Abstract
Met murine hepatocyte (MMH) lines were established from livers of transgenic mice expressing constitutively active human Met. These lines harbor two cell types: epithelial cells resembling the parental populations and flattened cells with multiple projections and a dispersed growth habit that are designated palmate. Epithelial cells express the liver-enriched transcription factors HNF4 and HNF1α, and proteins associated with epithelial cell differentiation. Treatments that modulate their differentiation state, including acidic FGF, induce hepatic functions. Palmate cells show none of these properties. However, they can differentiate along the hepatic cell lineage, giving rise to: (a) epithelial cells that express hepatic transcription factors and are competent to express hepatic functions; (b) bile duct-like structures in three-dimensional Matrigel cultures. Derivation of epithelial from palmate cells is confirmed by characterization of the progeny of individually fished cells. Furthermore, karyotype analysis confirms the direction of the phenotypic transition: palmate cells are diploid and the epithelial cells are hypotetraploid. The clonal isolation of the palmate cell, an immortalized nontransformed bipotential cell that does not yet express the liver-enriched transcription factors and is a precursor of the epithelial-hepatocyte in MMH lines, provides a new tool for the study of mechanisms controlling liver development.
Keywords: acidic FGF, epithelial morphogenesis, hepatic development and differentiation, HGF/SF, liver-enriched transcription factors
The liver contains two major differentiated cell types of endodermal origin: hepatocytes, located in the hepatic parenchymal plates, and biliary epithelial cells, located in the bile ducts. In addition, several cell types presumed to be of mesenchymal origin are present, including Kupffer cells, Ito/stellate cells, and endothelial cells (Desmet, 1994).
Hepatoblasts are the cells that have migrated from the liver diverticulum into the septum transversum at ∼9.5 d of development in the mouse, and they are already determined along the hepatic epithelial cell lineage (Le Douarin, 1975; Houssaint, 1980; Zaret, 1996). These bipotential progenitor cells give rise to mature hepatocytes and to intrahepatic bile ducts and portions of the extrahepatic ducts (Germain et al., 1988b ; Shiojiri et al., 1991). The hepatocytes assume their polarized definitive differentiation only just before birth (Stamatoglou et al., 1992).
An equivalent bipotential precursor cell (stem cell) in adult liver remains to be identified. Although the liver is a quiescent organ, terminally differentiated hepatocytes are able to undergo active proliferation after cell loss (Michalopoulos and DeFrances, 1997). In response to massive necrosis, when hepatocytes cannot respond to a growth stimulus and during some regimes of chemically induced hepatocarcinogenesis, a mixed cell type compartment emerges in the region of the portal triads, referred to as the “oval cell compartment” (Farber, 1956; Germain et al., 1988a ; Lemire et al., 1991; Evarts et al., 1993). A variable proportion of cells in the compartment share markers with fetal hepatocytes, including the expression of α-fetoprotein, albumin, γ-glutamyl transpeptidase, placental alkaline phosphatase, and the pattern of expression of cytokeratins (CK).1 On the basis of their localization in the liver cell plate and of their morphological features, the oval cells seem to correspond to the primitive intrahepatic bile duct elements descended from hepatoblasts (Fausto et al., 1993).
Although the different steps involved in liver development are well described from the hepatic specification of ventral endoderm to the generation of hepatoblasts and the differentiation of mature hepatocytes, the control mechanisms underlying these steps are still poorly understood. Recent experiments with primary tissue explants of foregut endoderm have suggested the influence of positive and negative extracellular signals, respectively provided by cardiac and dorsal mesoderm, during early hepatic specification (Gualdi et al., 1996). Gene disruption in mice has demonstrated that factors as diverse as c-Jun (Hilberg et al., 1993), RelA (Beg et al., 1995), and both hepatocyte growth factor/scatter factor (HGF/SF; Schmidt et al., 1995) and its receptor Met (Bladt et al., 1995) are implicated in hepatogenesis. It is generally accepted that particular combinations of members of four families of liver-enriched transcription factors (LETF), including hepatocyte nuclear factor (HNF)3 (Lai et al., 1991), HNF4 (Sladek et al., 1990), HNF1 (Frain et al., 1989), and CCAAT/ enhancer binding protein (C/EBP; Johnson et al., 1987) control critical steps in hepatic differentiation, and that a hierarchy of expression of these transcription factors exists (Cereghini, 1996). Among the possible LETF combinatorial mechanisms involved in hepatic cell differentiation the HNF4-HNF1α couple has a key role (Kuo et al., 1992; Spath and Weiss, 1997). Moreover the extracellular environment plays a critical role in regulating the activities of these transcription factors, thereby helping to maintain hepatocyte differentiation (Liu et al., 1991). Recently, a role for HNF4 in integrating the genetic programs of liver-specific gene expression and of epithelial morphogenesis of hepatocytes has been described (Spath and Weiss, 1998).
The lack of stable precursor cell lines that differentiate into hepatocytes in culture has hampered clarification of the events controlling liver development. Recently, we established ∼30 immortalized hepatocytic cell lines (Met murine hepatocyte [MMH]) from explants of embryonic, fetal, and new-born liver derived from transgenic mice expressing a constitutively active truncated human Met receptor (cyto-Met) under control of the human α1-antitrypsin transcription unit (Amicone et al., 1997). All of the MMH lines are untransformed and present a differentiated phenotype judging from the retention of epithelial cell polarity and the expression of LETF. In addition, many express hepatic functions. Although the immortalization of transgenic liver cells was reproducible, the event was rare: a small number of epithelial islands emerged and grew from a large number of cells in the primary cultures (Amicone et al., 1997).
With the goal of identifying an immortalized bipotential precursor cell within the MMH lines, several lines were cloned and the properties of the progeny clones determined. The results reveal that some of the MMH lines are composed of two distinct cell types: the expected epithelial cells as well as a cell type of spreading fibroblast-like morphology that we designate palmate. The epithelial cells express hepatocyte functions, or are competent to do so upon induction. In contrast, the palmate cells express neither LETF nor hepatic functions. However, they are bipotential, for they give rise to epithelial cells, spontaneously upon continuous culture, or precociously under the appropriate environmental conditions. Epithelial progeny of palmate cells express LETF and are competent to express hepatic functions. In addition, in three-dimensional cultures, palmate cells form hollow tubules lined with microvilli, reminiscent of bile ducts.
Unequivocal demonstration that palmate cells can give rise to epithelial-hepatocytes is provided by cloning of individually fished cells and characterization of their progeny. Moreover as true stem cells, palmate cells are diploid whereas their epithelial progeny are hypotetraploid. All of these findings demonstrate that palmate cells are the precursors of hepatocytes in MMH cell lines.
Materials and Methods
Cell Lines and Culture Conditions
The MMH lines are immortalized cell lines derived from livers of transgenic mice expressing a truncated human MET proto-oncogene. Details on the construction and on the tissue-specific and temporal expression of the transgene are published (Amicone et al., 1995).
The MMH lines were grown in RPMI 1640 supplemented with 10% FCS, 50 ng/ml EGF, 30 ng/ml IGF II, 10 μg/ml insulin (Boehringer Mannheim Corp., Indianapolis, IN), and antibiotics. Cells were usually plated on collagen I–coated (Sigma Chemical Co., St. Louis, MO) Petri dishes (Falcon Plastics, Cockeysville, MD). Each passage corresponds to five to six cell generations. Cell passages were calculated from initial thawing.
The differentiated rat hepatoma cell lines Fao and FGC4 were derived from clonal line H4IIEC3 (Pitot et al., 1964). Rat hepatoma cells were grown in modified Ham's F12 medium supplemented with 5% FCS (Coon and Weiss, 1969). All cell lines were incubated in a humidified atmosphere with 7% CO2 at 37°C.
Environmental Factors
For some experiments, the normal growth medium was supplemented with 2% DMSO (Sigma Chemical Co.) or 100 ng/ml aFGF (always supplied in association with 10 μg/ml Heparin; GIBCO BRL, Gaithersburg, MD). Where indicated, cells were grown on gelatin-coated dishes (0.1% gelatin in PBS; the sterile solution was left on the dishes for 20 min before aspiration and inoculation of cells). To prepare gels derived from the Engelbreth-Holm-Swarm (EHS) mouse tumor (Orkin et al., 1977) 0.5 ml of basement membrane (Matrigel; Collaborative Research, Inc., Waltham, MA) was poured into 60-mm dishes and permitted to set at 37°C for 1–2 h.
Cloning
Cloning of MMH lines was performed by inoculation of 100 or 200 cells into 10-cm Petri dishes. 1 wk later well isolated colonies were scraped and plated onto collagen I–coated 12-well microtiter plates (Falcon Plastics, Cockeysville, MD) and expanded.
Visually-verified single cell cloning of MMH lines was carried out by picking one cell from a diluted cell suspension of 102 cells/ml with a P-20 micropipette under a dissecting microscope. After verifying that one single cell had been picked, by expelling it into a drop of pure medium, the cell was recovered in the P-20 and seeded onto a feeder layer in a 12-well microtiter plate. Three types of feeder layers were used for the cloning experiments and were prepared as follows: MMH E14 cells and the MMH E14 epithelial clone cells were incubated over night with 10 μg/ml mytomicin (Sigma Chemicals Co.); a primary culture of mouse embryonic fibroblasts (EF) was incubated 3 h with 5 μg/ml mytomicin. After treatment with mytomicin cells were plated onto collagen I–coated 12-well microtiter plates (2 × 105 cells/well). In all experiments, feeder layers alone were incubated for several weeks to verify that no colony forming cells were present.
