Abstract
To identify components involved in the nuclear export of ribosomes in yeast, we developed an in vivo assay exploiting a green fluorescent protein (GFP)-tagged version of ribosomal protein L25. After its import into the nucleolus, L25-GFP assembles with 60S ribosomal subunits that are subsequently exported into the cytoplasm. In wild-type cells, GFP-labeled ribosomes are only detected by fluorescence in the cytoplasm. However, thermosensitive rna1-1 (Ran-GAP), prp20-1 (Ran-GEF), and nucleoporin nup49 and nsp1 mutants are impaired in ribosomal export as revealed by nuclear accumulation of L25-GFP. Furthermore, overexpression of dominant-negative RanGTP (Gsp1-G21V) and the tRNA exportin Los1p inhibits ribosomal export. The pattern of subnuclear accumulation of L25-GFP observed in different mutants is not identical, suggesting that transport can be blocked at different steps. Thus, nuclear export of ribosomes requires the nuclear/cytoplasmic Ran-cycle and distinct nucleoporins. This assay can be used to identify soluble transport factors required for nuclear exit of ribosomes.
Keywords: nuclear pore complex, ribosomal export, L25, GFP, nucleocytoplasmic transport
The nucleolus is the organelle in which ribosomal subunits are assembled from ribosomal RNA (rRNA)1 and ribosomal proteins (for review see Scheer and Weisenberger, 1994; Shaw and Jordan, 1995). In all eukaryotic cells, a large ribosomal precursor RNA (pre-rRNA) transcript is processed and modified to yield mature 18S, 5.8S, and 25S/28S rRNAs (Tollervey, 1996). During the steps of transcription, processing, and modification of rRNA, the ∼80 ribosomal proteins assemble inside the nucleolus with the different rRNA species including 5S rRNA to form preribosomal particles (Woolford, 1991). In the past, the analysis of yeast mutants defective in ribosome biogenesis yielded insight into the mechanism of rRNA transcription, processing, modification, and ribosomal assembly (for review see Tollervey, 1996). However, very little is known on how ribosomes are exported from the nucleolus into the cytoplasm.
Ribosomes are transported like any other nuclear export and import substrate through the nuclear pore complexes (NPC). It is assumed that the two ribosomal subunits are exported separately from the nucleus into the cytoplasm (Warner, 1990). The dimensions of ribosomal subunits are in the range of 20 nm, and, accordingly, they have to be actively transported through the central gated channel of the NPC which limits the functional size of passive diffusion to 9 nm (Dingwall and Laskey, 1986). It is known that the nuclear pore diameter can expand to 26 nm under transport conditions that would allow a ribosomal subunit to pass through the pore channel without unfolding (Feldherr and Akin, 1990). Larger RNP particles such as Balbiani ring RNPs pass through the NPCs in a partly unfolded configuration with a leading 5′ end of the mRNA (Skoglund et al., 1983).
Microinjection of radiolabeled eukaryotic ribosomes or ribosomal subunits into Xenopus oocytes and measurement of export kinetics has shown that nuclear export of ribosomes is an unidirectional and energy-dependent process (Khanna-Gupta and Ware, 1989). Coinjection of the two ribosomal subunits has been also shown to increase significantly the speed of translocation as compared with individually injected subunits. Interestingly, microinjected E. coli ribosomal subunits could not be exported from the nucleus, indicating that prokaryotic ribosomes lack a nuclear export signal (Khanna-Gupta and Ware, 1989). A kinetic competition assay has demonstrated that export kinetics between the 40S ribosomal subunit and tRNA are cooperative (Pokrywka and Goldfarb, 1995). Therefore, it has been suggested that nuclear retention of rRNA (as well as of other RNA classes) has to be overcome first in order for the rRNA to enter the export pathway. An in vitro assay for ribosomal export has recently been reported (Hassel et al., 1996), but it is not clear whether it reliably measures the in vivo ribosomal transport process.
Different transport routes through the NPC have been identified, and different classes of transport cargoes enter or leave the nucleus together with their specific transport factors (for review see Nigg, 1997). All the specific import and export receptors identified so far belong to the family of importin/karyopherin β–like transport factors (Görlich, 1998). The best-studied transport route is the nuclear uptake of cargo carrying a classical NLS (cNLS; Adam et al., 1989). This cNLS is recognized by the importin α and β complex (Görlich et al., 1994; Imamoto et al., 1995; Radu et al., 1995). Recently, importins or karyopherins for other import (karyophilic) cargoes, such as shuttling hnRNP proteins (Aitchison et al., 1996; Pollard et al., 1996; Pemberton et al., 1997; Senger et al., 1998), ribosomal proteins (Rout et al., 1997) and proteins involved in tRNA maturation pathways (Rosenblum et al., 1997) have been identified.
Although much less is known about nuclear export routes, it has recently become clear that a certain type of nuclear export signal (the prototype is the leucine-rich NES within HIV Rev protein), which occurs within exported or shuttling proteins, interacts with Crm1p/Xpo1p (the NES receptor), a member of the family of importin β–like transport factors (Fornerod et al., 1997; Fukuda et al., 1997; Kudo et al., 1997; Neville et al., 1997; Ossareh-Nazari et al., 1997; Stade et al., 1997). Several RNA-binding proteins that shuttle between nucleus and cytoplasm could also be shown to carry a nuclear export sequence. Accordingly, these NES-containing proteins may mediate the nuclear export of the RNA they associate with. This has indeed been shown to be the case for the HIV Rev protein that transports unspliced or partially spliced viral mRNA into the cytoplasm (Bogerd et al., 1995; Fischer et al., 1995; Stutz et al., 1995). Finally, the hnRNP A1 protein, which is thought to be involved in export of cellular mRNA, contains a different type of NES called M9, which can also function as a NLS (Michael et al., 1995).
Microinjection experiments have shown that the nuclear export of different classes of RNA (mRNA, snRNA, rRNA, tRNA) is mediated by different factors (Jarmolowski et al., 1994). One of these factors has recently been identified to be a member of the importin β protein family that functions as a tRNA exportin in both vertebrate cells (Arts et al., 1998; Kutay et al., 1998) and yeast (Los1p; Simos et al., 1996b; Hellmuth et al., 1998). Concerning nuclear export of rRNA, antibodies against the vertebrate nucleoporin Nup98, when microinjected into Xenopus nuclei, have been shown to cause a reduction in the export of rRNA besides the export defects for the other RNA species. Furthermore, in a thermosensitive RCC1 (Ran-GEF) mutant, rRNA export appeared to be also inhibited (Cheng et al., 1995). However, this mutant was also affected in the processing of the 45S rRNA precursor. Interestingly, the rRNA processing pathway is also affected in yeast mutants mapping in components of the Ran cycle such as Prp20p (Ran-GEF) or Rna1p (Ran-GAP) (Kadowaki et al., 1993). There is indeed experimental evidence that biogenesis (processing and modification) of the different classes of RNA is somehow coupled to transport. Examples include nuclear retention of unspliced mRNA (Legrain and Rosbash, 1989), splicing of tRNA by enzymes that are localized in close proximity to the NPCs (Clark and Abelson, 1987), coupling between tRNA pseudouridinylation and export (Simos et al., 1996b) and cap-methylation of snRNA as an important export signal (Hamm and Mattaj, 1990), whereas cap-hypermethylation of snRNP in the cytoplasm functions as a signal for nuclear import (Fischer and Lührmann, 1990).
Despite this increasing knowledge on transport routes through the NPC, very little is known about components involved in ribosomal export. It is clear that rRNA must first assemble into preribosomal particles before they can be exported from the nucleus (Trapman et al., 1975; Warner, 1990). It is possible that Ran somehow triggers for ribosomal export, ribosomal transport within the nucleolus, or release from intranucleolar retention sites. Particles seen in transit through the NPC were initially suspected to be ribosomes, but they were later thought to be a permanent structure of the NPC (Unwin and Milligan, 1982). Recently, EM revealed that ribosomes align on tracks before they are transported through the NPCs (Léger-Silvestre et al., 1997).
In the past, yeast served as a powerful genetic system to dissect the NPC into its numerous components and identify factors involved in nucleocytoplasmic transport reactions (Doye and Hurt, 1997). We have set up an in vivo assay that makes it possible to monitor nuclear export of the ribosomal protein L25 fused to GFP. This allows mutants known to be impaired in ribosome assembly, NPC structure and nucleocytoplasmic transport to be tested for defects in ribosomal subunit export. Furthermore, it should be possible to identify with this assay novel genes of the ribosomal export machinery.
Materials and Methods
Generation of L25-GFP and RPL25 Gene Disruption
A modified RPL25 (L25) gene in which an EcoRV restriction site was generated at the stop codon (Tollervey et al., 1993) was used to fuse its 3′ end to the GFP gene that was previously attached to the 3′-UTR of the MEX67 gene (Segref et al., 1997). In this ligation, the L25-containing BamHI-EcoRV restriction fragment was joined to a PCR amplified GFP-3′-UTRMEX67 construct that was cut with EcoRV at the 5′ end of GFP and with EcoRI at the 3′ end of the UTR. This ligation leads to an in-frame fusion between the L25 and GFP ORFs, generating an EcoRV site at the junction region. Finally, the L25-GFP construct that contains the authentic RPL25 5′-UTR (i.e., its authentic promoter) was inserted into the high copy vector YEplac112 and ARS/CEN plasmid pRS314, previously cut with BamHI-EcoRI. Later on, the L25-GFP gene was also inserted into the high copy number plasmid YEplac195 carrying both the URA3 and ADE2 gene.
