Abstract
The Saccharomyces cerevisiae Dnm1 protein is structurally related to dynamin, a GTPase required for membrane scission during endocytosis. Here we show that Dnm1p is essential for the maintenance of mitochondrial morphology. Disruption of the DNM1 gene causes the wild-type network of tubular mitochondrial membranes to collapse to one side of the cell but does not affect the morphology or distribution of other cytoplasmic organelles. Dnm1 proteins containing point mutations in the predicted GTP-binding domain or completely lacking the GTP-binding domain fail to rescue mitochondrial morphology defects in a dnm1 mutant and induce dominant mitochondrial morphology defects in wild-type cells. Indirect immunofluorescence reveals that Dnm1p is distributed in punctate structures at the cell cortex that colocalize with the mitochondrial compartment. These Dnm1p-containing structures remain associated with the spherical mitochondria found in an mdm10 mutant strain. In addition, a portion of Dnm1p cofractionates with mitochondrial membranes during differential sedimentation and sucrose gradient fractionation of wild-type cells. Our results demonstrate that Dnm1p is required for the cortical distribution of the mitochondrial network in yeast, a novel function for a dynamin-related protein.
Keywords: dynamin, GTPase, mitochondria, membrane morphology, S. cerevisiae
Virtually all eukaryotic cells contain mitochondria, which produce most of the ATP cells need to grow and divide. Both mitochondrial shape and mitochondrial distribution can be tailored to meet the specialized energy requirements of a cell. In muscle fibers, for example, mitochondria are organized into regular columns stacked between actin-myosin bundles (Bakeeva et al., 1978). During spermatogenesis in insects, mitochondria fuse into a few large organelles that later extend along the length of the growing flagellum (Fuller, 1993). Many other cell types contain a reticulum of interconnected, tubular mitochondrial membranes that are spread throughout the cytoplasm (Bereiter-Hahn, 1990; Bereiter-Hahn and Voth, 1994). The molecular mechanisms required to generate and/or maintain these diverse mitochondrial morphologies and distributions are not well understood.
The budding yeast Saccharomyces cerevisiae contains a branched network of mitochondrial tubules that are located near the plasma membrane and are distributed evenly in the peripheral cytoplasm (for review see Hermann and Shaw, 1998). Maintenance of this tubular network requires interactions between the mitochondrial compartment and cytoskeletal structures, including actin (Lazzarino et al., 1994; Simon et al., 1995; Hermann et al., 1997) and intermediate filaments (McConnell and Yaffe, 1992). Two mitochondrial outer membrane proteins, Mmm1p (mmm,1 mitochondrial morphology maintenance; Burgess et al., 1994) and Mdm10p (mdm, mitochondrial distribution and morphology; Sogo and Yaffe, 1994), appear to mediate the attachment of mitochondria to actin filaments (Boldogh et al., 1998), and in strains lacking these or another membrane protein called Mdm12p (Berger et al., 1997), the mitochondrial network is converted into giant, spherical organelles. Two other outer membrane proteins, the dynamin-related GTPase Mgm1p (mgm; mitochondrial genome maintenance) and the transmembrane GTPase Fzo1p (fzo; fuzzy onions), also regulate mitochondrial morphology. The function of Mgm1p is not known, but cells lacking this protein contain severely aggregated mitochondrial membranes (Jones and Fangman, 1992; Guan et al., 1993; Shepard, K.A., and M.P. Yaffe. 1997. Mol. Biol. Cell. 8:444a; Gorsich, S., and J.M. Shaw, unpublished). The Fzo1p GTPase regulates mitochondrial fusion, and loss of Fzo1p function causes the mitochondrial network to rapidly fragment (Hermann et al., 1998).
We previously reported the isolation of a collection of mdm mutants with defective mitochondrial distribution and morphology (Hermann et al., 1997). In one of these strains, mdm29-1, the mitochondrial network collapses to one side of the cell, forming an elongated mass of mitochondrial membranes that function normally, retain their mtDNA, and are efficiently transported into buds during division. MDM29 is identical to DNM1 and encodes one of three dynamin-related proteins in yeast (Gammie et al., 1995). The dynamins comprise a family of high–molecular weight GTPases that localize to a variety of different cellular structures (Chen et al., 1991; van der Bliek and Meyerowitz, 1991; De Camilli et al., 1995; Liu and Robinson, 1995; Warnock and Schmid, 1996; Schmid, 1997; Urrutia et al., 1997). Mammalian dynamin is the best characterized of the family members and has been shown to assemble into collars around the base of clathrin-coated pits (Takei et al., 1995). Subsequent GTP hydrolysis by dynamin in these collars is thought to stimulate a membrane scission activity that releases endocytic vesicles (Herskovits et al., 1993; van der Bliek et al., 1993; Damke et al., 1994, 1995; Hinshaw and Schmid, 1995; Takei et al., 1995; Sweitzer and Hinshaw, 1998). The yeast Dnm1 protein (Dnm1p) was originally thought to be the homologue of mammalian dynamin. However, mutations in DNM1 do not block endocytosis of pheromone receptors in Saccharomyces cerevisiae (Gammie et al., 1995). In addition, dnm1 mutations do not affect other cellular transport pathways, including the secretory pathway and the vacuole protein sorting pathway. Thus, the exact function of Dnm1p in vivo is unclear.
In this report, we present evidence that Dnm1p controls the morphology and cortical distribution of yeast mitochondrial membranes. Mutational analysis reveals that the predicted GTPase domain of Dnm1p is required for its activity in vivo. In wild-type cells, the Dnm1 protein is found in patches distributed uniformly at the cell cortex that colocalize with the mitochondrial network. These Dnm1p-containing structures could regulate mitochondrial branching or division, or could act as attachment sites to distribute the mitochondrial network evenly at the cell periphery.
Materials and Methods
Yeast Methods
Standard methods were used for growth, transformation, and genetic manipulation of S. cerevisiae. Rich medium (YP-Dextrose and YP-Glycerol), synthetic medium (synthetic dextrose [SD] and synthetic galactose [SGal]) and medias containing 5-fluoro-orotic acid (5-FOA) were prepared as described (Sherman et al., 1986).
Strains
Escherichia coli strains DH5α and JM109 (Promega Corp., Madison, WI) were used for bacterial manipulations of clones and plasmids. Yeast strains used in this study are listed in Table I. JSY1542 (containing genomic DNM1-HAc) was generated by integrating a 3×HA-URA3-3×HA (hemagglutinin) cassette between codons 756 and 757 of the DNM1 coding region in JSY1539 (Schneider et al., 1995). Cells that had undergone recombination and loss of the URA3 portion of the cassette were selected by growth on 5-FOA (confirmed by PCR). The resulting JSY1542 strain expresses a full-length Dnm1 protein containing 3×HA epitopes inserted in-frame before the last amino acid of the polypeptide chain (confirmed by Western blotting with a polyclonal anti-HA antibody; Berkeley Antibody Company, Richmond, CA). JSY1678 (containing genomic GAL1-DNM1) was also generated by integrating a GAL1-URA3-GAL1 cassette upstream of the DNM1 coding region in JSY1238 (Schneider et al., 1995) and selecting for URA3 loss on 5-FOA. The genomic region corresponding to −100 bp to −10 bp upstream of the start codon is replaced by the GAL1 promoter.
Table I.
Yeast Strains
| Strain | Genotype* | Source/Reference | ||
|---|---|---|---|---|
| FY10 | MATα ura3-52 leu2Δ1 | Winston et al., 1995 | ||
| FY250 | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 | Winston et al., 1995 | ||
| JSY1236 | MATa ura3-52 mdm29-1 | This study | ||
| JSY1943 | MATa ura3-52 leu2Δ1 his3Δ200 mdm29-2 | This study | ||
| MS3031 | MATα ura3-52 ade2-101 leu2-3,112 his3Δ200 dnm1::HIS3 | Gammie et al., 1995 | ||
| JSY1238 | MATa ura3-52 leu2Δ1 his3Δ200 | This study | ||
| JSY1361 | MATa ura3-52 leu2Δ1 his3Δ200 dnm1::HIS3 | This study | ||
| JSY1542 | MATa ura3-52 leu2Δ1 his3Δ200 DNM1-HAc | This study | ||
| JSY1678 | MATa ura3-52 leu2Δ1 his3Δ200 GAL1-DNM1 | This study | ||
| JSY1781 | MATa/MATα ura3-52/ura3-52 leu2Δ1/leu2Δ1 his3Δ200/HIS3 DNM1-HAc /DNM1-HA c | This study | ||
| JSY1729 | MATα his3 leu2 mdm10::URA3 DNM1-HAc | This study | ||
| SEY6210 | MATα leu2-3,112 his3Δ200 ura3-52 trp1-Δ901 lys2-801 suc2-Δ9 GAL | S. Emr | ||
| RH268-1C | MATa his4 leu2 ura3 bar1-1 end4 | Raths et al., 1993 | ||
| RH266-1D | MATa ura3 leu2 his4 bar1-1 end3 | Raths et al., 1993 | ||
| JSR18Δ1 | SEY6210; vps18Δ1::TRP1 | Robinson et al., 1991 | ||
| MBY3 | SEY6210; vps4::TRP1 | Babst et al., 1997 | ||
| BWY18 | MATa leu2-3,112 his3Δ200 ura3-52 trp1-Δ901 pan1-20 | Wendland et al., 1996 | ||
| JSY1914 | MATα ura3 his3 | This study | ||
| JSY1916 | MATa ura3 his3 mdm10::URA3 | This study |
All of the JSY strains used in this study are isogenic to FY10 (Winston et al., 1995) except JSY1914 and JSY1916, which were generated by crossing MYY503 (mdm10::URA3; Sogo and Yaffe, 1994) into the FY background, and JSY1729, which was generated by crossing MYY503 with an FY strain containing DNM1-HAc.
Plasmids
pRS415-DNM1 (pDO7) contains the DNM1 coding region flanked by 419 bp of upstream and 401 bp of downstream sequence and was prepared by cloning a PCR product into the BamHI site of pRS415 (CEN, LEU2; Stratagene, La Jolla, CA). The region of the plasmid containing DNM1 was sequenced. pRS425-DNM1 (pDO13) was constructed by cloning the BamHI fragment from pRS415-DNM1 into BamHI-digested pRS425 (2 micron, LEU2; Stratagene). pYEp213-DNM1 was constructed by cloning the BamHI fragment from pRS415-DNM1 into BamHI-digested pYEp213 (2 micron, LEU2). pRU1-DNM1 and pRL1-DNM1 were constructed by cloning the BamHI fragment from pRS415-DNM1 into BamHI-digested pRU1 and pRL1, respectively (see below). All of the plasmids described above complemented the mitochondrial morphology defect in the dnm1 null strain JSY1361 (see Table I).
Plasmids derived from the YCp50 (Rose et al., 1987) and p366 (P. Hieter, Johns Hopkins University, Baltimore, MD) shuttle vectors were modified to remove the unique BglII site in ARS1. YCp50 (CEN URA3) and p366 (CEN LEU2) were digested with BglII, end-filled to create blunt ends, and ligated to recircularize the vectors (a new ClaI site is created in the ligated vectors). The resulting plasmids were named pRhinoURA3/1 (pRU1) and pRhinoLEU2/1 (pRL1). These plasmids are maintained at low copy in both E. coli and yeast.
In some experiments, strains were transformed with the plasmid pDO10 to visualize mitochondrial compartments labeled with Cox4–GFP (green fluorescent protein). To construct pDO10, a NotI-XhoI restriction fragment from pOK29 (containing the mitochondrial targeting sequence of Cox4p fused in-frame to the NH2 terminus of GFP; provided by R. Jensen) was ligated into pRS415 (LEU2).
Mapping of the mdm29-1 Mutation to the DNM1 Locus
Genetic mapping of mdm29 was carried out in several steps. First, mdm29-1 was crossed to strains containing trp1 mutant alleles. Analysis of tetrads derived from this cross indicated that mdm29-1 was centromere-linked (1.35 cM, 37 tetrads). Second, mdm29-1 was localized to chromosome XII (linked to the asp5/aat2 locus; 14.29 cM, 28 tetrads) in crosses with a collection of mapping strains obtained from the American Type Culture Collection (Rockville, MD). Third, complementation analysis was performed by crossing mdm29-1 with strains harboring mutations in genes close to the centromere on chromosome XII. Subsequent studies showed that the mitochondrial morphology defect observed in mdm29-1 was not complemented in a diploid heterozygous for mutations in mdm29-1 and dnm1 (MS3031; Gammie et al., 1995). Meiotic segregation analysis with the mdm29-1/dnm1 diploid confirmed that mdm29-1 was closely linked to dnm1. The dnm1 null strain (MS3031) from the Rose laboratory exhibited mitochondrial morphology defects similar to those observed in the mdm29 mutant. Plasmids pDO7 and pDO13 (see above) containing the wild-type DNM1 gene fully complement the mitochondrial morphology defects observed in the mdm29-1 and dnm1 null mutant strains.
