Abstract
We expressed the γ-subspecies of protein kinase C (γ-PKC) fused with green fluorescent protein (GFP) in various cell lines and observed the movement of this fusion protein in living cells under a confocal laser scanning fluorescent microscope. γ-PKC–GFP fusion protein had enzymological properties very similar to that of native γ-PKC. The fluorescence of γ-PKC– GFP was observed throughout the cytoplasm in transiently transfected COS-7 cells. Stimulation by an active phorbol ester (12-O-tetradecanoylphorbol 13-acetate [TPA]) but not by an inactive phorbol ester (4α-phorbol 12, 13-didecanoate) induced a significant translocation of γ-PKC–GFP from cytoplasm to the plasma membrane. A23187, a Ca2+ ionophore, induced a more rapid translocation of γ-PKC–GFP than TPA. The A23187-induced translocation was abolished by elimination of extracellular and intracellular Ca2+. TPA- induced translocation of γ-PKC–GFP was unidirected, while Ca2+ ionophore–induced translocation was reversible; that is, γ-PKC–GFP translocated to the membrane returned to the cytosol and finally accumulated as patchy dots on the plasma membrane. To investigate the significance of C1 and C2 domains of γ-PKC in translocation, we expressed mutant γ-PKC–GFP fusion protein in which the two cysteine rich regions in the C1 region were disrupted (designated as BS 238) or the C2 region was deleted (BS 239). BS 238 mutant was translocated by Ca2+ ionophore but not by TPA. In contrast, BS 239 mutant was translocated by TPA but not by Ca2+ ionophore. To examine the translocation of γ-PKC–GFP under physiological conditions, we expressed it in NG-108 cells, N-methyl-d-aspartate (NMDA) receptor–transfected COS-7 cells, or CHO cells expressing metabotropic glutamate receptor 1 (CHO/mGluR1 cells). In NG-108 cells , K+ depolarization induced rapid translocation of γ-PKC–GFP. In NMDA receptor–transfected COS-7 cells, application of NMDA plus glycine also translocated γ-PKC–GFP. Furthermore, rapid translocation and sequential retranslocation of γ-PKC–GFP were observed in CHO/ mGluR1 cells on stimulation with the receptor. Neither cytochalasin D nor colchicine affected the translocation of γ-PKC–GFP, indicating that translocation of γ-PKC was independent of actin and microtubule. γ-PKC–GFP fusion protein is a useful tool for investigating the molecular mechanism of γ-PKC translocation and the role of γ-PKC in the central nervous system.
Protein kinase C (PKC),1 a family of phospholipid-dependent serine/threonine kinases and of which there are at least 12 subspecies, plays an important role in various cellular signal transductions (Nishizuka, 1984, 1988, 1992). Regardless of ubiquitous expression of PKCs in various tissues, the central nervous system abundantly contains several unique PKCs. In particular, the γ-subspecies of PKC (γ-PKC) is present only in the central nervous system and is thought to be involved in many neuronal functions including the formation of neural plasticity and memory (Nishizuka, 1986; Abeliovich et al., 1993a ,b; Tanaka and Nishizuka, 1994).
PKC isozymes are divided into three subfamilies based on differences in the regulatory domain: conventional PKC (cPKC), novel PKC (nPKC), and atypical PKC (aPKC). Conventional PKCs have two common regions in the regulatory domain, C1 and C2. The C1 region has two cysteine-rich loops (zinc finger–like motifs) that interact with diacylglycerol (DG) or phorbol esters (Nishizuka, 1988; Ono et al., 1989). The C2 region mediates calcium binding (Ono et al., 1989) and is only present in cPKCs (Ono et al., 1988b ), although a region related to C2 region was recently reported in nPKC, a calcium-independent PKC (Parker and Dekker, 1997). Full activation of cPKCs, including γ-PKC, requires DG and calcium. The C1 region is also present in nPKC, and one of the cysteine-rich loops is found in aPKCs.
Conventional PKCs and nPKCs, whose regulatory domains contain C1, are known to be translocated from the cytosol to particulate fraction when activated by DG or phorbol esters (Kraft et al., 1982). Therefore, the translocation of PKCs is a good marker of whether these enzymes are activated. Although this phenomenon is well known, the mechanism and physiological significance of PKC translocation have not yet been clarified. By conventional enzymological or immunohistochemical methods, it is impossible to observe the translocation of PKC in real time, in the same cells, and in living states, except in the investigation using fluorescent probes that directly bind PKC (Chen and Poenie, 1993). In addition, these fluorescent compounds are suggested to inhibit the activity of PKC itself at high concentration.
To resolve these problems and to directly observe the translocation of γ-PKC in living cells, we produced a fusion protein of γ-PKC and green fluorescent protein (GFP). The GFP, isolated from jellyfish Aequorea victoria, has fluorescence without additional substrates and cofactors (Cubitt et al., 1995). Recent studies have revealed that GFP is a good candidate as a molecular reporter protein to monitor the alternation of protein localization, gene expression, and protein trafficking in living cells (Cubitt et al., 1995). In this study, we visualized and analyzed the translocation of γ-PKC–GFP fusion protein with confocal laser scanning fluorescence microscopy, using various stimulations, such as phorbol esters, Ca2+ ionophore, K+ depolarization, and receptor-mediated stimulus.
Materials and Methods
Materials
A23187 was purchased from Calbiochem (La Jolla, CA). N-methyl-d-aspartate (NMDA), 12-O-tetradecanoylphorbol 13-acetate (TPA), and 4α-phorbol 12, 13-didecanoate (4α-PDD) were purchased from Sigma Chemical Co. (St. Louis, MO). (1S, 3R)-1-aminocyclopentane-1,3-dicarboxylic acid (trans-ACPD), 2-amino-8-phosphonopentanoic acid (AP-5), and (RS)-α-methyl-4-carboxyphenyl-glycine ([RS]-MCPG) were from Tocris (Bristol, UK). Nicardipine and thapsigargin were from Wako Pure Chemical Ltd. (Osaka, Japan). ω-Conotoxin GVIA was from Peninsula Laboratories Inc. (Belmont, CA). Cytochalasin D and colchicine were from nacalai tesque (Kyoto, Japan). All other chemicals were of analytical grade.
Cell Culture
COS-7 and NIH3T3 cells were purchased from Riken cell bank (Tsukuba, Japan). Mouse neuroblastoma × glioma hybrid cells (NG 108-15 cells) were obtained from Dr. T. Amano (The Mitsubishi-Kasei Institute of Life Science, Tokyo, Japan). The CHO cells, stably expressing metabotropic glutamate receptor 1 (CHO/mGluR1 cells) (Tanabe et al., 1992) were a gift from Dr. Nakanishi (Department of Biological Science, Kyoto University Faculty of Medicine, Kyoto, Japan). COS-7 cells were cultured in DME containing 25 mM glucose, which was buffered with 44 mM NaHCO3 and supplemented with 10% FBS, in a humidified atmosphere containing 5% CO2 at 37°C. NG 108-15 cells were cultured in the same conditions as for COS-7 cells. NIH3T3 cells were cultured in DME supplemented with 10% calf serum instead of FBS. CHO/mGluR1 cells were cultured as described (Tanabe et al., 1992). All media were supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml), and the FBS used was not heat-inactivated.
Construct of Plasmids Encoding γ-PKC–GFP Fusion Protein
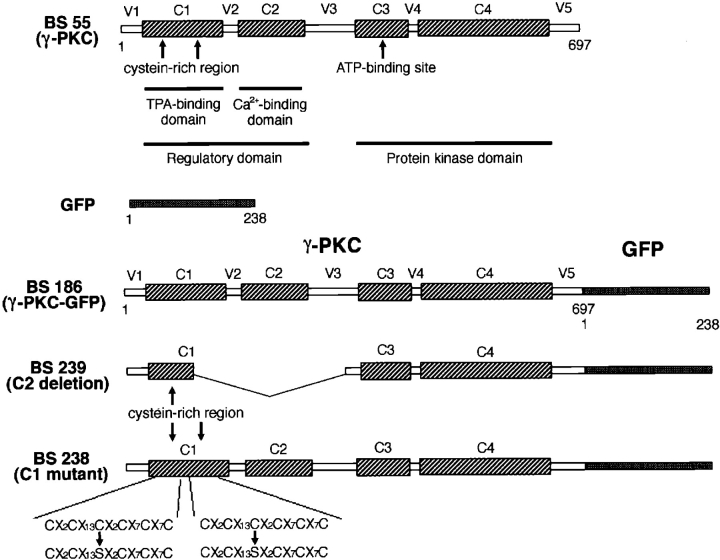
A plasmid containing GFP cDNA (pGFP 10.1) was donated by Dr. D.C. Prasher (Columbia University, New York) (Prasher et al., 1992). A cDNA fragment encoding GFP with a HindIII site in the 5′-terminal and an EcoRI site in the 3′-terminal end was obtained by PCR using pGFP 10.1 as a template. The sense and antisense primers used were 5′-TTAAGCTTATGGTGAGCAAGGGCCAGGAG-3′ and 5′-CCGAATTCTTACTTGTACAGCTCGTCCAT-3′, respectively. A rat γ-PKC cDNA was obtained from its cDNA clone of λCKRγ1 (Ono et al., 1988a ). After digestion with EcoRI, an insert fragment encoding rat γ-PKC was subcloned into an expression plasmid for mammalian cells, pTB 701 (Ono et al., 1988a ) (designated as BS 55) (Fig. 1). A cDNA fragment of γ-PKC with an EcoRI site in the 5′ terminus and a HindIII site in 3′ terminus was also produced by PCR using BS 55 as a template. The sense and antisense primers used were 5′-TTGAATTCATGGCGGGTCTGGGTCCTGG-3′ and 5′-TTAAGCTTATGGCGGGTCTGGGTCCTGG-3′, respectively. PCR products for both GFP and γ-PKC were together subcloned into the EcoRI site in pTB701 (BS 186) (Fig. 1).
