Abstract
The Drosophila MEI-S332 protein has been shown to be required for the maintenance of sister-chromatid cohesion in male and female meiosis. The protein localizes to the centromeres during male meiosis when the sister chromatids are attached, and it is no longer detectable after they separate. Drosophila melanogaster male meiosis is atypical in several respects, making it important to define MEI-S332 behavior during female meiosis, which better typifies meiosis in eukaryotes. We find that MEI-S332 localizes to the centromeres of prometaphase I chromosomes in oocytes, remaining there until it is delocalized at anaphase II. By using oocytes we were able to obtain sufficient material to investigate the fate of MEI-S332 after the metaphase II–anaphase II transition. The levels of MEI-S332 protein are unchanged after the completion of meiosis, even when translation is blocked, suggesting that the protein dissociates from the centromeres but is not degraded at the onset of anaphase II. Unexpectedly, MEI-S332 is present during embryogenesis, localizes onto the centromeres of mitotic chromosomes, and is delocalized from anaphase chromosomes. Thus, MEI-S332 associates with the centromeres of both meiotic and mitotic chromosomes and dissociates from them at anaphase.
Cohesion between sister chromatids is essential for proper segregation of chromosomes during mitosis and meiosis. By counteracting spindle forces pulling chromosomes towards the poles, cohesive forces between sister chromatids enable stable bipolar attachments to be established; these in turn allow the sister chromatids to be partitioned appropriately during anaphase. The consequences of inappropriate partitioning can be severe: aneuploidy is observed in many tumors and also in individuals with congenital disorders such as Down's syndrome. Defects in sister-chromatid cohesion have been suggested as an important factor that might be involved in oncogenesis or meiotic errors (Orr-Weaver, 1996; Lamb et al., 1996; Lengauer et al., 1997).
In both meiosis and mitosis, cohesion exists between the arms and the centromere regions of the sister chromatids after their replication, but release of sister-chromatid cohesion occurs differently in these two types of cell division (Moore and Orr-Weaver, 1998). In mitosis, the sister chromatids segregate from one another in a single cell division, and thus cohesion is released from both the chromosome arms and centromere regions at the same time, the onset of anaphase. Meiosis consists of two cell divisions that follow a single round of replication: the homologues segregate from one another in the first division, the sister chromatids in the second division. The homologues are typically connected at sites on their arms called chiasmata, and sister-chromatid cohesion along the chromosome arms is believed to be important for the maintenance of chiasmata (Maguire, 1974, 1993). With the onset of anaphase I, this arm cohesion is lost, but cohesion between the centromeric regions of the sister chromatids is maintained. This cohesion in the centromeric region is required to align the sister chromatids for metaphase II and is released at the beginning of anaphase II. Thus, meiosis is a specialized cell division that requires a two-step release of sister-chromatid cohesion.
The Drosophila protein MEI-S332 has been demonstrated both to be essential for cohesion between sister chromatids and to be localized to chromosomes (Goldstein, 1980; Kerrebrock et al., 1992, 1995). These cytological studies were performed in spermatocytes. In male meiosis, MEI-S332 localizes to the centromeric regions of meiotic chromosomes and is maintained there through the metaphase I–anaphase I transition (Kerrebrock et al., 1995). MEI-S332 is observed on chromosomes in metaphase II but is no longer detectable with the commencement of anaphase II, the time when cohesion between sister chromatids is released. The protein is required primarily for proper segregation during the second meiotic division, because by genetic assays, mei-S332 mutant males and females have nearly normal segregation during the first meiotic division and high levels of missegregation during the second meiotic division (Davis, 1971; Goldstein, 1980; Kerrebrock et al., 1992). Precociously separated sister chromatids are observed in mei-S332 spermatocytes in late anaphase I, suggesting that MEI-S332 is vital for centromeric cohesion after the metaphase I–anaphase I transition (Goldstein, 1980; Kerrebrock et al., 1992). Previous studies have not described the localization of MEI-S332 during female meiosis.
The structure of the meiotic chromatin and the meiotic spindle differs between the sexes in Drosophila melanogaster (for review see Orr-Weaver, 1995), so it cannot be assumed that localization of MEI-S332 is the same in both spermatocytes and oocytes. In females, but not in males, synaptonemal complex forms during prophase and reciprocal exchange occurs, resulting in the chiasmata that are assumed to hold homologues together. In males, pairing sites hold the homologues together without synaptonemal complex or reciprocal exchange between the homologues (for review see McKee, 1996). Another significant difference is that the oocyte metaphase I spindle is thought to be organized by the chromatin rather than by centrosomes (Theurkauf and Hawley, 1992), and this function could require that the meiotic chromosomes have a different structure in females. Finally, oocytes arrest during metaphase I, whereas spermatocytes normally do not, thus requiring cohesion to be maintained longer. Differences between meiosis in male and female Drosophila could impact MEI-S332 localization. Moreover, the existence of alleles that affect male and female meiosis with different severity suggests that there must be some differences in MEI-S332 mechanism between the sexes (Kerrebrock et al., 1992). Whereas Drosophila male meiosis has several unusual features, Drosophila female meiosis is more typical of meiosis in most eukaryotes; thus, localization of MEI-S332 in oocytes is of particular interest.
Sister chromatids are believed to be held together by proteins until anaphase (for review see Bickel and Orr-Weaver, 1996). The cohesive proteins that hold sister chromatids together could dissociate or could be degraded at the time when the chromatids separate. Studies in both yeast and Xenopus extracts have shown that release of cohesion is dependent on proteolysis of some substrates by the cyclin degradation machinery, the anaphase promoting complex (Holloway et al., 1993; Irniger et al., 1995; Funabiki et al., 1996). This complex could directly proteolyze the cohesive proteins at the chromosomes, or indirectly promote sister-chromatid separation by degrading inhibitors of anaphase. Recent work in budding yeast demonstrates that the Pds1p protein, which acts as an inhibitor of separation, is degraded by the anaphase promoting complex at the initiation of anaphase (Cohen-Fix et al., 1996; Yamamoto et al., 1996). A second protein more integrally involved in cohesion, the Mcd1p/Scc1p protein, has also been identified (Guacci et al., 1997; Michaelis et al., 1997). Mcd1p localizes to mitotic chromosomes and dissociates at the metaphase–anaphase transition, but its degradation is slow, and the protein persists after anaphase. Thus, both dissociation and degradation may play important roles in the release of sister-chromatid cohesion. Although the cohesion protein MEI-S332 is not observed on the chromatids after the sister chromatids separate during meiosis II, it is not known whether the protein simply dissociates or is degraded.
In this paper, we look at the localization of MEI-S332 during meiosis in females, and we find that, as in males, the protein disappears from centromeres at anaphase II. The fate of MEI-S332 at the metaphase II–anaphase II transition is examined using Western blots, and we find that MEI-S332 is not degraded detectably at that time. Because the protein is not degraded, we examine its localization during embryonic mitoses. Although centromeric cohesion also occurs in mitosis, mei-S332 is not essential for mitotic divisions (Kerrebrock et al., 1992, 1995). Strikingly, we find that the MEI-S332 protein is localized to the centromeric regions of mitotic chromosomes in the embryo.
Materials and Methods
Fly Strains
In the studies of MEI-S332—green fluorescent protein (GFP)1 localization in oocyte meiosis, females of genotype y w P( +mc 5.6KK mei-S332 + :: GFP = GrM)-13; P(w +mc 5.6KK mei-S332 + ::GFP = GrM)-1, containing four copies of the fusion transgene mei-S332 + ::GFP (Kerrebrock et al., 1995) and two endogenous copies of mei-S332 +, were used. (The insertion of the transgene on the X chromosome is named P(GrM)-13; the insertion on chromosome 2 is named P(GrM)-1.) For localization of MEI-S332-GFP in embryos, mothers of the genotype described above or mothers carrying only two copies of the fusion transgene mei-S332 + ::GFP in the y; mei-S3327/Df(2R)X58-6 background were used. The latter flies were generated by crossing y w P(GrM)-13; cn mei-S3327 px sp/SM1 females to y w P(GrM)-13/y + Y; Df(2R)X58-6 pr cn/SM1 males.
