Abstract
Recent studies with patients suffering from epidermolysis bullosa simplex associated with muscular dystrophy and the targeted gene disruption in mice suggested that plectin, a versatile cytoskeletal linker and intermediate filament-binding protein, may play an essential role in hemidesmosome integrity and stabilization. To define plectin's interactions with hemidesmosomal proteins on the molecular level, we studied its interaction with the uniquely long cytoplasmic tail domain of the β4 subunit of the basement membrane laminin receptor integrin α6β4 that has been implicated in connecting the transmembrane integrin complex with hemidesmosome-anchored cytokeratin filaments. In vitro binding and in vivo cotransfection assays, using recombinant mutant forms of both proteins, revealed their direct interaction via multiple molecular domains. Furthermore, we show in vitro self-interaction of integrin β4 cytoplasmic domains, as well as disruption of intermediate filament network arrays and dislocation of hemidesmosome-associated endogenous plectin upon ectopic overexpression of this domain in PtK2 and/or 804G cells. The close association of plectin molecules with hemidesmosomal structures and their apparent random orientation was indicated by gold immunoelectron microscopy using domain-specific antibodies. Our data support a model in which plectin stabilizes hemidesmosomes, via directly interlinking integrin β4 subunits and cytokeratin filaments.
Integrins comprise a large family of heterodimeric receptors that mediate the adhesion of cells to extracellular matrices and other cells (Buck and Horwitz, 1987; Hynes, 1987, 1992; Ruoslahti and Pierschbacher, 1987; Ginsberg et al., 1988; Hemler, 1990; Springer, 1990; Watt et al., 1993). In addition, they are involved in transducing extracellular signals into the cell (Hynes, 1992; Juliano and Haskill, 1993; Giancotti and Mainiero, 1994). Both the α and β subunits of integrins have a large extracellular portion, a transmembrane segment, and generally a short cytoplasmic domain. The cytoplasmic domains of integrins interact with the cytoskeleton and possibly with signaling molecules, but the molecular mechanisms of these interactions are not well understood.
The α6β4 integrin is a basement membrane receptor for laminins (Kajiji et al., 1989; De Luca et al., 1990; Sonnenberg et al., 1990a ,b; Lee et al., 1992) that engages in cytoplasmic interactions distinct from those of all other known integrins. Instead of being localized at adhesion plaques that serve as anchoring structures of actin filament networks, this receptor is part of hemidesmosomes (Carter et al., 1990; Stepp et al., 1990; Jones et al., 1991; Sonnenberg et al., 1991), junctional complexes that anchor cytokeratin intermediate filament (IF)1 networks and mediate adhesion of epithelial cells to the underlying basement membrane. The intracellular interactions of integrin α6β4 are mediated by the β4 subunit, the intracellular portion of which is much larger (∼1,000 amino acids) than that of all the other known β subunits (∼50 amino acids) and bears no apparent sequence homology to them (Hogervorst et al., 1990; Suzuki and Naitoh, 1990; Tamura et al., 1990). It contains four regions with homology to fibronectin type III (FNIII) repeats, arranged in two pairs separated by a 143–amino acid-long connecting segment. This large cytoplasmic tail of integrin β4 is required, and probably sufficient, for incorporation of the integrin into hemidesmosomes. Specifically, a minimal region on the integrin β4 subunit located in the linking segment between the second and third repeat has been reported to be critical to its localization in hemidesmosomes (Niessen et al., 1997a ). The targeted inactivation of the integrin β4 gene in mice convincingly demonstrated that hemidesmosome formation and proper skin attachment to the basal lamina are crucially dependent on the expression of this integrin subunit (Dowling et al., 1996; van der Neut et al., 1996).
Cytoplasmic components of the hemidesmosome that have been implicated in the attachment of IFs to this adhesion structure include the bullous pemphigoid antigen 1 (Stanley et al., 1981), also referred to as BP230 or BPAG1e (Yang et al., 1996), plectin (Wiche et al., 1984), the 200-kD 6A5 antigen (P200; Kurpakus and Jones, 1991), HD1 (Owaribe et al., 1991; Hieda et al., 1992), and the IF-associated protein 300 (Skalli et al., 1994). Among these, plectin, IF-associated protein 300, and HD1 have been shown to bind to IFs in vitro (Foisner et al., 1988; Skalli et al., 1994; Fontao et al., 1997), and the IF-binding site of plectin has recently been mapped to the fifth repeat domain within the carboxy-terminal globular region of the molecule (Nikolic et al., 1996). Also, BP230, which shares partial sequence homology with plectin and desmoplakin and is structurally related to these molecules (Tanaka et al., 1991; Green et al., 1992; Klatte and Jones, 1994), is likely to bind to IFs (Yang et al., 1996). A direct interaction of the intracellular domain of the integrin β4 subunit with any of the cytoplasmic hemidesmosome-associated proteins has not been demonstrated to date. There is evidence that integrin β4 interacts with HD1, based on coimmunoprecipitation of purified recombinant integrin β4 polypeptides expressed in bacteria with HD1 present in COS-7 cell lysates, and the observation that overexpression of integrin β4 leads to a redistribution of HD1 in transfected cells (Sánchez-Aparicio et al., 1997; Niessen et al., 1997a ,b), but the question whether this interaction was of direct or indirect nature remains unsolved.
Plectin, the most versatile cytoskeletal linker protein characterized to date, remains a strong candidate for bridging cytokeratin filament networks to hemidesmosomes. In fact, due to the very large size (>500 kD) of plectin molecules predicted on the basis of cDNA sequencing (Wiche et al., 1991; Liu et al., 1996; Elliott et al., 1997), and their extended (∼200-nm-long) multi-domain structure, as visualized by electron microscopy (Foisner and Wiche, 1987), plectin molecules would have the dimensions to span the entire tripartite structure characteristic of hemidesmosomes. In this way, they could extend from the plasma membrane, the location of integrin β4, to the filament anchorage site at the hemidesmosome inner plate structure. A role of plectin in hemidesmosome stabilization by physical linkage of proteins constituting this multi-component complex appears likely also in light of recent studies showing that defects in plectin expression lead to epidermolysis bullosa simplex (EBS)-MD, a severe hereditary skin blistering disease combined with muscular dystrophy (Chavanas et al., 1996; Gache et al., 1996; McLean et al., 1996; Pulkkinen et al., 1996; Smith et al., 1996). Moreover, plectin-deficient mice generated by targeted gene inactivation showed severe skin blistering and degeneration of keratinocytes, apparently caused by a significant reduction in number and mechanical stability of epidermal hemidesmosomes (Andrä et al., 1997).
The aim of the work presented here was to extend the human disease and plectin gene-knockout studies to the molecular level, and to investigate whether plectin directly interacts with integrin β4, and if so, to define the molecular domains of both proteins involved in this interaction. In addition, we were interested in the consequences that overexpression of integrin β4 mutant proteins containing putative plectin-binding sites might have on the integrity of cytokeratin filament networks and on the subcellular distribution of plectin. We show here that partially truncated versions of the integrin β4 cytoplasmic domain expressed in bacteria bind to recombinant carboxy- and amino-terminal domains of plectin in two independent in vitro binding assays, and that protein species shown to interact in vitro colocalized when ectopically coexpressed in living cells. In addition, we observed self-association of integrin β4 mutant proteins in vitro, and show that plectin proteins can modulate this effect. Furthermore, carboxy-terminal integrin β4 mutant proteins lacking plasma membrane anchoring sequences, but comprising plectin-binding region(s) in their tail domain, are shown to cause bundling and collapse of IF network arrays upon overexpression in PtK2 and 804G cells, and similar fragments dislocated endogenous plectin from hemidesmosomes in 804G cells.
Materials and Methods
cDNA Constructs
Integrin β4.
The integrin β4 subunit protein and cDNA sequences were numbered according to Suzuki and Naitoh (1990; Genbank/EMBL/DDBJ accession number X51841). Nucleotide and primer numbers refer to the nucleotide position (3′) relative to the first bp of the start codon (position 127 in X51841). For cloning, a part of the cytoplasmic tail region of integrin β4 was amplified by PCR from human placenta cDNA (Quick-Clone; CLONTECH Laboratories, Inc., Palo Alto, CA) using primers U3330 (5′-GAG CTT CAC GAG TCA GAT GTT GTC-3′) and L5250 (5′-GGG GCA GGG TGC GGT CAA GTT TGG-3′). A mixture of Klen-Taq (AB Peptides, St. Louis, MO) and PfuI (Stratagene, Heidelberg, Germany) polymerases was used under high fidelity conditions described by Barnes (1994). The 1944 bp PCR product obtained was used as template for nested PCR with EcoRI-tailed primers and the amplified fragments were subcloned into the unique EcoRI site of the bacterial expression vector pBN120 (Nikolic et al., 1996), a derivative of pET-15b (Novagen Inc., Madison, WI). Clones generated encoded the following domains of the integrin β4 subunit: β4-F1,2 (amino acid residues 1,126–1,315; plasmid construct pGR1), β4-L (1,316–1,457; pGR2), β4-F3,4C′(1,486–1,752; pGR3), β4-F2L (1,219–1,457; pGR4), β4-F1,2L′(1,126–1,485; pGR5), and β4-F1,2LF3,4C (1,126–1,752; pGR6). The clone encoding β4-F3,4 (1,457–1,662; pJP5) was generated by PCR from pGR6 (β4-F1,2LF3,4C; see Fig. 1 A for an overview). The correctness of all PCR-generated clones was verified by DNA sequencing. The clone encoding β4-F1,2L (1,126–1,457; pGR36) was obtained by exchanging the SmaI/PstI fragment from pGR5 with that of pGR4. Plasmids encoding β4-LF3,4C (1,316–1,752; pGR37) and β4-F3,4C (1,457–1,752; pGR44) were constructed by exchanging the SmaI/HindIII fragment of pGR2 with that of pGR6 and the XbaI/NotI fragment of pGR6 with that of pJP5, respectively. For expression of carboxy terminally c-myc–tagged proteins in mammalian cells, EcoRI fragments were subcloned into pAD29 (Nikolic et al., 1996), and excised XbaI/HindIII fragments were subcloned into the eukaryotic expression vector pRc/ CMV (Invitrogen Corp., San Diego, CA). This yielded the mammalian expression constructs encoding β4-F3,4C′myc (pGR13, derived from pGR3), β4-F1,2 myc (pGR16, derived from pGR1), β4-F1,2L′myc (pGR19, derived from pGR5), and β4-F1,2LF3,4Cmyc (pGR20, derived from pGR6). In addition, the EcoRI fragment from pGR6 was also subcloned into the eukaryotic expression vector pGR29, a modified version of pEGFP-N3 (CLONTECH Laboratories, Inc.). The resulting protein encoded by this construct (β4-F1,2LF3,4CGFP; pGR30) carried an enhanced version of the Aequorea victoria green fluorescent protein (GFP) at its carboxy terminus. Correct expression of proteins encoded by mammalian vector constructs was verified by Western blot analysis of lysates of transfected 804G cells.
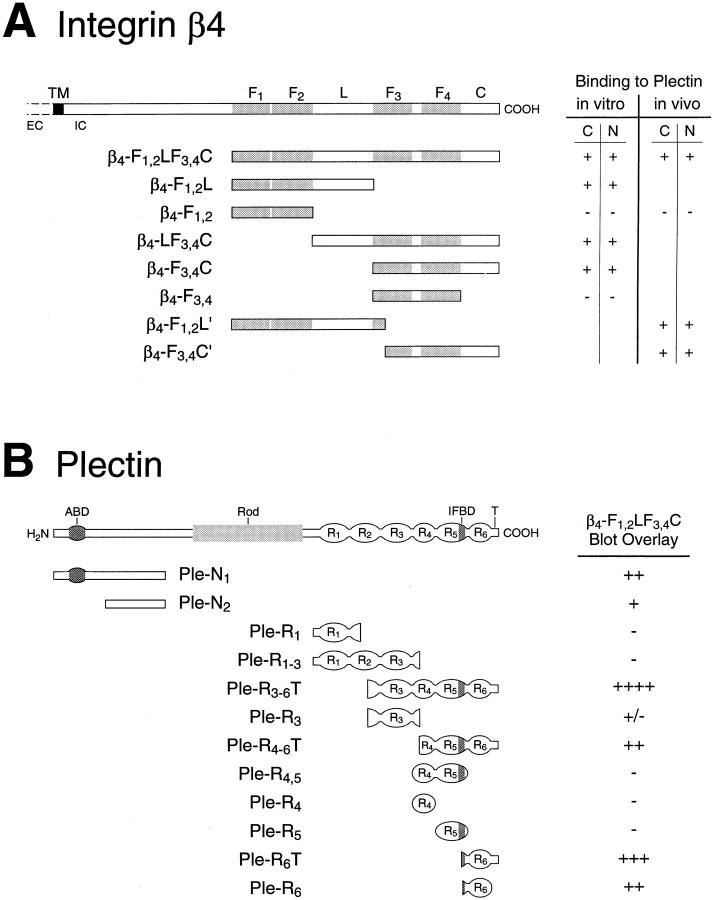
Figure 1.
(A) Schematic representation of human integrin β4 mutant proteins and summary of their plectin-binding phenotype in vitro and in vivo. Drawing on top shows major structural motifs contained in the integrin β4 subunit. EC and IC, extracellular part (not fully shown) and intracellular part, respectively; TM, transmembrane domain; F1–F4, fibronectin type III-like repeat domains; L, linking segment between the first and second repeat pair; C, carboxy-terminal domain after the second repeat pair. Details about generation and description of cDNA constructs and expressed proteins are given in the text. Table on right hand side indicates whether (+) or not (−) a given integrin mutant protein bound to the carboxy- (C = Ple-R3-6T) or amino-terminal (N = Ple-N1) plectin mutant protein in the Eu3+-overlay (in vitro) or transfection assays (in vivo). (B) Schematic representation of rat plectin mutant proteins and summary of their in vitro binding ability to the β4-F1,2LF3,4C polypeptide. Drawings include the major molecular domains of plectin: ABD, actin-binding domain; Rod, plectin rod domain; IFBD, IF binding domain; R1–R6, carboxy-terminal repeats domains 1–6; T, carboxy-terminal tail after repeat 6. Expression plasmids were generated as outlined in the text. Summary table at right shows semiquantitative assessment (strongest reaction: ++++, no binding: −) of the in vitro binding data shown in Fig. 3 (blot overlay of various plectin mutant proteins with the integrin β4 polypeptide β4-F1,2LF3,4C).
Plectin.
Rat plectin cDNA constructs were generated by PCR and/or other cloning techniques on the basis of the complete rat cDNA sequence (Genbank/EMBL/DDBJ database entry X59601). All constructs used were subcloned into the bacterial expression vector pBN120 (Nikolic et al., 1996), which enabled expression of amino terminally his-tagged proteins, except for the ones encoding Ple-N1 (pGR48) and Ple-R3-6T (pGR49), which were inserted into the unique EcoRI site of pFS23, a pET23a derivative driving the expression of carboxy terminally his-tagged proteins. Plectin constructs encoded the following segments of the protein (see Fig. 1 B for an overview): Ple-N1 (amino acids 1–1,128; construct pGR48), Ple-N2 (546–1,128; pMZ4), Ple-R1 (2,777–3,161; pJD11), Ple-R1-3 (2,777–3,851; pJD22), Ple-R3-6T (3,346–4,687; pGR49), Ple-R3 (3,346–3,851; pJD21), Ple-R4-6T (3,850–4,687; pJD23), Ple-R4,5 (3,780–4,367; pMZ3), Ple-R4 (3,780–4,024; pBN135), Ple-R5 (4,025–4,367; pBN132), Ple-R6T (4,262– 4,687; pMZ5), and Ple-R6 (4,277–4,620; pBN144). For expression in mammalian cells, EcoRI fragments of pGR48 (Ple-N1) and pGR49 (Ple-R3-6T) were subcloned into the eukaryotic expression vector pGR29, resulting in constructs encoding GFP-tagged Ple-N1 GFP (pGR31) and Ple-R3-6TGFP (pGR33). The latter plectin fragment was also expressed as a c-myc-tagged version (Ple-R3-6Tmyc), encoded by pBN72, a pRc/CMV derived plasmid.
