Abstract
Alcohol oxidase (AOX), the first enzyme in the yeast methanol utilization pathway is a homooctameric peroxisomal matrix protein. In peroxisome biogenesis-defective (pex) mutants of the yeast Pichia pastoris, AOX fails to assemble into active octamers and instead forms inactive cytoplasmic aggregates. The apparent inability of AOX to assemble in the cytoplasm contrasts with other peroxisomal proteins that are able to oligomerize before import. To further investigate the import of AOX, we first identified its peroxisomal targeting signal (PTS). We found that sequences essential for targeting AOX are primarily located within the four COOH-terminal amino acids of the protein leucine-alanine-arginine-phenylalanine COOH (LARF). To examine whether AOX can oligomerize before import, we coexpressed AOX without its PTS along with wild-type AOX and determined whether the mutant AOX could be coimported into peroxisomes. To identify the mutant form of AOX, the COOH-terminal LARF sequence of the protein was replaced with a hemagglutinin epitope tag (AOX–HA). Coexpression of AOX–HA with wild-type AOX (AOX-WT) did not result in an increase in the proportion of AOX–HA present in octameric active AOX, suggesting that newly synthesized AOX–HA cannot oligomerize with AOX-WT in the cytoplasm. Thus, AOX cannot initiate oligomerization in the cytoplasm, but must first be targeted to the organelle before assembly begins.
Alcohol oxidase (AOX)1 is a homooctameric flavoprotein consisting of eight identical subunits of ∼74 kD, each containing a flavin adenine dinucleotide molecule (FAD) as a prosthetic group (van der Klei et al., 1991). The protein catalyzes the oxidation of methanol to formaldehyde and hydrogen peroxide, the first step in the methanol utilization pathway of certain yeasts including Pichia pastoris, Hansenula polymorpha, and Candida boidinii (van der Klei et al., 1991). AOX is normally localized in the matrix of single membrane-bound organelles called peroxisomes. During methanol growth, the peroxisomes also contain large amounts of dihydroxyacetone synthase, the first enzyme in the methanol assimilatory pathway, and catalase (CAT), which converts the hydrogen peroxide generated by oxidases such as AOX into water and oxygen (Veenhuis and Harder, 1991). As a result, peroxisomes, which are small and few in number in glucose-grown cells, are massively induced in methanol-grown cells (Veenhuis and Harder, 1991). In previous studies, we have shown that functional peroxisomes are essential for growth of P. pastoris and H. polymorpha on methanol, but not on glucose, and have exploited this observation in the isolation of numerous mutants that are defective in the biogenesis/assembly of peroxisomes (pex mutants) (Cregg et al., 1990; Gould et al., 1992; Liu et al., 1992; Waterham et al., 1992; Tan et al., 1995a ).
Genes encoding AOX have been cloned from P. pastoris (AOX1 and AOX2; Ellis et al., 1985; Cregg et al., 1989; Koutz et al., 1989), H. polymorpha (MOX; Ledeboer et al., 1985), and C. boidinii (AOD1; Sakai and Tani, 1992). The predicted primary sequences of their products are 73– 85% identical. In all of these yeast species, the transcription of genes encoding AOX is highly repressed during growth on glucose or ethanol, and maximally induced during growth on methanol (Tschopp et al., 1987). Like other peroxisomal proteins, AOX is translated on free cytosolic ribosomes and posttranslationally imported into the peroxisomal matrix (Roa and Blobel, 1983). The four COOH-terminal amino acids of H. polymorpha AOX are capable of targeting a nonperoxisomal reporter protein to peroxisomes, suggesting that AOX is targeted by alanine-arginine- phenylalanineCOOH (ARF), an uncommon variant of the type 1 peroxisomal targeting signal (PTS1) motif (Hansen et al., 1992; Subramani, 1993). The typical PTS1 motif is a tripeptide of the sequence serine-lysine-leucineCOOH (SKL; and conserved variants) found at the extreme COOH terminus of many matrix proteins (Gould et al., 1989, 1990; Swinkels et al., 1992; Subramani, 1993). The motif is specifically recognized by a PTS1 receptor protein, Pex5p, as an early step in the peroxisomal protein import process (McCollum et al., 1993; Terlecky et al., 1995). Pex5p is thought to deliver PTS1-containing polypeptides to the surface of the peroxisome and then cycle back to the cytoplasm for further rounds of PTS1 protein binding and targeting (Dodt and Gould, 1996; Waterham and Cregg, 1997).
Recent observations indicate that the peroxisomal protein import mechanism may differ significantly from those known for other organelles (McNew and Goodman, 1996). In particular, newly synthesized peroxisomal proteins need not be in an extended monomeric conformation to be imported, but can assemble/oligomerize in the cytoplasm before import. For example, Saccharomyces cerevisiae thiolase, without its NH2-terminal PTS2 motif, is not imported into peroxisomes (Glover et al., 1994). However, if coexpressed with wild-type thiolase, it is efficiently imported. The interpretation of this result is that thiolase, a homodimer, must be capable of oligomerizing before import, and that PTS2-less thiolase monomers are capable of dimerizing with wild-type monomers in the cytoplasm, and being coimported (piggybacked) into the organelle. Similar results have been obtained with three PTS1-targeted proteins: a chloramphenicol acetyl transferase PTS1 fusion protein (CAT–PTS1; McNew and Goodman, 1994), human alanine/glyoxylate amino transferase 1 (AGT; Leiper et al., 1996), and S. cerevisiae malate dehydrogenase 3 (MDH3; Elgersma et al., 1996). Further evidence supporting the notion that peroxisomes can import preassembled structures has been provided from microinjection studies of human cell lines that show that colloidal gold particles ⩽9 nm and coated with a human serum albumin-PTS1 conjugate can be imported (Walton et al., 1995).
To investigate whether oligomeric import is a general characteristic of peroxisomal matrix protein import, we have examined the targeting, import, and assembly of P. pastoris AOX. Our results demonstrate that AOX is targeted by a COOH-terminal PTS1 motif and that assembled and active AOX is only found inside peroxisomes. In addition, we show that the efficiency of import and assembly of AOX without its PTS1 is not enhanced by coexpression with wild-type AOX. Thus, unlike other peroxisomal proteins, AOX cannot initiate assembly in the cytoplasm.
Materials and Methods
Strains, Media, and Microbial Techniques
Yeast strains used in this study are listed in Table I. Shake-flask cultures were incubated for 10–15 h at 30° (P. pastoris strains) or 37°C (H. polymorpha strains) in selective minimal YND or YNM medium (0.17% [wt/ vol] yeast nitrogen base without amino acids {Difco Laboratories Inc., Detroit, MI} supplemented with 0.5% [wt/vol] glucose [dextrose] or 0.5% [vol/vol] methanol). For growth of auxotrophic strains, amino acids were added to a final concentration of 50 μg/ml. Transformations of P. pastoris (Becker and Guarente, 1991) or H. polymorpha (Faber et al., 1994) were performed by electrotransformation. Cultivation of Escherichia coli strain DH5α and standard recombinant DNA techniques were performed essentially as described (Sambrook et al., 1989).
Table I.
Yeast Strains
| Strain | Genotype | Source or reference | ||
|---|---|---|---|---|
| P. pastoris | ||||
| Wild type | NRRL Y-11430 | |||
| GS115 | his4 | Cregg et al., 1985 | ||
| GS-HWOxx | GS115 transformed with plasmid pHWOxx. | This study | ||
| MC100-3 (aoxΔ) | aox1Δ::ScARG4 aox2Δ::Pphis4 arg4 his4 | Cregg et al., 1989 | ||
| MC-HWOxx | MC100-3 transformed with plasmid pHWOxx. | This study | ||
| MC200 (pex2Δ aoxΔ) | pex2Δ::ScHIS4 aox1Δ::ScARG4 aox2Δ::Pphis4 arg4 his4 | This study | ||
| MC200-MOX | aox1Δ::ScARG4 aox2Δ::Pphis4 pex2Δ::ScHIS4 arg4 his4 (pHWO56) | This study | ||
| JC116 (pex2-1) | pex2-1 his4 (formerly per6-1) | Waterham et al., 1996 | ||
| JC117 (pex2Δ) | pex2Δ::PpARG4 arg4 his4 | This study | ||
| JC120 (pex8-1) | pex8-1 his4 (formerly per3-1) | Liu et al., 1995 | ||
| JC125 (pex8Δ) | pex8Δ::ScARG4 arg4 his4 (formerly per3Δ) | Liu et al., 1995 | ||
| pex5Δ | pex5Δ::PpARG4 arg4 his4 (formerly pas8Δ) | McCollum et al., 1993 | ||
| pex5Δ-HWOxx | pex5Δ strain transformed with plasmid pHWOxx. | This study | ||
| H. polymorpha | ||||
| A16 | leu1.1 | Veale et al., 1992 | ||
| CT100 (pex10) | pex10-1 leu1.1 (formerly per8-1) | Tan et al., 1995a | ||
| CW110 (pex10 moxΔ) | pex10-1 moxΔ::ZEO leu 1.1 | This study | ||
| CW111 (pex10-1 moxΔ pex10-1 moxΔ [pHWO57] AOX) | This study | |||
| S. cerevisiae | ||||
| SFY526 | MATa, ura3-52, his3-200, ade2-101, lys2-801, | Bartel et al., 1993 | ||
| trp 1-901, leu2-3, 112, canr, gal4-542, gal80-538, | ||||
| URA3::GAL1-lacZ |
Construction of β-Lactamase Fusion Vector Strains
Plasmids encoding chimeric proteins composed of selected AOX amino acid sequences fused to a modified E. coli β-lactamase protein were constructed. The modified β-lactamase was composed of amino acid residues H24–W286 (Sutcliffe, 1978) preceded by the amino acids methione-serine-glycine (MSG) as previously described (Waterham et al., 1994, 1997). DNA sequences encoding the COOH termini of wild-type AOX (AOX-WT), AOX–RSC, and AOX–HA (see below) were ligated in reading frame to the 3′ end of the bla gene using either the StuI site located at 1,924 bp or the AgeI site (made blunt ended with the Klenow fragment of DNA polymerase I) located at 1,972-bp downstream of the AOX1 start codon (Koutz et al., 1989). The proper DNA sequence at each fusion junction was verified by DNA sequencing. The primary sequence of each fusion protein is shown in Table II. All β-lactamase fusion proteins were expressed under transcriptional control of the constitutive GAP promoter (PGAP) from the P. pastoris GAP gene (Waterham et al., 1997) in vector pHWO10K. This vector was created from the HIS4-based vector pHWO10 by replacing the ampicillin-resistance gene with the kanamycine-resistance gene. The vectors were integrated into the genomic HIS4 locus of P. pastoris strains GS115 or pex5Δ (Table I) after linearization with SalI, a unique site in the HIS4 gene of each vector.
