Abstract
We showed previously that protein kinase C, which is required to maintain cell integrity, negatively regulates cell fusion (Philips, J., and I. Herskowitz. 1997. J. Cell Biol. 138:961–974). To identify additional genes involved in cell fusion, we looked for genes whose overexpression relieved the defect caused by activated alleles of Pkc1p. This strategy led to the identification of a novel gene, KEL1, which encodes a protein composed of two domains, one containing six kelch repeats, a motif initially described in the Drosophila protein Kelch (Xue, F., and L. Cooley. 1993. Cell. 72:681– 693), and another domain predicted to form coiled coils. Overexpression of KEL1 also suppressed the defect in cell fusion of spa2Δ and fps1Δ mutants. KEL2, which corresponds to ORF YGR238c, encodes a protein highly similar to Kel1p. Its overexpression also suppressed the mating defect associated with activated Pkc1p. Mutants lacking KEL1 exhibited a moderate defect in cell fusion that was exacerbated by activated alleles of Pkc1p or loss of FUS1, FUS2, or FPS1, but not by loss of SPA2. kel1Δ mutants form cells that are elongated and heterogeneous in shape, indicating that Kel1p is also required for proper morphology during vegetative growth. In contrast, kel2Δ mutants were not impaired in cell fusion or morphology. Both Kel1p and Kel2p localized to the site where cell fusion occurs during mating and to regions of polarized growth during vegetative growth. Coimmunoprecipitation and two-hybrid analyses indicated that Kel1p and Kel2p physically interact. We conclude that Kel1p has a role in cell morphogenesis and cell fusion and may antagonize the Pkc1p pathway.
Keywords: Pkc1, kelch, cell fusion, yeast mating, morphology
Cell fusion occurs during a variety of biological processes—for example, in sperm–egg fusion during fertilization, in myoblast fusion during myotube formation, and in microorganisms such as yeast during mating. In yeast, this process is initiated when haploid cells respond to the peptide pheromone secreted by cells of the opposite mating type (a cells responding to α-factor, and α cells to a-factor). The pheromones bind a seven-transmembrane receptor, stimulating a signal transduction cascade that results in cell cycle arrest, transcriptional induction of genes required for mating and cell fusion (Trueheart et al., 1987; McCaffrey et al., 1987; Elion et al., 1995), and a morphological response called shmoo formation (for reviews see Bardwell et al., 1994; Herskowitz, 1995). Cells polarize their actin cytoskeleton towards their mating partner by detecting a pheromone gradient (Jackson and Hartwell, 1990; Madden and Snyder, 1992; Segall, 1993), resulting in polarized protein deposition to the region of future cell contact (Trueheart et al., 1987; Elion et al., 1995).
Once the mating partners come into contact, a series of events, poorly understood at the molecular level, must be executed successfully to produce a diploid zygote. First, the cell wall must be removed in the region separating the two cells. Since inappropriate removal of cell wall material could result in lysis, correct spatial and temporal regulation of this event is critical for maintaining cell integrity. Hence, cell wall degradation does not occur until a cell comes into contact with its mating partner. The cell wall is then degraded beginning in the center of the region of contact and proceeding towards the edges (Osumi et al., 1974; Gammie et al., 1998). Cell wall degradation normally occurs quickly, and thus few cells are observed that have adhered but failed to fuse (Trueheart et al., 1987). Once cell wall material is removed, plasma membranes fuse, and then intracellular organelles such as nuclei and mitochondria fuse to produce an a/α diploid zygote.
Our previous work demonstrated that Pkc1p and Fps1p regulate cell fusion (Philips and Herskowitz, 1997). Mutants defective in the FPS1 gene, predicted to encode a glycerol transporter (Luyten et al., 1995), accumulate intracellular glycerol and exhibit a defect in cell fusion (Luyten et al., 1995; Philips and Herskowitz, 1997). The fusion defect can be alleviated if intracellular glycerol concentration is restored to wild-type levels or if 1 M sorbitol is provided to osmotically stabilize the cells, suggesting that osmotic imbalance is the cause of the defect in cell fusion. Since the Pkc1p pathway responds to conditions that threaten cell integrity such as hypoosmotic shock (Davenport et al., 1995; Kamada et al., 1995), we proposed that Pkc1p inhibits cell fusion if cells are not osmotically balanced, a situation that makes cell fusion particularly dangerous (Philips and Herskowitz, 1997). Supporting the idea that Pkc1p negatively regulates cell fusion, cells expressing an activated allele of PKC1 (PKC1-R398P) exhibit a defect in cell fusion (Philips and Herskowitz, 1997). PKC1-R398P alters the pseudosubstrate binding site of Pkc1p, creating a dominant, activated allele (Nonaka et al., 1995). Pkc1p is proposed to function upstream of an MAP kinase module composed of the MEKK, Bck1p/ Slk1p (Costigan et al., 1992; Lee and Levin, 1992), two redundant MEKs, Mkk1p and Mkk2p (Irie et al., 1993), and the MAPK Mpk1p/Slt2p (Torres et al., 1991; Mazzoni et al., 1993; reviewed by Errede and Levin, 1993). Consistent with the idea that the Pkc1p pathway may inhibit cell wall degradation during mating, the pathway is activated in response to pheromone (Errede et al., 1995; Zarzov et al., 1996; Buehrer and Errede, 1997). Furthermore, mutants defective in MPK1 lyse in response to pheromone (Errede et al., 1995). These observations implicate Pkc1p in the regulation of cell fusion, but the nature of the fusion machinery and how it is controlled is unclear.
Numerous mutants have been identified that are defective in cell fusion (for review see Marsh and Rose, 1997). When such mutants mate, they adhere but fail to fuse, producing dumbbell-shaped structures called prezygotes. In prezygotes, the cell wall persists between mating partners, presenting a barrier to plasma membrane fusion, cytoplasmic mixing, and nuclear fusion. Proteins required for cell fusion fall into three classes. The first class is comprised of Fus1p and Fus2p, whose synthesis is highly induced by pheromone and seems to be specifically required for fusion (Trueheart et al., 1987; McCaffrey et al., 1987; Elion et al., 1995). fus1 fus2 double mutants are mildly compromised in mating to a wild-type strain but are severely defective in mating to fus1 or fus2 strains (Trueheart et al., 1987). Fig1p and Fig2p are also highly induced by pheromone, suggesting that they should be categorized with Fus1p and Fus2p, although they are also required for normal morphology during mating (Erdman et al., 1998). The other two classes include proteins that have functions during mating or vegetative growth in addition to their role in cell fusion. Axl1p, Ram1p, and Ste6p affect cell fusion in a but not in α cells (Elia and Marsh, 1996; Brizzio et al., 1996; Dorer et al., 1997). They were originally identified for their role in a-factor production and mating (Adames et al., 1995; Powers et al., 1986; Kuchler et al., 1989) and later shown to affect cell fusion. The third class of proteins is required in both cell types for fusion, and is also needed for processes in addition to cell fusion and mating. This class includes Rvs161p, Fps1p, Spa2p, Pea2p, Bni1p, and Chs5p (Brizzio et al., 1998; Philips and Herskowitz, 1997; Dorer et al., 1997; Gammie et al., 1998; Santos et al., 1997). These proteins regulate morphogenesis and cell integrity, which may be modulated during mating to bring about cell fusion. Rvs161p is required to stabilize Fus2p during mating, a function that is distinct from its role in actin organization and endocytosis (Brizzio et al., 1998).
Here we describe a novel protein, Kel1p, which is a member of this final class of fusion proteins. We identified Kel1p because its overexpression suppressed the mating defect associated with an activated allele of Pkc1p. Overexpression of Kel1p also suppressed the mating defect of fps1Δ and spa2Δ mutants but not fus1Δ or fus2Δ mutants. Kel1p localizes to regions of polarized growth during mating and vegetative growth, and mutants lacking Kel1p exhibit defects in cell fusion and morphology.
Materials and Methods
Yeast Strains and Media
Yeast strains are described in Table I. Standard yeast growth conditions and genetic manipulations are described in Rose et al. (1990). Cells were grown at 30°C in YEPD medium unless otherwise noted. DNA manipulations were performed as in Sambrook et al. (1989).
Table I.
Yeast Strains and Plasmids
| Strains | Genotype | Source | ||
|---|---|---|---|---|
| IH2350* | MATα ura3-52 his4-34 trp1Δ1 | IH collection | ||
| IH2351* | MATα ura3-52 trp1Δ1 fus1Δ1 fus2Δ3 | IH collection | ||
| IH1783** | MAT a | IH collection | ||
| IH3077** | MATa mpk1Δ::TRP1 | IH collection | ||
| IH3196 | MATa leu2Δ1 | Philips and Herskowitz (1997) | ||
| IH3197 | MATα leu2Δ1 | IH collection | ||
| IH3204 | MATa spa2Δ::TRP1 trp1Δ99 leu2Δ1 | IH collection | ||
| JP52 | MATa fus1Δ::TRP1 trp1Δ99 | Philips and Herskowitz (1997) | ||
| JP54 | MATa/MATα | This study | ||
| JP147 | MATa fps1Δ::URA3 leu2Δ1 | Philips and Herskowitz (1997) | ||
| JP317 | MATa PKC1::PKC1-R398P-URA3 leu2Δ1 | Philips and Herskowitz (1997) | ||
| JP338 | MATa ura3::URA3 leu2Δ1 | This study | ||
| JP358 | MATa kel1Δ::hisG-URA3-hisG leu2Δ1 | This study | ||
| JP363 | MATa kel1Δ::hisG leu2Δ1 | This study | ||
| JP371 | MATa kel2Δ::LEU2 leu2Δ1 | This study | ||
| JP385 | MATa kel1Δ::hisG kel2Δ::LEU2 leu2Δ1 | This study | ||
| JP410 | MATa kel1Δ::hisG-URA3-hisG fus1Δ::TRP1 trp1Δ99 | This study | ||
| JP412 | MATa kel2Δ::hisG-URA3-hisG fus1Δ::TRP1 trp1Δ99 | This study | ||
| JP414 | MATa fps1Δ::URA3 kel1Δ::hisG leu2Δ1 | This study | ||
| JP415 | MATa fps1Δ::URA3 kel2Δ::LEU2 leu2Δ1 | This study | ||
| JP416 | MATa fus2Δ::URA3 kel2Δ::LEU2 leu2Δ1 | This study | ||
| JP417 | MATa fus2Δ::URA3 kel1Δ::hisG leu2Δ1 | This study | ||
| JP418 | MATa fus2Δ::URA3 leu2Δ1 | This study | ||
| JP445A** | MATα kel1Δ::hisG-URA3-hisG | This study | ||
| JP446A** | MATα kel2Δ::hisG-URA3-hisG | This study | ||
| JP490** | MATa mpk1Δ::TRP1 kel1Δ::hisG-URA3-hisG | This study | ||
| JP491** | MATα mpk1Δ::TRP1 kel2Δ::hisG-URA3-hisG | This study | ||
| JP500 | MATa fps1Δ::URA3 kel1Δ::hisG kel2Δ::LEU2 leu2Δ1 | This study | ||
| JP502 | MATa spa2Δ::TRP1 kel1Δ::hisG-URA3-hisG trp1Δ99 leu2Δ1 | This study | ||
| Plasmid name | Description | |||
| pJP2 | fus1Δ::TRP1 | Philips and Herskowitz (1997) | ||
| pJP52 | fps1Δ::URA3 | Philips and Herskowitz (1997) | ||
| pJP67 (YCp50-DS1) | YCp50-PKC1-R398P | Nonaka et al. (1995) | ||
| pJP72 | pRS306-PKC1-R398P | Philips and Herskowitz (1997) | ||
| pJP81 | YEp351-KEL1 | This study | ||
| pJP92 | YEp351-KEL2 | This study | ||
| pJP94 | kel1Δ::hisG-URA3-hisG | This study | ||
| pJP113 | kel2Δ::LEU2 | This study | ||
| pJP123 | YEp351-KEL2 | This study | ||
| pJP126 | YEp351-KEL2-GFP | This study | ||
| pJP127 | YEp351-KEL1 | This study | ||
| pJP129 | YEp351-KEL1-GFP | This study | ||
| pJP131 | YEp351-KEL2-HA | This study | ||
| pJP138 | kel2Δ::hisG-URA3-hisG | This study | ||
| pJP139 | YIp5-KEL1-GFP | This study | ||
| pJP143 | YIp5-KEL1 | This study | ||
| pJP158 | pEG202-KEL2-DBD | This study | ||
| pJP160 | pRS426-GAL-KEL2 | This study | ||
| pJP167 | pJG4-5-KEL1-AD | This study | ||
| pJP168 | pJG4-5-KEL2-AD | This study | ||
| pJP202 | YEp351-KEL1-HA | This study | ||
| pJP207 | YCplac111-KEL1 | This study | ||
| pJP209 | YCplac111-KEL1-HA | This study | ||
| pJW192 | RAS2-GFP | J. Whistler | ||
| pKOFUS2 | fus2Δ::URA3 | Philips and Herskowitz (1997) | ||
| pRFHM1 | Bicoid-DBD | Gyuris et al. (1993) | ||
| pSH18-34 | LexAop-LacZ | Gyuris et al. (1993) | ||
| pRS306 | ||||
| YCplac111 | ||||
| YEp351 | ||||
| YIp5 | ||||
| pJG4-5 | ||||
| pRS426 |
pRS306, pRS426, YEp351, and YIp5 are described in Guthrie and Fink (1991). pEG202 and pJG4-5 are described in Gyuris et al. (1993). YCplac111 is described in Gietz and Sugino (1988).
