Abstract
Transfer of mitochondria to daughter cells during yeast cell division is essential for viable progeny. The actin cytoskeleton is required for this process, potentially as a track to direct mitochondrial movement into the bud. Sedimentation assays reveal two different components required for mitochondria–actin interactions: (1) mitochondrial actin binding protein(s) (mABP), a peripheral mitochondrial outer membrane protein(s) with ATP-sensitive actin binding activity, and (2) a salt-inextractable, presumably integral, membrane protein(s) required for docking of mABP on the organelle. mABP activity is abolished by treatment of mitochondria with high salt. Addition of either the salt-extracted mitochondrial peripheral membrane proteins (SE), or a protein fraction with ATP-sensitive actin-binding activity isolated from SE, to salt-washed mitochondria restores this activity. mABP docking activity is saturable, resistant to high salt, and inhibited by pre-treatment of salt-washed mitochondria with papain. Two integral mitochondrial outer membrane proteins, Mmm1p (Burgess, S.M., M. Delannoy, and R.E. Jensen. 1994. J.Cell Biol. 126:1375–1391) and Mdm10p, (Sogo, L.F., and M.P. Yaffe. 1994. J.Cell Biol. 126:1361– 1373) are required for these actin–mitochondria interactions. Mitochondria isolated from an mmm1-1 temperature-sensitive mutant or from an mdm10 deletion mutant show no mABP activity and no mABP docking activity. Consistent with this, mitochondrial motility in vivo in mmm1-1 and mdm10Δ mutants appears to be actin independent. Depolymerization of F-actin using latrunculin-A results in loss of long-distance, linear movement and a fivefold decrease in the velocity of mitochondrial movement. Mitochondrial motility in mmm1-1 and mdm10Δ mutants is indistinguishable from that in latrunculin-A–treated wild-type cells. We propose that Mmm1p and Mdm10p are required for docking of mABP on the surface of yeast mitochondria and coupling the organelle to the actin cytoskeleton.
Mitochondria are indispensable organelles for normal eukaryotic cell function. Since mitochondria cannot be synthesized de novo, these organelles are inherited, i.e., transferred from mother to daughter during cell division. In the yeast Saccharomyces cerevisiae, vegetative cell division occurs by budding, a form of proliferation in which growth is directed toward the developing bud. Previous studies indicate that mitochondria undergo a series of cell cycle–linked motility events during normal inheritance in yeast (Simon et al., 1997). These are: (a) polarization of mitochondria towards the site of bud emergence in G1 phase; (b) linear, polarized movement of mitochondria from mother cells to developing buds in S phase; (c) immobilization of newly inherited mitochondria in the bud tip during S and G2 phases; and (d) release of immobilized mitochondria from the bud tip during M phase.
There is mounting evidence that the actin cytoskeleton controls mitochondrial morphology and inheritance during vegetative yeast cell growth. The two major actin structures of yeast observed by light microscopy are patches and cables. Actin cables are bundles of actin filaments that extend from the mother into the bud. Mitochondria colocalize with these actin cables (Drubin et al., 1993; Lazzarino et al., 1994). Moreover, mutations such as deletion of the tropomyosin I gene, TPM1, or the mitochondrial distribution and morphology gene, MDM20, which selectively destabilize actin cables, result in the loss of polarized mitochondrial movement and reduce transfer of mitochondria into buds (Herman et al., 1997; Simon et al., 1997). Together, these studies indicate that normal mitochondrial inheritance in yeast requires association of mitochondria with actin cables.
Cell-free studies reveal a possible mechanism underlying actin control of mitochondrial inheritance. Sedimentation assays document binding of mitochondria to the lateral surface of F-actin. This mitochondrial actin-binding activity is ATP-sensitive, saturable, reversible, and mediated by protein(s) on the mitochondrial surface (Lazzarino et al., 1994). In addition, ATP-driven, actin-dependent motor activity has been identified on the surface of mitochondria (Simon et al., 1995). These observations support a model of mitochondrial inheritance whereby mitochondria use an actin-dependent motor to drive their movement from mother to daughter cells along actin cable tracks.
Yeast genetic screens have revealed several genes, collectively referred to as mdm (mitochondrial distribution and morphology) and mmm (maintenance of mitochondrial morphology), which are required for mitochondrial inheritance (McConnell et al., 1990; Burgess et al., 1994; Sogo and Yaffe, 1994). We have focused on two of these genes: MDM10 and MMM1. Deletion of MDM10 leads to the development of giant spherical mitochondria, presumably by the collapse of elongated mitochondria into a spherical mass (Sogo and Yaffe, 1994). Deletion of MMM1 (Burgess et al., 1994) produces a similar phenotype. In both mutants, the fraction of buds without mitochondria is high, indicating defective mitochondrial inheritance. The proteins encoded by these genes, Mdm10p and Mmm1p, appear to be integral membrane proteins in the mitochondrial outer membrane. Here, we report tests of the hypothesis that Mmm1p and Mdm10p are required to link mitochondria to the cytoskeleton.
Materials and Methods
Yeast Strains
Yeast strains used in this study are listed in Table I. Yeast cell growth was carried out according to Sherman (1991).
Table I.
Yeast Strains Used in This Study
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| D273-10B | MATα | 1 | ||
| YPH252 | MATα, leu2, his3, ade2, trp1, lys2, ura3 | 2 | ||
| YPH253 | MATα, mmm1-1, leu2, his3, ade2, trp1, lys2, ura3 | 2 | ||
| MYY291 | MATα, ura3, leu2, his3 | 3 | ||
| MYY504 | MATα, mdm10::URA3, ura3, leu2, his3 | 3 | ||
| AH216 | MATα, leu2, his3, phoE, phoC | 2 | ||
| AH217 | MATα, om45::LEU2, leu2, his3, phoE, phoC | 2 | ||
| DAUL1 | MATα, ade2, ura3Δ, lys2 | 4 |
Sources of strains: American Type Culture Collection: (1) ATCC25657 (Rockville, MD); (2) R. Jensen (Johns Hopkins University, Baltimore, MD); (3) M. Yaffe (University of California, San Diego, CA); and (4) T. Fox (Cornell University, Ithaca, NY).
Purification of Yeast Mitochondria
Mitochondria were isolated according to Lazzarino et al. (1994) with the following modifications. Since the mutant strains grow poorly on a nonfermentable carbon source such as lactate, mmm1-1 and mdm10Δ mutants and the corresponding parents were grown at 23°C in rich, raffinose-based media (YPR). All other strains were grown at 30°C in lactate medium. In all cases, mid-log phase cells were harvested, washed, and then converted to spheroplasts in zymolyase-catalyzed reactions as described previously (Daum et al., 1982). To restore metabolic activity, spheroplasts were “regenerated” using a modification of published methods (Baker and Schekman, 1989) by centrifugation (2,000 g, 10 min at 4°C) and incubation at a concentration of 106 cells/ml in 1.2 M sorbitol, 0.3% (wt/vol) yeast extract, 0.05% (wt/vol) CaCl2, 0.05% (wt/vol) NaCl, 0.06% (wt/vol) MgCl2, 0.1% (wt/vol) NH4Cl, and either 2% (vol/vol) lactate, pH 5.5, or 2% raffinose for 45 min. mdm10Δ, mmm1-1, and the corresponding wild-type strains were regenerated at 22°C. All other regenerations were carried out at 30°C. Regenerated spheroplasts were then isolated by centrifugation (2,000 g, 10 min at 4°C), resuspended in breaking buffer (0.6 M sorbitol, 20 mM Hepes-KOH, pH 7.4, 2 mM MgCl2, 1 mM EGTA, 1 mM PMSF, and protease inhibitor cocktail [10 mM benzamidine, 1 mg/ml 1,10-phenanthroline, 0.5 mg/ml antipain, 0.5 mg/ml chymostatin, 0.5 mg/ml leupeptin, 0.5 mg/ml pepstatin, and 0.5 mg/ml aprotinin]), and disrupted using a Dounce homogenizer. Crude mitochondria were isolated from the homogenate by differential centrifugation (Daum et al., 1982). Further purification of the crude mitochondrial fraction was carried out by isopycnic centrifugation using Nycodenz (5-N-2,3-dihydroxypropylacetamido-2,4,6-triiodo-N,N′-bis (2,3-dihydroxy-propyl) isophthal-amide; Sigma Chemical Co., St. Louis, MO) gradients (Glick and Pon, 1995). Purified mitochondria were stored at a concentration of 6–20 mg/ml in iso-osmotic Nycodenz/breaking buffer solutions in vials suspended in liquid nitrogen, and then thawed at 30°C immediately before use.
