Abstract
Titin is a giant elastic protein in vertebrate striated muscles with an unprecedented molecular mass of 3–4 megadaltons. Single molecules of titin extend from the Z-line to the M-line. Here, we define the molecular layout of titin within the Z-line; the most NH2-terminal 30 kD of titin is located at the periphery of the Z-line at the border of the adjacent sarcomere, whereas the subsequent 60 kD of titin spans the entire width of the Z-line. In vitro binding studies reveal that mammalian titins have at least four potential binding sites for α-actinin within their Z-line spanning region. Titin filaments may specify Z-line width and internal structure by varying the length of their NH2-terminal overlap and number of α-actinin binding sites that serve to cross-link the titin and thin filaments. Furthermore, we demonstrate that the NH2-terminal titin Ig repeats Z1 and Z2 in the periphery of the Z-line bind to a novel 19-kD protein, referred to as titin-cap. Using dominant-negative approaches in cardiac myocytes, both the titin Z1-Z2 domains and titin-cap are shown to be required for the structural integrity of sarcomeres, suggesting that their interaction is critical in titin filament–regulated sarcomeric assembly.
Keywords: titin, α-actinin, Z-disc, titin-cap (T-cap), sarcomere
The formation of perfectly aligned myofibrils, the structural units responsible for the production of active and passive forces in striated muscle, represents a dramatic example of supramolecular assembly in eukaryotic cells. The mechanisms by which this transpires are incompletely understood. During the last few years, the protein titin has increasingly been implicated in this process. Titin, also called connectin, is the largest protein known and is the third most abundant protein in striated muscle behind only myosin and actin (Maruyama et al., 1977; Wang et al., 1979). It is composed of a single strand of 27,000–34,000 residues with a molecular mass of 3.0–3.7 MD (Labeit and Kolmerer, 1995). Ultrastructural analysis has revealed that a single titin polypeptide spans half-sarcomeres (Maruyama et al., 1985; Fürst et al., 1988), with its NH2 terminus corresponding to the Z-line and COOH terminus at the M-line (Labeit et al., 1992). Thus, titin alone is long enough to provide half-sarcomere spacing for both thick and thin filaments (for recent reviews on the properties of titin in vitro see Trinick, 1996; Wang, 1996; Keller, 1997; Labeit et al., 1997; Maruyama, 1997).
In addition to titin's role as an elastic spring element during muscle contraction (Horowits et al., 1986; Funatsu et al., 1990; Granzier and Irving, 1995; Linke et al., 1996), this giant filamentous protein has been postulated to spatially define the positions of other contractile proteins during myofibril assembly by fulfilling a role as a molecular blueprint for the layout of the sarcomere, thereby directing the sequential events in sarcomere assembly (Whiting et al., 1989; Trinick, 1996; Wang, 1996). It has been demonstrated that titin interacts directly with several A band components, such as C protein, M protein, myomesin, and myosin at the level of the MHC rod (Maruyama et al., 1985a ; Labeit et al., 1992; Houmeida et al., 1995; Obermann et al., 1996). Furthermore, the modular arrangement of Ig-like and fibronectin-3–like domains within titin's A band region correlates remarkably with the ultrastructure of the sarcomere (Labeit et al., 1992; Labeit and Kolmerer, 1995). In the I band, titin binds the major thin filament component, actin, near the Z-line (Granzier et al., 1997; Linke et al., 1997; Trombitas and Granzier, 1997). Additionally, it binds to the major Z-line component, α-actinin (Ohtsuka et al., 1997; Sorimachi et al., 1997; Turnacioglu et al., 1997; Young et al., 1998).
The direct interaction of titin with numerous sarcomeric proteins is particularly pertinent with respect to its proposed role as a molecular template during myofibril assembly. Titin is one of the first myofibrillar proteins that assembles into nascent sarcomeres in in vivo studies and in most, but not all, studies using primary striated muscle cell cultures (for in vivo studies see Tokuyasu and Maher, 1987a ,b; Fürst et al., 1989a ; Shimada et al., 1996; for discussion of discrepancies in in vitro studies see Epstein and Fischman, 1991; Handel et al., 1991; Dabiri et al., 1997; Fulton and Alftine, 1997; Holtzer et al., 1997). During myogenesis, the integration of titin into nascent sarcomeres begins with the assembly of the Z-line–associated NH2 terminus and proceeds towards the COOH-terminal M band region in the middle of the sarcomere (Fürst et al., 1989a ). Together, these observations implicate titin in the assembly of I-Z-I bodies (containing α-actinin, actin, and other thin filament–associated proteins) and in the subsequent docking of the I-Z-I bodies to the muscle myosin-containing thick filaments to form colinear bundles and then mature periodic sarcomeres (Holtzer et al., 1997). Other studies emphasize the potential role of titin during myofibril assembly as a template for thick filament assembly (e.g., Trinick, 1996).
To understand the importance of titin in the Z-line, the identity of the sequences of the NH2-terminal domains of titin that correspond to particular Z-line locations and the functional significance of titin's interactions with other Z-line components need to be established. To date, different models for the layout of the 80-kD of the NH2-terminal titin in Z-discs have been proposed. Initial immunolocalization studies at the ultrastructural level demonstrated that ∼600–800 residues of the titin molecule are located within the Z-line (Yajima et al., 1996). Epitope-mapping studies of previously characterized titin antibodies T20 and T21 (Fürst et al., 1988) concluded that the NH2 terminus of titin is located in the center of the Z-disc, suggesting that two titin filaments from opposing half-sarcomeres do not share significant overlap in the Z-line (Gautel et al., 1996). Since the original submission of this manuscript, it was also suggested that titin filaments may overlap in Z-lines (Young et al., 1998). Clearly a clarification of titin's Z-line layout is needed.
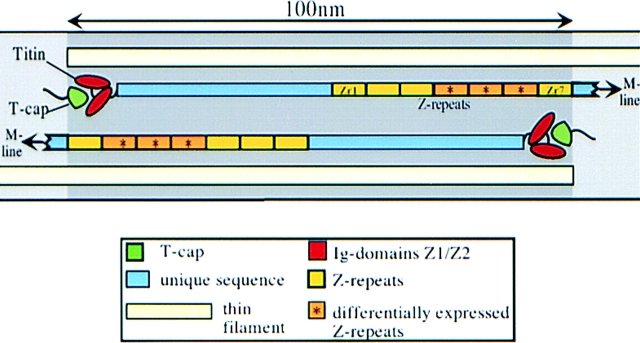
Identifying the molecular layout of Z-disc titin is also likely to be important for understanding why the morphology of Z-lines differs in different muscles and fiber types, and why the number of proteins incorporated into the Z-disc structure varies (Vigoreaux, 1994; Squire, 1997). The finding that a family of 45-residue titin Z-repeats is expressed in different copy numbers in different muscles gave rise to the proposal that differences in structure and protein composition of Z-lines result from the expression of different titin isoforms (Gautel et al., 1996). A detailed survey of the tissue-specific expression of Z-disc titin confirmed a tissue-specific variation of the Z repeat copy number and demonstrated the presence of additional Z-disc isoforms from different tissues (Sorimachi et al., 1997). Seven copies of Z-repeats are present in rabbit heart, referred from the NH2 terminus to the COOH terminus as Zr1 to Zr7 (Sorimachi et al., 1997). The repeats Zr1, 2, 3, and 7 are shared by all titins, whereas the repeats Zr4, 5, and 6 are differentially expressed in skeletal muscles, which express mixtures of titins with four and six Z-repeats. Comparison of Z-disc titins from different species suggests the presence of additional isoforms (Yajima et al., 1996). Therefore, the variability that exists among Z-lines appears to correlate with the presence of multiple differentially spliced Z-disc titin isoforms. However, a direct correlation of particular characteristics/functions (e.g., its width) of Z-lines with the titin motif family has not been established.
In this study, to increase our knowledge about Z-disc structure and the role that titin plays in this structure, we sought to (a) determine more precisely the molecular layout of the Z-disc region of titin and (b) identify, and determine the functional significance of, the interactions of titin with Z-line components. We found that the differentially expressed segment of titin is integral within Z-lines and, in fact, appears to span the entire width of the Z-line. The most NH2-terminal 200 residues, which are not differentially expressed and are shared by all titins, are located at the periphery of the Z-line (i.e., at the edge of the adjacent sarcomere). In vitro binding studies revealed that mammalian striated muscles contain at least four potential binding sites for the major Z-line component, α-actinin, within the region of titin that spans the Z-line. In addition, we have identified a novel, striated muscle–specific, 19-kD protein ligand of the extreme NH2-terminal region of titin; we refer to this protein as titin-cap (T-cap).1 Dominant-negative approaches in cardiac myocytes demonstrate that this interaction is critical for sarcomeric integrity. We hypothesize, based on our work and that of others, that the direct lateral association between titin and Z-line molecules imparts structural continuity to the sarcomere. Future studies will focus on understanding how the interaction of titin with titin-cap contributes to the dynamic process of myofibril assembly.
Materials and Methods
Antibodies
All antibodies were raised to recombinant peptides, which were obtained from the expression of the desired sequences in Escherichia coli. The insert sequences for the expression constructs were isolated by PCR (Saiki et al., 1985). For the titin Z1-Z2 antigen, the NH2-terminal 195 residues of the human cardiac titin (21 kD; sequence data available from EMBL under accession number X90568) were amplified. For the titin Zr5-6 antigen, the two Z-repeats Zr5 and Zr6 (residues 560–650; EMBL accession number Y18012), which are excluded in the rabbit psoas splice variant but included in rabbit heart (Sorimachi et al., 1997), were amplified from rabbit heart cDNA. For the T-cap antigen, a fragment coding for residues 1–138 and therefore the NH2-terminal 16 kD of the full-length 19-kD protein (accession number AJOOO491; Valle et al., 1997) was amplified from specific human cardiac cDNA clones. The fragments coding for the peptides titin Z1-Z2, titin Zr5-6, and T-cap were subcloned into modified pET9D vectors (Studier and Moffat, 1991), which expressed their insert sequences as fusions with NH2-terminal His6-tags. The titin Z1-Z2 and Zr5-6 peptides were purified from the soluble fraction, whereas the T-cap was purified under denaturing conditions in 8 M urea. All three peptides were purified by nickel chelate affinity chromatography on NTA (Ni-NTA) resins as specified by the manufacturer (QIAGEN, Valencia, CA), yielding ∼10 mg of each antigen from 2–4-liter cultures. Polyclonal antibodies were generated in rabbits (Eurogentec, Belgium), and the specific IgG fraction was isolated by affinity chromatography. Western blot analysis of extracts of rat cardiac muscle was used to verify the specificity of the antibodies (data not shown).
