Abstract
We have used gene disruption to isolate two talin (−/−) ES cell mutants that contain no intact talin. The undifferentiated cells (a) were unable to spread on gelatin or laminin and grew as rounded colonies, although they were able to spread on fibronectin (b) showed reduced adhesion to laminin, but not fibronectin (c) expressed much reduced levels of β1 integrin, although levels of α5 and αV were wild-type (d) were less polarized with increased membrane protrusions compared with a vinculin (−/−) ES cell mutant (e) were unable to assemble vinculin or paxillin-containing focal adhesions or actin stress fibers on fibronectin, whereas vinculin (−/−) ES cells were able to assemble talin-containing focal adhesions. Both talin (−/−) ES cell mutants formed embryoid bodies, but differentiation was restricted to two morphologically distinct cell types. Interestingly, these differentiated talin (−/−) ES cells were able to spread and form focal adhesion-like structures containing vinculin and paxillin on fibronectin. Moreover, the levels of the β1 integrin subunit were comparable to those in wild-type ES cells. We conclude that talin is essential for β1 integrin expression and focal adhesion assembly in undifferentiated ES cells, but that a subset of differentiated cells are talin independent for both characteristics.
Keywords: talin, integrins, focal adhesions, gene knockout, ES cells
The interaction between animal cells and the adhesive glycoproteins of the extracellular matrix is mediated by the integrin family of cell adhesion molecules, the cytoplasmic domains of which are thought to be linked in many, but not all cases, to the actin cytoskeleton (Burridge et al., 1996). Proteins that have been suggested to participate in this link include talin, vinculin and α-actinin (Jockusch et al., 1995) which colocalize with integrins in the specialized cell–matrix junctions or focal adhesions formed when cultured cells are grown on rigid supports. Initial in vitro biochemical studies based on equilibrium gel filtration indicated that talin can bind directly to integrins (Horwitz et al., 1986), and a putative talin-binding site in the cytoplasmic region of the β1-integrin subunit has been identified based on peptide inhibition experiments (Tapley et al., 1989). In a more recent study, purified αIIbβ3 integrin was shown to bind to talin deposited on plastic, and a monoclonal antibody raised against the cytoplasmic domain of the β3 subunit blocked binding (Knezevic et al., 1996). Talin was also found to bind to synthetic peptides corresponding to the cytoplasmic domain of the β3 subunit, but it also bound to an αIIb cytoplasmic domain peptide suggesting that integrin α-subunits might also participate in the interaction. In addition, talin (but also filamin) has been shown to bind to recombinant polypeptides containing the β1A cytoplasmic domain (Pfaff et al., 1998). Since talin is an actin-binding protein (Muguruma et al., 1992; Niggli et al., 1994; Hemmings et al., 1996; McCann and Craig, 1997), it may provide a direct link between integrins and the actin cytoskeleton. However, talin also binds to vinculin (Burridge and Mangeat, 1984; Gilmore et al., 1992, 1993) which has been shown to contain two F-actin binding sites (Huttelmaier et al., 1997) as well as a binding site for α-actinin (McGregor et al., 1994), a well-characterized actin bundling protein (Blanchard et al., 1989). α-Actinin has also been reported to bind directly to β1-integrin cytoplasmic domain peptides (Otey et al. 1990). Such data have lead to a model of the focal adhesion in which integrins are linked to F-actin either directly via talin or α-actinin, or indirectly via interactions between talin, vinculin and α-actinin (Burridge et al., 1996).
Evidence in support of this model has come from a variety of experimental approaches. Experiments with Caenorhabditis elegans mutants have clearly established that the localization of talin to focal adhesion-like structures requires a β-integrin, but not vinculin (Moulder et al., 1996). Similarly, studies with a chimeric molecule containing the extracellular domain of N-cadherin fused to the transmembrane and cytoplasmic domains of β1-integrin support the view that the β1-integrin cytoplasmic domain plays a key role in localizing talin specifically to cell– matrix rather than cell–cell junctions (Geiger et al., 1992). Microinjection of antibodies to vinculin (Westmeyer et al., 1990) and talin (Nuckolls et al., 1992; Bolton et al., 1997) into fibroblasts disrupts actin stress fibers, as do proteolytic fragments of α-actinin (Pavalko and Burridge, 1991) and recombinant talin polypeptides (Hemmings et al., 1996), presumably via a dominant negative effect. Antisense mRNAs to vinculin (Rodriguez Fernandez et al., 1993) and talin (Albiges-Rizo et al., 1995) have been found to reduce cell adhesion and cell spreading of BALB/c 3T3 cells and Hela cells, respectively, and a mouse F9 teratocarcinoma cell line in which the vinculin gene has been disrupted showed altered adhesive characteristics (Coll et al., 1995; Volberg et al., 1995). However, the vinculin (−/−) F9 mutants retained the capacity to assemble talin-containing focal adhesions suggesting that vinculin is not an essential component of cell–matrix junctions, at least in this cell type. In an attempt to define further the role of talin in the adhesion of cells to the extracellular matrix, we have used gene replacement vectors to isolate mouse ES cells1 in which both copies of the talin gene have been disrupted. The phenotypic properties of these cells are consistent with the hypothesis that talin plays a key role in cell–matrix interactions.
Materials and Methods
Isolation and Characterization of Mouse Talin and Vinculin Genomic Clones
A mouse talin cDNA spanning nucleotides 286–1,187 was generated by reverse transcription–PCR from mRNA purified from 4 × 107 mouse NIH 3T3 cells using acid guanidinium thiocyanate and oligo(dT)-cellulose. The PCR primers contained BamHI sites, and the PCR product was subcloned into the BamHI site in pBluescript SK+ (Stratagene, La Jolla, CA), and authenticated by sequencing. The cDNA was labeled with [32P]dCTP using the Quick Prime kit (Nycomed Amersham Inc., Princeton, NJ), and used to screen 1 × 106 recombinants of a C129 mouse ES (E14) cell genomic library in λ2001 (kindly provided by Dr. Andrew Smith, Centre for Genome Research, Edinburgh) plated onto Escherichia coli host strain Q358. Restriction enzyme mapping of clone 5Tλ2 with all combinations of BamHI, HindIII, SacI, and EcoRI, and Southern blotting using oligonucleotides based on 5′ talin cDNA sequence (nucleotides [nt] 163–206, 500–518, 940–957, and 1381–1398), and end labeled using an ECL kit (Nycomed Amersham Inc.), suggested that clone 5Tλ2 contained the first two coding exons. Clone 5Tλ2 was mapped in more detail by subcloning a 1-kb SacI–HindIII fragment and a 1.7-kb BamHI fragment into pBluescript. These constructs were sequenced with pBluescript reverse and −20 M13 primers, as well as with primers spanning nt 163–206, 287–271, and 294–314 of the mouse talin cDNA sequence (X56123). Comparison of the resulting genomic sequence with the cDNA sequence, and with consensus eukaryotic splice sites (Mount and Steitz, 1981) allowed definition of the boundaries of the first two coding exons. A mouse ES cell vinculin genomic clone containing exons 4–6 was isolated and characterized in a similar way.
Generation of Talin and Vinculin Gene Targeting Constructs
The talin genomic clone 5Tλ2 was digested with BamHI and a 2.4-kb fragment cloned into the BamHI site of plasmid pX53 (Ohno et al., 1994) in the opposite transcriptional orientation to the Neomycin (Neo) gene to give the vector pX53-5′arm. A 6.2-kb HindIII fragment from 5Tλ2 was cloned into the HindIII site of pBluescript SK+ such that the ClaI site in the multiple cloning site was 5′ with respect to the genomic DNA fragment. This fragment was then excised from pBluescript using ClaI–NotI and cloned into the pX53-5′ arm vector digested with ClaI–NotI. A vinculin gene targeting vector was created by subcloning a 7.5-kb BamHI–XbaI genomic fragment containing exons 4–6 into the BamHI–XbaI sites in pBluescript. A BamHI–XhoI Neo cassette from the pX53 vector was then blunt ended into the unique MscI site in exon 5. Equivalent constructs of both the talin and vinculin gene targeting vectors were also made in which the Neo gene was replaced with the Hygromycin (Hyg) gene. Plasmid DNA was prepared using maxiprep columns (QIAGEN Inc., Chatsworth, CA) and all constructs were linearized with NotI.
ES Cell Culture
Mouse ES cell clone HM1 (a generous gift of Dr. David Melton, University of Edinburgh) was cultured (37°C in 5% CO2 in air) on tissue culture plates coated with 0.1% gelatin in Dulbecco's modified Eagle's medium/ Ham's F12 nutrient mix (GIBCO BRL, Gaithersburg, MD) containing 0.1% NaHCO3, β-mercaptoethanol (0.001%), antibiotics/mycotics/kanamycin (GIBCO BRL), 15% batch-tested fetal calf serum, and sufficient leukemia inhibitory factor (LIF) to suppress differentiation. LIF was prepared by transiently expressing a plasmid containing a LIF cDNA construct (pXMT2) in monkey COS cells, and harvesting the culture medium. The ability of each LIF preparation to suppress ES cell differentiation was tested before use.
Electroporation of ES Cells with Targeting Constructs and Selection of Homologous Recombinants
Exponentially growing HM1 ES cells (1 × 108) were trypsinized, washed 2× with cold Hepes-buffered saline (2.5 mM Hepes buffer, pH 7.1, 0.75 mM Na2HPO4, 140 mM NaCl), and resuspended in 3.5 ml of Hepes-buffered saline on ice. The linearized (Not1) talin targeting construct (100 μg) containing the Neo selection marker was added in 0.5 ml Hepes-buffered saline, and 0.8-ml aliquots incubated in electroporation cuvettes for 10 min on ice. The cells were resuspended, then electroporated (240 volts, 500 μF) using a gene pulser (Bio-Rad Laboratories, Hercules, CA). The cells were kept for 10 min at room temperature, then resuspended in 200 ml of culture medium and distributed in 20 × 9-cm gelatin-coated dishes. After 24 h, the medium was changed and G418 (100 μg/ml) and 0.2 μM gancyclovir added. The medium was changed daily until surviving colonies were visible (∼10 d), and these were then picked using a Gilson micropipette into 96-well microtiter plates coated with gelatin, and dispersed by pipetting. When confluent, cells were trypsinized to produce two replica plates. Genomic DNA was prepared from one plate and digested with EcoRI using an “in well” procedure (Ramirez-Solis et al., 1992), before screening for homologous recombinants by Southern blotting. The second plate was stored at −70°C using 10% dimethyl sulfoxide as a cryoprotectant. Clones heterozygous at the talin locus were taken through a second round of electroporation using a targeting construct containing the Hyg selection marker to inactivate the remaining talin allele. Essentially identical methods were used to target the vinculin gene in ES cells, except that the construct used lacked the thymidine kinase gene (TK) negative selection marker, and therefore gancyclovir was not added to the culture medium.
