Abstract
The transport of newly synthesized proteins through the vacuolar protein sorting pathway in the budding yeast Saccharomyces cerevisiae requires two distinct target SNAP receptor (t-SNARE) proteins, Pep12p and Vam3p. Pep12p is localized to the pre-vacuolar endosome and its activity is required for transport of proteins from the Golgi to the vacuole through a well defined route, the carboxypeptidase Y (CPY) pathway. Vam3p is localized to the vacuole where it mediates delivery of cargoes from both the CPY and the recently described alkaline phosphatase (ALP) pathways. Surprisingly, despite their organelle-specific functions in sorting of vacuolar proteins, overexpression of VAM3 can suppress the protein sorting defects of pep12Δ cells. Based on this observation, we developed a genetic screen to identify domains in Vam3p (e.g., localization and/or specific protein–protein interaction domains) that allow it to efficiently substitute for Pep12p. Using this screen, we identified mutations in a 7–amino acid sequence in Vam3p that lead to missorting of Vam3p from the ALP pathway into the CPY pathway where it can substitute for Pep12p at the pre-vacuolar endosome. This region contains an acidic di-leucine sequence that is closely related to sorting signals required for AP-3 adaptor–dependent transport in both yeast and mammalian systems. Furthermore, disruption of AP-3 function also results in the ability of wild-type Vam3p to compensate for pep12 mutants, suggesting that AP-3 mediates the sorting of Vam3p via the di-leucine signal. Together, these data provide the first identification of an adaptor protein–specific sorting signal in a t-SNARE protein, and suggest that AP-3–dependent sorting of Vam3p acts to restrict its interaction with compartment-specific accessory proteins, thereby regulating its function. Regulated transport of cargoes such as Vam3p through the AP-3–dependent pathway may play an important role in maintaining the unique composition, function, and morphology of the vacuole.
Keywords: vacuole, SNARE, di-leucine, Saccharomyces cerevisiae, AP-3
In eukaryotic cells, accurate transport of proteins between intracellular compartments is essential to maintain the biochemical identity of each organelle. Proteins trafficking through the secretory pathway use a vesicular transport mechanism in which proteins are actively concentrated into budding vesicles and then delivered in a vectorial manner to subsequent compartments. Efficient sorting of proteins in this system depends upon both selective packaging of proteins into the appropriate vesicles and recognition and fusion of cargo-containing transport vesicles with the correct target organelle.
Formation of many of the vesicle populations that transport cargo through the endocytic and lysosomal pathways requires both the coat protein clathrin and distinct heterotetrameric adaptor protein complexes (Robinson, 1994). While clathrin acts to deform membranes into vesicles, adaptor proteins provide both a binding site for clathrin on the membrane, and act to select cargo for inclusion into vesicles by recognizing sorting signals contained within the cargo proteins themselves (Marks et al., 1997; Robinson, 1997). Three distinct adaptor protein complexes, AP-1, AP-2, and AP-3, have been identified in both mammalian cells and in yeast and are thought to direct transport of cargo proteins into the endocytic/lysosomal pathways (Phan et al., 1994; Stepp et al., 1995; Dell'Angelica et al., 1997; Simpson et al., 1997) by recognition of two main classes of sorting signals; tyrosine- (Chen et al., 1990) and di-leucine–based (Letourneur and Klausner, 1992; Marks et al., 1997) sorting motifs. In mammalian cells, AP-1 and AP-2 are associated with budding clathrin-coated vesicles at the TGN and the plasma membrane, respectively, although assigning sorting function to these proteins in yeast has been difficult as deletions of these genes do not result in detectable protein sorting phenotypes (Robinson, 1994). AP-3 has been assigned a sorting function in the process of pigment deposition in Drosophila (Simpson et al., 1997) and cargo selective transport of proteins to the vacuole in yeast (Cowles et al., 1997a ; Stepp et al., 1997).
Once vesicles have budded from a donor membrane, specific cognate interactions between SNAP receptor (SNARE)1 proteins, which are found on both vesicle (v)- SNARE and target (t)-SNARE membranes, are thought to provide the targeting specificity required for a transport vesicle to dock and fuse with the appropriate acceptor organelle (Bennett et al., 1993; Bennett and Scheller, 1993; Sollner et al., 1993). At each intracellular compartment, SNAREs act with members of several other protein families, including compartment-specific Sec1p and Rab proteins and the general factors N-ethylmaleimide–sensitive factor (NSF) (Sec18p) and soluble NSF attachment protein (SNAP) (Sec17p), which together are thought to regulate the formation and/or activity of SNARE complexes (for review see Novick and Brennwald, 1993; Rothman, 1994). In accordance with their targeting function, t-SNAREs are associated for the most part with individual compartments and become, in effect, markers for that compartment. Thus, t-SNAREs represent a large family of related proteins, each of which must be sorted to a distinct cellular location to maintain accurate intracellular protein trafficking.
In yeast, protein transport to the vacuole has proven to be a powerful model system for the study of protein sorting, as many of the required components (i.e., adaptor proteins, SNAREs, Sec1, and Rab proteins) are evolutionarily conserved (Bennett and Scheller, 1993; Stack and Emr, 1993). The characterization of a large collection of vacuolar protein sorting (VPS) genes has revealed two distinct routes by which proteins traffic from the TGN to the vacuole. These routes can be distinguished in temperature-sensitive mutants of transport components that display differential effects on the transport of two vacuolar hydrolases, carboxypeptidase Y (CPY) and alkaline phosphatase (ALP). Many vacuolar resident proteins, such as CPY, are delivered to the vacuole through a well defined route that requires the function of the VPS genes, one of which encodes the endosomal t-SNARE, Pep12p (Becherer et al., 1996). In contrast, the vacuolar integral membrane protein ALP is transported to the vacuole in a manner that is independent of PEP12 (Cowles et al., 1997b ), and is therefore presumed to bypass this endosomal compartment. Instead, ALP transport to the vacuole follows an AP-3 adaptor protein–dependent pathway (Cowles et al., 1997a ; Stepp et al., 1997). Vam3p, the vacuolar t-SNARE, is required for delivery of both CPY and ALP to the vacuole, indicating that these distinct pathways ultimately converge at this docking site (Darsow et al., 1997). Interestingly, localization studies indicate that Vam3p may also be transported to the vacuole via the ALP pathway, suggesting that it too may bypass the endosomal compartment defined by Pep12p (Cowles et al., 1997b ; Piper et al., 1997).
Although Vam3p and Pep12p perform analogous biochemical t-SNARE functions, these genes clearly have distinct sites of action (Becherer et al., 1996; Burd et al., 1997; Darsow et al., 1997). Surprisingly, however, overexpression of VAM3 can compensate for the loss of PEP12 function (Darsow et al., 1997; Gotte and Gallwitz, 1997), suggesting that under some conditions, Vam3p can substitute for Pep12p. We designed a random genetic screen to define the sequence determinants in Vam3p that allow it to replace Pep12p. This screen specifically identified a small region within the Vam3p sequence (NEQSPLL), which is similar to a region of the ALP cytosolic tail that contains a di-leucine sequence required for proper sorting (Cowles et al., 1997b ; Vowels and Payne, 1998). Furthermore, many of the mutations within the presumptive Vam3p sorting determinant caused mislocalization of Vam3p. Together, these results suggest that the Vam3p t-SNARE is likely to be transported to the vacuole through the AP-3/ALP pathway via recognition of an acidic di-leucine sorting signal. Thus, trafficking of Vam3p to the vacuole through the AP-3 pathway appears to play an important role in restricting its site of function and regulating its association with other components of the transport machinery.
Materials and Methods
Strains and Media
Saccharomyces cerevisiae strains used for these studies are listed in Table I. Yeast strains were grown in standard yeast extract-peptone-dextrose (YPD) or synthetic medium (YNB) supplemented with essential amino acids. Standard bacterial medium, containing 100 μg/ml ampicillin for plasmid selection, was used to propagate Escherichia coli. Transformation of S. cerevisiae was done by the lithium acetate method (Ito et al., 1983). E. coli transformations were done by the method of Hanahan (1983).
