Abstract
Yeast verprolin, encoded by VRP1, is implicated in cell growth, cytoskeletal organization, endocytosis and mitochondrial protein distribution and function. We show that verprolin is also required for bipolar bud-site selection. Previously we reported that additional actin suppresses the temperature-dependent growth defect caused by a mutation in VRP1. Here we show that additional actin suppresses all known defects caused by vrp1-1 and conclude that the defects relate to an abnormal cytoskeleton. Using the two-hybrid system, we show that verprolin binds actin. An actin-binding domain maps to the LKKAET hexapeptide located in the first 70 amino acids. A similar hexapeptide in other acting-binding proteins was previously shown to be necessary for actin-binding activity. The entire 70– amino acid motif is conserved in novel higher eukaryotic proteins that we predict to be actin-binding, and also in the actin-binding proteins, WASP and N-WASP. Verprolin-GFP in live cells has a cell cycle-dependent distribution similar to the actin cortical cytoskeleton. In fixed cells hemagglutinin-tagged Vrp1p often co-localizes with actin in cortical patches. However, disassembly of the actin cytoskeleton using Latrunculin-A does not alter verprolin's location, indicating that verprolin establishes and maintains its location independent of the actin cytoskeleton. Verprolin is a new member of the actin-binding protein family that serves as a polarity development protein, perhaps by anchoring actin. We speculate that the effects of verprolin upon the actin cytoskeleton might influence mitochondrial protein sorting/function via mRNA distribution.
The actin cytoskeleton plays crucial roles in fundamental processes such as cell growth, differentiation and migration, localized membrane growth, endocytosis, and cell division. Saccharomyces cerevisiae is an excellent organism for the study of the actin cytoskeleton because many of its components are conserved (Drubin and Nelson, 1996) and it is possible to probe the function of particular components in yeast via genetic approaches. S. cerevisiae cells display cytoskeletal asymmetry during the cell cycle. As cells initiate a cell cycle, the cytoskeleton becomes organized anisotropically to facilitate polarized growth. In G1, a ring of actin marks the site where the bud will grow. In S and G2, as the bud continues its growth, the actin cytoskeleton remains highly polarized toward the bud. Cortical patches are concentrated mainly in the bud and cables are visible converging toward the mother–bud axis. After cell division, the newly formed cells are unpolarized and another round of polarization starts with a new cell cycle (Adams and Pringle, 1984). During the cell cycle the actin cytoskeleton plays roles in bipolar bud-site selection and polarized secretion of components, as well as polarized movement of organelles such as mitochondria to the daughter cell (Smith et al., 1995; Wang and Bretscher, 1995; Yang et al., 1997).
The actin cytoskeleton is also important for processes not necessarily associated with the cell cycle, such as endocytosis (Munn et al., 1995). In our efforts to identify cellular components involved in the distribution of nucleus-encoded proteins to mitochondria, we identified mutations of VRP1, PAN1, (Zoladek et al., 1995) and RSP5 (Zoladek et al., 1997). Interestingly, all of these genes affect endocytosis and show genetic interactions with each other and/or with components of the actin cytoskeleton (Munn et al., 1995; Zoladek et al., 1995; Wendland et al., 1996; Zoladek et al., 1997). The current studies focus on deciphering the cellular function of the VRP1 gene product.
VRP1 codes for verprolin, a 817–amino acid protein very rich (22%) in proline. Besides a defect in the ability to grow at elevated temperatures, vrp1 mutants have aberrant cytoskeletons (Donnelly et al., 1993; Zoladek et al., 1995), defects in cytoplasmic/mitochondrial distribution of a mitochondrial protein (Zoladek et al., 1995), and defects in endocytosis (Munn et al., 1995; Zoladek et al., 1997). We provide evidence here that verprolin is also required for bipolar budding patterns. How verprolin is involved in these diverse processes is not understood. In particular, it is not known whether all the defects are related to the cytoskeletal disorganization or whether verprolin has multiple functions in yeast.
In attempts to clone VRP1, ACT1 encoding actin was uncovered as a suppressor of the vrp1-1 mutation. We showed that when supplied with actin encoded from a low copy centromere-containing vector or from a single additional integrated copy of ACT1, vrp1-1 mutant cells could grow at the nonpermissive temperature; however, ACT1 did not suppress the temperature-sensitive (ts)1 growth defect caused by a disruption allele of VRP1, vrp1::LEU2 (Zoladek et al., 1995). Therefore, an apparent increase in the intracellular concentration of actin caused allele-specific suppression of the ts growth defects of vrp1 mutant cells. To begin to understand the role of verprolin in yeast, we determined which of the defects of vrp1 mutant cells could be corrected by ACT1. Here we show that actin supplied from a low copy centromere-containing vector corrects all vrp1 defects.
We show that verprolin is an actin-binding protein located at positions coincident with the actin cortical cytoskeleton. Many other actin-binding proteins are localized similarly to verprolin (Doyle et al., 1996; Amberg et al., 1997; Waddle et al., 1997). The cytoskeletal location of many actin-binding proteins is dependent upon an appropriate cytoskeleton. However, the polarized locations of some actin-binding proteins can be actin cytoskeleton independent (Ayscough et al., 1997). Our results demonstrate that verprolin achieves its intracellular localization through an actin-independent pathway. The pattern of intracellular localization of verprolin, its independence from actin, its role in bud-site selection, as well as the phenotypes of vrp1 mutants provide evidence that verprolin is a polarity development protein required for the correct assembly of the actin cytoskeleton.
Materials and Methods
Strains and Genetic Techniques
Media for yeast growth were prepared according to Sherman (1991). Cells were transformed by the one-step method described by Chen et al. (1992). The strains used in this study are listed in Table I.
Table I.
S. cerevisiae Strains Used in This Study
| Strain | Genotype | Reference | ||
|---|---|---|---|---|
| T8.1D | MATα SUP11 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3,112 his4-519 | Zoladek et al., 1995 | ||
| TZ33 | MATα SUP11 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3,112 his4-519 vrp1-1 | Zoladek et al., 1995 | ||
| T65.1D | MATα leu2-3, 112 ade1 ura3-52 Ile− MEL1 vrp1::LEU2 | Zoladek et al., 1995 | ||
| Y190 | MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL- -> lacZ LYS2::GAL(UAS)- ->HIS3 cyhr | S.J. Elledge, gift (Baylor College of Medicine, Houston, TX) | ||
| Sc467 | MATα/MAT a leu2-3,112/leu2-3,112 ade1/ade1 ura3-52/ura3-52 Ile −/Ile − MEL1/MEL1 | Zoladek et al., 1995 | ||
| GVY1 | MATα/MAT a leu2-3,112/leu2-3,112 ade1/ade1 ura3-52/ura3-52 Ile −/Ile − MEL1/MEL1 vrp1::LEU2/vrp1::LEU2 | This laboratory |
General DNA Manipulations
DNA manipulations were done using standard techniques (Sambrook et al., 1989). Escherichia coli strains employed were DH5α and RR1. Restriction endonucleases and modifying enzymes were obtained from New England Biolabs Inc. (Beverly, MA) or from Promega (Madison, WI). Sequenase was obtained from United States Biochemical Corp., (Cleveland, Ohio).
Plasmid Construction
To construct the VRP1–green fluorescent protein (GFP) fusion, a 0.7-kb PCR product containing the GFP open reading frame (ORF) from pSEY18/GAL1-10/GFP (gift from Roger Y. Tsien, University of California, San Diego, CA) was obtained using the following primers: the 5′oligo, 5′GCCCCGGGCATGAGTAAAGGAGAAGAACTTTTC3′ and the 3′oligo, 5′CATTCCCGGGTATAGTTCATCCATGCCATG3′. The PCR fragment contained built-in SmaI ends and the amplified DNA was cloned into pGEM-T vectors. From pGEM-T vectors the 0.7-kb GFP was released by digesting with SmaI and further cloned in-frame at the carboxy terminus of VRP1 that had been digested at the unique BsaAI site, one codon before the stop codon. The ∼4.7-kb HindIII piece containing the VRP1–GFP fusion and the VRP1 regulatory sequences were transferred to the single-copy yeast shuttle vector YCplac33 generating YCpVRP1GFP.
For YCpVRP1 plasmid, a 4-kb piece containing the VRP1 gene was removed from the pT6 plasmid (Zoladek et al., 1995) by digesting with HindIII and cloned into pUC19. From pUC19, the 4-kb Hind III fragment containing VRP1 was further cloned into YCplac33 vector giving rise to YCpVRP1.
For all two-hybrid constructs, two vectors were used: pACT2, encoding the Gal4 DNA transcription activation domain, and pAS-CYH2, encoding the Gal4 DNA-binding domain. Both vectors were a gift from S.J. Elledge (Baylor College of Medicine, Houston, TX). To eliminate possible confusion related to the nomenclature of vector and genes we cloned in these vectors, we designated pACT2 as pDAD, for DNA Activating Domain. In the same way, pAS-CYH2 was replaced by pDBD for DNA- Binding Domain in the name of all constructs.
