Abstract
A small cell-binding proteoglycan for which we propose the name osteoadherin was extracted from bovine bone with guanidine hydrochloride–containing EDTA. It was purified to homogeneity using a combination of ion-exchange chromatography, hydroxyapatite chromatography, and gel filtration. The Mr of the proteoglycan was 85,000 as determined by SDS-PAGE. The protein is rich in aspartic acid, glutamic acid, and leucine. Two internal octapeptides from the proteoglycan contained the sequences Glu-Ile-Asn-Leu-Ser-His-Asn-Lys and Arg-Asp-Leu-Tyr-Phe-Asn-Lys-Ile. These sequences are not previously described, and support the notion that osteoadherin belongs to the family of leucine-rich repeat proteins. A monospecific antiserum was raised in rabbits. An enzyme-linked immunosorbent assay was developed, and showed the osteoadherin content of bone extracts to be 0.4 mg/g of tissue wet weight, whereas none was found in extracts of various other bovine tissues. Metabolic labeling of primary bovine osteoblasts followed by immunoprecipitation showed the cells to synthesize and secrete the proteoglycan. Digesting the immunoprecipitated osteoadherin with N-glycosidase reduced its apparent size to 47 kD, thus showing the presence of several N-linked oligosaccharides. Digestion with keratanase indicated some of the oligosaccharides to be extended to keratan sulfate chains. In immunohistochemical studies of the bovine fetal rib growth plate, osteoadherin was exclusively identified in the primary bone spongiosa. Osteoadherin binds to hydroxyapatite. A potential function of this proteoglycan is to bind cells, since we showed it to be as efficient as fibronectin in promoting osteoblast attachment in vitro. The binding appears to be mediated by the integrin αvβ3, since this was the only integrin isolated by osteoadherin affinity chromatography of surface-iodinated osteoblast extracts.
The extracellular matrix of bone is mineralized with crystals of hydroxyapatite. The spatial orientation of the crystals depends on the most abundant bone matrix protein, type I collagen (25, 48). Fibers of this collagen comprise 90% of the organic material in the mineralized bone matrix. They are highly insoluble becuase of intra- and intermolecular cross-links (9). During the last two decades a number of noncollagenous proteins have been isolated from bone tissue and characterized (17, 50). Examples are osteocalcin (38), matrix gla-protein (39), osteonectin (3, 8), osteopontin (33), bone sialoprotein (BSP; references 11 and 34),1 and the small bone proteoglycans decorin (27) and biglycan (10). However, in most cases very little is known about their function in the tissue, though it appears that osteopontin is crucially involved in anchoring osteoclasts to the mineral matrix of bone surfaces via the integrin αvβ3 (19, 40). Osteopontin is also enriched at the mineralization front (18), indicating its involvement in mineral deposition and growth, perhaps as an inhibitor (22) since it contains a polyaspartic acid sequence (33). BSP has been suggested to be involved in hydroxyapatite nucleation (21). In support, the protein has a predominant localization at the interface between mineralizing growth cartilage and bone (20). Decorin binds to collagen type I, modifying the properties of the completed fibril and potentially regulating collagen fibrillogenesis (15). Decorin also binds TGF-β (49) and may be involved in sequestering this factor in the bone matrix to be released upon bone remodeling. Somewhat surprisingly, little is known of the function of osteocalcin in bone, despite the fact that the protein was described early. However, a recently described inactivation of the gene gave a phenotype manifesting increased bone mineral density, and suggested osteocalcin involvement in bone remodeling (7).
Here we describe the isolation of a novel keratan sulfate proteoglycan from bovine long bone, and the structural and functional characteristics of this new bone component. The proteoglycan has strong integrin-dependant cell-binding ability. We propose the name osteoadherin, since it promotes cell attachment as efficiently as fibronectin in a manner dependent on the amino acid sequence RGD, and because of its high affinity to hydroxyapatite.
Materials and Methods
Bovine Bone Extraction
The diaphyseal part of the tibiae from 2-yr-old steers were carefully cleaned from adhering connective tissue and bone marrow. The bones were frozen in liquid nitrogen and crushed into small pieces with a hydraulic press, followed by grinding of the frozen bone pieces into powder. 100 g of frozen powdered bovine bone was extracted in sequence, first with 10 vol of 4 M guanidine hydrochloride in 50 mM sodium acetate, pH 5.8 (to remove non–mineral-associated proteins and cells), and then with 30 vol of 4 M guanidine hydrochloride containing 0.5 M disodium EDTA in 50 mM Tris/HCl buffer, pH 7.4 (to release proteins in the mineral compartment). Each extraction solution contained proteinase inhibitors as described in detail elsewhere (12). The EDTA extract was clarified by centrifugation at 10,000 g for 40 min. The supernatant of the extract was concentrated at 4°C by ultrafiltration (PM-10 filter; Amicon Corp., Easton, TX). The concentrate was transferred into 7 M urea, 0.1 M sodium acetate, 10 mM Tris/HCl buffer, pH 6.0, by diaflow with 10 vol of the urea solution.
Chromatographic Purification of Osteoadherin
The guanidine hydrochloride/EDTA extract from 100 g of bone was brought into the 7 M urea/Tris buffer (see above), chromatographed on a DEAE-cellulose (DE-52) ion-exchange column (4 × 15.0 cm) as described previously (13). The column was eluted with a linear gradient of sodium acetate (0.1–1.2 M) to a total volume of 1.5 liters in the urea/Tris buffer described above. A peak corresponding to 0.25–0.35 M sodium acetate was pooled and dialyzed against distilled water and freeze-dried. It was dissolved in 7 M urea, 20 mM sodium phosphate, pH 8.0. The sample was applied to a hydroxyapatite column (HTP, 4.0 × 5.5 cm; Bio-Rad Laboratories, Hercules, CA). The remaining bound material was eluted with a gradient of 0.02–0.2 M sodium phosphate (2 × 250 ml) in the same solvent. Fractions were collected and analyzed for protein content by measuring absorbance at 280 nm and by SDS-PAGE.
Fractions corresponding to 84–104 mM sodium phosphate from the hydroxyapatite chromatography were pooled and transferred by diaflow into 7 M urea, 20 mM bis Tris, 50 mM NaCl, pH 7.0. The sample was chromatographed on a Mono Q column HR 5/5 (Pharmacia Biotech Sverige, Uppsala, Sweden). Bound material was eluted from the column at a flow rate of 0.5 ml/min with a linear gradient of sodium chloride (0.050–0.5 M) in the same buffer as above. Fractions (0.5 ml) were collected and analyzed for protein content by measuring the absorbance at 280 nm and by SDS-PAGE.
Fractions 28–35 from the Mono Q column were pooled and transferred by diaflow into 4 M guanidine hydrochloride, 20 mM Tris, pH 8.0. The material was then reduced by adding 5 mM DTT (Merck, Darmstadt, Germany) and incubated at room temperature for 3 h. The sample was chromatographed on a Superdex 200 column (Pharmacia Biotech Sverige), using an HPLC system (Pharmacia Biotech Sverige). The column was eluted with the guanidine hydrochloride buffer at a flow rate of 0.25 ml/min, and 0.5-ml fractions were collected.
SDS-PAGE
Samples were prepared for electrophoresis by ethanol precipitation as described elsewhere (37), and dissolved in 5% SDS/sample buffer with or without mercaptoethanol, heated in a boiling water bath for 2 min, and electrophoresed on gradient polyacrylamide (4–16%) slab gels with a 3% stacking gel and the buffer system described by Laemmli (28). Gels were stained with Coomassie blue G 250 (Serva, Heidelberg, Germany) as described by Neuhoff et al. (32).
Amino Acid Analysis
The amino acid composition was determined (after hydrolysis of samples in 6 M HCl for 24 h at 110°C under argon) using an automatic amino acid analyzer (High Performance Analyzer System 6300™; Beckman Instruments, Inc., Fullerton, CA) equipped for ion exchange chromatography and detection with ninhydrin.
Protein Fragmentation and Amino Acid Sequence Determination
50 μg of pure osteoadherin was cleaved in 70% formic acid containing 1% cyanogen bromide (CNBr) for 24 h at room temperature (16). The peptides were separated by reversed phase chromatography on a μRPC C2/18 column (SC 2.1/10; Pharmacia Biotech Sverige) using a SMART system™ (Pharmacia Biotech Sverige). The CNBr peptides were eluted with a linear gradient of acetonitrile (0–40% in 160 min) in 0.065–0.05% trifluoroacetic acid at a flow rate of 100 μl/min. Amino acid sequences were determined with a protein/peptide sequencer (model 470A™; PE Applied Biosystems, Foster City, CA). The amino acid sequences obtained were used to search the Swiss-Prot protein sequence data base (Geneva University Hospital and University of Geneva, Geneva, Switzerland).
