Abstract
We have identified a Saccharomyces cerevisiae protein, Cyk1p, that exhibits sequence similarity to the mammalian IQGAPs. Gene disruption of Cyk1p results in a failure in cytokinesis without affecting other events in the cell cycle. Cyk1p is diffused throughout most of the cell cycle but localizes to a ring structure at the mother–bud junction after the initiation of anaphase. This ring contains filamentous actin and Myo1p, a myosin II homologue. In vivo observation with green fluorescent protein–tagged Myo1p showed that the ring decreases drastically in size during cell division and therefore may be contractile. These results indicate that cytokinesis in budding yeast is likely to involve an actomyosin-based contractile ring. The assembly of this ring occurs in temporally distinct steps: Myo1p localizes to a ring that overlaps the septins at the G1-S transition slightly before bud emergence; Cyk1p and actin then accumulate in this ring after the activation of the Cdc15 pathway late in mitosis. The localization of myosin is abolished by a mutation in Cdc12p, implicating a role for the septin filaments in the assembly of the actomyosin ring. The accumulation of actin in the cytokinetic ring was not observed in cells depleted of Cyk1p, suggesting that Cyk1p plays a role in the recruitment of actin filaments, perhaps through a filament-binding activity similar to that demonstrated for mammalian IQGAPs.
Cytokinesis in animal cells depends on drastic rearrangements of cortical actin filaments, leading to the assembly of an actomyosin-based contractile ring (for review see Schroeder, 1990; Satterwhite and Pollard, 1992; Fishkind and Wang, 1995). How actin and myosin are recruited to the cleavage site to form the contractile structure is not well understood. Proteins known to regulate actin cytoskeleton organization are likely to play important roles in contractile ring formation and function. Mutational or pharmacological inactivation of Rho-type GTPases, such as Rho and Cdc42, inhibits cell cleavage (Kishi et al., 1993; Drechsel et al., 1996; Larochelle et al., 1996). Calcium/calmodulin-dependent signaling events are also thought to regulate the onset and progression of actin ring contraction (for review see Schroeder, 1990; Walsh, 1994). The direct targets of the small GTP-binding proteins and calmodulin in cytokinesis have not yet been identified. Recently, a new family of proteins, termed IQGAP, have been implicated in connecting Cdc42 and calmodulin signaling to the remodeling of the actin cytoskeleton (Weissbach et al., 1994; Brill et al., 1996; Hart et al., 1996; McCallum et al., 1996; Bashour et al., 1997). These proteins contain a calponin homology domain and interact directly with calmodulin as well as the activated form of Cdc42. Furthermore, IQGAP1 has been shown to form a homodimer and bundle actin filaments with high affinity in vitro (Bashour et al., 1997). The precise in vivo function of IQGAP proteins has not yet been determined in animal cells.
The timing and positioning of the contractile ring are crucial for the faithful transmission of genetic and organelle materials to progeny cells. The mitotic spindle has long been thought to generate the spatial information for cleavage furrow formation, either through a diffusible signal or a direct interaction with the actin cortex (for review see Rappaport, 1986; Strome, 1993). Also, mitotic kinases, such as Cdc2 and Polo (for review see Fishkind and Wang, 1995; Glover et al., 1997), may be important for coordinating cell division with chromosome segregation. For example, phosphorylation of the regulatory light chain of myosin II by Cdc2/cyclinB kinase complex has been speculated to be a mechanism of delaying cell cleavage until the completion of chromosome segregation (Satterwhite et al., 1992; Yamakita et al., 1994).
Clearly, the mechanisms involved in the regulation of structural assembly during cytokinesis are highly complex. Studies in simple and genetically tractable organisms, such as yeast, could provide key insights into these mechanisms. However, a contractile ringlike structure was not known to exist in the budding yeast Saccharomyces cerevisiae, preventing the use of this organism as a general model for studying cytokinesis. The only filamentous ring previously found at the mother–bud junction is composed of septins, which include Cdc3p, Cdc10p, Cdc11p, and Cdc12p (Longtine et al., 1996). The septin ring is required for cytokinesis but does not appear to be contractile. The absence of an actomyosin ring and the morphology of the mother–bud junction have led to the view that budding yeast cell division may not involve constriction, as in animal cells, but instead may result from the growth of a septum and two plasma membranes across the neck, much as occurs in the phragmoplast during plant cytokinesis (Harold, 1990). Consistent with this notion, cortical actin patches, structures always associated with the growing surface area, accumulate at the site of cell division during cytokinesis (Adams and Pringle, 1984; Kilmartin and Adams, 1984). Actin cables, thought to direct vesicular transport (Liu and Bretscher, 1992; Bretscher et al., 1994), are oriented toward the forming septum (Adams and Pringle, 1984; Kilmartin and Adams, 1984).
Here we present evidence that cytokinesis in budding yeast does involve an actomyosin-based ring that exhibits a contractionlike change in shape during cytokinesis. This ring contains an IQGAP-like protein, gene disruption of which results in an inability to undergo cytokinesis. The assembly of the ring is controlled at various points in the cell cycle. The localization of one of the key components occurs early in the cell cycle and depends on the structure containing septin filaments, whereas the final maturation of the ring is triggered by a late mitotic pathway.
Materials and Methods
Media and Genetic Manipulations
Yeast cell culture and genetic techniques were carried out by methods described in (Sherman et al., 1974). Yeast extract, peptone, dextrose (YPD)1 contained 2% glucose, 1% yeast extract, and 2% Bactopeptone (Difco Laboratories Inc., Detroit, MI). YPG contained 2% galactose, 2% raffinose, 1% yeast extract, and 2% Bactopeptone.
Plasmid Construction
To construct Cyk1-expressing plasmids, a DNA fragment containing a 262-bp 5′ sequence and the entire open reading frame of CYK1 was obtained by PCR against yeast genomic DNA. This fragment was digested with XhoI (included in the 5′ primer) and EagI (included in the 3′ primer immediately following the coding sequence for the last amino acid), and then cloned between the XhoI and EagI sites in bluescript SK to yield pRL143. The same fragment was also blunted at the EagI site and cloned between the SalI and BamHI sites of pRL102, a vector for expressing COOH-terminal, myc-tagged proteins in yeast (Li, 1997). The Cyk1-myc expressing fragment of the resulting plasmid was further subcloned into the centromere-based plasmids, pRS313 and pRS316 (Sikorski and Hieter, 1989) to yield pTL12 and pTL13. A CYK1 gene disruption plasmid (pRL144) was constructed by replacing the segment corresponding to amino acids 23–1,342 with the LEU2 marker gene (Berben et al., 1991). To generate the plasmid that expresses Cyk1 under the GAL1 promoter, a fragment containing amino acids 25–1,495 (the last amino acid) of Cyk1, as well as the COOH-terminal myc tags was cloned into pRS306 (Sikorski and Hieter, 1989). A 670-bp fragment that contains the GAL1 promoter was cloned into the resulting plasmid at the 5′ end of the Cyk1 fragment to yield pRL170. An ATG is provided from an NcoI site immediately 5′ to amino acid 24 of Cyk1.
pRL72, a yeast vector for COOH-terminal 6-myc tagging was constructed by ligating the BamHI–SnaBI fragment that contains six copies of myc epitope from CS + MT (Roth et al., 1991) into vector pRS315 (Sikorski and Hieter, 1989), between BamHI and XbaI (blunt). pRL73, a yeast vector for COOH-terminal green fluorescent protein (GFP) tagging, was constructed by cloning an XbaI–BamHI fragment containing the coding sequence for GFP with S65T and V163A mutations between the XbaI– BamHI sites of pRS315, a centromere-based yeast expression vector (Sikorski and Hieter, 1989). The BamHI site is immediately 5′ to, and in frame with the ATG of GFP. A 5.75-kb fragment containing the MYO1 (Watts et al., 1987) coding region and a 226-bp 5′ flanking sequence was amplified by PCR using a sequence-specific 5′ primer containing a PstI site, and a sequence-specific 3′ primer containing a BamHI site. This fragment was cut with BamHI and PstI, and then cloned into the corresponding sites in pRL72 and pRL73 to generate pLP7 and pLP8.