Table II.
Plasmids
| Plasmid | Relevant properties | Reference | ||
|---|---|---|---|---|
| P. pastoris | ||||
| pHIL-A1 | Apr, PpHIS4, PAOX1,tAOX1 | Waterham et al., 1997 | ||
| pHWO40 | Apr, PpHIS4, PAOX1-AOX-WT (M1-F663)-tAOX1 | This study | ||
| PHWO41 | Apr, PpHIS4, PAOX1-AOX-RSC (M1-G659RSC)-tAOX1 | This study | ||
| pHWO42 | Apr, PpHIS4, PAOX1-AOX-HA (M1-G659YPYDVPDYAG)-tAOX1 | This study | ||
| pHWO43 | Apr, PpHIS4, PAOX1-AOX-Δ22 (M1 -A642)-tAOX1 | This study | ||
| pHWO10 | Apr, PpHIS4, PGAP, tAOX1 | Waterham et al., 1997 | ||
| pHWO10K | Kmr, PpHIS4, PGAP, tAOX1 | This study | ||
| pHWO19 | Kmr, PpHIS4, PGAP-β-lac (MSGH24-W286)-tAOX1 | Waterham et al., 1997 | ||
| pHWO44 | Kmr, PpHIS4, PGAP-β-lac-AOX642-663 (MSGH24-W286SRV-A642-F663)-tAOX1 | This study | ||
| pHWO45 | Kmr, PpHIS4, PGAP-β-lac-AOX660-663 (MSGH24-W286SRV-G659-F663)-tAOX1 | This study | ||
| pHWO46 | Kmr, PpHIS4, PGAP-β-lac-AOX642-659-RSC (MSGH24-W286SRV-A642-G659RSC)-tAOX1 | This study | ||
| pHWO47 | Kmr, PpHIS4, PGAP-β-lac-AOX642-659-HA (MSGH24-W286SRV-A642-G659YPYDVPDYAG)-tAOX1 | This study | ||
| pUZ12 | Apr, pex2Δ::ScHIS4 | Waterham et al., 1996 | ||
| pPICZ-B | ZEO, PAOX1, tAOX1 | Invitrogen | ||
| pHWO55 | ZEO, PAOX1, PMOX-MOX, tAOX1 | This study | ||
| pHWO56 | Apr, PpHIS4, PAOX1, PMOX-MOX, tAOX1 | This study | ||
| S. cerevisiae | ||||
| pGBT9 | Apr, ScTRP1, PADH1-GAL4B | Clontech | ||
| pHWO48 | Apr, ScTRP1, PADH1-GAL4B-AOX642-663 (E881IEFPA642-F663) | This study | ||
| pHWO49 | Apr, ScTRP1, PADH1-GAL4B-AOX642-663-RSC (E881IEFPA642-G659RSC) | This study | ||
| pHWO50 | Apr, ScTRP1, PADH1-GAL4B-AOX642-659-HA (E881IEFPA642-G659YPYDVPDYAG) | This study | ||
| pGAD424 | Apr, ScLEU2, PADH1-GAL4A | Clontech | ||
| pHWO51 | Apr, ScLEU2, PADH1-GAL4A-PEX5 (S147PEFPGIRG13-F576) | This study | ||
| pUT352 | Apr, ScURA3, ZEO | Wenzel et al., 1992 | ||
| H. polymorpha | ||||
| pMOX-BS | Apr, pUC18 + BamHI–SacI MOX fragment | Distel et al., 1987 | ||
| pET1 | Apr, ScLEU2, PMOX | Tan et al., 1995b | ||
| pHWO52 | Apr, moxΔ::ZEO | This study | ||
| pHWO57 | Apr, PpHIS4, ScLEU2, PAOX1-AOX-tAOX1 | This study | ||
| pHWO58 | Apr, ScLEU2, PMOX-AOX-tAOX1 | This study |
Bold, underline, and capital; PpAOX1 amino acid sequence numbered according to Koutz et al. (1989). Bold, italic, and capital; β-lactamase amino acid sequence numbered according to Sutcliffe (1978). Bold and capital; Pex5p amino acid sequence numbered according to McCollum et al. (1993).
Construction of Mutant AOX Expression Strains
The P. pastoris AOX1 gene was amplified from plasmid pPG5.4 (Cregg et al., 1989) by the PCR using a forward primer, composed of the first 18 bp of the AOX1 open reading frame preceded by an EcoRI site, and a reverse primer, composed of 19 bp located 247–266-bp downstream of the AOX1 stop codon which includes a genomic HindIII site (Koutz et al., 1989). After digestion with EcoRI and HindIII, the AOX1 gene was first subcloned in EcoRI–HindIII–digested pBS-SK (Stratagene, La Jolla, CA), and subsequently as an EcoRI–ClaI fragment under transcriptional control of the P. pastoris AOX1 promoter (PAOX1) into EcoRI- and ClaI-digested vector pHIL-AI (Invitrogen, San Diego, CA; Waterham et al., 1997). The resulting vector was named pHWO40. To modify sequences encoding the COOH terminus of AOX-WT, vector pHWO40 was digested with AgeI (located 1,972-bp downstream of the AOX1 start codon) and then the resulting termini were made blunt with Klenow fragment of DNA polymerase I and religated. This resulted in a frame shift changing the COOH-terminal residues of AOX-WT leucine-alanine-arginine-phenyl-alanineCOOH (LARF) to arginine-serine-cysteineCOOH (RSC). The resulting vector that expressed AOX–RSC was named pHWO41. A second vector encoding a COOH-terminal modified AOX was created by inserting two complementary oligonucleotides, which changed the four COOH-terminal residues from LARF to that for the human influenza virus epitope (HA) tag at the AgeI site of pHWO40. These residues are recognized by the mouse monoclonal antibody 12CA5 (Boehringer Mannheim Biochemicals, Indianapolis, IN). The resulting modified AOX protein is called AOX–HA (vector pHWO42). Finally, a vector was created that encodes a modified AOX (AOX-Δ22) in which the last 22 amino acids of the protein are deleted. This vector (pHWO43) was generated by inserting a blunt AgeI adaptor encoding an in-frame stop codon between the StuI and AgeI sites of AOX1 in vector pHWO40. The primary sequences of the COOH termini of the modified AOX proteins are shown in Table II. All constructs were verified by DNA sequencing. The constructs were integrated into the genomic HIS4 locus of P. pastoris strains MC100-3, GS115, and pex5Δ (Table I) by linearization at the unique EcoNI site in the HIS4 gene of each vector.
Two-hybrid System Experiments
Interactions between the P. pastoris PTS1 receptor protein Pex5p and selected sequences from AOX were studied with the yeast two-hybrid system (Matchmaker; Clontech Laboratories, Inc., Palo Alto, CA). The P. pastoris PEX5 open reading frame was released from pSP72 (a gift from S. Subramani, University of California at San Diego, San Diego, CA) by digestion at a SacII site located 42-bp downstream of the PEX5 start codon, treated with mung bean exonuclease, and then digested at a PstI site located downstream of the PEX5 stop codon. This fragment was ligated into SmaI–PstI–digested pGAD424 which resulted in the expression of Pex5p fused in reading frame with the GAL4 activation domain (pHWO51). The COOH termini of AOX-WT, AOX–RSC, and AOX– HA were released from vectors pHWO40, pHWO41, and pHWO42, respectively, by digestion with StuI (located 1,924-bp downstream of the AOX1 start codon) and PstI (located downstream of the AOX1 stop codon). These fragments were then ligated into pGBT9 that had been cut with XmaI, filled with Klenow, and then digested with PstI. The resulting vector expressed the GAL4–DNA binding domain protein fused in-frame to each of the AOX-derived peptides described above. The COOH-terminal sequences of the resulting constructs named pHWO48 through pHWO50 are shown in Table II. All constructs were verified by DNA sequencing. These constructs and control vectors were transformed by electrotransformation into the S. cerevisiae reporter strain SFY526 (Becker and Guarente, 1991). β-Galactosidase activity in each strain was determined qualitatively by the filter assay method and quantitatively by the cell-free activity assay method described in the Clontech technical manual.
Expression of Modified AOX1 Genes in P. pastoris
AOX–HA was coexpressed with AOX-WT by inserting vector pHWO42 in GS115 (GS-HWO42), and expressed alone by insertion of the vector into the AOX1 and AOX2 deletion strain MC100-3 (MS-HWO42). Octamerization of AOX proteins was examined using a modified version of the velocity sedimentation method described by Goodman et al. (1984). Cells (50 OD600 U) were lysed by vortexing for 15 min at 4°C with 1 vol of glass beads in 0.3 ml TENT buffer (10 mM Tris-HCl, pH 8, 5 mM EDTA, 50 mM NaCl, 1% Triton X-100) and then centrifuged for 10 min at maximum speed in a microcentrifuge (Eppendorf model 5415C; Brinkman Instruments, Westbury, NY).
Supernatant samples of 0.1 ml were separated through 5–30% (wt/vol) sucrose gradients (6 layers of 1.5 ml sucrose in TENT) by centrifugation for 5 h at 40,000 rpm and 2°C in a rotor (SW41Ti; Beckman Instruments, Palo Alto, CA). Fractions of 0.5 ml were removed from the top and analyzed for sucrose density, AOX, and CAT activities, and for AOX-WT or AOX–HA protein by immunoblotting using polyclonal antibodies against AOX or monoclonal antibodies against the HA epitope.
Heterologous Expression of AOX1 and MOX Genes
The H. polymorpha MOX gene was expressed under transcriptional control of its own promoter by integration of KasI-linearized vector pHWO56 into the HIS4 locus of the P. pastoris AOX1 and AOX2 deletion strain MC100-3 and by integration of SacI-linearized vector pHWO55 into the PAOX1 locus of the P. pastoris pex2Δ aox1Δ aox2Δ strain MC200. The MC200 strain was generated by disruption of the PEX2 gene in MC100-3 using BamHI-digested pUZ12 (Waterham et al., 1996). The P. pastoris AOX1 gene was expressed under transcriptional control of its own promoter (pHWO57) and the H. polymorpha PMOX (pHWO58) in a H. polymorpha moxΔ pex10-1 strain. This strain was constructed by disruption of the MOX gene in the H. polymorpha pex10-1 mutant strain using AlwNI– SalI–digested pHWO52.
