Abstract
Members of the Rho GTPase family regulate the organization of the actin cytoskeleton in response to extracellular growth factors. We have identified three proteins that form a distinct branch of the Rho family: Rnd1, expressed mostly in brain and liver; Rnd2, highly expressed in testis; and Rnd3/RhoE, showing a ubiquitous low expression. At the subcellular level, Rnd1 is concentrated at adherens junctions both in confluent fibroblasts and in epithelial cells. Rnd1 has a low affinity for GDP and spontaneously exchanges nucleotide rapidly in a physiological buffer. Furthermore, Rnd1 lacks intrinsic GTPase activity suggesting that in vivo, it might be constitutively in a GTP-bound form. Expression of Rnd1 or Rnd3/RhoE in fibroblasts inhibits the formation of actin stress fibers, membrane ruffles, and integrin-based focal adhesions and induces loss of cell–substrate adhesion leading to cell rounding (hence Rnd for “round”). We suggest that these proteins control rearrangements of the actin cytoskeleton and changes in cell adhesion.
Rho GTPases control rearrangements of the actin cytoskeleton in response to extracellular signals. Like Ras, members of the Rho family are thought to cycle between an inactive GDP-bound form and an active GTP-bound form and three major regulators controlling their activity have been identified: (a) guanine nucleotide dissociation inhibitors (RhoGDIs) interact with the GDP-bound form of Rho GTPases to keep them in a “resting” complex (Ueda et al., 1990; Van Aelst and D'Souza-Schorey, 1997); (b) guanine nucleotide exchange factors (GEFs),1 of the dbl family, promote GDP/GTP exchange leading to activation of Rho GTPases (Cerione and Zheng, 1996); and (c) GTPase-activating proteins (GAPs) stimulate GTP hydrolysis and return the activated Rho GTPase to the inactive GDP-bound form (Lamarche and Hall, 1994).
In mammalian cells, all members of the Rho family studied to date play important roles in regulating the dynamics of the actin cytoskeleton and its reorganization in response to extracellular stimuli (Van Aelst and D'Souza-Schorey, 1997). In Swiss 3T3 cells, Rho regulates a signal transduction pathway linking growth factor receptors to the assembly of focal adhesions and actin stress fibers, Rac regulates the formation of lamellipodia and membrane ruffles and Cdc42 controls the formation of filopodia (Ridley and Hall, 1992; Ridley et al., 1992; Kozma et al., 1995; Nobes and Hall, 1995). Rho, Rac, and Cdc42 have also been shown to regulate changes in gene transcription and in particular there have been numerous reports that Rac and Cdc42 activate the JNK and p38 MAP kinase pathways (Coso et al., 1995; Minden et al., 1995). It is likely, therefore that these three GTPases play a key regulatory role in cell movement and in morphogenetic processes where changes in the actin cytoskeleton are coordinated with changes in gene transcription (Hall, 1998).
Like all GTPases, the cellular effects of the Rho family are mediated through their GTP-dependent interaction with cellular targets or effectors and using yeast two hybrid and affinity chromatography techniques, numerous candidates have already been described (Van Aelst and D'Souza-Schorey, 1997). In the case of Rho, the Rho-associated kinases, p160ROKα and p160ROCK, have attracted much attention. p160 Rho kinases can directly phosphorylate myosin light chain (MLC) and can also phosphorylate and inhibit the regulatory subunit of myosin light chain phosphatase (Amano et al., 1996; Kimura et al., 1996). Furthermore, overexpression of p160 Rho kinase leads to the Rho-independent assembly of actin– myosin filaments (Leung et al., 1996). It appears, therefore, that Rho acting through Rho kinase and MLC phosphatase regulates the levels of phosphorylated MLC and thereby controls the bundling and assembly of actin filaments into stress fibers. Numerous candidate targets for Rac and Cdc42 have also been identified and of these p65PAK, IQGAP, WASP, POR1, and PI4-P5 kinases have been suggested to play a role in actin filament assembly (Van Aelst and D'Souza-Schorey, 1997). Other potential players that may be common to all three GTPase-dependent pathways are the ezrin/radixin/moesin (ERM) family of proteins. Rho has been suggested to regulate the interaction of actin with the plasma membrane by modifying the association of ERM proteins with plasma membrane proteins such as CD44 (Tsukita et al., 1997), whereas using a permeabilized cell assay, it has been shown that ERM proteins play an essential role in both the Rho- and Rac-dependent assembly of stress fibers and lamellipodia, respectively (Mackay et al., 1997).
Here we characterize three new proteins that form a distinct branch of the Rho family: Rnd1, Rnd2, and Rnd3/ RhoE. Although clearly related in sequence, these G proteins possess strikingly different functional properties from other members of the Rho family. We show here that these proteins are likely to exist constitutively in the activated, GTP-bound form. Rnd1 associates with adherens junctions in both confluent fibroblasts and in epithelial cells and overexpression in growing fibroblasts induces the loss of polymerized actin structures and focal adhesions and causes severe loss of cell–matrix adhesion. We conclude that these proteins act as negative regulators of actin assembly and of cell adhesion.
Materials and Methods
Cloning and Analysis of New Rho Family Members
A bovine brain cDNA library was screened with a mixed oligonucleotide probe derived from the Rho effector domain (YVPTVFENYVADIE) and 14 positive clones characterized. Nine were RhoA and three RhoC, none encoded RhoB although the probe gave a strong signal on a RhoB plasmid used as control. This frequency is representative of the level of expression of RhoA and RhoC in retina, similar frequencies were observed in different brain libraries. Two cDNAs coded for two new proteins closely related to Rho in their effector domains (Rnd1 and Rnd2). These bovine cDNAs were used as probes to screen a human fetal brain library and nine clones were isolated corresponding to human Rnd1 and 2 clones corresponding to human Rnd2, and a third clone encoded a closely related protein, Rnd3. Rnd3 corresponded to a previously published cDNA, RhoE. The Rnd3 cDNA was incomplete, but we noticed that a 5′ region of Rnd3/RhoE was present in the expressed sequence tag (EST) database (GenBank ID: T58202, I.M.A.G.E. Consortium clone ID: 74398). This cDNA was obtained from Research Genetics Inc. (Huntsville, AL) and sequenced. Rnd3/RhoE starts with the same amino acid sequence as Rnd1: MKERRA(P/S)Q. In Rnd1 the assignment of the initiating ATG is based on the presence of a GCAACCATG sequence that closely resembles the optimal “Kozak” sequence for the initiation of translation. Furthermore, there is no downstream ATG in the case of Rnd1.
12 partial sequences for the human Rnd1 cDNA 5′ end are present in the EST databank (Dec. 97; see Fig. 1), and all include 60–90 bp of 5′ noncoding sequence, without any additional upstream ATGs. Furthermore one mouse EST shows high homology with the human Rnd1 cDNA, starting at the position encoding the MKERRA... sequence. To confirm that the ATG of Rnd2 had been properly assigned as the start codon, we used the Marathon 5′ Race technique (CLONTECH, Palo Alto, CA) to obtain longer 5′ regions using testis cDNA. However, an upstream ATG could not be obtained for Rnd2, nor could a region coding for a sequence homologous to the MKERRA(P/S)Q sequence present at the NH2 terminus of Rnd1 and Rnd3/RhoE. In addition, two sequences for the 5′ end of mouse Rnd2 are present in the databases; like human Rnd2 they have the sequence encoding MEGQS... close to the 5′ end, but there is no homology upstream of this sequence, strongly suggesting that it is the authentic NH2 terminus of the protein. Rnd1, Rnd2, and Rnd3/RhoE were deposited in the databank originally as Rho6, Rho7, and Rho8, respectively.
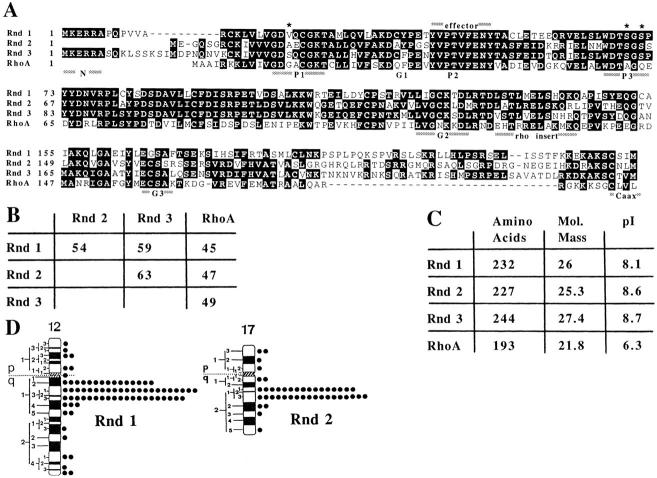
Figure 1.
