Abstract
Membrane fusion is required to establish the morphology and cellular distribution of the mitochondrial compartment. In Drosophila, mutations in the fuzzy onions (fzo) GTPase block a developmentally regulated mitochondrial fusion event during spermatogenesis. Here we report that the yeast orthologue of fuzzy onions, Fzo1p, plays a direct and conserved role in mitochondrial fusion. A conditional fzo1 mutation causes the mitochondrial reticulum to fragment and blocks mitochondrial fusion during yeast mating. Fzo1p is a mitochondrial integral membrane protein with its GTPase domain exposed to the cytoplasm. Point mutations that alter conserved residues in the GTPase domain do not affect Fzo1p localization but disrupt mitochondrial fusion. Suborganellar fractionation suggests that Fzo1p spans the outer and is tightly associated with the inner mitochondrial membrane. This topology may be required to coordinate the behavior of the two mitochondrial membranes during the fusion reaction. We propose that the fuzzy onions family of transmembrane GTPases act as molecular switches to regulate a key step in mitochondrial membrane docking and/or fusion.
Keywords: GTPase, membrane fusion, mitochondria, mtDNA, organelle morphology
The homotypic, or self-fusion, of mitochondrial membranes plays an important role in controlling the copy number and cellular distribution of these essential organelles (Bereiter-Hahn and Voth, 1994). In budding yeast and some mammalian cells, frequent mitochondrial fusion events are balanced by mitochondrial fission, maintaining the compartment as a highly branched, tubular network (Hoffman and Avers, 1973; Stevens, 1981; Koning et al., 1993; Nunnari et al., 1997; Hermann, 1998). Mitochondrial fusion also functions to remodel mitochondrial morphology during differentiation. Individual mitochondria in rat skeletal muscle cells undergo postembryonic fusion to generate a branched mitochondrial reticulum (Bakeeva et al., 1981). During spermatogenesis in insects, fusion generates giant mitochondria that associate with the axoneme of the sperm flagella (Lindsley and Tokuyasu, 1980; Fuller, 1993). The generation of such large, interconnected mitochondrial compartments is thought to facilitate the distribution of energy and metabolites as well as chemical and electrical signals throughout these cells (Bakeeva et al., 1978; Ichas et al., 1997). The genetic and molecular mechanisms that control mitochondrial fusion have not been defined.
Molecules that regulate the heterotypic and homotypic fusion of membranes in the secretory and endocytic pathways have been studied in some detail. Both types of fusion require the cytosolic factors NSF and SNAPs (or homologues) as well as compartment-specific integral membrane proteins termed v- and t-SNAREs (Denesvre and Malhotra, 1996; Pfeffer, 1996; Rothman, 1996; Hay and Scheller, 1997; Edwardson, 1998; Götte and Fischer von Mollard, 1998; Patel et al., 1998; Rabouille et al., 1998). During heterotypic fusion, v/t-SNARE pairing promotes the stable association of the vesicle with the target membrane and may be sufficient to catalyze bilayer mixing (Weber et al., 1998). In contrast, the homotypic fusion of ER and Golgi membranes depends on self-interactions among resident t-SNAREs (Patel et al., 1998; Rabouille et al., 1998). Yeast vacuole fusion requires a v/t-SNARE pairing for optimal fusion but can occur (with lower efficiency) between vacuoles containing only the t-SNARE partner (Nichols et al., 1997). Thus, in some cases, the recognition of “like” membranes in homotypic fusion reactions may be mediated by homomeric interactions between compartment-specific t-SNAREs. NSF and SNAPs appear to act generally during fusion to disrupt unproductive SNARE associations within a membrane before docking (Mayer et al., 1996; Otto et al., 1997) and may also regulate changes in SNARE conformation after docking but before membrane fusion (Söllner et al., 1993).
Although an isoform of the VAMP-1 SNARE has been reported to localize to mitochondria in transfected epithelial cells (Isenmann et al., 1998), molecules in the NSF, SNAP, and SNARE families have not been implicated in mitochondrial fusion. This is not entirely unexpected since mitochondrial fusion, unlike most other membrane fusion reactions in the cell, requires the sequential mixing of two distinct lipid bilayers. Moreover, the mitochondrial compartment is evolutionarily distinct from compartments of the secretory and endocytic pathways. Thus, the molecular machinery that mediates mitochondrial fusion is likely to be unique, even though the general mechanism of membrane fusion may be conserved. A novel gene required for mitochondrial fusion, fuzzy onions (fzo),1 was recently identified through the analysis of Drosophila mutants defective in spermatogenesis (Hales and Fuller, 1997). During spermatid differentiation in flies, individual mitochondria coalesce, fuse into two giant organelles, and coil tightly around one another forming the Nebenkern, a spherical structure that resembles an onion in cross section (Lindsley and Tokuyasu, 1980; Fuller, 1993). Mutations in the fzo gene disrupt mitochondrial fusion and result in disorganized aggregates of individual mitochondria that do not associate properly with the elongating axoneme (Hales and Fuller, 1997). fzo encodes a predicted transmembrane GTPase that is detected on the mitochondrial compartment just as fusion begins and disappears soon after fusion is complete. Together, the fzo mutant phenotype and the expression pattern of the Fzo protein suggest that this molecule either regulates, or is a direct mediator of, mitochondrial fusion.
The discovery of fzo homologues in yeasts, nematodes, and mammals defined a new family of multiple domain, high molecular weight GTPases, and raised the possibility that these molecules act generally to control mitochondrial fusion events in different organisms and cell types (Hales and Fuller, 1997). Fzo family members contain an amino-terminal GTPase domain, two adjacent carboxy-terminal transmembrane domains, and multiple heptad repeats (see Fig. 1 A) (Hales and Fuller, 1997). Here we show that the Saccharomyces cerevisiae orthologue of Drosophila Fzo, Fzo1p, is a mitochondrial integral membrane protein required to maintain a tubular mitochondrial reticulum during mitotic growth and demonstrate a direct role for this protein in mitochondrial fusion during yeast mating. Mutations in two conserved FZO1 GTPase motifs disrupt mitochondrial fusion but do not affect Fzo1p localization. Subcellular fractionation and protease protection experiments reveal that the amino terminus of the protein (containing the essential GTPase domain and multiple heptad repeats) extends into the cytoplasm where it could participate in organelle docking/fusion and interact with molecules that regulate GTP binding or hydrolysis. In addition, the distribution of Fzo1p in submitochondrial fractionation studies suggests that the carboxy-terminus of this protein may interact with both mitochondrial membranes. This topology is similar to that of SNARE molecules and viral fusion proteins and suggests that Fzo1p could play a direct role in the docking and fusion of mitochondrial compartments.
Figure 1.
S. cerevisiae FZO1 is required for mitochondrial function. (A) Domain structure of three Fzo family members showing the position of the predicted GTPase (GTPase), heptad repeat (Hep), and transmembrane (TM) domains. Amino acid identities between the entire protein sequences and the predicted GTPase domains of the S. cerevisiae Fzo1p, S. pombe Fzo1p (the newest family member), and D. melanogaster Fzo family members are indicated. GenBank/EMBL/DDBJ accession numbers are as follows: S. cerevisiae (Z36048), S. pombe (ALO23533), and D. melanogaster (U95821). (B) fzo1Δ cells exhibit growth defects on fermentable and nonfermentable carbon sources. Two fzo1Δ (top, JSY1809 and JSY1810) and two wild-type (bottom, JSY1811 and JSY1812) strains from a single tetrad were grown on YPDextrose medium for 2 d at 30°C (left) or YPGlycerol medium for 5 d at 30°C (right).
Materials and Methods
Strains and Genetic Techniques
All yeast strains (Table I) are isogenic to FY10 (Winston et al., 1995). All media including YPDextrose, YPGlycerol, SDextrose, SGalactose, and SRaffinose were prepared as described by Sherman et al. (1986). Strains were grown at 30°C unless otherwise noted. Genetic and molecular cloning manipulations followed standard techniques (Maniatis et al., 1982; Sherman et al., 1986; Baudin et al., 1993). The Escherichia coli host strain JM109 (Promega Corp., Madison, WI) was used for all bacterial manipulation of plasmids.
Table I.
