Abstract
Drosophila affords a genetically well-defined system to study apoptosis in vivo. It offers a powerful extension to in vitro models that have implicated a requirement for cytochrome c in caspase activation and apoptosis. We found that an overt alteration in cytochrome c anticipates programmed cell death (PCD) in Drosophila tissues, occurring at a time that considerably precedes other known indicators of apoptosis. The altered configuration is manifested by display of an otherwise hidden epitope and occurs without release of the protein into the cytosol. Conditional expression of the Drosophila death activators, reaper or grim, provoked apoptogenic cytochrome c display and, surprisingly, caspase activity was necessary and sufficient to induce this alteration. In cell-free studies, cytosolic caspase activation was triggered by mitochondria from apoptotic cells but identical preparations from healthy cells were inactive. Our observations provide compelling validation of an early role for altered cytochrome c in PCD and suggest propagation of apoptotic physiology through reciprocal, feed-forward amplification involving cytochrome c and caspases.
Keywords: apoptosis, programmed cell death, cytochrome c, Drosophila
Apoptosis, a genetically mediated suicide mechanism, is associated with distinctive morphological changes including membrane blebbing, nuclear condensation, chromatin aggregation, and formation of apoptotic bodies (Kerr et al., 1972; Wyllie et al., 1980). It is the predominant form of cell death associated with normal cell turnover and is also the basis of cell killing by some genotoxic agents (reviewed in Thompson, 1995; Smith and Fornace, 1996; Jacobson et al., 1997). Misregulated apoptosis is also implicated in a variety of human diseases including AIDS, cancer, and neurodegenerative disorders (reviewed in Hengartner, 1997; Rudin and Thompson, 1997).
The role of mitochondria and cytochrome c in apoptosis has drawn considerable attention from recent studies of in vitro models. Cytochrome c is an essential component of the mitochondrial respiratory chain. It is a soluble protein, localized in the intermembrane space, and is loosely attached to the surface of the inner mitochondrial membrane (Gonzales and Neupert, 1990). It has been observed that cytochrome c is released from mitochondria into the cytosol of cultured cells undergoing apoptosis, resulting in the activation of cysteine proteases, termed caspases (Liu et al., 1996; Kluck et al., 1997a; Reed, 1997a; Yang et al., 1997). Diverse stimuli which cause apoptosis result in the activation of caspases which cleave a variety of substrates including structural components, regulatory proteins, and other caspases (reviewed in Cohen, 1997; Nicholson and Thornberry, 1997; Salvesen and Dixit, 1997; Villa et al., 1997; Cryns and Yuan, 1998). More recent studies have shown that microinjection of cytochrome c is sufficient to induce apoptosis (Zhivotovsky et al., 1998) and that reconstitution of a purified caspase-3 activation complex requires cytochrome c as an obligate binding factor (Li et al., 1997). In addition, several reports suggest that bcl-2, located primarily on the outer membrane of mitochondria (Monaghan et al., 1992), may block apoptosis by preventing the release of cytochrome c from mitochondria (Kluck et al., 1997a; Yang et al., 1997). While several compelling lines of evidence favor an important role for cytochrome c in apoptosis, our knowledge in this area comes solely from in vitro models or cultured cells, where apoptosis is experimentally induced (reviewed in Reed, 1997a).
Drosophila melanogaster affords a powerful genetic context to investigate details of cellular and molecular events in apoptosis which otherwise remain obscure (reviewed in Rodriguez et al., 1998). In this animal model, we detected cytological changes associated with cytochrome c that precede overt signs of apoptosis, both in live tissues and in cultured cells. In late-stage egg chambers, nurse cells transfer their cytoplasmic contents to the developing oocytes and subsequently undergo apoptotic cell death (King, 1960; Tourmente et al., 1990; Foley and Cooley, 1998; McCall and Steller, 1998). Cytoplasmic dumping and the eventual demise of the nurse cells occurs through reproducible stages that provide an excellent model for the analysis of pre-apoptotic events in identifiable cells. We examined the status of cytochrome c in vivo using this model of programmed cell death. Our studies revealed that a pronounced change in this protein, evidenced by the exposure of an otherwise hidden epitope (i.e., apoptogenic display of cytochrome c), precedes overt signs of apoptosis. Display of a new cytochrome c epitope was highly specific for pre-apoptotic cells, and did not occur in cells destined to survive.
A similar alteration associated with cytochrome c was detected in the cultured Drosophila cells expressing the known apoptotic activators reaper (rpr)1 (White et al., 1994) or grim (Chen et al., 1996b). Consistent with our studies of ovarian tissues, the altered form of cytochrome c was specific for apoptotic cells and was not detected in a variety of controls. Moreover, this change was specific for apoptotic death because altered cytochrome c did not occur in cells killed by toxic challenge. However, in contrast to reports from studies of mammalian cells, the apoptogenic form of cytochrome c was not released from mitochondria during apoptosis, but instead remained localized to this organelle (as evidenced by cell fractionation and cytology). Since apoptosis induced by rpr and grim is caspase dependent (Chen et al., 1996b; Nordstrom et al., 1996; Pronk et al., 1996; White et al., 1996; Fraser et al., 1997; Kondo et al., 1997) we also examined the influence of caspase function upon the altered display of cytochrome c. The viral caspase inhibitor, p35, and peptide-based caspase inhibitors completely blocked display of the apoptogenic cytochrome c epitope and, conversely, expression of an activated Drosophila caspase was sufficient to trigger apoptogenic changes in cytochrome c. Finally, in cell-free studies, we found that mitochondria isolated from apoptotic cells could promote caspase activation whereas identical preparations from healthy cells did not.
Taken together, we demonstrate that the appearance of an altered form of cytochrome c selectively precedes the programmed death of cells in an intact, developing organ. This pre-apoptotic alteration uncovers an otherwise hidden epitope and was provoked in a caspase-dependent manner by the death activators, rpr or grim. In light of evidence that mitochondrial components such as cytochrome c can promote caspase activation in vitro (Liu et al., 1996; Kluck et al., 1997a; Zou et al., 1997), our data implicate a feed-forward amplification circuit involving an apoptogenic form of cytochrome c and caspase activation.
Materials and Methods
Anti–cytochrome c mAbs
The mouse anti–rat cytochrome c mAbs used in this study have been described (Goshorn et al., 1991; Mueller and Jemmerson, 1996). They were purified from ascites by affinity chromatography using rabbit cytochrome c–coupled Sepharose beads as in Urbanski and Margoliash (1977). Cytochrome c (Sigma Chemical Co.) was coupled to cyanogen bromide–activated Sepharose 4B (Sigma Chemical Co.).
Anti–cytochrome c mAb Staining on Ovaries
Ovaries were dissected into 1× PBS. Tips of the ovariole were separated from one another to facilitate adequate penetration of the fixative and mAbs. Fixation was done by agitating the ovaries in a mixture of 1 vol 4% paraformaldehyde in 1× PBT (1× PBS + 0.1% Tween 20) and 3 vol of heptane for 30 min at room temperature. Samples were then washed four times in 1× PBT for 10 min each, then preincubated for 1 h in 1× PBTB (PBT + 1.5% BSA) at room temperature. The ovaries were incubated overnight at 4°C with the mouse anti–cytochrome c mAb (22 μg/ml) in 1× PBTB, washed four times in 1× PBTB followed by a 2-h incubation in 1:200 dilution of goat anti–mouse IgG-FITC (Jackson ImmunoResearch Laboratories) in 1× PBTB, washed three times in 1× PBT, and mounted in 50% glycerol in 1× PBS.
Terminal Transferase Method (TUNEL) for Detection of Apoptotic Nuclei
Hand-dissected ovaries were fixed as before and washed four times in 1× PBT.