For the RPL25 gene disruption, 5′-UTR (520 bp; ending at the ATG start codon) and 3′-UTR (590 bp; starting at the stop codon) of the RPL25 gene were amplified by PCR as XbaI-BamHI and BamHI-HindIII fragments using the appropriate DNA primers, respectively. Both DNA fragments were ligated into pBluescript KS in a triple ligation and were joined at their common BamHI site. Next, the HIS3 gene was isolated as a BamHI fragment from plasmid YDp-H and inserted into the BamHI site previously generated at the junction between the 5′- and 3′-UTR of RPL25. The rpl25::HIS3 gene disruption construct was excised from this plasmid by restriction digestion with NotI-XhoI and used to transform the diploid strain RS453. Two types of HIS+ transformants were obtained. Those that grew significantly slower on SDC-his plates were shown to contain the rpl25::HIS3 gene disruption. Thus, RPL25/rpl25::HIS3 heterozygous diploids are impaired in cell growth due to the lack of one of the two RPL25 gene copies. Such slower growing heterozygous strains carrying the correct rpl25::HIS3 gene disruptions were sporulated and tetrad analysis was performed. A 2:2 segregation of viability was found and no growing progeny were HIS+, confirming earlier data that RPL25 is an essential gene in yeast (Rutgers et al., 1990). For complementation of the rpl25::HIS3 disrupted progeny with L25-GFP, one of the heterozygous diploid disruptants DSL25 (see Table I) was transformed with the plasmid YEplac195-ADE2-URA3-L25-GFP, and URA+/ADE+ transformants were sporulated and tetrads dissected. The L25-GFP complemented the lethal phenotype of haploid rpl25::HIS3 progeny and a 4:0 segregation for viability was observed. However, such haploid strains L25-GFP (Table I) grew slower than L25+ cells (hRS453).
Table I.
Yeast Strains
| Strain | Genotype | |
|---|---|---|
| CHRS 52 | MATa/α ade2/ade2,ade3/ade3,his3/his3,leu2/leu2,trp1/trp1,ura3/ura3 (this work) | |
| CHRS 1d | MATα,ade2,ade3,his3,leu2,trp1,ura3 (this work) | |
| RS453 | MATa/α,ade2/ade2,his3/his3,leu2/leu2,trp1/trp1,ura3/ura3 (Segref et al., 1997) | |
| DSL25 | MATa/α,ade2/ade2,his3/his3,leu2/leu2,trp1/trp1,ura3/ura3,L25::HIS3/L25 (RS453-derived) | |
| L25-GFP | MATa or α,ade2,his3,leu2,trp1,ura3,L25:HIS3 (pYEplac195-ADE2-URA3-L25-GFP) | |
| nup49-313/L25-GFP | ade2,his3,leu2,trp1,ura3,L25:HIS3, nup49::TRP1 (pYEplac195-ADE2-URA3-L25-GFP,pUN100LEU2-nup49-313) | |
| nup49-313 | MATα,ade2,ade3,his3,leu2,trp1,ura3,nup49::TRP1 (pUN100-LEU2-nup49-313) (Doye et al., 1994) | |
| nsp1-5/L25-GFP | ade2,his3,leu2,trp1,ura3,L25:HIS3,nsp1::HIS3 (pYEplac195-ADE2-URA3-L25-GFP,pSB32-LEU2-nsp1-5 [L640>S]) | |
| nsp1-5 | MATa,ade2,his3,leu2,trp1,ura3,nsp1::HIS3 (pSB32-LEU2-nsp1-5 [L640>S]) (Nehrbass et al., 1993) | |
| nop1-7 | nop1-7::HIS3,ura3,leu2 (Tollervey et al., 1993) | |
| nop1-7/L25-GFP | his3,leu2,ura3,L25:HIS3,nop1-7::HIS3 (pYEplac195-ADE2-URA3-L25-GFP) | |
| nop56-1 | MATa,ade2,trp1,leu2,ura3,HIS3::nop56 (pRS315-nop56-1) (Gautier et al., 1997) | |
| los1−/pus1− | MATα,ade2, leu2, ura3, his3, trp1,los1::HIS3,pus1::HIS3 (Simos et al., 1996b) | |
| nsp1-ala6 | ade2,his3,leu2,trp1,ura3,nsp1::HIS3 (pYEplac195-ADE2-URA3-L25-GFP,pSB32-LEU2-nsp1-ala6) | |
| nic96-1 | MATa,ade2,leu2,trp1,ura3,nic96::HIS3 (pUN100-nic96-1) | |
| nsp1-ala6/L25-GFP | ade2,his3,leu2,trp1,ura3,L25:HIS3,nsp1::HIS3 (pYEplac195-ADE2-URA3-L25-GFP,pSB32-LEU2-nsp1-ala6) | |
| xpo1-1 | ade2,his3,leu2,trp1,ura3,xpo1::LEU2 (pYEplac195-ADE2-URA3-L25-GFP,pHIS3-XPO1) | |
| (H. Santos-Rosa; derived from Stade et al., 1997) | ||
| xpo1-1/L25-GFP | ade2,his3,leu2,trp1,ura3,L25::HIS3,xpo1::LEU2 (pYEplac195-ADE2-URA3-L25-GFP,pHIS3-XPO1) | |
| nup85Δ | ade2,his3,leu2,trp1,ura3,nup85::HIS3 (nup85Δ) (Siniossoglou et al., 1996) | |
| nup85Δ/L25-GFP | ade2,his3,leu2,trp1,ura3,L25::HIS3, nup85::HIS3 (pYEplac195-ADE2-URA3-L25-GFP) | |
| nup84::HIS3 | ade2,his3,leu2,trp1,ura3,nup84::HIS3 (nup84) (Siniossoglou et al., 1996) | |
| nup84::HIS3/L25-GFP | ade2,his3,leu2,trp1,ura3,L25::HIS3, nup84::HIS3 (pYEplac195-ADE2-URA3-L25-GFP) | |
| mtr10::HIS3 | ade2,his3,leu2,trp1,ura3,mtr10::HIS3 (Senger et al., 1998) | |
| mtr10::HIS3/L25-GFP | ade2,his3,leu2,trp1,ura3,L25:HIS3,mtr10::HIS3 (pYEplac195-ADE2-URA3-L25-GFP) | |
| prp20-1 | ade2,his3,leu2,trp1,ura3,prp20-1 (pYEplac195-ADE2-URA3-L25-GFP) (M. Künzler; derived from Aebi et al., 1990) | |
| pse1-1/kap123::HIS3 | MATa,ura3,leu2,trp1,pse1-1,kap123::HIS3 (pYEplac195-ADE2-URA3-L25-GFP) (Seedorf and Silver, 1997) | |
| rna1-1 | ade2,his3,leu2,trp1,ura3,rna1-1 (H. Santos-Rosa; derived from Hopper et al., 1978) | |
| rnal-1/L25-GFP | ade2,his3,leu2,trp1,ura3,L25::HIS3,rna1-1 (pYEplac195-ADE2-URA3-L25-GFP) |
Generation and Growth of Double Mutants between L25-GFP and Nucleoporin/Nucleolar Mutants
Single mutant strains of opposite mating type and containing appropriate auxotrophic markers were mated in liquid YPD medium before diploids were selected on SDC minimal plates lacking the corresponding auxotrophic markers. Diploid progeny were sporulated on YPA plates and tetrad analysis was performed. Haploid progeny containing the rpl25::HIS3 and the nucleoporin/transport factor disruption gene plus plasmid-borne L25-GFP and mutant nucleoporin/transport factor genes were selected and analyzed for phenotypical defects (see also Table I). Cells were grown in YPD- or selective medium, either to logarithmic or stationary phase before transfer into fresh medium and shift for 1–14 h to 33°/37°C and further reshift to 20°C for 1–24 h. Samples were taken and inspected in the fluorescence microscope or ribosomes were isolated.
Overexpression of Dominant-negative LOS1, GSP1-G21V, KAP95, and YRB4 in L25-GFP Cells
The construction of pEMBLyex4-GAL-ProtA-LOS1 and pEMBLyex4-GAL-ProtA-LOS1ΔN was recently described (Hellmuth et al., 1998). To overexpress YRB4 and KAP95, plasmids YCpGAL-YRB4ΔN and YCpGAL-RSL1 (Schlenstedt et al., 1997) were used. Cells carrying these plasmids were allowed to grow in raffinose-containing medium before induction by dilution in medium containing 2% galactose. A GAL1-driven version of GSP1-G21V was used as described earlier (Hellmuth et al., 1998) and based on plasmid pGPCNR1 (Kadowaki et al., 1993). The L25-GFP strain was transformed with the plasmid YEp351-LEU2-GAL:: GSP1-G21V. A 20-ml culture of such a transformant was grown in synthetic medium containing raffinose, but lacking leucine (SRC-leu), at 30°C overnight to the stationary phase. Half of the culture (10 ml) was pelleted and resuspended in 1 ml SRC-leu and incubated on a plate with synthetic medium containing galactose. The plate was incubated at 30°C for 14 h. Cells were taken directly from the plate and observed in the fluorescence microscope.
Fluorescence Microscopy
To detect L25-GFP in vivo in the fluorescence microscope, the GFP-signal was examined in the fluorescein channel of a Zeiss Axioskop fluorescence microscope and pictures were obtained with a Xillix Microimager CCD camera. Digital pictures were processed by the software program Openlab (Improvision).
RNA Extraction and Hybridization
Steady state levels of rRNA and pre-rRNAs that were extracted from nup49-313/L25-GFP cells were analyzed by Northern hybridization using appropriate radiolabeled rRNA probes as described in (Tollervey, 1987).
DNA recombinant work such as restriction analysis, end filling, ligation and PCR amplification were performed according to Maniatis et al. (1982). Isolation of ribosomes under high salt (800 mM NaCl) and low salt conditions (100 mM NaCl) by sucrose gradient centrifugation was performed as described earlier (Tollervey et al., 1993). Poly(A)+ RNA export defects were analyzed by in situ hybridization (Segref et al., 1997). SDS-PAGE and Western blotting was done according to Siniossoglou et al. (1996). To detect L25-GFP, whole cell extracts were analyzed by Western blotting using commercially available anti-GFP antibodies and as previously described (Segref et al., 1997). Indirect immunofluorescence using anti-Nsp1p and anti-Nop1p antibodies was done essentially after Simos et al. (1996a).
