Abstract
A 130-kD protein that coimmunoprecipitates with the tight junction protein ZO-1 was bulk purified from Madin-Darby canine kidney (MDCK) cells and subjected to partial endopeptidase digestion and amino acid sequencing. A resulting 19–amino acid sequence provided the basis for screening canine cDNA libraries. Five overlapping clones contained a single open reading frame of 2,694 bp coding for a protein of 898 amino acids with a predicted molecular mass of 98,414 daltons. Sequence analysis showed that this protein contains three PSD-95/SAP90, discs-large, ZO-1 (PDZ) domains, a src homology (SH3) domain, and a region similar to guanylate kinase, making it homologous to ZO-1, ZO-2, the discs large tumor suppressor gene product of Drosophila, and other members of the MAGUK family of proteins. Like ZO-1 and ZO-2, the novel protein contains a COOH-terminal acidic domain and a basic region between the first and second PDZ domains. Unlike ZO-1 and ZO-2, this protein displays a proline-rich region between PDZ2 and PDZ3 and apparently contains no alternatively spliced domain. MDCK cells stably transfected with an epitope-tagged construct expressed the exogenous polypeptide at an apparent molecular mass of ∼130 kD. Moreover, this protein colocalized with ZO-1 at tight junctions by immunofluorescence and immunoelectron microscopy. In vitro affinity analyses demonstrated that recombinant 130-kD protein directly interacts with ZO-1 and the cytoplasmic domain of occludin, but not with ZO-2. We propose that this protein be named ZO-3.
The tight junction acts to limit movement of substances through the paracellular space and as a boundary between the compositionally distinct apical and basolateral plasma membrane domains of epithelial and endothelial cells. The molecular configuration of the tight junction has generated considerable interest in the last decade. Actin filaments (17, 29) and the peripheral membrane proteins ZO-1 (40), cingulin (10), ZO-2 (21), 7H6 (48), Rab3B (43), symplekin (22), and AF-6 (47) are now known to be found at the tight junction. Occludin, a transmembrane protein, is also localized to the tight junction (14). Limited information is available on the roles these elements play in tight junction physiology. For example, actin filaments are believed to function in the regulation of junction permeability (30), although the molecular linkage by which this occurs is unknown. Data from several laboratories indicate that occludin functions as part of the paracellular permeability barrier (6, 31, 45) and participates in intercellular adhesion (42); however, it remains possible that other, unidentified transmembrane components may also be involved.
ZO-1 and ZO-2 were among the first tight junction proteins identified (16, 21, 40). Cloning and sequence analysis of these proteins indicates that they are part of a larger family of proteins potentially involved in tumor suppression and/or signal transduction (21, 44). This family includes the discs large tumor suppressor gene product (dlg-A) of Drosophila (46); p55, an erythrocyte membrane protein (35); and PSD-95/SAP90, a synaptic membrane protein (9, 25). TamA, a second Drosophila protein homologous to ZO-1, has also been identified (41). Common to all family members is a region homologous to guanylate kinase (GUK)1, an src homology (SH3) domain, and variable numbers of PDZ domains. These latter domains have been shown to function in binding integral membrane proteins such as ion channels at synapses (23, 27). In addition, the GUK domain of PSD95/SAP90 has been shown to bind a novel synaptic protein (24), and the SH3 domain of ZO-1 binds a serine protein kinase that phosphorylates a region at the COOH-terminal end of the protein (4). The membrane association and presence of GUK domains in this collection of proteins has resulted in them being named the MAGUK family (1).
Information on direct interactions among tight junctional proteins is also limited. Affinity analyses indicate that ZO-1 binds to occludin (15) and the Ras target AF-6 (47). Additional observations suggest that ZO-1 has the capacity to bind spectrin (15, 20), although localization of this cytoskeletal protein to the tight junction has not been documented. These observations, together with the protein-binding capacities of the MAGUK domains, make it likely that the tight junction conforms to the architectural paradigm of the adherens junction and desmosome, that of transmembrane constituents linked to the cytoskeleton through a complex of peripheral membrane proteins.
Immunoprecipitation of ZO-1 performed under conditions designed to preserve protein–protein interactions indicates that an additional protein at 130 kD interacts with a complex containing ZO-1 and ZO-2 (5). Although this protein is known to be phosphorylated, it has not been further characterized to date. Here we report the isolation, identification, and partial characterization of the 130-kD protein. We find this protein to be a novel member of the MAGUK protein family that directly interacts with ZO-1 and occludin. As the protein localizes to the tight junction (zonula occludens), we name it ZO-3.
Materials and Methods
Cell Culture
MDCK strain II cells were grown and metabolically labeled as described previously (2, 37). The MDCK cell line MDCK/Z3 was generated by transfection of the parental line with a full-length ZO-3 construct in pBK-CMV (see below) using Lipofectin (GIBCO BRL, Gaithersburg, MD) and G418 selection according to established protocols (3). Passage and growth of MDCK/Z3 cells were identical to that of parental cells. Sf9 insect cells (GIBCO BRL) were grown in serum-free medium (Sf-900 II SFM; GIBCO BRL) at 28°C in a nonhumidified, ambient air incubator on an orbital platform rotating at 125 rpm. The cell density was maintained between 4 × 105 to 2 × 106 cells/ml with viability >90%.
Immunoprecipitation
Immunoprecipitations under conditions that maintain some protein-protein interactions (“low stringency”) were performed by a modification of previously published techniques (16, 21). Confluent monolayers of MDCK cells were rinsed twice in ice-cold TBS with 1 mM CaCl2, 0.5 mM MgCl2, 0.2 mM PMSF, and 1 μg/ml aprotinin. Cells were solubilized in 1% Triton X-100, 0.5% sodium deoxycholate, 0.2% SDS, 150 mM NaCl, 20 mM Hepes, pH 7.4, 2 mM EDTA, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 μg/ml chymostatin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin. Solubilized material was centrifuged at 13,000 g for 30 min at 4°C to remove cell debris. Rat anti–ZO-1 mAb R40.76 (2) was added to the supernatant, followed by goat anti–rat IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) coupled to cyanogen-bromide–activated Sepharose 4B (Pharmacia Biotech, Inc., Piscataway, NJ). As a negative control, R5.21, a rat mAb identical in isotype to R40.76 (40), was used. Sepharose beads were washed according to procedures of Pasdar and Nelson (1989) and solubilized in gel sample buffer.
High stringency immunoprecipitations of ZO-1, ZO-2, and ZO-3 (tagged with an 11–amino acid epitope from the cytoplasmic domain of vesicular stomatitis virus G protein [VSV-G]; see below) from cells solubilized in hot 1% SDS were done as described previously (39) using R40.76 anti–ZO-1/ anti-rat IgG–Sepharose, rabbit polyclonal anti-ZO-2 (reference 21; R9989, a gift from Lynne Jesaitis and Dan Goodenough, Harvard Medical School, Boston, MA)/protein A–Sepharose (Pharmacia Biotech, Inc.), and a mouse anti–VSV-G cytoplasmic domain mAb (clone P5D4; Boehringer Mannheim Corp., Indianapolis, IN)/protein G–Sepharose (Pharmacia Biotech, Inc.).
Isolation and Cloning of ZO-3
Bulk Immunoprecipitation and Purification.
ZO-1 was immunoprecipitated from 104 roller bottles (1,600 cm2 each) of MDCK cells under low stringency conditions. Immunoprecipitates solubilized in gel sample buffer were pooled, concentrated by centrifugation in filter concentrators (Centriprep 100; Amicon Corp., Danvers, MA) and run on preparative SDS-PAGE. Approximately 25 μg of ZO-3 (determined by comparison of Coomassie blue–stained standards) were transferred onto PVDF in a Hoeffer TE42 transfer apparatus operated at 90 volts for 6 h and maintained at 20°C by a temperature controlled water bath. Transfer buffer (25 mM Tris, pH 8.3, 190 mM glycine, 0.01% SDS) was changed after 4 h. Protein on the polyvinylidene difluoride (PVDF) was visualized by staining with amido black. Buffer and transfer conditions were optimized for ZO-3 by test transfers with metabolically labeled material.
Amino Acid Sequencing and RT-PCR.
The PVDF containing ZO-3 was submitted to the Rockefeller University Protein Sequencing Facility for Lys-C endopeptidase microdigestion (12) and amino acid (aa) sequencing. We obtained the following 19 aa sequence: TAEMPDQFGIADSVLRTDN. Degenerate primers corresponding to the 6 aa at each end were synthesized: forward, 5′-AC(AC) GC(CT) GAG ATG CC(CT) GA-3′; reverse, 5′-GTT GTC (GT)GT (GCT)CG (GC)AG (GC)AC-3′. RT-PCR was performed with these primers on total RNA isolated from MDCK cells with TRIZOL (GIBCO BRL) according to manufacturer directions. PCR products were subcloned into the TA vector (Invitrogen Corp., Carlsbad, CA). The authenticity of the PCR products was verified on Southern blots using a degenerate synthetic oligonucleotide (5′-GA(CT) CA(AG) TT(CT) GG(AGCT) AT(ACT) GC-3′) end-labeled with δ-[32P]ATP (ICN Pharmaceuticals Inc., Irvine, CA). This oligonucleotide corresponded to the internal aa of the original peptide. Positives on Southern blots were subjected to double-stranded sequencing. A 57-mer that verified the entire original 19-aa peptide was obtained.
Library Screening.
