Abstract
The classical adherens junction that holds epithelial cells together consists of a protein complex in which members of the cadherin family linked to various catenins are the principal components. δ-catenin is a mammalian brain protein in the Armadillo repeat superfamily with sequence similarity to the adherens junction protein p120ctn. We found that δ-catenin can be immunoprecipitated as a complex with other components of the adherens junction, including cadherin and β-catenin, from transfected cells and brain. The interaction with cadherin involves direct contact within the highly conserved juxtamembrane region of the COOH terminus, where p120ctn also binds. In developing mouse brain, staining with δ-catenin antibodies is prominent towards the apical boundary of the neuroepithelial cells in the ventricular zone. When transfected into Madin-Darby canine kidney (MDCK) epithelial cells δ-catenin colocalized with cadherin, p120ctn, and β-catenin. The Arm domain alone was sufficient for achieving localization and coimmunoprecipitation with cadherin. The ectopic expression of δ-catenin in MDCK cells altered their morphology, induced the elaboration of lamellipodia, interfered with monolayer formation, and increased scattering in response to hepatocyte growth factor treatment. We propose that δ-catenin can regulate adhesion molecules to implement the organization of large cellular arrays necessary for tissue morphogenesis.
Keywords: δ-catenin, armadillo, adhesive junctions, cell motility, neural development
The development of tissue boundaries and the maintenance of tissue integrity require highly dynamic cell–cell and cell–matrix adhesion. Various types of adhesive junctions represent key modulators of these relationships, and during cell migration these structures must be able to mediate highly dynamic interactions among neighboring cells and between cells and the substrate. Within the adherens junction, proteins of the cadherin family, single membrane-spanning glycoproteins, directly anchor cell–cell contacts via Ca2+-dependent homophilic interactions. Proteins with various numbers of cadherin extracellular tandem repeats comprise a superfamily which has achieved greatest diversity in the nervous system (reviewed in Uemura, 1998). The classical cadherins, which include E-, N-, and P-cadherin (Marrs and Nelson, 1996), all have a highly conserved cytoplasmic domain to which bind a set of associated proteins referred to as the catenins (Nagafuchi and Takeichi, 1988; Ozawa et al., 1989). The catenins form a cytoplasmic plaque complex, which links classical cadherins to the actin cytoskeleton (Takeichi, 1991, 1995; Yap et al., 1997). The adhesive activity of the junctions derives from both the cadherin ectodomain, which has weak intrinsic adhesive activity on its own (Brieher et al., 1996), and the cytoplasmic domain, which significantly strengthens the homophilic interaction. Thus, one role of the conserved cadherin cytoplasmic domain is to bind to β-catenin, which in turn binds to α-catenin, and anchors the complex to actin filaments (Aberle et al., 1994; Hoschuetzky et al., 1994; Funayama et al., 1995; Jou et al., 1995; Rimm et al., 1995).
Plakoglobin/γ-catenin also binds directly to E-cadherin, as well as to desmosomal cadherins, and may compete with β-catenin for binding (Mathur et al., 1994; Staddon et al., 1995). β-catenin and plakoglobin share a 42–amino acid repeated motif, the Arm repeat, originally described in the Drosophila segment polarity gene, armadillo (Riggleman et al., 1989). A more distantly related protein, p120ctn, also contains a series of Arm repeats and binds to cadherin (Reynolds et al., 1992, 1994; Shibamoto et al., 1995; Staddon et al., 1995) at a juxtamembrane site within the cytoplasmic domain of cadherin rather than the more distal site where β- and γ-catenin bind (Ozawa and Kemler, 1998; Yap et al., 1998). p120ctn does not bind to α-catenin (Daniel and Reynolds, 1995) and in nontransformed cells, only small amounts bind to cadherin (Shibamoto et al., 1995).
δ-catenin is a member of the p120ctn subfamily, defined as proteins with 10 Arm repeats (in contrast to the 13 Arm repeats of β-catenin) in a characteristic spacing and often with quite diverse NH2- and COOH-terminal sequences that flank the repeats (Peifer et al., 1994). p120ctn is the founding member of a subfamily of Arm repeat proteins (Peifer et al., 1994), which include p0071 and the plakophilins, both components of the desmosome (Kapprell et al., 1988; Heid et al., 1994; Hatzfeld and Nachtsheim, 1996; Mertens et al., 1996) and ARVCF, a protein known only on the basis of its sequence (Sirotkin et al., 1997). δ-catenin was discovered by its ability to bind to the loop region of presenilin-1 (Zhou et al., 1997), which is encoded by the gene most commonly mutated in familial Alzheimer's disease (Clark et al., 1995; Sherrington et al., 1995). Independently, the same gene was cloned from a human fetal brain library using oligonucleotides deduced from a plakophilin 1-related expressed sequence tag, or EST (Paffenholz and Franke, 1997; Zhou et al., 1997) and termed NPRAP. Like other members of the subfamily, δ-catenin has greatest similarity with the Arm repeats of p0071 (69.3% identical), a desmosomal protein, and is somewhat less related to p120ctn (48.0% identical). Both Northern blot and in situ hybridization studies showed that δ-catenin is almost exclusively expressed in the nervous system (Paffenholz and Franke, 1997; Zhou et al., 1997; Ho, C., Bhide, P., and Kosik, K.S., unpublished data).
We sought to determine whether δ-catenin is a cell junction–associated protein and may perform a morphoregulatory function. We demonstrated that δ-catenin colocalizes and interacts with adhesive junction proteins both in transfected cells and in mouse brain. Based on transfections in MDCK cells, the junctional targeting signal resides in the Arm repeats. Like p120ctn, δ-catenin binds to the juxtamembrane domain on cadherins. Functionally, δ-catenin can prime MDCK cells for growth factor–stimulated cell motility.
Materials and Methods
cDNA Cloning
In the region of the mouse δ-catenin start codon (Paffenholz and Franke, 1997), a homologous human EST of 150 base pairs is in the database (GenBank accession number AA670399). This short sequence was used to design a sense primer which, together with an antisense primer from the previously known human sequence (Paffenholz and Franke, 1997; Zhou et al., 1997), was used to amplify the human 5′ cDNA sequence by reverse transcriptase PCR. Total RNA from SH-SY5Y human neuroblastoma cells was isolated using the TRIzol Reagent system (GIBCO BRL). cDNA was synthesized from 1 μg of total RNA with random primers and reverse transcriptase (Perkin-Elmer Corp.) according to the manufacturer's instructions. This cDNA was then used as a template in a PCR reaction using primers DC8 (5′-GGTGCATGTTTGCGAGGAAGC-3′) and D1350-AS (5′-ATGGGCGAGCTGGTGCTGTAGGAC-3′) for 30 cycles (94°C for 30 s; 55°C for 30 s; 72°C for 1 min; and a final step at 72°C for 10 min) in the presence of 5% DMSO. A fragment of ∼950 bp was purified using Wizard PCR Preps (Promega Corp.) and then cloned directly into pCR-II using the TA Vector kit (Invitrogen Corp.) and sequenced.
δ-catenin cDNA was subcloned into the mammalian expression vector pcDNA containing the CMV promoter (Invitrogen Corp.). To visualize the δ-catenin distribution by direct fluorescent methods, δ-catenin and its truncated mutants were subcloned by PCR into pEGFP (CLONTECH Laboratories, Inc.). MDCK cells were transfected using Lipofectamine Plus (GIBCO BRL). The stable MDCK cells were selected and maintained in medium containing neomycin. Although the neomycin selection allowed us to obtain a population of cells, nearly all of which expressed δ-catenin, repeated passage of these cells resulted in a gradual reduction in δ-catenin expression.
Antibodies and Reagents
To generate antibodies specific for δ-catenin, a plasmid construct containing amino acids 434–530 from the NH2 terminus was fused to glutathione transferase in pGEX-4T1 (Pharmacia Biotech, Inc.). Fusion protein was purified according to the manufacturer's instruction. Rabbits were immunized with purified fusion protein (Charles River Laboratories) and collected and tested. Immune serum was affinity purified by the removal of glutathione transferase followed by the further purification against δ-catenin amino acids 434–530 cross-linked to Actigel A resin (Pharmacia Biotech, Inc.). The final purified rabbit anti–δ-catenin was designated rAb62. A second antibody, rAb64, was raised against amino acids 828–1022 of δ-catenin and was purified similarly. mAb specific for the intracellular domain of human N-cadherin was from Dr. K. Knudsen (Wynnewood, PA; Knudsen et al., 1995). mAbs against β-catenin, pp120, E-cadherin, and desmoglein were from Transduction Laboratories. Monoclonal and polyclonal antibodies against the COOH terminus of chick N-cadherin as well as monoclonal anti-uvomorulin (DECMA-1) were from Sigma Chemical Co. Unless otherwise indicated, all chemicals were from Sigma Chemical Co.
Cell Cultures and Immunohistochemistry
MDCK cells were grown on Nunclon plates (Fisher Scientific Co.) in MEM containing Earle's balanced salts solution and supplemented with 5% FCS, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Hepatocyte growth factor (HGF)1 was diluted to 100 ng/ml in serum free media (MEM supplemented with 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin) and added directly to cells for 24 h. Nontreated cells were incubated with serum free media without HGF. Cells used for fluorescent studies were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Following staining, the cells were mounted and photographed using a Zeiss Axioskop equipped with epifluorescence. Slides were also viewed with a Zeiss MC100 immunofluorescence microscope equipped with a Biorad MRC-1024 Confocal Imaging System. After z-axial collection of images, the vertical and other morphometric analyses were performed with MetaMorph Imaging software system (Universal Imaging Corp.).
Sample Preparation
Cultured Cells.
