Abstract
We show that a splice variant–derived cyclin B is produced in sea urchin oocytes and embryos. This splice variant protein lacks highly conserved sequences in the COOH terminus of the protein. It is found strikingly abundant in growing oocytes and cells committed to differentiation during embryogenesis. Cyclin B splice variant (CBsv) protein associates weakly in the cell with Xenopus cdc2 and with budding yeast CDC28p. In contrast to classical cyclin B, CBsv very poorly complements a triple CLN deletion in budding yeast, and its microinjection prevents an initial step in MPF activation, leading to an important delay in oocyte meiosis reinitiation. CBsv microinjection in fertilized eggs induces cell cycle delay and abnormal development. We assume that CBsv is produced in growing oocytes to keep them in prophase, and during embryogenesis to slow down cell cycle in cells that will be committed to differentiation.
Cyclins are a conserved family of proteins that play a central role in eukaryotic cell division cycle progression, as regulatory subunits of cyclin dependent kinases (CDKs, whose catalytic subunits are homologues of the fission yeast cdc2 protein).1 CDKs are downstream targets of convergent cascades of regulations at critical points of the cell cycle. M-phase–promoting factor (MPF, formerly maturation promoting factor, reference 21), the factor responsible for M-phase entry and progression in mitosis, has been purified three times by biochemical means (7, 19, 36). MPF from starfish, Xenopus, and carp oocytes has been found to be a heterodimer composed of one molecule of cdc2 and one molecule of cyclin B (CB). B type cyclins are archetypal mitotic cyclins, evolutively and functionally related to fission yeast cdc13p. Among CDKs, the regulation of MPF is by far the best understood today. Cyclin B is required for activity, as well for activation and for inhibition of MPF. The cdc2 monomer has never been found active. Its activation is conferred by the CAK-dependent T161-phosphorylation that requires cyclin B association (4, 28, 33). Inhibition of MPF during S- and G2-phases and also by the DNA replication checkpoint mechanism is achieved by wee1-catalyzed phosphorylation of the tyrosine 15 in cyclin B–bound molecules of cdc2 (9, 22). Cyclin B is also likely required for activation of the protein phosphatase cdc25p that specifically dephosphorylates tyrosine 15 and allows MPF amplification and entry into mitosis (5, 37). Finally, targeted proteolysis of cyclin B by an ubiquitin-dependent pathway is the mechanism by which MPF is inactivated and the cell returns to interphase (8). Therefore, the major part of MPF regulation is accounted for by cyclin B synthesis and proteolysis. This was emphasized in simplified early embryogenesis cycles that are composed of a succession of M- and S-phases without intervening G-phases. Cycles in acellular Xenopus egg extracts are driven by MPF as a basic oscillator, whose periodic activity is scheduled strictly by oscillating abundance of cyclin B (24). Accordingly, during the cleavage period of Xenopus embryogenesis, cdc2 tyrosine 15 is never found phosphorylated (3) and checkpoint mechanisms are downregulated.
Site-directed mutagenesis as well as protein crystallization have allowed the mapping of some sequences in cyclins involved in these regulations. Crystal structure of the homologous dimer cdk2–cyclin A showed that the cyclin interacts with the cdk via sequences distributed along the so-called cyclin box, a sequence well conserved among all cyclins (14). In the NH2 terminus of mitotic cyclins A and B, a destruction box is required to allow ubiquitination of the protein and its targeted proteolysis in anaphase (8). Mutants that are deleted for this box are stable in mitosis, and their overexpression triggers mitotic arrest. Also in the NH2-terminal region of B type cyclins, a cytoplasmic retention signal (CRS) is presumed to account for differential early prophase localization of nuclear cyclin A and cytoplasmic cyclin B (27). A chimeric cyclin A with the first amino acids of cyclin B remains cytoplasmic until early prophase. Further on, at the beginning of the cyclin box, conserved amino acids in the P-box are thought to be involved in the specific activation of cdc2 by cdc25 (37). Finally, two reports showed that a short COOH-terminal deletion of recombinant cyclins A or B abolished the binding to cdc2 (17, 34), although this region was not found to be directly involved in the physical interaction between cyclin A and cdk2 (14).
Here we show that such a COOH-terminal truncation, which removes universally conserved amino acids, is naturally realized in a splice variant of sea urchin cyclin B. Moreover, immunofluorescence experiments suggest this splice variant plays a role in embryogenesis and behaves like a marker of cell lineages in postcleavage embryos.
Materials and Methods
Handling of Animals
Sea urchins of the species Sphaerechinus granularis were collected by diving in the Baie de Banyuls (Banyuls, France). Eggs and sperm were collected in filtered natural sea water after injection of 0.2 M acetylcholine in the general cavity of the adults. After fertilization embryos were allowed to develop at 16°C under moderate agitation. The library was made with RNA from embryos taken after the first mitotic division (2-cell stage). RNA for Northern blots was prepared before fertilization (unfertilized stage) from 2-cell stage embryos and from swimming larvae collected at 24 h after fertilization. Hatching usually occurred at 18 h.
Library Construction
cDNA cloning procedure was derived from a primer-adapted method that has been described in detail (1). In brief, first strand synthesis was performed using 200 U of RNAseH–MoMuLV reverse transcriptase (GIBCO BRL, Gaithersburg, MD). Removal of the oligo(dT) primer was achieved through two 0.1% cetyl triethyl ammonium bromide (CTAB) precipitation steps, followed by two ethanol precipitations (35). Large-sized cDNA (>0.5 kb), size-fractionated on a S400 gel filtration column (Pharmacia Biotech., Inc., Piscataway, NJ), were then dG tailed, annealed to dC-tailed/BamH1–cut pT3T718U phagemid DNA (Pharmacia Biotech, Inc.) and to the oligo(dT)/BamHI adapter, and then DNA was ligated for 16 h at 16°C. After T4 DNA polymerase repair and ethanol precipitation, DNA was electroporated (Bio-Rad Laboratories, Hercules, CA) into TG1 bacteria. Yield was ∼105 recombinant clones per ng of cDNA. Total library was grown for 5 h at 37°C in LB medium supplemented with 40 μg/ml ampicillin and then frozen in 30% glycerol at −80°C.
Screening
Screening was performed with a heterologous probe comprising nucleotides (nt) 473–920 of the starfish Marthasterias glacialis cyclin B open reading frame (ORF). Hybridization (6× SSC, 42°C) and rinses (1× SSC, 50°C) were performed at low stringency. Three successive screenings were required to get pure clones.
PCR Analysis
The cyclin B cloned cDNA sequences were used to design the following primers required for sea urchin cDNA, RNA, and genomic DNA analysis (see Figs. 2 and 4 for the position of the primers): CB1, CCAATCGACCAGGATGTGA; CB5, GATGACACATTACAGCATG; CB64, ACCAAGGATGATCTGCGCACG; CB62, ACAAAACTGACCCA; CB8, TTATTATTCAATCGTTGATTGT; CB10, GTCAGAGAACAGGCTGGCGAT.
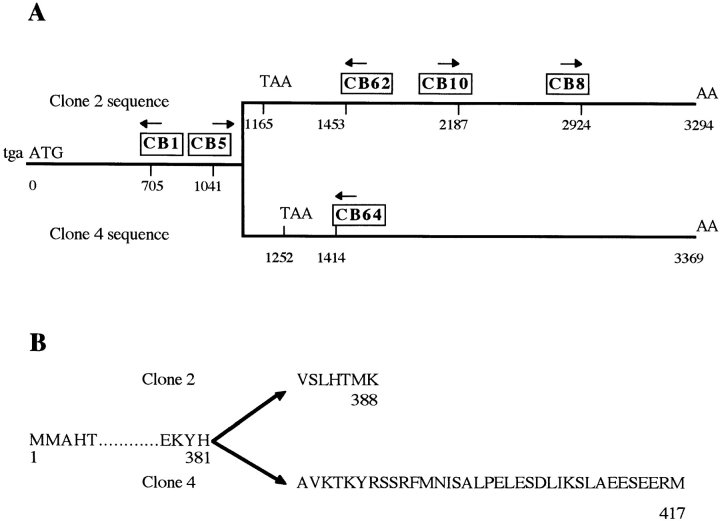
Figure 2.
The two different cyclin B mRNAs found in S. granularis. (A) schematic representation of the cDNAs. Nucleotide numbers from the beginning of ORF are indicated under the line representing the cDNA. Primers used in this work are positioned and boxed above the cDNA, with arrows indicating the direction of the primers. Both cDNAs are identical until nt 1143, and sequences downstream are specific for clone 2 (top) or clone 4 (bottom). (B) Amino acid sequences corresponding to clone 2 (top) or clone 4 (bottom) diverge downstream of histidine 381. Clone 2 stops after only seven specific residues, whereas specific sequence of clone 4 is 36 amino acids long.
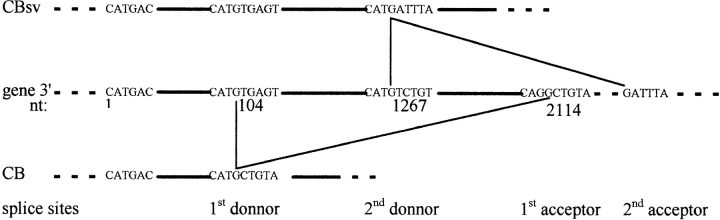
Figure 4.
Schematic representation of the 3′ part of the gene encoding cyclin B in S. granularis. nt 1 is the first nucleotide of CB5 primer (see Fig. 2). The two splice donor sites and the first acceptor site have been sequenced; the second acceptor site has not been sequenced and is deduced from CBsv cDNA sequence. CBsv mRNA is raised by splicing between donor 2 and acceptor 2, CB by splicing between donor 1 and acceptor 1. The corresponding spliced introns are thus overlapping.
Race PCR
To isolate 5′ ends of incomplete cDNA we used 5′RACE-PCR method with the Marathon™ cDNA Amplification Kit (CLONTECH Laboratories, Inc., Palo Alto, CA) according to the protocol of manufacturer.
Northern Blot Hybridization
Total RNA from unfertilized eggs, 2-cell embryos, and swimming blastulae were purified by the guanidinum thiocyanate/cesium chloride method (2). Poly(A)+ RNAs were isolated by oligo(dT)-cellulose chromatography. mRNA concentration was evaluated by densitometry and 1 μg mRNA was separated in a 1.2% formaldehyde–agarose gel and transferred to a hybond nitrocellulose membrane (Amersham Corp., Arlington Heights, IL). Hybridization and washing of the membrane were performed as described (30). The membrane was rehybridized with a new probe after soaking it twice in boiling 0.5% SDS.
