Abstract
In Saccharomyces cerevisiae, a single cyclin-dependent kinase, Cdc28, regulates both G1/S and G2/M phase transitions by associating with stage-specific cyclins. During progression through S phase and G2/M, Cdc28 is activated by the B-type cyclins Clb1–6. Because of functional redundancy, specific roles for individual Clbs have been difficult to assign. To help genetically define such roles, strains carrying a cdc28ts allele, combined with single CLB deletions were studied. We assumed that by limiting the activity of the kinase, these strains would be rendered more sensitive to loss of individual Clbs.
By this approach, a novel phenotype associated with CLB5 mutation was observed. Homozygous cdc28-4ts clb5 diploids were inviable at room temperature. Cells were defective in spindle positioning, leading to migration of undivided nuclei into the bud. Occasionally, misplaced spindles were observed in cdc28-4 clb5 haploids; additional deletion of CLB6 caused full penetrance. Thus, CLB5 effects proper preanaphase spindle positioning, yet the requirement differs in haploids and diploids. The execution point for the defect corresponded to the time of Clb5-dependent kinase activation. Nevertheless, lethality of cdc28-4 clb5 diploids was not rescued by CLB2 or CLB4 overexpression, indicating a specificity of Clb5 function beyond temporality of expression.
Keywords: cell cycle, nuclear migration, spindle dynamics
Cell cycle progression in all eukaryotes is controlled primarily at the G1/S and G2/M phase transitions by the activity of cyclin-dependent kinases (cdks).1 In the yeast Saccharomyces cerevisiae, a single cdk, Cdc28, regulates both transitions by associating with different stage-specific cyclins (Nasmyth, 1993). The G1/S transition, START, constitutes the point of commitment to a new round of cell division. Passage through START requires Cdc28 activation by the G1 cyclins Cln1, 2, and 3 (Richardson et al., 1989). During progression through S phase and G2/M, Cdc28 is activated by the B-type cyclins Clb1–6. Based on genetic evidence and the timing of expression, particular Clbs are likely to play specific roles either in S phase or G2/M. However, establishing these roles genetically has been difficult since B-type cyclins display partial functional redundancy. Thus, full penetrance of phenotypes associated with loss of Clb activity can only be observed in strains with multiple CLB deletions (Surana et al., 1991; Fitch et al., 1992; Richardson et al., 1992; Schwob and Nasmyth, 1993).
Successful cell duplication depends on a precise sequence of events in the cell cycle (Byers, 1981; Lew et al., 1997). Progression through START triggers initiation of DNA replication, bud emergence, and spindle pole body (SPB) duplication. After DNA replication is completed, one SPB migrates to the other side of the nucleus, resulting in generation of a short intranuclear spindle. Then, the nucleus migrates to the bud neck with the spindle oriented along the mother-bud axis in a process that requires cytoplasmic microtubules and motor proteins (Huffaker et al., 1988; Palmer et al., 1992; Sullivan and Huffaker, 1992; Eshel et al., 1993; Li et al., 1993; Yeh et al., 1995; Cottingham and Hoyt, 1997; DeZwaan et al., 1997). This process is followed by spindle elongation and nuclear division. The spindle finally disassembles coincident with cytokinesis.
Genetic analysis has implicated Clb function in spindle assembly. Strains containing multiple CLB deletions (e.g., clb1-4Δ, clb3-5Δ) fail to form a bipolar spindle, and arrest with a 2C DNA content (Surana et al., 1991; Fitch et al., 1992; Schwob and Nasmyth, 1993). The relative requirement of individual Clbs in the various aspects of the spindle pathway, however, has not been precisely determined because of the partial redundancy of Clb function.
As an alternative way to reveal specific roles for individual B-type cyclins, strains carrying cdc28ts alleles in combination with different clb− disruption alleles were studied. Since cdk activity is already compromised at the permissive temperature, the resulting strains were expected to be more sensitive to the absence of the individual Clbs. We chose to concentrate on CLB5 and CLB6, with the long-term goal of designing a genetic screen to identify targets of the S-phase form of the kinase. Clb5 and Clb6 are B-type cyclins required for efficient DNA replication. Expression of these cyclins peaks at the G1/S boundary and activation of Clb5-dependent kinase has been shown to occur as cells initiate DNA synthesis (Epstein and Cross, 1992; Schwob and Nasmyth, 1993; Schwob et al., 1994).
Here we describe a novel phenotype associated with loss of Clb5 function under limiting cdk activity. The phenotype implicates Clb5, and to lesser extent Clb6, in early events in the spindle pathway, ultimately ensuring proper coordination between preanaphase spindle positioning and nuclear division.
Materials and Methods
Yeast Strains, Genetic Procedures, Media, and Growth Conditions
All strains used in this study were isogenic derivatives of BF-264-15Daub: a, ade1, his2, leu2-3,112, trp1-1a, ura3Dns (Richardson et al., 1989) containing, in addition, a nonreverting allele of ARG4 (Epstein and Cross, 1992). Strains carrying a cdc28-4 allele (Lörincz and Reed, 1986) in combination with CLB deletions were obtained as progeny of a cross between MY1 (cdc28-4, ade1, his2, leu2-3,112, trp1-1a, ura3Dns, arg4) and one of the following: RBY2 (a, ade1, his2, leu2-3,112, trp1-1a, ura3Dns, arg4 clb5:: ARG4 clb6::ADE1; Basco et al., 1995), YH135 (a clb2::LEU2), or YS118 (a clb3::TRP1, clb4::HIS2; Richardson et al., 1992). The relevant genotypes of the strains used in this study are listed in Table I.
Table I.
Strains Used in This Study
| Strain | Relevant genotype | |
|---|---|---|
| MY101 | MATa cdc28-4 | |
| MY102 | MATα cdc28-4 | |
| MY104 | MATa cdc28-4 LEU2 | |
| MY105 | MATα cdc28-4 TRP1 | |
| MY126 | MATa cdc28-4 clb5::ARG4 | |
| MY124 | MATα cdc28-4 clb5::ARG4 | |
| MY127 | MATa cdc28-4 clb5::ARG4 LEU2 | |
| MY128 | MATα cdc28-4 clb5::ARG4 TRP1 | |
| MY1010 | MATa/α cdc28-4/cdc28-4 GAL1:CLB5(TRP1)/trp1 | |
| MY1416 | MATa/α cdc28-4/cdc28-4 clb5::ARG4/clb5::ARG4 GAL1:CLB5(TRP1)/trp1 | |
| MY1417 | MATa/α cdc28-4/cdc28-4 clb5::ARG4/clb5::ARG4 GAL1:CLB2(LEU2)/leu2 [CLB5-URA3] | |
| MY1418 | MATa/α cdc28-4/cdc28-4 clb5::ARG4/clb5::ARG4 GAL1:CLB4(LEU2)/leu2 [CLB5-URA3] | |
| MYT101 | MATa cdc28-4 HIS3:GFP:TUB1 (URA3) | |
| MYT102 | MATα cdc28-4 HIS3:GFP:TUB1 (URA3) | |
| MYT126 | MATa cdc28-4 clb5::ARG4 HIS3:GFP:TUB1 (URA3) | |
| MYT124 | MATα cdc28-4 clb5::ARG4 HIS3:GFP:TUB1 (URA3) | |
| MYT1010 | MATa/α cdc28-4/cdc28-4 GAL:CLB5(TRP1)/trp1 HIS3:GFP:TUB1 (URA3)/ura3 | |
| MYT1416 | MATa/α cdc28-4/cdc28-4 clb5::ARG4/clb5::ARG4 GAL1:CLB5(TRP1)/trp1 HIS3:GFP:TUB1 (URA3)/ura3 | |
| MY129 | MATa cdc28-4 clb5::ARG4 GAL:CLB5(TRP1) [MATa-LEU2] | |
| MY130 | MATa cdc28-4 clb5::ARG4 GAL:CLB5(TRP1) [MATα-LEU2] | |
| MY131 | MATα cdc28-4 clb5::ARG4 GAL:CLB5(TRP1) [MATa-LEU2] | |
| MY132 | MATα cdc28-4 clb5::ARG4 GAL:CLB5(TRP1) [MATα-LEU2] | |
| MY1263 | MATa cdc28-4 clb5::ARG4 GAL:CLB5(LEU2) bud3::URA3 |
Yeast cultures were grown at 23°C unless indicated. Yeast media and genetic procedures were according to Sherman et al. (1986). Gene disruptions were performed by the one-step method (Rothstein, 1983). Strains expressing the GFP:TUB1 fusion under the control of the HIS3 promoter were constructed by transformation with pAFS91 linearized at the unique StuI site (Straight et al., 1997).