Characterization of Rnd proteins and chromosomal localization of the Rnd1 and Rnd2 genes. (A) Alignment of Rnd1, 2, and 3 compared with RhoA. Identical amino acids are boxed; P1–3 and G1–3, the regions binding the phosphate and the guanine moieties of GTP, respectively. The region considered as the effector binding site including the asparagine (FENY) ADP ribosylated by the C3 transferase in Rho is overlined. Stars over the sequence indicate the positions corresponding to Ras-activating mutations. rho insert, an insertion of 12 amino acids that is characteristic of Rho family proteins. The COOH-terminal sequence for prenylation (CAAX) is underlined. These data are available from GenBank/EMBO/DDBJ under accession numbers Y07923 (Rnd1), X95456 (Rnd2), and X95282 (Rnd3/RhoE). (B) Percentage of identity between Rho family members. (C) Expected size and isoelectric point (pI)of Rnd1–3 proteins. (D) Chromosomal localization of Rnd1 and Rnd2. Idiograms of the human G-banded chromosomes 12 and 17, illustrating the distribution of grains with the Rnd1 and Rnd2 probes, respectively.
Northern blots of polyadenylated (poly[A]) RNA from human tissues (MTN blots) were obtained from CLONTECH and hybridized with Rnd cDNAs in stringent conditions.
Chromosomal Localizations
In situ hybridization was carried out on chromosome spreads from phytohemagglutinin-stimulated human lymphocytes cultures for 72 h. 5-Bromodeoxyuridine (60 μg/ml) was added for the final 7 h of culture to ensure a posthybridization chromosomal banding of high quality. pUC19 plasmids with ∼600 bp of Rnd1 or Rnd2 insert was tritium labeled by nick translation to specific activities of 1–2 × 108 dpm/μg. Radiolabeled probes were hybridized to metaphase spreads at a final concentration of 200 ng/ml of hybridization solution. After coating with nuclear track emulsion (NTB2; Eastman Kodak Co., Rochester, NY), the slides were exposed for 20 d at 4°C, and then developed. To avoid any slipping of silver grains during the banding procedure, chromosome spreads were first stained with buffered giemsa solution and metaphase photographed. R-banding was then performed by the fluorochrome-photolysis-giemsa method and metaphases re-photographed before analysis. In the 150 metaphase cells examined for Rnd1, 326 silver grains associated with chromosomes and 76 of these (23.3%) were located on chromosome 12. The distribution was not random, 57 of these (75%) mapped to the q12-q13 region of chromosome 12 long arm. In the 100 metaphase cells examined for Rnd2, 242 silver grains associated with chromosomes and 44 of these (18.2%) were located on chromosome 17. The distribution was not random, 34 of them (77.3%) mapped to the q21 region of chromosome 17 long arm.
Expression and Purification of Rnd Proteins
NdeI and BamHI sites were introduced by PCR at the 5′ and 3′ ends of Rnd1, 2, or 3 cDNAs, and cloned in the pET11a vector (Novagen Inc., Madison, WI) to express complete nonfusion proteins. Rnd1 was expressed at high levels and in a soluble form. The level of Rnd2 and Rnd3/ RhoE expression were lower so we focused on Rnd1. Using the most common lysozyme/EDTA protocol for bacterial lysis we were able to purify >20 mg of Rnd1 from the soluble fraction by a single-column procedure (>80% purity). This first protein preparation was used to raise antibodies against Rnd1. However the protein was relatively unstable and had a tendency to precipitate, even in the presence of GDP, suggesting that this protein bound GDP poorly. We have since avoided the use of EDTA and lysed the cells with a french press, directly in a buffer containing Mg2+ and GDP (or GTP). Unlike most Escherichia coli proteins Rnd1 bound to a SP-Sepharose Fast Flow (Pharmacia Biotechnology Inc., Piscataway, NJ) column and could be eluted at ∼200 mM NaCl, Rnd1 represented >70% of the total proteins in peak fractions. In these conditions, we were able to purify large amounts of functional protein (>10 mg/liter of bacterial culture).
The bacterial exoenzyme C3 transferase, ADP ribosylates Asn at codon 41 and inhibits the activity of RhoA, B, and C (Chardin et al., 1989; Aktories and Just, 1995). E. coli–expressed Rnd1, purified in the presence of GDP, GTP, or without added nucleotide, could not be ADP ribosylated by C3 in vitro. Since Rnd proteins have a molecular mass of ∼32 kD, they are easily separated from RhoA by SDS-PAGE. ADP ribosylation by C3 in the presence of [32P]NAD was performed on total tissue extracts. A strong band was observed at ∼25 kD (Rho A–C), but no labeling ∼30 kD could be observed, even though Western blot analysis using anti-Rnd antibodies revealed that these proteins were present. Finally, COS cells transfected with Rnd1 expression vectors did not show any additional ADP-ribosylatable proteins compared with untransfected COS cells. We conclude that Rnd proteins are not C3 substrates.
Exchange and GTP Hydrolysis Assays
For exchange assays, 1 μM Rnd1 was incubated with 10 μM [35S]GTPγS in 50 mM Hepes, pH 7.5, 100 mM KCl, 2 mM MgCl2, and 1 mM DTT (final volume of 200 μl), 25-μl aliquots containing 25 pmol of Rnd1 were taken at different times, added to cold “stop buffer,” and immediately filtered on 0.45 μm membranes (Schleicher & Schuell, Inc., Keene, NH). To measure GTP hydrolysis RhoA or Rnd1 were loaded with 10 μM [γ32P]GTP in the same buffer but including 5 mM Pi to inhibit a contaminant nucleotidase activity. Rnd1, which exchanges fast, was loaded in 1 mM Mg2+ at 37°C for 5 min, whereas RhoA was loaded in 1 μM Mg2+ at 37°C for 5 min, and then 1 mM Mg2+ added back to start GTP hydrolysis. About 0.1 μM of GST-RhoGAP COOH-terminal was added as indicated. 25-μl aliquots were removed at the indicated times, mixed with 975 μl of charcoal in 50 mM NaH2PO4, and then centrifuged. 0.8 ml of the supernatant was counted to measure 32Pi release.
Preparation of Membrane Pellets
Tissues or scraped cells were resuspended and washed at 4°C in fractionation buffer (250 mM sucrose, 20 mM Hepes, pH 6.8, 50 mM NaCl, 10 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM GTP) containing “complete” protease inhibitors (Boehringer Mannheim Corp., Indianapolis, IN) and then lysed by 50 strokes in a glass homogenizer (Kontes Glass Co., Vineland, NJ). Nuclei and unlysed cells were removed by low speed centrifugation, the supernatant was centrifuged at 100,000 g, and the P100 pellet was resuspended in the same buffer. Protein concentrations in the pellet or supernatant (S100) fractions were measured and Laemmli sample buffer added. Similar amounts of total proteins were loaded in each lane, electrophoresed on 12% polyacrylamide–SDS gels, and then transferred to nitrocellulose. The amounts of protein were checked by staining with Ponceau red and the membranes incubated with anti-Rnd1 antibodies.
In Situ Hybridization
Riboprobes were generated by in vitro transcription using [33Pα]UTP (3,000 Ci/mmol; ICN Pharmaceuticals, Costa Mesa, CA) with T3 (antisense probe) and T7 (sense probe) RNA polymerases (Boehringer Mannheim Corp.) from linearized plasmid containing a 750-bp DNA fragment of the Rnd1 cDNA. Brains from adult Wistar rats (Charles River, St. Aubin Les Elbeuf, France) were fixed overnight in a phosphate-buffered 4% paraformaldehyde solution and immersed in 20% sucrose overnight. Frozen tissues sections (12 μm) were cut on a cryostat (Leica AG, Heerbrugg, Switzerland). Prehybridization included treatment with 0.1 M glycine, 0.3% Triton X-100, (5 μg/ml, 15 min) proteinase K, and 0.1 M triethanolamine/0.25% acetic anhydride. Hybridization was carried out at 55°C overnight in a buffer containing 50% formamide, 5× Denhardt's solution, 20 mM NaPO4, 10% dextran sulfate, 5% sarcosyl, and 2× sodium saline citrate (SSC). After hybridization, sections were washed twice (45 min) in 1× SSC, treated 20 min with RNase A (10 μg/ml), and then washed in 0.1× SSC at room temperature (30 min). Specimens were dehydrated, air dried, and exposed to Hyperfilm β-max (Amersham Corp.) for 21 d.
