Abstract
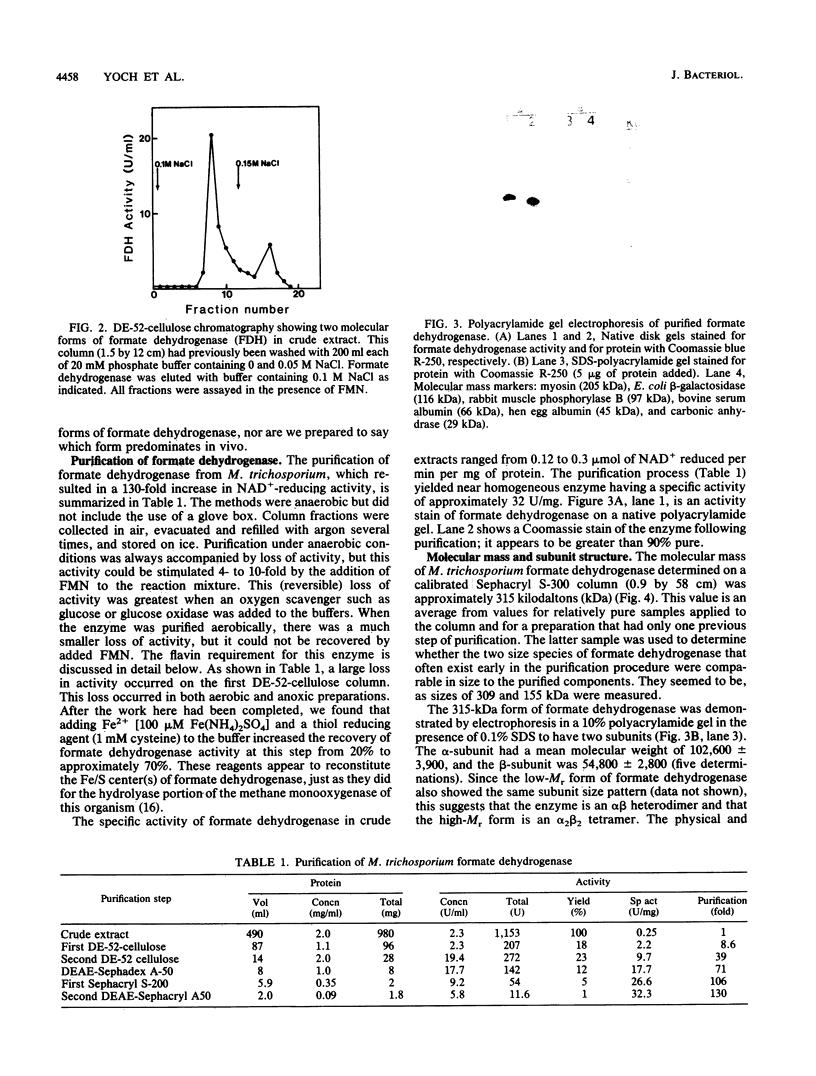
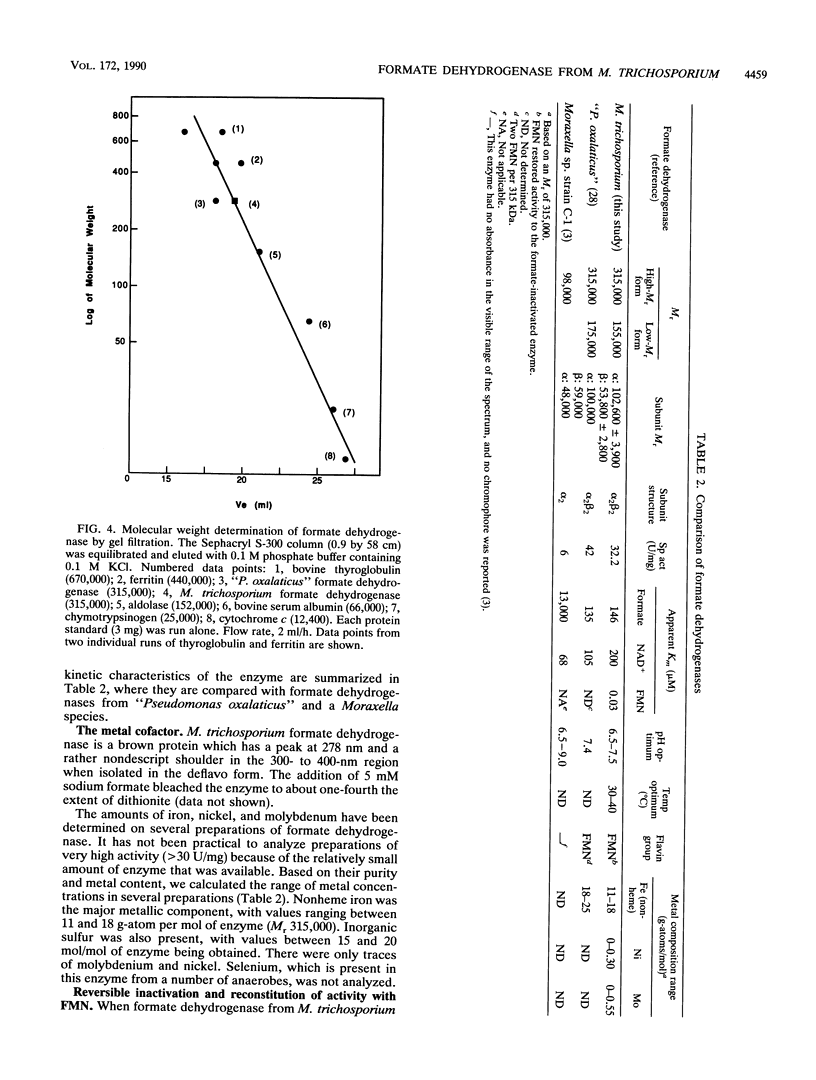
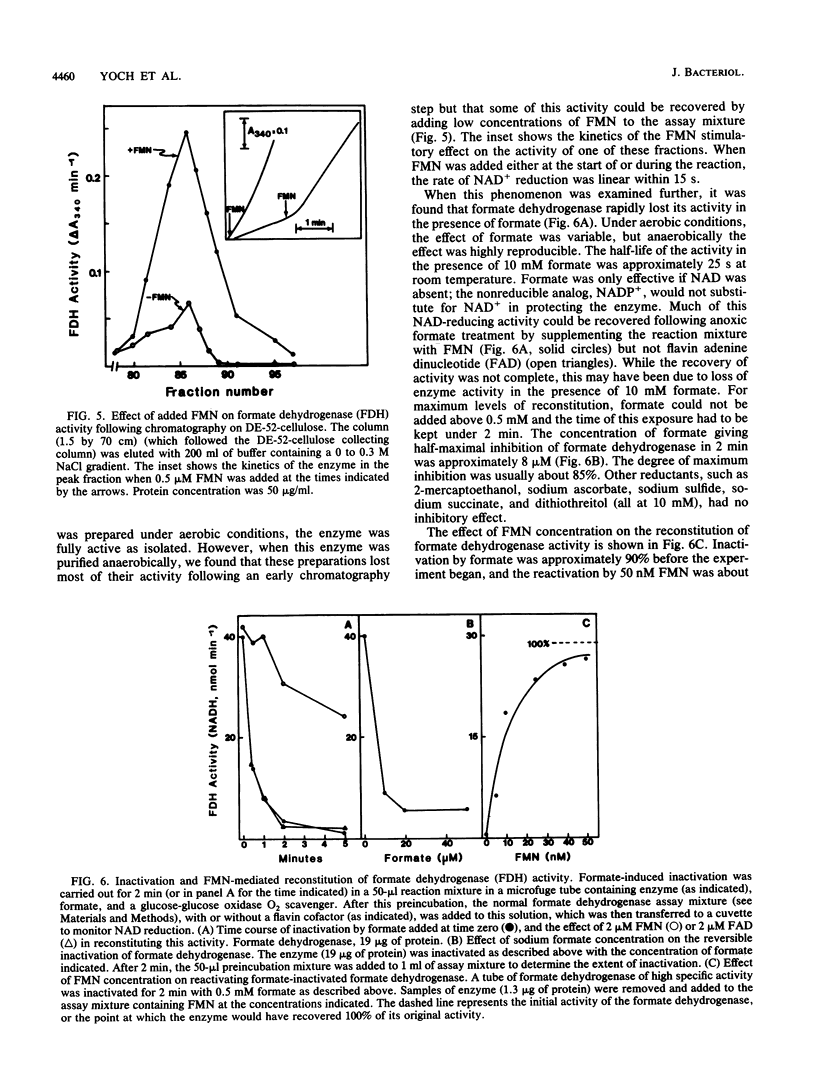
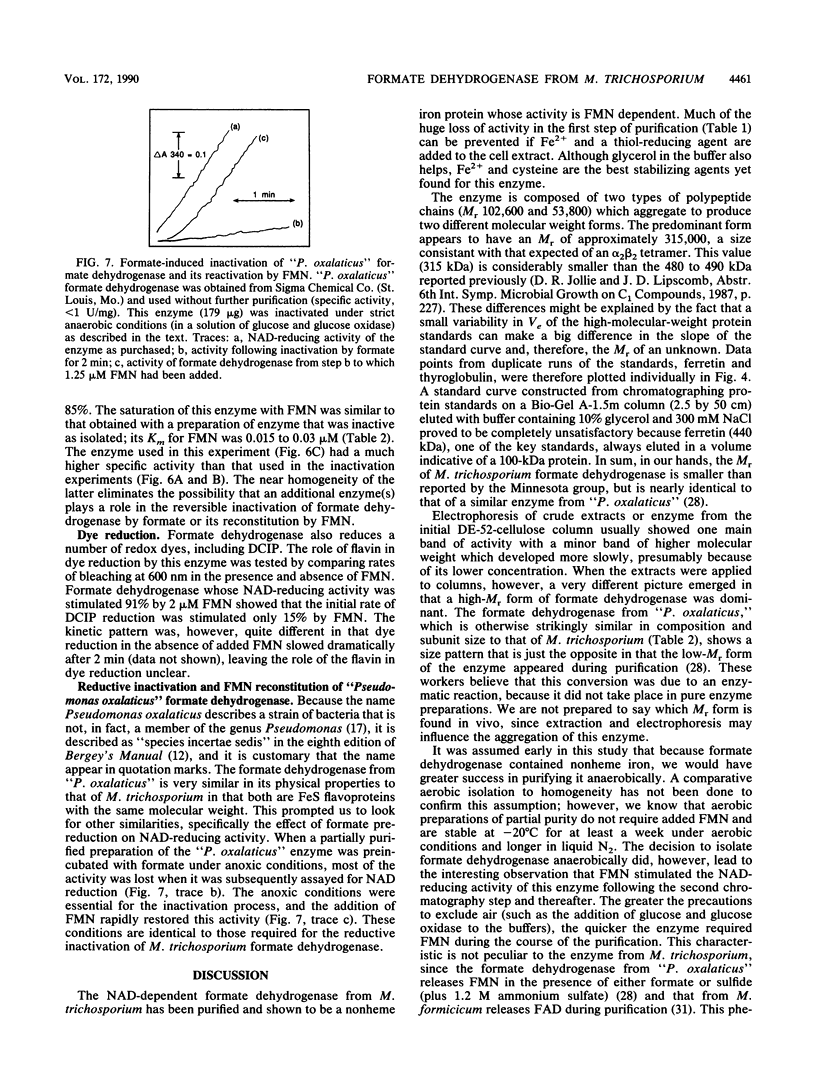
Formate dehydrogenase (NAD+ dependent) was isolated from the obligate methanotroph Methylosinus trichosporium OB3b. When the enzyme was isolated anaerobically, two forms of the enzyme were seen on native polyacrylamide gels, DE-52 cellulose and Sephacryl S-300 columns; they were approximately 315,000 and 155,000 daltons. The enzyme showed two subunits on sodium dodecyl sulfate-polyacrylamide gels. The Mr of the alpha-subunit was 53,800 +/- 2,800, and that of the beta-subunit was 102,600 +/- 3,900. The enzyme (Mr 315,000) was composed of these subunits in an apparent alpha 2 beta 2 arrangement. Nonheme iron was present at a concentration ranging from 11 to 18 g-atoms per mol of enzyme (Mr 315,000). Similar levels of acid-labile sulfide were detected. No other metals were found in stoichiometric amounts. When the enzyme was isolated aerobically, there was no cofactor requirement for NAD reduction; however, when isolated anaerobically, activity was 80 to 90% dependent on the addition of flavin mononucleotide (FMN) to the reaction mixture. Furthermore, the addition of formate to an active, anoxic solution of formate dehydrogenase rapidly inactivated it in the absence of an electron acceptor; this activity could be reconstituted approximately 85% by 50 nM FMN. Flavin adenine dinucleotide could not replace FMN in reconstituting enzyme activity. The Kms of formate dehydrogenase for formate, NAD, and FMN were 146, 200, and 0.02 microM, respectively. "Pseudomonas oxalaticus" formate dehydrogenase, which has physical characteristics nearly identical to those