Abstract
The budding yeast contains two type I myosins, Myo3p and Myo5p, with redundant functions. Deletion of both myosins results in growth defects, loss of actin polarity and polarized cell surface growth, and accumulation of intracellular membranes. Expression of myc-tagged Myo5p in myo3Δ myo5Δ cells fully restores wild-type characteristics. Myo5p is localized as punctate, cortical structures enriched at sites of polarized cell growth. We find that latrunculin-A–induced depolymerization of F-actin results in loss of Myo5p patches. Moreover, incubation of yeast cells at 37°C results in transient depolarization of both Myo5p patches and the actin cytoskeleton. Mutant Myo5 proteins with deletions in nonmotor domains were expressed in myo3Δ myo5Δ cells and the resulting strains were analyzed for Myo5p function. Deletion of the tail homology 2 (TH2) domain, previously implicated in ATP-insensitive actin binding, has no detectable effect on Myo5p function. In contrast, myo3Δ myo5Δ cells expressing mutant Myo5 proteins with deletions of the src homology domain 3 (SH3) or both TH2 and SH3 domains display defects including Myo5p patch depolarization, actin disorganization, and phenotypes associated with actin dysfunction. These findings support a role for the SH3 domain in Myo5p localization and function in budding yeast. The proline-rich protein verprolin (Vrp1p) binds to the SH3 domain of Myo3p or Myo5p in two-hybrid tests, coimmunoprecipitates with Myo5p, and colocalizes with Myo5p. Immunolocalization of the myc-tagged SH3 domain of Myo5p reveals diffuse cytoplasmic staining. Thus, the SH3 domain of Myo5p contributes to but is not sufficient for localization of Myo5p either to patches or to sites of polarized cell growth. Consistent with this, Myo5p patches assemble but do not localize to sites of polarized cell surface growth in a VRP1 deletion mutant. Our studies support a multistep model for Myo5p targeting in yeast. The first step, assembly of Myo5p patches, is dependent upon F-actin, and the second step, polarization of actin patches, requiresVrp1p and the SH3 domain of Myo5p.
The myosin superfamily of molecular motors has a crucial role in actin-dependent processes in all eukaryotic cells. This superfamily consists of conventional (type II or muscle-like) myosins and at least ten classes of unconventional myosins (for review see Cheney and Mooseker, 1995; Sellers and Goodson, 1995). All unconventional myosin proteins have conserved “head” and “neck” regions. The head or motor domain contains binding sites for ATP and actin. The necks of myosin proteins have a variable number of isoleucine- and glutamine-rich IQ motifs, possible binding sites for calmodulin or calmodulin-like light chains. The carboxy-terminal “tail” regions, which are highly divergent among classes of unconventional myosins, contain sequences implicated in protein–protein interactions, membrane binding, and intracellular signaling.
The type I myosin proteins were the first unconventional myosins to be discovered and remain the best studied class (Pollard and Korn, 1973). Representatives of this class are present in phylogenetically diverse organisms including filamentous fungi, ameba, yeast, and vertebrates. This points to a central role for these proteins in eukaryotic cell function (for review see Cheney and Mooseker, 1995). Phylogenetic sequence comparisons of the head regions of a host of novel myosins-I reveals at least four subclasses. Our study focuses on the subclass of myosins-I known as “classic” or “ameboid” myosins-I.
In Saccharomyces cerevisiae, there are two highly related classic type I myosin isoforms, MYO3 and MYO5, which appear to have overlapping activities (Goodson and Spudich, 1995; Goodson et al., 1996). Deletion of MYO3 or MYO5 alone leads to only subtle defects. In contrast, deletion of both MYO3 and MYO5 results in severe defects in actin organization and phenotypes associated with actin cytoskeletal dysfunction including slow growth, accumulation of intracellular membranes and vesicles, cell rounding, random bud site selection, and impaired secretion and endocytosis (Geli and Riezman, 1996; Goodson et al., 1996). Myo5p is localized as punctate, cortical structures that are enriched at sites of polarized growth and actin cytoskeletal polarization. These observations support a general role for Myo5p in organization of the yeast actin cytoskeleton.
Recent studies also support a role for the classic myosin I of Dictyostelium discoideum, myoB, in control of actin organization (Temesvari et al., 1996). Moreover, other classic myosin I proteins have been implicated in a variety of processes that are dependent upon actin cytoskeletal function (Jung and Hammer, 1990; Jung et al., 1993, 1996; Titus et al., 1993; Peterson et al., 1995; Novak et al., 1995; McGoldrick et al., 1995; Goodson et al., 1996). myoB, myoC, and myoD of Dictyostelium contribute to endocytosis, motility, streaming, and phagocytosis (Jung and Hammer, 1990; Titus et al., 1993; Jung et al., 1996). myoA, a classic type I myosin that is essential for viability in Aspergillus nidulans, is required for polarized growth and secretion (McGoldrick et al., 1995). Finally, myosin IC of Acanthamoeba castellani is essential for contractile vacuole function (Doberstein et al., 1993).
Consistent with this, classic myosin I proteins in yeast and other organisms (Fukui et al., 1989; Jung et al., 1993; Baines et al., 1995; McGoldrick et al., 1995; Goodson et al., 1996) are localized to actin-rich regions or sites of actin reorganization. First, many other myosin I proteins including myosin IC of Acanthamoeba, myoA and B of Dictyostelium, and myoA of Aspergillus display cortical localization. Second, classic myosin I proteins are localized at sites of actin organization in budding yeast, Aspergillus, and Dictyostelium. Third, the classic myosin I protein of Aspergillus occurs in punctate structures similar to cortical Myo5p patches of budding yeast.
Here, we report studies on the role of the tail homology domain 2 (TH2)1 and src homology domain 3 (SH3) domains of the Myo5p tail in Myo5p-control of actin organization and function in vivo. These regions are present in all classic myosin I proteins; however, their contribution to myosin I function within the context of the cell is not well understood (for review see Sellers and Goodson, 1995). The TH2 domain is a glycine-proline-alanine–rich region implicated in ATP-insensitive actin binding and actin cross-linking in vitro (Lynch et al., 1986; Pollard et al., 1991; Jung and Hammer, 1994; Rosenfeld and Rener, 1994). The SH3 domain, a region that binds proline-rich sequences, has been implicated in a variety of cell functions. Early studies indicate that the SH3 domain mediates protein–protein interactions that regulate enzymatic activity. For example, binding of p85 to the SH3 domain of phosphatidylinositol-3′ kinase (PI-3 kinase) increases PI-3 kinase activity five- to sevenfold (Pleiman et al., 1994). Since classic myosin I proteins contain proline-rich regions within their TH2 domain, it is possible that their SH3 domain participates in intramolecular links which regulate function (Goodson and Spudich, 1995). Other studies indicate that the SH3 domain contributes to protein targeting. For example, SH3 domains of the NADPH oxidase subunits, p47phox and p67phox, mediate protein–protein interactions required to recruit p47phox and p67phox to the membrane to form an activated oxidase (de Mendez et al., 1994, 1996). Moreover, microinjection studies with the SH3 domain of phospholipase C-γ indicate that the SH3 domain mediates localization of phospholipase C-γ to the microfilament network (Bar-Sagi et al., 1993). Indeed, many cytoskeleton-associated proteins including myosin I (Bement et al., 1994; Stoffler et al., 1995; Goodson and Spudich, 1995; Goodson et al., 1996), fodrin (Merilainen et al., 1993), nebulin (Wang et al., 1996), cortactin (Wu and Parsons, 1993), and the actin-associated proteins of yeast Abp1p (Drubin et al., 1990; Lila and Drubin, 1997), Rvs167p (Bauer et al., 1993), Bem1p (Chenevert et al., 1993), Boi1p and Boi2p (Bender et al., 1996; Matsui et al., 1996) contain SH3 domains, which underscores the importance of the SH3 domain in protein–protein interactions with the cytoskeleton.
We find that the SH3 domain is critical for Myo5p patch polarization and Myo5p-dependent actin organization, and identify Vrp1p as a likely binding partner for the SH3 domain of yeast myosin I proteins. Moreover, we find that targeting of Myo5p in yeast is a multistep process.
Materials and Methods
Yeast and Bacterial Manipulations
S. cerevisiae strains used in this study are the following: (a) HA10-1b (wild-type), MATα can1-100 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 (Goodson and Spudich, 1995); (b) HA31-9c (myo3Δ myo5Δ), MATa can1-100 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 myo3::HIS3 myo5::TRP1; (d) HA31-9a (myo3Δ), and (e) MATα can1-100 ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 myo3::HIS3 (Goodson et al., 1996). VHA-1 is an HA31-9c– derived strain in which the DNA encoding three copies of the hemagglutinin (HA) tag was inserted into the 3′ end of the chromosomal copy of the VRP1 gene (see below). Two-hybrid assays were carried out using the strain Y704 (MATa lexAop-LEU2 lex-Aop-lacZ sst1Δ his3 trp1 ura3-52 leu2). The effect of verprolin deletion on Myo5p localization was evaluated in the vrp1Δ strain (GVY1 - MATα/MATa leu2-3,112/leu2-3,112 ade1/ade1 ura3-52/ura3-52 Ile-/Ile- MEL1/MEL1 vrp1::LEU2/rrp1:: LEU2) and the corresponding wild-type strain (Sc467-MATα/MATa leu2-3,112/leu2-3,112 ade1/ade1 ura3-52/ura3-52 Ile-/Ile- MEL1/MEL1). Yeast manipulations including cell culture, transformation, and tetrad analysis were carried out according to Guthrie and Fink (1991). Bacterial manipulations were carried out according to Sambrook et al. (1989).