For pDAD-VRP1 or pDBD-VRP1, a 2.5-kb PCR product containing the VRP1 ORF and 71 bp from the trimer of the hemagglutinin (HA) epitope was cloned into pGEM-T giving rise to pGEM-VRP1-2Hy. The 5′oligo was 5′GTACGGATCCAAATGGCAGGTGCTCCA3′ and the 3′ oligo was 5′TCATATGGATAGGATCC3′. Both oligos have built-in BamHI ends. The ∼2.6-kb BamHI fragment was transferred to pDAD or pDBD vectors to generate in-frame fusions between Gal4p DNA-binding domain or the Gal4p transcription activation domain and Vrp1p.
pDAD-ACT1 and pDBD-ACT1 were constructed in the following way: a 1.1-kb PCR product containing the ACT1 ORF following the intron was cloned into pGEM-T vector giving rise to pGEM-ACTIN. For the PCR reaction, the template was pT3 (Zoladek et al., 1995). Use of the 5′ oligo, 5′GTACGGATCCAGGTTGCTGCTTTGGTT3′, eliminates the codons before the intron in the ACT1 gene. The 3′ oligo was 5′CAGTGGATCCACACAAAAGCAGAGATTA3′. Both oligos have built-in BamHI ends. From pGEM-ACTIN the BamHI fragment containing the ACT1 coding region was transferred to pDAD or pDBD.
For pDAD-PFY or pDBD-PFY the following procedure was used: a 0.35-kb PCR product containing the PFY1 ORF following the intron was cloned into pGEM-T giving rise to pGEM-PFY. For the PCR reaction the template was YCpPFY1 (a gift from S.S. Brown, University of Michigan, Ann Arbor, MI), the 5′ oligo was 5′GTACGGATCCCATACACTGATAACTTA3′ and the 3′ oligo was 5′GTACGGATCCAAATTAGTATTGAACACC3′. Both oligos have built-in BamHI ends. The BamHI fragment was transferred to pDAD or pDBD.
For constructing pDAD-VRP1ΔSalΔKpn1-2, the pGEM-VRP1-2Hy plasmid was digested with SalI and the 4.1-kb piece containing the plasmid backbone and amino terminus of VRP1 coding for 363 amino acids was ligated and further digested with KpnI. The release of an internal 0.35-kb KpnI fragment and further ligation at this KpnI site generated an out-of-frame fusion beyond codon 70, and generated pGEM-VRP1Δ SalΔKpn1-2. This construct encodes VRP1 amino acids 1–70. The ∼0.9-kb BamHI–SalI fragment was then cloned into pDAD digested BamHI–SalI giving rise to pDAD-VRP1ΔSalΔKpn1-2.
For pDAD-VRP1ΔKpn1-3, the pGEM-VRP1-2Hy plasmid was digested with KpnI and two pieces, 0.3 and 0.7 kb were released. The backbone plasmid was then closed to create an in-frame fusion between amino acids 70 and 717 of Vrp1p. This plasmid, pGEM-VRP1ΔKpn1-3, encodes the amino terminus (amino acids 1–70) and the carboxy end (amino acids 717–817) of verprolin fused in frame to the Gal4p DNA transcription activation domain. The ∼0.5-kb BamHI fragment was removed and cloned in frame into pDAD.
For pDAD-VRP1 71-817, a ∼2.2-kb PCR product of VRP1 coding for a protein missing the first 70 amino acids was first cloned into pGEM-T vectors. From pGEM-T, the 2.2-kb VRP1 fragment containing built-in BamHI ends was cut out and cloned into the pDAD vector. For PCR, the 5′ oligo was: 5′GGGATCCGTACCGTCAGTAGTAAAGGACCC3′ and the 3′ oligo was 5′TCATATGGATAGGATCC3′.
pDAD-VRP1ΔSalΔKpn1-2/EE contains a fusion between the Gal4-DNA–activating domain and the first 70 amino acids of verprolin in which the two lysines in the LKKAET motif have been mutated to glutamic acids. To construct this plasmid, pGEM-VRP1ΔSalΔKpn1-2 was used for reverse PCR (Hemsley et al., 1989). 5′GAGGAGGCTGAGACCAATGATAGAAGTGCCCCAATC3′ and 5′TAGTTTCATTCCCTTTCTA- ATGTCACC3′ were the primers used. The correct sequences of the 70– amino acids coding region was verified by DNA sequencing. After removing the A-tails of the PCR product with T4 polymerase and phosphorylation with T4 kinase, the PCR product was circularized with T4 ligase. The BamHI–SalI piece was removed and cloned in pDAD vector-digested BamHI–SalI to give rise to pDAD-VRP1ΔSalΔKpn1-2/EE.
YCpVRP1-HA was constructed as follows: a 4-kb HindIII VRP1 piece including the promoter region and the 3′UTR was cloned into pUC19, giving rise to pUCH. This plasmid was digested at the unique BsaAI site, one codon before the stop codon in VRP1 gene, and a HaeIII fragment containing roughly three copies of the hemmaglutinin trimer was placed in frame at the BsaAI site, creating the tagged version of VRP1. From pUC19 the HindIII piece containing the tagged VRP1 was digested and cloned in YCplac33 vector giving rise to YCpVRP1-HA.
pT3 is a plasmid from a library inserted into a centromere-containing vector that contains the ACT1 gene. Transposon mutagenesis of the genomic insert of this plasmid showed that ACT1 is responsible for complementation of the vrp1-1 mutation (Zoladek et al., 1995).
YCpLEUMod5p-I,KR6ET, a low copy plasmid bearing a hemagglutinin trimer-tagged version of Mod5p-I,KR6, was a gift from T. Zoladek (Polish Academy of Sciences, Warsaw, Poland).
Two-hybrid Analysis
Two-hybrid studies were done according to Chien et al. (1991). The host strain for two-hybrid studies was Y190, a derivative of Y153, and kindly provided by S.J. Elledge (Durfee et al., 1993). Strain Y190 was transformed sequentially with each of the pairs of plasmids tested. Nitrocellulose membrane lifts, assays of β-galactosidase activity, and growth on leu−trp−his− plates were done according to recommendations (Durfee et al., 1993). Growth on media-lacking histidine was tested in the presence of 25, 50, and 100 mM 3–amino-1,2,4-triazole.
Microscopic Imaging and Analysis
For verprolin-GFP localization studies, cells from exponentially growing cultures or from a one-day fresh plate were used. Cells were spotted on microscope slide, covered with a coverslip and visualized immediately under a 60× objective (1.4 numerical aperture) using either FITC-filtered illumination or differential interference contrast (DIC) optics. Pictures were taken on Kodak T-Max 400. A Nikon Microphot-FX equipped with B-1E filter cube/420–490 excitation filter/5151F barrier filter was used.
For verprolin-HA and actin staining in the same cells, indirect immunofluorescence was done as follows: 10 ml logarithmic cultures were fixed with 5% formaldehyde for 30 min, washed twice with 1× PBS, pH 6.5, treated with 0.2% Triton X-100 for 10 min, digested 45 min with zymolyase 20T in the presence of β-mercaptoethanol, washed once with sol. B (1.2 M sorbitol, 40 mM phosphate buffer, pH 6.5, 0.05 mM MgCl2). After incubating for an additional 10 min with 0.2% Triton X-100, cells were incubated with primary antibody against HA (12CA5; BAbCO, Richmond, CA) at a 1:750 dilution for 1 h in solution, washed once with PBS, and then incubated with secondary Cy3-conjugated goat anti–mouse antibody (Jackson Immunoresearch Laboratories, Inc., West Grove, PA) at a 1:600 dilution for 45 min. Cells were then washed two times with PBS, and stained with 3.3 μM Oregon green 488 phalloidin in the dark for 30 min. Cells were washed once and mounted on slides. Images were acquired and digitized directly with a SenSys (Photometrics Ltd., Tucson, AZ) CCD camera coupled to a Nikon Optiphot-2 microscope. Illumination was controlled by a Lambda 10-2C filter-wheel controller (Sutter Instrument Company, Novato, CA). Image processing was done using IPLab Spectrum software (Signal Analytics, Co., Vienna, VA) running on a 6500/225 Power Macintosh. Files were converted into and further processed using Adobe Photoshop 4.
Staining
For all experiments, except for the co-localization studies described above, rhodamine phalloidin was used. Cells were fixed and further processed as recommended (Adams and Pringle, 1991). The endocytosis test was done according to Dulic et al. (1991). Calcofluor staining was done on live cells as described (Pringle, 1991).
Latrunculin-A Studies
For growth of cells in the presence of Latrunculin-A (LAT-A), a 200-μl logarithmic culture was treated with either LAT-A (Molecular Probes Inc., Eugene, OR) at a concentration of 100 μM in DMSO for 15 min or with DMSO alone (control). The cells were visualized immediately. At the same time point, an aliquot from each culture was fixed and processed for immunofluorescence with rhodamine phalloidin. For cells released from G0, a culture enriched in unbudded cells was incubated with LAT-A at 100 μM as described for 4 h (Ayscough et al., 1997) and then visualized using the same conditions as for verprolin-GFP localization studies. Actin cytoskeleton disassembly was verified by staining with rhodamine phalloidin.
The halo assay to assess LAT-A sensitivity was done as described previously (Ayscough et al., 1997). Plates were incubated at 23°C for 48 h. Relative apparent LAT-A sensitivity was calculated according to Reneke et al., 1988.
Results
Additional Actin Causes Allele-specific Suppression of vrp1-1 Phenotypes
We previously showed that vrp1-1 cells transformed with a centromere-containing plasmid containing ACT1, pT3, or harboring an integrated copy of ACT1 are able to grow at nonpermissive temperatures (Zoladek et al., 1995). We analyzed actin levels in total cell extracts prepared from wild-type cells (strain T8.1D) or vrp1-1 cells (strain TZ33) transformed with vector alone or with pT3. When indexed against the cytoplasmic protein Rna1p, actin levels were two to fourfold greater in vrp1-1 cells bearing pT3 than in either parental (strain T8.1D) or vrp1-1 (strain TZ33) cells transformed with vector alone (data not shown).
