Abstract
In Sciara, unfertilized embryos initiate parthenogenetic development without centrosomes. By comparing these embryos with normal fertilized embryos, spindle assembly and other microtubule-based events can be examined in the presence and absence of centrosomes. In both cases, functional mitotic spindles are formed that successfully proceed through anaphase and telophase, forming two daughter nuclei separated by a midbody. The spindles assembled without centrosomes are anastral, and it is likely that their microtubules are nucleated at or near the chromosomes. These spindles undergo anaphase B and successfully segregate sister chromosomes. However, without centrosomes the distance between the daughter nuclei in the next interphase is greatly reduced. This suggests that centrosomes are required to maintain nuclear spacing during the telophase to interphase transition. As in Drosophila, the initial embryonic divisions of Sciara are synchronous and syncytial. The nuclei in fertilized centrosome-bearing embryos maintain an even distribution as they divide and migrate to the cortex. In contrast, as division proceeds in embryos lacking centrosomes, nuclei collide and form large irregularly shaped nuclear clusters. These nuclei are not evenly distributed and never successfully migrate to the cortex. This phenotype is probably a direct result of a failure to form astral microtubules in parthenogenetic embryos lacking centrosomes. These results indicate that the primary function of centrosomes is to provide astral microtubules for proper nuclear spacing and migration during the syncytial divisions. Fertilized Sciara embryos produce a large population of centrosomes not associated with nuclei. These free centrosomes do not form spindles or migrate to the cortex and replicate at a significantly reduced rate. This suggests that the centrosome must maintain a proper association with the nucleus for migration and normal replication to occur.
Centrosomes nucleate microtubule assembly and determine the number, length, and overall distribution of microtubules within the cell. Consequently, centrosomes influence the position and distribution of many cellular organelles including the nucleus. An animal centrosome typically contains a pair of centrioles formed from nine triplet microtubules arranged in a short cylinder surrounded by an electron-dense amorphous pericentriolar material (Kellogg et al., 1994). Centrosomes nucleate microtubules in a polarized array with their minus ends originating in the pericentriolar material and their fast growing plus ends directed outward. Centrosomes duplicate precisely once each cell cycle, and in mitosis the daughter centrosomes separate to define the poles of the mitotic spindle (for reviews see Kellogg et al., 1994; Balczon, 1996).
Although a number of studies demonstrate that the centrosomes nucleate and organize microtubule arrays, recent findings indicate that the cell maintains other means of nucleating microtubules and organizing spindles. For example, plasmid DNA attached to beads organizes a bipolar spindle in mitotic Xenopus extracts (Heald et al., 1996). These spindles assemble from microtubules nucleated near the chromatin, which are sorted into arrays. In a step requiring dynein, the minus ends of the microtubule arrays are bundled into focused poles, resulting in the formation of bipolar spindles with the plus end of the microtubules radiating toward the chromosomes.
In animal cells, examining centrosome function in vivo requires a means of completely eliminating centrosome function. One approach is to analyze specialized cell types that lack centrosomes. Female meiosis in Drosophila, for example, occurs without centrosomes (Theurkauf and Hawley, 1992; McKim and Hawley, 1995; Endow and Komma, 1997). To examine this issue in cells that normally possess centrosomes, an alternative approach is to remove the centrosome by micromanipulation. By this procedure, it was shown that intact centrosomes are required for spindle formation in grasshopper spermatocytes (Zhang and Nicklas, 1995a ,b, 1996). In the work presented here, we have taken advantage of the fact that unfertilized Sciara coprophila embryos are able to initiate development (Ruder et al., 1987). In addition, we found that sperm supplies the centrosomes in Sciara embryos. Therefore, the unfertilized embryos provide a means of analyzing mitosis, spindle assembly, and other early developmental events in the absence of a centrosome.
The early divisions in fertilized Sciara embryos are syncytial, and the nuclei divide without accompanying cytokinesis. During the initial five divisions, the nuclei divide synchronously and maintain an even distribution as they migrate to the cortex. Once at the cortex, the nuclei continue dividing synchronously, and nuclear cycles progressively lengthen until cellularization (de Saint Phalle and Sullivan, 1996). The fertilized embryo has two populations of centrosomes as a consequence of an unusual feature of the male germ-line. In the spermatogonial divisions, a giant centriole containing 60–90 singlet microtubules forms the basal body of the sperm flagellum (Phillips, 1967). We show that at fertilization, the giant basal body gives rise to multiple microtubule-organizing centers (MTOCs).1 These include the centrosomes associated with the dividing nuclei and a large population of free MTOCs.
Our studies demonstrate that unfertilized Sciara embryos initiate syncytial development without functional centrosomes. During mitosis, anastral spindles form from microtubule nucleation at or near the chromosomes. These spindles are functional, and sister chromosomes successfully segregate during anaphase. In contrast, fertilized Sciara embryos form spindles in a more conventional manner with microtubule nucleation initiating at the centrosomes. Although spindles formed without centrosomes undergo anaphase B, the distance between sister nuclei at the following interphase is much less than for spindles formed with centrosomes. Also in contrast to fertilized embryos, the syncytial nuclei in unfertilized embryos are not evenly distributed, undergo extensive fusions, and do not migrate to the cortex. It is likely that the astral microtubules, which do not form in unfertilized embryos lacking centrosomes, are essential for maintaining an even nuclear distribution and for cortical migration.
These results provide in vivo support for in vitro analysis demonstrating that a common cytoplasm can support two modes of spindle assembly (Heald et al., 1997; Merdes and Cleveland, 1997). In the absence of centrosomes, chromosomes serve as the primary organizer of the bipolar spindle. However, if centrosomes are present, they become the primary organizer of the spindle. Our results also demonstrate that the latter stages of mitosis, anaphase and telophase, can occur in the absence of centrosomes.
Materials and Methods
Strains and Egg Collection
Sciara coprophila strains 7298 and 6980 were kindly provided by Dr. Susan Gerbi (Brown University, Providence, RI). Unfertilized eggs were collected by decapitating virgin females, which promotes egg laying (Ruder et al., 1987). This procedure was successful with ∼15% of the females. The unfertilized eggs produced by decapitation have the same phenotype as a subset of eggs found in collections taken from live fertilized females, suggesting that decapitation produces normal unfertilized eggs.
Fluorescent Analysis
Embryos were fixed by a modification of the Mitchison and Sedat (1983) procedure and of procedures described elsewhere (Sullivan et al., 1990). After dechorionation with 50% Clorox for 90 s, embryos were washed in 0.4% NaCl and 0.03% Triton X-100 and placed in heptane to fenestrate the vitelline membrane (Karr and Alberts, 1986). Embryos were fixed by adding an equal volume of methanol to the heptane and shaking to fix and devitellinize. Fixed embryos were stored in methanol at 4°C for up to 20 wk before staining. Embryos were stained with 1 μg/ml propidium iodide (Sigma Chemical Co., St. Louis, MO) to visualize the DNA. Immunofluorescent analysis was used to visualize microtubules and performed according to the procedures described in Karr and Alberts (1986) and Sullivan et al. (1993).