In studying the MEI-S332 levels in oocytes before and after activation (see Fig. 3 C), y w females were used. Embryos and oocytes from y; pr cn mei-S3327 bw sp/Df(2R)X58-6, pr cn and y; pr cn mei-S3327 bw sp/cn mei-S3327 px sp females were used as negative controls for the anti–MEI-S332 peptide antibodies (Kerrebrock et al., 1992 and see below). Oregon-R (wild type) was used as the negative control for GFP fluorescence microscopy and positive control for Western blot analysis (see Fig. 3 B). For protein extracts from overexpressing oocytes, oocytes were obtained from females carrying six copies of the mei-S332 + gene (two endogenous copies and four copies from homozygous insertions of P(w +mc 5.6 KK mei-S332 +) on the second and third chromosomes; Kerrebrock et al., 1995). In all the mei-S332 transposons, the gene was expressed from the normal genomic regulatory regions.
Figure 3.
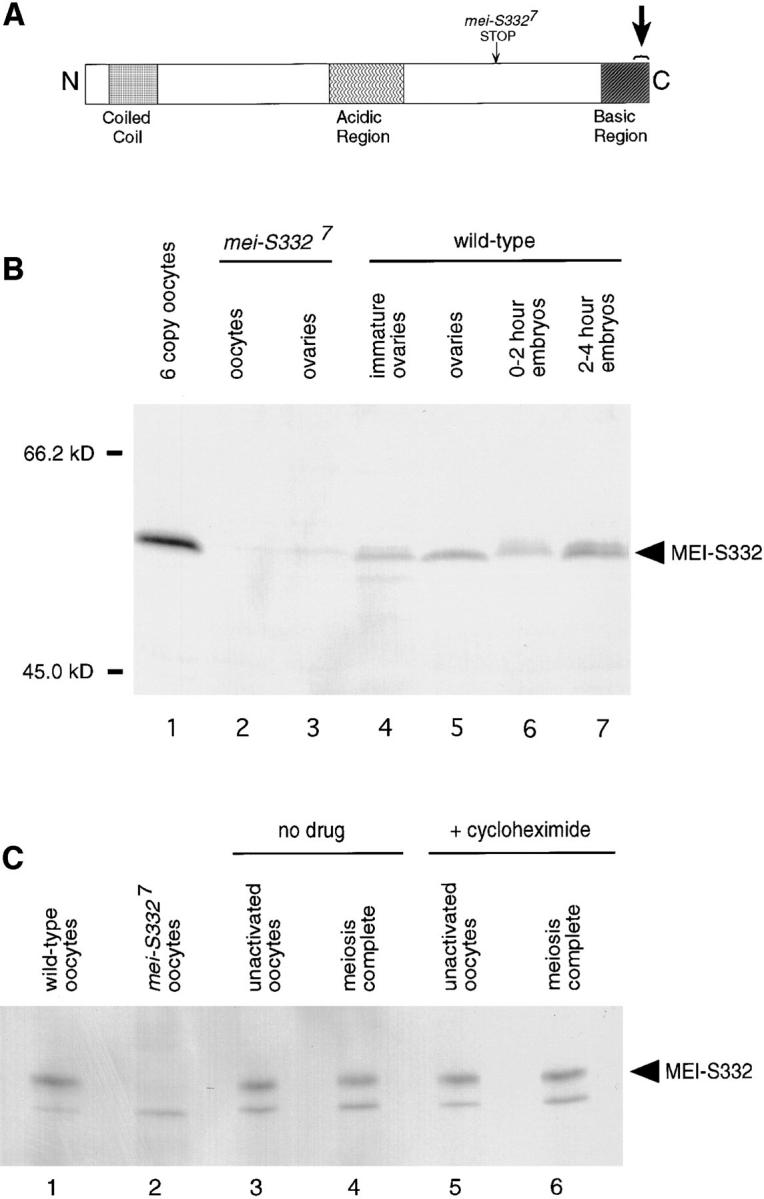
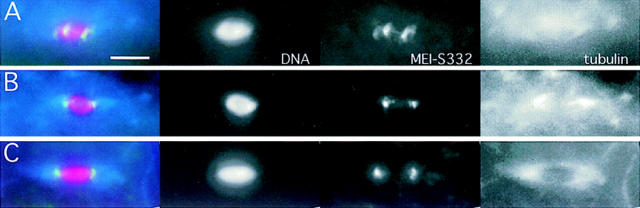
The MEI-S332 protein is present in embryos and is not globally degraded at the metaphase II–anaphase II transition. (A) A schematic of the MEI-S332 protein. Anti–MEI-S332 antibodies were generated against a COOH-terminal 15–amino acid peptide of MEI-S332 (large arrow). Tissues from mei-S3327 flies were used as negative controls for the antibodies because the mei-S3327 mutation generates a truncated form of the protein that lacks the epitope for the COOH-terminal peptide antibodies (small arrow). (B) The MEI-S332 protein, predicted to be 44 kD, is recognized as a 55-kD band on Western blots by affinity-purified anti–MEI-S332 peptide antibodies. Higher levels of MEI-S332 (lane 1) are seen in oocytes isolated from females carrying six copies of the mei-S332 + gene (two endogenous copies and four copies of a genomic fragment). MEI-S332 is present in previtellogenic ovaries (lane 4), mature ovaries (lane 5), 0–2 h embryos (lane 6), and 2–4 h embryos (lane 7). There appear to be different mobility forms of MEI-S332 in embryos. As expected, the 55-kD band is not detected in mei-S3327 oocytes and ovaries (lanes 2 and 3). (C) MEI-S332 protein levels remain essentially unchanged in activated eggs that have completed meiosis (compare lanes 3 and 4). Although MEI-S332 is no longer detectable on the chromosomes when sister-chromatid cohesion is lost, it is not degraded globally. Protein levels remain unchanged when meiosis is completed in the presence of the translational inhibitor cycloheximide (compare lanes 5 and 6). Oregon-R and mei-S3327 unactivated oocytes were used as positive and negative controls, respectively, for the antibodies (lanes 1 and 2). The lower nonspecific band, probably an artifact of this sample preparation, is not MEI-S332 as it is still present in extracts from mei-S3327 oocytes (lane 2).