Sequencing of mdm29 Mutant Alleles and Disruption of MDM29/DNM1
Overlapping segments of the DNM1 gene were PCR amplified from yeast genomic DNAs prepared from the MDM29/DNM1 (JSY1238), mdm29-1 (JSY1236), and mdm29-2 (JSY1943) strains. PCR products from triplicate reactions were pooled, purified (Qiagen, Chatsworth, CA), and sequenced on both strands (University of Utah Health Sciences Sequencing Facility). The wild-type DNM1 gene sequence from the FY strain background was identical to the sequence reported in the Saccharomyces Genome Database (SGD). However, our DNM1 sequence and the SGD DNM1 sequence differed from that reported by Gammie et al. (1995). Counting from the first (1) nucleotide of the methionine initiation codon, the sequence reported by Gammie et al. (1995) contains nine extra nucleotides (5′-TATCACCTG-3′) inserted between nucelotides 370 and 371 of the DNM1 sequence. In addition, nucleotide 370, reported as A in the Gammie et al. (1995) sequence, is a C in our sequence and in the SGD. These changes alter the predicted amino acid sequence in this region from F122LISPDIPG to F122LHIPG. All maps, plasmids, and strains used in this study contain the shorter DNM1 sequence listed in the SGD.
A dnm1 null mutation in the FY strain background (Winston et al., 1995) was generated by gene replacement using a PCR fragment containing the HIS3 gene (pRS403; Stratagene) contiguous with DNM1-flanking sequences. The resulting dnm1::HIS3 disruption (lacking all DNM1 coding sequences) was confirmed by Southern blotting of genomic DNA isolated from a His+ transformant.
Site-directed Mutagenesis
A 3.1-kb fragment containing the DNM1 coding region and flanking 5′ and 3′ sequence was subcloned into the BamHI site in the p-ALTER-1 vector (Promega Corp.). Site-directed mutagenesis was performed with oligonucleotides to generate the different point mutations as described by the manufacturer (Promega Corp.). The GTPase deletion construct was generated by adding a BglII site 1,014 nucleotides after the start codon. An additional BglII site is present in the wild-type DNM1 sequence 15 nucleotides after the Dnm1p start codon. Subsequent BglII digestion/religation removed DNM1 coding sequence corresponding to amino acids 8–338 and resulted in an in-frame fusion of sequences encoding amino acids 7–339 (GTPase deletion). Mutant dnm1 sequences confirmed by restriction digest and sequence analysis were subcloned into pRU1 (described above) or YEp213 (LEU2) and introduced into JSY1238 (wild-type), JSY1361 (dnm1Δ), and JSY1678 (GAL1-DNM1) strains. DNA sequencing and sequence analyses were performed as described for the mdm29 mutant alleles.
The effect of the GTP domain mutations on mitochondrial morphology was examined in strains grown in synthetic dextrose medium lacking uracil (to maintain the low copy pRU1 CEN plasmid) or synthetic dextrose medium lacking leucine (to maintain the high copy YEp213 2μ plasmid). Overexpression of wild-type Dnm1p in the GAL1-DNM1 strain was achieved by growing strains overnight in synthetic galactose medium lacking either uracil (pRU1) or leucine (YEp213).
Characterization of dnm1 Organellar Phenotypes
To characterize mitochondrial distribution and morphology, wild-type and dnm1 mutant cells were grown overnight in YPD or synthetic media at 30°C, diluted 1:10 in fresh media, and incubated at 30°C for an additional 3 h. 100 μl of culture was stained with 100 nM 3,3′ dihexyloxacarbocyanine (DiOC6) (Molecular Probes, Eugene, OR) as described previously (Hermann et al., 1997; Roeder et al., 1998). Alternatively, cells expressing the Cox4–GFP protein from the pOK29 or pDO10 plasmids were grown overnight under the same conditions and inspected microscopically. Mitochondrial morphology phenotypes were scored in n ≥ 100 cells in at least three independent experiments. Comparable results were obtained using both of these staining methods, indicating that the mitochondrial morphology phenotypes observed in dnm1Δ cells were not due to the presence of the Cox4–GFP fusion protein (see also Roeder et al., 1998).
Rhodamine-phalloidin staining of actin (Hermann et al., 1997; Roeder et al., 1998), antitubulin staining (Pringle et al., 1989), 4′,6-diamidino-2-phenylindole (DAPI) staining of nuclei and mtDNA (Hermann et al., 1997), FM 4-64 staining of vacuoles and kinetic analysis of FM 4-64 internalization (Roeder and Shaw, 1996; Vida and Emr, 1995), anti-Kar2p indirect immunofluorescence of ER membranes (Shamu and Walter, 1996), and Sec7p-EGFP staining of Golgi membranes (Séron et al., 1998; generously provided by O. Rossanese and B. Glick, University of Chicago, Chicago, IL) were performed as described.
Dnm1p Induction and Depletion Studies
To examine mitochondrial morphology changes during a Dnm1p induction time course, JSY1678 cells (harboring the genomic GAL1-DNM1 gene) were grown in synthetic medium containing 2% dextrose for 16 h at 30°C to deplete the cells of the endogenous Dnm1 protein. After diluting and growing for an additional 3 h to obtain log phase cultures, the cells were pelleted, washed with dH2O, and resuspended in fresh synthetic medium containing 2% galactose to induce expression of the GAL1-DNM1 gene. For the depletion time course, JSY1678 cells grown in synthetic medium containing 2% galactose for 16 h at 30°C were diluted into galactose-containing medium, grown to log phase, pelleted, washed in dH2O, and resuspended in synthetic dextrose-containing medium to turn off expression of the GAL1-DNM1 gene. Mitochondrial morphology was scored in 100-μl aliquots of DiOC6-stained cells removed at various times after transfer to the new medium. The density of the culture was maintained between 0.5–2.0 OD600 units during the course of the experiment. In control experiments, cells containing the DNM1 gene expressed from its own promoter (JSY1238) maintained wild-type mitochondrial networks throughout the same time course.
Protein extracts prepared from equivalent numbers of cells were analyzed by SDS-PAGE and Western blotting (Harlow and Lane, 1988) using a 1:2,500 dilution of the affinity-purified rabbit polyclonal antiserum generated against the COOH terminus of Dnm1p (described below). To control for differences in protein loading, the blots were stripped and reprobed with an antibody to the cytosolic marker 3-PGK (Molecular Probes, Eugene, OR). Detection was carried out using ECL (NEN™ Life Science Products, Boston, MA). Densitometry (NIH Image software) was used to determine the level of Dnm1p expression from the GAL1 promoter (JSY1678) relative to the endogenous DNM1 promoter (JSY1238).
Generation of Affinity-purified Anti-Dnm1p Antibodies
A 1-kb EcoRI fragment encoding the COOH-terminal amino acids 412– 757 of Dnm1p was subcloned into the pGEX-2T (128/129) expression vector (M. Blanar, Bristol-Myers Squibb, Princeton, NJ) to create the pGEX-2T-DNM1-C plasmid. E. coli BL21-(DE3) cells containing pGEX-2T-DNM1-C express a 65-kD glutathione-S-transferase (GST)–Dnm1412–757 fusion protein, which was purified on Glutathione Sepharose 4B beads (Pharmacia Biotech, Piscataway, NJ), run on 8% SDS-PAGE gels, excised, and used to immunize rabbits (Berkeley Antibody Co.). Soluble Dnm1p fused to 6× HIS residues at the COOH terminus (pET23a-DNM1-3) was expressed in E. coli BL21-(DE3) cells, purified by affinity chromatography on His-Bind resin (Novagen, Inc., Madison, WI), and coupled to cyanogen bromide–activated Sepharose 4B as recommended by the manufacturer (Pharmacia Biotech). Rabbit polyclonal anti–GST-Dnm1412–757 fusion protein antibody was affinity purified on the Dnm1-6XHISp-Sepharose column, equilibrated in PBS, pH 7.2, concentrated to 0.35 mg/ml (using a Centricon-30 filter unit; Amicon Corp., Easton, TX), and stored at −80°C after addition of 5 mg/ml BSA (Fraction V; Sigma Chemical Co., St. Louis, MO).
Immunolocalization of the Dnm1-HAC and Wild-Type Dnm1 Proteins
Indirect immunofluorescence was performed essentially as described (Roeder and Shaw, 1996) except that cells expressing Cox4–GFP (pDO10) were fixed in 4.4% formaldehyde for 2 h to preserve GFP fluorescence. To detect the HA epitope, cells were incubated with the following antibodies: (a) 1:1,000 dilution of a 1 mg/ml mouse monoclonal anti-HA antibody for 14 h (catalog No. MMS-101P-500; Berkeley Antibody Co.), (b) 1:10 dilution of a 2.4 mg/ml goat anti–mouse polyclonal antibody (catalog No. 115-005-003; Jackson ImmunoResearch, West Grove, PA), (c) 1:10 dilution of a 1.8 mg/ml mouse anti–goat polyclonal antibody (catalog No. 205-005-108; Jackson ImmunoResearch), and (d) 1:100 dilution of a 1.5 mg/ml CY5-conjugated goat anti–mouse polyclonal antibody (catalog No. 115-175-003; Jackson ImmunoResearch). To detect the wild-type Dnm1 protein (lacking the COOH-terminal HA epitope), cells were incubated with: (a) 1:50 dilution of the 0.345 mg/ml affinity-purified rabbit anti–GST-Dnm1412–757 antibody and (b) 1:25 dilution of a 1.5 mg/ml CY5-conjugated goat anti–rabbit polyclonal antibody (catalog No. 111-175-144; Jackson ImmunoResearch).
To quantify the degree of Dnm1p/mitochondrial colocalization, 1-μm optical sections were generated through fixed yeast cells costained for Dnm1p (CY5) and a mitochondrial matrix marker (Cox4–GFP). Three-dimensional projections of individual cells as well as separated and merged CY5 and Cox4–GFP fluorescence images were generated. Individual CY5 spots in 5–10 cells were scored relative to the GFP-stained mitochondrial network as localizing to a nonmitochondrial position or to the mitochondrial network, and the data were expressed as the percentage of total spots colocalizing with the mitochondrial network.
Differential Sedimentation and Sucrose Gradient Fractionation
Cells grown in SGal medium were fractionated by differential sedimentation as described previously (Daum et al., 1982; Zinser and Daum, 1995; Hermann et al., 1998), except that the P10,000 mitochondrial pellet was subjected to an additional wash step. In brief, P10,000 was resuspended in breaking buffer (0.6 M mannitol, 20 mM Hepes-KOH, pH 7.4, plus protease inhibitor cocktail), and spun at 1,500 g for 5 min to yield P1,500 and S1,500. S1,500 was spun at 10,000 g for 10 min to yield a washed P10,000 pellet and a second S10,000 supernatant. Protein concentrations were determined using the Bio-Rad Protein Assay (Hercules, CA), and 2 μg of protein from each fraction was analyzed by Western blotting with the following antibody dilutions: anti-Dnm1p (1:5,000), antiporin (1:1,000; Molecular Probes), and anti–3-PGK (1:1,000; Molecular Probes).
The P10,000 mitochondrial pellet was subjected to sucrose gradient fractionation as described by Walworth et al. (1989). The S10,000 supernatant was spun at 100,000 g for 1 h (model SW50.1 rotor; Beckman Instruments, Fullerton, CA) to generate S100,000 and P100,000 fractions. Equal volumes of gradient fractions or equal volumes of S100,000 and P100,000 fractions were assayed by Western blotting with the following antibody dilutions: anti-Dnm1p (1:1,000), antiporin (1:1,000), anti–3-PGK (1:1,000), anti– Dol-P-Man Synthase (1:1,000), anti-Mnn1p (1:400), anti-ALP (1:1,000), and anti-Gas1p (1:5,000). Na2CO3 and Triton X-100 treatments of the P10,000 fraction were performed essentially as described (Hermann et al., 1998).
Microscopy
Organellar phenotypes were quantified with a microscope (model Axioplan; Carl Zeiss, Inc., Thornwood, NY) equipped with differential interference contrast optics, epifluorescence capabilities, and a 100× (NA 1.3) objective (model Acroplan-Neofluar; Carl Zeiss, Inc.). In some experiments, cells were viewed with a confocal microscope (model MRC-600; Bio-Rad Microsciences, Cambridge, MA; 1-μm optical serial sections) attached to a microscope (model Optiphot; Nikon, Inc., Melville, NY) equipped with a 60× plan apochromat objective (NA 1.3; Carl Zeiss, Inc.). Images were collected using Bio-Rad image capture software, and projections were generated using Confocal Assistant software (copyright by Todd Clark Brelje). Electron microscopy was performed as described (Hermann et al., 1998).
Results
The Dynamin-like GTPase, Dnm1p, Is Required for the Maintenance of Mitochondrial Morphology in Yeast
The mdm29-1 and mdm29-2 mutations cause defective mitochondrial morphology in yeast. Genetic methods were used to map mdm29 to a region close to the centromere of chromosome XII. Analysis of open reading frames near this centromere revealed that mdm29 maps to a previously isolated gene called DNM1, one of three dynamin-related genes in yeast (Gammie et al., 1995). Sequence analysis showed that the mdm29-1 mutation causes a truncation after amino acid 120 within the conserved, NH2-terminal GTPase domain of the Dnm1 protein (Dnm1p). The mdm29-2 allele introduces a stop codon that removes the COOH-terminal 26 amino acids of Dnm1p. We also generated a dnm1 null allele by gene disruption (dnm1Δ). The phenotypes observed in dnm1Δ are identical to those found in mdm29-1 and mdm29-2, suggesting that both mutant alleles result in a complete loss of Dnm1p function.
