Abstract
Immediately before the transition from metaphase to anaphase, the protein kinase activity of maturation or M-phase promoting factor (MPF) is inactivated by a mechanism that involves the degradation of its regulatory subunit, cyclin B. The availability of biologically active goldfish cyclin B produced in Escherichia coli and purified goldfish proteasomes (a nonlysosomal large protease) has allowed the role of proteasomes in the regulation of cyclin degradation to be examined for the first time. The 26S, but not the 20S proteasome, digested recombinant 49-kD cyclin B at lysine 57 (K57), producing a 42-kD truncated form. The 42-kD cyclin was also produced by the digestion of native cyclin B forming a complex with cdc2, a catalytic subunit of MPF, and a fragment transiently appeared during cyclin degradation when eggs were released from metaphase II arrest by egg activation. Mutant cyclin at K57 was resistant to both digestion by the 26S proteasome and degradation at metaphase/anaphase transition in Xenopus egg extracts. The results of this study indicate that the destruction of cyclin B is initiated by the ATP-dependent and ubiquitin-independent proteolytic activity of 26S proteasome through the first cutting in the NH2 terminus of cyclin (at K57 in the case of goldfish cyclin B). We also surmise that this cut allows the cyclin to be ubiquitinated for further destruction by ubiquitin-dependent activity of the 26S proteasome that leads to MPF inactivation.
Proteolysis plays an important role in the regulation of the eukaryotic cell cycle. The termination of mitosis and meiosis, the transition from metaphase to anaphase, is induced by the degradation of cyclin B, a regulatory subunit of maturation or M phase promoting factor (MPF;1 Murray et al., 1989). Further, it is suggested that cyclin is degraded by a ubiquitin-dependent proteolytic pathway (Glotzer et al., 1991; Hershko et al., 1991; Sudakin et al., 1995). Proteins subject to ubiquitin-dependent proteolysis are ligated to ubiquitin through their lysine residues and then degraded by a nonlysosomal large protease called the proteasome (or the multicatalytic protease), which is found in all eukaryotes from yeast to humans (Armon et al., 1990; Driscol and Goldberg, 1990; Kanayama et al., 1992; for reviews see Hershko and Ciechanover, 1982; Orlowski, 1990). Thus proteasomes must play an important role in cyclin degradation. Indeed, a genetic approach has revealed that mutation of the gene encoding one proteasome subunit causes G2/M arrest (Ghislain et al., 1993; Gordon et al., 1993). To date however, direct biochemical evidence for the involvement of proteasomes in cyclin degradation is poor.
Fish oocytes provide an appropriate experimental system with which to investigate the molecular mechanisms controlling meiosis and the embryonic cell cycle. Several factors responsible for the regulation of meiotic maturation of fish oocytes have been identified. These include the isolation and characterization of a fish maturation-inducing hormone (17α,20β-dihydroxy-4-pregnen-3-one; Nagahama and Adachi, 1985) and the components of MPF (cdc2, the catalytic subunit and cyclin B, the regulatory subunit; Yamashita et al., 1992a ,b; Kajiura et al., 1993). Both truncated and full length recombinant goldfish cyclins are biologically active (Hirai et al., 1992; Katsu et al., 1993). To understand the role of the proteasome in meiotic maturation, particularly in cyclin degradation, we have purified and characterized SDS-dependent (20S) and -independent (26S) proteasomes from goldfish oocyte cytosol (Tokumoto et al., 1995a ,b). The availability of biologically active goldfish cyclin B produced in Escherichia coli and of purified goldfish proteasomes allows the role of proteasome in the regulation of cyclin degradation to be examined for the first time. Here we propose that the 26S proteasome initiates cyclin degradation through the first cut in its NH2 terminus.
Materials and Methods
Materials
Goldfish were purchased from a local supplier and maintained at 15°C until use. The 20S and 26S proteasomes were purified from immature goldfish oocytes by conventional column chromatography as described (Tokumoto et al., 1995a ,b). Xenopus laevis were obtained from a dealer and maintained at 20°C. Xenopus CSF-arrested egg extracts were prepared by the method of Murray et al. (1989).
Electrophoresis and Immunoblot Analysis
Electrophoresis proceeded as described by Laemmli (1970), using 12.5% gels under denaturing conditions. Cyclin B degradation was assessed by immunoblotting against anti-goldfish cyclin B (B63 and B112) monoclonal antibodies (Yamashita et al., 1992b ). Immunocomplexes were visualized using the ECL detection kit (Amersham Intl., Arlington Heights, IL).
Determination of the Digestion Site of Cyclin B
Electroblotted 42-kD fragments of cyclin B were prepared as described (LeGendre and Matsudaira, 1989). The NH2-terminal amino acids were determined using a protein sequencer (470A; Applied Biosystems, Chiba, Japan).
Production of Recombinant Cyclin Bs
Full length (Δ0) and NH2-terminal truncated (Δ41 and Δ68) goldfish cyclin Bs were produced as described (Hirai et al., 1992; Katsu et al., 1993; Yamashita et al., 1995). Mutant cyclin B in which lysine 57 was replaced by arginine (Δ0K57R) was produced as follows. A cDNA clone encoding full length goldfish cyclin B (Hirai et al., 1992) was mutated using a site- directed mutagenesis system (Mutan K; Takara, Tokyo, Japan), following a strategy based on the method of Kunkel (1985), according to manufacturer's instructions. Double-strand, mutated cDNA was prepared by T3 polymerase using single-strand cDNA and the following oligonucleotide: AAGAAGGAAGTGAGGGTGGCGCCCAAGGTGGAG. This oligonucleotide was designed to produce a mutation site (bold type) and a restriction enzyme site (underlined) as described (Yamashita et al., 1995). Mutant clones were screened by digestion with the restriction enzyme and confirmed by sequencing.
Recombinant proteins were produced in E. coli BL21 (DE3) and purified by SDS-PAGE followed by electroelution from the gel, as described previously (Hirai et al., 1992). 35S-labeled cyclins were produced using a TNT T7-coupled Reticulocyte Lysate System (Promega Biotech, Madison, WI) according to the manufacturer's instructions.
