Abstract
In plants, light perception by photoreceptors leads to differential expression of an enormous number of genes. An important step for differential gene expression is the regulation of transcription factor activities. To understand these processes in light signal transduction we analyzed the three well-known members of the common plant regulatory factor (CPRF) family from parsley (Petroselinum crispum). Here, we demonstrate that these CPRFs, which belong to the basic- region leucine-zipper (bZIP) domain-containing transcription factors, are differentially distributed within parsley cells, indicating different regulatory functions within the regulatory networks of the plant cell. In particular, we show by cell fractionation and immunolocalization approaches that CPRF2 is transported from the cytosol into the nucleus upon irradiation due to action of phytochrome photoreceptors. Two NH2-terminal domains responsible for cytoplasmic localization of CPRF2 in the dark were characterized by deletion analysis using a set of CPRF2-green fluorescent protein (GFP) gene fusion constructs transiently expressed in parsley protoplasts. We suggest that light-induced nuclear import of CPRF2 is an essential step in phytochrome signal transduction.
Keywords: light regulation, phytochrome, bZIP transcription factors, nucleocytoplasmic partitioning, retention domain
Light plays a central role in the morphogenesis of plants (Kendrick and Kronenberg, 1994). They have evolved a set of photoreceptor systems that include phytochromes, blue/UV-A, and UV-B receptors to percept the quality, quantity, direction, and duration of environmental light conditions to adapt optimal growth (Frankhauser and Chory, 1997). Many of the developmental changes occuring during photomorphogenesis are controlled by members of the phytochrome (phy)1 photoreceptor family encoded by five different genes in Arabidopsis thaliana (PHYA to PHYE). Phytochromes are photoreversible chromoproteins that are synthesized in their physiologically inactive, red light-absorbing forms (Pr) and converted to their active, far-red light-absorbing forms (Pfr) upon irradiation. The red/far-red photoreversibility (reversible Pfr/Pr conversion) of a given response (low fluence response) and the responsiveness to continuous far-red light (high irradiance response) are two well-established operational criteria for the involvement of phyB and phyA, respectively, in light perception and signaling (Quail et al., 1995; Whitelam and Devlin, 1997).
Regulatory expression of genes is essential for plant adaptation during photomorphogenesis (Frankhauser and Chory, 1997). Transcriptional regulation of gene expression is largely mediated through sequence-specific DNA-binding proteins that interact with cis-acting elements located in the promoter regions of the corresponding genes. The binding of transcription factors to relevant cis-acting elements alters the activity of the general transcription machinery stimulating or suppressing the expression of genes (for review see Tjian and Maniatis, 1994). Several cis-acting elements were identified within plant promotors (Terzaghi and Cashmore, 1995). One of the best characterized elements is the hexameric G-box (CACGTG) found in the promotors of light-regulated genes such as ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit from tomato and chalcon synthase (CHS) from parsley (for review see Menkens et al., 1995; Frankhauser and Chory, 1997). The G-box often functions together with other cis-elements as, for example, has been demonstrated for the light-regulated elements of the parsley CHS promotor (Schulze-Lefert et al., 1989a; Block et al., 1990). The proteins that bind to the G-box belong to the basic-region leucine-zipper (bZIP) transcription factor family (Foster et al., 1994). This domain comprises two structural motifs: a stretch rich in basic residues mediating DNA-binding (Weiss et al., 1990; O'Neil et al., 1991; O'Shea et al., 1991; Ellenberger et al., 1992) adjacent to a leucine-zipper responsible for homo- and heterodimer formation (Rasmussen et al., 1991). The basic stretch also contains the nuclear localization sequence (NLS) as shown exemplarily for opaque 2 (Varagona et al., 1992) and TGA-1 (van der Krol and Chua, 1991). In addition, with few exceptions, all plant bZIPs that have been cloned so far, such as the G-box–binding factors (GBF) from Arabidopsis and the CPRFs from parsley, contain a proline-rich NH2-terminal region that functions as a transcriptional activation or repression domain in animal and plant cells (Weisshaar et al., 1991; Schindler et al., 1992a,b; Feldbrügge et al., 1994). The specificity of homodimeric bZIP proteins for a G-box–like element depends on sequences flanking the ACGT core (for review see Foster et al., 1994; Meshi and Iwabuchi, 1995). For instance, the GBFs from Arabidopsis and CPRF1 and CPRF4 from parsley bind with high affinity to the classical G-box (CACGTG). In contrast, the bZIP factors TGA1, OBF4, and bA19 from Arabidopsis bind to GACGT(T/C) motifs but not to the G-box itself. Proteins with intermediate characteristics for DNA-binding specificity as for example maize opaque2 and parsley CPRF2 are known as well.
The capacity for heterodimer formation as well as for heterotypic interaction with members of other transcription factor families (Armstrong et al., 1992; Schindler et al., 1992b; Büttner and Singh, 1997; Vicente-Carbajosa et al., 1997) provides a wealth of possible transcription factor complexes with potentially distinct binding activities and activation abilities resulting in signal-specific regulation of a particular target gene. The regulation of the formation and activity of such transcription factor complexes and, thus, the ability to transactivate a certain gene is accomplished by several mechanisms on different regulatory levels. (a) Although most plant bZIP proteins are constitutively expressed, some are restricted to a particular tissue or differentially regulated by exogenic or endogenic signals (Schindler et al., 1992a; Menkens and Cashmore, 1994; Kircher et al., 1998). (b) The DNA-binding activity as well as the transactivation ability of bZIPs can be regulated by posttranslational modifications as, for instance, phosphorylation (Klimczak et al., 1992, 1995; Harter et al., 1994a). (c) The control of nuclear localization is also known to regulate transcription factor activity. The cytosolic retention in absence of the appropriate signal can be achieved by anchoring proteins or by retention factors (for reviews see Jans and Hübner, 1996; Ghosh et al., 1998). Cytoplasmic localization of some plant bZIP proteins was recently demonstrated for G-box binding proteins in dark-grown parsley cells (Harter et al., 1994a) and for Arabidopsis GBF1 and GBF2 in Arabidopsis and transiently transformed soybean protoplasts (Terzaghi et al., 1997). After irradiation with white light at least one of the cytosolic parsley bZIP factors is imported into the nucleus in vitro (Harter et al., 1994a). Similarly, during cultivation of transiently transformed soybean cells under blue light, the pool of GBF2, but not of GBF1 protein is now found in the nuclear compartment (Terzaghi et al., 1997).
Here we report that the bZIP factors CPRF1, CPRF2, and CPRF4 are differentially distributed within dark-cultivated parsley cell with CPRF1 localized in the nucleus, CPRF2 found in the cytosol, and CPRF4 present in both compartments. Using three different in vivo assays we are able to show that CPRF2 is transported from the cytosol into the nucleus upon light irradiation. Furthermore, immunolocalization studies reveal that phyA via high irradiance response and phyB via a low fluence response are the main photoreceptors involved in the nuclear translocation response of CPRF2. Mapping of the retention domains within the CPRF2 sequence reveals two functionally independent motifs responsible for cytoplasmic localization and let us propose two alternative hypothesis for the molecular mechanism of CPRF2 retention in darkness.
Materials and Methods
Light Sources
UV-containing white light, UV-A light, and blue light sources were used as described in Frohnmeyer et al. (1992). Standard red, far-red, and RG9 light conditions were used as described in Schäfer (1977). If not otherwise indicated, all experiments were carried out under dim-green safelight according to Schäfer (1977).
Suspension Culture, Preparation of Protoplasts, and Isolation of Cytosolic and Nuclear Extracts
Protoplasts were prepared from a dark-grown parsley (Petroselinum crispum) cell culture 6 d after subculturing (Dangl et al., 1987). Preparation of parsley protoplasts and evacuolated protoplasts was performed as described by Frohnmeyer et al. (1994). Isolation of cytosolic and nuclear extracts from evacuolated protoplasts was performed as described previously (Harter et al., 1994a). For technical reasons we used a newly established cell culture for immunostaining. A large part of these cells contained several small vacuoles instead of a big central one reducing the proportion of disintegrated cells which typically occurs during fixation of plant cells.
Plasmid Construction
All primers used in this study are shown in Table II. The green fluorescent protein (GFP) expression cassette was removed from the pBI121–GFP construct (Haseloff et al., 1997) by HindIII/EcoRI digest and transferred into pUC18 (GFP–pUC18). Afterwards a 5′ BamHI followed by an in-frame SmaI restriction site were introduced into GFP coding region by PCR using the primers P1 and P2. The resulting plasmid (mAV4) was used for the construction of COOH-terminal fusions of the CPRF1 and CPRF2 coding regions with GFP. For this purpose, BamHI-compatible restriction sites were introduced at the 5′ ends and SmaI sites at the 3′ ends into the full-length coding regions of common plant regulatory factors (CPRFs) by PCR using the gene-specific primers P3 to P6. For the construction of the 5′ deletions of CPRF2 the 5′ primers P7 (amino acid [aa]80CPRF2), P8 (aa159CPRF2), and P9 (aa178CPRF2) were used in combination with primer P6 introducing a BamHI-compatible site and a start ATG in front of the first codon. The ΔCPRF2–GFP construct was produced as follows: primers P5 and P10 were used to produce a PCR product encoding aa 1–177 that contains a BamHI restriction site at its 5′ end and a KpnI site at its 3′ end. Primers P11 and P6 were used to polymerize an additional PCR product encoding amino acid 193–403 that contains a KpnI site at its 5′ end and a SmaI site at its 3′ end. After cutting with BamHI and KpnI the PCR products were ligated into mAV4. The NLS–GFP and PHYA–GFP constructs were produced as follows: primer P12, P13, and P14 and P15, respectively, were used to polymerize 5′ BamHI and 3′ SmaI-containing PCR products that encode either the NLS of CPRF4 (aa306–332; according to Van der Krol and Chua, 1991 and Varagona et al., 1992) or phyA. The BamHI and SmaI-cut PCR products were ligated into mAV4 as described above. After the BamHI-compatible sites all 5′ primers also introduce the short sequence stretch 5′-AACA-3′ allowing efficient translation of RNA in plant cells.
