Abstract
Previously, we have suggested that vascular cell adhesion molecule-1 (VCAM-1) and its integrin receptor α4β1 mediate cell–cell interactions important for skeletal myogenesis. Expression of the receptors subsequently subsides in muscle after birth. Here, we examine the mechanism regulating VCAM-1 gene expression in muscle. An enhancer located between the TATA box and the transcriptional start site is responsible for VCAM-1 gene expression in muscle—this element is inactive in endothelial cells where VCAM-1 expression is dependent on nuclear factor κB sites and inflammatory cytokines. We identify interferon regulatory factor-2 (IRF-2), a member of the interferon regulatory factor family, as the enhancer-binding transcription factor and show that expression of IRF-2 parallels that of VCAM-1 during mouse skeletal myogenesis. IRF-2 is not dependent upon cytokines for expression or activity, and it has been shown to act as a repressor in other nonmuscle cell types. We show that the basic repressor motif located near the COOH-terminal of IRF-2 is not active in muscle cells, but instead an acidic region in the center of the molecule functions as a transactivating domain. Although IRF-2 and VCAM-1 expression diminishes on adult muscle fiber, they are retained on myogenic stem cells (satellite cells). These satellite cells proliferate and fuse to regenerate muscle fiber after injury or disease. We present evidence that VCAM-1 on satellite cells mediates their interaction with α4β1(+) leukocytes that invade the muscle after injury or disease. We propose that VCAM-1 on endothelium generally recruits leukocytes to muscle after injury, whereas subsequent interaction with VCAM-1 on regenerating muscle cells focuses the invading leukocytes specifically to the sites of regeneration.
Vascular cell adhesion molecule-1 (VCAM-1)1 is a member of the immunoglobulin gene superfamily that is expressed on the surface of endothelial cells in response to inflammation (Osborn et al., 1989; Elices et al., 1990; Rice et al., 1991). Through its interaction with the integrin receptors, α4β1 and α4β7, on T-cells, monocytes, and eosinophils, VCAM-1 targets these inflammatory cells to cytokine-stimulated endothelium (Erle et al., 1991; Freedman et al., 1991; Miyake et al., 1991; Scheeren et al., 1991; Schimizu et al., 1991; Ruegg et al., 1992).
In addition to its role in inflammation, VCAM-1-deficient mice demonstrated essential roles for VCAM-1 during embryonic development (Gurtner et al., 1995; Kwee et al., 1995). Mice lacking VCAM-1 died at two different developmental stages. Most VCAM-1-deficient embryos died before embryonic day (E) 11.5 due to a failure of the allantois to fuse with the chorion. In wild-type embryos, VCAM-1 and its receptor, α4β1, are expressed on the allantois and chorion, respectively (Kwee et al., 1995). Embryos that survived these defects in the extraembryonic membranes died by E12.5 due to cardiac abnormalities, including defects of the ventricular myocardium and intraventricular septum and a failure to form an epicardium. In wild-type embryos, VCAM-1 is expressed on the outer layer of the myocardium, which physically interacts with epicardial cells that express α4β1 (Kwee et al., 1995; Sheppard et al., 1994). Significantly, α4-deficient embryos exhibit similar defects in placental and cardiac development (Yang et al., 1995). These results suggest important roles for VCAM-1 and α4β1 in both placental and cardiac development.
Other studies have suggested that VCAM-1 also plays a role in the differentiation of skeletal muscle; however, VCAM-1-deficient embryos die before the role of VCAM-1 can be examined in this process. Mammalian skeletal muscle differentiation occurs in two stages, each represented by a distinct population of myoblasts (Ontell, 1977; Ross et al., 1987; Stockdale and Miller, 1987; Ontell et al., 1988). Around E12 in the mouse, an early-born population of myoblasts (primary myoblasts) fuse to form primary myotubes. Then, between E14 and E16, a second, distinct wave of myoblasts (secondary myoblasts) appear and align themselves along the primary myotubes, where they proliferate and appear to use primary myotubes as a template for fusion into secondary myotubes. These secondary myotubes comprise most adult muscle fiber. VCAM-1 is present on secondary myoblasts and portions of secondary myotubes that are apposed to primary myotubes, while α4β1 is present on primary myotubes (Rosen et al., 1992). This alignment suggests that these receptors play a role in secondary myogenesis. Antibodies that block the interaction between VCAM-1 and α4β1 inhibited myoblast fusion in culture (Rosen et al., 1992), and expression of antisense α4 RNA blocked myoblast fusion in vitro and prevented fusion into muscle in vivo (unpublished observations), providing further evidence of a role for these proteins in myogenesis. Expression of both VCAM-1 and α4β1 is developmental-specific since neither receptor is found in normal adult muscle fibers.
In endothelial cells where VCAM-1 expression is dependent upon inflammatory cytokines, silencer elements (identified as octamer binding sites) restrict VCAM-1 gene promoter activity in unstimulated endothelial cells (Iademarco et al., 1992, 1993). Inflammatory cytokines overcome the negative activity of the octamers and cause activation of the promoter through two adjacent nuclear factor κB sites located at positions −77 and −63 bp in the VCAM-1 gene promoter. In contrast to endothelial cells, where VCAM-1 expression is dependent upon cytokines, there is high basal expression of VCAM-1 on muscle cells. An element between the TATA box and the transcriptional start site appears to override the activity of other promoter elements causing constitutive VCAM-1 expression; this element is not active in endothelial cells. The nucleotide sequence of the VCAM-1 gene promoter in this region, between positions −17 and −5 bp, matches the consensus for an interferon (IFN) regulatory factor (IRF) binding element (Neish et al., 1995). The IRF family of transcription factors binds to these DNA sequences classically found in the IFN-α and IFN-β gene promoters. However, a number of other genes contain IRF elements, and not all of the IRF family members are regulated by cytokines. The IRF family members are characterized by a highly conserved amino-terminal DNA binding domain that contains a repeated tryptophan motif (Veals et al., 1992), but exhibit minimal sequence similarity outside this region. The IRF family contains both activators and repressors of transcription. Transfection studies using the IFN-β and other IFN- responsive gene promoters have shown that IRF-1 is a transcriptional activator while IRF-2 is a repressor (Harada et al., 1989, 1990; Reis et al., 1992). However, other studies have demonstrated that IRF-2 contains a latent activation domain (which becomes evident when the repressor domain is deleted; Yamamoto et al., 1994) that can activate transcription from some gene promoters (Vaughan et al., 1995).
Here, we demonstrate that IRF-2 interacts with the VCAM-1 gene promoter in muscle cells and is responsible for the transcriptional activation of the VCAM-1 gene promoter. We show that the repressor domain is inactive in muscle cells, and using a series of IRF-2 deletion mutants, we identify the transactivating domain in IRF-2 that is responsible for transcriptional activity during myogenesis. We show that IRF-2 expression closely parallels that of VCAM-1 in differentiating skeletal muscle. Additionally, we examined VCAM-1 expression during muscle regeneration, both in response to muscle injury and in a murine model of muscular dystrophy. Muscle regeneration differs from embryonic myogenesis in several ways. Myogenesis in the embryo occurs in two stages and involves distinct populations of myoblasts while regenerating muscle is derived from muscle satellite cells, which are stem cell–like precursors that are normally quiescent and do not express muscle transcription factors. In response to injury and disease, these satellite cells proliferate and fuse into muscle fibers. Muscle regeneration is intimately associated with inflammation, and in addition to their obvious role in inflammation, invading leukocytes may provide a trigger for proliferation of satellite cells that initiates the regeneration process. We present evidence that VCAM-1 is expressed on satellite cells and newly forming myotubes, where it plays a novel role in recruitment of leukocytes to specific sites of myogenic regeneration.
Materials and Methods
Cell Culture and DNA Transfection
C2C12 mouse myoblasts were maintained in DME supplemented with 13% FBS. For transient transfections, 20 μg of reporter plasmid were transfected by the calcium phosphate technique (Iademarco et al., 1992) followed by DMSO shocking (8 min in 12% DMSO in DME) 18 h later. Cells were maintained as myoblasts in 13% FBS, and cell extracts were prepared 36 h after transfection. 1 μg of TKluc, which contains the thymidine kinase promoter fused to the firefly luciferase gene, was cotransfected as an internal control and luciferase activity was used to correct for transfection efficiency. Luciferase and chloramphenicol acetyl transferase gene (CAT) assays were done as described previously (Iademarco et al., 1992). NIH 3T3 cells were maintained in DME supplemented with 10% FBS. P19 cells were maintained in α-MEM containing 10% FBS and differentiated with retinoic acid as described previously (Sheppard et al., 1995).
Cell Adhesion Assays
C2C12 cells were transferred to 96-well plates and allowed to adhere for 18 h. Ramos cells were washed twice with PBS and resuspended in DME with 10% FBS alone or with 2 μg/ml anti–α4 antisera (HP 2/1, a gift from Dr. F. Sanchez-Madrid, Hospital de la Princesa, Madrid, Spain) or control anti–myosin heavy chain antisera (a gift from Dr. R. Kopan, Washington University). Approximately 105 Ramos cells were allowed to attach to the C2C12 monolayers for 1 h. The plates were then washed three times with PBS and photographed.
Plasmid Construction
VCAM-1 promoter constructs have been described previously (Iademarco et al., 1992). Mutant −32 VCAMCAT plasmids were constructed by cloning double-stranded synthetic oligonucleotides corresponding to positions −32 to +8 bp of the VCAM-1 gene promoter into the PstI and HindIII sites of SkCAT-Pst/Bam (Rosen et al., 1991). IRF-1 and IRF-2 expression plasmids (Lin et al., 1994) were a gift from Dr. John Hiscott (McGill University, Montreal, Quebec, Canada).
