Abstract
We have determined the relationship between overall nuclear architecture, chromosome territories, and transcription sites within the nucleus, using three-dimensional confocal microscopy of well preserved tissue sections of wheat roots. Chromosome territories were visualized by GISH using rye genomic probe in wheat/rye translocation and addition lines. The chromosomes appeared as elongated regions and showed a clear centromere–telomere polarization, with the two visualized chromosomes lying approximately parallel to one another across the nucleus. Labeling with probes to telomeres and centromeres confirmed a striking Rabl configuration in all cells, with a clear clustering of the centromeres, and cell files often maintained a common polarity through several division cycles. Transcription sites were detected by BrUTP incorporation in unfixed tissue sections and revealed a pattern of numerous foci uniformly distributed throughout the nucleoplasm, as well as more intensely labeled foci in the nucleoli. It has been suggested that the gene-rich regions in wheat chromosomes are clustered towards the telomeres. However, we found no indication of a difference in concentration of transcription sites between telomere and centromere poles of the nucleus. Neither could we detect any evidence that the transcription sites were preferentially localized with respect to the chromosome territorial boundaries.
Keywords: BrUTP, transcription sites, chromosome territory, nuclear architecture, plant
There is accumulating evidence that interphase chromosomes occupy spatially distinct regions of the nucleus, often referred to as chromosome territories, separated by interchromosomal channels (for reviews see Strouboulis and Wolffe, 1996; Lamond and Earnshaw, 1998). Early evidence for this organization was provided by Cremer et al. (1982) who irradiated interphase nuclei with UV and showed that damage was localized to only a few chromosomes. The use of in situ hybridization with chromosome-specific DNA paints confirmed the arrangement of interphase chromosomes in distinct, non-overlapping territories (Cremer et al., 1988; Lichter et al., 1988). Since then, a territorial organization of chromosomes has been demonstrated in an increasing number of animal and plant species (e.g., Heslop-Harrison and Bennett, 1990). However, the way the interphase chromosomes are arranged within the nucleus and with respect to each other seems to vary from species to species. Many years ago, Rabl proposed that the centromere–telomere orientation established at mitotic anaphase would continue throughout the cell cycle (Rabl, 1885). This configuration, since referred to as the Rabl configuration, would imply that centromeres and telomeres were positioned at opposite poles of the nucleus. The Rabl configuration has been demonstrated in some species, including Drosophila polytene nuclei (Hochstrasser et al., 1986), Trypanosoma (Chung et al., 1990), fission yeast (Funabiki et al., 1993), and some plants (Heslop-Harrison et al., 1993; Noguchi and Fukui, 1995). However there is no evidence for a Rabl configuration in somatic cells of other studied species, such as humans and other mammals.
It has been proposed that the compartmentalization of the nucleus into chromosome territories and interchromosome channels is reflected in the spatial organization of the functional protein complexes responsible for nuclear processes such as transcription, splicing, replication, and repair (Cremer et al., 1993, 1995). Thus it has been postulated that transcription takes place at the surfaces of the chromosome territories, with transcript processing and export being directed through the three-dimensional (3D)1 network of interchromosome channels; interchromosome channels ending at a nuclear pore would provide an efficient exit route from the nucleus for mRNA complexes (Kurz et al., 1996). However, the experimental evidence for this hypothesis is based on the in situ localization of a few genes, and to our knowledge there has so far been no systematic study of the relation between all nuclear transcription sites and chromosome territorial organization.
Various groups have shown that BrUTP incorporation accurately localizes transcription sites in the nucleus (Jackson et al., 1993; Wansink et al., 1993, 1994). These studies have revealed several hundred distinct, punctate sites of labeling, showing that RNA polymerase II transcription takes place in numerous small domains dispersed throughout the nucleus. The labeled sites remained after most of the chromatin was digested away by nucleases, suggesting the transcription sites are attached to a resistant nuclear matrix. A similar pattern has been observed in plant cell nuclei (Straatman et al., 1996) and nucleoli (Hozak et al., 1994; Thompson et al., 1997). In confirmation of this distribution of transcription sites, RNA polymerase II and transcription factors were found distributed throughout the nucleoplasm in numerous small domains (Grande et al., 1997).
In the present work, we have examined the organization of transcription sites in relation to the arrangement of chromosomes in wheat root tissue. For this study we used well-preserved, intact tissue for whole-mount in situ hybridization and for BrUTP incorporation, and analyzed the labeling using 3D confocal microscopy. It has not so far been possible to produce chromosome paints to label individual wheat chromosomes, or indeed those of any other plant. For this reason we used a wheat line containing an extra pair of chromosomes from rye, as well as a translocation line containing a single pair of rye chromosome arms. The rye chromosomes and chromosome arms were detected using total rye genomic DNA as a probe for in situ hybridization. The arrangement of the chromosomes was confirmed by in situ hybridization with centromere and telomere probes. To study the pattern of distribution of transcription sites in the wheat nucleus we used BrUTP incorporation in unfixed root sections. By combining BrUTP incorporation with in situ hybridization we visualized transcription sites in relation to chromosome territories, and to telomeres and centromeres.