Isogenic to IH2350.
Isogenic derivatives in the EG123 strain background, whose full genotype is trp1 ura3 his4 leu2 can1. All other strains are isogenic derivatives of IH3160, whose full genotype is MATa ade2-101 ura3-52 met1-1 HMLa HMRa.
Yeast Plasmids and Transformations
Plasmids are listed in Table I. pJP72 is a pRS306-derived plasmid containing the PKC1-R398P allele as described in Nonaka et al. (1995) (Philips and Herskowitz, 1997). This plasmid was used to integrate PKC1-R398P at its genomic locus, generating strain JP317. Control strain JP338 was constructed by integrating pRS306 at ura3. Yeast transformations were performed by the lithium acetate method (Ito et al., 1983). pJW192 codes for a RAS2-GFP fusion protein under control of the GPD1 promoter (kindly provided by J. Whistler, University of California, Berkeley; Philips and Herskowitz, 1997).
Cloning of KEL1 and KEL2
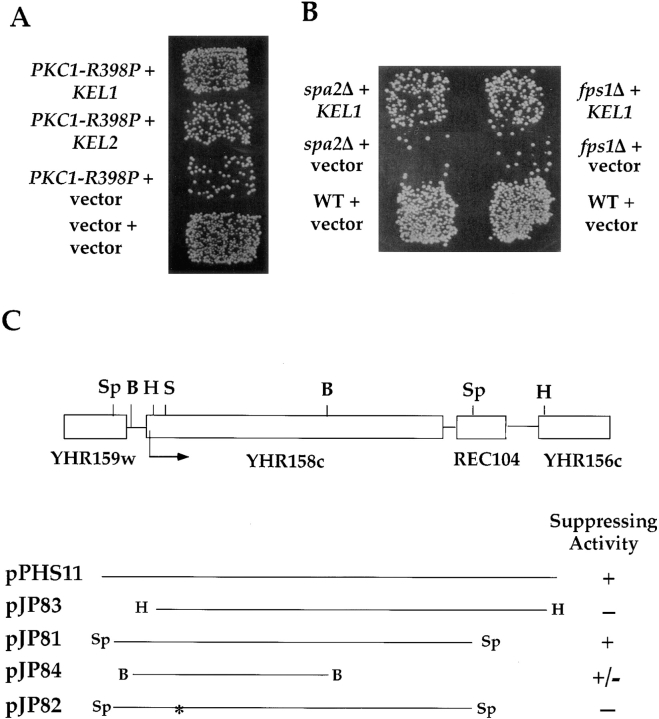
KEL1 was cloned by suppressing the mating defect of JP317. A plasmid (pPHS11) containing an ∼6-kb insert was isolated from a 2μ library (Nasmyth; see Fig. 1 C). A HindIII–HindIII fragment from pPHS11 was subcloned into the HindIII site of YEp351 to generate pJP83. An SphI– SphI fragment from pPHS11 was subcloned into the SphI site of YEp351 to generate pJP81. A BamHI–BamHI fragment from pPHS11 was subcloned into the BamHI site of YEp351 to generate pJP84. pJP81, which contains YHR158c, was indistinguishable from pPHS11 in its ability to suppress the mating defect of JP317 (data not shown). To confirm that the ORF responsible for suppression was YHR158c, YHR158c was disrupted by digesting pJP81 with SpeI, treating with Escherichia coli DNA polymerase I Klenow fragment and deoxynucleoside triphosphates, and ligating, to generate pJP82. pJP82 failed to suppress the mating defect exhibited by JP317. Additionally, a construct that contains approximately half of the YHR158c sequence (pJP84) partially suppressed the mating defect JP317, whereas a construct that removes the first 89 nucleotides of YHR158c (pJP83) failed to suppress the mating defect (Fig. 1 C).
Figure 1.
Identification of KEL1 and KEL2 as suppressors of PKC1-R398P. (A) High-copy KEL1 and KEL2 suppress the mating defect associated with PKC1-R398P. JP317 (MATa PKC1-R398P) carrying 2μ plasmids containing KEL1 (pJP81), KEL2 (pJP92), or vector (YEp351) were mated to a MATα fus1 fus2 strain (IH2351) as described in Materials and Methods. The wild-type control (JP338) harbored vector YEp351. (B) High-copy KEL1 suppresses the mating defect of spa2Δ and fps1Δ mutants. spa2Δ (IH3204) and fps1Δ (JP147) mutants harboring 2μ plasmids containing KEL1 (pJP81) or vector (YEp351) were mated to MATα fus1 fus2 strain (IH2351) as described in Materials and Methods. The wild-type strain (IH3196) contained vector (YEp351). (C) Restriction map and subcloning analysis of pPHS11. pPHS11 is the original plasmid obtained from the high-copy suppressor screen. pJP81-84 are YEp351-derived plasmids carrying the indicated segments. pJP82 was generated by digesting pJP81 with SpeI, filling in with Klenow (*), and religating to destroy the reading frame of YHR158c. The ability of each plasmid to suppress the mating defect of JP317 is indicated on the right: +, suppression; −, no suppression; ±, partial suppression. Restriction enzymes: S, SpeI; B, BamHI; H, HindIII; Sp, SphI. Additional information is provided in Materials and Methods and in the text.
Oligonucleotides OJP27 (5′-ACC TCT TGT AAC TAC TAC ATA CG) and OJP28 (5′-TCT TCT TGA CCT GAA CAT TGG) were used to amplify KEL2 from yeast genomic DNA by PCR. The amplified product was cut with PstI and cloned into the PstI site of YEp351 to generate pJP92.
Strain Construction
The KEL1 gene was deleted from yeast strains using pJP94, a construct that replaces KEL1 with hisG-URA3-hisG (Alani et al., 1987). This plasmid was constructed by ligating the SphI–SphI fragment containing KEL1 into the SphI site of pGEM11(F+). BglII sites were introduced at the start position and 31 nucleotides upstream of the stop codon of KEL1 by PCR. The BglII–BamHI fragment from pNKY51 (Alani et al., 1987) was ligated into the BglII sites to replace the KEL1 ORF with hisG-URA3-hisG, generating pJP94. kel1Δ strains were verified by PCR analysis.
kel2Δ deletion strains were generated using either pJP138 or pJP113. pJP113 replaces KEL2 with LEU2; pJP138 replaces KEL2 with hisG-URA3-hisG (Alani et al., 1987). These plasmids were generated by ligating a PstI–PstI fragment containing KEL2 into the PstI site of pBluescript. BglII sites were introduced 11 nucleotides upstream of the start site and 7 nucleotides downstream of the stop codon by PCR. A BamHI fragment containing LEU2 was ligated into the BglII sites to generate pJP113. A BglII–BamHI fragment containing hisG-URA3-hisG from pNKY51 was inserted into the BglII sites to generate pJP138. kel2Δ strains were confirmed by PCR analysis.
fus1Δ strains were constructed using pJP2 (Philips and Herskowitz, 1997) and confirmed by defective mating and PCR analysis. fus2Δ strains were constructed using pKOFUS2 (Philips and Herskowitz, 1997) and confirmed by mating ability and PCR analysis. fps1Δ strains were constructed using pJP52 (Philips and Herskowitz, 1997) and confirmed by PCR analysis.
Microscopic and Plate Mating Assays
Mating assays scored microscopically were performed as described in Philips and Herskowitz (1997). Prezygotes were defined as structures in which the nuclei of mating partners remained unfused, as evidenced by two distinct DAPI-staining structures, and in which a septum was visible between adherent mating partners. Percentage prezygotes was defined as prezygotes/(prezygotes + zygotes). At least 100 partnered cells (zygotes + prezygotes) were counted per sample. Numbers represent averages of at least three experiments. Assays in which one partner contained the RAS2-GFP plasmid were performed as described in Philips and Herskowitz (1997) except that cells were resuspended in Fluoromount G (Southern Biotechnology Associates, Inc., Birmingham, AL) before viewing. Mating assays scored on plates were performed as described in Philips and Herskowitz (1997) except that the spa2Δ and fps1Δ mutants were incubated on YEPD for ∼6 h before replica plating to selective medium.
Construction of Tagged Kel1p and Kel2p
GFP (S65T, F64L; Cormack et al., 1996) was fused to the carboxy terminus of both Kel1p and Kel2p. BglII sites were introduced just before the stop codon of KEL1 and KEL2 by oligonucleotide-mediated mutagenesis (Kunkel et al., 1987) using oligonucleotides OJP43 (5′-CAT TAC GCA TAT TGT CTT TTA AGA TCT ATC GCT GTC AGC ATC) for KEL1 and OJP46 (5′-AAC TAT ATA CTC TCG AAC AAA GAT CTT AGT CAT TGG AAG ACG) for KEL2. A SalI–SacII fragment of KEL1 that contained the introduced BglII site was cloned into the SalI–SacII sites of pJP81 to generate pJP127 (YEp351-KEL1). GFP was obtained on a BglII–BamHI fragment from pEBO413 (a gift from E. O'Shea, University of California, San Francisco) and ligated into the BglII site of pJP127 to generate pJP129 (YEp351-KEL1-GFP). To construct an integrating construct, the SphI–SphI fragment containing KEL1-GFP from pJP129 was inserted into the SphI site of YIp5 to generate pJP139 (YIp5-KEL1- GFP). The SphI–SphI fragment from pJP127 was cloned into the SphI site of YIp5 to generate pJP143 (YIp5-KEL1). pJP139 and pJP143 were used to integrate KEL1-GFP and untagged KEL1 at the KEL1 locus by digesting with XbaI before transformation. When integrated at the URA3 locus, Kel1p–GFP complemented the mating defect of kel1Δ mutants (data not shown). To construct an HA-tagged version of Kel1p, an oligonucleotide containing two copies of the HA epitope was inserted into pJP127 to generate pJP202 (YEp351-KEL1-HA). The SphI–SphI fragment from pJP202 containing KEL1-HA was cloned into the SphI site of YCplac111 to generate pJP209 (YCplac111-KEL1-HA). The SphI–SphI fragment from pJP127 containing the untagged version of KEL1 was ligated into the SphI site of YCplac111 to generate pJP207 (YCplac111-KEL1). For KEL2, the BglII–BamHI fragment from pEBO413 containing GFP was inserted into the BglII site generated in KEL2 by oligonucleotide-mediated mutagenesis (Kunkel et al., 1987). This fusion construct, expressed from the 2μ plasmid YEp351, was designated pJP126. To construct an HA-tagged version of Kel2p, an oligonucleotide containing two copies of the HA epitope was inserted into the BglII site to create pJP131. pJP123 encodes Kel2p on YEp351 lacking an epitope tag.