Table II.
Oligonucleotides Used as Primers in This Study
| Number | Sequence | PCR template* | ||
|---|---|---|---|---|
| P1 | 5′-CGGGATCCCGCCCGGGATGAGTAAAGGAGAAGAACTTTTCACTGG-3′ | GFP | ||
| P2 | 5′-GCGGAGCTCTTATTTGTATAGTTCATCCATCCATGCC-3′ | GFP | ||
| P3 | 5′-GAGGATCCAACAATGGGTAGCAACGAAGAAGGAAACCCC-3′ | CPRF1 | ||
| P4 | 5′-GAAGATCTCCCCGGGACCTGCAGCAACAGCATCTGTCCG-3′ | CPRF1 | ||
| P5 | 5′-GCGGATCCATAACAATGGATAGGGTGTTTTCAGTGG-3′ | CPRF2 | ||
| P6 | 5′-TACCCCGGGCTGCTTACCACTGACCTGGGCCTCGC-3′ | CPRF2 | ||
| P7 | 5′-GCGGATCCATGGAATTCTCGGAGGATTATCAAGCTTATCTC-3′ | CPRF2 | ||
| P8 | 5′-GCGGATCCATGGAATTCGCCTTGCAAAAGAAATCTGC-3′ | CPRF2 | ||
| P9 | 5′-GCGGATCCATGGAATTCGACCACTCAGACGATGACGATG-3′ | CPRF2 | ||
| P10 | 5′-GAGGTACCTCTAGATGAACCACTTGTTGATTT-3′ | CPRF2 | ||
| P11 | 5′-GAGAATTTCGCTACCACTCGGAACGGAGATCC-3′ | CPRF2 | ||
| P12 | 5′-GAGGATCCAACAATGGACGAAAGGGAGCTTAAAAGG-3′ | CPRF4 | ||
| P13 | 5′-TCGGATCCCCGGGCGCCTGCTTCCGTAACCTGGATCG-3′ | CPRF4 | ||
| P14 | 5′-GAAGATCTAACAATGTCATCTTCAAGACCTGCAAACTCTTCC-3′ | PHYA | ||
| P15 | 5′-GAAGATCTCCCCGGGAGATTCGCGTTTACTAGCTGCAGC-3′ | PHYA |
Transient Transformation of Protoplasts by Electroporation
Electroporation of protoplasts was performed in 0.4-cm cuvettes for 5 s at 320 V and 125 μF (Ω → ∞•, pulse time = 10 ms) using a Gene Pulser II (Bio-Rad) at room temperature. For transformation, 1 vol of protoplast suspension was diluted with 5 vol of electrode buffer (10 mM Tris/Hepes, pH 7.2, 15 mM MgCl2, 0.5 M sucrose). The suspension was centrifuged for 5 min at 500 g and the floating protoplasts were removed. 5 × 106 protoplasts were mixed with 75 μg of plasmid DNA in a final volume of 800 μl electrode buffer. After electroporation, protoplasts were diluted in 5 ml hemagglutinin and 0.4 M sucrose medium, transferred to Petri dishes, and then used for irradiation experiments.
Immunostaining and Confocal Microscopy
Immunofluorescence labeling of CPRF2 was performed following a protocol described in Petrášek et al. (1998). Parsley cells were settled on glass slides that had been coated with Meyer's adhesive (1% wt/vol sodium salicylate in 1:1 vol/vol egg-white/glycerol). Cells were fixed for 30 min in freshly prepared paraformaldehyde (3.7% wt/vol), solved in MS buffer (50 mM Pipes, pH 6.9, 1 mM MgSO 4, 5 mM EGTA, 1% vol/vol glycerol, 0.25% vol/vol Triton X-100), and then washed twice for 5 min in MS buffer. Before antibody incubation, the cells were incubated with a mixture of 1% wt/vol macerozyme (Yakuruto) and 0.1% wt/vol pectolyase (Yakuruto) in MS buffer for 5 min at 30°C. Afterwards the slides were blocked with 5% vol/vol goat normal serum (Sigma) in TBST (20 mM Tris-HCl, pH 7.3, 150 mM NaCl, 0.25% vol/vol Triton X-100) at 25°C and incubated with primary antibodies (diluted 1:300 in TBST). The slides were kept in a moist chamber at 37°C for 1 h, washed three times with TBST, and then incubated with anti mouse-IgG antibody conjugated to FITC (Sigma) diluted 1:100 in TBST. The cells were washed three times with TBST, sealed with a glass slide, and then stored at 4°C until microscopy. Each experiment was performed at least three times.
The fixed cells were visualized under a confocal laser microscope (model DM RBE, Leica), using a two-channel scan with an argon-krypton laser at 488 and 568 nm excitation, a beam splitter at 575 nm, and a 580- and 590-nm emission filter. To eliminate filter leakage, in some experiments, the cells were viewed by subsequent one-channel scans with identical scanning intervals. For this constellation, the FITC signal was analyzed using excitation at 488 nm, a beam splitter at 510 nm, and a 515-nm emission filter. Since our confocal microscope has no 4′,6-diamidino-2-phenylindole exciting laser, positioning of nuclei was assayed by transmission microscopy.
Antiserum Production, ELISA, Protein Assay, and Western Blot Analysis
Construction of expression plasmids, expression in Escherichia coli, purification of (His)6-tagged recombinant CPRF1, CPRF2, and CPRF4, and refolding of the bZIP factors as well as the production of antisera in mice are described in Kircher et al. (1998). For ELISA, 50 μl of purified antigen (concentration: 5 μg/ml) were bound for 2 h to M129 B high-affinity plate (Dynatech). After washing with PBST (8 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 0.14 M NaCl, 2.7 mM KCl, 0.05% vol/vol Tween 20) the plate was blocked for 2 h with PBST containing 3% wt/vol bovine serum albumin. After washing twice with PBST 50 μl of PBST containing a 1:1,000 dilution of antiserum was added and the plate incubated for 1 h at room temperature. The plate was washed twice with PBST. Afterwards, 50 μl of PBST containing a 1:1,200 dilution of horseradish peroxidase- labeled secondary antibody was added, and the plate again incubated for 1 h at room temperature. After washing twice with PBST the reaction was developed for 5 min in orthophenyldiamin/H2O2 solution, stopped by addition of 50 μl of 2 M H2SO4 (Harlow and Lane, 1988), and then the optical density was quantified at 490 nm. Protein assay and Western blot analysis were performed as described in Harter et al. (1994b). The polyclonal histone 2A/2B antibody was diluted 1:10,000 for use (Harter et al., 1994b). The monoclonal tubulin antibody (clone DM 1A; SERVA) was diluted 1:1,000 for use. The secondary anti-rabbit and anti-mouse sera were from Boehringer Mannheim and diluted according to the manufacturer's instruction.
Electrophoretic Mobility Supershift Assay
For electrophoretic mobility supershift assay (EMSSA), a monomeric DNA probe (5′-AATTCTCCCTTATTCCACGTGGCCATCCGG-3′) according to the G-box of the parsley CHS promotor (Schulze-Lefert et al., 1989a,b) or a monomeric C-box (5′-AATTCTCCCTTATCTGACGTCAGCATCCGG-3′) with a core-sequence (underline) according to Izawa et al. (1993) was used. Preparation of radioactive-labeled probes as well as experimental conditions for EMSSA were performed according to Harter et al. (1994a). For the assay 50 μg of cytoplasmic or 20 μg of nuclear protein in a total volume of 10 μl was incubated for 10 min on ice with 1 μl of CPRF antisera or the corresponding preimmunsera in the binding reaction mixture (Harter et al., 1994a).
Results
Characterization of CPRF-specific Antisera
To determine the intracellular distribution of members of the CPRF family from parsley we first had to produce CPRF-specific antisera and to test them for specifity against the refolded antigens. Therefore, recombinant (r) His-tagged CPRF1, CPRF2, and CPRF4 were overexpressed in E. coli, purified, and then used for antisera production (Kircher et al., 1998). As shown by ELISA, using refolded, functionally intact antigen, the antisera raised against rCPRF1 and rCPRF4 showed cross-reactivity not only with other CPRFs (Table I) but also with GBFs from Arabidopsis (data not shown). In contrast, the antiserum raised against rCPRF2 recognized only its antigen and did not cross-react with rCPRF1, rCPRF4 (Table I), or the GBFs from Arabidopsis (data not shown). As shown previously (Kircher et al., 1998), Western blot analysis of crude extracts from parsley cells demonstrated that the rCPRF2 antiserum detects a single band of 46 kD representing endogenous CPRF2, whereas the sera produced against rCPRF1 and rCPRF4 recognize two different proteins of 42 and 44 kD representing endogenous CPRF1 and CPRF4, respectively. These data are in good agreement with our ELISA results indicating that the serum produced against rCPRF2 is highly specific for its native antigen, whereas we had to expect a certain amount of cross-reactivity between CPRF1 and CPRF4 when we use the antisera raised against rCPRF1 and rCPRF4.
