Abstract
Muscle thick filaments are stable assemblies of myosin and associated proteins whose dimensions are precisely regulated. The mechanisms underlying the stability and regulation of the assembly are not understood. As an approach to these problems, we have studied the core proteins that, together with paramyosin, form the core structure of the thick filament backbone in the nematode Caenorhabditis elegans. We obtained partial peptide sequences from one of the core proteins, β-filagenin, and then identified a gene that encodes a novel protein of 201–amino acid residues from databases using these sequences. β-Filagenin has a calculated isoelectric point at 10.61 and a high percentage of aromatic amino acids. Secondary structure algorithms predict that it consists of four β-strands but no α-helices. Western blotting using an affinity-purified antibody showed that β-filagenin was associated with the cores. β-Filagenin was localized by immunofluorescence microscopy to the A bands of body–wall muscles, but not the pharynx. β-filagenin assembled with the myosin homologue paramyosin into the tubular cores of wild-type nematodes at a periodicity matching the 72-nm repeats of paramyosin, as revealed by immunoelectron microscopy. In CB1214 mutants where paramyosin is absent, β-filagenin assembled with myosin to form abnormal tubular filaments with a periodicity identical to wild type. These results verify that β-filagenin is a core protein that coassembles with either myosin or paramyosin in C. elegans to form tubular filaments.
The thick filaments of striated muscles are stable, highly differentiated supramolecular structures in contrast to the dynamic assemblies of the cytoskeleton. In the assembly of muscle thick filaments, myosin forms characteristic structures of uniform symmetry, length, and diameter. However, myosin alone, in the case of vertebrate thick filaments, and myosin and its homologous companion paramyosin, in invertebrates, do not assemble in this characteristic manner in the test tube. Furthermore, transgenic experiments in Drosophila melanogaster show that different myosin isoforms from muscles with structurally distinct thick filaments can be exchanged with one another without changes in the muscle-specific assembly (Wells et al., 1996). The cellular mechanisms for assembling these intricate and regularly organized structures of striated muscle, therefore, are still not understood, despite their significance in hereditary cardiac and neuromuscular diseases, protein metabolism in starvation and diabetes, and normal muscle development (Epstein and Fischman, 1991).
The nematode Caenorhabditis elegans provides a genetically, biochemically, and structurally tractable model for studying mechanisms of filament assembly in muscle. The thick filaments of C. elegans body–wall muscle contain two myosins with different myosin heavy chains. The two myosins are differentially located in the thick filaments with myosin A (its heavy chain encoded by myo-3) in the center, and myosin B (its heavy chain encoded by unc-54) in the polar regions (Miller et al., 1983). Furthermore, paramyosin (Waterston et al., 1974; Harris and Epstein, 1977), an α-helical coiled-coil protein encoded by unc-15 (Waterston et al., 1977), homologous to the rod domains of myosin heavy chains, is also found in the thick filaments (Epstein et al., 1985). Genetic studies have shown that myosin A can substitute for myosin B in the thick filaments in unc-54 mutants (Epstein et al., 1986). However, myo-3 null mutants do not assemble thick filaments at all, which leads to embryonic lethality (Waterston, 1989), whereas unc-15 null mutant worms produce abnormal filament-like structures with scrambled myosins A and B in the medial region, and myosin B in the hollow polar regions. Although viable, the unc-15 null worms appear very thin and paralyzed. Therefore, myosin A and paramyosin are essential for proper thick filament assembly. In addition to myosin and paramyosin, other proteins appear to be critical for thick filament assembly. For example, unc-45 and unc-82 mutants do not alter the amino acid sequences of myosin or paramyosin, but produce abnormal thick filaments (Epstein and Thomson, 1974; Waterston et al., 1980; Venolia and Waterston, 1990).
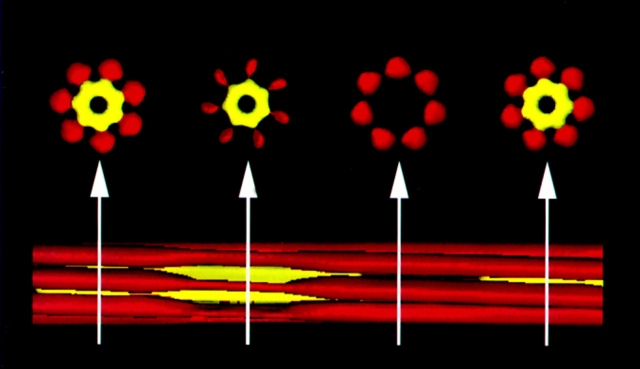
In the thick filaments of C. elegans body–wall muscle cells, a core substructure has been proposed as the template for the differential assembly of myosin heavy chains (Epstein et al., 1985). The cores are composed of a subpopulation of paramyosin molecules and at least three “core” proteins (Deitiker and Epstein, 1993). A three-dimensional model of the cores has been proposed based on the reconstruction of electron microscopy images of isolated cores (Fig. 1; Epstein et al., 1995). In this model, the core is composed of seven subfilaments of paramyosin that are cross-linked or coupled to form a tubule by the putative core proteins. We have named these proteins of 30-, 28-, and 20-kD α-, β-, and γ-filagenins (from the Latin filum; thread, and generare; to beget). The characterization of the filagenins should provide insights into the assembly of the cores and the subsequent assembly of native thick filaments.
Figure 1.
Three-dimensional model of C. elegans thick filament cores. In the longitudinal view (bottom), paramyosin subfilaments are shown in red whereas internal structures are shown in yellow. Not all seven subfilaments are visible in this projection. The internal structures are proposed to be composed of filagenins, and couple or cross-link the paramyosin subfilaments. Note that the internal structures are not continuous. Cross-sections of the core (top) are at the positions indicated by the arrows. All seven paramyosin subfilaments are visible. The internal tubule is missing in the cross-section second from the right, indicating the discontinuity. The cross-sections from left to right show the rotation of the subfilaments according to a seven-start helix. The paramyosin subfilaments are shown with different thicknesses to reflect the staggering of paramyosin.
