Abstract
The extracellular matrix glycosaminoglycan hyaluronan (HA) is an abundant component of skin and mesenchymal tissues where it facilitates cell migration during wound healing, inflammation, and em- bryonic morphogenesis. Both during normal tissue homeostasis and particularly after tissue injury, HA is mobilized from these sites through lymphatic vessels to the lymph nodes where it is degraded before entering the circulation for rapid uptake by the liver. Currently, however, the identities of HA binding molecules which control this pathway are unknown. Here we describe the first such molecule, LYVE-1, which we have identified as a major receptor for HA on the lymph vessel wall. The deduced amino acid sequence of LYVE-1 predicts a 322-residue type I integral membrane polypeptide 41% similar to the CD44 HA receptor with a 212-residue extracellular domain containing a single Link module the prototypic HA binding domain of the Link protein superfamily. Like CD44, the LYVE-1 molecule binds both soluble and immobilized HA. However, unlike CD44, the LYVE-1 molecule colocalizes with HA on the luminal face of the lymph vessel wall and is completely absent from blood vessels. Hence, LYVE-1 is the first lymph-specific HA receptor to be characterized and is a uniquely powerful marker for lymph vessels themselves.
Keywords: hyaluronic acid; endothelium, lymphatic; DNA, complementary; receptors, cell surface; recombinant fusion proteins
Hyaluronan (HA),1 a linear polymer of (1-β-4) d-glucuronic acid (1-β-3) N-acetyl-d-glucosamine, is a large glycosaminoglycan found in the tissue matrix and body fluids of all vertebrates which plays a fundamental role in regulating cell migration and differentiation (26). This role is first apparent during development, when changes in the levels of matrix HA induce condensation of mesenchymal cells and lead to the onset of chondrogenesis and myogenesis (reviewed in 19). In later life, HA facilitates cell migration in processes such as wound healing and inflammation by forming a pericellular matrix surrounding fibroblasts and epithelial cells, reducing the level of intercellular adhesion (20–22). The critical importance of HA in all of these roles is underlined by the observation that deletion of HA synthases in knockout mice results in early death of the embryo (31).
The largest concentrations of HA are found within the skin and musculo-skeletal system which account for >50% of total body HA. Here the rate of HA turnover is rapid (t 1/2 0.5 d) by comparison with other extracellular matrix components (26). Surprisingly, most degradation does not occur within the skin itself; rather HA is transported through the lymphatic system to distant lymph nodes where >90% of the glycosaminoglycan in afferent lymph is degraded or reenters the circulation to be rapidly endocytosed by liver endothelial HA receptors (11, 12). Hence, the lymphatic vessels are a major conduit for HA transport from tissues such as the skin and intestine, where flux measurements indicate they can remove as much as 10% of the total HA content within a 24-h period (36). This turnover can be further increased after either tissue injury or sepsis. Yet why HA should require transport through the lymphatics is not presently understood and little or nothing is known about the nature of any HA-binding molecules involved.
The majority of HA-binding proteins within the tissues belong to the Link protein superfamily. Members of this superfamily contain a conserved disulfide linked domain of ∼100 amino acid residues termed the Link module, which binds the minimal recognition unit HA6–8 and whose three-dimensional structure resembles the sugar-binding domain of C-type lectins. Other superfamily members in addition to the Link protein (34) include the large matrix proteoglycans Aggrecan (9), Versican (61), and Brevican (58), the tumor necrosis factor–inducible protein TSG-6 (28), and a single cell surface HA receptor, the CD44 molecule (1, 40). CD44 is widely expressed on epithelial, mesenchymal, and lymphoid cells, where it is thought to constitute the major receptor for HA (reviewed in 29). Interactions between CD44 and HA are implicated in several diverse functions including the maintenance of tissue structure within the epithelia (57), the extravasation of lymphocytes through inflamed vascular endothelium (8, 33), and the hematogenous dissemination of tumor cells (46, 47). The only other HA receptors characterized to date are RHAMM (receptor for HA-mediated motility), a 58-kD intracellular protein expressed transiently on the surface of transformed lymphocytes (15, 53), the cation-dependent liver endothelial cell (LEC) receptors responsible for the clearance of HA from the serum (59, 60), and the nuclear protein cdc37 (14). To date, however, there is no specific evidence that either CD44, RHAMM, or the LEC HA receptors are involved in the sequestration or transport of HA by lymphatic vessels, or in its uptake or degradation within the lymph nodes. Indeed, deletion of the CD44 gene in knockout mice produced no significant abnormalities, and resulted in no significant disruption of lymphatic function (43). Hence the identities of HA receptors involved in lymphatic HA homeostasis are completely unknown and we understand little about the regulation of this important physiological process.
Here we present the first identification of an HA receptor that is almost exclusively expressed on lymph vessels and is absent from blood vessels. This novel receptor which we have named LYVE-1 is a homologue of the CD44 HA receptor and a new member of the Link protein superfamily. We present evidence that LYVE-1 sequesters HA on lymph vessel endothelium in vivo and highlight the significance of this novel receptor in further defining the structure and function of the lymphatic system.
Materials and Methods
Cells, Antibodies, Chemicals, and Ig Fusion Proteins
Human umbilical vein endothelial cells (HUVEC) were kindly donated by Dr. Sue Adams (Molecular Parasitology Group, University of Oxford) and used at the third passage. mAbs to human CD31 (JC70), CD34 (QBEND 10), and von Willebrand factor (vWF, F8/86) were kindly donated by Dr. David Mason (Department of Cellular Science, University of Oxford). Texas red–conjugated goat anti–rabbit IgG and fluorescein-conjugated goat anti–mouse IgG reagents were obtained from Southern Biotechnologies. Biotinylated HA-binding complex (bHABC) was purified from bovine articular cartilage by extraction with 4 M guanidine-HCl and biotinylated as described previously (48). Biotin-LC-hydrazide was obtained from Pierce Chemical Co., and high molecular weight HA (rooster comb) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDAC) were from Sigma Chemical Co. Soluble Ig Fc (fragment-complement binding) fusion proteins corresponding to the extracellular domains of CD44 hematopoietic form (residues 1–200, CD44H Fc; see refs. 1, 37), ICAM-2 (complete extracellular domain, ICAM-2 Fc), and CD33 (complete extracellular domain, CD33 Fc) fused to the hinge, CH2, and CH3 domains of human IgG1 were donated, respectively, by Dr. Kelly Bennett (Bristol-Myers Squibb, Seattle, WA), Dr. Sue Adams (see above), and Dr. Regis Doyonnas (MRC Molecular Haematology Unit, Institute of Molecular Medicine, University of Oxford). A soluble truncated form of CD44H (CD44158his) comprising residues 1–158 of the ectodomain was expressed in Escherichia coli and refolded from urea-solubilized inclusion bodies (3). Details of the CD44158his protein which displays similar HA binding to CD44H Fc have been published recently (3).
Cloning and Identification of the LYVE-1 Receptor cDNA
A commercial cDNA database (Human Genome Sciences, The Institute for Genome Research) of >106 expression sequence tags (ESTs) obtained by random DNA sequencing of clones within 700 different human cDNA libraries was screened for ESTs bearing significant homology to the CD44 HA receptor using the program BlastSearch (Genetics Computer Group). Several overlapping ESTs with translated amino acid sequences that were at least 30% identical to that of full-length CD44 were identified in cDNA libraries constructed from tissues including umbilical vein, placenta, fetal liver, adipose tissue, lung, heart, prostate, embryo, spinal cord, bone marrow, bone, and ovarian tumor. The LYVE-1 EST described here was isolated from a HUVEC cDNA library, subcloned into the plasmid vector pBluescript, and sequenced in its entirety.
Northern Blot Hybridization
For Northern blot analysis, multiple tissue RNA blots (2 μg polyadenylated RNA per lane) were purchased from Clontech and hybridized (42°C, 50% formamide, 6× SSC) to a full-length LYVE-1 double-stranded DNA probe labeled with [32P]dCTP by random hexamer-priming (Rediprime DNA labeling system; Amersham International plc). Blots were washed at high stringency (0.1× SSC, 60°C, 15 min) before exposure to Kodak X-Omat RP x-ray film.
RT-PCR
For the detection of LYVE-1 and CD44 mRNAs in different cell lines, total RNA was isolated by extraction with guanidinium thiocyanate/sodium acetate, pH 4.0, and ethanol precipitation followed by first-strand cDNA synthesis using reverse transcriptase, and PCR using appropriate LYVE-1 or CD44 primers.
First-strand cDNA syntheses were carried out by oligo-dT priming in 50-μl reactions containing 5 μg total RNA, 0.5 μM dNTPs, 0.1 M Tris-HCl, pH 8.3, and 2.5 mU AMV reverse transcriptase for 3 h at 42°C. Samples (1 μl) of the final products were then committed to 50-μl PCR reactions (94°C, 1 min; 55°C, 1 min; 72°C, 1 min; 40 cycles) containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 1 mM dNTPs, 1 U Taq DNA polymerase, and the LYVE-1 primers LYVE-1F Hind/LYVE-1R Bam (see below), or the CD44 primers AMP1 (TCCCAGTATGACACATATTGC) and AMP2 (CCAAGATGATCAGCCATTCTGG). The integrity of the cDNA and input RNA was confirmed by PCR with the glyceraldehyde-3-phosphate dehydrogenase primers G3PF (TGGTCGTATTGGGCGCCTGG) and G3PR (CCAAATTCGTTGTCATACCAGG). Products were electrophoresed on 1.25% agarose gels, transferred to charged nylon membranes (Hybond N; Amersham International plc), and hybridized with 32P-end-labeled oligonucleotide probes to detect either LYVE-1 (CGCGGATCCCCATAAAAACTTCTGTGACAC) or CD44 (TGTACATCAGTCACAGACCTGCA). Blots were washed at 55°C in 6× SSC for 10 min before autoradiography.
Preparation of Full-Length LYVE-1 cDNA in pRcCMV and Transient Transfection of COS 1 Cells
For cell surface expression of full-length LYVE-1 molecules, the entire LYVE-1 coding sequence together with 11 bp of 5′ untranslated region and 40 bp of 3′ untranslated region was reamplified (94°C, 1 min; 55°C, 1 min; and 72°C, 1 min, 30 cycles) from the original pBluescript clone (see above) with the primers LYVE-1 F Hind (CGCGAAGCTTGGGTAGGCACGATGGCCAG) and LYVE-1 R Xba (GCTCTAGAGCCTCAGGTGTGTCTCCTC) using Pyrococcus furiosus (Pfu) DNA polymerase, and ligated into HindIII/XbaI cut pRcCMV. The pRcCMV construct was then used to transiently transfect COS 1 cells using DEAE dextran and chloroquine as described previously (17).