In Situ Hybridization
After dechorionation with 50% Clorox for 90 s, embryos were washed in 0.4% NaCl and 0.03% Triton X-100 and placed in heptane to fenestrate the vitelline membrane (Karr and Alberts, 1986). Embryos were fixed by adding an equal volume of 10-10-10 fixative (10% each of 37% formaldehyde, 5 M EGTA, and 10× PBS) to the heptane for 20 min, removing the fixative, and shaking with an equal volume of methanol to devitellinize. We used a DNA probe that hybridizes specifically to the Sciara rDNA on the X chromosome, immediately adjacent to the centromere (Renkawitz et al., 1979). Cloned Sciara rDNA was kindly provided by Dr. Susan Gerbi. This probe was used for whole mount in situ hybridization according to the procedure described by Hiraoka et al. (1993).
Microscopy
Microscopy was performed using an inverted photoscope (model IMT2; Olympus, Melville, NY) equipped with a laser confocal imaging system (model 600; Bio-Rad Labs, Hercules, CA). Confocal images were imported into Adobe Photoshop 3.0.5 (San Jose, CA), and composite plates were prepared. Images of embryos were not edited except in the preparation of Fig. 3, which shows the syncytial development of fertilized and unfertilized embryos. In this figure, images of fertilized embryos were edited by removing polar bodies and condensed debris left over from chromosome elimination, while images of unfertilized embryos had the contrast increased so interior nuclei were visible.
Figure 3.
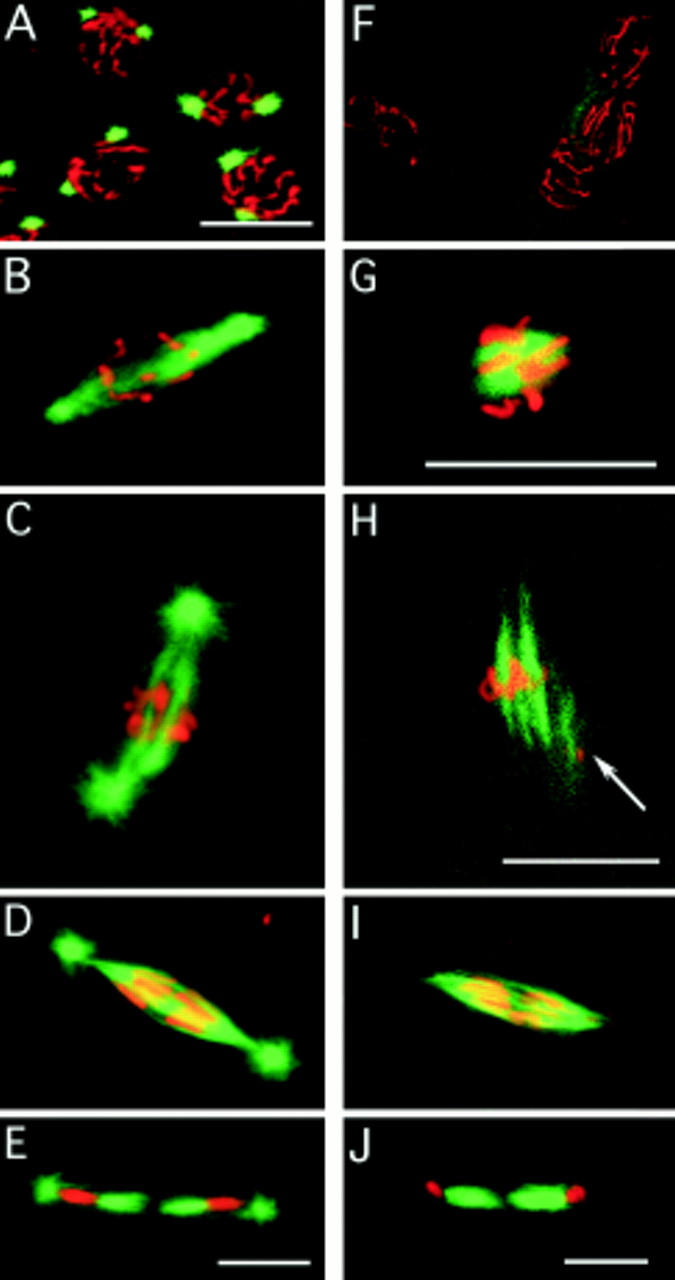
Spindle assembly in the absence of centrosomes. Embryos are double stained for their DNA (red) and microtubules (green). Fertilized embryos: (A) In prophase, the chromosomes are condensed on the nuclear membrane, and the centrosomes have migrated to approximately a 180 degree position. (B) After nuclear membrane breakdown, the centrosomes of fertilized embryos nucleate microtubules that are preferentially directed toward the chromosomes. (C) Metaphase fertilized embryos have a spindle between the two centrosomes. (D) In early anaphase, the spindle is sharply pointed, and the chromosomes are starting to segregate. (E) In telophase, the midbody (central spindle) has formed between the two daughter nuclei of the division. Unfertilized embryos: (F) In prophase, the chromosomes are condensed on the nuclear membrane, but there are no MTOCs. (G) Tubulin is apparent on and near the chromosomes as the spindle starts to assemble. (H) Arrays of microtubules arise from the chromatin. The arrow marks a second metaphase nucleus partly in the plane of focus with a spindle also assembling. The close apposition of nuclei is typical of unfertilized embryos. (I) The appearance of the anaphase spindle is very similar to the anaphase spindle of fertilized embryos, except that centrosomes with astral microtubules are conspicuously absent. (J) In telophase, the midbody (central spindle) has formed between the two daughter nuclei of the division, even though there are no centrosomes. Bars, 10 μm.
Free centrosomes were counted using optical sections of embryos double stained for their DNA and microtubules. Confocal images of medial sections on both the rhodamine (DNA) and fluorescein (tubulin) channels were taken at 5-μm intervals on the z-axis of each embryo and analyzed in Adobe Photoshop.
The three-dimensional location of the nuclei of fertilized and unfertilized embryos in cycles 3–4 were determined from confocal images of embryos stained for their DNA with propidium iodide. The x and y coordinates were obtained from the Comos Length/Profile function, and the z coordinate was obtained from the focus motor setting. For each embryo, the point at the center of the nuclei (CN) was found by averaging the coordinates. Embryos that did not have the proper number of nuclei (four in cycle 3, eight in cycle 4) because of clumping were not scored. Computations involving the coordinates were done using Excel 4.0 (Microsoft Corp., Redmond, WA).