Results
Large Subunit Ribosomal Protein L25 Tagged with GFP Assembles into 60S Ribosomes
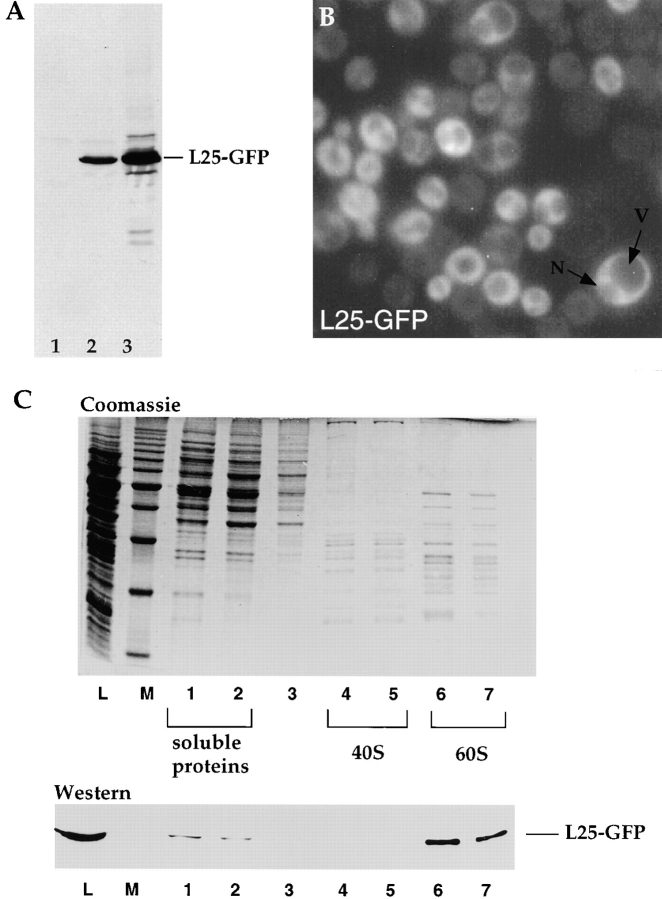
Our goal was to develop an in vivo assay that would allow us to monitor export of a ribosomal protein from the nucleus into the cytoplasm in living cells. Such an assay would facilitate the identification of mutants defective in ribosomal export. The ribosomal protein L25 was chosen as a reporter, because it contains a strong nuclear localization sequence (NLS) within its NH2-terminal half (Schaap et al., 1991; Nehrbass et al., 1993) that mediates efficient import into the nucleus and a COOH-terminal domain that binds to rRNA and facilitates assembly into ribosomes. L25 belongs to the group of r-proteins that bind to rRNA very early during rRNA transcription and processing (Kruiswijk et al., 1978) and thus could serve as reporter to measure ribosomal exit from the nucleus. In fact, L25 carrying a short peptide tag from the VSV G protein has been shown to assemble into 60S subunits and localizes in the cytoplasm under steady state conditions (Tollervey et al., 1993). This prompted us to test whether a larger tag, the GFP that allows in vivo localization studies, can be attached to L25 without disturbing assembly into 60S ribosomes.
L25 was GFP-tagged at its COOH-terminal end and the corresponding L25-GFP fusion construct was inserted into a single or high copy number yeast plasmid. When transformed into a wild-type yeast strain, expression of L25-GFP (calculated molecular mass is 15.6 kD for L25 plus 25 kD for GFP) could be shown by Western analysis using an antibody against the GFP moiety (Fig. 1 A). When yeast cells expressing L25-GFP were inspected in the fluorescence microscope, a cytoplasmic L25-GFP staining with vacuolar and nuclear exclusion was observed (Fig. 1 B). To show assembly of L25-GFP into 60S ribosomal subunits, ribosomes were isolated by sucrose gradient centrifugation from yeast cells that contained authentic L25 but also expressed L25-GFP from a high copy number plasmid. Clearly, L25-GFP is found in the fractions of the sucrose density gradient that contain the 60S large ribosomal subunits, but it is not detected in the adjacent 40S peak fractions (Fig. 1 C). A small portion of L25-GFP was also found on the top of the density gradient where soluble proteins fractionate. This fraction may represent nonassembled L25-GFP.
Figure 1.
Assembly of L25-GFP into 60S ribosomes. (A) Western blot analysis of whole cell extracts. 1, CHRS 52 strain; 2, CHRS 52 strain transformed with pRS314-L25-GFP (single copy plasmid); 3, CHRS 52 strain transformed with pYEplac112-L25-GFP (high copy plasmid). Equal amounts of cells were loaded on the SDS–polyacrylamide gel that was blotted onto nitrocellulose and probed with anti-GFP antibodies. The position of L25-GFP is shown. (B) Fluorescence microscopy of yeast cells (CHRS 1d) expressing L25-GFP. The vacuole (V) and nuclear compartment (N) is indicated in one of the L25-GFP-expressing cells. (C) Ribosome isolation by sucrose gradient centrifugation. Ribosomal proteins of the 60S and 40S subunits, which were obtained from CHRS 1d cells expressing L25-GFP, were separated by SDS-PAGE and visualized, respectively, by Coomassie-staining (upper panel) and Western blotting using anti-GFP antibodies (lower panel). L, load, i.e., whole cell extract of yeast strain CHRS 1d expressing L25-GFP; M, 10 kD protein ladder standard (the prominent band corresponds to 50 kD); 1–7, fractions from the sucrose gradient. The position of the 60S and 40S fractions, and of the soluble proteins, are indicated.
L25-GFP Can Complement the Lethal Phenotype of a rpl25::HIS3 Null Mutant
To find out whether L25-GFP can functionally replace authentic L25, the RPL25 gene that encodes the L25 ribosomal protein was disrupted in a diploid yeast strain. Tetrad analysis of sporulated heterozygous diploids (rpl25::HIS3/ RPL25) revealed a 2:2 segregation for viability (Fig. 2 A). This confirmed that RPL25 is essential for cell growth (Rutgers et al., 1990). The lethal phenotype of haploid rpl25:HIS3 progeny can be complemented by the expression of plasmid-borne L25-GFP, although L25-GFP cells grow slower than wild-type cells (Fig. 2 A). However, L25-GFP cells do not exhibit a particular heat- or cold-sensitive phenotype (Fig. 2 A, compare hRS453 and L25-GFP). This shows that the attachment of GFP to L25 only slightly impairs the L25 function. When haploid cells with disrupted RPL25 gene but complemented by plasmid-borne L25-GFP were inspected in the fluorescence microscope, the GFP signal increased and was still exclusively cytoplasmic (see also below). This suggests that L25-GFP assembles more efficiently into 60S ribosomal subunits when the endogenous L25 copy is not present as a competitor. This is consistent with the observation that no free L25-GFP can be detected in the upper part of a sucrose gradient when a whole cell extract derived from L25-GFP cells was analyzed (see also Fig. 1 C); instead L25-GFP was now found exclusively associated with 60S ribosomes (data not shown). In conclusion, L25 tagged with the GFP is functional in yeast, since it can replace authentic L25; accordingly, L25-GFP carrying ribosomes are exported from the nucleus into the cytoplasm.
Figure 2.
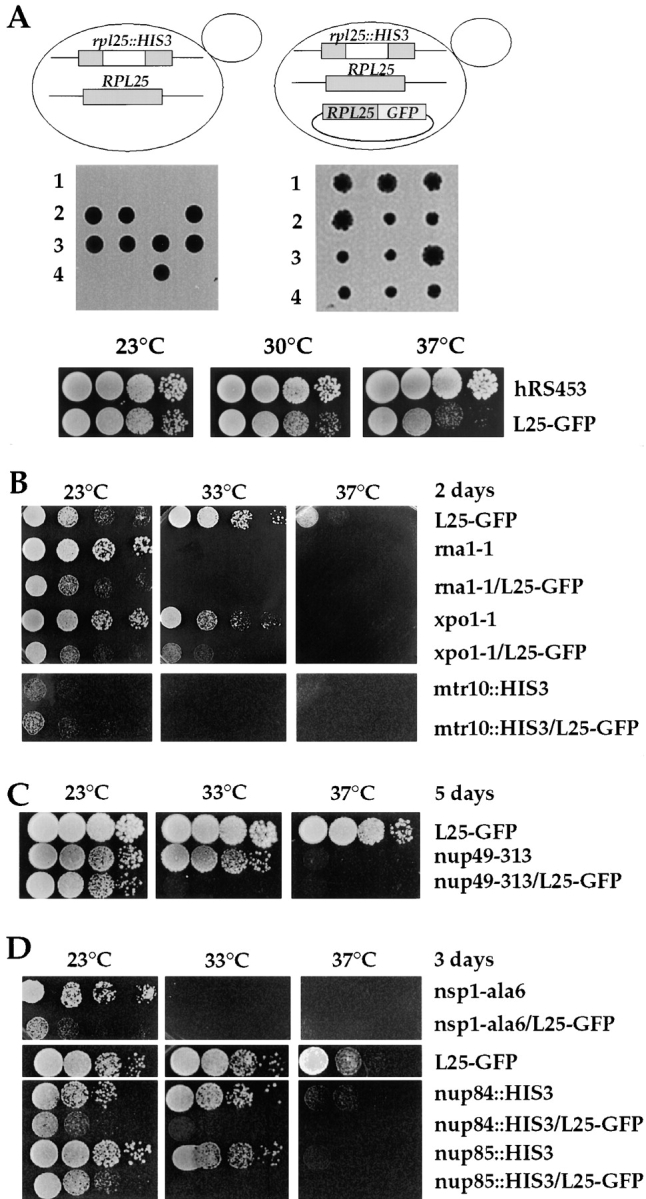
Growth of L25-GFP-expressing strains. (A) L25-GFP complements a rpl25::HIS3 null mutant. (Upper and middle panel) Tetrad analysis of the heterozygous rpl25::HIS3/RPL25 diploid strain reveals a 2:2 segregation, indicating that the RPL25 gene is essential for viability. Viability of the rpl25::HIS3 spores is rescued by cosegregation of the pYEplac195-ADE2-URA3-L25-GFP plasmid with chromosomal rpl25::HIS3. (Lower panel) Growth properties of haploid L25-GFP strain. Precultures were diluted in growth medium and equivalent amounts of cells (diluted in 10−1 steps) were spotted onto YPD plates. hRS453 is a haploid wild-type, L25-GFP a haploid strain disrupted for rlp25:: HIS3 and complemented by YEplac195-ADE2-URA3-L25-GFP. It was grown for 4 d. (B–D) Growth properties of different single and double mutant strains at different temperatures. Precultures were diluted in growth medium and equivalent amounts of cells (diluted in 10−1 steps) were spotted onto YPD plates. Cells were grown at the indicated temperatures and for the indicated days on YPD plates. For description of strains, see Materials and Methods and Table I.
