Abstract
Cdc42, activated with GTPγS, induces actin polymerization in supernatants of lysed neutrophils. This polymerization, like that induced by agonists, requires elongation at filament barbed ends. To determine if creation of free barbed ends was sufficient to induce actin polymerization, free barbed ends in the form of spectrin-actin seeds or sheared F-actin filaments were added to cell supernatants. Neither induced polymerization. Furthermore, the presence of spectrin-actin seeds did not increase the rate of Cdc42-induced polymerization, suggesting that the presence of Cdc42 did not facilitate polymerization from spectrin-actin seeds such as might have been the case if Cdc42 inhibited capping or released G-actin from a sequestered pool.
Electron microscopy revealed that Cdc42-induced filaments elongated rapidly, achieving a mean length greater than 1 μm in 15 s. The mean length of filaments formed from spectrin-actin seeds was <0.4 μm. Had spectrin-actin seeds elongated at comparable rates before they were capped, they would have induced longer filaments. There was little change in mean length of Cdc42-induced filaments between 15 s and 5 min, suggesting that the increase in F-actin over this time was due to an increase in filament number. These data suggest that Cdc42 induction of actin polymerization requires both creation of free barbed ends and facilitated elongation at these ends.
Keywords: actin polymerization, Cdc42, leukocytes, actin filament barbed ends, G proteins
The ability of agonists to induce actin polymerization is essential for cell motility and chemotaxis. In neutrophils, addition of chemoattractant causes a rapid twofold increase in F-actin. The chemoattractant-induced polymerization requires a pertussis toxin-sensitive G protein but downstream mediators of the response have remained obscure. Recently, members of the Rho family of G proteins have been implicated in this response. Thus, injection of Cdc42 or Rac into macrophages induces filopodia and lamellipodia, respectively (Allen et al., 1997), and expression of dominant-negative Cdc42 or Rac inhibited cytoskeletal responses to chemoattractants (Cox et al., 1997).
Cdc42 induces cytoskeletal changes in a wide range of organisms including mammals (Nobes et al., 1995), Drosophila (Eaton et al., 1996) and Saccharomyces cerevisiae (Adams et al., 1990; Johnson et al., 1990). The cytoskeletal functions affected include cytokinesis, cell polarity, formation of attachment sites, filopodial extension, motility, and phagocytosis (Kozma et al., 1995; Nobes et al., 1995; Stowers et al., 1995; Chen et al., 1996; Allen et al., 1997; Cox et al., 1997; Keely et al., 1997). Although functionally and morphologically distinct, each of these functions involves actin polymerization. Thus, a primary effect of Cdc42 appears to be the regulation of actin polymerization.
Stimulated actin polymerization in vivo is usually accomplished by elongation at the barbed end of an actin filament. The barbed end of an actin filament has a higher affinity than the pointed end for ATP–G-actin. Thus, if in the resting cell barbed ends are largely capped, agonist stimulation can shift the steady state toward more polymerized actin (F-actin) by freeing barbed ends. The difference in the affinity between the two ends only represents a small concentration difference of free G-actin, 0.1 versus 0.5 μM, and thus, in the absence of other factors, only that amount of actin would be polymerized on uncapping. However, in cells such as neutrophils, a large reservoir of G-actin bound to thymosin β4 exists, effectively buffering the free G-actin and allowing a large change in polymerization upon uncapping (Cassimeris et al., 1992; Fechheimer et al., 1993). Furthermore, the presence of profilin enhances barbed-end elongation and reduces the free [ATP–G-actin] at steady state (Pantaloni et al., 1993). Thus, although profilin sequesters free G-actin, in vivo this is buffered by the much larger thymosin β4 pool, and so profilin amplifies the polymerization seen on uncapping. Cofilin has the complementary effect of enhancing pointed-end depolymerization and decreasing its affinity (Carlier et al., 1997). Since these proteins also increase the rate of achieving a new steady state, their presence permits a cell to alter the level of polymerization rapidly and dramatically merely by regulating the availability of barbed ends (Carlier et al., 1997).
To define how Cdc42 affects actin polymerization, we developed a functional assay in which a GTPγS-charged insect cell produced recombinant Cdc42 (Cdc42) and induces actin polymerization in supernatants of lysed neutrophils (Zigmond et al., 1997). The Cdc42-induced polymerization is inhibited by cytochalasin and thus, like agonist-induced polymerization in vivo, requires barbed-end elongation (Zigmond et al., 1997). An increase in free barbed ends is detected in supernatants stimulated by Cdc42. The mechanism of creating free barbed ends is not known. Cdc42 could increase free barbed ends by creating them de novo from G-actin, by uncapping of existing filaments, or by cutting of filaments. It seems unlikely that they are created by cutting because the starting supernatants have little F-actin and the ability of Cdc42 to induce polymerization is not inhibited by phalloidin which inhibits cutting by the actin depolymerizing factor family of proteins (Yonezawa et al., 1988; Maciver, 1991). Creation de novo or the uncapping of small actin oligomers remain as possibilities.
However the free barbed ends are created, the question remains: is merely making barbed ends available sufficient to induce polymerization? We now report that addition of exogenous free barbed ends does not induce polymerization. Indeed, the exogenous free barbed ends do not contribute to polymerization even when Cdc42 is present. Furthermore, although both Cdc42-induced filaments and exogenous barbed ends appear to elongate for only a short period, the Cdc42-induced filaments elongate rapidly, within 15 s, attaining lengths greater 1 μm, whereas added free barbed ends do not elongate appreciably. It appears that Cdc42-induced polymerization depends on enhanced rates of elongation restricted to Cdc42-induced barbed ends.
Materials and Methods
Polymorphonuclear Leukocyte Lysates
Rabbit peritoneal exudate neutrophils were obtained and processed as described previously (Sullivan et al., 1980; Zigmond et al., 1997). Briefly, the neutrophils were suspended at 3–6 × 108 cells/ml in saline and incubated with 1 mM diisopropylfluorophosphate, DFP (Sigma Chemical Co., St. Louis, MO), for 5 min on ice. The cells were washed two times with cold saline and resuspended at 3 or 4 × 108 cells/ml in intracellular physiological buffer (IP): 135 mM KCl, 10 mM NaCl, 2 mM MgCl2, 2 mM EGTA, 10 mM Hepes, pH 7.1. Protease inhibitors (1 μg/ml leupeptin, 1 ug/ml benzamidine, 10 ug/ml aprotinin, 10 ug/ml TAME) were added. Cells were lysed either by nitrogen bombing (Zigmond et al., 1997) or by minimal sonication required to break the cells (three 1-s pulses on setting 40 of a Dyn probe sonicator [Dynatech Laboratories, Inc., Chantilly, VA]). Examination of F-actin levels, nucleation sites assayed by elongation of exogenous pyrenyl actin, and responses to GTPγS and Cdc42 indicated that the properties of lysates prepared by both methods were similar.
Cell Supernatant
Cell supernatant was the supernatant of lysate spun first at 4°C at 14,000 rpm for 5 min (∼1.5 ×105 g/min) and the supernatant of this spin was then spun at 80,000 rpm for 20 min (∼5.6 × 106 g/min) in a Beckman TL 100 centrifuge using a 100.3 rotor (both from Beckman Instrs., Palo Alto, CA). The protein concentration of the supernatant was determined and the supernatant was aliquoted at 200 μl, frozen in liquid nitrogen, and then stored at −80°C.
