Abstract
We previously described a kinesin-dependent movement of particles in the flagella of Chlamydomonas reinhardtii called intraflagellar transport (IFT) (Kozminski, K.G., K.A. Johnson, P. Forscher, and J.L. Rosenbaum. 1993. Proc. Natl. Acad. Sci. USA. 90:5519–5523). When IFT is inhibited by inactivation of a kinesin, FLA10, in the temperature-sensitive mutant, fla10, existing flagella resorb and new flagella cannot be assembled. We report here that: (a) the IFT-associated FLA10 protein is a subunit of a heterotrimeric kinesin; (b) IFT particles are composed of 15 polypeptides comprising two large complexes; (c) the FLA10 kinesin-II and IFT particle polypeptides, in addition to being found in flagella, are highly concentrated around the flagellar basal bodies; and, (d) mutations affecting homologs of two of the IFT particle polypeptides in Caenorhabditis elegans result in defects in the sensory cilia located on the dendritic processes of sensory neurons. In the accompanying report by Pazour, G.J., C.G. Wilkerson, and G.B. Witman (1998. J. Cell Biol. 141:979–992), a Chlamydomonas mutant (fla14) is described in which only the retrograde transport of IFT particles is disrupted, resulting in assembly-defective flagella filled with an excess of IFT particles. This microtubule- dependent transport process, IFT, defined by mutants in both the anterograde (fla10) and retrograde (fla14) transport of isolable particles, is probably essential for the maintenance and assembly of all eukaryotic motile flagella and nonmotile sensory cilia.
Assembly and maintenance of the eukaryotic flagellar axoneme presents some unique problems for the cell (Lefebvre and Rosenbaum, 1986; Johnson and Rosenbaum, 1993; Johnson, 1995). The organelle is composed of well over 200 different polypeptides (Piperno et al., 1977; Dutcher, 1995) that, after their synthesis in the cytoplasm (Lefebvre et al., 1978), must rapidly find their way to the distal tip of the flagellum where most of the assembly of the microtubular axoneme takes place (Rosenbaum et al., 1969; Witman, 1975; Johnson and Rosenbaum, 1992). Delivery of flagellar precursors to the base of the flagellum may be facilitated by the localization of mRNAs for these proteins near the basal bodies, as seen with tubulin mRNA in Naegleria gruberi (Han et al., 1997). Some of the axonemal polypeptides synthesized from these mRNAs are assembled in the cytoplasm into larger complexes, e.g., radial spokes (Diener, D.R., D.G. Cole, and J.L. Rosenbaum. 1996. Mol. Biol. Cell. 7:47a) and dynein arms (Fok et al., 1994; Mitchell, D.R., personal communication), which are then targeted to the flagellum. How these proteins are then transported to the flagellar tip during axonemal assembly has been one of our principal research concerns.
The use of fla10-1 (Huang et al., 1977; Adams et al., 1982), a mutant strain of the biflagellate alga Chlamydomonas that carries a temperature-sensitive (ts1) point mutation in the gene for a kinesin known as KHP1 (Walther et al., 1994; Vashishtha et al., 1996) or FLA10, has been pivotal to the study of transport of flagellar precursors. FLA10 is essential for flagellar assembly and maintenance; after shifting to the restrictive temperature, fla10 mutants are unable to assemble new flagella (Huang et al., 1977; Adams et al., 1982), and will resorb flagella assembled at the permissive temperature (Lux and Dutcher, 1991). Using a technique of in vivo complementation called dikaryon rescue, in which the cytoplasm of one cell supplies flagellar components to defective flagella of a second cell to which it is mated, FLA10 activity was shown to be necessary for the assembly of inner dynein arms at the flagellar tip (Piperno et al., 1996). The role of FLA10 in flagellar assembly and maintenance may, therefore, involve movement of precursors to the flagellar tip.
The mechanism by which FLA10 could transport axonemal components to the flagellar tip may involve a kinesin-driven motility called intraflagellar transport (IFT). IFT is observed by video-enhanced differential-interference contrast (DIC) microscopy as the movement of particles underneath the flagellar membrane distally at 2 μm/s and proximally at nearly 4 μm/s (Kozminski et al., 1993). The particles being moved by IFT have been shown by electron microscopy to consist of a varying number of electron opaque subunits (hereafter referred to as IFT particles) assembled into linear arrays called “rafts” (originally observed by Ringo, 1967), which appear to be linked to both the B tubules of the outer doublets and the overlying flagellar membrane (Kozminski et al., 1993, 1995). In fla10 cells incubated at restrictive temperatures, IFT gradually disappears just before the onset of flagellar resorption, concomitant with the reduction of rafts identified by electron microscopy (Kozminski et al., 1995). Together, these data suggest that anterograde (base to tip) IFT, powered by FLA10, delivers precursors to the flagellar tip for assembly.
Here we describe (a) the isolation, characterization, and subcellular localization of FLA10 and present further evidence that this kinesin is responsible for anterograde IFT, and (b) the isolation, initial characterization, and cellular localization of polypeptides that compose the IFT particles. Peptide sequence analysis shows that of the IFT particle polypeptides that have been analyzed so far, two are homologous to proteins that have been recently characterized in Caenorhabditis elegans. In keeping with the proposed role of IFT in Chlamydomonas flagellar assembly and maintenance, C. elegans strains with mutations affecting these proteins (Collet et al., 1998; Stone, S., and J. Shaw, personal communication) have both structural and functional defects in the sensory cilia of the sensory neurons (Perkins et al., 1986). In related work, Pazour et al. (1998) describe a new Chlamydomonas mutant in which retrograde IFT is absent, resulting in the accumulation of IFT particles in the flagella. Therefore, IFT in Chlamydomonas is one of the only microtubule-based motilities in which mutants are available in both the anterograde and retrograde motors responsible for transport of discrete isolable particles. Furthermore, this motility is essential for the assembly, maintenance, and function of motile cilia/flagella and nonmotile sensory cilia.
Materials and Methods
Cultures
Chlamydomonas reinhardtii wild-type strain cc125, cell wall–deficient strain cw92 (cc503), basal body–deficient strain bld2 (cc478), and ts flagellar assembly mutants fla1 (cc1389), fla2 (cc1390), fla3 (cc1391), fla8 (cc1396), fla9 (cc1918), fla10 (fla10-1 allele, cc1919), and fla13 (cc1922) were obtained from the Chlamydomonas Genetics Center (Duke University, Durham, NC). fla1, fla2, fla3, and fla10-1, were originally designated dd-a-6, dd-frag-1, dd-a-13, and dd-a-224, respectively, by Huang et al. (1977) and renamed by Adams et al. (1982). fla8, fla9, and fla13 have been previously described (Adams et al., 1982). Previous studies with fla mutants used 32°C as the restrictive temperature; in the studies presented here, fla mutants were incubated at a restrictive temperature of 33°C. Phenotypes were verified by monitoring flagellar loss after fla mutants with full-length flagella were shifted from the permissive temperature of 23°C to the restrictive temperature of 33°C. Cells were grown in liquid minimal medium, MI, (Sager and Granick, 1953) at 22°–23°C with a light/dark cycle of 14 h/10 h with constant aeration.
Flagellar Isolation
Flagella were isolated from Chlamydomonas by pH shock as described by Witman et al. (1972) with minor modifications as follows. Cells were harvested from 8 to 32 liters of liquid medium with a Pellicon tangential flow filtration device (Millipore, Bedford, MA) and/or low speed centrifugation of 400 g for 3 min. Cells were resuspended in fresh MI at ∼107 cells/ml and vigorously aerated under light for 1–4 h at 23°C. Restrictive temperature experiments were carried out under lights in a 33°C water bath with constant aeration of the cells. Cells were sedimented at 400 g for 3 min and then resuspended in 5% sucrose in 10 mM Hepes, pH 7.2. Immediately after placing the cells in an ice bath, the flagella were shed by reducing the pH to 4.6 with 0.5 M acetic acid. Deflagellation was confirmed using phase microscopy after 60 s, and the cells were neutralized with 0.5 M KOH before 2 min elapsed. EGTA was added to a final concentration of 0.1 mM and the following protease inhibitors were added to a final concentration of 1.0 mM PMSF, 50 μg/ml soybean trypsin inhibitor, 1 μg/ml pepstatin A, 2 μg/ml aprotinin, and 1 μg/ml leupeptin. All following steps were carried out at 4°C. Cell bodies were removed by centrifugation through 75-ml cushions of 25% sucrose in 10 mM Hepes, pH 7.2, in 250-ml conical bottles at 600 g for 5 min. The remaining cell bodies were removed from the top layer by centrifugation through 8-ml cushions of 25% sucrose in 10 mM Hepes, pH 7.2, in 50-ml conical tubes at 600 g for 10 min. The flagella were collected using an SS34 rotor (Sorvall Products, Newtown, CT) at 10,000 rpm for 20 min. The resulting pellet was thoroughly resuspended in 2–5 ml 5% sucrose in 10 mM Hepes, pH 7.2, with protease inhibitors, and then centrifuged through 2–4 ml 15% sucrose in 5 mM Hepes, pH 7.2, plus half-strength protease inhibitors in 10-ml conical tubes in an HB-4 rotor at 8,000 rpm for 20 min. This additional step usually produced a stratified pellet consisting of three layers. The bottom layer was small, tight, dark green, and contained primarily cell bodies. The middle layer was white and consisted primarily of flagella. The top layer was light green, flocculent, easily removed, and contained flagella and unidentified amorphous material. Only the white layer of flagella was recovered and resuspended in a minimal volume (0.1–1.0 ml) of HMDEK buffer (10 mM Hepes, pH 7.2, 5 mM MgSO4, 1 mM DTT, 0.5 M EDTA, and 25 mM KCl) containing protease inhibitors as described above. Flagella were extracted with NP-40 or flash frozen in liquid N2 within 20 min of final resuspension.
Isolation of FLA10 Kinesin-II
The purification of FLA10 kinesin-II was monitored using affinity-purified anti-FLA10T, a polyclonal antibody raised against the COOH-terminal portion (amino acids 416–688 containing the tail domain and most of the stalk domain) of the FLA10 gene product (Kozminski et al., 1995). The minimal concentration of NP-40 (Pierce Chemical Co., Rockford, IL) required to release FLA10 kinesin-II from flagella was found to be 0.02% (data not shown). Use of excess NP-40 (1–2% final concentration) frequently resulted in a 50% lower yield of extracted FLA10 kinesin-II, possibly due to irreversible aggregation. Therefore, to ensure maximum solubilization of FLA10 kinesin-II, extractions were routinely performed at 0.05% NP-40. Flagella were initially extracted with 0.05% NP-40 and 2 mM 5′-adenylylimidodiphosphate (AMPPNP) (Sigma Chemical Co., St. Louis, MO) in HMDEK buffer for 10 min before centrifugation in a microfuge at 12,000 rpm for 10 min. The supernatant, “membrane plus matrix,” was removed and saved for later analysis. The axonemal pellet was washed in 0.5 mM AMPPNP in HMDEK buffer and centrifuged again at 12,000 rpm for 10 min. The axonemal pellet was then extracted with 10 mM MgATP for 15 min on ice before centrifugation at 12,000 rpm for 10 min. The supernatant, termed the ATP eluate, was further clarified for 10 min in an Airfuge A-100/18 rotor (Beckman Instruments, Fullerton, CA) at 25 psi. The ATP eluate was then fractionated by gel filtration using a 1 × 90 cm S400 (Pharmacia Biotech Sevrage, Uppsala, Sweden) column equilibrated with HMDEK plus protease inhibitors. The FLA10 kinesin-II peak fractions were pooled, concentrated to 200 μl using a Centricon-10 (Amicon Corp., Beverly, MA), and further fractionated by sedimentation through 11 ml 5–20% sucrose density gradients in a SWTi41 rotor (Beckman Instruments) for 12 h at 41,000 rpm.
Preparation of IFT Complexes A and B
Freshly prepared flagella were extracted with 0.05% NP-40 and 10 mM MgATP in either HMDEK buffer or 10 mM Hepes, pH 7.2. Alternatively, fresh flagella in HMDEK buffer or 10 mM Hepes, pH 7.2, were flash frozen in liquid N2 and thawed to effect disruption of the flagellar membrane as described by Zhang and Snell (1995). The soluble proteins from freeze/ thaw preparations are believed to exclude both membrane and axonemal proteins and are referred to as the “matrix.” Detergent extractions and freeze/thaw extractions produced similar yields of complexes A and B. Axonemes and other insoluble matter were removed with a 10-min centrifugation in an Airfuge A-100/18 rotor at 25 psi. The supernatants, membrane plus matrix (with NP-40) or matrix (freeze/thaw) were fractionated through 11 ml 5–20% or 10–25% sucrose density gradients in a SWTi41 rotor for 12 h at 38,000 rpm. Gradients were typically fractionated into either 28 0.4-ml aliquots or 18 0.6-ml aliquots, and subsequently analyzed by SDS-PAGE and Western blotting.