Figure 1.
Constructs of γ-PKC–GFP fusion protein and its mutants. BS 186 was a γ-PKC–GFP fusion protein in which γ-PKC (BS 55) and GFP were bound at the COOH terminus of γ-PKC. In the C1 region of γ-PKC, there are two cysteine-rich sequences that interact with DG or phorbol esters, such as TPA. To produce BS 238 (C1 mutant), one cysteine residue in each of the two cysteine-rich regions was substituted with serine by site-directed mutagenesis. The C1 region of BS 238 no longer displayed activity for binding phorbol esters (Ono et al., 1989). The C2 region of γ-PKC, the Ca2+ binding domain, was deleted for the production of BS 239 (C2 deletion) as described in the text.
We next produced a construct encoding mutant γ-PKC–GFP, whose zinc finger–like motif in the C1 region was mutated. We used pTB966, a plasmid containing a cDNA for mutated C1 region, as a template for PCR (Ono et al., 1989). The protein derived from pTB966 was reported to have no binding activity for TPA (Ono et al., 1989). We also produced a mutant γ-PKC–GFP construct whose C2 region was deleted. For this, pTB971 was used as a template for PCR (Ono et al., 1989). The protein derived from pTB971 no longer had binding activity for Ca2+ (Ono et al., 1989). PCR was performed to obtain a cDNA fragment containing C1 and C2 regions with a sense primer (5′-TTGAATTCATGGCGGGTCTGGGTCCTGG-3′) and an antisense primer (5′-TTGGATCCTCTGCGCTCTGCCAG-3′) using pTB966 or pTB971 as templates. These PCR products were digested with EcoRI and BamHI and then subcloned into BS 186, whose EcoRI/ BamHI fragment in γ-PKC was removed. These constructs were designated as BS 238 (C1 mutant) and BS 239 (C2 deletion), respectively. All PCR products were verified by sequencing. The structures of three γ-PKC–GFP proteins (BS 186, BS 238, and BS 239) are summarized in Fig. 1.
Expression of γ-PKC–GFP Protein in Cultured Cells
Transient transfection into COS-7 cells was performed by electroporation. Plasmids (∼32 μg) encoding each type of γ-PKC–GFP or γ-PKC were transfected into 6 × 106 cells using a Gene Pulser (960 μF, 220 V; Bio-Rad Labs, Hercules, CA). Transfections into NG-108, NIH3T3, and CHO/ mGluR1 cells were carried out by lipofection using Tfx-TM-50 (Promega Corp., Madison, WI) according to the manufacturer's standard protocol. After the transfection, cells were cultured at 30°C to obtain the optimal fluorescence of GFP. The fluorescence of γ-PKC–GFP was detected 2 or 3 d after the transfection. Experiments were performed 3–5 d after the transfection.
Coexpression of NMDA Receptor and γ-PKC–GFP in COS-7 Cells
Mouse cDNAs encoding NMDARζ1 and NMDARε1 subunits, kindly donated by Dr. Mishina (Department of Pharmacology, Tokyo University, Faculty of Medicine, Tokyo, Japan) (Ikeda et al., 1992; Meguro et al., 1992; Yamazaki et al., 1992), were subcloned into an expression plasmid, pTB 701. The same amount of γ-PKC–GFP, NMDARζ1, and NMDARε1 plasmids (total 32 μg) was transfected into COS-7 cells by electroporation as described above. To prevent NMDA receptor–mediated cell death, transfected cells were cultured in the presence of 100 μM AP-5, an antagonist of NMDA receptor, until the experiment was performed.
Immunoblotting, Kinase Assay, and Immunoprecipitation of γ-PKC–GFP and γ-PKC
γ-PKC-GFP (BS 186) or γ-PKC (BS 55) cDNAs were transiently transfected into 6 × 106 COS-7 cells by electroporation. The same number of transfected cells were divided into two culture dishes 8 cm in diameter and cultured at 30°C for 3 d. After treatment with 5 μM TPA for 90 min at room temperature, cells were harvested with PBS(−) and centrifuged. The cell pellet/dish was resuspended in 200 μl homogenate buffer (250 mM sucrose, 10 mM EGTA, 2 mM EDTA, 50 mM Tris/HCl, 200 μg/ml leupeptin, 1 mM PMSF, pH 7.4). After the sonication (UD-210 TOMY SEIKO Co. Ltd., Tokyo, Japan; output 3, duty 50%, 10 times, at 4°C), samples were centrifuged at 19,000 g for 30 min at 4°C, and supernatant was collected as the cytosol fraction. The pellet was resuspended with 200 μl of homogenate buffer containing 0.5% Triton-X and used as particulate fraction after the sonication as described above.
For immunoblotting, the samples of each fraction were subjected to 10% SDS-PAGE in the same volume, and the separated proteins were electorophoretically transferred onto polyvinylidine difluoride (PVDF) filters (Millipore Corp., Bedford, MA). Nonspecific binding sites on the PVDF filters were blocked by incubation with 5% gelatine for 18 h. The PVDF filters were then incubated with anti–PKC-γ monoclonal antibody (diluted 1:2,000) (Hashimoto et al., 1988) or anti-GFP polyclonal antibody (diluted 1: 2,000) (CLONTECH Laboratories, Inc., Palo Alto, CA) for 30 min at 25°C. After washing with 0.01 M PBS containing 0.03% Triton X- 100, the filters were incubated with goat anti–mouse IgG (for γ-PKC antibody) or anti–rabbit IgG (for GFP antibody) for 15 min and then incubated for 15 min with rabbit peroxidase antiperoxidase complex. After three rinses, the immunoreactive bands were visualized with a chemiluminescence detection kit (ECL; Amersham, Buckinghamshire, UK).
Kinase assays of γ-PKC, γ-PKC–GFP, and its mutants expressed in COS-7 cells were performed as described previously (Kikkawa et al., 1983). The kinase activity in 10 μl of each fraction was assayed by measuring the incorporation of 32Pi into calf thymus H1 histone from [γ-32P]ATP in the presence of 8 μg/ml phosphatidylserine (PS), 0.8 μg/ml diolein (DO), and 0.5 mM Ca2+. Basal activity was measured in the presence of 0.5 mM EGTA instead of PS, DO, and Ca2+.
For immunoprecipitation of γ-PKC–GFP and γ-PKC, transfected cells in a dish 8 cm in diameter were harvested with 1 ml of homogenate buffer containing 1% Triton X-100 and were homogenized by pipetting. After a centrifuge at 19,000 g for 5 min at 4°C, the supernatant was rotated with anti–γ-PKC monoclonal antibody for 30 min at 4°C, then with protein A–Sepharose for an additional 30 min. Samples were centrifuged at 2,000 g for 5 min at 4°C, and pellets were washed three times with PBS(−). Finally, 10 μl of suspended pellet with 50 μl PBS(−) was used for kinase assay as described above.
Observation of γ-PKC–GFP Translocation
γ-PKC–GFP–transfected cells were spread onto the glass bottom culture dishes (MatTek Corp., Ashland, MA) and cultured for at least 16 h before the observation. For COS-7 cells and CHO/mGluR1 cells, the culture medium was replaced with normal Hepes buffer composed of: 135 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM Hepes, 10 mM glucose, pH 7.3. When necessary, CaCl2 or MgCl2 were eliminated. In the case of NG-108 cells, the buffer used was composed of: 165 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 5 mM Hepes, 10 mM glucose, pH 7.4.
The fluorescence of γ-PKC–GFP was monitored with a confocal laser scanning fluorescent microscope (model LSM 410 invert; Carl Zeiss, Jena, Germany) at 488-nm argon excitation using a 515-nm-long pass barrier filter. Translocation of γ-PKC–GFP was triggered by a direct application of various stimulants at high concentration into the Hepes buffer to obtain the appropriate final concentration. To observe NMDA-induced translocation, NMDA was applied into the dish in the presence of 10 μM glycine and absence of MgCl2. To induce K+ depolarization in NG-108 cells, the buffer was exchanged to a high K+-containing buffer composed of: 52 mM NaCl, 100 mM KCl, 1 mM MgCl2, 10 mM CaCl2, 5 mM Hepes, 10 mM glucose, pH 7.4. All experiments were performed at room temperature.