Meiosis in Activated Eggs
The cytology of activated eggs was performed essentially as described in Page and Orr-Weaver (1997) with changes in the fixation conditions to preserve the GFP fluorescence. 300 females of genotype y w P(GrM)-13; P(GrM)-1 were fattened on wet yeast for several days. Flies were disrupted in IB (55 mM NaOAc, 40 mM KOAc, 110 mM sucrose, 1.2 mM MgCl2, 1 mM CaCl2, 100 mM Hepes, final pH 7.4) in a blender, and oocytes were isolated by filtration and gravity settling. This isolation step took 10–11 min. Oocytes were activated by the addition of AB (3.3 mM NaH2PO4, 16.6 mM KH2PO4, 10 mM NaCl, 50 mM KCl, 5% PEG 8000, 2 mM CaCl2, final pH 6.4) for a 5-min incubation, and then the buffer was changed to ZAB (9 mM MgCl2, 10 mM MgSO4, 2.9 mM NaH2PO4, 0.22 mM NaOAc, 5 mM glucose, 27 mM glutamic acid, 33 mM glycine, 2 mM malic acid, 7 mM CaCl2, final pH 6.8) for an additional incubation of 10 min (for anaphase I and metaphase II) or 25 min (for anaphase II and the postmeiotic interphase). Eggs with cross-linked vitelline membranes, a hallmark of activation, were selected by a 3-min incubation in 50% Clorox bleach, and fixed in 8% EM-grade, MeOH-free formaldehyde (Ted Pella Inc., Irvine, CA) in cacodylate buffer (100 mM cacodylic acid, 100 mM sucrose, 40 mM KOAc, 10 mM NaOAc, 10 mM EGTA, pH to 7.2 with KOH; Theurkauf, 1994) for 10–15 min, and washed in PBST (PBS with 0.3% Triton X-100) containing ∼1% BSA to prevent sticking to glassware. Vitelline membranes were removed by rolling the fixed eggs between two microscope slides (Theurkauf, 1994), again using PBST–BSA as a lubricant. Eggs were incubated in 1% RNase A (boiled to destroy DNase activity) for 20 min, and then incubated with 1 μg/ml propidium iodide (Sigma Chemical Co., St. Louis, MO) for 30 min. Samples were mounted in Vectashield containing propidium iodide (Vector Labs Inc., Burlingame, CA).
Tubulin Immunofluorescence
Oocytes were prepared using the protocol described by Theurkauf (1994) for isolation and fixation of egg chambers. Tubulin was labeled using two anti-tubulin rat monoclonal antibodies, YL1/2 and YOL1/34 (Sera-Lab Ltd., Sussex, UK), overnight at room temperature at a dilution of 1:5 in 0.1% BSA in PBST, followed by a 3-h incubation with a Texas red-conjugated donkey anti–rat antibody (Jackson Immunoresearch Laboratories Inc., West Grove, PA) at room temperature at a dilution of 1:200. The oocytes were further stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma Chemical Co.) at 1 μg/ml in PBS for 10 min, followed by two 15-min rinses in PBS before mounting in 50% glycerol.
MEI-S332–GFP Localization in Embryos
Embryos were collected for 2.5 h from females of the genotype y w P(GrM)-13; P(GrM)-1. The embryo in Fig. 5 was from a 4-h collection from females of the genotype y w P(GrM)-13; cn mei-S3327 px sp/Df(2R)X58-6 pr cn. Oregon-R embryos were used as a control for background autofluorescence.
Figure 5.
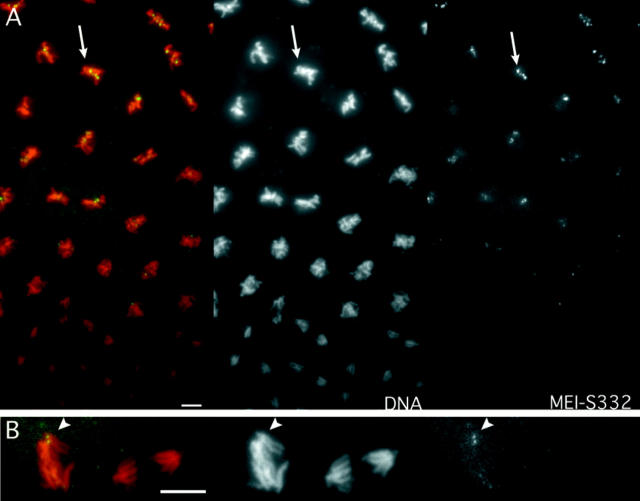
MEI-S332–GFP disappears from centromeres at the metaphase–anaphase transition in embryos. MEI-S332–GFP is shown in green, DNA in red. Images are also separated to show individual channels as labeled. (A) A field of syncytial nuclei in a cycle 12 embryo is in the process of mitosis. In each lane, metaphase figures are on the top, anaphase figures in the middle, and late anaphase figures on the bottom. MEI-S332– GFP localizes to discrete dots on the mitotic metaphase plates (arrow). MEI-S332–GFP is no longer detectable on mitotic chromosomes in late anaphase. (B) MEI-S332–GFP can be seen at the leading edge of the chromosomes in early anaphase (arrowhead), but it is no longer detectable on mid-anaphase chromosomes. Embryos were collected from mei-S3327 females carrying two copies of the mei-S332 + ::GFP transgene. Images were collected using a CCD camera. Bars, ∼5 μm.
Embryos were dechorionated in 50% bleach, and fixed for 30 min in 8% MeOH-free formaldehyde in cacodylate buffer (see above). After washing in PBS, embryos were rolled out of their vitelline membranes between two glass slides (Theurkauf, 1994). To stain for DNA, two methods were used. Embryos in Figs. 4 A, C, and D were treated with 1 mg/ml RNase A for 30 min, stained with 1 μg/ml propidium iodide for 30 min, and mounted in Vectashield with propidium iodide (Vector Labs Inc.). The embryos shown in Figs. 4 B and E, and 5 were stained with DAPI at 1 μg/ml in PBS for 10 min, followed by two 15-min rinses in PBS before mounting in 50% glycerol.
Figure 4.
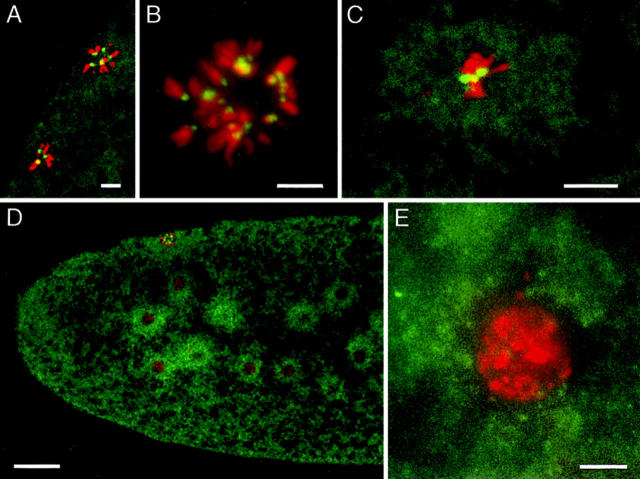
MEI-S332–GFP localizes to condensed chromosomes in embryos. MEI-S332– GFP is shown in green and DNA in red. (A) MEI-S332–GFP is present on the polar body rosettes. (B) A close-up image of a polar body rosette shows punctate MEI-S332–GFP localization on the inside ring of the rosette where centromeres are believed to be pulled to the center. 22 dots of MEI-S332–GFP can be counted in the single rosette found in this embryo. (C) MEI-S332–GFP localizes to discrete dots on a mitotic metaphase plate, resembling those on meiotic metaphase II chromosomes. In addition, a cloud of diffuse MEI-S332–GFP is observed around each mitotic nucleus. (D) MEI-S332–GFP is detected in clouds surrounding the interphase nuclei. The nuclei are not centered within the clouds. (E) A close-up image of the interphase nucleus demonstrates the absence of MEI-S332–GFP localization on the decondensed interphase chromatin. Embryos were collected from females carrying four copies of the mei-S332 + ::GFP transgene, fixed, and stained with either propidium iodide or DAPI. Images in A, C, and D were collected using confocal microscopy, and images in B and E were collected using a CCD camera. Bars: A–C and E ∼5 μm; (D) ∼30 μm.