Antibody Preparation
Antibodies were raised against osteoadherin by subcutaneous immunization of a rabbit with the protein (100 μg) dissolved in 0.15 M NaCl, 5 mM sodium phosphate, pH 7.4, and emulsified with an equal volume of Freund's complete adjuvant (Difco Laboratories Inc., Detroit, MI). A good titer level was obtained after two subsequent boosters (2 × 100 μg) with osteoadherin in Freund's incomplete adjuvant. Antibody titers were determined with an ELISA using polystyrene microtiter plates coated with 2 μg/ml of protein.
Western Blotting
Samples were transferred to nitrocellulose after electrophoresis on 4–16% gels, essentially as described by Towbin (47). The nitrocellulose sheet was immunoblotted with the primary antiserum (rabbit anti–bovine osteoadherin) using a 1:200 dilution in 0.15 M NaCl, 10 mM Tris, 0.2% Tween, pH 7.4, and a 1:500 dilution in the same buffer of the secondary antiserum (peroxidase-conjugated pig anti–rabbit IgG; DAKOPATTS, Copenhagen, Denmark). Bound antibodies were detected using DAB (Chemicon International, Inc., Temecula, CA) as the substrate.
ELISA
Tissues were extracted with 10 vol of guanidine hydrochloride and treated exactly as described previously (30). These tissues were bone, liver, trachea, tendon, articular cartilage, kidney cortex, heart, intestine, muscle, and cornea. The bone tissue was extracted a second time with guanidine hydrochloride/EDTA, and this extract was dialyzed against guanidine hydrochloride to remove the EDTA before ethanol precipitation and assay. The principles of the ELISA have been described elsewhere (30). Thus, plates were coated with 0.1 μg of osteoadherin in 4 M guanidine hydrochloride, 50 mM sodium carbonate, pH 10.0. Ethanol-precipitated extracts and isolated osteoadherin were dissolved and diluted in 0.8% SDS. An equal volume of antibody at appropriate dilution in 4% Triton was added before incubation in the coated plates. Bound antibodies were detected using a secondary antibody/alkaline phosphatase conjugate.
Metabolic Labeling of Cell Cultures and Immunoprecipitation
Primary bovine osteoblasts were prepared according to Robey (41). The cells were grown in Ham's F12 medium supplemented with 10% FBS, penicillin, and streptomycin. The osteoblasts were labeled with [3H]leucine (50 μCi/ml) and [35S]sulfate (0.1 mCi/ml) in Ham's F12 for 6 h. Conditioned media (3 ml) were precipitated with ethanol (37) and subsequently immunoprecipitated using the method of Oldberg et al. (35) with the specific antiserum. Precipitated radiolabeled proteins were identified by electrophoresis on a 4–16% gradient gel and fluorography (5).
Analysis of Carbohydrate Substituents
The osteoadherin protein was immunoprecipitated from [35S]sulfate- labeled cells (as described above) and digested with keratanase (keratan sulfate, 1,4-β-d-galactanohydrolase, EC 3.2.1.103; Boehringer Mannheim Corp., Indianapolis, IN) or with N-glycosidase F (Boehringer Mannheim Corp.) following the recommendations of the manufacturers. SDS-PAGE was performed using a 4–16% gradient polyacrylamide gel. Labeled proteins were detected by fluorography.
Immunohistochemical Staining
Ribs from a late third trimester bovine fetus were frozen in liquid N2 and mounted in OCT Compound (Tissue Tek II™; Miles Laboratories, Naperville, IL) on a cryostat stage. Frozen sections (5 μm) were prepared at −22°C and collected on gelatin-coated slides. Before immunostaining, the sections were dried at room temperature for 2 h, followed by incubation for 10 min in acetone and rehydration in PBS. Endogenous peroxidase was quenched by incubating in PBS containing 1% H2O2 for 20 min. To reduce nonspecific binding, each section was incubated with goat serum (1:70 in PBS-0.01% BSA) for 20 min. The sections were incubated with primary antibody against osteoadherin (diluted to 1:1,000 in PBS-0.01% BSA) or the preimmune serum (diluted to 1:1,000 in PBS 0.01% BSA) at 4°C overnight in a moist chamber. The sections were then treated with biotinylated secondary antibody (diluted 1:200) and avidin–peroxidase conjugate using the Vectastain ABC kit™ (Vector Labs, Burlingame, CA), following the recommendations of the manufacturer.
Cell Attachment to Osteoadherin
Primary osteoblasts isolated with the method of Robey et al. (41) were grown to near confluency in Ham's F12 medium supplemented with 10% FBS, 50 UI penicillin, and 50 μg/ml streptomycin. Wells in a 96-well polystyrene immunoplate (cat. no. 4-39454; Nunc, Roskilde, Denmark) were coated with 100 μl of 10 μg/ml bovine osteoadherin, 10 μg/ml fibronectin (Calbiochem-Novabiochem Corp., La Jolla, CA) or 10 μg/ml BSA (Sigma Chemical Co., St. Louis, MO) in 4 M guanidine hydrochloride, 50 mM Na2CO3, pH 10.0. In a separate experiment, wells in a 96-well immunoplate were coated with nonreduced bovine osteoadherin, bovine osteonectin, fibronectin, and BSA, respectively, at 10 μg/ml in 100 μl in the same coating buffer as above.
After coating overnight at room temperature, the wells were rinsed with PBS and aftercoated with 100 μl of 1 mg BSA/ml in PBS for 2 h. Plates were then rinsed with PBS and used directly for binding experiments performed as described elsewhere (45). Cultured primary osteoblasts were briefly treated with EDTA and rapidly washed three times in Ham's F12 medium, and resuspended in F12 at 0.2 × 106 cells per ml. An aliquot (100 μl) of the cell suspension was added to each well (20,000 cells). Some cell samples were incubated with 1 mM CaCl2 or 1 mM MgCl2. In some cases the cells were incubated with the combination of 1 mM CaCl2 and 1 mM MgCl2. After incubation for 1 h at 37°C, the wells were gently rinsed three times with PBS, and the number of attached cells was determined by measuring the N-acetyl-hexosaminidase activity as described by Landegren (29).
Inhibition of Cell Attachment
The RGD peptide inhibition experiment was performed in osteoadherin-coated wells (see above). The primary osteoblasts were allowed to attach in the presence of increasing concentrations of the synthetic peptides GRGDSL and GRGESL, respectively. The peptides were dissolved in Ham's F12, 1 mM CaCl2, 1 mM MgCl2, and 0.1% BSA at the following concentrations: l, 5, 25, 75, 100, and 400 μg/ml. Approximately 2 × 104 cells were allowed to attach to osteoadherin for 1 h in the presence of a competing or a noncompeting peptide. Bound cells were quantified as described above.
Receptor Characterization
Human osteosarcoma cells (MG 63) were grown in Ham's F12 medium plus 10% FBS, 50 UI penicillin, and 50 μg/ml streptomycin (Gibco Laboratories, Grand Island, NY). The cells were harvested by trypsinization and then washed once in PBS containing 1 mg/ml glucose (PBS-glucose).
The osteosarcoma cells (MG 63) were resuspended in 1 ml PBS-glucose. The cells were iodine-labeled on ice for 15 min with 1 mCi of carrier-free [125I]sodium iodide (Nycomed Amersham Inc., Princeton, NJ) including 3.8 U of lactoperoxidase (120 U/mg; Sigma Chemical Co.) and 0.04 U of glucose oxidase (1,000 U/ml, Sigma Chemical Co.) in PBS-glucose. The iodination was stopped by adding 10 ml of Ham's F12 culture medium. After washing three times in PBS, the cells were extracted for 1 h on ice with 2 ml extraction buffer containing 1% Triton-X100, 100 μg/ml aprotinin, 4 μg/ml leupeptin, 4 μg/ml pepstatin A, 1 mM PMSF (Sigma Chemical Co.), 1 mM MnCl2, 1 mM MgCl2, and 10 mM Tris-HCl, pH 7.4. Cell lysates were centrifuged at 10,000 rpm for 30 min at 4°C (Micromax centrifuge, rotor 851; International Equipment Co., Needham, MA).
An osteoadherin (OSAD) affinity column was prepared by coupling 1 mg of the proteoglycan to 1 ml of cyanogen bromide–activated Sepharose CL-4B (Pharmacia Biotech Sverige) according to the manufacturer. A control column was made by treating the agarose in exactly the same way but without protein.
Receptor isolation was accomplished by chromatographing detergent-solubilized osteosarcoma cell proteins on OSAD-agarose (1.0 ml) and control agarose (1.0 ml) in minicolumns (Bio-Rad Laboratories) essentially as described by Camper et al. (4). Samples of the affinity-purified proteins were precipitated with ethanol followed by separation on 4–12% SDS-PAGE and visualized by autoradiography or phosphor image analysis using the BioImaging Analyzer Bas2000 (Fuji Photo Film Co., Tokyo, Japan).