Immunoelectron Microscopy
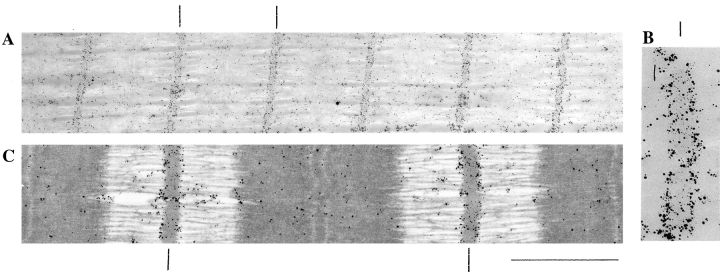
Single fibers were dissected from human soleus muscle (biopsies obtained in accordance with protocol “Role of titin in human muscle tissue 2,” Washington State University). Fibers were both chemically and mechanically skinned and stretched in relaxing solution that contained high levels of protease inhibitors (for details see Trombitas et al., 1998). Fibers were used within 48 h and immunolabeled with affinity-purified rabbit anti– titin Z1-Z2, anti–Zr5-6. or anti–T-cap antibodies. In brief, skeletal muscle fibers were fixed in 3% formaldehyde/PBS solution, blocked, and labeled with the primary antibodies (typically 50 μg/ml), followed by washing. To optimize labeling, the concentration of antibody and the duration of the blocking step were varied. Results shown were obtained by blocking for 60 min using 2% bovine serum albumin. Affinity-purified F(ab′) fragments, raised in goat against the whole rabbit IgG molecule were used as secondary antibodies (catalog No. 2004; Nanoprobes, Inc., Stonybrook, NY). These antibodies were conjugated with 1.4-nm gold particles that were intensified with HQ silver developer for 4 min (Trombitas et al., 1995). Labeled fibers were embedded in araldite and then sectioned. Some sections were stained with potassium permanganate and lead citrate (Figs. 1, C and F, and 6 C). To optimally visualize silver grains, some sections were unstained (Figs. 1, A, B, D, and E, and 6, A and B). Sections were visualized using a transmission electron microscope (model JEM 1200 IIE; Jeol, Tokyo, Japan). Z-disc–to–epitope distances were measured from electron micrographs after high-resolution scanning (UC-1260; UMAX Data System, Inc., Fremont, CA) and digital image processing using custom-written macros for the image analysis program NIH Image (v. 1.6, Wayne Rasband, National Institutes of Health, Bethesda, MD). For spatial calibration, the electron microscope's magnification was used.
Figure 1.
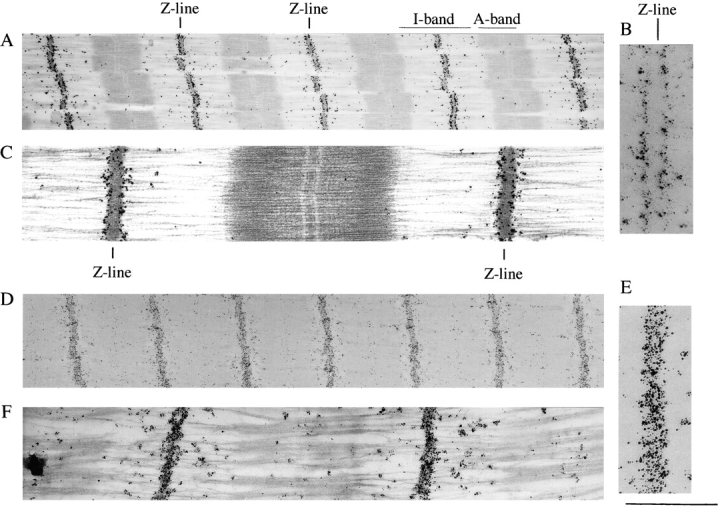
Immunoelectron microscopy of Z-disc titin. Fibers labeled with anti–titin Z1-Z2 antibodies (A–C). (A) Low-magnification image of a stained section. Darkly stained silver grains reveal epitopes at the edge of the Z-line. (B) High-magnification image of epitopes at edge of Z-line. Notice the absence of clear labeling inside the Z-disc. (C) High-magnification image of stained section. Fibers labeled with anti–titin Zr5-6 antibodies (D–F). (D) Low-magnification image of an unstained section. Darkly stained silver grains reveal epitopes throughout the width of the Z line. (E) High-magnification image. (F) High-magnification image of a stained section. Bar, 1 μm.
Figure 6.
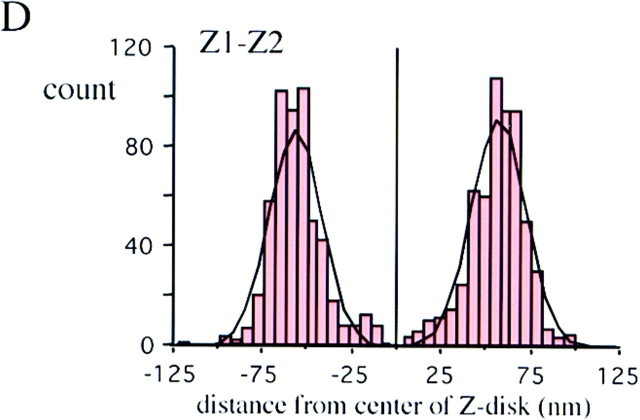
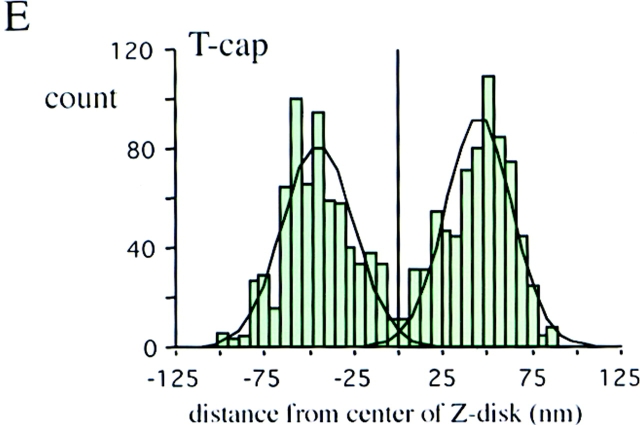
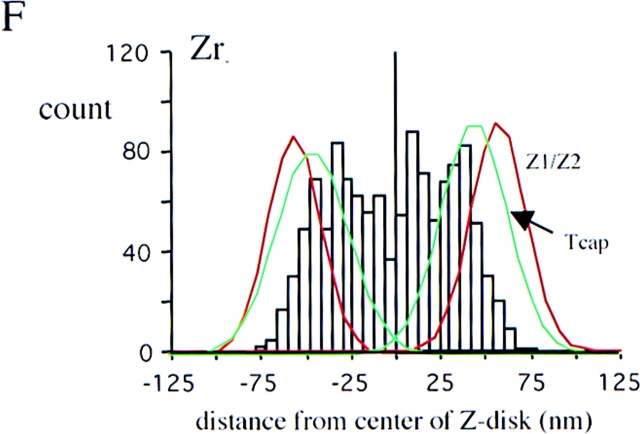
Immunoelectron microscopy of T-cap. Fibers labeled with anti–T-cap antibodies. (A) Low-magnification image of an unstained section. (B) High-magnification image. (C) High-magnification image of a stained section. The T-cap epitope is localized at the periphery of the Z-disc. (D–F) Location of epitopes as determined from the distance of silver grains to the center of Z-disc. Histograms of results with anti–titin Z1-Z2 (red) and anti–T-cap antibodies (green) reveal peaks 60 nm from the center of Z-disc, while the anti–Zr5-6 antibodies (purple) label within the Z-disc: the titin NH2 terminus and T-cap colocalize in the Z-disc periphery. Black lines mark Z-lines. Bar, 1 μm.
In Vitro Interaction Studies with Fusion Proteins
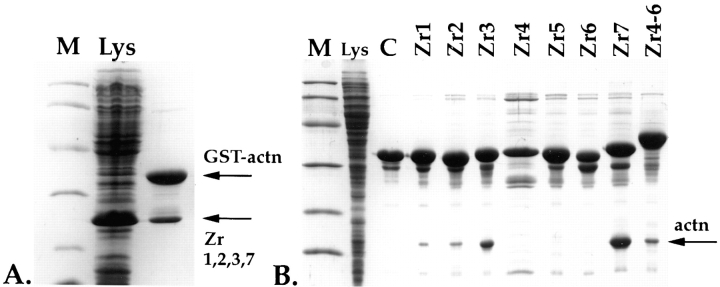
For the titin and α-actinin interaction studies, Z-repeat and α-actinin (ACTN2; Beggs et al., 1992) cDNA fragments were isolated by PCR and inserted into modified pET9D vectors (Studier and Moffat, 1991). Unmodified pET9D was used to express tagless proteins, whereas modified pET9D vectors were used to express NH2-terminal His6-tagged and His6-GST–double tagged fusion proteins. The glutathione-S-transferase (GST) fusion component was included to improve solubility of the expressed peptides, whereas the His6-tag was used to monitor interactions. Induction, expression, and cell lysis were carried out as previously described (Studier et al., 1990). E. coli cells expressing the four repeat fragment Zr1, 2, 3, and 7 were mixed with cells expressing His6-GST–tagged α-actinin (Fig. 2 A). Additionally, cells expressing the COOH-terminal 15 kD of α-actinin (containing residues 760–864) were mixed with His6-GST– tagged Z-repeats (Fig. 2 B). After cell lysis, proteins were passed over Ni-NTA resin.