The 57-mer was synthesized, end labeled with δ-[32P]ATP, and used to screen an oriented oligo(dT) primed MDCK cell cDNA library in λ-ZAP (provided by Marino Zerial, European Molecular Biology Laboratory, Heidelberg, Germany) using the method of Ausubel et al. (1992). 10 positively hybridizing phage were isolated after screening ∼500,000 plaques. pBluescript containing inserts were excised from plaque-purified phage as directed by the manufacturer (Stratagene, La Jolla, CA). The longest insert (A1, 1.6 kb) was subjected to double-stranded sequencing. Analysis of this sequence, together with the molecular mass of the protein and size of the mature ZO-3 mRNA determined by Northern blot (∼3.4 kb, see Fig. 4), indicated that the A1 clone contained the 3′ but not the 5′ end of the open reading frame. To extend the 5′ end of the ZO-3 sequence we constructed an oriented MDCK cDNA library specific for ZO-3 in Uni-ZAP XR (Stratagene). MDCK cell poly(A)+ RNA was isolated with PolyATract mRNA System III (Promega Corp., Madison, WI) and reverse transcribed with a ZO-3–specific antisense linker-primer (5′-GAG AGA GAG AGA GAG AGA GAA CTA GTC TCG AGC TGA TCT CCC TCC TGG ATG-3′) containing an XhoI site. This primer was situated 173 bp downstream of the 5′ end of clone A1. The cDNA was ligated with EcoRI adaptors, digested with XhoI, size fractionated, and ligated into the Uni-ZAP XR vector. The resulting library was screened with the oligonucleotide 5′-CAG CTA TGA CAT CTA CAG GGT GCC CAG CAG CCA GAG CGC AGA GGA CCG TG-3′ end-labeled with δ-[32P]ATP. This sequence is the complement of a known region of the A1 clone located in the 173 bp 5′ of the primer used to create the library. Approximately 200,000 plaques were screened and 6 positive clones plaque purified. Inserts were sized by restriction digestion and the 3′ 200–300 bp were sequenced to verify the clones were ZO-3 specific. The longest clones were selected for double strand sequencing. All sequencing was performed through the use of Sequenase Version 2.0 (United States Biochemical Corp., Cleveland, OH), Fidelity DNA Sequencing System (Oncor, Inc., Gaithersburg, MD), or the University of Alberta DNA Sequencing Facility.
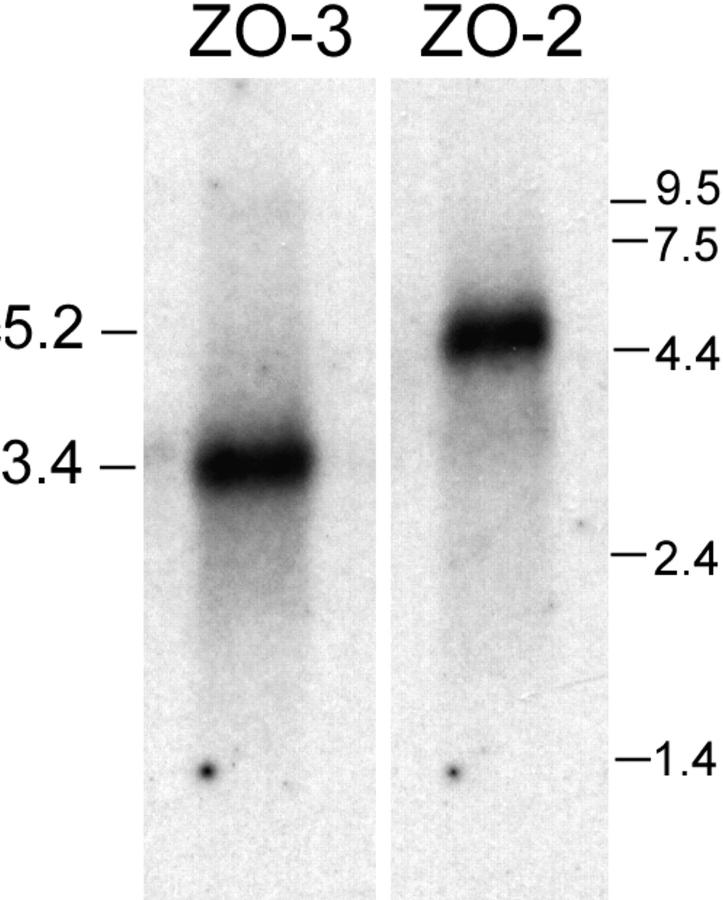
Figure 4.
Northern blots of ZO-3 and ZO-2. 5 μg of MDCK cell poly(A)+ RNA probed for ZO-3 shows a ∼3.4-kb transcript. Blot stripped and reprobed for ZO-2 shows the expected 5.2-kb transcript (21). RNA standards at right (kb).
Sequence Analysis.
Nucleotide sequences, aa compositions and isoelectric points of various domains, and the overall molecular mass of the protein were analyzed using the University of Wisconsin Genetics Computer Group (GCG) software package. Amino acid sequence comparisons to determine the percent identity and the percent similarity (percent identity + percent conserved substitutions) were done using the default parameters in the GCG GAP alignment program based on the algorithm of Needleman and Wunsch (1970). Sequence alignments were performed with GCG PILEUP. The amino acid numbers of compared sequences are shown in Table I.
Table I.
Amino Acid Numbers of Compared Sequences
| hZO-1 | cZO-2 | cZO-3 | TamA | dlg-A | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Start | 1–10 | 1–9 | 1–10 | |||||||
| PDZ1 | 11–97 | 10–96 | 11–92 | 23–110 | 40–122 | |||||
| Link | 98–173 | 97–290 | 93–185 | |||||||
| PDZ2 | 174–251 | 291–368 | 186–263 | 167–251 | 154–244 | |||||
| Link | 252–411 | 369–495 | 264–371 | |||||||
| PDZ3 | 412–490 | 496–574 | 372–450 | 408–486 | 486–567 | |||||
| Link | 491–507 | 575–591 | 451–467 | |||||||
| SH3 | 508–569 | 592–650 | 468–535 | 500–563 | 604–667 | |||||
| Link | 570–634 | 651–713 | 536–606 | |||||||
| GUK | 635–782 | 714–863 | 607–753 | 629–787 | 770–948 | |||||
| Link | 783–805 | 864–882 | 754–772 | |||||||
| Acid | 806–876 | 883–947 | 773–835 | |||||||
| End | 877–1736 | 948–1174 | 836–898 |
Northern and Southern Blotting
Total RNA was isolated from MDCK cells for Northern blots by the one step protocol of Ausubel et al. (1992), and poly(A)+ mRNA was generated by the PolyATract System III (Promega Corp.). RNA was transferred to Magna (MSI) nylon membrane and cross-linked with UV radiation (Stratalinker; Stratagene). A 230-bp fragment at the 5′ end of the ZO-3 A1 cDNA generated by EcoRI/PstI digestion (see Fig. 2) and an 850-bp segment of ZO-2 generated by PCR (primers: forward, 5′-GCACGAGAGAGACGGCAGC-3′; reverse, 5′-TTCTTCTTCATGCCAGATC-3′) were labeled by random-priming and used as probes. Blots were hybridized overnight at 68°C and washed according to previously published protocols (18). Southern blots were performed according to established techniques (3) using Duralon UV (Stratagene) and probed as described above.
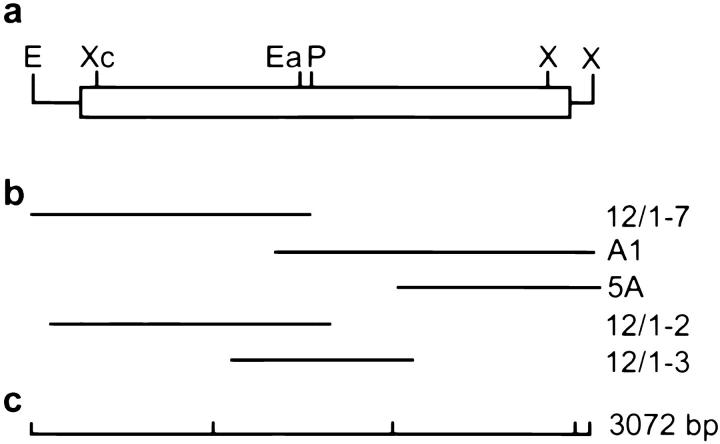
Figure 2.
Schematic representation of the full-length ZO-3 cDNA. (a) The 3,072-bp cDNA, with EcoRI (E) and XhoI (X) restriction sites at the 5′ and 3′ ends. (Solid lines) Untranslated regions. Open reading frame indicated by the box, with XcmI, EagI, PstI (unique to the A1 cDNA), and internal XhoI restriction sites. (b) Individual clones obtained from library screening. All partial cDNAs were subjected to double-stranded sequencing. (c) Scale bar in 1-kb segments.
Epitope Tagging of ZO-3
A full-length ZO-3 cDNA was assembled from overlapping clones and subcloned into the eukaryotic expression vector pBK-CMV (Stratagene). To remove the 241 bp 5′ untranslated region of the ZO-3 cDNA, the 5′ region was amplified by PCR with the following primers: forward, 5′-GGG AAT TCC AGG TGC TGG ACA TGG AGG A-3′; reverse 5′-CGG CAC CAC ATC GGA CAC GAC C-3′ (nucleotides 380–359 of Fig. 3). The forward primer contains an EcoRI restriction site and the 12 bp immediately upstream of the ZO-3 start codon. The amplified DNA fragment was used to replace the original 5′ untranslated region of ZO-3 by restriction digestion with EcoRI and XcmI (see Fig. 2). The COOH terminus of ZO-3 was tagged with the 11-aa VSV-G epitope by PCR using the following primers: reverse, 5′-CAA TCT AGA TTA CTT CTT AAG TCG GTT CAT CTC TAT GTC TGT ATA CAG GTC GGT GGC CGG GCC-3′, forward 5′-TAC GAG ACG GAC GGC GAG GG-3′ (nucleotides 2634–2653 of Fig. 3). The reverse primer codes for a XbaI restriction site, a stop codon and 11 aa of the VSV-G cytoplasmic domain immediately downstream of the terminal leucine of ZO-3. The amplified DNA fragment was used to replace the original 3′ end of ZO-3 by restriction digestion with XhoI and XbaI (see Fig. 2). All sequences of PCR- amplified fragments were verified by DNA sequencing.
