Abstract
Osteoclasts are multinucleated cells of hemopoietic origin that are responsible for bone resorption during physiological bone remodeling and in a variety of bone diseases. Osteoclast development requires direct heterotypic cell–cell interactions of the hemopoietic osteoclast precursors with the neighboring osteoblast/stromal cells. However, the molecular mechanisms underlying these heterotypic interactions are poorly understood. We isolated cadherin-6 isoform, denoted cadherin-6/2 from a cDNA library of human osteoclast-like cells. The isolated cadherin-6/2 is 3,423 bp in size consisting of an open reading frame of 2,115 bp, which encodes 705 amino acids. This isoform lacks 85 amino acids between positions 333 and 418 and contains 9 different amino acids in the extracellular domain compared with the previously described cadherin-6. The human osteoclast-like cells also expressed another isoform denoted cadherin-6/1 together with the cadherin-6. Introduction of cadherin-6/2 into L-cells that showed no cell–cell contact caused evident morphological changes accompanied with tight cell–cell association, indicating the cadherin-6/2 we isolated here is functional. Moreover, expression of dominant-negative or antisense cadherin-6/2 construct in bone marrow–derived mouse stromal ST2 cells, which express only cadherin-6/2, markedly impaired their ability to support osteoclast formation in a mouse coculture model of osteoclastogenesis. Our results suggest that cadherin-6 may be a contributory molecule to the heterotypic interactions between the hemopoietic osteoclast cell lineage and osteoblast/bone marrow stromal cells required for the osteoclast differentiation. Since both osteoclasts and osteoblasts/bone marrow stromal cells are the primary cells controlling physiological bone remodeling, expression of cadherin-6 isoforms in these two cell types of different origin suggests a critical role of these molecules in the relationship of osteoclast precursors and cells of osteoblastic lineage within the bone microenvironment.
Multinucleated osteoclasts are unique cells that are responsible for bone resorption during physiological bone remodeling and in a variety of bone diseases such as osteoporosis, Paget's disease, and osteolytic bone metastasis of cancer (for review see Roodman, 1996). Hemopoietic stem cells give rise to osteoclasts through a series of sequential steps that are under the influence of diverse hormones, cytokines, and growth factors. One critical step in osteoclast formation is the fusion of hemopoietic mononuclear precursors to form multinucleated osteoclasts. We have previously reported that this fusion step requires homotypic (between the same cells) cell–cell interactions between the osteoclast precursors and is mediated by E-cadherin (Mbalaviele et al., 1995). In addition to homotypic interactions, it has been demonstrated that osteoclast development also requires direct cell–cell interactions of the osteoclast precursors with neighboring cells of the osteoblast/stromal lineage (for review see Suda et al., 1992). These heterotypic (between two different cell types) interactions could be mediated through either homophilic (between the same molecules), heterophilic (between two different molecules) or both. However, the precise molecular mechanisms underlying these heterotypic cell–cell interactions are unknown.
Recently, a previously unrecognized property of cadherins has been reported. In the association between lymphocytes and intestinal mucosal epithelium, E-cadherin– expressed mucosal epithelial cells were found to bind with the integrin αEβ7 (human) (Cepek et al., 1994) or αM290β7 (mouse) (Karecla et al., 1994) expressed in the lymphocytes. These findings suggest that cadherins are also able to establish heterotypic or heterophilic cell–cell interactions. In the present study, we explored the possibility of mediation by cadherins of heterotypic cell-to-cell communication between the osteoclast precursors and osteoblast/stromal cells. Our previous study has demonstrated that none of the classical cadherins such as epithelial, neural, and placental (E-, N-, and P-) cadherin are expressed concomitantly in both cell types during osteoclastogenesis (Mbalaviele et al., 1995). The present study was, therefore, first aimed to identify a novel cadherin member that is expressed both in the osteoclast precursors and osteoblast/stromal cells. Second, we then determined its role in osteoclast development in a mouse coculture model of osteoclastogenesis.
Materials and Methods
Mouse Bone Marrow Cell Culture
We used a well-characterized mouse bone marrow cell culture technique (Takahashi et al., 1988) to obtain multinucleated osteoclast-like cells. Mouse bone marrow cells containing mononuclear hemopoietic cells and stromal cells were collected from femora and tibia of C57BL mice (male, 4–6-wk-old; Harlan Industries, Houston, TX). Cells were washed twice with serum-free αMEM (Hazleton Biologies Inc., Lenexa, KS) and cultured in αMEM supplemented with 10% FBS (Hyclone Laboratories, Logan, UT), and 10 nM 1,25-dihydroxyvitamin D3 (1,25D3)1 (BIOMOL, Plymouth Meeting, PA) in 48-well plates (Falcon, Becton Dickinson Labware, Lincoln Park, NJ) (106 cells/0.5 ml/well) for 6 d at 37°C in a humidified atmosphere of 5% CO2 in air. At day 2 of the culture when most of cells did not become adherent yet, medium change was not done but 0.5 ml fresh αMEM containing 10% FCS, 20 nM 1,25D3 was added to the cultures. At day 4 when majority of cells attached, 0.5 ml of the culture medium was gently aspirated without agitating non-adherent cells, and then 0.5 ml fresh αMEM-containing factors was added. At the end of the culture, cells were fixed in 60% acetone in citrate buffer, pH 5.4, for 30 s, washed twice with distilled water, air dried, and then stained for tartrate-resistant acid phosphatase (TRAP), a widely used cytochemical marker of mouse osteoclasts (Wijngaert and Burger, 1986), using a commercially available kit (Sigma Chemical Co., St. Louis, MO). Stained cultures were examined under light microscopy at a magnification of ×200. TRAP-positive (red-staining) multinucleated (three or more nuclei) cells (osteoclast-like cells) in each well were counted by manually scanning across the entire well in a systematic fashion (Mbalaviele et al., 1995). It should be noted that multinucleated osteoclast-like cells supported by stromal cells are developed only in the presence of 1,25D3 in this culture system. In the absence of 1,25D3, no multinucleated osteoclast-like cells are formed and there is only a mixture of mononuclear hemopoietic cells and stromal cells in the cultures.
RNA Preparation
Highly purified human mature osteoclast-like cells were isolated from giant cell tumors of bone. Highly purified human bone marrow hematopoietic CD34+ progenitors (purity >95%) were purchased from Poietic Technologies, Inc. (Gaithersburg, MD) and used straightforward for RNA isolation. CD34+ progenitors have been shown to contain pools of osteoclast precursors (Matayoshi et al., 1996; Pierelli et al., 1997). Primary mouse bone marrow cells (106 cells/cm2, 10-cm dish) containing hemopoietic mononuclear cells and stromal cells were cultured for 6 d in the presence of 10 nM 1,25D3 to generate osteoclast-like cells. Human osteoblastic cell lines (5 × 105 cells/well, 6-well plates) were cultured for 3–4 d until confluency. Total RNA was extracted with guanidinium thiocyanate using protocols based on the commercial product RNAzol according to the manufacturer's directions (Cinna/Biotecx Laboratories, Inc., Houston, TX).
PCR Techniques and DNA Analysis
To search for novel cadherins, PCR was performed on a cDNA library prepared from highly purified human osteoclast-like cells (Takahashi et al., 1994) using two degenerate oligonucleotide primers. These primers were designed based on the highly conserved cytoplasmic domain of known cadherins as previously described by Suzuki et al. (1991). PCR was performed for 35 cycles consisting of denaturation at 94°C for 10 min, annealing at 50°C for 2 min, and then polymerization at 72°C for 3 min. PCR products were size fractionated by agarose gel electrophoresis and components of ∼160 bp were extracted, ligated into PCR™II (Invitrogen, San Diego, CA), sequenced, and then used as probes to isolate full-length cDNA from the same library. Nucleotide sequencing of both strands of cloned PCR products and full cDNA clones was performed by the double-strand dideoxy-chain termination method, using T7 polymerase (Sequenase version 2.0; United States Biochemical Corp., Cleveland, OH). The nucleotide and amino acid sequences were analyzed using the Genepro™ software (Riverside Scientific Enterprises, Bainbridge Island, WA).