Immunostaining of γ-PKC–GFP–transfected Cells by Anti–γ-PKC Antibody
The γ-PKC–GFP–transfected COS-7 cells, cultured in glass bottom dishes, were stimulated by TPA or A23187 and it was confirmed that fluorescence was completely translocated. Then cells were fixed with 4% paraformaldehyde and 0.2% picric acid in 0.1 M phosphate buffer, pH 7.4, for 30 min. After two washes with 0.1 M PBS, pH 7.4, cells were treated with PBS containing 0.3% Triton X-100 and 5% normal goat serum for 10 min. Cells were sequentially incubated with anti–PKC-γ monoclonal antibody (Hashimoto et al., 1988) (diluted 1:1,000) for 40 min in PBS with 0.03% Triton X-100 (PBS-T) and 5% normal goat serum and then with Cy5-labeled goat anti–mouse IgG for 30 min at room temperature. The fluorescence of γ-PKC–like immunoreactivity was observed with a confocal laser scanning fluorescent microscope at 633-nm argon excitation and a 665-nm red glass filter.
Results
Characteristics of γ-PKC–GFP Fusion Protein
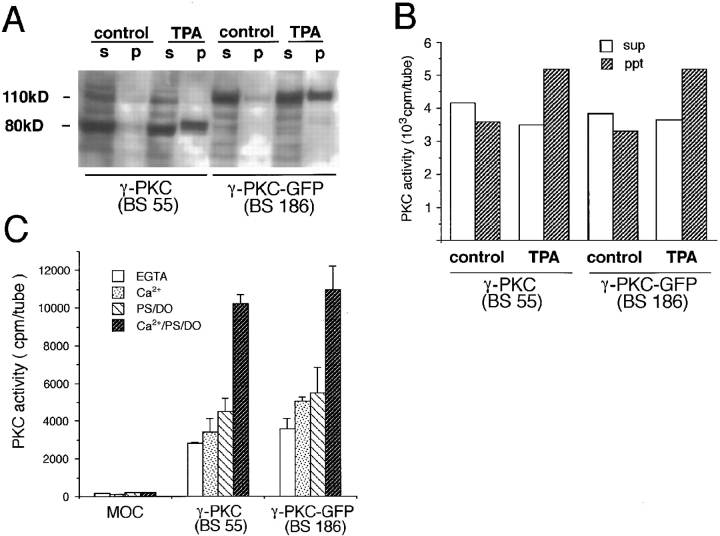
To examine whether γ-PKC–GFP had the characteristics of a phospholipid-dependent/calcium-activated protein kinase (PKC), we carried out immunoblotting and kinase assay of γ-PKC–GFP and γ-PKC expressed in COS-7 cells. As shown in Fig. 2 A, γ-PKC and γ-PKC–GFP were recognized as specific bands with the reasonable molecular masses of 80 and 110 kD, respectively. γ-PKC–GFP was also detected as a single 110-kD band by anti-GFP polyclonal antibody (data not shown). The amount of membrane-associated γ-PKC–GFP was increased after the treatment with TPA as seen in the case of γ-PKC (Fig. 2 A). The degradative products of γ-PKC–GFP and γ-PKC could not be detected by the antibodies against γ-PKC and GFP. Furthermore, kinase activities of γ-PKC–GFP and of γ-PKC were translocated from cytosol to membrane fractions (Fig. 2 B). The kinase assays revealed that both immunoprecipitated γ-PKC and γ-PKC–GFP were dependent on PS/DO and Ca2+ (Fig. 2 C). These results suggested that γ-PKC–GFP had similar enzymological properties to the native γ-PKC.
Figure 2.
Enzymological property of γ-PKC and γ-PKC–GFP transiently expressed in COS-7 cells. (A) Immunoblotting analysis by anti–γ-PKC antibody revealed that expressed γ-PKC (BS 55) and γ-PKC–GFP (BS 186) were proteins with molecular sizes of 80 and 110 kD, respectively. Treatment with 5 μM TPA for 90 min increased the amount of both γ-PKC and γ-PKC–GFP associated with the particulate fraction. p, pellet (particulate fraction); s, supernatant (cytosol fraction). (B) Kinase activity of expressed γ-PKC and γ-PKC–GFP in cytosol and particulate fractions. Kinase activities of both γ-PKC and γ-PKC–GFP were translocated from the cytosol to particulate fraction after treatment with 5 μM TPA for 90 min. ppt, pellet (particulate fraction) sup, supernatant (cytosol fraction). (C) Enzymological property of γ-PKC and γ-PKC–GFP immunoprecipitated by anti–γ-PKC antibody. Kinase activities of expressed γ-PKC and γ-PKC–GFP, which were immunoprecipitated by anti–γ-PKC antibody, were measured in the presence or absence of activators of γ-PKC. The kinase activity of γ-PKC–GFP maximized in the presence of PS, DO, and Ca2+, as did that of γ-PKC. The enzymological properties of γ-PKC and γ-PKC–GFP were very similar.
Translocation of γ-PKC–GFP Induced by TPA and A23187
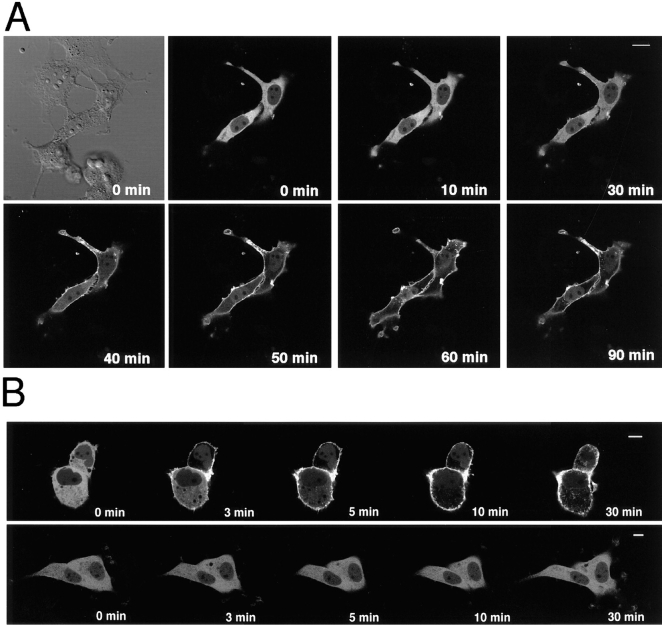
Intense fluorescence of γ-PKC–GFP was observed in the perikarya of the transfected COS-7 cells, and faint fluorescence was seen in the nuclei (Fig. 3, A and B). The activation of γ-PKC–GFP by 5 μM TPA induced the obvious translocation of the fluorescence from the cytoplasm to the membrane. Translocation began at 10 min and was completed by 60 min after the treatment with TPA (Fig. 3 A). The fluorescence remained on the plasma membrane for at least 90 min after TPA treatment and did not return to the cytoplasm in the cells tested. The γ-PKC–GFP in the nuclei did not appear to be translocated. The TPA-induced translocation of γ-PKC–GFP occurred more rapidly when the experiments were performed at 37°C (Fig. 3 B, upper trace). The translocation completed 10 min after the treatment with lower concentration of TPA (200 nM). In contrast, an inactive phorbol ester, 4α-PDD, at 500 nM failed to induce the translocation within 30 min (Fig. 3 B, lower trace).
Figure 3.
Phorbol ester– induced translocation of γ-PKC–GFP in COS-7 cells. (A) Change in the fluorescence of γ-PKC–GFP expressed in COS-7 cells by 5 μM TPA at room temperature. γ-PKC–GFP fusion protein was observed throughout the cytoplasm in transfected COS-7 cells. Activation of PKC by 5 μM TPA induced the obvious translocation of γ-PKC–GFP fluorescence from cytosol to membrane. Translocation was almost completed within 60 min after the treatment with TPA. The same view was taken before the stimulation under Nomarski interference microscope and shown at the upper left corner. (B) Change in the fluorescence of γ-PKC–GFP expressed in COS-7 cells by 200 nM TPA and 500 nM 4α-PDD at 37°C. Lower concentration of TPA (200 nM) induced the translocation of γ-PKC–GFP from cytosol to membrane when examined at 37°C (upper trace). The translocation occurred more rapidly, and the complete translocation was observed at 10 min after the treatment with TPA. In contrast, 4α-PDD, an inactive phorbol ester, at 500 nM failed to induce the translocation of γ-PKC–GFP even at 37°C. Bars, 10 μm.
