Abstract
Chloroplast subfractions were tested with a UV cross-linking assay for proteins that bind to the 5′ untranslated region of the chloroplast psbC mRNA of the green alga Chlamydomonas reinhardtii. These analyses revealed that RNA-binding proteins of 30–32, 46, 47, 60, and 80 kD are associated with chloroplast membranes. The buoyant density and the acyl lipid composition of these membranes are compatible with their origin being the inner chloroplast envelope membrane. However, unlike previously characterized inner envelope membranes, these membranes are associated with thylakoids. One of the membrane-associated RNA-binding proteins appears to be RB47, which has been reported to be a specific activator of psbA mRNA translation. These results suggest that translation of chloroplast mRNAs encoding thylakoid proteins occurs at either a subfraction of the chloroplast inner envelope membrane or a previously uncharacterized intra-chloroplast compartment, which is physically associated with thylakoids.
Plastids perform many diverse functions for plants and eukaryotic algae, e.g., photosynthesis, the assimilation of nitrogen and sulfur, and the synthesis of carbohydrates, amino acids, fatty acids, and glycerolipids. Plastids of plants and green algae are surrounded by two concentric envelope membranes, which regulate the trafficking of diverse molecules (e.g., amino acids, proteins, and metabolites) between the stroma and cytosol. In chloroplasts, one of the developmental fates of the plastids, the light-driven reactions of photosynthesis occur in the membranes of the disklike thylakoids. Multiple thylakoids are associated in stacks called grana.
Considerable progress has been made recently towards understanding the processes underlying the biogenesis of the thylakoid compartment. Thylakoid proteins encoded by chloroplast genomes are thought to be synthesized directly into thylakoids on membrane-bound ribosomes (for review see Harris et al., 1994). However, the glycerolipids, quinones, and pigments (i.e., carotenoids and chlorophyll) found in thylakoids are probably synthesized at the chloroplast envelope, and then transported to thylakoids by as yet unknown routes (for review see Joyard et al., 1991). Moreover, many thylakoid proteins are encoded by nuclear genes, synthesized on cytosolic ribosomes, and then imported through the chloroplast envelope and stroma to the thylakoid membrane or lumen (for review see Cline and Henry, 1996).
Translation in the chloroplast of the green alga Chlamydomonas reinhardtii, and in the mitochondria of Saccharomyces cerevisiae, requires genetic interactions between gene-specific, nucleus-encoded functions and sequences within the 5′ untranslated region (UTR)1 of the organellar mRNA (for review see Hinnebusch and Liebman, 1991; Gillham et al., 1994; Fox, 1996; Rochaix, 1996). Potential regulatory proteins of chloroplast mRNA translation have been identified by their ability to bind to the UTRs of chloroplast mRNAs in vitro (Hsu-Ching and Stern, 1991; Nickelsen et al., 1994; for review see Gillham et al., 1994; Zerges and Rochaix, 1994; Chen et al., 1995; Hauser et al., 1996). The most extensively studied protein in this class is a 47-kD protein in C. reinhardtii called RB47. A multiprotein complex containing RB47 has been postulated to specifically regulate translation of the psbA mRNA in response to light intensity (Danon and Mayfield, 1991) via stromal ADP levels (Danon and Mayfield, 1994a ) and redox potential (Danon and Mayfield, 1994b ). There is yet no direct evidence that RB47 is localized to the chloroplast. Other studies have suggested translational roles for RNA-binding proteins with molecular masses in the 46–47 kD molecular mass range (Zerges and Rochaix, 1994; Hauser et al., 1996). In this study we have used biochemical approaches to address the intra-chloroplast locations of chloroplast RNA-binding proteins.
Materials and Methods
Media and Growth Conditions
Cultures were grown in Tris-acetate-phosphate medium (Gorman and Levine, 1965), exposed to a light intensity of 100 microeinsteins (μE) m−2 s−1, to 2–5 × 106 cells/ml, and with aeration. Chloroplasts were isolated from a strain carrying the CW15 mutation, which affects the cell wall (Davies and Plaskitt, 1971), to allow breakage of cells but not chloroplasts.
Isolation and Fractionation of Chloroplasts
Cells were concentrated by centrifugation for 10 min at 4,000 g, resuspended in isotonic solution (0.3 M sorbitol, 10 mM tricine-HCl, pH 7.8, 5 mM MgCl2), at ∼5 × 107 cell/ml, and then broken by passage through a Nebulizer (Gascol Apparatus Co., Terre Haute, IN) at 22 bar. Samples were maintained at 0–4°C thereafter. Chloroplasts, other organelles, and unbroken cells were pelleted by centrifugation at 3,000 g for 1.0 min, gently resuspended in the same isotonic solution, loaded on a discontinuous (45/ 75%) Percoll gradient (with 0.3 M sorbitol, 10 mM tricine-HCl, pH 7.8, 5 mM MgCl2), and then centrifuged at 6,000 g for 20 min. Chloroplasts were collected from the interphase of the 45% and 75% Percoll solutions. Two volumes of the isotonic buffered solution were added to dilute the Percoll so that the chloroplasts could be pelleted by centrifugation for 5 min at 4,000 g. The chloroplast extract (see Fig. 1, lane 1) was prepared by lysis in 0.1% Triton X-100, 10 mM tricine, pH 7.8, 60 mM KCl, 5 mM β-mercaptoethanol, and 20% glycerol. The whole-cell extract (see Fig. 2) was prepared by lysing cells (carrying CW15) under the same conditions. To prepare the chloroplast fractions (Figs. 1 and 2), isolated chloroplasts were osmotically lysed in 2 ml of a hypotonic buffered solution (10 mM tricine, pH 7.8, 10 mM EDTA, 5 mM β-mercaptoethanol) by repeatedly pipetting (10–20 times) with a Pipetman p1000 (Gilson, Omnilab, Switzerland). For the stromal, thylakoid, and low density membranes analyzed in Figs. 1 and 2, this chloroplast lysate was fractionated on two sequential discontinuous sucrose gradients. The first gradient, which had 0.6 and 1.0 M sucrose phases (10 mM tricine pH 7.8, 10 mM EDTA, 5 mM β-mercaptoethanol), was centrifuged for 3 h at 105 g in an SW40 rotor (Beckman Instruments, Inc., Fullerton, CA). The stromal fraction was the material that did not enter the sucrose. The low buoyant density membranes were collected from the interphase. Thylakoids in the pellet were resuspended in 1.8 M sucrose, 10 mM tricine, pH 7.8, 10 mM EDTA, and a solution containing 1.3 M sucrose, 10 mM tricine, pH 7.8, and 10 mM EDTA was layered on top. This discontinuous sucrose gradient was centrifuged at 105 g for 3 h, and the thylakoid fraction was collected from the interphase. An equal volume of 10 mM tricine, pH 7.8, 10 mM EDTA was added to the low density and thylakoid membrane fractions to dilute the sucrose so that these membranes could be pelleted by centrifugation at 105 g for 1 h. Membranes were resuspended in 10 mM tricine, pH 7.8, 0.2 mM EDTA, 60 mM KCl, 5 mM β-mercaptoethanol, and 20% glycerol. The stromal extract was dialyzed against the same buffer, including also 20% (wt/vol) polyethylene glycol 8000 to concentrate it.
Figure 1.
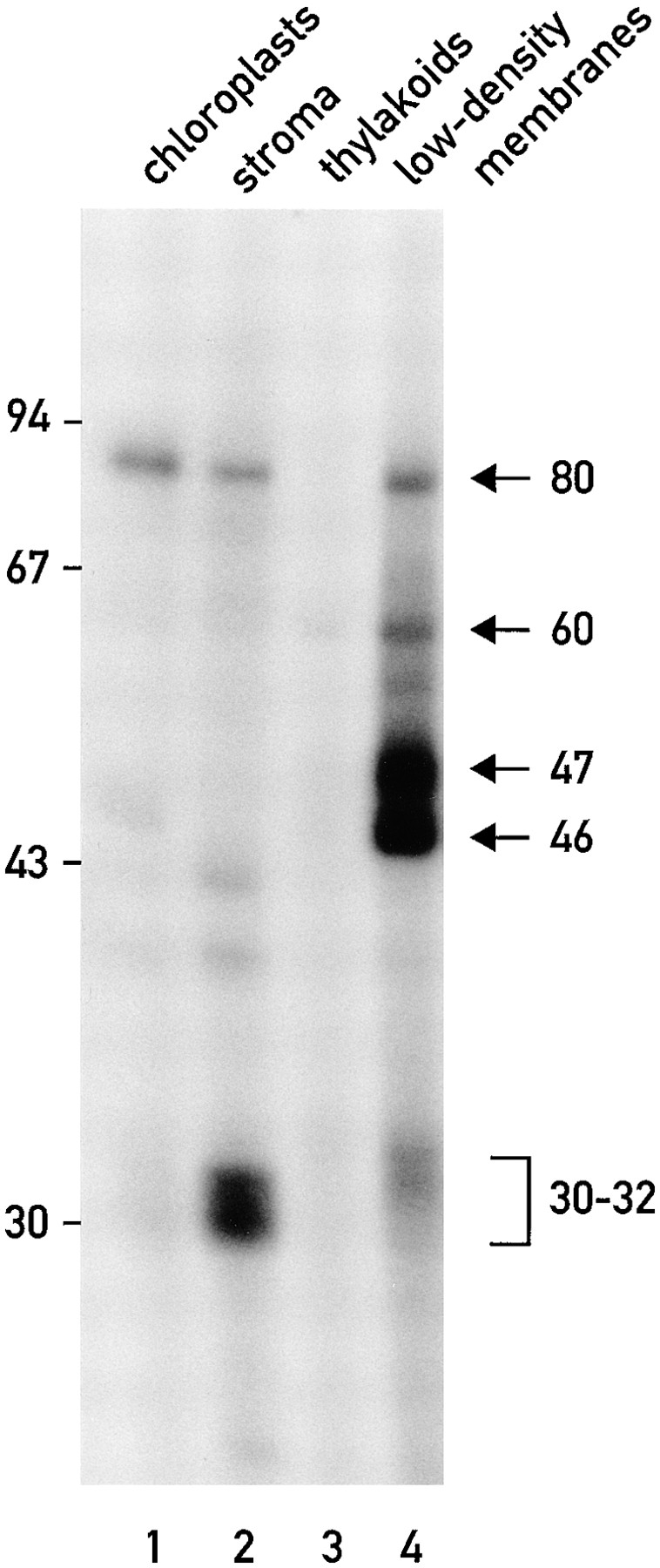
RNA-binding proteins (of ∼30, 32, 46, 47, 60, and 80 kD) cofractionate with the chloroplast low density membranes. UV light was used to cross-link the 32P-RNA probe derived from the psbC 5′ UTR to proteins in extracts from isolated chloroplasts (lane 1), chloroplast stroma (lane 2), thylakoids (lane 3), and the low density chloroplast membranes (lane 4). Each sample contained 20 μg of protein.
