Abstract
By the yeast two-hybrid screening of a rat brain cDNA library with the regulatory domain of protein kinase C ζ (PKCζ) as a bait, we have cloned a gene coding for a novel PKCζ-interacting protein homologous to the Caenorhabditis elegans UNC-76 protein involved in axonal outgrowth and fasciculation. The protein designated FEZ1 (fasciculation and elongation protein zeta-1) consisting of 393 amino acid residues shows a high Asp/Glu content and contains several regions predicted to form amphipathic helices. Northern blot analysis has revealed that FEZ1 mRNA is abundantly expressed in adult rat brain and throughout the developmental stages of mouse embryo. By the yeast two-hybrid assay with various deletion mutants of PKC, FEZ1 was shown to interact with the NH2-terminal variable region (V1) of PKCζ and weakly with that of PKCε. In the COS-7 cells coexpressing FEZ1 and PKCζ, FEZ1 was present mainly in the plasma membrane, associating with PKCζ and being phosphorylated. These results indicate that FEZ1 is a novel substrate of PKCζ. When the constitutively active mutant of PKCζ was used, FEZ1 was found in the cytoplasm of COS-7 cells. Upon treatment of the cells with a PKC inhibitor, staurosporin, FEZ1 was translocated from the cytoplasm to the plasma membrane, suggesting that the cytoplasmic translocation of FEZ1 is directly regulated by the PKCζ activity. Although expression of FEZ1 alone had no effect on PC12 cells, coexpression of FEZ1 and constitutively active PKCζ stimulated the neuronal differentiation of PC12 cells. Combined with the recent finding that a human FEZ1 protein is able to complement the function of UNC-76 necessary for normal axonal bundling and elongation within axon bundles in the nematode, these results suggest that FEZ1 plays a crucial role in the axon guidance machinery in mammals by interacting with PKCζ.
Keywords: neuropeptides, UNC-76 protein, phosphorylation, protein binding, protein kinase C
Protein kinase C (PKC)1 was originally isolated as a Ca2+- and phospholipid-dependent Ser/Thr protein kinase, exerting a wide range of physiological functions (Nishizuka, 1995). The PKC family consists of at least 10 isoforms and commonly possesses two functional domains, a regulatory domain in the NH2-terminal half and a catalytic domain in the COOH-terminal half. The PKC isoforms are widely distributed in many types of mammalian cells. Besides the conventional (α, βI, βII, and γ) and the novel (δ, ε, η, and θ) isoforms, the PKC family also comprises two atypical isoforms, ζ and λ/ι, that are distinguished structurally from the former isoforms by the presence of only a single PKC zinc finger motif in their regulatory domains (Nishizuka, 1995). PKCζ, one of the atypical isoforms, has been shown to be involved in a wide variety of important cellular functions: maturation of Xenopus laevis oocytes (Dominguez et al., 1992), enhancement of the NFκB-dependent promoter activity (Folgueira et al., 1996), activation of the mitogen-activated protein kinase (MAPK) (Berra et al., 1995), control of apoptosis (Diaz-Meco et al., 1996), regulation of neuronal differentiation (Wooten et al., 1994), and maintenance of the long-term potentiation in nervous systems (Sacktor et al., 1993). For exhibiting these cellular functions, it is essential that PKCζ interacts specifically with respective cellular substrates. Since all PKC isoforms show a subtle difference in the substrate specificity for synthetic peptides in the in vitro phosphorylation studies (Kazanietz et al., 1993; Nishikawa et al., 1997), it is conceivable that the regulatory domain of PKC isoforms contributes to the recognition of their own cellular substrates by protein–protein interaction. Recently, several proteins interacting with the regulatory domain of PKC isoforms have emerged to be the determinants for subcellular localization of PKC isoforms, cellular regulators for the activity of PKC isoforms, or cellular substrates specific for various PKC isoforms (Mochly-Rosen, 1995; Diaz-Meco et al., 1996; Faux and Scott, 1996; Jaken, 1996; Kuroda et al., 1996; Puls et al., 1997). For elucidating novel signaling pathways involving PKCζ, it is thus a prerequisite to identify the proteins interacting with the regulatory domain of PKCζ.
In this study, we have identified a novel PKCζ-interacting protein homologous to the nematode Caenorhabditis elegans UNC-76 protein necessary for axonal outgrowth (Bloom and Horvitz, 1997). We here demonstrate that the rat cDNA-derived protein, designated FEZ1 (fasciculation and elongation protein zeta-1) (Bloom and Horvitz, 1997), is a cellular substrate of PKCζ and is translocated from the plasma membrane to the cytoplasm by activation of PKCζ. We also show that FEZ1 mRNA is abundantly expressed in adult rat brain and throughout the developmental stages of mouse embryo. A human FEZ1 protein has been shown to rescue the defects caused by unc-76 mutations in the nematode (Bloom and Horvitz, 1997), indicating that both UNC-76 and FEZ1 are conserved evolutionarily on the functional and structural bases. Therefore, it is predicted that FEZ1 is involved in the axon guidance machinery in mammals by interacting with PKCζ.
Materials and Methods
Yeast Two-Hybrid Screening and Sequence Analysis
The yeast two-hybrid screening (Chien et al., 1991) of a rat brain cDNA library (Clontech) was conducted with a yeast strain CG-1945 [MATa ura3-52 his3-200 lys2-801 trp1-901 ade2-101 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1-HIS3 cyhr2 URA3::(GAL4 17-mers)3-CYC1-lacZ], a derivative of HF7c (Feilotter et al., 1994), by using the regulatory domain of rat PKCζ (residues 1–250) (Ono et al., 1988b) fused with the yeast GAL4 DNA-binding domain as a bait. The cDNA fragment of a positive clone obtained was sequenced with a DNA sequencer (model 373S; Perkin Elmer/Applied Biosystems). The full-length cDNA was obtained from a rat brain Marathon-Ready cDNA library (Clontech) by the method of rapid amplification of cDNA ends (RACE) (Frohman et al., 1988). Prediction for the coiled–coil structure (Lupas, 1996) was performed by software available at the web site of the Swiss Institute for Experimental Cancer Research (http://ulrec3.unil.ch/software/COILS_form.html).
Plate Assay for β-Galactosidase (β-Gal) Activity in Yeast Cells
β-Gal activity in yeast cells was measured by the plate assay method. Yeast transformants (Leu+, Trp+, His+) were transferred onto nylon membranes, permeabilized in liquid nitrogen, and placed on Whatman 3MM papers that had been soaked in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM MgCl2, 50 mM 2-mercaptoethanol, pH 7.0) containing 1 mg/ml 5-bromo-4-chloro-3-indoryl-β-d-galactoside (X-Gal). After developing at 37°C for 30 min, the yeast cells forming dark blue colonies were classified into a strongly positive group (+++). After developing at 37°C for 10 h, the yeast cells forming either dark blue or blue colonies were classified into a moderately positive (++) and a weakly positive group (+), respectively. Those forming white colonies were classified into a negative group (−). All measurements were repeated at least four times.
Northern Blot Analysis
Northern blots containing poly(A)+ RNA (∼2 μg/lane) from eight tissues of adult rats and mouse embryos in four different developmental stages were obtained from Clontech. The amount of poly(A)+ RNA in each lane was calibrated using the rat β-actin gene. The full-length FEZ1 cDNA fragment labeled with [α-32P]dCTP (∼110 TBq/mmol) by a Ready-To-Go DNA labeling kit (Pharmacia Biotech) was used as a probe. Hybridization was carried out under highly stringent conditions. The blots were autoradiographed by using a BAS-2000 bioimage analyzing system (Fuji).
Expression of Epitope-tagged Proteins in COS-7 Cells
For expression of the NH2-terminally FLAG-tagged FEZ1 protein (FEZ1-FLAG), a pTB701-FLAG-FEZ1 plasmid was constructed by placing in frame FEZ1 cDNA 3′ downstream of the FLAG epitope sequence of pTB701-FLAG (Kuroda et al., 1996). Similarly, for expression of the NH2-terminally HA-tagged PKCζ (PKCζ-HA), a pTB701-HA-PKCζ plasmid was constructed from pTB701-HA (Kuroda et al., 1996). An expression plasmid for a kinase-negative mutant protein of PKCζ-HA (K281M PKCζ-HA), pTB701-HA-K281M PKCζ, was prepared by replacing the ATP-binding Lys-281 residue by Met with a Quick-Change site-directed mutagenesis kit (Stratagene Cloning Systems). An expression plasmid for a constitutively active mutant of PKCζ-HA (caPKCζ-HA), pTB701-HA-caPKCζ, was prepared by deleting the pseudosubstrate region from Arg-116 to Trp-122 (Schonwasser et al., 1998). These plasmids were transferred into COS-7 cells by electroporation using a Gene Pulser II (Bio-Rad Laboratories).