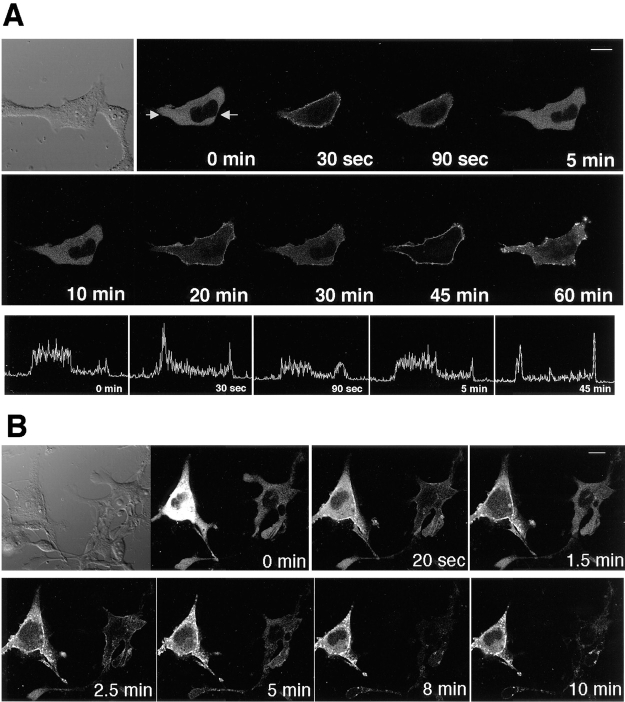
At 80 μM, A23187, the Ca2+ ionophore, also produced γ-PKC–GFP translocation, the time course of which was significantly different from the TPA-induced translocation. The Ca2+ ionophore–induced translocation was rapid and reversible (Fig. 4 A). The fluorescence of γ-PKC–GFP translocated transiently at only 30 s after the stimulation. The first phase of the translocation was quickly reversed. The γ-PKC–GFP was retranslocated to cytoplasm at 90 s after the stimulation. The second and third phase of the translocation was observed 20 and 45 min after the stimulation, and finally γ-PKC–GFP was accumulated as patchy dots at the plasma membrane 60 min after A23187 treatment (Fig. 4 A, upper and middle traces). In the lower trace of Fig. 4 A, the profiles of the GFP intensity on the same line across the cell at various time points were shown. The translocation of γ-PKC–GFP to the membrane was detected as the increase in the intensity at the fringe of the cell at 30 s and 45 min. Very similar profiles of the GFP intensity were obtained at 0 and 5 min, and the fading of the fluorescence is negligible between the two different time points.
Figure 4.
Ca2+ ionophore– induced translocation of γ-PKC–GFP in COS-7 cells. (A) Change in the fluorescence of γ-PKC–GFP expressed in COS-7 cells by 80 μM A23187, Ca2+ ionophore. A23187 also translocated the fluorescence of γ-PKC–GFP from cytosol to membrane; however, the time course of the translocation was significantly different from the TPA-induced one. Ca2+ ionophore–induced translocation was rapid and reversible (upper and middle traces). In the lower trace, profiles of the GFP intensity on the same line across the cell were shown. (The measured line is between the arrows in the upper left picture.) The translocation was expressed as the increase in the fluorescence at the fringe of the cell at 30 s and 45 min. Comparing the profile of the GFP intensity, very similar profiles were obtained at 0 and 5 min, and the fading of the fluorescence is negligible. (B) The γ-PKC– GFP translocation by A23187 was not always reversible. The left cell reveals unidirected translocation, while the right cell shows the reversible translocation, as seen in A. Unidirected translocation was more common than reversible translocation. γ-PKC–GFP eventually accumulated as patchy dots on the plasma membrane and in neighboring cytoplasm. Bars, 10 μm.
The γ-PKC–GFP translocation by A23187 was not always reversible (Fig. 4 B, left cells). The unidirected translocation (Fig. 4 B, left cells) was more common than the reversible translocation (Fig. 4 B, right cells). However, γ-PKC–GFP was always accumulated on plasma membrane as patchy dots (Fig. 4 B). A23187 at lower concentration (10 μM) also showed similar effects (data not shown).
Effects of Thapsigargin on Translocation of γ-PKC–GFP
To examine the influence of Ca2+ released from the intracellular Ca2+ store on γ-PKC–GFP translocation, we studied the effects of thapsigargin, which inhibits endoplasmic reticulum Ca2+-ATPase and increases the concentration of cytosolic Ca2+, on translocation of γ-PKC–GFP. Application of 5 μM thapsigargin induced a rapid translocation of fluorescence, which began within 1 min after the stimulation (Fig. 5). Finally, fluorescence was accumulated as patchy dots on the plasma membrane as seen in A23187-induced translocation. Accumulation of γ-PKC–GFP also occurred in the perikaryon.
Figure 5.
Thapsigargin- induced translocation of γ-PKC–GFP. 5 μM thapsigargin, an inhibitor of the endoplasmic reticulum Ca2+- ATPase, also induced rapid translocation of γ-PKC–GFP. Fluorescence of γ-PKC–GFP accumulated as patchy dots as in A23187-induced translocation. Bar, 10 μm.
Effects of Ca2+ Chelators on γ-PKC–GFP Translocation
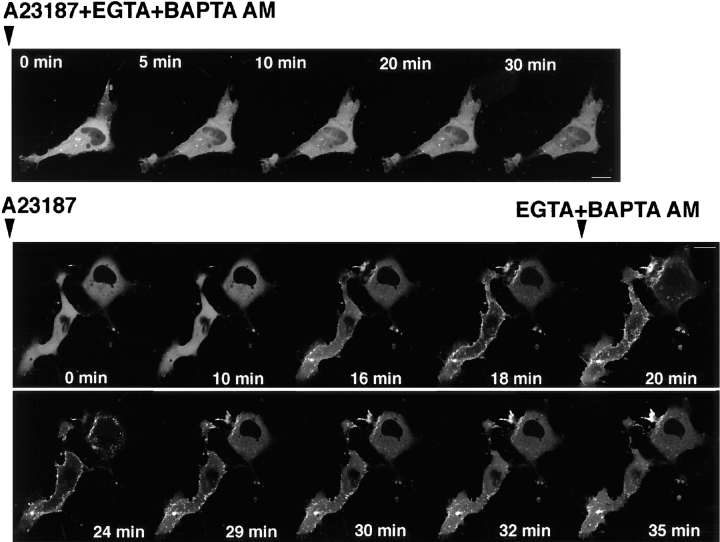
To elucidate whether Ca2+ ionophore–induced translocation depends on the increase in the intracellular Ca2+ concentration ([Ca2+]i), we examined the effects of Ca2+ chelators on A23187-induced translocation. Ca2+ ionophore–induced translocation was completely blocked by the pretreatment with Ca2+ chelators, 2.5 mM EGTA, and 15 μM BAPTA-AM (Fig. 6, upper trace). In addition, A23187-induced translocation was reversed by the treatment with Ca2+ chelators after the stimulation of A23187. The fluorescence of γ-PKC–GFP partially returned to the cytoplasm (Fig. 6, lower trace). TPA-induced translocation was not inhibited by either pre- or post-treatment with Ca2+ chelators (data not shown).
Figure 6.
Effects of Ca2+ chelators on A23187-induced translocation of γ-PKC–GFP. Pretreatment with 2.5 mM EGTA and 15 μM BAPTA-AM completely blocked A23187 (50 μM)–induced γ-PKC–GFP translocation (upper trace). Treatment with 2.5 mM EGTA and 15 μM BAPTA-AM retranslocated γ-PKC–GFP from membrane to cytosol, even after A23187- induced translocation had occurred (lower trace). Bars, 10 μm.
Immunostaining of Translocated γ-PKC–GFP
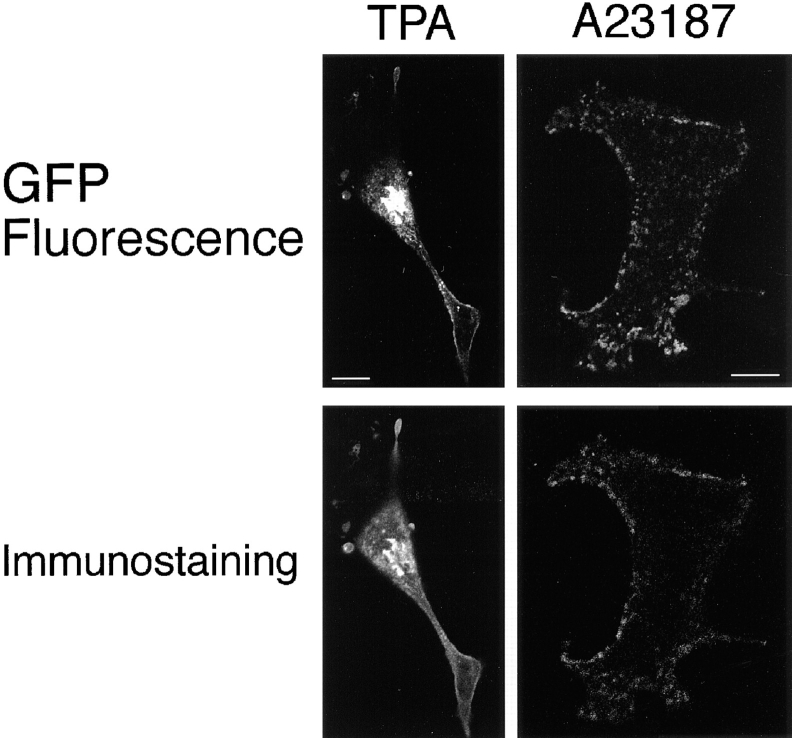
PKCs are known to be digested between the regulatory and catalytic domains by protease such as calpain (Kishimoto et al., 1983), suggesting that the fluorescence of translocated γ-PKC–GFP did not exactly reveal the localization of γ-PKC. Therefore, we immunostained the transfected cells using a monoclonal anti–γ-PKC antibody that recognizes the regulatory domain of γ-PKC after the translocation was completed. As shown in Fig. 7, γ-PKC-like Cy5 fluorescence was colocalized with the GFP fluorescence even after the TPA- or A23187-induced translocation was completed. These results suggested that fluorescence of GFP showed the localization of γ-PKC itself.