Cell Culture, DNA Transfection, and Immunofluorescence Microscopy
Rat kangaroo PtK2 (CCL 56; American Type Culture Collection, Rockville, MD) and rat bladder carcinoma 804G cells (Izumi et al., 1981) were cultured in DME, supplemented with 50 U/ml penicillin, 50 μg/ml streptomycin, and 10% of heat-inactivated fetal calf serum, at 37°C and 5% CO2. For transfection, cells were grown on glass coverslips and, while still subconfluent, transiently transfected with either 30 μg plasmid DNA per 10-cm plate using the calcium phosphate precipitation method (Graham and van der Eb, 1973) or 0.6 μg plasmid DNA per 13-mm coverslip in wells of 24-well tissue culture plates (Falcon Plastics, Cockeysville, MD) using the LipofectAMINE (GIBCO BRL, Paisley, Scotland) transfection method. In the latter case, the transfection medium (0.6 ml/coverslip) contained DNA and LipofectAMINE at final concentrations of 1 μg/ml and 8 μg/ml, respectively, in serum-free medium (Opti-MEM, GIBCO BRL); it was replaced with DME/10% FCS after 4.5 h of incubation at 37°C and 5% CO2. Cells were fixed 24–48 (PtK2) or 8–24 h (804G) after transfection using chilled (−20°C) methanol and processed for immunofluorescence microscopy as previously described (Wiche et al., 1993). The following immunoreagents were used: rabbit anti–human β4-antiserum (Giancotti et al., 1992; kindly provided by F.G. Giancotti), mouse monoclonal anti–rat plectin antibody (mAb) 5B3 (Foisner et al., 1994), guinea pig anti–mouse liver cytokeratin antibody (Denk et al., 1981; kindly provided by H. Denk), mouse monoclonal anti-vimentin antibody (clone V9; DAKOPATTS, Copenhagen, Denmark), and anti-myc monoclonal antibody 1-9E10.2 (American Type Culture Collection) as primary antibodies; and AMCA-, fluorescein (FITC)- or Texas red–conjugated AffiniPure donkey anti–mouse IgG (H+L), anti–rabbit IgG (H+L), and anti–guinea pig IgG (H+L; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) as secondary antibodies. Specimens were viewed in a Zeiss Axiophot fluorescence microscope (Carl Zeiss, Oberkochen, Germany) or using the Bio-Rad MRC600 confocal scanning laser microscope (Richmond, CA). Photographs were taken using Ilford ASA 400 black and white film, and digital images were processed using the NIH Image 1.59 and Adobe Photoshop 4.01 software packages.
Expression of Recombinant Proteins in Bacteria
His-tagged recombinant proteins encoded by pBN120 or pFS23 vector constructs were expressed in Escherichia coli BL21(DE3) and purified from inclusion bodies by solubilization in 6 M urea, 500 mM NaCl, 20 mM Tris-HCl, pH 7.9 (binding buffer) containing 5 mM imidazole, followed by affinity binding to His-Bind metal chelation resin, according to the manufacturer's (Novagen Inc.) protocol. Bound proteins were eluted from affinity columns using 250 mM imidazole in binding buffer and stored frozen at −20°C. Samples were dialyzed against desired buffers before use to remove urea.
Gel Electrophoresis, Immunoblotting, and Preparation of 804G Cell Lysates
SDS-polyacrylamide gel electrophoresis (PAGE) was carried out under reducing conditions (Laemmli, 1970). Proteins on gels were visualized by staining with Servablue-G (Serva, Heidelberg, Germany). For immunoblotting, proteins were transferred to nitrocellulose sheets (Schleicher & Schuell, Dassel, Germany) for 4 h at 35 V in 25 mM Tris, 191 mM glycine, 0.01% SDS. After blocking with 3% BSA in TBS-T (TBS, containing 0.1% Tween-20), membranes were incubated with mAb 5B3 (1 h, room temperature), washed with TBS-T, and incubated with alkaline phosphatase-conjugated secondary goat anti–mouse antibody (Promega, Heidelberg, Germany). After washing, antibody binding was visualized with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate in 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 5 mM MgCl2.
Lysates of 804G cells were prepared by boiling cells in Laemmli's sample buffer (400 μl per confluent 10-cm plate) for 5 min.
Eu3+ Labeling of Recombinant Proteins and Microtiter Plate Overlay Assay
Recombinant mutant proteins were labeled with Eu3+ following the recommendations provided by the manufacturer (Wallac, Turku, Finland). 50–200 μg of recombinant integrin (β4-F1,2, β4-F1,2LF3,4C, β4-F1,2L, β4-LF3,4C, β4-F3,4C, β4-F3,4) and plectin (Ple-N1, Ple-R3-6T) mutant proteins were dialyzed against labeling buffer (50 mM sodium carbonate, pH 8.5) overnight at 4°C, and subsequently incubated with the labeling reagent (Eu3+-chelate of N1-(p-isothiocyanatobenzyl)-DTTA) at room temperature for ∼30 h. Unreacted labeling reagent was removed by gel filtration through a P6 column (Bio-Rad) equilibrated with 50 mM Tris-HCl, pH 7.5, 0.9% NaCl, 0.01% NaN3. Proteins were stored at 4°C until use. Microtiter plates were coated with recombinant proteins expressed in bacteria (100 μl of a 100 nM solution in 25 mM sodium borate buffer, pH 9.2) overnight at 4°C. Blocking was carried out with 4% BSA in TBS for 1 h and then binding with Eu3+-labeled proteins in 100 μl overlay buffer (TBS, pH 7.5, containing 1 mM EGTA, 2 mM MgCl2, 1 mM DTT, and 0.1% Tween 20) for 90 min at room temperature. After extensive washing with overlay buffer, the amount of bound proteins was determined by releasing complexed Eu3+ with enhancement solution and measuring the fluorescence with a Delfia time-resolved fluorometer (Wallac). The fluorescence values were converted to concentrations by comparison with an Eu3+ standard.
125I Labeling of Proteins and Blot Overlay Assay
The recombinant integrin β4 mutant protein β4-F1,2LF3,4C (∼0.8 mg) was dialyzed against 20 mM Tris-HCl, pH 7.5, and incubated with one Iodo-Bead (Pierce, Rockford, IL) and 0.5 mCi of Na125I (DuPont-NEN, Boston, MA) at room temperature for 15 min. Free iodine was removed by gel filtration using a Sephadex G-25 (Pharmacia Biotech Sverige, Uppsala, Sweden) column (0.9 × 14 cm) equilibrated with 20 mM Tris-HCl, pH 7.5, 1 mM DTT. Labeled proteins were stored at 4°C until use.
Purified recombinant integrin and plectin mutant proteins and lysates of induced bacterial cultures expressing plectin protein fragments were analyzed by SDS-10% PAGE. Proteins were blotted onto 0.2-μm nitrocellulose membranes (Schleicher & Schuell), overnight in 25 mM Tris, pH 8.8, and 192 mM glycine, at 25 V using a wet mini-blot apparatus (Bio-Rad). Membranes were incubated with 0.1% gelatin (Merck, Darmstadt, Germany) in PBS for 4 h at room temperature, and rinsed once with TSDT (20 mM Tris-HCl, pH 7.5, 250 mM NaCl, 1 mM DTT, 1% (vol/vol) Triton X-100). After incubation with 10 μg/ml of 125I- β4-F1,2LF3,4C in TSDT containing 0.5% BSA for 4 h and washing with TSDT, sheets were dried, sealed in plastic bags, and X-Omat film (Eastman Kodak, Rochester, NY) was exposed to them.
Immunoelectron Microscopy
Vascular perfusion of Wistar rats was performed with 4% paraformaldehyde in PBS, pH 7.4. Skin samples were immersed in fresh paraformaldehyde solution for 30 min and embedded in lowicryl-HM20 (Agar Scientific Ltd., Stansted, UK) as described in detail by Villinger (1991). Small pieces (∼1–2 mm2) of fixed rat skin were rinsed in PBS and dehydrated in a series of ethanol dilutions at −20°C and ethanol finally replaced by overnight infiltration of pure lowicryl. Samples were transferred into transparent tubes (Eppendorf, Hamburg, Germany) that were filled with freshly prepared lowicryl. Polymerization was complete after 24 h of exposure to UV light −28°C.
Lowicryl thin sections (60–80 nm) cut with an ultramicrotome (Ultracut S; Reichert, Vienna, Austria) were mounted on formvar-coated nickel slot grids. Postembedding immunolabeling was performed as previously described (Polak and Varndell, 1984). Free aldehyde groups were reduced by incubating sections in 0.1 M glycine in PBS, three times for 5 min, and blocking was performed in normal goat serum (BioCell Res. Lab, Cardiff, UK) for 30 min. For immunolabeling, sections were incubated for 1 h at room temperature, or overnight at 4°C with one of the following domain-specific anti-plectin antibodies: a) anti-rod mAb 7A8 (Foisner et al., 1996), b) anti-amino terminus rabbit serum E1A raised against a synthetic peptide corresponding to a 12–amino acid residue-long sequence of human exon 1a, and c) anti-carboxy terminus mouse serum 135C raised against a recombinant rat plectin mutant protein corresponding to the carboxy-terminal repeat 4 domain (see Andrä et al., 1997), or with non- immune serum or PBS as controls. Secondary labeling using 5 nm gold-conjugated goat-anti–mouse IgG and 1 nm gold-conjugated goat-anti– rabbit IgG (BioCell Res. Lab) was performed for 1 h at room temperature. For visualization in the electron microscope, 1 nm or 5 nm colloidal gold particles were silver enhanced (Stierhof et al., 1991). Thin sections were counterstained with uranyl acetate and lead citrate and were viewed at 80 kV in a JEOL JEM-1210 electron microscope.
Results
Expression and Purification of Recombinant Forms of Integrin β4 and Plectin
Localization of plectin at hemidesmosomes (Wiche et al., 1984) and its function as an IF (cytokeratin)-binding protein make it a likely candidate to act as a linker between the cytokeratin filament network and the hemidesmosomal integrin α6β4. To test whether this proposed function of plectin involves its direct interaction with the uniquely long cytoplasmic tail of the β4 subunit of integrin α6β4, we generated a series of human integrin β4 and rat plectin truncation mutants (Fig. 1) for use in biochemical in vitro binding, as well as in vivo cell transfection assays.
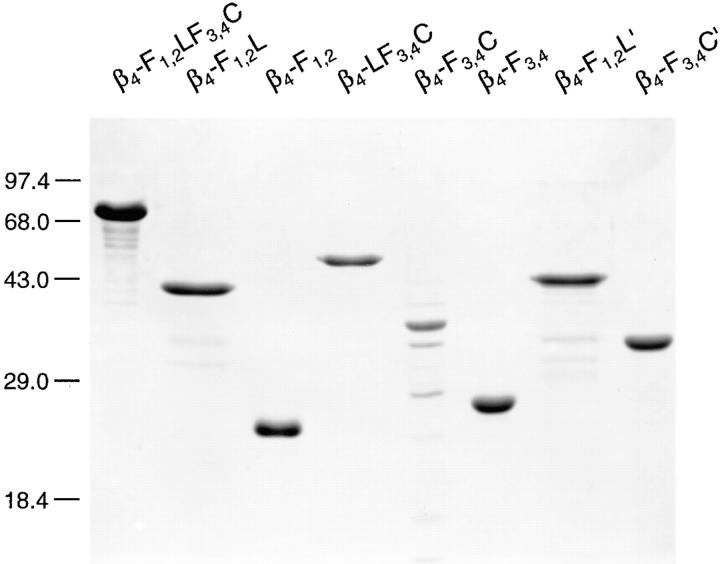
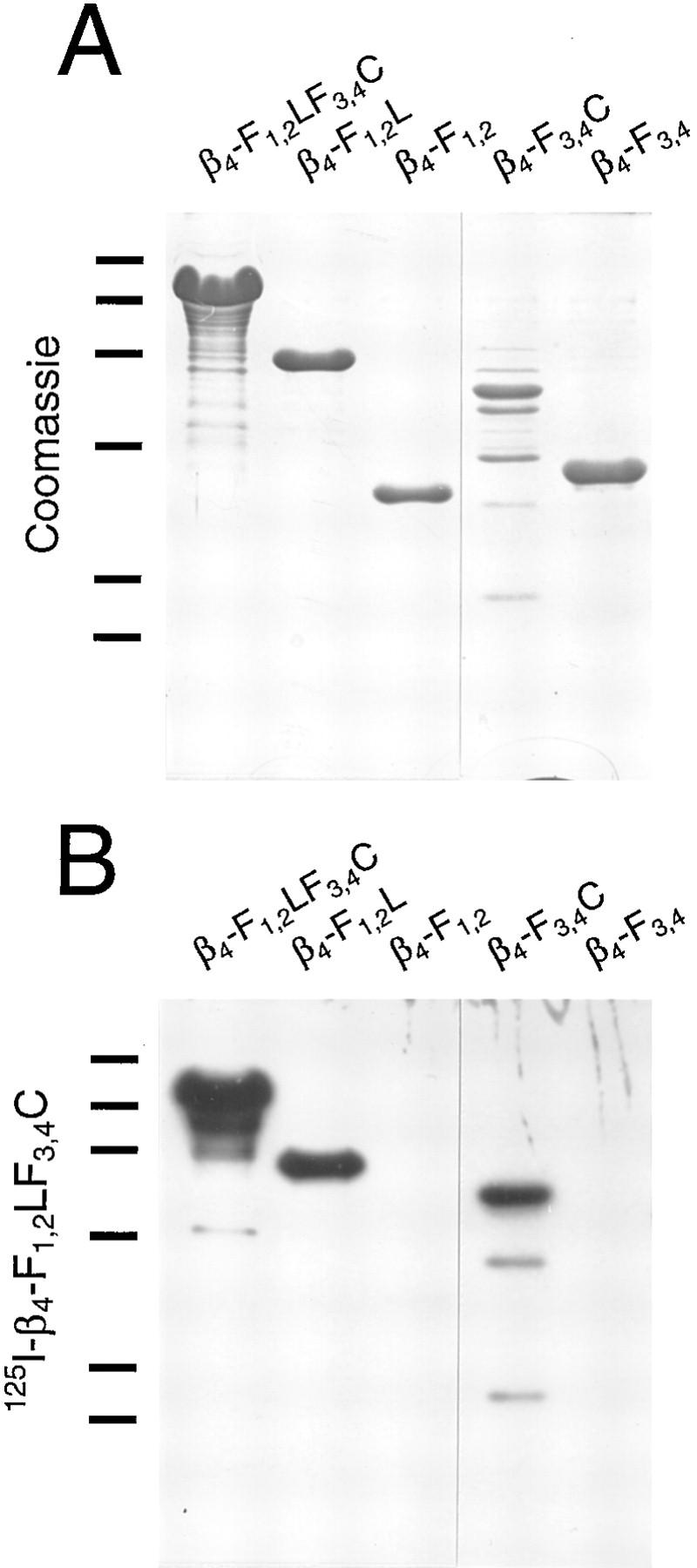
The original design of our integrin β4 expression constructs was based on a report by Spinardi et al. (1993), who showed that a 303–amino acid-long region of the cytoplasmic tail comprising the first of its two pairs of FNIII repeats and the following segment between the first and second pair was necessary for the recruitment of the integrin β4 subunit into hemidesmosomes. Integrin β4 subdomain-specific clones generated in the pET expression system are shown in Fig. 1 A. Encoded proteins contained the cytoplasmic tail starting at the beginning of the first FNIII repeat (β4-F1,2LF3,4C), the 303–amino acid region described above (β4-F1,2L′), and the rest of the tail, starting in the third FNIII repeat, close to its beginning (β4-F3,4C′). β4-F1,2L and β4-F3,4C were similar to β4-F1,2L′ and β4-F3,4C′, respectively, except for excluding (β4-F1,2L) and including (β4-F3,4C) the third FNIII repeat in its entirety. β4-F1,2 corresponded to the first FNIII repeat pair, β4-F3,4 to the second; β4-LF3,4C lacked the first FNIII repeat pair. SDS-PAGE analysis of purified recombinant integrin β4 polypeptides expressed in E. coli confirmed that they were of the sizes predicted on the basis of the corresponding cDNA sequences, including the 2.6-kD his-tags (Fig. 2).
