Abstract
A series of fusion protein constructs were designed to investigate the contribution of secretory nascent chains to regulation of the ribosome–membrane junction in the mammalian endoplasmic reticulum. As a component of these studies, the membrane topology of the signal sequence was determined at stages of protein translocation immediately after targeting and before signal sequence cleavage. Truncated translation products were used to delimit the analysis to defined stages of translocation.
In a study of secretory protein precursors, formation of a protease-resistant ribosome–membrane junction, currently thought to define the pathway of the translocating nascent chain, was observed to be precursor- and stage-dependent. Analysis of the binding of early intermediates indicated that the nascent chain was bound to the membrane independent of the ribosome, and that the binding was predominately electrostatic. The membrane topology of the signal sequence was determined as a function of the stage of translocation, and was found to be identical for all assayed intermediates. Unexpectedly, the hydrophobic core of the signal sequence was observed to be accessible to the cytosolic face of the membrane at stages of translocation immediately after targeting as well as stages before signal sequence cleavage. Removal of the ribosome from bound intermediates did not disrupt subsequent translocation, suggesting that the active state of the protein-conducting channel is maintained in the absence of the bound ribosome. A model describing a potential mode of regulation of the ribosome–membrane junction by the nascent chain is presented.
The structural requirements for signal sequence function in translocation across the ER membrane are well defined. Although signal sequences do not share a common primary sequence motif, they do display a strict conservation of physical characteristics. Such characteristics are paramount for function and consist of an amino-terminal positively charged domain (N-domain), a central hydrophobic core (H-domain), and a carboxy-terminal polar region containing, in nearly all precursor proteins, a consensus site for cleavage by the signal peptidase complex (von Heinje, 1984; von Heinje, 1985; Gierasch, 1989). The signal sequence performs at least two functions: it serves as a signal for targeting of ribosome-nascent chain complexes to the ER, an event mediated by the signal recognition particle (SRP)1, and is necessary for translocation initiation across the ER membrane (Blobel and Dobberstein, 1975; Walter et al., 1981a ,b).
It has been established that signal sequences physically interact with SRP, and that such interactions define the recognition stage of the ribosome/nascent chain targeting reaction (Walter et al., 1981a ; Kurzchalia et al., 1986). Recognition occurs via protein–protein interactions involving the methionine-rich domain of the 54-kD subunit of SRP and the hydrophobic core of the signal sequence (Romisch et al., 1990; Zopf et al., 1990). The mechanistic basis for signal sequence function after targeting is, however, unclear. That the recognition and targeting stages of translocation require direct physical interaction of the signal sequence with the 54-kD protein subunit of SRP suggests that subsequent functions of the signal sequence may also involve protein–protein interactions with membrane protein components of the ER (Wiedmann et al., 1987; Jungnickel and Rapoport, 1995; Voigt et al., 1996).
Isolated signal sequences spontaneously insert into phospholipid vesicles, and in doing so undergo conversion from a random to an α-helical conformation (Briggs et al., 1985; Batenburg et al., 1988; Gierasch, 1989; Jones and Gierasch, 1994a ,b). Of interest, studies with mutant signal peptides have indicated that the structural requirements for spontaneous insertion into membranes closely mimic the structural requirements for translocation in vivo (Briggs et al., 1985; Gierasch, 1989; Hoyt and Gierasch, 1991). These correlations have been extended in studies of mutant signal peptides to include the observations that translocation activity closely correlates with the mean central core hydrophobicity of the signal sequence, the capacity of the signal sequence to assume an α-helical conformation in a membrane-mimetic environment, and the capacity of the hydrophobic core to insert into the acyl chain region of the bilayer (Bird et al., 1987; Batenburg et al., 1988; Gierasch, 1989; Chou and Kendall, 1990; Hoyt and Gierasch, 1991). It has not yet been determined, however, whether such correlations are fortuitous, or rather representative of signal sequence function in vivo.
In addition to uncertainties regarding the site(s) of interaction of the membrane-bound signal peptide, the membrane topology of the ER-bound signal sequence remains to be established. Shaw et al. (1988) investigated this question through transfection of tissue culture cells with mutant forms of the vesicular stomatitis virus G-protein (VSV-G). The mutant proteins contained a point mutation that blocked signal peptide cleavage (as well as NH2-terminal extensions of the signal sequence) to allow signal sequence topology indentification by proteolysis assays. In protease protection studies of membranes isolated from infected cells, it was observed that the mutant constructs were retained in the ER in a topology consistent with the NH2 terminus remaining on the cytoplasmic side of the ER membrane (Shaw et al., 1988). Thus, the hydrophobic core and polar region of the signal sequence were observed to reside within the membrane, protected from digestion by exogenous proteases, whereas the NH2-terminal extensions were protease accessible (Shaw et al., 1988). These data provide insight into the topology of an uncleaved signal sequence after completion of the translocation reaction, and suggest that the signal sequence may assume an antiparallel loop topology throughout translocation.
Currently it is not understood how an antiparallel loop topology is conferred, be it through interaction with a protein receptor, the lipid bilayer, or both, or whether the antiparallel loop topology contributes mechanistically to translocation. Cross-linking studies have established that the signal sequence can reside in physical proximity to protein components of the ER membrane (Wiedmann et al., 1987; Krieg et al., 1989; Görlich et al., 1992a ,b; High et al., 1993; Nicchitta et al., 1995). Furthermore, it has been proposed that the signal sequence physically associates with the protein-conducting channel component Sec61p early in translocation, and thereby mediates formation of the tight ribosome–membrane junction thought to provide the pathway for the translocating nascent chain (Crowley et al., 1993, 1994; Jungnickel and Rapoport, 1995). Cross-linking studies have also demonstrated that the hydrophobic core of the signal sequence resides in direct physical proximity to membrane lipids, and thus the precise molecular architecture of the translocation site awaits further refinement (Martoglio et al., 1995).
In this communication, data are presented indicating that at all stages of translocation after signal cleavage, the signal sequence, including the hydrophobic domain, is accessible to the cis, or cytoplasmic face of the ER membrane. Analysis of staged translocation intermediate binding indicates that the association of secretory nascent chains with the ER membrane proceeds from an early salt-sensitive stage (in which the nascent chain can be extracted from the membrane after dissociation of the ribosome into its component subunits) to a later salt-insensitive stage in which the nascent chain is irreversibly assembled into the translocation site. Formation of a tight ribosome–membrane junction, as determined by proteolysis assays, was found to be precursor- and stage-dependent. After assembly of the nascent chain into the membrane, translocation can proceed in the absence of the intact ribosome, suggesting that the ribosome does not function to regulate continuously the activity state of the protein-conducting channel.
Materials and Methods
Construct Preparation
To experimentally determine the membrane topology of the signal sequence and mature domains of staged translocation intermediates, a construct was designed to allow isotope-specific labeling of either region of the nascent chain. This construct, termed TVG, contains the signal sequence of TRAPα (Hartmann et al., 1993) and leucine-free domains of the VSV-G protein and the ER chaperone GRP94. The construct was prepared as follows: By PCR, a cDNA encoding the TRAPα signal sequence was prepared using the oligonucleotide primers: sense: 5′-GCT-CTA-GAA-TGA-GGG-TCC-TCC-CGC-GC-3′ and antisense: 5′-GCG-GAT-CCA-TCT-GTG-GGT-TCA-TCT-TC-3′. Oligonucleotides were designed with 5′ add-on restriction enzyme sequences to facilitate subsequent cloning. A cDNA encoding a leucine-free domain of GRP94 (residues 247–313) was prepared by PCR using the oligonucleotide primers: sense: 5′-GCG-GAT-CCG-TCA-AGA-AAT-ATT-CAC-AG-3′ and antisense: 5′-GCG-AAT-TCC-TCC-CAA-TCC-CAG-AC. PCR products were gel-purified, digested with XbaI and BamHI (TRAPα) or BamHI and EcoRI (GRP94), and ligated into XbaI- and EcoRI-digested, gel-purified pGEMBP1 (Connolly and Gilmore, 1986). Ligation products were transformed into Escherichia coli strain DH5α, positive clones (pTG) were selected by ampicillin resistance, and sequences were confirmed by dideoxy sequencing of miniprep DNA. To allow labeling of the mature domain with [35S]methionine, a leucine-free domain of the VSV-G protein (residues 37–129), was prepared by PCR using the oligonucleotide primers: sense: 5′-GCA-GAT-CTC-ATA-GGC-ACA-GCC-ATA-3′ and antisense: 5′-GCG-GAT- CCA-GTT-CCT-TGT-TTC-GT-3′. The relevant PCR product was gel-purified, digested with BglII and BamHI, and ligated into BglII/BamHI-digested pTG. After transformation and antibiotic selection, positive colonies were selected, miniprep plasmid DNA was prepared, and positive clones (pTVG) were identified by dideoxy sequencing as described above. Using endogenous restriction sites, pTVG can be used to prepare transcripts encoding proteins of 60 (FokI) and 129 (NcoI) amino acids. During analysis of the binding and processing behavior of the 60- and 129-mer translation products, it was determined that a construct encoding an 80– amino acid form of TVG would prove useful. A TVG80 cDNA was thus prepared by PCR using the oligonucleotide primers: sense: 5′-GCT-CTA-GAA-TGA-GGG-TCC-TCC-CGC-GC-3′ and antisense: 5′-GCG-AAT-TCG-GAC-TGT-GTT-ATA-TAC-TT-3′ with the vector pTVG as template. The predicted PCR product was gel-purified, digested with XbaI and EcoRI, ligated into gel-purified, XbaI- and EcoRI-digested pTVG, and positive clones (pTVG80) were identified by antibiotic selection and dideoxy sequencing of miniprep plasmid DNA.