Construction of Plasmids Containing Mutant myo5 Constructs
Expression vectors for these studies will be referred to using the following convention: (a) pMYO5, plasmid-borne wild-type MYO5 gene bearing three copies of the myc epitope at its carboxy terminus; (b) pTH2Δ, plasmid-borne myc-tagged MYO5 gene bearing a deletion of the TH2 region; (c) pSH3Δ, plasmid-borne myc-tagged MYO5 gene bearing a deletion of the SH3 region; (d) pTH2Δ–SH3Δ, plasmid-borne myc-tagged MYO5 gene bearing deletions of the TH2 and SH3 regions; and (e) pSH3, plasmid-borne myc-tagged SH3 domain of the MYO5 gene.
All myo5 containing plasmids used in this study were generated using the pRS-Y2 MYO5-myc construct described in our previous work (Goodson et al., 1996). pTH2Δ was created using the Transformer Site-Directed Mutagenesis Kit from Clontech Laboratories, Inc. (Palo Alto, CA) according to the manufacturer's protocol. The selection oligonucleotide primer used for this construct (5′-GCGGTGGCGGTCGCTCTAGAAC-3′) destroys a unique NotI site in the noncoding region of the vector. The mutagenic primer used was 5′GCAGCCCAGCATGTTAAAGAACCCATGTTGAAGC-3′. The pSH3Δ and pTH2Δ–SH3Δ constructs were created using overlapping PCR products with site-directed mutations. For pSH3Δ and pTH2Δ–SH3Δ constructs, the outer primers used were 5′-CTCGAGGTCGACGGTATCGA-3′ and 5′-CAATATCAGTTCGTCGTGG-3′. The inner primers were 5′-CCAAAAGAACCCATGTTTGAAAAACCACATTCCGGAAATAATAATATC-3′ and 5′-GATATTATTATTTCCGGAATGTGGTTTTTCAAACATGGGTTCTTT- TGG-3′ for pSH3Δ and 5′-GCTGCAGCCCAGCATGTTAAACCACATTCCGGAAATAATAATATC-3′ and 5′-GATATTATTATTTCCGGAATGTGGTTTAACATGCTGGGCTGCAGC-3′ for pTH2Δ–SH3Δ. The full-length PCR products from the pSH3Δ and pTH2Δ–SH3Δ mutations were cloned in place of the endogenous BstEII–SalI fragment of pRS-Y2 MYO5-myc.
The plasmid containing the SH3 domain of MYO5 (pSH3) was constructed by PCR using the primers 5′-TTATGTGGCCAATACGAATTTAACCGCTTTATAGAAATGGCTATCCCAAAAGAACCCA-TGTTTGAAGCGGCTTACG-3′ and 5′-GACCATGATTACGCCAAGCTC-3′. The PCR product was digested with MscI and XhoI and cloned in place of the endogenous MscI–XhoI fragment of pRS-Y2 MYO5-myc. All constructs were confirmed by DNA sequencing.
Construction of Plasmids for Two-Hybrid Analysis
To create p1360, carrying a DNA fragment coding for Vrp1p (1–200), we amplified a 0.6-kb fragment of VRP1 by PCR with primers (5′-GGATCCAAATGGCAGGTGCTCCAGCTCCT-3′) and (5′-GCGGCCGCTCACCTTACTGAAGGCATATGCGG-3′) that incorporated BamHI and NotI sites. The product was ligated into pCR-Script/SK+ (Stratagene, La Jolla, CA). To create p1361, carrying a DNA fragment coding for Vrp1p (195–817), we amplified a 1.9-kb fragment of VRP1 by PCR with primers (5′-GGATCCATATGCCTTCAGTAAGGCCAGCAC-3′) and (5′-GCGGCCGCTCACGTAAATAATGTTAAGTCCAATGGCACACTACT-AC-3′) that incorporated BamHI and NotI sites. The product was ligated into pCR-Script/SK+ (Stratagene, La Jolla, CA).
To create p1645, carrying a DNA fragment coding for Myo3p (1,118–1,271), we amplified a 0.5-kb fragment of MYO3 by PCR with primers (5′-CAACCAAAGGATCCGAAATTCGAAGCT-3′) and (5′-TCGCGACCAGTCATCATCATC ATCGCCATCGT-3′) that used an internal BamHI site in the MYO3 sequence and incorporated an NruI site before the stop codon of the MYO3 sequence, and then the product was ligated into pCR 2.1 TOPO (Invitrogen, Carlsbad, CA). p1686 is similar to p1645 except that the NruI site is replaced by a stop codon followed by a NotI site. p1705 contains a BamHI to NotI fragment encoding for Myo3p (1,118– 1,271) cloned into KS+ (Stratagene). To create p1408, carrying a DNA fragment coding for Myo5p (1,082–1,219), we amplified a 0.4-kb fragment of MYO5 by PCR with primers (5′-TGGATCCGCGAATCAAAGCCAAAAGAACCCATGTTTGAAG-3′) and (5′-TGCGGCCGCTGATTACCAATCATCTTCCTCTTCATCT-3′) that incorporated BamHI and NotI sites and then the product was ligated into pCR-Script/SK+.
Two-hybrid constructs were based on pEG202 (Gyuris et al., 1991), which encodes the LexA DNA-binding domain, pJG4-5 (Gyuris et al., 1991), which encodes the B42 transcription activation domain, and pACT (Durfee et al., 1993), which encodes the Gal4p transcription activation domain. pEG202-derived plasmids used were p1386, which contains a BamHI to NotI fragment encoding Vrp1p (1–200); p1388, which contains a BamHI to NotI fragment encoding Vrp1p (195–817); p1707, which contains a BamHI to NotI fragment encoding Myo3p (1,118–1,271); and p1456, which contains a BamHI to NotI fragment encoding Myo5p (1,082–1,219). pJG4-5–derived plasmids used were p1387, which contains a BamHI to NotI fragment encoding Vrp1p (1–200); p1389, which contains a BamHI to NotI fragment encoding Vrp1p (195–817); p1706, which contains a BamHI to NotI fragment encoding Myo3p (1,118–1,271); p1457, which contains a BamHI to NotI fragment encoding Myo5p (1,082–1,219). p1962 is a pACT-derived plasmid that encodes full-length Vrp1p (Vaduva et al., 1997).
Epitope Tagging of Vrp1p
Vrp1p was tagged at its carboxy terminus with three copies of the peptide epitope from the HA protein of human influenza virus using a derivative of a gene disruption cassette (Wach, 1996; Longtine et al., 1998). This cassette codes for three copies of the HA epitope and the heterologous dominant resistance marker, kanMX, which confers geneticin (G418) resistance in yeast. The cassette with long-flanking VRP1 homology regions was produced using PCR amplification and the forward and reverse primers: 5′-AAAGGGTAGTAGTGTGCCATTGGACTTAACATTATTTACGCGGATCCCCGGGTTAATTAA-3′ and 5′-TGTTCTTCAGTGATTTATTGTAACCATGGAGAAATGCTCAGAATTCGAGCTCG TT T - AAAC-3. myo3Δ myo5Δ cells bearing pMYO5 were transformed with the PCR product. Cells bearing the integrated cassette were selected by growth on rich, glucose-based media (YPD) plates containing 200 mg/liter of G418 (Sigma Chemical Co., St. Louis, MO). HA tagging of Vrp1p was confirmed by Western blot analysis using an anti-HA antibody (clone 3F10; Boehringer Mannheim, Indianapolis, IN). HA-tagged Vrp1p is a 93-kD protein based on its amino acid content. However, HA-Vrp1p migrates on SDS gels as a 120-kD protein, presumably due to its high proline content.
Light Microscopy
Actin cytoskeletal structure and chitin deposition were visualized using rhodamine–phalloidin, a rabbit polyclonal antibody raised against yeast actin (Drubin et al., 1988), and calcofluor (Sigma Chemical Co.) according to published procedures (Adams et al., 1991; Pringle et al., 1991). Immunofluorescence detection of myc-tagged Myo5p was carried out as described previously (Goodson et al., 1996). HA-tagged Vrp1p was visualized using a rat monoclonal antibody (see above), and FITC-coupled goat anti–rat antibody (Kirkegaard and Perry Laboratories, Inc., Gaithersburg, MD). Other methods (fixation, mounting, and DAPI staining) were as described by Pringle et al. (1991). Photomicroscopy was performed on a Leitz Dialux microscope (Rockleigh, NJ) and a Zeiss Axioplan II microscope (Oberkochen, Germany). Images were collected using a cooled charge-coupled device camera (model Star-1; Photometrics, Tucson, AZ). Light output from the 100-W mercury arc lamp was controlled using a shutter driver (model Uniblitz D122; Vincent Associates, Rochester, NY) and attenuated using neutral density filters (Omega Optical Corporation, Brattleboro, VT). Image enhancement and analysis were performed on a Macintosh Quadra 800 computer (Cupertino, CA) using the public domain program NIH Image 1.60 (National Institutes of Health, Bethesda, MD). Images were stored on a magnetic optical disk drive (Peripheral Land Inc., Fremont, CA).
Electron Microscopy
Preparation of samples for transmission electron microscopy was carried out according to Stevens (1977). Yeast were fixed by addition of glutaraldehyde (Sigma Chemical Co.) to growth medium to a final concentration of 5%. After incubation for 3 h at room temperature (RT), cells were concentrated by centrifugation at 10,000 g for 10 min at RT and then washed two times with 0.9% NaCl. Samples were resuspended in 4% KMnO4 in 0.1 M Na-cacodylate, pH 7.4 (Electron Microscopy Sciences, Fort Washington, PA), and incubated at 4°C for 1 h with gentle rotation. After two washes with 0.9% NaCl, samples were resuspended in 2% uranyl acetate (Electron Microscopy Sciences) and then incubated for 1 h at RT. Samples were washed three times, dehydrated in a graded series of ethanol solutions, infiltrated with propylene oxide for 10 min, and then embedded in Epon-812 (Tousimis Research Co., Rockville, MD). Ultrathin sections were stained for 5 min with 1% lead citrate before viewing with a JEOL 1200 transmission electron microscope (Peabody, MA).