Western Blotting
Confluent dishes (9 cm) of ES cells were washed twice with PBS, the cells scraped into 0.5 ml lysis buffer (2% SDS in 75 mM Tris-HCl, pH 6.8, containing 10% glycerol, and 0.1 mM PMSF) and the sample heated at 100°C for 10 min. Aliquots of cell lysate containing equivalent amounts of protein were subjected to SDS-PAGE (7% gels) and the proteins electroblotted to Hybond C membranes using a semi-dry blot apparatus (Bio-Rad Laboratories). Excess protein binding sites were blocked by incubation in 5% dried milk in Tris-buffered saline, pH 7.4. Talin was detected using a mouse monoclonal antibody TD77 raised against human platelet talin (Bolton et al., 1997) and diluted 1:10,000 in blocking buffer. Vinculin was detected using the mouse monoclonal antibody F9 diluted 1:400 (a generous gift from Dr. V. Koteliansky, CNRS Ecole Normal Superieure, Paris). Antibodies to paxillin and the β3 integrin subunit were purchased from Transduction Laboratories (Lexington, KY) and antibodies to β1, αV, and α5 integrin subunits were purchased from Chemicon (Harrow, London, UK). An antibody to mouse pp125FAK was prepared by immunizing rabbits with a recombinant polypeptide derived from the COOH-terminal region of the protein. A rabbit polyclonal antibody raised against human platelet VASP was a generous gift from Dr. M. Reinhard (Medizinische Universitatsklinik, Wurzburg, Germany). Bound primary antibody was detected by adding anti–mouse or anti–rabbit antibodies coupled to HRP (1:3,500) in conjunction with an ECL kit (Nycomed Amersham Inc.).
Adhesion Assay
96-well Nunc-Immunoplates (MaxiSorp 439454A) were coated with either 25–50 μg /ml laminin or fibronectin (Sigma Chemical Co., St. Louis, MO) overnight at 4°C, and the wells washed three times with PBS. Excess protein-binding sites were blocked with 2% BSA (2 h at room temperature), and the wells washed with PBS. ES cells were trypsinized, the trypsin neutralized by addition of complete medium, and cells suspended (8 × 105/ ml) in serum-free medium. 100 μl of cell suspension was added to each well and the cells allowed to attach for 60 min at 37°C. Nonadherent cells were removed by washing 3× with PBS, and the attached cells were fixed with 10% formaldehyde (30 min) and stained with 1% toluidene blue for 60 min. The plates were washed extensively with water, air dried and the dye extracted with 2% SDS and the absorbance read at 630 nm using a microtiter plate reader. The assays were carried out in triplicate, and each experiment was repeated three times.
Immunofluorescence
ES cells cultured on glass coverslips coated with bovine fibronectin (50 μg/ml) for 24–48 h, were washed with PBS and fixed in 3.8% formaldehyde in PBS for 10 min at room temperature. Fixed cells were permeabilized with 0.2% Triton X-100 in PBS for 30 s, and incubated with 1% BSA in PBS for 1 h before staining for either talin or vinculin using mouse monoclonal antibodies TD77 and F9 respectively (1:50), and a Texas red– labeled anti–mouse Ig (Nycomed Amersham Inc.), all diluted in 0.2% BSA in PBS. Cells were counterstained for F-actin using FITC phalloidin (Sigma Chemical Co.) as per manufacturer's instructions.
Time-lapse Recording and Analysis of ES Cell Behavior Using Interference Microscopy
The phenotype of isolated, undifferentiated, ES cells cultured on fibronectin-coated coverslips was analyzed using the DRIMAPS method of interference microscopy for recording cell behavior (Dunn et al., 1997). This system produces digital images in which the intensity at each point is directly proportional to the mass of nonaqueous cellular material in the light path. Time-lapse recordings of wild-type cultures and of the two mutant types were made over 20-h periods at a lapse interval of 1 min. Individual cells in each sequence of images were next identified by tracking the cells with a mouse-operated cursor during playback of the sequence. Computer processing then yielded for each cell in each frame, its dry mass, its spread area, the coordinates of its mass centroid (center of gravity) and two parameters, elongation and dispersion, describing cell shape (Dunn and Brown, 1986). Comparison of consecutive frames yielded further parameters describing the protrusion and retraction of the cell margin. Protrusion is defined as the region of the substratum newly covered by the cell during the frame interval whereas retraction is the region from which the cell withdraws during the interval. The areas, positions of the area centroid and changes in dry mass of these regions were measured. For analysis of variance (ANOVA), data from each cell were partitioned into 1-h runs, discarding any remainder, and mean values of parameters were then calculated for each run. These means were then nested according to cells, cultures and genotypes and the significances of any differences between the genotypes were assessed using a nested ANOVA which prevents false significances arising due to differences between cells or between cultures (Dunn et al., 1997). Spectral analysis of rapid variations in protrusion and retraction was performed using the Time Series Pack issued as a supplement to the MathematicaR package. Covariances were obtained from runs of data up to a lag of 50 min. Then the covariances for all runs of one genotype were pooled by weighting according to run length and the smoothed power spectrum was obtained using the Tukey-Hanning lag window of width 50 (Dunn et al., 1997). Recordings from 16 wild-type cultures, 9 cultures of the talin (−/−) A28 mutant cells and 10 cultures of the vinculin (−/−) D7 mutant cells were analyzed. A total of 1,005 isolated cells were tracked and these yielded 3848 one hour runs.
ES Cell Differentiation
24-well tissue culture plates were coated with of 0.1% poly(2-hydroxyethylmethacrylate) (ICN) in 95% ethanol, and air dried at 37°C. 5 × 104 ES cells in 1 ml of complete medium plus LIF were added per well, and the following day, a further 1 ml of medium minus LIF was added to each well. On day three most of the medium was carefully aspirated from the wells while tilting the plate to allow the embryoid bodies to sink to the front of the well and fresh medium (1 ml) minus LIF was added to each well. On alternate days, the cells were either fed by addition of 1 ml of fresh medium (minus LIF) or the medium was replaced.
Results
The Use of Neo and Hyg Replacement Vectors to Target Both Copies of the Talin Gene in Mouse ES Cells
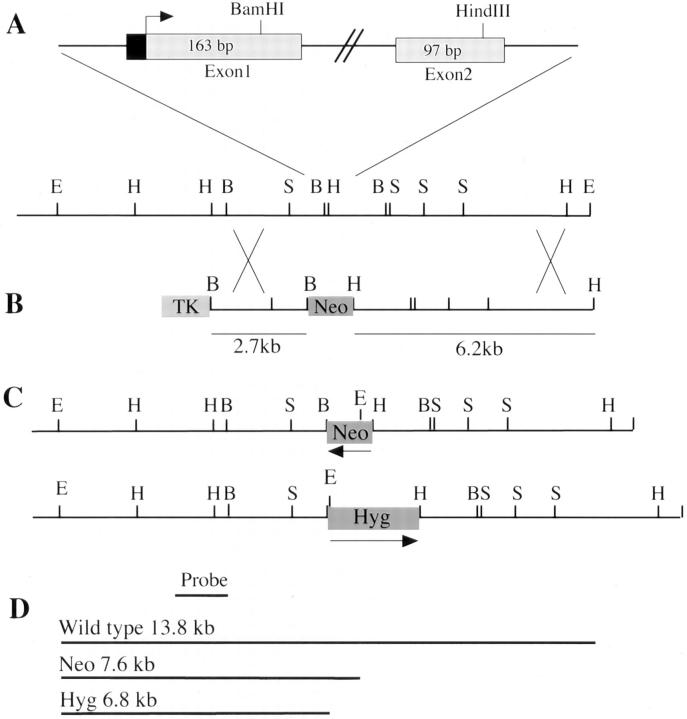
A mouse talin genomic clone (5Tλ2) was analyzed by restriction enzyme digestion and Southern blotting using oligonucleotide probes from the 5′ end of the talin cDNA. Sequencing of genomic fragments hybridizing to these probes allowed the boundaries of the first two coding exons to be identified (Fig. 1 A). The first coding exon is 163 bp long and contains 127 bp of coding sequence. Codon 44 is split by an intron between coding exons 1 and 2. The second coding exon contains 97 bp ending at codon 76. A 2.4-kb BamHI fragment containing the 5′ end of the first coding exon and a 6.2-kb HindIII fragment containing the 3′ end of the second coding exon were cloned either side of the Neo gene in the vector pX53, which also contains a TK negative selection marker (Fig. 1 B). Homologous recombination of this construct with the talin gene should lead to deletion of codons 37–66 and the fusion of the Neo gene with the residual parts of coding exons 1 and 2.
Figure 1.
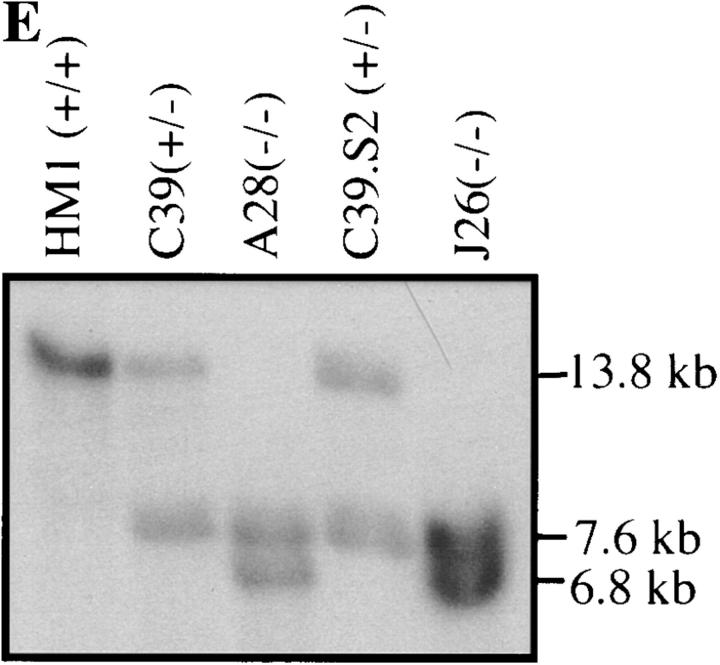
Targeting of the talin locus in ES cells. (A) Gene structure of talin locus around the targeting site. The position of the first two coding exons of talin is shown in relation to a restriction map of the mouse ES cell talin genomic DNA in that region. Exons are represented by rectangles (black being untranslated and grey being coding) and introns as solid lines. The arrow indicates the translation start. (B) Talin targeting construct with Neo replacing the sequence between the BamHI and HindIII sites found in exons 1 and 2, respectively. (C) Targeted talin alleles showing the replacement of BamHI–HindIII fragment with either Neo or Hyg and the location of the newly introduced EcoRI (E) sites. (D) Expected fragment sizes and positions when genomic DNA from either wild-type, Neo targeted, or Hyg targeted ES cells is digested with EcoRI and probed with the 5′ external probe shown. (E) Southern blots of targeted ES cell clones. EcoRI digested genomic DNA from wild-type ES cells (HM1), talin (+/−) ES cells with one allele targeted by the Neo vector (C39), and talin (−/−) ES cells with one allele targeted by the Neo vector, the other by the Hyg vector (A28 and J26). The clone C39.S2 which is (+/−) at the talin locus was isolated during attempts to inactivate the second allele with the Hyg targeting vector, and is a useful control for any phenotypic changes which might arise during Hyg selection. E, EcoRI; H, HindIII; B, BamHI; S, SacI.