Cell-free Transcription and Translation
pTVG was linearized within the coding region by digestion with FokI or NcoI, and was transcribed to yield mRNA transcripts encoding precursors of 60 and 129 amino acids, respectively. The plasmid pTVG80 was digested with EcoRI and transcribed to yield mRNA transcripts encoding a precursor of 80 amino acids. Truncated preprolactin transcripts were prepared as described previously (Nicchita et al., 1995). Transcription reactions were performed in a buffer containing 40 mM Tris/HCl (pH 8.0), 8 mM Mg(OAc)2, 25 mM NaCl, 2 mM spermidine, 10 mM dithiothreitol, 2.5 mM ATP, CTP, UTP, and GTP, 5 U/ml yeast inorganic pyrophosphatase, and 1 U/μl T7 RNA polymerase. Transcription reactions were extracted with phenol/chloroform, and mRNA was collected by ethanol precipitation at room temperature. mRNA pellets were resuspended in Tris/EDTA to a final concentration of 500 ng/μl and stored at −80°C.
Cell-free translations were performed in a rabbit reticulocyte lysate system as described in Nicchitta and Blobel (1989). Translations (20 μl) contained 8 μl of nuclease-treated rabbit reticulocyte lysate, 25 μCi of [35S]pro-Mix (methionine/cysteine), 0.05 U/μl RNasin, 1 mM DTT, 80 μM minus methionine amino acid mix and, where indicated, one equivalent of rough microsomes, as defined in Walter and Blobel (1983). In signal sequence–labeling reactions, 20 μCi of [3H]leucine was present, and a minus leucine amino acid mixture was substituted for the minus methionine mix. The ionic conditions of all translations were adjusted to 110 mM KOAc, 2.5 mM Mg(OAc)2. Nuclease-treated rabbit reticulocyte lysate was prepared by the method of Jackson and Hunt (1983), and canine pancreas rough microsomes were prepared by the method of Walter and Blobel (1983).
Analysis of Membrane Association
Cell-free translation reactions (20 μl) were chilled on ice and diluted to 150 μl in physiological salts buffer (110 mM KOAc, 20 mM K-Hepes, pH 7.2, 2.5 mM Mg[OAc]2), high-salt buffer (0.5 M KOAc, 20 mM K-Hepes, pH 7.2), neutral Tris buffer (0.5 M Tris/HCl, pH 7.2), or magnesium-free, physiological salts buffer (110 mM KOAc, 20 mM K-Hepes, pH 7.2, with or without the indicated concentrations of EDTA). After incubation on ice for 30 min, samples were overlaid onto a 70-μl cushion of 0.5 M sucrose, 20 mM K-Hepes, pH 7.2, and centrifuged in the Beckman TLA100 rotor (Fullerton, CA) for 5 min at 60 K rpm (4°C).The supernatant and cushion fraction was removed and processed as the supernatant. The pelleted membrane fraction was recovered by addition of 20 μl of 0.5 M Tris base, 5% SDS, 0.1 M DTT, and heating to 55°C for 15 min.
Protease Protection Studies
Completed translation reactions were chilled on ice and diluted to 50 μl in 110 mM KOAc, 20 mM K-Hepes, and 2 mM Mg(OAc)2. Proteinase K was added from freshly prepared stocks in water. Unless otherwise indicated, the final proteinase K concentration was 150 μg/ml. Where indicated, the detergent CHAPS was present at 0.5% (wt/vol) final concentration. Digestion reactions were performed for 30 min on ice and quenched by addition of PMSF to a final concentration of 2 mM.
Membrane Reconstitution
Lumenal protein-depleted RM were reconstituted by the procedures described in Nicchitta and Blobel (1990) and Nicchitta et al. (1991).
Sample Processing
After translation/translocation, reactions were placed on ice and diluted to 50 μl in 110 mM KOAc, 20 mM K-Hepes, 2 mM Mg(OAc)2. Two volumes of saturated ammonium sulfate solution were added and, after a 20-min incubation on ice, samples were centrifuged for 10 min at 15,000 g. In membrane centrifugation experiments, the supernatant fraction was diluted twofold with saturated ammonium sulfate and processed as described above. Supernatants were removed, and the pellets were washed in 10% TCA. Pellet fractions were resuspended in 35 μl of 0.5 M Tris base, 5% SDS, 0.1 M DTT, and heated to 55°C for 20 min. Samples were resolved on 12.5% Tris-Tricine gels by the protocol of Schägger and von Jagow (1987), fixed in 35% methanol, 10% acetic acid, and were either dried ([35S]methionine) or prepared for fluorography by the method of Bonner and Laskey (1974). Translations performed with [35S]methionine were analyzed and quantitated using a Fuji MacBAS1000 phosphorimager system (Tokyo, Japan) and version 2.2 software. Translations performed with [3H]leucine were subject to fluorography on Kodak XAR5 film (Rochester, NY) at −80°C. All phosphorimager files were exported as PICT images, and size and contrast was adjusted using Adobe Photoshop software (version 3.0; Adobe Systems, Inc., Mountain View, CA).
Results
Construct Design
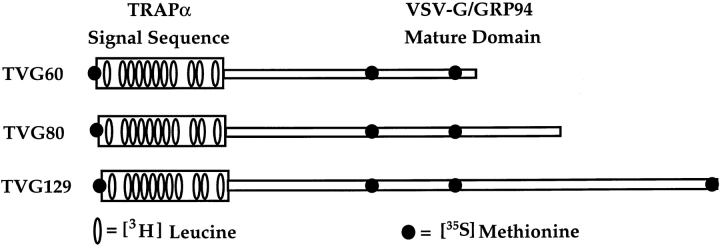
A series of constructs was designed to investigate the stage-specific topological disposition of the signal sequence during translocation in the endoplasmic reticulum and the role of the nascent chain in ribosome–membrane junction regulation. One family of constructs, termed TVG, contains the signal sequence of TRAPα, which contains 11 leucine residues and fragments of the mature domain of VSV-G and GRP94, chosen solely to provide a leucine-free mature domain that could be biosynthetically labeled with [35S]methionine. Restriction enzyme cleavage of the TVG cDNA and in vitro transcription was used to prepare transcripts encoding 60, 80, and 129 amino acid precursors. Precursor sizes were chosen to allow staging of the translation products from signal sequence binding (60-mer) to that phase of translocation immediately preceding signal sequence cleavage (129-mer). A schematic illustration of the domain-specific isotope-labeling patterns of the three translation products is presented in Fig. 1 A. Although not illustrated, all translation products lack termination codons and are expected to remain in association with the ribosome after synthesis.
Figure 1.
Schematic illustration of differential labeling constructs. A chimeric cDNA was designed to allow isotope-specific labeling of either the signal sequence or the mature portion of the protein. This construct, termed TVG, contains the signal sequence from TRAPα and a mature domain consisting of portions of the VSV-G and GRP94 coding sequence chosen solely on basis of amino acid distribution. With this construct, synthesis in the presence of [3H]leucine will yield isotopic labeling of the hydrophobic core of the signal sequence, whereas synthesis in the presence of [35S]methionine will yield products in which the majority of the labeling is limited to the mature domain.
Membrane Targeting and Physical Characteristics of Bound TVG Translation Products
To investigate the capacity of the TVG60, 80, and 129-mer constructs to target and bind to the ER membrane, in vitro translations were performed in the presence of pancreas rough microsomes (RM), the completed reactions were chilled on ice, and the interactions of the translation products with the membrane were evaluated by extraction and sedimentation assays (Gilmore and Blobel, 1985; Connolly and Gilmore, 1986). In these assays, translation/translocation reactions are diluted into buffers of varying salt composition, and the physical characteristics of the binding interactions are subsequently assessed by centrifugation of the membrane fraction free from the translation mix. Insensitivity of the translation product to extraction with EDTA or high salt, as well as resistance to digestion by exogenous proteases, are diagnostic of the formation of a translocation-competent ribosome–membrane junction (Gilmore and Blobel, 1985; Connolly and Gilmore, 1986; Connolly et al., 1989; Nicchitta and Blobel, 1989).