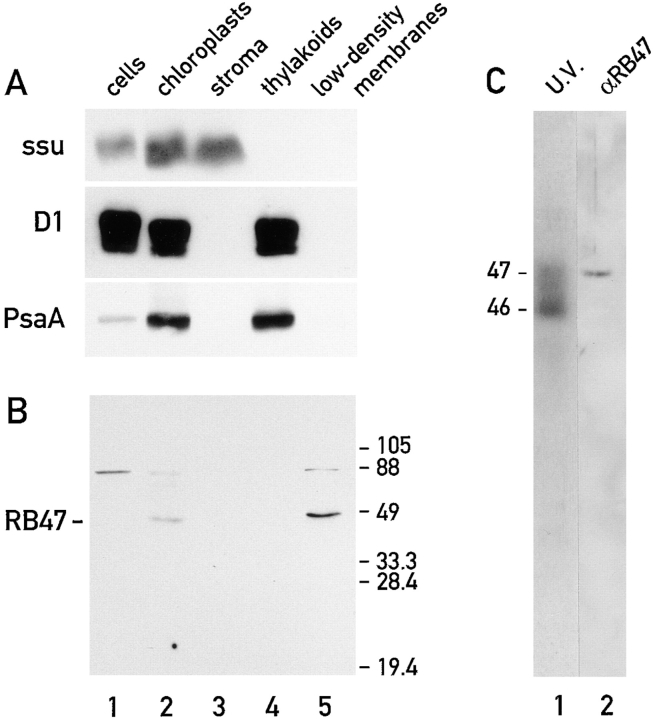
Figure 2.
The RNA-binding protein, RB47, cofractionates with low density chloroplast membranes. (A and B) The immunoblots analyze 20-μg samples of protein extracts from total cells (lane 1) isolated chloroplasts (lane 2), chloroplast stroma (lane 3), thylakoids (lane 4), and the low density chloroplast membranes (lane 5). The immunoblot in A was reacted with antisera against marker proteins for the chloroplast stroma, the small subunit of Rubisco (ssu); for the stacked (appressed) thylakoid membranes, the D1 subunit of PSII (D1), and for the nonstacked or stroma lamellae of thylakoid membranes, the PsaA subunit of PSI (PsaA). The immunoblot in B was reacted with antisera against the RNA-binding protein RB47 (Danon and Mayfield, 1991, 1994). (C) The 47-kD RNA-binding protein (radiolabeled by UV cross-linked to the 32P-RNA probe ) comigrates during SDS-PAGE (and in the same lane) with RB47 detected with the antiserum against it.
For the sucrose density gradients 1 and 3 shown in Fig. 4, osmotically lysed chloroplasts (see above) were fractionated on the basis of buoyant density by centrifugation at 105 g for 12–16 h on a continuous (0.3–1.8 M) sucrose gradient (with 10 mM tricine, pH 7.8, 5 mM MgCl2, 5 mM β-mercaptoethanol). Fractions were collected from the gradient, and then diluted with an equal volume of 10 mM tricine, pH 7.8, 5 mM MgCl2, 5 mM β-mercaptoethanol. Membranes in these fractions were concentrated by centrifugation at 105 g for 30 min. The resulting pellet fractions containing thylakoid membranes were resuspended in either 10 mM tricine, pH 7.8, 10 mM EDTA, or 10 mM tricine, pH 7.8, 5 mM MgCl2, and then fractionated on the gradients 2 and 4, respectively, which had 0.3–1.8 M sucrose gradient and either 10 mM tricine, pH 7.8, 10 mM EDTA, 5 mM β-mercaptoethanol (gradient 2), or 10 mM tricine, pH 7.8, 5 mM MgCl2, 5 mM β-mercaptoethanol (gradient 4) for at least 5 h at 105 g. Fractions were diluted twofold with either 10 mM tricine, pH 7.8, 10 mM EDTA (gradient 2 fractions), or 10 mM tricine, pH 7.8, 5 mM MgCl2 (gradient 4 fractions), and then centrifuged at 105 g for 30 min to pellet the membranes. The final membrane fractions were resuspended in 10 mM tricine, pH 7.8, 0.2 mM EDTA, 60 mM KCl, 5 mM β-mercaptoethanol, 20% glycerol and stored at −70°C. Each extract was standardized for protein concentration with the assay of Smith et al. (1985). The samples analyzed with the UV cross-linking assay represented equivalent proportions of each fraction. Polyribosomes were prepared by lysing cells carrying the CW15 mutation in 0.1% TritonX-100 and then immediately fractionating this lysate on a discontinuous sucrose gradient as described previously (Yohn et al., 1996). Chlorophyll and β-carotene were quantified spectrophotometrically (Porra et al., 1989).
Figure 4.