Materials and Methods
Root Sections
Seeds of Triticum aestivum (cv Chinese Spring/1R disomic addition and Chinese Spring 1A/1R translocation) were germinated on water-soaked filter paper. Roots were excised 3 d after germination. For in situ studies, root tips were fixed in 4% (wt/vol) PFA in PEM (50 mM Pipes, 5 mM EGTA, 5 mM MgSO4, pH 6.9, with KOH) for 1 h, followed by washing for 10 min in TBS (10 mM Tris, 140 mM NaCl, pH 7.4, with HCl). Root tips were sectioned under water into 30-μm-thick sections using a Vibratome Series 1000 (TAAB Laboratories Equipment Ltd., Aldermaston, UK). Sections were placed immediately on multi-well slides (ICN Biomedicals Inc., Costa Mesa, CA) coated with glutaraldehyde-activated γ-aminopropyl triethoxy silane (APTES; Sigma Chemical Co., St. Louis, MO) and left to air dry.
BrUTP Incorporation into Tissue Sections
For transcription studies, the method described by Thompson et al. (1997) was adapted. To improve nuclear transcription as opposed to nucleolar transcription, 1% BSA was added to the modified physiological buffer (MPB: 100 mM KAc, 20 mM KCl, 20 mM Hepes-KOH, pH 7.4, 1 mM MgCl2, 1 mM ATP in 50 mM Tris, pH 8, 1% thiodiglycol, 2 mg/ml aprotenin, 0.5 mM PMSF), and the permeabilization step with Triton X-100 was replaced by a very short (10 s) treatment with 0.05% Tween 20 in MPB. In vitro transcription was allowed to continue for 5 min.
In Situ Hybridization with Centromeric and Telomeric Probes
Tissue sections on slides were treated with 2% (wt/vol) cellulase (Onuzuka R-10) in TBS for 1 h at room temperature and washed in TBS followed by 0.1× SSC (SSC: 150 mM NaCl, 15 mM sodium citrate). Denaturation of both target DNA and probe was done in 0.1× SSC at 98°C for 5 min, followed by 5 min in ice-cold 0.1× SSC. The surface liquid was blotted off slides and 10 μl of ice-cold hybridization mixture was added. The hybridization mixture comprised 50% deionized formamide, 10% 100 mM Pipes/10 mM EDTA, pH 8, 20% dextran sulphate, 10% 3 M NaCl, 100 ng centromeric or telomeric probe, 50× excess blocking salmon sperm DNA. Probes were produced as described by Aragón-Alcaide et al. (1997). Hybridization was carried out overnight in a humid chamber at 37°C. Post-hybridization washes were performed in 0.1× SSC at 50°C for 1.5 h with two changes.
Centromere probes were detected using conjugated sheep anti-digoxigenin antibody-FITC (Boehringer Mannheim Corp., Indianapolis, IN) and the signal was sequentially amplified with rabbit anti–sheep-FITC (DAKO Corp., Carpinteria, CA) and sheep anti–rabbit-FITC (Sigma Chemical Co.). Telomere probes were detected using extravidin-Cy3 (Sigma Chemical Co.). Antibodies were diluted in TBS/3% BSA. Antibody incubations were carried out in a damp chamber for 45 min at room temperature and TBS washes were carried out between antibody incubations. Slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma Chemical Co.) (1 μg/ml) for 5 min, and then mounted in antifade solution (Vectashield; Vector Laboratories Inc., Burlingame, CA).
In Situ Hybridization with Rye Total Genomic Probe
Sections were treated with cellulase as described above. Probe preparation and hybridization procedures followed those described by Schwarzacher et al. (1992) with the following modifications: the hybridization mixture was denatured at 95°C for 5 min before addition to the preparations, and denaturation of slides with probe was performed at 78°C for 10 min using a modified thermocycler (Omnislide; Hybaid Ltd., Long Island, NY).
Combined BrUTP Incorporation and In Situ Hybridization on Root Sections
BrUTP incorporation was performed on sections as described above. After the antibody detection of BrUTP incorporation, the sections were fixed a second time with 4% PFA for 30 min. Then the sections were washed in TBS and in situ hybridization was performed as described above except in the case of centromere in situ labeling where the denaturation of sections was done for 5 min at 80°C in 70% formamide 2× SSC.
Microscopy, Photography, and Image Processing
Specimens were surveyed using a Zeiss Universal (Carl Zeiss, Inc., Oberkochen, Germany) or a Nikon Eclipse 800 (Nikon Corp., Tokyo, Japan) fluorescence microscope. Confocal optical section stacks were collected using a Biorad MRC-600 or a Biorad MRC-1,000 UV confocal scanning microscope (Bio-Rad Laboratories, Hercules, CA) as described previously (Beven et al., 1996). Images were transferred to a PC or a Macintosh computer and assembled into composite images using Photoshop (Adobe Systems Inc., Mountain View, CA) and NIH Image, a public domain program for the Macintosh available via anonymous ftp from zippy.nimh.nih.gov. Images were printed on a Pictrography P3000 printer.