Terminal Transferase Reaction.
Buffer (1× TdT) (Boehringer Mannheim Corp.) was mixed with 2.5 mM COCl2, 60 μM biotin-16dUTP, 200 nM dNTPs, and 15 U TdT to a final reaction volume of 50 μl. The ovaries were incubated in this mixture for 3 h at 37°C, rinsed and washed four times, 10 min each in 1× PBT.
Detection.
The ovaries were preincubated in 1× PBTB for 1 h at room temperature. Incorporated biotin-dUTP was detected by incubation in streptavidin-TRITC (Jackson ImmunoResearch Laboratories) in 1× PBTB (1:1,000) at room temperature in the dark for 2 h. Ovaries were mounted in 50% glycerol in 1× PBS and viewed under the rhodamine channel.
Rhodamine-phalloidin Staining
Rhodamine-phalloidin (Molecular Probes Inc.) was diluted (1:200) in 1× PBTB. Egg chambers were incubated in the solution for 20 min at room temperature in the dark, and then rinsed in 1× PBT. They were then mounted in 50% glycerol/50% 1× PBS.
Cell Culture and Treatments
Mt-rpr and Mt-grim are expression vectors that permit conditional expression of reaper or grim, respectively, in cell culture (Chen et al., 1996a,b, 1998; Nordstrom et al., 1996; Bose et al., 1998). The genes are placed downstream of the Drosophila metallothionein promoter and conditional expression in Schneider L2 (SL2) cells (Schneider, 1972) can be induced either in stably or transiently transfected cells. After transfection of Mt-rpr and Mt-grim, alone or in combination with the pMt-p35 plasmid (Nordstrom et al., 1996), induction was achieved by exposing the cells to 700 μM CuSO4.
Transient expression assays were done as in Chen et al. (1996a) and Nordstrom et al. (1996). Typical transfection efficiencies ranged from 40 to 60%. 48 h after transfection, cells from each well were split into two wells, and copper was added to one of the two wells. For ceramide treatment, SL2 cells were plated at 1 million/ml and incubated with 25 μM C2-ceramide (Biomol) for up to 24 h before being processed for antibody staining. Under these conditions, ∼50% cells were killed as measured by trypan blue exclusion (Pronk et al., 1996).
Construction of pMt-(Δ1–33)-dcp-1
A forward primer (5′ATCAGGGAGCTCGGATCCATATGGCCAAGGGCTGTACGCCG3′) and a reverse primer (5′GGGGTACCGTCGACTAATGATGATGATGATGATGGGATCCGCCAGCCTTATTGCCGTTCGG3′) were used together with a dcp-1 cDNA to produce a PCR product. This fragment was gel purified, digested with the appropriate enzymes (Sac1 and Kpn1), and then ligated to a Sac1/Kpn1 digested pRmHa.3 vector (Bunch et al., 1988). The resulting plasmid, pMt-(Δ1– 33)-dcp-1, expresses a truncated version of dcp-1 that is deleted for the prodomain (residues 1–33). Transient transfection was done as described above.
Anti–cytochrome c mAb Staining of Cultured Cells
Cells were plated at 25°C on coverslips, at a density of 106 cells/ml, and the following day apoptosis was induced. After an appropriate time of incubation in CuSO4 (∼6 h for rpr-transfected cells and ∼2.5 h for grim-transfected cells), cells were washed with 0.1 M sodium phosphate buffer (pH 7.5). Cells were then fixed at room temperature for 20 min with 3% paraformaldehyde in PBS containing 3 mM each of trinitrophenol, KCl, and MgCl2. Cells were then washed in PBS and incubated in 100 mM NH4Cl in PBS for 10 min. After washing three times with PBS for 5 min each, cells were permeabilized with 0.1% Triton X-100 for 5 min at room temperature.
Cells were then sequentially incubated with 0.8% BSA (Sigma Chemical Co.) at room temperature for 30 min, anti–cytochrome c mAb (22 μg/ml in 0.8% BSA) overnight at 4°C, and finally with 20 μg/ml goat anti–mouse Ig G-FITC (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Cells were washed twice with 0.4% BSA, twice with 0.1 M sodium phosphate buffer, and once in distilled water and mounted in fluoromount-G (Fisher Scientific Co.) containing 2.5% 1,4-diazabicyclo- (2.2.2)octane (DABCO; Sigma Chemical Co.). Photomicrographs were recorded on conventional and confocal microscopes using appropriate filters.
Rhodamine 123 Staining
Cells stably transfected with pMt-grim were induced for apoptosis as described above. After 2.5 h, samples were incubated in 50 nM rhodamine 123 for 15 min at 37°C to label mitochondria (Dumas et al., 1995). Cells were then rinsed twice in 1× PBS, fixed, and stained as described above.
Use of Caspase Inhibitors
Cells transfected stably with pMt-rpr and pMt-grim were treated with 50 μM each of caspase inhibitors DEVD-fmk and ZVAD-fmk (both purchased from Calbiochem). After appropriate times, induced and control cells were stained with anti–cytochrome c mAb as described before.
Immunoprecipitation
Immunoprecipitation was performed using protein A/G PLUS–Agarose (Santa Cruz Biotechnology) according to the manufacturer's instructions with a few modifications. Briefly, SL2 cells and apoptotic grim-expressing cells were pelleted, and lysed in lysis buffer (25 mM Hepes-KOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, 1 mM dithiothreitol, 5% glycerol, 0.1% Triton X-100, supplemented with 5 μg/ml pepstatin, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 0.1 mM PMSF) on ice for 30 min at a protein concentration of 0.5 mg/ml. The lysates were centrifuged at 3,000 g for 10 min, and 1 ml supernatant from each sample was precleared by incubating with 2 μg normal mouse IgG and 60 μl agarose conjugate for 30 min. Then 0.5 ml of precleared lysates was incubated with 2 μg anti–cytochrome c mAb for 2 h before 30 μl of agarose beads was added, and rotated at 4°C overnight. The agarose beads were washed three times with lysis buffer, resuspended in 30 μl 2× SDS sample buffer, resolved on a 15% polyacrylamide gel, transferred to PVDF membrane, and probed with anti–cytochrome c mAb, clone 7H8.2C12.
Subcellular Fractionation and Caspase Activity Assay
Subcellular fractionation was done as described (Yang et al., 1997). 8 × 107 cells were plated and, after 18 h, induced with copper as above. Cells were harvested 4 h later by centrifugation at 3,800 g for 5 min at room temperature. The pellets were washed once with ice-cold PBS and resuspended with 5 vol of buffer A (20 mM Hepes-KOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, 1 mM dithiothreitol, supplemented with 5 μg/ml pepstatin, 10 μg/ml leupeptin, 2 μg/ ml aprotinin, 0.1 mM PMSF) containing 250 mM sucrose for 15 min. Then cells were homogenized with 10 strokes in a Teflon homogenizer, and the homogenates were spun twice at 750 g for 10 min at 4°C. The supernatants (referred to as total cell lysates) were centrifuged at 10,000 g for 15 min at 4°C, and the resulting heavy membrane pellets were washed three times with cold buffer A containing sucrose, resuspended in the same buffer and stored at −80°C. The supernatants of the 10,000 spin were further centrifuged at 100,000 g for 1 h at 4°C, and the resulting supernatants (S-100) were aliquoted and stored at −80°C until use.
