Abstract
Several enzymes, including cytoplasmic and flagellar outer arm dynein, share an M r 8,000 light chain termed LC8. The function of this chain is unknown, but it is highly conserved between a wide variety of organisms. We have identified deletion alleles of the gene (fla14) encoding this protein in Chlamydomonas reinhardtii. These mutants have short, immotile flagella with deficiencies in radial spokes, in the inner and outer arms, and in the beak-like projections in the B tubule of the outer doublet microtubules. Most dramatically, the space between the doublet microtubules and the flagellar membrane contains an unusually high number of rafts, the particles translocated by intraflagellar transport (IFT) (Kozminski, K.G., P.L. Beech, and J.L. Rosenbaum. 1995. J. Cell Biol. 131:1517–1527). IFT is a rapid bidirectional movement of rafts under the flagellar membrane along axonemal microtubules. Anterograde IFT is dependent on a kinesin whereas the motor for retrograde IFT is unknown. Anterograde IFT is normal in the LC8 mutants but retrograde IFT is absent; this undoubtedly accounts for the accumulation of rafts in the flagellum. This is the first mutation shown to specifically affect retrograde IFT; the fact that LC8 loss affects retrograde IFT strongly suggests that cytoplasmic dynein is the motor that drives this process. Concomitant with the accumulation of rafts, LC8 mutants accumulate proteins that are components of the 15-16S IFT complexes (Cole, D.G., D.R. Deiner, A.L. Himelblau, P.L. Beech, J.C. Fuster, and J.L. Rosenbaum. 1998. J. Cell Biol. 141:993–1008), confirming that these complexes are subunits of the rafts. Polystyrene microbeads are still translocated on the surface of the flagella of LC8 mutants, indicating that the motor for flagellar surface motility is different than the motor for retrograde IFT.
An M r 8,000 polypeptide, first identified as a light chain LC8 of outer arm axonemal dynein in the green alga Chlamydomonas reinhardtii (Piperno and Luck, 1979; Pfister et al., 1982; King and Patel-King, 1995), has subsequently been shown to be associated with many different complexes, including cytoplasmic dynein (King et al., 1996), the Chlamydomonas inner arm dynein I1 (Harrison et al., 1998), myosin V (Espindola, F.S., R.E. Cheney, S.M. King, D.M. Suter, and M.S. Mooseker. 1996. Mol. Biol. Cell. 7:372a), neuronal nitric oxide synthase (Jaffrey and Snyder, 1996), and a complex within the tegument of the blood fluke Schistosoma (Hoffman and Strand, 1996). The protein has been highly conserved throughout evolution, with homologues from humans, Drosophila melanogaster, Caenorhabditis elegans, and Chlamydomonas having ∼90% sequence identity (King et al., 1996). The function of the polypeptide is not known, although it appears to have a negative effect on nitric oxide synthase activity (Jaffrey and Snyder, 1996). The fact that the protein is associated with many different enzymes raises the possibility that it has a common structural or functional role in diverse polypeptide complexes.
Because LC8 is associated with at least two complexes in Chlamydomonas, Chlamydomonas provides a potentially unique and valuable system for investigating the cellular distribution and roles of LC8. Of particular importance is the ability to transform the nuclear genome of Chlamydomonas (Kindle, 1990), which makes possible insertional mutagenesis (Tam and Lefebvre, 1993; Gumpel and Purton, 1994; Pazour et al., 1995). When Chlamydomonas is transformed, the transforming DNA inserts into the genome at random, and either disrupts any gene at the site of insertion or, more commonly, causes the deletion of a block of DNA flanking the site of insertion. In either case, the result is a mutation that is tagged by the exogenous DNA and can be identified as a restriction fragment length polymorphism. If a probe for a particular gene is available, insertional mutants can be screened by Southern blotting to identify those in which the gene is deleted or disrupted. An advantage of insertional mutagenesis in Chlamydomonas is that it usually results in a null mutant. We have used this strategy to identify null mutants for LC8 in Chlamydomonas. We used a cDNA encoding LC8 (King and Patel-King, 1995) to screen a large collection of insertional mutants originally selected for defects in cell or flagellar morphology, flagellar motility, or phototaxis (Pazour et al., 1995; Koutoulis et al., 1997), and found two strains that completely lack the LC8 gene, which we here term FLA14. The mutants grow normally but have short, paralyzed flagella that become progressively shorter during the light period. The mutants are rescued by transformation with the gene for LC8, confirming that the phenotype is due to loss of that polypeptide. Electron microscopy of the mutant flagella revealed deficiencies in the inner and outer arms, radial spokes, and beak-like projections that extend into the lumens of the B tubules of outer doublet microtubules numbers 1, 5, and 6 (Witman et al., 1972; Hoops and Witman, 1983). In addition, there is an abnormal accumulation of particles in the space between the flagellar membrane and doublet microtubules. These particles appear to be identical to the “rafts” that move rapidly to the tip of the flagellum and back again during intraflagellar transport (IFT)1 (Kozminski et al., 1993, 1995). Video-enhanced differential interference contrast (DIC) microscopy revealed that the mutants have normal anterograde IFT but lack retrograde IFT, accounting for the accumulation of rafts in the flagellum. The other structural abnormalities in the axoneme may reflect a direct requirement for LC8 in the assembly of the affected components, or in the transport of the components to the flagellum or within the flagellar shaft. In any case, the results show that LC8 is essential for normal flagellar morphogenesis and stability.
Because LC8 is a subunit of cytoplasmic dynein, our findings suggest that cytoplasmic dynein is the motor for retrograde IFT. The outer arm dynein, which also contains LC8 and is potentially in the correct location to generate IFT, can be ruled out as the retrograde motor because mutants lacking the outer arms have normal IFT (Kozminski et al., 1993). LC8 is not necessary for flagellar surface motililty (Bloodgood, 1989), indicating that retrograde IFT and retrograde surface motility are powered by different motors.
Recently, novel 15-16S particles have been identified in the cytoplasmic matrix fraction of Chlamydomonas flagella (Cole, D.G., and J.L. Rosenbaum. 1996. Mol. Biol. Cell. 7:47a; Piperno and Mead, 1997; Cole et al., 1998). Concomitant with the accumulation of rafts in the flagella of LC8 mutants, we find a massive accumulation of the polypeptides that are postulated to be components of these complexes. These results confirm that the 15-16S particles are subunits of the rafts (Cole et al., 1998).
Because Chlamydomonas is advantageous for both biochemical and molecular genetic studies, the null mutants described here should be very useful for definitive identification of the retrograde IFT motor, and for understanding the role of LC8 in the functioning of this motor. The mutants also should be useful for determining the full repertoire of complexes with which LC8 is associated in Chlamydomonas, and whether LC8 plays a direct role in the assembly of structures such as the inner and outer dynein arms.
Materials and Methods
Strains
Chlamydomonas reinhardtii strains used in this work included: g1 (nit1, NIT2, agg1, mt+) (Pazour et al., 1995), 137c (nit1, nit2, mt+), CC124 (nit1, nit2, mt−), ida1 (ida1, mt+) (obtained from R. Kamiya, University of Tokyo, Tokyo, Japan), and pf18 (pf18, mt−). Strains 137c, CC124, and pf18 can be obtained from the Chlamydomonas Culture Collection (Duke University, Durham, NC). Strains produced in the course of this study included: V64 (fla14-1::NIT1, nit1, mt+) and V101 (fla14-2::NIT1, nit1, mt+), both insertional mutants obtained by transforming g1 with cloned NIT1 DNA; F5 (FLA14, fla14-1::NIT1, nit1, mt+) obtained by transforming V64 with the cloned FLA14 gene; and 2782.1 (FLA14, fla14-1::NIT1, mt−) obtained by crossing F5 to 137c.
Growth Medium
Cells were grown in the following media: M (Sager and Granick [1953] medium I altered to have 0.0022 M KH2PO4 and 0.00171 M K2HPO4), M–N (M medium without nitrogen), R (M medium plus 0.0075 M sodium acetate), and SGII/NO3 (Sager and Granick [1953] medium II modified to have 0.003 M KNO3 as the nitrogen source).
Transformation
Transformation was performed using the glass bead method of Kindle (1990) as described in Pazour et al. (1995). Insertional mutants were obtained by transforming nit1 cells (strain g1) with plasmid pGP505 (Pazour et al., 1995) which contains the Chlamydomonas nitrate reductase gene (Fernandez et al., 1989). After transformation, the cells were plated on solid SGII/NO3 medium and allowed to grow into colonies. Individual colonies were picked into 5 ml of R medium and grown until the cultures were light green in color. The cells were then examined for photoaccumulation and motility (Pazour et al., 1995; Koutoulis et al., 1997). From 2,978 transformants screened, 55 lines with defects in motility were identified. DNA was isolated from the 55 lines and examined by Southern blotting using the LC8 cDNA clone as a probe.
For experiments in which fla14 cells were rescued by transformation with cloned FLA14 genomic DNA, transformants were selected using an enrichment technique based on the ability of transformants to swim. After vortexing in the presence of the exogenous DNA, cells were placed directly into flasks containing 125 ml of liquid R medium and then allowed to grow for ∼1 wk. Untransformed cells were unable to swim and remained on the bottoms of the culture flasks, whereas transformants could swim and were distributed throughout the flasks. Transformants were observed in all flasks. After a few rounds of enrichment obtained by removing the inoculi from the tops of the cultures, most of the cells were swimming. At this point, cells were plated on solid medium; a single colony was kept from each flask.
Genetic Analysis
Mating and tetrad analysis were performed as described by Levine and Ebersold (1960) and Harris (1989). Cells of each mating type were grown on solid R medium, resuspended in M–N liquid medium, and then mixed together. After pellicles became apparent in 1 or 2 d, the mixture was plated on solid M medium, allowed to dry, and then placed in the dark for 6–10 d. Zygotes were hatched on solid R medium and dissected using a glass needle. The meiotic progeny were allowed to grow for 3–5 d and then transferred to 5 ml of liquid R medium. Cells were allowed to grow for an additional 2–5 d and then scored for motility by microscopic observation of cells illuminated with dim red light.
Analysis of IFT and Flagellar Length
IFT was observed in living cells by video-enhanced DIC microscopy as described by Moss et al. (1992), except that a 20× projection lens was used, and images were recorded on a Toshiba (Japan) PCM VHS recorder. Cells were immobilized for observation by placing them between a slide coated with a thin layer of 0.5% agarose in M medium and a coverslip. Output was recorded as an AVI file using AV Master (Fast Multimedia, Munich, Germany) to capture the data. IFT was quantitated by manually counting the number of particles passing a point about halfway along the flagellum while also measuring elapsed time.