Expression of LYVE-1 as a Soluble IgFc Fusion Protein
A 684-bp LYVE-1 fragment encoding the predicted NH2-terminal leader and extracellular domain, truncating at Gly 232, was amplified (94°C 1 min; 60°C 5 min; and 72°C 10 min, 25 cycles) from the full-length LYVE-1 cDNA clone in pBluescript using Pfu polymerase and the primers LYVE-1F Hind (see above for sequence) and LYVE-1 R Bam (CGCGGATCCCCAGCAGCTTCATTCTTGAATG). After digestion with HindIII and BamH1, the product was cloned into BamH1/HindIII cut IgFc vector to yield a construct encoding amino acid residues 1–232 of the LYVE-1 sequence, fused at the COOH terminus with the 234-residue hinge, CH2, and CH3 region of human IgG1 (1). The absence of polymerase-induced errors was confirmed by sequencing the resulting construct on both strands. For expression, the LYVE-1 cDNA was transiently transfected into SV-40–transformed African green monkey kidney cells (COS 1) using DEAE dextran. After 72 h, culture supernatants were collected, supplemented with 0.1 M Tris-HCl, pH 8.0, and the fusion protein purified by protein A–Sepharose affinity chromatography. Purity was assessed by SDS-PAGE analysis under both reducing and nonreducing conditions, and by Western blot analysis with peroxidase-conjugated anti–human IgGFc antibody.
Western Blotting/Biosynthetic Labeling
For detection of LYVE-1 protein expressed in transfected COS 1 cell lysates, these were boiled in SDS-PAGE sample buffer, electrophoresed on 10% polyacrylamide SDS-PAGE gels, and transferred to nitrocellulose membranes (Hybond super; Amersham International plc) using a semi-dry transfer apparatus (Hoefer). Blots were incubated with LYVE-1 polyclonal serum (1:100 dilution in PBS, pH 7.5, 5% dried milk powder, 0.1% Tween 20), and developed with peroxidase-conjugated anti–rabbit Ig using chemiluminescent detection (ECL kit; Amersham International plc).
For biosynthetic labeling of LYVE-1 Fc fusion proteins, transiently transfected COS 1 cells were cultured (24 h, 37°C) in RPMI supplemented with 100 μCi/ml [35S]methionine/cysteine (Amersham International plc) followed by absorption of the labeled fusion proteins from the supernatants on protein A–Sepharose beads. Beads were then boiled in SDS-PAGE sample buffer and electrophoresed on 10% polyacrylamide SDS-PAGE gels. After electrophoresis, the gels were fixed, stained with Coomassie blue, and impregnated with AMPLIFY (Amersham International plc), before drying and exposing to Kodak X-Omat x-ray film at −70°C.
HA-binding Assays
Binding to Immobilized HA.
Binding of LYVE-1 IgFc fusion protein to immobilized HA was measured in a 96-well microtiter plate ELISA assay. In brief, microtiter plates (Nunc Maxisorp) were coated with HA (purified from rooster comb; Sigma) at a concentration of 2 mg/ml in coating buffer (15 mM sodium carbonate and 34 mM sodium bicarbonate, pH 9.3). After overnight incubation at 25°C, blocking (2 h) in PBS, pH 7.5, 0.25% (wt/ vol) BSA, 0.05% (wt/vol) Tween 20, and washing (three times, PBS, pH 7.5), plates were incubated with purified LYVE-1 IgFc or CD44H Fc, or with the control proteins ICAM-2 Fc or CD33 Fc (15–500 ng/well) for 1 h at 25°C. After removal of unbound protein by washing (three times) with PBS, bound fusion protein was detected by successive (1 h, 25°C) incubation with horseradish peroxidase–conjugated anti–human IgG1 antibody. Reactions were developed by the addition of O-phenylenediamine substrate and the absorbance measured at 490 nm in a Bio-Rad microplate reader. To determine the binding specificity of LYVE-1 for other glycosaminoglycans, the standard assay was modified to include either free chondroitin-4 sulfate, chondroitin-6 sulfate, or heparin (1.5–200 μg/ml) as competitor during the primary incubation step.
Binding to Soluble HA.
For binding assays with soluble HA, this was first biotinylated by a modification of the method of Yu and Toole (33). In brief, high molecular weight HA (5 mg/ml) in 0.1 M MES, pH 5.5, was derivatized with biotin-LC-hydrazide (1 mM) in the presence of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDAC, 0.16 mg/ml) in 1-ml stirred reactions overnight at room temperature, followed by dialysis against PBS, pH 7.5, to remove free biotin.
To perform the standard binding assay, microtiter plates were coated with LYVE-1 Fc, CD44H Fc, or the control fusion proteins CD33 Fc or ICAM-2 Fc (1.25–10 μg/ml, 1 h, 25°C) washed (three times) to remove unbound material, and incubated with biotinylated HA (5 μg/ml) for a further 1 h. Bound HA was then detected by incubation with horseradish peroxidase–conjugated streptavidin (Sigma) and O-phenylenediamine substrate. Absorbances were measured at 490 nm in a Bio-Rad microplate reader as described above.
Sandwich Assay for Binding to Preformed HA Receptor Complexes.
To measure binding of LYVE-1 to HA presented as a complex with CD44, microtiter plates were coated with bacterially expressed CD44158his (5 μg/ml), before successive incubations with HA (250 μg/ml) and either LYVE-1 (5 μg/ml) or CD33 Fc (5 μg/ml, negative control), followed by detection with peroxidase-conjugated anti–human IgFc (see above). To measure binding of CD44 to HA presented as a complex with LYVE-1, microtiter plates were coated with either LYVE-1 Fc or CD33 Fc (negative control), before successive incubations with HA (250 μg/ml) and CD44H Fc (5 μg/ml) and detection with the peroxidase-conjugated CD44 mAb A3D8 (1 μg/ml).
Production of LYVE-1 Polyclonal Serum
For the production of polyclonal antisera, rabbits were immunized with purified LYVE-1 Fc fusion protein (100 μg) in complete Freund's adjuvant, followed by three consecutive boosts (50 μg) in incomplete adjuvant, at intervals of 4 wk. Sera were then tested for reactivity with LYVE-1 Fc or with the irrelevant receptor-globulin controls CD33 Fc or ICAM-2 Fc, immobilized to 96-well microtiter dishes using an ELISA assay detected with peroxidase-conjugated goat anti–rabbit IgG and O-phenylenediamine. Antibodies reactive with the human IgG portion of the immunizing antigen were depleted by preabsorption on columns of human IgG Sepharose.
Immunoperoxidase and Immunofluorescent Antibody Staining of Cells and Tissues
For immunoperoxidase staining, normal human tissue samples were obtained from the Department of Histopathology, Oxford Radcliffe Hospital, as paraffin-embedded sections on silanized glass microscope slides. Slides were dewaxed and rehydrated by successive incubation in Citroclear (5 min), 100% ethanol (5 min), 70% ethanol (5 min), and water followed by microwave treatment (700 W, 8 min) and incubation (30 min, 25°C) with 1:100 diluted LYVE-1 serum in PBS, 5% human serum, 0.1% azide. Antigen was subsequently detected by incubation (30 min, 25°C) with an anti–rabbit Ig peroxidase conjugate, and diaminobenzidine (Envision kit; DAKO), before counterstaining (30 s) with hematoxylin and eosin.
For standard single/double immunofluorescent staining, tissue sections on glass microscope slides were dewaxed and rehydrated as described above before incubation (30 min, 5°C) with 1:100 diluted LYVE-1 serum in PBS, 5% human serum, 0.1% azide, either alone or in combination with 1:2 diluted CD31, CD34, or vWF hybridoma supernatants, or with CD44 mAb (10 μg/ml). After washing (twice) to remove unbound antibodies, samples were then stained (30 min, 5°C) with 1:25 diluted FITC-conjugated goat anti–rabbit Ig (to detect LYVE-1), either alone or in combination with 1–50 diluted Texas red–conjugated goat anti–rabbit Ig. Samples were fixed in 2% (wt/vol) formaldehyde and viewed under a Zeiss Axioskop fluorescence microscope using epifluorescent illumination.
Staining for HA and colocalization with LYVE-1 receptor was carried out using complexes of biotinylated bovine aggrecan G1 domain and bovine link protein as described previously (48). In brief, dewaxed and rehydrated tissue sections on glass microscope slides were blocked (30 min, room temperature) in PBS, pH 7.5, containing 50 mM glycine and 1% (wt/ vol) fat-free milk powder, before incubation (overnight, 5°C) with bHABC (5 μg/ml) with or without LYVE-1 serum (1:100 dilution) in 0.1 M sodium phosphate buffer, pH 7.4, supplemented with 1% BSA. After washing to remove unbound reagents, the sections were incubated (1 h, room temperature) with a mixture of Texas red–labeled anti–rabbit Ig and FITC-labeled avidin (Vector Laboratories, Inc.), diluted 1:50 and 1:500, respectively, in PBS, pH 7.5. Slides were mounted in Vectashield (Vector Laboratories, Inc.) for immunofluorescence microscopy.
Results
Identification of a Novel HA Receptor LYVE-1
To identify new HA receptors we searched recent databases of ESTs for cDNAs homologous to the CD44 molecule, currently thought to be the primary HA receptor on mammalian cells. A homology search of the combined Human Genome Sciences/TIGR EST databases with the amino acid sequence of full-length CD44H using the BLAST program identified a number of ESTs with predicted amino acid sequences >30% similar to CD44. 59 identical ESTs were identified in libraries prepared from fetal tissues including liver, spleen, brain, and heart; adult human placenta, bone marrow, lung, and spinal cord; glioblastoma, ovarian, and bone tumor; and HUVEC. The 2,313-bp EST from HUVEC which we have termed LYVE-1 (lymphatic vessel endothelial HA receptor) was subjected to complete nucleotide sequencing. Translation of the LYVE-1 cDNA (Fig. 1 A) revealed a large (966 bp) open reading frame starting with a Kozak (24) consensus ATG initiation codon at position 91 and terminating with the TAG stop codon at position 1057. The deduced amino acid sequence encodes a 322-residue polypeptide with the features characteristic of a type I integral membrane glycoprotein. These are: a sequence of 26 largely hydrophobic residues at the NH2 terminus most likely comprising the leader peptide; a hydrophilic sequence of 212 amino acids containing seven cysteine residues, a serine/threonine-rich region (residues 145–216) and two motifs for N-linked glycosylation (NFT and NSS, centered on residues 54 and 131, respectively) corresponding to an extracellular domain; a sequence of 21 hydrophobic residues immediately followed by the dibasic motif KR and a highly charged stretch of 63 residues predicted to form the transmembrane anchor and cytoplasmic tail, respectively (Fig. 1 A). The predicted transmembrane anchor is also apparent in Kyte-Doolittle (25) and Goldman hydropathy plots of the deduced amino acid sequence (Fig. 1 B). The position of the mature NH2 terminus is predicted to be Ala 27 based on the −3 +1 rule proposed by Von Heijne (56).
Figure 1.