The construct was linearized with NotI and electroporated into 1 × 108 ES cells (clone HM1). G418 was used to select for cells containing the plasmid, and gancyclovir was used to select against cells in which the construct had integrated randomly. Resistant colonies were transferred to four 96-well microtiter plates. The cells were then grown to confluence, replica plated, and genomic DNA from one set was analyzed by Southern blotting. The wild-type talin gene gives rise to a 13.8-kb EcoRI fragment that is detected by the probe indicated, whereas the Neo targeted allele gives rise to a novel 7.6-kb fragment due the presence of an EcoRI site within the Neo gene (Fig. 1, C and D). The result for one clone (C39) which is heterozygous at the talin locus is shown in Fig. 1 E. Gancylcovir lead to a twofold enrichment in homologous recombinants, and the talin gene was targeted in ∼1 in 3 of the G418/gancyclovir-resistant clones. To inactivate the second talin allele, an equivalent targeting vector was constructed in which the Neo gene was replaced by a Hyg selection marker. This was electroporated into the talin (+/−) ES cell clone C39, and EcoRI digests of genomic DNA from 384 hygromycin resistant clones screened for targeting of the second allele. The targeted allele should give rise to a novel 6.8-kb fragment, with corresponding loss of the remaining wild-type 13.8-kb fragment (Fig. 1, C and D). Only 1 clone (A28) was obtained which displayed this genotype (Fig. 1 E). This may be because the talin null cells adhere only weakly to gelatin-coated plates (see Fig. 3), and are lost during the selection procedures. The talin (+/−) ES cell clone C39 was used in a second experiment with the Hyg targeting vector, and an additional clone (J26) in which both talin alleles had been disrupted was isolated (Fig. 1 E).
Figure 3.
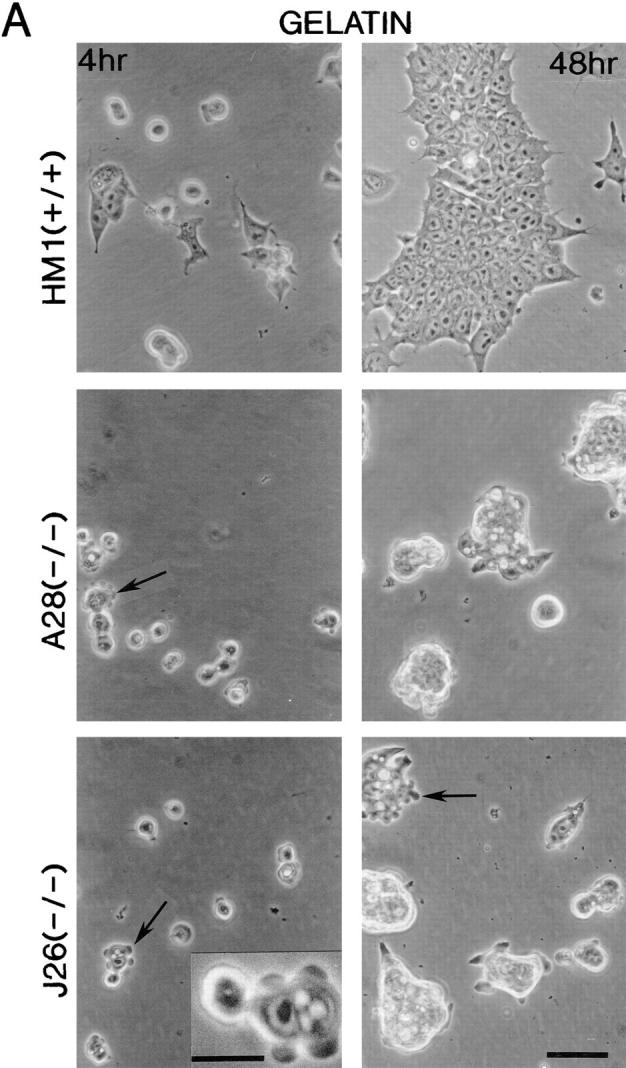
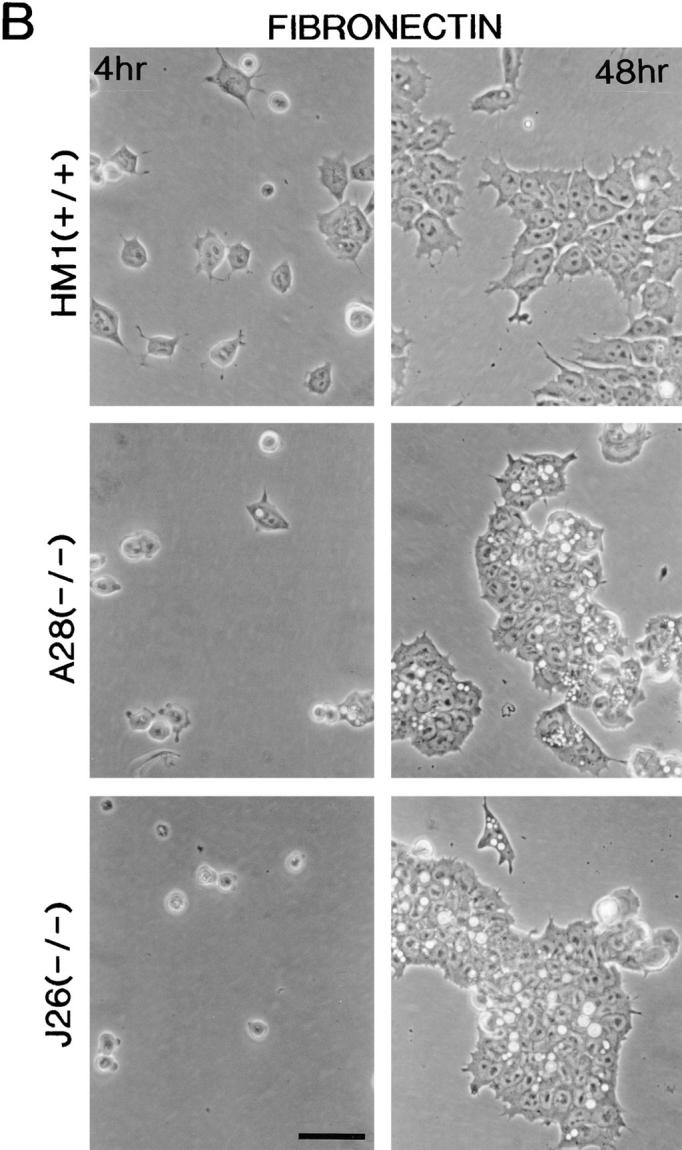
Morphology of the talin ES cell mutants plated on gelatin and fibronectin. Wild-type ES cells (clone HM1) and the two talin (−/−) ES cell lines A28 and J26, were trypsinized to obtain a single cell suspension and plated onto gelatin-coated (A) or fibronectin-coated (B) tissue culture plastic. These cells were then photographed after 4 and 48 h. Both talin (−/−) ES cell mutants spread more slowly than wild-type ES cells on gelatin and fibronectin (4 h time point), and both showed evidence of extensive membrane blebbing on gelatin (see arrows, and inset for high power view) and on fibronectin. After 48 h on gelatin, both talin (−/−) ES cell mutants formed colonies with a rounded, refractile appearance which was quite distinct from the well spread morphology of the colonies formed by wild-type ES cells. On fibronectin, the talin (−/−) ES cell mutants behaved much more like the wild-type ES cells, although the colonies formed were still less well spread than those formed by wild-type cells. Note the macropinocytic vesicles that occur at high frequency in the talin (−/−) ES cell mutants on both gelatin and fibronectin. Bars: (A and B) 50 μm; (inset) 20 μm.
Western Blot Analysis of ES Cell Talin Mutants
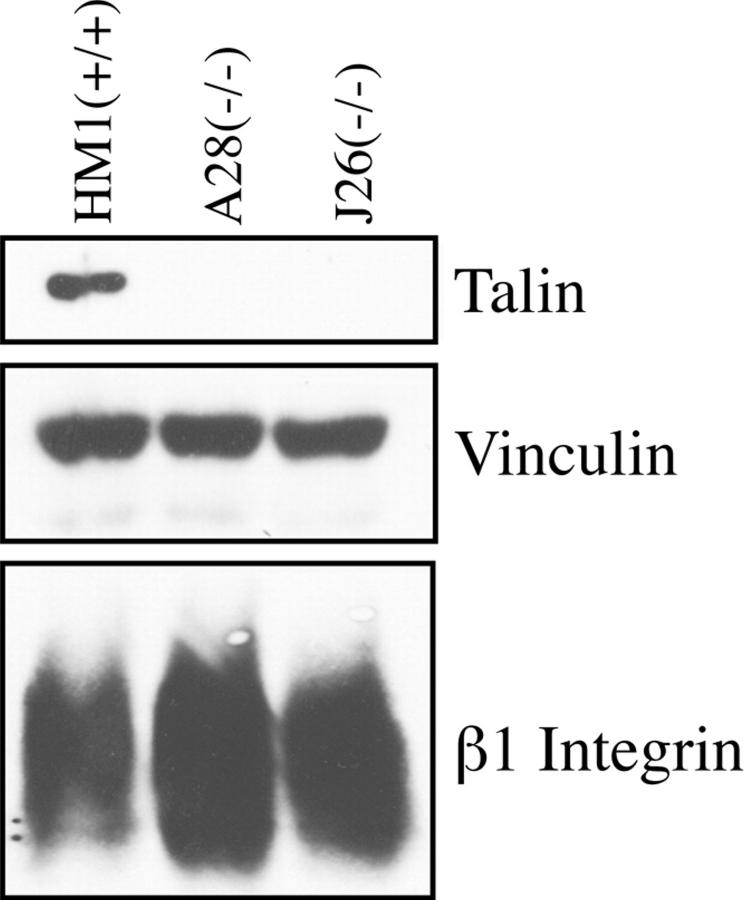
The expression of talin in these various ES cell clones, grown in the presence of LIF to suppress differentiation, was analyzed by Western blotting using a monoclonal antibody (TD77) raised against human platelet talin. This antibody, which recognizes an epitope at the extreme COOH-terminal region (residues 2464–2541) of talin (Bolton et al., 1997), detected a single talin polypeptide in the wild-type ES cells. The talin (+/−) ES cell clone C39 showed a significant reduction in talin immunoreactivity. However, these cells expressed two talin polypeptides, one which comigrated with the wild-type protein, the other migrating with slightly increased mobility (Fig. 2, A and B). The talin (−/−) ES cell mutants A28 and J26 showed almost complete loss of talin immunoreactivity (Fig. 2, A and B), although prolonged exposure of the blots showed that clone A28 (Fig. 2 A) and J26 (data not shown) expressed very low levels of a truncated talin polypeptide that comigrated with the truncated talin polypeptide expressed in the (+/−) C39 ES cell line. The results clearly establish that both the talin (−/−) A28 and J26 ES cell mutants express no intact talin. The levels of two other cytoskeletal proteins associated with the cytoplasmic face of focal adhesions, vinculin and α-actinin, were also somewhat reduced in the talin (−/−) A28 mutant, although the levels of paxillin and pp125FAK were similar to those in wild-type cells (Fig. 2 A). There was also a dramatic reduction in the levels of the β1 integrin subunit in both the talin (−/−) ES cell mutants (Fig. 2, A and B) whereas the levels of both α5 and αV integrin subunits were comparable to those in wild-type ES cells. The results show that loss of talin is associated with a reduction in the levels of a number of other focal adhesion proteins, most notably the β1 integrin subunit.