Table I.
Test of Different Polyclonal Antisera Raised against rCPRF1, rCPRF2, and rCPRF4 for Cross-reactivity in an ELISA
| Antiserum‡ | Antigen* | |||||
|---|---|---|---|---|---|---|
| rCPRF1 | rCPRF2 | rCPRF4 | ||||
| Anti-CPRF1 | 100§ | 1 | 46 | |||
| Anti-CPRF2 | 6 | 100 | 5 | |||
| Anti-CPRF4 | 22 | 5 | 100 | |||
Highly purified, recombinant and refolded (His)6-tagged CPRF1, CPRF2, and CPRF4 were used as antigens.
Antisera were produced in mice as described in Kircher et al. (1998).
The ELISA signal obtained from the antiserum incubated with the corresponding antigen was set to 100. The relative signals of the preimmunosera were between 0 and 5 (data not shown).
CPRF1, CPRF2, and CPRF4 Show Differential Intracellular Distribution in Dark-cultivated Parsley Cells
Recently, the involvement of bZIP proteins in G-box– binding activity in the cytosol of evacuolated dark-cultivated parsley protoplasts was demonstrated (Harter et al., 1994a). At least one of these bZIP factors was translocated into the nucleus in response to light, indicating that an inducible nuclear transport of a transcription factor is part of the light-modulated signal transduction network (Harter et al., 1994a). To identify this bZIP factor we used the CPRF antisera described in Table I in EMSSA using a G-box–containing sequence as DNA probe that allows detection of a wide spectrum of plant bZIP proteins (Foster et al., 1994; Menkens et al., 1995). EMSSA allows a sensitive monitoring of DNA-binding activities in combination with specific detection of the binding protein in crude cell extracts (Feldbrügge et al., 1994; Harter et al., 1994a). Dependent on the epitopes recognized within the aa sequence of its antigen, a polyclonal antiserum added to the binding reaction can inhibit the DNA/protein interaction resulting in the disappearance of a shifted band and/or can induce a supershifted complex with lower electrophoretic mobility (Harter et al., 1994a). The cytoplasmic and nuclear extracts for EMSSA were prepared from evacuolated parsley protoplast according to Harter et al. (1994b). These extracts were not contaminated by nuclear and cytoplasmic proteins, respectively, as demonstrated by Western blot analysis using histone 2A/2B- and tubulin-specific antibodies (Fig. 1 A). As shown in Fig. 1, B and C, lanes 3, 5, and 7, two major DNA–protein complexes could be detected in the cytoplasmic and one in the nuclear extract of dark-cultivated evacuolated parsley protoplasts in the presence of the preimmunosera. Addition of the CPRF2-specific antiserum to the DNA/cytosol-binding reaction caused the disappearance of the upper DNA–protein complex and a supershifted band (Fig. 1 B, lane 4). However, an EMSSA of the nuclear extract showed no disappearing CPRF2-containing complex (Fig. 1 C, lane 4). These data demonstrate that CPRF2 is present in the cytoplasmic compartment but not inside the nuclei of dark-cultivated, evacuolated parsley protoplasts. The use of the antiserum raised against rCPRF4 resulted in the disappearance of the lower cytoplasmic DNA–protein complex (Fig. 1 B, lane 6). An appearance of a supershifted DNA–protein complex as well as a weakening of a shifted band was also observed in nuclear extracts (Fig. 1 C, lane 6). In contrast, we obtained only a weak antibody activity in the cytoplasmic but a strong in the nuclear extracts when we tested the serum produced against rCPRF1 (Fig. 1, B and C, lanes 2). Since there is cross-reactivity of the CPRF1 antiserum with CPRF4 (see Table I), we conclude that the weak cytoplasmic antibody activity observed with the CPRF1 serum was due to cross-interaction with CPRF4, whereas the strong effect in the nuclear extract was due to true interaction with CPRF1.
Figure 1.
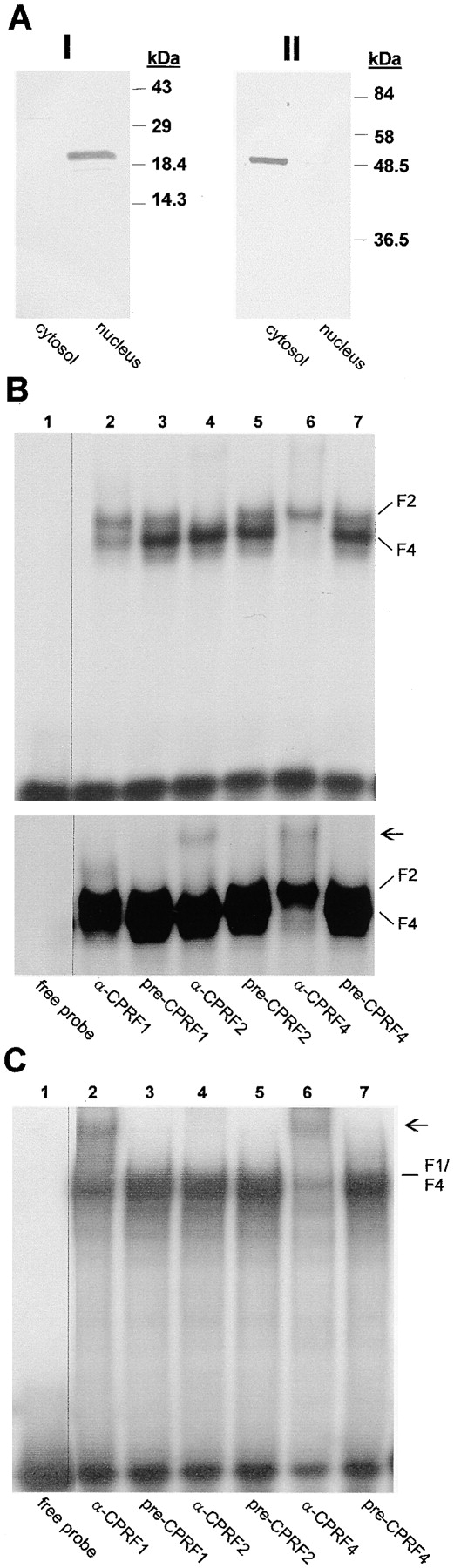
Analysis of the cytoplasmic and nuclear distribution of G-box–binding activities in dark-kept evacuolated parsley protoplasts. (A) Western blot analysis of cytoplasmic (cytosol) and nuclear (nucleus) extracts probed with histone 2A/2B (panel I) and tubulin (panel II) antibodies. In (I) 25 μg of protein per lane and in (II) 10 μg of protein per lane were loaded. In B and C autoradiograms of EMSSAs with 50 μg per lane of cytoplasmic and 20 μg per lane of nuclear extract are shown. For CPRF/antiserum interaction test the extracts were incubated for 10 min on ice with 1 μl of serum and the radioactive-labeled G-box probe before loading the samples on the gel (A and B, lanes 2–7). In B a 12- (top) and a 24-h (bottom) exposure of the identical shift gel is shown. In lanes 1, the binding reaction mix contained neither a serum nor protein (free probe). CPRF-containing DNA–protein complexes are marked (F1, CPRF1; F2, CPRF2; F4, CPRF4). Arrow, positions of supershifted DNA–CPRF complex.
Taken together, our data strongly indicate that the three members of the CPRF family exhibit a differential intracellular distribution in dark-kept cells with CPRF1 exclusively localized in the nucleus, CPRF4 found in both compartments, and CPRF2 retained in the cytosol. Furthermore, the absence from the nuclear compartment suggests that CPRF2 could be the bZIP factor that is translocated into the nucleus in response to light as described previously (Harter et al., 1994a). We therefore focused our further analysis on CPRF2 using its highly specific antiserum as a tool.