Microscopy
Two kinds of epifluorescence microscopy were used in our investigations. Conventional epifluorescence microscopy was performed using either a Nikon Optiphot-2 microscope or a Nikon Eclipse E800 equipped with a Nikon 60× oil objective (Garden City, NY). A Photometrics CE200A cooled CCD video camera was used to photograph images. The images were further processed with the CELLscan 2.0 system (Scanalytics) to create volume views from focal planes separated by 0.25 μm. 32 focal planes are shown for the oocyte images in Fig. 2, 45 focal planes for the rosette in Fig. 4 B, 20 focal planes for the mitotic interphase nucleus in Fig. 4 E, and 7 focal planes for the images in Fig. 5. Chromatin and MEI-S332-GFP in Figs. 1, A–G, 4 A, C, and D were visualized on a confocal laser scanning head (MRC 600; BioRad, Hercules, CA) equipped with a krypton/argon laser, mounted on a Zeiss Axioskop microscope (Oberkochen, Germany), with 20 and 40× oil Plan Neofluar objectives. In some cases, optical sections were taken and projected into a single plane. All images were further processed, colorized, and merged using Adobe Photoshop 3.0 on a Macintosh Power PC.
Figure 2.
MEI-S332–GFP assembly onto female meiotic chromosomes correlates with spindle formation. MEI-S332–GFP is shown in green, tubulin in blue, and chromatin in red. The images are also separated to show the individual channels. (A) MEI-S332–GFP is first observed on the meiotic chromosomes at multiple discrete sites before the formation of a bipolar spindle. (B) As the spindle becomes increasingly elongated and bipolar, the discrete dots of MEI-S332–GFP begin to cluster at opposite ends of the chromatin mass. (C) When the spindle is fully elongated, MEI-S332–GFP is observed in two caps at the opposite ends of the chromatin mass, aligned with the bipolar spindle. Oocytes were isolated from females carrying four copies of the mei-S332 + ::GFP transgene, fixed, and stained with anti-tubulin antibodies and DAPI. Images were collected using a CCD camera. Bar, ∼5 μm.
Figure 1.
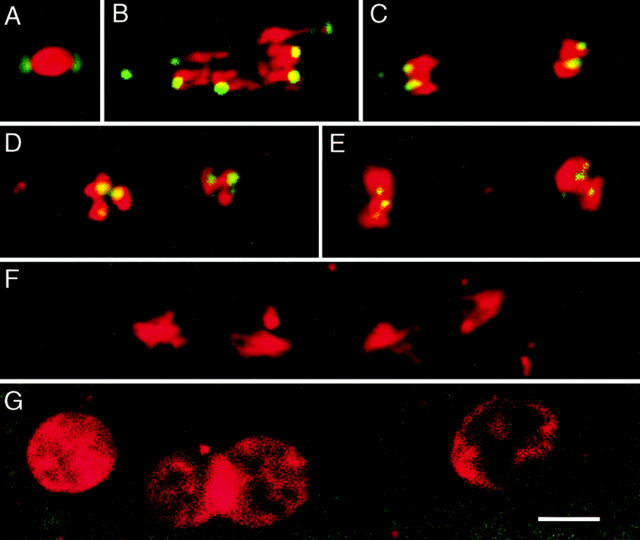
MEI-S332–GFP localizes to centromeric regions of female meiotic chromosomes until anaphase II. MEI-S332–GFP is shown in green and chromatin in red. (A) Unactivated stage 14 oocytes are arrested in metaphase I, with MEI-S332–GFP localized to two discrete sites on the opposite ends of the condensed meiotic chromosomes. (B) At the onset of anaphase I, eight dots of MEI-S332–GFP are visible at the leading edges of the separating anaphase chromosomes, one per pair of sister chromatids, with the fourth chromosomes closest to the poles. Chromosome 4 in Drosophila is very small and sometimes difficult to visualize. (C) In late anaphase I, MEI-S332–GFP is still detected at the leading edges of the chromosomes, which become shorter and rounder as they approach the poles. (D) Between the first and second meiotic divisions, nuclear decondensation does not occur. Rather, two clusters of 3–4 chromatin balls are observed. Each ball most likely represents a pair of sister chromatids and is associated with a dot of MEI-S332– GFP. (E) In metaphase II, the chromatin balls move together to form metaphase plates, and MEI-S322–GFP localizes to the middle of the chromatin. (F) When sister-chromatid cohesion is released at anaphase II, the sister chromatids separate, and MEI-S332–GFP is no longer detectable on the meiotic chromosomes. (G) During the postmeiotic interphase, MEI-S332–GFP is not visible on the decondensed chromosomes. Oocytes were isolated from females carrying four copies of the mei-S332 + ::GFP transgene, activated in vitro, fixed, and stained with propidium iodide. Images were collected using confocal microscopy. Bar, ∼5 μm.
Western Blot Analysis
The rabbit anti–MEI-S332 antibodies (Covance, Research Products Inc., Denver, PA) were generated against a COOH-terminal MEI-S332 peptide conjugated to keyhole limpet hemacyanin. This 15-mer peptide (residues 386–400), (C)KNKLRNGSKGKAKAK, was chosen as the antigen because of the availability of the mei-S3327 allele, which lacks the COOH-terminal region of the protein due to a nonsense mutation at residue arg293 (Kerrebrock et al., 1995) and, hence, provides a negative control for the antibodies. The anti-peptide antibodies were affinity purified from rabbit serum using GST–MEI-S332 fusion protein bound to immobilon-P strips. The antibodies were eluted from the strips by acid elution buffer (5 mM glycine-HCl, pH 2.5, 150 mM NaCl) and immediately neutralized by 1 M NaPO4 buffer, pH 8. The GST–MEI-S332 fusion protein was generated by cloning a 1.35-kb BamHI-EcoRI mei-S332 cDNA fragment in frame with GST in the pGEX-4T-3 expression vector (Pharmacia Biotechnology, Inc., Piscataway, NJ). The resulting pGEX.MEI plasmid allowed for expression of the full-length MEI-S332 protein, fused to GST at the NH2 terminus, in BL21 (λDE3)pLysS cells.
Embryonic extracts were made by dechorionating Oregon-R embryos in 50% Clorox bleach and homogenizing in urea sample buffer (USB: 8 M urea, 2% SDS, 5% β-mercaptoethanol, 100 mM Tris, pH 7.6, and 5% Ficoll) at 5:1 USB/embryo (vol/vol). Oocyte extracts were made from mature oocytes isolated as described in Page and Orr-Weaver (1997). Females were fattened for 3–5 d with yeast before blender isolation. Oocytes were homogenized in one urea sample buffer at 3:1 USB/oocyte (vol/vol). Ovary extracts were made by dissecting previtellogenic, immature ovaries, or mature ovaries in PBS from newly eclosed females or females that were fattened on yeast for 3 d, respectively, and homogenizing pooled ovaries in USB (∼1 μl buffer/ovary). All protein extracts were cleared by centrifugation, frozen in liquid nitrogen, and stored at −80°C.
For the analysis of MEI-S332 levels in oocytes before and after activation, oocytes were isolated in IB, in either the presence or absence of 100 μg/ml cycloheximide (Fluka), from 300 y w females fattened on wet yeast for 3 d, as described above. After isolation, half of the oocytes were fixed by immersion in MeOH (unactivated) and the other half were activated in AB and ZAB in either the presence or absence of 100 μg/ml cycloheximide, as described above. The total incubation time in AB + ZAB was 60 min. These activated eggs were then fixed by incubation in MeOH. After several hours of fixation in MeOH at room temperature, oocytes and eggs were rehydrated in PBS. Rehydrated samples were mixed with 1:1 EB/4 × Laemmli sample buffer (EB: 10 mM Tris 7.5, 80 mM Na β-glycerophosphate, pH 7.5, 20 mM EGTA, 15 mM MgCl2, 2 mM Na3VO4, 1 mM benzamidine, 1 mM sodium metabisulfite, 0.2 mM PMSF) by crushing with the melted tip of a glass pipette. The ratio of sample to buffer added was 1:4 (vol/vol). Samples were boiled for 15 min, cleared by centrifugation, and frozen in a dry ice/MeOH bath. Control extracts for this experiment were made by isolating and fixing unactivated oocytes from pr cn mei-S3327 bw sp/cn mei-S3327 px sp and Oregon-R females as above. A cross-reacting band on the Western blot, just below the MEI-S332 signal, is also present in the mei-S3327 negative control, and is perhaps an artifact of this sample preparation.
