Abstract
Cell polarity is fundamental to differentiation and function of most cells. Studies in mammalian epithelial cells have revealed that the establishment and maintenance of cell polarity depends upon cell adhesion, signaling networks, the cytoskeleton, and protein transport. Atypical protein kinase C (PKC) isotypes PKCζ and PKCλ have been implicated in signaling through lipid metabolites including phosphatidylinositol 3-phosphates, but their physiological role remains elusive. In the present study we report the identification of a protein, ASIP (atypical PKC isotype–specific interacting protein), that binds to aPKCs, and show that it colocalizes with PKCλ to the cell junctional complex in cultured epithelial MDCKII cells and rat intestinal epithelia. In addition, immunoelectron microscopy revealed that ASIP localizes to tight junctions in intestinal epithelial cells. Furthermore, ASIP shows significant sequence similarity to Caenorhabditis elegans PAR-3. PAR-3 protein is localized to the anterior periphery of the one-cell embryo, and is required for the establishment of cell polarity in early embryos. ASIP and PAR-3 share three PDZ domains, and can both bind to aPKCs. Taken together, our results suggest a role for a protein complex containing ASIP and aPKC in the establishment and/or maintenance of epithelial cell polarity. The evolutionary conservation of the protein complex and its asymmetric distribution in polarized cells from worm embryo to mammalian-differentiated cells may mean that the complex functions generally in the organization of cellular asymmetry.
Keywords: ASIP, atypical PKC, par, cell polarity, tight junction
The stimulus-coupled turnover of membrane lipids is an important event during cellular signal transduction (Nishizuka, 1995). Members of the protein kinase C (PKC)1 family are among the prominent candidates that transduce a variety of extracellular stimuli to the cellular signaling networks through lipid-derived second messenger molecules. PKC comprises at least 10 closely related isoforms that can be subdivided into three classes: cPKC (PKCα, PKCβI, PKCβII, and PKCγ), nPKC (PKCδ, PKCε, PKCη, and PKCθ), and aPKC (PKCζ and PKCλ/ι) (Nishizuka, 1995). The cPKC and nPKC isotypes can be positively regulated by a variety of lipid-derived messenger molecules including diacylglycerols, and are receptors for phorbol esters such as TPA. Unlike other PKC isotypes, aPKCs do not seem to be regulated directly by Ca2+, phorbol esters, or diacylglycerols (Nishizuka, 1995). Some studies have shown that ceramide regulates the activity of PKCζ (Lozano et al., 1994; Müller et al., 1995). Purified PKCζ is also regulated by phosphatidylinositol 3,4,5-trisphosphate (PI-3,4,5-P3), a product of phosphatidylinositol 3-kinase (PI3-kinase) (Nakanishi et al., 1993), and PKCλ is activated upon PDGF receptor stimulation through a pathway involving PI3-kinase (Akimoto et al., 1996; Toker and Cantley, 1997). Stimulation of insulin receptors also results in PKCζ activation through the PI3-kinase pathway (Standaert et al., 1997; Bandyopadhyay et al., 1997). A series of studies using a pseudosubstrate peptide or kinase-deficient dominant negative forms of PKCζ and PKCλ suggest that atypical PKC isotypes (aPKC) plays roles distinct from those of the cPKC and nPKC isotypes in a variety of cellular functions. These include Xenopus oocyte maturation (Dominguez et al., 1992; Berra et al., 1993), proliferation and survival of fibroblasts (Berra et al., 1993; Diaz-Meco et al., 1996), differentiation of PC12 (Wooten et al., 1994) and leukemic cells (Ways et al., 1994), activation of mitogen-activated protein kinase (MAPK) (Berra et al., 1995) and gene expression (Lozano et al., 1994; Akimoto et al., 1996; Xu et al., 1996), and insulin-induced glut4 translocation (Standaert et al., 1997). Furthermore, several proteins have been shown to interact directly with aPKC isotypes (Diaz-Meco et al., 1994; Diaz-Meco et al., 1996a ; Diaz-Meco et al., 1996b ; Puls et al., 1997; Sanchez et al., 1998). However, the downstream effectors and the exact cellular function of aPKCs remain elusive. In this paper we present evidence consistent with a hypothesis that atypical PKC isotype–specific interacting protein (ASIP) and aPKC complex is involved in establishment or maintenance of cell polarity.
Cell polarity is the reflection of complex mechanisms that establish and maintain functionally specialized domains in the plasma membrane and cytoplasm, and is fundamental to differentiation and function of most cells (Drubin and Nelson, 1996; Caplan, 1997). Cell polarity is being studied in model genetic systems like Caenorhabditis elegans and in tissue culture in epithelial cells. Studies of asymmetric cell division in embryogenesis have provided evidence that transient asymmetric distribution of proteins at the cell periphery is essential for cell polarity (Knoblich, 1997). In early C. elegans embryos, PAR proteins such as PAR-3 are required for embryonic polarity, and become localized asymmetrically at the periphery of the one-cell embryo (Etemad-Moghadam et al., 1995; Guo and Kemphues, 1996). The cue that triggers cell polarization and determines the axis of polarity is provided by the sperm (Goldstein and Hird, 1996). Mutations in the par-3 gene affect the asymmetric distribution of other proteins involved in cell fate determination and the orientation of mitotic spindles in successive cell cycle (Guo and Kemphues, 1996; Bowerman et al., 1997). How the sperm cue triggers asymmetric distribution of PAR proteins is not clear; neither is it clear how the asymmetric distribution of PAR proteins leads to other cellular asymmetries.
Mammalian epithelial cells provide an experimental system that has revealed essential features of cell polarity (Eaton and Simons, 1995; Drubin and Nelson, 1996; Gumbiner, 1996). Epithelial cells respond to asymmetric cell adhesion to organize cytoskeletal and membrane proteins into distinct apical and basal-lateral membrane domains; this apical/basal polarity provides a basis for directed transport across the epithelium. Tight junctions are specialized structures that play an essential role in epithelial cell polarity by creating a barrier to diffusion between cells in the epithelial sheet and forming an intramembrane diffusion fence that restricts intermixing of apical and basal-lateral membrane components (Balda and Matter, 1998). As in the C. elegans one-cell embryos, establishing of cell polarity in epithelial cells starts with a cortical spatial cue. The spatial cue in epithelial cells is cell adhesion. E-cadherin–mediated cell–cell contact and the contact between integrins and the extracellular matrix trigger the specialized assembly of actin-based cytoskeleton and signaling networks around the adhesion receptors and tight junctions, and position other cytoskeletal complexes and protein-sorting compartments (Eaton and Simons, 1995; Drubin and Nelson, 1996; Gumbiner, 1996). How adhesion receptors trigger the establishment of cellular asymmetry is not clear; neither is it clear how tight junctions reinforce and maintain the cellular asymmetry.
During experiments to clarify the role of aPKC isotypes, we searched for aPKC-interacting proteins using an interaction cloning approach using purified recombinant PKCζ as a probe. In the present study, we show that a novel protein, ASIP, interacts with aPKC isotypes, and that the interaction involves the kinase domain of aPKC and occurs within a region of 225 amino acids of ASIP. ASIP shows significant sequence similarity to a C. elegans polarity protein, PAR-3. Furthermore, the direct interaction with aPKC is conserved from worm PAR-3 to mammalian ASIP. Endogenous ASIP and PKCλ form a complex in NIH3T3 cells and epithelial MDCKII cells. In addition, ASIP colocalizes with PKCλ to the tight junction and adherens junction–containing junctional complex in MDCKII cells. ASIP also colocalizes with ZO-1, a tight junction protein, in MDCKII cells, and localizes to the tight junction in rat intestinal epithelium.
Materials and Methods
Materials and Chemicals
PKC expression vectors encoding PKCα (YK504), PKCδ (M241), PKCζ (M246), PKCλ (MLNP45), and PKCλRD (MLRD) have been described previously (Akimoto et al., 1994; Ohno et al., 1994; Akimoto et al., 1996; Izumi et al., 1997). PKCλKD encoding amino acid (a.a.) residues 191–586 of PKCλ was constructed by K. Akimoto. Tag-ASIP (SRHis-ASIP) encodes the entire ASIP sequence fused downstream of the six histidine residues and a 12–amino acid sequence from the T7 gene 10 leader sequence. Anti-PKCζ (ζRb2) was raised against the COOH-terminal peptide of mouse PKCζ (from residues 578–592). Anti-glutathione-S-transferase (GST) was raised against the purified protein for glutathione-S-transferase. Monoclonal antibodies (mAb) of anti-PKCα and anti-PKCδ were purchased from Transduction Laboratories (Lexington, KY), and anti-T7 tag was from Novagen. The mAb of PKCλ was purchased from Transduction Laboratories. This antibody was generated against the kinase domain of PKCι, a human homologue of mouse PKCλ, and specifically recognizes PKCλ, but not PKCζ. The anti-PKCλ (λ2) antibody is a polyclonal antibody against the D2/D3 region of mouse PKCλ. Both anti-PKCλ (mAb) and anti-PKCλ (λ2) recognize cellular PKCλ specifically, and not PKCζ (data not shown). The anti-ASIP antibody (C1 and C2) was raised against the GST fusion protein of aPKC-binding region (a.a. residues 712–936), and P1 was raised against the GST fusion protein of the third PDZ domain (a.a. residues 584–708). Anti-ZO-1 and MDCKII cells were kindly provided by Dr. S. Tsukita (Kyoto University, Kyoto, Japan). FITC-conjugated goat anti–rabbit and Cy3-conjugated goat anti–mouse antibodies were purchased from E.Y. Laboratories, Inc. (San Mateo, CA) and Amersham Life Science, Inc. (Arlington Heights, IL), respectively.
Purification and 32P-labeling of PKCζ
Mouse PKCζ (2.1 μg) purified from recombinant baculovirus-infected Sf21 cells (Fujise et al., 1994) was labeled with 32P by autophosphorylation in 155 μl of kinase buffer containing 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 10% glycerol, 0.6 μCi/ml [γ-32P]ATP, and 40 μg/ml phosphatidylserine at 30°C for 2 h. After the reaction, the [32P]-labeled PKCζ was separated from the unreacted ATP by gel filtration on Sephadex G-50 equilibrated with 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1 mM DTT, and 0.5% Triton X-100, and was used as a probe for screening a λEXlox cDNA expression library.