of the M. trichosporium enzyme, was also shown to be inactivated under anoxic conditions by formate and reactivated by FMN. The evolutionary significance of this similarity is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem J. 1965 Sep;96(3):595–606. doi: 10.1042/bj0960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y., Sekigawa T., Inukai H., Nakazawa A. Purification and properties of formate dehydrogenase from Moraxella sp. strain C-1. J Bacteriol. 1988 Jul;170(7):3189–3193. doi: 10.1128/jb.170.7.3189-3193.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilova T. V., Egorova O. A., Ioanesyan L. S., Egorov A. M. Biosynthesis, isolation and properties of NAD-dependent formate dehydrogenase from the yeast Candida methylica. Eur J Biochem. 1985 Nov 4;152(3):657–662. doi: 10.1111/j.1432-1033.1985.tb09245.x. [DOI] [PubMed] [Google Scholar]
- BRUMBY P. E., MILLER R. W., MASSEY V. THE CONTENT AND POSSIBLE CATALYTIC SIGNIFICANCE OF LABILE SULFIDE IN SOME METALLOFLAVOPROTEINS. J Biol Chem. 1965 May;240:2222–2228. [PubMed] [Google Scholar]
- Baron S. F., Ferry J. G. Purification and properties of the membrane-associated coenzyme F420-reducing hydrogenase from Methanobacterium formicicum. J Bacteriol. 1989 Jul;171(7):3846–3853. doi: 10.1128/jb.171.7.3846-3853.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S. F., Ferry J. G. Reconstitution and properties of a coenzyme F420-mediated formate hydrogenlyase system in Methanobacterium formicicum. J Bacteriol. 1989 Jul;171(7):3854–3859. doi: 10.1128/jb.171.7.3854-3859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. P., Yoch D. C. Isolation, characterization, and biological activity of ferredoxin-NAD+ reductase from the methane oxidizer Methylosinus trichosporium OB3b. J Bacteriol. 