Abstract
We have identified a new protein, Tim54p, located in the yeast mitochondrial inner membrane. Tim54p is an essential import component, required for the insertion of at least two polytopic proteins into the inner membrane, but not for the translocation of precursors into the matrix. Several observations suggest that Tim54p and Tim22p are part of a protein complex in the inner membrane distinct from the previously characterized Tim23p-Tim17p complex. First, multiple copies of the TIM22 gene, but not TIM23 or TIM17, suppress the growth defect of a tim54-1 temperature-sensitive mutant. Second, Tim22p can be coprecipitated with Tim54p from detergent-solubilized mitochondria, but Tim54p and Tim22p do not interact with either Tim23p or Tim17p. Finally, the tim54-1 mutation destabilizes the Tim22 protein, but not Tim23p or Tim17p. Our results support the idea that the mitochondrial inner membrane carries two independent import complexes: one required for the translocation of proteins across the inner membrane (Tim23p–Tim17p), and the other required for the insertion of proteins into the inner membrane (Tim54p–Tim22p).
Mitochondrial function depends on the import of hundreds of different proteins synthesized in the cytosol. Protein import is a multistep pathway which includes the binding of precursor proteins to surface receptors, translocation of the precursor across one or both mitochondrial membranes, and folding and assembly of the imported protein inside the mitochondrion (for review see Ryan and Jensen, 1995; Schatz and Dobberstein, 1996; Jensen and Kinnally, 1997; Pfanner and Meijer, 1997). Most precursor proteins carry amino-terminal targeting signals, called presequences (Hurt et al., 1984, a, b, 1985; Horwich et al., 1985; van Loon et al., 1986), and are imported into mitochondria via import complexes located in both the outer and the inner membrane (IM)1. The mitochondrial outer membrane of Saccharomyces cerevisiae contains at least four proteins, Tom70p, Tom37p, Tom22p, and Tom20p (Hase et al., 1983; Hines et al., 1990; Steger et al., 1990; Moczko et al., 1993; Ramage et al., 1993; Gratzer et al., 1995; Hönlinger et al., 1995), which have been proposed to recognize the targeting signal carried on imported mitochondrial proteins. These four receptors interact with at least six other polypeptides, and together comprise the TOM complex, which facilitate the movement of proteins across the outer membrane (Kiebler et al., 1990; Söllner et al., 1992).
In the IM, a TIM complex mediates the translocation of proteins into the matrix. The TIM complex consists of at least two integral IM proteins, Tim23p and Tim17p, which have been proposed to form part of a protein-translocating channel (Dekker et al., 1993; Emtage and Jensen, 1993; Maarse et al., 1994; Ryan et al., 1994). Tim23p, a 23-kD protein, was first identified as a temperature-sensitive mutant defective in the import of several different matrix- localized precursor proteins (Emtage and Jensen, 1993). TIM17 was identified as a multi-copy suppressor of the tim23 mutant, and was shown to encode a 17-kD protein homologous to the carboxyl-terminal domain of Tim23p (Ryan et al., 1994). The essential Tim23p and Tim17 proteins cooperate with Tim44p, located on the inside of the IM (Scherer et al., 1992; Rassow et al., 1994), and mt-Hsp70, a matrix-localized member of the 70-kD heat shock family (Kang et al., 1990; Scherer et al., 1992; Rassow et al., 1994). Tim44p and mt-Hsp70 are proposed to pull the imported precursor protein through the IM translocation channel into the matrix (Kronidou et al., 1994; Rassow et al., 1994; Schneider et al., 1994; Ungermann et al., 1994, 1996; Blom et al., 1995).
Tim23p, Tim17p, Tim44p, and mt-Hsp70 appear to form functional complexes in the IM. Tim23p and Tim17p cofractionate after detergent solubilization of the mitochondria (Berthold et al., 1995; Blom et al., 1995; Ryan et al., 1998). Both proteins can also be chemically cross-linked to each other in intact mitochondria (Berthold et al., 1995; Blom et al., 1995; Ryan et al., 1998). When a precursor destined for the matrix was arrested in transit across the IM, a large complex containing Tim44p, Tim23p, Tim17p, and mt-Hsp70 (and presumably other proteins) coimmunoprecipitated with the precursor (Berthold et al., 1995). It has recently been shown that Tim23p may exist in two subcomplexes, one complex consisting of Tim23p, Tim44p, and mt-Hsp70, and another complex with Tim23p, Tim17p, and mt-Hsp70 (Bömer et al., 1997). The function of the two subcomplexes in import is not known.
Recently, an essential IM protein, Tim22p, homologous to both Tim23p and Tim17p, has been identified (Sirrenberg et al., 1996). Tim22p is not required for the import of matrix-localized proteins, but Tim22p appears to be essential for the correct insertion of two membrane proteins, Aac1p, the ATP/ADP carrier, and PiC, the phosphate carrier, into the IM. Whether Tim22p is required for the import of other membrane proteins besides carrier proteins is not known. Tim22p is proposed to be in a separate complex from Tim23p and Tim17p in the IM, but other members of this new complex have not been identified. We have identified a new protein, Tim54p, which is essential for the IM insertion of the Aac1 carrier and Tim23 import proteins. Tim54p is not required for the translocation of precursors across the IM into the matrix. We find that Tim54p physically interacts with Tim22p, and forms an import complex in the IM separate from the Tim23p–Tim17p import machinery. Our results indicate that there are two import pathways in the IM. One pathway uses Tim23p and Tim17p in the translocation of precursors across the IM, and the other requires Tim54p and Tim22p for the insertion of proteins into the IM. Because Tim54p and Tim22p are both essential for yeast cell viability, we suggest that the Tim54p–Tim22p complex plays a key role in the insertion of many, if not all, proteins into the IM.
Materials and Methods
Strains and Relevant Genotypes
MATa gal4 cyh2 trp1-901 leu2-3 leu2-112 URA3::GAL1::lacZ LYS2:: GAL1::HIS3 strain Y190 (Bai and Elledge, 1996), MATa/MATα leu2-Δ1/ leu2-Δ1 strain YPH501 (Sikorski and Hieter, 1989), MATa/MATα ura3-52/ura3-52 trp1-Δ1/trp1-Δ1 strain YPH857/858 and MATa ura3-53 trp1-Δ1 strain YPH857 (Spencer et al., 1993), MATa/MATα trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 strain FY833/834 and MATa trp1-Δ63 his3-Δ200 strain FY833 (Winston et al., 1995), MATa tim23-1 ura3-52 strain 574 (Ryan et al., 1994), MATα TIM23 strain 55 and MATα tim23-1 strain 201 (Emtage and Jensen, 1993), MATa tim23::URA3 leu2 strain 474 (Emtage and Jensen, 1993), and MATa tim17::TRP1 leu2 strain 463 (Ryan et al., 1994) have been described. MATa/MATα ade2/ade2 ade3/ade3 trp1/TRP1 ura3/ura3 leu2/leu2 strain 534 was constructed by crossing strains 4795-202 and 4795-408 (gifts from D. Koshland, Carnegie Institute of Washington, Baltimore, MD). Yeast transformations were performed as described (Schiestl and Geitz, 1989). Standard yeast media and genetic techniques (Rose et al., 1988) were used.
Isolation of TIM54 and TIM22
Tim54p was identified as a potential interactor with the Mmm1 protein (Burgess et al., 1994) using the two-hybrid screen (Fields and Song, 1989; Bai and Elledge, 1996). pOK52, a TRP1 plasmid which expresses amino acids 123–426 of Mmm1p fused to the Gal4 DNA-binding domain, was constructed by inserting a 2.6-kbp NheI–NcoI MMM1–containing fragment from pSB75 into the NheI–NcoI sites of pAS1-CYH2 (Bai and Elledge, 1996). pOK52 was transformed into yeast strain Y190 along with a library of yeast cDNAs fused to the Gal4 activation domain carried on the LEU2 plasmid pACT (Bai and Elledge, 1996). Y190 carries both the Escherichia coli lacZ and the yeast HIS3 genes under the control of the GAL1 promoter region (Bai and Elledge, 1996). 106 transformants were plated onto medium containing 50mM 3-amino-triazole (Sigma Chemical Co., St. Louis, MO) to select for HIS3 expression, and ∼10,000 colonies were subsequently screened for β-galactosidase activity. 10 His+, β-gal+ colonies were identified and the plasmid from one transformant (pACT-15) was shown to specifically interact with pOK52. For example, pACT-15 did not interact with other proteins fused to the Gal4 DNA-binding domain, such as p53, yeast Snf1, and rat lamin. pACT-15 contained a cDNA of 750 bp, which was excised with BglII and subcloned into the BamHI site of Bluescript SK II+ (Stratagene, La Jolla, CA) to form pOK49.
The yeast DNA insert in pOK49 was sequenced completely and shown to encode the carboxyl-terminal region of a novel protein. Database analysis using the BLAST program (Altschul et al., 1990) showed the open reading frame was located upstream of the PEP8 gene (Bachhawat et al., 1994). Plasmid p7171, which carries a 7-kbp SpeI fragment in pRS316 (a gift from E. Jones, Carnegie Mellon University, Pittsburg, PA), was used to isolate a 3.1-kbp ClaI fragment, which was blunt-end ligated into the EcoRV site of Bluescript SK II+, forming pOK21A. Complete DNA sequencing of the insert in pOK21A (and subclones from the insert) identified an open reading frame encoding a 54-kD protein.
We identified the TIM22 gene as an open reading frame (YDL217C) on chromosome IV (Jacq et al., 1997) encoding a protein homologous to Tim23p and Tim17p. TIM22 was isolated by PCR-amplifying a 1.4-kbp fragment from yeast genomic DNA using oligonucleotide No. 178 (5′-GATCGAGCTCGTTACAGAGAAATGTC-3′) and oligonucleotide No. 180 (5′-CAGTCTCTAACGCCTCG-3′). The PCR fragment was digested with EcoRI and inserted into the EcoRI site of either the CEN6-URA3 plasmid, pRS316 (Sikorski and Hieter, 1989), or the 2μ-URA3 plasmid, pRS426 (Sikorski and Hieter, 1989), forming pJH201 and pJH202, respectively.
Disruption of the TIM54 and TIM22 Genes
Two different disruptions of the TIM54 open reading frame were constructed. First, tim54::URA3 was produced by digesting pOK49 with StyI, which removes nucleotides 1156–1409 from the TIM54 open reading frame, and the DNA ends were filled in with DNA polymerase. A 1.1-kbp SmaI fragment containing the URA3 gene was isolated from plasmid pSM32 (a gift from S. Michaelis, Johns Hopkins School of Medicine, Baltimore, MD) and blunt-end ligated into the StyI-cut pOK49 vector to form pOK3. For gene disruptions in yeast, a 1.5-kbp XhoI fragment containing tim54::URA3 was isolated from pOK3 and transformed into ura3/ura3 diploid strain YPH857/858. Southern analysis of this diploid confirmed that one of the two copies of TIM54 had been replaced by tim54::URA3.
A second TIM54 disruption was constructed by inserting the LEU2 gene into the NcoI site located at position 230 in the TIM54 open reading frame. pOK21A, which carries a TIM54 on a 3.1-kbp ClaI fragment blunt-end ligated into the EcoRV site of Bluescript SK II+ (Stratagene), was digested with NcoI and the ends filled-in with DNA polymerase. A 2.2-kbp XhoI–SalI fragment carrying the LEU2 gene was inserted into NcoI-cut pOK21A, forming pOK28. tim54::LEU2, carried on a 4.4-kbp NotI–HindIII fragment, was isolated from pOK28 and used to replace one of two copies of TIM54 in MATa/MATα leu2/leu2 diploid strains YPH501 and 534.
A complete disruption of the TIM22 open reading frame was constructed as described (Lorenz et al., 1995). A DNA fragment was amplified from pRS304 (Sikorski and Hieter, 1989) using oligonucleotides No. 181 (5′-TAAGATCAAGAAATTGTGATTTTAAATACTTTATACGAAGCTGTGCGGTATTTCACACCG-3′) and No. 182 (5′-AAATATAAAACATTCATCGTTCGTCGAAATTGGCTATTCAAGATTGTA-CTGAGAGTGCAC-3′) using the PCR (Saiki et al., 1985). This DNA fragment contained 40 bp of TIM22 5′ untranslated sequences joined to one end of the TRP1 gene and 40 bp of TIM22 3′ untranslated sequences joined to the other end of TRP1. Homologous recombination between the PCR product and the yeast chromosome produces a precise replacement of the TIM22 ORF by TRP1. The PCR product was transformed into trp1/ trp1 diploid strain FY833/834, and Trp+ transformants were selected. In one transformant (strain 777), one of the two copies of the TIM22 open reading frame was shown to be replaced by TRP1. Meiotic analysis of transformed diploids yielded only two viable spores in each tetrad. Similar to the TIM54 disruptions, tim22::TRP1 spores germinated and underwent six to eight cell divisions before arresting in their growth as unbudded cells.