Strain Construction
All strains used in this study are listed in Table I. To disrupt CYK1, pRL144 was digested with XhoI and EagI, and then transformed into RLY138, a Leu− diploid strain to yield RLY226. RLY226 cells transformed with pTL13 were also sporulated. CYK1 gene disruption was confirmed by PCR analysis of the genomic DNA prepared from the Leu+, pTL13-bearing colonies using a 5′ primer corresponding to a sequence within the CYK1 coding region that is replaced by the LEU2 marker gene in the disruption allele and a 3′ primer corresponding to a sequence 3′ to the CYK1 coding region, that is not present in pTL13 (data not shown). The Cyk1-myc expressing strain, RLY 230, which contains the Δcyk1 chromosomal mutation and pTL13, was obtained as a Leu+, Ura+ haploid colony after sporulation and tetrad dissection of RLY226 cells transformed with pTL13. To generate RLY277, the strain in which the expression of Cyk1p is controlled under GAL1 promoter, pRL170 was linearized with StuI and integrated into the URA3 locus of RLY261, a strain that grows well on galactose-containing media. The resulting strain was crossed with RLY230, and the resulting diploid was sporulated to yield Leu+ Ura+ spores that grow on galactose media but not on glucose-containing media. One such spore was then backcrossed two more times with RLY261 to generate RLY277. The original cdc15-2 and cdc12-6 strains were obtained from D. Pellman (Dana-Farber Cancer Institute, Boston, MA) and J.R. Pringle (University of North Carolina, Chapel Hill, NC), respectively, and backcrossed into the Li lab strain background (S288c).
Table I.
Yeast Strains
| Name | Genotype | Source | ||
|---|---|---|---|---|
| RLY8 | MATa ura3-52 his3-Δ200 leu2-3, 112 lys2-801Δbar1 | this work | ||
| RLY226 | MATa/α ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3,112/leu2-3, 112 lys2-801/lys2-801 CYK1/Δcyk1::LEU2 | this work | ||
| RLY230 | MATa ura3-52 his3-Δ200 leu2-3, 112 lys2-801 Δcyk1::LEU2 pCYK1-myc (pTL 13) | this work | ||
| RLY238 | MATa ura3-52 his3-Δ200 leu2-3, 112 trp1-1 ade2 cdc15-2 Δcyk1::LEU2 pCYK1-myc (pTL13) | this work | ||
| RLY261 | MATa ura3-52 his3-Δ200 leu2-3, 112 trp1-1 ade2 Δbar1 | Elion lab | ||
| RLY277 | MATa ura3-52 his3-Δ200 leu2-3, 112 lys2-801 trp1-1 Δbar1 Δcyk1::LEU2 pGAL1-CYK1-myc (pRL170) | this work | ||
| RLY301 | MATa ura3-52 his3-Δ200 leu2-3, 112 lys2-801Δbar1 pMYO1-myc (pLP7) | this work | ||
| RLY302 | MATa ura3-52 his3-Δ200 leu2-3, 112 lys2-801Δbar1 pMYO1-GFP (pLP8) | this work | ||
| RLY311 | MATa ura3-52 leu2-3, 112 cdc12-6 pMYO1-myc (pLP7) | this work |
Zymolyase Treatment
5 ml fixed cells were harvested, washed twice with PBS, and then once with 1 M sorbitol in 50 mM KPO4, pH 7.5. Cells were incubated with 0.2 mg/ml zymolyase 20T (Seikagaku Corporation, Tokyo, Japan) in the above sorbitol buffer containing 2 mM DTT for 10–20 min at 37°C. Typically >90% of the treated cells lost the refractile appearance when observed under a Labophot-2 microscope (Nikon Inc., Melville, NY) with a Plan40 0.5 ELWD objective, indicating that cell wall removal was efficient. Efficient cell wall removal by such treatment was also confirmed by electron microscopy (see cells in Fig. 3 E).
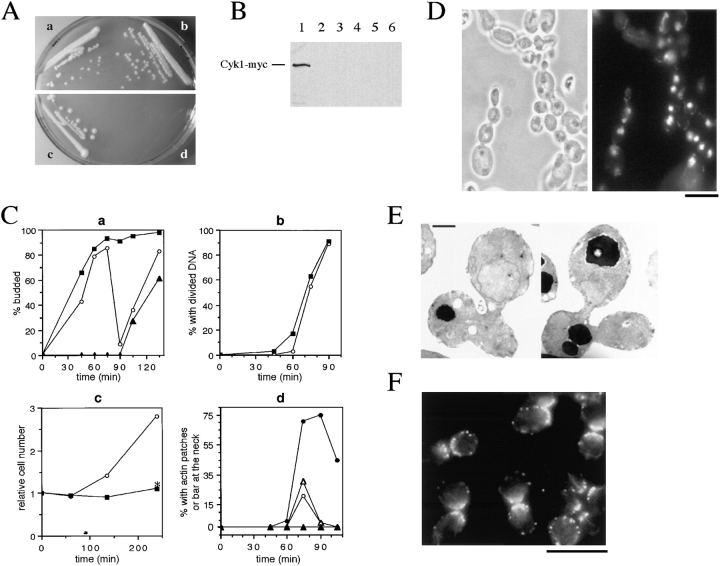
Figure 3.
Cyk1p is required for cytokinesis. (A) RLY261 (a and c; wild-type) and RLY277 (b and d; expressing Cyk1 under GAL1 promoter) were streaked onto a YPG plate (a and b) and a YPD plate (c and d), and then grown for 3 d at 30°C before photography. (B) RLY261 and RLY277 cells were cultured overnight in YPG at 30°C. A 10-ml sample of RLY277 culture was processed for immunoblot analysis (lane 1). The rest of the cells were shifted to YPD containing 0.1 μg/ml α factor (Sigma Chemical Co., St. Louis, MO) and grown for 3 h at 30°C. The cells were washed five times with water and transferred to YPD. 10-ml samples were taken at 0, 45, 60, 75, and 90 min (lane 2, 3, 4, 5, and 6, respectively) after the release. Immunoblot analysis using anti-myc antibody was carried out as described in Materials and Methods. (C) In the same experiment as in B, 5-ml samples were fixed with formaldehyde at each time point after the release from the G1 arrest (horizontal axis in each graph). A fraction of the cells were treated with zymolyase and stained with DAPI and rhodamine phalloidin. The number of budded cells or cells with divided DNA mass was counted and divided by total number of cells counted to give percent budded cells (% budded) (a), and percent with divided DNA (% with divided DNA) (b), respectively. The number of cells in which actin patches were concentrated at the mother–bud junction or with an actin bar at the mother–bud junction was divided by the total number of cells counted (typically 100–150) to give percent with patches at septum or percent with an actin bar at septum (d), respectively. Another fraction of the fixed cells from each time point was sonicated and counted on a hemocytometer. The resultant cell concentration at each time point was divided by that at time 0 to give relative cell number (c). In a, b, and c: ○, wild type; ▪, Cyk1− cells. ▴ in a, Cyk1− cells with more than one bud. Asterisk in c, relative cell number after zymolyase treatment. ○ in d, wild-type cells with actin patches at septum; ▵, wild-type cells with actin bar at septum; •, Cyk1− cells with actin patches at septum; ▴, Cyk1− cells with actin bar at septum. (D) Cyk1− cells after 4 h in glucose from the same experiment as in B and C were fixed, treated with zymolyase, and then stained with DAPI. A representative field of cells is shown. (E) Cyk1− cells from the 135-min time point were processed for thin section electron microscopy. Examples of cells with two buds connected to the mother cell are shown. (F) Cyk1− cells from the 75-min time point were stained with rhodamine phalloidin. A representative field of cells is shown. Bars: (D and F) 10 μm.; (E) 1 μm.
Fluorescence Staining of Yeast Cells
Cells grown in YPD were fixed directly in growth media by addition of 37% formaldehyde to 5% final concentration, and incubation at room temperature for 1 h with gentle agitation. Cells grown in synthetic media were shifted to growth in YPD for 1 h before fixation because the low pH of the synthetic media resulted in poor fixation. Phalloidin staining was performed as described (Guthrie and Fink, 1991) except that the cells were treated with zymolyase to remove the cell wall after fixation in all of the experiments in this study. Immunofluorescence staining of the Cyk1-myc–expressing strain (RLY230) with rat antitubulin (Kilmartin et al., 1982), mouse anti-myc (Evan et al., 1985), rabbit anti-Cof1 (Moon et al., 1993), rabbit anti-Sac6 (Drubin et al., 1988), and rabbit anti-Cdc11 (Ford and Pringle, 1991) primary antibodies and FITC-conjugated or rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) was carried out as described (Drubin et al., 1988). To double stain cells with rhodamine phalloidin and an antibody, the methanol/acetone treatment was omitted from the immunofluorescence staining procedure. After incubation with the secondary antibody, the cells were incubated with 0.05 μM rhodamine phalloidin (Molecular Probes, Inc., Eugene, OR) for 20–30 min in the dark. The cells were visualized on a Zeiss Axiophot microscope with a HB100 W/Z high pressure mercury lamp and a Zeiss ×63 Plan Neofluar oil immersion objective (Carl Zeiss, Inc., Thornwood, NY). Image acquisition was carried out using Northern Exposure (Phase 3 Imaging Systems, Milford, MA).
Electron Microscopy
Thin section electron microscopy was carried out as previously described (Li, 1997), except that the zymolyase-treated cells were filtered on to a piece of AcetatePlus membrane (Micron Separation Inc., Westboro, MA) before the incubation with osmium tetraoxide.