16 μg of S-100 cytosol from indicated cells was incubated with 150 ng of poly ADP-ribose polymerase (PARP) (Biomol) in a total volume of 25 μl in buffer A containing 250 mM sucrose for 1 h at room temperature. The reactions were stopped by adding 10 μl of 4× SDS sample buffer. PARP was resolved on an 8% SDS polyacrylamide gel, transferred to a PVDF membrane, and probed with an anti-PARP mAb (C-2-10 from Biomol). For mixing experiments, 20 μg of heavy membrane fractions from indicated cells were incubated with SL2 cytosol for 1 h at room temperature, then the heavy membrane fractions were spun down, and the supernatants were used for PARP cleavage activity as above. To ensure that caspase activation associated with mitochondrial fractions did not result from contaminating cytosolic components, heavy membranes were washed in a 20-fold excess volume of buffer A (with sucrose) at least five times. Also, in parallel experiments, we found that mitochondrial fractions from pre-apoptotic cells retained activity even after exposure to 50 μM irreversible caspase-inhibitor peptides (DEVD-fmk or zVAD-fmk) before washing.
Results
Immunodetection of Drosophila Cytochrome c
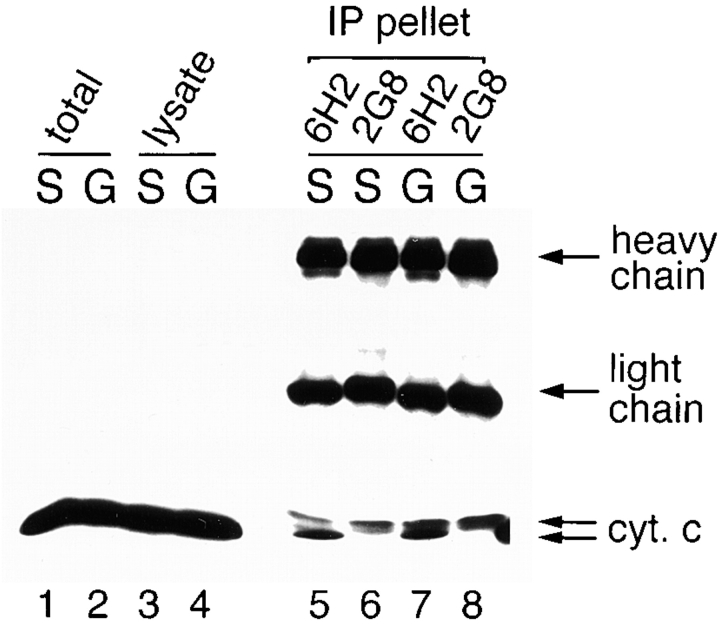
Although two cytochrome c genes, DC4 and DC3, have been described in Drosophila melanogaster (Limbach and Wu, 1985) studies at the level of protein (Inoue et al., 1986) and at the level of RNA (Limbach and Wu, 1985) suggest that DC4, which shows >86% identity with its rat counterpart, is either the predominant or only form of actively expressed product. We screened an existing panel of mAbs, directed against mammalian versions of cytochrome c, as possible probes for in situ analyses of the fly counterpart. The potential utility of these mAbs was assessed by immunoprecipitations of SL2 cell lysates, probed with a third anti–cytochrome c mAb, 7H8, to detect denatured cytochrome c in Western blots. The results in Fig. 1 show that two mAbs, 6H2 (Goshorn et al., 1991) and 2G8 (Mueller and Jemmerson, 1996), recognized Drosophila cytochrome c. Both antibodies detected a doublet that comigrated with mammalian cytochrome c at ∼13 kD. While mAb 2G8 preferentially precipitated the upper band, mAb 6H2 had about equal affinity for both forms of cytochrome c. No obvious correlation between the relative abundance of the two cytochrome c bands and apoptosis was observed (see below). The same banding pattern was also observed when immunoprecipitation was performed using purified Drosophila cytochrome c (Liu, J., and R. Jemmerson, unpublished observation).
Figure 1.
Immunoprecipita-tion of Drosophila cytochrome c. Western blot analyses were used to visualize immunoprecipitated proteins that were probed with anti–cytochrome c mAb 7H8. Lanes 1 and 2 show whole cell suspension of healthy SL2 cells (S) and grim-expressing cells 4 h after induction (G). Lanes 3 and 4 were precleared lysates used for immunoprecipitation (see Materials and Methods). Lanes 5–8 were immunoprecipitates from SL2 or grim-expressing cell lysates using two anti–cytochrome c mAbs, 6H2 and 2G8. The same two cytochrome c bands were also observed in lanes 1–4 on shorter exposures.
These bands clearly represent distinct cytochrome c species since they were immunoprecipitated with mAbs to the native protein, and were detected by Western blot using a different mAb that recognizes a distinct epitope on the denatured protein. While it is formally possible that these two bands correspond to the polypeptides encoded by the Drosophila cytochrome c genes, DC-3 and DC-4, earlier studies suggest that only DC-4 encodes a functional polypeptide (Inoue et al., 1986) and furthermore, the expected Drosophila cytochrome c products (DC-3 and DC-4) differ by only three amino acids, which is not likely to be resolved in our gels. Although the biochemical nature of these different forms is unresolved, our data indicate that mAb 2G8 preferentially recognized the higher molecular weight form of the doublet. This product occurs in relatively small amounts that were not overtly affected by the extent of apoptosis in the cultures (Fig. 1).
Altered Cytochrome c Anticipates Programmed Cell Death
Between stages 11 and 13 of Drosophila oogenesis, programmed cell death eliminates nurse cells which nourish the developing egg (Sang and King, 1959; Foley and Cooley, 1998; McCall and Steller, 1998). The apoptotic nature of nurse cell death is indicated by two distinct markers, acridine orange staining (Abrams et al., 1993) and TUNEL labeling (Gavrieli et al., 1992). These readily identifiable cells offer a unique opportunity to examine pre-apoptotic events before their eventual demise.
To directly demonstrate the involvement of cytochrome c during apoptosis, Drosophila ovaries were stained with the anti–cytochrome c mAbs described above and egg chambers at all stages of development were analyzed. In preliminary experiments, mAb 2G8 detected cytochrome c in apoptotic cells prompting extensive studies with this mAb. As shown in Fig. 2, A and B, only nurse cells at stage 10B exhibited pronounced cytochrome c immunoreactivity distributed as characteristically punctate labeling of the cytoplasm in a pattern consistent with localization to mitochondria (see Fig. 4 B). Such staining was not seen in nurse cells before this stage, nor was this overt immunoreactivity seen in any other cell type of the Drosophila egg chamber. Moreover, this staining was specific for cytochrome c immunoreactivity because no labeling of nurse cells (Fig. 2 C) was observed if the antibody was first preadsorbed with cytochrome c covalently bound to Sepharose 4B (Urbanski and Margoliash, 1977).
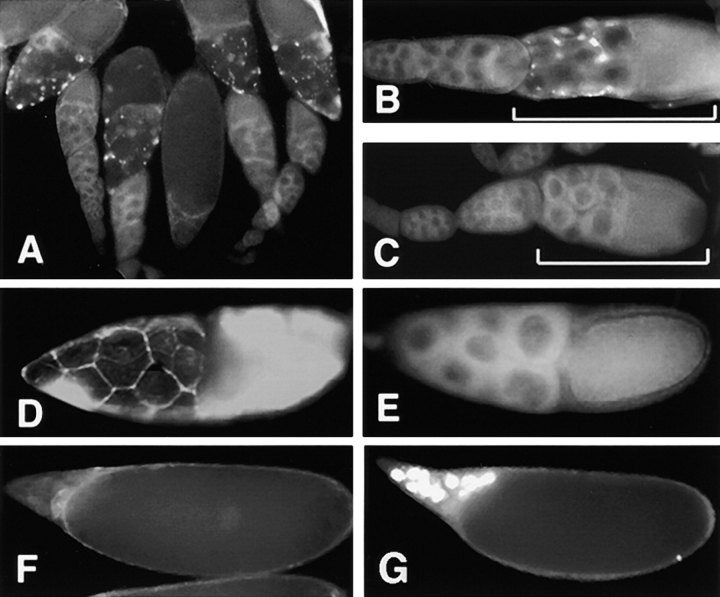
Figure 2.
