Abstract
Spore formation in yeast is an unusual form of cell division in which the daughter cells are formed within the mother cell cytoplasm. This division requires the de novo synthesis of a membrane compartment, termed the prospore membrane, which engulfs the daughter nuclei. The effect of mutations in late-acting genes on sporulation was investigated. Mutation of SEC1, SEC4, or SEC8 blocked spore formation, and electron microscopic analysis of the sec4-8 mutant indicated that this inability to produce spores was caused by a failure to form the prospore membrane. The soluble NSF attachment protein 25 (SNAP-25) homologue SEC9, by contrast, was not required for sporulation. The absence of a requirement for SEC9 was shown to be due to the sporulation-specific induction of a second, previously undescribed, SNAP-25 homologue, termed SPO20. These results define a developmentally regulated branch of the secretory pathway and suggest that spore morphogenesis in yeast proceeds by the targeting and fusion of secretory vesicles to form new plasma membranes in the interior of the mother cell. Consistent with this model, the extracellular proteins Gas1p and Cts1p were localized to an internal compartment in sporulating cells. Spore formation in yeast may be a useful model for understanding secretion-driven cell division events in a variety of plant and animal systems.
Sporulation in Saccharomyces cerevisiae is an unusual form of cell division in which the daughter cells are formed within the original mother cell (Esposito and Klapholz, 1981). In this process, a single round of DNA replication followed by two meiotic divisions occurs within a single, intact nuclear envelope. During the second meiotic division, an extension of the outer plaque is formed on the cytoplasmic side of each of the four spindle pole bodies (Moens, 1971; Moens and Rapport, 1971; Byers, 1981). A flattened membrane sac then forms on the cytoplasmic face of each of the outer plaques. This membrane, which is sometimes called the prospore wall, is referred to in this work as the prospore membrane. As meiosis II progresses, the chromatin segregates into four lobes of the nucleus and each prospore membrane grows larger to engulf the adjacent nuclear lobe. As meiosis II is completed and the nucleus divides, each prospore membrane fuses with itself so that every daughter nucleus (and associated cytoplasm) is captured inside two, now distinct, unit membranes of the prospore membrane, creating immature spores. The bilayer of the prospore membrane closest to the daughter nucleus now serves as the plasma membrane of the spore. In the final step of spore morphogenesis, the spore wall is formed in the lumenal space between the two membranes derived from the prospore membrane (Lynn and Magee, 1970).
Although the cytology of spore morphogenesis has been described, little is known about the molecular and genetic requirements for spore formation. A growing number of mutants have been identified that proceed through meiosis II but do not form proper spores (Moens et al., 1974; Briza et al., 1990; Friesen et al., 1994; Krisak et al., 1994; Rose et al., 1995; Tu et al., 1996). Some of the mutations disrupt proper assembly of the spore wall (Briza et al., 1990; Friesen et al., 1994; Krisak et al., 1994), whereas others seem to disrupt the coordination between prospore membrane growth and nuclear division. A spo3 mutant, for example, suffers from a delay in meiosis II, the consequence of which is premature closure (relative to nuclear division) of the prospore membrane and, therefore, the failure to package the daughter nuclei into spores (Moens et al., 1974).
It has been suggested on morphological grounds that the prospore membrane is a derivative of the ER (Lynn and Magee, 1970). However, this proposition has not been directly tested. One means to investigate the origin of the prospore membrane is to examine the effect of mutations in SEC genes on development of this membrane. A large number of sec mutants that block the transit of vesicles through the secretory pathway at distinct steps have been isolated in S. cerevisiae (Novick et al., 1981). One subset of SEC genes, for instance, are specifically required at the end of the secretory pathway for the fusion of post-Golgi vesicles with the plasma membrane (Novick et al., 1981). These late-acting SEC genes include SEC1, 2, 3, 4, 5, 6, 8, 9, 10, and 15, as well as two pairs of redundant genes SSO1 and 2 and SNC1 and 2. The molecular function of many of these gene products is understood. SEC4, for example, encodes a GTPase of the rab family (Salminen and Novick, 1987). SEC9 encodes a subunit of the plasma membrane t-SNARE complex (Aalto et al., 1993; Brennwald et al., 1994). Additionally, Sec6p, Sec8p, and Sec15p are subunits of a large, cytoplasmic protein complex that localizes to sites of vesicle fusion on the plasma membrane, and several of the other late-acting SEC genes are required for proper assembly of this complex (Terbush and Novick, 1995).
A large body of evidence supports a general model for vesicle fusion events in the secretory pathway, termed the soluble NSF attachment protein receptor (SNARE)1 hypothesis, in which fusion of transport vesicles with the appropriate target membrane is controlled by SNARE complexes (Söllner et al., 1993; Søgaard et al., 1994; Rothman, 1994). According to this model, every vesicle carries on its surface a vesicular (v-)SNARE that can only interact with target (t-)SNARE found on the correct acceptor membrane. Interaction of SNAREs is further regulated by the action of rab-family GTPases. The specificity of SNARE interactions controls the specificity of vesicular transport in the cell. In the case of secretion in S. cerevisiae, vesicles destined for the plasma membrane carry on their surface the v-SNARE proteins Snc1p or Snc2p (Protopopov et al., 1993). These Snc proteins are predicted to interact specifically with the t-SNARE proteins Sec9p and Sso1p or Sso2p, ensuring that Snc-containing vesicles fuse only with the plasma membrane and that no other vesicles fuse with the plasma membrane. The association of these proteins is expected to be regulated by Sec4p. Indeed, SEC4 has genetic interactions with SEC9 and, under appropriate conditions, Sso1p, Snc1p, and Sec9p can be coimmunoprecipitated (Brennwald et al., 1994).
Because the prospore membrane functions as the plasma membrane of the spore, it seemed possible that, like new plasma membrane, the prospore membrane was derived from post-Golgi vesicles, rather than directly from the ER as suggested by Lynn and Magee (1970). To differentiate between these possibilities, mutations in late-acting SEC genes, which effect only post-Golgi vesicular transport, were examined for their effect on spore formation. The results identify a previously unknown branch of the secretory pathway in yeast and suggest that spore morphogenesis begins by the de novo synthesis of a new plasma membrane in the interior of the mother cell.