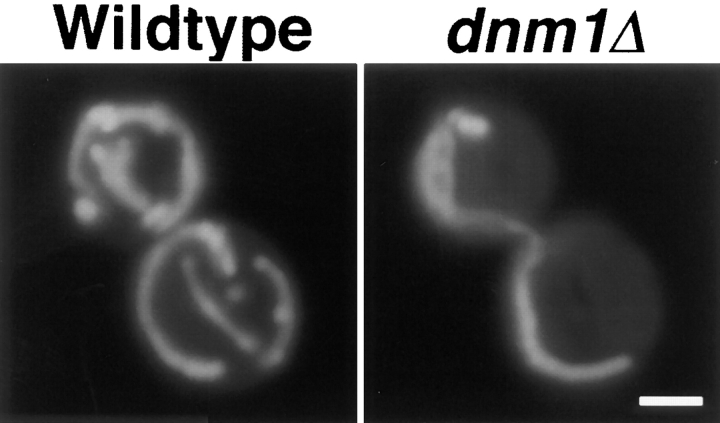
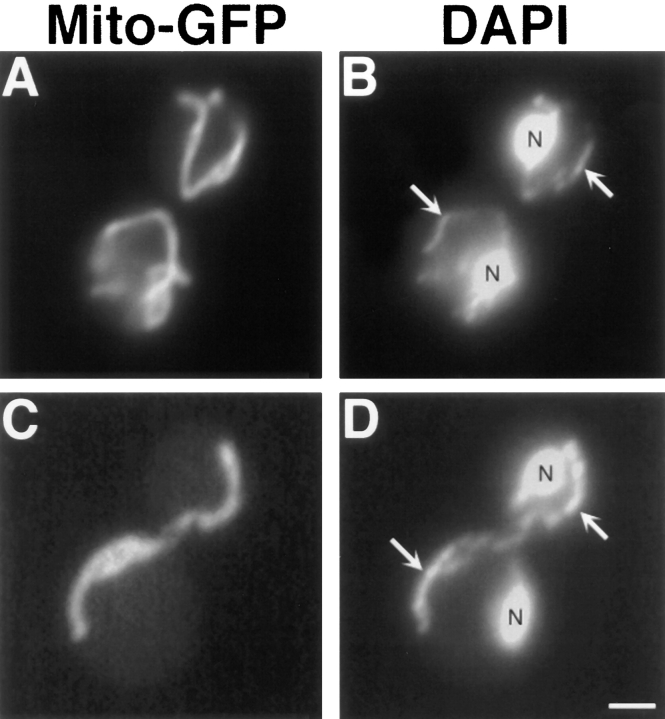
In dnm1Δ cells, the highly branched mitochondrial network observed in wild-type strains (Fig. 1, Wildtype) collapses to form a long tubular structure that is still localized to the cell cortex (Fig. 1, dnm1Δ). Staining with the DNA-specific dye DAPI (Pringle et al., 1989) indicates that these elongated organelles contain mitochondrial DNA nucleoids (Fig. 2 D). The mitochondrial phenotype caused by the dnm1Δ mutation is strikingly different from that caused by the previously isolated mmm1, mdm10, and mdm12 mutations, which convert mitochondrial networks into one or a few large spherical organelles (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997; for example see Fig. 12 G).
Figure 1.
Mitochondrial morphology in wild-type (DNM1) and dnm1Δ mutant strains. Mitochondrial morphology in the DNM1 (JSY1238, left) and dnm1Δ (JSY1361, right) strains were visualized using a matrix-targeted form of the green fluorescent protein (Cox4–GFP). Buds are in the upper left portion of each panel. Bar, 2 μm.
Figure 2.
Collapsed dnm1Δ mitochondria retain mtDNA nucleoids. DNM1 (JSY1238, A and B) and dnm1Δ (JSY1361, C and D) strains labeled with Cox4–GFP (A and C) to visualize mitochondrial compartments and DAPI (B and D) to visualize nuclei (N) and mtDNA (B and D, white arrows). Buds are in the upper right portion of each panel. Bar, 2 μm.
Figure 12.
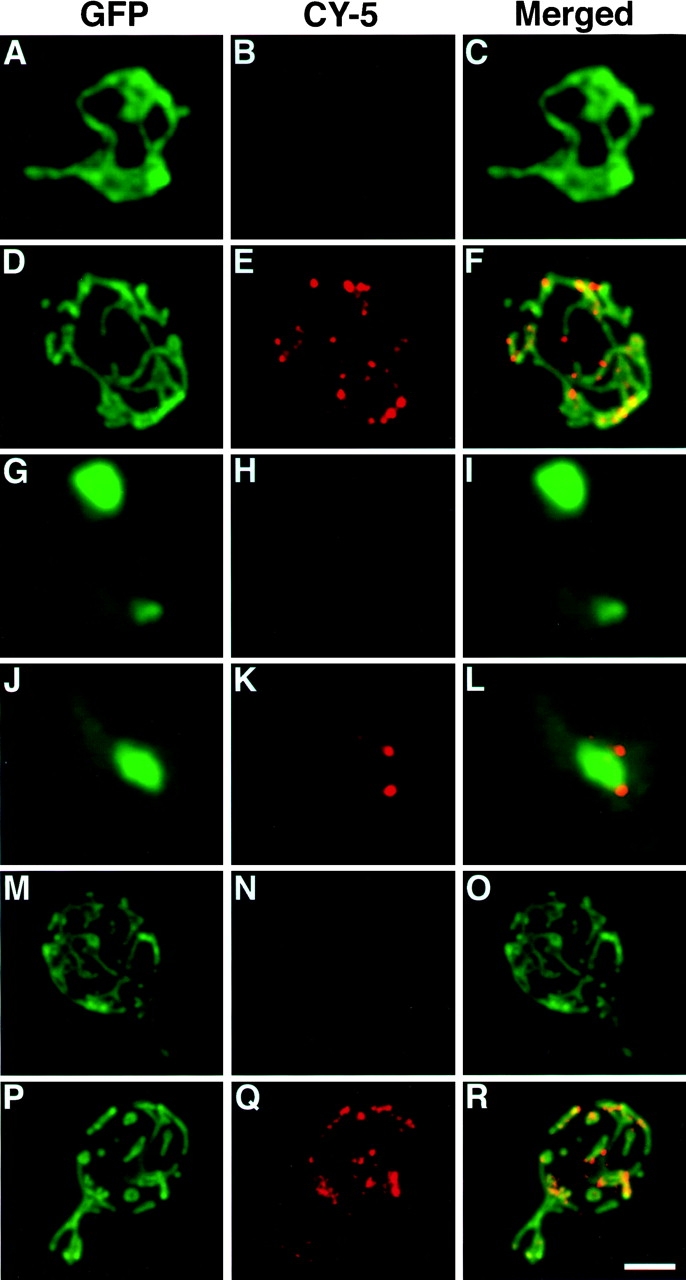
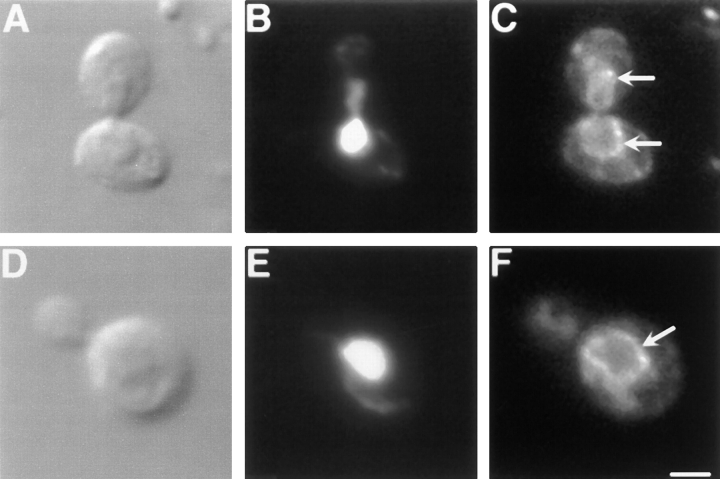
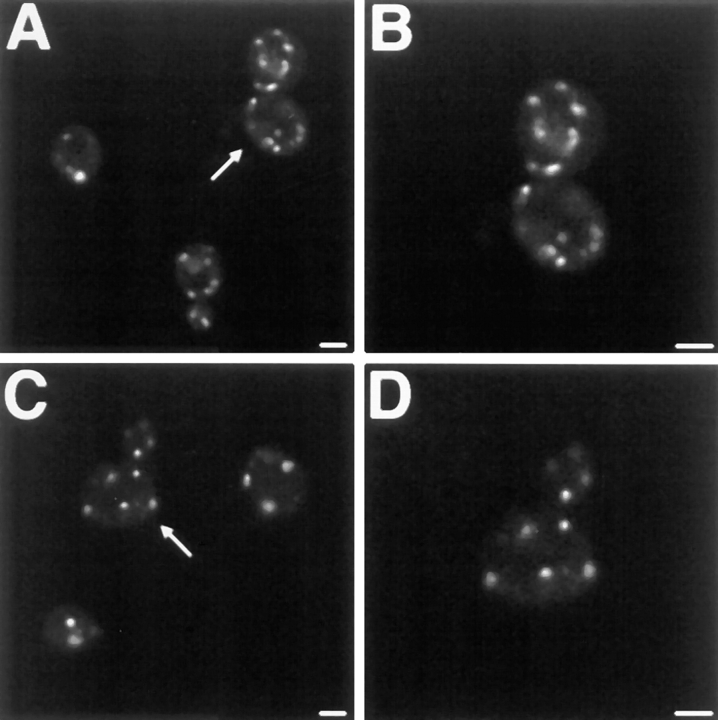
Dnm1p colocalizes with the yeast mitochondrial network. Wild-type (JSY1781, A–F; JSY1238, M–R) or mdm10 mutant (JSY1729, G–L) cells expressing Dnm1– HACp (A–L) or wild-type Dnm1p (M–R) were fixed and incubated with no primary antibody (A–C, G–I, and M–O), anti-HA antibody (D–F and J–L), or anti-Dnm1p412–757 antibody (P–R). A CY5-conjugated secondary antibody (red fluorescence in B, E, H, K, N, and Q) was used to visualize the Dnm1–HAc and Dnm1 proteins. Mitochondrial networks were visualized by expressing the matrix-targeted Cox4–GFP protein (green fluorescence in A, D, G, J, M, and P) in the same cells. The merged CY5 and GFP images are shown in C, F, I, L, O, and R. Bar, 2 μm.
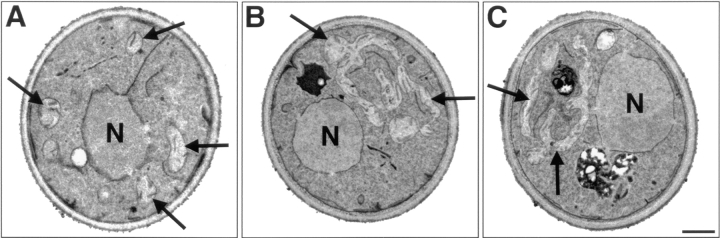
Transmission electron microscopy confirmed that mitochondrial distribution and morphology were altered in the dnm1Δ strain (Fig. 3). In wild-type cells, mitochondrial tubules in cross section appeared uniform in size, were distributed evenly in the peripheral cytoplasm, and contained distinct cristae (invaginations of the inner mitochondrial membrane) (Fig. 3 A). Although mitochondrial profiles in dnm1Δ cells contained normal cristae, the compartments often appeared as a series of larger tubules restricted to one half of the cell (Fig. 3, B and C). These mitochondrial tubules were usually interconnected or branched, indicating that dnm1Δ mitochondrial membranes still retain some features of a mitochondrial network or reticulum (Fig. 3, B and C) and had not fused to form a single, elongated mitochondrial compartment.
Figure 3.
Transmission electron microscopy of DNM1 and dnm1Δ cells. (A) Mitochondrial profiles (black arrows) in DNM1 cells (JSY1238) are uniformly dispersed in the peripheral cytoplasm and contain visible cristae. (B and C) Mitochondrial profiles (black arrows) in dnm1Δ cells (JSY1361) contain normal cristae but appear as enlarged tubules clustered at one side of the cell. Bar, 500 nm.
Despite their unusual morphology, a number of observations suggest that normal functions of dnm1Δ mitochondria are not severely compromised. First, the abnormal mitochondria in dnm1Δ cells are efficiently transported into buds during division (Fig. 1, dnm1Δ). Second, these mitochondria can be labeled with the potential-dependent dye DiOC6, which only stains actively respiring organelles (Pringle et al., 1989). Third, a Cox4–GFP fusion protein (targeted to the matrix; generously provided by R. Jensen) also stains collapsed dnm1Δ mitochondria (Figs. 1 and 2), indicating that the mitochondrial protein import machinery is not grossly affected. Fourth, in our strain background, cells harboring dnm1 mutations grow as well as wild-type on the nonfermentable carbon sources glycerol, lactate, and ethanol/acetate, which can only be metabolized in cells with functional mitochondria. In addition, the organelle morphology defect in dnm1Δ cells appears specific for mitochondria and does not affect the morphology of other membrane-bound structures, including nuclei (Fig. 2 D), the endoplasmic reticulum (Fig. 4 F), the Golgi apparatus (Fig. 5, C and D), and vacuoles (Fig. 6 D). In addition, indirect immunofluorescence studies indicate that the organization of the actin (Fig. 7 D) and tubulin (Fig. 7 H) cytoskeletons is not altered in dnm1Δ. Together, these results indicate that DNM1 plays an important and specific role in generating and/or distributing the branched mitochondrial network in yeast.
