Abstract
PML and Sp100 proteins are associated with nuclear domains, known as nuclear dots (NDs). They were discovered in the context of leukemic transformation and as an autoantigen in primary biliary cirrhosis, respectively. Both proteins are expressed in the form of many COOH-terminally spliced variants, and their expression is enhanced by interferons (IFN). The recent finding that PIC1/SUMO-1, a small ubiquitin-like protein, is covalently linked to the RanGAP1 protein of the nuclear pore complex and also binds PML in yeast cells led us to determine whether PML is covalently modified by PIC1/SUMO-1 and whether the same is true for Sp100. We found an immune reaction of PML and Sp100 proteins with a PIC1/SUMO-1–specific monoclonal antibody by immunoblotting when using cell extracts prepared from stably transfected cells inducibly expressing one isoform of each protein as well as from nontransfected cells. In contrast, both proteins did not react when synthesized in vitro. Immunofluorescence staining showed that PIC1/SUMO-1 colocalized with Sp100 and PML in NDs except in mitotic cells, in which PML and Sp100 are dissociated. Cell fractionation and immunoblotting demonstrated that PIC1/SUMO-1 immunoreactive Sp100 in IFN-treated and untreated cells was exclusively nuclear, whereas nonmodified Sp100 was also found in the cytoplasm. Taken together, these data strongly suggest covalent modification of specific nuclear isoforms of Sp100 and PML by PIC1/SUMO-1. This modification may play a regulatory role in ND structure, composition, and function.
Nuclear dots containing PML and Sp100 proteins (NDs)1 were originally discovered as targets of autoantibodies in patients suffering from the autoimmune disease primary biliary cirrhosis (PBC) (Bernstein et al., 1984; Powell et al., 1984). They belong to the heterogeneous group of nuclear bodies (Brasch and Ochs, 1992) and are distinct subnuclear organelles that do not colocalize with any of the other known nuclear substructures (Ascoli and Maul, 1991; Raska et al., 1992; Stuurman et al., 1992; Dyck et al., 1994; Weis et al., 1994). NDs gained major attention when their disruption in the hematopoietic malignancy acute promyelocytic leukemia (APL) was discovered (Daniel et al., 1993). NDs were also reported to play a role in cell transformation, growth control (Mu et al., 1994; Ahn et al., 1995; Koken et al., 1995; Le et al., 1996), and cellular stress response (Maul et al., 1995). Furthermore, several viral oncoproteins and transactivators were found to influence ND structure and composition (Maul et al., 1993; Carvalho et al., 1995; Desbois et al., 1996; Korioth et al., 1996; for review see Sternsdorf et al., 1997a ).
Among the currently known ND-associated proteins, the Sp100 and PML proteins are the best characterized so far. Although very different in primary structure, PML and Sp100 share many properties. Expression of both proteins is strongly enhanced by interferons (IFN), and with very similar kinetics (Guldner et al., 1992; Chelbi-Alix et al., 1995; Lavau et al., 1995; Grötzinger et al., 1996b ). A large number of variant proteins is expressed from both genes because of extensive COOH-terminal alternative splicing (Xie et al., 1993; Dent et al., 1996) (Guldner, H., C. Szostecki, and H. Will, manuscript submitted for publication). A further common feature of both proteins is their autoantigenicity in PBC (Szostecki et al., 1997). The Sp100 protein was originally identified as an autoantigen in PBC (Szostecki et al., 1987, 1990), whereas the PML protein was discovered as a fusion protein with the retinoic acid receptor α (RAR) in cells of APL patients (de Thé et al., 1990; Goddard et al., 1991; Kakizuka et al., 1991). The expression of the PML/RAR fusion protein in APL cells is due to a chromosomal translocation and is sufficient for transformation of these cells and the development of leukemia (Altabef et al., 1996; Brown et al., 1997; Grisolano et al., 1997). Unlike in normal cells, both PML and Sp100 were reported to be microdispersed into hundreds of tiny dots in the nucleus and the cytoplasm in APL cells (Dyck et al., 1994; Koken et al., 1994; Weis et al., 1994). After treatment of APL cells with all-trans retinoic acid, the normal ND pattern is restored in the nucleus (Daniel et al., 1993), and concomitantly, the differentiation block in the promyelocytes is released (Huang et al., 1988). Since treatment of patients has the same effect on APL cells and frequently leads to disease remission, disintegration of NDs is believed to play a key role in the development of APL. This is consistent with the observation that the normal PML protein has growth- and transformation-suppressing properties (Mu et al., 1994; Ahn et al., 1995; Koken et al., 1995). For the Sp100 protein, transcription transactivating properties were reported (Xie et al., 1993) (Guldner, H., C. Szostecki, and H. Will, manuscript submitted for publication).
Recently, a protein directly interacting with PML, termed PML interacting clone 1 (PIC1), was identified by using the yeast two-hybrid interaction assay (Boddy et al., 1996). In independent studies addressing questions ranging from nuclear pore complex function, apoptotic signaling, and DNA recombination/repair processes, four other research groups isolated cDNAs coding for the same protein and termed it SUMO-1 (small ubiquitin-related modifier) (Mahajan et al., 1997), GAP modifying protein 1 (GMP1) (Matunis et al., 1996), Sentrin (Okura et al., 1996), and ubiquitin-like 1 (UBL1 ) (Shen et al., 1996b ), respectively. This protein, designated here PIC1/SUMO-1 to stress the aspects studied in our report (see below), has a calculated molecular mass of 11.5 kD and migrates in SDS-PAGE at ∼17–22 kD (Boddy et al., 1996; Matunis et al., 1996). It has a considerable sequence homology with ubiquitin and is covalently linked to the nuclear pore complex-associated protein RanGAP1 (Matunis et al., 1996; Mahajan et al., 1997). In a very recent publication, conjugation of PIC1/SUMO-1 to uncharacterized nuclear proteins has been demonstrated (Kamitani et al., 1997). PIC1/SUMO-1 is a member of the so-called ubiquitin-homology family. For many of these proteins, functions different from ubiquitin-dependent proteolytic degradation were reported (Watkins et al., 1993; Nakamura et al., 1995; Aso et al., 1995; Garrett et al., 1995; Biggins et al., 1996).
The recent findings of an interaction of PIC1/SUMO-1 with PML, the sequence homology of PIC1/SUMO-1 with ubiquitin, as well as its function in covalent modification of one protein stimulated us to investigate whether PML does not only bind but is covalently modified by PIC1/ SUMO-1. We further tested whether Sp100, which colocalizes with PML and has many other common features, is also modified by PIC1/SUMO-1. In this report, we provide strong evidence for covalent linkage of specific nuclear but not cytoplasmic isoforms of PML and Sp100 by PIC1/ SUMO-1 or an immunologically cross-reactive protein.
Materials and Methods
Cell Extracts, Immunoblotting, and Antibodies
Preparation of total cellular protein extracts, SDS-PAGE (7.5% or 12.5% polyacrylamide), and protein transfer onto nitrocellulose filters were carried out according to standard protocols (Sambrook et al., 1989). Blots were blocked in 5% (wt/vol) dry milk in water and incubated with primary antibodies (Abs) in the same solution. After washing four times in TBS (10 mM Tris-HCl, pH 7.6, 150 mM NaCl) or PBS (8 mM Na2HPO4, 1.5 mM KHPO4, 140 mM NaCl, 2.6 mM KCl, pH 7.3), bound Abs were detected with HRP-conjugated secondary Abs and enhanced chemiluminescence. Rat anti-PML and rat anti-Sp100 Abs were diluted 1:1,000 and 1:2,000, respectively. The mAb 21C7 was diluted 1:1,000 in 5% dry milk (in water). Secondary Abs were diluted 1:10,000. Production of the rat and rabbit anti-Sp100 and rat anti-PML Abs was described elsewhere (Sternsdorf et al., 1995; Grötzinger et al., 1996b ). A ubiquitin-specific mAb (Ubi-1) was purchased from Zymed Laboratories Inc. (WAK-Chemie, Bad Homburg, Germany).
Subcellular Fractionation
Cells grown as monolayer were harvested, washed once with PBS, and the pellet was resuspended in two volumes (pellet volumes [PV]) hypotonic buffer A′ (10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 1% NP-40, complete® protease inhibitor cocktail [Boehringer Mannheim Corp., Indianapolis, IN]) (Dignam et al., 1983). After 15 min of incubation on ice, cells were pelleted at 800 g for 10 min (4°C). The supernatant was cleared by centrifugation at 10,000 g, mixed with an equal volume of 2× SDS sample buffer (Sambrook et al., 1989) and boiled for 10 min. The pellet containing crude nuclei was washed once or twice in a large volume of buffer A′. Optionally, the second washing step was performed in two PV of buffer A′ followed by 10 min of incubation on ice to check for leakiness of the nuclei. The resulting supernatant was mixed with an equal volume of 2× SDS sample buffer and boiled for 10 min. Washed nuclei were resuspended in two PV buffer A′, mixed with an equal volume of 2× SDS sample buffer, sonicated three times for 20 s, and boiled for 10 min. Alternatively, cells were fractionated using a detergent-free method described previously (Sternsdorf et al., 1997b ). Briefly, cells were resuspended in buffer A (buffer A′ without NP-40). After 15 min swelling on ice, cells were homogenized in a Kontes all glass Dounce homogenizer (Type B pestil). Proteins of the supernatant were precipitated by addition of ice-cold acetone.
Immunoprecipitation
Cell nuclei were isolated with one of the methods described above, and nuclei were resuspended in four PV. Buffer E (140 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl, pH 8.0, and protease inhibitor cocktail). Na-desoxycholate (DOC) was added to a final concentration of 1.5%. Benzonase (Merck, Darmstadt, Germany) was added to a final concentration of 1 U/ μl, and the nuclei were incubated for 20 min at room temperature for digestion of genomic DNA and RNA. After centrifugation at 2,000 g, the supernatant was removed, cleared by centrifugation at 10,000 g, and used for immunoprecipitation (DOC-supernatant). The supernatant contained most of the nuclear PML and Sp100 proteins as determined by control experiments. For quantitative immunoprecipitation, 50 μl serum were incubated with 60 μl protein G–Sepharose for 1 h at 4°C, washed three times with RIPA buffer (Sambrook et al., 1989), containing proteinase inhibitor cocktail (= washing buffer). The complexes were then incubated for 2 h at 4°C with 100–150 μl of the DOC-supernatant and washed five times for 10 min in washing buffer. The pellets were boiled for 5 min in an adequate volume of 2× SDS loading buffer. For immunoprecipitation, rabbit anti-PML and rabbit anti-Sp100 (anti-SpDF or anti-SpGH) Abs were used.