Isolation of cDNA Clones Encoding ASIP
Poly(A)+ RNA was isolated from NIH3T3 cells, and randomly primed cDNA was synthesized using a HindIII primer adapter (Novagen Inc., Madison, WI). cDNA was ligated to λEXlox vector arm (Novagen Inc.), and was packaged in phage particles using a Phage Marker System (Novagen Inc.). This library contains ∼0.95 × 106 independent clones. Approximately 1.8 × 105 clones were plated at 10,000 phages per plate using Escherichia coli strain BL21 (DE3) pLysE as host cells. After incubation for 6 h at 37°C, the plaques were covered with Hybond-C extra (Amersham Life Science, Inc.), impregnated with 10 mM IPTG, and allowed to grow for an additional 3.5 h at 37°C. The filters were then washed with TBS buffer (50 mM Tris-HCl, pH 7.5, 0.5 M NaCl), blocked with 5% skim milk at 4°C overnight, and incubated with 1 mM ATP solution (TBS buffer containing 1 mM ATP) at room temperature for 2 h to saturate any autophosphorylation sites or ATP-binding sites on the proteins. The filters were then incubated with [32P]-labeled PKCζ (106 cpm/ml) in 50 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 50 μg/ml phosphatidylserine, and 1% BSA for 5 h at room temperature. After washing in TBS buffer, the filters were exposed for autoradiography. Positive phage clones were converted to plasmids (pEXlox + cDNA insert) using E. coli strain BM25.5 according to the manufacturer's instructions. The initial isolate (clone I-1), which encodes amino acid residues 710–924, was used as a probe to screen a rat 3Y1-λZAP II cDNA library to obtain a cDNA clone encoding the full-length ASIP coding sequence. The nucleotide sequence of the ASIP cDNA has been submitted to the Genbank/EMBL databank with accession no. AB005549.
Northern Blot Analysis
Total RNA preparations from various mouse tissues were obtained as described (Osada et al., 1992). Poly(A)+ RNA preparations from various cell lines were isolated using a QuickPrep mRNA Purification Kit (Pharmacia Biotech, Inc., Piscataway, NJ). Northern blot analysis was performed according to the standard protocol.
FISH Mapping of ASIP on Chromosomes
Lymphocytes isolated from human blood were cultured in α-MEM supplemented with 10% FBS and phytohemagglutinin at 37°C for 68–72 h. The lymphocyte cultures were treated with BrdU (0.18 mg/ml: Sigma Chemical Co.) to synchronize the cell population. The synchronized cells were washed three times with serum-free medium to release the block, and were recultured at 37°C for 6 h in α-MEM containing thymidine (2.5 μg/ml; Sigma Chemical Co.). The cells were harvested, and slides were made by standard procedures including hypotonic treatment, fixation, and air-drying. For in situ hybridization and FISH detection, the rat ASIP cDNA probe was biotinylated with dATP using a BioNick labeling kit (15°C, 1 h; GIBCO BRL, Rockville, MD; Heng et al., 1992). The procedure for FISH detection was performed according to Heng et al. (1992) and Heng and Tsui (1993). In brief, the slides were baked at 55°C for 1 h. After RNase treatment, the slides were denatured in 70% formamide in 2× SSC for 2 min at 70°C, and were dehydrated with ethanol. Probes were denatured at 75°C for 5 min in a hybridization mix consisting of 50% formamide and 10% dextran sulphate. The probes were loaded on denatured chromosomal slides. After overnight hybridization, the slides were washed and amplified for detection. FISH signals and the DAPI banding pattern were recorded separately by photograph, and assignment of the FISH mapping data to the chromosomal bands was achieved by superimposing the FISH signals on the DAPI banded chromosomes (Heng and Tsui, 1993).
Cell Cultures
COS and MDCKII cells were grown in DME containing 10% FCS, penicillin, and streptomycin in an air-5% CO2 atmosphere at constant humidity. NIH3T3 cells were grown in DME containing 7% calf serum, penicillin, and streptomycin in an air-5% CO2 atmosphere at constant humidity. For the Ca2+ switch assay, subconfluent MDCKII cells were grown in normal growth medium, and were transferred to low Ca2+ medium (growth medium containing 4 mM EGTA) for 6 h and then transferred back to the normal Ca2+ medium.
Transfection, Cell Lysis, Immunoprecipitation, and Immunoblotting
COS cells were transfected with cDNA expression plasmids by electroporation (Gene Pulser; Bio-Rad Laboratories, Hercules, CA). Cells (NIH3T3, MDCKII, and cDNA-transfected COS cells) in 10-cm dishes were suspended in 200 μl of lysis buffer containing 20 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 10 μg/ml leupeptin, 1 mM PMSF, 1.8 μg/ml aprotinin, 1% Triton X-100, 0.1% deoxycholate, and 0.1% SDS. After a 30-min incubation on ice, the lysates were clarified by centrifugation at 14,000 rpm for 30 min, and were incubated with antibodies preabsorbed on Protein G-Sepharose (Pharmacia Biotech, Inc., Piscataway, NJ) for 1 h at 4°C. The immunocomplexes on Sepharose were washed six times with lysis buffer, after which the proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and probed with the indicated antibodies.
Gel Electrophoresis and Western Blot Analysis
SDS-PAGE was based on the discontinuous Tris-glycine system of Laemmli (1970). Western blot analysis was performed by one-dimensional electrophoresis, followed by electrophoretic transfer to PVDF membranes which were then incubated with various antibodies. Antibodies were detected by chemiluminescence ECL (Amersham Life Science, Inc., Arlington Heights, IL).
Blot Overlay Assay Using Recombinant aPKC, GST-ASIPs, and GST-PAR-3
GST-fusion proteins were produced in Escherichia coli, and were purified on Glutathione Sepharose 4B (Pharmacia Biotech, Inc.) by a standard procedure. GST-ASIP 584/708 and GST-ASIP 712/936 encode amino acid residues 584–708 and 712–936, respectively, of ASIP fused downstream of GST. GST-PAR-3 (p30-5) encodes amino acid residues 678–932 of PAR-3. These proteins were subjected to SDS-PAGE and blotted onto a PVDF membrane. After treatment with a 5% skim milk solution, the PVDF membrane was incubated with 4 μg/ml of purified mouse PKCζ in 50 mM Tris-HCl, pH 7.5, containing 0.5 M NaCl and 1% BSA for 3 h at room temperature. The membrane was washed with TBS buffer, and the bound proteins were detected with an anti-PKCζ antibody (ζRb2), an alkaline phosphatase-conjugated second antibody (Tago Inc., Burlingame, CA), and an artificial substrate for alkaline phosphatase (Vector Labs, Inc., Burlingame, CA).
Immunofluorescence Microscopy
MDCKII and NIH3T3 cells plated on glass coverslips were fixed with methanol-acetone (1:1) for 10 min at −20°C, and were then air-dried. The cells were incubated with TBST (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% Tween 20) containing 10% calf serum for 10 min at room temperature, incubated with primary antibodies for 45 min at 37°C, and washed three times for 5 min each with TBST. After the first incubation, the cells were incubated for 45 min at 37°C with secondary antibodies (FITC-conjugated goat anti–rabbit and Cy3-conjugated goat anti–mouse antibodies), and were washed three times for 5 min each with TBST. Samples were viewed using an Olympus BX50. Images were captured with a Princeton Instruments digital camera. Images were manipulated using Adobe Photoshop software.
Immunocytochemistry of Rat Intestinal Epithelium and Hepatic Bile Capillary
Rat (10-wk-old) tissue samples were frozen in liquid nitrogen, and the frozen sections (∼7 μM) were cut in a cryostat. The samples were mounted on glass slides, fixed with methanol/acetone (1:1) for 10 min at −20°C, and air-dried. After the samples were incubated with TBST containing 10% calf serum for 10 min at room temperature, they were incubated with primary antibodies for 45 min at 37°C, and were washed three times for 5 min each with TBST. After the first incubation, the cells were incubated for 45 min at 37°C with secondary antibodies (FITC-conjugated goat anti–rabbit and Cy3-conjugated goat anti–mouse antibodies) and washed three times for 5 min with TBST. Samples were viewed using an Olympus BX50. Images were captured with a Princeton Instruments digital camera (Princeton Instruments, Trenton, NJ). Images were manipulated using Adobe Photoshop software.
Immunoelectron Microscopy
Small pieces of rat small intestine were fixed in 2% formaldehyde (freshly depolymerized from paraformaldehyde; Merck & Co., Whitehouse Station, NJ) in 0.1 M sodium phosphate buffer, pH 7.4, and 5% sucrose for 30 min at 4°C. After rinsing, the tissue specimens were infused with a mixture of sucrose and polyvinylpyrrolidone (Sigma Chemical Co.; Tokuyasu, 1989) and frozen rapidly in liquid nitrogen. Ultrathin (50–70 nm) cryosections were prepared as described (Liou et al., 1996) and subjected to indirect immunolabeling with 10 nm colloidal gold-conjugated goat anti–rabbit IgG antibody (Amersham Life Science, Inc., Arlington Heights, IL). The specimens were embedded in a mixture of 2% methylcellulose (Nacalai Tesque, Kyoto, Japan) and 0.4% uranyl acetate (Griffiths et al., 1986).