Figure 7.
Comparison of the fluorescence of γ-PKC–GFP with the γ-PKC–like immunoreactivity. Immunostaining of γ-PKC– GFP–transfected COS-7 cells by anti–γ-PKC antibody showed that GFP fluorescence and γ-PKC–like immunoreactivity had very similar localizations, even after the TPA- or A23187-induced translocation was completed. Bar, 10 μm.
Translocation of Mutant γ-PKC–GFP Fusion Protein
To examine the significance of the C1 and C2 region of γ-PKC in the γ-PKC–GFP translocation, we constructed mutant γ-PKC–GFP, BS 238 (C1 mutant), and BS 239 (C2 deletion), as described above (Fig. 1). BS 238 is a mutant of the C1 region that binds TPA; it therefore was not expected to be activated by TPA. Indeed, TPA did not induce the translocation of BS 238 (C1 mutant), while A23187 did (Fig. 8 A). The Ca2+ ionophore–induced translocation of BS 238 was insufficient compared with that of wild-type γ-PKC–GFP (BS 186) (Fig. 8 A and 4 A). BS 239 (C2 deletion) is a deletion mutant of the C2 region that binds calcium ion. In contrast to BS 238, BS 239 was translocated by TPA but not by A23187 (Fig. 8 B).
Figure 8.
Translocation of mutant γ-PKC–GFPs (BS 238 and BS 239) expressed in COS-7 cells. (A) Translocation of BS 238 (C1 mutant γ-PKC–GFP). TPA at 5 μM did not induce the translocation of BS 238, while A23187 at 50 μM did. A23187- induced translocation of BS 238 was insufficient compared with that of BS 186 (control γ-PKC–GFP). (B) Translocation of BS 239 (C2 deletion γ-PKC–GFP). In contrast to BS 238, BS 239 was translocated by 5 μM TPA but not by 50 μM A23187. Bars, 10 μm.
We also examined the kinase character of BS 238 and BS 239. The kinase activity of BS 238 was dependent on Ca2+ (157.8 ± 9.1% of the control; control kinase activity was measured in the presence of EGTA), but was not activated by TPA (94.5 ± 5.6% of the control ). In contrast, the kinase activity of BS 239 was not activated by neither Ca2+ (100.4 ± 5.6% of the control ) nor TPA (81.2 ± 7.2% of the control). The kinase activity of BS239, however, was reduced to 27.2% ± 0.4% of the control by the treatment with 1 μM staurosporine, while the activity of BS238 was slightly inhibited by staurosporine (70.9 ± 14.5% of the control).
K+ Depolarization Induced Translocation of γ-PKC–GFP in Transfected NG 108-15 Cells
The translocation of γ-PKC–GFP protein was further examined under physiological conditions. To investigate whether depolarization and subsequent activation of the voltage-gated Ca2+ channel induce γ-PKC translocation, we expressed γ-PKC–GFP fusion protein in NG 108-15 cells, which have voltage-gated Ca2+ channels (Atlas and Adler, 1981). After the extracellular K+ was elevated from 5 to 100 mM, the fluorescence of γ-PKC–GFP was rapidly translocated from cytosol to membrane (Fig. 9). After the translocation was completed, patchy dotlike fluorescence accumulated in membrane and cytosol as seen in A23187- or thapsigargin-induced translocation (Fig. 9). Reversible translocation, as seen in Fig. 3 B, also occurred in the other cells tested (data not shown). K+ depolarization–induced translocation was abolished by 10 μM nicardipine or 10 μM ω-conotoxin GVIA, blockers of L- and N-type Ca2+ channels, respectively, but not by 2 μM tetrodotoxin, a Na+ channel blocker (data not shown). These findings suggested that translocation was triggered by Ca2+ influx through a certain type of voltage-gated Ca2+ channel expressed in NG 108-15 cells.
Figure 9.
K+ depolarization– induced translocation of γ-PKC–GFP expressed in NG 108-15 cells. Replacing the external solution with a high K+–containing one rapidly induced translocation of γ-PKC–GFP. The fluorescence of γ-PKC–GFP accumulated as patchy dots on the plasma membrane and in neighboring cytoplasm, as in Ca2+ ionophore–induced translocation. Bar, 10 μm.
NMDA Receptor–mediated Translocation of γ-PKC–GFP
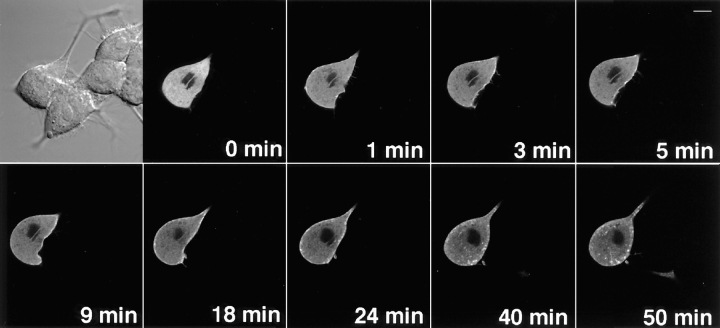
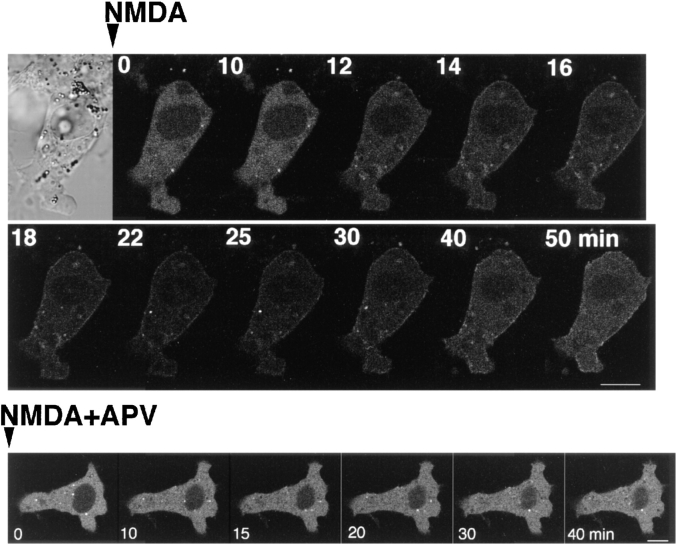
The translocation of γ-PKC–GFP by receptor-mediated stimulation was further examined. In COS-7 cells coexpressing NMDA receptor channels (ζ1 and ε1 subunits), the treatment with 1 mM NMDA plus 10 μM glycine induced faint but significant translocation of γ-PKC–GFP (Fig. 10). Simultaneous application of 500 μM AP-5, an antagonist of NMDA receptor, blocked NMDA-induced translocation (Fig. 10).
Figure 10.