Soft Agar Colony Formation
The soft agar assay was performed by inoculating 1 × 105 cells suspended in 1.5 ml of DME containing 10% FCS, 50 ng/ml EGF, 30 ng/ml IGF II, 10 μg/ml insulin, and supplemented with 0.34% agar (Sigma Chemical Co.) in 60-mm Petri dishes coated with a 0.68% agar underlay. Cultures were incubated for 20 d.
Karyotype and FACS® Analysis
Karyotype studies were performed on colcemid-arrested metaphases. Colcemid (0.1–0.2 μg/ml final; GIBCO BRL) was added to the cell culture for 45 min at 37°C. Cells were trypsinized, swollen with 75 mM KCl at 37°C for 10 min, fixed, and air dried. At least 25 metaphases of each clone were counted. FACS® analysis was used to verify and quantitate the karyology results: cells in the exponential phase were harvested, fixed in 70% ethanol, washed in PBS, treated with RNase A (100 μg/ml) for 30 min at 37°C and then stained in propidium iodide (40 μg/ml).
Immunofluorescence Analysis and Antisera
For indirect immunofluorescence staining, cells were grown on collagen I–coated glass coverslips, fixed, and treated as described (Mevel-Ninio and Weiss, 1981). The antibodies were obtained and diluted as follows: mouse monoclonal anti-pan-CK antibody from Sigma Chemical Co., diluted 1/100; rat monoclonal anti-zonula occludens (ZO)-1 antibody (Stevenson et al., 1986), supplied by Doris Cassio (Institut Curie, Orsay, France), used undiluted; mouse monoclonal anti-E-cadherin antibody from Signal Transduction Laboratories (Lexington, KY), diluted 1/100; rat monoclonal anti-CK 8, 18, and 19 antibodies (TROMA 1, 2, 3; Kemler et al., 1981) provided by R. Kemler (Max-Planck Institute of Immunobiology, Freiburg, Germany), used undiluted; sheep anti–mouse Ig FITC linked whole antibody from Amersham International, used 1/100; rabbit polyclonal anti–rat IgG FITC conjugate antibody from Sigma Chemical Co., diluted 1/100. Preparations were examined with a Zeiss axiophot.
Transmission Electron Microscopy
For thin sections, aggregates of cells grown on Matrigel-coated dishes were fixed with phosphate-buffered 2.5% glutaraldehyde for 12–18 h at 4°C and postfixed in 1% OsO4. Cells were then dehydrated and embedded in epoxy resin. Thin sections for electron microscopy were counterstained with uranil acetate and lead citrate and examined with a Zeiss 109 microscope.
Preparation of Nuclear Extracts and Gel Mobility Shift Assay
Preparation of nuclear extracts and gel shift assay were performed as described by Cereghini (Cereghini et al., 1988). The binding reaction mixture was incubated on ice for 30 min and consisted of a 14-μl reaction mix containing 0.6 ng of 32P end-labeled double-stranded oligonucleotide, 10 mM Hepes (pH 7.9), 0.125 mM EDTA (pH 8), 0.0625 mM EGTA, 0.5 mM dithiothreitol, 7 mM MgCl2/spermidine, 1 μg/μl poly(dI-dC) (Pharmacia Biotech, Inc., Piscataway, NJ), 1 μg/μl sonicated salmon sperm DNA (Pharmacia Biotech, Inc.), and 10 μg of nuclear extract. The double-stranded oligonucleotides used correspond to the binding sites for HNF4 in the human apolipoprotein CIII promoter (C3P 5′-GGTCAGCAGGTGACCTTTGCCCAGCG-3′), for HNF1α in the rat albumin proximal promoter (PE56 5′-TGGTTAATGATCTACAGTTA-3′), for HNF3 in the mouse transthyretin (TTR) promoter (TTR 5′-TTGACTAAGTCAATAATCAGAATCAG-3′). The protein-DNA complexes were loaded and electrophoresed on a 6% polyacrylamide gel in 0.25× Tris-borate EDTA at 12 V/cm, fixed, dried and developed by autoradiography. A 200-fold molar excess of unlabeled oligonucleotides was included in the reaction when competition experiments were performed unless otherwise indicated in the figure legend. The double-stranded oligonucleotides used in competition experiments are: for HNF3 the binding site for HNF3 in the rat L-type 6-phospho-fructo2-kinase promoter (PFK3 5′-GATCGTCTTTTATTTGCATACTCTA-3′), for HNF4 the C3P oligo. Supershifts of HNF1 complexes were generated with 3 μl of diluted antibody (1:10) for HNF1α (a gift from Tanguy Chouard, Institut Pasteur).
RNA Analysis
For Northern blot analysis total cellular RNA was extracted from cells according to standard protocols. RNA samples (20 μg per lane) were resolved by electrophoresis in 1.85 M formaldehyde-agarose gels and blotted onto nylon membranes (Nycomed Amersham Inc., Princeton, NJ) by vacuum blotting (Pharmacia Biotech, Inc.). Blots were hybridized with 32 P-labeled cDNA inserts corresponding to HNF1α (Rey Campos et al., 1991); HNF3 (a plasmid containing the HNF3α DNA-binding domain was a gift from Mario Zakin); HNF4 (Sladek et al., 1990); C/EBPα (Friedman et al., 1989); albumin (Kioussis et al., 1981); β-fibrinogen (Crabtree and Kant, 1981); TTR (Derman et al., 1981); cytokeratins endo A (CK 8; Singer et al., 1986) and endo B (CK 18; Oshima et al., 1988).
Results
Although all of the MMH lines characterized by Amicone et al. (1997) presented similar epithelial morphology at high density, some displayed morphological heterogeneity at low density. Indeed, at low density fibroblast-like migratory cells are visible, but at high density the cultures present a homogeneous appearing epithelial monolayer. To identify the cell types comprising the cultures, and to explore the possibility that precursors of hepatocytes were present, clones were isolated. This work focused on two such heterogeneous cell lines, E14 and D2, derived from primary liver cultures of a 14-d embryo or a 2-d-old mouse, respectively. In addition, a homogeneous epithelial line, D3 (from a 3-d-old mouse), was examined in parallel.
Clones of MMH Lines Can Be of Two Morphological Categories
Clones of the E14 and D2 lines were of two different types: epithelial and non-epithelial. In contrast, the D3 colonies were all of epithelial morphology (Table I). To have several representatives of the two classes of clones, we expanded and characterized two E14 clones, three clones from D2 and two from the D3 line. Most of the results presented below concern the E14 line and its clones; similar but less extensive analysis of the D2 clones revealed equivalent properties.
Table I.
Types of Progeny Colonies from Three MMH Lines
| MMH E14 | MMH D2 | MMH D3 | ||||
|---|---|---|---|---|---|---|
| PE | 26 | 12 | 28 | |||
| Epithelial colonies (%) | 61 | 22 | 100 | |||
| Non-epithelial colonies (%) | 39 | 78 | 0 | |||
| Clones expanded | 1 epith. | 2 epith. | 2 epith. | |||
| 1 non-epith. | 1 non-epith. |
The table summarizes the plating efficiency (PE) of the MMH lines 1 wk after inoculation of 100 or 200 cells into 10-cm Petri dishes, the percentage of epithelial and nonepithelial colonies obtained from each line, and the number of clones expanded and characterized for each line. The values given are averages of at least three independent experiments.
Fig. 1 shows the morphological characteristics of the two classes of clones. Although cells of both types of clones exhibit generation times of ∼24 h, striking differences concern: (a) the cell shape and (b) the growth habit. One class (referred to as epithelial) shows flattened epithelial morphology, similar to the parental lines (Amicone et al., 1997): the cells have a regular polygonal shape, with clear round nuclei, and grow in a tightly packed fashion. Cells of the second class show a flat fibroblast-like morphology: they are strongly attached to the dish, grow in a scattered and loosely packed fashion, and send out cytoplasmic extensions. To remain openminded concerning the origin of cells of this second type and to recall their characteristic form, we shall refer to them as palmate rather than fibroblast-like.
Figure 1.
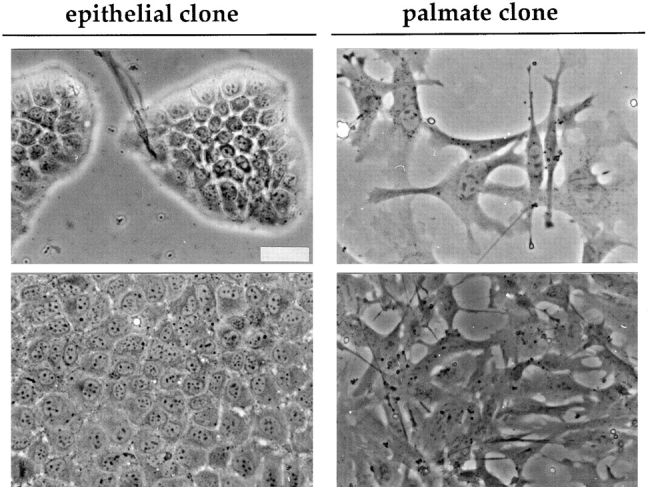
Morphology of clones derived from the MMH E14 line. Phase-contrast micrographs of the epithelial and the palmate clones at low (top) and high or intermediate (bottom) cell density, respectively. In the micrographs of the cultures at low density the differences in growth habit and cellular shape between the epithelial and the palmate clone are highlighted; those at higher density document that these differences are maintained. Bar, 40 μm.
