Abstract
A limited number of transport factors, or karyopherins, ferry particular substrates between the cytoplasm and nucleoplasm. We identified the Saccharomyces cerevisiae gene YDR395w/SXM1 as a potential karyopherin on the basis of limited sequence similarity to known karyopherins. From yeast cytosol, we isolated Sxm1p in complex with several potential import substrates. These substrates included Lhp1p, the yeast homologue of the human autoantigen La that has recently been shown to facilitate maturation of pre-tRNA, and three distinct ribosomal proteins, Rpl16p, Rpl25p, and Rpl34p. Further, we demonstrate that Lhp1p is specifically imported by Sxm1p. In the absence of Sxm1p, Lhp1p was mislocalized to the cytoplasm. Sxm1p and Lhp1p represent the karyopherin and a cognate substrate of a unique nuclear import pathway, one that operates upstream of a major pathway of pre-tRNA maturation, which itself is upstream of tRNA export in wild-type cells. In addition, through its association with ribosomal proteins, Sxm1p may have a role in coordinating ribosome biogenesis with tRNA processing.
The partitioning of nuclear from cytoplasmic matters is the defining feature of eukaryotes and separates the sites of transcription and translation. As a result, many proteins and RNAs need to be specifically transported into and out of the nucleoplasm. This transport takes place through the nuclear pore complex (NPC)1 and is mediated by a structurally related family of soluble factors known as karyopherins (also named transportins, importins, or exportins). The karyopherin of the most widely studied import pathway is a heterodimer, composed of karyopherin α (Kap60p in yeast) and karyopherin β1 (Kap95p in yeast) (for reviews see Moroianu, 1997; Nigg, 1997). The Kap95p/Kap60p heterodimer, via Kap60p, binds to basic nuclear localization sequences (NLSs) in the cytoplasm and, via Kap95p, to peptide repeat-containing nucleoporins at the NPC. In a complex series of events, the trimeric complex is dissociated through the actions of p10 and the small GTPase Ran and its modulators (Rexach and Blobel, 1995, Nehrbass and Blobel, 1996). Kap60p and the NLS-containing protein continue into the nucleus, whereas Kap95p rapidly returns to the cytoplasm. Finally, the Kap60p/NLS interaction is disrupted (Rexach and Blobel, 1995, Moroianu et al., 1996), and Kap60p is recycled to the cytoplasm (Kutay et al., 1997; Moroianu et al., 1997). As much as 50% of all nuclear proteins have putative NLSs that may mediate nuclear import through the karyopherin α/karyopherin β1 pathway (Makkerh et al., 1996).
Additional noncompeting pathways to and from the nucleus for both proteins and RNAs have been directly or indirectly observed (Fischer et al., 1991; Garcia-Bustos et al., 1991; Michaud and Goldfarb, 1991, 1992). The completion of the yeast genome project has clarified and intensified the search for the molecular components of these pathways. Thorough characterization of these pathways should provide profound insight into how eukaryotic cells function. At the present time, it is not clear which classes of nuclear proteins will need their own transporters and how all of the transport pathways will be coordinated to permit efficient cellular function. Characterization of these pathways is in its early stages, as the molecular components for two additional import pathways and an export pathway have recently been identified in yeast (Aitchison et al., 1996; Rout et al., 1997; Stade et al., 1997). Each of these pathways is mediated by a protein similar in sequence to Kap95p. Importantly, none of the additional pathways appears to use Kap60p. Further characterization of these newly discovered transport factors will reveal additional similarities and differences to the Kap60p/Kap95p pathway. A limited number of additional open reading frames with modest similarity to Kap95p have been noted (Fornerod et al., 1997). One of these open reading frames is YDR395w, which was recently renamed SXM1 (Seedorf and Silver, 1997). The following experiments were carried out to determine if Sxm1p is indeed a karyopherin, and if it is to determine its cargo.
Materials and Methods
Yeast Strains and Methods
Strain DF5 was the parent strain for all those in this study (Finley et al., 1987), with the exception of the LHP1 and lhp1 strains in Fig. 5, which were generously provided by Christopher Yoo and Sandra Wolin (Yale University, New Haven, CT). Strains were manipulated and maintained according to standard methods (Rose et al., 1990).
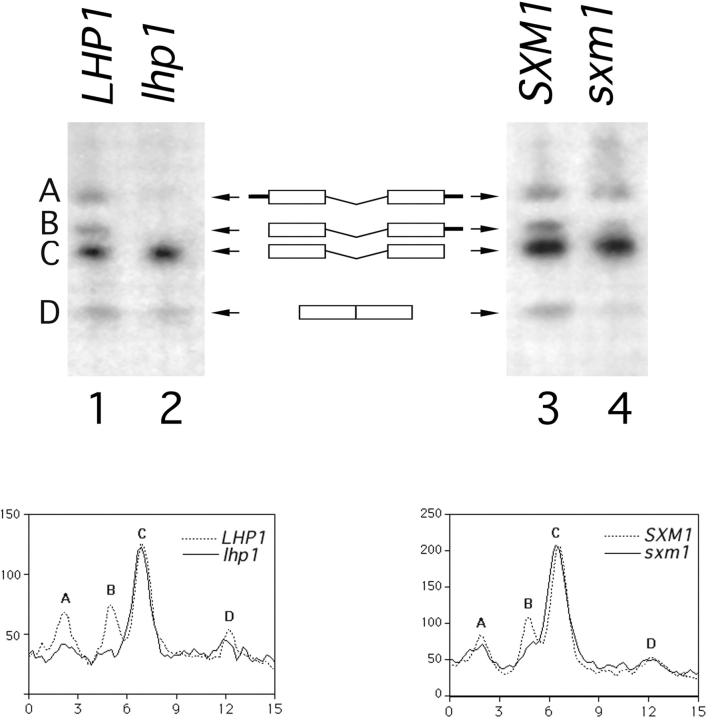
Figure 5.
