Abstract
The mammalian protein TAP/p115 and its yeast homologue Uso1p have an essential role in membrane traffic (Nakajima et al., 1991; Waters et al., 1992; Sztul et al., 1993; Rabouille et al., 1995). To inquire into the site and mechanism of TAP/p115 action, we aimed to localize it and to identify domains required for its function. We show that in interphase cells, TAP/p115 localizes predominantly to the Golgi and to peripheral structures that represent vesicular tubular clusters (VTCs) involved in ER to Golgi transport. Using BFA/ nocodazole treatments we confirm that TAP/p115 is present on ER to Golgi transport intermediates. TAP/ p115 redistributes to peripheral structures containing ERGIC-53 during a 15°C treatment, suggesting that it is a cycling protein. Within the Golgi, TAP/p115 is associated with pleiomorphic structures on the cis side of the cis-Golgi cisterna and the cis-most cisterna, but is not detected in more distal compartments of the Golgi.
TAP/p115 binds the cis-Golgi protein GM130, and the COOH-terminal acidic domain of TAP/p115 is required for this interaction. TAP/p115 interaction with GM130 occurs only in the Golgi and is not required for TAP/p115 association with peripheral VTCs. To examine whether interaction with GM130 is required to recruit TAP/p115 to the Golgi, TAP/p115 mutants lacking the acidic domain were expressed and localized in transfected cells. Mutants lacking the GM130-binding domain showed normal Golgi localization, indicating that TAP/p115 is recruited to the Golgi independently of its ability to bind GM130. Such mutants were also able to associate with peripheral VTCs. Interestingly, TAP/p115 mutants containing the GM130-binding domain but lacking portions of the NH2-terminal region were restricted from the Golgi and localized to the ER. The COOH-terminal domain required for GM130 binding and the NH2-terminal region required for Golgi localization appear functionally relevant since expression of TAP/p115 mutants lacking either of these domains leads to loss of normal Golgi morphology.
Keywords: TAP, p115, Golgi, GM 130, VTC
Movement of membrane and cargo through the secretory pathway proceeds via transport intermediates that originate from one compartment and fuse with the next compartment in the pathway. Through a combination of yeast genetics, neurochemistry, and cell-free transport assays, a number of proteins have been identified that regulate the fusion process. Prominent among these are the transmembrane proteins known as SNAREs. According to the SNARE1 (N-ethylmaleimide– sensitive factor attachment protein receptor) hypothesis, fusion of a transport vesicle with its target membrane is dependent on pairing of a compartment-specific vesicle (v)-SNARE with its cognate target (t)-SNARE (Söllner et al., 1993; Rothman, 1994). Consistent with the idea that SNAREs provide specificity in fusion of transport vesicles with their target, a growing number of v- and t-SNAREs have been identified and linked to specific stages of transport (Pfeffer, 1996; Hay and Scheller, 1997).
Distinct families of proteins are known to regulate fusion by modulating SNARE interactions. Proteins of the sec1/sly1 family associate with t-SNAREs before SNARE complex assembly and appear to regulate t-SNAREs in both a positive and negative manner (Pevsner, 1996; Halamachi and Lev, 1996). The rab/ypt1 group of ras related GTPases also regulate SNARE complex formation (Pfeffer, 1994; Novick and Zerial, 1997), and there is evidence that they associate with and activate t-SNAREs (Lupashin and Waters, 1997). Like SNAREs, the individual members of the sec1/sly1 and rab/ypt1 families of proteins act at specific transport stages.
Additional proteins have been shown to be required during events leading up to membrane fusion. Bovine p115 was initially purified as a soluble factor required in an intra-Golgi transport assay (Waters et al., 1992) and subsequently shown to be also essential in a cell-free assay reconstituting regrowth of Golgi cisternae from postmitotic Golgi fragments (Rabouille et al., 1995, 1998). Within that assay, binding of p115 to the cis-Golgi matrix protein GM130 is required for cisternal regrowth (Nakemura et al., 1997). The rat protein TAP was found to be necessary in an assay measuring fusion of transcytotic vesicles with the plasma membrane (Sztul et al., 1993). Dissection of this assay into docking and fusion steps showed that TAP is required for vesicle docking to the target membrane before fusion (Barroso et al., 1995). Cloning of bovine p115 and rat TAP revealed them to be 90% identical (Barroso et al., 1995; Sapperstein et al., 1995) with homology to yeast Uso1p (Nakajima et al., 1991). Sequence similarity between TAP/p115 and Uso1p is highest in three homology regions (H1, H2, and H3). H1 and H2 are found in the NH2-terminal approximately one-third of TAP/p115. H3 is at the COOH terminus and contains an acidic domain at the extreme COOH terminus. Analysis of TAP/p115 and Uso1p with the COILS program (Lupas et al., 1991) predicts extensive coiled-coil formation in the COOH-terminal region, which likely form the rod-like tail (Barroso et al., 1995; Sapperstein et al., 1995; Yamakawa et al., 1996). In agreement, electron micrographs of purified TAP/p115 show a homodimer, similar in overall appearance to myosin, with two globular heads and an extended rod-like tail (Sapperstein et al., 1995). Uso1p has a similar homodimeric structure, with a longer tail than TAP/p115 (Yamakawa et al., 1996). Uso1p has been shown to be involved in ER to Golgi transport (Nakajima et al., 1991), and more recently, for the formation of the ER to Golgi SNARE complex (Sapperstein et al., 1996), at a step before ATP hydrolysis by NSF (Lupashin et al., 1996). In agreement with the tethering function shown for TAP/ p115, Uso1p appears required for docking of ER-derived vesicles with Golgi membranes (Barlowe, 1997). The elongated structure of TAP/p115 and Uso1p seem suited to their proposed function in the docking of transport vesicles with target membrane (Barroso et al., 1995, Bajjalieh and Scheller, 1995; Sapperstein et al., 1996; Barlowe, 1997; Lowe et al., 1998; Orci et al., 1998).
To further investigate the role of TAP/p115 in membrane transport, we sought to identify subcellular compartments in which it is present and identify regions within TAP/p115 mediating its localization and its interactions with other proteins. In this report, we show that TAP/p115 is associated with peripheral vesicular tubular clusters (VTCs), with pleiomorphic structures adjoining the cis- side of the Golgi stack and the cis-most Golgi cisterna, and that it cycles between the Golgi and earlier compartments of the secretory pathway. Within the Golgi, TAP/p115 interacts with the cis-Golgi protein GM130. This interaction is not required for Golgi localization of TAP/p115 but is functionally essential since expression of TAP/p115 mutants lacking the GM130-binding domain lead to Golgi structure disruption. In addition, the combination of recombinant protein binding and transfection assays identified another relevant region within TAP/p115: the NH2-terminal H2 domain is required for TAP/p115 localization to the Golgi and expression of NH2 terminally truncated TAP/p115 results in a loss of compact Golgi morphology in transfected cells.
Materials and Methods
Antibodies
Rabbit polyclonal antibodies against TAP/p115 were generated by immunization with affinity-purified rat TAP/p115. The anti-TAP monoclonal antibody 5D6 has been described previously (Barroso et al., 1995). Mouse and rabbit polyclonal antibodies against GM130 were generated by immunization with inclusion bodies from bacteria expressing full-length GM130. Anti-giantin monoclonal G1/133 (Linstedt and Hauri, 1993) and anti–ERGIC-53 monoclonal G1/93 (Schweizer et al., 1988) were provided by H.-P. Hauri (University of Basel, Basel, Switzerland). Polyclonal antibodies against mannosidase II were kindly provided by M. Farquhar (University of California, San Diego, CA). Monoclonal antibodies against mannosidase II were purchased from Berkeley Antibody Co. (Berkeley, CA). Monoclonal antibodies against the Golgi marker 58K (Golgi 58K protein) were purchased from Sigma Biosciences (St. Louis, MO). Polyclonal antibodies against calnexin (SPA-860) were purchased from StressGen Biotechnologies (Victoria, BC, Canada). Goat anti–rabbit and anti– mouse antibodies conjugated with FITC or rhodamine were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Affinity Purification and Cross-linking of Antibodies
TAP/p115 was affinity purified from rat liver cytosol using the 5D6 monoclonal antibody as previously described (Barroso et al., 1995), run on 7.5% acrylamide gels, and then transferred to nitrocellulose. Strips of nitrocellulose containing TAP/p115 were incubated with immune serum in PBS, 5% dried milk, and 0.1% Tween 20 for 3 h at room temperature. Bound antibodies were eluted with 0.1 M glycine, pH 3.0, then neutralized with a 0.1 vol of 1 M Tris, pH 7.4. Anti-TAP/p115 antibodies were cross-linked to protein A–Sepharose 4 FF (Pharmacia Biotech, Piscataway, NJ) with dimethyl pimelimidate (Pierce Chemical Co., Rockford, IL) according to the manufacturer's protocol.