Stable MDCK cells were lysed in buffer A, 10 mM Tris, pH 7.4, 50 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.2% Triton X-100 containing protease and phosphatase inhibitors, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 10 μg/ml aprotinin, 10 μg/ml pepstatin A, 10 μg/ml of leupeptin, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 20 mM sodium fluoride. In some experiments, cell lysates were prepared in the absence of any detergent. The cell lysates were centrifuged at 100,000 g to remove cell debris. Cells were also lysed in buffer B (same as buffer A, but with 1% Triton X-100) and the supernatants were collected in the same way. The pellet was resuspended in RIPA buffer (10 mM Tris, 1 mM EDTA, 1 mM EGTA, 0.2% SDS, and 0.5% deoxycholate, pH 7.4). After removing cell debris with centrifugation, the lysates were either mixed with sample buffer for SDS-PAGE analysis, or for further immunoprecipitation.
Brain Tissue.
For the mouse brain samples, mice were killed and brains removed and rinsed in PBS with 1 mM orthovanadate and 2 mM PMSF. The washed brain tissues were homogenized in buffer containing 50 mM NaCl, 10 mM Pipes, pH 6.8, 3 mM MgCl2, 0.5% Triton X-100, 300 mM sucrose, 1.2 mM PMSF, and 10 μg/ml leupeptin, and spun at 5,000 g for 10 min at 4°C to remove nuclei and cell debris. The supernatant was then spun a second time at 100,000 g for 10 min. The supernatant from this spin was collected as the postnuclear homogenate supernatant (PnH) and the pellet was resuspended in a buffer containing 15 mM Tris, pH 7.5, 5 mM EDTA, 2.5 mM EGTA, and 0.2% SDS as postnuclear Triton X-100 insoluble pellet (PnP). To obtain brain synaptosomal fractions, the post nuclear homogenates were placed onto a sucrose gradient and different fractions were collected. Synaptosomes were identified by the enrichment of synaptophysin. In all cases the amount of protein was determined using the BCA Protein Assay Kit (Pierce Chemical Co.).
Immunoprecipitations
The lysates for immunoprecipitation (IP) were precleared by incubating with protein A–Sepharose beads (for mAb IP, protein G plus protein A–agarose beads was used instead) followed by centrifugation to remove the beads. The supernatants were immunoprecipitated with specific antibodies for 1 h at 4°C. The immunoprecipitates were captured by protein A–Sepharose (or protein G plus protein A–agarose beads for mAb IP) for an additional 2 h at 4°C. Samples were washed sequentially with IP buffer (15 mM Tris, pH 7.5, 5 mM EDTA, 2.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 120 mM NaCl, and 25 mM KCl), high salt buffer (15 mM Tris, pH 7.5, 5 mM EDTA, 2.5 mM, 1% Triton, 0.1% SDS, and 1 M NaCl), and a low salt buffer (15 mM Tris, pH 7.5, 50 mM NaCl, 5 mM EDTA). Immunoprecipitates were separated by SDS-PAGE and transferred to nitrocellulose membranes (PGC Scientifics) for immunoblotting. After blotting protein stainings were detected by enhanced chemiluminescence (Amersham Life Science) on X-OMAT film. When necessary, blots were stripped (100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl, pH 7.6) for 30 min at 55°C. The blots were then washed in TBS Tween 20 before reprobing with specific antibodies. Immunoblots were quantified with an AlphaImager™ 2000 Documentation & Analysis System (Alpha Innotech Corp.).
Cell Proliferation Analysis
To measure the growth rate of cells, two methods were used. In one, δ-catenin–transfected MDCK (MF) cells and mock-transfected MDCK cells were plated at equal densities (500,000 cells/plate), grown for 2 d (∼60% confluency), and trypsinized. An aliquot was taken for counting the number of cells with a hemacytometer. Each set, either δ-catenin– expressing or mock-transfected cells, was counted eight times, and the mean number of cells calculated. The second method used a 5-bromo-2′-deoxy-uridine (BrdU) Labeling and Detection kit (Boehringer Mannheim).
Cell Migration Analysis
To quantify the migration rate, a two-chambered system was used consisting of a smaller chamber placed inside a larger well from a 24-well plate (Costar Corp.). The inner, or upper, compartment was separated from the lower plate compartment by a 12-μm filter. 5,000 cells were plated in the upper chamber overnight, then either allowed to grow for another day or treated with 100 ng/ml HGF for 48 h (Balkovetz et al., 1997). Cells in the upper chamber were trypsinized and counted in a similar manner as used to measure the growth rate. Cells in the lower chamber were stained with Harris modified hematoxylin with acetic acid (Fisher Scientific Co.) and counted under the microscope using a hemacytometer. A ratio of the number of cells in the lower:upper chamber was calculated for each set to compare the migration rate of control cells to MF cells.
Yeast Two-Hybrid Analysis
These assays, as well as the vectors pCK2 and pCK4, are described in detail in Pai et al. (1996). Briefly, the indicated portions of mouse E-cadherin or DE-cadherin were PCR amplified and cloned into pCK4 to produce a Gal4 transactivation domain fusion. A murine OB-cadherin clone (Tao et al., 1996) in the pYP16 vector was provided by Pierre McCrea. The entire Arm repeat region of δ-catenin (amino acids 532–1013) was cloned into pCK2 to produce a lexA DNA binding domain fusion. All constructs were confirmed by sequencing. The yeast strain L40 was transformed and selected for both plasmids on synthetic media lacking tryptophan and leucine. Liquid β-galactosidase assays were carried out on at least six independent transformants in duplicate for each plasmid combination tested. β-galactosidase activity was calculated in Miller units as described in Pai et al. (1996).
Results
Structure of δ-catenin
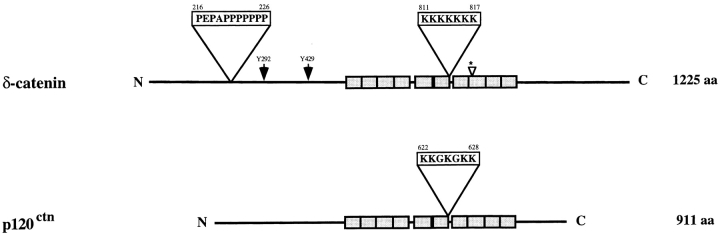
We determined the complete sequence of human δ-catenin. It encodes a 1,225–amino acid protein with a predicted molecular weight of 132544.86 and a pI of 7.94 (Fig. 1). Mouse δ-catenin encodes a 1,247–amino acid protein that is highly related with 95% identity and 98% similarity (Paffenholz and Franke, 1997). Neither initiator methionine fits very well with the Kozak consensus sequence (Kozak, 1986) for the initiation of translation. Within the eighth Arm repeat of the human sequence, the mouse has a 25–amino acid insert at position 879 which may represent alternative splicing. A significant portion of the molecular mass lies NH2- and COOH-terminal to the Arm domain of δ-catenin. These regions contain several potentially important consensus sequences. An analysis of the complete sequence was conducted using the software CANSITE 1.0 (http://himiris.bidmc.harvard.edu), which tests putative proteins against the results of a combinatorial peptide library for selectivity of phosphorylation by signal transduction kinases (Zhou et al., 1995). The analysis revealed two potential Abl sites (Fig. 1) and several potential Abl binding motifs with the sequence XPXXPP. We found that δ-catenin showed a very high probability of being phosphorylated by Abl at tyrosine residues 292 and 429, with a specificity predicted to be within the top 1% of >2 million peptides tested. A polylysine stretch from amino acid 811 to 817 resembles a nuclear localization signal (Kalderon et al., 1984) and a polyproline tract, which is not present in p120ctn, could serve as a profilin binding site (Perelroizen et al., 1994).
Figure 1.
Schematic representation of the domain structure of full-length human δ-catenin compared with p120ctn. Amino acids 216–226 contain a proline rich motif that is absent in p120ctn. For δ-catenin, the amino acids 532–1013 are boxed and correspond to the 10 Armadillo repeats. Two Abl tyrosine phosphorylation consensus sites (Y292 and Y429) are indicated by arrows. The small arrowhead with an asterisk indicates a 25–amino acid insertion site that was observed in murine δ-catenin but not in human clones. Amino acids 811–817 represent a lysine rich motif that is a potential nuclear localization signal (NLS) sequence. p120ctn has a similar, albeit somewhat weaker, potential NLS at amino acids 622–628. A partial human δ-catenin cDNA sequence was previously published (Zhou et al., 1997) and the full-length sequence is now updated (GenBank accession number U96136).
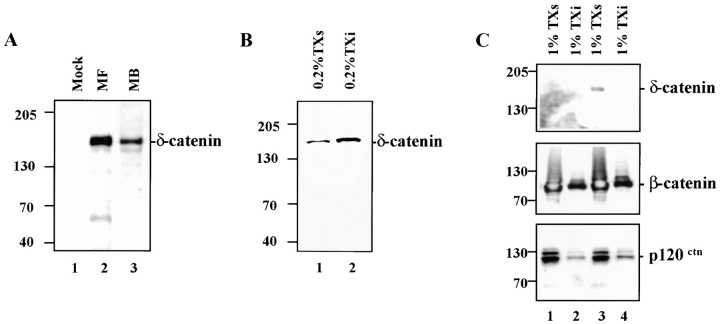
We raised two antibodies directed against different δ-catenin epitopes. Both specifically recognized a single band at 160 kD in MDCK cells transiently transfected with a full-length δ-catenin cDNA, but did not react with mock-transfected cells (Fig. 2 A). Because δ-catenin is specifically expressed in the nervous system (Zhou et al., 1997), postnatal day 2 mouse brain was also used for an immunoblot analysis. In this assay, δ-catenin migrated as a doublet at 160 kD (Fig. 2 A). The retarded mobility of δ-catenin relative to its predicted molecular weight may be due to the presence of charge groups, posttranslational modifications, or an extended structure which affects its electrophoretic migration.
Figure 2.
Expression of δ-catenin cDNA in transfected MDCK cells and endogenous δ-catenin in developing mouse brains. (A) Immunoblot showing δ-catenin expression in MDCK cells and developing mouse brains. (1) Mock-transfected MDCK cells; (2) MF (MDCK cells transfected with full-length δ-catenin cDNA); (3) MB (mouse brain lysate). (B) Immunoblot of transfected MDCK cells showing δ-catenin in the 0.2% Triton X-100 soluble (TXs, lane 1) and insoluble (Txi, lane 2) fractions. (C) Immunoblots of mock (1 and 2) and δ-catenin transfected (3 and 4) MDCK cells extracted in 1.0% Triton X-100. Soluble and insoluble fractions were blotted with the indicated antibodies. In all panels the molecular weight standard is indicated at the left.