ACT1 Restores the Cytoskeletal Polarization of vrp1-1 Cells.
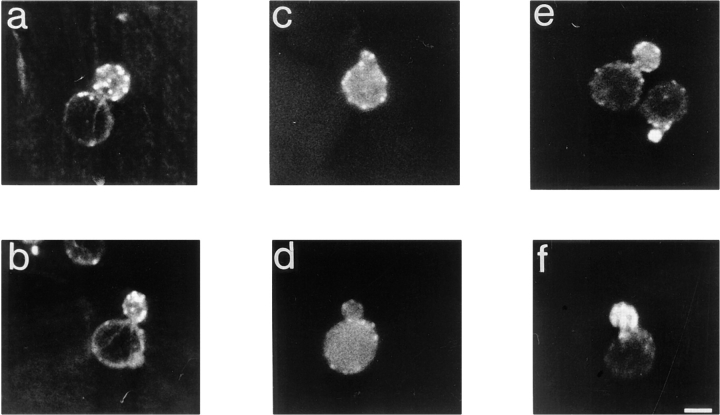
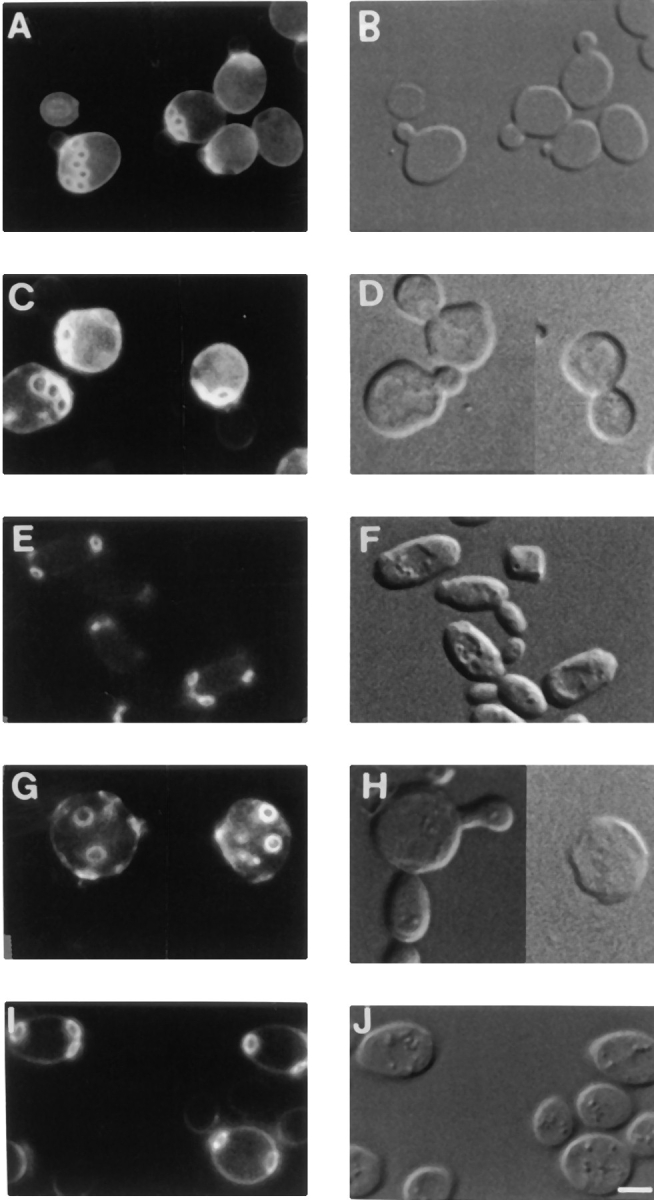
Mutant vrp1-1 cells have a disorganized cytoskeleton. Cortical patches are distributed randomly between mother and bud, no concentration of patches can be detected during any phase of the cell cycle, and no actin cables are visible. The defect is more severe when cells are grown on nonfermentable carbon sources (see mitochondrial defects below), even at permissive temperatures (Zoladek et al., 1995). To determine whether additional actin corrects the defects in actin cytoskeleton organization, cells were grown on lactate medium at the permissive temperature, fixed, and stained with rhodamine phalloidin. Wild-type cells transformed with vector alone have polarized cytoskeletons (Fig. 1, a and b), whereas mutant vrp1-1 cells bearing the vector alone have shape alterations and a random distribution of cortical patches between mother and bud (Fig. 1, c and d). In contrast, vrp1-1 cells bearing pT3, encoding ACT1, have a wild-type morphology and polarized distribution of cortical patches between the mother and the bud (Fig. 1, e and f). Therefore, a two to fourfold increase in the intracellular concentration of actin confers polarized cytoskeleton and normal shape to vrp1-1 cells.
Figure 1.
Cytoskeletal polarization is restored in vrp1-1 cells (strain TZ33) by two- to fourfold increases in the level of actin. Representative cells of wild-type strain T8.1D bearing an empty plasmid in a and b; vrp1-1 mutant cells (strain TZ33) bearing an empty plasmid in c and d; and vrp1-1 cells plus ACT1 on the centromere-containing plasmid pT3 in e and f. Cells were grown on 2% lactate medium at permissive temperature, fixed as described (Adams and Pringle, 1991) and then stained with rhodamine-phalloidin for 1 h. Bar, 3 μm.
ACT1 Corrects the Endocytosis Defects of vrp1-1 Cells.
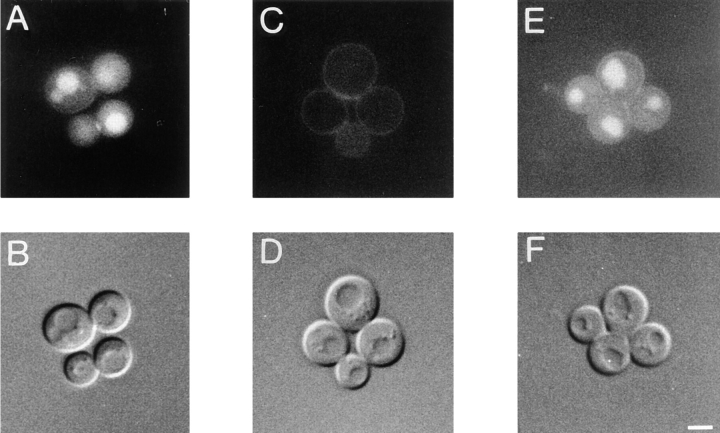
Cells with mutations in VRP1 are defective in endocytosis (Munn et al., 1995; Zoladek et al., 1997). It is known (Benedetti et al., 1994; Munn et al., 1995) that the actin cytoskeleton plays an essential role in the internalization step of endocytosis in yeast. Many cytoskeletal proteins, such as actin, Sac6p, calmodulin, End3p, End4p, and type I myosins (Geli and Riezman, 1996) are necessary for endocytic traffic. To determine whether the defect in endocytosis of vrp1 mutant cells is related to actin cytoskeleton defects, we studied fluid phase endocytosis (Dulic et al., 1991) of vrp1 cells in the presence or absence of the centromere-containing plasmid encoding actin. Parental cells (strain T8.1D) accumulate Lucifer yellow in vacuoles (Fig. 2, A and B), whereas vrp1-1 mutant cells do not accumulate any detectable levels of this compound (Fig. 2, C and D). However, mutant cells containing pT3 show accumulation of Lucifer yellow in vacuoles in a manner indistinguishable from wild-type cells (Fig. 2, E and F). Therefore, an increase in the intracellular quantity of actin suppresses the endocytosis defect caused by the vrp1-1 mutation.
Figure 2.
Fluid-phase endocytosis is restored in vrp1-1 cells (strain TZ33) by additional actin. Lucifer yellow uptake in wild-type (strain T8.1D) cells transformed with vector alone in A and B; vrp1-1 mutant cells bearing vector alone (C and D); and vrp1-1 cells bearing ACT1 on pT3 plasmid (E and F). Cells were grown at permissive temperature and then incubated with Lucifer yellow for 2 h. (A, C, and E), FITC channel. In B, D, and F, the same cells visualized under DIC. Bar, 3 μm.
ACT1 Partially Corrects the Mitochondrial Defects of vrp1-1 Cells.
Mutant vrp1-1 cells have defects in mitochondrial function as assessed by their inability to grow on nonfermentable carbon sources at temperatures exceeding 30°C (Table II). In contrast, vrp1-1 mutant cells grow well at 30°, 34°, and 35°C and poorly at 37° or 38°C on glucose-containing media. Vrp1-1 mutant cells provided with additional actin grow on glycerol-containing media at temperatures from 30° to 36°C, but not at 38°C (Table II). Therefore, a two- to fourfold increase in actin levels partially restores mitochondrial function to vrp1-1 mutants.
Table II.
Growth Characteristics of vrp1-1 Cells Bearing ACT1 on the Centromere-containing Plasmid pT3
| Glycerol | Glucose | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 23°C | 30°C | 36°C | 38°C | 23°C | 30°C | 36°C | 38°C | |||||||||
| VRP1 | + | + | + | + | + | + | + | + | ||||||||
| vrp1-1 | + | ± | − | − | + | + | + | − | ||||||||
| vrp1-1 and pT3 | + | + | + | − | + | + | + | + | ||||||||
Growth was assessed after incubating the plates for two days at the indicated temperature. Absence of growth is indicated by (−); wild-type growth indicated by (+), (±) designates poor growth.