Results
Restricted Digestion of Cyclin B by 26S Proteasome
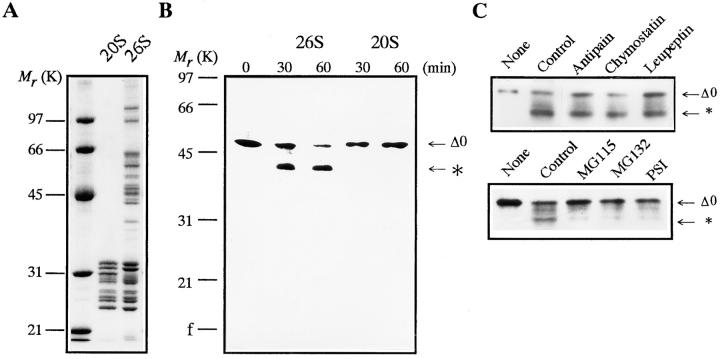
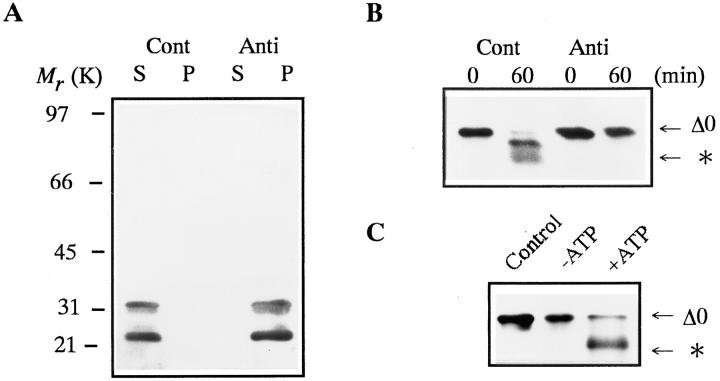
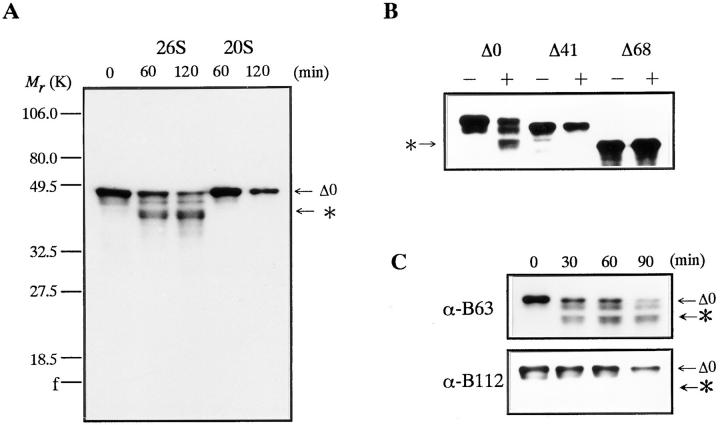
Although the 26S proteasome is a ubiquitin-dependent protease in general (Armon et al., 1990; Driscol and Goldberg, 1990; Kanayama et al., 1992), it also catalyzes an ATP-dependent and ubiquitin-independent proteolysis (Tanaka et al., 1983; Matthews et al., 1989; Murakami et al., 1992). Therefore, we initially investigated whether or not the 26S proteasome can degrade in vitro nonubiquitinated, full length goldfish cyclin B produced in E. coli (cyclin Δ0). Goldfish proteasomes (20S and 26S) were purified from immature goldfish oocytes by sequential chromatography (Tokumoto et al., 1995a ,b). SDS-PAGE has demonstrated that the 26S proteasome consists of multiple subunits with a molecular mass ranging from 23.5 to 140 kD, whereas the 20S proteasome includes subunits ranging from 23.5 to 31.5 kD (Fig. 1 A). The 26S, but not the 20S proteasome, digested 49-kD cyclin Δ0 and produced a 42-kD cyclin (Fig. 1 B). Protease inhibitors of microbial origin (antipain, chymostatin, and leupeptin) blocked the cyclin digestion at high (200 μM) but not at low concentrations (50 μM). Proteasome inhibitors (MG115, MG132, and PSI) that inhibit the chymotrypsin-like activity and ubiquitin-dependent protein degradation (Figueiredo-Pereira et al., 1994; Rock et al., 1994; Jensen et al., 1995), blocked the digestion at 50 μM (Fig. 1 C). No digestion proceeded when the 26S proteasome was depleted with an anti-proteasome antibody (Fig. 2, A and B). The reaction was also prevented when ATP was depleted from the reaction mixture (Fig. 2 C). These results indicate that the digestion of cyclin B is not due to a contaminating protease in the 26S proteasome fraction, but is catalyzed by the 26S proteasome itself.
Figure 1.
Digestion of E. coli-produced goldfish cyclin B by 26S proteasome purified from immature goldfish oocytes. (A) Subunit composition of 20S and 26S proteasomes. Purified 20S (10 μg) and 26S (27 μg) proteasomes were resolved by electrophoresis and stained with Coomassie brilliant blue R-250. (B) Digestion of full length cyclin B by purified proteasomes. Cyclin Δ0 (5 μg/ml) was incubated at room temperature with purified 20S or 26S proteasomes (60 μg/ml) in reaction buffer (100 mM Tris-HCl, 5 mM MgCl2, 0.04 mM ATP, pH 7.6). Samples were exposed to Laemmli's SDS sample buffer at the indicated times during incubation. Cyclin B was detected by immunoblotting against an anti–goldfish cyclin B (B63) monoclonal antibody. The position to which the digested cyclin B migrated is indicated by an asterisk. (C) Effect of protease and proteasome inhibitors on cyclin B digestion by the 26S proteasome. Cyclin Δ0 was incubated for 60 min without (None) or with the 26S proteasome in the absence (Control) or presence of various inhibitors at 50 μM. Cyclin B was detected by the B63 antibody. The position of the digested cyclin B is indicated by an asterisk.
Figure 2.
Inhibition of cyclin B digestion by proteasome or ATP depletion. Cyclin B was detected by the B63 antibody. The position to which the digested cyclin B migrated is indicated by an asterisk. (A) Immunodepletion of 26S proteasome from purified 26S proteasome fraction. 26S Proteasome was immunoprecipitated by affinity-purified anti-proteasome IgG (Anti) or control IgG (Cont), as described (Tokumoto and Ishikawa, 1993). Supernatants (S) and precipitates (P) were immunoblotted with a mixture of three monoclonal antibodies against goldfish 20S proteasome (GC4/5, 3α and 3β; Tokumoto et al., 1995a ). (B) Digestion of cyclin B in the mock- (Cont) or proteasome-depleted (Anti) 26S proteasome fraction. Cyclin Δ0 (5 μg/ml) was incubated at room temperature with the supernatant after immunoprecipitation with control or anti-proteasome IgG. Samples were exposed to Laemmli's SDS sample buffer at the indicated times during incubation. (C) Effect of ATP depletion on cyclin B digestion by the 26S proteasome. Cyclin Δ0 (5 μg/ml) was incubated at room temperature for 60 min without (Control) or with the 26S proteasome in the presence of an ATP-depleting system (10 mM glucose and 1 μg/ml hexokinase, −ATP) or 2 mM ATP (+ATP).