Gel Retardation Assays
Nuclear extracts were prepared from C2C12 cells using a modified Dignam protocol (Dignam et al., 1983). Briefly, cells were harvested and rinsed. The cells were then lysed through a syringe in hypotonic buffer (10 mM Hepes, pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 1 mM PMSF, and 1 mM DTT). 10 μg of nuclear extract was assayed for binding using 32P-labeled, double stranded oligonucleotides. The probe for the VCAM-1 IRF site spanned −32 to +8 bp of the VCAM-1 gene. For gel retardation assays, nuclear extract was incubated with probe on ice for 30 min in binding buffer (10 mM Tris-HCl, pH 7.9, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 5% glycerol, 1 μg nonspecific DNA, 2.5 mM MgCl2, 2.5 mM MnCl2). For supershift analysis, nuclear extract was incubated with 2.5 μl anti–IRF-2 or control anti–c-rel antisera (Santa Cruz Biotechnology, Santa Cruz, CA) for 45 min on ice before addition of binding buffer. The sequences of the oligonucleotides used are as follows: VCAM-1, 5′-TTTATAAAGCACAGACTTTCTATTTCACTCCGCGGTATCTGCA-3′; and HLA, 5′-ATTCCCCACTCCCCTGAGTTTCACTTCTTCTCCCAAC-3′.
Western Blot Analysis
Whole cell extracts were prepared from cultured cells in 10 mM Tris-HCl (pH 6.8) and 0.1% SDS. Muscle extracts were prepared by homogenization of embryonic limbs or adult hindlimb muscle as previously described (Andrew and Appel, 1973). Whole cell extracts were subjected to Western blot analysis using anti–IRF-1 and anti–IRF-2 (COOH-terminal) antibodies as described previously (Nguyen et al., 1995). Anti–IRF-1 and anti– IRF-2 antisera (Nguyen et al., 1995) were a gift from Dr. Hiscott.
Immunostaining and Muscle Injury
Immunostaining of embryonic mouse sections and sections of adult skeletal muscle was done as we have described (Rosen et al., 1992). Immunostaining for IRF-2 was confirmed with three different antisera, each to a distinct region of IRF-2. For muscle damage, the gastrocnemius muscle in the hindlimb of adult C3H mice was exposed surgically and damaged by either cutting or freezing (results were similar with both damaging techniques). At various times after muscle damage, the area of damage or a control region from the other undamaged hindlimb was dissected and frozen in O.C.T. (Miles-Yeda Inc., Elkhart, IN) as described previously (Rosen et al., 1992). Sections were immunostained for VCAM-1, NCAM, and CD45 as described previously (Rosen et al., 1992). Anti–CD45 was a gift from Dr. M. Thomas (Washington University).
Results
Expression of VCAM-1 in Skeletal Muscle Cells Is Controlled by an Element 3′ of the TATA Box
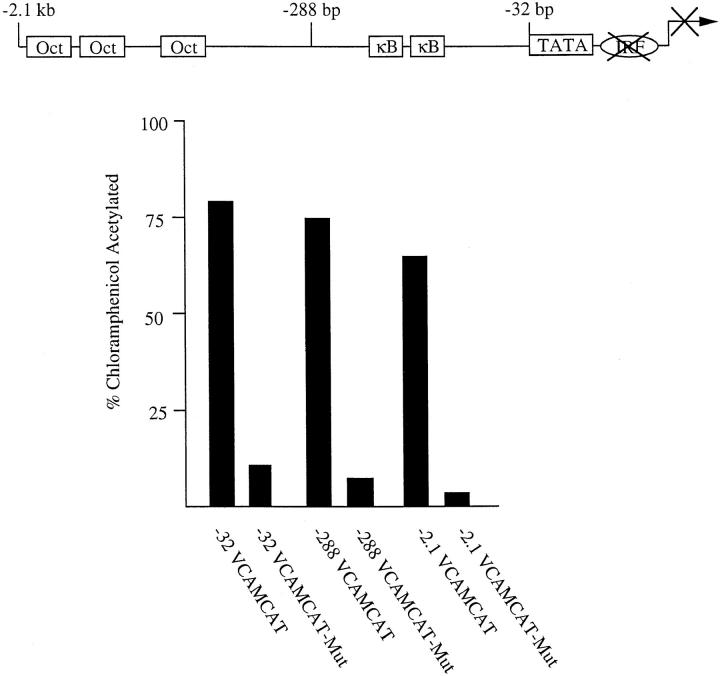
Previously, we found that a 2.1-kb fragment of the VCAM-1 gene promoter is transcriptionally active in C2C12 myoblasts and that deletion of the promoter to −32 bp, which is immediately upstream of the TATA box, did not inhibit promoter activity (Iademarco et al., 1993). Here, we demonstrate that mutation of the region 3′ of the TATA box, between positions −21 and −5 bp, in the context of the −32-, −288-, and −2.1-kb VCAMCAT reporter constructs inhibited VCAM-1 promoter activity in C2C12 myoblasts (Fig. 1). These results suggest that the region between −21 and −5 bp is necessary for the activity of the VCAM-1 gene promoter in skeletal myoblasts.
Figure 1.
An element between the TATA box and transcriptional start site controls VCAM-1 expression in skeletal muscle cells. Wild-type and mutant VCAM-1 promoter constructs (20 μg) were transfected into C2C12 myoblasts. Numbers in the name indicate the amount of VCAM-1 gene 5′ flanking region in each construct (−32, 32 bp; −288, 288 bp; −2.1, 2.1 kb). In mutant constructs, the IRF sequence between positions −21 and −5 bp was replaced with the following sequence, where mutated bases are indicated by lowercase letters: 5′-cCgGctcccgcgcTcCg-3′. The results with each construct are representative of at least five different assays done in duplicate, and using different preparations of plasmid DNAs. Mutation of the IRF site always resulted in at least a sevenfold decrease in reporter activity.
Mutations Identify an IRF Element in the VCAM-1 Gene Promoter
To more precisely identify regions critical for VCAM-1 gene promoter activity, a series of mutant constructs was created, and their activities were examined in transfection assays in C2C12 myoblasts (Fig. 2). This analysis identified an essential DNA sequence in the VCAM-1 gene promoter that matches the consensus binding site for the IRF family of transcription factors, G(A)AAANNGAAA(G/C) (T/C) (Tanaka et al., 1993). Mutations in either or both half-sites of the IRF-like DNA binding sequence decreased transcriptional activity. Additionally, mutations at positions −14 and −8 bp disrupted promoter activity; such mutations in the IRF binding sequence have previously been shown to disrupt IRF binding (Neish et al., 1995). However, mutations in the nonconserved spacer (NN) between the half-sites or in the sequences 5′ or 3′ of the IRF element had little or no effect. This mutational analysis suggests that the enhancer in the VCAM-1 gene promoter is an IRF site and that this site is responsible for VCAM-1 expression in muscle cells.
Figure 2.
The IRF element is necessary for VCAM-1 gene promoter activity in C2C12 myoblasts. Wild-type and mutant −32 VCAMCAT reporter constructs were transfected into C2C12 myoblasts. Black boxes indicate mutated regions in the VCAM-1 gene promoter. The wild-type VCAM-1 sequence between positions −32 and +5 bp is shown above. The sequence of the mutants between positions −23 and +1 bp, where mutated bases are indicated by lowercase letters, are as follows: 5′-cCcggcACTTTCTATTTCACTCCG-3′ (−23 to −18); 5′-GCcCgGctcccgcgcTcCgCTCCG-3′ (−21 to −5); 5′-GCcCgGctcccgTATTTCACTCCG-3′ (−21 to −12); 5′-GCACAGtaTTTCTATTTCACTCCG-3′ (−17 to −16); 5′-GCACAGACggTCTATTTCACTCCG-3′ (−15 to −14); 5′-GCACAGACTTgaTATTTCACTCCG-3′ (−13 to −12); 5′-GCACAGACTcTCTATgTCACTCCG-3′ (−14 and −8); 5′-GCACAGACTTTCTAcgcgcCTCCG-3′ (−9 to −5); 5′-GCACAGAggctcccgTCACTCCG-3′ (−15 to −8); 5′-GCACAGACTTTCccTTTCACTCCG-3′ (−11 to −10); 5′-GCACAGACTTTCTATTTCcCTCCG-3′ (−5); 5′-GCACAGACTTTCTATTTCgaTCCG-3′ (−5 to −4); 5′-GCACAGACTTTCTATTTCACTCgt-3′ (−1 to +1). The results with each construct are representative of at least five independent assays done in duplicate, with multiple preps of plasmid DNAs. There is not a significant difference between −21 to −5, −21 to −12, −15 to −14, −14 and −8, −9 to −5, or −15 to −8. Likewise, there is no significant difference between wild-type, −23 to −18, −17 to −16, −11 to −10, or −1 to +1. In contrast, −5 to −4 and −13 to −12 were reproducibly different from the other two groups.