Immunohistochemistry on Brain Sections
For immunohistochemistry, frozen tissues section (20 μm) were prepared as described for in situ hybridization. Endogenous peroxidase was inhibited in 3 mg/ml phenylhydrazine (Sigma Chemical Co., St. Louis, MO) for 30 min. The slides were treated for 10 min in Triton X-100 0.3% and incubated in 3% goat serum for 2h. Slides were then incubated overnight with a polyclonal antibody directed against Rnd1 (dilution 1:250) at 4°C. Immunoperoxidase revelation of the primary antibodies was carried out using the streptavidin/peroxidase method following the manufacturer's protocol (Vectastain Elite kit; Vector Labs, Inc., Burlingame, CA). Visualization of the signal was performed using 3,3′-diaminobenzidine as chromogen. In control procedures, anti-Rnd1 antibodies were preabsorbed by incubating in a solution containing a saturating concentration of the antigenic peptide.
Cell Microinjection and Immunofluorescence
Swiss 3T3, MDCK, and primary rat embryo fibroblast (REF) cells were cultured in DME/10% FCS. Confluent, quiescent Swiss 3T3 cells for microinjection were prepared by seeding 5 × 104 cells onto coverslips in DME/5% FCS followed by 6–10 d without medium change. Quiescent cells were serum starved overnight (16 h) in DME containing 2 g/liter NaHCO3. Swiss 3T3, MDCK, and REF cell nuclei were microinjected with eukaryotic expression vectors (pRK5) encoding wild-type Rnd1, N27Rnd1, A45Rnd1, L71Rnd1, NH2-terminal truncated Rnd1, CAAX-deleted Rnd1, myc-tagged Rnd2, or myc-tagged Rnd3/RhoE at a concentration of 100 μg/ml in PBSA. Cells were returned to the incubator for a further 1–3 h before fixation in 4% paraformaldehyde (wt/vol). MDCK cells were pre-permeabilized (2 min) in 80 mM potassium-Pipes buffer, 0.1% saponin, 5 mM EGTA, 1 mM MgCl2, at room temperature before fixation. To study the effects of Rnd proteins on actin structure assembly, Swiss 3T3 cells were further incubated in the presence of lysophosphatidic acid (LPA) (100 ng/ml) for 30 min or PDGF (5 ng/ml) for 20 min before fixation.
Fixed Swiss 3T3 cells on coverslips were stained for filamentous actin as described previously (Nobes and Hall, 1995), and for expressed Rnd protein by incubation in the presence of an affinity-purified anti-Rnd1 polyclonal antibody followed by a second antibody layer of FITC-conjugated donkey anti–rabbit (Jackson Immunoresearch Laboratories, Inc., West Grove, PA), or expressed myc-tagged Rnd3/RhoE, using an anti-myc antibody followed by FITC-conjugated goat anti–mouse (Pierce Chemical Co., Rockford, IL). Costaining for endogenous Rnd1 and cadherin localization was performed on Swiss 3T3 cells, using an anti-Rnd1 polyclonal antibody followed by a secondary antibody layer of FITC-conjugated goat anti–rabbit (Jackson Immunoresearch Laboratories) and a tertiary layer of FITC-conjugated donkey anti–goat along with mouse anti–pan cadherin antibodies (Sigma Chemical Co.) and a TRITC-conjugated goat anti–mouse antibody (Jackson Immunoresearch Laboratories). In the case of colocalization of expressed Rnd protein with cadherin, Swiss 3T3 cells were pre-permeabilized for 2 min before fixation. REF cells were costained for filamentous actin and vinculin as described in Nobes and Hall (1995).
Results
Characterization of Three New Members of the Rho Family
In a search for new Rho-related GTPases expressed in the retina, we cloned partial cDNAs for two new proteins (Rnd1 and Rnd2) closely related to Rho. The complete human cDNAs for Rnd1, Rnd2, and a third closely related member, Rnd3, were subsequently obtained (Fig. 1 A). Rnd3 is identical to the recently described RhoE (Foster et al., 1996), though it is clear from our sequence that the previously published RhoE sequence lacks 15 NH2-terminal amino acids. Rnd proteins share 54–63% identity pairwise, ∼45–49% identity with Rho (Fig. 1 B), and slightly less identity with other Rho family members, Rac or Cdc42. We conclude that the three Rnd proteins form a new branch of the Rho family.
Rnd proteins display striking differences from other members of the Rho family in their size, their charge, and biochemical properties. Their expected molecular weights are higher (Fig. 1 C) due to NH2-terminal extensions for Rnd1 and Rnd3/RhoE and COOH-terminal extensions of ∼30 amino acids for all three. Their apparent molecular mass on SDS-PAGE is ∼32 kD, whereas Rho migrates at ∼24 kD. They also have very different isoelectric points; whereas most Rho family proteins are negatively charged at neutral pH (pI = 5.5–6.8; Huber et al., 1994), Rnd proteins are positively charged with pI = 8.1–8.7 (Fig. 1 C). Rnd proteins end with a “CAAX box” motif for prenylation, but unlike Rho, Rac, and Cdc42, which have a COOH-terminal leucine and are predominantly geranyl-geranylated, Rnd proteins end with a methionine residue suggesting that the proteins are likely to be farnesylated.
The three guanine-binding motifs (Fig. 1 A, G1–3) are conserved in Rnd proteins, the two loops and a conserved threonine involved in phosphate binding (Fig. 1 A, P1–3) can also be recognized and the three major residues that coordinate magnesium in the GTP-bound form of Ras: T 17, T 35, and D 57 (Pai et al., 1990) are strictly conserved. However, three residues of the phosphate-binding site that are important for the intrinsic GTPase activity of Ras proteins differ in Rnd. The equivalent of Ras glycine 12 is replaced by valine, alanine, or serine in Rnd proteins, Ras glycine 13 is replaced by glutamine or glutamic acid and Ras alanine 59 and glutamine 61 are both replaced by serine. Any one of these substitutions in Ras decreases its intrinsic GTPase rate and prevents GAP-mediated GTPase stimulation, leading to constitutive activation of the protein. Furthermore, in viral Ras the presence of two substitutions at positions 12 and 59 (replaced by threonine) decreases GTPase activity more than individual substitutions (John et al., 1988). Based on the effects of these substitutions on Ras, the prediction is that Rnd proteins will have little or no intrinsic GTPase activity.
The chromosomal locations of Rnd1 and Rnd2 were determined (see Materials and Methods). Rnd1 maps to the q12-q13 region of chromosome 12 long arm and Rnd2 maps to the q21 region of chromosome 17 long arm (Fig. 1 D)
Biochemical Properties of Rnd1
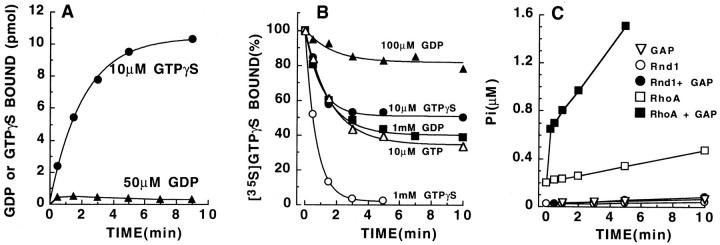
We have expressed Rnd1 in bacteria as a nonfusion protein using the pET11a vector and examined its biochemical properties. Fig. 2 A shows that GTPγS rapidly binds to Rnd1 (t 1/2 = 1.4 min at 37°C) in a physiological buffer containing 2 mM Mg2+. 10–15 pmol of nucleotide bound to 25 pmol of Rnd1 indicating that about half of the purified protein is properly folded and binds nucleotide. However, Fig. 2 A shows that GDP binds very poorly to Rnd1 under the same conditions. In Fig. 2 B, Rnd1 was first loaded with [35S]GTPγS and its dissociation in the presence of different concentrations of competing nucleotides was measured. At equilibrium, 10 μM GTP is about as efficient at promoting GTPγS dissociation as 1 mM GDP, indicating that the affinity for GDP is about 100 times lower than for GTP. The Rnd1 protein used in Fig. 2 A was prepared by lysing the bacteria in a buffer with 10 μM GTP; however, very similar kinetics were observed when the bacteria were lysed in a buffer with 10 μM GDP. Inside bacteria, Rnd1 proteins are most likely bound to GTP, which is present in high amounts, and when bacteria are lysed at 4°C, GTP does not exchange, even when exogenous GDP is added. At low magnesium (<1 μM), GTP dissociates extremely fast, but even when GTPγS is present the protein is unstable and it is unable to bind nucleotides if incubated for 10 min at 37°C with EDTA (data not shown). The precise dissociation rate at low magnesium could not, therefore, be measured reliably but is <30 s.