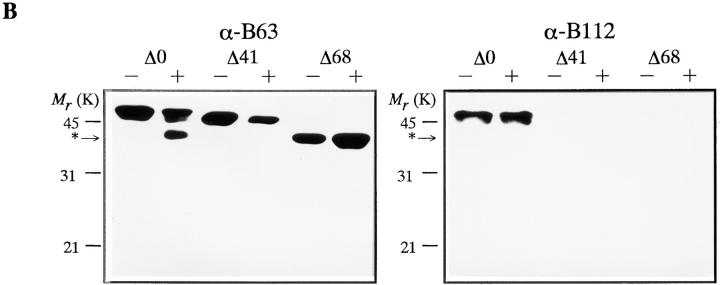
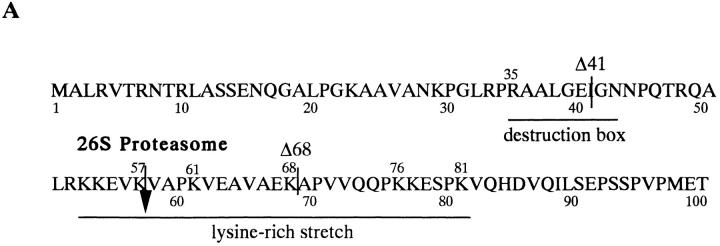
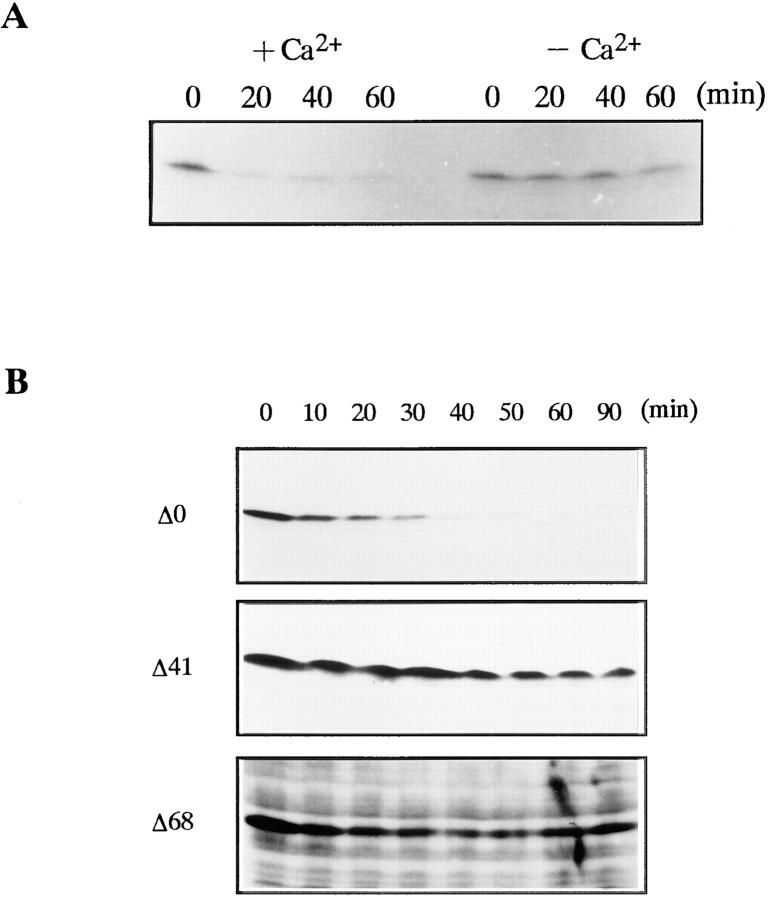
We determined whether the 26S proteasome digests the NH2- or COOH-terminal region of the cyclin B. We used monoclonal anti-cyclin B63, which recognizes the COOH terminus of cyclin B, and the anti-cyclin B112, which recognizes the NH2 terminus (Hirai et al., 1992; Katsu et al., 1993). The B63 antibody recognized the 42-kD intermediate and the NH2-terminal deletion mutants of cyclin B, while the B112 did not react with the intermediate but recognized the full length cyclin B (Fig. 3 B). Therefore, we concluded that cyclin B was digested in the NH2 terminus. To define the cleavage site, the NH2-terminal amino acid sequence of the electroblotted 42-kD cyclin was directly determined. The site of cyclin B cleavage by the 26S proteasome was the COOH-terminal peptide bond of lysine 57 (K57; Fig. 3 A).
Figure 3.
NH2-terminal sequence of goldfish cyclin B and digestion of recombinant cyclin B by the 26 S proteasome. (A) Amino acid sequence of the NH2-terminal region of goldfish cyclin B. The site digested by the 26S proteasome (COOH terminus of K57) and truncated sites of deletion mutants (Δ41, Δ68) are indicated. The destruction box and lysine-rich stretch are also indicated. (B) Digestion of full length and truncated cyclin Bs by the 26S proteasome. Cyclins Δ0, Δ41, and Δ68 were incubated in the absence (−) or presence (+) of the 26S proteasome for 120 min at room temperature. Cyclin degradation was assessed by immunoblotting against two kinds of anti- cyclin B (B63 and B112) monoclonal antibodies. B112 recognizes the NH2-terminal portion of goldfish cyclin B. The position of the digested cyclin B is indicated by an asterisk.
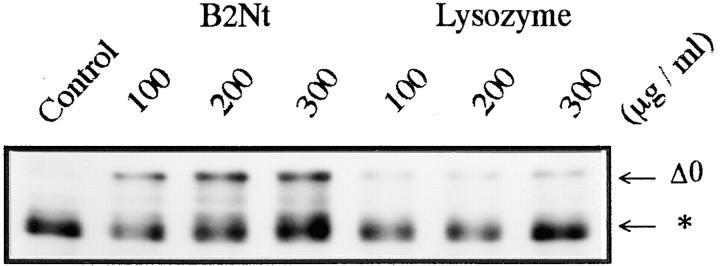
The NH2-terminal sequences of cyclins, including a consensus sequence called the destruction box, play a critical role in targeting cyclins for degradation (Glotzer et al., 1991; Lorca et al., 1991; Luca et al., 1991; Kobayashi et al., 1992). We therefore examined the role of NH2-terminal sequences in cyclin digestion by the 26S proteasome in vitro. We produced two NH2-terminal truncated cyclins: cyclin Δ41 lacking the destruction box and cyclin Δ68 lacking the destruction box and half of the lysine-rich stretch (Fig. 3 A). Neither cyclin Δ68 nor cyclin Δ41 were digested by 26S proteasome (Fig. 3 B). This finding suggests that the NH2-terminal region of cyclin B, including the destruction box, supplies an interaction site between cyclin B and 26S proteasome that is necessary for the subsequent cutting of cyclin B at K57. Interaction between the NH2 terminus of cyclin B and the 26S proteasome was also suggested by an experiment with a truncated protein containing the first 89 amino acids of Xenopus cyclin B2 (B2Nt, a gift from Dr. M.J. Lohka, University of Calgary, Calgary, Canada; Velden and Lohka, 1993). B2Nt inhibited the digestion of cyclin B by the 26S proteasome in a dose-dependent manner (Fig. 4). Control lysozyme, a basic and low molecular weight protein like B2Nt, did not inhibit the cyclin digestion (Fig. 4).
Figure 4.
Inhibition of 26S proteasome-catalyzed cyclin B digestion in vitro by the NH2-terminal fragment of Xenopus cyclin B2 (B2Nt). Cyclin Δ0 was incubated with purified 26S proteasome (60 μg/ml) for 120 min in the absence (Control) or presence of various concentrations of B2Nt or lysozyme. Cyclin B was detected with the B63 antibody. The migrating position of the digested cyclin B is indicated by an asterisk.
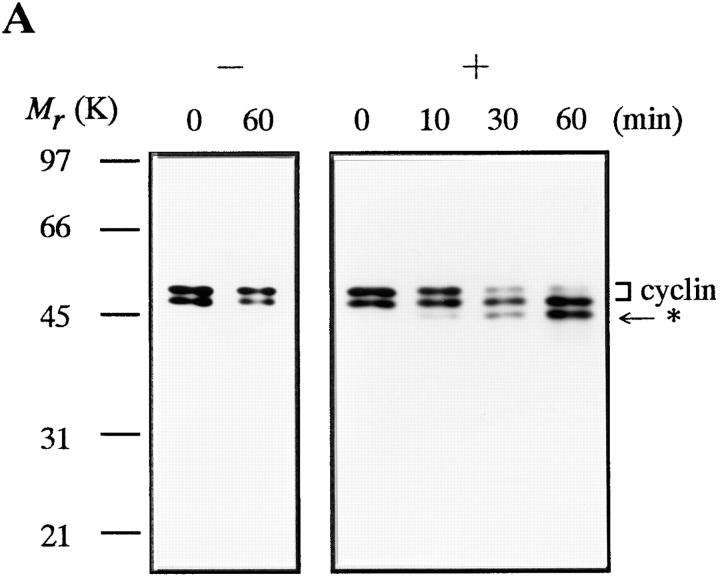
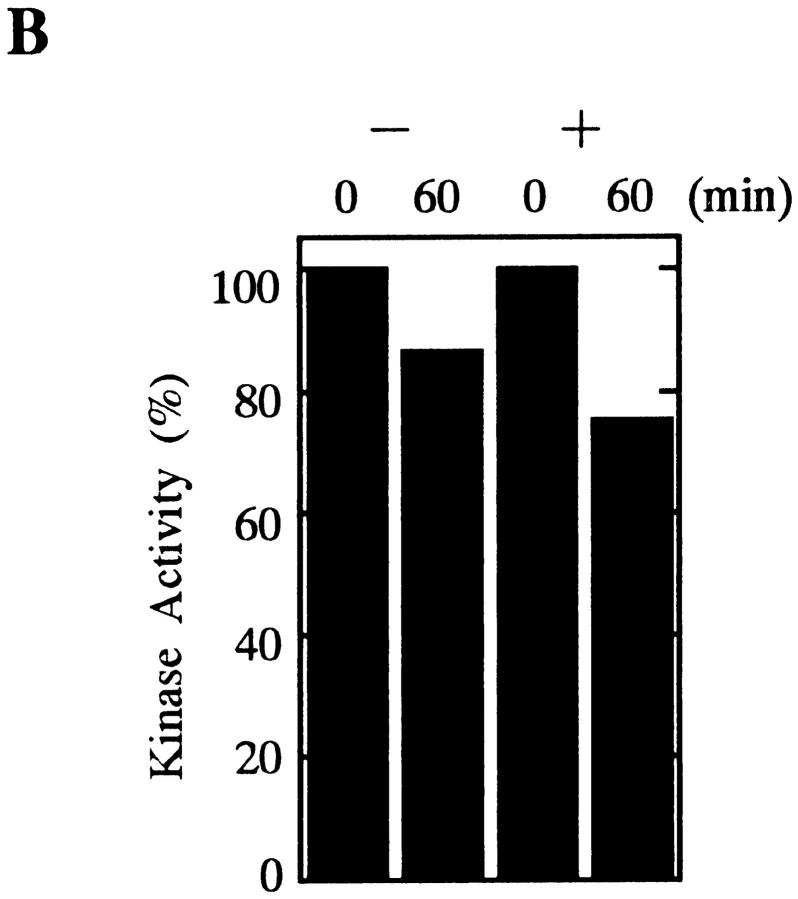
The 26S proteasome also digested native cyclin B that had been isolated as a complex with cdc2, yielding a truncated cyclin of ∼42-kD (Fig. 5 A). This was the same size as the fragment produced by digestion of recombinant, full length cyclin B with the 26S proteasome. The digestion of the cdc2–cyclin B complex with 26S proteasome, however, did not cause kinase inactivation of cdc2 (Fig. 5 B).