Negative Staining
The supernatant was incubated at a final concentration of 3 mg/ml with buffer or GTPγS-activated Cdc42 (see below) for various times at room temperature on a freshly carbon-coated EM grid. The grids were rinsed with water containing 0.4 μM phalloidin and then stained for 1 min with 1% uranyl acetate before draining and drying.
Photographs were taken of regions of the grid that contained filaments. For analysis of filament lengths, pictures at a final magnification of 18,500× were examined. Preliminary studies indicated that although prints at 48,000× enhanced recognition of filaments less than 0.3-μm long, a significant fraction of the filaments present extended off the edge of the photograph. The mean length of filaments with only one end in the photo was greater than the mean of filaments entirely within the photo. This was not true with prints at 18,500× and thus, the edge of the photo did not appreciably distort estimates of filament length distribution. Filaments greater than 0.25 μm were readily recognized and filament length distributions measured by one individual could be reproduced by a second observer. However, counts of filaments less that 0.25 μm were not reliable primarily because the supernatant contained numerous components in this size range. Thus, we did not attempt to count filaments less than 0.25 μm. It should be noted that even at the earliest measured times the great majority of Cdc42-induced filaments were not excluded by this. For example at 15 s, Model 1, which fits the data presented, predicts that 17% of all filaments were less than 0.25 μm. In experiments with phalloidin, 1–6 μM phalloidin was present in the supernatant during the incubation with the Cdc42.
S-1 Labeling
Samples activated with Cdc42 were applied to a grid and then washed with IP containing phalloidin. 10 μl of the S-1 fragment of myosin (gift of D. Safer, University of Pennsylvania, Philadelphia, PA) was added at 1 mg/ ml. The S-1 was added either in IP buffer or in 0.1 M K phosphate buffer, pH 6.8, both in the presence of 1 μM phalloidin. Grids were passed through two drops of S-1 and allowed to incubate for 5 min. The grid was then rinsed with water and stained with 1% uranyl acetate.
F-actin Determination
F-actin was quantified from TRITC-phalloidin staining of pelleted material as described originally by Howard and Oresajo (Howard et al., 1985) and modified slightly (Zigmond et al., 1997). Briefly, 60-μl aliquots of supernatant at a final concentration of ∼3 mg/ml protein incubated with various additions were stopped by dilution into 840 μl IP buffer containing 0.4 μM TRITC-phalloidin (Sigma Chemical Co.) and 0.1% Triton X-100. After staining for 1 h, the samples were spun in a 100.2 rotor at 80,000 rpm for 20 min in the Beckman TL100 ultracentrifuge (both from Beckman Instrs.). The pellets were extracted with 1 ml of methanol and after ∼20 h the fluorescence (Ex540/Em575) was read. To determine nonsaturable staining, ≥4 μM unlabeled phalloidin was included.
Spectrin-Actin Seeds
Spectrin-actin seeds were prepared according the method of Lin (Casella et al., 1986).
Pyrenyl-Actin Assays of Nucleation
Pyrenyl-actin assays of nucleation were performed as described previously (Cano et al., 1991). Briefly, supernatants were diluted 100-fold directly into 1.5 μM pyrenyl actin and the initial rate of polymerization was determined from the increase in pyrenyl fluorescence Ex370 /Em410. All samples had the same final concentration of seeds, supernatant, and pyrenyl-actin. The fact that the rates of polymerization decrease more rapidly with time in the presence of supernatant than in its absence indicates that even at this dilution of supernatant some capping activity is present (DiNubile et al., 1995). The presence of 2 μM cytochalasin B decreased the rate of polymerization by 90% (data not shown), indicating that the polymerization was due to barbed-end elongation.
Reagents
Recombinant Proteins.
Recombinant Cdc42 was expressed in a baculovirus insect cell expression system as described (Heyworth et al., 1993; Xu et al., 1994). The Cdc42 was a glutathione-S-transferase (GST)1 construct and was isolated on GST beads from a membrane detergent lysate (in order to obtain the isoprenylated protein only). The amount of proteins used in the assays was based on activity of the small GTPase proteins as determined by their ability to bind [35S]GTPγS. The purified G proteins were charged with GTPγS by incubation for 10 min at 30°C with a two- to threefold molar excess of GTPγS in EDTA/Mg to give a final Mg concentration between 100 and 1,000 nM (Knaus et al., 1992). The Mg concentration was then increased to 2 mM in excess of the EDTA present and the samples were stored on ice until use. In this manuscript, Cdc42, unless otherwise stated, means GTPγS-activated Cdc42.
Modeling
To test whether the filament length distributions observed at different times were compatible with the hypothesis that filaments elongated transiently, we investigated whether we could mimic the data with a simple model of Cdc42-induced nucleation and filament elongation, depolymerization, and termination of elongation, here modeled as capping. It should be noted that, when a model is found to fit the data, it merely shows that the schema used to generate the mathematical representation—or indeed any other of the infinitely many mechanisms that lead to the same or practically indistinguishable formulations—cannot be ruled out. If, however, a model can be shown not to be able to fit the data, then that model must deviate in at least some aspect from reality. The general elements of the model we used were as follows:
Filament Elongation, Depolymerization, and Capping.
An uncapped filament of n monomers (Fn) can elongate by adding G-actin [G] at the pointed or barbed ends and, in the presence of profilin [Pr], by the addition of profilin-actin [PrG] at the barbed end. It can depolymerize by loss of a monomer from either end or be capped at the barbed end, effectively irreversibly during the time course of the experiment, by a capping protein assumed to be present in large excess, leading to the differential equations (Eq. 1, n = 4, 5 . . .) (see Modeling Strategy below for an explanation of the factor fp):
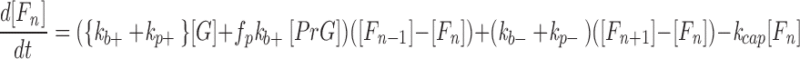 |
1 |
where [Fn] = number concentration of filaments with n monomers.
The capped filaments (FnC) also produced the following analogous equations with the barbed-end terms eliminated:
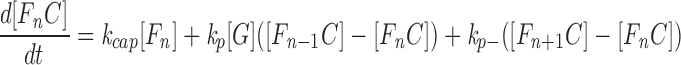 |
2 |
Nucleation and Filament Loss.
The formation of nuclei (Nuc) is presumed to occur at a constant net rate throughout the experiment (this reflects the observations, see Results). Nuclei are able to elongate at the barbed end with the normal elongation rate constants to form tetramers (F4); thus, they can be pictured as consisting of some facilitating entity bound to a trimer formed from association of three G-actins. In the model it is assumed that the facilitating entity dissociates as soon as the filament elongates; however, this is not a necessary feature: the slow kinetics of the pointed end and the fact that free [G] is not far from the pointed end critical concentration for much of the time course of the experiment make that end's status irrelevant to the outcome. Consequently, in the model (Nuc) substitutes for (F3) in the differential equations describing filament elongation. The depolymerization of a tetramer, capped or uncapped is, however, presumed to result in its breakup into the four component G-actins. These assumptions lead to the differential Eq. 3:
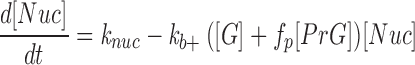 |
3 |
Free and Sequestered G-actin.