Results
Unfertilized Sciara Embryos Initiate Parthenogenetic Development
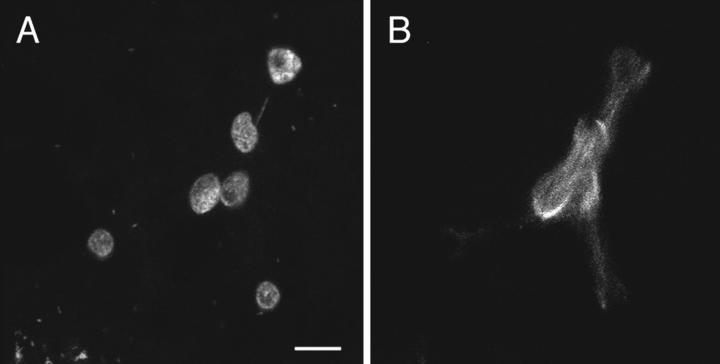
Unfertilized Sciara eggs complete meiosis and initiate rounds of mitotic divisions. As will be described, this parthenogenetic development is ultimately unsuccessful, and none of the embryos cellularize. In fertilized Sciara embryos, fusion of the male and female pronuclei occurs at 1.4 h after egg deposition. Embryos reach cycle 3 (four nuclei at interphase) and cycle 8 (128 nuclei at interphase) at ∼2.3 and 4.9 h after egg deposition (de Saint Phalle and Sullivan, 1996). The number of nuclei in 0–5-h collections of unfertilized embryos were scored to determine the percentage of Sciara embryos initiating parthenogenetic development. The results of this analysis are shown in Table I. 19% of the embryos had no detectable DNA staining and could not be classified. 9% of the embryos had one to four nuclei in the interior but no polar bodies at the cortex. In Table I, these are scored as undergoing meiosis. 19% of the embryos possessed one to four interior nuclei in addition to cortically localized polar bodies. 34% of the embryos possessed from 8 to greater than 32 nuclei. This class clearly demonstrates that the nuclei in the unfertilized embryos are undergoing multiple rounds of nuclear division. 13% of embryos had a highly disorganized pattern with patches of intense nuclear staining in the interior of the embryo. This phenotype is only observed in older collections, indicating that it is the terminal phenotype of unfertilized embryos that initiate parthenogenesis. Of 121 unfertilized embryos aged more than 5 h, 78% exhibited this terminal phenotype, 7% had one to four nuclei, and the remainder had no detectable DNA staining. This indicates that 78% of the unfertilized embryos undergo multiple rounds of nuclear division. The phenotypes are shown in Fig. 1 (right-hand panels).
Table I.
The Development of Unfertilized Embryos
| No stain | Meiosis | 1–4 nuclei | 6–8 nuclei | 8–32 nuclei | 32+ nuclei | Terminal (vesicles) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–5-h collection | 19 | 9 | 19 | 6 | 32 | 2 | 13 | |||||||
| (88 embryos) | ||||||||||||||
| 5-h plus collection | 15 | 0 | 7 | 0 | 0 | 0 | 78 | |||||||
| (121 embryos) |
Entries are percent of total embryos.
Figure 1.
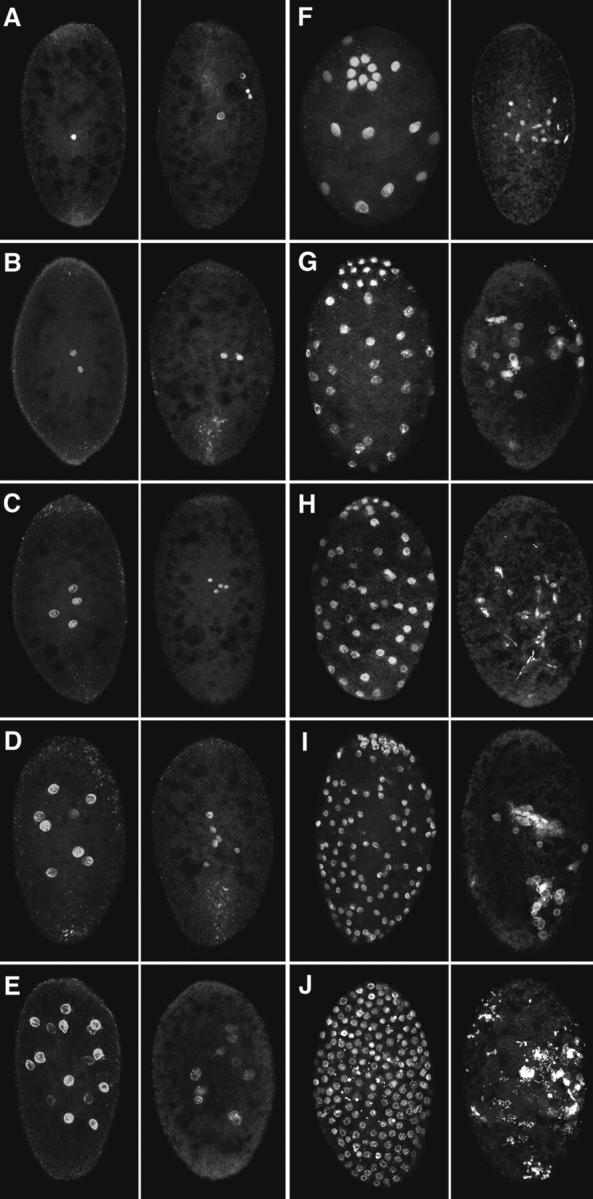
Unfertilized embryos undergo many rounds of nuclear division but exhibit an abnormal nuclear distribution. Syncytial development in fertilized (left-hand panels) and unfertilized (right-hand panels) embryos stained for their DNA. Cycles 1–10 are depicted in A–J. Because of extensive nuclear clumping, the cycles after nuclear cycle 5 in the unfertilized embryos are only estimates. Approximate width of the embryos is 150 μm, length 200 μm.