Figure 3.
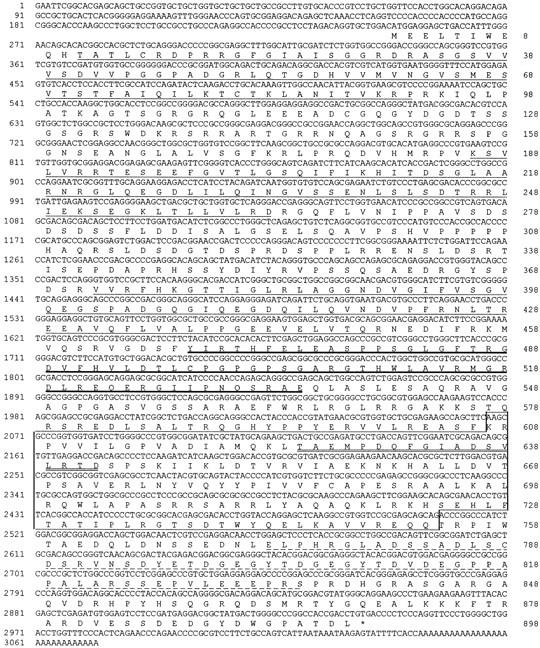
Full-length nucleotide and amino acid sequence of ZO-3. ZO-3 contains 3 PDZ domains (thin underlines), an SH3 domain (thick underline), and a GUK domain (box), demonstrating that it is another member of the MAGUK family of proteins. ZO-3 also contains a basic domain between PDZ1 and PDZ2, a proline-rich domain between PDZ2 and PDZ3 and an acidic domain (dashed underline). The double underline within the GUK box indicates the original peptide obtained by amino acid sequencing. The ZO-3 sequence is available from the GenBank database under accession number AF023617.
Immunoblotting
Immunoblots were performed using ECL (Amersham Corp., Arlington Heights, IL) according to manufacturer's protocols. Whole cell lysates were prepared according to Stevenson et al. (1994). The following primary antibodies were used: anti–ZO-1 mAb R40.76 (2); rabbit anti-ZO-2 (21); rabbit anti-VSV-G cytoplasmic domain (a gift from Carolyn Machamer, Johns Hopkins University, Baltimore, MD); an anti-human ZO-1 polyclonal antibody (Zymed Laboratories, Inc., South San Francisco, CA); and a partially characterized guinea pig antisera generated against a ZO-3 fusion protein corresponding to the linker region between PDZ1 and PDZ2 (aa 95–185). This anti–ZO-3 antisera shows strong reactivity to the fusion protein (data not shown), recombinant full-length ZO-3 and a 130-kD band present in whole MDCK cell lysate (see Fig. 9 c). However, it also shows faint reactivity on immunoblots of MDCK cell lysates with a band that comigrates with ZO-1. The nature of this reactivity has not been fully clarified but is irrelevant to the binding studies for which it was used (the antisera was not used for immunolocalizations; see below). Peroxidase-conjugated goat anti–rat IgG, goat anti–guinea pig IgG (Jackson ImmunoResearch Laboratories, Inc.) and goat anti–rabbit IgG (Bio-Rad Laboratories, Hercules, CA) were used as secondary antibodies.
Figure 9.
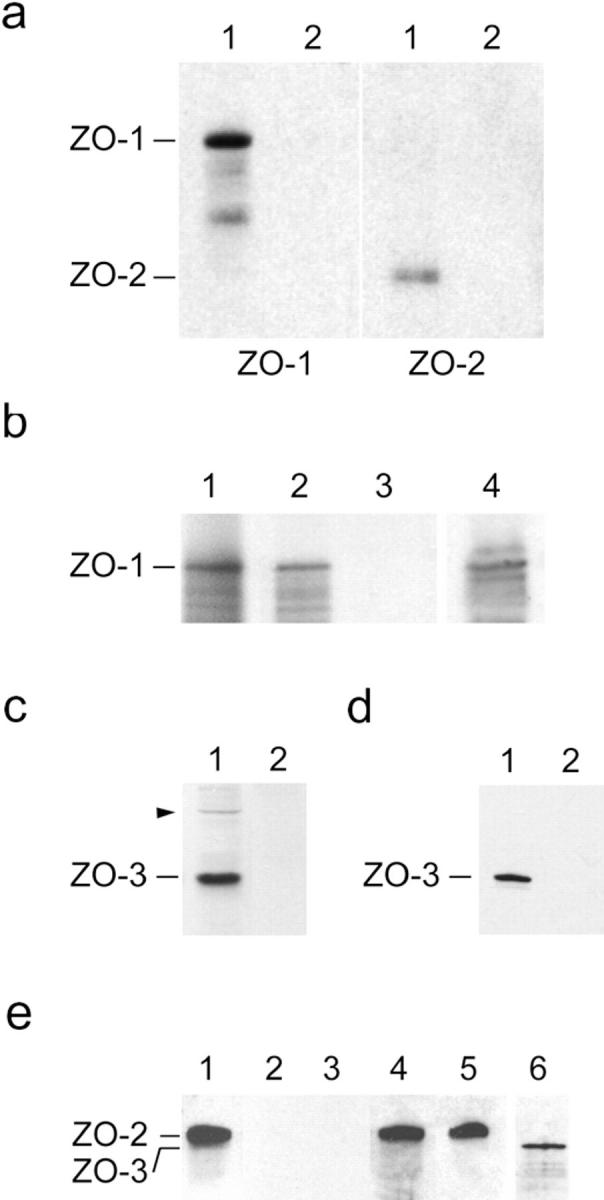
Binding of recombinant ZO-3 to tight junction proteins. (a) ZO-3 binds ZO-1 and ZO-2 from MDCK cell extracts. Affinity resin containing either full-length ZO-3 (lanes 1) or negative control peptide (lanes 2) were incubated with high salt extracts of MDCK cell membranes, washed, solubilized, and immunoblotted for either ZO-1 (left) or ZO-2 (right). The resin containing ZO-3 specifically retains both ZO-1 and ZO-2. (b) ZO-3 binds ZO-1 directly. Radioactively labeled ZO-1 generated by in vitro transcription/translation (lane 1) was incubated with affinity resin containing either full-length ZO-3 (lane 2) or negative control peptide (lane 3). Resin was washed, solubilized, and subjected to SDS-PAGE. The resin containing ZO-3 specifically retains a band which runs at 215 kD. This band was confirmed as ZO-1 by immunoblotting an identical aliquot of the bound material in lane 2 with anti-human ZO-1 antisera (lane 4). Lanes 1–3, autoradiograms; lane 4, ECL. (c) Partial characterization of an anti–ZO-3 antisera. Guinea pig antisera generated against a portion of ZO-3 reacts with a 130-kD band present in whole MDCK cell lysate (lane 1). It also shows faint reactivity with a band that comigrates with ZO-1 (arrowhead). No reaction with MDCK cell proteins was detected with preimmune sera (lane 2). (d) ZO-3 binds directly to the cytoplasmic tail of occludin. Recombinant ZO-3 was incubated with affinity resin containing either the COOH-terminal 148 aa of occludin (lane 1) or negative control peptide (lane 2). Bound material was eluted from washed resin and immunoblotted with anti–ZO-3 antisera. The resin containing occludin specifically retains ZO-3. (e) ZO-3 does not bind to ZO-2. Recombinant ZO-2 (lane 1) was incubated with affinity resin containing ZO-3 (lane 2) or negative control peptide (lane 3). Bound material was eluted from washed resin and immunoblotted with anti–ZO-2 antisera. No binding was detected in either case. Unbound fractions collected from resin containing ZO-3 (lane 4) or negative control peptide (lane 5) were also immunoblotted with anti–ZO-2 antisera. The presence of ZO-3 on the resin of lane 2 was verified by stripping the blot and reprobing with anti–ZO-3 antisera (lane 6).
Localization of ZO-3
Immunofluorescence was performed according to previously published protocols (18) using R40.76 anti-ZO-1 rat mAb/FITC-conjugated goat anti-rat IgG (Jackson ImmunoResearch Laboratories, Inc.) and rabbit anti-VSV-G/rhodamine-conjugated donkey anti–rabbit (Jackson ImmunoResearch Laboratories, Inc.). MDCK/Z3 cells were fixed with 2.5% paraformaldehyde and permeabilized with −20°C methanol before staining.
ImmunoEM was performed on isolated MDCK/Z3 cell membranes according to the technique of Jesaitis and Goodenough (1994) except that membranes were fixed with 2.5% paraformaldehyde for 15 min on ice followed by quenching in 50 mM glycine, 1% BSA in PBS for 20 min on ice before antibody incubations. Membranes were then colabeled with R40.76 anti–ZO-1/10 nm gold-conjugated goat anti–rat (British Biocell International, Cardiff, UK) and rabbit anti–VSV-G/5 nm gold-conjugated goat anti–rabbit (Sigma Chemical Co., St. Louis, MO) before processing for thin section EM (40).
Affinity Binding
Protein Expression.