Results
Chromosome Arrangement in the Wheat Interphase Nucleus
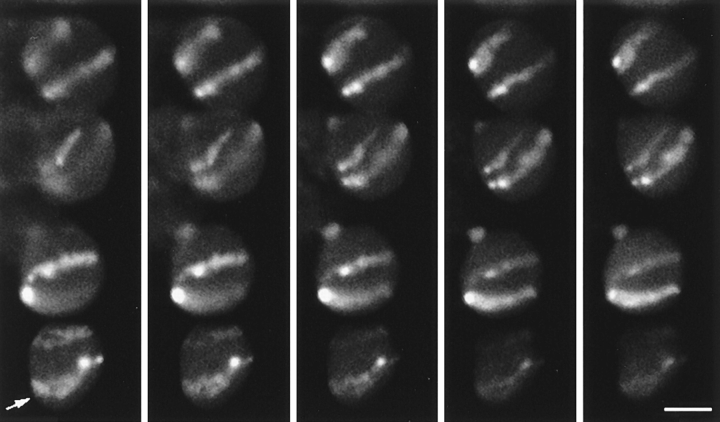
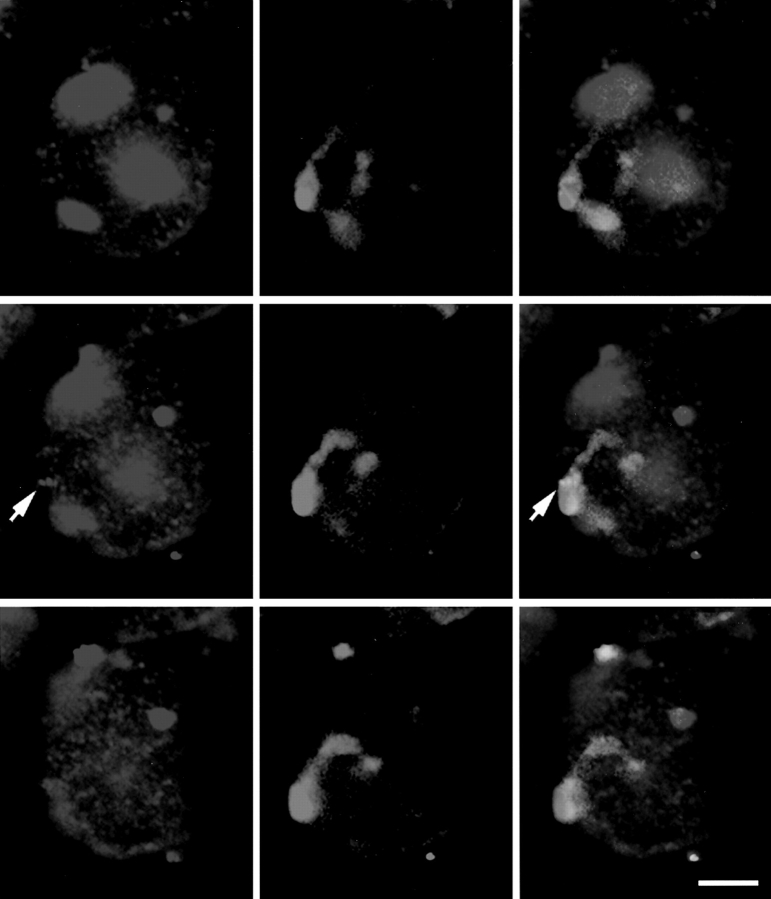
Wheat lines containing the addition of a pair of rye chromosomes (1R) or a 1A/1R translocation line, where one arm of wheat chromosome 1A is replaced by one arm of rye chromosome 1R, were used to visualize individual chromosome territories. In both lines the rye chromosomes or chromosome arms were labeled by in situ hybridization using fragmented total rye DNA into which digoxigenin had been incorporated. In the interphase nuclei the rye chromosomes appeared as elongated regions generally traversing the nucleus from one side to the other and they were usually parallel to each other (Fig. 1). Often, the labeled chromosomes in several nuclei in a cell file were in the same orientation (see Fig. 1). In the addition line, the two chromosome arms always lay alongside each other. Often the two arms were so close that only a single labeled region was seen; sometimes the two arms were distinguishable (Fig. 1, arrow). A similar pattern of chromosome labeling was seen, irrespective of the size of the nucleus, and thus the presumed phase of the cell cycle. We examined these specimens for evidence of somatic association of the homologues. We considered the two homologous chromosomes or chromosome arms to be associated if only one region of labeling, with no intervening space, could be seen for the two chromosomes or arms. In all, 20 out of 84 nuclei from the addition line, and 9 out of 99 nuclei from the translocation line showed evidence of homologous pairing (24% and 9%, respectively). We consider that this is probably not significant, although a detailed statistical analysis is beyond the scope of this paper.
Figure 1.
Root tissue from wheat 1R addition line, in which a pair of rye chromosomes (1R) is present. The rye chromosomes have been labeled by genomic fluorescence in situ hybridization using a total rye genomic DNA probe. A series of optical sections collected by confocal microscopy is shown. The chromosomes stretch across the nuclei, the two arms next to each other, and the two labeled chromosomes are usually parallel to one another. In some cases, the two arms can be distinguished (arrow). Focal distance between section = 1 μm. Bar, 10 μm.