Unfertilized Embryos Lack Functional Centrosomes
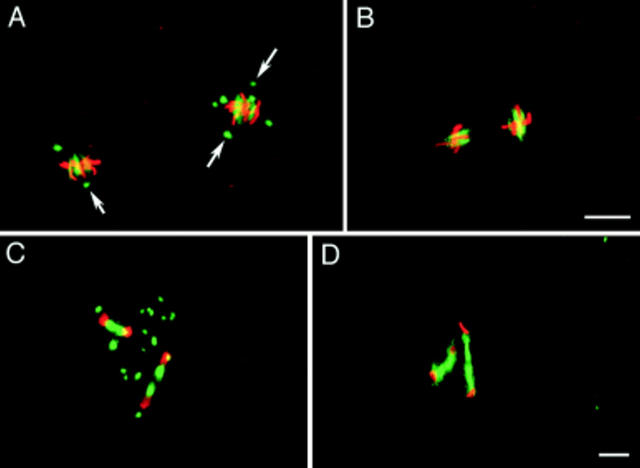
Fig. 2 A depicts a fertilized embryo in metaphase of cycle 2. The arrows indicate centrosomes located at the poles of the spindles. As will be described, fertilized Sciara embryos have a large population of MTOCs not associated with nuclei. These account for the additional MTOCs. Fig. 2 B depicts an unfertilized embryo. Although the chromosomes have condensed on the metaphase plate and spindles are beginning to form, no MTOCs are present. Fig. 2, C and D, depicts fertilized and unfertilized embryos in telophase of nuclear cycle 2. In both, the central spindle (midbody) is positioned between the daughter telophase nuclei. However, in the unfertilized embryo there are no MTOCs associated with the nuclei nor are there free MTOCs. Therefore, the centrosomes associated with the nuclei and free MTOCs are of paternal origin.
Figure 2.
Unfertilized embryos do not possess MTOCs. Nuclear cycle 2 embryos are stained for their DNA (red) and microtubules (green). (A) In a fertilized embryo, two of the MTOCs are associated with spindle poles while the others surround the metaphase plate. (B) In the equivalently staged unfertilized embryo, no MTOCs are present, and by counting the chromosome arms it can be seen that the nuclei are haploid. This may be why unfertilized nuclei seem smaller (see also Fig. 1). (C and D) Telophase of nuclear cycle 2 in fertilized and unfertilized embryos, respectively. While both exhibit robust midbodies, no MTOCs are present in the unfertilized embryo. Bars, 10 μm.
The Mitotic Spindle Is Assembled without Centrosomes in Unfertilized Embryos
The typical stages of spindle assembly in fertilized and unfertilized embryos double stained for their microtubules (green) and DNA (red) are shown in Fig. 3. Fig. 3, A–E, depicts spindle assembly in fertilized embryos, and Fig. 3, F–J, depicts spindle assembly in unfertilized embryos. Fig. 3 A depicts prophase in a fertilized embryo. The chromosomes are condensing, and the centrosomes are at opposite poles of the nucleus. During the initial stages of spindle assembly after nuclear envelope breakdown, the centrosomes nucleate microtubules that are preferentially directed toward the chromosomes before the chromosomes have fully aligned on the metaphase plate (Fig. 3 B). At metaphase, a robust spindle forms between the two opposing centrosomes (Fig. 3 C). In early anaphase, the chromosomes are observed segregating toward the spindle poles, marked by the presence of centrosomes with astral microtubules (Fig. 3 D). In telophase, the two daughter nuclei are separated by a robust midbody (Fig. 3 E).
In unfertilized embryos, mitotic spindles are anastral and appear to initiate at the chromosomes. During prophase, the chromosomes exhibit normal chromosome condensation but no centrosome-based MTOCs (Fig. 3 F). The first stage of spindle formation is marked by a concentration of microtubules around the chromosomes (Fig. 3 G). In the next stage of spindle formation, microtubule arrays are bundled into a bipolar spindle (Fig. 3 H). However, these spindles do not exhibit distinct MTOCs at the poles. Although these spindles lack functional centrosomes, they proceed into anaphase and successfully segregate sister chromosomes (Fig. 3 I), and a midbody forms between the two telophase daughter nuclei (Fig. 3 J).
To examine the fidelity of chromosome segregation in the absence of centrosomes, we analyzed the first division of unfertilized embryos, when there was no possibility of generating abnormal nuclei through collisions. We looked for DNA that might have been left at the metaphase plate. In 69 embryos examined, no chromatin fragments between the daughter nuclei were observed. With this assay, we found no evidence that chromosome segregation in mitosis is less accurate in spindles assembled without centrosomes than in normal spindles assembled with centrosomes. The last section in the results provides further support for this conclusion.
Spindle Length Increases as the Nuclei in Unfertilized Embryos Progress through Anaphase
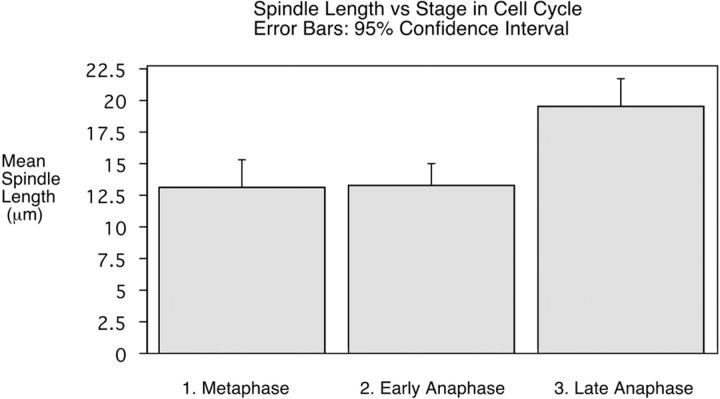
The average length of the spindle at metaphase in unfertilized embryos is 13.15 μm (n = 8, SD 2.65 μm). These are averages of measurements determined to 0.1 μm. In early anaphase, when the centromeres have separated but there is no gap between the segregating chromosomes, the average spindle length is 13.32 μm (n = 12, SD 2.58 μm). In late anaphase, when a gap has appeared between the segregating chromosomes, the average spindle length is 19.55 μm (n = 17, SD 4.20 μm). The difference in the mean length of the metaphase and early anaphase spindle is not significant. (The probability that it occurred by chance is 0.89 by an unpaired t test.) But the increase in mean length as the spindle progresses through anaphase is statistically highly significant (0.0001 by the unpaired t test). These data, shown in Fig. 4, indicate that anaphase B occurs in the absence of centrosomes in early Sciara embryos.
Figure 4.
Anaphase B elongation of the spindle in unfertilized embryos. Spindle length was measured in fixed embryos. The average length of the spindle at metaphase is 13.15 μm (n = 8, SD 2.65 μm). In early anaphase, when the centromeres have separated but there is no gap between the segregating chromosomes, the average spindle length is 13.32 μm (n = 12, SD 2.58 μm). In late anaphase, when a gap has appeared between the segregating chromosomes, the average spindle length is 19.55 μm (n = 17, SD 4.20 μm). The difference between the length of early anaphase and late anaphase spindles was statistically highly significant (P = 0.0001 by an unpaired t test). The mean length of the spindle in late anaphase is likely to be an underestimate because for the majority of the spindles measured, the chromosomes had not yet reached the poles.