Full-length canine ZO-2 (7) and ZO-3 cDNAs were subcloned into the pFastBac HT vector series of the Baculovirus eukaryotic expression system (GIBCO BRL, Gaithersburg, MD). The resulting constructs (pHTb/ZO-2 or pHTc/ZO-3) encode proteins with a 6-histidine tag at the NH2 terminus. pHTb/ZO-2 and pHTc/ZO-3 were transformed into Escherichia coli DH10Bac cells containing the Baculovirus genome. DNA isolated from the transformed DH10Bac bacteria was transfected into Sf9 insect cells using Lipofectin. Transfected Sf9 cells were harvested at 48 h (ZO-2) or 60 h (ZO-3) after infection. Harvested cells were centrifuged at 500 g for 5 min and cell pellets were kept at −70°C until needed. The parental vector pFastBac HTc alone (pHTc), coding for the 6-histidine tag plus 36 nonspecific aa, served as a negative control.
Full-length ZO-3 cDNA was also subcloned into prokaryotic expression vector pGEX-3X to generate GST/ZO-3 fusion protein. A cDNA encoding the cytoplasmic COOH-terminal tail (aa 358–505) of chicken occludin (14) in pGEX-2T was a gift from Alan Fanning and James Anderson (Yale University, New Haven, CT). GST fusion proteins were prepared according to manufacturer instruction (Pharmacia Biotech, Inc.). In brief, 50 ml of medium was inoculated with 5 ml of an overnight culture in the presence of 100 μg/ml ampicillin. The culture was incubated at 37°C for 1.0 h with shaking at 250 rpm followed by the addition of IPTG to a final concentration of 0.1 mM, and the incubation was continued for 3.0 h. Cells were harvested by centrifugation at 5,000 g for 10 min and resuspended in 2 ml of PBS containing 1.0 mM DTT, 1.0 mg/ml BSA, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 μg/ml chymostatin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin. Cells were sonicated briefly and then mixed with 10% Triton X-100 to a final concentration 1.0%. The cell lysate was clarified at 10,000 g for 30 min at 4°C to remove insoluble material. The supernatant was transferred to a new tube, mixed with 0.1 ml of a 1:1 slurry of glutathione/agarose (prewashed with PBS), and incubated for 30 min at room temperature. The beads were washed 4× 1.0 ml in cold PBS. Vector encoding GST alone served as negative control.
Recombinant ZO-1 was prepared by in vitro transcription/translation according to manufacturer's instructions (Promega Corp.) from a full-length human ZO-1 cDNA subcloned into pBluescript SK+ under control of T7 promoter (obtained from Drs. Anderson and Fanning). In brief, 25 μl TNT rabbit reticulocyte lysate, 2 μl TNT reaction buffer, 1 μl TNT T7 RNA polymerase, 1 μl amino acid mixture minus methionine (1 mM), 40 U RNasin, 1 μg template DNA, and 4 μl [35S]methionine (1,000 Ci/mMol, 10 mCi/ml; ICN Pharmaceuticals Inc., Irvine, CA) were used for each reaction in a total volume of 50 μl. The reaction mixtures were incubated at 30°C for 90 min and the protein product used immediately in binding assays.
Indirect Binding.
Binding of recombinant ZO-3 to ZO-1 and ZO-2 from whole MDCK cell extracts was assayed according to modifications of the techniques of Furuse et al. (1994). A cell pellet from a 50-ml culture of pHTc/ZO-3/Sf9 or pHTc/Sf9 cells was thawed on ice and resuspended in 2 ml lysis buffer containing 50 mM NaH2PO4, 300 mM NaCl, 10 mM MgCl2, 10 mM imidazole, pH 8.0, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 μg/ml chymostatin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin. Lysozyme was added to a final concentration of 1 mg/ml. The cell suspension was incubated on ice for 30 min, at which time DNase was added to 5 μg/ml and the suspension incubated on ice for another 15 min. After sonication, NP-40 was added to the cell lysate to a final concentration of 1%, and the mixture rotated at 4°C for 30 min to solubilize the over-expressed ZO-3 protein. The suspension was centrifuged at 10,000 g for 20 min and the pellet discarded. For each binding reaction, 50 μl of Probond beads (Invitrogen Corp.) were washed with 3 ml lysis buffer. 500 μl of pHTc/ZO-3/Sf9 or pHTc/Sf9 supernatant was added to the beads and incubated at 4°C for 30 min with gentle shaking. Beads were then washed with 3 ml wash buffer (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole, pH 8.0, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 μg/ml chymostatin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin) and used for binding assays.
Confluent MDCK cells in 2 × 15-cm dishes were scraped, pelleted and homogenized in 3 ml solution K (140 mM KCl, 10 mM Hepes, pH 7.5, 1 mM MgCl2, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 μg/ml chymostatin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin) using a Dounce homogenizer. The suspension was centrifuged at 100,000 g for 60 min at 4°C. The resulting pellet was resuspended in 1 ml high salt solution (1 M KCl, 10 mM Hepes, pH 7.5, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 μg/ml chymostatin, 1 μg/ml leupeptin and 1 μg/ml pepstatin) and incubated on ice for 60 min followed by a 100,000 g centrifugation for 60 min. Supernatant containing the cell extract was diluted with 4.65 ml of 10 mM Hepes, pH 7.5, 30 mM imidazole, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 μg/ml chymostatin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin. The protein aggregates formed during dilution into lower salt were removed by a 10,000 g, 10 min, 4°C centrifugation. 1 ml each of the resultant supernatant was made 1% in Triton X-100 for binding of ZO-1 or 1% in Brij 58 for binding of ZO-2. The 1-ml sample was then added to 50 μl of ZO-3– or negative control peptide-bound beads and incubated at 4°C overnight with gentle rotation. Beads were washed 3× with wash buffer with 0.2% Triton X-100 and 0.2% Tween 20. Beads were then resuspended in 45 μl of wash buffer plus 6 μl of 10× gel sample buffer and equivalent 5-μl samples run on SDS-PAGE and immunoblotted for ZO-1 and ZO-2 as described above.
Direct Binding.
In vitro transcribed/translated, [35S]methionine-labeled human ZO-1 was added to Baculovirus-expressed ZO-3 or negative control peptide bound to Probond resin in binding buffer (140 mM KCl, 25 mM imidazole, pH 8.0, 1.5% Brij 58, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 μg/ml chymostatin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin). Binding was allowed to proceed overnight at 4°C with rotation. Beads were washed 4× 1 ml with wash buffer, resuspended in an equal volume of 2× gel sample buffer and run on SDS-PAGE. The identity of ZO-1 bound to the ZO-3 beads was confirmed by immunoblot using the anti-human ZO-1 polyclonal antibody. Similar results were obtained with bacteria-expressed ZO-3 (data not shown). To assay ZO-3 binding to occludin, Baculovirus-expressed ZO-3 eluted from Probond resin with 300 mM imidazole was added to GST/occludin or GST alone bound to glutathione agarose in the presence of solution K. After overnight incubation at 4°C the glutathione agarose was washed with 4× 1 ml solution K. Bound protein was eluted with 25 mM glutathione, resolved on SDS-PAGE, transferred to nitrocellulose and immunoblotted with anti–ZO-3 antisera. Binding of ZO-3 to ZO-2 was assessed by adding Baculovirus-expressed ZO-2 eluted from Probond resin with 300 mM imidazole to GST/ZO-3 or GST alone bound to glutathione agarose in solution K. Beads were incubated 2 h at 4°C and washed with 4× 1 ml solution K. ZO-2 starting material, bound protein eluted with 25 mM glutathione and the unbound fractions were resolved on SDS-PAGE, transferred to nitrocellulose and immunoblotted with anti–ZO-2 antisera. The presence of ZO-3 on the beads was confirmed by stripping the blot and reprobing with anti–ZO-3 antisera.
Results
ZO-3 Is a Member of the MAGUK Family
Previous investigations showed that a protein of 130 kD specifically coimmunoprecipitates with ZO-1 and ZO-2 under conditions designed to preserve protein–protein interactions (5, 21). We repeated these observations and noted that the 130-kD protein is present in significantly smaller quantities than either ZO-1 or ZO-2 (Fig. 1). However, isolation by bulk immunoprecipitation and purification by gel electrophoresis provided enough of the 130-kD polypeptide for endopeptidase digestion (12) and aa sequencing. Degenerate primers based on the 6 aa at each end of a unique 19-aa fragment were synthesized, and RT-PCR from MDCK cell RNA generated a 57-bp oligonucleotide that confirmed the entire original 19 aa. This 57-mer was then used to screen an MDCK cDNA library. The first clone obtained, A1, was subsequently used to screen the same library. To obtain the 5′ end of the cDNA, an oriented MDCK cDNA library specific for the protein was created.
Figure 1.
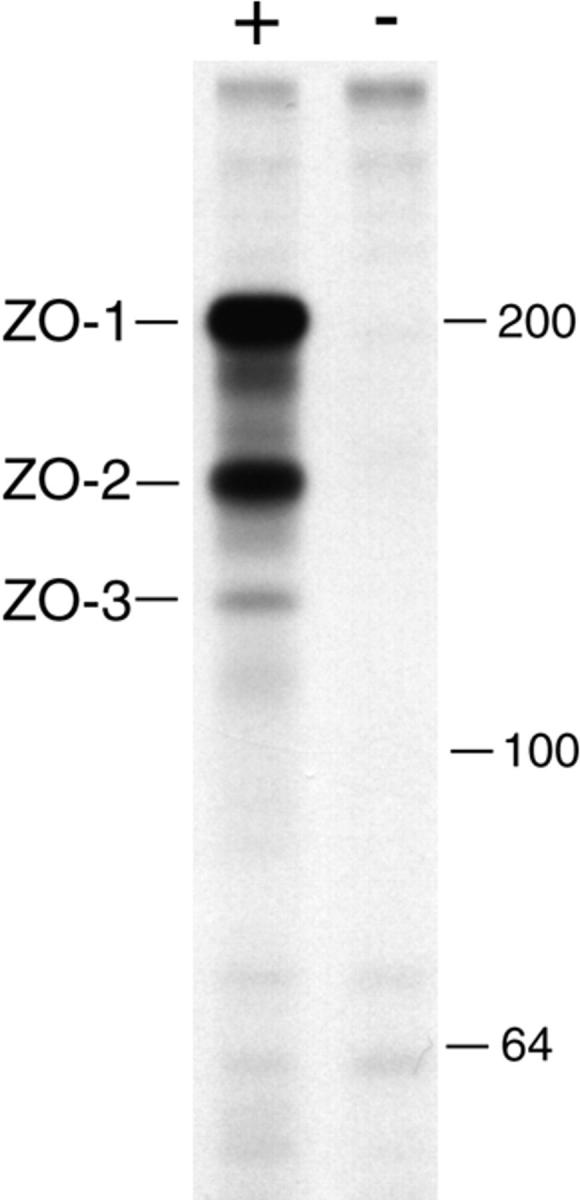
Immunoprecipitation from metabolically labeled MDCK cell extracts with an anti–ZO-1 mAb (+) or a nonspecific but identical subclass mAb (−) under conditions that preserve protein–protein interactions shows that both ZO-2 (160 kD) and ZO-3 (130 kD) specifically coprecipitate with ZO-1 (215 kD). Molecular mass markers at right (kD).