Results
Molecular Cloning, Primary Structure, and mRNA Expression of ASIP
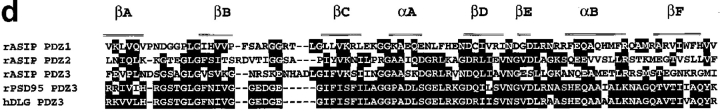
To identify proteins that interact directly with aPKC, we screened an NIH3T3 cDNA expression library using purified and autophosphorylated PKCζ as a probe. One cDNA found, clone I-1, encoded a truncated form of a novel protein, and we next screened a rat fibroblast 3Y1 cDNA library to obtain a full-length cDNA clone encoding the entire protein-coding sequence. This cDNA was 5,500 nucleotides in length, and contained a 1,337–amino acid open reading frame. The open reading frame is preceded by 259 nucleotides containing an in-frame termination codon at nucleotide position -96 (compared with A at the putative initiation codon), and is followed by a 1,227-nucleotide 3′ noncoding sequence. Since the encoded protein shows binding for aPKC isotypes of the PKC family (see below), we named it ASIP (aPKC-specific interacting protein). The sequence of ASIP does not contain any apparent structural motif suggesting catalytic activity, but contains three PDZ domains that are potentially involved in protein–protein interactions (Saras and Heldin, 1996; Sheng, 1996) in addition to the aPKC-binding region (see below; Fig. 1 d). A database search for sequence similarity revealed Caenorhabditis elegans PAR-3 as the only protein with a significant overall sequence similarity (Fig. 1, a and b). PAR-3 localizes asymmetrically to the periphery of the C. elegans one-cell embryo, and is required for embryonic cell polarity (Etemad-Moghadam et al., 1995). The similarity between ASIP and PAR-3 involves an NH2-terminal conserved region (CR1), the three PDZ domains (CR2), and a conserved sequence (CR3) in the aPKC-binding region. A Drosophila EST cDNA clone, AA439413, also contains a sequence that shows significant similarity to the sequence of CR1 (Fig. 1 c).
Figure 1.
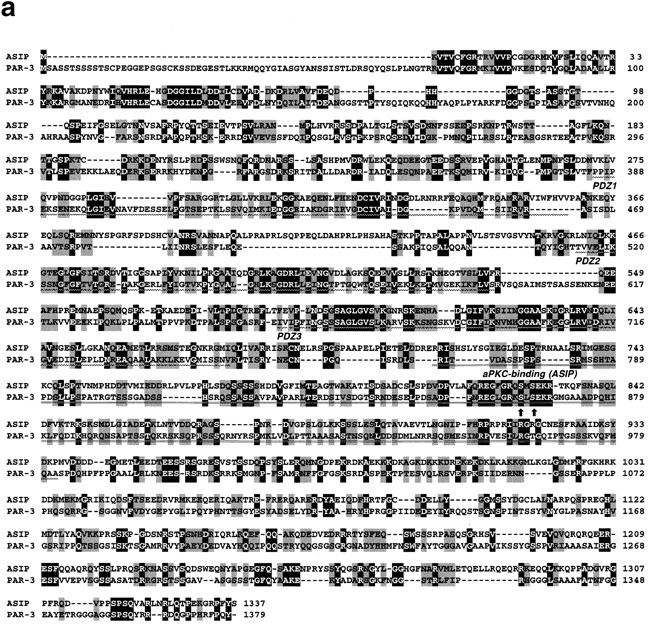
Sequence similarity between ASIP and PAR-3. (a) Amino acid sequence alignment of rat ASIP and C. elegans PAR-3. Alignment was done using the GeneStream align program (http: //genome.eerie.fr/bin/align-guess.cgl). Residues with black and shaded backgrounds indicate amino acid identity and similarity, respectively. PDZ domains (wavy underlining) and the aPKC-binding region (double underlining; see Fig. 5) are indicated. Arrows indicate potential PKC phosphorylation sites in the CR3. Dashes represent sequence gaps. (b) Schematic view of the ASIP and PAR-3 structures. Typical conserved regions—CR1, CR2, and CR3—are shown. The aPKC-binding region shown is that sufficient for a direct interaction with aPKC (see Fig. 5). (c) Amino acid sequence alignment of the NH2-terminal region of rat ASIP with C. elegans PAR-3 and Drosophila AA439413. Residues with a black background indicate amino acid identity. The amino acid sequence of Drosophila AA439413 is encoded by residues 137–401 of the nucleic acid sequence. (d) Amino acid sequence alignment of the PDZ domain of ASIP with the third PDZ domain of rat PSD95 and human DLG. Residues with a black background indicate amino acid identity. The six β sheets (βA–βF) and the two α helices (αA and αB) present in the structure of the third PDZ domain of rat PSD95 (Doyle et al., 1996) and human DLG (Cabral et al., 1996) are indicated in the upper alignment. r, rat; c, C. elegans; d, Drosophila; h, human.
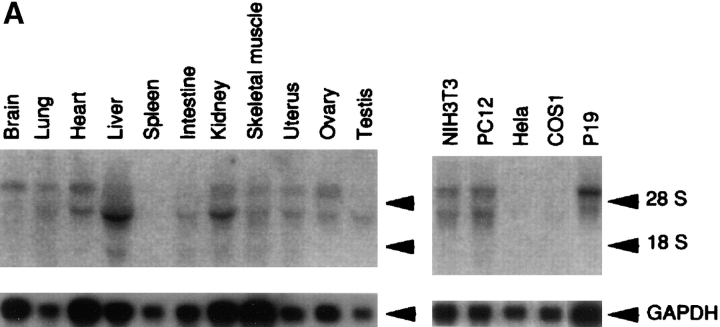
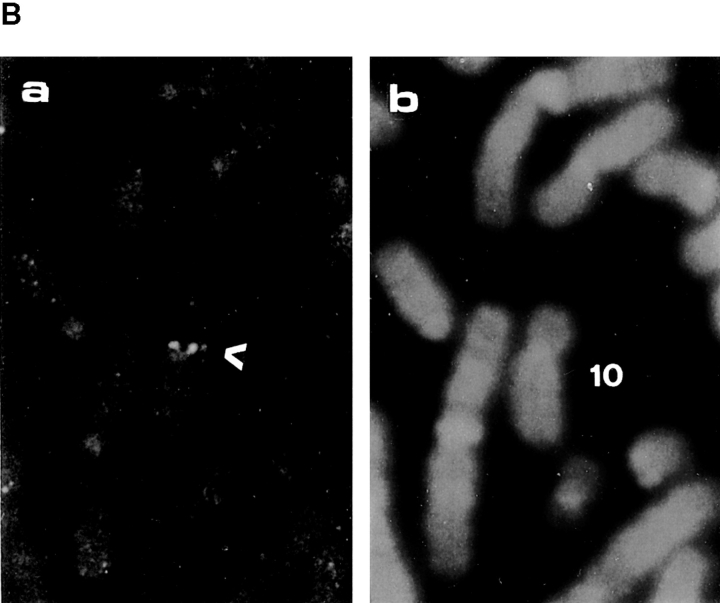
Northern analysis of the ASIP mRNA from mouse tissues and mammalian cell lines shows that there are at least two different mRNA sizes, and that both are expressed ubiquitously in adult tissues and cell lines in slightly different manners (Fig. 2 A). The two kinds of ASIP mRNAs might be products of the same gene produced by alternative splicing, promoter usage, poly(A) addition, or they may be products of a related gene. Human chromosome mapping of the gene for ASIP using rat ASIP cDNA as a probe (Fig. 2 B, a and b) shows a single chromosomal localization on chromosome 10, band p11.2. This result suggests that the human ASIP gene is a single gene, although the possibility of the presence of tandem aligned multiple genes can not be excluded.
Figure 2.
Expression of ASIP mRNA in mouse tissues and FISH mapping of ASIP on human chromosomes. (A) Total RNA (5 μg for brain; 7 μg for Hela; 10 μg for others) or poly(A)+ RNA (1.6 μg for p19) was analyzed using the original mouse cDNA isolate (clone I-1) of ASIP as a probe. The positions of the ribosomal RNAs are indicated. The same blot was reprobed with GAPDH cDNA (bottom). The natures of the two mRNAs (6 and 4 kb) remain to be clarified, although our cDNA clone (5.5 kb) corresponds to the longer mRNA. (B) FISH mapping of the ASIP probe (a) showing the FISH signals on the chromosome; and (b) showing the same mitotic figure stained with DAPI to identify chromosome 10.
Association Between ASIP and aPKC in COS Cells
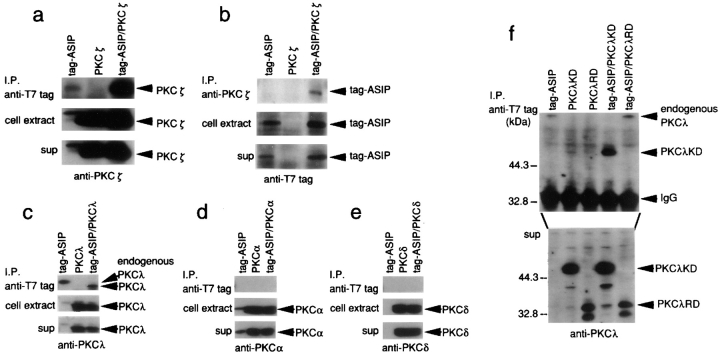
To confirm the association between aPKC and ASIP in intact cells and to examine the binding specificity of ASIP to various PKC isotypes, we coexpressed ASIP and PKC isotypes in COS cells, and performed immunoprecipitation experiments. When epitope-tagged-ASIP (tag-ASIP) is immunoprecipitated from cells transfected with tag-ASIP and PKCζ, the overexpressed exogenous PKCζ (lane tag-ASIP/PKCζ) coprecipitates with ASIP along with the endogenous PKCζ or PKCλ (lane tag-ASIP; Fig. 3 a). The reverse experiment, where PKCζ is first immunoprecipitated and the immunoprecipitates are then probed with anti-tag antibody, yields a tag-ASIP band (Fig. 3 b). Similar experiments using other PKC isotypes indicate that PKCλ can also interact with ASIP in COS cells (Fig. 3 c), while PKCα, a conventional PKC, and PKCδ, a novel PKC, do not (Fig. 3, d and e). Furthermore, the kinase domain of PKCλ coprecipitates with ASIP (Fig. 3 f). These results indicate that ASIP interacts with aPKC isotypes, and that the kinase domain of aPKC is sufficient for the interaction.
Figure 3.
