Abstract
CENP-E is a kinesin-like protein that binds to kinetochores and may provide functions that are critical for normal chromosome motility during mitosis. To directly test the in vivo function of CENP-E, we microinjected affinity-purified antibodies to block the assembly of CENP-E onto kinetochores and then examined the behavior of these chromosomes. Chromosomes lacking CENP-E at their kinetochores consistently exhibited two types of defects that blocked their alignment at the spindle equator. Chromosomes positioned near a pole remained mono-oriented as they were unable to establish bipolar microtubule connections with the opposite pole. Chromosomes within the spindle established bipolar connections that supported oscillations and normal velocities of kinetochore movement between the poles, but these bipolar connections were defective because they failed to align the chromosomes into a metaphase plate.
Overexpression of a mutant that lacked the amino-terminal 803 amino acids of CENP-E was found to saturate limiting binding sites on kinetochores and competitively blocked endogenous CENP-E from assembling onto kinetochores. Chromosomes saturated with the truncated CENP-E mutant were never found to be aligned but accumulated at the poles or were strewn within the spindle as was the case when cells were microinjected with CENP-E antibodies. As the motor domain was contained within the portion of CENP-E that was deleted, the chromosomal defect is likely attributed to the loss of motor function.
The combined data show that CENP-E provides kinetochore functions that are essential for monopolar chromosomes to establish bipolar connections and for chromosomes with connections to both spindle poles to align at the spindle equator. Both of these events rely on activities that are provided by CENP-E's motor domain.
The kinetochore establishes and maintains the connection between the chromosome and the dynamic plus ends of microtubules to generate force for accurately segregating chromosomes. After the nuclear envelope disassembles at the onset of mitosis, chromosomes establish connections to microtubules from each spindle pole, as each member of a kinetochore pair that resides on opposite sides of the centromere captures microtubules that are nucleated from the pole that they face. The bipolar attached chromosome congresses toward the spindle equator through the coordinated actions of the kinetochore pair. Net displacement of the chromosome occurs when one kinetochore moves towards its pole, while its sister kinetochore moves away from its pole. Concomitant with these motions, microtubules attached to the kinetochore that is moving poleward must shorten, and those that are attached to its sister elongate. Shrinkage and elongation of microtubules occur primarily at the kinetochore because this attachment site is where tubulin subunits are incorporated into or mostly lost from the microtubule (Mitchison et al., 1986; Gorbsky et al., 1987; for review see Inoue and Salmon, 1995; Yen and Schaar, 1996; Nicklas, 1997).
The discovery of various microtubule-based motors that reside at kinetochores suggest that these molecules generate force to move chromosomes and also maintain the mechanical link between kinetochores and the tips of shrinking and growing microtubules (Desai and Mitchison, 1995; Lombillo et al., 1995). Thus far, the motor composition of mammalian kinetochores includes cytoplasmic dynein and its associated dynactin complex (Pfarr et al., 1990; Steuer et al., 1990) and two kinesin-related proteins, CENP-E (Yen et al., 1992) and MCAK (XKCM1 is its frog homolog; Wordeman and Mitchison, 1995; Walczak et al., 1996). Video analysis and correlative electron microscopy of chromosome movements in newt lung cells show that if a kinetochore makes a lateral connection with a microtubule that glances past it, this interaction was sufficient to pull the chromosome polewards at a velocity that is consistent with the in vitro polarity and velocity of cytoplasmic dynein (Rieder et al., 1989; Hayden et al., 1990). In addition to a potential role in pulling chromosomes into the spindle proper, dynein and dynactin contribute to spindle morphogenesis, as disruption of their functions interferes with bipolar spindle formation (Vaisberg et al., 1993; Echeverri et al., 1996). XKCM1 (and perhaps MCAK) may also play a role in spindle morphogenesis as depletion of this protein from frog egg extracts disrupts spindle assembly (Walczak et al., 1996). However, the ability of XKCM1 to induce catastrophic depolymerization of microtubules in vitro suggests that this activity at the kinetochore may stimulate kinetochore microtubule depolymerization and facilitate poleward movement of chromosomes (Walczak et al., 1996).
CENP-E is a 312-kD protein (Yen et al., 1992) that assumes a highly elongated shape and was found to be associated with a minus end microtubule motor activity (Thrower et al., 1995). Recent immunogold EM data show that it is concentrated at the fibrous corona that occupies the surface of the outer kinetochore plate (Cooke et al., 1998). Early functional studies had shown that microinjection of an anti-CENP-E monoclonal antibody (mAb177) arrested cells in mitosis with chromosomes aligned in a metaphase plate at the spindle equator (Yen et al., 1991). The nature of this arrest was never clear, as the antibody was not directed towards obvious functional domains such as the motor or the carboxy-terminal microtubule binding domains (Liao et al., 1994), but rather, it recognized epitopes along the extended rod domain. Although the presence of this monoclonal antibody at kinetochores did not prevent chromosomes from aligning at the spindle equator, it interfered with critical events that were necessary to initiate chromosome separation and anaphase onset.
A role for CENP-E in chromosome motility has come from recent in vitro studies on the mechanism that is responsible for microtubule depolymerization-dependent movement of chromosomes (Lombillo et al., 1995). Chromosome movement in this in vitro system does not require ATP but relies simply on the ability of the kinetochore to remain attached to the shrinking end of a single microtubule induced to depolymerize by the dilution of free tubulin subunits. CENP-E was found to be important for coupling the kinetochore to the microtubule as antibodies directed against its neck region, which connects the motor domain of CENP-E to its stalk domain, disrupted chromosome movement by dissociating the kinetochore from a shrinking microtubule against a flow of buffer. This finding suggested that the kinetochore–microtubule interactions mediated by CENP-E in vitro might be important to chromosome attachment and alignment on the spindle in vivo.
The discrepancy between the in vitro data and the early microinjection studies necessitated a re-evaluation of the in vivo function of CENP-E. This renewed effort required that we develop strategies that directly test the contribution of CENP-E to kinetochore function. Optimally, this would require examining the behavior of chromosomes whose kinetochores lacked CENP-E. Our first strategy was to microinject highly specific polyclonal CENP-E antibodies to sterically block CENP-E from binding to kinetochores at mitosis. The second approach was to overexpress a motorless CENP-E mutant in transiently transfected cells to saturate limiting binding sites for CENP-E at kinetochores and thus competitively block endogenous CENP-E from binding. In both strategies, depletion of endogenous CENP-E from kinetochores to levels below our limits of detection blocked chromosome alignment but not bipolar spindle formation. Chromosome alignment was reproducibly blocked at very specific stages as they were found to be trapped very close to a pole (most likely with a monopolar connection), or chromosomes established defective bipolar connections that sustained normal velocities of oscillations but often failed to attain a stable position at the spindle equator. Thus, CENP-E and its motor domain provide functions that are essential for two events during prometaphase: monopolar chromosomes to establish bipolar connections and for bipolar chromosomes to align and form a stable metaphase plate at the spindle equator.