Immunoblot Analysis
Total yeast cell extract was prepared by glass bead lysis in SDS sample buffer. Immunoblot analysis was carried out using the enhanced chemiluminescence detection kit (Amersham Corp., Arlington Heights, IL).
Observation of Myo1-GFP–expressing Cells
RLY302 cells grown in selective media were placed on an agarose pad essentially as described (Waddle et al., 1996). Living cells were imaged at room temperature on a Diaphot 300 microscope with a 100/1.40 oil DIC objective (Nikon Inc.). Images were collected every minute with 0.1-s exposure to fluorescent light filtered through a Chroma 41018 filter using a CH250 cooled CCD camera (Photometric, Tucson, AZ). The shutter was controlled automatically using the UniBlitz D122 shutter driver and Metamorph 2.5 software (Universal Imaging Corp., West Chester, PA). Photobleaching was minimal and cells remained capable of budding and growing during a 3-h time course.
Results
An IQGAP-like Protein Is Specifically Required for Budding Yeast Cytokinesis
The original goal of this study was to identify downstream effectors of Cdc42 in regulating actin rearrangements. Recently, the IQGAP family proteins identified in mammalian organisms have been implicated in carrying out such function (Weissbach et al., 1994; Brill et al., 1996; Hart et al., 1996; McCallum et al., 1996; Bashour et al., 1997). As an attempt to study the in vivo role of IQGAP-like proteins through genetic analysis, we identified an open reading frame from the Saccharomyces cerevisiae genomic database that encodes a 165-kD polypeptide with significant structural similarity to IQGAP (Fig. 1). This polypeptide was named Cyk1 (cytokinesis) for its function demonstrated below. Like the mammalian IQGAPs, Cyk1p contains an NH2-terminal calponin homology domain, followed by eight IQ motifs and a COOH-terminal, GAP-related domain (GRD). Cyk1 does not appear to have a WW domain. Four consensus sites for phosphorylation by Cdc28 (S. cerevisiae homologue of cdc2) (Moreno and Nurse, 1990) or MPM-2 kinases (Westendorf et al., 1994) are present in the NH2-terminal part of Cyk1p (Ser 7, Thr 298, Ser 354, and Ser 404).
Figure 1.
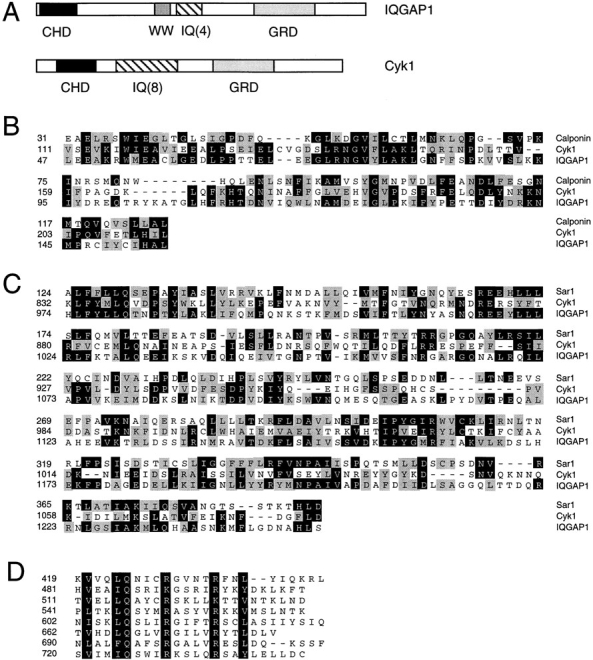
Comparison of structural domains of Cyk1p with human IQGAP1. The complete amino acid sequence of Cyk1 protein is not shown but these data are available from GenBank/ EMBL/DDBJ under accession number Z73598; and yeast genome database ORF name: YPL242c). (A) Schematic diagram comparing the domain organization of human IQGAP1 (these data are available from GenBank/EMBL/DDBJ under accession number L33075) and Cyk1p. CHD, calponin homology domain; WW, WW domain; IQ, IQ motifs; and GRD, GAP-related domain. (B) Alignment of the calponin homology domains of Cyk1p and IQGAP1 with the corresponding segment of mouse calponin (Swiss Protein database accession number: Q08093). Identical residues are shaded in black, conserved substitutions are shaded in gray. (C) Alignment of the GRDs of IQGAP1 and Cyk1p with the catalytic domain of Schizosaccharomyces pombe Ras GAP Sar1 (available from GenBank/EMBL/DDBJ accession number S37449). Identical residues are shaded in black, conserved substitutions are shaded in gray. (D) Alignment of the eight IQ motifs in Cyk1p. The consensus residues are shaded in black.
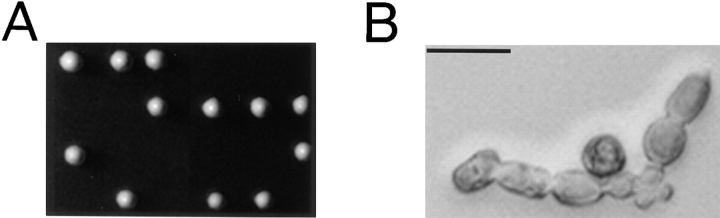
To determine the cellular function of Cyk1p, a null allele (Δcyk1) was constructed where 88% of the open reading frame was replaced by the LEU2 marker gene. Tetrad analysis of a diploid strain heterozygous for Δcyk1 showed 2:2 segregation for viability (Fig. 2 A), and all of the viable spores were Leu− (data not shown), indicating that CYK1 is an essential gene. Examination of the Δcyk1 microcolonies under a light microscope revealed a qualitatively uniform terminal morphology: a chain of large cells, sometimes with branches (Fig. 2 B) that could not be separated with a microdissection needle. This indicates a defect in cell division. The cells then became transparent and amorphous (an indication of lysis; data not shown).
Figure 2.
Phenotype of CYK1 gene disruption. (A) Tetrad analysis of a diploid strain (RLY226) heterozygous for the Δcyk1 mutation showing 2:2 segregation of viability. The plate was photographed after 5-d growth at 23°C. (B) The morphology of a typical Δcyk1 microcolony after 24-h growth at 23°C, after tetrad dissection. The microcolony did not further increase in size and the cells eventually lysed. The photograph was taken under a Zeiss Axiophot microscope with a ×40 Nikon 0.5 ELWD objective. Bar, 20 μm.
To determine if Cyk1p has a specific function in cytokinesis, a strain was constructed in which the sole expression of Cyk1p is controlled by the inducible GAL1 promoter. The Cyk1p in this strain is also tagged at the COOH terminus with six myc epitopes. This strain grows as well as the wild type in the presence of galactose but is unable to grow in glucose-containing media (Fig. 3 A). The Cyk1-myc– expressing cells cultured in the galactose-containing media were shifted to the glucose-containing media for 3 h. α-Mating factor was also present during this period to arrest the cells in G1. The cells were then released from the G1 arrest but maintained in the glucose media. The G1 arrested cells as well as those released from the arrest did not contain any detectable Cyk1-myc (Fig. 3 B), and will thus be referred to as Cyk1− cells. As shown in Fig. 3 C (a and b), Cyk1− cells budded and went through nuclear division at rates similar to those of the wild-type cells. The number of the wild-type cells had nearly tripled 4 h after release from the α-mating factor arrest, whereas Cyk1− cells did not increase in number and were therefore deficient in cell division (Fig. 3 C, c). At 135 min, while the second peak of budded cells appeared in the wild-type population, 60% of the Cyk1− cells had two buds that could not be separated from the mother by sonication (Fig. 3 C, a). After 4 h in the glucose media, >90% of the Cyk1− cells appeared as chains of cell bodies with many nuclei (Fig. 3 D). These results strongly suggest that Cyk1p is required for cell division but not for the progression of the budding or the nuclear cycle.
Two experiments were performed to determine if the cell division defect observed in Cyk1− cells was because of a block in cytokinesis (the division of the cytoplasm), or a failure in septum formation and cell separation. First, Cyk1− cells from the 240-min time point were fixed and treated with zymolyase to remove the cell wall. The cell number did not increase after such treatment (Fig. 3 C, c; asterisk), and the cells remained as chains of attached cells (Fig. 3 D). Thus, Cyk1− cells exhibited a cytokinesis defect defined by the criteria previously described (Hartwell, 1971). Second, Cyk1− cells from the 135-min time point were examined by thin section electron microscopy. At this time point, most of the cells had two buds attached to one mother. In cell sections that captured the mother and both of the buds, it was often apparent that the cytoplasm of the mother and both buds remained connected (Fig. 3 E), indicating that cytokinesis did not occur. Cells in which the mother had connected cytoplasm with two buds as shown in Fig. 3 E were never observed in sections of the wild-type cells (data not shown). The above results suggest that Cyk1p plays an essential role in cytokinesis.