Construction of HA Epitope-tagged Versions of Tim54p and Tim22p
pOK100, which contains a unique NotI site immediately preceding the termination codon of TIM54, was constructed as follows. Using pOK49, oligonucleotide No. 123 (5′-CTTGCGGCCGCCAATATCTGATTCTGGCTC-3′), and oligonucleotide No. 98 (5′-AACAGCTATGACCATG-3′), we isolated a 530-bp DNA fragment using PCR, and digested the fragment with XhoI and NotI. A 954-bp NotI–BamHI fragment containing a unique NotI site preceding the TIM23 termination codon and the 3′- untranslated region of TIM23 was isolated from pJE7 (Emtage and Jensen, 1993). Both fragments were inserted into BamHI–SalI digested pUC18 (Yanisch-Perron et al., 1985) to form pOK100. pOK101, which encodes Tim54p with the hemagglutinin (HA) epitope at its carboxyl terminus, was constructed by inserting a 114-bp NotI fragment containing three tandem copies of the HA epitope (Field et al., 1988; Tyers et al., 1992; a gift from B. Futcher, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) into pOK100. Because pOK101 lacks complete TIM54 sequences, we reconstituted TIM54 by integration into the yeast chromosome. A 3.6-kbp PvuI fragment from pOK101, carrying the TIM54–HA fusion, was inserted into the URA3 integrating vector pRS306 (Sikorski and Hieter, 1989) to form pOK102. pOK102 was transformed into ura3/ ura3 diploid strain YPH857/858, and we targeted the integration to the chromosomal TIM54 gene by digesting pOK102 with HpaI. Stable Ura+ transformants were isolated and shown to express Tim54–HA. Integration of the TIM54–HA construct at the chromosomal TIM54 gene was confirmed by Southern analysis. Isolation of meiotic products from sporulated diploid transformants yielded haploid cells containing the integrated TIM54–HA, which grew on both glucose and glycerol-containing medium at 23°, 30°, and 37°C. Because integration of the pOK102 plasmid inactivates the chromosomal copy of the essential TIM54 gene, our results indicate that the Tim54–HA fusion protein is fully functional.
pJH301, a LEU2 plasmid that carries the full-length TIM54–HA gene, was constructed as follows. First, a 1.5-kbp EcoRI fragment carrying the carboxyl-terminal region of Tim54p fused to HA was isolated from pOK102 and inserted into EcoRI-digested pOK21B. pOK21B carries the full-length TIM54 gene on a 3.1-kbp ClaI fragment blunt-end ligated into the EcoRV site of Bluescript SK II (Stratagene). Replacement of the 1.66-kbp EcoRI fragment of pOK21B with the fragment from pOK102 generated pOK48, which carries TIM54–HA with 376 bp of upstream sequences. pJH301 was constructed by inserting a PvuII fragment containing TIM54–HA from pOK48 into PvuII-digested pRS315 (Sikorski and Hieter, 1989).
An HA epitope-tagged version of Tim22p was constructed as follows. First, a NotI site was inserted proximal to the TIM22 stop codon by PCR-amplifying a 1.1-kbp fragment from genomic yeast DNA using oligonucleotide No. 178 (5′-GATCGAGCTCGTTACAGAGAAATGTC-3′) and oligonucleotide No. 179 (5′-GGAATTCCGCGGCCGCATTCTTTAAAATCGTTTTG-3′). This fragment was digested with SacI and NotI and inserted into SacI–NotI digested pJE7 (Emtage and Jensen, 1993) creating pJH101. pJH101 thus carries the TIM22 gene with 505 bp of its upstream sequences, a unique NotI site preceding the TIM22 stop codon, and 940 bp of 3′ untranslated sequences from the TIM23 gene. pJH102, which encodes Tim22p with the HA epitope at its carboxyl terminus (Tim22–HA), was constructed by inserting a 114-bp NotI fragment containing three tandem copies of the HA epitope (Field et al., 1988) into the NotI site of pJH101. The Tim22-HA protein expressed from pJH102 provides Tim22p function, as it complements the tim22::TRP1 disruption.
Indirect Immunofluorescence
pOK27, which expresses Tim54–HA from the GAL1 promoter was constructed as follows. First, TIM54 was PCR-amplified using oligonucleotide No. 134 (5′-CGCCTCGAGGGCAAAGACATCATACC-3′), oligonucleotide No. 99 (5′-AATACGACTCACTATAG-3′) and pOK21A. The PCR product was digested with XhoI and PstI and inserted into XhoI– PstI-cut pRS314GU (Nigro et al., 1992), forming pOK25. An EcoRI fragment containing TIM54–HA was isolated from pOK102, and inserted into EcoRI-cut pOK25, forming pOK27. trp1 strain FY833 was transformed with pOK27, and cells were grown to an OD600 of 0.7 in synthetic medium containing 2% galactose. Cells fixed in 4% paraformaldehyde were converted to spheroplasts, attached to glass cover slips, and permeabilized as described (Harlow and Lane, 1988). Samples were incubated with undiluted culture supernatant from 12CA5 cells followed by a 1:250 dilution of antiserum to the β subunit of the F1-ATPase. A 1:250 dilution of secondary antibodies (Texas red-conjugated goat anti–rabbit IgG and fluorescein-linked goat anti–mouse IgG; Molecular Probes, Inc., Eugene, OR) were then added. Samples were observed using a Zeiss Axiovert microscope with a 100× objective. Fluorescense and differential interference contrast (DIC) images were captured with a Photometrics PXL CCD camera using IP Lab software (Signal Analytics Co., Vienna, VA).
Subcellular and Submitochondrial Fractionation
Yeast cells were grown to OD600 of approximately two in either YPD medium (Rose et al., 1988) supplemented with 2% lactate, or with synthetic complete medium containing 2% galactose and supplemented with the appropriate amino acids. Cells were converted to spheroplasts, homogenized, and separated into mitochondrial pellet and a postmitochondrial supernatant by centrifugation at 9,600 g for 10 min as described (Daum et al., 1982). Preparation of mitochondrial membrane vesicles, and the separation of outer membrane and IM by sucrose step-gradients were as described (Emtage and Jensen, 1993). To test whether Tim54p was an integral membrane protein, we added 0.1 M sodium carbonate, pH 11, 1.5 M sodium chloride, or 7 M urea to mitochondrial membrane vesicles, and separated the membrane fraction from the supernatant by centrifugation at 40 psi in a Beckmann airfuge for 30 min. For analysis, sample buffer (125 mM tris[hydroxymethyl]aminomethane-HCl, pH 6.8, 2% SDS, 20% glycerol) containing 4% β-mercaptoethanol was added to proteins, the proteins were separated by SDS-PAGE (Laemmli, 1970; Haid and Suissa, 1983), and then transferred (Haid and Suissa, 1983) to Immobilon filters (Millipore Corp., Waters Chromatography, Milford, MA). HA fusion proteins were identified by incubation of filters with a 1:10,000 dilution of mouse ascites fluid prepared using 12CA5 cells (Niman et al., 1983; BAbCO, Berkeley, CA). Marker proteins were identified by incubation with antisera to the following proteins: the β subunit of the F1-ATPase (a gift from M. Yaffe, University of California, San Diego, CA), subunit IV of cytochrome oxidase (Jensen and Yaffe, 1988), hexokinase (a gift from M. Yaffe), and OM45p (Yaffe et al., 1989). Immune complexes were visualized using HRP-conjugated secondary antibody (Amersham Corp., Arlington Heights, IL) followed by chemiluminescence (SuperSignal; Pierce, Rockford, IL). We quantified immune blots using the ImageQuant Software (Molecular Dynamics Inc., Sunnyvale, CA).
Isolation of the Temperature-sensitive Tim54-1 Mutant
tim54-1 was produced by mutagenesis of the cloned TIM54 gene. pOK22, which carries TIM54 gene on a 3.1-kbp ClaI fragment blunt-end ligated into the EcoRV site of the TRP1-CHY2–containing plasmid, pKS1 was passed through bacterial mutator strain XL1-RED (Stratagene) according to manufacturer's instructions. Mutagenized pOK22 was transformed into ade2 ade3 trp1 ura3 leu2 tim54::LEU2 strain 551, which also carries the TIM54-ADE3-URA3 plasmid, pOK32. tim54 mutations carried on pOK22 were identified by a plasmid-shuffle scheme (Sikorski and Boeke, 1991). Specifically, if the TIM54 gene carried on pOK22 was defective, then transformants were unable to lose the TIM54-ADE3-URA3 pOK32 plasmid (and the ade2 ADE3 cells accumulated a red pigment). If TIM54 on pOK22 is functional, then transformants could lose the pOK32 plasmid (and the ade2 ade3 cells remained white). From 37,400 total transformants, 175 completely red colonies were identified as unable to lose the pOK32 plasmid at 34°C. One transformant was found to be temperature sensitive for TIM54 activity: pOK32 could be lost at 24°C, but not at 34°C. The majority of the other transformants were defective in TIM54 at both temperatures. Plasmid DNA (called pOK24) was isolated from this colony and shown to confer temperature-sensitive growth when reintroduced into strain 551. Subcloning experiments showed that the mutation was located in the TIM54 gene. MATa trp1 tim54-1 strain 809, which carries a chromosomal version of tim54-1, was constructed as follows. trp1 tim54:: URA3 strain 835, carrying plasmid pOK24, was patched onto medium containing 5-fluoro-orotic acid (Boeke et al., 1984) to select for cells in which the chromosomal tim54::URA3 gene was replaced with the plasmid-borne tim54-1 sequences by homologous recombination.
pOK32 was constructed by inserting TIM54 carried on a 3.5-kbp PvuII fragment isolated from pOK21A into the 2μ-URA3 plasmid pRS426 (Sikorski and Hieter, 1989), forming pOK30. Subsequently, the ADE3 gene was isolated as a 4.8-kbp BamHI–SalI fragment from pDK255 (a gift from D. Koshland) and inserted into BamHI–SalI-cut pOK30 to form pOK32. Yeast strain 551 was constructed as follows. First, a disruption of TIM54 in diploid strain 534 was produced by transformation with a 4.4-kbp NotI–HindIII fragment carrying tim54::LEU2 from pOK28. A diploid carrying one copy of tim54::LEU2 was transformed with pOK32, sporulated, and MATa tim54::LEU2 strain 551 was identified among the meiotic progeny.
Imports into Isolated Mitochondria
Mitochondria were isolated from yeast strains as described (Daum et al., 1982), except that SEH buffer (250 mM sucrose, 1 mM EDTA, 20 mM Hepes-KOH, pH 7.4) was sometimes used in place of breaking buffer, and temperature-sensitive strains were grown at 24°C. For import reactions, mitochondria were suspended in import buffer (Scherer et al., 1992) to a final concentration of 1-mg/ml protein. 80–200-μg mitochondria were used, and 10–15 μl of lysate containing the radiolabeled protein was added to each reaction. Imports were stopped by transferring tubes to ice and adding valinomycin (Sigma Chemical Co.) to a final concentration of 40 μM. Indicated reactions were treated with proteinase K (Sigma Chemical Co.) on ice for 30 min, followed by the addition of 1 mM PMSF (Sigma Chemical Co.). The outer membrane of mitochondria were disrupted to form mitoplasts by resuspending mitochondrial pellets in 20 mM Hepes-KOH, pH 7.4, followed by incubation at 0°C for 20 min. After all manipulations, mitochondria and mitoplasts were pelleted by centrifugation at 12,500 g for 10 min, resuspended in 1× sample buffer containing 4% β-mercaptoethanol, and analyzed by SDS-PAGE. Radiolabeled proteins were detected by fluorography (Bonner and Laskey, 1974) or by using a phosphorimager (Molecular Dynamics, Inc.). To quantitate data, we used the Molecular Dynamics ImageQuant Software.
For transcription/translations, pSP6-TIM23 plasmid pJE29 was constructed by inserting the 1.7-kbp HpaI–BamHI fragment containing TIM23 from pJE2 (Emtage and Jensen, 1993) into SmaI–BamHI-digested pSP65 (Promega Corp., Madison, WI). pSP6-AAC1 was constructed by inserting the HindIII–BamHI fragment containing AAC1 from pT7-AAC1 (a gift from M. Douglas, Sigma Biotech, St. Louis, MO) into the HindIII–BamHI sites of pSP64 (Promega Corp.). pSP6-COX4 was obtained from D. Allison (University of Washington, Seattle, WA), and pGEM-Su9-DHFR, which expresses a fusion protein between residues 1–69 of Neurospora crassa ATPase subunit 9 and mouse dihydrofolate reductase (Pfanner et al., 1987), was obtained from F.-U. Hartl (Max-Planck Institute, Munich, Germany). We produced radiolabeled proteins from the SP6-containing plasmids using 1.5-mCi/ml [35S]methionine (1,000 Ci/ mmol; Amersham Corp.) in a coupled transcription/translation system (SP6 TNT™ System, Promega Biotech, Madison, WI) according to manufacturer's instructions.
