The view that proteins are, in essence, nanoscale macromolecular machines is helping to foster a new paradigm in biology, with an interdisciplinary emphasis that incorporates strong elements of biophysics (1). Remarkable examples of cellular machinery abound, including ribosomes, polymerases, proteosomes, chaperonins, and spliceosomes. But perhaps no more spectacular example of proteins-as-machines exists than the motor proteins, also called mechanoenzymes. These molecular assemblies drive both rotary and linear motions in virtually all organisms. Rotary engines include the F1-F0 ATPase (ATP synthase) and the bacterial flagellar motor, both powered by currents of protons. Linear motors, which include members of the myosin, kinesin, and dynein superfamilies, are fueled by the hydrolysis of ATP.
Among the linear motors, kinesin is the only mechanoenzyme that is known to be processive—that is, it remains bound to its polymer substrate while undergoing multiple rounds of activity (23). Kinesin dimers, which consist of two identical heads attached to a common α-helical coiled-coil stalk, are widely believed to proceed by a “hand-over-hand” mechanism, involving a strict alternation of the two heads along the microtubule lattice. Three recent studies, two appearing in this issue (8, 19) and one in Biochemistry (31), shed new light on the mystery of kinesin processivity and lend additional experimental support to the hand-over-hand model.
Proceeding Processively
There is overwhelming evidence for kinesin processivity. Single molecules of kinesin typically travel for distances of a micrometer along microtubules, with a constant probability of dissociation per unit of distance traveled, corresponding to roughly 1% per molecular step (5, 28). This implies a processivity value of ∼100 enzymatic turnovers catalyzed before dissociation or termination. This value compares favorably with that calculated for many nucleic acid enzymes, which move processively but are not normally categorized as motor proteins, although it pales in comparison with DNA or RNA polymerase. Perhaps more impressively, single molecules of kinesin can pull tiny beads (∼0.5 μm in diameter) against loads up to ∼6 pN or more (24), all while taking steps measuring 8 nm (25). These numbers can be used to estimate an upper bound for the length of time any kinesin molecule might reasonably spend apart from its microtubule substrate, assuming it were to unbind at all during the mechanical cycle. A load of ∼3 pN (half of the stall force) can pull a dissociated bead rearwards through a distance equal to its forward stepsize, thereby precluding net progress, in ∼20 μs. Any time spent unbound must be shorter than this (perhaps 0). This is one thousandth of the time required for molecules to complete their mechanical cycle, which is ∼20 ms at ∼400 nm/s (half the peak velocity). The effective “duty ratio” for a single kinesin molecule, defined as that fraction of the total cycle during which it remains tightly attached to the microtubule during the translocation process, is thus >0.999, and possibly 1.0. Kinesin hangs on for dear life.
But does kinesin move hand-over-hand? Implicit in that scenario is a kind of molecular legerdemain whereby the trailing head can't advance until the leading head is already in position, and vice versa, so that strict alternation is accomplished. The first real indication for such coordination came from elegant biochemical work by Hackney (7), who showed that there is a fundamental asymmetry between the two heads of a kinesin dimer molecule bound to a microtubule in the absence of ATP: one head domain was directly attached to the microtubule in a rigor-like manner and carried no bound nucleotide, while the other head appeared to be tethered and carried ADP in its catalytic site. Moreover, the binding of ATP to the rigor head induced ADP release by the partner head. Subsequent work confirmed and extended this result and was consistent with a model in which ATP binding by one head is required for ADP release by the other (16). In this way, the two heads may be forced to run “out of phase” with one another.
The hand-over-hand model predicts that single-headed motors ought not to work very well, if at all, and they should lose their processivity. In ground-breaking work, Gelles and co-workers (2) demonstrated that a single-headed, monomeric construct based on the head domain of Drosophila kinesin did in fact support movement in vitro. As anticipated, however, the motion was slow and irregular, with motors wandering willy-nilly over the microtubule surface, and comparatively large surface densities of motors were required in assays. This was in contrast to native dimeric motors, which support movement at vanishingly low surface densities, down to the single-molecule level (11), and which track faithfully along paths parallel to single protofilaments of the microtubule lattice (18). All in all, this is consistent with the view that single-headed motors work in groups and that their heads don't hang on very well. But there are other possible explanations for the failure of single-headed kinesin, so it was important to go beyond these experiments.