NMDA-induced translocation of γ-PKC– GFP in COS-7 cells coexpressing NMDA receptors. NMDARζ1 and NMDARε1 subunits were cotransfected with γ-PKC–GFP into COS-7 cells by electroporation. NMDA at 1 mM was applied to cells in the absence of Mg2+ and presence of 10 μM glycine. NMDA induced faint but significant translocation of γ-PKC–GFP. Simultaneous application of 100 μM AP-5 with 1 mM NMDA blocked NMDA- induced γ-PKC–GFP translocation. Bars, 10 μm.
mGluR1–mediated Translocation of γ-PKC–GFP
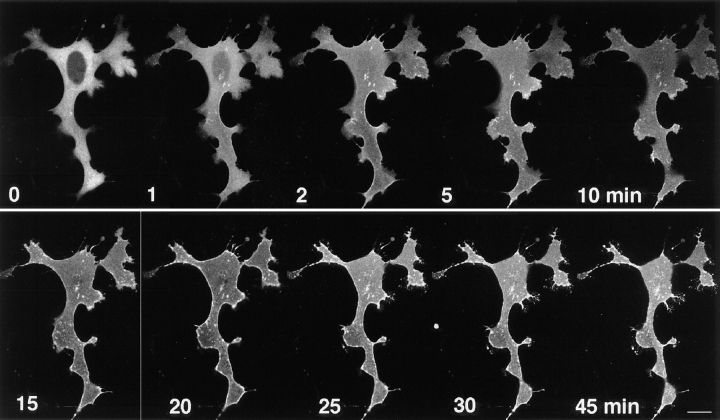
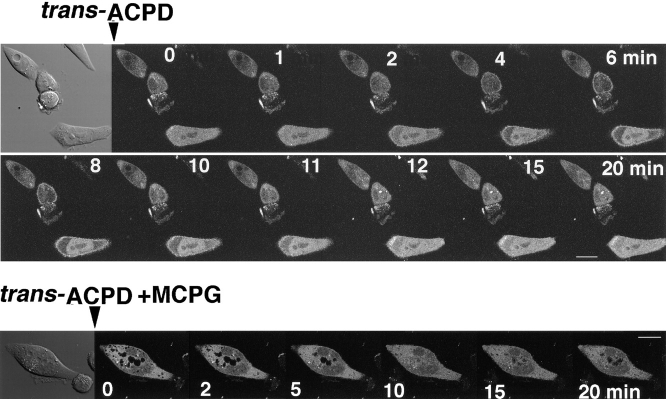
We transfected γ-PKC–GFP into the CHO cells stably expressing mGluR1 to investigate G-protein–coupled receptor-mediated translocation of γ-PKC. Activation of mGluR1 receptor by 1 mM trans-ACPD, an agonist of mGluR1, induced a rapid translocation of γ-PKC–GFP with re-translocation to the cytoplasm within 20 min (Fig. 11). Simultaneous application of 2.5 mM RS-MCPG, an antagonist of mGluR1, completely blocked mGluR1-mediated translocation of γ-PKC–GFP (Fig. 11).
Figure 11.
mGluR1-mediated translocation of γ-PKC–GFP. γ-PKC–GFP was transfected into CHO cells stably expressing mGluR1 by lipofection as described in the text. (Upper trace) Application of 1 mM trans-ACPD rapidly induced the translocation of γ-PKC–GFP from cytosol to membrane. The fluorescence was retranslocated from membrane to cytosol within 20 min. (Lower trace) Simultaneous application of 500 μM MCPG with 1 mM trans-ACPD completely blocked mGluR1-mediated translocation of γ-PKC–GFP. Bar, 10 μm.
Effects of Cytochalasin D and Colchicine on γ-PKC–GFP Translocation
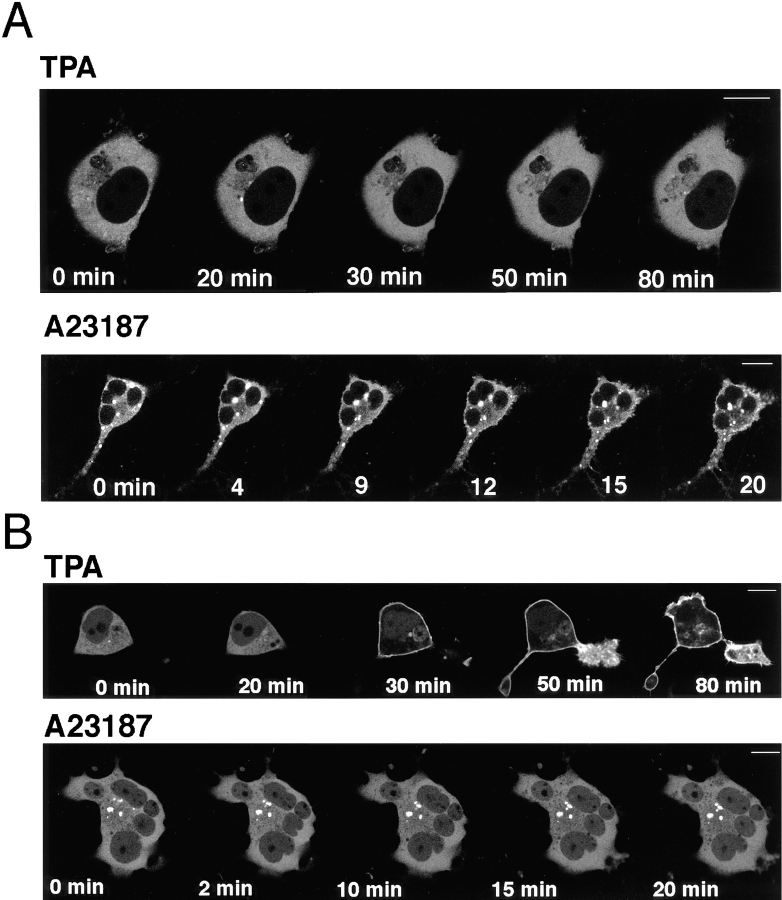
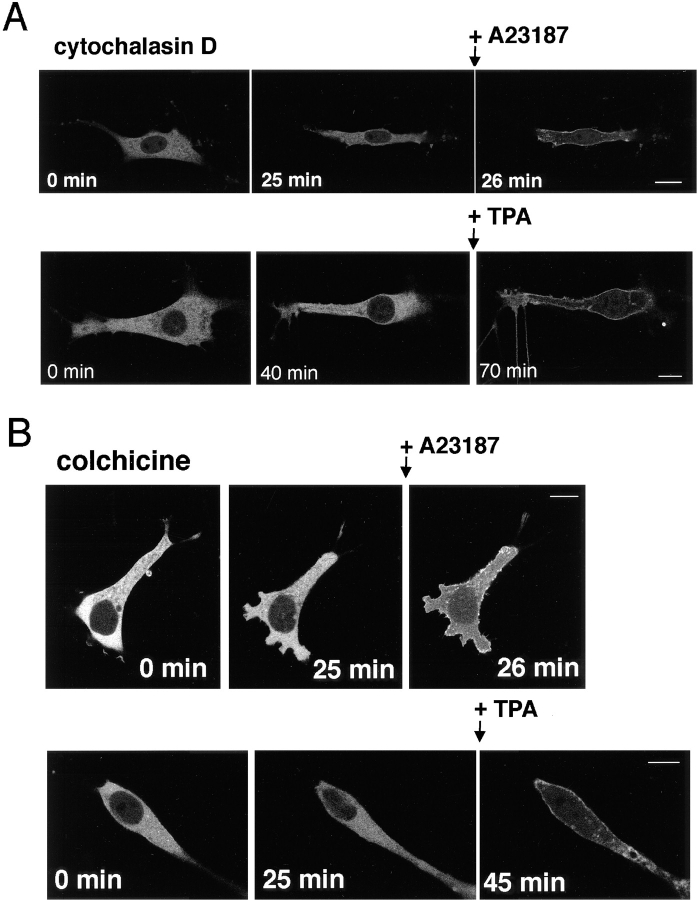
To investigate the involvement of filamentous actin in γ-PKC translocation, we studied the effects of cytochalasin D, an inhibitor of actin polymerization, on the translocation of γ-PKC–GFP. Pretreatment with 10 μM cytochalasin D for 25–40 min altered the shapes of cells; however, it did not inhibit the translocation of γ-PKC–GFP induced by either TPA or A23187 (Fig. 12 A). We also examined the effects of colchicine, an inhibitor of microtubule polymerization, to investigate the relationship between γ-PKC translocation and microtubules. Colchicine, like cytochalasin D, did not affect the TPA- or A23187-induced translocation of γ-PKC–GFP (Fig. 12 B). To assess whether the pretreatment with cytochalasin D or colchicine described above actually acted on cytoskeleton, we stained filamentous actin with rhodamine-phalloidin or microtubules with anti– α-tubulin antibody. Both 10 μM cytochalasin and 100 μM colchicine disrupted actin fibers and microtubules, respectively (data not shown).
Figure 12.
Involvement of γ-PKC–GFP translocation in cytoskeleton. (A) Effects of 10 μM cytochalasin D on γ-PKC–GFP translocation. Treatment with 10 μM γ-PKC–GFP affected neither TPA- nor A23187-induced translocation of γ-PKC–GFP. (B) Effects of 100 μM colchicine on γ-PKC–GFP translocation. Treatment with 100 μM colchicine did not affect TPA- or A23187-induced translocation. In these experiments, γ-PKC–GFP was transfected into NIH3T3 cells by lipofection as described in the text. The concentrations of TPA and A23187 applied were 1 and 50 μM, respectively. Bars, 10 μm.