Indirect Immunofluorescence
P. pastoris cells (25 OD600 U) were washed twice with water and fixed in 1 ml of 40 mM KPO4 buffer, pH 6.5, with 3.7% (vol/vol) formaldehyde for 1 h at room temperature. After two washes with 1 ml MOPS+ (5 mM 3-[morpholino]propane-sulfonic acid, 10 mM Na2SO3, and 0.5 M KCl), fixed cells were converted into spheroplasts in 0.5 ml MOPS+ supplemented to 85 mM β-mercaptoethanol and 0.08 mg/ml Zymolyase-100T for 1 h at 30°C. The spheroplasts were washed twice with 1 ml MOPS+, incubated for 5 min at −20°C in 100% methanol, washed twice with 1 ml PBS+ (PBS, pH 7.2, 1 M sorbitol, and 1% bovine serum albumin), and finally suspended in 1 ml PBS+. Suspended cells (0.2 ml) were incubated for 2 h at room temperature with 0.5 μl of polyclonal antibodies raised against CAT or β-lactamase (5 Prime, 3 Prime, Boulder, CO). After three washes with 1 ml PBS+, cells were suspended in 0.2 ml PBS+ and incubated for 1 h at room temperature in the dark with 0.5 μl FITC-conjugated goat anti–rabbit antiserum (Boehringer Mannheim, Indianapolis, IN). After three washes with 1 ml PBS+, cells were suspended in 0.1 ml PBS+. For microscopic observation, small samples of the cell suspensions were mixed with equal volumes of 0.1 M n-propyl gallate in 90% glycerol. Cells were examined with a microscope equipped for indirect immunofluorescence (Leitz Laborlux S; Wild Leitz, Rockleigh, NJ) at 1,000× and photographed (T-MAX 400 film; Eastman Kodak Co., Rochester, NY).
Biochemical Methods
Subcellular fractionation of P. pastoris and H. polymorpha cells was performed as described previously (Waterham et al., 1996) except that H. polymorpha cells were converted to spheroplasts in the presence of 3 M sorbitol and homogenized in the presence of 2 M sorbitol. Cell-free extracts were made using a glass bead method (Waterham et al., 1997). Peroxisomal AOX and CAT, mitochondrial cytochrome c oxidase, cytosolic glyceraldehyde-3-phosphate dehydrogenase, and β-lactamase activities were assayed as described previously (Waterham et al., 1996, 1997). Protein concentrations were determined with the bicinchoninic acid protein assay kit (Pierce Chemical Co., Rockford, IL) using bovine serum albumin as a standard. The transfer of proteins to nitrocellulose after SDS-PAGE electrophoresis was performed using the Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad Laboratories, Hercules, CA) as directed by the manufacturer. Immunoblotting experiments were performed using the Western Light Kit (Tropix Inc., Bedford, MA) with specific polyclonal antibodies against AOX and monoclonal antibody 12CA5 against the hemagglutinin epitope (Boehringer Mannheim). Immunoprecipitations were performed as described in Rehling et al. (1996) except that protein A–Sepharose (Pharmacia Biotech. Inc., Piscataway, NJ) was used instead of DynaBeads. Native (nonreducing nondenaturing discontinuous) gels were electrophoresed at 200 v for 2 h at 4°C through 5% polyacrylamide using a Mini-Protean II apparatus (BioRad Laboratories) as described in Ausubel et al. (1996).
Nucleotide Sequence Accession Numbers
Sequence data for the P. pastoris AOX1 and AOX2 genes, as published by Koutz et al. (1989), have been submitted to the databases and are available from GenBank/EMBL/DDBJ under accession numbers U96967 (AOX1) and U96968 (AOX2).
Results
Active AOX Is Present Only in Peroxisomes of P. pastoris
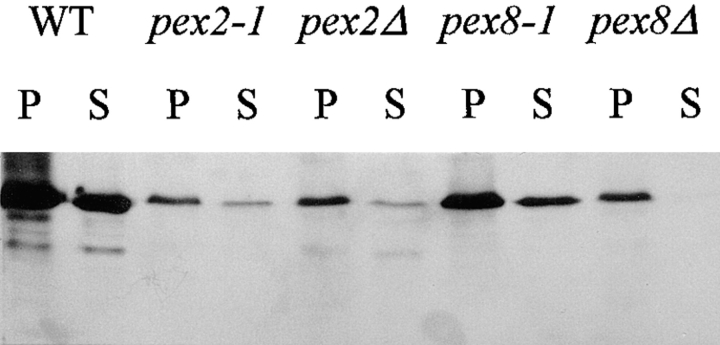
Our previous studies with P. pastoris pex mutants indicated that methanol-induced pex cells contain little or no AOX activity, although significant amounts of AOX protein are present in the cells (Liu et al., 1992, 1995; Waterham et al., 1996). The level of residual AOX activity in an individual pex mutant allele strongly correlated with the severity of the peroxisome-deficient phenotype of that strain. For example, in methanol-induced cells of pex2-1 and pex8-1 (two slightly leaky mutants generated by chemical mutagenesis), numerous small peroxisomal remnants were observed that retain the ability to import small amounts of peroxisomal enzymes (Liu et al., 1995; Waterham et al., 1996). In both of these mutants, small but significant amounts of active AOX were present and these appeared to be exclusively within the peroxisomal remnants as judged by subcellular fractionation studies (Table III). In contrast, methanol-induced cells of pex2 and pex8 deletion strains (pex2Δ and pex8Δ), which have a more severe peroxisome-deficient phenotype with few, if any, peroxisomal remnants and no measurable import ability, contained no active AOX, although AOX protein was present (Fig. 1). The inactive AOX protein was concentrated in cytoplasmic protein aggregates which sedimented with organelles during subcellular fractionation (Fig. 1; Liu et al., 1995; Waterham et al., 1996). These results indicate that active AOX exists only in the peroxisomal matrix and suggest that AOX precursors require the presence of functional peroxisomes to assemble into active octamers.
Table III.
Peroxisomal Enzyme Activities in Organelle Pellet and Cytosolic Supernatant Fractions of Methanol-induced P. pastoris pex strains
| Strain | Fraction* | AOX‡ | CAT§ | Cyt c ox.‡ | GAPDH‡ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | P | 0.35 | 33 | 1.9 | UD | |||||
| S | 0.33 | 32 | 0.0041 | 0.21 | ||||||
| %P§ | 51 | 51 | 99 | 0.0 | ||||||
| pex2-1 (JC116) | P | 0.0031 | 1.9 | 1.4 | 0.011 | |||||
| S | UD | 20 | 0.014 | 0.28 | ||||||
| %P | 100 | 8.7 | 99 | 3.8 | ||||||
| pex2Δ (JC117) | P | UD | 3.5 | 1.4 | 0.010 | |||||
| S | UD | 72 | UD | 0.32 | ||||||
| %P | — | 4.6 | 100 | 3.0 | ||||||
| pex8-1 (JC120) | P | 0.011 | 3.3 | 1.5 | 0.0050 | |||||
| S | 0.00070 | 45 | 0.0097 | 0.19 | ||||||
| %P | 94 | 6.8 | 99 | 2.6 | ||||||
| pex8Δ (JC125) | P | UD | 3.4 | 1.1 | 0.060 | |||||
| S | UD | 44 | UD | 0.32 | ||||||
| %P | — | 7.2 | 100 | 16 |
P and S are enzyme activities in organellar pellet and cytosolic supernatant fractions, respectively. %P is percentage of enzyme activity in the pellet fraction.
Activities are expressed as U/mg protein.
Activity is expressed as ΔE240/min per mg protein. UD indicates activity is undetectable.
Figure 1.
Location of AOX protein in P. pastoris wild-type (WT) and selected pex strains. Methanol-induced cells of each strain were spheroplasted, osmotically lysed, and subjected to differential centrifugation. Protein samples of 30 μg from the resulting organelle pellet (P) and cytosolic supernatant (S) fractions were subjected to SDS-PAGE and immunoblotting with anti-AOX antibodies.
AOX from H. polymorpha Is Active in the Cytoplasm of P. pastoris
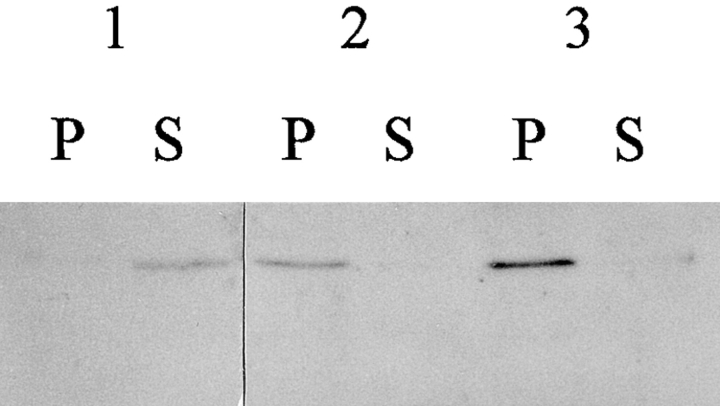
Unlike P. pastoris, AOX from H. polymorpha (MOX) efficiently assembles into an active octameric enzyme in the cytoplasm of H. polymorpha pex mutants (van der Klei et al., 1991). To determine whether this difference is a characteristic of the AOX proteins themselves or the cellular environment provided by their hosts, we constructed pex strains of each yeast species that expressed AOX/MOX from the heterologous species. For H. polymorpha, a vector containing the P. pastoris AOX1 gene was inserted into the genome of an H. polymorpha pex10 moxΔ strain. AOX activity could not be detected in the AOX-expressing pex10 moxΔ strain CW111 at either 30° or 37°C (Table IV). Immunoblot analysis performed on crude organelle pellet and cytosolic supernatant fractions from methanol-induced cells of this strain showed that AOX protein was synthesized and located primarily in pellet fractions (Fig. 2, group 2 and 3 lanes). The pellet location was most likely a consequence of AOX protein aggregation as seen in P. pastoris pex mutants (Fig. 1). Identical results were obtained when AOX1 was expressed under control of the H. polymorpha MOX promoter (data not shown). Thus, in contrast to endogenous H. polymorpha MOX, P. pastoris AOX is not assembled into active protein in the cytosol of an H. polymorpha pex mutant.
Table IV.
Peroxisomal Enzyme Activities in Organelle Pellet and Cytosolic Supernatant Fractions of Methanol-induced Strains
| Strain | Fraction* | AOX‡ | CAT§ | Cyt c ox‡ | GAPDH‡ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pp pex2 aoxΔ MOX | P | 0.0028 | 9.1 | 0.91 | 0.24 | |||||
| (MC200-MOX) | S | 0.050 | 52 | 0.0070 | 1.4 | |||||
| %P | 5.3 | 15 | 99 | 15 | ||||||
| Hp pex10 moxΔ AOX | P (37°C) | UD | 1.0 | 0.97 | 0.18 | |||||
| (CW111) | S (37°C) | UD | 17 | 0.018 | 6.4 | |||||
| %P | — | 5.6 | 98 | 2.7 | ||||||
| Hp pex10 moxΔ AOX | P (30°C) | UD | 1.4 | 1.1 | 0.20 | |||||
| (CW111) | S (30°C) | UD | 15 | 0.012 | 6.1 | |||||
| %P | — | 8.5 | 99 | 3.2 |
P and S are enzyme activities in organellar pellet and cytosolic supernatant fractions, respectively. %P is percentage of enzyme activity in the pellet fraction.