PCR detection of the alternatively spliced isoforms of cadherin-6 was performed either on human osteoclast-like cell cDNA library as described above or on reverse-transcribed single-strand cDNA from total RNA preparations (1 μg) using Moloney murine leukemia virus reverse transcriptase and random hexamers primers (Perkin Elmer Cetus, Norwalk, CT). The upstream primer 5′-CAGATGGGAGGATTATCTGGG-3′ (positions 760–781 bp; see Fig. 4) and the downstream primer 5′-GTACTCGACTACTTTGCTTTGGATT-3′ (positions 1,169–1,194 bp) were designed to amplify fragments encompassing the alternative splicing site. PCR was carried out as described above except that the denaturation was at 94°C for 1.5 min. The components of predicted size were extracted from agarose gel, ligated into PCR™II, and then sequenced.
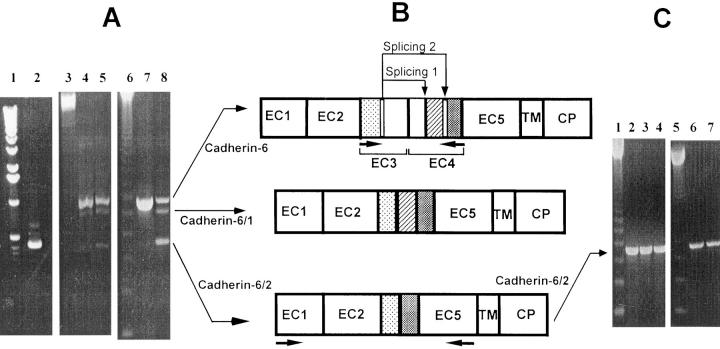
Figure 4.
(A) RT-PCR analysis of cadherin-6 gene expression. Human osteoblastic cell line MG-63 (lane 7); a cDNA library of human osteoclast-like cells (lane 8); highly purified human bone marrow CD34+ progenitors (lane 2); human mature osteoclast-like cells derived from giant cell tumors of bone (lane 5); and TE-85 (lane 4). 1-kb (lane 1) and 123-bp (lanes 3 and 6) DNA ladders, respectively. (B) Occurrence of alternative splicing in cadherin-6 gene. Cadherin-6/2 contains an extracellular domain (EC1 to EC5), a transmembrane domain (TM) and a cytoplasmic domain (CP). Comparison of amino acid sequences of PCR products to those of cadherin-6 suggested an alternative splicing reaction involving regions of EC3 and EC4 to generate cadherin-6/1 (Splicing 1). Another splicing involved the same sequences of EC3 and other sequences of EC4 to generate cadherin-6/2 (Splicing 2). Horizontal arrows below cadherin-6 and cadherin-6/2 indicate the location of primers used to analyze the alternative splicing and amplify the extracellular domain of cadherin-6/2 for polyclonal antibody production, respectively. (C) RT-PCR analysis of cadherin-6 gene expression in mouse and rat cells. A 123-bp DNA ladder (lanes 1 and 5); mouse stromal ST2 cells (lane 2); mouse bone marrow cells cultured in the presence (multinucleated osteoclast-like cells and supporting stromal cells were present in the cultures) (lane 3) or absence (no multinucleated osteoclast-like cells were formed and mononuclear hemopoietic cells and stromal cells were present in the cultures) (lane 4) of 1,25D3; mouse osteoblastic cell line MC3T3-E1 (lane 6); and rat osteoblastic cell line ROS 17/2.8 (lane 7).
Reverse transcription (RT)-PCR detection of cadherin-6/2 (see below for the designation of cadherin-6/2) antisense products from those of cadherin-6/2 sense was performed using the upstream primer 5′-CACTATAGGCTAGCCTCGAG-3′, which bound to the untranslated sequences of T7 promoter of pCIneo, and the downstream primer 5′-GAGTTCAGTGGAGAGGTTAGAGCC-3′ specific for cadherin-6/2.
Competitive RT-PCR on total RNA from ST2 cells transfected with truncated cadherin-6/2 (cadherin-6/2DE) (see below for the truncation of cadherin-6/2) was performed using SuperScript™ RNase H reverse transcriptase (Gibco Laboratories, Grand Island, NY). PCR was performed in the presence of different concentrations (0.00001–10 fmol) of a homologous competitor DNA using PCR SuperMix (Gibco Laboratories, Grand Island, NY). To distinguish amplified cadherin-6/2 fragments from those of cadherin-6/2DE, the primers were designed as follows: the downstream primer 5′-TGACCACAGGCAAGAGATAGG-3′ annealed both the endogenous cadherin-6/2 and cadherin-6/2DE, as well as the competitor. The upstream primers 5′-CAGATGGGAGGATTATCTGGG-3′ and 5′-CACTATAGGCTAGCCTCGAG-3′ in addition to the competitor, specifically annealed the endogenous cadherin-6/2 and the untranslated sequences of T7 promoter of pCIneo vector containing cadherin-6/2DE, respectively.
Expression Vectors
An expression vector for cadherin-6/2 (pCIneo-cadherin-6/2) was constructed by isolating the full-length cadherin-6/2 cDNA from pBlueScript by digestion with XhoI–NotI, which was then inserted into the XhoI/NotI sites of pCIneo vector (Promega Corp., Madison, WI).
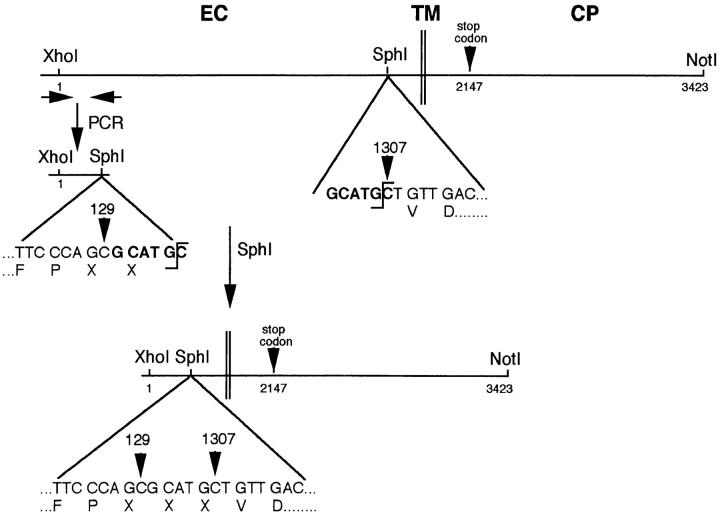
The expression vector for cadherin-6/2 truncated of its extracellular domain (pCIneo–cadherin-6/2DE) was made as follows: pCIneo–cadherin-6/2 was digested with XhoI–NotI to release full-length cadherin-6/2 and subsequently digested with SphI to generate a SphI–NotI fragment, 282-bp upstream of the transmembrane domain of cadherin-6/2 (see Fig. 3). In parallel, the upstream primer 5′-CACTATAGGCTAGCCTCGAG-3′ and the downstream primer 5′-GCATGCGCTGGGAAACCACTAGTCCT-3′ were designed to contain the sites of XhoI (at 5′ end) and SphI (at 3′ end), respectively, and amplify a fragment of 129 bp using the putative signal peptide and precursor sequences of cadherin-6/2 as a template. The fragment was then cloned into PCR™II vector, sequenced and released by XhoI–SphI digestion to generate a XhoI–SphI fragment. XhoI–SphI and SphI–NotI fragments that were in-frame were ligated into pCIneo vector, previously digested with XhoI–NotI.
Figure 3.
Truncation of the extracellular domain of cadherin-6/2 (cadherin-6/2DE). Cadherin-6/2DE contains SphI recognition site at position 1,933–1,938 bp (bold). To delete the most extracellular domain of cadherin-6/2, SphI site was created at position 130–135 bp (bold) of cadherin-6/2 by PCR as described in Materials and Methods. Note that the creation of SphI site generated 3 amino acids designated X, which are Ala-His-Ala. EC, extracellular domain; TM, transmembrane domain; CP, cytoplasmic domain.