In all, five overlapping clones were obtained (Fig. 2). Sequencing shows that these cDNAs contain a single open reading frame of 2,694 bp coding for 898 aa (Fig. 3). The overall size of the cDNAs obtained (∼3.1 kb) corresponds well with the ∼3.4 kb mRNA for the protein detected by Northern blot (Fig. 4). The predicted molecular mass for this aa sequence is 98,414 D, indicating that the protein runs anomalously on SDS-PAGE, as previously observed for other proteins including ZO-1 and ZO-2 (21, 44), or is posttranslationally modified. The 130-kD protein was previously shown to be phosphorylated (5). Sequence analysis demonstrates that the predicted aa sequence contains 3 PDZ domains, an SH3 domain and a GUK region (Fig. 3), indicating homology to ZO-1, ZO-2, the gene products from Drosophila discs large (dlg-A) and tamou (TamA), and other MAGUK family members. Amino acid sequence comparisons of the MAGUK domains from ZO-1, ZO-2, ZO-3, TamA, and dlg-A illustrate that ZO-3 has a high degree of similarity to ZO-1 and ZO-2 in all three domains, as well as in overall sequence (Table II, Fig. 5). Furthermore, ZO-3 shows approximately the same level of similarity to dlg-A as ZO-1. However, ZO-3 and especially ZO-1 show higher degrees of similarity to TamA than to dlg-A, suggesting that TamA is the closer Drosophila family member. We conclude that ZO-3 is a novel member of the MAGUK family of proteins.
Table II.
Amino Acid Comparison of MAGUK Domains of ZO-1, ZO-2, ZO-3, dlg-A, and TamA
| ZO-3 | ZO-1 | |||||
|---|---|---|---|---|---|---|
| PDZ1 | ZO-1 | 57(72) | — | |||
| ZO-2 | 61(76) | 71(82) | ||||
| TamA | 46(66) | 63(78) | ||||
| dlg-A | 35(60) | 44(68) | ||||
| PDZ2 | ZO-1 | 50(72) | — | |||
| ZO-2 | 55(74) | 65(86) | ||||
| TamA | 31(57) | 43(71) | ||||
| dlg-A | 22(30) | 44(78) | ||||
| PDZ3 | ZO-1 | 63(81) | — | |||
| ZO-2 | 59(75) | 70(86) | ||||
| TamA | 48(68) | 48(69) | ||||
| dlg-A | 42(66) | 42(69) | ||||
| SH3 | ZO-1 | 58(76) | — | |||
| ZO-2 | 59(80) | 69(85) | ||||
| TamA | 41(62) | 53(76) | ||||
| dlg-A | 34(58) | 29(58) | ||||
| GUK | ZO-1 | 52(69) | — | |||
| ZO-2 | 52(71) | 67(82) | ||||
| TamA | 40(66) | 43(68) | ||||
| dlg-A | 34(53) | 31(54) | ||||
| Overall | ZO-1 | 47(64) | — | |||
| ZO-2 | 47(65) | 51(67) | ||||
| TamA | 34(57) | 31(52) | ||||
| dlg-A | 27(48) | 26(47) |
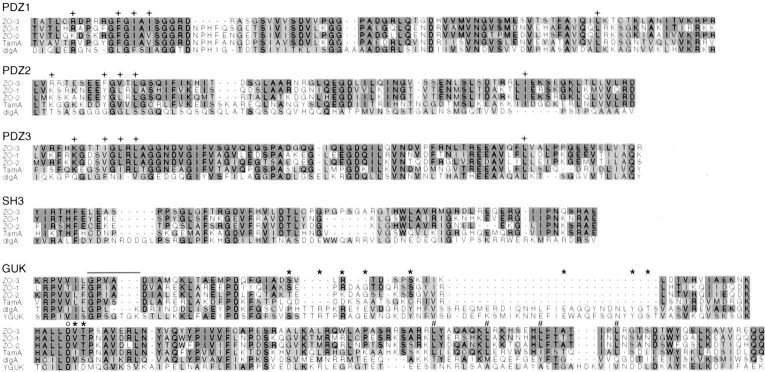
Figure 5.
Multiple sequence alignments of the MAGUK domains of canine ZO-3, human ZO-1 (44), canine ZO-2 (7), Drosophila TamA (41), and dlg-A (46), constructed with GCG PILEUP. The GUK domain is also compared with the yeast guanylate kinase sequence (8). Dark shaded regions indicate identity to the ZO-3 sequence, lighter shades are conserved aa substitutions. Amino acids in the PDZ domains believed to be important in protein–protein interactions (32) are marked (+), as are the aa involved in ATP (bar), GMP (*) and Mg2+ (o) binding in the GUK domain (26). # indicates the aa of the putative leucine zipper motif.
The aa believed to have a role in PDZ domain-mediated protein–protein interactions (32) are illustrated in Fig. 5. These aa are well conserved among ZO-1, ZO-2, ZO-3, TamA, and dlg-A, indicating a common functional importance. However, 8 of the 18 aa involved in ATP, GMP, and Mg2+ binding in yeast guanylate kinase (8, 26) are missing in ZO-1, ZO-2, ZO-3, and TamA (Fig. 5), making it unlikely that any of these proteins display GUK enzymatic activity. ZO-3, like ZO-1 (44) and ZO-2 (7, 21), contains leucine residues in a putative leucine zipper motif within the GUK domain (Fig. 5).
In addition to the MAGUK domains, ZO-3 shares with ZO-1 and ZO-2 the presence of a basic domain between PDZ1 and PDZ2 that is rich in arginine residues (ZO-3 pI = 11.92, 17% arg) and an acidic domain at the COOH-terminal end of the GUK domain (ZO-3 pI = 3.75). However, several features distinguish ZO-3 from ZO-1 and ZO-2. Both ZO-1 and ZO-2 contain proline-rich regions at the COOH-terminal end of the molecules (ZO-1 = 14% proline, ZO-2 = 13%; references 21, 44). Whereas ZO-3 is only 5% proline in this region, the link between PDZ2 and PDZ3 of ZO-3 is unique in that it contains 14% proline residues. Moreover, although the 3′ ends of both ZO-1 and ZO-2 contain alternatively spliced regions (7, 44), no such splicing could be detected by RT-PCR analysis of this region of ZO-3 using MDCK cell mRNA and primers that span from inside the acidic domain to near the COOH terminus (primers: forward, nucleotides 2633–2652; reverse, 2861–2841; Fig. 3; data not shown). Finally, aa sequence comparisons of all regions of ZO-1, ZO-2, and ZO-3 indicates the levels of similarity are generally highest in the MAGUK and acid domains, whereas most linker regions are less conserved (Table III). Such analysis also demonstrates that ZO-1 and ZO-2 are more closely related to each other in almost all regions than either is to ZO-3.
Table III.