Determination of Molecular Mass
Sedimentation coefficients were determined on both 5–20% and 10–25% sucrose density gradients with varying buffers, ionic strength, and pH. Sedimentation standards were added to the samples before centrifugation and included thyroglobulin (19.4 S), catalase (11.3 S), BSA (4.4 S), ovalbumin (3.66 S), and cytochrome c (2.1 S). Diffusion coefficients were determined on either an open column of S400 (1.0 × 90 cm) run by gravity or a prepacked S300 column (1.6 × 60 cm) using an FPLC system (Pharmacia Biotech Sevrage). The standards used were fibrinogen, 1.98 × 10−7 cm2/s; thyroglobulin, 2.6 × 10−7 cm2/s; catalase, 4.1 × 10−7 cm2/s; alcohol dehydrogenase, 4.76 × 10−7 cm2/s; and BSA, 6.3 × 10−7 cm2/s. The void volumes were determined with blue dextran and linear plasmid for the S400 and S300, respectively. The bed volume was determined with ATP. The apparent molecular mass of FLA10 kinesin-II and IFT complexes A and B were determined with the Svedberg equation,
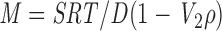 |
(Cantor and Schimmel, 1980), where R is the gas constant, T is temperature in Kelvin, V 2 is the partial specific volume of the protein, and ρ is the solution density, and S and D are the experimentally derived sedimentation and diffusion coefficients, respectively. The partial specific volume was assumed to be 0.72 cm3/g, and the solution density was assumed to be 1.00 g/cm3.
Electrophoresis
Isoelectric focusing was performed according to O'Farrell (1975) using a 1:4 mixture of 3–10 and 4–6 ampholines (Serva, Westbury, NY). Gels were focused for 8,000 V h followed by l h at 800 V. Second dimension SDS-PAGE was carried out on 5–20% acrylamide gradient gels (Laemmli, 1970).
Antibodies
The outer arm dynein intermediate chain mAb, 1869A (King et al., 1985), as purchased from Sigma Chemical Co. The pan-kinesin peptide antibodies, LAGSE and HIPYR (Cole et al., 1992; Sawin et al., 1992), were purchased from BABCO, Inc. (Berkeley, CA). Antibodies against the tail domain of FLA10 (anti-FLA10T, Kozminski et al., 1995) and RSP3 (Williams et al., 1989) were affinity purified (Olmsted, 1981).
Polyclonal anti-FLA10N antiserum was raised against the FLA10 neck domain as follows. A 390-bp SalI fragment encoding the putative neck domain and a portion of the stalk domain of FLA10, Ser-342 to Arg-472, was cloned into the SalI site of PGEX-5X2 expression vector (Pharmacia Biotech Sevrage) to produce a FLA10N-GST fusion protein. The fusion protein, expressed in XL1-Blue cells induced with 3 mM isopropylthio-β-d-galactoside (IPTG), was purified using glutathione–agarose resin (Frangioni and Neel, 1993). Two rabbits were injected subcutaneously with 500 μg FLA10N-GST fusion protein and boosted with 250 μg after 21 d and every 28 d thereafter. Antibody was affinity purified from serum taken at 10 d after the third and fourth boosts. All treatment and care of the rabbits was performed by Yale Biotechnology Services (New Haven, CT).
The anti–FLA10N-GST antiserum was affinity purified using a FLA10-His6 fusion protein that included most of the motor domain and all of the 130–amino acid neck/stalk fragment used for antigen, termed FLA10M/ N-His6. 1,122 bases of FLA10 encoding Met-97 to Arg-472 were cloned into the PQE-30 expression vector (Qiagen, Chatsworth, CA). The FLA10M/N-His6 fusion protein was expressed as described above and purified on Ni-NTA resin (Qiagen) according to the manufacturer's instructions. Purified FLA10M/N-His6 was dialyzed overnight against bicarbonate coupling buffer and coupled to cyanogen bromide–activated Sepharose 4B beads (Pharmacia Biotech Sevrage) as recommended by the manufacturer. Anti–FLA10N-GST antisera (20 ml) was incubated with 4 ml resin for 2 h at 25°C, washed twice with TBS, and then 1.0 M NaCl in TBS at 25°C. Affinity-purified anti-FLA10N antibody was eluted with 0.1 M glycine, pH 2.8, neutralized in 0.1 M Tris, pH 8.5, and then dialyzed overnight against 1× TBS.
mAbs were raised against select IFT particle polypeptides as follows. Sucrose density gradient–purified IFT complexes A and B (15–16S fractions) were injected subcutaneously into balbc mice (50 μg total protein per mouse) on days 0, 21, and 35. On day 49, one of the mice was given an intravenous injection of 40 μg total protein. On day 52, the mouse was killed and the cells from the spleen were fused with an sp2/0 myeloma cell line and plated out on 10 96-well microtiter plates. Between 7 and 14 d later, ∼300 hybridoma supernatants were tested against curtain blots of sucrose density gradient–purified IFT complexes A and B using a Miniblotter28 (Immunetics, Cambridge, MA). Seven positive clones produced antibodies named after the IFT particle polypeptide they recognize: 172.1, 139.1, 81.1, 81.2, 81.3, 81.4, and 57.1. In order, their isotypes as determined with the Sigma Immunotype Kit are IgG1, IgG2a, IgG1, could not be determined, IgG1, IgG1, and IgG2a. Immunofluorescence with these antibodies used tissue culture supernatants, whereas immunoprecipitation was carried out with antibodies purified from ascites (Yale Immunological Services; 172.1 ascites were raised by Maine Biotechnology Services, Portland, ME).
Indirect Immunofluorescence Microscopy
Two different protocols were used for immunofluorescence and both produced similar results. The first protocol was based on a procedure by Huang et al. (1988) in which 3% paraformaldehyde was used to fix cell wall–less mutants (cw92) in suspension before attachment to coverslips and labeling. cw92 cells from logarithmic phase liquid cultures were harvested by gentle centrifugation at 84 g for 6 min. The cells were resuspended in 3% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, PA) in 0.1 M KHPO4, pH 7.4, and then fixed for 30 min at room temperature. Fixed cells were washed three times in PBS by sedimenting 1.5-ml aliquots of cells in a microfuge at 4,000 rpm for 2 min. Washed cells were resuspended in 0.5 ml H2O and adsorbed onto No. 1 glass coverslips by thoroughly coating one side with the cell suspension and immediately removing any excess liquid. The cells were allowed to air dry on the coverslips, which were subsequently placed into Coplin jars for the following extractions. Cells were permeabilized in 0.5% NP-40 in PBS for 2 min and then washed several times with H2O. Chlorophyll was extracted with three changes of ice-cold acetone for 5 min each. After a brief rinse in distilled H2O, excess liquid was wicked away and the coverslips were placed cell side up in a humidity chamber. 150–250 μl primary antibody diluted in blocking buffer A (10 mM K2HPO4, pH 7.2, 5% normal goat serum [Jackson Immunoresearch, West Grove, PA], 5% glycerol, and 1% cold water fish gelatin [Sigma Chemical Co.]) was incubated on each coverslip at 37°C for 2 h. The coverslips were washed three times with PBS in Coplin jars for 5 min each and returned to the humidity chambers. 150–250 μl aliquots of secondary antibody in blocking buffer A were incubated on the coverslips at 37°C for 1 h. After secondary antibody incubation, the coverslips were washed three times with PBS as described above, rinsed briefly with distilled H2O, and then mounted onto slides in a medium containing 2% N-propyl gallate (Sigma Chemical Co.), 30% 0.1 M Tris, pH 9, and 70% glycerol.
The alternate protocol, used for staining wild-type (cc125), bld2, and fla10 mutants, was adapted from Sanders and Salisbury (1995). This procedure allowed simultaneous fixation and extraction of cells that possessed cell walls. Coverslips were coated with 1% poly-l-lysine (Sigma Chemical Co.) for 5 min, rinsed with distilled H2O, and then air dried for 30–60 min. Cells were placed on the coverslips for 5–10 min, rinsed briefly in fresh MI media, extracted in −20°C methanol in Coplin jars for 10 min, and then air dried for 10–15 min. The following steps were performed in humidity chambers at room temperature. The dried cells were rehydrated with PBS for 10 min and then treated for 30 min with 5× blocking buffer B containing 5% BSA, 1% cold water fish gelatin in PBS. The cells were incubated for another 30 min with 10% normal goat serum in 5× blocking buffer B in PBS. The cells were incubated with primary antibody in 1× blocking buffer B for 6–12 h, and then washed with six changes of 1× blocking buffer B before a 60-min incubation with secondary antibody in 1× blocking buffer B. The coverslips were washed six times with 1× blocking buffer B before mounting on slides with ProLong antifade reagent (Molecular Probes, Eugene, OR).
Stained cells were viewed on a Nikon Diaphot 300 inverted microscope. Images were collected with an Image Point CCD camera (Photometrics, Tucson, AZ) and Metamorph Imaging System software (Universal Imaging Corp., West Chester, PA) running on Windows 95 (Microsoft, Redmond, WA). Images were prepared for final publication using Photoshop (Adobe Microsystems, Mountain View, CA).
Electron Microscopy
Wild-type cells were fixed and embedded as described in Kozminski et al. (1995). Sections were cut with a diamond knife and quenched in blocking buffer C (2% BSA, 0.1% cold water fish gelatin, 0.05% Tween-20 in PBS, pH 7.2) for 30 min before incubation in mAb 172.1 tissue culture supernatant diluted 1:1 with blocking buffer C at 4°C, overnight. Grids were washed three times each in wash buffer (PBS plus 0.05% Tween-20) for 5 min each, incubated in 12-nm colloidal gold conjugated to goat anti–rabbit IgG (Jackson Immunologicals, West Grove, PA) for 60 min at room temperature, rinsed in wash buffer (three times for 5 min), PBS (5 min), water (twice for 5 min), and then stained with 2% aqueous uranyl acetate for 10 min before rinsing in water (three times for 5 min). Sections were viewed on a transmission electron microscope (CM10; Philips Electron Optics, Mahwah, NJ).
Results
Purification of Flagellar FLA10 Kinesin-II
IFT is the bidirectional movement of particles beneath the flagellar membrane along the outer doublet microtubules of the axoneme (Kozminski et al., 1993, 1995). The IFT particles, which are not bounded by membrane, exist in linear arrays or rafts of varying lengths that appear to be bridged to the B tubules of the outer doublets (Kozminski et al., 1993, 1995). The movement of the rafts is dependent on the kinesin-like protein FLA10: inactivation of this motor in the temperature-sensitive mutant fla10 results in cessation of IFT (Kozminski et al., 1995). FLA10 has also been localized by immunoelectron microscopy to the compartment where IFT occurs—between the outer doublets and the flagellar membrane (Kozminski et al., 1995).
In light of the role of FLA10 in IFT, this flagellar kinesin was isolated, not only to characterize the native motor complex, but also to identify polypeptides bound to the motor complex that might be components of the IFT particles/rafts. FLA10 was purified from flagella by use of standard kinesin purification procedures (see Materials and Methods): isolated flagella were first treated with detergent in the presence of AMPPNP to remove the flagellar membrane plus matrix while leaving the kinesin bound to the axonemal microtubules. The kinesin was then eluted from the axonemes with ATP and was further purified by gel filtration (Fig. 1) and sucrose gradient centrifugation (Fig. 2 A).
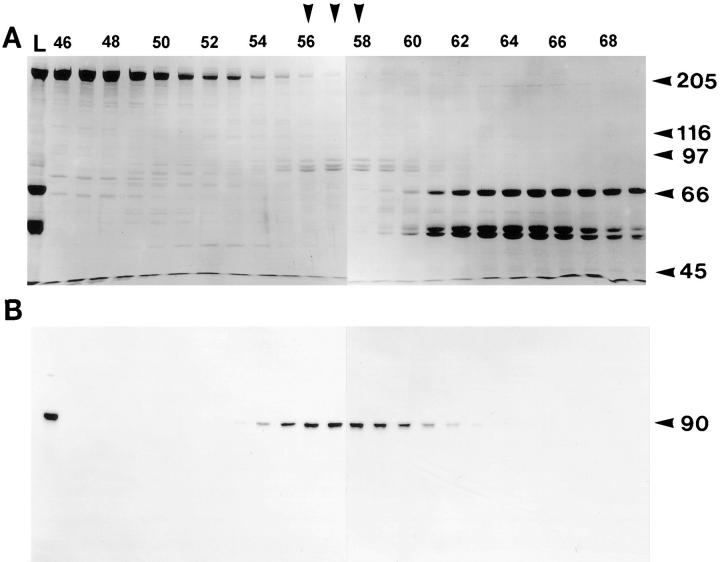
Figure 1.
Gel filtration profile of axonemal ATP eluate. Axonemes from wild-type cells were isolated in the presence of AMPPNP and then extracted with ATP. The resulting ATP eluate, shown in the first lane (L) was separated on an S400 sizing column. (A) Coomassie-stained gel (7.5%) of fractions 46–69 (left to right) shows the majority of eluted protein. The vertical arrowheads over lanes 56–58 indicate the peak fractions of three comigrating bands of M r 100, 90, and 85 kD. Molecular weight markers are indicated on the right. (B) Corresponding immunoblot probed with anti-FLA10T reveals that the 90-kD FLA10 gene product coelutes with the three comigrating bands, primarily between fractions 55 and 60.