Figure 2.
In vitro interaction of Z-repeats with α-actinin. (A) Binding of four Z-repeat fragments, Zr1, 2, 3, and 7 to an α-actinin fusion protein. A Coomassie blue–stained 15% Laemmli SDS gel of a whole-cell lysate of BL21 cells expressing the Z-repeat fragment Zr1, 2, 3, and 7, which corresponds to the splice variant of psoas muscle (lane Lys). These cells were mixed with cells expressing a GST-His double-tagged α-actinin (GST-actn) fusion peptide and lysed together, and lysates were passed over Ni-NTA columns. Interaction of α-actinin with the Z-repeats is indicated by also retaining the nontagged Z-repeats on an Ni-NTA column (arrow Zr1,2,3,7). (B) As in A, but here the Z-repeats were expressed as GST-His6 double-tagged fusions, and the expressed α-actinin COOH-terminal peptide was tagless. Interaction of the 15-kD from the α-actinin COOH terminus with the Z-repeats is indicated by retaining the α-actinin 15 kD band on the column (arrow actn). Here, Lys indicates a whole-cell lysate of BL21 cells overexpressing the α-actinin COOH terminus; control (C), Zr1-His6-GST fusion peptide alone loaded on the column, demonstrating that a 15-kD protein from E. coli does not bind nonspecifically on the column. Lanes Zr1–Zr7, interaction of each of the seven Z-repeats alone with the COOH terminus of α-actinin; lane Zr4-6, interaction of the three fused Z-repeats with the COOH terminus of α-actinin. The Z-repeats Zr1, 2, 3, and 7 alone are sufficient for binding to the α-actinin COOH terminus under the conditions used. The Zr4, 5, and 6 alone do not interact with the α-actinin COOH terminus, but as a fusion peptide, they also bind to the COOH terminus of α-actinin. Lane M, molecular mass markers corresponding to 97, 60, 40, 30, 20, and 15 kD.
For the titin Z1-Z2 and T-cap binding studies, the His6-tagged NH2-terminal 16-kD fragment comprising residues 1–138 of the full-length T-cap protein was expressed in E. coli. The T-cap fragment was purified under denaturing conditions in 8 M urea. The peptide was then purified by passing the solution over Ni-NTA agarose (QIAGEN), followed by dialysis against a buffer containing 20 mM Tris, pH 8.0, 50 mM NaCl. Approximately 100-μg aliquots of the solubilized T-cap NH2-terminal peptide were mixed with E. coli lysates from cells expressing tagless Z1-Z2 titin domains. After passing the lysates over Ni-NTA agarose, aliquots of the bound and unbound fractions were denatured in SDS sample buffer and subjected to 15 or 18% PAGE (Laemmli, 1970).
Yeast Two-Hybrid Studies
A 0.8-kb rabbit titin cDNA fragment (corresponding to bp 133–905 of the human entry; sequence data available from EMBL under accession number Y18102) was amplified from rabbit psoas muscle mRNA by reverse transcription–polymerase chain reaction (RT-PCR). The fragment was inserted into the pAS2-1 vector (CLONTECH Labs, Palo Alto, CA) to obtain a GAL4 reporter fusion protein for the detection of protein–protein interactions. The recombinant “bait” plasmid, referred to as pAS2-Z1-Z2-is1, was transformed into Saccharomyces cerevisiae, strain CG-1945. For screening, the bait pAS2-Z1-Z2-is1 was coexpressed in CG-1945 cells with human heart muscle cDNA library in a pGAD10 vector (HL4013AB; CLONTECH Labs). Candidate clones from a total of 500,000 colonies screened were rescued on minus (minimal media plates deficient in Leu, Trp, and His) plates, supplemented with 1.5 mM 3-amino-1, 2, 4-triazole (3-AT). Candidate clones were processed to lose the bait plasmid, and the DNA was prepared and sequenced. In addition, candidates were also retransformed with pAS2-Z1-Z2-is1 to confirm positive binding. pAS2-Z1-Z2 was constructed by introducing a termination codon at bp 717 by site-directed mutagenesis. The truncated versions pAS2-Z1-Z2, pAS2-Z1, and pAS2-Z2-is1 of the bait were constructed by digestion of pAS2-Z1-Z2-is1 with PstI, EcoRI, and Bpu1102I, respectively. Incompatible ends were filled with the Klenow fragment of DNA polymerase I, and the construct was religated. T-cap-Δ1 and -Δ2 were constructed by digestion of T-cap with BalI and PstI, respectively, after recircularization. All constructs were expressed in CG-1945 cells and examined for growth on minus plates with 3-AT and/or for β-galactosidase activity. β-galactosidase activity of the cells was measured using chlorophenol red-β-d-galactopyranoside (CPRG) as described by the manufacturer (CLONTECH Labs).
Detection of Transcripts for the Titin-Cap Protein
Whole-mount mouse embryos at different stages post coitum (pc) were hybridized in situ essentially as described (Conlon and Herrmann, 1993). Embryos were dissected free of extra-embryonic membranes in PBS where applicable and fixed overnight at 4°C in 4% paraformaldehyde/ PBS. Labeled antisense RNA probes were prepared by in vitro transcription of mouse titin-cap protein sequences in the presence of 11-digoxygenin-UTP (Boehringer Mannheim Corp., Indianapolis, IN). All embryos were cleared in 50% formamide. For RT-PCR analysis, total human cDNAs were either prepared from commercially available cDNA libraries by amplification, or by reverse-transcription of RNAs isolated from biopsies (MacDonald et al., 1987).
Construct Preparations
pCMVEGFP–T-cap.
A PCR fragment containing the entire T-cap open reading frame, except for the start-methionine, was generated using sense and antisense primers: 5′-tttagatctGCTACCTCAGAGCTGAGCTGCGAGGTGTCG-3′ and 5′-tttggtaccttaGCCCCCAACTCTGGGCAAACTACAAAGCAG-3′, respectively. (The BglII and KpnI sites, which were included on the primers to facilitate subcloning, are shown in small letters.) The pEGFP-C1 vector (CLONTECH Labs) was linearized by digestion with BglII and KpnI. The plasmid T-cap insert were ligated and bacteria transformed. Plasmids were verified by sequencing. Plasmid DNA were purified using QIAGEN columns.
pCMVmyc–T-cap and pCMVmyc–titin Z1-Z2.
For the pCMVmyc–T-cap construct, the insert was amplified similarly to the EGFP–T-cap construct, but the cloning sites included in the 5′ sense oligo were XbaI. The amplified fragment was digested with XbaI + KpnI and subcloned into the pCMVmyc vector, which was derived from the pBK-CMV vector (Stratagene, La Jolla, CA). The pCMVmyc vector was kindly provided by Dr. Naoji Toyota (Chiba University, Japan). For the pCMVmyc–titin Z1-Z2, a fragment encompassing base pairs 133–717 was amplified from the most NH2-terminal region of the human titin partial cDNA hh1 (Labeit and Kolmerer, 1995) and subcloned as a XbaI/KpnI fragment into the pCMVmyc vector. Plasmids were verified by sequencing. Plasmid DNA was purified using QIAGEN columns.
Cell Culture, Transfection, and Microinjection Procedures
Cardiac myocytes were prepared from day 6 embryonic chick hearts and cultured as described previously (Gregorio and Fowler, 1995), except that the cells were grown on CELLocate gridded coverslips (Eppendorf, Hamburg, Germany) that were coated with collagen according to the manufacturer's instructions (0.01% collagen type I; Sigma Chemical Co., St. Louis, MO). Approximately 15% of the cells in most of our primary cultures are fibroblasts.
For transient transfection experiments, cells were plated and maintained in antibiotic-free media, according to a procedure generously provided by Dr. Naoji Toyota (Chiba University, Japan). 24 h after plating, cultured myocytes were washed three times in OptiMEM, placed in 400 μl OptiMEM–10% FBS (Hyclone Laboratories Inc., Logan, UT), and returned to the incubator while DNA complexes were prepared. DNA liposome complexes were prepared by combining 2 μg plasmid with 2 μl LipofectAmine in 100 μl serum-free OptiMEM for each 12-mm coverslip. After 20 min, 100 μl of complexes was added dropwise to the 400 μl OptiMEM–10% FBS. After 24 h, coverslips were washed three times with Hank's Balanced Salt Solution and incubated in 1 ml of MEM–10% FBS. 24, 48, and 72 h later, cells were gently washed with PBS and fixed with 2% formaldehyde in PBS for 5 min. Coverslips were washed and stored in PBS at 4°C until staining. The percent of cardiac myocytes that were transfected was ∼5%; this is comparable to what others have found (e.g., Dabiri et al., 1998). Over 400 transfected cells were studied. All tissue culture reagents (except where noted) and the LipofectAmine were purchased from Life Technologies (Grand Island, NY).
For microinjection experiments, cells were injected with a 0.5–1.0 mg/ml solution of the titin Z1-Z2 fragment in 10 mM Tris, pH 7.4, using an Eppendorf micromanipulator/transjector system, fixed 1–8 h after injection, and processed for immunofluorescence microscopy as described below. To identify injected cells, cells were coinjected with 1 μg/ml of an antibody (MOPC21; Sigma Chemical Co.) that was previously found to have no effect on this cell type (Gregorio et al., 1995). Approximately 40 cells/ group were injected, stained, and observed per experiment. Each experiment was repeated three times. Note that although the disrupted myofibril distribution pattern detected in the titin Z1-Z2 and T-cap transfection/microinjection experiments is seen in ∼5–10% of the wide range of normal cardiomyocytes in primary cultures, 80–100% of cells that overexpressed these proteins displayed this distribution pattern.
