Abstract
The nuclear matrix is defined as the insoluble framework of the nucleus and has been implicated in the regulation of gene expression, the cell cycle, and nuclear structural integrity via linkage to intermediate filaments of the cytoskeleton. We have discovered a novel nuclear matrix protein, NRP/B (nuclear restricted protein/brain), which contains two major structural elements: a BTB domain–like structure in the predicted NH2 terminus, and a “kelch motif” in the predicted COOH-terminal domain. NRP/B mRNA (5.5 kb) is predominantly expressed in human fetal and adult brain with minor expression in kidney and pancreas. During mouse embryogenesis, NRP/B mRNA expression is upregulated in the nervous system. The NRP/B protein is expressed in rat primary hippocampal neurons, but not in primary astrocytes. NRP/B expression was upregulated during the differentiation of murine Neuro 2A and human SH-SY5Y neuroblastoma cells. Overexpression of NRP/B in these cells augmented neuronal process formation. Treatment with antisense NRP/B oligodeoxynucleotides inhibited the neurite development of rat primary hippocampal neurons as well as the neuronal process formation during neuronal differentiation of PC-12 cells. Since the hypophosphorylated form of retinoblastoma protein (p110RB) is found to be associated with the nuclear matrix and overexpression of p110RB induces neuronal differentiation, we investigated whether NRP/B is associated with p110RB. Both in vivo and in vitro experiments demonstrate that NRP/B can be phosphorylated and can bind to the functionally active hypophosphorylated form of the p110RB during neuronal differentiation of SH-SY5Y neuroblastoma cells induced by retinoic acid. Our studies indicate that NRP/B is a novel nuclear matrix protein, specifically expressed in primary neurons, that interacts with p110RB and participates in the regulation of neuronal process formation.
The nuclear matrix is formed by an ordered and highly compartmentalized protein structure consisting of a nuclear lamina, a residual nucleolus, and an internal matrix composed of a nonchromatin fibrogranular network associated with DNA (Berezney and Coffey, 1974; Berezney, 1984, 1991; Buttyan and Olsson, 1986; Schuchard et al., 1991; Carter et al., 1993; Xing et al., 1993). The nuclear matrix has been implicated in transcription, regulation of gene expression, cell cycle, primary transcription processing, and linkages to intermediate filaments of the cytoskeleton (He et al., 1995; Loidl and Eberharter, 1995; Penman, 1995; Mancini et al., 1996). The nuclear lamins and the nuclear matrins were identified as major nuclear matrix proteins (Hakes and Berezney, 1991a ,b; Nakayasu and Berezney, 1991).
The nuclear matrix, as an underlying nuclear framework, coordinates the different processes occurring at the chromatin sites, which are under stringent cell cycle control (Loidl and Eberharter, 1995). For example, the retinoblastoma gene product (p110RB) is associated with the nuclear matrix in a cell cycle–dependent manner (Mittnacht and Weinberg, 1991; Mancini et al., 1994; Riley et al., 1994). Isolated nuclear matrices from synchronized cultured cells contained a significant amount of hypophosphorylated retinoblastoma protein only during the G1 period (Goodrich et al., 1991). Some p110RB-binding proteins are themselves associated with the nuclear matrix, including several “inactivating” viral oncoproteins (SV-40 large T antigen, adenovirus E1a protein, and the human papilloma E7 protein), lamin A1C (Ozaki et al., 1994), and p84 (Durfee et al., 1994).
Neuronal differentiation involves migration, directional axon growth, synaptogenesis, and selective survival (Kaplan and Stephens, 1994; Diaznido et al., 1996). Recent evidence indicates the involvement of cell cycle regulatory molecules in neuronal differentiation (Keynes and Cook, 1995). During neuronal differentiation, cyclin-dependent kinase (CDK)1 activities decline, and phosphorylation of the p110RB is reduced, leading to the appearance of a p110RB-containing E2F DNA-binding complex (Dobashi et al., 1995; Kranenburg et al., 1995). Neuronal differentiation can be induced by overexpression of CDK inhibitor p27KIP or p110RB, suggesting that loss of p110RB phosphorylation is an important determinant for neuronal differentiation (Kranenburg et al., 1995). In addition, cyclin dependent kinase-2 (CDK2) overexpression inhibited the NGF-induced differentiation of PC-12 cells (Dobashi et al., 1995). CDK5 expression and kinase activity are correlated with the extent of differentiation of neuronal cells in the developing brain (Ohshima et al., 1996).
Nuclear actin and myosin are components of the nuclear matrix in neurons and are present in the interphase nuclei of intact dorsal root ganglia and PC-12 cells (Milankov and De Boni, 1993; Amankwash and De Boni, 1994). Neuronal nuclear matrix components might play an important role in neuronal development and in the dynamic positioning of specific chromatin domains in a tissue-specific nonrandom pattern in neurons (Amankwash and De Boni, 1994). Recent observations indicated that ataxin-1 (the protein encoded by the SCA1 gene), involved in the neurodegenerative disorder spinocerebellar ataxia, alters nuclear matrix–associated structures (Skinner et al., 1997). Despite the apparent importance of the nuclear matrix in the regulation of many biological processes, the roles of nuclear matrix in cell physiology are largely unknown. Thus, the discovery of a matrix constituent whose expression is tissue or cell specific offers a new area for research. We describe here the cloning, expression, and characterization of a novel neuronal nuclear matrix protein, NRP/B (nuclear restricted protein/brain), which binds to p110RB and is involved in the regulation of neuronal process formation.
Materials and Methods
Materials
Chemical reagents were purchased from Sigma Chemical Co. (St. Louis, MO). The human fetal λ-gtl0 cDNA library was obtained from Dr. Kunkel (Children's Hospital, Boston, MA). The λ-ZapII human hippocampus cDNA library was from Stratagene (La Jolla, CA). Restriction endonucleases, modifying enzymes, terminal deoxynucleotidyl transferase, random priming kits, and Sephadex G-25 quickspin columns were purchased from Pharmacia Biotech (Piscataway, NJ) and New England Biolabs (Beverly, MA). The primers for PCR, reverse transcriptase PCR, and sequencing were synthesized using an automated DNA synthesizer (model 394; PE Applied Biosystems, Foster City, CA). The PCR and RNA-PCR kits were obtained from Perkin-Elmer Corp. (Norwalk, CT). Sequenase and random priming kits were obtained from U.S. Biochemical Corp. (Cleveland, OH), and RNA isolation kits were from Stratagene. Antibodies for p110RB (X Z55, A431) were obtained from PharMingen (San Diego, CA).
Amplification of cDNA Fragments Encoding NRP/B
A novel cDNA sequence was identified using the protocol as described (Khan et al., 1992). The highest homology was found with the Drosophila ring canal kelch protein. Therefore, this fragment was designated RCR-1 (Ring canal–related protein). RCR-1 was radiolabeled by random priming and used as a probe to screen the human cDNA libraries.
Isolation and Characterization of cDNA Clones
The human fetal brain λ-gtl0 cDNA was screened (∼6 × 105 recombinants/screening) initially with the PCR fragment RCR-1 and further processed to obtain isolated cDNA clones as previously described (Avraham et al., 1995a ,b). A total of 12 clones was isolated, and two clones were sequenced on both strands. The human brain hippocampus λ-ZapII cDNA library (∼8 × 105 recombinants/screening) was also screened with the 32P-labeled RCR-1 fragment. A total of 12 clones was isolated, and two clones were further analyzed.
Northern Blot Analysis
Poly-A mRNA was extracted from cells and analyzed as described (Avraham et al., 1995b ). The level of expression for each mRNA was determined densitometrically (E-C Apparatus Corp., Holbrook, NY). The mRNA blots from the human adult and fetal tissue mRNAs and mRNAs from various human brain regions were obtained from CLONTECH Laboratories (Palo Alto, CA).
Cell Culture
Primary neurons and astrocytes were prepared from the hippocampal regions of Sprague-Dawley rats at gestational day 18, and cortex cultures were prepared from postnatal day 1 rats as described (Brewer et al., 1993; Grill and Pixley, 1993). PC-12, Neuro 2A, and SH-SY5Y neuroblastoma cells were grown as described in the American Tissue Cell Culture manual (Greene and Tischler, 1976; Leventhal and Feldman, 1996; Vignali et al., 1996; Cosgaya et al., 1997).
Cell Cycle Analysis
Cells were grown on six-well plates and were harvested by trypsinization after two washes with PBS. 1–2 × 106 cells/ml were fixed with 50% ice-cold methanol for 30 min on ice. After centrifugation at 300 g for 5 min, supernatants were aspirated, and cell pellets were resuspended with 500 μl staining solution containing propidium iodide and RNAse (100 U/ml). Samples were incubated at room temperature for 30 min and then analyzed by FACS® analysis (Dana Farber Cancer Institute, Boston, MA).
Effect of NRP/B Antisense Oligodeoxynucleotides on Neuronal Cell Growth and Differentiation
The S-modified 18-mer sense, antisense, and scrambled oligodeoxynucleotides of NRP/B were synthesized by Genosys Biotechnologies, Inc. (The Woodlands, TX) and dissolved in DME. NRP/B antisense oligodeoxynucleotides corresponded to nucleotides +4 to +24 and +221 to +224 from the first ATG. All experiments were carried out with the corresponding sense and scrambled sequence controls as described in Avraham et al. (1995b). Differentiation of Neuro 2A and SH-SY5Y cells was induced with dibutyryl cAMP (5 μM) and retinoic acid (10 μM), respectively, as described (Leventhal and Feldman, 1996).
Construction of FLAG-epitope–tagged NRP/B Expression Vector
The full-length NRP/B cDNA was subcloned in the pcDNA3neo expression vector (Invitrogen, Carlsbad, CA). The nucleotide sequence of the FLAG-epitope (DYKDDDDK) was introduced to the 5′-end of the coding sequence of NRP/B in the NRP/B-pcDNA3neo construct by PCR to obtain the FLAG-NRP/B-pcDNA3neo construct. The sequence and orientation were confirmed by the sequencing of both strands.
Transfections of NRP/B
Cells were grown in six-well plates with MEM or DME containing 10% FBS. Lipofectamine (GIBCO BRL, Gaithersburg, MD) was used for stable and transient transfections. The stable transfected cells were selected in DME (+ 10% FBS) with 400 μg/ml of geneticin (G418).
In Situ Hybridization
For preparation of the RNA probes, the murine homologue of NRP/B cDNA, Nrp/b, was isolated. A 390-bp XbaI-EcoRI 3′-fragment of Nrp/b was subcloned into multiple cloning sites of the pBluescript II SK+ vector (Stratagene). 35S-labeled RNA probes were transcribed in the antisense or sense orientation from the linearized pBluescript vector according to the manufacturer's protocol (Stratagene). For tissue preparation, hybrid-ready mouse brain tissue sections (7 μm) were mounted onto Vectabond-treated slides (purchased from Novagen, Madison, WI). The brain sections were either mid-coronal or mid-sagittal. Prehybridization, hybridization with a 35S-labeled radioactive probe, washes, autoradiography, and developing of the slides were performed according to the protocols described in the Sure Site II system manual using the hybridization reagent kit (Novagen).
Expression of His6-tagged NRP/B Protein in Sf-9 Insect Cells
The NRP/B-His6/pBlueBacIII recombinant plasmid was constructed by inserting a C-His6-NRP/B construct at BamHI and HindIII sites. To make the C-His6-NRP/B construct, which encodes six extra histidine residues added to the COOH-terminal end of the NRP/B protein, two sets of PCR reactions were performed using two flanking oligodeoxynucleotides. C-His6 NRP/B/pBlueBacIII recombinated baculovirus (AcMNPV) was obtained by using a baculovirus expression system (MaxBac; Invitrogen). To express the His6-tagged NRP/B protein, 2 × 106 cells/ml of Sf-9 cells were infected with the recombinant virus (MOI of 5) and harvested 48 h after infection. The cell pellet was resuspended in 5 ml of 6 M guanidine-HCl and incubated for 2 h on ice. After centrifugation at 25,000 g, the supernatant was applied to a 2-ml nickel column (Invitrogen) and incubated for 10 min at room temperature. The resin was washed according to the manufacturer's protocol, and the bound protein was eluted with elution buffer (8 M urea, 0.5 M NaCl in 20 mM phosphate buffer, pH 4.0). The eluted protein was then diluted slowly with 20 mM phosphate buffer, pH 7.4, and dialyzed against renaturation buffer (0.3 M arginine, 10% glycerol in 20 mM phosphate buffer, pH 6.0).