Table I.
S. cerevisiae Strains Used in This Study
| Strain | Genotype | Reference or source | ||
|---|---|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al., 1988 | ||
| TDY2 | SEY6210; vam3Δ::LEU2 | Darsow et al., 1997 | ||
| CBY31 | SEY6210; pep12Δ::HIS3 | Burd et al., 1997 | ||
| BWY102 | SEY6210; vps24Δ::HIS3 | Babst et al., 1998 | ||
| GOY8 | SEY6210; pep12Δ::LEU2 apm3Δ::HIS3 | Cowles et al., 1998 |
Plasmid Construction and Nucleic Acid Manipulations
Restriction and modification enzymes were purchased from Boehringer Mannheim (Indianapolis, IN), New England Biolabs (Beverly, MA), or U.S. Biochemical Corporation (Cleveland, OH). Alleles of vam3 were constructed in pVAM3.414 and were subcloned into pRS416 by excising the NotI–XhoI fragment and ligating into NotI–XhoI-digested pRS416 (Sikorski and Hieter, 1989). For construction of NH2-terminal green fluorescent protein (GFP) fusion constructs, a 900-bp fragment containing the VAM3 open reading frame (ORF) was amplified from either wild-type or mutant VAM3 template using primers that induced an in-frame BglII site at the start codon and an EcoRI site after the stop codon. BglII–EcoRI- digested PCR product was then ligated into BamHI–EcoRI-digested pGOGFP fusion vector (Cowles et al., 1997a ). Plasmids pVAM3.414 and pVAM3.424 were described previously (Darsow et al., 1997).
Alleles of VAM3 were constructed by PCR based mutagenesis (Muhlrad et al., 1992). Primers complimentary to chromosomal sequences immediately adjacent to the start and stop codons of VAM3 were used to amplify a 900-bp fragment (containing the entirety of the VAM3 ORF) under limiting dATP, dTTP, or dGTP conditions (20 μM) in separate reactions. The resulting PCR products were precipitated in 95% ethanol, combined, and then gel purified. A gapped plasmid was generated by digesting pVAM3.414 with BsmI and isolating the vector by gel purification. The mutagenized PCR product and gapped plasmid were co-transformed into CBY31 cells and transformants in which homologous recombination resulted in integration of mutagenized PCR product were selected by amino acid prototrophy. Transformants were replica plated onto YPD containing 50 μg/ml geneticin (Gibco Laboratories, Grand Island, NY) and grown at 30°C for 2 d. Presumptive mutant colonies were picked, retested, and then plasmid linkage of the G418 resistance phenotype was confirmed by retransformation of isolated plasmids into CBY31 cells.
Site-directed mutagenesis of Q156 was performed by amplifying the VAM3 locus by PCR using mutagenic primers in a gene SOE (splicing by overlap extension) reaction. Complimentary primers containing the appropriate mutation were used in conjunction with primers complimentary to sequences ∼300 bp both 5′ and 3′ of the VAM3 ORF for the initial amplification. A secondary amplification using the initial PCR products as template was performed using only the outside primers to amplify the full-length vam3 mutant. The resultant PCR product was cloned into TOPO TA vector (Invitrogen Corp., Carlsbad, CA). The EcoRI fragment of the TA clones was excised and ligated into EcoRI-digested pRS414 vector for yeast expression. The mutation introduced a unique SacI restriction site and positive insert-containing clones were confirmed to be point mutants by digestion with SacI.
Plasmids isolated from the G418 resistance screen were purified from E. coli using miniprep spin columns (QIAGEN Inc., Valencia, CA). Resultant plasmids were denatured, hybridized to sequencing primers, and then subjected to dideoxy chain termination sequence analysis using the Sequenase enzymes and protocol (U.S. Biochemical Corporation).
Metabolic Labeling and Immunoprecipitation
To analyze the transport of vacuolar proteins, yeast cells were grown at 26°C in synthetic medium supplemented with amino acids to an OD600 of 0.5–1.0. Cells were harvested and converted to spheroplasts as described previously (Paravicini et al., 1992). Spheroplasts were resuspended at a concentration of 3 OD600/ml in synthetic medium containing amino acids and supplemented with 100 μg/ml α2-macroglobulin and 1 mg/ml BSA to stabilize secreted proteins. Cultures were pre-incubated at the appropriate experimental temperature for 5 min, and then labeled with 60 μCi [35S]cysteine/methionine per ml of cell suspension. After labeling, cultures were chased with the addition of methionine, cysteine, yeast extract, and glucose to a final concentration of 5 mM, 1 mM, 0.4%, and 0.2%, respectively. After appropriate chase periods, samples were harvested and precipitated by addition of TCA (10% final concentration). For analysis of CPY in whole cells, metabolic labelings were done in a similar manner except BSA and α2-macroglobulin were excluded from the labeling medium. Whole cells lysates were generated by glass bead disruption in urea buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 1% SDS, and 6 M urea). In both spheroplast and whole cell experiments, immunoprecipitated proteins were resolved by SDS-PAGE and analyzed by autoradiography. Antibodies to CPY and Pep12p have been previously described (Klionsky and Emr, 1989; Becherer et al., 1996). mAbs to Vph1p and ALP were purchased from Molecular Probes (Eugene, OR).
Subcellular Fractionation and Gradient Analysis
For intracellular localization of proteins, cells were converted to spheroplasts and lysed in Hepes KOAc lysis buffer containing protease inhibitors to the following final concentrations: 20 μg/ml PMSF; 5 μg/ml antipain; 1 μg/ml aprotinin; 0.5 μg/ml leupeptin; 0.7 μg/ml pepstatin; and 10 μg/ml α2-macroglobulin (Gaynor et al., 1994). After a 5-min clearing spin at 300 g, the spheroplast lysate was sequentially centrifuged at 13,000 g (15 min) and 100,000 g (60 min) to generate both high and low speed pellet and supernatant fractions. Resulting samples were TCA precipitated and analyzed by immunoblotting and ECL detection as described previously (Babst et al., 1997). Gradient Accudenz (Accurate Chemical and Scientific Corporation, Westbury, NY) solutions were prepared (wt/vol) in 10 mM Hepes KOAc, pH 7.6, with protease inhibitors. The gradient was generated using the following Accudenz concentration steps from bottom to top: 0.5 ml 60%; 1 ml 50%; 1 ml 43%; 1 ml 37%; 1 ml 31%; 1 ml 27%; 1 ml 23%; 1 ml 20%; 1 ml 17%; 1 ml 13%; 1 ml 7%. Gradient analysis was performed on 15 OD600 equivalents of cleared spheroplast lysate in a volume of 1.5 ml loaded on top of the gradient. The gradient was subjected to centrifugation at 4°C in a Beckman SW41 rotor at 170,000 g for 20 h. 12 fractions were harvested manually from the top of the gradient, proteins were TCA precipitated and analyzed by immunoblotting. Quantitation of proteins on gels was done by densitometry using NIH Image.
Fluorescence Microscopy
To examine vacuolar and endocytic structures in live yeast cells, FM4-64 (Molecular Probes) labeling was done as previously described (Vida and Emr, 1995) except that the labeling was done at a concentration of 16 μM FM4-64 at 26°C for 1 h and the cells were chased for a period of 1.5 h. Visualization of FM4-64 and GFP was done either by confocal microscopy or fluorescence microscopy using rhodamine and fluorescein filters, respectively.