In Vitro Transcription/Translation
For in vitro transcription and translation of PML and Sp100, the coupled TNT® reticulocyte lysate system (Promega Corp., Madison, WI) was used. In vitro translation was carried out without addition of radiolabeled methionine, according to the manufacturer's instructions. Plasmids pSG5-Sp100, pSP65-SpAlt-C, and pSG5-PML were used, which contain cDNAs encoding Sp100 (GenBank/EMBL/DDBJ accession number M60618), SpAlt-C (GenBank/EMBL/DDBJ accession number L79986), and PML (GenBank/EMBL/DDBJ accession number S50913) proteins under control of the T7 (pSG5-derivative vectors) or the SP6 promoter (pSP65-derivative vector). 2–5 μl of a 50-μl reaction were used for SDS-PAGE and subsequent immunoblotting.
Cells, Cytokines, Transfections, and Indirect Immunofluorescence Microscopy
Human HEp-2, HeLa S3, HuH-7, and MG63 cells were maintained in DME (GIBCO-BRL, Gaithersburg, MD) supplemented with 10% FCS. The hematopoietic progenitor cell line TF-1 was maintained as described previously (Kitamura et al., 1989). Human IFN (1,000 U/ml) was added to the medium for 15–20 h. Plasmid DNAs were introduced into cells using the calcium phosphate precipitation procedure (Sambrook et al., 1989). 1–5 μg of pSG5-Sp100, pRK5-SpAlt-C, or pSG5-PML plasmids adjusted to 10 μg DNA with pUC19 were precipitated per 6-cm dish. For indirect immunofluorescence staining, cells were grown on coverslips and treated as indicated. Cells were fixed either in 1% freshly prepared paraformaldehyde in PBS for 5 min, followed by permeabilization in PBS containing 0.2% Triton X-100 and 20 mM glycine for 20 min (at room temperature), or at −20°C for 5 min in methanol, and for 20 s in acetone. Rabbit or rat anti-Sp100 (Sp26) and rat anti-PML (anti–PML-N) antibodies were diluted 1:400. mAb 21C7 was diluted 1:400. Cells were incubated with Abs for 30 min at room temperature. For detection, DTAF- or LRSC-conjugated donkey anti–mouse, goat anti–rabbit, or donkey anti–rat IgG Abs (DIANOVA, Hamburg, FRG) were diluted 1:200 in PBS.
Establishment of Cell Lines with Inducible High Levels of PML or SpAlt-C Protein
Establishment of the HeLa-PML++ cell line was described previously (Sternsdorf et al., 1995). Cell lines expressing high levels of the SpAlt-C protein upon induction were produced accordingly by transfection of HtTA-1 cells (kindly provided by M. Gossen and H. Bujard, Zentrum für Molekulare Biologie, Heidelberg, FRG) with an SpAlt-C expression vector. These cell lines allow inducible expression of PML or SpAlt-C from the integrated copies of the transfected plasmid upon removal of tetracycline. Cells were grown in DME supplemented with l-glutamine, vitamins, nonessential amino acids, 1 μg/ml tetracycline, 200 μg/ml hygromycine, 500 μg/ml neomycine, and 10% FCS. Induction was carried out for 24–48 h by removal of tetracycline. Correct expression of the cDNAs was verified by reverse transcriptase PCR using total RNA of induced HeLa-PML++ and HeLa-SpAltC++ cell lines and complete sequencing of the cDNA inserts from two independent clones per cell line.
Results
Evidence for Posttranslational Modification of Sp100 and PML Proteins
Differences in electrophoretic mobility of proteins synthesized by in vitro transcription/translation compared with the same ones synthesized in living cells can provide indirect evidence for post- or cotranslational modification. However, this type of evidence is hard to obtain when several splice variants are expressed in cells that give rise to many bands in immunoblots. To circumvent this problem, part of our analysis was focused on only one splice variant of each of both proteins. This was achieved by using extracts from HeLa-derived cell lines with low endogenous Sp100 and PML expression that were stably transfected with expression constructs containing cDNA of one Sp100 or one PML splice variant. Expression of the corresponding proteins can be switched off or on in these cell lines by using culture medium with or without tetracycline, respectively (for details see Material and Methods). The stably transfected cell lines are designated in the following with HeLa-SpAltC++ and HeLa-PML++, respectively. For comparison, the same proteins were synthesized by in vitro transcription/translation or by transient expression in the human hepatoma cell line HuH-7. Moreover, all PML and Sp100 proteins expressed from the endogenous genes were analyzed on immunoblots using extracts from IFN-treated and untreated HeLa S3 cells. A schematic representation of the PML and Sp100 splice variants used in these experiments is given in Fig. 1. Note that PML and, more drastically, Sp100 have aberrant electrophoretic mobility. In addition, we found that the relative mobility changes depending on the percentage of the polyacrylamide gel used. Apparent molecular masses of the corresponding isoforms are given for 7.5% gels.
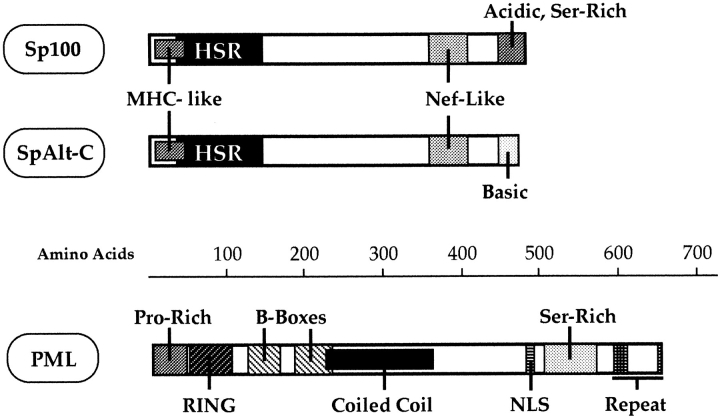
Figure 1.
Schematic representation of sequence motivs in the Sp100 and PML splice variants used. Sp100 represents the originally described major splice variant (Szostecki et al., 1990). In its NH2-terminal region, Sp100 harbors a region with homology to MHC class I molecules, followed by a domain that is conserved in its murine homologue (Grötzinger et al., 1996a ; Weichenhan et al., 1997) and in a lymphocyte-specific ND protein (Bloch et al., 1996; Dent et al., 1996), termed HSR domain (Sternsdorf et al., 1997a ). Also depicted is a region of sequence homology to the HIV I Nef-protein. The COOH terminus of Sp100 is acidic and contains a consensus phosphorylation site for casein kinase 2. This region (32 amino acids) is replaced by a basic stretch of 24 amino acids in SpAlt-C (Guldner, H., C. Szostecki, and H. Will, manuscript submitted for publication). The PML splice variant used in this study contains the proline-rich NH2 terminus, the RING-finger-B-Box motiv, followed by the predicted coiled coil region (de Thé et al., 1991). At its COOH terminus, the PML protein contains the nuclear localization signal (NLS), a serine-rich region, and five repeats of the amino acid motiv LASP(L).
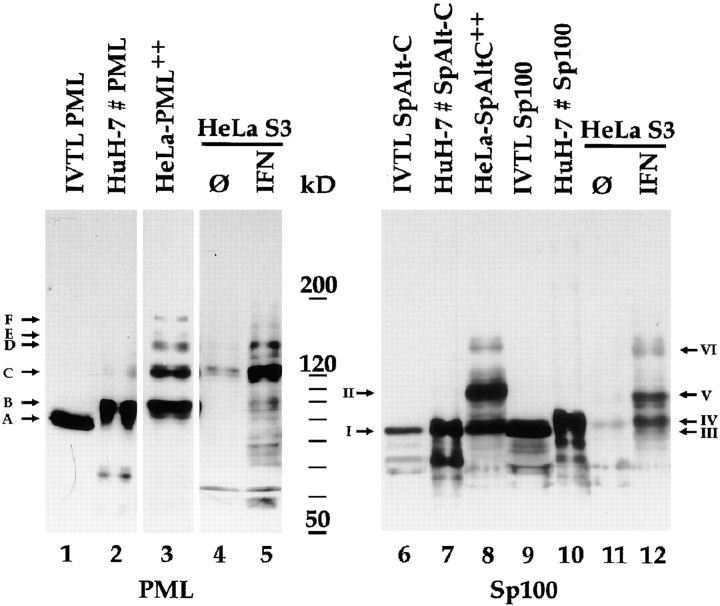
In vitro transcription/translation of PML resulted in a protein migrating at ∼85 kD as visualized by immunoblotting with a polyclonal rat anti-PML antibody (Fig. 2, lane IVTL PML, band A). When the same protein was expressed transiently in HuH-7 cells by short term transfection, a major band at the position of ∼95 kD and a minor band at ∼130 kD were visualized (Fig. 2, lane HuH-7 # PML, bands B and C). Interestingly, in cell line HeLa-PML++ three PML proteins with even slower electrophoretic mobility (Fig. 2, lane HeLa-PML ++, bands D–F) in addition to those seen in transiently transfected HuH-7 cells were observed. These data suggest that the PML protein analyzed is post- or cotranslationally modified by a covalently linked moiety. This modification was most prominent in the stably transfected cells, only weak in transiently transfected cells, and not detected when the protein was synthesized in vitro. The size differences between the PML proteins expressed in the different systems is roughly expected for PML covalently linked to one up to approximately four molecules of PIC1/SUMO-1, respectively.
Figure 2.
Immunoblot analysis of in vitro translated PML, SpAlt-C, and the Sp100 protein as well as of proteins of total cell extracts from various cells (HuH-7 cells transfected with PML, SpAlt-C and Sp100 expression vectors, induced HeLa-PML++ and HeLa-SpAltC++ cells, and HeLa S3 cells with and without prior treatment with IFN) separated by 7.5% SDS-PAGE. PML protein synthesized in vitro (lane 1, IVTL PML) has a higher electrophoretic mobility (band A) than the same PML protein expressed in transiently transfected HuH-7 (lane 2, band B) or in HeLa-PML++ cells (lane 3, band B), the latter showing additional bands of lower electrophoretic mobility (lane 3, bands C–F). PML proteins expressed from the endogenous gene are visualized as multiple bands in IFN-treated HeLa S3 cells (lane 5). In untreated cells, only the most prominent one is observed (lane 4). SpAlt-C protein comigrates when synthesized in vitro or in HuH-7 cells (lanes 6 and 7, band I). When expressed in HeLa-SpAltC++ cells (lane 8, band I), an additional band corresponding to a 110-kD protein is visualized (lane 8, band II). The Sp100 protein translated in vitro differs from that expressed transiently in HuH-7 cells by ∼3 kD (lane 10, bands III and IV). Sp100 proteins expressed from the endogenous gene in IFN-treated HeLa S3 cells are visualized as multiple bands; the major ones are indicated (lane 12, bands IV–VI). In untreated HeLa S3 cells, little Sp100 protein is detected (lane 11).