Subcellular Fractionation of COS-7 Cells
COS-7 cells (∼5.0 × 107 cells) expressing FEZ1-FLAG were suspended in 1 ml of PBS and sonicated on ice for 15 s. After centrifugation at 10,000 g at 4°C for 10 min, the supernatant was collected as a cytoplasmic fraction. The pellet was resuspended in 1 ml of PBS containing 1% (vol/vol) Triton X-100 and sonicated on ice for 15 s. After centrifugation at 10,000 g at 4°C for 10 min, the supernatant was collected as a membrane fraction. Samples (10 μl) derived from ∼5.0 × 105 cells were subjected to SDS-PAGE (12.5%) and analyzed by Western blotting using an anti-FLAG mAb M2 (Eastman Kodak).
In Vitro Transcription and Translation
In vitro synthesis of FEZ1-FLAG was performed with a Single Tube Protein System 2 (Novagen). In brief, the cDNA for FEZ1-FLAG was placed 3′ downstream of the T7 promoter and then was transcribed with T7 RNA polymerase in the presence of dNTPs at 30°C for 15 min. The synthesized mRNA was translated in the rabbit reticulocyte lysate containing 1.5 MBq of [35S]Met (∼37 TBq/mmol) at 30°C for 60 min. Samples were analyzed by SDS-PAGE (12.5%) and subsequent autoradiography.
Immunoprecipitation and Phosphorylation Assay
COS-7 cells (∼5.0 × 107 cells) coexpressing FEZ1-FLAG and either PKCζ-HA or K281M PKCζ-HA were suspended in 500 μl of the lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10 mM 2-mercaptoethanol, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and 1% [vol/vol] Triton X-100, pH 7.5) containing 1 tablet of the complete protease inhibitor cocktail (Boehringer Mannheim) per 50 ml of the buffer. After centrifugation at 10,000 g at 4°C for 10 min, the lysates (500 μl) were incubated on ice for 1 h with 2 μg of either an anti-FLAG or anti-HA 12CA5 (Boehringer Mannheim) mAb and then mixed with 20 μl of protein G–Sepharose 4 fast flow beads (50% slurry; Pharmacia Biotech). After shaking at 4°C for 1 h, the beads were washed four times with the lysis buffer. For Western blotting, the beads were subjected to SDS-PAGE (12.5%). Tagged proteins were detected with either an anti-FLAG or anti-HA mAb as a primary antibody and an alkaline phosphatase-conjugated anti–mouse IgG (Promega) as a secondary antibody. For phosphorylation assay, the beads were mixed with 25 μl of the reaction mixture containing 20 mM Tris, 10 mM MgCl2, 20 μM ATP, pH 7.5 (without PKC activators). After addition of 3.7 KBq of [γ-32P]ATP (∼220 TBq/ mmol), the beads were incubated at 30°C for 30 min. Samples were analyzed by SDS-PAGE (12.5%) and subsequent autoradiography.
In Vitro Phosphorylation Assay
The glutathione-S-transferase (GST)-fused FEZ1 protein was synthesized in Escherichia coli BL21 cells by using a pGEX6P-1 vector (Pharmacia Biotech) and purified by a glutathione-Sepharose 4B column (Pharmacia Biotech) according to the supplier's protocol. The phosphorylation reaction mixture (see above) (25 μl) and 50 ng of the conventional PKC isoforms (mixture of α, βI, βII, and γ) purified from the rat brain (Kikkawa et al., 1986) were mixed with 5 μg of the purified GST-fused FEZ1 protein and the reaction was started by addition of 3.7 KBq of [γ-32P]ATP (∼220 TBq/mmol). The mixture was incubated at 30°C until incorporation of phosphate was saturated (∼30 min). Samples were analyzed by SDS-PAGE (12.5%) and autoradiography, followed by measurement of radioactivities with a BAS-2000 image analyzer. Under the same conditions, ∼1.8 mol of phosphate was incorporated into each mole of H1 histone, a commonly used substrate of PKC (Kikkawa et al., 1986; Kuroda et al., 1996).
Deletion Analysis
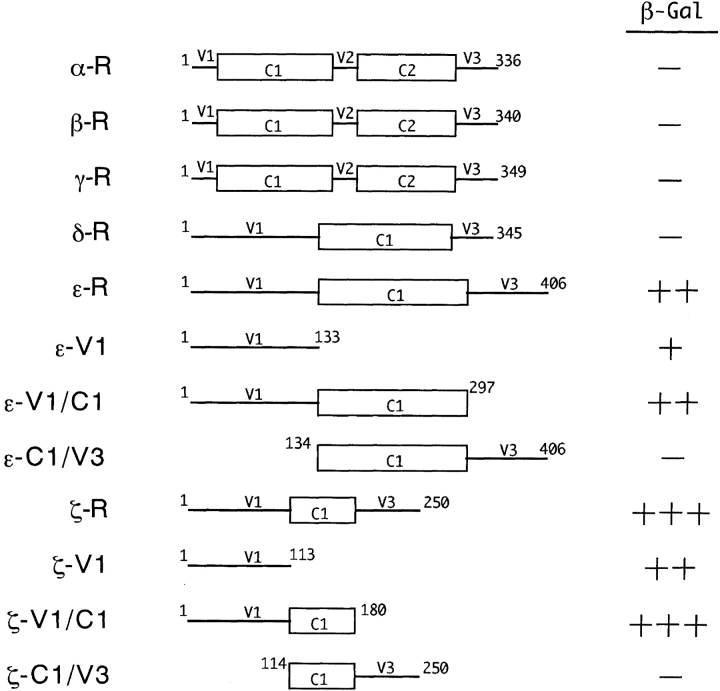
Essential regions in the PKC isoforms for interaction with FEZ1 were investigated by a yeast two-hybrid assay using the following 12 PKC-deletion mutants: α-R, residues 1–336 of rat PKCα (Ono et al., 1988a); β-R, residues 1–340 of rat PKCβI (Ono et al., 1986); γ-R, residues 1–349 of rat PKCγ (Ono et al., 1988a); δ-R, residues 1–345 of rat PKCδ (Ono et al., 1988b); ε-R, residues 1–406 of rat PKCε (similarly, ε-V1, residues 1–133; ε-V1/C1, residues 1–297; and ε-C1/V3, residues 134–406) (Ono et al., 1988b); and ζ-R, residues 1–250 of rat PKCζ (similarly, ζ-V1, residues 1–113; ζ-V1/C1, residues 1–180; and ζ-C1/V3, residues 114–250).
Immunocytochemical Observation
COS-7 cells coexpressing FEZ1-FLAG and either PKCζ-HA, K281M PKCζ-HA, or caPKCζ-HA were seeded in a 3.5-cm glass-bottom plate (MetTek Co.) at a concentration of ∼5.0 × 104 cells/plate. For experiments involving the treatment with a PKC inhibitor, cells were treated with 0.1 μM staurosporin (Wako Pure Chemical Ind., Ltd.) (Tamaoki et al., 1986) at 37°C for 2 h. Cells were fixed in 4% (wt/vol) paraformaldehyde at room temperature for 30 min, and then permeabilized and blocked in the mixture of 0.25% (vol/vol) Triton X-100, 5% (vol/vol) normal goat serum, and 5% (wt/vol) skim milk at room temperature for 30 min. The cells were incubated at room temperature for 2 h with 1 μg/ml of either an anti-FLAG or anti-HA mAb in PBS containing 0.03% (vol/vol) Triton X-100. Subsequently, the cells were incubated at room temperature for 30 min with 1 μg/ml of an FITC-conjugated anti–mouse IgG (Amersham International plc.). The fluorescence was visualized under a Zeiss LSM410 confocal laser scanning microscope (Carl Zeiss, Inc.). Parental COS-7 cells showed no fluorescence with either an anti-FLAG or anti-HA mAb.