Figure 2.
SDS-PAGE of integrin β4 mutant proteins expressed in bacteria. His-tagged recombinant proteins were purified from bacterial inclusion bodies as described in the text and analyzed on a 12% polyacrylamide gel. Proteins were visualized by staining the gel with Servablue-G. The predicted masses of recombinant proteins were: 71.4 kD (β4-F1,2LF3,4C), 39.4 kD (β4-F1,2L), 24.0 kD (β4-F1,2), 50.4 kD (β4-LF3,4C), 35.0 kD (β4-F3,4C), 25.4 kD (β4-F3,4), 42.6 kD (β4-F1,2L′), and 31.8 kD (β4-F3,4C′). Molecular weight markers are indicated (×10−3).
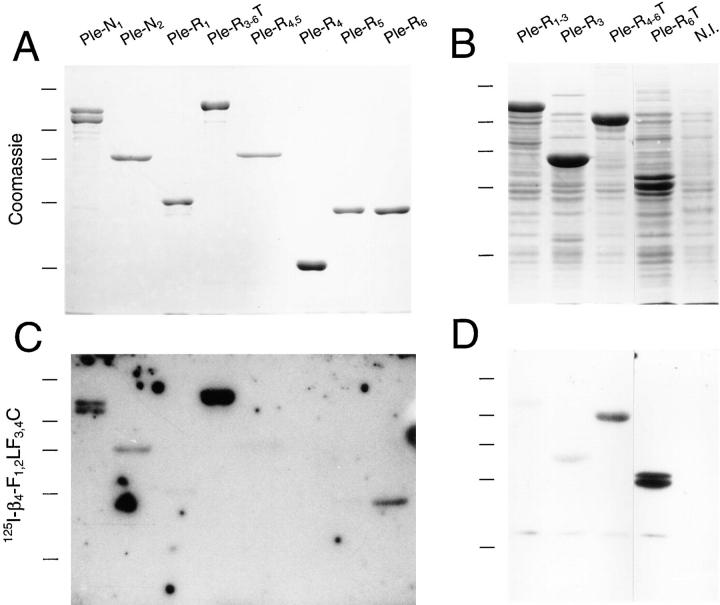
Plectin mutant proteins corresponding to different molecular regions of the amino- and carboxy-terminal globular domains of the molecule (Fig. 1 B) were generated by subcloning rat plectin cDNAs into the pET expression system. Two amino-terminal protein fragments used covered the amino acid sequence encoded by exons 1–24 (Ple-N1) and exons 12–24 (PleN2). The other fragments covered, partly overlapping, the complete carboxy-terminal globular domain with its repeat structure. Ple-R1 started within the hinge region after the rod domain and included the first and part of the second repeat domain; Ple-R3-6T contained the rest of plectin's carboxy terminus. Two constructs, Ple-R3 and Ple-R4-6T, split construct Ple-R3-6T into two shorter ones, one corresponding to repeat 3 and parts of the flanking repeats 2 and 4, the other to the carboxy terminus starting in repeat 4. Ple-R1-3 was a combination of Ple-R1 and Ple-R3, ranging from plectin's hinge region into repeat 4. Ple-R4,5 contained repeat 4 and 5, Ple-R4 repeat 4 alone, and Ple-R5 repeat 5 alone. Ple-R6T contained a small portion of repeat 5, repeat 6 and the carboxy-terminal tail. Ple-R6 was equivalent to Ple-R6T but lacked the tail. Plectin mutant proteins expressed from these plasmids were all of the expected sizes, as determined by SDS-PAGE (Fig. 3, A and B). The double band observed for Ple-N1 was likely due to an additional translation initiation event at an in-frame ATG codon, resulting in a ∼15-kD shorter his-tagged protein version.
Figure 3.
Blot overlay of recombinant plectin proteins with 125I-labeled β4-F1,2LF3,4C. Purified his-tagged recombinant plectin proteins Ple-N1, Ple-N2, Ple-R1, Ple-R3-6T, Ple-R4,5, Ple-R4, Ple-R5, and Ple-R6 (A and C), and whole lysates of induced bacterial cultures (B and D) expressing his-tagged plectin recombinant proteins Ple-R1-3, Ple-R3, Ple-R4-6T, Ple-R6T, and a non-induced bacterial culture, were subjected to SDS 10% PAGE in duplicate. Proteins on one set of gels were stained with Servablue-G (A and B), those on the other set were blotted onto nitrocellulose membranes, overlaid with 125I-β4-F1,2LF3,4C (his-tagged), and bound integrin β4 protein detected by autoradiography (C and D). Size markers of 200, 97, 68, 43, and 29 kD are indicated.
Different Subdomains of the Integrin β4 Cytoplasmic Tail Region Each Bind to Amino- and Carboxy-terminal Domains of Plectin
To detect direct interaction between the candidate integrin β4 cytoplasmic tail region and various subdomains of plectin, we first used a qualitative blot overlay assay in which recombinant plectin fragments immobilized on nitrocellulose membranes were overlaid with 125I-labeled purified recombinant integrin β4-F1,2LF3,4C (Fig. 3, C and D). The plectin fragments used in these experiments covered the complete cDNA sequence (exons 1–32; Liu et al., 1996) with the exception of the predicted rod region (exon 31), a small part of the preceding amino-terminal globular domain (exons 25–30), and the three additional alternative start exons (1a, 1b, and 1c) identified recently (Elliott et al., 1997). Among nearly a dozen recombinant plectin polypeptides tested, the one displaying highest binding activity to β4-F1,2LF3,4C was Ple-R3-6T, which contained most of plectin's carboxy-terminal globular domain. Plectin protein fragments corresponding to individual repeats (Ple-R3, Ple-R4, Ple-R5) showed no or hardly any binding activity, neither did fragments containing repeat 1 and part of the hinge region (Ple-R1), repeats 1–3 (Ple-R1-3), or repeats 4 and 5 combined (Ple-R4,5). Aside from Ple-R3-6T, the β4-F1,2LF3,4C polypeptide bound to all other plectin mutant proteins that contained repeat 6, such as Ple-R4-6T, Ple-R6T, and Ple-R6. β4-F1,2LF3,4C also reacted with the amino-terminal polypeptide fragments Ple-N1 and Ple-N2, suggesting that one or more additional interaction site(s) resided in this region of the plectin molecule.
To verify the blot overlay, and in order to assess plectin-integrin β4 interaction in a more quantitative way, we used an alternative, nonradioactive microtiter plate-binding assay involving Eu3+-labeled proteins, which had successfully been used in the molecular mapping of plectin's IF-binding site (Nikolic et al., 1996). The two plectin protein fragments representing amino- and carboxy-terminal domains and exhibiting highest binding efficiency in the blot overlay assay (Ple-N1 and Ple-R3-6T, Fig. 3) were used for this assay. β4-F1,2LF3,4C was coated onto 96-well microtiter plates and overlaid with increasing concentrations of the Eu3+-labeled plectin proteins (Fig. 4 A). The amounts of proteins bound to the coated integrin fragment were determined by measuring released Eu3+ by time-resolved fluorometry (Soini and Lövgren, 1987). Non-specific binding to coated BSA was also determined and subtracted to give results for specific binding to β4-F1,2LF3,4C. Binding of both plectin proteins was similar up to 500 nM, where Ple-N1 reached saturation, contrary to Ple-R3-6T, which exhibited approximately twofold higher binding at 1,000 nM, consistent with the apparently stronger binding of 125I-β4-F1,2LF3,4C to this protein in the blot overlay assay (Fig. 3).
Figure 4.
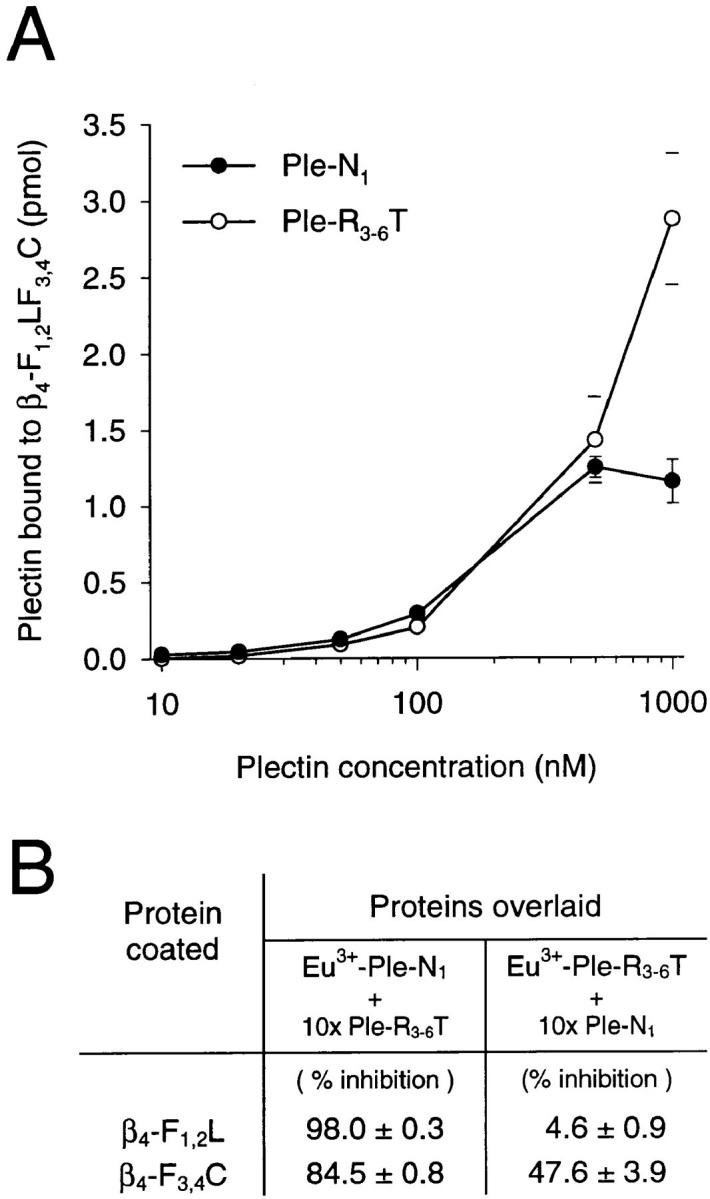
Concentration dependent (A) and competitive binding (B) of Eu3+- labeled recombinant plectin polypeptides to immobilized recombinant integrin β4 cytoplasmic tail polypeptide. (A) β4-F1,2LF3,4C (100 nM) was coated onto microtiter plates and overlaid with increasing concentrations (10–1,000 nM) of Eu3+-labeled Ple-N1 (•) or Ple-R3-6T (○). Data shown have been corrected for nonspecific binding to BSA. Note, for concentrations up to 500 nM error bars do not extend outside of symbols. (B) Table summarizing competition experiments: β4-F1,2L or β4-F3,4C were coated onto microtiter plates (100 nM) and overlaid with either Eu3+-labeled Ple-N1 (100 nM) in the absence and presence of a 10-fold molar excess (1 μM) of unlabeled Ple-R3-6T (column 2) or Eu3+-labeled Ple-R3-6T (100 nM) in the absence and presence of a 10-fold molar excess (1 μM) of unlabeled Ple-N1 (column 3). Extent of inhibition of binding was calculated as 100 − 100 × (binding with competitor/binding without competitor). Amounts of plectin proteins bound were determined by measuring released Eu3+ by time-resolved fluorometry after addition of enhancement solution as described in the text. All data (A and B) are presented as the mean ± SD of duplicate determinations.
To map the plectin-binding site(s) in the integrin β4 cytoplasmic domain more precisely, Ple-R3-6T and Ple-N1 were separately coated and overlaid with samples of Eu3+-labeled integrin β4 mutant proteins (Fig. 5, A and B). Consistent with the qualitative blot overlay assay (Fig. 3), strongest binding was observed between β4-F1,2LF3,4C and the carboxy-terminal plectin fragment Ple-R3-6T (Fig. 5 A, first bar). The same integrin β4 fragment bound to the amino-terminal plectin fragment Ple-N1 with just ∼30% lower efficiency (Fig. 5 B, first bar). Among a panel of truncated versions of β4-F1,2LF3,4C tested, only those containing either the region between the second and third FNIII repeat (β4-F1,2L and β4-LF3,4C) and/or the carboxy-terminal tail region following the last FNIII repeat (β4– LF3,4C and β4-F3,4C) showed significant binding to either one of the plectin fragments (Fig. 5, A and B). β4-F1,2 and β4-F3,4 hardly showed any detectable binding. These results suggested that at least two different plectin-binding domains were present in the cytoplasmic domain of the integrin β4 subunit, one between the two pairs of FNIII repeats, the other in the ultimate tail region trailing the second pair of FNIII repeats. Moreover, both binding domains apparently comprised interaction sites for amino- as well as carboxy-terminal domains of plectin.
Figure 5.
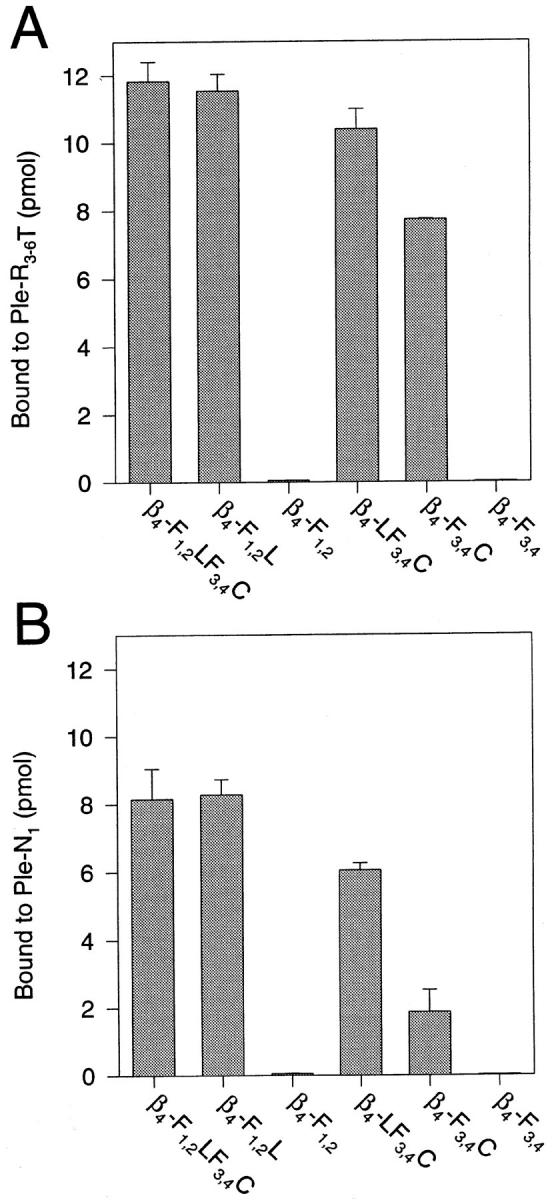
Binding of Eu3+- labeled recombinant integrin β4 polypeptides to plectin carboxy- (A) and amino-terminal (B) protein domains immobilized on microtiter plates. 100 nM purified his-tagged recombinant plectin mutant proteins Ple-R3-6T and Ple-N1 were coated onto microtiter plates and separately overlaid with Eu3+- labeled integrin β4 proteins β4-F1,2LF3,4C, β4-F1,2L, β4-F1,2, β4-LF3,4C, β4-F3,4C, and β4-F3,4 (2.5 μM each). Amounts of bound integrin β4 proteins were determined as described in Fig. 4. Data are presented as the mean ± SD of duplicate determinations.