For measurement of flagellar lengths, live cells were recorded at 2,000×. Subsequently, images of the flagella were displayed on a video monitor, traced onto transparent acetate overlays, and then measured with a flexible ruler. A slide micrometer was measured in the same way to establish the scale.
Isolation and Fractionation of Flagella
Flagella were isolated from the fla14 mutant strain V64 and from wild-type strain 137c by the dibucaine method (Witman, 1986). Isolated flagella were demembranated with 1% NP-40 and the membrane plus matrix fraction was separated from the axonemes by centrifugation (Witman, 1986). The axonemes were then washed with a solution containing 10 mM ATP in 30 mM Hepes pH 7.4, 10 mM MgSO4, 2 mM DTT, 0.5 mM EGTA, and 30 mM potassium acetate to remove any ATP-extractable components. Finally, the washed axonemes were resuspended in the same solution but without ATP. All three fractions from a particular preparation were equal in volume. Samples from different preparations were normalized by comparing the amounts of axonemal tubulin on Coomassie blue–stained gels.
Western Blotting
Flagellar fractions were separated by SDS-PAGE (Pfister et al., 1982) and electroblotted onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore Corp., Waters Chromatography, Milford, MA). Western blotting was carried out by standard procedures (Sambrook et al., 1987) using 5% nonfat dry milk in 10 mM Tris, pH 7.5, 166 mM NaCl, and 0.05% Tween to block the membrane. Primary antibodies were diluted in the blocking solution as recommended by the supplier and then hybridized for 2 h at room temperature. Horseradish peroxidase–conjugated secondary antibodies and enhanced chemiluminescence (Amersham Pharmacia Biotech. Inc., Piscataway, NJ) were used to detect the primary antibodies.
Primary antibodies used included: (a) R4058, specific for LC8 (King and Patel-King, 1995) (gift of S. King, University of Connecticut Health Center, Farmington, CT); (b) 1878β, specific for the outer arm dynein intermediate chain IC78 (King et al., 1985); (c) EU51, specific for the inner arm dynein intermediate chain IDA-IC140 (previously unpublished antibody gift of P. Yang and W. Sale, both from Emory University, Atlanta, GA); (d) 2-10-β6, specific for β tubulin (gift of G. Piperno, Mount Sinai School of Medicine, New York) (Piperno et al., 1987; King et al., 1991); (e) anti-FLA10N, specific for the anterograde IFT motor subunit FLA10 (Cole et al., 1998) (gift of D. Cole and J. Rosenbaum, both from Yale University, New Haven, CT); and (f) a mixture of mAB172.1, mAB139.1, mAB81.1, mAB81.2, mAB81.3, mAB81.4, and mAB57.1 specific for four of the raft proteins (Cole et al., 1998) (gift of D. Cole and J. Rosenbaum). Additional antibodies used were specific for radial spoke proteins RSP1 (Williams et al., 1986) and RSP3 (Williams et al., 1989) (gifts of D. Cole and J. Rosenbaum), the highly conserved kinesin sequences HIPYR and LAGSE (gift of C. Walczak, University of California, San Francisco, CA), and the molecular chaperone HSP70 (previously unpublished antibody gift of E. Savino and J. Rosenbaum, both from Yale University).
Electron Microscopy
Cells were fixed in glutaraldehyde (Hoops and Witman, 1983) and processed as described in Wilkerson et al. (1995) or Kozminski et al. (1995).
DNA Isolation and Analysis
DNA was isolated by digesting ∼0.3 ml of packed cells with 0.5 ml of proteinase K (1 mg/ml) in 5% sodium lauryl sulfate, 20 mM EDTA, and 20 mM Tris, pH 7.5, at 50°C for 12–16 h. Ammonium acetate was added to 1.5 M, the mixture extracted once with 50% phenol/50% chloroform, once with chloroform, and then precipitated with 0.7 vol of isopropyl alcohol. DNA was resuspended in a solution containing 10 mM Tris, pH 8.0, and 1 mM EDTA and then digested with PstI. Gel electrophoresis and Southern blotting were performed according to standard procedures (Sambrook et al., 1987).
Results
Identification of Mutants Deleted for the LC8 Gene
We previously used insertional mutagenesis to generate a large number of mutants with defective flagella (Koutoulis et al., 1997); these included slow-jerky swimmers, slow-smooth swimmers, uniflagellate cells, aflagellate cells, cells with paralyzed flagella, and cells with long flagella. In an effort to find mutations in the gene for LC8, we screened the slow-jerky swimming cell lines for a restriction fragment length polymorphism detectable using a cDNA clone encoding LC8. Slow-jerky swimming is indicative of outer dynein arm mutations (Kamiya, 1988), and it seemed reasonable that a defect in LC8 would affect the outer arm. However, we found no LC8 mutations in this collection, suggesting that the phenotype of an LC8 mutation could be quite different from that of other outer dynein arm mutations. We therefore expanded our search by screening all of the insertional mutants in our collection. This identified two cell lines, V64 and V101, that were completely deleted for LC8 (Fig. 1).
Figure 1.
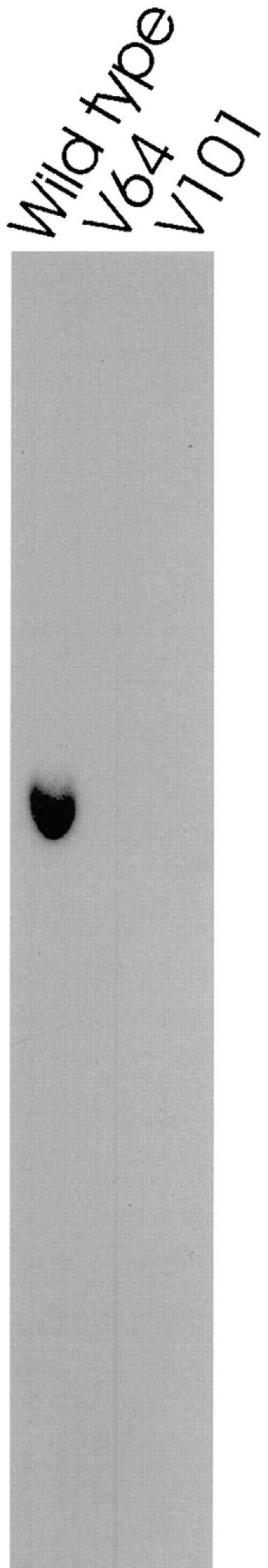
The gene encoding LC8 is deleted in two cell lines with flagellar defects. Chlamydomonas mutants with defects in motility were generated by insertional mutagenesis. DNA was isolated from each of the mutant cell lines, cleaved with PstI, and then analyzed by Southern blotting with the cDNA encoding LC8 as a probe. The probe hybridized with a single band in the lane of wild-type DNA, consistent with there being a single copy of this gene in the Chlamydomonas genome (King and Patel-King, 1995). All cell lines except V64 and V101 showed wild-type hybridization patterns. V64 and V101 had no hybridizing bands, indicating that the gene is completely deleted in these mutants.
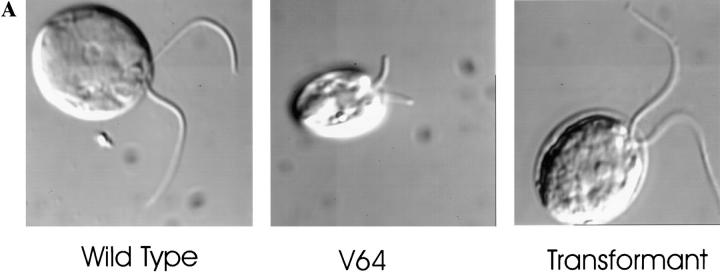
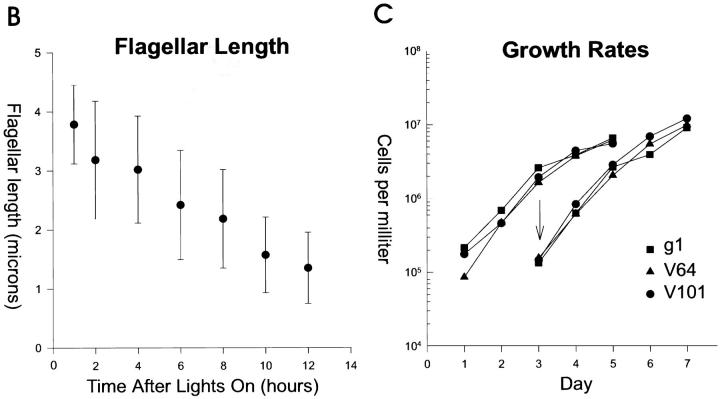
Both LC8 mutants have short, often unequal length flagella (Fig. 2 A) that sometimes have swellings on their sides or tip. The flagella are paralyzed and only rarely exhibit slight bending movements. They are longest at the beginning of the light cycle and shorten linearly at a rate of ∼0.2 μm/h as the day progresses (Fig. 2 B). The mutations do not appear to affect growth or division of the cells, since doubling times and cell body morphology are normal (Fig. 2, A and C).
Figure 2.
Phenotype of mutants lacking the LC8 gene. (A) Deletion of the LC8 gene in strain V64 (V64) causes flagella to be shorter than normal (Wild Type). After transformation of the mutant with wild-type genomic DNA encoding LC8, flagellar length is restored (Transformant). Cells were fixed with 0.1% glutaraldehyde in 10 mM Hepes and examined by DIC microscopy. (B) Deletion of the LC8 gene causes the flagella to shorten with time. V64 cells were grown synchronously on a 14-h light: 10-h dark cycle in liquid M medium with perfusion of 5% CO2. Starting 1 h after the beginning of the light cycle, samples were removed, hatched with autolysin, and then the length of one flagellum on each of 50–75 cells was measured. When the two flagella of a cell were of different lengths, the longer flagellum was measured. Data reported are mean and standard deviation. (C) Deletion of the LC8 gene does not affect growth rate. Wild-type (g1) or mutant (V64, V101) cells were grown in liquid M medium with perfusion of 5% CO2. Every 24 h a sample was removed and the cells counted with a hemocytometer. On day 3, a second set of cultures was inoculated by diluting cells from the first series to 105 cells/ml (arrow).
We attempted to use tetrad analysis to investigate if the short paralyzed flagella phenotype was linked to the LC8 deletion. The V64 and V101 cell lines did not mate well with wild-type cells and only two partial tetrads were obtained after many attempts. The mutant offspring of these tetrads were missing the LC8 gene, whereas the normal offspring contained the LC8 gene, suggesting that the phenotype was indeed linked to the LC8 deletion.