Centromeres and Telomeres Are Located at Opposite Poles of the Nucleus
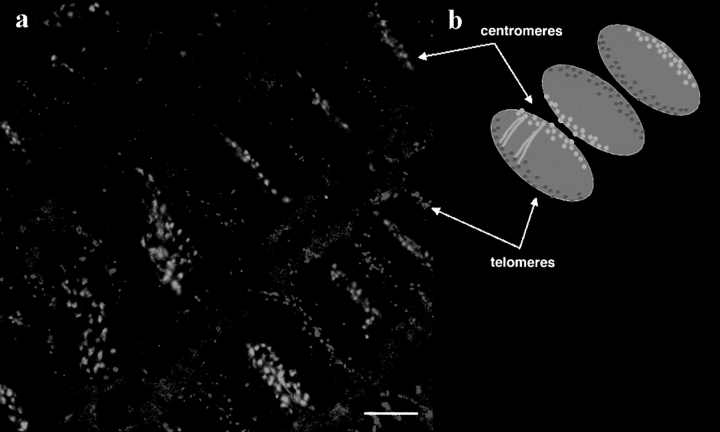
Double fluorescence in situ hybridization was carried out on root sections, using centromeric and telomeric probes. As shown in Fig. 2 a, there was a strong polarization of the sites labeled in the interphase nuclei, with the centromeres clustered at one side of the nucleus and the telomeres located at the opposite side—a clear Rabl configuration. Fig. 2 b shows a diagrammatic interpretation of the labeling pattern seen. It was striking that a common nuclear orientation was maintained for many adjacent cells in a given cell file, suggesting a strong conservation of overall chromosome order during several rounds of cell division, and confirming the previous observations of the orientation of the labeled chromosomes.
Figure 2.
(a) Wheat root tissue double labeled by fluorescence in situ hybridization with probes to the centromeres (green) and telomeres (red). Projection of five confocal optical sections (focal distance between original sections = 1 μm). (b) Diagram showing the interpretation of the labeling in (a) as the Rabl configuration. The chromosomes must all be parallel to one another, with the centromeres clustered on one side of the nuclear periphery, and the telomeres somewhat more dispersed on the other side of the nuclear periphery. A common (alternating) polarity is often maintained through the lines of cells as in this image. Bar, 10 μm.
Transcription Foci Are Scattered throughout the Nucleus
Transcription sites were visualized by incubating unfixed vibratome sections from wheat roots with a transcription mix containing BrUTP, fixing with formaldehyde, and then detecting incorporated BrUTP by immunofluorescence labeling. It was necessary to use very short permeabilization times (10 s) with 0.05% Tween to allow the transcription mix into the nuclei, while preserving nuclear transcriptional activity. Longer incubation with detergent gave solely nucleolar incorporation of BrUTP (Thompson et al., 1997), possibly because of disruption or inactivation of RNA polymerase II. All procedures were carried out at room temperature, and transcription was allowed to proceed for 5 min. Published methods for animal cells specify 4°C for permeabilization; we found that treatment at 4°C inactivated transcription in plant cells, and that it did not recover subsequently at room temperature. In control experiments, BrUTP was omitted from the transcription reaction, or actinomycin D, an inhibitor of RNA polymerases, was added. In these control experiments no fluorescence in the nucleus or nucleolus was seen.
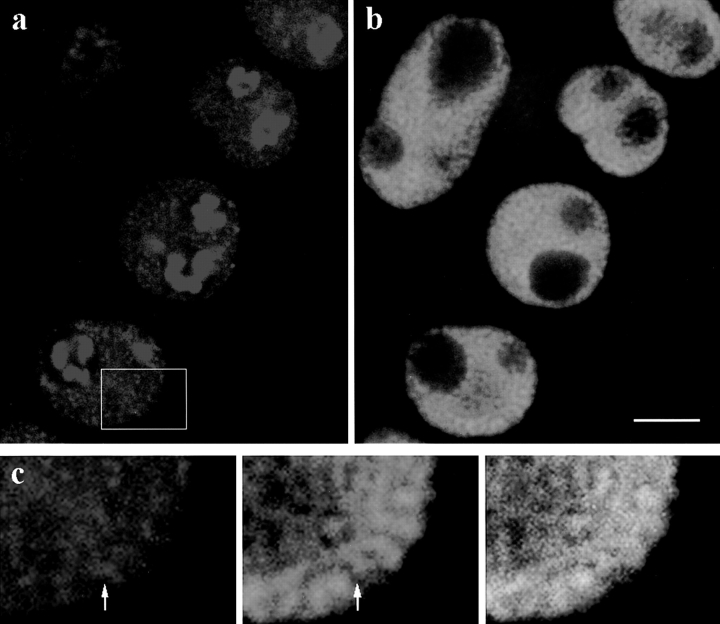
Fig. 3 shows an example of BrUTP incorporation (Fig. 3 a, red), compared with DAPI counterstain (Fig. 3 b, blue). The nucleoli are clearly visible in the DAPI-stained images as dark holes in the nuclei, and the nucleolar transcription sites are dispersed through a sub-region of the nucleoli. The nucleolar labeling is stronger than that in the nucleoplasm, and in these images the nucleolar labeling has been overexposed so as to show the fainter nucleoplasmic transcription sites. These are seen as many small foci, distributed fairly evenly throughout the nucleoplasm. Some sites are significantly brighter than the others. In this species the chromatin is arranged in a reticulate network, and there is some tendency for the transcription sites to be located in the darker interchromatin regions rather than in the bright DAPI-stained regions, but the pattern of transcription sites does not match the reticulate pattern of DAPI staining very closely (Fig. 3 c).
Figure 3.