Protein extracts were separated on 12% 150:1 (acrylamide/bis-acrylamide) gels and blotted onto immobilon-P membranes (Millipore Corp., Waters Chromatography, Milford, MA). About 200 μg of total protein was loaded per lane, and Ponceau S staining was used to verify equivalent protein loading before immunoblotting. Blots were blocked in 5% nonfat dry milk and 2% BSA in TBST (0.01 M Tris, pH 7.5, 0.9% NaCl, and 0.1% Tween 20) for 1 h at room temperature, and then incubated overnight at room temperature with affinity-purified anti–MEI-S332 peptide antibodies diluted at 1:40 in the block solution. Alkaline phosphatase-conjugated anti–rabbit secondary antibodies (Promega Corp., Madison, WI), diluted 1:7,500 in the block solution, were used to detect bound anti-peptide antibodies. The MEI-S332 protein was visualized using the BCIP/ NBT color development substrate (Promega). Although it is predicted to be 44 kD, the MEI-S332 protein migrates as a 55-kD band.
Results
MEI-S332 Localizes to Centromeric Regions in Oocytes
Although the localization of MEI-S332 has been determined in spermatocyte meiosis (Kerrebrock et al., 1995), the differences between male and female meiosis in D. melanogaster and the existence of mei-S332 alleles that affect the two sexes with different severity led us to ask where MEI-S332 is localized in oocyte meiotic divisions. Specifically, we asked whether it localizes to meiotic centromeres, and if so, what is the fate of the protein when the sisters separate at anaphase II.
To visualize the MEI-S332 protein in oocytes, we used a fusion of GFP to the NH2-terminal end of mei-S332 (mei- S332 + ::GFP) that has been shown to complement fully the mutant phenotype in both males and females (Kerrebrock et al., 1995). In Drosophila, mature oocytes arrest at metaphase I with a tapered spindle and an elongated nucleus. We examined fixed oocytes stained for DNA and observed that MEI-S332–GFP was present in two caps at opposite ends of the oocyte nucleus (Fig. 1 A). The orientation of the caps with respect to the morphology of the oocyte nucleus suggested that these caps were facing the poles of the metaphase I spindle, and tubulin staining later confirmed this interpretation (see below). Because it has been shown that the centromeric regions of chromosomes are positioned on opposite sides of the chromatin mass during the metaphase I arrest in Drosophila oocytes (Dernburg et al., 1996), it was likely that caps of MEI-S332–GFP represented centromeric localization.
We wanted to determine what happens to these caps of MEI-S332 when the meiotic cell cycle resumes after the oocyte arrest. In particular, we sought to observe the localization of the protein during anaphase I, when centromeric localization would be most apparent, and observe what happens to the protein at the metaphase II–anaphase II transition when the sister chromatids separate. Historically, it has been difficult to observe any of the stages of female meiosis that follow the metaphase I arrest in Drosophila oocytes, but recent advances in egg activation in vitro now allow all the stages of meiosis to be examined (Page and Orr-Weaver, 1997). Accordingly, oocytes from mothers carrying the mei-S332 + ::GFP transgene were activated in vitro to complete meiosis, and then fixed and stained for DNA. Oocytes in anaphase I had eight pairs of sister chromatids, four on each side, as is expected since the haploid chromosome number in Drosophila is four. Such oocytes also had eight dots of MEI-S332–GFP visible at the leading edges of the separating chromosomes, one per pair of sister chromatids (Fig. 1 B). The observation that each pair of sister chromatids had MEI-S332 at their leading edge argued strongly that MEI-S332 is localized at the centromeric regions of chromosomes in female meiosis. MEI-S332–GFP was continually visible on the chromosomes between anaphase I and metaphase II (Fig. 1, B–E and see below). When sister-chromatid cohesion was released at anaphase II, the sister chromatids separated, and for the first time during the meiotic divisions, MEI-S332 was not observed on the chromosomes (Fig. 1 F). After the meiotic divisions, the chromatin decondensed into four nuclei (three polar bodies and one pronucleus) in the postmeiotic interphase. MEI-S332–GFP was not detectably localized during the postmeiotic interphase (Fig. 1 G).
The cytology of nuclei between the meiotic divisions has been difficult to observe in oocytes. Indeed, even with the in vitro activation system, the lack of familiar cytological landmarks between anaphase I and metaphase II has meant that it was still unknown what happened to chromosome morphology between the divisions. Although it is known that in Drosophila male meiosis the telophase I nuclei decondense and then recondense for meiosis II (Cenci et al., 1994), it was unclear whether such decondensation occurred in Drosophila females. In experiments activating hundreds of mei-S332 + ::GFP transgenic oocytes, we never observed oocytes with only two decondensed nuclei, in agreement with our unpublished observations with oocytes from nontransgenic flies. Thus, it appears that Drosophila oocyte nuclei remain condensed throughout meiosis until telophase II.
Because it was clear from the early anaphase I figures that MEI-S332–GFP labels the centromeric regions of oocyte meiotic chromosomes (Fig. 1 B), we were able to use it as a tool in deducing the order of events in chromatin remodeling between anaphase I and metaphase II. We observed that late anaphase I chromosomes appear to become shorter and rounder as they approach the poles, but despite these morphological changes they could always be identified by the leading edge of MEI-S332–GFP at the centromere (Fig. 1 C). Between the divisions, the chromosomes rounded up and formed two clusters of three or four individual balls of chromatin (Fig. 1 D). Each ball was associated with a dot of MEI-S332–GFP, but the dots were no longer oriented at the leading (outside) edge of the chromosomes. We think it likely that each ball represents the sister chromatids of each of the three large chromosomes, with the small fourth chromosome only sometimes visible. Metaphase II was evident when the clusters of chromatin balls compacted to form metaphase plates, usually parallel to each other, with MEI-S332–GFP in the middle of the compacted chromatin (Fig. 1 E). Often, as in Fig. 1 E, the two nuclei were slightly out of synchrony. Even though there is no decondensation between the meiotic divisions, a series of interesting changes occurs in chromosome morphology between anaphase I and metaphase II.
When Does MEI-S332 Localize to Centromeres?
In spermatocytes, MEI-S332 protein is observed in the cytoplasm during prophase I, and it is localized to the chromosomes as they compact for prometaphase I (Kerrebrock et al., 1995). We examined when and how MEI-S332 is localized before metaphase I in oocytes, as there are marked differences between spermatocytes and oocytes during prophase I. The origin of the cytoplasm in oocytes differs from that in spermatocytes, because much of it is created in the nurse cells, and the volume of cytoplasm is much greater in oocytes than in spermatocytes. Another important difference is that synaptonemal complex is seen on oocyte chromosomes, but not on spermatocyte chromosomes. Sex-specific differences in the origin and amount of cytoplasm or in the structure of the meiotic chromosomes suggested that the timing of MEI-S332 localization should be examined in oocytes to see if it differed from spermatocytes.
To examine MEI-S332 localization in oocytes during early developmental stages, ovaries were dissected from females carrying the mei-S332 + ::GFP transgene, fixed, and stained for DNA (data not shown). MEI-S332–GFP was not observed in egg chambers during prophase I, corresponding to oocyte development through stage 12, either in the cytoplasm or on the condensed meiotic chromosomes in the karyosome. Multiple foci of MEI-S332–GFP were first observed on the meiotic chromatin after the chromatin compacted into the small round mass characteristic of prometaphase I. Using egg chamber morphology to judge developmental stage, we determined that these foci first appeared in stage 13. By stage 14, MEI-S332–GFP was observed in two caps on either side of the nucleus (Fig. 1 A and see below).
