Abstract
We examined the effect of cell cycle progression on various levels of chromosome organization in Drosophila. Using bromodeoxyuridine incorporation and DNA quantitation in combination with fluorescence in situ hybridization, we detected gross chromosomal movements in diploid interphase nuclei of larvae. At the onset of S-phase, an increased separation was seen between proximal and distal positions of a long chromsome arm. Progression through S-phase disrupted heterochromatic associations that have been correlated with gene silencing. Additionally, we have found that large-scale G1 nuclear architecture is continually dynamic. Nuclei display a Rabl configuration for only ∼2 h after mitosis, and with further progression of G1-phase can establish heterochromatic interactions between distal and proximal parts of the chromosome arm. We also find evidence that somatic pairing of homologous chromosomes is disrupted during S-phase more rapidly for a euchromatic than for a heterochromatic region. Such interphase chromosome movements suggest a possible mechanism that links gene regulation via nuclear positioning to the cell cycle: delayed maturation of heterochromatin during G1-phase delays establishment of a silent chromatin state.
Keywords: heterochromatin, chromosome movement, silencing, cell cycle, somatic pairing
The genome of eukaryotic organisms is subdivided into chromosomes, which are organized at multiple levels. First-order packaging into nucleosomes is well understood. However, very little is understood of packaging above the level of the 30-nm fiber (for reviews see van Driel and Otte, 1997). A description of the spatial arrangements of chromosomes within the whole of the interphase nucleus may well be as illuminating as the studies of nucleosome remodeling have been for understanding chromatin-mediated gene regulation.
Information concerning chromosome position during interphase comes from investigation of mammalian nuclei (Ferguson and Ward, 1992; Manuelidis, 1990), Drosophila embryos (Foe and Alberts, 1983; Marshall et al., 1996), and polytene nuclei (Hochstrasser et al., 1986). In mammalian cells, chromosomes do not range through the whole nucleus, but maintain discrete territories within a subset of the nuclear volume (Schardin et al., 1985). Additionally, there is some evidence that mammalian chromosomes are nonrandomly arranged with respect to each other (Nagele et al., 1995) and can undergo homologous associations at specific cell cycle stages (LaSalle and Lalande, 1996). Chromosomal territories have also been demonstrated in Drosophila early embryo and polytene nuclei, where most of the genome is contained on only five large chromosome arms. Unlike mammalian chromosomal territories, these span the length of the nucleus, giving a shape that is roughly similar to the segment of an orange (Marshall et al., 1996). This arrangement also results in a polar orientation of the centromeres and telomeres, referred to as a Rabl configuration.
Another general feature of interphase nuclear organization is the distinction seen between euchromatin and heterochromatin. Heterochromatin comprises the condensed regions of chromosomes, which are found primarily surrounding the centromere and remain condensed during interphase. Electron micrographic sections of interphase nuclei show much of the electron-dense heterochromatin near the nucleolus and nuclear periphery. All higher eukaryotic genomes contain heterochromatin. This portion of the genome consists mainly of middle repetitive sequences and highly repetitive simple satellite sequences, which in Drosophila can account for 15–30% of the genome (for reviews see Csink et al., 1997; John, 1988).
Heterochromatin can influence the relative position of a chromosomal region within the interphase nucleus (Csink and Henikoff, 1996; Dernburg et al., 1996; Talbert et al., 1994). The brown-Dominant (bwD) allele results from insertion of a large block (1–2 megabases) of heterochromatin into the brown (bw) eye color gene, which is located near the distal tip of the right arm of chromosome 2 (2R). In heterozygous bw + /bwD flies this insertion results in trans-inactivation of the wild-type gene on the homologue in a dominant variegating manner. Fluorescence in situ hybridization (FISH)1 to interphase nuclei from the third instar larval central nervous systems (CNS) demonstrated that the bwD insertion causes aberrant association of distal 2R euchromatin with 2R centric heterochromatin (2Rh) (Csink and Henikoff, 1996; Dernburg et al., 1996). Using various phenotypic modifiers, we showed that an increase or decrease of bw + trans-inactivation in the adult eye was correlated with the degree of bwD-2Rh association (Csink and Henikoff, 1996; Talbert et al., 1994).
While a significant level of bwD-2Rh heterochromatic association in the larval CNS nuclei was seen compared with wild-type nuclei, it was shown that between 40 and 60% of the bwD nuclei examined displayed no association (Csink and Henikoff, 1996; Dernburg et al., 1996). This lack of bwD-2Rh association was also seen in mitotic spreads and in the rapidly dividing nuclei of bwD syncytial blastoderm embryos. Therefore, it was speculated that the lack of association in a subset of nuclei resulted from disruption of bwD-2Rh association by mitosis.
Here we examine the cell cycle–dependent features of heterochromatic arrangements during interphase. We find that bwD-2Rh associations are broken down during early S-phase, and are not re-established until well into G1. Additionally, we examine the effect of cell cycle progression on two other organizational features of chromosomes in Drosophila: the Rabl configuration and pairing of homologous chromosomes. These three aspects of chromosome organization exhibit quite different postmitotic behavior. Our work indicates that, contrary to the notion that large-scale movements are confined to mitosis (Abney et al., 1997), chromosomes show dynamic behavior during interphase.
Materials and Methods
Fly Lines and Culture
Flies were maintained at 25°C on cornmeal-molasses media, except when being fed BrdU, in which case they were kept on Carolina Biological Supply Company (Burlington, NC) fly media. Flies were homozygotes from either a bwD;st or a wild-type (Amherst) line. Minute mutant lines were obtained from the Mid-America Stock Center (Bowling Green, OH).
Fluorescence In Situ Hybridization and Microscopy
All larvae in this study were female. Dissections, FISH, and measurements were done as previously described (Csink and Henikoff, 1996) except that anti-BrdU-fluorescein (Boehringer Mannheim Corp., Indianapolis, IN) and Cy5-streptavidin (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) were added to the final incubation mixture. Nuclei were treated briefly in a hypotonic solution, and were squashed slightly under a coverslip. This treatment resulted in nuclei with radii between 1.75 and 5.5 μm. Untreated nuclei from larval CNS cells have radii between 1.5 and 3 μm. All microscopy was performed using a Deltavision system (Applied Precision, Inc. Issaquah, WA) that records epifluorescence images on a CCD camera, allowing for quantitation of the fluorescence intensity signal (see below). A 60× 1.4 na PlanApo objective (Carl Zeiss Inc., Thornwood, NY) was used.
To describe the characteristics of a large population of nuclei, our study was limited by the amount of data that could be collected given finite computational and storage resources. For maximum efficiency and to avoid undersampling, we chose to analyze all data in two dimensions rather than to carry out 3D reconstructions. Whereas this procedure, along with slight flattening and hypotonic treatment, would be problematic in the context of absolute distance or size measurements, for statistical analysis of relative distance measurements required in this study, 2D projection data are sufficient. We note that in previous studies using hypotonically treated and squashed imaginal disc or CNS nuclei, quantitative measurements of heterochromatic associations (Dernburg et al., 1996) and homologous pairing (Fung et al., 1998) were very similar to those obtained in parallel using whole-mount preparations. Furthermore, measurements on heterochromatic associations (Dernburg et al., 1996) were indistinguishable from those collected after a milder hypotonic treatment (Csink and Henikoff, 1996), which was used in the present study. Thus, treatments that are needed to allow collection of large datasets of relative measurements should not affect conclusions concerning nuclear positioning. Furthermore, by avoiding weak fixations that have been used to obtain accurate absolute size measurements, but which might insufficiently fix a subset of the nuclei, we minimize differential effects that could bias relative distance measurements.