Indirect Immunofluorescence Microscopy
Cardiac myocytes were essentially stained as described (Gregorio et al., 1994; Gregorio and Fowler, 1995). In brief, coverslips were permeabilized in 0.1% Triton X-100/PBS for 15 min. To minimize nonspecific binding of antibodies, the coverslips were preincubated in 2% donkey serum, 2% BSA/PBS for 30 min. 12 well-characterized antibodies specific to various sarcomeric components and phalloidin were used to analyze the phenotypes. Details on the staining protocols used in the micrographs presented are described below. The green fluorescent protein (GFP)–T-cap–transfected cells were stained with anti–titin T12 antibody (1 μg/ml), followed by Texas red–conjugated F(ab′)2 fragments of donkey anti–mouse antibodies (1:600). Microinjected cells were stained with Texas red–conjugated phalloidin to stain for actin and Cy2-conjugated goat anti–mouse antibodies (1:600) to identify the injected cells. pCMVmyc–titin Zl-Z2– and pCMVmyc–T-cap–transfected cells were stained with monoclonal anti-cmyc antibodies (9E10), followed by FITC-conjugated donkey anti– mouse antibodies (1:600) and Texas red–conjugated phalloidin (1:400) or rabbit anti–α actinin antibodies (generously provided by Dr. S. Craig, Johns Hopkins University, Baltimore, MD), followed by Texas red–conjugated F(ab′)2 fragments of donkey anti–rabbit antibodies (1:600). pCMVmyc–titin Zl-Z2– and pCMVmyc–T-cap–transfected cells were also stained with polyclonal anti-cmyc antibodies (1:100), followed by fluorescein-conjugated donkey anti–rabbit antibodies (1:600), monoclonal anti-sarcomeric myosin antibodies (1:50) (F4G3; generously provided by Dr. F. Pepe, University of Pennsylvania, Philadelphia, PA), and Texas red–conjugated donkey anti–mouse antibodies (1:600). For the GFP-tagged experiments, cells were analyzed on a laser scanning confocal microscope (model TCS 4D; Leica, Inc.; Arizona Research Laboratory, Division of Biotechnology, University of Arizona, Tucson, AZ) using a 100× NA 1.4 oil immersion objective. Simultaneous two-channel recording was performed with a pinhole size of 85 μm. All figures are derived from a single optical section. Images were processed and merged using Adobe Photoshop software (San Jose, CA) and printed using a Codonics NP1600 dye sublimation printer (Middleburg Heights, OH). For all other transfection and microinjection experiments, stained cells were analyzed on a microscope (model Axiovert; Carl Zeiss, Inc., Thornwood, NY) using a 63× (NA 1.4) objective, and micrographs were recorded on Kodak TriX film (Rochester, NY) or as digital images on a SenSys cooled HCCD camera (Photometrics, Tucson AZ). All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA), except the Cy2-conjugated antibodies, which were purchased from Pierce Chemical Co. (Rockford, IL). Phalloidin was purchased from Molecular Probes, Inc. (Eugene, OR), and anti–titin T12 antibodies were purchased from Boehringer Mannheim Corp. Monoclonal anti-cmyc (9E10) antibodies (Evan et al., 1985) were produced from the hybridoma MYC 1-9E10.2, which was purchased from American Type Culture Collection (Rockville, MD), and polyclonal anti-cmyc antibodies were purchased from Upstate Biotechnology, Inc. (Lake Placid, NY).
Results
Ultrastructural Molecular Layout of Z-Line Titin
To ascertain the molecular layout of titin within the Z-line, we generated novel polyclonal antibodies against (a) the first two NH2-terminal Ig domains of titin (anti–Z1-Z2 antibodies), which are expressed in all striated muscles, and (b) the Zr5 and 6 titin repeats (anti–titin Zr5-6 antibodies) (Labeit and Kolmerer, 1995; Sorimachi et al., 1997). Under standard labeling and strong blocking (60 min with 2% BSA) conditions, antibodies that are specific to the titin Z1-Z2 domains stained the borders of the Z-lines only (Fig. 1, A–C). This result was surprising based on previous reports that predicted that the NH2 terminus of titin is found in the middle of the Z-line (Gautel et al., 1996). These new observations indicate that either the NH2 terminus of titin completely spans the Z-line to the adjacent sarcomere or that the NH2-terminal end of the titin molecule does not penetrate the Z-line. Next, to distinguish between these possibilities and to rule out that the absence of integral Z-line labeling was a result of the failure of the first antibody to penetrate the Z-line, labeling studies were also performed with the anti–titin Zr5-6 antibodies. Although these polyclonal antibodies were generated against the Zr5 and 6 repeats which are differentially expressed in striated muscle, they most likely recognize all Zr repeats because of the very high sequence similarity between them (Sorimachi et al., 1997); this is supported by the observation that the anti–titin Zr5-6 antibodies label the Z-lines in myofibrils from rat psoas muscle (data not shown) which only express the Z-repeats 1, 2, 3 and 7 (Sorimachi et al., 1997). Immunolabeling with the anti–titin Zr5-6 antibodies resulted in strong labeling throughout the Z-line, indicating that this structure can be penetrated by antibodies (Fig. 1, D–F). The labeling pattern for the Z-repeats, together with the labeling for the Z1-Z2 Ig domains, suggests that titin molecules from neighboring sarcomeres fully overlap and extend to the opposite ends of the Z-disc (see model: Fig. 11).
Figure 11.
Model for the insertion of the NH2-terminal region of titin into Z-discs. Titin's residues 1–200 are shared by titins from all muscle tissues and code for two Ig domains (Z1 and Z2). Together, these terminal domains interact with a 19-kD ligand, referred to as titin-cap (T-cap). The Z1-Z2/T-cap complex is located at the periphery of the Z-line. T-cap contains a 27-residue-long COOH-terminal serine/proline and basic residue-rich domain that codes for potential phosphorylation motifs and is not required for interaction with titin. Titin residues 200–430 are predicted to extend to the center of the Z-line. Titin residues 430–700 contain the Z-repeats that potentially bind to the COOH terminus of α-actinin. Titins from opposite half-sarcomeres are predicted to share full overlap in Z-lines (about 100 nm in soleus and cardiac muscle Z-lines).
Interaction of Titin Z-Repeats with α-Actinin
Many studies have suggested that titin and α-actinin have important roles in myofibril assembly (for example see Epstein and Fischman, 1991; Shimada et al., 1996; Holtzer et al., 1997; Turnacioglu et al., 1997). To explore further the interaction of Z-disc titin with the major Z-line component, α-actinin, an in vitro E. coli expression assay was used. Previously, the Zr7 repeat domain of titin, which is expressed in all striated muscle types, was shown to interact with the COOH-terminal region of the α-actinin isoform encoded by the ACTN2 gene (Ohtsuka et al., 1997; Sorimachi et al., 1997). First, in Fig. 2 A, an untagged titin Zr1, 2, 3, or 7 fusion protein, which corresponds to the splice variant seen in psoas muscle, was expressed in BL21 cells, and the whole-cell lysate was mixed with lysates from cells expressing a GST-His6 double-tagged α-actinin fusion protein and passed over a Ni-NTA column. The bound peptides that were eluted were run on a 15% polyacrylamide gel and visualized by Coomassie blue staining. Results from this experiment demonstrate that the fusion protein containing the titin repeats Zr1, 2, 3, and 7 binds to α-actinin. In Fig. 2 B, cells expressing all of the Z-repeats individually as GST-His6 double-tagged fusion proteins were mixed with cells expressing a tagless 15-kD COOH-terminal peptide of α-actinin. This experiment demonstrates that the titin Z-disc repeat domains Zr1, 2, 3, and 7 can individually bind to the COOH-terminal 15-kD region of α-actinin (Fig. 2 B). In contrast, the titin repeats Zr4, 5, and 6 can only bind to the COOH-terminal region of α-actinin when fused together (Fig. 2 B); this may suggest a weaker binding of the individual Z-repeats, but possibly only reflects the addition of more flanking residues or a change in domain folding, features important for in vitro binding assays using such short peptides as the Z-repeats. Taken together, the α-actinin/titin binding studies suggest that the titin Z-repeats provide at least four potential binding sites for the COOH terminus of α-actinin. We hypothesize that Z-lines with more titin Z-repeats and hence more connecting Z-filaments are mechanically stiffer and more resistant to stretch because of additional cross-linking of titin molecules and thin filaments (which are also cross-linked in the Z-line by α-actinin; Stromer and Goll, 1972).
Interaction of the NH2-terminal Titin Z1-Z2 Repeats with a Novel 19-kD Protein
The yeast two-hybrid system was used to identify new ligands that interact with the NH2-terminal Ig domains Z1 and Z2, which are shared by all titins. Our search for potential ligands revealed that the titin repeats Z1 and Z2 specifically bind to a 19-kD protein, which we refer to as T-cap. 500,000 clones from a human heart muscle cDNA library were screened with the recombinant bait, referred to as pAS2-Z1-Z2-is1. A total of seven colonies were identified on the plates and further analyzed. Six of the clones were confirmed by retransformation and β-galactosidase assays (CLONTECH Labs). One clone coded for a vimentin cDNA (data now shown). So far, we have not been able to confirm an interaction of titin Z1-Z2 and vimentin with independent approaches. Therefore, as of now, we consider the interaction of titin Z1-Z2 with vimentin as a possible false-positive. The remaining five clones encoded various portions of the same gene transcript, which we called “T-cap.” The screening of 400,000 cDNA clones from a human heart cDNA library with T-cap probes identified 40 T-cap cDNA clones. Sequence analysis indicates that the T-cap is coded for by a 950-bp cDNA, which contains a single open reading frame for a 19-kD protein. Inspection of this sequence revealed a 27-residue-long COOH-terminal serine/proline and basic residue-rich domain, which codes for potential phosphorylation motifs, and no obvious motifs for the NH2-terminal 140 residues. Data library searches with the 19-kD protein revealed sequence identity to the recently described cDNA sequence of the putative thick filament protein “telethonin,” which was obtained from the assembly of anonymous EST entries (sequence data available from EMBL under accession AJOOO491; Valle et al., 1997). No significant homology with other known protein sequences was found.