Figure 2.
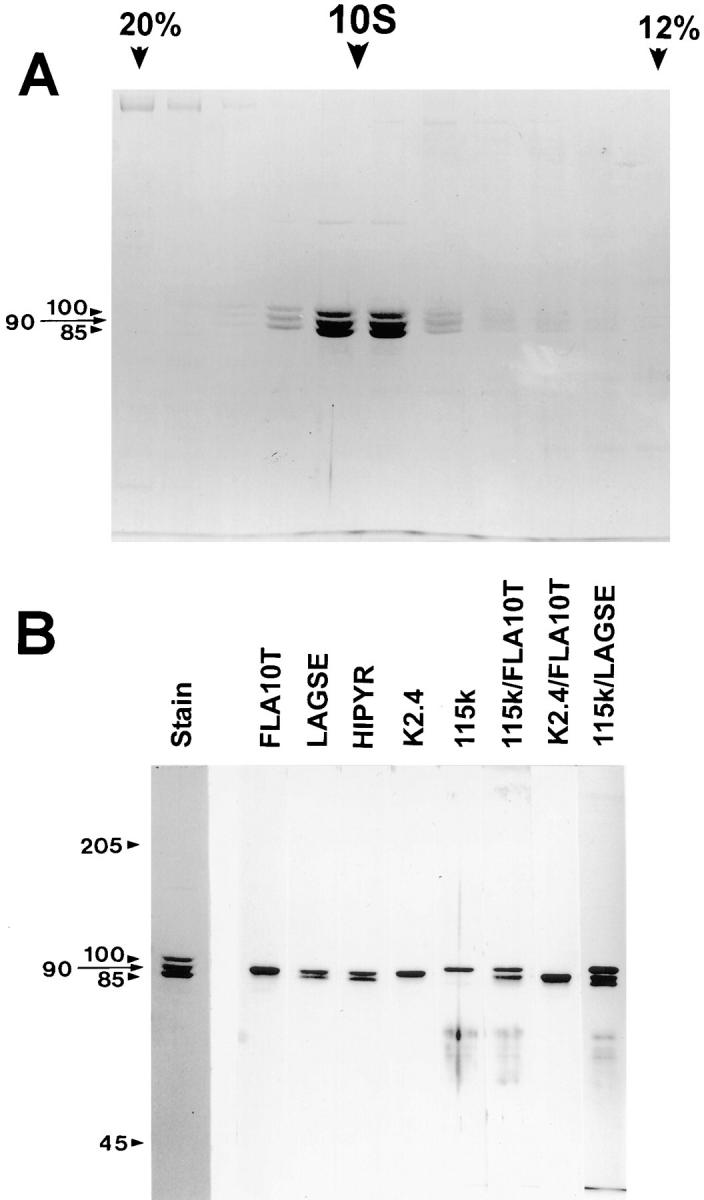
Sucrose density gradient profile of FLA10 kinesin-II and subsequent antibody analysis. The peak FLA10-containing fractions from S400 chromatography were pooled, concentrated, and further fractionated on a 5-ml, 5–20% sucrose gradient. (A) Coomassie-stained gel shows cosedimentation at 9.7 S of three polypeptides at 100, 90, and 85 kD with molar ratios of 0.8, 1.0, and 1.0, respectively. Only the portion of the gradient from 20% (left) to 12% (right) sucrose is shown. (B) Analysis of FLA10 kinesin-II subunits with antibodies. The left panel is a Coomassie-stained gel of sucrose density gradient–purified FLA10 kinesin-II. The right panel contains separate strips from corresponding immunoblots that have been probed with the antibodies listed above each strip. Both of the pan-kinesin peptide antibodies, LAGSE and HIPYR, reacted with both the 85- and 90-kD polypeptides, indicating a high probability that both subunits are kinesin-like proteins. Anti-FLA10T and K2.4, an mAb raised against the 85-kD subunit of sea urchin kinesin-II, reacted only with the 90-kD subunit (FLA10). The polyclonal anti-115k antisera, raised against the 115-kD nonmotor subunit of sea urchin kinesin-II, reacted only with the 100-kD subunit. The identities of the reactive bands were confirmed in lanes probed with mixtures of the antibodies. The mixture of K2.4 and anti-FLA10T reacted only with the 90-kD band, verifying that a monoclonal raised against the smaller of the two kinesin-like subunits (85 kD) of sea urchin kinesin-II recognizes the larger of the two kinesin-like subunits (90 kD) of Chlamydomonas kinesin-II.
The purified kinesin complex, similar to other previously described heterotrimeric kinesins (Cole et al., 1992, 1993; Kondo et al., 1994; Yamazaki et al., 1995, 1996; Wedaman et al., 1996), was composed of three polypeptides with molecular weights of 100, 90, and 85 kD (Fig. 2 A) in a molar ratio of 0.8:1:1. From the hydrodynamic properties of the protein measured by gel filtration (D = 3.1 × 10−7 cm2/s; Fig. 1; Table I) and sucrose density gradient centrifugation (S-value = 9.7 S; Fig. 2 A; Table I) the molecular mass of the complex was calculated to be 270 kD, in accord with the sum of the mass of the three subunits (275 kD). Using several anti-kinesin antibodies, including pan-kinesin antibodies, an anti-FLA10 antibody, and antibodies to motor and nonmotor subunits of sea urchin kinesin-II, the purified complex was determined to be a heterotrimeric kinesin with the 90-kD subunit being the FLA10 gene product (Fig. 2 B). We have named this Chlamydomonas heterotrimeric complex FLA10 kinesin-II in keeping with the nomenclature established for the sea urchin heterotrimeric kinesin-II (Scholey, 1996; Wedaman et al., 1996). The term FLA10, by itself, refers to the 90-kD subunit of FLA10 kinesin-II.
Table I.
Hydrodynamic Behavior and Predicted Stoichiometry of Chlamydomonas FLA10 Kinesin-II Complex
| Sedimentation | 9.7 S | |
| Diffusion coefficient | 3.1 × 10−7 cm2s−1 | |
| Molecular mass | 270 kD | |
| Stoichiometry | ||
| 1 × 100 kD | ||
| (FLA10 Subunit) | 1 × 90 kD | |
| 1 × 85 kD |
Regardless of the methods used to isolate the FLA10 kinesin-II from flagella (see Materials and Methods), it was always released as a 270-kD complex containing only three constituent polypeptides. Therefore, although IFT is dependent on FLA10 activity, the IFT particles did not copurify with the FLA10 kinesin-II heterotrimer under the conditions used.
Identification of Components of IFT Particles
Because the IFT particles did not appear to be bound to FLA10 kinesin-II, we took advantage of the observation that IFT particles are depleted from the flagella of the ts mutant fla10 to identify the components of the particles. When pf1 fla10 double mutants(the pf1 fla10 double mutant was used because it has paralyzed flagella, which greatly facilitates visualization of IFT using DIC microscopy) are incubated at the restrictive temperature for 1.5 h, IFT disappears, and very few rafts can be seen by electron microscopy (Kozminski et al., 1995). We therefore looked for proteins that disappeared from the flagella of fla10 cells at 33°C coincident with the loss of IFT particles.
Wild-type and fla10 cells were incubated at either 23° or 33°C, and their flagella were analyzed by SDS-PAGE. A set of prominent polypeptides was reduced in the flagella of fla10 cells following incubation at the restrictive temperature of 33°C. Although these changes could be observed on SDS-PAGE gels of intact flagella (data not shown), they were more obvious in membrane plus matrix extracts of these flagella (Fig. 3 A). The loss of a specific set of flagellar polypeptides concomitant with the reduction of flagellar rafts suggested that these polypeptides were components of the IFT particles.
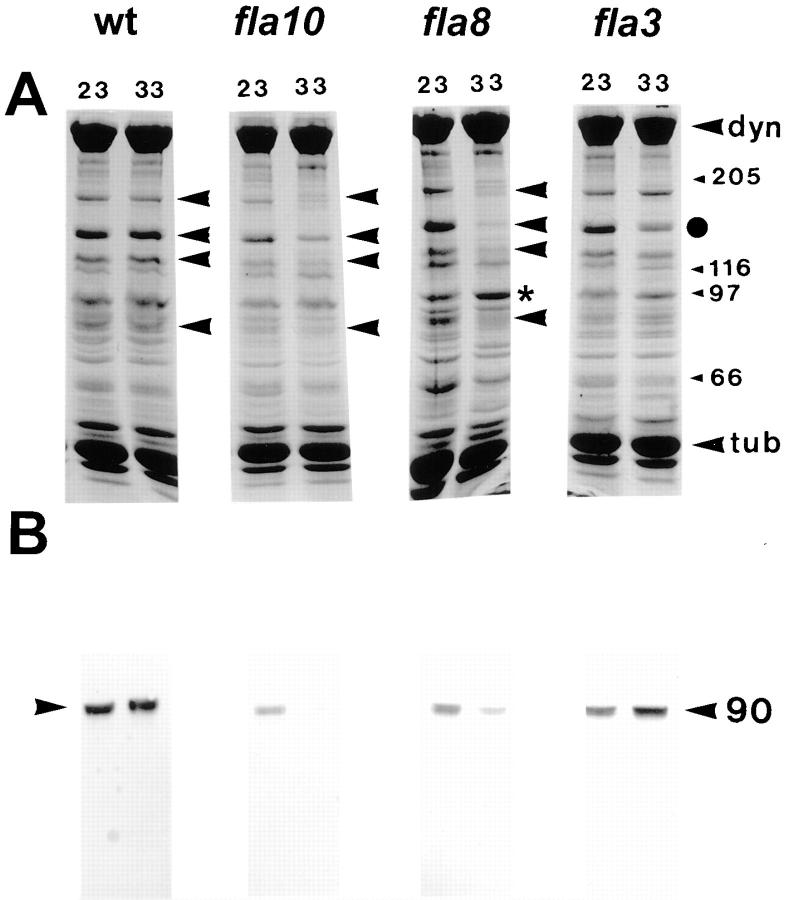
Figure 3.
Polypeptide analysis of flagellar extracts from wild-type and fla mutant cells. Flagella were isolated from cells incubated at either 23°C or the restrictive temperature of 33°C. Membrane plus matrix was isolated in the presence of 0.05% NP-40 and 10 mM ATP to optimize extraction of FLA10 kinesin-II. (A) Coomassie-stained 7.5% gels of membrane plus matrix. Arrowheads, prominent bands that are reduced after incubation of the fla10 and fla8 mutants at 33°C. • on the far right indicates a band, M r 140 kD, that is reduced in fla3 after heating, indicating that only a subset of the polypeptides lost in fla8 and fla10 appear to be reduced in the flagella of fla3 incubated at 33°C. An asterisk indicates a band at ∼100 kD that increases in fla8 after incubation at 33°C. Molecular weight standards, dynein, and tubulin are indicated on the right. (B) Corresponding immunoblots probed with anti-FLA10T. The FLA10 protein is significantly depleted in the flagellar extracts of the fla10 and fla8 mutants after incubation at 33°C.
Other ts fla mutants that, like fla10, resorb their flagella at 33°C, were likewise analyzed for changes in flagellar proteins at the restrictive temperature. The putative IFT particle polypeptides that were reduced in the flagella of fla10 cells at 33°C were also significantly reduced in the flagella of two additional ts fla mutants, fla1 and fla8, after incubation at 33°C (Fig. 3 A; Table II). In contrast, this same set of polypeptides was not similarly depleted in the flagella of the ts fla mutants fla2, 3, 4, 9, and 13 (Fig. 3 A; Table II) after incubation at 33°C. Indeed, in fla3 only a subset of the IFT particle polypeptides were reduced at 33°C (Fig. 3 A) and in fla2 the levels of all of the IFT particle polypeptides increased at 33°C (Table II).
Table II.
Flagellar Levels of FLA10 and IFT Particle Polypeptides in fla Mutants
| Cell line | Flagellar resorption at 33°C | FLA10 epitope | IFT polypeptides | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 23°C | 33°C | 23°C | 33°C | |||||||
| wild type | No loss | ++ | ++ | ++ | ++ | |||||
| fla1 | t1/2 = 5 h | + | − | + | − | |||||
| fla8 | t1/2 = 2 h | ± | − | + | − | |||||
| fla10-1 | t1/2 = 3 h | ± | − | + | − | |||||
| fla2 | t1/2 = 0.5 h | ++ | ++ | +++ | ++++ | |||||
| fla3 | t1/2 = 0.75 h | + | ++ | ++ | ++/+ | |||||
| fla4 | t1/2 > 6 h | + | + | + | + | |||||
| fla9 | t1/2 > 6 h | +++ | +++ | ++ | ++ | |||||
| fla13 | t1/2 = 4.5 h | ++ | +++ | +++ | ++ | |||||
t 1/2 is the time of incubation at 33°C when the average flagellar length is 50% preincubation length.