Immunoprecipitation
50-ml cultures of various yeast strains were grown to mid-log phase. Cells were collected and lysed by vortexing for 6 min at 4°C with 0.5-mm glass beads in a solution consisting of 10% glycerol, 10 mM EGTA, 1% Triton X-100, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM ATP, 2 mM PMSF, and protease inhibitor cocktail as described in Lazzarino et al. (1994). The protein extracts were clarified by centrifugation for 5 min at 10,000 g and the supernatant was used for immunoprecipitation experiments. 200 μl of 2 mg/ml protein extract were incubated with 6 μg of anti-myc antibody for 2 h at 4°C. Thereafter, 15 μl of protein–G Sepharose 4 Fast Flow (Pharmacia Biotech. Inc., Piscataway, NJ) pretreated with BSA was added. This mixture was further incubated for 1 h at 4°C. The Sepharose beads were then pelleted and washed two times with a buffer consisting of 20 mM Tris-HCl, pH 7.5, 10% glycerol, 1% Triton X-100, 150 mM NaCl, 1 mM PMSF, and protease inhibitor cocktail. Proteins recovered on Sepharose beads were separated by SDS-PAGE (Laemmli, 1970) and analyzed by immunoblots (Towbin et al., 1979).
Other Methods
To detect mutant and wild-type Myo5p protein expression levels, whole cell extracts were prepared by vortexing yeast cells with 0.5-mm glass beads in a solution consisting of 10% glycerol, 10 mM EGTA, 1% Triton X-100, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM PMSF, and protease inhibitor cocktail. Protein concentrations were determined using the bicinchoninic acid assay following the vendor's protocol (Pierce Chemical Co., Rockford, IL). Gel electrophoresis and immunoblots were performed as described above. Latrunculin-A (Molecular Probes, Eugene OR) treatment was carried out as described in Ayscough et al. (1997).
Results
Construction and Expression of myo5 Mutants
Expression of myc-tagged Myo5p on a low-copy number plasmid under control of the MYO5 promoter (pMYO5) complements slow growth rate and defects in actin organization and function produced by deletion of endogenous myosin I genes (Goodson et al., 1996). To investigate the functional roles of various domains within the tail of Myo5p, we created MYO5–myc constructs containing deletions of either the TH2, SH3, or TH2 and SH3 domains (Fig. 1). We defined the TH2 domain as amino acids (aa) 1,000–1,091, a region enriched in glycine, proline, and alanine that is amino-terminal to the SH3 domain. However, in some Acanthamoeba and Dictyostelium myosin I proteins, the TH2 region is believed to be bipartite: part of the TH2 region is amino-terminal to the SH3 domain, and part is carboxy-terminal to the SH3 domain. Since a proline- and alanine-rich region is present carboxy-terminal to the SH3 domain in Myo5p (aa 1,141–1,200), it is formally possible that the TH2 region of yeast myosin I proteins extends beyond the region deleted in this study. The SH3 domain of MYO5 encompasses aa 1,092–1,140. We expressed these mutant myosin I constructs using low-copy, centromere-based plasmids and the endogenous MYO5 promoter in cells bearing deletions of endogenous myosin I genes, and then examined the effect of mutant myosin I proteins on actin organization and function in vivo.
Figure 1.
Schematic diagram of Myo5p mutants. pMYO5, plasmid-borne wild-type MYO5 gene bearing three copies of the myc epitope at its carboxy terminus; pTH2Δ, plasmid-borne, myc-tagged MYO5 gene bearing a deletion of the TH2 region; pSH3Δ, plasmid-borne, myc-tagged MYO5 gene bearing a deletion of the SH3 region; pTH2Δ-SH3Δ, plasmid-borne, myc-tagged MYO5 gene bearing deletions of the TH2 and SH3 regions; pSH3, plasmid-borne, myc-tagged SH3 domain of MYO5 gene. With the exception of pSH3, all constructs were expressed in myo3Δ myo5Δ mutant cells. pSH3 was expressed in the myo3Δ strain. All proteins were expressed under control of the MYO5 promoter.
Expression of various epitope-tagged and untagged Myo5p in myo3Δ myo5Δ mutants was examined by Western blot analysis using an anti-myc antibody (Evan et al., 1985) and an antibody raised against a myosin I–specific peptide (Ruppert et al., 1993) (Fig. 2). Western blot analysis using a myosin I–specific antipeptide antibody indicated that the level of expression of plasmid-borne wild-type and mutant Myo5p in the myo3Δ myo5Δ cells is similar to that of endogenously expressed myosin I proteins in wild-type cells. Moreover, myc-tagged myosin I proteins of the expected electrophoretic mobility were detected in cells expressing wild-type and mutant MYO5 genes. The level of expression of Myo5p bearing deletions in both SH3 and TH2 domains is lower than that of wild-type Myo5p. However, the level of the other mutant proteins was comparable to that of protein expressed from pMYO5.
Figure 2.
Expression of myc-tagged myo5 mutant proteins. Western blot of whole cell protein extracts (200 μg/ lane) from wild-type (lane A), myo3Δ myo5Δ mutants containing empty vector (lane B), pMYO5 (lane C), pTH2Δ (lane D), pSH3Δ (lane E), and pTH2Δ-SH3Δ (lane F). The blot was probed using 9E10 anti-myc antibody (panel A) and myosin I protein-specific antibody G371 (panel B).
Effect of Mutant myo5 Constructs on Cell Growth
Deletion of MYO3 and MYO5 results in severe growth defects. The doubling time of the myosin I double deletion mutant in liquid culture at 30°C is two to three times greater than that of wild-type cells or myosin I single mutants (Goodson et al., 1996). In addition, myo3Δ myo5Δ cells do not grow on solid rich media at 37°C or at high osmotic strength (0.75 M KCl) (Goodson et al., 1996). Expression of pMYO5 in myo3Δ myo5Δ cells fully restores growth under all of the restrictive conditions (Goodson et al., 1996; Fig. 3 and Table I).
Figure 3.
Growth characteristics of myo3Δ,myo5Δ mutants expressing various myo5 constructs. Wild-type (row A), and myo3Δ myo5Δ mutants containing empty vector (row B), pMYO5 (row C), pTH2Δ (row D), pSH3Δ (row E), and pTH2Δ–SH3Δ (row F) were grown on YPD–containing solid media at 30° or 37°C, or in YPD supplemented with 0.75 M KCl at 30°C.
Table I.
Doubling Times of myo3Δ myo5Δ Cultures Expressing Mutant myo5 Constructs
| MYO5 construct | Doubling time | |||
|---|---|---|---|---|
| 30°C | 37°C | |||
| (min) | ||||
| Wild-type (n = 11) | 94 ± 6 | 97 ± 5 | ||
| myo3Δ myo5Δ + vector (n = 11) | 216 ± 22 | 368 ± 32 P < 0.001 | ||
| myo3Δ myo5Δ + pMYO5 (n = 11) | 98 ± 8 | 104 ± 10 | ||
| myo3Δ myo5Δ + pTH2Δ (n = 11) | 102 ± 5 | 108 ± 7 | ||
| myo3Δ myo5Δ + pSH3Δ (n = 10) | 163 ± 16 | 158 ± 14 P < 0.001 | ||
| myo3Δ myo5Δ + pTH2Δ-SH3Δ (n = 10) | 183 ± 14 | 180 ± 15 P < 0.001 | ||
Cultures of myo3Δ,myo5Δ mutant cells transformed with either wild-type or mutant myc-tagged MYO5 on a centromeric plasmid (pRS-Y2) were grown to early log phase in synthetic complete media lacking uracil at 30°C and at 37°C. Aliquots were removed periodically from all cultures and the cells were sonicated briefly to disrupt clumps. Cell densities were determined by OD600 measurements and apparent doubling times were calculated. The P values demonstrate significance levels compared to wild-type growth rates.
The growth of pTH2Δ-expressing cells is indistinguishable from that of wild-type cells or of pMYO5-rescued myosin double mutants (Fig. 3 and Table I). In contrast, deletion of the SH3 domain from MYO5 results in defects in Myo5p function (Fig. 3 and Table I). Although pSH3Δ- expressing cells grow under all conditions tested, the extent of growth on solid medium at 37°C or at high osmotic strength, and the rate of growth in liquid culture are significantly lower than those of wild-type cells or myo3Δ myo5Δ cells rescued with pMYO5. Unlike the myo3Δ myo5Δ cells, which exhibit a large increase in doubling time after temperature shift to 37°C, the pSH3Δ-expressing cells grow at 37°C as efficiently as at 30°C (Table I). Although deletion of the TH2 domain of Myo5p does not have any significant effect on growth, deletion of both the TH2 and SH3 domains produces a growth rate slightly lower than that observed after deletion of the SH3 domain alone (Fig. 3 and Table I). We attribute this decrease in growth rate to the fact that the expression level of mutant Myo5p bearing both TH2 and SH3 deletions is somewhat lower than that of all other expression constructs.