Marker Analysis Reveals that the Morphological Differences Attest to Distinct Phenotypic States
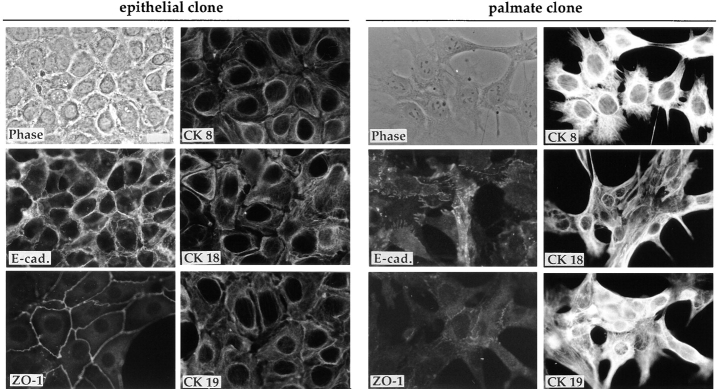
To explore possible differences in proteins that are implicated in defining cell morphology, immunofluorescence analysis was carried out to explore the localization and expression of several markers of differentiated epithelial cells in both epithelial and palmate clones. As shown in Fig. 2, in the epithelial cells there is strong expression of membrane-bound E-cadherin, an epithelium-specific adhesion molecule. ZO-1 is clearly visible as a fine network on the membrane areas of cell–cell contact. The palmate cells, as expected from their dispersed growth habit, show only weak and diffuse staining for E-cadherin and for the epithelial-polarity marker ZO-1. Interestingly, monoclonal antibodies directed against CK 8, CK 18, and CK 19, stage-specific markers of the hepatic cell lineage, reveal a difference in organization of the filaments in the two classes of clones, the epithelial cells showing weak staining of a dense network of filaments surrounding the nucleus, and the palmate cells' intense staining of a loosely organized network (Fig. 2).
Figure 2.
Immunofluores- cence analysis of expression of epithelial and stage-specific markers of the hepatic cell lineage (CKs 8, 18, and 19). Phase-contrast micrographs and immunofluorescence staining of the E14 epithelial and palmate clones for the expression and localization of E-cadherin (E-cad.) and ZO-1, and of the three CKs 8, 18, and 19. (The cytokeratin antibodies were used on liver sections as a control where they showed the expected staining patterns: CK 8 and 18 on hepatocytes, and CK 19 primarily on bile ducts). For the epithelial clone the E-cad. staining and the phase-contrast image are of the same field; for the palmate clone the CK 8 field corresponds to the phase-contrast image. Bar, 20 μm.
Our previous study (Amicone et al., 1997) of the MMH cell lines revealed that they failed to form colonies in soft agar, indicating that these cells do not have a transformed phenotype. We tested the ability of cells of both epithelial and palmate clones to form colonies in soft agar; both cell types failed to form colonies. This rules out the hypothesis that palmate cells correspond to transformed derivatives of the epithelial cells.
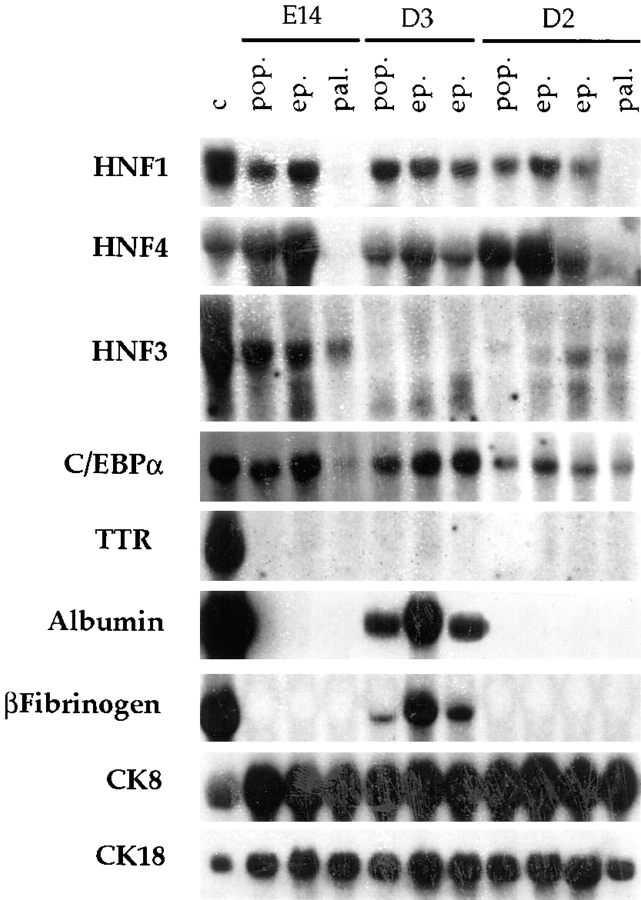
To document differences in gene expression underlying the respective morphological types, RNA was prepared from the three MMH parental lines (E14, D2, and D3) and the seven clones. Blots were prepared and hybridized with probes for LETF, for hepatic functions and for hepatocyte-specific CK 8 and 18 (Fig. 3).
Figure 3.
Characterization of the profile of liver-specific gene expression in MMH lines and their clones. Northern blot analysis of total RNA from the E14, D2, and D3 parental populations (pop.), from the epithelial (ep.) and palmate (pal.) clones, and from the control Fao rat hepatoma line (c). Each lane was loaded with 20 μg of total RNA. Probes used are indicated on the left.
The analysis of liver-specific gene expression confirms that the morphological differences correspond to the existence of two phenotypic classes of cells. Cells of the epithelial clones, as well as the three parental populations, express all of the LETF tested, with some variability in the forms of HNF3. By contrast, concerning the expression of liver-specific functions, only D3 cells and its two clones express the hepatic marker functions albumin, β-fibrinogen and α1-antitrypsin (Fig. 3 and data not shown). The palmate cells from both E14 and D2 lines are deficient for two major LETF (HNF1α and HNF4) and for the liver-specific functions. Data will be presented below to document that the presence and absence of LETF transcripts correlates with the presence and absence of the corresponding DNA-binding proteins, respectively. Transcripts for CK 8 and CK 18 are present in both the parental lines and the progeny clones of the two classes.
Palmate Cells Form Bile Ductular Structures on Matrigel
Because cell–matrix interactions play an important role in establishing and maintaining the differentiated cell phenotype, the capacity of both clones to undergo morphogenesis on Matrigel (a commercial derivative of matrix extracted from the EHS mouse sarcoma) was evaluated. Growth of cells on this three-dimensional matrix for 7–10 d permitted the cells to sink into the gel. Under these conditions a striking difference in intrinsic morphogenic activity between the two classes of clones was revealed. Fig. 4 A illustrates the spheroidal three-dimensional organization consistently observed with cultures of epithelial clones. In contrast, the palmate clone gave rise to spheroidal structures that send out tubules (Fig. 4 B). Electron microscopic examination of a thin section of a tubule reveals that cells in Matrigel become polarized, showing stabilized cell–cell contacts and circumscribing lumina densely decorated with microvilli, resembling bile ducts (Fig. 4 C).
Figure 4.
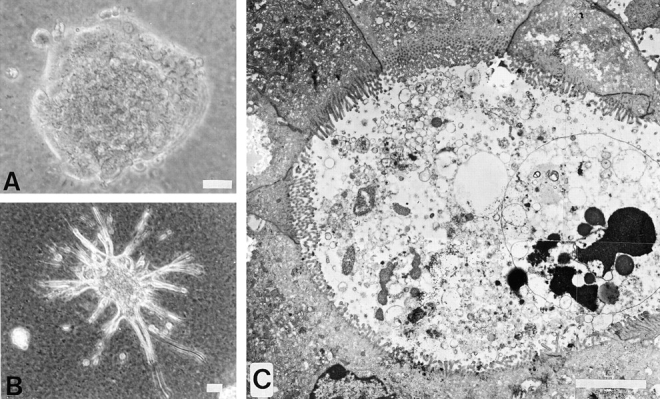
Differences in the intrinsic morphogenic activity of the epithelial and the palmate clones are revealed by culture on the three-dimensional matrix, Matrigel. Photographs taken from 10-d cultures on a thick layer of Matrigel. (A) Phase-contrast micrograph of the spheroidal organization of the epithelial clone; (B) palmate cells, showing a spheroidal colony sending out tubules. (C) composite of four overlapping electron micrographs of a thin section across a tubule of palmate cells highlighting a duct-like structure with a well-defined lumen and circumscribed by well-polarized cells with junctional complexes and luminal membranes covered with microvilli. Bars: (B) 40 μm; (C) 5 μm.
The tubules observed in Matrigel cultures of palmate but not epithelial cell clones are reminiscent of those observed by Montesano et al. (1997) upon the addition of HGF to cultures of several types of epithelial cells. Hence, it was appropriate to question whether the expression level of the cyto-MET transgene is different between epithelial and palmate cells. RT-PCR analysis of cyto-Met transcripts revealed similar signals for the two cell types, and in both cases the amounts of corresponding protein were below the level of detection by Western blot analysis of immunoprecipitated extracts (data not shown).