Because the meiotic spindle is organized shortly after the chromatin compacts, we further characterized the localization of MEI-S332–GFP with respect to formation of the spindle by isolating stage 13 and 14 oocytes, and labeling both the DNA and tubulin. After compaction of the chromatin in stage 12, the nuclear envelope breaks down and short microtubule fibers captured by the chromatin subsequently coalesce into a bipolar spindle during stage 13 (Theurkauf and Hawley, 1992). The earliest stage at which MEI-S332–GFP was observable was coincident with the beginning of spindle formation. A small number of dots of MEI-S332–GFP were distributed throughout the chromosomal mass (Fig. 2 A). When spindles appeared more bipolar and elongated, typical of late stage 13 and stage 14 oocytes, the MEI-S332–GFP foci were more clearly combined into caps on the ends of the chromatin mass that face the spindle poles (Fig. 2, B–C).
The Metaphase II–Anaphase II Transition
In both female and male meioses MEI-S332 was not visible on the sister chromatids after they separated at anaphase II, consequently we investigated what happened to the protein when sister-chromatid cohesion was released. In yeast and Xenopus mitosis, an inhibitor of sister-chromatid separation is degraded by the cyclin destruction machinery at the metaphase–anaphase transition (Holloway et al., 1993; Irniger et al., 1995; Cohen-Fix et al., 1996). Because MEI-S332 is essential for sister-chromatid cohesion, it seemed plausible that it might be degraded at the metaphase II–anaphase II transition.
To study protein levels directly, we generated polyclonal rabbit antibodies against a peptide corresponding to the COOH-terminal fragment of the MEI-S332 protein (Fig. 3 A). Affinity-purified antibodies recognized a band of ∼55 kD on a Western blot of ovary and oocyte extracts (Fig. 3 B). This band was absent in extracts made from mei-S3327 oocytes and ovaries (Fig. 3 B, lanes 2 and 3). Extracts from mei-S3327 homozygotes and hemizygotes provided a critical negative control, as this mutation creates a nonsense codon that prematurely truncates the protein so that it lacks the epitope for the COOH-terminal peptide antibodies (Fig. 3 A). As additional evidence that the identified band is MEI-S332, we probed extracts from transgenic ovaries that had four extra copies of a genomic mei-S332 + fragment, in addition to the two endogenous copies, and we found that the band was significantly more intense (Fig. 3 B, lane 1). These data lead us to conclude that the peptide antibodies recognize the MEI-S332 protein as a 55-kD band on Western blots. This protein migrates during electrophoresis as a 55-kD band even though its predicted size is 44 kD.
To determine whether MEI-S332 is degraded at the metaphase II–anaphase II transition, we analyzed in vitro activated oocytes. 60 min after activation, eggs can be selected so that 95–99% have completed meiosis (Page and Orr-Weaver, 1997). We compared MEI-S332 protein levels between extracts of unactivated oocytes, which have MEI-S332 localized to the chromosomes (Fig. 1 A), and extracts of eggs that have passed through the metaphase II–anaphase II transition after activation for 60 min. On Western blots, these protein levels remained essentially unchanged (Fig. 3 C, lanes 3 and 4), a result that was repeated several times. This suggests that although the protein dissociated from the chromosomes at anaphase II, it was not degraded.
Although the total levels of MEI-S332 remained constant before and after meiosis was completed, we were concerned that continuing translation of new MEI-S332 protein might mask protein degradation. To address this concern, we activated oocytes in the presence of the translational inhibitor cycloheximide. Metabolic labeling experiments have demonstrated that oocytes activated in the presence of cycloheximide have protein synthesis inhibited to about 5% of wild type levels, but that about 95% of oocytes still complete meiosis under these conditions, arresting at the postmeiotic interphase (Page and Orr-Weaver, 1997). Western blotting of extracts from arrested, unactivated oocytes incubated in cycloheximide, compared to extracts from oocytes activated in the presence of cycloheximide, further demonstrated that there was no detectable degradation of MEI-S332 during meiosis, suggesting that it instead delocalized (Fig. 3 C, lanes 3–6).
MEI-S332 during Mitosis
The phenotype of mei-S332 mutants was previously shown to be exclusively meiotic and not mitotic: no cytological defect has been detected in proliferating tissues, mutants are completely viable, and no increase in somatic clones from mitotic errors is observed (Kerrebrock et al., 1992, 1995). However, our finding that the protein was not degraded at anaphase II led us to ask whether the protein persisted in the developing embryo. We examined extracts from wild type oocytes and embryos by Western blotting, and we found significant amounts of MEI-S332 in a collection of embryos of ages 0–2 h (Fig. 3 B, lane 6). The protein level appeared to increase in populations of embryos of ages 2–4 h (Fig. 3 B, lane 7), suggesting that MEI-S332 did not merely persist into embryogenesis, but could be playing a role there. Additionally, we noted that there appeared to be different mobility forms of MEI-S332, an observation that is currently under investigation.
We used the mei-S332 + ::GFP transgene to determine whether MEI-S332 could localize onto chromosomes in the embryo, and we observed persistent localization of the protein on polar body rosettes (Fig. 4, A and B). Chromosomes from the unused meiotic products are pulled into a radial formation by a sphere of tubulin, after replicating and condensing into a metaphase-like state. These are found in the anterior dorsal quadrant of early embryos, typically fused so that there exist only one or two rosettes (Foe et al., 1993). MEI-S332–GFP localized to the condensed chromosomes facing the inside of the rosette, where centromeres are expected to be located (Foe et al., 1993). Moreover, when all the unused meiotic chromosomes have fused into a single rosette formation, the number of chromosomes should be 12, or after replication 24, and we count ∼24 foci of MEI-S332–GFP in a typical single rosette formation (Fig. 4 B). As in meiosis, MEI-S332 localized to the apparent centromeric regions of replicated sister chromatids.
MEI-S332–GFP also localized to condensed chromosomes in the early mitotic divisions. Drosophila embryos have 13 syncytial nuclear division cycles before gastrulation. On condensed prometaphase and metaphase chromosomes of these early cycles we observed MEI-S332–GFP in punctate dots resembling those on meiotic chromosomes, consistent with centromeric localization (Fig. 4 C). These punctate dots were not observed in interphase nuclei (Fig. 4, D and E). In addition to chromosome localization, diffuse clouds of fluorescence were observed in the vicinity of each mitotic nucleus (Fig. 4 C). Similar diffuse clouds of MEI-S332– GFP fluorescence were evident near interphase nuclei (Fig. 4, D and E) and produced a signal brighter than the background autofluorescence in embryos lacking the transgene (data not shown). These clouds of fluorescence may correspond to energids, regions of yolk-free cytoplasm that have been observed in the early cycles of Drosophila embryos (Foe et al., 1993). Immunofluorescence with anti-peptide antibodies confirmed the localization to polar body chromosomes and condensed mitotic chromosomes (data not shown).