Nuclear Migration and Spacing Is Defective in Unfertilized Embryos
The distribution of nuclei in fertilized and unfertilized embryos at equivalent stages of syncytial development is shown in Fig. 1. During nuclear cycles 1 and 2 (Fig. 1, A and B), there are no obvious differences between the fertilized and unfertilized embryos. However, during nuclear cycle 3, although there are the correct number of nuclei in the unfertilized embryo, they are abnormally distributed (Fig. 1 C). Relative to the fertilized embryos, the four nuclei are in a much tighter cluster. Abnormalities in the nuclear distribution become more extreme as nuclear division continues in the unfertilized embryos and large clusters of nuclei are observed (Fig. 1, F–J, right-hand panels). Irregularly sized and shaped nuclei result from the collision of the products of neighboring nuclear divisions.
In fertilized embryos, migration of the nuclei occurs from nuclear cycles 2 to 6. In the unfertilized embryos, nuclear migration never occurs. Fig. 5 represents an enlarged image of the unfertilized embryo depicted in Fig. 1 D (right). The figure shows three pairs of sister telophase nuclei. The products of each division are well separated, but the divisions are oriented so that one nucleus from each dividing pair will collide with one another. This pattern of nuclear division is never observed in fertilized embryos. The terminal phenotype of unfertilized embryos is characterized by large nuclear clusters irregularly distributed throughout the interior of the embryo with few or no nuclei at the cortex (Fig. 1 J, right).
Figure 5.
In unfertilized embryos, the spindles are improperly oriented and the products of neighboring nuclear divisions collide with one another. An unfertilized embryo in telophase of nuclear cycle 3, stained for both DNA (A) and tubulin (B). The spindles are in a single plane and are oriented so that the nuclear products from three separate divisions will collide with one another. This is a magnified view of the embryo depicted in Fig. 1 D. Bar, 10 μm.
To quantify these spacing defects, we determined the three-dimensional position of each nucleus with respect to its neighboring nuclei for 10 fertilized and 10 unfertilized embryos in nuclear cycles 2, 3, and 4. The average distances between the two nuclei in cycle 2 fertilized and unfertilized embryos were 31.2 μm (n = 10, SD 12.2 μm) and 17.4 μm (n = 10, SD 6.3 μm), respectively. This suggests that centrosomes influence the separation of the products of the first mitotic division.
For nuclear cycle 3 (four nuclei) and cycle 4 (eight nuclei), we quantified the overall nuclear distribution of each embryo by finding the point at the center of the group of nuclei and determining the average distance of the nuclei to this point. We call this point the “center of the nuclei” (CN), and its coordinates are the average of the x, y, and z coordinates of all the nuclei in the embryo. The average distance to the CN is a measure of the overall nuclear distribution in an embryo. For 10 fertilized and 10 unfertilized cycle 3 embryos, the averages of this value were 26.9 μm (n = 10, SD 6.6 μm) and 16.0 μm (n = 10, SD 3.5 μm), respectively. For 10 fertilized and 10 unfertilized cycle 4 embryos, the corresponding values were 35.8 μm (n = 10, SD 4.6 μm) and 19.3 μm (n = 10, SD 3.3 μm), respectively. There was some overlap in the range of these values between fertilized and unfertilized embryos in cycle 3, but in cycle 4 there was none. The nuclei of unfertilized embryos fail to spread out as much as the nuclei of fertilized embryos even in the first divisions.
Fertilized Sciara Embryos Have Extra MTOCs at Pronuclear Fusion
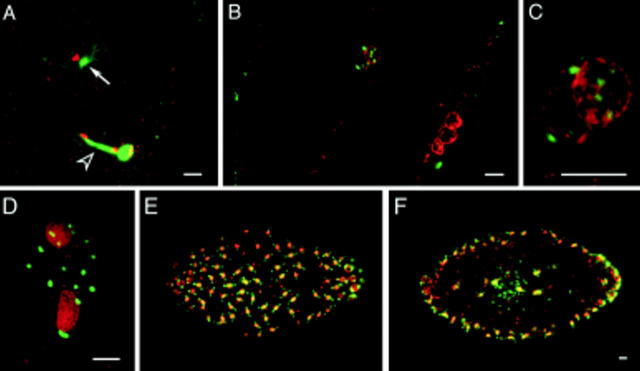
Oogenesis in Sciara coprophila and Drosophila melanogaster are similar in that mature but unactivated oocytes are arrested in metaphase of meiosis I. Upon fertilization in Sciara, the male pronucleus migrates toward the center of the embryo and retains a closely associated MTOC, indicating the presence of a centrosome (Fig. 6 A, solid arrow). At this time, telophase of female meiosis II is still in progress, and the midbody separating the meiotic products is prominent (Fig. 6 A, open arrowhead). The female meiotic products do not possess MTOCs. Sciara is unusual because multiple MTOCs are closely associated with the pronuclei during syngamy. These probably originate from the breakdown of the unusually large basal body of the Sciara sperm (see last section of Results). Fig. 6 B shows multiple MTOCs surrounding male and female pronuclei in the process of fusing. The fusing pronuclei are shown at higher magnification in Fig. 6 C with the asters of eight MTOCs clearly visible. There are no MTOCs associated with the polar bodies. By observing multiple focal planes, we find that 6–10 MTOCs surround the fused pronuclei. Directly after the first mitotic division, the two nuclei have numerous free MTOCs between them (Fig. 6 D). Free MTOCs also surround the nuclei at the second division, but by the third division the dividing nuclei migrate away from the free MTOCs (data not shown).
Figure 6.
Free MTOCs are present in fertilized Sciara embryos. These embryos are double stained for their DNA (red) and microtubules (green). (A) During telophase of female meiosis II, the centrosome associated with the decondensing male pronucleus is nucleating a large aster (arrow), and a large midbody separates the female pronucleus and the second polar body. (B) The asters of eight MTOCs are in the plane of focus surrounding the fusing pronuclei. Three polar bodies reside at the cortex. (C) Magnified view of pronuclei. (D) During interphase of nuclear cycle 2, the two nuclei are encompassed by 16 MTOCs. E and F depict surface and medial views of a nuclear cycle 8 embryo and demonstrate that the free MTOCs do not migrate to the cortex. Bars, 10 μm.