The flagellar membrane plus matrix fractions shown in Fig. 3 A were probed with anti-FLA10T for the presence of FLA10 protein (Fig. 3 B). As expected, incubation at 33°C did not affect FLA10 levels in wild-type cells; however, in fla1 (not shown), fla8, and fla10 the level of FLA10 decreased after the incubation. Even at the permissive temperature, the amount of FLA10 in these three ts mutants was reduced relative to wild-type cells, consistent with previous results showing little FLA10 in fla10 flagella at either the permissive or restrictive temperatures (Walther et al., 1994; Kozminski et al., 1995). The similarity of FLA10 behavior in fla10, fla1, and fla8, suggests that the gene products of fla1 and fla8 could be the 100- and 85-kD subunits of the heterotrimeric FLA10 kinesin-II complex. In the remaining fla mutants tested, the amounts of FLA10 remained relatively constant or increased after incubation at the restrictive temperature (Table II). Thus, the loss of the putative IFT particle polypeptides correlates with the loss of FLA10 in the flagella of fla1, fla8, and fla10 after incubation at the restrictive temperature. This correlation provided further evidence that the set of polypeptides described above are components of the IFT particles.
Membrane plus matrix extracts of flagella isolated from wild-type cells or fla mutants were analyzed by sucrose density gradient centrifugation to better characterize the IFT particle polypeptides. As seen in gradients of wild-type extracts (Fig. 4), the set of polypeptides that were reduced by incubating fla10 at 33°C (Fig. 3 A) cosedimented at ∼16 S. The loss of these putative IFT particle polypeptides was confirmed in gradients of flagellar extracts isolated from fla10 (data not shown) and fla1 (Fig. 5) mutants after incubation at 23° and 33°C. Note that, as with fla10 (Fig. 3 A), the concentration of the IFT particle polypeptides in fla1 flagellar extracts isolated at 23°C (Fig. 5 A) was low relative to wild-type extracts.
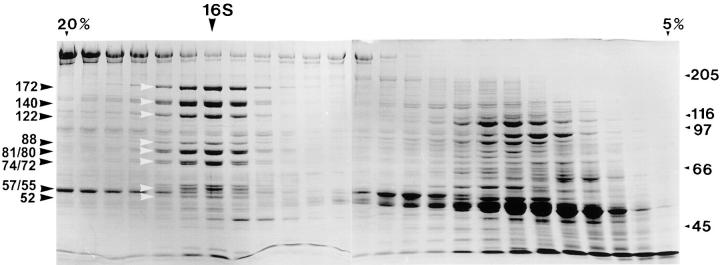
Figure 4.
Sucrose density gradient profile of membrane plus matrix. The membrane plus matrix fraction from flagella of wild-type cells was fractionated on an 11-ml 5–20% sucrose gradient in 10 mM Hepes, pH 7.2. The Coomassie-stained gel of the gradient profile shows that the putative IFT particle polypeptides cosediment at 16 S (highlighted by both sets of arrowheads with their apparent mobilities listed on the left).
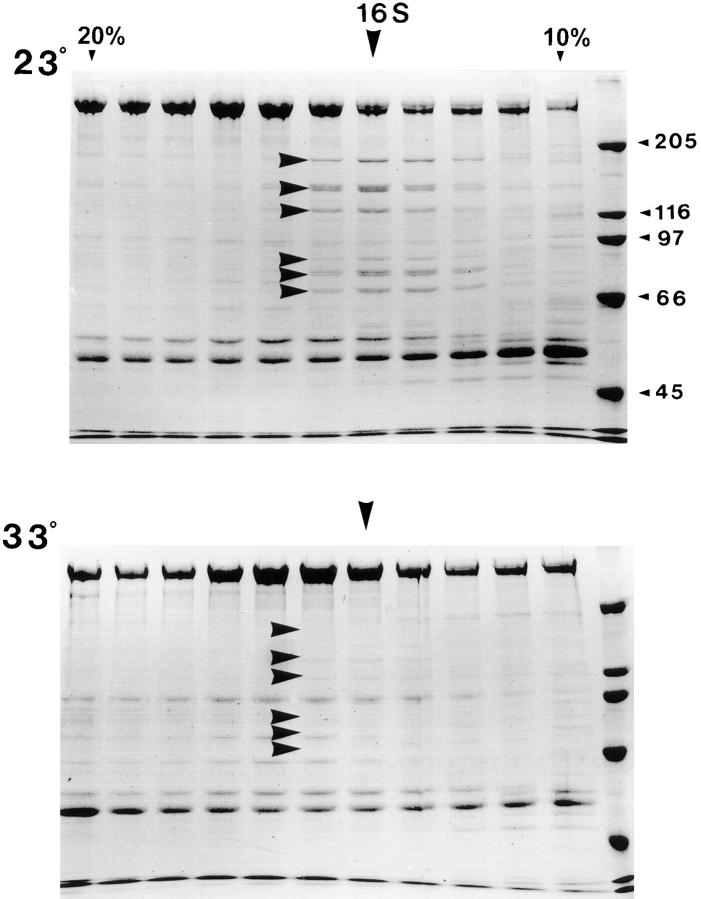
Figure 5.
Loss of 16 S polypeptides in the flagellar extracts of fla1 mutant cells at the restrictive temperature. Membrane plus matrix from fla1 mutant cells incubated at the permissive and restrictive temperatures of 23° and 33°C was fractionated on 11 ml 5–20% sucrose gradients in HMDEK buffer. Only the portion of the gradient from 20% (left) to 10% (right) is shown here on Coomassie-stained 7.5% gels. At 23°C, the IFT particle polypeptides, at 15–16 S, are reduced relative to wild type (arrowheads). These polypeptides are further depleted after incubation at 33°C (arrowheads). Similar results were obtained with membrane plus matrix isolated from fla8 and fla10 cells incubated at 23° and 33°C. Note that in the upper gel the polypeptides at 140 and 122 kD peak in the fraction highlighted by the 16 S arrow, whereas the other highlighted bands are sedimenting slightly slower.
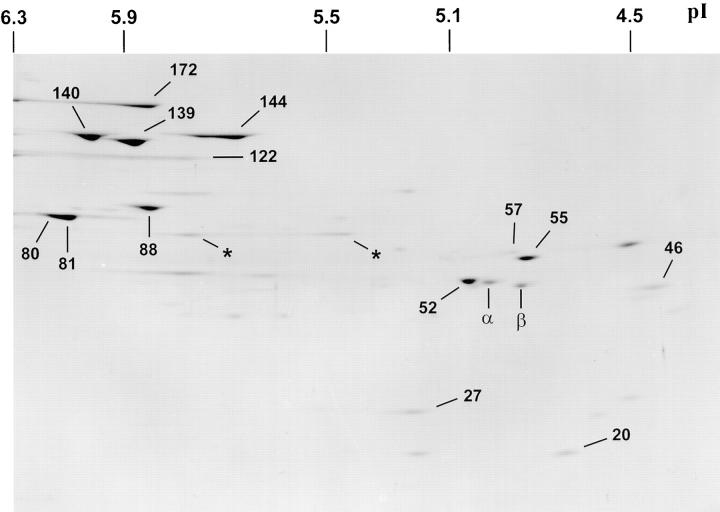
The IFT particle polypeptides were further characterized with two-dimensional gel analysis of 15–16 S fractions of wild type (Fig. 6), fla1, and fla10 flagellar extracts isolated from cells at 23° and 33°C. Comparative analysis of one- and two-dimensional gels of these extracts led to the identification of the following 15 IFT particle polypeptides: p172, p144, p140, p139, p122, p88, p81, p80, p74, p72, p57/55, p52, p46, p27, and p20. Routinely, all but the 74-/ 72-kD doublet could be identified on two dimensional gels as shown in Fig. 6. The lower band of the 140-kD doublet seen on one-dimensional gels (Figs. 4 and 5) was further resolved on two-dimensional gels into a doublet of p140 and p139 (Fig. 6). On immunoblots of one- and two-dimensional gels, both p57 and p55 reacted with the same mAb, 57.1, suggesting they are the product of the same gene (data not shown). p80 could be resolved from p81 on one-dimensional gels (Fig. 4) but was frequently not resolved from p81 on two-dimensional gels as illustrated in Fig. 6. Densitometric scans of Coomassie-stained one-dimensional gels suggested the polypeptide components of the IFT particles were present in a molar ratio of ∼1 (data not shown), with the exception of p88 and p46, which were frequently present at substoichiometric levels (for example, Fig. 4, p88; Fig. 6, p46).
Figure 6.
Two-dimensional gel of IFT particle polypeptides. The 15–16 S fraction from a sucrose density gradient of a wild-type flagellar extract was separated by two-dimensional gel electrophoresis and stained with Coomassie blue. Identifiable bands are labeled according to apparent molecular mobility in the second dimension. An asterisk may represent p74. p57/55 are resolved as two closely migrating bands. p172 and p122 frequently run as streaks. The pIs of two-dimensional gel standards are shown.
Hydrodynamic Analysis of the IFT Particle Polypeptides
The 15 IFT particle polypeptides appeared initially to cosediment at ∼16 S (Fig. 4). By varying the ionic strength, however, these polypeptides could be separated into two complexes: complex A, composed of p144, p140, p139, and p122, which sedimented at 16.1–16.4 S in both low and moderate ionic strength solutions; and complex B, composed of the remaining 11 polypeptides, which, at low ionic strength (10 mM Hepes, pH 7.2) sedimented at 16 S, but sedimented more slowly as the ionic strength was increased. The composition and hydrodynamic behavior of complexes A and B are listed in Table III. Fig. 7 A shows a profile of a sucrose density gradient (moderate ionic strength ≈125 mM) of membrane plus matrix isolated from fla2 (fla2 was used because flagella from fla2 cells contain two to three times as much complex A and complex B as flagella from wild-type cells). The salt-dependent shift in the sedimentation of complex B polypeptides was especially evident in a corresponding immunoblot of Fig. 7 A that was probed with mAbs raised against p172, p139, p81, and p57/55 (Fig. 7 B). The separation of p172 from the rest of complex B, most apparent on the immunoblot (Fig. 7 B), accounts for the variability in S-value and diffusion coefficient (see below) of complex B.
Table III.
Polypeptide Composition of IFT Complexes A and B
| Complex | Hydrodynamic behavior | Subunit polypeptides | ||||||
|---|---|---|---|---|---|---|---|---|
| Mass | pI | |||||||
| A | S-value | 16.1–16.4 S | p144 | 5.7–5.8 | ||||
| Diff. coef. | 2.5 × 10−7 cm2/s | p140 | 6.0 | |||||
| M w | 550 kD | p139 | 5.9 | |||||
| p122 | 5.8–6.0 | |||||||
| B | S-value | 15–16 S | p172 | 5.9 | ||||
| Diff. coef. | 1.8 × 10−7 cm2/s | p88 | 5.9 | |||||
| M w | 710–760 kD | p81 | 6.1 | |||||
| p80 | 6.0 | |||||||
| p74 | nd | |||||||
| p72 | nd | |||||||
| p57/55 | 4.9/4.8 | |||||||
| p52 | 5.0 | |||||||
| p46 | 4.4 | |||||||
| p27 | 5.2 | |||||||
| p20 | 4.7 | |||||||
Figure 7.