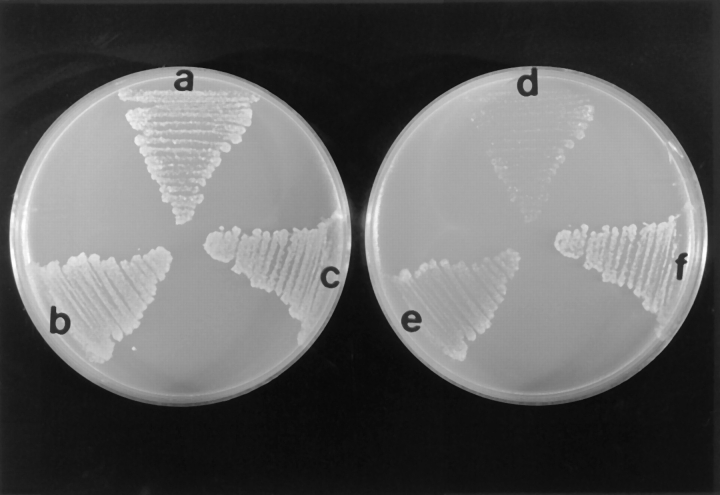
To determine if additional Act1p affects the delivery of proteins to mitochondria, we assessed the functional levels of cytosolic Mod5p-I. Mod5p is a tRNA isopentenyl transferase that modifies tRNAs. Cytosolic levels of Mod5p can be assessed by studying the levels of modified tRNA Tyr UAA, which in turn are monitored by assaying nonsense suppression of the lys2-1 allele (Zoladek et al., 1995). Mod5p-I,KR6 is a form of Mod5p-I, with an alteration that provides its quantitative delivery to mitochondria. Parental cells (strain T8.1D) harboring the plasmid-encoding Mod5p-I,KR6 deliver the great majority of Mod5p-I,KR6 to mitochondria. As these cells do not have sufficient cytosolic Mod5p-I,KR6 to modify the tRNA Tyr UAA encoded by SUP11, they can not grow on media lacking lysine (lys−). In contrast, vrp1-1 cells (strain TZ33) bearing the plasmid encoding Mod5p-I,KR6, grow on lys− media. We analyzed the growth on lys− media of vrp1-1 mutant cells with and without a centromere-containing plasmid encoding actin. Parental cells (strain T8.1D) bearing YCpLEUMod5p-I,KR6ET and a control centromere-containing plasmid encoding URA3 alone are able to grow on ura−leu− media (Fig. 3 a), but not on ura−leu−lys− (Fig. 3 d). By contrast, vrp1-1 cells bearing YCpLEUMod5p-I,KR6ET and the control URA3 plasmid are able to grow on both ura−leu− (Fig 3 c) and ura−leu−lys− media (Fig. 3 f). Although able to grow well on ura−leu− media (Fig. 3 b) vrp1-1 cells bearing YCp LEUMod5p-I,KR6ET and pT3 containing ACT1 show an intermediate phenotype on ura−leu−lys− media (Fig. 3 e). Thus, the partial suppression of the Lys+ phenotype in vrp1-1 cells indicates that excess actin causes a decrease in the functional cytoplasmic pool of Mod5p-I,KR6.
Figure 3.
Extra actin partially suppresses the Lys+ phenotype of vrp1-1 cells. The plate in the left is ura−leu− and the one in the right is ura−leu−lys−. Parental T8.1D cells (a and d); vrp1-1 cells (strain TZ33), (c and f); vrp1-1 cells bearing pT3 (encoding actin) (b and e). All strains have Mod5-I,KR6ET gene on a centromere-containing plasmid; the ura−leu− master plate was first replica plated to ura−leu−lys− plate and then to ura−leu− plate. For all strains, growth was assessed at 23°C.
Verprolin Is Required for Bipolar Budding But Not for Axial Budding
Because verprolin is associated with many actin-related functions and the actin cytoskeleton is required for diploid-specific bipolar budding pattern (Yang et al., 1997), we examined whether verprolin affects the budding pattern. Haploid vrp1::LEU2 cells (Fig. 4, C and D), like their wild-type counterparts (Fig. 4, A and B), bud axially. In contrast, diploid cells homozygous for vrp1::LEU2 (Fig. 4, G and H) are unable to bud bipolarly like wild-type cells do (Fig. 4, E and F); instead, they bud randomly. Introduction of the VRP1–GFP fusion on a low copy plasmid which complements the ts growth of the mutant cells leads to restoration of the bipolar budding pattern (Fig. 4, I and J). Thus, verprolin is required for bipolar budding pattern in diploid cells, but does not appear to influence the axial budding pattern of haploid cells. Although the budding pattern in diploid cells homozygous for vrp1::LEU2 is random, the cells are still able to form buds, indicating that verprolin is required for bud site-selection and not bud growth per se.
Figure 4.
Budding pattern phenotypes of haploid and diploid vrp1::LEU2 mutant strains and rescue of mutant phenotype by VRP1-GFP. Logarithmic cultures of each strain were grown in selective media at 23°C and then stained with Calcofluor to visualize bud scars. (A) vrp1::LEU2 (strain T65.1D) cells bearing YCpVRP1GFP plasmid show normal axial budding pattern; (C) vrp1::LEU2 (strain T65.1D) cells bearing vector alone show axial pattern; (E) wild-type diploid strain Sc467 bearing an empty plasmid shows bipolar budding pattern; (G) diploid vrp1::LEU2/vrp1:: LEU2 cells bearing vector alone show random budding pattern; (H) morphologically, a large proportion of these cells have multiple buds and shape defects; (I) restoration of bipolar budding in homozygous diploid vrp1::LEU2/vrp1::LEU2 cells by the VRP1-GFP gene on a centromere-containing plasmid. These cells have normal morphology (J). B, D, F, H, J are DIC images of A, C, E, G, I, respectively. Bar, 2.5 μm.
Verprolin Interacts with Actin in the Two-Hybrid System
ACT1 and VRP1 show allele-specific interactions as ACT1 can suppress the vrp1-1 mutation, but not the vrp1::LEU2 disruption. Single-copy suppression and allele-specific interactions provide genetic evidence that Act1p and Vrp1p may physically interact. We tested the genetic predictions by using the two-hybrid system (Fields and Song, 1989) to detect verprolin–actin physical interactions. Also, because verprolin has 12 stretches of three to nine polyprolines and polyproline stretches have been shown to bind another actin-binding protein, profilin, we tested whether there is an interaction between verprolin and profilin. Each gene was cloned into the vector encoding the Gal4p DNA-binding domain and the vector encoding the DNA-activating domain and all possible combinations were analyzed. Neither VRP1 nor ACT1 activated the reporter genes, LacZ and HIS3, when transformed alone in the Y190 tester strain. pDBD-PFY, encoding profilin appended to the Gal4p DNA-binding domain, activated the reporter genes by itself. However, pDAD-PFY, encoding profilin appended to the Gal4p transcription activation domain did not. Therefore, pDAD-PFY was used to test interactions between pDBD-ACT1 and pDBD-VRP1. We found that verprolin interacts with actin but not with profilin (Table III). Both LacZ and HIS3 transcription (at 100 mM 3-amino- 1,2,4-triazole) are activated when VRP1 and ACT1 are both present. However, the pDAD-VRP1 plasmid is mitotically unstable; the loss of plasmid correlates with the loss of blue color in these cells. The tendency to lose this plasmid may be related to our observations that overexpression of verprolin is detrimental to cells (data not shown). As expected (Amberg et al., 1995), we detected interactions between actin–profilin and actin–actin (Table III). We found no evidence for the dimerization of verprolin, since the pair pDAD-VRP1 and pDBD-VRP1 did not activate either reporter gene.
Table III.
Two-Hybrid Interactions between Verprolin, Actin, and Profilin
| pDBD | pDBD-ACT1 | pDBD-PFY | pDBD-VRP1 | |||||
|---|---|---|---|---|---|---|---|---|
| pDAD | − | − | + | − | ||||
| pDAD-ACT1 | − | + | + | + | ||||
| pDAD-PFY | − | + | + | − | ||||
| pDAD-VRP1* | − | ± | + | − |
The pDAD-VRP1 plasmid is mitoticallly unstable.
An Actin-binding Domain Maps at Amino Terminus of Verprolin
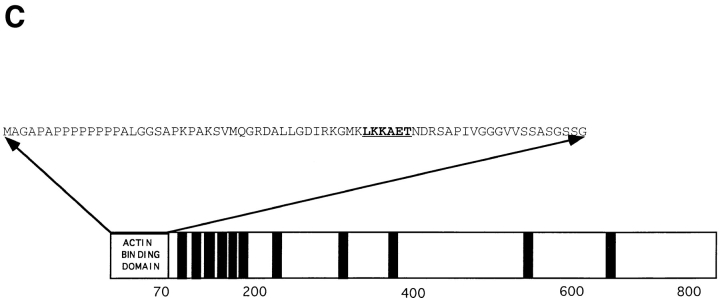
A computer search using the BEAUTY program (Worley et al., 1995) revealed that the amino terminal 70 amino acids of verprolin show strong homology with similarly located regions in a number of proteins from various species: Caenorhabditus elegans CELR144.4 gene product (GenBank/EMBL/DDBJ accession number U23515), Xenopus laevis proline rich protein (GenBank/EMBL/DDBJ accession number X68249) and rat SH3 domain-binding protein (GenBank/EMBL/DDBJ accession number U25281) (Fig. 5 A). All these genes code for proteins of unknown function. Further, inspection of the core of this extended motif revealed the presence of a hexapeptide, LKKAET, conserved in a variety of actin-binding proteins, including thymosin, actobindin, tropomyosin, villin, dematin, myosin heavy chain, fimbrin, α-actinin, and plastin (Fig. 5 B).