IRF-2 Is Expressed in Myoblasts
We examined expression of IRF family proteins in C2C12 myoblasts. Extracts of C2C12 myoblasts were tested for IRF-1, IRF-2, and IRF-4 expression by Western blot analysis. Other IRF family members, ICSBP and IGSF3γ, were not tested. (ICSBP is expressed only in macrophages and lymphocytes [Driggers et al., 1990; Nelson et al., 1993], and IGSF3γ is activated only in response to IFN treatment [Kessler et al., 1990].) IRF-2 was the only IRF protein detected in the C2C12 extracts (Fig. 3 A, and data not shown). As a control, IRF-2 expression was examined in P19 embryonic carcinoma cells. It has been demonstrated previously that IRF-2 is not expressed in undifferentiated P19 cells, but it is expressed when P19 cells are induced to differentiate into neural cells in response to retinoic acid treatment (Harada et al., 1990). Interestingly, we have found previously that VCAM-1 expression is also induced upon P19 cell differentiation (Sheppard et al., 1995).
Figure 3.
Expression of IRF proteins. (A) Extracts were prepared from undifferentiated and retinoic acid–treated P19 cells (15 μg), C2C12 myoblasts (15 μg), and embryonic and adult skeletal muscle (30 μg), separated by SDS-PAGE, transferred to nitrocellulose membrane, and immunoblotted with anti–IRF-2 antisera. (B) 15 μg of whole cell extracts prepared from untreated and IFN-γ-treated (3 h, 1000 U/ml) NIH 3T3 and C2C12 cells were separated by SDS-PAGE, transferred to nitrocellulose membrane, and immunoblotted with anti–IRF-1 antisera. NS denotes a nonspecific complex.
While IRF-1 was not normally expressed in C2C12 cells, it is expressed in response to treatment with IFN-γ (Fig. 3 B). Accordingly, VCAM-1 expression has been shown previously to be induced by IFN-γ (Thornhill et al., 1991; Sikorski et al., 1993; Gao and Issekutz, 1996; Seko et al., 1996; Prudhomme et al., 1996). As a control, we show that IRF-1 is also induced by IFN-γ in NIH 3T3 cells (Fig. 3 B). These patterns of expression suggest that IRF-2 is the IRF family member that normally regulates VCAM-1 gene transcription in muscle cells; however, other IRF proteins, such as IRF-1, may be able to substitute for IRF-2 (e.g., during inflammation).
IRF-2 Binds to the VCAM-1 IRF Element
Next, the ability of IRF-2 to bind to the IRF element from the VCAM-1 gene promoter was examined. Synthetic oligonucleotides containing the VCAM-1 gene IRF element were used in gel shift assays with extracts from 293 cells transfected with an IRF-2 expression vector. Two specific complexes were observed with extract from the IRF- 2-transfected cells, and mutation of the IRF site eliminated binding of these complexes to the probe; antisera against IRF-2 supershifted these complexes (data not shown). A control IRF binding element from the HLA-B7 gene promoter (Johnson and Pober, 1994) formed similar complexes and efficiently competed for binding to the VCAM-1 site. These results indicate that the VCAM-1 gene IRF element can bind IRF-2.
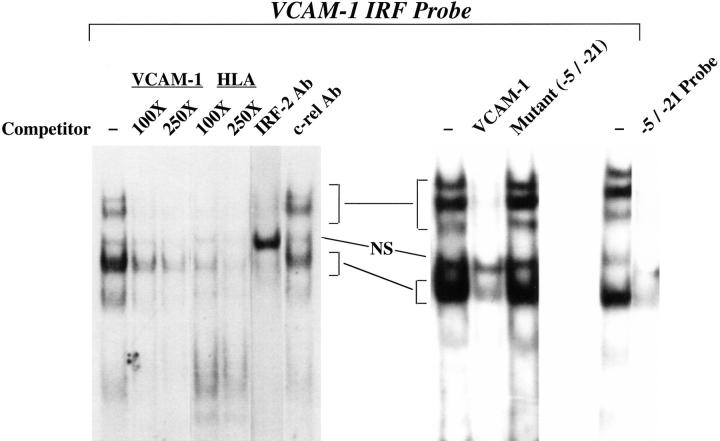
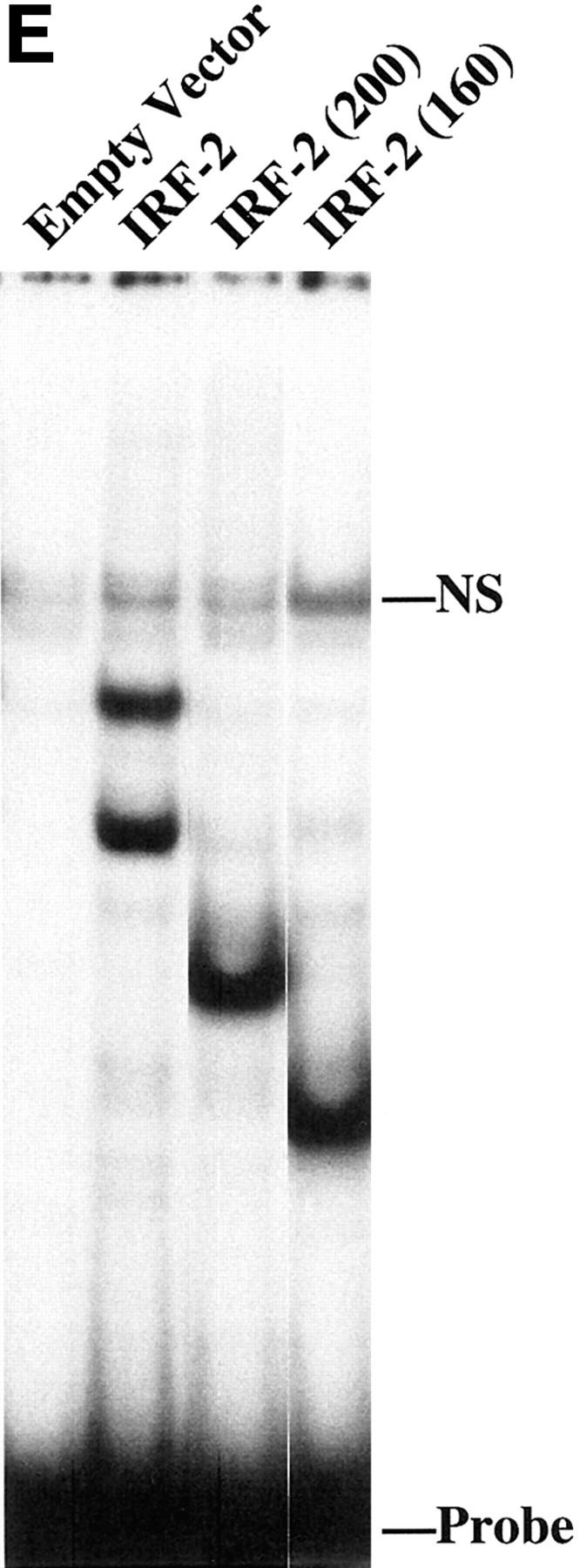
To determine which IRF proteins bind the VCAM-1 gene IRF element in C2C12 cells, the IRF element was used in gel retardation assays with nuclear extract from C2C12 cells (Fig. 4). A single major complex was observed, along with several minor, more slowly migrating complexes. The IRF element from the HLA-B7 probe efficiently competed for binding to the VCAM-1 IRF element. Mutation of the IRF site eliminates formation of the specific complexes and, accordingly, the mutant probe did not compete for binding of nuclear proteins to the wild-type probe. Formation of all specific complexes was blocked by the IRF-2 antibody, resulting in accumulation of the nonspecific complex; an irrelevant antibody had no effect. These results demonstrate that IRF-2 is responsible for all of the binding to the IRF site in the VCAM-1 gene promoter in myoblasts, and thus IRF-2 appears to be the IRF family member responsible for controlling VCAM-1 expression in C2C12 cells.
Figure 4.
IRF-2 binds to the VCAM-1 gene promoter in C2C12 myoblasts. A gel retardation assay is shown using C2C12 nuclear extract with a probe corresponding to positions −32 to +8 bp in the VCAM-1 gene promoter. A 100- or 250-fold molar excess of unlabeled competitor oligonucleotides or 2.5 μl of antisera (Ab) was included in the assays where indicated. HLA indicates a control IRF binding element from the HLA-B7 gene promoter (Johnson and Pober, 1994). NS denotes a nonspecific complex. −5/−21 indicates that the mutant sequence described in Fig. 1 was used as a competitor or a labeled probe. On the right-hand side of the figure, a 250-fold excess of competitor oligonucleotides was used.