Quantitative mating assays were performed as follows. In brief, 2 ml of 5 × 106 cell/ml a Leu+ and αTrp+ cultures were mixed and filtered onto 47-mm 0.45-μm GN-6 Metricel membranes (Gelman Sciences, Ann Arbor, MI). Filters were then placed on YEPD plates and incubated for 5 h. Cells were eluted from the filter into 1 M sorbitol, were briefly sonicated, and serial dilutions were plated by duplicates on single dropout plates (−LEU or −TRP) to score parental haploids, or double dropouts (−LEU −TRP) to score for diploid colonies. Dilutions yielding between 30 and 300 colonies were counted after 4 d at room temperature. Results were normalized to the frequency of zygote formation monitored microscopically.
Suppression of diploid cdc28-4 clb5 lethality by B-type cyclins expressed under the control of the GAL1 promoter was tested as follows. Homozygous cdc28-4 clb5 diploids were obtained in the presence of YEp24-CLB5 to ensure viability. After transformation with plasmids expressing each cyclin to be tested under the GAL1 promoter, transformants were grown overnight in rich galactose-containing medium to allow spontaneous loss of the CLB5 plasmid. Cells were then plated to obtain individual colonies on galactose plates, followed by replica-plating to selective plates to score the plasmid marker (URA3), YEPDextrose, and YEPGalactose to verify that viability was galactose-dependent for Ura− colonies.
Plasmids
YIpG2CLB2, YIpG2CLB4, and YIpG2CLB6 are integrative plasmids carrying CLB2, CLB4, or CLB6 open reading frames under the control of the GAL1 promoter, respectively (Lew and Reed, 1995). Plasmids were linearized at the unique BstEII site before transformation. pMST57 and pMSL42 contained the GAL1 promoter fused to the CLB5 open reading frame as a 2-kb EcoRI-SalI fragment into YIplac128 or YIplac204 (Gietz and Sugino, 1988), respectively. Plasmids were linearized using the unique EcoRV before transformation.
YEp24-CDC28, YEp24-CLB5 (not including CLB2), YEp24-CLB3, and YEp24-CLB2 (not including CLB5) are library clones isolated by screening a YEp24 library (Carlson and Botstein, 1982) for plasmids that could suppress the enhanced temperature sensitivity of a cdc28-4 clb5 strain (M. Segal and S.I. Reed, unpublished data). YEpCLB6/CLB1 contains a 12-kb SalI fragment spanning the CLB1-CLB6 loci in YEp351. pJH3-45 is a multicopy plasmid carrying the CLN2 gene (Hadwiger et al., 1989). Plasmid pAFS91 encoding a GFP:TUB1 fusion was obtained from A.F. Straight (University of California at San Francisco). pMATa, pMATα plasmids carry a 4.2-kb HindIII fragment of the MATa or MATα genes in pRS315. These plasmids were obtained from T. Galitski and A. Sherman (The Whitehead Institute for Biomedical Research, Cambridge, MA).
Plasmids for disruption of BUD3 and BUD5 were constructed as follows. A 2.7-kb fragment spanning the BUD3 gene was amplified by PCR and cloned into the pCRII vector (Invitrogen Corp., Carlsbad, CA). An internal 1.63-kb BglII–ClaI fragment was then replaced by the 1.1-kb BamHI–ClaI fragment containing the URA3 gene. A 2.86-kb fragment including BUD5 was amplified by PCR, and was cloned into pCRII. An internal 1.6-kb HpaI–HindIII fragment was then replaced by the HindIII– SmaI fragment of URA3. Plasmid pSK-URA3 containing the 1.1-kb SmaI–HindIII fragment of URA3 from YEp24 into the HindIII–SmaI sites of pBluescriptSK was the source of the URA3 fragments used for disruption. Disruptions were confirmed by PCR using primers flanking the site of integration.
Cell Biology Protocols
Differential interference contrast (DIC) and fluorescence microscopy of living or fixed cells was performed using an Eclipse E800 microscope (Nikon, Inc., Melville, NY) with a 60× objective. Cell images were captured with a Quantix CCD (Photometrics, Tucson, AZ) camera using Iplab Spectrum software (Signal Analytics Co., Vienna, VA). Cell measurements were performed on captured images using the IPlab cursor measuring tool calibrated with a stage micrometer. The distance between the spindle and the bud neck was scored by overlaying the DIC image onto the corresponding spindle image. For microscopy of living cells, parental (cdc28-4/cdc28-4 GAL1:CLB5) or mutant (cdc28-4/cdc28-4 clb5/clb5 GAL1:CLB5) diploids were grown to ∼5 × 106 cells/ml in synthetic galactose medium. Cells were collected by filtration, and were shifted to synthetic dextrose medium for 6 h at 23°C. The cells were then mounted in the same medium for microscopy at room temperature. Images were acquired using 500-ms exposures. Single cells were followed up to 60 min. The focus was manually adjusted throughout the imaging to keep the mitotic spindle fully in focus.
Cell cultures were analyzed for DNA content using flow cytometry as described previously (Lew et al., 1992) except that cells were stained using the green nucleic acid stain Sytox (Molecular Probes, Inc., Eugene, OR). Nuclei were visualized with DAPI as previously described (Mondésert and Reed, 1996). Chitin staining was performed as described by Pringle (1991).
Cytology of mating cells was carried out by eluting aliquots from a filter mating assay at 30-min intervals. Living cells were directly mounted for microscopy or fixed for nuclear staining by DAPI. Spindles, and in particular astral microtubules, were observed more effectively in live cells. For double staining, GFP-tubulin–expressing cells were fixed for 30 min at room temperature in 3.7% formaldehyde followed by DAPI staining.
Execution Point
Parental (a /α cdc28-4/cdc28-4 GAL:CLB5) or mutant (a/α cdc28-4/cdc28-4 clb5/clb5 GAL:CLB5) cells grown to early log phase (6 × 106 cells/ml) in YEPGalactose were synchronized by adding 15 μg/ml nocodazole to the culture and incubating for 4 h. Arrest was monitored microscopically. Cells were collected by centrifugation, washed twice in YEPR affinose, and released in the same medium at a density of 5 × 106 cells/ml. At 20-min intervals, aliquots were drawn from the YEPRaffinose culture for plating on YEPGalactose, FACS analysis, and cell counting. At least 1,000 microcolonies emerging on YEPGalactose plates were counted after 15 h at room temperature to score cell viability after reinduction of CLB5 expression. The percentage of cells that had entered S phase in the YEPR affinose culture was estimated by FACS analysis; the budding index and nuclear positioning were evaluated by scoring a minimum of 200 cells.