CPRF2 Is Transported In Vivo into the Nucleus in Response to Light
To test the possibility that CPRF2 is the target for a light-modulated nuclear import we performed an additional set of EMSSA with cytoplasmic and nuclear extracts from evacuolated parsley protoplasts that were either irradiated for 30 min with UV-containing white light exciting all plant photoreceptor systems or kept in darkness for the same time period before isolation of the compartments. To detect CPRF2 more clearly we switched from the G-box to the C-box as DNA probe. CPRF2 has a very high affinity to this sequence whereas the binding activities of CPRF1 and CPRF4 are low, reducing the signals of these two bZIP factors in EMSSA (Izawa et al., 1993; Foster et al., 1994). Furthermore, the C-box allows a better resolution of C-box–binding factors in EMSSA (Fig. 2) (Wellmer, F., S. Kircher, A. Rügner, H. Frohnmeyer, E. Schäfer, and K. Harter, manuscript submitted for publication). Using the highly specific CPRF2 antiserum we could not detect any CPRF2-dependent DNA-binding activity in the nuclear extract of dark-kept parsley cells demonstrating again the absence of the factor from this compartment under dark conditions (Fig. 2 A, lane 6). However, a supershifted CPRF2 signal can be observed in the corresponding cytosol (Fig. 2 A, lane 3). Irradiation of the cells caused the appearance of a supershifted CPRF2-containing DNA–protein complex in the nucleus (Fig. 2 B, lane 6). The use of the corresponding preimmunoserum showed no effect (Fig. 2, A and B, lanes 4 and 7). Note, that the amount of nuclear proteins used in the assays is 2.5 times lower than that of cytoplasmic one. In evacuolated parsley protoplasts, about 45%, each, of total protein was determined to be localized in the cytosol and the nucleus (Harter et al., 1994a). From this, we conclude that the amount of nucleus-imported CPRF2 is at least 50% of the total pool. We could not detect any changes in the intracellular distribution pattern of supershifted DNA–protein complexes when we used the antisera raised against rCPRF1 and rCPRF4 in EMSSA (data not shown). In conclusion, these results strongly indicate that CPRF2 is in fact the bZIP factor that is translocated from the cytosol into the nucleus in response to light. Note that there are additional C-box–binding activities in the cytoplasmic as well as in the nuclear extract that were not detected previously using the G-box as DNA probe (Fig. 2). Moreover, the pattern of these activities derived from unknown C-box binding proteins changed in the nuclear extract after light irradiation (Fig. 2).
Figure 2.
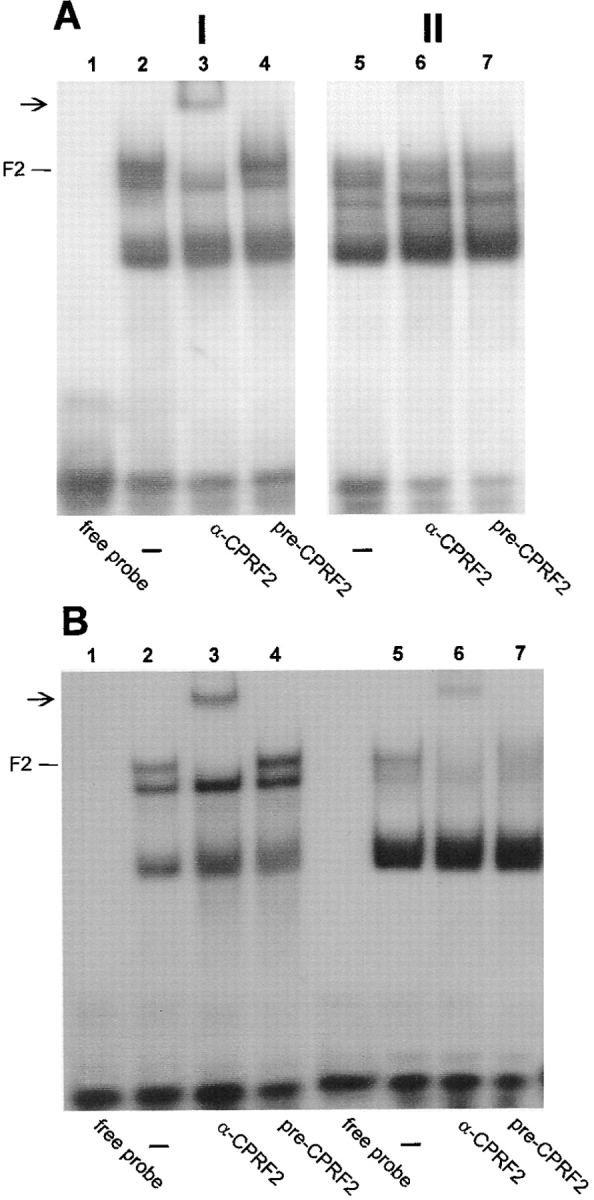
CPRF2 is imported into the nucleus of evacuolated parsley protoplasts in response to irradiation with UV-containing white light. Autoradiograms of EMSSAs with a radioactive-labeled C-box probe are shown. For EMSSA 50 μg per lane of cytosolic (panel I) and 20 μg per lane of nuclear (panel II) extracts isolated from dark-cultivated cells were used that were either further kept in continuous darkness (A) or irradiated for 30 min with UV-containing white light (B) before compartment separation. For CPRF–antiserum interaction tests, the extracts were incubated for 10 min on ice with 1 μl of serum and the radioactive-labeled C-box probe before loading the samples on the gel (A and B, lanes 3 and 4, and 6 and 7, respectively). In lanes 2 and 6, no serum was added (−). DNA–CPRF2 complexes are marked (F2). Arrows, positions of supershifted DNA–CPRF2 complex. For further abbreviations see Fig. 1.
Nuclear Import of CPRF2 Is Triggered by phyA and phyB
To confirm our biochemical data and to elucidate the role of the different plant photoreceptors in the induction of CPRF2 nuclear import, we analyzed the intracellular distribution of the factor under different light conditions by an immunolocalization assay combined with confocal microscopy. For this purpose, dark-grown parsley cells were irradiated for 2 h with light of different wavelengths or further kept in darkness, respectively. After treatment cells were fixed and then stained with anti-CPRF2 or preimmunoserum and FITC-labeled secondary antibodies. The rapid fixation protocol enables a semiquantitative analysis of the intracellular distribution of CPRF2 even if the cells disintegrate during the procedure as a result of the strong intracellular osmotic pressure (see Materials and Methods). Before scanning the cells the positioning of nuclei was confirmed by transmission microscopy. Whereas the preimmunoserum showed a very weak and constitutive background fluorescence (Fig. 3 A), the endogenous CPRF2 is detected in the cytosol of dark-incubated cells, but not in the nucleus (Fig. 3 B). However, after irradiation of the cells with continuous UV-A, blue, red, or far-red light, CPRF2 appeared in the nucleus and, dependent on the light quality, became less pronounced in the cytosol (Fig. 3, C–F). This shows that CPRF2 was actively moving from the cytosol into the nucleus. The efficiency of the nuclear translocation of CPRF2 seems to be dependent on the light quality with red light being most effective followed by blue, far-red, and UV-A light. These differences in the efficiency of the translocation response to the tested light qualities point to phyA and phyB to be the main photoreceptors involved in this photoresponse. To further test this hypothesis we treated dark-grown parsley cells with pulses of red and long wavelength far-red (RG9) light and then transferred them back into darkness for another 2 h before fixation. As shown in Fig. 3 G a red light pulse of 5 min induced an almost complete translocation of CPRF2 from the cytosol to the nucleus, whereas a 5-min RG9 pulse led to no effect (Fig. 3 H). If the red light pulse was followed by a RG9 pulse, the import of CPRF2 into the nucleus was less pronounced (Fig. 3 I) compared with the red light pulse alone (Fig. 3 G). The reversion of the red light effect by the RG9 pulse was incomplete, though, if one compares the cytoplasmic staining of CPRF2 in Fig. 3 I with that of Fig. 3 H. To test whether, in addition to light, other exogenic factors can induce the translocation of CPRF2, dark-grown cells were treated either with elicitor from Phytophtera megaspermum f. sp. glycinea (PMG; Somssich et al., 1986) or with heat (Walter, 1989). Neither the PMG elicitor nor the heat shock treatment produced any effect on the intracellular distribution of CPRF2 (Fig. 3, J and K). In conclusion, our irradiation data indicate a control of nuclear transport of CPRF2 by both phyA and phyB (see Discussion).
Figure 3.
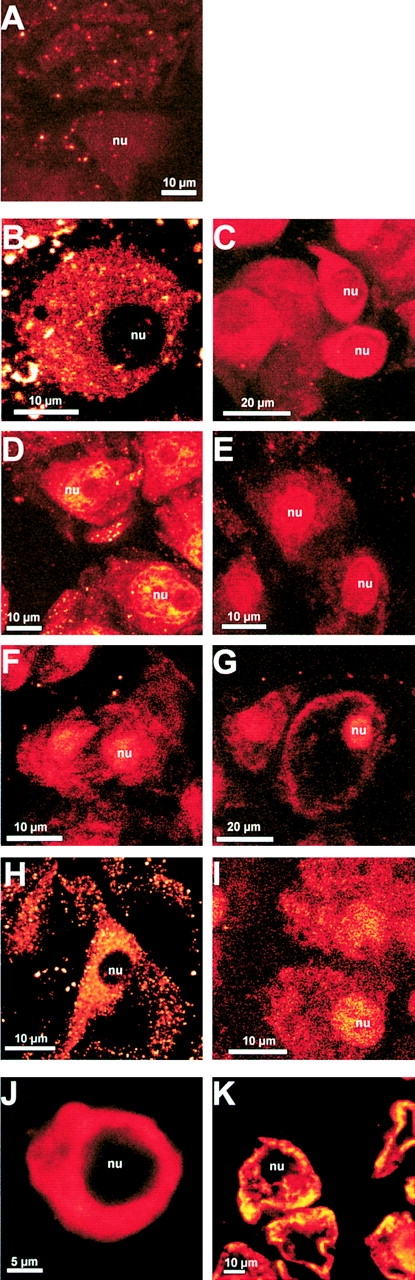
Intracellular distribution of CPRF2 is regulated by phytochrome. Confocal images of immunostained parsley cells probed with the preimmunoserum (A) and the CPRF2-specific antiserum (B–K). Before fixation cells had been kept either in darkness (B) or irradiated for 2 h with continuous UV-A (C), blue (D), red (E) or far-red light (F) or pulse-irradiated for 5 min with red light (G), RG9 light (H), or red light followed by a 5-min RG9 pulse (I). In J, cells incubated for 2 h in 0.1 mg/ml of PMG elicitor from Phytophtera megasperma and in K, cells treated by a 30-min heat shock (37°C) followed by 90 min of cultivation at 25°C in the dark are shown. After treatments cells were fixed and probed with a CPRF2-specific antiserum. nu, position of the nuclei.