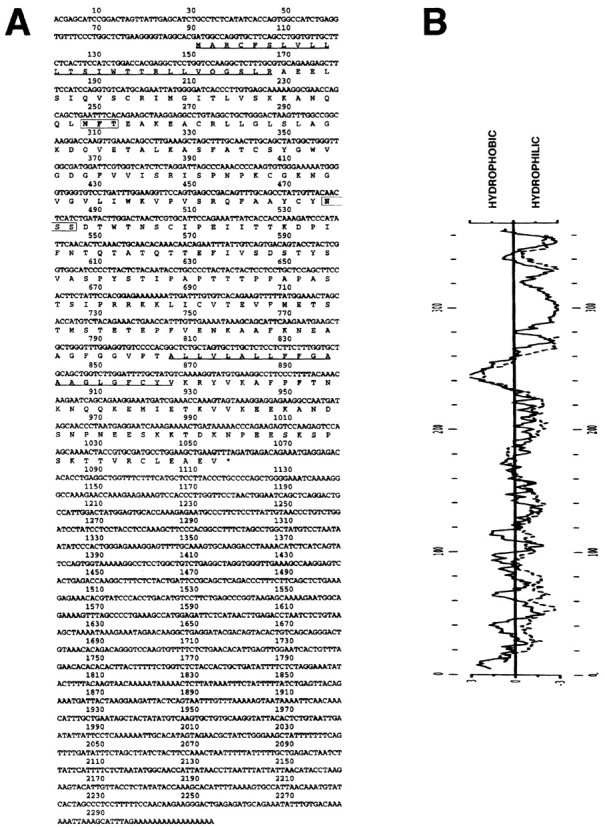
Nucleotide sequence and derived amino acid sequence of LYVE-1, a novel human HA receptor. A shows the complete nucleotide and derived amino acid sequence for the 2,313-bp LYVE-1 cDNA clone in pBluescript. A cleavable NH2-terminal leader predicted using the −3 +1 rule (56) and a probable transmembrane anchor are underlined. The positions of two putative N-glycosylation sites (N X S/T) are boxed. The sequence predicts a 322–amino acid residue polypeptide with a 26-residue leader peptide, a 21-residue hydrophobic membrane anchor, and a 63-residue cytoplasmic tail. B depicts Kyte-Doolittle (solid line) and Goldman (stippled line) hydropathy plots of the derived amino acid sequence, both of which confirm the position of the predicted hydrophobic transmembrane anchor. These sequence data are available from GenBank under accession number AF118108.
Similarities between the Link Modules in LYVE-1 and CD44
Alignment of the LYVE-1 amino acid sequence with that of the human CD44 HA receptor (Fig. 2) illustrates the degree of homology between the two molecules which have an overall similarity of 41%. Closer inspection reveals the region of maximal homology (57% similarity, 38% identity) to be contained within the area encompassing the extended CD44 Link domain and the corresponding region (residues 36–139) in the LYVE-1 sequence (Fig. 2 A). The location of the putative Link module in LYVE-1 is marked by four central cysteine residues (Cys 61, 85, 106, and 128), whose spacing (C1 - X23 - C2 - X20 - C3 - X20 - C4) is almost identical in CD44, and whose intervening sequences share 57% similarity. This similarity rises to 68% (57% identity) between C2 (Cys 85) and C3 (Cys 106). These four cysteine residues are conserved in all members of the Link superfamily where they form essential disulfide bridges that stabilize the HA-binding domain, a structure comprising two α helices and two anti–parallel β sheets surrounding a large hydrophobic core (23). Interestingly the two additional cysteine residues Cys 31 and Cys 133 that uniquely flank the CD44 Link module (13, 45) are also present at appropriately conserved locations in the LYVE-1 sequence (Cys 36 and Cys 139, Fig. 2 A). Downstream of the Link module, however, the region corresponding to the membrane proximal domain of LYVE-1 shows little similarity to CD44, with the exception of a moderately high proportion of serine and threonine residues (37% within residues 145–216).
Figure 2.
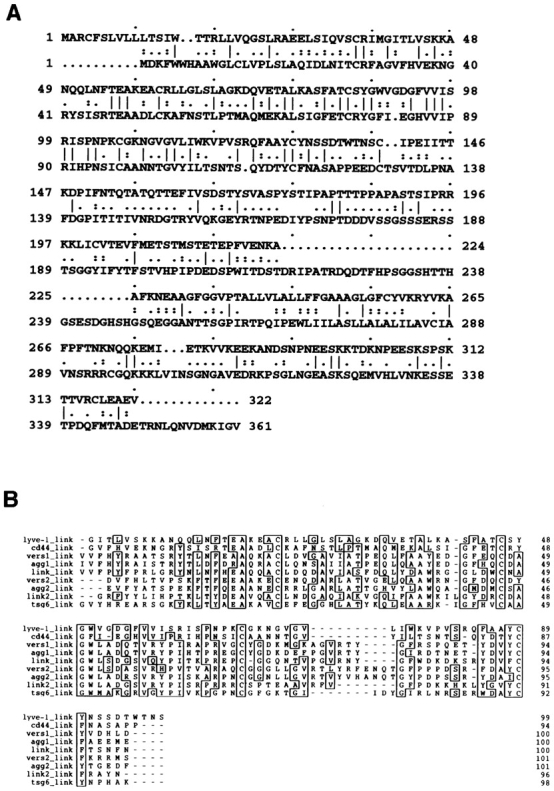
Comparison of the LYVE-1 receptor Link module with other Link superfamily members. In A, the derived amino acid sequence of LYVE-1 (top line) is shown aligned with that of the full-length human CD44 cDNA (bottom line) using the GCG program GAP. Positions where amino acid residues are identical are depicted with a line; semiconservative and conservative differences are depicted with one or two dots, respectively. The two sequences have an overall similarity of 41% and a similarity of 57% within the immediate Link homology unit (LYVE-1 residues 61–128). B shows a Prettyplot (GCG) comparison of sequence encompassing the “Link modules” (HA-binding domains) from LYVE-1 (corresponding to residues 40–138 in A), human CD44, the soluble human tumor necrosis factor–inducible TSG-6 molecule, and both tandem repeats (Links 1 and 2) of human Aggrecan, human Versican, and human Cartilage Link protein.
The presence of the LYVE-1 Link module is more clearly illustrated by sequence alignments with other human Link superfamily members including the cartilage matrix proteins Aggrecan, Versican, and Link protein itself, each of which contain tandem repeats of the Link homology unit (Fig. 2 B). A novel feature apparent from these alignments is that LYVE-1 and CD44 appear to define a subgroup within the Link superfamily. These findings suggest the divergence of the cell surface HA receptors as a separate branch from the HA-binding extracellular matrix proteins during evolution.
More detailed comparisons of individual amino acid residues within the putative HA-binding regions of the LYVE-1 and CD44 Link modules reveal a number of differences. For example, of the nine key residues identified by site directed mutagenesis (2, 37) as critical for HA binding within the human CD44 Link domain, Lys 38 + 68, Arg 41 + 78, Tyr 42, 79, + 105, and Asp 100 + 101, only three of these, Lys 38, Tyr 79, and Asp 100 are conserved within the LYVE-1 molecule (residues 46, 87, and 109, respectively, Fig. 2 A). Furthermore, the peripheral cluster of basic amino acids located downstream of the Link module in CD44, which includes Arg 158 and Lys 162 (Fig. 2 A) previously shown to be important for HA binding (37), is not conserved in the LYVE-1 sequence. Hence, despite overall similarities in Link module sequences, the specific residues involved in the predicted LYVE-1–carbohydrate interaction are likely to be different from those in the CD44 molecule.
The LYVE-1 Receptor Is Expressed on the Cell Surface
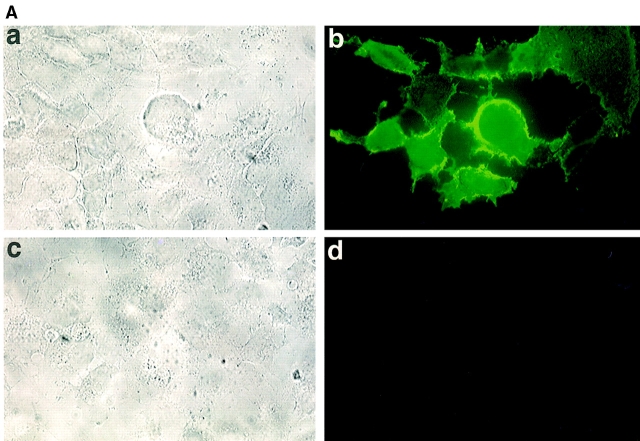
To confirm the identification of the LYVE-1 molecule as an integral membrane glycoprotein, COS 1 fibroblasts were transfected with LYVE-1 cDNA and stained with polyclonal LYVE-1 antibodies for immunofluorescence microscopy. The specificity of the LYVE-1 serum is characterized in detail below. As shown by the results in Fig. 3 A, the transfectants displayed intense surface staining. Western blotting of the LYVE-1 transfected COS cells using a polyclonal serum generated against soluble LYVE-1 revealed a major band which migrated with an apparent molecular mass of 60 kD, and which was absent from mock transfected cells. The discrepancy between the experimentally observed value and the size predicted from the primary sequence (35,158 D) is largely due to glycosylation, since treatment with either N-glycanase or O-glycanase/neuraminidase reduced the molecular mass by ∼20 kD (not shown).
Figure 3.
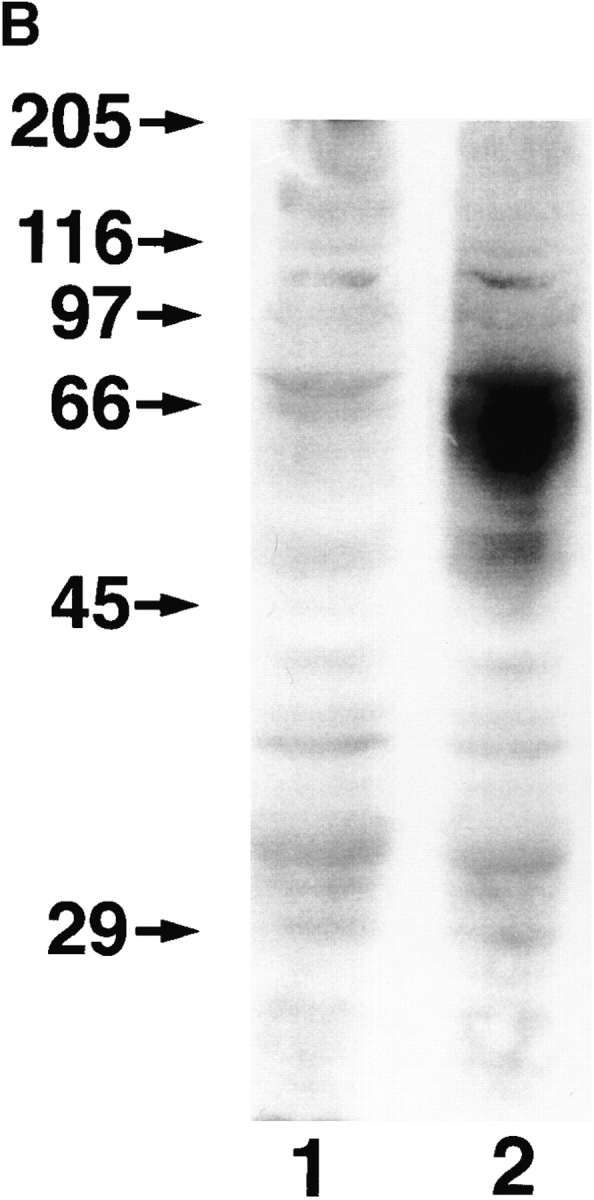
Cell surface expression of LYVE-1 receptor on transfected COS cells. In A, COS 1 cells were transiently transfected with either full-length LYVE-1 cDNA in the expression vector pRcCMV (a and b), or with a control empty pRcCMV vector (c and d) using DEAE dextran followed by surface immunofluorescent staining with rabbit polyclonal LYVE-1 antiserum (1:100 dilution) and FITC goat anti–rabbit IgG. In B, control and LYVE-1 transfected COS cells were electrophoresed on a 10% polyacrylamide SDS-PAGE gel, transferred to nitrocellulose, and Western blotted with LYVE-1 antiserum and peroxidase-conjugated goat anti–rabbit IgG (see Materials and Methods). Samples were control transfected COS (lane 1) and LYVE-1 transfected COS (lane 2). The positions and sizes in kilodaltons of the molecular mass calibration markers myosin (205 kD), β-galactosidase (116 kD), phosphorylase b (97 kD), BSA (66 kD), ovalbumin (45 kD), and carbonic anhydrase (29 kD) are indicated on the left.