Sequestration of G-actin by profilin and thymosin β4 [Tβ4] was assumed to be at equilibrium at all times. Thus, [PrG] and [Tβ4G] were given by Eqs. 4 and 5:
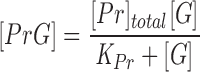 |
4 |
and
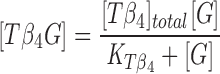 |
5 |
Furthermore, this assumption, combined with the elongation and depolymerization equations, leads to the differential Eq. 6 for [G]:
 |
6 |
Simplification of Equations for Numerical Integration.
Two simplifications were necessary to reduce the model equations to a finite and manageable number for numerical integration. (a) The elongation and depolymerization equations were only included up to a maximum filament length, usually 5.04 μm (F1,800). Longer filaments were accounted for by integrating the number concentration and [F-actin] present in all filaments of greater length. The only error introduced by this is that it fails to account explicitly for filaments that grow beyond the maximum length and then stochastically depolymerize below it. Systematic changes to the maximum length showed this error to be negligible. (b) To further reduce the number of equations, filaments below the maximum length were grouped into length ranges and the sums of concentrations of both capped and uncapped filaments in each length range were integrated severally under the assumption that their concentrations were uniformly distributed within a length range. For exploratory calculations, each range was 0.05-μm long. Referee calculations with shorter ranges showed that deviations from the uniformity assumption were typically less than 5%, and due to compensatory effects, the errors under the much wider binning used in presenting the length distributions were substantially less.
Initial Conditions and Rate Constants.
The following measured parameters were assumed based on the supernatants being twenty-fold dilutions of the cell cytoplasm: [G]total, t = 0 = 6 μM (Cassimeris et al., 1992); [Tβ4]total = 8.75 μM (Cassimeris et al., 1992), K Tβ4 = 0.6 μM (Weber et al., 1992); [Pr]total = 2 μM (Southwick et al., 1990), K Pr = 0.1 μM (Perelroizen et al., 1994). These lead to a calculated initial free G-actin [G]t = 0 = 0.48 μM.
Except where noted the following rate constants were used: elongation by free G-actin and depolymerization (Walsh et al., 1984)(numbers rounded), pointed-end k p+ = 106/M/s; k p− = 0.5/s; barbed-end k b+ = 107/ M/s; k b−= 1/s. Elongation by profilin-actin at the barbed end, as shown in the equations above, was assumed to be with the same rate constant as free G-actin (Pollard et al., 1984; Pring et al., 1992) multiplied by an adjustable factor, fp, as described below.
Filament capping (see Results): barbed-end k cap = 0.115/s (Model 1), within the range predicted for this supernatant concentration by values measured previously. Capper concentration = 50–100 nM, capper on rate, k on = 7 × 105 − 5 × 106 M−1s−1 (DiNubile et al., 1995); or for slowed rate of 0.025/s (Model 2).
Modeling Strategy.
In modeling the Cdc42-induced filament length distributions the elongation and nucleation (k nuc) rates were adjusted manually, the former via the factor fp determining the rate of profilin-actin addition, until the calculated average filament length and [F-actin] at 5 min matched those observed. The predicted and observed length distributions at 5 min and earlier times were then compared.
Results
Addition of Free Barbed Ends Does Not Induce Actin Polymerization
We showed previously (Zigmond et al., 1997) that recombinant Cdc42 charged with GTPγS (hereafter referred to as Cdc42) induces actin polymerization in supernatants of neutrophil lysates. The increase in F-actin is accompanied by an increase in free barbed ends detected as sites that, after dilution of supernatant, nucleate barbed-end elongation of labeled actin. The Cdc42-induced actin polymerization in supernatants is inhibited by cytochalasin, indicating that the increase in polymerization depends on free barbed ends. We now ask whether the ability of Cdc42 to produce free barbed ends is sufficient to account for the actin polymerization.
Free barbed ends, in the form of spectrin-actin seeds, added to cell supernatants caused no actin polymerization (Fig. 1 A). The spectrin-actin seeds contain some F-actin (seen as an increase in TRITC-phalloidin staining even at time = 0) which was maintained during incubation, suggesting that the seeds did not depolymerize. In the experiment shown in Fig. 1 A, the concentration of spectrin-actin seeds added had ∼20 times more barbed-end nucleating sites (determined from the initial rate of pyrenyl-actin polymerization) than those present in supernatant incubated for 5 min with 100 nM Cdc42. Addition of 10- or 100-fold lower concentrations of spectrin-actin seeds also did not induce polymerization (data not shown). To rule out the possibility that the failure to induce polymerization was unique to spectrin-actin seeds, we added barbed ends in the form of sheared actin filaments. These also did not induce detectable polymerization. Thus, it appeared that addition of free barbed ends is not sufficient to induce polymerization.
Figure 1.
Addition of spectrin-actin seeds to cell supernatants. (A) Effects of spectrin-actin seeds and/or Cdc42 on actin polymerization. Supernatants were incubated at 37°C with buffer (open circles), spectrin-actin seeds (open triangles), 100 nM GTPγS-charged Cdc42 (closed circles) or both spectrin-actin seeds and 100 GTPγS-Cdc42 (closed triangles), for 2 or 5 min before the F-actin levels were determined by the TRITC-phalloidin staining of pelletable material (refer to Materials and Methods). For t = 0, the seeds were added after a 15-fold dilution of supernatant. Data are from a single experiment representative of three. (B) Spectrin actin seeds become rapidly capped when incubated with supernatant. Spectrin-actin seeds were incubated in supernatants for 5, 10, or 60 s before the supernatant was diluted 100-fold into 1.5 μM pyrenyl-actin. Polymerization of the pyrenyl- actin was then followed over time from the pyrenyl fluorescence (refer to Materials and Methods). For the time 0 point, the seeds were added after supernatant to the pyrenyl-actin. Data shown are representative samples. (C) Time course of capping was determined from the decrease in initial rate of polymerization. The initial rate of increase in pyrenyl fluorescence (proportional to the number of elongating filaments) is plotted versus the duration of incubation of the seeds with the supernatant. The data, expressed as percent of seed-induced initial rate at the start of incubation, are from supernatants (closed squares) at 3 mg/ml protein (as used for most experiments) or 0.75 mg/ml (closed triangles). The half-time of capping, in supernatants at 3 mg/ml, ranged between 3 and 9 s in different supernatants.
Spectrin-Actin Seeds Are Rapidly Capped.
The failure of spectrin-actin seeds to increase F-actin levels might be due to rapid capping of the seeds by the cell supernatant. Indeed, incubation of spectrin-actin seeds for 1 min in supernatant before dilution into pyrenyl-actin decreased the rate of pyrenyl-actin polymerization induced to a steady (pointed-end) level ∼10% of that without incubation; the half-time of capping was 6 ± 3 s. (Fig. 1, B and C). This rapid capping is consistent with the kinetics of capping protein β2 present in neutrophil supernatants assayed previously (DiNubile et al., 1995).
Spectrin-Actin Seeds Did Not Enhance Cdc42-induced Polymerization.