Results
Genetic Interaction Between cdc28ts Alleles and clb5 or clb6 Mutations
Most cdc28ts strains undergo a G1 arrest before START when mutant asynchronous cultures are shifted to the restrictive temperature. To detect phenotypes specifically resulting from the absence of the S-phase cyclins Clb5 and 6, strains carrying a cdc28-4 temperature-sensitive allele in combination with deletions in CLB5 or CLB6 were constructed. It was anticipated that use of a temperature-sensitive allele that encodes a cdk already partially defective in kinase activity, even at the permissive temperature, would result in a higher sensitivity to the absence of Clb5 and Clb6 and render deletion of either or both genes more fully penetrant.
We observed that a clb5 mutation strongly interacted with cdc28-4 as well as a number of other cdc28ts alleles. This interaction was reflected by a 5°C increase in temperature sensitivity of the double mutant (data not shown). A clb6 mutation did not affect the temperature sensitivity of cdc28-4, however, it was lethal in combination with the cdc28-4 clb5 mutations. The additive effect of clb6 when in combination with the clb5 mutation was observed for all seven cdc28ts alleles tested (not shown).
Haploid cdc28-4 clb5 mutants were viable at the permissive temperature. However, when these mutants were mated with identical mutants of the opposite mating type, the resulting zygotes were inviable. A 1,000-fold decrease in formation of viable diploid colonies was observed between a cdc28-4 clb5 × cdc28-4 clb5 and a cdc28-4 × cdc28-4 clb5 cross, as determined by a quantitative mating assay at the permissive temperature (Fig. 1 A). Zygote formation, however, was not affected (see below). This phenotype was specific for a cdc28-4 clb5 mutant. Other mutant combinations such as cdc28-4 clb6, cdc28-4 clb3 clb4, or cdc28-4 clb2 formed viable homozygous diploid colonies as efficiently as did cdc28-4 parental cells (not shown).
Figure 1.
Viability of cdc28-4 clb5 mutant diploids. (A) Mating assay was carried out as described in Materials and Methods. In brief, opposite mating types were mixed and, after zygote formation, cells were plated to select for viable diploid colonies or total viable cells: (a) a cdc28-4 clb5 × α cdc28-4 (MY127 × MY105), dilution 1:1,000; (b) a cdc28-4 clb5 × α cdc28-4 clb5 (MY127 × MY128), dilution 1:100; and (c) a cdc28-4 × α cdc28-4 clb5 (MY104 × MY128), dilution 1:1,000. Zygote formation was comparable for the three crosses. Diploid plating efficiency was normalized to total plating efficiency in each cross (not shown), and was found to be 1,000-fold decreased for b. (B) Diploid cdc28-4 clb5 GAL1:CLB5 patching efficiency on dextrose and suppression by multicopy plasmids. Individual transformants of strain MY1416 were patched on synthetic galactose medium and replica-plated onto YEPDextrose or YEPGalactose medium. Plates were photographed after 2 d at 23°C.
To determine whether Clb5 is required for diploid viability after mating, a homozygous cdc28-4 clb5 diploid carrying CLB5 expressed under the control of the inducible GAL1 promoter was constructed. This strain grew on galactose-containing medium, but failed to form colonies on dextrose medium (when CLB5 expression is repressed). This result indicated that the CLB5 requirement for viability extended beyond the mating process, and reflected a defect in the diploid. Lethality was prevented by plasmids expressing CLB5 or CDC28, and was weakly suppressed by a multicopy plasmid containing CLB3. CLB2 or CLB6 multicopy plasmids failed to rescue the strain in dextrose medium (Fig. 1 B). A multicopy plasmid encoding the G1 cyclin CLN2 was unable to rescue the mutant diploid (not shown).
Nuclear Positioning in cdc28-4 clb5 Homozygous Diploids
To investigate the basis for the observed lethality, cdc28-4 clb5 homozygous zygotes were stained with the DNA-specific dye DAPI to observe nuclear behavior during mating. It was readily apparent that zygote formation per se was not defective in cdc28-4 clb5 mutants. Zygotes were produced with comparable efficiency to that observed with parental cdc28-4 strains (not shown). However, the dynamics of nuclear migration and proper nuclear positioning in the first mitotic division after karyogamy were seriously perturbed. In contrast to cdc28ts cells in which the nucleus initially remained at the bud neck, and then nuclear division proceeded to give rise to mother and daughter cells, each containing a diploid nucleus (Fig. 2 A; a–c), the double mutants contained a single nucleus that migrated completely into the bud in ∼80% of budded zygotes (Fig. 2 A; d–f). In cdc28-4 clb5 cells the nucleus rarely divided (once in the bud). The same defect was observed in cdc28-4 clb5 GAL1:CLB5 diploids after a 6-h shift of an asynchronous culture from galactose to dextrose medium: 35% of the cells contained a single nucleus fully positioned in the bud (Fig. 2 B; g–l). Parental cdc28-4 GAL1:CLB5 diploids positioned the nucleus at the neck (Fig. 2 B; a–b), resulting in correct nuclear division between the mother and daughter cells (Fig. 2 B; c–f).
Figure 2.

Nuclear position defect in cdc28-4 clb5Δ diploids. (A) Zygotes from a cross between a and α parental cdc28-4 (MY101 × MY102; a–c) or mutant cdc28-4 clb5 (MY126 × MY124, d–f) strains. Overlays of DIC and DAPI images are shown. Mating was followed in a filter assay by eluting and fixing aliquots for staining with DAPI. Large budded zygotes accumulated after approximately 3 h at 23°C. Representative stages for parental: G2 (a), early anaphase (b), late anaphase (c), or mutant nuclear migration into the bud (d–f) are shown. Bar, 5 μm. (B) Diploid cdc28-4 GAL1:CLB5 (MY1010) or cdc28-4 clb5Δ GAL1:CLB5 cells (MY1416) were fixed and stained with DAPI after a shift from galactose to dextrose containing medium at 23°C. Overlays of DIC and DAPI images are shown. Representative stages for parental early-budded (a), G2 (b), early and late anaphase (c–f); or mutant nuclear migration into the bud (g–l) are shown. Bar, 5 μm.
Spindle Orientation in cdc28-4 clb5 Homozygous Diploids Before Nuclear Division
In the unperturbed cell cycle, nuclear migration and nuclear division are coordinated to give rise to a mother and a daughter, each containing a single nucleus. The development of the mitotic spindle is central to these processes. As cells progress through START, the spindle pole bodies (SPB; the microtubule organizing center in yeast) are duplicated, and by the time DNA replication is complete, a short intranuclear spindle is generated by SPB separation. The spindle is then positioned at the bud neck, and orients along the mother-bud axis in a process that requires cytoplasmic microtubules and motor proteins (Byers, 1981; Palmer et al., 1992; Sullivan and Huffaker, 1992; Cottingham and Hoyt, 1997; DeZwaan et al., 1997). This is followed by spindle elongation concomitant with nuclear division. The spindle finally disassembles coincident with cytokinesis.