1989 Sep;171(9):5012–5016. doi: 10.1128/jb.171.9.5012-5016.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. P., Yoch D. C. Regulation of two nickel-requiring (inducible and constitutive) hydrogenases and their coupling to nitrogenase in Methylosinus trichosporium OB3b. J Bacteriol. 1987 Oct;169(10):4778–4783. doi: 10.1128/jb.169.10.4778-4783.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorov A. M., Avilova T. V., Dikov M. M., Popov V. O., Rodionov Y. V., Berezin I. V. NAD-dependent formate dehydrogenase from methylotrophic bacterium, strain 1. Purification and characterization. Eur J Biochem. 1979 Sep;99(3):569–576. doi: 10.1111/j.1432-1033.1979.tb13289.x. [DOI] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- Fox B. G., Froland W. A., Dege J. E., Lipscomb J. D. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J Biol Chem. 1989 Jun 15;264(17):10023–10033. [PubMed] [Google Scholar]
- Hou C. T., Patel R. N., Laskin A. I., Barnabe N. NAD-linked formate dehydrogenase from methanol-grown Pichia pastoris NRRL-Y-7556. Arch Biochem Biophys. 1982 Jun;216(1):296–305. doi: 10.1016/0003-9861(82)90214-4. [DOI] [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem. 1981 Jan 25;256(2):656–663. [PubMed] [Google Scholar]
- Karzanov V. V., Bogatsky YuA, Tishkov V. I., Egorov A. M. Evidence for the presence of a new NAD+-dependent formate dehydrogenase in Pseudomonas sp. 101 cells grown on a molybdenum-containing medium. FEMS Microbiol Lett. 1989 Jul 15;51(1):197–200. doi: 10.1016/0378-1097(89)90508-9. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Mayhew S. G., Massey V. Evidence for a novel flavin prosthetic group associated with NADH dehydrogenase from Peptostreptococcus elsdenii. Biochim Biophys Acta. 1971 May 12;235(2):303–310. doi: 10.1016/0005-2744(71)90208-7. [DOI] [PubMed] [Google Scholar]