3′ processing of pre-tRNA Ser CGA Ser CGA in LHP1- and SXM1-deletion strains. A schematic of the intron-containing pre-tRNAs, species A, B, and C and of mature tRNA, species D, is included for reference. Exons are represented by boxes, 5′ and 3′ extensions are represented by black bars, and the central intron is shown. A Northern blot of total RNA was probed for intermediates A, B, and C using an oligonucleotide from the intron, and for mature tRNA using a probe that spans the properly spliced exons. Lane 1, wild-type cells (LHP1); lane 2, lhp1 cells; lane 3, Lhp1-PrA cells, wild-type for SXM1 (SXM1); and lane 4, Lhp1-PrA/sxm1 cells (sxm1). The blot was quantified using a PhosphorImager with ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The graphs indicate radioactivity relative to distance, and peaks corresponding to intermediates are labeled. To account for minor loading discrepancies, the data in the graphs have been normalized to an internal control, precursor C, which was shown previously (Yoo and Wolin, 1997) to be independent of LHP1 genotype.
Protein A Tagging and Gene Replacement
Protein A tagging and gene replacement of SXM1 with HIS3 were performed using published procedures (Aitchison et al., 1995). For protein A tagging, a PCR product was generated that contained a protein A, HIS3, URA3 cassette flanked by the final 60 nucleotides of the coding region and 60 nucleotides downstream of the stop codon. SXM1 was replaced with HIS3 by generating a PCR product corresponding to the HIS3 gene flanked by 60 nucleotides directly upstream and 60 nucleotides directly downstream of the coding region of SXM1. Yeast cells were transformed by electroporation. Correct integration was verified by PCR. Heterozygous diploids were sporulated and dissected to generate haploid strains.
Whole Yeast Protein Extracts
Protein extracts were prepared using a method similar to a published procedure (Yaffe and Schatz, 1984). Briefly, 1 ml of a saturated, overnight YPD culture was added to 140 μl of 1.85 M NaOH/7.4% 2-mercaptoethanol. After 2 min at room temperature, 140 μl of 50% (wt/vol) trichloroacetic acid was added. The mixture was spun for 5 min at 4°C. After aspiration of supernatant, the pellet was washed with 95% ethanol and dried. The pellet was then resuspended in 100 μl of sample buffer and heated at 95°C for 5 min. After centrifugation, 1–5 μl were subjected to SDS-PAGE.
Immunofluorescence
Indirect immunofluorescence was carried out as described (Rout et al., 1997). Briefly, yeast cells were fixed with 3.7% formaldehyde for 5 and 20 min. After digestion of the cell wall, spheroplasts were attached to glass slides. Protein A moieties were visualized by probing with rabbit IgG that had been preadsorbed to wild-type yeast spheroplasts (Cappel Laboratories, Malvern, PA) followed by Cy3-conjugated donkey anti–mouse IgG (Jackson ImmunoResearch, West Grove, PA). Nuclear DNA in fixed cells was visualized with 4′,6-diamidino-2-phenylindole (DAPI). Direct immunofluorescence was used to visualize live, unfixed cells transformed with a plasmid encoding an SV-40 NLS–green fluorescent protein (GFP) fusion (Shulga et al., 1996).
Cell Fractionation and Immunoisolation
Fractionation and immunoisolation of Sxm1–protein A (PrA) were as described (Aitchison et al., 1996). Briefly, postnuclear, postribosomal cytosol from 160 ml of a YPD culture with an OD600 of 1.6 was prepared. Rabbit IgG–Sepharose (Cappel Laboratories) was added to the cytosol. After overnight incubation, the Sepharose was extensively washed, and bound proteins were eluted with a step gradient from 50 to 4,500 mM MgCl2. For mass spectrometry and microsequencing, eluted fractions were precipitated and resolved on a 10–20% acrylamide gel (Novex, San Diego, CA).
Northern Blot
Northern blot to analyze pre-tRNA processing was as described (Yoo and Wolin, 1997) with minor modifications. Briefly, total RNA from ∼3 × 107 cells was isolated using Tri Reagent (Sigma Chemical Co., St. Louis, MO) according to the manufacturer's instructions. This RNA was separated on a 10% TBE-Urea gel (Novex) and transferred in a tank apparatus to Hybond-N (Amersham Corp., Arlington, Heights). To assess overall RNA integrity, the samples were loaded in duplicate, and the half that was not transferred was stained with ethidium bromide and visualized with UV light. Oligonucleotides (Yoo and Wolin, 1997) and hybridization conditions (Tarn et al., 1995) have been described.
Overlay Assay
The overlay assay was carried out as previously described (Rout et al., 1997). Protein A fusion protein–containing cytosol was used analogously to antibodies, to probe blots for bands able to interact with the fusion protein. Briefly, cytosol was incubated overnight at 4°C with nitrocellulose blot. The blot was sequentially incubated with rabbit IgG (Cappel Laboratories) and HRP-conjugated goat anti–rabbit IgG (Amersham Corp.). Enhanced chemiluminescence was used for detection.
Results
Sxm1p Is Similar to Kap95p
Sxm1p is 16% identical and 50% similar to Kap95p (Fig. 1). When the two proteins are independently aligned, the similarity runs the length of the alignment. The common functions of the karyopherins are thought to be mediated by a more highly conserved amino-terminal domain; however, the boundaries of this domain are not obvious from our alignment. Although this similarity is modest, it is consistent with that seen for the similarity of other karyopherins, and proposed karyopherins, to Kap95p.