Materials and Methods
Production of Antibodies and Immunological Methods
A 1.3-kb Dra I CENP-E cDNA fragment was subcloned into pMAL expression vector (New England Biolabs, Beverly, MA) and purified maltose-binding (MBP)1 fusion protein was used to raise rabbit antibodies (Dra B) against the carboxy-terminal 360 amino acids. For HX-1 antibodies, an Xho/HindIII fragment that encodes amino acids 1,571–1,859 of CENP-E was subcloned into pGEX (Pharmacia Biotech, Piscataway, NJ), and the purified glutathione-S-transferase (GST) fusion protein was used to immunize rabbits. Antibodies were affinity purified by binding whole IgG to an Affigel-10 (Bio Rad, Richmond, CA) column that was covalently coupled with either the MBP:DraB or GST:HX-1 fusion proteins. Antibodies were eluted with 100 mM glycine, pH 2.5, neutralized, concentrated, and stored in PBS. Preimmune rabbit IgG was purified using protein A sepharose (Pierce, Rockland, IL). Mouse monoclonal antibody 177 was described previously (Yen et al., 1991). To detect the green fluorescent protein (GFP):CENP-E fusion protein by immunoblot, rat polyclonal antibodies were generated against bacterially expressed GFP. Affinity-purified GFP antibodies were used to probe filters at a final concentration of 1 μg/ml. Filters were washed, incubated with alkaline phosphatase–conjugated anti-rat secondary antibodies (Sigma Chemical Co., St. Louis, MO), and bound antibodies were visualized by chemiluminescent detection. Western blots were performed according to manufacturer's protocol (Tropix, Bedford, MA). HX-1 and DraB antibodies were used at 1 μg/ml final concentration. Immunofluorescence staining was performed as described (Yen et al., 1991) and visualized with a 100× 1.3 NA objective attached to a microscope (Microphot SA; Nikon, Inc., Melville, NY) equipped with epifluorescence optics. Photographs were taken with Kodak TMax 400 film.
Microinjections and Video Microscopy of Injected Cells
Microinjections were performed with a Nikon Diaphot that is equipped with an Eppendorf semi-automatic microinjector and micromanipulator. Antibodies were first filtered through a 0.2-μm membrane (Millipore, Bedford, MA), and injections were performed with Eppendorf femtotips. HeLa or U2OS cells that were grown to 30% confluence in Hepes buffered DME plus 10% FBS were injected in the cytoplasm with an antibody concentration of 0.5 mg/ml for the anti-carboxy terminus (DraB) antibody, 10 mg/ml for anti-rod domain (HX-1) antibody, and 20 mg/ml for preimmune antibodies. After injection, cells were returned to 37°C for ∼12 h before observation. For quantitative studies, HeLa cells were synchronized by double thymidine block as described (Yen et al.,1992). After release from the block, the tissue culture dish was placed into a TC-102 (Medical Systems Corp., Greenville, NY) microscope stage incubator that maintained the medium at 37°C. Cells were injected with antibody, the culture medium was overlaid with mineral oil, and the entire field of injected and uninjected cells were recorded using a CCD camera (Ultrachip; Javelin Electronics, Torrance, CA) and a time-lapse S-VHS video recorder (Panasonic, Secaucus, NJ). The duration of mitosis for each cell was determined by recording when each cell rounded up (entry into mitosis) and when they separated (cytokinesis).
Interphase U2OS cells were microinjected with DraB antibodies and incubated for ∼12 h at 37°C. When the cells were confirmed to be blocked in mitosis, the coverslip was removed from the culture dish and inverted onto a clean glass slide that had two strips of double stick tape. Hepes-buffered DME plus 10% FBS was added, and the observation chamber was sealed with a 1:1:1 mixture of Vaseline/lanolin/beeswax. The slide chamber was mounted on a microscope (Diaphot; Nikon, Inc.) and cells were observed under a 63× 1.4 NA differential interference contrast (DIC) objective and a 1.4 NA condenser. Images were captured with a Newvicon video camera (Hamamatsu Phototonics, Bridgewater, NJ), real-time background subtraction was performed with a DSP 2000 image processor (DAGE-MTI, Michigan City, IN), and the subtracted images were recorded onto time-lapse S-VHS VCR (Panasonic). Individual video frames were photographed directly from the monitor. To measure the velocities of the kinetochores, the videotape was analyzed as described (Skibbens et al., 1993).
Constructs and Transient Transfections
Construction of the motorless CENP-E was accomplished by fusing PCR fragments that contain either GFP (Chalfie et al., 1993) or the IgG-binding region of protein A (pRIT2T; Pharmacia Biotech) in the correct reading frame with a 5.6-kb cDNA fragment that encoded amino acids 880 to the carboxy terminus of CENP-E. The hybrid gene was subcloned into the expression vector pWS that uses a CMV promoter and a segment of the adenovirus 5′ tripartite leader sequence to achieve high level expression in mammalian cells (Sheay et al., 1993). CsCl purified plasmid DNA was used to transfect subconfluent HeLa cells according to Chen and Okayama (1987). Cells were harvested 30–40 h after transfection and lysates were prepared for immunoblot analysis. Immunofluorescence staining was performed as described above.
Results
Microinjection of CENP-E Antibodies Depletes CENP-E from Kinetochores
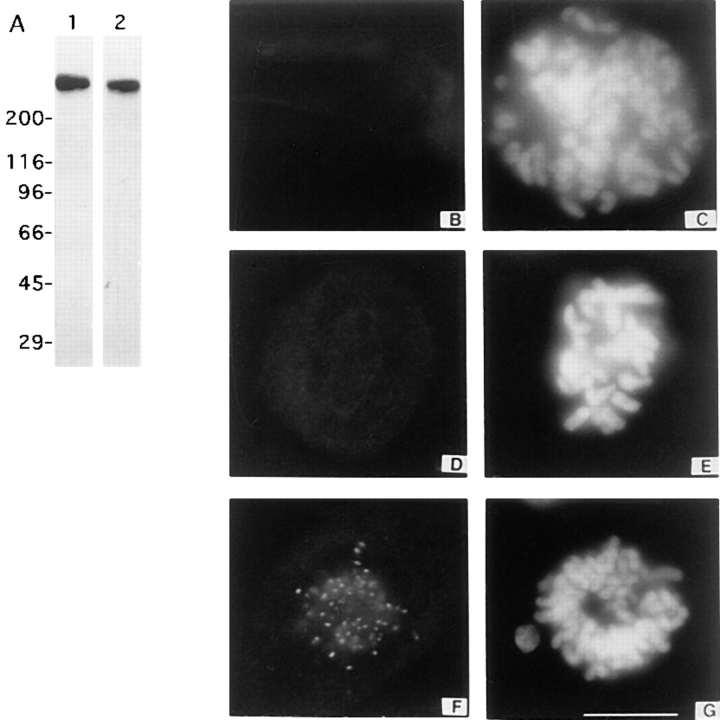
Our first strategy to deplete CENP-E from kinetochores took advantage of an earlier observation that CENP-E is a cytoplasmic protein during interphase and assembles onto kinetochores after nuclear envelope breakdown in mitosis (Yen et al., 1991). If microinjection of CENP-E–specific antibodies into interphase cells can block cytoplasmic CENP-E from assembling onto kinetochores after nuclear envelope breakdown, it would then be possible to examine the behavior of chromosomes that lacked CENP-E. Polyclonal antibodies were raised against either a segment of the stalk (amino acids 1,571–1,859) or the carboxy-terminal 360 amino acids that included a large portion of the kinetochore targeting domain (Chan, G.K.T., and T.J. Yen, unpublished observations). Both sets of antibodies were expected to form mutivalent complexes with CENP-E and thus sterically interfere with its ability to bind to kinetochores. In addition, the carboxy-terminal antibodies were expected to directly block CENP-E from binding to kinetochores by obscuring the kinetochore-targeting domain from its binding site. Both of the affinity-purified antibodies were highly specific as CENP-E was the only protein that was identified when they were used to probe filters that contained total protein from mitotic HeLa cells (Fig. 1 A).
Figure 1.