We next tested whether any of the putative modified forms of the single PML splice variant expressed in HeLa-PML++ cells corresponds in electrophoretic mobility to any of the multiple PML proteins expressed from the endogenous PML gene. This was done by immunoblot analysis of total extracts of untransfected, IFN-treated, and untreated HeLa S3 cells with anti-PML antibodies. As expected, upon long exposure of the blot a large number of immune reactive PML proteins corresponding to the many variants were observed in untreated cells and more intense and even more numerous in IFN-treated cells (data not shown). Upon short exposure of the gel, cell extracts from untreated cells showed a dominant signal corresponding to a protein of 130 kD (Fig. 2, lane 4), which corresponded to one of the slowly migrating, putatively modified PML variants previously detected in transiently transfected HuH-7 and stably transfected HeLa-PML++ cells (Fig. 2, compare lanes 1 to 4, band C). The same dominant signal, but much increased in intensity, was observed with extracts from IFN-treated cells (Fig. 2, lane 5). In addition to the dominant signal for the protein in band C, the second most intense signal appeared at the position of the modified PML splice variant located in band D. Taken together, these data strongly argue for co- or posttranslational modification of a subfraction of the major PML protein in HeLa S3 cells.
A similar conclusion was reached for Sp100 when an analogous study was performed with the major Sp100 protein and the minor splice variant SpAlt-C, as well as with Sp100 proteins expressed from the endogenous gene in IFN-treated and untreated HeLa S3 cells. SpAlt-C produced by in vitro translation or when expressed in transiently transfected HuH-7 cells migrated at a position corresponding to a protein of 85 kD (Fig 2, band I). In HeLa-SpAltC++ cells, the 85-kD SpAlt-C protein was also observed, but an additional strong signal corresponding to a SpAlt-C protein of 110 kD (Fig. 2, band III) and weak signals for proteins with even higher apparent molecular mass emerged. These data indicate that SpAlt-C is co- or posttranslationally modified, presumably by covalent linkage to one or several small proteins in cell line HeLa-SpAltC++ but not when synthesized in vitro or in transiently transfected HeLa S3 cells.
Since SpAlt-C is only a minor splice variant expressed from the endogenous gene, we also analyzed whether the major Sp100 protein synthesized by in vitro translation comigrates with Sp100 when expressed in transiently transfected cells. In contrast to SpAlt-C, a minor difference in electrophoretic mobility between the Sp100 proteins expressed in the two systems was observed (Fig. 2, lanes 9 and 10, bands III and IV). This may be due to co- or posttranslational modification of the major Sp100 protein by small molecules such as phosphate groups, when expressed by transient transfection in HuH-7 cells but not when synthesized in vitro. As performed for PML, we also used extracts from IFN-treated and untreated HeLa S3 cells to examine whether the co/posttranslationally modified forms of SpAlt-C identified in stably and transiently transfected cells comigrate with any of the Sp100 proteins expressed from the endogenous gene. Two major bands (Fig. 2, bands IV and V) corresponding to proteins of 88 and 105 kD, respectively, in addition to several minor ones were detected in IFN-treated cells (Fig. 2, lane 12). In untreated cells, only the signal corresponding to the 88-kD Sp100 protein (Fig. 2, lane 11, band IV) was observed at this exposition of the blot. This endogenous 88-kD protein migrates at exactly the same position as the major Sp100 when produced in transiently transfected cells. Therefore, it probably represents this Sp100 protein with the minor modification. In contrast, the most abundant endogenous Sp100 proteins with higher molecular mass did not comigrate with any of the modified forms of SpAlt-C, which are obviously only minor variants. This indicates that they are not modified SpAlt-C proteins but either correspond to modified forms of the major Sp100 protein or to additional Sp100 splice variant proteins that are most strongly expressed in IFN-treated cells. In summary, our data strongly suggest that at least some of the many PML and Sp100 proteins are co- or posttranslationally modified within cells but not when produced by in vitro translation. This discrepancy could well be due to the lack of components and enzymatic machinery from translation-competent reticulocyte lysates required for the attachment of the modification(s) or for proper ND protein folding, which may be a prerequisite for modification.
Evidence for Stable Modification of PML and Sp100 Proteins by PIC1/SUMO-1 or Immunologically Cross-reactive Polypeptides
The difference in apparent molecular mass between the in vivo– and in vitro–generated isoforms of PML and Sp100/ SpAlt-C described above could correspond to covalent modification by one or several molecules of PIC1/SUMO-1. This possibility and the previous report of covalent linkage of PIC1/SUMO-1 to RanGAP1 (Mahajan et al., 1997; Matunis et al., 1996) stimulated us to test whether PML and/ or Sp100 are also modified by this protein. If such modified proteins exist, they should immunologically react with PIC1/SUMO-1–specific antibodies. This was experimentally examined by using extracts of induced and noninduced HeLa-PML++ and HeLa-SpAltC++ cell lines and immunoblotting with the PIC1/SUMO-1–specific mouse mAb 21C7 (Matunis et al., 1996) and in addition with polyclonal rat or rabbit Abs against PML and Sp100, respectively.
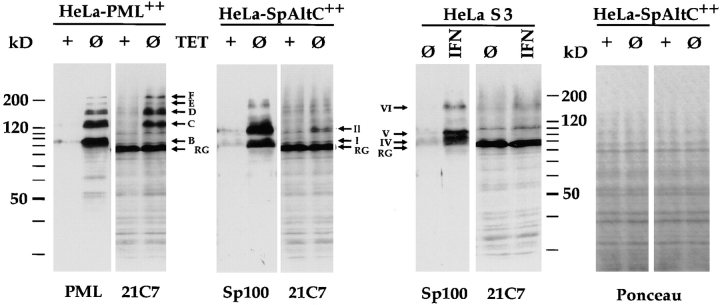
In immunoblots with extracts from induced and noninduced HeLa-PML++ cells or HeLa-SpAltC++ cells, a strong signal for a protein of ∼90 kD was observed with mAb 21C7 (Fig. 3, band RG). This protein corresponds in electrophoretic mobility to the previously described isoform of the Ran-GTPase–activating protein covalently modified by PIC1/SUMO-1 (Matunis et al., 1996; Mahajan et al., 1997). Four mAb 21C7–specific signals were observed on the blot with the cell extracts from the induced but not in extracts from noninduced HeLa-PML++ cells (Fig. 3, bands C–F). All four signals corresponded to proteins immune stained with anti-PML (Fig. 3, PML). This observation strongly suggests that bands C and D contain PIC1/SUMO-1–modified isoforms of the PML splice variant overexpressed in these cells. Note, however, band B, which most strongly stained with anti-PML (Fig. 3, second lane) and which was previously shown to contain modified PML (see Fig. 1), did not react with mAb 21C7 (Fig. 3, fourth lane). Therefore, the co- or posttranslationally modified overexpressed PML protein of 95 kD in band B is obviously not modified by PIC1/SUMO-1 and could be linked to a related protein that lacks the binding site of mAb 21C7. The analogous blots with the extracts from induced and noninduced HeLa-SpAltC++ cells revealed that only the SpAlt-C protein isoform of 110 kD but not the 88-kD isoform immune reacted with mAb 21C7 (Fig. 3, bands II and I, respectively). Therefore, the SpAlt-C variant protein tested is also presumably covalently modified by PIC1/SUMO-1 molecules or an immunologically cross- reactive polypeptide. The number of these peptides conjugated to the PML splice variant tested seems to vary and can be higher than those conjugated to SpAlt-C.
Figure 3.
Immunoblot with proteins of total extracts from noninduced (lanes + TET) and induced (lanes Ø TET) HeLa-PML++ and HeLa-SpAltC++ cells, and of untreated (lanes Ø) and IFN-treated (lanes IFN) HeLa S3 cells, separated by 12.5% SDS-PAGE. For detection, blots were incubated with polyclonal rat-anti-PML (PML), rat-anti-Sp100 (Sp100) Abs, and PIC1/SUMO-1–specific mAb 21C7 (21C7). In induced HeLa-PML++ cells, several strong bands were stained with anti-PML Abs (bands B–F) but only some of them (bands C–F) with anti-PIC1/SUMO-1 mAb. This indicates covalent linkage of PIC1/ SUMO-1 or a related polypeptide to proteins in the latter bands. In induced HeLa-SpAltC++, two major bands emerged when stained with anti-Sp100 Abs (bands I and II), whereas only the protein in band II, but not in band I, reacted with mAb 21C7. This indicates that band II represents the PIC1/SUMO-1–like modified form of the protein present in band I. In total extracts from untreated and IFN-treated HeLa S3 cells, a minor increase in staining by mAb 21C7 after IFN treatment in the range of 100–200 kD was observed. In all lanes, a 90-kD protein was recognized (band RG), which represents PIC1/SUMO-1–modified RanGAP1 protein (compare Fig. 4). Staining of the same blots with Ponceau S shows similar amounts of protein in all lanes (right; Ponceau S, shown for HeLa-SpAltC++).
To investigate whether Sp100 and PML proteins expressed from the endogenous genes are also modified by PIC1/SUMO-1, we performed immunoblots using extracts from untreated and IFN-treated HeLa S3 cells (Fig. 3, HeLa S3, lanes Ø and IFN, respectively). When total cell extracts were blotted with mAb 21C7, a strong signal for the PIC1/SUMO-1–modified RanGAP1 protein (band RG) among the large number of bands detected (Fig. 3, HeLa S3, panel 21C7) emerged, but only very faint IFN-enhanced signals, which may correspond to PIC1/SUMO-1–modified Sp100 or PML proteins, emerged (lane IFN). This suggests that PIC1/SUMO-1–modified proteins expressed from the endogenous PML and Sp100 genes (see below) are much less abundant than the modified RanGAP1 protein or the PML and SpAlt-C proteins expressed from the transgenes. In addition, they may be less well recognized by the mAb and therefore hardly detectable among the many other bands recognized.