IRF-2 Transactivates the VCAM-1 Gene Promoter
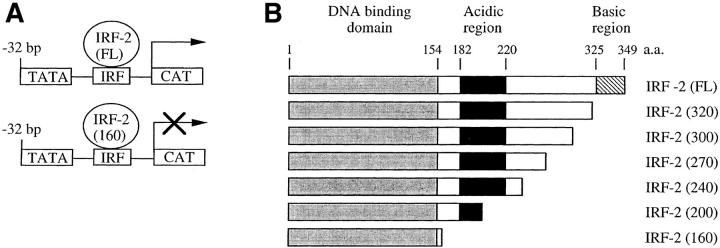
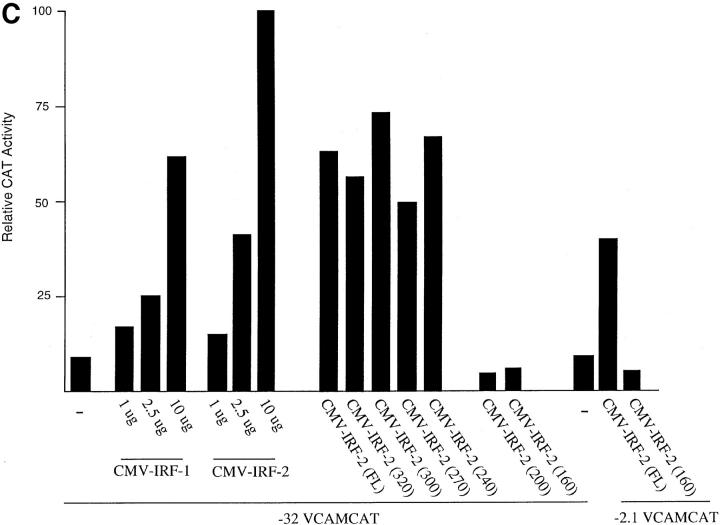
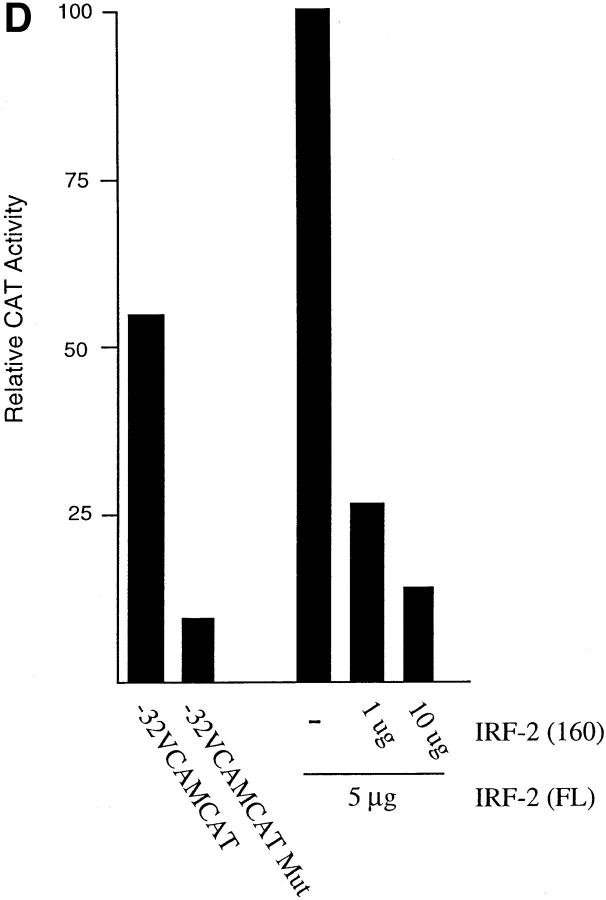
It was surprising that IRF-2 is the family member that binds to the VCAM-1 gene promoter in C2C12 cells since it has been demonstrated to be a transcriptional repressor in lymphoid cells. However, it has been demonstrated that IRF-2 contains a latent transcriptional activation domain that is apparent upon deletion or mutation of the transcriptional repression domain (Yamamoto et al., 1994). Recent studies have demonstrated that IRF-2 activates transcription, suggesting that the activation domain can function in the context of the wild-type protein (Vaughan et al., 1995; Luo and Skalnik, 1996). To demonstrate that IRF-2 activates transcription from the VCAM-1 gene promoter, a vector that overexpresses the protein was cotransfected with the −32 or −2.1 VCAMCAT reporter construct into C2C12 myoblasts (Fig. 5). Overexpression of IRF-2 significantly increased CAT activity (Fig. 5 C), suggesting that it activates the VCAM-1 gene promoter in myoblasts through interaction with the IRF element.
Figure 5.
A central acidic region of IRF-2 is responsible for transactivation of the VCAM-1 gene promoter. (A) Schematic representation of full-length IRF-2 (FL) activating the VCAM-1 gene promoter, and a dominant-negative IRF-2(160), which contains the DNA binding domain but lacks the acidic transcriptional activation domain, displacing wild-type IRF-2 and thereby inhibiting transcription. (B) Schematic representation of IRF-2 deletion proteins used in transfection studies. Numbers in parenthesis indicate the length in amino acids. (C) C2C12 myoblasts were cotransfected with 10 μg of reporter construct, either −32 VCAMCAT or −2.1 VCAMCAT, and the indicated amount of empty vector (−) or IRF expression vector. (D) C2C12 myoblasts were cotransfected with 10 μg of reporter construct, either −32 VCAMCAT or −32 VCAMCAT-Mut, which contains the mutated sequence described in Fig. 1 (−21/−5). (E) A gel retardation assay showing the binding of full-length IRF-2 and IRF-2 deletion mutants to the VCAM-1 IRF-2 site. CAT assays are all representative of at least five different assays, each done in duplicate with multiple preps of plasmid DNAs.
Although IRF-1 is not normally expressed in the C2C12 cells, it is expressed in response to IFN-γ treatment (see above). Therefore, the possibility that cotransfection of an IRF-1 expression vector would activate the VCAM-1 gene promoter was examined (Fig. 5 C). Overexpression of IRF-1 activated the promoter to a level similar to that observed with IRF-2, suggesting that other IRF proteins, if present, can substitute for IRF-2 in activation of the VCAM-1 gene promoter.
The Acidic Region of IRF-2 Is Required for Transcriptional Activation
Previous mutational studies of IRF-2 have identified the basic, COOH-terminal region of IRF-2 (amino acids 325– 349) as a transcriptional repressor motif (Yamamoto et al., 1994). Deletion or mutation of this inhibitory COOH-terminal region revealed a latent activation domain in the central region of the protein; this region (amino acids 182– 218) is relatively acidic. However, it is unclear whether this acidic activation domain is responsible for activation in the context of the full-length protein. To localize the region of IRF-2 required for transcriptional activation, a series of IRF-2 deletion mutants was tested in transfection assays for ability to transactivate the VCAM-1 gene promoter (Fig. 5, B and C). Deletion of the COOH-terminal region, which acted as a repressor in other systems, had no effect on transcriptional activity. However, deletions into the acidic central domain of the protein eliminated transcriptional activation of the VCAM-1 gene promoter. Therefore, this acidic domain is necessary for transcriptional activation in myoblasts, while the COOH-terminal repressor motif of the protein appears to be inactive in myoblasts.
IRF-2 deletion mutants that lack the acidic transcriptional activation domain, but contain the DNA binding domain, should act as a dominant negative and displace wild-type IRF-2 from the VCAM-1 gene promoter, thereby blocking transcription (Fig. 5, A and B). Expression of the DNA binding domain of IRF-2 indeed functioned as a dominant negative that blocked transactivation by full-length IRF-2 and inhibited transcriptional activity of the −32 and −2.1 VCAMCAT reporter constructs (Fig. 5, C and D). IRF-2 constructs were transfected into 293 cells, and nuclear extracts were examined in gel retardation assays for binding to an IRF binding site (Fig. 5 E). The results demonstrate that expression of full-length IRF-2 or the IRF-2 deletion mutants that contain only the DNA binding domain results in similar DNA binding activity in the nucleus. Taken together, the above results provide further evidence of an important role for IRF proteins in activating VCAM-1 gene expression in muscle cells.
Expression of IRF-2 Immediately Precedes VCAM-1 Expression in Differentiating Skeletal Muscle Cells in the Mouse
During skeletal myogenesis in the mouse, VCAM-1-positive cells appear relatively late, around E16 and remain present until at least postnatal day 2 (Rosen et al., 1992; Sheppard et al., 1994). However, no VCAM-1 is detectable on adult muscle fibers. Therefore, we examined whether IRF-2 expression correlates with VCAM-1 expression. Extracts from embryonic (E16 and E18) limb muscle and adult hindlimb muscle were examined for IRF-2 by Western blot analysis (Fig. 3 A). We found that IRF-2 is indeed expressed in limb muscle during embryogenesis, but is not expressed in adult skeletal muscle.
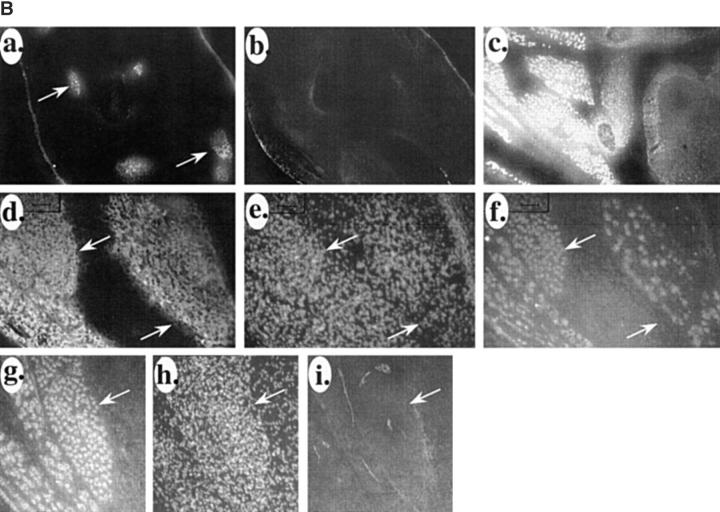
Next, we compared the patterns of VCAM-1, IRF-2, and the myogenic marker myosin heavy chain during mouse development. At E11, myosin was already evident in the differentiating somites (Fig. 6 A, a). No immunostaining for IRF-2 was evident at E11 in the muscle or anywhere else in the embryo (Fig. 6 A, b). We have demonstrated previously that VCAM-1 does not appear until relatively late in muscle differentiation process (well after myosin) (Rosen et al., 1992), and accordingly it is not yet evident at E11 (Fig. 6 A, c). At E15, expression of IRF-2 was evident on myogenic cells in skeletal muscle masses (Fig. 6 B, a and c)—no immunostaining was observed in other areas of the embryos. No VCAM-1 was evident on low power views of the muscle (Fig. 6 B, b). At this time, IRF-2 was only evident on a subset of the myosin-positive cells in the muscle masses (Fig. 6 B, d–f). Higher power views of the muscle masses showed very low levels of VCAM-1 expression in the muscle masses (Fig. 6 B, g–i). By E18, IRF-2 expression was evident on all skeletal muscle masses (Fig. 6 C, a and b). In contrast to E15, where IRF-2 was only expressed on a subset of myosin-positive cells within a muscle mass, by E18 all of the myosin-positive cells were IRF-2 positive (Fig. 6 C, c–e). Also by E18, VCAM-1 expression had increased and the protein was evident around the IRF-2-positive cells in the muscle masses (Fig. 6 C, f–h). These results suggest that IRF-2 expression precedes that of VCAM-1 on differentiating muscle cells. These patterns of IRF-2 immunostaining are consistent with the expression of IRF-2 detected by Western blot analysis described above. The pattern of IRF-2 expression in the embryos was surprisingly muscle specific, further suggesting an important role for the protein in the late stages of myogenesis.