Labeling for cytochrome c precedes programmed cell death in ovaries. Drosophila egg chambers were examined with mAb 2G8. (A) The mAb labels cytochrome c specifically in nurse cells at stage 10B. Characteristically punctate staining is found throughout the nurse cell cytoplasm only at this stage. (B) Single egg chamber magnified (brackets denote stage 10B egg chambers). Note that earlier stages are not stained. (C) With mAb 2G8 preadsorbed on cytochrome c–Sepharose 4B, this stage did not exhibit labeling. Cytoplasmic and nuclear integrity of nurse cells at this stage was substantiated by the cytoplasmic marker, rhodamine phalloidin, and nuclear staining technique, TUNEL (D and E), respectively (see text). Apoptotic nuclei, labeled by TUNEL, are evident at a later stage in oogenesis (G), when no cytochrome c labeling could be seen (F).
Figure 4.

Distribution of Drosophila cytochrome c during apoptosis. (A) Healthy SL2 cells and apoptotic rpr- or grim-expressing cells were fractionated 4 h after induction (see Materials and Methods). Cytochrome c in the total cell lysates (lanes 1–3), heavy membrane fractions (lanes 4–6), and S-100 cytosol (lanes 7–9) was visualized by Western blot analyses using mAb 7H8. Note that cytochrome c efflux into the cytosol of apoptotic cells was not observed, and that cytochrome c was substantially enriched in heavy membrane fractions which contain mitochondria. (B) Two separate panels of confocal immunofluorescence microscopy images showing the colocalization of mitochondria and 2G8 immunoreactivity. Cells were incubated in 50 nM rhodamine 123 before fixing (see Materials and Methods) and viewed for rhodamine and fluorescein. Coincidence is indicated by absence of green only labeling. (C) High magnification view of a single cell showing punctate 2G8 immunoreactivity, visualized by conventional microscopy.
To determine the chronology of cytochrome c display relative to other apoptotic changes, we compared the onset of mAb binding with other degenerative changes known to occur in these cells. Though nurse cells at stage 10B showed pronounced exposure of the epitope for mAb 2G8 (Fig. 2 B) no signs of apoptosis were apparent in either the cytoplasm or the nuclei of these cells (Fig. 2, D and E). Egg chambers were stained with rhodamine-conjugated phalloidin which detects filamentous actin at the cortices of the nurse cells. As shown in Fig. 2 D, this marker substantiates the integrity of the cytoplasmic membrane and shows that nurse cell dumping has not yet begun at a time when cytochrome c labeling is already very conspicuous. Similarly the nuclei of nurse cells at stage 10B were negative for TUNEL labeling (Fig. 2 E), a method that detects a nuclear hallmark of apoptosis. By stages 12–13 (at least 0.5 to 4 h later) the nuclei of the dying nurse cells adopt characteristic apoptotic features as evidenced by the TUNEL assay (Fig. 2 G) and acridine orange staining (data not shown). These results demonstrate that cytochrome c display precedes overt signs of apoptosis in intact organs.
Altered Cytochrome c Is Provoked by Apoptosis Activators
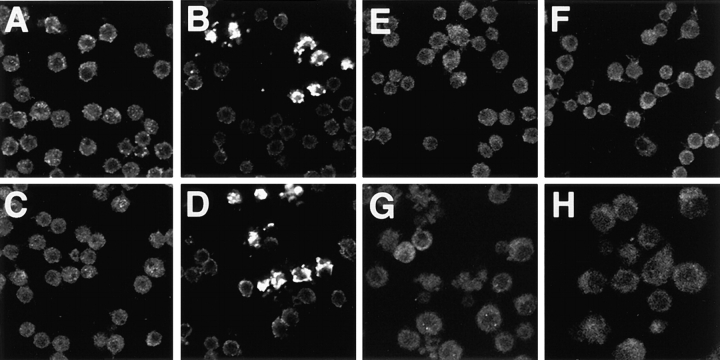
Previous studies on Drosophila SL2 cells have shown that conditional expression of rpr or grim triggers apoptosis in cultured cells and in transgenic animals (White et al., 1994, 1996; Chen et al., 1996b; Nordstrom et al., 1996). Transiently transfected SL2 cells were induced for rpr or grim and, at various time intervals after induction, the preparations were examined for cytochrome c immunoreactivity with mAb 2G8. As seen in Fig. 3, apoptotic cultures (Fig. 3, B and D) exhibited profound staining with the antibody. In contrast, uninduced healthy cells (Fig. 3, A and C) and cells transfected with the vector (pMt) alone in the absence or presence of the inducing agent copper (Fig. 3, E and F) showed no signs of cytochrome c display. As before, mAb 2G8 preadsorbed with cytochrome c–Sepharose 4B (Fig. 3, F and H) validated specificity of immunoreactivity for cytochrome c. It should also be emphasized that routine labeling of apoptotic SL2 cells (or egg chambers) with a variety of antibodies directed against different antigens does not behave like mAb 2G8, and we can readily exclude the possibility that the pre-apoptotic condition somehow promotes enhanced immunodetection.
Figure 3.
Altered cytochrome c display is provoked by expression of the apoptosis activators, rpr or grim. L2 cells were stably transfected with plasmids expressing rpr (A and B) and grim (C and D) driven by the metallothionein promoter. Upon activation of this conditional promoter by copper (B and D, respectively) cells exhibited pronounced labeling with mAb 2G8 and underwent apoptosis. Uninduced rpr (A) and grim (C) cells, or cells transfected with pMTAL vector alone uninduced (E) or induced (F) did not exhibit apoptotic morphology nor did they exhibit staining with the antibody. Specificity of this staining was further substantiated when rpr (G) and grim (H) expressing cells were stained with preadsorbed 2G8 mAb in parallel and did not exhibit cytochrome c immunoreactivity. The punctate and relatively large dimensions associated with immunoreactivity might reflect aggregations of mitochondria during apoptosis (see Li et al., 1998).
Cytochrome c Is Retained in Mitochondria during Apoptosis
To test the possibility that cytochrome c might be released into the cytosol during apoptosis, we fractionated healthy SL2 cells and apoptotic rpr- or grim-expressing cells, and assayed for cytochrome c in the mitochondrial (heavy membrane) and cytosolic fractions. Surprisingly, these cells showed no difference in cytochrome c distribution and we found no evidence for the transit of cytochrome c to the cytosol as a correlate to apoptosis (Fig. 4 A). It is worth mentioning that our experimental design is set up such that equal numbers of cells are plated ∼18 h before induction. Therefore, differences between parental and transfected samples in the levels of cytochrome c (in total and heavy membrane samples) result solely from differences in growth rate and cell number at the time of harvest (Bose et al., 1998). Note that reduced levels of immunogen detected (Fig. 4 A, lanes 5 and 6) are proportional to the reduced total levels of immunogen (Fig. 4 A, lanes 2 and 3). In addition, we should emphasize that nothing about our procedure would have prevented us from observing a genuine change in the subcellular location of this protein because in several preparations we could, in fact, cause cytochrome c to be released and detected in the cytosol simply by omitting sucrose from the preparation (see Materials and Methods).
Biochemical data indicating retention of cytochrome c in mitochondria during apoptosis is consistent with our cytological studies. For instance, under all circumstances the protein was compartmentalized in a punctate distribution which, in double labeling experiments, was coincident with the mitochondrial marker, rhodamine 123 (Fig. 4 B). These observations, together with our biochemical data, indicate that appreciable efflux of cytochrome c from mitochondria does not occur during apoptosis in Drosophila cells.