Recombinant Proteins
Cyclin B and variant cyclin B ORFs were amplified by reverse transcriptase (RT)-PCR with a common 5′ primer: GCGGCGGCCCATATGATGGCTCATACCACAAGAAATTCA; a 3′ primer specific for CB: GCGGCGGCCGGATCCTCACATCCGTTCTTCACTTTCTTCCGC; and a 3′ primer specific for CBsv: GCGGCGGCCGGATCCTTACTTCATGGTATGTAAACTCACATG. This sequence data information is available from GenBank/EMBL/DDBJ under accession numbers: Y08016 (CB); Y08017 (CBsv); and Y08018 (cyclin B genomic sequence). The PCR products were digested with Nde1 and BamH1 and cloned in frame in pET21A vector. The recombinant proteins were produced for 4 h in Bl 21. The major part of the recombinant protein was insoluble in inclusion bodies, which were prepared free of supernatant. The recombinant CB and CBsv proteins were purified by preparative SDS-PAGE (BRL Preparative Electrophoresis System; GIBCO BRL), dialyzed overnight, and concentrated at ∼1 mg/ml in TBS.
Polyclonal antibodies specific for CB and CBsv were raised in rabbit against synthetic COOH-terminal peptides (Genosys Co., Cambridge, UK) CSDLIKSLAEESEERM (CB) and CEKYHVSLHTMK (CBsv). Serums were affinity purified on Sepharose-coupled peptide, and resultant affinity-purified antibodies were kept frozen at 2 mg/ml concentration.
Immunofluorescence Experiments
Gametes, eggs, or embryos were fixed either directly in 75% methanol, 25% glycerol, 10 mM EGTA, or first in AT buffer (10 mM Mes, pH 6.7, 10 mM EGTA, 25% glycerol, and 1% NP-40) followed by fixation in methanol for at least 1 h. Eggs were incubated overnight in first antibody diluted 1/50 in TBS, plus 0.1% Triton X-100, and 10 μg/ml Hoechst dye. Texas red–coupled goat anti–rabbit second antibody was applied for 2 h. Cells were mounted in 10% polyvinyl alcohol (Sigma Chemical Co., St. Louis, MO).
Results
Cloning of S. granularis Cyclin B cDNA
1.2 × 105 clones of the cDNA library were screened. None of the five cyclin B cDNAs was full length. All sequences began approximately in the same region of the ORF, corresponding to amino acid (aa) 220 of the sea urchin Arbacia punctulata cyclin B, just after the beginning of cyclin B box. 5′ RACE-PCR with CB1 primer was performed to complete the 5′ end of the longest sized cDNA sequence (clone 4, 2.9 kbp long). Completed clone 4 sequence codes for a 417-aa protein highly homologous to the previously published A. punctulata cyclin B sequence (reference 26; Fig. 1).
Figure 1.

Alignment of amino acid sequences of cyclin B from the sea urchins Arbacia punctulata (26) and Sphaerechinus granularis (this paper). Residues identical in both sequences are boxed. A high degree of identity is noteworthy between cyclin B of these distant Echinid species.
Characterization of Alternative Splicing in Cyclin B mRNA
The sequence of one clone (clone 2) was found to be identical to others until nt 1143 (from ATG), corresponding to Histidine 381 of the clone 4 ORF, and to diverge abruptly after that until the end of 3′ untranslated sequence (Fig. 2). Coding sequence after the divergence ends after 36 amino acids in clone 4, and only after 7 amino acids in clone 2. An extensive search showed no significant homology between both 3′ UTR. Since sequence that surrounds the divergence in clone 2 (CAT/GTGAGT) corresponded to a canonical intron entry sequence, clone 2 could have resulted from a lack of intron removal in the premessenger RNA. RT-PCR with 3′ primers CB62 and CB64, specific respectively for sequences 2 and 4, and a common CB5 primer as a 5′ primer gave rise to the expected products (425 bp long with CB62 and 393 bp with CB64, respectively), showing that both splice variants were present in poly(A)+ mRNAs (Fig. 3, lanes 2 and 3). 5′-RACE-PCR with CB62 and CB64 confirmed that the whole 5′ ends of both sequences were identical, and that no other intron was retained in the 5′ end of clone 2 sequence (not shown).
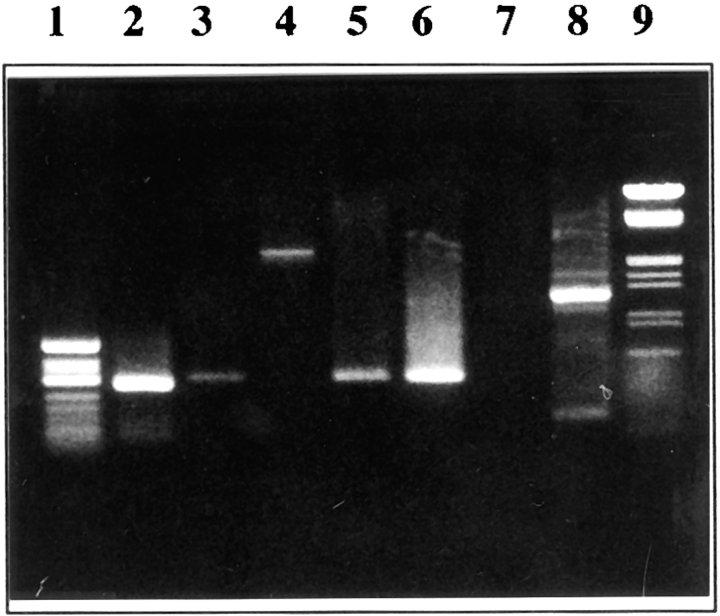
Figure 3.
PCR characterization of alternative splicing in cyclin B mRNA. (Lanes 1 and 9) Molecular mass markers (φ X 174/HaeIII and λ/EcoRI + HindIII, respectively). (Lanes 2 and 3) RT-PCR was done using poly(A)+ mRNA from unfertilized eggs as a template, and CB5-CB64 and CB5– CB62 primer pairs, respectively (see Fig. 2). Both pairs amplified fragments of the expected sizes. (Lanes 4 and 5) The same primer pairs were used, with genomic DNA as a template. While CB5-CB62 gave rise to the right-sized PCR product (compare lanes 3 and 5), CB5-CB64 amplified a 2.5-kbp PCR fragment (compare lanes 2 and 4). This last product was reamplified with CB5-CB62 and gave rise to a right-sized product (compare lanes 3, 5, and 6), showing part of the clone 2 cDNA sequence comprising primer CB62 was included in the 2.5-kbp genomic CB5-CB64 fragment. (Lanes 7 and 8) PCR amplification of genomic DNA with respectively primers CB8-CB64 and CB10-CB64 (see Fig. 2). Only CB10-CB64 amplified a fragment (1,300 bp, lane 8), whereas amplification with CB8-CB64 was unsuccessful, showing part of the clone 2 cDNA, comprising primer CB8, was not included in the 2.5-kbp genomic CB5-CB64 fragment. All PCR products showed in this figure were confirmed by cloning and partial sequencing (not shown).
The presence of an intron was confirmed by PCR using genomic DNA as a template and primer pairs CB5-CB64 or CB5-CB62. As expected, CB5-CB62 generated a 425-bp fragment, whereas CB5-CB64 PCR resulted in the amplification of an ∼2.5-kbp long fragment (Fig. 3, lanes 4 and 5). Further reamplification of the latter PCR product with CB5-CB62 gave rise to the expected 425-bp fragment (Fig. 3, lane 6). While CB5-primed sequencing of the 2.5-kbp product confirmed its identity with clone 2 sequence, CB64-primed sequence matched with clone 4 until a splice acceptor site (CAG/GCTGCA), but failed to match with the end of 3′ untranslated clone 2 sequence thereafter. Based on the NsiI restriction map we could localize the region of divergence and designed primers CB8 and CB10 to confirm this observation by PCR, using genomic DNA as a template. Although CB10-CB64–primed PCR gave rise to a 1,254-bp product, CB8-CB64–primed PCR was unsuccessful (Fig. 3, lanes 8 and 7, respectively). Finally, whole sequencing of the 2.5-kbp genomic fragment showed it contains another putative intron between canonical donor (CAT/GTCTGT) and acceptor sequences (Fig. 4).
Presently, in our work on the CB5-CB64 genomic PCR product, we have shown that clone 4 cDNA sequence (further reported as CB sequence) arises from removal of an intron between nt 104 and 2114 from the beginning of CB5. In contrast, in clone 2 cDNA sequence (further reported as CBsv sequence), the 5′ part of this intron is retained (alternative splicing), and another overlapping intron, comprising part or all of CB 3′ sequence, is removed downstream of nt 1267 (Fig. 4). Further work on the cyclin B gene will allow us to determine whether other splicings play a role in rising 3′ ends of both CB and CBsv mRNAs.