Cdc42 could induce polymerization by inhibiting capping proteins in the supernatant or by freeing G-actin from sequestration. Were either the case, the effects of Cdc42 would be expected to extend to spectrin-actin seeds after the creation of new barbed ends. Thus, we examined whether the presence of spectrin-actin seeds enhanced the rate of polymerization induced by Cdc42. Spectrin-actin seeds caused no increased rate of polymerization (Fig. 1 A). Thus, the effects of Cdc42 do not extend to the barbed ends of other filaments present.
Induced Filaments Can Be Monitored by Electron Microscopy.
Filaments induced by Cdc42 can be observed after negative staining in the electron microscope. Incubation of 100 nM Cdc42 with supernatant for 1 min resulted in the appearance of many filaments (Fig. 2 A). The lengths of the filaments were measured (refer to Materials and Methods) and the distribution of their lengths is shown in Fig 2 E. The mean length of filaments was 2 μm. Consistent with their inability to induce polymerization, spectrin-actin seeds incubated with supernatant produced only few short filaments (Fig. 2, B and F). This was not a result of the seeds being ineffective nucleators since after a 30-s incubation with 1.5 μM pure actin, they produced many filaments with a mean length of 1.1 μm (Fig. 2, C and G).
Figure 2.


Electron microscopy of Cdc42-induced and spectrin-actin seed-induced filaments. Representative electron micrograph of negative-stained sample (left column) for: (A) 100 nM Cdc42 incubated in supernatant for 1 min; (B) spectrin-actin seeds (1.5 nM) incubated for 2 min in supernatant; (C) spectrin-actin seeds incubated for 30 s in 1.5 μM pure actin; (D) spectrin-actin seeds (1.5 nM) incubated in supernatant for 5 min in the presence of 1 μM phalloidin. Filament length distributions (right column) E–H measured for samples illustrated in A–D, respectively. The lengths of all filaments present longer than 0.25 μm were measured from photographs (equal to 4.5 mm on the photo). The data are expressed as the number of filaments on the y axis with a length equal to the value ± 0.25 μm on the x axis. Thus, all filaments with lengths between 0.25 to 0.75 μm are represented by the bar labeled 0.5, those with lengths 0.75–1.25 μm are represented by the bar labeled 1.0, etc. In each case the total filament number has been normalized to 100. Actual counts for each sample were: E, 102; F , 59; G, 175; H, 67. The mean lengths were: E, 2.1 μm; F, 0.4 μm; G, 1.1 μm; and H, 0.5 μm.
What Accounts for the Differences between Spectrin-Actin Seeds and Cdc42-induced Filaments?
Clearly mere addition of barbed ends to cell supernatants was not sufficient to increase F-actin levels or to induce visible filaments. The difference between the Cdc42 and spectrin-actin seeds could be explained if Cdc42-induced filaments were: (a) capped at the same rate but elongated much faster than spectrin-actin seeds; and/or (b) capped at a slower rate than spectrin-actin seeds; and/or (c) inhibited from depolymerization whereas spectrin-actin seeds depolymerized.
Filaments Induced by Cdc42 Elongate Rapidly.
To estimate the rate of Cdc42-induced filament elongation, the lengths of filaments present in supernatants incubated for various times with Cdc42 were determined (Fig. 3). Cdc42-induced filaments elongated rapidly, reaching a mean length of 1.4 μm at 15 s. An effective G-actin concentration (i.e., the sum of all free actin and actin complexes able to contribute to elongation) of ∼3.3 μM would be required for a filament to elongate to 1.4 μm in 15 s (given an actin barbed-end on rate of 107 M−1s−1 and assuming no depolymerization or capping). This estimate is conservative since it assumes that all filaments were present at time 0 and continued to elongate throughout the 15 s. A higher concentration would be required if there is a lag before Cdc42 produces free barbed ends and if new filaments are produced continuously during incubation (see below). A higher concentration would also be required if elongation were transient, e.g. if some filaments were capped within 15 s. A more realistic estimate of effective G-actin concentration is at least double this, i.e., 6.6 μM.
Figure 3.
Length distribution of filaments induced by Cdc42. Supernatant was incubated in 100 nM Cdc42 for 15 s (top), 30 s (middle), or 5 min (bottom) before negative staining. Filament lengths are displayed as described in Fig. 2. For comparison between times the total number of filaments at each time was normalized to 76; actual number of filaments measured at 15 s was 76 in six photos, at 30 s was 64 filaments in three photos, and at 5 min was 406 filaments in four photos. The mean filament length at 15 s was 1.4 μm; at 30 s, 1.7 μm; and at 5 min, 1.8 μm.
The very short length of filaments nucleated by spectrin-actin seeds suggests that the G-actin concentration available to Cdc42-induced filaments was not available to the spectrin-actin seeds. With an effective G-actin concentration of 3.3 μM and a half-time of capping at 6 s, the mean filament length would be ∼1 μm; with an effective G-actin concentration of 6.6 μM, the mean filament length would be 2 μm. Rather, the few filaments long enough to be detected had a mean length less than 0.4 μm.
The Mean Length of Cdc42-induced Filaments Changes Little over Time.
A surprising feature of the filament length distribution was that it changed only slightly with incubation time, being similar after 15 s and 5-min incubations (Fig. 3). For the data shown in Fig. 3, the mean length at 15 s was 1.4 μm, at 30 s it was 1.7 μm, and at 5 min it was 1.8 μm. (by a Chi square test, the distribution at 15 s was not significantly different from that at 5 min). The similar length distributions at 30 s and 5 min were observed in two additional experiments although there was variation in the absolute filament length between experiments.
The rate of polymerization is a function of the Cdc42 concentration (Zigmond et al., 1997). Increasing the concentration of Cdc42 from 100 to 800 nM increased the number of filaments observed on the grid at early times; however, increasing the Cdc42 concentration had little effect on the distribution of filament lengths (Fig. 4). Some decrease in filament length after longer incubation in 800 nM Cdc42 might reflect the fact that a limited amount of G-actin must now be distributed in a larger number of filaments.
Figure 4.
Comparison of filaments induced by 800 versus 100 nM Cdc42. Supernatants were incubated with 800 or 100 nM Cdc42 for 30 s, 1 min, or 5 min before negative staining. Photographed filaments were measured and pooled as in Fig. 2. For comparison, the filament number at each time point was normalized to a total of 100 filaments. The number of photographs analyzed and filaments actually measured at each time point was: for 800 nM Cdc42 at 30 s, two photographs with total of 127 filaments; for 1 min, four photographs with 127 filaments; and for 5 min, three photographs with 198 filaments. The mean length at 30 s was 1.7 μm; at 1 min, 1.4 μm; and at 5 min, 1.6 μm. For 100 nM Cdc42 at 30 s, there were three photographs with total of 47 filaments; at 1 min, three photographs with 102 filaments; and at 5 min, four photographs with 196 filaments. The mean length at 30 s was 1.7 μm; at 1 min, 2.1 μm; and at 5 min, 1.9 μm.
Previous studies have shown that upon addition of Cdc42, F-actin increases as a function of time (Zigmond et al., 1997). Since the mean filament length does not change over time, the increase in F-actin must reflect an increase in filament number. Indeed, the number of filaments visible in the electron microscope did increase with time. Control supernatants incubated between 0 and 5 min with buffer have few actin filaments. Scanning the entire grid revealed an occasional region with 1–4 filaments per 100 μm2. In contrast, more than 150 filaments per 100 μm2 could be observed in supernatant incubated 5 min with 100 nM Cdc42. Filaments were seen after incubations as short as 15 s. Since the filament distribution on the grid was not uniform, filament number was not rigorously quantified as a function of time; rather, photographs of regions selected merely for the presence of filaments were analyzed. Nevertheless, in each of five experiments analyzed, the mean number of filaments present in photographs (at a standard magnification) increased with incubation time up to 5 min.