Effect of Mutant myo5 on Repolarization of the Actin Cytoskeleton after Temperature-induced Depolarization
Actin patches and cables are the two F-actin–containing structures detected by light microscopy in budding yeast. In wild-type yeast, patches are concentrated exclusively in buds and cables are aligned along the mother–bud axis (Kilmartin and Adams, 1984). This polarization of actin cytoskeletal structures is transiently disrupted by temperature shift. Transfer of wild-type yeast cultures from 30° to 37°C for 30 min causes loss of actin cables and delocalization of actin patches over both mother and bud cell surfaces. Polarization of actin cables and patches is restored within 90 min after the temperature shift (Lillie and Brown, 1994; Fig. 4 A, panels A, D, and G).
Figure 4.
The organization of the yeast actin cytoskeleton in myo3Δ myo5Δ cells rescued with various MYO5 constructs. Wild-type cells (A, panels A, D, and G), myo3Δ myo5Δ cells carrying empty vector (A, panels B, E, and H), pMYO5 (A, panels C, F, and I), pTH2Δ (B, panels A, D, and G), pSH3Δ (B, panels B, E, and H), or pTH2Δ–SH3Δ (B, panels C, F, and I) were fixed before shift to 37°C (A and B, panels A, B, and C), 30 min after a shift to 37°C (A and B, panels D, E, and F), and 90 min after a shift to 37°C (A and B, panels G, H, and I). Cells were converted to spheroplasts, stained with rhodamine– phalloidin, and then viewed as described in Materials and Methods. Bar, 1 μm.
Actin patches and cables are present in myo3Δ myo5Δ mutants. However, deletion of yeast myosin I genes results in loss of actin polarization in over 95% of the cells examined during vegetative growth at 30°C (Goodson et al., 1996; Fig. 5). This depolarization persists through the temperature shift (Fig. 4 A, panels B, E, and H). Expression of pMYO5 in these cells restores organization of the actin cytoskeleton during vegetative growth at 30°C (Goodson et al., 1996), and the normal time course of actin repolarization after temperature-induced depolarization (Fig. 4 A, panels C, F, and I).
Figure 5.
Actin cytoskeletal polarization and repolarization after shift to 37°C are defective in pSH3Δ- or pTH2Δ–SH3Δ-expressing cells. Wild-type cells (A), myo3Δ myo5Δ cells carrying empty vector (B) and pMYO5 (C), pTH2Δ (D), pSH3Δ (E), or pTH2Δ-SH3Δ (F) were removed from liquid culture at the indicated times after shift to 37°C, fixed, converted to spheroplasts, and then stained with rhodamine–phalloidin. Cells were scored as having a depolarized actin cytoskeleton if they contained more than six actin patches within the mother cell. The graphs indicate the percentage of cells that retained polarization of the actin cytoskeleton. These percentages are the average of values from three experiments. n > 100 for each experiment.
We examined the actin polarization and time course of actin repolarization in response to temperature shift in pTH2Δ-, pSH3Δ- and pTH2Δ–SH3Δ-expressing cells (Figs. 4 and 5). Actin organization during growth at 30°C, and the kinetics of actin repolarization after shift to 37°C, are not significantly altered in pTH2Δ-expressing cells compared with double deletion mutants expressing pMYO5 (Fig. 4 B, panels A, D, and G). In contrast, actin organization during vegetative growth at 30°C and the kinetics of actin reorganization are altered in pSH3Δ- and pTH2Δ– SH3Δ-expressing cells (Fig. 4 B, panels B, E, and H and C, F, and I, respectively). Depolarization of actin is observed in 20–30% of pSH3Δ- or pTH2Δ–SH3Δ-expressing cells even at 30°C (Fig. 5). Moreover, only 44–48% of these cells display repolarization of the actin cytoskeleton 90 min after temperature shift. For comparison, ∼85% of the cells expressing pMYO5 or pTH2Δ display actin repolarization under similar conditions. Therefore, the SH3 domain of Myo5p is required for full Myo5p function in actin organization.
Effect of Mutant myo5 on Actin-dependent Activities
Phenotypic analysis of yeast bearing mutations in actin (ACT1) or in the actin-binding proteins fimbrin (SAC6), capping protein (CAP1, 2), profilin (PFY1), conventional myosin (MYO1), and myosin V (MYO2) suggest a role for the actin cytoskeleton in control of cell shape, bud site selection, secretion, endocytosis, cell wall and chitin deposition, growth at high osmotic strength, and mitochondrial organization (Novick and Botstein, 1985; Haarer et al., 1990; Rodriguez and Patterson, 1990; Adams et al., 1991; Johnston et al., 1991; Amatruda et al., 1992; Chowdhury et al., 1992; Liu and Bretscher, 1992; Drubin et al., 1993; Kübler and Riezman, 1993; Lazzarino et al., 1994; Simon et al., 1995). To determine whether the defects in actin structure observed in cells expressing myo5 mutations also produce defects in actin-dependent functions, we studied the effect of myo5 mutations on mitochondrial, nuclear, and vesicular organization and on chitin and cell wall deposition (Table II).
Table II.
Effect of MYO5 Mutations on Actin Function
| Strain | % of cells displaying | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mitochondrial aggregation | Cell wall thickening | Membrane accumulation | Multinucleation | Chitin patches | ||||||
| Wild-type | <3 | 0 | 0 | 0 | 0 | |||||
| myo3Δ myo5Δ + vector | 21 | 34 | 61 | 58 | 60 | |||||
| myo3Δ myo5Δ + pMYO5 | <3 | 0 | 0 | 0 | 0 | |||||
| myo3Δ myo5Δ + pTH2Δ | <3 | 0 | <2 | 0 | 0 | |||||
| myo3Δ myo5Δ + pSH3Δ | 15 | 15 | 19 | 18 | 17 | |||||
| myo3Δ myo5Δ + pTH2Δ-SH3Δ | 15 | 11 | 19 | 11 | 15 | |||||
Mitochondria organization and multinucleation were determined in mid-log phase cells after fixation, staining of nuclear and mitochondrial DNA using the DNA-binding dye 4′,6-diamidino-2-phenylindole, and visualization by fluorescence microscopy as described previously (Goodson et al., 1996). Cell wall thickening and intracellular accumulation were visualized by transmission electron microscopy, as described previously (Goodson et al., 1996). In all cases, measurements were performed on fields of randomly selected cells. Chitin deposition was examined using calcofluor white staining and fluorescence microscopy (Goodson et al., 1996). The number of cells examined for each sample was 67–288.
Wild-type cells and myo3Δ myo5Δ mutants that express pMYO5, or pTH2Δ all display normal mitochondrial morphology: mitochondria in these cells are extended, tubular structures colocalizing with actin cables. These cells also display normal cell walls, normal chitin rings at bud sites, and no significant accumulation of intracellular membrane or vesicle. In contrast, pSH3Δ- or pTH2Δ–SH3Δ-expressing cells display (a) mitochondrial aggregation; (b) accumulation of small (50-nm) vesicles and large (200–500-nm diam) multilamellar structures that resemble the Berkeley bodies previously observed in late secretory mutants (Novick et al., 1981); (c) abnormal, delocalized chitin deposition over the surface of the mother cell, and abnormal, brightly stained patches of chitin irregularly deposited in the cell walls; and (d) thickened cell wall (Table II). The defects in actin-dependent processes in pSH3Δ- and pTH2Δ–SH3Δ-expressing cells are not as severe as those in myosin I double deletion cells expressing vector alone. Thus, the observed partial defects in actin organization described above correlate with partial defects in actin function.
Effect of Actin Depolarization and Depolymerization on Myo5p
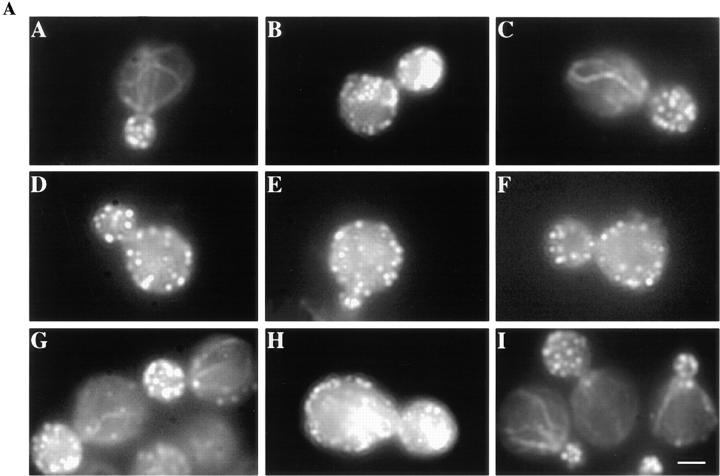
In wild-type cells grown at 30°C, immunostaining of myc-tagged Myo5p reveals punctate staining and polarization, i.e., enrichment in the bud at sites of polarized cell surface growth (Fig. 6 B). Myo5p patches show cell cycle dependent polarization that correlates with actin patch polarization. In cells bearing small- and medium-sized buds, actin patches (Fig. 6 A) and Myo5p patches (Fig. 6 B) are enriched in the bud. There is a correlation between actin patch depolarization and Myo5p depolarization in cells bearing large buds (Table III). 30 min after the temperature shift, concomitant with loss of actin polarization (Fig. 6 C), Myo5p patch localization is clearly altered. At this time point, Myo5p staining is still punctate; however, Myo5p patches are delocalized throughout both mother and daughter cells (Fig. 6 D). Finally, Myo5p patch repolarization occurs concomitant with repolarization of actin patches; both are complete within 90 min after the temperature shift (Fig. 6, E and F). This correlation between polarization of actin and Myo5p patches is consistent with a role of Myo5p in actin organization.
Figure 6.