More definitive evidence that loss of LC8 was responsible for the mutant phenotype was obtained by rescuing the mutants with the cloned LC8 gene. Genomic DNA encoding LC8 was isolated from a lambda phage library and used to transform mutant cells. The lambda phage clones and subclones as small as 3.1 kb complemented the mutation, restoring flagellar length (Fig. 2 A, Transformant) and the ability to swim. To rule out the possibility that the rescuing DNA contained genes in addition to LC8, the ends of the 3.1-kb complementing fragment were sequenced (data not shown). The LC8 gene occupied ∼700 bp beginning at ∼150 bp from one end. The sequence at the other end of the clone matched internal exons of the Arabidopsis propionyl-CoA carboxylase gene. The central ∼1,200 bp of the fragment were not sequenced but presumably contained the remaining exon(s) and the 3′ untranslated region of the propionyl-CoA carboxylase gene. This leaves very little room for any other gene, making it highly unlikely that this fragment contains any full-length genes except LC8.
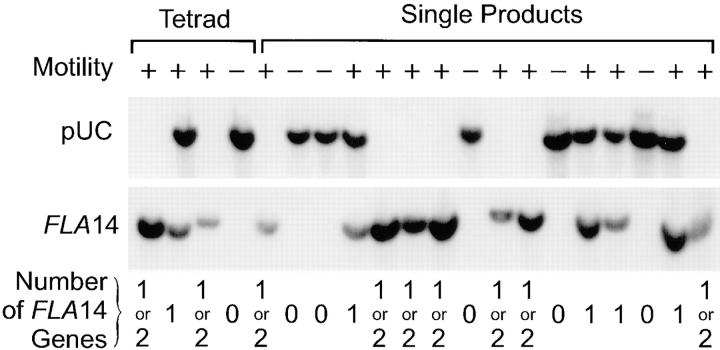
To confirm that rescue was due to integration of the LC8 gene as opposed to suppression of the phenotype by spontaneous mutation of some other gene, we examined the segregation pattern of the exogenous DNA in crosses involving the rescued mutants. One of these transformants was mated to wild-type cells and then tetrads were dissected. A motile meiotic product, deleted for the endogenous copy of the LC8 gene but carrying a transformed copy of this gene, was identified and mated to wild-type cells. Seventeen complete tetrads were obtained. The motility of the offspring was observed by light microscopy and the genotype was determined by Southern blotting (Fig. 3). Because the transformed copy of the LC8 gene is integrated at a site unlinked to the original locus, the inserted gene segregated independently of the original gene, resulting in offspring with zero, one, or two functional copies of the gene. Cells with one or two copies of the gene exhibited a wild-type phenotype, whereas those with no copies had a mutant phenotype. Thus, the defects in V64 and V101 are caused by deletion of the gene encoding LC8, and rescue was due to restoration of the LC8 gene. This gene will be termed FLA14, and the mutations have been named fla14-1 and fla14-2, respectively.
Figure 3.
Presence of the LC8 gene correlates with the wild-type phenotype in meiotic products. Strain V64 was transformed with a lambda phage clone containing the LC8 gene. Transformed cells recovered the ability to swim and were readily enriched by taking inoculi from the upper parts of unmixed cultures. After enrichment, a pure culture of one of the transformants was isolated and mated to a wild-type cell line of the opposite mating type. An offspring deleted for the endogenous LC8 gene, but carrying a transformed copy of this gene, was isolated. This was mated to a wild-type cell line of the opposite mating type. Tetrads were dissected and the offspring were scored for motility by light microscopy. DNA was isolated from each of the offspring and analyzed by Southern blotting using pUC119 (the cloning vector used in the original insertional mutagenesis) and the cDNA clone encoding LC8 as probes. pUC119 detects inserted exogenous DNA marking the deletion allele of this gene, whereas the cDNA detects the wild-type allele as well as the transformed copy. Offspring from all four of the products of one tetrad (marked in brackets) and a single product of 16 additional full tetrads were analyzed. All wild-type swimmers (+ Motility) had at least one copy of the LC8 gene, whereas the nonswimmers (− Motility) had no copies of this gene (bottom). Cells containing the deletion allele (top) were swimmers or nonswimmers depending on whether they inherited the transformed copy of the LC8 gene.
LC8 Deletion Mutants Have Severe Defects in Flagellar Ultrastructure
Electron microscopic analysis of V64 and V101 flagella revealed that the beak-like projections (Witman et al., 1972) in the B tubule of the outer doublet microtubules are missing, most of the radial spokes are missing or very defective, and the outer and inner dynein arms are reduced in number (Fig. 4). The central pair of microtubules and their projections appeared normal, although the central pair frequently is not centered in the axoneme, as previously observed in the absence of radial spokes (Witman et al., 1978).
Figure 4.
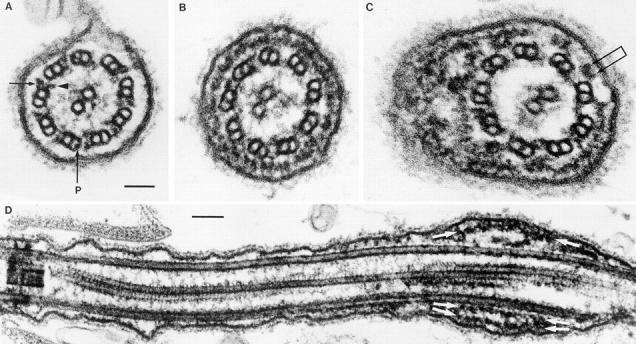
Electron micrographs of flagella from wild-type cells (A) and mutant V64 lacking the LC8 gene (B–D). Flagella of wild-type cells have prominent outer arms (arrow) and inner arms (arrowhead) and radial spokes, but the space between the axoneme and the flagellar membrane is usually devoid of material. In contrast, flagella of the LC8 mutants lack the radial spokes, most of the outer arms, and some inner arms. Due to the lack of radial spokes, the central pair of microtubules frequently is not centered in the axoneme. Mutant flagella also have a massive accumulation of material termed rafts (Kozminski et al., 1995) between the axoneme and membrane (B). In flagellar cross-sections, each raft appears as a pair of globular structures (C, conjoined arrows). Sometimes the rafts are piled up, forming a bulge on the flagellum (C and D). In D, the rafts (white arrows) are closely associated with either the flagellar membrane or the doublet microtubules; the base of the flagellum is to the left. Finally, LC8 flagella lack the projection into the lumen of the B tubule that is present in outer doublets number 1, 5, and 6 of wild type (P in panel A) (Witman et al., 1972; Hoops and Witman, 1983). Flagella were fixed for electron microscopy as described in Hoops and Witman (1983) (A–C) or Kozminski et al. (1995) (D). Bars: (A–C) 50 nm; (D) 100 nm.
The most striking feature of fla14 flagella is an unusual abundance of electron-dense material between the flagellar membrane and the outer doublet microtubules. This material looks very much like the rafts that have been correlated with the moving particles in IFT (Kozminski et al. 1993, 1995). When viewed in flagellar cross-sections, rafts appear as pairs of globular structures, each ∼20 nm in diameter (Fig. 4 C). These paired structures occasionally were observed in wild-type flagella, but in the mutant they were so abundant that they sometimes completely circumscribed the axoneme (Fig. 4, B and C). In longitudinal sections of flagella (Fig. 4 D) the rafts appeared as linear arrays of subunits, as reported by Kozminski et al. (1993). Frequently, especially at the distal tips of the flagella, the rafts were piled up, causing distension of the flagellar membrane; this presumably gives rise to the bulges seen by light microscopy. In such cases, some rafts were closely associated with the outer doublet microtubules, and others with the flagellar membrane; these associations appeared to be mediated by raft–microtubule or raft–membrane crossbridges, respectively (Fig. 5).
Figure 5.
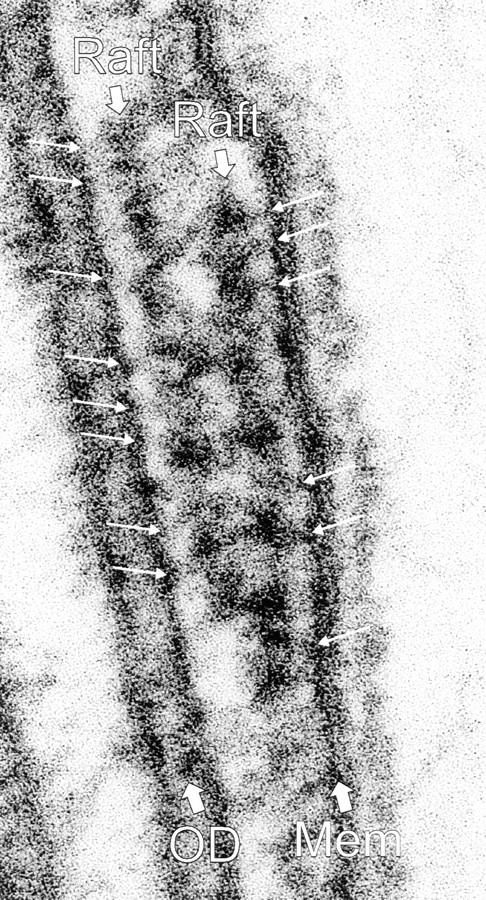
Two overlapping rafts (Raft, thick arrows) in a flagellum of an LC8 mutant. The raft on the right appears to be connected to the flagellar membrane (Mem) by links (right, thin arrows); the other appears to be connected to the outer doublet microtubule (OD) by slightly thinner crossbridges (left, thin arrows).
Retrograde IFT Is Defective in LC8 Mutants
It has been suggested that cytoplasmic dynein is the motor responsible for retrograde IFT (Kozminski et al., 1995). The fact that LC8 is a subunit of cytoplasmic dynein (King et al., 1996) prompted us to examine the flagella of the fla14 mutants to determine if IFT was affected in any way. IFT in flagella of control and mutant cells was assayed by video-enhanced DIC microscopy. In control cells, particles were observed moving in both the anterograde and retrograde directions (Fig. 6 A and Table I). Movement was smooth and continuous in each direction; the retrograde rate was slightly greater than the anterograde rate, as previously reported (Kozminski et al., 1995). In contrast, fla14 mutants almost completely lacked retrograde IFT, although anterograde IFT appeared to be normal (Fig. 6 B and Table I). No discrete particles were observed to move toward the cell body, although occasionally there appeared to be a peristaltic-like movement of material from tip to base. The rate of anterograde transport in the mutants was indistinguishable from that in control cells (Fig. 6, A and B).
Figure 6.