Results
Identification of VAM3 Mutants Capable of Substituting for PEP12
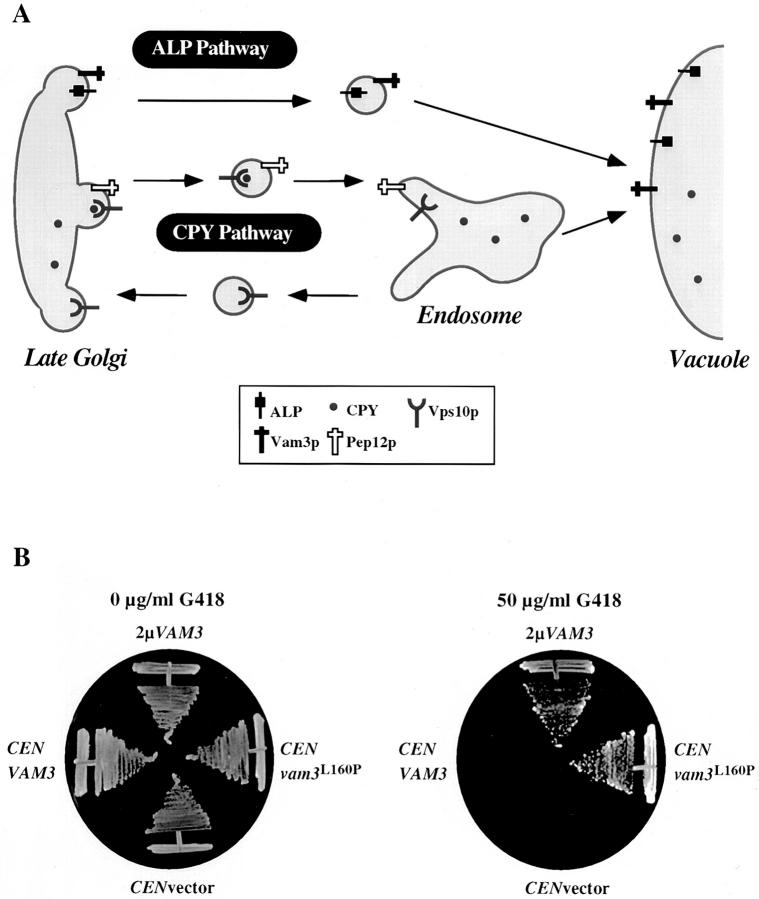
Vam3p and Pep12p are two homologous yeast t-SNARE proteins involved in protein transport to the vacuole in yeast. Although Vam3p and Pep12p have clearly distinguishable functions (i.e., Pep12p acts at the endosome and Vam3p acts at the vacuole) (Fig. 1 A), when overexpressed, these proteins are able to partially substitute for one another (Darsow et al., 1997; Gotte and Gallwitz, 1997). This suggests that the specificity of Vam3p and Pep12p functions may be dependent on interactions with other compartment-specific transport components (Sec1 homologues, Rab proteins). Based on this observation, we reasoned that mutations in Vam3p that allow it to substitute for Pep12p might uncover domains required for localization and/or specific protein–protein interactions of Vam3p with Pep12p-specific transport components.
Figure 1.
Novel genetic screen to identify regions of Vam3p required for function and localization at the vacuole. (A) In wild-type cells, Pep12p, the endosomal t-SNARE, directs traffic from the Golgi complex to the endosome through the CPY pathway. Vam3p, the vacuolar t-SNARE, is transported through the AP-3–dependent ALP pathway to the vacuole where it functions to receive incoming vesicular traffic from both the ALP and CPY pathways. We designed a G418-based selection scheme to identify VAM3 mutants that were able to compensate for the loss of PEP12 and thus identify regions of Vam3p sequence responsible for restricting Vam3p activity to the AP-3–dependent pathway. (B) Growth of both pep12Δ (CBY31) cells alone or pep12Δ (CBY31) cells expressing VAM3 from a single-copy plasmid (pVAM3.414) is compromised at concentrations of 50 μg/ml G418, while growth on media containing no G418 is normal. However, pep12Δ (CBY31) cells either overexpressing VAM3 from a 2μ plasmid (pVAM3.424) or expressing a mutant derived from the screen, vam3 L160P, on a single-copy plasmid (pVAM3L160P.414) results in growth at 50 μg/ml G418, as well as on media containing no G418.
Several vps mutants, including pep12Δ, are hypersensitive to geneticin (G418), an aminoglycoside antibiotic related to gentamicin (Fig. 1 B). The mechanism of this hypersensitivity is unknown but for pep12Δ mutants it appears to correlate with extent of vacuolar protein sorting defects. For example, overexpression of VAM3 from a multi-copy vector in pep12Δ cells restored viability to cells grown on media containing 50 μg/ml G418, while VAM3 expressed at single copy (which does not improve CPY sorting when expressed in pep12Δ cells) did not rescue the growth defects of pep12Δ cells on media containing 50 μg/ml G418 (Fig. 1 B). Therefore, selection by growth on G418-containing media presented an easily scoreable phenotype that could be used to select for vam3 mutants capable of suppressing the growth defects of pep12Δ cells on media containing G418.
The entire VAM3 ORF was randomly mutagenized by error-prone PCR-mediated mutagenesis and then co-transformed with a gapped, single-copy plasmid into pep12Δ mutant cells (Muhlrad et al., 1992). Transformants were replica plated onto media containing 50 μg/ml G418, and viable colonies were selected. Approximately 10,000 colonies were screened, and >100 vam3 mutant clones that had acquired the ability to suppress the growth defects of pep12Δ cells at 50 μg/ml G418 were recovered. We selected ∼50 representative clones that grew well at 50 μg/ml G418 and rescued plasmids from these strains. 30 mutants that showed plasmid linkage for the G418 resistance phenotype were selected for further analysis. As a secondary screen, we tested the growth of these mutants at elevated concentrations of G418. This criteria allowed us to separate the mutants into two general classes: (1) strong mutants that conferred resistance to 100 μg/ml G418, and (2) weaker mutants that conferred resistance to only 50 μg/ml G418.
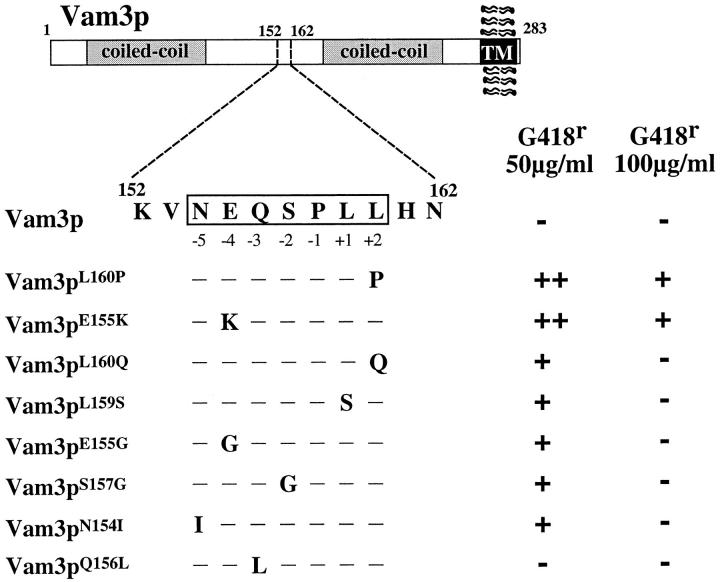
Sequence analysis revealed that each of the 30 mutants recovered in this screen contained at least one mutation within a 7–amino acid region of Vam3p, corresponding to amino acids 154–160 (Fig. 2). This sequence of Vam3p lies approximately in the middle of the protein, between the two coiled-coil domains, and it is distanced well away from the transmembrane domain in a region that does not exhibit high sequence similarity to Pep12p. However, the sequence closely resembles a region in the cytosolic tail of ALP that has been shown to be necessary for transport of ALP through the AP-3–dependent pathway to the vacuole (Cowles et al., 1997b ; Vowels and Payne, 1998). These characteristics, together with the presence of a leucine pair at positions 159–160, suggested that this region may represent a di-leucine–type sorting signal in Vam3p.
Figure 2.
Vam3p mutants define a di-leucine sorting signal. Vam3 protein domain structure is shown including the extreme COOH-terminal transmembrane domain and the two coiled-coil domains. The location of the region containing the mutations recovered from the G418 resistance screen is denoted by the dashed box. The wild-type sequence at the mutated region is shown in detail. Single mutations that were recovered from the G418 resistance screen are indicated in capital letters designating the amino acid changes. The glutamine 156 to leucine mutation was not recovered in the screen but was made by site-directed mutagenesis. The level of G418 resistance conferred by each of the mutants is as follows: ++, wild-type growth; +, slower growth to single colonies; and −, no growth.