Microscopy
Localization of Kel1p was analyzed in strains harboring pJP139 or pJP143 grown in SD-Ura. Localization of Kel2p was determined using strains harboring pJP126 or pJP123 grown in SD-Leu. Cells were grown to mid-log phase, sonicated, pelleted in a microcentrifuge, and resuspended in Fluoromount G (Southern Biotechnology Associates, Inc.). Samples were viewed with a BX50 microscope at 100× (Olympus America, Inc., Melville, NY).
Morphology of wild-type (IH3196), kel1Δ (JP363), kel2Δ (JP371), and kel1Δ kel2Δ (JP385) strains was determined by growing cells to mid-log phase in synthetic complete medium. IH3196 harboring YEp351 or pJP81 was grown to mid-log phase in SD-Leu. IH3196 harboring pRS426 or pJP160 was grown to mid-log phase in SGal-Ura. Cells were sonicated, pelleted in a microcentrifuge, and resuspended in 50% glycerol before viewing with an Axioskop microscope at 100× (Carl Zeiss, Inc., Thornwood, NY).
Northern Analysis
RNA preparations and sample analysis were performed as described previously (Cross and Tinkelenberg, 1991). The probes used were DNA restriction fragments that were gel-purified and labeled by random-prime labeling using a Prime-it kit (Stratagene, La Jolla, CA). The fragments used were a 1.7-kb PstI–HincII fragment containing coding sequence for FUS1, a 1.7-kb HindIII–BamHI fragment containing coding sequence for YHR158c, and a 0.8-kb HpaI–SalI fragment for TCM1 (Schultz and Friesen, 1983).
Coimmunoprecipitation Analysis
Yeast cell extracts were prepared by growing cells overnight in SD-Ura Leu media. Cells were inoculated into YEPD and allowed to grow for approximately two generations to an OD600 of ∼0.4. 100-ml cultures were harvested, resuspended in 250 μl ice-cold lysis buffer (50 mM Tris-HCl pH 8.0, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 50 mM NaF, 80 mM β-glycerophosphate, 1 mM Na3VO4, and a cocktail of protease inhibitors (Boehringer Mannheim Corp., Indianapolis, IN) and mixed with an equal volume of glass beads. After vortexing vigorously ten times with 10-s pulses, samples were centrifuged at 14,000 rpm for 15 min in a microfuge at 4°C. The supernatants were removed, and lysis buffer was added to a final volume of 0.5 ml. Samples were centrifuged again for 15 min at 4°C. 50 μl of supernatant was removed for detecting Kel1p-GFP in the extract (data not shown). Immunoprecipitations were performed on the remaining sample at 4°C for 2 h by incubating the remaining supernatant with 25 μl of a 50% slurry of protein G-sepharose bound to anti-HA antibodies (12CA5) with rocking. Precipitates were washed four times with 0.5 ml cold lysis buffer, resuspended in 50 μl loading buffer, and boiled for 5 min before resolving by 7.5% SDS-PAGE. Immunoblot analysis was performed with anti-HA (HA11; Berkeley Antibody Co., Inc., Richmond, CA) or anti-GFP antibodies as described below.
Immunoblot Analysis
Extracts that were not used for coimmunoprecipitations were prepared as follows. Cells were grown to an OD600 of ∼0.3. 15 ml of culture was pelleted in a microfuge and incubated on ice for 5 min. Pellets were resuspended in 150 μl ice-cold solution 1 (1.85 N NaOH, 7.4% β-mercaptoethanol) and incubated on ice for 10 min. 150 μl ice-cold 50% TCA was added. Cells were incubated on ice for 10 min, followed by centrifugation for 2 min at 4°C. Pellets were washed with 1 ml ice-cold acetone. Samples were centrifuged for 2 min at 4°C. The pellet was resuspended in sample buffer, boiled for 5 min, and resolved by 7.5% SDS-PAGE.
SDS-PAGE was followed by electroblotting onto nitrocellulose filters using the Minigel system (Bio-Rad Laboratories, Hercules, CA). Blots were incubated in TBST (TBS with 0.1% Triton X-100) with 10% nonfat dry milk for ∼1 h. Blots were incubated with primary antibody (mAb HA11 diluted 1:10,000 [Berkeley Antibody Co., Inc.]; mAb anti-GFP [C163; a gift from P. O'Farrell, University of California, San Francisco, CA] diluted 1:2) in TBST supplemented with 2% nonfat dry milk at 4°C overnight. Blots were washed three times for 5 min in TBST and incubated with peroxidase-linked secondary antibody (Bio-Rad Laboratories) diluted 1:2,000 in TBST with 2% nonfat dry milk for 1 h. Blots were washed twice for 5 min in TBS supplemented with 0.3% Triton X-100, followed by three washes in TBST. Blots were developed using an enhanced chemiluminescence detection kit (Amersham Corp., Arlington Heights, IL).
Two-Hybrid Analysis
Two-hybrid plasmids pJP158 and pJP168 were constructed by introducing a BamHI site before the start codon of KEL2 by oligonucleotide-mediated mutagenesis (Kunkel et al., 1987) using oligonucleotide OJP50 (5′-GG CAG GCA ACC CGG ATC CTA GCT ATG GTA CCT). A BamHI–NcoI fragment containing KEL2 was cloned into pEG202 to create pJP158. A BamHI–NotI fragment containing KEL2 from pJP158 was ligated into the BamHI–NotI sites of pJG4-5 to generate pJP168. pJP167 was generated by introducing a SalI site before the start codon of KEL1 by oligonucleotide-mediated mutagenesis (Kunkel et al., 1987) using oligonucleotide OJP49 (5′-GCT GAA TCC AGC CAT GTT TGG TCG ACT TTC TAG GTG C). A SalI–SalI fragment containing KEL1 was ligated into the SalI sites of pJG4-5 to create pJP167.
Two-hybrid assays were performed as described (Gyuris et al., 1993). Yeast strain EGY48 containing the LexAop–LacZ reporter plasmid pSH18-34 was cotransformed with pEG202-based plasmids expressing LexA DNA-binding domain fusions and pJG4-5–based plasmids containing transcriptional activation domain fusions (Gyuris et al., 1993). β-galactosidase activities were measured essentially as described by Stern et al. (1984). Cultures were grown overnight in S raffinose-His Ura Trp. Expression of Kel1p-AD was induced with galactose for ∼1 h. Data are expressed in Miller units (OD420 × 1000)/(OD600 × min × ml) as determined from at least three independent transformants, tested in duplicate.
Analysis of mpk1Δ kel1Δ and mpk1Δ kel2Δ Strains
An mpk1Δ strain (IH3077) was crossed with two independent kel1Δ strains (JP445A and JP445B) generated by gene replacement or two independent kel2Δ strains (JP446A and JP446B) generated by gene replacement. Segregants were allowed to germinate on YEPD containing 1 M sorbitol at 30°C. Cells were streaked for single colonies on either YEPD or YEPD + 1 M sorbitol and grown for 2 d at 34.5°C.
Results
Overexpression of KEL1 Suppresses Mutants Defective in Cell Fusion
Cells expressing an activated allele of PKC1 (PKC1-R398P) exhibit impaired mating due to a defect in cell fusion (Philips and Herskowitz, 1997). Expression of this allele under control of its own promoter resulted in a mating defect that was easily detected when cells were mated to a fus1 fus2 mutant strain, which is also partially defective in cell fusion (Fig. 1 A). To identify additional genes important for cell fusion, we performed a high-copy suppressor screen for improved mating of cells expressing PKC1-R398P. Strain JP317 (PKC1-R398P) was transformed with a high-copy library, and ∼4,500 transformants were screened by replica plating for mating with a fus1 fus2 mutant strain. Four plasmids containing distinct inserts were identified that restored mating when retransformed. One of them, designated pPHS11, is described here.
Sequencing revealed that pPHS11 contained two full-length ORFs, YHR158c and REC104, and part of YHR159w and YHR156c. Subcloning demonstrated that YHR158c was responsible for suppression (see Materials and Methods and Fig. 1 C). Fig. 1 A shows that a 2μ plasmid containing YHR158c (pJP81) suppressed the mating defect of JP317. To ascertain whether overexpression of YHR158c suppressed the fusion defect of JP317, we examined the matings microscopically. We found that pJP81 reduced the number of prezygotes that accumulated in a strain expressing PKC1-R398P by >40% (Table II, lines 2 and 3). To determine whether pJP81 could suppress the mating defect of additional cell fusion mutants, we examined its effect on the mating ability of fus1Δ, fus2Δ, spa2Δ, and fps1Δ mutants. Similar to what we observed in the PKC1-R398P mutant, pJP81 partially restored mating and cell fusion to spa2Δ and fps1Δ mutants (Fig. 1 B and Table II). In contrast, there was no significant effect of pJP81 on fus1Δ and fus2Δ mutants (data not shown). We conclude that overexpression of YHR158c partially suppresses the mating defect exhibited by a subset of mutants defective in cell fusion.
Table II.
The Effect of KEL1 Overexpression on Cell Fusion in spa2Δ, fps1Δ, and PKC1-R398P Mutants
| Genotype | Plasmid | Prezygotes* | ||
|---|---|---|---|---|
| % | ||||
| WT | vector | 4.7 ± 1.1 | ||
| PKC1-R398P | vector | 51 ± 0.5 | ||
| PKC1-R398P | KEL1 | 29 ± 7 | ||
| WT | vector | 2.3 ± 0.4 | ||
| spa2Δ | vector | 42 ± 2 | ||
| spa2Δ | KEL1 | 25 ± 4 | ||
| fps1Δ | vector | 84 ± 3 | ||
| fps1Δ | KEL1 | 67 ± 3 |
For lines 1–3, strains were: WT (JP338) and PKC1-R398P (JP317). Strains were transformed with YEp351 containing KEL1 (pJP81) or YEp351 alone as indicated. For lines 4–8, strains were: WT (IH3196), spa2Δ (IH3204), and fps1Δ (JP147). For lines 1–3, strains were grown overnight in SD-Leu Ura. For lines 4–8, strains were grown overnight in SD-Leu. All strains were mated to a wild-type α strain (IH2350). At least 100 partnered cells were counted in each experiment to determine % prezygotes. Values are means of three experiments ± SD.
Percent prezygotes represents the number of prezygotes/(zygotes + prezygotes).
YHR158c is predicted to encode an 1164–amino acid protein. Analysis of the sequence revealed six internal repeats in the amino-terminal half of the protein followed by a domain predicted to form coiled coils in the carboxy-terminal half of the protein (Fig. 2 A). The repeats of ∼50 amino acids each belong to the kelch family (Xue and Cooley, 1993; Bork and Doolittle, 1994), named after the Drosophila protein in which these repeats were first identified. The proteins most similar to YHR158c are a protein predicted in the S. cerevisiae database (YGR238c), Tea1p of Schizosaccharomyces pombe (Mata and Nurse, 1997), and a protein predicted by the S. pombe sequencing project (accession No. Z98603). All four proteins have the same domain structure: six kelch repeats in the amino-terminal half of the protein followed by a predicted coiled-coil domain in the carboxy terminus. Because YHR158c and YGR238c contain kelch repeats, we have named the two genes KEL1 and KEL2, respectively. The two S. cerevisiae genes are located in duplicated regions of the S. cerevisiae genome (chromosomal block 29), between SPO12 and SOL3 for KEL1, and between YGR230w and SOL4 for KEL2 (Wolfe and Shields, 1998). Kel1p and Kel2p are 62% identical in the region containing kelch repeats and 44% identical over their entire length. An alignment of the kelch repeats in Kel1p, Kel2p, and Tea1p is shown in Fig. 2 B. Tea1p is slightly more similar to the two S. cerevisiae proteins than it is to the second S. pombe protein (see dendrogram in Fig. 2 C).