Fractionation of Rat Liver
Rat liver fractions were prepared by modification of a procedure previously described (Taylor et al., 1997). 200–300-g Sprague-Dawley rats were anesthetized with sodium pentobarbital. Livers were excised and chilled and subsequent steps were performed on ice or at 4°C. Livers were minced, washed three times in PBS, and then resuspended in 3 vol of buffer containing 0.5 M sucrose, 0.1 M potassium phosphate, 5 mM MgCl2, 1 mM DTT, pH 7.0, 0.5 M SPMD. The SPMD buffer contained the protease inhibitors antipain (2 μg/ml), aprotinin (5 μg/ml), leupeptin (5 μg/ml), and pepstatin (1 μg/ml). The sample was homogenized by 15 strokes in a 40-ml Dounce tissue grinder (Wheaton, Millville, NJ) and spun at 1,000 g for 10 min. The postnuclear supernatant (PNS) was applied to a sucrose step gradient consisting of 1.3 M SPMD, 0.86 M SPMD, the PNS in 0.5 M SPMD, and 0.25 M SPMD. The gradient was centrifuged for 2 h at 25,000 rpm in a Beckman SW28 rotor (Beckman Instrs., Fullerton, CA). Fractions were removed from the top of the tube using a Pasteur pipette. The cytosolic fraction was collected from the 0.5 M SPMD layer. The stacked Golgi (SG) fraction was collected at the 0.5 M–0.86 M SPMD interface. The 0.86 M and 1.3 M SPMD layers were collected and pooled to make up the ER fraction.
Immunoprecipitation
Fractions were solubilized in a buffer containing 20 mM Hepes, 100 mM KCl, pH 7.4, 1 mM MgCl2, 0.1 mM EDTA, 1 mM DTT (HKMED) and 1% Triton X-100 or a combination of 1% Triton X-100, 0.5% deoxycholate, and 0.1% SDS. After incubation at 4°C for 30 min, samples were centrifuged at 16,000 g for 15 min. Supernatants were incubated at 4°C with anti-TAP/p115 antibodies for 2 h and protein A–Sepharose 4 FF for 1 h, and then washed four times with HKMED buffer containing the detergent used for solubilization. Precipitates were analyzed by SDS-PAGE followed by silver staining (Ansorge et al., 1985).
Purification of p135 for Peptide Sequencing
The SG fractions obtained from 10 rat livers were pooled and solubilized in HKMED 1% Triton X-100 for 30 min. p135 was purified from the SG fraction by coimmunoprecipitation with TAP/p115 using affinity-purified anti-TAP/p115 antibodies cross-linked to protein A–Sepharose 4 FF. Bound protein was eluted from beads with 0.1 M cyclohexylaminopropanesulfonic acid buffer, pH 10.5. The eluted protein was precipitated by addition of four parts 50:50 methanol/acetone solution followed by chilling overnight at −20°C and centrifugation at 16,000 g. The precipitated protein was analyzed on 7.5% SDS-PAGE and stained with Coomassie brilliant blue R-250. Approximately 8 μg of p135 were obtained. A gel slice containing p135 was excised and washed three times in 50:50 water/ acetonitrile. Preparation of p135 peptides from the gel slice by trypsin digestion and HPLC microbore separation, and peptide sequencing by Edman degradation was performed by Harvard University Microchemistry.
TAP/p115 and GM130 Constructs
NH2 terminally truncated GM130 (GM130-N75) was cloned by PCR from a rat brain cDNA library. A Bluescript plasmid encoding full-length GM130 was a gift of N. Nakamura and G. Warren. TAP/p115 mutants were constructed using restriction sites within the sequence, or restriction sites were introduced by PCR. Standard molecular techniques were used for growth of bacteria, plasmid purification, plasmid digestion, and transformation of bacteria (Sambrook et al., 1989).
TAP/p115 In Vitro Binding Experiments
To test binding of full-length and truncated forms of TAP/p115 and GM130, proteins encoded in pET21b or pET21c vectors were expressed in Escherichia coli BL21. Cells were lysed by sonication in 100 mM KCl, 20 mM Tris, pH 7.4, 1 mM DTT (100 KTD) and centrifuged at 100,000 g for 1 h, and the resulting supernatants were used for binding experiments. Bacterial supernatants containing TAP FL, TAP-C934, TAP-C708, TAP-N372, or TAP/p115 from rat liver cytosol were incubated with supernatant containing GM130-FL or GM130 N-75 in 100 KTD, 1% Triton X-100. Samples were immunoprecipitated with affinity purified polyclonal anti-TAP/p115 antibodies and analyzed by SDS-PAGE. The percent of GM130 in the binding reaction that coprecipitated with TAP/p115 was determined by scanning gels with a Bio-Rad GS-700 imaging densitometer (Bio-Rad Laboratories, Hercules, CA).
Immunofluorescence Microscopy
Cells grown on glass coverslips were washed three times in PBS and fixed in 3% paraformaldehyde in PBS for 10 min at room temperature. Paraformaldehyde was quenched with 10 mM ammonium chloride and cells were permeabilized with PBS, 0.1% Triton X-100 for 7 min at room temperature. The coverslips were washed three times for 5 min each with PBS, 0.2% Tween 20, then blocked in PBS, 0.4% fish skin gelatin, 0.2% Tween 20 for 5 min, followed by blocking in PBS, 2.5% goat serum, 0.2% Tween for 5 min. Cells were incubated with primary antibody diluted in PBS, 0.4% fish skin gelatin, 0.2% Tween 20 for 45 min at 37°C. Coverslips were washed five times for 5 min each with PBS, 0.2% Tween 20. Secondary antibodies coupled to FITC or rhodamine were diluted in 2.5% goat serum and incubated on coverslips for 30 min at 37°C. Coverslips were washed as above and mounted on slides in 9:1 glycerol/PBS with 0.1% q-phenylenediamine. Slides were examined on a Leitz Orthoplan microscope (Wetzlar, Germany) equipped for epifluorescence. Photographs were taken on Kodak Gold film (Eastman Kodak Co., Rochester, NY) at 1,000 ASA with an adjoining Leitz Orthomal camera system.
Transfection of COS-7 Cells
COS-7 cells growing on coverslips were transiently transfected using a calcium phosphate transfection system (GIBCO BRL, Gaithersburg, MD). 5 μg of DNA expressing the appropriate TAP/p115 construct was used per 10-mm dish of cells. 24 h after transfection, cells were lysed with 0.5 ml of 50 mM Tris-HC1, pH 7.5, 150 mM NaC1, 1% NP-40, 0.5% DOC, 0.1% SDS (RIPA) buffer, the lysate was centrifuged at 1,000 g for 10 min, and the resulting PNS was recovered. PNS was processed for SDS-PAGE, the gel transferred to NC, and the filter was immunoblotted with anti-rat TAP/p115 antibodies.
Results
TAP/p115 Cycles between the Golgi and Earlier Secretory Compartments
TAP/p115 has been localized to the Golgi by immunofluorescence microscopy (Waters et al., 1992; Nakamura et al., 1997), and more specifically to the cis-side of the Golgi by confocal microscopy (Shima et al., 1997). In agreement with these results, we found that TAP/p115 colocalizes with the Golgi marker giantin (Linstedt and Hauri, 1993) in a characteristic juxtanuclear pattern in normat rat kidney cells (Fig. 1, A–C). In addition, TAP/p115 was seen in peripheral punctate structures that did not contain giantin (Fig. 1 C, arrowheads). Distribution to both the Golgi and peripheral structures is suggestive of the pattern seen with proteins occupying the ER to Golgi intermediate compartment (ERGIC) and cycling between the ER and the Golgi (for review see Hauri and Schweizer, 1992; Pelham, 1997; Bannykh and Balch, 1997). The ERGIC is made up of pleiomorphic transport intermediates described as VTCs (Bannykh et al., 1998; Balch et al., 1994). VTCs form throughout the cell, translocate in a microtubule dependent manner to the cis-region of the Golgi, where they appear to merge with the Golgi (Presley et al., 1997; Scales et al., 1997).
Figure 1.