Specificity of the association between ASIP and aPKC. (a–e) The association of ASIP and PKC isotypes in COS cells. COS cells were transiently transfected with the expression vectors shown at the top. (a) Cell extracts were clarified by centrifugation (sup), and were immunoprecipitated with anti-T7 antibody. The immunoprecipitates were probed with anti-PKCζ (ζRb2) antibody. Overexpressed exogenous PKCζ (lane tag-ASIP/PKCζ) was coprecipitated with ASIP as well as endogenous PKCζ and PKCλ (lane tag-ASIP). The anti-PKCζ (ζRb2) antibody cross-reacts with PKCα as well as PKCλ. (b) The reverse experiment to a. Cell extracts were immunoprecipitated with anti-PKCζ (ζRb2) antibody. The immunoprecipitates were probed with anti-T7 tag antibody. Tag-ASIP is coprecipitated with PKCζ. (c–e) Cell extracts were immunoprecipitated with anti-T7 tag antibody followed by anti-PKCλ (mAb; c), PKCα (mAb; d), or PKCδ (mAb; e). PKCλ, but not PKCα or PKCδ, can interact with ASIP in COS cells. (f) The PKCλ-kinase domain (KD) coprecipitated with ASIP in COS cells. COS cells were transiently transfected with the expression vectors shown at the top, and were immunoprecipitated by anti-T7 tag antibody. The immunoprecipitates were probed with anti-PKCλ antibody (λ2) which recognizes both PKCλRD and PKCλKD.
Direct Association of ASIP and C. elegans PAR-3 with aPKC
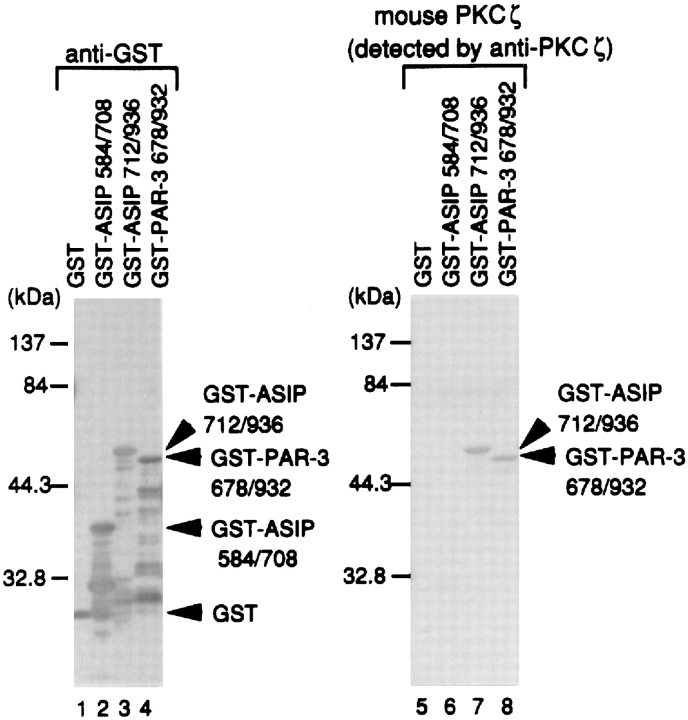
The original mouse ASIP cDNA (clone I-1), identified by screening for protein interactions, contains only a part of ASIP from amino acid residues 710–924; this region is sufficient for direct association with aPKC. An overlay assay where rat ASIP was expressed as a GST fusion protein, fixed on a filter, mixed with purified recombinant PKCζ, and probed with an anti-PKCζ antibody (Fig. 4), clearly indicates that the interaction between ASIP and aPKC is a direct one, and that the region from amino acids 712–936 of ASIP is sufficient for the interaction. Importantly, as suggested by the sequence similarity between ASIP and C. elegans PAR-3, the overlay assay also revealed that mouse PKCζ binds to GST-PAR-3 (a.a. residues 678–932; Fig. 4). These regions in the two proteins are in the same relative position with respect to the PDZ domains, and contain a small segment (CR3) with 73% amino acid identity that fits the consensus for phosphorylation by PKC (Fig. 1, a and b).
Figure 4.
Direct interaction between mouse PKCζ and rat ASIP or C. elegans PAR-3. The recombinant GST fusion proteins shown at the top were produced in E. coli. Purified proteins were subjected to SDS-PAGE and blotted onto PVDF membranes. The blots were then probed with anti-GST antibody (left) or overlaid with recombinant mouse PKCζ, followed by immunodetection with an anti-PKCζ (ζRb2) antibody and an alkaline phosphatase–conjugated secondary antibody (right).
In Vivo Association of ASIP with PKCλ in MDCKII Epithelial Cells and NIH3T3 Fibroblasts
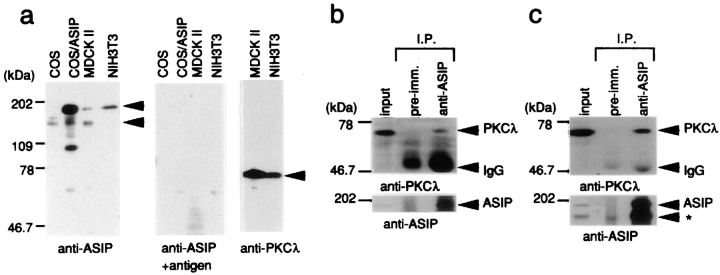
The experiments described above establish that ASIP and aPKC interact with each other directly in vitro and in COS cells. To evaluate the physiological significance of this interaction, we next examined the interaction in vivo. First, we raised anti-ASIP antibodies against two portions of the protein in three rabbits. One anti-serum, P1, was raised against the third PDZ domain (amino acid residues 584– 708), and the others, C1 and C2, were raised against the aPKC-binding region (712–936). All three antisera recognized bands of 180 and 150 kD on Western blots of proteins from MDCKII cells, a single band of 180 kD in NIH3T3 cells, and a major band of 150 kD in COS cells. The 180-kD band comigrates with the major band in COS cell extracts transfected with full-length ASIP cDNA (Fig. 5 a), indicating that the 180-kD band corresponds to ASIP encoded by the full-length cDNA sequence. Importantly, both the 180-kD and 150-kD bands disappear in the presence of excess amounts of antigen (Fig. 5 a). Although the present results strongly suggest that the 150-kD band observed in MDCKII and COS cells is a splicing variant of ASIP or the product of a related gene, its exact nature remains to be clarified in future experiments.
Figure 5.
ASIP expression and association with aPKC in NIH3T3 and MDCKII cells. (a) Western blot analysis with anti-ASIP (C2) or anti-PKCλ (mAb) antibodies. Total extracts of the indicated cells were subjected to Western blot analysis. The central panel shows the results obtained using anti-ASIP (C2) antibody preabsorbed with antigen. Arrowheads show two specific ASIP bands (180 and 150 kD) (left) and a PKCλ band (right). (b and c) Association of ASIP and aPKC in NIH3T3 (b) and MDCKII (c) cells. Cell extracts were clarified by centrifugation (input) and immunoprecipitated with anti-ASIP (C2) antibody. The immunoprecipitates were probed with anti-PKCλ (mAb) antibody (top) or anti-ASIP (C2) antibody (bottom). Asterisk indicates the 150-kD ASIP band (c).
For immunohistochemical determination of the distribution of different aPKC isoforms, we searched among available antibodies that can distinguish PKCλ from PKCζ. We selected anti-PKCλ mouse monoclonal antibody (mAb), which specifically detects PKCλ (data not shown). Unfortunately, in our hands all the available antibodies directed against PKCζ also recognize PKCλ (data not shown), so we have restricted our analysis to PKCλ, which complicates understanding of the distribution of PKCζ reported in previous publication. Western analysis using the anti-PKCλ (mAb) revealed a single band migrating at 70 kD in extracts of NIH3T3 and MDCKII cells, indicating that both cell lines express the PKCλ isoform of aPKC (Fig. 5 a).
To test the association of ASIP with PKCλ in vivo, we used the ASIP-specific antibody, C2, and PKCλ (mAb). Immunoprecipitation of NIH3T3 cell extracts with anti-ASIP antibodies followed by Western blotting using anti-PKCλ antibody detected a band corresponding to PKCλ (Fig. 5 b). In MDCKII cells, immunoprecipitates obtained using anti-ASIP antibodies (C2) contain PKCλ as well as the two ASIP bands (180 kD and 150 kD; Fig. 5 c). These results provide strong evidence that ASIP associates with PKCλ in vivo. Whether endogenous PKCζ also interacts with ASIP remains to be clarified because of the lack of PKCζ-specific antibodies.
Colocalization of ASIP with PKCλ at the Junctional Complexin MDCKII and NIH3T3 Cells
C. elegans PAR-3 becomes localized asymmetrically in the anterior periphery, and affects the asymmetric distribution of other cytoplasmic components. Conservation of the overall amino acid sequence and the aPKC-binding activity between ASIP and C. elegans PAR-3 led to our recent findings that a C. elegans homologue of aPKC, PKC-3, is required for establishing embryonic polarity, that PKC-3 localizes in the anterior periphery of the C. elegans one-cell embryo in a manner very similar to that of PAR-3, and that PKC-3 and PAR-3 are mutually required for their asymmetric distributions (Tabuse et al., 1998). These findings not only provide strong evidence that aPKC and PAR-3 cooperate together to determine cell polarity in the C. elegans embryo, but also suggest the physiological importance of the interaction between ASIP and aPKC. The conservation of the biochemical relationship between ASIP/PAR-3 and aPKC/PKC-3 led us to investigate whether ASIP and PKCλ are involved in regulating cell polarity as observed in C. elegans embryos.
In mammals, cell polarity has been studied intensively using cultured epithelial cells such as MDCKII cells. To evaluate the above possibility that the protein complex of ASIP and aPKC is involved in regulating epithelial cell polarity, and to test further whether ASIP and PKCλ physically interact in vivo, we examined the cellular distribution of ASIP and PKCλ in MDCKII cells by double-label immunofluorescence microscopy. The results were as expected and exciting in that ASIP and PKCλ showed overlapping localizations in vivo. Furthermore, they localize at junctional complexes including tight junctions and adherens junctions.