Figure 5.
The actin-binding domain of verprolin. (A) Homologies between the first 70 amino acids of verprolin and C. elegans CELR 144.4 protein (“CELR144.4”, GenBank/EMBL/DDBJ accession number U23515), X. laevis proline-rich protein (“Xlpro-rich”, GenBank/EMBL/DDBJ accession number X68249), and rat SH3-domain–binding protein (“RNU25281”, GenBank/EMBL/DDBJ accession number U25281). Alignment was done using BOXSHADE 3.21 program running on a WWW server at ISREC, Switzerland. Asterisks designate the position of the LKKAET hexapeptide. (B) LKKAET-like motifs in various cytoskeletal proteins. (C) Diagram of the localization of the actin-binding domain in verprolin. The LKKAET motif is in bold and underlined. The 11 black bars correspond to potential SH3-binding domains having the general pattern APPL/IP.
To determine whether there is an actin-binding domain at the amino terminus of verprolin (Fig. 5 C), VRP1 deletion and out-of-frame constructs were generated (Fig. 6). If the actin–verprolin interactions detected by the two- hybrid method result from the interaction of the region of verprolin containing the LKKAET motif, plasmids encoding this region of verprolin should then generate proteins that interact with actin. Deletion of the carboxy-terminal 455 amino acids generated pDAD-VRP1ΔSal coding for verprolin amino acids 1–362. This part of verprolin interacts with actin in the two-hybrid system (Fig. 6). pDAD-VRP1ΔSalΔKpn1-3, which fuses in-frame the first 70 with the last 100 codons of verprolin, generates a shorter version of verprolin that shows strong interaction with actin. Finally, pDAD-VRP1ΔSalΔKpn1-2 that has an out-of-frame fusion coding for only the first 70 amino acids of verprolin was found to code for a protein able to bind actin. Therefore, the first 70 amino acids of verprolin are sufficient for actin-verprolin interactions in the two-hybrid system.
Figure 6.
Mapping of the actin-binding domain of verprolin using two-hybrid analysis. Various DNA fragments encoding truncated Vrp1p were cloned in-frame with the Gal4 DNA-activation domain in vector pDAD and the resultant plasmids were transformed into Y190 strain bearing the pDBD–ACT1 construct encoding actin. Association of verprolin with actin was examined by the qualitative assay for β-galactosidase and by assessing growth on media-lacking histidine in the presence of 100 mM 3-amino-1,2,4-triazole. Numbers represent the length in amino acids. Asterisks designate mitotic instability of pDAD-VRP1 and pDAD-VRP1ΔSal plasmids. (N.D.) not done.
To test whether the LKKAET motif that resides within the 70–amino acid domain is necessary for the verprolin– actin interaction, we generated a mutant version of the sequence encoding this hexamer. Based on the alignment in Fig. 5 B and on mutagenesis studies of β-thymosin (Van Troys et al., 1996), we chose to mutate the two basic residues of this sequence to acidic residues. We found that the 70–amino acid motif encoding the sequence LEEAET was unable to interact with actin in the two-hybrid system. Therefore, as anticipated, the hexamer is necessary for the interaction of verprolin with actin.
If verprolin–actin interactions are solely dependent upon the 70–amino acid motif, constructs not containing this motif should then generate proteins that fail to interact with actin. To test this, a fusion between the Gal4p DNA-activating domain and a fragment of VRP1 encoding amino acids 71–817 (pDAD-VRP1 71–817) was constructed. pDAD-VRP1 71–817 shows less activation of LacZ or HIS3 reporter genes than pDAD-VRP1ΔSalΔKpn1-2. Therefore, whereas the first 70 amino acids of verprolin, containing the LKKAET motif, are sufficient for binding to actin, other actin-binding sequences might reside in verprolin and/or other sequences may influence the efficiency of actin-binding.
Suppression of vrp1-1 phenotypes by actin indicate that verprolin–actin interactions are important for the function of verprolin. If so, versions of verprolin lacking the actin-binding domain should fail to complement vrp1 mutations. Fortuitously, the plasmid encoding the entire verprolin appended to the Gal4p DNA-binding domain (pDAD-VRP1) constructed for the two-hybrid analyses generates sufficient hybrid protein to complement vrp1-1 (Fig. 6), even though most of the protein locates to the nucleus as assessed by indirect immunofluorescence (not shown). None of the other deletion constructs was able to complement vrp1-1. In particular, pDAD-VRP1 71–817, lacking the 70–amino acid actin-binding domain failed to code for a functional verprolin (Fig. 6). Provided that this shortened version of verprolin maintains its native conformation, we can conclude that the actin-binding domain of verprolin is important for its in vivo function.
Cellular Location of Verprolin
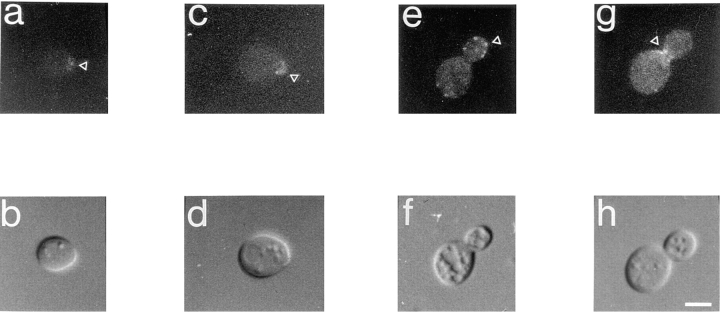
The allele-specific interactions described above and the results from the two-hybrid system provide evidence that verprolin and actin interact directly. To test the prediction that verprolin localizes with the actin cytoskeleton, we assessed the natural distribution of verprolin tagged with GFP (Chalfie et al., 1994) in live cells. Wild-type cells (strain T8.1D) as well as mutant vrp1-1 (strain TZ33) and vrp1:: LEU2 haploid (strain T65.1D) or diploid (strain GVY1) cells were transformed with low copy YCpVRP1GFP plasmid encoding verprolin–GFP. VRP1–GFP complements both the vrp1-1 mutation and the vrp1::LEU2 disruption. Verprolin–GFP expressed from YCpVRP1GFP plasmid has a similar distribution in wild-type, vrp1-1, vrp1::LEU2, and vrp1::LEU2/vrp1::LEU2 mutant cells. However, the signal is most easily visualized in vrp1::LEU2 (Fig. 7) and vrp1::LEU2/vrp1::LEU2 cells bearing YCpVRP1GFP plasmid. In cells preparing to bud, verprolin is localized at the presumptive budding site (Fig. 7, a and b). As the bud starts to grow, verprolin concentrates at the tip of the bud (Fig. 7, c and d). There is very little if any verprolin in the cytoplasm. In older buds verprolin has punctate staining, much like cortical patches (Fig. 7, e and f). Sometimes patches are at the tip of the buds and sometimes they are eccentric. When the cells prepare for cytokinesis, there are two rings of verprolin around the mother-bud neck (Fig. 7, g and h). Similar actin-containing rings are characteristic for this part of the yeast life cycle (Adams and Pringle, 1984) and at least one other actin-binding protein, Aip3p, locates at these rings during this part of the cell cycle (Amberg et al., 1997). In freshly divided cells, verprolin patches appear randomly distributed. The punctuate staining of verprolin-GFP is remarkably similar to that of actin-GFP (Doyle and Botstein, 1996).
Figure 7.
In vivo localization of verprolin-GFP fusion in vrp1::LEU2 disruptant (as shown by arrowheads). (a and c) A verprolin cap marks the site where the potential bud will form. (e) Verprolin localizes to the bud in patches-like structures. (g) Cells preparing for cytokinesis have two rings of verprolin-GFP around the neck. b, d, f, and h are DIC images of a, c, e, and g frames, respectively. Bar, 2 μm.
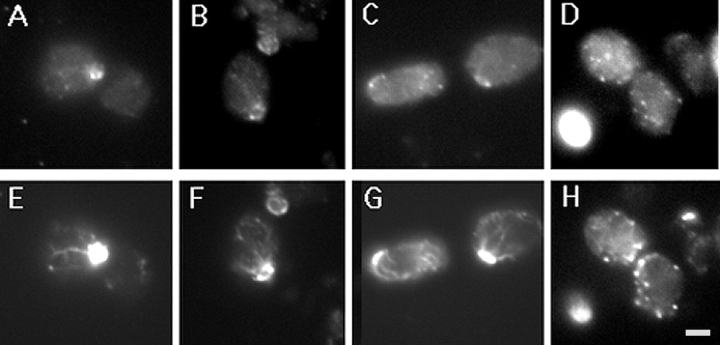
To determine whether verprolin and actin cortical patches co-localize, we used indirect immunofluorescence to locate a HA-tagged version of Vrp1p encoded by the low copy vector YCpVRP1-HA. This verprolin–HA fusion fully complements, in either single or multicopy, the vrp1-1 and the vrp1::LEU2 mutations in haploid or diploid cells. Actin was detected using fluorescent phalloidin. As it was not possible to stain the cytoskeleton with phalloidin on slides, the procedure was modified to perform all reactions in solution (see Materials and Methods). As was found in studies of live cells, verprolin in fixed cells is located in a cell cycle–dependent fashion to positions also occupied by the actin cortical cytoskeleton (Fig. 8). The verprolin patches overlap most often with actin patches stained with phalloidin. Interestingly, patches in mother cells not associated with the presumptive bud appear to locate at points of actin cable branch sites (Fig. 8). We conclude that verprolin is present during the whole cell cycle and is located at or very near the actin cortical patches.