Preparation of Anti-NRP/B Antibodies
NRP/B was expressed and purified from Sf-9 cells as described above, and polyclonal antibodies against the NRP/B protein were raised in rabbits, according to a standard protocol (Harlow and Lane, 1988). The titers of the antisera were analyzed by ELISA and Western blot analysis. Hybridoma-producing monoclonal anti-NRP/B antibodies were raised by the conventional PEG-fusion technique using 2 × 108 spleen cells and 1 × 108 PAI-O partner cells (generous gifts from Dr. J.A. Langer, Rutgers University School of Medicine and Dentistry, NJ). The clones that survived in the HAT medium supplemented with 20% FBS were screened by ELISA. Positive clones were subcloned three times by the limiting dilution method (Harlow and Lane, 1988).
Isolation of Nuclei
Nuclei from cells were isolated as described (Mirkovitch et al., 1994). Exponentially growing and SH-SY5Y cells (3 × 108 cells) were collected, washed, and resuspended in 10 ml isolation buffer containing 0.1% digitonin (Sigma Chemical Co.), 0.5 mM PMSF, and a protease inhibitor cocktail (Boehringer Mannheim Corp., Indianapolis, IN) and were disrupted in a 5-ml Dounce-type homogenizer (Cole-Parmer Instrument Co., Niles, IL) with 20 strokes. The nuclei were collected by centrifugation after washing with the isolation buffer. The isolated nuclei were resuspended in a 4-ml isolation buffer containing 0.1% digitonin but no EDTA. To digest chromosomal DNA, nuclei were supplemented with restriction buffer B (Boehringer Mannheim Corp.) and incubated with 250 U of EcoRI at 37°C for 4 h. After EcoRI digestion, the nuclei were extracted with a high-salt solution (2 M NaCl, 10 mM Pipes, 10 mM EDTA, pH 6.8, and protease inhibitors [Boehringer Mannheim Corp.]). The high-salt extracted nuclei were collected by centrifugation (10 min, 2,100 g) and washed three times with an isolation buffer supplemented with 0.1% digitonin, exonuclease restriction digestion buffer B, and protease inhibitors. The nuclei were again incubated at 37°C with 250 U EcoRI for 2 h and then extracted with the high-salt solution. The nuclear matrices were collected by centrifugation (10 min, 2,500 g), and the supernatants from the high-salt extractions were combined. Desalting was done with G-50 desalting columns (Boehringer Mannheim Corp.). For the in vivo phosphorylation experiments, nuclei and nuclear matrix protein were prepared from SH-SY5Y, based on modified procedures reported by Hakes and Berezney (1991a ,b) (Feuerstein and Randazzo, 1991). Briefly, purified nuclei were extracted with high ionic strength buffer (2 M NaCl, 10 mM Tris, pH 7.4, 0.2 mM MgCl2, 1 mM PMSF) and then digested with DNAse for 1 h. They were subsequently extracted with 0.5% NP-40 in low ionic strength buffer (10 mM Tris, pH 7.4, 0.2 mM MgCl2) and washed twice with low ionic strength buffer. The pellet was resuspended in low ionic strength buffer containing 9.5 M urea, 2% NP-40, 5% β-mercaptoethanol, and phosphatase inhibitors (10 mM NaF, 1 mM sodium pyrophosphate) and then centrifuged at 30,000 rpm for 1 h at 4°C. Supernatant was collected and subjected to two-dimensional gel electrophoresis.
Immunoprecipitation and Immunoblotting
The cells were lysed in modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM PMSF, 2 μg/ml aprotinin, leupeptin, and pepstatin, and 1 mM Na3VO4) (Hiregowdara et al., 1997). Total cell lysates were clarified by centrifugation at 10,000 g for 10 min. Identical amounts of protein from each sample were precleared by incubation with protein G–Sepharose CL-4B (Sigma Chemical Co.) for 10 min at 4°C. After the removal of protein G–Sepharose by brief centrifugation, protein concentrations were determined using a protein assay (Bio-Rad Laboratories, Hercules, CA), and cell lysates were incubated with NRP/B antibodies (as described for each experiment) overnight at 4°C. Immunoprecipitation of the antigen–antibody complex was accomplished by incubation for 1 h at 4°C with 30 μl of protein G–Sepharose. Bound proteins were solubilized in 20 μl of 2× Laemmli buffer. Samples were separated and analyzed by 7.5 or 10% SDS-PAGE and then transferred to Immobilon membranes. The membranes were blocked with 5% BSA (Boehringer Mannheim Corp.) and probed with primary antibody for 1 h at room temperature. Immunoreactive bands were visualized using horseradish peroxidase–conjugated secondary antibody and the ECL reagents (Amersham Corp., Arlington Heights, IL).
Far Western Blot Analysis
For binding experiments, total cell lysates were immunoprecipitated with control antibody or p110RB monoclonal antibody. The immunoprecipitates were analyzed on 7.5% SDS-PAGE and transferred to membranes. The blots were incubated at 4°C overnight in 5% milk in PBS containing 0.1% Tween 20 followed by incubation with 20 μg of purified tyrosine kinase Csk as a control or NRP/B protein for 2 h at 4°C. The blots were washed, and mouse anti-NRP/B monoclonal antibody was added for 1 h. After washing, horseradish peroxidase–conjugated anti–mouse IgG (Amersham Corp.) was added for 1 h. Immunoreactive bands were visualized using ECL reagents.
Confocal Microscopy
Cell Staining.
Primary hippocampal neurons and astrocytes were fixed for 10 min in 4% paraformaldehyde in PBS containing 0.1% BSA, and cell staining was performed as described (Li et al., 1996). Polyclonal NRP/B antiserum or preimmune rabbit serum (1:1,000 dilution in blocking solution) was used. Cells were incubated with FITC-conjugated goat anti–rabbit IgG (1:200) or rhodamine-conjugated goat anti–rabbit IgG for 1 h at room temperature, washed three times with PBS for 10 min, and mounted with Vectashield mount medium.
Microscopy.
Immunostained cells were examined using a Sarastro 2000 confocal laser scanning microscope (Molecular Dynamics, Sunnyvale, CA) fitted with a 25-mW argon-ion laser as described (Li et al., 1996). Images of neurons were recorded in 1,024 image size format. A 60× 1.4 numeric aperture objective was used to identify the distribution of NRP/B.
Electron Microscopy
Cells were fixed in 2.5% glutaraldehyde in a sodium cacodylate buffer and postfixed in 1% osmium tetroxide dehydrated through a graded series of ethanol. Cells were embedded in EPON 812 and thin-sectioned on RMC MT6000 ultramicrotone (Tucson, AZ). Treatment of NRP/B-specific polyclonal antibodies with 10 μM colloidal gold was performed, followed by staining with uranyl acetate and bead citrate. The thin sections were analyzed under an electron microscope (model 300; Philips Electron Optics, Mahwah, NJ).
In Vivo Phosphorylation
Cells were labeled for 4 h with 0.5 μCi/ml of 32P, supplied as orthophosphoric acid in 8 ml of phosphate-free DME. At the end of the pulse, the cells were harvested and lysed in RIPA buffer. 500 μg of total cell lysate was immunoprecipitated with anti-NRP/B monoclonal antibody as described above. After exposure, the blot was probed with anti-NRP/B antibody. In addition, the nuclei were isolated as described in the section regarding nuclear isolation procedures. 2 × 106 cpm were applied and analyzed by two-dimensional gel electrophoresis (Multipore System; Pharmacia Biotech).
Results
Identification and Isolation of the Full-Length Human NRP/B cDNA
Single pass sequencing was used to identify proteins of potential importance in brain development (Khan et al., 1992). One clone of 720 bp (designated RCR-1), representing a novel gene with no homology to known mammalian cDNAs, was selected and further characterized by Northern blot analysis. Using RCR-1 as a probe, expression of RCR-1 was observed only in fetal and adult brain tissues (data not shown). Therefore, RCR-1 was used as a probe to screen the human fetal brain and adult brain hippocampus cDNA libraries. Several independent cDNA clones were sequenced and appeared to encompass a complete reading frame. One of these clones, λ-ZapIIHFBLII5s, contained an open reading frame of 1,767 nucleotides, which encoded the entire protein of 589 amino acids (Fig. 1 A) with a predicted molecular mass of 67 kD. Human and mouse NRP/B cDNAs were highly conserved, sharing 99% sequence homology (unpublished data).
Figure 1.
Amino acid sequence analysis of NRP/B. (A) Deduced amino acid sequence of NRP/B. The BTB/ POZ homologous domain is underlined. (B) Sequence alignment of the 50–amino acid β-sheet repetitive domains. Sequence alignment indicates the 50–amino acid putative β-sheet repetitive domains compared with the most related protein, Drosophila kelch (d), with 28% identity (45% similarity). The consensus is indicated on the top line: capital letters, highly conserved residues; underlined italics, hydrophobic residues; #, hydrophilic residues; *, charged residues. Letters in shaded boxes denote hydrophobic or charged residues. White letters on black background represent conserved residues. Dashes indicate gaps in the alignment. The three digit numbers in the left column indicate the number of the first residue (or amino acid) in each repeat, and the numbers in the right column indicate the number of residues in a repeat.
Analysis of the NRP/B Protein Sequence
Analysis of NRP/B cDNA sequences and the predicted NRP/B protein showed that the amino terminus of the NRP/ B protein is predicted to be α-helical, while the carboxyl terminus is predicted to be a β-sheet and consists of an ∼50–amino acid residue motif repeated six times (“kelch motif”) (Fig. 1 B). Interestingly, sequence analysis indicates that the ∼113–amino acid NH2-terminal residues (amino acids 28–141) have significant homology (35% identity, ∼55% similarity) to the BTB/POZ domain–containing proteins (He et al., 1995) (Fig. 1 A). Although NRP/B has two basic residue-rich regions (amino acids 258–263 and 289–295), it lacks a classical nuclear localization signal. The kelch motif shares homology to several actin-associated proteins, including the actin-associated Drosophila kelch protein (Fig. 1 B) (Xue and Cooley, 1993), murine IAP-promoted placenta protein (MIPP) (Chang-Yeh et al., 1991), scruin (Way et al., 1995), protein Spe-26 of Caenorhabditis elegans (Varkey et al., 1995), calicin (von Bulow et al., 1995), and a large number of poxvirus proteins. This kelch motif contains a predicted structure of β-sheet repeats that form “superbarrel” structures (Bork and Doolittle, 1994).
NRP/B Is Highly Restricted in Tissue Expression
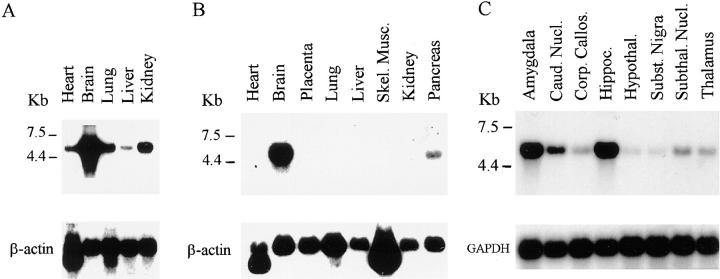
An extensive survey of human fetal and adult primary tissues was performed by Northern blot analysis. The 5.5-kb NRP/B mRNA was detected abundantly in human fetal brain tissue with moderate expression in fetal heart, lung, and kidney (Fig. 2 A). Probing of mRNAs derived from various human adult tissues revealed a very high level of NRP/B mRNA expression in the brain and a lower but detectable mRNA signal in the pancreas (Fig. 2 B). The expression of NRP/B mRNA was particularly high in the amygdala and hippocampus regions of human adult brain (Fig. 2 C).
Figure 2.
mRNA expression of NRP/B in human tissues. (A) Poly-A mRNA blot of human fetal tissues. (B) Expression of NRP/B mRNA in human adult primary tissues. (C) Expression of NRP/B mRNA in different regions of human brain. The probes used in these blots were the 3′-end gene-specific NRP/B cDNA probe (500-bp) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin cDNA probes as controls for the RNA amounts.