LYVE-1 Binds Both Immobilized and Soluble HA
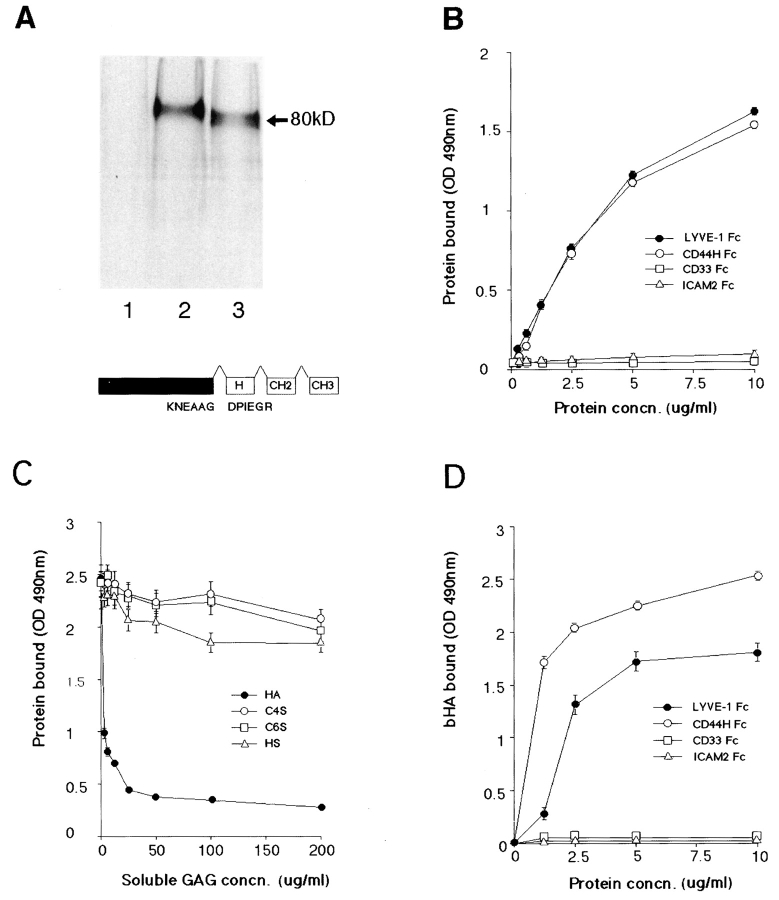
To confirm the presence of a functional HA-binding domain in the LYVE-1 receptor, we expressed the extracellular domain with its own NH2-terminal leader (residues 1–232) as a soluble fusion protein with the hinge, CH2, and CH3 domains of human IgG1 (total length 440 residues). The purified protein migrated as a single 80-kD band on SDS-PAGE, similar to a CD44H Fc fusion protein (total length 434 residues, Fig. 4 A). The discrepancy with the 46,900-D molecular mass predicted from the mature 440 residue primary sequence again is largely due to N- and O-glycosylation of the LYVE-1 core polypeptide (see above).
Figure 4.
LYVE-1 binds both immobilized and soluble HA. LYVE-1, expressed as a soluble IgFc fusion protein in transiently transfected human COS fibroblasts, was compared with CD44 for binding to HA and other glycosaminoglycans. A shows LYVE-1 and CD44H Fc fusion proteins isolated from the supernatants of [35S] methionine/cysteine-labeled transfectants and electrophoresed on a 7.5% polyacrylamide SDS-PAGE gel. Samples were the protein A–Sepharose adsorbed proteins from control untransfected cells (lane 1), CD44H Fc transfected cells (lane 2), and LYVE-1 transfected cells (lane 3). The LYVE-1 fusion protein comprises residues 1–232 of the extracellular domain fused to the hinge (H), CH2, and CH3 domains of human IgG1. Details of the CD44H Fc protein, which includes residues 1–200 of the extracellular domain, have been published previously (1). For ligand binding assays, LYVE-1 Fc was compared with CD44H Fc and the negative control fusion proteins CD33 Fc and ICAM-2 Fc for adhesion to immobilized and soluble HA in 96-well microtiter plates (see Materials and Methods). B shows binding of the fusion proteins to immobilized HA, in the absence of competing glycosaminoglycans; C shows binding (LYVE-1 Fc only) in the presence of free chondroitin-4-SO4, chondroitin-6-SO4, or heparin; and D shows binding to soluble biotinylated HA. Detection of bound fusion protein and biotinylated HA was carried out using peroxidase-conjugated anti–human IgFc antibody and peroxidase-conjugated streptavidin, respectively. Values are the mean ± SEM of at least three replicates.
Comparison of LYVE-1 Fc with CD44 Fc in a microtiter plate glycosaminoglycan binding assay (Fig. 4, B and C) revealed that LYVE-1 binds immobilized HA in a concentration-dependent manner that is similar if not identical to CD44. In contrast, neither of the two control fusion proteins CD33 Fc or ICAM-2 Fc displayed any significant binding over this range (0.25–10 μg/ml). The specificity of the LYVE-1 HA interaction was demonstrated by competition with free glycosaminoglycans, which showed that neither chondroitin-4-SO4, chondroitin-6-SO4, nor heparan sulfate blocked binding even at concentrations up to 200 μg/ml (Fig. 4 C). In contrast the LYVE-1 HA interaction was extremely sensitive to inhibition by free HA (IC50 ∼3 μg/ml). This indicates that LYVE-1 has a much higher specificity for glycosaminoglycan binding than either CD44 or the cation-dependent liver endothelial receptors. Importantly, this also distinguishes LYVE-1 from the lymph node receptors involved in the uptake and degradation of lymph fluid HA, since these are blocked by chondroitin sulfate and chondroitin sulfate proteoglycans (54). LYVE-1 Fc also bound soluble high molecular mass HA in a concentration-dependent fashion (Fig. 4 D), although the binding capacity decreased dramatically with increasing levels of HA biotinylation (data not shown), a feature that was not observed with CD44. This latter result suggests there is a significant difference in the size or geometry of the HA-binding surface within these two receptors.
LYVE-1 mRNA Has a Restricted Pattern of Tissue Expression
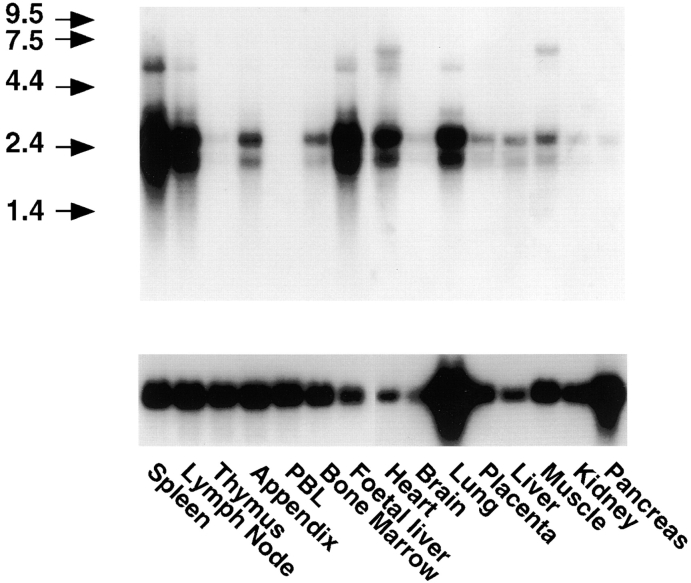
Expression of the LYVE-1 gene in vivo was investigated by Northern blot hybridization to poly(A)+ RNAs prepared from a range of human tissues (Fig. 5). Two major bands of ∼2 and 2.6 kb were abundant in spleen, lymph node, heart, lung, and fetal liver RNA and to a lesser degree in appendix, bone marrow, placenta, muscle, and adult liver RNA. These were largely if not completely absent from peripheral blood lymphocytes, thymus, brain, kidney, and pancreas RNA. In addition, minor bands of 6–7 kb were detected to a variable extent in spleen, lymph node, fetal liver, heart, lung, and muscle. The 2.6-kb band is predicted to correspond to the LYVE-1 transcript shown in Fig. 1, whereas the 2-kb band likely represents a transcript that is polyadenylated on one or other of the consensus AATAAA motifs at positions 1819 and 1905 within the 3′ untranslated region (see Fig. 1). The minor 6–7-kb bands have not yet been characterized. Hence it is possible they represent alternatively spliced LYVE-1 transcripts.
Figure 5.
Northern blot hybridization analysis of LYVE-1 receptor mRNA. RNA blots containing 2 μg poly(A)+ RNA per lane from each of the tissues shown were hybridized to a 32P-labeled full-length LYVE-1 DNA probe (top), or glyceraldehyde-3-phosphate dehydrogenase probe (bottom), and washed at high stringency before autoradiography (see Materials and Methods). The migration positions of RNA calibration markers (kilodaltons) are shown to the left of the figure.
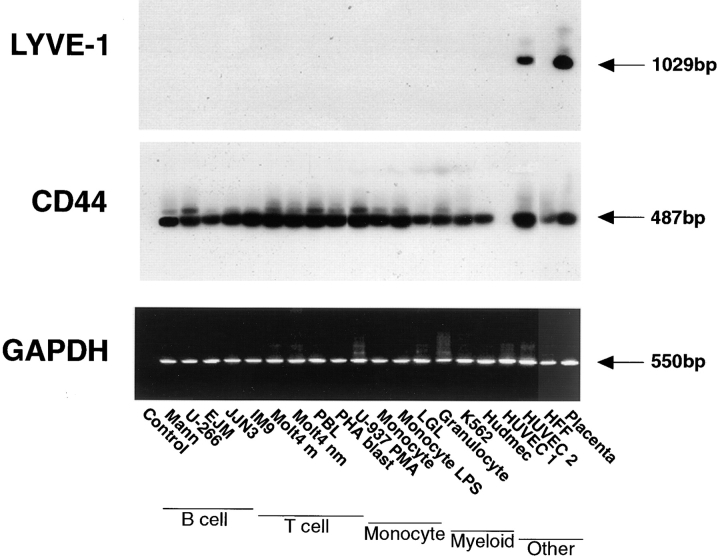
Intriguingly, RT-PCR analyses (Fig. 6) of individual cell lines derived from lymphocytes, myeloid cells, monocytes, microvascular endothelial cells, and fibroblasts yielded no products in any cell type with the exception of late passage HUVEC, and placental tissue, used as a positive control. In contrast, each cell line yielded abundant PCR products when amplified with primers to CD44, or the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase. These results suggested an unusually specific pattern of tissue expression for LYVE-1 that was clearly distinct from that of the CD44 molecule.