Materials and Methods
Yeast Strains and Media
S. cerevisiae strains used in this work are listed in Table I. To make strains AN79, AN81, AN126, and AN127, haploids AN63-2C, AN61-1A, HSF187, and AN65-1C (sec9-4, sec4-8, sec8-9, and sec1-1, respectively) were crossed to the SK1-related strain S2683 to create diploids AN74, AN75, AN124, and AN125. Several sec segregants from these diploids were crossed to each other to find diploids that sporulate efficiently at permissive temperature (AN79 AN81, AN126, and AN127). To create the spo20ΔURA3 diploid AN67, SPO20 was first disrupted in S2683 and RKY1145 by one-step gene replacement (Rothstein, 1983) using EcoRI- and XbaI-digested pspo20ΔURA3. The resulting haploids (AN1037 and AN1038) were then mated to produce AN67. AN80 was constructed in an identical manner except that the haploid strains used were AN74-3A and AN74-7C. Unless otherwise noted, standard media and growth conditions were used (Rose and Fink, 1990).
Table I.
Strains
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| AN61-1A | MATa ura3-52 sec4-8 | this work‡ | ||
| AN63-2C | MATa ura3-52 sec9-4 | this work§ | ||
| HSF187 | MATa sec8-9 | N. Dean | ||
| AN65-1C | MATα ura3 sec1-1 | this work‖ | ||
| RKY1145 | MATa ura3 leu2ΔhisG his4-x lys2 hoΔLYS2 | N. Hollingsworth | ||
| S2683 | MATα ura3 leu2-k arg4-NspI lys2 hoΔLYS2 | N. Hollingsworth | ||
| AN1037 | as RKY1145, plus spo20ΔURA3 | see text | ||
| AN1038 | as S2683, plus spo20ΔURA3 | see text | ||
| AN74-3A* | MATa ura3 arg4-NspI sec9-4 | segregant of AN63-2C × S2683 | ||
| AN74-7C* | MATα ura3 leu2-k sec9-4 | segregant of AN63-2C × S2683 | ||
| NH144 | MATa/MATα ura3/ura3/ura3 leu2ΔhisG/leu2-k his4-x/+ | RKY1145 × S2683 | ||
| his4-x/+ arg4-NspI/+ lys2/lys2 hoΔLYS2/hoΔLYS2 | ||||
| AN67 | as NH144 plus spo20ΔURA3/spo20ΔURA3 | AN1037 × AN1038 | ||
| AN79* | MATa/MATα ura3/ura3 leu2-k/+ | AN74-3A × AN74-7C | ||
| arg4-NspI/+ sec9-4/sec9-4 | ||||
| AN80* | as AN79, plus spo20ΔURA3/spo20ΔURA3 | see text | ||
| AN81* | MATa/MATα ura3/ura3 leu2-k/+ | see text | ||
| arg4-NspI/+ lys2/+ sec4-8/sec4-8 | ||||
| AN126* | MATa/MATα ura3/ura3 leu2-k/+ | see text | ||
| arg4-NspI/+ sec8-9/sec8-9 | ||||
| AN127* | MATa/MATα ura3/ura3 leu2-k/+ | see text | ||
| arg4-NspI/+ sec1-1/sec1-1 |
These strains are Lys+, which may be due to any of the following genotypes: LYS2 ho, lys2 ho::LYS2, or LYS2 ho::LYS2.
Segregant from an outcross of strain NY774 (sec4-8) from D. Terbush.
Segregant from an outcross of strain CKY31 (sec9-4) from C. Kaiser.
Segregant from an outcross of strain CKY23 (sec1-1) from C. Kaiser.
Sporulation Assays
Strains were sporulated by overnight growth in YPD followed by 1:300 dilution into YPAcetate medium. Cultures were then grown overnight to midlog phase, pelleted, washed once, and resuspended in 2% KOAc medium at a concentration of 3 × 107 cells/ml. For strains harboring temperature-sensitive mutations, cultures were incubated in sporulation medium at 23°C for ∼2 h before shift to 34°C. At intervals, aliquots were removed and fixed by addition of formaldehyde to a final concentration of 3.7%. Samples were then stained with 4′,6′-diamidino-2-phenylindoledihydrochloride (DAPI) and observed using a Zeiss Axiophot microscope (Thornwood, NY).
Plasmids
The SPO20 region was cloned directly from genomic DNA by PCR using the primers ANO85 (5′-GGC TGA ATT CTA GGC GCT TTC AAC C) and ANO86 (5′-GCC GTC TAG AGT GTA TAA CAG ATC ACC) and Taq polymerase (Boehringer Mannheim Corp., Indinapolis, IN). The resulting fragment was digested with EcoRI and XbaI and cloned into EcoRI-XbaI–cut Bluescript SK− (Promega Corp., Madison, WI) to create pSK-SEC9H. The spo20ΔURA3 allele was created by replacing a BamHI-BglII fragment of pSK-SEC9H carrying the central 0.7 kb of the SPO20 coding region with a 1.1-kb BamHI-BglII fragment of pMH3 (a gift of N. Hollingsworth, SUNY Stony Brook) carrying the URA3 gene. The resulting plasmid pspo20ΔURA3 was digested with EcoRI and XbaI before transformation to release the disruption fragment. The SPO20-HA fusion allele was constructed in three steps. First, the EcoRI-XbaI fragment of pSK-SEC9H carrying the entire SPO20 region was cloned into pRS315 (Sikorski and Hieter, 1989). Using the primers ANO95 (5′-GTG CTG AAT TCT ATA TAA TGG GGT TCA G) and ANO96 (5′-GTG TAT AAC TCA TAT GCA TCT TTT CCC G) and pSK-SEC9H as a template, PCR was then used to introduce EcoRI and NsiI sites 5′ of the ATG and at the stop codon of the SPO20 open reading frame, respectively. This PCR fragment was digested with EcoRI and NsiI and cloned into EcoRI-PstI– cut pSKHA3 (Neiman et al., 1997) to create an in-frame fusion of three copies of the influenza hemagglutinin epitope (YPYDVPDY) to the carboxy terminus of SPO20. Finally, a BglII-XbaI fragment coding for the COOH-terminal portion of the Spo20-HA fusion protein was swapped with a corresponding BglII-XbaI fragment of pRS315-SPO20 to create pRS315-SPO20-3xHA. A NotI-XhoI fragment of pRS315-SPO20-3×HA was cloned into similarly cut pRS425 to create pRS425-SPO20-3×HA. The original PCR cloning from the genome did not introduce deleterious mutations into the SPO20 coding region, as shown by the fact that pRS315-SPO20 completely complements the spo20Δ sporulation defect of strain AN67. The hemagglutinin (HA) tag may mildly impair Spo20p function, as a spo20ΔURA3 strain carrying pRS315-SPO20-3xHA is reduced in sporulation ∼50% relative to the same strain carrying pRS315-SPO20.