To explore the basis for the nuclear migration defect observed in cdc28-4 clb5 diploids, spindle position was determined in dividing zygotes or vegetatively growing diploids by expressing a fusion between the green fluorescent protein (GFP) and the major α tubulin (the product of the gene TUB1) to visualize microtubular structures by fluorescence microscopy in living cells (Straight et al., 1997).
In the mutant zygotes, the assembled spindle along with the nucleus migrated into the bud. Spindles were frequently misaligned with respect to the mother-bud axis (Fig. 3 A). The frequency of spindle migration into the bud correlated with the observed frequency of misplaced nuclei.
Figure 3.
Spindle positioning in cdc28-4 clb5Δ mutants. (A) Zygotes from parental or mutant crosses. Mating assays with cells expressing GFP:TUB1 were performed as described for Fig. 2 except that cells were visualized without fixation. Pairs of DIC and GFP fluorescence images are shown of representative stages for cdc28-4 cells: G2 (a), early anaphase (b), late anaphase (c), and cdc28-4 clb5 short and intermediate length spindles positioned in the bud (d–i). Bar, 5 μm. (B) Diploid cdc28-4 GAL1:CLB5 (MYT1010; a–c) or cdc28-4 clb5 GAL1:CLB5 (MYT1416; d–f) expressing GFP:TUB1 after a 6-h shift from galactose to dextrose containing medium at 23°C. Representative stages are shown for parental SPB duplication and G2 spindle (a), G2 and midanaphase spindles (b), late anaphase and spindle disassembly (c), and mutant (d–f) cells. Bar, 5 μm.
Homozygous diploid cdc28 clb5 GAL1:CLB5 cells expressing the GFP:TUB1 construct displayed normal spindle structures in galactose containing medium but, after a 6-h shift to dextrose, accumulated with spindles fully positioned in the bud corroborating the defect observed in mutant zygotes (Fig. 3 B).
Correlation between defects in nuclear and spindle positioning was confirmed by observation of cells expressing the GFP-tubulin construct and stained with DAPI (Table II).
Table II.
Cell Type Distribution of Parental and Mutant Cultures
Parental and mutant strains were grown in YEPGalactose followed by a 6-h shift to YEPDextrose at 23°C. Cells were fixed and prepared for microscopy as described in Materials and Methods. The result is the average of three independent counts of 200 cells. Nuclear distribution was essentially identical in the absence of the GFP-tubulin construct.
Astral Microtubular Function in cdc28-4 clb5 Diploid Cells
In addition to the defect in spindle positioning, fluorescence microscopy revealed anomalies in astral microtubules both in dividing zygotes and in vegetatively growing diploid cells. Compared with parental strains in which preanaphase spindles showed a proper position at the neck along with short, straight astral microtubules extending to sites at the cortex of the mother and daughter, the cdc28-4 clb5 mutants exhibited excessively long astral microtubules that often curved along the cortex surface. In addition, it was striking that deformation of microtubules was associated mostly with those oriented towards the mother cell cortex, while those oriented towards the cortex of the bud remained fairly straight and normal in appearance. Thus, an apparently aberrant microtubule–cortex interaction in the mother cell correlated with the nuclear positioning defect. Such behavior is reminiscent of mutations conferring defects in attachment to the cell cortex and/or processing of microtubules (e.g., kip3Δ, dhc1Δ, act5, jnm1). However, in those mutants, nuclei frequently fail to migrate properly, and orient at the bud neck before anaphase, correlating with long curving astral microtubules directed specifically towards the bud rather than the mother cell cortex (Carminati and Stearns, 1997; Cottingham and Hoyt, 1997).
The spindle defect was studied in more detail by time lapse photography of live cells (Fig. 4). Based on observations of at least 30 time lapse series, spindle behavior in the mutant diploids fell into two groups: those in which anaphase did not proceed (94%), and those in which anaphase did occur (Fig. 4 A). cdc28-4 clb5 mutant cells were occasionally capable of correcting spindle position and orientation. However, in most instances, spindles reached an intermediate size without achieving an apparently functional attachment in the mother cell, and remained mispositioned without further movement or elongation.
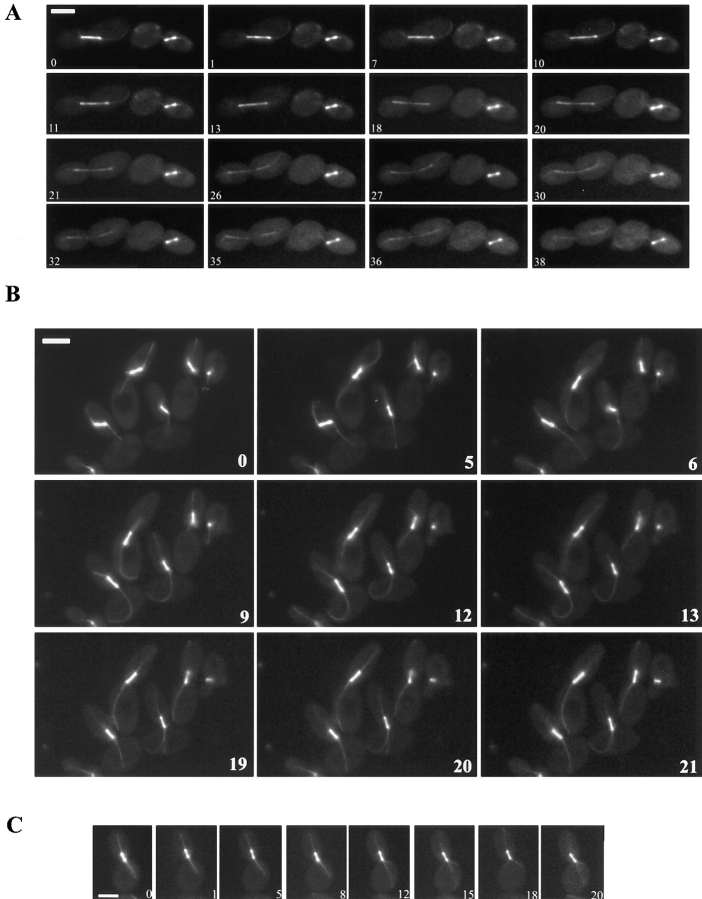
Figure 4.
Time-lapse series of spindle behavior in cdc28-4 or cdc28-4 clb5 diploids. Cells were grown in synthetic galactose medium followed by transfer to synthetic dextrose medium for 6 h at 23°C before mounting on the same medium for microscopy as described in Materials and Methods. Numbers correspond to the time elapsed in minutes. Representative series of the mutant (A– B) and parental (C) strains are shown. (A) The cell to the right contains a spindle positioned into the bud that underwent limited elongation. The cell to the left proceeded through anaphase. Selected frames from a 40-min time lapse experiment are shown. Bar, 5 μm. (B) Astral microtubular defects associated with spindle positioning and anaphase progression in cdc28-4 clb5 GAL1: CLB5 diploids. Cells improved alignment along the mother– daughter axis, but failed to restore position at the bud neck. Astral microtubules directed towards the mother cortex lengthened and curved. By an additional 20-min period after the last time point shown, cells had not proceeded through anaphase. Selected frames from a 40-min time lapse experiment are shown. Notice that the cell on the upper right side assembled a spindle throughout the time lapse. Bar, 5 μm. (C) Spindle positioning in a cdc28-4 diploid (MYT1010). Selected frames from a 20-min time lapse experiment to show parental behavior of astral microtubules and spindle positioning. Bar, 5 μm.
