Abstract
A single kinesin molecule can move for hundreds of steps along a microtubule without dissociating. One hypothesis to account for this processive movement is that the binding of kinesin's two heads is coordinated so that at least one head is always bound to the microtubule. To test this hypothesis, the motility of a full-length single-headed kinesin heterodimer was examined in the in vitro microtubule gliding assay. As the surface density of single-headed kinesin was lowered, there was a steep fall both in the rate at which microtubules landed and moved over the surface, and in the distance that microtubules moved, indicating that individual single-headed kinesin motors are not processive and that some four to six single-headed kinesin molecules are necessary and sufficient to move a microtubule continuously. At high ATP concentration, individual single-headed kinesin molecules detached from microtubules very slowly (at a rate less than one per second), 100-fold slower than the detachment during two-headed motility. This slow detachment directly supports a coordinated, hand-over-hand model in which the rapid detachment of one head in the dimer is contingent on the binding of the second head.
Conventional kinesin is a motor protein that transports membrane-bound vesicles and organelles along microtubules in neurons and other cells (for review see Bloom and Endow, 1995). Although the biochemical mechanism underlying this directed motility is not fully understood, it is thought that each time the motor hydrolyzes a molecule of ATP it steps through a distance of 8 nm to the next binding site lying closer to the plus end of the microtubule (Ray et al., 1993; Svoboda et al., 1993; Hua et al., 1997; Schnitzer and Block, 1997). An unusual and important property of kinesin is that it is processive: an individual molecule can move continuously for a distance up to several microns along the surface of a microtubule, taking hundreds of steps without dissociating (Howard et al., 1989; Block et al., 1990). Processivity has been confirmed by high resolution single molecule experiments showing that kinesin can move hundreds of nanometers even against loads up to several piconewtons (Svoboda and Block, 1994; Meyhöfer and Howard, 1995), and by biochemical experiments showing that kinesin hydrolyzes ∼100 ATP molecules per encounter with a microtubule (Hackney, 1995). Processive motility appears to be an adaptation to kinesin's function as an organelle transporter: since few motors can fit on the surface of a small organelle or vesicle, it is important that each motor remains bound to the microtubule for as long as possible to ensure that cargo is transported quickly and reliably over long distances.
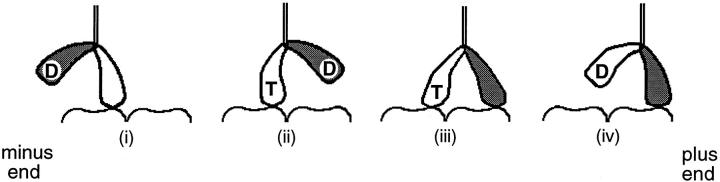
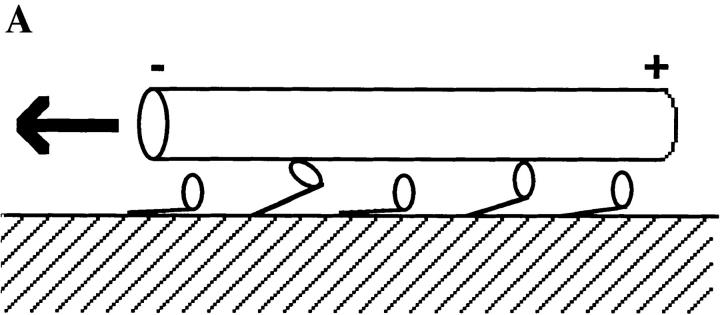
To move processively it is essential that kinesin remains bound to the microtubule throughout the motion, because if the motor detaches it will quickly diffuse away. The structure of kinesin suggests a possible mechanism by which kinesin can maintain attachment with the microtubule even while it steps. Kinesin is a multimeric protein with two identical globular “head” domains, a long coiled-coil rod responsible for dimerization, and a small “tail” domain (Bloom et al., 1988; Hirokawa et al., 1989; Yang et al., 1989). The heads are the motor domains: they contain the microtubule- and nucleotide-binding sites and they suffice for motility (Yang et al., 1990; Stewart et al., 1993; Berliner et al., 1995). The tail is thought to be an association domain that hitches the motor to its appropriate cargo (Coy and Howard, 1994). In view of kinesin's two heads, a simple model to account for processivity is that the motor moves in a hand-over-hand fashion so that the detachment of one head from the microtubule is contingent on the attachment of the other (see Fig. 1; Howard et al., 1989; Schnapp et al., 1990). In this way there is always one head bound to prevent the motor from dissociating. An additional feature of this model is that it provides a ready explanation for how kinesin can reach its next binding site, a distance 8 nm away, with heads that are themselves only ∼8-nm long (Kull et al., 1996); the second head may mechanically amplify a smaller motion of the attached head, thereby extending the motor's reach.
Figure 1.
The hand-over-hand model. It is postulated that the unbinding of the kinesin's first head (unshaded) in step iv occurs only after the binding of the second head (shaded) in step iii. In this way at least one head remains bound to the microtubule at all times. Also shown are the likely nucleotide states of the two heads during the motion (see text). The results presented in this paper indicate that the structure with both heads attached (iii) is highly strained and that this strain accelerates the release of the trailing head.
A hand-over-hand mechanism implies that the two heads move in a coordinated manner. One type of coordination has been observed in biochemical experiments that show the attachment of the second head to the microtubule is contingent upon the binding of ATP to the bound first head (Hackney, 1994; Ma and Taylor, 1997). These findings are incorporated into the model of Fig. 1. However, there is no direct evidence for the essential type of coordination required by the hand-over-hand model, namely that the release of the first head from the microtubule be contingent upon the binding of the second head.
Although the hand-over-hand model is attractive, it is difficult to reconcile with two findings. First, the atomic structure of dimeric kinesin with bound ADP shows that the two heads are rotated by ∼120° with respect to each other, and they cannot simultaneously bind to the polar microtubule unless the dimer is highly strained (Kozielski et al., 1997). This observation has led to the proposal that the motion is purely one headed and that the second head is a passenger or a crutch that plays no active role. Second, there are kinesin-related proteins, KIF1A and KIF1B, which, like conventional kinesin, are thought to be vesicle and organelle transporters, but which are monomeric (Nangaku et al., 1994; Okada et al., 1995). The existence of these motors shows that a two-headed structure is not essential for organelle motility, and raises the question of how monomeric motors could maintain contact with the microtubule while they move.
The hand-over-hand model makes several clear predictions. First, a single-headed molecule, in which one of the heads has been removed, should not be processive. Second, a single single-headed molecule should have a high affinity for the microtubule, even though it does not move, because there is no second head to induce its detachment. And third, we might expect single-headed molecules to move more slowly due to the loss of the mechanical amplification provided by the other head, just as myosin moves more slowly when its regulatory domain, also thought to act as a mechanical lever, is weakened (Lowey et al., 1993) or shortened (Anson et al., 1996; Uyeda et al., 1996).
In the present work, we have tested these predictions by examining the motility of single-headed kinesin in microtubule gliding assays. To facilitate direct comparison to the two-headed wild-type protein, we have used a construct that retains all the rod and tail domains, including the “neck” or dimerization domain comprising amino acids 345–380 adjacent to the motor domain (Huang et al., 1994). Because the rod and tail portions are the same as those in the wild-type protein, we presume that the single-headed protein attaches to the glass surfaces used in the motility assays in the same way that the wild-type protein attaches. A distinct advantage of our construct over its complement, the monomeric motor domain, is that the latter protein on its own shows no detectable motility (Vale et al., 1996). Thus, even though the motor domain is an ATPase (Huang and Hackney, 1994), it may not be a motor, perhaps due to the absence of a neck or even because it is monomeric; any discussion of its processivity is therefore moot. In all cases, the motility of the motor domain has been contingent upon the addition of an artificial tail (Stewart et al., 1993; Berliner et al., 1995; Vale et al., 1996; Inoue et al., 1997), which may substitute for the neck or may induce dimerization in solution or on the surface, especially when surface attachment is mediated by the multivalent protein streptavidin.
Our approach to measuring the number of single-headed molecules required for motility is to determine how the motor activity depends on the surface density of motor molecules in an in vitro motility assay. The rationale of the approach, which has been used widely in biochemistry and biophysics (Hill, 1913; Hecht et al., 1942), is that if only one molecule is necessary, then the activity is expected to decrease gradually as the density is reduced, whereas if a large number of molecules are required, then the activity is expected to decrease abruptly at a density that fails to provide the requisite number of motors. In this way we have determined the minimum number of single-headed molecules required for continuous motility.