Loss of actin patch polarity and Myo5p localization coincide after a shift to 37°C. myo3Δ myo5Δ cells expressing pMYO5 were grown to mid-log phase at 30°C (A and B) and shifted to 37°C. After 30 (C and D) or 90 min (E and F) of incubation, aliquots were removed, fixed, and then converted to spheroplasts. In this double-label experiment, actin (A, C, and E) and myc-tagged Myo5p (B, D, and F) were visualized as described above. Asymmetric Myo5p localization and actin patch polarization are lost after incubation at 37°C for 30 min (C and D) and restored after 90 min of incubation at 37°C (E and F). Bar, 1 μm.
Table III.
Myo5p Polarized Localization Is Coincident with Actin Polarization
| Strain | Bud size | % of cells with | ||||
|---|---|---|---|---|---|---|
| Polarized actin patches | Polarized Myo5p patches | |||||
| myo3Δ myo5Δ + pMYO5 | Small | 100 | 95 (n = 392) | |||
| Large | 1 | 1.5 (n = 135) | ||||
| myo3Δ + pMYO5 | Small | 98 | 95 (n = 318) | |||
| Large | 2 | 0 (n = 131) | ||||
Cultures of myo3Δ myo5Δ or myo3Δ mutant cells transformed with pMYO5 were grown to midlog phase. Cells were fixed and converted to spheroplasts and stained for actin and myc as described above. Cells were considered to be small-budded if their bud length was ≤1/3 of the length of the mother cell, and large budded if their bud length was >1/3 of the length of the mother cell. The actin cytoskeleton was scored as polarized, if the bud showed a significant concentration of actin patches with no more than five patches in the mother. Myo5p patches were scored as polarized if the bud showed a significant concentration of Myo5p patches. (n = number of cells observed).
To evaluate the role of F-actin depolymerization in Myo5p patch assembly and polarization, we treated cells with latrunculin-A (LAT-A; Spector et al., 1983; Ayscough et al., 1997). Treatment of myo3Δ myo5Δ cells expressing pMYO5 with LAT-A for 15 min causes complete loss of F-actin as assessed by rhodamine–phalloidin staining (Fig. 7, B and D). This treatment also causes a significant decrease in the intensity of Myo5p staining in patches, suggesting a defect in Myo5p patch assembly or stabilization. In addition, Myo5p patches which are present in LAT-A–treated cells show depolarization (Fig. 7, A and C). Over 90% of control cells treated with the same concentration of DMSO as that used for LAT-A treatment display enrichment of Myo5p patches in the bud of cells bearing small- and medium-sized buds. In contrast, only 12.5% of the LAT-A–treated cells examined display polarized Myo5p patches in cells bearing small- and medium-sized buds. Although polarization of Myo5p was observed in some LAT-A–treated cells, the level of enrichment of Myo5p in buds of LAT-A–treated cells was not as great as that observed in controls.
Figure 7.
Localization of Myo5p in LAT-A treated cells. myo3Δ myo5Δ cells expressing pMYO5 were grown to mid-log phase at 30°C. Aliquots were then removed and the cells were treated with 0.15 mM LAT-A dissolved in DMSO (C and D) or with equal volume of DMSO (A and B). After 15 min of incubation at 30°C cells were fixed, converted to spheroplasts, and then double labeled with anti-myc antibody (A and C) and rhodamine–phalloidin (B and D). Asymmetric Myo5p localization is lost in LAT-A– treated cells. Bar, 1 μm.
The SH3 Domain of Myo5p Is Required but Not Sufficient for Localization of Myo5p at Sites of Polarized Cell Surface Growth
To determine whether the TH2, SH3, or both domains have a role in Myo5p targeting, we examined the localization of proteins expressed from pMYO5, pTH2Δ, pSH3Δ, and pTH2Δ–SH3Δ by indirect immunofluorescence (Fig. 8). In all cases, staining of wild-type and mutant Myo5p was punctate and cortical (Fig. 8, B, D, F, and H). Moreover, patches containing TH2-deleted Myo5p resembled wild-type Myo5p patches and were enriched in the bud (n = 74). In contrast, patches containing Myo5p bearing deletions in the SH3 domain alone, or in the TH2 and SH3 domains, were evenly distributed in mother cells and bud: only 3% of cells examined showed enrichment of these proteins in the bud (n = 101 and n = 111, respectively; Fig. 8, F and H). Although partial depolarization of the actin cytoskeleton is observed in pSH3Δ- and pTH2Δ–SH3Δ-expressing cells, depolarization of Myo5p patches in these cells is not due to effects on actin organization. Patches containing mutant Myo5p protein are delocalized even in pSH3Δ- and pTH2Δ–SH3Δ-expressing cells that contain polarized actin patches and cables.
Figure 8.

Localization of wild-type and mutant MYO5 gene products. Mid-log phase myo3Δ myo5Δ cells expressing pMYO5 (A and B), pTH2Δ (C and D), pSH3Δ (E and F), or pTH2Δ– SH3Δ (G and H) were fixed, converted to spheroplasts, and then stained for actin (A, C, E, and G) and myc (B, D, F, and H) as described above. The asymmetric localization of wild-type Myo5p patches in the bud is lost after deletion of the SH3 or TH2 and SH3 domains, but not after deletion of the TH2 domain alone. Arrows point to examples of colocalization of Myo5p patches with actin patches. Bar, 1 μm.
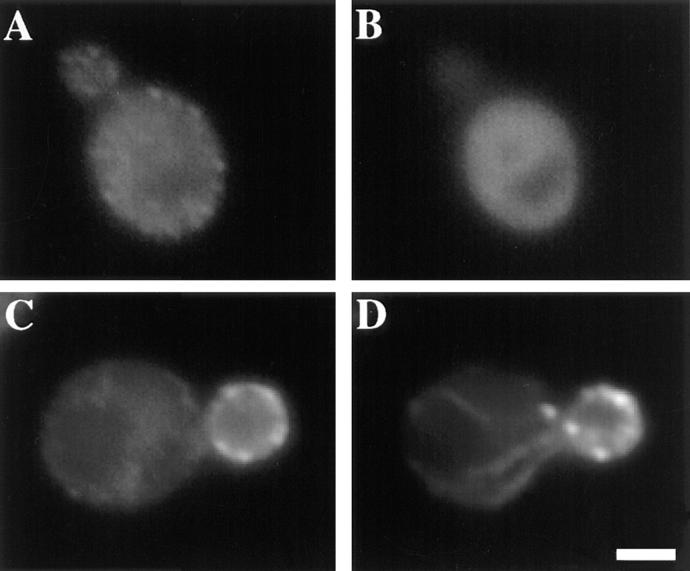
According to the results described above, the SH3 domain is required for targeting of Myo5p to growing buds. To examine the role of this domain in (a) assembly of Myo5p into patches, and (b) enrichment of Myo5p patches at sites of polarized cell surface growth, we constructed a plasmid, pSH3, carrying the SH3 domain and the subsequent 79-aa carboxy terminus of Myo5p (aa 1,086–1,219) fused to three copies of the myc epitope. This construct was expressed and localized in a MYO3 deletion mutant. myo3Δ cells were used for expression because they contain a polarized actin cytoskeleton, and only one of two endogenous myosin I genes. Western blot analysis using the anti-myc antibody indicates that expression of pSH3 using a low-copy plasmid and the endogenous MYO5 promoter produces a protein of the expected apparent molecular weight at levels comparable to that of protein expressed from pMYO5 (data not shown). Immunolocalization of this fusion protein using the anti-myc antibody reveals diffuse staining which is distributed throughout the cytoplasm (Fig. 9). Thus, the SH3 domain of Myo5p is not sufficient for either association of Myo5p with patches or for enrichment of Myo5p patches at sites of polarized cell surface growth.
Figure 9.
Localization of the SH3 domain of Myo5p. Mid-log phase myo3Δ cells expressing pMYO5 (A and B) or pSH3, the myc-tagged SH3 domain of Myo5p, (C and D) were fixed, converted to spheroplasts, and then stained for actin (A and C) and myc (B and D) as described above. The SH3 domain of Myo5p does not show the same localization as full-length Myo5p. Bar, 1 μm.
The Proline-rich, SH3-binding Protein, Verprolin, Is a Myo5p Binding Partner
In two-hybrid tests for protein–protein interaction, the SH3 domains of Myo3p and Myo5p bound a 10-residue polyproline sequence (Evangelista, M., and C. Boone, unpublished data). With a short polyproline sequence as a probe, we performed a BLAST search (Altschul et al., 1990) against the translated yeast genome to identify potential binding partners for the SH3 domain of yeast myosin I proteins. This search highlighted verprolin, Vrp1p, an actin-binding protein implicated in actin cytoskeletal organization and function (Donnelly et al., 1993; Vaduva et al., 1997). Vrp1p contains several short stretches of polyproline and 11 copies of a potential SH3-binding motif with the general sequence APPL/IP (Bar-Sagi et al., 1993). We therefore evaluated interactions between Vrp1p and the SH3 domain of yeast type I myosins using the two-hybrid system (Table IV). We find that both the Myo3p and Myo5p SH3 domains interact with full-length Vrp1p and with two smaller Vrp1p fragments, Vrp1p (1–200) and Vrp1p (195–817). Thus, the SH3 domains of the yeast myosin I proteins may bind to multiple sites within Vrp1p.
Table IV.