BrdU Incorporation
Larvae were used that were 95–105 h after egg deposition (AED), because larvae stop eating 10 h before pupariation (120 AED). BrdU-containing food was prepared in the following manner: 0.3 g of crushed blue fly media (Carolina Biological Supply Company) was placed in a 35-mm petri dish and hydrated with 1.6 ml of BrdU solution. BrdU concentrations were 1 mg/ml for the 1–4-h time points and 0.4 mg/ml for the longer time points and the pulse feed experiments. A stock solution of BrdU was made by dissolving 50 mg of BrdU in 2.5 ml 40% ethanol. This stock solution was then diluted to the desired concentration in water, and the solution was used to rehydrate the fly media. Larvae were placed on food for the desired period of time. Only larvae with blue food in their gut were dissected.
For the pulse feeding experiments, bromophenol blue was added to the BrdU solution at a concentration of 0.2 mg/ml. Larvae were placed on the dark blue BrdU-containing food mixture for 15–20 min, after which they were rinsed in PBS. Only those larvae with visible blue food in crop, but not in gut were selected and placed on BrdU-free crushed white food (Carolina Biological Supply Company). 2 h after the beginning of the pulse, larvae were rinsed in PBS, and only those with no visible blue food in their bodies were selected. The larvae were placed on BrdU-free crushed blue food and fed for an additional 5, 12, or 18 h. At the time of dissection, larvae from the 5 and 12-h chase periods had blue food in their gut, but larvae from the 18-h chase period had no food in their gut.
Controls were performed to insure that BrdU feeding did not affect the parameters measured in our study, and to determine the extent of BrdU toxicity. When larvae were fed 1 mg/ml BrdU from 95–105 h AED until pupation, 95% pupated and 12% emerged. However, when larvae were pulse-fed with 1 mg/ml BrdU as described above, all of the larvae emerged and all were fertile. bwD/bw + larvae were fed 0.4 mg/ml BrdU, or were pulsed with 1 mg/ml BrdU and chased with BrdU-free food, and the emergent adults examined for bwD trans-inactivation. No modification of trans-inactivation was observed.
To insure that bwD heterochromatic associations were not modified by BrdU incorporation, bwD larvae were placed on food either with or without 1 mg/ml BrdU and allowed to feed for 2.5–3 h. Data on the distances between heterologous probes (59E and AACAC) were collected as described above from fed and unfed larvae (n = 180 nuclei for each data set). There was no significant difference between the two data sets. Additionally, we determined the nuclei in both data sets that were in S- or G2-phase by quantifying the relative DAPI staining intensity. These nuclei would be the ones in the population most likely to display disruption by BrdU (as shown in this paper). Again, no significant difference was seen between the BrdU-fed and control nuclei.
Measuring Cell Cycle Timing in CNS Nuclei
The CNS of third instar larvae is a mixture of cell types including neuroblasts, ganglion mother cells, various neuronal precursor cells, and neurons. The reported cell cycle duration in these nuclei at 25°C is 8 h or more (Ashburner, 1989; Gatti et al., 1974). However, when different cell types of the CNS were examined, the cell cycle time was found to be variable (Ito and Hotta, 1992; Meinertzhagen and Hanson, 1993; Truman and Bate, 1988; Truman et al., 1993), and each of the cell types examined constituted only a small fraction of the CNS. Because of this heterogeneity, it was necessary to characterize the general cycling behavior of the bulk of CNS cells.
To determine the timing of cell cycle stages in CNS nuclei, we determined the proportion of labeled nuclei (Table I) and the number of labeled mitotic figures after various labeling times. Larvae ∼95–105 h AED were fed BrdU for 1.5 h (see below). All 20 randomly chosen mitotic figures from the CNS of these larvae were found to be unlabeled. This indicates that for most cells, the time from late S-phase to metaphase is at least 70 min because larvae must feed for ∼20 min before any BrdU label is detectable in nuclei. When we examined 60 mitotic figures from larvae ∼95–105 h AED that had been fed BrdU for 3–3.5 h, 23% were unlabeled, 43% were fully labeled, and 34% were partially labeled. Therefore, most nuclei have a G2-phase of <3.5 h, and for at least 43% of the nuclei, the time to complete S-phase and G2-phase is <3.5 h. When we examined 20 nuclei from larvae that had been labeled for 5 h, no unlabeled mitotic figures were found, so that none of the nuclei had a G2-phase longer than 5 h (Table II). From the proportion of nuclei in the larval CNS that entered S-phase during a given labeling period (Table I), a rate of 4–7% per hour was calculated.
Table I.
Percent Nuclei Incorporating BrdU
| Mean | SE | |||
|---|---|---|---|---|
| Continuous feed | ||||
| 1.5–1.75 | 9.9 | 1.0 | ||
| 2.25–2.5 | 10.5 | 1.1 | ||
| 3.25–3.5 | 17.2 | 1.7 | ||
| 4.75–5 | 25.8 | 2.1 | ||
| 6–7 | 28.6 | 2.1 | ||
| Pulse feed | ||||
| 7–7.5 | 8.4 | 2.0 | ||
| 14–14.5 | 7.3 | .9 | ||
| 20–20.5 | 9.7 | 2.2 |
Percent of nuclei from the CNS of third instar larvae labeled with BrdU using various feeding regimens. All of the nuclei (60–200) in nine microscope field were counted. DNA incorporated BrdU for 1.5–2.5 hr, as can be seen from the similarity of percent nuclei labeled in these experiments to the percent labeled in the 1.5–2.5 hr continuous-feed experiments. SE, standard error. The first column is hours after the start of BrdU feeding.
Table II.
Percentage of Mitotic Figures from the CNS of Drosophila Larvae Showing BrdU Labeling after Various Incorporation Times
| Time fed BrdU | n | Unlabeled | Partially labeled | Fully labeled | ||||
|---|---|---|---|---|---|---|---|---|
| h | % | % | % | |||||
| 1.5 | 20 | 100 | 0 | 0 | ||||
| 3–3.5 | 60 | 23 | 34 | 43 | ||||
| 5 | 20 | 0 | 0 | 100 |
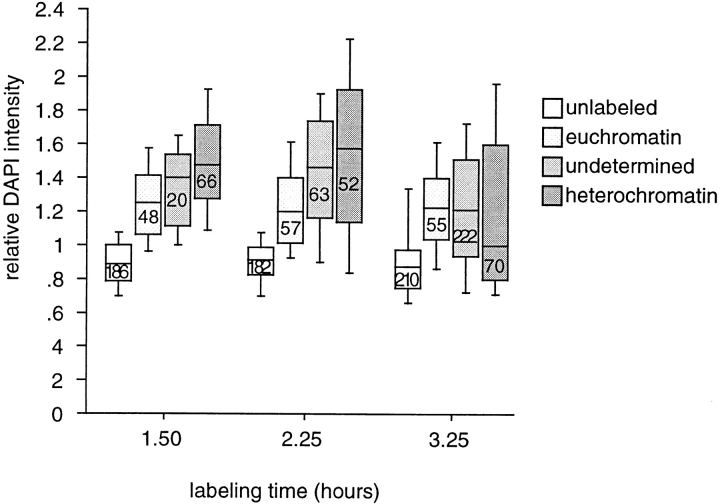
Further inferences concerning the length of the various parts of the cell cycle in CNS nuclei were made by determining if the labeled nuclei were in S-, G2- or G1-phase (postmitotic). We estimated the relative DNA content by quantitating intensity of the DAPI fluorescence, and noted the BrdU labeling pattern for each nucleus. Euchromatin labels early in S-phase with numerous origins of replication giving a fine grain pattern, while heterochromatin labels at the end of S-phase with fewer origins giving brighter spots (Yanishevsky and Prescott, 1978). We confirmed that heterochromatin is replicated late in larval CNS nuclei by observing that partially labeled mitotic figures had only pericentric incorporation of BrdU (data not shown). Interphase nuclei from larvae fed BrdU for 1.5, 2.25, or 3.25 h showed a mixture of heterochromatic, euchromatic, and undetermined labeling patterns. The progression of nuclei through the cell cycle can be seen in the box plots in Fig. 1, which shows the distribution of the relative DAPI intensity for each labeling class from larvae fed BrdU for three different intervals and the numbers of nuclei from each class. At 1.5 h, the median DAPI intensity of heterochromatically labeled nuclei is 1.7× the median for the unlabeled nuclei, but by 3.25 h the median is lower and the distribution is broader. At the earlier time point the heterochromatically labeled nuclei are primarily in G2-phase, while at the later time point this class contains a mixture of cells in G2-phase and those that are just postmitotic. This result indicates that for the bulk of CNS nuclei, G2-phase is >2 h. These data also show that the number of nuclei entering S-phase and the number in late S-phase is constant, whereas a substantial increase was seen in the number of nuclei in the undetermined class, which consists mainly of those nuclei that are completely labeled. The euchromatically and heterochromatically labeling classes did not increase, indicating that these classes include constant proportions of nuclei at the beginning of each feeding.