Figure 2.
Biochemical properties of E. coli–expressed Rnd1. (A) Kinetics of GDP and GTPγS binding on Rnd1 (25 pmol/point) at high magnesium. (B) Kinetics of GTPγS dissociation from Rnd1 with different concentrations of GDP, GTP, or GTPγS as competitors. (C) GTP hydrolysis by Rho and Rnd proteins.
Fig. 2 C shows that Rnd1 has no detectable GTPase activity, even in the presence of RhoGAP. The very small increase of Pi released when RhoGAP is added is because of trace amounts of phosphatase activity contaminating RhoGAP, since it is also observed when only RhoGAP is added to the [γ32P]GTP mix in the same conditions. As a positive control, the same amount of RhoGAP (0.1 μM) stimulates GTP hydrolysis very efficiently on RhoA (Fig. 2 C, squares). These biochemical results show that in physiological conditions, Rnd1 exchanges GTP faster and has a much lower affinity for GDP than most other small G proteins, and it has no intrinsic GTPase activity. This raises the possibility that in vivo, Rnd proteins are constitutively in the active, GTP-bound form.
Tissue Expression
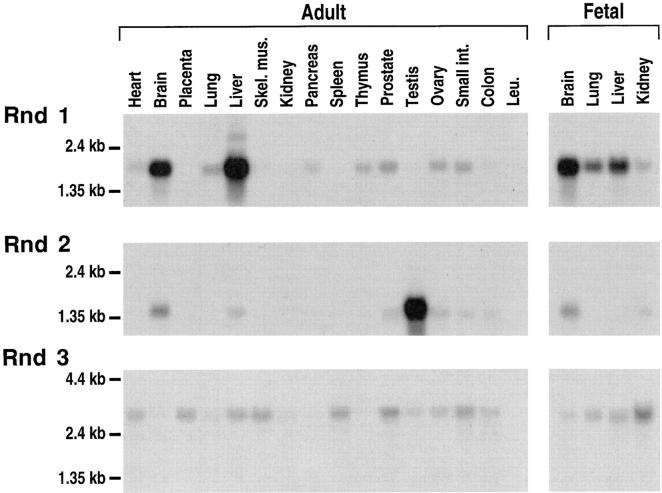
Northern blots show that Rnd1 is expressed as a major 1.9-kb mRNA, mostly in brain and liver, Rnd2 as a 1.5-kb mRNA abundant in testis, whereas Rnd3/RhoE is expressed at low levels as a 3.1-kb mRNA in all tissues studied (Fig. 3). Rnd3/RhoE expression is particularly low in brain, pancreas, thymus, testis, and peripheral blood leukocytes, but could be detected upon longer exposure. When the same blot was rehybridized with a ubiquitously expressed probe, the relative intensities of the signals were similar to those seen with the Rnd3/RhoE probe.
Figure 3.
Expression of Rnd1, 2, and 3 mRNAs in human adult and fetal tissues. Northern blots of human Poly(A+) RNAs from various tissues were hybridized with Rnd probes labeled to similar activities and autoradiographed for 1 wk, so that the intensities of the signals with the three probes could be compared. Hybridization of the same blots with an actin probe gave a much stronger signal, with comparable intensities for all organs. Expression of Rnd3/RhoE in brain, pancreas, thymus, testis, and peripheral blood leukocytes (Leu.) could only be detected upon longer exposure.
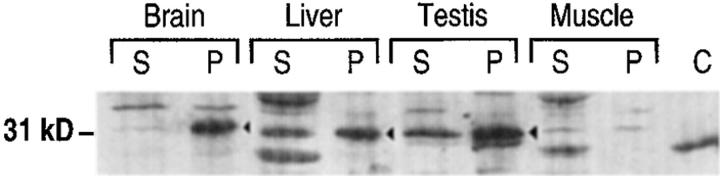
The expression of Rnd proteins (probably mainly Rnd1) in rat brain and liver could also be detected at the protein level, with antibodies raised against Rnd1. When these tissue extracts are centrifuged at 100,000 g the major portion of Rnd1 proteins are found in the pellet (P100) fraction (Fig. 4), whereas most Rho appears in the supernatant (S100) fraction, as described previously. In freshly prepared rat hepatocytes, the expression of the Rnd1 protein was similar to that found in total liver. We looked at the expression of the Rnd2 protein in testis of immature rats (6 d) with antibodies raised against bacterially expressed Rnd2 and found a level of expression similar to that in adult rats (data not shown).
Figure 4.
Expression and localization of Rnd proteins in rat tissues. Western blot analysis of supernatants (S) and pellets (P) after a 100,000 g centifugation of postnuclear supernatants from different rat tissues. Skeletal muscle was used as a negative control with undetectable Rnd expression (see mRNA expression on Fig. 3). C, 1 ng of bacterially expressed Rnd1, which runs slightly faster than tissue Rnd1. The polyclonal antiserum raised against bacterially expressed Rnd1 mostly detects the Rnd1 protein, but is also able to detect the high levels of Rnd2 expressed in testis. Arrowheads, Rnd proteins. Unrelated proteins of different molecular weights are detected in the supernatants.
Closer examination of Rnd1 expression in the brain using a Northern blot of mRNAs prepared from different regions of the brain revealed high levels in the cortex, the occipital pole and the frontal and temporal lobes (Fig. 5 A). Lower levels of expression are observed in most other regions of the brain. When the same blot was rehybridized with a ubiquitously expressed probe (β-actin), the intensity of the signal was much stronger and similar for all brain regions. Analysis of Rnd1 mRNA expression on rat brain sagittal sections was performed by in situ hybridization. The general expression of Rnd1 transcripts was low in most regions of the rat brain. Overall, the highest expression of Rnd1 mRNA was observed in the cerebral cortex including both the frontal, striate, and primary olfactory cortex (piriform cortex). Also the hippocampus, including the dentate gyrus, the cerebellar granular layer and pontine nuclei expressed relatively high levels. A lower expression was found in the inferior and superior colliculus, substantia nigra, ventral tegmental area (VTA), and several unidentified brain stem nuclei (Fig. 5 B). Hybridization with the sense riboprobe revealed no signal (Fig. 5 C).
Figure 5.