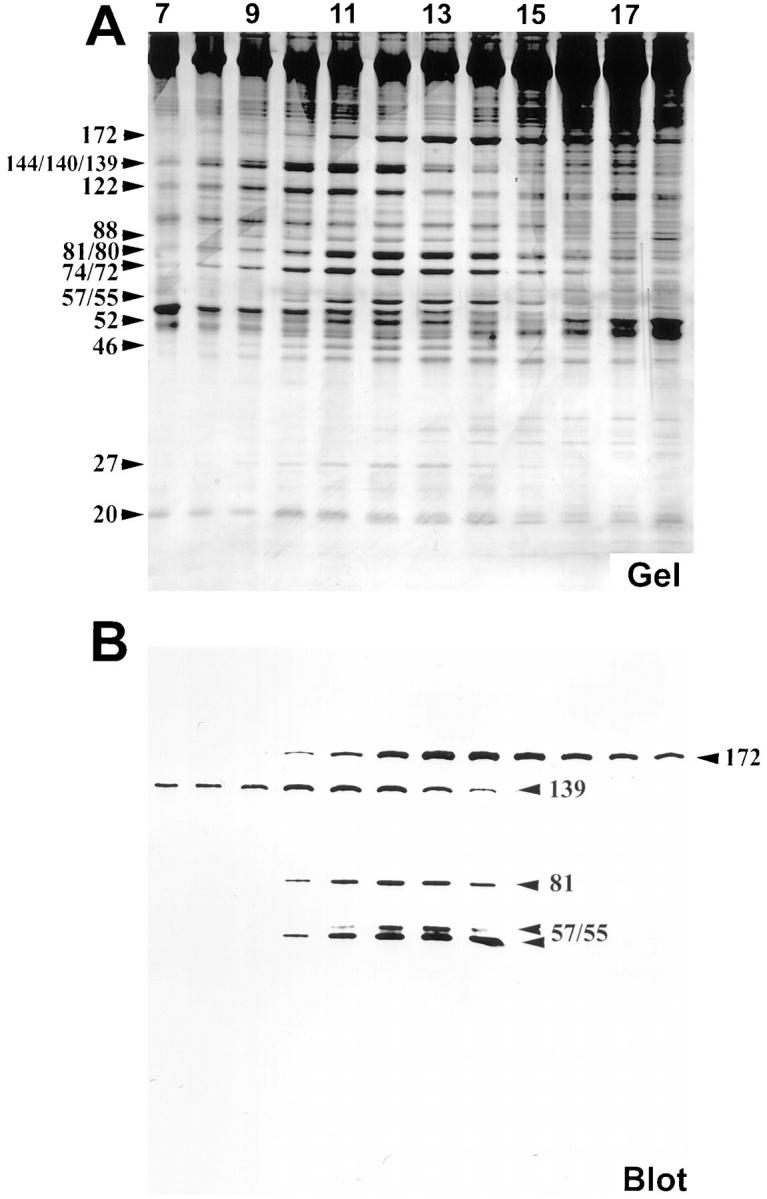
Resolution of 15 IFT particle polypeptides. Membrane plus matrix from fla2 cells incubated at 23°C was fractionated on an 11-ml 10–25% sucrose density gradient in HMDEK buffer + 50 mM NaCl. Only a portion of the gradient from fraction 7 (21% sucrose, left side) to fraction 18 (15% sucrose, right side) are shown here on 5–20% SDS-PAGE. (A) Silver-stained gel. A subset of the IFT particle polypeptides, p144, p140, p139, and p122 (complex A), peak at fraction 11. The wide band at ∼140 kD is a triplet of unresolved bands at 144, 140, and 139 kD. The p172 polypeptide peaks at fraction 14. The remaining 10 IFT particle polypeptides, p88, p81, p80, p74, p72, p57/55, p52, p46, p27, and p20 (complex B minus p172), peak between fractions 12 and 13. The bands at ∼80 and 74 kD are two sets of unresolved doublets at 81 and 80 kD, and 74 and 72 kD, respectively (these two sets of doublets were routinely resolved on 7.5% SDS-PAGE as seen in Fig. 4). Note that at this ionic strength, p172 is clearly sedimenting slower than the rest of complex B. (B) Corresponding immunoblot probed with mAbs 172.1, 139.1, 81.1, 57.1, raised against IFT particle polypeptides.
The IFT particle polypeptides also behaved as two complexes when fractionated by gel filtration: complex A exhibited a relatively constant diffusion coefficient of 2.5 × 10−7 cm2/s over a range of ionic strengths (75–375 mM). It was difficult to obtain an accurate diffusion coefficient for complex B by gel filtration because of its lability, especially at the moderate ionic strength (100 mM) needed to minimize interactions between proteins and the gel filtration support resin. Given this caveat, an approximate diffusion coefficient of 1.8 × 10−7 cm2/s was obtained for complex B at an ionic strength of 75 mM (Table III). From these hydrodynamic data, the molecular masses of complex A and complex B were calculated to be 550 kD and 710–760 kD, respectively. Assuming a stoichiometry of 1.0 for each subunit, the composite molecular masses of the complexes were estimated to be 545 and 769 kD for complexes A and B, respectively (Table III), in accord with the calculations made from the hydrodynamic data. These results suggest that complexes A and B each contain one each of the subunits as listed in Table III. We can not rule out, however, that one or more of the subunits may be present at a higher or lower stoichiometry.
Immunoprecipitation of Complexes A and B
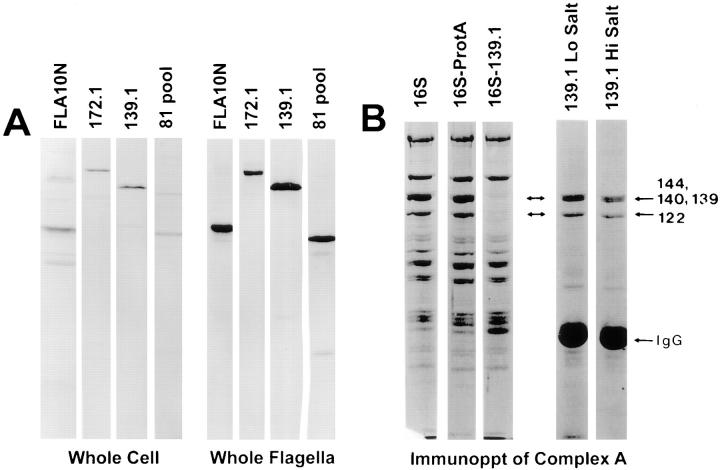
One-dimensional immunoblots of whole cells and whole flagella were probed with anti-FLA10N antibody and the complex A and complex B mAbs to demonstrate the specificity of the antibodies (Fig. 8 A) and to measure the relative distribution of FLA10 kinesin-II and the IFT particle polypeptides in flagella and the cell body (data not shown). Even though the majority (90%) of total FLA10 antigen was found in the cell body, the ratio of FLA10 antigen to total protein was enriched 10-fold in the flagella over that of the whole cell. The flagellar enrichment of FLA10 kinesin-II is in contrast to that reported for sea urchin blastula in which kinesin-II concentrations are 10– 20-fold lower in the cilia relative to the cell body (Morris and Scholey, 1997). Similar flagellar enrichments of the IFT particle polypeptides p172, p139, p81, and p57/55 relative to cell body levels were also observed (data not shown).
Figure 8.
Specificity of anti-FLA10N antibody and anti-IFT particle polypeptide antibodies and immunoprecipitation of complex A. (A) Whole wild-type cells and flagella were separately boiled in SDS-PAGE sample buffer, clarified, and then fractionated on 7.5% gels. Corresponding immunoblots were probed with antibodies as labeled. (B) Immunoprecipitation of complex A with 139.1. The membrane plus matrix from wild-type flagella was fractionated on a sucrose density gradient and fractions containing the 16 S proteins were pooled. An immunoprecipitate with the 139.1 antibody from this pool was analyzed on a Coomassie blue–stained 7.5% gel. Lane 1, 16 S pool; lane 2, 16 S pool after incubation with protein A–agarose resin; lane 3, 16 S pool immunodepleted with 139.1; lane 4, 139.1 immunoprecipitate of 16 S pool in HMDEK buffer; lane 5, 139.1 immunoprecipitate of 16 S pool after 0.5 M NaCl wash.
To identify other proteins weakly bound to complexes A and B, the complexes were rapidly purified by immunoprecipitation from both wild-type membrane plus matrix as well as with the 16 S fraction from sucrose gradients of membrane plus matrix. Antibody 139.1 coprecipitated primarily the four polypeptides previously described as comprising complex A: p144, p140, p139, and p122 from both membrane plus matrix (data not shown) and the 16 S fraction (Fig. 8 B). Antibody 172.1 immunoprecipitated the 172-kD polypeptide described previously and significant, albeit substoichiometric, amounts of the rest of the complex B polypeptides at an ionic strength of 100 mM. The lability of the interaction between p172 and the rest of complex B was evident by washing the resin with 0.2 M NaCl in 10 mM Hepes, pH 7.2, which released nearly all of the bound complex B polypeptides except for p172 (data not shown). Thus, the salt-sensitive immunoprecipitation of complex B with 172.1 agrees with the dissociation seen on sucrose gradients. We also found by Western blotting with anti-FLA10N that there was a small, but detectable, amount of FLA10 present in the 139.1 immunoprecipitate of membrane plus matrix (data not shown). Thus, in spite of clear separation between FLA10 kinesin-II and complexes A and B on sucrose gradients, coimmunoprecipitation of FLA10 and complex A raises the possibility that one or more polypeptides in complex A can bind directly, though weakly, to FLA10 kinesin-II. If such binding can occur, it appears to be weak under the conditions used and such weak binding may not be detected using long duration separation techniques such as the sucrose density gradient centrifugation and gel filtration procedures used to purify both FLA10 kinesin-II and complexes A and B.
Immunolocalization of FLA10 Kinesin-II and IFT Complexes A and B
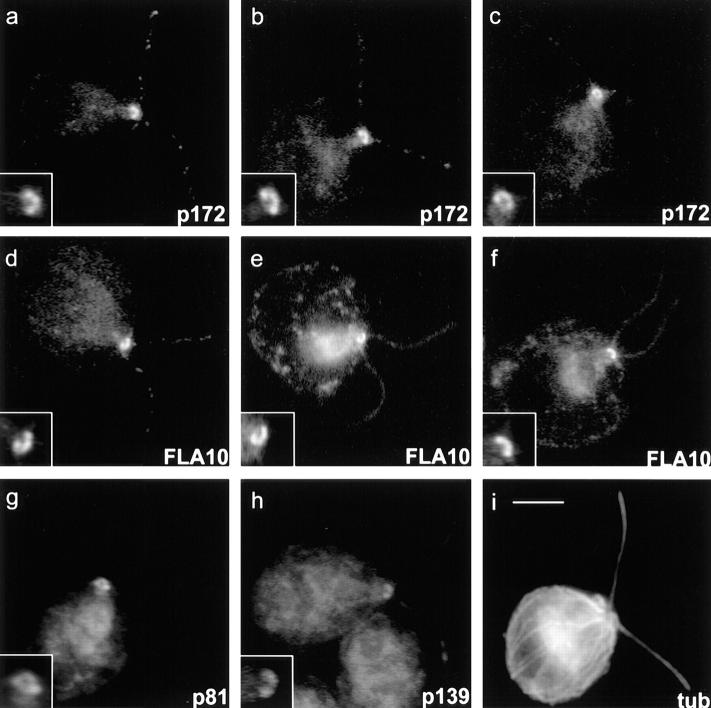
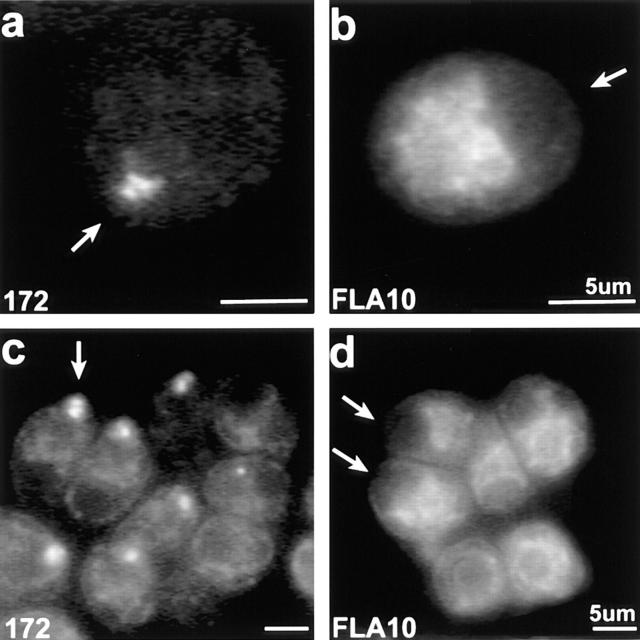
Immunofluorescence and immunogold electron microscopy were used to localize both FLA10 and complex A and B polypeptides (see Figs. 9–11). Immunofluorescence microscopy shows that in the cell body, FLA10 (Fig. 9, d–f), p172 (Fig. 9, a–c), p81 (Fig. 9 g), and p139 (Fig. 9 h) are highly enriched in the region around the basal bodies (Fig. 9, insets). This is in spite of the fact that the antigens were purified from isolated flagella. The basal body region staining appeared as a bi- or sometimes tri-lobed semi-circle with the concave side facing the nucleus. Unlike basal bodies, which are closest together at the point nearest the nucleus, FLA10 and IFT particle polypeptide staining was farthest apart nearest the nucleus. Weak, punctate staining of the flagella was also observed with these antibodies. It is probable, though not certain, that the punctate staining of the flagella represents the raft structures seen with DIC and electron microscopy.
Figure 9.
Immunofluorescent localization of FLA10, p172, p139, and p81 in cw92 cells (cell wall deficient but otherwise wild type). (a–c) 172.1. (d–f) Anti-FLA10N. (g) a pool of mAbs to p81: 81.1, 81.2, 81.3, and 81.4. (h) 139.1. Note the bi- or tri-lobed staining at the base of the flagella in a–h, insets. (i) Polyclonal anti–α-tubulin. Bar, 5 μm (inset, 10 μm).