Figure 2.

Relationship of Kel1p, Kel2p, and Tea1p. (A) Domain structure of Kel1p, Kel2p, and Tea1p. Grey boxes indicate kelch repeats. Striped boxes indicate predicted coiled-coil domains. (B) Alignment of the kelch repeats in Kel1p, Kel2p, and Tea1p. Consensus residues are indicated in bold (residues present in at least three of the six repeats). Consensus sequence is listed underneath repeats in bold. # Indicates presence of amino acids excluded from the alignment. (C) Dendrogram indicating the relationship among Kel1p, Kel2p, Tea1p, and the protein predicted by the S. pombe sequencing project (Z98603). Dendrogram was generated using Pileup and Cluster analysis.
When expressed from a 2μ plasmid, Kel2p, like Kel1p, suppressed the mating defect associated with activated Pkc1p (Fig. 1 A); however, the suppression was weaker and more variable from experiment to experiment than that seen with Kel1p. The inability of Kel2p to suppress as well as Kel1p may be due to lower protein levels. Western blot analysis of both HA-tagged and GFP-tagged versions indicated that, when expressed from 2μ or CEN-ARS plasmids, Kel1p was more abundant than Kel2p (Fig. 3 D and data not shown).
Figure 3.
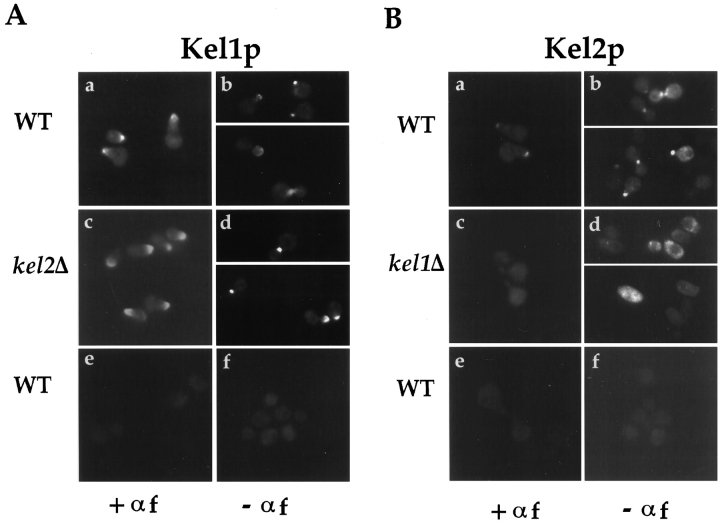
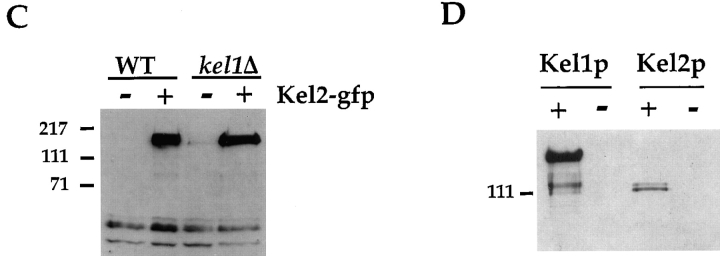
Localization of Kel1p and Kel2p. (A) An integrating plasmid containing Kel1p-GFP (pJP139) was used to localize Kel1p in wild-type cells (IH3196) (a and b) and in kel2Δ (JP370) cells (c and d). No signal was observed in wild-type cells expressing an untagged control plasmid (pJP143) (e and f). Cells in a, c, and e were treated with 25 μg/ml α-factor for ∼2 h. b, d, and f are composites from two different photographs. (B) YEp351 containing Kel2p-GFP (pJP126) was used to localize Kel2p in wild-type cells (a and b) and in kel1Δ cells (JP363; c and d). No signal was observed in wild-type cells expressing an untagged control plasmid (pJP123; e and f). Cells in a, c, and e were treated with 25 μg/ml α-factor for ∼2 h. b, d, and f are composites from two different photographs. (C) Kel2p is present in kel1Δ strains. Wild-type (IH3196) and kel1Δ (JP363) strains harboring plasmids encoding Kel2p-GFP (pJP126) or untagged control plasmid (pJP123) were analyzed by Western blot as described in Materials and Methods. Monoclonal antibodies recognizing GFP (C163) were a generous gift from P. O'Farrell. Molecular weight standards are indicated on the left. (D) Kel1p is more highly expressed than Kel2p. JP317 was transformed with YEp351-derived plasmids containing Kel1p-HA (pJP202), untagged Kel1p (pJP127), Kel2p-HA (pJP131), or untagged Kel2p (pJP123). +, HA-tagged versions; −, untagged versions. Extracts were prepared as described in Materials and Methods. Lanes were equally loaded, and Western blot analysis was performed with monoclonal antibodies recognizing the HA tag (HA11; Berkeley Antibody Co., Inc.). Molecular weight standards are indicated on the left.
Kel1p and Kel2p Localize to Regions of Polarized Growth
During mating, cells polarize the actin cytoskeleton and secretory apparatus towards their selected mating partner (Field and Schekman, 1980; Ford and Pringle, 1986; Hasek et al., 1987; Madden and Snyder, 1992; Read et al., 1992; Segall, 1993). Many proteins required for cell fusion, including Fus1p, Fus2p, and Spa2p, localize to the region of future cell contact (Trueheart et al., 1987; Elion et al., 1995; Gehrung and Snyder, 1990). To determine whether Kel1p is similarly localized, a fusion protein was constructed in which GFP was joined to the carboxy terminus of Kel1p (see Materials and Methods). Cells were treated with α-factor, and localization of Kel1p-GFP was examined by fluorescence microscopy. Kel1p-GFP expressed from 2μ, CEN-ARS, or integrating plasmids localized to shmoo tips (Fig. 3 A, a and data not shown). Focusing through sections of cells indicated that Kel1p-GFP was localized to the periphery of the shmoo tip. When overexpressed, Kel1p-GFP suppressed the fusion defect associated with PKC1-R398P. In this case, Kel1p-GFP remained localized to the shmoo tip, but the green fluorescent signal was more intense and more broadly localized than when the fusion construct was integrated (data not shown).
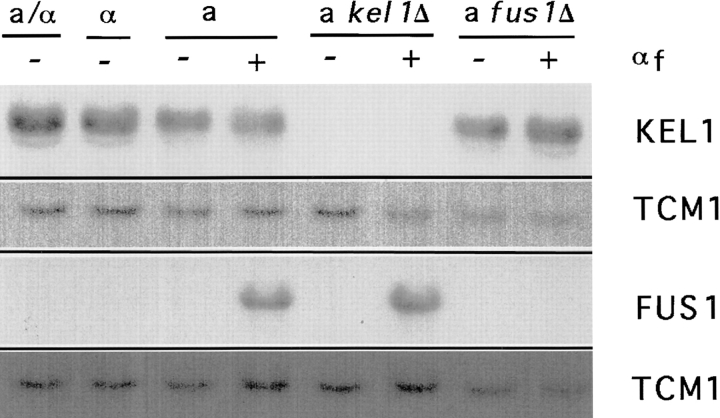
Northern blot analysis indicated that the KEL1 transcript was present in a, α, and a/α diploid cells, consistent with it having a function during vegetative growth, and was not induced by pheromone (Fig. 4). Hence, we examined its localization during vegetative growth. As with pheromone treatment, Kel1p-GFP localized to regions of polarized growth in vegetative cells (Fig. 3 A, b). Kel1p-GFP could be seen as a single spot in unbudded cells. In cells with small- or medium-sized buds, Kel1p localized to the bud tip. In large-budded cells, Kel1p-GFP was often localized to the neck region separating the mother and daughter cell.
Figure 4.
Northern analysis of KEL1 and FUS1 mRNA. mRNA was prepared as described in Materials and Methods from a /α wild-type (JP54), α wild-type (IH3197), a wild-type (IH3196), a kel1Δ (JP358), and a fus1Δ (JP52) strains after cells were treated with α-factor for 20 min (+, treated; −, untreated). Northern blots were hybridized with a probe to KEL1 (top) or FUS1 (third panel). Blots were stripped and hybridized with a probe to TCM1 to serve as a loading control.
To determine if Kel2p could function similarly to Kel1p, we examined its subcellular localization. GFP was fused to the carboxy terminus of Kel2p. We could only detect Kel2p-GFP signal when expressed from a 2μ plasmid, but not when expressed from a CEN-ARS plasmid. Like Kel1p-GFP, Kel2p-GFP localized to the shmoo tip in pheromone-treated cells (Fig. 3 B, a). Also, like Kel1p, Kel2p-GFP localized to regions of polarized growth during vegetative growth. Kel2p-GFP was found as a single spot in unbudded cells. In cells with small- or medium-sized buds, Kel2p-GFP localized to the bud tip. In large-budded cells, Kel2p-GFP was detected at the bud tip or at the neck between mother and bud (Fig. 3 B, b). We conclude that Kel1p and Kel2p localize to regions of polarized growth both during mating and budding.
Localization of Kel2p Depends Upon KEL1
Because Kel1p and Kel2p exhibit a similar pattern of localization, we determined whether their localization depends upon each other. In kel2Δ strains, localization of Kel1p was indistinguishable from that in wild-type strains (Fig. 3 A, c and d). However, Kel2p failed to localize in the absence of Kel1p both with and without pheromone treatment (Fig. 3 B, c and d). To determine whether Kel2p was present in the kel1Δ strain, we examined Kel2p-GFP by immunoblot and observed that the protein was present at equivalent levels in wild-type and kel1Δ strains (Fig. 3 C). We conclude that Kel1p is required to localize Kel2p to regions of polarized growth and that Kel2p is dispensable for Kel1p localization. Despite the considerable homology between Kel1p and Kel2p, Kel1p apparently interacts with a determinant at the site of polarized growth with which Kel2p fails to interact in the absence of Kel1p.