Developmental Regulation of CB and CBsv Messengers
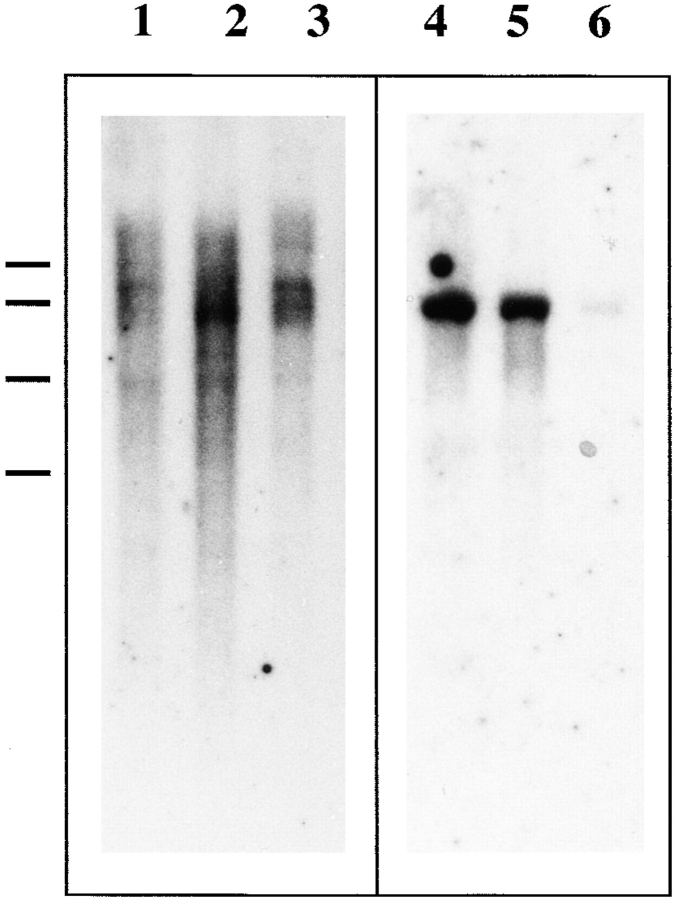
Northern blots were performed with 1 μg poly(A)+ RNA from unfertilized eggs, 2-cell embryos, and swimming blastulae. A CBsv-specific probe hybridized faintly but equally with three major bands (∼4, 5, and 5.5 kbp long) in mRNAs from all origins (Fig. 5, lanes 1–3). On the same blot after dehybridization, a CB-specific probe hybridized with a major 4.5-kbp mRNA, strongly in both unfertilized and stage 2 eggs, and faintly in larvae (Fig. 5, lanes 4–6). This result has two major implications: (a) both cDNAs, even completed by 5′ RACE-PCR, are not full length, since mRNAs are ∼5 kbp long; and (b) CB mRNA, which is the most homologous to known B cyclins, encodes the very major part of maternal cyclin B. After replacement of maternal mRNAs by embryonic ones the predominance of CB on CBsv mRNA is reduced.
Figure 5.
Relative abundance of CB and CBSV mRNAs varies with developmental stage. Northern blots of similar amounts (1 μg as estimated by densitometry) of poly(A)+ mRNAs from unfertilized eggs (lane 1), from 2-cell stage (lane 2), and from swimming late blastulae (lane 3) were probed with a probe specific for CBSV (lanes 1–3). After exposure for 50 h the blot was dehybridized and reprobed with a probe specific for CB and then reexposed for 18 h (lanes 4–6). CBSV mRNAs are essentially distributed in bands between 4 and 5.5 kbp, whereas a single prominent 4.5-kbp CB mRNA is evidenced. As we have used the same blot to visualize first CBSV and then CB mRNAs, we can state that the relative amount of CB mRNA with regard to CBSV mRNA decreases dramatically in swimming larvae, although no rigorous quantitation of poly(A)+ mRNAs was performed.
CBsv Lacks a Sequence Universally Conserved among Mitotic Cyclins
Visually we aligned the COOH-terminal amino acid sequences of 72 mitotic cyclins, including plant A and B types and the three known B3 cyclins, starting at the beginning of the last translated exon (AVKT... in sea urchin cyclin B). We chose a simple algorithm to estimate the degree of amino acid conservation at each given position (Fig. 6). In cyclin B, amino acids at positions 5 (K) and 6 (Y) are absolutely conserved, as they are in cyclins A with exception of the only sequenced Caenorhabditis elegans cyclin A (NY). Three other clusters display more or less good conservation: at position 10 (K), 14 (V in cyclin A) and 15 (S or A), and 20 (L in cyclin B, P in cyclin A). Accordingly, a search for homology with the COOH-terminal sequence of CB resulted in significant homology with various A and B type cyclins. In contrast, the CBsv COOH terminus displayed no significant homology with any known protein. These results support the view that, if CBsv displays a sequence unusual in other cyclins, most likely its meaningful peculiarity is the lack of the KY and amino acids conserved in all mitotic cyclins.
Figure 6.
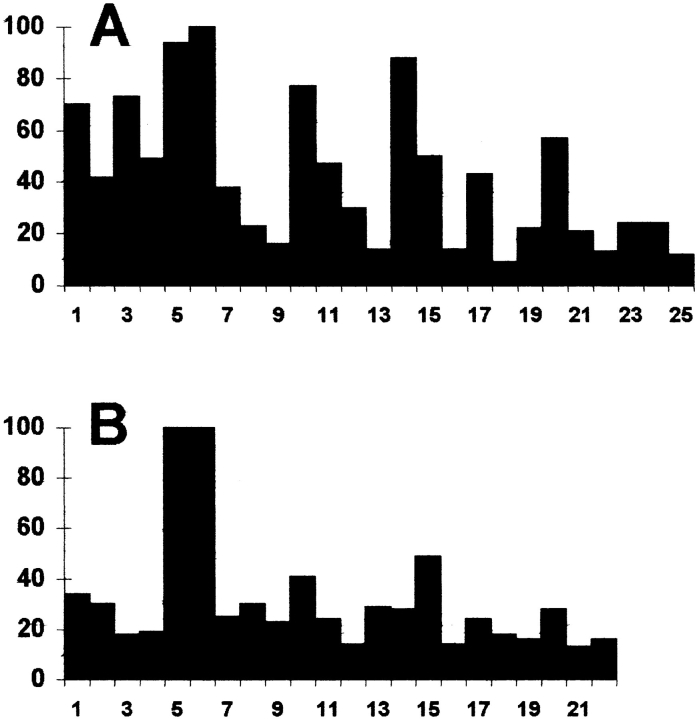
Conservation of amino acids at the COOH terminus of A type cyclins (A) and B type cyclins (B). Amino acid 1 is the first of the last translated exon in classical cyclin A and B. 33 cyclin A sequences and 39 cyclin B sequences were visually aligned and compared over the length of the shortest one (25 aa in A, 22 aa in B). The occurrences of each amino acid at a given position were cumulated, and the level of conservation was calculated as the summation of the squares of each score at a given position. The highest possible conservation (the same amino acid at a given position in overall sequences: 332 = 1089 for cyclin A, and 392 = 1521 for cyclin B) was taken as 100%. For example, for three sequences at a given position, the highest conservation will be obtained with three identical aminoacids at this position, thus 32 = 9, 9/9 = 100%; with two identical aminoacids and one divergent, 22 + 1 = 5, 5/9 = 56%; with three different aminoacids, 1 + 1 + 1 = 3, 3/9 = 33%. Given that (a + b)2 > a 2 + b 2, this method is simple and discriminates well the level of conservation. This figure shows that cyclin A COOH terminus is better conserved than cyclin B, and points out the high level of conservation of aminoacids 5 (K) and 6 (Y) in both families of mitotic cyclins.
Localization of CBsv
We raised antibodies specific for CBsv or CB COOH-terminal sequences and looked for the immunofluorescence pattern of both proteins during early embryogenesis. Sea urchin eggs are laid arrested in G1-phase of the first cell cycle after completion of meiosis. Fertilization then induces entry into early embryogenesis, and cell cycles occur synchronously in a given embryo until the fourth cell cycle (16-cell stage). At that time divisions in the vegetal half become unequal and essentially give rise to the precursors of both endoderm and mesoderm (12).
During the first steps of cleavage, CBsv staining was very low compared to CB staining. The most striking difference was that detergent treatment (1% NP-40) before methanol fixation removed most of CBsv staining that was found diffuse around the spindle area during mitosis (not shown), whereas CB-specific staining was, as already described, closely associated to microtubular structures. Thus, in the following results we used only direct methanol fixation to describe CBsv distribution.
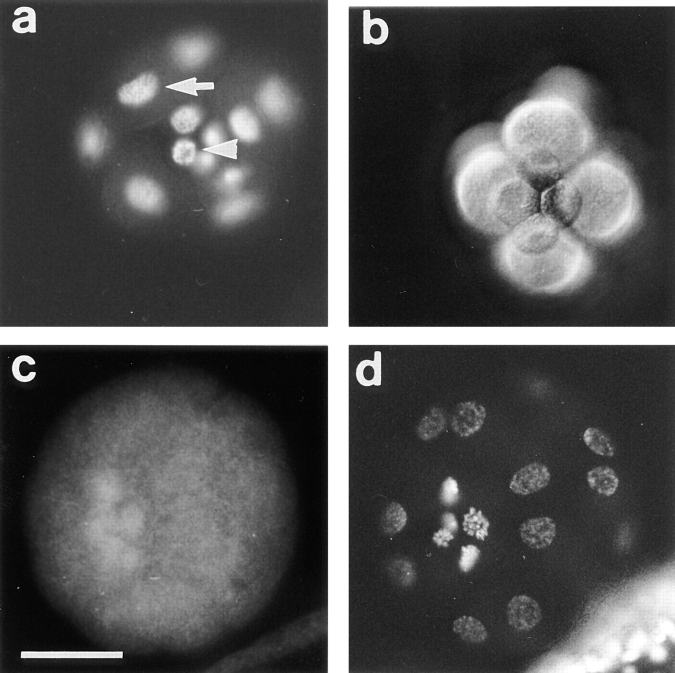
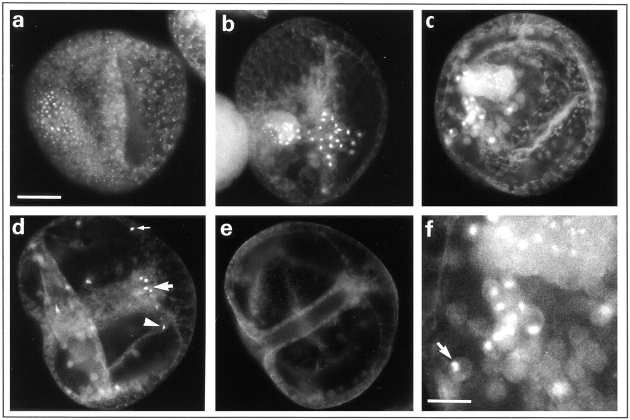
In sea urchin, the midblastula transition is not a one-step event occurring in all the cells at the same time, as it was shown in amphibians for example (25, 32). The first asymmetrical division occurs in vegetal blastomeres at the fourth cleavage and gives rise to micromeres and macromeres. In Sphaerechinus granularis, while divisions occur every 35 min in macromeres and animal mesomeres, the fifth cycle lasts ∼60 min in micromeres. Nevertheless, BrdU incorporation showed that the S-phase was not delayed in micromeres (Fig. 7 a). Thus, lengthening of the fifth cell cycle in micromeres was due to the acquisition of a G2-phase rather than a G1-phase. Furthermore, asymmetrical division and lengthening of the cell cycle occurred even in the presence of microinjected α-amanitin (200 μg/ml final concentration), a potent inhibitor of transcriptions (Fig. 7 b), showing that these events are under maternal control in the vegetal blastomeres and micromeres. We showed that micromeres overproduce CBsv, compared with macromeres and mesomeres (Fig. 7 c), and this holds true in the presence of α-amanitin (not shown). After hatching, which occurs at the late blastula stage (mesenchyme blastula), a strong cytoplasmic staining appeared in the bud of archenteron (Fig. 8 a). Occasionally at this stage a significant staining of almost all the cells of the embryo could be detected, as shown in Fig. 8 a. During invagination and differentiation of the archenteron, cells at the tip kept an intense staining, together with some blastocoelar cells (Fig. 8, b and c). Finally, archenteron cells, blastocoelar cells, and pigment cells of the late blastula were also strongly and specifically (see control in Fig. 8 e) stained by CBsv antibodies (Fig. 8 d). Staining inside individual cells could be clarified in large blastocoelar cells; CBsv accumulated in a single cytoplasmic structure near the nucleus (Fig. 8 f). Only immunogold labeling will confirm whether this structure is associated with the centrosome. As shown on Fig. 8 e, anti-CBsv antibodies are really specific for the CBsv peptide, but this did not rule out the possibility that the same sequence was shared by another protein, whose expression could be also developmentally controlled. For this reason, we compared the CBsv staining with the staining by a polyclonal antibody raised against the full-length Arbacia punctulata cyclin B in the presence of the CBsv peptide. The polyclonal antibody displayed the same strong staining as anti-CBsv in the bud of the archenteron and a more general staining (probably of CB) in all the cells of the embryo (Fig. 9, a and b). Moreover, we inferred from our own experience that the intronic sequence is not conserved between different species, and thus, that anti-CBsv should not recognize the splice variant in another sea urchin species. Accordingly, the polyclonal anti–Arbacia punctulata cyclin B strongly stained the bud of archenteron (Fig. 9 c), whereas anti-CBsv did not recognize any epitope (Fig. 9 d), in mesenchyme blastulae of the related species Paracentrotus lividus. This strongly supports the view that the pattern displayed by anti-CBsv represents really the distribution of CBsv protein in the embryo.