To examine whether SEC9 can rescue the spo20 sporulation defect, a genomic clone of SEC9 was isolated by complementation of the sec9-4 temperature-sensitive phenotype with a YEp13 based library and then transformed into strain AN67. For the reciprocal experiment, the PCR product carrying the SPO20 coding region (generated using primers AN95 and ANO96, see above) was cloned as a blunt end fragment into EcoRV-cut Bluescript KS+ and then subcloned as a 1.2-kb EcoRI-SalI fragment into plasmid pRD53 (R. Deshaies), placing SPO20 under the control of the GAL1 promoter. This plasmid, pRD53-SPO20, was then transformed into strain AN63-2C, and growth was assessed on galactose plates incubated at 36°C.
Electron Microscopy
Samples were prepared for the electron microscope by a modification of the procedure of Moens (1971). Samples were fixed by addition of glutaraldehyde (Sigma Chemical Co., St. Louis, MO) directly to the culture medium to a final concentration of 2.5%, mixed, and incubated on ice overnight. They were then washed twice in distilled water before being resuspended in 4% KMnO4 for 30 min. The KMnO4 was then removed by washing with distilled water and the samples were resuspended in a saturated uranyl acetate solution for 30 min. After one wash with distilled water, the samples were dehydrated by a series of ethanol washes and a final wash in 100% propylene oxide. The samples were then washed three times in Epon mix (5 ml Epon-812, 1.5 ml dodecenyl succinic anhydride [DDSA], 3.5 ml nadic methyl anhydride [NMA]). Finally, the samples were resuspended in Epon mix with DMP-30 (0.15 ml), left for 2 d at room temperature, and then shifted to 60°C for 24 h before sectioning. For all strains described here, at least 50 cells were analyzed that were, as judged by nuclear morphology, postmeiotic or in meiosis II.
Invertase and Carboxypeptidase Y Assays
Invertase activity was monitored as described (Pelham et al., 1988). Carboxypeptidase Y sorting and processing was monitored by Western analysis and colony lifts using anti-CPY antibodies (N. Dean, SUNY Stony Brook) as described previously (Rothman et al., 1986).
Western Analysis
Strain AN67 carrying pRS425-SPO20-3×HA was sporulated in liquid medium, and at intervals aliquots were removed. Extracts were made by disruption with glass beads in lysis buffer (10 mM Hepes, pH 7.5, 200 mM KCl, 0.1% NP-40) followed by 30 min of centrifugation in a microfuge at 13,000 rpm. Samples were separated by SDS-PAGE in a 10% gel and then blotted to nitrocellulose. Filters were probed with the anti-HA antibody 12CA5 (BAbCo, Berkeley, CA) at a 1:3,000 dilution and then HRP-conjugated anti–mouse secondary antibodies. Bands were visualized using ECL (Amersham Corp., Arlington Heights, IL).
Indirect Immunofluorescence
Immunofluorescence was performed essentially as described (Neiman et al., 1997). The anti-Gas1p and anti-Cts1p antibodies (N. Dean) were used at a 1:1 dilution. The secondary antibodies used were goat anti–rabbit coupled to Cy3 (Cappel Laboratories, Malvern, PA).
Results
Late-acting SEC Genes Are Required for Spore Formation
To determine if late-acting SEC genes play a role in spore morphogenesis, diploids homozygous for the temperature-sensitive mutations sec1-1, sec4-8, sec8-9, or sec9-4 were tested for their ability to form spores. Strains carrying these mutations were incubated briefly in sporulation medium at permissive temperature (23°C) and then shifted to nonpermissive temperature (34°C). Progression through meiosis was scored by staining the cells with DAPI and monitoring the appearance of tetranucleate staining by fluorescence microscopy, while formation of spores was assayed by phase microscopy. All four strains sporulated efficiently at permissive temperature (Table II). However, strains homozygous for sec1-1, sec4-8, or sec8-9 were severely impaired in sporulation at the restrictive temperature. DAPI staining indicated that the sec1-1/sec1-1, sec4-8/ sec4-8, and sec8-9/sec8-9 strains formed tetranucleate cells nearly as well as wild type (NH144) (Table II). Some tetranucleate staining may represent segregation of the chromatin within a single nucleus rather than the completion of nuclear division (Kupiec et al., 1997). Nonetheless, this result suggests that the failure of these mutants to sporulate is an inability to form spores, per se, rather than a defect in the completion of meiosis.
Table II.
Sporulation in sec Mutant Strains
| Strain | Relevant genotpye | Percent asci 23°C* | Percent asci 34°C* | Percent tetranucleate 34°C‡ | ||||
|---|---|---|---|---|---|---|---|---|
| NH144 | wild type | 96 | 95 | 61 | ||||
| AN127 | sec1-1/sec1-1 | 80 | <0.2 | 35 | ||||
| AN81 | sec4-8/sec4-8 | 74 | 0.4 | 46 | ||||
| AN126 | sec8-9/sec8-9 | 80 | <0.2 | 39 | ||||
| AN79 | sec9-4/sec9-4 | 63 | 69 | 23 | ||||
| AN67 | spo20Δ/spo20Δ | <0.2 | <0.2 | 32 |
Cultures were transfered to sporulation medium, incubated for 3 h at 23°C, and then split in two. One half was left at 23°C, and the other was shifted to 34°C. After overnight incubation, all cultures were examined by phase contrast microscopy. For each culture, at least 500 cells were counted.
Tetranucleate staining was unstable in strains AN67, AN81, AN126, and AN127, probably because of the failure to package nuclei (see text). Therefore cells were fixed 4 h after shift to 34°C.
In contrast, the sec9-4/sec9-4 strain (AN79) sporulated well at the restrictive temperature (Table II). This difference between sec9-4 and the other sec mutants is apparently not due to leakiness of the sec9-4 allele since in all sec strains tested the nonpermissive temperature used for sporulation was above the vegetative restrictive temperature (Salminen and Novick, 1987; Neiman, A.M., unpublished observations).