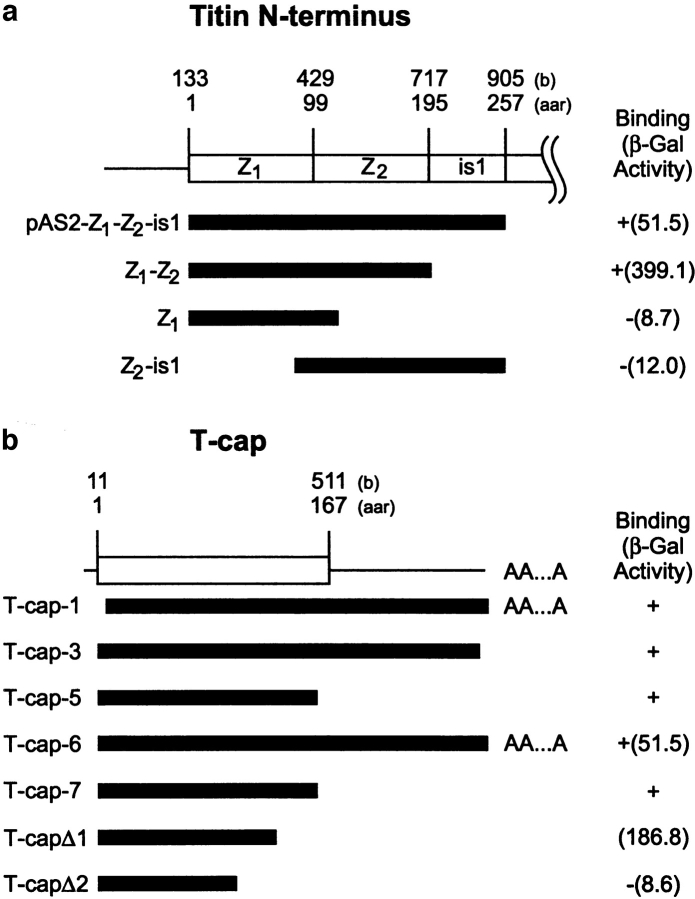
Next, we attempted to identify more precisely the binding sites for the titin/T-cap interaction. Truncation analysis of the bait plasmid, pAS2-Z1-Z2-is1, using the constructs pAS2-Z1-Z2 (containing the Z1 and Z2 domains), pAS2-Z1 (containing only the Z1 domain), and pAS2-Z2-is1 (containing the Z2 domain fused to residues 200-257) indicated that, individually, Z1 and Z2 are not capable of binding T-cap, but that both Z1 and Z2 Ig motifs are required for its binding activity to T-cap (Fig. 3 a). Truncation analyses of T-cap indicated that the COOH-terminal SP-rich domain is not involved in the binding of the Z1-Z2 domains (Fig. 3 b), whereas the deletion mutant T-cap-Δ2 has lost its interaction potential (representing nucleotide residues 19–246).
Figure 3.
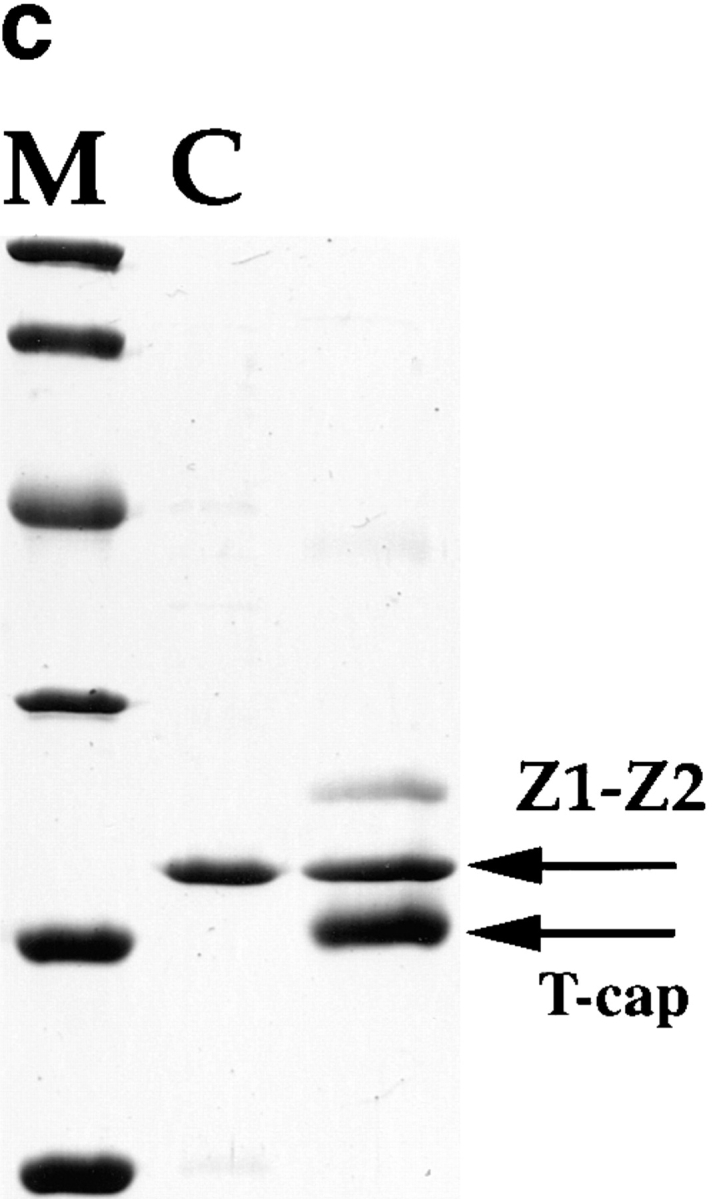
Interaction of T-cap and the titin Z1-Z2 domains. (a) Titin repeats Z1 and Z2 specifically interact with the 19-kD titin-cap protein by yeast two-hybrid analysis. Numbers above the schematic layout of the NH2-terminal titin region indicate the nucleotide residue numbers corresponding to the human entry (EMBL data library X90568) (top) and residue numbers of the deduced amino acid sequence (bottom). Two immunoglobulin domains at the NH2 terminus of titin are indicated as Z1 and Z2, respectively. Bars indicate positions of the cDNAs used for the bait plasmid (pAS2-Z1-Z2-is1), and the truncation constructs, pAS2-Z1-Z2 (containing the Z1 and Z2 domains), pAS2-Z1 (containing Z1 domain only), and pAS2Z2-is1 (containing the Z2 domain fused to residues 200– 257), used for the binding assay to T-cap. +/− signs refer to growth on minus (Leu, Trp, and His) plates supplemented with 3-AT, and numbers in parentheses refer to β-galactosidase activities (arbitrary units). (b) Interaction of the NH2-terminal 16 kD of T-cap with the titin Z1-Z2 Ig domains in the yeast two-hybrid system. Schematic of T-cap cDNA clone. Numbers above the titin-cap cDNA indicate the nucleotide residues of the human full-length cDNA sequence (sequence data available from EMBL under accession number AJ000491; from Valle et al., 1997) (top) and residue numbers of the deduced amino acid sequence (bottom). AA...A, a poly(A+) tail at the 3′ terminus of the corresponding cDNA clone. Bars indicate positions of cDNA inserts derived from the two-hybrid screening using pAS2-Z1-Z2-is1 (see a) as bait (T-cap-1, -3, -5, -6, and -7), and the truncation constructs of T-cap (T-cap-Δ1 and -Δ2) described in Materials and Methods. +/− signs refer to growth on minus (Leu, Trp, and His) plates supplemented with 3-AT, and numbers in parentheses refer to β-galactosidase activities (arbitrary units). (c) Interaction of expressed T-cap and titin Z1-Z2 peptides. A Coomassie blue– stained 18% Laemmli SDS gel of a whole-cell lysate of BL21 cells expressing tagless titin Z1-Z2 (lane C). These cells were mixed with a His6-tagged 16-kD fragment of T-cap and lysed. Lysates were passed over Ni-NTA agarose columns. Interaction of a stable complex of titin Z1-Z2 and T-cap is indicated by retaining also the nontagged titin Z1-Z2 on the column (arrow marks Z1-Z2 and T-cap in third lane). Lane M, molecular mass markers as in legend for Fig. 2.
To exclude the possibility of false-positive results from the yeast two-hybrid studies, the interaction of the T-cap with the titin Z1-Z2 domain was confirmed using an independent approach. E. coli cell lysates expressing the Z1-Z2 titin domains were mixed with a purified NH2-terminal fragment of T-cap. Mixing resulted in the formation of a T-cap/titin Z1-Z2 complex, and both proteins were copurified on Ni-NTA columns (Fig. 3 c). We conclude from the yeast two-hybrid and in vitro binding studies that the NH2-terminal 16-kD of T-cap specifically binds to titin, and both the Z1 and Z2 titin domains are required for this interaction.