Figure 2.

Western blot analysis of talin ES cell mutants. (A) 100 μg of total cellular protein from wild-type ES cells (HM1) and talin ES cell mutants, C39 (+/−) and A28 (−/−) was Western blotted onto Hybond C and probed for talin, vinculin, α-actinin, paxillin, pp125 focal adhesion kinase (FAK), β1 integrin, α5 integrin, and αV integrin. The panel labeled Talin* is a long exposure of a blot probed for talin. The β1, αV, and α5 integrin subunits were detected in nonreducing gels. (B) A second Western blot with similar amounts of total cellular protein from the above cell lines plus a second talin (−/−) ES line (J26), confirms that the reduction in β1 integrin levels is characteristic of talin (−/−) ES cell lines.
Morphology of the Undifferentiated Talin (−/−) ES Cell Mutants
Undifferentiated wild-type ES cells and the two talin (−/−) ES cell mutants were trypsinized and plated onto gelatin-coated tissue culture plates in the presence of LIF, and their morphology inspected after 4 or 48 h by phase contrast microscopy. After 4 h, many of the wild-type cells had begun to spread, whereas both of the talin (−/−) ES cell mutants remained rounded with evidence of extensive surface protrusions (Fig. 3 A). After 48 h, the wild-type ES cells formed large colonies in which the cells were well spread. In contrast, both the talin (−/−) ES cell mutants formed rounded colonies that were only loosely attached to the substrate, and contained very few spread cells. A notable feature of both talin (−/−) ES cell mutants was the higher frequency of macropinocytic vesicles compared with wild-type cells (Fig. 3 A). Whereas the talin (−/−) ES cell mutants also showed surface protrusions and macropinocytic vesicles when plated on fibronectin, they were able to spread, although spreading occurred more slowly, and was somewhat less extensive than the wild-type ES cells (Fig. 3 B). The morphology of the talin (−/−) ES cell mutants plated on laminin was similar to that on gelatin with a marked reduction in cell spreading compared with wild-type ES cells (data not shown).
Talin (−/−) ES Cell Mutants Show Reduced Adhesion to Laminin but Not Fibronectin
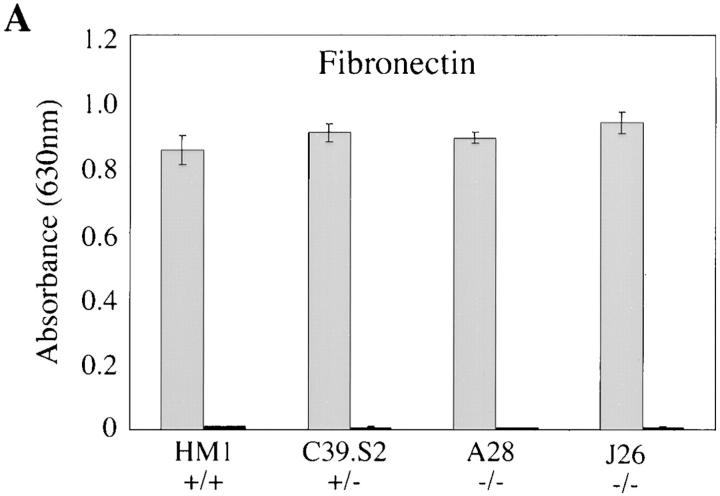
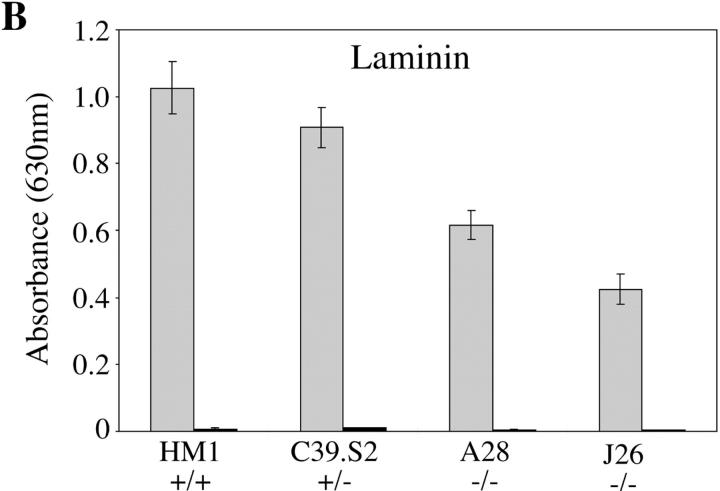
The ability of wild-type ES cells, and the various talin ES cell mutants to adhere to fibronectin and laminin is shown in Fig. 4. Wild-type ES cells adhered to both fibronectin and laminin to about the same degree over a 1-h period, whereas adhesion to gelatin or collagen was very low (OD630 < 0.1). Adhesion was abolished in the presence of 10 mM EDTA demonstrating a requirement for divalent cations, a characteristic of integrin-mediated adhesion. Although both talin (−/−) ES cell mutants expressed much reduced levels of β1 integrin (Fig. 2), their adhesion to fibronectin was identical to that of wild-type ES cells. In contrast, they showed a significant reduction in adhesion to laminin. The reduced adhesion of the talin (−/−) ES cell mutants to laminin is consistent with the fact that they spread less well on laminin than fibronectin.
Figure 4.
Adhesive properties of the talin (−/−) ES cell mutants. Wild-type ES cells (HM1, +/+) and the talin ES cell mutants C39.S2 (+/−) A28 (−/−) and J26 (−/−) were trypsinized to single cell suspension and then plated onto fibronectin (A) or laminin (B) coated wells for 1 h either in the presence (black bars) or absence (gray bars) of 10 mM EDTA. Cells that did not adhere in this time were washed off, and the number of adherent cells quantitated by staining with toluidine blue followed by the extraction of bound dye. Absorbance was read at 630 nm. The results are the mean of three separate experiments conducted in triplicate. Bars represent standard errors.
Undifferentiated Talin (−/−) ES Cell Mutants Fail to Form Focal Adhesions on Fibronectin
Undifferentiated wild-type ES cells (HM1) and the talin (−/−) A28 ES cell mutant grown in the presence of LIF were plated on coverslips coated with fibronectin and analyzed for their ability to form vinculin-containing focal adhesions and associated actin stress fibers after 48 h (Fig. 5). Wild-type ES cells adopted a well-spread morphology and assembled numerous actin stress fibers that terminated in focal adhesions that stained for talin, vinculin, and paxillin. In contrast, the talin (−/−) A28 ES cell mutant formed large rounded colonies with few if any spread cells. They failed to assemble any actin stress fibers, and actin staining was limited to the cell margins with diffuse cytoplasmic staining. There was no evidence of talin localized in focal adhesions, although the cells showed some diffuse staining for talin. Similarly, vinculin and paxillin staining was diffuse with only occasional punctate staining for vinculin at the margins of the cell aggregates. The results for the J26 talin (−/−) ES cell mutant were identical (data not shown). These findings establish that loss of talin compromises the ability of undifferentiated mouse ES cells to form focal adhesions and associated actin stress fibers on fibronectin, although talin is not required to support integrin-mediated adhesion to fibronectin.
Figure 5.

Localization of talin, vinculin, paxillin, and F-actin in undifferentiated talin (−/−) ES cell mutants by immunofluoresence. Wild-type (+/+) HM1 ES cells and the talin (−/−) A28 ES cell mutant were seeded onto fibronectin-coated glass coverslips. After 48 h, the cells were stained for talin and F-actin, vinculin and F-actin or paxillin and F-actin. Wild-type ES cells form focal adhesions containing talin, vinculin and associated actin filaments. The talin (−/−) A28 mutant failed to assemble vinculin or paxillin-containing focal adhesions and lacked actin stress fibers. Bar, 20 μm.
Phenotypic Properties of the Vinculin (−/−) D7 ES Cell Mutant
We have used a similar strategy to generate ES cells in which both copies of the vinculin gene have been disrupted (Fig. 6, A and B). The vinculin (−/−) D7 ES cell line expressed no intact vinculin (Fig. 6 C), although low levels of a truncated vinculin polypeptide could be detected on prolonged exposure of the Western blots. However, the levels of three vinculin-binding proteins, talin, α-actinin, and VASP (Fig. 6 C) as well as β1-integrin (data not shown) were comparable to those in wild-type ES cells. The vinculin (−/−) D7 ES cell line showed a tendency to form rounded colonies when grown under standard culture conditions on gelatin-coated tissue culture dishes, although on fibronectin, their morphology was similar to that of wild-type ES cells. These results are similar to those with the talin (−/−) ES cell mutants that also formed rounded colonies on gelatin, but were able to spread on fibronectin.
Figure 6.