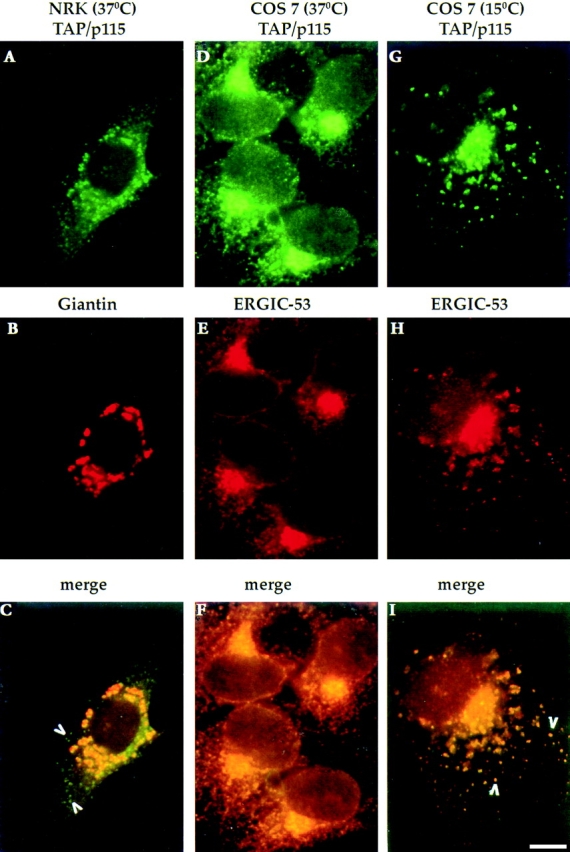
TAP/p115 is a cycling protein. NRK or COS-7 cells were incubated at 37° or at 15°C for 2 h, fixed, and then processed for IF. In NRK cells, TAP/p115 (A) colocalizes with the Golgi marker giantin (B) in a perinuclear region. TAP/p115 is also present in peripheral punctate structures that do not contain giantin (arrowheads in C). In COS-7 cells incubated at 37°C, TAP/ p115 (D) and ERGIC-53 (E) colocalize in a juxtanuclear region and in peripheral punctate structures (F). In COS-7 cells incubated at 15°C, TAP/p115 (G) and ERGIC-53 (H) colocalize in dispersed punctate structures (arrowheads in I). Bar, 10 μm.
To determine if TAP/p115 localizes to VTCs, the distribution of TAP/p115 and the VTC marker ERGIC-53 (Schweizer et al., 1988, 1991) were compared in COS-7 cells. As shown in Fig. 1 D, TAP/p115 was found in the Golgi and in peripheral punctate structures. ERGIC-53 (Fig. 1 E) was detected in the Golgi and in peripheral VTCs, in agreement with its localization in COS-1 cells (Itin et al., 1995). TAP/p115 and ERGIC-53 colocalized in the Golgi region and in the peripheral punctate structures (Fig. 1 F), suggesting that TAP/p115 is present in VTCs and is likely to cycle between the Golgi and earlier compartments of the secretory pathway.
ERGIC-53 is a cycling protein and to confirm that TAP/ p115 also cycles, its distribution was compared with that of ERGIC-53 in cells incubated at reduced temperature. Incubation of cells at 15°C inhibits the anterograde transport of VTCs from the periphery to the Golgi region, whereas retrograde transport appears less affected. The net effect on the localization of cycling proteins is a redistribution from the Golgi to enlarged peripheral VTCs (Lippincott-Schwartz et al., 1990; Schweizer et al., 1990; Saraste and Svensson, 1991). To determine if TAP/p115 exhibits such redistribution, cells were incubated at 15°C for 2 h, and TAP/p115 and ERGIC-53 were then localized. TAP/p115 (Fig. 1 G) and ERGIC-53 (Fig. 1 H) were present in enlarged peripheral elements scattered throughout the cell and colocalized in all structures (Fig. 1 I, arrowheads). These results suggest that TAP/p115 cycles between peripheral VTCs and the Golgi. It is possible that TAP/p115, like ERGIC-53, cycles through the ER, but this remains to be determined.
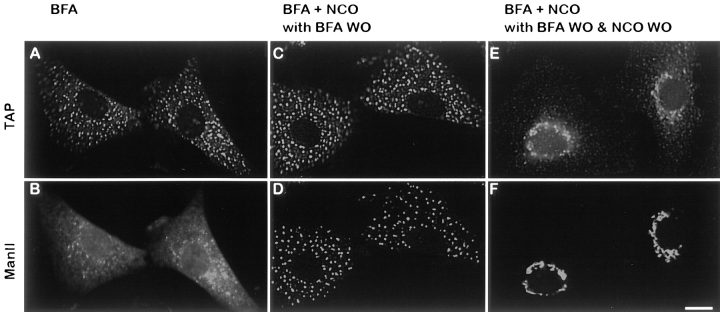
VTCs have been shown to mediate transport of cargo from the ER to the Golgi (Presley et al., 1997). To show directly that structures containing TAP/p115 are transport intermediates, their role in mannosidase II transport from the ER to the Golgi was analyzed. NRK cells were first treated with BFA to relocate mannosidase II from the medial-Golgi to the ER. After a 30-min BFA treatment, mannosidase II was detected in a characteristic ER pattern (Fig. 2 B), whereas TAP/p115 was found in punctate structures scattered throughout the cell (Fig. 2 A). Cells were then treated with nocodazole for 90 min to depolymerize microtubules. Addition of nocodazole did not cause a change in mannosidase II or TAP/p115 patterns (data not shown). BFA was then washed out during a 30-min incubation at 37°C in the presence of nocodazole to prevent movement of VTCs towards the microtubule organizing center (MTOC). As shown in Fig. 2, C and D, mannosidase II moved from the ER into the TAP/p115-containing structures, in agreement with previous results showing that traffic from the ER to VTCs is not microtubule dependent (Cole et al., 1996). Subsequent wash-out of nocodazole during a 45-min incubation at 37°C leads to migration of structures containing TAP/p115 and mannosidase II towards the MTOC and their coalescence to form a normal Golgi structure (Fig. 2, E and F). These results suggest that functional transport intermediates contain TAP/p115.
Figure 2.
Peripheral structures containing TAP/p115 are transport intermediates. NRK cells were treated with 5 μg/ml BFA for 30 min, fixed, and processed for IF. TAP/p115 (A) is present in punctate structures, whereas mannosidase II (B) is localized to the ER. Cells like in A and B were treated with 10 μg/ml nocodazole for 90 min. Cells were then incubated in medium without BFA but containing nocodazole (BFA + NCO with BFA WO) for 30 min, fixed, and processed for IF. Mannosidase II (D) redistributed from the ER into punctate structures containing TAP/p115 (C). Cells like in C and D were subjected to nocodazole wash-out (BFA + NCO with BFA WO & NCO WO) for 45 min, fixed, and then processed for IF. TAP/p115 (E) and mannosidase II (F) colocalized in a morphologically normal Golgi complexes. Bar, 10 μm.
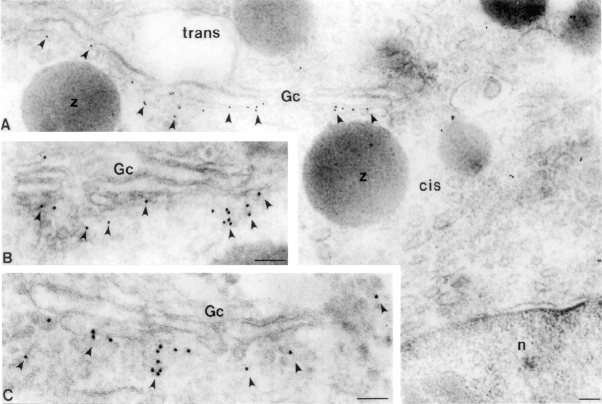
Within the Golgi, cycling proteins such as the ERGIC-53 homologue p58 can be detected in pleiomorphic structures adjoining the cis-side of the Golgi stack and the cis-cisterna (Lahtinen et al., 1992; Saraste and Svensson, 1991). To examine the localization of TAP/p115 in the Golgi, immunogold labeling was performed on frozen sections of rat pancreas. This tissue was selected because the morphology of pancreatic acinar cells has been well studied and allows one to distinguish between the cis and trans-sides of the Golgi complex (Farquhar, 1981; Farquhar and Palade, 1981). As shown in Fig. 3 A–C (arrowheads), gold particles were localized to cisternal and noncisternal elements on the cis-side of the Golgi stack. Cisternal labeling was almost entirely restricted to the highly fenestrated cis-most cisterna, with little labeling of more medial-cisternae (Fig. 3 A). Noncisternal labeling was found over structures with a vesicular appearance in close proximity to the cis-most cisternae (Fig. 3, B and C). Quantitation of TAP/p115 distribution close to the Golgi complex in multiple micrographs of distinct pancreatic cells, indicates that ∼60% of gold grains are associated with pleiomorphic structures and ∼40% with the cis-most cisterna.
Figure 3.
TAP/p115 associates with vesicular tubular structures and cis-Golgi membranes. Immunogold localization of TAP/p115 (arrowheads) in pancreatic acinar cells. Gc, Golgi complex; n, nucleus. Bar, 0.2 μm.
Taken together, the data indicate that TAP/p115 associates with VTCs in the periphery of cells and with the pleiomorphic structures in the Golgi region and that TAP/p115 cycles between the Golgi and the peripheral VTCs. The results suggest that TAP/p115 might move with VTCs as they translocate from cell periphery to the Golgi.