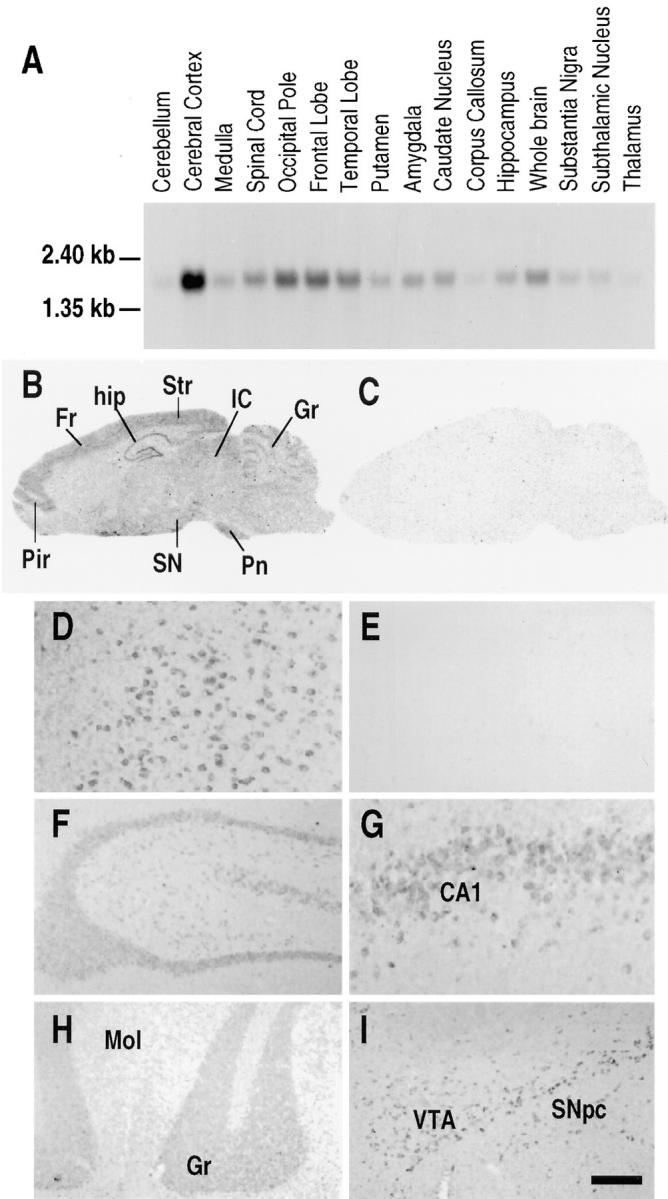
Expression of Rnd1 within human and rat brain. (A) Northern blot analysis of Rnd1 expression in different regions of the human brain. (B–C) Photomicrographs of the autoradiograms of sagittal rat brain sections after in situ hybridization histochemistry with (B) the antisense Rnd1 riboprobe and (C) the corresponding sense riboprobe. Exposure time is 21 d. (D–I) High power bright-field microphotographs of coronal rat brain sections immunostained with the (D and F–I) anti-Rnd1 antibodies, and (E) anti-Rnd1 antibodies after preincubation with the antigenic peptide. (D and E) cerebral cortex, (F) hippocampus, (G) CA1 pyramidal neurons, (H) granular cell layer of the cerebellar cortex, and (I) VTA and substantia nigra pars compacta. Cx, cerebral cortex; Fr, frontal cortex; Gr, cerebellar granular layer; hip, hippocampus; IC, inferior colliculus; Pir, piriform cortex; Pn, pontine nuclei; SN, substantia nigra; and Str, striate cortex. Bar: (D, E, and G) 65 μm; (F, H, and I) 200 μm.
The neuronal localization of Rnd1 revealed by immunohistochemistry was in good agreement with that of in situ hybridization. The highest density of immunohistochemical staining in the brain was observed in the cerebral cortex. Immunoreactivity was seen in all layers and throughout the whole cerebral cortex (Fig. 5 D). No signal was obtained when the serum was preincubated with an excess of the antigenic peptide (Fig. 5 E). In the hippocampal formation immunostaining was observed in all pyramidal cells and in the granule neurons of the dentate gyrus (Fig. 5, F and G). Within the cerebellum, highest expression of the Rnd1 protein was observed in the Purkinje cells, whereas there was a less intense staining of the granule cell layer (Fig. 5 H). Expression of Rnd1 was also found in the substantia nigra pars compacta and VTA (Fig. 5 I).
Subcellular Localization of Rnd1 in Swiss 3T3 Cells
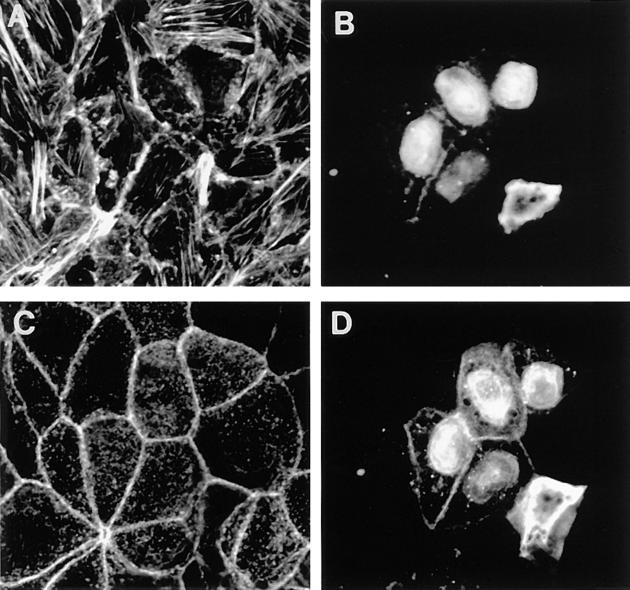
The subcellular localization of endogenous Rnd proteins was examined in Swiss 3T3 fibroblasts using polyclonal antibodies raised against Rnd1 protein. Fig. 6 A shows that endogenous Rnd is concentrated at the cell periphery at points of cell–cell contact in confluent monolayers of Swiss 3T3 fibroblasts. This peripheral staining is not observed with the preimmune serum (Fig. 6 C); the nuclear staining seen with anti-Rnd1 is also observed with the preimmune serum and is nonspecific. The peripheral staining for Rnd colocalized with an anti–pan cadherin antibody indicating that Rnd1 is localized in adherens junctions in confluent cells (Fig. 6 B).
Figure 6.
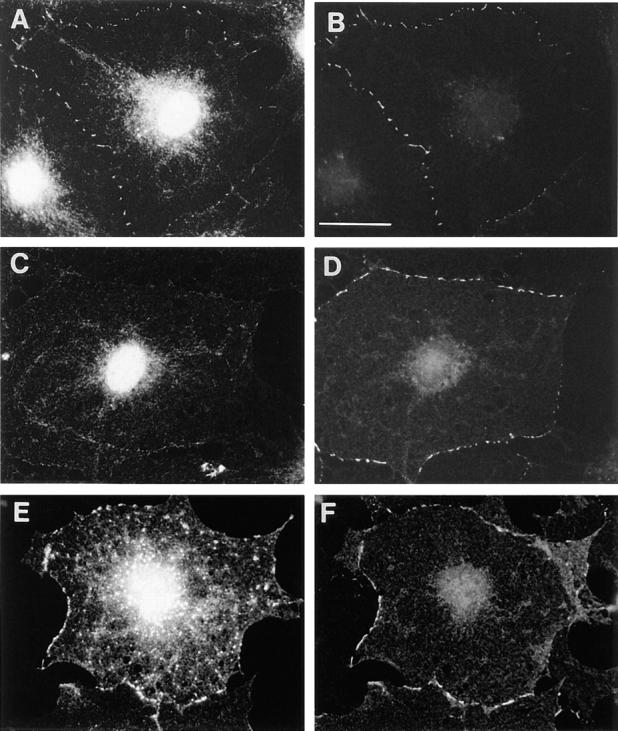
Localization of Rnd proteins in adherens junctions. Confluent, quiescent, serum-starved Swiss 3T3 cells were fixed and endogenous Rnd1 (A) localized using polyclonal antibodies raised against recombinant Rnd1. The control preimmune staining pattern is shown in C. These cells were costained for cadherin localization using a pan cadherin monoclonal antibody (B and D). Confluent, quiescent, serum-starved Swiss 3T3 cells were injected with a pRK5 vector expressing N27Rnd1 and fixed 3 h later. Expressed Rnd1 protein (E) and cadherin (F) were visualized by indirect immunofluorescence. Bar, 25 μm.
Since the Rnd1 polyclonal antibody cross-reacts with Rnd2 and Rnd3/RhoE on Western blots, we have looked at the subcellular localization of transiently expressed Rnd1. Overexpression of wild-type Rnd1 induces cell rounding and loss of cell–cell contacts (see below) and it was not possible, therefore, to localize the protein after microinjection of an expression vector into Swiss 3T3 cells. However, we observed that an N27 mutant (corresponding to the N17 dominant negative mutant of ras) and an A45 mutant of Rnd1 (predicted to destroy its effector site) do not affect cell morphology. When these constructs were expressed in confluent monolayers of Swiss 3T3 cells, both Rnd1 mutants predominantly localized at the cell periphery at points of cell–cell contact (shown for N27Rnd1 in Fig. 6 E). The peripheral staining for N27Rnd1 colocalized with an anti–pan cadherin antibody (Fig. 6 F), confirming that Rnd1 localizes to adherens junctions.