Generation and analysis of ES cell lines with disrupted vinculin alleles. (A) Targeting of the vinculin gene. On the top line is a schematic of the gene structure of vinculin with the introns as lines and exons as numbered boxes. The second line represents the generic vinculin positive/negative targeting vector where a Neo or Hyg selectable marker has been inserted into the unique MscI (M) site in exon 5. The bottom part represents the resulting targeted alleles with either Neo or Hyg inserted into the MscI site of exon 5. ES cell clones were screened by Southern blotting using EcoRI digests that were probed with the 5′ probe indicated. The wild-type band detected is 7.0-kb whereas targeted alleles containing either Neo or Hyg give rise to bands of 6.5 kb. E, EcoRI; B, BamHI; M, MscI; X, XbaI. An example of such a Southern blot is shown in B. (C) Western blot analysis of wild-type ES cells (HM1), and vinculin ES cell mutants ES2 (+/−) and D7 (−/−). (100 μg protein loaded per well). The blots were probed with antibodies to talin, vinculin, α-actinin and VASP. (D) Morphology of the vinculin (−/−) D7 ES cell mutant on gelatin and fibronectin coated dishes 48 h after plating. Note the more rounded morphology of the D7 colonies on gelatin. (E) Localization of talin and F-actin in wild-type ES cells (clone HM1) and the vinculin (−/−) D7 ES cell mutant by immunofluoresence. Bars: (D) 50 μm; (E) 5 μm.
To establish whether the vinculin (−/−) D7 ES cell mutant was able to form talin-containing focal adhesions, cells were plated on fibronectin-coated glass coverslips and stained for talin and F-actin. As expected, the wild-type ES cells formed prominent actin stress fibers that terminated in talin-containing focal adhesions (Fig. 6 E). Interestingly, the vinculin (−/−) D7 ES cell mutant retained the capacity to assemble prominent actin stress fibers, but these were invariably localized towards the outer cell margins. Talin was localized at the ends of these actin filaments, although the talin-containing structures were more elongated than in wild-type ES cells. Essentially identical results were obtained with a second ES cell clone (DK022) that also expressed no intact vinculin (data not shown). The results establish that vinculin is not essential for the assembly of talin-containing focal adhesions and associated actin stress fibers in undifferentiated ES cells, although the D7 ES cell clone that lacks vinculin, has a somewhat different actin cytoarchitecture compared with wild-type ES cells.
Analysis of the Behavior of Talin and Vinculin ES Cell Mutants
Wild-type ES cells and the talin (−/−) A28 and vinculin (−/−) D7 ES cell mutants were plated on fibronectin-coated coverslips and their behavior recorded over a period of 20 h using interference microscopy. The rate of increase in dry mass of individual cells served as a check on the viability of the cultures. All cultures grew at a mean rate of just over 5% h−1 with no significant differences due to genotype in an ANOVA test. Cell spreading and cell shape are all determined by the patterns of protrusion and retraction of the cell margin (Dunn et al., 1997) and, therefore, we examined these patterns directly. Fig. 7, a–c shows a sample of protrusions (green) and retractions (red) for cells of the three genotypes during a 2-min interval. Compared with the wild-type ES cells (Fig. 7 a), the talin (−/−) A28 ES cell mutants (Fig. 7 b) showed large bleb-like protrusions evenly interspersed with retractions around the cell periphery, whereas the vinculin (−/−) D7 ES cell mutant (Fig. 7 c) showed greater polarity, with the central cell displaying a large lamellar protrusion opposite an extensive retracting margin. Fig. 7 d is a summary of measurements of the mean polarity which we have previously defined as the distance in μm separating the centroids of protrusion and retraction over a 5-min period (Dunn et al., 1997). In this plot, the solid discs represent the median values and the distributions of the data are indicated by the rectangles that span 50% of data values, and the “bars” that span 80% of values. In ANOVA tests, the polarity of the talin (−/−) A28 ES cell mutant was significantly suppressed compared with wild-type ES cells (P < 0.05) whereas the polarity of the vinculin (−/−) D7 ES cell mutant was significantly increased (P < 0.01).
Figure 7.
Analysis of wild-type ES cells and the talin and vinculin mutants by time lapse video interference microscopy. (a) Wild-type ES cells (b) the talin (−/−) A28 ES cell mutant and (c) the vinculin (−/−) D7 ES cell mutant taken from DRIMAPS recordings processed to reveal the regions of protrusion (green) and retraction (red) of the cell margin during a 2-min interval. The grey levels inside the cells represent the dry mass distribution on an arbitrary scale. (d). Median values of cell polarity measured as the distance in μm between the centroids of the protrusion and retraction areas over a 5-min period. The rectangles and bars indicate the distributions of data values. (e). Power spectra of the variations in protrusion of the wild-type ES cells (black), talin (−/−) A28 ES cell mutant (red) and the vinculin (−/−) D7 ES cell mutant cells (green). The power (vertical axis) in arbitrary units reveals how much of the variance in the data is attributable to each frequency. Bars: (A–C) 20 μm.
To quantify the rapid marginal blebbing observed in recordings of the talin (−/−) A28 ES cell mutant, we performed a spectral analysis of the variations in protrusion. Compared with other protrusions, blebs tend to have small areas but high mass density, and the parameter we eventually chose for detecting rapid blebbing was therefore the areal mass density of the protrusion regions measured in units of pg/μm2. The power spectrum of variations in protrusion density for the three genotypes is shown in Fig. 7 e. A single strong peak in any of these three spectra would indicate a significant periodicity in the marginal activity of the corresponding cells. In fact there are several weak peaks in all three traces though their height in each case is only ∼20–30% of the total height of the spectrum. These weak peaks indicate that there are multiple minor periodicities in the marginal activity at different frequencies, but the net result is much closer to a random sequence than to a periodic one. It is clear from the spectra that the power of this random marginal activity is about seven times greater in the talin (−/−) A28 ES cell mutant than in the wild-type ES cells, whereas the vinculin (−/−) D7 mutant showed only about half the power of the wild-type.
Talin (−/−) ES Cell Mutants Form Embryoid Bodies, but Extensive Morphological Differentiation Is Inhibited
Both wild-type ES cells and the talin (−/−) A28 ES cell mutant form embryoid bodies of a similar size and appearance when grown in the absence of LIF for 8 d (Fig. 8). This is not unexpected as the process is dependent on cadherin-mediated cell–cell interactions (Larue et al., 1996), and talin is specifically localized to integrin-containing cell–matrix junctions (Geiger et al., 1985). When wild-type embryoid bodies were plated on gelatin-coated tissue culture dishes, cells spread out from the central cell mass as a continuous sheet over a period of 24–48 h (Fig. 8 A), and formed an extensive cell monolayer within 5 d. In contrast, only a few cells had begun to spread out from the margins of the embryoid bodies formed by the talin (−/−) A28 ES cell mutants after 24 h, and those cells that did migrate over a 48-h period migrated as individual cells rather than as a continuous cell sheet (Fig. 8 A). The leading edge of the migrating cells from the wild-type embryoid bodies always contained giant cells (Fig. 8 A, arrowheads). These were never seen in the cells emerging from the talin (−/−) A28 embryoid bodies. After 7 d on gelatin, wild-type embryoid bodies gave rise to a variety of cell types with distinct morphologies, including beating cardiomyocytes and giant cells (Fig. 8 B, arrowheads), as well as organized structures (Fig. 8 B, arrows). In contrast, the talin (−/−) A28 embryoid bodies typically gave rise to only two morphologically distinct cell types, and no organized structures were observed (Fig. 8 B). Essentially identical results were obtained with the talin (−/−) J26 ES cell mutant (data not shown). Deletion of β1-integrins in mouse F9 teratocarcinoma cells has also been reported to block morphological differentiation (Stephens et al., 1993).
Figure 8.
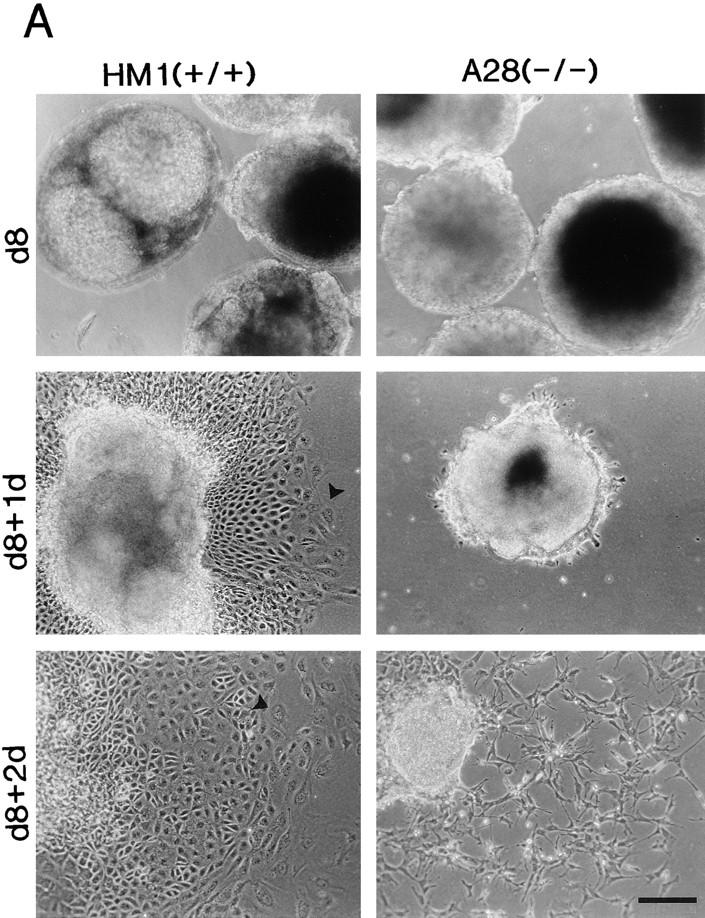
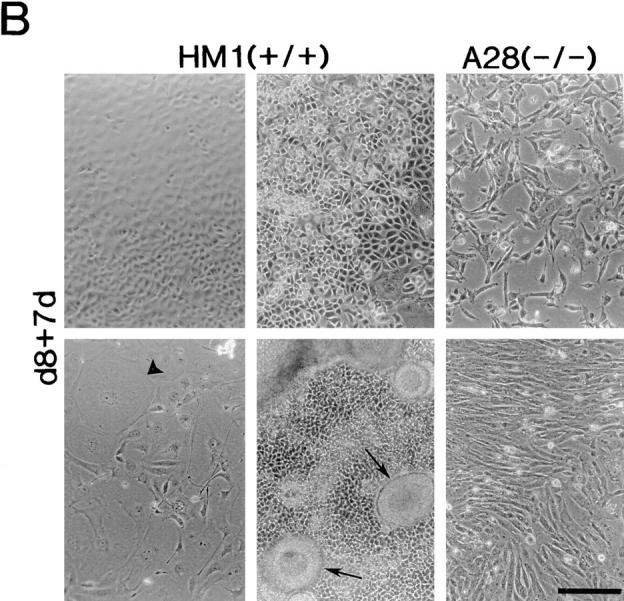
Differentiation capacity of talin (−/−) A28 ES cells. (A) Wild-type ES cells (HM1) and talin (−/−) A28 ES cells were used to generate embryoid bodies that were cultured in the absence of LIF for 8 d, and then were plated onto gelatin-coated tissue culture dishes. Embryoid bodies after 8 d in culture in the absence of LIF (d8), d8 embryoid bodies 1 d after plating onto gelatin-coated dishes (d8+1d), d8 embryoid bodies 2 d after plating onto gelatin-coated (d8+2d). (B) Examples of some of the morphologically distinct cell types (including giant cells; arrowheads) and structures (arrows) which were observed when d8 wild-type HM1 embryoid bodies were cultured on gelatin-coated plates for 7 d (d8+7d). The two right hand panels show the only two morphologically distinct cell types observed when the talin (−/−) A28 d8 embryoid bodies were cultured under identical conditions. Bars, 200 μm.