Immune Precipitations from Detergent-solubilized Mitochondria
Mitochondria were isolated from strain 494 that expresses Tim54–HA from the integrated pOK102 construct, tim22::TRP1 strain 800 carrying TIM22-HA plasmid pJH102, or from wild-type strain D273-10b (Sherman, 1964) as described above. For immune precipitations, mitochondria were solubilized as described (Berthold et al., 1995). Briefly, we added 0.5 ml solubilization buffer (0.5% digitonin, 50 mM NaCl, 30 mM Hepes-KOH, pH 7.4, 1 mM PMSF), 1 μg/ml aprotinin (CalBiochem Corp., La Jolla, CA), 1 μg/ml leupeptin (CalBiochem Corp.), 1 μg/ml chymostatin (Sigma Chemical Co.), 1 μg/ml antipain (Sigma Chemical Co.), 1 μg/ml pepstatin A (Sigma Chemical Co.) to 500 μg mitochondria, and incubated the suspension at 4°C with gentle agitation for 10 min. After centrifugation at 12,500 g for 10 min, we added 100 μl of a 1:1 slurry of anti–HA-protein A–Sepharose beads in solubilization buffer to a 1-ml aliquot of the supernatant. Samples were incubated at 4°C with gentle agitation at least 5 h. HA-protein A–Sepharose was prepared using IgG from 12CA5 cells (Niman et al., 1983) and an ImmunoPureTM IgG orientation kit (Pierce) according to manufacturer's instructions. Alternatively, 20 μl of antiserum against Tim23p (Emtage and Jensen, 1993) was added to the supernatant, followed by 150 μl of a 1:1 slurry of protein A–Sepharose beads (Sigma Chemical Co.). Beads were collected by centrifugation. We added 160 μl of 4× sample buffer to each supernatant and heated the supernatants at 65°C for 5 min. Pellets containing the protein A–Sepharose were washed four times with 1.0-ml solubilization buffer, and the bound proteins were eluted by two sequential extractions with 80 μl of 1× sample buffer. 500 μl of solubilization buffer was added to the pellet samples followed by heating at 65°C for 5 min. After separation by SDS-PAGE, proteins were immune blotted with antibodies to the HA epitope, or with antiserum to Aac1p (a gift from M. Douglas), mt-Hsp70 (a gift from M. Cumsky, University of California, Irvine, CA), Tim44p (a gift from G. Schatz, Biocenter, Basel, Switzerland), Tim54p, Tim22p, Tim23p, or Tim17p (Ryan et al., 1998).
Miscellaneous
To raise antiserum to Tim54p, a BglII fragment encoding the carboxyl-terminal region of Tim54p from pACT-15 was inserted into BamHI-cut pGEX-3X (Pharmacia LKB Biotechnology Inc., Piscataway, NJ). The GST–Tim54p fusion protein was expressed in bacteria and crude protein homogenates were isolated as per manufacturer's instructions. Proteins were separated by SDS-PAGE, stained with Coomassie blue R-250, and the bands containing the fusion protein were excised. Gel slices were frozen in liquid nitrogen, ground in a mortar and pestle, and lyophilized. To raise antiserum to Tim22p, a peptide based on the carboxyl-terminal region of Tim22p (CDGRPPQNDFKE; The Protein/Peptide/DNA Facility, Johns Hopkins School of Medicine, Baltimore, MD) was coupled to keyhole limpet hemocyanin (Sigma Chemical Co.) using m-maleimidobenzoic acid-N-hydroxy-succinimide (Pierce) as the crosslinking agent (Doolittle, 1986). Injection of antigens into rabbits and collection of antiserum were performed by Covance, Inc. (Denver, PA). pTB1 and pTB2, multi-copy plasmids which carry TIM17 and TIM23, respectively, were isolated from 2μ-URA3 genomic library (a gift from P. Hieter) by Southern hybridization.
Results
TIM54 Encodes a Novel 54-kD Protein that Potentially Interacts with Mmm1p
Mmm1p is a mitochondrial outer membrane protein required for the maintenance of mitochondrial morphology (Burgess et al., 1994). When Mmm1p function is lost, such as in the temperature-sensitive mmm1 mutant, the normal elongated mitochondria collapse into large, spherical organelles. We found that the Mmm1 protein resides in the mitochondrial outer membrane, but its role in mediating mitochondrial shape is unknown. To further investigate Mmm1p function, we identified potential Mmm1p-interacting proteins using the yeast two-hybrid screen (Fields and Song, 1989; Bai and Elledge, 1996). In particular, we fused the bulk of the Mmm1 protein, residues 123–426, to the DNA-binding domain of the yeast Gal4p transcriptional activator. This plasmid construct was cotransformed into Y190 yeast cells along with a cDNA library containing random fusions of yeast genes to the Gal4 activation domain (Bai and Elledge, 1996). Y190 cells contain both the yeast HIS3 gene and the E.coli lacZ gene under the control of Gal4 (Bai and Elledge, 1996). cDNAs that encode Mmm1p-interacting proteins (and therefore bring the activation and DNA-binding domains of Gal4 together) were identified as His+ lacZ+ transformants. From 10 6 transformants, we isolated 10 potential Mmm1p-interactors. To determine if the interaction with Mmm1p was specific in any of the cotransformants, we tested each of the 10 plasmids carrying cDNA–Gal4 activation domain fusions for their interaction with other proteins besides Mmm1p (p53, lamin, yeast Snf1) fused to the Gal4-DNA binding domain. One plasmid, pACT-15, contained a yeast cDNA-activation domain fusion that interacted specifically with Mmm1p.
DNA sequencing showed that pACT-15 contained a cDNA encoding the carboxyl-terminal 141 amino acids of a novel protein. Complete sequencing of genomic DNA revealed an open reading frame of 1,437 bp, which encodes a protein of 54.2 kD. Subsequent to our work, the yeast genome project revealed that the TIM54 gene is located on chromosome X, and corresponds to the YJLO54w open reading frame (Galibert et al., 1996). As described below, we found that the encoded protein is located in the mitochondrial IM and plays a pivotal role in protein import. We have named the new gene TIM54 and the protein Tim54p, consistent with the new nomenclature (Pfanner et al., 1996). Comparison of TIM54 to DNA and protein sequences in available databases revealed a potential cognate in Candida albicans, but no other significant homologies. An internal region of Tim54p, amino acids 133–154, is predicted to have a high propensity to form a coiled coil (Lupas, 1996).
TIM54 Is an Essential Gene
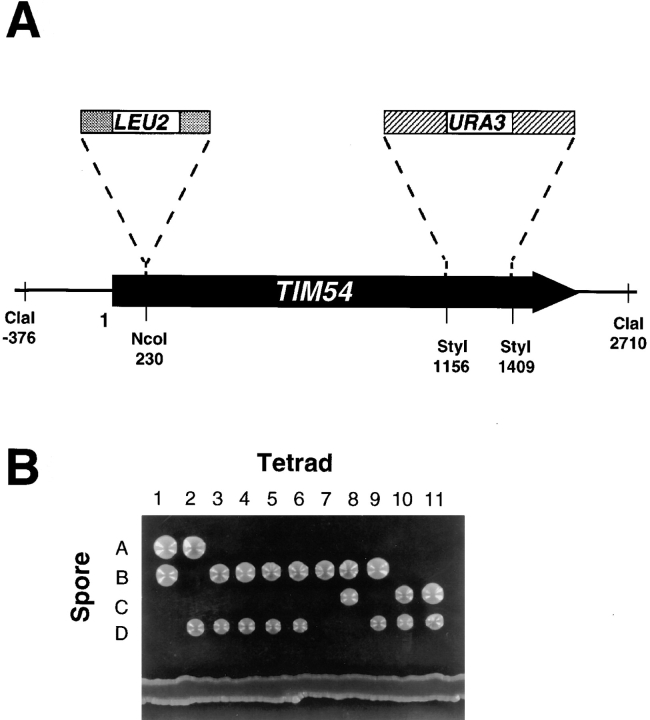
To investigate the function of the Tim54 protein, we constructed two disruptions of the TIM54 gene (Fig. 1 A). Briefly, we first inserted the yeast LEU2 gene into the NcoI site, which interrupts the TIM54 coding sequence after amino acid 77. We also inserted URA3 into the StyI sites of TIM54 (residues 386 and 470). We then transformed the tim54::LEU2 or tim54::URA3 constructs into diploid strains 410, 411, or 534, and stable Leu+ or Ura+ transformants were isolated. We sporulated these diploid cells and allowed the haploid progeny to grow at 24°, 30°, or 37°C. In more than 30 total tetrads, none gave rise to more than two viable spores, even after prolonged incubation (Fig. 1 B). All viable spores were shown to carry the wild-type TIM54 gene. TIM54 is thus an essential gene. Spores inferred to be tim54::LEU2 or tim54::URA3 germinated and underwent approximately eight cell divisions, and then arrested in their growth as unbudded cells. We consistently observed that tim54 spores formed microcolonies, never containing more than about 250 cells. This “delayed death” phenotype was similar to that seen in disruptions of the IM import protein, Tim22p (Holder, J., unpublished observations).
Figure 1.
TIM54 is an essential gene. (A) Restriction endonuclease map of the TIM54 gene. Relevant restriction sites in the cloned TIM54 gene, and their position in basepairs with respect to the amino terminus of the Tim54 protein (position 1) are shown. The arrow represents the TIM54 open reading frame, with the arrowhead indicating the 3′-end of TIM54. The location of the URA3 and LEU2 disruptions, which inactivate TIM54, are also shown. (B) Meiotic products from diploid strain 506, in which one of the two TIM54 genes was replaced by the tim54:: URA3 disruption, were separated by micromanipulation and allowed to grow at 30°C for ten days on YEP medium (Rose et al., 1988) containing 2% glucose. The dissection of eleven tetrads is shown, and the position where the four spores were initially placed is indicated (A–D).
Tim54p Is Located in the Mitochondrial IM, with its Carboxyl Terminus Facing the Intermembrane Space
To elucidate the function of Tim54p, we first identified its location within the yeast cell. We constructed an epitope-tagged version of Tim54p by inserting a segment from the influenza HA protein at the carboxyl terminus of Tim54p, and then integrated this construct into the yeast genome. The HA epitope is recognized by the monoclonal antibody 12CA5 (Niman et al., 1983). Cells expressing Tim54–HA contained a protein of ∼58 kD that reacted with the 12CA5 antibodies. The size of this protein was consistent with the addition of the 4-kD HA epitope to the 54-kD Tim54 protein. We also found that the Tim54–HA fusion protein was functional. Cells which only express Tim54–HA, and not wild-type Tim54p, grew on all media tested at 24°, 30°, or 37°C.
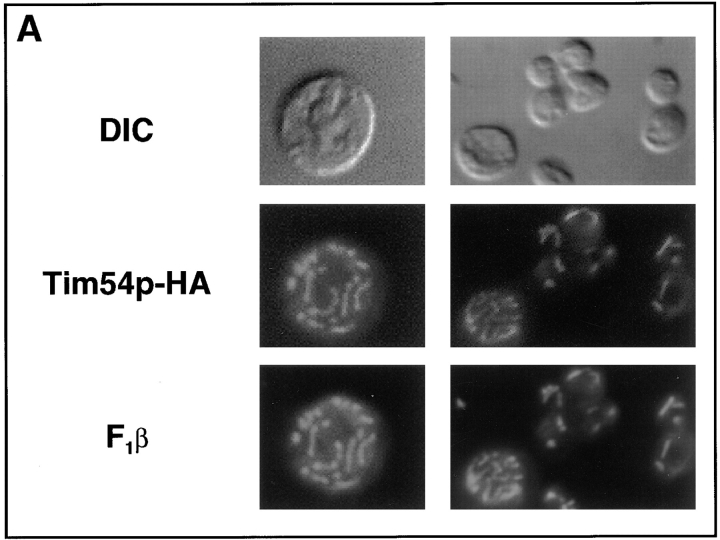
Immunofluorescence studies indicate that Tim54p is a mitochondrial protein (Fig. 2 A). Yeast cells containing plasmid pOK27, which expresses the Tim54–HA fusion protein, were fixed, permeabilized, and then incubated with antibodies to the HA-epitope and the mitochondrial ATPase β subunit (F1β) protein (Takeda et al., 1985). When immune complexes were visualized using fluorescence microscopy, we found that the Tim54–HA protein colocalized with the mitochondrial F1β protein. We also found in cell fractionation experiments that Tim54p was a mitochondrial protein (Fig. 2 B). Cells expressing the Tim54–HA fusion protein were homogenized, and separated into a mitochondrial fraction and a postmitochondrial supernatant. We found that Tim54p cofractionated with F1β, whereas little or no Tim54p was found in the supernatant along with the cytosolic hexokinase protein.
