Abstract
The flagellar basal apparatus comprises the basal bodies and the attached fibrous structures, which together form the organizing center for the cytoskeleton in many flagellated cells. Basal apparatus were isolated from the naked green flagellate Spermatozopsis similis and shown to be composed of several dozens of different polypeptides including a protein band of 95 kD. Screening of a cDNA library of S. similis with a polyclonal antibody raised against the 95-kD band resulted in a full-length clone coding for a novel protein of 834 amino acids (90.3 kD). Sequence analysis identified nonhelical NH2- and COOH-terminal domains flanking a central domain of ∼650 residues, which was predicted to form a series of coiled-coils interrupted by short spacer segments. Immunogold labeling using a polyclonal antibody raised against the bacterially expressed 95-kD protein exclusively decorated the striated, wedge-shaped fibers, termed sinister fibers (sf-fibers), attached to the basal bodies of S. similis. Striated fibers with a periodicity of 98 nm were assembled in vitro from the purified protein expressed from the cloned cDNA indicating that the 95-kD protein could be a major component of the sf-fibers. This structure interconnects specific triplets of the basal bodies with the microtubular bundles that emerge from the basal apparatus. The sf-fibers and similar structures, e.g., basal feet or satellites, described in various eukaryotes including vertebrates, may be representative for cytoskeletal elements involved in positioning of basal bodies/centrioles with respect to cytoskeletal microtubules and vice versa.
The base of eukaryotic flagella is formed by specialized microtubular structures, basal bodies. The structure of the basal bodies, consisting of nine microtubular triplets, is highly conserved and also characteristic for centrioles, which are part of the centrosome in many eukaryotic cells. During sexual and asexual reproduction, basal bodies are often converted into centrioles, indicating their functional similarity (Kellogg et al., 1994). It is generally accepted that basal bodies/centrioles function as a template for the structure of the axoneme (Lange and Gull, 1996b ). An open question is whether or not centrioles contribute to the centrosome of animal cells, for example, by organizing its microtubule nucleation sites or by controlling its duplication.
The basal bodies of many flagellated cells not only serve as assembly sites for the axoneme but also represent the central structures of the basal apparatus—a functional homologue of the centrosomal complex of animal cells (Baron and Salisbury, 1992). The basal apparatus consists of the basal bodies and a diverse array of filaments attached to them. Sets of flagellar root microtubules usually radiate from the basal apparatus and form the major part of the microtubular cytoskeleton (which is involved in the determination of polarity, shape, and internal organization of flagellated cells) (Moestrup, 1982; Melkonian, 1984; Holmes and Dutcher, 1989). Thus, analysis of the flagellar basal apparatus could provide information about the basal bodies/centrioles as centrosomal organizers.
In green flagellates like Chlamydomonas reinhardtii, two types of the filaments present in the basal apparatus have been analyzed in detail (Lechtreck and Melkonian, 1991a ). One is characterized by the presence of centrin, a 20-kD protein belonging to the EF-hand family of calcium-binding proteins (Huang et al., 1988). Centrin-based filaments (system II fibers; Melkonian, 1980) form a connection between the basal bodies, interconnect the basal bodies and the cell nucleus, and are part of the flagellar transitional region; they undergo calcium-induced contractions and are involved in various motile functions of the basal apparatus (Melkonian et al., 1992; Salisbury, 1995). The other well-characterized filaments of the basal apparatus are the noncontractile, striated microtubule-associated fibers (SMAFs,1 or system I fibers) (Melkonian, 1980) that are composed of striated fiber (SF)-assemblin, a highly α-helical 30-kD protein (Lechtreck and Melkonian, 1991b ; Weber et al., 1993). These are attached to the four bundles of flagellar root microtubules characteristic for Chlamydomonas-like flagellates and are thought to function as stabilizing elements in the basal apparatus (Lechtreck and Silflow, 1997). Electron microscopical studies have revealed additional filamentous structures attached to the basal bodies in the green algal basal apparatus (Ringo, 1967; Beech and Melkonian, 1993; Geimer et al., 1997a): transitional fibers (connectives between the basal bodies and the plasma membrane), proximal plates or connecting fibers, and linkers between basal bodies and flagellar root microtubules. The latter may establish the spatial relationship between the basal bodies and the flagellar root microtubules, as well as direct the microtubular roots to specific locations in the cell. In contrast to centrin- and SF-assemblin–based fibers, these other fibers and linkers are relatively small, making it more difficult to identify their biochemical composition.
We have recently developed a method to obtain homogeneous preparations of basal apparatus from the naked, biflagellate green alga Spermatozopsis similis (Geimer et al., 1997b ). The complex, filamentous system surrounding the two basal bodies in the isolated state has been studied in detail by serial thin section electron microscopy (Geimer et al., 1997a). Besides the major proteins of the basal apparatus (tubulin, centrin, and SF-assemblin) more than two dozen different polypeptides were present in our preparations. Here we identify a novel 95-kD protein as a component of the sinister fibers (sf-fibers), which are linkers between the basal bodies and two of the four microtubular flagellar roots in the basal apparatus of S. similis. The protein contains extended coiled-coil domains and assembles into striated fibers in vitro. The green algal sf-fibers resemble the basal feet that have been described in various eukaryotes (Gibbons, 1961), and may be representative of a particular type of basal body–microtubular flagellar root linker that is a common component of the cytoskeleton.
Materials and Methods
Strains and Culture Conditions
Spermatozopsis similis (Sammlung von Algenkulturen Göttingen [SAG] B 1.85; Schlösser, 1994) and Chlamydomonas reinhardtii cw 15+ (SAG 83.81) were maintained in aerated cultures (∼1 liter × min−1) at 15°C with a light-dark cycle of 14:10 h and a photon flux density of 20 μEinstein (E) × m−2 × s−1 (Osram 40W/25 universal white) in Waris medium (McFadden and Melkonian, 1986).
Isolation of a 95-kD Protein Band from the Basal Apparatus
The isolation and purification of basal apparatus of Spermatozopsis similis has been described (Snell et al., 1974; Geimer et al., 1997b ). Briefly, concentrated cells were lysed with 1.5% Triton X-100 in TE buffer (10 mM Tris-HCl, pH 7.4, 2 mM EDTA), disintegrated by homogenization, washed with TE buffer/0.5 % Triton X-100 (800 g, 30 min), and then the suspension was loaded onto a discontinuous sucrose gradient (6 ml 60%, 9 ml 50%, 9 ml 40%, and 6 ml 20% in TE buffer/0.1% Triton X-100) and centrifuged for 1 h at 12,500 g in a swing-out rotor (HB-4; Sorvall, Bad Homburg, Germany). Highly enriched basal apparatus were collected from a milky band below the 20–40% sucrose interphase, and the proteins were separated by preparative SDS-PAGE. A band of 95 kD was excised from Coomassie blue–stained gels, concentrated by electrophoresis, and then blotted onto nitrocellulose membrane. The membrane with ∼10 μg of the 95-kD band was dissolved in dimethyl sulfoxide, mixed with complete Freunds adjuvant, and subcutanously injected at several sites into a young, male rabbit. For subsequent booster injections on day 15 and 53, we used acrylamide gel pieces with ∼10 μg antigen, homogenized in PBS, and then mixed with incomplete Freunds adjuvant. An IgG fraction (anti-p95) was purified from the serum taken 10 d after the second injection using a fast protein liquid chromatography (FPLC) system (Pharmacia LKB, Uppsala, Sweden) and a protein A column (Pharmacia LKB).
Cloning and Sequencing of a cDNA Coding for the 95-kD Protein
Total RNA from S. similis was isolated by the phenol/SDS method basically following the protocol of Palmiter (1974). To purify mRNA, a self-packed oligo-dT column (Boehringer Mannheim GmbH, Mannheim, Germany) was used. A Uni-Zap XR cDNA library was constructed using a cDNA synthesis and cloning kit (Stratagene, Heidelberg, Germany) according to the manufacturer's instructions with the following modifications: first-strand synthesis was done for 15 min, 45°C; 20 min, 50°C; and 20 min, 55°C, with reverse transcriptase from GIBCO BRL (Eggenstein, Germany). The library was screened with anti-p95, and positive clones were analyzed by restriction mapping and partial sequencing after in vivo excision of the pBluescript phagemid. In addition, bacterial lysates obtained from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced cultures were analyzed with anti-p95 in Western blots. A 3.1-kb clone (clone 7/1) produced an immunoreactive band of 95 kD. The insert was sequenced completely in both directions (Abiprism, 310 Gene Analyzer; Perkin Elmer Applied Biosystems, Weiterstadt, Germany). The AutoAssembler 1.4.0 (Perkin Elmer Applied Biosystems) program was used for gene assembly and the sequence was analyzed using programs (COILS, FASTA, BLAST) provided by European Bioinformatics Institute (EBI; http://www.ebi.ac.uk/services/services.html) and Baylor College of Medicine (BCM; http://dot.imgen.bcm.tmc.edu:9331/seq-search/struc-predict.html) via the internet (Altschul et al., 1990; Lupas, 1996).