TAP/p115 Binds the cis-Golgi Protein GM130 Exclusively in the Golgi
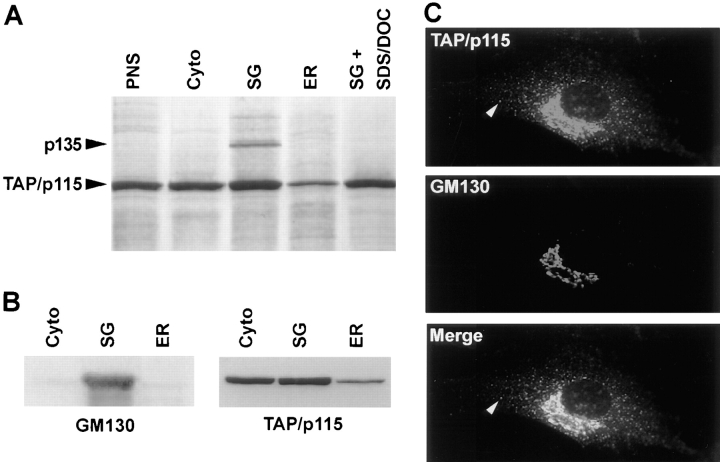
TAP/p115 is associated with Golgi and VTC membranes in vivo (Figs. 1–3) and since TAP/p115 is a soluble protein, it is likely that its binding to membranes is mediated by interactions with other proteins. To identify TAP/p115 associated protein(s), anti-TAP/p115 antibodies were used to immunoprecipitate rat liver fractions derived from a PNS (Fig. 4 A, lane PNS), and enriched in cytosolic (lane cyto), stacked Golgi (lane SG), or ER (lane ER) components. As shown in Fig. 4 A, approximately equal amounts of TAP/p115 were recovered from a Triton X-100–solubilized PNS, cytosolic, and SG fractions, with less TAP/p115 precipitating from the ER fraction. The amount of TAP/p115 in the cytosolic fraction reflects the peripheral nature of its membrane association. TAP/p115 recovered in the ER fraction could be present on VTCs or the ER since this fraction contained both, the VTC marker, p58, and the ER marker, calnexin (data not shown).
Figure 4.
TAP/p115 binds GM130 in the Golgi. (A) TAP/p115 was immunoprecipitated from PNS (lane PNS), cytosolic (lane cyto), stacked Golgi (lane SG), and ER (lane ER) fractions solubilized with Triton X-100 or from the SG fraction solubilized with Triton X-100/SDS/DOC (lane SG + SDS/DOC). A silver-stained gel is shown. p135 coimmunoprecipitates with TAP/ p115 only from the SG fraction. (B) Rat liver fractions were analyzed by immunoblot for the presence of GM130 and TAP/p115. GM130 was detected predominantly in the SG fraction whereas TAP/p115 was also present in cytosolic and ER fractions. (C) NRK cells were processed for IF using anti-TAP/p115 and anti-GM130 antibodies. TAP/p115 and GM130 colocalize in the Golgi region, but GM130 is not detected in peripheral punctate structures containing TAP/ p115 (arrowheads).
A 135-kD protein (p135) coimmunoprecipitated with TAP/p115 from the Golgi-enriched fraction (Fig. 4 A, lane SG). p135 did not coimmunoprecipitate when the anionic detergents SDS and deoxycholate (DOC) were added to the Golgi fraction (Fig. 4 A, lane SG + SDS/ DOC), or when preimmune serum was used (data not shown), suggesting a specific interaction with TAP/p115. p135 was purified by large scale coimmunoprecipitation with TAP/p115 and microsequenced. The sequences of two tryptic peptides were identical to sequences in the cis-Golgi protein GM130 (Nakamura et al., 1995). Peptide 1 was identical to the GM130 residues YQELAVALDSSYVTNK found at positions 147–162 and peptide 2 was identical to residues AQVQDNESLSHLNR at positions 473–487. Our independent finding that TAP/p115 binds GM130 is in agreement with the work from Warren and colleagues (Nakamura et al., 1997). GM130 is a peripheral membrane protein localized to multiple cisternae and pleiomorphic structures on the cis-side of the Golgi stack (Nakamura et al., 1995).
The finding that the TAP/p115–GM130 complex was only detected in the SG fraction, despite the fact that TAP/p115 was present in other fractions suggests that GM130 distribution might be restricted. To examine this, distribution of GM130 was compared by immunoblotting to that of TAP/p115 in cytosolic, SG, and ER fractions. As shown in Fig. 4 B, GM130 was concentrated in the SG fraction, with only small amounts detected in the cytosolic and ER fractions. In contrast, TAP/p115 was detected in all fractions, in a distribution similar to that seen after immunoprecipitation (Fig. 4 A). These results suggest that GM130 is present predominantly in the Golgi whereas TAP/p115 is expected to have a wider subcellular distribution. This was supported by double labeling of NRK cells for GM130 and TAP/p115. As shown in Fig. 4 C, the two proteins colocalize in the Golgi, but GM130 is not detected in peripheral structures that contain TAP/p115 (arrowheads).
The data suggest that TAP/p115–GM130 complex forms only in the Golgi. The previously published reports of GM130 immunogold localization (Nakamura et al., 1997) and our immunogold localization of TAP/p115 show that GM130 and TAP/p115 are detected in the cis-Golgi. These results suggest that the TAP/p115–GM130 complex might form in the cis-Golgi, but its exact spatial distribution remains to be determined. Significantly, the results indicate that TAP/p115 association with peripheral VTCs is GM130 independent, and that TAP/p115 binding to VTCs is likely to be mediated by a protein or proteins other than GM130.
COOH-terminal Acidic Domain of TAP/p115 Interacts with GM130
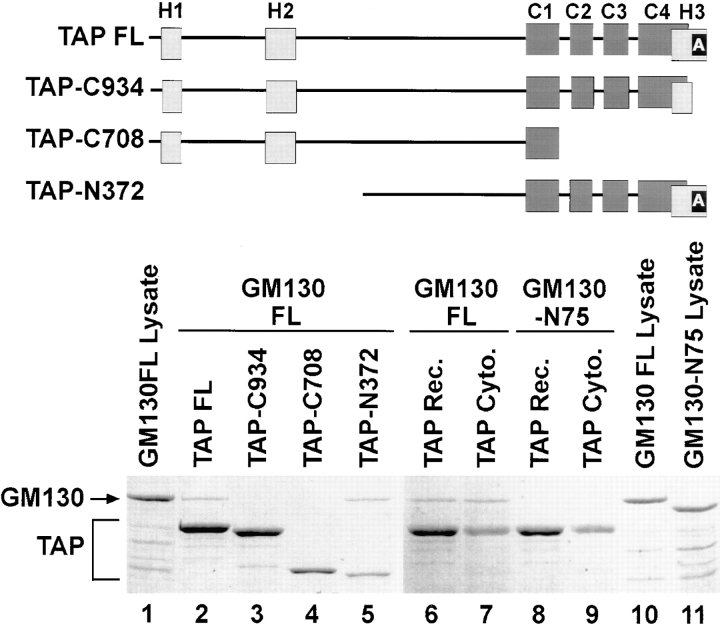
With the goal of testing the functional significance of TAP/p115 association with GM130, we first mapped their corresponding binding domains. To identify GM130 binding sequences within TAP/p115, truncated forms of the protein were expressed in bacteria and analyzed for their ability to interact with full-length recombinant GM130. Any formed complexes were recovered by immunoprecipitation with anti-TAP/p115 antibodies. As shown in Fig. 5, full-length TAP/p115 (lane 2) and NH2 terminally truncated TAP/p115 (lane 5, TAP-N372) bound GM130. Approximately 4% of the input recombinant GM130 was recovered in the TAP/p115 immunoprecipitate. In contrast, COOH terminally truncated TAP/p115 (Fig. 5, lane 4, TAP-C708) lacking the H3 region and three of the four coiled coils, failed to bind visible amounts of GM130. A TAP/ p115 construct with a smaller COOH-terminal truncation and lacking only the acidic domain (lane 3, TAP-C934), also failed to bind GM130, indicating that the COOH-terminal region has a critical role in binding GM130.
Figure 5.
The acidic COOH terminus of TAP/p115 and the basic NH2 terminus of GM130 are required for binding. Diagram of analyzed constructs. Light gray rectangles, Uso1p homology (H1, H2, and H3) regions. Dark gray rectangles, coiled-coil domains (C1–C4). Black rectangle, the acidic domain (A). (Lanes 1–5) Recombinant TAP/p115 constructs were incubated with recombinant full-length GM130 (20% of input is loaded in lane 1). Complexes were recovered by immunoprecipitation with anti-TAP/ p115 antibodies. Coomassie blue–stained gel is shown. Full-length TAP/p115 (lane 2) and TAP-N372 (lane 5) bound full-length GM130, whereas mutants lacking the COOH-terminal acidic domain, TAP-C934 (lane 3) and TAP-C708 (lane 4) did not bind GM130. (Lanes 6–11) Recombinant full-length TAP/p115 or cytosolic TAP/p115 were incubated with recombinant full-length GM130 (20% of input is loaded in lane 10) or GM130-N75 (20% of input is loaded in lane 11). Complexes were recovered by immunoprecipitation with anti-TAP/p115 antibodies. Coomassie blue–stained gel is shown. Recombinant (lane 6) and cytosolic (lane 7) TAP/p115 bound GM130, but neither bound GM130-N75 (lanes 8 and 9). The results indicate that the COOH-terminal acidic domain of TAP/p115 and the NH2-terminal basic domain of GM130 are required for binding.