Labeling of transcription sites in wheat root tissue by incorporation of BrUTP. A single confocal optical section is shown. (a) BrUTP labeling of several nuclei (red). The nucleolar transcription sites are more strongly labeled than the nucleoplasmic sites, and are overexposed in this image. The nucleoplasmic sites are fairly uniformly distributed throughout the nucleoplasm. There is some variability in the intensity of BrUTP labeling, so that, for example, the upper left nucleus is less strongly labeled, but the pattern of labeling is equivalent to that in the other cells. (b) Corresponding DAPI image (blue). (c) Enlargement of the boxed area in a, showing the correspondence between BrUTP sites and chromatin structure. In some cases transcription foci localize to DAPI-dark regions (e.g., arrows), but this is not universal. Bar, 10 μm.
The Distribution of Transcription Sites in Interphase Nuclei Is Not Polarized
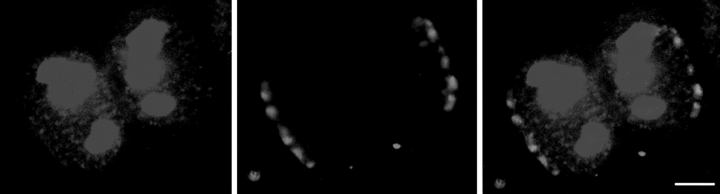
There is evidence that in the physical map of wheat chromosomes, the telomeric regions of all the chromosomes are significantly more gene-rich than the centromeric regions (Gill et al., 1996). Since we have shown above that wheat nuclei are highly polarized, with the centromeres clustered at one side of the nucleus, we might expect that transcription sites would be concentrated in the opposite half of the nucleus, and that the region of the nucleus near the centromeres would be relatively depleted in transcription sites. We therefore carried out double-labeling experiments to show transcription sites by BrUTP incorporation, followed by fluorescence in situ hybridization with centromere probe. Fig. 4 shows a confocal optical section through a pair of nuclei in G1. Transcription sites are shown in red in the left-hand panels, centromeres in green (clearly clustered on opposite sides of the sister nuclei) in the central panels, and the two probes superimposed in the right hand panels. There is no evidence for any polarization in the distribution of transcription sites; there seems to be as high a density near the centromeres as at the opposite pole of the nuclei.
Figure 4.
A single confocal image showing double labeling of transcription sites (red, left panel) and centromeres (green, central panel). The two labels are superimposed in the right panel. There is no indication of a polarized distribution of transcription sites, which appear as dense at the centromeric poles of the nuclei as at the opposite, telomeric poles. Bar, 10 μm.
Transcription Sites Are Not Preferentially Located at Chromosome Territory Boundaries
To compare the distribution of transcription sites with chromosome territories, we used a wheat line with an additional pair of rye chromosomes (1R addition), and a line containing a translocated rye chromosome arm (1A/1R translocation). Transcription was detected by BrUTP incorporation, and this was then followed by genomic in situ hybridization using total genomic rye probe. Three consecutive confocal optical sections from the translocation line are shown in Fig. 5; BrUTP incorporation is shown in red in the left-hand panels, the rye chromosomes in green in the central panels, and the two probes superimposed in the right hand panels. There is no correlation between the region occupied by the rye chromosome arms and the transcription sites. In fact, transcription sites are seen throughout the labeled chromosome territory. Two particularly strong transcription sites within this territory are indicated by an arrow. There is no sign that the interior regions of these chromosome territories are devoid of transcription sites. It should be noted that in places, including the position of the strong arrowed transcription sites, the labeled chromosome territories are at least 2-μm wide. Thus location of the transcription sites, which range in size from 0.5 μm to sizes at or below the resolution limit (0.25 μm), to sub-regions of the chromosome territory would be well within the resolution limit of this technique.
Figure 5.
Double labeling of the wheat 1A/1R translocation line, in which one arm of wheat chromosome 1A is substituted by an arm of rye chromosome 1. BrUTP incorporation is shown in red (left-hand panels), genomic in situ labeling in green (central panels), the two labels superimposed in the right hand panels. Three consecutive confocal sections are shown. The distribution of transcription sites shows no sign of being excluded from the interior of the labeled chromosome territory. In fact two prominent sites are clearly inside the chromosome territory (arrows). Section spacing = 1 μm. Bar, 5 μm.
Discussion
By applying fluorescence in situ hybridization combined with confocal 3D imaging to well-preserved wheat root tissue we have shown that the nuclei in these cells have a remarkably well ordered and consistent 3D architecture. We have shown previously that the in situ procedures we used cause minimal cellular disruption, at least at optical resolution (Shaw et al., 1995), in contrast to standard in situ methods which cause considerable structural distortion because of squashing and harsh denaturation conditions. We have also previously shown that the distribution of nucleolar transcription sites visualized by BrUTP incorporation into plant root tissue agrees well with that determined by in situ hybridization using an anti-sense probe to the external transcribed spacer of the rRNA (Thompson et al., 1997). Furthermore, in the present study, the distribution of transcription sites visualized by BrUTP incorporation was not affected by subsequent in situ labeling.
The interphase chromosomes occupy elongated regions, usually stretching right across the nucleus. The chromosomes are approximately parallel to one another and their arms lie next to each other. Both centromeres and telomeres are located at the nuclear periphery. The centromeres are highly clustered in one region of the nuclear periphery, whereas the telomeres are more dispersed around the opposite side of the nuclear periphery. This suggests that both centromeres and telomeres interact with peripheral nuclear structures, possibly the nuclear pore–lamina complex or another nuclear matrix component, whereas more specific interactions are also involved in the organization of the centromeres into clusters. This strong Rabl configuration gives a polarity to the nuclei, and the direction of this polarity can be maintained through several rounds of cell division.