Purification of the Recombinant 95-kD Protein and Antibody Production
For induction of gene expression, Escherichia coli XL-1Blue cells containing clone 7/1 were grown at 37°C to a density of OD600 0.8–1.0 and induced with 2 mM IPTG. After 4 h the cells were harvested (1,000 g, 15 min, 4°C), resuspended in 50 mM NaH2PO4, 300 mM NaCl, pH 7.8, and frozen at −80°C for 20 min. After thawing, the bacteria were sonicated five times for 10 s, and the insoluble material was pelleted by centrifugation (10,000 g, 10 min, 4°C). The pellet was washed three times (10,000 g, 10 min, 4°C) in washing buffer (50 mM Tris-HCl, 1 mM EDTA, 3% Triton X-100, 0.05% β-mercaptoethanol, pH 7.5) and finally twice in 50 mM Tris-HCl, pH 7.5. Then, the pellet was resuspended in 20 mM Tris-HCl, 2 mM EDTA, 1 mM DTT, pH 8.0, containing 8 M urea and extracted for 2 h at room temperature. To remove insoluble material the suspension was centrifuged at 18,000 g for 30 min and filtered (0.22 μm, Millex-GV; Millipore, Eschborn, Germany). The proteins were separated on a MonoQ column (HR 5/5, Pharmacia LKB) equlibrated with the urea buffer using a FPLC system (Pharmacia LKB). The column was eluted with a linear salt gradient (0–50 mM KCl) in the same solvent and fractions containing the 95-kD protein were identified by SDS-PAGE. MonoQ fractions of interest were applied to preparative SDS-PAGE. For internal or NH2-terminal amino acid sequencing, the protein was transferred to polyvinylidene difluoride membrane. For antibody production the recombinant 95-kD protein was cut out, reelectrophorized, and electroeluted. The protein concentration was determined using the method of Neuhoff et al. (1979), and four injections of 75 μg of homogenous 95-kD protein each were used to generate polyclonal antibodies (anti-rec95) in rabbit (as above or by BioGenes, Berlin, Germany) (day 1, intralymphatic; day 7, intramuscular; day 14, subcutan; day 29, intramuscular).
Reassembly of Striated Fibers from the Recombinant 95-kD Protein
MonoQ fractions containing the 95-kD protein (>95% pure, ∼0.1 μg/μl) were dialyzed using a floating membrane filter (type VS, pore size 0.025 μm; Millipore) against an excess amount of 10 mM Tris-HCl, 1 mM EDTA, 150 mM KCl, pH 7.0, with decreasing concentrations of urea (4 M, 2 M, 0.8 M, and 0 M) for 120 min at each urea concentration. An alternative protocol used dialysis tubes (12,000–14,000 D; Medicell, London, UK) and prolonged dialysis times (4 h) for each concentration. Reassembled fibers were analyzed by whole mount electron microscopy. The fibers were harvested by centrifugation (100,000 g, 30 min), the proteins in the supernatant were precipitated by the addition of acetone (1:9), and both fractions analyzed by SDS-PAGE.
Immunoblot Analysis
Whole cells, isolated cytoskeletons, or enriched basal apparatus of S. similis were separated by SDS-PAGE and transferred to PVDF membrane (Millipore Corp., Bedford, MA). After extensive blocking with PBS-T (PBS with 0.05% Tween 20) containing 3% gelatin from cold water fish (Sigma Chemical Co., St. Louis, MO) the membrane strips were incubated with the primary antibody (anti-p95, 1:500 or anti-rec95, 1:1,000 in PBS-T/3% gelatin) for 60 min. The membrane was washed five times for 15 min with PBS-T, and then blocked again for 30 min, before incubation with anti– rabbit IgG peroxidase conjugate (Sigma Chemical Co.; 1:1,000 in PBS-T/3% gelatin) for 60 min. Membrane strips were washed five times for 15 min in PBS and developed using 4-chloro-1-naphthol as the substrate.
Indirect Immunofluorescence
The preparation of isolated cytoskeletons of S. similis for indirect immunofluorescence and antibody staining has been described previously (Lechtreck and Melkonian, 1991b ). The anti-p95 and anti-rec95 sera were used in a 1:100 dilution; monoclonal anti–α-tubulin was applied simultaneously. Secondary antibodies (anti–rabbit IgG or anti–mouse IgG conjugated to FITC or TRITC, Sigma Chemical Co.) were diluted 1:50. Immunofluorescence images were observed using a Zeiss IM35 equiped with a 1,3/100× oil immersion objective (Carl Zeiss, Oberkochen, Germany), documented on HP5 (Ilford Ltd., Knutsford, UK), digitized, labeled, and assembled using Photoshop (Adobe Systems Inc., Mountain View, CA) and Powerpoint (Microsoft, Redmond, WA).
Preembedding Immunogold Electron Microscopy
Cells of S. similis were washed by centrifugation in MT (30 mM Hepes, 5 mM EDTA, 15 mM KCl, pH 7) supplemented with 5 mM MgSO4 (MT-Mg2+) and lysed by the addition of an equal volume MT-Mg2+/2% Triton X-100, immediately followed by fixation with 1% paraformaldehyde (final concentration in MT-Mg2+) for 5 min. The cytoskeletons were washed twice in the presence of the fixative (80 g, 10 min, 15°C). All subsequent steps were performed as described previously (McFadden et al., 1987). The anti-rec95 was used at a 1:200 final dilution and detected with protein A conjugated to 15-nm gold particles. Samples were transferred to agar, dehydrated, and then embedded in Epon 812 (Serva, Heidelberg, Germany) as previously described (McFadden and Melkonian, 1986).
Standard and Whole Mount Electron Microscopy
For whole mount electron microscopy, 5 μl of suspended particles were applied to copper grids coated with pioloform (Plano GmbH, Marburg, Germany) and allowed to adhere for 10–60 min. The grids were stained with 1% aqueous uranyl acetate for 1–2 min. Utrathin sections were cut with a MT-6000 microtome (RMC, Tucson, AZ) and stained with uranyl acetate and lead citrate (Reynolds, 1963). Micrographs were taken with a transmission electron microscope (CM 10; Philips Electron Optics, Mahwah, NJ) using Scientia EM Film (Agfa, Leverkusen, Germany).