Using in vitro transcribed/translated GM130 mutants, it has been shown that the NH2-terminal 74 amino acids of GM130 are required for binding TAP/p115 (Nakamura et al., 1997). To confirm this result in our system, we expressed NH2 terminally truncated GM130 (GM130-N75) in bacteria, and tested its ability to bind cytosolic TAP/p115 or full-length recombinant TAP/p115. Full-length GM130 bound both cytosolic TAP/p115 (Fig. 5, lane 7) and full-length recombinant TAP/p115 (lane 6). Approximately 4% of the input recombinant GM130 was recovered in the TAP/p115 immunoprecipitate. GM130-N75, lacking the NH2-terminal region did not bind TAP/p115 from either source (Fig. 5, lanes 8 and 9).
The NH2-terminal region of GM130 contains a stretch of 26 amino acids with 11 basic residues and only one acidic residue. The acidic domain of TAP/p115 contains 25 amino acids with 17 acidic residues. This suggests that the COOH-terminal acidic domain of TAP/p115 directly associates with the NH2-terminal basic region of GM130, probably through charge–charge interactions.
Interaction with GM130 Is Not Required for Golgi Localization of TAP/p115
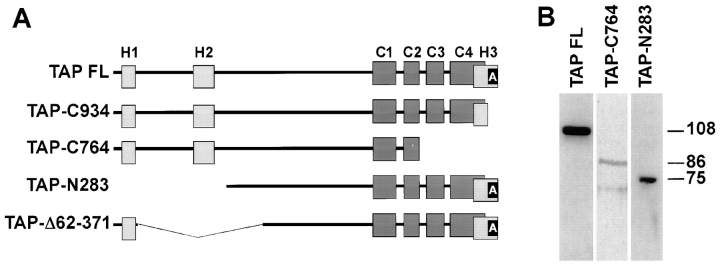
Because TAP/p115 and GM130 interact in the Golgi, binding to GM130 could have an important role in targeting TAP/p115 to the Golgi. To address this experimentally, COS-7 cells were transfected with full-length rat TAP/p115, COOH terminally truncated forms of TAP/p115 lacking the GM130-binding domain, or NH2 terminally truncated forms of TAP/p115 containing the GM130-binding domain, and their localization in the transfected cells analyzed by immunofluorescence. Diagrams of mutants used in this analysis are shown in Fig. 6 A. Two constructs lacking the acidic domain (TAP-C934 and TAP-C764) and two constructs containing the acidic domain but lacking an NH2-terminal region (TAP-N283 and TAP-Δ62-371) were analyzed.
Figure 6.
TAP/p115 mutants used in transfection studies. (A) Diagram of analyzed constructs. Light gray rectangles, Uso1p homology (H1, H2, and H3) regions. Dark gray rectangles, coiled-coil domains (C1–C4). Black rectangle, acidic domain (A). (B) Lysates of COS-7 cells tranfected with specific TAP/p115 constructs were analyzed by immunoblotting. Recombinant proteins of the appropriate molecular weight (∼108 kD for TAP-FL; ∼86 kD for TAP-C764, and ∼75 kD for TAP-N283) were detected.
To ensure that TAP/p115 mutant proteins of the appropriate size were produced in transfected cells, expression of selected constructs was monitored by SDS-PAGE and immunoblotting. COS-7 cells were transfected and cell lysates were prepared 24 h after transfection, subjected to SDS-PAGE, and then blotted with anti-rat TAP/p115 antibodies. These antibodies were used at low concentration and recognize the rat and not the simian TAP/p115. As shown in Fig. 6 B, the endogenous simian TAP/p115 is not detected on the immunoblot but recombinant rat proteins of the expected molecular weight are seen (108 kD for TAP-FL, 86 kD for TAP-C764, and 75 kD for TAP-N283). The t 1/2 of endogenous rat TAP/p115 in NRK cells is ∼22 h and the t 1/2 of endogenous simian TAP/p115 in COS-7 cells is ∼24 h (data not shown). The t 1/2 of rat TAP/p115 mutants in transfected COS-7 cells were not significantly different, ∼22 h for TAP-C764, and ∼18 h for TAP-Δ62-371 (data not shown), indicating that the recombinant proteins have normal turnover rates.
The localization of the wild-type rat TAP/p115 was explored first, to ensure that the recombinant protein localizes correctly in transfected COS-7 cells. In all transfection IF analyses, anti-TAP/p115 antibodies were used at a low concentration and detected the recombinant rat TAP/p115 and not the endogenous monkey TAP/p115. As shown in Fig. 7, A–C, full-length rat TAP/p115 was targeted to the COS-7 Golgi as evidenced by its colocalization with mannosidase II in a juxtanuclear structure with morphology characteristic of COS-7 Golgi. Recombinant TAP/p115 was also detected in peripheral punctate structures, in a pattern similar to that seen for endogenous TAP/p115 in COS-7 cells and likely to represent VTCs (compare Fig. 7 A with Fig. 1 D).
Figure 7.
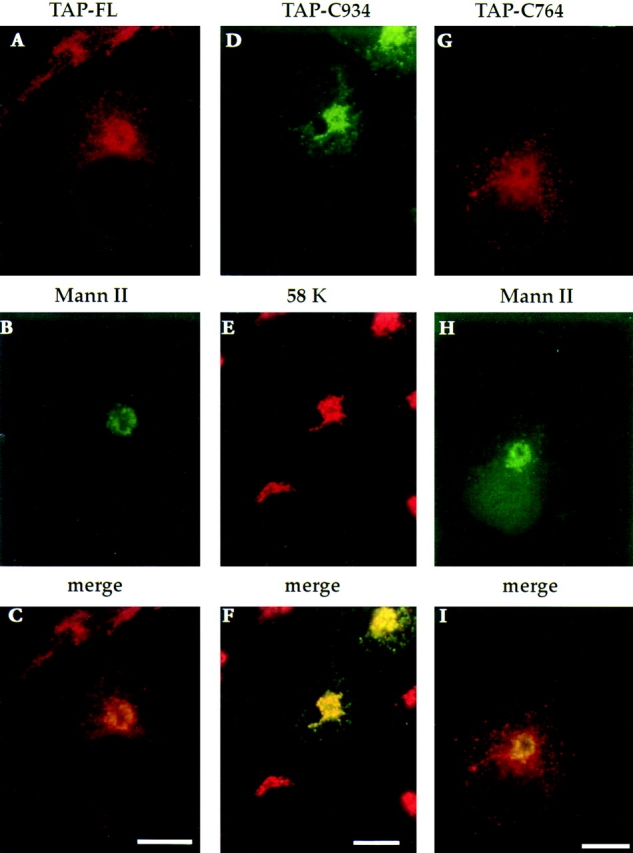
TAP/p115 mutants lacking the GM130-binding domain localize to the Golgi. Full-length TAP/p115 (A–C), TAP-C934 (D–F), and TAP-C764 (G–I) were transiently expressed in COS-7 cells. Cells were fixed and processed for IF using anti-TAP/ p115 and either anti–mannosidase II or anti-58K antibodies. Recombinant proteins were concentrated in the Golgi region and colocalized with the Golgi markers. Bars, 10 μm.
The localization of the COOH terminally truncated mutants was explored next. TAP-C934, lacking the acidic domain, was predominantly detected in the Golgi, where it colocalized with the Golgi marker protein 58K (Bloom and Brashear, 1989) (Fig. 7, D–F). In addition, TAP-C934 was also detected in peripheral punctate structures, TAP-C764, lacking the acidic domain and two of the four coiled coils was also concentrated in the Golgi, where it colocalized with mannosidase II (Fig. 7, G–I). TAP-C764 was also present in peripheral structures not containing mannosidase II, likely to represent VTCs.
The data show that TAP/p115 mutants lacking the GM130-binding domain localize efficiently to the Golgi and can associate with peripheral VTCs. Although the association of TAP/p115 with VTCs has been suggested by previous results to be GM130 independent (refer to Fig. 4 C), the finding that these mutants are efficiently localized to the Golgi, suggests that TAP/p115 localization to the Golgi also does not require association with GM130.