Yeast Strains Used in This Study
| Strain | Genotype + (Plasmid) | |
|---|---|---|
| JSY999 | MATα leu2Δ1 his3Δ200 ura3-52 rho + | |
| JSY1373 | MATa/α leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 ura3-52/ura3-52 rho + | |
| JSY1808 | MATa/α leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 ura3-52/ura3-52 fzo1Δ::HIS3/FZO1 rho + | |
| JSY1809 | MATα leu2Δ1 his3Δ200 ura3-52 fzo1Δ::HIS3 rho0 | |
| JSY1810 | MATa leu2Δ1 his3Δ200 ura3-52 fzo1Δ::HIS3 rho0 | |
| JSY1811 | MATα leu2Δ1 his3Δ200 ura3-52 FZO1 rho + | |
| JSY1812 | MATa leu2Δ1 his3Δ200 ura3-52 FZO1 rho + | |
| JSY2028 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 FZO1::N-3XMYC rho + | |
| JSY2034 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 FZO1::N-3XMYC rho + | |
| JSY2038 | MATa leu2Δ1 his3Δ200 ura3-52 fzo1Δ::HIS3 rho + + (pRS416-FZO1) | |
| JSY2270 | MATa leu2Δ1 his3Δ200 ura3-52 FZO1 rho + + (pGAL1-N-3XMYC-FZO1) | |
| JSY2273 | MATa leu2Δ1 his3Δ200 ura3-52 fzo1Δ::HIS3 rho + + (pGAL1-N-3XMYC-FZO1) | |
| JSY2287 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 rho + + (pRS416-FZO1) | |
| JSY2288 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 FZO1 rho + + (pRS416-FZO1) | |
| JSY2354 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 + (pRS414) | |
| JSY2355 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 + (pRS414-fzo1[T221A]) | |
| JSY2356 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 + (pRS414-fzo1[K371A]) | |
| JSY2357 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 + (pRS414-fzo1[K200A]) | |
| JSY2358 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 + (pRS414-fzo1[S201N]) | |
| JSY2392 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 + (pRS414-FZO1) | |
| JSY2394 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 rho + + (pRS414-N-9XMYC-FZO1) | |
| JSY2555 | MATα leu2Δ1 his3Δ200 ura3-52 rho0 | |
| JSY2579 | MATa leu2Δ1 his3Δ200 ura3-52 fzo1Δ::HIS3 rho0 + (pRS416-FZO1) | |
| JSY2634 | MATa leu2Δ1 his3Δ200 ura3-52 fzo1Δ::HIS3 rho + + (pGAL1-N-3XMYC-FZO1) + (pRS416-ADH-COXIVpre-GFP) | |
| JSY2793 | MATα leu2Δ1 his3Δ200 ura3-52 trp1Δ63 fzo1Δ::HIS3 rho + + (pRS414-FZO1) + (pRS416-ADH-COXIVpre-GFP) | |
| JSY2802 | MATα leu2Δ1 his3Δ200 ura3-52 trp1Δ63 lys2Δ-202 fzo1Δ::HIS3 rho + + (pRS414-fzo1-1) + (pRS416-ADH-COXIVpre-GFP) | |
| JSY2804 | MATa leu2Δ1 his3Δ200 ura3-52 trp1Δ63 fzo1Δ::HIS3 rho + + (pRS414-fzo1-1) + (pRS416-ADH-COXIVpre-GFP) | |
| JSY2926 | MATα leu2Δ1 his3Δ200 ura3-52 trp1Δ63 fzo1Δ::HIS3 rho + + (pRS414-FZO1) |
Plasmid Construction
For pRS416-FZO1, a genomic fragment containing the FZO1 coding region plus 500 bp of 5′and 3′ flanking sequence was PCR amplified from JSY999 using primers containing engineered EcoRI sites: P297 (5′-GGGGGAATTCCCAGGTGACAGAATGTCTGGGTTGAAAG-3′) and P298 (5′-GGGGGAATTCCTTGCTCCTTGTTGTCTTTTAAATGGAG-3′), and cloned into the unique EcoRI site in pRS416 (verified by sequencing; Stratagene, La Jolla, CA). An EcoRI fragment from pRS416-FZO1 was subcloned into pRS414 (Stratagene) to generate pRS414-FZO1. The pRS414-FZO1 and pRS416-FZO1 plasmids complemented both the mitochondrial morphology defects and the loss of mtDNA in fzo1Δ. To generate a GAL1 regulated form of FZO1, a PCR fragment containing a 3XMYC tag at the amino terminus of FZO1 (N-3XMYC-FZO1) was amplified from JSY2028 (see below) using primers with engineered SalI, P379 (5′-GGGGGTCGACTATCTAATCGATGTCTAAATTTATTTCTTC-3′), and XbaI, P380 (5′-GGGGTCTAGATTAACGATGTCTAGGGAACAAAAGCTGGAG-3′), sites. The PCR fragment was cloned into pRS415-GAL1 (Mumberg et al., 1994) (American Type Culture Collection, Rockville, MD) to generate pGAL1-N-3XMYC-FZO1 (pRS415- GAL1-N-3XMYC-FZO1, verified by sequencing). Growth in SRaffinose provided sufficient expression of N-3XMYC-Fzo1p from the GAL1 promoter to complement the mitochondrial morphology and growth defects of fzo1Δ cells. To generate mutations in the conserved FZO1 GTPase domain, a 3.5-kb EcoRI fragment from pRS416-FZO1 was cloned into the EcoRI site of pALTER-1 (Promega Corp.) to create pALTER-1-FZO1. Site-directed mutagenesis (Altered Sites II; Promega Corp.) was performed with the following mutagenic oligonucleotides: K200A (5′-GATGTAAATACTGGCGCCTCAGCTCTTTGCAAC-3′, introduces an EheI site); S201N (5′-CAGGTGATGTAAATACTGGTAAAAATGCATTATGCAACTCTCTATTAAAGCAGCG-3′, introduces an NsiI site); T221A (5′-GGATCAGCTACCATGCGCAAATGTATTTTCCGAA-3′, introduces an Fsp I site); K371A (5′-GTTTTTTGTTGTGAAA GCTTTTGACAAAATCAGGG-3′, introduces a HindIII site). The mutants were identified by restriction digest and confirmed by sequence analysis. The mutagenized fzo1 genes were subcloned into the EcoRI sites of pRS414 and pRS424.
Generation and Characterization of fzo1Δ::HIS3 Cells
The fzo1Δ::HIS3 mutation was generated by transforming the diploid strain JSY1373 with a PCR fragment containing 50 bp of FZO1 flanking sequence interrupted by the HIS3 gene (Baudin et al., 1993). The fzo1Δ:: HIS3 disruption precisely removed the entire FZO1 coding sequence and was verified by PCR analysis. In 39 tetrads from the sporulated heterozygous diploid (JSY1808), the HIS3 marker segregated 2:2 with a slow growth defect on YPDextrose and an inability to grow on YPGlycerol. Mitochondrial morphology was visualized in four tetrads using a primary anti-porin antiserum (1:200 dilution; Molecular Probes, Eugene OR) and a secondary goat anti–mouse FITC antibody (1:100 dilution) (Jackson ImmunoResearch Laboratories, West Grove, PA) (Pringle et al., 1991). DAPI (4′,6-diamidino-2-phenylindole; 25 ng/ml) was included in the mounting medium to visualize mtDNA. The loss of mtDNA was confirmed by mating JSY1810 (fzo1Δ) to a rhoo tester strain JSY2555 (FZO1, rhoo). pRS416-FZO1 was transformed into a fzo1Δ strain (JSY1810) to generate JSY2579 (wild-type mitochondrial morphology, no detectable mtDNA). To generate the rhoo strain JSY2555 (wild-type mitochondrial morphology, no detectable mtDNA), a rho+ strain (JSY999) was grown twice to saturation in synthetic minimal medium containing 25 μg/ml ethidium bromide (Fox et al., 1991).
Electron microscopy of wild-type (JSY1812) and fzo1Δ (JSY1810) cells was performed essentially as described with the following modifications (Yaffe, 1995). The strains were grown in YPDextrose before fixation, and two additional changes of anhydrous Spurr resin (Polysciences, Inc., Warrington, PA), followed by overnight incubation, were used to achieve maximum infiltration of the samples.
Depletion of N-3XMYC-Fzo1p
To deplete the N-3XMYC-Fzo1p, the pGAL1-N-3XMYC-FZO1 plasmid was transformed into JSY2038 (fzo1Δ rho+ + pRS416-FZO1) and cells that had lost the pRS416-FZO1 plasmid were selected on SRaffinose medium containing 5-FOA (5-fluoro-orotic acid) to yield JSY2273 (fzo1Δ rho+ + pGAL1-N-3XMYC-FZO1). JSY2273 was grown for 24 h in SRaffinose medium lacking leucine to select for pGAL1-N-3XMYC- FZO1. To block expression of N-3XMYC-Fzo1p, the cells were collected, rinsed, and grown in SDextrose minus leucine to a density of 2.5 × 106 cells/ml. Mitochondrial morphology and mtDNA nucleoid distribution were evaluated at the indicated time points by staining with DiOC6 (3,3′ dihexyloxacarbocyanine) (Molecular Probes Inc.) (Hermann et al., 1997) or DAPI (Pringle et al., 1991), respectively. JSY2270 (FZO1 rho+ + pGAL1-N-3XMYC-FZO1), containing the wild-type FZO1 gene, did not exhibit any changes in mitochondrial morphology or loss of mtDNA during the N-3XMYC-Fzo1p depletion (data not shown). N-3XMYC-Fzo1 protein levels in total cell extracts were analyzed by Western blotting (anti-MYC antibody) at the indicated time points (Harlow and Lane, 1988). Blots were stripped and reprobed with anti–3-PGK (3-phosphoglycerate kinase) (1:1,000 dilution) (Molecular Probes Inc.) to control for differences in protein loading. Mitochondrial morphologies were scored by GFP (green fluorescent protein) staining in JSY2273 cells containing the pRS416-ADH-COXIVpre-GFP plasmid (JSY2634; mito-GFP).
Generation and Characterization of the fzo1-1 Mutation
The fzo1-1 temperature-sensitive allele was generated by low-fidelity PCR (Muhlrad et al., 1992). Mutagenized pRS414-FZO1 plasmids were transformed into a strain in which fzo1Δ::HIS3 was covered with pRS416- FZO1 (JSY2287). The loss of pRS416-FZO1 from these cells was selected by growth on medium containing 5-FOA. Cells containing the pRS414- FZO1 mutagenized plasmids were tested for growth at 25° and 37°C on SGlycerol medium. One strain that was inviable at 37°C was identified (fzo1-1). The plasmid containing the mutant version of FZO1 was recovered, and when retransformed into a cell lacking FZO1 caused temperature-sensitive growth on SGlycerol medium. Transformation of the pRS414-fzo1-1 plasmid into a wild-type strain revealed that the fzo1-1 temperature-sensitive growth defect was recessive. Mitochondrial morphology was examined in fzo1-1 cells by staining with DiOC6 (not shown) or mito-GFP.
To examine mitochondrial fusion during mating, fzo1Δ + pRS414- fzo1-1 cells labeled with either mito-GFP (JSY2802) or Mitotracker red (JSY2804) (Molecular Probes Inc.), were mated at 25° or 37°C and analyzed as described by Nunnari et al. (1997).
Construction and Analysis of MYC-tagged Fzo1p
A 3XMYC epitope was introduced immediately downstream of the initiating Met in FZO1 (JSY2028) as described by Schneider et al. (1995). To add additional MYC epitope tags, the N-3XMYC-FZO1 coding region plus 500 bp of 5′ and 3′ flanking sequence was amplified from JSY2028 using P297 and P298 and cloned into the EcoRI site of pRS426 generating pRS426-N-3XMYC-FZO1 (verified by sequence analysis). N-3XMYC- FZO1 was subcloned into the EcoRI site of pALTER-1 (lacking a NotI site) and digested with NotI to release the 3XMYC tag. NotI fragments containing 3XMYC epitopes from the plasmid pMPY-3XMYC (Schneider et al., 1995) were inserted into the amino-terminal NotI site of FZO1. PCR screening and sequence analysis was used to identify a clone containing a 9XMYC insert (pALTER-1-N-9XMYC-FZO1). The N-9X-MYC-FZO1 was cloned into the EcoRI site of pRS414 to generate pRS414-N-9XMYC- FZO1. fzo1Δ cells expressing only the 3XMYC-tagged (pRS426-N-3XMYC- FZO1; JSY2028) or the 9XMYC-tagged (pRS414-N-9XMYC-FZO1; JSY2394) Fzo1 protein grew normally on nonfermentable carbon sources, retained mtDNA, and had wild-type mitochondrial morphology, indicating that the fusion proteins were functional. All MYC-tagged forms of Fzo1p were detected by Western blotting of total protein extracts using an anti-MYC mouse monoclonal antibody (9E10; 1:1,000) (Berkeley Antibody Co., Richmond, CA) (Hermann et al., 1997). The N-9XMYC-Fzo1p was localized in JSY2392 (fzo1Δ + pRS414-FZO1) and JSY2394 by indirect immunofluorescence with a primary anti-MYC antibody (1:100 dilution) and a goat anti–mouse FITC secondary antibody (1:100 dilution) (Jackson ImmunoResearch) (Pringle et al., 1991).