The Localization of δ-catenin Suggests an Association with Adhesive Junctions
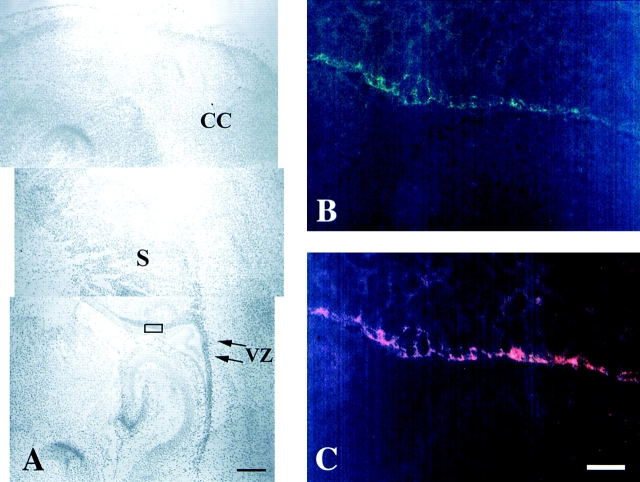
δ-catenin is preferentially expressed in brain, with the highest expression levels during brain development. We first sought to determine whether δ-catenin was localized to cell boundaries in the developing brain. Postnatal day 2 mouse brain sections were double labeled with rAb62 and mouse monoclonal anti–β-catenin (Fig. 3). We concentrated on the neuroepithelial precursor cell population in the cortical ventricular zone where δ-catenin staining was most intense and where junctional complexes are prominent. The staining was most intense at the apical end of these cells along the ventricular boundary. Consistent with the staining pattern, adherens junctions are known to be prominent along the lateral surface at the apical end of neuroepithelial cells (Hinds and Ruffett, 1971; Shoukimas and Hinds, 1978). Deeper in the ventricular zone, δ-catenin staining was less intense and outlined the cell periphery in a honeycomb pattern. Double labeled samples showed that δ-catenin colocalized with β-catenin at points of cell–cell contact (Fig. 3, B and C). Double labeling with anti–N-cadherin showed similar colocalization; anti–E-cadherin failed to demonstrate a neuroepithelial localization (data not shown). Although weaker than in the ventricular zone, neuronal staining was present throughout the nervous system as described in Paffenholz and Franke and Zhou et al. (1997).
Figure 3.
Double immunofluorescence microscopy showing the colocalization of δ-catenin with β-catenin in neocortical neuroepithelia of postnatal day 2 mouse brain. (A) Low magnification view in which the rectangle shows the location of the immunofluorescent images in B and C. CC, cerebral cortex. VZ, ventricular zone. S, striatum. Bar, 0.3 mm. (B) Frozen sagittal section stained by rAb62. (C) Same section immunostained by mouse monoclonal anti–β-catenin. Bar, 20 μm.
δ-catenin Forms Stable Complexes with Adhesive Junction Proteins
To characterize δ-catenin in a defined culture system where adhesive junctions are prominent, δ-catenin was transfected into MDCK epithelial cells and stable cell lines were established (see Materials and Methods). Anti–δ-catenin specifically labeled a single band at 160 kD (Fig. 2, A, lane 2, and B). The proportion of δ-catenin in the detergent-soluble pool increased when the Triton X-100 was increased from 0.2% to 1.0% (Fig. 2, B and C). Although δ-catenin was not soluble in cell lysates prepared in the absence of detergents, its solubility was nearly complete in 1.0% Triton X-100. In contrast, β-catenin was only partially solubilized under these conditions (Fig. 2 C). The extraction properties of δ-catenin resemble those of p120ctn which is also more soluble in 1.0% Triton X-100 than β-catenin as previously noted (Shibamoto et al., 1995), but nearly insoluble under aqueous conditions. Interestingly, the expression of δ-catenin did not drive more p120ctn either to the Triton soluble (Fig. 2 C) or aqueous compartments as quantified by densitometry (data not shown).
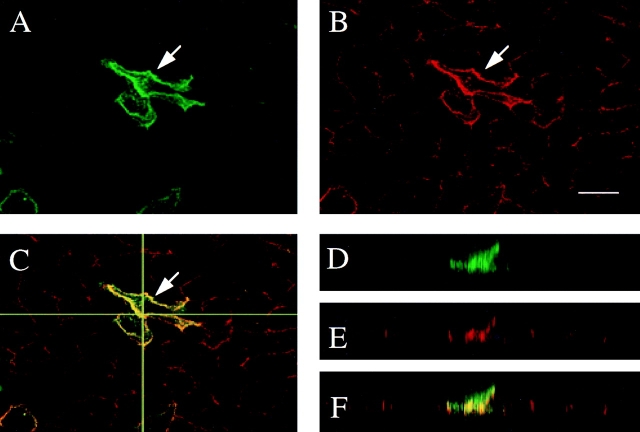
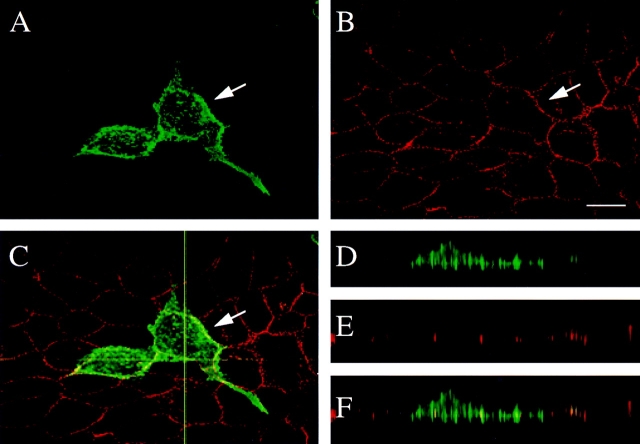
To study the subcellular distribution of δ-catenin, both transient and stably transfected MDCK cells were analyzed by immunofluorescent confocal light microscopy. rAb62 immunostaining showed that δ-catenin was localized to the periphery of transiently transfected cells in a pattern suggestive of cell–cell junctions (Figs. 4 A and 5 A). Although less intense than the peripheral staining, labeling of the cytoplasm was above background (Figs. 4 A and 5 A) suggesting that in transfected cells, a cytoplasmic pool of δ-catenin was also present. The pattern was reminiscent of the honeycomb pattern observed in the neuroepithelium. The immunofluorescent staining was specific because rAb62 did not stain cell–cell junctions in those cells in the untransfected dish. Double labeling immunofluorescence microscopy experiments showed that δ-catenin colocalized with E-cadherin (Fig. 4, A–C) and β-catenin (data not shown).
Figure 4.
Confocal immunofluorescence microscopy of MDCK cells transiently transfected with δ-catenin cDNA. The cells were double labeled with (A) δ-catenin antibodies and with (B) E-cadherin antibody. The arrow points to the transfected cell. (C) Merged fluorescent image showing colocalization of δ-catenin and E-cadherin. The horizontal line indicates where the XZ plane was selected for D–F. (D–F) Respective XZ vertical sections of A–C. Bar, 15 μm.
Because δ-catenin shares significant homology with p0071, a desmosomal protein (Hatzfeld and Nachtsheim, 1996), we compared the localization of δ-catenin with both desmoplakin and E-cadherin (Figs. 4 and 5). MDCK cells were transiently transfected with δ-catenin and double labeled with δ-catenin antibodies and either desmoplakin or E-cadherin antibodies. In transfected cells, δ-catenin codistributed with the adherens junction protein, E-cadherin in a thick band around the cell periphery, while neighboring untransfected cells retained a typical less intense junctional pattern of immunoreactivity (Fig. 4, A–C). Double labeling immunofluorescent microscopy showed that δ-catenin even colocalized with E-cadherin along cell processes (Fig. 4, A–C). Because δ-catenin–transfected cells are less flat than nontransfected MDCK cells, a z-series showed that δ-catenin and E-cadherin also codistributed in the vertical axis (Fig. 4, D–F). The greater intensity of E-cadherin staining in δ-catenin–transfected cells suggested that the concentration of E-cadherin in the transfected cells increased (Fig. 4, A and B). Densitometric measurement of protein profiles on immunoblots demonstrated a 30% increase in the E-cadherin level while p120ctn and desmoglein levels did not significantly change (data not shown). Double labeling immunofluorescence with δ-catenin and desmoplakin revealed a less close relationship (Fig. 5, A–F). In δ-catenin–transfected cells, the intensity of the desmoplakin immunoreactivity did not increase, and the pattern of desmoplakin labeling did not completely codistribute with δ-catenin. Desmoplakin labeling did not extend into cell processes (Fig. 5, A–C) and did not extend apically in the z-axis (Fig. 5, D–F). These findings do not exclude any colocalization between desmoplakin and δ-catenin, but they clearly point to a preferential cellular relationship to E-cadherin. δ-catenin also did not colocalize with the tight-junction associated protein, ZO-1 (data not shown). We therefore concluded that exogenous δ-catenin can be directed to the adherens junctions in MDCK cells.
Figure 5.
Confocal immunofluorescence microscopy of MDCK cells transiently transfected with δ-catenin. The cells were double labeled with (A) δ-catenin antibodies and with (B) desmoplakin antibody. The arrow points to the transfected cell. (C) Merged fluorescent image showing minimal colocalization of δ-catenin and desmoplakin. The horizontal line indicates where the XZ plane was selected for D–F. (D–F) Respective XZ vertical sections of A–C. Bar, 15 μm.