Transcription of T-cap during Development and in Adult
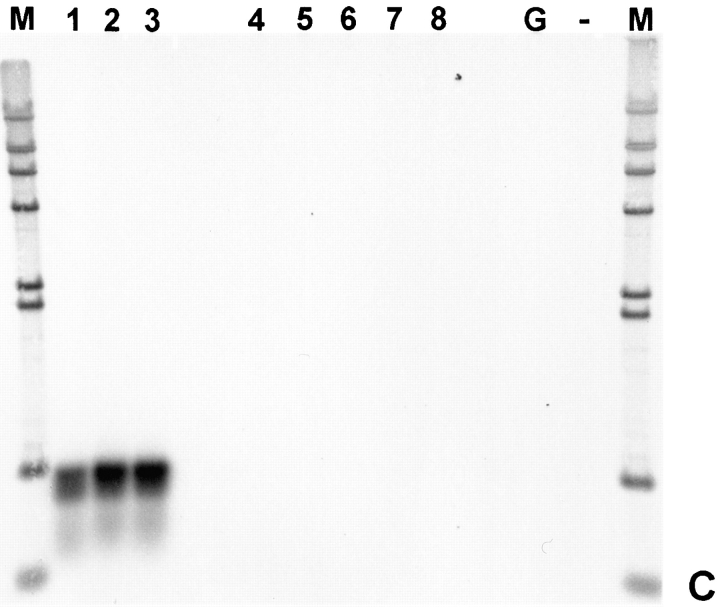
Titin has a characteristic tissue-specific and developmental expression pattern. In the mouse, titin transcripts first appear in the heart anlage at day 8 pc and remain abundantly expressed in the heart throughout development, whereas expression in the somites occurs about 1 d later (Kolmerer et al., 1996). To determine whether T-cap transcripts have the same characteristics, we performed in situ hybridization of whole-mount embryos using probes from the mouse titin-cap cDNA. The T-cap mouse cDNA sequence was obtained by cross-species PCR (sequence data available from EMBL under accession number Y14845). The comparison of the T-cap sequences from human and mouse predicted by their respective 950-bp full-length cDNAs revealed that the encoded 19-kD T-cap proteins share 90% sequence identity and 95% sequence similarity between species (Fig. 4). At day 9.5 pc, T-cap transcripts are detected in the developing heart and in the somites. At this stage, the transcription pattern of T-cap in the heart and somites is very similar to the transcripts of titin, except that transcripts for T-cap are also detected in the otic vesicle (Fig. 5, A and B). In a survey of adult tissues by RT-PCR, T-cap transcripts were found exclusively in striated muscles, including human fetal heart, adult heart, and skeletal muscles, but were absent in normal and pregnant uterus, fetal brain, liver, and spleen (Fig. 5 C). We conclude that the transcription pattern of titin and T-cap are similar at day 9.5 pc. The significance of the presence of T-cap (but not titin) transcripts in the otic vesicle and the observation that titin transcripts are detectable ∼1 d earlier in somites than T-cap transcripts will require further studies.
Figure 4.
Comparison of the T-cap protein sequences from human and mouse predicted by their respective 950-bp full-length cDNAs. The 19-kD T-cap proteins share 90% sequence identity and 95% sequence similarity between species. The human T-cap protein sequence is identical to the recently described telethonin entry (EMBL data library AJ000491) derived from anonymous EST entries (Valle et al., 1997); the mouse sequence data from skeletal muscle are available from GenBank/EMBL/DDBJ under accession number Y14845.
Figure 5.
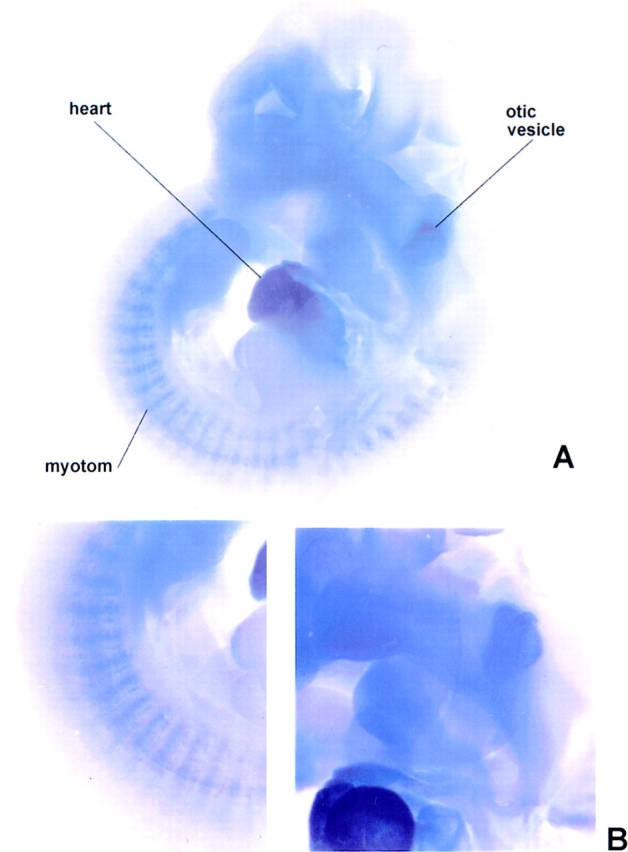
Developmental- and tissue-specific expression of T-cap transcripts. (A) In situ hybridization of whole-mount embryos at day 10.5 pc using probes from the mouse titin-cap cDNA. Transcripts are detected in the heart at 9.5 d of gestation (not shown). At day 10.5 pc, transcription has also commenced in the somites. The transcription pattern is very similar to that of titin (Kolmerer et al., 1996), except that transcripts for the titin-cap protein are also detected in the otic vesicle (B, selected details of A). (C) Presence of titin-cap protein transcripts in human cDNAs from different tissues, as detected by RT-PCR. T-cap protein is expressed exclusively in striated muscles. Lanes 1–3, fetal heart, adult heart, and adult skeletal muscles, respectively; lanes 4–8, normal and pregnant uterus, fetal brain, liver, and spleen, respectively. Controls: lane G, genomic DNA, “no template”; lane M, lambda HindIII size marker.
Localization and Assembly of T-cap in Striated Muscle
To ascertain the cellular localization of T-cap, we generated a polyclonal antibody against this molecule. This antibody specifically labeled Z-line regions by indirect immunofluorescence microscopy (data not shown). Immunoelectron microscopy with the anti–T-cap antibodies revealed that the labeling was confined to the edges of the Z-line (Fig. 6, A–C). Comparison of the histograms of the gold particle distribution obtained with the various antibodies (Fig. 6, D–F) support the hypothesis that the anti– Z1-Z2 and anti–T-cap antibodies both label the edge of the Z-line while the anti–Zr5-6 antibody labels inside the Z-line.
For functional studies on the assembly of T-cap, primary cultures of chick cardiac myocytes were chosen as a model system. Owing to their flat, well-spread morphology, these cells are ideally suited for detailed immunolocalization observations of the spatial relationships among assembling myofibrillar proteins. To determine the cellular localization of assembled T-cap, we transfected cardiac myocytes with a fusion construct of T-cap and GFP. Confocal analysis of T-cap–GFP–transfected cells, which were costained with the well-characterized monoclonal titin T12 antibody, which stains an epitope of titin in close proximity to the Z-line (Fürst et al., 1988), revealed overlap of the two fluorochromes at the level of resolution of the light microscope (identical results were obtained when T-cap–GFP– transfected cells were costained with anti–α-actinin antibodies; data not shown) (Fig. 7, a–c). This result indicates that newly synthesized T-cap is targeted to Z-lines in live cardiac muscle cells, which is consistent with the ultrastructural localization of this protein (Fig. 6) and its binding partner, titin Z1-Z2 (Fig. 1).
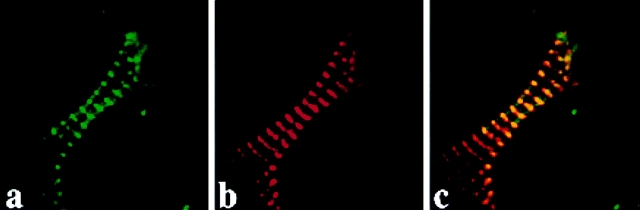
Figure 7.
T-cap assembles at the Z-disc in cardiac myocytes. Fluorescent confocal image of a cardiac myocyte expressing (a) T-cap–GFP that was fixed and stained with (b) anti–titin T12 antibodies followed by Texas red–conjugated donkey anti– mouse antibodies. (c) Merged image of a and b demonstrating overlap of the two fluorochromes. Bar, 10 μm.