Figure 4.
ER morphology is wild type in dnm1Δ cells. DIC (A and D), DAPI (B and E), and indirect immunofluorescence images (C and F) of DNM1 (JSY1238, A–C) and dnm1Δ (JSY1361, D–F) cells stained with anti-Kar2p antiserum. Buds are in the upper portion of each panel. White arrows in C and F mark the perinuclear ER stained with anti-Kar2p antiserum. Bar, 2 μm.
Figure 5.
Golgi apparatus morphology is wild type in dnm1Δ cells. Confocal microscopy images of DNM1 (JSY1238, A and B) and dnm1Δ (JSY1361, C and D) cells labeled with the peripheral Golgi membrane marker, Sec7p-EGFP. White arrows in A and C indicate magnified cells shown in B and D, respectively. Bars, 2 μm.
Figure 6.
Vacuole morphology is wild type in dnm1Δ cells. Differential interference contrast (A and C) and fluorescence images (B and D) of DNM1 (JSY1238, A and B) and dnm1Δ (JSY1361, C and D) cells labeled with the vacuole membrane–specific styryl dye, FM 4-64. Bar, 2 μm.
Figure 7.
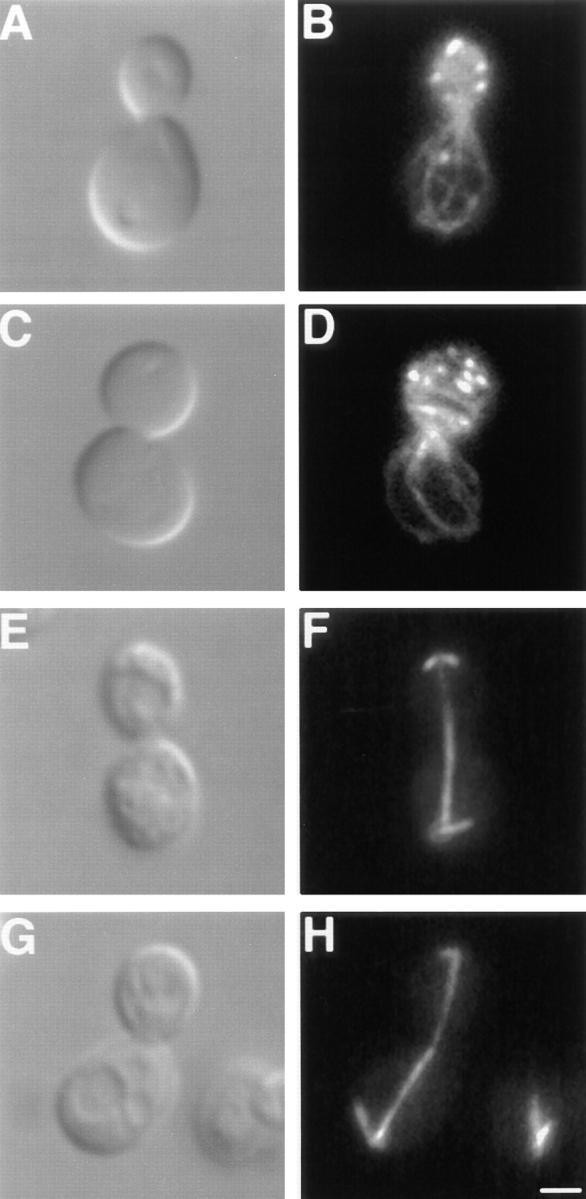
Actin and tubulin cytoskeleton organization are wild type in dnm1Δ cells. Differential interference contrast images (A, C, E, and G), rhodamine-phalloidin–stained actin (B and D), and antitubulin staining (F and H) of DNM1 (JSY1238; A, B, E, and F) and dnm1Δ (JSY1361; C, D, G, and H) cells. Buds are in the upper portion of each panel. Bar, 2 μm.
Mitochondrial Morphology Changes Dramatically when Dnm1p Expression Is Induced or Repressed in Yeast Cells
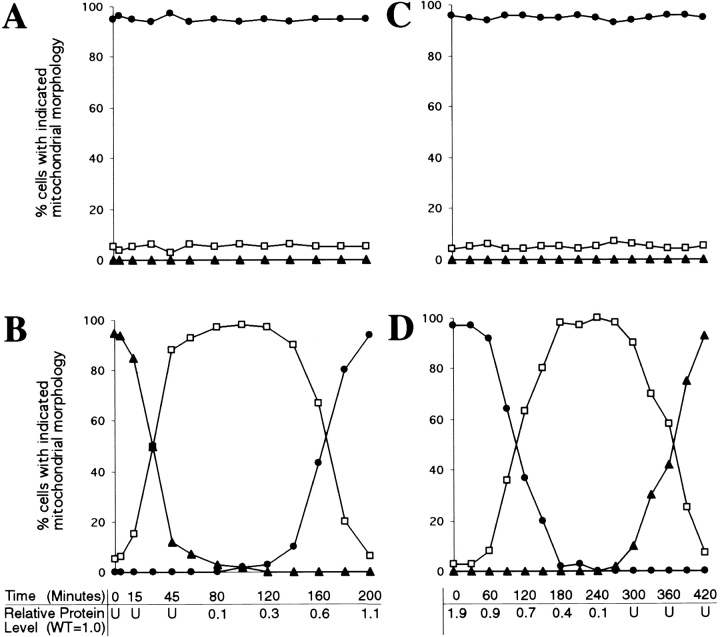
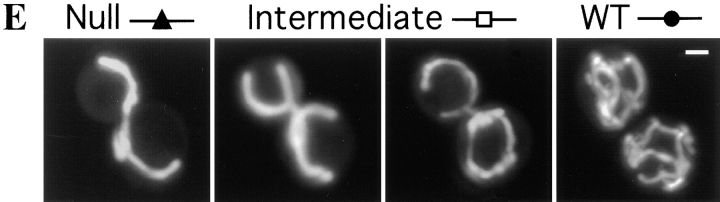
To further characterize the function of DNM1 in vivo, we examined mitochondrial morphology in cells carrying only a chromosomal copy of DNM1 under control of the GAL1 promoter (JSY1678). When this strain is grown overnight in dextrose-containing medium (which represses transcription from the GAL1 promoter), no Dnm1p can be detected (Fig. 8 B, 0 min; U, undetected), and >95% of the cells contain collapsed mitochondrial networks (Fig. 8 B, closed triangles; and 8 E, Null). 15 min after transfer to galactose-containing medium to induce expression of Dnm1p, the percentage of cells in the population exhibiting the null mitochondrial morphology begins to decline, and cells containing a more branched “intermediate” morphology appear (Fig. 8 B, open squares; and 8 E, Intermediate). As shown in the confocal microscope projections in Fig. 8 E, cells with this intermediate morphology appear to be extending tubules of mitochondrial membrane around the cell periphery to form a ring. By 140 min, the tubules extended around the cortex begin forming additional branches characteristic of the wild-type mitochondrial network, and at later time points (200 min), virtually 100% of the cells in the population exhibit wild-type mitochondrial morphology (Fig. 8 B, closed circles; and 8 E, WT). These changes in mitochondrial morphology are correlated with an increase in the intracellular level of the Dnm1 protein. As shown below the graph in Fig. 8 B, the steady-state level of Dnm1p in the induced GAL1-DNM1 strain increases from undetected (U, at time zero) to 1.1 (at 200 min) relative to the wild-type DNM1 strain (containing a single chromosomal copy of the DNM1 gene expressed from its own promoter). In control experiments, DNM1 cells maintain wild-type mitochondrial networks throughout the same time course (Fig. 8 A, closed circles).
Figure 8.
Mitochondrial morphology changes in response to changes in DNM1 gene expression. Null (closed triangles), intermediate (open squares), and wild-type (closed circles) mitochondrial morphologies were quantified by DiOC6 staining in cells containing a single genomic copy of the wild-type DNM1 (JSY1238; A and C) or GAL1-DNM1 (JSY1678; B and D) gene. (A) DNM1 cells transferred from dextrose to galactose. (B) GAL1-DNM1 cells transferred from dextrose to galactose. (C) DNM1 cells transferred from galactose to dextrose. (D) GAL1-DNM1 cells transferred from galactose to dextrose. The amount of Dnm1p expressed in the GAL1-DNM1 strain relative to the DNM1 strain is indicated for selected time points below graphs B and D. (WT = DNM1 = 1.0; U, undetected). (E) Representative examples of Null (closed triangles), intermediate (open squares), and WT (DNM1; closed circles) mitochondrial morphologies. Buds are in the upper portion of each panel. Bar, 2 μm.
We also found that the wild-type mitochondrial network collapses when cells are transferred from galactose to dextrose to turn off transcription of the GAL1-DNM1 gene (Fig. 8 D). In these experiments, >95% of the cells grown in galactose medium contain highly branched, wild-type mitochondrial networks (Fig. 8 D, closed circles; and 8 E, WT). Although the initial steady-state level of Dnm1p expressed from the chromosomal GAL1-DNM1 construct is 1.9-fold higher than that observed in the wild-type DNM1 strain (Fig. 8 D, relative protein level at time zero), this level of Dnm1p overexpression does not appear to produce additional mitochondrial phenotypes in otherwise wild-type cells. 60 min after transfer to dextrose, the intracellular level of Dnm1p falls below that observed in the DNM1 control strain (Fig. 8 D, relative protein level = 0.9), and branches begin to disappear from the mitochondrial network producing an intermediate mitochondrial morphology (Fig. 8 D, open squares; and 8 E, Intermediate). At later time points, mitochondrial morphology collapses further in these cells to produce the elongated mitochondrial structures characteristic of dnm1 null cells (Fig. 8 D, closed triangles; and 8 E, Null). Once again, control cells containing the DNM1 gene expressed from its own promoter maintain wild-type mitochondrial networks throughout an identical time course (Fig. 8 C, closed circles). These results demonstrate that changes in mitochondrial morphology can be directly correlated with changes in the level of Dnm1p expression.
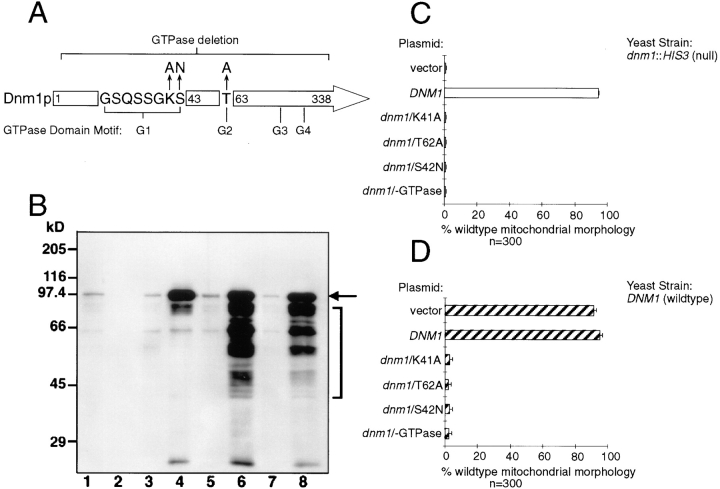
The Predicted GTPase Domain Is Required for Dnm1p Function in Mitochondrial Morphology Maintenance
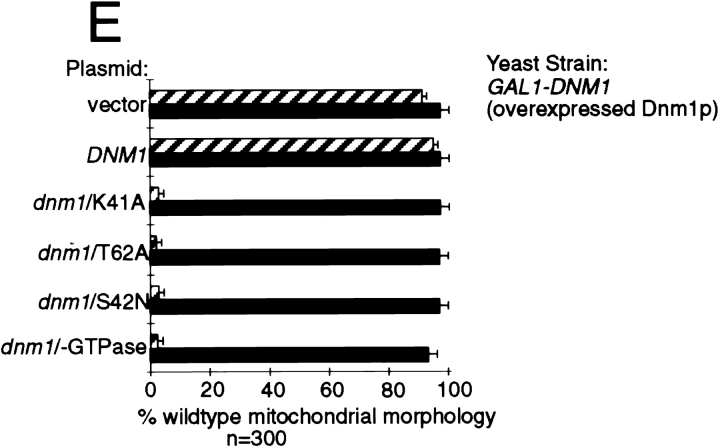
Like other members of the dynamin family, the NH2-terminal domain of Dnm1p contains a predicted GTP-binding motif with the conserved sequence elements GSQSSGKS42 (Fig. 9 A, G1), DLPG178 (Fig. 9 A, G3, sequence not shown), and TKLD247 (Fig. 9 A, G4, sequence not shown) (Bourne et al., 1991). Although a G2 motif consisting of a conserved threonine (Bourne et al., 1991) has not been experimentally defined in dynamins, Dnm1p contains a threonine at position 62 that has the potential to serve as a G2 domain (Fig. 9 A). To determine whether the predicted GTP-binding motif in DNM1 is important for function, point mutations that alter the nucleotide-binding and/or hydrolysis properties of GTPases (Fig. 9 A) were introduced into a DNM1 gene contained on a low-copy (pRU1) or high-copy (YEp213) (Rose and Broach, 1990) plasmid and expressed in yeast (Fig. 9 B). The G1 dnm1K41A mutation is predicted to abolish nucleotide binding (Bourne et al., 1991; van der Bliek et al., 1993), the G1 dnm1S42N mutation is predicted to have reduced affinity for GTP (Feig and Cooper, 1988; Herskovits et al., 1993), and the G2 dnm1T62A mutation is equivalent to a mutation in the p21ras GTPase that is thought to interfere with p21ras GAP binding (Adari et al., 1988; Cales et al., 1988) and with raf kinase interactions (Vojtek et al., 1993). A dnm1 mutant lacking the entire NH2-terminal GTP-binding domain was also constructed (dnm1−GTPase). None of these mutated dnm1 genes expressed from a low-copy (pRU1, not shown) or high-copy plasmid (YEp213) could rescue mitochondrial morphology defects in the dnm1 null strain (Fig. 9 C, dnm1::HIS3), suggesting that GTP binding and/or hydrolysis is required for Dnm1p function in vivo. In addition, all four mutant dnm1 constructs induced dominant mitochondrial morphology defects when expressed from a low-copy (pRU1, not shown) or high-copy number plasmid (YEp213) in cells containing a wild-type copy of the DNM1 gene (Fig. 9 D, hatched bars). These mutant constructs did not cause trans-dominant mitochondrial morphology defects in cells that were also overexpressing the wild-type Dnm1 protein. (Compare filled bars in Fig. 9 E with hatched bars from Fig. 9 D; wild-type Dnm1p is expressed from a genomic GAL1-DNM1 construct at a steady-state level approximately twofold that of wild-type DNM1 cells.) The ability of all four GTP-binding domain mutations to induce mitochondrial morphology defects in wild-type cells further supports the notion that Dnm1p plays a primary role in controlling mitochondrial morphology. Moreover, these dominant phenotypes suggest that Dnm1p associates with additional cellular components required for mitochondrial morphology maintenance.