One-headed Monsters
Hancock and Howard (8) produced a single-headed construct by coexpressing full-length kinesin heavy chains from Drosophila together with histidine-tagged, “decapitated” chains deleted for the motor domain. The decapitated chains consist largely of the rod domain (also called the stalk) that is responsible for kinesin dimerization via α-helical coiled-coil interactions. The his-tag on the headless chain was used to isolate the single-headed species from the two-headed one, generating a homogeneous population for testing. Consistent with earlier work, the motion produced by one-headed motors was slow and irregular, regardless of the surface density. However, the fact that these constructs contained the full-length rod, which under the right conditions can bind to surfaces without disrupting the motor activity in the heads, meant that it now became possible to titrate movement against motor surface density. This experiment is formally analogous to titrating the activity of an enzyme against its substrate concentration: when plotted appropriately, the result is a sigmoid curve whose steepness reveals the presence or absence of cooperative interactions—a kind of Hill curve. For dimeric motors, the effective mechanical “Hill coefficient” was unity, reproducing earlier results (11, 22). However, the new single-headed constructs gave a mechanical Hill coefficient somewhere between 4 and 6, implying that 4–6 kinesin heads must participate to generate continuous motion.
Perhaps the most interesting finding was that at the very lowest surface densities, isolated single-headed motors tended to bind microtubules for exceedingly long times (∼3 s, on average), leading to diffusional swiveling of the microtubules on surfaces about their points of attachment, a phenomenon known as “nodal point pivoting.” However, single-headed motors failed to move microtubules, in contrast to dimeric motors. This result poses something of a paradox. If single heads linger on microtubules for as long as 3 s, corresponding to a detachment rate of just ∼0.3 s−1, how can the two-headed species possibly drive movement at ∼800 nm/s, which requires an overall stepping rate of ∼100 s−1 given 8-nm steps? On this basis, Hancock and Howard reasoned that the binding of additional heads to a microtubule somehow accelerates the detachment of the first head, most likely because the first head senses an additional mechanical strain acting through the microtubule. This resolution seems plausible, although the evidence for it remains circumstantial, but it is the kind of coordination required for hand-over-hand motion.
Gelles and co-workers (31) took a somewhat different tack, using two different “tail-less” single-headed derivatives, K365-BIO and K340-BIO, which consist of slightly different sizes of the kinesin head domain from Drosophila fused to a biotin-accepting domain from Escherichia coli. These monomeric species were then bound at various densities to surfaces coated with streptavidin and used in microtubule gliding assays. By adding methycellulose to the motility buffer, they were able to prevent microtubules from diffusing away from the coverglass surface, and thereby to score movement at lower motor densities than previously (26). The logic of the experiment goes as follows: For motors with low duty ratios (those that are bound and stroking during only a fraction of the cycle), the effect of multiple motors is to produce faster motion than that of a single motor alone since the contribution from any one motor tends to occur at a time when all the others are unbound. Thus, the individual motions summate. In contrast, for motors with high duty ratios, the effect of several motors is roughly the same as for a single one (15), and there is little or no change in speed.
By carefully compiling histograms of speeds attained by both single-headed species and a double-headed control, Gelles and co-workers were able to demonstrate that single-headed motion is fundamentally different from double-headed motion. In particular, motions produced by the two-headed derivative tend to be biphasic: either microtubules are driven by motors near their peak speed, or they are not driven at all, whereupon the motion is purely diffusive. Episodes of movement at intermediate speeds are comparatively rare. This behavior is consistent with a processive motor having a high duty ratio. In contrast, both the single-headed species produce monophasic distributions, with a continuum of speeds that do not cluster near the maximal value. This is consistent with a nonprocessive motor having a low duty ratio. In addition, Gelles and colleagues were able to confirm reduced duty cycles for their single-headed derivatives by a clever autocorrelation analysis, which quantifies aspects of the persistence of the motion (31).
But is a low duty ratio for single-headed motors consistent with the observation that single heads remain bound for long periods (8), which would seem to imply the exact opposite? Curiously, yes. Assuming that the interpretation advanced by Hancock and Howard is right, i.e., that detachment rates for single heads are accelerated many-fold by the mechanical strain that develops when other heads bind, then assays that characterize the effects of multiple single heads (e.g., those used by Gelles and co-workers) would tend to “see” the detached forms, giving a low duty ratio. But more work remains to be done addressing this issue.
What's in a Name?