Amino Acid Comparisons of ZO-1, ZO-2, and ZO-3
| ZO-3/ZO-1 | ZO-3/ZO-2 | ZO-1/ZO-2 | ||||
|---|---|---|---|---|---|---|
| Start | 80(80) | 89(89) | 56(56) | |||
| PDZ1 | 57(72) | 61(76) | 71(82) | |||
| Link | 25(54) | 27(53) | 29(53) | |||
| PDZ2 | 50(72) | 55(74) | 65(86) | |||
| Link | 34(46) | 29(48) | 39(55) | |||
| PDZ3 | 63(81) | 59(75) | 70(86) | |||
| Link | 53(82) | 35(65) | 53(76) | |||
| SH3 | 58(76) | 59(80) | 69(85) | |||
| Link | 55(74) | 53(76) | 72(84) | |||
| GUK | 52(69) | 52(71) | 67(82) | |||
| Link | 26(42) | 16(47) | 47(58) | |||
| Acid | 43(60) | 51(61) | 60(71) | |||
| End | 17(41) | 24(41) | 25(43) |
ZO-3 Localizes to the Tight Junction
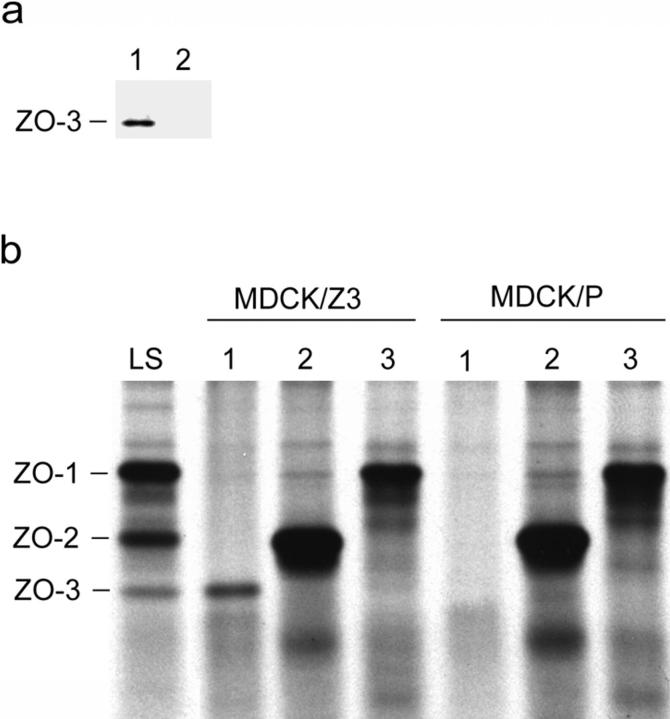
Generation of antibodies against ZO-3 has been compromised by the homology between ZO-3, ZO-1, and ZO-2. Injection of fusion proteins corresponding to even the low similarity linker domains has produced antibodies that cross-react with ZO-1 and/or ZO-2 (see Fig. 9 c). To circumvent these difficulties, a cDNA coding for full-length ZO-3 with a VSV-G cytoplasmic domain epitope tag at the COOH terminus was constructed. This construct was then transfected into MDCK cells to generate the stable MDCK/Z3 cell line. Immunoblot of a whole cell lysate from MDCK/Z3 cells with an anti–VSV-G antibody demonstrates the expression of an exogenous protein of ∼130 kD (Fig. 6 a, lane 1). No such band is detected in the parental, untransfected cell line (Fig. 6 a, lane 2) or in a cell line stably transfected with vector alone (data not shown). High stringency anti–VSV-G antibody immunoprecipitation from the MDCK/Z3 cell line likewise brings down a polypeptide that runs at approximately the same position as the original 130-kD band present in low-stringency ZO-1 immunoprecipitates (Fig. 6 b, compare MDCK/Z3 lane 1 with lane LS). The slightly higher molecular mass of the exogenous protein in the MDCK/Z3 cells can be accounted for by the additional 1,338 D of the epitope tag. No similar band is immunoprecipitated from the parental cell line with anti–VSV-G (Fig. 6 b, MDCK/P lane 1).
Figure 6.
An exogenous protein of ∼130 kD is expressed by the MDCK/Z3 cell line. (a) Immunoblot of whole cell lysates from the MDCK/Z3 line (lane 1) or parental MDCK cells (lane 2) probed with anti–VSV-G to detect the epitope-tagged ZO-3 construct. A reactive band, running at ∼130 kD, appears only in the MDCK/Z3 cell line. (b) Low-stringency immunoprecipitation of ZO-1 from metabolically labeled MDCK cells (LS) shows coprecipitation of ZO-2 and ZO-3 (see also Fig. 1). High-stringency immunoprecipitation from metabolically labeled MDCK/Z3 or parental (MDCK/P) cells using anti–VSV-G (lanes 1), anti–ZO-2 (lanes 2), or anti–ZO-1 (lanes 3) demonstrates the presence of a band only in the MDCK/Z3 cells that runs a fraction higher than the endogenous 130-kD band. This slight difference in molecular mass can be accounted for by the additional 1,338 D of the epitope tag.
To determine where ZO-3 is localized in epithelial cells, we immunofluorescently costained MDCK/Z3 monolayers for the epitope-tagged ZO-3 and endogenous ZO-1, a widely used marker of the tight junction in this cell type (6, 38, 40). Fig. 7 shows that ZO-3 is localized to sites of cell–cell interaction, identical to the distribution of ZO-1. Additional staining of the cytoplasm is visible in the ZO-3–stained cells (Fig. 7 a). It is not known if this is due to exogenous overexpression or a novel distribution of the protein.
Figure 7.
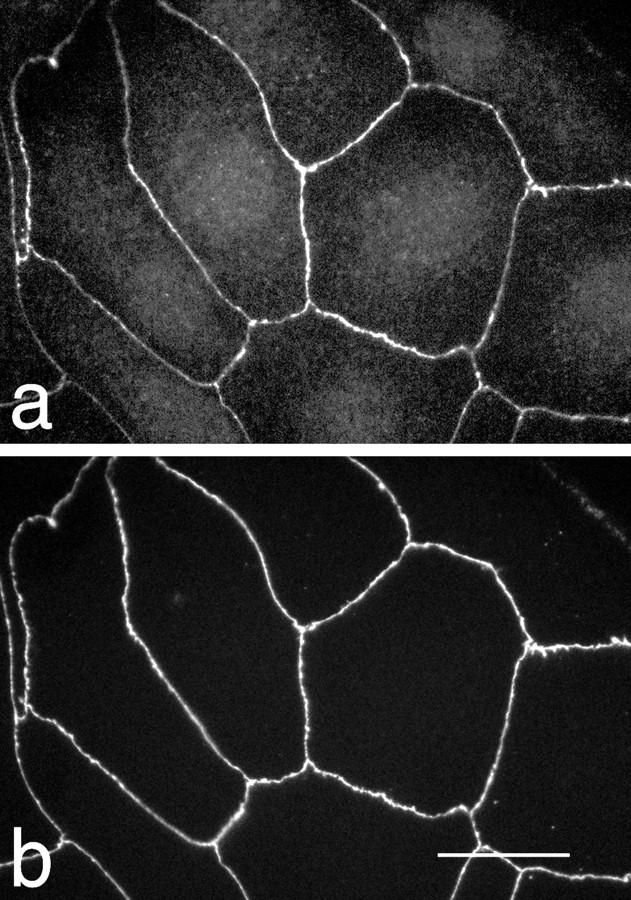
Immunofluorescent costaining of MDCK/Z3 cells for ZO-3 (a) and ZO-1 (b). ZO-3 and ZO-1 are identically distributed at cell borders. Additional staining of the cell cytoplasm is visible for ZO-3. Bar, 5 μm.
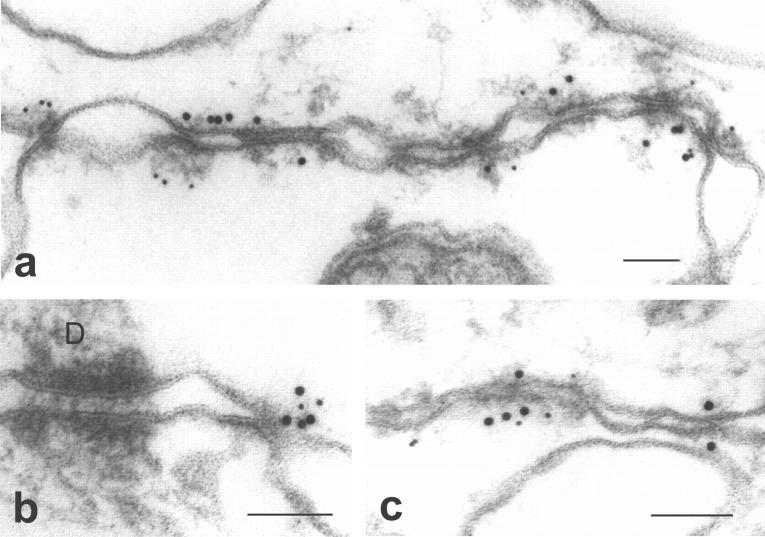
ImmunoEM of an isolated MDCK/Z3 cell membrane preparation was used to determine the ultrastructural colocalization of ZO-3 and ZO-1. It was previously demonstrated that ZO-1 is found exclusively at MDCK cell tight junctions by immunoEM (38, 40). As shown in Fig. 8, both ZO-1 and the ZO-3 are restricted to tight junctions in these membranes, whereas all other membranes, including other intercellular junctions, are free of label.
Figure 8.
ImmunoEM colocalization of ZO-1 (10 nm gold) and ZO-3 (5 nm gold) on membranes isolated from MDCK/Z3 cells. (a–c) Three different images of labeling are displayed, showing that ZO-3 and ZO-1 are colocalized at sites of tight junction membrane contact, while all other membranes, including a desmosome (D) in b, are free of label. Bars, 100 nm.
ZO-3 Directly Binds ZO-1 and Occludin, but Not ZO-2
We tested whether the ZO-3 cDNA codes for a protein capable of interacting with other proteins from the tight junction in in vitro binding analyses. First, either full-length recombinant ZO-3 or a negative control peptide was bound to affinity beads. Resin was then incubated with a high salt extract of MDCK cell membranes, washed, and the bound proteins subjected to SDS-PAGE and immunoblotting with either anti–ZO-1 or ZO-2 antibodies. As shown in Fig. 9 a, ZO-3, but not the control peptide, binds both ZO-1 and ZO-2 from MDCK cell extracts. Although this result indicates the presence of a protein complex that includes ZO-1, ZO-2, and ZO-3, it does not test for direct interactions among the proteins. Therefore, we tested for direct binding using recombinant ZO-1, ZO-2, and ZO-3. We also assayed for direct interaction between ZO-3 and the COOH-terminal 148 aa of occludin, the domain of this protein believed to be oriented in the cytoplasm (14, 15). As shown in Fig. 9 b, recombinant ZO-3 specifically binds in vitro transcribed/translated ZO-1. ZO-3 also binds directly to the cytoplasmic tail of occludin (Fig. 9 d). However, no binding was detected between recombinant ZO-3 and ZO-2 with our assay conditions (Fig. 9 e).