Depletion of CENP-E from kinetochores by microinjection of rabbit polyclonal antibodies directed against CENP-E. (A) Western blots of mitotic HeLa lysates probed with stalk HX-1 (lane 1) and carboxy-terminal DraB (lane 2) affinity-purified antibodies. (B, D, F, and I) Monoclonal antibody mAb177 (7) staining of Hela cells injected with 0.5 mg/ml of DraB antibodies (B and D), an uninjected prometaphase cell (F). mAb177 was visualized with FITC-conjugated anti–mouse antibodies. (C, E, and G) DAPI staining to visualize chromosomes. Images from B, D, and F were exposed for identical times. Bar, 10 μm.
Initial experiments showed that when interphase (asynchronous) HeLa or U2OS cells were microinjected with either of the two CENP-E antibodies, they were found to accumulate in mitosis when they were examined after an overnight incubation. These cells were probably blocked in mitosis as their mitotic index of injected cells was much higher than cells injected with nonimmune IgG. To examine the distribution of CENP-E in the mitotically blocked cells, the injected cells were stained with a murine monoclonal CENP-E antibody (mAb177) whose epitope did not overlap those recognized by the injected rabbit polyclonal antibodies. mAb177 staining did not reveal the presence of CENP-E at the kinetochores of these mitotic cells (Fig. 1, B and D) even though prominent levels of kinetochore staining were observed in adjacent uninjected prometaphase cells (Fig. 1, F and G). Direct visualization of the injected CENP-E antibodies also did not reveal specific localization at kinetochores (data not shown). The fate of CENP-E/antibody complexes in the injected cells is not known but most likely was released from the cytoplasm during extraction.
Inhibition of CENP-E binding to kinetochores was specific for the affinity-purified antibodies as injection of 40-fold more nonimmune rabbit IgG did not disrupt mitotic progression (Fig. 2, A, D, and G; and see below) or the normal distribution of CENP-E at kinetochores (Fig. 2, B, E, and H) at metaphase or at the interzonal microtubules at anaphase. As mAb177 was able to identify the normal subcellular distribution of CENP-E in the cells injected with control IgG, these results also show that the inability of mAb177 to stain kinetochores of cells injected with CENP-E polyclonal antibodies was not simply due to an overwhelming level of rabbit IgG in the cell. Microinjection of CENP-E antibodies was found to be a reliable method to deplete CENP-E from kinetochores and was therefore used to examine the in vivo function of CENP-E.
Figure 2.
Preimmune antibodies injected at a 40-fold higher concentration have no effect on mitotic progression or CENP-E localization. Preimmune HX-1 antibodies were injected in an identical manner to immune antibodies. DNA was detected with DAPI (A, D, and G) CENP-E was detected with mAb 177 (B, E, and H) and injected antibodies were detected with anti–rabbit Ig secondary antibodies (C, F, and I). No aberrations were seen in the organization of the chromosomes during alignment (A). CENP-E localized normally in these cells during anaphase (E and H). Anaphase cells as shown in D through F and G through I were never observed in immune HX-1 nor DraB antibody– injected cells. Bar, 10 μM.
Loss of CENP-E at Kinetochores Block Cells in Mitosis
To monitor the fates of the injected cells in a more quantitative way, 27 HeLa cells that were synchronized at the G1/S boundary were injected with the affinity-purified carboxy-terminal CENP-E antibodies shortly after they were released from the cell cycle block (Table I). Time-lapse videomicroscopy at low magnification (10× phase contrast) showed that the injection did not interfere with progression of the cells into mitosis as they took the same amount of time to enter mitosis as did adjacent uninjected cells. However, cells injected with the affinity-purified antibodies to CENP-E were arrested in mitosis in a period that varied between 4 to 17 h while mock-injected cells or cells injected with nonimmune IgG completed mitosis in ∼1 to 2 h. All of the cells arrested in mitosis by CENP-E antibodies eventually died by apoptosis. These results show that CENP-E is not required for progression of cells into mitosis but is essential for completion of mitosis.
Table I.
Time Course of Microinjected Cells
| No. cells | Time between cell rounding and anaphase onset (h:min) | |
|---|---|---|
| 21 Uninjected | 0:30–1:00 | |
| 8 Injected | 4:00–7:00 | |
| 19 Injected | 7:00–15:38* |
Injection of anti–CENP-E affinity-purified polyclonal antibodies causes mitotic arrest in HeLa cells synchronized at the G1/S boundary. HeLa cells were injected cytoplasmically with antibodies after release from a double thymidine block and then returned to the culture incubator. The culture was incubated for 7 h and then overlaid with mineral oil and observed under 10× phase microscopy for 24 h on a heated stage (37°C).
These cells underwent apoptosis rather than entering anaphase.
Chromosomes Lacking CENP-E at Kinetochores Fail to Align
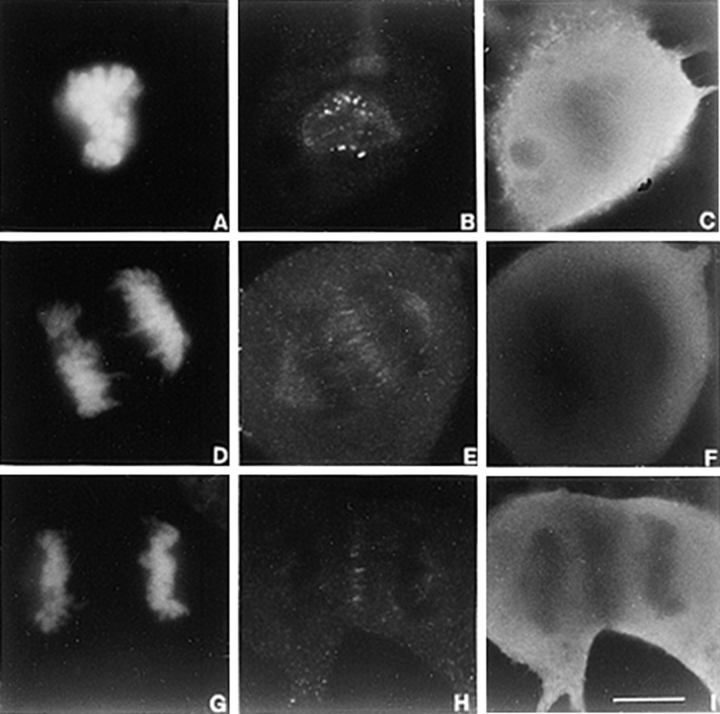
The nature of the mitotic defect that resulted from the depletion of CENP-E from kinetochores was next examined. Tubulin staining revealed that the mitotically blocked cells had established a bipolar spindle (Fig. 3 C). The overall morphology of the spindle in many cells took on a ragged appearance that is likely due to the prolonged mitotic block. Nevertheless, this result showed that CENP-E is not essential for bipolar spindle formation. Examination of the distribution of chromosomes in >100 blocked cells showed that they were either positioned very close to the poles or scattered between the poles (Fig. 3, D and F). The accumulation of large numbers of chromosomes at the poles in the mitotically blocked cells contrasts with a normal prometaphase cell where chromosomes are rarely seen so close to the poles but lie mostly within the spindle (Fig. 3, A and B). The chromosomes at the poles did not result from premature separation of the chromatids because immunofluorescence staining with anti-centromere autoimmune serum (ACA) produced the characteristic double dot staining pattern that is indicative of paired chromosomes (Fig. 3 E, open arrows). In addition to the accumulation of monopolar chromosomes in cells where CENP-E function was inhibited, there were also chromosomes that were strewn throughout the spindle. Normally, the chromosomes that are positioned in between the two poles at the time of nuclear envelope breakdown rapidly establish bipolar connections and immediately begin to congress towards the spindle equator (Fig. 3 B) (Alexander and Rieder, 1991; Rieder and Salmon, 1994) The loss of CENP-E affected their ability to align, as we never observed an injected cell with aligned chromosomes at the metaphase plate.