Transient Expression Assay for Neuronal Differentiation of PC12 Cells
PC12 cells (∼5.0 × 105 cells) were seeded in a 10-cm plate and cultured for 24 h in DME supplemented with 10% (vol/vol) horse serum (GIBCO BRL) and 5% (vol/vol) FCS. Cells were transfected with 9 μg of pTB701-FLAG-FEZ1, 3 μg of a pTB701-HA-PKCζ derivative, and 1 μg of a reporter plasmid, pRc-CMV-β-Gal (Higuchi et al., 1997) by the liposome method (SuperFect; QIAGEN GmbH). After 72 h, cells were washed with PBS and fixed with 1% (vol/vol) glutaraldehyde at 4°C for 5 min, followed by washing twice with PBS containing 5 mM MgCl2. Cells were stained by incubation at 37°C for 3 h in PBS containing 20 mM K3Fe(CN)6, 20 mM K4Fe(CN)6, 1 mM MgCl2, and 1 mg/ml X-Gal. The β-Gal–positive blue cells were scored by phase-contrast microscopy. Morphologically altered cells were judged from neurite outgrowth with a flattened shape and increased body mass, as typified previously (Higuchi et al., 1997).
Results
cDNA Cloning of PKCζ-interacting Protein
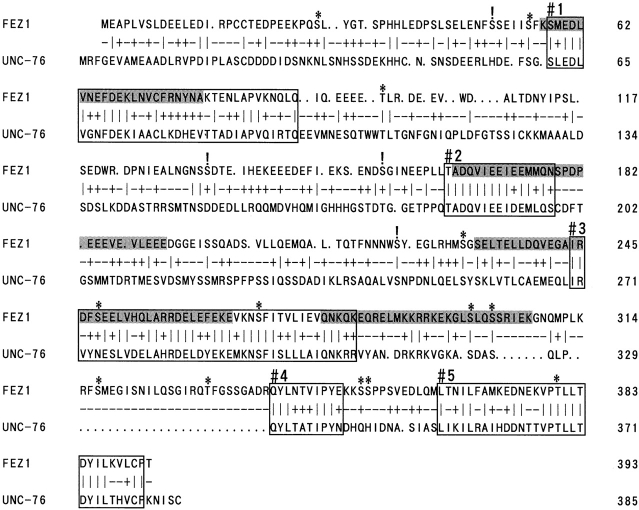
Using the regulatory domain of rat PKCζ (residues 1–250) fused with the yeast GAL4 DNA-binding domain as a bait, we screened a rat brain cDNA library by the two-hybrid method in yeast. Among ∼1.0 × 107 yeast transformants (Leu+, Trp+) expressing rat cDNA-derived proteins fused with the GAL4 activation domain, one positive clone that exhibited both β-Gal activity and His+ phenotype only in the presence of the bait plasmid was obtained. Since the clone was found to harbor a 5′-terminal truncated form of a cDNA fragment upon nucleotide sequencing (data not shown), RACE was performed with another rat brain cDNA library to obtain a full-length cDNA. Finally, a cDNA consisting of 1662 bp and encoding a polypeptide of 393 amino acid residues was isolated and named temporarily as zeta-1. By computer search of the protein sequences registered in the GenBank/EBI/DDBJ data bank, we have come across the C. elegans UNC-76 protein reported recently (Bloom and Horvitz, 1997), showing significant sequence homology with the rat zeta-1 protein (identity, 31%; similarity, 63%) (Fig. 1). The nematode gene unc-76 (unc, uncoordinated) is necessary for normal axonal bundling and elongation within fascicles in the nematode (Hedgecock et al., 1985; Desai et al., 1988; McIntire et al., 1992). A human gene coding for a protein homologous to the nematode UNC-76 protein has also been cloned and shown to rescue locomotory defects caused by unc-76 mutations in the nematode (Bloom and Horvitz, 1997). The rat zeta-1 protein shows a 96% sequence identity with the reported human homologue of UNC-76. Hence, these mammalian homologues of the nematode UNC-76 protein (human and rat zeta-1 proteins) have been designated FEZ1 on the basis of the presumed roles in axonal fasciculation and elongation in mammals (Bloom and Horvitz, 1997).
Figure 1.
Sequence alignment of rat FEZ1 and C. elegans UNC-76 proteins. The two sequences were aligned by using a BLOSUM 62–amino acid substitution matrix. Insertions (indicated by dots) are introduced to optimize the alignment. Identical and similar residues are indicated by vertical bars and plus marks, respectively, and the other residues by minus signs. The regions predicted to form amphipathic helices are shaded. Five conserved regions (1–5) are boxed. Potential sites for N-glycosylation and phosphorylation by PKC are indicated by ! and *, respectively. Amino acid residues are numbered on the right margin. The nucleotide sequence encoding rat FEZ1 protein has been submitted to the GenBank/EBI/DDBJ data bank with accession number U48249.
Rat FEZ1 is rich in acidic residues Asp and Glu (24% of the total residues), especially in the NH2-terminal half. Although the NH2-terminal half of UNC-76 (residues 13– 186) has been demonstrated to play as a signal for axonal localization in the nematode (Bloom and Horvitz, 1997), it is unknown whether the NH2-terminal half of FEZ1 possesses the similar signaling function. A computer analysis of the deduced amino acid sequence of FEZ1 has predicted that four regions (Lys-57 to Ala-85, Ala-165 to Glu-192, Ser-231 to Glu-266, and Gln-279 to Lys-307) have potentials to form amphipathic helices, which can mediate intra- and intermolecular interactions by constituting the coiled–coil structure (Lupas, 1996). Alignment of rat FEZ1 with the nematode UNC-76 (Fig. 1) reveals the presence of five well conserved regions (1–5), and three (1–3) of them approximately coincide with the regions predicted for forming amphipathic helical structures. In addition, FEZ1 has 4 potential sites for N-glycosylation (Asn-Xaa-Thr/Ser, where Xaa represents any amino acid except for Pro) and 13 putative sites for phosphorylation by PKC (Pearson and Kemp, 1991) (Fig. 1).
Expression of FEZ1 mRNA
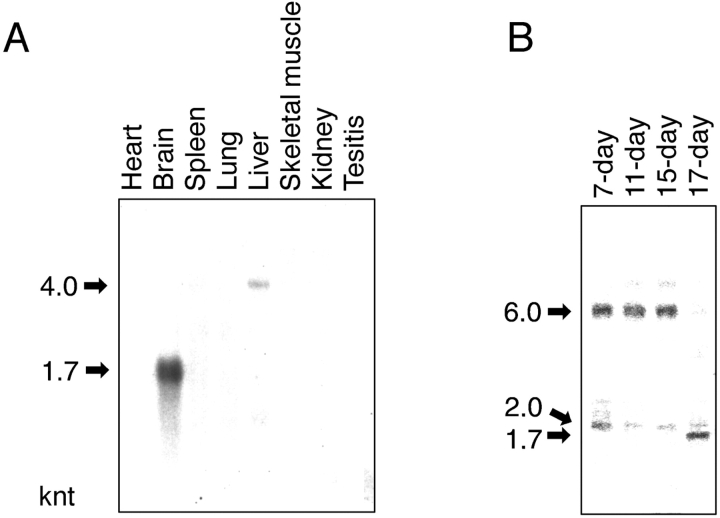
Northern blot analysis of eight tissues from adult rat has shown that FEZ1 mRNA with a size of ∼1700 nucleotides (nt) is expressed abundantly and exclusively in the brain, although faint expression of mRNA with a size of ∼4000 nt is also observed in the liver (Fig. 2 A). It is interesting to note that PKCζ mRNAs are also highly expressed in the brain (Ono et al., 1988b). The nematode UNC-76 is detected throughout the nervous system of the animals at all developmental stages from embryos (before outgrowth of the first axons) through adult worms (Bloom and Horvitz, 1997). Therefore, we investigated further the FEZ1 mRNA expression during development of mouse embryo with rat FEZ1 cDNA as a probe; the mouse FEZ1 gene deposited in the GenBank EST (Expressed Sequence Tag) database shows a very high sequence identity (>92%) with rat FEZ1. As shown in Fig. 2 B, a 6000-nt mRNA is expressed abundantly in the early to mid stage of development (from 7 d postcoitum [dpc] to 15 dpc). In the late stage of development (17 dpc), the 6000-nt mRNA disappears and instead a 1700-nt mRNA is expressed abundantly. Furthermore, a 2000-nt mRNA is expressed constantly, though slightly, in all developmental stages. It has been known that development of the nervous system in mouse embryo begins soon after 7 dpc with the neural plate formation and ends before 17 dpc. By cDNA cloning, all mRNAs observed here (1700-, 2000-, 4000-, and 6000-nt mRNAs) were confirmed to contain a 5′-untranslated sequence (∼100 nt), the same rat/mouse FEZ1 gene (∼1200 nt), a 3′-untranslated sequence of various lengths (from 340 to 4600 nt), and a poly (A)+ sequence (<100 nt).