Somewhat surprisingly, the mutations in the 30 plasmids represented only seven separate single mutations in the following five residues: asparagine 154, glutamate 155, serine 157, leucine 159, and leucine 160 (Fig. 2). No other amino acid substitutions in Vam3p were found outside of this region. The recovery of multiple identical mutants in each residue indicates that the screen was near saturation and that the most relevant residues in Vam3p that confer suppression of the pep12Δ mutation had likely been mutated. The mutants that conferred the highest level of G418 resistance (100 μg/ml) were glutamic acid 155 (at −4 relative to the di-leucine pair) to lysine (vam3 E155K), and leucine 160 to proline (vam3 L160P). However, when these same residues were mutated to either glycine (vam3 E155G) or glutamate (vam3 L160Q), respectively, the cells grew only at the lower concentration of G418 (50 μg/ml). Independently isolated clones containing mutations in asparagine 154 at the −5 position from the di-leucine (vam3 N154K), serine 157 at the −2 position (vam3 S157G), and leucine 159 (vam3 L159S) also resulted in maximum G418 resistance at 50 μg/ml G418 (Fig. 2).
The ALP cytoplasmic tail sequence and the Vam3p sequence both contain a conserved glutamine at the −3 position (Fig. 2). Since glutamine mutants were not recovered in the G418 selection, we wanted to determine whether it was required for the function of the Vam3p domain. Using site-directed mutagenesis, we changed the glutamine 156 to leucine (vam3 Q156L), transformed the mutant into pep12Δ cells and assayed for G418 resistance. The vam3 Q156L mutant did not confer G418 resistance to pep12Δ cells. However, when the vam3 Q156L point mutation was transformed into vam3Δ cells, it complemented both the morphological defects and the CPY sorting defects of the vam3Δ cells (data not shown), indicating that a full-length, functional protein was still produced. Therefore, the glutamine at the −3 position of the Vam3p motif is not required for normal Vam3p function.
Vam3p Mutants Partially Substitute for Pep12p Function in Vacuolar Protein Sorting
To confirm that the G418 resistance phenotype used to isolate vam3 mutants correlated with suppression of CPY defects of pep12Δ cells harboring these mutant forms of VAM3 we examined CPY sorting by pulse-chase/immunoprecipitation analysis. As expected, in pep12Δ cells CPY was recovered almost exclusively as the Golgi-modified p2 form, consistent with severe defects in Golgi-to-endosome transport. Similarly, pep12Δ cells expressing VAM3 from a single-copy plasmid did not show any significant improvement in CPY sorting. However, in all cases, pep12Δ cells expressing vam3 mutants from a single-copy plasmid that conferred G418 resistance also exhibited improved CPY sorting. Moreover, efficiency of CPY sorting in pep12Δ cells harboring the various vam3 mutants correlated with degree of G418 resistance as mutants that exhibited resistance to high concentrations of G418 sorted and matured more CPY than mutants resistant to only lower (50 μg/ml) G418 concentrations. For example, pep12Δ cells expressing the vam3 N154I mutant, which is resistant to G418 concentrations up to 50 μg/ml, converted ∼50% of p2 CPY to the mature form, whereas pep12Δ cells expressing the vam3 L160P mutant, which is resistant to 100 μg/ml G418, matured ∼80% of CPY (Fig. 3 A).
Figure 3.
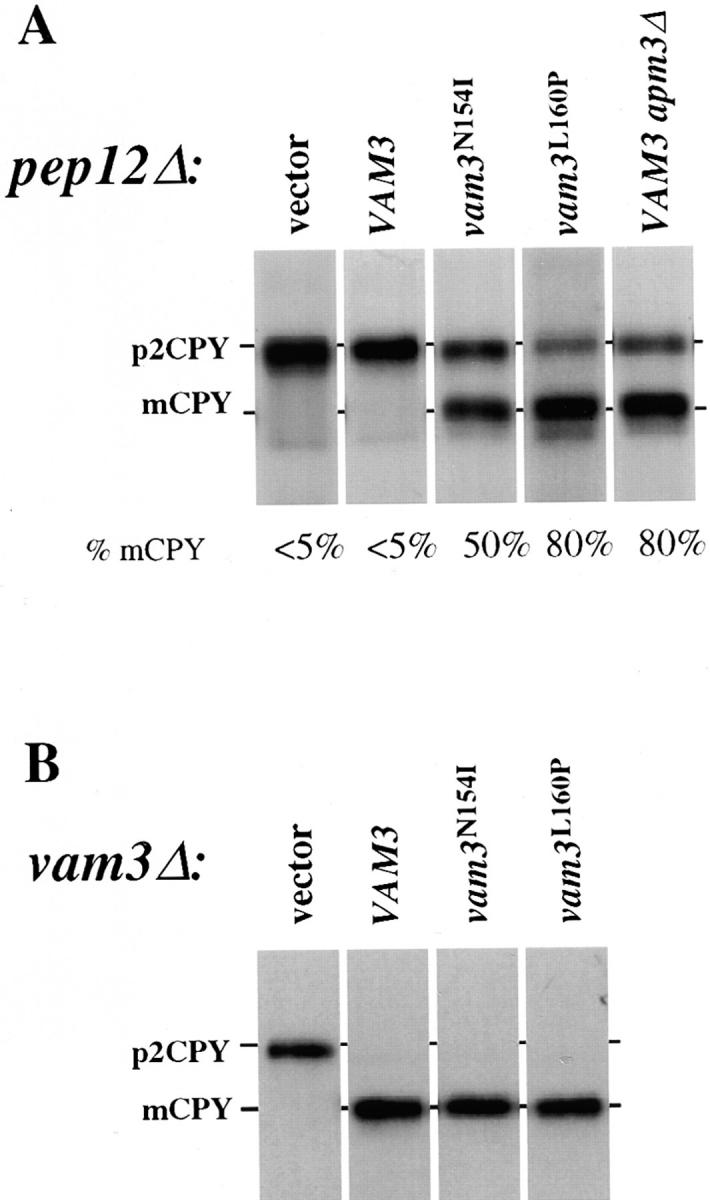
CPY sorting of vam3 mutants in both pep12Δ and vam3Δ mutant cells. (A) pep12Δ (CBY31) cells and pep12Δaps3Δ (GOY8) double-mutant cells transformed with a single-copy plasmid containing wild-type VAM3 (pVAM3.414), and pep12Δ (CBY31) cells transformed with vector (pRS414), vam3 L160P or vam3 N154I mutant isolates (pVAM3L160P. 414 and pVAM3N154I.414, respectively) were spheroplasted, and then metabolically labeled and chased for 45 min. (B) vam3Δ (TDY2) cells were transformed with the identical plasmids from A and were labeled and chased as whole cells for 30 min. CPY was immunoprecipitated and examined by autoradiography. For both A and B, mature and Golgi-modified precursor CPY are indicated as mCPY and p2CPY, respectively. In A, the percent of mature CPY is denoted beneath each individual lane.
As Vam3p requires AP-3 for its normal transport and localization to the vacuole, disruption of the AP-3 pathway may mimic the effects of the Vam3p sorting signal mutations. To examine this possibility, we analyzed CPY sorting in both pep12Δ cells and apm3Δpep12Δ double mutant cells, each expressing an additional copy of wild-type VAM3 to mimic the conditions of the mutant screen. In contrast to pep12Δ cells containing wild-type VAM3, in which CPY is blocked exclusively in the p2 form, ∼80% of CPY was converted to the mature vacuolar form in apm3Δpep12Δ double mutant cells (Fig. 3 A). This was remarkably similar to results obtained for Vam3p mutants such as vam3 L160P expressed in pep12Δ cells, and suggests that these mutations allow CPY maturation because they prevent normal recognition of Vam3p by the AP-3 adaptor complex and result in missorting of Vam3p to the pre-vacuolar endosome where it can partially substitute for Pep12p.