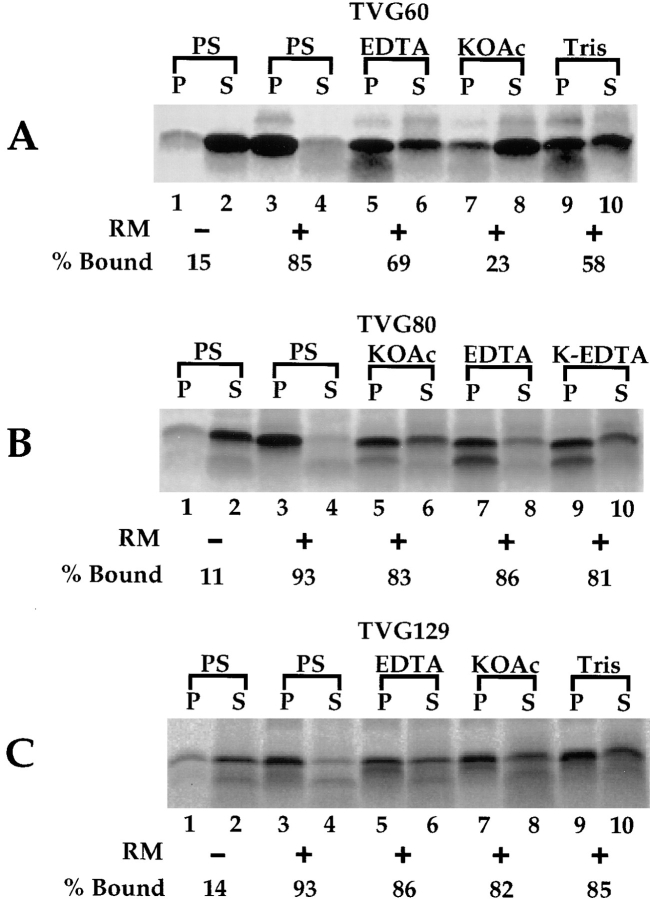
When translations were performed in the absence of RM, the TVG translation products were recovered predominately in the supernatant fraction, whereas in the presence of RM, all translation products were efficiently targeted to the RM fraction (Fig. 2, A–C; compare lanes 1 and 2 with lanes 3 and 4). In a centrifugation assay of targeting, then, the TVG60, 80, and 129-mer translation products were indistinguishable.
Figure 2.
Characterization of TVG translation product–membrane interactions. mRNAs encoding the TVG 60-, 80-, or 129-mer were translated in a reticulocyte lysate translation system in the presence or absence of canine pancreas rough microsomes (RM). After translation, reactions were chilled on ice and diluted eightfold in physiological salts buffer (PS), PS + 0.5 M KOAc (KOAc), PS + 7.5 mM EDTA, 0.5 M KOAc + 7.5 mM EDTA (K-EDTA) or 0.5 M Tris, pH 7.2. After a 30-min incubation on ice, translations were fractionated by centrifugation to yield membrane-bound (P) and supernatant (S) fractions. Samples were separated on 12.5% Tris-Tricine gels; a digital image derived from phosphorimager analysis is depicted. (A) TVG 60-mer; (B) TVG80-mer; and (C) TVG129-mer. Quantitation was based on both precursor and processed forms of the nascent chains.
Substantial differences in the physical characteristics of the bound TVG translation products were observed after extraction with buffers of high salt concentration. Most notably, when TVG60-mer translations were extracted with 0.5 M KOAc or 0.5 M Tris base, pH 7.2, a substantial fraction of the translation product was recovered in the supernatant fraction (Fig. 2 A, lanes 3 and 4 vs. 7–10). In contrast to the TVG60-mer translation products, TVG80- and TVG129-mers translated in the presence of RM were resistant to extraction under all conditions assayed (Fig. 2, A and B). In the experiment depicted in Fig. 2 A, high salt extractions were conducted in the absence of magnesium. Under these conditions, ribosomes dissociate into their component subunits (Noll and Noll, 1976), as is also observed after EDTA treatment (Sabatini et al., 1966). Substantial release of TVG60-mer nascent chains was not observed upon addition of EDTA, but was clearly apparent in the presence of 0.5 M KOAc, suggesting that at early stages of translocation the nascent chain is bound to the membrane, at least in part through electrostatic interactions between the nascent chain and components of the ER membrane, and furthermore, that such binding interactions occur independent of the ribosome.
Ribosome-independent Binding of Short Chain Intermediates at Early Stages of Translocation
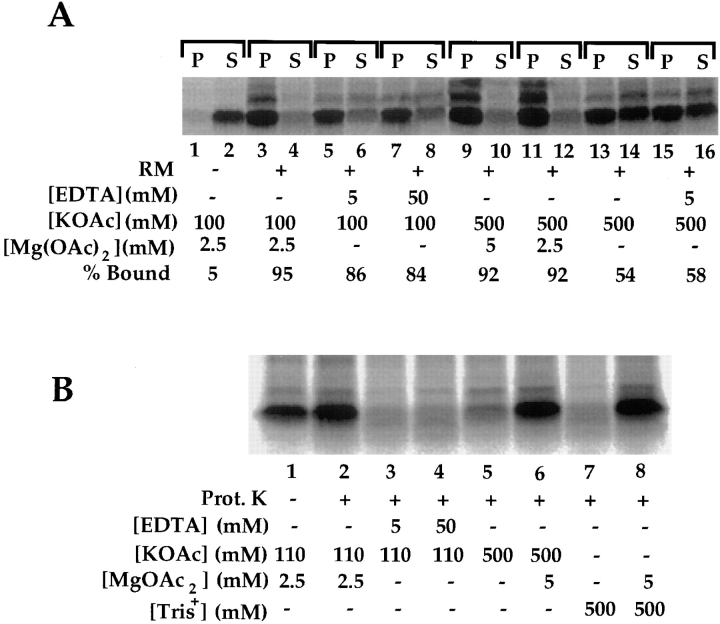
Previous investigations into the initial ribosome/nascent chain binding stage have indicated that salt- and protease-resistant nascent chain binding occur concommitantly, and thereby define a discrete stage of the insertion reaction in which the ribosome-nascent chain complex is bound to Sec61p (Jungnickel and Rapoport, 1995). In view of the salt-sensitive binding of TVG60-mer nascent chains depicted in Fig. 2, the characteristics of the TVG60-mer ribosome–membrane junction were further investigated. In the experiment depicted in Fig. 3 A, TVG60-mer nascent chains were synthesized in the presence or absence of RM, and the ribosome requirement for stable binding of the nascent chain to the membrane was determined. As shown in Fig. 2, TVG60-mer efficiently targeted to ER membranes (Fig. 3 A, lanes 1–4). Bound nascent chains remained in tight association with the ER membrane after addition of 5 or 50 mM EDTA (lanes 5–8). In the presence of 50 mM EDTA, the small ribosomal subunit is wholly, and the large ribosome subunit substantially, released from the membrane (Sabatini et al., 1966; Nicchitta, C.V., unpublished observations), a further indication that the binding of TVG60-mer nascent chains to the ER membrane is, in large part, independent of the ribosome. Interestingly, extraction with high salt, in the presence of either 2.5 or 5 mM magnesium, did not effect release of the nascent chain, whereas extraction with high salt in the absence of magnesium provoked release of a substantial fraction of the membrane-bound nascent chains (Fig. 3 A, lanes 9–16). These data are consistent with the interpretation that release of TVG60-mer nascent chains requires disassembly of the ribosome, which will occur after addition of 0.5 M salt in the absence, but not the presence, of magnesium (Wettenhall and Wool, 1974).
Figure 3.
Characterization of the ribosome–membrane junction of short chain intermediates. TVG60-mer nascent chains were synthesized in the presence or absence of rough microsomes, and, after translation, were subjected to extraction under differing salt and magnesium buffer conditions for 30 min on ice (A), or after addition of the indicated buffers and incubation on ice for 30 min, treated with proteinase K (150 μg/ml) for an additional 30 min on ice (B). In the experiment depicted in A, samples were resolved into pellet (membrane-bound) and supernatant fractions by centrifugation. Samples were prepared for SDS-PAGE as described in Materials and Methods. Digital images derived from phosphorimager analysis are depicted.
When the protease accessibility of the membrane-bound nascent chains was determined under the conditions used to assess nascent chain binding, it was evident that in the presence of EDTA (Fig. 3 B, lanes 3 and 4) or magnesium-free, high-salt buffers (Fig. 3 B, lanes 5 and 7), nascent chains were readily degraded by added protease, as would be predicted on the basis of the well-characterized effects of these reagents on ribosome structure. When ribosome structure is preserved by the continued presence of magnesium, the nascent chains are fully protected from proteolytic digestion (Fig. 3 B, lanes 6 and 8). Of particular interest are the observations that after addition of EDTA, the nascent chains are fully accessible to proteolytic digestion (Fig. 3 B, lanes 3 and 4), yet are in tight association with the ER membrane (Fig. 3 A, lanes 5–8). Therefore, it is evident that at early stages of translocation, the nascent chain associates with components of the ER membrane via, at least in part, electrostatic interactions. Such interactions are maintained in the absence of bound ribosomes or intact ribosomal subunits.
Salt-sensitive Nascent Chain Binding Correlates with Translocation Activity
The results presented in Figs. 2 and 3 define two stages of nascent chain interaction with the ER membrane. At an early binding stage, as displayed by the TVG60-mer, nascent chains are efficiently targeted to the ER membrane but can be extracted with magnesium-free high-salt buffers. At later stages of targeting, defined by the TVG80- and 129-mers, nascent chains bind to the ER membrane in a salt-, protonated amine (Tris)-, and EDTA-insensitive manner. To identify functional correlates to the different binding stages, the translocation activity of the different precursors was investigated.