Figure 6.
RT-PCR analysis of LYVE-1 mRNA expression in tissue culture cell lines. LYVE-1 and CD44 mRNA levels were compared within a panel of tissue culture cell lines representing different lineages. Samples of cDNA prepared from each of the cell lines shown were amplified with primers specific for either LYVE-1 (top), CD44 (middle), or glyceraldehyde-3-phosphate dehydrogenase (control, bottom) followed by electrophoresis on 1.25% agarose gels, staining with ethidium bromide (glyceraldehyde-3-phosphate dehydrogenase only), or Southern blotting and hybridization with the appropriate 32P-labeled detection oligonucleotides as described in Materials and Methods. Arrows depict the sizes of the PCR products in each case.
The LYVE-1 Molecule Is Present on the Walls of Lymphatic Vessels
To explore the expression pattern of the LYVE-1 protein in more detail, we generated a polyclonal serum by immunizing rabbits with purified LYVE-1 Fc fusion protein (see above). The serum was then preabsorbed on human IgG Sepharose to remove contaminating Fc-reactive antibodies, followed by affinity chromatography on a LYVE-1–Sepharose column. The specificity of the purified antiserum was established using immobilized LYVE-1 Fc fusion protein in an ELISA assay. As shown in the top panel of Fig. 7, the LYVE-1 antiserum was highly specific for LYVE-1 Fc at dilutions below 1:100 and reacted only weakly with the control ICAM-2 fusion protein, which in common with LYVE-1 Fc, contains the hinge, CH2, and CH3 domains of human IgG1. Importantly, the LYVE-1 antiserum was unreactive with a CD44H ectodomain construct CD44158his which contains a functional Link module (residues 1–158) expressed as a histidine-tagged bacterial fusion protein (3), with CD44H Fc and with CD44-transfected COS 1 cells (not shown). Finally, the LYVE-1 antiserum (1:100 dilution) had the capacity to completely block binding of soluble bHA (Fig. 7, bottom). These data indicate the polyclonal serum is specific for LYVE-1 and does not exhibit reactivity with CD44, its closest homologue.
Figure 7.
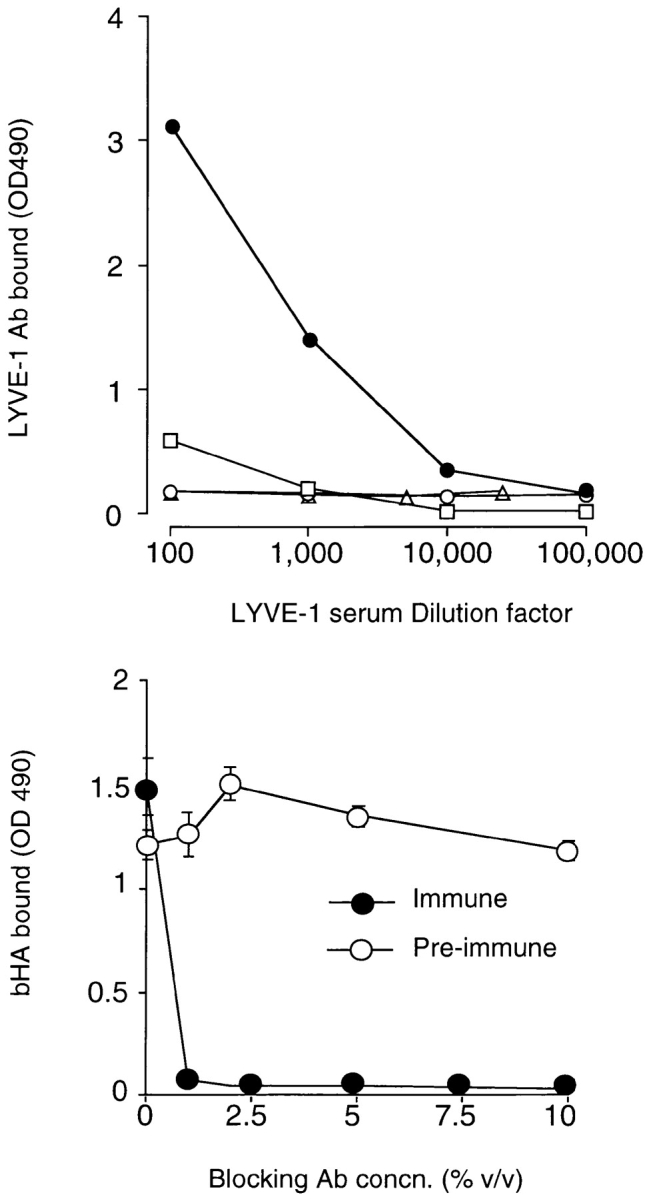
Specificity of a LYVE-1 receptor polyclonal serum. The specificity of an affinity-purified rabbit polyclonal antiserum generated against soluble LYVE-1 receptor and its capacity to block HA binding were assessed in microtiter plate binding assays. In the top panel, wells coated with either LYVE-1 Fc (filled circles), the CD44H ectodomain fragment CD44158his (triangles), or the control fusion protein ICAM-2 Fc (squares) were incubated with appropriately diluted LYVE-1 specific polyclonal antiserum, and binding was detected with peroxidase-conjugated anti–human Ig (see Materials and Methods). As a control, a second set of LYVE-1–coated wells was reacted with appropriately diluted preimmune serum (open circles). In the bottom panel, wells coated with LYVE-1 Fc were incubated with soluble biotinylated HA (5 μg/ml), in the presence of increasing concentrations of either LYVE-1 antiserum or control preimmune serum followed by detection of bound HA as described in Materials and Methods. Data in each case are the mean ± SEM of triplicate determinations.
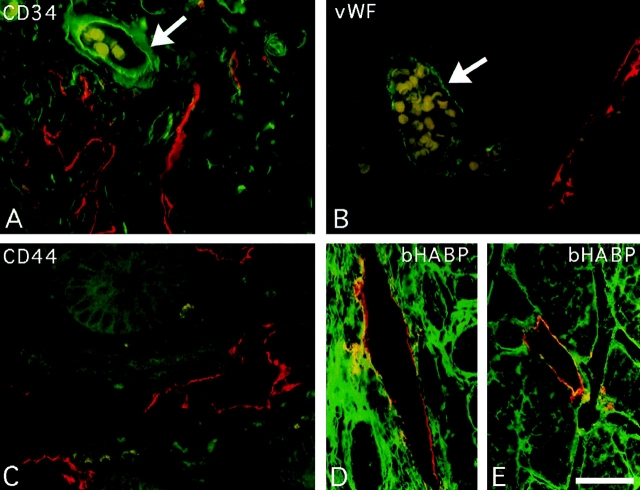
Immunoperoxidase staining of human tissues with the polyclonal LYVE-1 serum revealed an unusual and highly restricted expression pattern, as shown in Fig. 8 and summarized in Table I. Intriguingly the expression of LYVE-1 was mostly confined to endothelial cells lining vessels of the lymphatic system. The only other sites of prominent LYVE-1 expression were sinusoidal endothelial cells within the spleen and placental syncytiotrophoblasts (Table I and Fig. 8 k). In contrast with the expression pattern of CD44, no LYVE-1 was detected on lymphocytes or other hematopoietic cells within lymphoid organs such as the tonsil and thymus (Table I and Fig. 8 l). Localization of LYVE-1 within the lymphatics was most strikingly visible among the draining lymphatic vessels within the submucosae, underlying the intestinal crypts in colon and small intestine (Fig. 8, a–d). Many small lymph vessels, including some containing large numbers of lymphocytes, showed intense endothelial staining. High levels of LYVE-1 expression were also apparent on the lacteal vessels draining individual intestinal villi, on subdermal lymphatic vessels within the skin, and on lymphatic vessels within peripheral nodes (Fig. 8, e–j). A similar expression pattern was found in other tissues displaying prominent lymphatics, particularly the appendix and stomach (see Table I). In contrast, no LYVE-1 was detected on blood vessels within any of the tissues tested. Interestingly, LYVE-1 protein expression was detected on the surface of cultured HUVEC (Fig. 8 m), confirming the detection of LYVE-1 mRNA by RT-PCR (see above). This is most likely to be the result of culture induced de-differentiation as no vascular staining was observed in tissue sections of umbilical vein (not shown). The selectivity for lymphatic expression is underlined by a comparison of the two vessel types in sections of salivary gland (Fig. 8 g), where blood vessels that are clearly distinguishable by their erythrocyte content (visible through weak nonspecific staining with rabbit IgG) are negative for LYVE-1, while the adjacent empty lymph vessels stain intensely. This distinction was confirmed by double immunofluorescence staining of small intestine sections (Fig. 9) with antibodies to LYVE-1 and to the vascular endothelial markers CD34 (sgp90) and vWF (Factor VIII–related molecule). LYVE-1 staining was mutually exclusive with both of the other markers, and was confined to irregularly shaped vessels within the submucosae that were devoid of erythrocytes. Furthermore, double immunofluorescence staining revealed little or no CD44 expression on lymphatic vessels, indicating that LYVE-1 is likely to be the major receptor for HA on lymphatic endothelial cells (Fig. 9).
Figure 8.