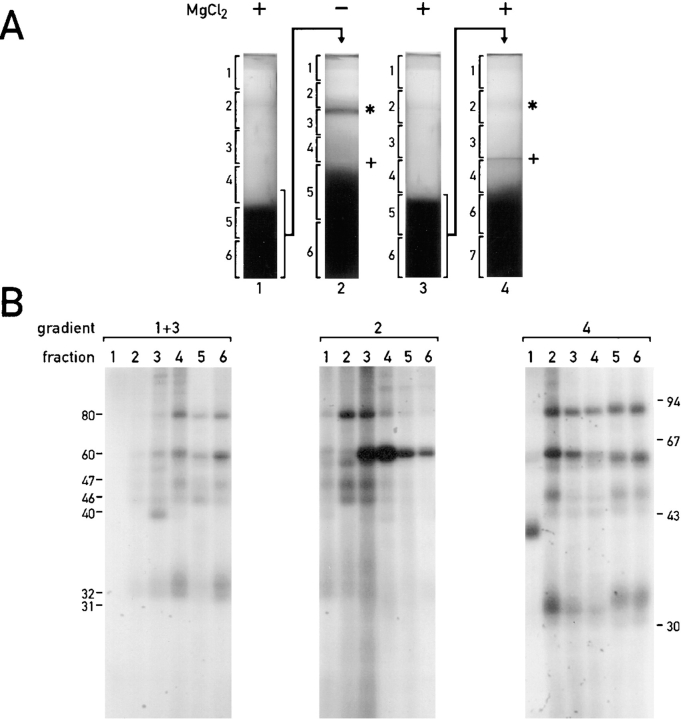
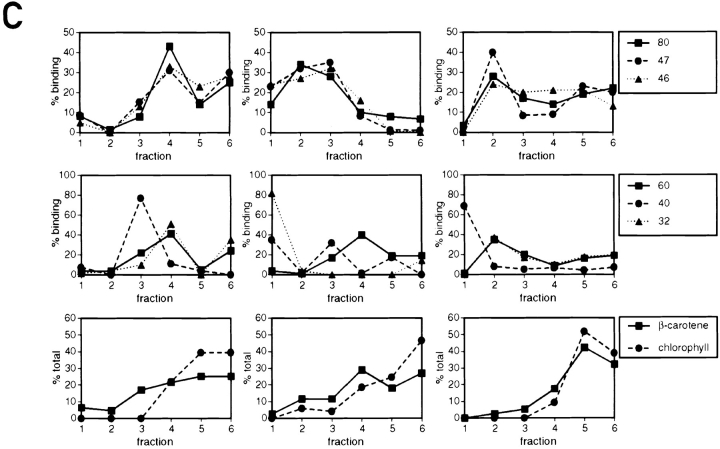
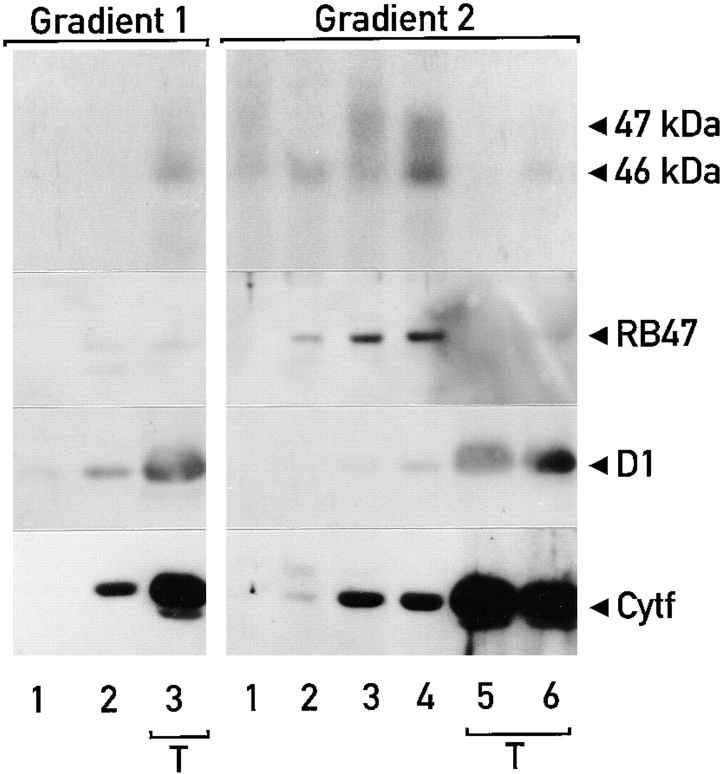
The low density membranes (with the RNA-binding proteins) are associated with thylakoid membranes. (A) Sequential sucrose density gradients were used to fractionate chloroplast membranes. On gradient 1 total chloroplast membranes were fractionated in the presence of 5 mM MgCl2. Thylakoids from this gradient (the dark band at the bottom) and the low density membranes associated with them were separated on gradient 2, which lacked MgCl2. The band of orange membranes observed on gradient 2 is indicated with an asterisk. When both gradients contained MgCl2 (gradients 3 and 4) considerably fewer low density membranes were observed above the thylakoid membranes. The lower density of the thylakoids in the absence of Mg2+ (indicated by their upward shift in gradient 2 and the +) reflects the destacking of the grana. (B) Binding of proteins to the 32P-RNA derived from the psbC 5′ UTR in the fractions from the gradients shown in A was measured with the UV cross-linking assay. Samples represented equivalent proportions of each fraction (i.e., were analyzed on a per chloroplast basis) and contained between 8 and 30 μg protein. (C) The binding signals for each protein were determined by phosphorimaging and graphed as percentage of the total signal for that protein in each gradient. Chlorophyll concentrations throughout each gradient are also graphed to indicate the distribution of thylakoid membranes.
To prepare the membranes used in Fig. 3, isolated chloroplasts (see above) were hypotonically lysed in 10 mM tricine, pH 7.8, 10 mM EDTA, 5 mM β-mercaptoethanol, and then fractionated on a discontinuous sucrose gradient, which had 0.3 and 0.8 M sucrose phases (10 mM tricine, pH 7.8, 10 mM EDTA, 5 mM β-mercaptoethanol) and was centrifuged for 3 h at 105 g in an SW40 rotor (Beckman Instruments, Inc.). The RNA-binding proteins in samples containing 10 μg protein were 32P labeled by UV cross-linking to a 32P-RNA probe (see below). Membranes were pelleted by centrifugation at 105 g for 60 min at 2°C. The resulting pellets were resuspended in the various solutions described in the text and incubated for 30 min on ice with intermittent vortexing. Membranes were pelleted by centrifugation at 105 g for 60 min at 2°C, and then resuspended in SDS-PAGE loading buffer (10% glycerol, 1% SDS, 100 mM DTT, 30 mM Tris-HCl, pH 6.8, 0.01% bromophenol blue). The supernatant fractions were extracted with the procedure of Wersel and Flügge (1984) to remove the Triton X-100, NaCl, or urea and concentrate the proteins. Fractions were analyzed by SDS-PAGE and autoradiography or phosphorimaging.
Figure 3.
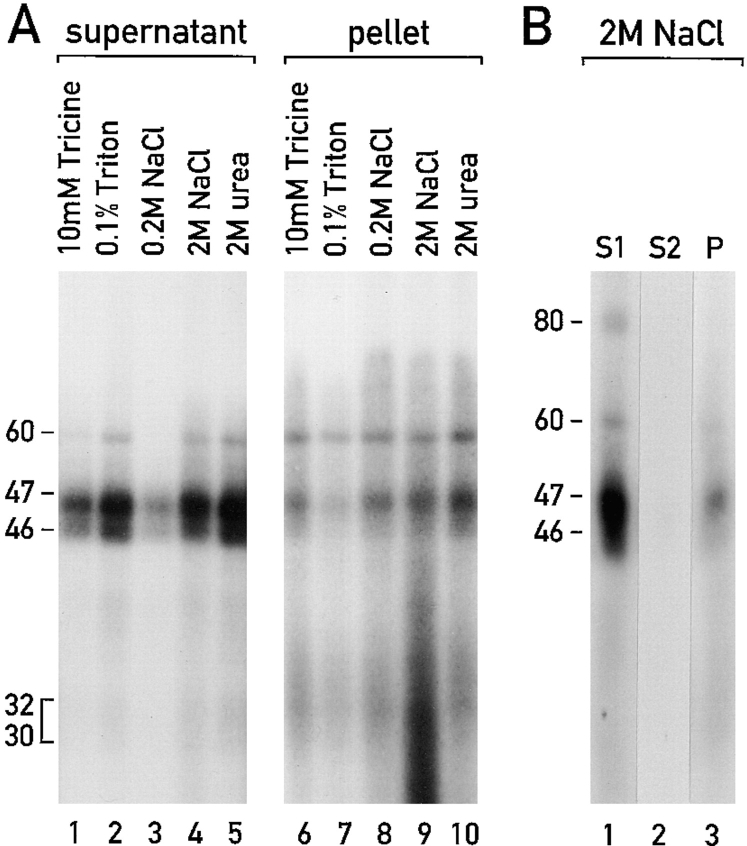
Membrane association of the RNA-binding proteins. (A) Samples (1.0 μg protein) of low density chloroplast membranes containing the RNA-binding proteins (which had been radiolabeled by the UV cross-linking to the 32P-RNA probe) were treated with 10 mM tricine, pH 7.8, 0.1% Triton X-100, 0.2 M NaCl, 2 M NaCl and 2 M urea. The samples were incubated 30 min at 0°C. After centrifugation of the extracts at 105 g for 60 min, supernatant and pellet fractions were analyzed by SDS-PAGE and autoradiography. (B) A sample of low density membranes with the 32P-RNA–binding proteins was treated for 30 min at 0°C with 2.0 M NaCl, and then centrifuged at 105 g for 60 min. The membrane pellet was resuspended in 2.0 M NaCl, and again incubated for 30 min at 0°C and then centrifuged at 105 g for 60 min. The first and second supernatant fractions (S1 and S2) and the final membrane pellet fraction (P) were analyzed by SDS-PAGE and autoradiography. The UV crosslinking of the 80-kD protein was often not detected for unknown reasons (see A and B). However, in other experiments, extraction of this protein paralleled the extraction of the 46-, 47- and 60-kD proteins.
RNA-binding
The psbC 5′ UTR RNA probe was transcribed in vitro from pDH245 (Zerges and Rochaix, 1994) in a 10-μl reaction containing 0.5 μg of linear DNA template, 40 mM Tris-HCl, pH 7.5, 6 mM MgCl2, 2 mM spermidine, 10 mM DTT, 25 U porcine RNase inhibitor (Biofinex, Praroman, Switzerland), 60 μCi of [α-32P]UTP (800 mCi/mmol; Amersham International, Little Chalfont, UK), 12 μM non-radiolabeled UTP, 0.3 mM each of ATP, CTP, and GTP, and 8 U T7 RNA polymerase (Biolabs, Beverly, MA) for 45 min at 37°C. 1 U of RNase-free DNase was added and the reaction was incubated for an additional 10 min at 37°C. The reactions were extracted once with phenol-chloroform. The RNA probes were separated from the nonincorporated nucleotide-triphosphates on spin columns of G-50 (Sambrook et. al., 1989). Binding reactions (15–20 μl) were performed at 22– 25°C for 5 min and contained 30 mM Tris-HCl, pH 7.0, 5.0 mM MgCl2, 5 mM DTT, 50 mM KCl, and 3–80 μg protein (depending on the extract used and as described in the text). Each reaction contained ∼2 × 105 cpm of 32P-RNA probe. Subsequent exposure to a 254-nm UV irradiation of 1.0 J/cm2 and for a ∼7 min duration using a Stratalinker UV cross-linker (Stratagene, La Jolla, CA) covalently cross-linked the RNA probe and bound proteins. After irradiation, the nonbound 32P-RNA probe was digested by treatment with 10 μg RNase A (Sigma Chemical Co., St. Louis, MO) for 10 min at 37°C. Samples were incubated at 85°C for 5 min in protein loading buffer (10% glycerol, 1% SDS, 100 mM DTT, 30 mM Tris-HCl, pH 6.8, 0.01% bromophenol blue), fractionated by SDS-PAGE (11% acrylamide), and analyzed by autoradiography or phosphorimaging.