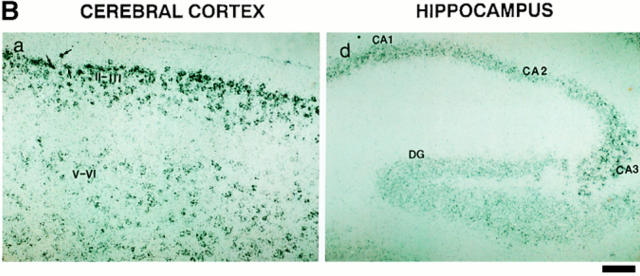
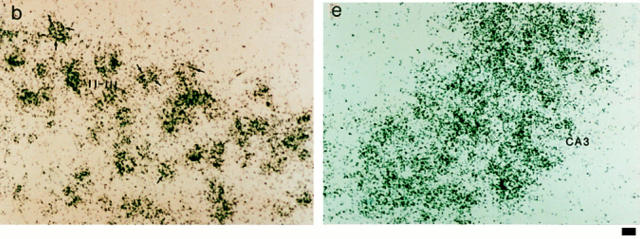
To assess the expression of NRP/B in tissues at different developmental stages, in situ hybridization was performed with murine Nrp/b cDNA probe(s) on mouse tissue sections. An in situ hybridization study of Nrp/b expression in early postimplantation mouse embryos (8–10 d post coitum [pc]) showed no expression (data not shown). However, in day 12 pc mouse embryos, NRP/B showed an interesting pattern of expression. It was mainly present in the cerebrum, spinal cord, and peripheral ganglia (Fig. 3 A). In the brain, Nrp/b expression was found in the diencephalon, mesencephalon, rhombencephalon, and on the floor of the third and fourth ventricles. In the spinal cord, Nrp/b expression was found in the dorsal and ventral regions and in the peripheral ganglia. In situ hybridization was also performed on the sagittal sections of adult mouse brain. Nrp/b mRNA expression was found abundantly in the neuronal layers of the cerebral cortex, hippocampus, and amygdala (Fig. 3 B). In the hippocampus, significant levels of Nrp/b mRNA were observed in the pyramidal cells of the CA1–CA3 subfields (Fig. 3 B, d). A strong signal was also observed in the CA3 region of the hippocampus, and a moderate level of expression was seen in the CA1 and CA2 subfields (Fig. 3 B, d and e). In the dentate gyrus, granule cells showed a strong signal (Fig. 3 B, d). Nrp/b expression was also abundant in the amygdala and basal ganglia. In the cerebral cortex and the piriform cortex, high levels of Nrp/b expression in layers II–III and moderate levels in layers V–VI were observed (Fig. 3 B, a and b). No signal was detected in the cerebral cortex (Fig. 3 B, c) or hippocampus (Fig. 3 B, f) when the brain sections were probed with the sense strand of Nrp/b.
Figure 3.
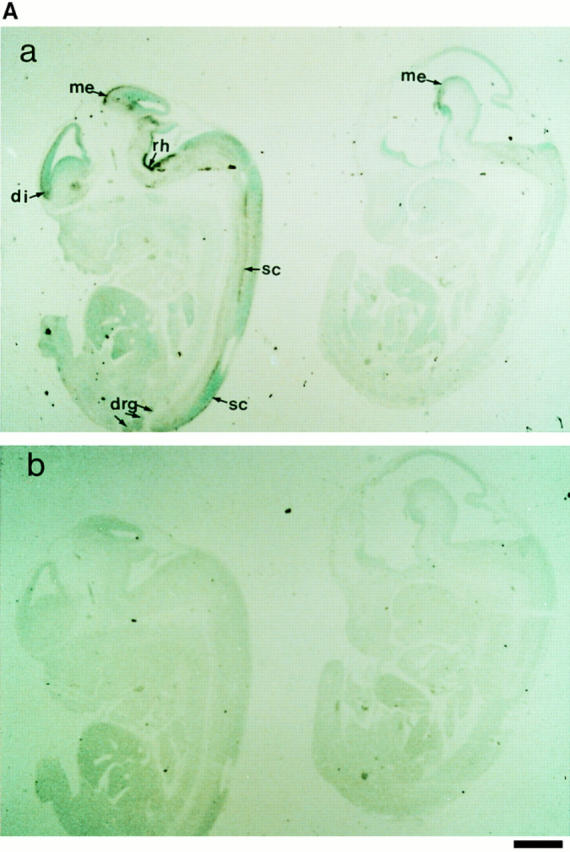
In situ hybridization of NRP/B. (A) NRP/B expression in day 12 pc mouse embryos: Sagittal sections of day 12 mouse embryos were probed with 35S-labeled antisense (a) and sense (b) RNA probes. (a) Bright-field photomicrograph of day 12 pc mouse embryo. di, diencephalon; me, mesencephalon; rh, rhombencephalon; sc, spinal cord; drg, dorsal root ganglia. (b) Serial sections (shown in a) hybridized with the sense probe show no signal. (B) Expression of NRP/B mRNA in adult mouse brain: Sagittal sections of mouse brain were probed with 35S-labeled antisense (a, b, d, and e) and sense (c and f) RNA probes. (a) Bright-field photomicrograph of cerebral cortex showing hybridization in layers II–III and V–VI. (b) High magnification of layer II–III shown in a. Arrows point to the signal in neuronal cells. (c) Serial section shown in a, hybridized with the sense probe. (d) Bright-field photomicrograph of hippocampal region showing hybridization in CA1–CA3 subfields and dentate gyrus. (e) Higher magnification of CA3 subfield of hippocampus. (f) Serial section shown in d hybridized with the sense probe shows no signal in the hippocampus region. DG, dentate gyrus. Bars: (A, a and b) 10 μm; (B, a, c, d, and f) 100 μm; (B, b and e) 400 μm.
NRP/B Is Localized to the Nucleus of Primary Hippocampal Neurons
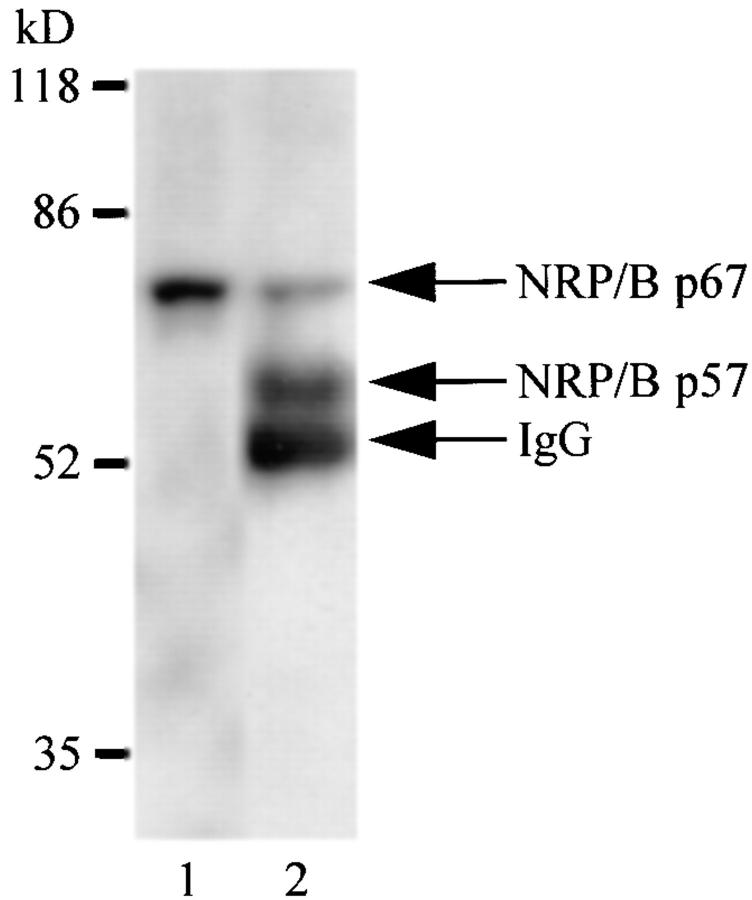
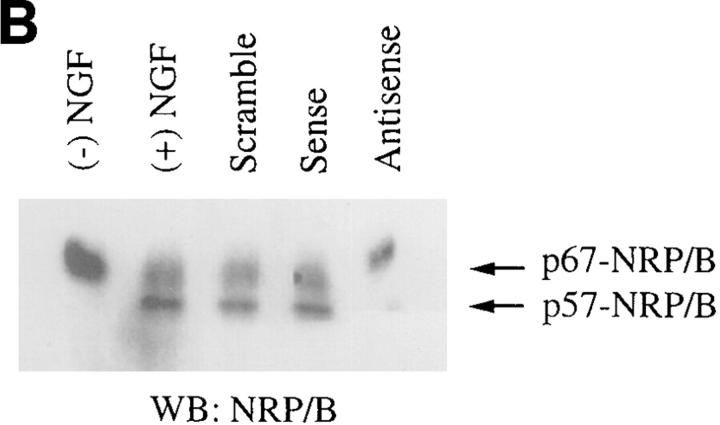
To identify NRP/B protein expression, we have generated specific anti-NRP/B antibodies: NRP/B polyclonal antibodies raised in rabbits immunized with the full-length NRP/B protein expressed in the baculovirus system, and NRP/B monoclonal antibodies raised in mice that were also immunized with the full-length NRP/B protein expressed in the baculovirus system. 24 hybridoma clones were characterized as NRP/B-specific monoclonal antibodies and recognized the full-length 67-kD NRP/B-purified protein expressed in the baculovirus system (data not shown). When total cell lysates were prepared from primary neurons (Fig. 4), PC-12 cells (Fig. 9 B), or SH-SY5Y cells (data not shown) and analyzed by immunoprecipitation followed by Western blot analysis, two forms of NRP/B protein (67 and 57 kD) were detected using NRP/B polyclonal and monoclonal antibodies (Fig. 4, lane 2). However, when the nuclear pellet of the primary neurons was analyzed by Western analysis, only the 67-kD form was identified using polyclonal or monoclonal antibodies (Fig. 4, lane 1). These analyses indicate that the NRP/B protein appears to have two forms: 57 and 67 kD.
Figure 4.
Characterization of two NRP/B forms. A nuclear pellet of neurons was prepared and resuspended in SDS loading buffer (lane 1). In addition, total cell lysates of primary neurons (500 μg) were immunoprecipitated with NRP/B monoclonal antibody and analyzed by SDS-PAGE. Both samples were blotted with VD2 monoclonal NRP/B antibody (dilution 1:1,000).
Figure 9.
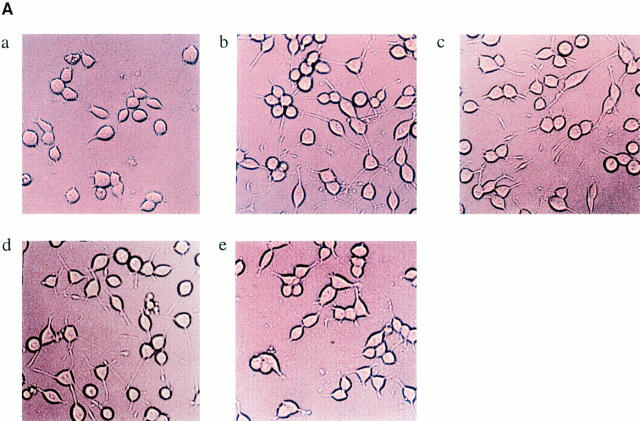
Effect of antisense oligodeoxynucleotides. (A) Effect of antisense oligodeoxynucleotides on cell morphology: PC-12 cells were plated at a density of 105 cells/ml and treated with 75 μg/ml oligodeoxynucleotide for 24 h in DME containing 1% horse serum, with or without NGF, 100 ng/ml as indicated: (a) Control PC-12 cells; (b) NGF (100 ng/ml)-treated PC-12 cells; (c) scramble oligodeoxynucleotides + NGF; (d) sense oligodeoxynucleotides + NGF; (e) antisense NRP/B oligodeoxynucleotides + NGF. (B) Western blot analysis of NRP/B protein from PC-12 cells treated with sense, scramble, or antisense oligodeoxynucleotides. Total cell lysates were obtained by the direct addition of prewarmed 2× Laemmli loading buffer to the cells. Equal amounts of cell equivalents (105 cells) were loaded. NRP/B monoclonal antibody was used (dilution 1:1,000) for Western blot analysis. The reactive proteins were detected using the ECL system (Amersham Corp.). (C) Quantitative analysis of neurite extension in PC-12 cells treated with oligodeoxynucleotides. A total of 200 cells from triplicate experiments were counted for the percentage of cells with neurites. The neurite length was measured under each condition. Data are the mean ± SE values.