Activities are expressed as U/mg protein.
Activity is expressed as ΔE240/min per mg protein. UD indicates activity is undetectable.
Figure 2.
Immunoblot analysis of AOX in organelle pellet (P) and cytosolic supernatant (S) fractions of methanol-induced P. pastoris and H. polymorpha strains expressing heterologous AOX. Group 1 lanes contain 20-μg samples from P. pastoris strain MC200 (pex2Δ aox1Δ aox2Δ) expressing the H. polymorpha MOX gene. Groups 2 and 3 contain 20-μg samples from H. polymorpha strain CW111 (pex10-1 moxΔ) expressing the P. pastoris AOX1 gene at 37° (2) or 30°C (3).
The converse result was obtained when H. polymorpha MOX was expressed in a P. pastoris pex mutant. As expected, MOX was able to complement for methanol growth in a P. pastoris strain on MC100-3 that is deleted for both AOX genes (aox1Δ aox2Δ). When the PAOX-MOX vector was then inserted into the genome of a P. pastoris pex2Δ aox1Δ aox2Δ strain (MC200), methanol-induced cells of the transformed strain contained active MOX (Table IV). Upon subcellular fractionation, both MOX activity and protein were found in the cytosolic supernatant (Fig. 2, 1 lanes). The same result was obtained when MOX was expressed in a P. pastoris pex5Δ aox1Δ aox2Δ strain (data not shown). Since H. polymorpha MOX assembles into an active enzyme in the cytoplasm of either H. polymorpha or P. pastoris pex strains, the inability of P. pastoris AOX to properly assemble in these yeasts must be a characteristic of the protein itself and not of the environment provided by the P. pastoris cytoplasm.
A Peroxisomal Targeting Signal Is Located at the COOH Terminus of AOX
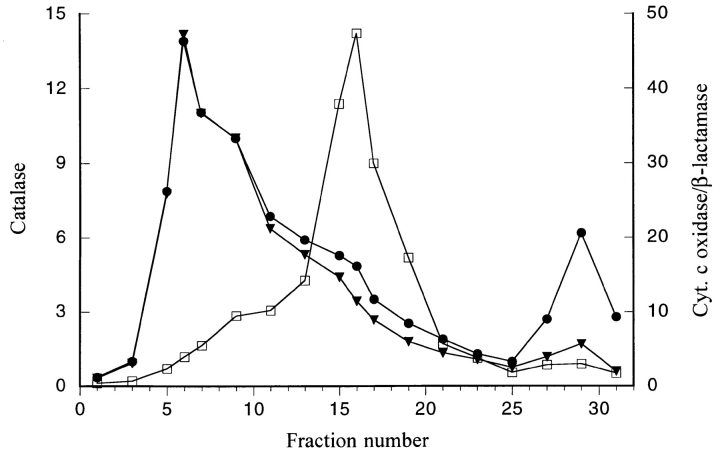
To further investigate the targeting and import of AOX, it was first necessary to define the AOX PTS. The three COOH-terminal amino acids of AOX, ARF, are similar in sequence to the PTS1 motif and, therefore, were a good candidate for the AOX PTS. Preliminary evidence in support of this was provided by Hansen et al. (1992) who showed that the last four amino acids of H. polymorpha MOX (LARF) are capable of targeting a nonperoxisomal protein to peroxisomes. To examine targeting of P. pastoris AOX, we first expressed a protein composed of the bacterial reporter enzyme β-lactamase fused to the four COOH-terminal amino acids of P. pastoris AOX (LARF) and determined whether this fusion protein (β-lac–LARF) was targeted to P. pastoris peroxisomes. Methanol-grown cells expressing β-lac–LARF were fractionated into an organelle pellet, consisting mainly of peroxisomes and mitochondria, and a cytosolic supernatant (Fig. 3 B). Biochemical analysis of these fractions revealed that a significant portion of β-lactamase activity colocalized with CAT activity in the organelle pellet fraction, suggesting that β-lac– LARF was targeted to peroxisomes. (Typically, a significant amount of peroxisomal matrix protein is also found in the supernatant fraction due to breakage of these fragile organelles.) Peroxisomal targeting of β-lac–LARF was confirmed by indirect immunofluorescence using β-lactamase– and CAT-specific antibodies (Fig. 3 B) and by further fractionation of the organelle pellet through sucrose density gradients (Fig. 4). In these gradients, β-lac–LARF was present at the same density as CAT and not with mitochondrial cytochrome c oxidase. As a control, β-lactamase without LARF expressed in P. pastoris fractionated to the cytosolic supernatant along with the cytoplasmic marker enzyme glyceraldehyde-3-phosphate dehydrogenase and appeared to be cytoplasmic in immunofluorescence assays (Fig. 3 A). We concluded a PTS is located within the LARF sequence.
Figure 3.
Location of β-lactamase fusion proteins in P. pastoris as determined by immunofluorescence and subcellular fractionation. The photomicrographs contain cells of the strains listed in the left column processed for immunofluorescence using anti-catalase (anti-CAT) and anti–β-lactamase (anti-βLAC) antibodies. The right column shows histograms of the percentage of activity for selected marker enzymes present in crude organelle pellet fractions from the same strains. Cat, Catalase; Lac, β-lactamase; Cyt c ox, Cytochrome c oxidase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Figure 4.
Sucrose density gradient of organelle pellet from a methanol-grown P. pastoris strain expressing β-lactamase–LARF. Catalase activity (•) is presented as ΔE240 U/ml; cytochrome c oxidase activity (□) as U/ml × 10−2, and β-lactamase activity (▾) as U/ml × 10−3.
LARF Is Necessary for Efficient Targeting of AOX
We next investigated the necessity of LARF for targeting and import of AOX. In one set of experiments, the 22 COOH-terminal amino acids of AOX were fused to the COOH terminus of β-lactamase (β-lac–AOX). As expected, this fusion protein was efficiently targeted to peroxisomes of P. pastoris (Fig. 3 D). We then deleted the four COOH-terminal residues from this construct and replaced them with either the non-PTS amino acid sequence RSC (β-lac–AOX–RSC) or the hemagglutinin–epitope-tag sequence (β-lac–AOX–HA). When expressed in methanol-grown P. pastoris cells, both of these chimeric proteins were localized to the cytoplasm as judged by indirect immunofluorescence and subcellular fractionation assays (Fig. 3, E and F).
The necessity of LARF for AOX targeting was further investigated by expressing mutant versions of AOX in which the tetrapeptide sequence had been removed and replaced with either RSC (AOX–RSC) or the HA tag (AOX–HA). Each mutant protein was expressed under control of the P. pastoris AOX1 promoter in a P. pastoris aox1Δ aox2Δ strain. Both the AOX–RSC and AOX–HA constructs partially complemented the strain for growth on methanol (18 and 22 h/generation, respectively, versus 3.5 h/generation for wild type), and both contained low but significant levels of AOX activity (16 and 3% of wild type; Table V). Subcellular fractionation studies of the AOX–RSC and AOX–HA expression strains showed that the majority of AOX activity was in the organelle pellet (Table V), suggesting that the active portions of the mutant AOXs were properly targeted to peroxisomes. Further fractionation of the organelle pellet from the AOX–HA strain through a sucrose density gradient confirmed that the AOX activity was peroxisomal (Fig. 5 B). Thus, a portion of both AOX–RSC and AOX–HA synthesized in P. pastoris cells is properly imported into peroxisomes where it assembles into an active enzyme.
Table V.
Peroxisomal Enzyme Activities in Organelle Pellet and Cytosolic Supernatant Fractions of Methanol-induced P. pastoris Strains Expressing Mutant AOX1 Genes
| Strain | t 1/2 | Fraction* | AOX‡ | CAT§ | Cyt c ox‡ | GAPDH‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AOX-WT | 3.5 | P | 0.54 | 74 | 5.3 | 0.077 | ||||||
| (MC-HWO40) | S | 0.38 | 65 | 0.51 | 2.3 | |||||||
| %P | 59 | 53 | 99 | 3.2 | ||||||||
| AOXΔ22 | >100 | P | UD | 56 | 0.59 | 0.067 | ||||||
| (MC-HWO43) | S | UD | 19 | 0.0052 | 2.0 | |||||||
| %P | — | 86 | 99 | 3.2 | ||||||||
| AOX–RSC | 18 | P | 0.56 | 71 | 4.5 | 0.36 | ||||||
| (MC-HWO41) | S | 0.025 | 29 | 0.055 | 2.4 | |||||||
| %P | 69 | 71 | 99 | 13 | ||||||||
| AOX–HA | 22 | P | 0.010 | 47 | 7.0 | 0.60 | ||||||
| (MC-HWO42) | S | UD | 20 | 0.043 | 4.9 | |||||||
| %P | 100 | 70 | 99 | 11 | ||||||||
| AOX+AOX–HA | 4.3 | P | 0.38 | 94 | 7.5 | 0.083 | ||||||
| (GS-HWO42) | S | 0.28 | 74 | 0.030 | 1.5 | |||||||
| %P | 58 | 56 | 99 | 5.2 |
P and S are enzyme activities in organellar pellet and cytosolic supernatant fractions, respectively. %P is percentage of enzyme activity in the pellet fraction.
Activities are expressed as U/mg protein.
Activity is expressed as ΔE240/min per mg protein. UD indicates activity is undetectable.
Figure 5.