Immunoblot Analyses
Samples of the various extracts (each containing 20 μg of protein) were incubated in 10% glycerol, 1% SDS, 100 mM DTT, 30 mM Tris-HCl, pH 6.8, and 0.01% bromophenol blue for 1–2 h at room temperature, electrophoresed through an 11% polyacrylamide gel (Sambrook et al., 1989), and then electroblotted to nitrocellulose membranes (Protran, 0.45 μm; Schleicher and Schuell, Inc., Keene, NH). Protein transfer was verified by staining the filter with Ponceau S (Sambrook et al., 1989). Filters were blocked in 5% nonfat dry milk, 0.02% Tween-20, PBS (Sambrook et al., 1989), and then incubated with primary antisera for 1 h, washed three times for 10 min in PBS (0.02% Tween-20), reacted with peroxidase-linked anti–rabbit Ig or anti–mouse Ig (Amersham International) for 1 h, and washed three times for 10 min with PBS containing 0.02% Tween-20. Signals were revealed with a chemiluminescence detection system (Durrant, 1990).
Acyl Lipid Analysis
Lipids were extracted with the procedure of Wersel and Flügge (1984) from fractions. Samples were all derived from an equal number of cells (∼1010). The final organic phase was dried under vacuum, and the remaining lipids were resuspended in 40 μl of chloroform/methanol (53:37). These samples were applied to silica gel 10 thin layer plates (Merck, Darmstadt, Germany). Chromatograms were developed with the solvent system; chloroform/methanol/glacial acidic acid/water (85:15:10:3; Goldberg and Ohad, 1970). The galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) were revealed by spraying the chromatogram with 50% (vol/vol) H2SO4 followed by a 10-min incubation at 120°C. Phosphatidylinositol (PI) and phosphatidylglycerol (PG) were revealed on the same thin layer chromatogram with phosphomolybdic acid spray (Sigma Chemical Co.), which is specific for phospholipids (Ryu and MacCoss, 1979). Acyl lipid classes were also identified from their reported R f values (Goldberg and Ohad, 1970)
Results
RNA-binding Proteins Cofractionate with the Low Density Chloroplast Membranes
Isolated chloroplasts were fractionated by sucrose density gradient centrifugation as described in Materials and Methods. When the resulting fractions were tested with an in vitro, UV cross-linking assay (see Materials and Methods for experimental details), we detected several proteins that bind to a 32P-RNA probe derived from the psbC 5′ UTR (Fig. 1). All of these proteins appear to be localized to the chloroplast because none were detected in a 20-μg sample (on the basis of protein content) of whole cells (data not shown). In addition, five of these proteins (of 30, 32, 40, 46, and 80 kD) were detected in a 20-μg sample of isolated chloroplasts (Fig. 1, lane 1). Enrichment of at least four RNA-binding proteins (of 32, 46, 47, and 60 kD) in the preparations of low density chloroplast membranes can be seen by comparing the binding signals of these proteins produced by a 20-μg sample of these membranes (Fig. 1, lane 4) and 20-μg samples of the stroma (Fig. 1, lane 2) or thylakoid membranes (Fig. 1, lane 3). In several similar experiments, in which the levels of binding of these proteins to the 32P-RNA probe were compared for 20-μg samples of stroma or thylakoids and 1-μg samples of the low density membranes, we found that these proteins are enriched over 100-fold in the low density membranes (data not shown). Although, these low density membranes have been reported to be derived from the chloroplast envelope (Douce et al., 1973; Clemetson et al., 1992), for reasons presented in the Discussion they will not be referred to as such in this report. An 80-kD RNA-binding protein was detected in both the stroma and the low density membrane fractions. Binding of a 30-kD protein to the 32P-RNA probe was detected in the stromal subfraction. The binding of one or more 31-kD protein(s) to the 32P-RNA probe was detected in the stroma and low density membranes.
To ascertain the purity of the stroma and thylakoid membrane fractions, each was tested for the enrichment of one or more marker protein(s) from the desired compartment and the depletion of marker protein(s) from the other. Separation of the proteins in these fractions by SDS-PAGE, followed by immunoblot analyses (Fig. 2 A), revealed that the stromal fraction was highly enriched for the small subunit of ribulose bisphosphate carboxylase (Rubisco; Fig. 2 A, lane 3). This stromal fraction lacked detectable levels of two marker proteins of thylakoid membranes, PsaA and D1, core subunits of photosystem I (PSI) and photosystem II (PSII), respectively. Similarly, the thylakoid membrane fraction was highly enriched for PsaA and D1, but contained neither of the Rubisco subunits (Fig. 2 A, lane 4; only the small subunit is shown). In the absence of reliable marker proteins for the chloroplast envelope of C. reinhardtii, we were unable to determine by immunoblot analyses whether the low density membranes are enriched for envelope membranes. That we detected neither Rubisco nor the PSI and PSII core subunits in this fraction demonstrates that the low density membranes are not significantly contaminated with stroma or thylakoids, including the stroma lamellae thylakoid membranes (see Discussion).
Stability of the Membrane Association of the RNA-binding Proteins
To further test whether these RNA-binding proteins are associated with membranes, and to determine the stability of these associations, we incubated samples of the low density membranes (corresponding to 1 μg of protein) under various conditions, and then tested whether the RNA-binding proteins still fractionated with the membranes. The RNA-binding proteins in a sample of low density membranes were labeled with 32P by UV cross-linking to the 32P-RNA probe derived from the psbC 5′ UTR (see Materials and Methods). These membranes and their associated 32P-RNA–binding proteins were incubated under various conditions for 30 min at 0°C, with intermittent vortexing. After solubilization of membranes with 0.1% (wt/ vol) Triton X-100 and centrifugation at 105 g for 30 min, the 46-, 47-, and 60-kD RNA-binding proteins were detected predominantly in the supernatant fraction (Fig. 3 A, lane 2), and only a small fraction of each protein remained in the pellet (Fig. 3 A, lane 7). To examine further this protein–membrane association we incubated samples of the low density membranes and their associated 32P-RNA– binding proteins (again containing 1.0 μg total protein) in the presence of various reagents including, as a control, 10 mM tricine, 0.2 M NaCl, 2 M NaCl, and 2 M urea. Peripheral membrane proteins should be extracted by some of these treatments, whereas integral membrane proteins remain membrane associated (Hatefi and Hanstein, 1974; Fujiki et al., 1982). After incubation, the samples were centrifuged at 105 g for 60 min, and the 32P-RNA–binding proteins in the pellet fractions were analyzed with SDS-PAGE followed by autoradiography and phosphorimaging. Membranes treated with Tricine retained ∼75% or more of the 60-kD protein and ∼25% of the 46- and 47-kD 32P-RNA–binding proteins (Fig. 3 A, lanes 1 and 6). A higher fraction of these proteins was retained on the membranes after treatment with 0.2 M NaCl (Fig. 3 A, lanes 3 and 8). Incubation in 2 M NaCl or 2 M urea resulted in an increased dissociation of the 32P-RNA–binding proteins from the membranes (Fig. 3 A, lanes 4, 5, 9, and 10). The retention of 32P-RNA–binding proteins by the low density membranes after the treatment with 2.0 M NaCl probably does not result from an inaccessibility of these proteins because to be 32P labeled they had to be accessible to the 32P-RNA probe. Most of the RNA-binding proteins in the 30–32-kD molecular mass range remained membrane associated after each of these treatments, suggesting a greater stability of these associations than we observed for the larger RNA-binding proteins. The 80-kD protein could not be identified reproducibly in these UV cross-linking experiments. However, in other experiments, extraction of this protein paralleled the extraction of the 46-, 47- and 60-kD proteins.
These results suggested that there are two populations of the 46-, 47- and 60-kD RNA-binding proteins; one that is weakly associated with the low density membranes and is extracted by treatment with tricine, NaCl, or urea, and another population that is stably associated and is not extracted by these treatments. To test this hypothesis, we asked whether any additional 32P-RNA–binding proteins could be extracted from the low density membranes by a second incubation with 2.0 M NaCl. After the first treatment with 2.0 M NaCl and centrifugation at 105 g, the pellet fraction was again treated with 2.0 M NaCl, and the membranes were pelleted by a second centrifugation at 105 g for 60 min. The first and second supernatant fractions and the final pellet fraction were tested for the 32P-RNA– binding proteins by SDS-PAGE and autoradiography (Fig. 3 B). Whereas the first treatment with 2.0 M NaCl extracted a fraction of the 46-, 47-, and 60-kD 32P-RNA– binding proteins from the low density membranes (Fig. 3 B, S1), only trace amounts of these proteins were extracted by the second treatment (Fig. 3 B, S2). The presence of these RNA-binding proteins in the pellet fraction after two extractions with 2.0 M NaCl strongly suggests that they are stably associated with the low density membranes. The majority of the 80-kD protein was extracted by the first treatment with 2.0 M NaCl in this experiment. In other trials, a subfraction of this protein appeared to be stably associated with the low density membranes.