For the cadherin-6/2 antisense construct, pCIneo-cadherin-6/2 was first digested with XhoI–NotI, the vector and the insert was filled in, and then the blunt end was ligated. The antisense orientation was checked by digestion with appropriate enzymes.
To generate glutathione-S-transferase (GST)–cadherin-6/2DC fusion protein for antibody production, the extracellular domain of cadherin-6/2 (see Fig. 4 B) was amplified using two oligonucleotide primers. The upstream primer 5′-CCCCAAGCTTTGGAATCAGTTCTTTCTCCTGGAA-3′ overlapped the BamHI site of cadherin-6/2 and the same restriction site was included in the downstream primer 5′-CCCCAAGCTTAGATAGGTGCTCATCTCGTGTC-3′. The amplified fragment was digested with BamHI and inserted into the BamHI site of pGSTag in frame with GST to generate the recombinant protein designated GST–cadherin-6/2DC. GST–cadherin-6/2DC was expressed in BL-21 (Stratagene, La Jolla, CA). The bacteria were grown in 2YT medium supplemented with 50 μg/ml ampicillin. Expression of the recombinant protein was induced with 0.3 mM IPTG for 1 h at 37°C at OD600 of 1. The bacteria were pelleted and resuspended in PBS containing 1 mM PMSF and 10 mM EDTA. The bacteria were lysed by sonication at 4°C for 20 s. Cell debris was removed by centrifugation (10 min, 14,000 g). GST fusion protein was purified by chromatography on glutathione-agarose (Sigma Chemical Co.), eluted from the beads with 10 mM of reduced glutathione in 50 mM Tris-HCl, pH 8.0.
DNA Transfection
L929 cells (American Type Culture Collection, Rockville, MD) or stromal ST2 cells were grown in DME and αMEM supplemented with 10% FBS and 1% penicillin-streptomycin, respectively. The cells were transfected with DNA by the standard calcium phosphate coprecipitation method. After 24 h of incubation, cells were fed with fresh medium, grown for further 24 h, split and seeded into 100-mm dishes, and then cultured in the presence of 250 μg/ml (ST2 cells) or 400 μg/ml (L929 cells) antibiotic G418 (Gibco Laboratories, Grand Island, NY). G418-resistant colonies were isolated and analyzed for recombinant protein expression.
Antibody Production
The purified recombinant protein GST–cadherin-6/2DC bound to glutathione-agarose beads was digested with human plasma thrombin (Sigma Chemical Co.) (ratio 1,000:1) at room temperature for 4 h. The thrombin digestion site is located between GST and the cloning site of pGSTag. The released cadherin-6/2DC fragment was separated from GST–glutathione-agarose by centrifugation at 14,000 g for 10 min, and then used to immunize rabbits for the generation of polyclonal antibody to cadherin-6/2DC.
Mouse mAb to rat K-cadherin (also known as cadherin-6) and β-catenin were purchased from Transduction Laboratories (Lexington, KY). The mouse mAb recognizing N-cadherin (13A9), but not P-, E-, or muscular (M)-cadherin (Donalies et al., 1991) was kindly provided by Dr. Wheelock (University of Toledo, Toledo, OH). Mouse mAb to pan cadherin was purchased from Sigma Chemical Co. This antibody recognizes cadherins containing the COOH-terminal cadherin tail, regardless of their cadherin type.
Immunoprecipitation and Immunoblotting
Cells were lysed in a buffer containing 20 mM Tris-HCl, pH 8.0, 2 mM CaCl2, 150 mM NaCl, 1% NP-40, 0.1% SDS, and protease inhibitors (20 mM leupeptin, 1 mM PMSF, 1% aprotinin solution) at 4°C, and then centrifuged at 10,000 g for 10 min at 4°C. The protein content of the supernatants was determined using the DC protein assay (Bio-Rad Laboratories, Hercules, CA), and the samples were then incubated with antibody for 2 h at 4°C. Protein A–Sepharose beads (Zymed Laboratories, South San Francisco, CA) were added to the mixtures and incubated for an additional 2 h. Beads and associated immune complexes were washed three times with lysis buffer. The immunoprecipitates were resuspended in SDS-PAGE sample buffer, boiled for 5 min, resolved on 7.5% Laemmli gels, and then processed for immunoblotting as follows.
The proteins were transferred onto polyvinyldine difluoride membranes (Immobilon-P; Millipore Corp., Bedford, MA) in transblotting buffer containing 20 mM Tris, 150 mM glycine, and 20% methanol, pH 8.0. The membranes were incubated overnight with blocking buffer consisting of 5% nonfat dry milk (Carnation Company, Los Angeles, CA) in TBS (50 mM Tris-HCl, 150 mM NaCl), and then with antibody diluted in the same buffer for 2 h, washed once with TBS containing 0.05% Tween 20 (TBST), and then twice with TBS, incubated for 1 h with HRP-conjugated secondary antibody or HRP-conjugated protein A, diluted in TBS containing 5% nonfat dry milk. After the membranes were washed three times with TBST, and then three times with TBS, the signal was visualized with ECL detection system (DuPont NEN, Boston, MA).
Immunocytochemistry
Mouse marrow cells (106 cells/cm2) were cultured in the presence of 10 nM 1,25D3 for 6 d to generate osteoclast-like cells, washed twice with PBS, fixed with a fresh solution of 3.7% (wt/vol) formaldehyde in PBS at room temperature for 30 min, blocked with 1% goat serum, incubated with preimmune or immune serum, at 1:500 dilution in PBS containing 0.1% goat serum at 37°C for 1.5 h in humidified atmosphere, washed once with 0.05% Tween 20 in PBS, and then twice in PBS. Cells were then incubated with goat secondary antibody and stained using the Vectastain ABC kit (Vector Laboratories, Inc., Burlingame, CA).
Coculture of Mouse ST2 Cells and Mouse Bone Marrow Cells
Mouse bone marrow cells (100,000 cells) and ST2 cells (5,000 cells) transfected with either empty vector, dominant-negative, antisense, or sense constructs were mixed in suspension, plated onto 48-well plates, and then cultured in αMEM supplemented with 10% FBS, 10 nM 1,25D3, and 100 nM dexamethasone. After 6 d of culture, number of multinucleated osteoclast-like cells formed was counted as described above.
Statistical Analysis
All data were analyzed by analysis of variance, followed by a paired t test.
Results
Cloning and Structural Analysis
cDNA Cloning.
To clone cadherins expressed by the osteoclast-like cells, PCR was performed on a cDNA library derived from highly purified human osteoclast-like cells. PCR products of predicted size of 160 bp were purified, cloned, and sequenced. One clone was 100% identical to E-cadherin, confirming our previous finding of E-cadherin expression by osteoclasts (Mbalaviele et al., 1995). Another clone that showed ∼60% homology with known cadherins was used as a probe to isolate a 3,423-bp cDNA from a cDNA library derived from highly purified human osteoclast-like cells. The cDNA consisted of an open reading frame of 2,115 bases (coding for 705 amino acids) flanked by 31 bases of untranslated sequence at the 5′ end and 1,277 bases at the 3′ end including a stop codon, a polyadenylation sequence and a polyadenylate tail (Fig. 1). The assigned initiator Met residue was followed by a typical signal sequence and a precursor peptide.
Figure 1.