Extracellular Matrix and Soluble Factors Modulate the Differentiation State of Cells of Both Epithelial and Palmate Clones
It was unexpected that epithelial clones from the E14 and D2 lines and those from D3 do not display the same pattern of expression of liver marker genes, even though they all express the LETF HNF1α and HNF4. Indeed, only the D3 cells and its clones express some liver-specific functions (Fig. 3). To examine whether the other epithelial clones possess the potential to express these functions, various factors that could affect their state of differentiation were tested. Cells were grown on a gelatin two-dimensional matrix or in the presence of the well-known differentiating agent DMSO or aFGF, a factor that has been suggested to play a role in the early steps of hepatic differentiation (Zaret, 1996).
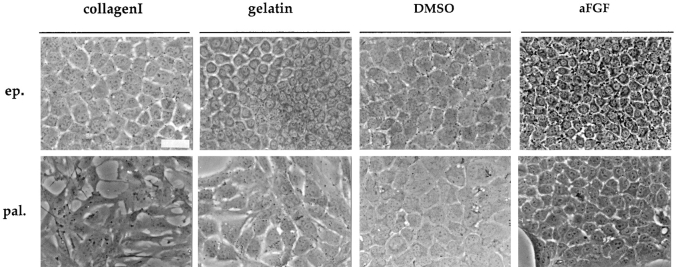
The growth of the E14 epithelial cells on gelatin-coated (rather than collagen-coated) dishes, or the addition of DMSO or aFGF leads to a more differentiated hepatocyte phenotype. After these treatments, the cells show a more regular shape with more phase dense and granular cytoplasm (Fig. 5). Significantly, they express albumin and transthyretin (TTR, Fig. 6 C). This induced phenotype resembles that presented by the D3 cells in the standard growth conditions.
Figure 5.
Morphological changes of epithelial (ep.) and palmate (pal.) clones after environmental modifications. Phase-contrast micrographs of epithelial and palmate clones in the standard growth conditions (on collagen I); grown on gelatin-coated dishes; treated with DMSO or aFGF. Photographs were taken 7–10 d after initiation of the different treatments. Bar, 40 μm.
Figure 6.
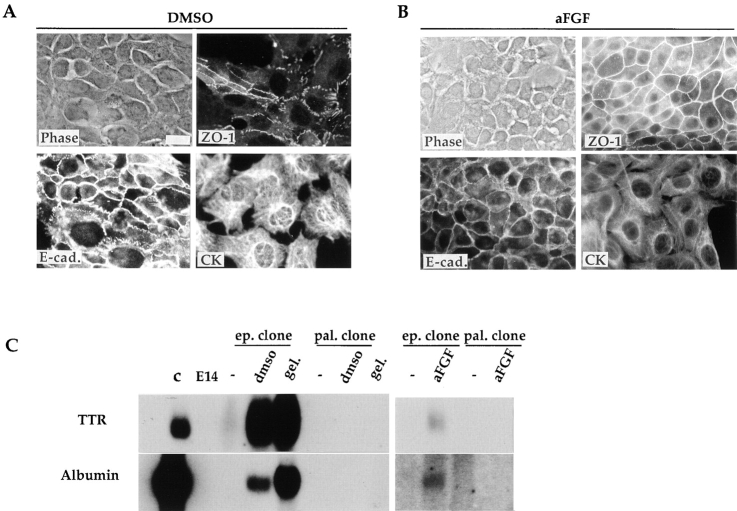
Characterization of the epithelial and the palmate cell clone responses to environmental treatments. Gelatin, DMSO, and aFGF treatments lead to epithelial morphogenesis in the palmate cells. (A and B) Immunofluorescence analysis of palmate cells treated for 1 wk with DMSO (A) or aFGF (B): expression and localization of the epithelial differentiation markers E-cadherin (E-cad.), ZO-1, and CK, using an anti-pan-CK antibody. In A, E-cadherin and the phase-contrast image are of the same field; in B, the ZO-1 and the phase-contrast image are of the same field. (C) Gelatin, DMSO and aFGF modulate the epithelial clone towards a differentiated hepatocyte phenotype. Northern blot analysis of RNA extracted from the E14 parental line (E14), the epithelial (ep.) and the palmate (pal.) clones as well as control (c) Fao rat hepatoma cells. Cultured cells were examined in the standard growth conditions (−), and 1 wk after growth on gelatin (gel.), or addition of DMSO or aFGF. Each lane was loaded with 20 μg of total RNA. Probes used are indicated on the left. Bar, 20 μm.
Cells of the palmate clone in the presence of gelatin, DMSO or aFGF acquire epithelial-like morphology, displaying a flat polygonal shape and growing in a tightly packed fashion (Fig. 5). These morphological changes reflect a reorganization of the cells as attested by the membrane localization of E-cadherin and ZO-1 (Fig. 6, A and B; Amicone et al., 1997). Concomitantly, expression of HNF1α and HNF4 transcripts was detected (data not shown), but neither albumin nor TTR transcripts were present (Fig. 6 C). The phenotypic modulation induced by gelatin and DMSO is reversible when the usual growth conditions are restored, whereas the effects of aFGF are heritable (data not shown).
The Palmate Clones Generate Epithelial Progeny Competent to Express Hepatocyte Functions
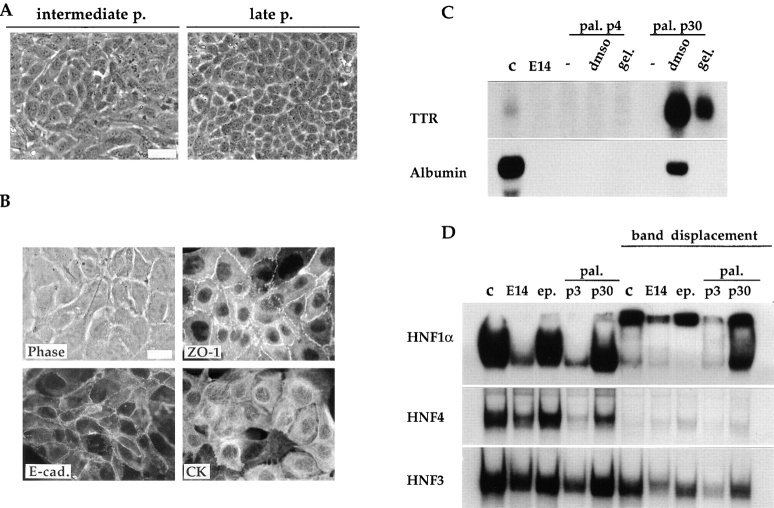
As described in the preceding paragraph, the palmate cells can evolve towards an epithelial phenotype after modification of the growth conditions. In addition, these cells undergo spontaneously the same morphological changes when maintained in continuous culture. As illustrated in Fig. 7 A, at 11 passages the E14 palmate clone showed both cell morphologies: among the palmate cells tightly packed epithelial islands appear. At 30 passages, cultures were essentially composed of cells of regular and polygonal shape. Concomitantly a well-organized compartmentalization of ZO-1 and E-cadherin can be observed (Fig. 7 B). Furthermore, in this clone after 30 passages treatment with DMSO or growth on gelatin-coated dishes is sufficient to lead to expression of albumin and TTR (Fig. 7 C).
Figure 7.
Spontaneous phenotypic transition of the palmate cells toward an epithelial-hepatocytic phenotype. (A) Phase-contrast micrographs showing the morphology of the palmate cells at intermediate (p11) and late passages (p30). (B) Appropriate re-organization of the epithelial polarity markers (E-cad., ZO-1, and CK) in the palmate cells at passage 30. The phase-contrast and immunofluorescence micrographs of the ZO-1 visualization is of the same field. (C) Reexpression of hepatic functions after environmental treatments in the palmate cells late passage. Northern blot analysis of RNA extracted from the E14 parental line (E14), the palmate clone (pal.) at early (p4) and late (p30) passages as well as control Fao rat hepatoma cell line (c). Cultured cells were examined in the usual growth conditions (−), and 1 wk after addition of DMSO or gelatin. Each lane was loaded with 20 μg of total RNA. Probes used are indicated on the left. (D) Activation of LETF (HNF1α and HNF4) expression in palmate cells continuously kept in culture. The gel shift assays were performed with labeled oligonucleotides corresponding to the HNF1 site PE56, the HNF4 site C3P and the HNF3 site of the mouse TTR promoter, as detailed in Materials and Methods. The displacement of the protein-DNA complexes was obtained with anti-HNF1α or with an excess of unlabeled oligonucleotides for HNF4 and HNF3. Nuclear extracts of FGC4, as positive control (c), of E14 parental line (E14), of the epithelial (ep.) and palmate (pal.) clones at early (p3) and late (p30) passage were examined. Bar, 20 μm.
As would be expected because albumin and TTR transcripts are present, cells of the palmate clones after 30 passages express HNF1α and HNF4 (Fig. 7 D). The gel mobility shift assays demonstrate that the epithelial cells do and the palmate cells at early passage do not produce HNF1α and HNF4, already indicated by the Northern blot analysis (Fig. 3). Nuclear extract of well-differentiated rat hepatoma cells was used as a positive control. The HNF1α and HNF1β homodimer and heterodimer complexes are present in the E14 parental line, in the epithelial and in the late passage palmate clone nuclear extracts and in the control extract (Fig. 7 D). The HNF1α homo- and heterodimer complexes undergo a supershift upon incubation with the anti-HNF1α antiserum. In the nuclear extract of the palmate clone at early passage only HNF1β is detected.