Sedimentation Assay for ATP-sensitive Binding of F-Actin to Mitochondria
Binding assays were carried out as described previously (Lazzarino et al., 1994) using F-actin prepared from blocks of baker's yeast according to Kron et al. (1992). Briefly, isolated yeast mitochondria (2.5 mg/ml) were incubated with phalloidin-stabilized F-actin (4.4 μM) for 10 min at 30°C in 80 μl of RM buffer (0.6 M sorbitol, 20 mM Hepes-KOH, pH 7.4, 100 mM KCl, 2 mM MgCl2, 1 mM DTT, 1 mg/ml fatty acid–free BSA, 1 mM PMSF and protease inhibitor cocktail). In ATP-treated samples, ATP concentration was maintained at 2 mM by addition of ATP and an ATP-regenerating system (0.1 mg/ml creatine kinase and 10 mM creatine phosphate). To deplete the system of ATP, apyrase (type VI; Sigma Chemical Company) was added to a final concentration of 12.5 U/ml (Pon et al., 1989). In all cases, ATP was either added to or depleted from the reaction mixture before the addition of mitochondria. Subsequent to binding, mitochondria were separated from unbound actin filaments by centrifugation through a sucrose cushion (25% sucrose, 20 mM Hepes-KOH, pH 7.4, 1 mM PMSF, and protease inhibitor cocktail) at 12,500 g for 10 min. Proteins recovered in the mitochondrial pellet were separated by SDS-PAGE (Laemmli, 1970) and identified by Western blot analysis (Towbin et al., 1979) using polyclonal antibodies raised against the mitochondrial marker proteins cytochrome b2 and porin (gifts from G. Schatz, University of Basel, Basel, Switzerland) and monoclonal antibody raised against actin (c4d6) (Lessard, 1988).
Preparation of Salt-washed Mitochondria and Mitochondrial Salt Extract
To remove salt-sensitive peripheral proteins from the outer membrane, mitochondria (30 mg/ml) were incubated in SM1 buffer (0.6 M sorbitol, 20 mM Hepes-KOH, pH 7.4, 1 M KCl, 2 mM MgCl2, 1 mM PMSF, and protease inhibitor cocktail) for 15 min on ice. Thereafter, the mixture was centrifuged at 10,000 g for 5 min at 4°C. Salt-washed mitochondria (SW)1 were recovered in the pellet. The salt-extracted mitochondrial peripheral membrane proteins (SE; 0.5–0.6 mg/ml) were recovered in the supernatant. To regenerate the actin binding activity of salt washed mitochondria, the KCl concentration of SE was diluted to 0.2 M and 0.03 mg of SE was incubated with 0.2 mg salt-washed mitochondria for 15 min on ice. Mitochondria were then separated from the mixture by centrifugation at 10,000 g at 4°C, resuspended in RM buffer, and then used for the sedimentation assay.
To test whether mABP is a protein, SE were treated with trypsin (5 μg/ml) and chymotrypsin (5 μg/ml) for 30 min at room temperature. Proteolysis was stopped by adding protease inhibitor cocktail supplemented with soybean trypsin inhibitor (100 μg/ml) and Pefabloc SC (1 mM; Boehringer Mannheim Corp., Indianapolis, IN). The ability of protease-treated SE to restore F-actin binding activity to SW was determined as described above.
To degrade salt insensitive, presumably integral membrane proteins, salt-washed mitochondria were treated with proteolytic enzymes. SW (6 mg/ml) were incubated on ice with papain (0.7 μg/ml) in protease inhibitor-free RM buffer for 15 min. The reaction was stopped by adding 1 mM PMSF and 2.5 mM DFP. The mitochondria were recovered by centrifugation at 10,000 g for 5 min at 4°C. SW or protease treated salt-washed mitochondria were assayed for ATP-sensitive actin-binding activity as described above.
Actin Affinity Chromatography
Preparation of F-actin affinity columns was carried out according to Miller et al. (1991). 0.5 ml Affi-Gel 10 (Bio-Rad Laboratories, Hercules, CA) and 0.5 ml Sepharose CL-6B (Pharmacia Biotechnology Inc., Piscataway, NJ) were mixed in a plastic syringe with 0.5 ml 2 mg/ml F-actin in 50 mM Hepes, pH 7.4, 0.1 M KCl, 0.2 mM CaCl2, 0.2 mM ATP, 5 mM MgCl2, 10% glycerol, and 10 μg/ml phalloidin, and then kept overnight at 4°C. After washing, 0.5–0.7 mg F-actin remained on the column. For affinity chromatography of the mitochondrial salt extract, the F-actin column was equilibrated with 5 vol of column buffer (0.6 M sorbitol, 50 mM Hepes, pH 7.4, 0.2 M KCl, 1 mM EGTA, 3 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail). Thereafter, salt extract from 50 mg Nycodenz-purified mitochondria was applied to the column. The salt extract used was prepared as described above and diluted with column buffer until the final concentration of KCl was 0.2 M before loading onto the column. To remove nonspecifically bound proteins, the column was washed twice with 2 vol of column buffer. Elution of ATP-sensitive actin-binding proteins was carried out in two steps by applying 2 vol of the column buffer containing first 0.1 mM ATP, and then 1 mM ATP. The flow rate during chromatography was maintained at 0.05 ml/min. The fractions from each step were collected.
To test the ability of the fractions to restore actin-binding activity of SW, 5% (vol/vol) of the salt extract and the column flow through, and 12% (vol/vol) of the eluates from the second wash, the 0.1 mM ATP wash, and the 1 mM ATP wash were incubated with SW. Mitochondria were then separated from the mixture by centrifugation at 10,000 g at 4°C, dissolved in RM buffer, and then used in the sedimentation assay.
Visualization of Mitochondria and the Actin Cytoskeleton
MDM10 parent (MYY291) and mdm10Δ (MYY504) mutant cells were grown to mid-log phase overnight in YPR at 22°C. MMM1 parent (YPH252) and mmm1-1 mutant (YPH253) cells were grown to mid-log phase in YPR at 22°C, and then shifted to 37°C for 3 h. Indirect immunofluorescence to stain mitochondria in fixed cells was carried out as described previously (Smith et al., 1995). In some cases, mitochondria were visualized in living cells using a fusion protein consisting of the mitochondrial signal sequence of citrate synthase 1 fused to green fluorescent protein (CS1-GFP). CS1-GFP was expressed in DAUL1 cells using a centromere-based plasmid under control of the Gal1-10 promoter (Zelenaya-Troitskaya et al., 1997). DAUL1 cells were transformed using the lithium acetate method (Ito et al., 1983). Cells expressing CS1-GFP were grown to mid-log phase in a synthetic, galactose-based liquid media (SGal-ura+4× ade) to mid-log phase at 30°C. Samples were mounted on microscope slides and visualized by fluorescence microscopy as described below. CS1-GFP labeling of mitochondria is specific and has no detectable effect on mitochondrial morphology or movement under our experimental conditions.
The actin cytoskeleton was counterstained in fixed DAUL1 cells expressing CS1-GFP using rhodamine–phalloidin (Molecular Probes, Eugene, OR), a ligand that binds specifically to actin polymers (Cooper, 1987). The fluorescent ligand was added to fixed samples to a final concentration of 2.5 mM in a solution consisting of a 4:1 ratio of NS (20 mM Tris-HCl, pH 7.6, 0.25 M sucrose, 1 mM EDTA, 1 mM MgCl2, 0.1 mM ZnCl2, 0.1 mM CaCl2, 0.8 mM PMSF, 0.05% [vol/vol] 2-mercaptoethanol) to methanol, and samples were allowed to stand in the dark at 4°C for 16 h. Stained cells were mounted onto microscope slides, and visualized by fluorescence microscopy.