To examine whether the different binding sites on integrin and plectin molecules have differential affinities for each other, integrin fragments β4-F1,2L and β4-F3,4C, each containing just one of the two putative plectin-binding regions, were coated onto microtiter plates and overlaid with either Eu3+-labeled Ple-N1 with or without a 10-fold molar excess of unlabeled Ple-R3-6T (Fig. 4 B, first column) or, vice versa, with Eu3+-labeled Ple-R3-6T with or without a 10-fold molar excess of unlabeled Ple-N1 (Fig. 4 B, second column). The results of these competition experiments, documented as the relative percentage of inhibition of binding in the presence of the competitor, are summarized in Fig. 4 B. Ple-R3-6T was able to efficiently displace Ple-N1 from both, β4-F1,2L and β4-F3,4C, whereas reversely, Ple-N1 hardly could displace Ple-R3-6T from β4-F1,2L and inhibited its binding to β4-F3,4C by only ∼50%. The ability of the plectin mutant proteins to displace each other from the more carboxy-terminal integrin β4 plectin-binding domain (contained in β4-F3,4C) indicated that both exhibited affinities for the same binding site. Apparently, however, Ple-R3-6T bound to this site with higher affinity compared to Ple-N1, as indicated by its higher competition efficiency. This was consistent with the more efficient binding of β4-F3,4C to immobilized Ple-R3-6T, as compared to Ple-N1 in the microtiter plate assay (compare bars β4-F3,4C in Fig. 5, A and B). The inability of Ple-N1 to displace Ple-R3-6T from β4-F1,2L indicated also a significantly higher affinity of Ple-R3-6T, compared to Ple-N1, for the second, more amino-terminal plectin-binding site contained in this integrin β4 fragment (β4-F1,2L).
Integrin β4 Mutant Proteins Exhibit Self-association upon Immobilization
Competition experiments similar to those shown above (Fig. 4 B) using immobilized plectin proteins and Eu3+- labeled integrin fragments in solution failed due to excessive self-interaction of integrin β4 mutant proteins upon immobilization, a situation which also prevented the determination of dissociation constants. The ability of integrin β4 cytoplasmic tail domains to self-associate potentially could have implications for certain functions of the protein, such as integrin clustering in hemidesmosomal complex formation and/or hemidesmosomal plaque-cytoskeleton anchorage. To obtain some information on possible molecular mechanisms and to identify the domain(s) mediating self-association, purified recombinant integrin β4 polypeptides representing various molecular domains were resolved by SDS-PAGE, blotted onto nitrocellulose, and overlaid with 125I-β4-F1,2LF3,4C (Fig. 6, A and B). β4-F1,2LF3,4C reacted strongly with itself, β4-F1,2L, and β4-F3,4C, but not detectably with β4-F1,2 or β4-F3,4. This indicated that the domains involved in the aggregation of integrin β4 were similar to those mediating integrin β4-plectin interactions. In solution, no such aggregation of integrin β4 proteins was found (data not shown). When β4-F1,2LF3,4C was coated and overlaid with Eu3+-β4-F1,2LF3,4C (Fig. 6 C, filled bars) in either the standard overlay assay buffer (150 mM NaCl), buffer without NaCl, or buffer with increased NaCl concentration (500 mM), binding under high-salt conditions was fivefold increased over standard condition and about 10-fold reduced at low-ionic strength. Non-specific binding to BSA (Fig. 6 C, open bars) was low and even decreased with increasing ionic strength. This experiment indicated that self-interaction of β4-F1,2LF3,4C was of hydrophobic nature. In a further experiment, increasing concentrations of Ple-R3-6T were added to a constant amount of Eu3+- β4-F1,2LF3,4C. As seen in Fig. 6 D, plectin fragments decreased the extent of integrin β4 self-association in a concentration dependent manner. Presumably by binding to coated integrin β4 molecules, plectin fragments blocked integrin β4 self-association sites, which reside near or are even overlapping with the plectin-binding sites, as indicated by the blot overlay assay (Fig. 6, A and B).
Figure 6.
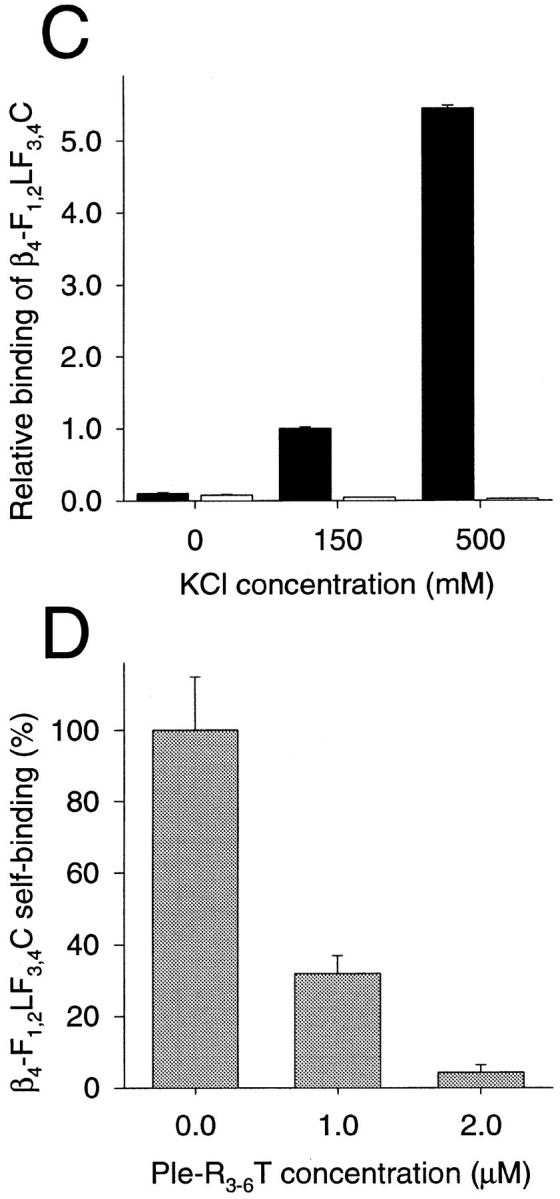
Self-association of recombinant integrin β4 cytoplasmic tail fragments. (A and B) Blot overlay: purified his-tagged recombinant integrin β4 proteins β4-F1,2LF3,4C, β4-F1,2L, β4-F1,2, β4-F3,4C, and β4-F3,4 were subjected to SDS 10% PAGE in duplicate. Proteins on one gel were stained with Servablue-G (A), those on the other were blotted onto nitrocellulose membrane, overlaid with 125I-β4-F1,2LF3,4C, and bound integrin β4 protein detected by autoradiography (B). Size markers of 97, 68, 43, 29, 18.4, and 14.3 kD are indicated. (C) Ionic strength dependence: β4-F1,2LF3,4C (filled bars) and BSA (open bars) were coated (both at 100 nM) and overlaid with Eu3+-labeled β4-F1,2LF3,4C (50 nM) in solutions of different ionic strengths (0, 150, and 500 mM NaCl). Scale is normalized for extent of binding under standard conditions (150 nM NaCl). Data are presented as the mean ± SD of duplicate determinations. (D) Competition experiment: β4-F1,2LF3,4C was coated (100 nM) and overlaid with 100 nM Eu3+-labeled β4-F1,2LF3,4C and different concentrations (0–2 μM) of Ple-R3-6T. Scale is normalized for extent of binding in the absence (0 μM) of Ple-R3-6T (100%). Data are presented as the mean ± SD of duplicate determinations.
Colocalization of Integrin β4 and Plectin Mutant Proteins Ectopically Expressed in Transfected PtK2 Cells
To test whether integrin β4 and plectin mutant proteins shown to interact in vitro would also interact when ectopically expressed in living cells, we transiently cotransfected PtK2 cells using various plectin and integrin β4 cDNA vector constructs driving the expression of c-myc–tagged and GFP-tagged proteins. Based on the in vitro binding data, we chose to express two plectin fragments, Ple-N1 GFP, containing the amino-terminal integrin β4-binding site(s) identified above, and Ple-R3-6Tmyc (Ple-R3-6TGFP), the carboxy-terminal polypeptide exhibiting the strongest binding to integrin β4 among all the recombinant plectin mutant proteins tested. In a first series, PtK2 cells were cotransfected with plasmids encoding the carboxy-terminal plectin fragment and one of the plasmids encoding correspondingly tagged integrin β4 fragments β4-F1,2LF3,4CGFP, β4-F1,2LF3,4Cmyc, β4-F1,2L′myc, β4-F1,2 myc, and β4-F3,4C′myc (Fig. 7). Most transfected cells were double-transfected, expressing both the plectin and integrin proteins. A few cells bearing only one plasmid served as controls for single-transfected cells.
Figure 7.
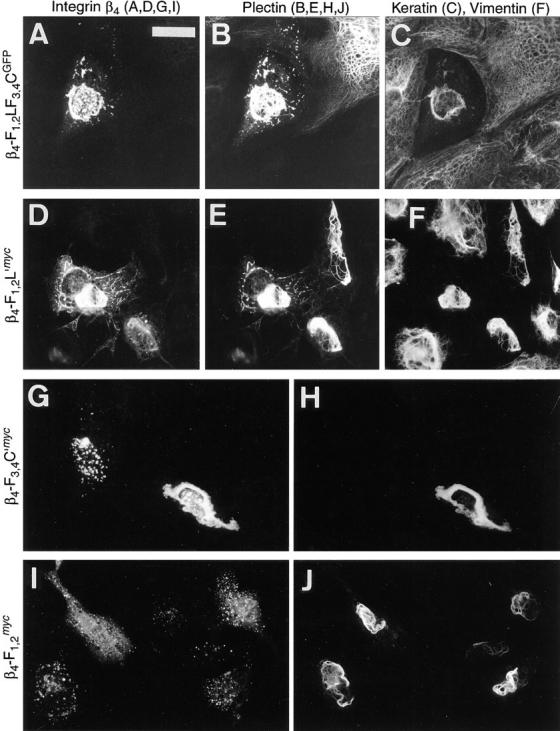
Coexpression of the carboxy-terminal plectin protein Ple-R3-6T and various integrin β4 mutant proteins in PtK2 cells. Cells plated on glass coverslips were cotransfected with a plectin construct encoding Ple-R3-6T (with either c-myc-tag or GFP linked to the carboxy terminus) and one of the integrin β4 constructs encoding β4-F1,2LF3,4CGFP (A–C), β4-F1,2L′myc (D–F), β4-F3,4C′myc (G and H), or β4-F1,2 myc (I and J). After ∼48 h, cells were fixed and double- or triple-stained. (A, D, G, and I) Ectopically expressed integrin β4 proteins, tagged with GFP (A; FITC optics) or c-myc (D, G, and I; Texas red optics). (B, E, H, and J) Ectopically expressed plectin protein tagged with c-myc (B; Texas red optics) or GFP (E, H, and J; FITC optics). (C and F), cytokeratins and vimentin, respectively, visualized by UV optics (AMCA). Note colocalization of β4-F1,2LF3,4CGFP/myc, β4-F1,2L′myc, and β4-F3,4C′myc, but not β4-F1,2 myc, with the carboxy-terminal plectin polypeptide (Ple-R3-6TGPF/myc). Bar, 15 μm.
In cells coexpressing Ple-R3-6Tmyc and β4-F1,2LF3,4CGFP, the bulk of both ectopically expressed proteins showed codistribution and association with dense structures surrounding the nucleus; in addition, both were associated with small dot- or patch-like structures of unknown nature throughout the cytosol in superimposable patterns (Fig. 7, A and B). In rarely observed single-transfected cells expressing the plectin mutant protein alone, similar dot- and patch-like structures were not observed; such cells rather displayed filament association of mutant proteins (e.g., single-transfected cell in upper right-hand corner of Fig. 7 B), or bundling and collapse of IFs, as previously reported (Nikolic et al., 1996). As revealed by triple-staining, the cytokeratin filaments of such cotransfected cells seemed to have completely collapsed onto the nuclei, rendering the remainder of the cytoplasmic space a cytokeratin-free zone, clearly outlined by adjacent cytokeratin-positive cells (Fig. 7 C).
β4-F1,2L′myc, containing the first pair of FNIII repeats plus the region between the second and third FNIII repeat, colocalized with the carboxy-terminal plectin protein (Ple-R3-6TGFP) in dense aggregates as well as in more delicate filamentous structures (Fig. 7, D and E). These delicate structures were not seen in cells expressing Ple-R3-6TGFP (see e.g., single-transfected cell in upper right-hand part of Fig. 7 E) or β4-F1,2L′myc alone (see e.g., transfected cell in center of Fig. 10 I), suggesting that their appearance was dependent on the coexpression of both the plectin and integrin β4 mutant proteins. This notion was supported by the observation that in double-transfected cells, expressing comparatively low levels of β4-F1,2L′myc compared to Ple-R3-6TGFP, delicate structures were hardly observed, but integrin-specific staining was confined to dense structures typical of collapsed filaments (see e.g., double-transfected cell in lower right-hand part of Fig. 7, D–F).
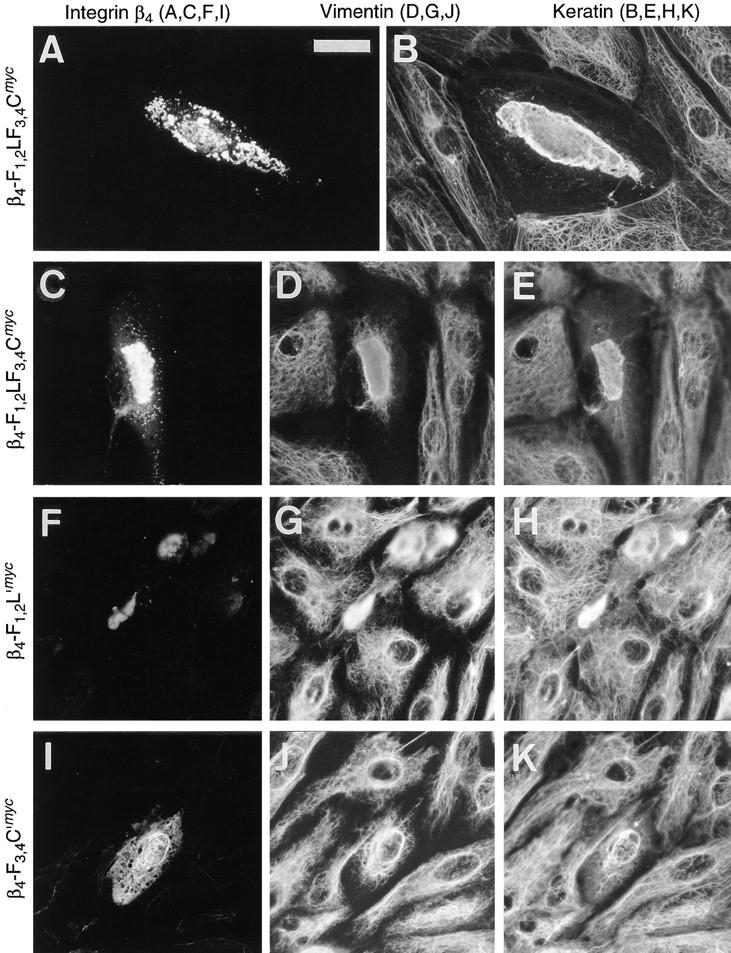
Figure 10.