The increase in filament number as a function of concentration and duration of incubation with Cdc42 was also detected as an increase in barbed-end nucleation sites for pyrenyl actin. Nucleation sites increased with Cdc42 concentration until a plateau level was reached (Fig. 5 A). The number of nucleation sites induced by 100 nM Cdc42 increased with time reaching a peak between 3 and 5 min (Fig. 5 B). With 100 nM Cdc42, there often was a lag before nucleation sites could be detected.
Figure 5.
Effect of Cdc42 on nucleation sites for pyrenyl actin. (A) Dose response of Cdc42-induced increase of nucleation sites. The supernatant was incubated for 5 min at 37°C with varying concentrations of GTPγS-charged Cdc42 before dilution into 1.5 μM pyrenyl-actin. The initial rate of polymerization of the pyrenyl-actin was determined from the pyrenyl fluorescence (refer to Materials and Methods). Data shown is from a representative experiment. The nucleation sites increase with concentration eventually reaching a plateau. The absolute levels of nucleation and the concentration of Cdc42 at the plateau vary somewhat with different supernatants and Cdc42 preparations. (B) Time course of Cdc42-induced increase in nucleation sites. The supernatant was incubated at 37°C with 100 nM GTPγS-charged Cdc42 for various times before dilution into 1.5 μM pyrenyl-actin. The initial rate of polymerization of the pyrenyl-actin was determined from the pyrenyl fluorescence (refer to Materials and Methods). Data shown is from a representative experiment.
Effect of Phalloidin on Filament Length Distributions.
It seemed unlikely that spectrin-actin seeds were depolymerizing in the supernatants since they contributed F-actin to the samples and this increase in F-actin was maintained during the incubations (refer to Fig. 1 A). Nevertheless, the filament length distribution of Cdc42-induced filaments could be affected by depolymerization. To examine the effects of depolymerization, supernatants were incubated in the presence of phalloidin to inhibit depolymerization (Cdc42 induced increases in F-actin were not inhibited by phalloidin [data not shown]). Even in the presence of phalloidin, few filaments were detected after 30 s or 1 min of incubation with spectrin-actin seeds. However, after 5 min, filaments with a mean length of ∼0.5 μm (n = 66) were present (refer to Fig. 2, D and H). Phalloidin, by inhibiting depolymerization, lowers the critical concentration at both ends of the filament. Thus, in the presence of phalloidin, filaments seen after 5 min may reflect pointed-end elongation from the spectrin-actin seeds or a partial inhibition/reversal of the barbed-end capping.
The length distributions of Cdc42-induced filaments present after 30 s, 1 min, or 5 min of cubation in supernatants containing 100 nM Cdc42 and phalloidin are shown in Fig. 6. As in the absence of phalloidin, the filament length distributions after incubation for 30 s and 1 min were similar. A direct comparison of filament length distributions induced by Cdc42 in the same supernatant with or without phalloidin confirmed that after 1 min of incubation, there was little difference between the length distributions of Cdc42-induced filaments in the presence or absence of phalloidin (data not shown). This indicates that during the first minute, Cdc42-induced filaments, even in the absence of phalloidin, are not depolymerizing (or being cut by cofilin since this is also inhibited by phalloidin). However, after a 5-min incubation, the presence of phalloidin causes a shift toward longer filaments: the mean length went from 1.5 μm (n = 231) at 1 min to 3.6 μm (n = 143) at 5 min. Thus, in the presence of phalloidin, Cdc42-induced filaments did elongate between 1 and 5 min.
Figure 6.
Time course of filament length distributions formed in the presence of phalloidin. Supernatants were incubated with Cdc42 as in Fig. 2 but 1 μM phalloidin was present during the incubation. After negative staining and photographing, filaments were measured and pooled as in Fig 3. For comparison, the filament number at each time point was normalized to a total of 231 filaments. The number of photographs analyzed and filaments actually measured at each time point was: at 30 s, four photographs with a total of 137 filaments; at 1 min, seven photographs with 231 filaments; and at 5 min, four photographs with 143 filaments. The mean length at 30 s was 1.5 μm; at 1 min, 1.5 μm; and at 5 min, 3.7 μm.
Clumps in the Cdc42 Preparation Serve As Foci for Polymerization and Are Preferentially Associated with the Barbed Ends of Filaments
The fact that enhanced elongation occurred selectively on the Cdc42-induced filaments suggested that Cdc42 or some Cdc42 target might remain associated with the barbed end during elongation. Geranylgeranylated Cdc42 often aggregates and then both the Cdc42 (as assayed by Western blots) and ability to induce polymerization are lost upon pelleting the Cdc42 preparation at high speed (Zigmond et al., 1997). Actin polymerization induced by Cdc42, when viewed by either fluorescence or electron microscopy occasionally emanated from foci. By S-1 labeling, the filaments emanating from these foci were observed to have their barbed ends preferentially (∼75% of the filaments) associated with the focus (Fig. 7). Although we have no direct evidence that Cdc42 is in these clumps, this filament orientation is similar to that of filaments induced at the leading edge of cells and by cytoplasmic Listeria and is consistent with the observation that, like Listeria, Cdc42 can mediate movement of lipid vesicles (Ma et al., 1998). Since filaments elongate at their barbed ends, it appears that the barbed end might for a time remain associated with factors that could affect their rate of elongation and/ or capping.
Figure 7.
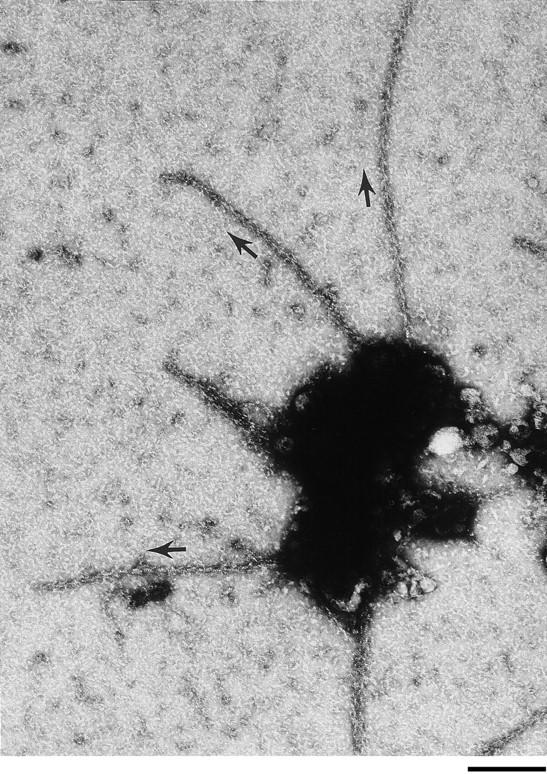
The barbed ends of actin filaments point toward foci of polymerization. Cdc42 was incubated in supernatant for 1 min before dilution into buffer containing phalloidin followed by incubation in S-1 fragment of myosin (refer to Materials and Methods). Samples were rinsed in water before negative staining. Arrows parallel to some of the filaments indicate the orientation of the arrowheads on the filaments. The orientation of 143 filaments was determined in 34 photographs of foci from two different experiments. 75% of the filaments emanating from a focus and whose orientation could be determined had their barbed ends at the foci. Bar, 0.25 μm.