In parallel, radiolabeled proteins were immunoprecipitated from the affinity-purified material. Thus, 5 μg/ml sample of antiintegrin αv-cyto (polyclonal peptide antiserum against the cytoplasmic tail of αv), antiintegrin αv (polyclonal antiserum against the whole αv-subunit), antiintegrin β3-cyto (polyclonal peptide antiserum against the cytoplasmic tail of β3; kind gifts by Dr. Erkki Ruoslahti; reference 14), or a monoclonal antiintegrin β3 (CD61, Clone RUU-PL 7F12; Becton Dickinson, Bedford, MA) were incubated overnight with antibodies followed by incubation with 100 μl protein A-Sepharose (Pharmacia Biotech Sverige) or with 75 μl of antimouse IgG-agarose (Sigma Chemical Co.) for 2 h. The agarose beads were centrifuged for 4 min at 4,000 rpm (Micromax centrifuge, rotor 851; International Equipment Co.) and washed three times with 1% Triton X-100, 0.5 M NaCl, and 10 mM Tris-HCl, pH 7.4. All steps were performed at 4°C. SDS-PAGE sample buffer was added to the washed immunoprecipitates, and the samples were boiled for 5 min with or without 2-mercaptoethanol (5%). The immunoprecipitates were electrophoresed along with aliquots of the total medium and eluate on SDS polyacrylamide gradient (4–16%) gels and visualized by autoradiography or by phosphor image analysis.
Results
Purification of Osteoadherin
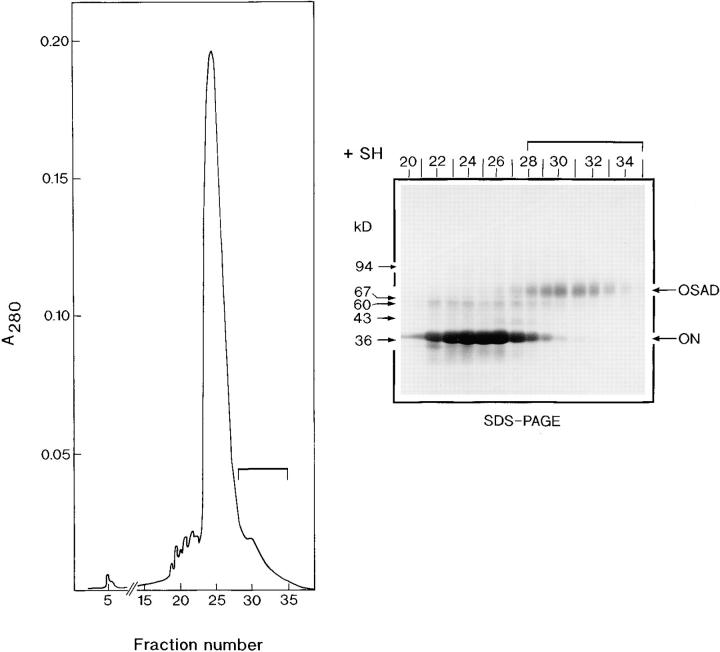
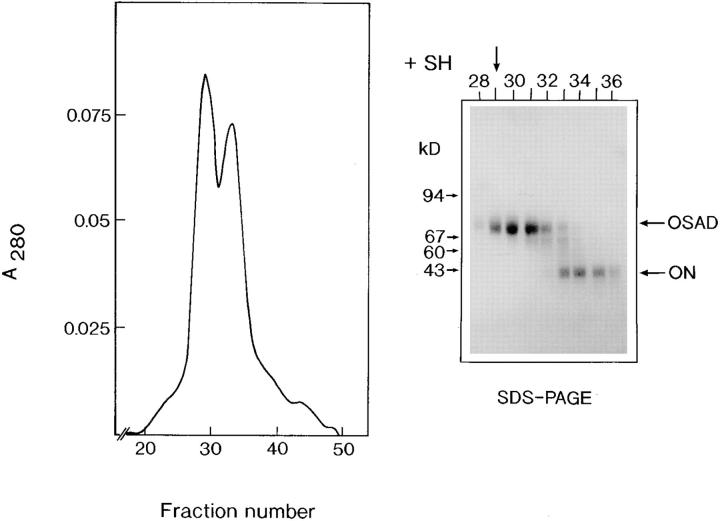
A novel keratan sulfate proteoglycan, which we named osteoadherin, was extracted from the mineral compartment of bovine diaphyseal bone and purified to homogeneity by means of four chromatographic steps. In the first step, the protein's acidic character was used to separate it from other more anionic bone matrix components by using an anion-exchange column. Osteoadherin has a M r of 85,000 under reducing conditions on SDS-PAGE. A complicating factor during purification was the presence of a component with the same charge and size, which comigrated in several chromatographic procedures we tried. As we found osteoadherin to have strong affinity for hydroxyapatite, we were able to separate osteoadherin from the comigrating contaminant in the second preparation step by using hydroxyapatite chromatography. However, as the preparation still contained large amounts of osteonectin, a subsequent ion-exchange chromatography step was performed on a Mono Q column (Fig. 1). Osteonectin elutes in the main peak in the chromatogram, while osteoadherin trails. The preparation still contained small amounts of osteonectin that could be completely removed only after reduction of the sample, final purification thus being achieved by gel filtration (Superdex 200) under reducing conditions (Fig. 2, inset).
Figure 1.
Mono Q chromatography at pH 7.0. A sample of semipurified osteoadherin, obtained by ion exchange chromatography of a bone extract followed by hydroxyapatite chromatography, was dialyzed against 7 M urea, 20 mM bis-Tris, 50 mM NaCl, pH 7.0. The sample was chromatographed on a Mono Q column to separate osteoadherin from osteonectin. The column was eluted with a sodium chloride gradient of 0.050–0.50 M in the same buffer. Samples of the fractions were taken to SDS-PAGE under reducing conditions after precipitation with ethanol, as outlined by Laemmli (28). Osteonectin (ON) elutes as a main peak with an M r of 39,000, while OSAD elutes later with an M r of 85,000. Osteoadherin-containing fractions 28–35, indicated by a bar, were pooled and taken to the final purification step.
Figure 2.
Superdex 200 chromatography of the reduced protein. The partially purified osteoadherin from the Mono Q column (Fig. 1) was dissolved in 4 M guanidine hydrochloride, 20 mM Tris, pH 8.0. The sample was reduced with DTT to break intermolecular disulfide bridges between OSAD and osteonectin (ON), followed by chromatography on a Superdex 200 column. 0.5-ml fractions were collected. Samples were precipitated with ethanol, and subjected to SDS-PAGE under reducing conditions. Fraction 29, indicated by an arrow, contains pure osteoadherin.
Chemical Composition of Osteoadherin
Determination of the amino acid composition of osteoadherin showed it to have very high content of glutamic acid/ glutamine and aspartic acid/asparagine, together accounting for more than one fourth of the residues (Table I). Another predominant amino acid in osteoadherin is leucine.
Table I.
Total Amino Acid Composition of Osteoadherin
| Amino acid | Residues/1,000 | |
|---|---|---|
| Aspartic acid/asparagine | 129 | |
| Threonine | 45 | |
| Serine | 58 | |
| Glutamic acid/glutamine | 145 | |
| Proline | 65 | |
| Glycine | 67 | |
| Alanine | 62 | |
| Cysteine | 9 | |
| Valine | 52 | |
| Methionine | 12 | |
| Isoleucine | 45 | |
| Leucine | 96 | |
| Tyrosine | 36 | |
| Phenylalanine | 40 | |
| Tryptophan | 5 | |
| Histidine | 30 | |
| Lysine | 50 | |
| Arginine | 54 |
Internal Peptide Sequences
The protein was cleaved by CNBr, and the peptides were separated by reversed phase chromatography on a μRPC column using the SMART™ system (data not shown). Two components eluting as symmetrical peaks were selected for sequencing. They yielded the following sequences: Glu-Ile-Asn-Leu-Ser-His-Asn-Lys and Arg-Asp-Leu-Tyr-Phe-Asn-Lys-Ile. A search of the databases available through GenBank identified no protein that contained these sequences or that was closely similar. Interestingly, in view of the high leucine content of the protein, both peptides contain sequences that would fit into the leucine-rich repeat consensus; i.e., LXXLXLXXNXL (where X is any amino acid and L is leucine or another hydrophobic amino acid).
Tissue Distribution of Osteoadherin
A specific antiserum against purified bovine bone osteoadherin was raised in rabbits. The antibody specificity was demonstrated in a Western blot of a bone extract (Fig. 3 B). The purified proteoglycan is well recognized by the antiserum (Fig. 3 B, right). Only one immunoreactive band was detected in the extract of demineralized bovine bone (Fig. 3 B, left).