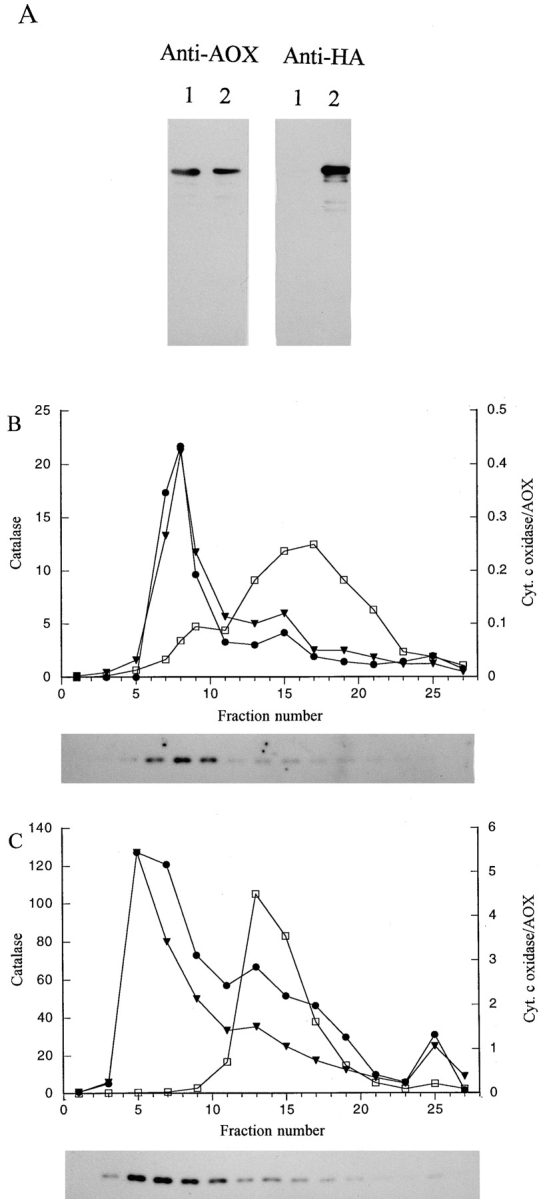
Sucrose density gradient analysis of P. pastoris strains expressing AOX–HA. (A) Total cell extracts (10 μg/lane) prepared from a strain expressing only AOX-WT (lane 1) or AOX-HA (lane 2) and immunoblotted with antibodies against AOX and the HA tag. Sucrose density gradient profiles of strain MC-HWO42 expressing only AOX–HA (B) or strain GS–HWO42 coexpressing both AOX–HA and AOX-WT (C). Symbols in the gradients are: AOX (•), catalase (▾), and cyt c oxidase (□). In B, catalase activities are presented as ΔE240 U/ml, cytochrome c oxidase activities as U/ml, and alcohol oxidase activities as U/ml × 10−2. In C, catalase activities are presented as ΔE240 U/ml, cytochrome c oxidase activities as U/ml, and alcohol oxidase activities as U/ml × 10−1. The immunoblots shown below each graph contained either 5 (B) or 1 μl (C) of each indicated fraction and were reacted with anti-HA monoclonal antibodies.
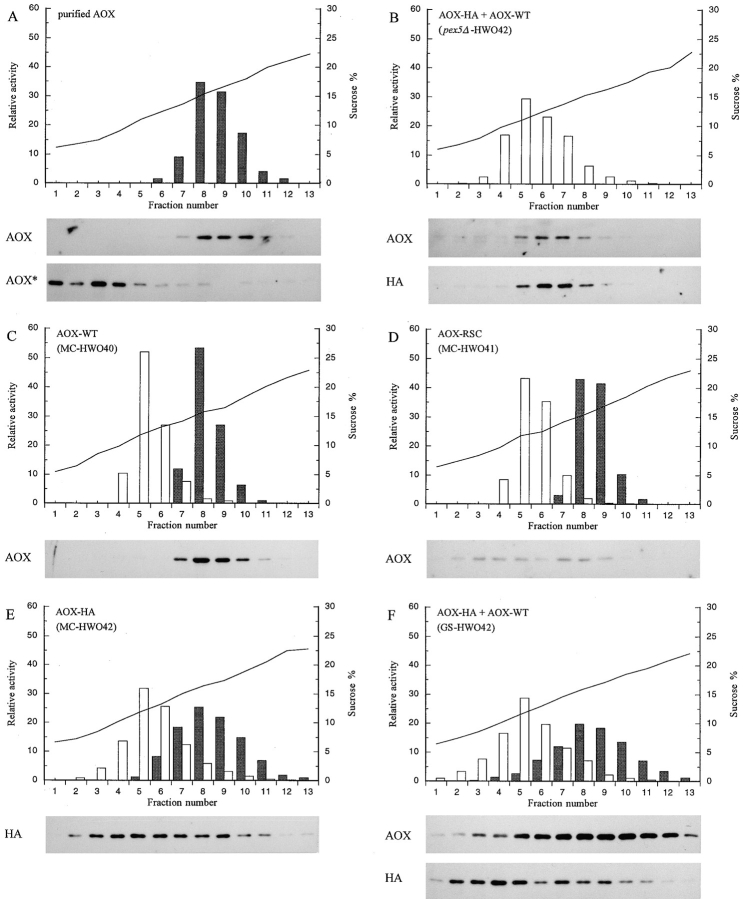
To estimate the proportion of total AOX–RSC and AOX–HA protein that assembled into an active enzyme, total cell extracts from methanol-induced cells of the AOX–RSC and AOX–HA-expressing strains were prepared by mechanical disruption and subjected to sucrose velocity gradient sedimentation (Goodman et al., 1984). This glass bead-based procedure was chosen because it avoids the long incubation period needed to prepare yeast protoplasts for subcellular fractionation, during which unstable inactive AOX protein is rapidly degraded relative to stable active AOX octamers. As a marker for the velocity gradients, we assayed activity for CAT, a tetrameric protein of ∼300 kD. In a control gradient with total lysates prepared from strains expressing AOX-WT (Fig. 6 C), AOX activity and protein comigrated at one position through the velocity gradient, the same position as purified octameric AOX with a molecular mass of ∼600 kD (Fig. 6 A). Gradients prepared from cells of strains expressing either AOX–RSC or AOX–HA alone contained small amounts of AOX activity at the normal octameric AOX position (Fig. 6, D and E). However, most of the AOX protein was spread over fractions representing lower molecular masses. Control gradients containing chemically denatured (mainly monomeric) AOX (Fig. 6 A, AOX*) or extracts prepared from a pex5Δ mutant strain expressing AOX–HA (Fig. 6 B) indicated that the inactive AOX proteins at these lower mass positions were most likely monomeric and aggregated forms of AOX.
Figure 6.
Sucrose velocity gradient analysis of AOX–HA expressing P. pastoris strains. Histograms show activity in fractions for tetrameric catalase (∼300 kD; white bars) and octameric AOX (∼600 kD; dark bars) as a percentage of the total activity in the gradient for each enzyme. Immunoblots containing 10-μl samples of gradient fractions were reacted with either anti-AOX or anti-HA antibodies. (A) shows purified active AOX and denatured AOX (AOX*). (B) shows extract prepared from a pex5Δ strain. C–F gradient results from strains expressing the following forms of AOX: C, AOX-WT; D, AOX–RSC; E, AOX–HA; F, both AOX–HA and AOX-WT. Fractions 14–19 from each gradient are not shown since they did not contain significant amounts of protein.
We concluded from these studies that a PTS sufficient for targeting proteins to peroxisomes is present within LARF (most likely ARF), and that this PTS is essential for the efficient targeting and import of AOX. However, since small but significant amounts of AOX are imported in the absence of LARF, a second less efficient PTS exists in another part of the protein. This second PTS may be located within the next 18 COOH-terminal amino acids of AOX, since a mutant protein in which the 22 COOH-terminal amino acid residues were deleted was completely inactive (Table V).
The COOH Terminus of AOX Interacts with the PTS1 Receptor
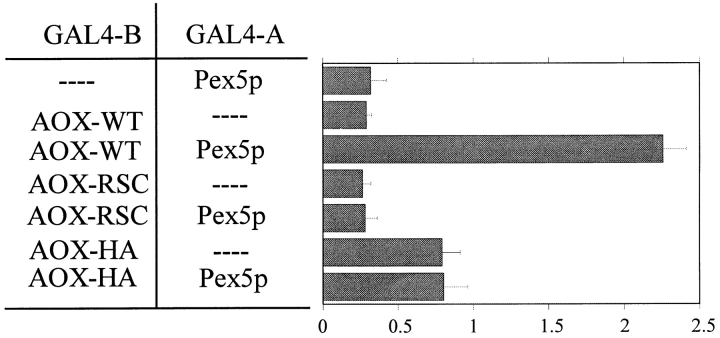
The similarity of ARF to the consensus PTS1 motif, SKL, suggests that AOX may be imported via the PTS1 pathway. Furthermore, import of AOX or β-lac–AOX is blocked in a pex5Δ mutant which is specifically defective in the PTS1 receptor Pex5p (Fig. 3 C; McCollum et al., 1993). Additional evidence that AOX is targeted to peroxisomes by the PTS1 pathway was obtained in yeast two- hybrid system assays in which the 22 COOH-terminal amino acids of AOX were expressed as a fusion with the GAL4 DNA-binding domain (GAL4B–AOX) in combination with the P. pastoris PTS1 receptor protein Pex5p fused to the GAL4 activation domain (GAL4A–Pex5p). This combination produced a strong response in the system (Fig. 7). The interaction was specific since expression of GAL4B–AOX with unfused GAL4A or GAL4A–Pex5p with unfused GAL4B produced little response. We then tested GAL4B–AOX variants in which the COOH-terminal LARF sequence was replaced by either RSC or the HA tag. Neither of these constructs produced a specific response in combination with GAL4A–Pex5p.
Figure 7.
Histogram of two hybrid system assays between Pex5p and the COOH-terminal 22 amino acids of AOX (AOX-WT). Data shown are the mean of three independent experiments. Units indicate β-galactosidase activity expressed as U/mg protein.
The Import Efficiency of AOX–HA Is Not Improved by Coexpression with AOX-WT
We examined whether the import efficiency of AOX without its PTS1 motif was increased when coexpressed with AOX-WT. To distinguish between AOX without its PTS1 motif and AOX-WT, we used the AOX–HA construct in which the four COOH-terminal amino acids had been replaced with the HA-epitope tag. Immunoblot analysis of total extracts prepared from P. pastoris strains expressing either AOX-WT or AOX–HA alone demonstrated that the anti-HA monoclonal antibodies exclusively recognized the AOX–HA protein, whereas polyclonal antibodies against AOX protein recognized both AOX-WT and AOX–HA (Fig. 5 A). To estimate the proportion of imported versus cytoplasmic AOX–HA, we again made use of the sucrose velocity sedimentation method to separate imported, assembled, and active octameric AOX from cytoplasmic inactive monomers and aggregated forms of the protein (Goodman et al., 1984). Velocity gradients prepared with lysates from a strain expressing only AOX–HA showed a small amount of AOX activity at the position expected for octameric AOX (Fig. 6 E). However, immunoblot analysis revealed that most AOX–HA protein migrated to lower molecular mass positions in the gradient. This distribution was not because of the HA tag since a similar result was obtained with a strain expressing AOX–RSC (Fig. 6 D). As described above, control gradients performed with purified and then chemically denatured AOX (Fig. 6 A, AOX*), or with a lysate prepared from a pex5Δ strain expressing AOX–HA (Fig. 6 B), indicated that the AOX protein at the lower positions represented monomeric and/or aggregated forms of AOX.