Figure 5.
Digestion of native cyclin B by 26S proteasome. The truncated cyclin B produced by the 26S proteasome digestion is indicated by an asterisk. (A) Digestion of cyclin B in MPF complex by the 26S proteasome. The MPF complex in mature carp oocytes was prepared using suc1 beads (Yamashita et al., 1992b ). The beads were washed with buffer (50 mM Tris-HCl, 20% glycerol, 10 mM 2-mercaptoethanol, 0.1 mM ATP, pH 7.5) and shaken in the absence (−) or presence (+) of 60 μg/ml of the 26S proteasome at room temperature with agitation. Samples were treated with SDS sample buffer at the indicated times and immunoblotted against the B63 antibody. Two cyclin bands were detected, and only the upper band was digested by the 26S proteasome. It is unlikely that these two bands correspond to different phosphorylation states of cyclin B (Yamashita et al., 1992b ). In C, only a single band of cyclin B was detected when oocytes were directly exposed to SDS sample buffer. Therefore, the lower band is probably produced by undesirable proteolysis during treatment with the suc1 beads. (B) Protein kinase activity of suc1 precipitates before and after the digestion with 26S proteasome. The kinase activity of suc1 precipitates incubated for 60 min in the absence (−) or presence (+) of 26S proteasome was measured with a synthetic peptide substrate for cdc2, as described (Yamashita et al., 1992a ). Activities are indicated as a percentage of the activity at 0 min for each condition. (C) Detection of a truncated cyclin B during goldfish egg activation. Ovulated eggs (2 ml) were placed in 3 ml goldfish Ringer's solution (Yamashita et al., 1992b ) and immediately homogenized in 5 ml SDS sample buffer at the indicated times. Before detecting truncated cyclin B by immunoblotting with B63 or B112, proteins with a molecular mass of 40–50 kD were separated by SDS-PAGE (Prep Cell Model 491; Bio Rad, Richmond, CA) and concentrated.
Detection of Intermediate Cyclin B during Egg Activation
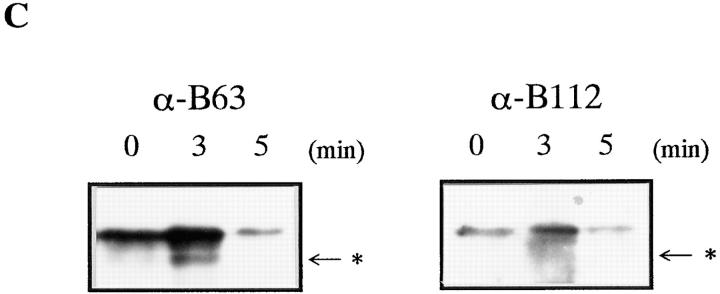
Mature goldfish oocytes are arrested at metaphase of meiosis II (metaphase II) and can be activated by the contact with water (Yamashita et al., 1992a ,b). Cyclin B was degraded within a few minutes after egg activation (Nagahama et al., 1995). A 42-kD fragment of cyclin B transiently appeared during the initial phase of goldfish egg activation. In a partially purified and highly concentrated fraction from egg extracts, intermediate cyclin B was detected 3 min after egg activation (Fig. 5 C). The monoclonal antibody B112, which recognizes the NH2-terminal region of cyclin B, did not react with the intermediate (Fig. 6 C). These results suggest that NH2-terminal digestion of cyclin B by the 26S proteasome is not an artifact of in vitro proteolysis but an initial reaction of cyclin B degradation that proceeds upon egg activation.
Figure 6.
Digestion of goldfish cyclin B by the Xenopus 26S proteasome. Cyclin was visualized by immunoblotting with B63 (A–C) and B112 (C). The position of the digested cyclin B is indicated by asterisks. (A) Digestion of full length cyclin B. Cyclin Δ0 (5 μg/ml) was incubated at room temperature with purified 20S or 26S proteasomes (60 μg/ml) in the reaction buffer (100 mM Tris-HCl, 5 mM MgCl2, 0.04 mM ATP, pH 7.6). Samples were exposed to Laemmli's SDS sample buffer at the indicated times during incubation. (B) Digestion of truncated cyclin B. Cyclins Δ0, Δ41, and Δ68 were incubated in the absence (−) or presence (+) of 60 μg/ml of 26S proteasome for 120 min at room temperature. (C) Cyclin was digested by Xenopus 26S proteasomes at the NH2 terminus. Goldfish cyclin Δ0 was digested by 26S proteasome for the indicated times and then stained with B63 or B112 antibody.
Destruction Analysis in Xenopus Egg Extracts
Although the results so far suggested that the 26S proteasome interacts with the NH2-terminal region of cyclin B and then cuts it at K57, digestion of cyclin B by the 26S proteasome in vitro is limited to cleavage of a single peptide bond and does not induce kinase inactivation of MPF (Fig. 5 B). We believe that the incomplete digestion of cyclin B by the 26S proteasome in vitro is due to the absence of factors responsible for further degradation of cyclin B. The most likely candidate is ubiquitin and its ligase. In fact, goldfish oocytes contain high levels of free ubiquitins (Tokumoto et al., 1993b ).
We purified and characterized the 26S proteasome from immature Xenopus oocyte extracts (Tokumoto and Ishikawa, 1995). We then examined whether or not this proteasome digests goldfish cyclin B, like the goldfish proteasome can. Xenopus 26S, but not 20S proteasomes, digested the NH2 terminus of goldfish cyclin B and produced the 42-kD intermediate (Fig. 6, A and C). NH2-terminal truncated cyclins were not digested by the Xenopus 26S proteasome (Fig. 6 B). These results indicate that the Xenopus 26S proteasome can digest goldfish cyclin B, suggesting a similar role of goldfish and Xenopus proteasomes in the regulation of cyclin degradation. We then examined the involvement of 26S proteasome in cyclin B degradation using a Xenopus cell-free system widely used for cell cycle studies, which contains the complete system necessary for cyclin degradation (Murray et al., 1989).