Effects of Rnd1 Expression in Fibroblasts
To examine the possible biological function of Rnd proteins, pRK5 vectors expressing wild-type Rnd1 or the different mutants were microinjected into the nuclei of quiescent serum-starved Swiss 3T3 fibroblasts and the cells left for 1–3 h. No induction of filamentous actin could be observed in wild-type Rnd1–expressing cells, rather we observed a dramatic loss of staining for filamentous actin (Fig. 7, A and B) and Rnd1–expressing cells retracted and severely rounded up to produce a dendritic-like phenotype. When microinjected cells were stimulated with LPA we observed that cells overexpressing wild-type Rnd1 did not form actin stress fibers (Fig. 7, C and D). Addition of PDGF to quiescent cells induces peripheral filamentous actin in lamellipodia and membrane ruffles as seen in uninjected cell in bottom right corner of Fig. 7, E and F. However, the cell in the top left corner of Fig. 7, E and F, which is expressing Rnd1, remains flat after PDGF addition and displays no filamentous actin in membrane ruffles at its periphery. We conclude that Rnd1 interferes with both Rho- and Rac-mediated reorganization of the actin cytoskeleton and in addition causes loss of cell–matrix adhesion. Expression of myc-tagged Rnd3/RhoE also blocked LPA-induced stress fiber formation (Fig. 7, G and H), similar to Rnd1 (Fig. 7, C and D), but expression of Rnd2 had no observable effects on actin (data not shown).
Figure 7.
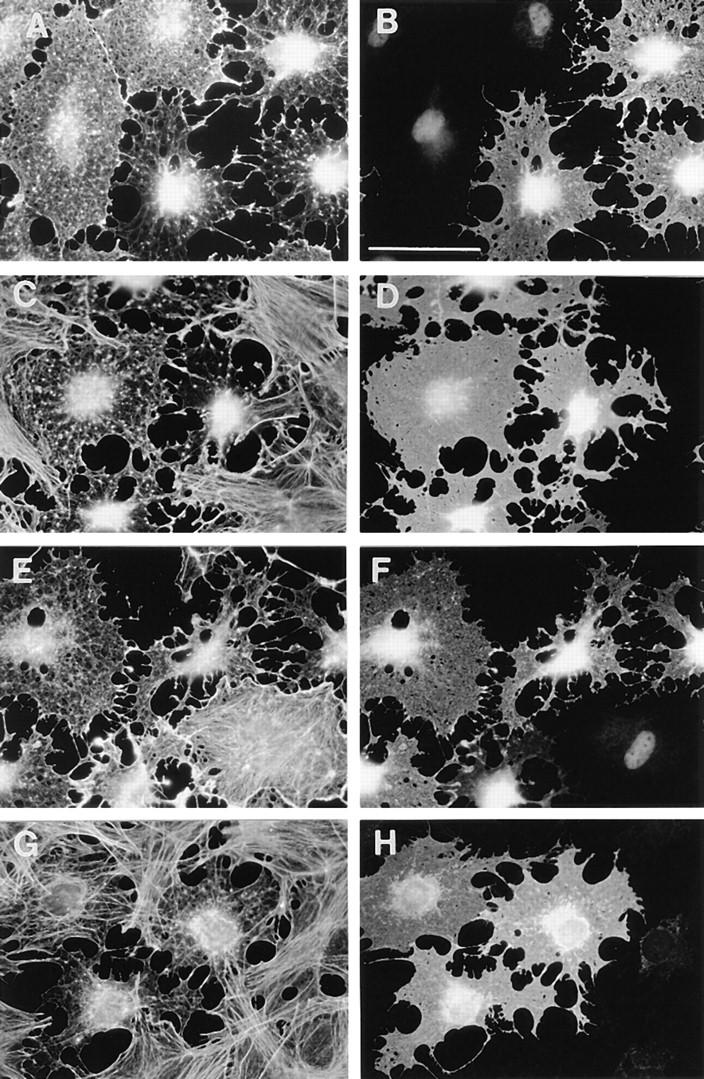
Effects of Rnd1 expression on actin filament assembly. Serum-starved confluent Swiss 3T3 fibroblasts were injected with a pRK5 vector expressing Rnd1 (A–F) or myc-tagged Rnd3/ RhoE (G and H). 1.5–2.5 h later, cells were either left unstimulated for a further 30 min (A and B), stimulated with LPA (100 ng/ml) for 30 min (C, D, G, and H), or stimulated with PDGF (5 ng/ml) for 20 min (E and F) before fixation. Permeabilized cells were stained to show Rnd1 expression (B, D, and F), myc-tagged Rnd3/RhoE expression (H), and actin filaments (A, C, E, and G). Bar, 50 μm.
In cells expressing N27Rnd1 (equivalent to a dominant negative N17 mutation in Ras) (Fig. 8, A and B) or A45Rnd1 (equivalent to an A35 effector site mutation in Ras; data not shown) LPA-induced stress fiber formation was not inhibited. Thus the binding of GTP and a functional effector domain are both required for Rnd1 to block stress fiber formation. Interestingly, Rnd1 deleted of its first six amino acids was also unable to block stress fiber formation (Fig. 8, C and D). We observed that a Rnd1 mutant deleted of its CAAX box was less potent than wild-type Rnd1 at preventing actin filament assembly (data not shown).
Figure 8.
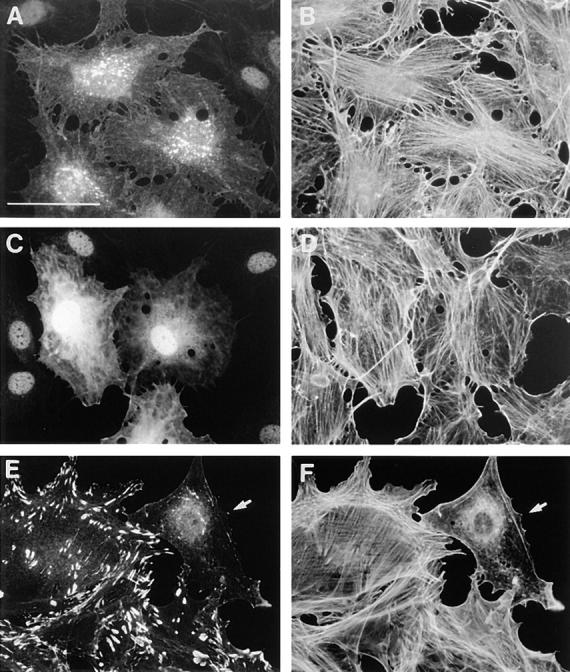
Effects of mutant Rnd1 expression on actin filament assembly. Serum-starved, confluent Swiss 3T3 fibroblasts were injected with pRK5 vectors expressing N27Rnd1 (A and B) or NH2-terminal truncated Rnd1 (C and D) and 2–3 h later, cells were stimulated with LPA (100 ng/ml) for 30 min before fixation. Permeabilized cells were stained to show Rnd1 expression (A and C) and actin filaments (B and D). Growing primary REFs were microinjected with a pRK5 vector expressing wild-type Rnd1 and fixed 2 h later. Permeabilized cells were costained to show vinculin localization (E) and actin filaments (F). Cells were coinjected with dextran-biotin (lysinated) and visualized with Cascade blue–conjugated avidin. At least 90% of injected cells expressed Rnd1 and showed loss of actin stress fibers and focal adhesions. The REF cell microinjected with Rnd1 is indicated by arrows in E and F. Bar, 50 μm.
We have introduced a leucine in position 71 of Rnd1 (L71, equivalent to codon 61 in Ras) instead of the serine. In Ras, the substitution of glutamine 61 by leucine gives a higher transforming potential than the substitution by threonine (serine has not been studied; Der et al., 1986) and blocks GAP stimulation of GTP hydrolysis. Thus we reasoned that the leucine 71 mutant of Rnd1 would have a higher activity than wild type if there was an endogenous RndGAP. We observed that L71Rnd1 has the same activity as wild-type Rnd1 in blocking LPA-induced stress fiber formation (data not shown), suggesting that fibroblasts do not contain a RndGAP.
Since expression of Rnd1 inhibits the assembly of actin stress fibers and membrane ruffles, we wanted to look at the effect of Rnd1 on actin structures and focal adhesions in growing cells. Expression of Rnd1 in growing Swiss 3T3 cells (data not shown) or in primary REFs (Fig. 8, E and F) rapidly (within 2 h) leads to the loss of both actin stress fibers and focal adhesions as visualized with antivinculin antibodies. It can also be seen in Fig. 8, E and F that Rnd1 overexpression inhibits lamellipodia and membrane ruffles in agreement with PDGF results in Fig. 7, E and F.