Evidence for PIC1/SUMO-1–modified Sp100 Proteins by Immunoprecipitation
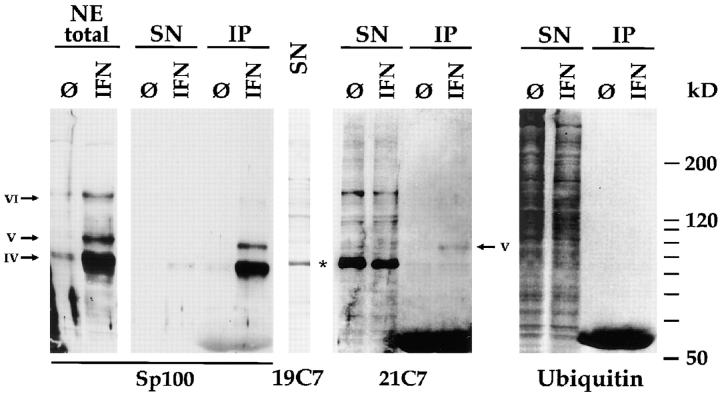
To increase the sensitivity and specificity of our detection, we performed immunoprecipitation with anti-PML and anti-Sp100 Abs using extracts of untreated and IFN-treated HuH-7 (Fig. 4) or HeLa S3 (not shown) cells. Since none of the various anti-PML Abs available to us efficiently immunoprecipitated PML (data not shown), we demonstrate the result of such an immunoprecipitation experiment with rabbit anti-Sp100 Abs only. In the protein extracts from IFN-treated HuH-7 cells, high Sp100 protein levels compared with untreated cells were detected as revealed by immunoblotting of total nuclear extracts (Fig. 4, lanes NE total; Sp100-specific bands are marked with IV, V, and VI as in Fig. 2). After precipitation of Sp100 proteins with an Sp100-specific polyclonal Ab and blotting of the precipitated proteins and, as a control, of the corresponding supernatants, Sp100 proteins were detected as strong signals in the precipitate of IFN-treated cells but only very faintly in the precipitate of untreated cells (Fig. 4, Sp100, lanes IP). Hardly any Sp100 was found in the supernatants (Fig. 4, Sp100, lanes SN), indicating that Sp100 proteins were precipitated almost quantitatively. Immunoblotting of the same extracts with mAb 21C7 revealed a band corresponding to Sp100 band V only in the precipitate of IFN-treated cells but not in untreated cells (Fig. 4, 21C7, lanes IP). On the contrary, the Sp100 band IV was not recognized by the antibody supporting the interpretation that band IV corresponds to unmodified and band V to the PIC1/SUMO-1–modified form of the major cellular Sp100 protein. A faint signal corresponding to the 90-kD PIC1/SUMO-1–modified form of the RanGAP1 protein was found unspecifically in equal amounts both in the precipitate of untreated and IFN-treated cells, confirming that equal amounts of protein were used for precipitation. All other 21C7-reactive bands, including most of the 90-kD RanGAP1 protein, were found in the supernatant (Fig. 4, 21C7, lanes SN). To confirm the identity of the 90-kD protein detected by mAb 21C7, a control incubation of a parallel blot with the RanGAP1-specific mAb 19C7 (Matunis et al., 1996) was performed (Fig. 4, 19C7, shown for supernatant of IFN-treated cells; position of RanGAP1 marked by asterisk). Note, band IV Sp100 protein has in 12.5% gels a slightly lower electrophoretic mobility than the RanGAP1 protein (Fig. 3) but migrates faster than RanGAP1 in 7.5% gels (Fig. 4). This is probably a consequence of the unusual distribution of charged amino acids in the Sp100 protein.
Figure 4.
Detection of endogenous Sp100 carrying the PIC1/ SUMO1–like modification after immunoprecipitation from total nuclear extracts of HuH-7 cells with rabbit anti-Sp100 Abs (SpDF). Immunoblot of total nuclear extracts (lanes NE total), supernatants (lanes SN), and precipitates (lanes IP) from untreated (lanes Ø) and IFN-treated (lanes IFN) cells, separated on a 7.5% SDS gel. Blots were incubated with rat anti-Sp100 Abs, mAb 19C7 (specific for RanGAP1), 21C7 (specific for PIC1/ SUMO-1), or Ubi-1 (specific for ubiquitin). After immunoprecipitation, Sp100 proteins are detected in the precipitate of IFN-treated cells but hardly in the precipitate of untreated cells or in the supernatant. When incubated with mAb 21C7, a band corresponding to Sp100 band V became visible only in the precipitate of IFN-treated cells but not of untreated cells, indicating that band V corresponds to a PIC1/SUMO-1–modified form of Sp100. A large number of bands became visible in the supernatant, with the most prominent band at a relative mobility of 90 kD. Control staining of a parallel blot with mAb 19C7 confirms that this band represents the PIC1/SUMO-1–modified form of the RanGAP protein (shown for supernatant of IFN-treated cells; position of RanGAP marked by asterisk). A blot with the same extracts stained with a ubiquitin-specific mAb showed no detectable reactive protein in the precipitate, even after long exposure of the blot. In contrast, many bands corresponding to ubiquitinylated cellular proteins were observed in the supernatant.
Since the PIC1/SUMO-1 protein shows homology with ubiquitin, we determined whether any of the Sp100 proteins present in the precipitate also react with a mAb specific for ubiquitin. However, even after long exposure of the blot, we could not visualize any bands in the precipitate that reacted with this mAb (Fig. 4, Ubiquitin, lanes IP). In contrast, a large number of proteins was detected in the supernatant (Fig. 4, Ubiquitin, lanes SN), again emphasizing the high specificity of the immunoprecipitation. In a parallel experiment using total extracts of induced and noninduced overexpressing cell lines, reactivity of this mAb with the modified PML- or SpAlt-C proteins was again not observed (data not shown). These results also indicate that endogenous cellular Sp100 proteins can be modified by PIC1/SUMO-1 or a related polypeptide, which must be different from ubiquitin.
Detection of PIC1/SUMO-1–associated Sp100, SpAlt-C, and PML by Immunofluorescence Staining of Cells
To analyze whether PIC1/SUMO-1 colocalizes with SpAlt-C and the PML splice variant proteins expressed in induced and noninduced HeLa-SpAltC++ and HeLa-PML++ cells, double immunostaining for PIC1/SUMO-1 with mAb 21C7 and for Sp100 or PML with rabbit anti-Sp100 or rat anti-PML Abs, respectively, was performed (Fig. 5). The cells were fixed with methanol/acetone, which makes ND complexes accessible for antibodies.
Figure 5.
Immunofluorescence staining of induced and noninduced HeLa-SpAltC++ and HeLa-PML++ cells fixed with methanol/acetone. In induced HeLa-PML++ cells, rat anti-PML Ab shows bright and enlarged ND staining in nonmitotic cells and large aggregates in a mitotic cell (A, arrow), whereas only a few small dots were stained in the noninduced cells (D). Double staining of the same slides with mAb 21C7 shows overlapping ND staining in induced nonmitotic cells (D) and a diffuse homogeneous pattern in noninduced cells (E). In mitosis, induced HeLa-PML++ cells resulted in no staining of PML aggregates with mAb 21C7 (B, arrow). For comparison, phase contrast images are given in C and F. In induced HeLa-SpAltC++ cells, rabbit anti-Sp100 Abs resulted in a microspeckled pattern with some large dots (G), whereas only a few small dots were stained in the noninduced cells (K). Additional incubation of the same slides with mAb 21C7 led to homogenous staining both in induced (H) and noninduced cells (L). For comparison, phase contrast images are given in I and M. Bar, 10 μm.
In induced, nonmitotic HeLa-PML++ cells, both the PML Ab (Fig. 5 A) and mAb 21C7 (Fig. 5 B) stained predominantly identical large ND-like structures. In addition, mAb 21C7 showed diffuse nuclear staining, which might be due to staining of modified RanGAP1. Interestingly, the staining pattern with both antibodies was drastically different in mitotic cells. PIC1/SUMO-1 was homogeneously distributed, and the PML splice variant protein was localized in large irregular aggregates (Fig. 5, A and B, mitotic cell marked by an arrow). In noninduced HeLa-PML++ cells, very few tiny dots were stained with anti-PML Abs (Fig. 5 D), whereas staining with mAb 21C7 (Fig. 5 E) was mainly homogeneous and nuclear. These data strongly support our conclusion that a large fraction of the PML splice variant is modified by PIC1/SUMO-1 in induced HeLa-PML++ cells. Moreover, the data indicate that the PIC1/SUMO-1–modified PML splice variant localizes predominantly in NDs in nonmitotic cells, whereas PIC1/SUMO-1 modification is absent from aggregated PML in mitotic cells.
The analogous experiment performed with HeLa-SpAltC++ cells showed the following. In induced HeLa-SpAltC++ cells, SpAlt-C localized in a microspeckled nuclear staining pattern with some additional larger dots, whereas PIC1/SUMO-1 staining was homogeneous with only a few additional dotlike structures (Fig. 5, G and H). In noninduced cells, Sp100 protein staining was weak and only present in few NDs in nuclei, whereas PIC1/SUMO-1 staining was homogeneously nuclear (Fig. 5, K and L). Taking into account our previous demonstration of PIC1/ SUMO-1–modified SpAlt-C in these cells (see Fig. 3), our findings suggest that the epitope recognized by mAb 21C7 is either not at all or only weakly accessible on PIC1/ SUMO-1–modified SpAlt-C fixed by methanol/acetone, or that the PIC1/SUMO-1–modified SpAlt-C is mainly homogeneously distributed in nuclei.
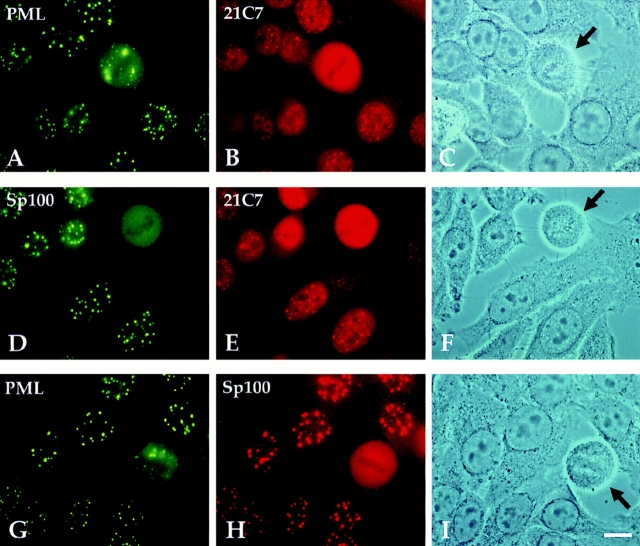
We next tested whether PIC1/SUMO-1 localizes in NDs of IFN-treated HeLa S3 cells, in which all splice variants of Sp100 and PML are most strongly expressed. Consistent with the previous colocalization of PIC1/SUMO-1 in non– IFN-treated cells, mAb 21C7 stained NDs brightly in nonmitotic IFN-treated HeLa S3 cells (Fig. 6, B and E). The same cells double stained with anti-PML and anti-Sp100 polyclonal Abs, respectively, showed largely overlapping ND staining (Fig. 6, A and B). Interestingly, the localization of the three proteins was drastically different in mitotic compared with nonmitotic cells (Fig. 6, mitotic cells marked by arrows). None localized in NDs, PIC1/SUMO-1 and Sp100 were homogeneously distributed, and PML localized in large aggregates. Three major conclusions can be drawn from these studies. First, Sp100 and PML proteins are clearly separated in mitotic cells. Second, PIC1/ SUMO-1 is either displaced from PML in mitosis, or the PIC1/SUMO-1 mAb 21C7–specific epitope is not accessible. Third, PIC1/SUMO-1–modified Sp100 and PML proteins are presumably localized primarily in the nucleus.