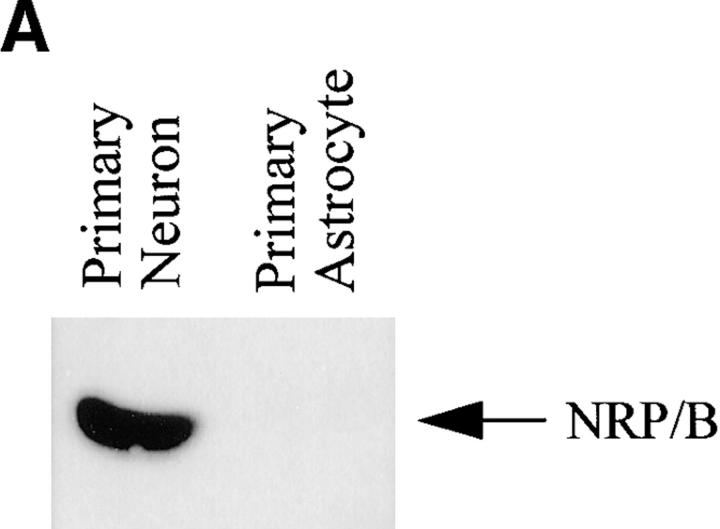
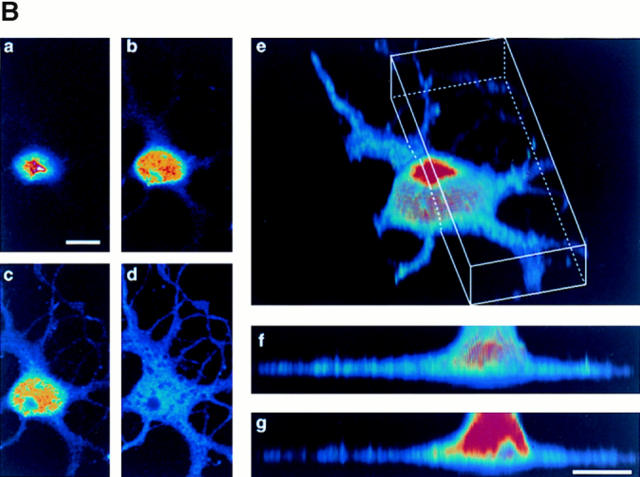
The expression of NRP/B protein in rat primary hippocampal neurons and astrocytes was further evaluated by Western blot analysis using NRP/B polyclonal and monoclonal (SA5) antibodies. NRP/B protein (67 kD) was expressed in primary hippocampal neurons but was absent in primary astrocytes (Fig. 5 A). In addition, primary hippocampal neurons were analyzed by immunostaining using polyclonal and monoclonal NRP/B antibodies, followed by immunofluorescent microscopy and confocal microscopy. NRP/B was observed in the nucleus of these neurons (Fig. 5 B). Microscopic imaging of rat primary hippocampal neurons revealed a nuclear-specific distribution of NRP/B (Fig. 5 B). Serial optical sections (3-μm intervals) recorded from the near apical surface (a) of the cell body toward the basal surface (d) revealed the NRP/B-labeled nuclear matrix. Three-dimensional volume rendering of entire cells indicates that NRP/B expression is limited to the nuclear matrix. In the cell shown (Fig. 5 B, e), over 95% of the NRP/B was localized to the cell body region. Volumetric analysis revealed this cell to be 2,866 μm3, with 390 μm3 occupied by the nucleus. Immunogold staining and transmission electron microscopy of rat primary hippocampal neurons revealed NRP/B antigen deposition in the nuclear matrix of the nucleoplasm, the peripheral heterochromatin, and the nucleolus (Fig. 5 C). No immunogold staining of NRP/B was seen in primary astrocytes. These results suggest that NRP/B is a nuclear protein associated with the nuclear matrix in neuronal cells.
Figure 5.
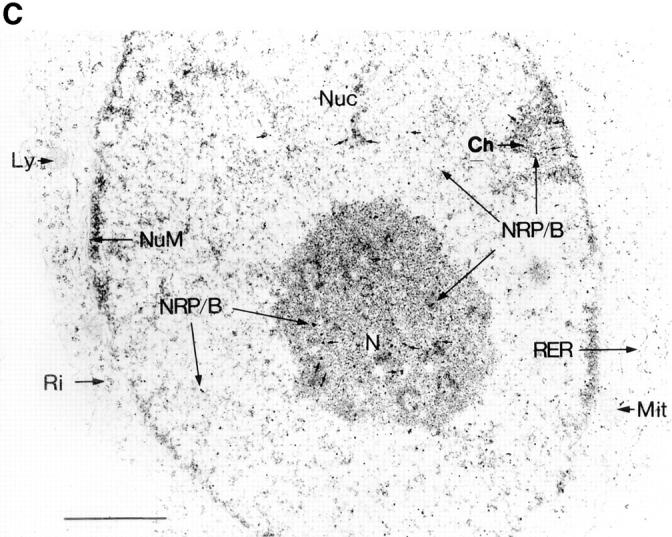
Subcellular localization of pNRP/B in primary hippocampal neurons. (A) Expression of NRP/B in primary neurons and astrocytes. Equal amounts of total cell lysate were applied for Western blot analysis using anti-NRP/B antibody (SA5 monoclonal antibody, 1:1,000 dilution). (B) Confocal micrographs of primary neurons isolated from the hippocampus of rat brain immunolabeled with NRP/B antibody (rabbit FITC secondary antibody) show nuclear staining. (a–d) Serial optical sections recorded 0.3 μm from the apical surface (a) of the cell body toward the basal surface (d) at 3-μm intervals. (e–g) Three-dimensional rendering of rat primary neuron of serial section represented in a–d. (e) A surface view of the neuron in an oblique orientation with the uppermost apical sections removed to reveal the greater intensity of the NRP/B labeling within the nuclear matrix of the cell body. The front surface of the open box indicates the position of an orthogonal view projection through the cell volume shown in g. (f) Side view three-dimensional projection of the reconstructed neuron seen in e. (C) Electron microscopic localization of NRP/B protein in primary neurons. Small arrows indicate the localization of the NRP/B protein. NuM, nuclear membrane; N, nucleolus; Mit, mitochondria; Ch, peripheral heterochromatin; Ri, ribosomes; Ly, Lysosome; RER, rough endoplasmic reticulum; Nuc, nucleus. Bar, 1 μm.
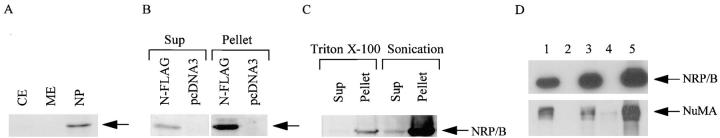
In addition to immunolocalization studies, nuclear localization of NRP/B was confirmed by two biochemical methods (Mirkovitch et al., 1994; Hakes and Berezney, 1991a ,b). Subcellular fractionation was performed on the COS-7 cells transfected with the FLAG-NRP/B-pcDNA3neo construct. Each fraction was processed for SDS-PAGE and Western blot analyses. NRP/B protein was detected only in the nuclear pellet using M5 antibody (Fig. 6 A). Subnuclear fractions from the COS-7 cells transfected with this construct contained insoluble NRP/B protein predominantly in the nuclear pellets (Fig. 6 B). Even after detergent solubilization (1% Triton X-100) or mechanical shear (sonication without detergent) of the nuclei, the majority of NRP/B protein was still found in the pellets (Fig. 6 C). These results suggest that NRP/B is localized to the nucleus and possesses the biochemical features of a nuclear matrix protein.
Figure 6.
Biochemical analysis of subcellular localization of NRP/B protein. Fractions were prepared from transfected COS-7 cells and SH-SY5Y neuroblastoma cells. (A) NRP/B expression in subcellular fractions of transiently transfected COS-7 cells. Pellet and supernatant were separated, and equal amounts of proteins were subjected to Western blot analysis using M5 anti-FLAG antibody. CE, cytoplasmic extract; ME, membrane extract; NP, nuclear pellet. (B) Nuclear fractionation from COS-7 cells transfected with either the N-FLAG-NRP/B-pcDNA3 neo construct or pcDNA3neo vector. Sup, supernatant from nuclei. (C) Fractionations of purified nuclei by detergent (Triton X-100, 1%) or sonication without detergent. (D) Fractionation of SH-SY5Y neuroblastoma cells; total lysates (lane 1), supernatant fraction after cell homogenization (lane 2), nuclear pellet (lane 3), high-salt extract nuclei after EcoRI digestion of the nuclei (lane 4), and the nuclear matrix protein (lane 5). A comparable amount of protein from each fraction was loaded onto a 10% SDS–polyacrylamide gel and separated. The NRP/B protein and the nuclear matrix protein NuMA were visualized using a murine monoclonal anti-NRP/B antibody (VD2) and monoclonal antibody for NuMA (Calbiochem, La Jolla, CA), respectively. The reactive proteins were detected using an ECL system (Amersham Corp.).
The biochemical properties of endogenous NRP/B were further characterized on actively growing SH-SY5Y human neuroblastoma cells (Fig. 6). Cells were collected and fractionated. After homogenization, a supernatant fraction (Fig. 6 D, lane 2) and a nuclear pellet (lane 3) were obtained. We further fractionated the nuclear pellet into soluble (Fig. 6 D, lane 4) and insoluble nuclear matrix fractions (lane 5) after EcoRI digestion and high-salt treatments. Western blot analysis of these fractions using monoclonal anti-NRP/B antibody indicated that NRP/B is a nuclear matrix protein (Fig. 6 D, lanes 3 and 5), as observed for the FLAG-NRP/B construct as described above. The molecular mass of the recognized protein was 67 kD. The nuclear matrix purification procedure was monitored and confirmed by using a monoclonal antibody to a nuclear matrix protein, NuMA, with a molecular mass of ∼240 kD (Compton et al., 1991, 1992; Yang et al., 1992).
NRP/B Is Involved in Neuronal Differentiation
To characterize the possible function(s) of the NRP/B protein in neurons, we have analyzed its expression in Neuro 2A cells upon dibutyryl cAMP treatment (Fig. 7) and in SH-SY5Y cells upon retinoic acid (RA) treatment (see Fig. 11). RA and dibutyryl cAMP are known to induce neuronal cell differentiation (Vignali et al., 1996; Cosgaya et al., 1997). The expression level of NRP/B was evaluated by Western blot analyses before and after induction by RA and dibutyryl cAMP treatment. In particular, Neuro 2A cells showed an increase in their level of NRP/B expression in response to dibutyryl cAMP (Fig. 7 B). Similar results were obtained with SH-SY5Y neuroblastoma cells in response to RA treatment (Fig. 11). These results indicate that NRP/B was upregulated during neuronal differentiation of the Neuro 2A and SH-SY5Y neuroblastoma cells.
Figure 7.
Effects of dibutyryl-cAMP on NRP/B expression and overexpression of NRP/B in Neuro 2A neuroblastoma cells. (A) Dibutyryl cAMP–induced neuronal differentiation of Neuro 2A cells: mouse Neuro 2A neuroblastoma cells were grown on MEM containing 10% fetal bovine serum as a control (a) or were induced by 5 μM dibutyryl-cAMP for 48 h (b). (B) Expression of NRP/B protein during neuronal differentiation of Neuro 2A cells: 1 × 105 cells were directly lysed with Laemmli loading buffer and subjected to 8% SDS-PAGE. Immunoblot analyses were performed using monoclonal NRP/B antibody: lane 1, cells without dibutyryl-cAMP treatment; lane 2, cells treated with dibutyryl-cAMP. (C) Morphological changes in Neuro 2A cells: stable Neuro 2A cells transfected with vector alone (pcDNA3) (a) and with FLAG-NRP/B-pcDNA3 (b) were examined for cell morphology under the same magnification using light microscopy. (D) Expression of NRP/B protein in transfected Neuro 2A cells by Western blot analysis: equal amounts of proteins were loaded and analyzed using FLAG monoclonal antibody M5; lane 1, total cell lysates prepared from FLAG-NRP/B-pcDNA3–transfected Neuro 2A cells; lane 2, total cell lysates prepared from pcDNA3-transfected cells.