VAM3 Mutants Still Maintain Vam3p Function
We were interested in determining whether the vam3 mutants were capable of normal Vam3p t-SNARE function at the vacuole. Since the screen was performed in a strain containing a wild-type VAM3 gene, it was possible that mutations allowing for suppression of pep12Δ sorting defects would also result in loss of the ability of these proteins to function as Vam3p. Therefore, we examined CPY sorting by pulse-chase analysis in vam3Δ cells transformed with the mutants recovered from the screen. As expected, vam3Δ cells accumulated only p2 precursor CPY, indicating that transport of CPY to the vacuole was blocked. However, vam3 mutants that resulted in both strong (vam3 L106P) and weak (vam3 N154I) suppression of pep12Δ mutant phenotypes completely complemented the CPY sorting defects of the vam3Δ strain (Fig. 3 B), indicating that although these mutants can substitute for the Pep12p t-SNARE, they also retain Vam3p t-SNARE function. These results suggest that the mutant Vam3 proteins must at least partially localize to the vacuole, the normal site of Vam3p function.
Mutant Vam3 Proteins Are Missorted into the CPY Pathway
The ability of the vam3 mutants to suppress pep12Δ vacuolar protein sorting defects suggests that mutant Vam3 proteins are at least partially localized to the pre-vacuolar endosome, the site of Pep12p function. To examine the localization of the Vam3 mutant proteins, we fractionated cells and examined the distribution of the Vam3 mutant proteins relative to wild-type Vam3p by differential centrifugation. Vam3p fractionates exclusively in a low speed (P13) pellet fraction by differential centrifugation, which is consistent with its vacuolar localization (Darsow et al., 1997). However, the mutant forms of Vam3p fractionated differently than the wild-type protein, with a significant portion of the proteins localized to a high speed P100 pellet, (data not shown) which indicated that at least a portion of the mutant proteins localized to a non-vacuolar fraction. Although the P13 fraction is enriched in vacuoles, other compartments also fractionate in the P13. For example, the endosomal t-SNARE Pep12p typically exhibits a 40% P13/60% P100 fractionation pattern, even though this protein is not localized to the vacuole (Becherer et al., 1996). To further resolve vacuoles away from other P13 material, vam3Δ cells harboring either single-copy wild-type VAM3 or the vam3 L160P mutant were analyzed by equilibrium density gradient fractionation. Cleared spheroplast lysates were applied to the top of Accudenz step gradients, which were then centrifuged to equilibrium. Fractions were collected from the top of the gradient and analyzed for the presence of Vam3p, Vph1p, and the endosomal t-SNARE, Pep12p. In both gradients, Vph1p fractionated primarily in the low density fractions (1–4) of the gradient, as is typical for vacuolar membrane proteins (Darsow et al., 1997) (Fig. 4 A). In contrast, Pep12p fractionated exclusively in more dense regions of the gradient (fractions 5–7), indicating that distinct separation of vacuoles and endosomes was achieved in these gradients (Fig. 4 B). As expected, wild-type Vam3p primarily co-fractionated with Vph1p in the first four fractions of the gradient. However, while a small percentage of Vam3pL160P also fractionated in the top (vacuolar) region of the gradient, the majority of the mutant Vam3 protein (∼80%) was recovered in more dense fractions (5–7), which also contained Pep12p (Fig. 4 B). Thus, while some Vam3pL160P is localized to the vacuole, most of the protein appears to reside in a non-vacuolar compartment that co-fractionates with Pep12p-containing endosomes.
Figure 4.
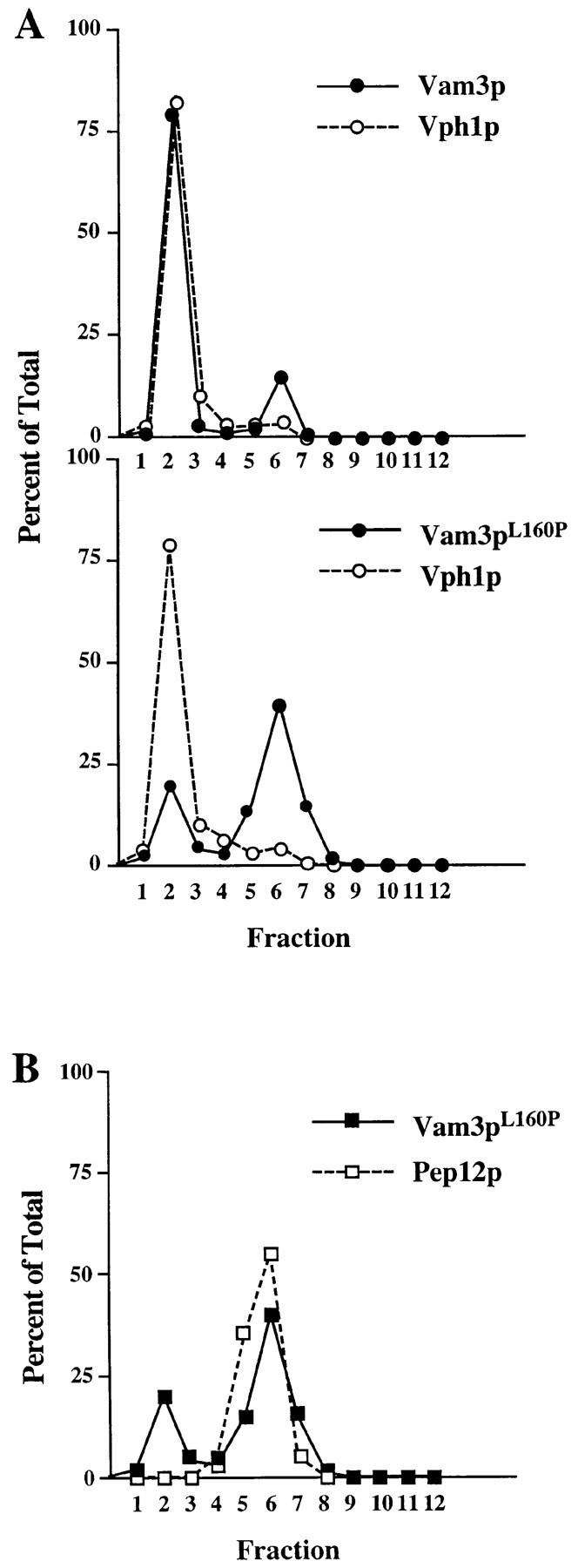
Localization of Vam3 mutant proteins. Cleared cell lysates generated from vam3Δ (TDY2) cells containing either wild-type VAM3 (pVAM3.414) or the vam3 L160P mutant (pVAM3L160P.414) were loaded onto the top of an Accudenz step gradient and centrifuged to equilibrium. Fractions 1–12 were collected from the top of the gradient. Proteins were precipitated from the fractions and then separated by SDS-PAGE and transferred to nitrocellulose. Vam3p, Vph1p, and Pep12p were detected by immunoblotting and visualized by ECL fluorography. The distribution of both wild-type and mutant (Vam3pL160P) Vam3p and Vph1p are shown graphically in A and the colocalization of Vam3pL160P and Pep12p are shown in B.
The cell fractionation data suggested that a significant portion of the mutant Vam3 proteins were mislocalized to compartments other than the vacuole. To visualize the compartments in which the mislocalized proteins were residing, we constructed GFP fusions with wild-type and mutant Vam3 proteins. We transformed the plasmid constructs into vam3Δ cells and examined the cells by fluorescence microscopy. Both the wild-type and mutant Vam3 fusion proteins complemented the protein sorting (data not shown) and morphology defects associated with vam3Δ cells (Fig. 5 A) and thus functioned as the native proteins. Consistent with the previous localization studies, wild-type GFP-Vam3 fusion protein localized almost exclusively to vacuolar membranes (Fig. 5 A). While GFP-Vam3L160P still accumulated in vacuolar membranes, a large portion of the protein was also seen in tubular and punctate structures throughout the cell (Fig. 5 A). GFP fusion proteins of the other Vam3p mutants also behaved in a similar manner (data not shown), indicating that these mutant proteins accumulate in non-vacuolar structures.
Figure 5.