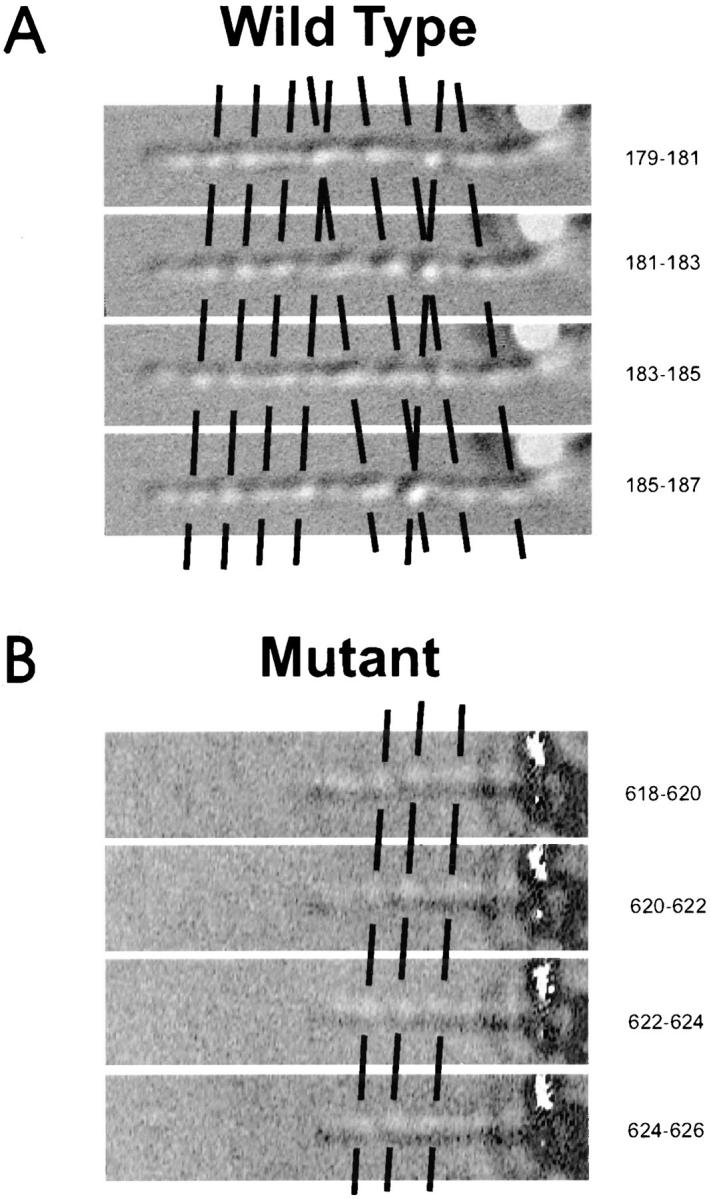
IFT examined in living cells by video-enhanced DIC microscopy. Output was recorded as an AVI file using AV Master (Fast Multimedia, Munich, Germany) to capture the data. Each series presented in this figure was produced by capturing five frames, each separated by 0.067 s. To enhance moving particles, each image (first number to right) had the subsequent image (second number to right) subtracted from it using the Adobe Photoshop subtract function with an opacity of 90%; this causes the moving particles to appear as bright objects. The flagellar tips are to the left; selected particles moving anterogradely or retrogradely are marked by diagonal lines having positive or negative slope, respectively. (A) Anterograde and retrograde IFT are observed in cells carrying a wild-type copy of the LC8 gene. The flagellum shown is from strain ida1, which lacks the inner dynein arm I1, but is wild-type for the LC8 gene and has normal IFT. The straightness of the lines indicates that movement in each direction occurred at a constant rate; the greater slope of the lines marking the retrogradely moving particles indicates that retrograde movement is faster than anterograde movement, as reported by Kozminski et al. (1995). (B) fla14 mutant (strain V64). Anterograde, but not retrograde, IFT was observed in flagella of mutants lacking the LC8 gene. Particles move anterogradely at the same rate in these cells as in ida1 cells, as indicated by the fact that the lines marking the anterogradely moving particles of both strains have the same slope.
Table I.
Loss of Retrograde IFT in Mutant V64 Lacking LC8
| Number of particles observed/min | ||||
|---|---|---|---|---|
| Anterograde | Retrograde | |||
| pf 18* | 59 ± 14 | 50 ± 10 | ||
| V64 | 52 ± 13 | 0.7 ± 1.3 | ||
Particles were counted as they moved past a point approximately halfway out on the flagellum. Values are mean ± SD.
Control. pf18 lacks the central pair of microtubules (Witman et al., 1972) but has normal IFT (Kozminski et al., 1993).
fla14 Mutations Do Not Abolish Surface Motility
In addition to beating its flagella to swim through liquid medium, Chlamydomonas also uses its flagella to glide along solid surfaces. This occurs when the flagella bind to the substrate and then glide over the substrate surface. If the flagella are oriented in opposite directions, one of the two becomes dominant and pulls the cell body and other flagellum along after it. In a process that may be related to gliding motility, Chlamydomonas flagella translocate exogenous microbeads that become bound to the outside of their flagella (Bloodgood, 1989). However, in contrast to gliding motility, where the apparent forces are unidirectional, bead movement occurs in both directions, indicating bidirectional forces.
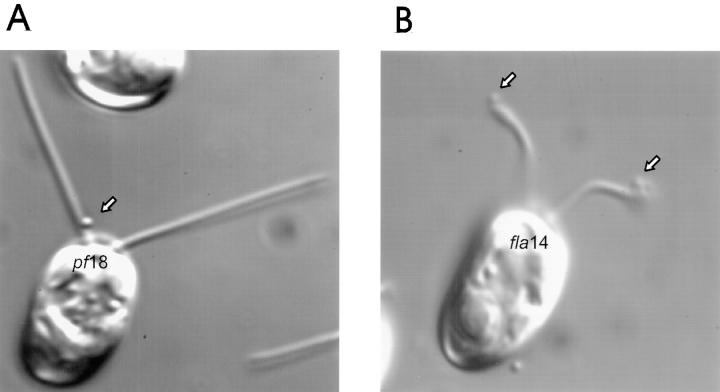
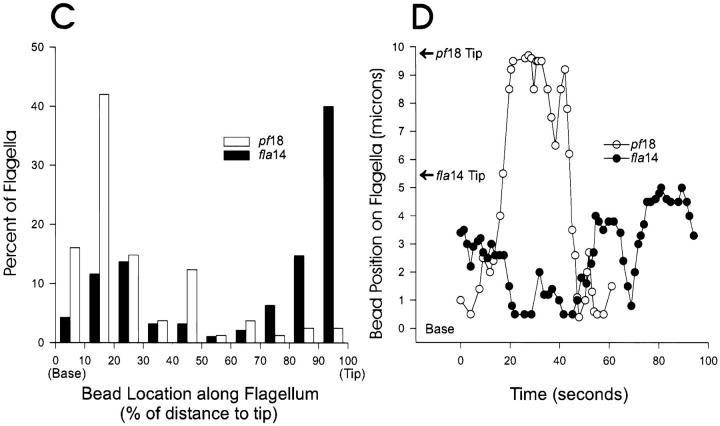
The flagella of fla14 cells do not adhere to glass and so we were unable to determine if they are capable of gliding motility. However, polystyrene microspheres bind to fla14 flagella and are translocated (Fig. 7). In pf18 cells, which have normal IFT (Kozminski et al., 1993) but have paralyzed flagella and thus facilitate observation of bead movement, the beads are seen moving along the flagella or pausing near the base of the flagella (Fig. 7, A and C, open bars). In contrast, the beads on fla14 flagella are much more likely to be found at the tips of the flagella (Fig. 7, B and C, closed bars). This is not due to a lack of retrograde movement of beads, since beads can be observed moving in both directions on fla14 flagella (Fig. 7 D, closed circles). The rates of both anterograde and retrograde bead movement (as indicated by the slopes of the lines in Fig. 7 D) are generally slower in the mutant than in wild type. It is not clear if the slower rate is directly due to the lack of LC8 or is an indirect effect related to the flagella being filled with rafts.
Figure 7.
Beads move in both directions but accumulate at the tips of fla14 mutant flagella and at the base of pf18 flagella. Polystyrene beads (Polysciences, Warrington, PA, 0.3 μm in diameter) were added to pf18 and fla14 (V64) cells. In general, beads (arrows) tended to be located at the bases of pf18 flagella (A) but at the distal tips of fla14 flagella (B). (C) Bead position was quantitated by continuously recording while moving through microscope fields and then measuring the position of bound beads with respect to the ends of the flagella. As soon as a flagellum with attached bead came into view, the position of the bead on the flagellum was measured; only one measurement was made per flagellum. On average, pf18 flagella were 9.8-μm long and fla14 flagella were 4.9-μm long. To compare data from different length flagella, the bead position was converted to percent of length and plotted as a histogram. (D) Beads bound to flagella of pf18 (open circles) and fla14 (closed circles) are actively translocated in both directions. Moving beads were observed by DIC microscopy and videotaped. In each case, the position of the bead on the flagellum was measured and plotted as a function of time. Note that beads bound to both pf18 and fla14 flagella make long, smooth runs in both directions. The pf18 sequence was atypical in that the bead was at the flagellar tip during much of the recording time.