Collapse of IF network arrays in PtK2 cells ectopically expressing different integrin β4 cytoplasmic tail domain fragments. Cells plated on glass coverslips were transfected with cDNA constructs encoding mutant integrin β4-F1,2LF3,4Cmyc (A–E), β4-F3,4C′myc (F–H), or β4-F1,2L′myc (I–K). After ∼48 h, cells were fixed and double- or triple-stained. (A, C, F, and I), c-myc-tagged integrin β4 proteins. (D, G, and J), vimentin, FITC optics. (B, E, H, and K), cytokeratins, UV optics (AMCA). Focus was set for the Texas red channel. Mutant proteins containing the carboxy-terminal 267 amino acids of the β4 cytoplasmic tail (β4-F1,2LF3,4Cmyc and β4-F3,4C′myc) led to the collapse of both IF systems (B, D, E, G, and H), whereas the protein lacking this domain (β4-F1,2L′myc) did not (J and K). Bar, 15 μm.
The expression product β4-F3,4C′myc, similar to β4-F1,2LF3,4CGFP and β4-F1,2L′myc, colocalized with the coexpressed carboxy-terminal plectin polypeptide in dense perinuclear structures (Fig. 7, G and H), different from the dispersed patches observed when this integrin construct was expressed alone (Fig. 7 G). In contrast, β4-F1,2 myc, which lacked plectin-binding activity in vitro, was diffusely localized in small dots throughout the cell, regardless of whether or not the cell was cotransfected with the plectin construct (Fig. 7, I and J). As expected, plectin staining was independent of integrin β4 localization and its characteristics were similar to those of plectin only expression (Fig. 7 J).
The plasmid encoding the amino-terminal plectin protein Ple-N1 GFP was used in a second set of similar cotransfection experiments using PtK2 cells (Fig. 8). Cells, cotransfected with the plasmid encoding β4-F1,2LF3,4Cmyc, displayed colocalization of both ectopically expressed polypeptides in brightly stained patches (Fig. 8, A and B). Colocalization was also observed with β4-F1,2L′myc (Fig. 8, C and D), but the dots and patches double-stained in this case were smaller and distributed more uniformly throughout the cell as compared to the structures formed with β4-F1,2LF3,4Cmyc. A similar confinement of both mutant proteins within distinct dots and patches was also found in cells coexpressing β4-F3,4C′myc and Ple-N1 GFP (Fig. 8, E and F). In a single-transfected cell the weakly expressed β4-F3,4C′myc was found diffusely distributed within the nuclear compartment and in form of dots and patches in the cytosol (Fig. 8 E). In contrast, β4-F1,2 myc, which lacks both putative plectin-binding sites, remained diffusely distributed throughout the cell (Fig. 8 G), without showing the dotty staining pattern of the plectin protein (Fig. 8 H).
Figure 8.
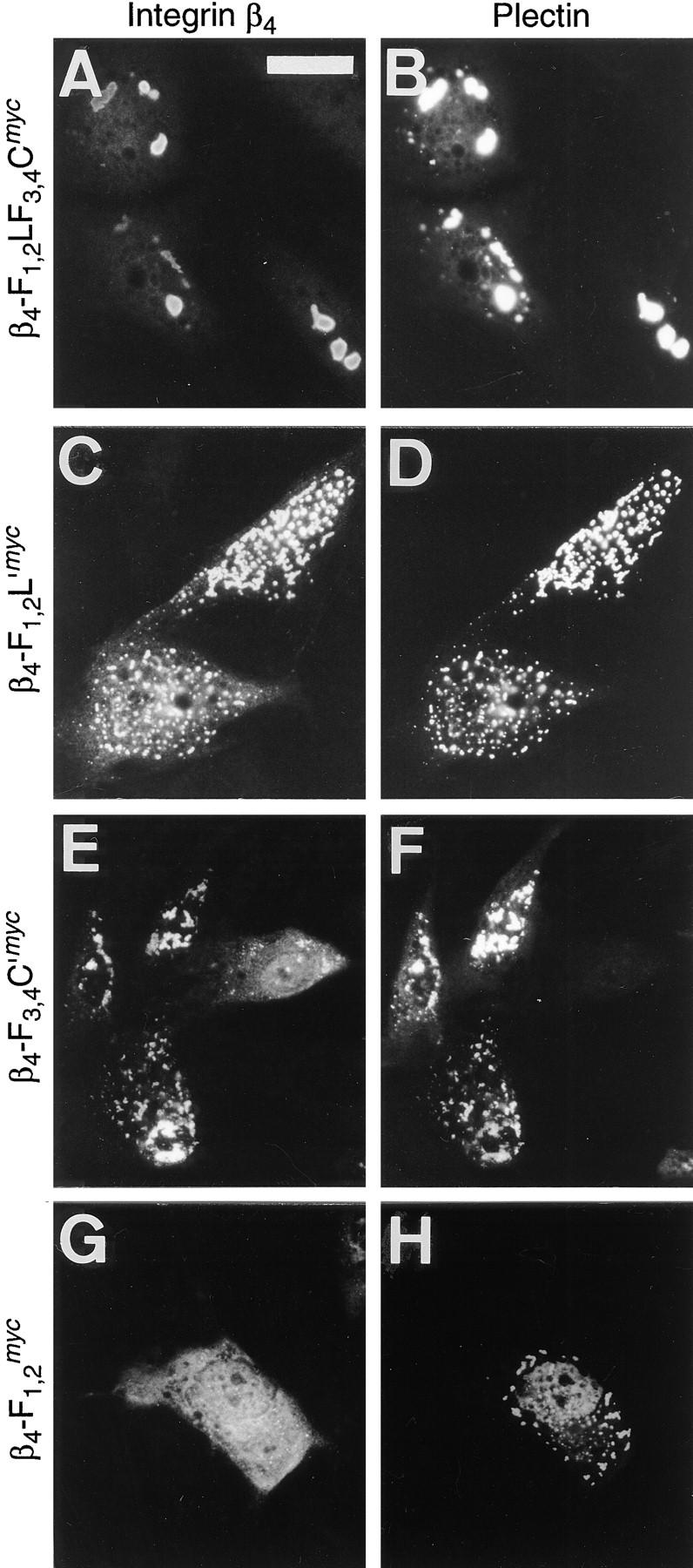
Coexpression of the amino-terminal plectin protein Ple-N1 and various integrin β4 mutant proteins in PtK2 cells. Cells plated on glass coverslips were cotransfected with a cDNA construct encoding Ple-N1 GFP and one of the plasmids encoding the integrin β4 mutants β4-F1,2LF3,4Cmyc (A and B), β4-F1,2L′myc (C and D), β4-F3,4C′myc (E and F), or β4-F1,2 myc (G and H). After ∼48 h, cells were fixed and double-stained for ectopically expressed c-myc-tagged integrin β4 proteins (A, C, E, and G; Texas red optics) and the GFP-tagged plectin protein (B, D, F, and H; FITC optics). Note colocalization of β4-F1,2LF3,4Cmyc, β4-F1,2L′myc, and β4-F3,4C′myc, but not β4-F1,2 myc, with Ple-N1 GFP. Bar, 15 μm.
Ultrastructural Localization of Plectin at Hemidesmosomes Using Domain-specific Antibodies
To examine whether plectin molecules found at hemidesmosomal junctions in tissues are located within distances short enough to enable their interaction with plasma membrane-anchored integrin β4, immunogold electron microscopy of acrylic resin-embedded rat skin samples was performed. Embedding in lowicryl HM20 was found to provide excellent conditions for thin sectioning of rat skin specimens (below 100 nm), and enabled adequate hemidesmosomal fine structure resolution, comparable to other protocols (Rappersberger et al., 1990; Shimizu et al., 1992). Considering that individual plectin molecules have been visualized as structures 200 nm in length, and probably can extend over even longer distances (Foisner and Wiche, 1987; Foisner et al., 1995; Svitkina et al., 1996), we used antibodies raised against epitopes of plectin residing in three different domains of the molecule, the amino terminus (serum E1A), the rod (mAb 7A8), and the carboxy terminus (serum 135C). All three of these domain-specific immunoreagents were found to be reactive with epitopes at the periphery as well as in the cytoplasmic interior of keratinocytes. Labeling was most intense near basolateral plasma membrane domains, in particular at the locations of hemidesmosomes (Fig. 9, A–C) and desmosomes (not shown; see also Eger et al., 1997), and along cytoplasmic cytokeratin filament bundles. In the area of hemidesmosomes, label was found at the plaque, at the inner plate, and in association with cytokeratin filaments (Fig. 9, A–C).
Figure 9.
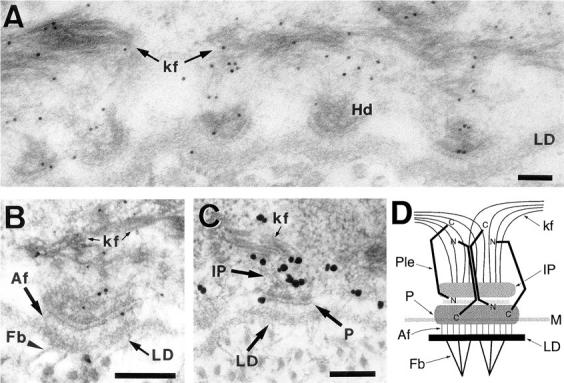
Postembedding immunogold electron microscopy of plectin in rat skin (A–C) and schematic model highlighting plectin's putative role as a stabilizer of hemidesmosomes (D). (A–C) Indirect immunogold labeling of plectin molecules using antibodies specific to the rod domain (A; mAb 7A8, 5 nm gold), the amino terminus (B; antiserum E1A, 1 nm gold silver enhanced), and the carboxy terminus (C; antiserum 135C, 5 nm gold silver enhanced). An overview over the interface between the basal keratinocyte layer and the basement membrane showing five regularly aligned hemidesmosomes is presented in A, and details of the hemidesmosomal ultrastructure comprising the inner plate linked to keratin filaments, plaque, lamina lucida, lamina densa, and extracellular anchoring filaments and fibrils can be identified in the ∼60-nm-thick sections shown in B and C. Note that all three domain-specific immunoreagents were found to be reactive with basolateral components of hemidesmosomes as well as cytoplasmic keratin filaments. Also note that specific immunolabeling using antiserum E1A was weak compared to mAb 7A8 and antiserum 135C and was observed only in combination with 1 nm, but not 5 or 10 nm, colloidal gold-conjugated secondary antibodies, indicating low accessibility of amino-terminal plectin epitopes, probably because of steric hindrance. Bars, 100 nM. (D) In this schematic drawing of a hemidesmosome, plectin molecules are depicted to connect the plaque structure with cytokeratin filaments anchored at the inner plate in a clamp-like fashion via interaction sites located at their opposite ends. Note that plectin molecules in dimeric (two polypeptide chains arranged parallel) as well as tetrameric forms (two dimers arranged anti-parallel) could provide IF- and integrin β4-binding sites at opposite ends and would be able to bridge the entire cytoplasmic domain of the hemidesmosome via their ∼200-nm-long rod domains. Hd, hemidesmosome; LD, lamina densa; kf, keratin filaments; Af, anchoring filaments; Fb, anchoring fibrils; IP, inner plate; P, plaque; Ple, plectin; M, cell membrane; N, plectin amino terminus; C, plectin carboxy terminus.
None of the domain-specific anti-plectin antibodies showed noticeable preference in labeling of any hemidesmosomal substructures. Furthermore, based on the resolution of the indirect immunogold labeling used (20–30 nm), it was unlikely that plectin molecules in hemidesmosomal basolateral regions were oriented in the same way and arranged in equidistal positions. These data were consistent with a model, in which randomly oriented plectin molecules can span over the entire distance between plasma membrane-associated hemidesmosomal integrin receptor complexes and hemidesmosomal inner plate-anchored cytokeratin filament bundles (Fig. 9 D).
Overexpression of Integrin β4 Cytoplasmic Tail Domain Leads to Intermediate Filament Collapse in PtK2 and 804G Cells
To assess whether the cytoplasmic domains of integrin β4 involved in plectin interaction would have any influence on cytoskeleton integrity, specifically IF-anchorage at plasma membrane sites, we monitored vimentin and cytokeratin network arrays in transfected PtK2 cells using double and triple immunofluorescence microscopy. Upon expression of β4-F1,2LF3,4CGFP, cytokeratin arrays were found aggregated and partially codistributed with the integrin mutant protein in perinuclear areas (Fig. 10, A and B). In fact, this phenotype was very similar to that observed after cotransfection of PtK2 cells using integrin β4 and plectin vector constructs in combination (compare to Fig. 7 C). Triple immunofluorescence microscopy revealed that both the vimentin and cytokeratin network arrays had undergone collapse in such cells (Fig. 10, C–E). Similar observations were made when PtK2 cells were transfected using the integrin β4 tail construct β4-F3,4C′myc (Fig. 7, F–H). In contrast, ectopic expression of β4-F1,2L′myc, which lacked the carboxy-terminal part contained in β4-F1,2LF3,4CGFP/myc and β4-F3,4C′myc, did not lead to such a drastic collapse of IF networks (Fig. 10, I–K). Instead, a partial association of the expressed protein with the IF network, specifically with a dense perinuclear ring-like structure, was observed. This ring-like structure, which was also present in untransfected cells, appeared more pronounced in transfected cells; and cytoplasmic IFs, especially the cytokeratin network, appeared diminished (Fig. 10, J and K). Because both, β4-F1,2LF3,4CGFP and β4-F3,4C′myc, but not β4-F1,2L′myc contained the carboxy-terminal region shown to exhibit plectin-binding activity in vitro, we conclude that this tail domain was sufficient to cause the collapse of both IF systems in PtK2 cells.
To confirm these studies, similar transfection experiments were carried out using rat bladder carcinoma 804G cells, a cell line that forms hemidesmosome-like structures when grown on uncoated glass coverslips or plastic tissue culture dishes (Riddelle et al., 1991). Similar to PtK2 cells, transfection of 804G cells with the plasmid encoding β4-F1,2LF3,4Cmyc led to bundling of cytokeratin filaments and their partial collapse in perinuclear areas; the ectopically expressed protein was again visualized within dense, patchy structures, and partly colocalized with the collapsed cytokeratin filaments (Fig. 11, A and B). Complete network collapse was observed in cells expressing β4-F3,4C′myc; in this case, the mutant protein was found colocalized with cytokeratins in dense, ring-like structures around the nucleus (Fig. 11, C and D). In cells transfected with plasmids encoding β4-F1,2L′myc (Fig. 11, E and F), the expressed protein was found more or less diffusely distributed throughout the cell, with an apparent association with dense, perinuclear cytokeratin structures. β4-F1,2 myc (Fig. 11, G and H) was diffusely distributed throughout the cytoplasm with no apparent effect on the cytokeratin network array. Thus, the phenotypes of transfected 804G cells were fully consistent with those of PtK2 cells.