Figure 2.
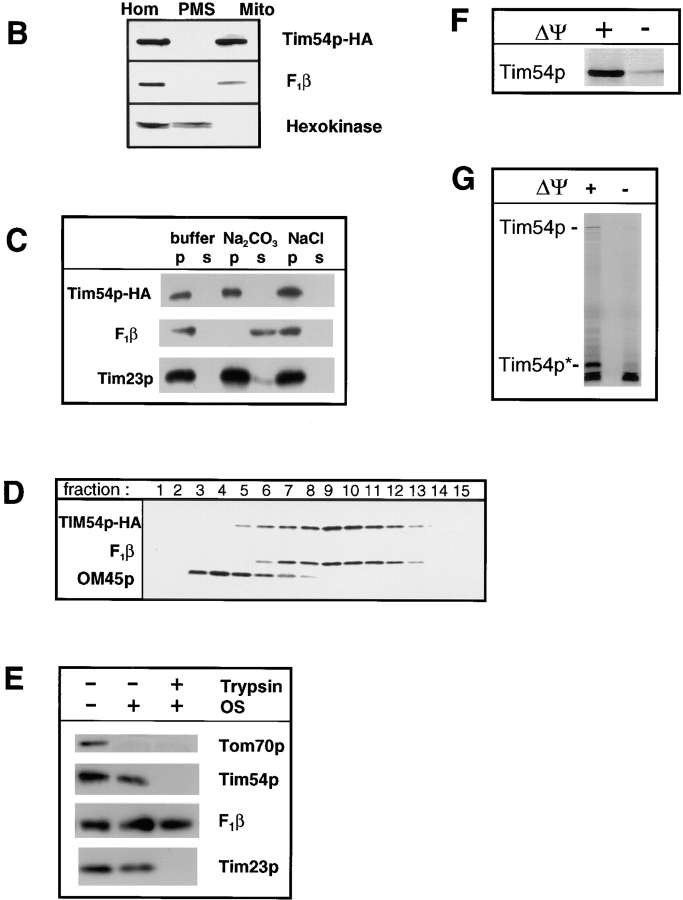
Tim54p is located in the mitochondrial inner membrane. (A) Tim54p colocalizes with mitochondria. Yeast strain 501 containing plasmid pOK27, which expresses the Tim54 protein tagged with the HA epitope (Tim54– HA), were fixed, permeabilized, and incubated with mouse antibodies to the HA epitope, or with rabbit antiserum to the β-subunit of the F1-ATPase (F1β). Cells were then incubated with Texas red–conjugated goat anti–rabbit IgG and fluorescein-linked goat anti–mouse IgG, and examined under the microscope at a magnification of 100. The three images in the left column show the same cell visualized by DIC illumination, or by fluorescence using the fluorescein (Tim54–HA) or rhodamine (F1β) channels. The three images in the right column show a group of eight cells examined by DIC and fluorescence. (B) Tim54p cofractionates with a mitochondrial marker. Strain 494, in which the integrated pOK102 construct expresses the Tim54–HA protein, were grown, converted to spheroplasts and homogenized. The homogenate (Hom) was separated into a mitochondrial pellet (Mito) and a postmitochondrial supernatant (PMS) by centrifugation. Aliquots of homogenate, mitochondria, and PMS representing equivalent numbers of cells were subjected to SDS-PAGE, blotted and proteins decorated with 12CA5 antibodies to the HA epitope (Tim54–HA), the β subunit of the F1-ATPase (F1β), or hexokinase. Immune complexes were visualized by chemiluminescence. (C) Tim54p is an integral membrane protein. 150-μg mitochondria isolated from strain 494 were sonicated, treated with either 0.1 M sodium carbonate, 1.5 M sodium chloride, or no additions (buffer), and centrifuged. Pellets (p) and supernatants (s) were analyzed by immune blotting with antibodies to the HA epitope (Tim54–HA), F1β, a peripheral membrane protein, and Tim23p, an integral membrane protein. (D) Tim54p is located in the inner membrane. Mitochondria from strain 494 were sonicated and the resulting vesicles were loaded onto sucrose step-gradients. After centrifugation, fractions were collected and analyzed by immune blotting with antibodies to the HA epitope (Tim54–HA), the inner membrane protein, F1β, or the outer membrane protein, OM45. Fraction 1 represents the top of the gradient. (E) The carboxyl terminus of Tim54 faces the intermembrane space. Mitochondria isolated from strain 494 were digested with 150 μg/ml trypsin for 20 min on ice, followed by the addition of 2 mg/ml soybean trypsin inhibitor. Mitochondria were reisolated by centrifugation and analyzed by immune blotting with antiserum to Tom70p, F1β, Tim23p, and Tim54p. To expose proteins located in the intermembrane space, the mitochondrial outer membrane was ruptured by osmotic shock (OS), and proteins were digested with 150 μg/ml trypsin as above. (F) Import of Tim54p into mitochondria requires an inner membrane potential. Mitochondria were isolated from wild-type strain D273-10B and incubated with the 35S-labeled Tim54 protein. In one sample, the inner membrane potential was dissipated (− Δψ) by the addition of 50 μM carbonyl cyanide m-chlorphenylhydrazone and 1 μM valinomycin prior to the import reaction. After 15 min at 30°C, imports were stopped by incubation at 0°C. Mitochondria were treated with 50-μg/ml proteinase K for 20 min at 0°C, isolated by centrifugation, and solubilized in SDS-sample buffer. Proteins were separated on SDS-polyacrylamide gels, and the radiolabeled Tim54p was identified by fluorography. (G) The bulk of the Tim54 protein faces the intermembrane space. Tim54p was imported into wild-type mitochondria in the presence (+ Δψ) or absence (− Δψ) of inner membrane potential as described above. After import, the mitochondrial outer membrane was disrupted by osmotic shock, and the resulting mitoplasts were digested with 50 μg/ml proteinase K for 20 min. Mitoplasts were recovered by centrifugation and subjected to SDS-PAGE and fluorography. The full-length Tim54 protein (Tim54p) and a fragment of Tim54p (Tim54p*) protected from protease digestion are indicated.
Tim54p could not be extracted from mitochondrial membranes following treatment with 1.5 M sodium chloride, 0.1 M sodium carbonate (Fig. 2 C), or 7 M urea (not shown), and is therefore an integral membrane protein. Hydropathy analysis (Kyte and Doolittle, 1982) suggests that Tim54p may contain one or two potential membrane-spanning segments (residues 37–54 and 358–386 in the Tim54 protein). To determine in which of the two mitochondrial membranes Tim54p resides, we isolated mitochondria from cells expressing the Tim54–HA protein and prepared membrane vesicles by sonication. As shown in Fig. 2 D, when the mitochondrial outer membrane vesicles were separated from IM vesicles on sucrose gradients, Tim54p cofractionated with the mitochondrial F1β protein, and not with the outer membrane OM45 protein (Yaffe et al., 1989). Tim54p, like several other IM proteins, does not appear to contain an amino-terminal, cleavable presequence typical of proteins that are imported into the mitochondrial matrix. Supporting this view, we found that Tim54p was imported into isolated mitochondria, and that its import required an IM potential (Fig. 2 F). We also observed no change in the molecular mass of Tim54p after its import. Thus, the mitochondrial targeting information of Tim54p appears to reside within the mature protein.
We found that the carboxyl terminus of the Tim54 protein faces the intermembrane space. Mitochondria were isolated from wild-type yeast strains and treated with trypsin and immune blots were probed with antiserum to Tom70p, Tim54p, F1β, and Tim23p. As shown in Fig. 2 E, in intact mitochondria, only the outer membrane Tom70 protein, which contains a large domain facing the cytosol (Hase et al., 1983), was removed by trypsin treatment. The IM proteins, Tim23p, F1β, and Tim54p, were not digested in intact mitochondria. When the mitochondrial outer membrane was disrupted by osmotic shock forming mitoplasts, both the Tim23 and Tim54 proteins were removed, while the matrix-localized F1β protein was not digested. Our Tim23p antibodies recognize the amino-terminal domain of the Tim23 protein (Ryan et al., 1998), which has been shown to face the intermembrane space (Bauer et al., 1996; Bömer et. al., 1997; Dekker et al., 1997; Hauke and Schatz, 1997; Emtage, J., O. Kerscher, and R.E. Jensen, manuscript submitted for publication). Since our antiserum to Tim54p was raised to the carboxyl-terminal third of the Tim54 protein, we conclude that the carboxyl terminus of Tim54p faces the intermembrane space. Supporting this view, we found that the bulk of the radiolabeled Tim54 protein that was imported into isolated mitochondria was located in the intermembrane space (Fig. 2 G). After import into energized mitochondria, trypsin digestion of mitoplasts yielded only a 5–6-kD fragment of Tim54p (Tim54p*). This protected fragment was not seen if the mitochondrial IM potential (Δψ) was dissipated prior to the import reaction.
Tim54p Is Required for the Insertion of Proteins into the IM, but Not for the Translocation of Proteins into the Matrix
All essential mitochondrial IM proteins to date have been shown to play a crucial role in protein import (Blom et al., 1993; Dekker et al., 1993; Emtage and Jensen, 1993; Maarse et al., 1994; Ryan et al., 1994). Because we found that Tim54p is an essential IM protein, we examined its potential role in import. To facilitate these studies, we first isolated a temperature-sensitive tim54 mutant by mutagenizing the cloned TIM54 gene. As described in Materials and Methods, we passed a plasmid carrying TIM54 through a bacterial strain defective in DNA repair (Greener and Callahan, 1994), and identified a temperature-sensitive tim54 mutant, tim54-1, using a plasmid-shuffle scheme (Sikorski and Boeke, 1991). Cells that contain the tim54-1 mutation as the sole source of TIM54 grew at normal rates at 24°C, but failed to grow at 35 or 37°C. We subsequently found that mitochondria isolated from the tim54-1 mutant grown at 24°C contained reduced levels of the altered Tim54 protein (see Fig. 6).
Figure 6.
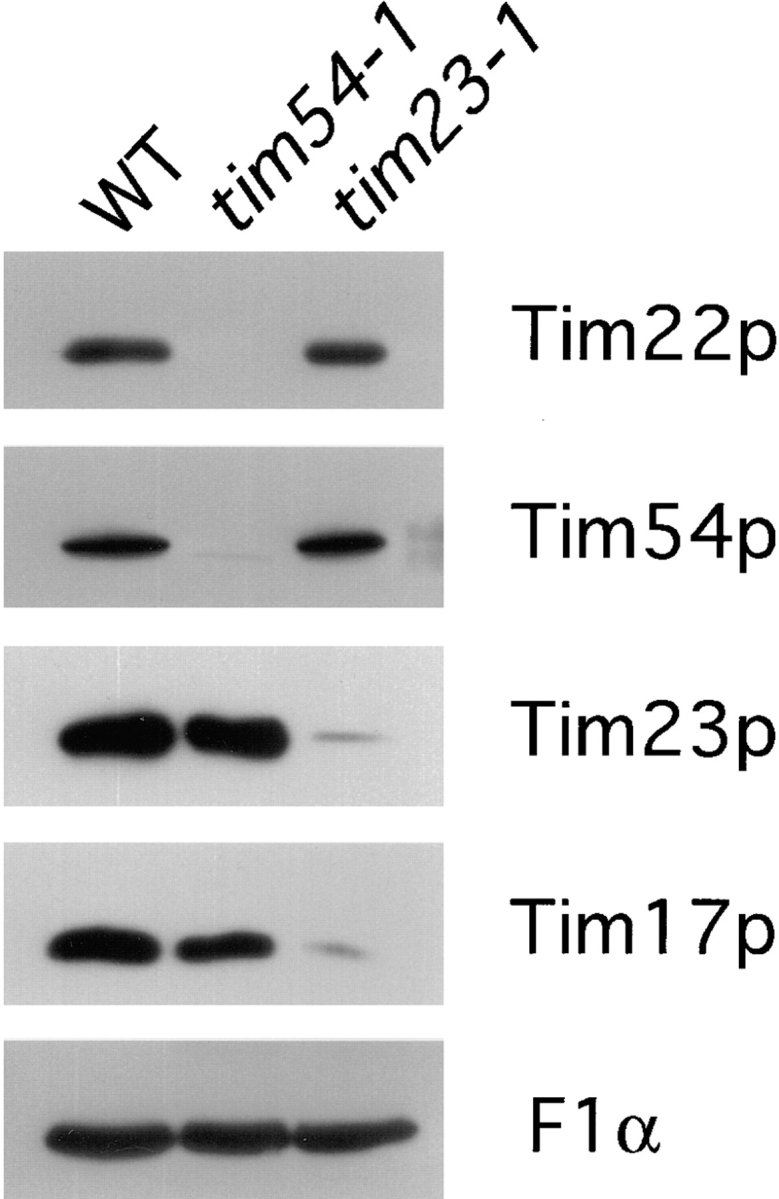
Mitochondria isolated from the tim54-1 mutant contain reduced amounts of both Tim54p and Tim22p. Mitochondria were isolated from wild-type strain 836, tim54-1 strain 835 and tim23-1 strain 201 grown at 24°C. 50 μg of mitochondria were analyzed by SDS-PAGE and immune blotting with antibodies to the α subunit of the F1-ATPase (F1α), Tim54p, Tim23p, Tim22p, Tim17p.