Mitochondria Isolated from Apoptotic Cells Trigger Caspase Activation In Vitro
Studies described in the previous section established that, although clearly altered, Drosophila cytochrome c is retained in the mitochondrial compartment during apoptosis. Since cell-free studies in vertebrate systems implicate mitochondrial factors (cytochrome c and/or other proteins) in the activation of some caspase enzymes (Liu et al., 1996; Zamzami et al., 1996; Evans et al., 1997; Kluck et al., 1997b; Kroemer et al., 1997) we sought to determine whether mitochondria from our insect model might exhibit similar properties. To test this possibility, we measured caspase activation in L2 cell cytosol that had been coincubated with mitochondria isolated from parental L2 cells or from pre-apoptotic cells (induced either for rpr or grim). Fig. 5 illustrates detection of caspase activation, as measured by signature cleavage of a bovine substrate, PARP. Cleavage of PARP in this assay (Bose et al., 1998) is indistinguishable from the signature activity reported in many mammalian systems (Lazebnik et al., 1994; Nicholson and Thornberry, 1997; Villa et al., 1997) and is readily detected in the cytosol of pre-apoptotic cells (Fig. 5, lane 2 induced for rpr; lane 3 induced for grim) but not in cytosol from parental L2 (Fig. 5, lane 1). Lanes 5 and 6 of Fig. 5 show that mitochondria isolated from rpr- or grim-expressing cells trigger the appearance of PARP cleavage activity in the otherwise silent L2 cell cytosol. In contrast, mitochondria from parental L2 cells did not provoke similar cleavage of PARP (Fig. 5, lane 4). These observations emphasize the importance of one or more mitochondrial factors in the activation of caspase function triggered by rpr or grim.
Figure 5.
Mitochondria from rpr- or grim-expressing cells promote caspase activation in vitro. Expression of rpr or grim triggers activation of caspase activity in the cytosol (S-100 fraction). Activity is assayed by cleavage of PARP and detection of a signature 85-kD fragment (lanes 2 and 3). PARP-cleavage activity is not detected in cytosolic preparations from parental SL2 cells (lane 1). After incubation with mitochondrial fractions from rpr- or grim-expressing cells (see Materials and Methods) the otherwise silent cytosolic S-100 fraction from SL2 cells now shows pronounced PARP cleavage activity (lanes 5 and 6). In contrast, mitochondria from nonapoptotic SL2 cells, tested in parallel, failed to promote caspase activation of the same cytosolic fraction (lane 4). L, SL2 cells; R, rpr-expressing cells; G, grim-expressing cells.
Altered Cytochrome c Display Is Blocked by Caspase Inhibitors In Vivo
The Drosophila death activators, rpr and grim, activate one or more caspases to elicit apoptosis (Chen et al., 1996b; Nordstrom et al., 1996; White et al., 1996; Fraser et al., 1997). To study the temporal relation of cytochrome c display with respect to caspase activity, we cotransfected SL2 cells with pMt-rpr and pMt-p35. 6 h after induction, cells induced for rpr alone showed pronounced labeling with mAb 2G8 (Fig. 6 A) whereas cells expressing rpr together with p35 were prevented from apoptosis and did not bind the mAb (Fig. 6 B). These observations suggested that apoptogenic cytochrome c display required caspase activity, a presumption that was further substantiated when rpr-expressing cells were treated with the peptide caspase inhibitors zDEVD-fmk and zVAD-fmk. As seen for p35-blocked cells, these inhibitors similarly prevented mAb 2G8 labeling and subsequent apoptosis (Fig. 6, C and D). Parallel results were observed in grim-expressing cells (not shown).
Figure 6.
p35 and peptide-based caspase inhibitors block altered cytochrome c display. Cells stably transfected and induced for Mt-rpr exhibited binding of mAb 2G8 as seen in A while cells cotransfected with Mt-rpr and Mt-p35 did not label with mAb 2G8 (B). Similarly, Mt-rpr cells induced with copper and treated with caspase inhibitors, DEVD-fmk (C) and zVAD-fmk (D), did not stain with mAb 2G8. Expression of rpr and p35 proteins in these cells was verified in separate studies.
Altered Cytochrome c Display Is Provoked by the Drosophila Caspase, dcp-1
The data above demonstrate that caspase activity is required for apoptogenic cytochrome c display. To determine if caspase function is sufficient to trigger this change, we induced apoptosis in SL2 cells by conditional expression of an activated version of the Drosophila caspase, dcp-1. If deleted for its prodomain, this caspase provokes considerable apoptosis in mammalian cells (Song et al., 1997) and SL2 cells (this paper). When labeled with mAb 2G8, cells transfected and induced for pMt-(Δ1–33)-dcp-1 expression exhibited profound punctate cytochrome c staining (Fig. 7 B) with features indistinguishable from those associated with expression of the death activators.
Figure 7.
Altered cytochrome c display is provoked by caspase action. Drosophila cells transiently transfected with pMt-(Δ1–33)- dcp-1, a vector which expresses dcp-1 lacking its prodomain, exhibited characteristically punctate labeling with mAb 2G8. B illustrates apoptogenic cytochrome c display induced by expression of the activated dcp-1 caspase. A shows uninduced control cells.
Discussion
Several compelling lines of evidence implicate a role for cytochrome c in apoptotic physiology. First, release of cytochrome c from mitochondria occurs as an early response to a variety of death stimuli, and this release is prevented by overexpression of bcl-2 (Kim et al., 1997; Kluck et al., 1997a; Kroemer et al., 1997; Reed, 1997b; Yang et al., 1997). Second, cytochrome c triggers activation of caspases in several in vitro models (Liu et al., 1996; Kluck et al., 1997a,b; Vaux, 1997; Yang et al., 1997; Zou et al., 1997). Third, direct injection of cytochrome c into cultured cells promotes death by apoptosis (Zhivotovsky et al., 1998). Finally, reconstitution of a purified caspase-3 activation complex was found to require cytochrome c as an obligate binding factor (Li et al., 1997).
We examined the status of cytochrome c during programmed cell death in an animal model. As part of this effort, we characterized mAbs directed against rat cytochrome c for binding Drosophila cytochrome c in situ. One mAb, 2G8, was of particular interest because it exhibited specific immunoreactivity to cytochrome c only in the mitochondria of cells that are committed to undergo apoptosis. For instance, pronounced labeling for cytochrome c was observed in mitochondria of cells expressing the apoptosis activators, rpr or grim, whereas healthy control cells were not labeled. Similar results were observed in intact ovarian tissues where cytochrome c display occurred specifically in nurse cells at a stage before any overt sign of apoptosis. To our knowledge, apoptogenic cytochrome c is the earliest reported molecular indicator of pre-apoptotic events in these cells and could reflect the point at which nurse cells become irreversibly committed to programmed cell death.
Although mAb 2G8 was originally produced against rat cytochrome c, we conclude that the immunoreactive signal corresponds to a specific configuration of the Drosophila counterpart for the following reasons. First, the immunogen for mAb 2G8 shares >86% identity to Drosophila cytochrome c (DC4) including the immunodominant aspartic acid at position 62 (Mueller and Jemmerson, 1996). Second, all labeling was abolished by pre-absorption with antigen. Therefore, mAb 2G8, rather than a contaminant, is responsible for the in situ signal we have observed. Third, in biochemical fractionation assays and immunofluorescence studies, the signal was localized to the mitochondrial compartment where cytochrome c resides. Fourth, mAb 2G8 immunoprecipitated products of the correct size (∼13 kD), which cross-reacted with a different cytochrome c antibody specific for a distinct epitope. These studies establish the ability of mAb 2G8 to recognize cytochrome c in crude fly cell extracts and were validated in similar assays showing that mAb 2G8 could immunoprecipitate purified Drosophila cytochrome c (not shown).