Two-hybrid Interactions between Verprolin and the SH3 Domains of Myo3p and Myo5p
| DNA-binding domain fusion | Activation domain fusion | lacZ activity | ||
|---|---|---|---|---|
| (Miller units) | ||||
| Vrp1p (1–200) | Vector | 0.2 ± 0.0 | ||
| Myo3p (1,118–1,271) | 1606.8 ± 263.8 | |||
| Myo5p (1,082–1,219) | 1056.0 ± 149.2 | |||
| Vrp1p (195–817) | Vector | 0.3 ± 0.2 | ||
| Myo3p (1,118–1,271) | 96.3 ± 21.8 | |||
| Myo5p (1,082–1,219) | 345.1 ± 7.3 | |||
| Myo3p (1,118–1,271) | Vector | 0.5 ± 0.1 | ||
| Vrp1p (1–200) | 253.8 ± 27.1 | |||
| Vrp1p (195–817) | 74.5 ± 4.3 | |||
| Vrp1p (1–817) | 7.3 ± 0.4 | |||
| Myo5p (1,082–1,219) | Vector | 0.3 ± 0.0 | ||
| Vrp1p (1–200) | 140.3 ± 2.3 | |||
| Vrp1p (195–817) | 28.5 ± 2.0 | |||
| Vrp1p (1–817) | 22.1 ± 0.8 |
Assays were done as described previously (Phizicky and Fields, 1995) using the strain Y704; pJG4-5 was the vector control. Assays involving Vrp1p (1–817) transformants were carried out in strain Y1003, a diploid W303 derivative carrying URA3: :lexAop-lacZ/URA3::lexAop-ADE2 reporters. The lacZ activity associated with the vector control in Y1003 was indistinguishable from that of Y704.
If Vrp1p and Myo5p interact in vivo, then these two proteins should colocalize. Localization of Vrp1p was performed by tagging the chromosomal copy of the VRP1 gene at its carboxy terminus with three copies of the HA epitope. We find that carboxy-terminal tagging has no effect on Vrp1p function; growth rates (data not shown) and actin cytoskeletal organization (Fig. 10 B) of the HA-Vrp1p expressing cells are indistinguishable from those of cells expressing untagged Vrp1p. This observation is consistent with the report that addition of GFP to the carboxy-terminus of Vrp1p does not affect Vrp1p function (Vaduva et al., 1997). Visualization of HA-tagged Vrp1p confirmed the observation that Vrp1p is localized to punctate structures enriched at sites of polarized cell surface growth (Vaduva et al., 1997). In addition, we find that Myo5p patches colocalize with Vrp1p patches (Fig. 10, C and D).
Figure 10.

Myo5p colocalizes with Vrp1p and mislocalizes in a vrp1 deletion mutant. myo3Δ myo5Δ cells expressing HA-Vrp1p and Myc-Myo5p (A–D), were grown to mid-log phase, fixed, converted to spheroplasts, and then stained for HA-tagged Vrp1p (A and C), actin (B), and myc-tagged Myo5p (D). Arrows point to examples of Myo5p andVrp1p colocalization. To evaluate the effect of VRP1 deletion on Myo5p localization, a vrp1Δ mutant (G and H) and the corresponding wild-type strain (E and F), which express myc-Myo5p, were grown to mid-log phase, fixed, and stained for myc-Myo5p (E and G) and actin (F and H). Bar, 1 μm.
Consistent with this, we find that Vrp1p is required for polarization of both Myo5p patches and the actin cytoskeleton. A vrp1Δ strain and the corresponding wild-type parent which express myc-Myo5p were fixed and stained for Myo5p and the actin cytoskeleton. As described previously, VRP1 deletion causes actin cytoskeletal depolarization (Vaduva et al., 1997). Our studies confirm this observation, and show that Myo5p patches are polarized in the wild-type strain (Fig. 10, E and F) and depolarized in the vrp1Δ cell (Fig. 10, G and H).
Finally, to test for direct interactions between Vrp1p and Myo5p in yeast, myc-tagged Myo5p was immunoprecipitated from extracts of cells that express myc-tagged Myo5p and HA-tagged Vrp1p. The immunoprecipitates were analyzed by Western immunoblotting using probes for myc and HA (Fig. 11). These reveal that Vrp1p coimmunoprecipitates with Myo5p. This coimmunoprecipitation is specific; it occurs only in cells that express both tagged constructs, and is antibody dependent. Our findings that Vrp1p (a) has the capacity to bind to the SH3 domain of Myo5p, (b) colocalizes with Myo5p, (c) is required for polarization of both Myo5p patches and the actin cytoskeleton, and (d) coimmunoprecipitates with Myo5p support a role for Vrp1p as a Myo5p binding partner.
Figure 11.
Vrp1p coimmunoprecipitates with Myo5p. Yeast strains that express no tagged constructs (HA10-1b), myc-tagged Myo5p (HA31-9c bearing pMYO5), or myc-tagged Myo5p and HA-tagged Vrp1p (VHA-1 bearing pMYO5) were grown to mid-log phase, disrupted, and then subjected to immunoprecipitation in the presence and absence of anti-myc antibody. Recovery of myc-tagged Myo5p was confirmed by Western blot analysis using anti-myc antibodies (left). Coimmunoprecipitation of HA-tagged Vrp1p with myc-tagged Myo5p was evaluated by decorating the same blot with anti-HA antibodies (right). The first lanes of each blot are whole cell lysates from VHA-1 bearing pMYO5 that illustrate the presence and electrophoretic mobility of tagged proteins.
Discussion
Type I myosin proteins have been identified in every eukaryotic cell studied. However, we are only beginning to understand the roles of these proteins in actin-dependent processes. In the budding yeast Saccharomyces cerevisiae, we have evidence that a primary function of classic type I myosins is the control of actin cytoskeletal organization (Goodson et al., 1996). Consistent with this, we observe a correlation between polarization of the actin cytoskeleton and that of Myo5p patches. Incubation of yeast cells at 37°C produces transient depolarization of both the actin cytoskeleton and Myo5p patches. Moreover, subsequent repolarization of the actin cytoskeleton occurs concomitant with Myo5p patch repolarization.
In the present study, we also examined the role of myosin I tail domains in the physiological function of the protein. To do so, we expressed mutated forms of the yeast type I myosin MYO5 in myo3Δ myo5Δ cells and examined the extent to which these mutant myosins can rescue wild-type phenotypes. From this, we have drawn additional conclusions concerning the nature of type I myosin function, and the role of tail domains in myosin I complex assembly and targeting.
In vitro data suggest that classic myosin I proteins have the capacity to bind and cross-link actin using the ATP- insensitive actin binding site in the TH2 domain of their tail and the ATP-sensitive actin binding site in their head, and may therefore organize microfilament networks by translocating actin structures in relation to other actin filaments (refer to Introduction). However, we find that a mutant Myo5p containing a deletion of the putative ATP-insensitive actin binding site in the TH2 domain is fully functional under the experimental conditions tested: growth rates, actin organization and repolarization subsequent to temperature shift, and actin-dependent processes including mitochondrial organization, chitin deposition, cell shape, and cell wall deposition are all unaffected. Additionally, TH2-deleted Myo5p is correctly assembled into patches and targeted to sites of polarized cell surface growth. Apparently, partial truncation of the tail alone does not have any functional consequences and the TH2 domain, as defined, is dispensable for the myosin I function in actin organization. Alternatively, since a proline- and alanine-rich region is present carboxy-terminal to the SH3 domain in Myo5p (see above), it cannot be excluded that deletion of this region might lead to alteration in Myo5p behavior.
In contrast to our observations with the TH2 domain, we find that the SH3 domain of the myosin I tail is required for optimal Myo5p function in yeast: expression of mutant MYO5 genes bearing deletions in either the SH3 domain or the SH3 and TH2 domains do not fully complement loss of MYO3 and MYO5. pSH3Δ- and pTH2Δ– SH3Δ-expressing cells have defects in actin organization and reorganization in response to temperature stress. In addition, these cells grow more slowly in liquid culture and are sensitive to high temperature (37°C) or to high osmotic strength. Finally, these cells display phenotypes of actin disorganization including aggregation of mitochondria, multinucleation, thickened cell walls, and accumulation of intracellular membranes. The extent of defects in pTH2Δ– SH3Δ-expressing cells is similar to that in pSH3Δ-expressing cells. We attribute the modest difference in the growth rates of these cells to the fact that the level of Myo5p expressed from pTH2Δ-SH3Δ is slightly lower than that from pSH3Δ. These findings support the model that the TH2 domain is dispensable for myosin I function, and argue against a role for intramolecular interactions between the SH3 and TH2 domains of Myo5p in Myo5p regulation. Moreover, our data indicate that the SH3 domain is required for normal Myo5p function, consistent with the finding that the SH3 domain of the classic myosin I of Dictyostelium, myoB, may be required for myoB function in cell migration (Novak and Titus, 1997).
The SH3 domain, a module which mediates protein– protein interactions, has been postulated to have diverse functions including control of protein activity, localization, and association with the cytoskeleton (refer to Introduction). Our studies indicate that SH3 contributes to Myo5p function through control of Myo5p localization. In budding yeast and in other organisms, classic myosin I proteins are resolved in patch-like structures, consistent with the assumption that these proteins are associated with some macromolecular complex. These myosin I–containing patches are localized at the cell cortex and are enriched at sites of actin organization and/or polarization. Here, we show that deletion of the SH3 domain of Myo5p produces defects in Myo5p localization. Wild-type and SH3-deleted forms of Myo5p are resolved as punctate structures localized at the cell periphery. Deletion of the SH3 domain results in depolarization of Myo5p patches. Thus, the SH3 domain of Myo5p is not required for Myo5p patch assembly. However, our findings indicate that the SH3 domain contributes to enrichment of Myo5p at sites of polarized cell surface growth. This observation is consistent with previous findings that a classic myosin I of Acanthamoeba, myosin IC, contains an SH3 domain and colocalizes with the proline-rich myosin IC-binding protein Acan125 (Xu et al., 1995). Since Myo5p patches do not colocalize with the actin cytoskeleton, our findings support a role for the SH3 domain in targeting events which are not strictly directed to the cytoskeleton.