Results
Identification of 95-kD Components in the Basal Apparatus
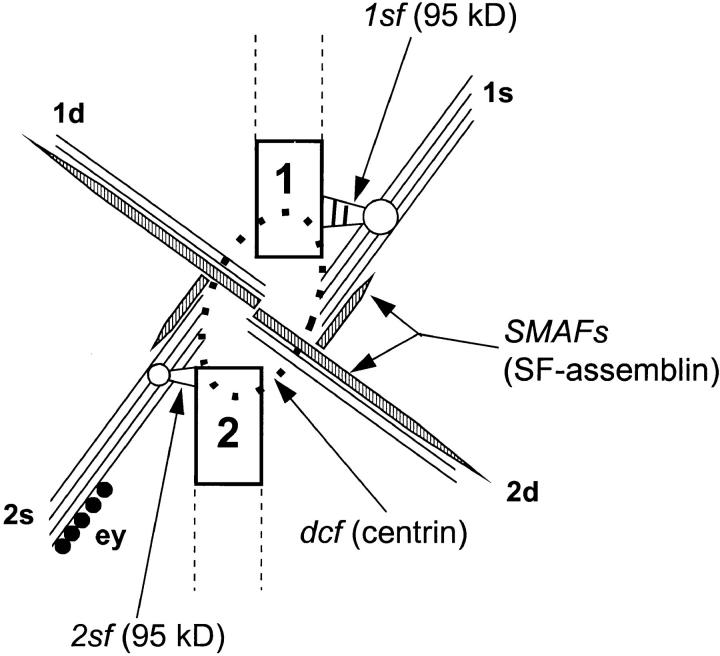
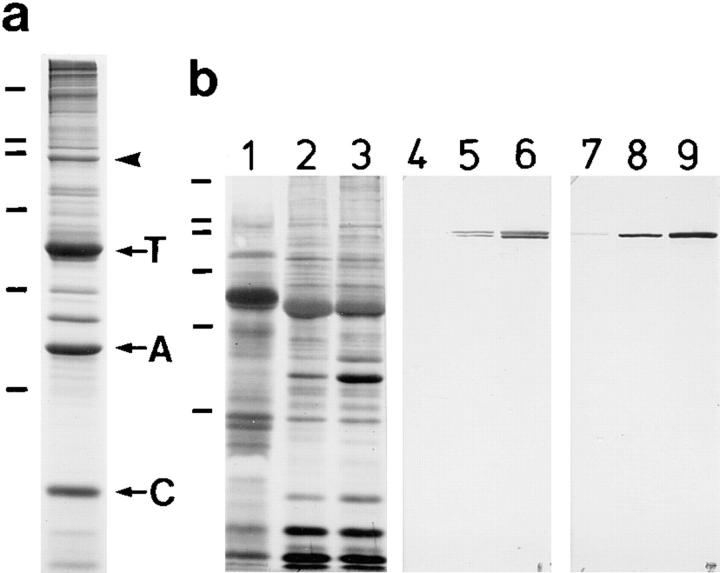
Spermatozopsis similis is a naked, biflagellate green alga that resembles Chlamydomonas reinhardtii in its major ultrastructural features. Detailed descriptions of the cells, isolated cytoskeletons, and basal apparatus are given in Preisig and Melkonian (1984), Lechtreck et al. (1989), and Geimer et al. (1997a). A uniform terminology for the structures in the basal apparatus of green flagellates has now been established (Moestrup and Hori, 1989; Beech et al., 1991), and the developmentally oldest basal body is designated as basal body No. 1, and the suffixes “s” (for sinister (Latin) = left) and “d” (for dexter = right) are used for the associated structures according to their position relative to basal body No. 1 (see Fig. 9). The four microtubular roots of the cruciate x-2-x-2 flagellar root system (where x = 3–6 microtubules), which are characteristic for Chlamydomonas-like flagellates, are called 1s (x-root of basal body No. 1) and 1d (two-stranded microtubular root of basal body No. 1), or 2s and 2d if associated with basal body No. 2. The fibrous structures that connect the basal bodies to the microtubular roots are called 1sf, 1df, 2sf, and 2df, respectively (Beech and Melkonian, 1993). The dominant proteins in preparations of isolated basal apparatus of S. similis were tubulin, centrin, and SF-assemblin (Geimer et al., 1997b ; Fig. 1 a). Several proteins with molecular weights >50 kD were enriched in fractions of basal apparatus purified by sucrose gradient centrifugation in comparison to crude fractions. Most of these proteins constitute only 1% or less of the total protein; however, a band of 95 kD was present comprising 2–4% of the proteins in purified basal apparatus (Fig. 1 a). The 95-kD band was purified by SDS-PAGE and used to raise a polyclonal antibody (anti-p95). When isolated cytoskeletons of S. similis were stained with anti-p95 for indirect immunofluorescence, the region of the basal apparatus was labeled (Fig. 6, a and b). In Western blotting experiments, this antibody labeled two closely spaced bands of 95 kD, which were notably enriched in the basal apparatus in comparison to whole cells or isolated cytoskeletons (Fig. 1 b).
Figure 9.
Cartoon of the basal apparatus of S. similis in top view. 1, older basal body; 2, younger basal body; 1d, 1s, 2d, and 2s, microtubular roots with attached SMAFs. dcf, distal connecting fiber indicated by a dotted line. ey, eyespot apparatus. Three biochemically distinct filament systems (centrin, SF-assemblin, and 95-kD protein) have been identified in the basal apparatus of flagellate green algae.
Figure 1.
(a) SDS-PAGE analysis (11%) of isolated basal apparatus of S. similis. Protein pattern characteristic for basal apparatus of S. similis purified by sucrose gradient centrifugation. Centrin (C), SF-assemblin (A), tubulin (T), and the 95-kD band (arrowhead) are marked. A polyclonal antibody (anti-p95) was raised against the electrophoretically purified 95-kD band. (b) Western blot analysis of S. similis with anti-p95 and anti-rec95. Membrane strips were stained with amido black (lanes 1–3), anti-p95 (lanes 4–6), or anti-rec95 (lanes 7–9), an antibody directed against bacterial expressed 95-kD protein (see also Fig. 4). 10 μg of whole cells (1, 4, and 7), isolated cytoskeletons (2, 5, and 8), or enriched basal apparatus (3, 6, and 9) were loaded per lane. Anti-p95 labeled two closely spaced bands, but only the upper one also cross-reacts with anti-rec95. The antibody staining reveals that the 95-kD proteins were highly enriched in the isolated basal apparatus of S. similis. Preimmune controls are shown in Fig. 4 b. The positions of standard proteins are indicated on the left (from top for a/b: 205, 116, 97, 66, 45, and 29 kD).
Figure 6.
Indirect immunofluorescence of S. similis. (a) Isolated cytoskeletons of S. similis stained with monoclonal anti–α-tubulin. (b) Anti-p95 immunofluorescence corresponding to a. In addition to the prominent dot-like staining in the basal apparatus, we often observed a faint labeling at the posterior end of the cell, where the microtubules are interconnected by a cap-like structure. (c and d) Isolated cytoskeletons of S. similis stained with anti-rec95, which labels two dots of unequal size. (e) Anti-tubulin staining of isolated cytoskeletons of S. similis. The corresponding signal obtained by anti-rec95 (f) consists of two dots in the region of the basal apparatus. Bars, 10 μm.
Cloning of the 95-kD Protein
SDS-PAGE analysis, indirect immunofluorescence and Western blotting analysis indicated that the antigens recognized by anti-p95 were components of the basal apparatus of S. similis (Figs. 1 and 6). Screening of a S. similis cDNA expression library with anti-p95 resulted in three clones with inserts of 3.1 kb and two shorter variants (1.9 and 2.3 kb), all with identical 3′ sequences. Both strands of one of the 3.1-kb clones (clone 7/1) were completely sequenced. The sequence contained a large open reading frame of 2,505 bp that had a GC content of 75%; the putative initiation codon was preceded by an in-frame stop codon (Fig. 2). An immunoreactive 95-kD protein was present in bacteria containing clone 7/1 after induction with IPTG (see Fig. 4, a, lane 2, and b), demonstrating that the clone is full length. Southern blot analysis using the insert of clone 7/1 as a probe indicated that the protein is encoded by a single copy gene in S. similis (data not shown).
Figure 2.
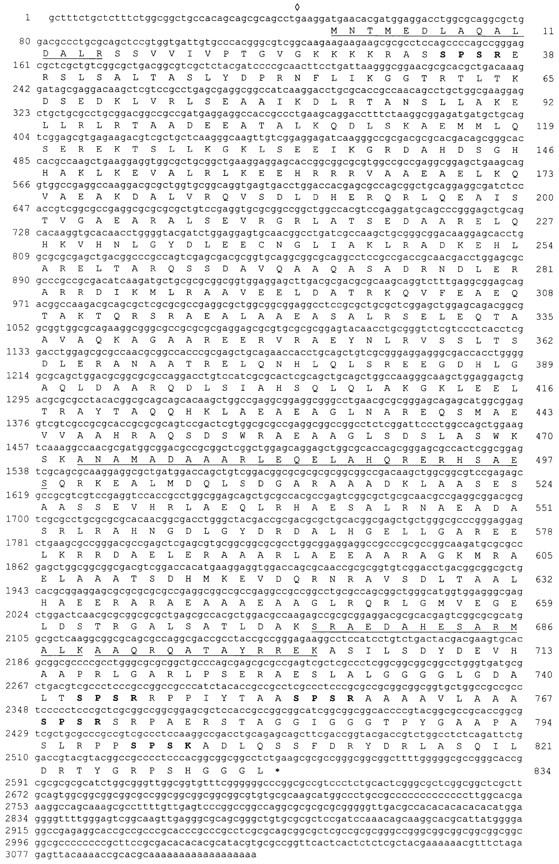
Sequence of the 95-kD protein. The cDNA sequence of clone 7/1 and the deduced amino acid sequence of the longest open reading frame are shown. The start codon is preceded by a stop codon (⋄). Peptide sequences determined by NH2-terminal or internal sequencing of the recombinant 95-kD protein are underlined. The SPS(R/K) motifs present in the NH2- and COOH-terminal domains are marked by bold letters. These sequence data are available from GenBank/EMBL/DDBJ under accession number AJ001438.