rna1-1 and prp20-1 Mutants Are Defective in Ribosomal Export
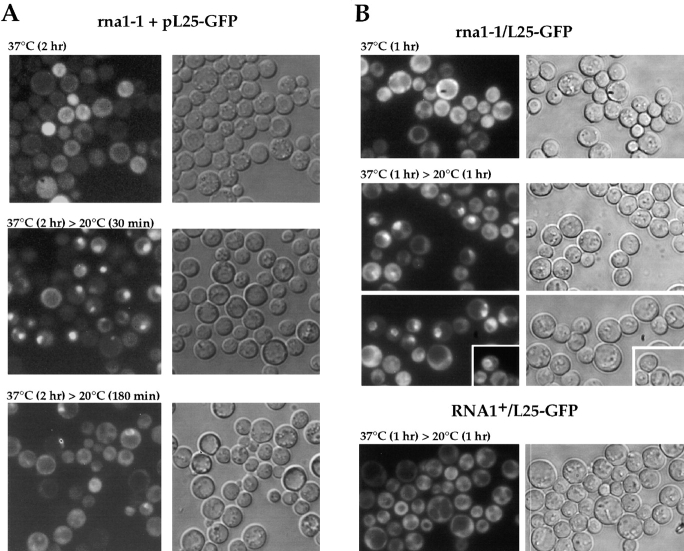
The small GTPase Ran has been suggested to be a universal regulator of nucleocytoplasmic transport (Koepp and Silver, 1996). By exploiting the L25-GFP assay, we tested whether ribosomal export is sensitive to perturbations of the Ran-GTP/GDP cycle. Therefore, the Ran-GAP mutant rna1-1 (Hopper et al., 1978; Bischoff et al., 1995) and the Ran-GEF mutant prp20-1 (Aebi et al., 1990; Amberg et al., 1993; Kadowaki et al., 1993) were transformed with the L25-GFP reporter construct. However, no nuclear accumulation of L25-GFP was seen when the rna1-1 cells were grown at the permissive temperature (23°C) and shifted for various time points to 37°C (Fig. 3 A). Since it is known that rRNA transcription and ribosome biogenesis is downregulated in a very complex way in yeast upon nutritional, mutational, and temperature stress (Warner, 1989; Woolford, 1991), the in vivo assay was performed differently. Ts mutants derived from prestationary or stationary culture conditions were first shifted to the restrictive temperature to induce the mutant phenotype and then incubated at the permissive condition to reinduce de novo ribosome biogenesis. Under these conditions, nuclear accumulation of the L25-GFP reporter was seen in many rna1-1 mutant cells (Fig. 3 A). The nuclear L25-GFP signal became visible already after 1 h shift to 37°C and 30 min regrowth at 20°C (data not shown), and developed stronger after 2 h at 37°C and 30 min induction at 20°C (Fig. 3 A). After prolonged permissive growth conditions (180 min), nuclear accumulation of L25-GFP vanished in most of the cells; interestingly, some cells showed a distinct nuclear envelope staining at this later time point (Fig. 3 A). A RNA1+ control strain exhibits under all tested conditions an exclusive cytoplasmic L25-GFP location with nuclear exclusion (see also later).
Figure 3.
Nuclear accumulation of L25-GFP in rna1-1 mutant. Thermosensitive mutant rna1-1 transformed with YEplac195-ADE2-URA3-L25-GFP (A), and the rpl25::HIS3 disruption mutant rna1-1/L25-GFP (B) were used. Cells were grown at 23°C on selective SDC-ura plates (A) and YPD plates (B) to stationary phase (3–4 d on plate) before they were inoculated in liquid YPD-medium. It was then shifted for the indicated time points to 37°C, before cells were further incubated at room temperature (20°C). After centrifugation, cells were resuspended in water, mounted on a slide and inspected in the fluorescence microscope.
Since the L25-GFP signal becomes stronger in cells lacking endogenous L25 (see above), a L25-GFP expressing haploid yeast was constructed in which the rna1-1 ts allele was combined with the chromosomal rpl25::HIS3 gene disruption (see Table I for strain construction, and Fig. 2 B for growth properties). When this rna1-1/L25-GFP strain was shifted for 1 h to 37°C and further incubated for 1 h at 20°C, a strong nuclear L25-GFP accumulation was noticed in many cells. Interestingly, individual cells stained differently showing L25-GFP in the entire nucleus, the nuclear envelope and distinct intranuclear spots, respectively (Fig. 3 B). Again, we noticed that nuclear accumulation of L25-GFP is best seen in the rna1-1 cells grown to prestationary or stationary phase (between OD600nm 4–8) before inoculation in fresh YPD-medium and shift to the restrictive temperature. A lower percentage of cells showed this defect if they were coming from a logarithmically growing culture in YPD-medium (data not shown). It is known that ribosome biogenesis is strongly induced when cells are shifted from stationary/starving conditions into fresh glucose-containing medium (Warner, 1989). This may also enhance nuclear accumulation of L25-GFP. As control served a RNA1+/L25-GFP strain, which did not show under identical conditions any nuclear accumulation of ribosomes after 1 h shift to 37°C and 1 h regrowth at 20°C (Fig. 3 B). The prp20-1 mutant that is defective in nuclear Ran-GDP/ GTP exchange (Kadowaki et al., 1993) was analyzed in a similar way. L25-GFP also accumulates in the nucleus in prp20-1 cells when shifted for 2 h to 37°C followed by regrowth for 40 min at 20°C (Fig. 4). Interestingly, the L25-GFP signal was not evenly distributed in the nucleus, but accumulated in a single or several spots, suggesting a segregation of L25-GFP into distinct subnuclear structures. It was reported earlier that prp20-1 cells are not only altered in mRNA metabolism, but also maintenance of the nuclear structure (Aebi et al., 1990). When the yeast Ran mutants gsp1-1 and gsp1-2 (Wong et al., 1997) expressing L25-GFP were tested, nuclear accumulation of L25-GFP was less prominent as compared with rna1-1 and prp20-1 mutant cells (data not shown). This could mean that not only export, but also nuclear uptake of L25-GFP is significantly impaired in Ran mutants, thus not allowing to detect a clear nuclear accumulation.
Figure 4.
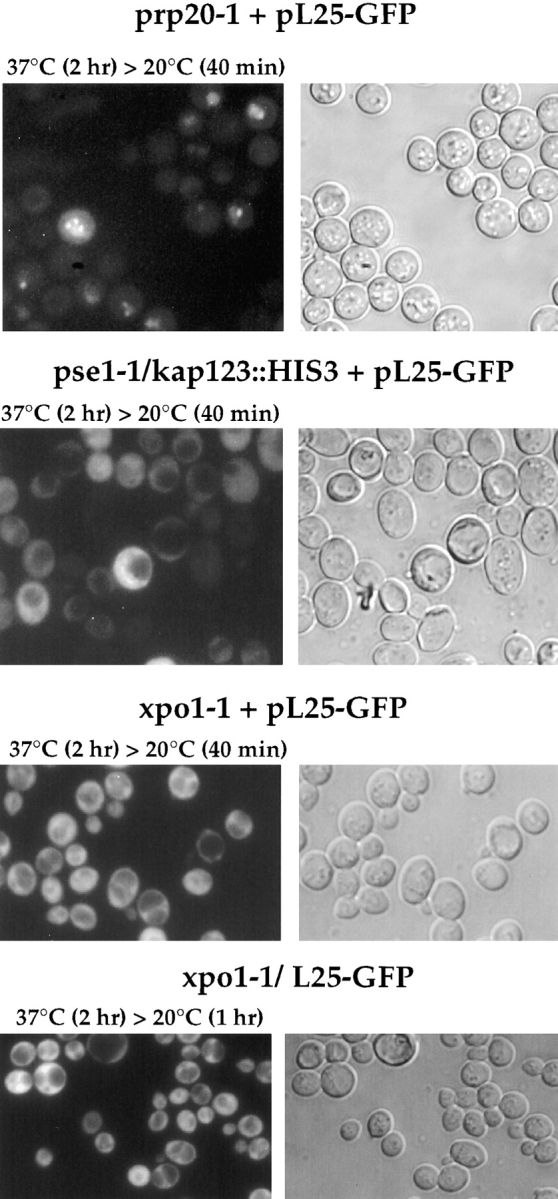
Ribosomal export in prp20-1 and mutants mapping in importin/karyopherin β–like transport factors. Thermosensitive mutants prp20-1, pse1-1/kap123::HIS3, and xpo1-1 were transformed with YEplac195-ADE2-URA3-L25-GFP (pL25-GFP), or the double xpo1-1/L25-GFP strain was constructed (see Table I). Cells were grown at 23°C on selective SDC-ura and YPD plates, respectively, to stationary phase, before they were inoculated in liquid YPD-medium. It was shifted for the indicated time points to 37°C, before cells were further incubated at room temperature (20°C). After centrifugation, cells were resuspended in water, mounted on a slide and inspected in the fluorescence microscope.
Next, mutants defective in nucleocytoplasmic transport factors belonging to the importin/karyopherin β-family were tested in the in vivo ribosomal export assay. Neither the pse1-1/kap123::HIS3 mutant (Fig. 4) that is impaired in ribosomal protein import and mRNA export (Rout et al., 1997; Schlenstedt et al., 1997; Seedorf and Silver, 1997), nor the xpo1-1 mutant (Fig. 4) that accumulates NES-containing nuclear proteins and mRNA inside the nucleus (Stade et al., 1997), nor the thermosensitive los1−/pus1− mutant (Simos et al., 1996b; data not shown) were impaired in nuclear exit of ribosomes. We also tested the double mutants xpo1-1/L25-GFP (Fig. 4) and mtr10::HIS3/ L25-GFP (data not shown; see Table I for strain construction, and Fig. 2 B for growth properties), but again could not see nuclear accumulation of L25-GFP. As shown earlier, mtr10::HIS3 cells are defective in nuclear import of the hnRNP-like protein Npl3p (Senger et al., 1998).