When short spindles (1–2 μm) are positioned at the bud neck in wild-type preanaphase cells, short movements within the neck are normally seen until alignment is completed (Kahana et al., 1995). Such movements were observed in parental cdc28-4 diploids, and were restricted to the vicinity of the neck (Fig. 4 C); on occasion, short spindles moved into the bud, but always remained close to the neck. Longer spindles (>3 μm) of cdc28-4 cells always extended across the neck with one SPB in the mother and the other in the bud, typical behavior of spindles in wild-type cells. In contrast, cdc28-4 clb5 diploids exhibited exaggerated spindle movement accompanied by lengthening and curving of astral microtubules extending into the mother cell (Fig. 4 B).
Defects in spindle dynamics were also apparent when spindle length and spindle position were correlated in cell populations. In log-phase cultures transferred to dextrose-containing medium for 6 h, parental diploids (cdc28-4 GAL1:CLB5) mostly had either short metaphase or long late anaphase spindles (Fig. 5). Mean sizes in each class indicated that spindles distributed mainly around 2 μm for short spindles, 4–5 μm for midanaphase, or 10 μm for late anaphase. Instead, diploid cdc28-4 clb5 GAL1:CLB5 cells accumulated with spindles of an intermediate length (3 μm); cells with late anaphase spindles were rare (Fig. 5 A). Most spindles in the mutant were positioned well into the bud with a high percentage >2 μm away from the neck, and were misaligned with respect to the main mother– daughter axis (Fig. 5, B and C).
Figure 5.

Spindle distribution by size and positioning in budded cells. Asynchronous parental MYT1010 (cdc28-4 GAL1: CLB5) or mutant MYT1416 (cdc28-4 clb5 GAL1:CLB5) diploids expressing the GFP:TUB1 fusion were shifted from galactose to dextrose-containing medium for 6 h before microscopy. (A) Distribution by size. At least 100 digitized images were captured and used for spindle measurements as described in Materials and Methods. Size intervals were as follows: <2 μm (black bars), 2–3 μm (white striped bars), 3–5 μm (black striped bars) and >5 μm (gray bars). Mean length and standard deviation for each size interval is indicated. (B) Distribution by position. At least 200 cells containing spindles shorter than 3 μm were scored by fluorescence microscopy. The proportion of cells containing the spindle in the mother cell (white bars), across the bud-neck (light gray bars), in the bud within 1 μm from the neck (dark gray bars), and in the bud farther than 1 μm away from the neck (black bars) are shown. The percentage of aligned spindles (as defined in C) with respect to the mother-bud axis is indicated for each position category. (C) Operative definition of spindle alignment. Aligned spindles were those within an angle of 35° (broken lines) with respect to the main mother–daughter axis (solid line).
Relative Penetrance of the Nuclear Migration Defect in Haploids and Diploids
As described above, cdc28-4 clb5 haploids were viable and displayed an increased temperature sensitivity compared with parental cdc28-4 cells. Since Clb5 and Clb6 are usually regarded as a pair of redundant S-phase cyclins, the relative contribution of Clb6 for haploid viability was tested by constructing a haploid cdc28-4 clb5 clb6 GAL1: CLB5 strain. Deletion of CLB6 resulted in lethality on dextrose medium and accumulation of large budded cells with 58% of spindles positioned in the bud (not shown), indicating that CLB6 becomes essential for this role in haploid cells in the absence of CLB5.
Nevertheless, with a low but significant frequency, aberrant spindle position and orientation could be also observed in haploid cdc28-4ts clb5 cells, suggesting weak penetrance of the phenotype seen in cdc28-4 clb5 diploids. To elucidate the basis for this difference, mating type was manipulated in haploid cells. Haploid cdc28-4 clb5 cells carrying a GAL1:CLB5 construct were transformed with plasmids encoding either MATa or MATα, and were tested for the ability to grow on dextrose-containing medium that repressed CLB5 expression. a cells carrying a MATa containing plasmid or α cells carrying a MATα plasmid, could grow in the presence of dextrose. In contrast, a/α haploids failed to form patches after replica-plating onto dextrose-containing medium (Fig. 6 A). The viability of parental cdc28-4 cells was not affected by either MAT plasmid (not shown). DAPI staining confirmed that nuclear positioning was defective in a/α haploids (Fig. 6 B).
Figure 6.

Penetrance of the positioning defect in a /α cdc28-4 clb5 haploid cells. (A) Patching efficiency of a/α cdc28-4 clb5 GAL1: CLB5 and a cdc28-4 clb5 bud3 GAL1:CLB5 haploids on galactose and dextrose media. The indicated strains were patched on galactose synthetic medium and replica-plated onto dextrose or galactose synthetic medium. (a) a cdc28-4 clb5 GAL1:CLB5 [MATα], (b) a cdc28-4 clb5 GAL1:CLB5 [MATa], (c) α cdc28-4 clb5 GAL1:CLB5 [MATa], (d) α cdc28-4 clb5 GAL1:CLB5 [MATα], (e) a cdc28-4 clb5 bud3 GAL1:CLB5. Plates were photographed after 3 d at 23°C. (B) Nuclear staining of a /α cdc28-4 clb5 GAL1:CLB5 haploids. Transformants were grown in galactose synthetic medium followed by a 6-h shift to dextrose synthetic medium. DIC images were overlaid onto the corresponding DAPI images. Bar, 5 μm. (C) Nuclear staining of a cdc28-4 clb5 bud3 GAL1:CLB5 haploids. Cells were grown to early log in YEPGalactose followed by a 4-h shift to YEPDextrose. Bar, 5 μm.
Budding patterns differ in haploid and diploid cells (Madden et al., 1992). Haploids form a new bud adjacent to the previous budding site (axial pattern). In contrast, diploid mother cells can either choose to form a new bud adjacent to the previous site or at the opposite end of the cell, whereas diploid daughters bud distal to the previous birth site (bipolar pattern). Budding pattern can be altered by mutations in BUD genes that can render the pattern bipolar in haploids (e.g., bud3) or random in haploids and homozygous diploids (e.g., bud5). Since lethality of the cdc28-4 clb5 cells correlated with the genotype at the MAT locus that controls budding pattern, the penetrance of the cdc28-4 clb5 defect in haploids was examined in mutants that are defective in bud site selection. Haploid a or α cdc28-4 clb5 bud3Δ GAL1:CLB5 cells were constructed and tested for defects in nuclear positioning. Staining of bud scars by calcofluor confirmed the change in budding pattern caused by the bud3Δ mutation (not shown). This strain had poor viability in dextrose medium (Fig. 6 A) and nuclear positioning defects comparable with those of a/α haploids (Fig. 6 C), consistent with the idea that budding pattern constitutes a factor in cdc28-4 clb5 diploid lethality. Nevertheless, a random pattern conferred by a bud5Δ mutation in cdc28-4 clb5 GAL1:CLB5 homozygous diploids could not suppress the lethality upon shift to dextrose medium suggesting that random budding is not equivalent to an axial pattern.