Figure 3.
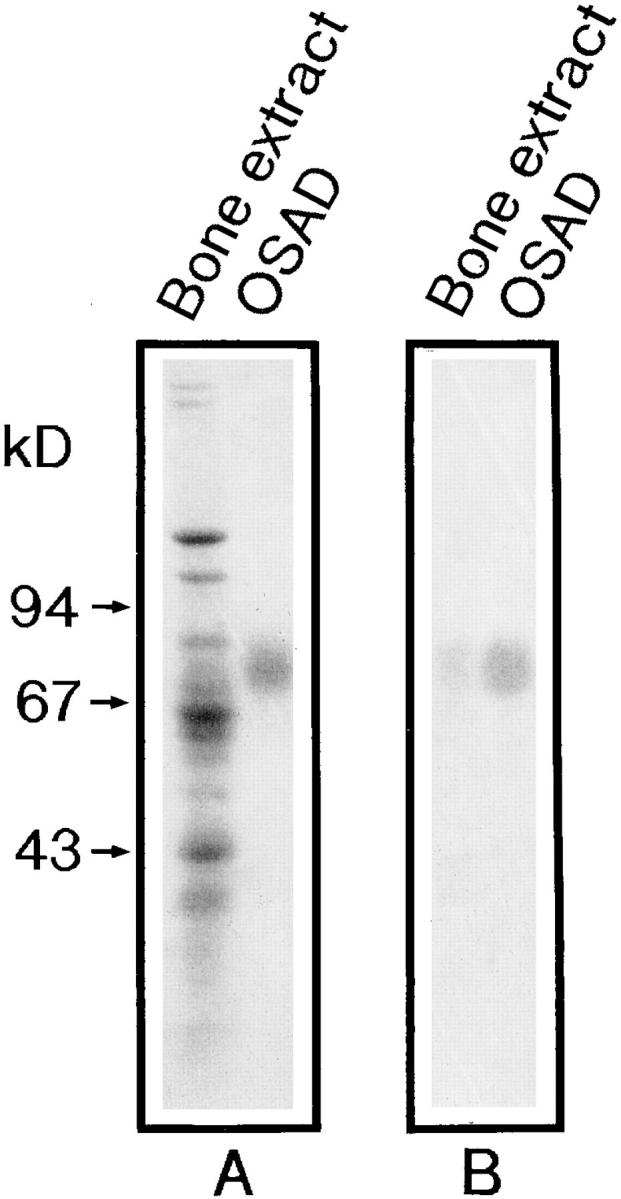
SDS-PAGE and immunoblotting of purified osteoadherin. An extract of the bone mineral compartment, i.e., with guanidine hydrochloride/EDTA (A, left lane) and osteoadherin purified from bovine bone (A, right lane) were applied to an SDS 4–12% polyacrylamide gel after reduction with mercaptoethanol. The gel was stained with Coomassie blue (A). An identical gel was transferred to nitrocellulose (47) and immunoblotted with a rabbit antiserum against the purified bovine osteoadherin (B). One band was detected in the guanidine hydrochloride EDTA extract (B, left lane) corresponding to the purified osteoadherin (B, right lane).
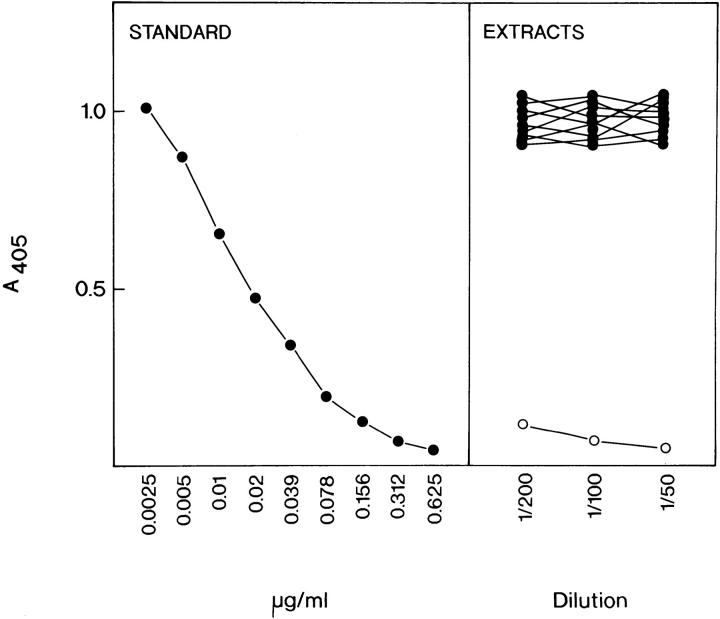
An ELISA was developed to determine the amount of osteoadherin in 4 M guanidine hydrochloride extracts of different bovine tissues (Fig. 4). The protein was detected only in the extract of bone. Extracts of liver, trachea, tendon, articular cartilage, kidney cortex, heart, intestine, muscle, and cornea did not contain detectable levels of osteoadherin. The concentration of osteoadherin in bone extracts was calculated to be 0.4 mg/g of tissue wet weight.
Figure 4.
Detection of osteoadherin in extracts of bovine tissues by inhibition ELISA. Various bovine tissues were extracted with 20 vol of 4 M guanidine hydrochloride, precipitated with ethanol, and dissolved in SDS buffer. Dilutions were prepared, and ELISA was performed as described in Materials and Methods section. The left panel shows an inhibition curve obtained with a standard of purified osteoadherin. The right panel shows that none of the non-bone tissues (•; liver, trachea, tendon, articular cartilage, kidney cortex, heart, intestine, muscle, or cornea) manifested significant inhibition, thus indicating the absence of osteoadherin at the level of detection limit (<0.3 μg/mg of original tissue weight). The bone tissue extract (○) manifested strong inhibition at corresponding dilutions.
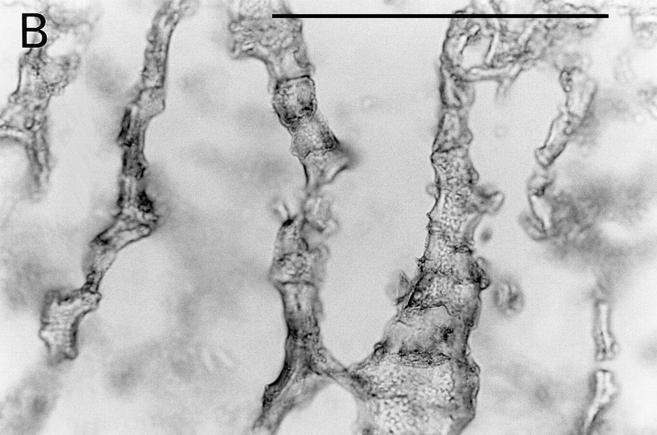
The distribution of osteoadherin in bovine fetal rib growth plate was investigated by immunohistochemistry (Fig. 5). Strong immunoreactivity with the antiosteoadherin antiserum was localized to the primary bone spongiosa (Fig. 5, A and B). No staining could be seen either in the cartilage or in the control with preimmune serum (Fig. 5 C).
Figure 5.
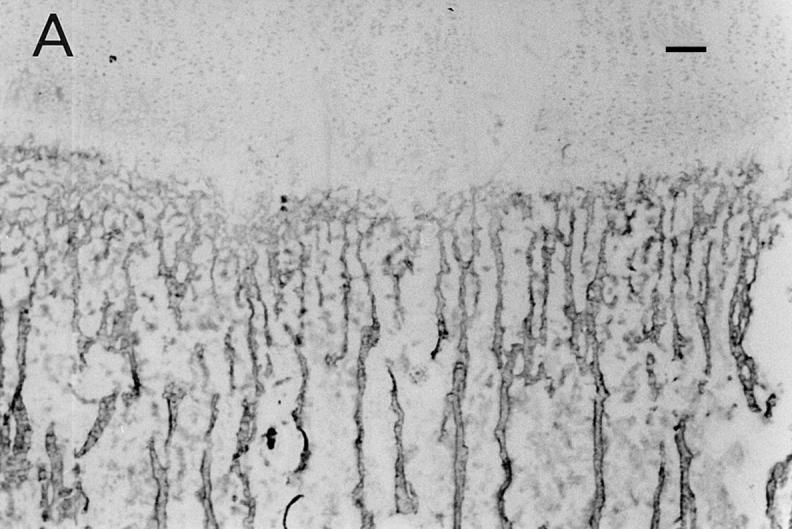
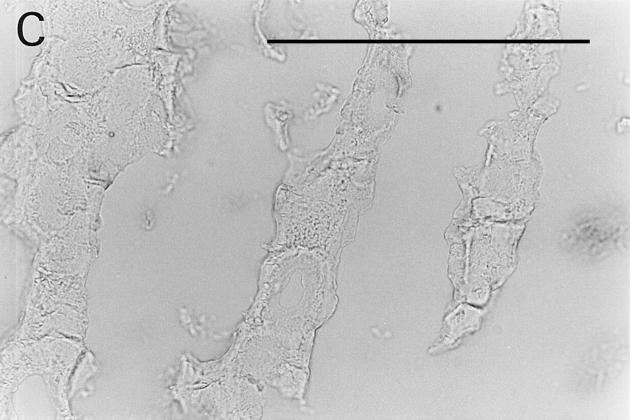
Immunolocalization of osteoadherin in the bovine fetal rib growth plate. The chondrocostal growth plate of a bovine fetus was sectioned on a cryostat in 5-μm sections without prior decalcification. An affinity-purified antiserum against bovine osteoadherin was used for immunostaining. The sections were incubated with antibodies against osteoadherin (A–B) or with preimmune serum (C) followed by a biotinylated second antibody. The sections were incubated with Vectastain ABC reagent and developed in peroxidase solution. (A) Strong specific staining for the osteoadherin in the primary spongiosa in the fetal rib growth plate. (B) Staining of bone trabeculae from the same section as in A with higher magnification. (C) The control with the preimmune serum where no staining can be detected. Bar, 200 μm.