Figure 2.
Northern blot analysis of FEZ1 mRNA. (A) Detection of FEZ1 mRNA in adult rat tissues. Northern blots containing 2 μg of poly(A)+ RNA from various adult rat tissues per lane were incubated with the full-length FEZ1 cDNA fragment labeled with [α-32P]dCTP. (B) Detection of FEZ1 mRNA in developing mouse embryos. Northern blots containing 2 μg of poly(A)+ RNA from mouse embryos in different developmental stages (7, 11, 15, and 17 dpc) per lane were used. The positions of FEZ1 mRNA are indicated by arrows with their approximate sizes (knt, kilonucleotides) on the left margin of each blot.
Expression of FEZ1 in COS-7 Cells
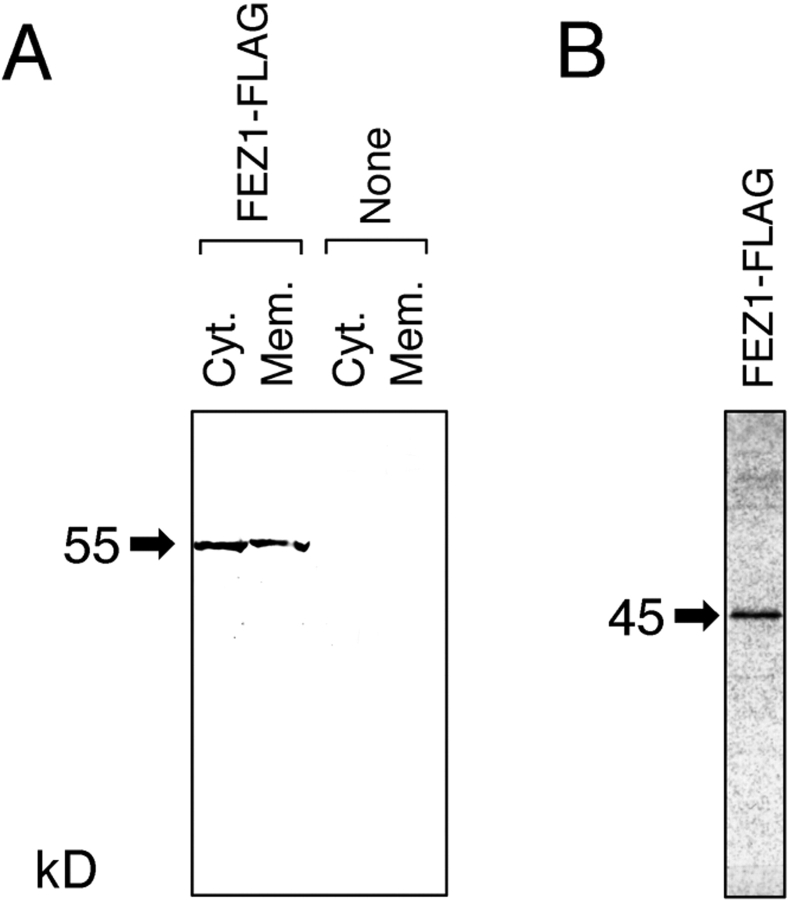
The lysate of COS-7 cells expressing FEZ1-FLAG was separated into the cytoplasmic and membrane fractions. Western blotting with an anti-FLAG mAb indicated that FEZ1-FLAG (∼55 kD) was equally present in both the cytoplasmic and membrane fractions (Fig. 3 A). The molecular mass of FEZ1-FLAG produced in COS-7 cells was ∼10 kD larger than that of the protein synthesized by in vitro transcription and translation (Fig. 3 B), which agreed well with the value calculated from the deduced amino acid sequence of FEZ1 (45,207). As described above, FEZ1 contains four potential sites for N-glycosylation. In addition, FEZ1-FLAG was found to be phosphorylated by in vivo labeling of COS-7 cells with [32P]H3PO4 (data not shown), which could also be a cause for the retarded migration on SDS-PAGE. These results strongly suggest that FEZ1 undergoes posttranslational modification (N-glycosylation and/or phosphorylation) in the mammalian cells.
Figure 3.
Expression of FEZ1 protein. (A) Subcellular localization of FEZ1-FLAG protein in COS-7 cells. The lysates of COS-7 cells expressing FEZ1-FLAG were separated into the cytoplasmic (Cyt.) and membrane fractions (Mem.) and analyzed by Western blotting with an anti-FLAG mAb. As a control, untransfected COS-7 cells were used (None). Each lane contained the sample derived from ∼5.0 × 105 cells. (B) In vitro synthesis of FEZ1-FLAG protein. FEZ1-FLAG protein labeled with [35S]Met was synthesized as described in Materials and Methods. The reaction mixture was subjected to SDS-PAGE (12.5%) and then autoradiographed. The molecular mass of FEZ1-FLAG protein is indicated on the left margin of each gel with an arrow (in kD).
Association of FEZ1 and PKCζ in COS-7 Cells
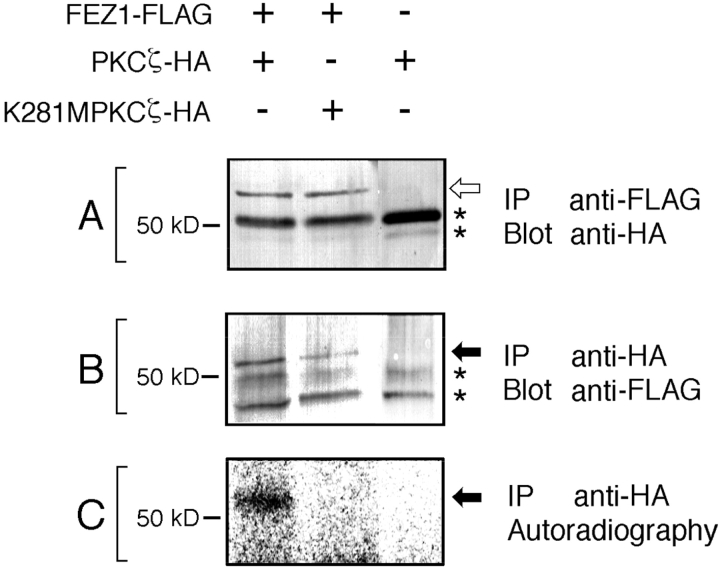
To examine in vivo association of FEZ1 and PKCζ, the lysates of COS-7 cells coexpressing FEZ1-FLAG and PKCζ-HA were analyzed by the immunoprecipitation assay. By Western blotting with the anti-HA mAb, PKCζ-HA with an approximate M r of 72,000 was detected in the anti-FLAG immunoprecipitates (Fig. 4 A), indicating coprecipitation of PKCζ-HA with FEZ1-FLAG. Similarly, by Western blotting with an anti-FLAG mAb, FEZ1-FLAG with an approximate M r of 55,000 was also detected in the anti-HA immunoprecipitates (Fig. 4 B), indicating coprecipitation of FEZ1-FLAG with PKCζ-HA. These results clearly show that FEZ1-FLAG associates with PKCζ-HA in vivo. Under the same conditions, FEZ1-FLAG did not associate with other HA-conjugated conventional and novel PKC isoforms (α, βI, γ, δ, and ε) (data not shown). We then analyzed the anti-HA immunoprecipitates by the phosphorylation assay as described in Materials and Methods and found that FEZ1 in the FEZ1-FLAG/PKCζ-HA complex could be phosphorylated (Fig. 4 C). When a kinase-negative mutant enzyme of PKCζ-HA (K281M PKCζ-HA) was coexpressed with FEZ1-FLAG in COS-7 cells, FEZ1-FLAG was found to be associated with K281M PKCζ-HA (Fig. 4, A and B) but not phosphorylated (Fig. 4 C). Collectively, these results indicate that FEZ1 is a novel cellular substrate of PKCζ and its association with PKCζ is independent of the activity of PKCζ. On the other hand, when GST-FEZ1 expressed in bacterial cells was incubated with the conventional PKC isoforms (mixture of α, βI, βII, and γ) purified from the rat brain in the presence of [γ-32P]ATP, ∼0.4 mol of phosphate was incorporated per mole of GST-FEZ1. The GST protein alone could not be phosphorylated under the same conditions (Kuroda et al., 1996). This result shows that FEZ1 can also be phosphorylated in vitro by the conventional PKC isoforms. The discrepancy between in vivo and in vitro phosphorylation assays is discussed later (see Discussion).