Materials and Methods
Construction of Coexpression Plasmid
Single-headed kinesin heterodimers were made by coexpressing full-length Drosophila kinesin heavy chain (Yang et al., 1989) with a decapitated kinesin heavy chain consisting of the rod and tail domains but lacking the 340-residue head domain (see Fig. 2). To increase expression levels of wild-type kinesin, the 5′ end of the gene (pET-kin [Yang et al., 1990]), generously provided by L.S.B. Goldstein (University of California, San Diego, CA) was altered to restore the correct NH2-terminal amino acid sequence and reduce possible secondary structure of the message. The DNA sequence and corresponding amino acids were changed from:
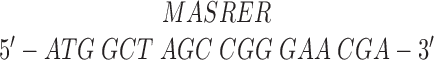 |
to:
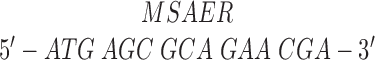 |
The decapitated kinesin gene was created using PCR-based mutagenesis. The sequence coding the NH2-terminal 340 residues was deleted and then a COOH-terminal sequence coding hexa-histidine (his)1 was added. The atomic structure of the dimeric rat kinesin shows that the coiled-coil neck domain of Drosophila kinesin begins at Ala345 (corresponding to Ala339 in the rat; Kozielski et al., 1997), consistent with secondary structure predictions (Howard, 1996). Thus, our single-headed heterodimer contains the entire neck domain. Physical measurements show that the 340-amino acid head domain does not dimerize in solution (Huang et al., 1994; Correia et al., 1995; Young et al., 1995); because the decapitated heavy chain contains all the dimerization regions, it is unlikely that our heterodimers form higher order oligomers via uncomplemented dimerization sequences. The forward primer, consisting of a GC clamp, NheI restriction site, and sequence corresponding to residues 341–347 of the kinesin sequence was 5′-GCGCGCGCTAGCGAGGAGCTTACTGCCGAGGAA-3′. The reverse primer, consisting of the reverse compliment of the sequence coding for residues 969–975 of the kinesin sequence, six histidine residues, two stop codons, as well as a BamHI site and a GC clamp was 5′-GCCGCGGATCCTCATCAGTGGTGGTGGTGGTGGTGCGAGTTGACAGGATTAACCTG-3′.
After PCR mutagenesis, the decapitated kinesin gene was digested with NheI and BamHI (molecular biology enzymes were purchased from New England Biolabs, Inc., Beverly MA; Life Technologies, Inc., Gaithersburg, MD; or Boehringer Mannheim Biochemicals, Indianapolis, IN; all other chemicals were purchased from Sigma Chemical Co., St. Louis, MO, unless otherwise noted), and then ligated into a pET5 plasmid (Novagen, Inc., Madison, WI). The sequences of the 5′-234 bases and the 3′-115 bases were confirmed using a sequencer (model ABI 373A; Perkin-Elmer Corp., Foster City, CA) and the intervening sequence between the SfiI and RsrII restriction sites was replaced by the corresponding sequence from the full-length kinesin gene to avoid any errors introduced by the Taq polymerase used in the PCR reaction.
To generate a coexpression plasmid (see Fig. 2 A), the decapitated kinesin gene, along with its T7 promoter and ribosome binding site, was cut out of the pET5 plasmid using SphI and EcoRI. The pET-kin plasmid was then linearized using SphI and then the decapitated gene ligated into the vector using a SphI to EcoRI adapter. This ligation resulted in a coexpression plasmid with the decapitated kinesin gene upstream of the full-length kinesin gene, with each gene having its own T7 promoter and ribosome binding site. Neither gene contained a transcriptional stop sequence, so the message was probably polycistronic.
Figure 2.
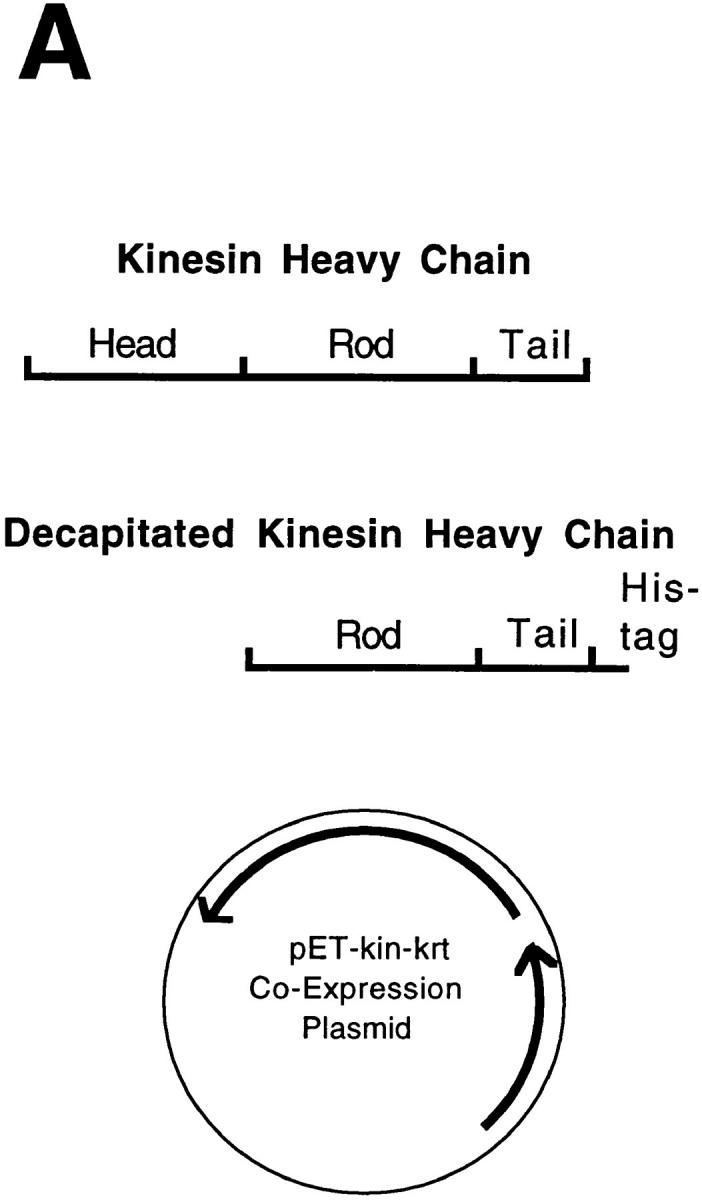
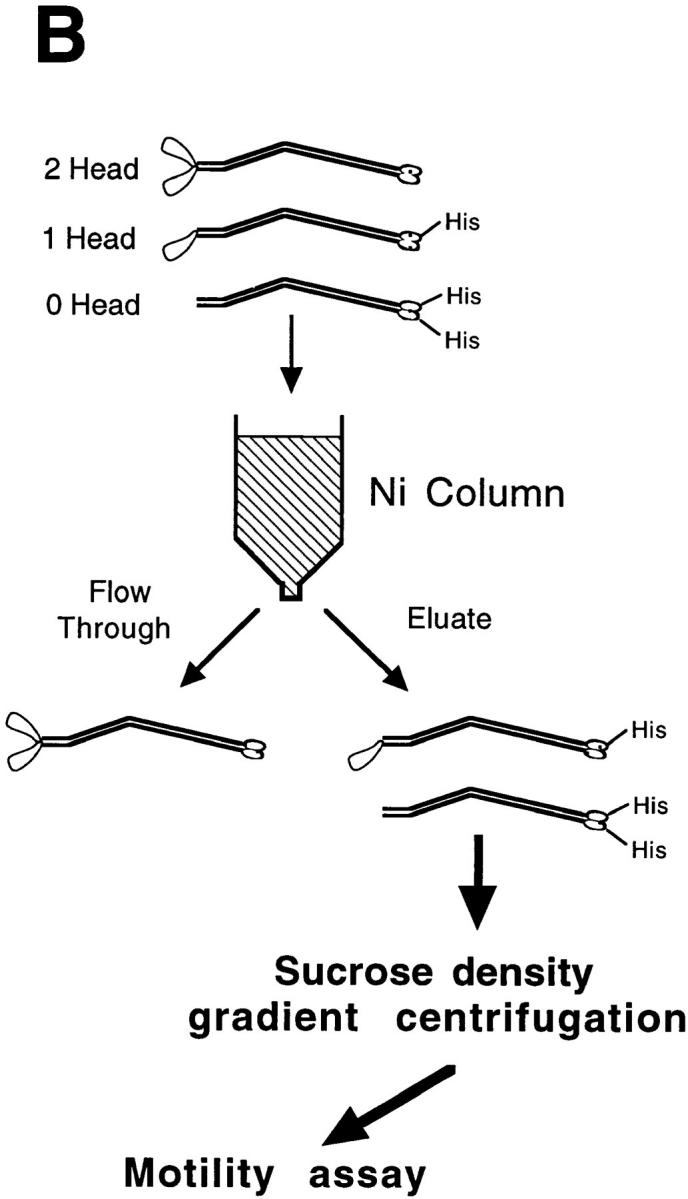
Protocol for generating single-headed kinesin heterodimers. (A) Coexpression vector. (B) Protein expression and purification strategy in which the single-headed heterodimers were separated from the two-headed homodimers by chromatography and centrifugation.