Many Flagellar Proteins Are Affected by Deletion of the LC8 Gene
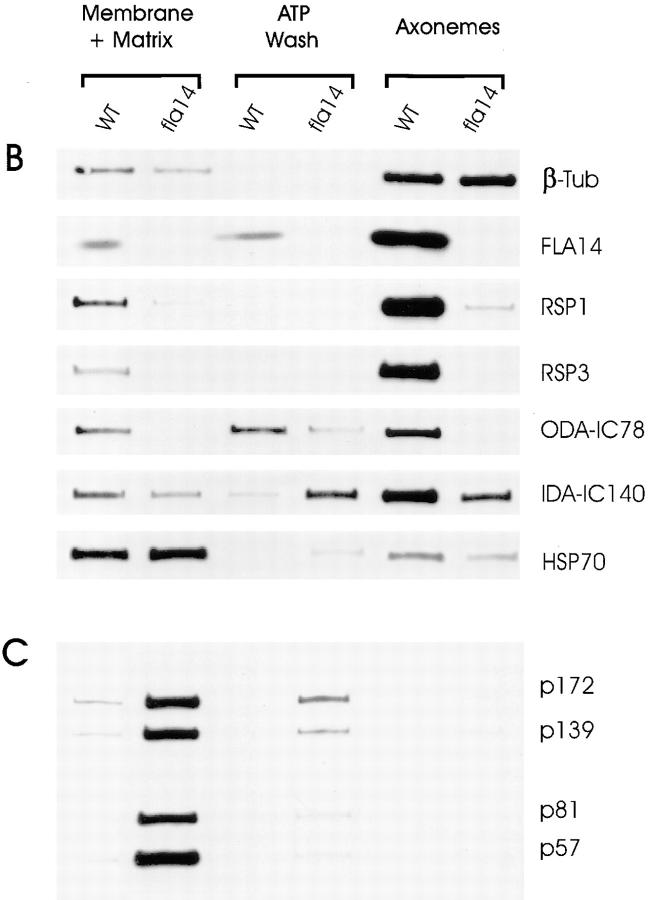
The defects in axonemal structures and the accumulation of raft-like material beneath the flagellar membrane suggested that there might be differences in the content of proteins in the various flagellar fractions of fla14 mutants as compared with wild-type cells. To investigate this, flagella were isolated from wild-type and fla14-1 cells and separated into the following three fractions: (a) “membrane plus matrix,” the detergent-soluble membrane proteins and the soluble proteins within the flagellum; (b) “ATP wash,” proteins that are bound to the axoneme only by an ATP-sensitive bond, as might be expected for active IFT motors and proteins associated with them; and (c) “axoneme,” the insoluble proteins not released by detergent or ATP. For each cell type (wild-type or mutant), the volumes of the fractions were adjusted so that equal aliquots of each fraction contained protein from an equivalent number of flagella. The proteins in these fractions were separated by SDS-PAGE and analyzed by silver staining or immunoblotting; because of the difference in flagellar length, loadings of fractions from wild-type versus mutant cells were normalized based on β tubulin content in the respective axonemal fractions (Fig. 8 B). In the membrane plus matrix fraction, a number of polypeptides were elevated in fla14 flagella (Fig. 8 A, white dots), whereas only two prominent proteins, migrating just behind tubulin in wild type, were missing from this fraction in the mutant (Fig. 8 A, black dots). In contrast, in the axoneme fraction, many proteins were decreased by the mutation.
Figure 8.
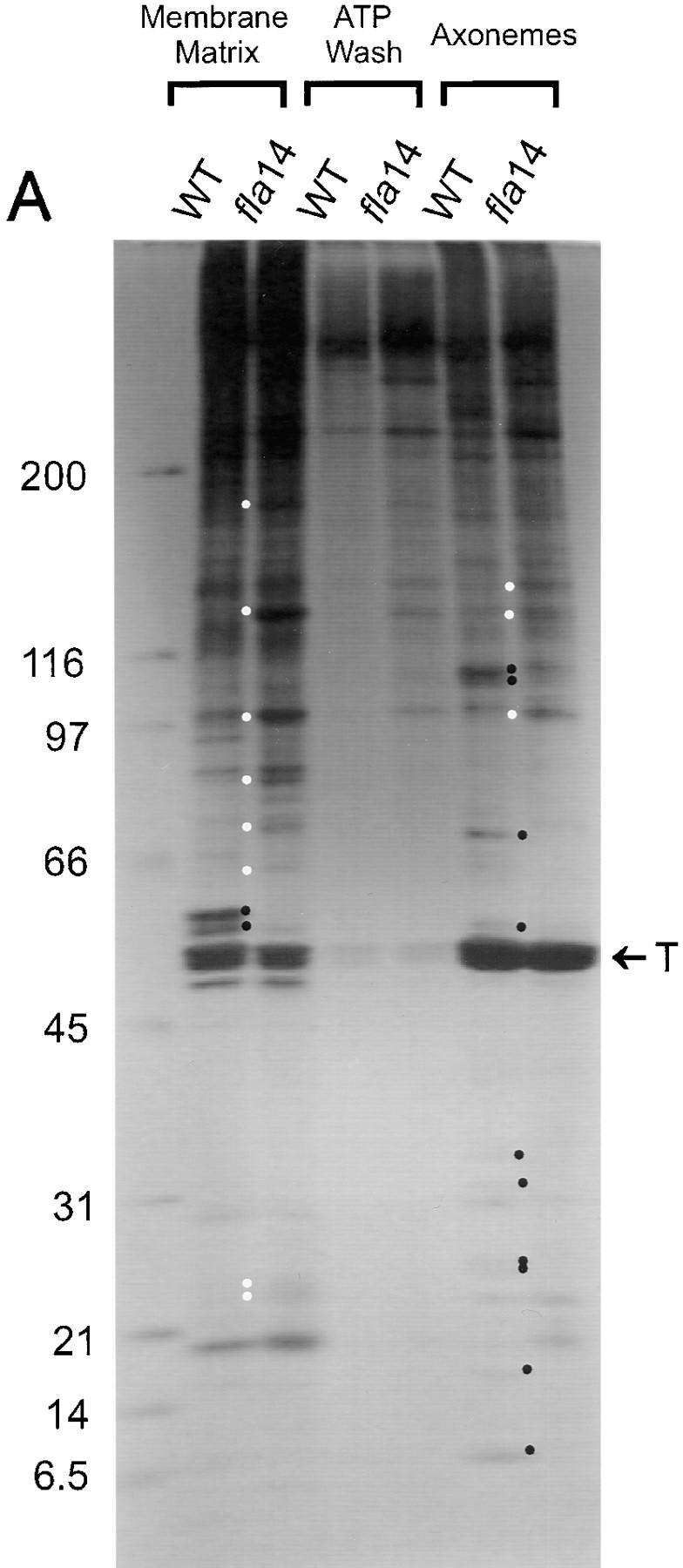
Many flagellar proteins are affected by deletion of the LC8 gene. Flagella were isolated from 137c (WT) and V64 (fla14) cells and separated into the NP-40–soluble fraction (Membrane + Matrix), the fraction of proteins released from the axoneme by 10 mM ATP (ATP Wash), and the remaining axonemal proteins (Axonemes). The proteins within these fractions were separated electrophoretically on SDS 5–15% gradient polyacrylamide gels. Loadings of wild-type and fla14 axonemes were normalized based on β tubulin content (see Fig. 8 B); for each cell type, membrane plus matrix, ATP wash, and axoneme fractions were loaded with protein from an equivalent number of flagella. (A) Silver-stained gel. Proteins that are more abundant or less abundant in fractions of fla14 as compared with wild-type flagella are marked by white or black dots, respectively. In general, the fla14 membrane plus matrix fraction had increased amounts of several proteins, whereas the fla14 axonemes exhibited reduced amounts of several other proteins. (B) Immunoblots probed with antibodies specific for LC8 (FLA14), the outer dynein arm intermediate chain IC78 (ODA-IC78), the I1 inner dynein arm intermediate chain IC140 (IDA-IC140), radial spoke stalk protein RSP1 (RSP1), radial spoke head protein RSP3 (RSP3), the molecular chaperone HSP70 (HSP70), and β tubulin (β-tub). (C) Immunoblot probed with a mixture of monoclonal antibodies against putative raft proteins p172, p139, p81, and p57 (Cole et al., 1998).
Western blotting with an antibody to LC8 showed that this protein was present in all fractions of the wild-type cells but in none of the fla14 fractions (Fig. 8 B, FLA14). This is expected as the fla14-1 allele is a complete deletion of the gene (refer to Fig. 1).
By EM, radial spokes appeared to be missing from fla14 axonemes. To confirm this result and to determine if the radial spokes had accumulated in an unassembled state in the fla14 membrane plus matrix fraction, blots were probed with antibodies to the radial spoke stalk protein RSP1 and the radial spoke head protein RSP3 (Fig. 8 B, RSP1 and RSP3). The blots revealed that these proteins are almost completely absent from the axonemes of the mutants, in agreement with the EM observations. The radial spoke proteins do not accumulate in an unassembled state in fla14 flagella, since these proteins also were reduced in the membrane plus matrix fraction of the mutant.
EM analysis indicated that the inner and outer dynein arms are reduced in number in the mutant flagella. Inasmuch as LC8 is a subunit of both outer arm dynein (King and Patel-King, 1995) and of the major inner arm dynein species I1 (Harrison et al., 1998), the absence of this subunit may affect the ability of the arms to assemble, be transported to, or attach to their normal binding sites on the doublet microtubules. Western blots using an antibody to the outer arm dynein intermediate chain IC78 showed that this protein was absent from fla14 axonemes and greatly reduced in the fla14 membrane plus matrix and ATP wash fractions (Fig. 8 B, ODA-IC78). These results are in agreement with the EM observations. It is likely that the few outer arms observed in fla14 flagella by EM are weakly attached to the axoneme and released by ATP. ATP-dependent release of at least some of the outer arms from wild-type axonemes has been described previously (Goodenough and Heuser, 1984), but the mechanism of this release is not understood. Western blots using an antibody to the inner arm I1 intermediate chain IC140 revealed a decrease in the amount of this chain in fla14 axonemes, nearly equivalent amounts of the chain in wild-type and fla14 membrane plus matrix fractions, but an increase in the amount of the chain in the ATP wash from fla14 axonemes (Fig. 8 B, IDA-IC140). The former result is consistent with our EM observations; the latter result suggests that more of the I1 inner arms are attached to the axoneme solely by ATP-dependent bonds in fla14 than in wild-type cells. The results also indicate that the deficiency for inner arm I1 is not as severe as that for the outer arm.
The large amount of raft-like material seen in the mutant flagella by EM suggested that proteins of the rafts should be elevated in the membrane plus matrix and/or ATP wash fractions of fla14 cells. These rafts are thought to be composed of two complexes (complex A and complex B) sedimenting at 15-16S and together containing at least 15 different polypeptides (Cole, D.G., and J.L. Rosenbaum. 1996. Mol. Biol. Cell. 7:47a; Piperno and Mead, 1997; Cole et al., 1998). Western blotting using monoclonal antibodies raised against one polypeptide (p139) of complex A and three polypeptides (p172, p81, and p57) of complex B (Cole et al., 1998) showed that all four proteins are greatly elevated in both the membrane plus matrix and ATP wash fractions of fla14 cells (Fig. 8 C). These findings provide further evidence that complexes A and B make up at least part of the rafts. The fact that these proteins were elevated in both fractions is consistent with the EM observation that rafts are closely associated with both flagellar outer doublet microtubules and membranes.