Figure 1.
Distribution of BrdU labeling patterns and DNA content in larval CNS nuclei during the cell cycle. Box plots of the relative DAPI intensity, wherein each horizontal line represents the 10th, 25th, 50th (median), 75th and 90th percentiles. The numbers in the boxes are the total number of nuclei in each plot. Larvae were fed BrdU for three different time periods, and the nuclei were analyzed for BrdU labeling pattern. A euchromatic pattern consists of fine-grained labeling over most of the nucleus, while a heterochromatic pattern consists of labeling in only a few bright spots. Undetermined nuclei were either entirely labeled or had a pattern that was not identifiable as heterochromatic or euchromatic.
Determination of DNA Content by Quantitation of DAPI Fluorescence
Accurate quantitation is possible because of software (Deltavision; Applied Precision, Inc.) that is able to calibrate the light field to compensate for variation in excitation light intensity across the field and for irregularities in the image path between the microscope and camera. For each data set (genotype and time point), at least nine microscope fields of nuclei from at least three different larvae were analyzed, and from each individual a maximum of three separate fields were collected. The total DAPI fluorescence intensity of each nucleus was measured using NIH Image (Wayne Rasband, National Institute of Mental Health) on a Macintosh computer, and a background value was subtracted. In each field, the intensity of >80% nuclei is very similar, with the remaining nuclei showing about twice the intensity of the bulk of the nuclei (G2-phase) or showing intermediate intensities (S-phase). From this we conclude that most nuclei are in G1-phase. Although most of the nuclei from each field were similar in DAPI fluorescence intensity, the intensity of each field differed greatly. Therefore, each intensity measurement was divided by the median density from its field, giving the relative intensity; this allowed data from different fields to be pooled.
Data Analysis
In each microscope field all labeled nuclei and 10 randomly selected unlabeled nuclei were scored. For each timepoint at least nine separate slide preparations from different larvae of a single genotype were used to collect data. Slides from the two genotypes were concurrently prepared using the same reagents in order to insure that there was no variation introduced and that any difference between the distances of the two probes was due to the presence of the bwD insertion.
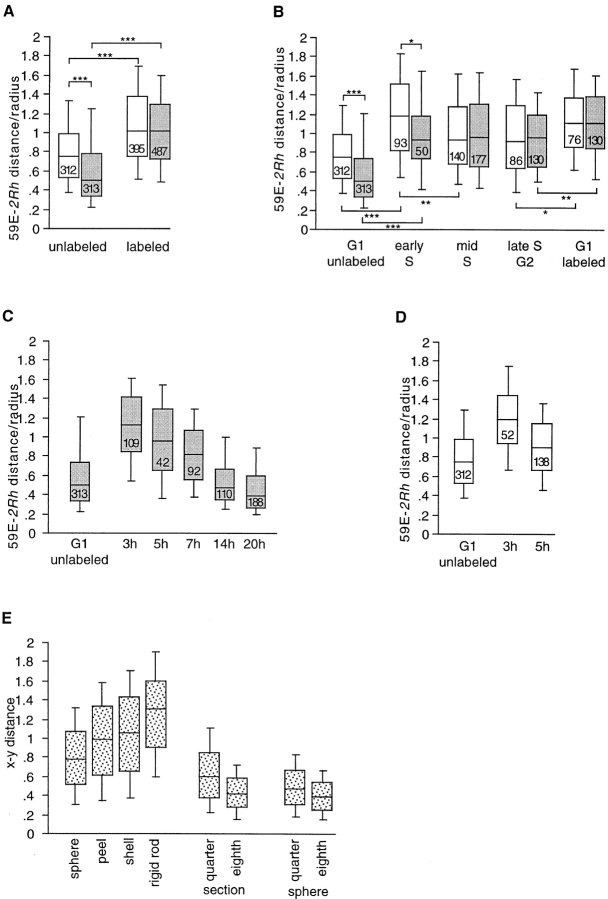
The partitioning of nuclei in Fig. 3 b was done in the following manner. All labeled nuclei were assigned to an early S-, a mid S-, or a late S/G2 class, except those assigned to the G1-labeled class, which had a G1-phase DNA content and either heterochromatic or undetermined labeling patterns. Those nuclei with a relative DAPI intensity <1.1 were considered to be in G1-phase. Labeled nuclei with a relative DAPI intensity between 1.1 and 1.2 or those with a G1 content and a euchromatic labeling pattern were considered to be in early S-phase. Nuclei with a relative DAPI fluorescence intensity between 1.2 and 1.6 were considered to be in mid S-phase, and those with a DNA content >1.6 were considered to be in late SG2-phase. Data in Fig. 3, a and b and the G1 unlabeled sets in Fig. 3, c and d, and in Fig. 4 were pooled from the three time points in Fig. 1. The G1-early-S boundary of 1.1 was chosen based on a test run, where 1.1 was close to half the maximum value (2.25), was the end point of the last bin of the histogram that contained >10% of the data, and was close to the 90th percentile of the unlabeled nuclei (1.12, see Fig. 1). The ES-MS boundary was chosen to reveal differences between early S- and mid S-phase. The choice of the MS-LSG2 boundary was arbitrary. Movement of the boundaries ± 0.1 did not change the direction or significance of the results.
Figure 3.
Cell cycle–dependent chromosome movements. Box plots of the distance between the 2Rh probe (AACAC) and the 59E probe divided by the radius of the nucleus. Data from wild-type nuclei are shown in white boxes, and bwD nuclei are shown in gray boxes. Simulated distributions are shown in spotted boxes. The numbers in the boxes are the total number of measurements in each plot. The asterisks on the brackets indicated the level of significance for the bracketed pair as determined using the two-tailed Mann-Whitney U test for nonparametric comparison of two unpaired groups. *P < 0.05; **P < 0.01; ***P < 0.001. (A) Comparison of BrdU-labeled and -unlabeled nuclei. Data from the first three time points in Table I are pooled. (B) Same data as in A categorized by cell cycle stage based on DAPI intensity and BrdU labeling pattern as described in Materials and Methods. G1 labeled nuclei are those that are just postmitotic. (C) Box plots showing reassociation of bwD and 2Rh after mitosis. The first box plot is for the unlabeled bwD nuclei from B. The second and third boxes are for post mitotic nuclei (G1-labeled class) from data collected from larvae fed BrdU for 3.5 or 5 h, respectively. The last three boxes are for labeled nuclei from larvae that have been pulse-fed BrdU. The time from initial feeding is given on the x axis, so the hours that these nuclei are postmitotic is roughly this time minus 2–4 h. All of the data sets were significantly different from each other (at least P < 0.05), with the exception of the G1 unlabeled set and the 14-hr set. (D) Box plots showing loss of Rabl orientation after mitosis in wild-type nuclei. The first box plot is for the unlabeled wild-type nuclei from B. The second and third boxes are for postmitotic nuclei (G1-labeled class) from data collected using larvae fed BrdU for 3.5 or 5 h, respectively. All of the data sets were significantly different from each other (at least P < 0.05). (E) Box plots drawn from the simulated xy distances for the models indicated.