Cyk1p Is Not Required for the Accumulation of Actin Patches at the Septum
The landmark for budding yeast cytokinesis has been considered as the accumulation of cortical actin patches around the mother–bud junction (Adams and Pringle, 1984; Kilmartin and Adams, 1984). We thus tested if Cyk1− cells are defective in such actin organization. Cells from each time point in the synchronized release experiment described above were fixed and stained with rhodamine phalloidin (Fig. 3 C, d). In small-budded Cyk1− cells, actin patches and cables were properly polarized as in wild-type cells (data not shown). In the wild-type cell culture, a small peak (21% of the total) of cells with actin patches accumulated at the septum was observed 75 min after release from the G1 arrest. At the same time point, 31% of the cells displayed an actin bar across the mother–bud junction, similar in appearance as that described below and shown in Fig. 4 D, c. In the Cyk1− cell culture, by contrast, >70% of the cells had accumulated actin patches around the septum (Fig. 3 F) and actin cables were oriented toward the septum. This population continued to increase until the 90-min time point. The actin bar at the mother–bud junction was not apparent in Cyk1− cells (only in 1 out of the 227 cells counted for the sample from the 75-min time point, we could not rule out the presence of an actin bar). This result suggests that Cyk1p is not required for the accumulation of actin patches to the septum, and that actin patches and cables are not sufficient for cytokinesis.
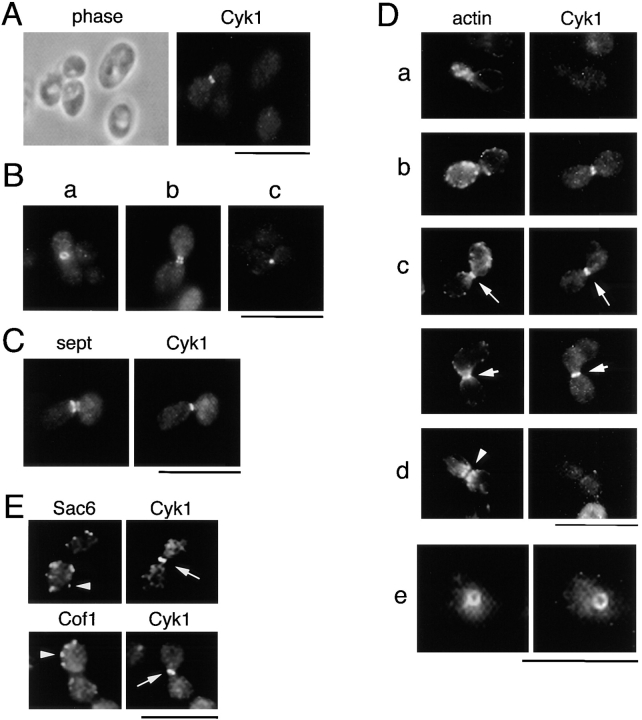
Figure 4.
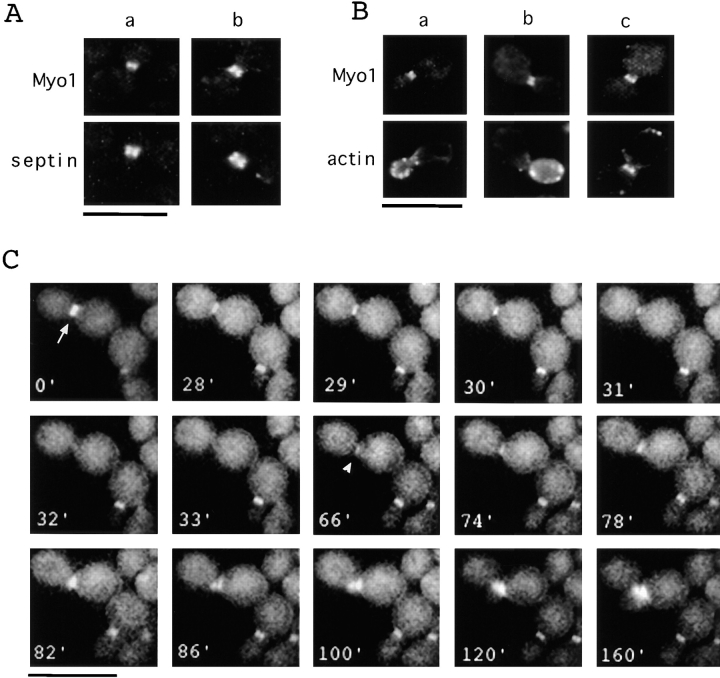
Cyk1p colocalizes with an actin ring. RLY230 (Cyk1-myc–expressing) cells were grown in YPD and fixed with formaldehyde. (A) A representative field of cells stained with mouse anti-myc antibody. (B) Examples of cells that (a) show a ring at the mother–bud junction when tilted at an angle from the glass surface; (b) show a double band at the mother–bud junction; and c show a dot at the mother–bud junction. (C) The fixed cells were double stained with mouse anti-myc and rabbit anti-Cdc11 (septin) primary antibodies and rhodamine-conjugated anti-mouse, and FITC-conjugated anti-rabbit secondary antibodies. (D) Double staining of fixed RLY230 cells with rhodamine phalloidin and anti-myc antibody. Arrows in c point to the actin or Cyk1 band in two large-budded cells. The arrowhead in d points to the actin-free zone in a representative cell with actin patches concentrated around the septum. e shows the ringlike appearance of the actin and Cyk1p containing structure in a cell that was tilted at an angle from the cover glass. Bars, 10 mm.
Cyk1p Localizes to a Ring at the Mother–Bud Junction
To gain further insights into the function of Cyk1p, a strain was constructed that expresses COOH-terminal, myc-tagged Cyk1p (Cyk1-myc) as the sole source of Cyk1p. This strain grows as well as the wild type (data not shown), and was used for determining the cellular location of Cyk1p. Immunofluorescence staining using a monoclonal anti-myc antibody showed that the distribution of Cyk1p was diffuse or slightly punctate in the majority of an exponentially growing cell population. In a small fraction of the cells (∼5% of the total population, all were large-budded), Cyk1-myc staining appeared as a single band across the mother–bud junction (Fig. 4 A). Frequently, in cells that were properly angled or floating in the mounting solution, the Cyk1p-containing structure appeared as a ring (Fig. 4 B, a and D, e). In ∼3% of the cells that had localized Cyk1 staining, the Cyk1 structure appeared as a double ring (Fig. 4 B, b). In rare occasions, Cyk1-myc staining appeared as a dot at the mother–bud junction (Fig. 4 B, c). These staining patterns were never seen in a wild-type strain bearing the vector alone or in any of the previous unrelated immunolocalization studies using the same primary and secondary antibodies (data not shown). Therefore, the staining is specific for Cyk1p, and the neck location supports a direct role in cytokinesis.
In budding yeast, the septins are known to form a ring at the mother–bud junction and are required for cytokinesis (Longtine et al., 1996). We thus determined the relationship of the Cyk1 ring with the septin ring. By immunofluorescence staining, the septin-containing structure appeared as double rings around the bud neck (Fig. 4 C). Very often, the Cyk1 single ring appeared to be “sandwiched” between the septin double ring (Fig. 4 C), although in a small population of cells the Cyk1 ring was asymmetrically located between the septin rings (data not shown). Interestingly, the Cyk1 ring was usually smaller in diameter (as judged by the maximum length of the Cyk1 band) than the septin ring, suggesting that it might be located inside the septin ring.
The Cyk1 Ring Colocalizes with an Actin Ring That Is Distinct From Actin Patches or Cables
Because human IQGAP1 has been shown to colocalize with actin structures (Bashour et al., 1997), the relationship between the Cyk1 ring with actin structures was determined. Double staining of Cyk1-myc–expressing cells with anti-myc antibody and rhodamine-phalloidin showed that in the majority (∼90%) of cells that contained the Cyk1 ring at the neck, a superimposable actin ring was also present (Fig. 4 D, c and e). At the point shown in Fig. 4 D (c), actin patches were randomly distributed in both the mother and the bud. Occasionally, the Cyk1 ring was observed in cells that still exhibited polarized actin patch distribution and without an apparent actin ring at the neck (Fig. 4 D, b). In small budded cells as shown in Fig. 4 D, a, it was difficult to tell if the actin ring was present because of the high concentration of actin patches and because the Cyk1 ring was not present to serve as a marker. In large budded cells where actin patches were less dense, an overlapping Cyk1 ring was also present whenever a clear actin ring as shown in Fig. 4 D (c) was observed. At the stage where actin patches were concentrated near the septum, the Cyk1 ring was not observed (Fig. 4 D, d). The actin ring also seemed to be absent, because there was usually an apparent actin-free zone (Fig. 4 D, d, arrowhead) separating the actin patches in the mother from those in the bud. To test whether the actin ring that overlaps with the Cyk1 ring is simply a ring of actin patches, Cyk1-myc– expressing cells were stained with antibodies against cofilin (Cof1) (Moon et al., 1993) and fimbrin (Sac6) (Drubin et al., 1988), two actin patch components. As shown in Fig. 4 E, the staining with these antibodies was observed only in patches and not in the Cyk1-containing ring, suggesting that the Cyk1/actin ring and actin patches may have different structural organization.