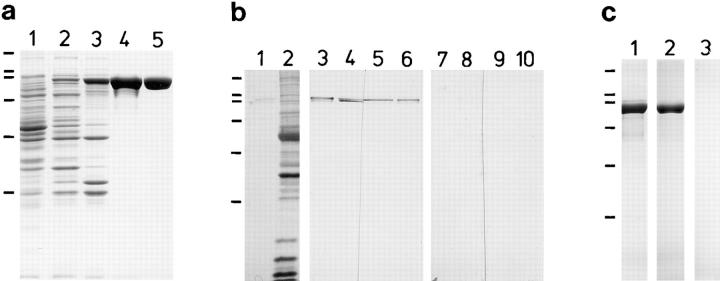
Figure 4.
(a) Purification of the recombinant 95-kD protein (SDS-PAGE 11%). Lane 1, bacteria without plasmid; lane 2, bacteria induced with IPTG for 2 h. Note the additional band of 95 kD. Lane 3, inclusion bodies enriched in the 95-kD protein; lane 4, MonoQ fraction containing the 95-kD protein; and lane 5, 95-kD protein purified by preparative electrophoresis from fractions as shown in lane 4. MonoQ-purified 95-kD protein was used for reassembly experiments, SDS-PAGE–purified protein was used for antibody (anti-rec95) production. (b) Western blot analysis of recombinant 95-kD protein (1, 3, 6, 7, and 10) and isolated basal apparatus of S. similis (2, 4, 5, 8, and 9). Membrane strips were stained with amido black (1 and 2), anti-p95 (3 and 4), anti-rec95 (5 and 6), and preimmune sera of both animals (7 and 8, and 9 and 10), respectively. To allow an exact comparison of the antigenic bands recognized by anti-p95 and anti-rec95, “double slots” were loaded with basal apparatus proteins (4/5 and 8/9), and the blot cut before antibody staining. Only the upper one of the two bands labeled with anti-p95 is also stained with anti-rec95 (compare lanes 4 and 5), demonstrating that anti-rec95 is monospecific. The additional bands visible in lanes 3 and 6 are caused by the presence of degradation products of the 95-kD protein (see also lane 1). (c) SDS-PAGE of in vitro reassembly experiments with recombinant 95-kD protein. Lane 1, MonoQ fraction; lane 2, pellet (100,000 g) after stepwise dialysis against urea-free buffer; and lane 3, supernatant corresponding to lane 2. Similar amounts of the samples were loaded in each lane. Almost the entire amount of the 95-kD protein is found in the pellet.
Sequence Analysis
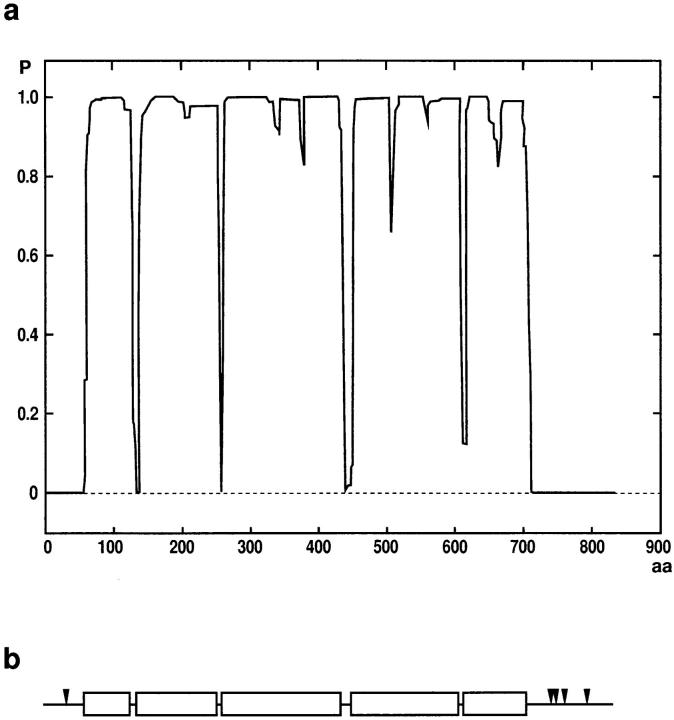
The ORF of clone 7/1 encodes a protein of 834 amino acids that has a predicted isoelectric point of 7.4 and a mass of 90.3 kD (Fig. 2). Sequence analysis using the algorithm of Garnier et al. (1978) identified a central domain of ∼650 residues that is predominantly α-helical (∼88%), flanked by nonhelical NH2- and COOH-terminal domains with lengths of ∼65 and 120 residues, respectively. The program COILS (Lupas, 1996) predicted a high probability of the central domain forming coiled-coil structures separated by short spacers around residues 140, 270, 450, and 620 (Fig. 3 a). Because of the presence of heptad repeats, two leucine zipper motifs (L-6X-L-6X-L-6X-L in position 392–413 and 572–593) were identified by PROSITE. The head and tail domains contained all prolines (14 of 17 located in the COOH-terminal domain) and five SPS(K/R) motifs (four in the COOH-terminal domain), which resemble the known consensus sequences for p34cdc2 kinase (Moreno and Nurse, 1990; Figs. 2 and 3 b). Database searches using the complete amino acid sequence of the 95-kD protein detected only low (<25%) homologies to other proteins with large coiled-coil domains (e.g., myosin, paramyosin, and intermediate filament proteins). In comparison with those proteins, the 95-kD protein displays a high content of alanine (19.7%), whereas isoleucine (1.4%) is underrepresented. This might reflect the high GC-content of the cDNA of the 95-kD protein (e.g., isoleucine has the codons AT[A/T/C]; 161 alanines are encoded by GC[C/G], but only three by GC[A/T]).
Figure 3.
Structure of the 95-kD protein. (a) Coiled-coil–forming probability (P) of the 95-kD protein as calculated with the program COILS (parameters: MTIDK matrix, window = 28, weighting of positions a and d = 2.5; Lupas, 1996). The central part of the 95-kD protein is formed by a series of predicted coiled-coils interrupted by short spacers. (b) Schematic presentation of the 95-kD protein. Boxes, coiled coil regions; arrowheads, position of SPS(R/K) motifs.
Purification of Recombinant 95-kD Protein and Antibody Production
Electrophoretic analysis of bacterial lysates showed that large quantities of recombinant 95-kD protein were present in the insoluble fraction (Fig. 4 a, lanes 2 and 3). The 95-kD protein was further purified by anion exchange chromatography and used for amino acid sequencing and reassembly experiments (Fig. 4 a, lane 4). NH2-terminal sequencing of the 95-kD protein identified two staggered sequences starting with methionine 1 or 4 (Fig. 2). Recombinant 95-kD protein (Fig. 4 a, lane 5) was used to raise a polyclonal antibody (anti-rec95). In Western blotting experiments this antibody recognized only the upper of the two bands labeled by the original anti-p95 (Fig. 4 b, lanes 4 and 5). In the meantime, immunoscreening of a cDNA library with anti-p95 yielded a clone different from clone 7/1 as revealed by partial sequences. Bacteria transformed with this clone expressed a protein of 95-kD that cross- reacts with anti-p95 but not with anti-rec95 (data not shown). We conclude that anti-p95 was heterospecific and that clone 7/1 encodes one of the two antigens of anti-p95.