An NH2-terminal Region Is Required for Golgi Localization of TAP/p115
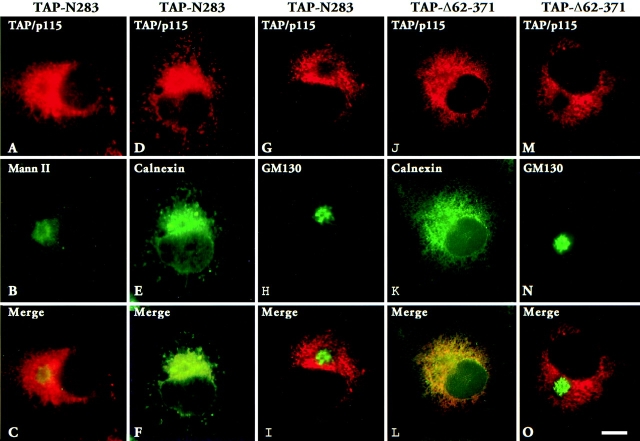
Localization of two TAP/p115 mutants containing the GM130 binding domain but lacking portions of the NH2 terminus were analyzed next. As shown in Fig. 8, A–C, TAP-N283 lacking the NH2-terminal 283 amino acids, was not concentrated in the Golgi region and did not colocalize well with mannosidase II. Instead, TAP-N283 had a rather diffuse distribution, resembling the pattern seen for ER proteins. Indeed, as shown in Fig. 8, D–F, TAP-N283 colocalized with the ER marker calnexin in an extensive reticular network meandering throughout the cell. Since TAP-N283 contains the GM130-binding acidic domain, the distribution of GM130 was analyzed in transfected cells to determine if GM130 was mislocalized to the ER. As shown in Fig. 8, G–I, GM130 was localized in a morphologically normal Golgi complex, in a region of the cell devoid of TAP-N283.
Figure 8.
TAP/p115 mutants with NH2-terminal deletions do not localize to the Golgi. TAP-N283 (A–I) or TAP-Δ62–371 (J–O) were transiently expressed in COS-7 cells. Cells were fixed and processed for IF using anti-TAP/p115 and either anti–mannosidase II, anti-calnexin, or anti-GM130 antibodies. Mutant TAP/p115 did not concentrate with mannosidase II in the Golgi (C), but colocalized with calnexin in the ER (F and L). Mutants did not cause the redistribution of GM130 from the Golgi to the ER (I and O). Bar, 10 μm.
The NH2-terminal region deleted in TAP-N283 contains two domains, H1 (amino acids 21–54) and H2 (amino acids 201–252) that are 62% identical to homologous regions in Uso1p. This high level of conservation suggests an important role in TAP/p115 function. To examine the importance of the H2 domain in TAP/p115 localization, an internal deletion mutant lacking amino acids 62–371 was generated (TAP-Δ62–371). This construct contains the H1 domain but lacks the H2 domain and flanking sequences. As shown in Fig. 8, J–L, TAP-Δ62–371 did not appear concentrated in the Golgi and was distributed in a reticular pattern, where it colocalized with calnexin. Since TAP-Δ62–371 contains the GM130-binding acidic domain, the distribution of GM130 was analyzed in transfected cells. GM130 was detected in morphologically normal Golgi structures from which TAP-Δ62–371 seemed restricted (Fig. 8, M–O).
These results show that the presence of the GM130-binding domain is not sufficient for Golgi localization of TAP-N283 and TAP-Δ62–371. Instead, the NH2-terminal region and specifically the H2 domain are required for Golgi localization.
Expression of TAP/p115 Mutants Affects Golgi Morphology
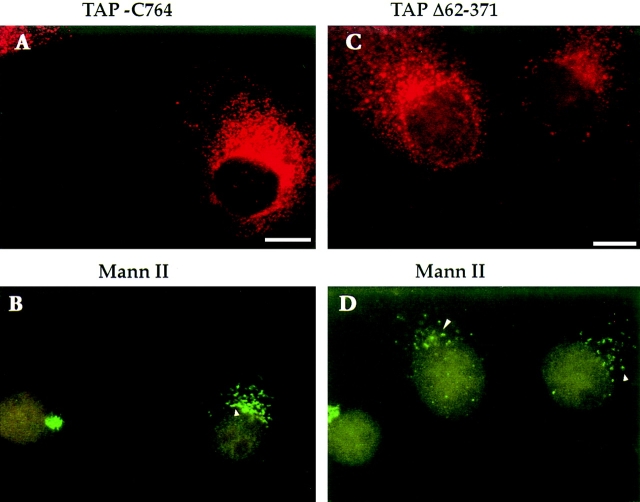
Expression of mutant forms of cycling proteins involved in ER to Golgi transport, such as rab1 and arf1, has been shown to perturb Golgi morphology (Wilson et al., 1994; Zhang et al., 1994). To determine the effect of overexpressing mutant TAP/p115 on Golgi morphology, localization of mannosidase II was monitored in cells transfected with TAP-C764 (mutant lacking the GM130-binding domain and localized to the Golgi) or TAP-Δ62–371 (mutant containing the GM130-binding domain and localized to the ER).
When TAP-C764–transfected cells were analyzed 36 h after transfection, mannosidase II was present in morphologically normal Golgi complexes (refer to Fig. 7 H). However, when cells were examined 63 h after transfection, cells containing high levels of recombinant TAP-C764 (Fig. 9 A) showed altered distribution of mannosidase II (Fig. 9 B). Instead of the normal compact Golgi seen in the untransfected cell in this field, mannosidase II was found in dispersed structures clustered in the perinuclear region (Fig. 9 B, arrowhead).
Figure 9.
Expression of TAP/p115 mutants leads to loss of characteristic Golgi morphology. TAP-C764 (A and B) and TAP-Δ62–371 (C and D) were transiently expressed in COS-7 cells, and 63 h after transfection cells were fixed and processed for IF using anti–mannosidase II and anti-TAP/p115 antibodies. In cells expressing high levels of mutant TAP/ p115 (A and C), mannosidase II is found in punctate structures clustered in the perinuclear region (arrowheads in B and D). Bar, 10 μm.
Similar changes in Golgi morphology were observed with prolonged expression of TAP-Δ62–371. At early time points after transfection the Golgi structure appeared morphologically unperturbed as evidenced by the localization of GM130 (refer to Fig. 8 N). However, 63 h after transfection in cells expressing high levels of recombinant TAP-Δ62-371 (Fig. 9 C), mannosidase II was detected in dispersed punctate structures concentrated in a perinuclear region (Fig. 9 D, arrowheads). The mannosidase II– containing structures in cells expressing the Golgi localized mutant are not readily distinguishable from those in cells expressing the ER localized mutant (Fig. 9, compare B with D).
The data suggest that TAP/p115 is required for processes involved in maintaining Golgi morphology and that the COOH-terminal (GM130 binding) and the NH2-terminal (GM130 independent) domains are relevant to this function.
Discussion
Mammalian TAP/p115 and its yeast homologue Uso1p have been shown to play an essential role in membrane traffic. TAP/p115 is required in cell-free assays designed to measure fusion of transcytotic vesicles with the plasma membrane (Sztul et al., 1993), intra-Golgi transport (Waters et al., 1992), and regrowth of Golgi cisternae from mitotic Golgi fragments (Rabouille et al., 1995, 1998). In yeast, Uso1p is essential for ER to Golgi transport (Nakajima et al., 1991). TAP/p115 has been shown to be required in the transcytotic assay for docking of transport intermediates to acceptor membranes, before ATP hydrolysis by NSF (Barroso et al., 1995). In agreement with the proposed role of TAP/p115 in vesicle tethering, it has been shown that TAP/p115 promotes docking of GTP-γS generated COP I vesicles to Golgi membrane remnants (Sönnischen et al., 1998). Similarly, Uso1p appears to perform a tethering role in ER to Golgi traffic (Barlowe et al., 1997), acting before the assembly of the ER to Golgi SNARE complex (Sapperstein et al., 1996) and ATP hydrolysis by NSF (Lupashin et al., 1996). To inquire into the mechanism of TAP/p115 action, we aimed to explore the compartments in which TAP/p115 is present, characterize TAP/p115 protein–protein interactions, and define the functional significance of such interactions.
Localization of TAP/p115
The subcellular localization of TAP/p115 was examined at the light and electron microscopy levels. In agreement with previous results (Waters et al., 1992; Nakamura et al., 1997; Shima et al., 1997), TAP/p115 was found to colocalize with Golgi marker proteins by immunofluorescence. By EM immunogold labeling, TAP/p115 was detected in association with pleiomorphic structures adjacent to the cis-side of the Golgi stack, and the fenestrated cis-most cisterna of the stack. TAP/p115 was not detected to any significant amount in medial- or trans-cisternae. The localization of TAP/p115 to the cis-Golgi was somewhat surprising considering the previously described role for TAP/p115 in cis- to medial–intra-Golgi transport (Waters et al., 1992). Proteins that localize exclusively to the cis- part of the Golgi stack (for example: rab1 [Saraste et al., 1995], syntaxin 5 [Banfield et al., 1994] and ERGIC-53 [Itin et al., 1995]) have been predominantly those involved in ER to Golgi traffic rather than in subsequent steps of transport. This finding is consistent with the possibility that TAP/p115 might function in ER to Golgi traffic.