The Cyk1/Actin Ring Assembles in Anaphase
To better define the cell cycle stage at which the Cyk1/actin ring is present, the Cyk1-myc–expressing cells were simultaneously stained with anti-myc and antitubulin antibodies and with 4',6-diamidino-2-phenylindole dihydrochloride (DAPI; a DNA stain). Spindle and nuclear morphology are well characterized and provide excellent markers for different cell cycle stages (Adams and Pringle, 1984; Kilmartin and Adams, 1984). The Cyk1 ring was predominantly observed in cells with an elongated spindle and two separated lobes of DNA (Fig. 5 A). Much less frequently, the Cyk1 ring was observed in a cell with a short spindle, but the rings in these cells were always much fainter than those in cells with a long spindle (Fig. 5 B, compare the intensity of the Cyk1 ring in cells with a long spindle [a and b] to that of the cell with a short spindle [c]). The distribution of cells in cell cycle stages characterized by different spindle and budding morphologies was compared to that of cells that displayed a Cyk1 ring. The data shown in Fig. 5 C suggest that the formation of the Cyk1 ring occurs predominantly after spindle elongation and disassembles after the completion of anaphase.
Figure 5.
The Cyk1 ring assembles in anaphase. Fixed RLY230 cells were triple stained with mouse anti-myc, rat antitubulin antibodies, and DAPI. (A) A representative cell that displays a Cyk1 ring and an elongated spindle. (B) Comparison of the intensity of the Cyk1 ring in cells with an elongated spindle (cells a and b) with that in a cell with a short spindle (cell c). The intensities of the Cyk1 rings shown are representative. The ends of the long spindles of cells a and b are out of focus. (C) White bars, cells in an exponentially growing RLY230 population that displayed the spindle and nuclear morphology as diagrammed below the histogram were counted and the resultant numbers were divided by the total number of cells counted (220) to yield the percentages. Solid bars, in the same population, cells that had a Cyk1 ring and the spindle and nuclear morphology as diagrammed were counted. The obtained numbers were divided by the total number of cells with a Cyk1 ring counted (145) to yield the percentages. Bars, 10 μm.
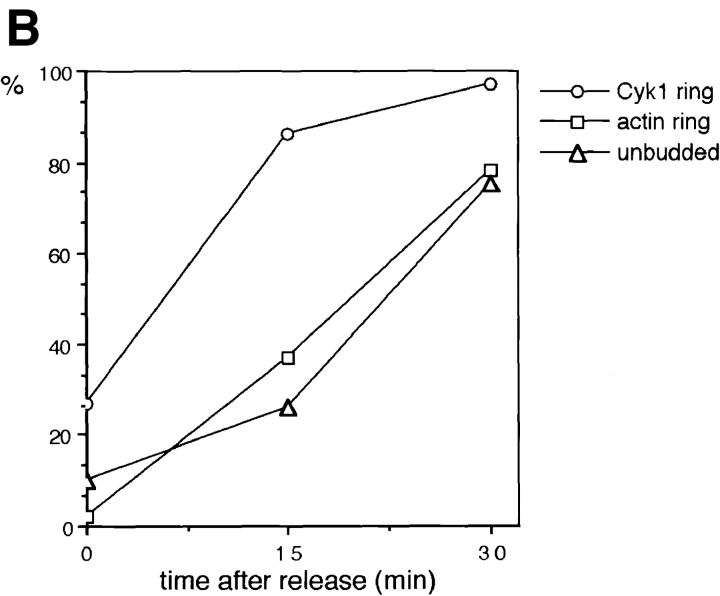
In budding yeast, the late events in mitosis are regulated by a set of genetically interacting proteins, including Cdc15p, Cdc5p, and Cdc14p, etc. (Hartwell et al., 1973; Schweitzer and Philippsen, 1991; Kitada et al., 1993; Morishita et al., 1995; Shirayama et al., 1996). Temperature-sensitive cdc15 cells grown at 37°C arrest with anaphase spindle and nuclear morphologies and high MPF activity (Schweitzer and Philippsen, 1991; Surana et al., 1993). We thus asked whether the assembly of the Cyk1/actin ring occurs downstream from the pathway constituted by these proteins. A temperature-sensitive cdc15 strain that expresses the myc-tagged Cyk1p was constructed. The cells were arrested at 37°C, and then released from the arrest to growth at 23°C. Samples were fixed at various time points after the release and stained with anti-myc antibody and rhodamine phalloidin. A small fraction (∼27% of the total) of the arrested cells displayed a weakly stained Cyk1 ring without an apparent actin ring (Fig. 6 A). 15 min after the cells were shifted to the permissive temperature, a bright Cyk1 ring was observed in 87% of the large budded cells, 34% of which displayed a clear actin ring. At 30 min, the majority of the large-budded cells had a Cyk1/actin ring. Concomitant with the appearance of the actin ring, the number of unbudded cells increased (Fig. 6 B), suggesting that cell division was occurring. Immunoblot analysis indicated that the level of Cyk1p did not change after the 37° to 23°C temperature shift (Fig. 6 C). These results suggest that the assembly of the Cyk1/actin ring is regulated by a posttranslational mechanism downstream of the Cdc15-dependent pathway. Cyk1p assembles to the ring probably before the accumulation of actin filaments, although we cannot rule out the presence of a small amount of actin in the ring before the accumulation of Cyk1p that was not detected by our staining method.
Figure 6.
The assembly of the actin ring occurs downstream of Cdc15. RLY238 cells (cdc15-2 CYK1-myc) were grown at 37°C for 3 h. 5 ml of cells were fixed as the 0 time point sample. The rest of the cells were harvested and resuspended in 23°C YPD and grown at 23°C for 15 and 30 min before fixation or processing for immunoblot analysis. The fixed cells were double stained with anti-myc antibody and with rhodamine phalloidin. (A) Representative cells from the 0 time point that had a Cyk1 ring (top left). Representative cells from the 30-min time point that had a Cyk1 ring (bottom left). (B) The number of budded cells (mostly large-budded) that had a Cyk1 ring or an actin ring was counted and divided by the total number of budded cells counted (∼150) to yield the percentage. The number of unbudded cells were also counted and divided by the total number counted to yield the percentage. All cells that had an obvious actin ring had a Cyk1 ring. (C) Cells from each time point were processed for immunoblot analysis using anti-myc antibody. Bar, 10 μm.
Myosin II Is a Component of the Actin Ring
The results presented above raise the possibility that a contractile ringlike structure may exist in budding yeast. One of the key components of the contractile ring in metazoan cells is myosin II (for review see Schroeder, 1990; Satterwhite and Pollard, 1992; Fishkind and Wang, 1995). Myo1p, a myosin II–like molecule was previously identified in S. cerevisiae, and its null mutation gives rise to chains of attached cells (Watts et al., 1987; Rodriguez and Paterson, 1990). Consistent with a role in cytokinesis, Myo1p was reported to localize to the bud neck (Watts et al., 1985). To identify the relationship between the Myo1 ring and the Cyk1/actin ring, we constructed a strain that expresses COOH-terminal, myc-tagged Myo1p under the control of the native MYO1 promoter. Immunofluorescence staining with anti-myc antibody showed that the tagged Myo1 protein localized to a ring or sometimes a double ring at the mother–bud junction. The Myo1 ring overlapped partially with the septin ring and was usually smaller in diameter and in width than the latter (Fig. 7 A). However, unlike the Cyk1/actin ring, the Myo1 ring appeared to be present in cells with buds of all sizes (see below). In small-budded cells where actin patches were concentrated in the bud, the Myo1 ring did not colocalize with an actin ring (Fig. 7 B, a and b). In anaphase cells, however, when an actin ring was observed, a superimposable Myo1 ring was also present (Fig. 7 B, c), suggesting that Myo1p is a component of the ring containing actin and Cyk1p. This result, together with the phenotype of MYO1-disrupted cells, supports the idea that an actomyosin-based ring is involved in cytokinesis in budding yeast.