Biochemical Analysis of N-9XMYC-Fzo1p
A strain expressing N-9XMYC-Fzo1p (JSY2394) was grown in SGalactose medium lacking tryptophan to select for the pRS414-N-9XMYC- FZO1 plasmid. Cell lysates (cytosol) were fractionated by differential sedimentation to generated a mitochondrial pellet and a postmitochondrial supernatant (Daum et al., 1982; Zinser and Daum, 1995). Samples (cell equivalents) from each fraction were analyzed by Western blotting. To determine the membrane association of N-9XMYC-Fzo1p, 100 μg of mitochondria purified from JSY2394 were pelleted at 12,000 g for 10 min, resuspended in 75μl of 100 mM Na2CO3, pH 11.5, or 1% Triton X-100 containing 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin, incubated on ice for 30 min, and then sedimented at 100,000 g for 30 min. Equal volumes of pellet and supernatant fractions were analyzed by Western blotting. Mitochondrial inner and outer membranes (Daum et al., 1982) were loaded onto 5 ml of 0.85–1.6 M sucrose step gradients (0.8, 1.1, 1.35, and 1.6 M) and then centrifuged for 16 h at 30,000 rpm in a Beckman SW50.1 rotor at 2°C (Fullerton, CA). The gradient was partitioned into 13 individual 400-μl fractions that were analyzed by Western blotting. Comparison of protein levels was performed using NIH Image 1.60 (National Institutes of Health, Bethesda, MD). The orientation of N-9XMYC-Fzo1p on mitochondria was determined by incubating 100 μg of purified mitochondria (from JSY2394) in breaking buffer (0.6 M mannitol, 20 mM Hepes-KOH, pH 7.4) containing 100 μg/ml trypsin (Sigma Chemical Co., St. Louis, MO) on ice. To disrupt the outer membrane, mitochondria were diluted with nine volumes of OS buffer (20 mM Hepes-KOH, pH 7.4) and trypsin was added to a final concentration of 100 μg/ml. After 20 min, the reaction was stopped by the addition of soybean trypsin inhibitor (2.5 mg/ ml, Sigma Chemical Co.) and 1 mM PMSF. Samples were analyzed by Western blotting. Western blots were performed with the indicated antibodies at the following dilutions: anti-MYC (1:1,000), anti-porin (1:1,000), anti–3-PGK (1:1,000), anti-CoxIV (cytochrome oxidase subunit IV; 1:20,000; Molecular Probes Inc.), and anti–cytochrome b 2 (1:10,000).
Analysis of FZO1 GTPase Point Mutants
pRS414, pRS414-FZO1, and pRS414 containing the FZO1 GTPase point mutations were shuffled into JSY2287 (fzo1Δ rho+ + pRS416-FZO1), to generate the following strains: (a) JSY2354 (fzo1Δ + pRS414); (b) JSY2392 (fzo1Δ + pRS414-FZO1); (c) JSY2355 (fzo1Δ + pRS414- fzo1[T221A]); (d) JSY2356 (fzo1Δ + pRS414-fzo1[K371A]); (e) JSY2357 (fzo1Δ + pRS414-fzo1[K200A]); and (f) JSY2358 (fzo1Δ + pRS414- fzo1[S201N]). None of the mutant Fzo1 proteins caused defects in mitochondrial morphology (anti-porin staining), mtDNA nucleoid retention (DAPI staining) or function (growth on glycerol) when introduced on low or high copy plasmids into a wild-type FZO1 strain (JSY2288) (data not shown). Subcellular fractionation (see above) and Western blotting with an anti-Fzo1p antibody confirmed that the mutant proteins were expressed and localized to mitochondrial membranes in fzo1Δ cells (data not shown).
Microscopic Techniques
Cells were viewed on a Zeiss Axioplan microscope (1.25× optivar setting; Carl Zeiss Inc., Thornwood, NY) as described in Roeder et al. (1998). Images were captured using a Hamamatsu C5810 color-chilled 3CCD camera (Hamamatsu Photonics, Hamamatsu City, Japan) interfaced to a Macintosh Quadra 840AV computer. For three-dimensional fluorescence microscopy, data collection was carried out using a Leica confocal microscope and a 100× 1.4 N.A. objective (Leica Inc., St. Gallen, Switzerland). All shutters, stage motion, and image acquisition were under computer control. Images were acquired by moving the stage in ∼0.2-μm intervals. Thin sections were viewed with a Hitachi H-700 electron microscope (Tokyo, Japan) and images were captured using the Kodak 161 digital camera system v1.55b (Eastman Kodak Co., Rochester, NY). Digital images were assembled into figures and printed as described in Roeder and Shaw (1996).
Results
S. cerevisiae FZO1 Is Required for Maintenance of Mitochondrial Morphology and Retention of Mitochondrial DNA
To determine whether Fzo family members act generally to control mitochondrial fusion, we disrupted one copy of S. cerevisiae FZO1 in a diploid strain. After sporulation and dissection, fzo1Δ haploid cells exhibited a significant growth defect relative to wild type on medium containing the fermentable carbon source dextrose and were unable to grow on medium containing the nonfermentable carbon source glycerol (Fig. 1 B). This growth pattern, also referred to as a “petite” phenotype (Dujon, 1981), is characteristic of strains with defective mitochondrial respiration and indicates that the loss of FZO1 disrupts normal mitochondrial function.
Indirect immunofluorescence revealed that Fzo1p is required for normal organization of the mitochondrial network in vegetatively growing cells (Fig. 2). In a wild-type strain, porin-stained mitochondrial membranes appeared as branched, tubular networks distributed at the cell surface (Fig. 2, A and B). In contrast, less than 1% of fzo1Δ cells contained a wild-type mitochondrial network. Instead, fzo1Δ mutants contained between one and five spherical or slightly elongated mitochondrial structures that were localized to the cell cortex but were not distributed evenly around the cell periphery (Fig. 2, G and H). In addition, numerous small mitochondria were occasionally observed scattered throughout the cytoplasm in the mutant (data not shown). The mitochondrial morphology in fzo1Δ cells resembles that in mutants with abnormal actin cytoskeletons (Drubin et al., 1993; Lazzarino et al., 1994). However, we did not observe defects in the organization of the actin and microtubule cytoskeletons in the fzo1Δ strain (data not shown). Although many of the yeast mitochondrial morphology mutants identified to date also have an associated mitochondrial inheritance defect (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997; Hermann and Shaw, 1998), our analysis did not reveal a mitochondrial inheritance defect in fzo1Δ cells relative to wild type (data not shown). Thus, Fzo1p controls mitochondrial network organization in yeast but is not required for mitochondrial transport into daughter cells during division.
Figure 2.
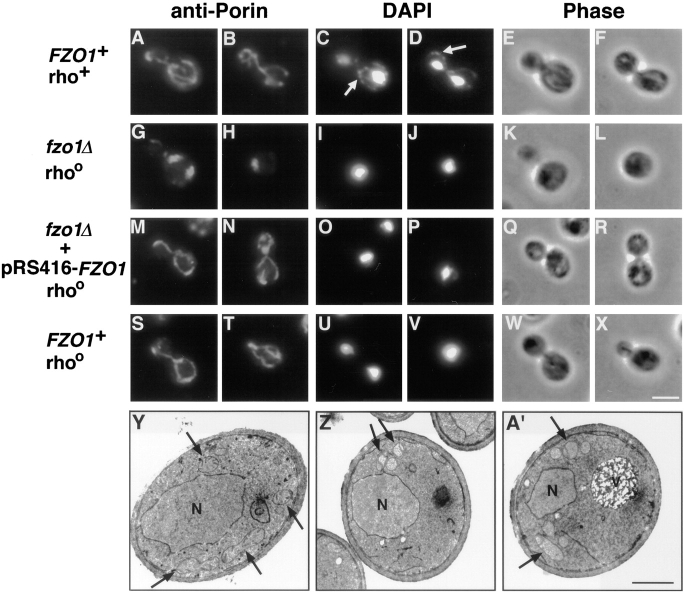
fzo1Δ cells exhibit defective mitochondrial morphology and lose their mtDNA. (A–X) Indirect immunofluorescence images of yeast cells stained with anti-porin antiserum to visualize mitochondrial membranes (leftmost columns), and DAPI to visualize nuclei and mtDNA (middle columns; white arrows in C and D indicate mtDNA nucleoids). Phase-contrast images of the same cells are shown in the rightmost columns. (A–F) Normal mitochondrial morphology and distribution of mtDNA nucleoids in a wild-type rho+ strain (JSY1812). (G–L) Mutant fzo1Δ rhoo cells (JSY1810) contained spherical or slightly elongated mitochondrial structures that lack DAPI-stained mtDNA. (M–R) Wild-type mitochondrial morphology, but not mtDNA, was restored in an fzo1Δ rhoo mutant strain (JSY2579) upon reintroduction of the wild-type FZO1 gene. (S–X) Mitochondrial morphology is normal in a wild-type rhoo strain (JSY2555) that lacks mtDNA. (Y–A′) Transmission electron micrographs of wild-type and fzo1Δ cells. (Y) Mitochondrial profiles in wild-type cells (JSY1812) are dispersed equally throughout the cell cortex and display numerous cristae (black arrows). (Z and A′) Mitochondrial profiles in fzo1Δ cells (JSY1810) are clustered together near the cell periphery and contain fewer cristae. N, nuclei; V, vacuoles; rho +, containing mtDNA; rhoo, lacking mtDNA. Bars: (A–X) 5 μm; (Y–A′) 1μm.