The NH2-terminal Domain Is Responsible for the Retention of CPRF2 in Dark-grown Cells
The observation that CPRF2 is found in the cytosol of dark-grown parsley cells prompted us to map the amino acid stretch of the CPRF2 molecule that could be responsible for cytoplasmic retention. For this purpose, we translationally fused the cDNA coding for CPRF2 to a 35 S promotor-driven GFP gene (Haseloff et al., 1997) resulting in a GFP fusion protein upon transient transformation into parsley protoplasts. A parsley PHYA–GFP, a nuclear targeting NLS–GFP, and a CPRF1–GFP fusion construct were used as controls for cytoplasmic (phyA–GFP; Speth et al., 1986, 1987) and nuclear localization, respectively (NLS–GFP, CPRF1–GFP; van der Krol and Chua, 1991; Varagona et al., 1992) (data in Fig. 1). After transformation, the protoplasts that were derived from dark-grown parsley cells were incubated for 16 h in darkness or irradiated with continuous UV-containing white light. The intracellular distribution of the GFP fusion proteins was then analyzed by confocal microscopy. Before scanning the cells the positioning of the nuclei was confirmed by transmission microscopy. As shown in Fig. 4 A, phyA–GFP was exclusively localized in the cytosol from dark-cultivated protoplasts. The NLS–GFP as well as the CPRF1–GFP fusion, in contrast, were always confined to the nucleus (Fig. 4, B and C). Irradiation of the protoplasts had no influence on the GFP fluorescence intensity indicating that the used light sources do not induce any bleaching effect. Furthermore, we observed no changes in the subcellular distribution of the control fusion proteins in response to light treatment (data not shown). In contrast, the CPRF2–GFP is observed in both compartments of dark-kept protoplasts (Fig. 4 D). Irradiation of the protoplasts expressing CPRF2–GFP resulted in the disappearance of the fusion protein from the cytosol indicating its nuclear import (Fig. 4 E). Due to a strong nuclear overreflection signal, a clear additional accumulation of CPRF2–GFP inside the nucleus could not be observed. However, as shown previously (Kircher et al., 1998), the total amount of CPRF2 is not reduced in response to irradiation excluding a compartment-specific degradation as reason for the disappearance of the bZIP factor from the cytosol.
Figure 4.
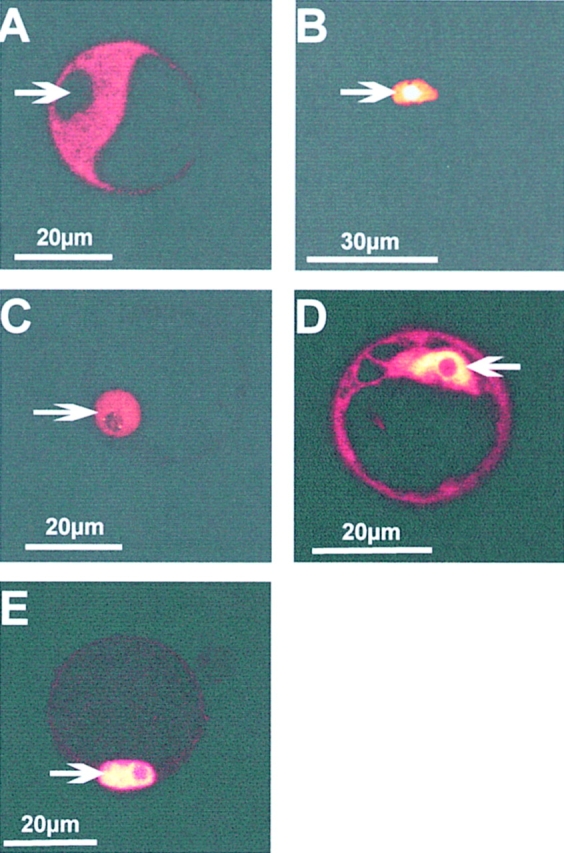
Intracellular sorting of GFP fusion proteins within parsley protoplasts. Confocal sections of parsley protoplasts transiently transformed with fusion constructs expressing phyA–GFP (A), NLS–GFP (B), CPRF1–GFP (C), and CPRF2–GFP (D and E). After transformation by electroporation protoplasts were kept for 16 h in darkness (A–D) or continuously irradiated with UV-containing white light (E) for the same time period before microscopical analysis. Arrows, positions of the nuclei.
Taken together, our data indicate that transiently transformed parsley protoplasts are capable of maintaining the correct intracellular sorting as demonstrated by the phyA– GFP, NLS–GFP, and CPRF1–GFP controls. Secondly, the observation that CPRF2–GFP is at least in part retained in the cytosol in darkness but transported into the nucleus in response to light indicates that the involved molecular mechanism is also functionally maintained. Since we never observed endogenous CPRF2 to be present in the nucleus of dark-grown cells (see Fig. 3 A), the accumulation of CPRF2–GFP in the nucleus of dark-kept protoplasts is most likely caused by overtitration of the cytoplasmic retention machinery due to strong overexpression of the fusion protein. Similar effects were found for GFP fusion proteins in other systems (Fukuda et al., 1997).
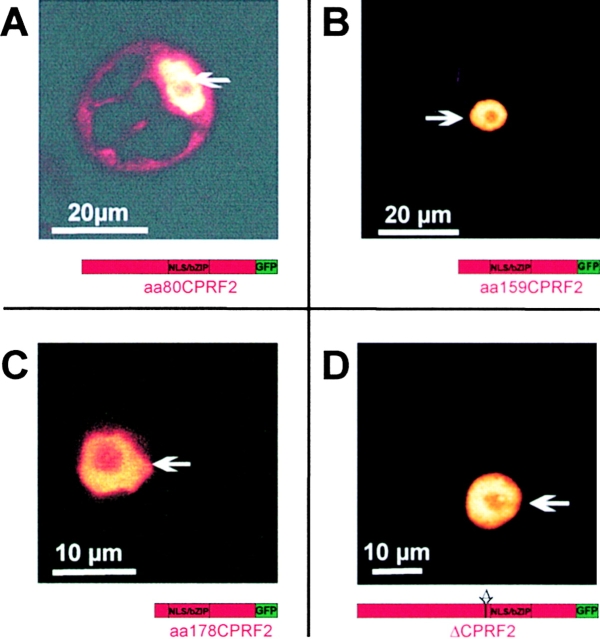
For our mapping purposes we now produced a set of CPRF2 constructs bearing NH2-terminal deletions fused to the GFP gene. These constructs were again transiently transformed into parsley protoplasts and the intracellular distribution of the corresponding fusion proteins determined by confocal microscopy after 16 h of cultivation in darkness. As shown in Fig. 5 A the fusion protein, where the first NH2-terminal 80 aa (aa80CPRF2–GFP) have been deleted, is observed in both the cytosol and the nucleus like it was shown for full-length CPRF2–GFP (compare to Fig. 4 D). The removal of additional 79 aa (aa15CPRF2– GFP) resulted in the loss of the cytoplasmic pool of the truncated CPRF2 (Fig. 5 B). The fusion peptide that misses the whole NH2 terminus up to aa 178 (aa178CPRF2–GFP) is exclusively found in the nuclear compartment as well (Fig. 5 C). In addition to these NH2-terminal deletions we determined the intracellular distribution of a fusion peptide from which an acidic amino acid stretch was removed (ΔCPRF2– GFP). This stretch (aa 178-DHSDDDDELEGETET-aa 192) is localized NH2-terminal of the NLS-containing basic DNA-binding motif of CPRF2. As shown in Fig. 5 D the fusion protein encoded by the ΔCPRF2–GFP construct is only observed in the nuclear compartment of dark-cultivated parsley protoplasts.
Figure 5.
CPRF2 contains two separable domains responsible for cytoplasmic retention in the dark. Confocal microscopy of parsley protoplasts transiently transformed with GFP fusion constructs expressing either 5′-deletions (A–C) or an internal deletion (D) of CPRF2. After transformation, protoplasts were kept in darkness for 16 h before microscopical analysis. The relative size and composition of the expressed GFP fusion protein is shown schematically below each photograph. aa80CPRF2, NH2-terminal deletion of CPRF2 starting with aa 80; aa159CPRF2, NH2-terminal deletion of CPRF2 starting with aa 159; aa178CPRF2, NH2-terminal deletion of CPRF2 starting with aa 178; ΔCPRF2, internal deletion of CPRF2 (aa178–192); NLS/ bZIP, nuclear localization sequence-containing basic-region leucine-zipper domain; GFP, green fluorescent protein. Arrows, positions of the nuclei.