Reassembly Experiments with Recombinant 95-kD Protein
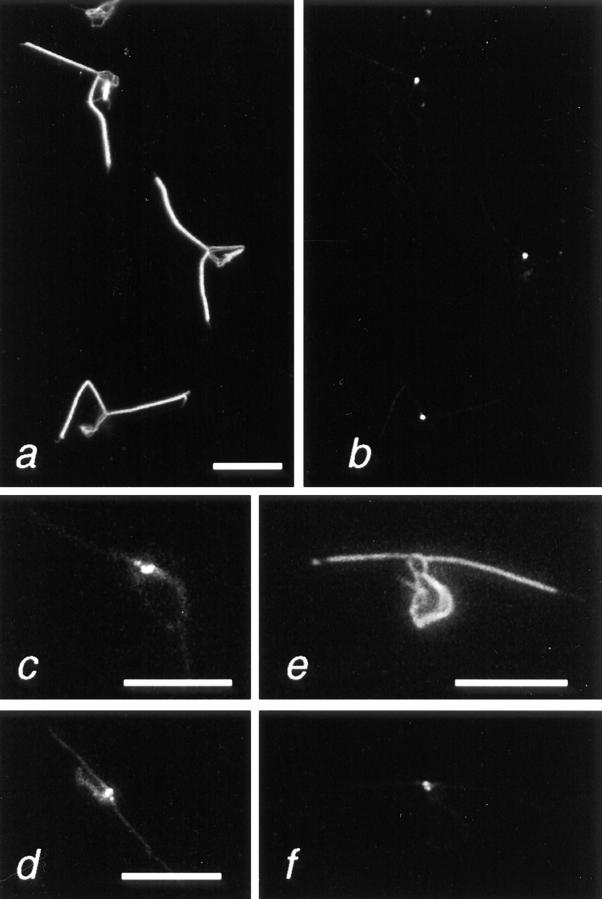
Purified recombinant 95-kD protein (>95%) dissolved in 8 M urea was dialyzed successively against buffers with 4, 2, 0.8, and 0 M urea. EM analysis of the final dialysate showed the formation of striated fibers with a periodicity of 98.2 ± 5.6 nm (n = 20; Fig. 5, a and b). Correspondingly, the post-centrifugation pellet was almost entirely composed of 95-kD protein (Fig. 4 c, lane 2). Structurally, the reassembled fibers showed repeats consisting of a broad electron-translucent band (∼60 nm) divided by an electron-opaque line of ∼6 nm and an electron-opaque band (∼40 nm; Fig. 5 b). The 95-kD protein formed thick, short fibers (length 643 ± 358 nm, n = 16; width 106 ± 61 nm, n = 15) often with tapered ends (Fig. 5 a). We sometimes observed ladder-like fibers with a periodicity of ∼100 nm, consisting of alternating loose regions formed by thin filaments and condensed regions (Fig. 5, c and d); these may represent incompletely assembled intermediates. We also observed clusters of rod-like particles (average length of 81 nm [± 18 nm, n = 16] and a diameter of 2–3 nm; Fig. 5 e), which may represent oligomeric precursors of the striated fibers and also reflect the elongated shape of the 95-kD molecules. Reassembly experiments using dialysis tubing instead of microdialysis membrane (see Materials and Methods) or higher protein concentrations (>0.4 μg/μl instead of 0.1 μg/μl) resulted in the formation of electron-dense, spindle-shaped protein aggregates (length 557 ± 55 nm, n = 15; width 243 ± 77 nm, n = 12), which were sometimes cross-striated with a periodicity of 16 nm (n = 3; Fig. 5, f and g).
Figure 5.
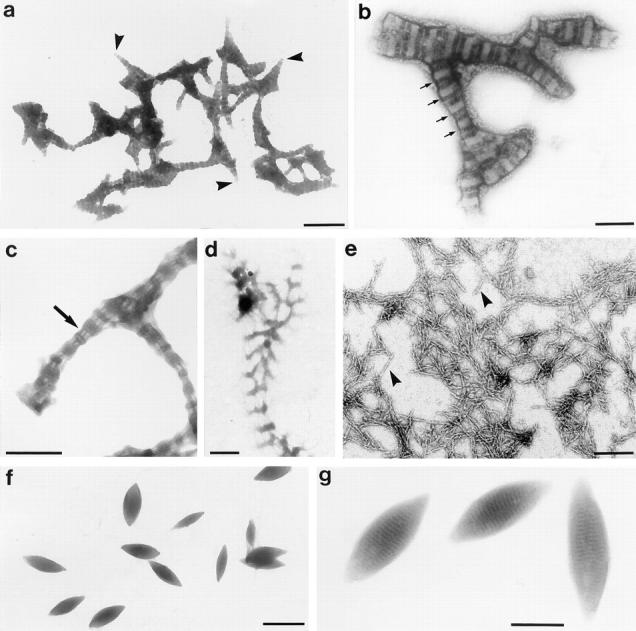
Negatively stained preparations of fibers assembled in vitro from the recombinant 95-kD protein. (a–e) Fibers from reassembly experiment using floating microdialysis membrane. (a) Overview. Note the tapering ends of several filaments (arrowheads). (b) Detail revealing the cross-striation pattern with a periodicity of 98 nm (indicated by arrows). (c and d) Fibers with alternating filamentous (arrow) and condensed regions. Similar filamentous parts are also present in the sf-fiber (compare Fig. 8, a and c). (e) Rod-like particles (arrowheads) and small filaments. These structures were observed frequently in reassembly experiments in addition to the fibers shown in a–d. Overview (f) and detail (g) of fibers formed by the 95-kD protein at high protein concentrations (>0.4 μg/μl) or using dialysis tubes. These spindle-shaped fibers often exhibit a 16-nm cross-striation. Bars: (a, d, and f) 500 nm; (b, c, e, and g) 200 nm.
Localization of the 95-kD Protein
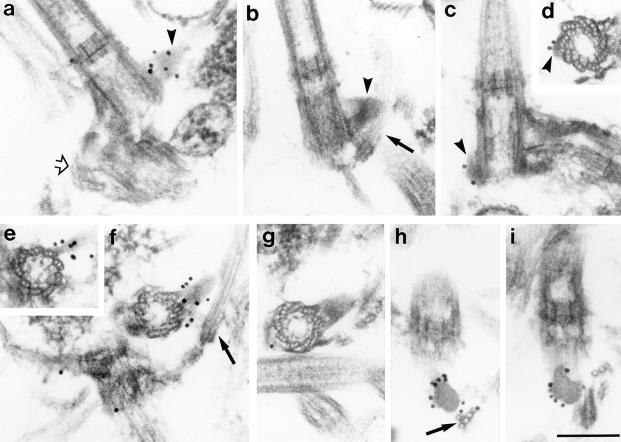
Immunofluorescence staining of isolated cytoskeletons of S. similis with anti-rec95 resulted in comparatively weak labeling of two dots, frequently of unequal size, in the region of the basal apparatus (Fig. 6, c, d, and f). To localize the 95-kD protein more precisely, we performed immunogold electron microscopy (preembedding technique) on isolated cytoskeletons from S. similis (Fig. 7). In samples treated with anti-rec95, cone-like structures associated with the basal bodies were decorated (Fig. 7, a and f). The immunoreactive structures were identified as the sf-fibers that link the basal bodies with the five-stranded microtubular roots (s-roots). The intense labeling of the exposed surface was most clearly seen in cross-sections through the sf-fibers (Fig. 7, h and i). Previously, we have shown that the two sf-fibers present in a basal apparatus differ in size depending on their association with the developmentally older (basal body No. 1) or younger (No. 2) basal body (Geimer et al., 1997a; Fig. 8). Immunogold labeling of the small 2sf-fiber attached to basal body No. 2 is depicted in Fig. 7, c and d; sections through the prominent 1sf-fiber are shown in Fig. 7, a, b, and e–i. Extensive analyses of immunolabeled cytoskeletons revealed no specific reaction of anti-rec95 with any other component of the basal apparatus and preimmune controls showed no specific labeling of the sf-fibers or any other parts of the cytoskeleton (Fig. 7, b and g). The sf-fibers were also decorated by immunogold labeling using anti-p95 (data not shown). We conclude that the sf-fibers are the primary location of the 95-kD protein in the basal apparatus of S. similis.
Figure 7.