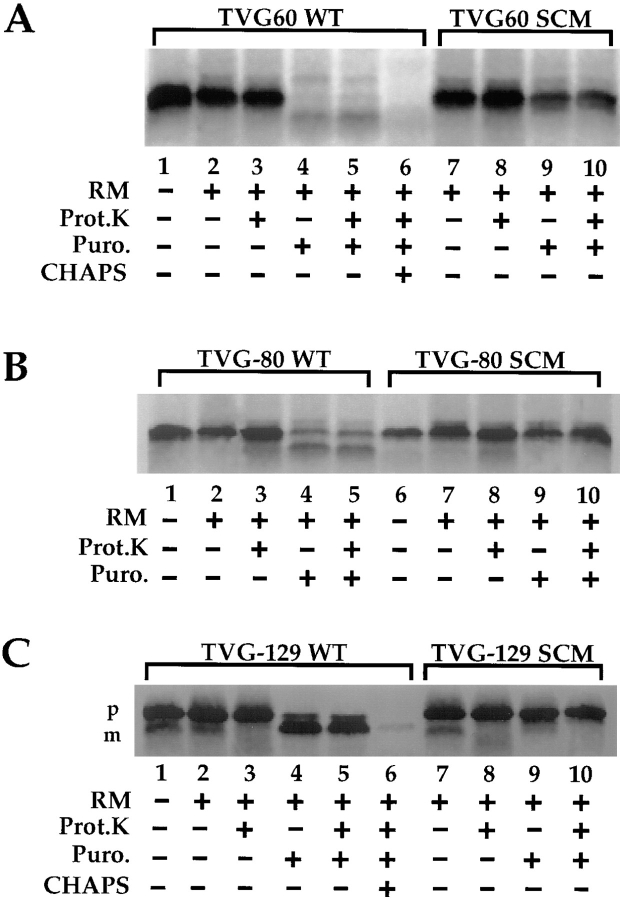
In the experiments depicted in Fig. 4, A–C, both wild-type (WT) and signal cleavage mutant (SCM) forms of the TVG60, 80, and 129-mers were investigated. The SCM forms contain a point mutation in the (-3,-1) signal peptidase cleavage site that prevents cleavage of the signal sequence by the signal peptidase complex. Although the TVG60-mer could readily be distinguished from the TVG80- and 129-mers in extraction assays of nascent chain interaction with the ER membrane, the membrane-bound translocation intermediates were markedly resistant to degradation by exogenous proteases (Fig. 3, A–C, lanes 2, 3, 7, and 8). Thus, in both centrifugation (Fig. 2) and proteolysis (Fig. 4) assays of ribosome/nascent chain targeting, all targeted intermediates appear to bind to the ER membrane in an identical manner and/or to identical sites. Depending upon the stage of translocation accessed by the nascent chain, however, the precursor may or may not be bound to the membrane in a salt-resistant manner. These data are consistent with findings from a recent study demonstrating that when membrane binding of the preprolactin 86-mer is performed at saturation (i.e., limiting membrane concentrations), membrane bound 86-mer nascent chains are protease-resistant, yet partially salt extractable (Murphy et al., 1997), and indicate that protease-resistant binding of ribosome/nascent chain complexes to the ER membrane is distinct from the process that yields a stably bound nascent chain.
Figure 4.
Translocation activity of TVG60-, 80-, and 129-mer translation products. Wild-type (WT) and signal cleavage mutant (SCM) forms of TVG60-, 80-, and 129-mer mRNAs were translated in the presence or absence of RM for 30 min at 25°C, and translocation was initiated by addition of 0.5 mM puromycin and incubation for 10 min at 25°C. Samples were chilled on ice, and translocation was assayed by protease protection assays with proteinase K (150 μg/ml) for 30 min on ice. Samples were processed as described in Materials and Methods, and were separated on 12.5% Tris-Tricine gels. A digital image derived from phosphorimager analysis is depicted.
After puromycin-dependent release of the nascent chain from the ribosome, the TVG60-, 80-, and 129-mer precursors displayed significant differences in translocation activity. Whereas the TVG129-mer was efficiently converted to a signal-cleaved mature form, which was protected from proteolytic degradation in the absence, but not the presence, of detergent (Fig. 4 C, lanes 2–6), the TVG60-mer was significantly, and the TVG80-mer partially, degraded (Fig. 4, A and B, lanes 3–5). These data suggest that the translocation competence of the membrane-bound precursors is markedly influenced by the size of the nascent chain. Degradation was reduced with the SCM forms, indicating that signal sequence cleavage enhances access to the relevant protease(s) (Fig. 4 A, lanes 2–4 vs. lanes 8–10). It is important to note that the translation products were biosynthetically labeled with [35S]methionine, and thus the disappearance of the precursor represents degradation of the signal sequence and mature regions of the protein. The degradation process was ATP- and proteasome inhibitor– independent, and required an intact membrane (data not shown). Further characterization of this degradation process will be reported elsewhere.
The Hydrophobic Core of the Signal Sequence Accesses the Cytosolic Domain of the Membrane
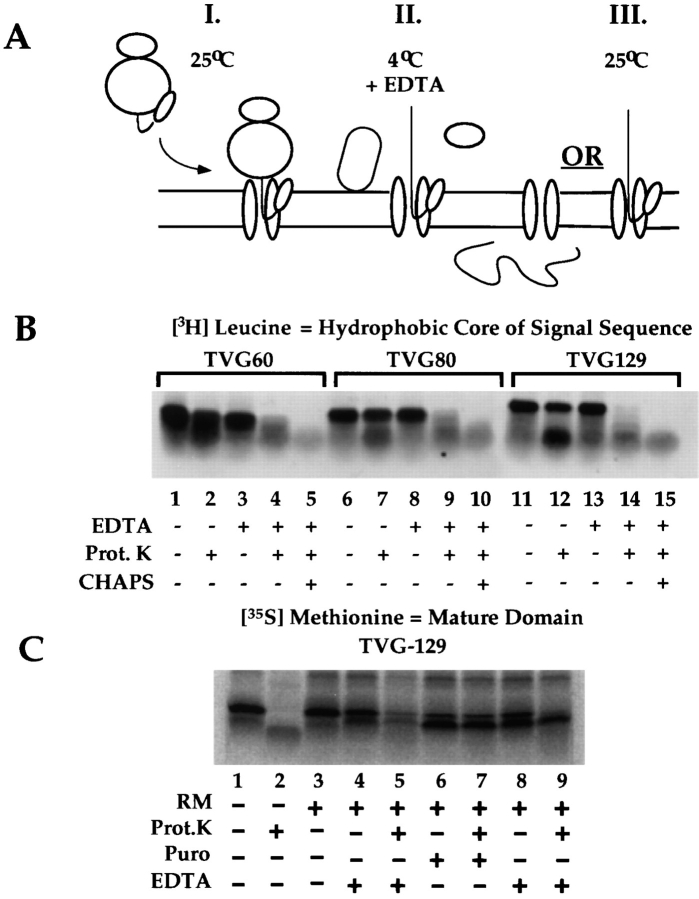
As noted previously, available experimental evidence suggests that the signal sequence assumes an antiparallel loop topology during translocation (Shaw et al., 1988). On the basis of this report, and with regard to the observations presented in Figs. 2–4, we postulated that the differences in membrane association and translocation behavior observed for the TVG60-, 80-, and 129-mers may reflect a scenario in which short chain (60-mer) precursors are of insufficient length to form a loop topology, and thus cannot associate with components of the ER membrane in a stable manner. If this explanation is correct, it would be predicted that the signal sequence membrane topology of the 60-, 80-, and 129-mer nascent chains would differ. Of relevance to this proposal, Shaw et al. (1988) demonstrated in proteolysis assays of completed translation products that the polar domain and hydrophobic core of the signal peptide were buried within the membrane, and were thus inaccessible to proteolytic digestion, whereas the NH2-terminal domain of the signal peptide extended into the cytosol.
To determine if the signal sequences of the TVG60-, 80-, and 129-mer nascent chains displayed any topological differences, precursors were synthesized in the presence of [3H]leucine specifically to limit incorporation of isotope into the hydrophobic core of the signal sequence, and membrane-bound nascent chains were isolated by centrifugation. The targeted nascent chains were then subjected to proteolysis on ice to determine the accessibility of the signal sequence (see schematic, Fig. 5 A, I). Consistent with previous data indicating that 35S-labeled nascent chains are protected from proteolytic degradation (Fig. 3), the majority of the ribosome-associated membrane-bound 3H-labeled signal sequences were protected from proteolytic degradation (Fig. 5 B, lanes 1, 2, 6, 7, 11, and 12). A small but reproducible increase in the nascent chain protease sensitivity as a function of chain length, however, was observed (Fig. 5 B, lanes 2, 7, and 12). Because of technical difficulties in accurately quantitating tritium- labeled translation products in excised PAGE gel fragments, we have been unable to precisely define the degree of sensitivity. In quantitating similar experiments performed with [35S]methionine labeling, however, the protease-sensitive fraction did not exceed 18 ± 7% (n = 5) of the total. We assume, then, that the protease-sensitive fraction seen for ribosome-associated, membrane-bound translation products may represent up to 18% of the total (Fig. 5 B, lanes 2, 7, and 12). In addition, because the tritium-labeled limit digestion products seen in Fig. 5 B, lanes 7 and 12, represent signal sequence labeling, it is clear that the observed proteolysis occurs at a site between the ribosome-protected domain and the signal sequence, suggesting that the characteristics of the ribosome–membrane junction may vary as a function of nascent chain length.