The molecular chaperone, HSP70, is found within the flagellum and has been suggested to play a role in targeting tubulin and other axonemal components to the tip of the flagellum where they are incorporated into the growing structure (Bloch and Johnson, 1995). Within the flagellum, HSP70 has been reported to be in both the membrane plus matrix fraction and bound to the axoneme; furthermore, part of the axonemal fraction is extractable by ATP (Bloch and Johnson, 1995). In our fractionation (Fig. 8 B), HSP70 had a distribution similar to that seen by Bloch and Johnson (1995). HSP70 also has been suggested to be a component of the 15-16S complexes believed to constitute the rafts (Piperno and Mead, 1997). We found that Hsp70 was slightly elevated in fla14 flagella, but much less so than the p172, p139, p81, and p57 components of the 15-16S complexes. Therefore, if Hsp70 is a component of either complex A or complex B, it is associated with only a subset of those particles.
Kinesin-related Proteins in fla14 Flagella
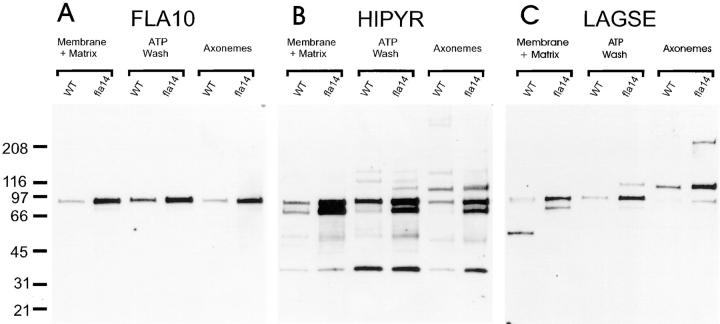
If retrograde transport is not functioning, anterograde motors may not be recycled back to the cell body and might accumulate in the flagella. The anterograde motor is thought to be a heterotrimeric kinesin, FLA10 kinesin-II (Kozminski et al., 1995; Cole et al., 1998). FLA10 kinesin-II contains two different kinesin-like motor subunits, one of which is the ∼90-kD FLA10 protein, and a slightly larger nonmotor subunit. We found that in wild-type flagella, FLA10 was most abundant in the ATP wash fraction, although it was present in the other two fractions as well (Fig. 9 A, FLA10). This protein was elevated in all fractions of fla14 flagella, consistent with the idea that the anterograde motors are not being returned to the cell body.
Figure 9.
FLA10 and several other kinesin-like proteins are affected by the deletion of LC8. Immunoblots prepared as described in Fig. 8 were probed with antisera to the anterograde IFT motor subunit FLA10 (A), and to the highly conserved kinesin sequences HIPYR (B) and LAGSE (C).
The pan-kinesin antisera, LAGSE and HIPYR, detect several other proteins within the Chlamydomonas flagella (Fox et al., 1994; Johnson et al., 1994). To determine how these are affected by fla14, Western blots were probed with these sera. Both LAGSE and HIPYR detect a FLA10-sized protein and a slightly smaller protein (Fig. 9, B and C) that are greatly elevated in fla14 flagella. This smaller protein may be the 85-kD motor subunit of the heterotrimeric FLA10 kinesin-II (Cole et al., 1998). However, the relative amounts of the two bands varied in the wild-type fractions, suggesting that more than one species is represented. There also is a >200-kD axonemal protein (detected by LAGSE), a ∼110-kD protein (detected by both LAGSE and HIPYR in the ATP wash and the axoneme), and a ∼40-kD protein (detected by HIPYR in the ATP wash and axoneme fractions) that are elevated by the mutation. Other proteins at ∼150 and ∼125 kD (detected by HIPYR in the ATP wash) are not greatly affected by the fla14 mutation. Interestingly, one protein at ∼55 kD (detected by LAGSE) in the membrane plus matrix fraction of wild type is completely missing in the mutant. This may correspond to one of the two proteins indicated by black dots in the membrane plus matrix fraction of Fig. 8 A.
Discussion
Phenotype of LC8 Deletion Mutants
We have isolated mutant cell lines of Chlamydomonas that are missing the gene encoding LC8, first identified as a subunit of outer arm dynein (Piperno and Luck, 1979; Pfister et al., 1982; King and Patel-King, 1995). Although this chain also is a subunit of cytoplasmic dynein (King and Patel-King, 1995), and has been reported to be associated with mammalian myosin V (Espindola, F.S., R.E. Cheney, S.M. King, D.M. Suter, and M.S. Mooseker. 1996. Mol. Biol. Cell. 7:37a) and neuronal nitric oxide synthase (Jaffrey and Snyder, 1996), deletion of the gene from Chlamydomonas does not have a detectable effect on cell growth or gross cell morphology. Therefore, the gene is not required for any critical cellular process in Chlamydomonas. However, loss of the gene, which we have named FLA14, does cause severe flagellar defects. Flagella of the mutant cells are short, unstable, and exhibit only slight bending movements. They are defective in retrograde IFT and are missing or have deficiencies in several axonemal structures. In addition, the space between the axonemal microtubules and the flagellar membrane of the mutants is filled with an abnormal amount of electron-dense material. This material appears to be identical to the rafts, described by Kozminski et al. (1993, 1995), that have been correlated with the moving particles of IFT. Consistent with the EM observations, there is an increase in many proteins in the fla14 membrane and matrix fraction, and a decrease in other proteins in the fla14 axonemal fraction, relative to wild-type.
LC8 Is Required for Assembly of Multiple Axonemal Components
Both EM and Western blotting of flagellar fractions demonstrate that fla14 mutants have greatly reduced numbers of outer dynein arms. This is most likely a direct result of the loss of LC8, which is an outer arm subunit and, like the outer arm dynein heavy and intermediate chains (Witman et al., 1994), may be required for arm assembly or attachment to the doublet microtubule.
LC8 also has been reported to be a subunit of inner arm I1 (Harrison et al., 1998). As with the outer arm, both EM and Western blotting indicate that there is a reduced number of inner arms in fla14 axonemes, although the deficiency is not as severe as that for the outer arm. The 140-kD intermediate chain of inner arm I1 is unique among the axonemal proteins that we examined in that it is elevated in the ATP-wash fraction of fla14 mutants relative to that of wild type. This could be because a substantial portion of I1 arms are more weakly attached than normal to their correct sites on the doublet microtubules of fla14 flagella, perhaps due to their deficiency in LC8. Alternatively, the I1 arms extracted by ATP could have been unassembled arms which were in transit to the tip of the flagellum (see below) and became bound directly or indirectly to the axonemal microtubules by rigor bonds after flagellar detachment.
Particularly intriguing is the nearly total loss of the radial spokes from fla14 axonemes. Similarly, fla14 axonemes are missing the beak-like projections present in the lumens of the B tubules of doublet microtubules 1, 5, and 6 (Witman et al., 1972; Hoops and Witman, 1983). A possible explanation is that LC8 also is a subunit of these components, and is directly required for their assembly and/or attachment to the doublet microtubules. In this context, it may be relevant that axonemes contain a pool of LC8 not associated with either the outer or inner dynein arms (Benashski, S.E., A. Harrison, and S.M. King, unpublished data, cited in Benashski et al., 1997).
The above explanations presume that LC8 is a subunit of four different axonemal structures, and is necessary for all four structures to assemble or bind to the doublet microtubules. An alternative explanation for the loss of inner arms, radial spokes, and beak-like projections in fla14 mutants is that the absence of LC8 affects transport of these structures in the flagellum. Both the radial spokes (Johnson and Rosenbaum, 1992) and the inner arms (Piperno et al., 1996) must be transported to the tip of the flagellum before binding to their normal sites on the doublet microtubules. In dikaryon rescue experiments (Segal et al., 1984), the beak-like projections are restored to fully formed flagella of mutants specifically lacking the projections, raising the possibility that these structures also are transported to the tip of the flagellum and then move back down the lumen of the B tubule to their binding sites. At least for the inner arms, movement to the tip of the flagellum is dependent upon FLA10 (Piperno et al., 1996), one of the subunits of the anterograde IFT motor (Walther et al., 1994; Kozminski et al., 1995). If defects in the fla14 flagellum prevented this transport, deficiencies in assembly of inner arms, radial spokes, and perhaps the beak-like projections would result. Although anterograde movement of rafts appears to proceed unimpaired in fla14 flagella, the movement of other components may be impeded.
It also is possible that loss of the structures from fla14 axonemes is due to a defect in the movement of axonemal precursors into the flagellum, or to the base of the flagellum before their transport into the flagellum. In the accompanying paper, Cole et al. (1998) show that FLA10 kinesin-II and the IFT particle proteins are highly concentrated in the cell body near the basal bodies, and point out that, inasmuch as the minus ends of the cytoplasmic microtubules terminate near the basal bodies, this accumulation may involve a retrograde microtubule motor such as cytoplasmic dynein. If the loss of LC8 impairs transport of axonemal precursors to the base of the flagellum, the complexes may not enter the flagellum in sufficient quantities to build a complete axoneme. Indeed, neither outer arm nor radial spoke proteins accumulate in fla14 flagella (refer to Fig. 8 B), consistent with there being a block to entry of these structures into the flagellum.
Retrograde IFT Is Dependent on LC8
A dramatic difference between wild-type and fla14 flagella is the presence of massive amounts of raft material at the tip of the fla14 flagella. This undoubtedly is due to the loss of retrograde IFT in the mutant; because anterograde IFT is present, the rafts are transported to the distal end of the flagellum where they accumulate.
Recently, two new complexes, sedimenting at 15-16S and together containing ∼15 different polypeptides, have been found in the membrane and matrix fraction of Chlamydomonas flagella (Cole, D.G., and J.L. Rosenbaum. 1996. Mol. Biol. Cell. 7:47a; Piperno and Mead, 1997; Cole et al., 1998). These complexes are lost from the flagella concomitantly with the anterograde motor FLA10 kinesin-II when the temperature-sensitive mutant fla10 is shifted to nonpermissive temperatures. As a result, it has been proposed that the complexes are the cargo of the FLA10 kinesin-II motor. Because the rafts are moved by IFT (Kozminski et al., 1995), the 15-16S complexes may be subunits of the rafts.
The fact that the rafts are highly enriched in fla14 flagella provided an opportunity to test this hypothesis. Using antibodies specific for four different polypeptides of complexes A and B, we found that all four of these proteins were greatly elevated in fla14 flagella. As expected, the proteins were present primarily in the membrane plus matrix fraction. These results strongly support the notion that the 15-16S complexes are raft components (Cole et al., 1998). That anterograde IFT continues unabated in the fla14 mutants despite the accumulation of large numbers of rafts at the tip of the flagella suggests that the cell body contains a large pool of raft components. This is consistent with immunolocalization studies showing that proteins of the 15-16S complexes are highly enriched in the basal body region (Cole et al., 1998).