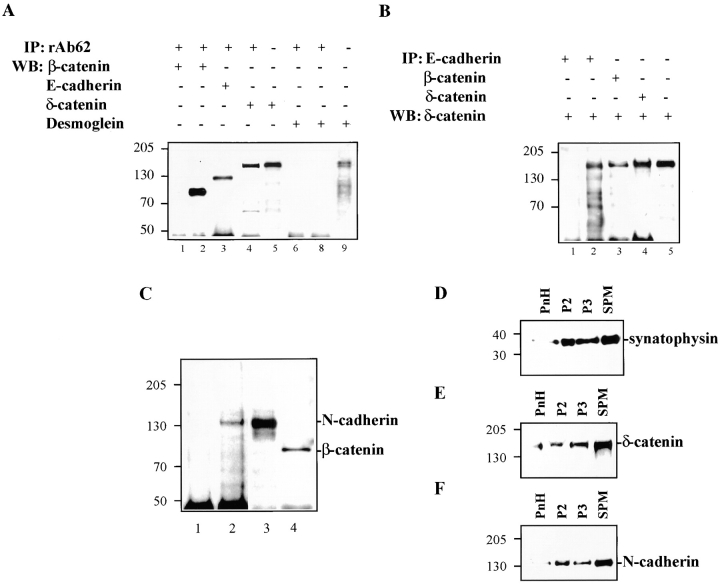
Based on colocalization data we asked whether δ-catenin interacts with the adherens junction proteins E-cadherin and β-catenin. In MDCK cells stably expressing δ-catenin, coimmunoprecipitation experiments indicated that δ-catenin coimmunoprecipitated both E-cadherin, β-catenin (Fig. 6 A), and p120ctn efficiently (not shown), but did not bring down desmoglein, which is abundantly present in MDCK cells and reacted with the desmoglein antibody (Fig. 6 A, lane 9). We concluded that at most, only minimal amounts of δ-catenin are associated with desmosomes. In reverse immunoprecipitation experiments, both β-catenin and E-cadherin coprecipitated δ-catenin (Fig. 6 B, lanes 2 and 3). The interaction between δ-catenin and E-cadherin/β-catenin was abolished in RIPA buffer that contained 0.2% SDS, although the coimmunoprecipitation of E-cadherin with β-catenin was retained (data not shown).
Figure 6.
Association of δ-catenin with adhesive junction proteins in brain and in MDCK cells stably expressing δ-catenin. (A) δ-catenin coimmunoprecipitates E-cadherin and β-catenin, but not desmoglein. (1 and 6) Mock-transfected MDCK cells. (2–5, 8, and 9) MF cells. (B) Reverse immunoprecipitation showing δ-catenin coprecipitated with E-cadherin and β-catenin. (1) Mock-transfected MDCK cells. (2–5) MF cells. (C) δ-catenin coimmunoprecipitates N-cadherin and β-catenin in brains. (1) N-cadherin immunoblot of brain fractions immunoprecipitated using nonimmune rabbit IgG. (2) N-cadherin immunoblot of brain fractions immunoprecipitated using rAb62. (3) N-cadherin immunoblot of brain lysate. (4) β-catenin immunoblot of brain fractions immunoprecipitated using rAb62. (D–F) Cofractionation of brain δ-catenin with N-cadherin and synaptophysin. PnH, postnuclear homogenates. P2, heavy membranes consisting of myelin, mitochondria, and crude synaptosomes. P3, mostly microsomes. SPM, synaptic plasma membranes. (D) Immunoblot showing brain fractionation profile of synaptophysin. (E) Immunoblot showing brain fractionation profile of δ-catenin. (F) Immunoblot showing brain fractionation profile of N-cadherin. Molecular weight markers are indicated at the left of each panel.
To determine whether endogenous δ-catenin associated with cadherins, postnatal day 2 mouse brain was used for similar coimmunoprecipitation experiments. Mouse brain lysates were immunoprecipitated with rAb62. Both N-cadherin and β-catenin were present in δ-catenin immunoprecipitants (Fig. 6 C, lanes 2 and 4). δ-catenin also cofractionated with N-cadherin when human brain tissue was homogenized and fractionated on a sucrose gradient to enrich for synaptosomes (Fig. 6, D–F). Therefore, δ-catenin is complexed with N-cadherin in a brain compartment that appears to be involved in cell–cell adhesion.
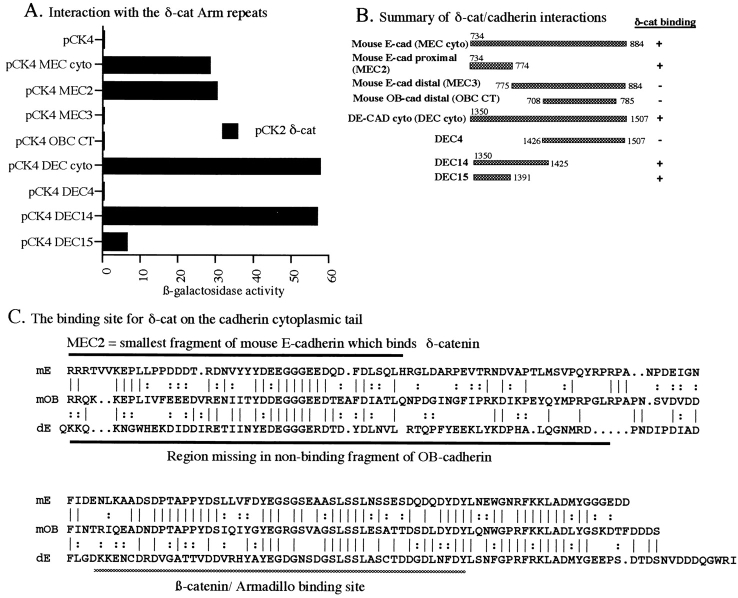
To determine whether the interaction between δ-catenin and the cytoplasmic domain of cadherin involves direct binding, their interaction was assayed in the yeast two-hybrid system. A construct was prepared containing the full Arm repeat region of δ-catenin, but lacking the NH2- and COOH-terminal domains. This construct specifically interacted with the full-length cytoplasmic tail of mouse E-cadherin (Fig. 7, A and B). Different classic cadherins in both mammals and flies carry two blocks of conserved sequence in their cytoplasmic tails (Fig. 7 C), a distal block which serves as the β-catenin/Armadillo binding site (Aberle et al., 1996; Pai et al., 1997) and a proximal region, which was recently discovered to bind p120ctn (Ozawa and Kemler, 1998; Yap et al., 1998). To begin to define the region required for δ-catenin binding, we tested two additional constructs, one containing the membrane-proximal region of mouse E-cadherin and the more distal region. δ-catenin specifically bound the membrane-proximal fragment, and failed to bind the more distal fragment. Also, δ-catenin did not bind to a truncated version of mouse OB-cadherin, which binds β-catenin (Tao et al., 1996), but lacks the 66 amino acids immediately following the transmembrane region that include the conserved proximal region (Fig. 7). We also tested whether δ-catenin could bind to Drosophila E-cadherin, to see whether the conserved sequences were sufficient for interaction. The full-length cytoplasmic tail of Drosophila E-cadherin binds δ-catenin, as do fragments containing the juxtamembrane region. The smallest interacting fragments of mouse E-cadherin and of Drosophila E-cadherin contain only the proximal 41 amino acids that include the highly conserved sequence YD(or E)D(or E)EGGGE (Fig. 7 C). This suggests that like p120ctn (Ozawa and Kemler, 1998; Yap et al., 1998), δ-catenin interacts with the conserved proximal region of the cadherin tail rather than the more distal site to which β-catenin and its fly homologue Armadillo bind (Aberle et al., 1996; Pai et al., 1997).
Figure 7.
δ-catenin interacts directly with the juxtamembrane region of cadherins in the yeast two-hybrid system. (A) Interaction of δ-catenin with various cadherin fragments. The Arm repeat region of human δ-catenin in the pCK2 “bait” vector was tested against pCK4 vector alone or with pCK4 fusions encoding the full-length murine E-cadherin cytoplasmic domain (pCK4 ME-CAD cyto), two complementary fragments of murine E-cadherin carrying either the membrane-proximal region (pCK4 MEC2) or the distal region with the β-catenin binding site (pCK4 MEC3), a COOH-terminal fragment of OB-cadherin not containing the juxtamembrane region (pCK4 OB-CAD CT), the entire cytoplasmic domain of Drosophila E-cadherin (pCK4 DEC), and smaller fragments of Drosophila E-cadherin as shown in B (pCK4 DEC 4, 14, and 15). (B) Schematic summary of the cadherin fragments and their interaction with δ-catenin. (C) Sequence alignment of the cytoplasmic tails of mouse E-cadherin, mouse OB-cadherin, and Drosophila DE-cadherin. Above the sequences are shown the smallest fragment of mouse E-cadherin which bound δ-catenin in the yeast two-hybrid assay, while below are diagrammed the amino acids missing from the OB-cadherin clone which does not bind to δ-catenin. The β-catenin/Armadillo binding site is also indicated.