We report here the molecular identification and characterization of β-filagenin, a core protein of novel amino acid sequence and predicted properties. Using an affinity-purified antipeptide antibody, we have localized β-filagenin to the A bands in C. elegans body–wall muscles by immunofluorescence microscopy, and to the cores by Western blotting. Using wild-type and paramyosin-deficient mutant strains, we have shown by immunoelectron microscopy that β-filagenin can coassemble with either myosin or paramyosin into tubular substructures of thick filaments in C. elegans.
Materials and Methods
Nematode Growth and Strains
For thick filament preparation, N2 (wild-type) C. elegans were grown on the peptone-enriched plates with a lawn of Escherichia coli strain NA22 at 20°C (Schachat et al., 1978). The nematodes were synchronized by starvation at L1 stage to obtain a relatively homogeneous population, and then harvested as L4 larvae. The nematodes were then cleaned and stored by mixing with two volumes of O.C.T. compound (Miles, Inc., Elkhart, IN) as described (Deitiker and Epstein, 1993). The paramyosin-deficient strain CB1214 was grown without starvation. For whole-mount immunofluorescence microscopy, nematodes were grown on nematode growth medium plates seeded with E. coli strain OP50 (Brenner, 1974).
Purification of Thick Filaments
The isolation of thick filaments was in accordance with the previously described procedures (Deitiker and Epstein, 1993). The 15K pellets made from 6 g of nematodes were resuspended in 3 ml of buffer used for extracting the thick filaments, and then loaded to a 34 ml 19–38% sucrose gradient. The gradient was centrifuged for 17 h at 5,000 rpm in a swing rotor (model SW 28; Beckman Instrs., Fullerton, CA). The gradient was divided into 12 fractions. Fractions 2–6 from the bottom of the gradient were pooled for filament protein precipitation. Because of the low abundance of the filagenins, multiple preparations were performed to accumulate enough thick filament proteins.
Ethanol Precipitation of Thick Filament Proteins
The pooled gradient fractions enriched with purified thick filaments were dialyzed against 10 mM sodium phosphate buffer, pH 6.36. Three volumes of precooled 95% ethanol were added slowly to the dialyzed fractions while stirring. After all ethanol was added, the precipitation was allowed to continue for 2 h by slow stirring at room temperature. Precipitated proteins were collected by centrifugation at 12,000 g for 20 min. The protein pellet was air-dried and stored at −80°C before being separated by SDS-PAGE.
Protein Separation, Digestion, and Sequencing
The precipitated thick filament proteins were separated by SDS-PAGE containing 11% acrylamide. Proteins were transferred to polyvinylidene difluoride (PVDF)1 membrane and stained with Coomassie blue as described previously. Because >90% of thick filament proteins by mass is myosin and paramyosin, large quantities of thick filament proteins were loaded in each lane to visualize the relatively scarce filagenins. Multiple lanes were required to obtain sufficient amounts of β-filagenin. The membranes containing β-filagenin were pooled and then digested with endoprotease Lys-C. Peptide fragments were separated by high performance liquid chromatography and sequenced on a protein sequencer (model 477A or 473A; Applied Biosystems, Foster City, CA).
Sequence Analysis
Most of the sequence information and homology searches were obtained through the C. elegans Genome Sequencing Project (The Sanger Centre, Hinxton Hall, Cambridge, UK; and the Washington University School of Medicine, St. Louis, MO). Further searches were done over the World Wide Web with the BCM Search Launcher (Smith et al., 1996). The cDNA clones were located using the BCM Search Launcher to the dbEST databases. The cDNA sequences originated from the C. elegans cDNA Sequencing Project in Japan (National Institute of Genetics, Mishima, Japan). Secondary structure prediction used the Type-1 Discrete State-Space Models from the Boston University BioMolecular Engineering Research Center PSA server (http://bmerc-www.bu.edu/psa/; Stultz et al., 1993; White et al., 1994). Other algorithms that we used are found in the Peptidestructure program of the Genetics Computer Group (GCG) Package, Wisconsin Package Version 9.0, GCG, Madison, Wisconsin; Chou and Fasman (1978), and Garnier et al. (1978). Isoelectric point calculation was obtained from the Isoelectric Program in the GCG Package.
Cloning of β-Filagenin cDNA
Primers P28gstup (35 nucleotide NH2-terminal) 5′GTGGATCCATGCCTTCGAGTCTTTCAGAGCC3′ and P28gstdn (35 nucleotide COOH-terminal) 5′CGGAATTCTTAAGAGAAAGAGTAGAAGTAGCGATG3′ were selected to amplify the complete predicted open reading frame of the β-filagenin gene by reverse transcriptase (RT)-PCR from total RNA isolated from nematodes of mixed stages. A single band of 630 bp was obtained, subcloned, and sequenced.
Antibodies
Rabbit antibody against β-filagenin was generated against the selected synthetic peptide YSSTLHKYRRDYDTL conjugated to the multiple antigenic peptide carrier protein core. The peptide was chosen based on its predicted high accessibility and antigenicity using the Surface Probability and Antigenicity subprograms of the Peptidestructure program in the GCG Package. The antiserum was affinity purified using the antigen as the ligand coupled to the matrix (model Affi-gel 10; Bio-Rad Laboratories, Hercules, CA). Coupling of the matrix and peptide was carried out mainly according to the manufacturer's instructions. 1 ml of the Affi-gel was drained of isopropanol and washed with 5 vol of cold water (4°C). The gel was then mixed with 4 ml of the peptide (2.5 mg/ml in 0.1 M morpholinepropanesulfonic acid, pH 7.5) by gentle rocking for 4 h at 4°C to allow coupling reaction to occur. Active esters left in the gel matrix were blocked by reacting the gel slurry with 0.1 ml of 1.0 M ethanolamine HCl, pH 8.0, for 1 h. The gel was then packed in a column, washed extensively with doubly distilled H2O, and then equilibrated with PBS (137 mM NaCl, 3 mM KCl, 10 mM Na2PO4, 2 mM KH2PO4, pH 7.2). For antibody binding, 5 ml of antiserum was dialyzed against PBS overnight, brought to 10 ml in total volume, and then passed through the column three times. Bound antibody was eluted with 0.1 M glycine-HCl, pH 2.5, and 1-ml fractions of the eluate were immediately neutralized, each with 0.3 ml of 1.0 M Tris-HCl, pH 8.2. The purified antibody was characterized by Western blotting using thick filament-enriched 6,200 g supernatants of C. elegans (Deitiker and Epstein, 1993) separated by 11% SDS-PAGE. Western blotting followed a previously described procedure using an affinity-purified IgG secondary antibody conjugated with alkaline phosphatase (Liu et al., 1997). The affinity-purified anti–β-filagenin antibody was used at 2.5 μg/ml. The antibodies against myosin heavy chains and paramyosin have been described (Miller et al., 1983).