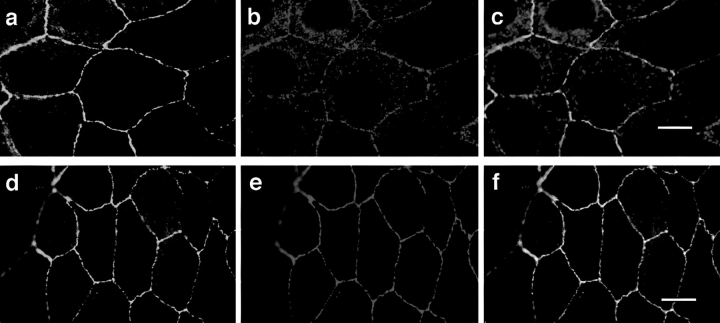
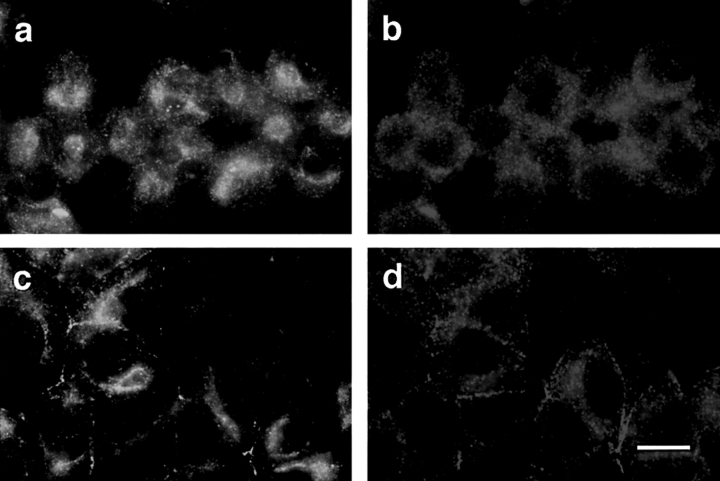
As shown in Fig. 6, ASIP staining (green) appears as a noncontinuous belt-like structure along the cell–cell contact region (Fig. 6 a). PKCλ staining (red) appears in a very similar pattern in addition to punctate staining throughout the cytoplasm (Fig. 6 b). Importantly, there is considerable overlap in the staining patterns for ASIP and PKCλ at the cell–cell contact region (orange/yellow) (Fig. 6 c). Furthermore, ASIP staining (green; Fig. 6 d) almost completely overlaps in double immunostaining with ZO-1 (red; Fig. 6, e and f), a protein known to localize at the tight junctions of epithelial cells. Confocal dissection revealed that signals for both antibodies are concentrated in the cell–cell contact region facing the apical membrane domain (data not shown). These results not only strongly support the in vivo association of ASIP and PKCλ, but also suggest that both proteins are components of the epithelial junctional complex. Light microscopy doesn't allow us to distinguish whether this staining reflects localization to the tight junction, the adherens junction, or to both. As described above, the anti-ASIP antibodies used in the present study all detect two ASIP bands, at 180 kD and 150 kD in MDCKII cell extracts. Overexpression of the 180-kD band by introducing full-length ASIP cDNA results in an increase in the staining at the cell–cell contact region (data not shown), clearly indicating that the 180-kD ASIP localizes to the junctional complex in MDCKII cells.
Figure 6.
Colocalization of ASIP with aPKC and ZO-1 at cell junctions in MDCKII cells. Confluent MDCKII cells were doubly stained with anti-ASIP (C2) antibody (a and d, green) and anti-PKCλ (mAb) antibody (b, red) or anti-ZO-1 (e, red) followed by FITC-conjugated anti-rabbit IgG and Cy3-conjugated anti-mouse IgG antibodies. The yellow and orange staining in c and f indicates the colocalization of ASIP and PKCλ or ZO-1. Bars (c and f), 10 μm.
Essentially similar results are obtained for confluent cultures of NIH3T3 cells (Fig. 7), where adherens junctions are formed at the cell–cell contact region (Reynolds et al., 1996). In NIH3T3 cells, however, localization of ASIP and PKCλ to the cell–cell contact region is not so complete as observed for MDCKII cells. More precisely, PKCλ staining (red) mostly appears as a punctate pattern throughout the cytoplasm (Fig. 7 b), whereas localization of ASIP is mostly concentrated in the cell–cell contact region (Fig. 7 a). Importantly, there is again considerable overlap in the staining patterns for ASIP and PKCλ in the cell–cell contact region (orange/yellow; Fig. 7 c). In addition, the ASIP staining (green; Fig. 7 d) also overlaps with that of ZO-1 (Fig. 7, e and f) in the cell–cell contact region in NIH3T3 cells. ZO-1 has been reported to localize at adherens junctions in confluent fibroblast cultures (Howarth et al., 1992; Itoh et al., 1993; Yonemura et al., 1995). Since NIH3T3 cells express only the 180-kD ASIP, the ASIP staining in NIH3T3 cells is derived from this isoform. ASIP staining in nonconfluent cultures of NIH3T3 cells appears as punctate structures distributed in the cytoplasm in a manner similar to that of PKCλ (data not shown).
Figure 7.
Colocalization of ASIP with aPKC and ZO-1 at cell junctions in NIH3T3 cells. Confluent NIH3T3 cells were doubly stained with anti-ASIP (C2) antibody (a and d, green) and anti-PKCλ antibody (mAb) (b, red) or anti-ZO-1 (e, red) followed by FITC-conjugated anti-rabbit IgG and Cy3-conjugated anti-mouse IgG antibodies. The yellow and orange staining in c and f indicates the colocalization of ASIP and PKCλ or ZO-1. Bars (c and f), 10 μm.
Ca2+-dependent Distribution of PKCλ and ASIP at the Junctional Complex of MDCKII Cells
To examine the requirement of cell–cell contact for the localization of PKCλ and ASIP at the junctional complex in MDCKII cells, we monitored the localization of PKCλ and ASIP during the formation and disappearance of cell– cell contacts. Previous experiments have shown that Ca2+ depletion from the culture medium of MDCKII cells results in the disappearance of cell–cell contacts and loss of tight junctions and adherens junctions (Gonzalez-Mariscal et al., 1985; Rajasekaran et al., 1996; Yamamoto et al., 1997). Furthermore, adding Ca2+ to the culture medium causes formation of cell–cell contacts with functional junctional complexes. As shown in Fig. 8, a and b, Ca2+ depletion from the culture medium results in cell rounding and disappearance of the specific staining for ASIP (green) and PKCλ (red) at the cell surface. Most of the staining shows a cytoplasmic distribution that is slightly different for PKCλ and ASIP. These results suggest that colocalization of ASIP and PKCλ require the presence of cell–cell contacts. Fig. 8, c and d, show that adding Ca2+ results in reconcentration of PKCλ and ASIP in the cell–cell contact region within 2 h. These properties of ASIP and PKCλ are very similar to those of ZO-1 and E-cadherin (data not shown; Rajasekaran et al., 1996), and support the notion that ASIP and PKCλ are peripheral components of cell– cell junctions.
Figure 8.
Immunofluorescence localization of ASIP and aPKC in Ca2+ switch experiments with MDCKII cells. Subconfluent MDCKII cells were transferred to low Ca2+ medium (growth medium containing 4 mM EGTA) for 6 h (a and b), and were then transferred back to normal Ca2+ medium for 2 h (c and d). The cells were doubly stained with anti-ASIP (C2) antibody (a and c) and anti-PKCλ (mAb) antibody (b and d), followed by FITC-conjugated anti-rabbit IgG and Cy3-conjugated anti-mouse IgG antibodies. Bar (d), 10 μm.
Localization of ASIP and PKCλ at Tight Junctions in Intestinal Epithelial Cells
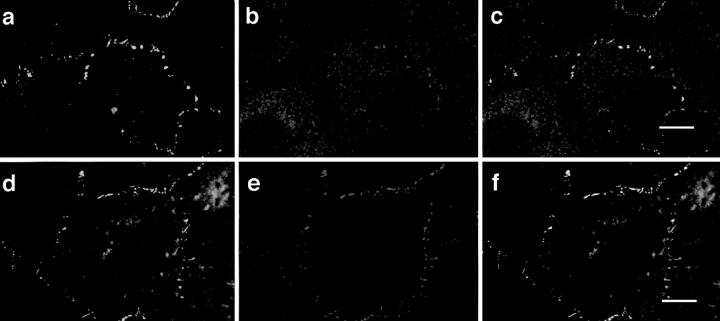
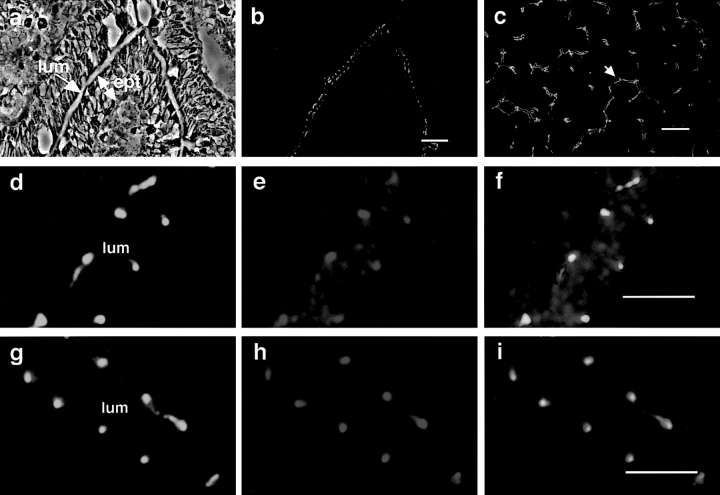
To analyze the subcellular localization of the ASIP-PKCλ complex in epithelial cells, we examined the distribution of ASIP and PKCλ in several rat tissues with epithelial junctional complexes. Small intestine is a typical tissue with differentiated epithelial cells containing well-developed cell–cell junctions. As shown in Fig. 9, a and b, ASIP staining (green) is concentrated at the lumen side of the epithelial cells where tight junctions and adherens junctions are concentrated. Fig. 9 c shows another example of well-differentiated cell–cell junctions at the bile capillary formed from hepatocytes. Again, ASIP staining is concentrated at the cell–cell junctions. Fig. 9, d, e, and f show enlarged views of the intestinal epithelium, clearly indicating localization of both ASIP (green) and PKCλ (red) staining at the cell–cell junctions; double staining indicates colocalization of the two proteins (Fig. 9 f). Fig. 9, g, h, and i show the results of similar experiments using antibodies for ASIP (green) and ZO-1 (red), indicating colocalization of ASIP and ZO-1. These results not only confirm the association between ASIP and PKCλ in vivo, but also demonstrate the strong correlation between the ASIP-PKCλ complex and the junctional complex.