For some experiments, mitochondria were visualized in living cells using the membrane potential–sensing dye, 3,3′-dihexyloxacarbocyanine iodide (DiOC6; Molecular Probes). The cell density of mid-log phase samples was adjusted to 107 cells/ml. Thereafter, the sample was incubated in medium containing 20 ng/ml of DiOC6 for 5 min at room temperature. Cells were washed once and resuspended to a final concentration of 2 × 108 cells/ml in medium. Samples were mounted on microscope slides and visualized by fluorescence microscopy. At the concentrations used, DiOC6 is specific for mitochondria and has no detectable effect on cell viability (Simon et al., 1995). In addition, since DiOC6 is a membrane potential– sensing dye, normal mitochondrial staining with this dye indicates that the organelle is intact and has normal respiratory activity. Therefore, although mmm1-1 and mdm10Δ mutants have a high incidence of loss of mitochondrial DNA and accompanying respiratory defects, all mutant and wild-type cells used in these experiments contain mtDNA.
Light Microscopy
Images were collected with a Zeiss Axioplan II microscope (Carl Zeiss, Oberkochen, Germany) using a Plan-Apochromat 100×, 1.4 NA objective lens, and a cooled CCD camera (Star-1; Photometrics, Tucson, AZ). Illumination with a 100-W mercury arc lamp was controlled with a shutter (Uniblitz D122; Vincent Associates, Rochester, NY). Image enhancement and analysis was performed on a Macintosh Quadra 800 computer (Apple Computer Co., Cupertino, CA) using the public domain program, NIH Image v1.60 (National Institutes of Health, Bethesda, MD).
Velocity Measurements
Velocities of mitochondrial movement were determined from images recorded at 20-s intervals >10 min of real time. In wild-type cells (MMM1, MDM10, and DAUL1), only velocities of organelles undergoing linear movement for at least three consecutive frames (1 min of real time) were measured. In the mmm1-1 and mdm10Δ mutants, and in DAUL1 cells treated with latrunculin-A, linear movements were not observed; therefore, the velocities of random samples of organelles were measured. For all velocity measurements, NIH Image v1.60 was used to determine the change in position (x and y coordinates) of mitochondria per unit time, and these were averaged to give a mean velocity. Polarized movement is defined as that which achieves a net displacement toward the bud in budding cells and is expressed as the percentage of all motile organelles exhibiting polarized movement over the time-lapse course.
Results
Latrunculin-A Treatment Produces Defects in Mitochondrial Morphology and Movement
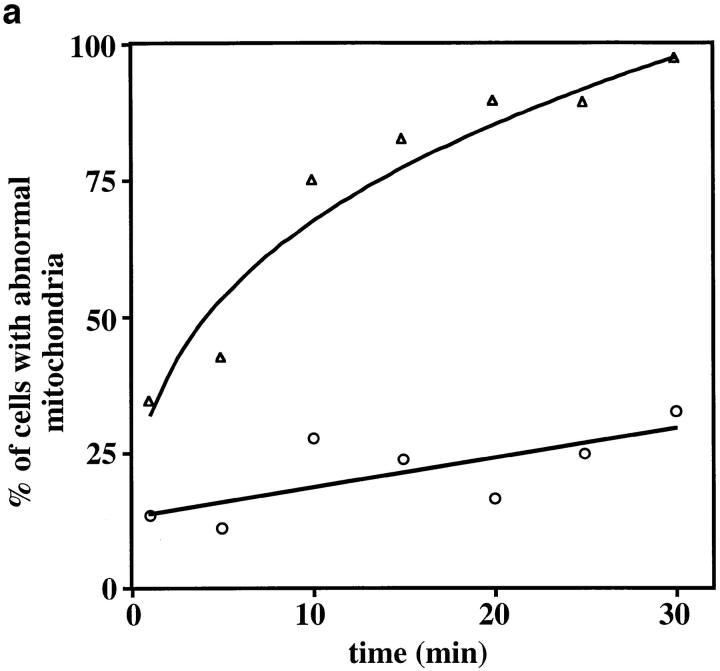
Latrunculin-A (Lat-A) treatment results in rapid and quantitative loss of F-actin in yeast and other eukaryotes (Spector et al., 1983; Ayscough et al., 1997). We used this agent to examine the acute effect of actin depolymerization on mitochondrial morphology and movement in vivo. Yeast containing green fluorescent protein (GFP)-labeled mitochondria were treated with a concentration of Lat-A that inhibits growth in the halo assay (Ayscough et al., 1997). This results in rapid depolymerization of F-actin, as assessed by rhodamine phalloidin staining, and fragmentation of tubular mitochondria into short tubular and small spherical structures (Fig. 1). There is a correlation between the time course of actin depolymerization and loss of normal mitochondrial morphology. Within 2 min of Lat-A addition, when most actin patches and cables are absent, we observe defects in mitochondrial morphology and position. Specifically, we observe loss of mitochondrial polarization along the mother–bud axis, and fragmentation of mitochondria to form short tubular or spherical structures. Moreover, we observe an increase in the extent of mitochondrial morphology and position defects with Lat-A treatment over a period of 30 min (Fig. 2 a).
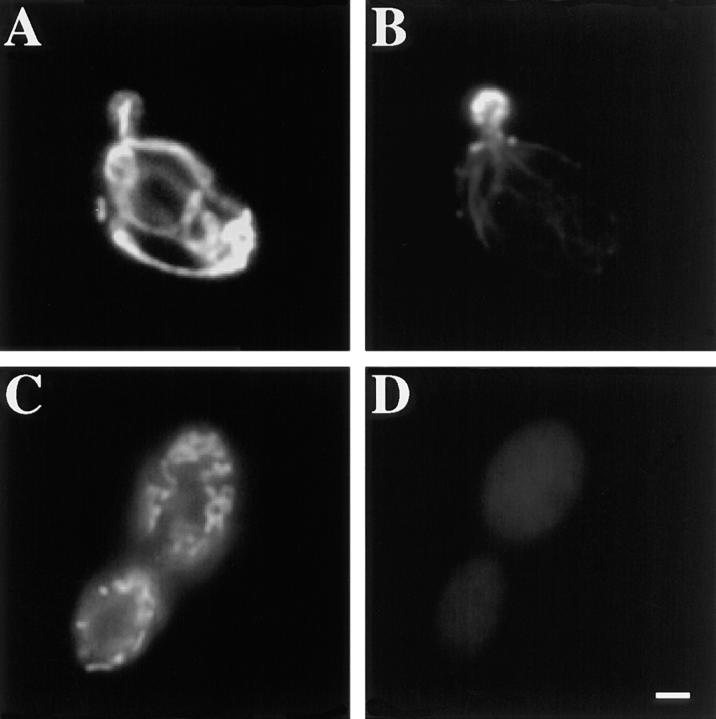
Figure 1.
Mitochondrial morphology is defective after short-term treatment with latrunculin-A. Wild-type cells expressing CS1-GFP were grown at 30°C in a synthetic galactose-containing media lacking uracil. 0.25 mM latrunculin-A dissolved in DMSO (C–D) or equal volume of DMSO alone (A–B) was added to mid-log phase cultures. After a 20-min incubation cells were fixed and stained with rhodamine phalloidin to visualize actin structures (B and D). Mitochondria are labeled with GFP (A and C). Bar, 1 μm.
Figure 2.