Figure 9.
GTPase domain mutations in DNM1 cause dominant interfering phenotypes in vivo. (A) The position of the four, NH2-terminal GTP-binding consensus elements (G1–G4) conserved among different dynamin family members is shown. The point mutations (arrows) and the deletion mutation (bracket) used in this study are indicated. The G1 dnm1K41A mutation and the deletion mutation are predicted to abolish nucleotide binding, the G1 dnm1S42N mutation is predicted to bind GDP more tightly than GTP, and the G2 dnm1T62A mutation is equivalent to a mutation in the ras GTPase that abolishes effector interactions (see text for references). (B) Endogenous and plasmid expression levels of wild-type and GTPase mutant forms of Dnm1p. A total cell extract from each strain was analyzed by Western blotting with anti-Dnm1p antiserum. Lane 1, DNM1 (JSY1238); lane 2, dnm1Δ (dnm1:: HIS3, JSY1361); lane 3, dnm1Δ + pRU1-dnm1K41A (CEN, single copy); lane 4, dnm1Δ + YEP213-dnm1K41A (2μ, multicopy); lane 5, dnm1Δ + pRU1-dnm1T62A; lane 6, dnm1Δ + YEP213-dnm1T62A; lane 7, dnm1Δ + pRU1-dnm1S42N; lane 8, dnm1Δ + YEP213-dnm1S42N. The black arrow marks the full-length form of wild-type and mutant Dnm1 proteins. Dnm1p breakdown products (bracket) accumulate in cells overexpressing Dnm1 proteins. Expression of the dnm1/-GTPase deletion mutant protein from the pRU1 and YEP213 plasmids was similar to that shown for the other mutants (not shown). (C) DiOC6 staining was used to quantify the ability of a multicopy vector alone (YEP213; LEU2), or the vector containing wild-type (DNM1) or mutated (dnm1/K41A, dnm1/T62A, dnm1/S42N, dnm1/-GTPase) forms of the DNM1 gene to restore wild-type mitochondrial morphology in dnm1::HIS3 null cells (JSY1361). The percentage of cells with wild-type mitochondrial morphology is indicated in each case (open bars). (D) The ability of the constructs described in C to induce mitochondrial morphology phenotypes in a wild-type DNM1 strain (JSY1238) was quantified by DiOC6 staining. The percentage of cells with wild-type mitochondrial morphology is indicated in each case (hatched bars). (E) The mutant constructs described in C (dnm1/K41A, dnm1/T62A, dnm1/S42N, and dnm1/-GTPase) failed to induce mitochondrial morphology defects in cells overexpressing Dnm1p from a genomic GAL1-DNM1 gene (JSY1678) (closed bars; overexpressed Dnm1p levels are approximately twofold that of the wildtype DNM1 strain). The dominant interfering phenotypes caused by these mutations in a strain expressing Dnm1p at wild-type levels (D; DNM1, JSY1238; hatched bars) are shown for comparison.
Endocytosis of FM 4-64 Occurs with Wild-Type Kinetics in dnm1Δ Cells
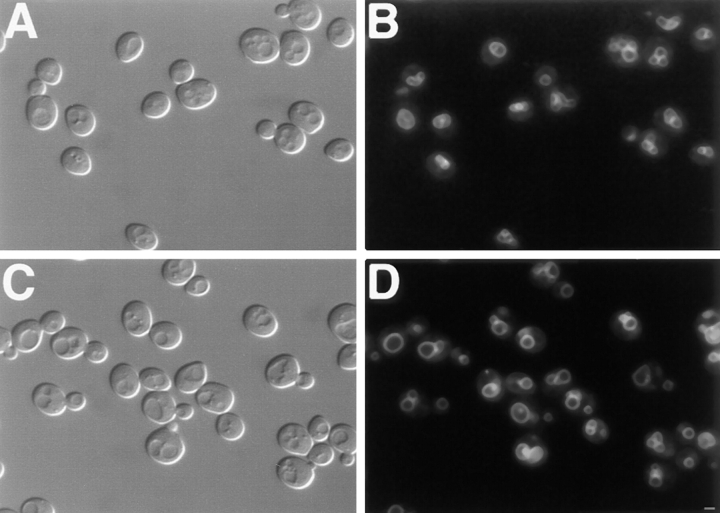
Dynamin mutations have been shown to cause endocytosis defects in both Drosophila melanogaster and mammalian cells (Poodry and Edgar, 1979; Herskovits et al., 1993; van der Bliek et al., 1993). In contrast, cells lacking DNM1 did not exhibit defects in the internalization of FM 4-64, a lipophilic styryl dye that has been used as a marker for endocytosis in yeast (Vida and Emr, 1995). Wild-type and dnm1Δ cells were labeled with FM 4-64 for 30 min at 0°C to allow the dye to accumulate in the plasma membrane (Table II; Fig. 10, A and B). The cells were subsequently washed to remove free dye and shifted to 30°C, and the internalization of FM 4-64 was monitored over a 60-min time course. As shown in Table II and Fig. 10, the kinetics of FM 4-64 uptake in the dnm1Δ strain and an isogenic wild-type strain were identical. After 10 min at 30°C, the FM 4-64 signal in the plasma membrane (Table II, PM) was still visible, and small fluorescent endosome-like intermediates (E) began to accumulate in the cytoplasm (Fig. 10, C and D). After 20 min, the PM-associated signal had decreased, and the majority of FM 4-64 fluorescence was concentrated in these endosome-like compartments (Table II; Fig. 10, E and F). At the 40-min time point, the FM 4-64 signal had begun to accumulate in the vacuole membrane (Table II, V), although some punctate cytoplasmic structures were still visible (Fig. 10, G and H). By 60 min, the majority of FM 4-64 signal was associated with the vacuole membrane (Table II; Fig. 10, I and J). The finding that the kinetics of FM 4-64 internalization in dnm1Δ cells are indistinguishable from wild type indicates that Dnm1p is not required for bulk endocytosis of the plasma membrane in yeast.
Table II.
Kinetics of FM 4-64 Internalization
| FM 4-64 Localization | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Time | Strain | PM | PM + E | E + V | V | |||||
| min | ||||||||||
| 0* | WT | 50/50 (100%) | 0/50 (0%) | 0/50 (0%) | 0/50 (0%) | |||||
| dnm1Δ | 50/50 (100%) | 0/50 (0%) | 0/50 (0%) | 0/50 (0%) | ||||||
| 10 | WT | 0/68 (0%) | 68/68 (100%) | 0/68 (0%) | 0/68 (0%) | |||||
| dnm1Δ | 0/55 (0%) | 55/55 (100%) | 0/55 (0%) | 0/55 (0%) | ||||||
| 20 | WT | 0/67 (0%) | 67/67 (100%) | 0/67 (0%) | 0/67 (0%) | |||||
| dnm1Δ | 0/58 (0%) | 58/58 (100%) | 0/58 (0%) | 0/58 (0%) | ||||||
| 40 | WT | 0/56 (0%) | 0/56 (0%) | 56/56 (100%) | 0/56 (0%) | |||||
| dnm1Δ | 0/50 (0%) | 0/50 (0%) | 50/50 (100%) | 0/50 (0%) | ||||||
| 60 | WT | 0/57 (0%) | 0/57 (0%) | 0/57 (0%) | 57/57 (100%) | |||||
| dnm1Δ | 0/60 (0%) | 0/60 (0%) | 0/60 (0%) | 60/60 (100%) | ||||||
PM, plasma membrane; E, endosome-like intermediate; V, vacuole.
0 min time point scored after incubation and wash performed at 0°C. Remaining time points were scored during the 30°C chase (see text).
Figure 10.
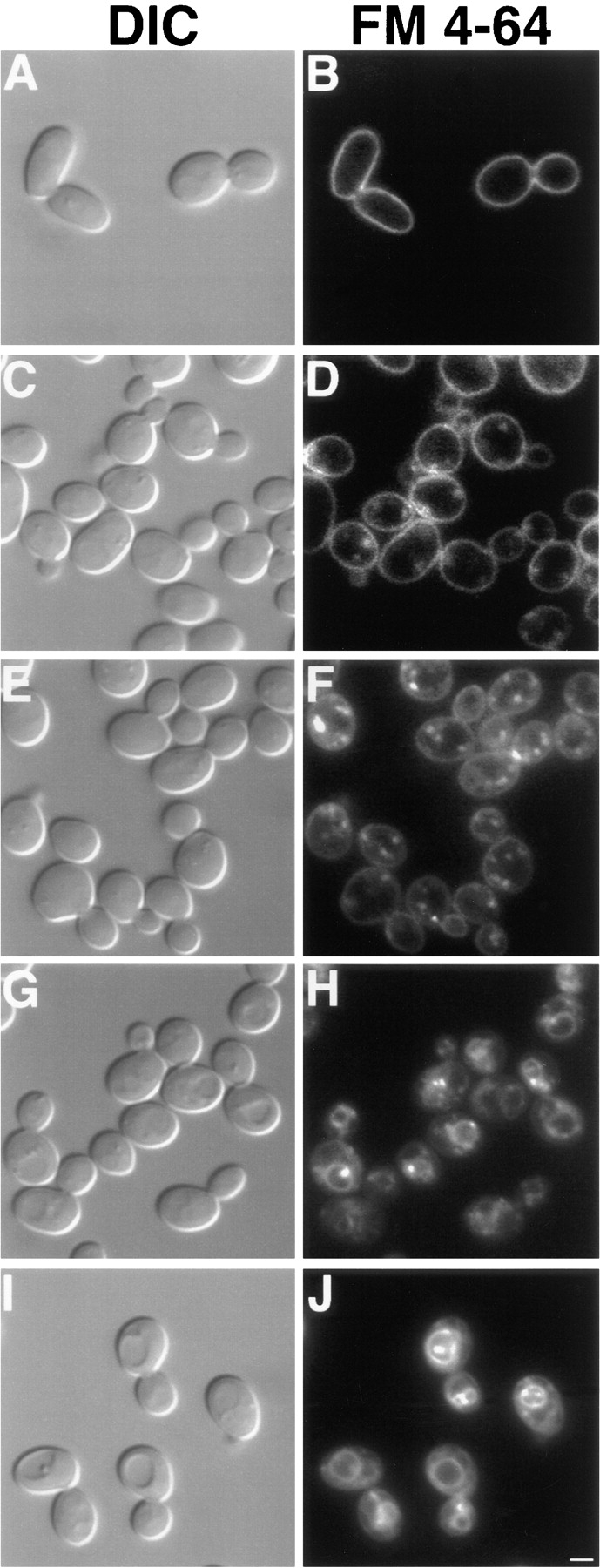
Kinetics of FM 4-64 internalization. DNM1 (JSY1238) and dnm1Δ (JSY1361) cells grown in YPD medium were labeled with FM 4-64 for 30 min at 0°C, washed, and chased in fresh medium at 30°C for 60 min. Aliquots removed at 0 (A and B), 10 (C and D), 20 (E and F), 40 (G and H), and 60 (I and J) min were scored for FM 4-64 distribution (see Table II) and photographed. Representative differential interference contrast (A, C, E, G, and I) and fluorescence (B, D, F, H, and J) images are shown for the dnm1Δ strain and were identical to those observed in wild type. Bar, 2 μm.