Figure 7.
Acquisition of a G2-phase in micromeres is under maternal control and coincides with accumulation of CBsv. (a) BrdU (1 mg/ml) was added during the fourth M-phase to sea urchin embryos. Embryos were directly fixed in 4 N HCl 10 min after the cytodieresis and then processed for BrdU detection, which showed that replication occurred in micromeres (arrowhead) to the same extent as in macromeres (arrow). As cell cycle is much longer in micromeres, this shows that the first G-phase appearing in micromeres is a G2-phase. (b) 200 μg/ml (final concentration) α-amanitin was microinjected in eggs before fertilization. This concentration was far higher than necessary (50 μg/ml) to inhibit hatching and gastrulation (not shown); nevertheless, in microinjected embryos, normal micromeres formed in schedule with control embryos. (c and d) Embryos were fixed after the fifth cell cycle in macromeres and mesomeres and processed for CBsv detection (c) and DNA Hoechst staining (d). While macromeres and mesomeres were in S-phase of the 6th cell cycle, micromeres were in early prophase of the fifth cycle and overexpressed CBsv. Bar, 35 μm.
Figure 8.
CBsv staining along embryogenesis. (a) Mesenchyme–blastula stage (late blastula). On the left a set of cells, representing the bud of archenteron, are heavily stained. Moreover, note that all the cells of this embryo are more or less stained (compare with the following pictures). (b) Young gastrula with a just growing archenteron. CBsv is localized in the cells of the archenteron, and in some secondary mesenchyme cells that have migrated inside the blastocoel. (c) Young gastrula at a later stage. The same categories of cells are stained as in Fig. 9 b. (d) Late gastrula. The tip of the archenteron has reached the apex of the embryo. A few cells (∼50 per embryo at a given time) are heavily stained by CBsv immunofluorescence: at the tip of the archenteron (large arrow), inside the blastocoel (filopodial cells, arrowhead), and at the periphery (presumably pigment cells, tiny arrow). (e) Control staining with anti-CBsv antibody loaded with recombinant CBsv. f is an enlargement of part of c showing that CBsv is concentrated in the cytoplasm as a dot near the nucleus of blastocoelar cells (arrow). The nucleus is seen as a black hole, whereas the surrounding cytoplasm is background stained. Bars: (a–e) 50 μm; (f) 20 μm.
Figure 9.
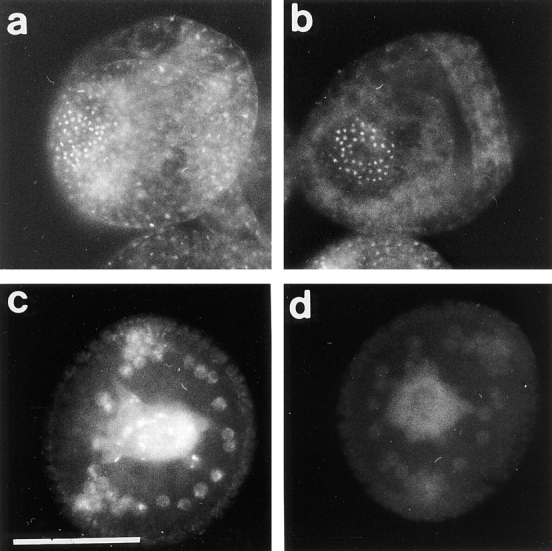
Anti-CBsv antibodies are specific for the Sphaerechinus granularis splice variant of cyclin B. 20-h-old embryos of S. granularis (a and b) or Paracentrotus lividus, another species of regular sea urchin, were methanol-fixed and processed for cyclin B detection with a polyclonal antibody raised against full-length Arbacia punctulata cyclin B (a and c) or for CBsv detection with the anti-CBsv antibody (b and d). Anti–cyclin B antibody clearly detected the overexpressed CBsv with a similar pattern in both species, even in the presence of 1 mg/ml CBsv peptide (a and c). In contrast, anti-CBsv antibody, which is directed against a specific intronic sequence in S. granularis, did not recognize any epitope in the related species P. lividus (b and d). Bar, 50 μm.
The staining of micromeres after the fourth division showed that CBsv was overproduced, by specific translation of maternal RNAs, in cells that were committed to stay for a long time in G2-phase. This prompted us to search for the expression of CBsv in growing oocytes, in which G2-phase is also long lasting. The process of meiosis reinitiation of sea urchin oocyte is not known. Growing oocytes are arrested in prophase of the first meiotic division, as in all animal species, and large oocytes quite often undergo spontaneous meiosis reinitiation when released in seawater by gonad dissection or artificial spawning induction.
Small oocytes were heavily stained by CBsv antibodies compared with eggs or early embryos (Fig. 10, a and b). CBsv was distributed essentially as dots in a thick cortical layer, and a few (1– 3) discrete dots in the germinal vesicle, without a clear association with nucleolus or chromatin (Fig. 10, c and d). Large oocytes displayed the same pattern, although the overall fluorescence intensity was lower than in small oocytes (not shown). After resumption of meiosis only a background staining was observed in G1 arrested eggs.
Figure 10.
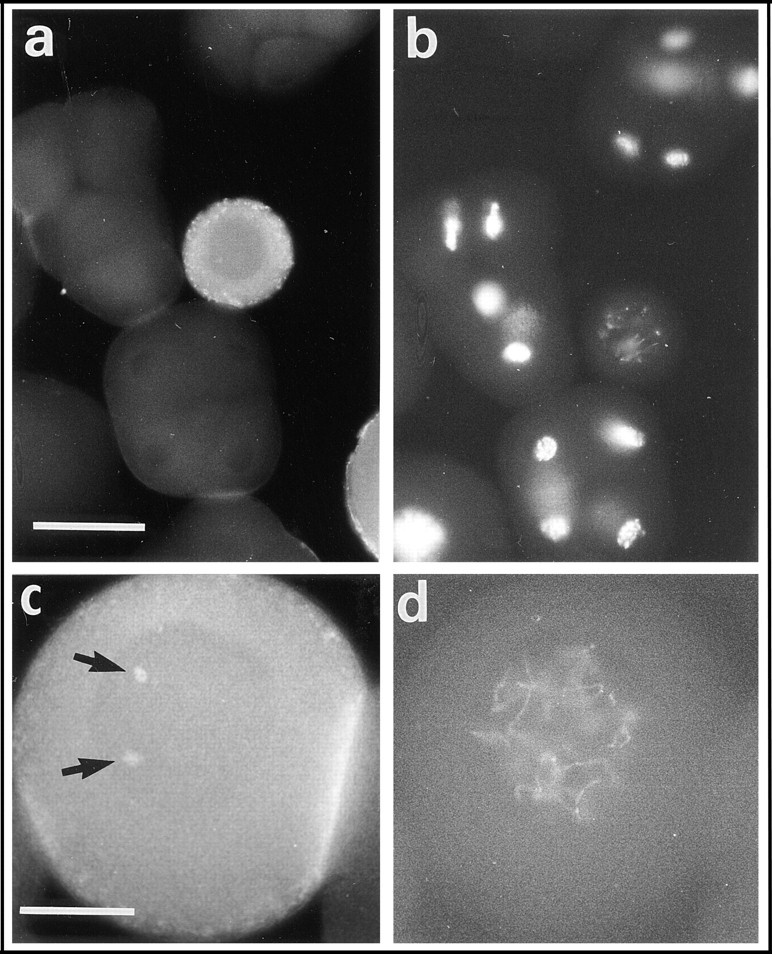
CBsv distribution in prophase-arrested oocytes (left) and corresponding Hoechst DNA staining (right). (a and b) View at small scale of a field comprising a small oocyte between various embryos, showing CBsv is prominently concentrated in the cortical layer of growing oocytes. (c and d) A near full-grown oocyte that also displays a cortical accumulation of CBsv and two intranuclear dots that do not seem to correspond to any chromatin structure (arrows). Such a nuclear staining in one to three dots was constantly observed in oocytes. Bars: (a and b) 55 μm; (c and d) 20 μm.