A P. pastoris strain coexpressing both AOX–HA and AOX-WT was first examined by subcellular fractionation. After the organelle pellet from methanol-grown cells of the strain were centrifuged through a sucrose density gradient, a significant portion of AOX–HA was found to be peroxisome-bound as indicated by the comigration of AOX–HA protein with activities for AOX and CAT (Fig. 5 C). When a lysate prepared from this strain was analyzed by sucrose velocity gradient sedimentation, large amounts of AOX activity migrated to the position of octameric protein (Fig. 6 F). Immunoblot analysis with anti-AOX antibodies that recognize both AOX-WT and AOX–HA confirmed that the major portion of AOX protein comigrated with the AOX activity. However, immunoblot analysis with the anti-HA antibodies did not show a shift of AOX–HA protein to the position of octameric AOX (compare the distribution of AOX–HA in Fig. 6, F and E). Thus, the import efficiency of AOX–HA, as measured by octamerization, was not improved by coexpression with AOX-WT, indicating that AOX–HA could not oligomerize with AOX-WT in the cytoplasm before import into peroxisomes.
AOX–HA Is Not Impaired in Ability to Assemble with AOX-WT
An alternative interpretation of the coexpression results was that AOX–HA could not oligomerize with AOX-WT in the cytoplasm because of interference by the HA tag in AOX–HA/AOX-WT assembly or to the rapid assembly kinetics of AOX-WT, as suggested for MDH by Elgersma et al. (1996). If this were true, then the small amount of AOX–HA that does assemble into active enzyme would be expected to be present as AOX–HA homooctamers and not as AOX–HA/AOX-WT heterooctamers. To examine whether or not active AOX–HA oligomerized with AOX-WT in cells coexpressing the two proteins, two experiments were performed. In the first, sucrose velocity gradient fractions containing octameric AOX from the coexpression strain (Fig. 6 F, fraction 9) and from the strain expressing AOX–HA alone (Fig. 6 E, fraction 9) were immunoprecipitated using anti–HA antibodies. The immunoprecipitates were subsequently examined via immunoblot for the relative amounts of HA and AOX cross- reacting protein (Fig. 8 A). If active AOX–HA in the coexpression strain was a homooctamer, the relative amount of HA- and AOX-reacting protein should be the same as in the AOX–HA fractions. In contrast, results showed that the HA antibodies precipitated significantly more AOX protein from the coexpression strain fractions, indicating that AOX-WT protein was coprecipitated along with AOX–HA. As a control for the specificity of the HA antibody preparation, no AOX protein was immunoprecipitated from AOX-WT gradient fractions (Fig. 6 C, fraction 9; Fig. 8 A, lane 4). These results suggested that AOX-WT and AOX–HA subunits exist as heteromers in octameric fractions from the coexpression strain.
Figure 8.
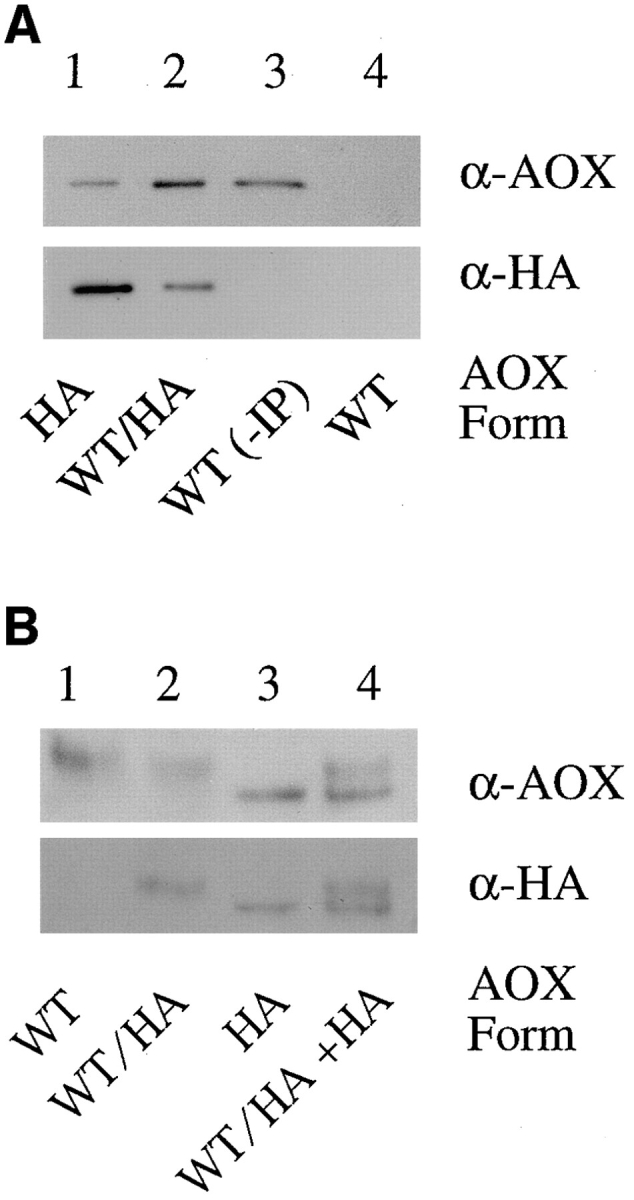
Analysis of octameric AOX in the P. pastoris strain coexpressing AOX–HA and AOX-WT. (A) Octameric AOX (fraction 9 from the sucrose velocity gradients shown in Fig. 6) was immunoprecipitated using HA-tag monoclonal antibodies, subjected to SDS-PAGE, and immunoblotted using antibodies against AOX (top) and HA (bottom). The SDS-PAGE lanes were loaded with equal ratios (relative to top and bottom) of sample from the following strains: Lane 1, AOX–HA; lane 2, AOX-WT/AOX–HA; lane 3, nonimmunoprecipitated AOX-WT, and lane 4, AOX-WT. (B) Octameric AOX from fraction 9 of the gradients in Fig. 6 were subjected to electrophoresis on a 5% native (nonreducing, nondenaturing) polyacrylamide gel and immunoblotted using antibodies against AOX (top) and HA (bottom). Lane 1, AOX-WT; lane 2, AOX-WT/AOX-HA; lane 3, AOX–HA; and lane 4, a mixture of fractions from both the AOX–HA strain and the AOX-WT/AOX–HA strain.
In the second experiment, the presence of AOX–HA and AOX-WT protein in the same species of AOX molecule was examined directly by subjecting active octameric AOX (Fig. 6, from the gradients in fraction 9) to native PAGE and immunoblotting (Fig. 8 B). In these native gels, AOX-WT homooctamers migrate slower than AOX–HA homooctamers (despite the fact that AOX–HA is slightly higher in predicted molecular mass than AOX-WT). In fractions from the coexpression strain, AOX–HA migrated at approximately the same rate as homooctameric AOX-WT. Thus, most of the AOX–HA was present as heterooctamers with AOX-WT. Furthermore, the fact that heteromers migrated at about the same rate as AOX-WT homomers indicated that the heteromers were composed mostly of AOX-WT with only one or two AOX–HA subunits, indicating that the small amount of AOX–HA that reaches the peroxisomal matrix had no difficulty oligomerizing with the large amount of AOX-WT present in the organelle.
Discussion
In this paper, we examined the targeting and assembly of peroxisomal AOX in the yeast P. pastoris. In particular, we were interested in determining whether newly synthesized AOX is capable of oligomerizing in the cytoplasm before import, as recently reported for certain other peroxisomal proteins. Previous results from our laboratory suggested that P. pastoris AOX could not assemble outside the peroxisome. Methanol-induced pex mutants of this yeast contain little or no activity for AOX, although substantial amounts of AOX protein are present in the cytoplasm (Liu et al., 1992, 1995; Waterham et al., 1996). Here we show that the amount of residual AOX activity in the pex mutants closely correlates with the severity of the peroxisomal biogenesis defect with chemically induced (and slightly leaky) pex mutants typically containing some residual AOX activity, while most pexΔ strains contain no detectable AOX activity. In addition, we show that residual active AOX in the pex mutants is located inside the few small peroxisomes or peroxisomal remnants present in these cells. Although these results do not eliminate the possibility that small amounts of AOX can octamerize into an active enzyme in the cytoplasm of pex mutants, they strongly suggest that the bulk of AOX protein must reach the peroxisome to efficiently assemble.
The inability of AOX to properly assemble in the cytoplasm of pex mutants contrasts with other peroxisomal enzymes which appear to be fully active in the cytoplasm and, therefore, have little difficulty assembling there. These enzymes include CAT from P. pastoris, an enzyme that must properly fold, incorporate a heme cofactor, and tetramerize to become active (Gould et al., 1992; Liu et al., 1992). Thus, the inability of AOX to assemble is not the result of some general inability of peroxisomal enzymes to incorporate cofactors or form homooligomers in the cytoplasm. The assembly problem does not appear to be because of the inability of AOX to oligomerize outside the peroxisome, since Evers et al. (1995) were able to efficiently reassemble active octameric AOX in vitro from FAD-containing monomeric subunits obtained from either P. pastoris or the related yeast H. polymorpha. They further showed that, in a riboflavin auxotrophic mutant of H. polymorpha, AOX protein is found in misfolded aggregates, indicating that insertion of FAD into AOX is an essential step in the assembly of H. polymorpha AOX (Evers et al., 1994, 1996). They postulated that the FAD insertion step may be mediated by an unknown assembly factor since, once removed, FAD could not be reinserted into AOX in vitro (Evers et al., 1995). Thus, in the absence of peroxisomes, the inability of P. pastoris AOX to assemble in the cytoplasm may also be related to a failure of FAD insertion (e.g., FAD and/or FAD insertion factor concentration in the cytoplasm may be too low to support efficient FAD binding).
Interestingly, AOX (MOX) from H. polymorpha efficiently assembles into an active enzyme in the cytoplasm of H. polymorpha pex mutants (Cregg et al., 1990; van der Klei et al., 1991). We found that MOX is active in the cytoplasm of P. pastoris pex mutants while AOX is inactive in the cytoplasm of pex mutants of H. polymorpha. Thus, this distinction between P. pastoris and H. polymorpha AOXs must be due to differences between their AOX polypeptides and not to the environment provided by their hosts. This is a surprising result given the high degree of sequence similarity shared by these polypeptides (∼85% identical). Perhaps, for proper assembly, P. pastoris AOX requires a higher concentration of FAD or FAD-insertion factor, or is more dependent on the acidic environment of the peroxisome than H. polymorpha AOX.
Recent studies suggest that at least some peroxisomal proteins are not only capable of cytoplasmic assembly but are also imported into peroxisomes in a preassembled oligomeric state. The key experiment in these studies is the coexpression of the peroxisomal protein without its PTS along with the wild-type protein. In each case, although the PTS-less protein expressed alone is not imported, coexpression resulted in the efficient coimport or piggybacking of the PTS-less polypeptides. Coimport has been reported for the human PTS1 enzyme, alanine/glyoxylate amino transferase (Leiper et al., 1996), a chloramphenicol acetyl transferase PTS1 fusion protein (McNew and Goodman, 1994), a yeast PTS1 enzyme, MDH3 (Elgermsa et al., 1996), and the yeast PTS2 enzyme thiolase (Glover et al., 1994). The ability of each of these proteins to piggyback into peroxisomes strongly suggests that these proteins can oligomerize in the cytoplasm before import.