Immunofluorescence Microscopy
Nematodes were freeze fractured (Liu et al., 1997) and fixed immediately in −20°C methanol for 5 min, and then followed by another 5 min in −20°C acetone. The fixed nematodes were rehydrated through a serial dilution of methanol, and then reacted with the antibodies as described (Epstein et al., 1993). The anti–β-filagenin antibody (2.5 μg/ml) was mixed with the rhodamine-conjugated monoclonal antiparamyosin antibody 5-23 (5 μg/ml). The mixed antibodies were used for the primary antibody reaction. The rabbit anti–β-filagenin antibody was reacted with affinity-purified, fluorescein isothiocyanate–conjugated goat anti–rabbit IgG secondary antibody.
Dissociation of Thick Filaments and Western Blotting
The 6.2K supernatant was dissociated with 450 mM NaCl (Deitiker and Epstein, 1993), and then centrifuged at 100,000 g for 40 min in an ultracentrifuge (model TL-100; Beckman Instrs., Inc.) using the microcentrifuge PA rotor. The pellets (cores) were resuspended and brought to the same volume as the supernatant. Equal volumes of 6.2K supernatant (thick filaments), dissociated supernatant, and resuspended pellet (cores) were separated by 11% polyacrylamide SDS-PAGE for Coomassie blue staining and Western blotting.
Immunoelectron Microscopy
Procedures for electron microscopy were as described (Miller et al., 1983; Epstein et al., 1985). Both monoclonal antibody 5-23 against paramyosin and anti–β-filagenin antibody were used at 50 μg/ml. The high concentration of both antibodies was necessary to visualize labeling in the electron microscope by the antibodies at relatively long repeats, observed in contrast to the lower concentration of antibody used in labeling myosin at shorter intervals (Levine et al., 1982; 1983; Woodhead, J.L., R.J.C. Levine, and H.A. King. 1986. J. Cell Biol. 109:267a; Deitiker and Epstein, 1993). Affinity-purified goat anti–mouse and goat anti–rabbit IgG secondary antibodies were used at 20 μg/ml.
Results
Purification and Peptide Sequencing of β-Filagenin
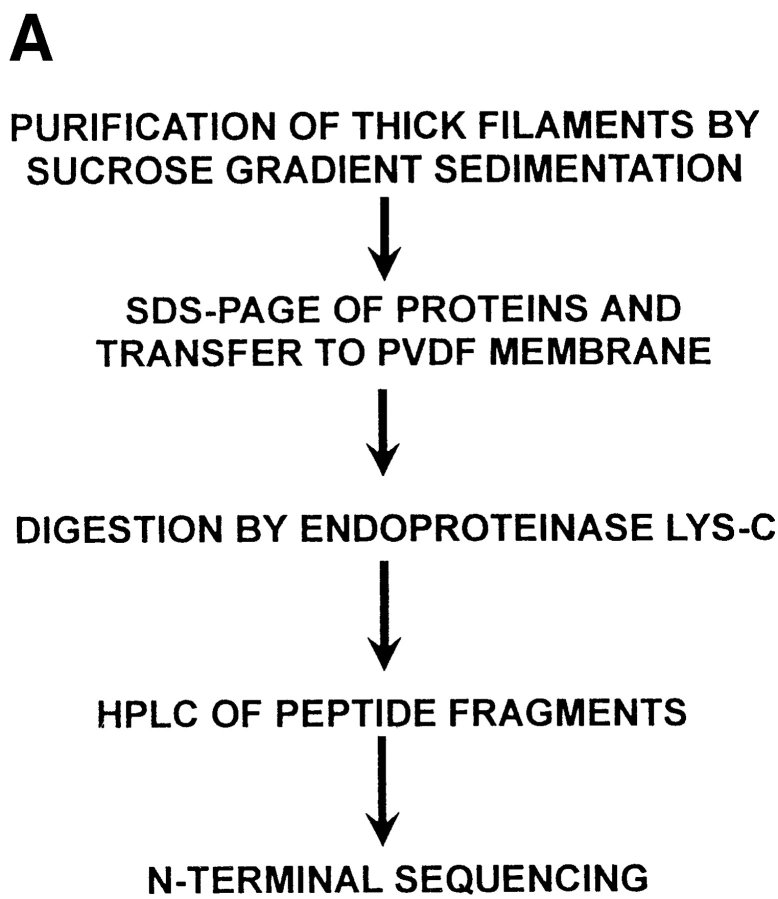
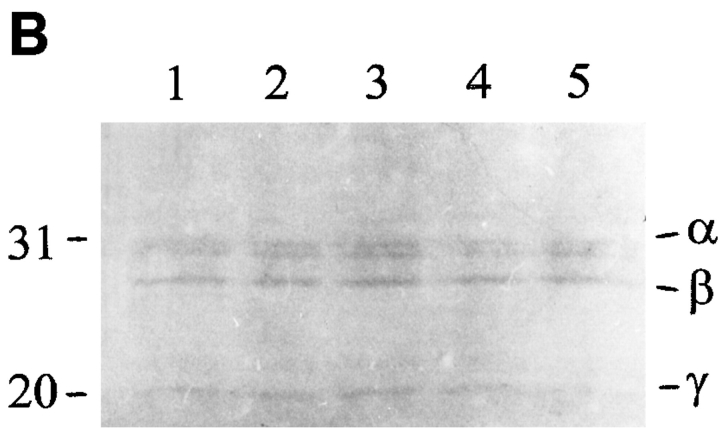
Because no previous examples of proteins that couple myosin or paramyosin subfilaments were known at the start of our experiments, we had to rely on direct biochemical methods to purify and initially characterize β-filagenin based upon our previous structural studies, rather than on more functional or genetic approaches. The biochemical approach was made more difficult by its low abundance of ∼0.01% of total protein (Deitiker and Epstein, 1993). Fig. 2 A outlines our procedure for isolating β-filagenin and characterizing it by peptide analysis and amino acid sequencing. Previously, purified thick filaments from the sucrose gradients were collected by sedimentation at high speed, a process that lowers the yield of thick filaments probably as the result of depolymerization (Deitiker and Epstein, 1993). One critical step that enabled us to obtain a sufficient amount of filagenins for protease digestion and amino acid sequencing was the ethanol precipitation of the proteins of thick filament preparation. Fig. 2 B shows another key step in this procedure, the Commassie blue– stained PVDF membrane to which the filagenins and other thick filament proteins were transferred after ethanol precipitation and SDS-PAGE. The β-filagenin band was proteolytically digested so that internal peptides could be purified and sequenced.