Figure 4.
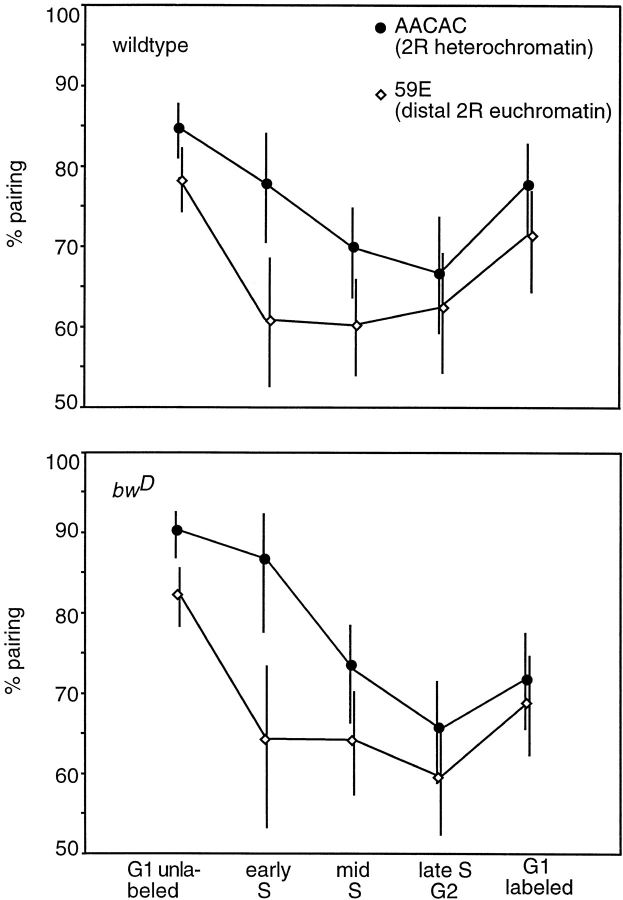
Somatic pairing of homologues through the cell cycle. Percentage of nuclei paired at the 59E and AACAC regions in bwD and wild-type nuclei as detected by the presence of only one dot of hybridization in each nucleus. Bars show 95% confidence interval of the percentages.
Distance measurement and intensity data were imported into Statview (Abacus Concepts, Inc., Berkeley, CA), where the box plots were drawn and the Mann-Whitney U test for significance was performed. The confidence intervals of the percentages in Fig. 4 were calculated as described by Sokal and Rohlf (1981), and are based on the binomial distribution.
Simulations
For a sphere of given radius r, a three-dimensional coordinate system was assumed with origin at the center of the sphere. A random point within the sphere was selected by repeatedly picking each of its three coordinates (x, y, z) at random between −r and +r using a uniform probability distribution until it was found that the point fell within the sphere that satisfies:
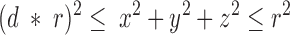 |
where 0.0 ≤ d ≤ 1.0. When d = 0, points can lie anywhere in the sphere, and when d = 1, points can lie only on its surface. A point could also be restricted to a quarter section of a sphere by specifying that two coordinates be nonnegative, and to an eighth section of a sphere by specifying that all three coordinates be nonnegative.
A second random point was selected in the same manner, and the pair of points was projected onto the x-y plane through the equator of the sphere. The Euclidean distance between the projected points was then computed. Each simulation reported consisted of 5,000 such distances.
Results
Cell Cycle Stage and bwD Heterochromatic Associations
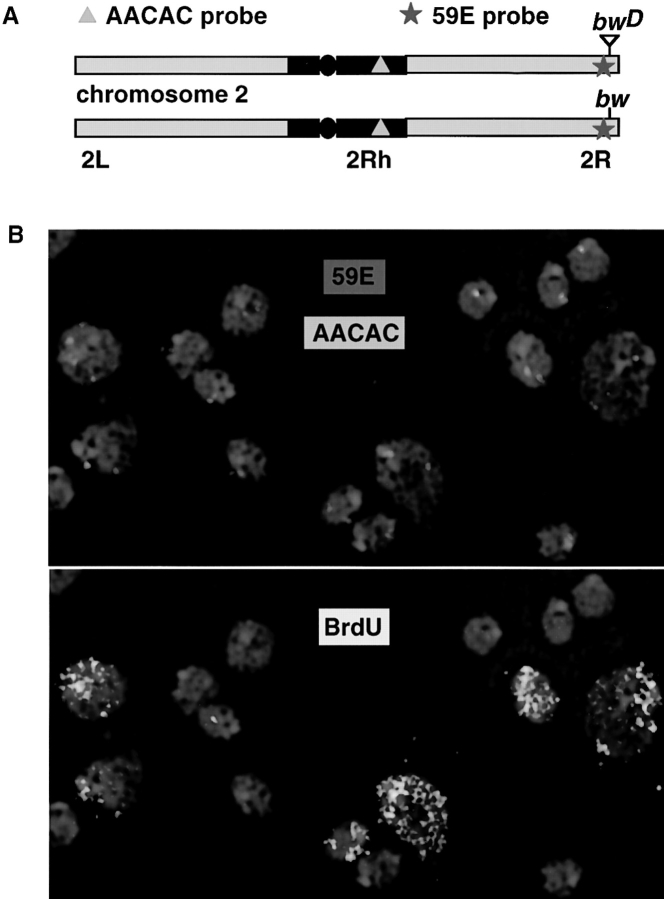
To detect possible cell cycle–dependent changes in bwD-heterochromatic associations, we used multicolored FISH to nuclei from the larval CNS. The experimental design used two probes: one for a satellite sequence (AACAC) specific to the centric heterochromatin of 2R, and a second probe consisting of a unique genomic clone of ∼80 kb located just proximal to the bw gene (59E) (Fig. 2 a; Csink and Henikoff, 1996). Third instar larvae 95–105 h AED were placed on food containing bromodeoxyuridine (BrdU), which is incorporated into the DNA and can be detected by an anti-BrdU antibody conjugated to fluorescein. Fig. 2 b shows CNS nuclei from bwD homozygous larvae that were fed BrdU for 4–5 h, fixed, hybridized with the 59E and 2Rh probes, and labeled with anti-BrdU. An examination of the bwD nuclei in Fig. 2 b reveals that those labeled with BrdU (lower panel) tend to display a greater distance between the signals from the distal and proximal probes than those that do not label.
Figure 2.
Concurrent FISH and fluorescent BrdU detection to measure chromosome position. (A) Diagram of chromosome 2 from bwD and wild type with the locations of the genomic probes indicated. Black region is heterochromatic. The circle represents the centromere. (B) Two panels of interphase nuclei from the CNS of a bwD larva. DAPI staining is in blue. (Top) Hybridization of the two probes in A. (Bottom) The same nuclei with the signal from anti-BrdU fluorescein indicating those nuclei that have incorporated DNA within the last 4–5 h. Differential and weak DAPI staining (relative to anti-BrdU staining) are responsible for the seeming discordance of DNA localization, FISH signal and BrdU incorporation.