sec4/sec4 Diploids Do Not Form Prospore Membranes
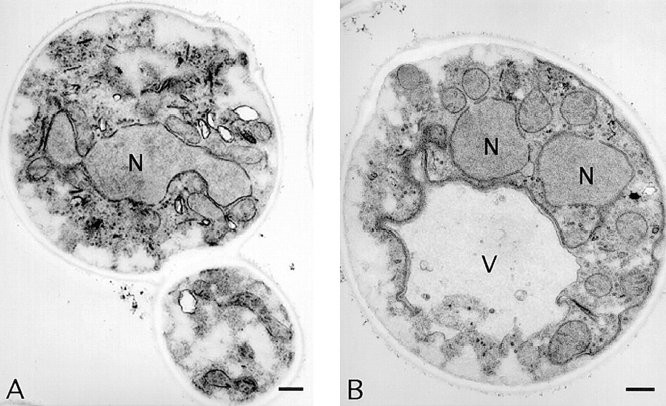
The failure of sec1-1, sec4-8, and sec8-9 mutants to form visible spores could be due to a defect at any point in the development of refractile spores. For instance, these mutant strains might form the prospore membrane normally but be unable to form a mature spore wall. To examine where in the process of spore morphogenesis the sec4 mutant defect occurs, the sec4-8 homozygous diploid AN81 was compared with a SEC4 diploid by transmission electron microscopy. Cultures of AN81 and NH144 were incubated in sporulation medium at 23°C for 3 h and then shifted to 34°C. At intervals, cells were removed and prepared for electron microscopy as described in Materials and Methods. Fig. 1 A shows a typical wild-type cell at a late stage of prospore membrane growth. The prospore membrane is a semicircular structure centered near the spindle pole body and curving down around a lobe of the nucleus. At a later stage (Fig. 1 B), the prospore membrane has closed around a daughter nucleus and a spore wall is forming. By contrast, in the sec4-8/sec4-8 diploid, membranes resembling the prospore membrane were never observed. In cells that appeared to be in meiosis II, as judged by their nuclear morphology (Fig. 1 C), no semicircular membranes were evident, and at later stages, cells appeared to have multiple, unpackaged daughter nuclei (Fig. 1 D). These results indicate that in the sec4-8 mutant, the prospore membrane does not form. Given the known function of SEC4 in the fusion of post-Golgi secretory vesicles with the plasma membrane, these results suggest that the prospore membrane is formed by the fusion of secretory vesicles within the cytoplasm of the cell.
Figure 1.
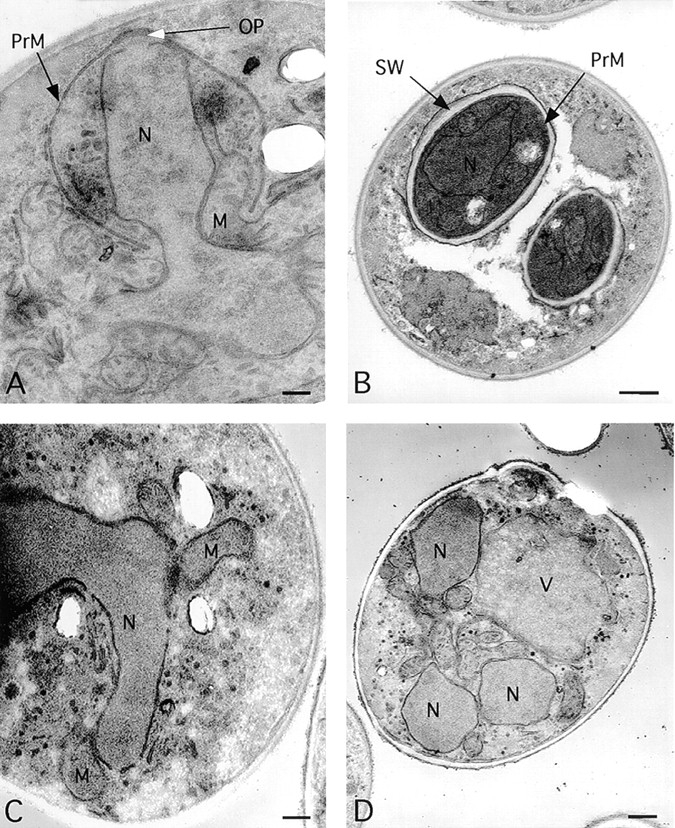
Prospore membranes are absent in a sec4/sec4 mutant. (A) SEC4 cell (NH144) late in prospore membrane formation. The gap in the nuclear envelope underneath the outer plaque is the position of the spindle pole body. (B) NH144, after closure. (C and D) Cells of strain AN81 (sec4-8/sec4-8) at stages comparable to those in A and B, respectively. N, nucleus; M, mitochondrion; V, vacuole; OP, outer plaque; PrM, prospore membrane; SW, spore wall. All cells were sporulated at 23°C for 3 h and then shifted to the restrictive temperature for sec4-8 (34°C). Bars: (A and C) 200 nm; (B and D) 500 nm.
A Second Soluble NSF Attachment Protein (SNAP-25) Homologue Functions in Spore Morphogenesis
A number of genetic interactions between SEC4 and SEC9 have led to the suggestion that Sec9p is the effector for Sec4p in secretory vesicle fusion with the plasma membrane (Brennwald et al., 1994). Therefore, the observation that SEC4 but not SEC9 is required for prospore membrane formation might suggest that fusion of vesicles with the prospore membrane is mechanistically different from fusion with the plasma membrane. Alternatively, there may be some sporulation-specific protein that can replace Sec9p in prospore membrane formation. To address this second possibility, the yeast genome was screened for SEC9-related sequences.
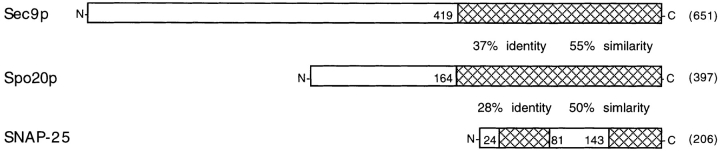
The S. cerevisiae genome contains one open reading frame, YMR017w, with significant sequence similarity to SEC9. For reasons described below, this open reading frame has been designated SPO20. The predicted protein encoded by SPO20 is 397 amino acids in length with 37% identity and 55% similarity to Sec9p over the carboxy-terminal 223 residues (Fig. 2). The amino-terminal region of Spo20p has no similarity to Sec9p or any other sequence in GenBank. In the domain conserved between Spo20p and Sec9p, Spo20p contains two 60 amino acid regions that together show 28% identity and 50% similarity to the mammalian t-SNARE subunit SNAP-25. Thus, SPO20 encodes a second S. cerevisiae SNAP-25 homologue.
Figure 2.
SPO20 encodes a second yeast SNAP-25 homologue. Alignment of domains in Sec9p, Spo20p, and mouse SNAP-25. Amino acid numbers are indicated adjacent to hatched areas, and numbers in parentheses are total amino acids. These sequence data are available from GenBank/EMBL/DDBJ under accession numbers Z49211 (SPO20), L34336 (SEC9), and M22012 (SNAP-25).
The SPO20 gene was cloned by PCR from yeast genomic DNA, and a diploid strain homozygous for a deletion of SPO20 was constructed (see Materials and Methods). This deletion replaces the coding region for amino acids 41–268 of Spo20p with the URA3 gene, presumably creating a null allele. The deletion strain, AN67, was examined for secretory phenotypes. No defect was found either in the secretion of invertase or in the sorting of carboxypeptidase Y to the vacuole (Neiman, A.M., unpublished observations); however, AN67 failed to sporulate. Specifically, DAPI staining indicated that the spo20ΔURA3 mutant strain proceeded through meiosis but failed to form spores, similar to the sec1, sec4, and sec8 mutants (Table II).