Protein Expression and Purification
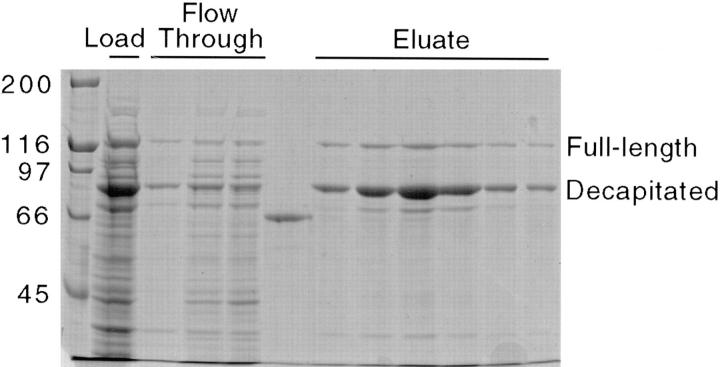
The coexpression plasmid was transfected into Escherichia coli strain BL21(DE3) (Novagen, Inc.), a bacterial stock was grown in Luria-Bertani medium supplemented with 100 μg/ml ampicillin, and then 2-ml aliquots were frozen and stored at −80°C. 1- to 2-liter vol of Luria-Bertani medium were inoculated with a stock aliquot, grown at 37°C in the presence of 100 μg/ml ampicillin to an OD of 0.5 per cm at 600 nm and then protein expression was induced by adding 0.4 mM isopropyl B-p-thiogalactopyranoside and incubating for 3 h at 20°C. Bacteria were pelleted by centrifuging for 6 min at 6,000 g, and were then resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 40 mM imidazole, 5 mM β-mercaptoethanol, 10% glycerol, pH 8.0), frozen in liquid nitrogen, and then stored at −80°C. After thawing, bacteria were lysed by twice passing through a French press (model FAO73; Spectronics Aminco, Inc., Rochester, NY) at 19,000 psi. in the presence of 1 mM PMSF. The bacterial lysate was then centrifuged for 30 min at 100,000 g to remove cell debris and insoluble protein. As seen in Fig. 3, lane 2, the expression level of the decapitated kinesin peptide was approximately fivefold higher than the full-length kinesin heavy chain; ideally this difference will maximize the formation of single-headed heterodimer over two-headed homodimer.
Figure 3.
SDS-PAGE of bacterial supernatant and Ni column fractions. Lane 1, molecular weight markers; lane 2, bacterial lysate supernatant loaded onto the Ni column (diluted 1:4); lanes 3–5, column flow-through fractions (diluted 1:4), lane 6: 50 μg/ml BSA standard; lanes 7–12, elution fractions.
His-tagged kinesin was purified on a 40-ml Ni-nitrilotriacetic acid (NTA) agarose column (QIAGEN, Inc., Santa Clarita, CA). Column buffers contained the following: 50 mM sodium phosphate, 300 mM NaCl, 1 mM MgCl2, 100 μM MgATP, 5 mM β-mercaptoethanol (added just before using), and either 60 mM (wash buffer) or 500 mM (elution buffer) imidazole, pH 7.0. To avoid loss of activity and prevent aggregation, kinesin was kept in 100 μM MgATP at all times.
The column was preincubated with wash buffer and then the bacterial supernatant was loaded onto the column, followed by five column vol of wash buffer to remove contaminating bacterial proteins. The protein absorbance at 280 nm was monitored to confirm the wash was complete. Single-headed kinesin was eluted from the column by a step elution. Gradient elutions were also found to work, with the his-tagged kinesin eluting at ∼150 mM imidazole. A gel of the elution fractions is seen in Fig. 3.
Sucrose Density–Gradient Centrifugation
Motility experiments on the Ni column elution fractions showed that despite the lack of a his tag on the two-headed kinesin species, there was a small contamination of two-headed kinesin in the elution fractions. To separate single-headed kinesin from the contaminating two-headed motors, the three different kinesin species (zero-head, one-head, and two-head with predicted molecular weights of 148, 185, and 222 kD, respectively) were separated by sedimentation rate using sucrose density–gradient centrifugation.
In preparation for the centrifugation step, the Ni column elution peak was concentrated by filling a 30–40 cm length of dialysis tubing (50,000 kD cutoff, 7.5 mm diameter; model Spectra/Por 6, Spectrum, Houston, TX) with 10–15 ml of column elution, and then covering with dry carboxymethyl cellulose. A 5–10 fold concentration was obtained after two or three h at 4°C with new dry powder added every 30–60 min. After concentration, the protein sample was spun for 5 min in an airfuge (model E4; Beckman Instrs., Palo Alto, CA) to remove any insoluble components. Using a 1.1-ml Sephadex G25 M column (Pharmacia Diagnostics AB, Uppsala, Sweden), the supernatant was exchanged into 25A25 buffer (Huang and Hackney, 1994) (25 mM N-2[acetamido]-2-amino-ethanesulphonic acid [ACES], 25 mM KCl, 2 mM MgOAc, 2 mM EGTA, 0.1 mM EDTA, 1 mM β-mercaptoethanol, pH 6.9) and then supplemented with 100 μM MgATP and 1 mM MgCl2.
Sucrose was dissolved in 25A25 buffer supplemented with 100 μM MgATP, 1 mM MgCl2 and 1 mM dithiothreitol. After degassing the solutions and cooling to 4°C, a 5–20% (wt/wt) sucrose density gradient was formed in a 12-ml Ultra-Clear tube (Beckman, Instrs.). To calculate sedimentation rates, proteins with known sedimentation values (carbonic anhydrase, 3.2; BSA, 4.4; alcohol dehydrogenase, 7.6; β-amylase, 8.9 S, respectively) were run either as internal standards or in a parallel tube. After the gradient was formed, 150–200 μl of the concentrated protein sample was carefully layered onto the gradient and then the sample was spun in an ultracentrifuge (model L8-80M; Beckman Instrs.) for 20–30 h at 41,000 rpm in a swinging bucket rotor (model SW-41, Beckman Instrs., Fullerton, CA).
The gradient was drained by positive displacement using 50% (wt/wt) sucrose solution. This method achieved significantly better results than draining the gradient by gravity through the bottom of the tube. An adapter, made from the tip of a 5-ml syringe, was fit to the top of the tube, connected to a fraction collector, and then ∼40 fractions of vol. of 300-μl each were collected and analyzed by SDS-PAGE and motility assay. To quantitate the concentration of full-length and decapitated kinesin peptide in each fraction, the Coomassie blue–stained gels were scanned using a 0.2-OD filter to reach the linear range of the scanner (model PS-2400X; UMAX Data Systems, Inc., Hsinchu, Taiwan), and then the band intensity was digitally integrated using National Institutes of Health (NIH) Image version 1.57 (Bethesda, MD) and compared to a BSA standard on the same gel.
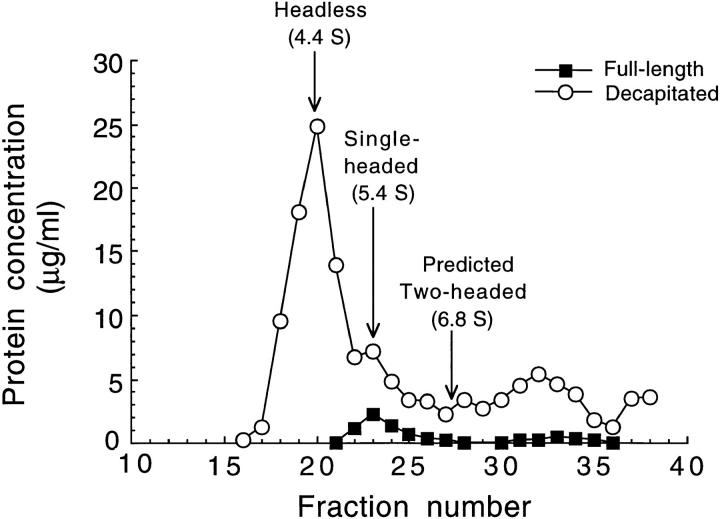
The separation of the various kinesin species as determined by SDS-PAGE is seen in Fig. 4. A prominent peak of decapitated kinesin heavy chain is seen followed by a smaller peak that coincides with a peak of full-length kinesin heavy chain. These two peaks are interpreted as the headless homodimer and single-headed heterodimer, respectively. By comparing the sedimentation rate to the protein standards, the values of 4.4 for headless kinesin, 5.4 for single-headed kinesin, and 6.8 S for two-headed kinesin, respectively, were calculated. Results from motility assays were consistent with the gel results: in the peak headless fraction (see Fig. 4, fraction 20) no microtubules stuck to the surface; in the peak single-headed fraction (see Fig. 4, fraction 23) many microtubules landed on the surface and moved at slow velocities; and in the expected peak two-headed fraction (see Fig. 4, fractions 26 and above) there was considerable movement at speeds similar to the wild-type speed.