Differentiated Talin (−/−) ES Cell Mutants Can Form Focal Adhesion-like Structures and Express Normal Levels of β1 Integrin
Because the differentiated talin (−/−) A28 and J26 ES cells that migrate out of embryoid bodies are able to adopt a spread morphology on gelatin-coated dishes, we investigated whether these cells could form focal adhesions and actin stress fibers when plated on fibronectin. The differentiated cells derived from wild-type embryoid bodies showed a variety of morphologies, and the organization of the actin cytoarchitecture within these cells took on a variety of forms. However, in all cases there was prominent staining for talin, vinculin and paxillin at the ends of actin filaments (Fig. 9). Interestingly, the differentiated talin (−/−) A28 cells also contained actin filaments, and in some cells, the ends of these filaments showed staining for vinculin and paxillin, but not talin. Identical results were obtained with the talin (−/−) J26 ES cell mutant (data not shown).
Figure 9.

Localization of talin, vinculin, paxillin, and F-actin in differentiated talin (−/−) ES cell mutants by immunofluoresence. Cells derived from the embryoid bodies formed by wild-type (+/+) ES cells (HM1) and the talin (−/−) A28 ES cell mutant were seeded onto fibronectin-coated glass coverslips. After 48 h, the cells were stained for talin and F-actin, vinculin and F-actin or paxillin and F-actin. Differentiated HM1 cells displayed a variety of morphologies, but all showed strong staining for talin, vinculin and paxillin in focal adhesions at the ends of actin filaments. The differentiated talin (−/−) ES cell mutants adopted a spread morphology with clear evidence of vinculin and paxillin-containing focal adhesion-like structures at the ends of actin filaments. However, the structures formed were less extensive than those seen in wild-type ES cells. Bar, 20 μm.
Western blot analysis of both the differentiated talin (−/−) ES cell mutants showed that although these cells still expressed no intact talin, the levels of β1 integrin subunit as well as vinculin were comparable with those in differentiated wild-type ES cells (Fig. 10). Therefore, it is apparent that differentiation of the talin (−/−) ES cell mutants in vitro generates cells in which neither β1 integrin subunit levels or the assembly of focal adhesions and associated actin filaments is dependent on talin.
Figure 10.
Western blot analysis of differentiated ES cells. 100 μg of total cellular protein from d8 embryoid bodies made from wild-type (HM1) ES cells or talin (−/−) mutant ES cells (A28 and J26) was resolved by SDS-PAGE, blotted onto Hybond C and probed for talin, vinculin, and β1 integrin.
Discussion
We have used gene disruption technology to isolate two talin (−/−) ES cell mutants (A28 and J26) that contain no intact talin, although both express low levels of a truncated talin polypeptide. The most notable features of the undifferentiated talin (−/−) ES cell mutants are extensive membrane blebbing, an accumulation of macropinocytic vesicles and an inability to spread on gelatin or laminin on which the cells grow as loosely attached colonies. Adhesion to laminin was significantly reduced, but adhesion to fibronectin was unaffected despite the fact that the cells showed a dramatic reduction in levels of the β1 integrin subunit. However, the talin (−/−) ES cell mutants were unable to assemble focal adhesions or associated actin stress fibers on fibronectin-coated coverslips, whereas a vinculin (−/−) mutant (D7) was able to do so. These results provide compelling evidence that talin plays a crucial role in the assembly of focal adhesions in undifferentiated ES cells. Interestingly, the talin (−/−) ES cell mutants were able to form apparently normal embryoid bodies, but when these were cultured on gelatin, cell migration from the central cell mass was much slower than wild-type, and differentiation was limited to just two morphologically distinct cell types. These expressed normal levels of β1 integrin, and were able to spread and assemble focal adhesion-like structures with associated actin filaments on fibronectin. These results raise the possibility that loss of talin restricts ES cell differentiation to a subset of cell types in which β1-integrin levels and focal adhesion assembly are talin independent.
The increase in membrane protrusions or blebs shown by the undifferentiated talin (−/−) ES cell mutants was observed in cells plated on both gelatin and fibronectin. Whether this reflects a failure of the cells to develop stable adhesions with the extracellular matrix, or is due to membrane instability per se remains to be established. Increased membrane blebbing activity has been noted in melanoma cells lacking the actin-binding protein ABP-280 (Cunningham et al., 1992), and cleavage of the actin-binding proteins talin, α-actinin (Miyoshi et al., 1996), and fodrin (Vanaga et al., 1996) has been implicated in membrane blebbing in apoptosis. Myosin light chain phosphorylation and activation of actomyosin contraction has also been shown to be involved in the membrane blebbing associated with apoptosis in PC12 cells (Mills et al., 1998). The authors speculate that actomyosin contraction in cells that lack the structural proteins that normally couple cortical actin to the plasma membrane may lead to cytoplasmic extrusion and membrane blebbing. Despite the much increased membrane blebbing activity displayed by the talin (−/−) ES cell mutants, there was no evidence of an increase in apoptosis, and they showed no obvious difference in growth rate compared with wild-type ES cells (data not shown).
The fact that the undifferentiated talin (−/−) ES cell mutants grew as poorly spread colonies on gelatin- and laminin-coated substrates, and showed reduced adhesion to laminin, is probably linked to the dramatic reduction in β1 integrin subunit expression, although we have not analyzed the levels of β1 integrins on the cell surface. Laminin receptors are all members of the β1 integrin family (Newham and Humphries, 1996), and reduced adhesion of the talin (−/−) ES cell mutants to laminin is not unexpected therefore. Why adhesion to fibronectin was not reduced is unclear as it has been reported that ES cells lacking the β1 integrin subunit fail to adhere to both laminin and fibronectin (Fassler et al., 1995). ES cells express the αV integrin subunit (see Fig. 2 and Fassler et al., 1995), and those used in this study also expressed a protein of 90 kD that reacted with an anti-β3 integrin antibody (data not shown). αVβ3 can act as a receptor for fibronectin, and may compensate for the effects of any reduction in surface expression β1 integrins in the talin (−/−) ES cell mutants. However, others have failed to detect β3 integrin in ES cells, although β1 integrin (−/−) ES cell mutants were able to adhere to vitronectin (Fassler et al., 1995). Although loss of talin did not affect adhesion of the undifferentiated talin (−/−) ES cell mutants to fibronectin, the rate of cell spreading was reduced, and the cells were completely unable to assemble vinculin or paxillin-containing focal adhesions or actin stress fibers. We conclude that talin plays a key role in the assembly of focal adhesions and the organization of the actin cytoarchitecture in undifferentiated ES cells. However, the fact that β1 integrin subunit levels were dramatically reduced in both talin (−/−) ES cell lines suggests that loss of talin may compromise focal adhesion assembly by more than one mechanism.
A number of observations suggest that talin is important in the early stages of cell adhesion and spreading including the formation of filopodia and lamellipodia. In Dictyostelium discoidum, talin accumulates at the tips of filopodia formed in response to cAMP (Kreitmeier et al., 1995), and microscale chromophore-assisted laser inactivation of talin in neuronal growth cones results in temporary cessation of filopodial extensions (Sydor et al., 1996). In chick embryo fibroblasts, talin is found in membrane ruffles as well as focal adhesions (Burridge and Connell, 1983; Bolton et al., 1997), and it is recruited to nascent focal adhesions before vinculin (DePasquale and Izzard, 1991). Moreover, microinjection of antibodies to talin disrupt focal adhesions (Nuckolls et al., 1992; Bolton et al., 1997), and downregulation of talin expression in Hela cells slowed down the rate of cell spreading and lead to a reduction in the size of focal adhesions (Albiges-Rizo et al., 1995).
Analysis of the sequence of mouse (Rees et al., 1990) and chicken (Hemmings et al., 1996) talin (2541 amino acids), and the biochemical properties of the protein shows that it has a number of features that make it highly suited to act as a key element in the assembly of focal adhesions. Talin is a dimeric elongated flexible protein (Goldmann et al., 1994; Winkler et al., 1997) that binds β1- and β3-integrins (Horwitz et al., 1986; Tapley et al., 1989; Kenzevic et al., 1996; Pfaff et al., 1998), vinculin (Burridge and Mangeat, 1984; Gilmore et al., 1992, 1993), and F-actin (Muguruma et al., 1992; Hemmings et al., 1996; McCann and Craig, 1997), and is reported to possess actin nucleation (Niggli et al., 1994) and bundling activity (Zhang et al., 1996). The NH2-terminal 433 residues of talin, which can be liberated as a 47-kD polypeptide by calpain II, contain sequences that target the protein specifically to cell– matrix junctions (Nuckolls et al., 1990), and are capable of binding acidic phospholipids (Niggli et al., 1994). Interestingly, residues 165–363 show homology with the ERM family of proteins that also act as linkers between the membrane and the actin cytoskeleton (Vaheri et al., 1997). The 190-kD COOH-terminal talin polypeptide liberated by calpain II, which is composed of multiple α-helical alanine-rich repeats (McLachlan et al., 1994), contains the dimerization site (Winkler et al., 1997), three vinculin-binding sites (Gilmore et al., 1993), and several actin-binding sites (Hemmings et al., 1996) including an actin nucleation site (Niggli et al., 1994). Although the 190-kD talin polypeptide is reported to bind integrins (Horwitz et al., 1986), the binding site has yet to be defined. However, the finding that differentiated talin (−/−) ES cell can form focal adhesion-like structures, and express normal levels of the β1 integrin subunit, suggests that talin is not required for focal adhesion assembly in all cell types, and that there are alternative pathways for the assembly of such structures. The actin-binding proteins α-actinin (Otey et al., 1990, 1993) and filamin (Pfaff et al., 1998) have been shown to bind to the cytoplasmic face of β1 integrins, and may provide a mechanism for linking integrins to F-actin.