Effects of Titin Z1-Z2 and T-cap Overexpression in Cardiac Myocytes
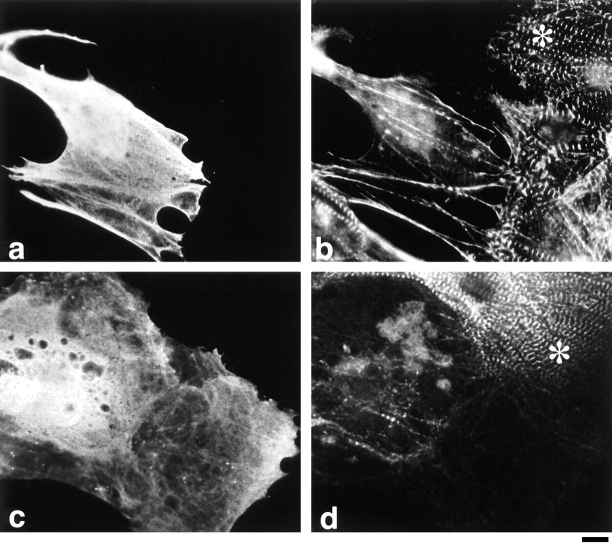
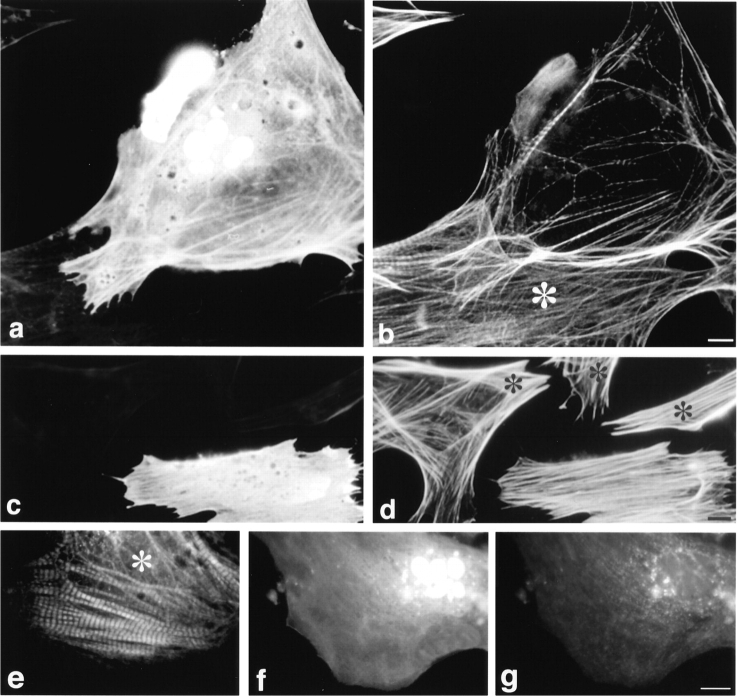
To gain insight into the functional significance of the NH2 terminus of titin in cells, we overexpressed titin Z1-Z2 under the control of a strong viral promoter (CMV) in the hope that the expressed protein would compete with T-cap to generate dominant-negative effects on sarcomere formation. First, we transfected cells with a plasmid encoding a myc-epitope–tagged titin Z1-Z2 fragment. A myc epitope was inserted at the NH2 terminus to distinguish the recombinant protein from endogenous protein. Severe disruption of Z-disc structure was observed in all transfected cells where titin Z1-Z2 was overexpressed (24, 48, and 72 h after transfection) as seen by a marked loss in α-actinin staining, as compared with nontransfected cells, which demonstrate a strong periodic Z-line staining with anti–α-actinin antibodies (Fig. 8, identical cells stained for [a] myc and [b] α-actinin; compare staining patterns in transfected cells that stain positively with anti-myc antibodies with nontransfected cells marked with *). As a complementary approach, to support the idea that the effects we were observing were a result of disrupting the interaction between titin and T-cap, cardiac myocytes were also transfected with a plasmid encoding a myc–T-cap fusion protein. Overexpression of T-cap resulted in an identical phenotype: severe disruption of Z-lines was discerned by staining for α-actinin (Fig. 8, staining for [c] myc and [d] for α-actinin; compare staining patterns in transfected cells with nontransfected cells marked with *). The severe phenotype observed suggested that other sarcomeric components might also be disrupted by the overexpression of these peptides. To study this, the distribution of the major thin filament protein, actin; the major thick filament protein, myosin; and different epitopes of titin along the length of the molecule were also observed in both pCMVmyc-titin Z1-Z2 and pCMVmyc–T-cap–transfected cells. Results from this experiment revealed that overexpression of either titin Z1-Z2 or T-cap resulted in disruption of all sarcomeric proteins studied. (See Fig. 9 for cardiac myocytes transfected with pCMVmyc titin Z1-Z2 and double stained for [a] myc and [b] actin, and [f] myc and [g] myosin; compare staining patterns in transfected cells that stain positively with anti-myc antibodies with nontransfected cells marked with * in b and e. Staining for other contractile protein constituents studied is not shown.) Similarly, when the T-cap–GFP COOH-terminal fusion protein was overexpressed, a similar disruption was observed, while overexpression of GFP alone did not lead to the disruption of myofibrils in the transfected cells (data not shown). This suggests that the observed phenotype was not due to overexpressing too much protein in this cell type (i.e., by using such a strong promoter; others have also observed no effect in the same cell type for example, Turnacioglu et al., 1997).
Figure 8.
Overexpression of titin Z1-Z2 or T-cap results in marked disruption of Z-discs. Cardiac myocytes transfected with a plasmid encoding the fusion proteins: (a and b) myc-titin Z1-Z2 and (c and d) myc–T-cap. Cells were fixed 48 h after transfection and double stained with (a and c) monoclonal anti-myc antibodies followed by FITC-conjugated donkey anti–mouse antibodies and (b and d) polyclonal anti–α-actinin antibodies followed by Texas red–conjugated F(ab′)2 fragments of donkey anti– rabbit antibodies. * marks cells that were not transfected. Bar, 10 μm.
Figure 9.
Overexpression of titin Z1-Z2 results in marked disruption of thin and thick filaments in cardiac myocytes: stress fibers in fibroblasts are unaffected. Cardiac myocytes (a, b, and e–g) and fibroblasts (c and d) transfected with a plasmid encoding myc-titin Z1-Z2. Cells were fixed 48 h after transfection and double stained with (a and c) monoclonal anti-myc antibodies followed by FITC-conjugated donkey anti-mouse antibodies and (b and d) Texas red–conjugated phalloidin to stain F-actin, or with (f) polyclonal anti-myc antibodies, followed by FITC-conjugated donkey anti–rabbit antibodies and (e and g) monoclonal antistriated muscle myosin antibodies, followed by Texas red–conjugated donkey anti– mouse antibodies. Staining for myc was not detected in the cell shown in e (data not shown). * marks cells that were not transfected. Bars, 10 μm.
It is important to note that the process of myofibril assembly in primary cultures of cardiac myocytes is temporarily irregular; thus numerous structures representing different stages in assembly are observed within the same cell, as well as within different cells in a culture dish (Rhee et al., 1994; Gregorio and Fowler, 1995 and references therein). Although this variability exists, variability was not seen in the phenotype in both the transfection and microinjection experiments. Myofibrils, whether in early, intermediate, or mature stages of assembly, were disrupted. Interestingly, there was also a correlation between the levels of expression and the integrity of the sarcomere. When the level of either the myc or GFP fusion protein expression was low, as resolved by a low level of unincorporated fluorescence in the cytoplasm, the fusion proteins were observed to assemble at the Z-line (e.g., Fig. 7). In all myocytes with a high level of expression of the fusion proteins (i.e., a high level of unincorporated fluorescence), a dominant-negative phenotype of myofibril disassembly and loss of physiological function (cell beating) occurred (e.g., Figs. 8 and 9). This was also confirmed in live cells when existing myofibrils were visualized disassembling as the level (intensity) of the GFP fluorescence increased in individual T-cap–GFP–expressing cells (data not shown). We conclude that both the titin Z1-Z2 domains and T-cap are required for sarcomerogenesis in cardiac myocytes.
As another complementary approach to rule out the possibility that the observed phenotype was due to overexpressing too much protein and to rule out effects due to the fusion proteins used, varying amounts of purified titin Z1-Z2 fragments were microinjected directly into the cytoplasm of the cells; again, a severe disruption of the myofibrils was observed (Fig. 10, cells microinjected with titin Z1-Z2 and double stained for [c and e] the anti–mouse Ig antibodies used to identify injected cells and [d and f] actin; compare with control microinjected cells [a and b]). Approximately, 80% of the cardiac myocytes that were positively identified as being injected expressed this phenotype.
Figure 10.
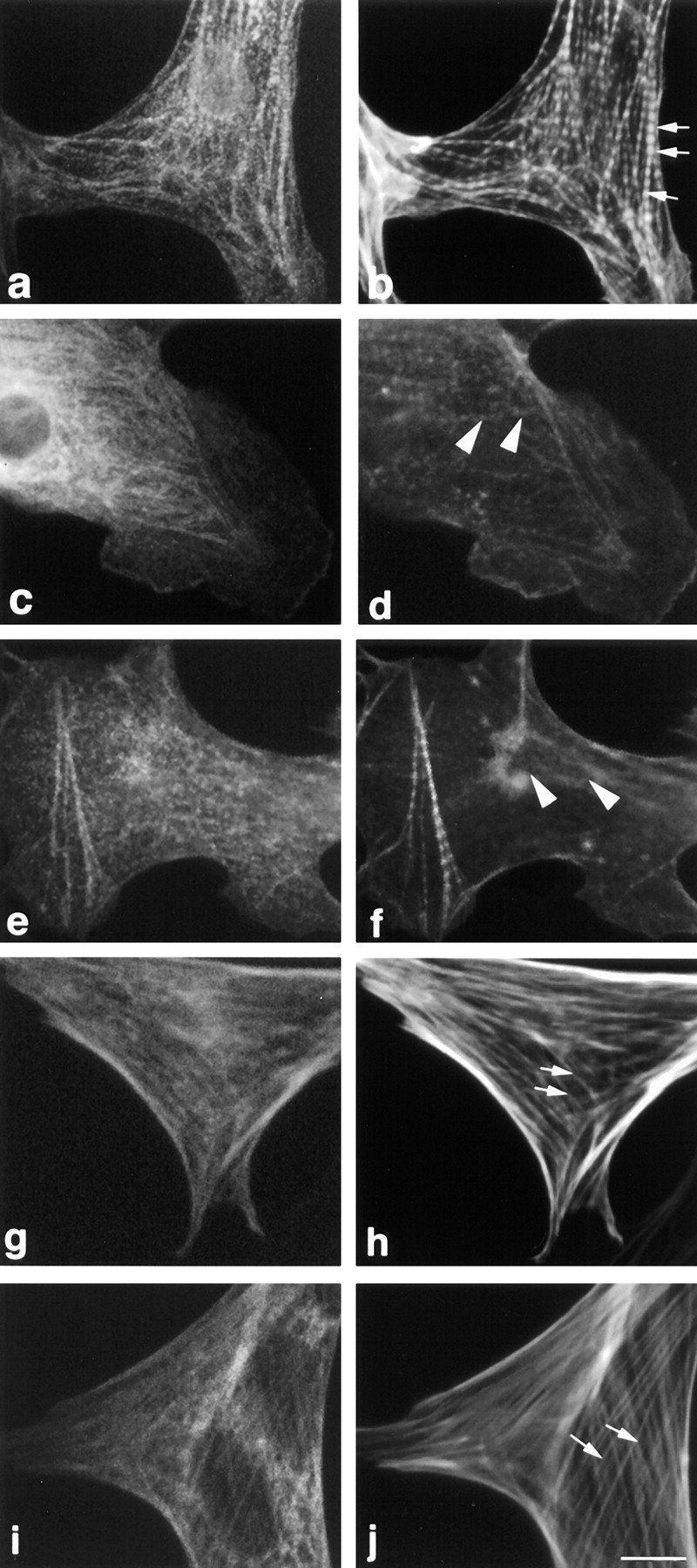
Microinjection of titin Z1-Z2 into the cytoplasm of cardiac myocytes results in disruption of myofibrils: stress fibers in fibroblasts are unaffected. Cells were microinjected with (a, b, g, and h) a nonspecific monoclonal antibody (MOPC-21) to identify injected cells and (c–f, i, and j) with the nonspecific antibody plus purified titin Z1-Z2. 2 h after injection, the cells were fixed and stained with (a) Cy2-conjugated anti–mouse antibodies and (b) Texas red–conjugated phalloidin. Note that the control microinjected antibody used to mark the injected cells appears to localize to some myofibrils in the cardiomyocytes and to some stress fibers in fibroblasts. Arrows point to the typical striated and stress fiber staining seen with Texas red–phalloidin (b) in cardiac myocytes and (h and j) in fibroblasts, respectively; arrowheads point to disrupted myofibrils in cardiac myocytes (d and f). Bar, 10 μm.