Free MTOCs Remain in the Interior of the Embryo and Undergo at Least One Round of Duplication or Splitting
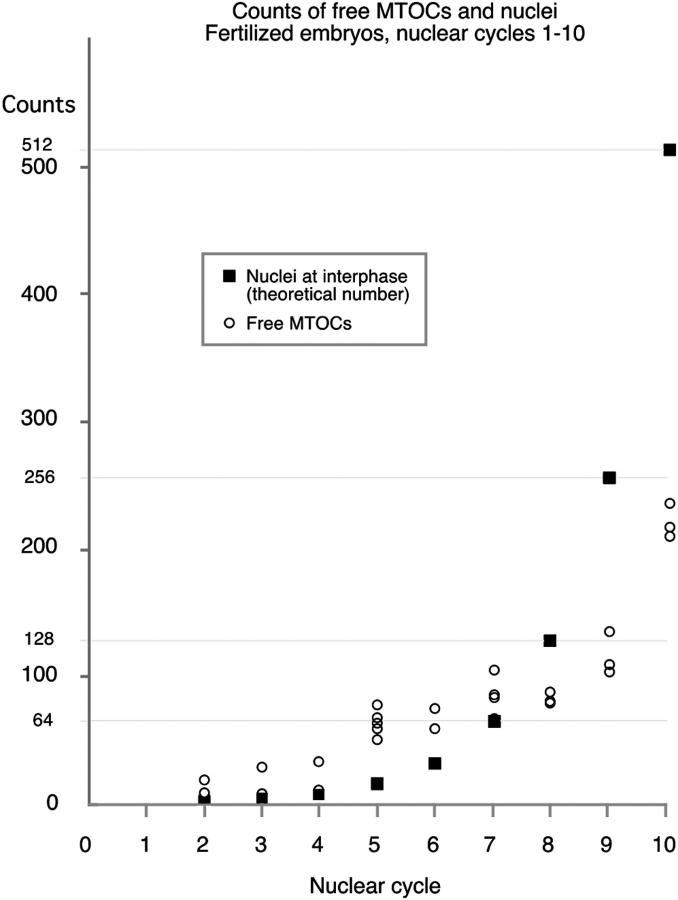
In fertilized embryos, the extra MTOCs remain in the interior of the embryos throughout the syncytial divisions. Fig. 6, E and F, shows surface and interior views of an embryo in which the nuclei have migrated to the cortex. Numerous free MTOCs are present in the interior of the embryo. We estimated the number of free MTOCs by taking a series of 5-μm optical sections and counting the MTOCs not associated with the yolk nuclei. As shown in Fig. 7, between nuclear cycles 2 and 10 the number of free MTOCs increases from 20 to ∼200. Unlike the increase in the number of nuclei, this increase is not exponential. At interphase of cycle 5, when the nuclei have almost completed their migration to the cortex, there are ∼60 free MTOCs in the interior of the embryo. At nuclear cycle 10, the MTOCs have increased to ∼200. This suggests that individual MTOCs undergo at most two rounds of replication after the dividing nuclei migrate to the cortex. The number of free MTOCs may be underestimated because undetected free MTOCs could lie between the 5-μm sections used to perform the analysis. The clumping of the free MTOCs also makes precise counting difficult.
Figure 7.
Free MTOC duplication during nuclear cycles 1–10. The number of free MTOCs, on the y axis, plotted against the nuclear cycle of the embryo on the x axis. The theoretical number of nuclei (2n −1, where n is the nuclear cycle) is also shown for each cycle. MTOCs not associated with nuclei (free MTOCs) were counted in a series of optical sections taken at 5-μm increments in mitotic embryos. The number of free MTOCs does not increase exponentially, but it does increase from ∼20 in cycle 1 to ∼200 in cycle 10.
Spindle Formation and Function in the First Mitotic Division of Fertilized Embryos Does Not Rely on Centrosome-based MTOCs
During the first mitotic division in fertilized embryos, numerous MTOCs surround the metaphase plate, but none are positioned at the spindle poles. The most likely origin of these MTOCs is from the breakdown of the giant basal body consisting of 60–90 singlet microtubules that is brought in with the sperm during fertilization (Phillips, 1967). These MTOCs do not appear to nucleate spindle microtubules. Instead, the microtubules are concentrated around the chromatin. During the first mitotic division in fertilized embryos, spindle formation appears to be similar to spindle formation in unfertilized embryos and seems to be independent of the MTOCs. This is often the case in the second division as well (data not shown). By nuclear cycle 3, however, spindles have centrosomes at the poles. These centrosomes may originate through the capture of free MTOCs during the second and third nuclear cycles.
The first two nuclear cycles in fertilized embryos are not marked by any special problems. The daughter nuclei separate without incident, appear to be of the same size, and have properly segregated their chromosomes as determined by fluorescent in situ hybridization with an X chromosome probe, which showed that the nuclei in the syncytial blastoderm at cycles 5–6 had the correct number of X chromosomes (data not shown).
Discussion
Sciara Embryos Maintain Both a Centrosome-based and Centrosome-independent Pathway of Assembling a Functional Spindle
The dominant model for spindle assembly in animal cells with centrosomes has been the search-and-capture model in which two centrosomes at opposite sides of the nucleus define the poles of the spindle by nucleating microtubules whose plus ends are directed outward (Kirschner and Mitchison, 1986; Nicklas, 1997). These dynamic microtubules are captured and stabilized if they encounter a kinetochore. Chromosomes align on the metaphase plate through the balance of forces mediated by pole to kinetochore microtubules. These forces are generated by microtubule depolymerization and motor proteins at the kinetochore (Cassimeris et al., 1994; Rieder and Salmon, 1994; Inoue and Salmon, 1995; Skibbens et al., 1995). In this model, the centrosome plays a key role in spindle assembly, and one would predict that centrosomes are essential for proper formation of the spindle. In grasshopper spermatocytes, this prediction has been verified by micromanipulation. When centrosomes are removed in prophase, the mitotic spindle does not form (Zhang and Nicklas, 1995a ,b).
However, there is evidence that in some cell types, spindles can assemble in the absence of centrosomes. For example, the meiocytes of Drosophila females assemble spindles without centrosomes (Theurkauf and Hawley, 1992; McKim and Hawley, 1995). In addition, in vitro studies demonstrate that if plasmid DNA attached to beads is placed in Xenopus extracts, a bipolar spindle forms (Heald et al., 1996). The spindle assembles from microtubules nucleated near the chromatin.
Our work complements and extends these studies by demonstrating in vivo that the cytoplasm of early Sciara embryos supports centrosome-based and centrosome-independent mitosis. In Sciara embryos, the sperm supplies the centrosomes, but development is initiated whether or not the egg has been fertilized. Therefore, early Sciara embryogenesis provides the opportunity to examine spindle assembly and mitosis in a common cytoplasm with or without centrosomes. Our data demonstrate that in the absence of centrosomes, spindles are assembled from microtubules nucleated near the chromosomes, while the presence of centrosomes in fertilized embryos redirects the nucleation of spindle microtubules to the centrosomes. Perhaps the most striking result is that the spindles formed in the absence of centrosomes are fully functional and proceed normally through anaphase.
Although functional spindles assembled in the absence of centrosomes, the presence of centrosomes may increase the fidelity of chromosome segregation. To measure this, we assayed for bridges and chromosome fragments in the first division and found none. At this level of analysis, we did not observe any defect in the fidelity of chromosome segregation in unfertilized embryos. In addition, the first division in fertilized embryos is very accurate, and the spindle is assembled without centrosomes.