Cytoplasmic Dynein Is Likely To Be the Motor for Retrograde IFT
Retrograde IFT moves particles toward the base of the flagellum, where the minus ends of the flagellar microtubules are located. The motor driving retrograde IFT is unknown, but has been suggested to be either cytoplasmic dynein or a kinesin that moves towards the minus ends of microtubules (Kozminski et al., 1995). Inasmuch as cytoplasmic dynein contains LC8 (King et al., 1996), our finding that deletion of the LC8 gene disrupts retrograde IFT points to cytoplasmic dynein as the most likely motor. It should be noted that loss of the outer arms and inner arms in the LC8 mutants cannot be responsible for the loss of retrograde IFT, since IFT is normal in mutants that specifically lack either or both of these structures (Kozminski, 1993, and Fig. 6 A of this article).
The involvement of cytoplasmic dynein in retrograde IFT is further supported by studies of C. elegans mutants. In the nematode, the ends of the sensory neurons are modified cilia (Perkins et al., 1986; White et al., 1986). Defects in the C. elegans CHE-3 gene, which encodes a dynein heavy chain (DHC) (Grant, W., personal communication, and see below), cause the tips of the ciliary neurons to become swollen and packed with electron-dense material (Lewis and Hodgkin, 1977; Albert et al., 1981). The OSM-6 gene product, which is homologous to the p52 polypeptide of the Chlamydomonas 15-16S complex B (Cole et al., 1998), is massively accumulated at the tips of the che-3 neurons (Collet et al., 1998). Therefore, the electron-dense material in the tips of the C. elegans che-3 neurons is likely to correspond to the rafts of Chlamydomonas flagella. The ciliated tips of the neurons also are much shorter in che-3 than in wild-type worms (Lewis and Hodgkin, 1977). Thus, the che-3 mutation in C. elegans appears to cause a phenotype closely related to that reported here for the Chlamydomonas fla14 mutant.
The occurrence of similar defects in organisms as evolutionarily distant as Chlamydomonas and Caenorhabditis suggest that the underlying process—presumably retrograde IFT—is widely distributed among organisms. Likewise, the occurrence of similar defects in organelles as different as the motile flagellum and the sensory neuron suggests that IFT is of importance in a wide range of cilia-based structures. This is likely to include the rods and cones of the vertebrate retina, which are modified cilia and contain a FLA10 kinesin-II homologue (Beech et al., 1996).
Because C. elegans has no motile cilia and the doublet microtubules of the ciliary neurons lack dynein arms, the CHE-3 gene product must be a component of cytoplasmic dynein. In a phylogenetic tree based on the predicted amino acid sequences of members of the DHC family, the C. elegans CHE-3 gene product clustered with the DHC1b isoform from human, rat, mouse, sea urchin, and Tetrahymena (Pazour, G., and G. Witman, unpublished data). DHC1b is more closely related to the conventional cytoplasmic DHC isoform (DHC1a) than to known axonemal DHCs (Gibbons et al., 1994; Tanaka et al., 1995; Porter et al., 1996), yet its expression is induced by deciliation (Gibbons et al., 1994) and it accumulates at the apical ends of ciliated cells (Criswell et al., 1996). A likely explanation for these observations is that DHC1b is a cytoplasmic dynein that operates in cilia and flagella. Retrograde IFT may be powered specifically by DHC1b.
Evidence that defects in LC8 cause defects in cytoplasmic dynein function is provided by a comparison of LC8 mutants and cytoplasmic dynein mutants in the fly Drosophila melanogaster and in the fungus Aspergillus nidulans. In Drosophila, strong LC8 alleles are embryonic lethals, whereas weak alleles cause sterility and defects in the wings and bristles (Dick et al., 1996; Phillis et al., 1996). Similarly, DHC1a deletions are homozygous lethals, whereas weaker alleles result in sterility and bristle and eye defects (Gepner et al., 1996). In Aspergillus, LC8 (nudG) and cytoplasmic DHC (nudA) mutations both affect nuclear migration (Xiang et al., 1994; Beckwith, S.M., and N.R. Morris. 1995. Mol. Biol. Cell. 6:5a). Thus, in both Drosophila and Aspergillus, the phenotypes of LC8 mutations closely resemble those of cytoplasmic DHC mutations. Assuming that LC8 is associated with both DHC1a and DHC1b, defects in LC8 are likely to affect the functions of both isoforms.
The above arguments notwithstanding, one cannot yet rule out a role for a minus-end–directed kinesin in retrograde IFT. Chlamydomonas flagella contain several proteins that react with antisera against highly conserved kinesin sequences. One of these is an ∼55-kD membrane plus matrix protein missing in fla14 flagella (Fig. 9 C), a pattern that might be expected for the IFT motor. Further study will be necessary to determine if this is a motor protein.
Retrograde IFT and Retrograde Surface Motility Are Powered by Different Motors
The discovery of IFT (Kozminski et al., 1993) raised the possibility that the movement of beads in flagellar surface motility (Bloodgood, 1989) simply reflected the movement of rafts to which the beads had become attached via a transmembrane protein. Previous experiments to resolve this have yielded ambiguous results. Both IFT and bead movement ceased together within 60–90 min after elevation of fla10 cells to nonpermissive temperature, indicating that both were dependent upon the anterograde IFT motor (Kozminski et al., 1995); however, this dependency might be due to a requirement for IFT to deliver the surface motor to the tip of the flagellum. Both IFT and bead movement were inhibited by NaCl and sucrose (Kozminski et al., 1993); this inhibition most likely was due to an osmotic affect, which could have inhibited more than one process. Rafts and polystyrene beads are transported at different rates (Kozminski et al., 1993), but this may be due to differences in the viscous drag acting on particles of vastly different sizes. Low Ca2+ and EGTA blocked bead movement without affecting IFT (Kozminski et al., 1993), although this might have been due to uncoupling of beads from rafts. Our observation that retrograde bead movement continues in fla14 flagella even in the absence of retrograde IFT provides the strongest evidence to date that these two types of movement are powered by different motors. Possible candidates for the retrograde surface motility motor include the kinesin-like proteins that are not affected by the loss of LC8 (Fig. 9).
The loss of LC8 does result in an abnormal accumulation of beads at the distal tip of the fla14 flagellum. Assuming that the motor responsible for bead movement is microtubule-based, this may occur because the accumulation of rafts at the tip of the flagellum pushes the membrane so far away from the axoneme that the surface motility motor can no longer span the gap between membrane and microtubules.
The Role of IFT
Observations on Chlamydomonas defective in the anterograde IFT motor FLA10 kinesin-II have shown that IFT is essential for flagellar assembly. As discussed above, anterograde IFT may be necessary for transporting specific axonemal components to the flagellar tip before their assembly into the axoneme. Whether retrograde IFT itself has a role in flagellar assembly is less clear. Although the flagella of fla14 mutants are short and paralyzed, this could be a secondary consequence of loss of the radial spokes and inner and outer arms. Mutants with specific defects in the radial spokes have paralyzed flagella (Witman et al., 1978), and double mutants with defects in both inner and outer arms have short flagella and are nonmotile (Brokaw and Kamiya, 1987; Kamiya et al., 1989; Kurimoto and Kamiya, 1991). It also is possible that axonemal assembly and flagellar growth in fla14 strains are not affected by loss of retrograde IFT per se, but by a defect in the transport of axonemal precursors in the cytoplasm to the region of the basal body, or from there into the flagellum (see above).
In related work, Morris and Scholey (1997) showed that microinjection of kinesin-II antibodies into fertilized sea urchin eggs blocked ciliogenesis. The injected embryos assembled only short, immotile flagella that lacked a central pair. It is likely that this is due to an effect of the antibody on IFT, and that IFT also is necessary for cilia formation in sea urchin cells.
Although the importance of IFT in flagellar assembly is established, the purpose of this process in nongrowing flagella is less well understood. When fla10 cells are shifted to nonpermissive temperatures, their flagella slowly shorten. Similarly, the flagella of fla14 cells shorten throughout the day. Therefore, both anterograde and retrograde IFT are necessary for maintaining flagellar length. IFT may supply some component essential for axonemal stability or maintenance. Perhaps the continuous active movement of large numbers of rafts to and fro in the confined space between the doublet microtubules and the flagellar membrane stirs the flagellar matrix, facilitating mass transport of molecules along the flagellar shaft.
It is not clear why the flagella of fla14 cells form within the mother cell wall, but then shorten after hatching. The findings reported here indicate that retrograde IFT is necessary for returning the rafts to the base of the flagellum. By extension, retrograde IFT is likely to be involved in recycling of other components that are transported to the tip of the flagellum. The lack of recycling of certain components may result in a deficiency that accumulates with time, eventually affecting flagellar stability.
IFT also is likely to have an important role in the maintenance of the flagellar membrane. A 65-kD membrane protein is one of the major species transported to the flagellum of nonregenerating Chlamydomonas cells (Remillard and Witman, 1982). Turnover of membrane proteins probably represents replacement of proteins lost by blebbing of membrane vesicles from the flagellar tip (Bergman et al., 1975), and failure to maintain the membrane could have deleterious effects on axonemal stability.
Future Directions
Because LC8 is associated with multiple polypeptide complexes, it is not surprising that deletion of the LC8 gene has pleiotrophic effects. Importantly, loss of the chain is not lethal in Chlamydomonas. As a result, it should be quite feasible to learn more about the role of LC8 and the complexes of which it is a part in this organism. It also should now be possible to isolate mutants with defects in other polypeptides of the retrograde motor; these should permit the effects of loss of retrograde IFT per se to be distinguished from other effects due to loss of LC8.
Acknowledgments
We thank J. Aghajanian (Worcester Foundation for Biomedical Research, Shrewsbury, MA) for electron microscopy, and D. Cole and J. Rosenbaum for discussion and for sharing unpublished results. Antibodies were graciously provided by D. Cole and J. Rosenbaum, P. Yang and W. Sale, S. King, G. Piperno, and C. Walczak.
These studies were supported by grants from the National Institutes of Health (GM30626) and the Campbell and Hall Charity Fund.
Abbreviations used in this paper
- DHC
dynein heavy chain
- DIC
differential interference contrast
- IFT
intraflagellar transport
Note Added in Proof
We recently have identified an insertional mutant of Chlamydomonas in which the gene for the dynein heavy chain isoform DHC1b is deleted. The mutant has short flagella, confirming that DHC1b is important for flagellar assembly.
Footnotes
Address all correspondence to George Witman, Department of Cell Biology, UMMC, Worcester Foundation Campus, 222 Maple Ave., Shrewsbury, MA 01545. Tel.: (508) 842-8921 Ext. 344. Fax: (508) 842-3915. E-mail: witman@sci.wfbr.edu
C. Wilkerson's present address is Michigan State University, Department of Energy Plant Research Laboratory and Department of Botany and Plant Pathology, Michigan State University, East Lansing, MI 48824-1312.