Biosynthesis of Osteoadherin by Primary Osteoblasts
Immunoprecipitation of radiolabeled medium from cultures of calf bone primary osteoblasts showed osteoadherin to be a bone cell–derived protein (Fig. 6 A).
Figure 6.
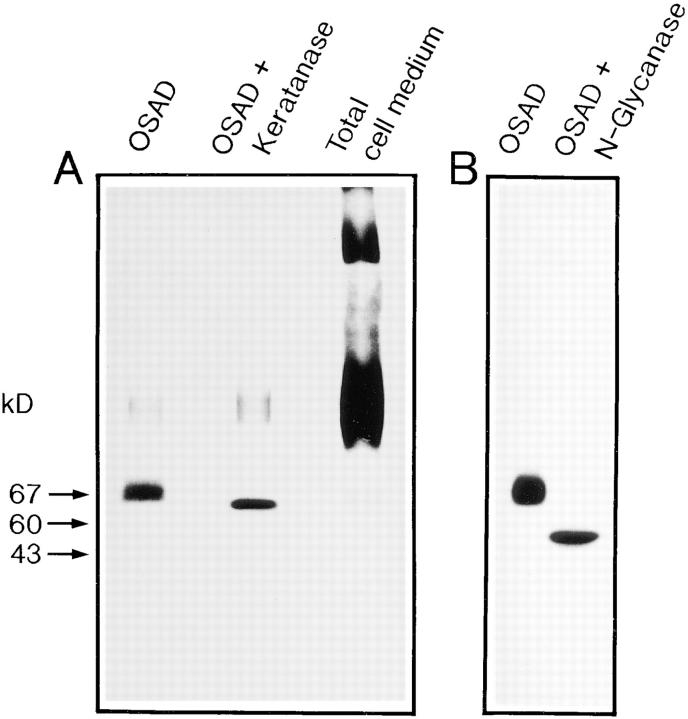
Synthesis of osteoadherin by primary bovine osteoblasts and digestion of the immunoprecipitated osteoadherin with keratanase and N-glycosidase F. Osteoblasts were prepared from bovine bone using the method of Robey (41) and grown to confluency in Ham's F12 medium containing 10% calf serum. The cells were metabolically labeled in the presence of [35S]sulfate for 6 h. Radiolabeled proteins in the culture medium were immunoprecipitated with the osteoadherin antiserum. The precipitates were digested with two different glycosidases. Digested samples, undigested control, and the whole cell medium were electrophoresed on 4–16% gradient polyacrylamide gels and stained with Coomassie blue. Labeled proteins were detected by fluorography. (A) Immunoprecipitated osteoadherin in the left lane, immunoprecipitated osteoadherin digested with keratanase in the middle lane, and nonprecipitated whole culture medium in the right lane. (B) Immunoprecipitated osteoadherin in the left lane, and immunoprecipitated osteoadherin digested with N-glycosidase F (N-glycanase) in the right lane. Both digestions were performed according to the recommendations of the manufacturers. The positions of molecular mass markers are indicated by arrows.
Confluent bovine osteoblasts were labeled with either [3H]leucine or [35S]sulfate, and the medium was immunoprecipitated with the specific antiosteoadherin antiserum. The immunoprecipitated material was analyzed by SDS-PAGE and fluorography. The protein could be demonstrated by [3H]leucine labeling (data not shown). Since the protein manifested some polydispersity typical of proteoglycans, we also tried [35S]sulfate labeling, which gave clearer results than [3H]leucine labeling (Fig. 6 A). Thus, osteoadherin is produced by the osteoblasts and contains sulfate.
Carbohydrate Constituents on Osteoadherin
To determine whether osteoadherin is glycosylated, we analyzed [35S]sulfate-labeled osteoadherin by SDS-PAGE (described above) after treatment with glycosidases. The size of the immunoprecipitated [35S]sulfate-labeled osteoadherin from primary osteoblasts decreased upon digestion with keratanase, suggesting osteoadherin to be substituted with keratan sulfate (Fig. 6 A). Digestion with N-glycanase decreased its mobility more extensively from 85 to 47 kD (Fig. 6 B). However, labeled osteoadherin was still detectable, suggesting the presence of other sulfate-containing groups such as tyrosine sulfate. Digestion of the protein with chondroitinase did not affect its mobility (data not shown).
Cell Attachment to Osteoadherin
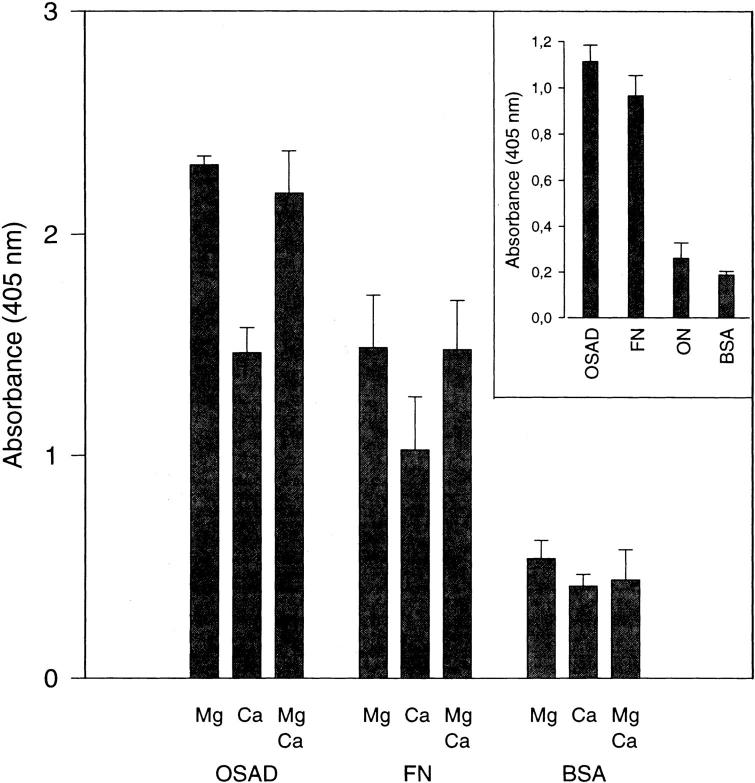
Primary osteoblasts were shown to attach quite well to osteoadherin in a cell attachment assay using reduced (Fig. 7) or nonreduced protein (Fig. 7, inset). As mentioned above, preparations of nonreduced osteoadherin contain some osteonectin. Therefore, in a separate experiment with pure nonreduced osteonectin, it was confirmed that osteonectin does not promote cell attachment (Fig. 7, inset). The effect of ligand binding of two different divalent cations—magnesium and calcium—was tested. Magnesium ions promoted binding to a higher degree than did calcium ions. A combination of magnesium and calcium ions did not increase the level of cell binding, as compared with Mg2+ alone. The use of fibronectin as a positive control yielded the same result, i.e., showed Mg2+ to be the most efficient promotor of cell binding. When BSA was used as a negative control, no binding was obtained. Similar results were obtained in experiments using rat osteosarcoma cells (data not shown).
Figure 7.