Lat-A treatment results in loss of linear, long-distance mitochondrial movement. (a) Cells containing GFP-labeled mitochondria were incubated in the presence (▵) or absence (○) of 0.25 mM Lat-A, and mitochondrial morphology and position were evaluated as a function of time of incubation. Cells containing fragmented mitochondria, spherical mitochondria, or mitochondria that did not align along the mother–bud axis were scored as abnormal (n > 100 for each time point). (b) Paths of GFP-labeled mitochondria were obtained from cells incubated in the absence (A) or presence (B) of 0.25 mM Lat-A for 20 min. Images were acquired at 20-s intervals over a period of 10 min. Tracings of the movements of individual mitochondria were made by marking the tip of motile organelles during the time in which they remained in the plane of focus. The points denote the position of organelles at 20-s intervals. Tracings are shown relative to the boundary of the dividing yeast cell. Bar, 1 μm.
The effect of Lat-A–induced F-actin depolymerization on mitochondrial motility was evaluated further using time-lapse fluorescence microscopy. Movement of GFP-labeled mitochondria in untreated control cells is similar to that observed in several other wild-type strains (Simon et al., 1995, 1997). This movement is linear and long distance (Fig. 2 b, panel A), with an average velocity of 27.5 ± 4.7 nm/s. In addition, mitochondrial movement in control cells is polarized: 51.5% of motile mitochondria move toward the bud (n = 100). Within 2–5 min of Lat-A addition we also observe loss of the normal pattern of mitochondrial movement. Mitochondria in Lat-A–treated cells undergo short oscillatory movements which resemble Brownian motion (Fig. 2 b, panel B); only 2.5% of the observed movements are directed towards the bud (n = 100). In addition, the average velocity of nonlinear movement produced by treatment with Lat-A for 2–5 min is 5.7 ± 1.7 nm/s, a velocity at the limit of detection in our system. Thus, actin-independent mitochondrial movements are nonlinear and nonpolarized with a velocity at least fivefold less than that of actin-dependent mitochondrial movement.
Mitochondrial–Actin Interactions In Vivo Are Disrupted in mmm1 and mdm10 Mutants
Mdm10p and Mmm1p are integral mitochondrial outer membrane proteins required for normal mitochondrial morphology and inheritance (Burgess et al., 1994; Sogo and Yaffe, 1994). Consistent with previous reports, we found that the efficiency of mitochondrial inheritance is decreased in an mdm10 deletion mutant (mdm10Δ) and an mmm1 temperature-sensitive mutant (mmm1-1). Analysis of mdm10Δ at 22°C, and mmm1-1 at restrictive temperature, reveals that 30% of mutant cells with small buds contain mitochondria in the bud compared with >90% for wild-type cells (Table II). In addition, buds of these mutants display defects in mitochondrial localization. Whereas 65% of wild-type cells bearing large buds show accumulation of mitochondria in the bud tip, accumulation of mitochondria in the bud tip is observed in only 10–12% of large-budded mmm1-1 and mdm10Δ mutants (Table II).
Table II.
Characteristics of Mitochondrial Motility and Inheritance in mmm1-1 and mdm10Δ Mutants
| Velocity | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Linear movements | Non-linear movements | % Polarized movement | % Inheritance | % Bud tip accumulation | ||||||||||||
| nm/s | ||||||||||||||||
| MMM1 | 32.4 ± 7.1 | ND | 52 | (n = 51) | 97.0 | (n = 600) | 64.7 | (n = 371) | ||||||||
| mmm1-1 | 0 | 9.27 ± 3.3 | 2.1 | (n = 54) | 30.5 | (n = 600) | 10.7 | (n = 343) | ||||||||
| MDM10 | 25.1 ± 6.6 | ND | 51 | (n = 56) | 95.8 | (n = 400) | 65.1 | (n = 347) | ||||||||
| mdm10Δ | 0 | 8.1 ± 3.1 | 1.8 | (n = 54) | 27.5 | (n = 400) | 12.9 | (n = 322) | ||||||||
Parent and mutant cells were grown and visualized as described for Fig. 3. Mitochondria were visualized with the membrane potential–sensing dye DiOC6. Mitochondrial velocities were measured as described in Materials and Methods. Polarized movement refers to the percentage of motile mitochondria that show net displacement toward the bud. Inheritance represents the percentage of small buds that contain mitochondria. Bud tip accumulation refers to the percentage of large buds that show immobilization of mitochondria in the tip of the bud. ND, not determined.
Bud-directed mitochondrial movement during inheritance in vegetative yeast requires actin cables (Simon et al., 1997). Since mitochondria coalign with actin cables in vivo (Drubin et al., 1993; Lazzarino et al., 1994; Fig. 3, arrows), and support F-actin filament sliding on their surface in vitro (Simon et al., 1995), it is likely that mitochondria use actin cables as tracks for movement into the bud during vegetative growth. We examined the organization of mitochondria and the actin cytoskeleton in mmm1-1 and mdm10Δ mutants (Fig. 3). Although mitochondrial morphology and inheritance are abnormal in these mutants (Burgess et al., 1994; Sogo and Yaffe, 1994) (Table II), these mutants contain actin cables with normal polarization along the mother–bud axis.
Figure 3.

The organization of mitochondria and actin in mmm1-1 and mdm10Δ strains. MMM1 parent (YPH252; A–B) and mmm1-1 mutant (YPH253; C–D) cells were grown overnight at 22°C, and then shifted to 37°C for 3 h. MDM10 parent (MYY291; E–F) and mdm10Δ mutant (MYY504; G–H) cells were grown at 22°C overnight. Fixed cells were stained for mitochondria by indirect immunofluorescence using an antibody raised against mitochondrial outer membrane proteins (A, C, E, and G) and for actin using rhodamine phalloidin (B, D, F, and H). Arrows point to examples of alignment of mitochondria with actin cables in wild-type cells. Bar, 1 μm.
In wild-type yeast, actin–mitochondria interactions control mitochondrial movement. Therefore, we studied the effect of mdm10Δ and mmm1-1 mutations on mitochondrial motility using time-lapse fluorescence microscopy and the membrane potential–sensing dye, DiOC6. Mitochondrial movement in MMM1 and MDM10 wild-type strains is similar to that in other wild-type cells: we observe linear, long-distance, bud-directed mitochondrial movement with an average velocity of 25–30 nm/s (Fig. 4, A and C; Table II). In contrast, mitochondrial movement in the mmm1-1 (at restrictive temperatures) and mdm10Δ mutants is abnormal, and indistinguishable from the actin- independent movement observed with Lat-A. Mitochondrial movement in mmm1-1 and mdm10Δ mutants is short, oscillatory, and nonpolarized with a velocity that is at least threefold lower than that of mitochondria in wild-type cells (Fig. 4, B and D; Table II). Thus, mitochondrial movement in mdm10Δ and mmm1-1 mutants is indistinguishable from the actin-independent movements observed in Lat-A–treated cells. This supports the interpretation that MMM1 and MDM10 are required for association of mitochondria with the actin cytoskeleton.
Figure 4.
Mitochondrial motility is defective in mmm1 and mdm10 mutants. Tracings of mitochondrial movement in MMM1 (A), mmm1-1 (B), MDM10 (C), and mdm10Δ cells (D) were performed as for Fig. 2. The points denote the position of organelles at 20-s intervals. Tracings are shown relative to the boundary of the dividing yeast cell. Bar, 1 μm.