Kel1p and Kel2p Are Present in a Complex
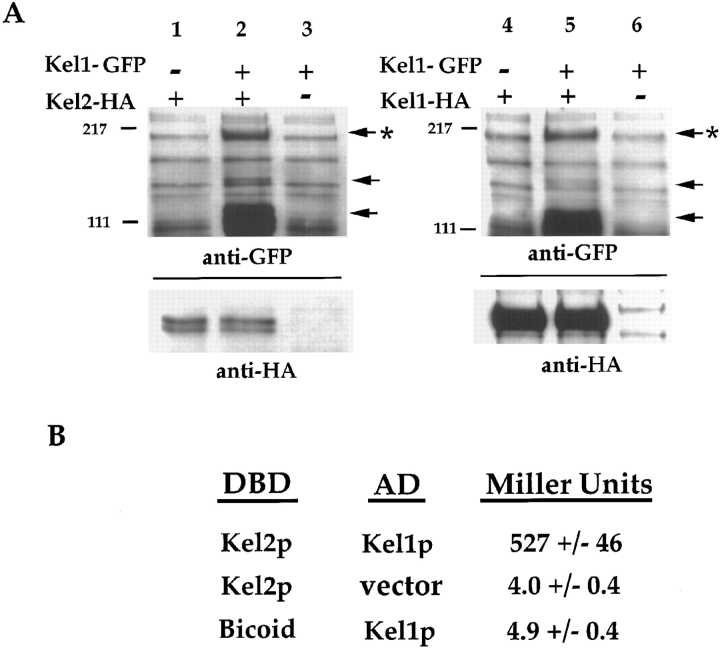
Since localization of Kel2p was dependent upon Kel1p, we wondered whether the two proteins physically interact. To address this question, coimmunoprecipitation and two-hybrid analyses were performed. An epitope-tagged version of Kel2p (Kel2p-HA) was constructed in which a double hemagglutinin (HA)1 tag was fused to the carboxy terminus of Kel2p as described in Materials and Methods. Kel2p-HA was expressed in a strain that harbored a GFP-tagged version of Kel1p or an untagged version. In cells expressing Kel2p-HA, we were able to immunoprecipitate Kel2p with anti-HA antibodies (Fig. 5 A, bottom panel, lanes 1 and 2). If the cells coexpressed Kel1p-GFP, Kel1p co-immunoprecipitated with Kel2p-HA as detected by immunoblot with anti-GFP antibodies (Fig. 5 A, top panel, lane 2). To determine whether Kel1p-GFP was specifically coimmunoprecipitated by Kel2p-HA, immunoprecipitations were performed from cells expressing the untagged version of Kel2p. We were unable to immunoprecipitate Kel1p-GFP with anti-HA antibodies in such extracts (Fig. 5 A, top panel, lane 3).
Figure 5.
Kel1p and Kel2p physically interact. (A) Coimmunoprecipitation of Kel2p and Kel1p. HA-tagged Kel1p or Kel2p was expressed in cells (JP363) that coexpressed Kel1p-GFP. HA-tagged proteins were immunoprecipitated as described in Materials and Methods, and the ability to coimmunoprecipitate Kel1p-GFP was assessed. In lanes 1–3, cells expressed Kel2p-HA (pJP131), indicated by +, or untagged Kel2p (pJP123) indicated by −. In lanes 4–6, cells expressed Kel1p-HA (pJP209), indicated by +, or untagged Kel1p (pJP207), indicated by −. Immunoprecipitated HA-tagged proteins were detected in the lower blots using antibodies that recognize the HA epitope (HA11; Berkeley Antibody Co., Inc.). Equal amounts of Kel2p-HA were immunoprecipitated in lanes 1 and 2, whereas no band was detected in extracts prepared from strains expressing the untagged control (lane 3). Equal amounts of Kel1p-HA were immunoprecipitated in lanes 4 and 5, whereas no band was detected in extracts prepared from strains expressing the untagged control (lane 6). Cells coexpressed Kel1p-GFP (pJP139), indicated by +, or untagged Kel1p (pJP143), indicated by −. Monoclonal antibodies recognizing GFP were used to detect coimmunoprecipitated Kel1p-GFP in the upper blots. Arrowhead with an asterisk indicates full-length Kel1p-GFP. Other arrowheads indicate breakdown products of Kel1p-GFP that also coimmunoprecipitate. Molecular weight standards are indicated on the left of each gel. (B) Two-hybrid analysis of Kel1p and Kel2p. Cells expressed Kel2p or Bicoid fused to the lexA DNA-binding domain (DBD; pJP158 and pRFHM1, respectively), and Kel1p fused to a transcriptional activation domain (AD; pJP167) or vector. Ability to activate transcription from a reporter construct was determined as described in Materials and Methods. Miller Units were determined as described in Materials and Methods. Values are means ± SD.
Similar experiments were performed to determine whether Kel1p can associate with itself. An HA-tagged version of Kel1p was expressed in a cell that also harbored an integrated copy of a GFP-tagged version of Kel1p. Kel1p-HA was immunoprecipitated using antibodies that recognize the HA tag. Kel1p-GFP coimmunoprecipitated as determined by Western blotting (Fig. 5 A, top panel, lane 5). In control experiments, we were unable to immunoprecipitate Kel1p-GFP if an untagged version of Kel1p rather than an HA-tagged version was coexpressed (Fig. 5 A, top panel, lane 6). These data indicate that, in addition to being able to detect Kel1p in a complex with Kel2p, we are able to detect Kel1p in a complex with itself.
Additional evidence that Kel1p and Kel2p physically interact was obtained by two-hybrid analysis. Full-length Kel2p was fused to the lexA DNA-binding domain (Kel2-DBD) to generate pJP158 (see Materials and Methods). On its own, this fusion protein did not activate transcription from a reporter construct in which lexA operator sites lie upstream of the lacZ gene (pSH18-34; Gyuris et al., 1993). Full-length Kel1p was fused to a transcriptional activation domain (Kel1-AD; see Materials and Methods) and expressed under control of the GAL1,10 promoter (pJP167). When Kel1-AD and Kel2-DBD were coexpressed, β-galactosidase activity increased more than 100-fold compared with cells that expressed either Kel1-AD or Kel2-DBD alone (Fig. 5 B). When cells were grown on glucose to repress expression of Kel1-AD, β-galactosidase was not produced (data not shown). When coexpressed with a Bicoid-lexA DNA-binding domain fusion (pRFHM1; Gyuris et al., 1993), Kel1-AD did not activate transcription of lacZ, indicating that interaction with the Kel2-DBD fusion is specific. To test whether Kel2p interacts with itself, Kel2-DBD and Kel2-AD constructs were examined in the two-hybrid system. When expression of Kel2-AD was induced by galactose for 1 h or less, no interaction was detectable. Longer inductions resulted in an interaction that was ∼10% that seen for Kel1p-AD and Kel2-DBD (data not shown). Based upon the coimmunoprecipitation and two-hybrid analyses, we conclude that Kel1p physically interacts with Kel2p and itself. Our data indicate, moreover, that the Kel1p–Kel2p interaction is significant in vivo, as localization of Kel2p depends upon Kel1p.
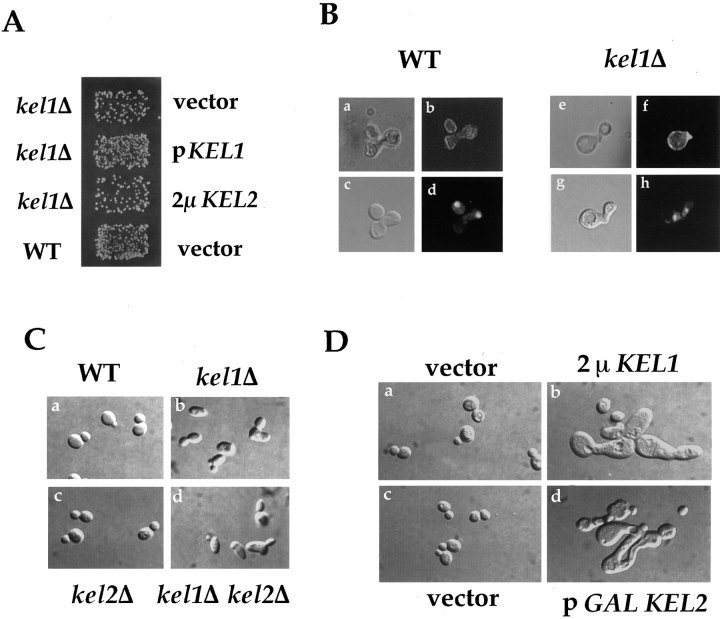
kel1Δ Mutants Are Defective in Cell Fusion
To determine whether KEL1 has a role in cell fusion, we examined the phenotype of a strain in which KEL1 was deleted. In a diploid strain, one copy of the KEL1 open reading frame was replaced with the URA3 gene (see Materials and Methods). Examination of haploid segregants indicated that deletion of KEL1 had no effect on growth rate, budding pattern (in kel1Δ haploids and in a/α kel1Δ/ kel1Δ diploids), or sensitivity to high and low temperatures. kel1Δ mutants did exhibit a mating defect (Fig. 6 A). This defect was not due to inability to produce or respond to pheromone normally, as determined by halo assays and shmoo formation (Fig. 3 B, c and data not shown). Additionally, kel1Δ mutants signaled in response to pheromone as evidenced by normal transcriptional induction of FUS1 (Fig. 4). kel1Δ mutants were examined microscopically to ascertain if prezygotes accumulated when they were mated to a wild-type strain. Prezygotes were scored as structures in which the nuclei of mating partners remained unfused, as evidenced by two distinct DAPI-staining structures, and in which a septum was visible between adherent mating partners. kel1Δ mutants did exhibit a defect in cell fusion, displaying a four- to fivefold increase in the number of prezygotes when compared with wild-type cells (Table III, lines 1 and 2; Table IV, lines 1 and 2). The defect was not exacerbated by mating to kel1Δ cells (data not shown). Similarly, the mating defect of fps1Δ mutants is not exacerbated by mating to fps1Δ cells (Philips and Herskowitz, 1997). In contrast, a number of other mutants defective in cell fusion, such as fus1Δ and fus2Δ mutants, exhibit a severe defect when mated to a mutant partner (Trueheart et al., 1987).
Figure 6.
Kel1p and Kel2p are involved in mating and morphology. (A) kel1Δ mutants are defective in mating. kel1Δ strain JP363 was transformed with a CEN-ARS plasmid containing KEL1 (pJP207), a 2μ plasmid expressing Kel2p (pJP92), or vector (YCplac111). The wild-type control (IH3196) was transformed with vector (YCplac111). Strains were mated to an α fus1 fus2 strain (IH2351) as described in Materials and Methods. (B) Wild-type (a–d) and kel1Δ (JP363; e–h) mutants were mated on filters to wild-type strain IH2350 carrying the RAS2-GFP fusion plasmid (pJW192) as described in Materials and Methods. Nuclei were visualized by DAPI staining (d and h). RAS2-GFP was visualized by fluorescence microscopy (b and f). (C) kel1Δ and kel1Δ kel2Δ mutants exhibit altered morphology. Morphology of isogenic (a) wild-type (IH3196), (b) kel1Δ (JP363), (c) kel2Δ (JP371), and (d) kel1Δ kel2Δ (JP385) strains. (D) Morphological phenotype exhibited by cells overexpressing KEL1 or KEL2. Morphology exhibited by a wild-type strain (IH3196) transformed with (a) vector (YEp351), (b) 2μ KEL1 (pJP81), (c) vector (pRS426), or (d) pGAL KEL2 (pJP160) grown in SD-Ura (a and b) or S galactose-Ura (c and d).
Table III.
The Effect of Deletion of KEL1 and KEL2 on Cell Fusion
| Genotype | Prezygotes* | |
|---|---|---|
| % | ||
| WT | 2.6 ± 0.7 | |
| kel1Δ | 9.4 ± 1.9 | |
| kel2Δ | 3.7 ± 0.8 | |
| kel1Δ kel2Δ | 7.1 ± 1.4 | |
| fus1Δ | 22.4 ± 3.8 | |
| fus1Δ kel1Δ | 54.0 ± 10.6 | |
| fus1Δ kel2Δ | 31.9 ± 3.5 | |
| fus2Δ | 44 ± 7 | |
| fus2Δ kel1Δ | 70 ± 3 | |
| fus2Δ kel2Δ | 38 ± 2 | |
| fps1Δ | 62 ± 7 | |
| fps1Δ kel1Δ | 93 ± 4 | |
| fps1Δ kel2Δ | 66 ± 13 | |
| fps1Δ kel1Δ kel2Δ | 99.3 ± 0.2 | |
| spa2Δ | 39 ± 12 | |
| spa2Δ kel1Δ | 42 ± 10 |
Analysis of lines 1–7, 8–10, 11–14, and 15–16 were carried out separately, in each case with wild-type and kel1Δ controls that behaved similarly to lines 1 and 2, respectively. Strains were WT (IH3196), kel1Δ (JP363), kel2Δ (JP371), fus1Δ (JP52), fus1Δ kel1Δ (JP410), fus1Δ kel2Δ (JP412), kel1Δ kel2Δ (JP385), fus2Δ (JP418), fus2Δ kel1Δ (JP417), fus2Δ kel2Δ (JP416), fps1Δ (JP147), fps1Δ kel1Δ (JP414), fps1Δ kel2Δ (JP415), fps1Δ kel1Δ kel2Δ (JP500), spa2Δ (IH3204), and spa2Δ kel1Δ (JP502). Cells were grown in YEPD and were mated to a wild-type α strain (IH2350). For lines 1–7, values are means of at least four experiments in which a total of at least 900 partnered cells were counted ± SD. For lines 8–14, values are the means of three experiments in which more than 100 partnered cells were counted per experiment. For lines 15 and 16, more than 200 partnered cells were counted in three experiments.