Ultrastructural localization of the 95-kD protein in S. similis by immunogold labeling of isolated cytoskeletons using the preembedding technique. Cytoskeletons treated with anti-rec95 (a, c–f, h, and i) or preimmune serum (b and g). (a and b) Longitudinal section through No. 1 basal bodies with attached 1sf-fiber (arrowheads). The sf-fiber in a is decorated with 15-nm gold particles, no labeling is visible in b (preimmune control). Open arrow in a, distal connecting fiber. (c and d) the 2sf-fiber (arrowheads) in longitudinal section (c) or cross-section (d). (e and f) Two consecutive cross-sections through the basal body No. 1 and the 1sf-fiber. Arrow, five-stranded root. Note that the sf-fiber is attached to only one triplet in the more distal section (e). (g) Similar section as in f, but the preimmune control. (h and i) Cross-sections through the 1sf-fiber revealing an intense labeling of the exposed surface. Arrow in h, five-stranded roots with characteristic four over one-pattern of the microtubules. Bar, 200 nm.
Figure 8.
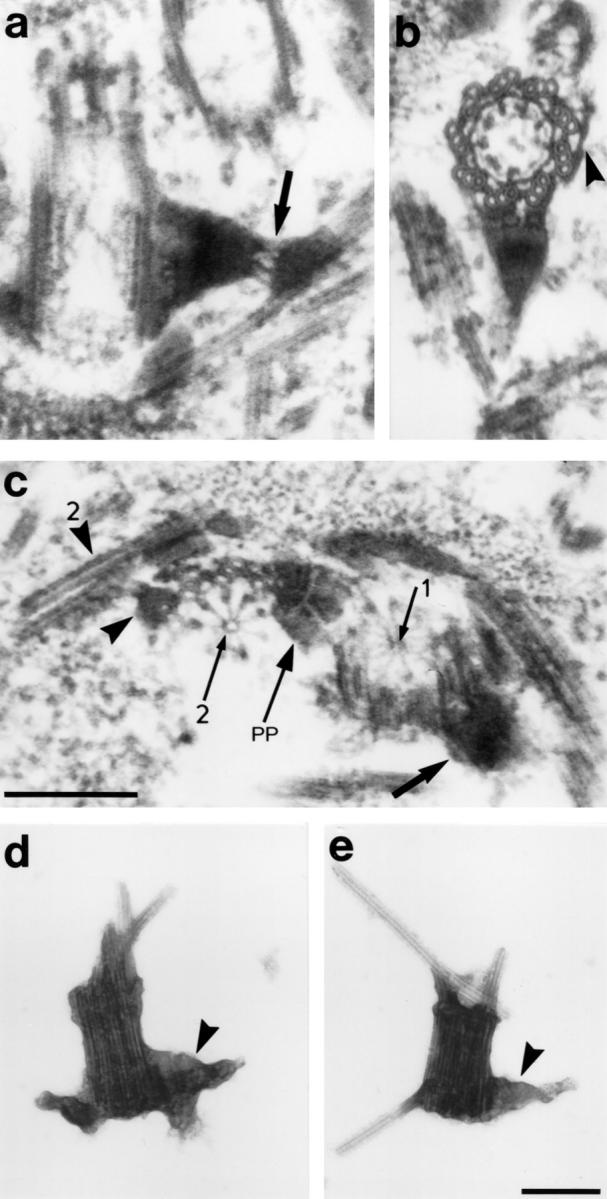
Structure of the sf-fibers. (a) Longitudinal section through basal body No. 1 with 1sf-fiber. Arrow, filamentous part of the 1sf-fiber. (b) Basal body and 1sf-fiber in cross-section. Note that the 1sf-fiber is attached to three triplets. Arrowhead, attachment sites of the df-fiber that links the basal body to the two-stranded microtubular root. (c) Cross-section through the proximal parts of the basal bodies in antiparallel configuration (the left basal body [No. 2] is pointing towards the observer, the No. 1 basal body away from the observer). The 2sf-fiber (arrowhead), the 2s-root (arrowhead with 2), the 1sf-fiber (large arrow), one of the proximal plates (arrow with PP), and the cartwheels (arrows with 1 and 2, respectively) are marked. (d and e) Isolated basal bodies obtained by mechanical disintegration of basal apparatus with a French press at 300, 250, and 200 bar, consecutively (French Pressure Cell Press, 40,000 Psi Pressure cell; SLM Instruments Inc., Urbana, IL). The distal end of the basal bodies contains remnants of the axonemal microtubules. The 1sf-fibers are labeled with arrowheads. Bar, 200 nm.
Structure of the sf-Fibers
The sf-fibers connect each of the two basal bodies with the corresponding bundle of five microtubules. The latter are part of the cruciate microtubular flagellar root system characteristic for Chlamydomonas-like flagellates, which in the case of S. similis, consist of two two-stranded (d-type) and two five-stranded (s-type) microtubular bundles (Preisig and Melkonian, 1984). The 1sf-fiber is attached to basal body No. 1 in a subdistal position and extends about 200 nm towards the s-type root (Fig. 8, a and b). The connection to the basal body involves 1–3 triplets depending on the level of section (Fig. 7, e and f; Fig. 8 b), and the junctions with at least some of the basal body triplets are formed by several thin filaments (Geimer et al., 1997a). Similar filaments are present between the part of the sf-fiber associated with the basal body and its distal part which overlies the flagellar root microtubules, suggesting a flexible junction (Fig. 8 a). The 1sf-fiber exhibits a cross-striation consisting of broad electron-translucent and electron-opaque bands (Fig. 8, a and b). The 2sf-fiber resides in a proximal position on basal body No. 2 and is only ∼80-nm long (Fig. 8 c). For a detailed analysis of the structure of the sf-fibers by serial ultrathin sections, see Geimer et al. (1997a). After intensive mechanical disintegration of basal apparatus, we still observed complexes of basal bodies and attached sf-fibers, indicating that the 1sf-fibers are linked firmly to the basal bodies (Fig. 8, d and e).
Discussion
We have identified a 95-kD protein as a component of the basal apparatus of the green flagellate Spermatozopsis similis. Analysis of the deduced sequence of the 95-kD protein showed a high probability for the formation of extended coiled-coils. The 95-kD protein forms rod-shaped particles in vitro and is located in the sf-fibers, which connect the basal bodies with the two five-stranded microtubular roots. Furthermore, fibers reconstituted in vitro from the recombinant 95-kD protein resembled the sf-fibers in that they had a cross-striation pattern consisting of broad electron-translucent and electron-opaque bands. We conclude that the 95-kD protein is the major structural component of the sf-fibers in S. similis.
Until now there are only very limited data about the distribution in other organisms of proteins homologous to the 95-kD protein from S. similis: no specific labeling was observed in various green algal (Dunaliella bioculata, Polytomella agilis), protist (Tetrahymena), or animal cells (sea urchin sperm, 3T3 cells, rod cells of bovine retina, mouse trachea). However, the antisera raised against the 95-kD protein decorate two dots in the basal apparatus of C. reinhardtii in indirect immunofluorescence (not shown). A possible explanation for the low species cross-reactivity of the antibodies to the 95-kD protein could be the low sequence conservation characteristic for many coiled-coil proteins. This is also true for SF-assemblin, the major protein of the noncontractile striated roots in green flagellates, which has only 54% identity of the amino acid sequence between S. similis and C. reinhardtii (Lechtreck and Silflow, 1997), but obviously has homologues in organisms as phylogenetically distant as Giardia lamblia (Weber et al., 1993).