Figure 5.
The hydrophobic core of the signal sequence is accesible to the cytoplasmic domain of the ER membrane. (A) Schematic illustration of experimental protocol. TVG translation products were synthesized in the presence of RM at 25°C, to yield membrane-targeted intermediates. Reactions were chilled on ice, and EDTA was added to dissociate the bound ribosome. Under these conditions (4°C), translocation is blocked. For the experiment depicted in C, samples were warmed to 25°C, and translocation was assayed by signal sequence cleavage and protease protection experiments. (B) Stage-specific accessibility of the signal sequence to proteolytic degradation. TVG60-, 80-, and 129-mers were translated in the presence of RM and [3H]leucine to label the signal sequence. After translation, the membrane-bound translation products were isolated by centrifugation, resuspended, and the accessibility of the bound translation products to digestion by exogenous protease was assayed. In lanes 2, 7, and 12, the accessibility of the ribosome-associated, membrane-bound precursors was assayed, whereas in lanes 4, 5, 9, 10, 14, and 15, the ribosome was disassembled by addition of 7.5 mM EDTA before proteolysis. Protease digestions were performed on ice for 30 min. Samples were processed and analyzed by SDS-PAGE. Translation products were detected by fluorography of PPO impregnated gels. (C) Effects of ribosome dissociation on translocation of targeted TVG129-mer. TVG129-mer was translated in the presence of [35S]methionine and RM. Bound translation products were recovered by centrifugation, resuspended in membrane buffer, and placed on ice. Aliquots were treated on ice for 15 min with either 7.5 mM EDTA (lanes 4, 5, 8, and 9) or 0.5 mM puromycin (lanes 6 and 7) and subsequently treated with protease (lane 5) or warmed to 25°C for 10 min (lanes 6–9). Samples 6–9 were subsequently chilled and, where indicated, subjected to proteolysis as described above. Samples were processed and analyzed by SDS-PAGE. A digital image derived from phosphorimager analysis is depicted.
To determine the membrane topology of the signal sequence at the stages of translocation represented by the TVG60-, 80-, and 129-mer translation products, ribosome/ nascent chain/membrane complexes were chilled on ice, and the ribosomes were extracted by EDTA addition (see schematic Fig. 5 A, II). Subsequently, the accessibility of the signal sequence to the cytosolic compartment was assayed by proteolysis. Under these conditions, the signal sequences of the three precursors were readily accessible to protease digestion, indicating that at early stages of translocation, the hydrophobic core of the signal sequence resides in, or reversibly populates, an environment that includes the cytosolic domain of the ER bilayer (Fig. 5 B, lanes 3–5, 8–10, and 13–15).
The protease accessibility data depicted in Fig. 5 B suggest that the antiparallel topology identified for completed signal cleavage mutant forms of the VSV-G signal peptide may not be conferred until late stages of translocation, perhaps those stages associated with protein synthesis termination. To address the possibility that the experimental conditions used to obtain this result (EDTA-mediated disassembly of the membrane-bound ribosome) yield an altered signal sequence topology, however, functional studies with 35S-labeled TVG129-mer nascent chains were performed. These studies were designed to determine the activity of the signal sequence after ribosome/nascent chain targeting and removal of the membrane-bound ribosome. Thus, TVG129-mers were translated in the presence of RM, the reactions were chilled on ice, treated with EDTA, and the samples were subsequently rewarmed to 25°C (see schematic Fig. 5 A, II and III). In the experiment depicted in Fig. 5 C, protease accessibility of the TVG129-mer was assayed in the absence of membranes (lanes 1 and 2), the presence of membranes (lanes 3 and 4), after disassembly of the ribosome on ice (lane 5), after addition of puromycin at 25°C (lanes 6 and 7), or after EDTA-dependent disassembly of the ribosome on ice and subsequent warming of the reaction to 25°C (lanes 8 and 9). To summarize the data, the TVG129-mer was, as expected, protease-accessible in the absence of RM and after disassembly of the membrane-bound ribosome/nascent chain complexes on ice. Because the nascent chains depicted in Fig. 5 C were labeled with [35S]methionine, these data indicate that the mature domain of the membrane-bound TVG129-mer nascent chain resides in the cytosol upon disassembly of the ribosome. TVG129-mer nascent chains, released from the ribosome by addition of EDTA on ice, were translocated upon warming of the sample to 25°C (Fig. 5 C, lanes 8 and 9). These data indicate that after targeting and salt-resistant membrane binding of the nascent chain, protein translocation can occur in the absence of the intact, bound ribosome. Therefore, the continued presence of the bound ribosome is not itself essential for channel function. Furthermore, the combined data indicate that the hydrophobic core of the signal sequence can reside, or reversibly populate, the cytosolic domain of the ER membrane, and yet be fully competent to mediate protein translocation.
Analysis of the Ribosome–membrane Junction in Native and Reconstituted Membranes
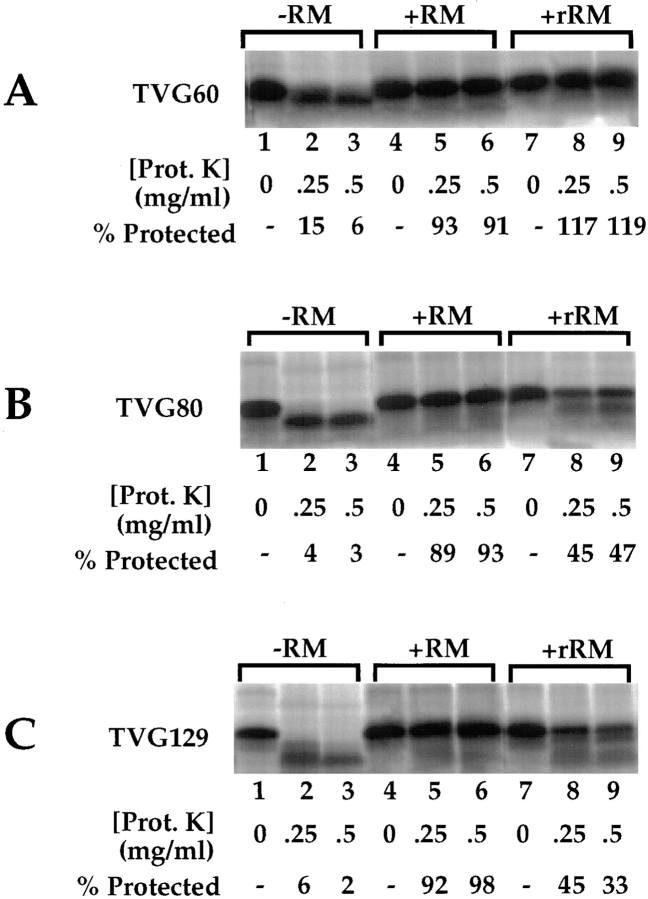
The data presented in Figs. 2–5 indicate that membrane-bound early (TVG60-mer) intermediates can be distinguished from late (TVG80-, 129-mer) intermediates by their sensitivity to extraction with high salt, yet the topology of the signal sequence for the different intermediates is identical, and all intermediates are bound to the membrane in a protease-resistant environment. To test the hypothesis that the differences in early- and late-stage intermediates may occur through late-stage specific interactions of translocation intermediates with components of the ER membrane, the ribosome–membrane junction for the TVG60-, 80-, and 129-mer intermediates was investigated in native and reconstituted membranes (Nicchitta and Blobel, 1990; Nicchitta et al., 1991; Görlich et al., 1992a ). Previously we have reported that reconstituted membranes, which lack the complement of lumenal chaperone/protein-folding enzymes, efficiently translocate nascent chains up to the stage of signal peptide cleavage, yet display reduced activity for net transfer of the nascent chain into the vesicle lumen (Nicchitta and Blobel, 1990). This reduction in activity is thought to represent the contribution of lumenal proteins to translocation of late-stage intermediates, as it is mimicked by depletion of the lumenal proteins (Nicchitta and Blobel, 1993) and, in yeast, by mutations in either the lumenal hsp70 protein Kar2p (BiP) (Sanders et al., 1992) or by its membrane regulatory partner Sec63p (Lyman and Schekman, 1995).
In these experiments, TVG60-, 80-, and 129-mer nascent chains were synthesized in the absence of RM, or in the presence of native or reconstituted RM (rRM). The ribosome–membrane junction was subsequently investigated by protease protection assays. As illustrated in Fig. 6 A, lanes 1–3, TVG60-mer nascent chains synthesized in the absence of membranes were readily degraded by proteases to yield the ribosome-protected fragment (Malkin and Rich, 1967). When synthesized in the presence of native (Fig. 6 A, lanes 4–6), or reconstituted membranes (Fig. 6 A, lanes 7–9), the TVG60-mer nascent chains were completely protected from digestion with proteases present at concentrations up to 0.5 mg/ml. The TVG80- and 129-mer nascent chains behaved identically to the TVG60-mer nascent chains when proteolysis assays were conducted on nascent chains synthesized in the absence of membrane, or in the presence of native membranes (Fig. 6, B and C, lanes 1–6). When synthesized in the presence of rRM, however, TVG80- and TVG129-mer nascent chains were partially degraded by exogenous protease (Fig. 6, B and C, lanes 7–9). These data suggest that the ribosome–membrane junction is, at least in part, regulated by the nascent chain. Thus, although the ribosome–membrane junction is identical in native and reconstituted membranes with regard to early secretory translocation intermediates, the characteristics of the junction in native and reconstituted membranes differ with later stage intermediates.