The RNA-binding Protein RB47 Cofractionates with the Low Density Chloroplast Membranes
To determine whether the RNA-binding protein RB47 is associated with chloroplast membranes (Danon and Mayfield, 1991, 1994a ,b; Yohn et al., 1996; see Introduction), an immunoblot with equal mass amounts (20 μg) of extracts of whole cells, isolated chloroplasts, and the three chloroplast subfractions (described above and in Materials and Methods) was reacted with a polyclonal antiserum raised against this protein (Fig. 2 B). Although RB47 was not detected in an extract derived from whole cells (Fig. 2 B, lane 1), it was detected in an extract of isolated chloroplasts (Fig. 2 B, lane 2). This provides the first direct evidence that this protein is located in the chloroplast. Comparison of the levels of RB47 in the three chloroplast subfractions on this immunoblot revealed an enrichment of this protein in the low density membrane fraction (Fig. 2 B, lane 5) and that it was not detected in the stroma (Fig. 2 B, lane 3) or thylakoid fractions (Fig. 2 B, lane 4). Therefore, RB47 appears to be associated with the low density membranes.
To determine whether the RNA-binding protein RB47 (Danon and Mayfield, 1991, 1994a ,b) could be one of the RNA-binding proteins in the 46–47 kD molecular mass range detected in preparations of the low density chloroplast membranes with the 32P-RNA probe derived from the psbC 5′ UTR, we asked whether it comigrates with either of these proteins during SDS-PAGE. The 46- and 47-kD RNA-binding proteins in a sample of low density membranes were 32P labeled by UV cross-linking to the psbC 5′ UTR 32P-RNA probe, subjected to SDS-PAGE, and then transferred to a nitrocellulose filter. As seen in Fig. 2 C, the 47-kD RNA-binding protein, but not the 46-kD species, comigrated (in the same lane) with the RB47 detected with the antiserum against it. (The exposure time of this autoradiograph was too short to reveal the weaker signals from the 31-, 32-, 60- and 80-kD RNA-binding proteins.) These results are consistent with RB47 being the 47-kD protein that binds to the psbC 5′ UTR 32P-RNA probe. The 46-kD RNA-binding protein associated with the low density membranes comigrates with RB46 (Zerges and Rochaix, 1994) and the 47-kD RNA-binding protein described by Nickelsen et al. 1994 (data not shown).
A protein of ∼80 kD, which cross-reacted with the antiserum against RB47, is more abundant in the extract of whole cells (Fig.2 B, lane 1) than in the extract of isolated chloroplasts (Fig. 2 B, lane 2) and therefore appears not to be a chloroplast protein. This protein was not detected in either the extracts of stroma (Fig. 2 B, lane 3) or thylakoids (Fig. 2 B, lane 4). The presence of trace amounts of this protein in the low density membrane extract (Fig. 2 B, lane 5) may be because of a low level of contamination of this fraction by extra-chloroplast proteins. The RNA-binding protein with a similar molecular mass was detected in stroma extracts (Fig. 1, lane 2), and not in extracts of whole cells (data not shown), as well as in the extract of low density membranes. Therefore, these two proteins are probably distinct.
The Low Density Membranes with the RNA-binding Proteins Are Physically Associated with Thylakoid Membranes
The composition of RNA-binding proteins in different preparations of low density membranes purified by discontinuous sucrose gradient centrifugation appeared to be highly dependent on the sucrose concentrations used, whether or not MgCl2 was present during the purification of chloroplasts and membrane fractionation steps, and/or other factors (data not shown). When chloroplast membranes were fractionated on the continuous sucrose gradient 1 in Fig. 4 A and Mg2+ ions were present throughout the preparative procedures, the dark green thylakoid membranes (Fig. 4 A, in fractions 4–6) separated from orange membranes of lower density (Fig. 4 A, fractions 1–3), which have been shown to be derived from the chloroplast envelope (Douce et al., 1973; Clemetson et al., 1992; see below). The presence of thylakoids in fractions 4–6 is evidenced by the presence of all the detectable chlorophyll in these fractions (Fig. 4 C). Similarly, the orange color of the low density membranes is because of the absence of chlorophyll, which reveals the β-carotene present throughout the gradient (Fig. 4 C, fractions 1–3). When the fractions of this gradient were tested with the UV cross-linking assay, the RNA-binding proteins of 31, 32, 46, 47, 60, and 80 kD were detected in the fractions containing thylakoid membranes (fractions 4–6). A 40-kD RNA-binding protein was detected predominantly in fraction 3, which contains low density membranes. Although these results would seem to contradict the cofractionation of the 31-, 32-, 46-, 47-, 60- and 80-kD RNA-binding proteins with the low density membranes seen in Figs. 1 and 2, these differences appear to have resulted from an association of the low density membranes with thylakoids, which is stabilized by Mg2+ ions. For example, when the RNA-binding proteins cofractionated with the low density membranes, and not with thylakoid membranes (Figs. 1 and 2), the membranes had been isolated in the absence of Mg2+ ions (see Materials and Methods). This issue was clarified when the membranes in fractions 5 and 6 from gradient 1 were depleted of Mg2+ ions by centrifugation at 105 g followed by vigorous resuspension in a solution lacking MgCl2 (and containing 10 mM EDTA). When these membranes were centrifuged on a sucrose density gradient 2 (lacking Mg2+ ions), orange low density membranes were observed that appeared to have been released from the thylakoids obtained from the first gradient. This was indicated by the presence of β-carotene and, to a lesser extent, chlorophyll in fractions 1–3 (Fig. 4 C). In addition, some of these low density membranes formed a discrete band (asterisk) that was not detected in gradient 1. In addition, the thylakoids were less dense than in gradient 1, due to their being unstacked in the absence of Mg2+ ions (for review see Barber, 1980). Fractions 2 and 3 of gradient 2, which are above the thylakoid membranes in fractions 4–6, contained the majority of the RNA-binding proteins of 31-, 32-, 46-, 47- and 80-kD. The majority of the 60-kD RNA-binding protein was detected in fractions 3 and 4, also above the thylakoid membranes. In other trials of this dual-gradient fractionation scheme, the 60-kD protein fractionated with thylakoids on gradient 1 and with low density membranes on gradient 2 (data not shown). In summary, the difference in the buoyant densities of the membranes associated with the RNA-binding proteins in the presence and absence of Mg2+ ions appears to result from an association of these membranes with the denser thylakoid membranes.
To determine the extent to which depletion of Mg2+ ions caused the release of the low density membranes associated with the RNA-binding proteins from thylakoids on gradient 2, we fractionated thylakoid membranes obtained from a gradient containing MgCl2 (Fig. 4 A, gradient 3) on a second gradient that also contained MgCl2 (Fig. 4 A, gradient 4). Note that gradients 1 and 3 are identical, whereas gradients 2 and 4 differ only by the absence and presence of MgCl2, respectively. After centrifugation, gradients 2 and 4 differed drastically in appearance; on gradient 4 only a minor fraction of the thylakoids became unstacked, as indicated by the presence of green material (+) above the denser band of stacked thylakoids. No chlorophyll and low amounts of β-carotene (2–5% total) were detected in the first three fractions. In addition, the band of low density membranes seen in gradient 2 was only faintly visible in gradient 4 (asterisk). Analysis of the gradient 4 fractions with the UV cross-linking assay revealed that the membranes associated with the RNA-binding proteins of 30, 32, 46, 47, and 80 kD were retained by thylakoids to a much greater extent than they were in the absence of Mg2+ ions on gradient 2. The partial release of the low density membranes associated with the RNA-binding proteins from the thylakoid membranes may have resulted from mechanical forces produced by repeated pipetting that was used to resuspend these membranes before they were loaded on gradient 4. Consistent with this possibility, we observed that when thylakoid membranes from gradients similar to gradients 1 and 3 were gently resuspended (with a paint brush), all of the RNA-binding proteins cofractionated again with thylakoid membranes both in the presence and absence of Mg2+ ions. Therefore, whereas the removal of Mg2+ ions appears to be required for the release of all of these low density membranes from thylakoid membranes, mechanical forces also appear to facilitate this dissociation.
The fractionation behavior of the RNA-binding proteins shown in Fig. 4 is not because of their association with ribosomes because these proteins were not detected in several preparations of polyribosomes (data not shown). Also, chloroplast ribosomes (detected by RNA-gel blot analysis using a probe complementary to the chloroplast 16-S rRNA) did not cofractionate with the RNA-binding proteins in the gradients 1 and 2 shown in Fig. 4 A, they were detected only in fractions 5 and 6 of both gradients (data not shown).