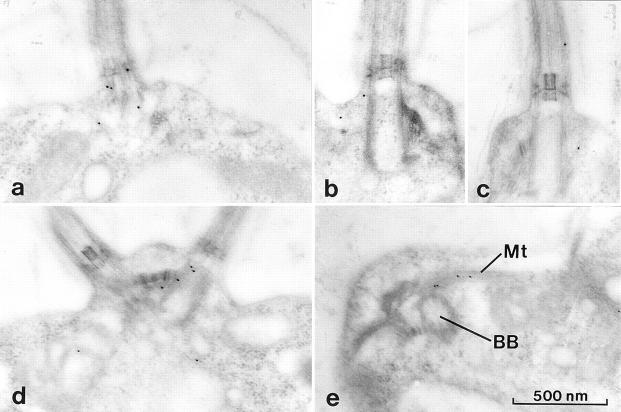
Figure 11.
Immunogold electron micrographs of p172 and FLA10 in wild-type cells. All micrographs illustrate the basal body region. (a–c) 172.1 staining was limited to the basal bodies and surrounding area with occasional gold particles seen between the flagellar membrane and outer doublet MTs. (d and e) Anti-FLA10N staining is similar to 172.1 staining. (e) Occasional labeling of the rootlet MTs (Mt) was seen in the vicinity of the basal bodies (BB).
Although FLA10 kinesin-II and complexes A and B appear to be highly concentrated near the basal bodies, their precise relationship to these organelles is difficult to resolve. To further elucidate this relationship, the distribution of these proteins in a mutant cell line that lacks basal bodies, bld2 (Goodenough and St. Clair, 1975; Ehler et al., 1995) was examined. In bld2 cells, as in wild type, p172 was concentrated at the anterior of the cell as judged by the position of staining relative to the nucleus and pyrenoid body (Fig. 10, a and c). However, the bi- or tri-lobed structure often seen in wild-type cells was not resolved, with the staining more variable in shape and somewhat more diffuse than in wild-type cells. In contrast to p172, FLA10 was dispersed in bld2 cells, showing no localization to the anterior of the cell (Fig. 10, b and d) as observed in wild-type (Fig. 9) and fla10 cells. Thus, FLA10 kinesin-II is either not targeted to, or cannot accumulate at the base of the flagella unless the basal bodies themselves are present; whereas p172 does not require the basal bodies for targeting or accumulation in this region. Similar results were obtained with fla10 cells incubated at 33°C for 24 h. During this incubation, the total cellular FLA10 protein in the fla10 cells was reduced from 10% (t = 0 h) to 3% (t = 24 h) of wild-type levels, which resulted in weak anti-FLA10N staining around the basal bodies. Concomitant with the reduction of FLA10 protein, p172 levels were reduced from 100% (t = 0 h) to 50% (t = 24 h) of wild-type levels and 172.1 staining continued to display the tripartite basal body region localization characteristic of cells maintained at 23°C (data not shown). In both cells lacking basal bodies (bld2) and cells where the FLA10 kinesin-II has been inactivated (fla10 at 33°C), p172 and presumably complex B still accumulate in the same cellular region as in wild-type cells.
Figure 10.
Immunofluorescent staining of p172 and FLA10 in bld2 cells. (a and c) 172.1 staining. (b and d) Anti-FLA10N staining. The background chloroplast labeling of anti-FLA10N is intentionally bright to indicate the cells' orientation and to demonstrate that FLA10 kinesin-II is not concentrated at what should be the basal body region of the cell (highlighted by arrows).
Immunogold electron microscopy using both 172.1 and anti-FLA10N was performed to increase our resolution of the distribution of p172 and FLA10 relative to the basal bodies (Fig. 11). As often seen with thin section immunogold labeling, the labeling was not intense, but was specific and reproducible. Consistent with immunofluorescence, most of the 172.1 label was found near the basal bodies with no labeling of the chloroplast or nucleus (data not shown). Sometimes the label was scattered up to a micron away from the basal bodies, but more typically, one to three gold particles were seen on, or within 100 nm of the basal bodies (Fig. 11, a–c). Labeling was also seen infrequently along the flagella between the flagellar membrane and axoneme (Fig. 11 c). Similarly, anti-FLA10N labeling was concentrated around the basal bodies (Fig. 11, d and e), and in rare fortuitous sections labeling of the rootlet microtubules near the basal bodies was seen (Fig. 11 e).
Analysis of Complex A and B Polypeptides
To describe the components of the IFT complexes, microsequences were obtained from complex A and B polypeptides that were purified on two-dimensional gels (as shown in Fig. 6). Database searches with most of these peptides failed to reveal any matches of significant homology with characterized proteins. However, two of the complex B polypeptides, p52 and p172, appear to be homologous to proteins of known biological function. Three peptide sequences from p52 share significant homology with OSM-6 (Collet et al., 1998), a protein whose expression is limited to sensory neurons in C. elegans and NGD5 (Wick et al., 1995), a protein of unknown function expressed in neural tissue of the mouse (Fig. 12). OSM-6, NGD5, and p52 are all of similar size and possess similar isoelectric point (pI) values, as might be expected for closely related proteins. One peptide from p172 shares significant homology with a repeated sequence found in the recently identified C. elegans OSM-1 protein (Fig. 12; Stone, S., and J. Shaw, personal communication). These findings are particularly significant because osm-1 and osm-6 share a common ultrastructural defect; these mutants fail to properly assemble the nonmotile sensory cilia located at the distal ends of the dendritic processes of sensory neurons (Perkins et al., 1986). This phenotype is very similar to that seen with osm-3, which encodes a KIF3-like kinesin (FLA10 homologue) whose expression is limited to a subset of sensory neurons in C. elegans (Shakir et al., 1993; Tabish et al., 1995). These parallels suggest a common mechanism exists for controlling the proper assembly and maintenance of both motile and nonmotile cilia and that FLA10 kinesin-II and the IFT complexes identified in Chlamydomonas play essential roles in this mechanism.
Figure 12.
(A) Alignment of three tryptic peptide sequences derived from p52 of complex B with deduced amino acids sequences from C. elegans OSM-6 and mouse neuronal NGD5. (B) Alignment of endoproteinase Lys-C peptide sequence derived from p172 with deduced amino acid sequence from C. elegans OSM-1.
Discussion
FLA10 Is a Subunit of a Heterotrimeric Kinesin
Chlamydomonas FLA10 was previously found to belong to the kinesin superfamily of microtubule (MT)-based motor proteins (Walther et al., 1994), and was classified as a member of the heterotrimeric kinesins (Moore and Endow, 1996; Scholey, 1996). These kinesins are characterized by the presence of two distinct, albeit similar, kinesin-like motor subunits, and a third, slightly larger, nonmotor subunit (for review see Scholey, 1996; Cole, 1998). More than 10 genes related to heterotrimeric kinesin motor subunits have been identified in various organisms (see the kinesin homepage on the world wide web at www.blocks. fhrc.org/∼http://kinesin/index.html), but only the sea urchin (Cole et al., 1992, 1993) and mouse (Yamazaki et al., 1995, 1996) proteins have been isolated and shown to belong to heterotrimeric complexes. The Chlamydomonas FLA10 polypeptide (90 kD) is also purified as a component of a 9.7 S heterotrimeric complex (270 kD) with a second motor subunit (85 kD), and a third nonmotor subunit (100 kD) (Figs. 1 and 2). The two motor subunits were probably previously identified (by pan-kinesin peptide antibodies) in partially purified Chlamydomonas flagellar extracts as a kinesin-related 97-kD doublet sedimenting at ∼9 S (Fox et al., 1994). We refer to the Chlamydomonas heterotrimeric complex as FLA10 kinesin-II using the nomenclature suggested by Scholey and coworkers (Scholey, 1996; Wedaman et al., 1996).
IFT Particles Are Composed of Two Large Protein Complexes
To identify components of the IFT particles, we looked for membrane plus matrix proteins that were reduced in the flagella of the ts fla10 mutant after incubation at the restrictive temperature of 33°C (Fig. 3), in which both IFT and the number of flagellar rafts, composed of IFT particles, were dramatically reduced (Kozminski et al., 1995). Two protein complexes, termed complex A (550 kD) and complex B (710–760 kD), disappeared from fla10 flagella at the restrictive temperature. At low ionic strength, both complexes sedimented at ∼16 S (Fig. 4); at higher ionic strength, complex A continued to sediment at 16 S while complex B dissociated into two or more species with lower S-values (Fig. 7). Because of their large size, presence in the membrane plus matrix fraction, relative abundance, and FLA10-dependent presence in the flagella, these complexes were excellent candidates for components of the IFT particles.
Additional evidence that complexes A and B compose the IFT particles comes from analysis of a retrograde IFT mutant described in the accompanying report (Pazour et al., 1998). This mutant, fla14, is a null mutant for the dynein light chain LC8, a component of flagellar outer arm dynein (King and Patel-King, 1995), which is also associated with cytoplasmic dynein (King et al., 1996) and myosin-V (Espindola, F.S., R.E. Cheney, S.M. King, D.M. Suter, and M.S. Mooseker. 1996. Mol. Biol. Cell. 7:372a). fla14 cells have short, nonmotile flagella, often exhibiting bulges on the side or tip of the flagellum (Pazour et al., 1998). DIC microscopic observations of these flagella showed that anterograde IFT continued while retrograde IFT stopped. Ultrastructural analysis revealed that large numbers of IFT rafts accumulated in the space between the axoneme and the flagellar membrane of the mutant because anterograde IFT continued to transport IFT particles into the flagellum, but without retrograde IFT, both IFT particles (complexes A and B) and the anterograde motor (FLA10 kinesin-II) lacked a mechanism by which to leave the flagellum. The levels of FLA10 kinesin-II and IFT particle polypeptides, but not other flagellar proteins, increased dramatically in fla14 flagella over that found in wild-type flagella (Pazour et al., 1998), providing convincing evidence that complexes A and B are major components of the IFT particles that compose the rafts.
Composition of IFT Complexes A and B
IFT complex A is a 550-kD tetramer containing subunits of 144, 140, 139, and 122 kD, whereas IFT complex B is a 750-kD complex containing 11 subunits ranging from 20 to 172 kD (Table III). Recently, a set of 13 polypeptides, including HSP70, were identified as a single 17 S complex whose presence in Chlamydomonas flagella required FLA10 activity (Piperno and Mead, 1997). It is probable that these 13 polypeptides are a subset of those described here, despite discrepancies in their apparent molecular weights. However, although we detected flagellar HSP70 sedimenting at 16 S (data not shown), the level of the 16 S fraction of HSP70 increased while the IFT particle polypeptides disappeared from the flagella of fla10 cells incubated at 33°C, indicating that HSP70 is not a subunit of complex A or complex B.
A small amount of complex A and to a much lesser extent, complex B, were also observed sedimenting at higher S-values (>18 S; see first four fractions of Fig. 4), suggesting the presence of larger complexes, perhaps bound to flagellar precursors. Indeed, a fraction of inner arm dynein, which sediments at 11 S when extracted from axonemes, was found to sediment at 18 S when isolated from membrane plus matrix, as if bound to IFT particles (Piperno and Mead, 1997). We also observed varying amounts of the following flagellar antigens sedimenting in unexpectedly heavy fractions (15–20 S) on immunoblots of sucrose density gradients of membrane plus matrix (data not shown) probed with antibodies to: (a) inner arm dynein intermediate chain IC140 (Yang, P., and W.S. Sale. 1996. Mol. Biol. Cell. 7:568a); (b) outer arm dynein intermediate chains IC78 (King et al., 1991) and IC69 (King et al., 1985); (c) radial spoke proteins 3 (Williams et al., 1989) and 6 (Diener, D.R., D.G. Cole, and J.L. Rosenbaum, manuscript in preparation); (d) actin (Kirk, D., personal communication); and (e) HSP70 (Savino, E., and J.L. Rosenbaum, unpublished observations). These proteins, however, did not disappear in parallel with complexes A and B in fla10 cells incubated at 33°C. Neither did immunoprecipitation of either complex A or complex B from membrane plus matrix coprecipitate detectable amounts of either I1 inner arm dynein or radial spoke complexes, suggesting that these axonemal proteins are not directly associated with the IFT complexes in our preparations. Though the presence of complexes A and B at >18 S remains unexplained, the higher sedimentation is consistent with weak self-association of complex A and a weak association between complex A and B.
Colocalization of FLA10 Kinesin-II and p172 at the Basal Bodies
Immunofluorescence microscopy revealed that complex A polypeptide p139 and the complex B polypeptides p172 and p81 were most concentrated in a semi-circular bi- or tri-lobed arc in the region surrounding the basal bodies. The similarity in the staining patterns of these antibodies suggest that complexes A and B are in close proximity to, if not associated with, each other in a unique subcellular domain. The precise relationship between the IFT complexes and the basal bodies themselves or the microtubule rootlets that emanate from the basal bodies remains unclear.