Figure 3.
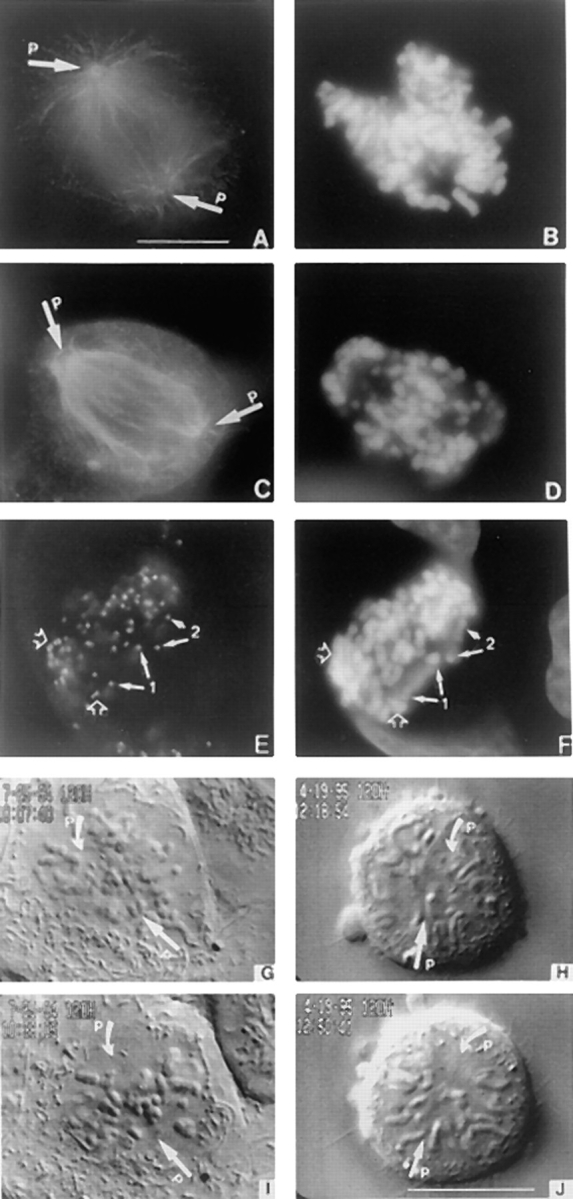
Loss of CENP-E disrupts chromosome alignment. (A) Tubulin staining of a normal prometaphase U2OS cell and (C) one blocked in mitosis by injection of carboxy-terminal DraB antibodies. (E) Mitotically blocked U2OS cell stained with anti-centromere autoimmune serum to reveal presence of paired centromeres (open arrows) and separated centromeres (chromosomes 1 and 2, solid arrows). (B, D, and F) Chromosomes visualized by DAPI. (G through J) Selected VE-DIC images from a videotape showing a normal U2OS cell aligns its chromosomes in ∼30 min (G and I) at room temperature. Chromosomes that lacked CENP-E failed to align during the same period (H and J) and never aligned. Arrows (P) denote positions of the separated poles. Bars, 10 μm.
To further characterize the behavior of chromosomes whose kinetochores were depleted of CENP-E, the motions of chromosomes in normal and antibody-injected U2OS cells were monitored by time-lapse video enhanced (VE)-DIC microscopy. In a normal cell, chromosomes were found to be completely aligned into a metaphase plate at the spindle equator ∼30 min after nuclear envelope breakdown (Fig. 3, G and I). In an injected cell that was already blocked in mitosis for 2 h before viewing, the mono-oriented chromosomes never moved away from their pole to establish bipolar connections during a 2-h period of continuous observation. In contrast, chromosomes within the spindle were connected to both poles as they oscillated back and forth between the poles at velocities typical of chromosomes in uninjected cells (1.7 vs. 1.6 μm/ min). However, these bipolar connections are defective as the chromosomes failed to align into a metaphase plate during the 2-h period of observation (Fig. 3, H and J). As these chromosomes retained the ability to oscillate between the poles during the extended period of the block, the loss of CENP-E from the kinetochores does not appear to affect their ability to move with normal velocities and maintain a bipolar connection. However, the inability of these chromosomes to align into a metaphase plate demonstrates that kinetochores that lack CENP-E are defective in the mechanisms that govern their movements.
Bipolar Attached Centromeres Lacking CENP-E Separate
In ∼25% of the mitotically blocked cells that were injected with CENP-E antibodies, some of the chromosomes within the spindle appeared to have separated at their centromeres because ACA produced single dots of staining instead of the typical double dot pattern that was indicative of paired centromeres (Fig. 3 E, solid arrows). The failure to detect a double dot pattern was not because one of the centromeres was obscured as a second dot of ACA staining was not found within close proximity in any focal plane. In all cases, the single foci of ACA staining were located at the extreme ends of what appeared to be centromeres that had separated from each other and were pulled towards opposite poles. However the chromosomes located at the poles remain paired based on the double dot ACA staining at their centromeres (Fig. 3 E, open arrows).
An example of a cell that was examined by time-lapse VE-DIC microscopy (Fig. 4, A–N) showed that the chromosomes within the spindle were indeed separated at their centromeres (Fig. 4, B and I, solid arrows) and along the length of their arms but remained connected near the telomeres (Fig. 4, B and I, open arrows). It is noteworthy that all separated metacentric chromosomes remained connected at both telomeres. However, acrocentric chromosomes were invariably found to be connected only by the telomere region of the long arm as the connections at the telomeres of the short arm were never observed.
Figure 4.