Figure 11.
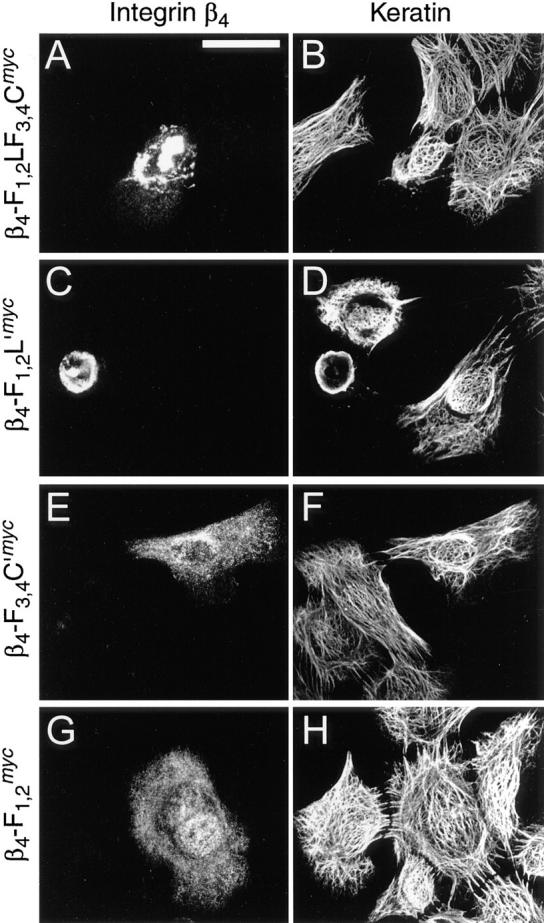
Collapse of the cytokeratin filament networks in 804G cells. Cells plated on glass coverslips were transfected with cDNA constructs encoding mutant integrin β4-F1,2LF3,4Cmyc (A and B), β4-F3,4C′myc (C and D), β4-F1,2L′myc (E and F), or β4-F1,2 myc (G and H). After 24 h, cells were fixed and double-stained. (A, C, E, and G), c-myc-tagged (Texas red optics) integrin β4 proteins. (B, D, F, and H), cytokeratin (FITC optics). Note cytokeratin filament network collapse in cells expressing β4-F1,2LF3,4Cmyc or β4-F3,4C′myc (B and D), whereas no such collapse is observed with β4-F1,2L′myc or β4-F1,2 myc (F and H). Bar, 25 μm.
Expression of Integrin β4 Cytoplasmic Tail Domain Prevents Hemidesmosome Association of Plectin in 804G Cells
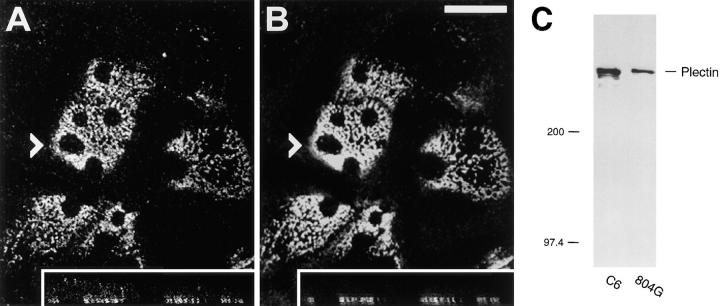
Because some of the integrin β4 mutant proteins overexpressed in 804G and PtK2 cells had a dramatic impact on cytoplasmic IF network organization, and were able to bind to recombinant plectin fragments, we wanted to test whether overexpression of these polypeptides in such cells would also affect the distribution of endogenous cytoplasmic plectin structures. As revealed by double immunofluorescence microscopy of untransfected 804G cells using antiserum raised against the 17 most carboxy-terminal amino acids of the integrin β4 subunit (Giancotti et al., 1992), and mAb 5B3 reacting with plectin's central rod domain (Foisner et al., 1994), plectin and integrin β4 showed colocalization in a pattern previously described as Swiss cheese-like (Jones et al., 1991) and characteristic of the hemidesmosome-like structures abundantly expressed by these cells (Fig. 12, A and B). Both antigens were predominantly located at the basal membrane of these cells, as confirmed by confocal laser scanning microscopy of vertical sections (Fig. 12, A and B, insets). This hemidesmosome-associated 5B3-immunoreactive plectin species represented the major form of the protein in 804G cells, as confirmed by Western blot analysis of 804G cell lysates. Using mAb5B3, a single major immunoreactive band of apparent high molecular weight (>300,000) comigrating with plectin isolated from rat glioma C6 cells, the original source of the immunogen (Foisner et al., 1994), was observed (Fig. 12 C).
Figure 12.
Immunolocalization (A and B) and Western blotting (C) of endogenous plectin in 804G cells. Cells were grown on glass coverslips to medium density, fixed, and double-immunostained using rabbit antibodies to integrin β4 (A) and mAb 5B3 to plectin (B). Note colocalization of plectin with integrin β4 in Swiss cheese-like patterns typical of hemidesmosomes formed by this cell line (plane of focus is just above substrate level). Insets show optical vertical sections with their starting positions indicated by arrowheads. Note localization of the bulk of integrin β4 as well as plectin at the basal side of the cells. Bar, 25 μm. Purified plectin from the rat glioma cell line C6 (C, lane C6) and a 804G cell lysate (C, lane 804G) were analyzed by SDS-4.5% PAGE and transferred onto a nitrocellulose membrane. Plectin was detected using mAb 5B3. Molecular weight markers were as indicated (×10−3).
Upon transient expression of integrin β4 mutant proteins containing one or both of the putative plectin-binding regions, the distribution of endogenous plectin was significantly altered. In cells expressing mutant proteins that caused the collapse of cytokeratin network arrays, such as β4-F1,2LF3,4Cmyc and β4-F3,4C′myc, the punctuate plectin-staining pattern typical of hemidesmosomes was greatly diminished (Fig. 13, A–D). Instead, remaining plectin-specific staining appeared rather diffuse, with the possible exception of peripheral areas, where a few positively stained filamentous structures became apparent. Similarly, upon expression of β4-F1,2L′myc, the punctuate plectin-specific signal associated with hemidesmosomes was hardly detectable anymore (Fig. 13 F). In fact, such cells seemed almost completely depleted of plectin. Expression of the integrin mutant protein corresponding to the first and second FNIII repeat (β4-F1,2 myc), which lacked a plectin-binding site and was diffusely distributed over the entire cell (Fig. 13 G), left the punctuate plectin-staining pattern intact, indicating no alteration of plectin's localization at hemidesmosomes (Fig. 13 H).
Figure 13.
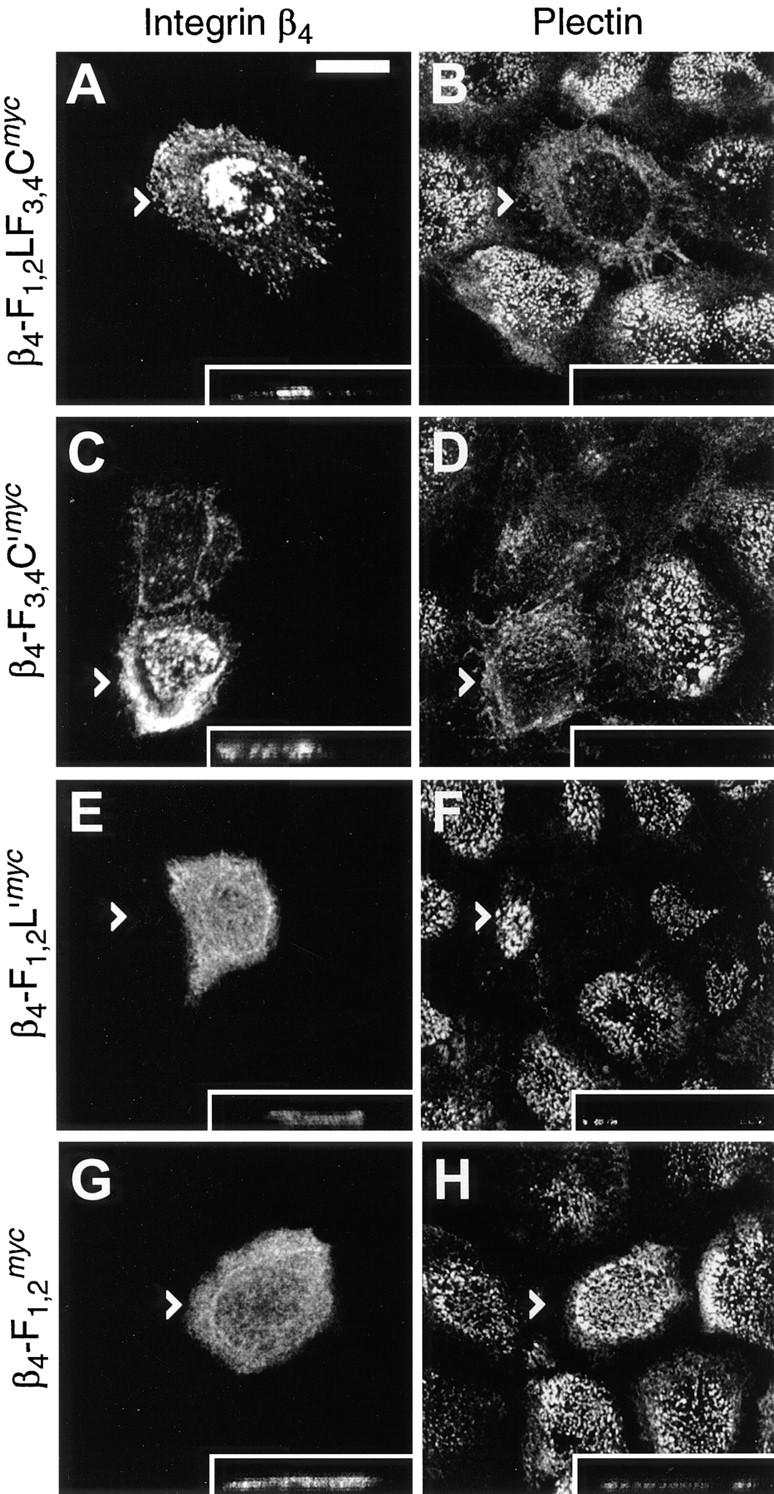
Immunolocalization of plectin in 804G cells overexpressing integrin β4 cytoplasmic tail domain fragments. Cells grown on glass coverslips were transfected with cDNA constructs encoding integrin β4 mutant proteins β4-F1,2LF3,4Cmyc (A and B), β4-F3,4C′myc (C and D), β4-F1,2L′myc (E and F), or β4-F1,2 myc (G and H). Cells were fixed 8 h posttransfection and double-stained using anti-c-myc antibodies (A, C, E, and G) and anti-plectin mAb 5B3 (B, D, F, and H). Confocal laser scanning microscopy was used to generate extended focus images over the total height of the cells. Arrowheads indicate positions of optical vertical sections shown in insets (tips of arrowheads correspond to left hand margins of insets). Note, plectin staining is substantially diminished at hemidesmosomes of cells overexpressing β4-F1,2LF3,4Cmyc, β4-F3,4C′myc, or β4-F1,2L′myc (B, D, and F), but still present at such structures in cells overexpressing β4-F1,2 myc (H), as judged by the absence or presence of the typical hemidesmosomal staining pattern. Bar, 10 μm.
Discussion
The uniquely long cytoplasmic domain of the β4 subunit protein has been implicated in connecting the α6β4 integrin complex with cytoskeletal elements (Schwarz et al., 1990; Spinardi et al., 1993; Borradori and Sonnenberg, 1996; Green and Jones, 1996), likely via cytoplasmic components of hemidesmosomes that are capable of cytokeratin filament interaction. Plectin, as a versatile IF-binding and cytoskeletal linker protein that has been shown to occur at hemidesmosomes (Wiche et al., 1984), was a strong candidate for mediating this interaction. In this report, we demonstrate a direct interaction between recombinant integrin β4 and plectin proteins using two independent in vitro binding assays and in vivo cotransfection experiments. Furthermore, we show that integrin β4 polypeptides containing a ∼250–amino acid-long carboxy-terminal sequence led to bundling and collapse of IF network arrays, possibly through interference with IF-membrane association, and affected the subcellular distribution of cellular plectin when ectopically expressed in a nonmembrane- associated state.
The integrin β4 cDNA constructs used in our study were designed such that they encoded the entire or truncated forms of the cytoplasmic tail domain starting with the first FNIII repeat. One of the objectives pursued in this way was to express, in cells, integrin β4 proteins that would not associate with the cell membrane but would remain soluble in the cytoplasm. In this form they would also resemble fragments of ∼70 kD resulting from proteolytic processing of cellular integrin β4, which has been investigated by Giancotti et al. (1992). As suggested by these authors, formation of these cytoplasmic, nonmembrane-attached β4 subunit fragments is likely to be functional and related to the remodeling of adhesive and cytoskeletal structures occurring in the skin when basal cells detach from the basement membrane at the onset of differentiation and movement to upper cell layers. In addition, we used plectin cDNA constructs that encoded parts of the amino- and carboxy-terminal globular domains, both of which have been shown to harbor functional sites, including a highly conserved amino-terminal actin-binding domain (Elliott et al., 1997), and an IF-binding site within the carboxy-terminal repeat 5 domain (Nikolic et al., 1996).
We used affinity blotting, a commonly used method to detect direct interaction between molecules (for review see Phizicky and Fields, 1995), to demonstrate that plectin's carboxy-terminal repeat 6 domain is essential for binding to integrin β4. However, our data also indicated that other synergy sites contained in domains flanking repeat 6 facilitated binding, because larger fragments extending beyond repeat 6 showed stronger binding. Additional integrin β4-binding sites were found to be contained in the amino-terminal globular domain of plectin. These binding data were confirmed by a microtiter plate-binding assay using Eu3+-labeled proteins as soluble reagents. Moreover, at least two separate plectin-binding sites were found within the cytoplasmic domain of integrin β4, one within the 142–amino acid-long segment connecting the two pairs of FNIII repeats, the other within the ultimate tail domain of the molecule. Both showed affinities for the same amino- as well as carboxy-terminal parts of plectin, as indicated by competition assays. Cotransfection of PtK2 cells with plasmids encoding mutant integrin β4 and plectin polypeptides fully confirmed these in vitro results. In fact, the in vivo expression experiments can be considered essentially as a third binding assay, as interacting proteins were found codistributed in patterns distinctly different from those observed when only one of the mutant proteins was ectopically expressed in cells. Thus, in vitro and in vivo binding data both suggested that plectin-integrin β4 binding was complex, involving multiple interfaces of the proteins.
Sequences in the linker segment between the first and second FNIII repeat pair have been shown to be important for integrin β4 functions. Spinardi et al. (1993) showed that a 303–amino acid-long region of the integrin β4 cytoplasmic tail domain starting with the first FNIII repeat was essential for association of the molecule with hemidesmosomes. The necessary sequences were later narrowed down to the second FNIII repeat and a stretch of 27 amino acids (1,329–1,355) of the linker segment (Niessen et al., 1997a ). Furthermore, the regions responsible for hemidesmosome localization have also been implicated in regulating the distribution of HD1 upon overexpression in COS-7 cells (Niessen et al., 1997a ). We show here for the first time that this linker segment apparently is also involved in integrin β4 self-interaction, aside from harboring one of the two plectin-binding regions identified in our study. Self-association of integrin β4 may allow for efficient, cooperative clustering of these molecules in the dense hemidesmosomal plaque. Interestingly, our competition binding experiments indicated that plectin fragments containing integrin-binding domains attenuated the self-association of integrin β4, suggesting a potential role of plectin in the regulation of this process. Whether integrin β4 and plectin molecules compete for binding sites located close enough to sterically hinder binding of the second ligand, or even compete for the same sites, remains to be established. Further functional diversity of the segment between the FNIII repeat pairs is further underlined by the complexity of the gene locus encoding it and the identification of variants generated by alternative splicing of exons 33 and 35 within this locus (Hogervorst et al., 1990; Tamura et al., 1990; Pulkkinen et al., 1997).