To quantitate this observation, we measured the distance between the distal and proximal hybridization signals and divided each measurement by the nuclear radius. Larvae were fed BrdU for 1.5 to 3.5 h (Fig. 1). The results for >1,000 nuclei, divided into BrdU-labeled and -unlabeled classes, are presented in the box plots in Fig. 3 a. With this labeling schedule most of the labeled nuclei would be premitotic (See Materials and Methods). We found significant differences between the wild-type and bwD data sets for only the unlabeled nuclei. There was no significant difference between wild-type and bwD in labeled nuclei, implying that cells that were progressing or had recently progressed through S-phase lacked bwD-2Rh associations. Therefore, contrary to expectations, breakdown of heterochromatic associations occurs before mitosis.
To determine more precisely when the breakdown of heterochromatic associations occurs, each nucleus was classified as G1-unlabeled, S-phase (early or mid), late SG2-phase, or G1-labeled (postmitotic), based on DAPI staining intensity and BrdU labeling patterns (Fig. 1 and Materials and Methods). This subdivision of the labeled class (Fig. 3 b) revealed further interesting points. In early S-phase, the distance between the proximal and distal probes increased significantly in both wild-type and bwD nuclei, although the distance was less in the bwD nuclei, indicating that there were still some bwD-2Rh associations at early S-phase. By mid S-phase, however, there was no difference between bwD and wild type. Even in the G1-labeled postmitotic nuclei, we found no evidence of heterochromatic associations (Fig. 3 b).
The unlabeled nuclei in Fig. 3 would be a mixture of nuclei from postmitotic neurons and nuclei destined to reenter mitosis at some point in larval development. From our analysis of labeling kinetics (Table I), we can predict that 15–21% of the unlabeled class will enter S-phase within 4–5 h. If the G1 unlabeled class were depleted preferentially of nuclei that lacked bwD-2Rh associations, then we would expect the median of that class to decrease with longer labeling times. No such decrease was observed (data not shown). Therefore, the G1 unlabeled class behaves as if it were the precursor for the S-phase classes, so that the individual nuclei entering S-phase would consist of those that display bwD-2Rh associations and those that do not.
The lack of any apparent association of 2Rh-bwD in the newly postmitotic nuclei indicates that this association may take a long time to form, perhaps well into G1, and may require large-scale reorganization of chromosomes. Additionally, bwD trans-inactivation is correlated with association. Therefore, we wished to determine how long it takes to set up these associations, because this could provide a window for escape of bw + from the influence of heterochromatin. By providing a pulse of BrdU, we could follow a subset of nuclei through mitosis and beyond. Effective pulse labeling procedures for larval CNS require dissection and subsequent incubation in BrdU-containing media. However, this protocol may affect nuclear organization or the cell cycle due to anoxia (Foe and Alberts, 1985) or disassociation with neighboring structures (Selleck et al., 1992). Therefore, we developed a pulse-feed method for labeling nuclei from larvae, which allowed us to directly compare data from continuously fed larvae.
This pulse-feed method allowed us to examine later events after mitosis. Fig. 3 c shows the distributions of 59E-2Rh distances in bwD of unlabeled nuclei, of postmitotic nuclei from larvae fed BrdU for 3 or 5 h, and of nuclei from pulse-fed larvae at 7, 14, and 20 h after the beginning pulse. Although there may be some reassociation in the 7-h data set, the distribution does not reach the unlabeled level until the 14-h time point (10–12 h after mitosis). The 20-h data set shows even greater association than the unlabeled data set. Therefore, we conclude that bwD- 2Rh associations do not form soon after mitosis, but form gradually over a period >5 h. In addition, these data show that heterochromatic associations occur within the population of cycling cells and are not merely a feature of a terminally differentiated subset of cells.
Cell Cycle–dependent Movement of Wild-type Chromosomes
An unexpected feature of the data presented in Fig. 3, a–b is the increase in distance between proximal 2Rh and distal 59E in wild-type nuclei when the labeled and unlabeled classes were compared. Below we consider how these observations conform with various models of chromosomal arrangement.
To determine the expected distributions of two probes in a nucleus under various constraints, computer simulations were carried out. Fig. 3 e shows box plots for simulated distributions. The first box plot shows the distribution of distances between two independent random points in the full volume of a sphere of radius 1, projected onto a two-dimensional surface. As has been previously noted in the context of genomic distances in an interphase nucleus, such data are approximated by a Rayleigh distribution (median .78), which is consistent with a random walk polymer model of chromatin conformation in the interphase nucleus (Yokota et al., 1995). The distributions of simulated data from other models are also presented. The peel allows the points to be distributed randomly though a surface that is 0.3× the radius (like a thick orange peel). The shell allows the points to be distributed randomly over the surface of the sphere. The rigid rod distribution was also previously described (Yokota et al., 1995), and models the distribution of the projected distances of two points on the tips of a randomly oriented rod when the length of the rod is twice the radius.
To model a territory consisting of chromosome 2 crudely, we simulated a one-quarter orthogonal section of the sphere. To model a territory encompassing only the distal and proximal 2R probes, we simulated a one-eighth orthogonal section, which is equivalent to the proportion of the genome between these two probes. The quarter-sphere and eighth-sphere, respectively, model radially symmetrical territories of the same volumes as quarter- and eighth-sections. Simple models of these types have been used to illustrate chromosomal territories in Drosophila CNS and imaginal disc nuclei (Dernburg et al., 1996). However, these models for territories (Fig. 3 f) do not resemble any distributions seen for wild-type CNS nuclei in our study (Fig. 3, a, b, and d).
Generally, a median greater than .78 indicates that the probes are nonrandomly separated, or that they are confined to an irregular volume. A median of <.78 indicates that the probes are confined to a subset of the nucleus or are interacting with each other. The medians and distributions of the simulated distances (Fig. 3 e) can be compared with those of actual distances between the proximal and distal probes in wild-type nuclei (Fig. 3, b and d) at various stages of the cell cycle. The data from the unlabeled nuclei most closely resemble the sphere distribution. During S-phase, this distribution shifts: the median is close to or greater than 1, and the distribution is more dispersed. In the case of early S-phase, the distribution approaches that of a rigid rod (compare Fig. 3 b, wild type, early-S, and Fig. 3 e), implying that the chromosome moves in a coordinated fashion. Later in S-phase the distribution more closely resembles that of a shell or a peel.
Postmitotic nuclei display a distribution that is similar to that of the rigid rod, which suggests a polar Rabl configuration with the centromeres at one pole and the telomeres at the other. To determine the length of time this arrangement persists, we examined nuclei another 2 h after mitosis. Because the larvae were fed BrdU for the full course of this experiment, this distribution is a mixture of those nuclei that are just postmitotic and those that are older. At 5 h, the median distance decreases to a level close to the G1-unlabeled nuclei (Fig. 3 b) and more closely resembles that of two points in a sphere than that of a randomly oriented rigid rod (Fig. 3 e). This result indicates that the Rabl configuration is breaking down within 2 h of mitosis.