Cycling of TAP/p115
In addition to the Golgi localization, TAP/p115 was also detected in peripheral punctate structures, likely to represent VTCs (Bannykh and Balch, 1997). Morphologically, VTCs are composed of clusters of tubular and vesicular profiles that form throughout the cell, close to ER exit sites (Bannykh and Balch, 1997). VTCs translocate by a microtubule dependent mechanism towards the Golgi and then fuse (Presley et al., 1997). Whether VTCs fuse with preexisting structures or with each other is currently unresolved (for review see Lippincott-Schwartz et al., 1998). VTCs are transient structures and proteins detected therein have been shown to cycle between the Golgi and earlier compartments of the secretory pathway. TAP/p115 localization to VTCs suggested that TAP/p115 might also cycle. One of the hallmarks of cycling proteins is their redistribution in cells incubated at reduced temperatures. Incubation at 15°C inhibits the translocation of VTCs from the periphery to the Golgi region without significantly affecting the retrograde cycling of the proteins, resulting in the net redistribution of these proteins from the Golgi region to peripheral structures. In addition to ERGIC-53, proteins known to exhibit this behavior include the cycling transmembrane proteins the KDEL receptor (Tang et al., 1993), mammalian Sec13 (Tang et al., 1997), p23 (Rojo et al., 1997), and rbet1 (Zhang et al., 1997); and the peripherally associated proteins, β-COP (Griffiths et al., 1995), and rab1 (Saraste et al., 1995). Consistent with the behavior of other cycling proteins, incubation of cells at 15°C caused redistribution of TAP/p115 from the Golgi to peripheral punctate structures containing ERGIC-53. The redistribution of TAP/p115 indicates that it cycles at least through VTCs and the Golgi. Many cycling proteins have been shown to transit through the ER (for review see Pelham, 1991), but whether TAP/p115 also cycles through the ER remains to be determined.
TAP/p115 localization to peripheral VTCs and its extensive cycling between the Golgi and earlier compartments of the secretory pathway suggests that TAP/p115 might associate with peripheral VTCs and be transported with them as they move from cell periphery to the Golgi region. Such a delivery mechanism for TAP/p115 would require the specific association of TAP/p115 with peripheral VTCs, but the subsequent transport to the Golgi would occur by default. Therefore, Golgi localization of TAP/p115 would not require a specific Golgi receptor.
Interaction between TAP/p115 and GM130
Previous reports showed that TAP/p115 interact with the tightly associated Golgi protein GM130 and mapped the TAP/p115 interacting domain within the GM130 to the basic NH2-terminal region (Nakamura et al., 1997). We defined the GM130-binding domain within TAP/p115 using an in vitro binding assay, and showed that the highly conserved COOH-terminal acidic domain of TAP/p115 is required to bind the NH2-terminal basic region of GM130. The results suggest a possible linear tail to head binding of TAP/p115 and GM130. TAP/p115 is a homodimer with two globular heads (∼9 nm) composed of the NH2-terminal regions of each monomer and a tail (∼45 nm) formed by the dimerization and close parallel apposition of four independent coiled-coil regions from each TAP/p115 monomer (Sapperstein et al., 1995). The acidic domain is found at the COOH terminus following the last coiled-coil region. GM130 is predicted (based on amino acid sequence) to exist as a long (∼100 nm) rod-like homodimer composed of parallel apposition of seven distinct coiled-coil regions in each GM130 monomer. Binding of the COOH terminus of TAP/p115 to the NH2-terminus of GM130 would result in a ∼150-nm-long structure, an arrangement that might be functionally important.
What is the relevance of TAP/p115–GM130 complex formation? Based on the finding that microinjection of peptide corresponding to the TAP/p115 binding domain of GM130 blocked TAP/p115 association with Golgi membranes in vivo, it was proposed that GM130 acts as a Golgi receptor for TAP/p115 (Nakamura et al., 1997). To test this hypothesis, we examined the localization of TAP/p115 mutants lacking the GM130-binding domain in transfected cells. Two mutants (TAP-C934 and TAP-C764), were shown to efficiently localize to the Golgi and to associate with peripheral VTCs. These results indicate that targeting of TAP/p115 to the Golgi and the VTCs does not require binding to GM130. The data are consistant with the hypothesis that TAP/p115 might be delivered to the Golgi by VTCs and that the ability to associate with VTCs might be sufficient for Golgi localization. However, real time observation of TAP/p115 movement on VTCs will be required to unequivocally confirm this hypothesis.
Interestingly, the presence of the GM130-binding domain is not sufficient to target TAP/p115 to the Golgi. Two mutants (TAP-N283 and TAP-Δ62-371) with deletions in the NH2-terminal region but containing the GM130-binding domain were not localized to the Golgi. Instead, they were restricted to the ER, where they colocalized with calnexin. It is possible that these mutants could not associate with peripheral VTCs and thus could not be transported to the Golgi.
Earlier studies showed that TAP/p115 is required for cis to medial step of intra-Golgi transport in an in vitro assay (Waters et al., 1992), but the exact molecular interactions were not defined. Recently, Sönnischen et al. (1998) have shown that TAP/p115 binds to giantin on COP I vesicles derived from GTP-γS–treated Golgi membranes and to GM130 present on the remaining membranes, and thereby might bridge the COP I vesicles and the Golgi membranes. However, since the vesicles generated after GTP-γS treatment are not functional transport intermediates (Weidman et al., 1993) and their exact origin and destination are uncertain, the significance of the proposed GM130–TAP/ p115–giantin interaction remains unclear. It will be important to determine what Golgi process is dependent on the GM130–TAP/p115–giantin interaction and what is the nature and dynamics of complex formation between these proteins.
Although the exact site of TAP/p115–GM130 complex formation within the Golgi is unknown, our results suggest the possibility that the interaction might occur between TAP/p115 on the surface of incoming VTCs and GM130 on the acceptor cis-Golgi membrane. If TAP/p115 is delivered to the Golgi on VTCs, the complex might form as part of a docking interaction before membrane fusion. The elongated structure of TAP/p115 and GM130, and the possible tail to head association of these molecules during binding seem well suited for a tethering interaction. Such a model is consistent with all available data but remains to be confirmed.
Acknowledgments
We wish to thank M. Farquhar and L. Woodward (University of California, San Diego, CA) for expert immunogold labeling of TAP/p115. We thank K. Howell (University of Colorado, Denver, CO), M. Barroso (University of Virginia, Charlottesville, VA), and J. Lippincott-Schwartz (National Institutes of Health, Bethesda, MD) for critical reading of the manuscript, and N. Nakamura and G. Warren for helpful discussion. We thank A. Tousson (University of Alabama, Birmingham, AL) for expert assistance with immunofluorescence microscopy and C. Beckers and T.H. Thai (University of Alabama) for helpful discussions.
This work was supported by a grant from the National Institutes of Health (RO1-DK46058).
Abbreviations used in this paper
- DOC
deoxycholate
- ERGIC
ER to Golgi intermediate compartment
- IF
immunofluorescence
- NSF
N-ethylmaleimide–sensitive factor
- PNS
postnuclear supernatant
- SG
stacked Golgi
- SNAP
soluble NSF atttachment protein
- SNARE
N-ethylmaleimide–sensitive factor attachment protein
- t-SNARE
target SNARE
- v-SNARE
vesicle SNARE
- VTC
vesicular tubular cluster
Footnotes
D.S. Nelson and C. Alvarez contributed equally to this work.
Address all correspondence to E. Sztul, Department of Cell Biology, University of Alabama at Birmingham, Birmingham, AL 35294. Tel.: (205) 934-1465. Fax: (205) 975-9131. E-mail: esztul@bmg.bhs.uab.edu
D.S. Nelson's present address is Department of Physiology and Biophysics, University of Alabama at Birmingham, Birmingham, AL 35294.