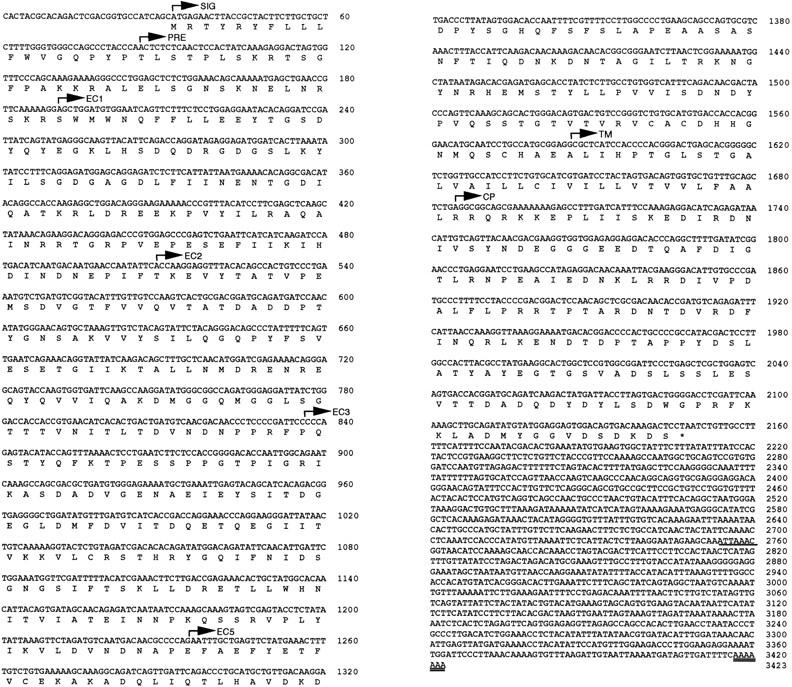
Nucleotide and deduced amino acid sequence. The cDNA contained non-coding 31 bp at the 5′ end (non-coding) followed by an open reading frame of 2,115 bases. The protein contained a signal peptide (SIG), a precursor peptide (PRE), an extracellular domain (EC1–EC5), a transmembrane domain (TM) and cytoplasmic domain (CP). The polyadenylation signal and the polyadenylate tail are shown by single and double underlining, respectively.
Structure of the Cadherin Protein.
Comparison of the deduced full-length amino acid sequence revealed that the isolated molecule belonged to the cadherin family. The mature protein structure predicted an extracellular domain of 498 amino acids that could be subdivided into five subdomains, and a hydrophobic transmembrane domain of 33 amino acids followed by an intracellular domain of 154 amino acids (Fig. 1). When the sequence of the isolated molecule was aligned to that of cadherin family members, it showed high homology with cadherin-6 (Shimoyama et al., 1995), the human counterpart of the rat K-cadherin (Xiang et al., 1994). However, the newly cloned molecule lacked 85 amino acids (255 bp) at positions 333–418 and had different amino acid sequence at positions 283–291, compared with the previously reported cadherin-6 sequence (Figs. 2 and 3), suggesting a polymorphism in the cadherin-6 gene.
Figure 2.
Comparison of the amino acid sequences of mature proteins. The sequence of cadherin-6/2 (bottom) was 100% identical to that of cadherin-6 (top) except that the new cadherin was depleted of 85 amino acids from position 282–367 in cadherin-6. In addition, 9 amino acids at position 283–291 (CRSTHRYGQ) in cadherin-6/2 were different to those of cadherin-6. EC, extracellular domain; TM, transmembrane domain; CP, cytoplasmic domain.
Expression Analysis
mRNA.
Northern analysis of K-cadherin (Xiang et al., 1994) and cadherin-6 (Shimoyama et al., 1995; Paul et al., 1997) demonstrated the presence of three transcripts. It is unknown if some of these transcripts arise by alternative splicing, alternate 3′ noncoding regions or different genes. We therefore chose a PCR approach to address this issue. PCR was carried out using oligonucleotide primers that allowed the amplification of fragments encompassing the splicing site. The cDNA library from human osteoclast-like cells was used as a template. We found an expected spliced fragment of 433 bp (Fig. 4 A, lane 8), which corresponded to our cDNA clone. In addition, a 688-bp fragment and a novel 615-bp fragment were obtained. The 688-bp fragment was identical to the previously identified unspliced isoform of cadherin-6 (Shimoyama et al., 1995). The 433- and 615-bp fragments were likely the alternatively spliced isoforms of cadherin-6. We denoted the unspliced isoform as cadherin-6 and the isoforms depleted of 73 and 255 bp as cadherin-6/1 and cadherin-6/2, respectively (Fig. 4, A and B). Both cadherin-6/1 and cadherin-6/2 lacked regions of the extracellular subdomains 3 and 4. We also confirmed that the three cadherin-6 isoforms were expressed by highly purified human CD34+ progenitors which contain pools of osteoclast precursors (Fig. 4 A, lane 2) and osteoclast-like cells derived from giant cell tumors of bone (Fig. 4 A, lane 5). Human osteoblastic cell lines MG-63 and TE-85 also expressed these three isoforms (Fig. 4 A, lanes 7 and 4, respectively). In contrast, we found that mouse stromal ST2 cells expressed only cadherin-6/2 (Fig. 4 C, lane 2). Similarly, mouse bone marrow cells that were cultured in the presence (multinucleated osteoclast-like cells and supporting stromal cells were present in the cultures) (Fig. 4 C, lane 3) or absence (no multinucleated osteoclast-like cells were formed but there was a mixture of mononuclear hemopoietic cells and stromal cells in the cultures) (Fig. 4 C, lane 4) of 1,25D3 also showed an expression of only cadherin-6/2. Moreover, we found that mouse osteoblastic cell line MC3T3-E1 and rat osteoblastic cell line ROS 17/2.8 also expressed only cadherin-6/2 (Fig. 4 C, lanes 6 and 7, respectively).
Protein.
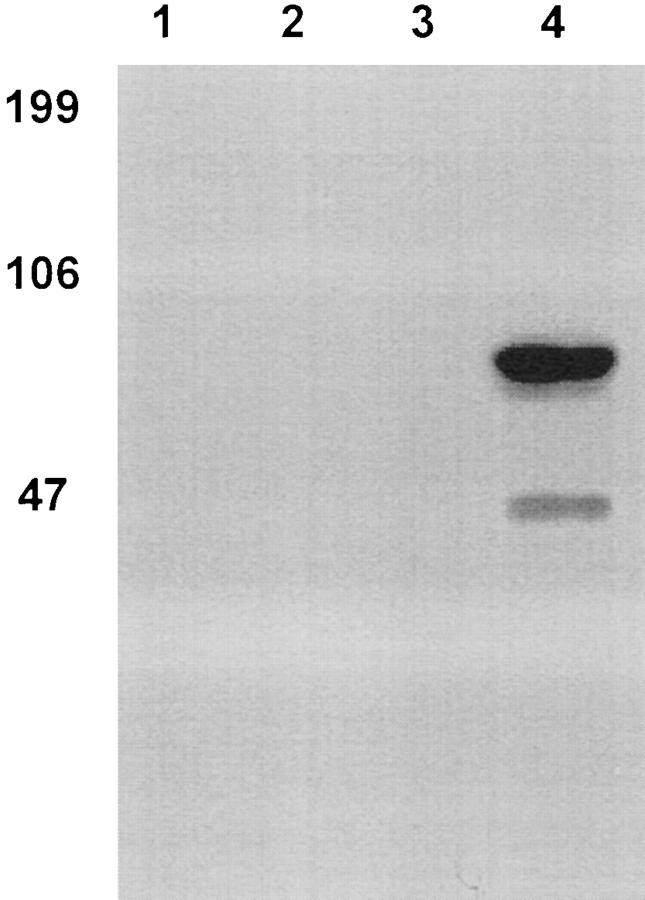
To determine if the polyclonal antibody recognized cadherin-6/2DC, against which it was raised, recombinant GST–cadherin-6/2DC was subjected to SDS-PAGE and immunoblotting. Fig. 5 shows that the antibody recognized the expected fusion protein of 69 kD (Fig. 5, lane 4) but not the GST protein (lane 3). The preimmune serum did not recognize either GST–cadherin-6/2DC (Fig. 5, lane 2) or GST (lane 1).
Figure 5.
Expression of GST–cadherin-6DC fusion protein. The extracellular domain of human cadherin-6/2 (cadherin-6/2DC) (Fig. 4) was used to generate polyclonal antibody. GST (lanes 1 and 3) or GST–cadherin-6/2DC fusion protein (lanes 2 and 4) were expressed in Escherichia coli and purified by affinity chromatography. After SDS-PAGE and transblotting, the membrane was incubated with the preimmune serum (lanes 1 and 2) or polyclonal antibody to cadherin-6/2DC (lanes 3 and 4). An expected 6-kD fusion protein was recognized by the antibody but not by the preimmune serum. The antibody showed no cross-reaction with the GST protein. The smaller fragment was likely a proteolytic product.