By using wheat lines containing the addition of pairs of rye chromosomes or translocations of single rye arms, we were able to visualize individual chromosome territories by genomic in situ hybridization. This confirmed the strong Rabl configuration in these nuclei, with all the chromosomes lying parallel to one another across the nucleus, the two chromosome arms next to each other. We found no evidence for significant association of homologues, in contrast to previous observations on squashed preparations of nuclei in wheat and other plants (Avivi and Feldman, 1980). This confirms previous observations on pre-meiotic wheat nuclei (Aragón-Alcaide et al., 1997), using these and other similar addition lines. In the developmental stages leading up to meiosis in wheat anthers no association of the homologues was found until shortly before meiosis. However, in the pre-meiotic interphase a high level (90%) of homologue pairing occurred in both the meiocytes and the surrounding somatic tapetal cells (Aragón-Alcaide et al., 1997). In some species, notably in Drosophila polytene salivary gland nuclei (Hochstrasser et al., 1986) there is a very clear somatic association of all the homologues. On the other hand, in most species, including mammals, there is no evidence for association of homologues, except in meiotic prophase. It appears that wheat shows an intermediate behavior, with little or no homologous pairing in somatic cells until shortly before meiosis. Whether the homologous pairing observed in wheat anthers should be regarded as part of the process of meiosis, or as a switching on of a mechanism for somatic pairing in the developmental pathway leading to meiocytes (and associated somatic cells) is still unknown.
The fact that wheat root nuclei, along with the other wheat somatic cell types we have examined, show such a high degree of structural organization makes them a very good system to analyze the organization of transcription sites, and to test previous hypotheses relating transcription sites to chromosome territories. Such a well-ordered and reproducible interphase chromosomal organization should clearly reveal a systematic organization of transcription sites if the location of transcription sites is related in any obvious way to chromosome territories. BrUTP incorporation shows many small foci distributed in the nucleoplasm, in addition to the strong incorporation we previously showed in the nucleolus (Thompson et al., 1997). This nuclear distribution of transcription foci is very similar to published images of human HeLa and T24 cultured cells (Jackson et al., 1993; Wansink et al., 1993). It has been shown previously that BrUTP incorporation faithfully represents transcription sites (e.g., Wansink et al. [1993] used microinjection of BrUTP precursors to verify results using permeabilization of cells). In plants, we have shown that BrUTP is incorporated into the same nucleolar sites as are labeled by an in situ probe to nascent rRNA transcripts (Thompson et al., 1997). It was not possible to quantify accurately the number of nucleoplasmic foci, but most nuclei contained of the order of a few hundred. This is consistent with the numbers of sites observed in mammalian cells, and is significantly less than the estimated number of active genes. This may mean that only the most active genes were seen, with many other transcription sites below the detection limit. An alternative explanation is that each site represents transcription of several genes— either a group of smaller sites too close together to be resolved, or more than one gene being transcribed at a single cluster of many polymerase molecules. Iborra et al. (1996) have provided evidence that all the transcription sites are in fact visualized in similar experiments in mammalian cells, and have suggested that each transcription site represents a “factory” where more than one gene is transcribed, and where other nuclear activities such as DNA replication take place (Jackson and Cook, 1995; Jackson, 1995).
It has been reported that in the physical map of wheat chromosomes, the distal, telomeric regions of all the chromosomes are gene-rich compared with the proximal, centromeric region (Gill et al., 1996, and references therein). This was based on the analysis of a number of markers in several series of deletion lines, and contrasts with the genetic map based on recombination frequencies, particularly in the proximal region. Given the chromosome arrangement we have shown for the wheat nucleus, we might expect that there would be a significant polarity in the arrangement of transcription sites, the volume of the nucleus nearer the telomeres containing a higher concentration of transcription sites than that nearer the centromeres. We did not observe this; in fact, the transcription foci were distributed fairly homogeneously throughout the volume of the nucleoplasm. One possible explanation could be that the fully condensed metaphase chromosomes decondense unevenly on reinitiation of transcription, the gene-rich regions decondensing more than the gene-poor regions. This would imply a gradient or polarity in chromatin decondensation levels along the chromosomes. We found no sign of such a difference in chromatin density, on the basis of staining intensity with DAPI. We stained with several different concentrations of DAPI to check this point, but did not observe any polarity in DAPI staining intensity. A second possibility is that the limited number of markers used by Gill et al. (1996) in constructing their physical maps is not representative of all the transcribed genes. In particular, the markers used in the physical maps may have been biased towards non-conserved genes and away from conserved housekeeping genes, which may be more likely to be located in recombination-poor chromosome regions such as the proximal, centromere regions. In addition to this, it may be that the genes in these proximal regions, even if they are sparsely distributed, may be highly expressed, as would be expected for housekeeping genes. Although we cannot exclude this possibility, it seems unlikely in view of the comparison of wheat and rice chromosome maps which show considerable synteny and confirm that the distal halves of the wheat chromosome arms are gene-rich compared with the proximal halves (Kurata et al., 1994). A third possibility is that there is not a strict relationship between chromosomal location of a gene and the site at which it is transcribed. In support of this possibility, Toledo et al. (1992) showed that two amplified markers that alternated in multiple repeats on a single chromosome often clustered together in two distinct regions of the interphase nucleus. This implied that the linear DNA carrying the successive copies of the two markers could be arranged in a complex way, with the successive gene copies looping back and forth. There is also recent evidence that long-range interactions can alter the nuclear positioning of genes and lead to gene silencing. Dernburg et al. (1996) have shown that the insertion of heterochromatin at the brown locus in Drosophila caused this gene to associate specifically with the centromeric heterochromatin at a particular developmental stage. Brown et al. (1997) have recently demonstrated that the transcriptional regulator protein ikaros is localized to domains containing centromeric heterochromatin, and that inactive, but not active genes are recruited to these domains. Thus there is accumulating evidence that genes can be moved quite large distances through the nucleus depending on their transcriptional state. Such an organization is consistent with the idea of groups of genes being transcribed at factories (Jackson, 1995), to which DNA loops carrying transcriptionally active genes can move. In this way a gene could be transcribed at a site some distance from its physical map position along the chromosome. In this hypothesis, transcription factories might be assembled throughout the nucleus at locations on a nuclear matrix, rather than directly organized on the interphase chromosomes. If this is the case, it would appear that one of the organizing principles, at least in this species, for such factories is a fairly homogeneous distribution throughout the nucleoplasm.