The Arm Domain Is Necessary and Sufficient for Targeting δ-catenin to the Adherens Junction
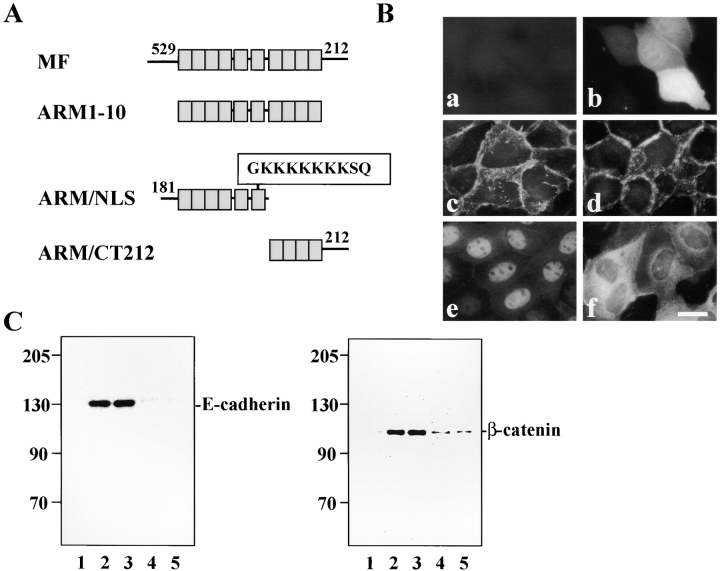
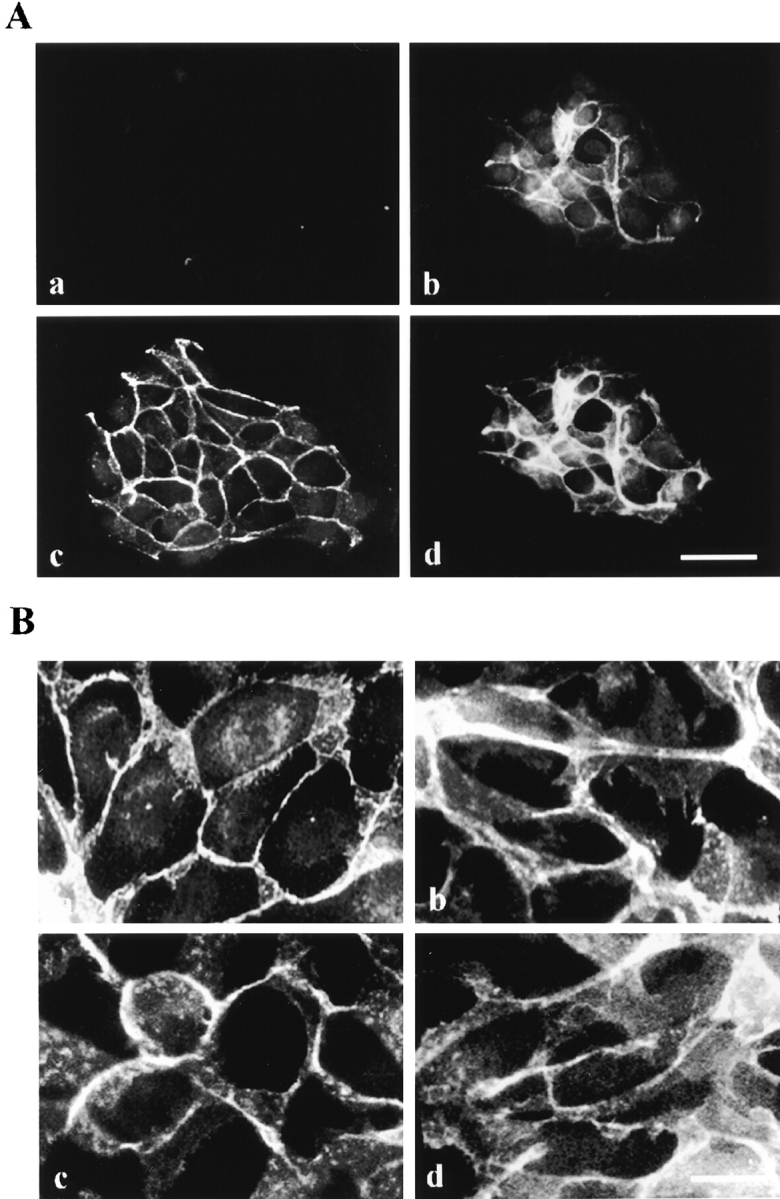
We generated a number of δ-catenin deletion mutants to define the domains necessary for targeting δ-catenin to the adherens junction (Fig. 8 A). To visualize these deletion mutants directly following transfection, full-length and various mutant constructs were fused with green fluorescent protein (GFP) and subcloned into the pEGFP vector. Full-length δ-catenin fused to EGFP showed a fluorescence distribution pattern that was identical to the antibody-labeling pattern of cells transfected with δ-catenin alone (Fig. 8 B, panel c). Although the primary site of δ-catenin immunoreactivity is the cell–cell junction, immunolabeling of cells also suggested a cytoplasmic pool. When both the NH2- and COOH-terminal extensions were deleted, leaving only the arm repeats, δ-catenin maintained its localization to cell–cell junctions based on detection of the GFP marker (Fig. 8 B, panel d) and on coimmunoprecipitation with E-cadherin and β-catenin (Fig. 8 C, lane 3). A significantly reduced interaction was observed for those constructs with truncations in the Arm repeat region (Fig. 8 C, lanes 4 and 5). Thus the armadillo repeat domain alone was sufficient for δ-catenin localization to cell–cell junctions as is the case for β-catenin which requires only a portion of the Arm domain to bind cadherin and localize to adherens junctions (Funayama et al., 1995; Orsulic and Peifer, 1996). These data are consistent with the two-hybrid analysis described above which also demonstrated that the δ-catenin Arm domain was sufficient for interacting directly with cadherin.
Figure 8.
Deletion analysis of δ-catenin localization and interaction with adhesive junction proteins. (A) Schematic drawings show the design of deletion mutants. MF, full-length δ-catenin. ARM1-10, δ-catenin sequence containing only the Arm repeats. ARM/NLS, δ-catenin containing partial NH2 terminus including six Arm repeats. Within Arm repeat 6, the boxed sequence indicates a putative nuclear localization signal. ARM/CT212: δ-catenin containing the complete COOH terminus but with only four Arm repeats from the COOH-terminal end. (B) Localization of mutant δ-catenin in MDCK cells. (a) Mock transfection. (b) GFP reporter. (c) Full-length δ-catenin. (d) Armadillo domain alone. (e) ARM/NLS. (f) ARM/CT212. Bar, 10 μm. (C) Coimmunoprecipitation of δ-catenin deletion mutants from transfected MDCK cells. Fractions were immunoprecipitated with GFP antibody and labeled with either E-cadherin or β-catenin antibody. (1) Mock-transfected cells. (2) MF. (3) ARM1-10. (4) ARM/ CT212. (5) ARM/NLS. Molecular weight markers are indicated at the left of each panel.
Two additional GFP fusion constructs, both of which interrupted the arm repeats, were transfected. One of these (ARM/CT212) contained four COOH-terminal arm repeats and the more COOH-terminal sequence (Fig. 8 A). This construct represents the fragment of δ-catenin originally cloned from the yeast two-hybrid system using the presenilin loop region as the bait (Zhou et al., 1997). This fragment, which can interact with presenilin, neither forms an immunocomplex with cadherin efficiently (Fig. 8 C, lane 4), nor demonstrates a primary localization to cell– cell junctions (Fig. 8 B, panel f). Instead, it appears to have a more diffuse cytoplasmic distribution similar to that in the endoplasmic reticulum observed for presenilin (Kovacs et al., 1996). A construct with the first six arm repeats plus the 181 residues NH2-terminal to the repeats (ARM/ NLS) localizes to nuclei (Fig. 8 B, panel e). This construct has a putative nuclear localization signal within the sixth repeat. However, we have not yet identified physiologic circumstances that drive full-length δ-catenin to the nucleus. Neither of these truncated proteins effectively coimmunoprecipitates with E-cadherin or β-catenin (Fig. 8 C, lanes 4 and 5).
δ-catenin Induces Morphological Alterations
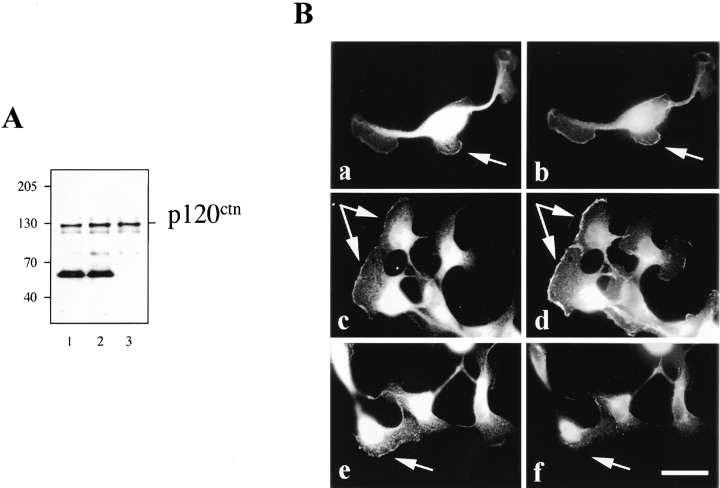
MDCK cells transfected with δ-catenin displayed an altered morphology. Transfected cells tended to lose their polygonal morphology and assumed either irregular shapes or an elongated fibroblastic appearance, sometimes with cell processes (Figs. 4 A and 5 A). Cells stably transfected with δ-catenin lost their organization as a regularly packed monolayer (compare Fig. 9 A, panels c and d). Immunolabeling of the δ-catenin–transfected cells showed that other major proteins of the adherens junction remained predominantly localized to cell–cell junctions (Fig. 9 A, c and d; B, a–d). Because δ-catenin and p120ctn both bind to the juxtamembrane region of cadherin, one might expect that their interaction with E-cadherin is competitive. However, in δ-catenin–expressing cells p120ctn retained its localization at the cell boundary (Reynolds et al., 1992; Fig. 9 B, c and d). Although a few cells showed increased cytoplasmic p120ctn staining after δ-catenin transfection, the differences from controls were insignificant, and there was no increase in the soluble pool of p120ctn (Fig. 2 C). Furthermore, after δ-catenin transfection there was no change in the amount of p120ctn coimmunoprecipitated with E-cadherin compared to mock-transfected cells (see Fig. 11 A, lanes 1 and 2).
Figure 9.
δ-catenin transfection altered MDCK cell morphology. (A) Double immunofluorescent labeling showing colocalization of δ-catenin with E-cadherin in MDCK cells stably expressing δ-catenin cDNA. (a and b) Anti–δ-catenin immunofluorescent microscopy. (c and d) Monoclonal anti–E-cadherin immunofluorescent microscopy. (a and c) Mock-transfected MDCK cells. (b and d) MDCK cells stably expressing δ-catenin cDNA. Note in b and d multilayers of MDCK cells can be observed while in a and c the monolayer is intact. Bar, 20 μm. (B) Double immunofluorescent microscopy showing the localization of adherens junction–associated proteins β-catenin and p120ctn in mock (a and c) and δ-catenin–transfected MDCK cells (b and d). (a and b) Anti– β-catenin. (c and d) Anti-p120ctn. Note p120ctn localization to cell–cell contact in mock- and δ-catenin–transfected MDCK cells. Bar, 5 μM.
Figure 11.