The sf-fibers are the third nontubulin filament system identified in the green algal basal apparatus (Fig. 9). They are distinct from the previously characterized centrin- and SF-assemblin filaments in their biochemical composition and putative function (Salisbury et al., 1984; Lechtreck and Melkonian, 1991a ). Centrin fibers maintain the spatial orientation of the basal bodies to each other and to the nucleus. Further, centrin may be involved in controlling basal body replication (Taillon et al., 1992). Because centrin fibers are contractile, they also serve to reorientate the basal bodies (e.g., during reversal of swimming direction), to decrease the distance between the basal apparatus and the nucleus, or to sever microtubules during flagellar autotomy (McFadden et al., 1987; Salisbury et al., 1987; Sanders and Salisbury, 1994). SMAFs run parallel to the microtubular roots and may stabilize the flagellar root microtubules and absorb any mechanical stress caused by the flagellar beat (Lechtreck and Melkonian, 1991b ). The sf-fibers, finally, link the basal bodies to the five-stranded microtubular roots (s-roots; Beech and Melkonian, 1993). Functionally, the sf-fibers could determine the position of the microtubular roots with respect to the basal bodies and guide the s-roots into a specific direction. This seems important with respect to the stereotyped internal order characteristic for most flagellate cells that is thought to be maintained by the microtubular flagellar roots (Melkonian, 1984). Previous studies in green algae have shown that the 2s-root connects the eyespot apparatus with the corresponding basal body (Holmes and Dutcher, 1989; Lechtreck et al., 1997). Thus, the eyespot is in a defined position with respect to the plane of flagellar beat, a situation important for phototaxis (Kreimer, 1994). Further, the two s-roots often persist during mitosis and form a specialized structure, which in Chlamydomonas is known as the metaphase band (Johnson and Porter, 1968). The s-root microtubules interconnect the dividing basal apparatus in an antiparallel fashion and arc over the mitotic spindle (Gaffal and el-Gammal, 1990). Thus, the metaphase band and homologous structures present in other green flagellates (Segaar and Gerritsen, 1989; Lechtreck et al., 1997) may be involved in the separation of the duplicated basal apparatus and the positioning of the spindle apparatus and the cleavage furrow (Ehler et al., 1995). Another important feature of the sf-fibers is the difference in size between the two fibers present in the basal apparatus of S. similis. The 1sf-fiber is much more prominent than its counterpart at the younger basal body resulting in a rather unequal distribution of the 95-kD protein to the two basal bodies. A similar distribution has been described for cenexin, a 96-kD protein of unknown primary structure, that is located exclusively in the pericentriolar material surrounding one of the two centrioles in the centrosome of mammalian cells (Lange and Gull, 1995). Interestingly, as described for cenexin, the prominent sf-fiber is a marker for the older basal body/ centriole. Furthermore, monoclonal antibodies directed against Spc110p from the yeast spindle pole body identified an immunologically related 100-kD protein at the proximal end of centrioles of mammalian cells, sometimes more intensely decorating the parental centriole (Tassin et al., 1997). In summary, the link formed by the sf-fibers between the basal bodies and the microtubular s-roots may establish the spatial relationship of these microtubular roots to other cellular structures and the plane of flagellar beat. In general, the decoration of specific microtubular triplets of the basal body with fibrous structures like the sf-fibers could enable the basal bodies to act as the organizing center for the cytoskeleton of flagellate cells.
Fibrous structures (e.g., satellites, basal feet, appendices) that are similar to the sf-fibers have been described in association with basal bodies and centrioles in various metazoan cells (Gibbons, 1961; Reese, 1965; Wolfe, 1972; Dentler, 1987; Sandoz et al., 1988; Paintrand et al., 1992; Lange and Gull, 1996a ). Basal feet are striated, champagne cork–shaped structures attached to two or three triplets of the basal bodies in a subdistal, median, or proximal position. Basal feet as sf-fibers are pointing in the direction of the effective flagellar stroke, indicating that they may be attached to a similar if not identical group of triplets (triplet Nos. 4, 5, and 6 according to the nomenclature of Afzelius, 1959; and Hoops and Witman, 1983) of the basal body, and thus could be markers for the lateral asymmetry of basal bodies (Gibbons, 1961). In diseases resulting in the immotile cilia syndrome, the basal feet of ciliated epithelia show a random disposition (Afzelius, 1980). Basal feet, sf-fibers, and satellites connect the basal bodies/centrioles with cytoskeletal microtubules (Vorobjev and Chentsov, 1982; Baron and Salisbury, 1988). Whereas most of the microtubules radiating from the mammalian centrosome are nucleated in the pericentriolar material, some are attached with their minus ends to the centriole-associated satellites or the centriole itself (Rieder and Borisy, 1982; Rattner, 1992). It has been speculated that these centriolar microtubules may function in anchoring and positioning of the centrosomal complex (Rattner, 1992). Only a few studies deal with the biochemical nature of basal feet or satellites. Recently, basonuclin, a zinc finger protein located in the nuclei of epidermal basal cells, has been localized to satellites of the flagellated centriole/ basal body in spermatocytes/spermatids (Yang et al., 1997). In PtK2 cells antibodies directed against algal centrin decorated the basal feet in addition to other centrosomal structures but cross-reacted with a protein of 165 kD in Western blotting (Baron and Salisbury, 1988). The 95-kD protein from S. similis described in this study is specifically located in a linker between basal bodies and cytoplasmic microtubules. Future analysis of the 95-kD protein may provide insights how the spatial relation between microtubules and basal bodies/centrioles is established and maintained.
Acknowledgments
We thank K. Weber (Max-Planck Institute for Biophysical Chemistry, Göttingen, Germany) for the determination of peptide sequences, and M. Haring (BioCentrum, University of Amsterdam, Netherlands) for advice during the preparation of the cDNA library.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG; Bonn, Germany).
Abbreviations used in this paper
- IPTG
isopropyl-β-d-thiogalactopyranoside
- sf-fibers
sinister fibers
- SMAF
striated microtubule-associated fibers
Footnotes
This work is dedicated to the memory of Annette Teltenkötter (2 May 1997).
Address all correspondence to Karl-F. Lechtreck, Botanisches Institut, Gyrhofstrasse 15, Universität zu Köln, D-50931 Köln, Germany. Tel.: 49-221-470-3795. Fax: 49-221-470-5181. E-mail: kflecht@novell.biolan.uni-koeln.de
J. Clees' present address is Universitätskrankenhaus Eppendorf, Zentrum für Molekulare Neurobiologie, Institut für Biosynthese Neuraler Strukturen, Martinistrasse 85, D-20251 Hamburg, Germany.
References
- Afzelius B. Electron microscopy of the sperm tail. Results obtained with a new fixative. J Biophys Biochem Cytol. 1959;5:269–278. doi: 10.1083/jcb.5.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzelius, B.A. 1980. Genetic disorders of cilia. In International Cell Biology 1980-81, H.G. Schweiger, editor. Springer-Verlag, Berlin. 440–447.
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic alignment search tool. J Mol Biol. 1990;215:69–76. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baron AT, Salisbury JL. Identification and localization of a novel, cytoskeletal, centrosome-associated protein in PtK2cells. J Cell Biol. 1988;107:2669–2678. doi: 10.1083/jcb.107.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, A.T., and J.L. Salisbury. 1992. Role of centrin in spindle pole dynamics. In The Centrosome. V.I. Kalnins, editor. Academic Press, San Diego. 167–195.