Figure 6.
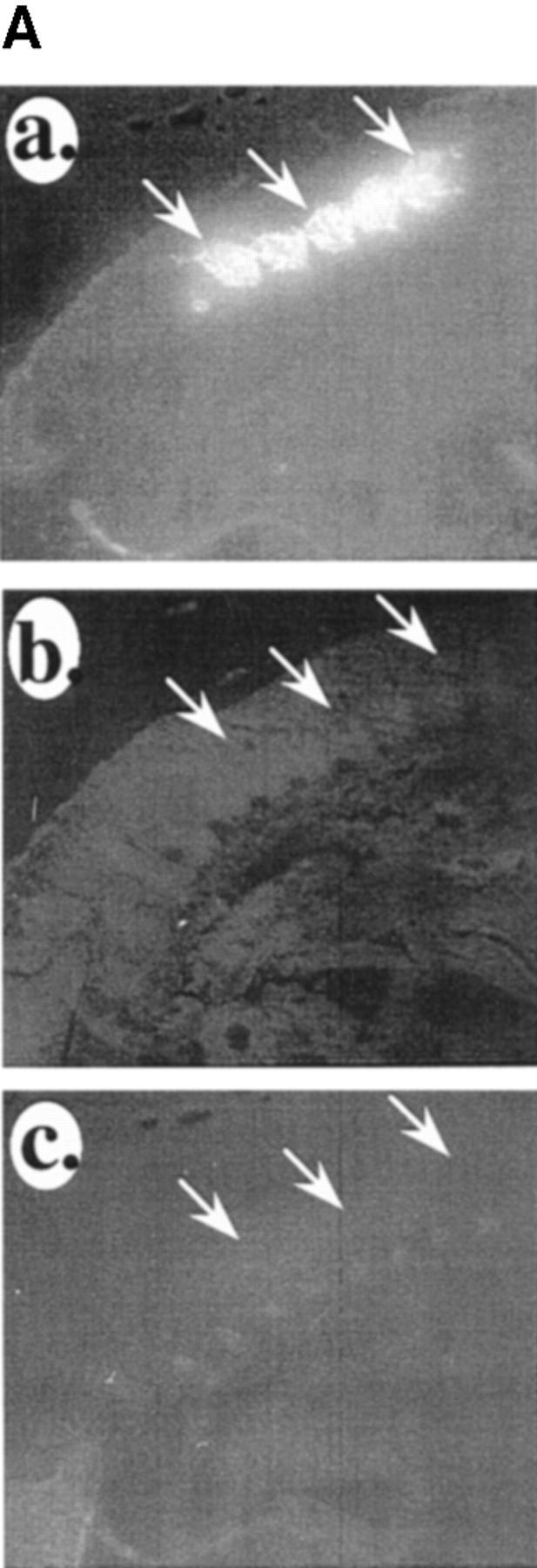
Expression of VCAM-1 closely follows that of IRF-2 in skeletal muscle differentiation. (A) IRF-2 and VCAM-1 expression follow that of the muscle differentiation marker myosin heavy chain during myogenesis in the mouse. (a) Myosin heavy chain immunostaining in differentiating somites in the mouse at E11. A sagittal section is shown. (b) Double immunostaining of the section in A for IRF-2. (c) Immunostaining of an adjacent section for VCAM-1. Arrows indicate differentiating somites. (B) Expression of IRF-2 precedes expression of VCAM-1 in differentiating mouse skeletal muscle. (a) Low power view (5×) of a section of E15 mouse immunostained for IRF-2. Arrows indicate IRF-2-positive muscle masses. (b) Double immunostaining of the section in a for VCAM-1. (c) A 10× magnification of a section of E15 mouse immunostained for IRF-2. Intense staining is evident only on some muscle masses. (d) High power view of muscle masses from E15 mouse immunostained for myosin heavy chain. (e) The section in e was treated with bis-benzimide and nuclei were viewed by UV illumination. (f) The section in d and e was double immunostained for IRF-2. Note that IRF-2 is only expressed on a subset of the myosin-positive cells in the muscle mass. (g) High power view of another E15 mouse muscle mass immunostained for IRF-2. (h) Bis-benzimide nuclear staining for the section in g. (i) Double immunostaining of the section in g and h for VCAM-1. Note that a very low level of VCAM-1 immunostaining is evident around a small subset of the cells in the muscle mass. Arrows indicate the same location in d–f and g–i, respectively. (C) Expression of VCAM-1 and IRF-2 coincide in mouse skeletal muscle at E18. (a) Low power view of IRF-2 immunostaining of muscle masses in a section of an E18 mouse. R indicates a rib. (b) Bis-benzimide nuclear staining of the section in a. Arrows in a and b indicate the same location. (c) High power view of immunostaining of E18 muscle mass for myosin heavy chain. (d) Bis-benzimide nuclear staining of the section in c. (e) Double immunostaining of the section in c and d for IRF-2. Note that in contrast to E15 (B, d–f), all of the myosin-positive cells in the muscle mass are now IRF-2 positive. Arrows in c–e indicate the same location. (f) High power view of immunostaining of another E18 muscle mass for IRF-2. (g) Bis-benzimide nuclear staining of the section in f. (h) Double immunostaining of the section in f and g for VCAM-1. Note that VCAM-1 is now expressed around the IRF-2-positive cells in the muscle mass. Arrows indicate the same location in f–h. Similar patterns of IRF-2 immunostaining were observed with three different antibodies.
IRF-2 and VCAM-1 Are Downregulated in Adult Skeletal Muscle Fibers, but Are Retained on Satellite Stem Cells
We have found previously that VCAM-1 expression diminishes in adult muscle fibers; however, the protein is retained on myogenic stem cells known as satellite cells (Rosen et al., 1992). Therefore, we wondered whether IRF-2 expression also resembled that of VCAM-1 in adult mice. Indeed, as with VCAM-1, we found that expression of IRF-1 diminished on adult myofibers, but it persisted on VCAM-1-positive cells that appeared to be scattered among the muscle fibers and located under the basement membrane surrounding the muscle fibers (Fig. 7, A–E). As we have shown previously, these cells also express NCAM, which is a marker for satellite cells (Rosen et al., 1992; Fig. 7, F and G). The above results suggest that lack of IRF-2 on adult muscle fibers may be responsible for the loss of VCAM-1 expression in adult muscle. Additionally, we conclude that IRF-2 is expressed in satellite cells and that this expression may be responsible for the presence of VCAM-1 on these cells.
Figure 7.
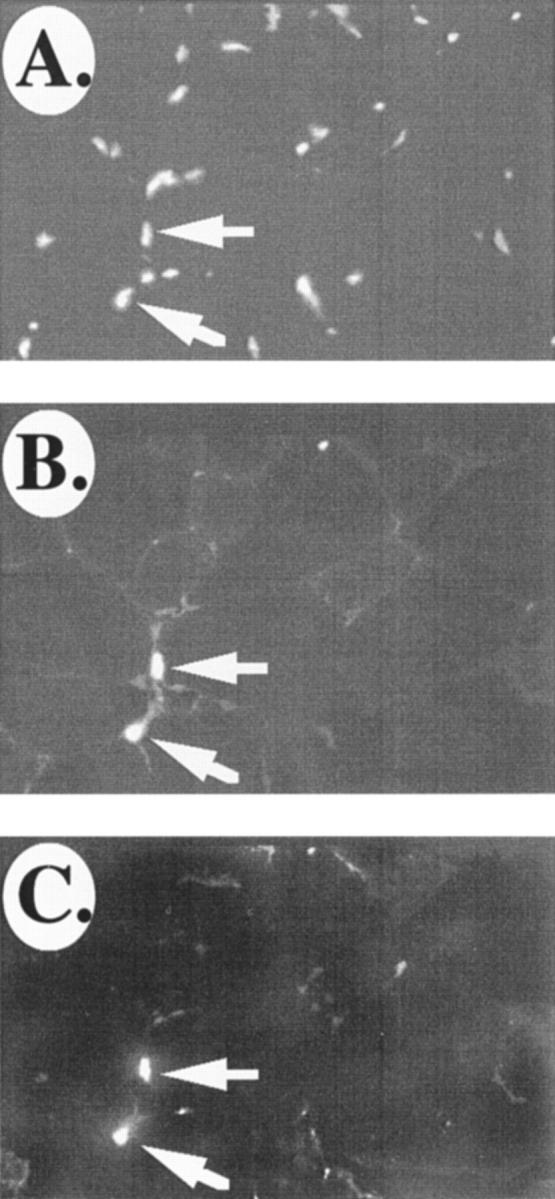


IRF-2 and VCAM-1 are restricted to satellite cells in adult skeletal muscle. (A) Bis-benzimide nuclear staining of a section of mouse adult skeletal muscle. (B) Immunostaining of the section in A for VCAM-1. (C) Double immunostaining of the section in A and B for IRF-2. Arrows indicate IRF-2/VCAM-1-positive cells. (D) High power view of laminin immunostaining of a section of mouse adult skeletal muscle. (E) The section in D was double immunostained for VCAM-1. Note that the VCAM-1-positive cells in the adult muscle are wedged between the basement membrane and the muscle fiber, a location indicative of satellite cells. Arrows in D and E indicate the same location. (F and G) VCAM-1-positive cells in adult skeletal muscle are also positive for the satellite cell marker NCAM. G and F show double immunostaining of a section of adult mouse skeletal muscle for VCAM-1 and NCAM, respectively. Bar, = 25 μm.