Execution Point for the CLB5 Requirement
The specific role of CLB5 in spindle positioning could reflect a temporal requirement for Clb-kinase met by the cell cycle–regulated expression of CLB5. Alternatively, Clb5 may have a unique specific ability to promote functions required to coordinate spindle dynamics and nuclear migration. To address this issue, the execution point for the spindle positioning defect was determined in cdc28-4 clb5 GAL1:CLB5 diploids. Cells were synchronized by a nocodazole block in YEPGalactose medium and released into YEPRaffinose. Paired aliquots were removed from this culture at set intervals: one to monitor cell cycle progression of the synchronous population, and the second to determine cell viability resulting from reinduction of CLB5 in galactose-containing medium. Viability was estimated by plating cells on YEPGalactose and scoring viable microcolonies arising after 15 h (Fig. 7 B). The time at which cells in the YEPRaffinose culture were no longer rescued by reinduction of CLB5 corresponded to entry into S phase, as assessed by FACS analysis (Fig. 7 A). A similar result was obtained when the experiment was performed in a synchronized population of small G1 cells obtained by elutriation (not shown). The apparent execution point corresponds to the point in the cell cycle at which Clb5-dependent kinase is normally active. However, the appearance of cells containing the nucleus positioned in the bud occurred later as cells accumulated in G2. Parental cdc28-4 GAL1:CLB5 diploids subjected to a similar protocol maintained constant viability throughout the time course (not shown).
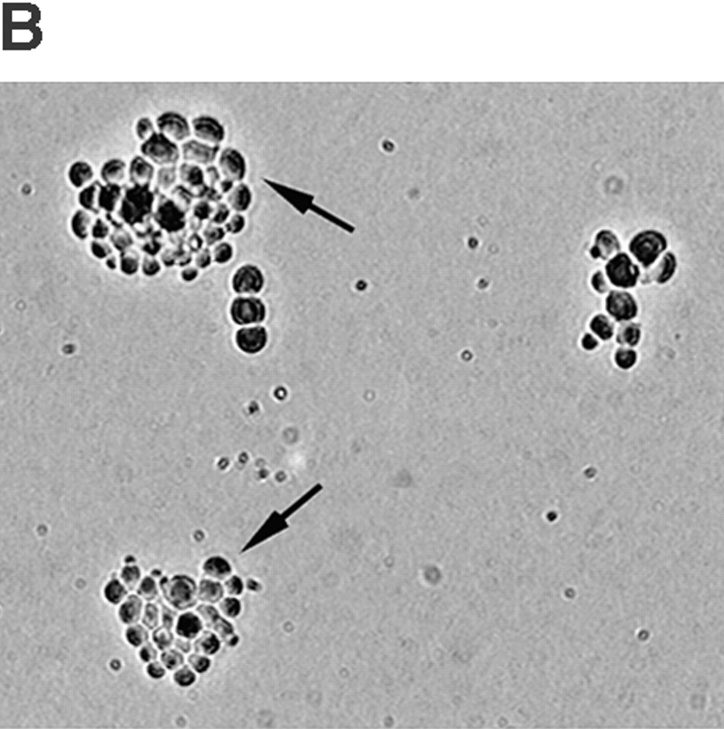
Figure 7.
Induction of GAL1: CLB5 before S phase rescues the cdc28-4 clb5 diploid lethality. (A) cdc28-4 clb5 GAL1: CLB5 diploids (MY1416) were released from a nocodazole block (YEPGalactose + 15 μg/ ml nocodazole) into YEPRaffinose. At 20-min intervals, two aliquots were removed for plating on galactose medium and cytology studies (to assess cell cycle progression in the YEPRaffinose culture). After 15 h, viability was estimated by counting the proportion of normal microcolonies emerging on YEPGalactose (open circles). Entry into S phase (open squares) was estimated from FACS analysis. At least 200 cells were scored for bud emergence (crossed squares) and nuclear positioning in the bud (crossed circles). (B) Viability on YEPGalactose. Arrows indicate examples of viable colonies. Microcolonies on a plate were photographed at the time of scoring.
Based on this result, it remained possible that other B-type cyclins could substitute for Clb5 if expressed ectopically at the G1/S boundary. To test this possibility, a homozygous diploid cdc28-4 clb5 GAL1:CLB2 or cdc28-4 clb5 GAL1:CLB4 strain carrying YEp24-CLB5 was constructed. After growth in rich galactose medium, the rate of spontaneous loss of the CLB5 plasmid was determined. 100% of the viable colonies retained the CLB5 plasmid in both cases, indicating that neither CLB2 nor CLB4 overexpression could replace CLB5. This result was further confirmed by the failure of these strains to grow on GAL-FOA plates (to apply negative selection for cells carrying the CLB5 plasmid). In contrast, haploid cdc28-4 clb5 GAL1:CLB2 or GAL1:CLB4 strains are viable both in dextrose- or galactose-containing media. It is therefore unlikely that a high dosage of CLB5 is essential to tolerate CLB2 or CLB4 overexpression.
Thus, the specific role of CLB5 under these conditions did not solely rely on the temporal pattern of CLB5 expression, since constitutive CLB2 or CLB4 could not substitute for the CLB5 requirement. Although unlikely, it nevertheless remains formally possible that Clb2 or Clb4 proteins cannot accumulate to the required level within the narrow early S-phase time window suggested by the execution point experiment.
In contrast, constitutive overexpression of CLB6 from the inducible GAL1 promoter was sufficient for viability of cdc28-4 clb5 diploids (although wild-type levels of Clb6 or even moderate overexpression from a multicopy plasmid are not sufficiently high to promote the required function in combination with the cdc28ts allele, see Fig. 1). The fact that highly elevated expression of Clb6 can substitute for Clb5 indicates that these two cyclins have overlapping functional potential. To some extent, this is also the case for Clb3, which can weakly suppress the lethality of cdc28-4 clb5 diploids from a multicopy plasmid (Fig. 1 B). Such overlap is presumably more manifest in the presence of the wild-type CDC28. In this case, deletion of CLB5 is not lethal, but results in a pronounced delay of DNA replication.
Discussion
Clb5-dependent Kinase is Limiting for Proper Preanaphase Nuclear Positioning
A genetic approach to reveal more specific functions of individual B-type cyclins led to the observation of a novel phenotype resulting from loss of Clb5-dependent kinase activity under conditions where the cyclin-dependent kinase Cdc28 is impaired. This strategy relied on the fact that a temperature-sensitive allele of CDC28, cdc28-4, encodes a protein that is already compromised for kinase activity at room temperature, creating a sensitized genetic environment for the loss of individual cyclins. The results presented here reveal that Clb5 under limiting kinase conditions is essential for early events of the spindle pathway that ultimately affect spindle dynamics at the G2/M boundary. Presumably, other cyclins can carry out this function when Cdc28 kinase activity is more robust (CDC28 clb5 homozygous diploids are viable).
A cdc28-4 clb5 homozygous diploid is inviable at room temperature as a result of lack of coordination of nuclear migration with other cell cycle events. This defect was first noticed by the failure of haploid cdc28-4 clb5 cells to produce viable diploids upon mating, but was finally ascribed to a requirement for Clb5 in mitotically dividing diploids (Fig. 1). Lethality correlated with the observation that 80% of zygotes and 35% of dividing diploids positioned an undivided nucleus in the bud per cell cycle (Fig. 2). Restoring either CLB5 or CDC28 corrected nuclear movement defects. The specificity of CLB5 was further confirmed by the fact that a sublethal dose of hydroxyurea that causes a delay in DNA replication comparable to a clb5Δ mutation cannot induce the phenotype. This result, however, cannot exclude that a delayed S phase in combination with the absence of Clb5 compromises coordination of the spindle pathway with the cell cycle. However, since other Clbs can reverse the S-phase defects associated with CLB5 deletion but not the nuclear migration defect, this scenario is unlikely to be correct.