Actin–Mitochondrial Interactions In Vitro Are Mediated by Salt-extractable Peripheral Mitochondrial Outer Membrane Protein(s) with ATP-sensitive Actin-binding Activity
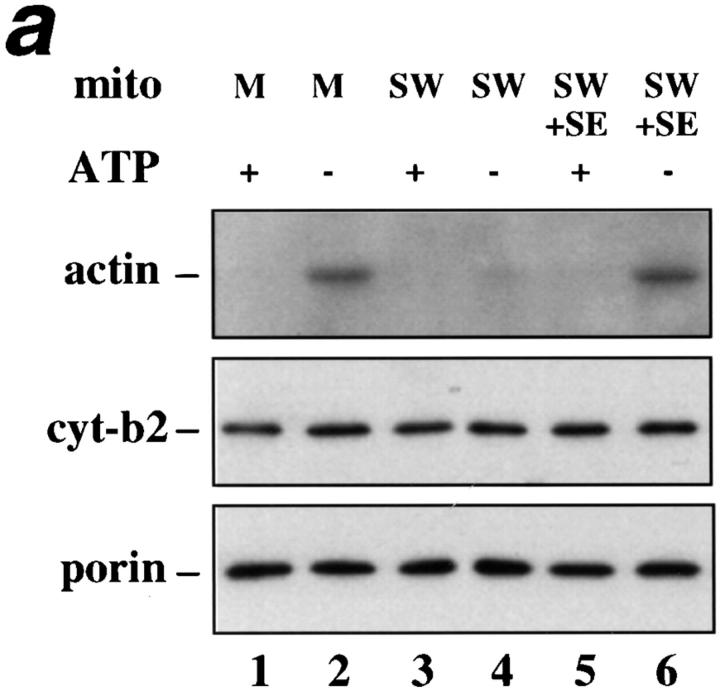
We studied the mechanism of coupling mitochondria to the actin cytoskeleton of yeast using a sedimentation assay that measures binding of isolated mitochondria to F-actin (Lazzarino et al., 1994). Using this assay, we find that mitochondrial actin binding is ATP sensitive, saturable, reversible, and mediated by a mitochondrial surface protein or proteins. In a typical assay, mitochondria are incubated with phalloidin-stabilized yeast actin filaments in the presence or absence of ATP, and actin recovered with the mitochondrial pellet after low speed centrifugation is interpreted as mitochondria-bound actin. As shown in Fig. 5 a (lanes 1 and 2) actin–mitochondria interactions are ATP sensitive: F-actin sediments with mitochondria only in the absence of ATP. Protease sensitivity experiments (see below) show that this activity is protein mediated. For simplicity, we will refer to the proteins that mediate ATP-sensitive actin-binding activity on mitochondria as mABP (mitochondrial actin binding protein[s]).
Figure 5.
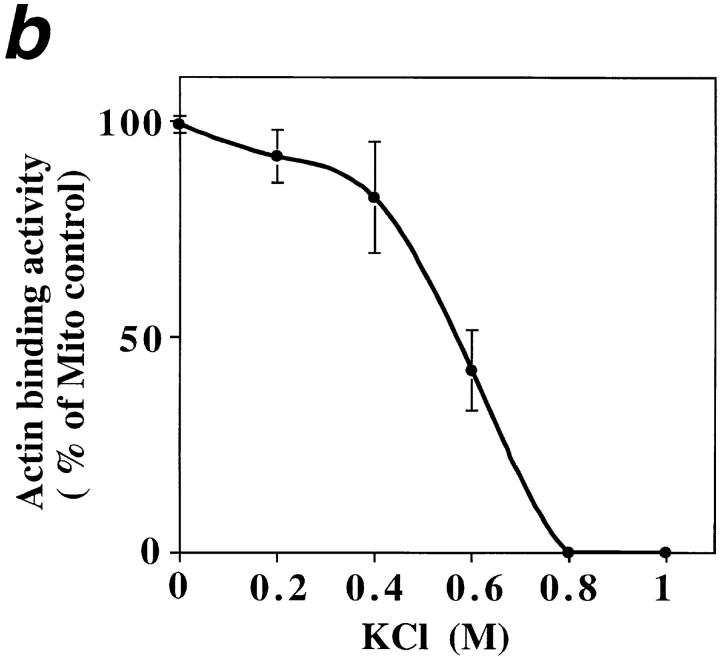
ATP-sensitive actin-binding activity of mitochondria requires a peripheral membrane protein which binds reversibly to mitochondria. (a) Sedimentation assays of isolated mitochondria from the D273-10B strain with phalloidin-stabilized F-actin in the presence or absence of ATP were carried out as described in Materials and Methods. The mitochondrial pellets of the sedimented reaction mixtures were evaluated for the levels of actin and of two mitochondrial marker proteins (cytochrome b2 and porin) by Western blot analysis. (1 and 2) Untreated (control) mitochondria (M); (3 and 4) salt-washed mitochondria (SW); and (5 and 6) salt-washed mitochondria (SW) pretreated with mitochondrial salt extract (SE). (b) The effect of KCl concentration on isolating SE. Isolated mitochondria were incubated in increasing amounts of salt. KCl was removed and the level of ATP-sensitive actin-binding activity of SW was tested using the sedimentation assay. The relative ATP-sensitive actin binding is shown here. 100% was defined as the amount of actin-binding activity in control, untreated mitochondria.
To evaluate the disposition of mABP activity on mitochondrial membranes, isolated mitochondria were pre-treated with 1 M KCl. This high salt treatment releases peripheral membrane proteins, but has no effect on the recovery of either an integral mitochondrial outer membrane marker (porin) or an intermembrane space marker protein (cytochrome b2; Fig. 5 a). Thus, 1 M KCl treatment selectively releases peripheral membrane proteins from the mitochondrial outer membrane without affecting the integrity of the organelle. This high salt pre-treatment of mitochondria blocks all detectable mABP activity (Fig. 5 a, lanes 3 and 4).
One interpretation of this finding is that salt treatment releases a peripheral mitochondrial membrane protein or proteins required for mABP activity. It is also possible that salt treatment inhibits mABP activity by some other mechanism. To distinguish between these possibilities, the KCl concentration of the SE was diluted to 200 mM, and the mixture was incubated with SW for 10 min at 4°C (SE + SW). This treatment restores mABP activity to SW (Fig. 5 a, lanes 5 and 6). Thus, mABP is or requires a peripheral mitochondrial membrane protein that binds reversibly to the mitochondrial surface.
To characterize mABP activity further, we evaluated the effect of KCl titration on actin–mitochondrial interactions (Fig. 5 b). Pre-treatment with ≤200 mM KCl had no significant effect on the level of mABP activity. However, pre-treatment with increasing concentrations of KCl from 0.4–0.8 M resulted in a KCl concentration-dependent loss of this activity. 0.8 M KCl pre-treatment abolishes all detectable activity. Thus, mABP activity resides in or requires a protein that is tightly associated with the mitochondrial outer membrane.
Finally, to distinguish between the possibilities that mABP either is or requires a peripheral mitochondrial outer membrane protein(s), we tested (a) for ATP-sensitive actin-binding activity in SE, and (b) whether these actin binding protein(s) could restore mABP activity to SW. Mitochondria were extracted with 1 M KCl, and salt- extracted proteins were passed over an F-actin affinity column. After washes to remove nonspecifically bound material, proteins with ATP-sensitive actin-binding activity were eluted by sequential washes with buffers containing 0.1 mM and 1 mM ATP. Approximately 0.1% of the protein loaded was recovered from the actin column upon elution with 0.1 mM ATP; virtually no protein was recovered after subsequent elution with 1 mM ATP. Thus, there are proteins in SE with ATP-sensitive actin-binding activity.
Next, we tested the ability of this actin binding fraction to restore mABP activity to salt-washed mitochondria (Fig. 6). mABP activity was restored to SW with (a) the flow through from the F-actin column, and (b) the fraction eluted from the F-actin column with 0.1 mM ATP. No other fraction showed mABP restoration activity. These observations indicate that mABP is a peripheral mitochondrial membrane protein with actin-binding activity that shows reversible association with the organelle.