Selective labeling of pre-apoptotic cells by mAb 2G8 indicates that apoptotic signaling triggers specific alterations in the configuration of cytochrome c that uncover an otherwise hidden epitope. Moreover, routine labeling of apoptotic SL2 cells with a variety of antibodies directed against different antigens does not behave like mAb 2G8, and we can readily exclude the possibility that the pre-apoptotic condition somehow promotes enhanced immunodetection. Based on numerous precedents, exposure of an otherwise hidden epitope through immunodetection could pave the way for developing reagents that probe specific protein conformations (Kapoor et al., 1988; Cumber et al., 1991; Yewdell et al., 1993; Cebolla et al., 1996). In the case of fly cytochrome c, the precise nature of this change is not yet clear, but the alteration could reflect either a modified conformation and/or changes in associated proteins that permit immunoreactivity with the 2G8 antibody. In contrast to several mammalian models of apoptosis (Reed, 1997a), the Drosophila protein was not released to the cytosol during apoptosis but instead continued to cofractionate with mitochondria. Therefore, the fly version of this protein could be more tightly tethered to its resident organelle than its mammalian counterpart. Also, since no covalent differences between healthy and pre-apoptotic samples of cytochrome c have yet been detected (either in our system or in mammalian models), our studies implicate a conformational and/or contextual change in cytochrome c which specifically occurs in cells that are committed to apoptotic death.
One possibility is that antibody binding to the reactive epitope could be hindered by mitochondrial components in live cells. In cells committed to apoptosis, fly cytochrome c might translocate from the intermembrane space, but remain tethered to the outer membrane, and in the process, also expose the epitope for mAb 2G8. Except for the fact that the protein is not physically released, this scenario suggests features in common with mammalian models, where cytochrome c is liberated from mitochondria. Alternatively the polypeptide may simply undergo a conformational change detected by mAb 2G8 that does not affect its location. Consistent with this possibility, Jemmerson et al. (1999) have obtained evidence that mouse cytochrome c does assume a different conformation early in apoptosis of a T cell hybridoma. The altered conform was detected using yet a different mAb than the one used here, which does not recognize native cytochrome c but does recognize nonnative forms of the protein, such as large peptide fragments. These observations are entirely consistent with at least some mammalian cell models of apoptosis where cytochrome c remains in the mitochondria but is inactivated by death signals (Krippner et al., 1996; Adachi et al., 1997). While the precise nature of the change remains to be determined, the alteration is evidently specific for apoptotic forms of cell death. For instance, 2G8 immunostaining was not observed when SL2 cultures were killed by exposure to levels of ceramide that provoke a mode of cell death that is distinctly necrotic (see Materials and Methods).
We authenticated a role for cytochrome c in apoptosis by examining staged ovarian tissues where the programmed death of identifiable cell types is well described. In a single egg chamber, one oocyte is connected through cytoplasmic bridges to 15 sister nurse cells. Over a period of slightly >3 d, the egg chamber develops through a sequence of well-defined stages before reaching maturity at the final stage of oogenesis, stage 14. Between stages 10B and 13 of oogenesis, the nurse cells undergo considerable changes that include a reorganization of the actin cytoskeleton (Gutzeit, 1986; Cooley et al., 1992) and a permeabilization of nuclear membranes (Okada and Waddington, 1959; Giorgi and Deri, 1976; Cooley et al., 1992). These changes are thought to facilitate the transfer of nurse cell components into the developing oocyte (King, 1960; Tourmente et al., 1990) and once completed, this process is quickly followed by the programmed death of the nurse cells themselves (Sang and King, 1959). Dying nurse cells display physiological changes common to apoptotic cells, such as elevated caspase RNAs (Chen et al., 1998) and a requirement for caspase function (McCall and Steller, 1998). Similarly, diverse morphological criteria such as TUNEL labeling and acridine orange staining (Foley and Cooley, 1998; McCall and Steller, 1998; see Fig. 2) and ultrastructural features (Giorgi and Deri, 1976) all point toward an apoptotic mode of death for late staged nurse cells.
In our studies with the 2G8 antibody, only pre-apoptotic nurse cells were positive for cytochrome c staining. Furthermore, in double-label analyses the immunoreactive signal was detected long before overt signs of apoptosis were visualized either with a cytoplasmic marker (cortical actin visualized with phalloidin) or with a nuclear marker (TUNEL labeling). To our knowledge, these observations are the first demonstration that changes in cytochrome c actually anticipate programmed cell death within intact tissues and, since these observations occur in an invertebrate model, they also argue for widespread conservation of cytochrome c–associated functions in apoptotic physiology.
In Drosophila, at least three genes, rpr, grim, and hid function as potent activators of the apoptotic pathway and, collectively, these genes are required for cell death in the embryo (reviewed in Rodriguez et al., 1998). Generally, expression of each precedes programmed cell death, and each is sufficient to elicit apoptosis in cultured cells and in transgenic animals (White et al., 1994; Grether et al., 1995; Chen et al., 1996b). Work presented here offers a cytological demonstration that signaling by rpr and grim trigger events which ultimately engage cytochrome c and is consistent with recent observations that rpr can provoke cytochrome c release in heterologous extracts from Xenopus (Evans et al., 1997). The Drosophila death activators engage caspase function (reviewed in Rodriguez et al., 1998) and therefore we examined the effects of the viral caspase inhibitor, p35, and peptide-based inhibitors, ZVAD-fmk and DEVD-fmk, upon cytochrome c display. We and others had shown previously that caspase inhibitors prevent apoptosis induced by rpr and grim (Grether et al., 1995; Chen et al., 1996b; Nordstrom et al., 1996; Pronk et al., 1996; White et al., 1996) and, as demonstrated here, coexpression of p35 or incubation with anticaspase peptides similarly prevented immunodetection of cytochrome c that would otherwise occur in the presence of rpr or grim alone. These results demonstrate that display of apoptogenic cytochrome c requires caspase activity. To further test this idea, we examined the status of cytochrome c in cells where apoptosis was directly triggered by expression of an activated caspase, dcp-1. As expected, this protein promoted apoptogenic cytochrome c display, with features that were indistinguishable from labeling provoked by the death activators. Taken together, our data establish that caspase action is both necessary and sufficient to elicit pre-apoptotic alterations in cytochrome c.
Cell-free models of apoptosis from vertebrate sources have implicated an important role for mitochondrial factors in the activation of some caspase enzymes (Liu et al., 1996; Zamzami et al., 1996; Evans et al., 1997; Kluck et al., 1997b; Kroemer et al., 1997). The cell-free experiments presented here are consistent with these reports, and with our in vivo studies indicating that signaling by rpr and grim engage one or more resident mitochondrial proteins. Is the apoptogenic form of cytochrome c solely responsible for caspase activation in our cell-free system? We tested this possibility in two ways, both of which were inconclusive. First, we found that the 2G8 anti–cytochrome c antibody did not interfere with the ability of apoptotic mitochondria to provoke caspase activation and second, we found that purified cytochrome c (from Drosophila, horse, and rabbit) was not sufficient on its own to promote caspase activity when added to cytosol from L2 cells. These data suggest that the 2G8 mAb might recognize, but not inhibit, cytochrome c in association with other proteins as component of a preformed apoptosome mitochondrial complex (Green, 1998; Green and Reed, 1998; Hengartner, 1998). It is also entirely possible that other mitochondrial factors (perhaps a Drosophila equivalent of AIF; Kroemer et al., 1997, a mitochondrial caspase, or a novel molecule) might play important roles during caspase amplification in this system. Further studies will be required to address this issue.