Figure 2.
Purification and sequencing of β-filagenin. (A) The purification procedure; (B) thick filament proteins by Coommassie blue staining on PVDF membrane were separated by and transferred from a 11% SDS-PAGE (all five lanes). Numbers on the left show molecular masses of 31 and 20 kD. Symbols on the right indicate positions of α-, β-, and γ-filagenins. The band just below α-filagenin is ATP translocase, which is not a core protein and does not copurify with cores.
The Primary and Secondary Structures of β-Filagenin
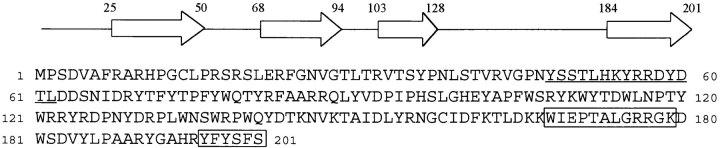
Two peptides were purified from protease digestion of β-filagenin and sequenced from their NH2 terminals. Computer-based searches with the peptide sequences obtained led to perfect matches within the predicted open reading frame of an X-linked genomic DNA sequence that had been determined by the C. elegans Genome Project, cosmid T14G12. The full predicted sequence of 201–amino acid residues is shown in Fig. 3. The predicted exons were verified by a partial cDNA sequence in the dbEST database, the cDNA clone yk102c12, and by our sequencing of a full-length cDNA generated by reverse transcriptase-PCR (data not shown). The amino acid sequence shows that β-filagenin may represent a novel group of proteins. The only homologous protein detected by a computer-based search to date is another C. elegans protein predicted from a genomic sequence, which shares 32% identity with β-filagenin. This predicted protein is also small, with only 221–amino acid residues, and has no known function. The gene encoding this protein is located physically on chromosome V (EMBL/GenBank/DDBJ accession number Z81579). Analysis of the β-filagenin sequence predicts four isolated β-strands separated by turns and loops as the only regular secondary structural motifs using the Discrete State-Space Models. Several other programs were also used, but none of them indicated any strong α-helical structures for β-filagenin unlike myosin or paramyosin. The protein is very basic with an isoelectric point of 10.61. Interestingly, tryptophan and tyrosine comprise ∼15% of the total amino acids, an unusually high fraction. These results may have functional significance for β-filagenin, as tryptophan and tyrosine are found in many protein binding sites, and the strongly basic character may be related to its putative interactions with the polyanionic subfilaments (Kagawa et al., 1989). It is unlikely that β-filagenin interacts with myosin or paramyosin through formation of α-helical coiled coils.
Figure 3.
Amino acid sequence and predicted secondary structure of β-filagenin. (Top) Predicted secondary structure. Each open arrow represents a β-strand with the numbers indicating its start and end. (Bottom) Predicted amino acid sequence of β-filaenin from the third open reading frame of cosmid T14G12 on chromosome X. The boxed sequences were obtained by peptide sequencing (Fig. 2) and used for database searches. The underlined 15–amino acid peptide was chosen as an immunogen.
β-Filagenin Localizes to A Bands of Body–Wall Muscle
To demonstrate that β-filagenin is a true component of thick filaments and their cores in C. elegans, we produced a specific antibody to β-filagenin in rabbits with a synthetic peptide (underlined sequence in Fig. 3) for localization experiments. The antiserum was purified by affinity chromatography against the same peptide, and the specificity of the resulting antibody was verified by Western blotting of thick filament-enriched proteins (Fig. 4). The purified antibody shows high affinity and specificity to β-filagenin.
Figure 4.