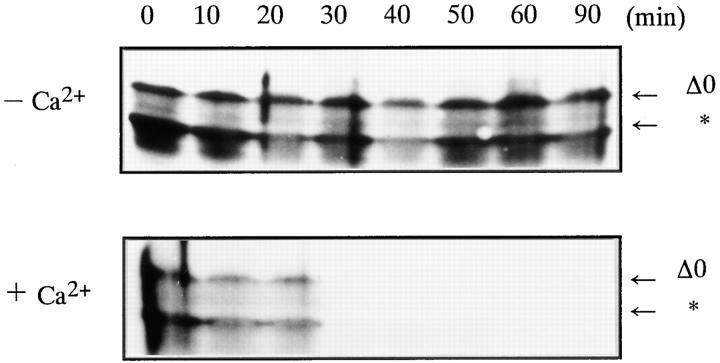
Cyclin Δ0 was completely degraded within 30 min after adding Ca2+ to Xenopus egg extracts, although it was stable in the absence of Ca2+ (Fig. 7 A). NH2-terminal truncated cyclins Δ41 and Δ68 were not degraded in Xenopus egg extracts even after activation with Ca2+ (Fig. 7 B). In contrast to cyclins Δ41 and Δ68, the 42-kD cyclin fragment (cyclin Δ57), which had been produced by the prior digestion of cyclin Δ0 with the purified 26S proteasome, was degraded in Xenopus extracts after adding Ca2+ (Fig. 8). To exclude the possibility that the 42-kD cyclin fragment remained in complex with the NH2-terminal portion after digestion with the 26S proteasome, we performed gel chromatography under the buffer conditions that were used for cyclin digestion. When cyclin treated with 26S proteasome was chromatographed, 42-kD cyclin and NH2-terminal fragment (∼9 kD) were clearly separated (Fig. 9). This result indicates that there is no significant interaction between 42-kD cyclin and the NH2-terminal fragment after digestion.
Figure 7.
Degradation of goldfish cyclin B in Xenopus egg extracts. Cyclin B was detected with B63 antibody. (A) E. coli-produced goldfish cyclin Δ0 was added to Xenopus egg extract at a final concentration of 5 μg/ml. Incubations proceeded in the absence (−Ca2 +) or presence (+Ca2 +) of 0.4 mM CaCl2 for the indicated times. (B) E. coli-produced cyclin Δ0, Δ41, and Δ68 were added to Xenopus extracts at the final concentration of 5 μg/ml. Cyclin degradation was induced by 0.4 mM Ca2+ and terminated by adding SDS sample buffer at the indicated times.
Figure 8.
Degradation of intermediate cyclin B (cyclin Δ57) in Xenopus egg extracts. Cyclin Δ57 was obtained by digesting cyclin Δ0 with 26S proteasome. 1/20 vol of the digestion mixture containing cyclins Δ0 and Δ57 was added to Xenopus extracts, and cyclin degradation was examined in the absence (−Ca2 +) or presence (+Ca2 +) of 0.4 mM Ca2+. Samples were exposed to SDS sample buffer at the indicated times. Cyclin degradation was assessed by immunoblotting with B63 antibody. The position of cyclin Δ57 is indicated by an asterisk.
Figure 9.
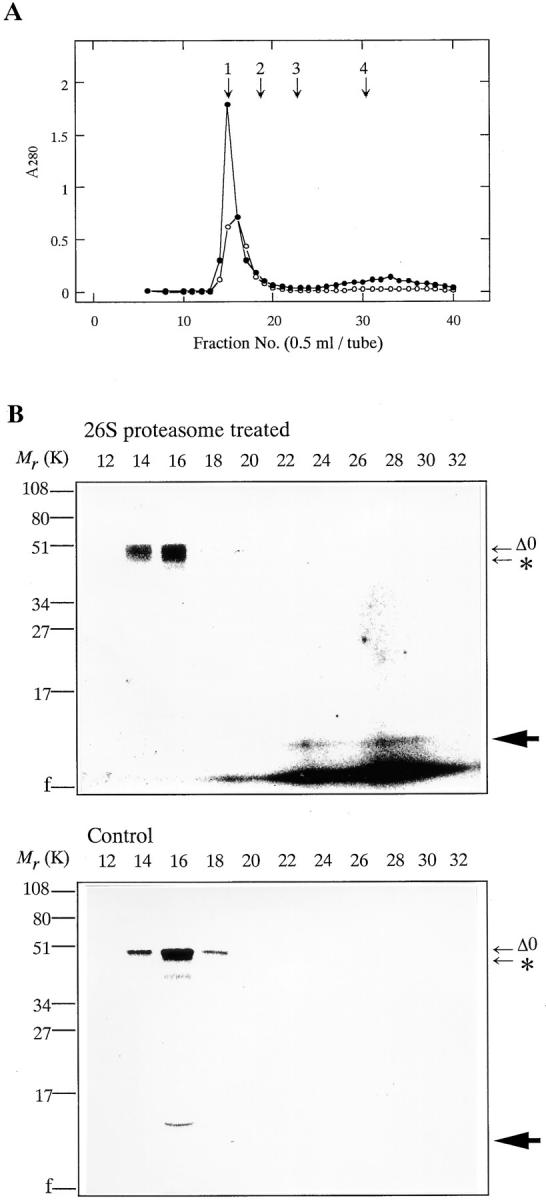
Separation of intermediate cyclin B and NH2-terminal fragment by gel chromatography. (A) Sephadex G-50 column chromatography. The 35S-labeled cyclin Δ0 was produced in vitro in rabbit reticulocyte lysate. Digestion of 35S-labeled cyclin Δ0 was performed for 60 min at room temperature with (•) or without (○) 26S proteasome. Samples were then separated on Sephadex G-50 column (1.0 × 19.0 cm) in 100 mM Tris-HCl, 5 mM MgCl2, pH 7.6. Fractions of 0.5 ml were collected. Arrows indicate the eluted positions of molecular weight standards as follows: 1, bovine serum albumin; 2, myoglobin; 3, ubiquitin; 4, total column volume (Vt). (B) SDS-PAGE analysis of gel chromatography fractions. Sephadex G-50 column chromatography fractions from 26S proteasome-treated cyclin Δ0 and untreated (Control) were separated by SDS-PAGE (15% gel) followed by autoradiography on Imaging plates (Fuji Film). The positions of the digested cyclin B is indicated by an asterisk, and the positions of NH2-terminal portion of cyclin B is indicated by an arrowhead.