Transmission electron microscopy indicated that mitochondrial morphology and distribution were also abnormal at the ultrastructural level in fzo1Δ. In wild-type cells, mitochondrial profiles were distributed throughout the peripheral cytoplasm and contained elaborate cristae (invaginations of the inner mitochondrial membrane) (Fig. 2 Y). In contrast, mitochondrial profiles in fzo1Δ cells appeared to cluster together in one or two regions near the plasma membrane (Fig. 2, Z and A′), similar to the mitochondrial distribution observed by indirect immunofluorescence (Fig. 2, G and H). The clustered mitochondrial profiles in fzo1Δ could represent closely opposed tubules of a collapsed, but still interconnected, mitochondrial reticulum. Alternatively, the clusters could be composed of individual unfused mitochondrial fragments (see below). Cross sections also revealed that fzo1Δ mitochondria often contained fewer cristae or lacked cristae altogether (Fig. 2, Z and A′).
The wild-type function of Fzo1p is also required for the maintenance of mtDNA (mitochondrial DNA) nucleoids. Wild-type cells labeled with the DNA-specific dye DAPI always contained brightly stained nuclei as well as 25–50 punctate mtDNA nucleoids localized at the cell periphery (Fig. 2, C and D). In contrast, mitochondrial nucleoids were never detected in fzo1Δ cells (Fig. 2, I and J). Crosses of fzo1Δ with a known rhoo strain (lacking mtDNA) confirmed that mitochondrial genomes were absent in fzo1Δ cells (data not shown). Since mtDNA encodes RNAs and proteins essential for mitochondrial respiratory function (Pon and Schatz, 1991), these results raised the possibility that fzo1Δ mitochondrial morphology defects were an indirect consequence of mtDNA loss. However, reintroduction of the wild-type FZO1 gene was sufficient to restore elongated and branched mitochondrial networks in fzo1Δ cells (Fig. 2, M and N) in the absence of mtDNA (Fig. 2, O and P). The morphology of the restored mitochondrial networks was identical to that observed in an isogenic rhoo strain containing a wild-type FZO1 gene (Fig. 2, S–V). These results demonstrate that the loss of mtDNA, and the resulting defects in mitochondrial respiration, do not cause mitochondrial morphology defects in wild-type cells (Guan et al., 1993) or in fzo1Δ mutants and suggest that Fzo1p is directly required for the maintenance of normal mitochondrial morphology.
Fzo1p depletion studies revealed that changes in mitochondrial morphology preceded the loss of mtDNA nucleoids. A GAL1 regulated, N-3XMYC-tagged form of Fzo1p on a plasmid was introduced into the fzo1Δ mutant strain using a plasmid shuffling technique to prevent mtDNA loss (refer to Materials and Methods). Growth in raffinose-containing medium (which neither induces nor represses transcription from the GAL1 promoter) resulted in sufficient expression of N-3XMYC-Fzo1p to fully support wild-type growth and restore wild-type mitochondrial morphology in fzo1Δ cells (Fig. 3, A, solid squares, t = 0, and D, Wildtype). After transfer to glucose-containing medium to block expression of N-3XMYC-Fzo1p from the GAL1 promoter, aliquots were harvested at the indicated times and mitochondrial morphology (DiOC6 staining) and mtDNA distribution (DAPI staining) were examined. During the 8 h after transfer, wild-type mitochondrial networks broke down into smaller segments which eventually collapsed, forming between one and five spherical or slightly elongated membrane clusters per cell (Fig. 3, A and D). The number, shape, and size of these mitochondrial clusters at the 8-h time point were identical to those visualized in the fzo1Δ mutant by anti-porin staining (Fig. 2, G and H). The defects in mitochondrial morphology observed 8 h after the shift did not severely affect mitochondrial function, since these misshapen organelles were still able to accumulated the membrane potential-sensitive dye DiOC6 (Chen, 1988). Western blotting with an anti-MYC antiserum showed a 10-fold reduction in N-3XMYC-Fzo1 protein levels during the course of the depletion experiment (Fig. 3 C). Cells lacking DAPI-stained mtDNA nucleoids were not detected, even after 8 h when the majority of cells exhibited defective mitochondrial morphology (Fig. 3 B). Moreover, 24 h after the initial N-3XMYC-Fzo1p expression block (11 doublings), only 25% of the cells lacked nucleoids, making it unlikely that Fzo1p is required for mtDNA partitioning into buds (data not shown). Together, these results indicate that the loss of Fzo1p leads first to defects in mitochondrial membrane morphology and only later to defects in mtDNA maintenance.
Figure 3.
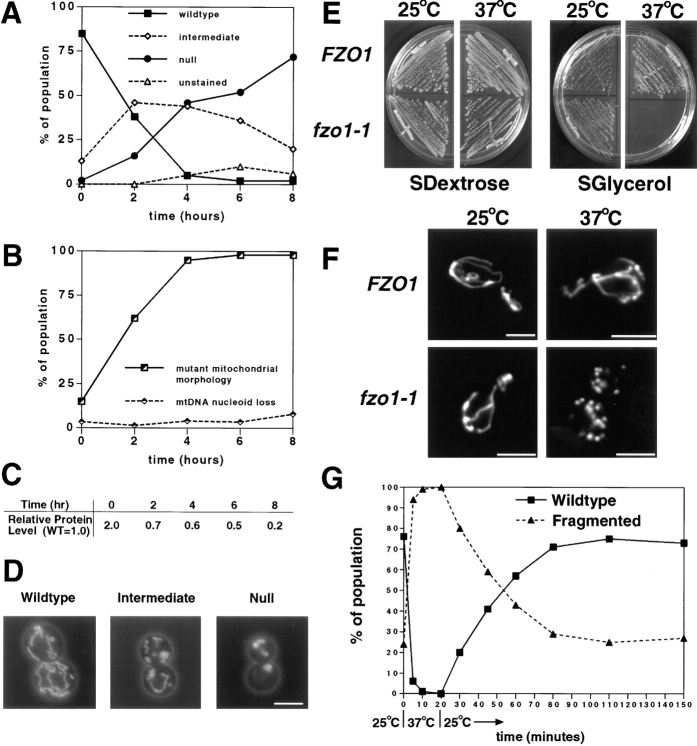
Depletion or loss of Fzo1p function leads to defects in mitochondrial morphology and mtDNA maintenance. (A–D) Depletion of Fzo1p causes defects in mitochondrial morphology that precede the loss of mtDNA nucleoids. fzo1Δ cells containing pGAL1-N-3XMYC-FZO1 (JSY2273) were shifted from raffinose medium, which allows nearly wild-type levels of Fzo1p expression, to dextrose medium, which represses expression from the GAL1 promoter. (A) Mitochondrial morphology scored by DiOC6 staining after shift to depletion medium (n = 100): mitochondrial morphologies were classified as wild type (branched tubular network), intermediate (tubular condensed network), null (spherical or partially elongated structures), and unstained (nonrespiring mitochondria do not accumulate DiOC6). (B) Distribution of mtDNA nucleoids scored by DAPI staining during the Fzo1p depletion shown in A (n = 200). The mutant mitochondrial morphology class included the intermediate, null, and unstained categories scored in A. (C) N-3XMYC-Fzo1p levels during the depletion experiment. Protein extracts prepared from equivalent numbers of cells were analyzed by Western blotting with anti-MYC antiserum. N-3XMYC-Fzo1 protein levels during the depletion were normalized to the level of N-3XMYC-Fzo1p expressed from the wild-type FZO1 promoter (WT = 1.0; JSY2034). (D) Representative wild-type, intermediate, and null mitochondrial morphologies visualized with a matrix-targeted form of the GFP (mito-GFP). (E and F) The temperature-sensitive fzo1-1 mutation causes rapid and reversible fragmentation of the mitochondrial network. (E) fzo1-1 cells cannot grow on a nonfermentable carbon source at 37°C. fzo1Δ strains containing pRS414-FZO1 (JSY2926) or pRS414-fzo1-1 (JSY2793) were grown on SDextrose media (left panels) for 2 d at 25° or 37°C and SGlycerol media (right panels) for 6 d at 25° or 37°C. (F) Mitochondrial morphology in FZO1 (JSY2793) and fzo1-1(JSY2802) cells grown at 25°C and shifted to 37°C for 10 min. Mitochondrial compartments were visualized with mito-GFP. Representative cells are shown. (G) Log-phase fzo1-1 cells (JSY2804) grown at 25°C (t = 0) were shifted to 37°C. After 20 min at 37°C, the cells were returned to 25°C. Mitochondrial morphology was quantified at the indicated times using mito-GFP (n ≥ 200). A representative experiment is shown. Bar, 2.5 μm.