Endocytosis Defects Do Not Induce Changes in Yeast Mitochondrial Morphology
To further examine the possibility that the mitochondrial morphology defect in dnm1 was a secondary consequence of an endocytic defect, we examined the mitochondrial network in well-characterized yeast mutants blocked at distinct steps in endocytosis. The end3 and end4 mutations block the internalization step of endocytosis (Raths et al., 1993; Benedetti et al., 1994). end13/vps4 does not affect internalization but does affect delivery of internalized material to the vacuole (Munn and Riezman, 1994; Babst et al., 1997). vps18 mutants completely lack a structure resembling the vacuole (the terminal endocytic compartment) (Robinson et al., 1991), and pan1 mutants accumulate compartments similar to early endosomes (Wendland et al., 1996). We visualized mitochondrial morphology in these mutants (Fig. 11, B–F) and their isogenic wild-type parents (Fig. 11 A and data not shown) and detected no defects in mitochondrial morphology. Thus, the mitochondrial morphology phenotype observed in the dnm1 mutant is not an indirect consequence of an endocytosis defect.
Figure 11.
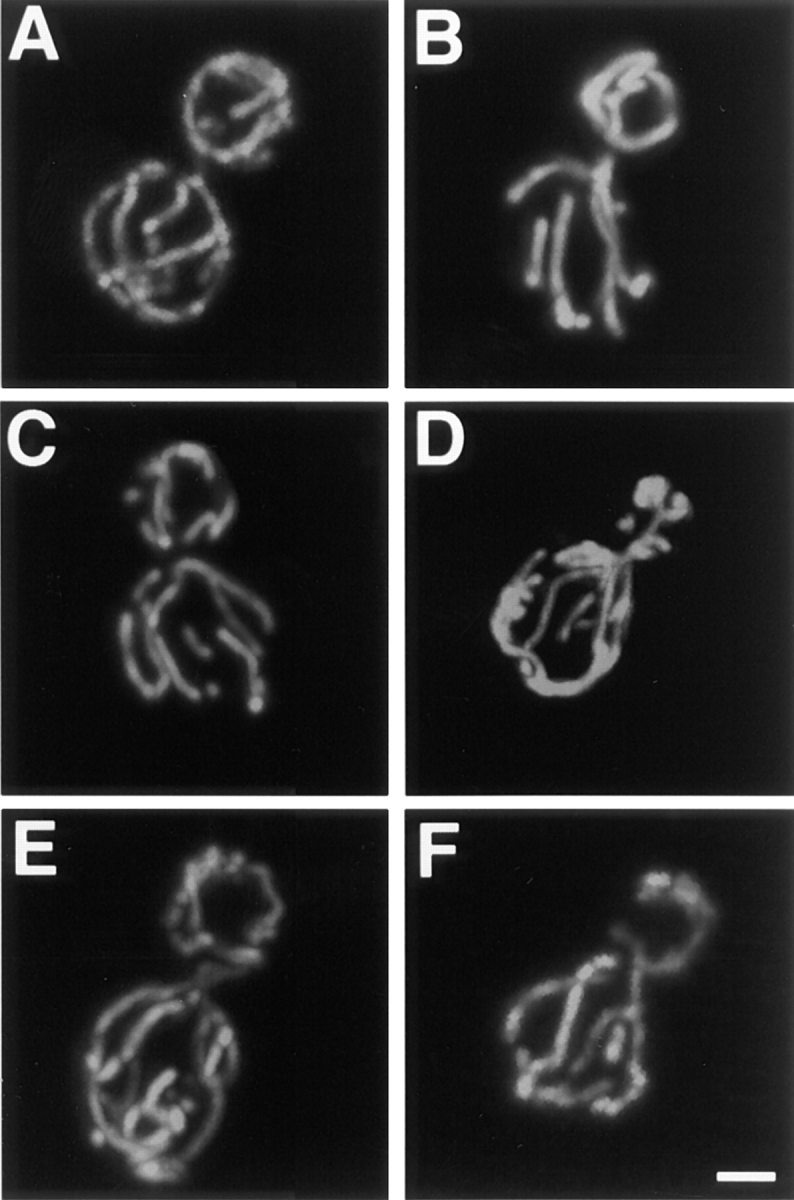
Mitochondrial morphology is wild type in mutants that block endocytosis. Mitochondrial morphology was visualized with Cox4–GFP in wild-type (A), end3 (B), end4 (C), vps4 (D), vps18 (E), and pan1 (F) cells under conditions that produce an endocytosis defect in the mutants. The wild-type mitochondrial morphology observed in A was also observed in >95% of the mutant cells. Buds are in the upper portion of each panel. Bar, 2 μm.
Dnm1p Is Found in Punctate Structures that Colocalize with the Mitochondrial Network at the Cell Cortex
The localization of Dnm1p in wild-type cells is consistent with its role in mitochondrial morphology maintenance. We performed indirect immunofluorescence studies to localize a Dnm1-HA–tagged protein (Dnm1–HACp) in yeast (Fig. 12, B, E, H, and K). Mitochondrial networks in the same cells were simultaneously visualized with the Cox4–GFP fusion protein (Fig. 12, A, D, G, and J). Confocal microscopy was used to evaluate the degree of Dnm1– HACp colocalization with mitochondrial networks in merged images (Fig. 12, C, F, I, and L). In a wild-type strain, yeast mitochondrial networks visualized with Cox4–GFP were localized at the cell cortex and distributed evenly around the cell circumference (Fig. 12, A and D, green fluorescence, Cox4–GFP). In cells expressing the Dnm1–HACp protein, monoclonal antibodies specific for the HA epitope (Fig. 12 E, red fluorescence, CY5-conjugated secondary antibody) stained punctate structures that also localized to the cell cortex. In addition, a haze of fluorescence was observed throughout the cell, suggesting that there was also a pool of cytoplasmic Dnm1–HAC protein. Dnm1–HACp staining was not detected in control experiments performed in the absence of the primary anti-HA antibody (Fig. 12 B). When the images in Fig. 12, D and E, were merged (Fig. 12 F), the majority of the Dnm1–HACp spots visualized with CY5 (85%) appeared on the sides of mitochondrial tubules, at mitochondrial network branch points, or at the tips of mitochondrial tubules. (Yellow areas correspond to regions where the red and green signals are superimposed.) Similar results were obtained when antibodies raised against the Dnm1 protein were used to stain wild-type cells (Fig. 12, P–R). Once again, these antibodies stained punctate structures at the cell cortex (Fig. 12 Q) that colocalized with the mitochondrial network (Fig. 12 P) in a merged image (Fig. 12 R; 75% colocalization).
We were concerned that some of the colocalization might be coincidental since mitochondrial morphology is complex and both the mitochondrial network and Dnm1p are found at the cell periphery. For this reason, we also localized Dnm1–HACp in an mdm10 mutant strain (Sogo and Yaffe, 1994) that lacks mitochondrial networks and, instead, contains one or a few giant spherical mitochondria (Fig. 12, G and J). A significant fraction (57%) of the Dnm1–HACp fusion protein (Fig. 12 K) still localized at or near the spherical mitochondria in mdm10 cells (Fig. 12 L), suggesting that these Dnm1–HACp-containing structures are bound directly or indirectly to the mitochondrial compartment. In addition, a portion of the Dnm1–HACp (43%) remained in punctate spots at the cortex in mdm10 cells, indicating that Dnm1p may be localized to the cell cortex independent of the mitochondrial network.
Subcellular Localization of Dnm1p
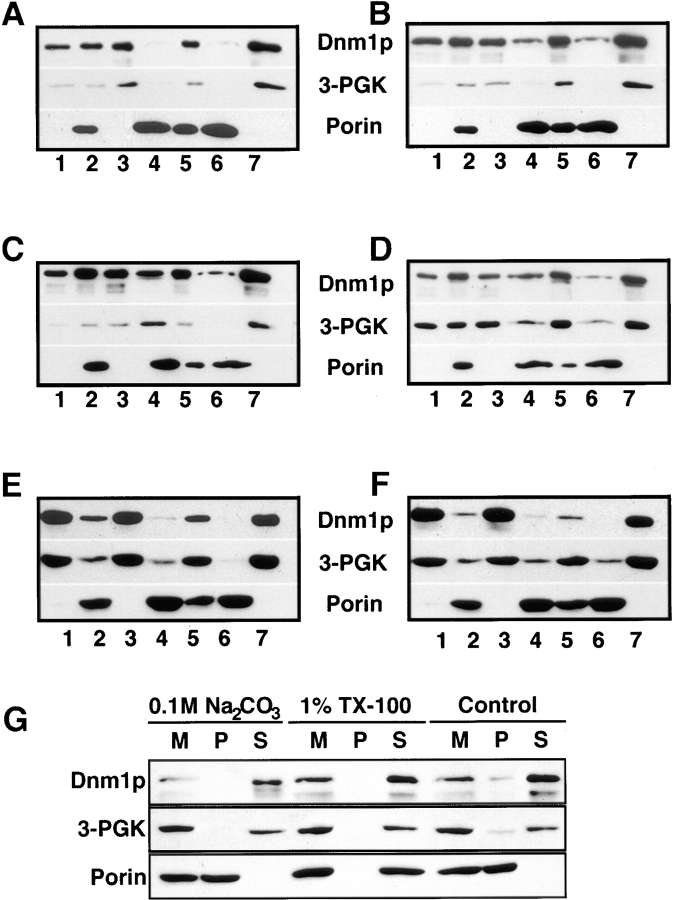
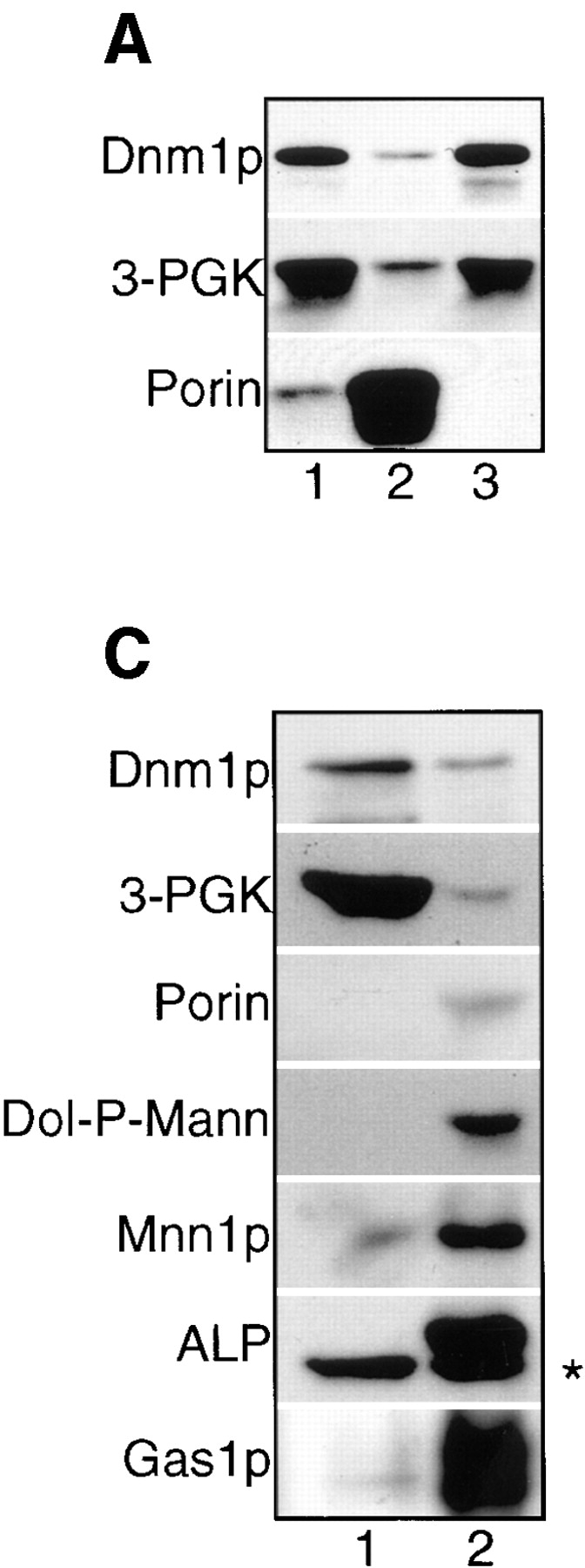
Immunoblot analysis of yeast subcellular fractions suggested that a portion of Dnm1p was associated with an enriched mitochondrial pellet. A total cell extract from a wild-type strain (JSY1238; Fig. 13 A, lane 1) was subjected to differential sedimentation to generate a P10,000 pellet (Fig. 13 A, lane 2) and an S10,000 supernatant (Fig. 13 A, lane 3). Characterization of these fractions by immunoblotting with anti-Dnm1p, anti–3-PGK (cytosol), and antiporin (mitochondrial outer membrane) antisera revealed that a substantial amount of Dnm1p was either soluble or associated with a nonmitochondrial membrane fraction that could not be pelleted at 10,000 g (Fig. 13 A, lane 3). However, a portion of Dnm1p cofractionated with the P10,000 pellet, a fraction enriched in mitochondrial membranes (Fig. 13 A, lane 2). Washing the P10,000 pellet and sedimenting a second time at 10,000 g released additional Dnm1p into the supernatant, suggesting that the protein was not tightly associated with membranes in this fraction (Fig. 13 A, compare washed P10,000 in lane 6 with the S10,000 wash in lane 7). Indeed, treatment of the initial P10,000 fraction with 0.1 M Na2CO3, pH 11.5, to release peripheral membrane proteins completely disrupted the association of Dnm1p with the membrane pellet (Fig. 13 G).