Modeling of the Data
Examination of the length distributions of Cdc42-induced filaments suggested that these filaments elongated rapidly and transiently. To determine if the data were compatible with the hypothesis that Cdc42-induced filaments elongated more rapidly than spectrin-actin seeds but were stopped at the same rate, we developed a mathematical model (Model 1) of the system. Alternatively, to determine if our data were compatible with the hypothesis that Cdc42-induced filaments elongated at a normal rate but were stopped at a slower rate than for spectrin-actin seeds, we developed a second model (Model 2). The starting conditions for both models were based on measured concentrations of various components in neutrophil supernatants (refer to Materials and Methods). Both models assumed that Cdc42 created new filaments at a constant rate. This was compatible with the observed increase in filament number as a function of time.
As shown in Fig. 8, the filament length distribution of Cdc42-induced filaments at various times in the absence of phalloidin could be fit by Model 1. The fit required enhancing the rate of polymerization over that expected from homogenous concentrations of G-actin and profilin-actin (see Fig. 8 legend). This demonstrated that the small changes in filament length distribution over time did not require modification of the capping rate. The same parameters allowed a fit of the spectrin-actin seed filament length distribution (refer to Fig. 2 F) if the rate of elongation was assumed to be due to free G-actin alone, without any contribution by profilin-actin (data not shown). In contrast, it was not possible to fit both the early and late Cdc42-induced filament length distributions with Model 2. If the lengths of the filament population at later times was determined by capping, the lengths at earlier times would need to be shorter. In fact, increasing the half-time of capping beyond 6 s resulted in a less good fit to these data. If rapid elongation of Cdc42-induced filaments has a half-life longer than 6 s, a more complicated model is required to generate the distribution.
Figure 8.

Match of models to data. The Cdc42-induced filaments that contribute to Fig. 3 are replotted here as the absolute number of filaments falling in different length categories (bar graph). Closed triangles, numbers predicted by Model 1 (refer to Materials and Methods), assuming that Cdc42 enhances the rate of profilin-actin mediated elongation by a factor fp = 4.5, compared to that seen with spectrin-actin seeds, but elongation is terminated at a rate similar to that of spectrin-actin seeds. The nucleation rate used was 0.026 nM filaments/s. A chi-square test of goodness of fit gave P > 0.1. Open circles, numbers predicted by Model 2, with no acceleration of elongation (fp = 1.0), but an increase in the duration of elongation by a factor of 4.6 (k cap = 0.025/s) compared to that of spectrin-actin seeds (k cap = 0.115/s). The nucleation rate used was 0.029 nM filaments/s. No meaningful chi-square could be calculated due to the very small predicted numbers of longer filaments at the earlier times.
The presence of phalloidin resulted in an increase in long filaments by 5 min. This shift could not be fit merely by eliminating all depolymerization in Model 1. Thus, some means of enhancing filament length via barbed-end elongation or filament annealing is needed. The situation may be complex. For example, it would be possible to explain the results shown in Fig. 5 by the presence of two populations of filaments: one that predominates at 30 s and grows as much as do filaments in the absence of phalloidin, but that is slowly converted to the second population, that can grow to substantially greater lengths and supplies the majority of the filaments at 5 min. The latter population could both be protected from capping and subject to enhanced rates of elongation, either permanently or transiently.
Exactly how Cdc42 promotes polymerization is not clear. However, a model consistent with our data is shown in Fig. 9: Cdc42 activates a complex which both creates a free barbed end (Nuc) and promotes elongation (EP) of that end. We suggest that Cdc42 remains with the elongation-promoting complex, thus acting in manner parallel to ActA in Listeria movement. Alternatively, Cdc42 could merely activate the elongation-promoting complex. The promotion of elongation involves enhanced rates of G-actin addition (we cannot rule out the possibility that it also allows prolonged elongation). After a short time the rapid elongation is terminated either by release of the elongation promoting complex and/or by capping. The activated Cdc42 can then assemble a new active complex.
Figure 9.
A Model. Cdc42 is depicted as interacting with several factors including Nuc that creates free barbed ends and an elongation promoting factor (EP) that facilitates rapid elongation. Filament elongation is terminated by release of EP and/or addition of a capper.
Discussion
The current studies demonstrate that: (a) the ability of Cdc42 to induce actin polymerization in cell supernatant cannot be explained merely through its ability to create free barbed ends, since exogenous barbed ends do not induce measurable polymerization in cell supernatant; (b) Cdc42 facilitates rapid filament elongation producing filaments greater than 1 μm in length whereas spectrin-actin seeds elongate little in supernatant. Had spectrin-actin seeds elongated at comparable rates before they were capped, they would have induced longer filaments; (c) there was little change in mean filament length between 15 s and 5 min, suggesting that the increase in F-actin over this time was due to an increase in filament number; and (d) whatever Cdc42 does to facilitate elongation, its effects are processive, i.e., restricted to Cdc42-induced filaments as demonstrated by the fact that the presence of Cdc42 did not allow spectrin-actin seeds to contribute to polymerization.
Free barbed ends are required for polymerization in vivo. But, it appears that barbed ends alone are not sufficient. Thus, addition of barbed ends did not increase the F-actin level in cell supernatants. Interestingly, the inability of F-actin seeds to elongate and induce polymerization has also been observed after their injection into live cells (Sanders et al., 1990). Only when enough barbed ends were added to adsorb out all of the capper was polymerization stimulated (Handel et al., 1990). In preliminary experiments we also find that if the spectrin-actin seed concentration in the supernatants exceeds the capper concentration (200 nM seeds added to supernatant with an estimated capper concentration of 100 nM), polymerization is induced. It seems unlikely that Cdc42 acts by adsorbing out, or inactivating, capper since: (a) even if all the G-actin present in supernatants were used to create filaments with a mean length of 2 μm (as measured), the concentration of filaments would be only 7 nM, much less than the concentration of capping protein, 100 nM (DiNubile et al., 1995); (b) if Cdc42 inactivated all the capper, spectrin-actin seeds should have increased the rate of polymerization but they did not; and (c) supernatant that had been activated by lipid plus GTPγS to polymerize actin (Zigmond et al., 1997) was able to cap spectrin-actin seeds with a time course similar to control supernatants (data not shown). This indicates that activation of polymerization does not require the inhibition of capper.
The Cdc42-induced increase in F-actin level is associated with an increase in filament number. The observations are consistent with the hypothesis that GTPγS-activated Cdc42 continuously creates new filaments until the G-actin level is depleted (refer to Fig. 4) (Zigmond et al., 1997). If, at this point more G-actin is added, further polymerization is observed (data not shown). Since Cdc42 does not stimulate polymerization of pure actin, additional factors in the supernatant must be involved. Whether new filaments are created by Cdc42 activating a complex like Arp2/3 that then nucleates new filaments (Mullins et al., 1998) or removes a capper from actin oligomers remains to be determined.