Discussion
Differential Subcellular Localization of the CPRF Family Members
Our data show that CPRF1, CPRF2, and CPRF4 are differentially distributed within dark-cultivated parsley cells. Whereas CPRF1 is exclusively localized in the nucleus and CPRF2 is only found in the cytosol, CPRF4 is present in both cell compartments (Fig. 1). This differential distribution suggests that these three bZIP factors exhibit different functional duties within the regulatory networks of the plant cell. For instance, the constitutive localization of CPRF1 inside the nucleus may indicate that this factor is constitutively active or is posttranscriptionally regulated on the level of DNA-binding or transcriptional activity rather than on intracellular partitioning. In contrast, the more uniform distribution of CPRF4 and the absence of CPRF2 from the nucleus point to inducible redistribution of these bZIP factors as a regulatory mechanism, though the inducing signal for CPRF4 is unknown. However, we could show for CPRF2 that light is the inducing signal for its import from the cytosol into the nucleus. We observed quantitative differences in the extent of nuclear-imported CPRF2 that is most likely due to the different parsley cell systems (evacuolated protoplasts versus cell suspension versus protoplasts). However, the existence of the light- induced nuclear import of CPRF2 was confirmed by following three in vivo assays: (a) in EMSSA, CPRF2 can be detected as supershifted DNA–protein complex in the nuclear extract of irradiated evacuolated parsley protoplasts that is absent in the corresponding extract of dark-kept cell culture (Fig. 2). (b) Immunolocalization technique combined with confocal microscopy showed that, in dark-grown parsley cells, CPRF2 is only found in the cytosol and is imported into the nucleus in response to irradiation (see Fig. 3). (c) Using a transient transformation system we demonstrated that a pool of CPRF2–GFP exists in the cytosol of dark-kept parsley protoplasts that is moved into the nucleus in response to light (see Fig. 4).
Induction of Nuclear Import of CPRF2 Is Light-specific and Regulated by Phytochrome
The induction of nuclear transport of CPRF2 is specific for light. Neither heat shock treatment nor treatment with the PMG elicitor from Phytophtera megaspermum had any effect on the intracellular distribution of CPRF2 (Fig. 3, J and K). This excludes that the nuclear import of CPRF2 directly participates in elicitor- or stress-induced signal transduction. The irradiation of cells with continuous light of different wavelengths suggests that the translocation response of CPRF2 is triggered by phytochrome. The strong import response achieved by irradiation of the cells with continuous far-red light indicate the involvement of phyA via a high irradiance response. Although the effects of irradiation with continuous blue and UV-A light can also be attributed to a phyA action, minor contributions of other photoreceptor systems besides phy cannot be entirely excluded. To test the participation of phyB we treated the cells with pulses of red and far-red light to disclose a classical phyB-dependent low fluence response. A red light pulse induced an almost complete translocation of CPRF2, whereas a RG9 pulse had no effect. When the RG9 pulse was given immediately after an inducing red light pulse, this partially “reverted” the translocation of the factor (see Fig. 3). A partial reversion of the effect of an inducing red light pulse by a subsequent RG9 pulse is described for several phy-dependent photoresponses (Schäfer and Briggs, 1986) and can be explained by a very fast coupling of the excited photoreceptor to downstream signal transducing elements (e.g., phosphorylation). Taken together, our data suggest that phyA and phyB trigger the nuclear import of the plant bZIP factor CPRF2. Recently, Terzaghi et al. (1997) described a blue/UV light-specific cytoplasmic/nuclear relocalization of a GUS–GBF2 fusion protein in soybean cell cultures. Since GBF2 from Arabidopsis and CPRF2 from parsley belong to two different subclasses of bZIP proteins with different characteristics for DNA-binding and homotypic dimerization (for review see Armstrong et al., 1992; Meier and Gruissem, 1994; Meshi and Iwabuchi, 1995), it is an attractive hypothesis that different photoreceptors may mediate wavelength-specific gene expression by the release of different transcription factors from cytosolic retention.
The incomplete “reversion” by RG9 of the translocation response triggered by a red light pulse indicate that the nuclear import of CPRF2 is rapid and is initiated immediately upon onset of irradiation, whereas the following RG9 terminates any further accumulation of CPRF2 in the nucleus. Similar rapid nuclear transport kinetics have been reported for several nucleophilic transcription factors retained in the cytosol in the absence of the inducing stimulus (Zabel and Baeuerle, 1990; Carey et al., 1996; Ghosh et al., 1998).
CPRF2 was initially isolated by Southwestern screening with a DNA probe derived from the parsley CHS promotor (Weisshaar et al., 1991). This motif was functionally characterized as an element mediating the light responsiveness of the CHS gene (Schulze-Lefert et al., 1989a,b; Block et al., 1990; Weisshaar et al., 1991; Frohnmeyer et al., 1992, 1994) consisting of two domains (box 1 and box 2) that are necessary and sufficient for light regulation. Box 2 represents a classical G-box (Schulze-Lefert et al., 1989a,b). CPRF2 was shown to bind to this G-box suggesting that it is involved in the light regulation of CHS gene expression (Weisshaar et al., 1991; Armstrong et al., 1992). Detailed physiological analysis in cultured parsley cells and protoplasts demonstrated, however, that the expression of the CHS gene is regulated by the activation of the UV photoreceptors but not by phytochrome (Ohl et al., 1989; Frohnmeyer et al., 1992). Since the nuclear import of CPRF2 is induced by phyA and phyB, it is very unlikely that the G-box of the CHS promotor is the functional target of CPRF2 in this system. However, no other genes are known to be regulated by phytochrome in parsley cell culture and protoplasts. Future research must be directed toward an investigation on which genes are the in vivo targets of CPRF2.
Localization of Retention Domains within the CPRF2 Protein
The demonstration that the disappearance of CPRF2– GFP from the cytosol of parsley cells in response to light is due to active transport into the nucleus allowed us to perform a deletion analysis to map the retention domain(s). The removal of the entire NH2 terminus up to aa 159 abolished cytosolic retention in darkness whereas the aa80CPRF2–GFP fusion protein was still detectable in the cytosol. A similar loss of cytosolic localization was found when a short internal stretch between aa 178 and 192 (ΔCPRF2–GFP) was removed from the full-length protein. The deletion analysis suggests that CPRF2 contains two separable domains for cytoplasmic retention (see Fig. 6 A). In addition, the aa stretch containing both retention domains can be functionally transferred to heterologous bZIP factors (e.g., CPRF1) not found in the cytoplasmic compartment of evacuolated parsley protoplasts (Kircher, S., unpublished data). It is noteworthy that neither CPRF1 nor CPRF4 contain the sequence motifs that could be homologous to the retention domains of CPRF2.
Figure 6.
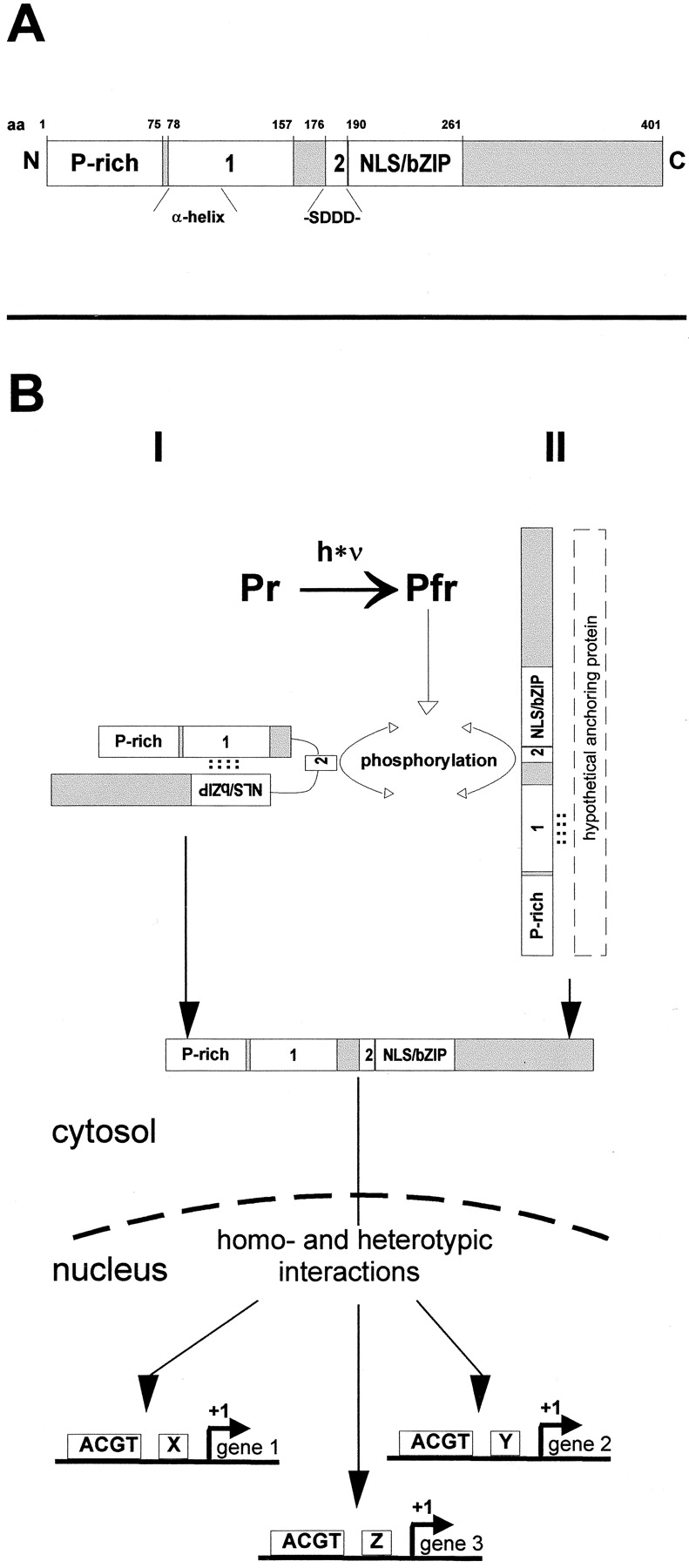
(A) Schematic representation of functional domains within CPRF2. P-rich, prolin-rich transactivation domain (Rügner, A., and E. Schäfer, unpublished data). 1, retention domain 1 that has strong homology to the α-helical cytoplasmic retention domain (α-helix) of the mammalian heat shock factor 2. 2, acidic retention domain 2 that contains a CKII phosphorylation site as indicated (-SDDD-). NLS/bZIP, nuclear localization sequence-containing basic-region leucine-zipper domain. N, NH2 terminus; C, COOH terminus; aa, amino acid. (B) Alternative models of phy-regulated nuclear import of CPRF2 in parsley cells. Pfr formation induces phosphorylation of the CKII site within retention domain 2 of CPRF2. This modification either abolishes intracellular masking of the NLS (model I) or results in the release of CPRF2 from a cytoplasmic anchoring protein (model II). In both cases, accession to the NLS results in nuclear import, homo- and/or heterotypic CPRF2–protein interactions, and binding of CPRF2-containing transcription factor complexes to promoters of light-regulated genes. Further explanation in the text. Pr, inactive form of phytochrome; Pfr, active form of phytochrome; ACGT, G-box like elements; x, y, z, cis-elements other than G-box like motifs; see A for other abbreviations and notes.