Digestion by the 26S Proteasome and Degradation of In Vitro Translated Cyclin Bs in Xenopus Egg Extract
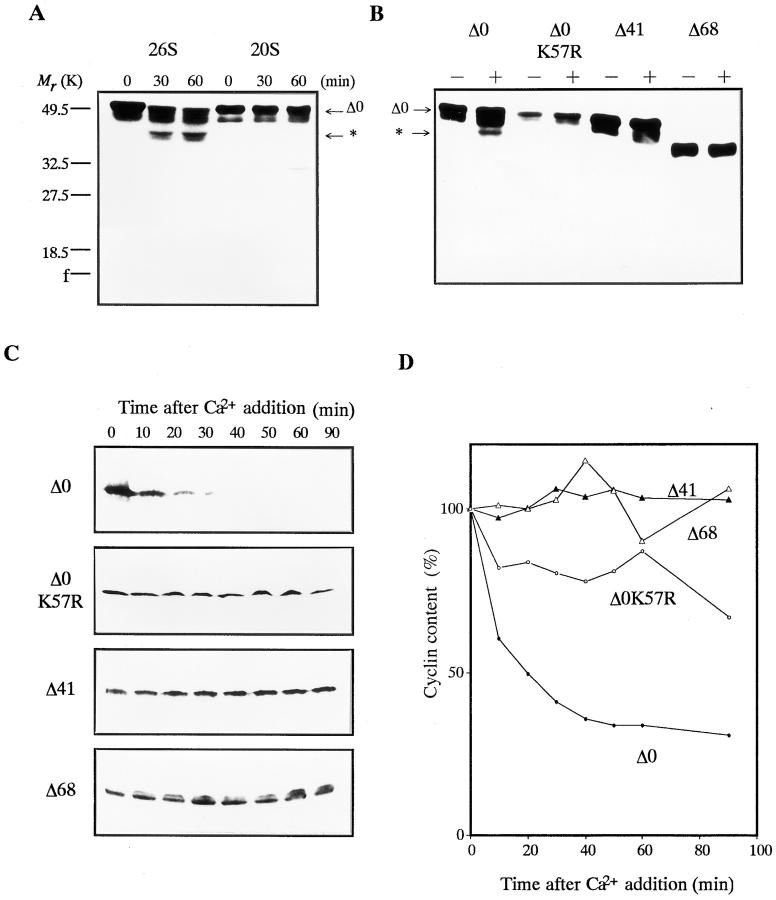
Our experiment using E. coli-produced cyclins demonstrated that the 26S proteasome restrictively digests the NH2 terminus of cyclin B and that only cyclins digestible or digested by 26S proteasome are degradable in Xenopus egg extracts activated with Ca2+. To confirm that these results are not artifacts derived from the incorrect form of cyclins produced in E. coli, we performed a similar experiment using cyclin B proteins translated in vitro in rabbit reticulocyte lysate. Cyclin B proteins produced in E. coli and translated in vitro gave the same results. Cyclin Δ0 translated in vitro was digested by the 26S , but not the 20S proteasome (Fig. 10 A), and degraded in Xenopus egg extracts (Fig. 10, C and D). NH2-terminal truncated cyclins Δ41 and Δ68 were resistant to 26S proteasome digestion (Fig. 10 B) and to degradation in Xenopus egg extracts (Fig. 10, C and D). Furthermore, the point mutant, cyclin Δ0K57R (in which the position of cleavage by the 26S proteasome, lysine 57, was converted to arginine), was neither digested by the 26S proteasome nor degraded in Xenopus extracts (Fig. 10, B–D).
Figure 10.
Digestion and degradation of in vitro translated cyclin B. The 35S- labeled cyclins Δ0, Δ0K57R, Δ41, and Δ68 were produced in vitro in rabbit reticulocyte lysate. After the translation of each cyclin, the lysate was incubated in the presence of 100 μg/ml of cycloheximide at room temperature under the indicated conditions. The 35S-labeled proteins were resolved by SDS-PAGE followed by autoradiography on Imaging plates (Fuji Film). The position of the digested cyclin B is indicated by an asterisk. (A) Digestion of full length cyclin B by purified 20S and 26S proteasomes. The reticulocyte lysate containing cyclin Δ0 was incubated with 60 μg/ml of proteasomes. (B) Digestion of full length, point mutated, and NH2-terminal truncated cyclin Bs by purified 26S proteasome. The reticulocyte lysate containing cyclin Δ0, Δ0K57R, Δ41, or Δ68 was incubated in the absence (−) or presence (+) of 60 μg/ ml of the 26S proteasome for 60 min. (C) Degradation of cyclin B in Xenopus egg extracts. One ninetieth of the lysate containing cyclin Δ0, Δ0K57R, Δ41, or Δ68 was added to the Xenopus egg extracts, and its degradation was induced by 0.4 mM Ca2+. At the indicated times, the reaction was terminated by adding SDS sample buffer. (D) The same sample as in C. Cyclin contents were quantified using an image analyzer (BAS2000; Fuji Film).
Discussion
Immediately before the transition from metaphase to anaphase, the kinase activity of MPF is inactivated through the degradation of its cyclin B subunit. The mechanism of cyclin degradation, which must be a highly selective process since few other proteins are degraded only at this time, is poorly understood. Using recombinant goldfish cyclin B and purified 26S proteasomes, we investigated the role of proteasomes in the regulation of cyclin degradation during egg activation. We found that purified 26S proteasome digests not only recombinant cyclin B, but also native cyclin B at its NH2-terminal portion, producing a 42-kD intermediate form. Since the 42-kD cyclin B appears transiently during the initial phase of normal egg activation, this digestion should not be an artifact but rather an initial step in cyclin B degradation upon egg activation. Using Xenopus egg extracts, we also showed that the initial digestion and further degradation of cyclin B are tightly linked. Cyclins Δ41 and Δ68 that are indigestible by the 26S proteasome are not degraded, whereas cyclins Δ0 and Δ57 digested by the 26S proteasome, are degraded. These findings strongly suggest that the initial cutting of the NH2-terminal region of cyclin B by 26S proteasome is a prerequisite for the subsequent degradation that leads to the inactivation of MPF at the metaphase/anaphase transition.
The NH2-terminal sequences of cyclin B, including a consensus sequence that is called the destruction box, play a critical role in targeting cyclins for degradation, since truncated sea urchin (Murray et al., 1989), human (Lorca et al., 1991), and clam (Luca et al., 1991) B-type cyclins missing the first 90, 72, or 97 amino acids, respectively, and clam (Luca et al., 1991) and Xenopus (Kobayashi et al., 1992) A-type cyclins missing the NH2-terminal 60 or 62 amino acids are resistant to degradation. Each of these truncated cyclins continuously activates cdc2, which prevents cells or cellular extracts from leaving mitosis. A truncated protein containing only the first 89 amino acids of Xenopus cyclin B2 (B2Nt), including sequences essential for cyclin degradation in other species, also inhibited cyclin degradation (Velden and Lohka, 1993). These results indicate interaction of the NH2-terminal portion of cyclin with the destruction machinery.
In this study, we proposed that the initial reaction of cyclin B destruction is the restricted cleavage of its NH2-terminal portion (K57 in the case of goldfish cyclin B) by the 26S proteasome. As well as cyclin Δ68 that lacks the cutting site K57, cyclin Δ41 containing K57 was not cleaved, indicating that the NH2-terminal region affords not only the cutting site but also the interaction site necessary for digestion by the 26S proteasome. This notion was confirmed by inhibiting the cyclin digestion with B2Nt that consists of the first 89 amino acid of Xenopus cyclin B2. Cyclins Δ41 and Δ68 were neither digested by the 26S proteasome nor degraded in Xenopus egg extracts activated by Ca2+, whereas cyclin Δ0 was digested at K57 by 26S proteasome and degraded in the extracts. In addition, cyclin Δ57 produced by digesting cyclin Δ0 with the 26S proteasome, was degraded in the extracts. These results suggest that only cyclins that have undergone 26S proteasome digestion at K57 can be degraded upon egg activation.
The mechanism of cyclin B degradation after initial cleavage by the 26S proteasome remains to be determined. The most likely candidate will be ubiquitin-dependent proteolysis. Proteins to be degraded by the ubiquitin pathway are ligated to ubiquitin through their lysine amino acid groups and then degraded by the 26S proteolytic complex (Hershko and Ciechanover, 1982). The first evidence that cyclin B degradation is mediated by ubiquitin-dependent proteolysis was provided by Glotzer et al. (1991). Other support for the involvement of a ubiquitin-dependent pathway in the cyclin degradation arises from the observation that methylated ubiquitin, which prevents the polyubiquitination of proteins destined for degradation, delays cyclin degradation in an extract from clam embryos (Hershko et al., 1991). A complex containing cyclin-selective ubiquitin ligase activity has been identified in clam oocytes (Sudakin et al., 1995). These findings suggest that the cell cycle-specific cyclin degradation is mediated by a ubiquitin-dependent proteolytic system.