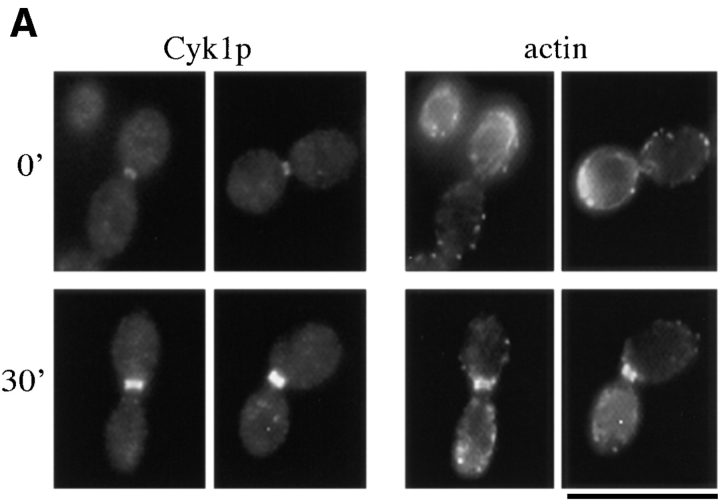
Figure 7.
The localization of Myo1p. (A and B) RLY301 (Myo1-myc–expressing) cells were fixed and double stained with mouse anti-myc and rabbit anti-Cdc11 (A) or rhodamine phalloidin (B). (C) RLY302 (Myo1-GFP–expressing) cells were observed by video microscopy as described in Materials and Methods. Arrow in the 0′ panel indicates the cell discussed in the text. Arrowhead in the 66′ panel indicates the site where a new Myo1 structure started to appear. Bar, 10 μm.
Observation of Myo1 Ring Dynamics In Vivo
To further determine the timing of assembly and the activity of the Myo1p-containing ring, a strain was constructed that expresses Myo1p tagged at the COOH terminus with GFP under the control of the native MYO1 promoter. These cells grow at a wild-type rate (data not shown) and display a green fluorescent ring similar to that observed by immunofluorescence staining of fixed cells except that it is clearly a single ring (Fig. 7 C), suggesting that the double ring appearance by immunofluorescence staining may be artifactual. The Myo1-GFP–expressing cells were observed by time-lapse video microscopy. Fig. 7 C shows one representative series that was recorded. The Myo1 ring in the large-budded cell (Fig. 7 C, 0′, arrow) stayed quiescent for >28 min after the start of recording. At 30 min the ring started to get smaller, and at 31 min, it had the appearance of a dot at the center of the mother–bud junction. The dot disappeared completely 2 min later. The ring to dot change and the disappearance of the dot were not because of the bleaching of the fluorescence because the Myo1 ring in other cells in the same field did not change in shape or intensity during the same period (the budded cell near the bottom right-hand corner of each panel in Fig. 7 C is such an example). At 66 min, a small fluorescent patch appeared next to the previous Myo1 ring. Within 4 min, a newborn bud of the right daughter cell was clearly visible in the same region when observed under Nomarski optics (data not shown; the new bud was not apparent in the fluorescence channel until the 120-min time point). The new Myo1-GFP patch became brighter and wider as the new bud grew. At 82 min, another fluorescent patch also appeared and correlated with the bud of the left daughter cell. After examination of several such series, we can summarize the observations as: (a) the assembly of the Myo1 ring occurs 4–6 min before the time of bud emergence; (b) the ring does not change significantly in appearance for most of the cell cycle, and then abruptly reduces to a dot; and (c) the dot disappears completely within 1–2 min. A septum can be observed at all focal planes under Nomarski optics 2–4 min after the ring-to-dot transition (data not shown), indicating that cytokinesis has occurred.
The Myo1 Ring Is Disrupted by a Septin Mutation
The observation that the Myo1 ring appears slightly before bud emergence and overlaps with the septin ring raised the possibility that the septins may be required for the assembly and maintenance of the Myo1 ring. To test this possibility, the Myo1-myc–expressing construct was introduced into a cdc12-6 (Kim et al., 1993) mutant strain. The cells were cultured at 23°C, and then shifted to growth at 37°C, the nonpermissive temperature. As expected, the septin ring was lost in >99% of the mutant cells 30 min after the temperature shift (Table II). The Myo1 ring was also absent in these cells. The Myo1 ring was only seen in 2 of the 500 cells counted, but both cells also maintained the septin ring. There was only a small drop in the fraction of the septin ring–containing or Myo1 ring–containing cells in the wild-type culture shifted to 37°C, suggesting that disappearance of the septin and Myo1p rings in the mutant culture was not simply because of the temperature shift. This result suggests that the septins play an important role in the assembly and maintenance of the Myo1 ring.
Table II.
The Myo1 Ring Is Abolished by a Septin Mutation
| Percent with a septin ring | Percent with a Myo1 ring | |||
|---|---|---|---|---|
| CDC12 at 23°C | 68 | 28 | ||
| CDC12 at 37°C | 56 | 23 | ||
| cdc12-6 at 23°C | 67 | 37 | ||
| cdc12-6 at 37°C | 0 | 0 |
RLY301 (Myo1-myc–expressing; CDC12) and RLY311 (myo1-myc–expressing; cdc12-6) cells were grown overnight at 23°C. Half of the cells were maintained at 23°C, and the other half were shifted to growth at 37°C for 30 min, fixed, and then stained with anti-myc and anti-Cdc11 antibodies. Fields of cells were first counted in the phase channel. The number of cells within the same field that had a septin ring or a Myo1 ring was counted in the appropriate fluorescence channel. The resultant numbers from the counting in the fluorescence channels were divided by the total cell number counted in the phase channel to give percent of cells with a septin ring or percent of cells with a Myo1 ring. 250–500 cells were counted for each sample.
Discussion
An Actomyosin Ring Involved in Budding Yeast Cytokinesis
Cell division in budding yeast appears to occur in a manner different from that in metazoan cells or fission yeast. S. cerevisiae cells bud early in the cell cycle at the G1–S transition and set up the division site at the bud neck upon the assembly of a septin ring (Byers, 1981). Because the neck is already constricted throughout the cell cycle, the activity of a contractile ring does not seem necessary for the completion of cytokinesis. Studies on actin-based structures in budding yeast have been focused on cortically associated actin patches and cytoplasmic actin cables. A significant body of evidence suggests that these structures play an important role in directing plasma membrane growth and cell wall synthesis (for review see Drubin, 1991; Bretscher et al., 1994). Because actin patches accumulate at and actin cables are oriented toward the mother–bud junction around the time of cytokinesis (Adams and Pringle, 1984; Kilmartin and Adams, 1984), and because it is known that cell division involves the formation of a cell wall septum (Byers, 1981), it is conceivable that local membrane insertion and cell wall formation are sufficient for the closure of the mother–bud connection.
In the present study, we first identified a mutation, Δcyk1, that inhibits cytokinesis but does not affect the organization of actin patches and cables at the predicted cell division site, suggesting that additional factors may be required for the completion of cytokinesis. Immunolocalization of Cyk1p revealed a ring that contains actin and occurs transiently around the time of cell division. In an earlier study on actin organization in S. cerevisiae cells (Adams and Pringle, 1984), it was reported that “cells with very large buds often show a concentration of fluorescence (actin) in the neck region, sometimes apparently as clusters of spots, sometimes apparently as a band.” The significance of the actin band was not further studied, probably because it was only observed in a small fraction of an exponentially growing culture and might not be distinct from other actin structures. In this study, the specific localization of Cyk1p to the actin ring provided a molecular marker for this structure and allowed further dissection of its assembly and function in the cell cycle .
Several properties of this structure are consistent with those of a contractile ring. First, this structure is composed of filamentous actin and a myosin II–like protein, whose mammalian counterparts are thought to generate the contractile force in the cleavage furrow (for review see Schroeder, 1990; Satterwhite and Pollard, 1992; Fishkind and Wang, 1995). Second, Cyk1p, a protein that concentrates solely to this but not other known structures, plays an essential and specific role in cytokinesis, suggesting that the structure it composes is likely to be critical for the same process. Third, the Cyk1/actin ring assembles in anaphase after spindle elongation and chromosome segregation, the same morphological stage when contractile rings in animal cells are formed. Fourth, time-lapse recording of living cells that expressed Myo1-GFP showed that the ring reduces to a dot at the end of the cell cycle. A similar change in ring appearance was also observed in GFP-Cyk1p expressing cells (Li, R., unpublished result). It is unlikely that this change is because of breakage of the ring followed by disassembly from the ends, because the dot always ends up at the center of the bud neck. We thus suspect that this change in shape is due to contraction of the ring during cytokinesis. Interestingly, the Myo1p-containing structure immediately disappears after the ring-to-dot change. This might be similar to the observation in marine eggs that as the cleavage furrow ingresses, the width of the contractile ring decreases abruptly and vanishes as the cleavage completes (Schroeder, 1972).