tim54-1 mutants are defective in the insertion of proteins into the mitochondrial inner membrane, but not in the translocation of proteins across the IM into the matrix. We isolated mitochondria from wild-type cells and the tim54-1 mutant, and examined their ability to import 35S-labeled precursor proteins in vitro. As shown in Fig. 3 A, we first examined the import of two proteins: Su9-DHFR, a fusion protein consisting of the amino-terminal mitochondrial targeting sequence of the matrix-localized N. crassa ATPase subunit 9 protein fused to mouse dihydrofolate reductase (Pfanner et al., 1987), and Aac1p, the yeast ATP/ADP carrier protein which resides in the mitochondrial IM (Lawson and Douglas, 1988). We found that Su9-DHFR was efficiently imported and processed to the mature form in mitochondria isolated from both wild-type and tim54-1 cells. This import of Su9-DHFR into tim54-1 mitochondria appeared to be complete. Sonication of wild-type and tim54-1 mitochondria after Su9-DHFR import liberated similar amounts of the mature form of Su9-DHFR into a supernatant fraction (see Fig. 4 B). In contrast to the matrix-localized Su9-DHFR protein, the import of IM-destined Aac1 protein into tim54-1 mitochondria was reduced at least fivefold relative to wild-type mitochondria. As shown in Fig. 3 C, we also found that tim54-1 mitochondria were defective in the import of another inner membrane protein, Tim23p. The import of Tim23p into the IM can be assessed by protease digestion: a 14-kD fragment of Tim23p (Tim23p*), which is resistant to protease digestion when the outer membrane is disrupted, is diagnostic of its correct insertion into the IM (Bömer et al., 1997; Dekker et al., 1997; Hauke and Schatz, 1997; Emtage et al., manuscript submitted for publication). We found that the rate of insertion of Tim23p into the IM, as measured by the appearance of Tim23p*) was at least 40% reduced in tim54-1 mitochondria as compared to wild-type. The import of Tim23p*, was dependent upon inner membrane potential (Δψ): the 14-kD Tim23p fragment protected from protease digestion was not seen in either wild-type or tim54-1 imports when the inner membrane potential was dissipated by valinomycin.
Figure 3.
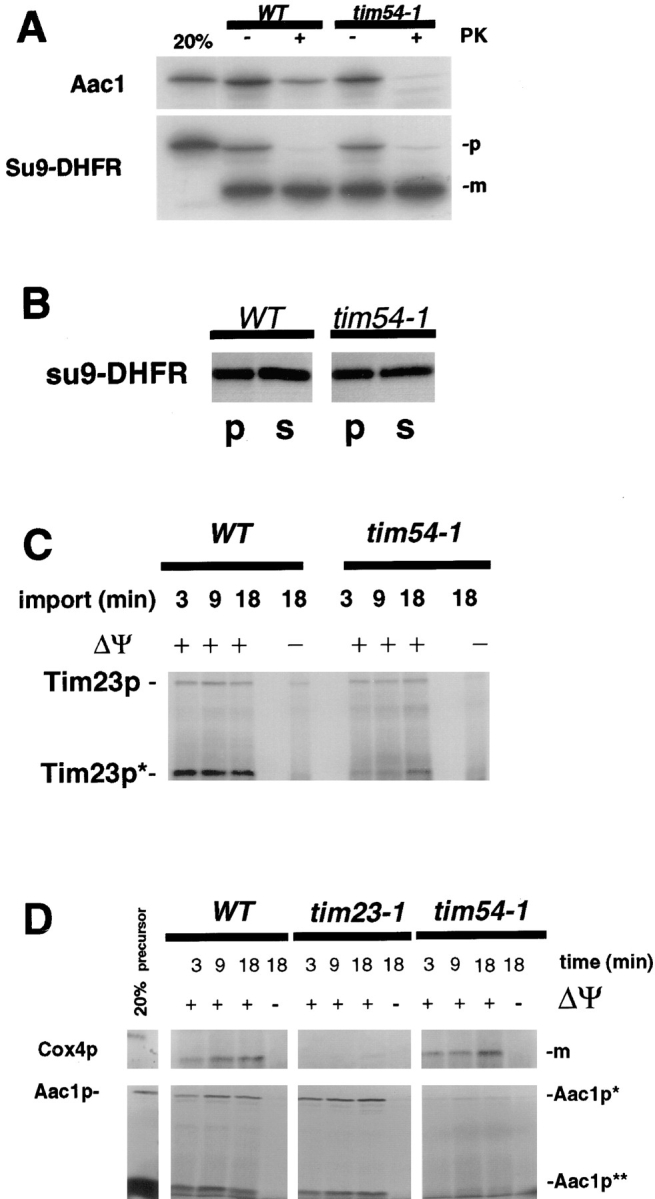
Mitochondria isolated from the tim54-1 mutant is defective in the insertion of proteins into the inner membrane, but not in the import of matrix proteins. (A) Su9-DHFR and Aac1p imports: Mitochondria were isolated from tim54-1 stain 809 and wild-type strain YPH857, and incubated with the 35S-labeled Aac1p or Su9-DHFR proteins. After 20 min at 30°C, import was stopped by the addition of 40 μM valinomycin and incubation at 0°C. An aliquot of the mitochondria was treated with 200 μg/ml proteinase K for 20 min at 0°C (+ PK). Samples were isolated by centrifugation, and pellets were solubilized in SDS-sample buffer. Proteins were separated on SDS-polyacrylamide gels, and the radiolabeled proteins were identified by fluorography. Precursor (p) and mature (m) forms of the Su9-DHFR are indicated. 20% of the precursor added to each import reaction is also shown. (B) Su9-DHFR is imported into the matrix in tim54-1 mitochondria. Mitochondria were isolated from tim54-1 stain 809 and wild-type strain YPH857, incubated with the 35S-labeled Su9-DHFR protein, and treated with proteinase K as described above. Mitochondria were sonicated and then centrifuged at 200,000 g for 45 min. Equal aliquots of the pellet (p) and supernatant (s) fractions were subjected to SDS-PAGE and fluorography. The mature form of Su9-DHFR is shown. (C) Tim23p imports: 35S-labeled Tim23 protein was imported into either wild-type or tim54-1 mitochondria for 3, 9, or 18 min at 30°C. Import was stopped by the addition of valinomycin, and mitochondria were treated with 200 μg/ml trypsin for 20 min at 0°C. After the addition of 1 mg/ml soybean trypsin inhibitor and centrifugation, the outer membrane of the mitochondria was disrupted by resuspending the mitochondrial pellet in 20 mM Hepes-KOH; pH 7.4, and incubation for 20 min at 0°C. Proteins were digested by treatment with 100 μg/ml proteinase K for 20 min 0°C. Samples were then isolated by centrifugation, and analyzed by SDS-PAGE and fluorography. Tim23* indicates the 14-kD protease-protected fragment of Tim23p indicative of correct insertion into the inner membrane. In some reactions, the inner membrane potential (Δψ) was dissipated prior to import by the addition of 40 μM valinomycin. (D) Import of Aac1p and Cox4p into wild-type, tim23-1, and tim54-1 mitochondria: Mitochondria were isolated from wild-type strain 55, tim23-1 strain 201, and tim54-1 strain 809 and incubated with the 35S-labeled Aac1 or Cox4 proteins for the indicated times at 30°C. After import, mitochondria were reisolated by centrifugation, converted to mitoplasts, and treated with protease as described above.
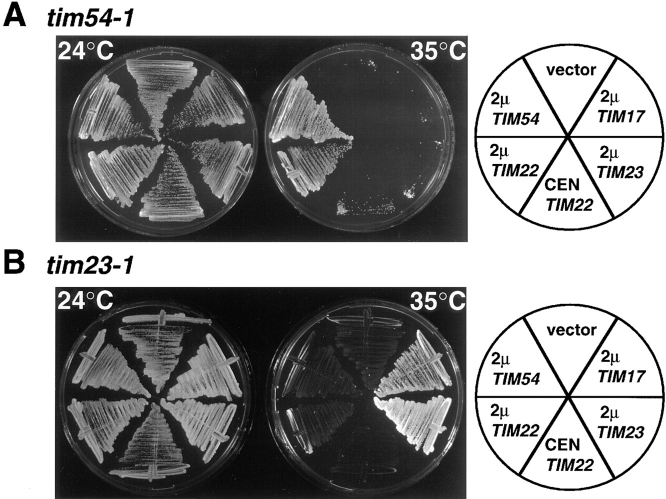
Figure 4.
Multiple copies of TIM22 suppress tim54-1, but not tim23-1. (A) tim54-1 ura3 strain 723 was transformed with the following multi-copy, URA3-containing plasmids: 2μ-TIM17 plasmid pTB1, 2μ-TIM23 plasmid pTB2, 2μ-TIM22 plasmid pJH202, 2μ-TIM54 plasmid pOK32, or the empty vector pRS426 (Sikorski and Hieter, 1989). tim54-1 was also transformed with CEN- TIM22 plasmid pJH201, which carries TIM22 on a centromere-containing plasmid. Ura+ transformants were streaked onto YEP medium containing 2% glycerol and ethanol, and incubated at 24° or 35°C for 5 d. (B) tim23-1 ura3 strain 574 was transformed with the same set of plasmids described above. Transformants were streaked onto YPglycerol/ethanol medium and incubated at 24 or 35°C for 5 d.
We previously showed that Tim23p is essential for the import of proteins into the matrix (Emtage and Jensen, 1993). To determine if Tim23p also played a role in IM protein import, we isolated mitochondria from the temperature-sensitive tim23-1 mutant, and compared their ability to import different precursor proteins to mitochondria isolated from wild-type and tim54-1 strains. Consistent with our earlier results, we found that the matrix-localized Cox4 precursor was efficiently imported and processed to the mature form in wild-type mitochondria, but was imported at greatly reduced rates into tim23-1 mitochondria (Fig. 3 D). The tim23-1 mutation had no effect on the import of the Aac1 protein: Aac1p was imported into wild-type and tim23-1 mitochondria at virtually identical rates. We found that the imported Aac1p was correctly inserted into the inner membrane. Import of Aac1p was dependent upon the inner membrane potential, and protease treatment of mitoplasts after import yielding characteristic protected fragments of Aac1p (Aac1p* and Aac1p**; Sirrenberg et al., 1996; Dekker et al., 1997). In contrast to the inner membrane Aac1 protein, tim54-1 mitochondria imported Cox4p into the matrix at a rate similar to wild-type mitochondria (Fig. 3 D). We found that Cox4p was completely translocated across the IM into the matrix of tim54-1 mitochondria. Similar amounts of the mature form of Cox4p were protected from protease digestion after disruption of the mitochondrial outer membrane (Fig. 3 D). Our results thus suggest that there are two separate import pathways in the mitochondrial inner membrane. Tim23p mediates the translocation of precursors into the matrix, whereas Tim54p is required for the insertion of at least some proteins into the IM, including at least one inner membrane component (Tim23p) of the alternative pathway.
TIM54 and TIM22 Genetically Interact with Each Other, but Not with TIM23 or TIM17
To further explore the role of Tim54p in the import of inner membrane proteins, we looked for genetic interactions between TIM54 and other IM import components. In particular, we examined the ability of TIM23, TIM17, and TIM22 to suppress the temperature-sensitive tim54-1 mutant. We transformed tim54-1 ura3 strain 723 with URA3-2μ–containing plasmids carrying either TIM54, TIM23, TIM22, TIM17, or an empty vector. We then grew these strains at either 24 or 35°C (Fig. 4 A). Plasmids carrying the 2μ origin of replication are present in multiple copies, resulting in overexpression of genes carried on the plasmid (Armstrong et al., 1989). All transformants grew equally well at 24°C, but only cells that contained either TIM54 or the 2μ-TIM22 plasmid, were able to grow at 35°C. Thus, multiple copies of TIM22 can suppress the growth defect of the tim54-1 mutant. TIM22 present on a low-copy, centromere-containing plasmid (CEN-TIM22), only weakly suppressed the tim54-1 growth defect. Multiple copies of TIM23, TIM17, or an empty vector failed to suppress the tim54-1 defect and did not grow at 34°C.