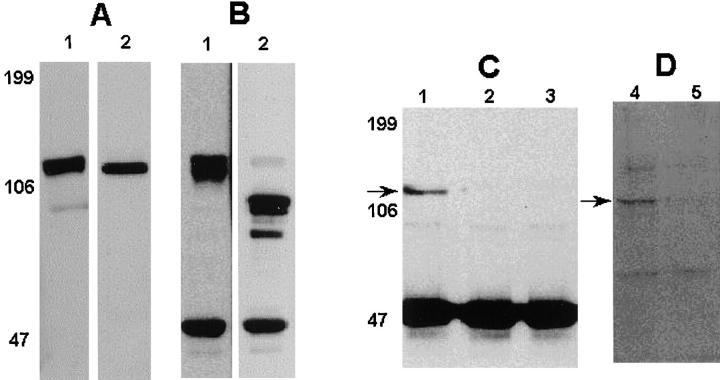
Immunoblotting analysis using the commercially available mAb to cadherin-6 showed an expression of ∼110– 120 kD protein in the human osteoblastic cell line MG-63 and mouse stromal ST2 cells (Fig. 6 A, lanes 1 and 2, respectively). The band at 110–120 kD was also the only one detected using polyclonal antibody to cadherin-6/2 or pan cadherin antibody that recognizes most of the cadherins described so far (data not shown). Although these antibodies recognize all three cadherin-6 isoforms, the band visualized in the ST2 cell lysates likely represents cadherin6/2, since ST2 showed only cadherin6/2 expression by RT-PCR (Fig. 4 C, lane 2).
Figure 6.
(A) Immunoblotting analysis of cadherin-6 protein expression. Confluent MG-63 (lane 1) or ST2 (lane 2) were lysed and subjected to immunoblotting. An an ∼110–120 kD protein was recognized by the mAb to cadherin-6. (B) Coimmunoprecipitation of cadherin-6 and β-catenin in human osteoblastic cell line MG-63. MG-63 lysates were immunoprecipitated using β-catenin antibody and immunoblotted using the mAb to cadherin-6. The expected 110–120 kD protein was detected (lane 1). The same membrane was stripped off and then reblotted with the β-catenin antibody. Note a protein at 85 kD corresponding to β-catenin (lane 2). (C and D) Effects of cadherin-6/2DE on the association of cadherin-6 and β-catenin. Mouse stromal ST2 cells transfected with empty vector (lanes 1 and 4) or cadherin-6/2DE (lanes 2, 3, and 5) were lysed and immunoprecipitated using β-catenin antibody, subjected to SDS-PAGE, and then blotted with monoclonal cadherin-6 antibody (C) or polyclonal antibody to cadherin-6/2 (D).
To determine whether cadherin-6 or its isoforms, like other cadherins, associate with catenins, the lysates of MG-63 were immunoprecipitated using the antibody to β-catenin. The antibody to cadherin-6 could not be used for direct immunoprecipitation experiments. Proteins were then subjected to SDS-PAGE and immunoblotted with the mAb to cadherin-6. The result shows that the 110–120 kD protein coimmunoprecipitated with β-catenin (Fig. 6 B, lane 1). When the same membrane was stripped off and re-blotted with the mAb to β-catenin, an expected band at 85 kD, which corresponded with the molecular size of β-catenin was observed (Fig. 6 B, lane 2). In ST2 cells that expressed only cadherin-6/2 (Fig. 4 C, lane 2), the cadherin-6/2 also coimmunoprecipitated with β-catenin (Fig. 6 C, lane 1, and D, lane 4). To confirm cadherin-6 protein expression in osteoclast cell lineage and stromal cells, mouse bone marrow cells were cultured for 6 d in the presence of 10 nM 1,25D3 to form multinucleated osteoclast-like cells. The polyclonal antibody to cadherin-6 was used in this experiment because the mAb to cadherin-6 could not be used for immunocytochemical analysis of mouse cells because of high background. As shown in Fig. 7, multinucleated osteoclast-like cells as well as mononuclear cells with round (hemopoietic cells) and spindle (stromal cells) shapes exhibited the reactivity to the cadherin-6 antibody. Although this polyclonal antibody recognizes all three cadherin-6 isoforms, it is likely that the immunocytochemical stain displayed in these cells represents cadherin-6/2, since RT-PCR data showed that only cadherin-6/2 was expressed in mouse marrow cultures (Fig. 4 C, lane 2). The result indicates that both cells of the osteoclast lineage and stromal cells in the mouse bone marrow cultures express cadherin-6/2.
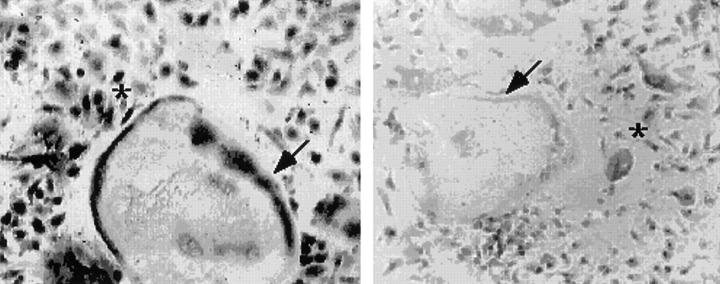
Figure 7.
Immunocytochemical analysis of cadherin-6 gene expression. Mouse bone marrow cells containing mononuclear hemopoietic cells and stromal cells were treated with 10 nM 1,25D3 and 100 nM dexamethasone for 6 d. The cultures containing multinucleated osteoclast-like cells, mononuclear hemopoietic cells and stromal sells were then incubated with the polyclonal antibody to cadherin-6/2. Multinucleated osteoclast-like cells (arrows) and mononuclear cells (asterisks) with round (hemopoietic cells) or spindle (stromal cells) were positively stained by the cadherin-6/2 antibody (left) but not by the preimmune serum (right).
Functional Analysis
Cell–Cell Adhesion Function of Cadherin-6/2.
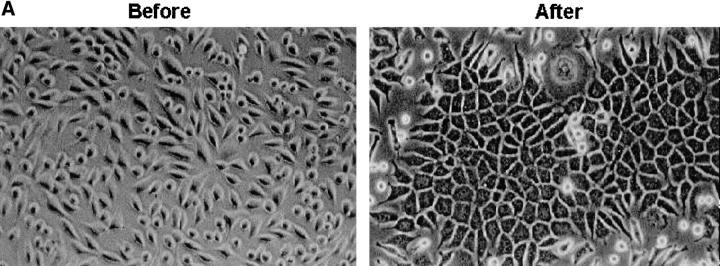
To examine whether the cadherin-6/2 we identified induces cell–cell adhesion, mouse fibroblasts L cells that normally do not express endogenous cadherins and do not display tight cell–cell contact (Nakagawa et al., 1995) were stably transfected with cadherin-6/2 and determined for cadherin-6/2 expression and cell–cell contact. Several G418-resistant clones of L-cells that expressed cadherin-6/2 (Fig. 8 B, After) showed polygonal shape and tight cell–cell contact (Fig. 8 A, After).
Figure 8.