The length of a filament, in the absence of depolymerization, depends on the rate and duration of elongation. If both ends of the filament are free, filaments can also anneal and thus increase their length. However, since the total filament concentration, especially at early times, is low (nM), annealing would be infrequent. Thus, unless enhanced by Cdc42, annealing is unlikely to account for the lengths observed. Rather, since Cdc42-induced filaments are approximately five times longer than spectrin-actin seed-induced filaments (mean length of 2 μm after incubation in Cdc42 for 1 min versus 0.4 μm for spectrin-actin seeds), Cdc42-induced filaments probably elongate five times faster and/or longer than spectrin-actin–induced filaments. If both seeds and Cdc42-induced filaments elongate for a half-time of 6 s as illustrated in Model 1 of Fig. 8, all of the length difference would be achieved by the rate of elongation. On the other hand, the outer limits of our data, i.e., spectrin-actin seeds capped with a half-time of 3 s and Cdc42-induced filaments elongating with a half-time of 12 s, would allow a fourfold difference in duration of elongation. Thus, from existing data, we cannot rule out that such a difference in duration of elongation exists and contributes the differences in filament length.
Nevertheless, our data suggest that elongation of Cdc42-induced filaments is more rapid than that of spectrin-actin seeds. Even if seeds were capped with a half -time of 3 s, had they elongated at the rate observed for Cdc42-induced filaments, longer filaments would have been seen. A spatially restricted concentration of effective G-actin is also suggested from estimates of the concentrations of free G-actin and profilin-actin in the supernatants (Kang et al., 1997). Given measured concentrations of actin, Tβ4 and profilin in neutrophil supernatants, we estimate the sum of the free G-actin concentration and the profilin-actin concentration to be approximately 2.1 μM. Although each concentration could be somewhat in error and vary from preparation to preparation, it is unlikely, (given currently known factors), that the supernatant provides a homogeneous effective G-actin concentration of 6.6 μM. Our data are compatible with Cdc42-induced elongation being terminated by either dissociation of an elongation-promoting factor followed by capping or by capping itself terminating the elongation. Further studies will be required to distinguish between these alternatives.
The form of the effective G-actin and the means of localizing it to Cdc42-induced filaments will require further investigation. Profilin-actin can contribute to barbed-end elongation and several Cdc42-associated proteins bind profilin either directly, i.e., Bni1p (Evangelista et al., 1997; Imamura et al., 1997) or indirectly, i.e., Wiscott-Aldrich syndrome protein, WASP (Burbelo et al., 1995; Symons et al., 1996; Miki et al., 1998) which binds WIP, a profilin-binding protein (Ramesh et al., 1997). The association of Cdc42 with profilin-binding proteins could locally enhance polymerization at the barbed ends of the Cdc42-induced filaments just as profilin bound to VASP is believed to enhance the rate of Listeria induced polymerization (Smith et al., 1996; Kang et al. 1997). The observation that filaments emanating from clumps in the Cdc42 preparation are preferentially oriented with their barbed ends at the clump and the observation that Cdc42 mediates movement of lipid vesicles in Xenopus extracts (Ma et al., 1998) support the idea that Cdc42, or a Cdc42-activated complex, is transiently associated with the barbed end. In this regard, the Cdc42-induced polymerization resembles ActA-induced polymerization which results in movement of Listeria. The filaments are oriented with their barbed ends toward ActA which is known to bind VASP which in turn binds profilin (Chakraborty et al., 1995; Reinhard et al., 1995). However, it appears that profilin is not required for movement of Listeria in Xenopus extracts (Marchand et al., 1995).
Induction of actin polymerization by Cdc42 appears to require both production of free barbed ends and enhancement of their elongation. The concentration of capping protein in neutrophils and the estimated on rate of capping suggest that a free barbed end would have a half-life of only 0.6 s in vivo (DiNubile et al., 1995). Thus, it seems unlikely that Cdc42 first activates a process to create a barbed end and then activates a process to promote the elongation of the end. Rather, the two processes are likely to be coupled. They may be activated simultaneously as parts of a complex activated by Cdc42. Alternatively, the elongation-promoting complex forms first since its action would not be triggered until the barbed end is available. Interestingly, a single protein, ActA, of Listeria has separate regions that promote nucleation and elongation.
Cells have likely evolved several mechanisms for regulating actin polymerization including production of free barbed ends, enhanced rates of elongation, and inhibition of capping. Different agonists may vary with regard to which mechanism they use. Thus, for reasons not yet clear, Cdc42 but not Rac can induce polymerization in broken cells (Ma et al., 1998). Rac is a better stimulator of PIP2 synthesis (Zigmond et al., 1997), and thus potentially may be more effective at locally inhibiting capping when the polymerization occurs near a phosphatidylinositol-rich membrane (Hartwig et al., 1995). Possibly when membrane associated, Cdc42 may also inhibit capping. Listeria, when inducing membrane protrusions, must inhibit capping since filaments associated with Listeria in pseudopodial extensions are very long. On the other hand, when in the cell center, many Listeria-induced filaments are rather short, perhaps because capping has not been inhibited (Tilney et al., 1992; Sechi et al., 1997).
Comparison with In Vivo Systems
The Cdc42-induced polymerization in supernatants shares a number of properties with actin polymerization induced in neutrophils in vivo by chemoattractant. In each case, polymerization is associated with an increase in the number of filaments with little change in the filament length distribution (Cano et al., 1991). Future studies are aimed at defining the molecular mechanisms mediating these actin changes.
Acknowledgments
We are grateful to G. Bokoch (The Scripps Research Institute, La Jolla, CA) for generously supplying us with recombinant Cdc42. We thank D. Safer (University of Pennsylvania, Philadelphia, PA) for the myosin fragment S-1. We are indebted to L. Tilney and P. Connelly (both from University of Pennsylvania) as well as P. Sterling and S. Shrom (both from University of Pennsylvania Medical School) for use of electron microscopes, darkrooms, and important advice regarding the electron microscopy. We thank summer students J. Lartique and H. Sun for filament measurements. Finally, we have been greatly aided by our Penn colleagues: A. Weber, V. Nachmias, L. Tilney, M. Ostap, and E. De La Cruz for conversations and suggestions on the manuscript.
This work was supported by a National Institutes of Health grant (AI-19883) to S.H. Zigmond.
Footnotes
Address all correspondence to Sally H. Zigmond, Biology Department, University of Pennsylvania, Philadelphia, PA 19104-6018. Tel.: (215) 898-4559. Fax: (215) 898-8780. E-mail: szigmond@sas.upenn.edu