To perform the coexpression study with P. pastoris AOX, it was first necessary to identify and characterize its PTS. We found that critical information for efficient peroxisomal targeting of P. pastoris AOX is located within its four COOH-terminal amino acids, LARF. Previously, a similar conclusion was drawn for the same four COOH-terminal amino acids of H. polymorpha MOX (Hansen et al., 1992). This conclusion was based on immunocytochemical data which indicated that these four amino acids were capable of targeting β-lactamase to peroxisomes in H. polymorpha. We show that these same four COOH-terminal amino acids are also sufficient to target β-lactamase to P. pastoris peroxisomes. In addition, we show that these four amino acids are critical for targeting since their removal and substitution with either RSC or an HA-epitope tag results in proteins that are only inefficiently imported into peroxisomes. Interestingly, a small portion of both AOX– RSC and AOX–HA is properly targeted to peroxisomes. Cell fractionation studies confirmed that these activities represent AOX protein that is correctly imported in peroxisomes, although the efficiency of import is not sufficient to support normal methanol-growth rates. One explanation for the residual import of LARF-less AOX is that, in addition to a COOH-terminal PTS, AOX contains a second independent but less efficient PTS. A second PTS has been reported for other peroxisomal matrix proteins including S. cerevisiae catalase A (Kragler et al., 1993) and H. polymorpha Per1p (Waterham et al., 1994). This second PTS may be located within the 18 amino acids immediately adjacent to LARF in AOX, since a mutant AOX deleted for the 22 COOH-terminal amino acids is not imported at all. Furthermore, a β-lactamase fusion containing the 22 COOH-terminal amino acids of AOX is targeted more efficiently than one fused to just the LARF sequence. A second explanation for the residual import of AOX without LARF is that the protein oligomerizes in the cytoplasm with another methanol pathway protein, such as dihydroxyacetone synthase, and is coimported. The preimport coassembly of AOX and dihydroxyacetone synthase was previously suggested from the results of ionophore experiments with the yeast C. boidinii (Bellion and Goodman, 1987). We observed that, in glucose-grown cells that do not synthesize dihydroxyacetone synthase, AOX– RSC (expressed under control of the constitutive GAP promoter) is not imported, although AOX-WT is imported (not shown), a result that is consistent with the notion that residual import of AOX without LARF may be dependent on another methanol pathway protein.
AOX is clearly targeted to peroxisomes via the PTS1 import pathway. Previous results demonstrated that AOX behaves in a manner similar to luciferase, the prototypical PTS1 protein, in P. pastoris wild-type and pex mutant cells (McCollum et al., 1993; Spong et al., 1993; Liu et al., 1995; Waterham et al., 1996). Here, we extend these results by demonstrating that neither AOX nor β-lactamase fused to LARF is targeted to peroxisomes in a pex5Δ strain that is specifically defective in the import of PTS1 proteins (Spong et al., 1993). In addition, two-hybrid system results indicate that AOX strongly interacts with the PTS1 receptor Pex5p, and that this interaction is dependent upon the last four amino acids of AOX. We conclude that the primary targeting signal for AOX is a PTS1 located within LARF and that the PTS most probably consists of the COOH-terminal tripeptide sequence ARF. The first two amino acids of the motif in AOX, alanine and arginine, are known functional variants of the prototypical PTS1 sequence (SKL; Gould et al., 1989; Swinkels et al., 1992). The substitution of a phenylalanine for leucine at the ultimate position was shown to abolish peroxisomal targeting in mammalian cells (Swinkels et al., 1992). However, while this work was in progress, Elgersma et al. (1996) reported that PTS1 motifs ending in phenylalanine were imported in S. cerevisiae.
To investigate the effect of coexpressing AOX-WT and AOX without its primary PTS, it was not possible to examine samples by differential or sucrose density gradient centrifugations, as done in previous studies of this kind. These techniques require prolonged incubation of cells to remove cell walls, conditions that result in degradation of improperly folded AOX (Liu et al., 1992, 1995; and Waterham et al., 1996). Furthermore, improperly folded AOX forms into protein aggregates which sediment upon differential centrifugation and, thus, appear in organelle pellets along with properly imported and assembled AOX. To observe import of AOX, cell extracts were prepared by the glass bead disruption method at a low temperature and were subjected to sucrose velocity sedimentation (Goodman et al., 1984). The proportion of AOX protein present at the position of fully assembled AOX octamers was then noted in the gradients. Previous studies (Liu et al., 1992, 1995) as well as those presented here indicate that active octameric AOX is only found in the peroxisomal matrix. Thus, the presence of AOX–HA at the position of octameric AOX in velocity gradients represents AOX–HA that was properly imported into the organelle.
The coexpression studies showed that the efficiency of AOX–HA import is not improved by AOX-WT. Thus, it appears that AOX–HA cannot initiate cytoplasmic assembly with AOX-WT in the cytoplasm and be piggybacked into the peroxisome. An alternative explanation for the failure of AOX–HA to efficiently oligomerize with AOX-WT in the coexpression experiment was that the HA tag somehow interfered with its assembly. Elgersma et al. (1996) proposed that the apparent inability of S. cerevisiae PTS1 enzyme MDH to coimport PTS1-less MDH was because of a reduction in the assembly kinetics of the mutant polypeptide. As a result, the mutant subunits were outcompeted by the wild-type subunits that rapidly homodimerize in the cytoplasm. Our results indicate that AOX–HA is not impaired in its ability to assemble with AOX-WT. When we examined the small portion of AOX–HA in the coexpression strain that does reach the peroxisomal matrix and assemble into active octameric enzyme, we found that virtually all of the mutant protein was present as heterooligomers with AOX-WT. Thus, it appears that despite the presence of large amounts of AOX-WT, most AOX–HA remains in the cytoplasm because AOX cannot initiate oligomerization in this compartment and, without its primary PTS1, the mutant AOX is inefficiently targeted to and imported into the peroxisome. In summary, although peroxisomes are capable of importing some proteins as preassembled structures, AOX must be targeted to peroxisomes as monomers for import and assembly to occur properly.
Despite recent progress, basic features of protein import into peroxisomes remain largely unknown (Waterham and Cregg, 1997). Although the organelle appears to be morphologically simple with a single membrane and uncomplicated matrix, the import mechanism is unexpectedly complex. Peroxisomes have evolved at least two matrix protein import pathways and a third independent pathway specific for peroxisomal integral membrane proteins. As shown by several recent studies, the matrix protein import machinery is capable of importing large preassembled structures ⩾9 nm in diameter (Walton et al., 1995), a size much greater than AOX octamers at ∼600 kD. Therefore, the necessity of importing AOX as unassembled monomeric polypeptides is probably not related to its size but may reflect the necessity to incorporate FAD before octamerization, a process proposed to occur in the peroxisomal matrix (Evers et al., 1996).
Although this is the first description of a yeast peroxisomal protein that cannot assemble in the absence of functional peroxisomes, this phenomenon may be relatively common in patients afflicted with the lethal peroxisomal biogenesis disorder Zellweger syndrome (Lazarow and Moser, 1994). In cells of these patients, peroxisomes are absent and peroxisomal matrix enzymes are left in the cytoplasm. Some of these enzymes are stable and active, whereas several others (including the plasmalogen biosynthetic enzymes, acyl-CoA:dihydroxyacetone phosphate acyltransferase and alkyl-dihydroxyacetone phosphate synthase, and the peroxisomal β-oxidation pathway enzymes, acyl-CoA oxidase, bifunctional enzyme, and 3-oxoacyl-CoA thiolase) are deficient (Lazarow and Moser, 1994). Pulse chase studies on acyl-CoA oxidase and thiolase demonstrated that the proteins are synthesized normally but remain largely inactive in the cytosol where they are rapidly degraded (Schram et al., 1986; Suzuki et al., 1986). The inability of these and other enzymes to assemble into stable active enzymes may be the reason that specific peroxisomal metabolic pathways are defective in Zellweger patients. Further investigations into the fate of AOX in P. pastoris mutants may shed light on the molecular details of this phenomenon and the molecular etiology of this aspect of Zellweger syndrome.
Acknowledgments
We thank S. Subramani (University of California at San Diego) for the gift of vector pSP72 containing the P. pastoris PEX5 gene and the pex5 deletion strain. We thank T. Hadfield (Oregon Graduate Institute, Portland, OR) for assistance in preparing the manuscript.
Abbreviations used in this paper
- AOX
alcohol oxidase
- AOX-WT
wild-type AOX
- ARF
alanine-arginine-phenylalanineCOOH
- CAT
catalase
- FAD
flavine adenine dinucleotide
- LARF
leucine-alanine-arginine-phenylalanineCOOH
- MDHC
malate dehydrogenase 3
- MOX; AOX from Hansenula polymorpha; MSG; methione-serine-glysine; pex
peroxisome biogenesis defective
- PTS
proximal targeting signal
- SKL
arginine-serine-cysteineCOOH
Footnotes
This research was supported by a National Institutes of Health grant (DK-43698) and a National Science Foundation grant (MCB-9514289) to J.M. Cregg.
H.R. Waterham's present address is Department of Pediatrics, Academic Medical Centre, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands.
Y. de Vries' present address is Department of Bacterial Genetics, Intervet B.V., P.O. Box 31, 5830 AA Boxmeer, The Netherlands.
Address all correspondence to James M. Cregg, Department of Biochemistry and Molecular Biology, Oregon Graduate Institute of Science and Technology, P.O. Box 91000, Portland, OR 97291-1000. Tel.: (503) 690-1217. Fax: (503) 690-1464. E-mail: cregg@bmb.ogi.edu
References
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1996. Analysis of proteins. In Current Protocols in Molecular Biology. Vol. 2. John Wiley and Sons, New York. pp. 10.1–10.20.