Overexpression of the Dominant-negative Ran-GTP Mutant or the tRNA Exportin Homologue Los1p Causes Nuclear Accumulation of L25-GFP
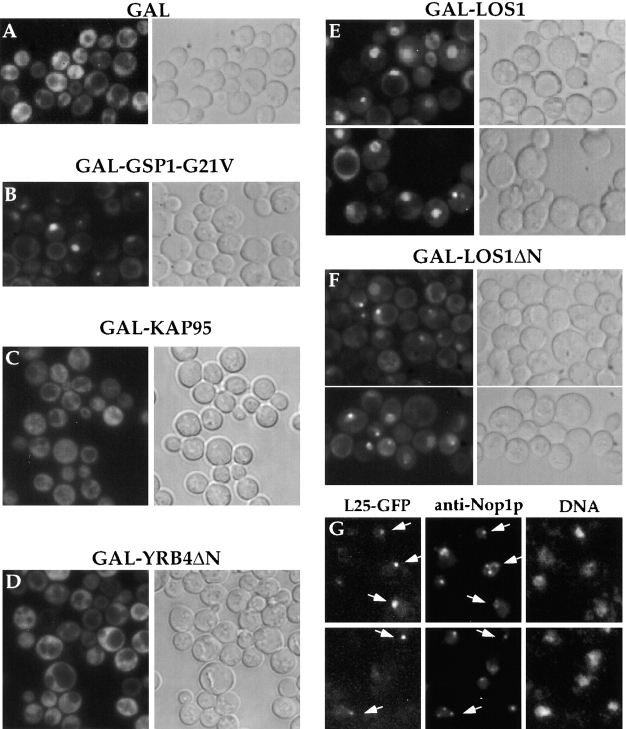
Since overexpression of a dominant-negative yeast Ran mutant that is stabilized in its GTP-bound form inhibits nucleocytoplasmic transport (Schlenstedt et al., 1995), we tested how this allele affects L25-GFP import and export reactions. Strain L25-GFP was transformed with a plasmid that expresses dominant-negative Gsp1p-G21V under the control of a galactose-inducible promoter. Transformed cells were first grown in repressive conditions, i.e., raffinose-containing medium before shifting them to galactose-containing medium to induce the expression of Gsp1p-G21V. Under repressed conditions, L25-GFP shows an exclusive cytoplasmic location; however, after induction of Gsp1p-G21V, a significant number of cells exhibit nuclear accumulation of L25-GFP that can be distributed throughout the entire nucleus or restricted to a distinct nuclear spot (Fig. 5, compare A with B).
Figure 5.
Accumulation of L25-GFP inside the nucleus in strains overexpressing Los1p and Gsp1p-G21V. L25-GFP cells carrying the plasmids (A) pEMBLyex4 (GAL), (B) pGAL-GSP1-G21V, (C) pGAL-KAP95, (D) pGAL-YRB4ΔN, (E) pEMBLyex4-ProtA-LOS1 (GAL-LOS1), (F and G) pEMBLyex4-ProtA-LOS1ΔN (GAL-LOS1ΔN) were first grown in raffinose-containing medium before transferring them into galactose-containing medium/ plate (see in Materials and Methods). After growth in galactose for 12 h, the localization of L25-GFP was determined in the fluorescence microscope (A–F). GAL-LOS1ΔN cells were also fixed in 3.7% formaldehyde for 30 min before spheroplasting and indirect immunofluorescence microscopy (G). The L25-GFP signal (fluorescein channel; indicated by arrows), the Nop1p signal (rhodamin channel) and the DNA signal (Hoechst channel) were recorded in a Zeiss Axioskop fluorescence microscope. L25-GFP and Nop1p colocation is indicated by arrows.
We also analyzed whether overproduction of importin β–like proteins, which can interfere with nucleocytoplasmatic transport mechanisms (Schlenstedt et al., 1997), affects ribosomal export. Overexpression of import receptors such as Kap95p (Enenkel et al., 1995) and the ΔN mutant of Yrb4p/Kap123p (Rout et al., 1997; Schlenstedt et al., 1997), known to block both nuclear protein import and mRNA export (Schlenstedt et al., 1997), did not reveal any intranuclear accumulation of L25-GFP (Fig. 5, C and D). It is possible that nuclear uptake of L25-GFP is impaired, therefore not allowing to detect a L25-GFP nuclear export defect. Los1p is another member of the importin β–like protein family involved in tRNA nuclear export, and its overexpression causes both a dominant-negative growth defect and inhibition of predominantly nuclear export reactions (Hellmuth et al., 1998). LOS1 and LOS1ΔN (a mutant lacking the Ran-GTP-binding domain) constructs were placed under the control of the GAL10 promoter and transformed into the L25-GFP strain. Overexpression of Los1p, which was induced by adding galactose to cultures pregrown in raffinose, caused a strong intranuclear accumulation of GFP-tagged L25 (Fig. 5 E); some cells also showed a nuclear envelope staining or a single spot inside the nucleus. This spot-like staining was particularly evident in cells that overexpressed Los1ΔNp (Fig. 5 F). To find out whether this presents nucleolar staining, indirect immunofluorescence microscopy using antibodies against the nucleolar marker Nop1p was performed (Fig. 5 G). Indeed, colocalization of the L25-GFP nuclear spot with Nop1p was seen in a significant number of Los1ΔNp overproducing cells (Fig. 5 G); however, the nucleolus disintegrates in Los1p-overproducing cells (Hellmuth et al., 1998) causing the appearance of more than one of Nop1p-staining nuclear spots, which do not necessarily contain L25-GFP. In conclusion, overproduction of the tRNA-exportin homologue Los1p interferes with nuclear export of ribosomes.
Nucleoporin Mutants Defective in Ribosomal Export
By exploiting the L25-GFP assay, we sought for nuclear accumulation of ribosomes in nucleoporin and ribosomal assembly mutants. To achieve a stronger L25-GFP signal in cells, haploid rpl25::HIS3 progeny expressing L25-GFP were generated that carried the nup49-313, nsp1-5, nsp1-ala6, nup85::HIS3, nup84::HIS3, and nop1-7 alleles, respectively (see also Table I). When the growth of these strains was analyzed, many of them showed an enhanced, i.e., synergistic inhibition of growth as compared with the single mutant strains. In particular, double mutants often did not grow at 33°C and above, whereas the single mutant strains could grow well at this intermediate temperature (Fig. 2, C and D). Therefore, 33°C was taken as restrictive temperature in the following experiments. Microscopic inspection revealed that these double mutants could go through several cell divisions at 33°C (data not shown).
Since the onset of the ts phenotype in the various nucleoporin and nop1 mutants requires several hours of incubation at the nonpermissive temperature (Nehrbass et al., 1993; Doye et al., 1994; Siniossoglou et al., 1996), ts strains were shifted for longer than 6 h to the restrictive growth condition (see also Doye et al., 1994; Nehrbass et al., 1993), before regrowth at the permissive temperature to allow for de novo ribosome biogenesis (see also Fig. 3). A time course was performed to follow the onset of a ribosomal export defect in the nup49-313/L25-GFP strain (Fig. 6 A). The minimal time required to observe L25-GFP nuclear mislocation in nup49-313 cells was to shift for 7 h to 33°C, but strongest accumulation was seen between 10– 14 h shift to 33°C. For reasons of convenience, we incubated over night at 33°C (i.e., 14 h), before regrowth at 20°C was induced. Already after 1 h incubation of nup49-313/L25-GFP cells at permissive conditions, nuclear L25-GFP accumulation became apparent, which increased within the next 3 h of incubation (Fig. 6 A). Upon longer growth at 20°C (>10 h), cells progressively lost their nuclear L25-GFP signal and cytoplasmic labeling became stronger. During the first 10 h of growth at 20°C, the optical density of the nup49-313/L25-GFP cell culture increased continuously showing that the cells indeed regrew (data not shown). Such an incubation scheme did not impair ribosomal export at all in the L25-GFP control strain that exhibits an exclusive cytoplasmic location of L25-GFP with no nuclear accumulation under all tested conditions (data not shown). To verify that L25-GFP accumulated inside the nucleus in nup49-313 cells, indirect immunofluorescence microscopy using anti-Pus1p antibodies (a nuclear marker protein; Simos et al., 1996b), and DNA staining was performed. Clearly, the L25-GFP signal colocalized with the Pus1p immunolabeling and partly with the DNA staining (Fig. 6 B).
Figure 6.