The cyclin B subunit of MPF must be ubiquitinated immediately before the onset of its destruction at the metaphase/anaphase transition. The findings of the present study indicate that the restricted cleavage of cyclin B triggers its ubiquitination. It is likely that the digestion of the NH2-terminal restricted portion by the 26S proteasome (K57 in goldfish cyclin B) changes the cyclin structure available for further chemical modifications including ubiquitination, which leads to the complete destruction of the cyclin at metaphase/anaphase transition. Since cyclin Δ57 was destroyed, whereas cyclin Δ68 was not, the lysine residues between amino acids 58 to 68 constitute the most likely ubiquitination site, and cutting by the 26S proteasome at K57 might be necessary to expose them to ubiquitinating enzymes. This notion should be verified by investigating the difference in the three-dimensional structure of cyclins Δ0, Δ41, Δ57, and Δ68.
Cyclin B is absent in immature (prophase I-arrested) goldfish oocytes. Cyclin B is de novo synthesized during oocyte maturation and forms an MPF complex with extant cdc2, which drives the prophase I-arrested oocytes to metaphase II (mature oocytes; Hirai et al., 1992; Katsu et al., 1993; Yamashita et al., 1995). As shown in this study however, the 26S proteasome purified from immature goldfish oocytes can digest cyclin B. If the initial cleave of cyclin B by the 26S proteasome triggers cyclin destruction as proposed in this study, the question remains why cyclin B is stable during oocyte maturation and in mature oocytes but destroyed upon egg activation. Based on the results obtained from clam oocytes, Sudakin et al. (1995) have suggested that the initiation of cyclin degradation is triggered by ubiquitination caused by the activation of cyclin-selective ubiquitin ligase near the end of M-phase, which targets cyclin B for destruction by the 26S proteasome that is constitutively active during the cell cycle. Contrary to this, we found that the 26S proteasome purified from mature goldfish oocytes cannot digest cyclin B and that at least two subunits in 26S proteasomes from immature and mature oocytes differ (Tokumoto, T., Horiguchi, R., Nagahama, Y., unpublished results). These findings suggest that some inhibitory mechanisms preventing cyclin B degradation proceed on the proteasome itself at least during metaphase II arrest. The amount of the proteasome in egg cytosol also changes dramatically during oocyte maturation and egg activation; the lowest level is in mature metaphase II-arrested oocytes, and there is a transient increase between the first and second meiotic cell cycles and upon egg activation in goldfish (Tokumoto et al., 1993a ). Further studies should reveal how the subunit composition of proteasomes and their contents during oocyte maturation and egg activation are involved in controlling the cell cycle by regulating cyclin stability.
Stewart et al. (1994) have shown that binding with cdc2 is necessary for the degradation of Xenopus cyclins A and B2 but not for that of cyclin B1. This implies that the mechanisms of cyclin degradation vary according to the types of cyclins. Since goldfish cyclin B exhibits higher homology to Xenopus cyclin B1 (66%) than to cyclin B2 (50%), the mechanism of cyclin degradation by the 26S proteasome proposed in this study may be specific to cyclin B1.
Our present data suggest that the NH2-terminal restricted cleavage of cyclin B by 26S proteasome allows cyclin to be ubiquitinated. Compared with the idea that the initiation of cyclin destruction is dependent on the activity of ubiquitin ligase, irrespective of 26S proteasome activity (Sudakin et al., 1995), we propose that cyclin destruction is primarily controlled by the activity of 26S proteasome.
Ubiquitination of cyclins has been well studied genetically and biochemically (for review see Murray, 1995). A cyclin-specific ubiquitin ligase complex, the cyclosome, or APC complex, has been characterized in clam and Xenopus, respectively (King et al., 1995; Sudakin et al., 1995). These ubiquitin ligases (E3) catalyze ubiquitination using a specialized ubiquitin carrier protein (E2). Among the multiple species of E2s, UBC9 is required for cell cycle progression in late G2 or early M-phase (Seufert et al., 1995). UBC4 protein can ubiquitinate cyclins in Xenopus egg extracts (King et al., 1995). Recently, a novel cyclin- selective UBC family member, E2-C, was reported which can ubiquitinate cyclin B(13-91)/protein A fusion protein in cyclosome-dependent manner (Aristarkhov et al., 1996). These reports have shown that destruction box mutants cannot be ubiquitinated or degraded after extract activation, suggesting that the destruction box is a recognition sequence for the ubiquitinating system. However, we have shown that mutants that lack the proteasome cleavage site cannot be degraded after extract activation. The relative importance of these two processes is unclear because of the discrepancy between in vivo and in vitro results. In vitro, proteasome cleavage and ubiquitination of cyclin seem independent of each other. Destruction box-dependent ubiquitination of cyclin by purified proteins does not depend on the previous proteasome cleavage of cyclin B, and cyclin cleavage by purified proteasome does not depend on previous ubiquitination of cyclin B. But, since mutations that block in vitro ubiquitination and mutations that block proteasome cleavage also block destruction in extracts, the simple conclusion is that the ubiquitination and proteasome cleavage are both necessary for cyclin destruction in extracts. There are, however, no experimental data available at present that provide information on the relative order of these two steps in vivo. Therefore, there are three possible in vivo scenarios. (a) Proteasome cleavage precedes ubiquitination to expose an NH2-terminal lysine that is a good substrate for ubiquitination. An extreme view would be that destruction box-dependent ubiquitination is an artifact that plays no role in vivo and that the purpose of the destruction box is solely to induce the initial ubiquitin-independent cleavage of cyclin. (b) Destruction box-dependent ubiquitination precedes proteasome cleavage to recruit cyclin to the proteasome by virtue of the proteasome's polyubiquitin binding subunit. An extreme view would be that proteasome cleavage is not necessary in vitro, and mutants like K57R are having a direct effect on destruction box-dependent ubiquitination. (c) There is no required order of proteasome cleavage and destruction box-dependent ubiquitination, although both events would be necessary for efficient cyclin destruction, they could occur in either order. Further studies are necessary to understand the molecular mechanism of cyclin degradation, especially identification of the lysine residue that is destined to be ubiquitinated.
Acknowledgments
We are grateful to Dr. M.J. Lohka for the B2Nt and to Drs. H.A. Bern and A.W. Murray for critical reading of the manuscript.
This study was supported in part by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (07283104 to Y. Nagahama) and by a Grant-in-Aid (Bio Media Program) from the Ministry of Agriculture, Forestry, and Fisheries (BMP 96-II-2-6). This study was performed under the National Institute for Basic Biology Cooperative Research Program (95-112 to T. Tokumoto).