The fact that limiting Cdk activity is important for this phenotype was confirmed using a less-severe temperature-sensitive allele of CDC28. Homozygous cdc28-13 clb5 diploids were viable at room temperature, but an increased frequency of nuclear migration into the bud was observed when cells were shifted to 35°C (data not shown).
Nuclear Positioning and Astral Microtubule Disfunction in cdc28-4 clb5 Cells
Nuclear migration and spindle positioning at the bud neck is a prerequisite for proper nuclear division between mother and daughter cells. Genetic analysis has demonstrated that astral microtubular function as well as a delicate balance of motor activities first contribute to spindle alignment at the bud neck and then to establish forces operating on the mitotic spindle that ensure timely elongation at the onset of anaphase (Huffaker et al., 1988; Palmer et al., 1992; Sullivan and Huffaker, 1992; Roof et al., 1992; Hoyt et al., 1993; Yeh et al., 1995; Carminati and Stearns, 1997; Saunders et al., 1995; Palmer et al., 1989; Endow et al., 1994; DeZwaan et al., 1997; Cottingham and Hoyt, 1997; Roof et al., 1992; Saunders et al., 1997; Yang et al., 1997; Huyett et al., 1998).
Genes encoding motor proteins implicated in astral microtubular function have been characterized in S. cerevisiae. Mutations in DHC1 (dynein heavy chain; Eshel et al., 1993; Li et al., 1993), KIP2 or KIP3 (Cottingham and Hoyt, 1997) lead to failure to position the spindle at the bud neck, which translates into an increased frequency of spindle elongation within mother cells. A similar phenotype arises in mutants defective in presumptive astral microtubule attachment components such as act5 (dynactin; Muhua et al., 1994), jnm1 (McMillan and Tatchell, 1994), num1 (Farkasovsky and Kuntzel, 1995), or kar9 (Miller and Rose, 1998). Another kinesin-like protein, Kar3, has been implicated in the control of astral microtubule number and length at the spindle pole body (Page et al., 1994; Saunders et al., 1997). kar3Δ causes a G2 delay with excessive astral microtubules emanating from the SPBs. Such delay is alleviated by microtubule depolimerizing agents.
cdc28-4 clb5 homozygous diploids exhibited a distinct behavior not observed in any of the mutants mentioned above. Cells accumulated with nuclei fully positioned in the bud. Spindle behavior at the G2/M transition was most consistent with a defect in maintenance of functional attachment sites and/or proper astral microtubule processing towards the mother cortex. Consistent with these observations, mutations in DHC1 or KIP3 as well as a low dose of nocodazole (3 μg/ml) could not relieve the lethality (not shown).
In contrast to the known mutants affecting nuclear migration in which anaphase proceeds in the mother, spindles of cdc28-4 clb5 cells once positioned in the bud rarely elongated to permit nuclear division. As shown by the execution point experiment (Fig. 7), this situation is essentially lethal. The penetrance of the defect was not absolute in a single cell cycle, but accounted for the inability of cells to give observable growth on solid medium.
The data presented here cannot conclusively demonstrate a causative relationship between spindle positioning in the bud and lack of spindle elongation. It remains possible that the latter constitutes the primary defect, or that both positioning and elongation are direct consequences of the clb5 mutation. Still, kinase inactivation in a clb1 clb2 mutant causes arrest with a short spindle positioned at the neck, indicating that impeding spindle elongation does not necessarily lead to nuclear positioning into the bud.
It has been proposed that cortical regions at the extremities of mother and daughter cells are specialized for microtubular interactions (Schroer, 1994; Carminati and Stearns, 1997). A prominent role has been proposed for dynactin as part of the attachment complex, which in turn interacts with cytoplasmic dynein molecules on the microtubules. To establish this interaction initially, cellular factors might partition asymmetrically to the bud cortex, a process that could accompany the polarized growth characteristic of yeast cells entering S phase. Consistent with this notion, proteins associated specifically with the mother (Num1; Farkasovsky and Kuntzel, 1995) or daughter (Kar9; Miller and Rose, 1998) cell cortex have been described. Once established, the microtubule–cortex interactions would determine spindle movement, and perhaps be required for normal spindle elongation. The dynamic properties of these interactions, in fact, have been found to be subjected to cell cycle regulation in the mother and daughter cells (Carminati and Stearns, 1997). Interestingly, novel temperature-sensitive alleles of MDM1, which encode an intermediate filament-like protein required for nuclear and mitochondrial inheritance, result in nuclear migration into the bud while the original temperature-sensitive alleles behave like motor mutants at the restrictive temperature (i.e., the nucleus divides in the mother cell; Fisk and Yaffe, 1997). Based on the genetic evidence, Mdm1 has been proposed to act in a scaffold-like network to position and orient cellular structures, including the spindle. Still, the possible relationship between this protein and determinants of spatial cues at the cortex remains an open question.
Several lines of evidence suggest the asymmetric nature of the spindle pathway. It is the newly synthesized spindle pole that is destined to the daughter cell (Vallen et al., 1992), and insertion of the spindle into the bud involves a microtubule-based searching mechanism that is accompanied by asymmetric distribution of dynein on microtubules directed towards the bud (Shaw et al., 1997). The behavior of the cdc28-4 clb5 mutant described in this study points to the existence of cell cycle–regulated mechanisms to promote maintenance of functional astral microtubule attachments towards the mother cell cortex as well. The assignment of possible molecular targets for this function based on genetic analysis, however, is not straightforward. Single mutations leading to defective components presumed to participate in either attachment to the mother cortex (num1) or to impose pulling forces towards the mother (kip2) paradoxically prevent nuclear migration to the bud neck, and also result in an increased frequency of binucleate mother cells.
Several models to interpret the phenotype observed may be proposed. These models must accommodate the fact that the essential function performed by the Clb5-dependent kinase, to enable proper spindle positioning before anaphase, occurs in early S-phase. In a first model, Clb5-dependent kinase may participate in an essential definition of functional microtubule attachment sites as cells progress through START. This event may involve proteins associating with the cortex such as Kar9 or Num1. Definition of such sites may impact the distribution of docking molecules on the one hand, and motor proteins on the other that later in the cell cycle will be required for proper spindle and nuclear positioning. Alternatively, in the absence of strong evidence supporting the docking of motors at the cell cortex, it can be proposed that control of astral microtubule dynamics reflects the differential association of motors with the old and newly synthesized SPB during the cell cycle. The polarity of the spindle may be imparted at the time of SPB duplication and separation by cyclin-dependent kinases. In this context, the contribution of the observed cell cycle–dependent localization of dynein and Kar3p (Yeh et al., 1995; Shaw et al., 1997; Saunders et al., 1997) to the dynamic properties of astral microtubules interacting with the mother and daughter cortex remains to be assessed. In a second model, Clb5-dependent kinase might differentially control motor activities involved in astral microtubule stability at the mother cell proximal spindle pole body, possibly in concert with other B-type cyclins regulating additional aspects of the spindle pathway. Deregulation of such activities by the combination of cdc28-4 and clb5 mutations (rather than loss of function of a single motor) may lead to an imbalance of forces, favoring spindle positioning in the bud. This type of model would be less consistent with the observation of an execution point in early S-phase for the defect.