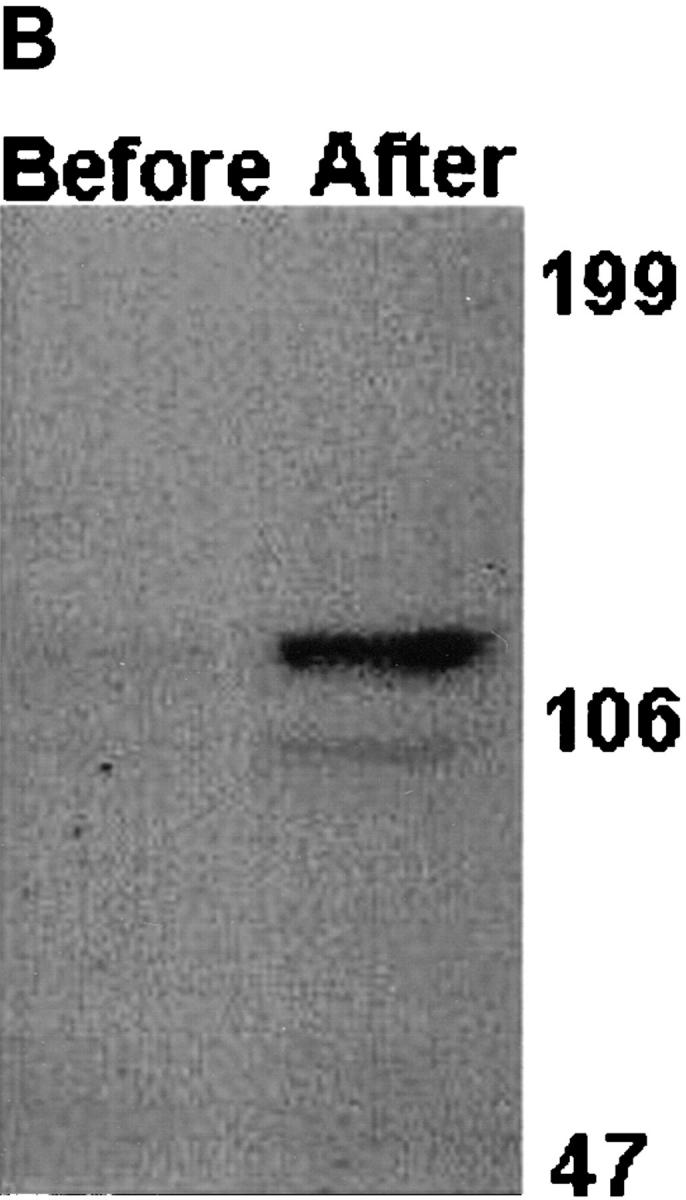
Effect of cadherin-6/2 transfection into mouse fibroblast L929 cells. L929 cells were stably transfected either with empty vector or cadherin-6/2 expression vector. Transfected cells expressed cadherin-6/2 detected by immunoblotting (B, After) and showed strong cell–cell adhesiveness (A, After) compared with non-transfected cells (A, Before).
Role of Cadherin-6/2 in Osteoclast Differentiation; the Effect of Dominant-Negative Cadherin-6/2 (Cadherin-6/2DE).
Mouse stromal ST2 cells are well characterized for their ability to support osteoclast differentiation in coculture with the mononuclear osteoclast precursors (Takahashi et al., 1988). As shown above, ST2 cells expressed cadherin-6/2 by RT-PCR (Fig. 4 C, lane 2) and immunoblotting (Fig. 6). We, therefore, attempted to determine the role of cadherin-6/2 in osteoclastogenesis through interfering with the function of cadherin-6/2 in ST2 cells. ST2 cells were stably transfected with cadherin-6/2 depleted of its extracellular domain (cadherin-6/2DE), and then examined for the ability to support osteoclast differentiation. Because all antibodies to cadherin-6 were raised against its extracellular domain, competitive RT-PCR analysis was used to select ST2 clones overexpressing cadherin-6/2DE. We isolated several clones in which the levels of cadherin-6/2DE was >100-fold higher compared with those of endogenous cadherin-6/2 (Fig. 9).
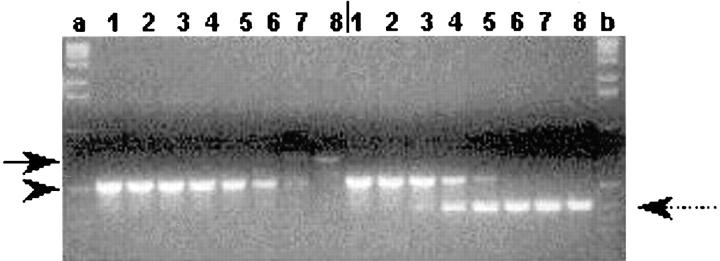
Figure 9.
Competitive RT-PCR analysis of cadherin-6/2 expression. Lanes 1–8 on left and on right of the vertical bar, represent empty vector- and cadherin-6/2DE–transfected ST2 cells, respectively. PCR was performed in the presence of decreasing concentrations of the competitor DNA (arrowhead) (10; 1; 0.1; 0.01; 0.001; 0.0001; 0.00001; 0 fmol). Lanes a and b, 123-bp DNA ladder. Note that endogenous cadherin-6/2 (arrow) was no longer amplified in the presence of 0.001 fmol of competitor versus 1 fmol in the case of cadherin-6/2DE (spotted arrow).
Overexpressed cadherin-6/2DE, which is truncated of its extracellular domain should compete with endogenous cadherin-6/2 for the binding to catenins as has been described for other truncated cadherins (Kintner, 1992). To confirm this, the cell lysates of ST2 cells overexpressing either empty vector or cadherin-6/2DE were coimmunoprecipitated with the mAb to β-catenin. The membranes were blotted with the mAb to cadherin-6, which recognizes only endogenous cadherin-6/2 but not transfected truncated cadherin-6/2DE since this antibody was raised against the extracellular domain of cadherin-6. As shown above, endogenous cadherin-6/2 coimmunoprecipitated with β-catenin in ST2 cells transfected with empty vector (Fig. 6 C, lane 1), whereas it failed to bind with β-catenin in the two different clones of ST2 cells overexpressing cadherin-6/2DE (Fig. 6 C, lanes 2 and 3). The polyclonal antibody to cadherin-6/2 that we generated, produced identical results (Fig. 6 D, lane 4, empty vector; and lane 5, cadherin-6/2DE). Because the cadherin-6/2DE most likely competes for the binding to β-catenin with all cadherin-6 isoforms, these data indicate that transfected cadherin-6/2DE had a dominant-negative effect on the binding of the endogenous cadherin-6. However, since ST2 cells express only cadherin-6/2 based on the RT-PCR data (Fig. 4 C, lane 2), it is probable that cadherin6/2 binding to β-catenin was interfered by the cadherin6/2DE in case of ST2 cells.
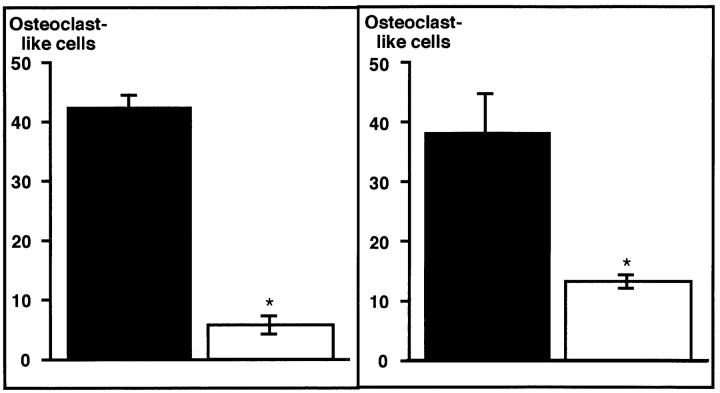
Transfection of cadherin-6/2DE did not alter the morphology and growth of ST2 cells. Furthermore, ST2 cells overexpressing cadherin-6/2DE showed an increase in alkaline phosphatase expression in response to retinoic acid as did nontransfected ST2 cells (data not shown). Stimulation of alkaline phosphatase activity by retinoic acid has been shown to be a critical property of ST2 cells that retain the ability to support osteoclast differentiation (Takahashi et al., 1988). These results suggest that, at least, the phenotype of ST2 cells required for a support of osteoclastogenesis is not altered by cadherin-6/2DE transfection. When ST2 cells overexpressing cadherin-6/2DE were cocultured with mouse bone marrow cells containing mononuclear osteoclast precursors for 6 d in the presence of 10 nM 1,25D3, multinucleated osteoclast-like cell formation was decreased by >70% (Fig. 10, left). Three different clones overexpressing cadherin-6/2DE showed identical effects (data not shown). These data suggest that cadherin-6/2, which is the only cadherin-6 isoform expressed in ST2 cells (Fig. 4 C, lane 2), is involved in interactions between stromal ST2 cells and osteoclast precursors in the formation of osteoclast-like cells in this coculture model.
Figure 10.