Previous studies indicate that proteins which act early in the polarity establishment pathway (Vrp1p, Bem1p, and Cdc42p) and proteins which act during cytokinesis (septin proteins Cdc10p and Cdc11p) do not require actin for localization. In contrast, actin-binding proteins (Abp1p and cofilin), and secretory proteins known to interact with actin (Sec4p and Sec8p) require actin for their localization (Ayscough et al., 1997; Vaduva et al., 1997). We find that Vrp1p, a proline-rich protein implicated in establishment of cell polarity, is a possible Myo5p binding partner. Two-hybrid tests document specific binding between the SH3 domains of both myosin I proteins of yeast and Vrp1p. Indeed, this assay for protein–protein interaction indicates that the SH3 domains of Myo3p and Myo5p can bind to multiple sites on Vrp1p. Since mutation of a conserved tryptophan residue conserved in the Myo3p SH3 domains abolished this interaction (data not shown), two-hybrid interaction between Vrp1p and the myosin I SH3 domain appears to be specific.
Several findings indicate that Vrp1p has functionally significant interactions with yeast myosin I proteins. First, we find that Myo5p colocalizes and coimmunoprecipitates with Vrp1p. Second, deletion of the VRP1 gene results in phenotypes that are similar to those of myosin I double deletion mutants: both mutants show actin cytoskeletal depolarization and defects in actin dependent functions including endocytosis, bud site selection, and mitochondrial organization (Vaduva et al., 1997). Third, deletion of VRP1 results in depolarization of both actin patches (Vaduva et al., 1997) and Myo5p patches. These observations support the model that Vrp1p binding to the SH3 domain of myosin I proteins contributes to (a) targeting of myosin I proteins to sites of polarized cell surface growth, and (b) myosin I function in control of actin organization. We propose that Vrp1p functions as a proline-rich scaffold that binds to multiple myosin I proteins and concentrates these proteins at sites of polarized cell surface growth. Since multiple proline-rich proteins including Srv2p and Bni1p are localized at sites of polarized cell surface growth (Freeman et al., 1996; Evangelista et al., 1997) it is possible that multiple SH3-mediated protein–protein interactions contribute to myosin I targeting in yeast.
Several findings indicate that the SH3 domain contributes to, but is not sufficient for, Myo5p targeting. First, pSH3Δ-expressing cells show only partial loss of actin organization and only partial defects in actin dependent processes. That is, deletion of the SH3 domain results in a decrease in the efficiency of Myo5p function in actin organization. Thus, some fraction of the SH3-deleted Myo5p must reach the correct site of action. Indeed, we find that patches containing SH3-deleted Myo5p are present but are not enriched at sites of polarized cell surface growth. Second, we find that a fusion protein containing of the SH3 domain of Myo5p shows diffuse cytosolic staining when expressed in yeast at levels comparable to wild-type, endogenous Myo5p. Thus, the SH3 domain is sufficient neither for the assembly of Myo5p patches, nor for Myo5p patch polarization. Together, these observations indicate that multiple factors in addition to the SH3 domain are required for Myo5p assembly and targeting. Indeed, since the SH3 domain of myoB, a myosin I protein of Dictyostelium, is not required for myoB targeting (Novak and Titus, 1997), it is possible that the relative contribution of the SH3 domain to myosin I targeting may vary among cell types.
One region that may participate in Myo5p targeting is its TH1 sequence, a highly basic region found in all myosin I subclasses. Previous studies indicate that the TH1 domain of myosin proteins can bind to acidic phospholipids (Adams and Pollard, 1989; Doberstein and Pollard, 1992). Moreover, the basic TH1 region of brush border myosin I protein is sufficient to target myosin molecules to actin-rich apical structures (Footer and Bretscher, 1994). We find that deletion of the hyperbasic segment of the Myo5p TH1 region, or deletion of the motor domain of Myo5p, impairs protein expression and/or stability (Swayne, T., and L. Pon, unpublished results). Therefore, we cannot draw conclusions regarding the role of the TH1 region in Myo5p targeting from this line of experiments.
Another region that may participate in Myo5p targeting is the actin-binding motor domain. We find that Myo5p patch assembly requires F-actin. LAT-A treatment results in rapid, quantitative actin depolymerization in yeast and also produces a decrease in total staining intensity of myc-tagged Myo5p in punctate structures and an increase in diffuse, cytosolic staining. Myo5p-containing patches that are detected are cortical, but delocalized and equally distributed in mother and daughter cells. Thus, a drug that causes F-actin depolymerization produces partial defects in Myo5p patch assembly and defects in enrichment of Myo5p patches at sites of polarized cell surface growth. Since myosins are actin-binding proteins, the simplest interpretation of this finding is that Myo5p patch assembly requires association of Myo5p with F-actin, presumably through its high-affinity F-actin binding site in the motor domain. Current studies focus on analysis of the actin-binding activity of the Myo5p motor domain, and the role of this activity in Myo5p targeting.
The data we present supports a model for myosin I localization and function in yeast. In this model, Myo5p is assembled into a protein complex at the cell cortex. Assembly of cortical Myo5p patches does not require either the TH2 domain or the SH3 domain of the Myo5p tail. However, Myo5p patch assembly is at least partially dependent upon F-actin. Cortical Myo5p patches are ultimately transported to and concentrated at sites of polarized cell surface growth where they function in polarization of the actin cytoskeleton. This final targeting event requires the SH3 domain of the Myo5p tail, a region that binds to Vrp1p. We propose that Vrp1p serves as a scaffold to bind and concentrate Myo5p patches within the bud. This is the first example of protein-mediated localization of a myosin I protein. Moreover, since Vrp1p has been implicated in actin polarization, our finding provides (a) additional support for a role of myosin I in actin polarization, and (b) an important link between the cell polarity machinery and a mediator of actin organization.
Acknowledgments
We thank F. Chang and P. Brandt (both from Columbia University, New York), and members of the Pon laboratory for critical evaluation of the manuscript; G. Vaduva and A. Hopper for yeast strains and helpful discussion regarding verprolin; S. Swamy for assistance in immunofluorescence microscopy; K. Brown for expert assistance with the electron microscopy; T. Leeuw for helpful discussion regarding Myo3p localization, and I. Pot for two-hybrid analysis of Myo3p protein–protein interactions.
This work was supported by research grants from the American Cancer Society (RPG-97-163-01-C) and National Institutes of Health (GM45735) to L.A. Pon, a National Institutes of Health Grant (NS16036) to L.A. Greene, a Medical Scientists Training Program Award (5T32 GM07367) to B.L. Anderson, and grants from the Natural Sciences and Engineering Research Council of Canada, and the National Cancer Institute of Canada to C. Boone.