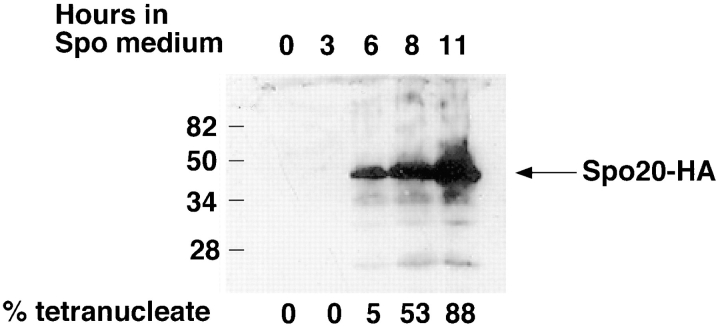
To examine the SPO20 gene product further, an epitope-tagged allele was constructed by fusing a sequence encoding three copies of the influenza hemagglutinin epitope (HA) to the 3′ end of the coding region. This fusion gene partially complemented the spore formation defect of the spo20ΔURA3 mutant (see Materials and Methods). To examine the expression of SPO20, AN67 carrying the SPO20-HA fusion gene under the SPO20 promoter on a high copy plasmid was sporulated, and cells were removed at various times after transfer to sporulation medium. Extracts were made from these cells, and the proteins were analyzed by Western blot using anti-HA antibodies. Consistent with the lack of any obvious phenotype of the spo20ΔURA3 mutant in vegetative cells, the protein was not detectable in vegetative cells and was first detected between 3 and 6 h after shift to sporulation medium (Fig. 3). The induction of Spo20p is coincident with or just before the appearance of tetranucleate cells and thus corresponds to the time of the second meiotic division and the onset of prospore membrane formation (Fig. 3). Northern analysis of transcripts from a meiotic time course using SPO20 as a probe indicates that the transcript is seen only in sporulating cells (Conrad, M., personal communication). Therefore, induction of Spo20p seems to be regulated at the level of transcription.
Figure 3.
Time course of Spo20p induction. At the times indicated, samples of strain AN67 carrying pRS425-SPO20HA were removed from sporulation medium, and extracts were made and analyzed by Western blot using anti-HA antibodies. Equal cell equivalents were loaded in each lane. Marks at left indicate the position of molecular size standards. The fraction of cells at each time point (at least 200 cells counted) that were tetranucleate, as determined by DAPI staining, is shown below each lane. The predicted molecular mass for the Spo20-HA fusion protein is 49,800 D.
spo20 Mutants Display a Defect in the Capture of Nuclei by the Prospore Membrane
A simple model based on the sporulation phenotypes of sec9 and spo20 is that Sec9p and Spo20p have separate functions. Sec9p is a component of a t-SNARE specifically required for vesicle fusion at the plasma membrane, and Spo20p could play the analogous role in a prospore membrane-specific t-SNARE complex. The remaining fusion machinery (e.g., Sec1p, Sec4p, and Sec8p) may be used for both membranes. If so, then during sporulation spo20 mutants should have the same cytological phenotype as sec4 mutants; namely, no prospore membranes should be formed. To examine the spo20ΔURA3 phenotype more closely, strain AN67 was sporulated, and at various times samples were removed and examined by electron microscopy (Fig. 4).
Figure 4.
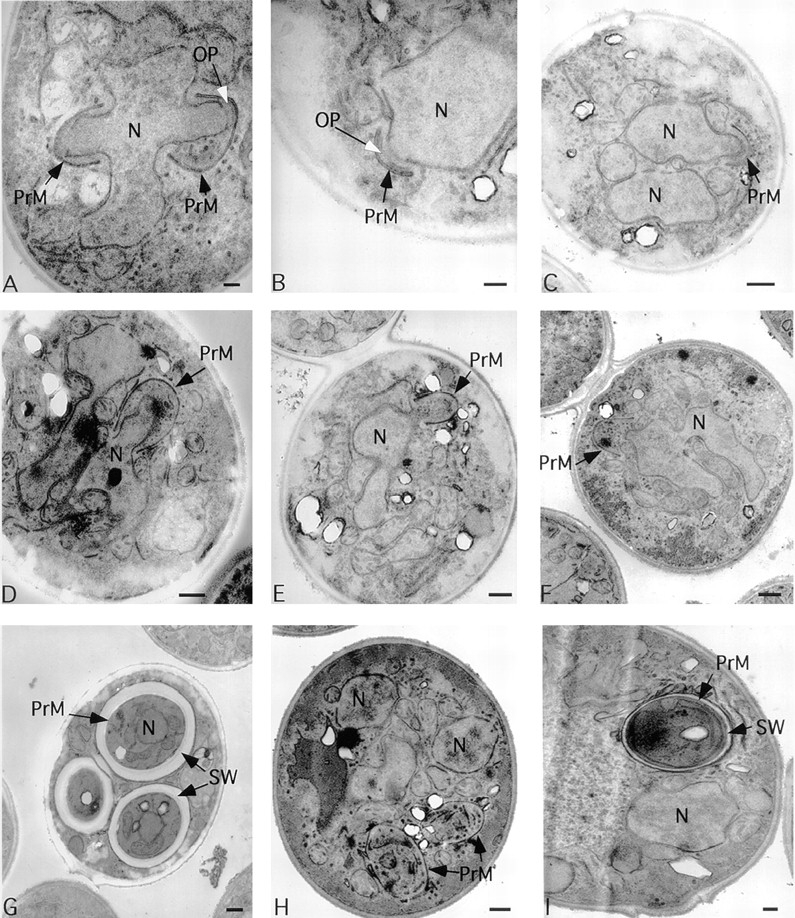
spo20 mutants fail to package daughter nuclei into spores. (A, D, and G) Isogenic SPO20 (NH144) or (B, C, E, F, H, and I) spo20ΔURA3/spo20ΔUR A3 (AN67) cells at different stages of prospore membrane formation: early (A– C); middle (D–F); or after closure (G–I). (A and B) The gap in the nuclear envelope underneath the outer plaque is the position of the spindle pole body. N, nucleus; OP, outer plaque; PrM, prospore membrane; SW, spore wall. Bars: (A, B, and I) 200 nm; (C–H) 500 nm.