Figure 4.
Separation of kinesin species by sucrose density–gradient centrifugation. The concentrations of full-length kinesin heavy chain and decapitated kinesin heavy chain peptides in each fraction were estimated from scans of Coomassie blue–stained SDS-PAGE gels. Inferred positions of the peak headless and single-headed fractions are highlighted. There was no evidence for a two-headed kinesin peak; the expected position of the two-headed peak is based on separate centrifugation experiments with pure wild-type kinesin. Fractions 22–24 were used as single-headed kinesin samples for motility assays.
The single-headed fractions (see Fig. 4, fractions 22–24) were divided into 50-μl aliquots, augmented with 0.5 mg/ml BSA and 0.5 mg/ml casein to prevent protein adsorption to the sides of the tubes (in some experiments this was done immediately after thawing on the day of the motility experiment), frozen in liquid nitrogen, and then stored at −80°C.
Wild-type (two-headed) kinesin was expressed and purified in an identical manner as single-headed kinesin. Briefly, a full-length Drosophila kinesin construct containing a COOH-terminal his tag was bacterially expressed, purified on an identical Ni column, further isolated by sucrose density–gradient centrifugation, and then quantified by gel scanning.
In Vitro Motility Assays
Motility assays were performed as previously described (Howard et al., 1993) using BRB80 buffer (80 mM Pipes, 1 mM EGTA, 1 mM MgCl2, pH 6.9) for all solutions. Briefly, the glass surfaces of the flow cell were blocked by 0.5 mg/ml casein solution for 3 min, and then the motor-containing solution (that included 100 μg/ml MgATP and 0.2 mg/ml casein) was flowed into the cell and the motors were allowed to adhere to the glass for 5 min. After this, motility solution (rhodamine-labeled bovine brain microtubules at 32 nM tubulin dimer, 1 mM ATP, 10 μM Taxol™ (Bristol-Meyers Squibb, New York, NY), 0.2 mg/ml casein, 20 mM d-glucose, 0.02 mg/ml glucose oxidase, 0.008 mg/ml catalase, and 0.5% β-mercaptoethanol) was added. Microtubules in the motility solution were sheared by twice passing through a 30-gauge needle at a flow rate of 100 μl/sec, resulting in a mean length of 2.05 ± 0.92 μm (mean ± SD, n = 75).
Motor Densities and Their Uncertainties
The surface density of motors was calculated from the motor concentration and the geometry of the flow cell. Control experiments demonstrated that >90% of the motors adsorbed to the glass surface during the 5-min incubation. Flow cells were 100-μm deep (made with number zero coverslips as spacers), and then the motors were assumed to distribute evenly between the top and bottom glass surfaces. Densities are expressed as the number of motor molecules per squared micrometer of surface area, where one wild-type molecule has two heads and one single-headed molecule has one head. To vary the motor density, motor stock solution was diluted into BRB80 supplemented with 100 μM MgATP and 0.2 mg/ml casein immediately before adding to the flow cell.
Uncertainty in the motor density was due to several factors, the largest of which was uncertainty in protein concentration as determined by gel scanning. For two-headed kinesin, the relative SEM was 25%, whereas for single-headed kinesin it was 30%. In addition to these random errors, there was a systematic error due to inactive protein. When the concentration of wild-type kinesin adsorbed to glass surfaces was measured from the number of nucleotide binding sites (Coy, D., personal communication), the concentration was only ∼50% of that determined by comparison to BSA standards as described above. Since this latter measure has been found to be a good estimate of the protein concentration of native kinesin (Hackney, 1988), it is possible that only about half of our recombinant, wild-type protein is active. If the single-headed protein has a higher proportion of active protein, then the true density of single-headed kinesin might be twice that of wild-type kinesin. These uncertainties are taken into account when the motor activities of the two proteins are compared in the Results.
Video Analysis
Motility experiments were recorded on standard VHS videotape for later analysis. Displacement and velocity data were collected by recording the digitized position of the front end of moving microtubules using MEASURE hardware (M. Walsh Electronics, San Dimas, CA) and software generously provided by S. Block (Princeton University, Princeton, NJ). Microtubule lengths and distances moved were measured directly from the video monitor by tracing the microtubule position on a transparent plastic sheet placed over the video screen. The threshold of detectable movement from the video was 0.3 μm.
Theory
To quantitate the minimum number of single-headed motors required for motility, we considered models of microtubules landing on and moving across kinesin-coated surfaces. Suppose that the motors are adsorbed at random locations on a planar surface and that for a microtubule to successfully land on the surface and move, at least n motors must be engaged. We assume that the landing is a sequential process with the microtubule first binding to one motor on the surface, then to another, and so on until n motors are engaged and the microtubule is able to move continuously. The landing rate, R n, is expected to depend on the density of motors, ρ, according to
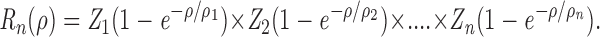 |
1 |
The individual bracketed terms reflect the probability that there is at least one motor within each area 1/ρi that characterizes the ith step in the landing process, and the Z i's are constants that do not depend on density. The product is formed because each of the n steps must be successful for the landing to be successful. For example, the first step corresponds to the collision with the surface, 1-exp(-ρ/ρ1) is the probability that there is at least one motor in the area 1/ρ1 (expected to be of the order of the length of the microtubule times twice the “reach” of the motor), and then Z 1 depends on the collision rate and the fraction of collisions that are successful. In simplifying the model, it can be shown that when all the ρi's are equal, the landing rate depends most steeply on the density (Howard, J., manuscript in preparation). Hence, a lower limit for n will be obtained when the landing rate data are fit with the equation
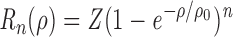 |
2 |
where Z is a parameter that incorporates collision of microtubules with the surface and ρ0 corresponds to the surface area over which motors interact with a microtubule. Eq. 2 yields a lower bound for n that is independent of the precise details of the landing process.
Suppose that to continue to move, there must always be at least n motors interacting with the microtubule. Because of the random placement of motors on the surface, it is possible that the microtubule, though initially moving, will reach a place on the coverglass that has too few motors to support motility and will stop. The probability that it will move a distance greater than or equal to its own length before it reaches such a place on the surface and stops is
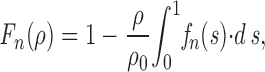 |
3 |
where
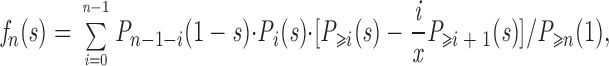 |
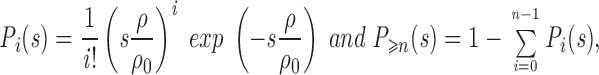 |
(Howard, J., manuscript in preparation).
The models of Eqs. 2 and 3 were fit to the landing rate and distance moved curves, respectively, using the Levenberg–Marquardt algorithm (Igor; Wavemetrics, Lake Oswego, OR). The data points were weighted by the SEM. For the landing rate curves, the standard errors were calculated as the sum of the errors associated with the counting statistics (SEM1 = mean/√ n, n = number of microtubules observed), together with an estimate of the variability from flow cell to flow cell, attributed to other factors, and derived from the landing rates at the highest densities (SEM2 = mean × 0.18 for single-headed and SEM2 = mean × 0.16 for wild-type). The sum was performed by adding the variances. For the distance-moved curves, the error bars were calculated from counting statistics alone: SEM = [f(1−f)/n]1/2 where f (≠ 0 or 1) is the measured fraction of microtubules that moved more than their own length and n is the number of observations; SEM = 1/n if f = 0 or 1.
Results
To investigate the coordination between the two heads of the native kinesin dimer, the motor activities of wild-type and single-headed kinesin were compared at a series of motor surface densities in the in vitro microtubule gliding assay.