It has been suggested that nuclei contain a three-dimensional network of intra-chromosomal channels where the machinery for transcription, splicing, and other essential nuclear functions are located. According to this model, the channels would be maintained between adjacent chromosome surfaces, possibly by repulsive electrostatic forces between the chromosome surfaces, and would enable transport of proteins and RNAs either by channeled diffusion or via matrix filaments. Kurz et al. (1996) provided evidence that active and inactive genes were localized preferentially at the periphery of chromosome territories, whereas non-expressed fragments were randomly distributed or localized preferentially in the interior of the chromosome territory. However, an objection to this evidence is that only a few genes were examined, rather than the full range of transcribed genes. Zirbel et al. (1993) also showed that viral RNA concentration was generally localized at the territory surface of the chromosome harboring the viral genes. The latter data certainly supports the idea that RNA processing or export is somehow related to the chromosome territory surface, but does not clearly demonstrate where transcription takes place. Other observations that might support such a model are the differences observed in the surface shape of active and inactive human X interphase chromosomes (Eils et al., 1996).
A prediction of this model would be that transcription sites would be located at a series of surfaces bounding the chromosome territories within the nucleus, and that the interior of the chromosome territories would be devoid of transcription sites. We used both the addition line and the translocation line to test this prediction in double labeling experiments, and obtained very similar results. We present the results from the translocation line here because they are more unequivocal. The two chromosome arms lie next to each other in the nuclei, and are almost invariably visualized as a single region. However, there is a territorial boundary between the two arms. In the addition line, it would be impossible to exclude the possibility that transcription sites inside the chromosome region visualized were located at this inter-arm boundary. For this reason we have shown results from the translocation line, where only a single arm is labeled, and this problem does not arise. It is quite clear that the prediction is not borne out by our results. The rye chromosome territories are well differentiated by genomic in situ hybridization, and there are clear transcription foci throughout the volume of the chromosome territories. There is no sign of chromosome-shaped regions devoid of transcriptional activity. This result also demonstrates that the rye chromosomes and chromosome arms are transcriptionally active. Given the highly organized structure of these wheat nuclei, with all the chromosomes parallel to each other lying across the nucleus in a Rabl configuration, we should expect to see some clear indication of this in the distribution of transcription sites. In fact we could not detect any polarity, density gradient, or systematic departures from homogeneity in the distribution of BrUTP foci. Our conclusion is, therefore, that the distribution of transcription sites in wheat nuclei is not restricted to chromosome territorial boundaries, at least as revealed by total chromosome in situ labeling. It might be argued that the real chromosome territory surfaces might be much more convoluted than shown by the in situ labeling, and that the transcription sites apparently in the chromosome interior in fact lie on such a convoluted surface (e.g., Wansink et al., 1996). However, in our view, this reduces the strength of the territorial surface hypothesis to the extent that it makes no testable predictions.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council of the UK; by a fellowship from the Gulbenkian Foundation and Program PRAXIS XXI Portugal (R. Abranches), and by the John Innes Foundation (L. Aragón-Alcaide).
Abbreviations used in this paper
- BrUTP
bromouridine triphosphate
- DAPI
4′,6-diamidino-2-phenylindole
- MPB
modified physiological buffer
- 3D
three-dimensional
Footnotes
Address all correspondence to Peter J. Shaw, John Innes Centre, Colney, Norwich NR4 7UH, UK. Tel.: (44) 1603-452571. Fax: (44) 1603-501771. E-mail: peter.shaw@bbsrc.ac.uk
References
- Aragón-Alcaide L, Reader S, Beven A, Shaw P, Miller T, Moore G. Association of homologous chromosomes during floral development. Curr Biol. 1997;7:905–908. doi: 10.1016/s0960-9822(06)00383-6. [DOI] [PubMed] [Google Scholar]
- Avivi L, Feldman M. Arrangement of chromosomes in the interphase nucleus of plants. Hum Genet. 1980;55:281–295. doi: 10.1007/BF00290206. [DOI] [PubMed] [Google Scholar]
- Beven AF, Lee R, Razaz M, Leader DJ, Brown JWS, Shaw PJ. The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J Cell Sci. 1996;109:1241–1251. doi: 10.1242/jcs.109.6.1241. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes ar centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Chung H-MM, Shea C, Fields S, Taub RN, van de Ploeg HT. Architectural organization in the interphase nucleus of the protozoan trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO (Eur Mol Biol Organ) J. 1990;9:2611–2619. doi: 10.1002/j.1460-2075.1990.tb07443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M. Analysis of chromosome positions in the interphase nucleus of Chinese hamsters cells by laser UV microirradiation experiments. Hum Genet. 1982;62:201–209. doi: 10.1007/BF00333519. [DOI] [PubMed] [Google Scholar]
- Cremer T, Lichter P, Borden J, Ward DC, Manuelidis L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Hum Gen. 1988;80:235–246. doi: 10.1007/BF01790091. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schrock E, Speicher MR, Mathieu U, Jauch A, Emmerich P, et al. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp Quant Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- Cremer, T., S. Dietzel, R. Eils, P. Lichter, and C. Cremer. 1995. Chromosome territories, nuclear matrix filaments and interchromatin channels: a topological view on nuclear architecture and function. In Kew Chromosome Conference IV. P.E. Brandham and M.D. Bennett, editors. Royal Botanical Gardens, Kew. 63–81.