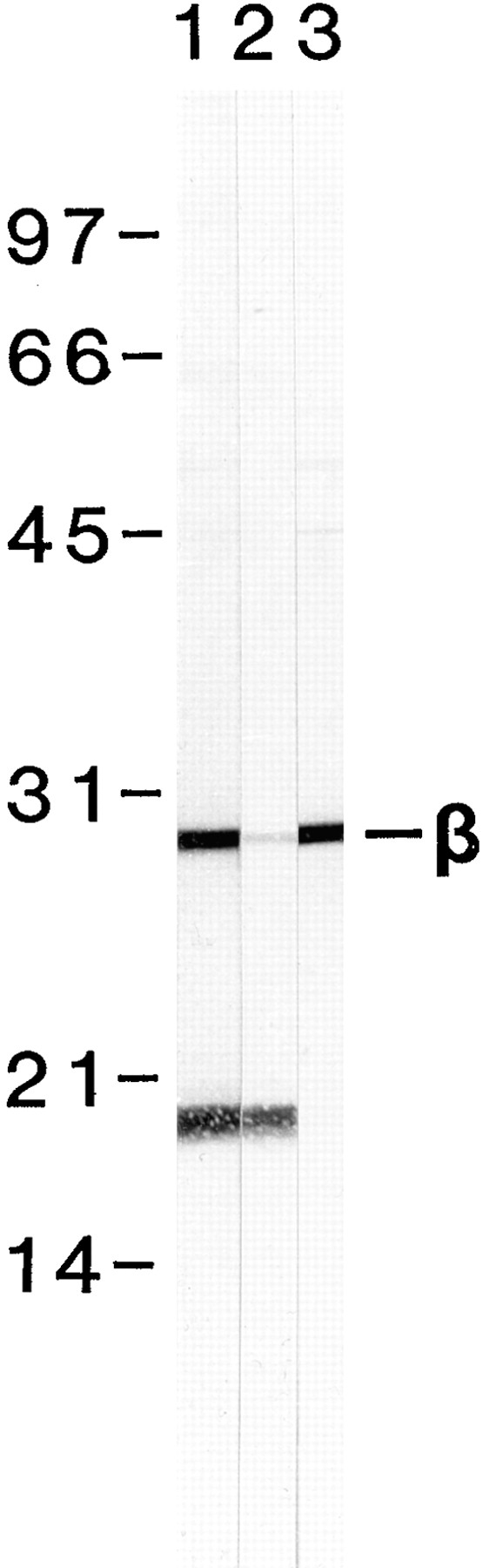
Antibody production, affinity purification, and characterization by Western blotting. Thick filament- enriched 6,200 g supernatants were separated by SDS-PAGE, transferred to nitrocellulose membranes, and reacted with different fractions of the affinity-purified anti–β-filagenin antiserum. Lane 1, antiserum before affinity purification at 1:100 dilution. Lane 2, antiserum after passing through the affinity column (flowthrough) at 1:100 dilution. Lane 3, affinity-purified antibody at 2.5 μg/ml. Numbers on the left are molecular weight markers in kilodaltons. Reaction of β-filagenin with the antibody is indicated by the β symbol on the right.
To confirm that β-filagenin is associated with thick filaments, C. elegans whole mounts were reacted with the affinity-purified antibody and viewed by immunofluorescence microscopy. The antibody labeled the thick filament-containing A bands of the body–wall muscles and the anal– intestinal muscles, but not the pharyngeal muscles (Fig. 5 B). This distribution is identical to the localization of myosin A and B (Miller et al., 1983; Ardizzi and Epstein, 1987). In contrast, antibody to paramyosin labeled all C. elegans muscles (Fig. 5 A), and served as a control for the more restricted localization of β-filagenin.
Figure 5.
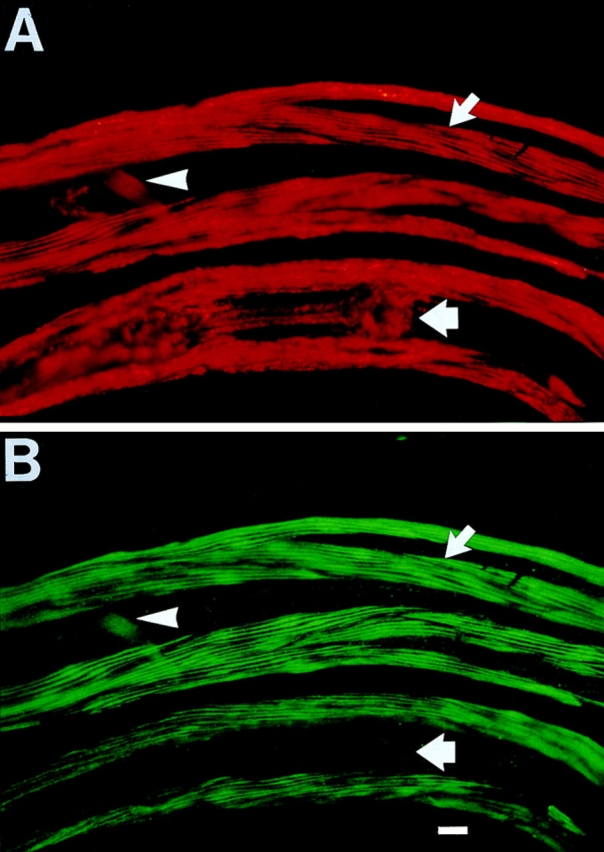
Immunolocalization of β-filagenin in a C. elegans whole mount. (A) A late larva was labeled with monoclonal antibody 5–23 against paramyosin. The antibody reacted with the body–wall muscle (narrow arrow), the pharynx (wide arrow), and the anal-intestinal muscle (arrowhead). (B) The same larva was labeled with the anti–β-filagenin polyclonal antibody. Note that this antibody also reacted with the body–wall and anal–intestinal muscles, but not the pharynx. Bar, 10 μm.