Figure 8.
Co-localization of verprolin-HA and actin. GVY1 cells bearing hemagglutinin-tagged VRP1 on a centromere-containing vector (YCpVRP1-HA) were fixed and stained with 12CA5 antibody against the hemagglutinin tag and then co-stained with Oregon green phalloidin as described in Methods. In A–D representative cells bearing the verprolin-HA fusion are shown. The same cells, visualized in the FITC channel to detect F-actin, are shown in E, F, G, and H. The two proteins co-localize in many of the cortical patches. However, apparently there are actin patches which do not seem to contain verprolin and vice versa. Bar, 1.5 μm.
Verprolin Localization is Actin Independent
Verprolin exhibits a cell cycle–dependent pattern of localization which closely resembles that of its interactor, actin. To explore the co-dependence of the subcellular distribution of Act1p and Vrp1p we used a pharmacological tool which recently has been shown to allow the temporal dissection of events involved in bud morphogenesis. LAT-A, a macrolide isolated from a marine sponge, is a potent inhibitor of actin cytoskeleton assembly in vertebrates (Spector et al., 1989) and yeast (Ayscough et al., 1997). We examined the consequences that disruption of actin cytoskeleton with LAT-A have on verprolin-GFP localization. Cultures of vrp1::LEU2/vrp1::LEU2 diploid cells (strain GVY1) containing the VRP1–GFP fusion on a low copy vector were grown for 15 minutes in the presence of 100 μM LAT-A or in the presence of DMSO diluent alone and visualized immediately by microscopy. At the same time point, an aliquot was withdrawn from each culture, fixed, and processed for rhodamine phalloidin staining. Whereas the actin cytoskeleton of LAT-A treated cells is virtually absent (compare Fig. 9, G and H to Fig. 9, C and D), the localization of verprolin–GFP is unaffected (compare Fig. 9, E and F, to Fig. 9, A and B). Thus, maintenance of the intracellular polarized location of verprolin is not dependent upon the integrity of actin cytoskeleton.
Figure 9.
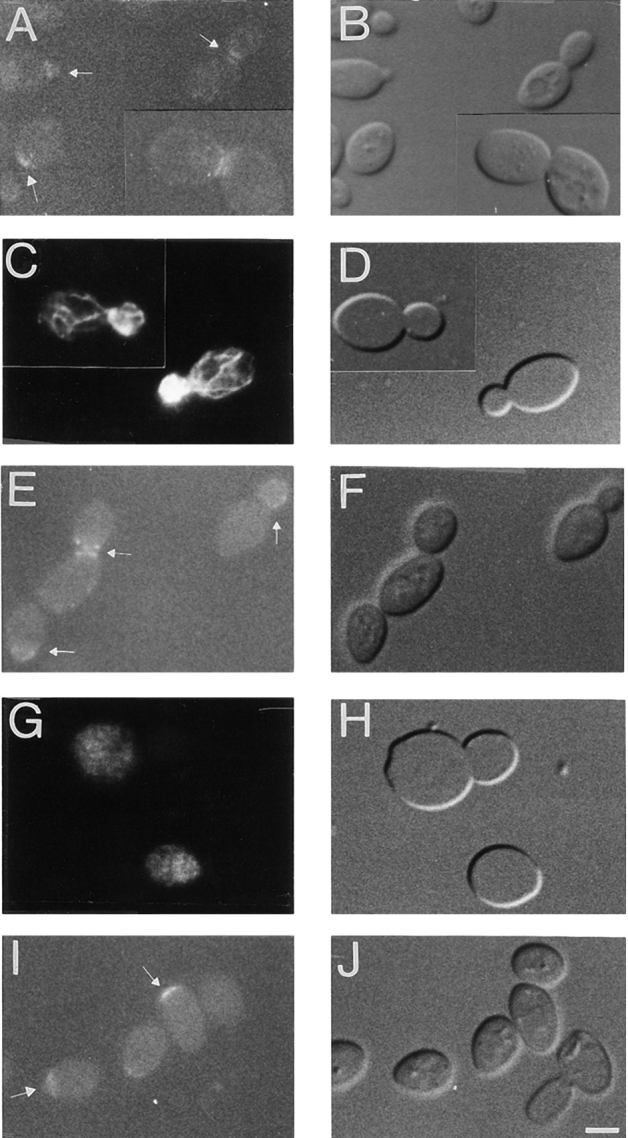
Localization of verprolin-GFP in the absence or presence of LAT-A in diploid vrp1::LEU2/vrp1::LEU2 cells (strain GVY1) containing YCpVRP1GFP plasmid. (A–H) Cells were grown to logarithmic phase, the culture was then divided in two: half was treated with DMSO alone for 15 min (A–D) and half with LAT-A solubilized in DMSO for 15 min (E–H). (I and J), a separate culture of cells enriched for unbudded cells was treated for 4 h with LAT-A solubilized in DMSO. (A) Localization of verprolin-GFP in the absence of LAT-A shows the polarized localization of verprolin at the presumptive budding site, in small buds, and at the mother-bud neck; (C) An aliquot of the culture used in (A) was fixed and further stained with rhodamine phalloidin to confirm the normal morphology of actin cytoskeletons; (E) Localization of verprolin-GFP in a logarithmic culture after incubation with LAT-A for 15 min. Verprolin-GFP maintains its intracellular localization at the mother-bud neck or at the tip of buds. (G) An aliquot of the culture used in E was fixed and stained with rhodamine phalloidin. Actin cytoskeletons of these cells are disassembled. (I) Localization of verprolin-GFP in the presence of LAT-A (4 h) in cells exiting G0. The verprolin-GFP signal is present at the presumptive budding site while the actin cytoskeletons of these cells are virtually absent (not shown). B, D, F, H, and J are DIC images of A, C, E, G, and I, respectively. Bar, 2 μm.
To determine whether verprolin requires the actin cytoskeleton to achieve its location, cultures enriched in unbudded cells (Ayscough et al., 1997) were grown for 4 h in 100 μM LAT-A and then visualized by microscopy. Under these conditions, while the actin cytoskeleton is disassembled (data not shown), the majority of the cells have verprolin-GFP at the presumptive budding site (Fig. 9, I and J). This indicates that verprolin achieves its location inside the cell through an actin-independent pathway.
Effect of Verprolin on Actin Cytoskeleton Stability
Sensitivity to LAT-A varies among actin cytoskeleton-associated mutants and can be used as an indicator of the effect the respective proteins have on the stability of the actin cytoskeleton (Ayscough et al., 1997). To test the sensitivity of vrp1 mutant strains to LAT-A we used a halo assay. The vrp1::LEU2 strain harboring the VRP1 gene on a centromeric plasmid is more resistant to LAT-A than is the same strain containing vector alone (Fig. 10, compare A and B). Therefore, disruption of VRP1 gene confers sensitivity to LAT-A. The relative apparent sensitivity (Reneke et al., 1988) for vrp1::LEU2 compared to vrp1::LEU2 bearing YCpVRP1 is 2.5, a value comparable to that of other cytoskeletal proteins (Ayscough et al., 1997). This result suggests that verprolin acts to increase actin cytoskeleton stability. Also, we compared the sensitivity of vrp1-1 cells bearing pT3 to that of vrp1-1 cells bearing YCpVRP1 or an empty vector as control. Although no difference is apparent between vrp1-1 cells bearing VRP1 or the empty vector at the permissive temperature, vrp1-1 cells bearing ACT1 on pT3 show resistance to LAT-A even at a concentration of 2 mM (Fig. 10 E).
Figure 10.
Halo assay testing the sensitivity of vrp1 mutant strains to LAT-A. (A) vrp1::LEU2 cells (strain T65.1D) bearing the VRP1 gene on the centromeric plasmid YCpVRP1. (B) vrp1:: LEU2 cells bearing a vector alone. (C) vrp1-1 cells (strain TZ33) bearing VRP1 gene on the centromeric plasmid YCpVRP1. (D) vrp1-1 cells (strain TZ33) with vector alone. (E) vrp1-1 cells (strain TZ33) bearing ACT1 gene on plasmid pT3. Concentrations of LAT-A are: 0 (top), 0.5 mM (right), 1 mM (bottom), and 2mM (left). Strains were grown on ura− selective media at 23°C for 48 h.