References
- Ansorge W. Fast and sensitive detection of protein and DNA bands by treatment with potassium permanganate. J Biochem Biophys Methods. 1985;11:13–20. doi: 10.1016/0165-022x(85)90037-5. [DOI] [PubMed] [Google Scholar]
- Bajjalieh SM, Scheller RH. The biochemistry of neurotransmitter secretion. J Biol Chem. 1995;270:1971–1974. doi: 10.1074/jbc.270.5.1971. [DOI] [PubMed] [Google Scholar]
- Balch WE, McCaffery JM, Plutner H, Farquhar MG. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell. 1994;76:841–852. doi: 10.1016/0092-8674(94)90359-x. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Rabouille C, Warren G, Pelham HR. Localization of Sed5, a putative vesicle targetingmolecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannykh SI, Balch W. Membrane dynamics at the endoplasmic reticulum–Golgi interface. J Cell Biol. 1997;138:1–4. doi: 10.1083/jcb.138.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannykh SI, Nisihmura N, Balch W. Getting into the Golgi. Trends Cell Biol. 1998;8:21–25. doi: 10.1016/s0962-8924(97)01184-7. [DOI] [PubMed] [Google Scholar]
- Barlowe C. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J Cell Biol. 1997;139:1097–1108. doi: 10.1083/jcb.139.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso M, Nelson DS, Sztul E. Transcytosis-associated protein (TAP)/p115 is a general fusion factor required for binding of vesicles to acceptor membranes. Proc Natl Acad Sci USA. 1995;92:527–533. doi: 10.1073/pnas.92.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption:regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar MG. Membrane recycling in secretory cells: implications for traffic of products and specialized membranes within the Golgi complex. Methods Cell Biol. 1981;23:399–427. doi: 10.1016/s0091-679x(08)61511-3. [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. The Golgi apparatus (complex) (1954– 1981)—from artifact to center stage. J Cell Biol. 1981;91:77–103. doi: 10.1083/jcb.91.3.77s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths GR, Pepperkok R, Krijnse-Locker J, Kreis TE. Immunocytochemical localization of b-COP to the ER-Golgi boundary and the TGN. J Cell Sci. 1995;108:2839–2856. doi: 10.1242/jcs.108.8.2839. [DOI] [PubMed] [Google Scholar]
- Halachmi N, Lev Z. The Sec1 family: a novel family of proteins involved in synaptic transmission and general secretion. J Neurochem. 1996;66:889–897. doi: 10.1046/j.1471-4159.1996.66030889.x. [DOI] [PubMed] [Google Scholar]
- Hauri HP, Schweizer A. The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol. 1992;4:600–608. doi: 10.1016/0955-0674(92)90078-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Scheller RH. Snares and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- Itin C, Schindler R, Hauri HP. Targeting of protein ERGIC-53 to the ER/ERGIC/cis-Golgi recycling pathway. J Cell Biol. 1995;131:57–67. doi: 10.1083/jcb.131.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Donaldson JG, Schweizer A, Berger EG, Hauri HP, Yuan LC, Klausner RD. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Cole N, Presley J. Unraveling Golgi membrane traffic with green fluorescent protein chimeras. Trends Cell Biol. 1998;8:16–20. doi: 10.1016/s0962-8924(97)01199-9. [DOI] [PubMed] [Google Scholar]
- Lowe M, Nakamura N, Warren G. Golgi division and membrane traffic. Trends Cell Biol. 1998;8:40–44. doi: 10.1016/s0962-8924(97)01189-6. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein secuences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Hamamoto S, Schekman RW. Biochemical requirements for the targeting and fusion of ER-derived transport vesicles with purified yeast Golgi membranes. J Cell Biol. 1996;132:277–289. doi: 10.1083/jcb.132.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin VV, Waters MG. t-SNARE activation through transient interaction with a rab-like guanosine triphosphatase. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Hirata A, Ogawa Y, Yonehara T, Yoda K, Yamasaki MA. Cytoskeleton-related gene, uso1, is required for intracellular protein transport in Saccharomyces cerevisiae. . J Cell Biol. 1991;113:245–260. doi: 10.1083/jcb.113.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui P, Slusarewicz N, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- Orci L, Perrelet A, Rothman JE. Vesicles on strings—morphological evidence for processive transport within the Golgi stack. Proc Natl Acad Sci USA. 1998;95:2279–2283. doi: 10.1073/pnas.95.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB. Recycling of proteins between the endoplasmic reticulum and the Golgi complex. Curr Biol. 1991;3:585–591. doi: 10.1016/0955-0674(91)90027-v. [DOI] [PubMed] [Google Scholar]
- Pelham HRB. Membrane transport—green light for golgi traffic. Nature. 1997;389:17. doi: 10.1038/37870. [DOI] [PubMed] [Google Scholar]
- Pevsner J. The role of Sec1p-related proteins in vesicle trafficking in the nerve terminal. J Neurosci Res. 1996;45:89–95. doi: 10.1002/(SICI)1097-4547(19960715)45:2<89::AID-JNR1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPases: master regulators of membrane trafficking. Curr Opin Cell Biol. 1994;6:522–556. doi: 10.1016/0955-0674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Transport vesicle docking: SNAREs and associates. Annu Rev Cell Dev Biol. 1996;12:441–461. doi: 10.1146/annurev.cellbio.12.1.441. [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJM, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Rabouille CT, Misteli R, Watson R, Warren G, Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system J Cell Biol. 1995;129:605–618. doi: 10.1083/jcb.129.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Rojo M, Pepperkok R, Emery G, Kellner R, Stang E, Parton RG, Gruenberg J. Involvement of the transmembrane protein p23 in biosynthetic protein transport. J Cell Biol. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatas. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Labortatory Press, Cold Spring Harbor, NY. 546 pp.
- Sapperstein SK, Walter DM, Grosvenor AR, Heuser JE, Waters MG. p115 is a general vesicular transport factor related to the yeast endoplasmic reticulum to Golgi transport factor Uso1p. Proc Natl Acad Sci USA. 1995;92:522–526. doi: 10.1073/pnas.92.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J, Svensson K. Distribution of the intermediate elements operating in ER to Golgi transport. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- Saraste J, Lahtinen U, Goud B. Localization of the small GTP-binding protein rab1p to early compartments of the secretory pathway. J Cell Sci. 1995;108:1541–1552. doi: 10.1242/jcs.108.4.1541. [DOI] [PubMed] [Google Scholar]
- Scales S, Pepperkok R, Kreis T. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Fransen JA, Bachi T, Ginsel L, Hauri HP. Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J Cell Biol. 1988;107:1643–1653. doi: 10.1083/jcb.107.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Fransen JA, Matter K, Kreis TE, Ginsel L, Hauri HP. Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. Eur J Cell Biol. 1990;53:185–196. [PubMed] [Google Scholar]
- Schweizer A, Matter K, Ketcham CM, Hauri HP. The isolated ER-Golgi intermediate compartment exhibits properties that are different from ER and cis-Golgi. J Cell Biol. 1991;113:45–54. doi: 10.1083/jcb.113.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living Hela cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993a;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993b;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Sönnischen B, Lowe M, Levine T, Jämsä E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul ES, Colombo M, Stahl P, Samanta R. Control of protein traffic between distinct plasma membrane domains. Requirement for a novel 108,000 protein in the fusion of transcytotic vesicles with the apical plasma membrane. J Biol Chem. 1993;268:1876–1885. [PubMed] [Google Scholar]
- Tang BL, Wong SH, Qi XL, Low SH, Hong W. Molecular cloning, characterization, subcellular localization and dynamics of p23, the mammalian KDEL receptor. J Cell Biol. 1993;120:325–328. doi: 10.1083/jcb.120.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL, Peter F, Krijnse-Locker J, Low SH, Griffiths G, Hong W. The mammalian homolog of yeast Sec13p is enriched in the intermediate compartment and is essential for protein transport from the endoplasmic reticulum to the Golgi apparatus. Mol Cell Biol. 1997;17:256–266. doi: 10.1128/mcb.17.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RS, Jones SM, Dahl RH, Nordeen MH, Howell KE. Characterization of the Golgi complex cleared of proteins in transit and examination of Calcium uptake activities. Mol Biol Cell. 1997;8:1911–1931. doi: 10.1091/mbc.8.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MG, Clary DO, Rothman JE. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidman P, Roth R, Heuser J. Golgi membrane dynamics imaged by freeze-etch electron microscopy: views of different membrane coatings involved in tubulation versus vesiculation. Cell. 1993;75:123–133. [PubMed] [Google Scholar]
- Wilson BS, Nuoffer C, Meinkoth J, McCaffery M, Feramisco JR, Balch W, Farquhar MG. A Rab1 mutant Affecting guanine nucleotide exchange promotes disassembly of the Golgi apparatus. J Cell Biol. 1994;125:557–571. doi: 10.1083/jcb.125.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Seog DH, Yoda K, Yamasaki M, Wakabayashi T. Uso1 protein is a dimer with two globular heads and a long coiled-coil tail. J Struct Biol. 1996;116:356–365. doi: 10.1006/jsbi.1996.0053. [DOI] [PubMed] [Google Scholar]
- Zhang CJ, Rosenwald AG, Willingham MC, Skuntz S, Clark J, Kahn RA. Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J Cell Biol. 1994;124:289–300. doi: 10.1083/jcb.124.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wong SH, Tang BL, Xu Y, Peter F, Subramaniam VN, Hong W. The nammalian protein (rbet1) homologous to yeast Bet1p is primarily associated with the pre-Golgi intermediate compartment and is involved in vesicular transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1997;139:1157–1168. doi: 10.1083/jcb.139.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]