Association of β-Filagenin with Cores by Western Blotting
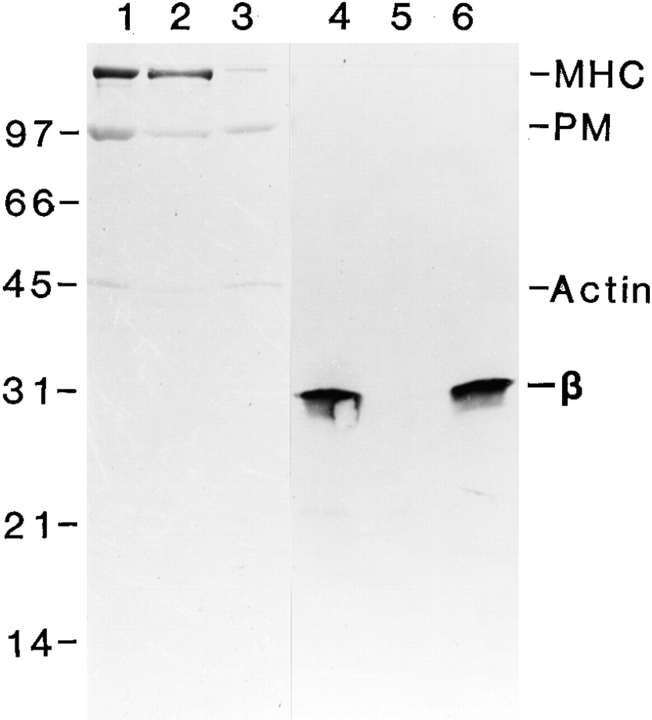
To verify that β-filagenin is a component of the cores, 6,200 g supernatants that are enriched for muscle filaments (Deitiker and Epstein, 1993) were dissociated with 450 mM NaCl, and then β-filagenin was traced by Western blotting using the affinity-purified antibody. Western blotting of thick filament- and core-enriched fractions reacted with the anti–β-filagenin antibody, whereas the fraction containing dissociated, soluble myosin and paramyosin did not (Fig. 6). This result is consistent with β-filagenin being a component of body–wall thick filaments and their core substructures in C. elegans.
Figure 6.
Localization of β-filagenin to the cores by Western blotting. Lanes 1 and 4, undissociated thick filaments; lanes 2 and 5, supernatants containing dissociated proteins; lanes 3 and 6, pellets containing remaining cores. Lanes 1–3, Coommassie blue staining; lanes 4–6, Western blotting. MHC represents myosin heavy chain; PM represents paramyosin; β represents β-filagenin. A small amount of actin filaments was also pelleted with the cores. Numbers on the left are molecular weight markers in kilodaltons.
Localization of β-Filagenin to Cores by Electron Microscopy
The localization of β-filagenin was further confirmed by immunofluorescence (data not shown) and immunoelectron microscopy of isolated tubular core structures after reaction with the specific antibody. The isolated cores showed a periodicity of 72 nm either by negative staining with uranyl acetate (Fig. 7 A), or by labeling with monoclonal antiparamyosin antibody (Fig. 7 B). The 72-nm repeats are also observed in paracrystals formed with purified paramyosin from either molluscan muscles (Cohen et al., 1971), or C. elegans (Waterston et al., 1974; Harris and Epstein, 1977). When isolated cores were reacted with the affinity-purified anti–β-filagenin antibody (Fig. 7 C), a repeating pattern matching the 72-nm repeats in the negatively stained and the antiparamyosin antibody-labeled cores was observed. This periodic nature of β-filagenin localization was consistent with the predictions of the model for the core structure (Epstein et al., 1995), and the coassembly of β-filagenin and paramyosin.
Figure 7.
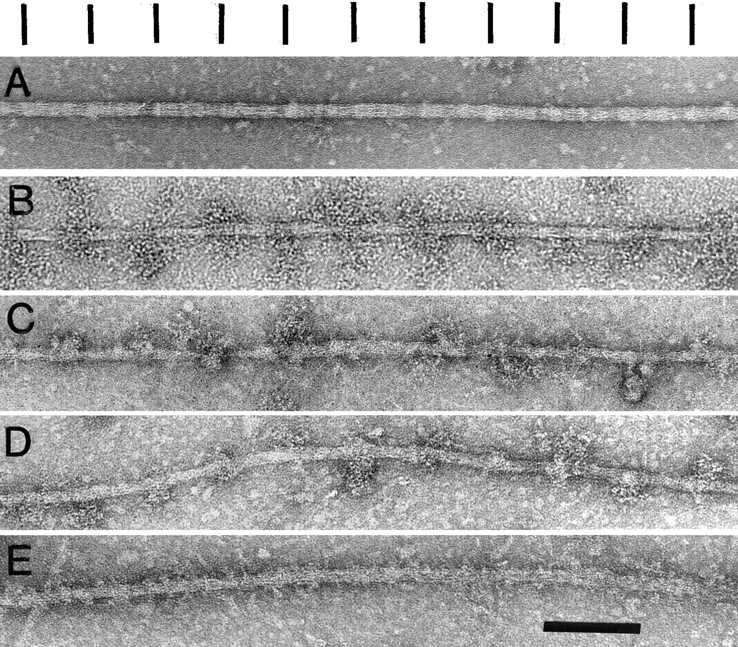
Localization of β-filagenin by immunoelectron microscopy. (A) Negatively stained wild-type core; (B) monoclonal antibody 5-23 labeling of wild-type core localizing a paramyosin epitope; (C) anti–β-filagenin antibody labeling of wild-type core; (D) anti-β-filagenin antibody labeling of the filament-like structure from the paramyosin-deficient strain CB1214; (E) negatively stained structure as in D. Note the different periodic structures have been aligned in register for easy comparison. Although the major periodicities of these different structures appear to be very similar, they are not in register with one another, which is consistent with the model of Epstein et al. (1995). Bar, 100 nm.
Localization of β-Filagenin in the Filaments of Paramyosin-null Mutant CB1214
To test whether β-filagenin functions in assembly, we examined the thick filament-like structures produced in the paramyosin-deficient mutant CB1214 (Fig. 7 E; Waterston et al., 1977; Waterston and Brenner, 1978; Mackenzie and Epstein, 1980; Gengyo-Ando and Kagawa, 1991). In these mutant filaments, tubular core-like substructures that contain myosin B protrude from medial filament-like regions containing myosin A and myosin B (Epstein et al., 1986). Since purified myosin from C. elegans does not assemble into tubular structures (Harris and Epstein, 1977), the behavior of myosin B in CB1214 was likely the result of interactions with β-filagenin and the other core proteins. The anti–β-filagenin antibody reacted with the CB1214 structures (Fig. 7 D), matching the periodicity observed in wild-type cores. This result is consistent with β-filagenin being necessary for the assembly of tubular substructures, whether of wild-type cores with paramyosin or of CB1214 mutant core-like structures with myosin B.