For comparison, we transformed the temperature sensitive tim23-1 ura3 strain 574 with URA3-2μ–containing plasmids carrying either TIM54, TIM23, TIM22, TIM17, or empty vector (Fig. 4 B). Although all transformants grew at 24°C, cells that contained either TIM23 or 2μ- TIM17 also grew at 35°C. Consistent with our previous results (Ryan et al., 1994), increased levels of TIM17 suppressed the growth defect of the tim23-1 mutant. TIM54 and TIM22, in contrast, did not allow tim23-1 cells to grow at 35°C. Thus, there is a genetic interplay between TIM54 and TIM22 distinct from that between TIM23 and TIM17. Our observations suggested the hypothesis that there are two separate IM import complexes, one containing Tim54p and Tim22p, and the other with Tim23p and Tim17p.
Tim54p and Tim22p Are Part of a Protein Complex Separate from the Tim23p–Tim17p Complex
To test the possibility that the Tim54 and Tim22 proteins interact with each other, but are not part of the Tim23p– Tim17p complex, we asked whether Tim54p and Tim22p could be coimmune precipitated from detergent-solubilized mitochondria. We isolated mitochondria from cells expressing the Tim54–HA protein as the sole source of Tim54p, and solubilized the mitochondria in buffer containing 0.5% digitonin. We then immune precipitated Tim54–HA using antibodies to the HA epitope and analyzed the pellet and supernatant fractions by immune blotting. As shown in Fig. 5 A, all of the Tim54–HA protein was found in the pellet fraction, and little or no Tim54–HA was seen in the supernatant. When we examined our fractions with antibodies to Tim22p, we found that all of the Tim22 protein coprecipitated along with Tim54–HA, indicating that all of the Tim22p in the mitochondria is associated with the Tim54 protein. Other IM proteins not involved in import, such as the ATP/ADP carrier protein Aac1p (Fig. 5 A), or the F1β and Cox4 proteins (not shown), did not precipitate with Tim54–HA. Thus the interaction of Tim54p and Tim22p was specific. Furthermore, proteins previously shown to be part of an IM complex mediating the translocation of proteins into the mitochondrial matrix did not coprecipitate with Tim54–HA: Tim23p, Tim17p, Tim44p, and mt-Hsp70 all remained in the supernatant fraction after Tim54–HA precipitation.
Figure 5.
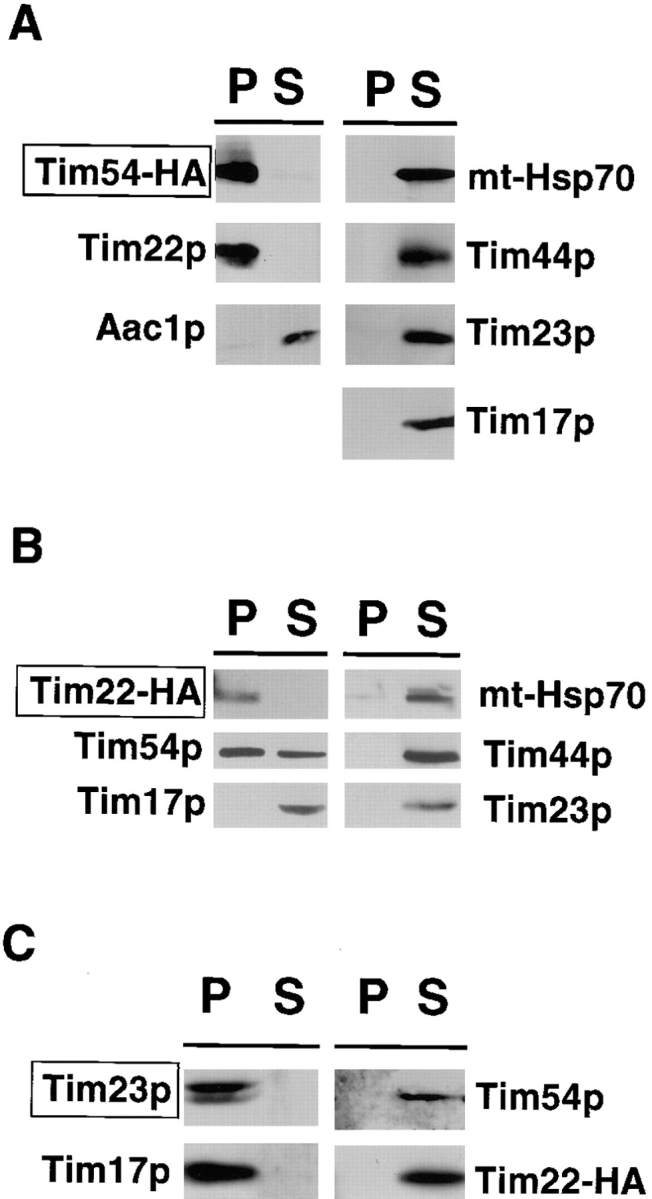
Tim54p and Tim22p physically interact, but are not part of the Tim23p– Tim17p complex. Mitochondria were isolated from strain 494, in which the integrated pOK102 construct expresses the Tim54–HA protein (A) or tim22::TRP1 strain 800, which expresses Tim22–HA from plasmid pJH102 (B and C), and solubilized in 0.5% digitonin. Extracts were immune precipitated with antibodies against the HA epitope (A and B), or antiserum to the Tim23 protein (C). Immunoprecipitates (P) and supernatants (S) were analyzed by SDS-PAGE and duplicate samples were immune blotted with antibodies to the HA epitope, Tim22p, Aac1p, Tim23p, Tim17p, Tim44p, or mt-Hsp70. A box in the upper left of each figure highlights the protein that was directly immune precipitated in each experiment.
When we immune precipitated the Tim22 protein, we found that part, but not all of the Tim54 protein, associated with Tim22p (Fig. 5 B). We isolated mitochondria from cells expressing Tim22–HA, solubilized the mitochondria in digitonin-containing buffer, and precipitated the Tim22–HA protein using anti–HA antibodies. Immune blots showed that while all of the Tim22–HA could be precipitated, only about half of Tim54p was in the pellet and the other half was present in the supernatant. Similar to our precipitations using Tim54–HA, the Tim23, Tim17, Tim44, and mt-Hsp70 proteins were found only in the supernatant fraction after Tim22–HA precipitation. Thus, neither Tim54p nor Tim22p interact with the Tim23p/Tim17p complex using our assay conditions.
As a control, mitochondria isolated from cells expressing Tim22–HA were solubilized and immune precipitated with antiserum to the Tim23 protein (Fig. 5 C). Consistent with previous studies we found a tight association between Tim23p and Tim17p (Berthold et al., 1995; Blom et al., 1995; Ryan et al., 1998): both Tim23p and Tim17p were found in the pellet fraction after Tim23p immune precipitation. In contrast, all of the Tim54 and Tim22 proteins were found in the supernatant fraction and thus did not associate with Tim23p.
When we precipitated Tim54–HA, all of the Tim22 protein coprecipitated (Fig. 5 A), but precipitation of Tim22–HA, on the other hand, brings down only half of the Tim54 protein (Fig. 5 B). We conclude that not all of the Tim54 protein in the inner membrane is interacting with Tim22p. We suggest that there may be two pools of Tim54p, one interacting with Tim22p, the other playing an unknown function. Supporting this possibility, we found that the Tim54–HA protein was approximately two to three times more abundant than Tim22–HA when both proteins were expressed from identical centromere-containing plasmids (Holder, J., and R. Jensen, unpublished observations).
Mitochondria Isolated from the tim54-1 Mutant Contain Reduced Amounts of the Tim54 and Tim22 Proteins, but Normal Levels of Tim23p and Tim17p
Mutations in one subunit of a multi-subunit complex often destabilize the complex and cause the rapid turnover of other subunits (Dowhan et al., 1985; Fang and Green, 1994; Shani et al., 1996). Consequently, if Tim54p and Tim22p are part of the same IM complex then tim54-1 may have an effect on the steady-state level of the Tim22 protein. To test this possibility, we isolated mitochondria from wild-type cells and the tim54-1 mutant grown at the permissive temperature, 24°C, and examined the level of several mitochondrial proteins by immune blotting. As shown in Fig. 6, the α-subunit of the F1-ATPase, Tim17p, and Tim23p were present in similar amounts in wild-type and tim54-1 mitochondria. In contrast, the levels of the Tim54 and Tim22 proteins were at least 10-fold reduced in tim54-1 mitochondria as compared to wild-type. The tim54-1 mutation thus appears to destabilize the Tim54p–Tim22p complex and leads to the turnover of the Tim22 protein in yeast cells. Since tim54-1 has no effect on the steady-state levels of the Tim23 and Tim17 proteins, our results argue that Tim23p and Tim17p are not part of the Tim54p– Tim22p complex. Consistent with this idea, we find that mitochondria isolated from tim23-1 mutants have lower levels of the Tim23 and Tim17 proteins, but normal levels of Tim54p and Tim22p (Fig. 6). Similar amounts of the outer membrane protein, Tom70, and the inner membrane protein, Aac1p, were found in all three mitochondria (not shown).
Discussion
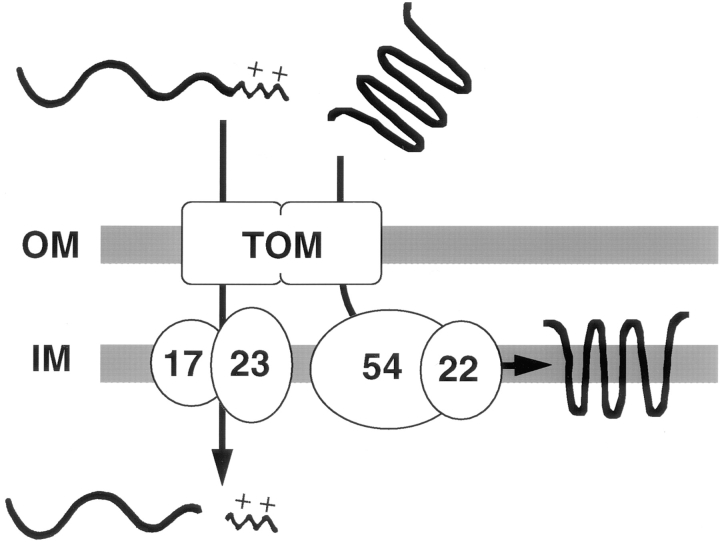
We have identified a new mitochondrial protein, Tim54p, which is required for the insertion of proteins into the inner membrane, but not for the translocation of proteins across the IM into the matrix. When mitochondria were isolated from the temperature-sensitive tim54-1 mutant, they efficiently translocated precursor proteins across the inner membrane, but were defective in the insertion of two inner membrane proteins, Aac1p and Tim23p. In contrast, tim23-1 mitochondria failed to import precursor proteins into the matrix, but efficiently inserted Aac1p and Tim23p into the IM. Our surprising results suggest that there are two distinct import pathways in the inner membrane: a Tim54p-dependent pathway for inserting membrane proteins, and a Tim23p-dependent pathway for translocating precursors across the IM into the matrix (Fig. 7).
Figure 7.
A model diagramming the role of two import complexes in the mitochondrial inner membrane. The pathway of two imported proteins is shown. Precursor proteins destined for the matrix carry a positively charged, amino-terminal presequence (indicated by the wavy line with + +), which is removed by the matrix-localized processing protease. After their import through the outer membrane machinery (TOM complex), these precursors are translocated across the IM through the Tim23p–Tim17p complex. Polytopic IM proteins carry internal targeting information. After their translocation through the outer membrane, the Tim54p–Tim22p complex mediates their insertion into the IM.
The role of Tim54p appears similar to that of Tim22p, which has recently been shown to mediate the IM import of the Aac1p and PiC carrier proteins (Sirrenberg et al., 1996). However, in addition to Aac1p, tim54-1 mitochondria are defective in the import of the Tim23 protein. In preliminary studies, we have also found that the import of the Tim22 protein is defective in tim54-1 mitochondria (Kerscher, O., and R. Jensen, manuscript in preparation). Because Tim23p and Tim22p are not obvious members of the carrier family, our results raise the possibility that Tim54p and Tim22p may be required for the insertion of many, if not all, inner membrane proteins. Supporting this idea, we find that both TIM54 and TIM22 are essential genes. The Aac1p and PiC proteins, on the other hand, are not essential for yeast cell viability, but are only required for growth on nonfermentable medium (Lawson and Douglas, 1988; Zara et al., 1991). It is therefore likely that both Tim54p and Tim22p mediate the import of essential proteins, including the Tim23p and Tim22p import components. To determine the complete repertoire of substrates for Tim54p and Tim22p, we are currently examining the import of additional inner membrane proteins, as well as proteins sorted to other mitochondrial locations.