As would be expected for a motor binding its cargo, localizations of FLA10 and IFT particle polypeptides were largely coincident (Fig. 9). FLA10 is found in association with the basal bodies in interphase cells and migrates with these organelles as they become centrioles during mitosis (Vashishtha et al., 1996). A direct interaction between FLA10 and basal bodies was further suggested by the failure of FLA10 to concentrate at the anterior of bld2 cells, which lack basal bodies (Goodenough and St. Clair, 1975; Ehler et al., 1995). Unlike FLA10, however, IFT polypeptide p172 accumulates at the anterior of bld2 cells, though its distribution is more diffuse than in wild-type cells. The targeting of p172, and presumably complex B, to the basal body region appears, therefore, to be independent of FLA10 kinesin-II. Likewise, after 24 h at the restrictive temperature, p172 accumulated at the basal body region of fla10 cells even though the small amount of FLA10 present was presumably inactive. FLA10 activity, therefore, is required for transport of IFT particles into the flagella, but not to the basal body region. Since the minus ends of cytoplasmic MTs are in close proximity to the basal bodies, a minus end–directed MT motor such as cytoplasmic dynein may function to deliver IFT complexes or their constitutive polypeptides to the basal body region before their entry into the flagellum.
Model of FLA10 Kinesin-II–mediated IFT
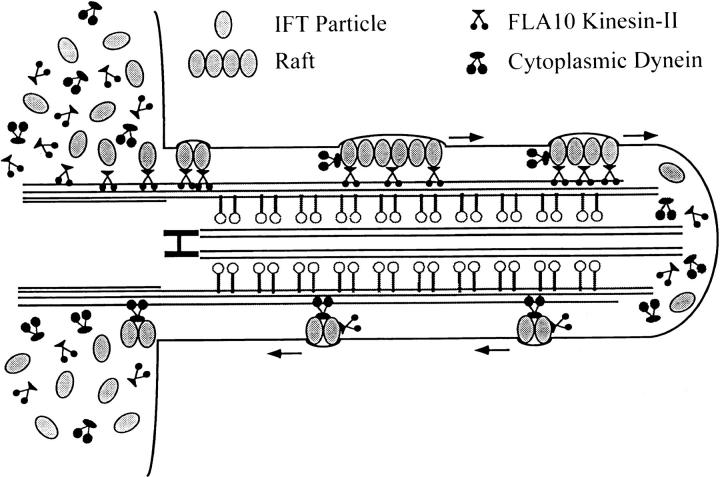
A working model of FLA10 kinesin-II–mediated IFT is shown in Fig. 13. FLA10 kinesin-II, complex A, and complex B accumulate in the region around the basal bodies perhaps resulting from localized synthesis or transport to this region. Multiple copies of each complex associate, possibly along with other still-undetermined molecules, to form IFT particles. The basal bodies are likely to serve as a docking site for FLA10 kinesin-II, which in turn binds to IFT particles. At this site, other molecules that require transport toward the flagellar tip may be bound. This cargo may include flagellar precursors (e.g., radial spoke complexes and inner dynein arms), integral membrane proteins (Bloodgood, 1992), the retrograde IFT motor (Pazour et al., 1998), and ATP. The large size and complexity of the IFT particles may be necessary to sequester flagellar precursors so they do not associate with each other or bind the axonemal MTs during transport. Association of the IFT complexes to form linear arrays or rafts may occur on the basal body or along the transition region of the axoneme. The rafts are then transported by FLA10 kinesin-II into the flagellum along the B tubules of the outer doublets (Kozminski et al., 1995). Once the precursors are delivered to the tip, the rafts, which appear to dissociate into smaller oligomers, are transported by the retrograde IFT motor, thought to be cytoplasmic dynein (Pazour et al., 1998), back to the base of the flagellum.
Figure 13.
Model of IFT in a Chlamydomonas flagellum. Fla10 kinesin-II and IFT complexes A and B are highly concentrated around the basal body region of the cell. Multiple copies of complexes A and B associate to form IFT particles. FLA10 kinesin-II binds to the basal bodies and then IFT particles bind to FLA10 kinesin-II. IFT particles may also bind to the kinesin before docking to the basal bodies. After oligomerization of IFT particles to form rafts, FLA10 kinesin-II transports IFT particles toward the flagellar tip. At the tip, the rafts dissociate into smaller oligomers of IFT particles and are returned to the base of the flagellum by cytoplasmic dynein.
If IFT delivers precursors to the tip during flagellar assembly, why does it operate continually even in full-length flagella? Several lines of evidence suggest that protein turnover occurs in full-length flagella. Metabolically labeled protein rapidly incorporates into full-length cilia of sea urchins and scallops even though these cilia have ceased to elongate (Stephens 1994, 1996). In Chlamydomonas, dikaryon rescue experiments also suggest that precursors are continually entering the flagella. Within minutes of cell fusion during mating, cytoplasmic components from one cell can enter immotile flagella of the other cell and restore motility (Lewin, 1954; Starling and Randall, 1971; Luck et al., 1977; Johnson and Rosenbaum, 1992), presumably transported by a mechanism that operates continuously. Finally, in dikaryons formed from two wild-type cells with full-length flagella, in which one cell expresses an epitope-tagged tubulin, the epitope-tagged tubulin incorporates into the full-length flagella beginning at the tip and moves toward the base at a rate consistent with microtubule treadmilling in vitro (Marshall, W., and J.L. Rosenbaum, unpublished observations). It is possible that, like many other microtubule structures once thought to be stable, axonemes constantly turn over and IFT is needed to continually deliver axonemal precursors to the flagellar tip.
IFT may also serve to transport proteins and/or lipids within the flagellar membrane. Evidence supporting this hypothesis comes from electron micrographs showing periodic associations of the IFT particles with the flagellar membrane (Kozminski et al., 1993, 1995; Pazour et al., 1998) and from studies showing that fla10 gametes lose the ability to mate 30–60 min after being shifted to the restrictive temperature of 32°C (Piperno et al., 1996). Mating in Chlamydomonas depends on flagellar adhesion (Bergman et al., 1975; Snell, 1976) and the movement of external flagellar membrane glycoproteins (agglutinins) to the flagellar tip (Mesland, 1976; Forest et al., 1978; Goodenough and Jurivich, 1978). It is possible that heat inactivation of FLA10 and the concomitant loss of IFT in fla10 mutants held at 33°C compromises one or more of the flagellar membrane-dependent processes required for successful mating.
C. elegans Homologues of p172, p52, and FLA10 Are Involved in Ciliogenesis
Sequence of internal peptides from 13 IFT particle polypeptides reveal that at least 6 of these have homologues in higher organisms. Peptide sequences from p172 and p52 are very similar to sequences of the nematode proteins, OSM-1 (Stone, S., and J. Shaw, personal communication) and OSM-6 (Collet et al., 1998), respectively (Fig. 12). osm-1 and osm-6 phenotypes are similar to that of osm-3 (Shakir et al., 1993; Tabish et al., 1995), a mutation affecting a kinesin-like protein closely related to FLA10. Like osm-3, osm-1 and osm-6 were identified as mutants unable to respond to changes in external osmotic gradients (Culotti and Russell, 1978) due to defects in the formation and function of sensory cilia located on the distal ends of the dendritic processes of sensory neurons (Perkins et al., 1986). Although the ultrastructure of the nematode sensory cilium varies, the typical cilium is a membrane-bound nonmotile axonemal structure containing nine outer doublet MTs and a variable number of central MTs, but lacking many of the proteins, such as dynein and radial spoke complexes, present in motile cilia. In osm-1 and osm-6 mutants, all sensory cilia are much shorter than in wild-type, but osm-3 affects only sensory cilia exposed to the external environment, suggesting that while IFT particles function in all sensory cilia, the role of OSM-3 may be assumed by alternate kinesins-II in assembly of some sensory cilia. Thus, in C. elegans sensory neurons, homologues of FLA10 kinesin-II and complex B polypeptides p52 and p172 are all required for formation of normal sensory cilia (Table IV).
Table IV.
Comparison of Proteins Required for Assembly of Chlamydomonas Flagella and Caenorhabditis elegans Sensory Cilia
| Chlamydomonas | Caenorhabditis | |||||
|---|---|---|---|---|---|---|
| Protein | Comments | Protein | Comments | |||
| FLA10 | 87 kD KIF3-like kinesin. | OSM-3 | 75 kD KIF3-like kinesin. | |||
| Required for IFT and | Required for assembly | |||||
| assembly of flagella | of sensory cilia | |||||
| p52 | 52 kD, pI = 5.0 | OSM-6 | 53 kD, pI = 4.8 | |||
| IFT particle protein | Required for assembly | |||||
| of sensory cilia | ||||||
| p172 | 172 kD, pI = 5.9 | OSM-1 | 196 kD, pI = 5.0 | |||
| IFT particle protein | Required for assembly | |||||
| of sensory cilia | ||||||
The phenotypes seen in C. elegans with mutations in IFT particle polypeptide homologues suggest that these particles have roles in the nematode that do not involve ciliary beating, since these cilia are immotile, nor do the particles deliver proteins that bind to the center of the axoneme, e.g. radial spokes and inner dynein arms, since these proteins are not present in sensory cilia. One possible function of IFT is suggested from sequence analysis of the p52 homologues, OSM-6 and NGD5; both OSM-6 and NGD5 contain PxxP motifs (Wick et al., 1995; Collet et al., 1998), that might interact with SH3 domains found in a variety of proteins, including signal transduction and cytoskeletal proteins (Cohen et al., 1995). One could speculate that, in addition to ciliary assembly, OSM-6 and p52 might also be involved with signal transduction, e.g. the transport of external signals originating on the flagellar membrane to the cell body.
Association of Kinesin-II with Ciliary Structures in Other Organisms
Although there is immunological evidence for the presence of at least five different kinesins in Chlamydomonas flagella (for review see Bernstein and Rosenbaum, 1994; Bernstein et al., 1994; Fox et al., 1994; Johnson et al., 1994), only FLA10 kinesin-II is known to be required for flagellar assembly and maintenance. There is evidence that kinesin-II homologues in organisms besides Chlamydomonas may be similarly associated with ciliary structures. Like OSM-3 in C. elegans (described above), the vertebrate kinesin-II, KIF3A/3B-KAP3, is also found in neurons but is thought to serve as a fast axonal transport motor in mice (Aizawa et al., 1992; Kondo et al., 1994; Yamazaki et al., 1995, 1996). KIF3A/3B-KAP3 may also function in ciliogenesis, since the expression of the separate subunits is very high in ciliated tissues like testes and lungs (Kondo et al., 1994; Yamazaki et al., 1995, 1996). In the sunfish, antibodies raised against KIF3A (Kondo et al., 1994) were used to localize a kinesin-II homologue to the connecting cilium of the photoreceptors (Beech et al., 1996). Vertebrate rod photoreceptors are specialized sensory neurons consisting of a photosensory outer segment connected to the cell body, or inner segment, by a membrane-bound “connecting cilium” (Besharse and Horst, 1990), which contains a 9+0 axonemal structure. The KIF3-like antigen in these cells is most concentrated around the basal bodies with some antigen being found along the connecting cilium (Beech et al., 1996), strikingly similar to the distribution of Chlamydomonas FLA10 kinesin-II reported here (Fig. 10). A similar distribution also occurs in echinoderm sperm, where kinesin-II (formerly KRP85/95) is concentrated around the midpiece that contains the basal body and mitochondria, and is found to a lesser extent throughout the flagellum (Henson et al., 1997). These distributions demonstrate that kinesin-II is associated with cilia in diverse organisms.
Direct evidence for participation of kinesin-II in ciliogenesis in echinoderms was shown by microinjection of anti–kinesin-II antibodies into sea urchin embryos (Morris and Scholey, 1997). Injected embryos developed normally until reaching the ciliated blastula stage, when the cells could only assemble short, immobile cilia lacking the central pair microtubules and varying amounts of other axonemal components. Thus, like Chlamydomonas FLA10 kinesin-II and C. elegans OSM-3, sea urchin kinesin-II appears to be directly involved in ciliogenesis.
Conclusion
Kinesin-II and IFT particle polypeptide homologues may be involved with the assembly and maintenance of all cilia found in eukaryotic cells. Like kinesin-II, IFT particle polypeptides appear to be widespread, as both nematode and vertebrate homologues of six of these polypeptides have been identified. Mutations affecting two of the nematode polypeptides, OSM-1 and OSM-6, cause defects in ciliogenesis; whether or not the vertebrate homologues also participate in ciliogenesis has yet to be determined. IFT particle polypeptides also play a role in axonemal assembly in both Chlamydomonas and C. elegans. Thus, the process of IFT, involving both kinesin-II and the IFT particle polypeptides, may be a universal mechanism required for the assembly, maintenance, and function of all eukaryotic motile cilia/flagella and sensory cilia.
Acknowledgments
We thank T. Ferguson for superb technical assistance, Dr. J. Leszyk at the Worcester Foundation for Biomedical Research Microsequencing Facility, Dr. J. Wolenski (MCDB, Yale University) for assistance in light microscopic imaging, Drs. P. Yang and W. Sale (Emory University), D. Kirk (Washington University), E. Savino, C. Wilkerson, and G. Witman (Worcester Foundation) for generously sharing antibodies, and Drs. G. Pazour and G. Witman (Worcester Foundation), and R. Herman and J. Shaw (University of Minnesota), and R. Morris and J. Scholey (University of California) for helpful discussions and sharing unpublished results.
This work was supported by National Institutes of Health grant GM14642 and National Science Foundation grant 45147 to J.L. Rosenbaum.