Figure 1.
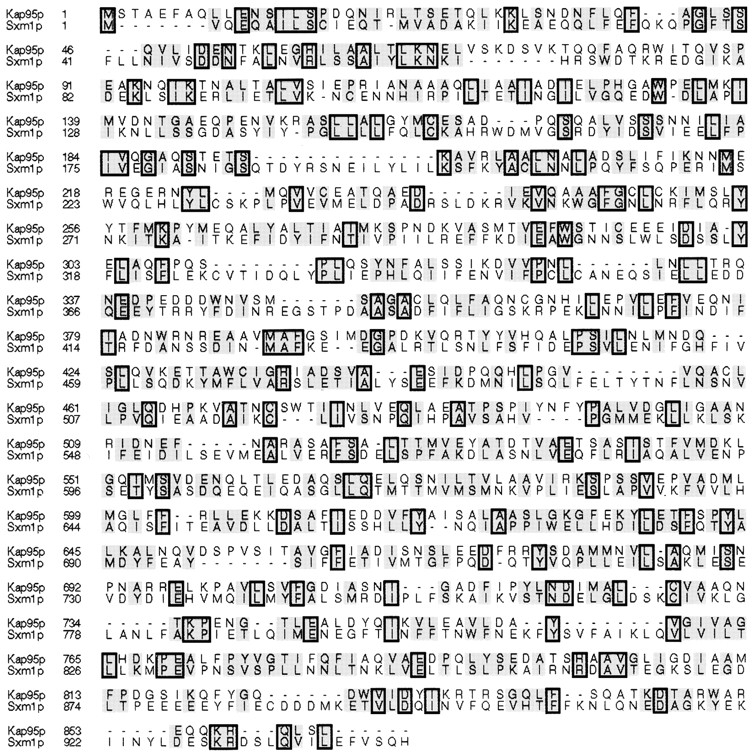
Kap95p and Sxm1p are homologous. The sequences were aligned by the Clustal method in MegAlign (DNA Star, Inc., Madison, WI). Similar amino acids are shaded, and identical amino acids are boxed and shaded.
Genomic Tagging of SXM1 and Localization of Tagged Sxm1p
The carboxy terminus of the genomic copy of SXM1 was directly fused, in frame, to the IgG-binding domains of Staphylococcus aureus PrA via integrative transformation into diploid cells. Sporulation and tetrad dissection yielded four viable colonies on YPD plates. The four strains produced grew at the same rate at 16, 30, and 37°C. Two of the four strains grew on plates lacking His and Ura. These strains also expressed a 140-kD protein reactive with rabbit IgG by immunoblotting, as expected for a fusion protein composed of Sxm1p and protein A.
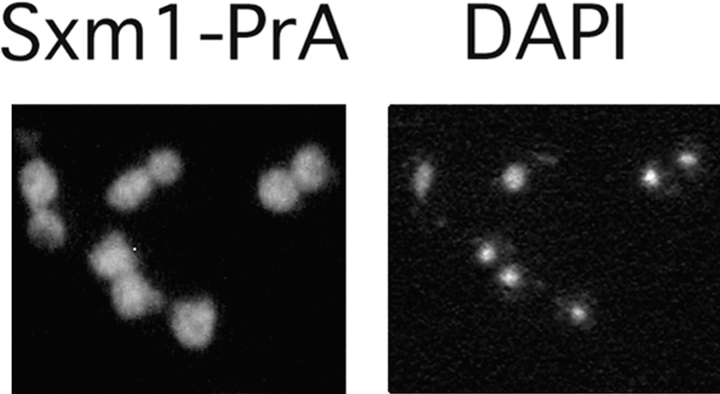
The resulting fusion protein (Sxm1–PrA) was used to determine the subcellular distribution of Sxm1p by indirect immunofluorescence microscopy. Using a fixation time of 5 min, Sxm1–PrA appeared predominantly in the nucleus (data not shown). This nuclear concentration is consistent with that seen for an Sxm1p–GFP fusion as reported by Seedorf and Silver (1997). After more prolonged fixation, however, the cytoplasmic pool of Sxm1–PrA appeared at least as concentrated as that in the nucleus (Fig. 2). Localization of Sxm1p in both the nucleus and cytoplasm is consistent with a role for it in nucleocytoplasmic transport.
Figure 2.
Sxm1–PrA is localized throughout the cell. The PrA moiety was visualized by indirect immunofluorescence. For reference, the right panel represents the nuclei of the same field of cells by staining for DNA with DAPI.