What might be the function of a contractile ring during cytokinesis, if the neck region is already constricted early in the cell cycle? One possibility is that the actomyosin ring drives the membrane closure at the mother–bud junction and this provides the docking sites for the vesicles that deliver the cell wall material for the generation of the septum. Alternatively, contraction might occur in a coordinated fashion with vesicle fusion along the plasma membrane around the bud neck, such that the new membrane growth generates an invagination rather than a longer or broader neck. Consistent with this second possibility, many of the Cyk1− cells observed by electron microscopy appeared to have an unusually long or wide neck (Fig. 3 E).
Assembly of the Cytokinetic Ring in the Cell Cycle
In animal cells, cleavage furrow formation and cell cleavage occur as a continuous sequence of events (Schroeder, 1990). In budding yeast, on the other hand, cell division occurs in two steps: first, bud emergence at the G1–S transition, which creates a partially advanced “cleavage furrow;” and second, completion of cytokinesis and septum formation at the end of anaphase. This unique feature implies that the assembly of the cytokinesis apparatus in yeast may also occur in temporally separated steps. Indeed, localization of four different proteins or a protein complex (septins) involved in cytokinesis to the mother–bud junction occurs at different stages in the cell cycle (Fig. 8). The assembly of the septin filaments to form a ring at the presumptive bud site occurs ∼15 min before bud emergence (Kim et al., 1991), and is likely to be dependent on Cdc28 activation by G1 cyclins (Lew and Reed, 1995). Electron microscopy studies showed that the septin ring, which contains Cdc3, 10, 11, and 12 proteins, consists of 10-nm filaments aligned with constant spacing encircling the bud neck (for review see Byers and Goetsch, 1976; Longtine et al., 1996). Mutations in any of the septin proteins results in a cytokinesis defect as well as an inability to depolarize cell growth later in the cell cycle (for review see Pringle et al., 1995). The precise role of the septins in cytokinesis is not clear. We showed that disruption of the septin ring by the cdc12-6 mutation results in a loss of the Myo1 ring, suggesting that at least one of the important functions of the septins might be to recruit components of the actomyosin ring to the division site.
Figure 8.
A model for the pathway of cytokinetic ring assembly in budding yeast cell cycle. Activation of Cdc28 kinase activity by the G1 cyclins (Cln) triggers the assembly of the septin ring and the myosin ring a few minutes before the bud becomes visible. During mitosis, activation of the Cdc15/Cdc5 pathway induces the localization of Cyk1p to the Myo1 ring, which subsequently recruits actin filaments. The septin filaments may disassemble at this point, as suggested by EM studies (Byers and Goetsch, 1976). This could be a trigger for actin ring contraction. After the completion of cytokinesis, the actin ring disassembles immediately by an unknown mechanism. Septum formation then occurs to complete cell division.
The assembly of Myo1p to a ring that overlaps part of the septin ring also occurs slightly before bud emergence. The Myo1 ring does not appear to colocalize with any distinct actin structure until the appearance of the actin ring during anaphase. Therefore, the function of Myo1p before anaphase may not be dependent on its motor activity. Since in vitro studies have shown that myosin II forms filaments under physiological salt conditions (Niederman and Pollard, 1975), it is possible that a filamentous ring of Myo1p acts together with the septin ring to provide the constriction at the bud neck during bud growth. Myo1p may also play a direct role in recruiting or organizing actin filaments in the ring during anaphase, since myosin filaments in vitro are able to assemble actin filaments of opposite polarities (Hayashi et al., 1977). A role for Myo1p in cytokinesis is supported by the reported phenotype of MYO1 gene–disrupted cells: long chains of undivided cells (Watts et al., 1987; Rodriguez and Paterson, 1990). However, the Δmyo1 mutation also seems to result in a number of other defects, including those in budding pattern, chitin deposition, and cell wall organization (Rodriguez and Paterson, 1990). Further investigation using conditional myo1 mutations should lead to a better understanding of the primary function of myosin II in yeast.
The assembly of Cyk1p to the ring at the mother–bud junction occurs predominantly in anaphase, and is stimulated by Cdc15 activation. This step seems to be immediately followed by the accumulation of actin filaments in the ring. Genetic analysis indicated that Cdc15p functions in the same pathway as Cdc5p (Hartwell et al., 1973; Schweitzer and Philippsen, 1991; Kitada et al., 1993; Morishita et al., 1995; Shirayama et al., 1996), the budding yeast polo– like kinase (Kitada et al., 1993; Lee and Erikson, 1997), which, in animal cells, is thought to regulate various mitotic events (Glover et al., 1997). Plo1, the fission yeast polo–like kinase is required for actin ring formation and its overexpression induces actin ring assembly in interphase cells (Okura et al., 1995). The relevant target of Plo1 is not known. The human polo–like kinase has been shown to phosphorylate a class of mitotically phosphorylated sites that are recognized by the MPM-2 antibody (Kumagai and Dunphy, 1996). Cyk1p contains four sites that match the consensus sequences found in MPM-2 antigens (Westendorf et al., 1994), and may be a direct target of Cdc5p in the induction of the actin ring.
The Role of Cyk1p in Cytokinesis
We have characterized the role of Cyk1p in the cell cycle by turning off its expression in G1 and allowing the cell cycle to proceed in the absence of Cyk1p. At least in the initial cycle, we detected no defects in cell polarization, budding or nuclear division, but only a block in cytokinesis. Therefore, Cyk1p appears to be required specifically for cytokinesis but not directly for other morphogenetic events. The fact that localized staining of Cyk1p is observed only in the ring that eventually contains both myosin and actin further supports this conclusion. Only after several cell cycles, some Cyk1− cells in the long chains that formed appeared aberrant in morphology and size, or contain none or more than one nuclei. We suspect that the sustained accumulation of actin patches at the mother– bud junction in Cyk1− cells caused a slight delay in the next round of budding (Fig. 3 C). After several generations, the accumulated defects further impaired budding and the coordination between the nuclear cycle and the budding cycle.
Two observations suggest that Cyk1p may play a role in recruiting actin filaments to the ring in anaphase. First, the appearance of the Cyk1 ring occurs slightly earlier than that of the actin ring, after the release from the cdc15-2 block. Although a small amount of actin might have been obscured by the background fluorescence at the mother– bud junction, a distinguishable actin ring was only observed where there was a Cyk1 ring even when cells were observed first in the rhodamine (phalloidin) channel. Second, when the wild-type cells were released from the mating factor–induced G1 arrest, a peak of cells with an apparent actin ring was observed during anaphase, but this peak was not observed in the Cyk1− cell culture. Bovine IQGAP1 has recently been shown to bind to actin filaments with high affinity, inducing the formation of actin bundles in vitro (Bashour et al., 1997). A similar activity may allow Cyk1p to recruit actin filaments into the ring.
It was recently reported that an IQGAP-related protein, GapA, is required for cytokinesis in Dictyostelium discoideum (Adachi et al., 1997). The GapA mutation does not affect the assembly or contraction of the actin ring but seems to block a late step in the completion of cytokinesis, possibly the cleavage of the midbody. It is unlikely that GapA and Cyk1p function by the same mechanism, because GapA contains only the GRD but not the calponin homology domain or IQ motifs. Cyk1p, on the other hand, contains all three domains and is therefore more likely to be functionally related to IQGAPs. IQGAP1 has been shown to colocalize with actin-rich structures at the cell margin, but it was not reported whether this protein is also enriched in the cleavage furrow. It is possible that IQGAPs are involved in actin assembly in contractile structures, which are not restricted to the cleavage furrow in mammalian cells. In yeast, contractile structures may only exist during cytokinesis, which could explain the specific association of Cyk1p to the cytokinetic ring.
IQGAP family proteins have the ability to interact with at least two signaling proteins, Cdc42 and calmodulin (Weissbach et al., 1994; Brill et al., 1996; Hart et al., 1996; McCallum et al., 1996; Bashour et al., 1997). This feature has led to the speculation that IQGAPs may mediate the role of Cdc42 in regulating actin rearrangements. Modulation of Cdc42 activity in Xenopus laevis eggs blocks cytokinesis by affecting cleavage furrow ingression (Drechsel et al., 1996). In yeast, Cdc42 localizes to the bud tip during polarized growth (Ziman et al., 1993), but its involvement in actin ring formation has not been investigated. The IQ motifs in IQGAP1 have been shown to mediate an interaction with calmodulin (Brill et al., 1996; Hart et al., 1996), and calmodulin binding seems to reduce the affinity of IQGAP1 with actin (Bashour et al., 1997). It remains to be determined whether calmodulin is a binding partner with Cyk1p and the effect of this interaction on actin ring formation.
Acknowledgments
We are grateful to T. Lechler for the generation of pTL12 and pTL13; L. Trakimas and M. Ericsson for preparing sections for electron microscopy; D. Sun for assistance in photography of EM images; D. Winter, J. Kahana, and the Silver lab for help in video microscopy; D. Drubin for providing the anti-Cof1 and Sac6 antibodies; J. Pringle for the anti-Cdc11 antibody; and E. Elion, D. Pellman, and J. Pringle for providing yeast strains.