The difference in behavior between single- and double-headed kinesin species raises anew a frequently posed question: Which parts of the kinesin molecule are responsible for its various properties? Is it possible to assign specific roles to one or more functional domains, such as force production, ATP hydrolysis, stepping distance, elastic compliance, processivity, or directionality? A search for functional domains generally begins with the identification of structural domains. Unfortunately, the kinesin field developed an obsolete and/or misleading nomenclature by assigning colorful structural names to regions of the molecule before their structures were actually known, based on a combination of electron microscopy, biochemistry, sequence gazing, and wishful thinking (14, 27, 29, 30). Originally, the NH2-terminal ∼430 amino acids (aa)1 of the kinesin heavy chain, which contain the ATPase and microtubule-binding activities, were called the “head,” and assumed to form a globular unit. The middle region of the heavy chain, responsible for dimerization through α-helical coiled-coil interactions, was termed the “stalk,” while the COOH-terminal ∼50–90 aa formed a third globular region called the “tail.” This created a problem right from the start, because of the structural analogy with myosin, where the globular NH2-terminal domain is also called the “head,” but where the coiled-coil portion is named the “tail.” To avoid confusion, many prefer to call the kinesin “stalk” a “rod,” which is a term also used for myosin.
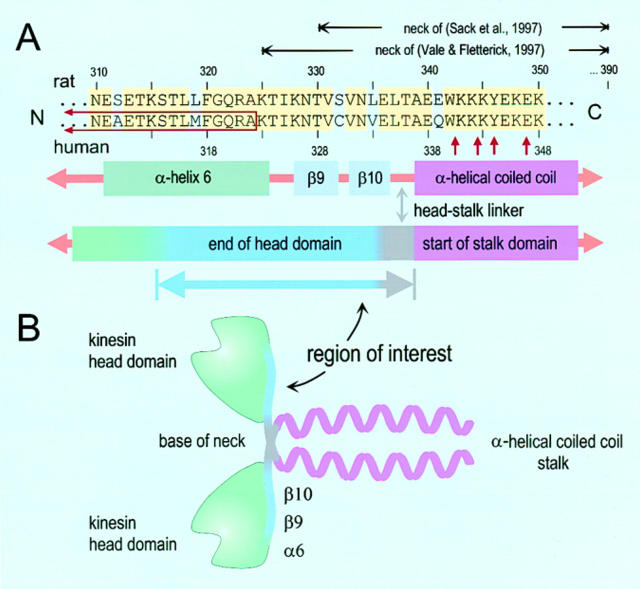
Alas, the problems didn't end there. The NH2-terminal ∼430 aa of kinesin constitute rather more than a globular head; they stretch well into the rod domain. The head fragment whose structure was originally determined was much smaller than this, comprising the first ∼350 aa, of which ∼325 aa were visible in the crystal structure (14). The latter portion was dubbed the kinesin “motor domain,” despite the fact that fragments truncated at this point did not retain motor activity. The name for this fragment was later revised to the “catalytic domain” (6), or to the “catalytic core” (27). The sequence of 20–40 aa immediately following the catalytic domain is evolutionarily conserved, in a class-specific way, across the whole superfamily of kinesin proteins, and is thus of considerable interest. Tendentiously, this sequence, whose structure was unknown at the time, was given the name “neck” (20, 27) (although the term “neck” had previously been used in a different context [12]), and the “motor domain” was apparently redefined to be the catalytic domain plus this neck (6). Soon thereafter, the crystal structures of both kinesin monomers and dimers containing portions of the neck were solved, comprising the NH2-terminal ∼350 and ∼370 aa, respectively (13, 21). These structures reveal that “neck” was an unfortunate choice of words. First, the neck isn't located at the base of the head; it starts near the midzone of the structural head domain, and its first dozen aa are closely associated with the rest of the head, forming two short β strands (β-9, β-10) packed against the core of the crystal structure. Second, the stretch of polypeptide linking the head to the “body” (stalk), that is, the part one might be inclined to call a “neck” in common parlance, is exceedingly short, comprising just 3–4 aa. The remaining ∼50 aa of the neck region, from ∼340–390, dimerize into a coiled-coil. Thus, the neck defined by Vale and Fletterick (27) spans at least three structural subdomains: the head, the short head–stalk linker, and the start of the rod, or stalk (Fig. 1). Now that crystal structures are available, it would seem to be a good time for people to get together and reach a consensus on a structurally consistent set of names for the parts of the kinesin protein.
Figure 1.
Sequence alignment for a portion of the kinesin heavy chain and corresponding elements in the kinesin structure. (A) Alignment of the rat (top) and human (bottom) sequences spanning the region ∼310–350 aa. Identical amino acids are colored (yellow). The numbering scheme for rat and human sequences differs by two. Neck regions, as defined by Sack et al. (21) and Vale and Fletterick (27), are indicated (horizontal black arrows). The sequence replaced in the ncd-kinesin chimera of Case et al. (6) is indicated on the left (red open box, left), as well as the sites of various mutations introduced by Romberg et al. (19) (vertical red arrows, right). Immediately under the alignment are structural elements of the dimer: α-helix 6 (olive), β-strands 9 and 10 (lavender), and the start of the coiled-coil (violet). Below these are the structural domains, consisting of the head (green, COOH terminus blue), head-stalk linker (gray), and initial portion of the stalk (violet). A new “region of interest” begins in α-helix 6 and continues until the start of the stalk (see text). (B) The corresponding structural domains are shown mapped onto a cartoon of the kinesin dimer, using the same coloring scheme.