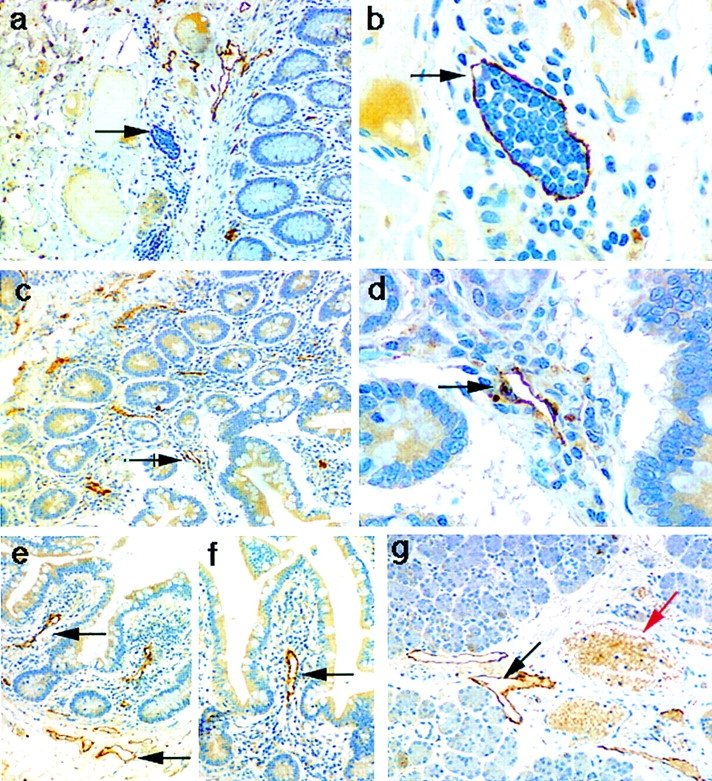
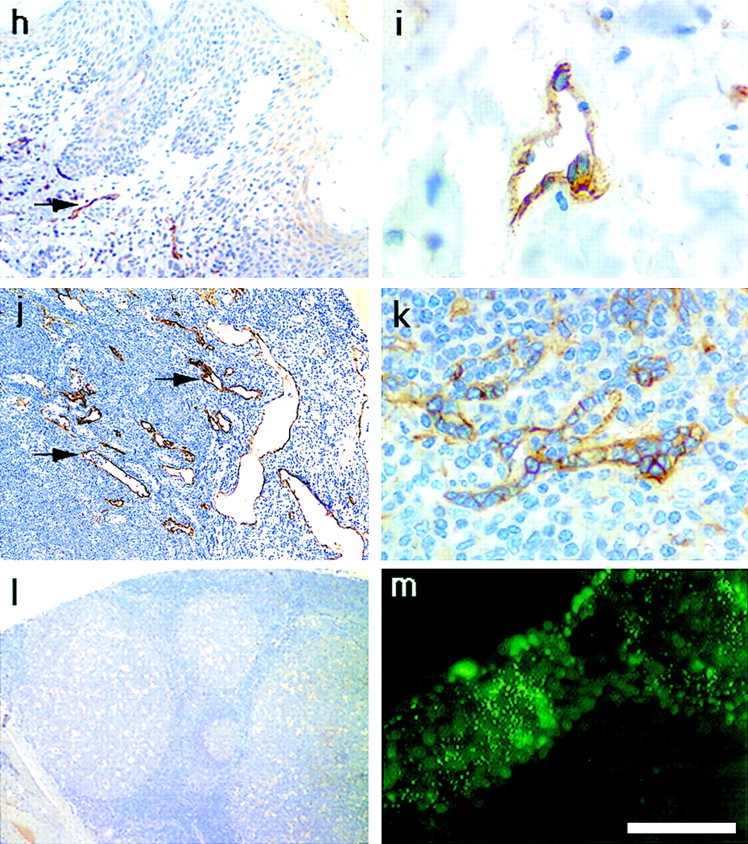
Localization of the LYVE-1 HA receptor in human tissues. Paraffin-embedded human tissue sections were stained with rabbit polyclonal LYVE-1 antiserum (1:100 dilution), followed by peroxidase-conjugated goat anti–rabbit IgG as described in Materials and Methods. Tissues shown are colon (a and b), small intestine (c–f), salivary gland (g), skin (h and i), lymph node (j), spleen (k), and tonsil (l). m shows cultured HUVEC stained with LYVE-1 and FITC-conjugated anti–rabbit Ig (see Materials and Methods). Bars, j and l, 500 μm; a, c, and e–h, 200 μm; b, d, and k, 50 μm; and i and m, 20 μm. Black arrows depict lymphatic vessels, both empty (d–g) and containing lymphocytes (b), and red arrows depict blood vessels clearly identified by their content of weakly stained erythrocytes (g and i).
Table I.
Tissue Distribution of the LYVE-1 Molecule
| Tissue type* | Staining intensity and description‡ | |||
|---|---|---|---|---|
| Hematopoietic | ||||
| Lymph node | ++ | Lymphatic vessels | ||
| Spleen | ++ | Sinusoidal cells | ||
| Tonsil | − | |||
| PBMC | − | |||
| Endocrine | ||||
| Adrenal | + | Zona reticularis cells | ||
| Breast | ++ | Lymphatic vessels | ||
| Pancreas | + | Exocrine cells/islets of Langerhans | ||
| Thryroid | + | |||
| Prostate | − | |||
| Salivary gland | + | Lymphatic vessels | ||
| Reproductive | ||||
| Cervix | − | |||
| Ovary | − | |||
| Placenta | + | Syncytiotrophoblasts | ||
| Testis | − | |||
| Uterus | +/− | |||
| Gastrointestinal | ||||
| Appendix | + | Lymphatic vessels | ||
| Colon | ++ | Lymphatic vessels | ||
| Omentum | + | Lymphatic vessels | ||
| Small intestine | ++ | Lymphatic vessels | ||
| Stomach | + | Lymphatic vessels | ||
| Cardiorespiratory | ||||
| Heart | − | |||
| Lung | +/− | Lymphatic vessels | ||
| Other | ||||
| Brain | + | Cerebral cortex neurons | ||
| Cerebellum | − | |||
| Bone marrow | + | |||
| Bladder | − | |||
| Kidney | ++ | Tubular cuboidal epithelium | ||
| Liver | +/− | |||
| Skin | + | Lymphatic vessels |
Paraffin-embedded human tissue sections were incubated with a polyclonal rabbit anti–human LYVE-1 protein antiserum and stained with horseradish peroxidase–conjugated anti–rabbit antibody as described in Materials and Methods (see also Fig. 8).
Staining intensity was assessed semiquantitatively using the following indices: +++, strong staining; ++, moderate staining; +, weak staining; +/−, equivocal staining; −, no staining. PBMC, peripheral blood mononuclear cell.
Figure 9.
Immunofluorescent staining of LYVE-1 in lymphatic vessels and colocalization with HA. Sections of human small intestine were double stained for LYVE-1, CD44, the vascular endothelial molecules CD34 and vWF, and for HA, by indirect immunofluorescence microscopy, using Texas red or fluorescein-conjugated secondary antibodies or bHABC as described in Materials and Methods. The antibody combinations were as follows: A, LYVE-1 (red) and CD34 (green); B, LYVE-1 (red) and vWF (green); C, LYVE-1 (red) and CD44 (green); and D and E, LYVE-1 (red) and bHABC (green). Arrows depict small blood vessels containing erythrocytes (pale yellow/ green), which are LYVE-1-ve. Lymph vessel endothelium positive for both LYVE-1 and HA in D and E is stained yellow/orange. Bar, 50 μm.
Physiological Function of the LYVE-1 HA Receptor
The lymphatic system is the main conduit for transport of HA from the tissues and degradation within lymph nodes. However, the molecules within lymphatic vessels which interact with lymph fluid HA have not been identified, and the mechanisms driving HA transport to the lymph nodes have yet to be elucidated. Therefore, we considered the possibilities that LYVE-1 might either sequester HA on the lymph vessel wall or promote HA-mediated rolling of leukocytes present within the lymph fluid. In the first instance, we looked for an association between LYVE-1 and its ligand HA on the luminal face of tissue lymphatics by double immunofluorescent staining, using bHABC composed of bovine Aggrecan and Link protein (Fig. 9). Abundant quantities of HA were detected throughout the parenchyma and lining the walls of numerous vessels in all the tissues tested (data not shown). A significant amount of this HA indeed colocalized with LYVE-1 in numerous patches (visible as yellow staining), around the luminal surface of lymph vessels, particularly those in the small intestines (Fig. 9). Although relatively high levels of HA (50 μg/ml) are normally found within lymph vessels in vivo (49), we observed some variation in the amounts retained on LYVE-1 stained vessels between and within different tissues. It is not yet clear whether this reflects variation in the amount of HA, or its recovery during the preparation of different tissues, or variation in the binding affinity of LYVE-1 in different locations.
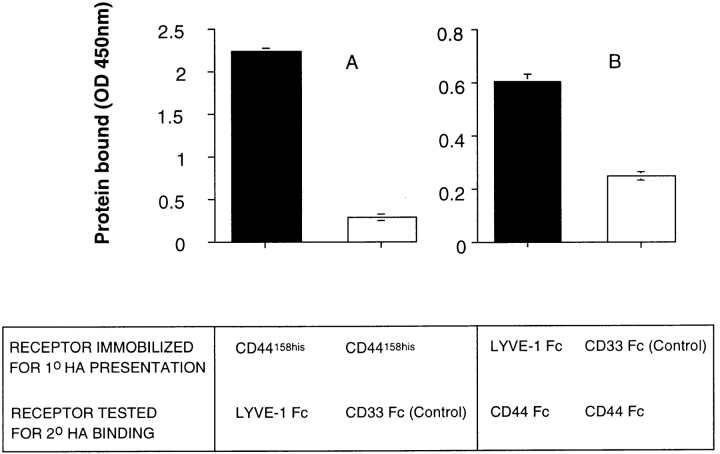
Finally, we looked for evidence that leukocytes adhere to HA presented by LYVE-1 present on the lymph vessel wall. Our initial attempts to demonstrate such adhesion using CD44 transfected Namalwa B lymphoma cells and frozen sections of gut lymphatics in Stamper-Woodruff type assays were difficult to interpret, because of the high levels of binding to HA within the surrounding parenchyma (not shown). As an alternative, we tested the ability of LYVE-1 Fc immobilized on plastic microtiter dishes to support HA-mediated binding to CD44. As shown in Fig. 10, LYVE-1 indeed supported HA-mediated binding of CD44, regardless of the order used for primary presentation or secondary HA binding. In conclusion, our results indicate that LYVE-1, in addition to binding HA within the lymphatics, may also bind HA-coated lymphocytes in transit to the lymph nodes.
Figure 10.
LYVE-1 molecules support HA-mediated binding to CD44. The capacity of LYVE-1 and CD44 to form ternary complexes with HA was tested using a modification of the HA-binding assay in Fig. 4 B. A shows binding of LYVE-1 Fc and CD33 Fc (control) to HA, presented by immobilized CD44158his protein. B shows binding of CD44 Fc to HA, presented by immobilized LYVE-1 Fc or CD33 Fc (control). Bound fusion proteins were detected with peroxidase-conjugated anti–human IgFc antibody, or with peroxidase-conjugated CD44 mAb A3D8, respectively, as described in Materials and Methods. No binding was observed in the absence of HA (not shown). Data are the mean ± SEM (n = 3).
Discussion
The interaction between cells and extracellular matrix HA is clearly of fundamental importance both during embryonic limb development and in processes such as wound healing and inflammation in later adult life (19–22). Such importance predicts that many receptors must be involved in HA adhesion, and that complex mechanisms must exist to regulate both their expression and their ligand-binding affinity. Yet at present, the list of professional cell surface HA receptors is small and includes only the CD44 molecule, RHAMM, and the LEC HA receptors. Furthermore, deletion of the gene for CD44, the most abundant and widely expressed of these, has little if any deleterious effects on the embryo (43). Thus, it has become increasingly clear to many workers in the field that additional receptors for HA are likely to exist within the genome.
In this paper we have described the primary structure and biological function of one such receptor, termed LYVE-1, which is present within vessels of the lymphatic system. This new receptor is a type I integral membrane polypeptide whose extracellular domain encodes a single cartilage Link module, the prototypic HA-binding domain conserved within all members of the Link or hyaladherin superfamily (51). The central core of the LYVE-1 Link module (C2-C3) is 57% identical to that of the human CD44 HA receptor, the only other Link superfamily HA receptor to be described to date and the closest homologue of LYVE-1. Nevertheless, there are distinct differences between LYVE-1 and CD44 that suggest the two homologues differ either in the mode of HA binding or in its regulation.