Figure 6.
mABP is a peripheral mitochondrial membrane protein. Fractions recovered from F-actin affinity chromatography of SE (see Materials and Methods) were incubated with SW for 10 min at 4°C. mABP activity in these organelles was then assayed as for Fig. 5. Mito, control mitochondria; SW, salt-washed mitochondria; FT, flow through from the actin affinity column; W, eluate after washing the column with 0.2 M KCl.
mABP Association with Mitochondria Is Saturable and Mediated by a Salt-inextractable Mitochondrial Membrane Protein(s)
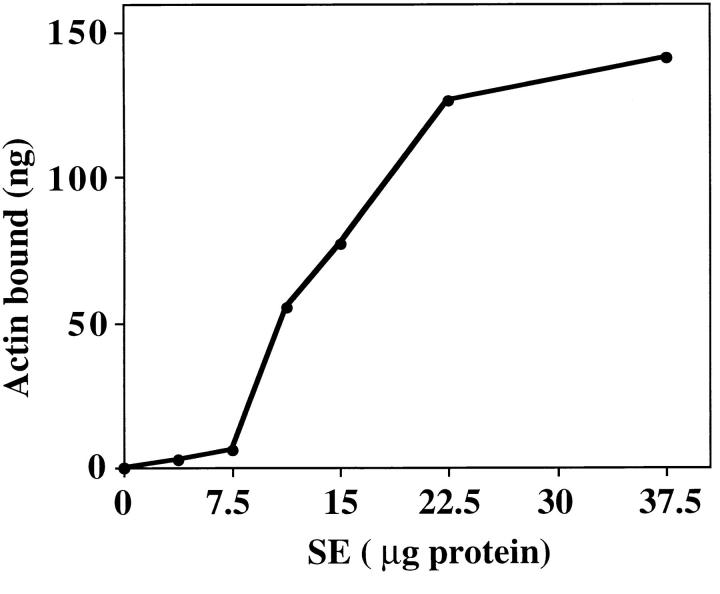
To investigate mABP docking on mitochondria, we incubated SW with increasing amounts of SE (Fig. 7). Low levels of SE do not restore mABP activity to SW. Upon incubation with increasing amounts of SE, the amount of restored mABP activity in SW increases linearly and then plateaus. This indicates that binding of mABP to the organelle is both saturable and dependent on SE concentration, suggesting a limited number of binding sites on the surface of SW. The maximum level of restored mABP activity is greater than that of control, untreated mitochondria. Thus, the salt-inextractable mABP docking protein is present in excess over mABP in our organelle preparation.
Figure 7.
The effect of increasing amount of SE on restoration of mABP activity in SW. SW were pre-treated with increasing amounts of salt-extractable membrane proteins (SE). Mitochondria were re-isolated and their ATP-sensitive binding activity was determined by sedimentation assay. The level of actin cosedimenting with mitochondria was measured by comparing the band densities of organelle-associated actin with those of known quantities of actin.
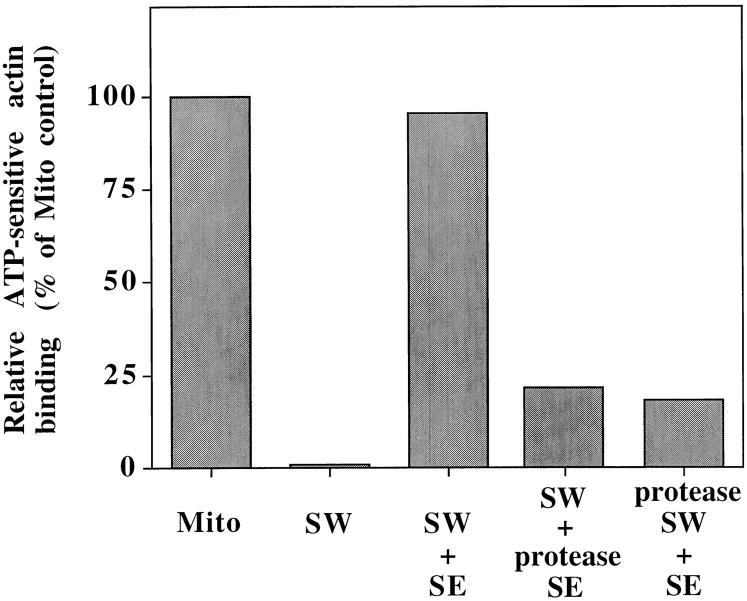
Finally, to test whether mABP and the salt-inextractable component required for mABP association with mitochondria are proteins, we tested the effect of protease treatment on mABP activity and mABP docking activity. For the former, we find that protease treatment of salt- extracted mitochondrial peripheral membrane proteins blocks their ability to restore mABP activity to SW (Fig. 8). Thus, mABP is a protein. For the latter, SW were treated with protease before incubation with saturating levels of SE. This treatment results in degradation of mitochondrial surface proteins without affecting the integrity of the organelle, and inhibits binding of mABP to SW (Fig. 8). Thus, mABP docking on mitochondria is also protein mediated. Together our findings reveal two components required for actin–mitochondria interactions: one is mABP, a salt-extractable peripheral mitochondrial membrane protein(s); the other is a salt-inextractable mitochondrial membrane protein(s) needed for mABP docking on mitochondria.
Figure 8.
Salt-inextractable mitochondrial membrane protein(s) are required for binding of mABP to mitochondria. To confirm that mABP is a protein, salt-extracted mitochondrial proteins (SE) were treated with trypsin and chymotrypsin (protease SE). The ability of protease-treated SE to restore F-actin–binding activity to SW was determined as for Fig. 5. To confirm that mABP docking is protein mediated, salt-washed mitochondria (SW) were incubated with papain (protease SW). Protease-treated mitochondria were recovered by centrifugation at 10,000 g for 5 min at 4°C. Actin-binding activity of each sample was determined as in Fig. 5, and the relative ATP-sensitive actin-binding activities are presented. 100% was defined as the amount of binding activity in the control mitochondria sample. In all cases, the recovery of actin with the mitochondrial pellet was normalized for equal recovery of porin.
Mmm1p and Mdm10p Are Required for mABP Association with Mitochondria
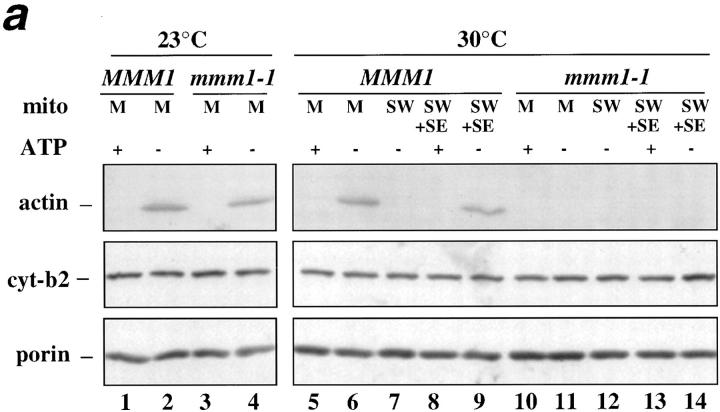
In light of this finding, we tested whether the mitochondrial integral outer membrane proteins, Mdm10p and Mmm1p, are required for (a) mABP activity, and (b) association of mABP with salt-washed mitochondria. Assays were performed using mitochondria isolated from the mdm10Δ mutant, the temperature-sensitive mmm1-1 mutant, and corresponding wild-type parent strains. The temperature-sensitive mmm1-1 mutant displays normal morphology at 23°C, and defects in mitochondrial morphology at both 30° and 37°C. Consistent with this, we find that mitochondria from the mmm1-1 strain show mABP activity at 23°C (Fig. 9 a, lanes 3 and 4), but no mABP activity at 30°C (Fig. 9 a, lanes 10 and 11). Thus, we observe a correlation between the temperature sensitivity of mABP activity in vitro and mitochondrial morphology in vivo of mmm1-1 mutants. In addition, we find that SE from wild-type MMM1 mitochondria restores mABP activity in salt-washed MMM1 mitochondria at 30°C (Fig. 9 a, lanes 8 and 9). However, this SE from wild-type MMM1 mitochondria does not restore mABP activity to salt-washed mmm1-1 mitochondria (Fig. 9 a, lanes 13 and 14). Thus, Mmm1p is required for binding of mABP with the mitochondrial surface.
Figure 9.