References
- Albert PS, Brown S, Riddle DL. Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol. 1981;198:435–451. doi: 10.1002/cne.901980305. [DOI] [PubMed] [Google Scholar]
- Beech PS, Pagh-Roehl K, Noda Y, Hirokawa N, Burnside B, Rosenbaum JL. Localization of kinesin superfamily proteins to the connecting cilium of fish photoreceptors. J Cell Sci. 1996;109:889–897. doi: 10.1242/jcs.109.4.889. [DOI] [PubMed] [Google Scholar]
- Benashski SE, Harrison A, Patel-King RS, King SM. Dimerization of the highly conserved light chain shared by dynein and myosin V. J Biol Chem. 1997;272:20929–20935. doi: 10.1074/jbc.272.33.20929. [DOI] [PubMed] [Google Scholar]
- Bergman K, Goodenough UW, Goodenough DA, Jawitz J, Martin H. Gametic differentiation in Chlamydomonas reinhardtii. II. Flagellar membranes and the agglutination reaction. J Cell Biol. 1975;67:606–622. doi: 10.1083/jcb.67.3.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch M, Johnson KA. Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J Cell Sci. 1995;108:3541–3545. doi: 10.1242/jcs.108.11.3541. [DOI] [PubMed] [Google Scholar]
- Bloodgood, R.A. 1989. Gliding Motility and Flagellar Glycoprotein Dynamics in Chlamydomonas. In Ciliary and Flagellar Membranes. R. Bloodgood, editor. Plenum Press, New York. 91–128.
- Brokaw C, Kamiya R. Bending patterns of Chlamydomonasflagella. IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskel. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II–dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis eleganssensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. . Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell PS, Ostrowski LE, Asai DJ. A novel cytoplasmic dynein heavy chain: expression of DHC1b in mammalian ciliated epithelial cells. J Cell Sci. 1996;109:1891–1898. doi: 10.1242/jcs.109.7.1891. [DOI] [PubMed] [Google Scholar]
- Dick T, Ray K, Salz HS, Chia W. Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. . Mol Cell Biol. 1996;16:1966–1977. doi: 10.1128/mcb.16.5.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez E, Schnell R, Ranum LPW, Hussey SC, Silflow CD, Lefebvre PA. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. . Proc Natl Acad Sci USA. 1989;86:6449–6453. doi: 10.1073/pnas.86.17.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox LA, Sawin K, Sale WS. Kinesin-related proteins in eukaryotic flagella. J Cell Sci. 1994;107:1545–1550. doi: 10.1242/jcs.107.6.1545. [DOI] [PubMed] [Google Scholar]
- Gepner J, Li M, Ludmann S, Kortas C, Boylan K, Iyadurai SJP, McGrail M, Hays TS. Cytoplasmic dynein function is essential in Drosophila melanogaster. . Genetics. 1996;142:865–878. doi: 10.1093/genetics/142.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BH, Asai DJ, Tang W-JY, Hays TS, Gibbons IR. Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol Biol Cell. 1994;5:57–70. doi: 10.1091/mbc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Heuser J. Structural comparison of purified dynein proteins with in situdynein arms. J Mol Biol. 1984;180:1083–1118. doi: 10.1016/0022-2836(84)90272-9. [DOI] [PubMed] [Google Scholar]
- Gumpel NJ, Purton S. Playing tag with Chlamydomonas. . Trends Cell Biol. 1994;4:299–301. doi: 10.1016/0962-8924(94)90222-4. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. 1989. The Chlamydomonas Sourcebook. Academic Press, Inc., San Diego, CA. 780 pp. [DOI] [PubMed]
- Harrison A, Olds-Clarke P, King SM. Identification of the tcomplex–encoded cytoplasmic dynein light chain Tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J Cell Biol. 1998;140:1137–1147. doi: 10.1083/jcb.140.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann KF, Strand M. Molecular identification of a Schistosoma mansonitegumental protein with similarity to cytoplasmic dynein light chains. J Biol Chem. 1996;271:26117–26123. doi: 10.1074/jbc.271.42.26117. [DOI] [PubMed] [Google Scholar]
- Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonasflagella. J Cell Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. PIN: An associated protein inhibitor of neuronal nitric oxide synthase. Science. 1996;274:774–777. doi: 10.1126/science.274.5288.774. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. . J Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Haas MA, Rosenbaum JL. Localization of a kinesin-related protein to the central pair apparatus of the Chlamydomonas reinhardtiiflagellum. J Cell Sci. 1994;107:1551–1556. doi: 10.1242/jcs.107.6.1551. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. . J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, R., E. Kurimoto, H. Sakakibara, and T. Okagaki. 1989. A genetic approach to the function of inner and outer arm dynein. In Cell Movement. Vol. 1. The Dynein ATPases. F. Warner, P. Satir, and I. Gibbons, editors. Alan R. Liss, Inc., New York. 209–218.
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. . Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Patel-King RS. The M r = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonasflagella have cytoplasmic homologues. J Biol Chem. 1995;270:11445–11452. doi: 10.1074/jbc.270.19.11445. [DOI] [PubMed] [Google Scholar]
- King SM, Otter T, Witman GB. Characterization of monoclonal antibodies against Chlamydomonasflagellar dyneins by high-resolution protein blotting. Proc Natl Acad Sci USA. 1985;82:4717–4721. doi: 10.1073/pnas.82.14.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Wilkerson CG, Witman GB. The M r 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with α-tubulin in situ. . J Biol Chem. 1991;266:8401–8407. [PubMed] [Google Scholar]
- King SM, Barbarese E, Dillman JF, III, Patel-King RS, Carson JH, Pfister KK. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved M r8,000 light chain. J Biol Chem. 1996;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- Koutoulis A, Pazour GJ, Wilkerson CG, Inaba K, Sheng H, Takada S, Witman GB. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J Cell Biol. 1997;137:1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonaskinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto E, Kamiya R. Microtubule sliding in flagellar axonemes of Chlamydomonasmutants missing inner- or outer-arm dynein: Velocity measurements on new types of mutants by an improved method. Cell Motil Cytoskel. 1991;19:275–281. doi: 10.1002/cm.970190406. [DOI] [PubMed] [Google Scholar]
- Levine RP, Ebersold WT. The genetics and cytology of Chlamydomonas. . Annu Rev Microbiol. 1960;14:197–216. doi: 10.1146/annurev.mi.14.100160.001213. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Hodgkin JA. Specific neuroanatomical changes in chemosensory mutants of the nematode Caenorhabditis elegans. . Comp Neur. 1977;172:489–510. doi: 10.1002/cne.901720306. [DOI] [PubMed] [Google Scholar]
- Morris RL, Scholey JM. Heterotrimeric kinesin-II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss AG, Gatti J-L, Witman GB. The motile β/IC1 subunit of sea urchin sperm outer arm dynein does not form a rigor bond. J Cell Biol. 1992;118:1177–1188. doi: 10.1083/jcb.118.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Sineshchekov OA, Witman GB. Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. . J Cell Biol. 1995;131:427–440. doi: 10.1083/jcb.131.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. . Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Pfister KK, Fay RB, Witman GB. Purification and polypeptide composition of dynein ATPases from Chlamydomonasflagella. Cell Motil. 1982;2:525–547. doi: 10.1002/cm.970020604. [DOI] [PubMed] [Google Scholar]
- Phillis R, Statton D, Caruccio P, Murphey RK. Mutations in the 8 kDa dynein light chain gene disrupt sensory axon projection in the Drosophilaimaginal CNS. Development. 1996;122:2955–2963. doi: 10.1242/dev.122.10.2955. [DOI] [PubMed] [Google Scholar]
- Piperno G, Luck DJL. Axonemal adenosine triphosphatases from flagella of Chlamydomonas reinhardtii. Purification of two dyneins. J Biol Chem. 1979;254:3084–3090. [PubMed] [Google Scholar]
- Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonasflagella. Proc Natl Acad Sci USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang X-j. Microtubules containing acetylated α-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1Fla10 to reach the distal part of the flagella in Chlamydomonas. . J Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M, Knott JA, Myster SH, Farlow SJ. The dynein gene family in Chlamydomonas reinhardtii. . Genetics. 1996;144:569–585. doi: 10.1093/genetics/144.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remillard SP, Witman GB. Synthesis, transport, and use of specific flagellar proteins during flagellar regeneration in Chlamydomonas. . J Cell Biol. 1982;93:615–631. doi: 10.1083/jcb.93.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies with Chlamydomonas reinhardtii. . Annu NY Acad Sci. 1953;56:831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1987. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 545 pp.
- Segal RA, Huang B, Ramanis Z, Luck DJL. Mutant strains of Chlamydomonas reinhardtiithat move backwards only. J Cell Biol. 1984;98:2026–2034. doi: 10.1083/jcb.98.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L-W, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtiiby DNA insertional mutagenesis. Genetics. 1993;135:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Zhang Z, Hirokawa N. Identification and molecular evolution of new dynein-like protein sequences in rat brain. J Cell Sci. 1995;108:1883–1893. doi: 10.1242/jcs.108.5.1883. [DOI] [PubMed] [Google Scholar]
- Walther Z, Vashishtha M, Hall JL. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J.G., E. Southgate, J.N. Thomson, and S. Brenner. 1986. The nervous system of Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. 314B:1–340. [DOI] [PubMed]
- Wilkerson CG, King SM, Koutoulis A, Pazour GJ, Witman GB. The 78,000 M r intermediate chain of Chlamydomonasouter arm dynein is a WD-repeat protein required for arm assembly. J Cell Biol. 1995;129:169–178. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Mitchell DR, Rosenbaum JL. Molecular cloning and expression of flagellar radial spoke and dynein genes of Chlamydomonas. . J Cell Biol. 1986;103:1–11. doi: 10.1083/jcb.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Velleca MA, Curry AM, Rosenbaum JL. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: Flagellar mutation pf-14is an ochre allele. J Cell Biol. 1989;109:235–245. doi: 10.1083/jcb.109.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonasflagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Witman GB, Carlson K, Berliner J, Rosenbaum JL. Chlamydomonasflagella. J Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB, Plummer J, Sander G. Chlamydomonasflagellar mutants lacking radial spokes and central tubules. J Cell Biol. 1978;76:729–747. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B., C.G. Wilkerson, and S.M. King. 1994. The biochemistry, genetics, and molecular biology of flagellar dynein. In Microtubules. J. Hyams and C. Lloyd, editors. Wiley-Liss, Inc., New York. 229–249.
- Xiang X, Beckwith S, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. . Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]