- Beech PL, Melkonian M. The basal apparatus of the quadriflagellate Spermatozopsis exsultans(Chlorophyceae): numbering of basal body triplets reveals triplet individuality and developmental modifications. J Phycol. 1993;29:191–202. [Google Scholar]
- Beech PL, Heimann K, Melkonian M. Development of the flagellar apparatus during the cell cycle in unicellular algae. Protoplasma. 1991;164:23–37. [Google Scholar]
- Dentler WL. Cilia and Flagella. Int Rev Cytol (Suppl) 1987;17:391–456. [Google Scholar]
- Ehler LL, Holmes JA, Dutcher SK. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtiimutant strain lacking basal bodies. Genetics. 1995;141:945–960. doi: 10.1093/genetics/141.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffal KP, el-Gammal S. Elucidation of the enigma of the “metaphase band” of Chlamydomonas reinhardtii. . Protoplasma. 1990;156:139–148. [Google Scholar]
- Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Geimer S, Lechtreck K-F, Melkonian M. The cytoskeleton of the naked green flagellate Spermatozopsis similis(Chlorophyta). Analysis of isolated basal apparatuses in the parallel configuration. J Phycol. 1997a;33:241–253. [Google Scholar]
- Geimer G, Teltenkötter A, Plessmann U, Weber K, Lechtreck K-F. Purification and characterization of basal apparatuses from a flagellate green alga. Cell Motil Cytoskeleton. 1997b;37:72–85. doi: 10.1002/(SICI)1097-0169(1997)37:1<72::AID-CM7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gibbons IR. The relationship between the fine structure and the direction of the beat in gill cilia of a lamellibranch mollusc. J Biophys Biochem Cytol. 1961;11:179–205. doi: 10.1083/jcb.11.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes JA, Dutcher SK. Cellular asymmetry in Chlamydomonas reinhardtii. . J Cell Sci. 1989;94:273–285. doi: 10.1242/jcs.94.2.273. [DOI] [PubMed] [Google Scholar]
- Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonasflagella. J Cell Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Mengersen A, Lee VD. Molecular Cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: Homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J Cell Biol. 1988;107:133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson UG, Porter KR. Fine structure of cell division in Chlamydomonas reinhardii.Basal bodies and microtubules. J Cell Biol. 1968;38:403–425. doi: 10.1083/jcb.38.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiaespindle pole body whose transcript is cell cycle regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer G. Cell biology of phototaxis in flagellate algae. Int Rev Cytol. 1994;148:229–310. [Google Scholar]
- Lange BMH, Gull K. A molecular marker for centriole maturation in the mammalian cell cycle. J Cell Biol. 1995;130:919–927. doi: 10.1083/jcb.130.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BMH, Gull K. A structural study of isolated mammalian centrioles using negative staining electron microscopy. J Struct Biol. 1996a;117:222–226. doi: 10.1006/jsbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- Lange BMH, Gull K. Structure and function of centrioles in animal cells: progress and questions. Trends Cell Biol. 1996b;6:348–352. doi: 10.1016/0962-8924(96)10033-7. [DOI] [PubMed] [Google Scholar]
- Lechtreck K-F, Melkonian M. An update on fibrous flagellar roots in green algae. Protoplasma. 1991a;164:38–44. [Google Scholar]
- Lechtreck K-F, Melkonian M. Striated microtubule-associated fibers: Identification of assemblin, a novel 34-kD protein that forms paracrystals of 2-nm filaments in vitro. . J Cell Biol. 1991b;115:705–716. doi: 10.1083/jcb.115.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K-F, Silflow CD. SF-assemblin in Chlamydomonas: sequence conservation and localization during the cell cycle. Cell Motil Cytoskeleton. 1997;36:190–201. doi: 10.1002/(SICI)1097-0169(1997)36:2<190::AID-CM8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Lechtreck K-F, McFadden GI, Melkonian M. The cytoskeleton of the naked green flagellate Spermatozopsis similis: isolation, whole mount electron microscopy, and preliminary biochemical and immunological characterization. Cell Motil Cytoskeleton. 1989;14:552–561. [Google Scholar]
- Lechtreck K-F, Reize IB, Melkonian M. The cytoskeleton of the naked green flagellate Spermatozopsis similis: flagellar and basal body developmental cycle. J Phycol. 1997;33:254–265. [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- McFadden GI, Melkonian M. Use of Hepes buffer for microalgal culture media and fixation for electron microscopy. Phycologia. 1986;25:551–557. [Google Scholar]
- McFadden GI, Schulze D, Surek B, Salisbury JL, Melkonian M. Basal body reorientation mediated by a Ca2+-modulated contractile protein. J Cell Biol. 1987;105:903–912. doi: 10.1083/jcb.105.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian M. Ultrastructural aspects of basal body associated fibrous structures in green algae: a critical review. Biosystems. 1980;12:85–104. doi: 10.1016/0303-2647(80)90040-4. [DOI] [PubMed] [Google Scholar]
- Melkonian, M. 1984. Flagellar root-mediated interactions between the flagellar apparatus and cell organelles in green algae. In Compartments in Algal Cells and Their Interactions. W. Wiessner, D. G. Robinson, and R.C. Starr, editors. Springer-Verlag, Berlin. 96–108.
- Melkonian, M., P.L. Beech, C. Katsaros, and D. Schulze. 1992. Centrin-mediated cell motility in algae. In Algal Cell Motility. M. Melkonian, editor. Chapman & Hall, New York. 179–221.
- Moestrup Ø. Flagellar structure in algae: a critical review, with new observations particulary on the Chrysophyceae, Phaeophyceae (Fucophyceae), Euglenophyceae, and Reckertia. . Phycologia. 1982;21:427–528. [Google Scholar]
- Moestrup Ø, Hori T. Ultrastructure of the flagellar apparatus in Pyramimonas octopus(Prasinophyceae) II. Flagellar root, connecting fibers, and numbering of individual flagella in green algae. Protoplasma. 1989;148:41–56. [Google Scholar]
- Moreno S, Nurse P. Substrates for p34cdc2; in vitro veritas? . Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- Neuhoff V, Philipp K, Zimmer H-G, Mesecke S. A simple, versatile and volume-independent method for quantitative protein determination which is independent of other external influence. Hoppe-Seyler's Z Physiol Chem. 1979;360:1657–1670. doi: 10.1515/bchm2.1979.360.2.1657. [DOI] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- Palmiter RD. Magnesium precipitation of ribonucleoprotein complexes: expedient techniques for the isolation of undegraded polysomes and messenger ribonucleic acid. Biochemistry. 1974;13:3606. doi: 10.1021/bi00714a032. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Preisig HR, Melkonian M. A light and electron microscopical study of the green flagellate Spermatozopsis similisspec. nova. Plant Syst Evol. 1984;146:57–74. [Google Scholar]
- Rattner, J.B. 1992. Ultrastructure of centrosome domains and identification of protein components. In The centrosome. V.I. Kalnins, editor. Academic Press, San Diego. 45–68.
- Reese TS. Olfactory cilia in the frog. J Cell Biol. 1965;25:209–230. doi: 10.1083/jcb.25.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Borisy GG. The centrosome cycle in PtK2 cells: asymmetric distribution and structural changes in the pericentriolar materiel. Biol Cell. 1982;44:117–132. [Google Scholar]
- Ringo DL. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. . J Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL. Centrin, centrosomes, and mitotic spindle poles. Curr Opin Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- Salisbury JL, Baron AT, Surek B, Melkonian M. Striated flagellar roots: Isolation and partial characterization of a calcium-modulated organelle. J Cell Biol. 1984;99:962–970. doi: 10.1083/jcb.99.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL, Sanders MA, Harpst L. Flagellar root contraction and nuclear movement during flagellar regeneration in Chlamydomonas reinhardtii. . J Cell Biol. 1987;105:1799–1805. doi: 10.1083/jcb.105.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MA, Salisbury JL. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. . J Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz D, Chailley B, Boisvieux-Ulrich E, Lemullois M, Laine M-C, Bautista-Harris G. Organization and function of the cytoskeleton in metazoan ciliated cells. Biol Cell. 1988;63:183–193. doi: 10.1016/0248-4900(88)90057-3. [DOI] [PubMed] [Google Scholar]
- Schiebel E, Bornens M. In search of a function for centrins. Trends Cell Biol. 1995;5:197–201. doi: 10.1016/s0962-8924(00)88999-0. [DOI] [PubMed] [Google Scholar]
- Schlösser UG. SAG - Sammlung von Algenkulturen at the University of Göttingen: Catalogue of strains. Bot Acta. 1994;107:113–186. [Google Scholar]
- Segaar PJ, Gerritsen AF. Flagellar roots as vital instruments in cellular morphogenesis during multiple fission (sporulation) in the unicellular green flagellate Brachiomonas submarina(Chlamydomonadales, Chlorophyta) Crypt Bot. 1989;1:249–274. [Google Scholar]
- Snell WJ, Dentler WL, Haimo LT, Binder LI, Rosenbaum JL. Assembly of chick brain tubulin onto isolated basal bodies of Chlamydomonas reinhardtii. . Science. 1974;185:357–360. doi: 10.1126/science.185.4148.357. [DOI] [PubMed] [Google Scholar]
- Taillon BE, Adler SA, Suhan JP, Jarvik JW. Mutational analysis of centrin: An EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. . J Cell Biol. 1992;119:1613–1624. doi: 10.1083/jcb.119.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Celati C, Paintrand M, Bornens M. Identification of an Spc110p-related protein in vertebrates. J Cell Sci. 1997;110:2533–2545. doi: 10.1242/jcs.110.20.2533. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;98:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Geisler N, Plessmann U, Bremerich A, Lechtreck K-F, Melkonian M. SF-assemblin, the structural protein of the 2-nm filaments from striated microtubule associated fibers of algal flagellar roots, forms a segmented coiled coil. J Cell Biol. 1993;121:837–845. doi: 10.1083/jcb.121.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, J. 1972. Basal body fine structure and chemistry. In Advances in Cell and Molecular Biology. Vol. 2. E.J. DuPraw, editor. Academic Press, New York/London. 151–192.
- Yang Z-h, Gallicano I, Yu Q-C, Fuchs E. An unexpected localization of basonuclin in the centrosome, mitochondria, and acrosome of developing spermatids. J Cell Biol. 1997;137:657–669. doi: 10.1083/jcb.137.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]