Figure 11.
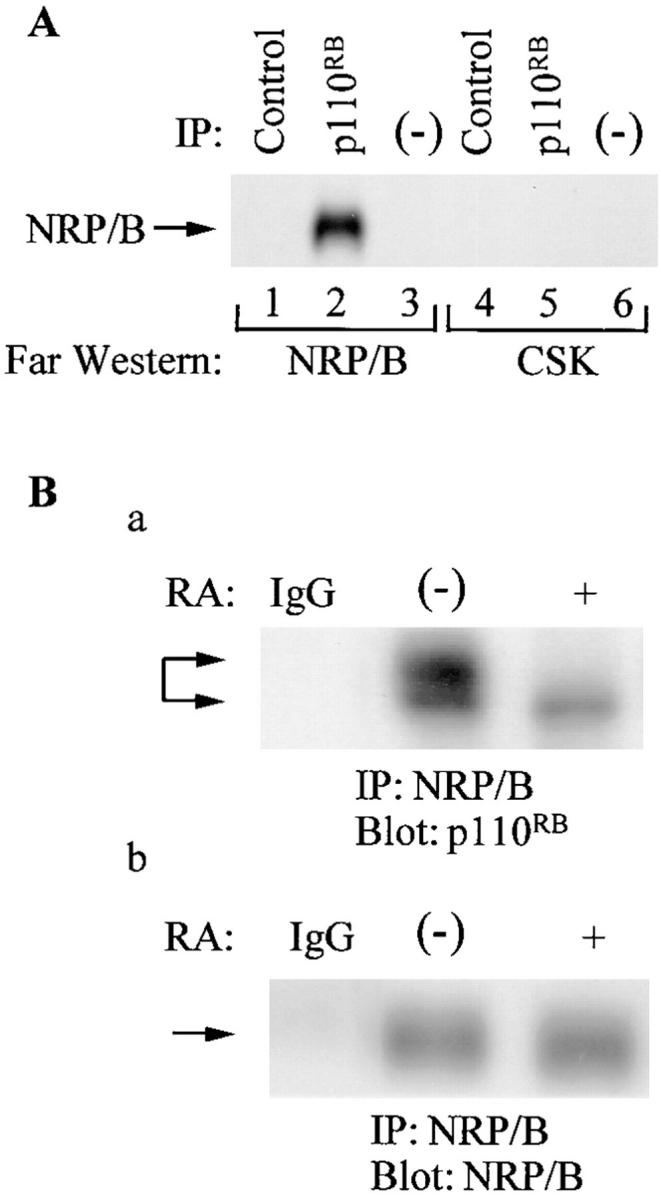
In vitro and in vivo association of NRP/B with p110RB. (A) Lysates of SH-SY5Y cells were immunoprecipitated with anti-p110RB antibody (lanes 2 and 5), analyzed by 7.5% SDS-PAGE, and transferred to Immobilon-P membranes. The membranes were processed for Far Western blotting with the Csk kinase (as a negative control) and NRP/B-purified proteins. Lanes 1 and 4, immunoprecipitations were performed with control antibody; lanes 3 and 6, control Sepharose beads alone. (B) In vivo association of NRP/B with p110RB: (a) 500 μg of total cell lysates obtained from retinoic acid (10 μM) treated and untreated SH-SY5Y cells were subjected to immunoprecipitation with NRP/B and blotted with monoclonal p110RB antibody. Two different migrated forms were observed: The slower migrated form (lane 1) represents the hyperphosphorylated p110RB, and the faster migrated form (lane 2) represents the hypophosphorylated p110RB. (b) After deprobing the blot in a, the membrane was probed with polyclonal NRP/B antibody.
In addition, we investigated the effects of NRP/B overexpression in murine Neuro 2A cells. We transfected these cells with FLAG-NRP/B-pcDNA3neo cDNA and generated a stable NRP/B-transfected cell line (Neuro 2A– NRP) that overexpressed the NRP/B protein (Fig. 7 D). The Neuro 2A–NRP demonstrated a significant augmentation of neuronal process formation, similar to that observed in Neuro 2A cells upon treatment with dibutyryl cAMP (Fig. 7 A, Table I). No morphological changes were observed in either the untreated Neuro 2A cells or in the Neuro 2A cells transfected with the pcDNA3neo vector alone (Fig. 7). The expression of FLAG-NRP/B in the Neuro 2A–NRP cells was observed by Western blot analysis using monoclonal antibodies for the FLAG-tagged protein (Fig. 7 D). Furthermore, the data summarized in Table I indicate that there was a significant increase in the number and length of neurites of the Neuro 2A cells stably transfected with NRP/B cDNA (fivefold), results similar to that obtained after stimulation with dibutyryl cAMP (fivefold).
Table I.
Neurite Outgrowth in NRP/B-transfected Neuro 2A Cells
| Transfected Neuro 2A cells | Percentage of cells with neurites | Neurite length | ||
|---|---|---|---|---|
| μm | ||||
| Control (+ dibutyryl cAMP) | 30 ± 3 | 82 ± 7 | ||
| NRP/B | 32 ± 3 | 81 ± 8* | ||
| pcDNA3 (Mock) | 6 ± 1 | 16 ± 1 |
Neuro 2A (mouse neuroblastoma) cells were stably transfected with 10 mg NRP/B-pcDNA3 as described in Materials and Methods. After G418 selection, cells were cultured on poly-d-lysine–treated plastic dishes. The percentages of cells with neurites were calculated from three experiments. Neurite lengths were measured from 50 neurites per condition in a representative experiment repeated three times. Data are the means ± SE values.
Significantly enhanced compared to control transfection with pcDNA3 (Mock) (P < 0.005).
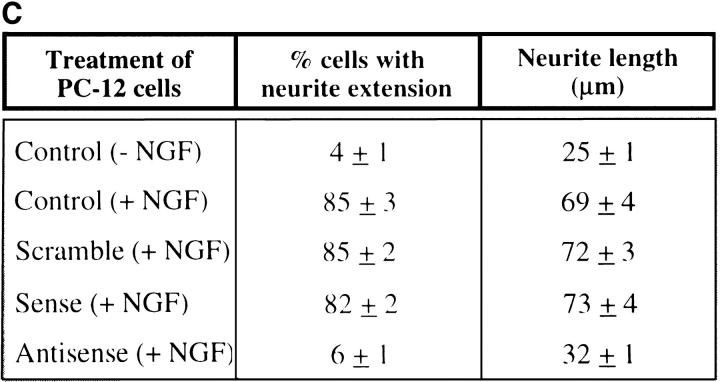
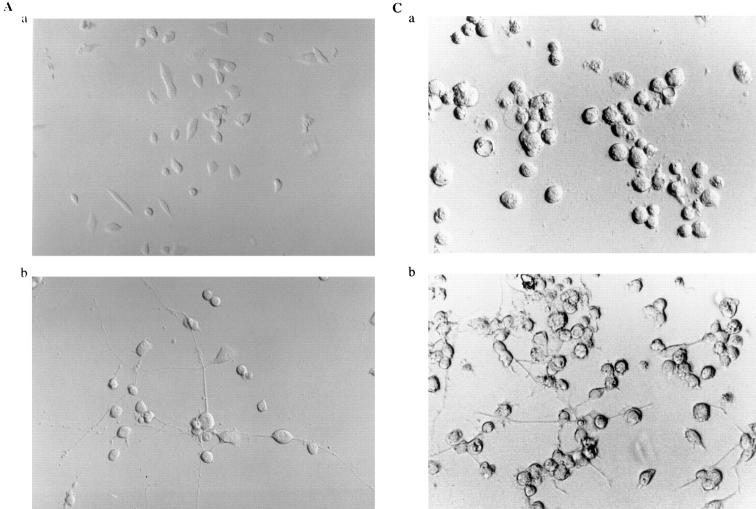
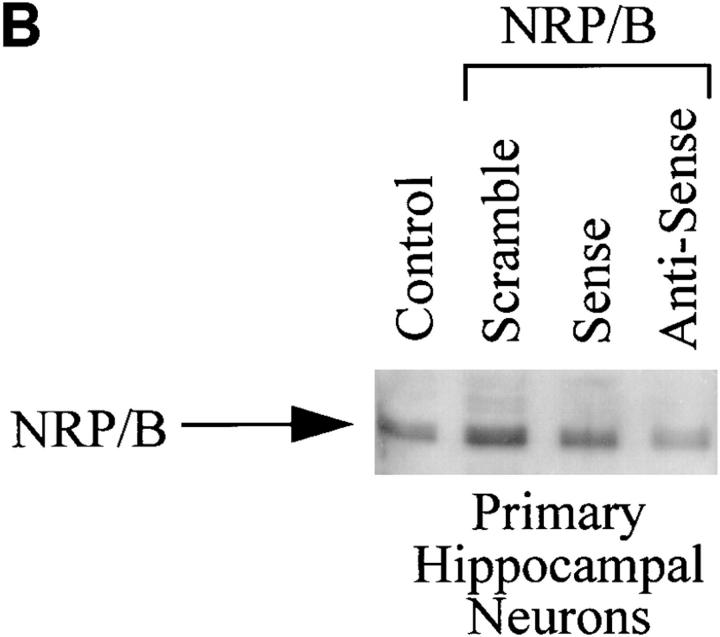
In addition, we used a sense/antisense oligodeoxynucleotide approach against NRP/B mRNA in cultures of rat primary hippocampal neurons. Over 90% of the cells in culture were primary neurons, as shown by immunostaining with neuronal specific markers such as MAP-2 (Binder et al., 1984; Geisert et al., 1990) (data not shown). Kinetics and dose response studies using NRP/B oligodeoxynucleotides were first performed at various densities of primary neurons to determine the optimal assay conditions. Antisense treatment (25 μg/ml) of primary neurons (1 × 105 cells/ml) resulted in a reduction in neuronal process formation (Fig. 8 A, d). This reduction could be observed even in low-density cultures (1 × 104 cells/ml) treated with NRP/B antisense oligodeoxynucleotides (data not shown). As shown in Table II, antisense treatment of rat primary hippocampal neurons resulted in a significant decrease (77%) in neuronal process length. No significant changes were observed when primary astrocytes were treated with the antisense oligodeoxynucleotides (Fig. 8 A, h), suggesting that the effect of NRP/B antisense oligodeoxynucleotides was specific to primary neurons (Fig. 8 A). In conjunction with these antisense effects on cell morphology, we observed decreased levels of NRP/B protein to ∼50% of baseline, supporting the specificity of these biological changes (Fig. 8 B). NRP/B sense and scrambled oligodeoxynucleotide treatments had no effect on primary neuronal cells (Fig. 8, A and B).
Figure 8.