The integrin β4 tail segment following the second FNIII repeat pair apparently is another subdomain of integrin β4 involved in multiple functions. Similar to the segment between the first and second FNIII repeat pair, this region exhibited binding to the integrin β4 cytoplasmic domain itself, as well as plectin amino- and carboxy-terminal domains. In addition, fragments containing this segment were highly efficient in causing IF collapse upon overexpression in PtK2 and 804G cells. In none of the previous studies we are aware of were specific functions assigned to, or binding partners identified in this region of integrin β4. A possible reason for this may be the predominant usage of carboxy terminally truncated mutants of integrin β4, because functions of carboxy-terminal fragments may easily escape detection, particularly when they are partially redundant, such as in the case of integrin β4 self-interaction and binding to plectin.
The role of the FNIII repeats remains unclear. We have shown that the pairs of FNIII repeats present in the integrin β4 cytoplasmic tail do not interact directly with plectin. However, they may be important for providing a structural framework allowing other parts to function efficiently. Supporting this point, integrin β4-LF3,4C, which lacked the first FNIII repeat pair, showed reduced binding to plectin in vitro, compared to β4-F1,2LF3,4C. Alternatively, some of the repeats may directly take part in interaction with other components. In a recent study, the second FNIII repeat, in addition to parts of the linker segment between the two FNIII pairs, has been shown to be crucial for the targeting of recombinant integrin β4 mutants to hemidesmosomes and interaction with HD1 (Niessen et al., 1997a ). Additionally, the FNIII repeats may also interact with elements of the signal transduction pathway(s) coupled to the α6β4 integrin (Mainiero et al., 1995).
In view of plectin's putative major role as a cytoskeletal linker and stabilizer molecule, one may expect that overexpression of a plectin-binding protein would exert some influence on plectin's cellular function(s) by competitive binding to plectin, possibly leading to blocking of, or interference with, some of the endogenous binding partners. This could result in dominant negative phenotypes, detectable in structures, the integrity of which is dependent on a particular plectin linkage function. In line with this notion, we found that overexpression of integrin β4 mutant polypeptides containing the second putative plectin-binding site located in the carboxy-terminal region (β4-F1,2LF3,4Cmyc /GFP and β4-F3,4C′myc) led to the complete collapse of vimentin and cytokeratin IF network systems in PtK2 and 804G cells. Because ectopically expressed β4-F1,2L′myc, which lacked this domain but still contained the more amino-terminal plectin-binding region, was much less effective, we conclude the carboxy-terminal tail segment of integrin β4, starting with the second FNIII repeat pair, is the dominant effector of this phenotype.
Niessen et al. (1997a ,b), who expressed membrane- associated integrin β4 molecules comprising the complete or parts of the cytoplasmic domain in COS-7 cells, contrary to our results, did not observe any cytoskeletal phenotype. There are a number of possible explanations for this difference. First, integrin mutant effector molecules need to be freely available (soluble) in the cytoplasm, because in a membrane-bound state they may be too distant from target plectin molecules. Second, the membrane-bound mutant proteins do in fact interact with plectin, but keep the IF network bound to the membrane, and thus, in a noncollapsed state. Third, though unlikely, COS-7 cells are phenotypically different from PtK2 and 804G cells. We favor the second explanation, which allows for a mechanism, in which cytosolic integrin β4 mutant proteins are in competition with their wild-type counterparts as well as other membrane-associated plectin-binding proteins, such as fodrin and α-spectrin (Herrmann and Wiche, 1987). Prohibiting the attachment of IFs to the plasma membrane, this situation could eventually lead to a collapse of IFs toward the perinuclear region. This interpretation is supported by the observation that endogenous 804G cell plectin, the localization of which was restricted largely to basal hemidesmosome-like structures in untransfected cells, upon expression of the plectin-binding sites containing integrin β4 polypeptides (β4-F1,2LF3,4Cmyc and β4-F3,4Cmyc), showed a diffuse cytoplasmic localization. Downregulation of plectin expression, in addition to relocalization of the membrane-bound species, may have been responsible for the even more drastic phenotype in the case of β4-F1,2Lmyc, where endogenous plectin was hardly detectable any more (Fig. 13 F). Similar phenomena involving IF collapse and/or relocalization of endogenous proteins have been observed after overexpression of wild-type or mutant versions of IF-associated proteins, including desmoplakin (Stappenbeck and Green, 1992), plectin (Wiche et al., 1993), and various IF subunit proteins themselves (Albers and Fuchs, 1987; for review also see Fuchs and Weber, 1994), and after microinjection of antibodies to a whole range of antigens, including IF-associated and other proteins, into cultured cells.
In all, the plectin phenotypes of cells, transfected with various truncated versions of the integrin β4 cytoplasmic domain, were fully compatible with the data-characterizing plectin as a direct interaction partner of hemidesmosomal integrin β4. Notably, mutant integrin β4 proteins leading to filament collapse also caused relocalization of plectin from hemidesmosomes in 804G cells, suggesting that the collapse may in fact be indirect, mediated through plectin. Furthermore, overexpression of integrin β4 polypeptides, containing the different plectin-binding sites identified individually, resulted in distinct phenotypes, consistent in two different cell lines. Hence, our findings are in favor of a hypothesis put forward by Giancotti et al. (1992), that proteolytic processing of the β4 subunit of the basement membrane receptor integrin α6β4, resulting in a number of defined cleavage products is related to the remodeling of adhesive and cytoskeletal structures. Additional support for this proposal is provided by our unpublished observation that transfected 804G cells overexpressing integrin β4 polypeptides, representing such proteolytic fragments, frequently had a rounder morphology, were apparently in the process of detachment from the substrate, and could be found in a second cell layer in dense cultures.
Recent studies with patients suffering from EBS-MD linked this disease to defects in plectin expression (McLean et al., 1996). Furthermore, in plectin (−/−) mice with a similar phenotype, the hemidesmosomes in epidermal keratinocytes were found to be ultrastructurally intact and numerous filaments emanated from their cytoplasmic face, indicating that their ability to serve as filament anchorage sites was at least to a certain extent preserved (Andrä et al., 1997). In contrast, BPAG1-deficient mice clearly lacked the hemidesmosomal inner plate, and consequently did not show keratin filament anchorage at the basal cell surface membrane (Guo et al., 1995). Thus, plectin is probably dispensable for the formation of hemidesmosomes. However, there is mounting evidence that this protein is important for the integrity and stability of hemidesmosomal junctions. First, the expression of integrin β4 was found to be significantly reduced in plectin-deficient mice (Andrä et al., 1997), as has been reported for mice deficient in another binding partner of integrin β4, the integrin subunit α6 (Georges-Labouesse et al., 1996), and second, hemidesmosome structures of plectin-deficient mice seemed to be fragile, as a peculiar, bipartite distribution of BPAG1 was observed in a discontinuous pattern both at the bottom and the top of blisters in the skin of these animals; this suggested a mechanism of cell breakage in which rupture can occur irregularly on both sides (apical and basal) of the hemidesmosome inner plate, or within the plate itself.
Based on a) the results reported in this study, in particular, plectin's ability to interact via carboxy- as well as amino-terminal domains with the cytoplasmic tail of integrin β4 and its close association with hemidesmosomes as demonstrated by immunoelectron microscopy, b) our previous identification of a highly conserved IF (cytokeratin)- binding site in the carboxy-terminal globular domain of plectin molecules (Nikolic et al., 1996), and c) the skin phenotype observed in plectin (−/−) mice (Andrä et al., 1997) and EBS-MD patients, we propose that plectin molecules act as stabilizers of hemidesmosomes according to the schematic model depicted in Fig. 9 D. In this model, plectin molecules form clamps bridging hemidesmosomal components with the IF networks by directly interacting with both plasma membrane-associated integrin β4 and cytoplasmic cytokeratin filaments. Binding sites for integrin β4 in both the amino- and carboxy-terminal globular domains of plectin would allow parallel as well as antiparallel plectin oligomeres to form interactions on both ends (see Foisner and Wiche, 1987, and Wiche et al., 1991, for discussions of putative dimeric and tetrameric plectin molecule structures). Multiple mutual binding sites on both integrin β4 and plectin may afford stronger resistance towards the great mechanical stress hemidesmosomes must be exposed to, yet retain the flexibility needed for such dynamic structures as they have shown to be (Kitajima et al., 1992; Riddelle et al., 1992).
In conclusion, we have identified the β4 subunit of the hemidesmosome-associated laminin receptor complex integrin α6β4 as a novel interaction partner of plectin and we have mapped interacting domains on both molecules. In a more global picture our results lend further support to the proposed role of plectin as a versatile and essential cytoskeletal linker protein that may contribute to the cytoarchitectural organization and mechanical reinforcement of cells by cross-linking IFs, interlinking them with other cytoskeletal network systems, and participating in their anchorage at the plasma membrane at specialized structures.
Acknowledgments
We wish to thank K. Andrä, J. Dubendorff, B. Nikolic, and M. Zachlederova, all from this laboratory, for providing some of the plectin cDNA constructs and antibodies used in this study. B. Nikolic performed the Western blot analysis shown in Fig. 12 C. We also wish to thank F.G. Giancotti (New York University School of Medicine, New York, NY) for kindly exchanging antibodies with us.
Abbreviations used in this paper
- EBS-MD
epidermolysis bullosa simplex combined with muscular dystrophy
- FNIII
fibronectin type III repeat
- GFP
green fluorescent protein
- IF
intermediate filament
Footnotes
J.M. de Pereda and S. Reipert were recipients of postdoctoral fellowships from the European Community Human Capital and Mobility Program, and the Wellcome Trust, U.K., respectively. This work was supported by grants from the Austrian Science Research Fund.
Address all correspondence to Gerhard Wiche, Vienna Biocenter, Institute of Biochemistry and Molecular Cell Biology, University of Vienna, Dr. Bohr-Gasse 9, 1030 Vienna, Austria. Tel.: 43 (1) 79515-5119. Fax: 43 (1) 79515-5121. E-mail: wiche@abc.univie.ac.at
References
- Albers K, Fuchs E. The expression of mutant epidermal keratin cDNAs transfected in simple epithelial and squamous cell carcinoma lines. J Cell Biol. 1987;105:791–806. doi: 10.1083/jcb.105.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrä K, Lassmann H, Bittner R, Shorny S, Fässler R, Propst F, Wiche G. Targeted inactivation of plectin reveals essential function in maintaining the integrity of skin, muscle, and heart cytoarchitecture. Genes Dev. 1997;11:3143–3156. doi: 10.1101/gad.11.23.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes WM. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradori L, Sonnenberg A. Hemidesmosomes: roles in adhesion, signaling and human diseases. Curr Opin Cell Biol. 1996;8:647–656. doi: 10.1016/s0955-0674(96)80106-2. [DOI] [PubMed] [Google Scholar]
- Buck CA, Horwitz AF. Cell surface receptors for extracellular matrix molecules. Annu Rev Cell Biol. 1987;3:179–205. doi: 10.1146/annurev.cb.03.110187.001143. [DOI] [PubMed] [Google Scholar]
- Carter WG, Kaur P, Gil SG, Gahr PJ, Wayner EA. Distinct functions for integrins α3β1 in focal adhesions and α6β4/bullous pemphigoid antigen in a new stable anchoring contact (SAC) of keratinocytes: relation to hemidesmosomes. J Cell Biol. 1990;111:3141–3154. doi: 10.1083/jcb.111.6.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanas S, Pulkkinen L, Gache Y, Smith FJD, McLean WHI, Uitto J, Ortonne JP, Meneguzzi G. A homozygous nonsense mutation in the PLEC1 gene in patients with epidermolysis bullosa simplex with muscular dystrophy. J Clin Invest. 1996;98:2196–2200. doi: 10.1172/JCI119028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Tamura RN, Kajiji S, Bondanza S, Rossino P, Cancedda R, Marchisio PC, Quaranta V. Polarized integrin mediated human keratinocyte adhesion to basal lamina. Proc Natl Acad Sci USA. 1990;87:6888–6892. doi: 10.1073/pnas.87.17.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk H, Franke WW, Dragosics B, Zeiler I. Pathology of cytoskeleton of liver cells: demonstration of Mallory bodies (alcoholic hyalin) in murine and human hepatocytes by immunofluorescence microscopy using antibodies to cytokeratin polypeptides from hepatocytes. Hepatology. 1981;1:9–20. doi: 10.1002/hep.1840010103. [DOI] [PubMed] [Google Scholar]
- Dowling J, Yu Q-C, Fuchs E. β4integrin is required for hemidesmosome formation, cell adhesion and cell survival. J Cell Biol. 1996;134:559–572. doi: 10.1083/jcb.134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eger A, Stockinger A, Wiche G, Foisner R. Polarisation-dependent association of plectin with desmoplakin and the lateral submembrane skeleton in MDCK cells. J Cell Sci. 1997;110:1307–1316. doi: 10.1242/jcs.110.11.1307. [DOI] [PubMed] [Google Scholar]
- Elliott CE, Becker B, Oehler S, Castañón MJ, Hauptmann R, Wiche G. Plectin transcript diversity: identification and tissue distribution of variants with distinct first coding exons and rodless isoforms. Genomics. 1997;42:115–125. doi: 10.1006/geno.1997.4724. [DOI] [PubMed] [Google Scholar]
- Foisner R, Wiche G. Structure and hydrodynamic properties of plectin molecules. J Mol Biol. 1987;198:515–531. doi: 10.1016/0022-2836(87)90297-x. [DOI] [PubMed] [Google Scholar]
- Foisner R, Leichtfried FE, Herrmann H, Small JV, Lawson D, Wiche G. Cytoskeleton-associated plectin: in situ localization, in vitro reconstitution, and binding to immobilized intermediate filament proteins. J Cell Biol. 1988;106:723–733. doi: 10.1083/jcb.106.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R, Feldman B, Sander L, Seifert G, Artlieb U, Wiche G. A panel of monoclonal antibodies to rat plectin: distinction by epitope mapping and immunoreactivity with different tissues and cell lines. Acta Histochem. 1994;96:421–438. doi: 10.1016/S0065-1281(11)80029-2. [DOI] [PubMed] [Google Scholar]