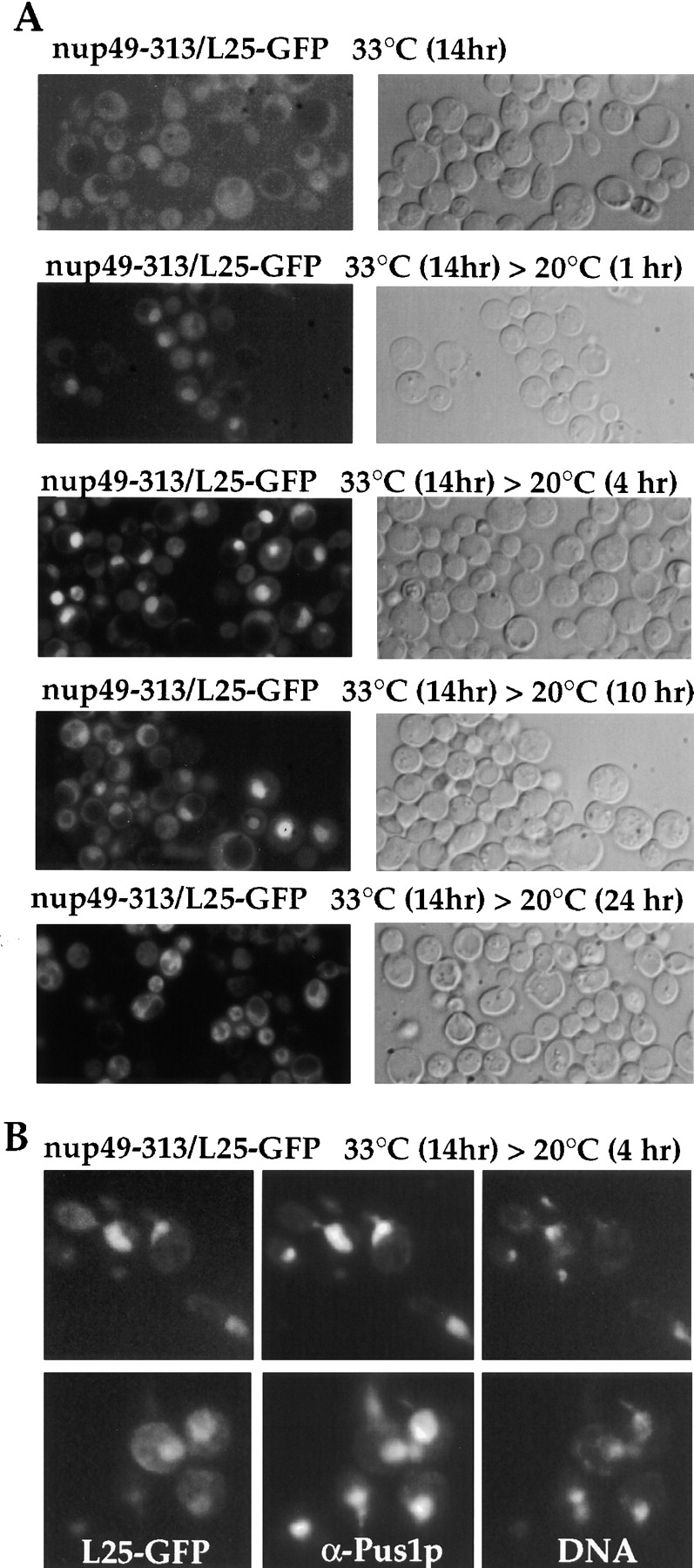
Nuclear accumulation of L25-GFP in nup49-313 ts mutant. (A) Time-course of nuclear accumulation of L25-GFP in nup49-313/L25-GFP cells. Cells were first grown at 23°C before shift for 14 h to 33°C. After the restrictive growth condition, cells were incubated at the permissive temperature (20°C) and inspected in the fluorescence microscope after 0, 1, 4, 10, and 24 h. (B) Indirect immunofluorescence microscopy of nup49-313/L25-GFP cells. After fixation of cells in 3.7% formaldehyde, spheroplasted cells were processed for indirect immunofluorescence using anti-Pus1p antibodies. Cells were also stained for DNA with Hoechst 33258. The L25-GFP signal was recorded in the fluorescein channel, the Pus1p immunostaining in the rhodamin channel of a Zeiss Axioskop fluorescence microscope.
According to these findings, additional nucleoporin mutants were tested in the in vivo ribosomal export assay. Similarly, nsp1-ala6/L25-GFP cells (Fig. 7) and to a lesser extent also nsp1-5/L25-GFP cells (data not shown) revealed an intranuclear accumulation of L25-GFP under the same conditions of incubation as described above. In contrast, neither nup85::HIS3/L25-GFP nor nup84::HIS3/ L25-GFP cells are defective in ribosomal export (Fig. 7), although these double mutant cells are also impaired in growth at 33°C (see also Fig. 2 D). Surprisingly, the nop1-7/ L25-GFP thermosensitive mutant that is inhibited in ribosome biogenesis (Tollervey et al., 1993) did not display a significant nuclear accumulation of L25-GFP; only in a very small number of cells, a spot-like L25-GFP signal was noticed (Fig. 7).
Figure 7.
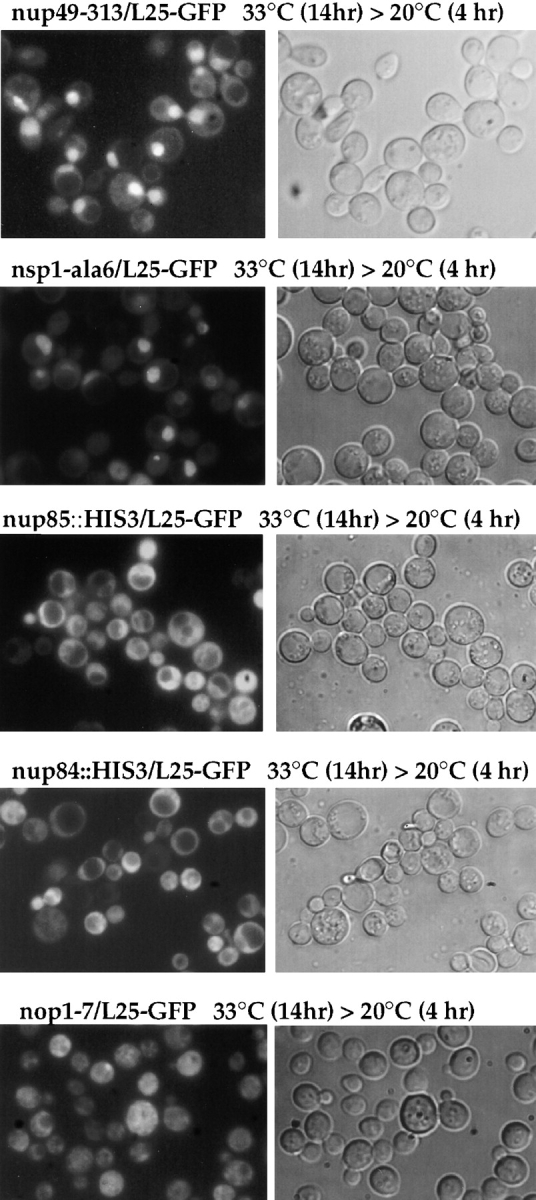
Nuclear accumulation of L25-GFP in nucleoporin mutants. Analysis of L25-GFP nuclear accumulation in nucleoporin and ribosome assembly mutants. Haploid double mutants (see Table I) nup49-313/L25-GFP, nsp1-ala6/L25-GFP nup85::HIS3/ L25-GFP, nup84::HIS3/L25-GFP, and nop1-7/L25-GFP were grown as described in Materials and Methods in YPD-medium to OD (600 nm) of 0.2 OD before shift for 14 h to 33°C and reshift for further 4 h to 20°C. Cells were viewed in the fluorescence microscope to see L25-GFP and under Nomarski optics.
When the original nup49-313 mutant still harboring the wild-type RLP25 gene was transformed with a plasmid carrying the L25-GFP reporter and analyzed as described above, L25-GFP also accumulated inside the nucleus when cells were shifted for more than 7 h to 37°C followed by a 4-h incubation at 20°C (data not shown). However, the intensity of labeling was reduced as compared with the nup49-313/L25-GFP strain (see also above). Accordingly, another thermosensitive nucleoporin mutant nic96-1, but not the ribosomal assembly mutant nop56-1 (Gautier et al., 1997), showed nuclear accumulation of L25-GFP in our in vivo assay (data not shown).
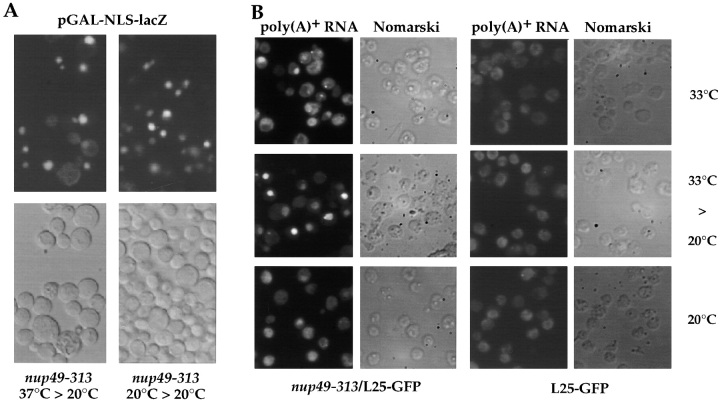
Apparently, nuclear uptake of L25-GFP still proceeds in nup49-313 cells (as well as in nsp1-ala6, rna1-1, and prp20-1 cells) after shift from restrictive to permissive growth conditions. Therefore, we tested other nucleocytoplasmic transport reactions. A galactose-inducible GAL-NLS-GFP-lacZ construct was transformed into the nup49-313 mutant and the culture was shifted for 11 h to 37°C under conditions of promoter repression (raffinose medium). Then, cells were transferred into galactose-containing medium to induce pGAL-NLS-GFP-lacZ and incubated for 5 h at 20°C. When cells were inspected in the fluorescence microscope, NLS-GFP-lacZ still accumulated inside the nucleus, although also a weak cytoplasmic staining could be seen in a few cells (Fig. 8 A). This shows that nuclear accumulation of this NLS-reporter can occur under conditions in which ribosomal export is apparently impaired (see also Figs. 6 and 7). When the distribution of poly(A)+ RNA was examined in the nup49-313/L25-GFP mutant, nuclear mRNA accumulation was seen in ∼10–20% of the cells when incubated at 33°C, and this percentage further increased when cells were brought back from the restrictive to permissive condition (Fig. 8 B). No mRNA export defect was observed in the L25-GFP strain grown at the various temperatures (Fig. 8 B).
Figure 8.
NLS-mediated nuclear protein uptake and poly(A)+ RNA export in thermosensitive nup49-313 cells. (A) The thermosensitive nup49-313 mutant was transformed with pGAL-NLS-GFP-lacZ (classical NLS of SV-40 large T antigen) and transformants were selected on SDC (−ura) plates. Transformants were grown in selective synthetic raffinose (−ura) medium at 20°C. One-half of the culture was left at 20°C (right panel), the other half was shifted for 11 h to 37°C (left panel). Cells from both cultures were collected by centrifugation, transfered into in SGal (−ura) medium and incubated for 5 h at 20°C to induce pGAL-NLS-GFP-lacZ expression. Cells were finally inspected in the fluorescence microscope. Note that at both temperatures the nuclear import reporter accumulates in the nucleus. (B) Nuclear export of poly(A)+ RNA in nup49-313/L25-GFP and L25-GFP cells. Cells were grown at the indicated temperatures (14 h at 33°C; 14 h at 33°C, and 4 h at 20°C; 18 h at 20°C) before they were fixed and processed for in situ hybridization using a fluorescently labeled oligo dT-probe.