Abbreviations used in this paper
- AMPPNP
5′-adenylylimidodiphosphate
- FLA10
the fla10 gene product
- FLA10 kinesin-II
heterotrimeric complex containing FLA10 and two additional subunits
- IFT
intraflagellar transport
- MT
microtubule
- ts
temperature-sensitive
Footnotes
Address all correspondence to Joel Rosenbaum, MCDB KBT 310, Yale University, P.O. Box 208103, New Haven, CT 06520-8103. Tel.: (203) 432-3472. Fax: (203) 432-5059. E-mail: joel.rosenbaum@yale.edu
Brief reports of this work were presented at the International Congress on Chlamydomonas in Regensburg, Germany on May 27–June 1, 1996, and appeared in abstract form (Cole, D.G., and J.L. Rosenbaum. 1996. Mol. Biol. Cell. 7S:47a; Cole, D.G., D.R. Diener, J. Fuster, A. Himelblau, and J.L. Rosenbaum. 1997. Mol. Biol. Cell. 8S:54a).
References
- Adams GMW, Huang B, Luck DJL. Temperature-sensitive, assembly-defective flagella mutants of Chlamydomonas reinhardtii. . Genetics. 1982;100:579–586. doi: 10.1093/genetics/100.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N. Kinesin family in murine central nervous system. J Cell Biol. 1992;119:1287–1296. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech PL, Pagh-Roehl K, Noda Y, Hirokawa N, Burnside B, Rosenbaum JL. Localization of kinesin superfamily proteins to the connecting cilium of fish photoreceptors. J Cell Sci. 1996;109:889–897. doi: 10.1242/jcs.109.4.889. [DOI] [PubMed] [Google Scholar]
- Bergman K, Goodenough UW, Goodenough DA, Jawitz J, Martin H. Gametic differentiation in Chlamydomonas reinhardtii. II. Flagellar membranes and the agglutination reaction. J Cell Biol. 1975;67:606–622. doi: 10.1083/jcb.67.3.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein M, Rosenbaum JL. Kinesin-like proteins in the flagella of Chlamydomonas. . Trends Cell Biol. 1994;4:236–240. doi: 10.1016/0962-8924(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Bernstein M, Beech PL, Katz SG, Rosenbaum JL. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonasflagellum. J Cell Biol. 1994;125:1313–1326. doi: 10.1083/jcb.125.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besharse, J.C., and C.J. Horst. 1990. The photoreceptor connecting cilium. A model for the transition zone. In Ciliary and Flagellar Membranes. R.A. Bloodgood, editor. Plenum Press, New York. 389–417.
- Bloodgood RA. Directed movements of ciliary and flagellar membrane components: a review. Biol Cell. 1992;76:291–301. doi: 10.1016/0248-4900(92)90431-y. [DOI] [PubMed] [Google Scholar]
- Cantor, C.R., and P.R. Schimmel. 1980. Biophysical Chemistry. Part II. Techniques for the Study of Biological Structure and Function. W.H. Freeman and Company, San Francisco. 846 pp.
- Cohen GB, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- Cole, D.G. 1998. Kinesin-II, the heterotrimeric kinesin. Cell Mol. Life Sci. In press. [DOI] [PMC free article] [PubMed]
- Cole DG, Cande WZ, Baskin RJ, Skoufias DA, Hogan CJ, Scholey JM. Isolation of a sea urchin egg kinesin-related protein using peptide antibodies. J Cell Sci. 1992;101:291–301. doi: 10.1242/jcs.101.2.291. [DOI] [PubMed] [Google Scholar]
- Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. . Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotti JG, Russell RL. Osmotic avoidance defective mutants of the nematode Caenorhabditis elegans. . Genetics. 1978;90:243–256. doi: 10.1093/genetics/90.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 1995;11:398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- Ehler LL, Holmes JA, Dutcher SK. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtiimutant strain lacking basal bodies. Genetics. 1995;141:945–960. doi: 10.1093/genetics/141.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok AK, Wang H, Katayama A, Aihara MS, Allen RD. 22S Axonemal dynein is preassembled and functional prior to being transported to and attached on the axonemes. Cell Motil Cytoskeleton. 1994;29:215–224. doi: 10.1002/cm.970290304. [DOI] [PubMed] [Google Scholar]
- Forest CL, Goodenough DA, Goodenough UW. Flagellar membrane agglutination and sexual signaling in the conditional gam-1 mutant of Chlamydomonas. . J Cell Biol. 1978;79:74–84. doi: 10.1083/jcb.79.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox LA, Sawin KE, Sale WS. Kinesin-related proteins in eukaryotic flagella. J Cell Sci. 1994;107:1545–1550. doi: 10.1242/jcs.107.6.1545. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Jurivich D. Tipping and mating structure activation induced in Chlamydomonasgametes by flagellar membrane antisera. J Cell Biol. 1978;79:680–693. doi: 10.1083/jcb.79.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, St. Clair HS. Bald-2: A mutant affecting the formation of doublet and triplet sets of microtubules in Chlamydomonas reinhardtii. J Cell Biol. 1975;66:480–491. doi: 10.1083/jcb.66.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JW, Park JH, Kim M, Lee J. mRNAs for microtubule proteins are specifically colocalized during the sequential formation of basal body, flagella, and cytoskeletal microtubules in the differentiation of Naegleria gruberi. . J Cell Biol. 1997;137:871–879. doi: 10.1083/jcb.137.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Cole DG, Roesener CD, Capuano S, Mendola RJ, Scholey JM. The heterotrimeric motor protein kinesin-II localizes to the midpiece and flagellum of sea urchin and sand dollar sperm. Cell Motil Cytoskeleton. 1997;38:29–37. doi: 10.1002/(SICI)1097-0169(1997)38:1<29::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Huang B, Rifkin MR, Luck DJL. Temperature-sensitive mutations affecting flagellar assembly and function in Chlamydomonas reinhardtii. . J Cell Biol. 1977;72:67–85. doi: 10.1083/jcb.72.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Watterson DM, Lee VD, Schibler MJ. Purification and characterization of a basal body-associated Ca2+-binding protein. J Cell Biol. 1988;107:121–131. doi: 10.1083/jcb.107.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA. Keeping the beat: form meets function in the Chlamydomonasflagellum. Bioessays. 1995;17:847–854. [Google Scholar]
- Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. . J Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. Flagellar regeneration in Chlamydomonas: a model system for studying organelle assembly. Trends Cell Biol. 1993;3:156–161. doi: 10.1016/0962-8924(93)90136-o. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Haas MA, Rosenbaum JL. Localization of a kinesin-related protein to the central pair apparatus of the Chlamydomonas reinhardtiiflagellum. J Cell Sci. 1994;107:1551–1556. doi: 10.1242/jcs.107.6.1551. [DOI] [PubMed] [Google Scholar]
- King SM, Patel-King RS. The M r = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonasflagella have cytoplasmic homologues. J Biol Chem. 1995;270:11445–11452. doi: 10.1074/jbc.270.19.11445. [DOI] [PubMed] [Google Scholar]
- King SM, Otter T, Witman GB. Characterization of monoclonal antibodies against Chlamydomonasflagellar dyneins by high-resolution protein blotting. Proc Natl Acad Sci USA. 1985;82:4717–4721. doi: 10.1073/pnas.82.14.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Wilkerson CG, Witman GB. The M r 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with α-tubulin in situ. . J Biol Chem. 1991;266:8401–8407. [PubMed] [Google Scholar]
- King SM, Barbarese E, Dillman JF, III, Patel-King RS, Carson JH, Pfister KK. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved M r8,000 light chain. J Biol Chem. 1996;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonaskinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lefebvre PA, Rosenbaum JL. Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Annu Rev Cell Biol. 1986;2:517–546. doi: 10.1146/annurev.cb.02.110186.002505. [DOI] [PubMed] [Google Scholar]
- Lefebvre PA, Nordstrom SA, Moulder JE, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas.IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J Cell Biol. 1978;78:8–27. doi: 10.1083/jcb.78.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin RA. Mutants of Chlamydomonas moewusiiwith impaired motility. J Gen Microbiol. 1954;11:358–363. doi: 10.1099/00221287-11-3-358. [DOI] [PubMed] [Google Scholar]
- Luck D, Piperno G, Ramanis Z, Huang B. Flagellar mutants of Chlamydomonas: Studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc Natl Acad Sci USA. 1977;74:3456–3460. doi: 10.1073/pnas.74.8.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux III, F.G., and S.K. Dutcher. Genetic interactions at the FLA10 locus: Suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. . Genetics. 1991;128:549–561. doi: 10.1093/genetics/128.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesland DAM. Mating in Chlamydomonas eugametos. A scanning electron microscopical study. Arch Microbiol. 1976;109:31–35. doi: 10.1007/BF00425109. [DOI] [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Morris RL, Scholey JM. Heterotrimeric kinesin-II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Olmsted JB. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981;256:11955–11957. [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. . Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Piperno G, Huang B, Luck DJL. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. . Proc Natl Acad Sci USA. 1977;74:1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonasflagella. Proc Natl Acad Sci USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1FLA10 to reach the distal part of flagella in Chlamydomonas. . J Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringo DL. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. . J Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas:The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies with Chlamydomonas reinhardtii. . Ann NY Acad Sci. 1953;56:831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Sanders MA, Salisbury JL. Immunofluorescence microscopy of cilia and flagella. Methods Cell Biol. 1995;47:163–169. doi: 10.1016/s0091-679x(08)60805-5. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ, Wordeman LG. Evidence for kinesin-related proteins in the mitotic apparatus using peptide antibodies. J Cell Sci. 1992;101:303–313. doi: 10.1242/jcs.101.2.303. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Kinesin-II, a membrane traffic motor in axons, axonemes, and spindles. J Cell Biol. 1996;133:1–4. doi: 10.1083/jcb.133.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir MA, Fukushige T, Yasuda H, Miwa J, Siddiqui SS. C. elegans osm-3gene mediating osmotic avoidance behaviour encodes a kinesin-like protein. Neuroreport. 1993;4:891–894. doi: 10.1097/00001756-199307000-00013. [DOI] [PubMed] [Google Scholar]
- Snell WJ. Mating in Chlamydomonas:A system for the study of specific cell adhesion. II. A radioactive flagella-binding assay for quantitation of adhesion. J Cell Biol. 1976;68:70–79. doi: 10.1083/jcb.68.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starling D, Randall J. The flagella of temporary dikaryons of Chlamydomonas reinhardtii. . Genet Res. 1971;18:107–113. [Google Scholar]
- Stephens RE. Tubulin and tektin in sea urchin embryonic cilia: pathways of protein incorporation during turnover and regeneration. J Cell Sci. 1994;107:683–692. [PubMed] [Google Scholar]
- Stephens RE. Selective incorporation of architectural proteins into terminally differentiated molluscan gill cilia. J Exp Zool. 1996;274:300–309. doi: 10.1002/(SICI)1097-010X(19960401)274:5<300::AID-JEZ5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Tabish M, Siddiqui ZK, Nishikawa K, Siddiqui SS. Exclusive expression of C. elegans osm-3kinesin gene in chemosensory neurons open to the external environment. J Mol Biol. 1995;247:377–389. doi: 10.1006/jmbi.1994.0146. [DOI] [PubMed] [Google Scholar]
- Vashishtha M, Walther Z, Hall JL. The kinesin-homologous protein encoded by the Chlamydomonas FLA10gene is associated with basal bodies and centrioles. J Cell Sci. 1996;109:541–549. doi: 10.1242/jcs.109.3.541. [DOI] [PubMed] [Google Scholar]
- Walther Z, Vashishtha M, Hall JL. The Chlamydomonas FLA10gene encodes a novel kinesin-homologous protein. J Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman KP, Meyer DW, Rashid DJ, Cole DG, Scholey JM. Sequence and submolecular localization of the 115-kD accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J Cell Biol. 1996;132:371–380. doi: 10.1083/jcb.132.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick MJ, Ann DK, Loh HH. Molecular cloning of a novel protein regulated by opioid treatment of NG108-15 cells. Brain Res Mol Brain Res. 1995;32:171–175. doi: 10.1016/0169-328x(95)00090-f. [DOI] [PubMed] [Google Scholar]
- Williams BD, Velleca MA, Curry AM, Rosenbaum JL. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14is an ochre allele. J Cell Biol. 1989;109:235–245. doi: 10.1083/jcb.109.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB, Carlson K, Berliner J, Rosenbaum JL. Chlamydomonasflagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB. The site of in vivoassembly of flagellar microtubules. Ann NY Acad Sci. 1975;253:178–191. doi: 10.1111/j.1749-6632.1975.tb19199.x. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc Natl Acad Sci USA. 1996;93:8443–8448. doi: 10.1073/pnas.93.16.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Snell WJ. Flagellar adenylyl cyclases in Chlamydomonas. . Methods Cell Biol. 1995;47:459–465. doi: 10.1016/s0091-679x(08)60845-6. [DOI] [PubMed] [Google Scholar]