Figure 6.
Immunofluorescence staining of methanol/ acetone-fixed, IFN-treated HeLa S3 cells. Cells were either double-labeled with polyclonal rat anti-PML (green) and mAb 21C7 (red) (A–C), with polyclonal rabbit anti-Sp100 antibodies (green) and mAb 21C7 (red) (D to F), or with anti-PML Abs and anti-Sp100 Abs (G–I). In a mitotic cell, PML is aggregated at the periphery of the cell (A and G, arrow), whereas Sp100 and PIC1/ SUMO-1 are diffusely distributed (B, D, E, and H), indicating that PIC1/SUMO-1 and Sp100 dissociate from PML in mitosis. Bar, 10 μm.
Predominant Nuclear Localization of PIC1/SUMO-1–modified PML and Sp100 Splice Variant Proteins
Three major reasons led us to examine by subcellular fractionation in which cellular compartment PIC1/SUMO- 1–modified Sp100 and PML proteins are present. First, PIC1/SUMO-1 modification was reported for the cytoplasmic part of a receptor protein, for a nuclear membrane-associated as well as for nuclear proteins (Boddy et al., 1996; Matunis et al., 1996; Okura et al., 1996; Shen et al., 1996; Mahajan et al., 1997). Second, evidence that ND-associated proteins are not always localized exclusively in nuclei (Koken et al., 1995; Gambacorta et al., 1996) are compatible with their modification in the cytoplasm and/ or nuclei. Third, the predominant nuclear localization of PIC1/SUMO-1 by immunofluorescence staining suggested but did not prove modification of PML and SpAlt-C predominantly or exclusively in nuclei. Moreover, we also wondered whether IFN treatment has any effect on the ratio and subcellular localization of PIC1/SUMO-1–modified and nonmodified Sp100 and PML proteins.
To address these questions and to examine further whether there are cell type–specific differences, subcellular fractions were prepared from untreated and IFN-treated cells of different origins: the hematopoietic progenitor cell line TF-1, the hepatoma cell line HuH-7, the osteosarcoma cell line MG63, and the fibroblastoid cell lines HEp-2 and HeLa S3 (for fractionation procedure see Materials and Methods). In addition, protein extracts from induced and noninduced HeLa-SpAltC++ and HeLa-PML++ cell lines were analyzed to investigate the localization of PIC1/SUMO-1–modified forms of a single, specific Sp100 and PML splice variant protein expressed in stable cell lines from transgenes instead of from the endogenous genes.
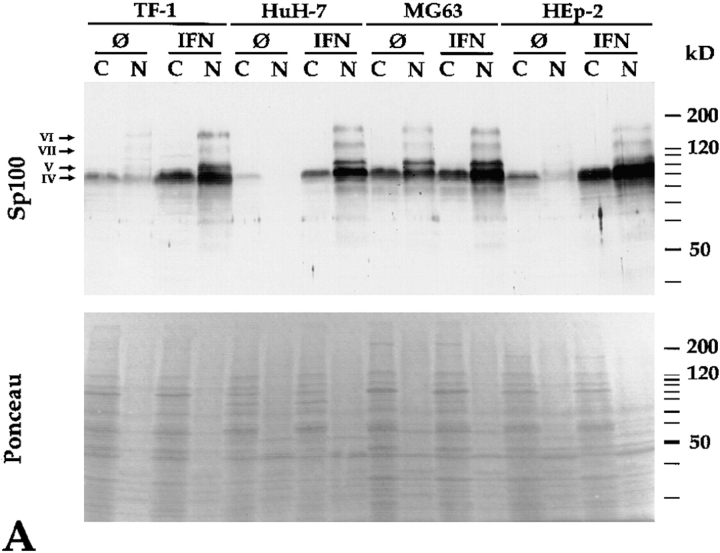
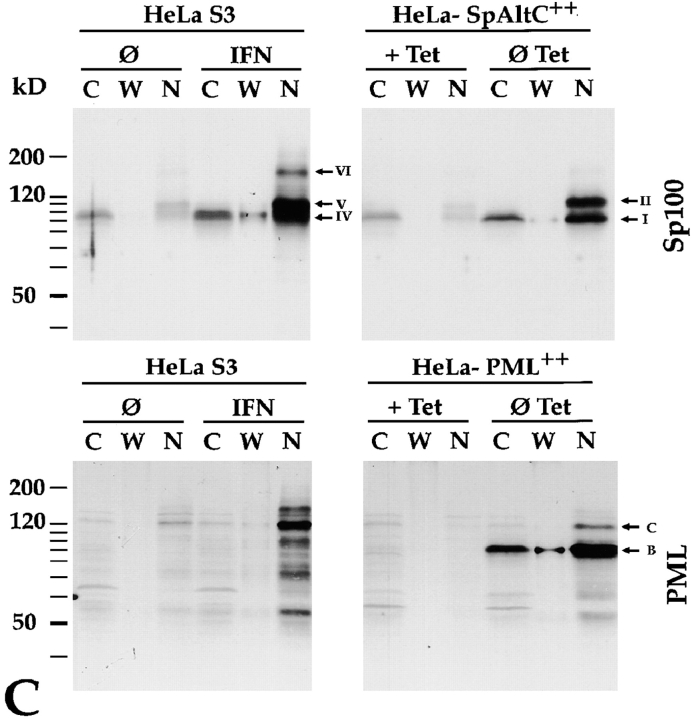
Interestingly, in these fractionation experiments we observed cytoplasmic forms of Sp100 in all cell lines tested so far. Immunoblot analysis of protein extracts from untreated (Fig. 7 A, lanes Ø) TF-1 and MG63 cells with anti-Sp100 antibodies revealed predominantly an Sp100 protein of 88 kD (Fig. 7 A, band IV) in the cytoplasmic (lanes C) and nuclear (lanes N) fraction. Intriguingly, in extracts from untreated HEp-2 and HuH-7 cells the same protein was detected mainly or even exclusively in cytoplasmic extracts. These data demonstrate not only that Sp100 proteins are present in nuclei but also that a substantial fraction is always, and in some cells predominantly, in the cytoplasm of untreated cells. After IFN treatment (Fig. 7 A, lanes IFN), the amount and number of immunoreactive Sp100 proteins did not only increase in all cell lines as expected, but the ratio between nuclear and cytoplasmic Sp100 proteins drastically changed towards dominant nuclear localization. Most notably, the Sp100 proteins previously shown to immunoreact with anti–PIC1/SUMO-1 mAb 21C7 (compare Figs. 3 and 4, bands V and VI) as well as one additional band with low electrophoretic mobility (Fig. 7, band VII) were detected exclusively in the nuclear fractions.
Figure 7.
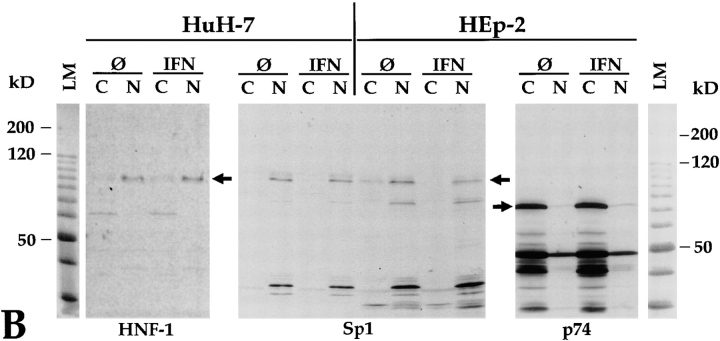
(A) Differentiation of nuclear and cytoplasmic forms of Sp100 proteins from various untreated (Ø) and IFN-treated (IFN) cell lines on immunoblots (12.5% SDS-PAGE) incubated with anti-Sp100 antibodies. With the exception of HuH-7, an Sp100 protein of ∼88 kD (band IV) is detected not only in the nuclear (lanes N) but also in the cytoplasmic fractions (lanes C) of all cell lines tested. In untreated HuH-7 cells, this protein is detected exclusively in the cytoplasmic fraction. With the exception of MG63 cells, IFN treatment strongly increases Sp100 levels in the nuclear fractions and only moderately in the cytoplasmic fractions. Isoforms of Sp100 with slower electrophoretic mobility (bands V–VII, arrow) are detectable only in the nuclear fractions. A similar amount of proteins of the respective fractions was loaded as documented by Ponceau S staining (Ponceau). (B) Immunoblot with nuclear and cytoplasmic extracts of HuH-7 and HEp-2 cells and polyclonal rabbit anti-HNF-1 (HNF-1) and monoclonal anti-Sp1 (Sp1) antibodies, and a human autoimmune serum, recognizing among other proteins the mitochondrial 74-kD dihydrolipoamide acetyltransferase subunit (p74). The transcription factors HNF-1 and Sp1 are detected predominantly in the nuclear fraction (lanes N) of HuH-7 or HuH-7 and HEp-2, cells, respectively (arrows). In contrast, the 74-kD mitochondrial protein was detected exclusively in the cytoplasmic fraction of HEp-2 cells (p74, lanes C, arrow). Treatment of the cells with interferon does not alter the subcellular localization of these proteins (lanes IFN). Lanes LM, Ponceau S staining of the molecular mass marker of the corresponding blots. (C) Differentiation of nuclear and cytoplasmic forms of Sp100 proteins in untreated (Ø) and IFN-treated (IFN) HeLa S3 cells, in induced (ØTet) and noninduced (+Tet) HeLa-PML++ as well as in HeLa-SpAlt-C++ cells by immunoblotting (12.5% SDS-PAGE) with anti-Sp100 or anti-PML antibodies. Similar to A, Sp100 as well as PML proteins are both in the nuclear (lanes N) and cytoplasmic (lanes C) fractions. IFN treatment as well as induction of HeLa-PML++ or HeLa-SpAltC++ cells increase their amounts predominantly in the nuclear fraction. Modified Sp100 proteins (bands II, V, and VI) are exclusively nuclear. Modified PML protein also is detected exclusively in nuclei of HeLa-PML++ cells. This is not evident in nontransfected HeLa S3 cells. Little Sp100 and PML proteins are detected in a nuclear wash fraction (lanes W).