Finally, rRNA processing was analyzed in the nup49-313/L25-GPF cells by Northern blotting. No striking changes in rRNA processing were observed (data not shown). Pre-rRNA processing in the strains expressing L25 was not clearly different from a wild-type control strain. In the nup49-313/L25-GFP strain, the levels of the mature 18S and 25S rRNAs were not detectably altered. The levels of the 35S pre-rRNA were marginally elevated, but this was observed at all temperatures. The 23S RNA, an aberrant processing intermediate that is indicative of a delay in the early pre-rRNA cleavages, was detected in the nup49-313/L25-GFP strain. Accumulation was strongest at 33°C, probably reflecting a mild delay in pre-rRNA processing due to reduced ribosomal protein import. The levels of the 27SA and 27SB pre-rRNAs, the precursors to the 25S rRNA with which L25 is associated, were unaltered in the nup49-313/L25-GFP strain during the temperature shift experiment. We conclude that nup49-313/L25-GFP cells are not significantly impaired in pre-rRNA processing under conditions that lead to strong nuclear accumulation of L25-GFP, and that the accumulated nuclear L25-GFP is therefore very likely to be associated with the mature 25S rRNA.
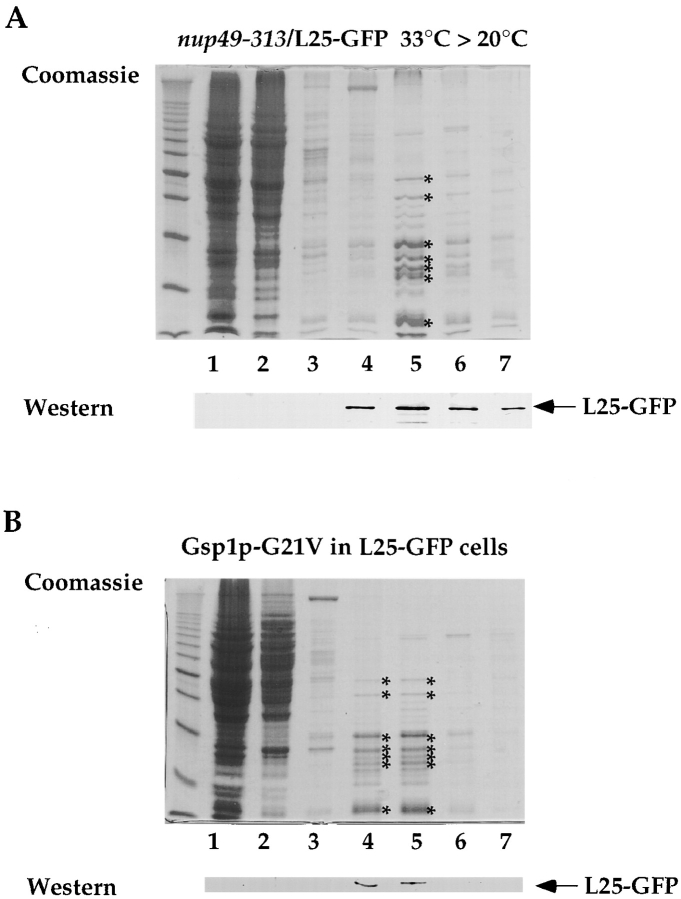
Isolation of Ribosomes from Yeast Mutants Accumulating L25-GFP Inside the Nucleus
To analyze whether L25-GFP is associated with ribosomes under conditions in which it accumulates inside the nucleus (see also Figs. 6 and 7), a whole cell extract was prepared from nup49-313/L25-GFP cells and ribosomes and soluble proteins were separated by sucrose gradient centrifugation. When the various sucrose gradient fractions were tested by Western blotting using anti-GFP antibodies, no free L25-GFP was detected in the upper part of the gradient, but L25-GFP was exclusively recovered in the lower gradient fractions in which the ribosomes migrated (Fig. 9 A, compare the Coomassie pattern of the major ribosomal proteins with the L25-GFP signal). This suggests that the L25-GFP that has accumulated in the nucleus is part of assembled ribosomes. As described above, free L25-GFP could be detected in the upper fractions of the sucrose density gradient, when authentic L25 was coexpressed (see Fig. 1 C). Ribosomes were also isolated from the strain L25-GFP that expresses dominant-negative Gsp1p-G21V (see also Fig. 4) under galactose-inducing conditions. All L25-GFP cofractioned with the ribosomal peak fractions and no free L25-GFP was found in the upper part of the sucrose gradient (Fig. 9 B).
Figure 9.
L25-GFP containing ribosomes isolated from nup49-313/L25-GFP and Gsp1p-G21V/L25-GFP cells. Ribosomes were isolated under low salt conditions (100 mM KCl) from (A) nup49-313/L25-GFP cells shifted for 14 h to 33°C and incubated for another 4 h at 20°C and (B) L25-GFP cells expressing Gsp1p-G21V under restrictive conditions (i.e., galactose medium) as described in the legend of Fig. 5 and in Materials and Methods. Whole cell extracts were prepared, loaded on a 10–40% sucrose gradient and centrifuged for 12 h at 150,000 g. The fractions from the sucrose density gradient were TCA-precipitated, resuspended in SDS-sample buffer and analyzed by SDS-PAGE and Coomassie-staining (upper part) or Western blotting using anti-GFP antibodies (lower part). The position of L25-GFP is indicated. The asterisks mark the position of prominent ribosomal proteins. Marker proteins (10-kD ladder with a stronger 50-kD band) are also depicted.
Discussion
We describe here a novel in vivo assay to monitor ribosomal export based on the GFP-tagged ribosomal protein L25 that assembles into 60S ribosomal subunits and forms green fluorescent ribosomes. This enabled us to test whether mutants defective in ribosomal assembly, NPC organization or nucleocytoplasmic transport are impaired in nuclear exit of ribosomes.
Our findings suggest that Ran is involved in nuclear export of ribosomes, since mutations affecting the Ran-GTP/ GDP cycle such as in Rna1p (Ran-GAP) and Prp20p (Ran-GEF), or overproduction of dominant negative Ran-GTP (Gsp1-G21V), inhibit nuclear export of L25-GFP. However, at present we cannot exclude that the rna1-1 and prp20-1 mutants also affect nuclear import of so far unknown specific ribosomal export factors. A Ran requirement for ribosomal export is also indicated from studies in the mammalian system in which the RCC1 (Ran-GEF) thermosensitive mutant was shown to be impaired in ribosomal export (Cheng et al., 1995). How could Ran be involved in ribosomal export? Ribosomes may contain NES (either in ribosomal proteins, rRNA or adaptor proteins) that are recognized by ribosomal export receptors. Whether these transport receptors belong to the importin/karyopherin β–like protein family remains to be shown. Recently, it has been found that two members of the importin β–like family, tRNA-exportin as well as its yeast homologue Los1p, bind cooperatively to tRNA in the presence of Ran-GTP and are involved in nuclear export of tRNA (Kutay et al., 1998; Arts et al., 1998; Hellmuth et al., 1998). According to current models, the cooperative binding of (a) putative ribosome exportin(s) to ribosomes is predicted to require Ran-GTP, explaining why disturbing the Ran-cycle also inhibits export of ribosomes.
Furthermore, overproduction of the tRNA exportin Los1p inhibits ribosomal export. How Los1p can bring upon this impairment is not clear, but excess of Los1p which interacts with both nucleoporins and Ran-GTP (Hellmuth et al., 1998) may saturate sites at the NPC or titrate Ran-GTP, thereby blocking ribosomal export. It is also possible that NES within newly assembled ribosomes compete with the nuclear export machinery in which Ran and Los1p are integrated. On the opposite, nuclear export of ribosomes is not inhibited when import factors belonging to the importin/karyopherin β–like family (e.g., Kap95p and Yrb4p/Kap123p, Pse1p/Kap121p, and Mtr10p) are overproduced or mutated.
By applying our L25-GFP assay, we found a distinct requirement of nucleoporins Nup49p, Nsp1p, and Nic96p for nuclear exit of ribosomes. Although we cannot yet exclude that these NPC proteins participate in the import of specific ribosomal export factors, it is likely that Nup49p, Nsp1p, and Nic96p are directly involved in ribosomal export. It was recently shown that the Nsp1p/Nup57p/ Nup49p/Nic96p complex is located at both sites of the central gated channel and thus could play a crucial role in the bi-directional translocation through the pore channel (Fahrenkrog et al., 1998). In contrast, Nup85p and Nup84p nucleoporin mutants are not impaired in the nuclear export of L25-GFP. Nup85p and Nup84p are members of a large nucleoporin complex that plays a dual role in NPC distribution and poly(A)+ RNA export (Siniossoglou et al., 1996). Interestingly, L25-GFP accumulates predominantly throughout the entire nucleus in nup49-313, nsp1-ala6, and nic96-1 cells, but has the tendency to concentrate in one or a few intranuclear spots or at the nuclear periphery in Ran-cycle mutants or when Los1p is overproduced. This suggests, that depending on the mutant, ribosomes are impaired in leaving the nucleolus, exiting the nucleus or passing through the nuclear pores. Whether the observed different locations correspond to bona fide intermediates during ribosomal export from the nucleolus to the cytoplasm remains to be shown.
How can one explain why nuclear export of L25-GFP containing ribosomes is more strongly inhibited than nuclear uptake of the import substrate L25-GFP, as observed in many mutants tested here? It is conceivable that transport of huge RNP particles such as ribosomes through the nuclear pores is more sensitive to alterations of the NPC transporter and the nucleocytoplasmic transport machinery than transport of smaller cargo such as the L25-GFP import substrate. Along these lines, it was recently suggested that the exit of the large ribosomal subunits (as well as larger messenger RNP particles) from the nucleus into the cytoplasm would be significantly slower in stage-1 versus vitellogenic oocytes that depends on the functional size of the NPC and which varies during oogenesis (Feldherr et al., 1998). Alternatively, the ribosomal export machinery may have requirements that are distinct from the protein import machinery.
Finally, the nop1-7 and nop56-1 mutants that are defective in rRNA processing and ribosome assembly (Tollervey et al., 1993; Gautier et al., 1997) did not show nuclear accumulation of L25-GFP, under conditions where nup49-313 and nsp1-ala6 cells clearly show this defect. This could mean that L25-GFP no longer assembles into ribosomes in nop1-7 or nop56-1 cells, either because its expression is downregulated or L25-GFP is degraded. As shown earlier, nop1-7 (as well as nop1-4) are strongly impaired in later steps of ribosome assembly yielding ribosomal particles that lack many ribosomal proteins (which are most likely degraded) and show strongly aberrant mobilities on sucrose gradients even at permissive temperatures (Tollervey et al., 1993).