Discussion
Characterization of the tight junction is proceeding rapidly and has stimulated advances in the fields of cell–cell interactions and epithelial cell biology. In addition to the list of proteins now found at the tight junction, we have learned that two previously characterized tight junction components, ZO-1 and ZO-2, are members of a larger protein family that appear to function in the organization of specific areas of the cell surface (11, 21, 23, 24, 27, 41, 44). Moreover, data suggest that some members of this family are involved in signal transduction and/or tumor suppression (1, 41, 46, 47), highlighting the importance of analyzing these molecules.
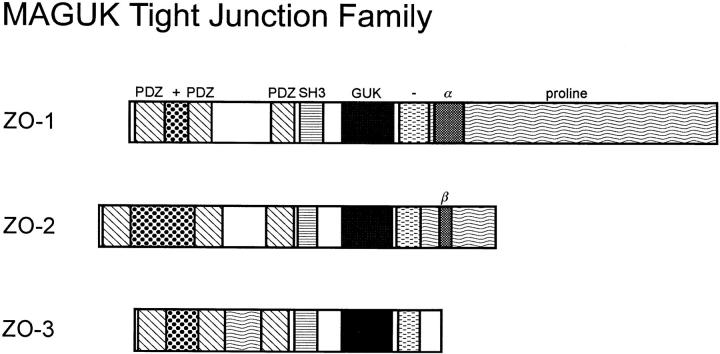
Here we present evidence of a novel member of the MAGUK family of proteins found at the tight junction. This 130-kD polypeptide, named ZO-3 because of homology to ZO-1 and ZO-2 (Figs. 5, Tables II and III) and localization at the tight junction (Figs. 7 and 8), contains 3 PDZ domains, an SH3 domain and a GUK region (Fig. 3). The arrangement of these domains along the length of the molecule is identical to that of ZO-1 and ZO-2, as well as to the Drosophila proteins dlg-A and TamA. In common with ZO-1 and ZO-2, but distinct from other MAGUK members, ZO-3 contains an acidic domain at the COOH-terminal end of the molecule. All three proteins (previously unnoted in ZO-1) also contain a basic region between PDZ1 and PDZ2. Although all three tight junction molecules contain proline-rich regions, those in ZO-1 and ZO-2 are located at the COOH-terminal ends of the molecules whereas that in ZO-3 is found between PDZ2 and PDZ3. Finally, using primers from the 3′ end of the ZO-3 cDNA, no evidence was obtained for an alternative splice site in this region (data not shown). Alternative splice regions in the COOH-terminal regions of ZO-1 and ZO-2 have been reported (7, 44).
ZO-3 was originally identified on the basis of coimmunoprecipitation with ZO-1 under specific conditions where some protein–protein interactions are preserved (reference 5; Fig. 1). We isolated and purified enough of this protein from MDCK cells for aa sequencing and subsequent cloning. This is similar to the approach used to identify ZO-2 (21), although our task was somewhat more difficult due to the smaller quantities of the 130-kD band that coprecipitate under these conditions (Fig. 1). It is unclear whether this reflects a smaller amount of ZO-3 present in cells or a reduced efficiency of coprecipitation. Despite the obvious identification of ZO-3 as another MAGUK element by sequence analysis, the continued characterization of the protein was complicated by our inability to generate anti–ZO-3 antisera that did not cross-react to ZO-1 and/or ZO-2 (Fig. 9 c). This problem extended through the injection of multiple fusion proteins corresponding to regions with lowest similarity to ZO-1 or ZO-2 into various antibody generating species. While our efforts to this end are ongoing, we constructed a full-length version of ZO-3 that contains an epitope tag at the COOH terminus. This version allowed us to determine that MDCK cells stably transfected with the construct express the exogenous protein at the predicted molecular mass by both immunoblot and immunoprecipitation (Fig. 6). Furthermore, this approach permitted us to localize ZO-3 using immunohistochemical techniques with an antiepitope antibody. Both immunofluorescence (Fig. 7) and immunoEM (Fig. 8) show that ZO-3 colocalizes with ZO-1 at the tight junction.
To provide additional evidence that ZO-3 was a junctional protein, and to clarify binding interactions at the tight junction, we explored interactions between recombinant ZO-3 and tight junction proteins using in vitro affinity binding analyses (Fig. 9). Our results show that ZO-3 binds both ZO-1 and ZO-2 from MDCK cell extracts (Fig. 9 a), as predicted from the original coimmunoprecipitation observations. This indicates that our ZO-3 construct binds these proteins when they are present at low concentrations in a diverse protein mixture. However, these results do not address whether ZO-3 binds both proteins directly or through intermediary components. To that end, using purified recombinant proteins we determined that ZO-3 binds directly to ZO-1 but not to ZO-2. This indicates that the presence of ZO-2 in the coprecipitating ZO-1/ZO-2/ ZO-3 complex from MDCK cell extracts is due to a direct interaction between ZO-2 and ZO-1 or another unidentified intermediary protein. However, the absence of binding between ZO-3 and ZO-2 should be interpreted with caution. It is possible that missfolding occurring during recombinant protein expression or the binding conditions themselves prevents detection of interaction. We also demonstrated that ZO-3 binds directly to the cytoplasmic 148-aa tail of occludin. Our results, combined with previous studies, indicate that occludin interacts directly with both ZO-1 (15) and ZO-3 (Fig. 9 d). ZO-2 is likely bound to this complex via ZO-1 (5, 16, 21; Fig. 1). ZO-1 also directly interacts with AF-6 (47). It has not been determined if ZO-2 binds directly to occludin or if AF-6 interacts with any other junctional component.
Limited information is available on the function of any of the tight junction proteins. Data clearly indicate that occludin has a role in the paracellular permeability barrier (6, 31, 45), as expected from a transmembrane element localized to the fibrils visible in freeze-fractured tight junctions (13). Evidence also suggests that additional transmembrane components may be present at the tight junction (6). The function of the cluster of peripheral membrane proteins found at the tight junction is less evident. The presence of actin filaments at the tight junction (29) indicates that some of these proteins may be involved in linking transmembrane elements to the cytoskeleton, and evidence suggesting that ZO-1 may function in this regard has recently been published (19). However, it is the MAGUK protein family members found at the tight junction, now totaling three with the data presented in this paper, that are the focus of much current investigation.
The founding member of the MAGUK family is the lethal(1)discs-large-1 (dlg) tumor suppressor gene product (dlg-A) of Drosophila, and information has been derived from genetic analysis of this gene. Mutations in the GUK domain of Drosophila dlg-A result in loss of normal epithelial cell polarity and neoplastic overgrowth in the larval imaginal disc, suggesting that this domain is important for normal protein function and that the protein plays a role in tumor suppression (46). Mutations in the PDZ, SH3, or GUK domains of dlg also disrupt normal synaptic structure at the neuromuscular junction (28), consistent with evidence that the PDZ domains bind synaptic ion channels (23, 27). Binding interactions have also been defined for the SH3 (4) and GUK domains (24), as well as other regions (19) of MAGUK proteins. Taken as a whole, the presence of multiple domains capable of specific protein– protein interactions suggests that MAGUK proteins at the tight junction act as connector molecules on the cytoplasmic surface of the plasma membrane.
In addition to the MAGUK domains, ZO-1, ZO-2, and ZO-3 have other common and distinguishing features (Fig. 10). The presence of three homologous proteins at the tight junction raises several questions. Do these proteins have unique functions or is their homology indicative of functional redundancy related to an overall critical role? If there are unique aspects to the function of these proteins, it is likely to fall within the regions linking the MAGUK domains or the COOH-terminal tails where aa similarity between them is the lowest (Table III). Will these molecules, if structurally altered or ablated, result in the neoplastic transformation observed for dlg-A? Genetic analysis in mammalian systems, although more difficult than in Drosophila, will clearly be instructive. What protein domains are responsible for the specific binding interactions among tight junction proteins? Investigations in this regard are ongoing in several laboratories, with obvious focus on the PDZ domains of the tight junction MAGUK subfamily. Finally, what roles might these proteins play in normal tight junction gate and fence physiology? There is now evidence linking paracellular permeability to tyrosine phosphorylation of ZO-1 (36), but otherwise there is no direct information on the function of ZO-1, ZO-2, or ZO-3. Although the molecular composition of the tight junction is now becoming understood, the understanding of tight junction molecular biology is still in its infancy.
Figure 10.
Schematic diagram showing the domain arrangement of the three MAGUK family members found at the tight junction. PDZ, diagonal stripes; SH3, horizontal stripes; GUK, black; basic domain, dots (+); acidic domain, horizontal dashes (−); proline-rich, wavy horizontal lines, alternative splices, α (ZO-1) and β (ZO-2).
Acknowledgments
We thank James Anderson, Honey Chan, Alan Fanning, Joe Fernandez, Dan Goodenough, Tom Hobman, Lynne Jesaitis, Brigitte Keon, Carolyn Machamer, Sheena Miche, David Paul, Jim Stone, Mandeep Tamber, Tom Turner, Dwayne Weber, Rick Wozniak, and Marino Zerial for their generous advice and technical support.
Abbreviations used in this paper
- aa
amino acid
- GUK
guanylate kinase
- PVDF
polyvinylidene difluoride
- SH3
src homology
- VSV-G
vesicular stomatitis virus G protein
Footnotes
The Rockefeller University Protein Sequencing Facility is supported in part by National Institutes of Health shared instrumentation grants and by funds provided by the US Army and Navy for purchase of equipment. This work was supported by operating grants to B.R. Stevenson from the Medical Research Council of Canada and the Kidney Foundation of Canada. B.R. Stevenson is a Senior Scholar of the Alberta Heritage Foundation for Medical Research.