Fractions of sequential sucrose density gradients similar to gradients 1 and 2 in Fig. 4, were assayed for the 46- and 47-kD RNA-binding proteins with the UV cross-linking assay, and for RB47 and various thylakoid proteins by immunoblot analyses (Fig. 5). RB47 fractionated with the 46- and 47-kD RNA-binding proteins. The enrichment of thylakoid membranes at the bottom of the gradients is evidenced by the abundance of the known thylakoid proteins (the D1 subunit of the PSII reaction center, cytochrome f) in fractions 5 and 6. Trace amounts of these proteins were detected in the fractions containing the low density membranes (fractions 2 and 3 of gradient 2). The distributions of P6, the PSII subunit encoded by psbC, and the PsaA subunit of PSI were similar to those of D1 and cytochrome f (data not shown).
Figure 5.
Immunoblot and UV cross-linking analyses of fractions from gradients 1 and 2. The 46-, 47-, and 60-kD RNA-binding proteins, which were found to bind to the 32P-RNA probe in the UV cross-linking assay, cofractionate with thylakoids (T) in the presence of magnesium ions (Gradient 1). In the absence of magnesium ions (Gradient 2), these proteins and the low density membranes associated with them were separated from thylakoid membranes (T). Thylakoid membranes were followed by their enrichment for the PSII reaction center subunit D1, and cytochrome f detected by immunoblot analysis. RB47 (detected by immunoblot analysis) also cofractionates with the low density membranes that are associated with thylakoid membranes in the presence of magnesium ions. Samples contained 20 μg of protein.
A Sub-fraction of the Chloroplast Envelope is Associated with Thylakoids
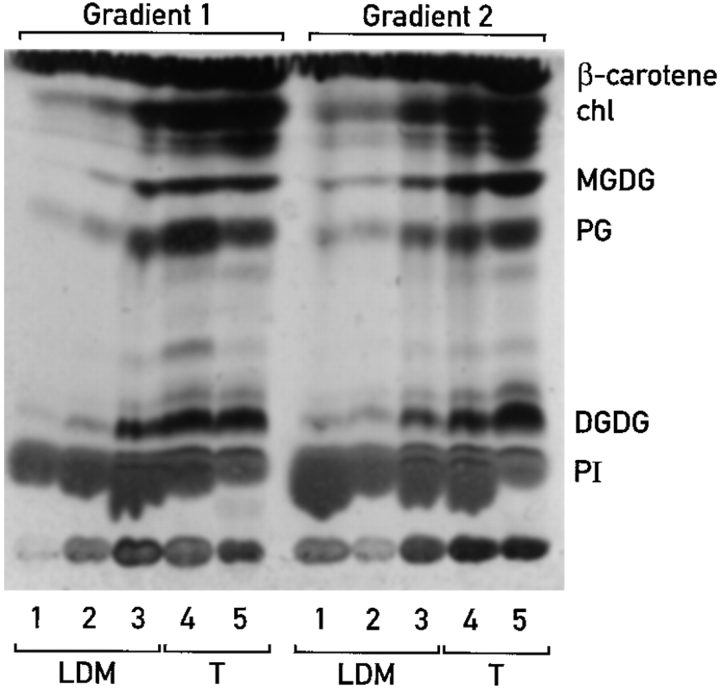
To test the hypothesis that low density membranes and the associated RNA-binding proteins are released from thylakoids upon the removal of magnesium ions, the acyl lipid compositions of fractions from gradients similar to those shown in Fig. 4 A were determined by thin layer chromatography. Inspection of the thin layer chromatogram in Fig. 6 revealed that the distributions of the various acyl lipid classes in gradients 1 and 2 are similar. The thylakoid membranes on both gradients (fractions 4 and 5) are rich in the galactolipids DGDG and MGDG, and also contain the phospholipids PG and PI (see Materials and Methods for experimental details). The membranes above the thylakoids in gradient 1 have been reported to be derived from the chloroplast envelope (Douce et al., 1973; for review see Joyard et al., 1991; Clemetson et al., 1992). The outer envelope membrane is less dense than the inner membrane, enriched in PI, but also contains low amounts of MGDG, DGDG, and PG (Joyard et al., 1991). On the thin layer chromatogram in Fig. 6, the least dense fractions from both gradients (i.e., fractions 1) contain predominantly PI. Therefore, these fractions appear to be enriched for the outer envelope membrane. In buoyant density, the inner envelope membrane is intermediate to the outer envelope and thylakoid membranes and therefore, should be found in fractions 2 and 3 of each gradient. The inner envelope membrane has an acyl lipid composition that is close to that of thylakoid membranes; both are rich in MGDG and DGDG and contain lower amounts (1%) of PI than does the outer envelope membrane (for review see Joyard et al., 1991). In fractions 2 and 3, the presence of MGDG and DGDG, and the lack of chlorophyll are consistent with an enrichment in the inner envelope membrane.
Figure 6.
The low density membranes that are associated with thylakoids in the presence of magnesium have acyl lipid compositions that are similar to the chloroplast envelope membranes. The thin layer chromatogram resolves acyl lipid classes in fractions from gradients similar to those shown in Fig. 4. Indicated are the pigments β-carotene and chlorophyll (chl), the galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), and the phospholipids phosphatidylglycerol (PG) and phosphatidylinositol (PI). LDM, the low density membranes. T, thylakoid membranes. Samples represent equivalent fractions of each fraction (i.e., were analyzed on a per chloroplast basis).
The most important conclusion that can be drawn from these data is that membranes were released from the thylakoid containing fractions from gradient 1 (Fig. 4) after the removal of Mg2+ and vigorous resuspension. This is evidenced by the presence of acyl lipids in fractions 1–3 of gradient 2. The 46- and 47-kD RNA-binding proteins were detected in fractions 2 and 3 of gradient 2, and not in the corresponding fractions of gradient 1 (data not shown; but see Fig. 4 B and C). Thus, the fractions from gradient 2 that contain the RNA-binding proteins also contain membranes that are similar to the inner envelope membrane in their acyl lipid composition and buoyant density.
Discussion
This study has revealed that RNA-binding proteins are associated with chloroplast membranes in C. reinhardtii. From our data, low density membranes in the chloroplast can be subdivided into two general classes. The first class separates from the thylakoids in the presence of magnesium ions and cofractionates with an RNA-binding protein of 40 kD (Fig. 4, gradient 1). The second class is associated with thylakoids and six RNA-binding proteins of 31, 32, 46, 47, 60, and 80 kD (Fig. 4 and data not shown), and RB47 (Fig. 5). The stability of the association of the second class of membranes with thylakoid membranes is evidenced by their cofractionation during a 16 h sucrose density gradient centrifugation (Fig. 4, gradient 1). The association of these RNA-binding proteins with the low density membranes was only revealed when thylakoid membranes were pelleted by centrifugation and harshly resuspended in the absence of Mg2+ ions. After these steps, the RNA-binding proteins and the low density membranes separated from thylakoid membranes on a second sucrose density gradient lacking Mg2+ ions (Fig. 4, gradient 2). On a second sucrose density gradient containing Mg2+ ions (Fig. 4, gradient 4), considerably greater amounts of low density membranes with the RNA-binding proteins remained associated with thylakoids than was observed on gradient 2 (i.e. in the presence of Mg2+ ions). As the stacking of thylakoids requires divalent cations (for review see Barber, 1980), these low density membranes may be retained by thylakoid grana. However, the Mg-dependent association between thylakoids in grana differs from the association of the low density membranes with thylakoid membranes because the latter association is maintained in the absence of Mg2+ ions if the membranes are gently resuspended (data not shown).
A fraction of each of the RNA-binding proteins, comprising ∼25% of the total, were found to be stably associated with the second class of low density membranes because they were not extracted with 2 M NaCl or 2 M urea (Fig. 3). NaCl concentrations >0.7 M have been shown to remove the OE23 subunit from the oxygen-evolving complex of PSII, which is embedded in thylakoid membranes (Ghanotakis and Yocum, 1990; Hashimoto et al., 1997). However, another fraction of the 46- and 47-kD RNA-binding proteins, comprising ∼75% of the total amount, was extracted by vortexing several times in tricine and, therefore, is weakly associated with membranes (Fig. 3 A). As these RNA-binding proteins did not appear to have been liberated to the chloroplast stroma during the preparation of the low density membranes analyzed in Figs. 1 and 2, we assume that the extraction of the low density membrane from thylakoids, binding of the 32P-RNA probe, or vortexing weakened the association of the RNA-binding proteins with the low density membranes. The distinction between these different subpopulations of the RNA-binding proteins on the basis of the strength of their membrane association was supported by the observation that a second incubation in 2.0 M NaCl did not result in a further release of the tightly associated 32P-RNA–binding proteins (Fig, 3B, S2). Thus, these tightly associated 32P-RNA–binding proteins were detected in the pellet fraction (P) after two sequential treatments with 2 M NaCl.