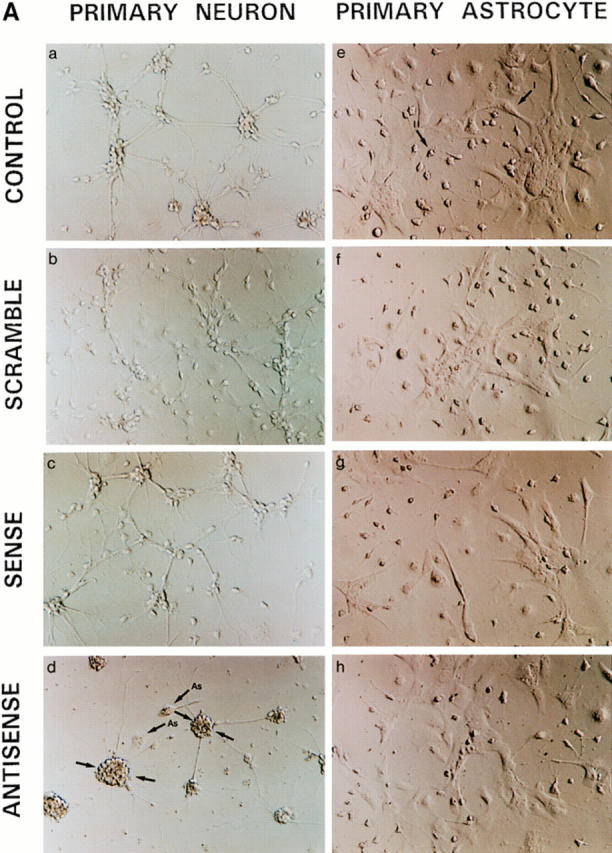
Effect of antisense oligodeoxynucleotides against NRP/B on rat primary hippocampal neurons and primary astrocytes. (A) Effect of antisense oligodeoxynucleotides on cell morphology: Primary neurons (a–d) and astrocytes (e–h) were plated at a density of 3 × 105 cells/ml and treated with 25 μg/ml oligodeoxynucleotide for 48 h. Two different types of primary astrocyte are indicated as I and II (e). Cells were treated with oligodeoxynucleotides against NRP/B as follows: a and e indicate controls without treatment; b and f indicate treatment with scramble oligodeoxynucleotides; c and g indicate treatment with NRP/B sense oligodeoxynucleotides; d and h indicate treatment with NRP/B antisense oligodeoxynucleotides. As, astrocytes. (B) Western blot analysis of NRP/B protein from primary neurons treated with sense, scramble, or antisense oligodeoxynucleotides. Total cell lysates were obtained by the direct addition of prewarmed 2× Laemmli-loading buffer to the cells. Equal amounts of cell equivalents (106 cells/ml) were loaded. NRP/B monoclonal antibody was used (dilution 1:1,000) for Western blot analysis. The reactive proteins were detected using the ECL system (Amersham Corp.).
Table II.
Quantitative Analysis of Neurite Outgrowth in Sense- and Antisense-treated Hippocampal Neuron Cultures
| Oligonucleotide | Percentage of cells with neurites | Neurite length | ||
|---|---|---|---|---|
| μm | ||||
| Control | 92 ± 4 | 115 ± 11 | ||
| Sense | 90 ± 4 | 116 ± 9 | ||
| Scramble | 88 ± 3 | 120 ± 7 | ||
| Antisense | 21 ± 2* | 44 ± 5 |
Embryonic day 18 hippocampal neuron cultures were prepared as described in Materials and Methods. Neurite lengths and the percentages of cells with neurites were measured 48 h after treatment with sense, scramble, and antisense oligonucleotides against NRP/B mRNA. A total of 150 cells from three repeated experiments for each condition were counted. Data are the means ± SE values.
Significantly inhibited neurite outgrowth compared to sense treatment (P < 0.005).
Since we observed a significant reduction in neuronal process formation in rat primary hippocampal neuronal cultures upon NRP/B antisense oligodeoxynucleotide treatment, we next studied PC-12 cells, which are known to differentiate upon treatment with the growth factor NGF (Greene and Tischler, 1976). Treatment of PC-12 cells with NGF (100 ng/ml) induced neuronal differentiation (which includes neuronal process formation) in more than 85% of the cell population as compared with the untreated cells (Fig. 9, A and C). Treatment of these cells with NRP/B antisense oligodeoxynucleotides (75 μg/ml) inhibited neuronal process formation in response to NGF (Fig. 9 C). No effect was observed in PC-12 cells treated with NRP/B sense or scrambled oligodeoxynucleotides. A significant decrease in neurite outgrowth and neurite length was observed in PC-12 cells treated with NRP/B antisense oligodeoxynucleotides (Fig. 9 C). Antisense oligodeoxynucleotide treatment completely inhibited p57-NRP/B expression and decreased the expression of p67 NRP/B (Fig. 9 B). These studies further indicated that NRP/B is involved in neuronal differentiation. Thus, the inhibition of NRP/B expression reduced neuronal process formation and overexpression of NRP/B enhanced neuronal process formation of neuronal cells.
In Vivo Phosphorylation of NRP/B and Its Binding to p110RB
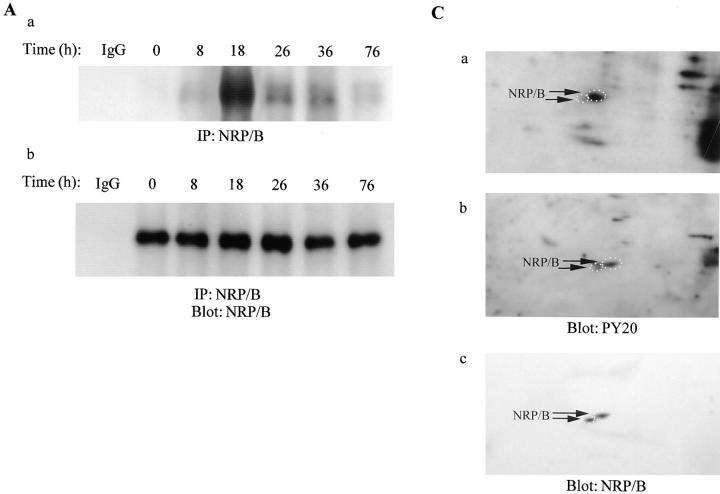
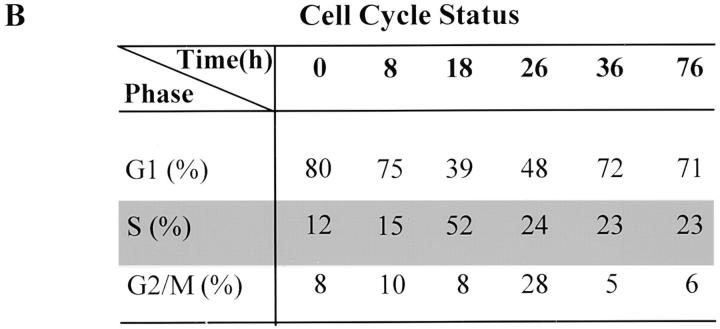
Evidence suggests that the hypophosphorylated form of p110RB is active in growth suppression (Riley et al., 1994). The hypophosphorylated form of p110RB protein is found to be associated with the nuclear matrix and “tethered” to the nuclear structure, whereas hyperphosphorylated and mutated forms of the protein are not (Mittnacht and Weinberg, 1991). Since NRP/B is found to be expressed in cells that are in the mitotic-active ventricular zone (unpublished data) as well as in postmitotic neurons in other brain regions (Fig. 3), we investigated whether NRP/B is associated with p110RB and its in vivo phosphorylation during cell cycle progression. To study in vivo phosphorylation of NRP/B during cell cycle progression, we used synchronized SH-SY5Y cells. Postconfluent SH-SY5Y cells were arrested in the G1 phase by serum deprivation (Fig. 10 B). 8 h after cells were stimulated by the addition of 10% fetal bovine serum, NRP/B became phosphorylated (Fig. 10 A). As cells were entered in S-phase, NRP/B became highly phosphorylated (Fig. 10 A, Time = 18 h). To further analyze NRP/B phosphorylation, in vivo 32P-labeled nuclear extract was analyzed by Isoelectric-Focusing (IEF) two-dimensional gel electrophoresis. Interestingly, two forms of in vivo phosphorylated NRP/B were detected (Fig. 10 C, a). These phosphorylated NRP/B forms are tyrosine-phosphorylated as shown in Fig. 10 C, b. These results suggest that phosphorylation of NRP/B is regulated during cell cycle progression.
Figure 10.
In vivo phosphorylation of NRP/B during cell cycle progression. (A) Postconfluent SH-SY5Y cells were cultured in serum-free medium for 24 h. (a) Cells were stimulated in phosphate-free DME with 10% dialyzed fetal bovine serum for the indicated times. For metabolic labeling, [32P]orthophosphoric acid (500 μCi) was added for 4 h. Total cell lysates (500 μg) were immunoprecipitated with monoclonal NRP/B antibody, and the blot was exposed overnight. (b) Western blot analysis of the same blot in a using NRP/B antibody (1:1,000). (B) Cells parallel to experiment A were harvested and stained with propidium iodide for flow cytometric analysis. (C) Analysis of in vivo 32P-labeled nuclear extract on two-dimensional gel electrophoresis. (a) Nuclear extract from metabolically labeled cells were resuspended in 9.5 M urea. 2 × 106 cpm were subjected to IEF two-dimensional gel electrophoresis. Blot was exposed overnight. (b) Immunoblot with PY-20 monoclonal antibody. (c) After deprobing the membrane in b, the blot was probed with NRP/B monoclonal antibody. Arrows indicate two forms of the phosphorylated NRP/B.
In an attempt to explore the interaction between NRP/B and p110RB during neuronal differentiation, the ability of NRP/B to bind native p110RB was tested using an in vitro binding assay. Total cell lysates were prepared from untreated or RA-treated SH-SY5Y cells and immunoprecipitated with anti-p110RB antibody. Immunoprecipitates were washed, separated on a 7.5% SDS-PAGE gel, and subjected to Western blotting. While the control-purified Csk protein did not bind to p110RB, the full-length recombinant NRP/B protein did bind directly to p110RB (Fig. 11 A). Thus, these results indicate that NRP/B can specifically interact with p110RB in vitro. To determine the nature of this NRP/B association to p110RB during neuronal differentiation, SH-SY5Y cells were treated with RA (10 μM) in the presence of 10% fetal bovine serum for 2 d, during which time cells underwent differentiation. p110RB was coimmunoprecipitated with NRP/B (Fig. 11 B). Although, NRP/B expression was upregulated during neuronal differentiation (Figs. 7 and 11), the association of NRP/B with p110RB was decreased compared with untreated cells (Fig. 11 B). These results demonstrate that the interaction of NRP/B with p110RB is specific and suggest that the hypophosphorylated form of p110RB associates with NRP/B during neuronal differentiation, while the hyperphosphorylated form of p110RB associates with NRP/B in nondifferentiating cells.
Discussion
In this study, we have described for the first time a novel neuronal nuclear matrix protein, designated NRP/B (for nuclear-restricted protein in brain), which appears to play a role in neuronal differentiation. This determination is based on four types of experimental and analytical sets of findings: (a) structural elements, (b) developmental studies, (c) localization, and (d) functional studies.
Structural Elements
Structural analysis of both the nucleotide sequences and the predicted secondary structure of NRP/B revealed two major structural elements, the α-helix NH2-terminal BTB domain–like structure (35% identity, 59% similarity) and the β-sheet COOH-terminal kelch motif. The BTB domain has been proposed to mediate protein–protein interactions that are associated with higher order structures involved in chromatin folding or cytoskeleton organization (Geisert et al., 1990). This domain is found in diverse molecules: in several developmentally regulated zinc finger– type transcription factors of Drosophila and mammals, in several open-reading frames of the poxviruses, and in the cytoskeletal protein calicin from the sperm head. Similar to WT1 and the Koriippel-type zinc finger protein TFIIIA (Caricasole et al., 1996), the NRP/B protein, through its BTB domain, might function as a regulatory domain in protein–protein interactions at the transcriptional and/or posttranscriptional level. In addition, the BTB homologous domain might be responsible for the insolubility of the 67-kD NRP/B protein (unpublished data). This characteristic of insolubility is probably due to oligomerization of the NRP/B protein. Similar data have been shown with the BTB domain of the bab protein (Albagli et al., 1995).
A consensus sequence derived from the six repeats of NRP/B showed ∼28% identity (45% similarity) with several actin-associated proteins that contain 50–amino acid “kelch-like motifs” (Fig. 1 B). These motifs are found in the actin cross-linking protein scruin (Way et al., 1995) and in a series of other proteins (Chang-Yeh et al., 1991; Xue and Cooley, 1993). Although the role of the β-sheet repeat in the kelch family of proteins is not yet known, the predicted structure of β-sheet repeat motifs may have functional significance in binding actin, protein folding, or protein–protein interactions. Structural analysis suggested that the β-sheet motifs form “superbarrel” structures (Bork and Doolittle, 1994; Albagli et al., 1995; Chen et al., 1995). Interestingly, a recent crystallographic study on the β-subunit of the G protein (Ito et al., 1991; Robinson et al., 1994) revealed that its β-stranded repeat motifs form a superbarrel structure (Wall et al., 1995) and might play an important role in intra- and intermolecular interactions. By analogy, NRP/B, with its six-repeat structure, may communicate directly with structural elements and may recruit other nuclear matrix proteins to form a nuclear scaffold structure.