Immunoisolation and Identification of Sxm1p-associated Proteins
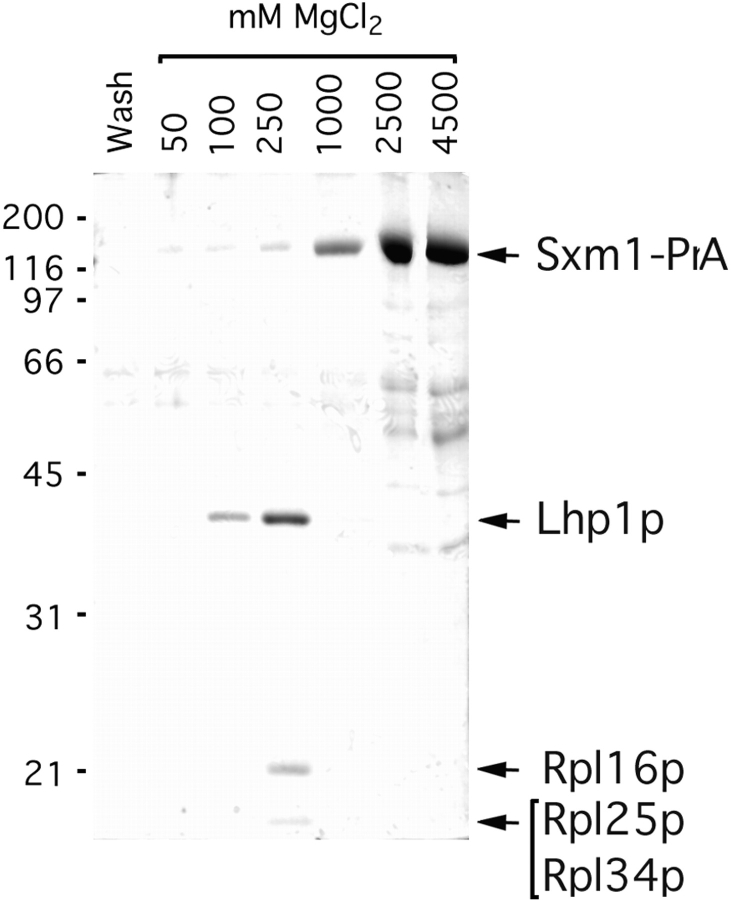
If Sxm1p is indeed a factor involved in transporting newly synthesized proteins from the cytoplasm to the nucleus, some portion of the cytoplasmic pool of Sxm1p can be expected to be complexed with its nucleus-bound cargo. As such, we should be able to immunoisolate this transport complex from the cytoplasm of our Sxm1–PrA strain by virtue of the high affinity of PrA for IgG (Aitchison et al., 1996; Rout et al., 1997). We fractionated the Sxm1–PrA strain into crude nuclear and cytoplasmic fractions and subjected the cytoplasmic fraction to IgG-Sepharose chromatography. Four major Sxm1–PrA–associated proteins were eluted using either low pH (data not shown) or a magnesium chloride gradient (Fig. 3). The associated proteins eluted in the 100 and 250 mM MgCl2 fractions, whereas the Sxm1–PrA/IgG interaction was not disrupted until ∼1,000 mM MgCl2. By Coomassie blue staining, there appeared to be similar concentrations of the associated proteins as compared with Sxm1–PrA, indicating that much of the cytosolic Sxm1–PrA exists complexed to these proteins.
Figure 3.
Immunoisolation of cytosolic Sxm1–PrA interacting proteins. Whole cytosol from an Sxm1–PrA strain was subjected to IgG-Sepharose chromatography. Fractions from the final wash and MgCl2 gradient were separated by SDS-PAGE. Molecular mass standards are indicated to the left in kilodaltons. The gel was stained with Coomassie blue R.
After SDS-PAGE and staining with Coomassie blue, associated protein bands were individually excised from the gel and digested with endoprotease Lys-C. The resulting peptides were subjected to microsequencing (Fernandez et al., 1994) and/or mass spectrometry (Gharahdaghi et al., 1996). The 38-kD band was identified as Lhp1p, which is the yeast homologue of the human autoantigen La. This assignment was made by sequencing the peptide VIEALRSSEILEVSADGENV, which corresponds to amino acids 83–102 of Lhp1p. From the 22-kD band, the peptide EQLSGQTPVQ was sequenced, corresponding to amino acids 38–47 of the 60S ribosomal protein Rpl16p. Two other ribosomal proteins, Rpl25p and Rpl34p, were identified through mass spectrometry of the doublet centered at 15 kD.
Lhp1p, Unlike Other Nuclear Proteins, Is Mislocalized in a Strain Missing Sxm1p
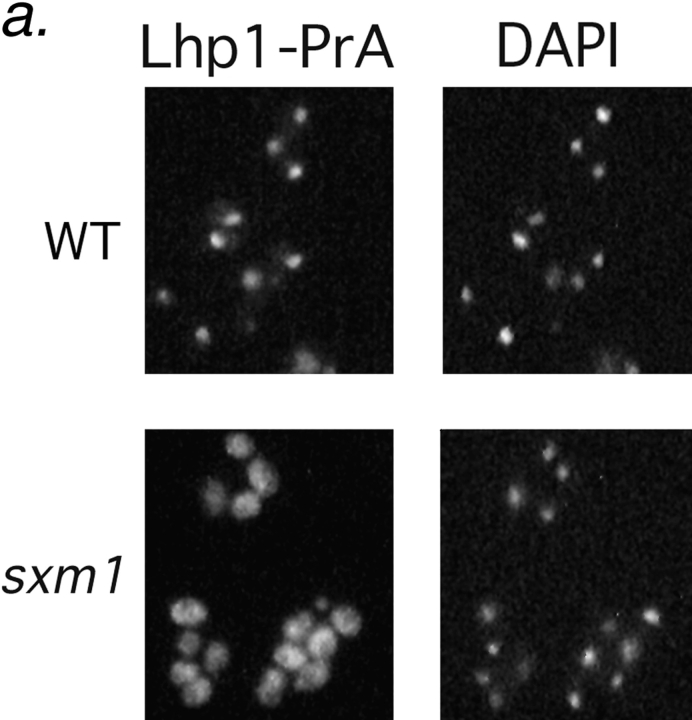
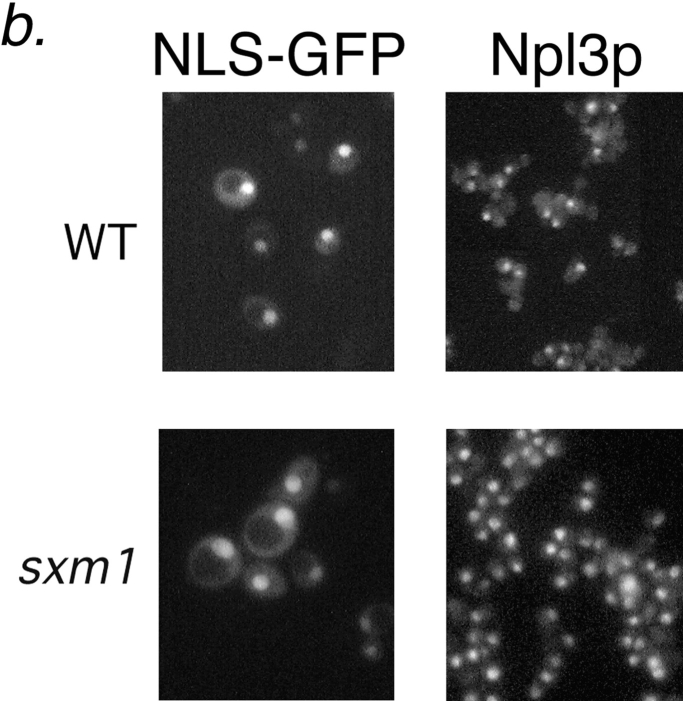
An Sxm1p-deficient strain was generated by replacement of SXM1 by the HIS3 gene. After sporulation and tetrad dissection, all four spores were viable on YPD plates. No growth phenotype was observed for the sxm1::HIS3 strain at any temperature. The absence of a growth phenotype is consistent with a previous study (Seedorf and Silver, 1997). Lhp1p, like the La protein of vertebrates (Simons et al., 1996), is concentrated in the nucleus at steady state. Like SXM1, LHP1 is nonessential for growth (Yoo and Wolin, 1994; Lin-Marq and Clarkson, 1995). We characterized the impact of SXM1 deletion on nuclear import of Lhp1p. As we had identified Lhp1p as a potential import substrate for Sxm1p, a dependence on Sxm1p for nuclear localization of Lhp1p would secure the definition of Sxm1p as a karyopherin. To examine the localization of Lhp1p, we constructed genomic Lhp1–PrA fusions in both a wild-type strain and in our sxm1::HIS3 strain. As