Expression of VCAM-1 during Muscle Regeneration
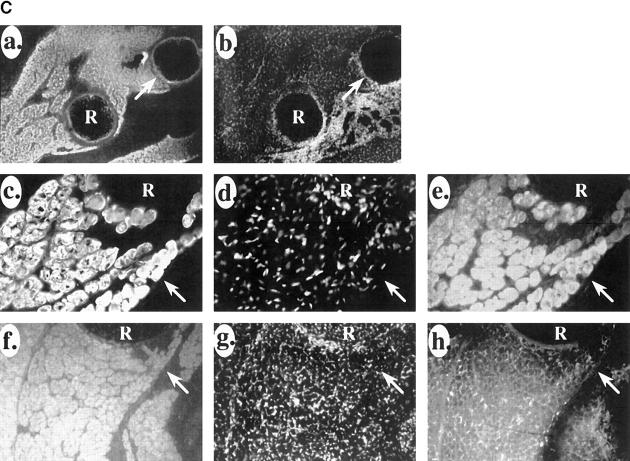
In the adult mouse, the myogenic program can be reactivated after muscle injury or in diseases such as muscular dystrophy. However, myogenesis after muscle injury or disease is distinct from embryonic myogenesis in that stem cell–like satellite cells proliferate and fuse to form muscle fibers. VCAM-1 is expressed on resting satellite cells in adult muscle; however, to determine whether VCAM-1 continues to be expressed on proliferating satellite cells and newly forming myotubes after muscle injury, the gastrocnemius muscle in the mouse hindlimb was exposed surgically and cut. 6 d after the surgery, mice were killed and muscle in the damaged region and control muscle from the undamaged hindlimb were removed, and frozen sections were prepared for immunostaining. Fig. 8, A and B, shows immunostaining for laminin, which is a component of the basement membrane surrounding individual muscle fibers, in normal and damaged muscle. Note the presence of small, newly forming myotubes in the area of muscle damage. NCAM is a marker of the proliferating satellite cells and immature myotubes (Rosen et al., 1992). Neither VCAM-1 nor NCAM were evident on undamaged adult muscle fibers (Fig. 8, C and D). However, the proteins were coexpressed on what appear to be proliferating satellite cells and immature myotubes formed by the fusion of these satellite cells (Fig. 8, E and F). Together, these results indicate that VCAM-1 is expressed during regeneration of myogenic tissue.
Figure 8.
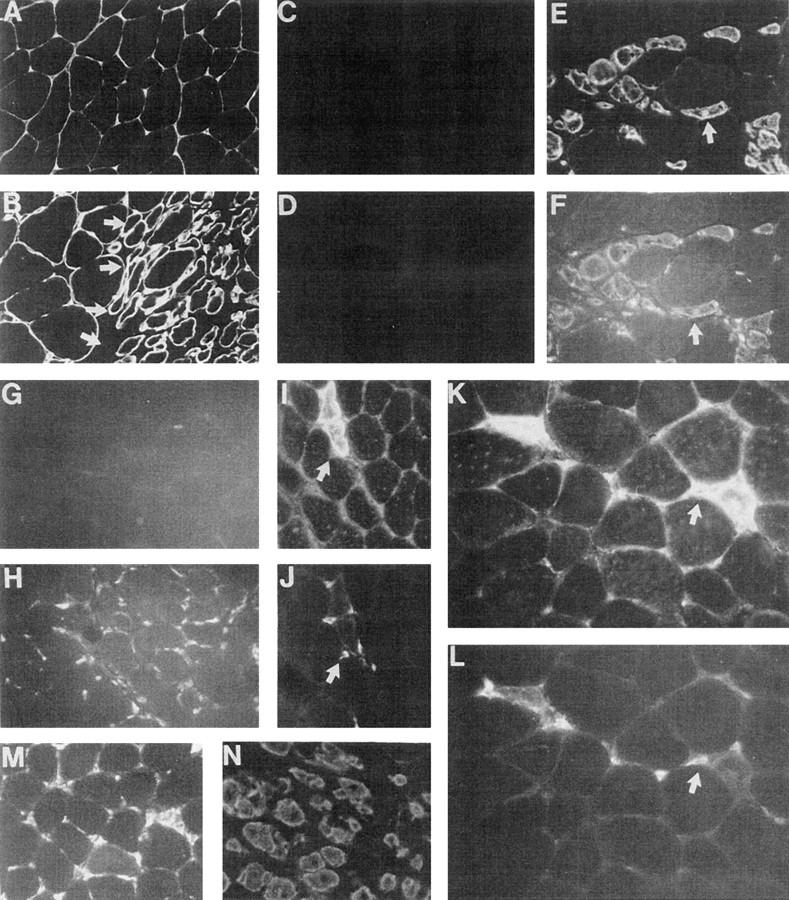
VCAM-1 is expressed after muscle injury and during muscle disease in adult mice. The gastrocnemius hindlimb muscle of adult mice was exposed surgically and damaged by cutting. 6 d after surgery, the mice were killed and the region of damaged muscle along with muscle from a corresponding site in the control hindlimb muscle were removed, and frozen sections were prepared. Immunostaining for laminin in control and damaged muscle is shown in A and B, respectively. Arrows in B denote the site of damage. Small, newly forming myotubes are evident in the damaged area (to the right of the arrows). C and D show immunostaining for NCAM and VCAM-1, respectively, in control muscle. E and F show double immunostaining for NCAM and VCAM-1, respectively, in damaged muscle. Arrows indicate the same location in the two panels. Note the presence of groups of NCAM(+)/ VCAM-1(+) small cells that appear to be proliferating satellite cells and larger cells that appear to be the myotubes that form from the satellite cell fusion. G and H show immunostaining for leukocytes (anti–CD45) in normal and damaged muscle, respectively. I and K show immunostaining for VCAM-1 in damaged muscle, whereas J and L show the same sections double immunostained for CD45. Arrows indicate the same location in the panels. Note the close association of the CD45(+) leukocytes and the VCAM-1(+) muscle cells. M and N show immunostaining for CD45 and VCAM-1, respectively, in sections of limb muscle from dystrophin-deficient mice. Results are all representative of at least four separate experiments.
Expression of VCAM-1 in Regenerating Muscle Tissue Is Closely Associated with Infiltrating Inflammatory Cells
Are IRF-2 and α4β1, the VCAM-1 ligand, also expressed on proliferating satellite cells and the new myotubes formed from the fusion of these cells? This question has proven difficult to address because the muscle injury is accompanied by a large influx of IRF-2-positive, α4β1-positive leukocytes (data not shown), and the number of these leukocytes makes it difficult to determine whether the regenerating muscle cells themselves also express these proteins. IRF-2 was originally identified in lymphocytes, where it functions as a repressor, and α4β1 was originally characterized on leukocytes, where it plays an important role in targeting the leukocytes to the endothelium, which expresses VCAM-1 in response to inflammatory cytokines. It is then difficult to determine whether an interaction between VCAM-1 and α4β1 has a role in muscle regeneration as we propose during embryonic development. However, we noticed that the infiltrating lymphocytes (which were followed by expression of the leukocyte markers CD45, CD3, and α4β1) were selectively and very tightly associated with VCAM-1(+) cells (Fig. 8, I–L and data not shown). None of the lymphocyte markers were evident in undamaged muscle.
VCAM-1 Is Expressed on Adult Muscle in Dystrophin-deficient Mice, Where It Is also Associated with Infiltrating Leukocytes
Dystrophin-deficient mice are a model for human muscular dystrophy and show continuous muscle damage and regeneration. Sections of gastrocnemius muscle from dystrophin-deficient mice were immunostained for VCAM-1 and CD45 to detect infiltrating leukocytes. As in muscle damage, VCAM-1 was expressed on what appear to be proliferating satellite cells and newly forming myotubes, and this expression was closely associated with infiltrating leukocytes (Fig. 8, M and N; results not shown).