Speed and Character of Microtubule Movement
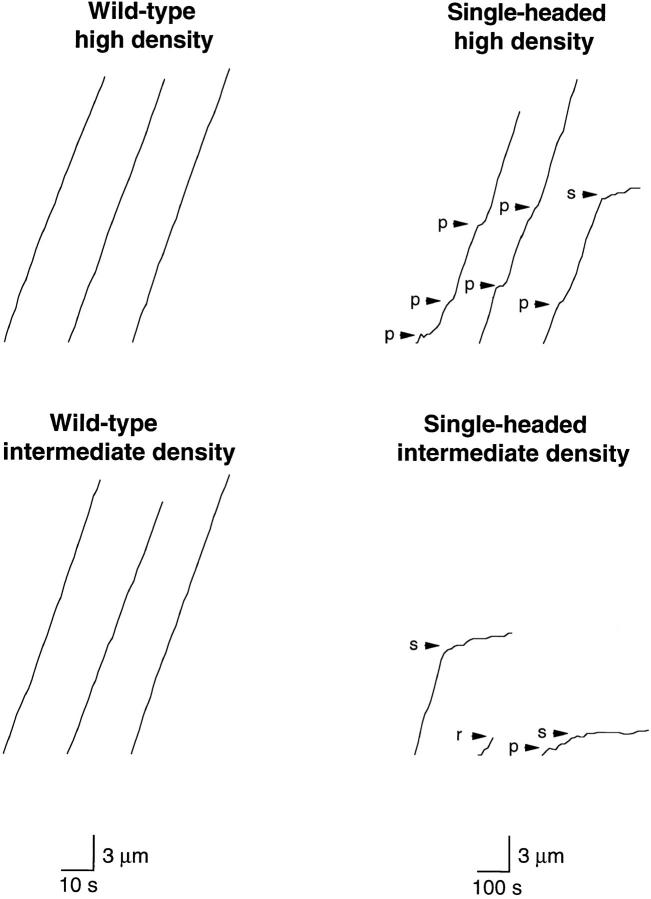
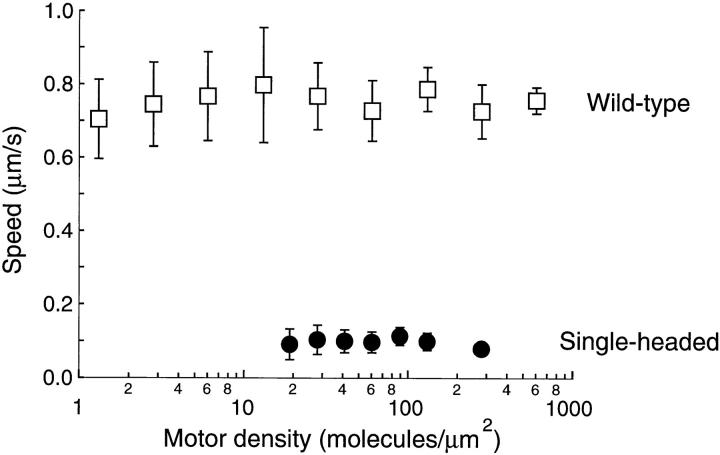
When adsorbed to glass surfaces at medium or high density, single-headed kinesin induced microtubule gliding, though the speed was lower and the motion less smooth than that induced by wild-type kinesin (Fig. 5). During periods of smooth movement, single-headed kinesin moved microtubules at 0.096 ± 0.030 μm/s (mean ± SD, n = 86), whereas wild-type kinesin moved at 0.76 ± 0.10 μm/s (mean ± SD, n = 92) (Fig. 6). Similar to two-headed kinesin, the gliding speed was independent of motor density over the ranges measured.
Figure 5.
Displacement versus time traces for microtubules moving over surfaces coated with wild-type kinesin (left panels) and single-headed kinesin (right panels) at high motor density (280 motors/μm2; top panels) and intermediate motor density (60 motors/μm2, bottom panels). The tracks of 12 microtubules are shown. The position of the leading end of each microtubule was measured every second for wild-type kinesin or every 10 s for single-headed kinesin. Note that microtubules often paused (p) or stalled (s) when moving over surfaces coated with single-headed kinesin. The short trace in the bottom right panel corresponds to a microtubule that released from the surface and diffused away (r). Note that the scale bars correspond to 10 s for wild-type kinesin and 100 s for single-headed kinesin, so the similar slopes of the traces indicates that wild-type kinesin moves about ten times faster than single-headed kinesin.
Figure 6.
Microtubule gliding speeds for wild-type kinesin (open squares) and single-headed kinesin (closed circles). Each point is the average of 7–18 observations. Error bars are standard deviations. For single-headed kinesin, speeds were determined during windows of smooth movement.
Single-headed kinesin, like wild-type kinesin, is a plus end-directed motor. When gliding assays were performed with polarity-marked microtubules (Howard and Hyman, 1993), the marked end almost always led: for wild-type kinesin 45 out of 47 microtubules moved with the marked end leading, and for single-headed kinesin the fraction was 28 out of 30. Because the bright marker is usually located at the minus end, these results indicate that both motors move toward the plus end of the microtubule.
The uneven motion shown in Fig. 5, with frequent pausing and stalling, was an inherent property of single-headed kinesin. It was observed in all four preparations and was not due to a subpopulation of motors that bound to microtubules but could not move: when such hypothetical “dead heads” were removed by incubating the kinesin with microtubules (0.1 μM tubulin dimer) in the presence of 1 mM ATP and 300 mM KCl followed by an airfuge spin to remove any irreversibly microtubule-bound motors, the quality of the movement was unaffected.
Single-headed Motility Is Not Due to Wild-Type Contamination
It is important to establish that the observed microtubule movement is generated solely by single-headed kinesin heterodimer and not by some other kinesin species. Artifactual movement could arise from at least three possible sources: (a) wild-type motors that were not eliminated during the purification process; (b) wild-type motors formed by recombination of single-headed heterodimers in solution; or (c) the formation of two-headed, four-tailed kinesin tetramers in solution or on the surface.
The first experiment used to rule out these possibilities was the physical characterization by sucrose density–gradient centrifugation (Fig. 4). Simple wild-type contamination was ruled out because the single-headed kinesin peak in the sedimentation assay was clearly separated from the sedimentation rate of two-headed kinesin; had the two-headed contamination been large enough to obtain the high landing rates seen in Fig. 7, it would have been detectable in the sedimentation data in Fig. 4. The second possibility, contamination by recombination, is also not supported by the sedimentation data: if the zero-, one-, and two-headed species exist in a dynamic equilibrium through the 30-h centrifugation, one would expect a broad sedimentation peak encompassing the three predicted sedimentation values instead of the discrete peaks observed in Fig. 4. Finally, two-headed, four-tailed tetramers formed from two single-headed dimers are expected to have a sedimentation coefficient even larger than the wild-type dimer; hence, they would have been observable if present and, in any case, would have been well separated from the heterodimers in the centrifugation step. The possibility that motility requires the formation of tetramers on the surface is ruled out because either their formation would be too slow (Howard et al., 1989) or their density would be very low, in which case the landing rate and distance curves of Figs. 7 and 8 would have been shifted much farther to the right.
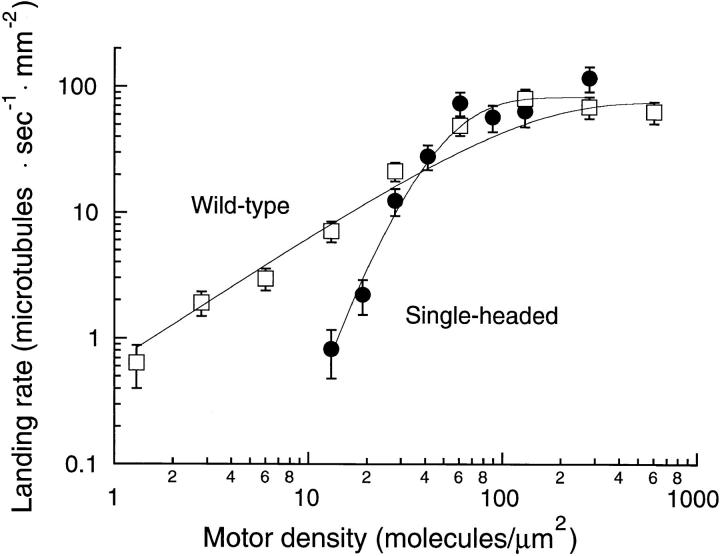
Figure 7.
Landing rate profile for wild-type (open squares) and single-headed kinesin (closed circles). The landing rate was measured by counting the number of microtubules that landed and moved ⩾0.3 μm in a 4000-μm2 video screen. Each data point is derived from two to six video screens taken from each of one or two flow cells. Standard error bars were calculated according to Materials and Methods. The continuous curves are the fits to the model of Equation 2 with n = 1 (wild-type) and n = 5 (single-headed).
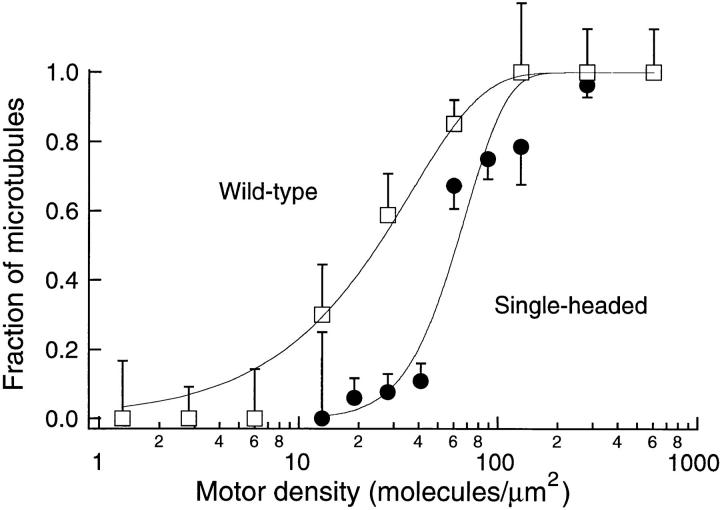
Figure 8.