Neck and Neck
Nomenclature notwithstanding, the neck is turning out to be a very interesting portion of the kinesin molecule. Two recent studies of genetically engineered kinesin-ncd chimeras, in which the head of kinesin (a plus end–directed motor) was replaced by the head of its relative, ncd (a minus end–directed motor), have furnished some insights into the possible origins of directional polarity (6, 9). When the kinesin catalytic domain was replaced by the corresponding domain of ncd, with the chimeric junction chosen to coincide with the NH2-terminal start of the kinesin neck sequence (Fig. 1), the resulting constructs moved towards the plus ends of microtubules, despite their ncd heads. In its simplest interpretation, this result suggests that the secret of directionality may reside somewhere in the neck and is therefore located “outside of the core catalytic domain” (6). Once again, though, the nomenclature may serve to mislead because the “neck” starts within the globular head domain.
The paper by Romberg et al. (19) investigates the effect of directed mutations elsewhere in the neck region of the human kinesin heavy chain. The mutations they introduced all turned out to be located towards the base of the neck, beyond the start of the α-helical coiled-coil stalk, which begins at Ala339 in the rat structure (Ala337 of the human sequence; Fig. 1). It had been hypothesized that some degree of “melting” or unwinding of this coiled-coil region might be required to separate the two kinesin heads by a distance equal to the step size (4, 27). Romberg and co-workers produced a number of sequence variants of the kinesin heavy chain designed to test this notion. Chimeric molecules were created, consisting of the NH2-terminal portion of the kinesin heavy chain sequence out to ∼aa 560, which is midway through the long stalk. Beyond this point, the kinesin sequence was fused to the sequence for green fluorescent protein equipped with a his-tag. These constructs could be scored for motor activity at the single-molecule level, using the powerful new tool of total internal reflection microscopy (17, 28).
The working hypothesis was that stabilizing the coiled-coil region to prevent any hypothetical unraveling might destroy kinesin processivity altogether. Interestingly, however, processivity was never completely abolished in any of the mutants scored. In one, a complete turn of the coiled-coil was duplicated (7 aa, starting at roughly the second heptad repeat of the coiled-coil, based on the rat structure). In a second, 3 aa in the second and third heptad repeats of the coiled-coil region were changed to hydrophobic residues in an attempt to make their coil interactions more “ideal.” In a third, roughly the second through fifth heptad repeats were replaced by a coiled-coil consensus sequence in an effort to further stabilize interactions. In all three cases, kinesin processivity, as assayed by single-molecule run length, was comparable to that of wild type. If anything, the duplication mutant had a greater-than-normal processivity, which seems hard to rationalize in this kind of model. Two additional mutants were tested. In one, a Gly-Gly-Gly “swivel” was inserted into the sequence between positions 342 and 343 (human numbering), which was expected to facilitate head motion. This mutation had about half the processivity of wild type. Unfortunately, the swivel may have been introduced at a less-than-perfect position in the human sequence, in light of the rat structure. Ideally, it would be placed in the head– stalk linker (aa ∼335, Fig. 1) and not after the first heptad of the coiled-coil. Finally, a ∼30-aa deletion was made at the start of the coiled-coil, beginning at human residue 341. This mutation affected kinesin processivity the most, reducing it to ∼14% of wild type, but nevertheless failed to destroy it altogether.
It's hard to know exactly what to make of these results, except that kinesin is a hard molecule to keep down! The initial coiled-coil region of kinesin dimers may well still unravel during motion, despite the attempts to stabilize it by mutagenesis. The fact that the mutations were placed mainly in the second heptad repeat or beyond, and not in the first, may compromise the strength of any conclusions one might draw. But equally plausibly, unwinding of the coiled-coil region may not be required for processivity after all, which is precisely the conclusion favored by Romberg et al. (19). This raises the question of what, exactly, does provide the means by which the kinesin heads separate by 8 nm. By a process of elimination, it is intriguing to speculate that the region from the middle of α-helix 6 of the head domain (aa ∼310) to the start of the coiled-coil (aa ∼340) might be the site of the requisite structural changes (Fig. 1, region of interest). Romberg et al. (19) show with computer modeling that swiveling of one of the two kinesin heads, in combination with restructuring of the peptide in this region of interest, might, in principle, allow the kinesin dimer to dock with a microtubule in such a way that both heads are bound (their Fig. 4). As explained above, the hand-over-hand model requires both kinesin heads to bind, at least transiently, during the mechanical cycle.