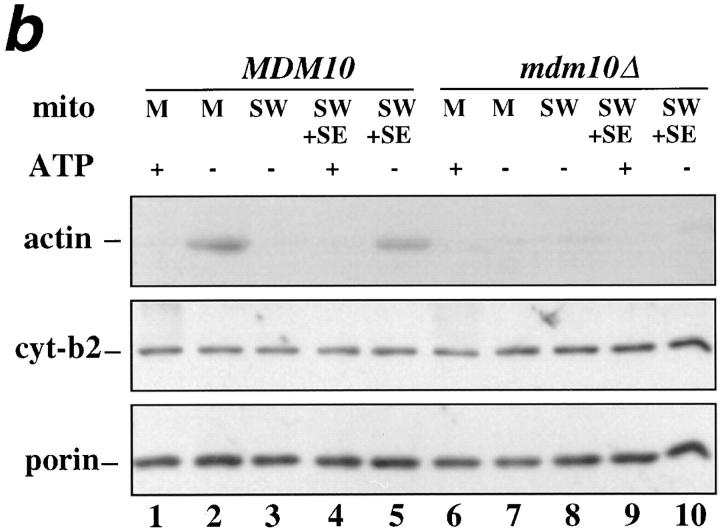
mmm1-1 and mdm10Δ mitochondria, but not om45Δ mitochondria, are defective in ATP-sensitive actin-binding activity. Mitochondria were isolated from wild-type parent and the corresponding mutant strains: MMM1, mmm1-1 (a), MDM10, mdm10Δ (b), and OM45, om45Δ (c). Sedimentation assays of isolated mitochondria were carried out with phalloidin-stabilized F-actin in the presence and absence of ATP (+/− ATP) at 30°C and also at 23°C in the case of MMM1 and mmm1-1 mitochondria. The mitochondrial pellets of the sedimented reaction mixtures were evaluated for the levels of actin, cytochrome b2, and porin by Western blot analysis. M, untreated mitochondria incubated with F-actin. SW, salt-washed mitochondria incubated with F-actin. SW+SE, salt-washed mitochondria from the strains indicated pretreated with the corresponding wild-type mitochondrial salt extract before incubation with F-actin.
We observe similar results with mitochondria isolated from mdm10Δ cells. MDM10 wild-type mitochondria show mABP activity, and mABP-binding activity (Fig. 9 b, lanes 1–5). In contrast, neither activity is observed in mitochondria isolated from mdm10Δ cells (Fig. 9 b, lanes 6–10). Based on these experiments, both Mmm1p and Mdm10p are required for the association of mABP to the mitochondrial surface in vitro.
To test whether deletion of other mitochondrial membrane proteins affects mABP docking activity, we measured actin–mitochondria interactions in organelles lacking OM45p. OM45p is a major integral mitochondrial outer membrane protein, facing the cytoplasm. Deletion of this protein has no discernible effect on cell viability or mitochondrial function (Yaffe et al., 1989). The absence of OM45p also did not alter either mABP activity or mABP docking activity (Fig. 9 c, lanes 6–10).
Discussion
Interactions between organelles and the cytoskeleton can provide a means for localization and transport of organelles in the cell. According to the simplest model, force-generating molecules on the surface of organelles use the cytoskeleton as tracks for this motility. Although numerous observations document microtubule-based transport systems in mitochondrial motility, increasing evidence supports the idea that mitochondrial motility can be actin based. Microfilament-dependent mitochondrial movement has been observed in axons of mammalian neurons (Morris and Hollenbeck, 1995), in the photoreceptor cells of the locust Schistocerca gregaria (Sturmer et al., 1995) and in Malpighian tubule cells of the insect Rhodnius prolixus (Bradley and Satir, 1979). Here, we focus on the role of the actin cytoskeleton in mitochondrial movement in the budding yeast, Saccharomyces cerevisiae.
Previous studies in yeast indicate that mitochondria colocalize with actin cables, and that mutations that cause actin disorganization or selective destabilization of actin cables affect mitochondrial morphology, motility, and inheritance. These studies support functional interactions between yeast mitochondria and the actin cytoskeleton. We present evidence that Lat-A, an agent that causes rapid, quantitative actin depolymerization, also produces rapid, quantitative defects in mitochondrial motility. Within minutes of Lat-A application, concomitant with actin depolymerization, mitochondria undergo fragmentation, loss of polarized, linear, long-distance movement, and show severely reduced velocity of movement. Indeed, mitochondria display random, apparently Brownian motions in the absence of F-actin. Thus, these studies provide further support for the model that mitochondria interact directly with the actin cytoskeleton in vegetative yeast, and support a role for the yeast actin cytoskeleton as the sole mediator of directed mitochondrial movement.
Our studies on the mechanism underlying actin–mitochondria interactions focus on mABP, a mitochondrial surface protein or proteins that mediates ATP-sensitive, saturable, and reversible binding of the organelle to F-actin in vitro. Here, we demonstrate that mABP is a peripheral mitochondrial outer membrane protein or proteins. We find that mABP (a) is released from the mitochondrial surface with high salt, (b) binds reversibly and saturably to the mitochondrial surface, and (c) requires a salt-inextractable, presumably integral membrane protein or proteins for association with mitochondria.
We present evidence that Mmm1p and Mdm10p, two integral mitochondrial outer membrane proteins, are required for mABP binding to mitochondria in vitro, for colocalization of mitochondria with actin cables, and for actin-dependent, directed mitochondrial movement in vivo. These studies used mmm1-1, a temperature-sensitive mutant that shows wild-type mitochondrial characteristics at permissive temperature, and defects in mitochondrial morphology and inheritance at restrictive temperatures (Burgess et al., 1994). We observe temperature-sensitive actin– mitochondrial interactions in vitro: mitochondria isolated from mmm1-1 mutants grown at permissive temperature show mABP activity only at permissive temperature. Moreover, mmm1-1 mitochondria have no mABP binding or mABP docking activity upon shift to restrictive temperatures. Similar results were obtained with an mdm10 deletion mutant. Mitochondria isolated from mdm10Δ cells show no mABP activity and no mABP docking activity. This requirement for Mdm10p and Mmm1p in mABP binding appears to be specific. Mitochondria isolated from cells bearing a deletion in OM45p, another integral mitochondrial outer membrane protein, have full mABP and mABP docking activities.
In complementary studies, we find that mitochondrial motility in mmm1-1 and mdm10Δ mutants is similar to the actin-independent, random motility observed in Lat-A–treated cells. In these mutants, mitochondria display nonpolarized, low velocity movements with abrupt directional changes. Indeed, mitochondria in both mutants and in Lat-A–treated wild-type cells appear to flail randomly. Our interpretation of these observations is that Mmm1p and Mdm10p are required for mABP binding to mitochondria, and that mABP–mitochondrial association is required for actin-directed mitochondrial motility in vivo.
Mmm1p, a 49-kD protein, resides in the mitochondrial outer membrane and has a putative membrane-spanning domain close to its NH2 terminus. Biochemical studies indicate that the COOH terminus of Mmm1p faces the cytosol (Burgess et al., 1994). Mdm10p, a 56-kD protein, also resides in the mitochondrial outer membrane (Sogo and Yaffe, 1994). Although the predicted amino acid sequence of Mdm10p does not reveal a membrane-spanning domain, Mdm10p has the characteristics of an integral membrane protein. Since both Mmm1p and Mdm10p are required for mABP binding, the simplest interpretation of our results and previous phenotypic analysis of mdm10 and mmm1 mutants is that both proteins are components of a multimeric mABP receptor complex. Consistent with this, Yaffe and colleagues find that a mmm1/mdm10Δ double mutant has the same phenotype as either single mutant (Yaffe, M.P, and K.H. Berger, personal communication). Thus, there is genetic evidence that Mmm1p and Mdm10p contribute to the same function.