- Fontao L, Dirrig S, Owaribe K, Kedinger M, Launay JF. Polarized expression of HD1: relationship with the cytoskeleton in cultured human colonic carcinoma cells. Exp Cell Res. 1997;231:319–327. doi: 10.1006/excr.1996.3465. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- Gache Y, Chavanas S, Lacour J-P, Wiche G, Owaribe K, Meneguzzi G, Ortonne JP. Defective expression of plectin/HD1 in epidermolysis bullosa simplex with muscular dystrophy. J Clin Invest. 1996;97:2289–2298. doi: 10.1172/JCI118671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges-Labouesse E, Messaddeq N, Yehia G, Cadalbert L, Dierich A, Le-Meur M. Absence of integrin alpha 6 leads to epidermolysis bullosa and neonatal death in mice. Nat Genet. 1996;13:370–373. doi: 10.1038/ng0796-370. [DOI] [PubMed] [Google Scholar]
- Giancotti FG, Mainiero F. Integrin-mediated adhesion and signaling in tumorigenesis. Biochim Biophys Acta. 1994;1198:47–64. doi: 10.1016/0304-419x(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Giancotti FG, Stepp MA, Suzuki S, Engvall E, Ruoslahti E. Proteolytic processing of endogenous and recombinant β4 integrin subunit. J Cell Biol. 1992;118:951–959. doi: 10.1083/jcb.118.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg MH, Loftus JC, Plow EF. Cytoadhesins, integrins and platelets. Thromb Haemostasis. 1988;59:1–6. [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green KJ, Jones JCR. Desmosomes and hemidesmosomes: structure and function of molecular components. FASEB (Fed Am Soc Exp Biol) J. 1996;10:871–881. doi: 10.1096/fasebj.10.8.8666164. [DOI] [PubMed] [Google Scholar]
- Green KJ, Virata MLA, Elgart GW, Stanley JR, Parry DAD. Comparative structural analysis of desmoplakin, bullous pemphigoid antigen and plectin: members of a new gene family involved in organization of intermediate filaments. Int J Biol Macromol. 1992;14:145–153. doi: 10.1016/s0141-8130(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Guo L, Degenstein L, Dowling J, Yu QC, Wollmann R, Perman B, Fuchs E. Gene targeting of BPAG1: abnormalities in mechanical strength and cell migration in stratified epithelia and neurological degeneration. Cell. 1995;81:233–243. doi: 10.1016/0092-8674(95)90333-x. [DOI] [PubMed] [Google Scholar]
- Hemler ME. VLA proteins in the integrin family: structures, functions, and their role in leukocytes. Annu Rev Immunol. 1990;8:365–400. doi: 10.1146/annurev.iy.08.040190.002053. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Wiche G. Plectin and IFAP-300K are homologous proteins binding to microtubule-associated proteins 1 and 2 and to the 240-kilodalton subunit of spectrin. J Biol Chem. 1987;262:1320–1325. [PubMed] [Google Scholar]
- Hieda Y, Nishizawa Y, Uematsu J, Owaribe K. Identification of a new hemidesmosomal protein HD1: a major high molecular mass component of isolated hemidesmosomes. J Cell Biol. 1992;116:1497–1506. doi: 10.1083/jcb.116.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogervorst F, Kuikman I, Kr AEG, von dem Borne, Sonnenberg A. Cloning and sequence analysis of β4cDNA: an integrin subunit that contains a unique 118-kd cytoplasmic domain. EMBO (Eur Mol Biol Organ) J. 1990;9:745–770. doi: 10.1002/j.1460-2075.1990.tb08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Integrins: a family of cell surface receptors. Cell. 1987;48:549–554. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: versatility, modulation and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Izumi K, Hirao Y, Hopp L, Oyasu R. In vitro induction of ornithine decarboxylase in urinary bladder carcinoma cells. Cancer Res. 1981;41:405–409. [PubMed] [Google Scholar]
- Jones JCR, Kurpakus MA, Cooper HM, Quaranta V. A function for the integrin α6β4in the hemidesmosome. Cell Regul. 1991;2:427–438. doi: 10.1091/mbc.2.6.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano RL, Haskill S. Signal transduction from the extracellular matrix. J Cell Biol. 1993;120:577–585. doi: 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiji S, Tamura RN, Quaranta V. A novel integrin (αEβ4) from human epithelial cells suggests a fourth family of integrin adhesion receptors. EMBO (Eur Mol Biol Organ) J. 1989;8:673–680. doi: 10.1002/j.1460-2075.1989.tb03425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima Y, Owaribe K, Nishizawa Y, Jokura Y, Yaoita H. Phorbol ester- and calcium-induced reorganization of 180-kDa bullous pemphigoid antigen on the ventral surface of cultured human keratinocytes as studied by immunofluorescence and immunoelectron microscopy. Exp Cell Res. 1992;203:17–24. doi: 10.1016/0014-4827(92)90034-6. [DOI] [PubMed] [Google Scholar]
- Klatte DH, Jones JCR. Purification of the 230-kD bullous pemphigoid antigen (BP230) from bovine tongue mucosa: structural analysis and assessment of BP230 tissue distribution using a new monoclonal antibody. J Invest Derm. 1994;102:39–44. doi: 10.1111/1523-1747.ep12371728. [DOI] [PubMed] [Google Scholar]
- Kurpakus MA, Jones JCR. A novel hemidesmosomal plaque component: tissue distribution and incorporation into assembling hemidesmosomes. Exp Cell Res. 1991;194:139–146. doi: 10.1016/0014-4827(91)90143-i. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee EC, Lotz MM, Steele GD, Jr, Mercurio AM. The integrin α6β4is a laminin receptor. J Cell Biol. 1992;117:671–678. doi: 10.1083/jcb.117.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CG, Maercker C, Castañón MJ, Hauptmann R, Wiche G. Human plectin: organization of the gene, sequence analysis, and chromosome localization (8q24) Proc Natl Acad Sci USA. 1996;93:4278–4283. doi: 10.1073/pnas.93.9.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainiero F, Pepe A, Wary KK, Spinardi L, Mohammadi M, Schlessinger J, Giancotti FG. Signal transduction by the α6β4 integrin: distinct β4subunit sites mediate recruitment of Shc/Grb2 and association with the cytoskeleton of hemidesmosomes. EMBO (Eur Mol Biol Organ) J. 1995;14:4470–4481. doi: 10.1002/j.1460-2075.1995.tb00126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean WHI, Pulkkinen L, Smith FJ, Rugg EL, Lane EB, Bullrich F, Burgeson RE, Amano S, Hudson DL, Owaribe K, et al. Loss of plectin causes epidermolysis bullosa with muscular dystrophy: cDNA cloning and genomic organization. Genes Dev. 1996;10:1724–1735. doi: 10.1101/gad.10.14.1724. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Hulsman EHM, Lauran CJM, Oomen I, Kuikman I, Sonnenberg A. A minimal region of the integrin β4subunit that is critical to its localization in hemidesmosomes regulates the distribution of HD1/ plectin in COS-7 cells. J Cell Sci. 1997a;110:1705–1716. doi: 10.1242/jcs.110.15.1705. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Hulsman EHM, Rots ES, Sánchez-Aparicio P, Sonnenberg A. Integrin α6β4forms a complex with the cytoskeletal protein HD1 and induces its redistribution in transfected COS-7 cells. Mol Biol Cell. 1997b;8:555–566. doi: 10.1091/mbc.8.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic B, Mac E, Nulty, Mir B, Wiche G. Basic amino acid residue cluster within nuclear targeting sequence motif is essential for cytoplasmic plectin-vimentin network junctions. J Cell Biol. 1996;134:1455–1467. doi: 10.1083/jcb.134.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owaribe K, Nishizawa Y, Franke WW. Isolation and characterization of hemidesmosomes from bovine corneal epithelial cells. Exp Cell Res. 1991;192:622–630. doi: 10.1016/0014-4827(91)90084-8. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak, J.M., I.M. Varndell. 1984. Immunolabeling for Electron Microscopy. Elsevier, Amsterdam/New York/Oxford.
- Pulkkinen L, Smith FJD, Shimizu H, Murata S, Yaoita H, Hachisuka H, Nishikawa T, McLean WHI, Uitto J. Homozygous deletion mutations in the plectin gene (PLEC1) in patients with epidermolysis bullosa simplex associated with late-onset muscular dystrophy. Hum Mol Genet. 1996;5:1539–1546. doi: 10.1093/hmg/5.10.1539. [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Kurtz K, Xu Y, Bruckner-Tuderman L, Uitto J. Genomic organization of the integrin β4gene (ITGB4): a homozygous splice-site mutation in a patient with junctional epidermolysis bullosa associated with pyloric atresia. Lab Invest. 1997;76:823–833. [PubMed] [Google Scholar]
- Rappersberger K, Binder M, Zonzits E, Hornick U, Wolff K. Immunogold staining of intermediate-sized filaments of the vimentin type in human skin: a postembedding immunoelectron microscopic study. J Invest Dermatol. 1990;94:700–705. doi: 10.1111/1523-1747.ep12876275. [DOI] [PubMed] [Google Scholar]
- Riddelle KS, Green KJ, Jones JCR. Formation of hemidesmosomes in vitro by a transformed rat bladder cell line. J Cell Biol. 1991;112:159–168. doi: 10.1083/jcb.112.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddelle KS, Hopkinson SB, Jones JCR. Hemidesmosomes in the epithelial cell line 804G: their fate during wound closure, mitosis and drug induced reorganization of the cytoskeleton. J Cell Sci. 1992;103:475–490. doi: 10.1242/jcs.103.2.475. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- Sánchez-Aparicio P, Martínez de Velasco AM, Niessen CM, Borradori L, Kuikman I, Hulsman EHM, Fässler R, Owaribe K, Sonnenberg A. The subcellular distribution of the high molecular mass protein, HD1, is determined by the cytoplasmic domain of the integrin β4subunit. J Cell Sci. 1997;110:169–178. doi: 10.1242/jcs.110.2.169. [DOI] [PubMed] [Google Scholar]
- Schwarz MA, Owaribe K, Kartenbeck J, Franke WW. Desmosomes and hemidesmosomes: constitutive molecular components. Annu Rev Cell Biol. 1990;6:461–491. doi: 10.1146/annurev.cb.06.110190.002333. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Ishida-Yamamoto A, Eady RA. The use of silver- enhanced 1-nm gold probes for light and electron microscopic localization of intra- and extracellular antigens in skin. J Histochem Cytochem. 1992;40:883–888. doi: 10.1177/40.6.1375240. [DOI] [PubMed] [Google Scholar]
- Skalli O, Jones JCR, Gagescu R, Goldman RD. IFAP300 is common to desmosomes and hemidesmosomes and is a possible linker of intermediate filaments to these junctions. J Cell Biol. 1994;125:159–170. doi: 10.1083/jcb.125.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FJD, Eady RAJ, Leigh IM, McMillan JR, Rugg EL, Kelsell DP, Bryant SP, Spurr NK, Geddes JF, Kirtschig G, et al. Plectin deficiency results in muscular dystrophy with epidermolysis bullosa. Nat Genet. 1996;13:450–457. doi: 10.1038/ng0896-450. [DOI] [PubMed] [Google Scholar]
- Soini E, Lövgren T. Time-resolved fluorescence of lanthanide probes and applications in biotechnology. CRC Crit Rev Anal Chem. 1987;18:105–154. [Google Scholar]
- Sonnenberg A, Linders CJT, Daams JH, Kennel SJ. The α6β1 (VLA-6) and α6β4protein complexes: tissue distribution and biochemical properties. J Cell Sci. 1990a;96:207–217. doi: 10.1242/jcs.96.2.207. [DOI] [PubMed] [Google Scholar]
- Sonnenberg A, Linders CJT, Modderman PW, Damsky CH, Aumailley M, Timpl T. Integrin recognition of different cell-binding fragments of laminin (P1, E3, E8) and evidence that α6β1 but not α6β4functions as a major receptor for fragment E8. J Cell Biol. 1990b;110:2145–2155. doi: 10.1083/jcb.110.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Calafat J, Janssen H, Daams H, van der Raaij-Helmer LMH, Falcioni R, Kennel SJ, Aplin JD, Baker J, Loizidou M, Garrod D. Integrin α6β4complex is located in hemidesmosomes, suggesting a major role in epidermal cell-basement membrane adhesion. J Cell Biol. 1991;113:907–917. doi: 10.1083/jcb.113.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinardi L, Ren Y-L, Sanders R, Giancotti FG. The β4 subunit cytoplasmic domain mediates the interaction of α6β4integrin with the cytoskeleton of hemidesmosomes. Mol Biol Cell. 1993;4:871–884. doi: 10.1091/mbc.4.9.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA. Adhesion receptors of the immune system. Nature. 1990;346:425–434. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- Stanley JR, Hawley-Nelson P, Yuspa SH, Shevach EM, Katz SI. Characterization of bullous pemphigoid antigen: a unique basement membrane protein of stratified squamous epithelia. Cell. 1981;24:897–903. doi: 10.1016/0092-8674(81)90115-x. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Green KJ. The desmoplakin carboxyl terminus coaligns with and specifically disrupts intermediate filament networks when expressed in cultured cells. J Cell Biol. 1992;116:1197–209. doi: 10.1083/jcb.116.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp MA, Spurr-Michaud S, Tisdale A, Elwell J, Gipson IK. Alpha 6 beta 4 integrin heterodimer is a component of hemidesmosomes. Proc Natl Acad Sci USA. 1990;87:8970–8974. doi: 10.1073/pnas.87.22.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierhof YD, Humbel BM, Schwarz H. Suitability of different silver enhancement methods applied to 1 nm colloidal gold particles: an immunoelectron microscopic study. J Electron Microsc Tech. 1991;17:336–343. doi: 10.1002/jemt.1060170307. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Naitoh Y. Amino acid sequence of a novel integrin β4subunit and primary expression of the mRNA in epithelial cells. EMBO (Eur Mol Biol Organ) J. 1990;9:757–763. doi: 10.1002/j.1460-2075.1990.tb08170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, Borisy GG. Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J Cell Biol. 1996;135:991–1007. doi: 10.1083/jcb.135.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura RN, Rozzo C, Starr L, Chambers J, Reichardt L, Cooper HM, Quaranta V. Epithelial integrin α6β4: complete primary structure of α6 and variant forms of β4 . J Cell Biol. 1990;111:1593–1604. doi: 10.1083/jcb.111.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Parry DAD, Klaus-Kovtun V, Steinert PM, Stanley JR. Comparison of molecularly cloned bullous pemphigoid antigen to desmoplakin I confirms that they define a new family of cell adhesion junction plaque proteins. J Biol Chem. 1991;266:12555–12559. [PubMed] [Google Scholar]
- Van der Neut R, Krimpenfort P, Calafat J, Niessen CM, Sonnenberg A. Epithelial detachment due to absence of hemidesmosomes in integrin beta 4 null mice. Nat Genet. 1996;13:366–369. doi: 10.1038/ng0796-366. [DOI] [PubMed] [Google Scholar]
- Villinger, W. 1991. Lowicryl resins. In Colloidal Gold: Principles, Methods, and Applications, Vol. 3. M.A. Hayat, editor. Academic Press, New York. 59–71.
- Watt FM, Kubler MD, Hotchin NA, Nicholson LJ, Adams JC. Regulation of keratinocyte terminal differentiation by integrin-extracellular matrix interactions. J Cell Sci. 1993;106:175–182. doi: 10.1242/jcs.106.1.175. [DOI] [PubMed] [Google Scholar]
- Wiche G, Krepler R, Artlieb U, Pytela R, Aberer W. Identification of plectin in different human cell types and immuno-localization at the epithelial basal cell surface membranes. Exp Cell Res. 1984;155:43–49. doi: 10.1016/0014-4827(84)90766-3. [DOI] [PubMed] [Google Scholar]
- Wiche G, Becker B, Luber K, Weitzer G, Castañón MJ, Hauptmann R, Stratowa C, Stewart M. Cloning and sequencing of rat plectin indicates a 466-kD polypeptide chain with a three-domain structure based on a central alpha-helical coiled coil. J Cell Biol. 1991;114:83–99. doi: 10.1083/jcb.114.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche G, Gromov D, Donovan A, Castañón MJ, Fuchs E. Expression of plectin mutant cDNA in cultured cells indicates a role of COOH-terminal domain in intermediate filament association. J Cell Biol. 1993;121:607–619. doi: 10.1083/jcb.121.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Dowling J, Yu Q-C, Kouklis P, Cleveland DW, Fuchs E. An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell. 1996;86:655–665. doi: 10.1016/s0092-8674(00)80138-5. [DOI] [PubMed] [Google Scholar]