In later syncytial divisions, the nuclei migrate to the surface of the embryo, and mitosis proceeds in a wave across the embryo. We examined mitotic chromosomes in these easily visualized nuclei to analyze localization of MEI-S332 during the metaphase–anaphase transition in mitosis. To simulate the same mei-S332 gene dosage as that of wild type oocytes, embryos from mothers hemizygous for mei- S3327 and carrying two copies of the mei-S332 + ::GFP transgene were examined after fixation in formaldehyde and DNA staining. MEI-S332–GFP was observed in bright dots aligned on the metaphase plates with the chromatin (Fig. 5 A; see arrow for one example). Sometimes much dimmer dots of MEI-S332–GFP were observed on chromosomes in early anaphase (Fig. 5 B, arrowhead). The residual MEI-S332–GFP was found on the leading edge of chromosomes. By late anaphase and telophase, no MEI-S332–GFP was observed on any of the chromatin. Thus, the metaphase–anaphase transition begins a process of delocalization of MEI-S332. The alignment of the dots on the metaphase plate and the association of residual MEI-S332–GFP with the leading edges of chromosomes strongly suggests that MEI-S332 is localized to the centromeric regions of mitotic chromosomes. Thus, in mitosis as in meiosis, MEI-S332 is localized to the centromeric regions of chromosomes condensed for metaphase, and MEI-S332 begins to dissociate from the chromatin when cohesion is lost and the sister chromatids segregate.
Discussion
In this study we examined the expression and localization of MEI-S332 in Drosophila oocytes and embryos. We found that in oocytes, MEI-S332 localizes to the centromeric region of condensed meiotic chromosomes from prometaphase I until the metaphase II–anaphase II transition, when sister chromatids separate. This is essentially the same localization pattern as has been observed in spermatocyte meiosis (Kerrebrock et al., 1992). It is striking that although no mitotic phenotype has been observed in mei-S332 mutants (Kerrebrock et al., 1995), MEI-S332 protein has a similar localization pattern in the early mitotic divisions in the embryo, where it appears bound to condensed chromosomes until the sister chromatids separate at anaphase. On the chromosomes of polar bodies, which are constitutively condensed in a configuration analogous to metaphase, MEI-S332 is consistently observed at the expected centromeric regions. Thus MEI-S332 appears localized to centromeres of metaphase chromosomes in each of these three different cell cycles, and it is dispersed each time sister chromatids separate.
MEI-S332 and the Metaphase–Anaphase Transition
Precisely what happens to MEI-S332 when sister chromatids separate at anaphase is a question of great interest. One possibility is that the protein is degraded at the metaphase–anaphase transition. To test this idea, we examined the levels of MEI-S332 in oocytes before and after the completion of meiosis. We found that even in the presence of cycloheximide to prevent new protein synthesis, the levels of MEI-S332 appeared unchanged before and after the metaphase II–anaphase II transition. This result demonstrates that on a global level MEI-S332 is not degraded at anaphase II. Although we have not directly examined the question of degradation in mitosis, the observation that MEI-S332 protein is visible in clouds around interphase nuclei strongly supports the idea that it is not degraded on a global level in the developing embryo during the syncytial divisions. Still, we cannot exclude the possibility that centromere-localized protein is locally degraded at either the metaphase II–anaphase II transition in oocytes or at the mitotic metaphase–anaphase transition. If a subpopulation of MEI-S332 was degraded at anaphase II, however, the amount degraded would have to be insignificant compared to the persisting fraction, as we do not observe any decrease in protein levels by Western blotting.
A second possibility is that dissociation of MEI-S332 from the centromeric regions triggers sister-chromatid separation. An analogous mechanism may occur in the yeast S. cerevisiae, because the Mcd1p/Scc1p cohesion protein localized on the chromosomes is not degraded until after anaphase. Instead it is removed from the chromosomes beginning at anaphase (Guacci et al., 1997; Michaelis et al., 1997). Noting that MEI-S332 appears to run as a doublet on Western blots, we speculate that dissociation of MEI-S332 may be regulated at some level by phosphorylation. Consistent with this speculation, the MEI-S332 protein has 30 possible phosphorylation sites recognized by protein kinase C, casein kinase II, cAMP-dependent protein kinase, and tyrosine protein kinase.
There is a third possibility, however, that MEI-S332 may first be inactivated to permit anaphase movement and subsequently dissociate from the chromosomes. This model is supported by our detection of MEI-S332 on the centromeres of chromosomes in early anaphase, although the levels are reduced compared to metaphase. Similarly, some Mcd1p/Scc1p remains localized to the chromosomes in anaphase (Guacci et al., 1997; Michaelis et al., 1997). We cannot distinguish between these two latter models at this point, because it is possible that sufficient amounts of MEI-S332 or Mcd1p/Scc1p dissociate at the metaphase– anaphase transition to permit sister-chromatid separation. Residual levels may then be removed subsequently.
Establishment Versus Maintenance of Sister-Chromatid Cohesion
In spermatocytes, oocytes, and early embryos, MEI-S332 is not detectable on the chromosomes until prometaphase. It is possible that sister-chromatid cohesion is not fully established until this point and that the localization of MEI-S332 marks the establishment of cohesion. It may be the case, however, that cohesion is established immediately after DNA replication. In FISH studies done in yeast, separate signals from the two sister chromatids were not observed until anaphase, indicating that sister chromatids are tightly associated from the time of their replication (Guacci et al., 1993, 1994). This suggests that cohesion is established during S phase. If this is true, then MEI-S332 may be required to maintain or augment cohesion when spindle forces come into play, rather than to establish cohesion. For example, it may serve to protect and preserve proteins directly attaching the sister chromatids until anaphase.
A Mitotic Role for MEI-S332?
We were surprised to find that MEI-S332 localizes to mitotic chromosomes in much the same way it localizes to meiotic chromosomes in spermatocytes and oocytes because no function has been ascribed to MEI-S332 in mitosis. The presence of MEI-S332 on mitotic chromosomes is not unique to the early embryonic cycles. MEI-S332 protein is present in dividing larval tissues and can localize to the chromosomes during mitosis (LeBlanc, H., T.T. Tang, and T.L. Orr-Weaver, unpublished results). We and our colleagues have undertaken careful phenotypic analyses of mei-S332 mutants in order to determine whether the protein is required for mitosis. Viability studies have demonstrated that mei-S332 homozygotes and their heterozygous siblings survive equally (Kerrebrock et al., 1992), even when the maternal mei-S332 contribution is eliminated (LeBlanc, H., and T.L. Orr-Weaver, unpublished data). Examinations of large numbers of larval brains, a mitotically active tissue that when squashed flat gives excellent mitotic cytology, demonstrated no significant difference in mitotic index or premature sister–chromatid separation between mei-S332 hemizygous (mei-S332/Df) and wild type larval brains (Kerrebrock et al., 1995). Furthermore, experiments testing the frequency of chromosome missegregation in the developing wing demonstrated no significant difference between mei-S332 hemizygotes and their heterozygous siblings (Kerrebrock et al., 1995).
If MEI-S332 is localized to mitotic centromeres, why do we not see a phenotype in mei-S332 mutants? One possibility is that in mitosis there is redundancy in the mechanisms that hold sister chromatids together. The simplest model for redundancy is that both MEI-S332 and another protein act independently to bind sister chromatids together at the centromeric regions in mitosis, and therefore no phenotype is observed when mei-S332 is mutated. Currently there are no candidates for such a protein. Although mutations have been characterized in three genes that encode Drosophila centromere–binding proteins, none appear to promote sister-chromatid cohesion. The HP1 and PROD proteins affect centromere condensation and presumably kinetochore function (Kellum and Alberts, 1995; Torok et al., 1997), whereas ZW10 may monitor spindle attachment to the kinetochore (Williams et al., 1996). Another version of this redundancy model is that whereas MEI-S332 acts at mitotic centromeres to attach sister chromatids, other proteins act along the lengths of the chromatid arms to ensure cohesion and proper orientation with respect to the mitotic spindle. The loss of MEI-S332 would result in the loss of centromeric cohesion, but this would not have phenotypic consequences in mitosis because arm cohesion would be sufficient to hold the chromatids together. This redundancy is not provided solely by ORD, a Drosophila protein required for arm cohesion in meiosis, because flies lacking both mei-S332 and ord have demonstrated no abnormalities in somatic mitoses (Bickel, S.E., D.P. Moore, C. Lai, and T.L. Orr-Weaver, manuscript submitted for publication).