Several studies support our results indicating that Tim23p is not required for the insertion of at least some inner membrane proteins. For example, Dekker et al. (1997) recently found that mitochondria isolated from a different tim23 mutant, tim23-2, were not defective in the insertion of Aac1p, Tim23p, or Tim17p into the inner membrane, but were defective in the import of several precursors into the matrix. Dekker et al. (1997) also found that chemical amounts of a matrix-destined precursor blocked the import of additional matrix proteins, but not the import of the inner membrane Tim23 and Aac1 proteins. Similarly, while we showed that Tim23p was required for the import of several different matrix proteins (Emtage and Jensen, 1993), we have recently found that the tim23-1 mutation and antibodies to Tim23p do not block the insertion of the Aac1, Tim23, or Tim17 proteins into the IM (Emtage, J., O. Kerscher, R.E. Jensen, manuscript submitted for publication). In contrast to the above studies, Volker and Schatz (1997) have reconstituted the protein insertion machinery of the mitochondrial inner membrane, and find that Tim23p is required for the insertion of Aac1p and Tim23p into the IM. Why the reconstituted system apparently differs from studies with intact mitochondria awaits further analyses.
We find that Tim54p and Tim22p physically interact in the inner membrane, but that Tim54p and Tim22p are not part of the previously-characterized Tim23p–Tim17p complex. Tim22p can be coprecipitated from detergent-solubilized mitochondria along with Tim54p, but the Tim23 and Tim17 proteins do not associate with Tim54p. In addition, Tim44p and mt-Hsp70, two members of the Tim23p– Tim17p complex (Berthold et al., 1995; Blom et al., 1995; Bömer et al., 1997; Ryan et al., 1998), fail to interact with either Tim54p or Tim22p. This observation may not be surprising since Aac1p and Tim23p, both imported via the Tim54p-dependent pathway, do not require mt-Hsp70 function for their import (Bömer et al., 1997; Emtage, J., O. Kerscher, R.E. Jensen, manuscript submitted for publication). Our genetic studies support the possibility that Tim54p and Tim22p are part of a complex separate from Tim23p–Tim17p. Multiple copies of the TIM22 gene, but not TIM23 or TIM17, suppress the growth defect of a tim54-1 strain. Moreover, TIM22 and TIM54 do not suppress the tim23-1 mutant. Our observations are also strengthened by recent findings that TIM22 is one of the genes identified in a library screen for multi-copy suppressors of the tim54-1 mutant (Leung, R., and R. Jensen, unpublished observations).
Our immune precipitation results suggest that there may be two populations of the Tim54 protein. When we precipitate Tim54p, all of the Tim22 protein coprecipitates. Precipitating Tim22p brings down only about half of the Tim54 protein. Furthermore, in preliminary experiments, the Tim54 protein in mitochondria appears to be present in two to threefold greater amounts than the Tim22 protein (Holder, J., and R. Jensen, unpublished observations). We speculate that there may be two Tim54p-containing complexes in the inner membrane: one containing Tim22p and the other containing unknown components. Tim54p may be similar to Tim23p, which is also found in two sub-complexes (Bömer et al., 1997). One Tim23p-containing subcomplex contains Tim17p, Tim44p, and mt-Hsp70; the other contains Tim17p and mt-Hsp70. Alternatively, the population of Tim54p that does not interact with Tim22p may represent an unassembled pool of Tim54p in the inner membrane. Thus, why Tim54p is present in more than one location awaits further studies.
If separate import pathways exist in the inner membrane, specific signals must then direct proteins to a given pathway. Virtually all matrix-localized proteins carry amino- terminal presequences, which target the protein to receptors on the mitochondrial surface. We propose that the presequence also directs the protein to the Tim23p–Tim17p machinery. For example, we found that the addition of the Cox4 presequence to the amino terminus of Tim23p, caused the mislocalization of Tim23p to the matrix (Emtage et al., manuscript submitted for publication). Because the import of Cox4-Tim23p was defective in tim23-1 mitochondria, we concluded that Cox4-Tim23p was imported via the Tim23p–Tim17p pathway, instead of Tim23p's normal route via the Tim54p–Tim22p pathway. In similar studies, the addition of a presequence to another IM protein, the mammalian uncoupling protein, UCP, caused its mislocalization to the matrix (Liu et al., 1990). Whereas the presequence appears to target proteins to the Tim23p– Tim17p complex, it is not clear what signal directs membrane proteins to the Tim54p–Tim22p pathway. Inner membrane proteins, such as Aac1p and Tim23p, do not contain amino-terminal presequences, and their import signal has not yet been identified.
Interestingly, when Tim23p and UCP were mislocalized to the matrix by the addition of a presequence, neither protein was inserted into the membrane and both could be readily extracted by alkali (Liu et al., 1990; Emtage, J., manuscript submitted for publication). Mislocalization to the Tim23p–Tim17p pathway prevents membrane insertion, even though the Tim23 and UCP proteins contain multiple hydrophobic transmembrane segments. It is possible that the Tim23p–Tim17p machinery does not have the capacity to recognize the membrane-spanning segments in at least some proteins, and instead behaves like a passive channel (Ungermann et al., 1994; Berthold et al., 1995). Only when Tim23p and UCP are imported through their normal route, the Tim54p–Tim22p pathway, are they correctly inserted into the bilayer.
Why was Tim54p, an inner membrane import protein, identified as a potential protein–protein interactor with Mmm1p, an outer membrane protein required to maintain normal mitochondrial shape (Burgess et al., 1994)? Since the inner membrane appears to contain separate machinery for the import of IM proteins, it is possible that the outer membrane similarly has distinct import machinery for different proteins. Mmm1p may function in a specific import pathway for IM proteins (which includes the Tim54 protein), and may transiently interact with these proteins during their import. We, however, consider this possibility unlikely since we have not been able to detect an import defect in mmm1 mutants or mmm1 mitochondria (Srinivasan, M., S. Burgess, and R. Jensen, manuscript in preparation). Furthermore, IM proteins and matrix proteins use many of the same outer membrane machinery for their import (Pfanner and Neupert, 1987; Pfaller et al., 1988; Hines et al., 1990; Kiebler et al., 1990; Dietmeier et al., 1997; Emtage, J., and R. Jensen, unpublished observations). It is also possible that the Mmm1p and Tim54p interaction is indirect. For example, we have preliminary evidence that Mmm1p is located in contact sites between the inner and outer membrane (Srinivasan, M., S. Burgess and R. Jensen, manuscript in preparation). The interaction between Tim54p and Mmm1p may occur since the protein import machinery is present, at least transiently, in these contact sites (Horst et al., 1995). We have found that the carboxyl terminus of Tim54p faces the intermembrane space and is therefore potentially accessible to outer membrane proteins. Further studies, however, are needed to clarify the relationship between Tim54p and Mmm1p.
Our studies clearly show that Tim54p is required for the insertion of IM proteins, but we have not shown that Tim54p plays a direct role in this import pathway. Since mitochondria isolated from the tim54-1 mutant have lowered amounts of Tim22p, it is possible that the primary function of Tim54p is to stabilize or localize the Tim22 protein in the mitochondrial inner membrane, and that Tim22p directly mediates IM protein insertion. Our observations that TIM54 is an essential gene and that Tim54p forms a stable complex with Tim22p argue that regardless of whether Tim54p's role in import is direct or indirect, the function of Tim54p is very important. It is also interesting to note that tim23-1 mitochondria have lowered amounts of both Tim23p and Tim17p, and both proteins have been shown to play direct roles in import of matrix proteins (Kübrich et al., 1994; Ryan et al., 1994; Berthold et al., 1995; Dekker et al., 1997). Nonetheless, further experiments are needed to pinpoint the roles of both Tim54p and Tim22p in the insertion of IM proteins.
In summary, the mitochondrial inner membrane contains distinct machinery for the translocation of proteins across the bilayer (Tim23p–Tim17p), and for the insertion of proteins into the membrane (Tim54p–Tim22p). It is possible that the mechanism of the Tim23p–Tim17p machinery differs significantly from that of the Tim54p– Tim22p complex. Tim22p, however, is homologous to both Tim23p and Tim17p, with over 50% similar residues shared between the three proteins. We therefore suggest that the membrane insertion machinery and the translocation machinery in the mitochondrial IM share at least some common activities. Supporting this idea, a single translocon in the ER mediates the transport of both soluble and membrane proteins (Kehry et al., 1980; McCune et al., 1980; Do et al., 1996; Mothes et al., 1997). It is tempting to speculate whether the two import complexes in mitochondria represent a more primitive or advanced evolutionary state compared to the ER machinery.
Acknowledgments
We wish to thank Fred Winston, Doug Koshland, Phil Hieter, Jef Boeke, and Steve Elledge for strains. We also thank Thad Baker for the 2μ- TIM17 and 2μ-TIM23 plasmids, Steve Elledge for the cDNA library, Doug Koshland for pDK255, Elizabeth Jones for the p7171 plasmid, Doug Murphy for use of his microscope, Susan Michaelis for pSM32, S. Burgess for pSB75, Jeff Schatz for antiserum to Tim44p, and Mike Yaffe for antiserum to F1β, hexokinase, and Tom70p. We thank Carolyn Machamer, Kathy Wilson, Alison Davis, and Susan Michaelis for critical comments on the manuscript.
This work was supported by grant R01-GM54021 from the United States Public Health Service to R.E. Jensen.
Abbreviations used in this paper
- CEN
centome-containing plasmid
- DIC
differential interference contrast
- HA
hemagglutinin
- IM
inner membrane
- Tim
translocase inner membrane
- Tom
translocase outer membrane
Footnotes
Address all correspondence to Robert E. Jensen, Department of Cell Biology and Anatomy, The Johns Hopkins University School of Medicine, 725 N. Wolfe St., Baltimore, MD 21205. Tel.: (410) 955-7291; Fax: (410) 955-4129; E-mail: rob_jensen@qmail.bs.jhu.edu
O. Kerscher and J. Holder contributed equally to this work.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Armstrong KA, Som T, Volkert FC, Rose A, Broach JR. Propagation and expression of genes in yeast using 2-micron circle vectors. Biotechnology. 1989;13:165–192. [PubMed] [Google Scholar]
- Bachhawat AK, Suhan J, Jones EW. The yeast homolog of H<β>58, a mouse gene essential for embryogenesis, performs a role in the delivery of proteins to the vacuole. Genes Dev. 1994;8:1379–1387. doi: 10.1101/gad.8.12.1379. [DOI] [PubMed] [Google Scholar]
- Bai C, Elledge SJ. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- Bauer M, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- Berthold J, Bauer MF, Schneider HC, Klaus C, Dietmeier K, Neupert W, Brunner M. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ ATP driving system. Cell. 1995;81:1085–1093. doi: 10.1016/s0092-8674(05)80013-3. [DOI] [PubMed] [Google Scholar]
- Blom J, Dekker PJT, Meijer M. Functional and physical interactions of components of the mitochondrial inner-membrane import machinery (MIM) Eur J Biochem. 1995;232:309–314. doi: 10.1111/j.1432-1033.1995.tb20813.x. [DOI] [PubMed] [Google Scholar]
- Blom J, Kübrich M, Rassow J, Voos W, Dekker PJT, Maarse A, Meijer M, Pfanner N. The essential yeast protein MIM44 (encoded by MPI1)is involved in an early step of preprotein translocation across the mitochondrial inner membrane. Mol Cell Biol. 1993;13:7364–7371. doi: 10.1128/mcb.13.12.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bömer U, Meijer M, Maarse AC, Hönlinger A, Dekker PJT, Pfanner N, Rassow J. Multiple interactions of components mediating translocation across the inner membrane. EMBO (Eur Mol Biol Organ) J. 1997;16:2205–2216. doi: 10.1093/emboj/16.9.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Laskey RA. A film detection method for tritium- labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen R E. MMM1encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Cell Biol. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Dekker P, Keil P, Rassow J, Maarse AC, Pfanner N, Meijer M. Identification of MIM23, a putative component of the protein import machinery of the mitochondrial inner membrane. FEBS Lett. 1993;330:66–70. doi: 10.1016/0014-5793(93)80921-g. [DOI] [PubMed] [Google Scholar]
- Dekker PJT, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO (Eur Mol Biol Organ) J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmeier K, Hönlinger A, Bömer U, Dekker PJT, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- Do H, Falcone D, Lin J, Andrews DW, Johnson AE. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Doolittle, R.F. 1986. Of Urfs and Orfs. University Science Books, Mill Valley, CA. 103 pp.