The E14 parental line and epithelial clone that we have shown to produce HNF4 transcripts do contain the corresponding protein, and the complex formation with the C3P-labeled oligonucleotide can be competed with unlabeled oligonucleotide. In the nuclear extract of palmate cells at early passage a very weak HNF4 complex was detected, that became stronger in late passage cells. Gel mobility shift assays were performed also with an oligonucleotide probe for HNF3, and they revealed a single HNF3 complex in all the extracts. The addition of an excess of unlabeled oligonucleotide PFK-3 corresponding to HNF3 binding sequence from the PFK2/FBPase-2 L-promoter failed to prevent HNF3 complex formation. It is likely that factors other than HNF3 can bind to the HNF3 site from the mouse TTR promoter, used as a probe. Further studies are required to evaluate the contribution of the different HNF3 isoforms to the cell phenotype.
The Progeny of Single Palmate Cells Become Epithelial at a Frequency That Is Modulated by the Environment
An unequivocal demonstration of cell lineage relationships is clonal analysis, and the most convincing clonings involve inoculation of individually fished cells. Therefore, to verify the occurrence of the transition from palmate to epithelial-hepatocytic cell we carried out cloning of individually fished cells. Since this procedure is most efficient when coupled with inoculation onto a mitomycin-treated non-proliferating feeder layer, we could address in the same experiments the role of cell environment in the transition from palmate to epithelial cell. Three types of feeder layers were used: the E14 parental cells, known to be composed of a mixture of palmate and epithelial cells, embryonic mouse fibroblasts (EF), traditionally used for culture of embryonic stem cells, and the epithelial clone derived from E14. These three cell types were chosen as feeders because they provided a mixed cell environment (E14), or a purely fibroblastic (EF) or epithelial (E14 ep.) one. If the environment were critical either to maintain the palmate phenotype or to insure the transition from palmate to epithelial cell, one would expect to obtain from a given cell line a different ratio of palmate to epithelial colonies as a function of the characteristics of the feeder layer.
The E14 parental line, an E14 palmate clone at early passage, and an E14 epithelial clone were all cloned on the three types of feeder layers. Cell suspensions were prepared and single cells were seeded onto a feeder layer in a 12-well dish. The results of the cloning operations are compiled in Table II according to the cell line cloned (left-most column) and the nature of the feeder layer. Plating efficiencies (PE) are given, as well as the morphological type of each progeny colony: epithelial or palmate. For each cell line and type of feeder layer, the results obtained with early and later passage cells are averaged in Table II since no consistent differences were observed between early and late passage cells. In all cases, each colony observed could be classified as palmate or epithelial; mixed colonies, or colonies presenting an alternative morphology, were not observed.
Table II.
Clones Derived from Individually Fished Cells
| Starting cell lines | Feeder layers | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E14 | EF | E14 ep. | ||||||||||||||||
| PE | ep. | pal. | PE | ep. | pal. | PE | ep. | pal. | ||||||||||
| E14 | 59 | 871 * | 122 | 32 | 29 | 712 | 46 | 50 | 502 | |||||||||
| (16/27) | (14) | (2) | (7/22) | (2) | (5) | (12/26) | (6) | (6) | ||||||||||
| E14 pal. | 71 | 85 | 15 | 41 | 221 | 781 | 56 | 601 | 403 | |||||||||
| (20/28) | (17) | (3) | (9/22) | (2) | (7) | (15/27) | (9) | (6) | ||||||||||
| E14 ep. | 82 | 100 | 0 | 45 | 100 | 0 | 45 | 100 | 0 | |||||||||
| (9/11) | (9) | (0) | (5/11) | (5) | (0) | (5/11) | (5) | (0) | ||||||||||
The table summarizes the results of 15 individual fished cell cloning operations. All the values given are percents, except for the numbers in italics and parentheses, which give the total number of clones observed over the number of individually plated cells, and the total number of epithelial or palmate colonies obtained. The E14 line was analyzed at passages 8 and 16; E14 pal. at passages 2 and 6. No differences were observed and averages are presented. PE, plating efficiency; ep., epithelial; pal., palmate; EF, embryonic fibroblasts.
The exponents refer to the numbers of clones from fished cells that were established in culture and analyzed (see Table III).
These experiments permitted us to pose two simple questions. Do palmate cells give rise to epithelial cells? Is the ratio of palmate to epithelial cells constant for a cell line, or does it vary depending upon the nature of the feeder layer?
The results in Table II clearly demonstrate that palmate cells do indeed give rise to epithelial ones. In addition, as anticipated from the results presented above, the purely epithelial clone was unable to give rise to palmate progeny. Surprisingly, the proportions of epithelial and palmate colonies from the E14 parental and E14 palmate lines were very similar on a given feeder layer, in spite of the facts that (a) epithelial cells were not detectable in the palmate clone until late passage, and (b) palmate cells were not visible in the E14 line except when cultures were seeded at low density.
A clear effect of the feeder layer on the colony types generated concerns the progeny of E14 palmate cells. At passages 2 and 6, epithelial cells are not yet visible in the cultures used to prepare the suspensions, and indeed the epithelial islands are apparent only after passage 10. (We recall here that each passage corresponds to about 5 cell generations, and that colonies are classified as epithelial or palmate after ∼8–10 generations, e.g., the equivalent of two passages). Nevertheless, a significant fraction of the colonies generated from E14 palmate cells were epithelial, especially when E14 parental or E14 epithelial cells were used as feeder layer. Moreover, the similarities in the colony type ratios for the palmate clone and E14 parental line as a function of the nature of the feeder layer strongly supports the conclusion that the nature of the feeder layer influences the colony type. Thus, the environment created by the E14 parental cells favors the transition from palmate to epithelial cell, and that of the EF cells favors the proliferation of palmate cells to the detriment of epithelial cells (Table II). These observations encourage us to attempt to identify diffusible factors or extracellular matrix molecules that could be implicated in the transition from palmate to epithelial cells.
The characterization of a number of clones from these experiments (Table III) permits us to affirm that they conform to the phenotypic states already described: (a) robust expression of the LETF HNF1α and HNF4 was observed only for epithelial clones; (b) palmate clones are deficient for both HNF1α and HNF4 (6 clones) or express the two factors only very weakly (4 clones), reflecting perhaps the existence of epithelial cells within these 4 clones even at early passage. Furthermore, 10 palmate clones were maintained in continuous culture to confirm their ability to generate epithelial-like cells. The spontaneous phenotypic transition from palmate cells to epithelial-like cells occurred consistently in all 10 of the palmate clones analyzed (Table III), indicating that the potential to undergo this transition is a heritable and consistent property of palmate cells. Logically, the MMH should eventually become depleted of palmate cells in favor of epithelial ones. However, our analyses of the E14 parental line failed to document such a depletion: when cloned at 8 or 16 passages, corresponding to ∼40 and 80 cell generations, the ratio of epithelial to palmate colonies observed was constant for a given type of feeder layer (Table II).
Table III.
Analysis of Fished Clones*
| Fished clones | HNF1α HNF4 at p2-3 | Pal.→ ep. transition‡ | ||
|---|---|---|---|---|
| E14(E14) pal.1§ | − | +at p10 | ||
| E14(E14) pal.2 | − | +at p12 | ||
| E14(E14) ep. | ++ | |||
| E14(EF) pal.1 | +/− | +at p16 | ||
| E14(EF) pal.2 | − | +at p5 | ||
| E14(ep.) pal.1 | − | +at p9 | ||
| E14(ep.) pal.2 | +/− | +at p10 | ||
| Pal.(EF) pal. | − | +at p18 | ||
| Pal.(EF) ep. | ++ | |||
| Pal.(ep.) pal.1 | +/− | +at p4 | ||
| Pal.(ep.) pal.2 | +/− | +at p9 | ||
| Pal.(ep.) pal.3 | − | +at p15 | ||
| Pal.(ep.) ep. | ++ |
Abbreviations are the same as in Table II except p (culture passage).
The passage at which the first epithelial cell islands are visible in the culture, probably corresponding to 10–25% of the population.
The identifying name of each fished clone refers to, in order: the parental line, the feeder layer (in parenthesis) and the morphological features of the clone, e.g., epithelial or palmate. When more than one clone, displaying exactly the same morphotype, was isolated the abbreviation is followed by progressive numbers.
Karyotype Analysis Confirms the Direction of the Phenotypic Transition
To confirm the filiation of the two morphological cell types, karyotypes were determined and FACS® analysis of the DNA content was carried out. The results of the karyology are summarized in Table IV, and the results of FACS® analysis (data not shown) confirmed the conclusions. The parental E14 line was examined at different passages; in all cases two populations were present. Three quarters of the metaphases were diploid or pseudodiploid and one quarter hypotetraploid. To determine whether the diploid versus hypotetraploid karyotype corresponds to one or the other of the distinct cellular phenotypes, both palmate and epithelial clones were analyzed. The palmate clone of the E14 line was analyzed at early and late passages. At early passage, 100% of the cells were diploid or pseudodiploid, with a clear mode at 40 chromosomes per metaphase. 20 passages later, corresponding to the time when epithelial cells were majority, the population had become bi-modal: most of the population remained diploid or pseudodiploid but with a weaker mode at 40 chromosomes, and a small fraction had become hypotetraploid. The epithelial clone derived from the E14 parental line was entirely hypotetraploid. The D2 line, which is composed of both palmate and epithelial cells, presents a bimodal karyotype, whereas the D3 line, whose progeny are uniquely epithelial, is uniformly hypotetraploid.
Table IV.