During Early Sciara Development, the Primary Function of the Centrosome May Be to Provide Astral Microtubules for Proper Migration and Spacing of the Syncytial Nuclei
Sciara embryos lacking centrosomes form pole-to-pole microtubules, pole-to-kinetochore microtubules, and midbodies. Astral microtubules appear to be the only class of microtubules missing in these embryos. We believe the defective nuclear distribution and migration in unfertilized Sciara embryos is a direct consequence of the lack of astral microtubules. Our data suggest that there are two microtubule-dependent processes affecting nuclear distribution in the early embryo. The first is mitosis, which produces daughter nuclei separated by a midbody. This occurs in both fertilized and unfertilized embryos. The second relies on astral microtubules and is probably responsible for nuclear spacing and nuclear migration to the cortex. In syncytial Drosophila embryos, astral microtubules have been implicated in the migration of the nuclei to the cortex (Baker et al., 1993). Because astral microtubule formation requires centrosomes, this latter process does not occur in unfertilized Sciara embryos. Therefore, the abnormal nuclear distribution in unfertilized embryos may be the net result of the movement of nuclei due to mitosis in the absence of astral microtubule-mediated movement of the nuclei.
An alternative interpretation of the abnormal nuclear distribution in unfertilized Sciara embryos is that they are haploid and exhibit altered gene expression. This is unlikely because, as with Drosophila, during the syncytial nuclear divisions development is controlled through maternally supplied gene products (Edgar et al., 1986; Merrill et al., 1988; Wieschaus and Sweeton, 1988). In Sciara, the T1 translocation, which is lethal in homozygous male embryos because of incorrect ploidy, exhibits normal syncytial development (de Saint Phalle and Sullivan, 1996).
Plus End–directed Motor Proteins May Mediate the Distribution of Nuclei by Acting upon the Astral Microtubules
Where astral microtubules from the centrosomes of neighboring nuclei overlap, plus end–directed motors could push the nuclei apart by sliding opposing astral microtubules past one another. In a group of nuclei, the force exerted on each nucleus would be balanced when nuclei were spaced to minimize the overlap of their astral microtubules. There are known motor proteins that slide microtubules relative to one another and that are cell cycle regulated. The BimC subfamily of kinesin-related motor proteins are plus end–directed motors that function in centrosome separation and spindle assembly (Sawin and Endow, 1993; Boleti et al., 1996; Karsenti et al., 1996; Vernos and Karsenti, 1996). The Drosophila KRP130 BimC subfamily kinesin is a bipolar motor that cross-links microtubules and slides them relative to one another (Kashina et al., 1996). Similar motors could act during interphase when astral microtubules are more stable, and consequently longer, than in mitosis (Gliksman et al., 1993; Inoue and Salmon, 1995). Therefore, the orientation of neighboring spindles may be mediated by the interaction of astral microtubules. In unfertilized Sciara embryos, the orientation of the spindle axis may be completely random with respect to neighboring spindles. In support of this, spindle orientations that do not occur in fertilized embryos are observed in unfertilized embryos (Fig. 5).
Anaphase B Occurs in Spindles Assembled without Centrosomes
Astral microtubule-based pulling forces and spindle-based pushing forces have both been proposed as potential mechanisms of anaphase B separation of the spindle poles. The pulling force of astral microtubules was demonstrated by creating asymmetric astral arrays. The centrosome always moved in the direction of the more concentrated astral array (Hiramoto et al., 1986). A single aster has been shown to move an entire mitotic apparatus in the fungus Nectria haematococca (Aist et al., 1991). Observations of spindles exerting pressure on the cortex in Caenorhabditis elegans (Hyman and White, 1987) and natural bending of spindles provide evidence for the pushing of overlap spindle microtubules as a mechanism of anaphase B (Aist et al., 1991). This is further supported by in vitro observations of elongating diatom spindles (Hogan and Cande, 1990, and references therein). In addition, studies in Saccharomyces cerevisiae demonstrate that astral microtubules are not required for anaphase B (Sullivan and Huffaker, 1992). In fact, both forces can act in the same organism, and studies by Aist et al. (1991) suggest that the spindle acts as governor on the rate of anaphase B spindle pole separation mediated by astral forces in Nectria haematococca.
In Sciara embryos, we find that anaphase B does occur in the absence of centrosomes and astral microtubules. This suggests that spindle-based pushing contributes to the separation of poles during anaphase B. It is unlikely that astral-based pulling also contributes to the separation of the spindle poles during anaphase B in fertilized embryos. The initial divisions in fertilized Sciara embryos occur in the interior of the embryo with large (>50 μm) and unequal distances between each spindle pole and the plasma membrane. Therefore, astral-based microtubule pulling would not be a viable mechanism to achieve anaphase B in these embryos.
Centrosomes Not Associated with Nuclei Do Not Replicate in Synchrony with the Nuclei and Do Not Migrate
In Sciara, free centrosomes do not form spindles and do not migrate to the cortex. The number of free centrosomes doubles twice between nuclear cycles 5 and 10, but they do not divide in synchrony with the nuclei. It is possible that after one or two rounds of replication the free centrosomes cease duplicating. These results suggest that continued replication and migration require a close nuclear association. Studies in syncytial Drosophila embryos in which free centrosomes were experimentally induced indicated that they could undergo multiple rounds of duplication provided nuclear division cycles were normal. Aphidicolin injection, which prevented normal nuclear replication, also prevented multiple rounds of centrosome duplication (Debec et al., 1996). These studies, combined with our observations in Sciara embryos, suggest that during the syncytial divisions, centrosome duplication requires signals generated from normally dividing nuclei. In the Sciara embryo, even though the free centrosomes are competent to respond to the signal, they may be too far away from the nuclei to receive it.
Acknowledgments
We are indebted to Susan Gerbi for kindly providing Sciara stocks, the rDNA clone, technical expertise, helpful discussions, and encouragement. William Rice's support and encouragement made this work possible. Finally, we are especially grateful to Helen Crouse for her encouragement, enthusiasm, and pioneering work with Sciara.
This work was supported with a National Institutes of Health (NIH) grant to W. Sullivan (R2961446409), a supplemental grant to B. de Saint Phalle from NIH (R01GM46409-06), and further assistance to B. de Saint Phalle from the Marilyn C. Davis Scholarship Fund.