Excess CBsv Delays MPF Activation in Starfish Oocytes
In prophase-blocked oocytes (germinal vesicle stage), cyclin B in most species is associated with tyrosine 15– and threonine 14–phosphorylated cdc2 in an inactive pre-MPF complex. Meiosis reinitiation is achieved by dephosphorylation of T14-Y15, upon stimuli varying according to material. A natural hormone, 1-methyl-adenine (1-MeAde) at micromolar concentration, triggers starfish oocytes maturation in a very well-known and reproducible way (10, 15, 16). Thus, we used this convenient model to compare the effect of CB and CBsv protein microinjection. We microinjected CB and CBsv recombinant proteins (∼1 μM final concentration) and searched for changes in the process of meiosis reinitiation.
Compared to control (buffer microinjection), CBsv microinjection considerably delayed breakdown of the germinal vesicle (GVBD), whereas CB microinjection significantly advanced it (Table I). Both recombinant cyclin isoforms delayed exit from meiosis I as shown by late first polar body emission (Table I). Accordingly, microinjection of fivefold diluted solutions had only reduced effects (Table I).
Table I.
CB Microinjection Advances, and CBsv Microinjection Delays Initial Steps of Hormone-induced Meiosis Reinitiation in Starfish Oocytes
| Microinjection (final concentration) | Time of 50% GVBD | Time of 50% pb1 emission | ||
|---|---|---|---|---|
| Min post-hormone addition | ||||
| Control buffer | 27 | 90 | ||
| CB (1 μM) | 22 | 123 | ||
| CB (0.2 μM) | 25 | 95 | ||
| CBsv (1 μM) | 50 | 145 | ||
| CBsv (0.2 μM) | 32 | 95 | ||
Each series of microinjections (lines) concerned 10 oocytes, except for controls that were 20. The experiment was made in two parts (one for CB, one for CBsv). The protein was first microinjected at two concentrations, then ten controls were microinjected with the buffer (TBS). 30 min after the last microinjection all the samples of the series were allowed to mature with 1 μM 1-MeAde. GVBD and completion of first polar body (pb1) emission, which are obvious events of meiosis, were scored every 2 min. The times of 50% GVBD and 50% pb1 were estimated from the obtained cumulative curves.
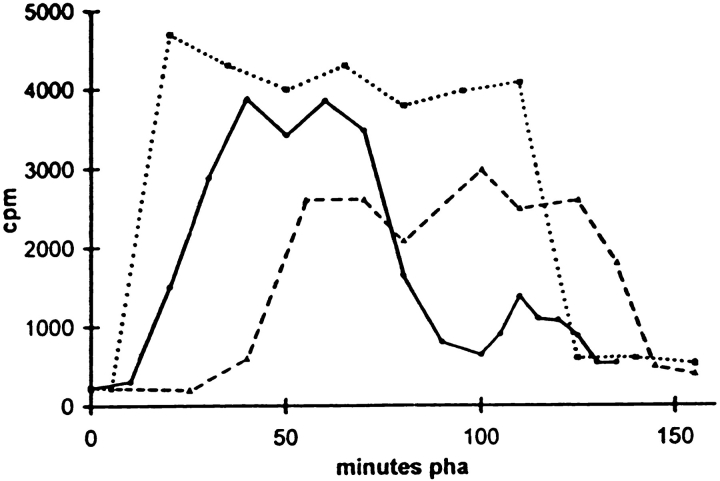
During starfish oocyte meiosis, MPF is the only cycling H1 kinase (20). Whole H1 kinase activity in homogenates, reflecting cyclin B–dependent kinase activity, can be easily measured on individual oocytes in Astropecten aranciacus. The striking changes in cytological events were confirmed and completed by time course measurements of H1 kinase activity in microinjected oocytes. Compared with controls, H1 kinase raised in advance and dropped late in oocytes microinjected with CB (Fig. 11). On the contrary, in CBsv-microinjected oocytes H1 kinase raised and dropped later than in controls (Fig. 11). The average level of kinase activity, measured on 10 oocytes at 60 min after hormone addition, was 10% higher in CB microinjected, and 20% lower in CBsv microinjected, compared to buffer microinjected control oocytes (not shown).
Figure 11.
CBsv microinjection delays activation and reduces activity of the mitotic H1 kinase during hormone-stimulated meiosis reinitiation of starfish oocytes. A. aranciacus oocytes were microinjected with 1 μM (final concentration) CB or CBsv and then allowed to mature. Individual oocytes of each batch, comprising one batch of uninjected control oocytes, were periodically sampled and frozen in 2 μl of distilled water. H1 kinase assay was performed by addition of 8 μl of the assay buffer containing 5 mM MgCl2, 50 μM ATP, 0.5 mg/ml histone H1, 5 mM Hepes, pH 7.0, and 50 μCi γ-[32P]ATP, for 10 min. After SDS-PAGE, the band containing histone H1 in each sample was excised and scintillation counted. This experiment was partially reproduced three times more with essentially the same results. In controls (solid line) H1 kinase reached maximal activity at 40 min, dropped almost completely at 100 min, and rose again for the second meiotic cell cycle. CB microinjection (dotted line) resulted in a significant advance in activation of the kinase, since the maximal activity was reached at 20 min, in an increase of the maximal activity, and a delay for mitosis exit. In contrast, CBsv microinjection (broken line) resulted in a large delay for activation of the kinase (maximal value at 55 min) and a lowering of its activity.
The way by which 1-MeAde triggers MPF activation is not yet well understood. Former experiments on oocytes maturation showed that one can experimentally discriminate two steps. During the first one, a little amount of active MPF is produced (MPF initiation), and during the second one this small MPF activity induces activation of the whole MPF in the cell (MPF amplification). MPF-containing cytoplasm microinjection, which is supposed to mimic MPF initiation, directly triggers MPF amplification, provided a sufficient amount of cytoplasm is transferred. This allowed us to discriminate between a CBsv-induced inhibition of MPF initiation or amplification: cytoplasmic transfer showed that the sensitivity to microinjected MPF-containing cytoplasm was not altered by a previous CBsv microinjection (Table II). Moreover, the mean time of GVBD after cytoplasmic transfer (31 min) did not vary significantly in this experiment. This shows that only the early step of MPF initiation is concerned by CBsv-induced inhibition. In contrast, MPF amplification was greatly facilitated by CB microinjection (Table II).
Table II.
Previous Microinjection of CBsv Does Not Reduce Efficiency of Mitotic Cytoplasm Transfer in Triggering Meiosis Reinitiation in Starfish Oocytes
| Recipient oocytes | Volume of cytoplasmic transfert | GVBD % (nb GVBD/nb microinjected) | ||
|---|---|---|---|---|
| pl | ||||
| Buffer microinjected | 600 | 50% (5/11) | ||
| Buffer microinjected | 400 | 10% (1/9) | ||
| CB microinjected (1 μM final) | 600 | 100% (11/11) | ||
| CB microinjected (1 μM final) | 400 | 90% (9/10) | ||
| CBsv microinjected (1 μM final) | 600 | 50% (5/10) | ||
| CBsv microinjected (1 μM final) | 400 | 13% (2/15) |
Astropecten aranciacus oocytes (∼10 nl in volume) were microinjected first with buffer (controls), CB, or CBsv, and then with cytoplasm taken from donor maturing oocytes. In brackets are the number of oocytes with GVBD/number of microinjected oocytes in each experiment. Clearly, CB microinjection facilitates meiosis reinitiation, whereas CBsv microinjection has no obvious effect.
Excess CBsv Slows Down and Disturbs Early Embryogenesis in Sea Urchin
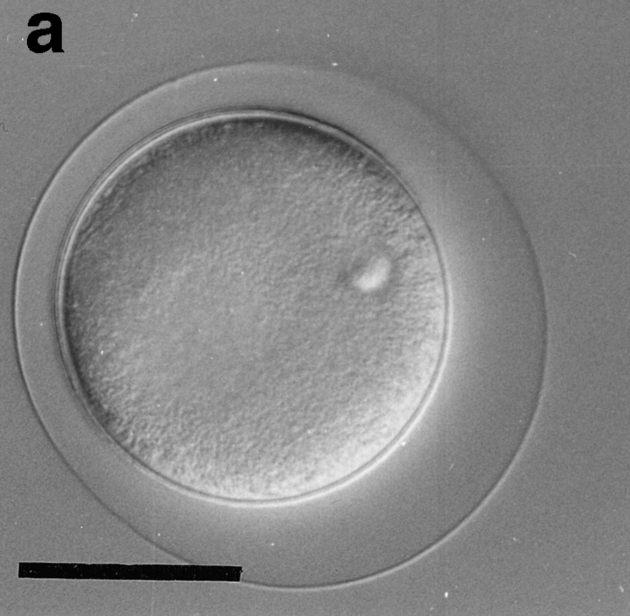
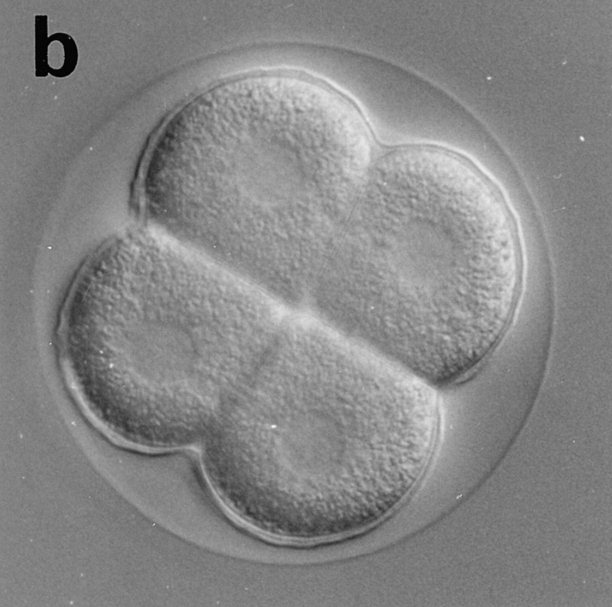
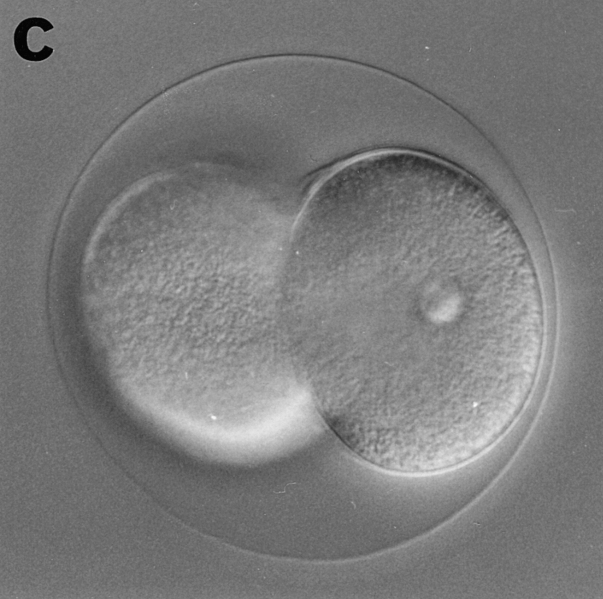
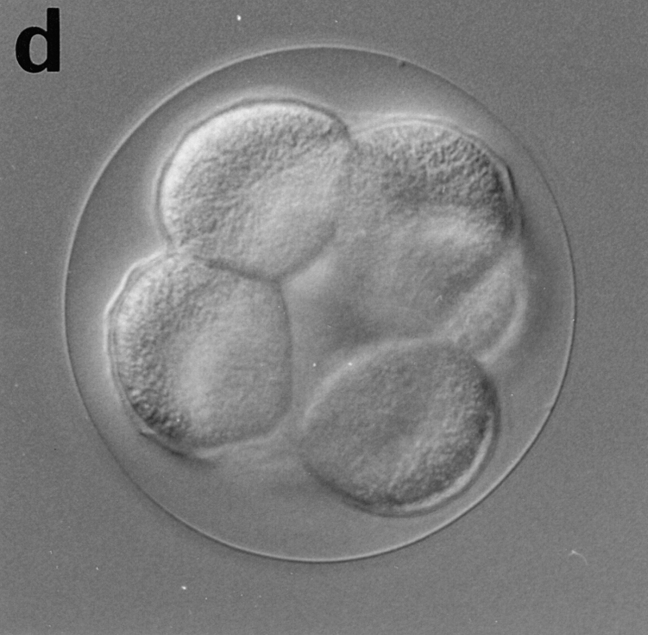
Eggs of Sphaerechinus granularis were microinjected with recombinant CBsv protein (1 μM final), then fertilized, and development of microinjected embryos was compared with controls. Entry and exit of the first M-phase were both considerably delayed, since microinjected eggs were in metaphase of the first cell cycle when controls were in metaphase of the third cell cycle (Fig. 12, a and b). BrdU incorporation showed that S-phase entry was only slightly delayed by ∼15 min (45 min against 30 min in controls), which could be explained, at least partially, by the injury due to microinjection. After completion of the first cell cycle, the rhythm of cleavages became more rapid (Fig. 12 c), but disorders in the synchronism and in the size of the cells were observed later (Fig. 12 e), and generally cytolysis occurred in morulae containing less than 100 cells (not shown).