Firstly, in terms of HA binding, only three of nine residues identified as essential within the CD44 Link module are conserved in LYVE-1 (Lys 38, Tyr 79, and Asp 100) and none of the downstream COOH-terminal basic residues of CD44 are present. Furthermore, unlike CD44, the affinity of LYVE-1 for bHA is drastically reduced with increasing degrees of biotinylation. These features suggest HA is orientated differently within the LYVE-1 Link module and suggest it may bind a larger HA unit than the minimal HA6 unit bound by CD44 (18, 55). Competition experiments with defined HA fragment sizes will be required to distinguish between these possibilities. A further distinction between LYVE-1 and CD44 is their dramatic difference in substrate specificity. Unlike CD44, LYVE-1 displays exquisite specificity for binding HA and no affinity for the glycosaminoglycans chondroitin-4 sulfate, chondroitin-6 sulfate, or heparan sulfate. This remarkable ligand specificity also distinguishes LYVE-1 from the lymph node HA receptors which mediate the uptake and degradation of HA from afferent lymph, a process that is blocked by chondroitin sulfate and chondroitin sulfate proteoglycans (54).
Secondly, the regulation of HA binding and its functional consequences are likely to differ significantly from CD44. Excluding the Link domain and its immediate boundaries, the LYVE-1 extracellular domain, transmembrane anchor, and cytoplasmic tail bear no similarity to those of the CD44 molecule. In the CD44 ectodomain, alternative splicing of up to 10 exons within the membrane-proximal region regulates HA binding in some cell types (5, 44), and introduces new ligand-binding specificities (4, 17). Although we have detected high molecular weight LYVE-1 transcripts in some tissues by Northern blotting (see Fig. 5), we have so far not observed alternatively spliced variants by RT-PCR. In the CD44 transmembrane anchor, a central cysteine residue (30) regulates HA binding in a cell-specific manner, through the formation of lipid domains within the bilayer (35, 39). No such residue is present in the LYVE-1 transmembrane anchor. Lastly, in the CD44 molecule, the 71-residue cytoplasmic tail binds the cytoskeletal components ankyrin and ezrin (52), and is subject to serine phosphorylation that regulates cell motility on HA substrata (16, 38). Although the lack of sequence conservation does not rule out the possibility that LYVE-1 interacts with these cytoskeletal components via different residues or that the LYVE-1 cytoplasmic tail interacts with other as yet undefined molecules, it seems unlikely that the mechanisms which regulate CD44-HA interactions are duplicated for LYVE-1, its closest known homologue.
Perhaps the most striking distinction between LYVE-1 and CD44 is in their patterns of tissue expression. Intriguingly, the LYVE-1 molecule is largely if not exclusively restricted to lymph vessel endothelial cells from which CD44 is almost completely absent. Comprehensive analyses of a large panel of human tissues by immunoperoxidase staining with specific antisera revealed LYVE-1 lining the lymphatics in virtually every tissue where these structures could be distinguished, and included vessels draining gastrointestinal tissues, skin, lymph nodes, breast, and salivary gland. The greatest concentration of LYVE-1 expression was seen in submucosal lymph vessels underlying smooth muscle in the colon, and the Lacteal vessels of intestinal villi that transport dietary lipid absorbed from the small intestine. Lymphatic vessels are morphologically distinguishable from blood vessels by their reduced or missing basement membrane and lack of erythrocyte content. There are currently no specific immunochemical markers that allow identification of lymphatics, although limited studies with endothelial markers indicate differential expression of the L-selectin ligand CD34 and the coagulation Factor VIII–related protein vWF (32, 42). Our own results using double immunofluorescence indicate that LYVE-1 defines a set of lymphatic vessels which express neither CD34 nor vWF and identify LYVE-1 as a powerful new marker for future studies of lymphatic structure and function.
The absence of LYVE-1 from vascular endothelial cells, in addition to its absence from lymphocytes, macrophages, fibroblasts, and epithelial cells, implies a rather different physiological role from that of the CD44 molecule, which is expressed abundantly in each of these locations (41) but is largely absent from lymphatic vessels. The primary role of the CD44 HA receptor appears to be the regulation of cell motility (50), as evidenced by its association with the migration of mesenchymal cells during differentiation, and with the hematogenous spread of tumor cells in experimental neoplasia (47). This is also apparent in inflammation, where CD44 expression on lymphocytes promotes migration through HA-coated vascular endothelium (7, 8), as well as the migration of lymphocytes through HA-coated reticular networks within peripheral lymph nodes (6). LYVE-1 by contrast appears to be restricted to relatively nonmotile lymphatic endothelial cells, where it seems more likely to function in the immobilization of HA in the vessel lumen, rather than in its recognition as a positional cue for migration.
A prominent function of the lymphatic system is the provision of fluid drainage of immune cells and foreign antigens from the tissues to the peripheral lymph nodes, where the latter are sampled by professional antigen-presenting cells and phagocytes. Lymph fluid contains high levels (40–50 μg/ml) of HA (see ref. 12 and reviewed in ref. 10), and its flux through the lymphatics is known to increase in inflammatory diseases such as rheumatoid arthritis and psoriasis and in response to bacterial infection (10, 27). The lymph vessels thus act as a conduit both for migrating inflammatory cells and for HA, although the precise relationship between the two is not clear. One possibility is that LYVE-1 functions as an endocytic receptor in an analogous fashion to CD44 on macrophages and the LEC HA receptors on liver endothelium. The specificity of LYVE-1 for HA does, however, appear to rule out any likelihood that it is the receptor involved in HA uptake and degradation within the lymph node. Alternatively, since inflammatory cells express CD44, we are tempted to speculate that LYVE-1 molecules may promote their adhesion to lymphatic endothelium via HA. Such a role is supported by our finding that LYVE-1 and HA colocalize to the luminal face of the lymph vessels, and that LYVE-1 can support HA-mediated adhesion to CD44. We are currently performing experiments to distinguish between these and other interesting possibilities.
Acknowledgments
We thank Professor David Y. Mason and Professor Kevin Gatter (Department of Cellular Science, University of Oxford) for helpful discussions and for the generous donation of multiple tissue sections and reagents for immunohistochemistry. We also gratefully acknowledge Mr. Simon Biddulph (Paediatric Pathology Laboratory, Oxford Radcliffe Hospital) for preparing paraffin-embedded sections for microscopy.
This study was supported by a Medical Research Council Senior Fellowship and a Cancer Research Campaign Project Grant to D. Jackson.
Abbreviations used in this paper
- bHABC
biotinylated HA-binding complex
- EST
expression sequence tag
- HA
hyaluronan
- HUVEC
human umbilical vein endothelial cells
- LEC
liver endothelial cell
- LYVE-1
lymphatic vessel endothelial HA receptor 1
- RHAMM
receptor for hyaluronan mediated motility
- vWF
von Willebrand factor
References
- 1.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 2.Bajorath J, Greenfield B, Munro SB, Day AJ, Aruffo A. Identification of CD44 residues important for hyaluronan binding and delineation of the binding site. J Biol Chem. 1998;273:338–343. doi: 10.1074/jbc.273.1.338. [DOI] [PubMed] [Google Scholar]
- 3.Banerji S, Day AJ, Kahmann JD, Jackson DG. Characterization of a functional hyaluronan-binding domain from the human CD44 molecule expressed in Escherichia coli. . Protein Exp Purification. 1998;14:371–381. doi: 10.1006/prep.1998.0971. [DOI] [PubMed] [Google Scholar]
- 4.Bennett K, Jackson DG, Simon JC, Tanczos E, Peach R, Modrell B, Stamenkovic I, Plowman G, Aruffo A. CD44 isoforms containing exon v3 are responsible for the presentation of heparin binding growth factor. J Cell Biol. 1995;128:687–698. doi: 10.1083/jcb.128.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett K, Modrell B, Greenfield B, Bartolazzi A, Stamenkovic I, Peach R, Jackson D, Spring F, Aruffo A. Regulation of CD44 binding to hyaluronan by glycosylation of variably spliced exons. J Cell Biol. 1995;131:1623–1633. doi: 10.1083/jcb.131.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark RA, Alon R, Springer TA. CD44 and hyaluronan-dependent rolling interactions of lymphocytes on tonsillar stroma. J Cell Biol. 1996;134:1075–1087. doi: 10.1083/jcb.134.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Grendele HC, Estess P, Picker LJ, Siegelman MH. CD44 and its ligand hyaluronate mediate rolling under physiological flow: a novel lymphocyte–endothelial cell primary adhesion pathway. J Exp Med. 1996;183:1119–1130. doi: 10.1084/jem.183.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Grendele HC, Estess P, Siegelman MH. Requirement for CD44 in activated T cell extravasation into an inflammatory site. Science. 1997;278:672–675. doi: 10.1126/science.278.5338.672. [DOI] [PubMed] [Google Scholar]
- 9.Doege KJ, Sasaki M, Kimura T, Yamada Y. Complete coding sequence and deduced primary structure of the human cartilage large aggregating proteoglycan, aggrecan. J Biol Chem. 1991;266:894–902. [PubMed] [Google Scholar]
- 10.Engstrom-Laurent A. Changes in hyaluronan concentration in tissues and body fluids in disease states. CIBA Found Symp. 1989;143:233–247. doi: 10.1002/9780470513774.ch14. [DOI] [PubMed] [Google Scholar]
- 11.Fraser JR, Laurent TC. Turnover and metabolism of hyaluronan. CIBA Found Symp. 1989;143:41–53. [PubMed] [Google Scholar]
- 12.Fraser JR, Kimpton WG, Laurent TC, Cahill RN, Vakakis N. Uptake and degradation of hyaluronan in lymphatic tissue. Biochem J. 1988;256:153–158. doi: 10.1042/bj2560153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein LA, Zhou DF, Picker LJ, Minty CN, Bargatze RF, Ding JF, Butcher EC. A human lymphocyte homing receptor, the hermes antigen, is related to cartilage proteoglycan core and link proteins. Cell. 1989;56:1063–1072. doi: 10.1016/0092-8674(89)90639-9. [DOI] [PubMed] [Google Scholar]
- 14.Grammatikakis N, Grammatikakis A, Yoneda M, Yu Q, Banerjee SD, Toole BP. A novel glycosaminoglycan-binding protein is the vertebrate homologue of the cell cycle control protein, Cdc37. J Biol Chem. 1995;270:16198–16205. doi: 10.1074/jbc.270.27.16198. [DOI] [PubMed] [Google Scholar]