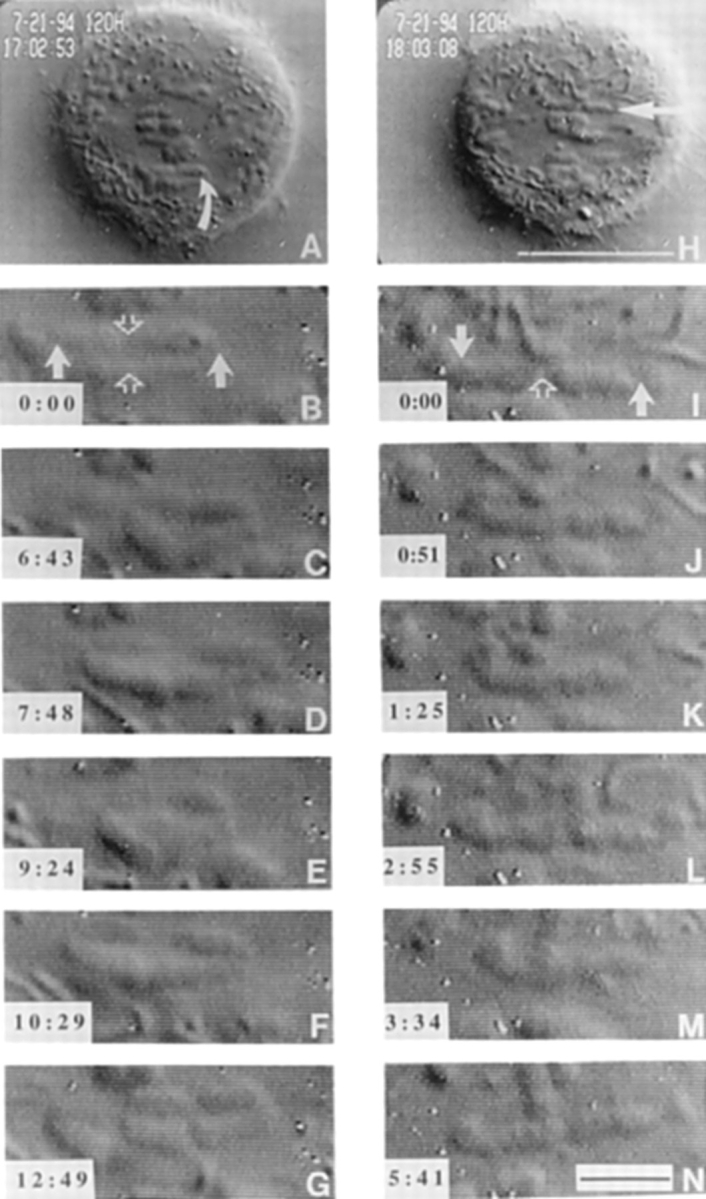
VE-DIC microscopy of chromosome dynamics in microinjected cells. The metacentric chromosome in A (curved arrow) exhibited uncoordinated movements shown in B through G. Centromeres of this chromosome are indicated with solid arrows, and the telomere connections are indicated by an open arrow in B. The centromeres of this chromosome stretch and compress the chromosome to show separation between the centromeres and sister chromatid connections (C and E) Time of observation is indicated in the lower left hand corner (min:s). The acrocentric chromosome in H, indicated by an arrow, was also followed. This chromosome only has a single telomere holding the two sister chromatids together (open arrow in I). The uncoordinated movements of the centromeres (solid arrows in I) stretch and bend the chromosome into a “u” shape (K and M) twice during the time of observation. Bars: (H) 10 μm; (N) 2 μm.
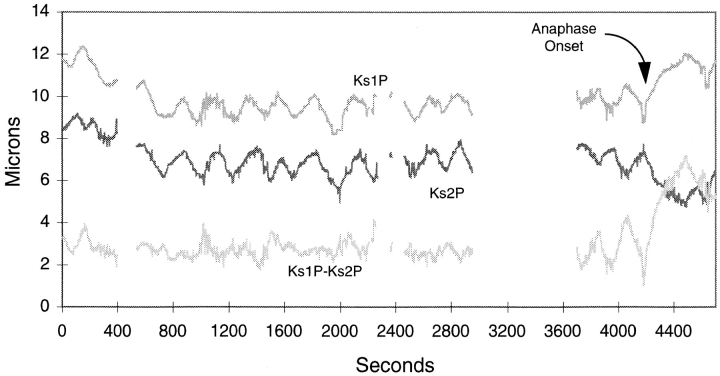
To search for clues that would help us understand how depletion of CENP-E might cause the bipolar chromosomes to prematurely separate, several parameters of kinetochore movement were quantitated. We were able to track the movements of sister kinetochores of partially separated chromatids in cells like that shown in Fig. 4 for nearly 50 min before the cell went into anaphase or the chromosomes went out of focus. First, we determined that the separated chromosomes were still connected to both poles as they were found to oscillate between the poles at rates that were not significantly different than for a normal bipolar chromosome. The kinetochores on partially separated sister chromatids (Fig. 5, Ks1P and Ks2P) oscillated back and forth between the poles with average velocities (0.65 ± 0.2 μm/min, n = 12) similar to those exhibited by kinetochores on bipolar-oriented chromosomes in uninjected cells (0.8 ± 0.2 μm/min, n = 5). In addition, the plots (Fig. 5, Ks1P and Ks2P) showed that sister kinetochores on the partially separated sister chromatids oscillated at frequencies that were similar to normal paired kinetochores; the duration of kinetochore movement in one direction of oscillation was 1.7 ± 0.7 min for both injected and uninjected cells. We also quantitated the distance that separated the two kinetochores from one of the poles and plotted these measurements over time. While both kinetochores were separated from each other by the partially separated arms, they were able to maintain a relatively constant distance of 3 μm between them (Fig. 5, Ks1P-Ks2P). This indicated that for the majority of time (63%), the two kinetochores exhibit coordinated motions where one sister is able to sense the direction of motion of the other sister (Skibbens et al., 1993). There were discrete points in time when the sister kinetochores moved away from each other, stretching their chromatid arms (Fig. 5, peaks in Ks1P-Ks2P plot) and also towards each other so that the chromosome appeared compressed (Fig. 5, valleys in Ks1P-Ks2P plot; and Fig. 4, B–E, and I–L). The overall similarity in velocity of movement, frequency of oscillation and frequency of coordinated kinetochore movements between the prematurely separated chromosomes and a normal bipolar chromosome (Skibbens et al., 1993) indicates that substantial depletion of CENP-E does not impair these kinetochore motility parameters.
Figure 5.
Plot of kinetochore to pole distance over time for two kinetochores (Ks1P and Ks2P) of a stretched chromosome in a cell that was injected with anti–CENP-E antibodies. Gaps in the plot indicate when kinetochores went out of the focal plane. Ks1P-Ks2P indicated the distance between the two kinetochores. Also indicated is the time when anaphase onset occurred and sister chromatids separated at their telomeres.
Unlike the bipolar chromosomes that oscillated for extended periods of time, the video images also showed that chromosomes that were positioned near the poles never exhibited any motion (data not shown). Separation of the bipolar chromosomes in these cells was not due to premature anaphase, because in rare instances, a cell overcame its checkpoint arrest and entered anaphase (Figs. 5 and 6). The telomeric connections that once held the separated chromosomes together were synchronously dissolved and the completely separated chromatids moved towards opposite poles at rates that were typical of that seen in normal anaphase (Figs. 5 and 6, A–E).
Figure 6.
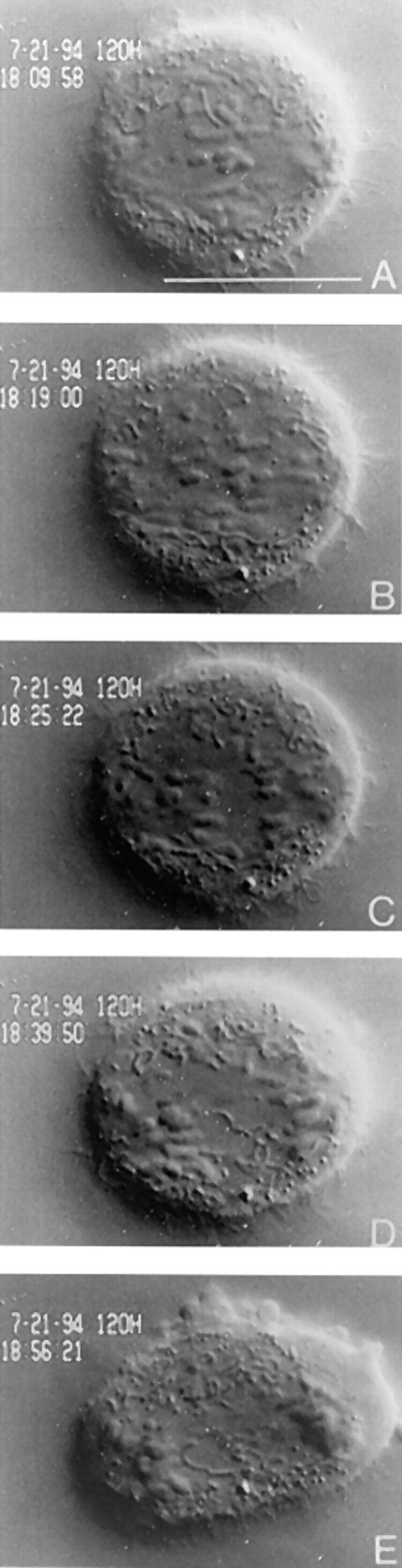
Separation of bipolar chromosomes depleted of CENP-E is not due to a precocious anaphase. The cell viewed in Fig. 4 was followed for 2 h after which the telomere connections of unzipped chromosomes synchronously disjoined, and sister chromatids moved towards opposite poles in a manner indistinguishable from anaphase in an uninjected cell. The cell also underwent normal anaphase B events (E) and cytokinesis. Bar, 10 μm.