The fraction of microtubules that moved a distance greater than their length as a function of motor density. Microtubules of length between two and three micrometers were observed for 30 s for wild-type kinesin (open squares) or for 300 s for single-headed kinesin (closed symbols). Standard error bars were calculated according to Materials and Methods. The continuous curves are the fits to the model of Eq. 3 with n = 1 (wild-type) and n = 4 (single-headed).
The second experiment used to rule out artifactual motility relied on our ability to detect small contaminations of wild-type kinesin in the single-headed fraction. This ability was demonstrated by adding known amounts of wild-type motors to the single-headed fraction; in these mixtures (or in the early stages of the project when separation of single- and two-headed kinesin was still a problem), high-speed, single-motor events were observed at low motor density. Because none of these events were seen in any of the preparations of pure single-headed kinesin, we conclude that the motility is generated solely by single-headed heterodimers and not by wild-type motors present either from simple contamination or by recombination.
Rates at Which Microtubules Attach to and Move across Surfaces Coated with Wild-Type and Single-headed Motors
To determine the minimum number of single-headed motors required to move a microtubule, the rates at which microtubules bound to and moved at least 0.3 μm across surfaces coated with wild-type and single-headed kinesin were measured over a range of motor densities (Fig. 7).
For surface densities greater than 100 motors/μm2, the landing rate for both single-headed and wild-type kinesin was constant at ∼75 microtubules · s−1 · mm−2. This maximum rate is presumably limited by the rate at which microtubules diffuse from solution to the motor-coated surface.
As the surface density of wild-type motors was decreased, the landing rate also decreased. When the landing rate was plotted against the density on a log–log plot (Fig. 7), the slope was approximately equal to one, consistent with just one wild-type molecule being sufficient for motility. Below a surface density of 10 motors/μm2, virtually every moving microtubule swiveled around a single point on the surface, moved until its trailing end reached this nodal point, and then released and diffused back into solution. This swiveling behavior is also consistent with single-motor motility. These results are very similar to those obtained using bovine brain kinesin (Howard et al., 1989), except that the recombinant Drosophila kinesin used in this study had a 10-fold higher motor activity, meaning that similar landing rates were obtained when the recombinant kinesin was at only one-tenth the surface density of the bovine kinesin. Presumably a large fraction of bovine brain kinesin is in an inactive form perhaps due to posttranslational modification.
As the surface density of single-headed kinesin was decreased, the landing rate decreased steeply. Below 100 motors/μm2, the slope was approximately equal to three in the log–log plot, indicating that more than one single-headed molecule is required for movement. Below a motor density of 10 motors/μm2, where individual wild-type motors were seen moving microtubules, no moving microtubules were detected. To determine the minimum number of single-headed motors necessary to move a microtubule, the landing rate data were fit to a model in which the motors are assumed to be bound randomly on the surface and the microtubules assumed to bind and move only if they encounter at least n motors (refer to Materials and Methods). Whereas the wild-type landing rate data were best fit by the model with n = 1, the single-headed data were well fit with n ⩾4. The chi-squared values for n = 2, 3, 4, 5, and 6 were 28.5, 15.8, 10.7, 8.2, 7.3, respectively, and the corresponding P values with six degrees of freedom were <0.001, 0.015, 0.1, 0.2, and >0.2, respectively. Hence, the shape of the landing rate curve indicates that the minimum number of single-headed motor molecules required for motility is at least four. This conclusion remains valid even if the detailed mechanism describing the landing and movement is more complex than assumed in the model. For example, the association of microtubules with surfaces sparsely coated with single-headed kinesin might be cooperative: the microtubule might first bind to one motor, then it might swivel around until a second motor is engaged. However, such cooperativity leads to shallower curves and an underestimation of the minimum number of molecules required (refer to Materials and Methods). Thus, the lower bound is not affected.
The slope of the landing rate curve does not provide a reliable upper bound because several experimental factors, such as the dispersion of microtubule length, might make the curve shallower. However, an upper bound on the minimum number of single-headed motors required for motility can be obtained from the relative positions of the two landing rate curves along the density axis. If a large number of single-headed motors were required for motility, then the single-headed landing rate curve would be notably rightward shifted with respect to the two-headed curve. However, the data show very little shift, indicating that the minimum number is modest. To calculate an upper bound, we estimated the largest possible shift between the curves that is consistent with the errors associated with measuring the densities of the single-headed and wild-type molecules. These errors, expressed as relative SEMs, include uncertainties in measuring the protein concentration (30% for single-headed and 25% for wild-type kinesin), and uncertainties in measuring the positions of the curves (6% for the single-headed and 18% for the wild-type curve). Furthermore, the fraction of active wild-type kinesin on the glass surfaces was estimated to be ∼50% (refer to Materials and Methods), leading to an additional potential rightward shift of the single-headed curve by a factor of two. Combining these uncertainties, values for an upper limit, with their corresponding P values, are n ⩽ 4 (P > 0.05), n ⩽ 5 (P > 0.01), or n ⩽ 6 (P > 0.005). However, the estimate of the upper bound, unlike the estimate of the lower bound, does depend on the detailed mechanism of the landing process, so these numbers might need to be increased slightly. For example, if the swiveling of a microtubule on one single-headed molecule increases the possibility of finding a second molecule, this would increase the upper bound by one.
Combining our upper and lower limits, we deduce that four to six single-headed motors are necessary and sufficient for continuous motility.
Distances Moved by Wild-Type and Single-headed Motors
A second, independent estimate of the number of single-headed kinesin motor molecules required to move a microtubule was obtained from measurements of the distances moved by microtubules over surfaces coated with single-headed or wild-type kinesin at various densities. Microtubules two to three micrometers in length were followed for 30 s over wild-type kinesin or for 300 s over single-headed kinesin, and then the proportion of microtubules that moved a distance greater than their length was plotted as a function of motor density (Fig. 8).
For wild-type kinesin, every microtubule moved a distance greater than its length when the motor density was high. The fraction of microtubules that moved more than their own length gradually decreased as the motor density decreased, until at the lowest densities (< 10 motors/μm2), no microtubules moved more than their length (Fig. 8, open squares). At these low densities microtubules land on single motors, move until the motor reaches the end of the microtubule, and then diffuse away.
For single-headed kinesin, the curve was both steeper and shifted to the right of the wild-type curve (Fig. 8, closed circles). At high densities, almost all the microtubules moved greater than their own length but as the density decreased, the fraction that moved more than their own length fell abruptly. The abrupt drop implies that multiple single-headed motor molecules are required to move a microtubule. Microtubule movement at lower densities of single-headed motors failed both because the microtubules stopped moving (stalled) and because they released from the surface and diffused away as in the wild-type assay. To estimate the minimum number of single-headed motors required for motility, the data were fit to a model in which the motors are assumed to be randomly located on the surface but the microtubule can only move if more than a minimum number, n, of motors can interact with the microtubule (refer to Materials and Methods). The best fit to the wild-type data was with n = 1, consistent with wild-type kinesin being processive. In contrast, the best fit for the single-headed data was with n = 4. The chi-squared values for n = 1, 2, 3, and 4 were 48.6, 26.0, 19.6, and 17.9, respectively, and the corresponding P values with seven degrees of freedom were <0.001, <0.001, 0.007, and 0.012, respectively. These P values are small for all values of n, possibly indicating that the uncertainties were underestimated. However, it is clear that the fit for n = 2 is significantly worse than the fits for n ⩾ 3, and so we conclude that at least three single-headed molecules are required for continuous motility.
As in the case of the landing rate curves, an upper bound on the minimum number of single-headed motors was estimated from the relative positions of wild-type and single-headed curves along the log (density) axis. We calculate that n ⩽ 10 with 95% confidence.