Ectopic expression of δ-catenin does not lead to displacement of p120ctn from adhesive junction proteins. (A) Immunoblot showing p120ctn coimmunoprecipitation with E-cadherin in mock (1), and MF cells (2). (3) MDCK cell lysate. (B) Double immunofluorescent microscopy showing redistribution of junctional proteins in δ-catenin–transfected MDCK cells after HGF stimulation. (a, c, and e) Anti–δ-catenin. (b) Anti– β-catenin. (d) Anti-p120ctn. (f) Anti-desmoplakin. Note the colocalization of δ-catenin with β-catenin and p120ctn, but not desmoplakin, at the leading edges of cells treated with HGF. Arrows point to the lamellipodia formation. Bar, 5 μm.
One explanation for the morphological changes was that δ-catenin altered MDCK cell proliferation. To test this hypothesis, we measured growth rates for control and δ-catenin–expressing cells. The growth rates of control and MDCK cells stably expressing δ-catenin did not significantly differ (Table I). To confirm this result BrdU incorporation into dividing cell nuclei was detected by a mAb specific for BrdU. Again, no significant difference was observed (Table II). Therefore, we concluded that the expression of δ-catenin does not significantly alter cell proliferation.
Table I.
Growth Rate Comparison between Control and MF Cells
| Cell type | No. of cells 104 | SEM | ||
|---|---|---|---|---|
| Control | 24.6 | ±2.26 | ||
| MF | 19.8 | ±3.47 |
Table II.
Nuclear Incorporation of BrdU in Control and MF Cells
| Cell type | Positive/total | SEM | ||
|---|---|---|---|---|
| Control | 37.5% | ±6.78 | ||
| MF | 36.6% | ±10.4 |
δ-catenin Primes Growth Factor–Induced Cell Spreading
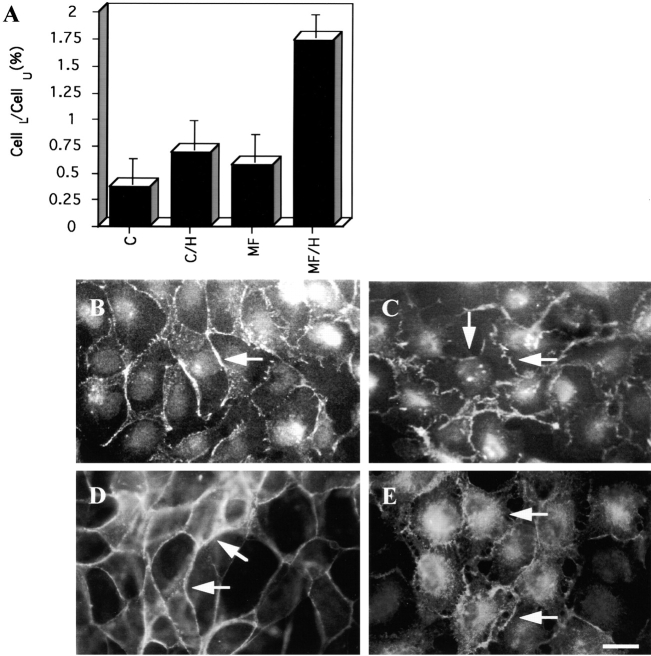
Another explanation for the altered cell–cell relationships in transfected cells was that δ-catenin conferred enhanced motility on the cells, perhaps by altering the composition of the wild-type junctions. To explore this possibility the transfected cells were treated with hepatocyte growth factor/scatter factor (HGF/SF) (Rosen et al., 1994; Balkovetz et al., 1997) in two cell dispersion assays. In one assay, 5,000 control or transfected MDCK cells were plated in the upper chambers of a two-chamber system overnight. The upper chambers were then transferred to another chamber in the presence or absence of HGF for an additional 2 d. HGF treatment induced the migration of control MDCK cells to the lower chamber. When those cells expressed δ-catenin, a significantly greater number of cells migrated to the lower chamber compared to controls (Fig. 10 A). In a second assay, the effect of HGF on the scattering of islands of MDCK cells was measured. In this dispersion assay, islands of δ-catenin–expressing cells and control cells were treated overnight with HGF and immunostained with rAb62. δ-catenin–expressing cells showed a greater degree of dispersion than control cells. Indeed, after HGF stimulation >78% of the δ-catenin–expressing cells completely detached from each other, while only 17% of the untransfected cells reached the same stage of dissociation. Thus, δ-catenin further stimulated HGF-induced cell scattering.
Figure 10.
Ectopic expression of δ-catenin promotes HGF-stimulated cell spreading and scattering. (A) Two-chamber system showing the enhanced migration of δ-catenin–expressing cells. CellL/CellU is the ratio of cells which migrated from the upper chamber to the lower chamber. C, control MDCK cells. C/H, control cells treated with HGF. MF, δ-catenin–transfected MDCK cells. MF/H, δ-catenin–transfected MDCK cells treated with HGF. (B and C) Monoclonal anti–E-cadherin immunofluorescence after overnight treatment with HGF. (B) Mock-transfected MDCK cells. (C) MDCK cells stably expressing δ-catenin. In B, cell–cell contact is still largely intact while in C cell–cell contact points appear disrupted (see arrows). (D and E) Anti–δ-catenin immunofluorescent microscopy showing the effect of HGF on δ-catenin distribution. (D) δ-catenin–expressing cells before HGF treatment. (E) δ-catenin–expressing cells after HGF treatment sometimes remain in clusters but show disruptions at points of cell–cell contact (see arrow) and show a redistribution of δ-catenin to the intracellular compartment. Bar, 5 μm.
To determine whether HGF-induced a redistribution of junctional proteins including δ-catenin, we treated mock-transfected and δ-catenin–transfected MDCK cells with HGF. When δ-catenin–transfected MDCK cells were stained with E-cadherin antibodies, the smooth honeycomb pattern of labeling in the periphery was lost, and the cell junctions appeared disrupted even among those cells which remained clustered (Fig. 10 C). Comparable groups of mock-transfected MDCK cells retained their junctional relationships as visualized by E-cadherin antibody labeling (Fig. 10 B). The disruption of the cell–cell junctions in this setting may be due to a reorganization of δ-catenin induced by HGF. To determine whether δ-catenin undergoes a shift in its localization following HGF treatment, we compared MDCK cells transfected with δ-catenin in the presence or absence of HGF (Fig. 10, D and E). Compared to untreated cells (Fig. 10 D), δ-catenin appeared to shift from a predominantly peripheral localization to a diffuse cytoplasmic distribution (Fig. 10 E). Clustered cells appeared to lose the integrity of their cell–cell contacts as judged by a less smooth appearance of the pool of δ-catenin which remained peripheral (Fig. 10 E). Other cells which completely dissociated from their neighbors underwent radical morphological changes that included the elaboration of processes and flattened lamellar veils (Fig. 11 B). In these dissociated cells δ-catenin remained colocalized with β-catenin (Fig. 11 B, a and b) and p120ctn (Fig. 11 B, c and d), even at the leading edge of the lamellae (see arrows). However, HGF made the dissociation from desmoplakin more apparent (Fig. 11 B, e and f).
Discussion
We have examined the cell biological and biochemical properties of a novel p120ctn family member, δ-catenin. Based on coimmunoprecipitations, immunolocalization, and expression studies, we show that there is a pool of δ-catenin associated with adherens junctions. In contrast to the related protein, p0071 (Hatzfeld and Nachtsheim, 1996), it appears that δ-catenin is not a component of the desmosome. Although it was previously claimed that δ-catenin (referred to as NPRAP, for neural plakophilin-related arm-repeat protein) did not label cell junctions, no data were presented on this point (Paffenholz and Franke, 1997; Zhou et al., 1997). Like p120ctn (Ozawa and Kemler, 1998; Yap et al., 1998), δ-catenin binds to a site within a 41–amino acid juxtamembrane region on cadherins. This region contains the sequence DEGGGE conserved among mouse E-cadherin, OB-cadherin, N-cadherin, Xenopus C-cadherin, and Drosophila E-cadherin. β-catenin binds to a more distal site at the extreme COOH terminus. δ-catenin can coimmunoprecipitate with β-catenin, as well as with cadherins, suggesting that all are part of the same higher order complex, but it is unknown whether a single cadherin molecule can simultaneously bind β-catenin and a p120ctn family member such as δ-catenin.
Components of cell–cell junctions may also serve as determinants of cell shape and contribute to motile behavior (Reynolds et al., 1996; Barth et al., 1997b). p120ctn, originally defined as a tyrosine kinase substrate phosphorylated both by the activated form of Src kinase and rapidly phosphorylated in response to ligand-induced signaling (reviewed in Daniel and Reynolds, 1997), also associates with classical cadherins (Reynolds et al., 1992, 1994; Shibamoto et al., 1995; Staddon et al., 1995). Because p120ctn is a prominent target of both nonreceptor and receptor tyrosine kinases (Daniel and Reynolds, 1997), it has been suggested that p120ctn may mediate some of the effects of oncogenic tyrosine kinases on cell behavior during transformation and growth factor receptors on cell morphology. Overexpression of p120ctn can alter the morphology of some cells (Reynolds et al., 1996; Barth et al., 1997a). Based upon consensus motifs, δ-catenin is also predicted to be a tyrosine kinase substrate and δ-catenin can alter cell morphology when coupled to a trophic stimulus. In our studies, the expression of δ-catenin also significantly enhanced the scattering response to the receptor tyrosine kinase ligand HGF. Thus δ-catenin appears to prime cells for morphogenic and motogenic behaviors. This observation suggests that an active form of δ-catenin can be induced in cells and this active form may be a phosphorylated isoform.