Figure 12.
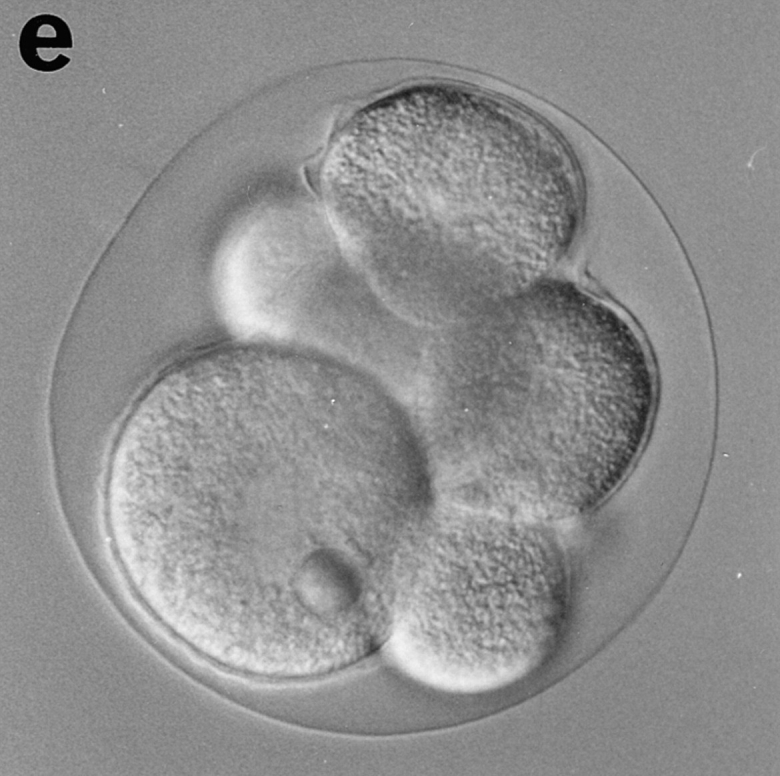
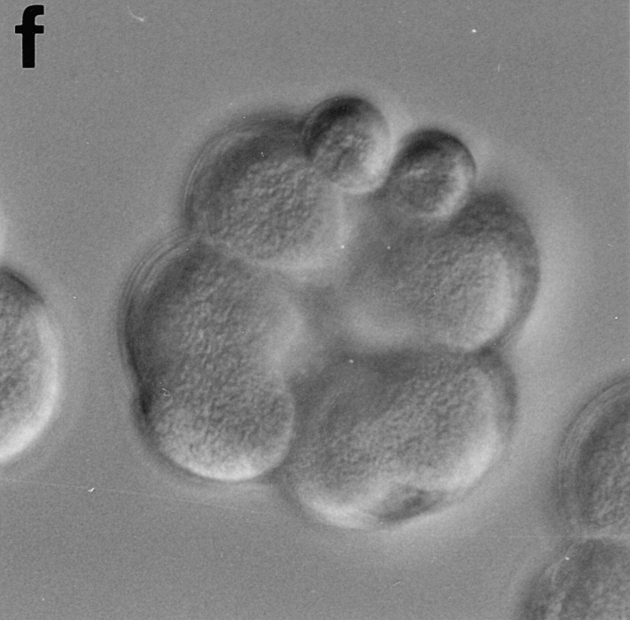
CBsv microinjection induces first cell cycle lengthening and developmental disorders in sea urchin embryos. CBsv (1 μM final concentration) was microinjected before fertilization in sea urchin eggs. Pictures were then taken during cleavage of microinjected (a, c, and e) or control uninjected (b, d, and f) embryos. (a and b) At 140 min controls were in metaphase of the third cycle, whereas CBsv microinjected just initiated anaphase of the first cycle. (c and d) At 170 min, controls were at the 8-cell stage, and microinjected eggs at the 2-cell stage. (e) At 240 min, injected embryos displayed various numbers of blastomeres irregular in size, here six blastomeres, one large and five small. (f) At 205 min, control embryos formed micromeres. a, c, and e are successive pictures of the same microinjected embryo. Bar, 50 μm.
Both CB and CBsv Bind to cdc2 with Differential Affinity and Rescue a Triple CLN Deletion in Budding Yeast with Differential Efficiency
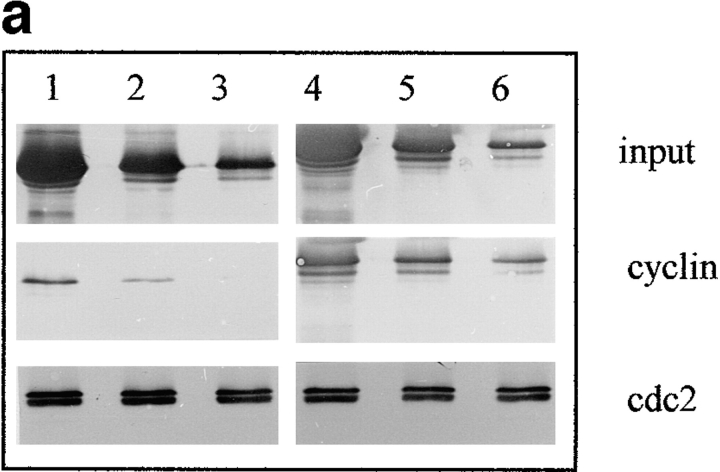
Previous reports showed that a recombinant COOH terminally deleted cyclin A does not bind cdk2 in vitro (17). This holds true for cyclin B2-cdc2 complex in interphase acellular extracts, in which a recombinant COOH terminally truncated GST-cyclin B2 was unable to bind efficiently endogenous cdc2 (34). To confirm these results we microinjected similar amounts of recombinant CB and CBsv in 10 Xenopus oocytes at different concentrations (1, 0.2, and 0.04 μM final concentration). After 2 h these oocytes were frozen in 1 ml EB (15 mM EGTA, 60 mM Na-β-glycerophosphate, 250 mM KCl, pH 7.4), and the soluble material processed for p13suc1 affinity purification. Western blot analysis of the material retained on p13 beads showed that CB was recovered much more efficiently than CBsv, whereas similar amounts of cdc2 were recovered (Fig. 13 a).
Figure 13.