(Left) Effects of the expression of cadherin-6/2DE on multinucleated osteoclast-like cell formation. Mouse bone marrow cells containing mononuclear hemopoietic cells and stromal cells were cocultured with stromal ST2 cells stably transfected with empty vector or cadherin-6/2DE for 6 d in the presence of 10 nM 1,25D3 and 100 nM dexamethasone. ST2 cells expressing cadherin-6/2DE (open bar) showed decreased ability to support multinucleated osteoclast-like cell formation compared with empty vector–transfected control cells (closed bar). Data shown are mean + SE (n = 4, for three experiments). Asterisk, significantly different from control (P < 0.05). (Right) Effect of cadherin-6/2 antisense on multinucleated osteoclast-like cell formation. Mouse bone marrow cells containing mononuclear hemopoietic cells and stromal cells were cocultured with stromal ST2 cells stably transfected with sense or cadherin-6/2 antisense for 6 days in the presence of 10 nM 1,25D3 and 100 nM dexamethasone. ST2 cells expressing cadherin-6 antisense (open bar) showed decreased ability to support multinucleated osteoclast-like cell formation compared with sense-transfected control cells (closed bar). Data shown are mean + SE (n = 4, for three experiments). Asterisk, significantly different from control (P < 0.05).
Effects of Cadherin-6/2 Antisense on Osteoclast-like Cell Formation.
It seems likely that cadherin-6/2DE shows a dominant-negative effect on all the endogenous cadherins that associate with catenins. Therefore, the effect observed above may not be due to a dominant-negative effect specific for cadherin-6/2. To specifically target cadherin-6/2, ST2 cells were next transfected with a cadherin-6/2 antisense construct. Positive clones for antisense expression were identified by RT-PCR as described in Materials and Methods (data not shown). Cadherin-6/2 antisense expression had no effect on ST2 cell morphology, growth, and alkaline phosphatase expression (data not shown). However, when ST2 cells expressing the cadherin-6/2 antisense construct were cocultured with mouse bone marrow cells containing mononuclear osteoclast precursors, the ability of ST2 cells to support multinucleated osteoclast-like cell formation was decreased by >50% (Fig. 10, right). These data strongly suggest that cadherin-6/2 plays a critical role in osteoclast-like cell formation in this coculture model.
Discussion
In this study we report the isolation of cadherin-6 isoform from the cDNA library of human osteoclast-like cells, the principal cells responsible for bone resorption during physiological bone resorption and a variety of bone diseases (for review see Roodman, 1996). Although isolation of cadherin-6 in human cancer (Shimoyama et al., 1995; Shimazu et al., 1996), chick embryo (Nakagawa et al., 1995; Inoue et al., 1997), and its rat homologue K-cadherin (Xiang et al., 1994) has been reported previously, here we describe the isolation of the identical gene in the human normal mature osteoclast-like cells that have distinctive physiological and pathological functions. Our RT-PCR data showed that cadherin-6 isoforms (cadherin-6, cadherin-6/1, and cadherin-6/2) are expressed in the human mature osteoclast-like giant cells as well as in human CD34+ progenitors that contain pools of osteoclast precursors. We then studied the biological functions of the cadherin-6 in a well-characterized murine co-culture model in which osteoclast-like cells form under the heterotypic cooperation between the hemopoietic osteoclast precursors and osteoblasts/stromal cells (Takahashi et al., 1988). Our immunohistochemical result together with RT-PCR data suggests that both osteoclast-like cells and osteoblasts/stromal cells express cadherin-6/2 in this murine culture model. Of note, however, cadherin-6 and cadherin-6/1 were not detected by RT-PCR in these two types of mouse cells. The sole expression of cadherin-6/2 in both hemopoietic cells of osteoclast lineage and bone marrow stromal cells raises a possibility that cadherin-6/2 may be a critical cell adhesion molecule (CAM) in osteoclastogenesis in mouse. It has been shown that osteoclast generation requires heterotypic cell–cell contact between the hemopoietic osteoclast precursors and bone marrow–derived stromal cells such as ST2 cells using the similar coculture model to that used here (Takahashi et al., 1988). ST2 cells are known to possess the ability to support osteoclast generation through cell–cell contact and, thus, have been frequently used in the coculture (Takahashi et al., 1988). Consistent with these earlier results, disruption of cadherin-6/2 function or expression with a dominant-negative or an antisense cadherin-6/2 construct, respectively, in the ST2 cells suppressed osteoclast formation in the coculture. It is, therefore, likely that cadherin-6/2 is the primary cadherin-6 responsible for establishing the heterotypic interaction in this mouse culture model. We did not study the role of other cadherin-6 isoforms, namely cadherin-6 and cadherin6/1 in the present study, since these cadherin-6 isoforms were not detected in mouse cells. However, it should be noted that these isoforms were isolated from a human osteoclast-like cell cDNA library and detected in highly purified human CD34+ progenitors, which contain pools of osteoclast precursors and mature osteoclast-like cells derived from giant cell tumors of bone and human osteoblastic cell lines MG-63 and TE-85. Thus, it is probable that these isoforms also play a critical role in mediating heterotypic cell–cell interactions in osteoclastogenesis in human. An establishment of human osteoclast-like cell line will greatly facilitate to approach this issue. We have already reported that E-cadherin mediation of homotypic interactions between hemopoietic mononuclear osteoclast precursors is a prerequisite for the fusion to form multinucleated osteoclasts (Mbalaviele et al., 1995). Since osteoclasts and osteoblasts are the key cellular component in physiological bone remodeling, our data suggest that cadherins including E-cadherin and cadherin-6 may play an important role in the maintenance of the bone microenvironment through mediation of homotypic and heterotypic cell–cell interactions. The result, thus, suggests that cadherins are one of CAMs that modulate cellular cross-talk between bone cells of different origin in the bone marrow cavity. It is most likely that heterophilic mediation between hemopoietic osteoclast precursors and osteoblasts/stromal cells also plays an important role in the generation of osteoclasts. Importance of heterophilic interactions remains to be elucidated.
The molecular mechanisms by which cadherin-6/2 mediates the interactions necessary for osteoclast differentiation are unclear at the present time. However, since we found that cadherin-6/2 associated with β-catenin, it is conceivable that the interactions of the hemopoietic osteoclast precursors with the stromal ST2 cells through cadherin-6/2 may induce activation of intracellular signaling pathways that involve β-catenin. Recent studies have revealed that β-catenin, which was originally identified as a bridge between cadherins and cytoskeleton, associates with DNA binding proteins of the T cell factor–lymphoid enhancer factor family, and thereby alters gene transcription (for review see Miller and Moon, 1996; Korinek et al., 1997; Morin et al., 1997; Rubinfeld et al., 1997). It is possible that cadherin-6/2–induced intracellular signaling cascades involving β-catenin may regulate transcription of genes which encode (a) cytokines in ST2 cells that are required for the differentiation and survival of the osteoclasts. In this context, ST2 cells are known to produce macrophage colony–stimulating factor (Perkins and Kling, 1995), which is an essential cytokine for osteoclast differentiation (for review see Roodman, 1996); or (b) autocrine/intracrine factors in the osteoclast precursors, which are important for their differentiation. Consistent with this notion, we found that cadherin-6/2 that was depleted of its extracellular domain, which inhibits osteoclast formation, prevented endogenous cadherin-6/2 from binding to β-catenin in ST2 cells. In agreement with our finding, Kintner (1992) has also shown the impairment of the association of endogenous cadherins with catenins in Xenopus embryos in which N-cadherin depleted of its extracellular domain was overexpressed.
By immunoblotting analysis using cadherin-6 antibodies of different sources, we consistently observed only a single broad band of ∼110–120 kD in MG-63 cells despite the fact that three cadherin-6 transcripts are present in these cells. Similar observation has been recently reported by Paul et al. (1997) using cadherin-6 antibody of another source. It is most likely that cadherin-6 and cadherin-6/1 may not be distinguishable by immunoblotting because they only differ by 24 amino acids. Thus, the broad single band ∼120–110 kD may represent cadherin-6, cadherin-6/1, and cadherin-6/2.