Amino-Terminal Truncated CENP-E Behaves as a Dominant-Negative Mutant
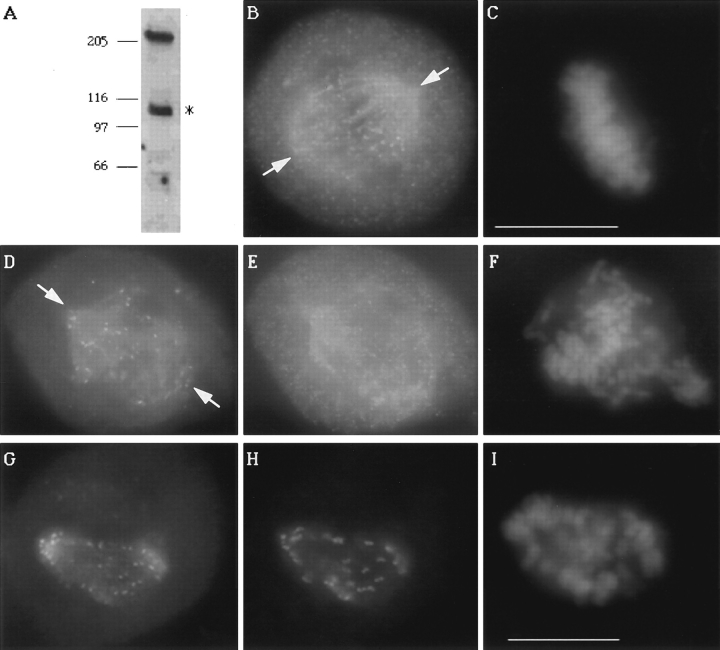
The disruption of kinetochore motility when CENP-E was depleted from kinetochores led us to adopt a second approach to directly test the contribution of its motor domain to kinetochore function. This method relied on the ability of an overexpressed CENP-E mutant to saturate the binding sites on kinetochores and competitively block endogenous CENP-E from assembling onto kinetochores. We deleted the amino-terminal 803 amino acids from CENP-E that included the kinesin-like motor domain and replaced the deleted portion with green fluorescent protein (GFP; Chalfie et al., 1993). Western blots of lysates prepared from transiently transfected HeLa cells showed that the GFP: CENP-E NΔ 803 mutant expressed a protein of the expected size (Fig. 7 A). Examination of the distribution of the GFP:CENP-E NΔ 803 (Fig. 7, D and G) in transfected mitotic cells showed that it accumulated at kinetochores as confirmed by colocalization with ACA staining (Fig. 7 H). Because the mutant CENP-E lacked its amino-terminal 803 amino acids, an antibody that specifically recognized this domain was used to detect the distribution of endogenous CENP-E. This antibody was able to detect endogenous CENP-E at the kinetochores of an untransfected metaphase cell (Fig. 7, B and C). In contrast, overexpression of the CENP-E mutant saturated the kinetochore binding sites (Fig. 7 D) as endogenous CENP-E was not detected at kinetochores (Fig. 7 E).
Figure 7.
Kinetochores saturated with a truncated CENP-E mutant that lacked its motor domain fail to align chromosomes. (A) Western blot of a transfected HeLa lysate probed with rat anti-GFP antibody to detect the 220-kD GFP: CENP-E NΔ 803. The asterisk denotes an endogenous 110-kD protein that crossreacts with the GFP antibody that is also present in untransfected lysates (not shown). (B) Localization of CENP-E on metaphase chromosomes of an untransfected cell by rabbit anti–CENP-E neck antibodies. (D) Direct visualization of GFP:CENP-E distribution in a transfected mitotic cell, (E) that was also stained with rabbit anti–CENP-E neck antibodies to detect endogenous CENP-E. (G) GFP:CENP-E colocalizes with ACA staining (H). (C, F, and I) DAPI staining. Arrows in B, D, and G denote the two separated spindle poles (P). Cells in B and E are from the same coverslip, and exposure times were identical. Bar, 10 μm.
The reproducible increase in the mitotic index of cells that expressed the CENP-E NΔ 803 mutant relative to untransfected or mock-transfected cells suggested that the accumulation of the mutant protein blocked cells in mitosis. In all cases (>100 cells), the occupation of the kinetochore by the mutant did not disrupt bipolar spindle formation but selectively disrupted chromosome alignment. Inspection of the chromosomes in fixed preparations of cells that were saturated with the CENP-E mutant at their kinetochores showed that chromosomes were found at the poles as well as in between the poles (Fig. 7, F and I). This data provides independent confirmation of the microinjection data. As the region that was deleted contains the motor domain that was shown to exhibit ATP-sensitive microtubule binding in vitro (Liao et al., 1994), the results strongly suggest that the motor domain of CENP-E is critical for proper kinetochore function. It is formally possible that the ∼350 amino acids that extend beyond the motor domain might contribute to some non-motor function of CENP-E that is also important for kinetochore function. The ability of the CENP-E mutant to compete effectively with the endogenous CENP-E for kinetochore-binding sites suggests that the mutant protein retains the same interactions with other proteins at the kinetochore as with the endogenous CENP-E. Thus, the chromosome defects observed when CENP-E is depleted from kinetochores by microinjecting antibodies is unlikely the result of compromising the overall structural integrity of the kinetochore. The collective data obtained by antibody injection and overexpressing the CENP-E NΔ803 mutant show that we have specifically disrupted CENP-E function at kinetochores and demonstrated that this molecule is critical for monopolar chromosomes to establish bipolar connections and for bipolar chromosomes to align into a metaphase plate.
Discussion
CENP-E Functions to Align Chromosomes
In this study, we have directly examined the kinetochore function of CENP-E in vivo by characterizing the behavior of chromosomes whose kinetochores were depleted of CENP-E or were saturated with a CENP-E mutant that lacked its motor domain. In both approaches, cells arrested in mitosis as they failed to align their chromosomes at the spindle equator. Our current findings differ from early experiments, which showed that cells microinjected with a CENP-E monoclonal antibody became blocked at metaphase with aligned chromosomes. The disparity between the data can be accounted for by differences in experimental approaches. In the earlier study (Yen et al., 1991), microinjection of the monoclonal antibody did not block assembly of CENP-E at kinetochores, as was the case here. Instead, the monoclonal antibody was bound to the kinetochore-associated CENP-E at what we now know to be a segment of the central rod domain. When the rod domain of CENP-E was bound by this antibody, it did not interfere with its normal function, which is to align chromosomes at the spindle equator. However, the presence of this antibody at the kinetochores was sufficient to prevent the aligned chromosomes from separating. Similar outcomes have been reported for other antibodies, which when bound to kinetochores or centromeres arrested cells in mitosis with aligned chromosomes (Bernat et al., 1989; Tomkiel et al., 1994).