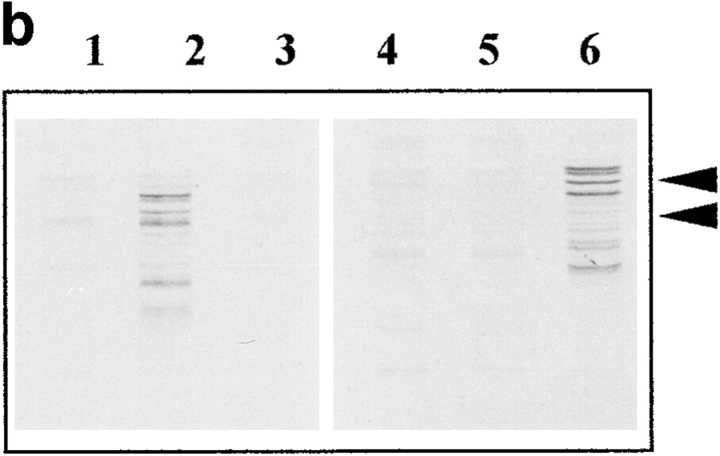
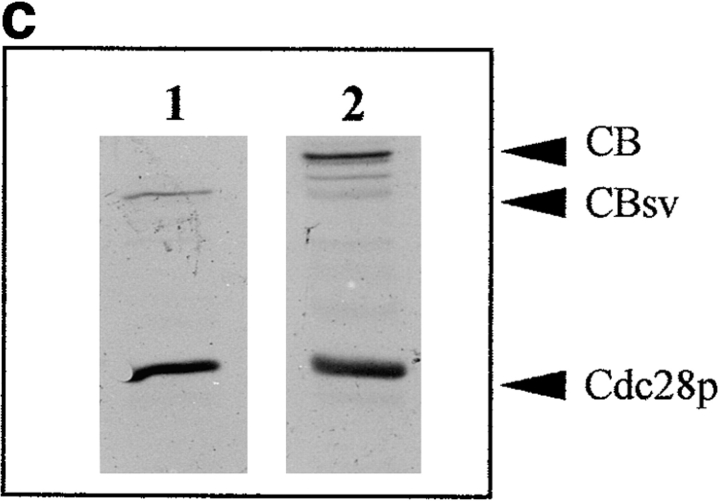
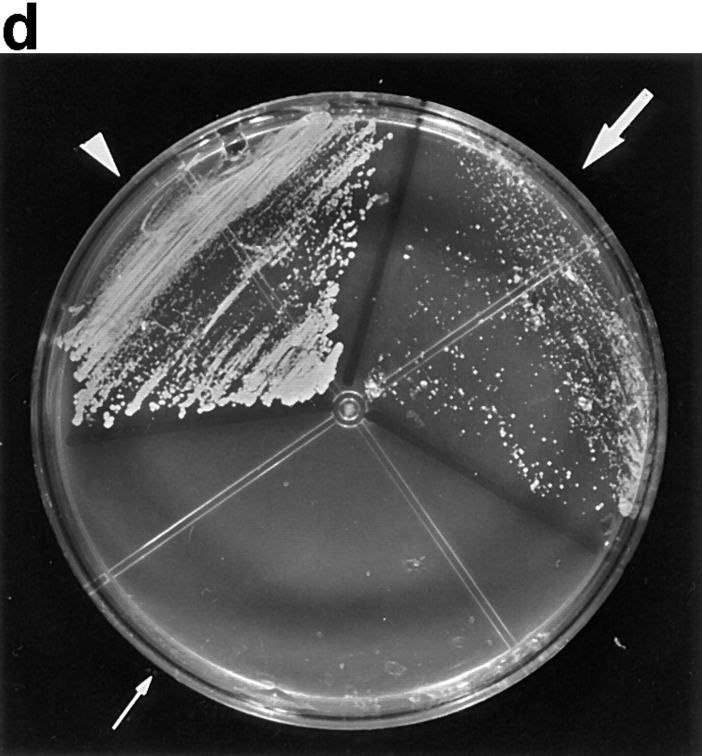
CBsv binds to cdc2 with a reduced affinity and is less efficient than CB for rescuing a CLN1-3 triple deletion in budding yeast. (a) Western blot showing binding of CBsv (lanes 1–3) and CB (lanes 4–6) after microinjection of the recombinant proteins in Xenopus laevis oocytes: (input) total amount of microinjected material; (cyclin) amount of cyclin (CB or CBsv) recovered on p13suc1 beads after a 2-h incubation in the cells: CB (right) was better recovered than CBsv (left), whereas the amount of cdc2 in the purified material was constant (cdc2). (b) The triple CLN deleted strain 589-5 was transformed with either the vector pADNS alone (lanes 1 and 4), pADNS containing CBsv ORF (lanes 2 and 5), or pADNS containing CB (lanes 3 and 6). Homogenates of the three transformed strains grown on galactose were subjected to detection with CBsv antibody (lanes 1–3, lower arrowhead), or with CB antibody (lanes 4–6, upper arrowhead). This panel shows that similar amounts of both proteins were efficiently produced, together with homologous degradation products. (c) Supernatants of the same homogenates were subjected to p13suc1 affinity purification, then the purified material detected with antibodies specific for CB (lane 2, upper arrowhead), or CBsv (lane 1, lower arrowhead), showing that the binding of CBsv to endogenous CDC28p (in similar amounts on p13 beads, Cdc28p) was reduced compared with the binding of CB. (d) Accordingly, when grown on dextrose, CB-transformed cells (arrowheads) were efficiently rescued, whereas CBsv-transformed cells grew only hardly (arrow). Cells transformed with vector alone (tiny arrow) did not grow at all.
Several previous studies (see reference 18 and our unpublished observations) have shown that mitotic cyclins (A and B types) are able to functionally complement the deletion of the three G1-S cyclins (CLN1 to 3) in budding yeast, where the same catalytic subunit, CDC28p, drives both G1-S and G2-M transitions. We introduced the ORFs of CB and CBsv behind the constitutive yeast ADH promoter in the vector pADNS. This construct was introduced in the yeast strain 589-5, in which CLN1 to 3 are deleted, and CLN3 is conditionally expressed on galactose by the GAL1 promoter (18). Rescue of this strain thus allows the colonies to grow on glucose. Western blots of homogenates and p13-purified material showed that both proteins were efficiently and similarly expressed (Fig. 13 b), but that CBsv did bind to CDC28p, the yeast homologue of cdc2, with a reduced affinity compared with CB (Fig. 13 c). Accordingly, CBsv was only hardly able to complement the yeast strain (Fig. 13 d): whereas the doubling time in the strain complemented by CB was 4.5 h, it was 10 h in the strain complemented by CBsv.
Discussion
In this work we have given evidence for the first time that two variants of the cyclin B protein can be generated by alternative splicing of a premessenger RNA. Although we failed to detect the rare mRNA corresponding to CBsv cDNA in a purified polysomal fraction (not shown), we cloned both sequences from oligo(dT)-primed cDNA and amplified both sequences in RT-PCR with poly(A)+ mRNA. Moreover, the structure of part of the cyclin B gene shows that CBsv sequence cannot result from an incomplete maturation of the premessenger, in which only one intron could have been retained, since another intron has to be removed to give rise to the 3′-untranslated CBsv sequence.
Only rare reports have mentioned alternative splicing affecting the translated sequence of cyclins. A chicken cyclin B3 cDNA isoform has been cloned, in which an in-frame intron was retained in the 5′ part of the ORF (6). However, in this case RT-PCR amplification of a corresponding product with appropriate primers was unsuccessful. According to the authors, a tempting significance of this alternative splicing in cyclin B3 is that the unspliced variant could have an altered destruction behavior. However, the functional role of cyclin B3 itself is not yet known. Finally, the only well-characterized alternative splicings in a cyclin ORF have been found in human cyclin E (23, 31). In the first report, a cyclin E splice variant (cyclin Es) that lacks part (49 aa) of the cyclin box was evidenced in Hela cell extracts. As expected, cyclin Es does not bind to cdk2, and thus is unable to activate it. However, the significance of cyclin Es remains an enigma, since it was shown unable to compete with cyclin E for developing histone H1 kinase activity in vitro. Probably, the role of cyclin Es will be explained by its association with other components of the cell cycle machinery. The second variant, cyclin ET, has an intact cyclin box, but has been found unable to complement a triple CLN mutant strain of S. cerevisiae, or to impede complementation of this strain by cyclin E. Once more, the role of cyclin ET remains elusive today.
Thus, our report is the first one describing alternative splicing in a B type cyclin ORF. Given the universal role of cyclin B in mitosis regulation and generally in proliferation, the comparative study of biochemical properties of both variants will be of crucial interest. As the specific amino acid sequence of CBsv is very short (7 aa) and does not share homology with other cyclins, CBsv most probably represents a shorter cyclin that lacks significant specific sequence. Although the COOH terminus is not usually considered as belonging to cyclin box, its sequence is well conserved among B type cyclins. In particular, a KY (lysine386–tyrosine387 in human cyclin B1 sequence) is absolutely conserved in mitotic cyclins, even in yeast and plant B cyclins, cyclin B3, or almost all known A cyclins, but is lacking in CBsv.
This study confirms previous observations that the COOH-terminal sequence of cyclin B is required for a correct cyclin–cdk association, at least in vivo, although in Xenopus oocytes as well as in yeast, CBsv was found to associate with cdc2, but with a reduced efficiency. Accordingly, the splice variant CBsv was hardly able to complement a triple CLN deletion in budding yeast. The fact that CBsv associates poorly with cdc28, whereas CB associates well enough to allow a correct functioning of the kinase, raises a puzzling problem: in contrast to higher eukaryotes cdc2, CDC28 in yeast is able to associate in the cell with mitotic and G1 cyclins, and in particular human cyclin E has been cloned by complementation of the CLN-deleted yeast strain (18). Yet, G1 cyclins do not share the COOH-terminal conserved sequences of mitotic cyclins, nor even similar motifs. This, and the fact that in crystallized cyclin A–cdk2 complex this sequence does not interact directly with cdk2 (14), suggests that it is required in mitotic cyclins only to allow a correct folding of parts of the molecule implicated in the interaction.
The second observation is that an excess of CBsv severely delays G2-M transition in hormone-stimulated starfish oocytes, as well as in fertilized sea urchin eggs. The low affinity of CBsv for cdc2 makes unlikely that the variant dimers could constitute dominant negative molecules. Moreover, yeast complementation showed that the dimer CBsv-CDC28 is probably active, since the strain 589-5 was effectively complemented. For the same reason, and also because cyclin B–cdc2 inactive complexes are already present in starfish oocytes, CBsv cannot be considered as a competitor of CB for binding to cdc2. Most likely, the CBsv monomer could activate inhibitors, or inhibit activators of early steps in MPF initiation. Monomeric cyclin B was found to interfere with, and activate, in some conditions, the phosphatase activity of cdc25 in the absence of any cyclin B–cdc2 dimer (5). One possible interpretation of our experimental data is that CBsv could trap cdc25 or other activator molecules apart from the preformed inactive cyclin B–cdc2 dimers in the cytoplasm, and thus competitively inhibit MPF activation. In this view, CBsv monomers can play a role of dominant negative only at two conditions: first, the protein should have a reduced affinity for cdc2, which is observed, without losing affinity for other ligands of cyclin B; then, CBsv has to be produced in excess in the target cell. Indeed, immunofluorescence experiments showed that CBsv is overrepresented in growing oocytes, which are arrested in G2 during vitellogenesis. This further suggests that the reversible inhibition of MPF initiation by CBsv is a physiological mechanism.