Karyotype Analysis
| Diploid chromosome number | Hypotetraploid chromosome number | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % Population | Mode | Range | % Population | Range | ||||||
| Parental lines and first generation clones* | ||||||||||
| E14 | ||||||||||
| p8 | 74 | 40 (67)‡ | 35-41; 48-55 | 21 | 59-64; 68-78 | |||||
| p13 | 74 | 40 (64) | 39-48 | 26 | 60-80 | |||||
| p19 | 76 | 40 (45) | 36-44 | 24 | 58-77 | |||||
| E14 pal. | ||||||||||
| p5 | 100 | 40 (72) | 39-45 | 0 | ||||||
| p25 | 94 | 40 (35) | 35-47 | 6 | 70-78 | |||||
| E14 ep. | 0 | 100 | 60-79 | |||||||
| mode 69-70 | ||||||||||
| D2 p17 | 40 | 40 (80) | 39-47 | 60 | 62-81 | |||||
| mode 74-77 | ||||||||||
| D3 p20 | 0 | 100 | 72-82 | |||||||
| mode 76-79 | ||||||||||
| Clones derived from fished cells | ||||||||||
| E14(E14)pal.2 p7 | 83 | 40 (64) | 40-47 | 17 | 50-65 | |||||
| Pal.(EF)pal. p6 | 100 | 40 (55) | 40-42 | 0 | ||||||
| Pal.(EF)pal. p20 | 32 | 40 (75) | 39-40 | 68 | 61-80 | |||||
| E14(E14)ep. p2 | 56 | 40 (36) | 39-47 | 44 | 58-82 | |||||
| Pal.(EF)ep. p3 | 0 | 100 | 51-65 | |||||||
| mode 60-65 | ||||||||||
The table summarizes the data of the karyotype analysis carried out on: the three MMH parental lines, the first generation clones (epithelial and palmate clones), and some fished clones representative of both colony types.
Abbreviations are the same as those in Tables II and III. Based upon the analysis of at least 25 metaphases for each cell line.
In parenthesis the percentage of the population with 40 chromosomes.
The karyological analysis of additional clones, including some obtained by cloning of individually fished cells, confirmed this pattern. Clones composed of palmate cells are diploid, whereas the epithelial ones are hypotetraploid. Moreover a fished clone of palmate cells at late passage, which had undergone the phenotypic transition, presents a bimodal karyotype (Table IV, bottom). These observations are compatible with the idea advanced above, that palmate cells give rise to epithelial cells.
Taken together, the results obtained from the characterization of clones and subclones, as well as the karyotype analysis of parental lines and of first and second generation clones, provide strong support to the hypothesis that a single immortalization event, most likely of a palmate cell, underlies the emergence of the MMH lines. The palmate cells possess morphogenic potential intrinsically different from the epithelial cells and they seem to be the precursors of the epithelial-hepatocytic cells.
Discussion
Analysis of MMH lines has revealed that two of the three examined (E14 and D2) are composed of two distinct cell types: epithelial cells and a spreading cell type with multiple projections that we refer to as palmate.
Clones of the two morphological types were isolated and indeed they represent distinct phenotypes, differing in cytoskeletal organization, in the capacity to form bile duct-like tubules in three-dimensional culture, in the expression of LETF, in their potential to respond to environmental stimuli by the expression of hepatocyte specific functions, and finally by their karyotypes. Most importantly, we demonstrate that palmate cells have the potential to undergo a transition to an epithelial cell morphology, and these cells are competent to express hepatocyte functions. An extensive study of the progeny of palmate clones reveals that they express the full spectrum of properties of either palmate or epithelial MMH cells. These findings demonstrate that in MMH lines palmate cells are the bipotential precursors of hepatocytes.
One of the two potentials of palmate cells is the formation of bile duct-like tubules on Matrigel, similar to the branching tubules formed in three-dimensional cultures of kidney and mammary epithelial cells in response to HGF (Montesano et al., 1997). In contrast, Matrigel cultures of palmate cells gave rise to tubules without addition of exogenous HGF. Under the same culture conditions, the epithelial cells formed only simple spheroids. The morphotype of palmate cells in two-dimensional culture, with the extension of multiple finger-like structures and the absence of junctional complexes, is indicative of the “scatter” response, and indeed is reminiscent of the effect of the addition of HGF/SF to epithelial cultures (Medico et al., 1996; Balkovetz et al., 1997; Pasdar et al., 1997; Royal et al., 1997). When these spreading cells are given the opportunity to form three-dimensional relationships with the substratum, as on Matrigel, they establish a new organizational pattern, forming well organized tubules composed of polarized cells and a lumen that is decorated with microvilli. Therefore, cyto-Met seems to confer constitutive morphogenic activity to the palmate cells, probably via the motogenic/scatter response. It is noteworthy to recall here that no difference in expression of the transgene was observed between palmate and epithelial clones. Consequently, intrinsic differences must exist between palmate and epithelial cells that are responsible for their different response to the product of the transgene; the scatter activity of HGF signaling is effective only in the palmate cells.
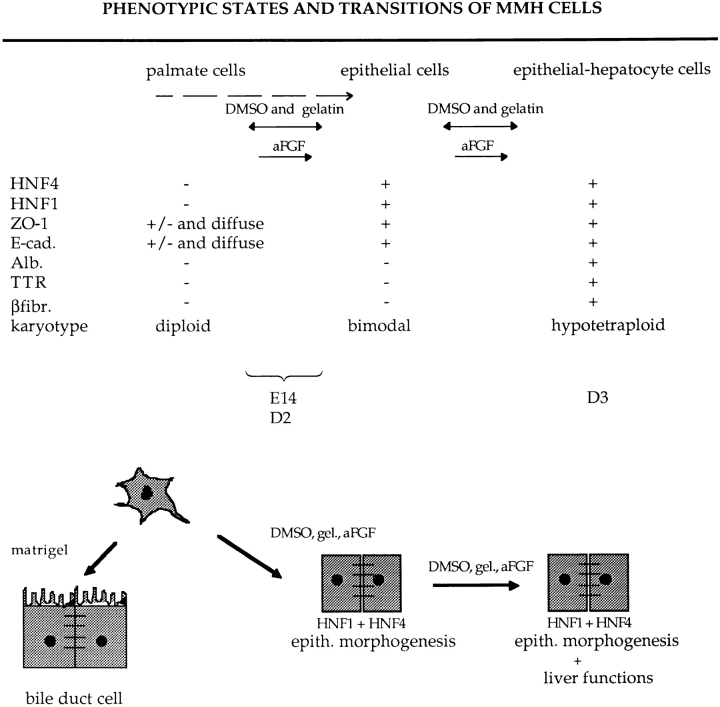
A limitation to the study of liver development has been the absence of lines of nontransformed precursor cells that can differentiate into hepatocytes in culture, and that possesses sufficient proliferative potential that clonal progeny can be isolated and characterized. The immortalized palmate cell may fill this gap. The palmate cell goes through at least two stages when differentiating in vitro, whether the transition is spontaneous or induced. Fig. 8 presents in a schematic fashion these transitions, the factors that induce them, and the properties of cells of the different stages. Characterization of the MMH clones revealed that there must exist at least two, and possibly three, classes of epithelial cells, going from the most to the least differentiated: (a) the most differentiated cells are hypotetraploid, polarized, and express the LETF as well as the liver functions in the absence of any stimulus. This is the only phenotype observed for the D3 cells and its clones. (b) An intermediate class is composed of hypotetraploid cells that display a well-polarized epithelial phenotype and express HNF1α and HNF4 but fail to express hepatic functions unless stimulated to do so. Cells of this class present an uncoupled phenotype (expression of HNF1α and HNF4 concomitant with absence of hepatic functions), recently described for some rat hepatoma variants (Chaya et al., 1997). (c) Finally, there is probably a transitory epithelial cell population that still retains a diploid chromosome number. Existence of this hypothetical third class is suggested by the facts that (a) 75% of the E14 parental population is diploid while more than half of the progeny clones are epithelial, and (b) some of the epithelial clones present bimodal karyotypes. Among the three MMH lines analyzed, only the D3 line is devoid of diploid cells and fails to give rise to palmate colonies upon cloning. A possible explanation for the absence of palmate cells from the D3 line is their depletion in favor of epithelial progeny. Alternatively, the initial immortalized D3 cell could have been epithelial rather than palmate. Further study of the events underlying emergence of immortalized transgenic liver cells should provide an answer to this question.
Figure 8.
Schematic representation of the different phenotypic states of the MMH lines and their clones, and spontaneous or induced transitions. The discontinuous arrow indicates a spontaneous transition; continuous two- and one-headed arrows indicate reversible and irreversible phenotypic modulations, respectively. Abbreviations used are the same as those in Fig. 6.
Although the environment (soluble factors, extracellular matrix, and cell–cell contacts) plays a key role in cell determination and differentiation (Ekblom, 1989; Hay, 1993; Gullberg and Ekblom, 1995; Cantley, 1996), the critical factors for the hepatocyte are only beginning to be defined (Michalopoulos and Pitot, 1975; Isom et al., 1985; Ben-Ze'ev et al., 1988; Germain et al., 1988a ,b; Caron, 1990; DiPersio et al., 1991; Coleman et al., 1994). Indeed, Block et al. (1996) have defined some of the factors that influence the differentiation state of hepatocytes in culture. We have shown that the differentiation state of both the palmate and the epithelial cells is modulated by extracellular matrix proteins and general differentiating agents, such as DMSO, or more developmentally relevant factors, such as aFGF. These factors are required to permit the MMH epithelial cells, which appear to be blocked at an early stage of differentiation, to progress within the hepatic program to express liver functions. The differentiating effects of gelatin and DMSO are entirely reversible, whereas those of aFGF are not, as though this stimulus were sufficient to push the cells to a new and heritable state of differentiation. Indeed, growth factors are known to act as embryonic inducers, setting off new reaction cascades, whereas ECM proteins may provoke reversible modulation (Di Persio et al., 1991). This is in line with the observations that aFGF is expressed at high levels during cardiac development, and its receptors are present in the endoderm cells (Stark et al., 1991; Mima et al., 1995). Therefore, a signaling role of aFGF from the cardiac mesoderm to the endoderm would not be surprising (Zaret, 1996).