Figure 4.
In vivo association of FEZ1 and PKCζ. Either PKCζ-HA or its kinase-negative mutant K281M PKCζ-HA was coexpressed with FEZ1-FLAG in COS-7 cells. The lysates were immunoprecipitated (IP) and then blotted (Blot) with either an anti-FLAG or anti-HA mAb. (A) Detection of PKCζ-HA and K281M PKCζ-HA (open arrow) in the anti-FLAG immunoprecipitates with an anti-HA mAb. (B) Detection of FEZ1-FLAG protein (closed arrow) in the anti-HA immunoprecipitates with an anti-FLAG mAb. (C) In vitro phosphorylation assay of the anti-HA immunoprecipitates. Phosphorylated FEZ1-FLAG protein is indicated by a closed arrow. IgG heavy chains derived from the mAb used for immunoprecipitation are indicated by asterisks.
Mapping of Regions Involved in the FEZ1/PKCζ Association
To investigate the specificity of FEZ1 for PKC isoforms and also to identify the region(s) in the PKC molecules involved in the association with FEZ1, the regulatory domains of various PKC isoforms were used as a bait in the yeast two-hybrid assay. As shown in Fig. 5, FEZ1 interacted strongly with the regulatory domain of PKCζ (ζ-R, residues 1–250) and moderately with that of PKCε (ε-R, residues 1–406) but not with those of other PKC isoforms. Based on the sequence comparison of PKC isoforms, the regulatory domains of PKCζ and PKCε are further divided into a conserved region (C1) and two variable regions (V1 and V3) with a considerable sequence diversity (Nishizuka, 1988). Therefore, various deletion mutants containing a part(s) of these regions of PKCζ and PKCε were then constructed to map the region interacting with FEZ1. The yeast two-hybrid assays indicated that the V1 regions of PKCζ and PKCε were essential for interaction with FEZ1 (Fig. 5). However, in the case of full-length PKC isoforms, only PKCζ interacted with FEZ1 in the two-hybrid assay (data not shown), indicating that PKCζ but not PKCε could form a stable complex with FEZ1.
Figure 5.
Delineated structures of the regulatory domains of PKC isoforms and interaction with FEZ1 in the yeast two-hybrid system. β-Gal activity of yeast transformants was assayed by the plate method as described in Materials and Methods (+++, strongly positive; ++, moderately positive; +, weakly positive; and −, negative). Based on the sequence comparison, the primary structures of the regulatory domains of PKC isoforms are divided into conserved regions (C1 and C2) (boxes) and variable regions (V1–V3) (lines).
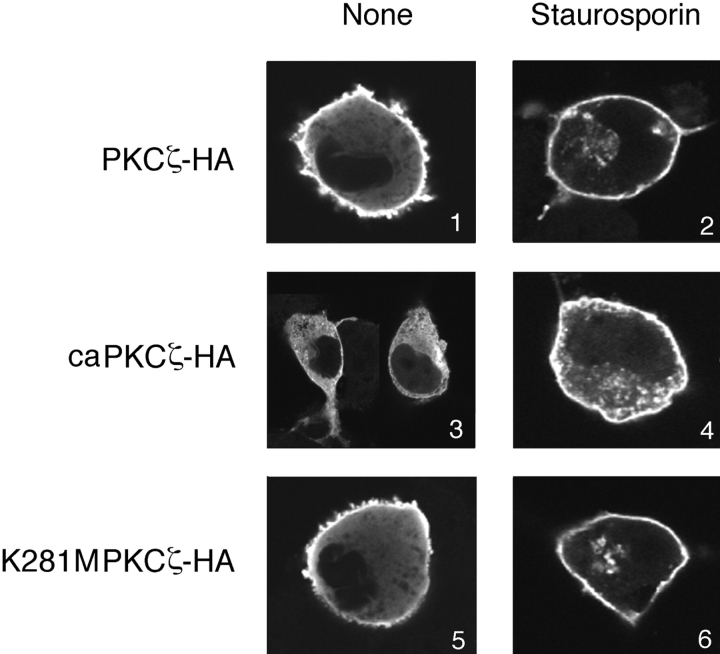
Cytoplasmic Translocation of FEZ1 in Response to the PKCζ Activity
A confocal laser scanning microscope was used to observe the intracellular localization of FEZ1 expressed in COS-7 cells. When the cells coexpressing FEZ1-FLAG and PKCζ-HA were examined, the plasma membrane was stained strongly with an anti-FLAG mAb and its cytoplasmic peripheries were also stained weakly (Fig. 6, panel 1). This indicates that FEZ1-FLAG is present predominantly in the plasma membrane of COS-7 cells, in which the expressed PKCζ is usually in an inactive form. On the other hand, in the cells expressing caPKCζ-HA (constitutively active mutant) (Schonwasser et al., 1998) instead of PKCζ-HA, FEZ1-FLAG was detected uniformly in the cytoplasm (Fig. 6, panel 3). In the cells expressing K281M PKCζ-HA (kinase-negative mutant), however, FEZ1-FLAG was again localized mostly in the plasma membrane (Fig. 6, panel 5). When these cells were treated with staurosporin, a PKC inhibitor common to all isoforms (Tamaoki et al., 1986), FEZ1-FLAG was present in the plasma membrane (Fig. 6, panels 2, 4, and 6). Particularly, it should be noted that FEZ1-FLAG present in the cytoplasm of the caPKCζ-HA–expressing cells was dynamically translocated into the plasma membrane by the staurosporin treatment (Fig. 6, panel 4). These results demonstrate that the translocation of FEZ1 from the plasma membrane, where it is normally localized, to the cytoplasm is regulated directly by the PKCζ activity.
Figure 6.
Intracellular localization of FEZ1 protein in COS-7 cells expressing various PKCζ. Either the wild-type PKCζ-HA, a constitutively active mutant (caPKCζ-HA), or a kinase-negative mutant (K281M PKCζ-HA) was coexpressed with FEZ1-FLAG protein in COS-7 cells. The cells were untreated (panels 1, 3, and 5) or treated with 0.1 μM staurosporin for 2 h (panels 2, 4, and 6), stained with an anti-FLAG mAb, and observed under a confocal laser scanning microscope.
Expression of FEZ1 Protein Stimulates the Constitutively Active PKCζ-induced Neuronal Differentiation of PC12 Cells
PC12 cells are known to differentiate into neuron-like cells in response to NGF or ectopic expression of an activated form of either Ras or other signaling molecules (Raf, MAPK kinase, MAPK) (Marshall, 1995). Because PKCζ is involved in the MAPK activation (Berra et al., 1995) and activation of PKCζ is required for NGF-induced neuronal differentiation of PC12 cells (Wooten et al., 1994; Zhou et al., 1997), we first examined the effects of PKCζ on morphology of PC12 cells by the transient expression assays. When PC12 cells were transfected with pTB701-HA-caPKCζ and the reporter plasmid pRc-CMV- β-Gal in a ratio of 9:1 (wt/wt), morphological changes (including extension of neurites, enlargement of cell mass, and flattened shapes) were induced in ∼18% of β-Gal– positive cells (Table I). No apparent morphological changes were observed in PC12 cells transfected with pTB701-HA-PKCζ and pRc-CMV-β-Gal. Under the same conditions, expression of the oncogenically activated form of Ras (RasG12V) significantly stimulated neuronal differentiation in ∼71% of transfected PC12 cells. These results suggest that caPKCζ can induce neuronal differentiation of PC12 cells, though weakly, presumably by activation of MAPK. On the other hand, expression of FEZ1 alone had no effect on PC12 cells.