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Eils R, Dietzel S, Bertin E, Schrock E, Speicher MR, Ried T, Robert-Nicoud M, Cremer C, Cremer T. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill KS, Gill BS, Endo TR, Taylor T. Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics. 1996;144:1883–1891. doi: 10.1093/genetics/144.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande M, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J, Bennett M. Nuclear architecture in plants. Trends Genet. 1990;6:401–405. doi: 10.1016/0168-9525(90)90300-u. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison, J.S., A.R. Leitch, and T. Schwarzacher. 1993. The physical organization of interphase nuclei. In The Chromosome. J.S. Heslop-Harrison and R.B. Flavell, editors. Bios Scientific Publishers Ltd., Oxford. 221–232.
- Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber HA. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. . J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozak P, Cook PR, Schofer C, Mosgoller W, Wachtler F. Site of transcription of ribosomal-RNA and intranucleolar structure in HeLa cells. J Cell Sci. 1994;107:639–648. doi: 10.1242/jcs.107.2.639. [DOI] [PubMed] [Google Scholar]
- Iborra F, Pombo A, Jackson D, Cook P. Active RNA polymerases are localized within discrete transcription factories in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Jackson DA. Nuclear organization: uniting replication foci, chromatin domains and chromosome structure. Bioessays. 1995;17:587–591. doi: 10.1002/bies.950170704. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. The structural basis of nuclear function. Intl Rev Cytol. 1995;162:125–149. doi: 10.1016/s0074-7696(08)61230-9. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO (Eur Mol Biol Organ) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata N, Moore G, Nagamura Y, Foote T, Yano M, Minobe Y, Gale M. Conservation of genome structure between rice and wheat. Biotechnology. 1994;12:276–278. [Google Scholar]
- Kurz A, Lampel S, Nickolenko JE, Bradl J, Benner A, Zirbel RM, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual human-chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Gen. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Noguchi J, Fukui K. Chromatin arrangement in intact interphase nuclei examined by laser confocal microscopy. J Plant Res. 1995;108:209–216. [Google Scholar]
- Rabl C. Uber Zelltheilung. Morphol Jahrbuch. 1885;10:214–330. [Google Scholar]
- Schwarzacher T, Anamthawat-Jonsson K, Harrison GE, Islam AKMR, Jia JZ, King IP, Leitch AR, Miller TE, Reader SM, Rogers WJ, et al. Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theor Appl Genet. 1992;84:778–786. doi: 10.1007/BF00227384. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Highett MI, Beven AF, Jordan EG. The nucleolar architecture of polymerase I transcription and processing. EMBO (Eur Mol Biol Organ) J. 1995;14:2896–2906. doi: 10.1002/j.1460-2075.1995.tb07289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straatman KR, Trompeter CM, Schul W, Schel JHN. Fluorescent labelling of nascent RNA reveals nuclear transcription domains throughout plant cell nuclei. Protoplasma. 1996;192:145–149. [Google Scholar]
- Strouboulis J, Wolffe AP. Functional compartmentalization of the nucleus. J Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Thompson WF, Beven AF, Wells B, Shaw PJ. Sites of rDNA transcription are widely dispersed through the nucleolus in Pisum sativum and can comprise single genes. Plant J. 1997;12:571–582. doi: 10.1046/j.1365-313x.1997.00571.x. [DOI] [PubMed] [Google Scholar]
- Toledo F, Le Roscouet D, Buttin G, Debatisse M. Co-amplified markers alternate in megabase long chromosomal inverted repeats and cluster independently in interphase nuclei at early steps of mammalian gene amplification. EMBO (Eur Mol Biol Organ) J. 1992;11:2665–2673. doi: 10.1002/j.1460-2075.1992.tb05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Manders EEM, van der Kraan I, Aten JA, van Driel R, de Jong L. RNA polymerase II transcription is concentrated outside replication domains throughout S-phase. J Cell Sci. 1994;107:1449–1456. doi: 10.1242/jcs.107.6.1449. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of a nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Sibon OCM, Cremers FFM, van Driel R, de Jong L. Ultrastructural localization of active genes in nuclei of A431 cells. J Cell Biochem. 1996;62:10–18. doi: 10.1002/(sici)1097-4644(199607)62:1<10::aid-jcb2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]