Palmate cells show an original immortalized phenotype, grow indefinitely while maintaining a diploid karyotype, and give rise to epithelial cells expressing LETF and hepatic functions. Now that their existence has been demonstrated, two critical questions must be considered. First, do they exist in vivo and where do they come from; second, what can they become?
Whether or not palmate cells exist in vivo cannot yet be answered. It is important to recall that although the palmate cells seem to express an activated met gene (cyto-MET) that is transforming in fibroblasts (Zhen et al., 1994), they do not express any characteristics of transformation. Expression of the transgene would mimic HGF/SF signaling, a state that is certainly physiological during liver development, when HGF/SF secretion by neighboring mesenchymal cells is high. The continuous signaling conferred by the expression of cyto-Met could prolong the survival of hepatoblasts in the liver. Indeed, the major effect of the transgene could be to stabilize a phenotypic state that is normally only transitory. A characteristic that is shared between palmate cells and fetal hepatoblasts in situ is an absence of epithelial cell polarity (Stamatoglou et al., 1992). Another obvious candidate for a palmate cell in the mature liver is the oval cell, whose presence becomes obvious only after appropriate stimuli, and which can differentiate into both hepatocytes and bile duct cells. However, the palmate cell is different from oval cells in failing to express LETF, and the early hepatic function albumin. Thus although the palmate cell appears to share properties with both hepatoblasts and oval cells, it is a perfect match with neither.
Established notions of embryology would lead us to argue that palmate cells must be of endodermal origin since they express the hepatic CKs and can give rise to hepatocytes. However, the morphology of palmate cells is strikingly reminiscent of monolayer cultures of Ito/stellate cells, presumed to be of mesenchymal origin. Future experiments will test the expression by palmate cells of markers that are specific to Ito/stellate cells. At present, we prefer to remain openminded concerning the possibility that the origin of the Ito cell has not yet been definitively established (Enzan et al., 1997).
What can the palmate cells become? In culture, they can express at least some traits of hepatocytes and they can form bile duct-like structures. The possibility that they can become functional hepatocytes capable of repopulating the liver is a subject for future experimentation. The existence in the mouse of a true hepatic stem cell capable of serial repopulation of six generations of livers, has been unequivocally demonstrated (Overturf et al., 1997). Identification of this stem cells is an important challenge for better understanding of liver development and regulation of its homeostasis. Oval cells are already candidates for hepatic stem cells; the palmate cell is now one as well.
The analysis presented here has provided hints of a relationship between HGF/SF signaling, the acquisition of epithelial morphology and the expression of LETF. The product of the constitutively active cyto-MET transgene, if it is expressed at all, is present at equivalent levels in palmate cells and their epithelial progeny, yet it is only in the palmate cells that evidence of a scatter response is observed. If the scatter response to cyto-Met signaling is a consequence of cytoskeletal components and organization, it can be suggested that the underlying intrinsic difference between palmate and epithelial cells is in the cytoskeleton. Moreover, since loss of the scatter response correlates with acquisition of epithelial cell polarity and of expression of HNF1α and HNF4, it can be further suggested either that (a) HGF/SF signaling inhibits expression of the LETF, and this inhibition can be alleviated by aFGF signaling and reinforced by factors that control the scatter response; or (b) the differentiative signals leading to the expression of the LETF constrain the HGF-mediated scattering response. Further studies will be needed to test these predictions.
Acknowledgments
We are grateful to Jean Claude Benichou for the electron microscopy analysis; to Latifa Bakiri and Moshe Yaniv for the help in the FACS® analysis; to Rolf Kemler for the gift of the TROMA 1, 2, and 3 antibodies; to Tanguy Chouard for the HNF1α antiserum; to Doris Cassio for the ZO-1 antibody. We thank Catherine Fougère-Deschatrette, Dina Chaya, and Giulio Cossu for critical reading of the manuscript and for helpful discussions.
Abbreviations used in this paper
- aFGF
acidic FGF
- C/EBP
CAAT/enhancer binding protein
- CK
cytokeratin
- EF
embryonic fibroblast
- EHS
Engelbreth-Holm-Swarm
- HGF/SF
hepatocyte growth factor/scatter factor
- HNF
hepatocyte nuclear factor
- LETF
liver-enriched transcription factor
- MMH
Met murine hepatocyte
- TTR
transthyretin
- ZO
zonula occludens
Footnotes
This work in the Paris laboratory was supported in part by grants from the Biotechnology Programme of the European Economic Community under contract number BIOT CT 93-0103 from the Association pour la Recherche sur le Cancer and La Ligue Nationale contre le Cancer. Work in the Rome laboratory was supported by the AIRC (Associazione Italiana Ricerca sul Cancro), Ministero dell' Universita e Ricerca Scientifica e Tecnologica (MURST), and Centro Nazionale delle Ricerche (CNR) Target project on Biotechnology. A NATO Collaborative Research Grant facilitated the collaboration.
Address all correspondence to Mary C. Weiss, Unité de Génétique de la Différenciation, URA 1773 du CNRS, Institut Pasteur, 25 rue du Dr. Roux, 75724 Paris Cedex 15, France. Tel.: 33 1 45 68 85 00. Fax: 33 1 40 61 32 31. E-mail: mweiss@pasteur.fr
References
- Amicone L, Galimi MA, Spagnoli FM, Tommasini C, De Luca V, Tripodi M. Temporal and tissue-specific expression of the MET ORF driven by the complete transcriptional unit of human A1AT gene in transgenic mice. Gene. 1995;162:323–329. doi: 10.1016/0378-1119(95)00277-d. [DOI] [PubMed] [Google Scholar]
- Amicone L, Spagnoli FM, Spath G, Giordano S, Tommasini C, Bernardini S, De Luca V, Della C, Rocca, Weiss MC, Comoglio PM, Tripodi M. Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. EMBO (Eur Mol Biol Organ) J. 1997;16:495–503. doi: 10.1093/emboj/16.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkovetz DF, Pollack AL, Mostov KE. Hepatocyte Growth Factor alters the polarity of Madin-Darby Canine Kidney cell monolayers. J Biol Chem. 1997;7:3471–3477. doi: 10.1074/jbc.272.6.3471. [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- Ben-Ze'ev A, Robinson GS, Bucher NL, Farmer SR. Cell-cell and cell-matrix interactions differentially regulate the expression of hepatic and cytoskeletal genes in primary cultures of rat hepatocytes. Proc Natl Acad Sci USA. 1988;85:2161–2165. doi: 10.1073/pnas.85.7.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursors cells into the limb bud. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, Howard TA, Michalopoulos GK. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LG. Growth factors and the kidney: regulation of cell movement and morphogenesis. Am J Physiol. 1996;271:F1103–F1113. doi: 10.1152/ajprenal.1996.271.6.F1103. [DOI] [PubMed] [Google Scholar]
- Caron JM. Induction of albumin gene transcription in hepatocytes by extracellular matrix proteins. Mol Cell Biol. 1990;10:1239–1243. doi: 10.1128/mcb.10.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghini S, Blumenfeld M, Yaniv M. A liver-specific factor essential for albumin transcription differs between differentiated and dedifferentiated rat hepatoma cells. Genes Dev. 1988;2:957–974. doi: 10.1101/gad.2.8.957. [DOI] [PubMed] [Google Scholar]
- Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- Chaya D, Fougère-Deschatrette C, Weiss MC. Liver-enriched transcription factors uncoupled from expression of hepatic functions in hepatoma cell lines. Mol Cell Biol. 1997;17:6311–6320. doi: 10.1128/mcb.17.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman WB, Smith GJ, Grisham JW. Development of dexamethasone-inducible tyrosine aminotransferase activity in WB-F344 rat liver epithelial stemlike cells cultured in the presence of sodium butyrate. J Cell Physiol. 1994;161:463–469. doi: 10.1002/jcp.1041610309. [DOI] [PubMed] [Google Scholar]
- Coon HG, Weiss MC. A quantitative comparison of formation of spontaneous and virus-produced viable hybrids. Proc Natl Acad Sci USA. 1969;62:852–859. doi: 10.1073/pnas.62.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR, Kant JA. Molecular cloning of cDNA for the alpha, beta, and gamma chains of rat fibrinogen. A family of coordinately regulated genes. J Biol Chem. 1981;256:9718–9723. [PubMed] [Google Scholar]
- Derman E, Krauter K, Walling L, Weinberg C, Ray M, Darnell JJ. Transcriptional control in the production of liver-specific mRNAs. Cell. 1981;23:731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Desmet, V.J. 1994. Organizational principles. In The Liver: Biology and Pathology. I.M. Arias, J.L. Boyer, N. Fausto, W.B. Jakoby, D.A. Schachter, and D.A. Shafritz, editors. Raven Press, Ltd., New York. 3–14.