expected, Lhp1–PrA colocalized with DAPI-stained DNA in the nucleus of the wild-type strain (Fig. 4 a, top). In marked contrast, in the sxm1::HIS3 strain, Lhp1-PrA was mislocalized to the cytoplasm, and in fact appears to be excluded from the nucleus in many cells (Fig. 4 a, bottom). Importantly, our sxm1 strain retained its ability to import the endogenous nuclear protein Npl3p as well as a reporter bearing the SV-40 large T nuclear localization sequence (Fig. 4 b). From these results, it is clear that Sxm1p is necessary for the nuclear import of Lhp1p and that this import operates in parallel to other nuclear import pathways.
Figure 4.
Deletion of SXM1 leads to mislocalization of Lhp1-PrA but not other nuclear proteins. (a) Wild-type haploid cells (top row) and sxm1 cells (bottom row), both with a genomic Lhp1-PrA fusion, are probed for Lhp1-PrA (left) and stained for DNA with DAPI (right). (b) Wild-type cells (top row) and sxm1 cells (bottom row) were probed for either an NLS-GFP reporter (left) or the endogenous protein Npl3p (right).
Deletion of SXM1 Does Not Markedly Affect tRNA Processing
By virtue of their in vivo interactions with small RNAs, La and Lhp1p have been suggested to have a number of functions in RNA biogenesis (Gottlieb and Steitz, 1989; Bachmann et al., 1990; Meerovitch et al., 1993; Yoo and Wolin, 1997). The first direct effect of Lhp1p on small RNA biogenesis has recently been described. Lhp1p was shown to be essential in vivo for 3' endonucleolytic maturation of pre-tRNA (Yoo and Wolin, 1997). In the absence of Lhp1p, pre-tRNA was matured by an exonucleolytic pathway that does not appear to contribute to pre-tRNA maturation in wild-type cells (Yoo and Wolin, 1997). Maturation is important not only for the biogenesis of tRNA, but it is also a prerequisite for tRNA export, as unprocessed RNAs are retained in the nucleus (Haselbeck and Greer, 1993). To determine if our Lhp1–PrA fusion is functional and to characterize further phenotypes of the SXM1 deletion, we analyzed maturation of tRNA Ser CGA in our Lhp1–PrA strains that are wild type or deleted for SXM1. By Northern analysis of total RNA, it is possible to discern three distinct, intron-containing pre-tRNA Ser CGA species in wild-type cells (Fig. 5, A–C; D is properly processed tRNA Ser CGA). The intermediates have 5′ and 3′ extensions (Fig. 5 A), only a 3′ extension (B), and no extension (C).
As previously described (Yoo and Wolin, 1997), deletion of LHP1 leads to an alteration in precursors A and B; in our assay, both are dramatically reduced in abundance (Fig 5, compare lanes 1 and 2). When the data are quantitated and normalized for intermediate C, whose abundance is independent of LHP1 genotype (Yoo and Wolin, 1997) (Fig. 5, bottom left) the peaks representing intermediates A and B are both clearly smaller. Deletion of SXM1 did not drastically affect the appearance of these intermediates for this pre-tRNA, although a minor effect was seen in precursor B (Fig. 5, compare lanes 3 and 4; see also Fig. 5, bottom right). It is possible that even in the absence of Sxm1p, some Lhp1p is brought into the nucleus with low efficiency by another pathway. As Lhp1p is likely to act catalytically, inefficient transport or diffusion of Lhp1p into the nucleus could account for the appearance of intermediates A and B in the SXM1-deficient strain.