Figure 6.
Stage-dependent variations in the ribosome–membrane junction in native and reconstituted RM. TVG60-, 80-, and 129-mers were translated in the presence of native membrane (RM) or reconstituted membranes lacking lumenal contents. After translation, samples were chilled on ice and subject to proteolysis as described in Materials and Methods. After proteolysis, samples were processed for SDS-PAGE and analyzed by phosphorimager analysis. Quantitation was performed using MacBAS v2.2 software. Digital images derived from phosphorimager analysis are depicted.
The Tight Ribosome–Membrane Junction Is Stage and Precursor Specific
The data presented in Fig. 6 can be interpreted to indicate that the ribosome–membrane junction, as assayed by proteolysis, is regulated by the nascent chain at early stages of presecretory protein translocation. Compelling evidence for sequence-specific regulation of the ribosome–membrane junction has been demonstrated for pause-transfer sequences (Hegde and Lingappa, 1996) and single-spanning transmembrane domains (Liao et al., 1997). In view of the fact that the data presented in Fig. 6 was obtained with a chimeric protein, this conclusion was further examined in studies of preprolactin (pPl) intermediates in native and reconstituted membranes.
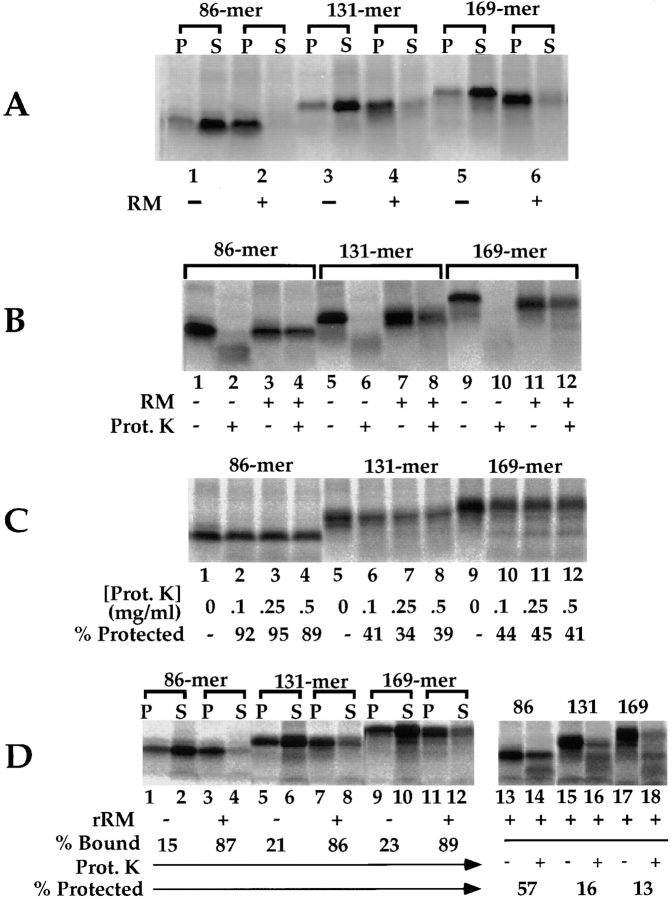
In these experiments, truncated pPl nascent chains of 86, 131, and 169 amino acids were translated in the presence or absence of native or reconstituted membranes, and the ribosome–membrane junction was investigated by proteolysis. The results of these experiments are depicted in Fig. 7 (A–D). To insure that the analysis was limited to membrane-targeted nascent chains, conditions were first defined in which the ribosome/nascent chain complexes were efficiently targeted to the RM. As shown in Fig. 7 A, under the described assay conditions, preprolactin 86-, 131-, and 169-mer nascent chain complexes were targeted to the RM with high efficiency. When membrane-bound 86–, 131–, and 169–amino acid precursors were subject to proteolysis in the absence of RM, all assayed precursors were protease-sensitive (Fig. 7 B, lanes 1, 2, 5, 6, 9, and 10). In contrast, only the 86-mer was efficiently protected from proteolytic degradation when translation was performed in the presence of RM (Fig. 7 B, lanes 3, 4, 7, 8, 11, and 12). These observations were further extended in the experiment depicted in Fig. 7 C, which depicts the relative protease-sensitive fraction of the three membrane-bound pPl intermediates, assayed as a function of protease concentration. In this experiment, 86-mer nascent chains were resistant to digestion by proteinase K concentrations of up to 0.5 mg/ml, whereas ∼55–65% of the 131- and 169-mer nascent chains were digested at all protease concentrations assayed. These data, although they do not corroborate the data of Mothes et al. (1997), are in agreement with the study of Connolly et al. (1989).
Figure 7.
Stage-dependent variations in the ribosome–membrane junction with native preprolactin: comparison of native and reconstituted membranes. Preprolactin 86-, 131-, and 169-mer translation products were synthesized in the presence of native or reconstituted lumenal protein–deficient membranes. In A, the efficiency of targeting was determined by centrifugation of the completed translations to yield membrane (P) and supernatant (S) fractions. In B, the ribosome–membrane junction was evaluated by proteolysis assays. Completed translation reactions, performed in the presence or absence of RM, were chilled on ice and treated with proteinase K (150 μg/ml) for 30 min on ice. In C, the dependence of the observed precursor protection was evaluated by titration of protease concentration in the digestion stage of the experiment. Protease concentrations of 0–500 μg/ml were used. In D, translations were performed in the presence or absence of reconstituted RM (rRM). Membrane targeting and the ribosome–membrane junction were analyzed as described for A–C.
When the ribosome–membrane junction for the truncated pPl intermediates was assayed with rRM as the target membrane, results similar to those depicted for the TVG nascent chains (Fig. 6) were obtained. Thus, as illustrated in Fig. 7 D, pPl 86-, 131-, and 169-mer precursors were efficiently recovered in the pellet fraction in the presence, but not the absence of rRM (Fig. 7 D, lanes 1–12). Analysis of the ribosome–membrane junction by proteolysis indicated that rRM, lacking lumenal proteins, afforded consistently lower protection of the 86–169–amino acid translocation intermediates to exogenous protease than that oberved with native RM (Fig. 7 D, lanes 13–18). Such differences are not an artifact of reconstitution, for in direct paired experiments with pPl 86-mer, the rRM-bound TVG60-mer is insensitive to proteolytic digestion (data not shown). These data further support the conclusion that the characteristics of the ribosome–membrane junction, in particular, the physical intimacy of the ribosome–membrane seal, is significantly influenced by the translocation stage and the sequence, or perhaps the structure of the precursor protein.
Discussion
The role of the nascent chain in ribosome–membrane junction regulation was analyzed through the use of truncated secretory protein translocation intermediates. As assessed by protease protection studies, the existence of a tight ribosome–membrane junction was observed to be stage- and precursor-specific. At early stages of translocation, operationally defined as the stage in which the signal sequence has emerged from the membrane-bound ribosome, the nascent chain is completely shielded from digestion with exogenous proteases. At later stages of translocation, in particular those stages occurring after signal sequence cleavage, the membrane-bound secretory nascent chains displayed enhanced sensitivity to digestion with exogenous proteases. These observations are consistent with a scenario in which the ribosome–membrane junction undergoes structural changes during of protein translocation. A dynamic ribosome–membrane junction has been previously identified in the mechanism of pause-transfer sequence recognition (Hedge and Lingappa, 1997), and as a component of the mechanism of integral membrane protein assembly (Mothes et al., 1997; Liao et al., 1997). We propose that at early stages of secretory protein translocation, the signal sequence–bearing nascent chain contributes substantially to the regulation of the ribosome–membrane junction. Such interactions are likely limited to stages of translocation occurring before signal sequence cleavage and furthermore, are distinct from the sequence-specific, regulated conformational changes in the ribosome–membrane junction that accompany recognition of pause-transfer and stop-transfer signals (Hedge and Lingappa, 1997; Mothes et al., 1997; Liao et al., 1997).
It has been proposed that the appearance of a protease-resistant ribosome–membrane junction occurs coincident with salt-insensitive binding of the nascent chain, and represents insertion of the nascent chain into the protein-conducting channel (Jungnickel and Rapoport, 1995). We have observed that membrane-bound short-chain intermediates (60-mer) were highly resistant to proteolysis, yet were extractable with magneisum-free, high-salt buffers. Interestingly, salt-sensitive extraction of the TVG60-mer was only observed under conditions that yielded ribosome disassembly; ribosome disassembly alone was insufficient for TVG60-mer release. It appears, therefore, that at early stages of translocation, the ribosome and the nascent chain make independent contributions to the binding reaction, and that protease-resistant and salt-resistant nascent chain binding represent two distinct processes. In all likelihood, it is the use of magnesium-free extraction buffers that have allowed us to resolve this novel stage of ribosome/nascent chain interaction with the ER membrane. When extractions were conducted in the presence of magnesium, results identical to those of Jungnickel and Rapoport (1995) were obtained.