Figure 13.
Distribution of Dnm1p in wild-type and mutant yeast cells after differential sedimentation. Yeast cells were spheroplasted, lysed, and sedimented at 1,500 g to remove unlysed cells and debris. The soluble extracts were sedimented at 10,000 g generating (lane 1) extract, (lane 2) P10,000 (contains mitochondria), and (lane 3) S10,000. The P10,000 pellet was resuspended and sedimented again at 1,500 g to remove aggregates and debris generating (lane 4) P1,500 and (lane 5) S1,500. S1,500 was spun at 10,000 g generating (lane 6) washed P10,000 and (lane 7) S10,000 wash. Fractions (2 μg protein) were separated by SDS-PAGE and analyzed by Western blotting with anti-Dnm1p, anti–3-PGK (cytoplasm), and antiporin (mitochondria) serum. Strains: (A) DNM1 (JSY1238), (B) DNM1 + pRU1-DNM1, (C) DNM1 + pRU1- dnm1K41A, (D) dnm1Δ (JSY1361) + pRU1-dnm1K41A, (E) MDM10 (JSY1914), (F) mdm10Δ (JSY1916). (G) P10,000 pellets (M) containing mitochondria were treated to dissociate peripheral membrane proteins (0.1 M Na2CO3) or solubilize integral membrane proteins (1% Triton X-100), separated into pellet (P) and supernatant (S) fractions, and analyzed by SDS-PAGE and Western blotting with anti-Dnm1p, anti–3-PGK, and antiporin serum. The release of soluble cytochrome b 2 and the peripheral F1β ATPase subunit into the supernatant fraction after 0.1 M Na2CO3 treatment was confirmed by Western blotting (data not shown).
The cellular distribution of Dnm1p was not dramatically affected by overexpressing wild-type Dnm1p, or expressing mutant forms of Dnm1p, in wild-type and dnm1Δ strains. The relative amount of Dnm1p associated with the P10,000 fraction during differential sedimentation did not significantly increase or decrease when an extra copy of the wild-type DNM1 (Fig. 13 B) or the mutated dnm1K41A (Fig. 13 C) gene was introduced into wild-type cells on a low-copy plasmid. Similar fractionation results were obtained when low-copy plasmids harboring mutated dnm1K41A (Fig. 13 D), dnm1S42N(not shown), or dnm1T62A (not shown) genes were introduced into a dnm1Δ strain. In addition, the Dnm1p fractionation pattern did not change significantly when differential sedimentation studies were performed on wild-type or dnm1Δ cells expressing the DNM1, dnm1K41A, dnm1S42N, and dnm1T62A genes from high-copy plasmids (data not shown). Finally, the subcellular distribution of Dnm1p in an mdm10 mutant strain (Fig. 13 F) did not differ significantly from wild type (Fig. 13 E).
The fractionation studies described above did not reveal whether Dnm1p was associated with mitochondrial membranes or with other cellular membranes in the P10,000 pellet. In addition, these studies did not address the possibility that Dnm1p forms aggregates that nonspecifically sediment with or are trapped by membranes during centrifugation. To distinguish between these possibilities, we separated membranes in the P10,000 pellet (Fig. 14 A, lane 2) by flotation in a sucrose density gradient (Fig. 14; Walworth et al., 1989). As shown in Fig. 14 B, the majority of Dnm1p in P10,000 was soluble and remained at the bottom of the gradient (graph ii, fractions 18–20) with the cytoplasmic protein 3-PGK (graph iii, fraction 20). However, a portion of Dnm1p cofractionated with the mitochondrial outer membrane protein porin in fraction 15 (Fig. 14 B, graph ii). The Dnm1p peak in fraction 15 was not completely coincident with marker peaks for the Golgi apparatus (graph iv, Mnn1p), the vacuole (graph v, ALP), or the plasma membrane (graph v, Gas1p). Although the ER marker Dol-P-Man synthase was found in two well separated peaks in the gradient (Fig. 14 B, graph iv, peaks at fractions 7 and 15), Dnm1p only localized with the ER peak in fraction 15. Since the porin mitochondrial marker was also found in fraction 15, and since Dnm1p colocalized with intact mitochondrial networks in indirect immunofluorescence experiments, it seems likely that Dnm1p associates with mitochondrial membranes in this fraction.
Figure 14.
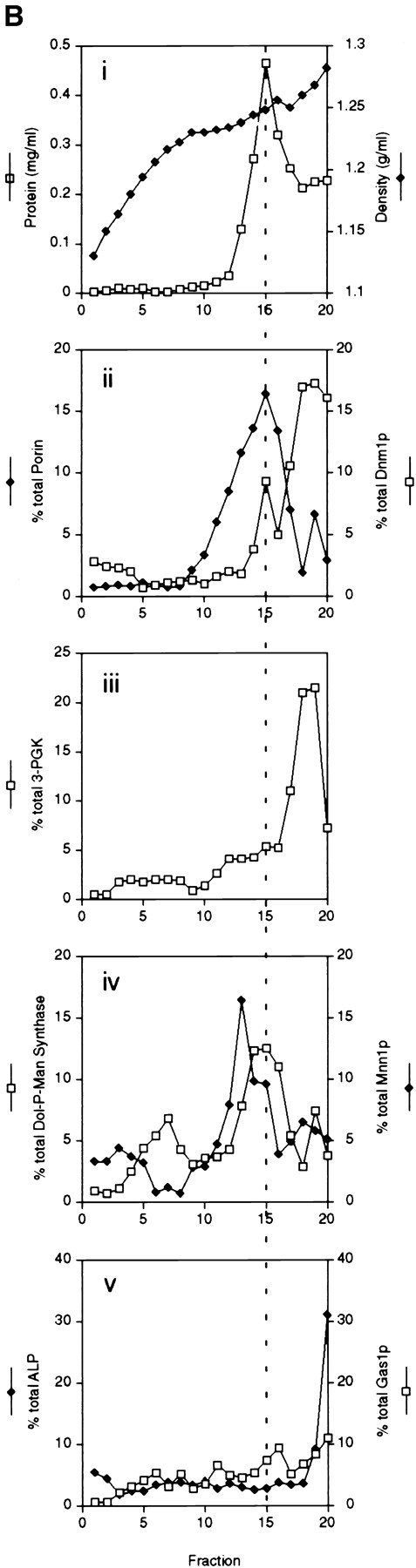
Elution profile of Dnm1p and organelle membrane markers after sucrose density fractionation of P10,000 from wild-type cells. (A) (lane 1) Extract, (lane 2) P10,000, and (lane 3) S10,000 fractions were generated and analyzed as described in the legend to Fig. 13. (B) The P10,000 pellet from A was resuspended in 60% sucrose and loaded on the bottom of a 35–60% sucrose gradient. The gradient was spun to equilibrium and fractionated, and equal volumes of each fraction were analyzed by Western blotting and densitometry. Results are expressed as the percent total signal per fraction. Density (g/ml); protein (μg/ml). Fraction 1 is the top of the gradient and fraction 20 is the gradient pellet. The dotted line transects fraction 15 in each graph. (C) S10,000 supernatant from A was spun at 100,000 g and equivalent volumes of the resulting S100,000 (lane 1) and P100,000 (lane 2) fractions were analyzed by SDS-PAGE and Western blotting with anti-Dnm1p, anti– 3-PGK, antiporin, anti–Dol-P-Man (endoplasmic reticulum), anti- Mnn1p (Golgi apparatus), anti-ALP (vacuole), and anti-Gas1p (plasma membrane) serum. The asterisk (*) marks a soluble, ALP breakdown product in the vacuole lumen that is partially released during fractionation (Stepp et al., 1997; Vowels and Payne, 1998).
To determine whether the Dnm1p found in the initial S10,000 supernatant (Fig. 14 A, lane 3) was associated with additional cellular membranes, this supernatant was further fractionated at 100,000 g to yield S100,000 and P100,000. Although the ER (Dol-P-Man), Golgi apparatus (Mnn1p), vacuole (ALP), and plasma membrane (Gas1p) markers sedimented under these conditions and were found in the P100,000 pellet (Fig. 14 C, lane 2), the majority of Dnm1p was soluble and remained in the S100,000 supernatant along with the soluble cytoplasmic protein 3-PGK (Fig. 14 C, lane 1).
Discussion
The studies described here establish Dnm1p as an important regulator of mitochondrial morphology in yeast. In dnm1 mutants, the mitochondrial membranes fail to be distributed properly at the cortex and collapse to one side of the cell. In contrast, the loss of Dnm1p function has no apparent effect on the morphology and distribution of other cytoplasmic organelles, including nuclei, the endoplasmic reticulum, the Golgi apparatus, and vacuoles. Despite their unusual morphology, dnm1 mitochondria are able to generate a membrane potential and import a Cox4–GFP marker protein into the mitochondrial matrix, suggesting that basic organelle functions are not severely impaired. Unlike other yeast mitochondrial morphology mutants, which rapidly lose mtDNA and exhibit defects in mitochondrial inheritance (Hermann and Shaw, 1998), dnm1 mitochondria retain their mtDNA and are efficiently transported into buds during division. Together, our results indicate that Dnm1p plays a very specific role in defining the shape and distribution of yeast mitochondrial membranes.
The immunolocalization of Dnm1p in intact cells is consistent with a role for this protein in controlling mitochondrial morphology and distribution. In wild-type cells, Dnm1p is found in punctate structures at the cell cortex that colocalize with branch points in the mitochondrial network and the tips and sides of mitochondrial tubules. A portion of these Dnm1p-containing structures remain associated with the spherical mitochondria found in the mdm10 mutant, suggesting that Dnm1p associates directly or indirectly with the organelle. This interpretation is supported by the observation that a small amount of Dnm1p reproducibly fractionates with mitochondrial membranes in sucrose density gradients. The proportion of total cellular Dnm1p associated with mitochondria in these gradients (and in the differential sedimentation studies) may be underrepresented because of fragmentation of the mitochondrial network during cell lysis. Our subcellular fractionation studies suggest that there is also a large cytoplasmic pool of Dnm1p. Although it is tempting to speculate that cycles of GTP binding and hydrolysis regulate the reversible association of Dnm1p with mitochondria, we did not detect differences in the subcellar distribution of wild-type and GTPase mutant Dnm1 proteins. However, we cannot rule out the possibility that the GTP-bound form of Dnm1p associates with mitochondrial membranes since we were unable to make mutations predicted to bind but not hydrolyze GTP. Further experiments are needed to determine the effect of GTP binding and hydrolysis on Dnm1p–mitochondrial interactions.
A deletion mutation and three different amino acid substitutions in the predicted GTP-binding domain of Dnm1p failed to rescue mitochondrial morphology defects in a dnm1 strain. Two of these mutations were generated in the G1 motif and are predicted to either reduce the affinity of the protein for both GTP and GDP (dnm1K41A; Bourne et al., 1991; van der Bliek et al., 1993), or are predicted to have reduced affinity for GTP (dnm1S42N; Feig et al., 1988; Herskovits et al., 1993). The failure of these mutant proteins to rescue the dnm1 mutant phenotype demonstrates that a functional Dnm1p GTPase domain is required for maintenance of the mitochondrial network in yeast. In support of this interpretation, preliminary studies in our lab indicate that a bacterially-expressed form of wild-type Dnm1p can bind and hydrolyze GTP in an in vitro assay (Brisch, E., and J.M. Shaw, unpublished data).
The Dnm1p GTPase domain mutations also cause dominant mitochondrial morphology defects in wild-type cells. These defects can be completely rescued by overexpressing the wild-type Dnm1 protein in the same strain. Analogous mutations in mammalian dynamin (Herskovits et al., 1993; van der Bliek et al., 1993; Damke et al., 1994) and the yeast dynamin-like protein Vps1p (Vater et al., 1992) cause similar dominant negative defects in endocytosis and vacuolar protein sorting, respectively. In the case of mammalian dynamin, the dominant negative endocytosis phenotype is thought to be related to the self assembly properties of this protein (Warnock et al., 1996). It is possible that coassembly of mutant and wild-type Dnm1 proteins produces oligomers or higher order structures that cannot be disassembled and block the function of Dnm1p in wild-type cells. Alternatively, mutant Dnm1 proteins could produce dominant phenotypes in wild-type cells by titrating out other physiologically important binding partners. Studies are currently underway to determine whether Dnm1p homooligomerizes and/or interacts with additional protein-binding partners.