Attachment of primary osteoblasts to osteoadherin. OSAD, fibronectin (FN), and BSA were coated in a 96-well immunoplate. Subconfluent primary osteoblasts were briefly EDTA-treated, washed, and resuspended in Ham's F12 medium. Approximately 20,000 cells were added to each well. The cell samples were incubated with 1 mM CaCl2, 1 mM MgCl2, or both. After incubation for 1 h at 37°C, the wells were washed with PBS, and the number of attached cells was determined by measuring the cellular N-acetyl-hexosaminidase activity. Maximal attachment was 30% of added cells. The data represent mean values for triplicate wells with a standard error of <10% of the mean. (Inset) In a separate experiment, nonreduced OSAD, fibronectin (FN), osteonectin (ON), and BSA were coated in a 96-well immunoplate. The cell attachment experiment was performed under the same conditions as above, except that the primary osteoblasts were incubated in 1 mM CaCl2 only.
RGD Inhibition of Cell Attachment to Osteoadherin
The synthetic peptide GRGDSL, containing the classical first-described cell integrin–binding RGD (42) sequence (in this case derived from osteopontin) inhibited adhesion of primary osteoblasts to osteoadherin (Fig. 8). The negative control peptide GRGESL did not have any inhibitory effect on the cell adhesion to osteoadherin (Fig. 8).
Figure 8.
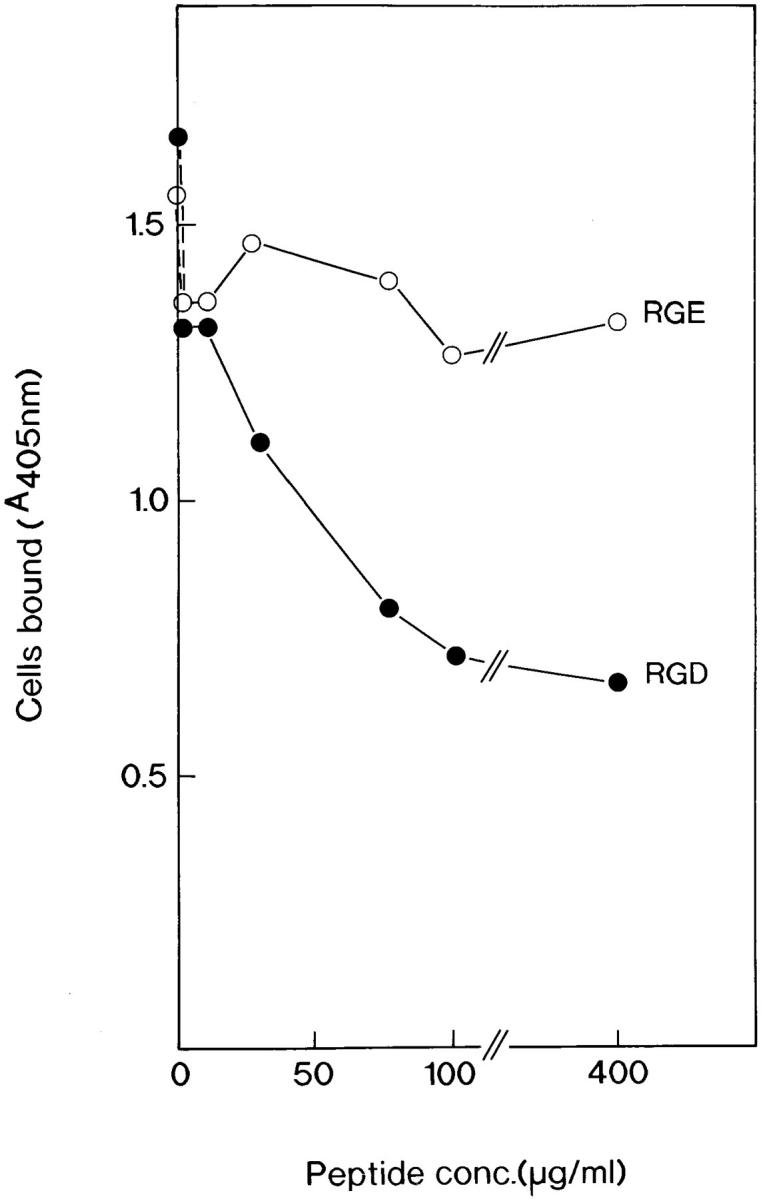
RGD-mediated inhibition of cell attachment to osteoadherin. The ROS cells were allowed to attach to osteoadherin-coated wells in the presence of increasing concentrations of the peptides indicated. The data represent mean values for triplicate wells, with a standard error of <10% of the mean.
Identity of an Osteoadherin-binding Integrin from Osteoblasts
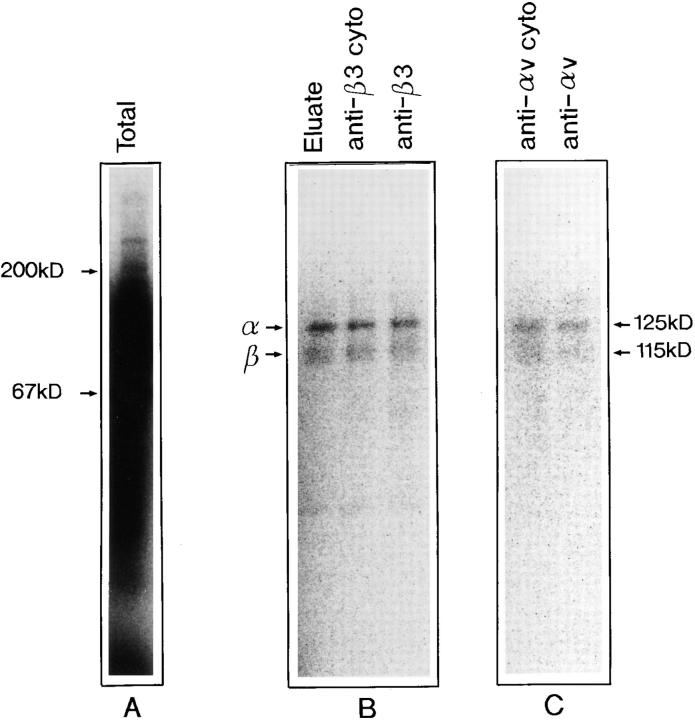
Affinity purification on immobilized osteoadherin of an extract of surface-iodinated osteoblasts demonstrated the presence of two binding proteins of an apparent M r of 125,000 and 115,000 (Fig. 9). These bands corresponded in mobility to αv and β3, respectively. Identification of these proteins as the αv and β3 integrin subunits, respectively, was verified by immunoprecipitation with the corresponding subunit-specific antibodies (Fig. 9). In a separate experiment (not shown), the antibodies to the cytoplasmic part of β3 precipitated both subunits while the antibodies to the cytoplasmic part of the αv subunit only precipitated this chain. At the same time, none of these subunits were precipitated by a control rabbit serum under the same condition.
Figure 9.
Affinity purification and immune precipitation of integrins binding to osteoadherin. A detergent extract of surface- iodinated MG 63 osteosarcoma cells was applied to a control agarose followed by OSAD agarose. Consecutive fractions were collected after adding EDTA to the elution buffer. The receptor-containing fractions were pooled. Aliquots of the pooled fractions were immunoprecipitated with polyclonal antibodies against the integrin subunits αv and β3, respectively. The pooled eluate and precipitated proteins were electrophoresed on SDS-PAGE (4–12%) under reducing conditions, and were visualized with the phosphor image analyzer Bas2000 (Fuji). (A) Total cell extract. (B) The left lane shows the EDTA eluate from the OSAD column. The middle lane shows immunoprecipiation with a peptide antiserum against the cytoplasmic tail of β3. The right lane presents precipitation with a monoclonal antiserum specific for the β3 subunit. (C) The left lane shows precipitation with a polyclonal antiserum against the cytoplasmic tail of αv. The right lane shows immunoprecipitation with a polyclonal antiserum against the αv subunit.
Discussion
A purification scheme was developed for isolation of osteoadherin from bovine bone. In the final purification step, the sample has to be reduced to achieve a complete separation of osteoadherin from osteonectin on a gel filtration column.
It is very likely that some of the osteonectin had actually been bound to osteoadherin via disulfide bonds. It is not likely that this had occurred via disulfide exchange during extraction in view of the use of N-ethyl maleimide to block free sulphydryl groups. Thus, it is possible that the protein has the capacity for specific interaction with osteonectin.
The protein constitutes a novel entity since the amino acid sequence of two internal octapeptides could not be found in the Swiss-Prot or the GenBank database. Unfortunately, the amino terminus of osteoadherin appears to be blocked, which precluded amino acid NH2-terminal sequencing. Some information on the properties of the protein could, however, be derived from its amino acid composition, which shows it to be an acidic protein rich in aspartic acid/asparagine and glutamic acid/glutamine. The acidic nature of the protein is consistent with its tight binding to hydroxyapatite. A high content of acidic amino acids and of leucine has been found in many extracellular matrix proteins (17), in particular in a family of proteins with leucine-rich repeats (LRR; for review see reference 24), such as decorin (27) and biglycan (10), which are found in bone. Other members of the family are fibromodulin (36), lumican (2), PG-Lb (44), chondroadherin (31), PRELP (1), and keratocan (6). Support of the notion that osteoadherin belongs to the family of leucine-rich repeat proteins was obtained from the amino acid sequences of the two internal peptides. However, one characteristic that sets osteoadherin apart is the size of its core protein (47 kD), which is larger than that of any LRR proteins/proteoglycans described previously.