Binding of VCAM-1 on C2C12 Satellite Cells to α4β1 on Lymphocytes Mediates Cell–Cell Interactions
Our results in vivo suggest that infiltrating leukocytes are being targeted selectively to satellite cells and newly forming myotubes arising from satellite cell fusion at sites of muscle damage. To determine whether this interaction might be mediated by VCAM-1 on the satellite cells and α4β1 on infiltrating leukocytes (all except neutrophils), we asked whether antibodies to α4β1 would block interaction of lymphocytes with C2C12 satellite cells. C2C12 cells were grown on tissue culture dishes as monolayers, and then the T-cell line Ramos was allowed to adhere to the C2C12 cells. The Ramos cells bound efficiently to the C2C12 cells, and addition of blocking antibodies α4β1 on the lymphocytes blocked the cell–cell interactions, whereas a control antibody to myosin heavy chain has no effect (Fig. 9, A–E). These results demonstrate that VCAM-1 expression can mediate the adhesion of leukocytes to satellite cells through interaction with α4β1 on the leukocytes.
Figure 9.
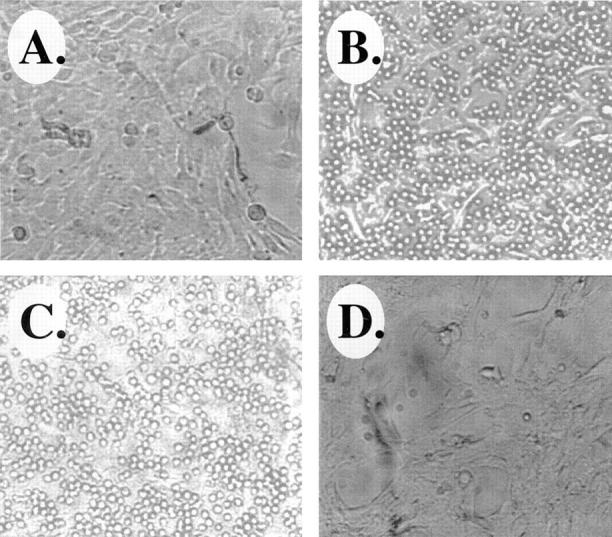
VCAM-1/α4β1 interactions mediate binding of lymphocytes to satellite cells. (A) Photomicrograph of C2C12 satellite cells growing as a monolayer on a tissue culture dish. (B) Photomicrograph showing adhesion of the Ramos T-cell line to monolayers of C2C12 cells. (C) Addition of a control antimyosin heavy chain antibody does not inhibit interaction of Ramos cells with C2C12 cells. (D) Addition of an antibody that specifically blocks the VCAM-1/α4β1 interaction disrupts binding of Ramos cells to the C2C12 cells (see Materials and Methods).
Discussion
We show that IRF-2 regulates transcription of the VCAM-1 gene in C2C12 cells. In vivo expression of IRF-2 is remarkably muscle specific during embryogenesis and is expressed in a pattern consistent with a regulator of VCAM-1 expression in muscle. IRF-2 appears relatively late in muscle differentiation, after myosin heavy chain expression and just preceding VCAM-1 expression. As with VCAM-1, IRF-2 expression is lost in adult muscle fibers; however, IFR-2 is maintained on adult satellite cells, which we propose is responsible for the expression of VCAM-1 in these muscle stem cells. These studies not only provide a mechanism for how VCAM-1 expression is controlled during myogenesis, they also provide the first evidence of a role for IRF-2 in muscle differentiation and regeneration.
Most transcription factors associated with skeletal myogenesis (i.e., myoD, myf-5, etc.) are expressed early in myoblasts and are thought to drive expression of genes such as myosin that are important for the differentiation process. We present evidence here that IRF-2 is expressed after myosin, and is thus a marker of late stages of myogenesis. IRF-2 is then the first transcription factor that we are aware of that specifically marks these late myogenic stages. We propose that it regulates expression of VCAM-1 (and perhaps other late stage myogenic genes). Myogenesis in the mouse and human occur in two waves (see references in Rosen et al., 1992). Initially, a primary generation of myoblasts appear and fuse to form primary myotubes. These primary myotubes are α4β1 positive, but VCAM-1 negative. Then, a second generation of myoblasts appears. These myoblasts are VCAM-1 positive (and α4β1 negative) and appear to use the interaction with α4β1 on primary myotubes to align themselves along the primary myotubes where they, in turn, fuse into myotubes. Most of the muscle fibers are then comprised of such secondary myotubes. It is thus conceivable that IRF-2 is a marker of the second generation myoblasts.
IRF-2 contains both transcriptional activation and repression domains. In lymphocytes, IRF-2 appears to function primarily as a transcriptional repressor (Harada et al., 1989, 1990). However, we demonstrate here that in muscle cells the protein functions as a transcriptional activator. In L929 cells, it has been shown that deletion of the COOH-terminal repressor domain converts IRF-2 into a transcriptional activator (Yamamoto et al., 1990). These results suggest that the repressor domain may be dominant over the transactivation domain. Conceivably, a tissue-specific corepressor molecule may determine whether the repressor domain is active, and in the absence of repressor activity, the molecule functions as a transcriptional activator by default.
We have previously found that VCAM-1 and α4β1 appear to have a role in myogenesis in culture and in vivo (Rosen et al., 1992; unpublished observations). Additionally, here we present evidence of a novel role for VCAM-1 in muscle regeneration. We show that leukocytes are targeted to proliferating satellite cells and newly forming myotubes after muscle injury and disease. In culture, we demonstrate that this interaction is mediated by VCAM-1 on the satellite cells and α4β1 on leukocytes. We propose a model where VCAM-1 appears on endothelial cells in response to muscle damage, and this is responsible at least in part for general recruitment of leukocytes to muscle after injury or disease. After the leukocytes invade the muscle tissue, they are then focused to sites of regeneration by their interaction with VCAM-1 on the regenerating muscle cells.
Hepatocyte growth factor (HGF)/scatter factor interacts with the c-met tyrosine kinase receptor and this can signal satellite cell proliferation (Anastasi et al., 1996). It is likely that c-met is responsible for expansion of the myogenic cell population during development and for stimulating proliferation of resting satellite cells in response to muscle injury. c-met also stimulates hepatocyte proliferation during liver regeneration (Ishiki et al., 1992) and keratinocyte proliferation during wound healing (Dunsmore et al., 1996). Although nothing is known of how the c-met pathway is triggered in response to injury and disease, one common aspect of muscle damage, liver regeneration, and wound healing is the influx of leukocytes into the injury/ disease site. It is possible that cytokines or proteases released from the infiltrating leukocytes play a role in triggering expression/activation of HGF/scatter factor or c-met in the region of VCAM-1-positive satellite cells that proliferate and fuse into myotubes after muscle damage. We suggest that cytokines released by these leukocytes may directly activate HGF/scatter factor or cause its release, which in turn triggers the satellite cell proliferation. One role of VCAM-1 in such a process would be to target the infiltrating leukocytes to the sites of muscle regeneration. These conclusions are supported in tissue culture by the finding that VCAM-1 on C2C12 satellite cells can efficiently mediate interaction with α4β1-positive lymphocytes. Additionally, although VCAM-1 does not appear to have any signaling capabilities, its ligand α4β1 on leukocytes does. It has been demonstrated that binding of α4β1 to VCAM-1 can activate a signal transduction cascade in leukocytes and this interaction, in conjunction with cross-linking of the T-cell receptor, can provide an efficient coactivator role. Therefore, interaction of leukocytes with VCAM-1 not only provides a mechanism for targeting leukocytes to specific areas for muscle regeneration, it also provides a trigger for leukocyte activation and the resulting release of cytokines and proteases into the tissue.
Acknowledgments
We thank Dr. J. Hiscott for anti–IRF-1 and anti–IRF-2 antisera, and IRF-1 and IRF-2 expression plasmids, Dr. M. Thomas for anti–CD45 antisera, Dr. F. Sanchez-Madrid for anti–α4 antisera, and Dr. R. Kopan for antimyosin antisera.
These studies were supported by grants to D.C. Dean from the National Institutes of Health.