Percent prezygotes represents the number of prezygotes/(zygotes + prezytotes).
Table IV.
The Effect of PKC1-R398P on Cell Fusion in kel1Δ and kel2Δ Mutants
| Genotype | Prezygotes* | |||
|---|---|---|---|---|
| Vector | PKC1-R398P | |||
| WT | 5.5 ± 1.0 | 47.9 ± 2.6 | ||
| kel1Δ | 30.2 ± 2.4 | 79.2 ± 0.3 | ||
| kel2Δ | 8.7 ± 2.8 | 53.9 ± 4.8 | ||
| fus1Δ | 47.3 ± 3.7 | 87.6 ± 3.6 | ||
| kel1Δ kel2Δ | 19.9 ± 2.0 | 69.6 ± 1.8 | ||
Strains were: WT (IH3196), kel1Δ (JP363), kel2Δ (JP371), fus1Δ (JP52), and kel1Δ kel2Δ (JP385). Strains were transformed with either YCp50 containing PKC1-R398P (pJP67) or YCp50 alone as indicated. Strains were grown in SD-Ura, and were mated to a wild-type α strain (IH2350). More than 200 partnered cells were counted in each experiment to determine % prezygotes. Values are means of three experiments ± SD.
Percent prezygotes represents the number of prezygotes/(zygotes + prezygotes).
To examine further the defect in cell fusion, strains were mated to a partner that produces RAS2-GFP fusion protein (kindly provided by J. Whistler, University of California, Berkeley) that localized green fluorescence around the cell periphery (Philips and Herskowitz, 1997; Dorer et al., 1997). In matings between two wild-type partners, the green fluorescent signal was seen throughout the entire zygote (Fig. 6 B, b). In contrast, in matings between kel1Δ mutants and a wild-type partner containing RAS2-GFP, prezygotes were found in which the green fluorescent signal remained restricted to one cell, indicating a failure in plasma membrane fusion and cytoplasmic mixing (Fig. 6 B, f). These structures could also be observed in wild-type matings, but were found at an elevated frequency in matings in which one partner was lacking KEL1. The increased frequency of prezygotes as determined by RAS2-GFP staining was similar to that quantitated by Nomarski optics and DAPI staining (data not shown).
To investigate further the role of KEL1 in cell fusion, we examined genetic interactions between kel1Δ mutants and other mutants defective in cell fusion. Because fus1Δ and fus2Δ mutants display only mild defects in cell fusion, whereas fus1Δ fus2Δ double mutants exhibit a more severe defect than either single mutant alone, it has been suggested that FUS1 and FUS2 function in parallel pathways (Trueheart et al., 1987; Brizzio et al., 1998). To determine whether KEL1 functions in either of these pathways, we examined the fusion competence of kel1Δ fus1Δ and the kel1Δ fus2Δ double mutants. In matings between a kel1Δ fus1Δ double mutant and a wild-type mating partner, ∼54% of the partnered cells were prezygotes, a defect significantly worse than that seen with kel1Δ or fus1Δ single mutants (9.4% and 22.4%, respectively; Table III). Loss of KEL1 exacerbated the defect in cell fusion of a fus1Δ and α fus1Δ cells, indicating that KEL1 is required in both cell types for efficient cell fusion (Table III and data not shown). Similar to the fus1Δ mutant, a fus2Δ single mutant exhibited 44% prezygotes, whereas the kel1Δ fus2Δ double mutant exhibited 70% prezygotes (Table III). These results demonstrate that loss of KEL1 exacerbates the defect in cell fusion of fus1Δ and fus2Δ mutants, suggesting that KEL1 is not in either the FUS1 or FUS2 pathways. In both fus1Δ and fus2Δ mutants, loss of KEL1 caused a 30% increase in the number of prezygotes. A 30% increase in the number of prezygotes was also seen in fps1Δ mutants, in which the number of prezygotes increased from 62% for fps1Δ mutants to 93% for fps1Δ kel1Δ double mutants (Table III). In contrast, loss of KEL1 did not significantly affect the ability of spa2Δ mutants to fuse. In this case, spa2Δ single mutants exhibited 39% prezygotes, whereas the spa2Δ kel1Δ double mutant exhibited 42% prezygotes, suggesting that Kel1p may function in the same pathway as Spa2p. We conclude that loss of KEL1 causes a mild defect in cell fusion, which is exacerbated by loss of FUS1, FUS2, or FPS1, but not by loss of SPA2.
kel2Δ Mutants Do Not Exhibit a Defect in Cell Fusion
To determine whether KEL2 was also required for cell fusion, we constructed a strain in which KEL2 was deleted, and analyzed its phenotype. One copy of the KEL2 open reading frame was replaced with the LEU2 gene (see Materials and Methods) in a diploid strain. Haploid segregants were examined for their ability to mate. Unlike kel1Δ mutants, kel2Δ mutants exhibited normal mating and cell fusion (Tables III and IV. In fus1Δ, fus2Δ, fps1Δ, and PKC1-R398P mutant strains, loss of Kel2p had little effect on cell fusion (Table III and IV). Since there is no detectable requirement for Kel2p in cell fusion in a wild-type strain, the primary function of Kel1p during fusion does not appear to be for localization of Kel2p.
Because kel1Δ mutants fail to localize Kel2p, one possible cause of the defect in cell fusion in such strains is mislocalization of Kel2p, as opposed to the absence of Kel1p or Kel2p at the shmoo tip. If such an explanation were correct, then deletion of KEL2 in the kel1Δ mutant should suppress the defect in cell fusion. When grown in rich medium and mated to a wild-type partner, kel1Δ mutants accumulated 9.4 ± 1.9% prezygotes, whereas kel1Δ kel2Δ double mutants accumulated 7.1 ± 1.4% prezygotes compared with 2.6 ± 0.7% prezygotes for wild-type cells (Table III). When cells were grown in minimal medium, there was a higher background level of prezygotes. In this case, kel1Δ strains accumulated 30.2 ± 2.4% prezygotes, whereas kel1Δ kel2Δ double mutants accumulated 19.9 ± 2.0% prezygotes compared with 5.5 ± 1.0% for wild-type cells (Table IV, compare lines 2, 5, and 1, respectively). We conclude that loss of KEL2 may modestly suppress the fusion defect of kel1Δ strains, but that mislocalized Kel2p does not completely account for the fusion defect of kel1Δ strains. Consistent with this interpretation, overexpression of KEL2 did not significantly alter the mating ability of kel1Δ mutants (Fig. 6 A) Taken together, these data indicate that Kel1p has a role in cell fusion that is not solely to localize Kel2p. In contrast, there is little requirement for Kel2p in cell fusion.
kel1Δ Mutants Have a Defect in Cell Morphology
In addition to the defect in cell fusion, kel1Δ mutants exhibited a defect in morphology, appearing slightly elongated and heterogeneous in shape when compared with wild-type control cells (Fig. 6 C, b). This morphological defect was observed in two different strain backgrounds (data not shown). kel2Δ mutants exhibited normal morphology (Fig. 6 C, c). Examination of the kel1Δ kel2Δ double mutant revealed the same elongated morphology as seen in the kel1Δ single mutant (Fig. 6 C, d), suggesting that, like the fusion defect, the altered morphology of kel1Δ mutants is not due to mislocalized Kel2p.
Although loss of Kel2p did not alter morphology, overexpressing Kel2p from the GAL1,10 promoter caused ∼5% of cells to display a grossly aberrant morphology (Fig. 6 D, d). When Kel1p was overexpressed from a 2μ plasmid, an equivalent fraction of cells displayed a similar morphology (Fig. 6 D, b). We did not observe this morphology in cells containing vector when grown in galactose (Fig. 6 D, c), or when cells containing pGAL-KEL2 were grown in glucose (data not shown). These data show that although only Kel1p is required for proper morphology, overexpression of either Kel1p or Kel2p disrupts normal cellular morphology.
Because of the potential role of kelch repeats in mediating interaction with cytoskeletal components (see Discussion), we examined actin in kel1Δ strains by staining with rhodamin–phalloidin. We did not detect any obvious differences between wild-type and kel1Δ mutants. Microtubules were visualized in kel1Δ, kel2Δ, and kel1Δ kel2Δ mutants by indirect immunofluorescence both with and without pheromone treatment. Again, we did not detect any obvious differences (data not shown), but subtle defects cannot be excluded. Additionally, loss of Kel1p or Kel2p did not affect sensitivity to the microtubule-depolymerizing drug benomyl, and kel1Δ mutants did not exhibit genetic interactions with tub1-1 mutants (Carminati, J., personal communication). Hence, it is currently unclear whether Kel1p interacts with the actin or microtubule cytoskeleton.
Relationship of Kel1p to the Pkc1p Pathway during Mating and Vegetative Growth
KEL1 was identified by its ability to suppress an activated allele of Pkc1p. In principle, Kel1p could be a target of Pkc1p, a negative regulator or Pkc1p, or could act in parallel to promote fusion. To examine the relationship between Kel1p and Pkc1p, we analyzed the phenotype of a kel1Δ mutant expressing PKC1-R398P. If Kel1p were the sole downstream target inhibited by Pkc1p to prevent cell fusion, then we would expect the PKC1-R398P kel1Δ double mutant to behave like the kel1Δ and PKC1-R398P single mutants. Examination of a kel1Δ PKC1-R398P double mutant revealed a defect in cell fusion that was worse than that seen in a kel1Δ mutant or in a wild-type cell expressing PKC1-R398P (Table IV). Matings between kel1Δ mutants and wild-type partners yielded ∼30% prezygotes, whereas the kel1Δ PKC1-R398P strain exhibited 79% prezygotes, higher than that seen with wild-type strains carrying the PKC1-R398P allele (48%). These data suggest that Kel1p is not the sole target of Pkc1p. Kel1p could be one of several Pkc1p targets or could function upstream or parallel to Pkc1p to promote cell fusion.