1. Abbreviation used in this paper: GST, glutathione-S-transferase.
References
- Adams AEM, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen WE, Jones GE, Pollard JW, Ridley AJ. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J Cell Sci. 1997;110:707–720. doi: 10.1242/jcs.110.6.707. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- Cano M, Lauffenburger DA, Zigmond SH. Kinetic analysis of F-actin depolymerization in polymorphonuclear leukocyte lysates indicates that chemoattractant stimulations increases actin filament number without altering filament length distribution. J Cell Biol. 1991;115:677–687. doi: 10.1083/jcb.115.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M-F, Laurent V, Santolini J, Melki R, Didry D, Xia G-X, Hong Y, Chua N-H, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1323. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M-F, Pantaloni D. Control of actin dynamics in cell motility. J Mol Biol. 1997;269:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- Casella JF, Maack DJ, Lin S. Purification and initial characterization of a protein from skeletal muscle that caps the barbed ends of actin filaments. J Biol Chem. 1986;261:10915–10921. [PubMed] [Google Scholar]
- Cassimeris L, Safer D, Nachmias VT, Zigmond SH. Thymosin β4 sequesters the majority of G-actin in resting human polymrophonuclear leukocytes. J Cell Biol. 1992;119:1261–1270. doi: 10.1083/jcb.119.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T, Ebel F, Domann E, Niebuhr K, Gerstel B, Pistor S, Temm-Grove CJ, Jockusch BM, Reinhard M, Walter U, Wehland J. A focal adhesion factor directly linking intracellularly motile Listeria monocytogenes and Listeria ivanoviito the actin-based cytoskeleton of mammalian cells. EMBO (Eur Mol Biol Organ) J. 1995;14:101–108. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-M, Hobbie S, Galan JE. Requirement for CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science. 1996;274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNubile MJ, Cassimeris LU, Joyce M, Zigmond SH. Actin filament barbed-end cappng activity in neutrophil lysates: the role of capping protein β2. Mol Biol Cell. 1995;12:1659–1671. doi: 10.1091/mbc.6.12.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Wepf R, Simons K. Roles for Rac1 and Cdc42 in planar polarization and hair outgrowth in the wing of Drosophila. . J Cell Biol. 1996;135:1277–1289. doi: 10.1083/jcb.135.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtime MS, Chow CJ, Adams N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fechheimer M, Zigmond SH. Focusing on unpolymerized actin. J Cell Biol. 1993;123:173–181. doi: 10.1083/jcb.123.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel SE, Hendry KAK, Sheterline P. Microinjection of covalently cross-linked actin oligomers causes disruption of existing actin filament architecture in PtK2 cells. J Cell Sci. 1990;97:325–333. doi: 10.1242/jcs.97.2.325. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Bokoch GM, Carpenter CL, Janmey PA, Taylor LA, Toker A, Stossel TP. Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis in permeabilized human platelets. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- Heyworth PG, Knaus UG, Xu X, Uhlinger DJ, Conroy L, Bokoch GM, Curnutte JT. Requirement for postranslational processing of Rac GTP-binding proteins for activation of human neutrophil NADPH oxidase. Mol Biol Cell. 1993;4:261–269. doi: 10.1091/mbc.4.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, T.H., and C.O. Oresajo. 1985. A method for quantifying F-actin in chemotactic peptide activated neutrophils: study of the effects of tBOC peptide. Cell Motil. Cytoskeleton. 5:545–557. [DOI] [PubMed]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family of small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiaegene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F, Laine RO, Rubb MR, Southwick FS, Purich DL. Profilin interacts with the Gly-Pro-Pro-Pro-Pro-Pro sequences of vasodilator-stimulated phosphoprotein (VASP): implications for actin-based Listeriamotility. Biochemistry. 1997;36:8384–8392. doi: 10.1021/bi970065n. [DOI] [PubMed] [Google Scholar]
- Keely PJ, Westwick JK, Whitehead IP, Der CD, Parise LV. Cdc42 and Rac1 induced integrin-mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390:632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- Knaus UG, Heyworth PG, Kinsella BT, Curnutte JT, Bokoch GM. Purification and characterization of Rac 2: A cytosolic GTP-binding protein that regulates human neutrophil NADPH oxidase. J Biol Chem. 1992;267:23575–23582. [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Cantley LC, Janmey PA, Kirschner MW. Corequirement of specific phosphoinositide and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopusegg extracts. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba canstellanii. . J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand J-B, Moreau P, Paoletti A, Cossart P, Carlier M-F, Pantaloni D. Actin-based movement of Listeria monocytogenes: actin assembly results from the local maintenance of uncapped filament barbed ends at the bacterium surface. J Cell Biol. 1995;130:331–343. doi: 10.1083/jcb.130.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N_WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation high affinity pointed end capping and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Carlier M-F. How profilin promotes actin filament assembly in the presence of thymosin β4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Perelroizen I, Marchand J-B, Blanchoin L, Didry D, Carlier M-F. Interaction of profiin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profiinon actin filament nucleation and elongation. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Pring M, Weber A, Bubb MR. Profilin-actin complexes directly elongate actin filaments at the barbed end. Biochemistry. 1992;31:1827–1836. doi: 10.1021/bi00121a035. [DOI] [PubMed] [Google Scholar]
- Ramesh N, Anton IM, Hartwig JH, Geha RS. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilin. EMBO (Eur Mol Biol Organ) J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MC, Wang Y-L. Exogenous nucleation sites fail to induce detectable polymerization of actin in living cells. J Cell Biol. 1990;110:359–365. doi: 10.1083/jcb.110.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi AS, Wehland J, Small JV. The isolated comet tail of pseudopodium of Listeria monocytogenes: a tail of two actin filament populations, long and axial and short and random. J Cell Biol. 1997;139:155–167. doi: 10.1083/jcb.137.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Theriot JA, Portnoy DA. The tandem repeat domain in the Listeria monocytogenesActA protein control the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J Cell Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwick FS, Young CL. The actin released from profilin-actin complexes is insufficient to account for the increase in F-actin in chemoattractant-stimulated polymorphonuclear leukocytes. J Cell Biol. 1990;110:1965–973. doi: 10.1083/jcb.110.6.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, Yelon D, Berg LJ, Chant J. Regulation of the polarization of T cells toward antigen-presenting cells by Ras-related GTPase Cdc42. Proc Natl Acad Sci USA. 1995;92:5027–5031. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SJ, Zigmond SH. Chemotactic receptor modulation in polymorphonuclear leukocytes. J Cell Biol. 1980;85:703–711. doi: 10.1083/jcb.85.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M, Derry JMJ, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiscott-Aldrich Syndrome Protein, a novel effect for the GTPase Cdc42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Tilney LG, DeRosier DJ, Tilney MS. How Listeriaexploits host cell actin to form its own cytoskeleton. I. Formation of a tail and how that tail might be involved in movement. J Cell Biol. 1992;118:71–81. doi: 10.1083/jcb.118.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TP, Weber A, Higgins J, Bonder EM, Mooseker M S. Effect of villin on the kinetics of actin polymerization. Biochemistry. 1984;23:2613–2621. doi: 10.1021/bi00307a012. [DOI] [PubMed] [Google Scholar]
- Weber A, Nachmias VT, Pennise CR, Pring M, Safer D. Interaction of thymosin β4 with muscle and platelet actin: implications for actin sequestration in resting platelets. Biochemistry. 1992;31:6179–6185. doi: 10.1021/bi00142a002. [DOI] [PubMed] [Google Scholar]
- Xu X, Barry DC, Settleman J, Schwartz MA, Bokoch GM. Differing structural requirements for GTPase-activating protein responsiveness and NADPH oxidase activation by Rac. J Biol Chem. 1994;269:23569–23574. [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Maekawa S, Sadai H. Studies on the interaction between actin and cofilin purified by a new method. Biochem J. 1988;251:121–127. doi: 10.1042/bj2510121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Joyce M, Borleis J, Bokoch GM, Devreotes PN. Regulation of actin polymerization in cell-free systems by GTPgS and Cdc42. J Cell Biol. 1997;138:363–374. doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]