As shown for the rel/NF-κB and the glucocorticoid receptor family of transcription factors cytoplasmic localization is achieved by retention factors such as IκB and HSP90, respectively (for review see Muller and Renkawitz, 1991; Jans and Hübner, 1996; Ghosh et al., 1998). Matsui et al. (1995) recently identified a possible retention factor of COP1 named CIP1. However, a detailed sequence comparison of the CPRF2-specific retention domains with those of the mentioned protein families does not reveal any sequence relationships. This indicates that the cytosolic compartmentalization of CPRF2 is probably not achieved by homologues of IκB, HSP90, or CIP1. Similarly, the retention domain of Arabidopsis GBF1 does not show any relationship to the CPRF2 sequences characterized here (Terzaghi et al., 1997). However, the NH2-terminal amino acid stretch of CPRF2 between aa 80 and 159 (Fig. 6 A) exhibits a significant sequence homology (degree of 25% identity and of 53% similarity) and especially structural homology to a well-characterized α-helical domain within the mammalian heat shock factor 2 (Sheldon and Kingston, 1993). The corresponding domain of this heat shock factor is discussed to mediate cytoplasmic retention under nonstress conditions by either intramolecular masking of the NLS or by acquiring a hypothetical NLS-masking protein.
Acidic motifs similar to that of CPRF2 (aa 178–192) are found in several nucleophilic proteins like, for instance, Nopp 140 (Meier and Blobel, 1992) and nucleolins from plants (Bögre et al., 1996) that shuttle between the cytosol and the nucleus. The exact molecular mechanisms responsible for regulating NLS activity have remained enigmatic, though. However, one possibility to modulate NLS activity is based on phosphorylation (Jans and Hübner, 1996). For example, casein kinase II (CKII) increases the nuclear import of the SV-40 T antigen through phosphorylation of a CKII-site 13 amino acids NH2-terminal to the NLS (Rihs et al., 1991; Jans and Jans, 1994). Interestingly, the acidic domain of CPRF2 contains a CKII phosphorylation site 20 amino acids NH2-terminal to the NLS (see Fig. 6 A) that might modulate NLS activity upon phosphorylation. Recent data (Wellmer, F., S. Kircher, A. Rügner, H. Frohnmeyer, E. Schäfer, and K. Harter, manuscript submitted for publication) indicate that phytochrome activation causes rapid phosphorylation of CPRF2 in vivo. This phosphorylation does not interfere with the DNA-binding activity of CPRF2 and might therefore be involved in the regulation of nuclear import.
The results presented in this work can be summarized in two alternative models of phytochrome-regulated nuclear import of CPRF2 (Fig. 6 B): CPRF2 is retarded in the cytosol of dark-grown parsley cells via retention domains 1 and 2. Irradiation photoconverts phyA and phyB from the inactive Pr to the active Pfr form that in turn induces the phosphorylation of the CKII-site within the acidic retention domain 2 of CPRF2. This modification either abolishes intracellular masking of the NLS (Fig. 6 B, I) or results in the release of CPRF2 from a cytoplasmic anchoring protein (Fig. 6 B, II). In both cases, accession of the NLS for the components of the nuclear import machinery results in the nuclear import of CPRF2. Inside the nucleus CPRF2 binds to promotors of certain light-regulated genes. The specificity to bind to a certain promotor element could be achieved by homo- or heterotypic protein-protein interactions of CPRF2 with members of the bZIP or of other transcription factor families (see examples in Vicente-Carbajosa et al., 1997; Büttner and Singh, 1997). One possible candidate for homotypic interaction could be the parsley homologue of the bZIP factor HY5 that has not been shown to have transcriptional activity and likely needs a transcriptionally active bZIP as a partner for function (Ang et al., 1998). HY5 was shown to be sequestered and kept inactive by interaction with the nuclear, general repressor of photomorphogenesis COP1 in dark-grown plants and released upon irradiation (Ang et al., 1998). The demonstration of a potential CPRF2/HY5 heterodimerization would point to how a photoreceptor-specific signaling pathway could interconnect with the general transcriptional repression mechanism consisting of the COP1 and HY5 gene products.
Acknowledgments
We are grateful to H. Frohnmeyer for methodical support, I. Abel (both from University of Freiburg, Freiburg, Germany) for technical assistance, and J. Haseloff (University of Cambridge, Cambridge, UK) for the GFP plasmid.
Abbreviations used in this paper
- aa
amino acid
- bZIP
basic-region leucine-zipper domain
- CHS
chalcon synthase
- CKII
casein kinase II
- CPRF
common plant regulatory factor
- EMSSA
electrophoretic mobility supershift assay
- GBF
G-box–binding factor
- GFP
green fluorescent protein
- NLS
nuclear localization sequence
- Pfr
far-red light absorbing form of phytochrome
- phy
phytochrome
- Pr
red light-absorbing form of phytochrome
- r
recombinant
- RG9
long wavelength far-red light
Footnotes
S. Kircher was supported by the Konrad Adenauer Stiftung and F. Wellmer was supported by the Graduiertenkolleg “Molekulare Mechanismen pflanzlicher Differenzierung.” The work was supported by grants to E. Schäfer from the Deutsche Forschungsgemeinschaft (SFB 388) and from the Human Frontier Science Program.
S. Kircher and F. Wellmer contributed equally to this work.
References
- Ang L-H, Chattopadhyay NW, Oyama T, Okada K, Batschauer A, Deng X-W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsisdevelopment. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Armstrong GA, Weisshaar B, Hahlbrock K. Homodimeric and heterodimeric leucine zipper proteins and nuclear factors from parsley recognize diverse promotor elements with ACGT cores. Plant Cell. 1992;4:525–537. doi: 10.1105/tpc.4.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Dangl JL, Hahlbrock K, Schulze-Lefert P. Functional borders, genetic fine structure, and distance requirements of cis elements mediating light responsiveness of the parsley chalcone synthase promotor. Proc Natl Acad Sci USA. 1990;87:5387–5391. doi: 10.1073/pnas.87.14.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Jonak C, Mink M, Meskiene I, Traas J, Ha DTC, Swoboda I, Plank C, Wagner E, Heberle-Bors E, Hirt H. Developmental and cell cycle regulation of alfalfa nucMs1, a plant homolog of yeast Nsr1 and mammalian nucleolin. Plant Cell. 1996;8:417–428. [PMC free article] [PubMed] [Google Scholar]
- Büttner M, Singh KB. Arabidopsis thalianaethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey KL, Richards S, Lounsbury KM, Macara IG. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the RAN/TC4 GTPase mediates an essential function independent of nuclear protein import. J Cell Biol. 1996;133:985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Hauffe KD, Lipphardt S, Hahlbrock K, Scheel D. Parsley protoplasts retain differential responsiveness to UV light and fungal elicitor. EMBO (Eur Mol Biol Organ) J. 1987;6:2551–2556. doi: 10.1002/j.1460-2075.1987.tb02543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: Crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Feldbrügge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B. Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell. 1994;6:1607–1621. doi: 10.1105/tpc.6.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua N-H. Plant bZIP proteins gather at ACGT elements. FASEB (Fed Am Soc Exp Biol) J. 1994;8:192–200. doi: 10.1096/fasebj.8.2.8119490. [DOI] [PubMed] [Google Scholar]
- Frankenhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H, Ehmann B, Kretsch T, Rocholl M, Harter K, Nagatani A, Furuya M, Batschauer A, Hahlbrock K, Schäfer E. Differential usage of photoreceptors for chalcon synthase gene expression during plant development. Plant J. 1992;2:899–906. [Google Scholar]
- Frohnmeyer H, Hahlbrock K, Schäfer E. A light inducible in vitrotranscription system from evacuolated parsley protoplasts. Plant J. 1994;5:437–449. doi: 10.1111/j.1365-313x.1994.00437.x. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO (Eur Mol Biol Organ) J. 1997;16:1901–1908. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-κB and rel proteins: evolutionarily conserved mediators of immune response. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 511–552.