Discussion
Previous work from this laboratory has shown that two myosin isoforms, A and B, are assembled about a core containing paramyosin within the thick filaments of body– wall muscle cells in C. elegans (Epstein et al., 1985, 1995). The core has been proposed to serve as a template for the assembly of the myosins. A major question remains as to what mechanism regulates the assembly of paramyosin to yield core structures of precise length and, thereby, thick filaments of precisely regulated dimensions. The role of additional core proteins in this process has been suggested (Epstein et al., 1985, 1988, 1995; Deitiker and Epstein, 1993), and we have described here the first identification and characterization of such a protein, β-filagenin.
Using several antibody-based methods, we have shown that β-filagenin is a core protein. The direct localization of β-filagenin in cores isolated from wild-type C. elegans was observed at the electron microscope level. Its labeling with specific antibody produced a repeating pattern consistent with the predictions of our structural model as shown in Fig. 1. This result is consistent with the proposal that paramyosin strands in the cores are cross-linked or coupled by β-filagenin. The filagenins may be essential, therefore, for the assembly and stabilization of the paramyosin cores and, consequently, the assembly of thick filaments.
β-Filagenin is also localized to the mutant thick filaments of CB1214, which is produced by the premature chain termination e1214 mutation in the unc-15 locus, and leads to a paramyosin-deficient state. The CB1214 thick filaments are core-like tubular structures where myosin B assembles in place of paramyosin. Importantly, β-filagenin localized to these myosin tubules with the same repeat pattern as in wild-type cores. Since myosin itself does not form tubules or assemble into wild-type cores, these results suggest that β-filagenin and other possible core proteins (α-, γ-filagenins) may be capable of directing the assembly of myosin as well as of paramyosin. Although apparently not homologous at the amino acid sequence level to β-filagenin, the myosin binding protein C family of cardiac and skeletal muscles may perform analogous functions during the assembly of thick filaments in vertebrates (Moos et al., 1975; Einheber and Fischman, 1990; Okagaki et al., 1993).
Analysis of the amino acid sequence of β-filagenin suggests that it is a protein of unusual biochemical characteristics. The protein is very basic with a predicted isoelectric point of 10.61. The protein also has a high percentage of tryptophan and tyrosine, two amino acids often found at the binding sites of protein–protein interactions. Whether these characteristics are of special importance to the function of β-filagenin is unknown. Biochemical studies using purified protein should provide further information on the relationships between the structure and function of β-filagenin.
Our studies also showed that β-filagenin was present in body–wall muscle cells and the specialized anal–intestinal muscles, but not in the pharynx. The pharyngeal thick filaments contain myosin C and D instead of myosin A and B (Ardizzi and Epstein, 1987). The thick filaments of the pharynx are also characteristically shorter and more electronlucent than those of the body–wall (Epstein et al., 1974; Albertson and Thomson, 1976). However, they share the only paramyosin (Ardizzi and Epstein, 1987). What protein performs the putative functions of β-filagenin in the pharynx is not known. We have identified another protein in C. elegans with a predicted sequence of 221–amino acid residues that shares 32% identity in sequence as well as other characteristics with β-filagenin (Liu, F., C.C. Bauer, and H.F. Epstein, unpublished results). The possibility that β-filagenin and this putative homologue could, in part, be responsible for the different properties of body– wall and pharyngeal muscle thick filaments will be tested in future experiments. The potential interactions of β-filagenin with proteins operating in thick filament assembly other than myosin and paramyosin, including such likely candidates as the α- and γ- filagenins and the UNC-45 protein (Epstein and Thomson, 1974; Venolia and Waterston, 1990), are also under study.
Acknowledgments
We thank our colleagues at Baylor, D.L. Casey for excellent assistance and J.M. Barral for comments.
Supported by grants from the Muscular Dystrophy Association and National Science Foundation.
Abbreviations used in this paper
- GCG
Genetics Computer Group
- PVDF
polyvinylidene difluoride
Footnotes
Address all correspondence to Henry F. Epstein, Department of Neurology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030. Tel.: (713) 798-4629. Fax: (713) 798-3771. E-mail: hepstein@bcm.tmc.edu
References
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. . Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Ardizzi JP, Epstein HF. Immunochemical localization of myosin heavy chain isoforms and paramyosin in developmentally and structurally diverse muscle cell types of the nematode Caenorhabditis elegans. . J Cell Biol. 1987;105:2763–2770. doi: 10.1083/jcb.105.6.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Empirical prediction of protein conformation. Annu Rev Biochem. 1978;49:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Cohen C, Szent-Gyorgyi AG, Kendrick J, Jones Paramyosin and the filaments of the molluscan “catch” muscles. I. Paramyosin: structure and assembly. J Mol Biol. 1971;56:223–237. doi: 10.1016/0022-2836(71)90461-x. [DOI] [PubMed] [Google Scholar]