CENP-E Is Essential for Two Steps during Chromosome Alignment
Our observation that chromosomes that lacked CENP-E at their kinetochores were reproducibly trapped at the poles or within the spindle reveal two discrete steps during chromosome alignment, when CENP-E function is critical. In normal cells, chromosomes that are positioned near a pole at the onset of nuclear envelope breakdown establish a transient monopolar connection that is converted to a bipolar connection when its unoccupied kinetochore captures a microtubule from the opposite pole (Nicklas, 1988; Nicklas and Ward, 1994). As the frequency that the unoccupied kinetochore encounters a microtubule that traverses across the entire spindle is low, it is critical that a stable connection is made when the kinetochore comes in contact with a microtubule (Nicklas and Kubai, 1985). One likely kinetochore function of CENP-E is to use its motor domain to capture and stabilize kinetochore–microtubule interactions. This possibility is supported by in vitro data that show the motor domain of CENP-E directly binds to microtubules (Liao et al., 1994). When CENP-E is depleted from kinetochores, the ability of the unoccupied kinetochore of a monopolar chromosome to capture microtubules and establish a bipolar connection is reduced (Fig. 8, A and B).
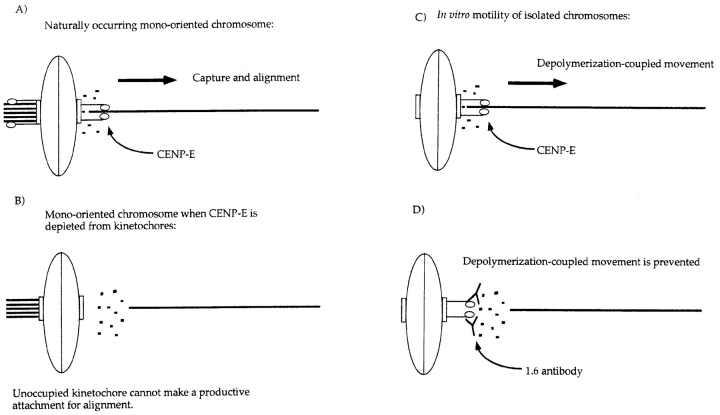
Figure 8.
Diagram illustrating the similar effects of inhibiting CENP-E function in vivo (this study) and in vitro (Lombillo et al., 1995). (A) Mono-oriented chromosomes (depicted as an oval) occur as a natural consequence of the random positioning of chromosomes after nuclear envelope breakdown. A chromosome that is captured and drawn close to one pole has one kinetochore (rectangle) saturated by microtubules from the pole it faces due to its proximity to that pole. The sister kinetochore must attain a microtubule connection from the opposite pole to congress the chromosome to the spindle equator. Because of the increase of catastrophic shrinkage during mitosis, a microtubule spanning the full length of the spindle (15–20 μm in the cells used in this study) is a rare event. We postulate that this creates a situation in which CENP-E (drawn as tadpoles sticking out of the kinetochore) is necessary to stabilize this connection by applying a poleward vector of force to the newly captured microtubule, creating tension along the microtubule as well as across the centromere. Once the microtubule is stabilized, other kinetochore proteins and motors engage the microtubule to induce movement. This model is based on the following observations: (a) when CENP-E is experimentally depleted from the kinetochore, mono-oriented chromosomes are abundant at both poles, a phenomenon not seen in control cells, and these mono-oriented chromosomes do not move from the attached pole to the metaphase plate; and (b) CENP-E–depleted chromosomes move with velocities comparable to controls.A role for CENP-E in maintaining attachment to a shrinking microtubule is also seen in the in vitro studies of Lombillo et al. (1995). (C) In the in vitro system, isolated chromosomes (ovals) capture the plus ends of microtubules nucleated from Tetrahymena pellicles at their kinetochores (rectangles) and mediate depolymerization-coupled movement when the tubulin is diluted in the absence of nucleotides. This movement is believed to be mediated in part by the motor domain of CENP-E (tadpoles), because if the chromosomes are first incubated with a polyclonal that spans a portion of the neck and stalk region of CENP-E, the chromosome detaches from the microtubule when buffer is perfused into the chamber to dilute the tubulin in (D). This is analogous to the situation that the unoccupied kinetochore of a mono-oriented chromosome that is depleted of CENP-E faces. Transient microtubules might be made but they cannot be stabilized in the absence of CENP-E.
In normal cells, chromosomes that are positioned within the spindle after nuclear envelope breakdown encounter microtubules at higher frequencies than the unoccupied kinetochore of a monopolar chromosome and therefore establish rapid bipolar connections (Cassimeris et al., 1994; Rieder and Salmon, 1994). Even when CENP-E is depleted from these kinetochores, the higher frequency of microtubule encounters within this region of the spindle appears to be sufficient to establish and maintain bipolar connections. These connections may be mediated by other kinetochore proteins or by a residual amount of endogenous CENP-E that is below the limits of our detection. Although these bipolar connections are sufficient to sustain relatively normal rates of oscillations, the CENP-E depleted kinetochores are unable to align at the spindle equator. The reason for this very specific defect is unclear, but it may be due to the inability to establish critical kinetochore microtubule connections that are essential for alignment. A normal mammalian kinetochore can be attached to as many as 20 to 30 microtubules (McIntosh, 1991; McEwen, 1997). These microtubule connections are not static, as they undergo cycles of release and capture at the kinetochores even after the chromosomes have aligned. At metaphase, a microtubule is estimated to have a half-life of ∼5 min so that it spends ∼80% of its time attached to the kinetochore (Zhai et al., 1995). Whether this represents the critical number of kinetochore microtubules that are required for chromosome alignment is unknown. If the role of CENP-E is to maintain stable kinetochore microtubule connections, kinetochores that lack CENP-E may be unable to establish the connections that are essential for alignment at the equator. This explanation is consistent with our interpretation of why monopolar chromosomes that lack CENP-E fail to establish bipolar connections as well as in vitro data that suggest that CENP-E is important for maintaining stable kinetochore connections (Lombillo et al., 1995). On the other hand, the failure to align is also consistent with the argument that kinetochores that lack CENP-E exhibit an unregulated poleward force that sustains prolonged oscillations but cannot maintain chromosomes at a steady position at the equator.
Bipolar Chromosomes Can Partially Separate in the Absence of CENP-E
The separation of centromeres and chromatid arms when CENP-E was depleted from the kinetochores were only observed for chromosomes that established bipolar attachments as monopolar chromosomes that accumulated in the same cell remained paired. This aberration is not due to a general consequence of a prolonged mitotic block for two reasons. First, chromosomes that are arrested at the metaphase plate by microinjection of centromere-specific antibodies remain paired even after several hours of the block. It is perhaps noteworthy that in the two cases that were previously examined, CENP-E was still present at the kinetochores of the metaphase-arrested chromosomes (Yen et al., 1991; Tomkiel et al., 1994). Second, when cells are blocked in mitosis for extended periods with microtubule-destabilizing drugs, the connections between the chromsome arms separate but the centromeres remain paired (Rieder and Palazzo, 1992).