Abbreviations used in this paper
- CN
center of the nuclei
- MTOC
microtubule-organizing center
Footnotes
Address all correspondence to William Sullivan, Department of Biology, 225 Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064. Tel.: (408) 459-4295. Fax: (408) 459-3139. E-mail: sullivan@biology.ucsc.edu, saint@darwin.ucsc.edu
References
- Aist JR, Bayles CJ, Tao W, Berns MW. Direct evidence for the existence, structural basis and function of astral forces during anaphase B in vivo. J Cell Sci. 1991;100:279–288. doi: 10.1242/jcs.100.2.279. [DOI] [PubMed] [Google Scholar]
- Baker J, Theurkauf WE, Schubiger G. Dynamic changes in microtubule configuration correlate with nuclear migration in the preblastoderm Drosophilaembryo. J Cell Biol. 1993;122:113–121. doi: 10.1083/jcb.122.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R. The centrosome in animal cells and its functional homologs in plant and yeast cells. Int Rev Cytol. 1996;169:25–82. doi: 10.1016/s0074-7696(08)61984-1. [DOI] [PubMed] [Google Scholar]
- Boleti H, Karsenti E, Vernos I. Xklp2, a novel Xenopuscentrosomal kinesin-like protein required for centrosome separation during mitosis. Cell. 1996;84:49–59. doi: 10.1016/s0092-8674(00)80992-7. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Rieder CL, Salmon ED. Microtubule assembly and kinetochore directional instability in vertebrate monopolar spindles: implications for the mechanism of chromosome congression. J Cell Sci. 1994;107:285–297. doi: 10.1242/jcs.107.1.285. [DOI] [PubMed] [Google Scholar]
- de Saint Phalle, B., and W. Sullivan. Incomplete sister chromatid separation is the mechanism of programmed chromosome elimination during early Sciara coprophilaembryogenesis. Development (Camb) 1996;122:3775–3784. doi: 10.1242/dev.122.12.3775. [DOI] [PubMed] [Google Scholar]
- Debec A, Kalpin RF, Daily DR, McCallum PD, Rothwell WF, Sullivan W. Live analysis of free centrosomes in normal and aphidicolin-treated Drosophilaembryos. J Cell Biol. 1996;134:103–115. doi: 10.1083/jcb.134.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophiladevelopment. Cell. 1986;44:365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- Endow SA, Komma DJ. Spindle dynamics during meiosis in Drosophilaoocytes. J Cell Biol. 1997;137:1321–1336. doi: 10.1083/jcb.137.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliksman NR, Skibbens RV, Salmon ED. How the transition frequencies of microtubule dynamic instability (nucleation, catastrophe, and rescue) regulate microtubule dynamics in interphase and mitosis: analysis using a Monte Carlo computer simulation. Mol Biol Cell. 1993;4:1035–1050. doi: 10.1091/mbc.4.10.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopusegg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopusegg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW. The onset of homologous chromosome pairing during Drosophila melanogasterembryogenesis. J Cell Biol. 1993;120:591–600. doi: 10.1083/jcb.120.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto, Y., Y. Hamaguchi, M.S. Hamaguchi, and Y. Nakano. 1986. Roles of microtubules in pronuclear migration and spindle elongation in sand dollar eggs. In Cell Motility: Mechanisms and Regulation. H. Ishikawa, S. Hatano, and H. Sato, editors. Alan R. Liss, Inc., New York. 349–356.
- Hogan CJ, Cande WZ. Antiparallel microtubule interactions— spindle formation and anaphase-B. Cell Motil Cytoskel. 1990;16:99–103. doi: 10.1002/cm.970160203. [DOI] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. . J Cell Biol. 1987;105:2123–2136. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly-disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr TL, Alberts BM. Organization of the cytoskeleton in early Drosophilaembryos. J Cell Biol. 1986;102:1494–1509. doi: 10.1083/jcb.102.4.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Boleti H, Vernos I. The role of microtubule dependent motors in centrosome movements and spindle pole organization during mitosis. Semin Cell Dev Biol. 1996;7:367–378. [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- McKim KS, Hawley RS. Chromosomal control of meiotic cell division. Science. 1995;270:1595–1601. doi: 10.1126/science.270.5242.1595. [DOI] [PubMed] [Google Scholar]
- Merdes A, Cleveland DW. Pathways of spindle pole formation: different mechanisms; conserved components. J Cell Biol. 1997;138:953–956. doi: 10.1083/jcb.138.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. . Development (Camb) 1988;104:495–510. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Sedat JW. Localization of antigenic determinants in whole Drosophilaembryos. Dev Biol. 1983;99:261–264. doi: 10.1016/0012-1606(83)90275-0. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Phillips DM. Observations on spermiogenesis in the fungus gnat Sciara coprophila. . J Cell Biol. 1966;30:477–497. doi: 10.1083/jcb.30.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DM. Giant centriole formation in Sciara. J Cell Biol. 1967;33:73–92. doi: 10.1083/jcb.33.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz R, Gerbi SA, Glatzer KH. Ribosomal DNA of fly Sciara coprophilahas a very small and homogeneous repeat unit. Mol Gen Genet. 1979;173:1–13. doi: 10.1007/BF00267685. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruder FJ, Frasch M, Mettrenleiter TC, Buesen W. Appearance of two maternally directed histone H2A variants precedes zygotic ubiquitination of H2A in early embryogenesis of Sciara coprophila(Diptera) Dev Biol. 1987;122:568–576. doi: 10.1016/0012-1606(87)90320-4. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Endow SA. Meiosis mitosis and microtubule motors. Bioessays. 1993;15:399–407. doi: 10.1002/bies.950150606. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Rieder CL, Salmon ED. Kinetochore motility after severing between sister centromeres using laser microsurgery: evidence that kinetochore directional instability and position is regulated by tension. J Cell Sci. 1995;108:2537–2548. doi: 10.1242/jcs.108.7.2537. [DOI] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W, Minden JS, Alberts BM. Daughterless-abo-like, a Drosophilamaternal-effect mutation that exhibits abnormal centrosome separation during the late blastoderm divisions. Development (Camb) 1990;110:311–324. doi: 10.1242/dev.110.2.311. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Daily DR, Fogarty P, Yook KJ, Pimpinelli S. Delays in anaphase initiation occur in individual nuclei of the syncytial Drosophilaembryo. Mol Biol Cell. 1993;4:885–896. doi: 10.1091/mbc.4.9.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophilafemales: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernos I, Karsenti E. Motors involved in spindle assembly and chromosome segregation. Curr Opin Cell Biol. 1996;8:4–9. doi: 10.1016/s0955-0674(96)80041-x. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Sweeton D. Requirements for X-linked zygotic gene activity during cellularization of early Drosophilaembryos. Development (Camb) 1988;104:483–494. doi: 10.1242/dev.104.3.483. [DOI] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. Chromosomes initiate spindle assembly upon experimental dissolution of the nuclear envelope in grasshopper spermatocytes. J Cell Biol. 1995a;131:1125–1131. doi: 10.1083/jcb.131.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. The impact of chromosomes and centrosomes on spindle assembly as observed in living cells. J Cell Biol. 1995b;129:1287–1300. doi: 10.1083/jcb.129.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. “Anaphase” and cytokinesis in the absence of chromosomes. Nature. 1996;382:466–468. doi: 10.1038/382466a0. [DOI] [PubMed] [Google Scholar]