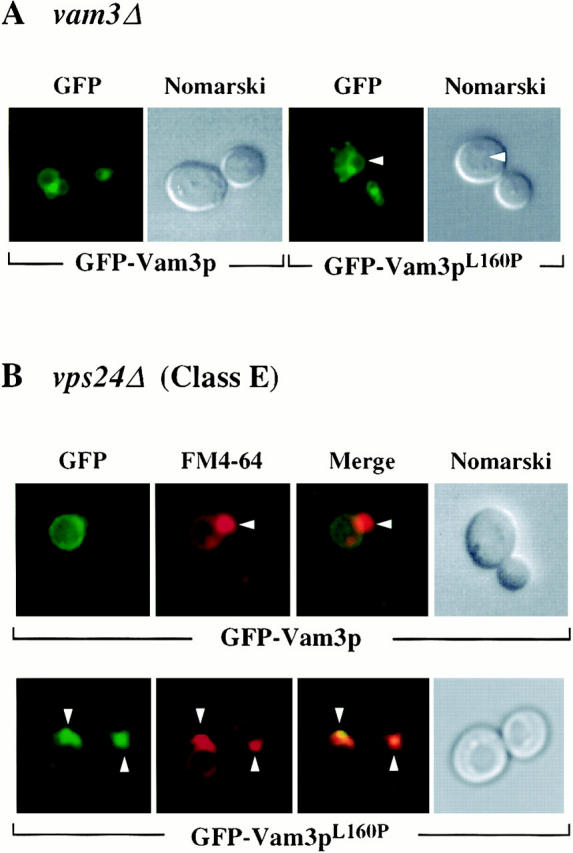
Localization of Vam3 mutant proteins in vps24Δ cells. vam3Δ (TDY2) and vps24Δ (BW102) cells were transformed with plasmids containing both wild-type (pGFPVAM3.426) and mutant (pGFPVAM3L160P.426) GFP-Vam3 fusion proteins. These strains were grown in selective media to exponential phase, harvested, and then resuspended in YNB for examination by microscopy. vps24Δ cultures were further labeled with 16 μM FM4-64 for a period of 1 h at 26°C in YPD. Labeled cells were then harvested, resuspended in YPD, and then chased for 1.5 h. After chase, cells were examined by Nomarski and fluorescence/ confocal microscopy for both GFP and FM4-64 fluorescence. Arrows in A indicate vacuoles and in B indicate class E compartments.
Class E vps mutants contain large aberrant endosomal structures (class E compartments), which accumulate both biosynthetic cargoes (e.g., CPY) as well as endocytic cargoes (e.g., the lipophilic dye FM4-64) (Rieder et al., 1996). However, proteins that travel through the AP-3–dependent pathway (e.g., ALP, Vam3p) do not accumulate in the class E compartment, as this pathway bypasses the endosome (Babst et al., 1997; Piper et al., 1997). Unlike wild-type Vam3p, if the mutant Vam3 proteins travel through the CPY pathway to the vacuole, they should accumulate in the class E compartment. We transformed class E mutant vps24Δ cells with plasmids encoding either GFP-Vam3 or GFP-Vam3L160P mutant fusion protein, labeled the cells with FM4-64, and examined the cells for both markers by fluorescence microscopy. Wild-type GFP-Vam3 fusion protein accumulated primarily on vacuolar membranes and was excluded from the class E compartment (Fig. 5 B). In contrast, a large percentage of the cells expressing GFP-Vam3pL160P displayed brightly fluorescent signal in a perivacuolar compartment, in addition to a lower intensity fluorescent signal on the vacuolar membrane (Fig. 5 B). FM4-64 counterstaining showed that this perivacuolar compartment was coincident with the class E compartment. GFP fusion proteins made with the other Vam3p mutants also accumulated in the E compartment in a similar manner (data not shown), indicating that mislocalization through the CPY pathway occurred in all of the mutants. Together, these localization data provide compelling evidence that the mutant proteins do not traffic to the vacuole through the AP-3–dependent pathway but instead transit the CPY pathway and accumulate in pre-vacuolar compartments.
Discussion
We have described a novel genetic screen for mutations in the vacuolar t-SNARE Vam3p that allow it to functionally replace the endosomal t-SNARE, Pep12p. SNARE proteins would appear to be one of the most important cargoes to be selectively sorted, as they represent a core component of the vesicle docking/fusion machinery. The correct sorting of many other cargo proteins depends on proper localization of these t-SNARE molecules. Here we provide the first identification of a defined sorting signal in a t-SNARE protein. Previous studies have indicated that both transmembrane and cytosolic domains of t-SNAREs contain information that influences their localization (Banfield et al., 1994; Rayner and Pelham, 1997). However, these studies did not define primary sequence sorting signals, nor did they identify specific transport components that may be involved in the sorting of these proteins (e.g., sorting receptors). This study provides significant, in vivo evidence that Vam3p, like ALP, is sorted by recognition of an acidic di-leucine motif through an AP-3–dependent pathway. Furthermore, disruption of the di-leucine sorting signal in Vam3p results in mislocalization of Vam3p to the pre-vacuolar endosome where it functionally substitutes for the endosomal t-SNARE Pep12p. These results suggest that the trafficking route of a SNARE acts to restrict its site of function and that localization plays an important role in determining the functional specificity of a given SNARE protein. Therefore, trafficking of Vam3p through the AP-3/ALP pathway plays a vital role in maintaining the unique composition and function of the vacuole. Furthermore, these observations provide insight into the requirement for the AP-3–dependent alternative Golgi-to-vacuole transport pathway.
Vam3p Contains a Putative Di-leucine–type Sorting Signal That Directs It into the ALP Pathway
The only known cargo of the AP-3–dependent sorting pathway to the vacuole in yeast is ALP (Cowles et al., 1997a ; Stepp et al., 1997). However, several lines of evidence suggest that Vam3p also traffics through this route. First, Vam3p localizes to the vacuole in a manner independent of the function of Pep12p (Cowles et al., 1997b ) and the class E Vps proteins Vps4p and Vps27p, and is specifically excluded from the class E compartment (Babst et al., 1997; Piper et al., 1997), suggesting that it does not transit through the pre-vacuolar endosome in the CPY pathway en route to the vacuole. Furthermore, in an AP-3 mutant, Vam3p is mislocalized to a non-vacuolar compartment in the same manner as ALP (Cowles et al., 1997a ). A di-leucine sorting signal in the cytosolic tail of ALP has been shown to be necessary for AP-3–dependent sorting of ALP to the vacuole (Cowles et al., 1997b ; Vowels and Payne, 1998). While Vam3p contains multiple putative di-leucine sequences within its cytoplasmic domain, we have found through a random mutagenesis screen that disruption of only one of the potential di-leucine signals in Vam3p alters its trafficking route and allows it to function at a non-vacuolar compartment. Furthermore, deletion of AP-3 complex components results in similar phenotypes as disruption of the di-leucine sequence of Vam3p. Together, these data strongly suggest that Vam3p traffics through the AP-3–dependent ALP pathway via recognition of a di-leucine motif. It was recently shown that di-leucine sorting signals in the mammalian proteins LIMP II, a lysosomal membrane protein, and tyrosinase, a melanosomal membrane protein that functions in melanin synthesis, bind to AP-3–enriched fractions in an in vitro surface plasmon resonance (SPR) assay, which is suggestive of direct interactions between di-leucine motifs and AP-3 (Honing et al., 1998). Our work provides strong in vivo evidence that di-leucine motifs are required for directing cargo into the AP-3–mediated transport pathway in yeast.