The GTPase domains of many proteins include a G2 motif with a conserved threonine residue (Bourne et al., 1991). In p21ras, this threonine interacts directly with a catalytic Mg2+ ion and the γ-phosphate of GTP (Pai et al., 1990). p21ras mutations that affect this residue prevent GTP-dependent interactions with the p21ras-GAP and the raf kinase (Adari et al., 1988; Cales et al., 1988; Bourne et al., 1991; Vojtek et al., 1993). Although dynamins contain a conserved threonine 20 residues downstream of the G1 motif, this region has not been defined as a G2 motif. In this study, we show that a T62A mutation in Dnm1p fails to rescue mitochondrial morphology defects in dnm1 cells and induces the mitochondrial network to collapse in wild-type cells. These results demonstrate that threonine 62 constitutes a bona fide G2 element in the Dnm1p GTPase domain. In keeping with the nomenclature originally used to define the domain structure of GTPases, we have renamed the conserved threonine in Dnm1p “G2,” and the more carboxy-terminal elements G3 and G4 (see Fig. 9 A).
An earlier study suggested that endosomal trafficking was delayed two- to threefold in cells lacking the DNM1 gene (Gammie et al., 1995). Although we observed mitochondrial morphology defects in the Gammie et al. (1995) dnm1Δ mutant, we did not detect endocytosis defects in our dnm1Δ strain. The different endocytic phenotypes observed for these two dnm1Δ mutants may reflect differences in the strain backgrounds or assays used by the two labs. It is also formally possible that the Dnm1 protein has dual roles in endosomal trafficking and mitochondrial morphology maintenance. Alternatively, endosomal trafficking defects in the Gammie et al. (1995) strain may be a secondary consequence of defects in mitochondrial morphology. In this study, we showed that mitochondrial morphology is wild type in five mutants blocked at distinct steps in the endocytic pathway. These data indicate that defects in endocytosis do not lead to secondary defects in mitochondrial membrane shape or distribution and provide further support that the mitochondrial morphology defects observed in dnm1 mutant cells are a direct result of the loss of Dnm1 protein function.
Exactly how the Dnm1 GTPase acts to control mitochondrial morphology and distribution in cells is not yet clear. It is possible that assembly of Dnm1p-containing structures at specific sites on the mitochondrial double membrane is responsible for initiating or regulating the formation of mitochondrial tubules or branches. Our TEM analysis of dnm1 mitochondria suggests that some tubular and branched membranes are still present in these organelles. Thus, while Dnm1p could play a role in generating mitochondrial tubules and branches, it is probably not the only molecule required for this process. An alternative possibility is that Dnm1p-containing structures are required to spread out and anchor portions of the mitochondrial network at the cell periphery. This latter model is consistent with the localization of Dnm1p to discrete patches at the cell cortex, the colocalization of these Dnm1p patches with the mitochondrial network, and the observation that the peripheral mitochondrial reticulum collapses to a restricted region of the cell cortex when Dnm1p is absent. Finally, it is also possible that, like mammalian dynamin, the Dnm1p GTPase plays a role in regulating membrane scission events at discrete positions on the mitochondrial reticulum. These models are not mutually exclusive, and some combination of these events could be simultaneously regulated by Dnm1p in vivo. Regardless of the mechanism of Dnm1p function, our results indicate that this protein plays a central role in controlling the shape and distribution of mitochondrial membranes.
A human gene closely related to Dnm1p was recently identified and termed variously DVLP (Dnm1p/Vps1p-like protein; Shin et al., 1997), Dymple (dynamin family member proline-rich carboxy-terminal domain less; Kamimoto et al., 1998), DLP1 (dynamin-like protein 1; Yoon et al., 1998), and DRP1 (dynamin-related protein 1; see related article by Smirnova et al., 1998; Imoto et al., 1998). Interestingly, the transient expression of Drp1 mutant proteins with altered GTP-binding domains causes aggregation of mitochondrial tubules in COS-7 cells but does not affect the morphology of other organelles or the transport of proteins through the secretory and endocytic pathways (see related article by Smirnova et al., 1998). While it is not yet clear whether mammalian Drp1 is a functional homologue of S. cerevisiae Dnm1p, the ability of mutant Drp1 to induce mitochondrial phenotypes in mammalian cells raises the intriguing possibility that Dnm1p-like GTPases play an evolutionarily conserved role in regulating mitochondrial distribution and morphology.
Dnm1p joins a growing family of dynamin-related proteins that localize to different organelles in mammals (Henley and McNiven, 1996; Jones et al., 1998; Yoon et al., 1998), yeast (Rothman et al., 1990; Jones and Fangman, 1992; Guan et al., 1993), and plants (Gu and Verma, 1996; Park et al., 1998). Future studies should reveal the role these different GTPases play in remodeling cellular membranes.
Acknowledgments
We are grateful to E. Bensen, S. Emr, A. Gammie, B. Glick, T. Graham, R. Jensen, J. Nunnari, G. Payne, H. Riezman, M. Rose, O. Rossanese, R. Schekman, B. Schneider, D. Stillman, and B. Wendland for providing strains, plasmids, and antibodies. We thank N. Fukushima for generating the HIS-tagged DNM1 construct used for affinity purification of the anti– GST-Dnm1412–757 antibody, E. King for technical assistance with the confocal microscopy, S. Perry for performing the original mdm29 × trp1 cross, and the staff of the Research Microscopy Facility (K.H. Albertine and N.B. Chandler) at the University of Utah Health Sciences Center for assistance with the TEM analysis. We also thank A.M. van der Bliek at the University of California, Los Angeles for sharing unpublished results and for many interesting discussions and G. Payne and members of the Shaw lab for their careful review of the manuscript.
This work was supported by grants from the American Cancer Society (CB-97) and the National Institutes of Health (NIH) (GM53466) awarded to J.M. Shaw, an NIH Predoctoral Genetics Training Grant (5 T32 GM07464) awarded to G.J. Hermann, a research grant from the Primary Children's Foundation at the University of Utah (PID 9802038) awarded to E. Brisch, and grants from the Hughes Undergraduate Research Program (Biology Department, University of Utah) awarded to W. Bleazard. The University of Utah Research Microscopy Facility is supported by a grant from the NIH (S10-RR-10489). The University of Utah Health Sciences Sequencing Facility is supported by a National Cancer Institute Grant (5-P30CA42014).
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole
- DiOC6
3,3′ dihexyloxacarbocyanine
- 5-FOA
5-fluoro-orotic acid
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- HA
hemagglutinin
- mmm
mitochondrial morphology maintenance
- mdm
mitochondrial distribution and morphology
Footnotes
D. Otsuga and B.R. Keegan contributed equally to this work.
Address all correspondence to Janet M. Shaw, Department of Biology, University of Utah, Salt Lake City, UT 84112. Tel.: (801) 585-6205. Fax: (801) 581-4668. E-mail: shaw@bioscience.utah.edu
References
- Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 raseffector binding domain. Science. 1988;240:518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO (Eur Mol Biol Organ) J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakeeva LE, Chentsov YS, Skulachev VP. Mitochondrial framework (reticulum mitochondriale) in rat diaphragm muscle. Biochim Biophys Acta. 1978;501:349–369. doi: 10.1016/0005-2728(78)90104-4. [DOI] [PubMed] [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. The END3gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I, Vojtov N, Karmon S, Pon LA. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol. 1998;141:1371–1381. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cales C, Hancock JF, Marshall CJ, Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the rasgene product. Nature. 1988;332:548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- Chen MS, Obar RA, Schroeder CC, Austin TW, Poodry CA, Wadsworth SC, Vallee RB. Multiple forms of dynamin are encoded by shibire, a Drosophilagene involved in endocytosis. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, van der Bliek AM, Schmid SL. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- De Camilli P, Takei K, McPherson PS. The function of dynamin in endocytosis. Curr Opin Neurobiol. 1995;5:559–565. doi: 10.1016/0959-4388(95)80059-x. [DOI] [PubMed] [Google Scholar]
- Feig LA, Cooper GM. Inhibition of NIH 3T3 cell proliferation by a mutant rasprotein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, M.T. 1993. Spermatogenesis. In The Development of Drosophila melanogaster. M. Bate and A. Martinez-Arias, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 71–147.
- Gammie AE, Kurihara LJ, Vallee RB, Rose MD. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Verma DPS. Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell. 1997;9:157–169. doi: 10.1105/tpc.9.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Farh L, Marshall TK, Deschenes RJ. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1gene. Curr Genet. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Henley JR, McNiven MA. Association of a dynamin-like protein with the Golgi apparatus in mammalian cells. J Cell Biol. 1996;133:761–775. doi: 10.1083/jcb.133.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Shaw JM. Mitochondrial dynamics in yeast. Annu Rev Cell Dev Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher J, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits JS, Burgess CC, Obar RA, Vallee RB. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Imoto M, Tachibana I, Urrutia R. Identification and functional characterization of a novel human protein highly related to the yeast dynamin-like GTPase Vps1p. J Cell Sci. 1998;111:1341–1349. doi: 10.1242/jcs.111.10.1341. [DOI] [PubMed] [Google Scholar]
- Jones BA, Fangman WL. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- Jones SM, Howell KE, Henley JR, Cao H, McNiven MA. Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science. 1988;279:573–577. doi: 10.1126/science.279.5350.573. [DOI] [PubMed] [Google Scholar]
- Kamimoto T, Nagai Y, Onogi H, Muro Y, Wakabayashi T, Hagiwara M. Dymple, a novel dynamin-like high molecular weight GTPase lacking a proline-rich carboxyl terminal domain in mammalian cells. J Biol Chem. 1998;273:1044–1051. doi: 10.1074/jbc.273.2.1044. [DOI] [PubMed] [Google Scholar]
- Lazzarino DA, Boldogh I, Smith MG, Rosand J, Pon LA. Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol Biol Cell. 1994;5:807–818. doi: 10.1091/mbc.5.7.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Robinson PJ. Dynamin and endocytosis. Endocrine Rev. 1995;16:590–607. doi: 10.1210/edrv-16-5-590. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J Cell Biol. 1992;118:385–395. doi: 10.1083/jcb.118.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H+-ATPase defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 angstrom resolution: implications for the mechanism of GTP hydrolysis. EMBO (Eur Mol Biol Organ) J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Cho JH, Kang SG, Jang HJ, Pih KT, Piao HL, Cho MJ, Hwang I. A dynamin-like protein in Arabidopsis thalianais involved in biogenesis of thylakoid membranes. EMBO (Eur Mol Biol Organ) J. 1998;17:859–867. doi: 10.1093/emboj/17.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poodry CA, Edgar L. Reversible alterations in the neuromuscular junctions of Drosophila melanogaster bearing a temperature-sensitive mutation, shibire. . J Cell Biol. 1979;81:520–527. doi: 10.1083/jcb.81.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in the receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. . J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Graham TR, Emr SD. A putative zinc finger protein, Saccharomyces cerevisiaeVps18p, affects late Golgi functions required for vacuolar protein sorting and efficient α-factor prohormone maturation. Mol Cell Biol. 1991;11:5813–5824. doi: 10.1128/mcb.11.12.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AD, Hermann GJ, Keegan BR, Thatcher SA, Shaw JM. Mitochondrial inheritance is delayed in Saccharomyces cerevisiae cells lacking the serine/threonine phosphatase, PTC1. . Mol Biol Cell. 1998;9:917–930. doi: 10.1091/mbc.9.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AD, Shaw JM. Vacuole partitioning during meiotic division in yeast. Genetics. 1996;144:445–458. doi: 10.1093/genetics/144.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Raymond CK, Gilbert T, O'Hara PJ, Stevens TH. A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell. 1990;61:1063–1074. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- Rose AB, Broach JR. Propagation and expression of cloned genes in yeast: 2 micron circle based vectors. Methods Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiaegenomic plasmid bank based on a centromere-containing shuttle vector. Gene (Amst) 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Schmid SL. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. . Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Séron K, Tieaho V, Prescianotto-Baschong C, Aust T, Blondel M-O, Guillaud P, Devilliers G, Rossanese OW, Glick BS, Riezman H, et al. A yeast t-SNARE involved in endocytosis. Mol Biol Cell. 1998;9:2878–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO (Eur Mol Biol Organ) J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY. 186 pp.
- Shin H-W, Shinotsuka C, Torii S, Murakami K, Nakayama K. Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J Biochem. 1997;122:525–530. doi: 10.1093/oxfordjournals.jbchem.a021784. [DOI] [PubMed] [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Huang K, Lemmon SK. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTPγS in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family of related GTPases? . Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibiregene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Vowels JJ, Payne GS. A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO (Eur Mol Biol Organ) J. 1998;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Goud B, Ruohola H, Novick PJ. Fractionation of yeast organelles. Methods Cell Biol. 1989;31:335–354. doi: 10.1016/s0091-679x(08)61618-0. [DOI] [PubMed] [Google Scholar]
- Warnock DE, Hinshaw JE, Schmid SL. Dynamin self-assembly stimulates its GTPase activity. J Biol Chem. 1996;271:22310–22314. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]
- Warnock DE, Schmid SL. Dynamin GTPase, a force generating molecular switch. Bioessays. 1996;18:885–893. doi: 10.1002/bies.950181107. [DOI] [PubMed] [Google Scholar]
- Wendland B, McCaffery MJ, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiaestrains that are isogenic to S228C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, Dahan S, McNiven MA. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J Cell Biol. 1998;140:779–793. doi: 10.1083/jcb.140.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. . Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]