- Harter K, Kircher S, Frohnmeyer H, Krenz M, Nagy F, Schäfer E. Light-regulated modification and nuclear translocation of cytoplasmic G-box binding factors (GBFs) in parsley (Petroselinum crispumL.) Plant Cell. 1994a;6:545–559. doi: 10.1105/tpc.6.4.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harter K, Frohnmeyer H, Kircher S, Kunkel T, Mühlbauer S, Schäfer E. Light induces rapid changes of the phosphorylation pattern in the cytosol of evacuolated parsley protoplasts. Proc Natl Acad Sci USA. 1994b;91:5038–5042. doi: 10.1073/pnas.91.11.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsisplants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua N-H. Plant bZIP protein DNA binding specifity. J Mol Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Jans DA, Jans P. Negative charge at the casein kinase II site flanking nuclear localization signal of the SV40 large T-antigen is mechanistically important for enhanced nuclear import. Oncogene. 1994;9:2961–2968. [PubMed] [Google Scholar]
- Jans DA, Hübner S. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol Rev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E., and G.H.M. Kronenberg. 1994. Photomorphogenesis in Plants. Martinus MNijhoff/W. Junk, Dordrecht, The Netherlands. 828 pp.
- Kircher S, Ledger S, Hayashi H, Weisshaar B, Schäfer E, Frohnmeyer H. CPRF4a, a novel plant bZIP protein of the CPRF family: comparative analyses of their light-dependent expression, post-transcriptional regulation, nuclear import and heterodimerisation. Mol Gen Genet. 1998;257:595–605. doi: 10.1007/s004380050687. [DOI] [PubMed] [Google Scholar]
- Klimczak LJ, Schindler U, Cashmore AR. DNA binding activity of the ArabidopsisG-box binding factor GBF-1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell. 1992;4:87–98. doi: 10.1105/tpc.4.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak LJ, Collinge MA, Farini D, Giuliano G, Walker JC, Cashmore AR. Reconstitution of Arabidopsiscasein kinase II from recombinant subunits and phosphorylation of transcription factor GBF1. Plant Cell. 1995;7:105–115. doi: 10.1105/tpc.7.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Stoop CD, von Arnim AG, Wei N, Deng X-W. Arabidopsis COP1 protein specifically interacts in vitrowith a cytoskeleton- associated protein, CIP1. Proc Natl Acad Sci USA. 1995;92:4239–4243. doi: 10.1073/pnas.92.10.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Gruissem W. Novel conserved sequence motifs in plant G-box binding proteins and implications for interactive domains. Nucl Acids Res. 1994;22:470–478. doi: 10.1093/nar/22.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Menkens AE, Cashmore AR. Isolation and characterization of a fourth Arabidopsis thalianaG-box-binding factor, which has similarities to Fos oncoprotein. Proc Natl Acad Sci USA. 1994;91:2522–2526. doi: 10.1073/pnas.91.7.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Meshi T, Iwabuchi M. Plant transcription factors. Plant Cell Physiol. 1995;36:1405–1420. [PubMed] [Google Scholar]
- Muller M, Renkawitz R. The glucocorticoid receptor. Biochim Biophys Acta. 1991;1088:171–182. doi: 10.1016/0167-4781(91)90052-n. [DOI] [PubMed] [Google Scholar]
- Ohl S, Hahlbrock K, Schäfer E. A blue-light derived signal modulates ultraviolet-light-induced activation of the chalcone synthase gene in cultured parsley cells. Planta. 1989;177:228–236. doi: 10.1007/BF00392811. [DOI] [PubMed] [Google Scholar]
- O'Neil KT, Shuman JD, Ampe C, DeGrado WF. DNA-induced increase in the α-helical content of C/EBP and GCN4. Biochemistry. 1991;30:9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Petrášek J, Freudenreich A, Heuing A, Opatrny Z, Nick P. Heat-shock protein 90 is associated with microtubules in tobacco cells. Protoplasma. 1998;202:161–174. [Google Scholar]
- Poppe C, Ehmann B, Frohnmeyer H, Furuya M, Schäfer E. Regulation of phytochrome A mRNA abundance in parsley seedings and cell-suspension culture. Plant Mol Biol. 1994;26:481–486. doi: 10.1007/BF00039558. [DOI] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short T, Xu Y, Wagner D. Phytochromes: Photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Rasmussen R, Benvegny D, O'Shea E, Kim PS, Alber T. X-ray scattering indicates that the leucine zipper is a coiled coil. Proc Natl Acad Sci USA. 1991;88:561–564. doi: 10.1073/pnas.88.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihs H-P, Jans DA, Fan H, Peters R. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO (Eur Mol Biol Organ) J. 1991;10:633–639. doi: 10.1002/j.1460-2075.1991.tb07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, E. 1977. Kunstlicht und Pflanzenzucht. Lexika-Verlag, Grafenau, Germany. 249–266.
- Schäfer E, Briggs WR. Photomorphogenesis from signal perception to gene expression. Photobiochem Photophys. 1986;12:305–320. [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitously expressed ArabidopsisGBF bZIP proteins. EMBO (Eur Mol Biol Organ) J. 1992a;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Terzaghi WB, Beckmann H, Kadesch T, Cashmore AR. DNA binding site preferences and transcriptional activation properties of the Arabidopsistranscription factor GBF-1. EMBO (Eur Mol Biol Organ) J. 1992b;11:1275–1289. doi: 10.1002/j.1460-2075.1992.tb05171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P, Dangl JL, Becker-André M, Hahlbrock K, Schulz W. Inducible in vivo footprints define sequences necessary for UV light activation of the parsley chalcone synthase gene. EMBO (Eur Mol Biol Organ) J. 1989a;8:651–656. doi: 10.1002/j.1460-2075.1989.tb03422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P, Becker-André M, Schulz W, Hahlbrock K, Dangl JL. Functional architecture of the light-responsive chalcone synthase promotor from parsley. Plant Cell. 1989b;1:707–714. doi: 10.1105/tpc.1.7.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon LA, Kingston RE. Hydrophobic coiled-coil domains regulate the subcellular localization of the human heat shock factor 2. Genes Dev. 1993;7:1549–1558. doi: 10.1101/gad.7.8.1549. [DOI] [PubMed] [Google Scholar]
- Somssich IE, Schmelzer E, Bollmann J, Hahlbrock K. Rapid activation by fungal elicitors of genes encoding “pathogenesis-related” proteins in cultured parsley cells. Proc Natl Acad Sci USA. 1986;83:2427–2430. doi: 10.1073/pnas.83.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth V, Otto V, Schäfer E. Intracellular localization of phytochrome in oat coleoptiles by electron microscopy. Planta. 1986;168:299–304. doi: 10.1007/BF00392353. [DOI] [PubMed] [Google Scholar]
- Speth V, Otto V, Schäfer E. Intracellular localization of phytochrome and ubiquitin in red-light-irradiated oat coleoptiles by electron microscopy. Planta. 1987;171:332–338. doi: 10.1007/BF00398678. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Terzaghi WB, Bertekap RL, Cashmore AR. Intracellular localisation of GBF proteins and blue light-induced import of GBF2 fusion proteins into the nucleus of cultures Arabidopsisand soybean cells. Plant J. 1997;11:967–982. doi: 10.1046/j.1365-313x.1997.11050967.x. [DOI] [PubMed] [Google Scholar]
- Tijan R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- van der Krol AR, Chua N-H. The basic domain of plant b-ZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell. 1991;3:667–675. doi: 10.1105/tpc.3.7.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt R, Raikhel NV. Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein opaque-2. Plant Cell. 1992;4:1213–1227. doi: 10.1105/tpc.4.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger protein binds the prolamine box in zein gene promotors and interacts with the basic leucine zipper transcriptional activator opaque2. Proc Natl Acad Sci USA. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MH. The induction of phenylpropanoid biosynthetic enzymes by ultraviolet light or fungal elicitor in cultured parsley cells is overridden by a heat-shock treatment. Planta. 1989;177:1–8. doi: 10.1007/BF00392148. [DOI] [PubMed] [Google Scholar]
- Weiss MA, Ellenberger T, Wobbe CR, Lee JP, Harrison SC, Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- Weisshaar B, Armstrong GA, Block A, da Costa e Silva O, Hahlbrock K. Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promotor element with functional relevance in light responsivness. EMBO (Eur Mol Biol Organ) J. 1991;10:1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF. Roles of different phytochromes in Arabidopsisdevelopment. Plant Cell Environ. 1997;20:752–758. [Google Scholar]
- Zabel U, Baeuerle P. Purified human IκB can rapidly dissociate the complex of the NFκB transcription factor with its cognate DNA. Cell. 1990;61:255–265. doi: 10.1016/0092-8674(90)90806-p. [DOI] [PubMed] [Google Scholar]