Two potential explanations reconcile our in vivo observations on apoptogenic cytochrome c with reports from mammalian cell-free systems that cytochrome c can trigger caspase activation. One possibility is that the order and/or nature of cytochrome c apoptotic function is not conserved between mammals and insects and thus, relative to caspase action, cytochrome c is upstream in the former case and downstream in the latter case. This scenario, however, seems unlikely given the widespread conservation of apoptotic components, the fact that display of fly cytochrome c in the animal significantly precedes all signs of programmed cell death, and reports from mammalian systems that upstream caspases can trigger cytochrome c release (Reed, 1997a; Li et al., 1998; Luo et al., 1998; Scaffidi et al., 1998). Therefore, a more likely interpretation of our results is that cytochrome c propagates apoptotic physiology by functioning together with caspases in a feed-forward amplification loop. In this scenario, altered cytochrome c and caspase activity exert positive and reciprocal feedback upon each other, similar to observations recently reported for caspase 8 (Kuwana et al., 1998). Thus, agents that restrain caspase action (p35) are also predicted to suppress pro-apoptotic display of cytochrome c, which behaves as an amplifier of caspase function. This interpretation is also consistent with recent studies on Fas signaling in type II cells, where molecular ordering studies found that activation of an initiator caspase (caspase 8/Flice) occurs upstream of changes associated with cytochrome c (Scaffidi et al., 1998).
Acknowledgments
We thank J. Chapo, S.I. Ho, and other members of the Abrams lab for their assistance. We also thank Dr. E. Margoliash for the gift of purified Drosophila cytochrome c and J. Liu for performing immunoprecipitation and Western blots. We are grateful to Dr. H. Steller for the dcp-1 clone.
This work was supported by grants from the American Cancer Society (CB-80362) and from the National Institutes of Health (NIH) (AG12466) to J.M. Abrams and, in part, by a grant from the National Science Foundation to R. Jemmerson (MCB-9630412). P. Chen was supported, in part, by a postdoctoral fellowship from NIH (GM18215-02).
Abbreviations used in this paper
- PARP
poly ADP-ribose polymerase
- rpr
reaper
- SL2
Schneider L2 cells
References
- Abrams JM, White K, Fessler L, Steller H. Programmed cell death during Drosophilaembryogenesis. Development. 1993;117:29–44. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- Adachi S, Cross AR, Babior BM, Gottlieb RA. Bcl-2 and the outer mitochondrial membrane in the inactivation of cytochrome c during fas-mediated apoptosis. J Biol Chem. 1997;272:21878–21882. doi: 10.1074/jbc.272.35.21878. [DOI] [PubMed] [Google Scholar]
- Bose R, Chen P, Loconti A, Grullich C, Abrams JM, Kolesnick RN. Ceramide generation by the reaper protein is not blocked by the caspase inhibitor, p35. J Biol Chem. 1998;273:28852–28859. doi: 10.1074/jbc.273.44.28852. [DOI] [PubMed] [Google Scholar]
- Bunch TA, Grinblat Y, Goldstein LS. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogastercells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A, Guzman C, de Lorenzo V. Nondisruptive detection of activity of catabolic promoters of Pseudomonas putidawith an antigenic surface reporter system. Appl Environ Microbiol. 1996;62:214–220. doi: 10.1128/aem.62.1.214-220.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Lee P, Otto L, Abrams JM. Apoptotic activity of REAPER is distinct from signalling by the tumor necrosis factor receptor 1 death domain. J Biol Chem. 1996a;271:25735–25737. doi: 10.1074/jbc.271.42.25735. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. Grim, a novel cell death gene in Drosophila. . Genes Dev. 1996b;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Chen P, Rodriguez A, Erskine R, Thach T, Abrams JM. Dredd, a novel effector of the apoptosis activators reaper, grim, and hid in Drosophila. . Dev Biol. 1998;201:202–216. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Verheyen E, Ayers K. chickadee encodes a profilin required for intercellular transport during Drosophilaoogenesis. Cell. 1992;69:173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Cryns V, Yuan J. Proteases to die for. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- Cumber PM, Jacobs A, Hoy T, Whittaker JA, Tsuruo T, Padua RA. Increased drug accumulation ex vivo with cyclosporin in chronic lymphatic leukemia and its relationship to epitope masking of P-glycoprotein. Leukemia. 1991;5:1050–1053. [PubMed] [Google Scholar]
- Dumas M, Maftah A, Bonte F, Ratinaud MH, Meybeck A, Julien R. Flow cytometric analysis of human epidermal cell aging using two fluorescent mitochondrial probes. C R Acad Sci III. 1995;318:191–197. [PubMed] [Google Scholar]
- Evans EK, Kuwana T, Strum SL, Smith JJ, Newmeyer DD, Kornbluth S. Reaper-induced apoptosis in a vertebrate system. EMBO (Eur Mol Biol Organ) J. 1997;16:7372–7381. doi: 10.1093/emboj/16.24.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley K, Cooley L. Apoptosis in the late stage Drosophilanurse cells does not require genes within the H99 deficiency. Development. 1998;125:1075–1082. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- Fraser AG, McCarthy NJ, Evan GI. Drlce is an essential caspase required for apoptotic activity in Drosophilacells. EMBO (Eur Mol Biol Organ) J. 1997;16:6192–6199. doi: 10.1093/emboj/16.20.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi F, Deri P. Cell death in ovarian chambers of Drosophila melanogaster. . J Embryol Exp Morphol. 1976;35:521–533. [PubMed] [Google Scholar]
- Gonzales DH, Neupert W. Biogenesis of mitochondrial c-type cytochromes. J Bioenerg Biomembr. 1990;22:753–768. doi: 10.1007/BF00786929. [DOI] [PubMed] [Google Scholar]
- Goshorn SC, Retzel E, Jemmerson R. Common structural features among monoclonal antibodies binding the same antigenic region of cytochrome c. J Biol Chem. 1991;266:2134–2142. [PubMed] [Google Scholar]
- Green DR. Apoptotic pathways: the roads to ruin. Cell. 1998;94:695–698. doi: 10.1016/s0092-8674(00)81728-6. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogasterfunctions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Gutzeit HO. The role of microfilaments in cytoplasmic streaming in Drosophilafollicles. J Cell Sci. 1986;80:159–169. doi: 10.1242/jcs.80.1.159. [DOI] [PubMed] [Google Scholar]
- Hengartner MO. Apoptosis and the shape of death. Dev Genet. 1997;21:245–248. doi: 10.1002/(SICI)1520-6408(1997)21:4<245::AID-DVG1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Hengartner MO. Apoptosis. Death cycle and Swiss army knives. Nature. 1998;391:441–442. doi: 10.1038/35036. [DOI] [PubMed] [Google Scholar]
- Inoue S, Inoue H, Hiroyoshi T, Matsubara H, Yamanaka T. Developmental variation and amino acid sequences of cytochrome c of the fruit fly Drosophila melanogaster and the Flesh Fly Boettcherisca peregrina. . J Biochem. 1986;100:955–965. doi: 10.1093/oxfordjournals.jbchem.a121808. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Jemmerson, R., J. Liu, D. Hausauer, K.-P. Lam, A. Mondino, and R.D. Nelson. 1999. A conformational charge in cytochome c of apoptotic and necrotic cells is detected by monoclonal antibody binding and mimicked by association of the native antigen with synthetic phospholipid vesicles. Biochemistry. In press. [DOI] [PubMed]