The effects of δ-catenin on cell morphology and motility support this view. The cytoplasmic domain of cadherin may regulate morphogenic events by selective binding at the distal site to β-catenin and at the juxtamembrane site to δ-catenin or p120ctn. The effects of each of these proteins when bound to cadherin, either alone or in combination, need to be systematically studied because a shift in their binding affinities can distort the phenotype. For example, an NH2-terminal deletion of β-catenin in MDCK cells results in a more fibroblastic appearance that is similar to the changes we observed when δ-catenin was expressed in MDCK cells (Barth et al., 1997b; Pollack et al., 1997). This mutant β-catenin is more stable in E-cadherin and adenomatous polyposis coli (APC) complexes. The regulation of cadherin cytoplasmic domain binding probably contributes to the requirement for a migrating cell to become motile while retaining dynamic adhesive interactions with other cells and with the substrate. For example, the ras-induced tyrosine phosphorylation state of MCF-10A breast epithelial cells is thought to determine whether β-catenin or p120ctn bind to cadherin (Kinch et al., 1995). In this model, ras transformation increased tyrosine phosphorylation in the cells, bound p120ctn to cadherin, and induced the loss of adherens junctions and the formation of focal adhesions. Further supporting a reciprocal balance between the expression of adherens junction proteins and proteins in focal adhesions is the increase in the β-catenin and cadherin concentrations of pheochromocytoma cells after suppression of the extracellular signal regulated kinases, which are downstream of ras (Lu et al., 1998).
In the case of E-cadherin, the juxtamembrane region negatively regulates adhesion by preventing lateral dimerization of the extracellular domain (Ozawa and Kemler, 1998). Therefore, molecules which bind to this site such as δ-catenin are candidates for negatively regulating cell–cell adhesion. The effects of δ-catenin on cell scattering meet the functional predictions for a molecule with this role in that cells detach and disperse. This response required HGF and therefore, δ-catenin activation depends on upstream signaling elements. p120ctn is also a candidate for a negative regulator of cell adhesion; however, there are some contradictory findings in the literature on this point. p120ctn binds to the juxtamembrane region of C-cadherin, but in the experiments with C-cadherin, p120ctn was thought to increase adhesive strength (Yap et al., 1998). On the other hand, p120ctn may not bind specifically to the juxtamembrane region, but to several sites on the E-cadherin COOH terminus (Ozawa and Kemler, 1998).
Because δ-catenin and p120ctn both bind to the juxtamembrane region, it is possible that the phenotypic changes we have observed are due to displacement of p120ctn from cadherin by δ-catenin. We believe this is unlikely for the following reasons: (a) after δ-catenin transfection we did not detect increased soluble p120ctn (Fig. 2 C); (b) after δ-catenin transfection immunoprecipitable p120ctn did not change (Fig. 11 A); and (c) after δ-catenin transfection the immunolocalization of p120ctn did not change (Fig. 9 B, panels c and d). What may explain how the two molecules bind to a nearly identical site, and yet not appear to displace each other? The most interesting explanation is that the expression of δ-catenin upregulates the expression of cadherin making more binding sites available. The evidence for this is apparent in Fig. 4 where the δ-catenin–transfected cells have increased cadherin-labeling relative to the nontransfected cells. Another possibility assumes that binding to sites in the juxtamembrane region is more difficult when cadherin is dimerized and δ-catenin interferes with dimer formation, thus increasing the number of binding sites. Finally, p120ctn may not bind exclusively to the juxtamembrane region (Ozawa and Kemler, 1998) and therefore, it may not directly compete for binding with δ-catenin.
Even if δ-catenin did displace p120ctn, the MDCK cell may not be ideal for observing phenotypic changes due to p120ctn overexpression. The phenotypic effects of p120ctn are most apparent in cells of mesenchymal origin (Kinch et al., 1995; Reynolds et al., 1996; Barth et al., 1997a,b), whereas the effects of δ-catenin are most apparent in epithelial cells. In contrast to our observations in MDCK cells, transfections of δ-catenin into fibroblasts showed only minimal effects on morphology (data not shown). Members of the Arm subfamily which bind to the juxtamembrane region of cadherin may represent a site where a cell implements its own specific morphological determinants. δ-catenin shows neuronal specificity, p120ctn is expressed more generally, and other members of this class with their own morphological determinants may emerge. Predicted from p120ctn and δ-catenin is the source of diversity which may arise from differences in the sequences that lie on either side of the Arm repeats. One function of the adherens junction is to link the cell surface with the actin cytoskeleton. β-catenin can serve this purpose by binding α-catenin which in turn interacts with actin either directly or indirectly (Jou et al., 1995; Nieset et al., 1997). The relationship of the p120ctn subfamily proteins to actin is more problematic because p120ctn does not bind to α-catenin (Daniel and Reynolds, 1995). δ-catenin, however, contains a poly-l-proline stretch (Fig. 1), not present in p120ctn, which may represent a profilin-binding site and a link to actin filaments. Thus, the downstream links of p120ctn and δ-catenin may differ greatly.
A novel interaction of both β-catenin and δ-catenin is with presenilin 1 (Zhou et al., 1997; Yu et al., 1998), which is encoded by the gene most commonly mutated in Alzheimer's disease (Clark et al., 1995; Sherrington et al., 1995), and is localized to the endoplasmic reticulum (Kovacs et al., 1996). The significance of this interaction in brain development or in Alzheimer's disease is unknown. Although full-length δ-catenin does not primarily reside in association with the endoplasmic reticulum where presenilin is resident, a potential site of residence may be revealed by the expression of a truncated protein. For example, when NH2-terminal deletions of β-catenin were expressed in MDCK cells the protein colocalized with APC, whereas full-length β-catenin failed to show this colocalization (Barth et al., 1997b). In the case of δ-catenin, NH2-terminal deletions revealed a potential for codistribution with the endoplasmic reticulum (Fig. 8 B, f). Just as the β-catenin binding partners, which include cadherins, fascin (Tao et al., 1996), APC (Rubinfeld et al., 1996), and certain tyrosine kinase receptors (Hoschuetzky et al., 1994; Shibata et al., 1996) suggest a versatility of function, so δ-catenin may engage in a variety of interactions through its rich content of functional domains.
To maintain tissue integrity, cells utilize cell–cell and cell–matrix junctions to create a highly ordered space-filling array. During development and in response to environmental stimuli, cells can alter their interactions with other cells and the substrate by detaching from their neighbors and migrating to other sites. In so doing adhesive junctions can accommodate both stable cell–cell interactions within established epithelia while accommodating cell rearrangement and cell migration. Ultimately, changes in either the composition of the proteins that make up the adherens junction or their posttranslational modifications are likely to play a pivotal role. δ-catenin is a neuronal specific protein expressed at high levels in the ventricular zone (Fig. 3), a neuroepithelial cell population destined to migrate tangentially and form the mature brain. A cautionary note is that δ-catenin is a neuronal cell protein and there may be some functional differences in the epithelial cells used here. Within δ-catenin is a consensus sequence for Abl phosphorylation, which functions as an in vitro substrate for Bcr/Abl (Lu, Q., and K.S. Kosik, unpublished observations). Among the genes associated with migration is the mouse disabled-1 gene (Howell et al., 1997b; Sheldon et al., 1997; Ware et al., 1997) which can associate with the nonreceptor tyrosine kinases, Src, Abl, and Fyn (Howell et al., 1997a). Given the localization of δ-catenin, its effects on cell scattering, and its putative relationship to nonreceptor tyrosine kinases, we hypothesize that δ-catenin functions as a regulator of neuronal migration.
Acknowledgments
We thank T.W. Kim for brain samples, Y.H. Chen and B. Keon for antibodies against Occludin, ZO-1, and desmoplakin, R. Paffenholz and W. Franke for a mouse δ-catenin cDNA and antibody against desmoplakin, K. Knudsen for antibodies against human N-cadherin, P. McCrea for the OB-cadherin cDNA, Mei Lu for help with image processing, and Kosik lab members for stimulating discussions.
This work was supported by National Institutes of Health grants AG06601 to K.S. Kosik and GM47857 to M. Peifer. R. Cavallo is supported by a United States Army Breast Cancer Research Program Pre-doctoral fellowship.