Figure 9.
Colocalization of ASIP with aPKC and ZO-1 at cell junctions in rat intestinal epithelium and hepatic bile capillaries. (a) Phase contrast images of frozen cryosections of rat small intestine. lum, lumen; ept, epithelium. (b and c) The localization of ASIP immunofluorescence in frozen sections of rat small intestine (b) and liver (c). b shows the same fields as a. The arrowhead in c shows a bile capillary. (d–i) Enlarged view of the intestinal epithelia. The samples were doubly stained with anti-ASIP (C2) antibody (d and g, green) and anti-PKCλ (mAb) antibody (e, red) or anti-ZO-1 (h, red) followed by FITC-conjugated anti-rabbit IgG and Cy3-conjugated anti-mouse IgG antibodies. The yellow and orange staining in f and i indicates colocalization of ASIP and PKCλ or ZO-1. Bars (b and c), 20 μm; bars (f and i), 5 μm.
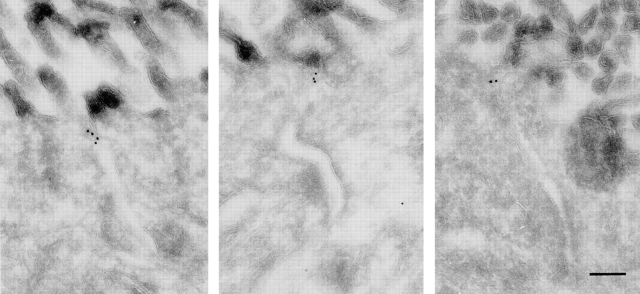
We next used immuno-gold EM to examine whether ASIP localizes to tight junctions or adherens junctions in rat epithelia. Fig. 10 shows examples indicating that ASIP localizes to tight junctions, but not to adherens junctions in intestinal epithelia. This feature is very similar to that of ZO-1. These results provide strong evidence that ASIP and aPKC are peripheral components of tight junctions in epithelial cells.
Figure 10.
Ultrastructural localization of ASIP in rat small intestine. Immunogold EM of the small intestinal epithelium. The labeling for ASIP is localized in the tight junction. The adherens junction, nonjunctional plasma membrane, and cytoplasm are not labeled. Bar, 200 nm.
Discussion
ASIP is a Physiological Partner of PKCλ in Differentiated Mammalian Cells
Protein kinase C plays a major role in intracellular signaling through lipid-derived second messengers (Nishizuka, 1995). However, the existence of multiple protein kinase C isoforms, their different sensitivities to lipids and calcium, and their diverse tissue and subcellular distribution suggest that different isoforms may have quite different physiological roles (Nishizuka, 1995; Mochly-Rosen and Gordon, 1998). Although overexpression studies and the use of pseudosubstrate peptides or dominant negative mutants of PKC isotypes have provided evidence consistent with the involvement of specific PKC isotypes in distinct cellular functions, their roles remain mostly elusive. As an alternative way to sort out these roles, efforts are being made to identify and study proteins that interact directly with different PKC isotypes. For example, attempts to understand the role of the atypical class of PKCs have led to the identification of several interacting proteins. These include p62/ZIP (PKCζ interacting protein), LIP (PKCλ interacting protein), PAR-4 (which is induced upon cell apoptosis and is completely different from C. elegans polarity gene par-4), and Ras (Puls et al., 1997; Diaz-Meco et al., 1996a ; Diaz-Meco et al., 1996b ; Diaz-Meco et al., 1994). Demonstrating that these interactions are physiologically relevant is the next step.
In this study, we identified a novel protein, ASIP, that interacts with aPKC isotypes, PKCζ and PKCλ, but not with other PKC isotypes including cPKC (PKCα) and nPKC (PKCδ). This interaction is direct and involves the kinase domain of aPKC binding to ASIP within a 225– amino acid region. Importantly, this interaction is shown for endogenous proteins in NIH3T3 cells and in MDCKII cells. Furthermore, we provide evidence that a protein complex containing ASIP and PKCλ is a peripheral component of tight junctions in epithelial cells. Quite interestingly, ASIP shows strong overall sequence similarity to the C. elegans polarity protein PAR-3, and the interaction between ASIP and aPKC is conserved for PAR-3 and aPKC. As described above, this led to our recent findings that a C. elegans homologue of aPKC, PKC-3, is required for establishing embryonic polarity, binds to PAR-3 in COS cells, and colocalizes with PAR-3 at the anterior periphery of the one-cell embryo (Tabuse et al., 1998).
ASIP as well as PAR-3 does not contain any sequence motifs, suggesting catalytic activity, but does contain three PDZ domains in CR2. PDZ domains (also called DHR or GLGF) are found in many proteins such as the Drosophila Dlg, PSD-95, and ZO-1, leading to the current understanding that PDZ domains serve as protein–protein interaction domains to localize protein complexes to the cell periphery (Saras and Heldin, 1996; Sheng, 1996). Furthermore, analysis of the InaD protein, which contains five PDZ domains each interacting with a different protein in the Drosophila phototransduction pathway, suggested that PDZ domain–containing proteins may serve as scaffolds upon which components of signaling complexes are assembled (Tsunoda et al., 1997). In addition to CR2, the CR1 region is conserved from worm to mammals, and may promote self-association of ASIP since the CR1 motif can interact with itself in the yeast two-hybrid system (T. Hirose, Y. Izumi, and S. Ohno, unpublished results). Thus, we propose that ASIP and PAR-3 serve as scaffolds for aPKC and other proteins to organize a signaling complex. Candidates for additional components in the ASIP/PAR-3 signaling complex include the previously reported aPKC-binding proteins, proteins localizing at tight junctions (see below), and the putative mammalian homologues of C. elegans of par-5 and par-6, genes required for the proper localization of PAR-3 (Watts et al., 1996; Guo and Kemphues, 1996).
ASIP and aPKC are Peripheral Components of Cell–Cell Junctions
Immunofluorescence microscopy reveals that ASIP and PKCλ colocalize to the most apical end of the cell–cell contact region in MDCKII cells, and to the apical junction in intact epithelial cells in rat intestine. Evidence that this distribution reflects association with the tight junction comes from colocalization of ASIP with the tight junction component ZO-1 in both cases examined, the requirement for Ca2+ to maintain this localization in MDCKII cells, and immunoelectron microscopy in intestinal epithelia. The localization of PKCλ to the cell–cell contact region is consistent with the previous observation that the signals obtained using anti-PKCζ antibodies overlap with those of ZO-1 in MDCKII cells (Saxon et al., 1994; Stuart and Nigam, 1995; Dodane and Kachar, 1996).
Tight junctions are the most apical intercellular junctions of epithelial and endothelial cells, and create a regulatable semipermeable diffusion barrier between individual cells (Balda and Matter, 1998). On a cellular level, they form an intramembrane diffusion fence that restricts the intermixing of apical and basolateral membrane components, leading to the asymmetric distribution of proteins and other cytoplasmic structures. These functions of tight junctions are dynamically regulated, but the molecular mechanism remains elusive. Identification of proteins localizing at tight junctions is starting to help us to understand the function of and regulation of tight junctions. These include proteins localizing at tight junctions such as ZO-1 (Stevenson et al., 1986; Willott et al., 1993; Itoh et al., 1993), ZO-2 (Gumbiner et al., 1991; Jesaitis and Goodenough, 1994), and p130/ZO-3 (Balda et al., 1993; Haskins et al., 1998), which share sequence similarities to Drosophila tumor suppressor DlgA, a tumor suppressor and peripheral component of septate junctions, the Drosophila version of mammalian tight junction (Woods et al., 1996). In addition, cingulin (Citi et al., 1988), 7H6 (Zhong et al., 1993), Rab3B (Weber et al., 1994), symplekin (Keon et al., 1996), and AF-6 (Yamamoto et al., 1997) are known to be present at tight junctions. However, limited information is available concerning the roles these proteins play in the function and regulation of the tight junction.
Although some tight junction proteins such as ZO-1, ZO-2, p130/ZO-3, and cingulin are phosphorylated, there are no correlations between phosphorylation level and tight junction assembly (Balda et al., 1993; Citi and Denisenko, 1995). The presence of the consensus PKC phosphorylation sequence in the CR3 of ASIP and PAR-3 suggests that PKCλ regulates this complex through phosphorylation. Consistent with this notion, ASIP domains, expressed as GST-fusion proteins in E. coli, are efficiently phosphorylated by recombinant mouse PKCζ (data not shown). The identification of ASIP and PKCλ as proteins locating at tight junctions has the potential to accelerate understanding of tight junction regulation or function.
We have shown that ASIP colocalizes with ZO-1 to the cell–cell contact region in confluent cultures of NIH3T3 cells, which do not form tight junctions. Since ZO-1 localizes to the adherens junction in nonepithelial cells (Howarth et al., 1992; Itoh et al., 1993; Yonemura et al., 1995), it appears that ASIP and PKCλ are components of adherens junctions in cells that do not form tight junctions. Not all of the PKCλ colocalizes with ASIP at putative junctional complexes. Punctate staining is detectable in the cytoplasm of MDCKII cells and confluent NIH3T3 cells. One possibility is that this cytoplasmic staining represents PKCλ localization to the lysosome-targeted endosomal compartment as was recently described for PKCλ/ι and its binding partner ZIP/p62 (Sanchez et al., 1998).