- Müller U., Willnow P., Ruschig U., Höpner T. Formate dehydrogenase from Pseudomonas oxalaticus. Eur J Biochem. 1978 Feb;83(2):485–498. doi: 10.1111/j.1432-1033.1978.tb12115.x. [DOI] [PubMed] [Google Scholar]
- Petrov R. R., Utkin I. B., Popov V. O. Redox-dependent inactivation of the NAD-dependent hydrogenase from Alcaligenes eutrophus Z1. Arch Biochem Biophys. 1989 Jan;268(1):298–305. doi: 10.1016/0003-9861(89)90591-2. [DOI] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Composition of the coenzyme F420-dependent formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1986 Feb;165(2):405–411. doi: 10.1128/jb.165.2.405-411.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. FAD requirement for the reduction of coenzyme F420 by formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1983 Aug;155(2):467–472. doi: 10.1128/jb.155.2.467-472.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D. I., Dalton H. Properties of the methane mono-oxygenase from extracts of Methylosinus trichosporium OB3b and evidence for its similarity to the enzyme from Methylococcus capsulatus (Bath). Eur J Biochem. 1979 May 2;96(1):205–212. doi: 10.1111/j.1432-1033.1979.tb13030.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto I., Saiki T., Liu S. M., Ljungdahl L. G. Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J Biol Chem. 1983 Feb 10;258(3):1826–1832. [PubMed] [Google Scholar]
- Yoch D. C., Lindstrom E. S. Nicotinamide adenine dinucleotide-dependent formate dehydrogenase from Rhodopseudomonas palustris. Arch Mikrobiol. 1969;67(2):182–188. doi: 10.1007/BF00409684. [DOI] [PubMed] [Google Scholar]