Abbreviations used in this paper
- APC
adenomatous polyposis coli
- BrdU
5-bromo-2′-deoxy-uridine
- GFP
green fluorescent protein
- HGF
hepatocyte growth factor
- IP
immunoprecipitation
- MF
δ-catenin–transfected MDCK
References
- Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- Aberle H, Schwartz H, Kemler R. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61:514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Balkovetz DF, Pollack AL, Mostov KE. Hepatocyte growth factor alters the polarity of Madin-Darby canine kidney cell monolayers. J Biol Chem. 1997;272:3471–3477. doi: 10.1074/jbc.272.6.3471. [DOI] [PubMed] [Google Scholar]
- Barth AI, Nathke IS, Nelson WJ. Cadherins, catenins and APC protein: interplay between cytoskeletal complexes and signaling pathways. Curr Opin Cell Biol. 1997a;9:683–690. doi: 10.1016/s0955-0674(97)80122-6. [DOI] [PubMed] [Google Scholar]
- Barth AI, Pollack AL, Altschuler Y, Mostov KE, Nelson WJ. NH2-terminal deletion of beta-catenin results in stable colocalization of mutant beta-catenin with adenomatous polyposis coli protein and altered MDCK cell adhesion. J Cell Biol. 1997b;136:693–706. doi: 10.1083/jcb.136.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher WM, Yap AS, Gumbiner BM. Lateral dimerization is required for the homophilic binding activity of C-cadherin. J Cell Biol. 1996;135:487–496. doi: 10.1083/jcb.135.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RF, Hutton M, Fuldner RA, Froelich S, Karran E, Talbot C, Crook R, Lendon C, Prihar G, He C, et al. The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nat Genet. 1995;11:219–222. doi: 10.1038/ng1095-219. [DOI] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. The tyrosine kinase substrate p120cas binds directly to E-cadherin but not to the adenomatous polyposis coli protein or alpha-catenin. Mol Cell Biol. 1995;15:4819–4824. doi: 10.1128/mcb.15.9.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. Tyrosine phosphorylation and cadherin/ catenin function. Bioessays. 1997;19:883–891. doi: 10.1002/bies.950191008. [DOI] [PubMed] [Google Scholar]
- Funayama N, Fagotto F, McCrea P, Gumbiner BM. Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M, Nachtsheim C. Cloning and characterization of a new armadillo family member, P0071, associated with the junctional plaque-evidence for a subfamily of closely related proteins. J Cell Sci. 1996;109:2767–2778. doi: 10.1242/jcs.109.11.2767. [DOI] [PubMed] [Google Scholar]
- Heid HW, Schmidt A, Zimbelmann R, Schafer S, Winter-Simanowski S, Stumpp S, Keith M, Figge U, Schnolzer M, Franke WW. Cell type-specific desmosomal plaque proteins of the plakoglobin family: plakophilin 1 (band 6 protein) Differentiation. 1994;58:113–131. doi: 10.1046/j.1432-0436.1995.5820113.x. [DOI] [PubMed] [Google Scholar]
- Hinds JW, Ruffett TL. Cell proliferation in the neural tube: an electron microscopic and Golgi analysis in the mouse cerebral vesicle. Z Zellforsch Mikrosk Anat. 1971;115:226–264. doi: 10.1007/BF00391127. [DOI] [PubMed] [Google Scholar]
- Hoschuetzky H, Aberle H, Kemler R. Beta-catenin mediates the interaction of the cadherin-catenin complex with epidermal growth factor receptor. J Cell Biol. 1994;127:1375–1380. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BW, Gertler FB, Cooper JA. Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO (Eur Mol Biol Organ) J. 1997a;16:121–132. doi: 10.1093/emboj/16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997b;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- Jou T, Steward D, Stappert J, Nelson W, Marrs J. Genetic and biochemical dissection of protein linkages in the cadherin-catenin complex. Proc Natl Acad Sci USA. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kapprell HP, Owaribe K, Franke WW. Identification of a basic protein of M r75,000 as an accessory desmosomal plaque protein in stratified and complex epithelia. J Cell Biol. 1988;106:1679–1691. doi: 10.1083/jcb.106.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinch MS, Clark GH, Der CJ, Burridge K. Tyrosine phosphorylation regulates the adhesions of ras-transformed breast epithelia. J Cell Biol. 1995;130:461–471. doi: 10.1083/jcb.130.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of alpha-actinin with the cadherin/catenin cell–cell adhesion complex via alpha-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs DM, Fausett HJ, Page KJ, Kim T-W, Moir RD, Merriam DE, Hollister RD, Hallmark OG, Mancini R, Felsensetin KM, et al. Alzheimer-associated presenilins 1 and 2: neuronal expression in brain and localization to intracellular membranes in mammalian cells. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Lu Q, Paredes M, Zhang J, Kosik K. Basal extracellular signal-regulated kinase activity modulates cell–cell and cell-matrix interactions. Mol Cell Biol. 1998;18:3257–3265. doi: 10.1128/mcb.18.6.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs JA, Nelson WJ. Cadherin cell adhesion molecules in differentiation and embryogenesis. Int Rev Cytol. 1996;165:159–205. doi: 10.1016/s0074-7696(08)62222-6. [DOI] [PubMed] [Google Scholar]
- Mathur M, Goodwin L, Cowin P. Interactions of the cytoplasmic domain of the desmosomal cadherin Dsg1 with plakoglobin. J Biol Chem. 1994;269:14075–14080. [PubMed] [Google Scholar]
- Mertens C, Kuhn C, Franke WW. Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J Cell Biol. 1996;135:1009–1025. doi: 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A, Takeichi M. Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO (Eur Mol Biol Organ) J. 1988;7:3679–3684. doi: 10.1002/j.1460-2075.1988.tb03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieset JE, Redfield AR, Jin F, Knudsen KA, Johnson KR, Wheelock MJ. Characterization of the interactions of alpha-catenin with alpha-actinin and beta-catenin/plakoglobin. J Cell Sci. 1997;110:1013–1022. doi: 10.1242/jcs.110.8.1013. [DOI] [PubMed] [Google Scholar]
- Orsulic S, Peifer M. An in vivo structure-function study of armadillo, the beta-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Kemler R. The membrane-proximal region of the E-cadherin cytoplasmic domain prevents dimerization and negatively regulates adhesion activity. J Cell Biol. 1998;142:1605–1613. doi: 10.1083/jcb.142.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO (Eur Mol Biol Organ) J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffenholz R, Franke WW. Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation. 1997;61:293–304. doi: 10.1046/j.1432-0436.1997.6150293.x. [DOI] [PubMed] [Google Scholar]
- Pai LM, Kirkpatrick C, Blanton J, Oda H, Takeichi M, Peifer M. Drosophila alpha-catenin and E-cadherin bind to distinct regions of Drosophila Armadillo. J Biol Chem. 1996;271:32411–32420. doi: 10.1074/jbc.271.50.32411. [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development (Camb) 1997;124:2255–2266. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- Pollack AL, Barth AIM, Altschuler Y, Nelson WJ, Mostov KE. Dynamics of beta-catenin interactions with APC protein regulate epithelial tubulogenesis. J Cell Biol. 1997;137:1651–1662. doi: 10.1083/jcb.137.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Herbert L, Cleveland JL, Berg ST, Gaut JR. p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors β-catenin, plakoglobin and armadillo. . Oncogene. 1992;7:2439–2445. [PubMed] [Google Scholar]
- Reynolds AB, Daniel J, McCrea PD, Wheelock MJ, Wu J, Zhang Z. Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol Cell Biol. 1994;14:8333–8342. doi: 10.1128/mcb.14.12.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Daniel JM, Mo YY, Wu J, Zhang Z. The novel catenin p120cas binds classical cadherins and induces an unusual morphological phenotype in NIH3T3 fibroblasts. Exp Cell Res. 1996;225:328–337. doi: 10.1006/excr.1996.0183. [DOI] [PubMed] [Google Scholar]
- Riggleman B, Wieschaus E, Schedl P. Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev. 1989;3:96–113. doi: 10.1101/gad.3.1.96. [DOI] [PubMed] [Google Scholar]
- Rimm DL, Koslov ER, Kebraiaei P, Cianci CD, Morrow JS. Alpha 1 (E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen EM, Nigam SK, Goldberg ID. Scatter factor and the c-met receptor: a paradigm for mesenchymal/epithelial interaction. J Cell Biol. 1994;127:1783–1787. doi: 10.1083/jcb.127.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3 beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- Sheldon M, Rice DS, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Cloning of a novel gene bearing missense mutations in early onset familial Alzheimer disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Shibamoto S, Hayakawa M, Takeuchi K, Hori T, Miyazawa K, Kitamura N, Johnson KR, Wheelock MJ, Matsuyoshi N, Takeichi M, et al. Association of p120, a tyrosine kinase substrate, with E-cadherin/catenin complexes. J Cell Biol. 1995;128:949–957. doi: 10.1083/jcb.128.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Ochiai A, Kanai Y, Akimoto S, Gotoh M, Yasui N, Machinami R, Hirohashi S. Dominant negative inhibition of the association between beta-catenin and c-erbB-2 by N-terminally deleted beta-catenin suppresses the invasion and metastasis of cancer cells. Oncogene. 1996;13:883–889. [PubMed] [Google Scholar]
- Shoukimas GM, Hinds JW. The development of the cerebral cortex in the embryonic mouse: an electron microscopic serial section analysis. J Comp Neurol. 1978;179:795–830. doi: 10.1002/cne.901790407. [DOI] [PubMed] [Google Scholar]
- Sirotkin H, O'Donnell H, DasGupta R, Halford S, St. Jore B, Puech A, Parimoo S, Morrow B, Skoultchi A, Weissman SM, et al. Identification of a new human catenin gene family member (ARVCF) from the region deleted in velo-cardio-facial syndrome. Genomics. 1997;41:75–83. doi: 10.1006/geno.1997.4627. [DOI] [PubMed] [Google Scholar]
- Staddon JM, Smales C, Schulze C, Esch F, Rubin LL. p120, a p120-related protein (p100), and the cadherin/catenin complex. J Cell Biol. 1995;130:369–381. doi: 10.1083/jcb.130.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- Tao YS, Edwards RA, Tubb B, Wang S, Bryan J, McCrea PD. Beta-catenin associates with the actin-bundling protein fascin in a noncadherin complex. J Cell Biol. 1996;134:1271–1281. doi: 10.1083/jcb.134.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T. The cadherin superfamily at the synapse: more members, more missions. Cell. 1998;93:1095–1098. doi: 10.1016/s0092-8674(00)81452-x. [DOI] [PubMed] [Google Scholar]
- Ware ML, Fox JW, Gonzalez JL, Davis NM, Lambert de Rouvroit C, Russo CJ, Chua SC, Jr, Goffinet AM, Walsh CA. Aberrant splicing of a mouse disabled homolog, mdab1, in the scrambler mouse. Neuron. 1997;19:239–249. doi: 10.1016/s0896-6273(00)80936-8. [DOI] [PubMed] [Google Scholar]
- Xia Y, Rohan de Silva HA, Rosi BL, Yamaoka LH, Rimmler JB, Pericak-Vance MA, Roses AD, Chen X, Masliah E, DeTeresa R, et al. Genetic studies in Alzheimer's disease with an NACP/α-synuclein polymorphism. Ann Neurol. 1996;40:50–58. doi: 10.1002/ana.410400212. [DOI] [PubMed] [Google Scholar]
- Yap AS, Brieher WM, Gumbiner BM. Molecular and functional analysis of cadherin-based adherens junctions. Annu Rev Cell Dev Biol. 1997;13:119–146. doi: 10.1146/annurev.cellbio.13.1.119. [DOI] [PubMed] [Google Scholar]
- Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120. J Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Chen F, Levesque G, Nishimura M, Zhang DM, Levesque L, Rogaeva E, Xu D, Liang Y, Duthie M, et al. The presenilin 1 protein is a component of a high molecular weight intracellular complex that contains beta-catenin. J Biol Chem. 1998;273:16470–16475. doi: 10.1074/jbc.273.26.16470. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liyanage U, Medina M, Ho C, Simmons AD, Lovett M, Kosik KS. Presenilin 1 interacts in brain with a novel member of the Armadillo family. Neuroreport. 1997;8:1489–1494. doi: 10.1097/00001756-199704140-00033. [DOI] [PubMed] [Google Scholar]
- Zhou S, Carraway KL, III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]