Analogy Between Epithelial Cell Polarity and That in Worm Embryogenesis
The conservation of a physical interaction between aPKC and ASIP/PAR-3 in worms and mammals raises the intriguing possibility that they are components of a widely used signaling complex whose role might include the determination of cell polarity. Another component of the C. elegans polarity system, PAR-1, is a member of a conserved family of kinases with several mammalian members. Rat family members MARK1 and MARK2 have been shown to phosphorylate microtubule-associated proteins, and the overexpression of MARK1 or MARK2 disrupts microtubule arrays in CHO cells (Drewes et al., 1997). Two other family members, a human protein, Kp78, and a mouse protein, EMK, localize asymmetrically in polarized epithelial cells (Parsa, 1988; Böhm et al., 1997). Interestingly, the mouse homologue of MARK2, EMK, is distributed on the lateral surface of MDCK cells, and this distribution ends at the tight junction where ZO-1 localizes (Böhm et al., 1997). Taken together with the findings on C. elegans PKC-3 (Tabuse et al., 1998), the present study supports earlier speculation about a possible parallel between C. elegans embryos and mammalian epithelial cells (Fig. 11; Böhm et al., 1997).
Figure 11.
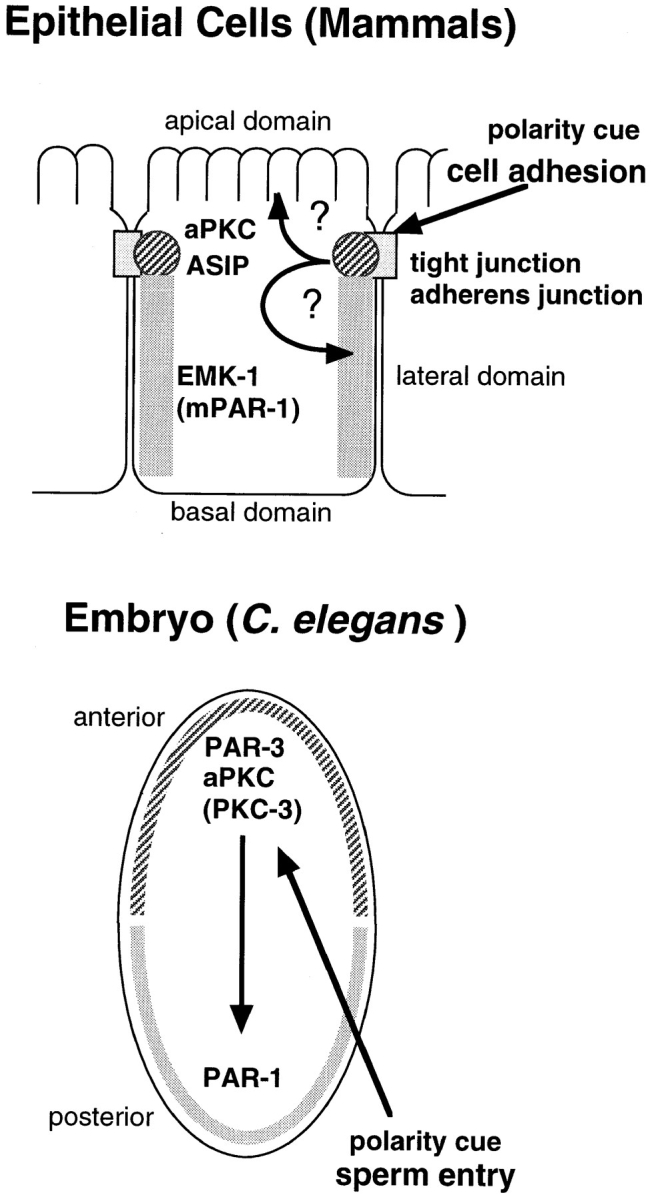
Comparison of the polarized asymmetric distribution of the protein complex of aPKC and PAR-3/ASIP between the C. elegans one-cell embryo and differentiated mammalian epithelial cells. In the C. elegans one-cell embryo, PAR-3 localizes at the anterior periphery with aPKC and determines the distribution of PAR-1, whereas PAR-1 localizes at the posterior periphery in a reciprocal manner. In mammalian epithelial cells, ASIP and PKCλ colocalize at tight junctions, whereas EMK (m-PAR-1) localizes in the lateral domain.
As clearly demonstrated in the present study, ASIP and aPKC are components of tight junctions in epithelial cells, and may be involved in the function of tight junctions. They also localize to adherens junctions in cells without tight junctions, and are expressed in cells without cell–cell junctions. These results support the hypothesis that the protein complex of ASIP and PKCλ is involved in the organization of cell polarity in epithelial cells. Alternatively, the protein complex might be a type of conserved biochemical machine used for diverse cellular functions. Clarification of the nature of the ASIP-aPKC protein complex will greatly enhance our understanding of the function of cell–cell junctions in the organization of cells, and hence the mechanism regulating cell growth and differentiation in multicellular organisms.
Acknowledgments
We thank Dr. Shoichiro Tsukita and Dr. Masahiko Ito (Kyoto University) for the anti-ZO-1 monoclonal antibody, MDCKII cells, and discussion, Dr. Hiroshi Nojima (Osaka University) for the rat 3Y1-cDNA library, and Kumi Noda for technical help. This work was supported by grants to S. Ohno and Y. Izumi from the Ministry of Education, Science, Sports, and Culture of Japan and Japanese Society for the Promotion of Science.
Abbreviations used in this paper
- ASIP
atypical PKC isotype–specific interacting protein
- GST
glutathione-S-transferase
- PKC
protein kinase C
- PVDF
polyvinylidene difluoride
- tag-ASIP
epitope-tagged-ASIP
Footnotes
Address all correspondence to Dr. Shigeo Ohno, Department of Molecular Biology, Yokohama City University School of Medicine, Kanazawa-ku, Yokohama 236-0004, Japan. Tel.: 81-45-787-2596. Fax: 81-45-785-4140. E-mail: ohnos@med.yokohama-cu.ac.jp
References
- Akimoto K, Mizuno K, Osada S, Hirai S, Tanuma S, Suzuki K, Ohno S. A new member of the third class in the protein kinase C family, PKCλ, expressed dominantly in an undifferentiated mouse embryonal carcinoma cell line and also in many tissues and cells. J Biol Chem. 1994;269:12677–12683. [PubMed] [Google Scholar]
- Akimoto K, Takahashi R, Moriya S, Nishioka N, Takayanagi J, Kimura K, Fukui Y, Osada S, Mizuno K, Hirai S, et al. EGF or PDGF receptors activate atypical PKCλ through phosphatidylinositol 3-kinase. EMBO (Eur Mol Biol Organ) J. 1996;15:788–798. [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Gonzalez-Mariscal L, Matter K, Cereijido M, Anderson JM. Assembly of the tight junction: the role of diacylglycerol. J Cell Biol. 1993;123:293–302. doi: 10.1083/jcb.123.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Matter K. Tight junctions. J Cell Sci. 1998;111:541–547. doi: 10.1242/jcs.111.5.541. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay G, Standaert ML, Galloway L, Moscat J, Farese RV. Evidence for involvement of protein kinase C (PKC)-ζ and noninvolvement of diacylglycerol-sensitive PKCs in insulin-stimulated glucose transport in L6 myotubes. Endocrinology. 1997;138:4721–4731. doi: 10.1210/endo.138.11.5473. [DOI] [PubMed] [Google Scholar]
- Berra E, Diaz-Meco MT, Dominguez I, Municio MM, Sanz L, Lozano J, Chapkin RS, Moscat J. Protein kinase Cζ isoform is critical for mitogenic signal transduction. Cell. 1993;74:555–563. doi: 10.1016/0092-8674(93)80056-k. [DOI] [PubMed] [Google Scholar]
- Berra E, Diaz-Meco MT, Lozano J, Frutos S, Municio MM, Sanchez P, Sanz L, Moscat J. Evidence for a role of MEK and MAPK during signal transduction by protein kinase Cζ. EMBO (Eur Mol Biol Organ) J. 1995;14:6157–6163. doi: 10.1002/j.1460-2075.1995.tb00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H, Brinkmann V, Drab M, Henske A, Kurzchalia TV. Mammalian homologues of C. elegansPAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Curr Biol. 1997;7:603–606. doi: 10.1016/s0960-9822(06)00260-0. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Ingram MK, Hunter CP. The maternal par genes and the segregation of cell fate specification activities in early Caenorhabditis elegansembryos. Development. 1997;124:3815–3826. doi: 10.1242/dev.124.19.3815. [DOI] [PubMed] [Google Scholar]
- Cabral JH, Petosa C, Sutcliffe MJ, Raza S, Byron O, Poy F, Marfatia SM, Chishti AH, Liddington RC. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- Caplan MJ. Membrane polarity in epithelial cells: protein sorting and establishment of polarized domains. Am J Physiol. 1997;272:F425–F429. doi: 10.1152/ajprenal.1997.272.4.F425. [DOI] [PubMed] [Google Scholar]
- Citi S, Denisenko N. Phosphorylation of the tight junction protein cingulin and the effects of protein kinase inhibitors and activators in MDCK epithelial cells. J Cell Sci. 1995;108:2917–2926. doi: 10.1242/jcs.108.8.2917. [DOI] [PubMed] [Google Scholar]
- Citi S, Sabanay H, Jakes R, Geiger B, Kendrick-Jones J. Cingulin, a new peripheral component of tight junctions. Nature. 1988;333:272–276. doi: 10.1038/333272a0. [DOI] [PubMed] [Google Scholar]
- Diaz-Meco MT, Lozano J, Municio MM, Berra E, Frutos S, Sanz L, Moscat J. Evidence for the in vitro and in vivo interaction of Ras with protein kinase Cζ. J Biol Chem. 1994;269:31706–31710. [PubMed] [Google Scholar]
- Diaz-Meco MT, Municio MM, Frutos S, Sanchez P, Lozano J, Sanz L, Moscat J. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell. 1996a;86:777–786. doi: 10.1016/s0092-8674(00)80152-x. [DOI] [PubMed] [Google Scholar]
- Diaz-Meco MT, Municio MM, Sanchez P, Lozano J, Moscat J. Lambda-interacting protein, a novel protein that specifically interacts with the zinc finger domain of the atypical protein kinase C isotype λ/ι and stimulates its kinase activity in vitro and in vivo. Mol Cell Biol. 1996b;16:105–114. doi: 10.1128/mcb.16.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodane V, Kachar B. Identification of isoforms of G proteins and PKC that colocalize with tight junctions. J Membr Biol. 1996;149:199–209. doi: 10.1007/s002329900020. [DOI] [PubMed] [Google Scholar]
- Dominguez I, Diaz-Meco MT, Municio MM, Berra E, Garcia de Herreros A, Cornet ME, Sanz L, Moscat J. Evidence for a role of protein kinase Cζ subspecies in maturation of Xenopus laevisoocytes. Mol Cell Biol. 1992;12:3776–3783. doi: 10.1128/mcb.12.9.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Eaton S, Simons K. Apical, basal, and lateral cues for epithelial polarization. Cell. 1995;82:5–8. doi: 10.1016/0092-8674(95)90045-4. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegansembryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Fujise A, Mizuno K, Ueda Y, Osada S, Hirai S, Takayanagi A, Shimizu N, Owada MK, Nakajima H, Ohno S. Specificity of the high affinity interaction of protein kinase C with a physiological substrate, myristoylated alanine-rich protein kinase C substrate. J Biol Chem. 1994;269:31642–31648. [PubMed] [Google Scholar]