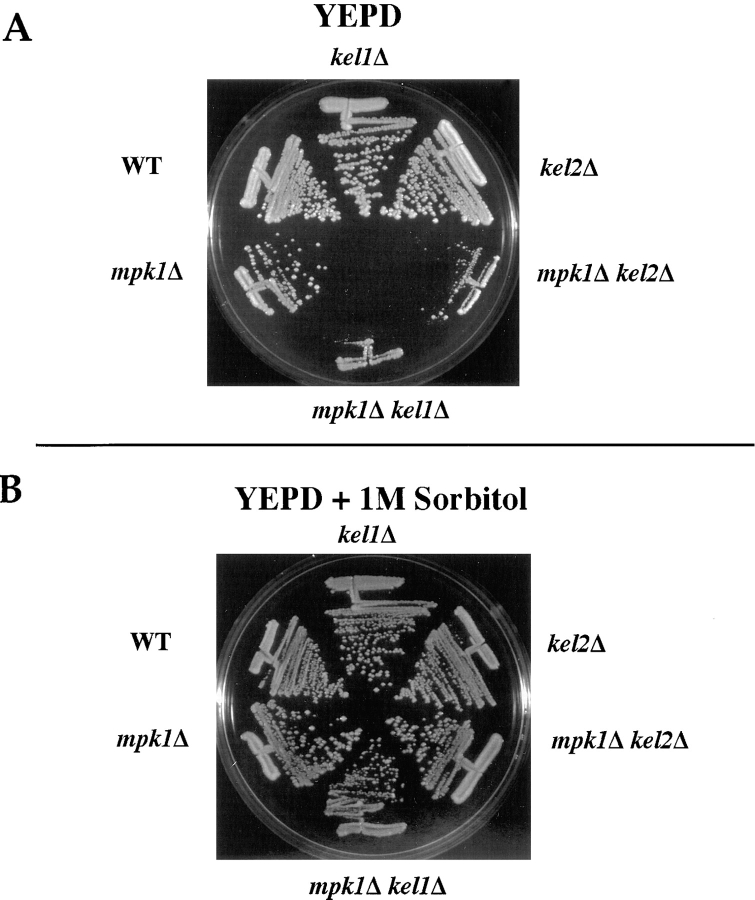
To examine the relationship between Kel1p and Kel2p and the Pkc1p pathway during vegetative growth, we analyzed the phenotype of strains lacking MPK1 and KEL1 or KEL2. MPK1 encodes a MAP kinase that appears to function downstream of Pkc1p (Lee and Levin, 1992; Kamada et al., 1995). mpk1Δ mutants grow slowly at 34.5°C and fail to grow at 37°C; the growth defect at high temperature is suppressed by 1 M sorbitol. To determine whether loss of KEL1 or KEL2 affects growth of an mpk1Δ mutant, kel1Δ mpk1Δ and kel2Δ mpk1Δ double mutant strains were constructed (see Materials and Methods). All 18 kel1Δ mpk1Δ and 13 kel2Δ mpk1Δ double mutants grew more poorly than mpk1Δ single mutants at 34.5°C (Fig. 7 A), and the growth defect was suppressed by 1 M sorbitol (Fig. 7 B). We conclude that loss of KEL1 or KEL2 exacerbates the growth defect of mpk1Δ mutants.
Figure 7.
Deletion of KEL1 or KEL2 exacerbates the growth defect of mpk1Δ mutants. Wild-type (IH1783), kel1Δ (JP445A), kel2Δ (JP446A), mpk1Δ (IH3077), mpk1Δ kel1Δ (JP490), and mpk1Δ kel2Δ (JP491) strains were grown on YEPD (A) or YEPD supplemented with 1 M sorbitol (B) at 34.5°C for 2 d.
Discussion
Identification and Analysis of Kel1p and Kel2p
We identified a novel gene, KEL1, whose overexpression can partially relieve the defect in cell fusion caused by PKC1-R398P. We found that overexpression of Kel1p could partially reduce the defect in cell fusion exhibited by spa2Δ and fps1Δ mutants. Kel1p localizes to the region of the cell where fusion is initiated during mating and to regions of polarized growth in vegetative cells. Mutants lacking Kel1p exhibit defects in cell fusion that are exacerbated by loss of FUS1, FUS2, or FPS1, or by expression of PKC1-R398P. kel1Δ mutants are also elongated and heterogeneous in shape, indicating that Kel1p has a role not only in cell fusion but also in morphogenesis.
KEL1 and KEL2 are present in duplicated regions of the genome, encoding proteins that are approximately 44% identical. The function of Kel2p is related to that of Kel1p in several respects: overexpression of Kel2p suppresses the mating defect caused by an activated form of Pkc1p, localization of Kel2p is indistinguishable from that of Kel1p, and Kel2p is in a complex with Kel1p. In addition, when Kel1p or Kel2p are overexpressed, a fraction of cells display an aberrant morphology that is strikingly similar in both cases. Despite their considerable homology, Kel1p and Kel2p exhibit some differences. First, Kel2p is unable to localize in the absence of Kel1p, suggesting that Kel1p, but not Kel2p, can interact with at least one factor at the cell cortex. Second, only kel1Δ mutants have detectable phenotypes. In an otherwise wild-type cell, Kel2p is not required for cell fusion or normal cellular morphology, nor does loss of KEL2 significantly affect the fusion ability of fus1Δ, fus2Δ, fps1Δ, or PKC1-R398P mutants. Kel1p and Kel2p may have diverged such that Kel2p does not play a role in these processes. Another possibility is that Kel1p and Kel2p have a similar function but that Kel1p is the major contributor. Coimmunoprecipitation and two-hybrid analyses demonstrate that Kel1p can associate with itself and with Kel2p (Fig. 5). If there are distinct Kel1p– Kel1p and Kel1p–Kel2p complexes, then loss of Kel1p would disrupt both types of complexes. In the absence of Kel2p, Kel1p–Kel1p complexes would remain, perhaps explaining the lesser role that Kel2p plays in cell fusion and morphogenesis.
Kel1p and Kel2p Are Members of the Kelch Family of Proteins
Kelch repeats, a motif of approximately fifty amino acids, are found in two to seven copies in proteins from diverse organisms including poxviruses, Drosophila, C. elegans, and mouse. Based upon sequence similarity to a superfamily of proteins that includes galactose oxidase (Ito et al., 1994) and neuraminidase (Varghese and Colman, 1991), these repeats are thought to form the blades of a β-propeller structure (Bork and Doolittle, 1994). Several kelch-containing proteins are implicated in actin interactions. For example, the Drosophila protein Kelch localizes to ring canals, actin-containing structures that separate nurse cells in the developing egg chamber. In the absence of Kelch, actin in the ring canals becomes disorganized (Xue and Cooley, 1993; Robinson and Cooley, 1997). The Limulus protein α scruin bundles actin filaments in the acrosomal process of sperm, and is thought to be important at fertilization when the filaments undergo a conformational change (Sanders et al., 1996). Additional kelch-containing proteins implicated in actin interactions include the actin-fragmin kinase from Physarum (Eichinger et al., 1996), SPE-26 from C. elegans (Varkey et al., 1995), and ENC-1 from the mammalian nervous system (Hernandez et al., 1997).
It is not clear whether all kelch domains mediate interaction with actin. β-scruin from Limulus and calicin from bovine sperm are thought to be localized to regions of the cell that lack actin structures (Way et al., 1995; von Bulow et al., 1995). Tea1p, the closest homologue to Kel1p and Kel2p, influences microtubule dynamics rather than actin dynamics and is required in S. pombe cells for proper rod-like morphology (Mata and Nurse, 1997). In the absence of Tea1p or when Tea1p is overexpressed, cells exhibit bent and T-shaped morphologies. Kel1p is also required for proper cell shape, but there is little evidence in S. cerevisiae that microtubules play a role in morphology, whereas actin is thought to be important (for review see Botstein et al., 1997). Hence, it is currently unclear whether Kel1p interacts with the actin or microtubule cytoskeleton, or whether it is involved in some other process.
Relationship of Kel1p to the Pkc1p Pathway
The observation that overexpression of Kel1p can partially reverse the fusion defect caused by hyperactive Pkc1p suggests that Kel1p functions in opposition to Pkc1p to activate cell fusion, perhaps by acting upstream of Pkc1p (where it might affect Rho1p or other potential regulators of Pkc1p; see Kamada et al., 1996; Gray et al., 1997; Zarzov et al., 1996) or in parallel to the Pkc1 pathway. Kel1p also plays a role in vegetative cells that has some functional connection to the Pkc1p pathway. A role in vegetative cells is inferred from the observation that loss of either KEL1 or KEL2 exacerbates the growth defect of mpk1Δ mutants, which is suppressed by 1 M sorbitol. Formally, these results indicate that both Kel1p (or Kel2p) and Mpk1p promote cell integrity. Although various explanations are possible for the reduced viability of the mpk1Δ kel1Δ and mpk1Δ kel2Δ strains, one possibility is that cells respond to loss of KEL1 or KEL2 by increasing Mpk1p activity, which is necessary to maintain cell integrity under these conditions. A notable feature of this explanation is that Kel1p would act in vegetative cells as it is proposed to act in mating cells, antagonistically to the Pkc1p pathway.
Toda et al. (1993) made the striking observation that overexpression of pck2 +,which codes for a Pkc1p-like protein of S. pombe, results in production of branched cells similar to those seen in tea1 mutants (Mata and Nurse, 1997). This relationship is analogous to what we have observed for cell fusion in budding yeast: hyperactivation of protein kinase C leads to a phenotype similar to that due to loss of a kelch protein (Kel1p). Kelch proteins in other organisms may also function along with protein kinase C pathways. The relationship between other kelch proteins and protein kinase C could be explored in organisms without facile genetics using dominant negative forms of the kelch proteins and dominant activated forms of protein kinase C.
The Role of Kel1p in Cell Fusion
Kel1p belongs to a class of proteins including Spa2p and Fps1p that is required for cell fusion during mating (Dorer et al., 1997; Gammie et al., 1998; Philips and Herskowitz, 1997) and that also functions during vegetative growth for morphogenesis and cell integrity (Snyder, 1989; Snyder et al., 1991; Evangelista et al., 1997; Luyten et al., 1995). The defect in cell fusion exhibited by mutants lacking Fps1p, Spa2p, or Kel1p could result from increased activity of the Pkc1p pathway. Our observation that overexpression of KEL1 partially restored mating ability to spa2Δ, fps1Δ, and PKC1-R398P mutants raises the possibility that spa2Δ and fps1Δ mutants have elevated Pkc1p activity, which may account for their defect in cell fusion. Additional data support this possibility. In particular, we have proposed earlier that the fusion defect of fps1Δ mutants results from high levels of intracellular glycerol, which activates the Pkc1p pathway (Philips and Herskowitz, 1997). Several observations suggest that the Pkc1p pathway may be activated in spa2Δ and kel1Δ mutants as well. Loss of Spa2p results in increased phosphorylation of Swi6p (Sheu et al., 1998), an apparent substrate of Mpk1p (Madden et al., 1997). Spa2p exhibits two-hybrid interactions with components of the Pkc1p pathway (Mkk1p/ Mkk2p; Sheu et al., 1998). Loss of Spa2p as well as Fps1p or Kel1p exacerbates the growth defect of mutants defective in the Mpk1p MAP kinase pathway (Costigan et al., 1992; Philips and Herskowitz, 1997; Fig. 7 A). These observations lead us to suggest that Spa2p, Kel1p, and Fps1p negatively regulate Pkc1p activity. In their absence, we propose that the Pkc1p pathway becomes activated, which may be essential for cells to remain viable and which leads to inhibition of cell fusion during mating. We suggest that during mating of wild-type cells, Pkc1p monitors the osmotic and morphological integrity of the cell. If cells are not osmotically stable, as in fps1Δ mutants, or exhibit disrupted morphology, as in spa2Δ and kel1Δ mutants, Pkc1p inhibits cell fusion.
It is also possible that Kel1p does not act by inhibiting the Pkc1p pathway but may act, for example, on the actin cytoskeleton. Kel1p, as well as proteins such as Spa2p and Bni1p, might be important for directing or maintaining vesicles at a cell fusion zone through interactions with the cytoskeleton (Evangelista et al., 1997; Gehrung and Snyder, 1990; Sheu et al., 1998). Consistent with such a possibility, EM analysis of mating cells shows the presence of clustered vesicles at the zone of cell fusion (Osumi et al., 1974; Gammie et al., 1998), which in the absence of Spa2p are more dispersed than in wild-type strains (Gammie et al., 1998). Cytoskeletal components play a possible role in myoblast fusion as well. In Drosophila, mutants defective in myoblast city (mbc) exhibit defects in cytoskeletal organization and myoblast fusion (Rushton et al., 1995; Erickson et al., 1997), failing to localize paired vesicles to the site of cell fusion (Doberstein et al., 1997). Similarly, overexpression of mutant forms of Drac1, a small GTPase known to affect the actin cytoskeleton, disrupts myoblast fusion and morphology (Luo et al., 1994; Doberstein et al., 1997). In vertebrates, cytochalasin B, which disrupts actin filaments, interferes with myoblast fusion (Sanger et al., 1971). It is unclear in myoblast fusion, as in yeast fusion, exactly what role the cytoskeleton plays in the fusion process. As more components become identified, we are in a better position to determine how proteins involved in cell fusion interact with cytoskeletal machinery, and the ways in which the protein kinase C pathway governs fusion.