Effects of Rnd1 Expression in MDCK Epithelial Cells
The above results indicate that integrin-associated actin filaments are sensitive to expression of wild-type Rnd1 protein. To look at the effects of Rnd1 on actin filaments in epithelial cells, MDCK cells were microinjected with a Rnd1 expression vector. Fig. 9, A and B reveals that as with fibroblasts, Rnd1 induces loss of actin stress fibers found juxtaposed to the basal membrane. However, injected cells did not dissociate or round up and further confocal analysis revealed that actin filaments associated with adherens junctions in lateral membranes remained intact up to 3 h after expression of Rnd1 (Fig. 9, C and D). Furthermore, there was no disruption of cadherin-based junctional complexes as visualized with an anti–E-cadherin antibody (data not shown). As with confluent fibroblasts, Rnd1 was found to associate predominantly at adherens junctions in the lateral membrane (Fig. 9 D).
Figure 9.
Effects of Rnd1 expression on actin filaments in epithelial cells. Confocal analysis of MDCK cells microinjected with a pRK5 vector expressing wild-type Rnd1. Cells were prepermeabilized, fixed 3 h after injection, and then stained to show Rnd1 expression (B and D) and actin filaments (A and C). Images A and B show a confocal section through a region juxtaposed to the basal membrane, and images C and D represent a section through the adherens junction lateral membranes. We are unable to assess whether there is any significance to the nuclear staining pattern of overexpressed Rnd1. It has often been observed that overexpression of other small G proteins leads to nuclear localization of the posttranslationally unprocessed protein.
Discussion
Rnd1, Rnd2, and Rnd3/RhoE form a distinct branch of the mammalian Rho family, which includes Rho (A, B, and C isoforms), Rac (1, 2, and other isoforms), Cdc42 (Cdc42Hs and G25K isoforms), TC10, RhoG, RhoD, and TTF. Rnd3 is identical to the recently described RhoE (Foster et al., 1996), except that in Rnd3 we have found an upstream ATG not reported in the RhoE cDNA. The first five amino acids of this new NH2-terminal sequence (MKERRA) of Rnd3/RhoE are identical to those of Rnd1. At least five EST sequences encoding the NH2 terminus of Rnd3 are present in the databases and they each show this longer NH2-terminal sequence. We have shown that in Rnd1, this sequence is essential for the intracellular localization and functional activity of the protein (see below). The NH2- and COOH-terminal extensions found in Rnd proteins show no close sequence similarity to other sequences in the databases.
Rnd1, Rnd2, and Rnd3/RhoE proteins expressed in bacteria, or in intact rat tissues, are not substrates for ADP ribosylation by C3 transferase (data not shown), despite having an asparagine residue at the equivalent of RhoA asparagine 41, which is modified by C3 (Chardin et al., 1989; Aktories and Just, 1995). Rnd1, Rnd2, and Rnd3/ RhoE each have substitutions at three positions known to be involved in GTPase activity and conserved in most of the 70 or so other small GTPases in the Ras superfamily. We also show here that Rnd1 exchanges GTP faster than most other small GTPases and has a much lower affinity for GDP than for GTP. Furthermore, Rnd1 has no detectable GTPase activity and Rho GAP, which stimulates GTP hydrolysis on all other Rho family proteins tested so far, does not stimulate GTP hydrolysis on Rnd1. Foster et al. (1996), also reported that a tagged version of RhoE expressed in COS cells was constitutively GTP bound. We cannot formally rule out the presence of a GAP specific for Rnd proteins, but our observation that Rnd1 with a leucine to serine substitution at codon 71, a mutation that would presumably block the effect of any GAP, has the same activity as wild-type Rnd1 in inhibiting LPA-induced stress fiber formation, argues strongly against the presence of a Rnd GAP. Taken together, our results and those of Foster et al. (1996) suggest that Rnd proteins are constitutively in the activated GTP-bound form.
These observations raise the important question as to how the cellular activities of Rnd proteins are regulated. One possibility is that Rnd function is attenuated by inhibitory binding proteins or by sequestration at specific intracellular locations. An alternative possibility is that Rnd activity is determined by expression. Rnd1 is expressed at high levels in liver and brain, where it is predominantly in the cortex, whereas Rnd2 is expressed at high levels in the testis. These results were confirmed by Western blots: Rnd1 is mostly found in the particulate (P100) fraction of brain and liver, with little or no expression in skeletal muscle and other tissues; Rnd2 is specifically expressed in testis, mostly in the particulate (P100) fraction; Rnd3/RhoE, however, appears to be ubiquitously expressed as judged by Northern analysis, at least. The expression of Rnd1 in freshly prepared hepatocytes is similar to that in total liver, indicating that hepatocytes are the major liver cells expressing Rnd1. In situ hybridization and immunocytochemistry on brain sections show that Rnd1 is mostly expressed in specialized neurons in the cortex, in the hippocampus, in CA1 pyramidal neurons, in the granular cell layer of the cerebellar cortex, and in specialized neurons of substantia nigra. We would like to propose that the high levels of Rnd1 expression in these cells are associated with specific properties of the actin cytoskeleton.
We have looked at the possible function of Rnd proteins by overexpressing wild-type Rnd 1 in cells. In Swiss 3T3 fibroblasts, Rnd1 blocks LPA-induced stress fiber formation and PDGF-stimulated lamellipodia protrusions. Rnd1 also promotes the disassembly of actin stress fibers and focal adhesions in growing REFs and in epithelial (MDCK) cells. It appears therefore, that Rnds may be general negative regulators of actin filament assembly. Interestingly, however, even after 3 h of expression, Rnd1 did not lead to loss of cortical actin filaments associated with adherens junctions in MDCK cells. This suggests that integrin-linked actin filaments might be more susceptible to Rnd-induced disassembly than cadherin-linked actin filaments. It is possible, however, that long-term expression of Rnd1 might also lead to disruption of lateral actin filaments in epithelial cells. These effects on the actin cytoskeleton are specific for the wild-type form of Rnd1; mutations that would be predicted to decrease its affinity for GTP (N27) or to disrupt its “effector” region are unable to affect actin filaments in either fibroblasts or epithelial cells. Furthermore, deletion of six NH2-terminal amino acids from Rnd1 leads to loss of both functional activity and subcellular localization. Interestingly, these residues are strictly conserved in Rnd3/RhoE and this could explain the lack of effects on the cytoskeleton originally reported for RhoE, since the clone used was not full length (Foster et al., 1996). We have observed that wild-type Rnd3/RhoE blocks LPA-induced stress fiber formation similar to wild-type Rnd1. Rnd2 on the other hand lacks this activity.
Our results show that expression of wild-type Rnd1, which may be constitutively in the GTP form, inhibits the formation and promotes disassembly of both actin stress fibers and focal adhesion complexes. Rnd1 appears, therefore, to have an antagonistic effect to both Rho- and Rac-regulated signaling pathways perhaps by interfering with a common element in the Rho and Rac pathways. We have attempted to determine if Rnd1 acts upstream or downstream of Rho and Rac. Coinjection of constitutively active Rho or Rac prevents Rnd1-induced actin disassembly (data not shown) and this might suggest that Rnd1 interferes with the activation of Rho and Rac (i.e., acts upstream) rather than competing with Rho and Rac for a downstream target. However, the experiment is not easy to interpret since it may depend on the relative levels of expression of Rho/Rac and Rnd.
Expression of Rnd1 in quiescent Swiss 3T3 cells leads not only to loss of actin filaments and focal adhesions, but also to loss of cell adhesion leading to cell rounding. The cell takes on a dendritic appearance and we have seen similar effects after injection of the Rho inhibitor C3 transferase. It appears, therefore, that Rnd1 also inhibits another Rho function related to cell adhesion. We do not know the nature of this function, it is not clear that it is integrin dependent because it may involve other surface adhesion proteins.