- Goldstein B, Hird SN. Specification of the anteroposterior axis in Caenorhabditis elegans. . Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L, Chavez de Ramirez B, Cereijido M. Tight junction formation in cultured epithelial cells (MDCK) J Membr Biol. 1985;86:113–125. doi: 10.1007/BF01870778. [DOI] [PubMed] [Google Scholar]
- Griffiths G, Simons K, Warren G, Tokuyasu KT. Immunoelectron microscopy using thin frozen section: application to studies of the intracellular transport of Semliki forest virus spike glycoproteins. Methods Enzymol. 1986;96:466–483. doi: 10.1016/s0076-6879(83)96041-x. [DOI] [PubMed] [Google Scholar]
- Gumbiner B, Lowenkopf T, Apatira D. Identification of a 160-kDa polypeptide that binds to the tight junction protein ZO-1. Proc Natl Acad Sci USA. 1991;88:3460–3464. doi: 10.1073/pnas.88.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegansembryo. Curr Opin Genet Dev. 1996;6:408–415. doi: 10.1016/s0959-437x(96)80061-x. [DOI] [PubMed] [Google Scholar]
- Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng HH, Squire J, Tsui LC. High-resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc Natl Acad Sci USA. 1992;89:9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng HH, Tsui LC. Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma. 1993;102:325–332. doi: 10.1007/BF00661275. [DOI] [PubMed] [Google Scholar]
- Howarth AG, Hughes MR, Stevenson BR. Detection of the tight junction-associated protein ZO-1 in astrocytes and other nonepithelial cell types. Am J Physiol. 1992;262:C461–C469. doi: 10.1152/ajpcell.1992.262.2.C461. [DOI] [PubMed] [Google Scholar]
- Itoh M, Nagafuchi A, Yonemura S, Kitani-Yasuda T, Tsukita S, Tsukita S. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Hirai S, Tamai Y, Fujise-Matsuoka A, Nishimura Y, Ohno S. A protein kinase Cδ-binding protein SRBC whose expression is induced by serum starvation. J Biol Chem. 1997;272:7381–7389. doi: 10.1074/jbc.272.11.7381. [DOI] [PubMed] [Google Scholar]
- Jesaitis LA, Goodenough DA. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophiladiscs-large tumor suppressor protein. J Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keon BH, Schafer S, Kuhn C, Grund C, Franke WW. Symplekin, a novel type of tight junction plaque protein. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric cell division during animal development. Curr Opin Cell Biol. 1997;9:833–841. doi: 10.1016/s0955-0674(97)80085-3. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- Lozano J, Berra E, Municio MM, Diaz-Meco MT, Dominguez I, Sanz L, Moscat J. Protein kinase Cζ isoform is critical for κB-dependent promoter activation by sphingomyelinase. J Biol Chem. 1994;269:19200–19202. [PubMed] [Google Scholar]
- Mochly-Rosen D, Gordon AS. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J. 1998;12:35–42. [PubMed] [Google Scholar]
- Müller G, Ayoub M, Storz P, Rennecke J, Fabbro D, Pfizenmaier K. PKCζ is a molecular switch in signal transduction of TNF-α, bifunctionally regulated by ceramide and arachidonic acid. EMBO (Eur Mol Biol Organ) J. 1995;14:1961–1969. doi: 10.1002/j.1460-2075.1995.tb07188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Brewer KA, Exton JH. Activation of the ζ isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1993;268:13–16. [PubMed] [Google Scholar]
- Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;9:484–496. [PubMed] [Google Scholar]
- Ohno S, Mizuno K, Adachi Y, Hata A, Akita Y, Akimoto K, Osada S, Hirai S, Suzuki K. Activation of novel protein kinases Cδ and ε upon mitogenic stimulation of quiescent rat 3Y1 fibroblasts. J Biol Chem. 1994;269:17495–17501. [PubMed] [Google Scholar]
- Osada S, Mizuno K, Saido TC, Suzuki K, Kuroki T, Ohno S. A new member of the protein kinase C family, nPKCθ, predominantly expressed in skeletal muscle. Mol Cell Biol. 1992;12:3930–3938. doi: 10.1128/mcb.12.9.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa I. Loss of a Mr 78,000 marker in chemically induced transplantable carcinomas and primary carcinoma of human pancreas. Cancer Res. 1988;48:2265–2272. [PubMed] [Google Scholar]
- Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase Cζ with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci USA. 1997;94:6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran AK, Hojo M, Huima T, Rodriguez-Boulan E. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J Cell Biol. 1996;132:451–463. doi: 10.1083/jcb.132.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Daniel JM, Mo YY, Wu J, Zhang Z. The novel catenin p120cas binds classical cadherins and induces an unusual morphological phenotype in NIH3T3 fibroblasts. Exp Cell Res. 1996;225:328–337. doi: 10.1006/excr.1996.0183. [DOI] [PubMed] [Google Scholar]
- Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18:3069–3080. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saras J, Heldin CH. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- Saxon ML, Zhao X, Black JD. Activation of protein kinase C isozymes is associated with post-mitotic events in intestinal epithelial cells in situ. J Cell Biol. 1994;126:747–763. doi: 10.1083/jcb.126.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M. PDZs and receptor/channel clustering: rounding up the latest suspects. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Standaert ML, Galloway L, Karnam P, Bandyopadhyay G, Moscat J, Farese RV. Protein kinase C-ζ as a downstream effector of phosphatidylinositol 3-kinase during insulin stimulation in rat adipocytes. Potential role in glucose transport. J Biol Chem. 1997;272:30075–30082. doi: 10.1074/jbc.272.48.30075. [DOI] [PubMed] [Google Scholar]
- Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RO, Nigam SK. Regulated assembly of tight junctions by protein kinase C. Proc Natl Acad Sci USA. 1995;92:6072–6076. doi: 10.1073/pnas.92.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical Protein Kinase C Cooperates with PAR-3 to Establish Embryonic Polarity In Caenorhabditis elegans. . Development. 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Toker A, Cantley LC. Signaling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT. Use of poly(vinylpyrrolidone) and poly(vinyl alcohol) for cryoultramicrotomy. Histochem J. 1989;21:163–171. doi: 10.1007/BF01007491. [DOI] [PubMed] [Google Scholar]
- Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker CS. A multivalent PDZ-domain protein assembles signaling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ. par-6, a gene involved in the establishment of asymmetry in early C. elegansembryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- Ways DK, Posekany K, deVente J, Garris T, Chen J, Hooker J, Qin W, Cook P, Fletcher D, Parker P. Overexpression of protein kinase C-ζ stimulates leukemic cell differentiation. Cell Growth Differ. 1994;5:1195–1203. [PubMed] [Google Scholar]
- Weber E, Berta G, Tousson A, St. John P, Green MW, Gopalokrishnan U, Jilling T, Sorscher EJ, Elton TS, Abrahamson DR, et al. Expression and polarized targeting of a rab3 isoform in epithelial cells. J Cell Biol. 1994;125:583–594. doi: 10.1083/jcb.125.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willott E, Balda MS, Fanning AS, Jameson B, Van Itallie C, Anderson JM. The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppressor protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DF, Hough C, Peel D, Callaini G, Bryant PJ. Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophilaepithelia. J Cell Biol. 1996;134:1469–1482. doi: 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten MW, Zhou G, Seibenhener ML, Coleman ES. A role for ζ protein kinase C in nerve growth factor-induced differentiation of PC12 cells. Cell Growth Differ. 1994;5:395–403. [PubMed] [Google Scholar]
- Xu J, Zutter MM, Santoro SA, Clark RA. PDGF induction of α2 integrin gene expression is mediated by protein kinase C-ζ. J Cell Biol. 1996;134:1301–1311. doi: 10.1083/jcb.134.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Harada N, Kano K, Taya S, Canaani E, Matsuura Y, Mizoguchi A, Ide C, Kaibuchi K. The ras target AF-6 interacts with ZO-1 and serves as a peripheral component of tight junctions in epithelial cells. J Cell Biol. 1997;139:785–795. doi: 10.1083/jcb.139.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Itoh M, Nagafuchi A, Tsukita S. Cell-to-cell adherens junction formation and actin filament organization: similarities and differences between non-polarized fibroblasts and polarized epithelial cells. J Cell Sci. 1995;108:127–142. doi: 10.1242/jcs.108.1.127. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Saitoh T, Minase T, Sawada N, Enomoto K, Mori M. Monoclonal antibody 7H6 reacts with a novel tight junction-associated protein distinct from ZO-1, cingulin and ZO-2. J Cell Biol. 1993;120:477–483. doi: 10.1083/jcb.120.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]