Antibodies raised against the purified protein were used in studies of its biosynthesis in order to ascertain whether primary bovine osteoblasts produce and secrete osteoadherin. Indeed, we were able to immunoprecipitate osteoadherin from medium of cell cultures labeled with [35S]sulfate or [3H]leucine. The sulfate label may have been incorporated into tyrosine sulfate or carbohydrate structures, e.g., glycosaminoglycans. However, the protein appears not to contain chondroitin sulfate, since digestion with chondroitinase did not affect its electrophoretic mobility (data not shown). In contrast, digestion of the immunoprecipitated osteoadherin with N-glycosidase reduced its apparent size to 47 kD, demonstrating the presence of several N-linked oligosaccharides. Digestion with keratanase indicated some of these oligosaccharides to be extended to keratan sulfate chains. The presence of highly sulfated keratan sulfate appears to be unlikely, since immunoblotting of osteoadherin with a monoclonal antibody (5D4) raised against such epitopes was negative (data not shown). The only keratan sulfate–containing molecule previously identified in bone is BSP, which only in rabbit bone appears as a keratan sulfate proteoglycan (26). However, BSP is quite distinct from osteoadherin, and antibodies manifest no cross-reactivity. Other proteins with a core protein of similar size to that of osteoadherin and containing KS chains are fibromodulin, lumican, and keratocan, but these molecules manifest no cross-reactivity with the osteoadherin antibodies. The fact that N-glycanase– digested osteoadherin still contains [35S]sulfate indicates the presence of other sulfated residues. Since digestion with O-glycanase did not affect the mobility of the protein, it appears unlikely that it contains O-linked oligosaccharides. Thus, it is possible that osteoadherin contains sulfated tyrosine residues.
The antiserum was shown to be specific for osteoadherin in Western blots of total bone extracts. In addition, pure osteoadherin was shown not to react with antisera against other acidic glycoproteins from bone matrix such as osteonectin, osteopontin, bone sialoprotein, decorin, or biglycan.
Osteoadherin seems to be restricted to bone as assayed by an inhibition ELISA. Other proteins exclusively restricted to bone include osteocalcin and BSP. In this context, it is noteworthy that BSP is also a cell-binding protein.
Antibodies against osteoadherin were used in immunohistochemical studies of the protein's distribution in bovine fetal rib growth plate. Strong reactivity was found in newly deposited bone, but staining could also be detected throughout the mineralized matrix. Bone sialoprotein is characterized by a similar tissue distribution, occurring predominantly in the newly formed osteoid seam (20). Osteopontin manifests a different tissue distribution pattern, occurring predominantly in bone resorption areas (20) and at the mineralization front.
We showed osteoadherin to have strong cation-dependent cell-binding properties, and to promote osteoblast attachment in vitro. Its cell-binding efficiency is comparable to that of fibronectin. Initially, reduced osteoadherin was used in the cell-binding assay since reduction is a prerequisite for complete separation of osteoadherin from osteonectin in the last step of the purification process using gel filtration. Reduction, however, may unfold the protein to expose cryptic sites for cell binding. In control experiments, we found the nonreduced protein also to bind cells. Using the same assay protocol, the only detectable contaminant in this preparation, osteonectin, was not found to mediate cell attachment. This result is consistent with earlier findings suggesting that osteonectin does not promote cell attachment in vitro (43).
Cell adhesion is usually mediated by integrins, a family of transmembrane receptors composed of two subunits: α and β (23). In several cases the RGD peptide motif has been shown to mediate cell adhesion in proteins including fibronectin, vitronectin, fibrinogen, osteopontin, and BSP (42). That an integrin is involved in osteoadherin binding was indicated in studies of the dependence of adhesion in the presence of divalent ions. Thus, magnesium ions are more potent than calcium ions in promoting binding of primary osteoblasts to osteoadherin. A further indication that this cell-binding is dependent on an integrin is that an RGD (Arg-Gly-Asp)-containing peptide was found to inhibit attachment of primary osteoblasts to osteoadherin. The control RGE (Arg-Gly-Glu) peptides did not affect cell binding. To probe further the nature of the integrin, we used affinity chromatography of selectively labeled cell surface proteins followed by immune precipitation with the respective α chain–specific and β chain–specific antibodies. The results showed the integrin to be αvβ3. This integrin was initially purified as a vitronectin receptor (46), but has since been shown to bind a large number of extracellular matrix molecules including osteopontin (40) and BSP (35). Like other ligands for this integrin, osteoadherin may function to act as an initial cell attachment factor for osteoblasts as well as an inducer of integrin-mediated signals.
It is noteworthy that bone contains several ligands for αvβ3. Furthermore, the distribution of BSP (also αvβ3 binding) visualized by light microscopy resembles that of osteoadherin. In neither case are the functional consequences yet known.
We propose the name osteoadherin in view of the proteoglycan's high affinity to hydroxyapatite, and of its cell-binding activity. The aim of future work will be its further characterization by functional studies, including elucidation of its putative involvement in mineral deposition.
Acknowledgments
We are grateful to Viveka Nilsson, Ros-Marie Sandfalk, and Lelis Zunino for their skillful experimental work.
This work was supported by grants from the Swedish Medical Research Council, the Axel and Margareta Ax:son Johnson's Trust, the King Gustav V 80th Birthday Foundation, the Greta and Johan Kock Trust, the Alfred Österlund Trust, and the Medical and Odontological Faculties, University of Lund.
Abbreviations used in this paper
- BSP
bone sialoprotein
- OSAD
osteoadherin
Footnotes
Address all correspondence to Mikael Wendel, Department of Cell and Molecular Biology, University of Lund, P.O. Box 94, S-221 00 Lund, Sweden. Tel.: +46 46 222 95 87. Fax: +46 46 211 34 17. E-mail: mikael.wendel@medkem.lu.se
References
- 1.Bengtsson E, Neame PJ, Heinegard D, Sommarin Y. The primary structure of a basic leucine-rich repeat protein, PRELP, found in connective tissues. J Biol Chem. 1995;270:25639–25644. doi: 10.1074/jbc.270.43.25639. [DOI] [PubMed] [Google Scholar]
- 2.Blochberger TC, Vergnes JP, Hempel J, Hassell JR. cDNA to chick lumican (corneal keratan sulfate proteoglycan) reveals homology to the small interstitial proteoglycan gene family and expression in muscle and intestine. J Biol Chem. 1992;267:347–352. [PubMed] [Google Scholar]
- 3.Bolander ME, Young MF, Fisher LW, Yamada Y, Termine JD. Osteonectin cDNA sequence reveals potential binding regions for calcium and hydroxyapatite and shows homologies with both a basement membrane protein (SPARC) and a serine proteinase inhibitor (ovomucoid) Proc Natl Acad Sci USA. 1988;85:2919–2923. doi: 10.1073/pnas.85.9.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camper L, Heinegård D, Lundgren-Åkerlund E. Integrin alpha2beta1 is a receptor for the cartilage matrix protein chondroadherin. J Cell Biol. 1997;138:1159–1167. doi: 10.1083/jcb.138.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chamberlain JP. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 6.Corpuz LM, Funderburgh JL, Funderburgh ML, Bottomley GS, Prakash S, Conrad GW. Molecular cloning and tissue distribution of keratocan. Bovine corneal keratan sulfate proteoglycan 37A. J Biol Chem. 1996;271:9759–9763. doi: 10.1074/jbc.271.16.9759. [DOI] [PubMed] [Google Scholar]
- 7.Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, et al. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- 8.Engel J, Taylor W, Paulsson M, Sage H, Hogan B. Calcium binding domains and calcium-induced conformational transition of SPARC/BM-40/osteonectin, an extracellular glycoprotein expressed in mineralized and nonmineralized tissues. Biochemistry. 1987;26:6958–6965. doi: 10.1021/bi00396a015. [DOI] [PubMed] [Google Scholar]
- 9.Eyre DR, Dickson IR, Van Ness K. Collagen cross-linking in human bone and articular cartilage. Age-related changes in the content of mature hydroxypyridinium residues. Biochem J. 1988;252:495–500. doi: 10.1042/bj2520495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher LW, Termine JD, Young MF. Deduced protein sequence of bone small proteoglycan I (biglycan) shows homology with proteoglycan II (decorin) and several nonconnective tissue proteins in a variety of species. J Biol Chem. 1989;264:4571–4576. [PubMed] [Google Scholar]
- 11.Fisher LW, McBride OW, Termine JD, Young MF. Human bone sialoprotein. Deduced protein sequence and chromosomal localization. J Biol Chem. 1990;265:2347–2351. [PubMed] [Google Scholar]
- 12.Franzen A, Heinegård D. Extraction and purification of proteoglycans from mature bovine bone. Biochem J. 1984;224:47–58. doi: 10.1042/bj2240047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzen A, Heinegård D. Isolation and characterization of two sialoproteins present only in bone calcified matrix. Biochem J. 1985;232:715–724. doi: 10.1042/bj2320715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed E, Gailit J, van der Geer P, Ruoslahti E, Hunter T. A novel integrin beta subunit is associated with the vitronectin receptor alpha subunit (alpha v) in a human osteosarcoma cell line and is a substrate for protein kinase C. EMBO (Eur Mol Biol Organ) J. 1989;8:2955–2965. doi: 10.1002/j.1460-2075.1989.tb08445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedbom E, Heinegård D. Interaction of a 59-kDa connective tissue matrix protein with collagen I and collagen II. J Biol Chem. 1989;264:6898–6905. [PubMed] [Google Scholar]
- 16.Heinegård D. Polydispersity of cartilage proteoglycans. Structural variations with size and buoyant density of the molecules. J Biol Chem. 1977;252:1980–1989. [PubMed] [Google Scholar]
- 17.Heinegård, D., and Å. Oldberg. 1993. Glycosylated matrix proteins. In Connective Tissue and Its Heritable Disorders. P.M. Royce and B. Steinmann, editors. Wiley-Liss, Inc., New York. 189–209.