Vam3p Mutants Reveal a Role for Additional Residues in the Di-leucine Motif
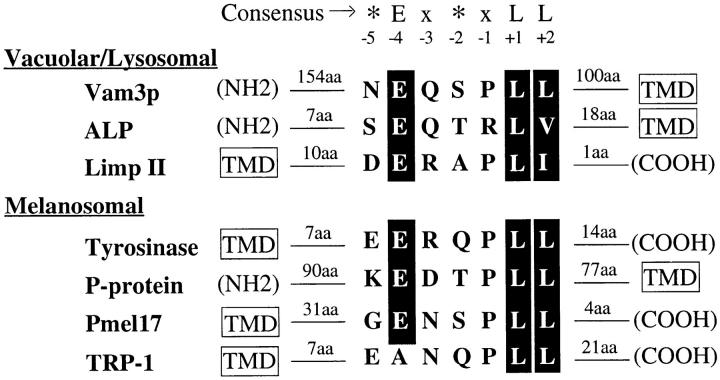
Random mutagenesis of Vam3p identified a small 7–amino acid region in the middle of the cytoplasmic domain of Vam3p that includes a leucine pair as well as several amino acids just to the NH2-terminal side of the di-leucine sequence. Because di-leucine sorting signals have been implicated in endocytosis from the plasma membrane (Letourneur and Klausner, 1992; Pond et al., 1995) and also have been shown to be capable of direct binding to both AP-1 and AP-2 in vitro (Heilker et al., 1996; Dietrich et al., 1997; Rapoport et al., 1998), it is likely that only a subset of di-leucine motifs interact with AP-3. Mutations in the glutamate at position −4 relative to the leucine pair in Vam3p resulted in particularly strong phenotypes, comparable to mutations in either of the leucine residues. These results are consistent with in vitro studies that have shown that mutation of the acidic residues in the di-leucine sorting signals in LIMP II and tyrosinase abrogate binding to AP-3–enriched fractions (Honing et al., 1998). However, acidic residues in addition to the di-leucine pair do not seem to be sufficient to direct AP-3 binding, since many mammalian proteins containing similar sequences do not seem to be sorted through AP-3–dependent pathways (Honing et al., 1998). Our analysis has identified several additional residues that also contribute to the function of the di-leucine motif. In addition to the acidic residue at −4 and the leucine pair, the Vam3p motif requires an upstream polar residue at −5 and a hydrophilic amino acid at the −3 position for optimal sorting activity. In fact, the sorting determinant in ALP contains conserved residues at the same positions to each of the additional residues that were mutated in Vam3p (Fig. 6).
Figure 6.
Vam3p di-leucine sequence alignment with putative AP-3 cargoes. The Vam3p di-leucine sequence shares significant sequence similarity to di-leucine sequences in both lysosomal and melanosomal proteins. The consensus motif (*Ex*xLL) is derived from mutagenesis data. An asterisk denotes a bias toward charged, polar amino acids, while x can be any amino acid. Residues that were mutated in the Vam3p sequence and are also conserved in other cargo proteins are indicated by shaded regions. ALP shares sequence similarity with Vam3p at every base that was recovered in our Vam3p mutagenesis. The acidic amino acid at −4, the polar amino acids at −5 and −3, and the proline at −1 as well as the di-leucine sequence, are conserved in the majority of the proteins that have been defined as potential AP-3 cargoes.
Similar sequence characteristics can also be found in many of the other candidate AP-3 cargoes that have been identified in mammalian systems. Melanosomal resident enzymes have been implicated as cargo for AP-3–directed sorting, as mutant mice in both AP-3 components and melanosomal proteins result in similar aberrant coat color phenotypes, suggesting that these pigmentation defects may be due to improper sorting of proteins to melanosomes (Odorizzi et al., 1998). Di-leucine motifs in many melanosomal proteins also contain a conserved hydrophilic amino acid at the −3 position (Fig. 6). In addition, mutation of residues at the same positions in proteins such as invariant chain, which do not bind to AP-3, have been shown to have no effect on the transport of these proteins (Motta et al., 1995; Pond et al., 1995; Honing et al., 1998). Both the Vam3p di-leucine sequence and the di-leucine sequences in many of the other potential AP-3 cargoes contain a conserved proline at the −1 position. However, we did not recover mutations in this residue and it is not conserved in the ALP sequence, suggesting that this proline may not be required for the function di-leucine sorting signals, at least in yeast. Together, these data point to a conserved sorting motif for most AP-3–dependent cargoes and a common mechanism for the recognition and packaging of these cargoes. Further mutational analysis of conserved amino acids in other vacuolar/lysosomal and melanosomal protein sequences will confirm the importance of these residues in AP-3–directed sorting. Sequence search algorithms derived from such analysis may also be useful in identifying new potential cargo proteins in the AP-3 pathway and may help to further define the pathway's biological significance.
Disruption of the Vam3p Di-leucine Motif Causes Mislocalization of Vam3p into the CPY Pathway
The CPY pathway seems to be the “default” route to the vacuole, as no sorting signals required for transport of membrane proteins into this pathway have been defined and proteins that normally transit through this pathway can be diverted into other pathways by the addition of positive sorting signals (Cowles et al., 1997b ). Furthermore, overexpression of either ALP or Vam3p results in their overflow into the CPY pathway, consistent with saturation of the signal recognition/packaging machinery in the AP-3 pathway (Cowles et al., 1997b ; Darsow et al., 1997). We have shown that disruption of the di-leucine sorting signal in Vam3p results in the mislocalization of the protein into the CPY pathway. It has been previously shown that Vam3p is missorted in AP-3 mutants and co-fractionates with late Golgi and endosomal markers (Cowles et al., 1997a ). Consistent with these observations, we found that Vam3p sorting signal mutants accumulate in a Pep12p-containing intermediate compartment, most likely a pre-vacuolar endosome. The following observations support this model: (a) the Vam3 mutant proteins are capable of suppressing the defects of pep12Δ cells, which argues that the proteins are reaching a compartment where Pep12p normally operates, (b) the mutant proteins co-fractionate with Pep12p and are in a distinct location from vacuolar markers in fractionation experiments and finally, and (c) the accumulation of the Vam3 mutant proteins in a class E mutant endosome implies that the proteins are not being specifically retained in the Golgi complex but instead are being transported forward at least until the class E block is initiated, which is thought to be at the point of exit from the endosomal compartment (Babst et al., 1997). Together, these data suggest that a large pool of the Vam3 mutant proteins reside at steady state in a pre-vacuolar endosome.
The localization data also indicate that only a minor fraction of the mutant proteins arrive at the vacuole (∼20%), and thus, that transport of Vam3p to the vacuole through the CPY pathway is inefficient. It is therefore possible that mechanisms may exist which retain the majority of Vam3 mutant protein in a stable, pre-vacuolar compartment. In wild-type cells, Pep12p is also retained within the pre-vacuolar endosomal compartment, possibly by a similar mechanism. Perhaps within their highly conserved primary sequence, Pep12p and Vam3p contain retention motifs that maintain their localization in the endosome, either by preventing forward transport or through recycling. Alternately, interactions with compartment-specific SNARE accessory proteins that function at the pre-vacuolar endosome, such as Vps45p, Vps21p, or Vac1p (Cowles et al., 1994; Horazdovsky et al., 1994; Burd et al., 1997), could act to retain both Pep12p and the mutant Vam3 proteins in the endosome. This would be consistent with our previous findings that Vam3p-specific suppression of pep12Δ requires expression of endosomal transport components (Darsow et al., 1997).
AP-3–mediated Localization Restricts the Function of the Vam3p t-SNARE Protein
It has previously been shown that overexpression of VAM3 can partially suppress the vacuolar protein sorting defects of a pep12 null mutant and that this activity may be dependent on mislocalization of Vam3p to the Pep12p compartment (Darsow et al., 1997; Gotte and Gallwitz, 1997). Our results presented here confirm that mislocalization of Vam3p allows it to efficiently substitute for Pep12p. While we can not rule out that Vam3 mutant proteins have increased affinity for Pep12p-specific components (e.g., Vps45p), the data are most consistent with a model in which these mutations result in quantitative missorting of Vam3p to the pre-vacuolar endosome where it can efficiently substitute for Pep12p.