Surprisingly, the spo20ΔURA3 mutant forms prospore membranes. Examination of a large number of cells revealed that the spo20 diploid does display a defect in prospore membrane growth, but a much more subtle one than in sec4 mutants. At early stages in prospore membrane development in the spo20 strain, the membranes appear normal and, as in wild type, are juxtaposed to the outer plaque of the spindle pole body (Fig. 4, A–C). In wild-type cells, as the membrane grows it surrounds a lobe of the nucleus (Fig. 4 D). However, in the spo20 strain, the connection between prospore membrane and nucleus seems to be lost. Prospore membranes are found in the cytoplasm, no longer adjacent to the nucleus (Fig. 4, E and F). These detached prospore membranes continue to grow and eventually fuse with themselves without enclosing a daughter nucleus, thus forming anucleate, immature spores (Fig. 4 H). In some cases, these anucleate spores appear to initiate spore wall development (Fig. 4 I), although no mature spore walls were ever seen.
Spo20p and Sec9p Have Partially Overlapping Functions
This cytological analysis demonstrated that, although abnormal, the prospore membrane does form in spo20 mutants. Analysis of sec9-4/sec9-4 diploids in the electron microscope showed that spores form normally (data not shown). Taken together, these two results suggest either that fusion of vesicles with the prospore membrane can occur without the function of a SNAP-25 homologue or that SEC9 and SPO20 have overlapping functions. In the latter case, one would expect that prospore membrane formation in the spo20ΔURA3 diploid would be dependent on SEC9 function. To test this possibility, a spo20ΔURA3/ spo20ΔURA3 sec9-4/sec9-4 strain (AN80) was constructed. This strain was sporulated at 34°C to inactivate SEC9 and examined in the electron microscope. The sporulation defect in the double mutant resembled the sec4-8/sec4-8 mutant in that no prospore membranes were observed (Fig. 5). It seemed possible that the cytological differences seen between AN67 (spo20ΔURA3/spo20ΔURA3) and AN80 (sec9-4/ sec9-4 spo20ΔURA3/spo20ΔURA3) were due to differences in strain background and not the presence of the sec9-4 mutation. To examine this possibility, a sec9-4 allele in AN80 was converted to wild type by transformation with a linear DNA fragment carrying the wild-type SEC9 gene and direct selection for growth at 36°C. The resulting strain (sec9-4/SEC9 spo20ΔURA3/spo20ΔURA3) was analyzed by electron microscopy and displayed the same prospore membrane defect described above for AN67 (data not shown). Thus, the lack of prospore membrane development is a direct consequence of losing both Sec9p and Spo20p function. These results demonstrate that Sec9p plays a role in prospore membrane growth and provide additional evidence that the late acting SEC genes are required for fusion of vesicles with the prospore membrane.
Figure 5.
Prospore membranes are absent in a spo20/spo20 sec9/ sec9 double mutant. Cells of strain AN80 (spo20ΔURA3/spo20ΔURA3sec9-4/sec9-4) at times corresponding to middle (A) and postclosure stages (B) of prospore membrane formation. N, nucleus; V, vacuole. Cells were sporulated at the restrictive temperature for sec9-4 (34°C). Bars, 500 nm.
To further examine the functional overlap between Sec9p and Spo20p, we tested whether overexpression of one gene could rescue a mutation in the other. Expression of SPO20 from the GAL promoter failed to rescue the sec9-4 temperature-sensitive phenotype (data not shown). Similarly, expression of SEC9 from a high copy plasmid could not rescue the spo20 sporulation defect (data not shown). Thus, though the two proteins have some functional overlap, they cannot completely substitute for each other.
The Cell Surface Protein Gas1p Localizes to the Growing Prospore Membrane
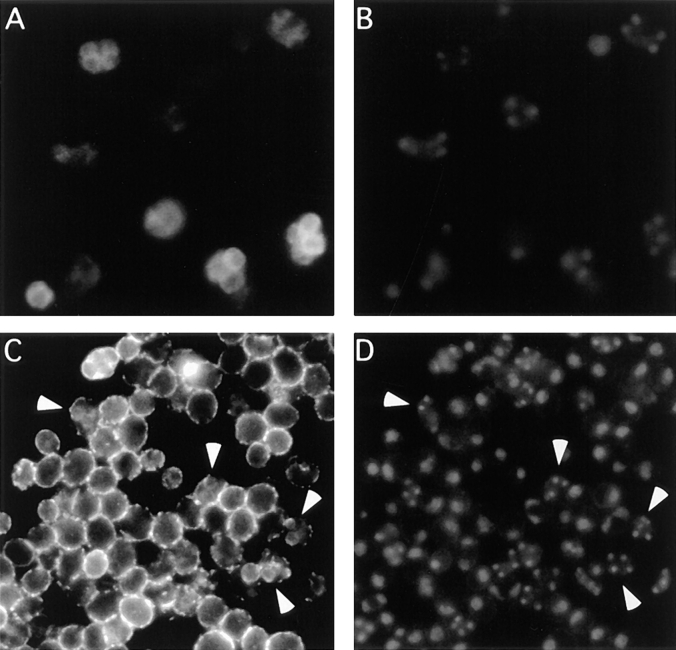
If the prospore membrane is formed by the redirection of secretory vesicles from the plasma membrane to an intracellular site, then the prospore membrane may also contain the usual cargo of those vesicles. One might expect, for example, to find plasma membrane proteins localized to the growing prospore membrane. To test this hypothesis we have examined the localization of the GPI-anchored cell surface protein Gas1p (Nuoffer et al., 1991) during sporulation (Fig. 6). In tetranucleate cells, Gas1p appears to be localized in an internal membrane that surrounds the nucleus (Fig. 6 A) consistent with a prospore membrane localization. This localization of Gas1p is disrupted in a sec1-1 mutant strain (data not shown), indicating that this internal membrane is the prospore membrane.
Figure 6.
Gas1p and Cts1p localization in sporulating cells. (A) Indirect immunofluorescence of Gas1p in sporulating cells of strain NH144. (B) DAPI staining of cells in A. (C) Indirect immunofluorescence of Cts1p in sporulating cells of strain NH144. (D) DAPI staining of cells in C. Arrowheads (C and D) indicate tetranucleate cells.
The localization of the secreted chitinase Cts1p (Kuranda and Robbins, 1987) was also examined in sporulating cells. Though the signal is fainter than for Gas1p, Cts1p displays an intracellular staining pattern similar to Gas1p in tetranucleate cells (Fig. 6, C and D, arrowheads). This staining may represent localization to the lumen of the prospore membrane.
Discussion
This report describes evidence for a role of the late-acting SEC genes in formation of the prospore membrane. Several late-acting SEC genes were examined. SEC4 and, in a spo20 background, SEC9 are required for the formation of this intracellular compartment. sec1 and sec8 mutants also fail to sporulate at restrictive temperature, presumably because of a similar defect in prospore membrane formation. These are the first genes shown to be specifically required for formation of the prospore membrane.