Discussion
Verprolin Is an Actin-binding Protein Important for Numerous Actin-associated Processes
Verprolin was identified as a protein important for both sorting of Mod5p-I between mitochondria and cytosol (Zoladek et al., 1995) and endocytosis (Munn et al., 1995). vrp1-1 mutant cells have an aberrant actin cytoskeleton which is most apparent at permissive temperatures if the cells are grown on nonfermentable carbon sources (Zoladek et al., 1995). Allele-specific suppression of vrp1-1 by a single additional copy of ACT1 (Zoladek et al., 1995) indicated that verprolin and actin physically interact. To confirm the genetic predictions and to understand the nature of these interactions, we examined vrp1-1 phenotypes, in addition to temperature sensitive growth, for suppression by ACT1. We found that ACT1 suppresses vrp1-1 defects in cytoskeletal polarization, endocytosis, and mitochondrial function. Therefore, it is likely that all phenotypes of vrp1 mutants relate to an abnormal cytoskeleton.
Our studies of fluid phase endocytosis are consistent with other observations that some, but not all, cytoskeletal mutations show defects in endocytosis (Munn et al., 1995; Wendland et al., 1996; Zoladek et al., 1997;). Our data support the involvement of the actin cytoskeleton and, more probably, cortical actin patches as components of the endocytic machinery. Restoration of endocytosis in vrp1-1 mutant cells by additional actin indicates that their endocytosis defect is most likely a direct consequence of a defective cytoskeleton.
Actin supplied from a centromere-containing plasmid also partially corrects the vrp1-1 mitochondrial deficiency as assessed by growth on nonfermentable carbon sources, and restores near parental cytoplasmic levels of Mod5p-I,KR6 as assessed by nonsense suppression studies. The involvement of the actin cytoskeleton with delivery of proteins to mitochondria and with mitochondrial function implicates a novel function for the actin cytoskeleton. Ideas as to how mitochondria and the cytoskeleton might interrelate are outlined below.
Because the actin cytoskeleton has been shown to be important for bud-site selection (Chant and Pringle, 1995; Yang et al., 1997), we examined the budding pattern of diploid vrp1::LEU2/vrp1::LEU2 and haploid vrp1::LEU2 cells. Verprolin is required for the bipolar budding pattern, but not for the axial budding pattern. The randomization of budding pattern in vrp1::LEU2/vrp1::LEU2 diploids indicates that verprolin is among the proteins required for bipolar site selection in these cells.
We used the two-hybrid system to test the genetic prediction that actin and verprolin interact. We found that actin and verprolin are able to bind each other. The data indicate that verprolin is a new member of the family of actin-binding proteins. The first 70 amino acids of verprolin are homologous with a similar region of a group of proteins of unknown function from three higher eukaryotes. Using the two-hybrid system and deletion analysis we show that the 70 amino acid conserved domain in verprolin is sufficient for interaction with actin.
The hexapeptide motif LKKAET, found in a variety of actin-binding proteins, is in the middle of this conserved domain. The conservation of the 70–amino acid domain and the possession of a motif similar to the LKKAET hexapeptide within this domain leads us to predict that these C. elegans, Xenopus, and rat proteins of unknown function will subsequently be shown to interact with actin. The hexapeptide motif has been shown to directly bind actin. Actobindin, a small actin-binding protein from Acanthamoeba, contains two LKHAET hexapeptides, between amino acids 15 to 20 and 51 to 56. Lys16 and Lys52 of actobindin can be cross-linked to particular amino acids of actin (Vandekerckhove et al., 1990; Vancompernolle et al., 1991). The LKKEKG hexapeptide is crucial for the morphogenic activity of the actin-bundling protein villin. In vitro studies with a 22–amino acid synthetic peptide including the villin LKKEKG hexapeptide showed that it is part of an F-actin binding site involved in the conversion of G-actin to a polymerization-competent form. The Ser809-Lys825 fragment of villin, containing the first five amino acids of the LKKEKG hexapeptide, is important for binding to preformed actin filaments, but insufficient for their induction (Friederich et al., 1992). Moreover, conversion of KKEK to KEEE in villin significantly decreases villin's morphogenic activity. Finally, the LKKTET hexapeptide in thymosin is essential for binding to actin; mutation of Lys18-19 to Glu drastically abolishes cross-linking with monomeric actin (Van Troys et al., 1996). By two-hybrid analysis we showed that the analogous alteration of the verprolin LKKAET motif to LEEAET destroys the ability of the 70–amino acid domain to interact with actin. Thus it appears that the function of this motif is conserved from lower to higher eukaryotes.
A similarity also exists between the 70–amino acid homology domain of verprolin and COOH-terminal regions of N-WASP and WASP, both of which contain less conserved versions of the LKKAET motif (Miki et al., 1996; Symons et al., 1996). Both WASP and N-WASP influence the organization of the actin cytoskeleton. WASP colocalizes with actin. N-WASP contains a tripartite actin-binding motif of two shorter verprolin-like domains, one of which terminates right in the middle of the LKKAET motif followed by a cofilin-like actin-binding motif. When only the verprolin-like part was tested for actin binding in vitro, there was very poor binding of N-WASP to actin. When the whole motif (including the cofilin-like part) was included, actin binding occured (Miki et al., 1996). The data suggest that part of the 70–amino acid region of verprolin, in addition to LKKAET, participates in actin binding.
Verprolin Localization Is Independent of Actin Cytoskeleton
The changes in the cellular distribution of verprolin-GFP during the cell cycle are very similar to the pattern of changes that the actin cortical patches undergo during the cell cycle. However, whereas the actin cytoskeleton is disassembled using the actin assembly inhibitor LAT-A, verprolin's localization is unaffected. Therefore, verprolin does not require actin to achieve or maintain its polarized cellular localization. This is not a unique case. Other proteins involved in cell polarity development, including Myo2p, calmodulin, and Bud6/Aip3p (Ayscough et al., 1997), achieve polarized localization by an actin-independent pathway. Also, polarity establishment proteins Cdc42p and Bem1p become polarized even though the actin cytoskeleton is totally disassembled (Ayscough et al., 1997). Thus, it appears that yeast cells have evolved an actin-independent pathway for localizing polarity development proteins. By contrast, other actin-binding proteins including Arp2p, proteins involved in secretion such as Sec4p, Sec8p, and the kinesin-like protein Smy1p require actin to achieve their polarized localization (Ayscough et al., 1997).
The ability of verprolin to localize independently from actin as well as its role in bud-site selection indicate that it functions in cell polarity development and maintenance. As supported by data regarding verprolin's actin-binding properties, its spatial co-localization with actin and its downstream effects on cytoskeleton morphogenesis and stability, verprolin might convey the polarity development information to the actin cytoskeleton via acting as an anchor for the actin cytoskeleton.
Possible Roles of Verprolin in Mitochondrial Function and Mitochondrial Protein Sorting
How would verprolin affect cytoskeletal morphology and, at the same time, be involved in mitochondrial function and Mod5p-I mitochondria/cytosol sorting? A priori, three mechanisms could account for the mitochondrial phenotypes: defective mitochondrial movement, altered protein delivery, and disruption of mRNA sorting. Although it is known that movement of mitochondria is impaired by ACT1 mutations that perturb the myosin-binding site (Smith et al., 1995) and it is known that many, but not all, thermal sensitive act1 alleles cause abnormal mitochondrial distribution and morphology (Drubin et al., 1993), we do not favor the model that altered mitochondrial distribution/morphology accounts for the mitochondrial defects observed in vrp1 mutant cells. This is because even on lactate-containing media when the cytoskeleton is clearly depolarized, vrp1 mutants show no obvious defects in mitochondrial distribution or morphology (Zoladek et al., 1995 and these studies). Recently, it has been shown that Mft52p, which affects the delivery of particular fusion proteins to yeast mitochondria, is a component of a greater than 5,000-kD cytosolic particle. Mft52p interacts with mitochondrial targeting sequences and it has been proposed that Mft52p plays a role in co-translational delivery of proteins to mitochondria (Cartwright et al., 1997; for review see Lithgow et al., 1997). If such a co-translational mechanism required a role of the actin cytoskeleton, then the mitochondrial defects we detect in vrp1 mutant cells could be accounted for by defects in this putative co-translational mechanism of mitochondrial import.
Alternatively, the actin cytoskeleton could be involved in the delivery of mRNAs encoding mitochondrial proteins to the vicinity of mitochondria. In higher eukaryotes, the actin cytoskeleton has a role in mRNA sorting (Singer et al., 1992; Bassell, 1993). mRNA sorting plays an important role in the cellular distribution of cytoskeletal proteins (Lawrence and Singer, 1986; Kislauskis et al., 1994) and numerous morphogens like Vg-1 (Yisraeli et al., 1990), nanos (Gavis and Lehmann, 1992) and oskar (Kim-Ha et al., 1993). The cellular distribution of actin itself has been shown to occur via sorting of actin mRNA to appropriate subcellular locations (Sundell and Singer, 1991). Moreover, there are reports implicating mRNA sorting in the delivery of proteins to mitochondria in higher eukaryotes (Rings et al., 1992; Ricart et al., 1997; for review see Lithgow et al., 1997).