Developmental Studies
We have observed NRP/B expression in fetal brain to be about 50-fold higher than in other tissues during fetal development (embryogenesis). Northern blot analysis demonstrated that NRP/B mRNA was highly expressed in the human adult brain. These different patterns of NRP/B expression in fetal and adult human tissues suggest that NRP/B is regulated during embryogenesis and may have tissue-specific functions at different developmental stages. Using in situ hybridization, NRP/B expression was not detected in the early embryos (8–10 d pc), while in the day 12 pc mouse embryos, NRP/B mRNA was readily detected in the brain and spinal cord (Fig. 3). There were particularly high NRP/B mRNA levels in the hippocampus and amygdala cortex, which are major components of the limbic system. NRP/B expression was also found in the piriform cortex, which is also a component of the limbic system. The expression pattern of NRP/B suggests the possible involvement of NRP/B in neuronal development.
Localization
The localization of the NRP/B protein is also suggestive of its regulatory activity. This protein is expressed abundantly in brain (Fig. 2) and appears to be expressed specifically in primary neurons (Fig. 3 A). Confocal micrographs and electron microscopy using NRP/B-specific antibodies indicated that NRP/B is highly expressed in the nucleus of neurons but not in primary astrocytes. Cellular commitment and differentiation of the cell lineages in the nervous system, like all developing tissues, depend ultimately on intricate programs establishing specific patterns of gene expression (Maniatis et al., 1987; Ptashne, 1988; Mitchell and Tjian, 1989). NRP/B gene expression in a tissue-specific manner (in neurons but not in astrocytes) might be the result of an interplay of ubiquitous as well as tissue-specific factors (Maniatis et al., 1987). Furthermore, the distribution of NRP/B was condensed in peripheral heterochromatin and in the nucleolus, a subcellular organelle known to harbor cyclin B–cdc2 complex, annexin V, angiogenin, and HSP70, proteins involved in cell cycle regulation and cell differentiation.
Our biochemical studies using two known methods for biochemical analysis of nuclear matrix proteins indicate that NRP/B is a nuclear matrix protein (Mirkovitch et al., 1994; Hakes and Berezney, 1991a ,b). It was localized to the nuclei in the subcellular fractions, and it was tightly associated to the insoluble matrix. Although NRP/B contains several motifs that are rich in positively charged amino acids, no classical nuclear localization signal (Sondek et al., 1996) was predicted from the primary sequence. The NRP/B protein might be localized through a nuclear localization signal generated by the interaction of two “half nuclear localization signal” sequences contributed by different parts of the NRP/B protein, or alternatively, NRP/B protein may be cotranslocated by another protein to the nucleus as a complex. Further studies are necessary to establish how the NRP/B protein is targeted into the nucleus and integrated into the nuclear matrix.
Two forms of NRP/B (67 and 57 kD) were detected by immunoprecipitation of total cell lysates of primary neurons, SH-SY5Y cells, and PC-12 cells, while in the neuronal nuclear pellet, only one form of NRP/B (p67) was detected, using specific monoclonal and polyclonal NRP/B antibodies (Fig. 4). Interestingly, the nuclear-mitotic apparatus (NuMA) protein undergoes dynamic changes during the cell cycle, and truncated forms of NuMA appear during apoptosis (Hsu and Yeh, 1996). NuMA behaves solely as a 220-kD nuclear matrix protein during interphase. However, during mitosis NuMA is phosphorylated by Cdc2 kinase into a 240-kD form. The 240-kD form of NuMA either becomes a 180-kD truncated form or a 220-kD form during the metaphase–anaphase. The 220-kD form relocates to the daughter nuclei and remains throughout interphase (Hsu and Yeh, 1996). Future studies will determine the function of both NRP/B forms and whether NRP/B undergoes dynamic changes, similar to that of NuMA.
Functional Studies
Neuronal differentiation involves migration, directional axon growth, synaptogenesis, and selective survival (Kaplan and Stephens, 1994; Diaznido et al., 1996). Neuronal growth factors and cell adhesion molecules stimulate and guide neuronal differentiation. Development of neurons from neuroblasts involves the extension of cytoplasmic processes (neurites) that mature into axons and dendrites in response to distinct extracellular signals (Henderson, 1996). Tyrosine phosphorylation has been implicated in neuronal differentiation, by which neurite outgrowth is regulated (Keegan and Halegoua, 1993). In addition, changes in the cytoskeleton are crucial for neurite outgrowth and maturation (Diaznido et al., 1996). The interaction of neuronal cells with the extracellular matrix and soluble neurotrophic factors regulates the formation and guidance of neurites by directing changes in the microtubules and the actin cytoskeleton (Sanes, 1989) and by triggering the protein tyrosine phosphorylation of key signaling proteins (Sanes, 1989; Bixby and Jhabvala, 1992; Ingber, 1993; Smith, 1994; Williams et al., 1994; Helmke and Pfenninger, 1995). Furthermore, the reorganization of the actin cytoskeleton is necessary for neurite formation (Ingber, 1993) and cell spreading (Smith, 1994), while microtubule polymerization is required for the elongation of neurites (Sanes, 1989; Smith, 1994).
To elucidate the function(s) of NRP/B in neuronal development, NRP/B expression in response to RA or dibutyryl cAMP was studied in neuronal cells. The NRP/B protein was upregulated during neuronal differentiation (Fig. 7). To further demonstrate the possible involvement of NRP/B in neuronal differentiation, two experimental approaches were taken. Overexpression of NRP/B in Neuro 2A neuroblastoma cells was used and was shown to significantly enhance the extension of neuronal processes during neuronal differentiation (Fig. 7, Table I). In the second experimental approach, the antisense and sense strategy was used, which has been used successfully to address the function of several important regulatory genes, including those expressed in neuronal cells (Wagner, 1994, 1995). Antisense oligodeoxynucleotide treatment of dynamin 1, a microtubule-activated GTPase, prevented neurite formation in cultured hippocampal neurons (Torre et al., 1994). NRP/B antisense treatment of rat primary hippocampal neurons resulted in a significant reduction in neuronal process formation (Fig. 8). Similarly, NRP/B antisense treatment inhibited NGF-induced differentiation of PC-12 cells, resulting in the inhibition of neuronal process formation (Fig. 9).
These results suggest that the NRP/B protein plays a role in neuronal differentiation. As suggested in the “Tensegrity” model (Sanes, 1989), nuclear morphology and the cytoskeletal structure are dynamically associated, and if changes occur in the nuclear structure, then the cytoskeletal structure and gene expression may also be altered. The consequences of the inhibition of neuronal process formation by NRP/B antisense treatment and the extension of neuronal processes by NRP/B protein overexpression are in accordance with this model.
Cell cycle control mechanisms can affect the nuclear matrix and its complex constituents at several levels. The p110RB is a nuclear matrix–associated protein that is cell cycle dependent (Loidl and Eberharter, 1995). This protein is distributed widely throughout the matrix (Mancini et al., 1994) and is associated with the major nuclear matrix protein, lamin A (Ozaki et al., 1994). p110RB regulates progression of the cell cycle and is important for cell survival and differentiation (Riley et al., 1994). Mutants or hyperphosphorylated forms of p110RB are less firmly attached to the nuclear structures, while the hypophosphorylated forms of p110RB become tightly associated with them (Mittnacht and Weinberg, 1991). In addition, a novel 84-kD nuclear protein that localizes to subnuclear regions associated with RNA processing was found to bind preferentially to the functionally active hypophosphorylated form of p110RB (Durfee et al., 1994). Neuronal differentiation can be induced by overexpression of p27KIP or p110RB, suggesting that inhibition of cyclin-dependent kinase activity leading to loss of p110RB phosphorylation is a major determinant for neuronal differentiation (Kranenburg et al., 1995). In vivo phosphorylation of NRP/B is regulated during cell cycle progression (Fig. 10). Both in vivo and in vitro experiments indicated that NRP/B is associated with p110RB (Fig. 11) and that the association of p110RB to NRP/B is decreased during neuronal differentiation (Fig. 11). These observations suggest that NRP/B is involved in neuronal differentiation through its interaction with the hypophosphorylated form of p110RB. The kinase that is responsible for NRP/B phosphorylation is not yet identified. These data suggest strongly that NRP/B interacts with p110RB and that NRP/B is involved in neuronal cell differentiation.
Therefore, to our knowledge NRP/B is the first nuclear matrix protein identified to be specifically expressed in primary neurons, to interact with p110RB, and to participate in the regulation of neuronal process formation. The results presented in this study might suggest that NRP/B is involved in cell cycle withdrawal after commitment to differentiation, or alternatively, NRP/B might be involved in the regulation of neuronal cell differentiation by interfering with the function of cell cycle regulatory proteins such as p110RB. Future studies will address these models of NRP/B function in neuronal cells.
Acknowledgments
We thank Dr. Shuxian Jiang for performing the RNA blot experiments, and Dr. Bijia Deng for her help in establishing rat primary hippocampal cultures and performing some of the sense/antisense treatment experiments. We are grateful to Roanna London for the computer analysis of the NRP/B sequences. We would like to thank Janet Delahanty for her help in editing the manuscript and preparing the art work, Evelyn Gould for her assistance with the figures, and Tee Trac for her typing assistance. The authors wish to thank Dr. Larry Benowitz (Children's Hospital, Boston, MA) for his help in analyzing the in situ hybridization results, Boney Meek (Harvard School of Public Health, Boston, MA) for electron microscopy analyses, and Dr. Jerome E. Groopman and Dr. Soner Altiok for their critical reading of this manuscript.
Abbreviations used in this paper
- CDK
cyclin-dependent kinase
- NRP/B
nuclear restricted protein/brain
- NuMA
nuclear-mitotic apparatus
- p110RB
retinoblastoma protein
- pc
post coitum
- RA
retinoic acid
- RCR-1
Ring canal–related protein
Footnotes
This paper is supported in part by National Institutes of Health grants HL55445, HL51456, and HL43510-06A.
This paper is dedicated to Raphael Recanati and his family for their friendship and support for our research program.
Address all correspondence to Dr. Shalom Avraham, Divisions of Experimental Medicine and Hematology/Oncology, Beth Israel Deaconess Medical Center, Harvard Institutes of Medicine, 4 Blackfan Circle, 3rd Floor, Boston, MA 02115. Tel.: (617) 667-0063. Fax: (617) 975-5240.
Sequence data are available from GenBank/EMBL/DDBJ under accession number AF059611.
References
- Albagli O, Dhordain P, Deweindt C, Lecoq G, Deprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Diff. 1995;6:1193–1198. [PubMed] [Google Scholar]
- Amankwash K, De Boni U. Ultrastructural localization of filamentous actin within neuronal interphase nuclei in situ. Exp Cell Res. 1994;210:315–325. doi: 10.1006/excr.1994.1044. [DOI] [PubMed] [Google Scholar]
- Avraham S, London R, Fu Y, Ota S, Hiregowdara D, Li J, Jiang S, Pasztor LM, White RA, Groopman JE, Avraham H. Identification and characterization of a novel related adhesion focal tyrosine kinase (RAFTK) from megakaryocytes and brain. J Biol Chem. 1995a;270:27742–27751. doi: 10.1074/jbc.270.46.27742. [DOI] [PubMed] [Google Scholar]
- Avraham S, Jiang S, Ota S, Fu Y, Deng B, Dowler LL, White RA, Avraham H. Structural and functional studies of the intracellular tyrosine kinase MATK gene and its translated product. J Biol Chem. 1995b;270:1833–1842. doi: 10.1074/jbc.270.4.1833. [DOI] [PubMed] [Google Scholar]
- Berezney, R. 1984. Organization and functions of the nuclear matrix. In Chromosomal Non-histone Proteins—Structural Associations. 4:119–180.