- Di Persio CM, Jackson DA, Zaret KS. The extracellular matrix coordinately modulates liver transcription factors and hepatocyte morphology. Mol Cell Biol. 1991;11:4405–4414. doi: 10.1128/mcb.11.9.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom P. Developmentally regulated conversion of mesenchyme to epithelium. FASEB J. 1989;3:2141–2150. doi: 10.1096/fasebj.3.10.2666230. [DOI] [PubMed] [Google Scholar]
- Enzan H, Himeno H, Hiroi M, Kiyoku H, Saibara T, Onishi S. Development of hepatic sinusoidal structure with special reference to the Ito cells. Micr Res Technique. 1997;39:336–349. doi: 10.1002/(SICI)1097-0029(19971115)39:4<336::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Evarts RP, Hu Z, Fujio K, Marsden ER, Thorgeirsson SS. Activation of hepatic stem cell compartment in the rat: role of transforming growth factor α, hepatocyte growth factor, and acidic fibroblast growth factor in early proliferation. Cell Growth Differ. 1993;4:555–561. [PubMed] [Google Scholar]
- Farber E. Similarities in the sequence of early histological changes induced in the liver of the rat by ethionine, 2-acetylaminofluorene and 3′-methyl-4-dimethylaminoazobenzene. Cancer Res. 1956;16:142–155. [PubMed] [Google Scholar]
- Fausto N, Lemire JM, Shiojiri N. Cell lineages in hepatic development and the identification of progenitor cells in normal and injured liver. Proc Soc Exp Biol Med. 1993;204:237–241. doi: 10.3181/00379727-204-43659. [DOI] [PubMed] [Google Scholar]
- Frain M, Swart G, Monaci P, Nicosia A, Stampfli S, Frank R, Cortese R. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell. 1989;59:145–157. doi: 10.1016/0092-8674(89)90877-5. [DOI] [PubMed] [Google Scholar]
- Friedman AD, Landschulz WH, McKnight SL. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- Germain L, Noel M, Gourdeau H, Marceau N. Promotion of growth and differentiation of rat ductular oval cells in primary culture. Cancer Res. 1988a;48:368–378. [PubMed] [Google Scholar]
- Germain L, Blouin MJ, Marceau N. Biliary epithelial and hepatocytic cell lineage relationships in embryonic rat liver as determined by the differential expression of cytokeratins, alpha-fetoprotein, albumin and cell surface-exposed components. Cancer Res. 1988b;48:4909–4918. [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Gullberg D, Ekblom P. Extracellular matrix and its receptors during development. Int J Dev Biol. 1995;39:845–854. [PubMed] [Google Scholar]
- Hay ED. Extracellular matrix alters epithelial differentiation. Curr Opin Cell Biol. 1993;5:1029–1035. doi: 10.1016/0955-0674(93)90088-8. [DOI] [PubMed] [Google Scholar]
- Hilberg F, Aguzzi A, Howells N, Wagner EF. c-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- Houssaint E. Differentiation of the mouse hepatic primordium. I. An analysis of tissue interactions in hepatocyte differentiation. Cell Differ. 1980;9:269–279. doi: 10.1016/0045-6039(80)90026-3. [DOI] [PubMed] [Google Scholar]
- Isom HC, Secott T, Georgoff I, Woodworth C, Mummaw J. Maintenance of differentiated rat hepatocytes in primary culture. Proc Natl Acad Sci USA. 1985;82:3252–3256. doi: 10.1073/pnas.82.10.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PF, Landschulz WH, Graves BJ, McKnight SL. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987;1:133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Kemler R, Brulet P, Schnebelen MT, Gaillard J, Jacob F. Reactivity of monoclonal antibodies against intermediate filament proteins during embryonic development. J Embryol Exp Morph. 1981;64:45–60. [PubMed] [Google Scholar]
- Kioussis D, Eiferman F, Van de Rijn P, Gorin MB, Ingram RS, Tilghman SM. The evolution of alpha-fetoprotein and albumin. II. The structures of the alpha-fetoprotein and albumin genes in the mouse. J Biol Chem. 1981;256:1960–1967. [PubMed] [Google Scholar]
- Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JJ, Crabtree GR. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- Lai E, Prezioso VR, Tao WF, Chen WS, Darnell JJ. Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. An experimental analysis of liver development. Med Biol. 1975;53:427–455. [PubMed] [Google Scholar]
- Lemire JM, Shiojiri N, Fausto N. Oval cell proliferation and the origin of small hepatocytes in liver injury induced by D-galactosamine. Am J Pathol. 1991;139:535–552. [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Di Persio CM, Zareth KS. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol Cell Biol. 1991;11:773–784. doi: 10.1128/mcb.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medico E, Mongiovi A, Huff J, Jelinek M-A, Follenzi A, Gaudino G, Parsons JT, Comoglio PM. The tyrosine kinase receptors Ron and Sea control “scattering” and morphogenesis of liver progenitor cells in vitro. Mol Biol Cell. 1996;7:495–504. doi: 10.1091/mbc.7.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio M, Weiss MC. Immunofluorescence analysis of the time-course of extinction, reexpression, and activation of albumin production in rat hepatoma-mouse fibroblast heterokaryons and hybrids. J Cell Biol. 1981;90:339–350. doi: 10.1083/jcb.90.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, Pitot HC. Primary culture of parenchymal liver cells on collagen membranes. Morphological and biochemical observations. Exp Cell Res. 1975;94:70–78. doi: 10.1016/0014-4827(75)90532-7. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Mima T, Ueno H, Fischman DA, Williams LT. Fibroblast growth factor receptor is required for in vivo myocyte proliferation at early embryonic stages of heart development. Proc Natl Acad Sci USA. 1995;92:467–471. doi: 10.1073/pnas.92.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano R, Soriano JV, Pepper MS, Orci L. Induction of epithelial branching tubulogenesis in vitro. J Cell Physiol. 1997;173:152–161. doi: 10.1002/(SICI)1097-4652(199711)173:2<152::AID-JCP14>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Orkin RW, Gehron P, McGoodwin EB, Martin GR, Valentine T, Swarm R. A murine tumor producing a matrix of basement membrane. J Exp Med. 1977;145:204–220. doi: 10.1084/jem.145.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima RG, Trevor K, Shevinsky LH, Ryder OA, Cecena G. Identification of the gene coding for the Endo B murine cytokeratin and its methylated, stable inactive state in mouse nonepithelial cells. Genes Dev. 1988;2:505–516. doi: 10.1101/gad.2.5.505. [DOI] [PubMed] [Google Scholar]
- Overturf K, Al-Dhalimy M, Ou C, Finegold M, Grompe M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol. 1997;151:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- Pasdar M, Li Z, Marreli M, Nguyen B, Park M, Wong K. Inhibition of junction assembly in cultured epithelial cells by hepatocyte growth factor/scatter factor is concomitant with increased stability and altered phosphorylation of the soluble junctional molecules. Cell Growth Differ. 1997;8:451–462. [PubMed] [Google Scholar]
- Pitot HC, Peraino C, Morse PA, Potter VA. Hepatoma in tissue culture compared with adapting liver in vitro. Natl Cancer Inst Monogr. 1964;13:229–242. [PubMed] [Google Scholar]
- Rey Campos, J., T. Chouard, M. Yaniv, and S. Cereghini. vHNF1 is a homeoprotein that activates transcription and forms heterodimers with HNF1. EMBO (Eur Mol Biol Organ) J. 1991;10:1445–1457. doi: 10.1002/j.1460-2075.1991.tb07665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal I, Fournier TM, Park M. Differential requirement of Grb2 and PI3-kinase in HGF/SF-induced cell motility and tubulogenesis. J Cell Physiol. 1997;173:196–201. doi: 10.1002/(SICI)1097-4652(199711)173:2<196::AID-JCP20>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- Shiojiri N, Lemire JM, Fausto N. Cell lineages and oval cell progenitors in rat development. Cancer Res. 1991;51:2611–2620. [PubMed] [Google Scholar]
- Singer PA, Trevor K, Oshima RG. Molecular cloning and characterization of the Endo B cytokeratin expressed in preimplantation mouse embryos. J Biol Chem. 1986;261:538–547. [PubMed] [Google Scholar]
- Sladek FM, Zhong WM, Lai E, Darnell JJ. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- Spath GF, Weiss MC. Hepatocyte nuclear factor 4 expression overcomes repression of the hepatic phenotype in dedifferentiated hepatoma cells. Mol Cell Biol. 1997;17:1913–1922. doi: 10.1128/mcb.17.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spath GF, Weiss MC. Hepatocyte Nuclear Factor 4 provokes expression of epithelial marker genes, acting as morphogen in dedifferentiated hepatoma cells. J Cell Biol. 1998;140:935–946. doi: 10.1083/jcb.140.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoglou SC, Enrich C, Manson MM, Hughes RC. Temporal changes in the expression and distribution of adhesion molecules during liver development and regeneration. J Cell Biol. 1992;116:1507–1515. doi: 10.1083/jcb.116.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, McMahon JA, McMahon AP. FGFR-4, a new member of the fibroblast growth factor receptor family, expressed in the definitive endoderm and skeletal muscle lineages of the mouse. Development. 1991;113:641–651. doi: 10.1242/dev.113.2.641. [DOI] [PubMed] [Google Scholar]
- Stevenson BR, Siciliano J, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. Molecular genetics of early liver development. Ann Rev Physiol. 1996;58:231–251. doi: 10.1146/annurev.ph.58.030196.001311. [DOI] [PubMed] [Google Scholar]
- Zhen Z, Giordano S, Longati P, Medico E, Campiglio M, Comoglio PM. Structural and functional domains critical for constitutive activation of the HGF-receptor (Met) Oncogene. 1994;9:1691–1697. [PubMed] [Google Scholar]