Effects of the Cell Cycle on Somatic Pairing
Pairing of homologous chromosomes has been found in most interphase nuclei of Drosophila. Using the same nuclei and classes scored for heterologous probe distances in Fig. 3 b, we determined the percentage of nuclei that showed only one fluorescent dot. Nuclei in which two dots could be distinguished were scored as unpaired, regardless of distance between the signals. It is unlikely that the presence of two dots resulted from separation of sister chromatids. If this were the case, we would expect to see three or four probe signals occasionally, but fewer than 0.5% of all nuclei examined ever showed more than two signals. In G1 unlabeled cells, ∼80% of the homologous chromosomes were found to be paired, and this percentage decreased during S-phase (Fig. 4). Interestingly, the pairing of the 59E euchromatic probe was disrupted in early S-phase in both bwD and wild-type nuclei, while the 2R heterochromatic probe was not disrupted until mid or late S-phase. Pairing did not recover during G2-phase, but did begin to recover shortly after mitosis. There is a direct correlation between pairing disruption and timing of replication, early S-phase for euchromatin, and later in S-phase for heterochromatin. In neither of the genotypes do we see full unpairing of the homologues, i.e., the least amount of pairing seen is 60% (Fig. 4). Since we see no recovery in G2-phase, unpairing is not likely to be transient. Rather, pairing may be disrupted in only a subset of nuclei. Another possibility is that we only detect the subset of disruptions with distance that exceed our limit of resolution.
Minute mutations prolong the development in Drosophila. Developmental delay is speculated to be the result of an increase in the length of the cell cycle (Morata and Ripoll, 1975). If such delay is caused by a lengthening of the cell cycle, this could allow somatic pairing to increase (Golic and Golic, 1996). From our results, we would predict that lengthening of the cell cycle gives more time both for homologues to pair and for bwD-2Rh associations to form. A Minute line, M(2)60E (Lindsley and Zimm, 1992), was crossed to bwD and wild type, and was examined for pairing at both 59E and AACAC. No significant differences were detected in the degree of homologous pairing in the Minute line, although any such effect of the Minute mutation may have been undetectable in our assay. Moreover, neither this mutation nor the M(2)53 Minute mutation (Lindsley and Zimm, 1992) was found to modify the trans-inactivation phenotype of bwD (data not shown). The lack of phenotypic modification also parallels a lack of modification of heterochromatic association. The 59E- 2Rh distance measurements in interphase nuclei from the CNS of wandering third instar larvae indicated a lack of significant difference (P < .05, n = 90) between M/bwD and +/bwD larvae or between M/+ and +/+ larvae.
The lack of effect of Minutes in our system could be due to a number of reasons. Homologue pairing and heterochromatic associations might be cued into events of the cell cycle that change proportionally in Minutes. For instance, if the deposition of heterochromatic proteins is also delayed in Minutes, one would not expect to see an overall change in heterochromatic associations. It is also possible that Minutes do not slow down the cell cycle. Cell cycle length has only been inferred by observations that somatic clones of M/+ cells grow more slowly than their wild-type neighbors; however, this observation is equally well explained by an increase in cell death. An early study of Minutes acknowledges that the data can have this alternative interpretation (see footnote 2 in Morata and Ripoll, 1975). To our knowledge, no direct measurement of Minute cell cycle time has been published.
Discussion
We have shown that heterochromatic associations, overall chromosomal positioning, and somatic pairing are disrupted during S-phase in Drosophila tissues. Large-scale interphase movements were unexpected because most such movements were assumed to be confined to mitosis. Our findings have implications for heterochromatic interactions for chromosome dynamics after mitosis and for the concept of chromosomal territories.
Territories can be unambiguously visualized in mammalian interphase nuclei using single-chromosome painting probes. A territory appears as an amorphous but compact mass encompassing only a fraction of the volume of the interphase nucleus (Manuelidis, 1990; Schardin et al., 1985; Zink et al., 1998). This fraction is roughly similar to the proportion of the genome contained within the specific chromosome. In Drosophila melanogaster, the second chromosome is one-third of the full complement (Ashburner, 1989), and so it would encompass a larger proportion of the interphase volume than is encompassed by a typical mammalian chromosome. This makes the chromosomal territories of Drosophila more difficult to detect. By simulating the distances between probes that lie near opposite ends of a long chromosome arm, we showed that confinement of these two probes to a compact subnuclear territory should detectably decrease the distribution of probe distances (Fig. 3 e). We do not observe such a decrease (compare Fig. 3 a, wild-type, unlabeled nuclei, and Fig. 3 e). Additionally, we have examined other sets of distant intrachromosomal probes and have failed to see such a decrease (data not shown). These observations suggest that if chromosome arm 2R is confined to a subnuclear territory, then it must have a complex shape.
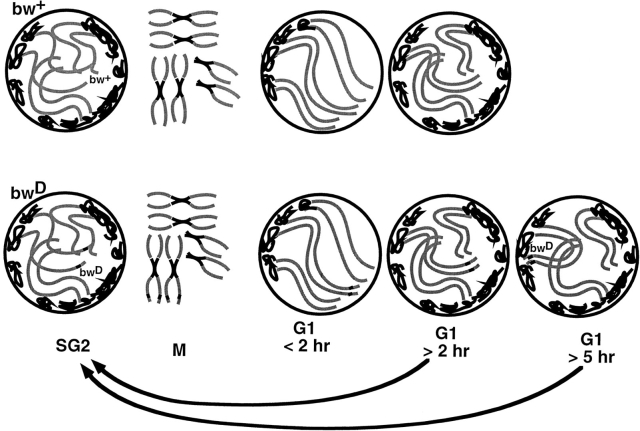
In larval CNS nuclei, probes that hybridize in situ to pericentric heterochromatin from different chromosomes are found in various locations in interphase diploid nuclei, not all in one place as would be expected for a chromocenter (Lifschytz and Hareven, 1982; Lohe and Roberts, 1988; Lohe et al., 1993; Dernburg et al., 1996). Indeed this fact is illustrated in Fig. 1 b, where the DAPI bright regions, which indicate the location of certain AT-rich heterochromatic blocks (Gatti et al., 1994), generally do not overlap the AACAC satellite probe. This result is in contrast to the chromocentral polar organization extensively described in interphase nuclei from precellular blastoderm embryos (Foe and Alberts, 1985; Hiraoka et al., 1993) and polytene larval tissues (Hochstrasser et al., 1986). These two types of nuclei are atypical; embryonic nuclei have no G1 phase and polytene nuclei have no mitosis: rather the diploid nuclei of the larval CNS are more representative of nuclei found in most organisms. While we detect a non-Rabl configuration during most of interphase in diploid nuclei, a polar chromosomal arrangement formed by the act of chromosome segregation at anaphase persists for ∼2 h after mitosis, leading to the conclusion that a redistribution of pericentric heterochromatin must take place in the hours after mitosis. A diagrammatic summary of the chromosome movements detected in this study is shown in Fig. 5.
Figure 5.
Summary of chromosomal movements detected in this study. The nuclei are between 3 and 6 μm in diameter. Some of the movements bring together a chromosome tip and pericentric heterochromatin, which at certain times are located at opposite poles of the nucleus. Therefore, the scale of these movements could exceed 3 μm. Circles represent interphase nuclei. Black lines indicate heterochromatin, and gray lines indicate various euchromatic arms.
Our results describe chromosomal movements associated with the onset of DNA replication (Fig. 3 b and Fig. 5). It is unlikely that any of these movements are influenced by experimental procedures. Heterochromatic associations involving bwD are measured relative to bw + controls, large-scale movements are deduced by comparison of nuclei that differ only by their cell cycle phase, and pairing measurements are additionally controlled by using probes from different parts of the chromosome. Moreover, evidence for chromosomal movements during S-phase has been reported in other studies. In cultured mouse and human lymphocytes, centromeres were found to change position during S- and G2-phase (Ferguson and Ward, 1992; Vourc'h et al., 1993). In Drosophila CNS nuclei, examination of telomere associations in a ubiquitin-conjugating enzyme mutant has led to the proposal that chromosome ends disperse in S- or G2-phase nuclei (Cenci et al., 1997). We see quite pronounced increases in the distances between the proximal and distal 2R in early S-phase, indicating that these regions may be confined to different relative positions within S-phase nuclei. Other hypothetical influences on nuclear positioning, such as nuclear matrix disorganization, would lead to randomization, and this would only have reduced our ability to detect these increases in distance.