- Bartel PL, Chien C-T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- Becker DM, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- Bellion E, Goodman JM. Proton ionophores prevent assembly of a peroxisomal protein. Cell. 1987;48:165–173. doi: 10.1016/0092-8674(87)90367-9. [DOI] [PubMed] [Google Scholar]
- Cregg JM, Barringer KJ, Hessler AY, Madden KR. Pichia pastorisas a host system for transformations. Mol Cell Biol. 1985;9:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, Madden KR, Barringer KJ, Thill G, Stillman CA. Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris. . Mol Cell Biol. 1989;9:1316–1323. doi: 10.1128/mcb.9.3.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, van der Klei IJ, Sulter GJ, Veenhuis M, Harder W. Peroxisome-deficient mutants of Hansenula polymorpha. . Yeast. 1990;6:87–97. [Google Scholar]
- Distel B, Veenhuis M, Tabak HF. Import of alcohol oxidase into peroxisomes of Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1987;6:3111–3116. doi: 10.1002/j.1460-2075.1987.tb02620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Gould SJ. Multiple PEXgenes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol. 1996;135:1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Vos A, van den Berg M, van Roermund CW, van der Swijs P, Distel B, Tabak HF. Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. . J Biol Chem. 1996;271:26375–26382. doi: 10.1074/jbc.271.42.26375. [DOI] [PubMed] [Google Scholar]
- Ellis SB, Brust PF, Koutz PJ, Waters AF, Harphold MM, Gingeras TR. Isolation of alcohol oxidase and two other methanol regulatable genes from the yeast Pichia pastoris. . Mol Cell Biol. 1985;5:1111–1121. doi: 10.1128/mcb.5.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers ME, Titorenko VI, van der Klei IJ, Harder W, Veenhuis M. Assembly of alcohol oxidase in peroxisomes of the yeast Hansenula polymorpharequires the cofactor flavin adenine dinucleotide. Mol Biol Cell. 1994;5:829–837. doi: 10.1091/mbc.5.8.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers ME, Harder W, Veenhuis M. In vitro dissociation and re- assembly of peroxisomal alcohol oxidases of Hansenula polymorpha and Pichia pastoris. . FEBS (Fed Eur Biochem Soc) Lett. 1995;368:293–296. doi: 10.1016/0014-5793(95)00653-q. [DOI] [PubMed] [Google Scholar]
- Evers ME, Titorenko V, Harder W, van der Klei IJ, Veenhuis M. Flavin adenine dinucleotide binding is the crucial step in alcohol oxidase assembly in the yeast Hansenula polymorpha. . Yeast. 1996;12:917–923. doi: 10.1002/(SICI)1097-0061(199608)12:10%3C917::AID-YEA984%3E3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Faber KN, Haima P, Harder W, Veenhuis M, AB G. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. . Curr Genet. 1994;25:305–310. doi: 10.1007/BF00351482. [DOI] [PubMed] [Google Scholar]
- Glover JR, Andrews DW, Rachubinski RA. Saccharomyces cerevisiaeperoxisomal thiolase is imported as a dimer. Proc Natl Acad Sci USA. 1994;91:10541–10545. doi: 10.1073/pnas.91.22.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JM, Scott CW, Donahue PN, Atherton JP. Alcohol oxidase assembles post-translationally into the peroxisome of Candida boidinii. . J Biol Chem. 1984;259:8485–8493. [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Scheider M, Howell SH, Garrard LJ, Goodman JM, Distel B, Tabak HF, Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO (Eur Mol Biol Organ) J. 1990;9:85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, McCollum D, Spong AP, Heyman JA, Subramani S. Development of the yeast Pichia pastorisas a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;6:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Hansen H, Didion T, Thiemann A, Veenhuis M, Roggenkamp R. Targeting signals of the major peroxisomal proteins in the methylotrophic yeast Hansenula polymorpha. . Mol Gen Genet. 1992;235:269–278. doi: 10.1007/BF00279370. [DOI] [PubMed] [Google Scholar]
- Koutz P, Davis GR, Stillman C, Barringer K, Cregg JM, Thill G. Structural comparison of the Pichia pastorisalcohol oxidase genes. Yeast. 1989;5:167–177. doi: 10.1002/yea.320050306. [DOI] [PubMed] [Google Scholar]
- Kragler F, Langeder A, Raupachova J, Binder M, Hartig A. Two independent peroxisomal targeting signals in catalase A of Saccharomyces cerevisiae. . J Cell Biol. 1993;120:665–673. doi: 10.1083/jcb.120.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow, P.B., and H.W. Moser. 1994. Disorders in peroxisome biogenesis. In The Metabolic Basis of Inherited Disease. 7th ed. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. McGraw Hill, New York. pp. 2287–2324.
- Ledeboer AM, Edens L, Maat J, Visser C, Bos JW, Janowicz Z, Eckart M, Roggenkamp R, Hollenberg CP. Molecular cloning and characterization of a gene coding for methanol oxidase in Hansenula polymorpha. . Nucleic Acids Res. 1985;13:3063–3082. doi: 10.1093/nar/13.9.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiper JM, Oatey PB, Danpure CJ. Inhibition of alanine:glyoxylate aminotransferase 1 dimerization is a prerequisite for its peroxisome- to-mitochondrial mistargeting in primary hyperoxaluria type 1. J Cell Biol. 1996;135:939–951. doi: 10.1083/jcb.135.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Tan X, Veenhuis M, McCollum D, Cregg JM. An efficient screen for peroxisome-deficient mutants of Pichia pastoris. . J Bacteriol. 1992;174:4943–4951. doi: 10.1128/jb.174.15.4943-4951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Tan X, Russell KA, Veenhuis M, Cregg JM. PER3, a gene required for peroxisome biogenesis in Pichia pastoris, encodes a peroxisomal membrane protein involved in PTS1- and PTS2-protein import. J Biol Chem. 1995;270:10940–10951. doi: 10.1074/jbc.270.18.10940. [DOI] [PubMed] [Google Scholar]
- McCollum D, Monosov E, Subramani S. The pas8 mutant of Pichia pastorisexhibits the peroxisomal import deficiencies of Zellweger syndrome cells the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J Cell Biol. 1993;121:761–774. doi: 10.1083/jcb.121.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Goodman JM. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Goodman JM. The targeting and assembly of peroxisomal proteins: some old rules do not apply. TIBS (Trends Biochem Sci) 1996;21:54–58. [PubMed] [Google Scholar]
- Rehling P, Marzioch M, Niesen F, Wittke E, Veenhuis M, Kunau W-H. The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7gene. EMBO (Eur Mol Biol Organ) J. 1996;15:2901–2913. [PMC free article] [PubMed] [Google Scholar]
- Roa M, Blobel G. Biosynthesis of peroxisomal enzymes in the methylotrophic yeast Hansenula polymorpha. . Proc Natl Acad Sci USA. 1983;80:6872–6876. doi: 10.1073/pnas.80.22.6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Tani Y. Cloning and sequencing of the alcohol oxidase- encoding gene (AOD1) from the formaldehyde-producing asporogenous methylotrophic yeast, Candida boidiniiS2. Gene. 1992;114:67–73. doi: 10.1016/0378-1119(92)90708-w. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schram AW, Strijland A, Hashimoto T, Wanders RJA, Schutgens RBH, van den Bosch H, Tager JM. Biosynthesis and maturation of peroxisomal β-oxidation enzymes in fibroblasts in relation to the Zellweger syndrome and infantile Refsum disease. Proc Natl Acad Sci USA. 1986;83:6156–6158. doi: 10.1073/pnas.83.16.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spong AP, Subramani S. Cloning and characterization of PAS5: a gene required for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. . J Cell Biol. 1993;123:535–548. doi: 10.1083/jcb.123.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JG. Nucleotide sequence of the ampicillin resistance gene of Escherichia coliplasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Orii T, Mori M, Tatibana T, Hashimoto T. Deficient activities and proteins of peroxisomal beta oxidation enzymes in infants with Zellweger syndrome. Clin Chim Acta. 1986;156:191–196. doi: 10.1016/0009-8981(86)90152-x. [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Subramani S. Targeting efficiencies of various permutations of the consensus C-terminal tripeptide peroxisomal targeting signal. FEBS (Fed Eur Biochem Soc) Lett. 1992;305:133–136. doi: 10.1016/0014-5793(92)80880-p. [DOI] [PubMed] [Google Scholar]
- Tan X, Titorenko VI, van der Klei IJ, Sulter GJ, Haima P, Waterham HR, Evers M, Harder W, Veenhuis M, Cregg JM. Characterization of peroxisome-deficient mutants of Hansenula polymorpha. . Curr Genet. 1995a;28:248–257. doi: 10.1007/BF00309784. [DOI] [PubMed] [Google Scholar]
- Tan X, Waterham HR, Veenhuis M, Cregg JM. The Hansenula polymorpha PER8gene encodes a novel peroxisomal integral membrane protein involved in proliferation. J Cell Biol. 1995b;128:307–319. doi: 10.1083/jcb.128.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlecky SR, Nuttley WM, McCollum D, Sock E, Subramani S. The Pichia pastorisperoxisomal protein Pas8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO (Eur Mol Biol Organ) J. 1995;14:3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp JF, Brust PF, Cregg JM, Stillman CA, Gingeras TR. Expression of the lacZ gene from two methanol-regulated promoters in Pichia pastoris. . Nucleic Acids Res. 1987;15:3859–3876. doi: 10.1093/nar/15.9.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei IJ, Harder W, Veenhuis M. Biosynthesis and assembly of alcohol oxidase, a peroxisomal matrix protein in methylotrophic yeasts: a review. Yeast. 1991;7:195–209. doi: 10.1002/yea.320070302. [DOI] [PubMed] [Google Scholar]
- Veale RA, Guiseppin MLF, van Eijk HMJ, Sudbery PE, Verrips CT. Development of a strain of Hansenula polymorphafor the efficient expression of guar α-galactosidase. Yeast. 1992;8:361–372. doi: 10.1002/yea.320080504. [DOI] [PubMed] [Google Scholar]
- Veenhuis, M., and W. Harder. 1991. Microbodies. In The Yeasts. 2nd ed., Vol. 4. A.H. Rose and J.S. Harrison, editors. Academic Press, London. pp. 601–653.
- Walton PA, Hill PE, Subramani S. Import of stably folded proteins into peroxisomes. Mol Biol Cell. 1995;6:675–683. doi: 10.1091/mbc.6.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Cregg JM. Peroxisome biogenesis. Bioessays. 1997;19:57–66. doi: 10.1002/bies.950190110. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Titorenko VI, van der Klei IJ, Harder W, Veenhuis M. Isolation and characterization of peroxisomal protein import (Pim−) mutants of Hansenula polymorpha. . Yeast. 1992;8:961–972. [Google Scholar]
- Waterham HR, Titorenko VI, Haima P, Cregg JM, Harder W, Veenhuis M. The Hansenula polymorpha PER1gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J Cell Biol. 1994;127:737–749. doi: 10.1083/jcb.127.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, de Vries Y, Russell KA, Xie W, Veenhuis M, Cregg JM. The Pichia pastoris PER6gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger Syndrome protein PAF-1. Mol Cell Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Digan ME, Koutz PJ, Lair SV, Cregg JM. Isolation of the Pichia pastorisglyceraldehyde-3-phosphate dehydrogenase gene and use and regulation of its promoter. Gene. 1997;186:37–44. doi: 10.1016/s0378-1119(96)00675-0. [DOI] [PubMed] [Google Scholar]
- Wenzel TJ, Migliazza A, Steensma HY, van den Berg JA. Efficient selection of phleomycin-resistant Saccharomyces cerevisiaetransformants. Yeast. 1992;8:667–668. doi: 10.1002/yea.320080810. [DOI] [PubMed] [Google Scholar]