- Deitiker PR, Epstein HF. Thick filament substructures in Caenorhabditis elegans: evidence for two populations of paramyosin. J Cell Biol. 1993;123:303–311. doi: 10.1083/jcb.123.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einheber S, Fischman DA. Isolation and characterization of a cDNA clone encoding avian skeletal muscle C-protein: an intracellular member of the immunoglobulin superfamily. Proc Natl Acad Sci USA. 1990;87:2157–2161. doi: 10.1073/pnas.87.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Waterston RH, Brenner S. A mutant affecting the heavy chain of myosin in Caenorhabditis elegans. . J Mol Biol. 1974;90:291–300. doi: 10.1016/0022-2836(74)90374-x. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Thomson JN. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. . Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- Epstein, H.F., and D.A. Fischman. 1991. Molecular analysis of protein assembly in muscle development. Science (Wash. DC). 251:1039–1044. [DOI] [PubMed]
- Epstein HF, Miller DM, Ortiz I, Berliner GC. Myosin and paramyosin are organized around a newly identified core structure. J Cell Biol. 1985;100:904–915. doi: 10.1083/jcb.100.3.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Ortiz I, Traeger LA, Mackinnon The alteration of myosin isoform compartmentation in specific mutants of Caenorhabditis elegans. . J Cell Biol. 1986;103:985–993. doi: 10.1083/jcb.103.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Berliner GC, Casey DL, Ortiz I. Purified thick filaments from the nematode Caenorhabditis elegans: evidence for multiple proteins associated with core structures. J Cell Biol. 1988;106:1985–1995. doi: 10.1083/jcb.106.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Casey DL, Ortiz I. Myosin and paramyosin of Caenorhabditis elegansembryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J Cell Biol. 1993;122:845–858. doi: 10.1083/jcb.122.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Lu GY, Deitiker PR, Ortiz I, Schmid MF. Preliminary three-dimensional model for nematode thick filament core. J Struct Biol. 1995;115:163–174. doi: 10.1006/jsbi.1995.1041. [DOI] [PubMed] [Google Scholar]
- Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gengyo-Ando K, Kagawa H. Single charge change on the helical surface of the paramyosin rod dramatically disrupts thick filament assembly in Caenorhabditis elegans. . J Mol Biol. 1991;291:429–441. doi: 10.1016/0022-2836(91)90184-8. [DOI] [PubMed] [Google Scholar]
- Harris HE, Epstein HF. Myosin and paramyosin of Caenorhabditis elegans: biochemical and structural properties of wild type and mutant proteins. Cell. 1977;10:709–719. doi: 10.1016/0092-8674(77)90105-2. [DOI] [PubMed] [Google Scholar]
- Kagawa H, Gengyo K, McLachlan AD, Brenner S, Karn J. Paramyosin gene (unc-15) of Caenorhabditis elegans.Molecular cloning, nucleotide sequence and models for thick filament structure. J Mol Biol. 1989;207:311–333. doi: 10.1016/0022-2836(89)90257-x. [DOI] [PubMed] [Google Scholar]
- Levine, R.J.C., R.W. Kensler, M. Stewart, and J.C. Haselgrove. 1982. Molecular organization of Limulus thick filaments. In Basic Biology of Muscles: A Comparative Approach. B.M. Twarog, R.J. Levine, and M.M. Dewey, editors. Raven Press. New York. 37–52. [PubMed]
- Levine RJC, Kensler RW, Reedy M, Hofmann W, King HA. Structure and paramyosin content of tarantula thick filaments. J Cell Biol. 1983;97:186–195. doi: 10.1083/jcb.97.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Thatcher JD, Epstein HF. Induction of glyoxylate cycle expression in Caenorhabditis elegans: a fasting response throughout larval development. Biochemistry. 1997;36:255–260. doi: 10.1021/bi9623800. [DOI] [PubMed] [Google Scholar]
- Mackenzie JM, Epstein HF. Paramyosin is necessary for determination of nematode thick filament length in vivo. . Cell. 1980;22:747–755. doi: 10.1016/0092-8674(80)90551-6. [DOI] [PubMed] [Google Scholar]
- Miller DM, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Moos C, Offer G, Starr R, Bennett P. Interaction of C-protein with myosin, myosin rod and light meromyosin. J Mol Biol. 1975;97:1–9. doi: 10.1016/s0022-2836(75)80017-9. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Weber FE, Fischman DA, Vaughan KT, Mikawa T, Reinach FC. The major myosin-binding domain of skeletal muscle MyBP-C (C-protein) resides in the COOH-terminal, immunoglobulin C2 motif. J Cell Biol. 1993;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachat F, Garcea RL, Epstein HF. Myosins exist as homodimers of heavy chains: demonstration with specific antibody purified by nematode mutant myosin affinity chromatography. Cell. 1978;15:405–411. doi: 10.1016/0092-8674(78)90009-0. [DOI] [PubMed] [Google Scholar]
- Smith RF, Wiese BA, Wojzynski MK, Davison DB, Worley K C. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- Stultz CM, White JV, Smith TF. Structural analysis based on state-space modeling. Protein Sci. 1993;2:305–314. doi: 10.1002/pro.5560020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L, Waterston RH. The unc-45 gene of Caenorhabditis elegansis an essential muscle-affecting gene with maternal expression. Genetics. 1990;126:345–353. doi: 10.1093/genetics/126.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH. The minor myosin heavy chain, mhc A, of Caenorhabditis elegansis necessary for the initiation of thick filament assembly. EMBO (Eur Mol Biol Organ) J. 1989;8:3429–3436. doi: 10.1002/j.1460-2075.1989.tb08507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Brenner S. A supressor mutation in the nematode acting on specific alleles of many genes. Nature. 1978;275:715–719. doi: 10.1038/275715a0. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Epstein HF, Brenner S. Paramyosin in Caenorhabditis elegans. . J Mol Biol. 1974;90:285–290. doi: 10.1016/0022-2836(74)90373-8. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Fishpool RM, Brenner S. Mutants affecting paramyosin in Caenorhabditis elegans. . J Mol Biol. 1977;117:679–697. doi: 10.1016/0022-2836(77)90064-x. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure in Caenorhabditis elegans. . Dev Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Wells L, Edwards KA, Bernstein SI. Myosin heavy chain isoforms regulate muscle function but not myofibril assembly. EMBO (Eur Mol Biol Organ) J. 1996;15:4454–4459. [PMC free article] [PubMed] [Google Scholar]
- White JV, Stultz CM, Smith TF. Protein classification by stochastic modeling and optimal filtering of amino-acid sequences. Mathemat Biosci. 1994;119:35–75. doi: 10.1016/0025-5564(94)90004-3. [DOI] [PubMed] [Google Scholar]