As we have not observed chromosomes that lack CENP-E in the process of separating, we can only speculate about their origin. We favor an explanation where we consider CENP-E's role in kinetochore motility although it could also contribute in an unknown way to the structural glue that holds the centromeres together. The glue hypothesis alone is inconsistent with our observation that only some chromosomes in a cell become separated while others, mostly at the poles, remain paired. Separation is likely due to secondary effects that resulted from the prolonged periods of oscillations exhibited by the chromosomes that achieved bipolar connections. This explanation is consistent with the lower frequency at which cells with separated chromosomes are seen. Our observations are consistent with experiments in which cells blocked in metaphase by a low dose of taxol show a transient increase in the distance between sister kinetochores (Waters et al., 1996a ,b). Separation of these sister kinetochores is thought to occur as a consequence of the poleward force that is simultaneously exerted on both kinetochores as a result of microtubule flux. If kinetochores lacking CENP-E tended to move towards opposite poles at higher frequency, the extended periods of oscillations due to a mitotic block might provide sufficient time for the centromeres to come apart. Another possibility is that CENP-E–deficient kinetochores produce an unchecked poleward force that increases the frequency or magnitude of force at which both kinetochores are simultaneously pulling towards their respective poles. The inappropriate application of poleward force would initially pull apart the connections holding together the centromeres, which would then propagate along the length of the chromosome arms.
Proposed Mechanism of CENP-E Function
We propose that CENP-E plays a critical role in maintaining stable kinetochore–microtubule connections that allows chromosomes to establish bipolar connections and to align at the spindle equator. This possibility is supported by in vitro experiments that showed the importance of CENP-E in microtubule depolymerization–dependent motility of kinetochores (Lombillo et al., 1995). In this assay, depolymerization of a single microtubule that was attached at the kinetochore of an isolated chromosome was sufficient to induce movement of the chromosome in the direction of the shrinking microtubule. Although this assay was performed in the absence of detectable ATP and low tubulin concentrations, there are interesting parallels that can be drawn between the in vitro and in vivo data. Both the in vitro and in vivo data revealed the importance of CENP-E's motor domain in kinetochore function. The ability of an isolated chromosome to move while remaining attached to a depolymerizing microtubule was completely inhibited if chromosomes were preincubated with CENP-E antibodies that were directed against the neck region that connected the motor domain to the stalk. The antibodies disrupted motility most likely by interfering with CENP-E's motor domain. Likewise, kinetochores saturated with a motorless CENP-E mutant were defective in their ability to align chromosomes. Both studies also showed that CENP-E was not critical for kinetochores to bind microtubules. Inhibition of CENP-E function with antibodies in vitro did not prevent microtubule capture by kinetochores. Similarly, kinetochores depleted of CENP-E in vivo were able to bind microtubules, although the connections were defective.
The basis of the in vivo defects observed for kinetochores that lack CENP-E may be explained by the in vitro finding that CENP-E is essential for coupling the kinetochore to a shrinking microtubule. Specifically, the in vitro studies showed that CENP-E was essential for kinetochores to remain attached to a shrinking microtubule or against a counterflow of buffer (i.e., under tension). This suggests that the connection between the kinetochore that was defective in CENP-E function and its microtubule was disrupted when tension was exerted on it. Perhaps, the critical function of CENP-E in vivo is to maintain and stabilize the connections between the kinetochore and microtubules. As tension is known to stabilize kinetochore microtubule connections in vivo (Nicklas and Ward, 1994), CENP-E could fulfill its role as a kinesin-like protein generating motive force at the kinetochore.
Acknowledgments
The authors are grateful to S. Jablonski, D. Gately, E. Golemis, and T. Coleman for critical reading of the manuscript.
This work was supported by National Institutes of Health grant GM24364 to E.D. Salmon. T.J. Yen was supported by grants GM44762, Leukemia Society Scholar's Award, core grant CA06927, Council for Tobacco Research, and an Appropriation from the Commonwealth of Pennsylvania.
Abbreviations used in this paper
- ACA
anti-centromere autoimmune serum
- DIC
differential interference contrast
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- MBP
maltose-binding protein
- VE
video enhanced
Footnotes
Address all correspondence to Tim J. Yen, Fox Chase Cancer Center, 770 Burholme Avenue, Philadelphia, PA 19111. Tel.: (215) 728-2590. Fax: (215) 728-3616.
References
- Alexander SP, Rieder CL. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol. 1991;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ault JG, Nicklas RB. Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma (Berl) 1989;98:33–39. doi: 10.1007/BF00293332. [DOI] [PubMed] [Google Scholar]
- Bernat RL, Borisy GG, Rothfield NF, Earnshaw WC. Injection of anticentromere antibodies in interphase disrupts events required for chromosome movements at mitosis. J Cell Biol. 1990;111:1519–1533. doi: 10.1083/jcb.111.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L, Rieder CL, Salmon ED. Microtubule assembly and kinetochore directional instability in vertebrate monopolar spindles: implications for the mechanism of chromosome congression. J Cell Sci. 1994;107:285–297. doi: 10.1242/jcs.107.1.285. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirschen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1993;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High efficiency transformation of mammalian cells with plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, C.A., B. Schaar, T.J. Yen, and W.C. Earnshaw. 1998. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma (Berl.). In press. [DOI] [PubMed]
- Desai A, Mitchison TJ. A new role for motor proteins as couplers to depolymerizing microtubules. J Cell Biol. 1995;128:1–4. doi: 10.1083/jcb.128.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ, Sammack PJ, Borisy GG. Chromosomes move poleward in anaphase along stationary microtubules that coordinately disassemble from their kinetochore ends. J Cell Biol. 1987;104:9–18. doi: 10.1083/jcb.104.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden JH, Bowser SS, Rieder CL. Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells. J Cell Biol. 1990;111:1039–1045. doi: 10.1083/jcb.111.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly/ disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Li G, Yen TJ. Mitotic regulation of microtubule crosslinking activity of CENP-E kinetochore protein. Science. 1994;265:394–398. doi: 10.1126/science.8023161. [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1cells and its implications for the mechanism of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR. Structural and mechanical control of mitotic progression. Cold Spring Harbor Symp Quant Biol. 1991;56:613–619. doi: 10.1101/sqb.1991.056.01.070. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Evans L, Schultze E, Kirschner MW. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45:515–527. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Kubai DF. Microtubules, chromosome movement, and reorientation after chromosomes are detached from the spindle by micromanipulation. Chromosoma (Berl) 1985;92:313–324. doi: 10.1007/BF00329815. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Ward SC. Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Palazzo RE. Colcemid and the mitotic cycle. J Cell Sci. 1992;102:387–392. doi: 10.1242/jcs.102.3.387. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP, Rupp G. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1989;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheay W, Nelson S, Martinez I, Chu TH, Bhatia S, Dornburg R. Downstream insertion of the adenovirus tripartite leader sequence enhances expression in universal eukaryotic vectors. Biotechniques. 1993;15:856–862. [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation of mitotic newt lung cell: a push–pull mechanism. J Cell Biol. 1993;122:859–876. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer ER, Wordemann L, Schroer TA, Sheetz MP. Localization of cyoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Thrower DA, Jordan MA, Schaar BT, Yen TJ, Wilson L. Mitotic HeLa cells contain a CENP-E-associated minus-end directed microtubule motor. EMBO (Eur Mol Biol Organ) J. 1995;14:918–926. doi: 10.1002/j.1460-2075.1995.tb07073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiel J, Cooke CA, Saitoh H, Bernat RL, Earnshaw WC. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: A Xenopuskinesin related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Waters JC, Mitchison TJ, Rieder CL, Salmon ED. Kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol Biol Cell. 1996a;7:154–1558. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Skibbens RV, Salmon ED. Oscillating mitotic newt lung cell kinetochores are, on average, under tension. J Cell Sci. 1996b;109:2823–2831. doi: 10.1242/jcs.109.12.2823. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–105. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Schaar BT. Kinetochore function: molecular motors, switches and gates. Curr Opin Cell Biol. 1996;8:381–388. doi: 10.1016/s0955-0674(96)80014-7. [DOI] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO (Eur Mol Biol Organ) J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase–anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]