Like Other Karyopherins, Sxm1p Binds Components of the Nuclear Pore Complex
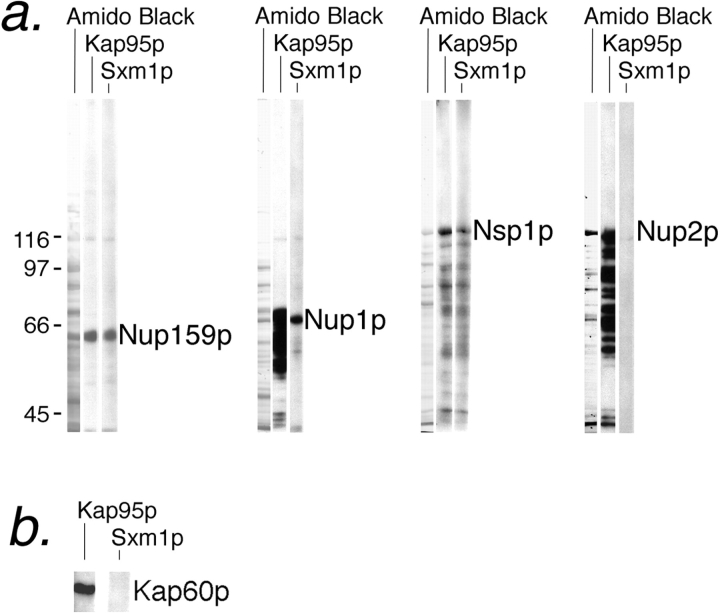
Having demonstrated in vivo that Sxm1p is necessary for both nuclear import and maximal activity of Lhp1p, we next tested the ability of Sxm1p to bind to known components of the yeast nuclear import machinery. As docking of transport complexes to the NPC is an essential step for import, we assayed the ability of Sxm1–PrA to bind either full-length or peptide-repeat regions of nucleoporins by a blot overlay assay. Bacterially expressed nucleoporins or nucleoporin fragments were separated by SDS-PAGE and transferred to nitrocellulose. Strips were overlaid with crude cytosol from our Sxm1–PrA strain. As a control, an adjacent strip was probed with cytosol from a Kap95–PrA strain. Like Kap95–PrA, Sxm1–PrA bound to full-length Nsp1p and the peptide-repeat domains of Nup159p and Nup1p. In contrast to Kap95–PrA, no binding of Sxm1– PrA to full-length Nup2p was seen (Fig. 6 a). As such, it appears that although both Sxm1p and Kap95p carry substrates through the NPC into the nucleus, they may each have distinct interactions along the way. In further experiments, Sxm1–PrA did not bind to either purified Kap60p (Fig. 6 b) or to purified Kap95p (data not shown) by overlay blot. As Sxm1–PrA does not appear to bind nucleoporins via Kap95p or substrates via Kap60p, Sxm1p most likely falls into the class of β karyopherins that includes Kap104p and Kap123p. The members of this class of karyopherins bind directly to both nucleoporins and to transport substrates.
Figure 6.
Sxm1–PrA binds to several nucleoporins but not to Kap60p. (a) Cytosol from either Kap95–PrA or Sxm1–PrA was overlaid on a nitrocellulose strip of electrophoretically separated crude E. coli lysate expressing repeat regions of Nup159p (amino acids 441–876; Kraemer et al., 1995) or Nup1p (amino acids 432– 816; Rexach and Blobel, 1995) or full-length Nsp1p or Nup2p. A strip stained for total protein with amido black is included for reference. (b) Purified Kap60p was overlaid with Kap95-PrA cytosol or Sxm1–PrA cytosol.
Discussion
In this study, we have shown that Sxm1p is a karyopherin and identified one of its major transport substrates. Lhp1p was shown to bind to Sxm1p in cytosol, a compartment where the complex between an import substrate and its karyopherin is expected to be stable by virtue of a low concentration of Ran-GTP. Furthermore, we observed that in a strain deleted for SXM1, Lhp1p was mislocalized to the cytoplasm, apparently unable to enter the nucleus. Significantly, the localization of other nuclear proteins was unaffected, suggesting that the Sxm1p pathway operates in parallel to previously characterized pathways. In addition to Lhp1p, three ribosomal proteins were isolated in complex with Sxm1p. It is possible that Sxm1p is able to mediate the import of these ribosomal proteins. As for Lhp1p, the import of the ribosomal proteins may be the result of either direct or indirect interaction with Sxm1p. Finally, we showed that Sxm1p is able to bind in vitro to peptide-repeat–containing nucleoporins, but not to Kap60p. Sxm1p, therefore, is a β karyopherin able to bind transport substrates and to dock at the NPC.
A Nuclear Import Pathway for Lhp1p
The La autoantigen directly binds all nascent RNA polymerase III transcripts (Stefano, 1984). In vivo, La is found in ribonucleoprotein particles with many classes of small RNAs, including pre-tRNA and U6 RNA, as well as the 5S rRNP (Hendrick et al., 1981; Rinke and Steitz, 1982, 1985). Specific nuclear retention and export of several classes of RNAs have been suggested to be mediated by La (Guddat et al., 1990; Boelens et al., 1995; Grimm et al., 1997). Lhp1p, the yeast homologue of La (Lin-Marq and Clarkson, 1995, Yoo and Wolin, 1994), was recently shown to be essential for accurate pre-tRNA processing (Yoo and Wolin, 1997). In wild-type cells, the 3′ end of tRNA is determined by an endonucleolytic cleavage facilitated by Lhp1p (Yoo and Wolin, 1997). As LHP1 is nonessential, it is evident that an alternative, exonucleolytic pathway can also produce mature, exportable tRNA. The relative kinetics and downstream events from each of these pathways is unknown. As Lhp1p binds to all RNA polymerase III transcripts and not just pre-tRNAs, it is likely to have additional roles in RNA biogenesis (Yoo and Wolin, 1997). Many of these other functions of Lhp1p are likely to be carried out in the nucleus as well. As a result, factors influencing the import of Lhp1p may be used to modulate its activities.