Relative Penetrance of the cdc28 clb5 Defect in Haploids and Diploids
The frequency of nuclear positioning in the bud was maximal for the first mitotic division in cdc28-4/cdc28-4 clb5/ clb5 zygotes (80%), intermediate in mitotically dividing diploids (35%), and very low in cdc28-4 clb5 haploids (0.3%). The fact that a/α cdc28-4 clb5 haploids also displayed the phenotype at high frequency excluded ploidy as the reason for the difference, and prompted us to search for other features distinguishing haploids and a/α diploids. A bud3 mutation increased the frequency of nuclei positioned into the bud in cdc28-4 clb5 haploids, pointing to a strong interaction between bipolar budding pattern and the underlying defect caused by the cdc28-4 clb5 double mutation. Little is known about the establishment of spindle polarity in axial vs. bipolar pattern, even in wild-type cells. However, the surprising finding that the relative penetrance of the defect is affected by the budding pattern indicates that spatial cues, particularly in the mother cell, may become more limiting when a change of 180° in polarity takes place. This result might also be consistent with the fact that the maximal penetrance of the phenotype was observed in the first division after mating. Under these conditions, karyogamy occurs along an axis defined by the shmoo projections of mating partners followed by the first mitotic division, generally at a perpendicular axis. This extraordinary situation may be most sensitive to a deficiency in the establishment of attachment sites at the mother cortex, while not having an impact on the microtubular functions required for karyogamy.
Specificity of Clb5 Function vs. Redundancy of B-type Cyclins
Migration of nuclei into the bud was not observed when other CLB deletions were combined with the cdc28-4 allele, underscoring a specific role of the Clb5-dependent kinase in this functional context. In addition, the inability of various cyclins to suppress the lethality of cdc28-4 clb5 diploids efficiently further indicated the uniqueness of Clb5 for this role.
Clb5-dependent kinase normally peaks as cells enter S-phase (Schwob et al., 1994), and is necessary for efficient DNA replication in otherwise wild-type cells. This role can be performed by a number of B-type cyclins when expressed under control of the GAL1 promoter. In addition, a previous genetic analysis has implicated Clb5 in spindle formation. clb3 clb4 clb5 mutants are defective in spindle assembly, and arrest with unseparated SPBs and a 2N DNA content (Schwob and Nasmyth, 1993). In other words, Clb3, Clb4, and Clb5 are to some extent redundant for an essential function in the spindle pathway after SPB duplication.
The situation is somewhat different under limiting cdk conditions. A GAL1:CLB2 construct can suppress the first cycle arrest in S-phase of haploid cdc28-4 clb5 cells at the restrictive temperature. The same construct, however, could not override the requirement of Clb5 for viability of homozygous cdc28-4 clb5 diploids. Thus, the involvement of Clb5 in the spindle pathway takes precedence in diploids over the requirement to promote DNA replication, and is ultimately responsible for the lethality observed in cdc28-4 clb5 mutants. It follows that B-type cyclins are quite redundant in DNA replication function, while presumably Clb5 plays a more specific role in early steps of the spindle pathway, a function for which Clb2 cannot substitute.
On the other hand, the limited ability of CLB6 to suppress the defect in cdc28-4 clb5 cells appears to be mostly a consequence of an insufficient dosage of this cyclin. It is important to point out that multicopy CLB6 has proven insufficient to suppress various defects associated with a clb5 mutation, even in haploids (not shown), suggesting that these two cyclins may not play overlapping physiological roles at normal expression levels.
Acknowledgments
We thank A.F. Straight, T. Galitski, A. Sherman, and D. Stuart for the generous gift of plasmids and strains, M. Smeets for her help with FACS analysis and comments on the manuscript, and N. Rhind and P. Russell for their help with microscopy techniques. Thanks to members of the Reed, Wittenberg and Russell labs for stimulating discussions.
Abbreviations used in this paper
- cdk
cyclin-dependent kinase
- DIC
differential interference contrast
- SPB
spindle pole body
Footnotes
M. Segal acknowledges fellowships from the European Molecular Biology Organization and the Human Frontiers in Science Program. D.J. Clarke was supported by a fellowship from the European Molecular Biology Organization. This work was supported by United States Public Health Service grant GM38328 to S.I. Reed.
Address all correspondence to Steven I. Reed, The Scripps Research Institute, 10666 North Torrey Pines Road, La Jolla, CA 92037. Tel.: (619) 784-9836. Fax: (619) 784-2781. E-mail: sreed@scripps.edu
References
- Basco RD, Segal MD, Reed SI. Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5030–5042. doi: 10.1128/mcb.15.9.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 59–96.
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiaeis accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM, Kinesin -related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Kang LJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO (Eur Mol Biol Organ) J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, Van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;909:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Kuntzel H. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J Cell Biol. 1995;131:1003–1014. doi: 10.1083/jcb.131.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk HA, Yaffe MP. Mutational analysis of Mdm1p function in nuclear and mitochondrial inheritance. J Cell Biol. 1997;138:485–494. doi: 10.1083/jcb.138.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. . Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia colishuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Hadwiger JA, Wittenberg C, Richardson HE, de Barros M, Lopes, Reed SI. A family of cyclin homologues that control the G1 phase in yeast. Proc Natl Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Totis L, Saunders WS. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3motor domain mutations. Genetics. 1993;135:35–44. doi: 10.1093/genetics/135.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyett A, Kahana J, Silver P, Zeng X, Saunders WS. The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J Cell Sci. 1998;111:295–301. doi: 10.1242/jcs.111.3.295. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Marini NJ, Reed SI. Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells in the budding yeast Saccharomyces cerevisiae. . Cell. 1992;69:317–327. doi: 10.1016/0092-8674(92)90412-6. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., T. Weinert, and J.R. Pringle. 1997. Cell cycle control in Saccharomyces cerevisiae. In The molecular and cellular biology of the yeast Saccharomyces. J.R. Pringle, J.R. Broach, and E.W. Jones, editors. Cold Spring Harbor Laboratory Press. Plainview, NY. 607–695.
- Li Y-Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lörincz AT, Reed SI. Sequence analysis of temperature sensitive mutations in the Saccharomyces cerevisiae gene CDC28. . Mol Cell Biol. 1986;6:4099–4103. doi: 10.1128/mcb.6.11.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Costingan C, Snyder M. Cell polarity and morphogenesis in Saccharomyces cerevisiae. . Trends Cell Biol. 1992;2:22–29. doi: 10.1016/0962-8924(92)90140-i. [DOI] [PubMed] [Google Scholar]
- McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiaeis required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondésert G, Reed SI. BED1, a gene encoding a galactosyltransferase homologue, is required for polarized growth and efficient bud emergence in Saccharomyces cerevisiae. . J Cell Biol. 1996;132:137–151. doi: 10.1083/jcb.132.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–589. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- Richardson HE, Wittenberg C, Cross FR, Reed SI. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- Richardson H, Lew DJ, Henze M, Sugimoto K, Reed SI. Cyclin-B homologues in Saccharomyces cerevisiaefunction in S phase and in G2. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hornack D, Lengyel V, Deng D. The Saccharomyces cerevisiaekinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiaekinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. New insights into the interaction of cytoplasmic dynein with the actin-related protein Arp1. J Cell Biol. 1994;128:617–624. doi: 10.1083/jcb.127.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. . Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. . Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G. Fink, and J.B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. . Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Vallen EA, Scherson TY, Roberts T, van Zee K, Rose MD. Asymmetric mitotic segregation of the yeast spindle pole body. Cell. 1992;69:505–515. doi: 10.1016/0092-8674(92)90451-h. [DOI] [PubMed] [Google Scholar]
- Yang SS, Yeh E, Salmon ED, Bloom K. Identification of a mid-anaphase checkpoint in budding yeast. J Cell Biol. 1997;136:345–354. doi: 10.1083/jcb.136.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]