The L25-GFP in vivo may be exploited for a detailed genetic analysis of a so far poorly characterized nucleocytoplasmic transport route. The analysis of other transport pathways has been facilitated by the availability of appropriate assays and reporters in yeast, including NLS-mediated nuclear uptake of reporter proteins carrying a classical NLS (Silver et al., 1989), ribosomal NLS (Moreland et al., 1985; Nehrbass et al., 1993) and the RGG-box NLS of the hnRNP protein Npl3p (Senger et al., 1998), shuttling of proteins between the nucleus and cytoplasm (Koepp et al., 1996), NES-mediated nuclear export (Stade et al., 1997) and in situ hybridization using oligo-dT probes to analyze nuclear export of poly(A)+ RNA (Amberg et al., 1992; Kadowaki et al., 1992). In principle, the in vivo assay based on the intracellular L25-GFP location could be exploited to identify novel components of the ribosomal export machinery. Yeast served in the past as an excellent system to dissect the NPC into its numerous components and identify nucleocytoplasmic transport factors (Doye and Hurt, 1997). In particular, an assay based on in situ hybridization in combination with screening random ts mutants for defects in poly(A)+ RNA export led to the identification of a large number of genes involved in mRNA export (Amberg et al., 1992; Kadowaki et al., 1992). A related genetic screen may be performed exploiting the L25-GFP reporter protein to identify genes involved in ribosomal export, e.g., by transforming a collection of thermosensitive mutants (heat- or cold-sensitive) with the L25-GFP construct and isolate those mutants that are defective in ribosomal export under restrictive conditions followed by a permissive growth pulse.
In summary, a novel assay based on the intracellular location of L25-GFP allowed us to monitor whether ribosomes are efficiently exported into the cytoplasm. This revealed that the Ran-GTP/GDP cycle and distinct nucleoporins participate in ribosomal export. The identification of new and specific factors mediating ribosomal export should become possible using this assay.
Acknowledgments
We would like to thank Dr. H. Santos-Rosa for providing the strains CHRS 52 and CHRS 1d, Dr. P. Silver (Dana-Farber-Institute, Boston, MA) for providing the pse1-1/kap123::HIS3 strain, Dr. M. Aebi (ETH Zürich, Switzerland) for prp20-1, Dr. A. Hopper (Pennsylvania State University, College of Medicine, Hershey, PA) for rna1-1, Dr. K. Weis (UCSF, San Francisco, CA) for xpo1-1, and Elisabeth Petfalski and Karina Deinert for expert technical assistance. The plasmids expressing NLS-GFP-lacZ, GAL::YRB4ΔN, and GAL-KAP95 were kindly provided by Dr. G. Schlenstedt (University of Homburg, Germany).
Abbreviations used in this paper
- GFP
green fluorescent protein
- NES
nuclear export signal
- NLS
nuclear localization sequence
- NPC
nuclear pore complex
- rRNA
ribosomal RNA
Footnotes
Address correspondence to Ed Hurt, Biochemie-Zentrum Heidelberg, Im Neuenheimer Feld 328, D-69120 Heidelberg, Germany. Tel.: 49 6221 54 41 73. Fax: 49 6221 54 43 69. E-mail: cg5@ix.urz.uni-heidelberg.de
E. Hurt was recipient of a grant from the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm “Funktionelle Architektur des Zellkerns”).
References
- Adam SA, Lobl TJ, Mitchell MA, Gerace L. Identification of specific binding proteins for a nuclear location sequence. Nature. 1989;337:276–279. doi: 10.1038/337276a0. [DOI] [PubMed] [Google Scholar]
- Aebi M, Clark MW, Vijayraghavan U, Abelson J. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1, which is involved in the control of chromosome condensation. Mol Gen Genet. 1990;224:72–80. doi: 10.1007/BF00259453. [DOI] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiaerequired for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Fleischmann M, Stagljar I, Cole CN, Aebi M. Nuclear PRP20 protein is required for mRNA export. EMBO (Eur Mol Biol Organ) J. 1993;12:233–241. doi: 10.1002/j.1460-2075.1993.tb05649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts G-J, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Madore S, Cullen BR. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dahlberg JE, Lund E. Diverse effects of the guanine nucleotide exchange factor RCC1 on RNA transport. Science. 1995;267:1807–1810. doi: 10.1126/science.7534442. [DOI] [PubMed] [Google Scholar]
- Clark MW, Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987;105:1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Protein import into the nucleus. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt EC. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly (A)+RNA transport and nuclear pore distribution. EMBO (Eur Mol Biol Organ) J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Hurt EC, Aebi U, Panté N. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J Cell Biol. 1998;143:577–588. doi: 10.1083/jcb.143.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr CM, Akin D. The permeability of the nuclear envelope in dividing and nondividing cell cultures. J Cell Biol. 1990;111:1–8. doi: 10.1083/jcb.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr C, Akin D, Moore MS. The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J Cell Sci. 1998;111:1889–1896. doi: 10.1242/jcs.111.13.1889. [DOI] [PubMed] [Google Scholar]
- Fischer U, Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. Transport into and out of the cell nucleus. EMBO (Eur Mol Biol Organ) J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Hamm J, Mattaj IW. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Hassel I, Cézanne V, Treviño C, Schlatter H, Romero-Matuschek I, Schmidt A, Fasold H. Export of ribosomal subunits from resealed rat liver nuclear envelopes. Eur J Biochem. 1996;241:32–37. doi: 10.1111/j.1432-1033.1996.0032t.x. [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Künzler M, Hurt EC, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6364–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Banks F, Evangelidis V. A yeast mutant which accumulates precursor tRNAs. Cell. 1978;14:211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Imamoto N, Tachibana T, Matsubae M, Yoneda Y. A karyophilic protein forms a stable complex with cytoplasmatic components prior to the nuclear pore binding. J Biol Chem. 1995;270:8559–8565. doi: 10.1074/jbc.270.15.8559. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Zhao Y, Tartakoff AM. A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc Natl Acad Sci USA. 1992;89:2312–2316. doi: 10.1073/pnas.89.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Goldfarb D, Spitz LM, Tartakoff AM, Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO (Eur Mol Biol Organ) J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Gupta A, Ware VC. Nucleocytoplasmic transport of ribosomes in a eukaryotic system: is there a facilitated transport process? . Proc Natl Acad Sci USA. 1989;86:1791–1795. doi: 10.1073/pnas.86.6.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Silver PA. A GTPase controlling nuclear trafficking: running the right way or walking RANdomly? . Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T, Planta RJ, Knop JM. The course of the assembly of ribosomal subunits. Biochim Biophys Acta. 1978;517:378–389. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- Léger-Silvestre I, Noaillac-Depeyre J, Faubladier M, Gas N. Structural and functional analysis of the nucleolus of the fission yeast Schizosaccharomyces pombe. . Eur J Cell Biol. 1997;72:13–23. [PubMed] [Google Scholar]
- Maniatis, T., E.T. Fritsch, and J. Sambrook. 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Michael WM, Choi MY, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Moreland RB, Nam HG, Hereford LM, Fried HM. Identification of a nuclear localization signal of a yeast ribosomal protein. Proc Natl Acad Sci USA. 1985;82:6561–6565. doi: 10.1073/pnas.82.19.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Fabre E, Dihlmann S, Herth W, Hurt EC. Analysis of nucleo-cytoplasmic transport in a thermosensitive mutant of the nuclear pore protein NSP1. Eur J Cell Biol. 1993;62:1–12. [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum JS, Blobel G. A distinct and parallel pathway for nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrywka NJ, Goldfarb DS. Nuclear export pathways of tRNA and 40 S ribosomes include both common and specific intermediates. J Biol Chem. 1995;270:3619–3624. doi: 10.1074/jbc.270.8.3619. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Rutgers CA, Schaap PJ, Van't Riet J, Woldringh CL, Raué HA. In vivo and in vitro of structure-function relationships in ribosomal protein L25 from Saccharomyces cerevisiae. . Biochim Biophys Acta. 1990;1050:74–79. doi: 10.1016/0167-4781(90)90144-q. [DOI] [PubMed] [Google Scholar]
- Schaap PJ, Van't Riet J, Woldringh CL, Raué HA. Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. . J Mol Biol. 1991;221:225–237. doi: 10.1016/0022-2836(91)80216-h. [DOI] [PubMed] [Google Scholar]
- Scheer U, Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Saavedra C, Loeb JDJ, Cole CN, Silver PA. The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and appearance of poly(A)+RNA in the cytoplasm. Proc Natl Acad Sci USA. 1995;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff FR. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO (Eur Mol Biol Organ) J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M, Silver PA. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Hurt EC. Mex67p which is an essential factor for nuclear mrna export binds to both poly(A)+RNA and nuclear pores. EMBO (Eur Mol Biol Organ) J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov AV, Mann M, Hurt EC. Mtr10p functions as a nuclear import receptor for the mRNA binding protein Npl3p. EMBO (Eur Mol Biol Organ) J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Silver P, Sadler I, Osborne MA. Yeast proteins that recognize nuclear localization sequences. J Cell Biol. 1989;109:983–989. doi: 10.1083/jcb.109.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. The yeast protein Arc1p binds to tRNA and functions as a cofactor for methionyl- and glutamyl-tRNA synthetases. EMBO (Eur Mol Biol Organ) J. 1996a;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO (Eur Mol Biol Organ) J. 1996b;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Björkroth B, Lamb MM, Daneholt B. Visualization of the formation and transport of a specific hnRNP particle. Cell. 1983;34:847–855. doi: 10.1016/0092-8674(83)90542-1. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO (Eur Mol Biol Organ) J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. trans-acting factors in ribosome synthesis. Exp Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Jansen RP, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Trapman J, Retèl J, Planta RJ. Ribosomal precursor particles from yeast. Exp Cell Res. 1975;90:95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- Unwin PN, Milligan RA. A large particle associated with the perimeter of the nuclear pore complex. J Cell Biol. 1982;93:63–75. doi: 10.1083/jcb.93.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. Synthesis of ribosomes in Saccharomyces cerevisiae. . Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990;2:521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Wong DH, Corbett AH, Kent HM, Stewart M, Silver PA. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL., Jr The structure and biogenesis of yeast ribosomes. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]