This work was supported by the Funds for Discovery Award to R. Li.
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride
- GFP
green fluorescent protein
- GRD
Gap-related domain
- YPD
yeast extract, peptone, dextrose
- YPG
yeast extract, peptone, galactose
Footnotes
Address all correspondence to R. Li, Department of Cell Biology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115. Tel.: (617) 432-0640. Fax: (617) 975-0524. E-mail: RLi@warren.med.harvard.edu
References
- Adachi H, Takahashi Y, Hasebe T, Shirouzu M, Yokoyama S, Sutoh K. DictyosteliumIQGAP-related protein specifically involved in the completion of cytokinesis. J Cell Biol. 1997;137:891–898. doi: 10.1083/jcb.137.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AEM, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic mutant Saccharomyces cerevisiae. . J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashour AM, Fullerton AT, Hart MJ, Bloom GS. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and crosslinks microfilaments. J Cell Biol. 1997;137:1555–1566. doi: 10.1083/jcb.137.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. The YDp plasmid: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. . Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Drees B, Harsay E, Schott D, Wang T. What are the basic functions of microfilaments? Insights from studies in budding yeast. J Cell Biol. 1994;126:821–825. doi: 10.1083/jcb.126.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S, Li S, Lyman CW, Church DM, Wasmuth JJ, Weissbach L, Bernards A, Snijders AJ. The Ras GTPase-activating protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory. Cold Spring Harbor, New York. 59–96.
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel DN, Hyman AA, Hall A, Glotzer M. A requirement for Rho and Cdc42 during cytokinesis in Xenopusembryos. Curr Biol. 1996;7:12–23. doi: 10.1016/s0960-9822(06)00023-6. [DOI] [PubMed] [Google Scholar]
- Drubin DG. Development of cell polarity in budding yeast. Cell. 1991;65:1093–1096. doi: 10.1016/0092-8674(91)90001-f. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-mycproto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishkind DJ, Wang YL. New horizons for cytokinesis. Curr Opin Cell Biol. 1995;7:23–31. doi: 10.1016/0955-0674(95)80041-7. [DOI] [PubMed] [Google Scholar]
- Ford SK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11gene product and the timing of events at the budding site. Dev Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Glover DM, Ohkura H, Tavares A. Polo kinase: The choreographer of the mitotic stage? . J Cell Biol. 1997;135:1681–1684. doi: 10.1083/jcb.135.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Methods Enzymol. 1991;194:729–731. [PubMed] [Google Scholar]
- Harold FM. To shape a cell: and inquiry into the causes of morphogenesis of microorganisms. Microbiol Rev. 1990;54:381–431. doi: 10.1128/mr.54.4.381-431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO (Eur Mol Biol Organ) J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of cell division cycle in yeast. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast: V. genetic analysis of cdcmutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Silver RB, Ip W, Cayer ML, Smith DS. Actin-myosin interaction. Self-assembly into a bipolar “contractile unit.” . J Mol Biol. 1977;111:159–171. doi: 10.1016/s0022-2836(77)80120-4. [DOI] [PubMed] [Google Scholar]
- Kilmartin J, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. . J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J, Wright B, Milstein C. Rat monoclonal anti-tubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Haarer BK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: Localization of the CDC3gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Kim Y, An G. Molecular cloning and characterization of anther-preferential cDNA encoding a putative actin-depolymerizing factor. Plant Mol Biol. 1993;21:39–45. doi: 10.1007/BF00039616. [DOI] [PubMed] [Google Scholar]
- Kishi K, Sasaki T, Kuroda S, Itoh T, Takai Y. Regulation of cytoplasmic division of Xenopusembryo by Rho p21 and its inhibitory GDP/ GTP exchange protein (Rho GDI) J Cell Biol. 1993;120:1187–1195. doi: 10.1083/jcb.120.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K, Johnson AL, Johnson LH, Sugino A. A multicopy suppresser gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. . Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopusegg extracts. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- Larochelle DA, Vithalani KK, De Lozanne A. A novel member of the rho family of small GTP-binding proteins is specifically required for cytokinesis. J Cell Biol. 1996;133:1321–1329. doi: 10.1083/jcb.133.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Erikson RL. Plk is a functional homolog of Saccharomyces cerevisiaeCdc5, and elevated Plk activity induces multiple septation structures. Mol Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Li R. Bee1, a yeast protein with homology to Wiskott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bretscher A. Characterization of TPM1disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- McCallum SJ, Wu WJ, Cerione RA. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol Chem. 1996;271:21732–21737. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Nurse P. Substrates for p34cdc2: in vivo veritas? . Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- Morishita T, Mitsuzawa H, Nakafuku M, Nakamura S, Hattori S, Anraku Y. Requirement of Saccharomyces cerevisiaeRas for completion of mitosis. Science. 1995;270:1213–1215. doi: 10.1126/science.270.5239.1213. [DOI] [PubMed] [Google Scholar]
- Niederman R, Pollard TD. Human platelet myosin. II. In vitroassembly of myosin and structure of myosin filaments. J Cell Biol. 1975;67:72–92. doi: 10.1083/jcb.67.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura H, Hagan I, Glover D. The conserved Schizosaccharomyces pombekinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., E. Bi, H.A. Harkins, J.E. Zahner, C. De Virgilio, J. Chant, K. Corrado, and H. Fares. 1995. Establishment of cell polarity in yeast. Cold Spring Harbor Symp. Quanto. Biol. [DOI] [PubMed]
- Rappaport R. Establishment of the mechanism of cytokinesis in animal cells. Int Rev Cytol. 1986;105:245–281. doi: 10.1016/s0074-7696(08)61065-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez JR, Paterson BM. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskeleton. 1990;17:301–308. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterwhite LL, Pollard TD. Cytokinesis. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- Satterwhite LL, Lohka MJ, Wilson KL, Scherson TY, Cisek LJ, Corden JL, Pollard TD. Phosphorylation of myosin-II regulatory light chain by cyclin-p34cdc2: A mechanism for the timing of cytokinesis. J Cell Biol. 1992;118:595–605. doi: 10.1083/jcb.118.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. The contractile ring. II. Determining its brief existence, volumetric changes, and vital role in cleaving Arbaciaeggs. J Cell Biol. 1972;53:419–434. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. The contractile ring and furrowing in dividing cells. Ann NY Acad Sci. 1990;582:78–87. doi: 10.1111/j.1749-6632.1990.tb21669.x. [DOI] [PubMed] [Google Scholar]
- Schweitzer B, Philippsen P. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 1991;7:265–273. doi: 10.1002/yea.320070308. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. Fink, and C. Lawrence. 1974. Methods in Yeast Genetics. Cold Spring Harbor Laboratory. Cold Spring Harbor, New York. 186 pp.
- Shirayama M, Matsui Y, Toh-e A. Dominant mutant alleles of yeast protein kinase gene CDC15 suppress the lte1 defect in termination of M phase and genetically interact with CDC14. . Mol Gen Genet. 1996;251:176–185. doi: 10.1007/BF02172916. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. Determination of cleavage planes. Cell. 1993;72:3–6. doi: 10.1016/0092-8674(93)90041-n. [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLBmitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO (Eur Mol Biol Organ) J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddle JA, Karpova TS, Waterson RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MP. Calmodulin and the regulation of smooth muscle contraction. Mol Cell Biochem. 1994;135:21–41. doi: 10.1007/BF00925958. [DOI] [PubMed] [Google Scholar]
- Watts FZ, Miller DM, Orr E. Identification of myosin heavy chain in Saccharomyces cerevisiae. . Nature. 1985;316:83–85. doi: 10.1038/316083a0. [DOI] [PubMed] [Google Scholar]
- Watts FZ, Shiels G, Orr E. The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO (Eur Mol Biol Organ) J. 1987;6:3499–3505. doi: 10.1002/j.1460-2075.1987.tb02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach L, Settleman J, Kalady MF, Snijders AJ, Murthy AE, Yan YX, Bernards A. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269:4869–4878. [PubMed] [Google Scholar]
- Westendorf JM, Rao PN, Gerace L. Cloning of cDNAs for M-phase phosphoproteins recognized by the MPM2 monoclonal antibody and determination of the phosphorylated epitope. Proc Natl Acad Sci USA. 1994;91:714–718. doi: 10.1073/pnas.91.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakita Y, Yamashiro S, Matsumura F. In vivo phosphorylation of regulatory light chain of myosin II during mitosis of cultured cells. J Cell Biol. 1994;124:129–137. doi: 10.1083/jcb.124.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Preuss D, Mulholland J, O'Brien JM, Botstein D, Johnson DI. Subcellular localization of Cdc42p, a Saccharomyces cerevisiaeGTP-binding protein involved in the control of cell polarity. Mol Biol Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]