We have previously reported that protease-resistant salt-sensitive binding of a native 86–amino acid–truncated preprolactin is observed when ribosome/nascent chain binding is conducted at limiting membrane concentrations (binding saturation; Murphy et al., 1997). Under standard assay conditions, however, pPl86-mer binds to RM in a salt- and protease-resistant manner (Connolly and Gilmore, 1986; Murphy et al., 1997). In context of this study, the phenomena of protease-resistant, salt-sensitive nascent chain binding can be identified under at least two experimental conditions: (a) through use of short-chain truncated translocation intermediates of length sufficient to make the signal sequence available for targeting (TVG60-mer); or (b) with longer (86-mer) nascent chains translated in the presence of limiting membrane concentrations. On the basis of these observations, we propose that salt-resistant binding of the nascent chain to the ER membrane represents the stage of productive insertion into the translocation site. Implicit in this proposal is the hypothesis that ribosome/nascent chain complexes can bind to the ER membrane and form a protease-resistant junction, yet because of limiting access to translocation sites, are unable to achieve stable insertion (Murphy et al., 1997).
It is generally thought that the signal sequence inserts into the ER membrane in an antiparallel loop topology. In a study in which this hypothesis was directly investigated, clear evidence for an antiparallel loop topology was obtained (Shaw et al., 1988). The loop conformation may provide a structural rationale for a chain-length requirement for stable (salt-resistant) nascent chain binding. Quite simply, the relative stability of membrane-bound early translocation intermediates would be expected to vary as a function of chain length, with short chain intermediates being of insufficient length to yield an antiparallel loop structure. As has been demonstrated, when present as an antiparallel loop, the hydrophobic core of the signal sequence is protected from digestion with exogenous proteases (Shaw et al., 1988). We tested this prediction through use of a domain-specific isotope-labeling protocol. A construct was designed to allow differential, biosynthetic labeling of either the hydrophobic core of the signal sequence or the mature region of the nascent chain. Using this construct, the topology of the signal sequence and mature domains of secretory nascent chains were investigated at different stages of translocation. Construct sizes were chosen such that the smallest (60–amino acid) precursor would extend an extraribosomal domain of approximately 30 amino acids, which represents the entirety of the TRAPα signal sequence (Hartmann et al., 1989). The longest construct (129 amino acids) is of sufficient length to engage fully the translocation machinery, but is of insufficient length to enter the ER lumen and undergo signal sequence cleavage.
The results from these studies were unexpected. As assayed by a protease protection assay, the hydrophobic core of the signal sequence was accessible to the cytoplasmic face of the ER membrane at all stages of translocation preceding signal peptide cleavage. These data are consistent with reports that the signal sequence resides in an aqueous environment throughout translocation (Crowley et al., 1993, 1994), but are in apparent contradiction to the report of Shaw et al. (1988). To reconcile this contradiction, we propose that the signal sequence is structurally dynamic, and when bound to the ER membrane, may interconvert between a cytoplasmic and membrane-inserted loop orientation. In this proposal, a stable antiparallel loop topology would require release of the nascent chain from the ribosome and translocation of the flanking domain.
Because the protease accessibility experiments indicate that the relative topology of the signal sequence is identical at stages of translocation immediately after targeting (60-mer) and preceding signal sequence cleavage (129-mer), it is of value to identify the processes that are responsible for stable insertion of the signal sequence. To begin investigations into this question, we addressed the hypothesis that lumenal chaperones would contribute, either directly or indirectly, to stable insertion of the nascent chain. The rationale for this prediction was based on the observations that in reconstituted membranes lacking lumenal proteins, the translocation stages after signal sequence cleavage are compromised (Nicchitta and Blobel, 1990) and that, in yeast, the lumenal hsp70 Kar2p (BiP) participates in the early signal sequence insertion reactions (Sanders et al., 1992), and performs a direct function in the lumenal translocation reaction (Sanders et al., 1992; Brodsky et al., 1995; Lyman and Schekman, 1995). In comparing the ribosome–membrane junction in native and lumenal protein–depleted (reconstituted) RM, it was observed that although both membrane populations were highly active in the targeting reaction, the ribosome–membrane junction varied as a function of chain length. Thus, at early stages of translocation (60-mer), the ribosome–membrane junction achieved by native and lumenal protein– depleted RM was identical. That is, membrane-bound TVG60-mer nascent chains were highly resistant to digestion by exogenous proteases. At later stages of translocation, however, nascent chains were considerably more sensitive to protease digestion when bound to lumenal protein–depleted versus native RM. This phenomena was observed for both the TVG construct, which is a chimera, and a native protein, preprolactin. These data suggest, but as yet do not prove, that in mammalian microsomes, lumenal proteins may contribute either directly or indirectly, to insertion and stabilization of the nascent chain. This proposal mirrors that previously made regarding the function of Kar2p in the yeast ER (Sanders et al., 1992).
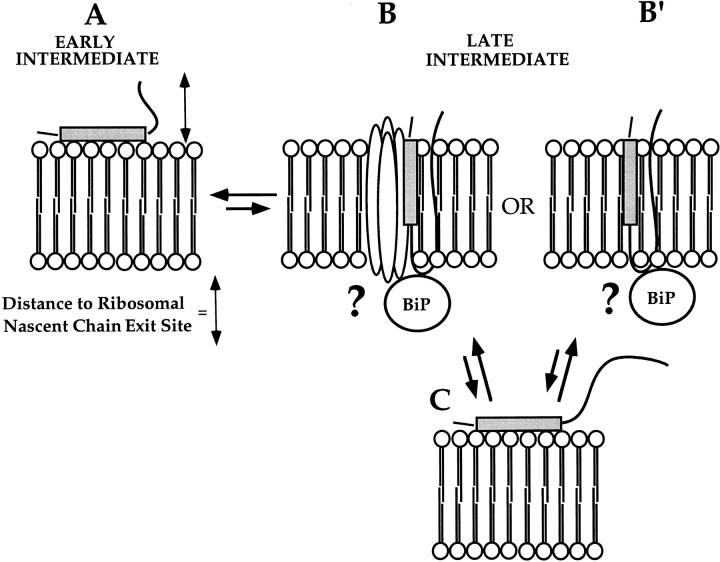
The observations presented in this communication are summarized in the model depicted in Fig. 8. In this model, the early translocation events are viewed as a dynamic continuum in which the conformation and the topology of the signal sequence vary as a function of the stage of translocation. Beginning at early stages of translocation (60-mer), the signal sequence is bound in a salt-sensitive protease-resistant manner to the ER membrane (Fig. 8 A). The ribosome–membrane topology that provides the protease-insensitive environment for the nascent chain is a direct consequence of the binding of the ribosome/nascent chain complex to the ER membrane, and likely reflects a stage-specific close apposition of the ribosomal nascent chain exit site to the membrane. In this view, the tight junction is predicted to exist in the interval between signal sequence binding and signal sequence cleavage. Because of the short length of the early intermediates, and, experimentally, the fact that if present as a truncated intermediate, such intermediates remain bound to the ribosome, the signal sequence is depicted as being topologically incapable of either insertion into the lipid bilayer (Fig. 8 B) or interaction with a signal sequence receptor protein (B′). Late-stage intermediates, in contrast, would be of sufficient length to undergo dynamic interactions with the bilayer interior, or receptor protein, and thus would be resistant to salt extraction. Necessarily, such interactions need be energetically favorable and thus would involve either a structural change in the signal sequence and/or an interaction with a receptor protein. To explain the signal sequence hydrophobic domain protease accessibility data, we propose that the TVG80- and 129-mer signal sequences can reversibly sample the membrane topography illustrated in Fig. 8 A. In sampling this topology, the hydrophobic core of the signal sequence would be predicted to become accessible to proteolytic digestion. Should the insertion event be accompanied by presentation of the mature domain to the ER lumen, interactions of this domain with either lumenal or integral membrane chaperones may act to further stabilize the loop topology. These predictions are currently under investigation.
Figure 8.
Model of the stage-dependent topological dynamics of the membrane-bound signal sequence. In this model, postulated interactions of the signal sequence with components of the ER membrane are illustrated. In A, represented by the TVG60-mer, the signal sequence is bound to the ER membrane but, because of the short length of the nascent chain and the fact that truncated intermediates remain ribosome-associated, the signal sequence is predicted to be topologically constrained, and therefore unable to interact with the membrane components that mediate signal sequence recognition. In the case of a truncated intermediate of longer length (i.e., TVG129-mer), the dynamics of signal sequence interaction with the ER membrane would be relatively unrestricted, thereby allowing formation of a loop topology (B or B′). Although the loop topology would be favored, the nascent chain is depicted as remaining capable of sampling the topology indicated for the newly targeted intermediate (A), and thus exhibiting protease accessibility after release from the ribosome (C). The relative size of the arrows is intended to suggest the propensity for sampling of a given topology. In B and B′, the lumenal protein BiP is depicted as interacting with the nascent chain and/or components of the translocation apparatus. These interactions are postulated to stabilize a translocation competent topology of the signal sequence. The speculative nature of this proposal is indicated by the question marks. For clarity, the membrane-bound ribosome is not depicted.