Mitochondrial Networks Fragment at 37°C in a Conditional fzo1-1 Mutant
Mitochondrial morphology is likely to be regulated, in part, by opposing membrane fusion and membrane fission reactions (Nunnari et al., 1997). If the primary effect of an fzo1 mutation is to block membrane fusion, we predicted that ongoing mitochondrial fission would initially cause the mitochondrial network to fragment. To test this, a conditional FZO1 allele (fzo1-1) was identified by replacing a wild-type FZO1 plasmid with mutagenized FZO1 plasmids in an fzo1Δ strain (refer to Materials and Methods). fzo1-1 allowed growth of the fzo1Δ strain at 25° but not 37°C on SGlycerol medium (Fig. 3 E). The fzo1-1 sequence contained three different nucleotide changes, resulting in amino acid substitutions K538I, N543I, and P553Q in the predicted polypeptide (refer to Materials and Methods). Staining with DiOC6 (data not shown) or GFP (Fig. 3, F and G) demonstrated that mitochondrial network morphology was wild type in the majority (76%) of fzo1-1 cells grown at the permissive temperature (Fig. 3, F, fzo1-1, 25°C, and G, t = 0 min). 10 min after shifting to 37°C, however, the networks in 99% of the fzo1-1 cells had fragmented into many small, uniformly distributed, mitochondrial compartments (Fig. 3, F, fzo1-1, 37°C, and G, t = 10 min). Upon extended incubation at 37°C, these mitochondrial fragments clustered together forming large membrane aggregates similar to those observed in the fzo1Δ strain (data not shown). If after 20 min at 37°C the temperature was reduced to 25°C, the mitochondrial network regained its wild-type morphology within 60 min (Fig. 3 G). In control experiments, mitochondrial fragmentation was never observed in wild-type strains grown at 25° or 37°C (Fig. 3 F, FZO1, 25°C and 37°C). The rapid fragmentation of the mitochondrial network observed in the conditional fzo1-1 strain provides a direct demonstration that Fzo1p controls mitochondrial morphology in yeast and is consistent with a role for this protein in mitochondrial membrane fusion.
fzo1-1 Blocks Mitochondrial Fusion during Mating
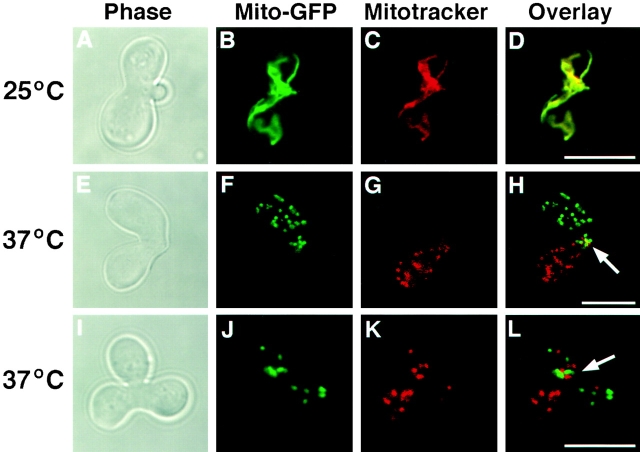
Mitochondrial fusion and content mixing has been shown to occur in yeast zygotes soon after cell fusion (Azpiroz and Butow, 1993; Nunnari et al., 1997). To test directly the requirement of Fzo1p in mitochondrial fusion, we examined the effect of the fzo1-1 mutation on mitochondrial content mixing during mating. Mitochondrial networks were visualized by labeling one haploid parent with the fluorescent vital dye Mitotracker red and the other haploid parent with a matrix targeted form of GFP (mito-GFP) (Nunnari et al., 1997). After mating and zygote formation, the distribution of the fluorophores was examined by fluorescence confocal microscopy. In matings between wild-type cells performed at 25° or 37°C, the two fluorescent markers rapidly and completely colocalized in zygotes, indicating that the parental mitochondrial membranes had fused and mitochondrial contents had mixed (Nunnari et al., 1997) (Table II). Mitochondrial fusion also occurred efficiently in fzo1-1 × fzo1-1 zygotes formed at 25°C (Fig. 4, A–D; Table II). In contrast, mitochondrial networks fragmented and failed to fuse in fzo1-1 × fzo1-1 zygotes formed at 37°C (Fig. 4, E–L; Table II). When fzo1-1 zygotes were optically sectioned using the confocal microscope, mitochondrial membranes containing the haploid-derived green and red fluorescent markers never colocalized (Fig. 4, E–L). Moreover, fzo1-1 × fzo1-1 zygotes formed at the restrictive temperature contained unfused mitochondria even after they had completed karyogamy and formed a new bud (Fig. 4, I–L). Thus, fzo1-1 causes a block, and not simply a delay, in mitochondrial docking and/or fusion. Finally, mitochondrial fusion was also reduced in matings between fzo1-1 and wild-type parents at 37°C (Table II), suggesting that the function of Fzo1p is required in both haploid parents for efficient mitochondrial fusion.
Table II.
Mitochondrial Fusion in Zygotes
| Fused mitochondria | ||||||
|---|---|---|---|---|---|---|
| Strains crossed | °C | Unbudded zygotes | Budded zygotes | |||
| FZO1 + × FZO1 + | 25 | 40/50 (80%) | 50/50 (100%) | |||
| 37 | 18/50 (36%) | 51/51 (100%) | ||||
| FZO1 + × fzo1-1 | 25 | 40/55 (73%) | 53/56 (95%) | |||
| 37 | 1/49 (2%) | 32/60 (53%) | ||||
| fzo1-1 × fzo1-1 | 25 | 27/50 (54%) | 51/55 (93%) | |||
| 37 | 0/51 (0%) | 4/53 (8%) | ||||
Figure 4.
FZO1 is required for mitochondrial fusion during mating. fzo1-1 cells of opposite mating type (JSY2802 and JSY2804) were labeled with mito-GFP or Mitotracker red and mated at 25° (A–D) and 37°C (E–H and I–L). Confocal microscopy was used to score the distribution of mito-GFP (green in B, F, and J) and Mitotracker (red in C, G, and K) in serial optical sections (representative single optical section are shown). Fusion and mixing of mitochondrial contents (yellow in D) was evaluated in merged mito-GFP and Mitotracker red images (D, H, and L). Zygote morphology was visualized by phase-contrast microscopy (A, E, and I). The zygote bud is on the right in A and on the top in I. White arrows, regions where parental mitochondrial membranes have intermixed. Bar, 5 μm.
We think it is unlikely that the fzo1-1 mutation prevents mitochondrial fusion indirectly by interfering with mitochondrial motility and/or distribution. First, although mitochondrial networks fragment at 37°C in fzo1-1 × fzo1-1 zygotes, these membrane fragments were segregated normally into daughter cells, indicating that mitochondrial motility was not severely impaired (Fig. 4, I–L, arrow). Second, the fragmentation of mitochondria in fzo1-1 × fzo1-1 zygotes does not appear to block their association. We often observed red and green mitochondrial compartments in close apposition near the zygote neck or in the diploid daughter cell (Fig. 4, H and L, arrows). Together, these results provide compelling evidence that Fzo1p plays an essential and direct role in mitochondrial fusion.
Fzo1p Is a Mitochondrial Outer Membrane Protein with Its GTPase Domain Facing the Cytoplasm
The Fzo1 protein is localized on the mitochondrial network in vegetatively growing yeast cells (Fig. 5). A plasmid encoding Fzo1p tagged near the amino terminus with nine MYC epitopes (N-9XMYC-Fzo1p) was constructed and shown to rescue the mitochondrial morphology defect, the mtDNA loss phenotype, and the glycerol growth defect in the fzo1Δ strain (data not shown). Anti-MYC antibodies recognized a 116-kD protein in extracts prepared from wild-type cells expressing the N-9XMYC-Fzo1p (Fig. 6 A, lane 2) but not the native Fzo1 protein (Fig. 6 A, lane 1). Indirect immunofluorescence with anti-MYC antibodies revealed that N-9XMYC-Fzo1p localized to the mitochondrial network in wild-type cells and was uniformly distributed on this compartment (Fig. 5, A–D). The pattern of DAPI-stained mtDNA nucleoids in these cells overlapped with the fluorescent signal confirming that N-9XMYC-Fzo1p was located on the mitochondrial network (Fig. 5, B and D; compare white arrows in A and C with B and C). Similar results were obtained when cells were stained with polyclonal antibodies generated against the native Fzo1 protein (data not shown). In control experiments, no signal was detected in cells expressing only the wild-type Fzo1p (Fig. 5 E).
Figure 5.
Fzo1p localizes to the mitochondrial network. (A–D) fzo1Δ cells (JSY2394) expressing the N-9XMYC-Fzo1 protein were stained with anti-MYC antiserum (A and C) and DAPI (B and D). The N-9XMYC-Fzo1p staining was completely coincident with mitochondria as marked by mtDNA nucleoids (compare white arrows in A and C with B and D). (E and F) Anti-MYC serum did not stain fzo1Δ cells (JSY2392) expressing the Fzo1p lacking the MYC tag. Bar, 5 μm.
Figure 6.
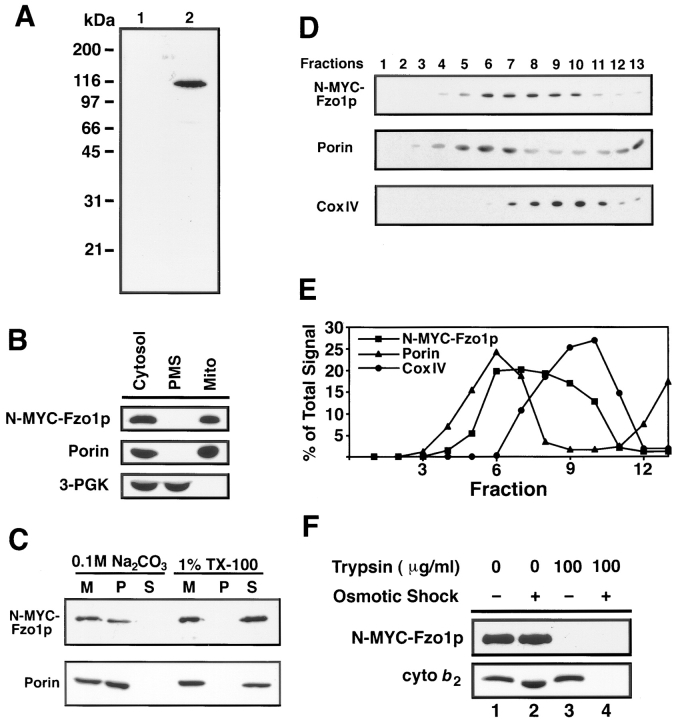
Fzo1p is a mitochondrial integral membrane protein with its GTPase domain exposed to the cytoplasm. (A) Total cell extracts from fzo1Δ cells containing either the wild-type pRS414-FZO1 (lane 1; JSY2392) or pRS414-N-9XMYC- FZO1 plasmids (lane 2; JSY2394) analyzed by Western blotting with an anti-MYC antiserum. The same fzo1Δ strain harboring pRS414-N-9XMYC-FZO1 (JSY2394) was used for all of the experiments described in 6B-F. (B) N-9XMYC-Fzo1p cofractionated with the mitochondrial protein porin. Protein extracts from equivalent numbers of cells were fractionated by differential centrifugation, separated by SDS-PAGE, and analyzed by Western blotting with anti-MYC, anti-porin, and anti–3-PGK serum. Cytosol, postnuclear cytoplasmic extract; Mito, crude mitochondrial pellet; PMS, supernatant depleted of mitochondria. (C) N-9XMYC-Fzo1p is an integral membrane protein. Purified mitochondria (M) were treated to solubilize peripheral membrane proteins (0.1 M Na2CO3) or integral membrane proteins (1% Triton X-100), separated into pellet (P) and supernatant (S) fractions, and analyzed by SDS-PAGE and Western blotting. The release of soluble cytochrome b 2 and the peripheral F1β ATPase subunit into the supernatant fraction after 0.1M Na2CO3 treatment was confirmed by Western blotting (data not shown). (D and E) N-9XMYC-Fzo1p fractionates in an intermediate density mitochondrial membrane fraction. Fractions from a 30–50% sucrose step gradient (top, fraction 1) analyzed by Western blotting with anti-MYC, anti-porin, and anti–CoxIV serum. (E) The percentage of total N-MYC-Fzo1p, porin, and CoxIV per fraction. (F) The GTPase domain of Fzo1p is exposed on the cytoplasmic surface of mitochondria. Untreated (lane 1), trypsin-treated (lanes 3 and 4, 100 μg/ml), and osmotically shocked (lanes 2 and 4) mitochondria were analyzed by SDS-PAGE and Western blotting with anti-MYC and anti-cytochrome b 2 serum.