Discussion
We first examined the enzymological property of γ-PKC– GFP protein by measuring kinase activity with or without activators of PKC. As shown in Fig. 2 C, γ-PKC–GFP protein expressed in COS-7 cells had similar enzymological character to the native γ-PKC. In addition, immunoblotting analysis revealed that γ-PKC–GFP of reasonable molecular size, but not the degradation product of γ-PKC–GFP, was present as a donor of GFP fluorescence, even after the translocation from cytosol to membrane by the stimulation with TPA (Fig. 2 A). Kinase activity of γ-PKC–GFP was also translocated from cytosol to membrane (Fig. 2 B). These results suggest that γ-PKC–GFP protein did not lose its enzymological character as a γ-PKC, even though a GFP, a protein with 238 amino acids, was added to the COOH terminus of γ-PKC. When the GFP was added to the NH2 terminus of γ-PKC (GFP–γ-PKC), the distribution of GFP–γ-PKC within a cell was similar to the present observation of γ-PKC–GFP (data not shown). The NH2-terminal methionine of cPKC, however, is known to be posttranslationally cleaved and replaced with an acetyl group (Tsutakawa et al., 1995). Therefore, it is suggested that the fusion protein γ-PKC–GFP, rather than GFP-γ-PKC, is better to monitor the exact localization of γ-PKC itself.
PKCs are reported to be proteolysed by protease such as calpain, a Ca2+-dependent neutral protease (Kishimoto et al., 1983). If γ-PKC–GFP was proteolysed during its translocation, GFP fluorescence did not reveal the exact localization of γ-PKC. Therefore, we also carried out the immunostaining of γ-PKC–GFP with the antibody that recognizes the regulatory domain of γ-PKC when the translocation was completed. As shown in Fig. 7, the immunoreactivity of γ-PKC–GFP coincided with the fluorescence of GFP. Furthermore, the fact that the line intensity profiles across the cell were similar before and after a transient translocation to the membrane (Fig. 4 A) suggests that the fluorescence of γ-PKC–GFP exactly revealed the localization of γ-PKC–GFP itself, and the time-dependent movement of GFP fluorescence showed the translocation events of γ-PKC in real time.
Immunohistochemical and enzymological analyses revealed that TPA-induced translocation was completed within 5 min (Kraft et al., 1982), although γ-PKC–GFP was translocated more slowly and a high dose of TPA was necessary for the translocation at room temperature. The discrepancy was due to the temperature since 200 nM TPA was enough to translocate γ-PKC–GFP to the membrane, and the translocation was completed within 10 min when the experiment was performed at 37°C (Fig. 3 B).
In contrast with TPA-induced translocation, Ca2+ ionophore–induced translocation was rapid and reversible. Even the TPA-induced translocation of γ-PKC–GFP at 37°C was still slower than that induced by Ca2+ ionophore. Ca2+ chelators blocked the translocation induced by Ca2+ ionophore, suggesting that Ca2+-induced translocation depended on the intracellular Ca2+ concentration ([Ca2+]i). In this regard, the wavelike translocation by A23187 may reflect the alternation of [Ca2+]i . To prove this, we observed the A23187-induced change in [Ca2+]i by loading cells with calcium green-1-AM, a fluorescent Ca2+ indicator, using a confocal laser fluorescent microscope. A transient elevation of fluorescence was observed; however, no wavelike phenomenon of the [Ca2+]i could be detected (data not shown). In addition, A23187 commonly induced the unidirected translocation of γ-PKC–GFP, and the typical reversible translocation as shown in Fig. 3 was infrequent. The reason why Ca2+ ionophore induced the wavelike translocation of γ-PKC–GFP is unclear at present; however, possible explanations can be proposed. First, a wavelike alternation of [Ca2+]i is induced by A23187 under certain conditions only, and the experiments using calcium green-1-AM were not performed under such conditions or the calcium green-1-AM was not a suitable drug with which to fine-tune change in [Ca2+]i becuase of its effect as a Ca2+ chelator. Alternatively, when the A23187-induced elevation of [Ca2+]i was insufficient, the γ-PKC–GFP translocation was incomplete and transient, but subsequent production of phospholipids by various Ca2+-activated phospholipases induced a second or third translocation of γ-PKC– GFP. The results of the kinase assay of γ-PKC–GFP that both phospholipids and Ca2+ were needed for the full activation of γ-PKC–GFP support this idea. Thapsigargin also induced rapid translocation of γ-PKC–GFP, indicating that Ca2+ influx from intracellular Ca2+ stores could translocate γ-PKC–GFP.
When translocation was completed, the localization of γ-PKC in TPA-induced translocation was different from in the Ca2+-induced one. Fluorescence of γ-PKC–GFP was accumulated in plasma membrane in TPA-induced translocation, whereas it was accumulated in plasma membrane and submembrane cytoplasm as patchy dots in Ca2+-induced translocation. These findings suggest that TPA-induced and Ca2+-induced translocation are mediated by different pathways and that the final localization of γ-PKC is distinct after each stimulation.
The treatment with TPA caused subtype-specific subcellular distribution in cardiac myocytes (Disatnik et al., 1994). γ-PKC was also reported to be translocated to Golgi organelle by TPA in NIH3T3 cells stably overexpressing γ-PKC (Goodnight et al., 1995). In contrast, γ-PKC–GFP was mainly translocated from cytosol to membrane by TPA in our study. As far as we could determine in various cells (COS-7, CHO, NG-108, etc.), γ-PKC was not translocated to the Golgi complex. Transiently expressed γ-PKC may have different properties of translocation from native or stably expressed γ-PKC. Otherwise, an addition of GFP may alter the translocation nature of γ-PKC.
In our present study, γ-PKC–GFP was translocated by various physiological stimuli. The findings showed that γ-PKC is activated and translocated in living cells. In addition, receptor mediated-translocation of γ-PKC–GFP were rapid and reversible. In particular, mGluR1-mediated translocation was transient, indicating that γ-PKC translocation induced by receptor-mediated breakdown of phosphatidylinositol was not sustained in living cells. As mGluR1-mediated elevation of Ca2+ was found to be transient by measuring fluorescence of intracellular-loaded Ca2+ green-1-AM (data not shown), mGluR1-mediated translocation of γ-PKC–GFP may depend on intracellular Ca2+.
To elucidate whether the translocation of γ-PKC is associated with the activation mechanism, we examined the translocation of mutant γ-PKC–GFPs. TPA induced translocation of BS 239 (C2 mutant) but not BS 238 (C1 mutant). In contrast, Ca2+ ionophore translocated BS 238 but not BS 239. The kinase activity of BS 238 was dependent on Ca2+, in parallel with the results of BS 238 translocation, while the kinase activity of BS 239 depended on neither Ca2+ nor TPA but was blocked by the staurosporine, a potent kinase inhibitor, suggesting that BS 239 may become a constitutively active form by a mutation. TPA-induced translocation of γ-PKC may not be coupled with its kinase activity, as the staurosporine did not block TPA-induced translocation (data not shown). As shown in Fig. 8 A, Ca2+ ionophore–induced translocation of BS 238 (C1 mutant) was insufficient in all cells tested. Based on the results of the kinase assay (Fig. 2 C), Ca2+ alone appears not to induce the complete translocation of γ-PKC. As Ca2+ has been reported to activate the production of phospholipids, including DG, both calcium and phospholipids were needed to induce complete translocation of γ-PKC.
Although PKC translocation is a well-known phenomenon, the molecular mechanism and significance of PKC translocation have not yet been clarified. Phorbol esters translocated some subtypes of PKCs from cytosol to particulate fractions including cytoskeleton (Zalewski et al., 1988; Jaken et al., 1989; Papadopoulos and Hall, 1989; Kiley and Jaken, 1990; Mochly-Rosen et al., 1990). In βII-PKC, which was reported to be translocated to actin fiber (Goodnight et al., 1995), it was proposed that receptors for activated C-kinase (RACK), present in the detergent- insoluble fraction and bound activated βII-PKC, play a role in the mechanism of PKC translocation (Mochly-Rosen et al., 1991; Ron and Mochly-Rosen, 1995). To elucidate whether cytoskeleton proteins such as actin or microtubule are involved in PKC translocation, the effects of cytochalasin D, an inhibitor of actin polymerization, and colchicine, an inhibitor of microtubule polymerization, on the translocation of γ-PKC–GFP were examined. As shown in Fig. 12, pretreatment with neither cytochalasin D nor colchicine affected γ-PKC–GFP translocation. Furthermore, γ-PKC–GFP translocation occurred even when the glucose in the external solution was eliminated (data not shown). These results indicated that PKC translocation did not need the cytoskeleton and glucose-dependent motor protein that are essential for some types of protein trafficking, such as an axonal flow or vesicle transport (Bloom, 1992; Cheney et al., 1993). In addition, based on the findings that the C2 deletion mutant of γ-PKC (BS 239) could be translocated by TPA, phorbol ester–induced translocation of γ-PKC from cytosol to plasma membrane is probably not necessary for the association of RACK because RACK was reported to bind the C2 region of PKC (Ron and Mochly-Rosen, 1995).