In addition to large-scale S-phase movements described above, we detected a dramatic disruption of heterochromatic associations in bwD nuclei. This S-phase disruption was unexpected, because the forces exerted during chromosome condensation and mitosis have been assumed to cause the most vigorous movements. The most parsimonious explanation is that this disruption results from the same process of S-phase rearrangement as in wild-type nuclei. For example, DNA replication of 2R euchromatin could cause an overall stiffening that would push apart the proximal and distal ends of the arm, which in turn would force apart bwD and 2R heterochromatin. Alternatively, there may be a heterochromatin-specific change at early S-phase causing the breakdown of bwD-2Rh associations, perhaps an initial chromatin decondensation in preparation for DNA replication. Interestingly, Li et al. (1998) have recently described the in vivo dynamics of a region of a highly repetitive transgene-induced array that shows heterochromatic properties. They found that this array decondenses and moves towards the center of the nucleus at mid S-phase where it is then replicated. Perhaps such movement towards the nuclear interior is necessary for DNA replication, and in the case of the brown locus, the movement takes place in early S-phase because of its euchromatic location. The consequence of this movement would be the breaking apart of bwD heterochromatic associations.
We were also surprised by the long time required for bwD to reassociate with pericentric 2R heterochromatin after mitosis. We had expected that heterochromatic associations would be set up shortly after mitosis, perhaps in concert with dispersion of the centromeres and breakdown of the Rabl configuration (∼2 h). However, examination of these time points showed no apparent bwD-2Rh heterochromatic association. Not until cells were ∼5–12 h postmitotic did we begin to detect levels of bwD-2Rh heterochromatic association close to the levels seen in unlabeled nuclei. A longer time might be required for the distal tip of 2R to traverse the nucleus and contact 2Rh than for similarly large-scale movements of centromere dispersion and Rabl configuration breakdown. The tip-to-base movement is somewhat slower than what would be predicted from direct measurements of diffusion of Drosophila embryonic chromatin (Marshall et al., 1997). However, those diffusion measurements were over shorter distances than are traversed by our markers, and other factors might be impeding movement.
The association of bwD and 2Rh seems inconsistent with dispersion of the centromeres after mitosis. Heterochromatin appears to show a general self-stickiness that is sequence-independent and is mediated by proteins that recognize the repetitive nature of heterochromatin (Csink and Henikoff, 1996; Dorer and Henikoff, 1997). If heterochromatic regions from opposite ends of a chromosome arm eventually can become stably associated, then why don't pericentric regions associate to form a chromocenter when they are close together in the Rabl configuration? One possibility is that there is a discrete postmitotic event, perhaps associated with exit from the cell cycle or differentiation, that allows cohesion of heterochromatin or chromosome movement. However, some nuclei displaying bwD-2Rh associations enter S-phase (Fig. 5). Moreover, an examination of the 7-, 14-, and 20-h distributions shows a gradual increase in bwD heterochromatic interactions, which means that any such event would differ temporally from cell to cell. Alternatively, there may be a property of the nucleus that is gradually built up after mitosis and promotes bwD-2Rh heterochromatic associations. We propose that this property is the maturing of heterochromatin, including the deposition of heterochromatic proteins. Some proteins associate with heterochromatin during mitosis, but appear to be lost from heterochromatin gradually (on the order of hours) during interphase (Platero et al., 1998). In contrast, most of Heterochromatin Protein 1 (HP1) is removed from chromosomes during mitosis, and is redeposited at an undetermined rate in G1 (Kellum et al., 1995). Indeed, our previous work has shown that decreasing HP1 will reduce heterochromatic associations (Csink and Henikoff, 1996). It is possible that removing mitosis-specific proteins is necessary for depositing the HP1, and it is not until a proteinaceous glue is of sufficient tackiness that chance encounters of bwD and centric heterochromatin will persist. The lack of centric cohesion in the early G1 Rabl configuration would thus be explained by the lack of tackiness of immature heterochromatin. This scenario predicts that the differential kinetics of release of mitosis-specific proteins from their target heterochromatic sequences (Platero et al., 1998) will result in differential kinetics of heterochromatin maturation.
Recent studies examining chromatin movements during interphase have concluded that most movement can be described as constrained Brownian motion (Marshall et al., 1997; Abney et al., 1997; Shelby et al., 1996; Zink et al., 1998). Rare large-scale movements have also been detected (Shelby et al., 1996; Zink et al., 1998; Buchenau et al., 1997). Perhaps these occasional large-scale movements allow the chromosome to escape the usual constraints and establish associations of bwD with pericentric heterochromatin.
Another nuclear interaction that we encountered is the somatic pairing of homologous chromosomes. While homologue pairing is seen during prophase of meiosis I of all eukaryotes, it is only rarely seen elsewhere. The exception is the Dipteran insects, in which almost all nuclei examined display somatic pairing (Kopczynski and Muskavitch, 1992; Lifschytz and Hareven, 1982; Metz, 1916). More than 75% of the nuclei that we examined showed only one spot of hybridization for a given probe, indicating that the signals from each homologue are too close to distinguish. Only short postmitotic periods are required for homologous pairing, in contrast to the long times required for heterochromatic associations within these nuclei. Somatic pairing does not approach this level in nuclei undergoing the initial divisions in the syncytial embryo until the cell cycle is >20 min at nuclear division 14 (Hiraoka et al., 1993), probably because this is the minimal amount of time necessary for pairing to occur.
We find that somatic pairing is partially disrupted during the course of S-phase. The disruption of the 59E euchromatic region occurs before the disruption of the heterochromatic AACAC satellite. Since euchromatin replicates early and heterochromatin replicates late, this difference between the two probes could indicate that this pairing disruption is caused by the passage of the replication fork. It should be noted that these pairing disruptions are not necessarily large-scale. Close juxtaposition of homologues is to some extent maintained during G2- and M-phases, because in metaphase figures homologous chromosomes, though separated, are closely apposed (Metz, 1916). Pairing does not recover immediately after passage of the fork, but only begins to recover after the completion of mitosis. This provides an interval of at least 1.5 h, which in embryos is more than sufficient for pairing to occur. Pairing may be inhibited during this postreplicative interval. Understanding this inhibition may provide insight into the mechanism of somatic pairing.
The expression of some Drosophila genes can be affected by pairing (for review see Henikoff, 1997). Our work showing that pairing is disrupted postreplicatively implies that pairing-dependent effects would be at least partially disrupted in G2-phase. Similar sensitivity to cell-cycle progression might be a feature of regulation of gene expression by heterochromatic associations. The idea that a heterochromatic compartment can sequester and thus regulate a gene (Eberl et al., 1993; Wakimoto and Hearn, 1990) has been a recent topic of much discussion (Maillet et al., 1996; Marcand et al., 1996). Gene regulation by heterochromatin may also be occurring in certain mouse lymphoid cells: a correlation was found between contact of genes with heterochromatin in the interphase nucleus and downregulation of those genes' transcripts (Brown et al., 1997). If this contact is indeed causative and analogous to bwD heterochromatic silencing, we speculate that the mode of transcriptional regulation would not be available until well into G1 when heterochromatin becomes sticky. Hence, silencing of a gene by heterochromatic associations offers a potential mechanism that ties gene regulation directly to mitosis. Silencing may become a regulatory option for cells during development as the length of G1-phase increases to allow heterochromatic associations to occur.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute.