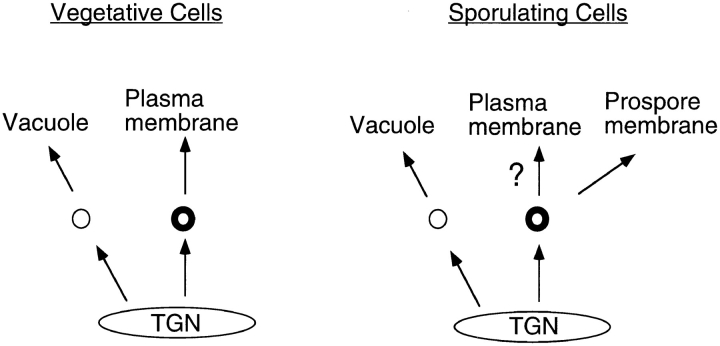
In vegetative cells, these SEC gene products are required solely for the fusion of post-Golgi vesicles with the plasma membrane. Therefore, the simplest interpretation of these results is that these gene products are required during sporulation for fusion of vesicles to form the prospore membrane. Furthermore, the vesicles that fuse to form the prospore membrane are likely the same ones regulated by late-acting SEC genes in vegetative cells, i.e., secretory vesicles. Consistent with this interpretation, the cell surface protein Gas1p and the secreted protein Cts1p are localized to the prospore membrane compartment in sporulating cells. Thus, trafficking to the prospore membrane defines a new, post-Golgi branch of the yeast secretory pathway (Fig. 7).
Figure 7.
A sporulation-specific branch of the yeast secretory pathway. Routes for vesicular traffic out of the TGN in vegetative and sporulating cells are shown. Thick circles indicate vesicles dependent on SEC4 function for fusion with their target membrane. In both situations, SEC4-independent vesicles (thin circles) are targeted to the vacuole. “?” indicates that it is unclear whether trafficking to the plasma membrane continues in cells developing a prospore membrane (see text).
During vegetative growth, proteins sorted at the TGN can be packaged into vesicles destined for either the vacuole or the plasma membrane. Only the latter class of vesicles is dependent on Sec4p function for fusion with their target membrane. In sporulating cells, the Sec4p-dependent vesicles could potentially fuse with either the plasma membrane or the prospore membrane (Fig. 7). The presence of two potential target membranes raises the possibility that secretory vesicles are differentially targeted to these two membranes during sporulation.
Recent studies have shown that at least two different classes of secretory vesicle can be distinguished in yeast on the basis of their density (Harsay and Bretscher, 1995). Though all of these vesicles require late-acting SEC genes to fuse with the plasma membrane, the different classes contain different cargo molecules. The results presented here do not, therefore, indicate whether all secretion is directed into the prospore membrane or if only a subset of secretory vesicles are directed there. Related to the question of which vesicles are delivered to the prospore membrane is the question of how secretory vesicles are targeted to this intracellular compartment. In vegetative yeast, fusion of secretory vesicles occurs only at particular sites on the cell surface (Winsor and Scheibel, 1997). The restriction of secretion to specified regions of the plasma membrane is mirrored in the polarized organization of the actin cytoskeleton, and mutations that disrupt the cytoskeleton are often associated with a delocalization of secretion (Winsor and Scheibel, 1997). Presumably, the targeting of secretory vesicles to the interior of the cell is accompanied by changes in cell polarity and the cytoskeleton. An illustration of this is the redistribution of septins (Fares et al., 1996). In vegetative cells, septins form a ring of 10-nm filaments around the mother/bud neck, whereas in sporulating cells the septins are found in a band (presumably) underlying the growing prospore membrane. It will be of interest to examine a variety of mutations in genes affecting cell polarity and the cytoskeleton to determine their effects on sporulation.
Distinct Functions for Sec9p and Spo20p
SEC9 and SPO20 are partially redundant in that the presence of one or the other of these gene products is required for formation of a prospore membrane. However, the single mutations differ in their effects. Mutations in sec9 have no effect on spore formation, whereas no spores form in spo20 mutants because the growing prospore membranes lose their attachment to the nucleus and subsequently fail to capture daughter nuclei. One possible explanation for this difference is that Sec9p and Spo20p are completely redundant but in the absence of Spo20p, the level of Sec9p during sporulation is insufficient to support proper spore formation. However, overexpression of SEC9 cannot rescue the spo20 sporulation defect. Therefore, Spo20p appears to have a function, which it does not share with Sec9p, that is required to maintain the position of the prospore membrane adjacent to the nucleus.
Similarly, ectopic expression of SPO20 in vegetative cells cannot suppress the temperature-sensitive phenotype of a sec9-4 mutant, suggesting that Sec9p has some essential function that Spo20p cannot provide. This result is surprising given that Spo20p can replace Sec9p during sporulation. It may be that there are additional sporulation-specific proteins required for Spo20p to function in vesicle fusion. Alternatively, Sec9p and Spo20p may mediate the fusion of overlapping but distinct sets of vesicles so that neither protein can completely compensate for a defect in the other.
Secretion-driven Cell Division in Other Systems
Spore formation is a process in which cell division occurs not by a classical cytokinetic mechanism but rather by the growth of new plasma membranes, discontinuous from the mother cell plasma membrane, to give rise to daughter cells within the cytoplasm of the mother cell. This form of cell division has features in common with “nonclassical” division events in a number of systems. In Drosophila, for example, during both cellularization of the syncytial blastoderm and spermatogenesis, polynucleate cells are divided into mononucleate cells by processes that involve the coalescence of vesicles to form new plasma membranes (Tokuyasu et al., 1972; Loncar and Singer, 1995). Similarly, in higher plants cell plate formation occurs first by the coalescence of vesicles in the interior of the cell between the daughter nuclei (Wick, 1991; Staehelin and Hepler, 1996). The vesicles then fuse to form a flattened sheet, which then grows outward by vesicle addition until it fuses with the mother cell plasma membrane to complete division.
Perhaps the most striking parallel to ascospore morphogenesis is also found in plants: the formation of the generative cell during pollen development (McCormick, 1993). The four haploid products of the male meiosis are termed microspores. Each of these microspores then undergoes a mitotic division. One of the daughter nuclei from this division will remain in the mother cell cytoplasm and is termed the vegetative nucleus. The second is enclosed in a double membrane within the mother cell cytoplasm. This smaller cell is termed the generative cell. The generative cell will eventually undergo a mitotic division to give rise to two sperm cells. Together, these three cells (two sperm and one vegetative) form the mature pollen grain. The double membrane around the generative cell is formed by the coalescence of vesicles around the generative cell nucleus (Gimenez-Martin et al., 1969; Ledbetter and Porter, 1970) and appears directly analogous to the formation of spores in S. cerevisiae. A cell wall even forms between the two membranes surrounding the generative cell (Angold, 1968; Heslop-Harrison, 1968), similarly to spore wall maturation. It will be of interest to learn if these different events in plants, yeast, and metazoans share some common molecular mechanisms, such as the use of alternative SNAP-25 proteins.