Alternatively, it is possible that MEI-S332 does play a nonredundant role in mitosis, but it is required only in response to perturbations of the cell cycle. For example, if it were necessary for a cell to delay the onset of anaphase, persistence of MEI-S332 at the centromeric regions could, in principle, restrain the sister chromatids from separating. The discovery of a mitotic phenotype, under any conditions, would greatly enhance our understanding of the mitotic function of MEI-S332.
Meiotic Cytology
Because MEI-S332 localizes to centromeres throughout meiosis until anaphase II, we were able to use it as a tool to examine meiotic chromosome morphology. In metaphase I arrested oocytes, the two caps of MEI-S332–GFP demonstrate that the centromeric regions of homologues are closest to the spindle poles during metaphase I, as would be anticipated if homologues are connected by chiasmata on the chromosome arms. Again using MEI-S332 to identify centromeres and chromosome orientation, we were able to infer an order of events after anaphase I. We found that in Drosophila, as in Xenopus and other organisms, oocyte chromosomes do not decondense between the two meiotic divisions (Murray and Hunt, 1993), in contrast to spermatocyte chromosomes that do decondense between the divisions (Cenci et al., 1994).
Conclusions
Our finding that MEI-S332 is present on both meiotic and mitotic chromosomes reinforces the idea that meiosis and mitosis are highly conserved processes, even at the molecular level. In both types of divisions, it is localized to the centromeric regions of sister chromatids aligned on a bipolar spindle, and it is no longer present on the sister chromatids when they segregate from one another in anaphase. The function of MEI-S332 is essential during meiosis and not mitosis probably because of the meiosis-specific requirement that sister chromatids remain attached in the centromeric region during the first meiotic cell division. It is ironic that MEI-S332 is now implicated in mitosis, because if it had a strong mitotic phenotype, lethality would have hindered the genetic and cytological analyses that defined its role in sister-chromatid cohesion. Our findings indicate that the analysis of meiosis will lead to a deeper understanding of chromosome segregation mechanisms in general.
Acknowledgments
We thank Sharon Bickel for advice on DNA stains and antibody affinity purification, Lisa Elfring for help with Western blots, and Ya-Huei Tu for assistance with microscopy (all from Whitehead Institute for Biomedical Research, Cambridge, MA). We are grateful to Abby Dernburg (Stanford University, Palo Alto, CA), Doug Koshland (Carnegie Institute of Washington, Baltimore, MD), and Kim Nasymth (Institute for Molecular Pathology, Vienna, Austria) for communicating results before publication, and to Sharon Bickel and Heidi LeBlanc (Whitehead) for permission to cite unpublished results. Angelika Amon (Whitehead), Dean Dawson (Tufts University, Boston, MA), Lisa Elfring, Heidi LeBlanc, and Jacqueline Lopez (Whitehead) provided critical comments on the manuscript. We thank an anonymous reviewer for insightful comments and suggestions.
Footnotes
A.W. Page and T.T. Tang were supported by predoctoral fellowships from the National Science Foundation. This research was funded by a grant from the National Science Foundation (MCB-9604135).
Address all correspondence to Terry L. Orr-Weaver, Whitehead Institute for Biomedical Research and the Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02142. Tel.: (617) 258-5245. Fax: (617) 258-9872.
D.P. Moore and A.W. Page contributed equally to this work.
1. Abbreviation used in this paper: GFP, green fluorescent protein.
References
- Bickel SE, Orr-Weaver TL. Holding chromatids together to ensure they go their separate ways. Bioessays. 1996;18:293–300. doi: 10.1002/bies.950180407. [DOI] [PubMed] [Google Scholar]
- Cenci G, Bonaccorsi S, Pisano C, Verni F, Gatti M. Chromatin and microtubule organization during premeiotic, meiotic, and early postmeiotic stages of Drosophila melanogasterspermatogenesis. J Cell Sci. 1994;107:3521–3534. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters J-M, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiaeis controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes and Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Davis B. Genetic analysis of a meiotic mutant resulting in precocious sister-centromere separation in Drosophila melanogaster. . Mol Gen Genet. 1971;113:251–272. doi: 10.1007/BF00339546. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- Foe, V.E., G.M. Odell, and B.A. Edgar. 1993. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. In The development of Drosophila melanogaster. M. Bate and A. Martinez Arias, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 149–300.
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Goldstein LSB. Mechanisms of chromosome orientation revealed by two meiotic mutants in Drosophila melanogaster. . Chromosoma (Berl) 1980;78:79–111. doi: 10.1007/BF00291909. [DOI] [PubMed] [Google Scholar]
- Guacci V, Yamamoto A, Strunnikov A, Kingsbury J, Hogan E, Meluh P, Koshland D. Structure and function of chromosomes in mitosis of budding yeast. Cold Spring Harbor Symp Quant Biol. 1993;58:677–685. doi: 10.1101/sqb.1993.058.01.075. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. . Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108:1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Miyazaki WY, Birnby D, Orr-Weaver TL. The Drosophila mei–S332gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130:827–841. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. MEI-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Lamb N, Freeman SB, Savage-Austin A, Pettay D, Taft L, Hersey J, Gu Y, Shen J, Saker D, May KM, et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–405. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Maguire MP. The need for a chiasma binder. J Theor Biol. 1974;48:485–487. doi: 10.1016/s0022-5193(74)80017-2. [DOI] [PubMed] [Google Scholar]
- Maguire M P. Sister chromatid association at meiosis. Maydica. 1993;38:93–106. [Google Scholar]
- McKee BD. The license to pair: identification of meiotic pairing sites in Drosophila. . Chromosoma. 1996;105:135–141. doi: 10.1007/BF02509494. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Condensins and cohesins: proteins that hold sister chromatids together. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Moore, D.P., and T.L. Orr-Weaver. 1998. Chromosome segregation during meiosis: building an unambivalent bivalent. In Meiosis and Gametogenesis. M.A. Handel, editor. Academic Press, San Diego, CA. 263–299. [DOI] [PubMed]
- Murray, A., and T. Hunt. 1993. The Cell Cycle: An Introduction. Freeman, New York. 251 pp.
- Orr-Weaver TL. Meiosis in Drosophila: seeing is believing. Proc Natl Acad Sci USA. 1995;92:10443–10449. doi: 10.1073/pnas.92.23.10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL. Meiotic nondisjunction does the two-step. Nat Genet. 1996;14:374–376. doi: 10.1038/ng1296-374. [DOI] [PubMed] [Google Scholar]
- Page AW, Orr-Weaver TL. Activation of the meiotic divisions in Drosophilaoocytes. Dev Biol. 1997;183:195–207. doi: 10.1006/dbio.1997.8506. [DOI] [PubMed] [Google Scholar]
- Theurkauf, W.E. 1994. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. In Methods in Cell Biology. Vol 44. L.S.B. Goldstein, and E.A. Fyrberg, editors. Academic Press, New York. 489–505. [DOI] [PubMed]
- Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: Behavior of nonexchange chromosomes and the effects of mutations in the nodkinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok T, Harvie PD, Buratovich M, Bryant PJ. The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila. . Genes and Dev. 1997;11:213–225. doi: 10.1101/gad.11.2.213. [DOI] [PubMed] [Google Scholar]
- Williams BC, Gatti M, Goldberg ML. Bipolar spindle attachments affect redistribution of ZW10, a Drosophila centromere/kinetochore component required for accurate chromosome segregation. J Cell Biol. 1996;134:1127–1140. doi: 10.1083/jcb.134.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. . J Cell Biol. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]