Individual Single-headed Motors Bind to but Do Not Move Microtubules
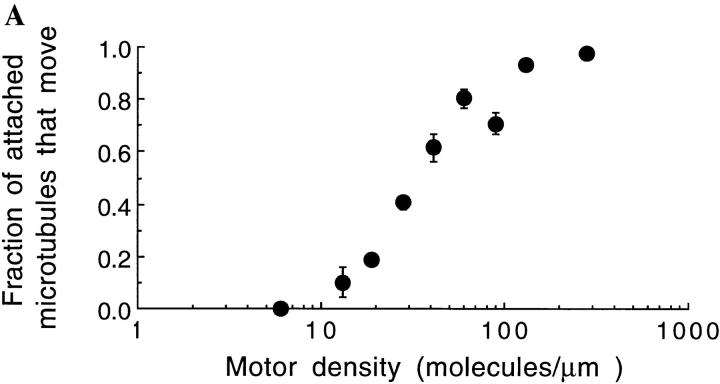
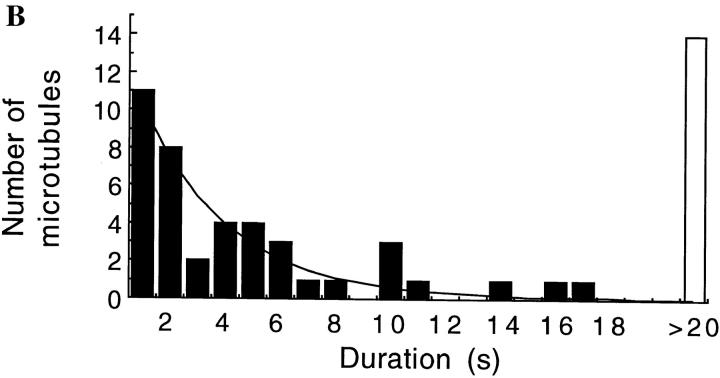
The abrupt decrease in the motor activity of single-headed kinesin is not due to its failure to bind to microtubules at low densities. On the contrary, even at 1 mM ATP concentration, microtubules did bind to surfaces sparsely coated with single-headed kinesin (though they failed to move). The fraction of microtubules that moved more than 0.3 μm decreased from nearly one at high motor density, to zero as the surface density of motors was decreased (Fig. 9 A). At the lowest motor densities where no movement was detected, microtubules swiveled around single points indicating that individual single-headed kinesin can bind microtubules but not move them. At higher densities, moving microtubules never swiveled, although sometimes they paused, transiently released from the surface, and then continued in a different direction. A histogram of bound times for microtubules bound to individual single-headed motors was fit by a single exponential with a time constant of 3.1 s (Fig. 9 B). Hence, individual single-headed motors can bind to microtubules for many seconds, but multiple single-headed motors are required to move a microtubule.
Figure 9.
The fraction of attached microtubules that moved ⩾0.3 μm as a function of motor density. Only microtubules of length 1 μm or longer that landed on the surface for 2 s were counted. The bars represent standard errors of the mean. (B): The duration of binding events for microtubules that landed on a surface coated with a low density of single-headed kinesin and swiveled, indicating single motor interactions. Although every event of duration 2 s or longer was counted, the count for 1-s events is likely to be an underestimate due to missed events. A fit of the data to a single exponential curve with a time constant of 3.1 s is shown. The ATP concentration was 1 mM.
Discussion
By analyzing the movement of microtubules over surfaces coated with single-headed kinesin at various densities, we conclude that single-headed kinesin is not a processive motor. Instead, some four to six single-headed molecules are necessary and sufficient for continuous motility, and when an insufficient number of motors are present, single-headed kinesin binds to microtubules and releases only very slowly.
The motility of single-headed kinesin molecules implies that there is a directed conformational change induced by ATP hydrolysis within the head domain itself, and that directed movement does not require a two-headed structure. This result confirms previous experiments using one-headed, tailless kinesin fragments (Stewart et al., 1993; Berliner et al., 1995; Vale et al., 1996), although, as mentioned in the Introduction there was the possibility that these other single-headed proteins could have oligomerized in solution or on the surface.
Single-headed Kinesin Is Not Processive
Several lines of evidence show that single-headed kinesin is not processive: (a) There is no movement at low density; (b) When movement does occur at intermediate densities, the microtubules do not swivel: since kinesin molecules have high torsional flexibility (Hunt and Howard, 1993), the failure to swivel indicates that the moving microtubules must be attached to motors located at two or more points on the surface; (c) The landing rate falls abruptly as the density is decreased; and (d) The probability of a microtubule moving more than its length also decreases abruptly as the density is decreased. The last two arguments are especially strong because they do not rely on knowledge of the absolute concentration of active protein, since the presence of inactive protein does not change the slope of the dilution curves when they are plotted against the logarithm of density. This was a major rationale for our approach.
Taken together, the landing rate and distance data indicate that a minimum of four to six single-headed motors are necessary for motility. Importantly, these data clearly rule out the possibility that two single-headed heterodimers suffice for motility. Therefore, uncoupling the two heads of kinesin, which are normally tightly associated through the coiled-coil dimerization domain, destroys the coordination necessary for processive motility.
Evidence for Mechanical Communication between Individual Single-headed Kinesin Motors
The finding that individual single-headed kinesin molecules remain bound to microtubules for several seconds accords with a key prediction of the hand-over-hand model, namely that the binding of the second head accelerates the detachment of the bound head. A wild-type motor moving at 800 nm/s takes 100 steps/s, and each head must detach 50 times/s on average. Given that the mean bound time of single-headed kinesin is 3 s, corresponding to a detachment rate of 0.3/s, the second head in the wild-type dimer must accelerate the unbinding of the first head by at least 100-fold. We do not believe that this acceleration is simply due to direct head-to-head contacts within the dimer, since comparable acceleration occurs in the case of single-headed molecules where there is no possibility of direct contact between heads (see below). Instead, we believe that the acceleration is due to strain arising from the binding of the second head or from a subsequent conformational change in the second head after it binds (refer to Fig. 1, iii). Therefore, the finding that simultaneous binding of the two heads to the polar microtubule may require considerable strain (Kozielski et al., 1997) is not inconsistent with the hand-over-hand model. Indeed, the approximate twofold symmetry of the dimer may be the structural basis for the mechanical communication between kinesin's two heads (Howard, 1996).
The notion of mechanical strain provides a simple insight into the directionality and load dependence of the motor. If the nucelotide-free head (the second) has a longer attached time than the ATP head (the first), then the first head will usually release before the second (refer to Fig. 1 [iii]), thereby imparting directionality to the process. If the motor is placed under a load that opposes the movement toward the plus end of the microtubule, then this load is expected to partially relieve the strain and therefore decelerate the release of the first head, accounting for the decrease in speed (Svoboda and Block, 1994; Meyhöfer and Howard, 1995). Thus, the notion of strain leads to a specific prediction as to which structural step is slowed by load. Conversely, a negative load is expected to accelerate release and lead to an increase in speed, as observed (Coppin et al., 1997).
Interestingly, some sort of mechanical communication must also take place between single-headed molecules that are moving microtubules at intermediate and high densities (Fig. 10 A). This is because the single-headed speed of 100 nm/s still requires a release rate of 12 s−1, or even higher if the single-headed steps are smaller than 8 nm, as is likely (see below). Thus, the release rate must also be accelerated during motility by single-headed motors, implying that they are also coordinated according to the hand-over-hand mechanism. Such coordination between single-headed molecules is possible because they are mechanically coupled via their tails through the glass substrate and via their heads through the microtubule; in this way strain developed in one head after it binds to the microtubule can be felt by the other attached heads. Because the moving microtubules are several microns in length, allostery must be occurring over distances of one micron! Is a microtubule rigid enough to mediate such mechanical coupling? The maximum work generated by a single kinesin molecule is ∼40 pN·nm (Svoboda and Block, 1994; Meyhöfer and Howard, 1995), so the maximum distortion of a 1-μm-long microtubule, whose stiffness is ∼400 pN/nm (Gittes et al., 1993), is only ∼0.45 nm (=[2 × 40/400]1/2). Because this distance is smaller than the 8-nm step size (or even the ∼1-nm step of the single-headed motor, see below) this argument shows that the microtubule is rigid enough to transmit such mechanical signals without significant loss.
Figure 10.
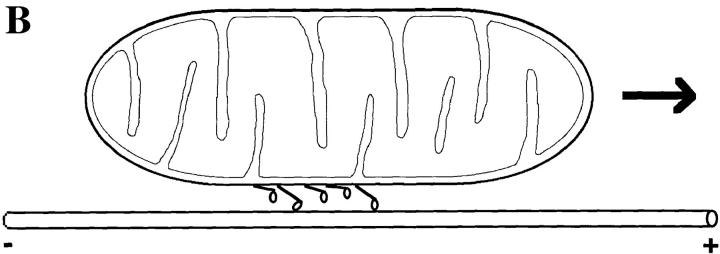
(A) Coordination between single-headed molecules moving a microtubule. In this example, the binding of the fourth head and its subsequent conformational change is hypothesized to accelerate the unbinding of the second, attached head and to move the microtubule a small distance to a position that will allow one of the other detached heads to bind. (B) Model of organelle transport by monomeric KIF1 molecules. If these kinesin-related proteins are coordinated like single-headed kinesin, then continuous motility would require only four to six molecules on the surface of a mitochondrion.