Table I.
Morphology of Transfected PC12 Cells
| Plasmid DNA* | Morphological change‡ | |
|---|---|---|
| (%, mean ± SD) | ||
| (A) | ||
| pTB701 (negative control) | 3.2 ± 1.5 | |
| pTB701-HA-PKCζ | 3.7 ± 1.9 | |
| pTB701-HA-caPKCζ | 17.6 ± 2.9 | |
| pTB701-FLAG-FEZ1 | 4.2 ± 1.8 | |
| pTB701-RasG12V§ (positive control) | 70.8 ± 4.5 | |
| (B) | ||
| pTB701-FLAG-FEZ1 + pTB701-HA-PKCζ | 4.9 ± 2.2 | |
| pTB701-FLAG-FEZ1 + pTB701-HA-caPKCζ | 48.3 ± 3.6 |
PC12 cells were transfected with a 9:1 (wt/wt) ratio of the indicated plasmid:pRc-CMV-β-Gal (reporter plasmid) in A, and with a 9:3:1 (wt/wt/wt) ratio of pTB701-FLAG-FEZ1:pTB701-HA-PKCζ derivatives:pRc-CMV-β-Gal in B.
X-Gal–positive blue cells were counted as transfected cells. Flattened cells with neurite longer than twice the soma diameter were judged as differentiated cells. The percentage of differentiated cells (neurite-bearing and flattened shape) was calculated from at least 300 transfected cells counted. An average percentage was calculated from three plates per transfection and three independent transfections. Differences from control cells (pTB701) are statistically significant (P < 0.001) according to t test.
The expression plasmid for an oncogenically active form of Ras (substituting Gly-12 by Val). RasG12V protein was used as a positive control for the neuronal differentiation of PC12 cells (Higuchi et al., 1997).
Since no FEZ1 protein was detected in PC12 cells by Western blotting using a polyclonal anti-FEZ1 mAb (data not shown), we next asked whether expression of FEZ1 would affect caPKCζ-induced neuronal differentiation of PC12 cells. Cells were cotransfected with pTB701-FLAG-FEZ1, pTB701-HA-caPKCζ, and pRc-CMV-β-Gal in a ratio of 9:3:1 (wt/wt/wt) and the number of morphologically changed cells in β-Gal–positive cells was scored. The percentage of differentiated cells was increased (∼48%), as compared with that (∼18%) observed in the case of expression of caPKCζ alone (Table I). When PC12 cells were transfected with pTB701-FLAG-FEZ1, pTB701-HA-PKCζ, and pRc-CMV-β-Gal, there were no apparent morphological changes. These results strongly suggest that FEZ1 is located downstream of PKCζ and stimulates the caPKCζ-induced neuronal differentiation of PC12 cells.
Discussion
Predicted Role of UNC-76 in the C. elegans Axon Guidance Machinery
In the developing nervous systems, axons outgrow along other axonal cell surfaces to reach their targets, and then most axons associate with other axons in specific fascicles. This axonal association in fascicles is crucial for the assembly of nervous systems (Jessell, 1988; Grenningloh and Goodman, 1992). By genetic analysis of the C. elegans unc mutants with locomotory defects, the axonal elongation in fascicles has been shown to require at least two groups of genes (Hedgecock et al., 1985; Desai et al., 1988; McIntire et al., 1992). One group of genes (unc-14, unc-33, unc-44, unc-51, and unc-73) is required for the axonal elongation along nonneuronal as well as neuronal cell surfaces, and is likely to encode molecules essentially required for the axonal elongation. The other group of genes (unc-34, unc-71, and unc-76) is necessary for the axonal elongation in fascicles but not for the axonal elongation along nonneuronal cell surfaces, suggesting that the products of these genes are involved in the interaction of axons with the neuronal cell surfaces. The C. elegans unc-76 mutants show two types of axonal defects; the axons in fascicles often do not reach their full lengths and fail to bundle tightly together. Nonetheless, the axons around the body wall elongate normally, which are not accompanied by other axons, showing that UNC-76 is required only for the recognition of adjacent neuronal cells (Hedgecock et al., 1985; Desai et al., 1988; McIntire et al., 1992; Bloom and Horvitz, 1997).
Many molecules related to the axon guidance are conserved among mammals and the nematode in both the structural and functional levels: netrin (Serafini et al., 1994), a mammalian homologue of UNC-6; CRMP-62 (Goshima et al., 1995), that of UNC-33; and transforming growth factor-β (Colavita et al., 1998), that of UNC-129. FEZ1 protein identified here as a PKCζ-interacting protein was shown recently to rescue the locomotory defects caused by unc-76 mutations (Bloom and Horvitz, 1997), indicating that both FEZ1 and UNC-76 are also conserved evolutionarily. In the nematode, the axon guidance machinery involves several signaling molecules, for instance, UNC-33 protein in the heterotrimeric GTP-binding protein cascade (Goshima et al., 1995), Dock protein in the Tyr kinase cascade (Garrity et al., 1996), and UNC-51 and UNC-14 proteins in the Ser/Thr kinase cascade (Ogura et al., 1994, 1997). In addition, various PKC isoforms similar to the mammalian PKC isoforms have been identified in the nematode (Land et al., 1994a,b; Sano et al., 1995; Islas-Trejo et al., 1997). Thus, it is likely that both mammals and the nematode share a common machinery for axon guidance using similar signaling molecules. To date, however, there has been no report describing the relationship between the nematode PKCs and the molecules involved in axon guidance such as UNC-76.
FEZ1 Protein as a Cellular Substrate for PKCζ
The findings presented herein demonstrate that FEZ1 protein, a mammalian homologue of the nematode UNC-76 protein, is a novel cellular substrate for PKCζ. Northern blot analysis has shown that FEZ1 mRNA is abundantly expressed in rat brain (see Fig. 2 A). PKCζ mRNA is also highly expressed in rat brain (Ono et al., 1988b). In mammalian cells including the neuronal cells, endogenous PKCζ is localized evenly in the cytoplasm and the plasma membrane (Wooten et al., 1994; Goodnight et al., 1995; Parrow et al., 1995). Therefore, it is likely that FEZ1 and PKCζ coexist in the cytoplasmic periphery of the plasma membrane and the activation of PKCζ induces the cytoplasmic translocation of FEZ1 in the mammalian neuronal cells (Fig. 6). Various FEZ1 mRNAs of different sizes are expressed throughout the developmental stages of mouse embryo (Fig. 2 B), whereas all mRNAs observed encode the same FEZ1 gene. Thus, we assume that these FEZ1 mRNAs may be expressed in different cell types and/or may possess distinct in vivo stabilities. The 6000-nt FEZ1 mRNA coordinately expressed in embryos during 7–15 dpc is suggested to play an important role in the development of the mouse nervous system, whereas the 1700-nt FEZ1 mRNA likely bears a continual role in the neuronal tissues already developed. However, further studies by in situ hybridization analysis of the mouse embryos and construction of the FEZ1 gene–deficient mice are needed to clarify the role of FEZ1 protein during development.
A mixture of conventional PKC isoforms (PKCα, βI, βII, and γ) could phosphorylate FEZ1 in vitro, although these PKC isoforms did not form a stable complex with FEZ1 in vivo (Fig. 4). Recently, many proteins interacting with the regulatory domain of PKC isoforms were shown to regulate the in vivo PKC functions (Mochly-Rosen, 1995; Diaz-Meco et al., 1996; Faux and Scott, 1996; Jaken, 1996; Kuroda et al., 1996; Puls et al., 1997). Therefore, it is suggested that the absence of an unidentified cellular protein(s) determining the specificity of PKC isoforms to FEZ1 may cause the in vitro phosphorylation of FEZ1 by conventional PKC isoforms. Attempts to identify the cellular factor(s) interacting with the PKCζ/FEZ1 complex are underway. Alternatively, in the phosphorylation by PKC, the substrate protein may not necessarily form a stable complex with PKC. Indeed, H1 histone and myelin basic protein, both of which are good substrates for PKC and are often used in the in vitro phosphorylation assay, could not form a stable complex with PKC in vivo (data not shown).