Abbreviations used in this paper
- AED
after egg deposition
- CNS
central nervous system
- FISH
fluorescence in situ hybridization
Footnotes
Zheng Fan performed the distance measurements. Jorja Henikoff wrote the computer program for the simulations and advised on statistical analysis. The possibility that cell death could play a roll in Minute developmental delay was brought to our attention by Thomas Neufeld. Georgette Sass provided helpful comments on earlier versions of this manuscript.
The present address of Amy K. Csink is Department of Biological Sciences, Carnegie Mellon University, 4400 Fifth Avenue, Pittsburgh, Pennsylvania 15213-2683.
Address all correspondence to Amy K. Csink or Steven Henikoff, Howard Hughes Medical Institute, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue N, Seattle, WA 98109-1024. Tel.: 206-667-4515. Fax: 206-667-5889. E-mail: csink@andrew.cmu.edu or steveh@muller.fhcrc.org
References
- Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Buchenau P, Saumweber H, Arndt-Jovin DJ. The dynamic nuclear redistribution of an hnRNP K-homologous protein during Drosophilaembryo development and heat shock. Flexibility of transcription sites in vivo. J Cell Biol. 1997;137:291–303. doi: 10.1083/jcb.137.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G, Rawson RB, Belloni G, Castrillon DH, Tudor M, Petrucci R, Goldberg ML, Wasserman SA, Gatti M. UbcD1, a Drosophilaubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev. 1997;11:863–875. doi: 10.1101/gad.11.7.863. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. . Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- Csink, A.K., G.L. Sass, and S. Henikoff. 1997. Drosophila heterochromatin: retreats for repeats. In Nuclear organization, chromatin structure and gene expression. R. van Driel and A.P. Otte, editors. Oxford University Press, New York. 223–235.
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl DF, Duyf BJ, Hilliker AJ. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster. Genetics. 1993;134:277–292. doi: 10.1093/genetics/134.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Ward DC. Cell cycle dependent chromosomal movement in pre-mitotic human T-lymphocyte nuclei. Chromosoma. 1992;101:557–565. doi: 10.1007/BF00660315. [DOI] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Fung JC, Marshall WF, Dernburg A, Agard DA, Sedat JW. Homologous chromosome pairing in Drosophila melanogasterproceeds through multiple independent initiations. J Cell Biol. 1998;100:1623–1636. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Bonaccorsi S, Pimpinelli S. Looking at Drosophilamitotic chromosomes. Methods Cell Biol. 1994;44:371–391. doi: 10.1016/s0091-679x(08)60924-3. [DOI] [PubMed] [Google Scholar]
- Gatti M, Tanzarella C, Olivieri G. Analysis of the chromosome aberrations induced by x-rays in somatic cells of Drosophila melanogaster. Genetics. 1974;77:701–719. doi: 10.1093/genetics/77.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic MM, Golic KG. A quantitative measure of the mitotic pairing of alleles in Drosophila melanogaster and the influence of structural heterozygosity. Genetics. 1996;143:385–400. doi: 10.1093/genetics/143.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW. The onset of homologous chromosome pairing during Drosophilamelanogaster embryogenesis. J Cell Biol. 1993;120:591–600. doi: 10.1083/jcb.120.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M, Mathog D, Gruenbaum H, Saumweber H, Sedat JW. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. . J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- John, B. 1988. The biology of heterochromatin. In Heterochromatin molecular and structural aspects. R.S. Verma, editor. Cambridge University Press, Cambridge, United Kingdom. 1–147.
- Kellum R, Raff JW, Alberts BM. Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J Cell Sci. 1995;108:1407–1418. doi: 10.1242/jcs.108.4.1407. [DOI] [PubMed] [Google Scholar]
- Kopczynski CC, Muskavitch MA. Introns excised from the Delta primary transcript are localized near sites of Delta transcription. J Cell Biol. 1992;119:503–512. doi: 10.1083/jcb.119.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- Li G, Sudlow G, Belmont AS. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J Cell Biol. 1998;140:975–989. doi: 10.1083/jcb.140.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E, Hareven D. Heterochromatin markers: arrangement of obligatory heterochromatin, histone genes and multisite gene families in the interphase nucleus of D. melanogaster. . Chromosoma. 1982;86:443–455. doi: 10.1007/BF00330120. [DOI] [PubMed] [Google Scholar]
- Lindsley, D.L., and G.G. Zimm. 1992. The Genome of Drosophila melanogaster. Academic Press, Inc., San Diego.
- Lohe, A., and P. Roberts. 1988. Evolution of satellite DNA sequences in Drosophila. In Heterochromatin molecular and structural aspects. R.S. Verma, editor. Cambridge University Press, Cambridge, United Kingdom. 148–186.
- Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. . Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A view of interphase chromosomes. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- Marcand S, Gasser SM, Gilson E. Chromatin: a sticky silence. Curr Biol. 1996;6:1222–1225. doi: 10.1016/s0960-9822(96)00701-4. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. . Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen, I.A., and T.E. Hanson. 1993. The development of the optic lobe. In The Development of Drosophila melanogaster. M. Bate and A.M. Arian, editors. Cold Spring Harbor Laboratory Press, Plainview, New York. 1327–1492.
- Metz CW. Chromosome studies on the Diptera. II. The paired association of chromosomes in the Diptera, and its significance. J Exp Zool. 1916;21:213–279. [Google Scholar]
- Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Nagele R, Freeman T, McMorrow L, Lee HY. Precise spatial positioning of chromosomes during prometaphase: evidence for chromosomal order. Science. 1995;270:1831–1835. doi: 10.1126/science.270.5243.1831. [DOI] [PubMed] [Google Scholar]
- Platero J, Csink A, Quintanilla A, Henikoff S. Changes in chromosomal localization of heterochromatin binding proteins during the cell cycle in Drosophila. J Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardin M, Cremer T, Hager HD, Lang M. Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories. Hum Genet. 1985;71:281–287. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- Selleck SB, Gonzalez C, Glover DM, White K. Regulation of the G1-S transition in postembryonic neuronal precursors by axon ingrowth. Nature. 1992;355:253–255. doi: 10.1038/355253a0. [DOI] [PubMed] [Google Scholar]
- Shelby RD, Hahn KM, Sullivan KF. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R.R., and F.J. Rohlf. 1981. Biometry. W.H. Freeman and Company, New York.
- Talbert PB, LeCiel CD, Henikoff S. Modification of the Drosophilaheterochromatic mutation brown Dominant by linkage alterations. Genetics. 1994;136:559–571. doi: 10.1093/genetics/136.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Truman, J.W., B.J. Taylor, and T.A. Awad. 1993. Formation of the adult nervous system. In The development of Drosophila melanogaster. M. Bate and A.M. Arian, editors. Cold Spring Harbor Laboratory Press, Plainview, New York. 1245–1275.
- van Driel, R., and A.P. Otte. 1997. Nuclear organization, chromatin structure and gene expression. Oxford University Press, New York.
- Vourc'h C, Taruscio D, Boyle AL, Ward DC. Cell cycle-dependent distribution of telomeres, centromeres and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytles. Exp Cell Res. 1993;205:142–151. doi: 10.1006/excr.1993.1068. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT, Hearn MG. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. . Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanishevsky RM, Prescott DM. Late S phase cells (Chinese hamster ovary) induce early S phase DNA labeling patterns in G1 phase nuclei. Proc Natl Acad Sci USA. 1978;75:3307–3311. doi: 10.1073/pnas.75.7.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota H, van den Engh G, Hearst JE, Sachs RK, Trask BJ. Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J Cell Biol. 1995;130:1239–1249. doi: 10.1083/jcb.130.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Cremer T, Saffrich R, Fischer R, Trendelenburg MF, Ansorge W, Stelzer EH. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]