Still unresolved is the role of the actin cytoskeleton in control of mitochondrial morphology. The finding that mitochondria undergo fragmentation and lose tubular form upon Lat-A–induced actin depolymerization supports a role for actin–mitochondrial interactions in maintaining mitochondrial shape. However, mitochondrial morphology in Lat-A–treated cells is not identical to that in mmm1-1 or mdm10Δ mutants. It is possible that the difference in mitochondrial morphology between Lat-A treatment and mmm1-1 or mdm10Δ mutations reflects acute versus steady-state uncoupling of mitochondria from the actin cytoskeleton. Thus, the spherical morphology of mmm1-1 and mdm10Δ mutant mitochondria may result from long-term dissociation of mitochondria from the actin cytoskeleton, together with other processes including ongoing mitochondrial biogenesis. Since long-term actin depolymerization with Lat-A produces pleiotropic effects including defects in cell shape and loss of cell integrity, it is difficult to assess the long-term effect of Lat-A treatment. Alternatively, it is possible that the actin cytoskeleton is not solely responsible for control of mitochondrial morphology. Indeed, other elements such as Mdm1p, a protein required for mitochondrial inheritance that shows homology to an intermediate filament protein (McConnell and Yaffe, 1993), may contribute to the maintenance of tubular shape of the organelle.
Previous studies defined an actin-dependent motor activity on yeast mitochondria capable of driving ATP- dependent F-actin sliding on the mitochondrial surface at velocities similar to that of actin-dependent mitochondrial movement in living yeast. The precise relationship between mABP and the mitochondrial motor is yet to be determined. However, preliminary evidence indicates that the mitochondrial motor is salt extractable, binds reversibly to the mitochondrial surface, and shows fractionation behavior on actin affinity chromatography similar, but not identical, to that of mABP (data not shown). Thus, it is possible that the mitochondrial motor activity observed in the microfilament sliding assay contributes to mABP activity observed in sedimentation assays. These observations raise the possibility that Mdm10p and Mmm1p are required for docking of an actin-dependent motor on mitochondria.
Although receptors for actin-dependent motors are not definitely known, there is a precedent for a microtubule motor receptor. Studies show that kinesin-dependent motility of lytic granules from T cells is sensitive to protease treatment of granules (Burkhardt et al., 1993) and that binding of kinesin to microsomal membranes is saturable and protease sensitive (Skoufias et al., 1994; Yu et al., 1995), consistent with the idea that a kinesin receptor is present on organelle membranes. A putative kinesin receptor, kinectin, has been identified in chick embryo microsomes (Toyoshima et al., 1992). Ongoing studies focus on the further characterization of the mitochondrial motor activity and the role of Mmm1p and Mdm10p in docking of the motor protein to the organelle surface.
Yeast genetic screens have revealed a requirement for at least 30 MDM and MMM genes in mitochondrial inheritance. With the exception of MDM20, a gene required for stabilization of actin cables (Herman et al., 1997), the mechanisms underlying the function of MDM and MMM genes in this process are not well understood. Our studies establish a role for Mmm1p and Mdm10p in linking mitochondria to the actin cytoskeleton of yeast. We find that loss of function of these proteins and uncoupling of mitochondria from actin results in loss of all polarized, directed mitochondrial movement, and severe defects in mitochondrial spatial arrangement and inheritance. These studies identify the first molecular components required for actin- dependent mitochondrial movement, and support the model that the actin cytoskeleton is the sole mediator of directed mitochondrial movement in vegetative yeast. It is possible that other proteins also contribute to mABP docking activity. Recent findings indicate that Mdm12p, an integral mitochondrial outer membrane protein, is required for normal mitochondrial morphology and inheritance (Berger et al., 1997). Ongoing studies focus on the role of Mdm12p and other proteins in mABP–mitochondrial interactions and mitochondrial motility.
Acknowledgments
We thank Dr. F. Chang, and Dr. P. Brandt, and members of the Pon Laboratory for critical evaluation of the manuscript; Drs. R. Jensen, M. Yaffe, and T. Fox for yeast strains; Dr. R. Butow for the mitochondria-targeted GFP; and Drs. J. Lessard and G. Schatz for antibodies.
This work was supported by a research grant from the National Institutes of Health (GM45735).
Abbreviations used in this paper
- mABP
mitochondrial actin binding protein
- SE
salt-extracted mitochondrial peripheral membrane proteins
- SW
salt-washed mitochondria
- Lat-A
latrunculin-A
Footnotes
Address all correspondence to Dr. L.A. Pon, Department of Anatomy and Cell Biology, Columbia University, P&S 12-425, 630 West 168th Street, New York, NY 10032. Tel.: (212) 305-1947. Fax: (212) 305-3970. E-mail: lap5@columbia.edu
References
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D, Schekman R. Reconstitution of protein transport using broken yeast spheroplasts. Methods Cell Biol. 1989;31:127–141. doi: 10.1016/s0091-679x(08)61605-2. [DOI] [PubMed] [Google Scholar]
- Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley TJ, Satir P. Evidence of microfilament-associated mitochondrial movement. J Supramol Struct. 1979;12:165–175. doi: 10.1002/jss.400120203. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt JK, McIlvain JM, Jr, Sheetz MP, Argon Y. Lytic granules from cytotoxic T cells exhibit kinesin-dependent motility on microtubules in vitro. J Cell Sci. 1993;104:151–162. doi: 10.1242/jcs.104.1.151. [DOI] [PubMed] [Google Scholar]
- Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Gasser SM, Schatz G. Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J Biol Chem. 1982;257:13075–13080. [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. . Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Drubin DG, Botstein D, Spudich JA. Yeast actin filaments display ATP-dependent sliding movement over surfaces coated with rabbit muscle myosin. Proc Natl Acad Sci USA. 1992;89:4466–4470. doi: 10.1073/pnas.89.10.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazzarino DA, Boldogh I, Smith MG, Rosand J, Pon LA. Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol Biol Cell. 1994;5:807–818. doi: 10.1091/mbc.5.7.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard JL. Two monoclonal antibodies to actin: one muscle selective and one generally reactive. Cell Motil Cytoskeleton. 1988;10:349–362. doi: 10.1002/cm.970100302. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Intermediate filament formation by a yeast protein essential for organelle inheritance. Science. 1993;260:687–689. doi: 10.1126/science.8480179. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Field CM, Alberts BM, Kellogg DR. Use of actin filament and microtubule affinity chromatography to identify proteins that bind to the cytoskeleton. Methods Enzymol. 1991;196:303–319. doi: 10.1016/0076-6879(91)96028-p. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon L, Moll T, Vestweber D, Marshallsay B, Schatz G. Protein import into mitochondria: ATP-dependent protein translocation activity in a submitochondrial fraction enriched in membrane contact sites and specific proteins. J Cell Biol. 1989;109:2603–2616. doi: 10.1083/jcb.109.6.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Simon VR, Karmon SL, Pon LA. Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. . Cell Motil Cytoskeleton. 1997;37:199–210. doi: 10.1002/(SICI)1097-0169(1997)37:3<199::AID-CM2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: Identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, Cole DG, Wedaman KP, Scholey JM. The carboxyl-terminal domain of kinesin heavy chain is important for membrane binding. J Biol Chem. 1994;269:1477–1485. [PubMed] [Google Scholar]
- Smith MG, Simon VR, O'Sullivan H, Pon LA. Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangement in the budding yeast Saccharomyces cerevisiae. . Mol Biol Cell. 1995;6:1381–1396. doi: 10.1091/mbc.6.10.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- Sturmer K, Baumann O, Walz B. Actin-dependent light-induced translocation of mitochondria and ER cisternae in the photoreceptor cells of the locust Schistocerca gregaria. . J Cell Sci. 1995;108:2273–2283. doi: 10.1242/jcs.108.6.2273. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima I, Yu H, Steuer ER, Sheetz MP. Kinectin, a major kinesin-binding protein on ER. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Jensen RE, Guido EC. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J Biol Chem. 1989;264:21091–21096. [PubMed] [Google Scholar]
- Yu H, Nicchitta CV, Kumar J, Becker M, Toyoshima I, Shetz MP. Characterization of kinectin, a kinesin-binding protein: primary sequence and N-terminal topogenic signal analysis. Mol Biol Cell. 1995;6:171–183. doi: 10.1091/mbc.6.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya O, Newman SM, Okamoto K, Perlman PS, Butow RA. Functions of the High Mobility Group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. . Genetics. 1997;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]