- Kapoor R, Sakai LY, Funk S, Roux E, Bornstein P, Sage EH. Type VIII collagen has a restricted distribution in specialized extracellular matrices. J Cell Biol. 1988;107:721–730. doi: 10.1083/jcb.107.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JFR, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Pettingell WH, Jung YK, Kovacs DM, Tanzi RE. Alternative cleavage of Alzheimer-associated presenilins during apoptosis by a caspase-3 family protease. Science. 1997;277:373–376. doi: 10.1126/science.277.5324.373. [DOI] [PubMed] [Google Scholar]
- King RC. Oogenesis in adult Drosophila melanogaster.IX. Studies on the cytochemistry and ultrastructure of developing oocytes. Growth. 1960;24:265–323. [Google Scholar]
- Kluck RM, Bossywetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for bcl-2 regulation of apoptosis. Science. 1997a;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- Kluck RM, Martin SJ, Hoffman BM, Zhou JS, Green DR, Newmeyer DD. Cytochrome c activation of cpp32-like proteolysis plays a critical role in a Xenopuscell-free apoptosis system. EMBO (Eur Mol Biol Organ) J. 1997b;16:4639–4649. doi: 10.1093/emboj/16.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Yokokura T, Nagata S. Activation of distinct caspase-like proteases by fas and reaper in Drosophilacells. Proc Natl Acad Sci USA. 1997;94:11951–11956. doi: 10.1073/pnas.94.22.11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krippner A, Matsunoyagi A, Gottlieb RA, Babior BM. Loss of function of cytochrome c in Jurkat cells undergoing fas-mediated apoptosis. J Biol Chem. 1996;271:21629–21636. doi: 10.1074/jbc.271.35.21629. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Zamzami N, Susin SA. Mitochondrial control of apoptosis. Immunol Today. 1997;18:44–51. doi: 10.1016/s0167-5699(97)80014-x. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Smith JJ, Muzio M, Dixit V, Newmeyer DD, Kornbluth S. Apoptosis induction by caspase-8 is amplified through the mitochondrial release of cytochrome c. J Biol Chem. 1998;273:16589–16594. doi: 10.1074/jbc.273.26.16589. [DOI] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- Li HL, Zhu H, Xu CJ, Yuan JY. Cleavage of bid by caspase 8 mediates the mitochondrial damage in the fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang XD. Cytochrome c and dATP-dependent formation of apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Limbach KJ, Wu R. Characterization of two Drosophila melanogastercytochrome c genes and their transcripts. Nucleic Acids Res. 1985;13:631–644. doi: 10.1093/nar/13.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang XD. Induction of apoptotic program in cell-free extracts: requirement for datp and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang XD. Bid, a bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- McCall K, Steller H. Requirement for DCP-1 caspase during Drosophilaoogenesis. Science. 1998;279:230–234. doi: 10.1126/science.279.5348.230. [DOI] [PubMed] [Google Scholar]
- Monaghan P, Robertson D, Amos TAS, Dyer MJS, Mason DY, Greaves MF. Ultrastructural localization of bcl-2 protein. J Histochem Cytochem. 1992;40:1819–1825. doi: 10.1177/40.12.1453000. [DOI] [PubMed] [Google Scholar]
- Mueller CM, Jemmerson R. Maturation of the antibody response to the major epitope on the self antigen mouse cytochrome c. Restricted V gene usage, selected mutations, and increased affinity. J Immunol. 1996;157:5329–5338. [PubMed] [Google Scholar]
- Nicholson DW, Thornberry NA. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- Nordstrom W, Chen P, Steller H, Abrams JM. Activation of the reaper gene during ectopic cell killing in Drosophila. . Dev Biol. 1996;180:213–226. doi: 10.1006/dbio.1996.0296. [DOI] [PubMed] [Google Scholar]
- Okada E, Waddington CH. The submicroscopic structure of the Drosophilaegg. J Embryol Exp Morphol. 1959;7:583–597. [PubMed] [Google Scholar]
- Pronk GJ, Ramer K, Amiri P, Williams LT. Requirement of an ICE-like protease for induction of apoptosis and ceramide generation by reaper. Science. 1996;271:808–810. doi: 10.1126/science.271.5250.808. [DOI] [PubMed] [Google Scholar]
- Reed JC. Cytochrome c: Can't live with it—can't live without it. Cell. 1997a;91:559–562. doi: 10.1016/s0092-8674(00)80442-0. [DOI] [PubMed] [Google Scholar]
- Reed JC. Double identity for proteins of the bcl-2 family. Nature. 1997b;387:773–776. doi: 10.1038/42867. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Chen P, Abrams JM. Molecular prophets of death in the fly. Am J Hum Genet. 1998;62:514–519. doi: 10.1086/301775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Thompson CB. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annu Rev Med. 1997;48:267–281. doi: 10.1146/annurev.med.48.1.267. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- Sang JH, King RC. Nutritional requirements for normal oogenesis in Drosophila melanogaster. . Drosophila Information Service. 1959;33:156–158. [Google Scholar]
- Scaffidi C, Fulda S, Srinivasan A, Friesen C, Feng L, Tomaselli KJ, Debatin K, Krammer PH, Peter ME. Two CD95 (APO-1/Fas) signaling pathways. EMBO (Eur Mol Biol Organ) J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from the late embryonic stages of Drosophila melanogaster. . J Embryol Exp Morphol. 1972;27:353–356. [PubMed] [Google Scholar]
- Smith ML, Fornace AJ. Mammalian DNA damage-inducible genes associated with growth arrest and apoptosis. Mut Res. 1996;340:109–124. doi: 10.1016/s0165-1110(96)90043-3. [DOI] [PubMed] [Google Scholar]
- Song ZW, McCall K, Steller H. Dcp-1, a drosophila cell death protease essential for development. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Tourmente S, Lecher P, Degroote F, Renaud M. Mitochondrial development during Drosophilaoogenesis: distribution, density and in situ RNA hybridizations. Biol Cell. 1990;68:119–127. doi: 10.1016/0248-4900(90)90296-f. [DOI] [PubMed] [Google Scholar]
- Urbanski GJ, Margoliash E. Topographic determinants on cytochrome c. I. The complete antigenic structures of rabbit, mouse, and guanaco cytochromes c in rabbits and mice1. J Immunol. 1977;118:1170–1180. [PubMed] [Google Scholar]
- Vaux DL. CED-4: the third horseman of apoptosis. Cell. 1997;90:389–390. doi: 10.1016/s0092-8674(00)80497-3. [DOI] [PubMed] [Google Scholar]
- Villa P, Kaufmann SH, Earnshaw WC. Caspases and caspase inhibitors. Trends Biochem Sci. 1997;22:388–393. doi: 10.1016/s0968-0004(97)01107-9. [DOI] [PubMed] [Google Scholar]
- White K, Grether M, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. . Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wyllie AH, Kerr JFR, Currie AR. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- Yang J, Liu XS, Bhalla K, Kim CN, Ibrado AM, Cai JY, Peng TI, Jones DP, Wang XD. Prevention of apoptosis by bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Taylor A, Yellen A, Caton A, Gerhard W, Bachi T. Mutations in or near the fusion peptide of the influenza virus hemagglutinin affect an antigenic site in the globular region. J Virol. 1993;67:933–942. doi: 10.1128/jvi.67.2.933-942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamzami N, Susin SA, Marchetti P, Hirsch T, Gomezmonterrey I, Castedo M, Kroemer G. Mitochondrial control of nuclear apoptosis. J Exp Med. 1996;183:1533–1544. doi: 10.1084/jem.183.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S, Brustugun OT, Doskeland SO. Injected cytochrome c induces apoptosis. Nature. 1998;391:449–450. doi: 10.1038/35060. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu XS, Lutschg A, Wang XD. Apaf-1, a human protein homologous to c-elegans ced-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]