Acknowledgments
We thank M. Peter and members of our laboratory, in particular L. Huang, M. Maxon, S. O'Rourke, and R. Tabtiang, for assistance and valuable discussion. We thank R. Tabtiang, A. Gammie, M. Maxon, M. Peter, S. O'Rourke, and L. Huang for helpful comments on the manuscript. We thank Y. Takai and E. O'Shea for plasmids, P. O'Farrell for anti-GFP antibodies, Chris Botka and Herve Recipon for assistance with sequence analysis, and J. Carminati, A. Gammie, V. Brizzio, and M. Snyder for communicating results before publication.
This work was supported by Research and Program Project Grants from the National Institutes of Health to I. Herskowitz. J. Philips was supported by the Julius Krevans Graduate Research Fellowship and by the Sussman Fund, supplemented by the Herbert W. Boyer Fund.
Footnotes
Address all correspondence to Dr. Jennifer Philips, Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94143-0448. Tel.: (415) 476-4985. Fax: (415) 502-5145. E-mail: philips@socrates.ucsf.edu
1. Abbreviation used in this paper: HA, hemagglutinin.
References
- Adames N, Blundell K, Ashby MN, Boone C. Role of yeast insulin-degrading enzyme homologues in propheromone processing and bud site selection. Science. 1995;270:464–467. doi: 10.1126/science.270.5235.464. [DOI] [PubMed] [Google Scholar]
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Inouye CJ, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. . Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- Bork P, Doolittle RF. Drosophilakelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Botstein, D., D. Amberg, and J. Mulholland. 1997. The yeast cytoskeleton. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. J.R. Pringle, J.R. Broach, and E.W. Jones, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 1–90.
- Brizzio V, Gammie AE, Nijbroek G, Michaelis S, Rose MD. Cell fusion during yeast mating requires high levels of a-factor mating pheromone. J Cell Biol. 1996;135:1727–1740. doi: 10.1083/jcb.135.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V, Gammie AE, Rose MD. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. . J Cell Biol. 1998;141:567–584. doi: 10.1083/jcb.141.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehrer BM, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. . Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homologue implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Davenport KR, Sohaskey M, Kamada Y, Levin DE, Gustin MC. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer R, Boone C, Kimbrough T, Kim J, Hartwell LH. Genetic analysis of default mating behavior in Saccharomyces cerevisiae. . Genetics. 1997;146:39–55. doi: 10.1093/genetics/146.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Bomblies L, Vandekerckhove J, Schleicher M, Gettemans J. A novel type of protein kinase phosphorylates actin in the actin-fragmin complex. EMBO (Eur Mol Biol Organ) J. 1996;15:5547–5556. [PMC free article] [PubMed] [Google Scholar]
- Elia L, Marsh L. Role of the ABC transporter Ste6 in cell fusion during yeast conjugation. J Cell Biol. 1996;135:741–752. doi: 10.1083/jcb.135.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA, Trueheart J, Fink GR. Fus2 localizes near the site of cell fusion and is required for both cell fusion and nuclear alignment during zygote formation. J Cell Biol. 1995;130:1283–1296. doi: 10.1083/jcb.130.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S, Lin L, Malczynski M, Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J Cell Biol. 1998;140:461–483. doi: 10.1083/jcb.140.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson MR, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B, Cade RM, Yashar BM, Kamada Y, Levin DE, Irie K, Matsumoto K. Dynamics and organization of MAP kinase signal pathways. Mol Reprod Dev. 1995;42:477–485. doi: 10.1002/mrd.1080420416. [DOI] [PubMed] [Google Scholar]
- Errede B, Levin DE. A conserved kinase cascade for MAP kinase activation in yeast. Curr Opin Cell Biol. 1993;5:254–260. doi: 10.1016/0955-0674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Field C, Schekman R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. . J Cell Biol. 1980;86:123–128. doi: 10.1083/jcb.86.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, S., and J.R. Pringle. 1986. Development of spatial organization during the formation of zygotes and shmoos in Saccharomyces cerevisiae. Yeast. S114.
- Gammie AE, Brizzio V, Rose MD. Distinct morphological phenotypes of cell fusion mutants. Mol Biol Cell. 1998;6:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrung S, Snyder M. The SPA2 gene of Saccharomyces cerevisiaeis important for pheromone-induced morphogenesis and efficient mating. J Cell Biol. 1990;111:1451–1464. doi: 10.1083/jcb.111.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia colishuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gray JV, Ogas JP, Kamada Y, Stone M, Levin DE, Herskowitz I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiaein bud emergence and identification of a putative upstream regulator. EMBO (Eur Mol Biol Organ) J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. Fink. 1991. Guide to yeast genetics and molecular biology. In Methods in Enzymology. Vol. 194. Academic Press, Inc., San Diego, CA. 933 pp. [PubMed]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Hasek J, Rupes I, Svobodova J, Streiblova E. Tubulin and actin topology during zygote formation of Saccharomyces cerevisiae. . J Gen Microbiol. 1987;133:3355–3363. doi: 10.1099/00221287-133-12-3355. [DOI] [PubMed] [Google Scholar]
- Hernandez MC, Andres-Barquin PJ, Martinez S, Bulfone A, Rubenstein JL, Israel MA. ENC-1: a novel mammalian kelch-related gene specifically expressed in the nervous system encodes an actin-binding protein. J Neurosci. 1997;17:3038–3051. doi: 10.1523/JNEUROSCI.17-09-03038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Irie K, Takase M, Lee KS, Levin DE, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiaemitogen-activated protein kinase-kinase homologues, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N, Phillips SE, Yadav KD, Knowles PF. Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol. 1994;238:794–814. doi: 10.1006/jmbi.1994.1335. [DOI] [PubMed] [Google Scholar]
- Jackson CL, Hartwell LH. Courtship in Saccharomyces cerevisiae: an early cell-cell interaction during mating. Mol Cell Biol. 1990;10:2203–2213. doi: 10.1128/mcb.10.5.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Qadota H, Python CP, Anraku Y, Ohya Y, Levin DE. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Jung US, Piotrowski J, Levin DE. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiaemediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- Kuchler K, Sterne RE, Thorner J. Saccharomyces cerevisiae STE6gene product: a novel pathway for protein export in eukaryotic cells. EMBO (Eur Mol Biol Organ) J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee KS, Levin DE. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiaeprotein kinase C homologue. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: DrosophilaDrac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Luyten K, Albertyn J, Skibbe WF, Prior BA, Ramos J, Thevelein JM, Hohmann S. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO (Eur Mol Biol Organ) J. 1995;14:1360–1371. doi: 10.1002/j.1460-2075.1995.tb07122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Sheu YJ, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- Madden K, Snyder M. Specification of sites for polarized growth in Saccharomyces cerevisiaeand the influence of external factors on site selection. Mol Biol Cell. 1992;3:1025–1035. doi: 10.1091/mbc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, L., and M.D. Rose. 1997. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. J.R. Pringle, J.R. Broach, and E.W. Jones, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 827–888.
- Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Mazzoni C, Zarov P, Rambourg A, Mann C. The SLT2 (MPK1) MAP kinase homologue is involved in polarized cell growth in Saccharomyces cerevisiae. . J Cell Biol. 1993;123:1821–1833. doi: 10.1083/jcb.123.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey G, Clay FJ, Kelsay K, Sprague GF. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. . Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homologue of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi M, Shimoda C, Yanagishima N. Mating reaction in Saccharomyces cerevisiae. V. Changes in fine structure during the mating reaction. Arch Microbiol. 1974;97:27–38. [PubMed] [Google Scholar]
- Philips J, Herskowitz I. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. . J Cell Biol. 1997;138:961–974. doi: 10.1083/jcb.138.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S, Michaelis S, Broek D, Santa S, Anna, Field J, Herskowitz I, Wigler M. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell. 1986;47:413–422. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- Read EB, Okamura HH, Drubin DG. Actin- and tubulin-dependent functions during Saccharomyces cerevisiaemating projection formation. Mol Biol Cell. 1992;3:429–444. doi: 10.1091/mbc.3.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Drosophilakelch is an oligomeric ring canal actin organizer. J Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 123 pp.
- Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophilamuscle development. Development. 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, New York. 545 pp.
- Sanders MC, Way M, Sakai J, Matsudaira P. Characterization of the actin cross-linking properties of the scruin-calmodulin complex from the acrosomal process of Limulussperm. J Biol Chem. 1996;271:2651–2657. doi: 10.1074/jbc.271.5.2651. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Holtzer S, Holtzer H. Effects of cytochalasin B on muscle cells in tissue culture. Nature. 1971;229:121–123. doi: 10.1038/newbio229121a0. [DOI] [PubMed] [Google Scholar]
- Santos B, Duran A, Valdivieso MH. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. . Mol Cell Biol. 1997;17:2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LD, Friesen JD. Nucleotide sequence of the TCM1 gene (ribosomal protein L3) of Saccharomyces cerevisiae. . J Bacteriol. 1983;155:8–14. doi: 10.1128/jb.155.1.8-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall JE. Polarization of yeast cells in spatial gradients of α-mating factor. Proc Natl Acad Sci USA. 1993;90:8332–8336. doi: 10.1073/pnas.90.18.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast morphogenesis. Mol Cell Biol. 1998;7:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Gehrung S, Page BD. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. . J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HOgene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Yanagida M. Two novel protein kinase C-related genes of fission yeast are essential for cell viability and implicated in cell shape control. EMBO (Eur Mol Biol Organ) J. 1993;12:1987–1995. doi: 10.1002/j.1460-2075.1993.tb05848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres L, Martin H, Garcia-Saez MI, Arroyo J, Molina M, Sanchez M, Nombela C. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2mutants. Mol Microbiol. 1991;5:2845–2854. doi: 10.1111/j.1365-2958.1991.tb01993.x. [DOI] [PubMed] [Google Scholar]
- Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese JN, Colman PM. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 Å resolution. J Mol Biol. 1991;221:473–486. doi: 10.1016/0022-2836(91)80068-6. [DOI] [PubMed] [Google Scholar]
- Varkey JP, Muhlrad PJ, Minniti AN, Do B, Ward S. The Caenorhabditis elegans spe-26gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- von Bulow M, Heid H, Hess H, Franke WW. Molecular nature of calicin, a major basic protein of the mammalian sperm head cytoskeleton. Exp Cell Res. 1995;219:407–413. doi: 10.1006/excr.1995.1246. [DOI] [PubMed] [Google Scholar]
- Way M, Sanders M, Chafel M, Tu YH, Knight A, Matsudaira P. β-Scruin, a homologue of the actin crosslinking protein scruin, is localized to the acrosomal vesicle of Limulussperm. J Cell Sci. 1995;108:3155–3162. doi: 10.1242/jcs.108.10.3155. [DOI] [PubMed] [Google Scholar]
- Wolfe, K., and D. Shields. 1998. Yeast Gene Duplications. http:://acer.gen. tcd.ie/∼khwolfe/yeast/topmenu.html
- Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophilaegg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- Zarzov P, Mazzoni C, Mann C. The SLT2 (MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO (Eur Mol Biol Organ) J. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]