The buoyant densities and the acyl lipid compositions of both classes of low density membranes are compatible with their origin being the inner chloroplast envelope membrane (Fig. 5). However, at present we cannot exclude the possibility that the second class of low density membranes, which has not been described previously, is derived from an intra-chloroplast compartment. The unavailability of a reliable marker protein for the inner envelope of C. reinhardtii chloroplasts has impeded the identification of these low density membranes. An antiserum against the 37-kD inner envelope protein E37 from spinach chloroplasts (Block et al., 1983) detected a 33-kD protein in the middle fractions of both gradients 1 and 2 (Fig. 4 A), which contain the RNA-binding proteins in gradient 2 (unpublished data). However, as this C. reinhardtii protein has not been shown to be homologous to E37 or localized at the inner chloroplast envelope membrane, it cannot be used as a definite marker for this compartment. Similarly, an antiserum raised against total envelope membranes from C. reinhardtii chloroplasts detected proteins with both classes of low density membranes, many of which were specific to one class. However, the membranes against which this antiserum was raised were prepared by similar fractionation procedures to those used in this study. Therefore, to use the proteins detected by this antiserum as criteria for chloroplast envelope would entail a circular argument. It is clear that the low density membranes were not derived from the stroma lamellae (non stacked) sub-fraction of thylakoid membranes because they contain only trace amounts of PSI (Fig. 2; and unpublished data) and the stroma lamellae have been shown to be enriched in PSI complexes (Andreasson et al., 1988).
The ability of the membrane-associated proteins to bind to RNA in vitro suggests that they function in chloroplast mRNA metabolism or translation. Previous studies have supported translational roles of 46- and 47-kD RNA-binding proteins (Danon and Mayfield, 1991, 1994a ,b; Hauser et al., 1996; Yohn et al., 1996; Zerges and Rochaix, 1994). We show here that RB47, an RNA-binding protein that has been proposed to activate the translation of the chloroplast psbA mRNA (RB47; Danon and Mayfield, 1991, 1994a ,b; Yohn et al., 1996), is associated with the second class of low density membranes. Although RB47 and the 47-kD RNA-binding protein detected in our UV cross-linking assay comigrate during SDS-PAGE (Fig. 2 C) and cofractionate on the two sequential sucrose density gradients shown in Fig. 5, the binding specificities of these proteins differ. The RNA-binding proteins described here bind to A- and U-rich RNAs; non-radiolabeled 5′ UTRs of various chloroplast mRNAs, including psbA, compete equally well (on a μg basis) for binding of the membrane-associated RNA-binding proteins to the psbC 32P-UTR probe, whereas no competition was observed by an RNA with a normal base composition (Zerges and Rochaix, 1994; and unpublished data). In addition, poly-A and poly-U competed for binding, whereas no competition was observed for poly-C or poly-G (Zerges and Rochaix, 1994; and unpublished data). In contrast, RB47 has been reported to bind specifically to the 5′ UTR of the psbA mRNA, based on the results of similar competition experiments (Danon and Mayfield, 1991; Yohn et al., 1996). Thus, the possibility remains that RB47 and the 47-kD RNA-binding protein described here are distinct. Alternatively, the psbA 5′ UTR affinity chromatography used in the purification of the multiprotein complex containing RB47 (Danon and Mayfield, 1991) may have selected for a subpopulation of the 47-kD RNA-binding protein described here that binds specifically to the psbA 5′ UTR. Consistent with this possibility, the binding of this protein to the psbC 32P-RNA probe, like the binding or RB47 to the psbA 5′ UTR (Danon and Mayfield, 1991), is light regulated and inhibited by ADP, and to a lesser extent by ATP, but not by other nucleotide-triphosphates (Zerges, W., and J-D. Rochaix, unpublished data).
In the mitochondria of S. cerevisiae, five gene-specific translational activator proteins (Michaelis et al., 1991; McMulin and Fox, 1993; for review see Fox, 1996), and other proteins involved in RNA processing and turnover (Dake et al., 1988; Wiesenberger and Fox, 1997) are associated with the inner mitochondrial membrane. This association has been postulated to reflect a compartmentalization of processing, translation, and turnover of mitochondrial RNAs at the inner mitochondrial membrane, where the proteins encoded by these RNAs function in the respiratory electron transport and the synthesis of ATP (McMulin and Fox, 1993). Whereas there is substantial genetic evidence that the activator proteins of COX3 mRNA translation physically interact with the COX3 5′ UTR (for review see Fox, 1996), the translational activator proteins of S. cerevisiae mitochondria have yet to be shown to interact with RNA in vitro. In chloroplasts, however, proteins known to function in RNA metabolism or translation have not yet been shown to be associated with membranes. Similar to the proposed roles of the translational activator proteins in S. cerevisiae mitochondria, the association of RNA-binding proteins with low density membranes in chloroplasts may reflect a compartmentalization of chloroplast RNA metabolism and translation.
Several previous observations suggest a role of the inner envelope membrane in chloroplast gene expression and thylakoid biogenesis. The galactolipids of thylakoid membranes are synthesized at the chloroplast envelope (Rawyler et al., 1995). Consequently, the acyl lipid compositions of the inner envelope and the thylakoid membranes are nearly identical (for review see Joyard et al., 1991). Extensions of the inner envelope membrane and membrane vesicles in the stroma have been observed by electron microscopy in the chloroplasts of C. reinhardtii (Hoober et al., 1991), tobacco, pea, soybean, and spinach (Morré et al., 1991), and in the chromoplasts of red pepper fruits (Hugueney et al., 1995). Moreover, steps in chlorophyll synthesis after Mg-protoporphyrin IX occur in the chloroplast envelope (for review see Joyard et al., 1991). How chlorophyll is transported to the thylakoids is unknown (for review see Reinbothe and Reinbothe, 1996). If the chlorophyll-binding proteins encoded by chloroplast mRNAs are synthesized on the inner envelope membrane, newly synthesized chlorophyll could bind directly to them there. Moreover, cocompartmentalization of the synthesis of chlorophyll and chloroplast genome–encoded, chlorophyll-binding proteins could facilitate a coordination of the these two processes. The plastid genome in the rapidly dividing chloroplasts of young pea leaves has been shown to be associated with the envelope (Sato et al., 1993). Lastly, a recent study has demonstrated the presence of a homologue of the Escherichia coli ribosome releasing factor associated with the chloroplast envelope in spinach. This protein was not detected in purified thylakoid membranes (Rolland, N., and J. Joyard, personal communication). Thus, many lines of evidence suggest that thylakoid biogenesis, and possibly translation of chloroplast mRNAs, occurs at the chloroplast inner envelope membrane. We should point out that other hypotheses are also compatible with our data. For example, the RNA-binding proteins might hold a pool of nontranslated or “silent” mRNAs at the chloroplast envelope as a reserve to sustain protein synthesis during conditions of limiting transcription.
A role of the low density membranes in chloroplast mRNA metabolism and translation would seem to be contradicted by many previous studies that have demonstrated that the synthesis of thylakoid membrane proteins occurs on thylakoid-associated ribosomes (Herrin et al., 1981; for review see Harris et al., 1994). However, magnesium ions were always present in the thylakoid preparations used in these studies. We find that low density membranes are associated with thylakoids (Fig. 4). Thus, these previous studies did not determine whether polyribosomes are associated with thylakoids directly, or with the membranes of lower buoyant density. The observation that, after the removal of magnesium ions, thylakoid membranes can be separated from the low density membranes and their associated RNA-binding proteins on the basis of buoyant density (Fig. 4) demonstrates that these membranes are distinct membrane compartments.
The hypothesis that a subfraction of the inner envelope membrane, or a previously uncharacterized internal chloroplast membrane compartment of similar buoyant density, is the site of thylakoid biogenesis necessitates a means by which the newly synthesized thylakoid proteins move from this membrane compartment to thylakoids. As mentioned above, some reports have suggested vesicular trafficking between the inner envelope and thylakoids (Hugueney et al., 1995; Hoober et al., 1991). However, it is also possible that this low density membrane compartment is contiguous with thylakoid membranes. Movement of the newly synthesized proteins between the two compartments, in this case, would occur laterally. We have made several attempts to detect the newly synthesized thylakoid proteins in the low density, thylakoid-associated membranes during short pulse-labeling experiments. These experiments showed that the proteins synthesized during the shortest possible pulse (1 min) were in thylakoid membranes (Zerges, W., and J.-D. Rochaix, unpublished data). However, this does not exclude the possibility that these proteins were synthesized in the low density membranes and then transported to thylakoids because known secretory pathways are typically too rapid to detect proteins in intermediate stages in the absence of inhibitors or mutations that result in a specific block in the pathway (for review see Gruenberg and Clague, 1992).
Acknowledgments
We thank B. Kohorn, J. Nickelsen, U. Oberholzer, N. Rolland, and the members of the laboratory (especially A. Auchincloss, M. Goldschmidt-Clermont, M. Hippler, O. Stampacchia, and K. Redding) for stimulating discussions and helpful comments; Y. Xu and P.-A. Siegenthaler for help with the lipid analyses and helpful comments; E. Boudreau for critical reading of the manuscript; N.H. Chua for the antisera against Rubisco; A. Danon for the antiserum against RB47; L. McIntosh for the antiserum against D1; S. Merchant for the antiserum against cytochrome f, K. Redding for antiserum against PsaA; and N. Roggli for preparing the figures.
This work was supported by grant 31-34014.92 from the Swiss National Fund.
Abbreviations used in this paper
- DGDG
digalactosyldiacylglycerol
- MGDG
monogalactosyldiacylglycerol
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- PSI
and PSII, photosystem I and II
- UTR
untranslated region
Footnotes
Address all correspondence to Jean-David Rochaix, Departments of Molecular Biology and Plant Biology, University of Geneva, 30 Quai Ernest-Ansermet, CH-1211, Geneva 4, Switzerland. Tel.: 41-22-702-6187. Fax: 41-22-702-6868.