Could this region of interest be in any way analogous to the so-called “lever arm” of the myosin motor, which is widely believed to be the site of conformational changes that power motion in muscle (3, 10)? Superficially at least, there is no structural resemblance whatsoever, and even if this region were to undergo large-scale conformational changes, it is not immediately apparent how it could produce either the requisite displacement (8 nm) or force (∼6 pN). And that, of course, has us all scratching our heads.
Footnotes
1. Abbreviation used in this paper: aa, amino acid(s).
Support by grants from the NIH and NSF is gratefully acknowledged.
References
- 1.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Berliner E, Young EC, Anderson K, Mahtani HK, Gelles J. Failure of single-headed kinesin to track parallel to microtubule protofilaments. Nature. 1995;373:718–721. doi: 10.1038/373718a0. [DOI] [PubMed] [Google Scholar]
- 3.Block SM. Fifty ways to love your lever: myosin motors. Cell. 1996;87:151–157. doi: 10.1016/s0092-8674(00)81332-x. [DOI] [PubMed] [Google Scholar]
- 4.Block, S.M. 1998. Kinesin molecular mechanics: what gives? Cell. In press.
- 5.Block SM, Goldstein LSB, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 6.Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- 7.Hackney DD. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock WO, Howard J. Processivity of the motor protein kinesin requires two heads. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henningsen U, Schliwa M. Reversal in the direction of movement of a molecular motor. Nature. 1997;389:93–95. doi: 10.1038/38022. [DOI] [PubMed] [Google Scholar]
- 10.Holmes KC. The swinging lever-arm hypothesis of muscle contraction. Curr Biol. 1997;7:R112–R118. doi: 10.1016/s0960-9822(06)00051-0. [DOI] [PubMed] [Google Scholar]
- 11.Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 12.Jiang W, Stock MF, Li X, Hackney DD. Influence of the kinesin neck domain on dimerization and ATPase kinetics . J Biol Chem. 1997;272:7626–7632. doi: 10.1074/jbc.272.12.7626. [DOI] [PubMed] [Google Scholar]
- 13.Kozielski F, Sack S, Marx A, Thormählen M, Schönbrunn E, Biou V, Thompson A, Mandelkow E-M, Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 14.Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leibler S, Huse DA. Porters vs. rowers: a unified stochastic model of motor proteins. J Cell Biol. 1993;121:1357–1368. doi: 10.1083/jcb.121.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma YZ, Taylor EW. Interacting head mechanism of microtubule-stimulated ATPase. J Biol Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 17.Pierce DW, Hom-Booher N, Vale RD. Imaging green fluorescent proteins. Nature. 1997;388:338. doi: 10.1038/41009. [DOI] [PubMed] [Google Scholar]
- 18.Ray S, Meyhöfer E, Milligan RA, Howard J. Kinesin follows the microtubule's protofilament axis. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romberg L, Pierce DW, Vale RD. Role of the kinesin neck region in processive microtubule-based motility. J Cell Biol. 1998;140:1407–1416. doi: 10.1083/jcb.140.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 21.Sack S, Müller J, Marx A, Thormählen M, Mandelkow E-M, Brady ST, Mandelkow E. X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry. 1997;36:16155–16165. doi: 10.1021/bi9722498. [DOI] [PubMed] [Google Scholar]
- 22.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 23.Stryer, L. 1995. Biochemistry. 4th Edition, Chap. 31. W.H. Freeman and Company, New York. 1,064 pp.
- 24.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 25.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 26.Uyeda TQP, Kron SJ, Spudich JA. Myosin step size: estimation from slow sliding movement of actin over low densities of heavy meromyosin. J Mol Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 27.Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;12:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 28.Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T. Direct observation of kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang JT, Laymon RA, Goldstein LSB. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analysis. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- 30.Yang JT, Saxton WM, Stewart RJ, Raff EC, Goldstein LSB. Evidence that the head of kinesin is sufficient for force generation and motility in vitro. Science. 1990;249:42–47. doi: 10.1126/science.2142332. [DOI] [PubMed] [Google Scholar]
- 31.Young, E.C., H.K. Mahtani, and J. Gelles. 1998. One-headed kinesin derivatives move by a non-processive, low duty ratio mechanism unlike that of two-headed kinesin. Biochemistry. In press. [DOI] [PubMed]