To exclude the possibility that the cytoplasmic localization of Sp100 observed in these experiments was due to leakage of the nuclei and release of a weakly bound nuclear form of Sp100, we performed immunoblotting experiments with the HuH-7 and HEp-2 extracts as in Fig. 7 A and used antibodies against the transcription factors HNF-1 (liver-specific, nuclear) and Sp1 (ubiquitous, nuclear) and an autoimmune serum containing autoantibodies against the dihydrolipoamide acetyltransferase (p74), which is a mitochondrial protein (Van de Water et al., 1988) (Fig. 7 B). Since these transcription factors can easily be extracted from nuclei by established procedures (Dignam et al., 1983), they were chosen as an adequate control for the quality of the fractionation. In these blots, HNF-1 (Fig. 7 B, HNF-1, shown for HuH-7 cells) and Sp1 (Fig. 7 B, Sp1, shown for HuH-7 and HEp-2 cells) were observed predominantly in the nuclear fraction (Fig. 7 B, lanes N), and the mitochondrial protein (Fig. 7 B, p74, shown for HEp-2 cells) was observed in the cytoplasm (Fig. 7 B, lanes C). This implies that cytoplasmic and nuclear proteins are efficiently separated by our fractionation method and rules out a major cross contamination by leakage of nuclear proteins into cytoplasm or vice versa. Taken together, we conclude from these experiments that IFN treatment greatly enhances association of Sp100 proteins with nuclei even in cells in which Sp100 proteins are predominantly or exclusively cytoplasmic before treatment. Most importantly, Sp100 proteins modified by PIC1/SUMO-1 as well as other Sp100 proteins with low electrophoretic mobility are predominantly or exclusively associated with nuclei. Analogous experiments performed with anti-PML yielded tentatively similar results (data not shown) but were less convincing because of the low sensitivity of PML protein detection by immunoblotting.
Fractionation experiments with induced and noninduced HeLa-SpAlt-C++ and HeLa-PML++ cells and, for comparison, with IFN-induced and noninduced HeLa S3 cells revealed the following. In untreated HeLa S3 and noninduced HeLa-SpAltC++ cells, a single signal corresponding to an Sp100 protein of 88 kD (Fig. 7 C, band IV) was observed with cytoplasmic extracts (Fig. 7 C, lanes C superscribed with Ø and +Tet). The same signal, but less intense, and a signal (band V) for an Sp100 protein of ∼105 kD (Fig. 7 C, lanes N superscribed with Ø and +Tet) were observed with nuclear extracts. This implies that Sp100 proteins expressed from the endogenous gene in both cell lines are cytoplasmic and nuclear, whereas modified Sp100 proteins are exclusively associated with the nucleus. Upon IFN treatment of HeLa S3 cells and induction of transgene expression in HeLa-SpAltC++ cells, the intensity and number of signals for immune reactive Sp100 proteins increased (Fig. 7 C, lanes IFN and ØTet). In the cytoplasm, only the Sp100 proteins of 88 and 85 kD (bands IV and I) were identified whereas additional ones corresponding to PIC1/SUMO-1–modified Sp100 (Fig. 7 C, bands V and VI) and SpAlt-C (band II) proteins were exclusively nucleus associated. The analogous experiment with anti-PML also showed that PML proteins are not only associated with nuclei but are also present at least in part in the cytoplasm (Fig. 7 C, PML). Similar to Sp100, the ratio of nuclear versus cytoplasmic PML proteins strongly increased upon IFN treatment. This was also apparent when extracts from induced HeLa-PML++ cells were blotted. However, in this case an additional immune reactive protein of 130 kD (Fig. 7 C, band C), previously shown to correspond to PIC1/SUMO-1–modified PML, was exclusively in the nuclear extract (Fig. 7 C, last lane).
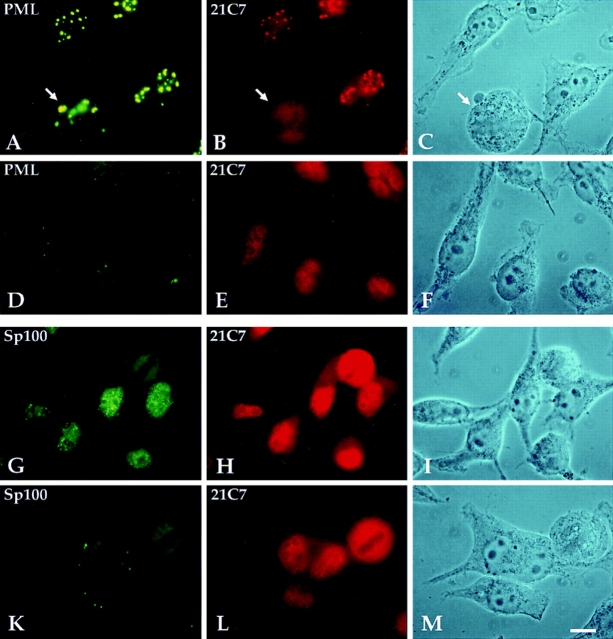
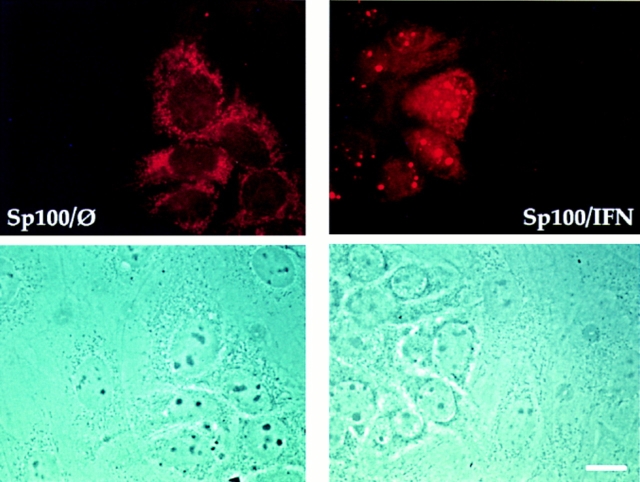
Since previous immunofluorescence-based localization studies only revealed nuclear Sp100 staining, we also tested whether Sp100 can be localized in the cytoplasm by immunofluorescence staining. For this experiment, we used the HuH-7 hepatoma cell line, which almost completely lacked nuclear Sp100 unless treated with IFN. Since a weak but specific cytoplasmic staining is hard to differentiate from nonspecific staining by antibodies, we have cocultivated HuH-7 cells with rat R1H cells. None of our anti-Sp100 antisera stained the corresponding rat Sp100 in these cells. Therefore, they were used as an internal negative control. In untreated HuH-7 cells, no ND staining was observed with polyclonal anti-Sp100 Abs (Fig. 8, Sp100/Ø). However, in these cells a diffuse or granular cytoplasmic staining was visualized. The adjacent R1H cells, however, at this exposition of the image were not visibly stained by the anti-Sp100 Abs. The corresponding phase contrast picture demonstrates that the two cell types can easily be discriminated by morphological criteria. Treatment of the cells with IFN led to clear ND staining in HuH-7 cells in addition to the cytoplasmic staining (Fig. 8, Sp100/IFN). The R1H cells visible on the same slide again did not visibly stain with the anti-Sp100 Abs. Therefore, in full agreement with the fractionation data, immunofluorescence analysis of HuH-7 cells shows exclusively cytoplasmic localization of Sp100 in untreated cells and after IFN treatment in addition in NDs.
Figure 8.
Detection of cytoplasmic Sp100 protein in a coculture of human HuH-7 and rat R1H cells by immunofluorescence analysis. Untreated HuH-7 cells when stained with polyclonal rabbit anti-Sp100 Abs (Sp26) lack ND staining but show cytoplasmic fluorescence (Sp100/Ø). After treatment with IFN, a clear ND staining became visible in addition to the cytoplasmic staining (Sp100/IFN). Both images were taken at the same exposure time. At this exposure, rat R1H cells are not visibly stained by the Abs and serve as an internal negative control. Localization of the rat cells is visible in the corresponding phase contrast pictures given below. Bar, 10 μm.
Taken together, these data indicate that PML and Sp100 proteins localize in a cell type–dependent manner either in part, predominantly, or even exclusively (Sp100 in HuH-7 cells) in the cytoplasm. Upon IFN treatment, both types of proteins predominantly fractionate with nuclei. Both PIC1/ SUMO-1–modified proteins are detectable exclusively in nuclei, regardless of prior IFN treatment. These findings may imply that PIC1/SUMO-1 conjugation enhances Sp100 and PML nuclear transport and/or matrix association.
Discussion
In this report, we provided several lines of evidence strongly suggesting that a fraction of some nuclear Sp100 and PML proteins is co- or posttranslationally modified by covalent linkage to the ubiquitin-like protein PIC1/SUMO-1 or an immunologically cross-reactive protein. Specific variants of both proteins, when expressed in vivo, had electrophoretic mobilities compatible with this modification but not when synthesized in vitro. A PIC1/SUMO-1–specific mAb but not an ubiquitin-specific mAb specifically reacted with some Sp100 and PML proteins when using total cell extracts or proteins immunoprecipitated with anti-Sp100 Ab. Using subcellular fractionation, the putative PIC1/SUMO-1–modified proteins were exclusively associated with nuclei. Moreover, our data indicate that IFN strongly enhances the amount of PIC1/SUMO-1–modified Sp100 proteins in the nucleus and suggest that PML is not modified by PIC1/SUMO-1 in mitotic cells. These findings imply a possible cell cycle–dependent role of PIC1/SUMO-1 modification of PML and Sp100 proteins in structure and function of NDs.
Our strongest argument for covalent linkage of Sp100 and PML proteins to PIC1/SUMO-1 is their immune reactivity with a PIC1/SUMO-1 mAb and appropriate electrophoretic mobility in SDS–gel electrophoresis. Although we have no definite proof for covalent modification, the immune reactivity of the proteins with mAb 21C7 after boiling in SDS under reducing conditions leaves hardly any other interpretation. However, strong binding of PIC1/SUMO-1 to Sp100 and PML, which resists this treatment, is not ruled out by our experiments. Additional and strong arguments for covalent linkage of PIC1/SUMO-1 to Sp100 and PML proteins are the sequence similarity of PIC1/SUMO-1 with ubiquitin and the recent demonstration of covalent modification of another nucleus-associated protein, RanGAP1, by PIC1/SUMO-1 (Matunis et al., 1996; Mahajan et al., 1997). Furthermore, preliminary evidence that the modification is reversible when nuclei are incubated at 37°C in the absence of ionic detergents (data not shown) argues also for covalent (de)modification, which depends on enzymatic processes, similar to what is known for PIC1-modified RanGAP1 (Matunis et al., 1996) and classical ubiquitinylation (Hochstrasser, 1996b ). Therefore, it is most reasonable to assume that PIC1/SUMO-1 is covalently linked to PML and Sp100, probably through an isopeptide bond formed between ε-amino groups of lysine residues in Sp100/PML and the COOH-terminal carboxyl group of PIC1/SUMO-1.