Subcellular fractionation confirmed the mitochondrial localization of Fzo1p (Fig. 6 B). N-9XMYC-Fzo1p cofractionated with the mitochondrial pellet, along with the outer mitochondrial membrane protein porin, during differential centrifugation of a postnuclear cell extract (Fig. 6 B, Mito). No N-9XMYC-Fzo1p was detected in the postmitochondrial supernatant fraction that contained the cytoplasmic protein 3-PGK (Fig. 6 B, PMS). Similar results were obtained from wild-type cells using anti-Fzo1p antiserum to follow fractionation of the native Fzo1 protein (data not shown).
Fzo1p behaved like an integral membrane protein, as predicted based on the two closely spaced and conserved hydrophobic domains near its carboxy terminus (refer to Fig. 1 A) (Hales and Fuller, 1997). When mitochondria containing N-9XMYC-Fzo1p were extracted with 100 mM Na2CO3, pH 11.5, to release peripheral membrane proteins, both the N-9XMYC-Fzo1p and the integral outer-membrane protein porin were resistant to sodium carbonate extraction and remained associated with the membrane pellet (Fig. 6 C). In contrast, Fzo1p and porin were released into the supernatant when mitochondrial membranes were solubilized with 1% Triton X-100 (Fig. 6 C). Fzo1p also remained associated with mitochondrial membranes after the organelles were disrupted by osmotic and mechanical methods to release soluble intermembrane space and matrix proteins (Fig. 6 D and data not shown).
Analysis of submitochondrial membrane fractions separated on sucrose density gradients indicated that Fzo1p associates with both the inner and outer mitochondrial membranes. N-9XMYC-Fzo1p fractionated at an intermediate density that overlapped with, but was distinct from, the distribution of both inner membrane vesicles containing the integral membrane protein CoxIV and outer membrane vesicles containing porin (Fig. 6, D and E). Although this result does not prove that Fzo1p crosses both mitochondrial membranes, this fractionation pattern is characteristic of “trapped” translocation intermediates that span both mitochondrial membranes at contact sites formed by the translocation pore (Pon et al., 1989). Proteins that fractionate with both inner and outer mitochondrial membranes and are enriched at contact sites have been observed previously (Pon et al., 1989).
Fzo1p is oriented with its amino-terminal GTPase domain and adjacent heptad repeats exposed on the cytoplasmic face of the mitochondrial compartment. Treatment of isolated, intact mitochondria with trypsin resulted in the complete digestion of the MYC tag on the amino terminus of Fzo1p (Fig. 6 F, lane 3). In contrast, cytochrome b 2, a protein in the intermembrane space, was resistant to proteolysis, indicating that the outer mitochondrial membrane remained intact (Fig. 6 F, lane 3). Cytochrome b 2 could be digested, however, if the mitochondrial outer membrane was disrupted by osmotic shock (Fig. 6 F, lane 4). Since Fzo1p behaves like an integral membrane protein, these data also indicate that at least one of the hydrophobic domains at the carboxy-terminus of Fzo1p is embedded in the mitochondrial outer membrane.
The signals and machinery that target Fzo proteins to mitochondria may be conserved. When a cDNA encoding the Drosophila melanogaster Fzo protein was introduced into wild-type cells on a GAL1 low copy plasmid, the Drosophila protein cofractionated with mitochondrial membranes as assayed by Western blotting with antibodies specific for the D. melanogaster homologue (data not shown). However, the Drosophila Fzo protein did not rescue mitochondrial morphology or mtDNA loss phenotypes in the S. cerevisiae fzo1Δ strain.
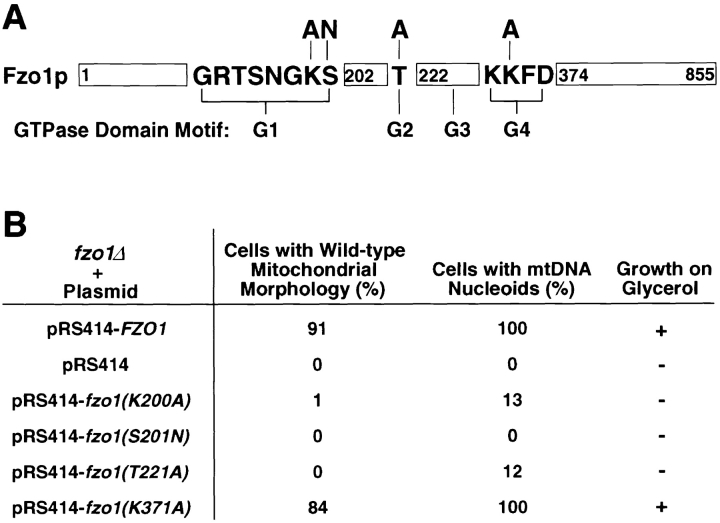
Fzo1p Function Requires the Conserved GTPase Domain
The Fzo1p GTPase domain contains four conserved motifs designated G1–G4 (Fig. 7 A). In most GTPases, these domains are required for GTP binding and hydrolysis as well as conformational changes elicited by nucleotide binding (Bourne et al., 1991). Conserved residues in three of the four G motifs in Fzo1p (K200A and S201N in G1, T221A in G2, and K371A in G4) were altered by site-directed mutagenesis (Fig. 7 A). All of these amino acid substitutions are known to disrupt either nucleotide binding or interactions with effector proteins in other GTPases (Sigal et al., 1986; Adari et al., 1988; Cales et al., 1988; Feig and Cooper, 1988; Farnsworth and Feig, 1991; Vojtek et al., 1993; Murphy et al., 1997). When low copy plasmids containing the mutated fzo1 genes were introduced into fzo1Δ cells, the mutant Fzo1 proteins were expressed at wild-type levels and were targeted to the mitochondrial compartment as assayed by differential centrifugation and Western blotting with anti-Fzo1p antiserum (data not shown). However, the fzo1(K200A), fzo1(S201N), and fzo1(T221A) mutant genes failed to rescue the glycerol growth defect or the mitochondrial morphology defects in the fzo1Δ strain (Fig. 7 B). Interestingly, a significant percentage (13 and 12%, respectively) of cells containing the fzo1(K200A) and fzo1(T221A) mutant genes contained detectable mtDNA nucleoids (Fig. 7 B), although the total number of nucleoids was reduced relative to wild type (1–5 instead of 25–50) (data not shown). It is possible that these mutant Fzo1 proteins retain some residual functions required for mtDNA maintenance. Alternatively, cells containing these mutant proteins may simply lose their mitochondrial genomes more slowly than fzo1-null cells. Mutation of a conserved residue in the G4 domain (K371A) did not disrupt the function of FZO1 (Fig. 7 B). This result is somewhat surprising, since lysine 371 is conserved throughout the GTPase superfamily and is known to be required for high-affinity GTP binding by Ras (Der et al., 1988; Bourne et al., 1991). In addition, the same mutation in Drosophila fzo had a modest but significant effect on the function of the protein (Hales and Fuller, 1997). Finally, wild-type cells overexpressing the mutant Fzo1 proteins (10–20 fold from a 2-μ vector) did not exhibit dominant growth or mitochondrial morphology defects when compared with cells overexpressing the wild-type Fzo1p (10–20 fold from a 2-μ vector) (data not shown). These observations indicate that GTP binding and/or hydrolysis is essential for Fzo1p function.
Figure 7.
GTPase domain mutations block Fzo1p function. (A) Schematic representation of Fzo1p illustrating the location and sequence of the GTPase domain motifs (G1–G4; not to scale) with mutations changing conserved residues indicated. The mutant amino acids are depicted above the original residues. (B) Mutations in the G1 and G2 motifs disrupt the function of Fzo1p. Glycerol growth, mitochondrial morphology (anti-porin, n = 400), and mtDNA nucleoid distribution (DAPI staining, n ≥ 100) analyzed in fzo1Δ cells (JSY2354) containing wild-type FZO1 (JSY2392) or mutated fzo1 genes (JSY2355-2358) carried on low copy plasmids (pRS414).
Discussion
Fzo1p Regulates Mitochondrial Fusion
We have shown that Fzo1p is a transmembrane GTPase required for mitochondrial fusion in yeast. Mitochondrial membranes rapidly fragment at the nonpermissive temperature in a conditional fzo1-1 strain. Since the opposing processes of mitochondrial fusion and fission are responsible for the reticular structure of the mitochondrial network in mitotically dividing cells (Nunnari et al., 1997), the simplest explanation for this result is that fzo1-1 causes a selective block in fusion and that fragmentation occurs as a result of continuing mitochondrial fission. fzo1-1 is the only yeast mitochondrial morphology mutant with a fragmentation phenotype, consistent with its novel role in mitochondrial fusion. Mitochondrial fusion and content mixing were also blocked in homozygous fzo1-1 × fzo1-1 zygotes and reduced in heterozygous fzo1-1 × FZO1 + matings. Functional Fzo1 protein could be required on opposing mitochondrial membranes for efficient fusion. If this is the case, then the residual mitochondrial fusion observed in fzo1-1 × FZO1 + matings could result from new protein synthesis and complementation in the heterozygous zygote. Alternatively, functional Fzo1 protein on only one of the fusion partners may allow fusion between Fzo1p+ and Fzo1p− mitochondrial membranes at lower efficiency.