Footnotes
Please address all correspondence to Dr. Y. Nagahama, Laboratory of Reproductive Biology, National Institute for Basic Biology, Okazaki 444, Japan. Tel.: (81) 564-55-7550; Fax: 81-564-55-7556; E-mail: nagahama@nibb.ac.jp
1. Abbreviation used in this paper: MPF, maturation or M-phase promoting factor.
References
- Aristarkhov A, Eytan MW, Moghe A, Admon A, Hershko A, Ruderman JV. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci USA. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armon T, Gonoth D, Hershko A. Assembly of the 26S complex that degrades proteins ligated to ubiquitin is accompanied by formation of ATPase activity. J Biol Chem. 1990;265:20723–20726. [PubMed] [Google Scholar]
- Driscol J, Goldberg A. The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J Biol Chem. 1990;265:4789–4792. [PubMed] [Google Scholar]
- Figueiredo-Pereira ME, Berg KA, Wilk S. A new inhibitor of the chymotrypsin-like activity of the multicatalytic proteinase complex (20S proteasome) induces accumulation of ubiquitin-protein conjugates in a neuronal cell. J Neurochem. 1994;63:1578–1581. doi: 10.1046/j.1471-4159.1994.63041578.x. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae26S protease mutants arrest cell division in G2/metaphase. Nature (Lond) 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature (Lond) 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Dillon P, Rosen C, Hastie ND. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature (Lond) 1993;366:355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. Mechanisms of intracellular protein breakdown. Annu Rev Biochem. 1982;51:335–364. doi: 10.1146/annurev.bi.51.070182.002003. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ganoth D, Pehrson J, Palazzo RE, Cohen LH. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J Biol Chem. 1991;266:16376–16379. [PubMed] [Google Scholar]
- Hirai T, Yamashita M, Yoshikuni M, Lou YH, Nagahama Y. Cyclin B in fish oocytes: its cDNA and amino acid sequences, appearance during maturation, and induction of p34cdc2activation. Mol Reprod Dev. 1992;33:131–140. doi: 10.1002/mrd.1080330204. [DOI] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Kajiura H, Yamashita M, Katsu Y, Nagahama Y. Isolation and characterization of goldfish cdc2, a catalytic component of maturation-promoting factor. Develop Growth Differ. 1993;35:647–654. doi: 10.1111/j.1440-169X.1993.00647.x. [DOI] [PubMed] [Google Scholar]
- Kanayama H, Tamura T, Ugai S, Kagawa S, Tanahashi N, Yoshimura T, Tanaka K, Ichihara A. Demonstration that a human 26S proteolytic complex consists of a proteasome and multiple associated protein components and hydrolyzes ATP and ubiquitin-ligated proteins by closely linked mechanisms. Eur J Biochem. 1992;206:567–578. doi: 10.1111/j.1432-1033.1992.tb16961.x. [DOI] [PubMed] [Google Scholar]
- Katsu Y, Yamashita M, Kajiura H, Nagahama Y. Behavior of the components of maturation-promoting factor, cdc2 kinase and cyclin B, during oocyte maturation of goldfish. Dev Biol. 1993;160:99–107. doi: 10.1006/dbio.1993.1289. [DOI] [PubMed] [Google Scholar]
- King RW, Peters J, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Stewart E, Poon R, Adamczewski JP, Gannon J, Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2protein kinase subunits. Mol Biol Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeGendre, N., and P. Matsudaira. 1989. Purification of proteins and peptides by SDS-PAGE. In A Practical Guide to Protein and Peptide Purification for Microsequencing. P.T. Matsudaira, editor. Academic Press, San Diego, CA. 49–69.
- Lorca T, Fesquet D, Zindy F, Bouffant F, Cerrui M, Brechot C, Devauchelle G, Doree M. An okadaic acid-sensitive phosphatase negatively controls the cyclin degradation pathway in amphibian eggs. Mol Cell Biol. 1991;11:1171–1175. doi: 10.1128/mcb.11.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca FC, Shibuya EK, Dahlmann CE, Ruderman JV. Both cyclin A Δ60 and B Δ97 are stable and arrest cells in M-phase, but only cyclin B Δ97 turns on cyclin destruction. EMBO (Eur Mol Biol Organ) J. 1991;10:4311–4320. doi: 10.1002/j.1460-2075.1991.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews W, Tanaka K, Driscoll J, Ichihara A, Goldberg AL. Involvement of the proteasome in various degradative processes in mammalian cells. Proc Natl Acad Sci USA. 1989;86:2597–2601. doi: 10.1073/pnas.86.8.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature (Lond) 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cyclin ubiquitination: the destructive end of mitosis. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature (Lond) 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- Nagahama Y, Adachi S. Identification of maturation-inducing steroid in a teleost, the amago salmon (Oncorhynchus rhodurus) . Dev Biol. 1985;109:428–435. doi: 10.1016/0012-1606(85)90469-5. [DOI] [PubMed] [Google Scholar]
- Nagahama, Y., M. Yoshikuni, M. Yamashita, T. Tokumoto, and Y. Katsu. 1995. Regulation of oocyte growth and maturation in fish. In Current Topics in Developmental Biology. Volume 30. R.A. Pederson and G.P. Schatten, editors. Academic Press, San Diego, CA. 103–145. [DOI] [PubMed]
- Orlowski M. The multicatalytic proteinase complex, a major extralysosomal proteolytic system. Biochemistry. 1990;29:10289–10297. doi: 10.1021/bi00497a001. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature (Lond) 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Stewart E, Kobayashi H, Harrison H, Hunt T. Destruction of Xenopus cyclins A and B2, but not B1, requires binding to p34 cdc2 . EMBO (Eur Mol Biol Organ) J. 1994;13:584–594. doi: 10.1002/j.1460-2075.1994.tb06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Waxman L, Goldberg AL. ATP serves two distinct roles in protein degradation in reticulocytes, one requiring and one independent of ubiquitin. J Cell Biol. 1983;96:1580–1585. doi: 10.1083/jcb.96.6.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumoto T, Ishikawa K. A novel “active” proteasomes from Xenopus laevisovary cytosol. Biochem Biophys Res Commun. 1993;192:1106–1114. doi: 10.1006/bbrc.1993.1531. [DOI] [PubMed] [Google Scholar]
- Tokumoto T, Ishikawa K. Characterization of active proteasome (26S proteasome) from Xenopus oocytes. . Biomed Res. 1995;16:295–302. [Google Scholar]
- Tokumoto T, Yamashita M, Yoshikuni M, Nagahama Y. Changes in the activity and protein levels of proteasomes during oocyte maturation in goldfish (Carassius auratus) . Biomed Res. 1993a;14:305–308. [Google Scholar]
- Tokumoto T, Kajiura H, Yoshikuni M, Yamashita M, Nagahama Y. Purification of ubiquitin from goldfish oocytes. Biomed Res. 1993b;14:309–311. [Google Scholar]
- Tokumoto T, Yamashita M, Yoshikuni M, Kajiura H, Nagahama Y. Purification of latent proteasome and demonstration of active proteasome in goldfish (Carassius auratus)oocyte cytosol. Biomed Res. 1995a;16:173–186. [Google Scholar]
- Tokumoto T, Yoshikuni M, Yamashita M, Kajiura H, Nagahama Y. Purification and characterization of active proteasome (26S proteasome) from goldfish ovaries. Biomed Res. 1995b;16:207–218. [Google Scholar]
- Velden HMW, Lohka MJ. Mitotic arrest caused by the amino terminus of Xenopuscyclin B. Mol Cell Biol. 1993;13:1480–1488. doi: 10.1128/mcb.13.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Fukada S, Yoshikuni M, Bulet P, Hirai T, Yamaguchi A, Yasuda H, Ohba Y, Nagahama Y. M-phase-specific histone H1 kinase in fish oocytes: its purification, components and biochemical properties. Eur J Biochem. 1992a;205:537–543. doi: 10.1111/j.1432-1033.1992.tb16810.x. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Fukada S, Yoshikuni M, Bulet P, Hirai T, Yamaguchi A, Lou YH, Zhao Z, Nagahama Y. Purification and characterization of maturation-promoting factor in fish. Dev Biol. 1992b;148:8–15. doi: 10.1016/0012-1606(92)90259-j. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Kajiura H, Tanaka T, Onoe S, Nagahama Y. Molecular mechanisms of the activation of maturation-promoting factor during goldfish oocyte maturation. Dev Biol. 1995;168:62–75. doi: 10.1006/dbio.1995.1061. [DOI] [PubMed] [Google Scholar]