We have identified three cadherin-6 isoforms by RT-PCR, an observation that is consistent with previous reports where three cadherin-6 transcripts were detected by Northern analysis (Xiang et al., 1994; Shimoyama et al., 1995; Shimazu et al., 1996; Paul et al., 1997). In contrast, only cadherin-6/2 was found in cells of mouse origin including bone marrow cells and ST2 stromal cells. The alternative splicing reaction generating three cadherin-6 isoforms is compatible with the genomic structure of other cadherins (Berx et al., 1995). In the present study, we only determined the functional role of cadherin-6/2 in the mouse model of osteoclastogenesis. It is not known whether cadherin-6, cadherin-6/1, or cadherin-6/2 has identical or different roles in the osteoclast development in human. Loss of 85 amino acids and a 9–amino acid difference in the extracellular domain may confer altered functions on cadherin-6/2. However, in a search of the data bank, neither the sequence of lacking 85 amino acids nor the 9 different amino acids showed homology with known molecules that are shown to have biological activity. This might indicate that cadherin-6 or cadherin-6/1 has no additional or different functions compared with those of cadherin-6/2. It remains, however, still possible that each isoform of cadherin-6 may possess its own unique function in the regulation of osteoclast formation in the bone microenvironment. These issues are beyond the scope of the present study, but need to be examined in the future experiments.
Finally, the regulatory mechanisms of the splicing reaction generating three variants of cadherin-6 in human osteoblastic cells and one variant in mouse osteoblastic and stromal cells are unknown at the present time. Several disparities have been reported between human and mouse models of osteoclastogenesis in vitro. Whether the profile of cadherin-6 gene expression described here contributes to these differences is not known but may be potentially an intriguing issue to be pursued.
Acknowledgments
We would like to thank Dr. X. Ji for his contributions to the data base search, Dr. M. Wheelock for providing us with N-cadherin antibody and Drs. X. Liu and L. Cheng for sharing some of their materials with us.
Abbreviations used in this paper
- 1,25D3
1,25-dihydroxyvitamin D3
- GST
glutathione-S-transferase
- RT
reverse transcription
Footnotes
Address all correspondence to Toshiyuki Yoneda, D.D.S., Ph.D., Department of Medicine/Endocrinology, University of Texas Health Science Center, 7703 Floyd Curl Drive, San Antonio, TX 78284-7877. Tel.: (210) 567-4900. Fax: (210) 567-6693. E-mail: yoneda@uthscsa.edu
References
- Berx G, Staes K, Hengel JV, Molemans F, Bussemakers MJG, Bokhoven AV, Roy FV. Cloning and characterization of the human invasion suppressor gene E-cadherin (CDH1) Genomics. 1995;26:281–289. doi: 10.1016/0888-7543(95)80212-5. [DOI] [PubMed] [Google Scholar]
- Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, Brenner MB. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the αEβ7 integrin. Nature. 1994;372:190–193. doi: 10.1038/372190a0. [DOI] [PubMed] [Google Scholar]
- Donalies M, Cramer M, Ringwald M, Starzinski-Powitz A. Expression of M-cadherin, a member of the cadherin multigene family, correlates with differentiation of skeletal muscle cells. Proc Natl Acad Sci USA. 1991;88:8024–8028. doi: 10.1073/pnas.88.18.8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Chisaka O, Matsunami H, Takeichi M. Cadherin-6 expression transiently delineates specific rhombomeres, other neural tube subdivisions, and neural crest subpopulations in mouse embryos. Dev Biol. 1997;183:183–194. doi: 10.1006/dbio.1996.8501. [DOI] [PubMed] [Google Scholar]
- Karecla PI, Bowden SJ, Green SJ, Kilshaw PJ. Recognition of E-cadherin on epithelial cells by the mucosal T cell integrin αM290β7 (αEβ7) Eur J Immunol. 1995;25:852–856. doi: 10.1002/eji.1830250333. [DOI] [PubMed] [Google Scholar]
- Kintner C. Regulation of embryonic cell adhesion by the cadherin cytoplasmic domain. Cell. 1992;69:225–236. doi: 10.1016/0092-8674(92)90404-z. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, Wichen DV, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin–Tcf complex in APC−/−colon carcinoma. Science. 1997;275:1784–1786. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Matayoshi A, Brown C, DiPersio JF, Haug J, Abu-Amer Y, Liapis H, Kuestner R, Pacifici R. Human blood-mobilized hematopoietic precursors differentiate into osteoclasts in the absence of stromal cells. Proc Natl Acad Sci USA. 1996;93:10785–10790. doi: 10.1073/pnas.93.20.10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbalaviele G, Chen H, Boyce FB, Mundy GR, Yoneda T. The role of cadherin in the generation of multinucleated osteoclasts from mononuclear precursors in murine marrow. J Clin Invest. 1995;95:2757–2765. doi: 10.1172/JCI117979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Moon RT. Signal transduction through β-catenin and specification of cell fate during embryogenesis. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler WK. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1789. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest cell–cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development (Camb) 1995;121:1321–1332. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- Paul R, Ewing CM, Robinson JC, Marshall FF, Johnson KR, Wheelock MJ, Isaacs WB. Cadherin-6, a cell adhesion molecule specifically expressed in the proximal renal tubule and renal cell carcinoma. Cancer Res. 1997;57:2741–2748. [PubMed] [Google Scholar]
- Perkins SL, Kling SJ. Local concentrations of macrophage colony-stimulating factor mediate osteoclastic differentiation. Am J Physiol. 1995;269:1024–1030. doi: 10.1152/ajpendo.1995.269.6.E1024. [DOI] [PubMed] [Google Scholar]
- Pierelli L, Scambia G, D'Onofrio G, Ciarli M, Fattorossi A, Bonanno G, Menichella G, Battaglia A, Panici PB, Tommasi M, et al. Generation of multinuclear tartrate-resistant acid phosphatase positive osteoclasts in liquid culture of purified human peripheral blood CD341 progenitors. Br J Haematol. 1997;96:64–69. doi: 10.1046/j.1365-2141.1997.8602490.x. [DOI] [PubMed] [Google Scholar]
- Roodman GD. Advances in bone biology: the osteoclast. Endocr Rev. 1996;17:308–332. doi: 10.1210/edrv-17-4-308. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Schalken JA, Giroldi LA, Jansen CFJ, Akaza H, Koiso K, Debruyne FMJ, Bringuier P. Pronostic value of cadherin-associated molecules (α, β, and γ-catenins and p120cas) in bladder tumors. Cancer Res. 1996;56:3234–3237. [PubMed] [Google Scholar]
- Shimoyama Y, Gotoh M, Terasaki T, Kitajima M, Hirohashi S. Isolation and sequence analysis of human cadherin-6 complementary DNA for the full coding sequence and its expression in human carcinoma cells. Cancer Res. 1995;5:2206–2211. [PubMed] [Google Scholar]
- Suda T, Takahashi N, Martin TJ. Modulation of osteoclast differentiation. Endocr Rev. 1992;13:66–80. doi: 10.1210/edrv-13-1-66. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Sano K, Tanihara H. Diversity of the cadherin family: evidence for eight new cadherins in nervous tissue. Cell Regul. 1991;2:261–270. doi: 10.1091/mbc.2.4.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Akatsu TA, Udagawa N, Sasaki T, Yamaguchi A, Moseley JM, Martin TJ, Suda T. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 1988;123:2600–2602. doi: 10.1210/endo-123-5-2600. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Reddy SV, Chirgwin JM, Devlin RD, Haipek C, Anderson J, Roodman GD. Cloning and identification of annexin II as an autocrine/paracrine factor that increases osteoclast formation and resorption. J Biol Chem. 1994;269:28696–28701. [PubMed] [Google Scholar]
- Wijngaert FB, Burger EH. Demonstration of tartrate-resistant acid phosphatase in undecalcified, glycomethacrylate-embedded mouse bone: a possible marker for (pre) osteoclast identification. J Histol Cytol. 1986;34:1317–1323. doi: 10.1177/34.10.3745910. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tanaka M, Suzuki M, Igarashi H, Kiyokawa E, Naito Y, Ohtawara Y, Shen Q, Sugimura H, Kino I. Isolation of complementary DNA encoding K-cadherin, a novel rat cadherin preferentially expressed in fetal kidney and kidney carcinoma. Cancer Res. 1994;54:3034–3041. [PubMed] [Google Scholar]