Disruption of the FZO1 gene, depletion of the Fzo1 protein, or prolonged incubation of the fzo1-1 strain at 37°C lead to the severe clumping and aggregation of mitochondrial membranes. It seems likely that a constitutive block in fusion is responsible for the dramatic defects in mitochondrial morphology we observed under all of these conditions. Although mitochondrial inheritance is often defective in yeast mutants with abnormal mitochondrial morphology (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997; Hermann, 1998), the loss of Fzo1p function did not affect the motility or transport of mitochondrial membranes during mitotic division and the abnormal mitochondrial compartments in fzo1-1 and fzo1Δ cells were efficiently transmitted to daughter buds (refer to Fig. 2 G). Defects in Fzo1p function also lead to the loss of mitochondrial genomes. Although we cannot rule out that Fzo1p participates directly in mtDNA replication and/or segregation, several lines of evidence suggest that the loss of mtDNA in fzo1Δ cells is a secondary consequence of changes in mitochondrial morphology. First, when Fzo1p is depleted from wild-type cells, defects in mitochondrial membrane structure are observed many hours before DAPI-stained mtDNA nucleoids are lost. Second, a number of studies indicate that mutations affecting mitochondrial morphology result in decreased mtDNA stability (Guan et al., 1993; Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997; Hermann, 1998). Further experiments are required to determine the mechanism by which mtDNA is lost from fzo1 mutant strains.
Fzo1 Mitochondrial Membrane Association and Topology
Immunolocalization, fractionation, and protease digestion studies indicated that Fzo1p is an integral membrane protein with its amino terminus displayed on the mitochondrial surface. This topology positions Fzo1p's GTPase domain and adjacent heptad repeats in the cytoplasm where they could interact with binding partners and/or regulatory molecules that might regulate Fzo1p function. Given the conservation of the domain structure and overall charge distribution of the different Fzo family members (Fig. 1 A; Hales and Fuller, 1997), we predict that Fzo homologues from other organisms will display a similar mitochondrial distribution and topology.
Mitochondrial fusion has been reported to initiate at stable contact sites between the inner and outer mitochondrial membranes (Bereiter-Hahn and Voth, 1994). The carboxy terminus of Fzo proteins could play an important role in coordinating the behavior of the two lipid bilayers at these sites during the fusion reaction. Protease digestion and protein solubilization studies suggested that at least one of Fzo1p's carboxy-terminal hydrophobic domains is embedded in the outer mitochondrial membrane. In addition, Fzo1p migrated in sucrose gradients with an intermediate density fraction that partially overlapped both inner and outer mitochondrial membrane markers. This fractionation pattern has been observed for proteins that are physically associated with mitochondrial contact sites (Pon et al., 1989), suggesting that Fzo1p is located in these structures. There are a number of topologies that could account for the ability of Fzo1p to fractionate with both mitochondrial membranes. It is possible that Fzo1p spans both membranes with its carboxy terminus in the matrix as originally proposed by Hales and Fuller (1997). Alternatively, the carboxy-terminal tail of Fzo1p could extend into the intermembrane space and interact with proteins in the inner membrane. Finally, Fzo1p might be exclusively associated with the outer membrane at contact sites formed by other proteins. Localization of the carboxy terminus of Fzo1p will help to distinguish between these models.
Although the Drosophila and yeast Fzo proteins are both required for mitochondrial fusion, several observations suggest that they may be regulated differently. First, our studies indicate that the Drosophila Fzo protein is efficiently targeted to mitochondrial membranes in wild-type yeast but cannot rescue mitochondrial phenotypes in the fzo1Δ strain (data not shown). Second, the Drosophila Fzo and yeast Fzo1 proteins exhibit distinct expression patterns in the two organisms. Drosophila Fzo was only detected on sperm mitochondria during a short period of time when mitochondrial fusion was occurring (Hales and Fuller, 1997), suggesting that the timing of mitochondrial fusion could be developmentally regulated by controlling Fzo expression, localization, and/or degradation. In contrast, yeast Fzo1p protein levels and mitochondrial localization did not change during mitotic growth, mating, or meiosis (data not shown), consistent with the observation that mitochondrial fusion occurs during all stages of the yeast life cycle (Pon and Schatz, 1991; Nunnari et al., 1997; Hermann and Shaw, 1998). In time-lapse studies, yeast mitochondrial fusion is observed when the tip of a mitochondrial tubule encounters the tip or side of another mitochondrial tubule (Nunnari et al., 1997). This has led to the suggestion that key fusion components are localized or specifically activated at the tips of mitochondrial tubules (Nunnari et al., 1997). Our observation that Fzo1p is uniformly distributed on the mitochondrial compartment suggests that the fusion machinery is not localized at tips. Instead, Fzo1p could be activated locally at sites of membrane contact.
Fzo1 Function Requires the Amino-terminal GTPase Domain
The GTPase domain of Fzo1p is essential for its function and could act as a molecular switch to regulate mitochondrial docking and/or fusion. The fzo1(K200A) and fzo1(S201N) mutations in the G1 motif are equivalent to mutations in the Ras GTPase that reduce guanine nucleotide binding (Sigal et al., 1986; Feig and Cooper, 1988; Farnsworth and Feig, 1991). The fzo1(T221A) substitution in the G2 motif is based on a Ras mutation which eliminates interactions with effector molecules (Adari et al., 1988; Cales et al., 1988; Vojtek et al., 1993; Murphy et al., 1997). None of these mutations disrupted Fzo1p localization suggesting that GTP binding and/or hydrolysis is not required to target Fzo1p to mitochondria.
In contrast, the fzo1(K371A) mutation in the G4 motif retained wild-type function and was able to rescue defects in mitochondrial fusion, mtDNA maintenance, and glycerol growth in fzo1 mutant cells. This lysine is highly conserved among the superfamily of GTPases (Bourne et al., 1991) and is required for efficient nucleotide binding and stablilization of the GTP binding pocket in Ras (Lys117) (Der et al., 1988; Pai et al., 1990). Although mutating Lys117 in Ras significantly reduces its affinity for GTP, it does not disrupt its oncogenic potential (Clanton et al., 1986; Der et al., 1988). In addition, the analogous mutation in the G4 motif of Drosophila Fzo did not completely disrupt mitochondrial fusion (Hales and Fuller, 1997). These results suggest that the yeast and fly Fzo proteins may have a high intrinsic affinity for GTP that is not completely compromised by alterations in the G4 lysine. Alternatively, this lysine may be completely (yeast Fzo1p) or partially (fly Fzo) dispensable with respect to nucleotide binding.
Mutant forms of the yeast dynamin-like proteins Vps1p and Dnm1p (Vater et al., 1992; Otsuga et al., 1998), mammalian dynamin (Herskovits et al., 1993; van der Bliek et al., 1993), and Ras (Sigal et al., 1986; Feig and Cooper, 1988; Farnsworth and Feig, 1991) induce dominant-interfering phenotypes when overexpressed in wild-type cells. These dominant phenotypes are thought to result because the mutant forms of the proteins either titrate out or block the activities of binding partners required for their function. In contrast, none of the disabled forms of Fzo1p we tested induced dominant mitochondrial phenotypes in wild-type cells (data not shown). The simplest interpretation of these results is that Fzo1p acts alone or that mutations in the GTPase domain of Fzo1p completely disrupt its ability to interact with itself or other proteins.
Models for the Mechanism of Fzo1p Action
Given what is known regarding the role of other integral membrane proteins in fusion, we propose that Fzo GTPases act as molecular switches to directly mediate mitochondrial docking and/or membrane fusion. Both the domain structure of the Fzo family members and the topology we have determined for Fzo1p are consistent with this model. Fzo molecules share structural features with two classes of integral membrane proteins that mediate membrane fusion events. The SNAREs regulate membrane docking and fusion during vesicle transport and during the homotypic fusion of organelle membranes (Denesvre and Malhotra, 1996; Hay and Scheller, 1997; Edwardson, 1998; Götte and Fischer von Mollard, 1998; Weber et al., 1998). The viral-type fusion proteins regulate extracellular membrane docking and fusion events (Hernandez et al., 1996; Huovila et al., 1996). Both SNAREs and viral fusion proteins contain multiple heptad repeats in their amino-terminal domains. These repeats form parallel coiled coils that are proposed to mediate the direct association of v- and t-SNAREs on opposing membranes and the oligomerization of viral fusion proteins within a membrane (Kee et al., 1995; Hernandez et al., 1996; Hanson et al., 1997; Hay and Scheller, 1997; Hughson, 1997). Our studies indicate that the two amino-terminal heptad repeats in Fzo1p extend into the cytoplasm. If Fzo1p serves a SNARE-like function, these repeats could mediate the direct or indirect association of Fzo1p with itself or with another protein binding partner on an opposing mitochondrial membrane. The cytoplasmic GTPase domain of Fzo1p might function to control the rate or fidelity of such Fzo1p interactions, similar to the manner in which Rab GTPases regulate SNARE–SNARE associations (Lupashin and Waters, 1997). The idea that Fzo1p functions as a novel type of mitochondrial SNARE is attractive, given that none of the v- or t-SNAREs encoded by the S. cerevisiae genome localize to the mitochondrial compartment (H.R.B. Pelham, personal communication).
Alternatively, Fzo1p may operate more like viral fusion proteins (White, 1990, 1992; Hernandez et al., 1996; Hughson, 1997; Qiao et al., 1998). These integral membrane proteins contain additional hydrophobic fusion peptides that normally remain masked. When activated, the fusion peptide inserts into the target membrane generating a docked state. Conformational changes in the protein pull the membranes together and promote fusion. Fzo1p contains two hydrophobic sequences in its cytoplasmic domain that resemble fusion peptides (Hermann, 1998). It is possible that Fzo1p uses its GTPase activity as a switch to stimulate the insertion of these putative fusion peptides into a neighboring mitochondrial membrane. The GTPase domain might also regulate conformational changes in Fzo1p that pull the two docked mitochondrial membranes together after peptide insertion. The observation that Fzo1p is tightly associated with both mitochondrial membranes suggests that it might regulate the fusion of the inner membrane as well. Whether the predicted transmembrane domain and heptad repeat closest to the carboxy terminus of Fzo1p are in a position to influence inner membrane behavior remains to be seen.