References
- 1.Anderson JM. Cell signaling: MAGUK magic. Curr Biol. 1996;6:382–384. doi: 10.1016/s0960-9822(02)00501-8. [DOI] [PubMed] [Google Scholar]
- 2.Anderson JM, Stevenson BR, Jesaitis LA, Goodenough DA, Mooseker MS. Characterization of ZO-1, a protein component of the tight junction from mouse liver and Madin-Darby canine kidney cells. J Cell Biol. 1988;106:1141–1149. doi: 10.1083/jcb.106.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1992. In Current Protocols in Molecular Biology. John Wiley & Sons, Inc., NY.
- 4.Balda MS, Anderson JM, Matter K. The SH3 domain of the tight junction protein ZO-1 binds to a serine protein kinase that phosphorylates a region C-terminal to this domain. FEBS Lett. 1996;399:326–332. doi: 10.1016/s0014-5793(96)01352-x. [DOI] [PubMed] [Google Scholar]
- 5.Balda MS, Gonzalez-Mariscal L, Matter K, Cereijido M, Anderson JM. Assembly of the tight junction: the role of diacylglycerol. J Cell Biol. 1993;123:293–302. doi: 10.1083/jcb.123.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balda MS, Whitney JA, Flores C, Gonzalez S, Cereijido M, Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatch M, Jesaitis LA, Gallin WJ, Goodenough DA, Stevenson BR. The tight junction protein ZO-2 contains three PDZ (PSD-95/ Discs-large/ZO-1) domains and an alternatively spliced region. J Biol Chem. 1996;271:25723–25726. doi: 10.1074/jbc.271.42.25723. [DOI] [PubMed] [Google Scholar]
- 8.Berger A, Schilts E, Schulz GE. Guanylate kinase from Saccharomyces cerevisiae. . Eur J Biochem. 1989;184:433–443. doi: 10.1111/j.1432-1033.1989.tb15035.x. [DOI] [PubMed] [Google Scholar]
- 9.Cho K-O, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 10.Citi S, Sabanay H, Jakes R, Geiger B, Kendrick-Jones J. Cingulin, a new peripheral component of tight junctions. Nature. 1988;333:272–276. doi: 10.1038/333272a0. [DOI] [PubMed] [Google Scholar]
- 11.Fanning AS, Anderson JM. Protein-protein interactions:PDZ domain networks. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez, J., M. DeMott, D. Atherton, and S.M. Mische. 1992. Internal protein sequence analysis: enzymatic digestion for less than 10 micrograms of protein bound to polyvinylidene difluoride or nitrocellulose membranes. Anal. Biochem 201:255-264. [DOI] [PubMed]
- 13.Furuse M, Fujimoto K, Sato N, Hirase T, Tsukita Sa, Sh, Tsukita Overexpression of occludin, a tight junction-associated integral membrane protein, induces the formation of intracellular multilamellar bodies bearing tight junction-like structure. J Cell Sci. 1996;109:429–435. doi: 10.1242/jcs.109.2.429. [DOI] [PubMed] [Google Scholar]
- 14.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita Sa, Sh, Tsukita Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita Sa, Sh, Tsukita Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gumbiner B, Lowenkopf T, Apatira D. Identification of a 160-kD polypeptide that binds to the tight junction protein ZO-1. Proc Natl Acad Sci USA. 1991;88:3460–3464. doi: 10.1073/pnas.88.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirokawa N, Tilney LG. Interactions between actin filaments and between actin filaments and membranes in quick-frozen and deeply etched hair cells of the chick ear. J Cell Biol. 1982;95:249–261. doi: 10.1083/jcb.95.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howarth AG, Hughes MR, Stevenson BR. Detection of the tight junction-associated protein ZO-1 in astrocytes and other nonepithelial cell types. Am J Physiol. 1992;262:C461–C469. doi: 10.1152/ajpcell.1992.262.2.C461. [DOI] [PubMed] [Google Scholar]
- 19.Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to a catenin and actin filaments. J Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh M, Yonemura S, Nagafuchi A, Tsukita Sa, Sh, Tsukita A 220-kD undercoat-constitutive protein: its specific localization at cadherin-based cell-cell adhesion sites. J Cell Biol. 1991;115:1449–1462. doi: 10.1083/jcb.115.5.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jesaitis LA, Goodenough DA. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the drosophila discs-large tumor suppressor protein. J Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keon BH, Schafer S, Kuhn C, Grund C, Franke WW. Symplekin, a novel type of tight junction plaque protein. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 24.Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kistner U, Wenzel BM, Veh RW, Cases-Langhoff C, Garner AM, Appeltauer U, Voss B, Gundelfinger EC, Garner CC. SAP90, a rat presynaptic protein related to the product of the Drosophila tumor suppressor gene Dlg-A. J Biol Chem. 1993;268:4580–4583. [PubMed] [Google Scholar]
- 26.Kistner U, Garner CC, Linial M. Nucleotide binding by the synapse associated protein SAP90. FEBS Lett. 1995;359:159–163. doi: 10.1016/0014-5793(95)00030-d. [DOI] [PubMed] [Google Scholar]
- 27.Kornau H-C, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 28.Lahey T, Gorczyca M, Jia XX, Budnik V. The drosophila tumor suppressor gene dlg is required for normal synaptic bouton structure. Neuron. 1994;13:823–835. doi: 10.1016/0896-6273(94)90249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madara JL. Intestinal absorptive cell tight junctions are linked to cytoskeleton. Am J Physiol. 1987;253:C171–C175. doi: 10.1152/ajpcell.1987.253.1.C171. [DOI] [PubMed] [Google Scholar]
- 30.Madara JL, Barenberg D, Carlson S. Effects of cytochalasin D on occluding junctions of intestinal absorptive cells: further evidence that the cytoskeleton may influence paracellular permeability and junctional charge selectivity. J Cell Biol. 1986;102:2125–2136. doi: 10.1083/jcb.102.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy KM, Skare IB, Stankewich MC, Furuse M, Tsukita S, Rogers RA, Lynch RD, Schneeberger EA. Occludin is a functional component of the tight junction. J Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- 32.Morais Cabral, J.J., C. Petosa, M.J. Sutcliffe, S. Raza, O. Byron, F. Poy, S.M. Marfatia, A.H. Chishti, and R.C. Liddington. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- 33.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 34.Pasdar M, Nelson WJ. Regulation of desmosome assembly in epithelial cells: kinetics of synthesis, transport, and stabilization of desmoglein I, a major protein of the membrane core domain. J Cell Biol. 1989;109:163–178. doi: 10.1083/jcb.109.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruff P, Speicher DW, Husain-Chishti A. Molecular identification of a major palmitoylated erythrocyte membrane protein containing the src homology 3 motif. Proc Natl Acad Sci USA. 1991;88:6595–6599. doi: 10.1073/pnas.88.15.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staddon JM, Herrenknecht K, Smales C, Rubin LL. Evidence that tyrosine phosphorylation may increase tight junction permeability. J Cell Sci. 1995;108:609–619. doi: 10.1242/jcs.108.2.609. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson BR, Anderson JM, Goodenough DA, Mooseker MS. Tight junction structure and ZO-1 content are identical in two strains of Madin-Darby canine kidney cells which differ in transepithelial resistance. J Cell Biol. 1988;107:2401–2408. doi: 10.1083/jcb.107.6.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson BR, Heintzelman MB, Anderson JM, Citi S, Mooseker MS. ZO-1 and cingulin: tight junction proteins with distinct identities and localizations. Am J Physiol. 1989;257:C621–C628. doi: 10.1152/ajpcell.1989.257.4.C621. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson BR, Richards CL, Howarth AG, Maraj VA, Hibbard JG. Quantitative immunoblot detection of rare proteins in whole cell extracts using biotin-strepavidin reagents. J Exp Zool. 1994;268:224–228. doi: 10.1002/jez.1402680307. [DOI] [PubMed] [Google Scholar]
- 40.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahisa M, Togashi S, Suzuki T, Kobayashi M, Murayama A, Kondo K, Miyake T, Ueda R. The Drosophilatamou gene, a component of the activating pathway of extramacrochaetae expression, encodes a protein homologous to mammalian cell-cell junction-associated protein ZO-1. Genes Dev. 1996;10:1783–1795. doi: 10.1101/gad.10.14.1783. [DOI] [PubMed] [Google Scholar]
- 42.Van Itallie CM, Anderson JM. Occludin confers adhesiveness when expressed in fibroblasts. J Cell Sci. 1997;110:1113–1121. doi: 10.1242/jcs.110.9.1113. [DOI] [PubMed] [Google Scholar]
- 43.Weber E, Berta G, Tousson A, St. John P, Green MW, Gopalokrishnan U, Jilling T, Sorscher EJ, Elton TS, Abrahamson DR, Kirk KL. Expression and polarized targeting of a rab3 isoform in epithelial cells. J Cell Biol. 1994;125:583–594. doi: 10.1083/jcb.125.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willott E, Balda MS, Fanning AS, Jameson B, Van Itallie C, Anderson JM. The tight junction protein ZO-1 is homologous to the drosophila discs-large tumor suppressor protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong V, Gumbiner BM. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J Cell Biol. 1997;136:399–409. doi: 10.1083/jcb.136.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woods DF, Bryant PJ. The discs-large tumor suppressor gene of drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto T, Harada N, Kano K, Taya S-I, Canaani E, Matsuura Y, Mizoguchi A, Ide C, Kaibuchi K. The ras target AF-6 interacts with ZO-1 and serves as a peripheral component of tight junctions in epithelial cells. J Cell Biol. 1997;139:785–795. doi: 10.1083/jcb.139.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong Y, Saitoh T, Minase T, Sawada N, Enomoto Y, Mori M. Monoclonal antibody 7H6 reacts with a novel tight junction-associated protein distinct from ZO-1, cingulin, and ZO-2. J Cell Biol. 1993;120:477–483. doi: 10.1083/jcb.120.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]