- Dowhan W, Bibus CR, Schatz G. The cytoplasmically-made subunit IV is necessary for assembly of cytochrome c oxidase in yeast. EMBO (Eur Mol Biol Organ) J. 1985;4:179–184. doi: 10.1002/j.1460-2075.1985.tb02334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage JLT, Jensen RE. MAS6encodes an essential inner membrane component of the yeast mitochondrial import pathway. J Cell Biol. 1993;122:1003–1012. doi: 10.1083/jcb.122.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Green N. Nonlethal sec71-1 and sec72-1 mutations eliminate proteins associated with the Sec63p–BiP complex from S. cerevisiae. . Mol Biol Cell. 1994;5:933–942. doi: 10.1091/mbc.5.9.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson IA, Lerner RA, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Galibert F, Alexandraki D, Baur A, Boles E, Chalwatzis N, Chuat JC, Coster F, Cziepluch C, De Haan M, Domdey H, et al. Complete nucleotide sequence of Saccharomyces cerevisiaechromosome X. EMBO (Eur Mol Biol Organ) J. 1996;15:2031–2049. [PMC free article] [PubMed] [Google Scholar]
- Gratzer S, Lithgow T, Bauer RE, Lamping E, Paltauf F, Kohlwein SD, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greener, A., and M. Callahan. 1994. XL1-Red: a highly efficient random mutagenesis strain. Stratagene Newsletter: Strategies in Molecular Biology. 7:32–34.
- Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor, Cold Spring Harbor, NY. 726 pp.
- Hase T, Riezman H, Suda K, Schatz G. Import of proteins into mitochondria: nucleotide sequence of the gene for a 70-kd protein of the yeast mitochondrial outer membrane. EMBO (Eur Mol Biol Organ) J. 1983;2:2169–2172. doi: 10.1002/j.1460-2075.1983.tb01718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauke V, Schatz G. Reconstitution of the protein insertion machinery of the mitochondrial inner membrane. EMBO (Eur Mol Biol Organ) J. 1997;15:4560–4567. doi: 10.1093/emboj/16.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines V, Brandt A, Griffiths G, Horstmann H, Brutsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO (Eur Mol Biol Organ) J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönlinger A, Kubrich M, Moczko M, Gartner F, Mallet L, Bussereau F, Eckerskorn C, Lottspeich F, Dietmeier K, Jacquet M, Pfanner N. The mitochondrial receptor complex - mom22 is essential for cell viability and directly interacts with preproteins. Mol Cell Biol. 1995;15:3382–3389. doi: 10.1128/mcb.15.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst M, Hilfiker-Rothenfluh S, Oppliger W, Schatz G. Dynamic interaction of the protein translocation systems in the inner and outer membranes of yeast mitochondria. EMBO (Eur Mol Biol Organ) J. 1995;14:2293–2297. doi: 10.1002/j.1460-2075.1995.tb07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Kalousek F, Mellman I, Rosenberg LE. A leader peptide is sufficient to direct mitochondrial import of a chimeric protein. EMBO (Eur Mol Biol Organ) J. 1985;4:1129–1135. doi: 10.1002/j.1460-2075.1985.tb03750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EC, Pesold-Hurt B, Schatz G. The amino-terminal region of an imported mitochondrial precursor polypeptide can direct cytoplasmic dihydrofolate reductase into the mitochondrial matrix. EMBO (Eur Mol Biol Organ) J. 1984a;3:3149–3156. doi: 10.1002/j.1460-2075.1984.tb02272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EC, Pesold-Hurt B, Schatz G. The cleavable prepiece of an imported mitochondrial protein is sufficient to direct cytosolic dihydrofolate reductase into the mitochondrial matrix. FEBS Lett. 1984b;178:306–310. doi: 10.1016/0014-5793(84)80622-5. [DOI] [PubMed] [Google Scholar]
- Hurt EC, Pesold-Hurt B, Suda K, Oppliger W, Schatz G. The first twelve amino acids (less than half of the pre-sequence) of an imported mitochondrial protein can direct mouse cytosolic dihydrofolate reductase into the yeast mitochondrial matrix. EMBO (Eur Mol Biol Organ) J. 1985;4:2061–2068. doi: 10.1002/j.1460-2075.1985.tb03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq C, Alt-Morbe J, Andre B, Arnold W, Bahr A, Ballesta JP, Bargues M, Baron L, Becker A, Biteau N, et al. The nucleotide sequence of Saccharomyces cerevisiaechromosome IV. Nature. 1997;387:75–78. [PubMed] [Google Scholar]
- Jensen RE, Kinnally KW. The mitochondrial import pathway: are precursors imported through membrane channels? . J Bioenerg Biomemb. 1997;29:3–10. doi: 10.1023/a:1022470303365. [DOI] [PubMed] [Google Scholar]
- Jensen RE, Yaffe MP. Import of proteins into yeast mitochondria: the nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MAS1-encoded subunit. EMBO (Eur Mol Biol Organ) J. 1988;7:3863–3871. doi: 10.1002/j.1460-2075.1988.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- Kehry M, Ewald S, Douglas R, Sibley C, Raschke W, Fambrough D, Hood L. The immunoglobulin mu chains of membrane-bound and secreted IgM molecules differ in their C-terminal segments. Cell. 1980;21:393–406. doi: 10.1016/0092-8674(80)90476-6. [DOI] [PubMed] [Google Scholar]
- Kiebler M, Pfaller R, Söllner T, Griffiths G, Horstmann H, Pfanner N, Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990;348:610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- Kronidou NG, Oppliger W, Bolliger L, Hannavy K, Glick BS, Schatz G, Horst M. Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1994;91:12818–12822. doi: 10.1073/pnas.91.26.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübrich M, Keil P, Rassow J, Dekker PJ, Blom J, Meijer M, Pfanner N. The polytopic mitochondrial inner membrane proteins MIM17 and MIM23 operate at the same preprotein import site. FEBS Lett. 1994;349:222–228. doi: 10.1016/0014-5793(94)00670-9. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawson JE, Douglas MG. Separate genes encode functionally equivalent ADP/ATP carrier proteins in Saccharomyces cerevisiae. Isolation and analysis of AAC2. J Biol Chem. 1988;263:14812–14818. [PubMed] [Google Scholar]
- Liu XQ, Freeman KB, Shore GC. An amino-terminal signal sequence abrogates the intrinsic membrane-targeting information of mitochondrial uncoupling protein. J Biol Chem. 1990;265:9–12. [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Maarse AC, Blom J, Keil P, Pfanner N, Meijer M. Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett. 1994;349:215–221. doi: 10.1016/0014-5793(94)00669-5. [DOI] [PubMed] [Google Scholar]
- McCune JM, Lingappa VR, Fu SM, Blobel G, Kunkel HG. Biogenesis of membrane-bound and secreted immunoglobulins. I. Two distinct translation products of human mu-chain, with identical N-termini and different C-termini. J Exp Med. 1980;152:463–468. doi: 10.1084/jem.152.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M, Gartner F, Pfanner N. The protein import receptor MOM19 of yeast mitochondria. FEBS Lett. 1993;326:251–254. doi: 10.1016/0014-5793(93)81801-6. [DOI] [PubMed] [Google Scholar]
- Mothes W, Heinrich SU, Graf R, Nilsson I, von Heijne G, Brunner J, Rapoport TA. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Sikorski R, Reed SI, Vogelstein B. Human p53 and CDC2Hs genes combine to inhibit the proliferation of Saccharomyces cerevisiae. . Mol Cell Biol. 1992;12:1357–1365. doi: 10.1128/mcb.12.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niman HL, Houghten RA, Walker LE, Reisfeld RA, Wilson IA, Hogle JM, Lerner RA. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller R, Steger HF, Rassow J, Pfanner N, Neupert W. Import pathways of precursor proteins into mitochondria: multiple receptor sites are followed by a common membrane insertion site. J Cell Biol. 1988;107:2483–2490. doi: 10.1083/jcb.107.6.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Douglas MG, Endo T, Hoogenraad NJ, Jensen RE, Meijer M, Neupert W, Schatz G, Schmitz UK, Shore G. Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem Sci. 1996;21:51–52. [PubMed] [Google Scholar]
- Pfanner N, Meijer M. Mitochondrial biogenesis: the Tom and Tim machine. Curr Biol. 1997;7:100–103. doi: 10.1016/s0960-9822(06)00048-0. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Müller HK, Harmey MA, Neupert W. Mitochondrial protein import: involvement of the mature part of a cleavable precursor protein in the binding to receptor sites. EMBO (Eur Mol Biol Organ) J. 1987;6:3449–3454. doi: 10.1002/j.1460-2075.1987.tb02668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Neupert W. Distinct steps in the import of ADP/ATP carrier into mitochondria. J Biol Chem. 1987;262:7528–7536. [PubMed] [Google Scholar]
- Ramage L, Junne T, Hahne K, Lithgow T, Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO (Eur Mol Biol Organ) J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Maarse AC, Krainer E, Kubrich M, Muller H, Meijer M, Craig EA, Pfanner N. Mitochondrial protein import - biochemical and genetic evidence for interaction of matrix Hsp70 and the inner membrane protein Mim44. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1988. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 164 pp.
- Ryan KR, Jensen RE. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Menold MM, Garrett S, Jensen RE. SMS1, a high-copy suppressor of the yeast mas6mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol Biol Cell. 1994;5:529–538. doi: 10.1091/mbc.5.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.R., R.S. Leung, and R.E. Jensen. 1998. Characterization of the mitochondrial inner membrane translocase (TIM) complex: the Tim23p hydrophobic domain interacts with Tim17p, but not other Tim23p molecules. Mol. Cell. Biol. In press. [DOI] [PMC free article] [PubMed]
- Saiki RK, Scharf S, Faloona KB, Mullis GT, Horn HA, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Scherer PE, Manning-Krieg UC, Jenö P, Schatz G, Horst M. Identification of a 45-kDa protein at the protein import site of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1992;89:11930–11934. doi: 10.1073/pnas.89.24.11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schneider H-C, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, Neupert W. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature. 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- Shani N, Sapag A, Watkins PA, Valle D. An S. cerevisiaeperoxisomal transporter, orthologous to the human adrenoleukodystrophy protein, appears to be a heterodimer of two half ABC transporters: Pxa1p and Pxa2p. Ann NY Acad Sci. 1996;804:770–772. doi: 10.1111/j.1749-6632.1996.tb18697.x. [DOI] [PubMed] [Google Scholar]
- Sherman F. Mutants of yeast deficient in cytochrome c. Genetics. 1964;49:39–48. doi: 10.1093/genetics/49.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R, Boeke JD. In vitromutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sikorski R, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–28. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22p. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- Söllner T, Rassow J, Wiedmann M, Schlossmann J, Keil P, Neupert W, Pfanner N. Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature. 1992;355:84–87. doi: 10.1038/355084a0. [DOI] [PubMed] [Google Scholar]
- Spencer F, Ketner G, Hieter P. Targeted recombination-based cloning and manipulation of large DNA segments in yeast. Methods. 1993;5:161–175. [Google Scholar]
- Steger HF, Sollner T, Kiebler M, Dietmeier KA, Pfaller R, Trulzsch KS, Tropschug M, Neupert W, Pfanner N. Import of ADP/ATP carrier into mitochondria: two receptors act in parallel. J Cell Biol. 1990;111:2353–2363. doi: 10.1083/jcb.111.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Vassarotti A, Douglas MG. Nuclear genes coding the yeast mitochondrial adenosine triphosphatase complex. Primary sequence analysis of ATP2 encoding the F1-ATPase beta-subunit precursor. J Biol Chem. 1985;260:15458–15465. [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiaeis regulated by proteolysis and phosphorylation. EMBO (Eur Mol Biol Organ) J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Guiard B, Neupert W, Cyr DM. The Δψ- and Hsp70-MIM44–dependent reaction cycle driving early steps of protein import into mitochondria. EMBO (Eur Mol Biol Organ) J. 1996;15:735–744. [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Neupert W, Cyr DM. The role of Hsp70 in conferring unidirectionality on protein translocation in mitochondria. Science. 1994;266:1250–1253. doi: 10.1126/science.7973708. [DOI] [PubMed] [Google Scholar]
- van Loon APGM, Brändli AW, Schatz G. The presequences of two imported mitochondrial proteins contain information for intracellular and intramitochondrial sorting. Cell. 1986;44:801–812. doi: 10.1016/0092-8674(86)90846-9. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Jensen RE, Guido EC. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J Biol Chem. 1989;264:21091–21096. [PubMed] [Google Scholar]
- Yanisch-Perron C, Viera J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zara V, Rassow J, Wachter E, Tropschug M, Palmieri F, Neupert W, Pfanner N. Biogenesis of the mitochondrial phosphate carrier. Eur J Biochem. 1991;198:405–410. doi: 10.1111/j.1432-1033.1991.tb16029.x. [DOI] [PubMed] [Google Scholar]