Acknowledgments
We thank R. Jensen (Johns Hopkins University, Baltimore, MD) for the mito-GFP plasmid and cytochrome b2 antibody, the staff of the Research Microscopy Facility (K.H. Albertine and N.B. Chandler) at the University of Utah Health Sciences Center (Salt Lake City, UT) for assistance with the ultrastructural studies, and Q. Tieu (University of California, Davis, CA) for technical assistance isolating the fzo1-1 allele.
This work was supported by grants from the American Cancer Society (CB-97) and the National Institutes of Health (NIH) (GM-53466) to J.M. Shaw, the National Science Foundation (MCB-9724143) to J. Nunnari, and the NIH (HD-29194) to M.T. Fuller. G.J. Hermann, K.G. Hales, and J.P. Mills were supported by NIH training grants (GM-07464, HG-00044, and GM-07790, respectively). The University of Utah Research Microscopy Facility is supported by a grant from the NIH (S10-RR-10489). The Utah Health Sciences Sequencing Facility is supported by a National Cancer Institute grant (5-P30CA42014).
Abbreviations used in this paper
- 3-PGK
3-phosphoglycerate kinase
- 5-FOA
5-fluoro-orotic acid
- CoxIV
cytochrome oxidase subunit IV
- DAPI
4′,6-diamidino-2-phenylindole
- DiOC6
3,3′ dihexyloxacarbocyanine
- fzo
fuzzy onions gene
- GFP
green fluorescent protein
- mtDNA
mitochondrial DNA
Footnotes
Address all correspondence to Janet M. Shaw, Department of Biology, University of Utah, Salt Lake City, UT 84112. Tel.: (801) 585-6205. Fax: (801) 581-4668. E-mail: shaw@bioscience.utah.edu, or Jodi Nunnari, Department of Molecular and Cellular Biology, University of California, Davis, CA 95616. Tel.: (530) 754-9774. Fax: (530) 752-7522. E-mail: fzmito@peseta.ucdavis.edu
References
- Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 raseffector binding domain. Science. 1988;240:518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Azpiroz R, Butow RA. Patterns of mitochondrial sorting in yeast zygotes. Mol Biol Cell. 1993;4:21–36. doi: 10.1091/mbc.4.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakeeva LE, Chentsov YS, Skulachev VP. Mitochondrial framework (reticulum mitochondriale) in rat diaphragm muscle. Biochim Biophys Acta. 1978;501:349–369. doi: 10.1016/0005-2728(78)90104-4. [DOI] [PubMed] [Google Scholar]
- Bakeeva LE, Chentsov YS, Skulachev VP. Ontogenesis of mitochondrial reticulum in rat diaphragm muscle. Eur J Cell Biol. 1981;25:175–181. [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. . Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cales C, Hancock JF, Marshall CJ, Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the rasgene product. Nature. 1988;332:548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- Chen LB. Mitochondrial membrane potential in living cells. Annu Rev Cell Biol. 1988;4:155–181. doi: 10.1146/annurev.cb.04.110188.001103. [DOI] [PubMed] [Google Scholar]
- Clanton DJ, Hattori S, Shih TY. Mutations of the rasgene product p21 that abolish guanine nucleotide binding. Proc Natl Acad Sci USA. 1986;83:5076–5080. doi: 10.1073/pnas.83.14.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Denesvre C, Malhotra V. Membrane fusion in organelle biogenesis. Curr Opin Cell Biol. 1996;8:519–523. doi: 10.1016/s0955-0674(96)80030-5. [DOI] [PubMed] [Google Scholar]
- Der CJ, Weissman B, MacDonald MJ. Altered guanine nucleotide binding and H-rastransforming and differentiating activities. Oncogene. 1988;3:105–112. [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B. 1981. Mitochondrial genetics and functions. In Molecular Biology of the Yeast Saccharomyces. J.M. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 505–635.
- Edwardson JM. Membrane fusion: All done with SNAREpins? . Curr Biol. 1998;8:R390–R393. doi: 10.1016/s0960-9822(98)70245-3. [DOI] [PubMed] [Google Scholar]
- Farnsworth CL, Feig LA. Dominant inhibitory mutations in the Mg2+-binding site of RasHprevent its activation by GTP. Mol Cell Biol. 1991;11:4822–4829. doi: 10.1128/mcb.11.10.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA, Cooper GM. Inhibition of NIH 3T3 cell proliferation by a mutant rasprotein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox TD, Folloy LS, Mulero JJ, McMullin TW, Thorsness PE, Hedin LO, Costanzo MC. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Fuller, M.T. 1993. Spermatogenesis. In The Development of Drosophila melanogaster. M. Bate and A. Martinez-Arias, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 71–147.
- Götte M, Fischer von Mollard G. A new beat for the SNARE drum. Trends Cell Biol. 1998;8:215–218. doi: 10.1016/s0962-8924(98)01272-0. [DOI] [PubMed] [Google Scholar]
- Guan K, Farh L, Marshall TK, Deschenes RJ. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1gene. Curr Genet. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane, editors. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 726 pp.
- Hay JC, Scheller RH. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G.J. 1998. Mitochondrial inheritance and morphology in yeast. Ph.D. thesis. University of Utah, Salt Lake City, UT. 242 pp.
- Hermann GJ, Shaw JM. Mitochondrial dynamics in yeast. Annu Rev Cell Dev Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:565–578. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Herskovits JS, Burgess CC, Obar RA, Vallee RB. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H, Avers CJ. Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science. 1973;181:749–751. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- Hughson FM. Enveloped viruses: a common mode of membrane fusion? . Curr Biol. 1997;7:R565–R569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- Huovila AJ, Almeida EAC, White JM. ADAMs and cell fusion. Curr Opin Cell Biol. 1996;8:692–699. doi: 10.1016/s0955-0674(96)80111-6. [DOI] [PubMed] [Google Scholar]
- Ichas F, Jouaville LS, Mazat JP. Mitochondria are excitable organelles capable of generating and conveying electrical and calcium signals. Cell. 1997;89:1145–1153. doi: 10.1016/s0092-8674(00)80301-3. [DOI] [PubMed] [Google Scholar]
- Isenmann S, Khew-Goodall Y, Gamble J, Vadas M, Wattenberg B. A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting signal. Mol Biol Cell. 1998;9:1649–1660. doi: 10.1091/mbc.9.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Lin RC, Hsu S, Scheller R. Distinct domains of syntaxin are required for synaptic vesicle fusion complex formation and dissociation. Cell. 1995;14:991–998. doi: 10.1016/0896-6273(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Koning AJ, Lum PY, Williams JM, Wright R. DiOC6staining reveals organelle structure and dynamics in living yeast cells. Cell Motil Cytoskeleton. 1993;25:111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Lazzarino DA, Boldogh I, Smith MG, Rosand J, Pon LA. Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol Biol Cell. 1994;5:807–818. doi: 10.1091/mbc.5.7.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D., and K.T. Tokuyasu. 1980. Spermatogenesis. In Genetics and biology of Drosophila. Vol. 2. M. Ashburner and T.R. Wright, editors. Academic Press, New York. 225–294.
- Lupashin VV, Waters MG. t-SNARE activation through transient interaction with a rab-like guanosine triphosphatase. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E.F. Fritsch, and J. Sambrook. 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 545 pp.
- Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GA, Moore MS, Drivas G, de la Ossa PP, Villamarin A, D'Eustachio P, Rush MG. A T42A Ran mutation: differential interactions with effectors and regulators, and defect in nuclear protein import. Mol Biol Cell. 1997;8:2591–2604. doi: 10.1091/mbc.8.12.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HRB, Wickner W, Haas A. Homotypic vacuolar fusion mediated by v- and t-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiaeis determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H, Hanson PI, Jahn R. Assembly and disassembly of a ternary complex of synaptobrevin, syntaxin, and SNAP-25 in the membrane of synaptic vesicles. Proc Natl Acad Sci USA. 1997;94:6197–6201. doi: 10.1073/pnas.94.12.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 angstrom resolution: implications for the mechanism of GTP hydrolysis. EMBO (Eur Mol Biol Organ) J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SK, Indig FE, Oliviera N, Levine ND, Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Transport vesicle docking: SNAREs and associates. Annu Rev Cell Biol. 1996;12:441–461. doi: 10.1146/annurev.cellbio.12.1.441. [DOI] [PubMed] [Google Scholar]
- Pon L, Moll T, Vestweber D, Marshallsay B, Schatz G. Protein import into mitochondria: ATP-dependent protein translocation activity in a submitochondrial fraction enriched in membrane contact sites and specific proteins. J Cell Biol. 1989;109:2603–2616. doi: 10.1083/jcb.109.6.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon, L., and G. Schatz. 1991. Biogenesis of Yeast Mitochondria. In The Molecular Biology of the Yeast Saccharomyces. J.R. Broach, J.R. Pringle, and E.W. Jones, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 334–406.
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Qiao H, Pelletier SL, Hoffman L, Hacker J, Armstrong RT, White JM. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J Cell Biol. 1998;141:1335–1347. doi: 10.1083/jcb.141.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Fremont P, Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Roeder AD, Shaw JM. Vacuole partitioning during meiotic division in yeast. Genetics. 1996;144:445–458. doi: 10.1093/genetics/144.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AD, Hermann GJ, Keegan BR, Thatcher SA, Shaw JM. Mitochondrial inheritance is delayed in Saccharomyces cerevisiae cells lacking the serine/threonine phosphatase, PTC1. . Mol Biol Cell. 1998;9:917–930. doi: 10.1091/mbc.9.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. The protein machinery of vesicle budding and fusion. Prot Sci. 1996;5:185–194. doi: 10.1002/pro.5560050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. . Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY. 186 pp.
- Sigal IS, Gibbs JB, D'Alonzo JS, Temeles GL, Wolanski BS, Socher SH, Scolnick EM. Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc Natl Acad Sci USA. 1986;83:952–956. doi: 10.1073/pnas.83.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T, Whitehart SW, Brunner M, Erdjumentbronage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stevens, B. 1981. Mitochondrial Structure. In The Molecular Biology of the Yeast Saccharomyces. J.M. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 471–504.
- van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homologue of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachi M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- White JM. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- White JM. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiaestrains that are isogenic to S228C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Yaffe MP. Isolation and analysis of mitochondrial inheritance mutants from Saccharomyces cerevisiae. . Methods Enzymol. 1995;260:447–453. doi: 10.1016/0076-6879(95)60157-0. [DOI] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. . Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]