The immune reactivity of some Sp100 and PML proteins with mAb 21C7 is also compatible with the assumption that both proteins are modified by a protein different from but immunologically cross-reactive with PIC1/SUMO-1. Modification of Sp100 and PML by one other ubiquitin-like protein, ISG15 (Loeb and Haas, 1992), was ruled out experimentally by performing immunoblots with polyclonal anti-ISG15 antiserum, similar to that done with mAb 21C7. On these blots, there was no indication for proteins reacting both with Sp100/PML and ISG15 (data not shown). One may also argue that the Sp100 and PML proteins reactive with mAb 21C7 may correspond to classically ubiquitinylated proteins. However, our data with the commercially available mAb Ubi-1, which reacts with ubiquitins from humans to Caenorhabditis elegans, are not consistent with this possibility.
The large number of proteins detected with mAb 21C7 on immunoblots with total cell lysates suggests that this mAb either cross-reacts immunologically with other ubiquitin-like proteins and/or with PIC1/SUMO-1 present on many cellular proteins in addition to those known so far and described here. Despite the most dominant reactivity of mAb 21C7 with PIC1/SUMO-1, additional cross-reaction of mAb 21C7 with PIC1/SUMO-1–related proteins different from ubiquitin is conceivable. This would be in line with the observation that the modified form of Sp100 reacts more weakly with the PIC1/SUMO-1–specific mAb 21C7 than the modified PML or especially the RanGAP1 protein (see Fig. 3). On the other hand, this could also be explained by a different accessibility of the corresponding 21C7 epitopes in the various conjugates. The 15-kD difference between the most dominant PML protein expressed in induced HeLa-PML++ cells and the same protein synthesized in vitro, however, is more likely due to covalent modification with proteins that do not cross-react with mAb 21C7. In any case, a final definite answer to the question as to whether PIC1/SUMO-1 and other ubiquitin-like proteins are covalently linked to PML and Sp100 protein requires protein sequence data.
When we analyzed PML in induced HeLa-PML++ cells, several proteins with slow electrophoretic mobility were observed (Fig. 2 A), which could represent covalent modification of this specific PML splice variant protein by more than one PIC1/SUMO-1 molecule. This could indicate the modification of more than one lysine residue in the PML protein. However, multiple modification of PML might also result from formation of multimeric PIC1/SUMO-1 conjugates. Formation of such multimeric conjugates is typical for modification of cellular proteins by polyubiquitin chains (Hochstrasser, 1996b ). For conventional polyubiquitinylation, the ε-amino group of lysine at position 48 of ubiquitin is most frequently used and was reported so far mostly in the context of protein degradation. However, other lysine residues are known to be used alternatively (Arnason and Ellison, 1994; Baboshina and Haas, 1996; Hodgins et al., 1996), though less frequently. In PIC1/ SUMO-1, one of these residues (corresponding to lysine 6 in ubiquitin) is conserved, suggesting that PIC1/SUMO-1 can potentially also form oligomers that are linked to a subpopulation of the PML protein(s). Since polyubiquitinylation at residues other than lysine 48 was speculated to play a role not only in proteolytic but also in nonproteolytic processes (Arnason and Ellison, 1994; Hochstrasser, 1996a ), the latter function is likely for PIC1/ SUMO-1 modification of ND proteins. This speculation is consistent with the absence of an analogous lysine at position 48 in PIC1/SUMO-1 and the fact that there is no evidence for increased degradation of modified Sp100 and PML proteins. In contrast to PML, there was no indication for modification of Sp100 proteins by oligomers of PIC1/ SUMO-1. This could imply that PML and Sp100 contain different variants of PIC1/SUMO-1–related molecules, one with and one without oligomerisation potential.
Our observation that PML is not modified by PIC1/ SUMO-1 when synthesized in vitro is consistent with the reported failure of PML/PIC1 coprecipitation when synthesized by in vitro translation and weak or no binding of PIC1 to PML–GST fusion protein (Boddy et al., 1996). However, in the same study, interaction of PIC1/SUMO-1 with a PML protein was observed when tested in the yeast two-hybrid system. To further characterize this interaction, we have performed analogous experiments in mammalian cells (HuH7 and HeLa S3) with various constructs expressing full-length and truncated fragments of PML and Sp100 fused to Gal4-DNA binding- or VP16-transactivation domains. Upon cotransfection of these constructs with constructs expressing PIC1/SUMO-1 fused to Gal-4 or VP16, we observed no interaction (data not shown). This finding is consistent with the lack or very inefficient modification of PML and Sp100 by PIC1 that we observed in transiently transfected cells. Moreover, these data suggest that the interaction of PIC1/SUMO-1 with PML in yeast is due to long-term expression of both proteins, yeast-specific PML and/or PIC1/SUMO-1 conformations, and possibly more abundant ubiquitin-conjugating enzymes.
An interesting observation in our study was the absence of detectable PIC1/SUMO-1 in large PML aggregates in mitotic cells, which may imply that this modification regulates ND protein function in a cell cycle–dependent manner as well as ND protein interaction. The known cell cycle–dependent variation in size and number of NDs is consistent with this hypothesis and is currently being examined experimentally by using extracts from cells in different stages of the cell cycle. In this context it is worth mentioning that the PML and Sp100 (SpAlt-C) variants used and overexpressed stably in the inducible cell lines have a remarkably different distribution within the nuclei. Whereas PML forms enlarged, brightly staining dots and therefore might use previously existing NDs to accumulate, SpAlt-C exhibits a more disperse and finely granular pattern of distribution. The apparent lack of staining of these fine granular structures by mAb 21C7 in immunofluorescence might be merely due to the lower concentration of antigen when compared with the PML overexpressing cells.
Interestingly, in a prior publication (Matunis et al., 1996) staining of the mitotic spindle by PIC1/SUMO-1–specific antibodies was demonstrated. The lack of such staining pattern in our experiments is presumably due to differences in the fixation procedure. With PML- and Sp100-specific antibodies, we never observed a staining of the mitotic spindle, regardless of the fixation conditions, indicating that a different PIC1/SUMO-1–modified protein might be responsible for this staining. Matunis et al. (1996) reported that this might be RanGAP1.
The coexistence of Sp100 and PML proteins with and without PIC1/SUMO-1 modification in the nucleus, the lack of this modification in mitotic cells (for PML) as suggested by immunofluorescence data, as well as the staining of the mitotic spindle apparatus with the mAb 21C7 (Matunis et al., 1996) may be an indication for cell cycle–dependent regulation of PIC1/SUMO-1 modification for other proteins as well. Moreover, if PIC1/SUMO-1 modification of PML and Sp100 occurs at the nuclear membrane, in a way similar to that described for RanGAP1, this could be the signal for nuclear pore association, subsequent nuclear transport, and/or attachment to the nuclear matrix. The latter is less likely since preliminary experiments with nuclear fraction incubated at 37°C showed that a large amount of Sp100 remained nuclear matrix-associated even though there was no more PIC1/SUMO-1–modified Sp100 detectable under these conditions (data not shown).
The covalent modification of RanGAP1 and Sp100/PML by PIC1/SUMO-1, the ubiquitin-like nature of PIC1/SUMO-1, and the cell stage specific modification of PML postulate the existence of enzymes that mediate modification and demodification, respectively. The most likely candidate enzyme that may mediate covalent linkage of PIC1/SUMO-1 to PML and Sp100 is UBE2I/HsUbc9, a human homolog of the yeast UBC9 ubiquitin-conjugating enzyme (Kovalenko et al., 1996; Yasugi and Howley, 1996). This enzyme is known to play a role in S- and M-phase cyclin degradation and mitotic control, but most importantly, it was recently shown to interact with PIC1/SUMO-1 (Shen et al., 1996a ) as well as with RAD51/52 proteins known to associate with PIC1/SUMO-1 (Shen et al., 1996b ). A very good candidate for removal of PIC1/SUMO-1 from ND proteins is the cellular HAUSP protein, a recently described herpes simplex virus–associated ubiquitin-specific protease that dynamically associates with NDs (Everett et al., 1997). The authors speculated that the viral transactivator protein ICP0/Vmw110 might use this protease to initiate the well-documented dispersal of Sp100 and PML proteins from NDs. The authors also speculated that PIC1/ SUMO-1 could be a target for HAUSP (Everett et al., 1997). Our observation that dissociation of PML and Sp100 happens concurrently with loss of PIC1/SUMO-1 modification of PML provides indirect support for these exciting speculations and furthermore argues for a regulatory role of this modification in ND composition, integrity, and function. The drastic increase in the amount of modified Sp100 only in the nucleus after IFN treatment argues in the same direction. The identification of PIC1/SUMO-1 as an interaction partner of proteins with very different functions points to a broad physiological relevance of this modification in cell biology. The modification of the RanGAP1 protein is known to mediate its association to the nuclear pore, whereas the nonmodified 70-kD isoform remains entirely cytoplasmic (Matunis et al., 1996; Mahajan et al., 1997). This strongly resembles the situation with—and may be functionally similar for—Sp100 and PML proteins. Furthermore, the association of PIC1/SUMO-1 with the death domains of Fas/APO-1 and tumor necrosis factor receptor 1 was speculated to have an antiapoptotic effect (Okura et al., 1996). Our study raises the question as to whether PIC1/SUMO-1–modified ND proteins contribute to this antiapoptotic effect. The association of PIC1/ SUMO-1 with DNA repair proteins RAD51/RAD52 proteins (Shen et al., 1996b ) may have consequences for several of their functions and of their interaction partners. Since the breast cancer susceptibility protein BRCA1 is one of the interaction partners (Scully et al., 1997), a possible role of PIC1/SUMO-1 modification of these and other proteins in tumorigenesis is therefore well conceivable. Consistent with this speculation is the key role of ND disruption in the development of acute promyelocytic leukemia (Dyck et al., 1994; Koken et al., 1994; Weis et al., 1994). This may be triggered by the possible loss of the PIC1/SUMO-1 modification site in the PML–RAR fusion protein, resulting in a functionally deregulated PML protein. Potent and specific inhibitors for PIC1/SUMO-1 modification would be useful to test this hypothesis and would also help to gain new information on the putative function of NDs in regulation of growth and differentiation, autoimmunity, and virus infection.