Abbreviations used in this paper
- aa
amino acids
- HA
hemagglutinin
- LAT-A
latrunculin-A
- RT
room temperature
- SH3
src homology domain 3
- TH1
tail homology domain 1
- TH2
tail homology domain 2
- YPD
rich, glucose-based media
Footnotes
Address all correspondence to Liza A. Pon, Department of Anatomy and Cell Biology, Columbia University P&S 12-425, 630 West 168th Street, New York, NY 10032. Tel.: (212) 305-1947. Fax: (212) 305-3970. E-mail: lap5@columbia.edu
References
- Adams RJ, Pollard TD. Binding of myosin I to membrane lipids. Nature. 1989;340:565–568. doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- Adams AE, Botstein D, Drubin DG. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. . Nature. 1991;354:404–408. doi: 10.1038/354404a0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amatruda JF, Gattermeir DJ, Karpova TS, Cooper JA. Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution, and polarized secretion in yeast. J Cell Biol. 1992;119:1151–1162. doi: 10.1083/jcb.119.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-a. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines IC, Corigliano-Murphy A, Korn ED. Quantification and localization of phosphorylated myosin I isoforms in Acanthamoeba castellani. . J Biol Chem. 1995;130:591–603. doi: 10.1083/jcb.130.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi D, Rotin D, Batzer A, Mandiyan V, Schlessinger J. SH3 domains direct cellular localization of signaling molecules. Cell. 1993;74:83–91. doi: 10.1016/0092-8674(93)90296-3. [DOI] [PubMed] [Google Scholar]
- Bauer F, Urdaci M, Aigle M, Crouzet M. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol Cell Biol. 1993;13:5070–5084. doi: 10.1128/mcb.13.8.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Wirth JA, Mooseker MS. Cloning and mRNA expression of human unconventional myosin 1C. A homologue of amoeboid myosins-I with a single IQ motif and an SH3 domain. J Mol Biol. 1994;243:356–363. doi: 10.1006/jmbi.1994.1662. [DOI] [PubMed] [Google Scholar]
- Bender L, Lo H, Lee H, Kokojan V, Peterson J, Bender A. Associations among PH and SH3-containing proteins and Rho-type GTPases in yeast. J Cell Biol. 1996;133:879–894. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert J, Corrado K, Bender A, Pringle J, Herskowitz I. A yeast gene (BEM1)necessary for cell polarization contains two SH3 domains. Nature. 1992;356:77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- Cheney RE, Mooseker MS. Unconventional myosins. Annu Rev Cell Dev Biol. 1995;11:633–675. doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Smith KW, Gustin MC. Osmotic stress and the actin cytoskeleton: phenotype-specific suppression of an actin mutation. J Cell Biol. 1992;118:561–571. doi: 10.1083/jcb.118.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendez I, Garrett MC, Adams AG, Leto TL. Role of the p67-phox SH3 domains in assembly of the NADPH oxidase system. J Biol Chem. 1994;269:16326–16332. [PubMed] [Google Scholar]
- de Mendez I, Adams AG, Sokolic RA, Malech HL, Leto TL. Multiple SH3 domain interactions regulate NADPH oxidase assembly in whole cells. EMBO (Eur Mol Biol Organ) J. 1996;15:1211–1220. [PMC free article] [PubMed] [Google Scholar]
- Doberstein SK, Pollard TD. Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoebamyosin IC. J Cell Biol. 1992;117:1241–1249. doi: 10.1083/jcb.117.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein SK, Baines IC, Wiegand G, Korn ED, Pollard TD. Inhibition of contractile vacuole function in vivoby antibodies against myosin-1. Nature. 1993;365:841–843. doi: 10.1038/365841a0. [DOI] [PubMed] [Google Scholar]
- Donnelly SFH, Pocklington MJ, Pallotta D, Orr E. A proline-rich protein, verprolin, involved in cytoskeletal organization and cellular growth in the yeast Saccharomyces cerevisiae. . Mol Microbiol. 1993;10:585–596. doi: 10.1111/j.1365-2958.1993.tb00930.x. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Mulholland J, Zhu Z, Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-1. Nature. 1990;343:288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Pete M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Footer M, Bretscher A. Brush border myosin I microinjected into cultured cells is targeted to actin-containing surface structures. J Cell Sci. 1994;107:1623–1631. doi: 10.1242/jcs.107.6.1623. [DOI] [PubMed] [Google Scholar]
- Freeman NL, Lila T, Mintzer KA, Chen Z, Pahk AJ, Ren R, Drubin DG, Field J. A conserved proline-rich region of the Saccharomyces cerevisiaecyclase-associated protein binds SH3 domains and modulates cytoskeletal localization. Mol Cell Biol. 1996;16:548–556. doi: 10.1128/mcb.16.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Lynch TJ, Brzeska H, Korn ED. Myosin I is located at the leading edge of locomoting Dictyostelium amoebae. Nature. 1989;341:328–331. doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Role of type I myosins in receptor-mediated endocytosis in yeast. Science. 1996;272:533–535. doi: 10.1126/science.272.5261.533. [DOI] [PubMed] [Google Scholar]
- Goodson HV, Spudich JA. Identification and molecular characterization of a yeast myosin I. Cell Motil Cytoskeleton. 1995;30:73–84. doi: 10.1002/cm.970300109. [DOI] [PubMed] [Google Scholar]
- Goodson HV, Anderson BL, Warrick HM, Pon LA, Spudich JA. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for organization of the actin cytoskeleton. J Cell Biol. 1996;133:1277–1291. doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to Yeast Genetics and Molecular Biology. In Methods in Enzymology Vol. 194. J.N. Abelson and M.I. Simon, editors. Academic Press, Inc., San Diego, CA. 931 pp. [PubMed]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Lillie SH, Adams AE, Magdolen V, Bandlow W, Brown SS. Purification of profilin from Saccharomyces cerevisiaeand analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Hammer JA. Generation and characterization of Dictyosteliumcells deficient in a myosin I heavy chain isoform. J Cell Biol. 1990;110:1955–1964. doi: 10.1083/jcb.110.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Hammer JA. The actin binding site in the tail domain of Dictyosteliummyosin IC (myoC) resides within the glycine- and proline-rich sequence (tail homology region 2) FEBS (Fed Eur Biochem Soc) Lett. 1994;342:197–202. doi: 10.1016/0014-5793(94)80500-8. [DOI] [PubMed] [Google Scholar]
- Jung G, Wu X, Hammer JA. Dictyosteliummutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J Cell Biol. 1996;133:305–323. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Fukui Y, Martin B, Hammer JA. Sequence, expression pattern, intracellular localization, and targeted disruption of the Dictyosteliummyosin ID heavy chain isoform. J Biol Chem. 1993;268:14981–14990. [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during cell cycle of the yeast Saccharomyces. . J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO (Eur Mol Biol Organ) J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazzarino D, Boldogh I, Smith MG, Rosand J, Pon LA. Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol Biol Cell. 1994;5:807–818. doi: 10.1091/mbc.5.7.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila T, Drubin DG. Evidence for physical and functional interactions among two Saccharomyces cerevisiaeSH3 domain protein, adenylyl cyclase-associated protein and the actin cytoskeleton. Mol Biol Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. . J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bretscher A. Characterization of TPM1disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. DeMarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. Pringle. 1998. Additional molecules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. In press. [DOI] [PubMed]
- Lynch T, Albanesi JP, Korn ED, Robinson EA, Bowers B, Fujisaki H. ATPase activities and actin-binding properties of subfragments of Acanthamoebamyosin IA. J Biol Chem. 1986;261:17156–17162. [PubMed] [Google Scholar]
- Matsui Y, Matsui R, Akada R, Toh-e A. Yeast src homology 3 domain binding proteins involved in bud formation. J Cell Biol. 1996;133:865–878. doi: 10.1083/jcb.133.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick CA, Gruver C, May GS. MyoA of Aspergillus nidulansencodes an essential myosin I required for secretion and polarized growth. J Cell Biol. 1995;128:577–587. doi: 10.1083/jcb.128.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merilainen J, Palvuori R, Sormunen R, Wasenius VM, Lehto VP. Binding of the alpha-fodrin SH3 domain to the leading lamellae of locomoting chicken fibroblasts. J Cell Sci. 1993;105:647–654. doi: 10.1242/jcs.105.3.647. [DOI] [PubMed] [Google Scholar]
- Novak KD, Titus MA. Myosin I overexpression impairs cell migration. J Cell Biol. 1997;136:633–647. doi: 10.1083/jcb.136.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Novak KD, Peterson MD, Reedy MC, Titus MA. Dictyosteliummyosin I double mutants exhibit conditional defects in pinocytosis. J Cell Biol. 1995;131:1205–1221. doi: 10.1083/jcb.131.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Peterson MD, Novak KD, Reedy MC, Ruman JL, Titus MA. Molecular genetic analysis of myoC, a Dictyosteliummyosin I. J Cell Sci. 1995;108:1093–1103. doi: 10.1242/jcs.108.3.1093. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Fields S. Protein–protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiman CM, Hertz WM, Cambier JC. Activation of phosphatidylinositol-3′ kinase by Src-family kinase SH3 binding to the p85 subunit. Science. 1994;263:1609–1612. doi: 10.1126/science.8128248. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Korn ED. Acanthamoeba myosin. I. Isolation from Acanthamoeba castellaniiof an enzyme similar to muscle myosin. J Biol Chem. 1973;248:4682–4690. [PubMed] [Google Scholar]
- Pollard TP, Doberstein SK, Zot HG. Myosin-I. Annu Rev Physiol. 1991;53:653–681. doi: 10.1146/annurev.ph.53.030191.003253. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–601. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rodriguez JR, Paterson BM. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskeleton. 1990;17:301–308. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Rener B. The GPQ-rich segment of Dictyosteliummyosin IB contains an actin binding site. Biochemistry. 1994;33:2322–2328. doi: 10.1021/bi00174a045. [DOI] [PubMed] [Google Scholar]
- Ruppert C, Kroschewski R, Bähler M. Identification, characterization and cloning of myr 1, a mammalian myosin-I. J Cell Biol. 1993;120:1393–1403. doi: 10.1083/jcb.120.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Second edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 545 pp.
- Sellers JR, Goodson HV. Motor proteins 2: myosins. Protein Profiles. 1995;2:1323–1423. [PubMed] [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- Stevens B. Variation in number and volume of the mitochondria in yeast according to growth conditions. A study based on serial sectioning and computer graphics reconstruction. Biologie Cellulaire. 1977;28:37–56. [Google Scholar]
- Stoffler HE, Ruppert C, Reinhard J, Bähler M. A novel mammalian myosin I from rat with an SH3 domain localizes to Con A-inducible, F-actin-rich structures at cell-cell contacts. J Cell Biol. 1995;129:819–830. doi: 10.1083/jcb.129.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temesvari LA, Bush JM, Peterson MD, Novak KD, Titus MA, Cardelli JA. Examination of the endosomal and lysosomal pathways in Dictyostelium discoideummyosin I mutants. J Cell Sci. 1996;109:663–673. doi: 10.1242/jcs.109.3.663. [DOI] [PubMed] [Google Scholar]
- Titus MA, Wessels D, Spudich JA, Soll D. The unconventional myosin encoded by the myoA gene plays a role in Dictyosteliummotility. Mol Biol Cell. 1993;4:233–246. doi: 10.1091/mbc.4.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaduva G, Martin NC, Hopper AK. Actin-binding verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J Cell Biol. 1997;139:1821–1833. doi: 10.1083/jcb.139.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruption in S. cerevisiae. . Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wang K, Knipfer M, Huang QQ, van Heerden A, Hsu LC, Gutierrex G, Quian XL, Stedman H. Human skeletal muscle nebulin sequence encodes a blueprint for thin filament architecture. Sequence motifs and affinity profiles of tandem repeats and terminal SH3. J Biol Chem. 1996;271:4304–4314. doi: 10.1074/jbc.271.8.4304. [DOI] [PubMed] [Google Scholar]
- Wu H, Parsons JT. Cortactin, an 80/85-kilodalton Pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zot AS, Zot H. Identification of Acan125 as a myosin- I-binding protein present with myosin I on cellular organelles in Acanthamoeba. . Proc Natl Acad Sci USA. 1995;270:25316–25319. doi: 10.1074/jbc.270.43.25316. [DOI] [PubMed] [Google Scholar]