Abbreviations used in this paper
- CAT
chloramphenicol acetyl transferase gene
- E
embryonic day
- IFN
interferon
- IRF
interferon regulatory factor
- VCAM-1
vascular cell adhesion molecule-1
References
- Anastasi S, Giordano S, Sthandier O, Gambarotta G, Maione R, Comoglio P, Amati P. A natural hepatocyte growth factor/scatter factor autocrine loop in myoblast cells and the effect of constitutive met kinase activation on myogenic differentiation. J Cell Biol. 1996;137:1057–1068. doi: 10.1083/jcb.137.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew CG, Appel SH. Macromolecular characterization of muscle membranes. I. Proteins and sialic acid of normal and denervated muscle. J Biol Chem. 1973;248:5156–5163. [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsmore SE, Rubin JS, Kovacs SO, Chedid M, Parks WC, Welgus HG. Mechanisms of hepatocyte growth factor stimulation of keratinocyte metalloproteinase production. J Biol Chem. 1996;271:24576–24582. doi: 10.1074/jbc.271.40.24576. [DOI] [PubMed] [Google Scholar]
- Driggers PH, Ennist DL, Gleason SL, Mak W, Marks MS, Levi B, Flanagan JR, Appella E, Ozato K. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1990;87:3743–3747. doi: 10.1073/pnas.87.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elices MJ, Osborn L, Takada Y, Crouse C, Luhowskyj S, Hemler ME, Lobb RR. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/fibronectin binding site. Cell. 1990;60:577–584. doi: 10.1016/0092-8674(90)90661-w. [DOI] [PubMed] [Google Scholar]
- Erle DJ, Ruegg C, Sheppard D, Pytela R. Complete amino acid sequence of an integrin beta subunit (beta 7) identified in leukocytes. J Biol Chem. 1991;266:11009–11016. [PubMed] [Google Scholar]
- Freedman AS, Munro JM, Rice GE, Bevilacqua MP, Morimoto C, McIntyre BW, Rhynhart K, Pober JS, Nadler LM. Adhesion of human B cells to germinal centers in vitro involves VLA-4 and INCAM-110. Science. 1991;249:1030–1033. doi: 10.1126/science.1697696. [DOI] [PubMed] [Google Scholar]
- Gao JX, Issekutz AC. Expression of VCAM-1 and VLA-4 dependent T-lymphocyte adhesion to dermal fibroblasts stimulated with proinflammatory cytokines. Immunology. 1996;89:375–383. doi: 10.1046/j.1365-2567.1996.d01-750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC, Davis V, Li H, McCoy MJ, Sharpe A, Cybulsky MI. Targeted disruption of the murine VCAM-1 gene: essential role of VCAM-1 in chorioallantoic fusion and placentation. Genes Dev. 1995;9:1–14. doi: 10.1101/gad.9.1.1. [DOI] [PubMed] [Google Scholar]
- Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- Iademarco MF, McQuillan JJ, Dean DC. Vascular cell adhesion molecule 1: contrasting transcriptional control mechanisms in muscle and endothelium. Proc Natl Acad Sci USA. 1993;90:3943–3947. doi: 10.1073/pnas.90.9.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iademarco MF, McQuillan JJ, Rosen GD, Dean DC. Characterization of the promoter for vascular cell adhesion molecule-1 (VCAM-1) J Biol Chem. 1992;267:16323–16329. [PubMed] [Google Scholar]
- Ishiki Y, Ohnishi H, Muto Y, Matsumoto K, Nakamara T. Direct evidence that hepatocye growth factor is hepatotropic for liver regeneration and has potential anti–hepatitis effect in vivo. Hepatology. 1992;16:1227–1235. [PubMed] [Google Scholar]
- Johnson DR, Pober JS. HLA class I heavy-chain gene promoter elements mediating synergy between tumor necrosis factor and interferons. Mol Cell Biol. 1994;14:1322–1332. doi: 10.1128/mcb.14.2.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler DS, Veals SA, Fu X, Levy DE. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 1990;4:1753–1765. doi: 10.1101/gad.4.10.1753. [DOI] [PubMed] [Google Scholar]
- Kwee L, Baldwin HS, Shen HM, Stewart CL, Buck C, Buck CA, Labow MA. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development (Camb) 1995;121:489–503. doi: 10.1242/dev.121.2.489. [DOI] [PubMed] [Google Scholar]
- Lin R, Mustafa A, Nguyen H, Gewert D, Hiscott J. Mutational analysis of interferon (IFN) regulatory factors 1 and 2. J Biol Chem. 1994;269:17542–17549. [PubMed] [Google Scholar]
- Miyake K, Medina K, Ishahara H, Kimoto R, Auerbach R, Kincade P. A VCAM-like adhesion molecule on murine bone marrow stromal cells mediates binding of lymphocyte precursors in culture. J Cell Biol. 1991;114:557–565. doi: 10.1083/jcb.114.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish AS, Read MA, Thanos D, Pine R, Maniatis T, Collins T. Endothelial interferon regulatory factor 1 cooperates with NF-kappa-B as a transcriptional activator of vascular cell adhesion molecule 1. Mol Cell Biol. 1995;15:2558–2569. doi: 10.1128/mcb.15.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Mustafa A, Hiscott J, Lin R. Transcription factor IRF-2 exerts its oncogenic phenotype through the DNA binding/transcription repression domain. Oncogene. 1995;11:537–544. [PubMed] [Google Scholar]
- Nelson N, Marks MS, Driggers PH, Ozato K. Interferon consensus sequence-binding protein, a member of the interferon regulatory factor family, suppresses interferon-induced gene transcription. Mol Cell Biol. 1993;13:588–599. doi: 10.1128/mcb.13.1.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontell M. Neonatal muscle: an electron microscopic study. Anat Rec. 1977;189:669–690. doi: 10.1002/ar.1091890410. [DOI] [PubMed] [Google Scholar]
- Ontell M, Bourke D, Hughes D. Cytoarchitecture of the fetal murine soleus muscle. Am J Anat. 1988;181:267–278. doi: 10.1002/aja.1001810305. [DOI] [PubMed] [Google Scholar]
- Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi RG, Lobb R. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell. 1989;59:1203–1211. doi: 10.1016/0092-8674(89)90775-7. [DOI] [PubMed] [Google Scholar]
- Prudhomme JG, Sherman IW, Land KM, Moses AV, Stenglein S, Nelson JA. Studies of Plasmodium falciparum cytoadherence using immortalized human brain capillary endothelial cells. Int J Parasitol. 1996;26:647–655. doi: 10.1016/0020-7519(96)00027-6. [DOI] [PubMed] [Google Scholar]
- Reis LFL, Harada H, Wolchok JD, Taniguchi T, Bilcek J. Critical role of a common transcription factor, IRF-1, in the regulation of IFN-beta and IFN-inducible genes. EMBO (Eur Mol Biol Organ) J. 1992;11:185–193. doi: 10.1002/j.1460-2075.1992.tb05041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GE, Munro JM, Corless C, Bevilacqua MP. Vascular and nonvascular expression of INCAM-110. A target for mononuclear leukocyte adhesion in normal and inflamed human tissues. Am J Pathol. 1991;138:385–393. [PMC free article] [PubMed] [Google Scholar]
- Rosen GD, Sanes JR, LaChance R, Cunningham JM, Roman J, Dean DC. Roles for the integrin VLA-4 and its counter receptor VCAM-1 in myogenesis. Cell. 1992;69:1107–1119. doi: 10.1016/0092-8674(92)90633-n. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Duxson MJ, Harris AJ. Neural determination of muscle fibre numbers in embryonic rat lumbrical muscles. Development (Camb) 1987;100:395–409. doi: 10.1242/dev.100.3.395. [DOI] [PubMed] [Google Scholar]
- Ruegg C, Postigo AA, Sikorski EE, Butcher EC, Pytela R, Erle DJ. Role of integrin alpha 4 beta 7/alpha 4 beta P in lymphocyte adherence to fibronectin and VCAM-1 and in homotypic cell clustering. J Cell Biol. 1992;117:179–189. doi: 10.1083/jcb.117.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeren RA, Koopman G, Van der Baan S, Meijer CJ, Pals ST. Adhesion receptors involved in clustering of blood dendritic cells and T lymphocytes. Eur J Immunol. 1991;21:1101–1105. doi: 10.1002/eji.1830210503. [DOI] [PubMed] [Google Scholar]
- Seko Y, Yagita H, Okumura K, Yazaki Y. Expression of vascular cell adhesion molecule-1 in murine hearts with acute myocarditis caused by coxsackievirus B3. J Pathol. 1996;180:450–454. doi: 10.1002/(SICI)1096-9896(199612)180:4<450::AID-PATH693>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Sheppard AM, McQuillan JJ, Iademarco MF, Dean DC. Control of vascular cell adhesion molecule-1 gene promoter activity during neural differentiation. J Biol Chem. 1995;270:3710–3719. doi: 10.1074/jbc.270.8.3710. [DOI] [PubMed] [Google Scholar]
- Sheppard AM, Onken MD, Rosen GD, Noakes PG, Dean DC. Expanding roles for alpha 4 integrin and its ligands in development. Cell Adhes Commun. 1994;2:27–43. doi: 10.3109/15419069409014200. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Newman W, Gopal TV, Horgan KJ, Graber N, Beall LD, van Seventer GA, Shaw S. Four molecular pathways of T cell adhesion to endothelial cells: roles of LFA-1, VCAM-1, and ELAM-1 and changes in pathway hierarchy under different activation conditions. J Cell Biol. 1991;113:1203–1212. doi: 10.1083/jcb.113.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski EE, Hallmann R, Berg EL, Butcher EC. The Peyer's patch high endothelial receptor for lymphocytes, the mucosal vascular addressin, is induced on a murine endothelial cell line by tumor necrosis factor-alpha and IL-1. J Immunol. 1993;151:5239–5250. [PubMed] [Google Scholar]
- Stockdale FE, Miller JB. The cellular basis of myosin heavy chain isoform expression during development of avian skeletal muscles. Dev Biol. 1987;123:1–9. doi: 10.1016/0012-1606(87)90420-9. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Kawakami T, Taniguchi T. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol Cell Biol. 1993;13:4531–4538. doi: 10.1128/mcb.13.8.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill MH, Wellicome SM, Mahiouz DL, Lanchbury JS, Kyan AU, Haskard DO. Tumor necrosis factor combines with IL-4 or IFN-gamma to selectively enhance endothelial cell adhesiveness for T cells. The contribution of vascular cell adhesion molecule-1-dependent and -independent binding mechanisms. J Immunol. 1991;146:592–598. [PubMed] [Google Scholar]
- Vaughan PS, Aziz F, Vanwijnen AJ, Wu SJ, Harada H, Taniguchi T, Soprano KJ, Stein JL, Stein GS. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- Veals SA, Schindler C, Leonard D, Fu XY, Aebersold R, Darnell JE, Levy DE. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Lamphier MS, Fujita T, Taniguchi T, Harada H. The oncogenic transcription factor IRF-2 possesses a transcriptional repression and a latent activation domain. Oncogene. 1994;9:1423–1428. [PubMed] [Google Scholar]
- Yang JT, Rayburn H, Hynes RO. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development (Camb) 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]