We also show here that CBsv is overrepresented in some cells during embryogenesis: mainly the cells of the archenteron, some blastocoelar, and some pigment cells. Previous studies on related species showed that these cells belong to the lineage of veg2 blastomeres, four blastomeres raised by the cleavage of macromeres after micromere formation (29). Additionally, a noteworthy, although less intense, staining was found occasionally in other cells, or even in all the cells of the embryo (see Fig. 8 a). This suggests that CBsv can be produced at some time in cell lineages to allow differentiation, or at least remodeling of cell cycle progression of these cells. Accordingly, the first cellular accumulation of CBsv was observed in micromeres, which are the first cells to desynchronize during embryogenesis and to be committed to differentiation. Moreover, we showed that the overproduction of CBsv in micromeres was under maternal control, as also were micromere formation and acquisition of a new G2-phase. This supports the view that accumulation of CBsv participates to the mechanism that induces cell cycle lengthening and appearance of G-phases and then allows starting zygotic transcriptions during midblastula transition. Recently, it has been shown that cyclin E degradation, which could be involved in appearance of a G1 phase and slowing down of S-phase at MBT of Xenopus embryos, was also under maternal control (11, 13). So far, nothing is known concerning the status of cyclin E in sea urchin embryos, but these results suggest that several mechanisms can cooperate to slow down the cycles, either simultaneously in the whole embryo, like in amphibian, or gradually in cell lineages, like in sea urchin. Our recent unpublished results show that the homologous splice variants also exist in both human cyclins B1 and B2, the genes of which contain an intron at exactly the same position, suggesting CBsv production is an universal mechanism for exit from early embryogenesis or induction of cell differentiation, at least in Deuterostomians.
Acknowledgments
We thank D. Caput (SANOFI Elf Bio-Recherches, Toulouse, France) for communication of the library construction method.
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and the University Paris VI, by grant from the Association pour la Recherche sur le Cancer (ARC), and by grants from the Ligue Nationale contre le Cancer and the Ligues Départementales contre le Cancer de l'Ardèche et du Gard.
Abbreviations used in this paper
- 1-MeAde
1-methyl-adenine
- aa
amino acid
- CB
cyclin B
- CBsv
CB splice variant
- CDK
cyclin-dependent kinase
- CRS
cytoplasmic retention signal
- GVBD
germinal vesicle breakdown
- MPF
M-phase–promoting factor
- nt
nucleotide
- ORF
open reading frame
- RT
reverse transcriptase
Footnotes
Jean-Claude Lozano and Philippe Schatt contributed equally to this work.
References
- 1.Belyavsky A, Vinogradova T, Rajewsky K. PCR-based cDNA library construction: general cDNA libraries at the level of a few cells. Nucleic Acids Res. 1989;17:2919–2932. doi: 10.1093/nar/17.8.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chirgwin JM, Przybyla AE, McDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 3.Ferrel JE, Wu M, Gerhart JC, Martin GS. Cell cycle tyrosine phosphorylation of p34cdc2 and a microtubule-associated protein kinase homolog in xenopus oocytes and eggs. Mol Cell Biol. 1991;11:1965–1975. doi: 10.1128/mcb.11.4.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fesquet D, Labbé JC, Derancourt J, Capony JP, Galas S, Girard F, Lorca T, Shuttleworth J, Dorée M, Cavadore JC. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of T161 and its homologs. EMBO (Eur Mol Biol Organ) J. 1993;12:3111–3121. doi: 10.1002/j.1460-2075.1993.tb05980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galaktionov K, Beach D. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclins. Cell. 1991;67:1181–1194. doi: 10.1016/0092-8674(91)90294-9. [DOI] [PubMed] [Google Scholar]
- 6.Gallant P, Nigg EA. Identification of a novel vertebrate cyclin: cyclin B3 shares properties with both A- and B-type cyclins. EMBO (Eur Mol Biol Organ) J. 1994;13:595–605. doi: 10.1002/j.1460-2075.1994.tb06297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautier J, Minshull J, Lohka M, Glotzer M, Hunt T, Maller JM. Cyclin is a component of maturation-promoting factor from xenopus. Cell. 1990;60:487–494. doi: 10.1016/0092-8674(90)90599-a. [DOI] [PubMed] [Google Scholar]
- 8.Glotzer M, Murray AW, Kirshner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 9.Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 10.Guerrier P, Dorée M. Hormonal control of meiosis in starfish. Dev Biol. 1975;47:341–348. doi: 10.1016/0012-1606(75)90288-2. [DOI] [PubMed] [Google Scholar]
- 11.Hartley RS, Sible JC, Levellyn AL, Maller J. A role for cyclin E/Cdk2 in the timing of the midblastula transition in Xenopus embryos. Dev Biol. 1997;188:312–321. doi: 10.1006/dbio.1997.8647. [DOI] [PubMed] [Google Scholar]
- 12.Hörstadius, S. 1973. Experimental Embryology of Echinoderms. Clarendon Press, Oxford. 41–91.
- 13.Howe JA, Newport JW. A developmental timer regulates degradation of cyclin E1 at the MBT during Xenopus embryogenesis. Proc Natl Acad Sci USA. 1996;93:2060–2064. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 15.Kanatani H, Shirai H, Nakanishi K, Kurosawa T. Isolation and identification of meiosis-inducing substance in starfish Asterias amurensis. . Nature. 1969;211:273–277. doi: 10.1038/221273a0. [DOI] [PubMed] [Google Scholar]
- 16.Kishimoto T, Kanatani H. Cytoplasmic factor responsible for germinal vesicle breakdown and meiotic maturation in starfish oocytes. Nature. 1976;260:321–322. doi: 10.1038/260321a0. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi H, Stewart E, Poon R, Adamczewski JP, Gannon J, Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdk2 protein kinase subunits. Mol Biol Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koff A, Cross F, Fisher A, Shumacher J, Leguellec K, Philippe M, Roberts JM. Human cyclin E, a new cyclin that interacts with two members of the cdc2 gene family. Cell. 1991;66:1217–1228. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- 19.Labbé JC, Capony JP, Caput D, Cavadore JC, Derancourt J, Kaghad M, Lelias JM, Picard A, Dorée M. MPF from starfish at first meiotic metaphase is a heterodimer containing one molecule of cdc2 and one molecule of cyclin B. EMBO (Eur Mol Biol Organ) J. 1989;8:3053–3058. doi: 10.1002/j.1460-2075.1989.tb08456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labbé JC, Picard A, Peaucellier G, Cavadore JC, Nurse P, Dorée M. Purification of MPF from starfish: identification as the H1 histone kinase p34cdc2 and a possible mechanism for its periodic activation. Cell. 1989;57:253–263. doi: 10.1016/0092-8674(89)90963-x. [DOI] [PubMed] [Google Scholar]
- 21.Masui Y, Markert CL. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J Exp Zool. 1971;177:129–145. doi: 10.1002/jez.1401770202. [DOI] [PubMed] [Google Scholar]
- 22.Meijer L, Azzi L, Wang JY. Cyclin B targets p34cdc2 for tyrosine phosphorylation. EMBO (Eur Mol Biol Organ) J. 1991;10:1545–1554. doi: 10.1002/j.1460-2075.1991.tb07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mumberg D, Wick M, Bürger C, Haas K, Funk M, Müller R. Cyclin ET, a new splice variant of human cyclin E with a unique expression pattern during cell cycle progression and differentiation. Nucleic Acids Res. 1997;25:2098–2105. doi: 10.1093/nar/25.11.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray AW, Kirshner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 25.Newport J, Kirshner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 26.Pines J, Hunt T. Molecular cloning and characterization of the mRNA for cyclin from sea urchin eggs. EMBO (Eur Mol Biol Organ) J. 1987;6:2987–2995. doi: 10.1002/j.1460-2075.1987.tb02604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pines J, Hunter T. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO (Eur Mol Biol Organ) J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon R, Yamashita K, Adamczewski JP, Hunt T, Shuttleworth J. The cdc2-related protein p40 (MO15) is the catalytic subunit of a protein kinase that can activate p33 (cdk2) and p34 (cdc2) EMBO (Eur Mol Biol Organ) J. 1993;12:3123–3132. doi: 10.1002/j.1460-2075.1993.tb05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruffins SW, Ettensohn CA. A fate map of the vegetal plate of the sea urchin (Lytechinus variegatus)mesenchyme blastula. Development. 1996;122:253–263. doi: 10.1242/dev.122.1.253. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning, A Laboratory Manual. Second edition. Cold Spring Harbor Laboratory Press, Plainview, NY. 7–52.
- 31.Sewing A, Rönicke V, Bürger C, Funk M, Müller R. Alternative splicing of human cyclin E. J Cell Sci. 1994;107:581–588. doi: 10.1242/jcs.107.2.581. [DOI] [PubMed] [Google Scholar]
- 32.Signoret J et J. Lefresne. Contribution à l'étude de la segmentation de l'oeuf d'Axolotl: I Définition de la transition blastuléenne. Ann Embryol Morphog. 1971;4:113–123. [Google Scholar]
- 33.Solomon M, Harper JW, Shuttleworth J. CAK, the p34 (cdc2) activating kinase, contains a protein identical or closely related to p40 (MO15) EMBO (Eur Mol Biol Organ) J. 1993;12:3133–3142. doi: 10.1002/j.1460-2075.1993.tb05982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van der Velden HM, Lohka MJ. Cell cycle-regulated degradation of xenopus cyclin B2 requires binding to p34cdc2. Mol Biol Cell. 1994;5:713–724. doi: 10.1091/mbc.5.7.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent S, Jeanteur P, Fort P. Growth-regulated expression of rhoG, a new member of the ras homolog gene family. Mol Cell Biol. 1992;12:3138–3148. doi: 10.1128/mcb.12.7.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita M, Fukada S, Yoshikuni M, Bulet P, Hirai T, Yamagushi A, Lou YH, Zhao Z, Nagahama Y. Purification and characterization of maturation-promoting factor in fish. Dev Biol. 1992;148:8–15. doi: 10.1016/0012-1606(92)90259-j. [DOI] [PubMed] [Google Scholar]
- 37.Zhend X-F, Ruderman JV. Functional analysis of the P-box, a domain in cyclin B required for the activation of cdc25. Cell. 1993;75:155–164. [PubMed] [Google Scholar]