Cell rounding and loss of stress fibers is known to occur during cytokinesis, whereas a more restricted loss of stress fibers occurs close to cell protrusions such as lamellipodia and filopodia seen during cell movement. We are currently examining whether Rnd1 might play a role in either of these processes using the putative dominant-negative version of Rnd1 (N27Rnd). Another possible area of interest is transformed cells, which are often devoid of stress fibers and have a rounded, “transformed” morphology. Microinjection of immortalized fibroblast cell lines with activated Ras rapidly induces spreading and increased ruffling (Bar-Sagi and Feramisco, 1986), but at later times morphological transformation occurs, characterized by cell rounding (Lloyd et al., 1989). It is unclear which biochemical activities induced by Ras contribute to cell rounding, some possibilities include suppression of integrin activity via the MAP kinase pathway, expression and secretion of cathepsins d and l (matrix proteases implicated in the acquisition of invasiveness), decreased expression of cytoskeletal-associated proteins such as α-actinin, vinculin, and tropomyosin 2 or 3, and inhibition of Rho GTPase signaling (Qiu et al., 1995; Hughes et al., 1997). We would like to propose that the activation or increased expression of Rnd proteins might contribute to the aquisition of a transformed morphology involving the loss of stress fibers and decreased cell adhesion. Interestingly and unlike other members of the Rho family (with the exception of RhoB), Rnd proteins are predicted to be farnesylated and for RhoE this has been experimentally confirmed (Foster et al., 1996). There is currently much interest in the use of farnesyltransferase inhibitors as potential anti-Ras, and therefore anti-cancer, agents. These compounds have been reported to revert the transformed morphology of some tumor cells leading to cell flattening, reassembly of actin stress fibers, and inhibition of anchorage-independent growth (Prendergast et al., 1994). If Rnd proteins do play a role in the induction of a transformed morphology, their inhibition by farnesyltransferase inhibitors might account for morphological reversion. We are currently looking into this possibility.
We determined the chromosomal localization of Rnd1 and Rnd2. Rnd2 maps to a region frequently rearranged in breast and ovarian tumors. Breast cancer is one of the most frequent malignancies in women, and rearrangements of chromosome 17 are the most frequent genetic alterations observed in breast and ovarian tumors. More than 100 kb of human genomic DNA containing the BRCA1 gene have recently been sequenced and Rnd2 is the closest gene found after BRCA1, in the opposite orientation (Smith et al., 1996). Mutations in BRCA1 increase susceptibility to breast and ovarian cancers, but other genetic rearrangements of this region are also frequent. The human plakoglobin (γ-catenin) gene also localizes to 17 q21 and is subject to loss of heterozygosity in breast and ovarian cancers (Aberle et al., 1995). In cells, plakoglobin (γ-catenin) associates with E-cadherin and is predominantly localized in adherens junctions, as Rnd proteins.
We are currently looking at the expression of Rnd2 in breast tissue, modifications of Rnd proteins expression in cancers, and a possible involvement of Rnd proteins in tumor invasion.
Acknowledgments
We thank E. van Obberghen-Schilling, V. Vouret-Craviari, P. Lenormand, M. Rassoulzadegan, F. Cuzin, N. Tapon, and N. Lamarche for their help and discussions; C. Bazenet for the gift of rat primary neurons; B. Durand for the gift of rat astrocytes primary cultures; D. Crenesse for the gift of rat hepatocytes; D. Lawson for the gift of primary rat embryo fibroblasts; G. Jarretou for her help with brain and liver sections; N. Leroudier for DNA sequencing; and M. Chabre for support.
Abbreviations used in this paper
- GEF
guanine nucleotide exchange factors
- GAP
GTPase-activating proteins
- LPA
lysophosphatidic acid
- REF
rat embryo fibroblast
Footnotes
C.D. Nobes and A. Hall were supported by the Cancer Research Campaign (UK). P. Chardin's work in A. Hall's laboratory was supported by a short term European Molecular Biological Organisation fellowship.
Address all correspondence to Pierre Chardin's present address, Cancer Center, University of California, 2340 Sutter Street, San Francisco, CA 94115. Fax: (415) 502-3179. E-mail: chardin@cc.ucsf.edu
References
- Aberle H, Bierkamp C, Torchard D, Serova O, Wagner T, Natt E, Wirshing J, Heidkamper C, Montagna M, Lynch HT, et al. The human plakoglobin gene localizes on chromosome 17 q21 and is subjected to loss of heterozygosity in breast and ovarian cancers. Proc Natl Acad Sci USA. 1995;92:6384–6388. doi: 10.1073/pnas.92.14.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktories K, Just I. In vitro ADP-ribosylation of Rho by bacterial ADP-ribosyl transferases. Methods Enzymol. 1995;256:184–195. doi: 10.1016/0076-6879(95)56023-8. [DOI] [PubMed] [Google Scholar]
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by Ras proteins. Science. 1986;233:1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- Chardin P, Boquet P, Madaule P, Popoff MR, Rubin EJ, Gill DM. The mammalian G protein RhoC is ADP-ribosylated by clostridium botulinum exoenzyme C3 and affects actin microfilaments in Vero cells. EMBO (Eur Mol Biol Organ) J. 1989;8:1087–1092. doi: 10.1002/j.1460-2075.1989.tb03477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coso OA, Chiariello M, Yu JC, Teramoto H, Crespo P, Xu N, Miki T, Gutkind JS. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- Der CJ, Finkel T, Cooper GM. Biological and biochemical properties of human Hras genes mutated at codon 61. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- Foster R, Hu K-Q, Lu Y, Nolan KM, Thissen J, Settleman J. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in-vivo farnesylation. Mol Cell Biol. 1996;16:2689–2699. doi: 10.1128/mcb.16.6.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Huber LA, Ullrich O, Takai Y, Lutcke A, Dupree P, Olkkonen V, Virtta H, Dehoop MJ, Alexandrov K, Peter M, Zerial M, Simons K. Mapping of Ras-related GTP-binding proteins by GTP overlay following two-dimensional gel electrophoresis. Proc Natl Acad Sci USA. 1994;91:7874–7878. doi: 10.1073/pnas.91.17.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes PE, Renshaw MW, Pfaff M, Forsyth J, Keivens VM, Schwartz MA, Ginsberg MH. Suppression of integrin activation: a novel function of a Ras/raf-initiated MAP kinase pathway. Cell. 1997;88:521–530. doi: 10.1016/s0092-8674(00)81892-9. [DOI] [PubMed] [Google Scholar]
- John J, Frech M, Wittinghofer A. Biochemical properties of H-Ras encoded p21 mutants and mechanism of the autophosphorylation reaction. J Biol Chem. 1988;263:11792–11799. [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuka M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase. Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche N, Hall A. GAPs for Rho-related GTPases. Trends Genet. 1994;10:436–440. doi: 10.1016/0168-9525(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Leung T, Chen XQ, Manser E, Lim L. The p160 RhoA-binding kinase ROK alpha is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol Cell Biol. 1996;16:5313–5327. doi: 10.1128/mcb.16.10.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AC, Paterson HF, Morris JD, Hall A, Marshall CJ. p21H-ras-induced morphological transformation and increases in c-myc expression are independent of protein kinase C. EMBO (Eur Mol Biol Organ) J. 1989;8:1099–1104. doi: 10.1002/j.1460-2075.1989.tb03479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJG, Esch F, Furthmayr H, Hall A. Rho- and Rac- dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: An essential role for ezrin/radixin/moesin proteins. J Cell Biol. 1997;138:927–938. doi: 10.1083/jcb.138.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO (Eur Mol Biol Organ) J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast GC, Davide JP, Desolms SJ, Guiliani EA, Graham SL, Gibbs JB, Oliff A, Kohl NE. Farnesyltransferase inhibition causes morphological reversion of Ras-transformed cells by a complex mechanism that involves regulation of the actin cytoskeleton. Mol Cell Biol. 1994;14:4193–4202. doi: 10.1128/mcb.14.6.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R-G, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Patterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Smith TM, Lee MK, Szabo CI, Jerome N, McEven M, Taylor M, Hood L, King MC. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Research. 1996;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- Tsukita Sa, Yonemura S, Sh, Tsukita ERM family: from cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997;9:70–75. doi: 10.1016/s0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- Ueda T, Kikuchi A, Ohga N, Yamamoto J, Takai Y. Purification and characterization from bovine brain cytosol of a novel regulatory protein inhibiting the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. J Biol Chem. 1990;265:9373–9380. [PubMed] [Google Scholar]
- Van Aelst L, D'Souza–Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]