Abbreviations used in this paper
- DAPI
4′,6′-diamidino-2-phenylindoledihydrochloride
- HA
hemagglutinin
- NSF
N-ethylmaleimide–sensitive factor
- SNAP
soluble NSF attachment protein
- SNARE
SNAP receptor
- v- and t-SNARE
vesicular and target SNARE
Footnotes
I thank N. Hollingsworth, R. Sternglanz, P. Pryciak (University of Massachusetts, Worcester, MA), N. Dean, and an anonymous reviewer for comments on the manuscript; N. Hollingsworth, C. Kaiser (Massachusetts Institute of Technology, Cambridge, MA), and D. Terbush (Yale University, New Haven, CT) for strains; and M. Conrad (Oklahoma Medical Research Foundation) for communication of results before publication. I am grateful to N. Dean for antibodies and to G. Rudomen for able assistance with the electron microscopy. I am also indebted to N. Hollingsworth, N. Dean, B. Aronson, and R. Sternglanz for discussion, advice, and encouragement throughout the course of this project.
A.M. Neiman was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation, DRG-1276 and National Institutes of Health grant GM-28220 to R. Sternglanz.
Address all correspondence to Aaron M. Neiman, Department of Biochemistry and Cell Biology, State University of New York, Stony Brook, Stony Brook, NY 11794. Tel.: (516) 632-8565. Fax: (516) 632-8575. E-mail: aneiman@mcbsgi.bio.sunysb.edu
References
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO (Eur Mol Biol Organ) J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angold RE. The formation of the generative cell in the pollen grain of Emdmyion non-scriptus(L) J Cell Sci. 1968;3:573–578. doi: 10.1242/jcs.3.4.573. [DOI] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Briza P, Breitenbach M, Ellinger A, Segall J. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. . Genes Dev. 1990;4:1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 59–96.
- Esposito, R.E., and S. Klapholz. 1981 Meiosis and ascospore development. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 211–287.
- Fares H, Goetsch L, Pringle JR. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. . J Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen H, Lunz R, Doyle S, Segall J. Mutation of the SPS1- encoded protein kinase of Saccharomyces cerevisiaeleads to defects in transcription and morphology during spore formation. Genes Dev. 1994;8:2162–2175. doi: 10.1101/gad.8.18.2162. [DOI] [PubMed] [Google Scholar]
- Gimenez-Martin G, Risueno MC, Lopez-Saez JF. Generative cell envelope in pollen grains as a secretion system, a postulate. Protoplasma. 1969;67:223–235. [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J. Synchronous pollen mitosis and the formation of generative cell in massulate orchids. J Cell Sci. 1968;3:457–466. [Google Scholar]
- Krisak L, Strich R, Winters RS, Hall JP, Mallory MJ, Kreitzer D, Tuan RS, Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. . Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- Kupiec, M., B. Byers, R.E. Esposito, and A.P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae. In The Molecular and Cellular Biology of the Yeast Saccharomyces Cell Cycle and Cell Biology J.R. Pringle, J.R. Broach, and E.W. Jones, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 889–1036.
- Kuranda MJ, Robbins PW. Cloning and heterologous expression of glycosidase genes from Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1987;84:2585–2589. doi: 10.1073/pnas.84.9.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter, M.C., and K.R. Porter. 1970. Introduction to the Fine Structure of Plant Cells. Springer-Verlag, New York. 270 pp.
- Loncar D, Singer SJ. Cell membrane formation during the cellularization of the syncytial blastoderm of Drosophila. . Proc Natl Acad Sci USA. 1995;92:2199–2203. doi: 10.1073/pnas.92.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn RR, Magee PT. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. . J Cell Biol. 1970;44:688–692. doi: 10.1083/jcb.44.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. Male gametophyte development. Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. . Can J Microbiol. 1971;17:507–510. doi: 10.1139/m71-084. [DOI] [PubMed] [Google Scholar]
- Moens PB, Rapport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae(Hansen) J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Esposito RE, Esposito MS. Aberrant nuclear behavior at meiosis and anucleate spore formation by sporulation-deficient (SPO) mutants of Saccharomyces cerevisiae. . Exp Cell Res. 1974;83:166–174. doi: 10.1016/0014-4827(74)90700-9. [DOI] [PubMed] [Google Scholar]
- Neiman AM, Mhaiskar V, Manus V, Galibert F, Dean N. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics. 1997;145:637–645. doi: 10.1093/genetics/145.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Nuoffer C, Jeno P, Conzelmann A, Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiaeGAS1 protein to the plasma membrane. Mol Cell Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB, Hardwick KG, Lewis MJ. Sorting of soluble ER proteins in yeast. EMBO (Eur Mol Biol Organ) J. 1988;7:1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, Gerst JE. Homologues of the synaptobrevin/VAMP family of synaptic vesicle proteins function in the late secretory pathway in S. cerevisiae. . Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci USA. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., and G.R. Fink. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY. 119–187.
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Hunter CP, Valls LA, Stevens TH. Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc Natl Acad Sci USA. 1986;83:3248–3252. doi: 10.1073/pnas.83.10.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T, Whitehearst SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Søgaard M, Tani K, Ruby ye R, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Söllner T. A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Hepler KP. Cytokinesis in higher plants. Cell. 1996;84:821–824. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. . J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT, Peacock WJ, Hardy RW. Dynamics of spermiogenesis in Drosophila melanogaster. I. Individualization process. Z Zellforsch. 1972;124:479–506. doi: 10.1007/BF00335253. [DOI] [PubMed] [Google Scholar]
- Tu J, Song W, Carlson M. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. . Mol Cell Biol. 1996;16:4199–4206. doi: 10.1128/mcb.16.8.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick SM. Spatial aspects of cytokinesis in plant cells. Curr Opin Cell Biol. 1991;3:253–260. doi: 10.1016/0955-0674(91)90149-s. [DOI] [PubMed] [Google Scholar]
- Winsor B, Schiebel E. An overview of the Saccharomyces cerevisiaemicrotubule and microfilament cytoskeleton. Yeast. 1997;13:399–434. doi: 10.1002/(SICI)1097-0061(199704)13:5<399::AID-YEA126>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]