The human La protein (hLa) has recently been shown to contain a nuclear localization sequence capable of leading to the import of a normally cytosolic reporter (Simons et al., 1996). The hLa NLS bears several similarities to the consensus bipartite NLS (Dingwall and Laskey, 1991; Makkerh et al., 1996). One major difference, however, is the presence of several acidic residues in the hLa NLS. This NLS may have been assumed to mediate import via the karyopherin α/β1pathway. However, extrapolating from our data in yeast, it may be assumed that mammalian cells also have a parallel import pathway for this highly conserved protein mediated by a Sxm1p-type karyopherin. In this light, it should be noted that all of the proteins that we isolated in complex with Sxm1p have homologues in higher eukaryotes.
A Backup Pathway for the Import of Ribosomal Proteins
Like KAP123, both SXM1 and LHP1 are nonessential in yeast. Even so, the benefits of the Sxm1p pathway have outweighed its metabolic cost. SXM1 was originally isolated in a screen for genes able to complement the conditional loss of Pse1p (Seedorf and Silver, 1997). Kap123p has recently been shown to be the primary carrier of ribosomal proteins into the nucleus, possibly backed up by Pse1p (Rout et al., 1997). Interestingly, Seedorf and Silver (1997) have generated a strain lacking Kap123p with a conditional allele of Pse1p that rapidly accumulates nuclear mRNA at 36°C. Overexpression of SXM1 in this strain complements its growth defect and partially restores normal mRNA export (Seedorf and Silver, 1997). On the basis of having a phenotype relating to a defect in the export of macromolecules from the nucleus, Sxm1p has been proposed to be an export factor (Ullman et al., 1997).
Our results do not address the possibility that Sxm1p may have a role in the export of macromolecules from the nucleus. However, from our data we can conclude that Sxm1p is necessary for the import of Lhp1p. In addition, it is possible that the Sxm1p pathway also is able to import ribosomal proteins Rpl16p, Rpl25p, and Rpl34p. It has recently been shown that various karyopherins overlap in their ability to import ribosomal proteins (Rout et al., 1997). In fact, one of the ribosomal proteins that we isolated in complex with Sxm1p, Rpl16p, has also been isolated in complex with Kap123p (Rout et al., 1997). It is therefore likely that in cells lacking a functional Sxm1p import pathway, Rpl16p, Rpl25p, and Rpl34p would be imported by Kap123p or Pse1p. Likewise, it is possible that the genetic interaction seen by Seedorf and Silver (1997) between KAP123/PSE1 and SXM1 results not from an overlap in their ability to export mRNA but from their ability to import ribosomal proteins. Indeed, the accumulation of mRNA in the nucleus is a common phenotype, shown in one study to affect 2% of all temperature-sensitive strains examined (Kadowaki et al., 1994). 16 complementation groups were found, and the screen does not appear to be saturated (Kadowaki et al., 1994). Various modulators of nucleocytoplasmic transport have been shown to be associated with mRNA export phenotypes, including components of the NPC and regulators of the small GTPase Ran (Kadowaki et al., 1994; Doye and Hurt, 1997). In many of these cases, the RNA export phenotype is not a primary phenotype.
The Sxm1p Pathway May Coordinate tRNA Processing/ Export with Ribosome Biogenesis
We have described a nuclear import pathway, one of whose substrates is a protein required for a step in pre-tRNA maturation in wild-type cells. This 3′ maturation occurs via an endonucleolytic cleavage event. As LHP1 is not an essential gene, it is clear that the exonucleolytic processing of pre-tRNAs that occurs in its absence also produces mature, exportable tRNA. As tRNA maturation is upstream from export, it is possible that the karyopherin that we have characterized is involved in this process in Lhp1p-containing cells as well. Lhp1p binds in vivo to several other classes of small RNAs and thus may also be involved in their processing or localization. The convergence of the pathway to import Lhp1p and that which may import several ribosomal proteins could not have been predicted. However, this convergence is likely to be physiologically relevant. In this light, it should be noted that studies in Xenopus have implicated a relay of La and a ribosomal protein in the export of 5S rRNA (Guddat et al., 1990). Newly transcribed, nuclear 5S rRNA was seen to transiently bind the Xenopus La, followed by binding to the ribosomal protein L5. The L5/5S RNP then exited the NPC and could be observed in the cytoplasm. It is possible that the ribosomal proteins that we have isolated in complex with Sxm1p perform a similar function in accompanying Lhp1p-interacting RNAs out of the nucleus.