- 15.Hardwick C, Hoare K, Owens R, Hohn HP, Hook M, Moore D, Cripps V, Austen L, Nance DM, Turley EA. Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility. J Cell Biol. 1992;117:1343–1350. doi: 10.1083/jcb.117.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isacke CM. The role of the cytoplasmic domain in regulating CD44 function. J Cell Sci. 1994;107:2353–2359. doi: 10.1242/jcs.107.9.2353. [DOI] [PubMed] [Google Scholar]
- 17.Jackson DG, Bell JI, Dickinson R, Timans J, Shields J, Whittle N. Proteoglycan forms of the lymphocyte homing receptor CD44 are alternatively spliced variants containing the v3 exon. J Cell Biol. 1995;128:673–686. doi: 10.1083/jcb.128.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knudson CB. Hyaluronan receptor–directed assembly of chondrocyte pericellular matrix. J Cell Biol. 1993;120:825–834. doi: 10.1083/jcb.120.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knudson CB, Knudson W. Hyaluronan-binding proteins in development, tissue homeostasis and disease. FASEB (Fed Am Soc Exp Biol) J. 1993;7:1233–1241. [PubMed] [Google Scholar]
- 20.Knudson W, Knudson CB. Assembly of a chondrocyte-like pericellular matrix on non-chondrogenic cells. J Cell Sci. 1991;99:227–235. doi: 10.1242/jcs.99.2.227. [DOI] [PubMed] [Google Scholar]
- 21.Knudson W, Biswas C, Li XQ, Nemec RE, Toole BP. The role and regulation of tumour-associated hyaluronan. CIBA Found Symp. 1989;143:150–169. doi: 10.1002/9780470513774.ch10. [DOI] [PubMed] [Google Scholar]
- 22.Knudson W, Bartnik E, Knudson CB. Assembly of pericellular matrices by COS-7 cells transfected with CD44 lymphocyte-homing receptor genes. Proc Natl Acad Sci USA. 1993;90:4003–4007. doi: 10.1073/pnas.90.9.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohda D, Morton CJ, Parkar AA, Hatanaka H, Inagaki FM, Campbell ID, Day AJ. Solution structure of the link module: a hyaluronan binding domain involved in extracellular matrix stability and cell migration. Cell. 1996;86:767–775. doi: 10.1016/s0092-8674(00)80151-8. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucl Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyte J, Doolittle RF. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Laurent TC, Fraser JR. Hyaluronan. FASEB (Fed Am Soc Exp Biol) J. 1992;6:2397–2404. [PubMed] [Google Scholar]
- 27.Lebel L, Smith L, Risberg B, Laurent TC, Gerdin B. Increased lymphatic elimination of interstitial hyaluronan during E. colisepsis in sheep. Am J Physiol. 1989;256:1524–1531. doi: 10.1152/ajpheart.1989.256.6.H1524. [DOI] [PubMed] [Google Scholar]
- 28.Lee TH, Wisniewski HG, Vilcek J. A novel secretory tumor necrosis factor–inducible protein (TSG-6) is a member of the family of hyaluronate binding proteins, closely related to the adhesion receptor CD44. J Cell Biol. 1992;116:545–557. doi: 10.1083/jcb.116.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesley J, Hyman R, Kincade PW. CD44 and its interaction with the extracellular matrix. Adv Immunol. 1994;54:271–335. doi: 10.1016/s0065-2776(08)60537-4. [DOI] [PubMed] [Google Scholar]
- 30.Liu D, Sy MS. A cysteine residue located in the transmembrane domain of CD44 is important in binding of CD44 to hyaluronic acid. J Exp Med. 1996;183:1987–1994. doi: 10.1084/jem.183.5.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald JA, Brehmgibson T, Cananisch T, Spicer AP. Molecular cloning and gene targeting of hyaluronan synthase 2 reveals a critical role in the development of the cardiovascular system in the mouse. Glycobiology. 1998;8:146–155. [Google Scholar]
- 32.Miettinen M, Lindenmayer AE, Chaubal A. Endothelial cell markers CD31, CD34, and BNH9 antibody to H- and Y-antigens: evaluation of their specificity and sensitivity in the diagnosis of vascular tumors and comparison with von Willebrand factor. Mod Pathol. 1994;7:82–90. [PubMed] [Google Scholar]
- 33.Mikecz K, Brennan FR, Kim JH, Glant TT. Anti-CD44 treatment abrogates tissue edema and leukocyte infiltration in murine arthritis. Nat Med. 1995;1:558–563. doi: 10.1038/nm0695-558. [DOI] [PubMed] [Google Scholar]
- 34.Neame PJ, Christner JE, Baker JR. The primary structure of Link protein from rat chondrosarcoma proteoglycan aggregate. J Biol Chem. 1986;261:3519–3535. [PubMed] [Google Scholar]
- 35.Neame SJ, Uff CR, Sheikh H, Wheatley SC, Isacke CM. CD44 exhibits a cell type dependent interaction with Triton X-100 insoluble, lipid rich, plasma membrane domains. J Cell Sci. 1995;108:3127–3135. doi: 10.1242/jcs.108.9.3127. [DOI] [PubMed] [Google Scholar]
- 36.Ostgaard G, Reed RK. Intravenous saline infusion in rat increases hyaluronan efflux in intestinal lymph by increasing lymph flow. Acta Physiol Scand. 1993;147:329–335. doi: 10.1111/j.1748-1716.1993.tb09506.x. [DOI] [PubMed] [Google Scholar]
- 37.Peach RJ, Hollenbaugh D, Stamenkovic I, Aruffo A. Identification of hyaluronic acid binding sites in the extracellular domain of CD44. J Cell Biol. 1993;122:257–264. doi: 10.1083/jcb.122.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peck D, Isacke CM. CD44 phosphorylation regulates melanoma cell and fibroblast migration on, but not attachment to, a hyaluronan substratum. Curr Biol. 1996;6:884–890. doi: 10.1016/s0960-9822(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 39.Perschl A, Lesley J, English N, Hyman R, Trowbridge IS. Transmembrane domain of CD44 is required for its detergent insolubility in fibroblasts. J Cell Sci. 1995;108:1033–1041. doi: 10.1242/jcs.108.3.1033. [DOI] [PubMed] [Google Scholar]
- 40.Picker LJ, De J los Toyos, M.J. Telen, B.F. Haynes, and E.C. Butcher. Monoclonal antibodies against the CD44 [In(Lu)-related p80], and Pgp-1 antigens in man recognize the Hermes class of lymphocyte homing receptors. J Immunol. 1989;142:2046–2051. [PubMed] [Google Scholar]
- 41.Picker LJ, Nakache M, Butcher EC. Monoclonal antibodies to human lymphocyte homing receptors define a novel class of adhesion molecules on diverse cell types. J Cell Biol. 1989;109:927–937. doi: 10.1083/jcb.109.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauter B, Foedinger D, Sterniczky B, Wolff K, Rappersberger K. Immunoelectron microscopic characterization of human dermal lymphatic microvascular endothelial cells. Differential expression of CD31, CD34, and type IV collagen with lymphatic endothelial cells vs. blood capillary endothelial cells in normal human skin, lymphangioma, and hemangioma in situ. J Histochem Cytochem. 1998;46:165–176. doi: 10.1177/002215549804600205. [DOI] [PubMed] [Google Scholar]
- 43.Schmits R, Filmus J, Gerwin N, Senaldi G, Kiefer F, Kundig T, Wakeham A, Shahinian A, Catzavelos C, Rak J, et al. CD44 regulates hematopoietic progenitor distribution, granuloma formation, and tumorigenicity. Blood. 1997;90:2217–2233. [PubMed] [Google Scholar]
- 44.Sleeman J, Rudy W, Hofmann M, Moll J, Herrlich P, Ponta H. Regulated clustering of variant CD44 proteins increases their hyaluronate binding capacity. J Cell Biol. 1996;135:1139–1150. doi: 10.1083/jcb.135.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamenkovic I, Amiot M, Pesando JM, Seed B. A lymphocyte molecule implicated in lymph node homing is a member of the cartilage link protein family. Cell. 1989;56:1057–1062. doi: 10.1016/0092-8674(89)90638-7. [DOI] [PubMed] [Google Scholar]
- 46.Sy MS, Guo YJ, Stamenkovic I. Distinct effects of two CD44 isoforms on tumor growth in vivo. J Exp Med. 1991;174:859–866. doi: 10.1084/jem.174.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sy MS, Guo YJ, Stamenkovic I. Inhibition of tumor growth in vivo with a soluble CD44-immunoglobulin fusion protein. J Exp Med. 1992;176:623–627. doi: 10.1084/jem.176.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tammi R, Ågren UM, Tuhkanen A-L, Tammi M. Hyaluronan metabolism in skin. Progress Histochem Cytochem. 1994;29:1–77. doi: 10.1016/s0079-6336(11)80023-9. [DOI] [PubMed] [Google Scholar]
- 49.Tengblad A, Laurent UB, Lilja K, Cahill RN, Engstrom-Laurent A, Fraser JR, Hansson HE, Laurent TC. Concentration and relative molecular mass of hyaluronate in lymph and blood. Biochem J. 1986;236:521–525. doi: 10.1042/bj2360521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas L, Byers HR, Vink J, Stamenkovic I. CD44H regulates tumor cell migration on hyaluronate-coated substrate. J Cell Biol. 1992;118:971–977. doi: 10.1083/jcb.118.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toole BP. Hyaluronan and its binding proteins, the hyaladherins. Curr Opin Cell Biol. 1990;2:839–844. doi: 10.1016/0955-0674(90)90081-o. [DOI] [PubMed] [Google Scholar]
- 52.Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turley EA, Austen L, Moore D, Hoare K. Ras-transformed cells express both CD44 and RHAMM hyaluronan receptors: only RHAMM is essential for hyaluronan-promoted locomotion. Exp Cell Res. 1993;207:277–282. doi: 10.1006/excr.1993.1194. [DOI] [PubMed] [Google Scholar]
- 54.Tzaicos C, Fraser JR, Tsotsis E, Kimpton WG. Inhibition of hyaluronan uptake in lymphatic tissue by chondroitin sulphate proteoglycan. Biochem J. 1989;264:823–828. doi: 10.1042/bj2640823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Underhill CB, Chi-Rosso G, Toole BP. Effects of detergent solubilization on the hyaluronate-binding protein from membranes of simian virus 40-transformed 3T3 cells. J Biol Chem. 1983;258:8086–8091. [PubMed] [Google Scholar]
- 56.Von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984;173:243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- 57.Weiss JM, Sleeman J, Renkl AC, Dittmar H, Termeer CC, Taxis S, Howells N, Hofmann M, Kohler G, Schopf E, et al. An essential role for CD44 variant isoforms in epidermal Langerhans cell and blood dendritic cell function. J Cell Biol. 1997;137:1137–1147. doi: 10.1083/jcb.137.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada H, Watanabe K, Shimonaka M, Yamaguchi Y. Molecular cloning of brevican, a novel brain proteoglycan of the aggrecan/ versican family. J Biol Chem. 1994;269:10119–10126. [PubMed] [Google Scholar]
- 59.Yannariello-Brown J, Frost SJ, Weigel PH. Identification of the Ca(2+)-independent endocytic hyaluronan receptor in rat liver sinusoidal endothelial cells using a photoaffinity cross-linking reagent. J Biol Chem. 1992;267:20451–20456. [PubMed] [Google Scholar]
- 60.Yannariello-Brown J, McGary CT, Weigel PH. The endocytic hyaluronan receptor in rat liver sinusoidal endothelial cells is Ca2+-independent and distinct from a Ca2+-dependent hyaluronan binding activity. J Cell Biochem. 1992;48:73–80. doi: 10.1002/jcb.240480111. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerman DR, Ruoslahti E. Multiple domains of the large fibroblast proteoglycan versican. EMBO (Eur Mol Biol Organ) J. 1989;8:2975–2981. doi: 10.1002/j.1460-2075.1989.tb08447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]