In conclusion, a GFP fusion protein with γ-PKC is a useful tool for investigating the mechanism and significance of γ-PKC translocation in living cells.
Acknowledgments
We thank Dr. Ushio Kikkawa for many useful discussions.
This work was supported by grants from the Ministry of Education, Science, Sports and Culture in Japan, the Yamanouchi Foundation for Research on Metabolic Disorders, and the Kato Memorial Bioscience Foundation.
Abbreviations used in this paper
- 4α-PDD
4α-phorbol 12, 13-didecanoate
- DG
diacylglycerol
- DO
diolein
- GFP
green fluorescent protein
- NMDA
N-methyl-d-aspartate
- MCPG
α-methyl-4-carboxyphenyl-glycine
- mGluR1
metabotropic glutamate receptor 1
- PKC
protein kinase C
- PS
phosphatidylserine
- TPA
12-O-tetradecanoylphorbol 13-acetate
- trans-ACPD
(1S, 3R)-1-aminocyclopentane-1,3-dicarboxylic acid
Footnotes
Address all correspondence to Naoaki Saito, Laboratory of Molecular Pharmacology, Biosignal Research Center, Kobe University, 1-1 Rokkodai-cho, Nada-ku, Kobe 657, Japan. Tel.: 81-78-803-1251. Fax: 81-78-803-0993. E-mail: naosaito@inherit.biosig.kobe-u.ac.jp
References
- Abeliovich A, Chen C, Goda Y, Silva AJ, Stevens CF, Tonegawa S. Modified hippocampal long-term potentiation in PKC γ-mutant mice. Cell. 1993a;75:1253–1262. doi: 10.1016/0092-8674(93)90613-u. [DOI] [PubMed] [Google Scholar]
- Abeliovich A, Paylor R, Chen C, Kim JJ, Wehner JM, Tonegawa S. PKCγ mutant mice exhibit mild deficits in spatial and contextual learning. Cell. 1993b;75:1263–1271. doi: 10.1016/0092-8674(93)90614-v. [DOI] [PubMed] [Google Scholar]
- Atlas D, Adler M. α-Adrenergic antagonist as possible calcium channel inhibitors. Proc Natl Acad Sci USA. 1981;78:1237–1241. doi: 10.1073/pnas.78.2.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom GS. Motor proteins for cytoplasmic microtubules. Curr Opin Cell Biol. 1992;4:66–73. doi: 10.1016/0955-0674(92)90060-p. [DOI] [PubMed] [Google Scholar]
- Chen CS, Poenie M. New fluorescent probes for protein kinase C. Synthesis, characterization, and application. J Biol Chem. 1993;268:15812–15822. [PubMed] [Google Scholar]
- Cheney RE, Rieley MA, Mooseker MS. Phylogenetic analysis of the myosin superfamily. Cell Motil Cytoskel. 1993;24:215–223. doi: 10.1002/cm.970240402. [DOI] [PubMed] [Google Scholar]
- Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- Disatnik MH, Buraggi G, Mochly-Rosen D. Localization of protein kinase C isozymes in cardiac myocytes. Exp Cell Res. 1994;210:287–297. doi: 10.1006/excr.1994.1041. [DOI] [PubMed] [Google Scholar]
- Goodnight JA, Mischak H, Kolch W, Mushinski JF. Immunocytochemical localization of eight protein kinase C isozymes overexpressed in NIH 3T3 fibroblasts. Isoform-specific association with microfilaments, Golgi, endoplasmic reticulum, and nuclear and cell membranes. J Biol Chem. 1995;270:9991–10001. doi: 10.1074/jbc.270.17.9991. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Ase K, Sawamura S, Kikkawa U, Saito N, Tanaka C, Nishizuka Y. Postnatal development of a brain-specific subspecies of protein kinase C in rat. J Neurosci. 1988;8:1678–1683. doi: 10.1523/JNEUROSCI.08-05-01678.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa M, Mori H, Araki K, Sakimura K, Watanabe M, Inoue Y, Mishina M. Cloning and expression of the epsilon 4 subunit of the NMDA receptor channel. FEBS Lett. 1992;313:34–38. doi: 10.1016/0014-5793(92)81178-o. [DOI] [PubMed] [Google Scholar]
- Jaken S, Leach K, Klauck T. Association of type 3 protein kinase C with focal contacts in rat embryo fibroblasts. J Cell Biol. 1989;109:697–704. doi: 10.1083/jcb.109.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa U, Minakuchi R, Takai Y, Nishizuka Y. Calcium-activated, phospholipid-dependent protein kinase (protein kinase C) from rat brain. Methods Enzymol. 1983;99:288–298. doi: 10.1016/0076-6879(83)99064-x. [DOI] [PubMed] [Google Scholar]
- Kiley SC, Jaken S. Activation of α-protein kinase C leads to association with detergent-insoluble components of GH4C1 cells. Mol Endocrinol. 1990;4:59–68. doi: 10.1210/mend-4-1-59. [DOI] [PubMed] [Google Scholar]
- Kishimoto A, Kajikawa N, Shiota M, Nishizuka Y. Proteolytic activation of calcium-activated, phospholipid-dependent protein kinase by calcium-dependent neutral protease. J Biol Chem. 1983;258:1156–1164. [PubMed] [Google Scholar]
- Kraft AS, Anderson WB, Cooper HL, Sando JJ. Decrease in cytosolic calcium/phospholipid-dependent protein kinase activity following phorbol ester treatment of ELA4 thymoma cells. J Biol Chem. 1982;257:13193–13196. [PubMed] [Google Scholar]
- Meguro H, Mori H, Araki K, Kushiya E, Kutsuwada T, Yamazaki M, Kumanishi T, Arakawa M, Sakimura K, Mishina M. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature. 1992;357:70–74. doi: 10.1038/357070a0. [DOI] [PubMed] [Google Scholar]
- Mochly-Rosen D, Henrich CJ, Cheever L, Khaner H, Simpson PC. A protein kinase C isozyme is translocated to cytoskeletal elements on activation. Cell Regul. 1990;1:693–706. doi: 10.1091/mbc.1.9.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D, Khaner H, Lopez J. Identification of intracellular receptor proteins for activated protein kinase C. Proc Natl Acad Sci USA. 1991;88:3997–4000. doi: 10.1073/pnas.88.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984;308:693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. Studies and perspectives of protein kinase C. Science. 1986;233:305–312. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. The molecular heterogeneity of protein kinase C and implications for cellular regulation. Nature. 1988;334:661–665. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- Ono Y, Fujii T, Igarashi K, Kikkawa U, Ogita K, Nishizuka Y. Nucleotide sequences of cDNAs for α and γ subspecies of rat brain protein kinase C. Nucleic Acids Res. 1988a;16:5199–5200. doi: 10.1093/nar/16.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Fujii T, Ogia K, Kikkawa U, Igarashi K, Nishizuka Y. The structure, expression, and properties of additional members of the protein kinase C family. J Biol Chem. 1988b;263:6927–6932. [PubMed] [Google Scholar]
- Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci USA. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos V, Hall PF. Isolation and characterization of protein kinase C from Y-1 adrenal cell cytoskeleton. J Cell Biol. 1989;108:553–567. doi: 10.1083/jcb.108.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, P.J., and L.V. Dekker. 1997. Introduction. In Protein Kinase C. P.J. Parker and L.V. Dekkers, editors. R.G. Landes Co., Austin, TX. 1–9.
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoriagreen-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- Ron D, Mochly–Rosen D. An autoregulatory region in protein kinase C: the pseudoanchoring site. Proc Natl Acad Sci USA. 1995;92:492–496. doi: 10.1073/pnas.92.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, Masu M, Ishii T, Shigemoto R, Nakanishi S. A family of metabotropic glutamate receptors. Neuron. 1992;8:169–179. doi: 10.1016/0896-6273(92)90118-w. [DOI] [PubMed] [Google Scholar]
- Tanaka C, Nishizuka Y. The protein kinase C family for neuronal signaling. Annu Rev Neurosci. 1994;17:551–567. doi: 10.1146/annurev.ne.17.030194.003003. [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Medzihradszky KF, Flint AJ, Burlingame AL, Koshland DJ. Determination of in vivo phosphorylation sites in protein kinase C. J Biol Chem. 1995;270:26807–26812. doi: 10.1074/jbc.270.45.26807. [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Mori H, Araki K, Mori KJ, Mishina M. Cloning, expression and modulation of a mouse NMDA receptor subunit. FEBS Lett. 1992;300:39–45. doi: 10.1016/0014-5793(92)80160-i. [DOI] [PubMed] [Google Scholar]
- Zalewski PD, Forbes IJ, Valente L, Apostolou S, Hurst NP. Translocation of protein kinase C to a Triton-insoluble sub-cellular compartment induced by the lipophilic gold compound auranofin. Biochem Pharmacol. 1988;37:1415–1417. doi: 10.1016/0006-2952(88)90802-7. [DOI] [PubMed] [Google Scholar]