The finding that in the absence of a functional di-leucine motif, Vam3p both localizes and functions at an earlier step in the CPY pathway has several interesting implications for the function of t-SNARE proteins. Both Vam3p and Pep12p act in what appear to be classical SNARE- mediated transport steps. They both require specific accessory proteins such as Rab and Sec1 homologues, as well as the general cytosolic fusion machinery, including Sec18p (NSF), for their activity (Burd et al., 1997; Darsow et al., 1997; Sato et al., 1998). By simply driving Vam3p into a different biosynthetic transport pathway, as may be accomplished either by Vam3p overexpression, disruption of the Vam3p sorting signal, or by disruption of the sorting machinery itself, Vam3p is able to function at a different compartment in the Golgi-to-vacuole transport pathway. These results suggest that a major distinguishing characteristic between Pep12p and Vam3p is their biosynthetic transport route and localization. Current models for SNARE function postulate that cognate interactions between t- and v-SNAREs define the primary specificity of each transport step. However, our observations are most consistent with a model in which SNARE proteins are much more promiscuous in their interactions with accessory proteins and, when localized improperly, are able to partially function at alternate sites. For example, although Vam3p normally acts in conjunction with the vacuolar Sec1 homologue, Vps33p, the suppression of pep12Δ by VAM3 overexpression also requires Vps45p, the endosomal Sec1 homologue (Darsow et al., 1997). In this case, Vam3p appears to use the docking and fusion machinery of the endosome rather than recruiting these accessory proteins from the vacuole. Clearly then, t-SNARE proteins do not define the sole component of specificity of vesicular transport, and other compartment specific proteins must also be present and act with the t-SNARE to accomplish this goal. It remains to be determined which proteins, or combination of proteins are ultimately responsible for transport specificity, but it seems likely that a set of complex interactions at each transport step may be required for accurate vesicular transport.
Acknowledgments
We thank both past and present members of the Emr lab, especially E. Gaynor, M. Babst, and B. Wendland for helpful comments and for critical reading of this manuscript. We would also like to thank G. Odorizzi for strains used in this study and D. Katzmann for assistance with the confocal microscopy.
This work was supported by grants GM32703 and CA58689 from the National Institutes of Health to S.D. Emr. S.D. Emr is an Investigator of the Howard Hughes Medical Institute.
Abbreviations used in this paper
- ALP
alkaline phosphatase
- CPY
carboxypeptidase Y
- GFP
green fluorescent protein
- NSF
N-ethylmaleimide–sensitive factor
- ORF
open reading frame
- SNAP
soluble NSF attachment protein
- SNARE
SNAP receptor
- VPS
vacuolar protein sorting
Footnotes
Address all correspondence to Scott D. Emr, Division of Cellular and Molecular Medicine and Department of Biology, Howard Hughes Medical Institute, University of California, San Diego, La Jolla, CA 92093-0668. Tel.: (619) 534-6462. Fax: (619) 534-6414. E-mail: semr@ucsd.edu
References
- Babst M, Sato TK, Banta LM, Emr SE. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO (Eur Mol Biol Organ) J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosomal function. EMBO (Eur Mol Biol Organ) J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Rabouille C, Warren G, Pelham HR. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer KA, Reider S, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Scheller RH. The molecular machinery for secretion is conserved from yeast to neurons. Proc Natl Acad Sci USA. 1993;90:2559–2563. doi: 10.1073/pnas.90.7.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Garcia-Arraras JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Burd CG, Peterson M, Cowles CR, Emr SD. A novel Sec18p/ NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol Biol Cell. 1997;8:1089–1104. doi: 10.1091/mbc.8.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- Cowles CR, Emr SD, Horazdovsky BF. Mutations in the VPS45 gene, a SEC1homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J Cell Sci. 1994;107:3449–3459. doi: 10.1242/jcs.107.12.3449. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997a;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Snyder WB, Burd CG, Emr SD. An alternative Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO (Eur Mol Biol Organ) J. 1997b;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor–like protein complex with ubiquitous expression. EMBO (Eur Mol Biol Organ) J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J, Kastrup J, Nielsen BL, Odum N, Geisler C. Regulation and function of the CD3gamma DxxxLL motif: A binding site for adaptor protein-1 and adaptor protein-2 in vitro. J Cell Biol. 1997;138:271–281. doi: 10.1083/jcb.138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte M, Gallwitz D. High expression of the yeast syntaxin-related Vam3 protein suppresses the protein transport defects of a pep12 null mutant. FEBS (Fed Eur Biochem Soc) Lett. 1997;411:48–52. doi: 10.1016/s0014-5793(97)00575-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Heilker R, Manning-Krieg U, Zuber JF, Spiess M. In vitro binding of clathrin adaptors to sorting signals correlates with endocytosis and basolateral sorting. EMBO (Eur Mol Biol Organ) J. 1996;15:2893–2899. [PMC free article] [PubMed] [Google Scholar]
- Honing S, Sandoval IV, von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO (Eur Mol Biol Organ) J. 1998;17:1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Busch GR, Emr SD. VPS21encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO (Eur Mol Biol Organ) J. 1994;13:1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO (Eur Mol Biol Organ) J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur F, Klausner RD. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- Marks MS, Ohno H, Kirchhausen T, Bonifacino JS. Protein sorting by tyrosine-based signals. Trends Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- Motta A, Bremnes B, Morelli MA, Frank RW, Saviano G, Bakke O. Structure-activity relationship of the leucine-based sorting motifs in the cytosolic tail of the major histocompatibility complex-associated invariant chain. J Biol Chem. 1995;270:27165–27171. doi: 10.1074/jbc.270.45.27165. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Novick P, Brennwald P. Friends and family: the role of Rab GTPases in vesicular traffic. Cell. 1993;75:597–601. doi: 10.1016/0092-8674(93)90478-9. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Cowles CR, Emr SD. The AP-3 complex: a coat of many colors. Trends Cell Biol. 1998;8:282–287. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- Paravicini G, Horazdovsky BF, Emr SD. Alternative pathways for the sorting of soluble vacuolar proteins in yeast: a vps35null mutant missorts and secretes only a subset of vacuolar hydrolases. Mol Biol Cell. 1992;3:415–427. doi: 10.1091/mbc.3.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HL, Finlay JA, Chu DS, Tan PK, Kirchhausen T, Payne GS. The Saccharomyces cerevisiae APS1 gene encodes a homolog of the small subunit of the mammalian clathrin AP-1 complex: evidence for functional interaction with clathrin at the Golgi complex. EMBO (Eur Mol Biol Organ) J. 1994;13:1706–1717. doi: 10.1002/j.1460-2075.1994.tb06435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Bryant NJ, Stevens TH. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond L, Kuhn LA, Teyton L, Schutze MP, Tainer JA, Jackson MR, Peterson PA. A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J Biol Chem. 1995;270:19989–19997. doi: 10.1074/jbc.270.34.19989. [DOI] [PubMed] [Google Scholar]
- Rapoport I, Chen YC, Cupers P, Shoelson SE, Kirchhausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO (Eur Mol Biol Organ) J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Pelham HR. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO (Eur Mol Biol Organ) J. 1997;16:1832–1841. doi: 10.1093/emboj/16.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome like compartment accumulates in the yeast vps28. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. The role of clathrin, adaptors, and dynamin in endocytosis. Curr Opin Cell Biol. 1994;6:538–544. doi: 10.1016/0955-0674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Robinson MS. Coats and vesicle budding. Trends Cell Biol. 1997;7:99–102. doi: 10.1016/S0962-8924(96)10048-9. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Sato, T.K., T. Darsow, and S.D. Emr. 1998. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homologue function together in vacuolar protein trafficking. Mol. Cell. Biol. In press. [DOI] [PMC free article] [PubMed]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F, Peden AA, Christopoulou L, Robinson MS. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stack JH, Emr SD. Genetic and biochemical studies of protein sorting to the yeast vacuole. Curr Opin Cell Biol. 1993;5:641–646. doi: 10.1016/0955-0674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- Stepp JD, Pellicena-Palle A, Hamilton S, Kirchhausen T, Lemmon SK. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain- related protein. Mol Biol Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Huang K, Lemmon SK. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels JJ, Payne GS. A dileucine-like sorting signal directs transport into an AP-3–dependent, clathrin-independent pathway to the yeast vacuole. EMBO (Eur Mol Biol Organ) J. 1998;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]