- 18.Hultenby K, Reinholt FP, Oldberg Å, Heinegård D. Ultrastructural immunolocalization of osteopontin in metaphyseal and cortical bone. Matrix. 1991;11:206–213. doi: 10.1016/s0934-8832(11)80160-5. [DOI] [PubMed] [Google Scholar]
- 19.Hultenby K, Reinholt FP, Heinegård D. Distribution of integrin subunits on rat metaphyseal osteoclasts and osteoblasts. Eur J Cell Biol. 1993;62:86–93. [PubMed] [Google Scholar]
- 20.Hultenby K, Reinholt FP, Norgård M, Oldberg Å, Wendel M, Heinegård D. Distribution and synthesis of bone sialoprotein in metaphyseal bone of young rats show a distinctly different pattern from that of osteopontin. Eur J Cell Biol. 1994;63:230–239. [PubMed] [Google Scholar]
- 21.Hunter GK, Goldberg HA. Nucleation of hydroxyapatite by bone sialoprotein. Proc Natl Acad Sci USA. 1993;90:8562–8565. doi: 10.1073/pnas.90.18.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter GK, Kyle CL, Goldberg HA. Modulation of crystal formation by bone phosphoproteins: structural specificity of the osteopontin-mediated inhibition of hydroxyapatite formation. Biochem J. 1994;300:723–728. doi: 10.1042/bj3000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 24.Iozzo RV, Murdoch AD. Proteoglycans of the extracellular environment: clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB (Fed Am Soc Exp Biol) J. 1996;10:598–614. [PubMed] [Google Scholar]
- 25.Jacenko O, Olsen BR, LuValle P. Organization and regulation of collagen genes. Crit Rev Eukaryotic Gene Expr. 1991;1:327–353. [PubMed] [Google Scholar]
- 26.Kinne RW, Fisher LW. Keratan sulfate proteoglycan in rabbit compact bone is bone sialoprotein II. J Biol Chem. 1987;262:10206–10211. [PubMed] [Google Scholar]
- 27.Krusius T, Ruoslahti E. Primary structure of an extracellular matrix proteoglycan core protein deduced from cloned cDNA. Proc Natl Acad Sci USA. 1986;83:7683–7687. doi: 10.1073/pnas.83.20.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Landegren U. Measurement of cell numbers by means of the endogenous enzyme hexosaminidase. Applications to detection of lymphokines and cell surface antigens. J Immunol Methods. 1984;67:379–388. doi: 10.1016/0022-1759(84)90477-0. [DOI] [PubMed] [Google Scholar]
- 30.Larsson T, Sommarin Y, Paulsson M, Antonsson P, Hedbom E, Wendel M, Heinegård D. Cartilage matrix proteins. A basic 36-kDa protein with a restricted distribution to cartilage and bone. J Biol Chem. 1991;266:20428–20433. [PubMed] [Google Scholar]
- 31.Neame PJ, Sommarin Y, Boynton RE, Heinegård D. The structure of a 38-kDa leucine-rich protein (chondroadherin) isolated from bovine cartilage. J Biol Chem. 1994;269:21547–21554. [PubMed] [Google Scholar]
- 32.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 33.Oldberg Å, Franzen A, Heinegård D. Cloning and sequence analysis of rat bone sialoprotein (osteopontin) cDNA reveals an Arg-Gly-Asp cell-binding sequence. Proc Natl Acad SciUSA. 1986;83:8819–8823. doi: 10.1073/pnas.83.23.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oldberg Å, Franzen A, Heinegård D. The primary structure of a cell-binding bone sialoprotein. J Biol Chem. 1988;263:19430–19432. [PubMed] [Google Scholar]
- 35.Oldberg Å, Franzen A, Heinegård D, Pierschbacher M, Ruoslahti E. Identification of a bone sialoprotein receptor in osteosarcoma cells. J Biol Chem. 1988;263:19433–19436. [PubMed] [Google Scholar]
- 36.Oldberg Å, Antonsson P, Lindblom K, Heinegård D. A collagen-binding 59-kd protein (fibromodulin) is structurally related to the small interstitial proteoglycans PG-S1 and PG-S2 (decorin) EMBO (Eur Mol Biol Organ) J. 1989;8:2601–2604. doi: 10.1002/j.1460-2075.1989.tb08399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulsson M, Sommarin Y, Heinegård D. Metabolism of cartilage proteins in cultured tissue sections. Biochem J. 1983;212:659–667. doi: 10.1042/bj2120659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poser JW, Esch FS, Ling NC, Price PA. Isolation and sequence of the vitamin K-dependent protein from human bone. Undercarboxylation of the first glutamic acid residue. J Biol Chem. 1980;255:8685–8691. [PubMed] [Google Scholar]
- 39.Price PA, Fraser JD, Metz G, Virca Molecular cloning of matrix Gla protein: implications for substrate recognition by the vitamin K-dependent gamma-carboxylase. Proc Natl Acad Sci USA. 1987;84:8335–8339. doi: 10.1073/pnas.84.23.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinholt FP, Hultenby K, Oldberg Å, Heinegård D. Osteopontin--a possible anchor of osteoclasts to bone. Proc Natl Acad Sci USA. 1990;87:4473–4475. doi: 10.1073/pnas.87.12.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robey PG, Termine JD. Human bone cells in vitro. Calcif Tissue Int. 1985;37:453–460. [PubMed] [Google Scholar]
- 42.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 43.Sage EH, Bornstein P. Extracellular proteins that modulate cell-matrix interactions. SPARC, tenascin, and thrombospondin. J Biol Chem. 1991;266:14831–14834. [PubMed] [Google Scholar]
- 44.Shinomura T, Kimata K. Proteoglycan-Lb, a small dermatan sulfate proteoglycan expressed in embryonic chick epiphyseal cartilage, is structurally related to osteoinductive factor. J Biol Chem. 1992;267:1265–1270. [PubMed] [Google Scholar]
- 45.Sommarin Y, Larsson T, Heinegård D. Chondrocyte-matrix interactions. Attachment to proteins isolated from cartilage. Exp Cell Res. 1989;184:181–192. doi: 10.1016/0014-4827(89)90376-5. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki S, Argraves WS, Pytela R, Arai H, Krusius T, Pierschbacher MD, Ruoslahti E. cDNA and amino acid sequences of the cell adhesion protein receptor recognizing vitronectin reveal a transmembrane domain and homologies with other adhesion protein receptors. Proc Natl Acad Sci USA. 1986;83:8614–8618. doi: 10.1073/pnas.83.22.8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Rest M, Garrone R. Collagen family of proteins. FASEB J. 1991;5:2814–2823. [PubMed] [Google Scholar]
- 49.Yamaguchi Y, Mann DM, Ruoslahti E. Negative regulation of transforming growth factor-beta by the proteoglycan decorin. Nature. 1990;346:281–284. doi: 10.1038/346281a0. [DOI] [PubMed] [Google Scholar]
- 50.Young, M.F., J.M. Kerr, K. Ibaraki, A.M. Heegaard, and P.G. Robey. 1992. Structure, expression, and regulation of the major noncollagenous matrix proteins of bone. Clin. Orthop. 275–294. [PubMed]