Little is known about sorting of mRNA in yeast. ASH1 mRNA is one example of a mRNA which is selectively transported to and expressed solely in daughter cells (Long et al., 1997). The actin cytoskeleton is actively involved in the localized transport of ASH1 transcript since mutations of cytoskeletal genes like ACT1, PFY1, TPM1, and BNI1 impair the transport of ASH1 mRNA to daughter cells (Long et al., 1997). If mRNAs encoding mitochondrial proteins are also sorted in yeast, as they are in higher eukaryotic cells, then lesions similar to those affecting the ability of yeast to distribute ASH1 mRNA might also affect the distribution of transcripts encoding mitochondrial proteins. The actin-binding protein Bni1p, required for correct sorting of ASH1 mRNA to daughter cells (Long et al., 1997), binds the polarity establishment protein Cdc42p and also Bud6/Aip3p (Evangelista et al., 1997), another actin-binding protein whose intracellular location (Amberg et al., 1997) is remarkably similar to that of verprolin. Like verprolin, Bud6/Aip3p is required for bipolar, but not for axial budding pattern (Amberg et al., 1997), and both verprolin and Bud6/Aip3p (Ayscough et al., 1997) can achieve their polarized location even though the actin cytoskeleton is disassembled. Of these three proteins, only Bni1p has been tested and shown to be responsible for sorting of ASH1 mRNA. The prediction is that verprolin will be found to play a role in mRNA sorting.
Acknowledgments
We thank S.S. Brown, S.J. Elledge, S.D. Emr, R.Y. Tsien, and T. Zoladek for plasmids and strains. We are grateful to D.R. Stanford for his generous help with computer data analysis and to A.L. Benko, L.A. Hunter, and D.R. Stanford for their comments on the manuscript. We thank D.C. Amberg for stimulating discussions, and R.M. Long and R.H. Singer for sharing data before publication. We are grateful to James E. Hopper for allowing us to use the CCD camera and computer facilities, Allison Adams for suggestions concerning double staining of cells with antibodies and phalloidin, and Sorin Vaduva for help with photographic and computer work.
This work was supported by grants from the American Cancer Society and the National Science Foundation to A.K. Hopper and N.C. Martin.
Abbreviations used in this paper
- DIC
differential interference contrast
- GFP
green fluorescent protein
- HA
hemagglutinin
- LAT-A
Latrunculin-A
- lys
lysine
- ORF
open reading frame
- ts
temperature sensitive
Footnotes
Address all correspondence to Anita K. Hopper, Department of Biochemistry and Molecular Biology, The Milton S. Hershey Medical Center, The Pennsylvania State University, Hershey, Pennsylvania 17033. Tel.: (717) 531-6008. Fax: (717) 531-7072.
References
- Adams AE, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenic-mutant Saccharomyces cerevisiae. . J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, A.E., and J.R. Pringle. 1991. Staining of actin with fluorochrome-conjugated phalloidin. In Methods Enzymol. Guide to Yeast Genetics and Molecular Biology; Vol. 194. C. Guthrie and G.R. Fink, editors. Academic Press Inc., San Diego, CA. 729–731. [DOI] [PubMed]
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. . Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol BiolCell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin D. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor Latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ. High resolution distribution of mRNA within the cytoskeleton. J Cell Biochem. 1993;52:127–133. doi: 10.1002/jcb.240520203. [DOI] [PubMed] [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. The END3gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, Beilharz T, Hansen P, Garrett J, Lithgow T. Mft52, an acid-bristle protein in the cytosol that delivers precursor proteins to yeast mitochondria. J Biol Chem. 1997;272:5320–5325. doi: 10.1074/jbc.272.8.5320. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D-C, Yang B-C, Kuo T-T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly SFH, Pocklington MJ, Pallotta D, Orr E. A proline-rich protein, verprolin, involved in cytoskeletal organization and cellular growth in the yeast Saccharomyces cerevisiae. . Mol Microbiol. 1993;10:585–596. doi: 10.1111/j.1365-2958.1993.tb00930.x. [DOI] [PubMed] [Google Scholar]
- Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. . Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Dulic, V., M. Egerton, I. Elguindi, S. Raths, B. Singer, and H. Riezman. 1991. Yeast endocytosis assays. In Methods Enzymol. Guide to Yeast Genetics and Molecular Biology; Vol. 194. C. Guthrie and G.R. Fink, editors. Academic Press Inc., San Diego, CA. 697–710. [DOI] [PubMed]
- Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn AE, Lee W-H, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O-K. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Friederich E, Vancompernolle K, Huet C, Goethals M, Finidori J, Vandekerckhove J, Louvard D. An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenic effect of villin. Cell. 1992;70:81–92. doi: 10.1016/0092-8674(92)90535-k. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R. Localization of nanosRNA controls embryonic polarity. Cell. 1992;71:301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Role of type I myosins in receptor-mediated endocytosis in yeast. Science. 1996;272:533–535. doi: 10.1126/science.272.5261.533. [DOI] [PubMed] [Google Scholar]
- Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Webster PJ, Smith JL, Macdonald PM. Multiple RNA regulatory elements mediate distinct steps in localization of oskarmRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer R. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. Intracellular localization of messenger RNA for cytoskeletal proteins. Cell. 1986;45:407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- Lithgow T, Cuezva JM, Silver P. Highways for protein delivery to the mitochondria. TIBS (Trends Biochem Sci) 1997;22:110–113. doi: 10.1016/s0968-0004(97)01007-4. [DOI] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Mating-type switching in yeast controlled by asymmetric localization of ASH1mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2- dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Stevenson BJ, Geli MI, Riezman H. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. . Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R. 1991. Staining of bud scars and other cell wall chitin with calcofluor. In Methods Enzymol. Guide to Yeast Genetics and Molecular Biology; Vol. 194. C. Guthrie and G.R.Fink, editors. Academic Press Inc., San Diego, CA. 732–735. [DOI] [PubMed]
- Reneke J, Blumer K, Courchesne W, Thorner J. The carboxy-terminal segment of the yeast a-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- Ricart J, Egea G, Izquierdo JM, San C, Martin, Cuezva JM. Subcellular structure containing mRNA for beta subunit of mitochondrial H+-ATP synthase in rat hepatocytes is translationally active. Biochem J. 1997;324:635–643. doi: 10.1042/bj3240635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rings EH, Buller HA, de Boer PA, Grand RJ, Montgomery RK, Lamers WH, Charles R, Moorman AF. Messenger RNA sorting in enterocytes. Co-localization with encoded proteins. FEBS Lett. 1992;300:183–187. doi: 10.1016/0014-5793(92)80192-j. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. A Laboratory Manual. Second edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sherman, F. 1991. Getting started with yeast. In Methods Enzymol. Guide to Yeast Genetics and Molecular Biology; Vol. 194. C. Guthrie and G.R. Fink, editors. Academic Press Inc., San Diego, CA. 3–21.
- Singer RH. The cytoskeleton and mRNA localization. Curr Opin Cell Biol. 1992;4:15–19. doi: 10.1016/0955-0674(92)90053-f. [DOI] [PubMed] [Google Scholar]
- Smith MG, Simon V, O'Sullivan H, Pon LA. Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangement in the budding yeast Saccharomyces cerevisiae. . Mol Biol Cell. 1995;6:1381–1396. doi: 10.1091/mbc.6.10.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I, Shochet N, Blasberger D, Kashman Y. Latrunculins-novel marine macrolides that disrupt microfilament organization and affect cell growth: 1. Comparison with cytochalasin D. Cell Motil. Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin messanger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Symons M, Derry JMJ, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Vancompernolle K, Vandekerckhove J, Bubb MR, Korn ED. The interfaces of actin and Acanthamoebaactobindin. J Biol Chem. 1991;266:15427–15431. [PubMed] [Google Scholar]
- Vandekerckhove J, Van Damme J, Vancompernolle K, Bubb MR, Lambooy PK, Korn ED. The covalent structure of Acanthamoebaactobindin. J Biol Chem. 1990;265:12801–12805. [PubMed] [Google Scholar]
- Van Troys M, Dewitte D, Goethals M, Carlier M-F, Vandekerckhove J, Ampe C. The actin binding site of thymosin β4 mapped by mutational analysis. EMBO J. 1996;15:201–210. [PMC free article] [PubMed] [Google Scholar]
- Waddle JA, Karpova TS, Waterson RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Bretscher A. The rho-GAP encoded by BEM2regulates cytoskeletal structure in budding yeast. Mol Biol Cell. 1995;6:1011–1024. doi: 10.1091/mbc.6.8.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley KC, Wiese BA, Smith RF. BEAUTY: an enhanced BLAST-based search tool that integrates multiple biological information resources into sequence similarity search results. Genome Res. 1995;5:173–184. doi: 10.1101/gr.5.2.173. [DOI] [PubMed] [Google Scholar]
- Yang S, Ayscough KR, Drubin DG. A role for the actin cytoskeleton of Saccharomyces cerevisiaein bipolar bud-site selection. J Cell Biol. 1997;136:111–123. doi: 10.1083/jcb.136.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli JK, Sokol S, Melton DA. A two-step model for the localization of maternal mRNA in Xenopusoocytes: involvment of microtubules and microfilaments in the translocation and anchoring of Vg-1 mRNA. Development. 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- Zoladek T, Vaduva G, Hunter LA, Boguta M, Go BD, Martin NC, Hopper AK. Mutations altering the mitochondrial-cytoplasmic distribution of Mod5p implicate the actin cytoskeleton and mRNA 3′ ends and/or protein synthesis in mitochondrial delivery. Mol Cell Biol. 1995;12:6884–6894. doi: 10.1128/mcb.15.12.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoladek T, Tobiasz A, Vaduva G, Boguta M, Martin NC, Hopper AK. MDP1, a Saccharomyces cerevisiae gene involved in mitochondrial/cytoplasmic protein distribution, is identical to the ubiquitin-protein ligase gene RSP5. . Genetics. 1997;145:595–603. doi: 10.1093/genetics/145.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]