We have yet to investigate the mechanisms involved in the downregulation of β1 integrins in talin (−/−) ES cells. However, our findings are reminiscent of those of Albiges-Rizo et al. (1995), who used an antisense RNA construct to downregulate talin in Hela cells. Cells in which talin levels were reduced by 38–60% showed loss of immunofluorescence staining for β1 integrins, although there was no reduction in β1 integrin mRNA levels. Surface labeling studies showed that both α5 and β1 integrin subunits appeared on the cell surface in an abnormal form, suggesting that there was a defect in integrin processing. Loss of talin might compromise integrin trafficking at several levels. A number of actin-binding proteins have recently been shown to be localized in the Golgi apparatus (reviewed in Stow et al., 1998) including myosin I which supports transport of vesicles along actin filaments towards the barbed membrane-attached end (Fath et al., 1994). Although there is no evidence that talin is localized in the Golgi, the fact that loss of talin leads to a marked change in actin cytoarchitecture might result in the inhibition of vesicular transport of integrins via such a pathway. It is also conceivable that talin plays a more specific role in β1 integrin transport by binding transport vesicles containing integrins to actin filaments. The actin cytoskeleton and myosin I have also been implicated in endocytic pathways (Durrbach et al., 1996), and loss of talin may also effect the endocytic pathways thought to be important in integrin recycling (Bretscher, 1996). An alternative explanation for the down regulation of the β1 integrin subunit in the talin (−/−) ES cell mutants is suggested by recent evidence that poly(A)+ RNA and ribosomes are recruited to the focal adhesion complexes formed in response to binding of fibronectin-coated beads to the cell surface (Chicurel et al., 1998). The authors speculate that certain mRNAs may be translated in focal adhesions. Whether the mRNA encoding β1 integrin falls into this category remains to be investigated, but were this the case, loss of talin and the consequent inability of undifferentiated ES cells to assemble such structures might reduce translation of the β1 integrin mRNA.
The origin of the truncated talin polypeptide expressed by both talin (−/−) ES cell mutants has not yet been established. The Neo gene (unlike the Hyg gene) was cloned into the targeting vector in the opposite transcriptional orientation to the talin gene. As a consequence, the Neo polyadenylation signal and translation stop codons would not be expected to halt transcription or translation from this allele. Translational initiation at an internal in frame methionine (codon 79) downstream of the Neo sequence would lead to a talin polypeptide lacking the first 78 amino acids and differing in apparent molecular mass by 8,708 D. Such a deletion is consistent with the size of the truncated talin polypeptide expressed by the talin ES (−/−) mutants. Alternatively, the targeted coding exons 1 and 2 could be spliced out of the primary transcript to give a novel talin mRNA lacking codons 1–76. Translation could be initiated internally at methionine codon 79 or at any number of in frame downstream AUG's, albeit with low efficiency. Interestingly, a mouse F9 teratocarcinoma vinculin (−/−) mutant, which was originally null for vinculin protein, was found to express a truncated vinculin polypeptide after a few passages in culture due to alternative splicing of the exon containing the Neo gene (Coll et al., 1997). The possibility that such a truncated talin polypeptide might act as a dominant negative mutant seems unlikely as it is expressed at very low levels, and the talin (+/−) ES cell mutants that also express this polypeptide are essentially indistinguishable from wild-type ES cells.
Interestingly, the undifferentiated vinculin (−/−) D7 null mutant was able to assemble talin-containing focal adhesions and stress fibers, although the morphology of the structures formed was distinctly different from those in wild-type ES cells. Others have previously reported that vinculin is not essential to focal adhesion assembly. A mouse F9 teratocarcinoma vinculin (−/−) mutant (Volberg et al., 1995), as well as immortalized cells from vinculin (−/−) mouse embryos (Xu et al., 1998) were both able to assemble focal adhesions in the absence of vinculin, although microinjection of antibodies to vinculin disrupted focal adhesions in chick embryo fibroblasts (Westmeyer et al., 1990), and antisense RNA downregulation of vinculin in BALB/c 3T3 cells reduced the number and size of focal adhesions (Rodriguez Fernandez et al., 1993). The finding that vinculin, which can bind both talin (Gilmore et al., 1992) and F-actin (Huttelmaier et al., 1997) is not essential to focal adhesion assembly, and the fact that cells expressing little (Rodriguez Fernandez et al., 1992) or no vinculin (Coll et al., 1995) are more motile, suggests a role in stabilising focal adhesions rather than in assembly. The proline-rich region in vinculin contains a binding site for VASP (Brindle et al., 1996), a protein that can bind profilin (Reinhard et al., 1995). Since profilin binds to G-actin and catalyses the exchange of ADP for ATP (Goldschmidt-Clermont et al., 1992), it is possible that vinculin also facilitates talin-mediated actin nucleation by recruiting a VASP/profilin/G-actin/ATP complex. We anticipate that the ability to express both wild-type and mutant talin and vinculin polypeptides in the ES cell mutants described here should allow us to address these various possibilities.
Acknowledgments
The authors are grateful to Mr. Paul E. Fraylich for technical assitance, to Dr. D. Melton (University of Edinburgh) for the HM1 clone of ES cells, to Dr. W. Colledge (University of Cambridge) and Dr. C. Pritchard (University of Leicester) for advice on the design of targeting constructs, and to Dr. R.O. Hynes (MIT, Cambridge, MA) for advice on ES cell electroporation.
Abbreviations used in this paper
- ANOVA
analysis of variance
- ES cell
embryonic stem cell
- Hyg
hygromycin
- LIF
Leukemia inhibitory factor
- Neo
neomycin
- nt
nucleotides
- TK
thymidine kinase
Footnotes
The work was supported by the Medical Research Council and the Wellcome Trust.
Address all correspondence to D.R. Critchley, Department of Biochemistry, University of Leicester, University Road, Leicester LE1 7RH, UK. Tel.: 0116 252 3477. Fax: 0116 252 5097. E-mail: drc@leicester.ac.uk
Dr. Priddle's current address is Centre for Genome Research, University of Edinburgh, The King's Buildings, West Mains Rd., Edinburgh EH9 3JQ, UK.
Dr. Hemmings current address is Perkin Elmer, Applied Biosystems Division, 7 Kingsland Grange, Woolston, Warrington, Cheshire WA1 4SR, UK.
References
- Albiges-Rizo C, Frachet P, Block MR. Down regulation of talin alters cell adhesion and the processing of the α5β1 integrin. J Cell Sci. 1995;108:3317–3329. doi: 10.1242/jcs.108.10.3317. [DOI] [PubMed] [Google Scholar]
- Blanchard A, Ohanian V, Critchley DR. The structure and function of α-actinin . J Muscle Res Cell Motility. 1989;10:280–289. doi: 10.1007/BF01758424. [DOI] [PubMed] [Google Scholar]
- Bolton SJ, Barry ST, Mosley H, Patel B, Jockusch BM, Wilkinson JM, Critchley DR. Monoclonal antibodies recognizing the N- and C-terminal regions of talin disrupt actin stress fibers when microinjected into human fibroblasts. Cell Motil Cytoskeleton. 1997;36:363–376. doi: 10.1002/(SICI)1097-0169(1997)36:4<363::AID-CM6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Bretscher MS. Moving membrane up to the front of migrating cells. Cell. 1996;85:465–467. doi: 10.1016/s0092-8674(00)81246-5. [DOI] [PubMed] [Google Scholar]
- Brindle NPJ, Holt MR, Davies JE, Price CJ, Critchley DR. The focal adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem J. 1996;318:753–757. doi: 10.1042/bj3180753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signalling. Annu Rev Cell Dev Biol. 1996;12:463–519. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- Burridge K, Connell L. Talin: a cytoskeletal component concentrated in adhesion plaques and other sites of actin-membrane interaction. J Cell Biol. 1983;97:359–367. doi: 10.1002/cm.970030509. [DOI] [PubMed] [Google Scholar]
- Burridge K, Mangeat P. An interaction between vinculin and talin. Nature. 1984;308:744–746. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- Chicurel ME, Singer RH, Meyer CJ, Ingber DE. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–733. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- Coll J-L, Ben A, Ze'ev, Ezzell RM, Rodriguez-Fernandez JL, Baribault H, Oshima RG, Adamson ED. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc Natl Acad Sci USA. 1995;92:9161–9165. doi: 10.1073/pnas.92.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll JL, Okamura D, Matheny W, Adamson ED. A unique RNA splicing event after the disruption of vinculin genes by homologous recombination. Transgenics. 1997;2:183. [Google Scholar]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers R, Stossel TP. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- DePasquale JA, Izzard CS. Accumulation of talin in nodes at the edge of the lamellipodium and separate incorporation into adhesion plaques at focal contacts in fibroblasts. JCell Biol. 1991;113:1351–1359. doi: 10.1083/jcb.113.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GA, Zicha D, Fraylich PE. Rapid, microtubule-dependent fluctuations of the cell margin. J Cell Sci. 1997;110:3091–3098. doi: 10.1242/jcs.110.24.3091. [DOI] [PubMed] [Google Scholar]
- Dunn GA, Brown AF. Alignment of fibroblasts on grooved surfaces described by a simple geometric transformation. J Cell Sci. 1986;83:313–340. doi: 10.1242/jcs.83.1.313. [DOI] [PubMed] [Google Scholar]
- Durrbach A, Collins K, Matsudaira P, Louvard D, Coudrier E. Brush-border myosin-I truncated in the motor domain impairs the distribution and the function of endocytic compartments in an hepatoma-cell line. Proc Nat Acad Sci USA. 1996;93:7053–7058. doi: 10.1073/pnas.93.14.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath KR, Trimbur GM, Burgess DR. Molecular motors are differentially distributed on golgi membranes from polarized epithelial-cells. J Cell Biol. 1994;126:661–675. doi: 10.1083/jcb.126.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler R, Pfaff M, Murphy J, Noegel AA, Johansson S, Timpl R, Albrecht R. Lack of β1 integrin gene in embryonic stem cells affects morphology, adhesion, and migration but not integration into the inner cell mass of blastocysts. J Cell Biol. 1995;128:979–988. doi: 10.1083/jcb.128.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Volk T, Volberg T. Molecular heterogeneity of adherens junctions. J Cell Biol. 1985;101:1523–1531. doi: 10.1083/jcb.101.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Salomon D, Takeichi M, Hynes RO. A chimeric N-cadherin/β1-integrin receptor which localizes to both cell-cell and cell-matrix adhesions. J Cell Sci. 1992;103:943–951. doi: 10.1242/jcs.103.4.943. [DOI] [PubMed] [Google Scholar]