Acknowledgments
We would like to thank Drs. G. Blobel and M.J. Matunis (Rockefeller University, New York) for providing mAbs 19C7 and 21C7 and Drs. H. de Thé (Hôpital St. Louis, Paris, France) and P. Staeheli (University of Freiburg, Freiburg, Germany) for polyclonal rabbit anti-PML and anti-ISG15 antisera, respectively. We thank Dr. K. Kühlcke (Heinrich-Pette-Institut) for providing the TF-1 cell line. We are indebted to Dr. P. Forster (Heinrich-Pette-Institut) for critical reading of the manuscript and Dr. G. Rutter (Heinrich-Pette-Institut) for fruitful discussion.
This work was supported by grants from the Fritz-Thyssen-Stiftung, the Deutsche Forschungsgemeinschaft, and the Deutsche Krebshilfe. The Heinrich-Pette-Institut is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Forschung und Gesundheit.
Abbreviations used in this paper
- Ab
antibody
- APL
acute promyelocytic leukemia
- IFN
interferon
- ND(s)
nuclear dot(s) (nuclear domain(s) containing PML and Sp100 proteins)
- PBC
primary biliary cirrhosis
- PIC1
PML interacting clone 1
- PV
pellet volumes
- RAR
retinoic acid receptor α
- snRNP
small nuclear ribonucleoprotein
- SUMO-1
small ubiquitin-related modifier
Footnotes
Address all correspondence to Hans Will, Heinrich-Pette-Institut für Experimentelle Virologie und Immunologie an der Universität Hamburg, Martinistraße 52, D-20251 Hamburg, FRG. Tel. and Fax: 0049 40 48051221. E-mail: will@hpi.uni-hamburg.de
References
- Ahn M-J, Nason-Burchenal K, Moasser MM, Dmitrovsky E. Growth suppression of acute promyelocytic leukemia cells having increased expression of the non-rearranged alleles: RARα or PML. Oncogene. 1995;10:2307–2314. [PubMed] [Google Scholar]
- Altabef M, Garcia M, Lavau C, Bae S-C, Dejean A, Samarut J. A retrovirus carrying the promyelocyte-retinoic acid receptor PML-RARα fusion gene transforms haematopoietic progenitors in vitroand induces acute leukaemias. EMBO (Eur Mol Biol Organ) J. 1996;15:2707–2716. [PMC free article] [PubMed] [Google Scholar]
- Arnason T, Ellison MJ. Stress resistance in Saccharomyces cerevisiaeis strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli CA, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Baboshina OV, Haas AL. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem. 1996;271:2823–2831. doi: 10.1074/jbc.271.5.2823. [DOI] [PubMed] [Google Scholar]
- Bernstein RM, Neuberger JM, Bunn CC, Callender ME, Hughes GR, Williams R. Diversity of autoantibodies in primary biliary cirrhosis and chronic active hepatitis. Clin Exp Immunol. 1984;55:553–560. [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Ivanovska I, Rose MD. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch DB, de la Monte SM, Guigaouri P, Filippov A, Bloch KD. Identification and characterization of a leukocyte-specific component of the nuclear body. J Biol Chem. 1996;271:29198–29204. doi: 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- Brasch K, Ochs RL. Nuclear bodies (NBs): a newly “rediscovered” organelle. Exp Cell Res. 1992;202:211–223. doi: 10.1016/0014-4827(92)90068-j. [DOI] [PubMed] [Google Scholar]
- Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci PG, Atwater S, Bishop JM. A PMLRARα transgene initiates murine acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T, Seeler JS, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix MK, Pelicano L, Quignon F, Koken MHM, Venturini L, Stadler M, Pavlovic J, Degos L, de Thé H. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027–2033. [PubMed] [Google Scholar]
- Daniel MT, Koken MHM, Romagne O, Barbey S, Bazarbachi A, Stadler M, Guillemin MC, Degos L, Chomienne C, de Thé H. PML protein expression in hematopoietic and acute promyelocytic leukemia cells. Blood. 1993;82:1858–1867. [PubMed] [Google Scholar]
- de Thé H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor α gene to a novel transcribed locus. Nature. 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- Dent AL, Yewdell J, Puviondutilleul F, Koken MHM, de Thé H, Staudt LM. LYSP100-associated nuclear domains (LANDs): description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood. 1996;88:1423–1436. [PubMed] [Google Scholar]
- Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I tax oncoprotein. Science. 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WJ, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO (Eur Mol Biol Organ) J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambacorta M, Flenghi L, Fagioli M, Pileri S, Leoncini L, Bigerna B, Pacini R, Tanci LN, Pasqualucci L, Ascani S, et al. Heterogeneous nuclear expression of the promyelocytic leukemia (PML) protein in normal and neoplastic human tissues. Am J Pathol. 1996;149:2023–2035. [PMC free article] [PubMed] [Google Scholar]
- Garrett KP, Aso T, Bradsher JN, Foundling SI, Lane WS, Conaway RC, Conaway JW. Positive regulation of general transcription factor SIII by a tailed ubiquitin homolog. Proc Natl Acad Sci USA. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard AD, Borrow J, Freemont PS, Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RARα under control of cathepsin G regulatory sequences. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- Grötzinger T, Jensen K, Guldner HH, Sternsdorf T, Szostecki C, Schwab M, Savelyeva L, Reich B, Will H. A highly amplified mouse gene is homologous to the human interferon-responsive Sp100 gene encoding an autoantigen associated with nuclear dots. Mol Cell Biol. 1996a;16:1150–1156. doi: 10.1128/mcb.16.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grötzinger T, Sternsdorf T, Jensen K, Will H. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML) Eur J Biochem. 1996b;238:554–560. doi: 10.1111/j.1432-1033.1996.0554z.x. [DOI] [PubMed] [Google Scholar]
- Guldner HH, Szostecki C, Grötzinger T, Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996a;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996b;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hodgins R, Gwozd C, Arnason T, Cummings M, Ellison MJ. The tail of a ubiquitin-conjugating enzyme redirects multi-ubiquitin chain synthesis from the lysine 48-linked configuration to a novel nonlysine-linked form. J Biol Chem. 1996;271:28766–28771. doi: 10.1074/jbc.271.46.28766. [DOI] [PubMed] [Google Scholar]
- Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, Gu LJ, Wang ZY. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–572. [PubMed] [Google Scholar]
- Kakizuka A, Miller WJ, Umesono K, Warrell RJ, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Nguyen HP, Yeh ET. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.22.14001. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, Piao YF, Miyazono K, Urabe A, Takaku F. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- Koken MHM, Puvion DF, Guillemin MC, Viron A, Linarez-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO (Eur Mol Biol Organ) J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken MHM, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thepot J, Juhlin L, Degos L, Calvo F, de Thé H. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- Kovalenko OV, Plug AW, Haaf T, Gonda DK, Ashley T, Ward DC, Radding CM, Golub EI. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc Natl Acad Sci USA. 1996;93:2958–2963. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi PP, Pelicci PG, Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–878. [PubMed] [Google Scholar]
- Le XF, Yang P, Chang KS. Analysis of the growth and transformation suppressor domains of promyelocytic leukemia gene, PML. J Biol Chem. 1996;271:130–135. doi: 10.1074/jbc.271.1.130. [DOI] [PubMed] [Google Scholar]
- Loeb KR, Haas AL. The interferon-inducible 15-kD ubiquitin homolog conjugates to intracellular proteins. J Biol Chem. 1992;267:7806–7813. [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Guldner HH, Spivack JG. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- Maul GG, Yu E, Ishov AM, Epstein AL. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- Mu ZM, Chin KV, Liu JH, Lozano G, Chang KS. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Xavier RM, Tsunematsu T, Tanigawa Y. Molecular cloning and characterization of a cDNA encoding monoclonal nonspecific suppressor factor. Proc Natl Acad Sci USA. 1995;92:3463–3467. doi: 10.1073/pnas.92.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei CF, Chang HM, Yeh ET. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- Powell F, Schroeter AL, Dickson ER. Anti-nuclear antibodies in primary biliary cirrhosis. Lancet. 1984;1:288–289. doi: 10.1016/s0140-6736(84)90164-8. [DOI] [PubMed] [Google Scholar]
- Raska I, Dundr M, Koberna K. Structure-function subcompartments of the mammalian cell nucleus as revealed by the electron microscopic affinity cytochemistry. Cell Biol Int Rep. 1992;16:771–789. doi: 10.1016/s0309-1651(05)80021-9. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Second Edition. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY. 545 pp.
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics. 1996a;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 1996b;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T, Guldner HH, Szostecki C, Grötzinger T, Will H. Two nuclear-dot associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand J Immunol. 1995;42:257–268. doi: 10.1111/j.1365-3083.1995.tb03652.x. [DOI] [PubMed] [Google Scholar]
- Sternsdorf, T., T. Grötzinger, K. Jensen, and H. Will. 1997a Nuclear dots: actors on many stages. Immunobiolology. In press. [DOI] [PubMed]
- Sternsdorf T, Jensen K, Züchner D, Will H. Cellular localization, expression, and structure of the nuclear dot protein 52. J Cell Biol. 1997b;138:435–448. doi: 10.1083/jcb.138.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Szostecki C, Krippner H, Penner E, Bautz FA. Autoimmune sera recognize a 100 kD nuclear protein antigen (Sp100) Clin Exp Immunol. 1987;68:108–116. [PMC free article] [PubMed] [Google Scholar]
- Szostecki C, Guldner HH, Netter HJ, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- Szostecki C, Guldner HH, Will H. Autoantibodies against “nuclear dots” in primary biliary cirrhosis. Semin Liver Dis. 1997;17:71–78. doi: 10.1055/s-2007-1007184. [DOI] [PubMed] [Google Scholar]
- Van de Water J, Fregeau D, Davis P, Ansari A, Danner D, Leung P, Coppel R, Gershwin ME. Autoantibodies of primary biliary cirrhosis recognize dihydrolipoamide acetyltransferase and inhibit enzyme function. J Immunol. 1988;141:2321–2324. [PubMed] [Google Scholar]
- Watkins JF, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiaeDNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenhan D, Kunze B, Zacker S, Traut W, Winking H. Structure and expression of the murine Sp100 nuclear dot gene. Genomics. 1997;43:298–306. doi: 10.1006/geno.1997.4834. [DOI] [PubMed] [Google Scholar]
- Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Xie K, Lambie EJ, Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T, Howley PM. Identification of the structural and functional human homolog of the yeast ubiquitin conjugating enzyme UBC9. Nucleic Acids Res. 1996;24:2005–2010. doi: 10.1093/nar/24.11.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]