W. Zerges' present address is Developmental, Cell and Molecular Biology, LSRC, Box 91000, Duke University, Durham, NC 27708-1000. E-mail: zerges@acpub.duke.edu
References
- Andreasson E, Svensson P, Weibull C, Albertsson P-A. Separation and characterization of stromal and grana membranes -evidence for heterogeneity in antenna size of both photosystem I and photosystem II. Biochim Biophys Acta. 1988;936:339–350. [Google Scholar]
- Barber J. Membrane surface charges and potentials in relation to photosynthesis. Biochim Biophys Acta. 1980;594:253–308. doi: 10.1016/0304-4173(80)90003-8. [DOI] [PubMed] [Google Scholar]
- Block, M.A., A-J. Dorne, J. Joyard, and R. Douce. 1983. Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. J. Biol. Chem. 258. 13273–13280. [PubMed]
- Chen Q, Adams CC, Usack L, Yang J, Monde R-A, Stern DB. An AU-rich element in the 3′ untranslated region of the spinach chloroplast petDgene participates in sequence-specific RNA-protein complex formation. Mol Cell Biol. 1995;15:2010–2018. doi: 10.1128/mcb.15.4.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemetson JM, Boschetti A, Clemetson KJ. Chloroplast envelope proteins are encoded by the chloroplast genome of Chlamydomonas reinhardtii. . J Biol Chem. 1992;267:19773–19779. [PubMed] [Google Scholar]
- Cline K, Henry R. Import and routing of nucleus-encoded chloroplast proteins. Annu Rev Cell Dev Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- Dake E, Hofmann TJ, McIntire S, Hudson A, Zassenhaus H. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. . J Biol Chem. 1988;263:7691–7702. [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light regulated translational activators: identification of chloroplatic gene specific mRNA binding proteins. EMBO (Eur Mol Biol Organ) J. 1991;10:3993–4001. doi: 10.1002/j.1460-2075.1991.tb04974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. ADP-dependent phosphorylation regulates RNA-binding in vitro: implications in light-modulated translation. EMBO (Eur Mol Biol Organ) J. 1994a;13:2227–2235. doi: 10.1002/j.1460-2075.1994.tb06500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994b;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Davies DR, Plaskitt A. Genetical and structural analyses of cell-wall formation in Chlamydomonas reinhardtii. . Genet Res. 1971;17:33–43. [Google Scholar]
- Douce R, Holtz RB, Benson AA. Isolation and properties of the envelope of Spinach chloroplasts. J Biol Chem. 1973;248:7215–7222. [PubMed] [Google Scholar]
- Durrant I. Light-based detection of biomolecules. Nature. 1990;346:297–298. doi: 10.1038/346297a0. [DOI] [PubMed] [Google Scholar]
- Fox, T.D. 1996. Genetics of mitochondrial translation. In Translational Control. J.W.B. Hershey, M.B. Matthews, and N. Sonenberg, editor. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 733–758.
- Fujiki Y, Hubbard AL, Fowler S, Lazrow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanotakis DF, Yocum CF. Photosystem II and the oxygen-evolving complex. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:255–276. [Google Scholar]
- Gillham NW, Boynton JE, Hauser CR. Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet. 1994;28:71–93. doi: 10.1146/annurev.ge.28.120194.000443. [DOI] [PubMed] [Google Scholar]
- Goldberg I, Ohad I. Biogenesis of chloroplast membranes: IV. Lipid and pigment changes during synthesis of chloroplast membranes in a mutant of Chlamydomonas reinhardtii y-1. . J Cell Biol. 1970;44:563–571. doi: 10.1083/jcb.44.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. . Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Clague MJ. Regulation of intracellular membrane transport. Curr Opin Cell Biol. 1992;4:593–599. doi: 10.1016/0955-0674(92)90077-p. [DOI] [PubMed] [Google Scholar]
- Harris EH, Boynton JE, Gillham N W. Chloroplast ribosomes and protein synthesis. Microbiol Rev. 1994;58:700–754. doi: 10.1128/mr.58.4.700-754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto A, Ettinger WF, Yamamoto Y, Theg S M. Assembly of newly imported oxygen-evolving complex subunits in isolated chloroplasts: sites of assembly and mechanism of binding. Plant Cell. 1997;9:441–452. doi: 10.1105/tpc.9.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatefi Y, Hanstien WG. Destabilization of membranes with chaotropic ions. Methods Enzymol. 1974;31:770–790. doi: 10.1016/0076-6879(74)31080-4. [DOI] [PubMed] [Google Scholar]
- Hauser CR, Gillham NW, Boynton J E. Translational regulation of chloroplast genes. J Biol Chem. 1996;271:1486–1497. doi: 10.1074/jbc.271.3.1486. [DOI] [PubMed] [Google Scholar]
- Herrin D, Michaels A, Hickey E. Synthesis of a chloroplast membrane polypeptides on thylakoid-bound ribosomes during the cell cycle of Chlamydomonas reinhardtii. . Biochim Biophys Acta. 1981;655:136–145. [Google Scholar]
- Hinnebusch, A.G., and S.W. Liebman. 1991. Protein synthesis and translational control in Saccharomyces. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics, Vol. 1. J.R. Broach, E.W. Jones and J.R. Pringle, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 627–735.
- Hoober JK, Boyd CO, Paavola L. Origin of thylakoid membranes in Chlamydomonas reinhardtii y-1at 38°C. Plant Physiol. 1991;96:1321–1328. doi: 10.1104/pp.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu-Ching C, Stern DB. Specific binding of chloroplast proteins in vitro to the 3′ untranslated region of spinach chloroplast petD mRNA. Mol Cell Biol. 1991;11:4380–4388. doi: 10.1128/mcb.11.9.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, D'Harlingue A, Kuntz M, Camara B. Identification of a plastid protein involved in vesicle fusion and/ or membrane protein translocation. Proc Natl Acad Sci USA. 1995;92:5630–5634. doi: 10.1073/pnas.92.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J, Block MA, Douce R. Molecular aspects of plastid envelope biochemistry. Eur J Biochem. 1991;199:489–509. doi: 10.1111/j.1432-1033.1991.tb16148.x. [DOI] [PubMed] [Google Scholar]
- Michaelis U, Körte A, Rödel G. Association of cytochrome btranslational activator proteins with the mitochondrial membrane: implications for cytochrome b expression in yeast. Mol Gen Genet. 1991;230:177–185. doi: 10.1007/BF00290666. [DOI] [PubMed] [Google Scholar]
- McMulin TW, Fox TD. COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. . J Biol Chem. 1993;268:11737–11741. [PubMed] [Google Scholar]
- Morré, D.J., Selldén, G., C. Sundqvist, and A.S. Sandelius. 1991. Stromal low temperature compartment derived from the inner membrane of the chloroplast envelope. Plant Physiol. 97:1558–1564. [DOI] [PMC free article] [PubMed]
- Nickelsen J, van Dillewijn J, Rahire M, Rochaix J-D. Determinants for stability of the chloroplast psbD mRNA are located within its short leader region in Chlamydomonas reinhardtii. . EMBO (Eur Mol Biol Organ) J. 1994;13:3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying the concentration of chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- Rawyler A, Meylan-Bettex M, Siegenthaler P-A. (Galacto)lipid export from envelope to thylakoid membranes in intact chloroplasts. II. A general process with a key role for the envelope in the establishment of lipid asymmetry in thylakoid membranes. Biochim Biophys Acta. 1995;1233:123–133. doi: 10.1016/0005-2736(94)00248-n. [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C. The regulation of enzymes involved in chlorophyll biosynthesis. Eur J Biochem. 1996;237:323–343. doi: 10.1111/j.1432-1033.1996.00323.x. [DOI] [PubMed] [Google Scholar]
- Rochaix J-D. Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. . Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- Ryu EK, MacCoss M. Modification of the Dittmer-Lester reagent for the detection of phospholipid derivatives on thin-layer chromatograms. J Lipid Res. 1979;20:561–563. [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Second edition. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 18.60–18.74.
- Sato N, Albrieux C, Joyard J, Douce R, Kuroiwa T. Detection and characterization of plastid envelope DNA-binding protein which may anchor plastid nucleoids. EMBO (Eur Mol Biol Organ) J. 1993;12:555–561. doi: 10.1002/j.1460-2075.1993.tb05687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Wersel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Wiesenberger G, Fox TD. Pet127p, a membrane-associated protein involved in stability and processing of Saccharomyces cerevisiaemitochondrial RNAs. Mol Cell Biol. 1997;17:2816–2824. doi: 10.1128/mcb.17.5.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CB, Cohen A, Danon A, Mayfield SP. Altered mRNA binding activity and decreased translation initiation in a nuclear mutant lacking translation of the chloroplast psbAmRNA. Mol Cell Biol. 1996;16:3560–3566. doi: 10.1128/mcb.16.7.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W, Rochaix J-D. The 5′ leader of a chloroplast mRNA mediates the translational requirements for two nucleus-encoded functions in Chlamydomonas reinhardtii. . Mol Cell Biol. 1994;14:5268–5277. doi: 10.1128/mcb.14.8.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]