- Gilmore AP, Jackson P, Waites GT, Critchley DR. Further characterisation of the talin binding site in the cytoskeletal protein vinculin. J Cell Sci. 1992;103:719–731. doi: 10.1242/jcs.103.3.719. [DOI] [PubMed] [Google Scholar]
- Gilmore AP, Wood C, Ohanian V, Jackson P, Patel B, Rees DJG, Hynes RO, Critchley DR. The cytoskeletal protein talin contains at least two distinct vinculin binding domains. J Cell Biol. 1993;122:337–347. doi: 10.1083/jcb.122.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann WH, Bremer A, Haner M, Aebi U, Isenberg G. Native talin is a dumbbell-shaped homodimer when it interacts with actin. J Struct Biol. 1994;112:3–10. doi: 10.1006/jsbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Furman FI, Wachsstock D, Safer DN, Nachmias VT, Pollard TD. The control of actin nucleotide exchange by thymosin-β4 and profilin—a potential regulatory mechanism for actin polymerisation in cells. Mol Biol Cell. 1992;3:1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings L, Rees DJG, Ohanian V, Bolton SJ, Gilmore AP, Patel B, Priddle H, Trevithick JE, Hynes RO, Critchley DR. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J Cell Sci. 1996;109:2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- Horwitz A, Duggan K, Buck C, Beckerle MC, Burridge K. Interaction of plasma-membrane fibronectin receptor with talin—a transmembrane linkage. Nature. 1986;320:531–533. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, Bubeck P, Rudiger M, Jockusch BM. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur J Biochem. 1997;247:1136–1142. doi: 10.1111/j.1432-1033.1997.01136.x. [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Bubeck P, Giehl K, Kroemaker M, Moschner J, Rothkegel M, Rudiger M, Schluter K, Stanke G, Winkler J. The molecular architecture of focal adhesions. Ann Rev Cell Dev Biol. 1995;11:379–416. doi: 10.1146/annurev.cb.11.110195.002115. [DOI] [PubMed] [Google Scholar]
- Knezevic I, Leisner TM, Lam SC-T. Direct binding of the platelet integrin αIIbβ3 (GPIIb-IIIa) to talin: evidence that interaction is mediated through the cytoplasmic domains of both aIIb and β3. J Biol Chem. 1996;271:16416–16421. doi: 10.1074/jbc.271.27.16416. [DOI] [PubMed] [Google Scholar]
- Kreitmeier M, Gerisch G, Heizer C, Muller-Taubenberger A. A talin homologue of Dictyosteliumrapidly assembles at the leading edge of cells in response to chemoattractant. J Cell Biol. 1995;129:179–188. doi: 10.1083/jcb.129.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Antos C, Butz S, Huber O, Delmas V, Dominis M, Kemler R. A role for cadherins in tissue formation. Development. 1996;122:3185–3194. doi: 10.1242/dev.122.10.3185. [DOI] [PubMed] [Google Scholar]
- McCann RO, Craig SW. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor A, Blanchard AD, Rowe AJ, Critchley DR. Identification of the vinculin-binding site in the cytoskeletal protein α-actinin. Biochem J. 1994;301:225–233. doi: 10.1042/bj3010225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclachlan AD, Stewart M, Hynes RO, Rees DJG. Analysis of repeated motifs in the talin rod. J Mol Biol. 1994;235:1278–1290. doi: 10.1006/jmbi.1994.1081. [DOI] [PubMed] [Google Scholar]
- Mills JC, Stone NL, Erhardt J, Pittman RN. Apoptotic membrane blebbing is regulated by myosin light chain phosphorylation. J Cell Biol. 1998;140:627–636. doi: 10.1083/jcb.140.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Umeshita K, Sakon M, Imajoh-Ohmi S, Fujitani K, Gotoh M, Oiki E, Kambayashi J, Monden M. Calpain activation in plasma membrane bleb formation during tert-butyl hydroperoxide-induced rat hepatocyte injury. Gastroenterology. 1996;110:1897–1904. doi: 10.1053/gast.1996.v110.pm8964416. [DOI] [PubMed] [Google Scholar]
- Mount SM, Steitz JA. Sequence of U1 RNA from Drosophila melanogaster: implications for U 1 secondary structure and possible involvement in splicing. Nucleic Acids Res. 1981;9:6351–6368. doi: 10.1093/nar/9.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder GL, Huang MM, Waterston RH, Barstead RJ. Talin requires β-Integrin, but not vinculin, for its assembly into focal adhesion-like structures in the nematode Caenorhabditis elegans. . Mol Biol Cell. 1996;7:1181–1193. doi: 10.1091/mbc.7.8.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muguruma M, Matsumura S, Fukazawa T. Direct interactions between talin and actin. Biochem Biophys Res Commun. 1990;171:1217–1223. doi: 10.1016/0006-291x(90)90815-5. [DOI] [PubMed] [Google Scholar]
- Muguruma M, Matsumura S, Fukazawa T. Augmentation of α-actinin-induced gelation of actin by talin. J Biol Chem. 1992;267:5621–5624. [PubMed] [Google Scholar]
- Newham, P., and M.J. Humphries. 1996. Integrin adhesion receptors: structure, function and implications for biomedicine. Mol. Med. Today. 304–313. [DOI] [PubMed]
- Niggli V, Kaufmann S, Goldmann WH, Weber T, Isenberg G. Identification of functional domains in the cytoskeletal protein talin. Eur J Biochem. 1994;224:951–957. doi: 10.1111/j.1432-1033.1994.00951.x. [DOI] [PubMed] [Google Scholar]
- Nuckolls GH, Turner CE, Burridge K. Functional-studies of the domains of talin. J Cell Biol. 1990;110:1635–1644. doi: 10.1083/jcb.110.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuckolls GH, Romer LH, Burridge K. Microinjection of antibodies against talin inhibits the spreading and migration of fibroblasts. J Cell Sci. 1992;102:753–762. doi: 10.1242/jcs.102.4.753. [DOI] [PubMed] [Google Scholar]
- Ohno H, Goto S, Taki S, Shirasawa T, Nakano H, Miyatake S, Aoe T, Ishida Y, Maeda H, Shirai T, Rajewsky K, Saito T. Targeted disruption of the CD3-ETA locus causes high lethality in mice—modulation of OCT-1 transcription on the opposite strand. EMBO (Eur Mol Biol Organ) J. 1994;13:1157–1165. doi: 10.1002/j.1460-2075.1994.tb06365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey CA, Pavalko FM, Burridge K. An interaction between α-actinin and the β-1 integrin subunit invitro. J Cell Biol. 1990;111:721–729. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey CA, Vasquez GB, Burridge K, Erickson BW. Mapping the α-actinin binding site within the β1 integrin cytoplasmic domain. J Biol Chem. 1993;268:21193–21197. [PubMed] [Google Scholar]
- Otto JJ. Detection of vinculin-binding proteins with an 125I-vinculin gel overlay technique. J Cell Biol. 1983;97:1283–1287. doi: 10.1083/jcb.97.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavalko FM, Burridge K. Disruption of the actin cytoskeleton after microinjection of proteolytic fragments of α-actinin. J Cell Biol. 1991;114:481–491. doi: 10.1083/jcb.114.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis R, Davis AC, Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1992;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- Pfaff M, Liu S, Erle DJ, Ginsberg MH. Integrin b cytoplasmic domains differentially bind to cytoskeletal proteins. J Biol Chem. 1998;273:6104–6109. doi: 10.1074/jbc.273.11.6104. [DOI] [PubMed] [Google Scholar]
- Rees D, Ades SE, Singer SJ, Hynes RO. Sequence and domain-structure of talin. Nature. 1990;347:685–689. doi: 10.1038/347685a0. [DOI] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilin. EMBO (Eur Mol Biol Organ) J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fernandez J, Geiger B, Salomon D, Benzeev A. Overexpression of vinculin suppresses cell motility in Balb/c 3T3 cells. Cell Motil Cytoskelet. 1992;22:127–134. doi: 10.1002/cm.970220206. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Fernandez JL, Geiger B, Salomon D, Benzeev A. Suppression of vinculin expression by antisense transfection confers changes in cell morphology, motility, and anchorage-dependent growth of 3T3-cells. J Cell Biol. 1993;122:1285–1294. doi: 10.1083/jcb.122.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens LE, Sonne JE, Fitzgerald ML, Damsky CH. Targeted deletion of β1-integrins in F9-embryonal carcinoma cells affects morphological differentiation but not tissue-specific gene expression. J Cell Biol. 1993;123:1607–1620. doi: 10.1083/jcb.123.6.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow JL, Fath KR, Burgess DR. Budding roles for myosin II on the Golgi. Trends Cell Biol. 1998;8:138–141. doi: 10.1016/s0962-8924(98)01238-0. [DOI] [PubMed] [Google Scholar]
- Sydor AM, Su AL, Wang F-S, Xu A, Jay DG. Talin and vinculin play distinct roles in filopodial motility in the neuronal growth cone. J Cell Biol. 1996;134:1197–1207. doi: 10.1083/jcb.134.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley P, Horwitz A, Buck C, Duggan K, Rohrschneider L. Integrins isolated from Rous sarcoma virus-transformed chicken embryo fibroblasts. Oncogene. 1989;4:325–333. [PubMed] [Google Scholar]
- Vaheri A, Carpen O, Heiska L, Helander TS, Jaaskelainen J, Majander-Nordenswan P, Sainio M, Timonen T, Turunen O. The ezrin protein family: membrane-cytoskeleton interactions and disease associations. Curr Opin Cell Biol. 1997;9:659–666. doi: 10.1016/s0955-0674(97)80119-6. [DOI] [PubMed] [Google Scholar]
- Vanaga DM, Pornares MI, Coppola S, Burgess DH, Orrenius S. Protease involvement in fodrin cleavage and phosphatidylserine exposure in apoptosis. J Biol Chem. 1996;271:31075–31085. doi: 10.1074/jbc.271.49.31075. [DOI] [PubMed] [Google Scholar]
- Volberg T, Geiger B, Kam Z, Pankov R, Simcha I, Sabanay H, Coll J-L, Adamson E, Ben-Ze'ev A. Focal adhesion formation by F9 embryonal carcinoma cells after vinculin gene disruption. J Cell Sci. 1995;108:2253–2260. doi: 10.1242/jcs.108.6.2253. [DOI] [PubMed] [Google Scholar]
- Weiming Xu, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- Westmeyer A, Ruhnau K, Wegner A, Jockusch BM. Antibody mapping of functional domains in vinculin. EMBO (Eur Mol Biol Organ) J. 1990;9:2071–2078. doi: 10.1002/j.1460-2075.1990.tb07374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Lunsdorf H, Jockusch BM. Energy-filtered electron microscopy reveals that talin is a highly flexible protein composed of a series of globular domains. Eur J Biochem. 1997;243:430–436. doi: 10.1111/j.1432-1033.1997.0430a.x. [DOI] [PubMed] [Google Scholar]
- Wood CK, Turner CE, Jackson P, Critchley DR. Characterisation of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin. J Cell Sci. 1994;107:709–717. [PubMed] [Google Scholar]
- Zhang J, Robson RM, Schmidt JM, Stromer MH. Talin can crosslink actin filaments into both networks and bundles. Biochem Biophys Res Commun. 1996;218:530–537. doi: 10.1006/bbrc.1996.0095. [DOI] [PubMed] [Google Scholar]