Mechanical coordination between single-headed kinesins might explain how the monomeric kinesin-related proteins KIF1A and KIF1B mentioned in the Introduction function as organelle transporters. If they have similar mechanical properties to single-headed kinesin, then only four to six such motors on the surface of an organelle would suffice for motility, since the mechanical coordination would ensure that the organelle remain associated with the microtubule (Fig. 10 B). Thus, a dimeric structure might not be essential for organelle transport; instead, our results suggest that it is mechanical communication between motor domains in a macromolecular assembly that is the essential requirement for continuous motility. Such mechanical coordination might occur in smooth muscle myosin or may contribute to the ability of unconventional myosins to mediate organelle transport along actin filaments (Howard, 1997).
The Working Stroke and the Duty Ratio
A simple interpretation of the low speed of single-headed kinesin is that removal of the second head reduces the mechanical amplification of a small conformational change that occurs in the remaining head. This is analogous to the reduction in speed of myosin when its “lever arm” is truncated (refer to Introduction). The speed of a motor is the working distance (the distance moved during the part of the ATP hydrolysis cycle that the head is attached), divided by the attached time (Howard, 1997). If the attached times of single- and two-headed kinesin are the same, then the lower speed suggests that single-headed kinesin has a smaller working distance of only ∼1 nm (= 8 nm × 0.096 nm/s ÷ 0.76 nm/s). Although this conclusion is tentative in the absence of a direct measurement of the working distance or the attached time, it does offer a simple reason why several single-headed kinesin molecules are required for continuous motility: a 1-nm working distance is not large enough to bring the head to its next binding site on the microtubule. If each head were to track a single protofilament, then the next site would be 8 nm away, the length of the tubulin dimer (Ray et al., 1993). However, if the heads are able to interact with several different protofilaments, as appears to be the case when dimerization is disrupted (Berliner et al., 1995), then the next binding site is only ∼6 nm away, since adjacent subunits in the B lattice are offset by ∼1 nm (Song and Mandelkow, 1993). Hence, this structural argument predicts a minimum number of single-headed motors required for motility of approximately six, in agreement with the measurements presented in this paper. Thus, we believe that single-headed kinesin is not processive because its working distance is too small to reach the next tubulin dimer.
Acknowledgments
The authors wish to thank D. Coy, D. Frank, A. Gordon, M. Wagenbach, and L. Wordeman (all from the University of Washington, Seattle, WA) for comments and suggestions.
This work was supported by a Muscular Dystrophy Association Research Fellowship to W.O. Hancock and grants from the National Institutes of Health (AR40593) and the Human Frontiers Science program to J. Howard.
Footnotes
Address all correspondence to Jonathon Howard, Department of Physiology and Biophysics, University of Washington, Box 357290, Seattle, WA 98195-7290. Tel.: (206) 685-3201. Fax: (206) 685-0619. E-mail: johoward@u.washington.edu
1. Abbreviation used in this paper: his, histidine.
References
- Anson M, Geeves MA, Kurzawa S, Manstein DJ. Myosin motors with artificial lever arms. EMBO (Eur Mol Biol Organ) J. 1996;15:6069–6074. [PMC free article] [PubMed] [Google Scholar]
- Berliner E, Young EC, Anderson K, Mahtani HK, Gelles J. Failure of a single-headed kinesin to track parallel to microtubule protofilaments. Nature. 1995;373:718–721. doi: 10.1038/373718a0. [DOI] [PubMed] [Google Scholar]
- Block SM, Goldstein LS, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- Bloom GS, Endow SA. Motor proteins 1: kinesins. Protein Profile. 1995;2:1105–1171. [PubMed] [Google Scholar]
- Bloom GS, Wagner MC, Pfister KK, Brady ST. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 1988;27:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- Coppin CM, Pierce DW, Hsu L, Vale RD. The load dependence of kinesin's mechanical cycle. Proc Natl Acad Sci USA. 1997;94:8539–8544. doi: 10.1073/pnas.94.16.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia JJ, Gilbert SP, Moyer ML, Johnson KA. Sedimentation studies on the kinesin motor domain constructs K401, K366, and K341. Biochemistry. 1995;34:4898–4907. doi: 10.1021/bi00014a047. [DOI] [PubMed] [Google Scholar]
- Coy DL, Howard J. Organelle transport in axons. Curr Opin Neurosci. 1994;4:662–667. doi: 10.1016/0959-4388(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Kinesin ATPase: Rate limiting ADP release. Proc Natl Acad Sci USA. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature. 1995;377:448–450. doi: 10.1038/377448a0. [DOI] [PubMed] [Google Scholar]
- Hecht S, Schlaer S, Pirenne MH. Energy, quanta, and vision. J Gen Physiol. 1942;25:819–840. doi: 10.1085/jgp.25.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AV. The combination of hemoglobin with oxygen and with carbon monoxide. Biochem J. 1913;7:471–480. doi: 10.1042/bj0070471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- Howard J. The movement of kinesin along microtubules. Annu Rev Physiol. 1996;58:703–729. doi: 10.1146/annurev.ph.58.030196.003415. [DOI] [PubMed] [Google Scholar]
- Howard J. Molecular motors: structural adaptations to cellular functions. Nature. 1997;389:561–567. doi: 10.1038/39247. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence microscopy. Methods Cell Biol. 1993;39:105–113. doi: 10.1016/s0091-679x(08)60164-8. [DOI] [PubMed] [Google Scholar]
- Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- Howard J, Hunt AJ, Baek S. Assay of microtubule movement driven by single kinesin molecules. Methods Cell Biol. 1993;39:137–147. doi: 10.1016/s0091-679x(08)60167-3. [DOI] [PubMed] [Google Scholar]
- Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388:390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- Huang T, Hackney DD. Drosophila kinesin minimal motor domain expressed in Escherichia coli. . J Biol Chem. 1994;269:16493–16501. [PubMed] [Google Scholar]
- Huang TG, Suhan J, Hackney DD. Drosophila kinesin motor domain extending to amino acid position 392 is dimeric when expressed in Escherichia coli. . J Biol Chem. 1994;269:16502–16507. [PubMed] [Google Scholar]
- Hunt AJ, Howard J. Kinesin swivels to permit microtubule movement in any direction. Proc Natl Acad Sci USA. 1993;90:11653–11657. doi: 10.1073/pnas.90.24.11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Toyoshima YY, Iwane AH, Miromoto S, Higuchi H, Yanagida T. Movements of truncated kinesin fragments with a short or an artificial flexible neck. Proc Natl Acad Sci USA. 1997;94:7275–7280. doi: 10.1073/pnas.94.14.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielski F, Sack S, Marx A, Thormählen M, Schönbrunn E, Biou V, Thompson A, Mandelkow E-M, Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey S, Waller GS, Trybus KM. Skeletal muscle myosin light chains are essential for physiological speeds of shortening. Nature. 1993;365:454–456. doi: 10.1038/365454a0. [DOI] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW. Interacting head mechanism of microtubule-kinesin ATPase. J Biol Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- Meyhöfer E, Howard J. The force generated by a single kinesin molecule against an elastic load. Proc Natl Acad Sci USA. 1995;92:574–578. doi: 10.1073/pnas.92.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M, Sato Y-R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine A-Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Ray S, Meyhöfer E, Milligan RA, Howard J. Kinesin follows the microtubule's protofilament axis. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp BJ, Crise B, Sheetz MP, Reese TS, Khan S. Delayed start-up of kinesin-driven microtubule gliding following inhibition by adenosine 5′-[β,γ-imido]triphosphate. Proc Natl Acad Sci USA. 1990;87:10053–10057. doi: 10.1073/pnas.87.24.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer MJ, Block SM. Kinesin hydrolyzes one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- Song Y-H, Mandelkow E. Recombinant kinesin motor domain binds to β tubulin and decorates microtubules with a B surface lattice. Proc Natl Acad Sci USA. 1993;90:1671–1675. doi: 10.1073/pnas.90.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RJ, Thaler JP, Goldstein LS. Direction of microtubule movement is an intrinsic property of the motor domains of kinesin heavy chain and Drosophilancd protein. Proc Natl Acad Sci USA. 1993;90:5209–5213. doi: 10.1073/pnas.90.11.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- Uyeda TQ, Abramson PD, Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:415–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Laymon RA, Goldstein LS. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analyses. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- Yang JT, Saxton WM, Stewart RJ, Raff EC, Goldstein LS. Evidence that the head of kinesin is sufficient for force generation and motility in vitro. Science. 1990;249:42–47. doi: 10.1126/science.2142332. [DOI] [PubMed] [Google Scholar]
- Young EC, Berliner E, Mahtani HK, Perez-Ramirez B, Gelles J. Subunit interactions in dimeric kinesin heavy chain derivatives that lack the kinesin rod. J Biol Chem. 1995;270:3926–3931. doi: 10.1074/jbc.270.8.3926. [DOI] [PubMed] [Google Scholar]