By the yeast two-hybrid assay, the NH2-terminal variable region (V1) of PKCζ was shown to interact with FEZ1 protein (Fig. 5). Since the V1 region of all PKC isoforms shows a considerable sequence diversity (Nishizuka, 1988), involvement of this region in the association with FEZ1 appears reasonable for displaying specific protein– protein interactions and is consistent with our recent finding that several PKC-interacting proteins containing the LIM domain recognize the similar region of PKC isoforms (Kuroda et al., 1996). As for the region(s) in FEZ1 protein involved in the association with PKCζ, the COOH-terminal half of FEZ1 (residues 185–393) likely participates in the association with the regulatory domain of PKCζ, because the clone containing a 5′-terminal truncated form of cDNA has been isolated in the first two-hybrid screening. Furthermore, the sequence homology between the COOH- terminal halves of FEZ1 and nematode UNC-76 proteins is higher than that between their NH2-terminal halves (see Fig. 1). Also, the COOH-terminal truncated forms of UNC-76 were observed in most of the nematode mutant alleles that failed to complement unc-76 (Bloom and Horvitz, 1997). Although further analyses with various deletion mutants of FEZ1 are needed to map the PKCζ-interacting region in more detail, it is suggested that interaction of the COOH-terminal half of FEZ1 with the regulatory domain of PKCζ is essential for the predicted cellular function(s) of FEZ1, as discussed below.
Possible Function of FEZ1 in the Neuronal Differentiation of PC12 Cells
In this study, we have shown that FEZ1 interacts with PKCζ in vivo and its intracellular localization is regulated by the PKCζ activity. By the transient expression assay, FEZ1 protein was found to stimulate the caPKCζ-induced neuronal differentiation of PC12 cells. On the other hand, in the NGF-induced neuronal differentiation of PC12 cells, a PKCζ substrate “nucleolin” was proposed recently to play a pivotal role in the connection between cell surface signaling and nucleus in PC12 cells (Zhou et al., 1997). Although it has been shown that the intracellular localization of nucleolin is changed by the activation of PKCζ in the NGF-induced neuronal differentiating PC12 cells, there is no structural similarity between nucleolin and FEZ1. Furthermore, no direct evidence is available showing that nucleolin enhances the caPKCζ-induced neuronal differentiation of PC12 cells. Thus, it is unclear whether or not there is any functional relationship between nucleolin and FEZ1.
Rho family GTP-binding proteins, which work downstream of the PKC activation (Tominaga et al., 1993), have also been shown to regulate the axon guidance in mammals and the nematode (Kozma et al., 1997; Luo et al., 1997; Zipkin et al., 1997). To examine the effects of Rho family proteins on the intracellular localization of FEZ1 protein (Fig. 6), we have coexpressed each of the constitutively active Rho family mutants (RhoA G14V, Rac1 G12V, and Cdc42Hs G12V/Q61L) with PKCζ-HA and FEZ1-FLAG in COS-7 cells. In all the cells examined, the localization of FEZ1 protein in the plasma membrane was unaffected by the ectopic expression of these Rho family mutants (data not shown), strongly suggesting that FEZ1 protein is not located downstream of the Rho family proteins but may be rather located upstream. As shown in Fig. 1, most of the FEZ1 protein is predicted to form coiled– coil structures (at least four amphipathic helices). These helices are considered to form a four-helix bundle, where the four helices are packed together by intramolecular interactions (Kohn et al., 1997), and the amphipathic property of these helices may contribute to the membrane localization of FEZ1 protein. More importantly, it has been demonstrated that the amphipathic helices (coiled–coil structures) often participate in the association with RhoA proteins, e.g., p116Rip (Gebbink et al., 1997), citron (Madaule et al., 1995), p160 ROCK (Ishizaki et al., 1996), and Rho-kinase (Leung et al., 1995). Thus, it is likely that the amphipathic helices of FEZ1 protein may interact with RhoA family proteins.
Concluding Remarks
Taken together, we propose that FEZ1 transduces the signals from unidentified receptors for neuronal cells to the axon guidance machinery by interacting with PKCζ through the following presumed pathway. First, cell-surface receptors receive the signals from adjacent neuronal cells. Second, receptors evoke an unknown molecule(s) that activates PKCζ. Third, FEZ1 proteins phosphorylated by PKCζ are translocated from the plasma membrane to the cytoplasm. Fourth, FEZ1 proteins in the cytoplasm transduce the signals to the intracellular machinery involving the Rho family GTP-binding proteins, which will finally induce axonal growth in fascicles. We expect that the FEZ1–PKCζ interaction demonstrated herein will be an important clue for elucidation of the signaling pathway and molecules involved in the axon guidance machinery in mammals.
Acknowledgments
We thank Dr. K. Mizuno for providing the reporter plasmid pRc-CMV-β-Gal. We also thank Drs. L. Bloom and H.R. Horvitz for sharing unpublished data. We are also grateful to Drs. U. Kikkawa and N. Saito for encouragement and helpful advice.
This study was supported in part by a Grant-in-Aid for Scientific Research (No. 09780576) from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations used in this paper
- β-Gal
β-galactosidase
- caPKCζ-HA
a constitutively active mutant of PKCζ-HA
- dpc
days postcoitum
- FEZ1
fasciculation and elongation protein zeta-1
- FEZ1-FLAG
NH2-terminally FLAG-tagged FEZ1 protein
- GST
glutathione-S-transferase
- K281M PKCζ-HA
a kinase-negative mutant protein of PKCζ-HA
- MAPK
mitogen-activated protein kinase
- nt
nucleotides
- PKC
protein kinase C
- PKCζ-HA
NH2-terminally HA-tagged PKCζ
- RACE
rapid amplification of cDNA ends
- X-Gal
5-bromo-4-chloro-3-indoryl-β-d-galactoside
References
- Berra E, Diaz-Meco MT, Lozano J, Frutos S, Municio MM, Sanchez P, Sanz L, Moscat J. Evidence for a role of MEK and MAPK during signal transduction by protein kinase C ζ. EMBO (Eur Mol Biol Organ) J. 1995;14:6157–6163. doi: 10.1002/j.1460-2075.1995.tb00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom L, Horvitz HR. The Caenorhabditis elegans gene unc-76and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc Natl Acad Sci USA. 1997;94:3414–3419. doi: 10.1073/pnas.94.7.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegansTGF-β. Science. 1998;281:706–709. doi: 10.1126/science.281.5377.706. [DOI] [PubMed] [Google Scholar]
- Desai C, Garriga G, McIntire SL, Horvitz HR. A genetic pathway for the development of the Caenorhabditis elegansHSN motor neurons. Nature. 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- Diaz-Meco MT, Municio MM, Frutos S, Sanchez P, Lozano J, Sanz L, Moscat J. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell. 1996;86:777–786. doi: 10.1016/s0092-8674(00)80152-x. [DOI] [PubMed] [Google Scholar]
- Dominguez I, Diaz-Meco MT, Municio MM, Berra E, Garcia de Herreros A, Cornet ME, Sanz L, Moscat J. Evidence for a role of protein kinase C ζ subspecies in maturation of Xenopus laevisoocytes. Mol Cell Biol. 1992;12:3776–3783. doi: 10.1128/mcb.12.9.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux MC, Scott JD. Molecular glue: kinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddel CJ, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueira L, McElhinny JA, Bren GD, MacMorran WS, Diaz-Meco MT, Moscat J, Paya CV. The Ras-Raf pathway is activated in human immunodeficiency virus-infected monocytes and participates in the activation of NF-κB. J Virol. 1996;70:223–231. doi: 10.1128/jvi.70.4.2332-2338.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity PA, Rao Y, Salecker I, McGlade J, Pawson T, Zipursky SL. Drosophilaphotoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- Gebbink MFBG, Kranenburg O, Poland M, van Horck FPG, Houssa B, Moolenaar WH. Identification of a novel, putative Rho-specific GDP/GTP exchange factor and a RhoA-binding protein: control of neuronal morphology. J Cell Biol. 1997;137:1603–1613. doi: 10.1083/jcb.137.7.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnight J, Mischak H, Kolch W, Mushinski JF. Immunocytochemical localization of eight protein kinase C isozymes overexpressed in NIH 3T3 fibroblasts. Isoform-specific association with microfilaments, Golgi, endoplasmic reticulum, and nuclear and cell membranes. J Biol Chem. 1995;270:9991–10001. doi: 10.1074/jbc.270.17.9991. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- Grenningloh G, Goodman CS. Pathway recognition by neuronal growth cones: genetic analysis of neural cell adhesion molecules in Drosophila. . Curr Opin Neurobiol. 1992;2:42–47. doi: 10.1016/0959-4388(92)90160-m. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegansidentified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Higuchi O, Amano T, Yang N, Mizuno K. Inhibition of activated Ras-induced neuronal differentiation of PC12 cells by the LIM domain of LIM-kinase 1. Oncogene. 1997;14:1819–1825. doi: 10.1038/sj.onc.1201020. [DOI] [PubMed] [Google Scholar]
- Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. The small GTP-binding protein Rho binds to and activates a 160 kD Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO (Eur Mol Biol Organ) J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- Islas-Trejo A, Land M, Tcherepanova I, Freedman JH, Rubin CS. Structure and expression of the Caenorhabditis elegans protein kinase C2 gene. Origins and regulated expression of a family of Ca2+-activated protein kinase C isoforms. J Biol Chem. 1997;271:6629–6640. doi: 10.1074/jbc.272.10.6629. [DOI] [PubMed] [Google Scholar]
- Jaken S. Protein kinase C isozymes and substrates. Curr Opin Cell Biol. 1996;8:168–173. doi: 10.1016/s0955-0674(96)80062-7. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Adhesion molecules and the hierarchy of neural development. Neuron. 1988;1:3–13. doi: 10.1016/0896-6273(88)90204-8. [DOI] [PubMed] [Google Scholar]
- Kazanietz MG, Areces LB, Bahador A, Mischak H, Goodnight J, Muschinski JF, Blumberg PM. Characterization of ligand and substrate specificity for the calcium-dependent and calcium-independent protein kinase C isozymes. Mol Pharmacol. 1993;44:298–307. [PubMed] [Google Scholar]
- Kikkawa U, Go M, Koumoto J, Nishizuka Y. Rapid purification of protein kinase C by high performance liquid chromatography. Biochem Biophys Res Commun. 1986;135:636–643. doi: 10.1016/0006-291x(86)90040-9. [DOI] [PubMed] [Google Scholar]
- Kohn WD, Mant CT, Hodges RS. α-Helical protein assembly motifs. J Biol Chem. 1997;272:2583–2586. doi: 10.1074/jbc.272.5.2583. [DOI] [PubMed] [Google Scholar]
- Kozma R, Sarner S, Ahmed S, Lim L. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol Cell Biol. 1997;17:1201–1211. doi: 10.1128/mcb.17.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill GN, Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271:31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- Land M, Islas-Trejo A, Freedman JH, Rubin CS. Structure and expression of a novel, neuronal protein kinase C (PKC1B) from Caenorhabditis elegans. PKC1B is expressed selectively in neurons that receive, transmit, and process environmental signals. J Biol Chem. 1994a;269:9234–9244. [PubMed] [Google Scholar]
- Land M, Islas-Trejo A, Rubin CS. Origin, properties, and regulated expression of multiple mRNAs encoded by the protein kinase C1 gene of Caenorhabditis elegans. . J Biol Chem. 1994b;269:14820–14827. [PubMed] [Google Scholar]
- Leung T, Manser E, Tan L, Lim L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- Luo L, Jan LY, Jan YN. Rho family GTP-binding proteins in growth cone signalling. Curr Opin Neurobiol. 1997;7:81–86. doi: 10.1016/s0959-4388(97)80124-9. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Madaule P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- McIntire SL, Garriga G, White J, Jacobson D, Horvitz HR. Genes necessary for directed axonal elongation or fasciculation in C. elegans. . Neuron. 1992;8:307–322. doi: 10.1016/0896-6273(92)90297-q. [DOI] [PubMed] [Google Scholar]
- Mochly-Rosen D. Localization of protein kinases by anchoring proteins: a theme in signal transduction. Science. 1995;268:247–251. doi: 10.1126/science.7716516. [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature. 1988;334:661–665. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB (Fed Am Soc Exp Biol) J. 1995;9:484–496. [PubMed] [Google Scholar]
- Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Muller F, Oshima Y. Caenorhabditis elegans unc-51gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- Ogura K, Shirakawa M, Barnes TM, Hekimi S, Oshima Y. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegansinteracts with the serine/threonine kinase UNC-51. Genes Dev. 1997;11:1801–1811. doi: 10.1101/gad.11.14.1801. [DOI] [PubMed] [Google Scholar]
- Ono Y, Kurokawa T, Fujii T, Kawahara K, Igarashi K, Kikkawa U, Ogita K, Nishizuka Y. Two types of complementary DNAs of rat brain protein kinase C. Heterogeneity determined by alternative splicing. FEBS Lett. 1986;206:347–352. doi: 10.1016/0014-5793(86)81010-9. [DOI] [PubMed] [Google Scholar]
- Ono Y, Fujii T, Ogita K, Kikkawa U, Igarashi K, Nishizuka Y. The structure, expression, and properties of additional members of the protein kinase C family. J Biol Chem. 1988a;263:6927–6932. [PubMed] [Google Scholar]
- Ono Y, Fujii T, Igarashi K, Kikkawa U, Ogita K, Nishizuka Y. Nucleotide sequences of cDNAs for α and γ subspecies of rat brain protein kinase C. Nucleic Acids Res. 1988b;16:5199–5200. doi: 10.1093/nar/16.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrow V, Fagerstrom S, Meyerson G, Nanberg E, Pahlman S. Protein kinase C-α and -ε are enriched in growth cones of differentiating SH-SY5Y human neuroblastoma cells. J Neurosci Res. 1995;41:782–791. doi: 10.1002/jnr.490410609. [DOI] [PubMed] [Google Scholar]
- Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase C ζ with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci USA. 1997;94:6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor TC, Osten P, Valsamis H, Jiang X, Naik MU, Sublette E. Persistent activation of the ζ isoform of protein kinase C in the maintenance of long-term potentiation. Proc Natl Acad Sci USA. 1993;90:8342–8346. doi: 10.1073/pnas.90.18.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Tabuse Y, Nishiwaki K, Miwa J. The tpa-1 gene of Caenorhabditis elegans encodes two proteins similar to Ca(2+)-independent protein kinase Cs: evidence by complete genomic and complementary DNA sequences of the tpa-1gene. J Mol Biol. 1995;251:477–485. doi: 10.1006/jmbi.1995.0449. [DOI] [PubMed] [Google Scholar]
- Schonwasser DC, Marais RM, Marshall CJ, Parker PJ. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol Cell Biol. 1998;18:790–798. doi: 10.1128/mcb.18.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T, Kennedy TE, Galko MJ, Mirzayan C, Jessell TM, Tessier-Lavigne M. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegansUNC-6. Cell. 1994;78:409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Staurosporine, a potent inhibitor of phospholipid/Ca++dependent protein kinase. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- Tominaga T, Sugie K, Hirata M, Morii N, Fukata J, Uchida A, Imura H, Narumiya S. Inhibition of PMA-induced, LFA-1–dependent lymphocyte aggregation by ADP ribosylation of the small molecular weight GTP binding protein, rho. J Cell Biol. 1993;120:1529–1537. doi: 10.1083/jcb.120.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten MW, Zhou G, Seibenhener ML, Coleman ES. A role for ζ protein kinase C in nerve growth factor-induced differentiation of PC12 cells. Cell Growth Diff. 1994;5:395–403. [PubMed] [Google Scholar]
- Zhou G, Seibenhener ML, Wooten MW. Nucleolin is a protein kinase C-ζ substrate: connection between cell surface signaling and nucleus in PC12 cells. J Biol Chem. 1997;272:31130–31137. doi: 10.1074/jbc.272.49.31130. [DOI] [PubMed] [Google Scholar]
- Zipkin ID, Kindt RM, Kenyon CJ. Role of a new Rho family member in cell migration and axon guidance in C. elegans. . Cell. 1997;90:883–894. doi: 10.1016/s0092-8674(00)80353-0. [DOI] [PubMed] [Google Scholar]