Acknowledgments
We are grateful to Dr. David Y. Thomas (McGill University, Montreal, Canada) for the gift of cDNA-encoding canine TRAPα (gp35), Dr. John Rose (Yale University, New Haven, CT) for the gift of VSV-G cDNA, and Dr. Steve Cala (Wayne State University, Detroit, MI) for the gift of GRP94 cDNA. We thank Edwin Murphy III, Matthew Potter, Robert Seiser, and Pamela Wearsch for helpful comments and criticism, and Dr. Tom Thompson (University of Virginia, Charlottesville, VA) for support, advice, and encouragement.
This work was supported by National Institutes of Health grant DK47897.
Abbreviations used in this paper
- pPl
preprolactin
- pTVG
fusion clone
- RM
rough microsomes
- rRM
reconstituted rough microsomes
- SRP
signal recognition protein
- VSV-G
vesicular stomatitis virus G-protein
Footnotes
Address all correspondence to Christopher V. Nicchitta, Department of Cell Biology, Box 3709, Duke University Medical Center, Durham, NC 27710. Tel.: 919-684-8948; Fax: 919-684-5481; E-mail: C.Nicchitta@cellbio.duke.edu
References
- Batenburg AM, Demel RA, Verkleij AJ, de Kruif B. Penetration of the signal sequence of the Eschericia coliPhoE protein into phospholipid model membranes leads to lipid specific changes in signal peptide structure and alterations of lipid structure. Biochemistry. 1988;27:5678–5685. doi: 10.1021/bi00415a043. [DOI] [PubMed] [Google Scholar]
- Bird P, Gething MJ, Sambrook J. Translocation in yeast and mammalian cells: not all signal sequences are functionally equivalent. J Cell Biol. 1987;105:2905–2914. doi: 10.1083/jcb.105.6.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes II. reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Laskey RA. A film detection method for tritium- labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Goeckler J, Schekman R. BiP and Sec63p are required for both cotranslational and posttranslational translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MS, Gierasch LM, Zlotnick A, Lear JD, DeGrado WF. In vivo function and membrane binding properties are correlated for Escherichia coliLamB signal peptides. Science. 1985;228:1096–1099. doi: 10.1126/science.3158076. [DOI] [PubMed] [Google Scholar]
- Chou MM, Kendall DA. Polymeric sequences reveal a functional interrelationship between hydrophobicity and length of signal peptides. J Biol Chem. 1990;265:2873–2880. [PubMed] [Google Scholar]
- Connolly T, Gilmore R. Formation of a functional ribosome-membrane junction during translocation requires the participation of a GTP-binding protein. J Cell Biol. 1986;129:2253–2261. doi: 10.1083/jcb.103.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Collins P, Gilmore R. Access of proteinase K to partially translocated nascent polypeptides in intact and detergent-solubilized membranes. J Cell Biol. 1989;108:299–307. doi: 10.1083/jcb.108.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Liao S, Worrell VW, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Crowley KS, Reinhart GD, Johnson AE. The signal sequence moves through a ribosomal tunnel into a non-cytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- Gierasch L. Signal sequences. Biochemistry. 1989;28:923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G. Translocation of secretory proteins across the microsomal membrane occurs through an environment accessible to aqueous perturbants. Cell. 1985;42:497–505. doi: 10.1016/0092-8674(85)90107-2. [DOI] [PubMed] [Google Scholar]
- Görlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in translocation. Nature. 1992a;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies K-U, Rapoport TA. A mammalian homolog of Sec61p and SecYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992b;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Hartmann E, Görlich D, Kostka S, Otto A, Kraft R, Knespel S, Burger E, Rapoport TA, Prehn S. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem. 1993;214:375–381. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- Hartmann E, Wiedmann M, Rapoport TA. A membrane component of the endoplasmic reticulum that may be essential for protein translocation. EMBO (Eur Mol Biol Organ) J. 1989;8:2225–2229. doi: 10.1002/j.1460-2075.1989.tb08346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Lingappa VR. Sequence-specific alteration of the ribosome-membrane junction exposes nascent secretory proteins to the cytosol. Cell. 1996;85:217–228. doi: 10.1016/s0092-8674(00)81098-3. [DOI] [PubMed] [Google Scholar]
- High, S., B. Martoglio, D. Gorlich, S.S. Andersen, A.J. Ashford, A. Giner, E. Hartmann, S. Prehn, T.A. Rapoport, and B. Dobberstein. 1993. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J. Biol. Chem., 268:26745–26751. [PubMed]
- Hoyt DW, Gierasch LM. Hydrophobic content and lipid interactions of wild-type and mutant OmpA signal peptides correlate with their in vivo function. Biochemistry. 1991;30:10156–10163. doi: 10.1021/bi00106a012. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–73. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Jones JD, Gierasch L. Effect of charged residue substitutions on the thermodynamics of signal peptide-lipid interactions for the Escherichia coliLamB signal sequence. Biophys J. 1994a;67:1546–1561. doi: 10.1016/S0006-3495(94)80628-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Gierasch LM. Effect of charged residue substitutions on the membrane-interactive properties of signal sequences of the Escherichia coliLamB protein. Biophys J. 1994b;67:1534–1545. doi: 10.1016/S0006-3495(94)80627-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Krieg UC, Johnson AE, Walter P. Protein translocation across the endoplasmic reticulum membrane: identification by photocross-linking of a 39-kD integral membrane glycoprotein as part of a putative translocation tunnel. J Cell Biol. 1989;109:2033–2043. doi: 10.1083/jcb.109.5.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia TV, Wiedmann M, Girshovich AS, Bochkareva ES, Bielka H, Rapoport TA. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- Liao S, Lin J, Do H, Johnson AW. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin LI, Rich A. Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosomal shielding. J Mol Biol. 1967;26:329–346. doi: 10.1016/0022-2836(67)90301-4. [DOI] [PubMed] [Google Scholar]
- Martoglio B, Hofmann MW, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:201–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Mothes W, Heinrich SU, Graf R, Nilsson I, von Heinje G, Brunner J, Rapoport TA. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Murphy EC, III, Zheng T, Nicchitta CV. Identification of a novel stage of ribosome/nascent chain association with the endoplasmic reticulum membrane. J Cell Biol. 1997;136:1213–1226. doi: 10.1083/jcb.136.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV, Blobel G. Nascent chain binding and translocation are distinct processes: differentiation by chemical alkylation. J Cell Biol. 1989;108:789–795. doi: 10.1083/jcb.108.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV, Blobel G. Assembly of translocation-competent proteoliposomes form detergent solubilized rough microsomes. Cell. 1990;60:259–269. doi: 10.1016/0092-8674(90)90741-v. [DOI] [PubMed] [Google Scholar]
- Nicchitta CV, Migliaccio G, Blobel G. Reconstitution of secretory protein translocation from detergent-solubilized rough microsomes. Methods Cell Biol. 1991;34:263–285. doi: 10.1016/s0091-679x(08)61685-4. [DOI] [PubMed] [Google Scholar]
- Nicchitta CV, Blobel G. Lumenal proteins of the endoplasmic reticulum are required to complete protein translocation. Cell. 1993;73:989–998. doi: 10.1016/0092-8674(93)90276-v. [DOI] [PubMed] [Google Scholar]
- Nicchitta CV, Murphy EC, Haynes R, Shelness GS. Stage and ribosome-specific alterations in nascent chain-Sec61p interactions accompany translocation across the ER membrane. J Cell Biol. 1995;129:957–970. doi: 10.1083/jcb.129.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M, Noll H. V. Magnesium-dependent dissociation of tight couples into subunits: measurements of dissociation constants and exchange rates. J Mol Biol. 1976;105:111–130. doi: 10.1016/0022-2836(76)90197-2. [DOI] [PubMed] [Google Scholar]
- Romisch K, Webb J, Lingelbach K, Gausepohl H, Dobberstein B. The 54K protein of signal recognition particle contains a methionine-rich RNA binding domain. J Cell Biol. 1990;111:1793–1802. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DD, Tashiro Y, Palade GE. On the attachment of ribosomes to microsomal membranes. J Mol Biol. 1966;19:503–524. doi: 10.1016/s0022-2836(66)80019-0. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- Schägger H, Von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shaw A, Rottier PJM, Rose JK. Evidence for the loop model of signal-sequence insertion into the endoplasmic reticulum. Proc Natl Acad Sci USA. 1988;85:7592–7596. doi: 10.1073/pnas.85.20.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heinje G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984;173:243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- von Heinje G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in vitro-assembled polysomes synthesizing secretory proteins. J Cell Biol. 1981a;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in vitro-assembled polysomes synthesizing secretory proteins. J Cell Biol. 1981b;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Wettenhall REH, Wool IG. Assay of reassociation of eukaryotic ribosomal subunits catalyzed by initiation factors. Methods Enzymol. 1974;30:186–197. doi: 10.1016/0076-6879(74)30021-3. [DOI] [PubMed] [Google Scholar]
- Wiedmann M, Kurzchalia TV, Hartmann E, Rapoport TA. A signal sequence receptor in the endoplasmic reticulum membrane. Nature. 1987;328:830–833. doi: 10.1038/328830a0. [DOI] [PubMed] [Google Scholar]
- Zopf D, Bernstein HD, Johnson AE, Walter P. The methionine-rich domain of the 54kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO (Eur Mol Biol Organ) J. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]