A further physiological role for the Sxm1p transport route may be to coordinate the processing and export of tRNA with ribosome biogenesis. As tRNA processing is an essential step that precedes export, the kinetics of processing may be capable of regulating export rate. Export of mature tRNA is extremely fast; in amphibians, about 2 × 109 molecules of a particular tRNA are exported per minute per nucleus (Zasloff, 1983). It is possible that regulation of the synthesis or activity of Sxm1p could affect the quantity of Lhp1p transported to the nucleus. Kinetic control of tRNA maturation could be achieved by controlling the contribution of two pathways, endonucleolytic and exonucleolytic, to 3′ tRNA processing, a process in which Lhp1p has been implicated (Yoo and Wolin, 1997). Furthermore, the potential Sxm1p substrate Rpl16p has a role in ribosome biogenesis (Rotenberg et al., 1988; Moritz et al., 1990, 1991). Therefore, Sxm1p may serve as a unifying factor in the coordination of tRNA export and ribosome assembly.
Acknowledgments
We thank Joe Fernandez and Farzin Gharahdaghi of the Rockefeller University Protein/DNA Technology Center for protein sequencing and mass spectral analysis. We are grateful to Christopher Yoo and Sandra Wolin for providing the lhp1 and LHP1 strains, John Aitchison and Mike Rout for Kap95-PrA cytosol, and Ulf Nehrbass, Monique Floer, and Michael Rexach for bacterially expressed proteins. The advice and suggestions of members of the Blobel lab are gratefully appreciated.
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- NLS
nuclear localization sequence
- NPC
nuclear pore complex
- PrA
protein A
Footnotes
J.S. Rosenblum was supported by a postdoctoral fellowship from the National Institutes of Health.
Address all correspondence to Günter Blobel, Laboratory of Cell Biology, Howard Hughes Medical Institute, 1230 York Avenue, Box 168, The Rockefeller University, New York, NY 10021. Tel.: (212) 327-8096. Fax: (212) 327-7880. E-mail: blobel@rockvax.rockefeller.edu
References
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Pfeifer K, Schroder HC, Muller WE. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell. 1990;60:85–93. doi: 10.1016/0092-8674(90)90718-t. [DOI] [PubMed] [Google Scholar]
- Boelens WC, Palacios I, Mattaj IW. Nuclear retention of RNA as a mechanism for localization. RNA. 1995;1:273–283. [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Biol Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Fernandez J, Andrews L, Mische SM. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Fischer U, Darzynkiewicz E, Tahara SM, Dathan NA, Luhrmann R, Mattaj IW. Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J Cell Biol. 1991;113:705–714. doi: 10.1083/jcb.113.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur Mol Biol Organ) J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos JF, Wagner P, Hall MN. Nuclear import substrates compete for a limited number of binding sites. Evidence for different classes of yeast nuclear import receptors. J Biol Chem. 1991;266:22303–22306. [PubMed] [Google Scholar]
- Gharahdaghi F, Kirchner M, Fernandez J, Mische SM. Peptide-mass profiles of polyvinylidene difluoride-bound proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in the presence of nonionic detergents. Anal Biochem. 1996;233:94–99. doi: 10.1006/abio.1996.0012. [DOI] [PubMed] [Google Scholar]
- Gottlieb E, Steitz JA. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO (Eur Mol Biol Organ) J. 1989;8:841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Lund E, Dahlberg JE. In vivo selection of RNAs that localize in the nucleus. EMBO (Eur Mol Biol Organ) J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guddat U, Bakken AH, Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- Haselbeck RC, Greer CL. Minimum intron requirements for tRNA splicing and nuclear transport in Xenopus oocytes. Biochemistry. 1993;32:8575–8581. doi: 10.1021/bi00084a026. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. 1981;1:1138–1149. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiaemRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Lin-Marq N, Clarkson SG. A yeast RNA binding protein that resembles the human autoantigen La. J Mol Biol. 1995;245:81–85. doi: 10.1006/jmbi.1994.0008. [DOI] [PubMed] [Google Scholar]
- Makkerh JPS, Dingwall C, Laskey RA. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud, N., and D. Goldfarb. 1992. Microinjected U snRNAs are imported to oocyte nuclei via the nuclear pore complex by three distinguishable targeting pathways. J. Cell Biol . 116:851–861. [DOI] [PMC free article] [PubMed]
- Michaud N, Goldfarb DS. Multiple pathways in nuclear transport: the import of U2 snRNP occurs by a novel kinetic pathway. J Cell Biol. 1991;112:215–223. doi: 10.1083/jcb.112.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Paulovich AG, Tsay Y-F, Woolford JL., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol. 1990;111:2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Pulaski BA, Woolford JL., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J. Molecular mechanisms of nuclear protein transport. Crit Rev Eukaryotic Gene Expr. 1997;7:61–72. doi: 10.1615/critreveukargeneexpr.v7.i1-2.40. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. The binding site of karyopherin α for karyopherin β overlaps with a nuclear localization sequence. Proc Natl Acad Sci USA. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. RanGTP-mediated nuclear export of karyopherin α involves its interaction with the nucleoporin Nup153. Proc Natl Acad Sci USA. 1997;94:9699–9704. doi: 10.1073/pnas.94.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rinke J, Steitz JA. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- Rinke J, Steitz JA. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 1985;13:2617–2629. doi: 10.1093/nar/13.7.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY. 198 pp.
- Rotenberg MO, Moritz M, Woolford JL., Jr Depletion of Saccharomyces cerevisiaeribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Silver PA. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons FH, Broers FJ, Van Venrooij WJ, Pruijn GJ. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. doi: 10.1006/excr.1996.0132. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- Tarn W-Y, Yario TA, Steitz JA. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of intron. RNA. 1995;1:644–656. [PMC free article] [PubMed] [Google Scholar]
- Ullman KS, Powers MA, Forbes DJ. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- Zasloff M. tRNA transport from the nucleus in a eukaryotic cell: carrier-mediated translocation process. Proc Natl Acad Sci USA. 1983;80:6436–6440. doi: 10.1073/pnas.80.21.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]