- Berezney R. Visualizing DNA replication sites in the cell nucleus. Semin Cell Biol. 1991;2:103–115. [PubMed] [Google Scholar]
- Berezney R, Coffey D. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60:1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Binder L, Frankfurter A, Kim H, Caceres A, Payne M, Rebhun L. Heterogeneity of microtubule-associated protein 2 during rat brain development. Proc Natl Acad Sci USA. 1984;81:5613–5617. doi: 10.1073/pnas.81.17.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixby JL, Jhabvala P. Inhibition of tyrosine phosphorylation potentiates substrate-induced neurite growth. J Neurobiol. 1992;23:468–480. doi: 10.1002/neu.480230503. [DOI] [PubMed] [Google Scholar]
- Bork P, Doolittle R. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Buttyan R, Olsson C. Prediction of transcriptional activity based on gene association with the nuclear matrix. Biochem Biophys Res Commun. 1986;138:1334–1340. doi: 10.1016/s0006-291x(86)80429-6. [DOI] [PubMed] [Google Scholar]
- Caricasole A, Duarte A, Larsson S, Hastie N, Little M, Holmes G, Todorov I, Ward A. RNA binding by the Wilms tumor suppressor zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:7562–7566. doi: 10.1073/pnas.93.15.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter K, Bowman D, Carrington W, Fogarty K, McNeil J, Fay F, Lawrence J. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Chang-Yeh A, Mold D, Huang R. Identification of a novel murine IAP-promoted placenta-expressed gene. Nucleic Acids Res. 1991;19:3667–3672. doi: 10.1093/nar/19.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zollman S, Couderc Z-L, Laski F. The BTB domain of bric a'brac mediates dimerization in vitro. Mol Cell Biol. 1995;15:3424–3429. doi: 10.1128/mcb.15.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Compton D, Yen T, Cleveland D. Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies generated against human mitotic chromosome scaffolds. J Cell Biol. 1991;112:1083–1097. doi: 10.1083/jcb.112.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D, Szilak I, Cleveland D. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J Cell Biol. 1992;116:1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgaya J, Recio J, Aranda A. Influence of Ras and retinoic acid on nerve growth factor induction of transin gene expression in PC12 cells. Oncogene. 1997;14:1687–1696. doi: 10.1038/sj.onc.1200997. [DOI] [PubMed] [Google Scholar]
- Diaznido J, Ulloa L, Sanchez C, Avila J. The role of the cytoskeleton in the morphological changes occurring during neuronal differentiation [review] Semin Cell Dev Biol. 1996;7:733–739. [Google Scholar]
- Dobashi Y, Kudoh T, Matsumine A, Toyoshima K, Akiyama T. Constitutive overexpression of CDK2 inhibits neuronal differentiation of rat pheochromocytoma PC12 cells. J Biol Chem. 1995;270:23031–23037. doi: 10.1074/jbc.270.39.23031. [DOI] [PubMed] [Google Scholar]
- Durfee T, Mancini M, Jones D, Elledge S, Lee W. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that colocalizes to centers for RNA processing. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisert E, Johnson H, Binder L. Expression of microtubule-associated protein 2 by reactive astrocytes. Proc Natl Acad Sci USA. 1990;87:3967–3971. doi: 10.1073/pnas.87.10.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich D, Wang N, Qian Y, Lee E, Lee W. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill RJ, Jr, Pixley SK. 2-Mercaptoethanol is a survival factor for olfactory, cortical and hippocampal neurons in short-term dissociated cell culture. Brain Res. 1993;613:168–172. doi: 10.1016/0006-8993(93)90470-8. [DOI] [PubMed] [Google Scholar]
- Hakes D, Berezney R. DNA binding properties of the nuclear matrix and individual nuclear matrix proteins. J Biol Chem. 1991a;266:11131–11140. [PubMed] [Google Scholar]
- Hakes D, Berezney R. Molecular cloning of matrin F/G: a DNA binding protein of the nuclear matrix that contains putative zinc-finger motifs. Proc Natl Acad Sci USA. 1991b;88:6186–6190. doi: 10.1073/pnas.88.14.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies. In A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1–726.
- He, D., C. Zeng, and B. Brinkley. 1995. Nuclear matrix proteins as structural and functional components of the mitotic apparatus. Int. Rev. Cytol. 162B: 1–74. [DOI] [PubMed]
- Helmke S, Pfenninger KH. Growth cone enrichment and cytoskeletal association of non-receptor tyrosine kinases. Cell Motil Cytoskel. 1995;30:194–207. doi: 10.1002/cm.970300304. [DOI] [PubMed] [Google Scholar]
- Henderson C. Role of neurotrophic factors in neuronal development. Curr Opin Neurobiol. 1996;6:64–70. doi: 10.1016/s0959-4388(96)80010-9. [DOI] [PubMed] [Google Scholar]
- Hiregowdara D, Avraham H, Fu Y, London R, Avraham S. Tyrosine phosphorylation of the related adhesion focal tyrosine kinase in megakaryocytes upon stem cell factor and phorbol myristate acetate stimulation and its association with paxillin. J Biol Chem. 1997;272:10804–10810. doi: 10.1074/jbc.272.16.10804. [DOI] [PubMed] [Google Scholar]
- Hsu H, Yeh N. Dynamic changes of NuMA during the cell cycle and possible appearance of a truncated form of NuMA during apoptosis. J Cell Sci. 1996;109:277–288. doi: 10.1242/jcs.109.2.277. [DOI] [PubMed] [Google Scholar]
- Ingber D. The riddle of morphogenesis: a question of solution chemistry or molecular cell engineering? . Cell. 1993;75:1249–1252. doi: 10.1016/0092-8674(93)90612-t. [DOI] [PubMed] [Google Scholar]
- Ito N, Phillips S, Stevens C, Ogel Z, McPherson M, Keen J, Yadav K, Knowles D. Novel thioether bond revealed by a 1.7 Å crystal structure of galactose oxidase. Nature. 1991;350:87–90. doi: 10.1038/350087a0. [DOI] [PubMed] [Google Scholar]
- Kaplan D, Stephens R. Neurotrophin signal transduction by the Trk receptor. J Neurobiol. 1994;25:1404–1417. doi: 10.1002/neu.480251108. [DOI] [PubMed] [Google Scholar]
- Keegan K, Halegoua S. Signal transduction pathways in neuronal differentiation. Curr Opin Neurobiol. 1993;3:14–19. doi: 10.1016/0959-4388(93)90029-x. [DOI] [PubMed] [Google Scholar]
- Keynes R, Cook G. Axon guidance molecules. Cell. 1995;83:161–169. doi: 10.1016/0092-8674(95)90157-4. [DOI] [PubMed] [Google Scholar]
- Khan A, Wilcox A, Polymeropoulos M, Hopkins J, Stevens T, Robinson M, Orpana A, Sikela J. Single pass sequencing and physical and genetic mapping of human brain cDNAs. Nat Genet. 1992;2:180–185. doi: 10.1038/ng1192-180. [DOI] [PubMed] [Google Scholar]
- Kranenburg O, Scharnhorst V, Van der Eb A, Zantema A. Inhibition of cyclin-dependent kinase activity triggers neuronal differentiation of mouse neuroblastoma cells. J Cell Biol. 1995;131:227–234. doi: 10.1083/jcb.131.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal P, Feldman E. Tyrosine phosphorylation and enhanced expression of paxillin during neuronal differentiation in vitro. J Biol Chem. 1996;271:5957–5960. doi: 10.1074/jbc.271.11.5957. [DOI] [PubMed] [Google Scholar]
- Li J, Avraham H, Rogers RA, Raja S, Avraham S. Characterization of RAFTK, a novel focal adhesion kinase and its integrin-dependent phosphorylation and activation in megakaryocytes. Blood. 1996;88:417–428. [PubMed] [Google Scholar]
- Loidl, P., and A. Eberharter. 1995. Nuclear matrix and the cell cycle. Int. Rev. Cytol. 162B:377–403. [DOI] [PubMed]
- Mancini MA, Shan B, Nickerson J, Penman S, Lee W. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc Natl Acad Sci USA. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini MA, He D, Ouspenski D, Brinkley B. Dynamic continuity of nuclear and mitotic matrix protein in the cell cycle. J Cell Biochem. 1996;62:158–164. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C158::AID-JCB3%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Goodbourn S, Fischer J. Regulation of inducible and tissue-specific gene expression. Science. 1987;236:1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Milankov K, De Boni U. Cytochemical localization of actin and myosin aggregates in interphase nuclei in situ. Exp Cell Res. 1993;209:189–199. doi: 10.1006/excr.1993.1301. [DOI] [PubMed] [Google Scholar]
- Mirkovitch S, Mirault M-E, Laemmli U. Organization of the high-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1994;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Mittnacht S, Weinberg R. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 1991;65:381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Nuclear matrins: identification of the major nuclear matrix proteins. Proc Natl Acad Sci USA. 1991;88:10312–10316. doi: 10.1073/pnas.88.22.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T, Ward J, Huh C, Longenecker G, Veeranna, Pant H, Brady R, Martin L, Kulkarni A. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci USA. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T, Saijo M, Murakami K, Enomoto H, Taya Y, Saikiyama S. Complex formation between lamin A and the retinoblastoma gene product: identification of the domain on lamin A required for its interaction. Oncogene. 1994;9:2649–2653. [PubMed] [Google Scholar]
- Penman S. Rethinking cell structure. Proc Natl Acad Sci USA. 1995;92:5251–5257. doi: 10.1073/pnas.92.12.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Riley D, Lee E, Lee W. The retinoblastoma protein: more than a tumor suppressor. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- Robinson D, Cant K, Cooley L. Morphogenesis of Drosophila ovarian ring canals. Development (Camb) 1994;120:2015–2025. doi: 10.1242/dev.120.7.2015. [DOI] [PubMed] [Google Scholar]
- Sanes JR. Extracellular matrix molecules that influence neural development. Annu Rev Neurosci. 1989;12:491–516. doi: 10.1146/annurev.ne.12.030189.002423. [DOI] [PubMed] [Google Scholar]
- Schuchard M, Subramanian M, Ruesink T, Spelsberg T. Nuclear matrix localization and specific matrix DNA binding receptor binding factor 1 of the avian oviduct progesterone receptor. Biochemistry. 1991;30:9516–9522. doi: 10.1021/bi00103a019. [DOI] [PubMed] [Google Scholar]
- Skinner PJ, Koshy BT, Cummings CJ, Klement IA, Helin K, Servadio A, Zoghbi HY, Orr HT. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- Smith CL. Cytoskeletal movements and substrate interactions during initiation of neurite outgrowth by sympathetic neurons in vitro. J Neurosci. 1994;14:384–398. doi: 10.1523/JNEUROSCI.14-01-00384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Bohrn A, Lambright D, Hamm H, Sigler P. Crystal structure of a G protein βγ dimer at 2.1 A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Torre E, McNiven M, Urrutia R. Dynamin 1 antisense oligodeoxynucleotide treatment prevents neurite formation in cultured hippocampal neurons. J Biol Chem. 1994;269:82411–82417. [PubMed] [Google Scholar]
- Varkey JP, Muhlrad PJ, Minniti AN, Do B, Ward S. The Caenorhabditis elegans spe-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Gen Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- Vignali G, Niclas J, Sprocati M, Vale R, Sirtori C, Navone F. Differential expression of ubiquitous and neuronal kinesin heavy chains during differentiation of human neuroblastoma and PC12 cells. Eur J Neurosci. 1996;8:536–544. doi: 10.1111/j.1460-9568.1996.tb01238.x. [DOI] [PubMed] [Google Scholar]
- von Bulow M, Heid H, Hess H, Franke WW. Molecular nature of calicin, a major basic protein of the mammalian sperm head cytoskeleton. Exp Cell Res. 1995;219:407–413. doi: 10.1006/excr.1995.1246. [DOI] [PubMed] [Google Scholar]
- Wagner R. Gene inhibition using antisense oligodeoxynucleotide. Nature. 1994;392:333–335. doi: 10.1038/372333a0. [DOI] [PubMed] [Google Scholar]
- Wagner R. The status of the art in antisense research. Nat Med. 1995;11:1116–1118. doi: 10.1038/nm1195-1116. [DOI] [PubMed] [Google Scholar]
- Wall M, Coleman D, Lee E, Iniguez-Lluhi J, Posner B, Gilman A, Sprang S. The structure of the G protein heterotrimer G αβγ. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Way M, Sanders M, Garcia C, Sakai J, Matsudaira P. Sequence and domain organization of scruin, an actin-cross-linking protein in the acrosomal process of Limulus sperm. J Cell Biol. 1995;128:51–60. doi: 10.1083/jcb.128.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EJ, Walsh FS, Doherty P. Tyrosine kinase inhibitors can differentially inhibit integrin-dependent and CAM-stimulated neurite outgrowth. J Cell Biol. 1994;124:1029–1037. doi: 10.1083/jcb.124.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Johnson C, Dobner P, Lawrence J. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xue F, Cooley L. Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- Yang C, Lambie E, Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J Cell Biol. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]