It is also interesting to note that high levels of expression of titin Z1-Z2 or T-cap in transfected or microinjected fibroblasts resulted in no observable alteration to stress fibers as determined by staining for F-actin. (Figs. 9, c and d, and 10, i and j, data shown for overexpression of titin Z1-Z2. Compare with fibroblasts not expressing titin Z1-Z2 in Fig. 9, c and d, marked with an * and in Fig. 10, g and h.) For T-cap, this might be related to the observation that this protein appears to be striated muscle specific (Valle et al., 1997; this report, Fig. 5). For titin, a nonmuscle isoform of titin has been suggested to be involved in the assembly of stress fibers (Eilertsen et al., 1994). Using similar approaches, Sanger and colleagues observed that expressing a fragment of titin encompassing the NH2-terminal Z1-Z2 domains in fibroblasts results in disruption of stress fibers (Turnacioglu et al., 1997). One possibility is that the stress fiber disruption phenotype they observed may reflect the fact that they overexpressed a larger fragment of titin than was used in this study. We conclude that the nature of nonmuscle isoforms and their roles in stress fiber structure will require further studies.
Discussion
Vertebrate Z-lines link actin filaments in one sarcomere to actin filaments with opposite polarity, in the adjacent sarcomere. Although there are substantial variations in Z-lines from different vertebrate muscles and fiber types, all vertebrate Z-lines share the packaging of the antiparallel thin filaments from opposite half-sarcomeres into a tetragonal lattice, which is cross-linked by α-actinin (otherwise referred to as Z-filaments: Yamaguchi et al., 1985; Vigoreaux, 1994; Luther et al., 1995; Schroeter et al., 1996; Squire, 1997). Other less characterized components of Z-lines, including titin, are likely to be important in the regulation of Z-line assembly. Thus, a detailed knowledge of titin's layout and of its molecular interactions in Z-lines is necessary.
The Z1 and Z2 Ig domains (residues 1–200) are expressed in all striated muscles, whereas the subsequent residues 200–700, including the Z-repeats, are differentially expressed (Sorimachi et al., 1997). We sought to define the titin Z-disc layout more precisely by generating antibodies to the constitutively expressed titin Z1-Z2 domains, and to the differentially expressed residues 560– 650. Immunolocalization at the ultrastructural level revealed that the NH2-terminal Z1-Z2 domains localized to the periphery of the Z-disc; this finding suggests that titin molecules from opposite sarcomeres fully cross each other within the Z-disc (Fig. 1). A possible explanation for the conflicting Z-line layout models that have been proposed by others (Gautel et al., 1996; Young et al., 1998) is that the antibodies used in these studies were generated against titin repeats which may cross-react with other Z repeats (e.g., Fürst et al., 1989b ; Gautel et al., 1996; anti– Zr5-6 antibodies used in this study). Here, we also used immunoelectron microscopy to localize the nonrepetitive T-cap. As expected from our localization of titin Z1-Z2, T-cap epitopes were also detected at the edges of the Z-disc (Fig. 6). Furthermore, the Z-repeat epitopes were present throughout the Z-line (Fig. 1). This may suggest that the Z-repeat family longitudinally extends within the Z-disc lattice (Fig. 11). Taken together, the immunolocalization studies with the anti–Z1-Z2, anti-Zr5/6, and anti– T-cap antibodies demonstrate that the titin NH2-terminal region spans the entire Z-line with the titin Z1-Z2 (residues 1–200)/T-cap protein complex extending to the I-Z junction of the adjacent sarcomere. In combination with other immunoelectron microscopy data (Yajima et al., 1996; Gautel et al., 1996; Young et al., 1998), it can be concluded that ∼200–700 residues of titin correspond to titin's Z-line–spanning region. Therefore, we suggest the term “Z-disc integrative domain” for this segment of titin.
In order for the ∼500 residues to span the 110-nm-wide Z-line of rabbit soleus muscle, each residue of Z-disc titin would have a mean axial extension of ∼2.2 Å per residue. This figure is intermediate between the values predicted for an extended α-helix (1.5 Å per residue) or an extended β-sheet structure (3.2–3.4 Å per residue). Sequence analysis of the titin residues 200–700 reveals segments with both predicted β-sheet and α-helical secondary structural folds. Therefore, we speculate that titin in the interior of the Z-line is composed of extended β-sheet and α-helical secondary structure elements connected in series. More highly folded domains in Z-disc titin, such as globular folds, appear unlikely. Based on our model, we would predict that residues 200–700 of the titin gene contain differentially expressed exons that are skipped in tissues and fibers that have narrower Z-lines. In support of this hypothesis, many differential splicing events have been observed to occur in this region (Gautel et al., 1996; Sorimachi et al., 1997). Furthermore, studies on chick breast muscles that have narrow Z-lines revealed that the differentially expressed Z-line segment of titin is very short and contains only ∼450 residues (Yajima et al., 1996). It will be interesting to also determine Z-disc titin sequences from teleost bony fish muscles, which have ∼50-nm-wide Z-lines (Luther et al., 1995), in contrast to the ∼70–110-nm-wide Z-lines of mammalian muscles.
Rabbit cardiac Z-disc titin contains seven copies of the 45-residue Z-repeats, soleus muscle expresses isoforms of four and six repeats, and rabbit psoas muscle contains four repeats (Sorimachi et al., 1997). Previously the Z-repeat, Zr7, was shown to interact with α-actinin (Ohtsuka et al., 1997; Sorimachi et al., 1997). Here, in vitro binding studies show that although Zr1, 2, 3, and 7 (repeats expressed in psoas muscle) together bind to the COOH terminus of α-actinin, each of these single Z-repeats are alone sufficient for this interaction. In contrast, the single repeats Zr4, 5, and 6 from soleus and heart muscles failed to bind in our assay; however, the triple repeat construct Zr4, 5, 6 interacted with α-actinin. This may suggest that the individual repeats Zr4, 5, and 6 also bind, although possibly more weakly, to the COOH terminus of α-actinin. Earlier studies using the yeast two-hybrid system detected titin/ α-actinin interactions only within the Z-repeat region (Ohtsuku et al., 1997; Sorimachi et al., 1997). However, after the original submission of this manuscript, it was reported that specific titin Z-repeats also bind to the central spectrin repeats within α-actinin, and that another non– Z-repeat α-actinin binding site (residues 760–826 of the human cardiac titin) is present in titin (Young et al., 1998). It was concluded from these findings that two distinct types of titin interactions lead to an asymmetrical sorting of α-actinin. Further studies will be required to resolve the range of possible in vivo titin/α-actinin interactions. At present, we speculate, based upon our data, that the titin Z-repeats may regulate the number and distribution of Z-filaments in Z-lines by their binding to the COOH-terminal α-actinin domains. In cardiac titin, Z-repeats may provide up to seven attachment sites for Z-filaments per titin molecule, whereas soleus and psoas muscle titins may provide six and four, respectively (Sorimachi et al., 1997). The differential expression of the titin Z-repeats may explain the presence of different numbers of Z-filament layers in the Z-lines (Rowe, 1973). As for the functional significance, we hypothesize that the differential expression of Z-repeats modulates the α-actinin/titin/actin network of the Z-lines and that this allows the mechanical strength of the Z-line to be varied.
The NH2-terminal immunoglobulin domains Z1 and Z2 are shared by all titins. We searched for a potential function(s) of these domains. Using the yeast two-hybrid approach and by subsequent studies with expressed recombinant proteins, we found that titin Z1 and Z2 domains bind to a 19-kD sarcomeric protein. Deletion studies indicated that both the Z1 and Z2 domains are required for this interaction, suggesting that the binding site for the 19-kD protein contains residues from both domains. Surprisingly, the identified sequence of the human 19-kD protein is identical to a recently determined cDNA sequence from a putative thick filament–associated protein, telethonin (Valle et al., 1997). Note that thick filament association in this study was determined by a single immunofluorescence study. To clarify this, we analyzed further if the cloned 19-kD protein is indeed a Z-disc protein interacting with titin, or rather a thick filament–associated protein. Initially, we investigated the targeting of the 19-kD titin ligand into myofibrils by expressing it in cardiac myocytes as a green fluorescent fusion protein. This experiment demonstrated that the 19-kD protein in cardiac myocytes is targeted to the Z-line, but not to the A band. Moreover, localization studies using a polyclonal antibody that specifically recognizes the 19-kD protein demonstrated staining at the Z-line, but not the A band. These data are consistent with the finding that the 19-kD protein is a ligand of the Z1-Z2 titin domains. Based on the 19-kD protein's localization, assembly, and interaction with the extreme NH2-terminal domains of titin, we propose the name “titin-cap” (T-cap) for the 19-kD protein, rather than the initial suggestion, Telethonin, which referred to a putative thick filament protein (Valle et al., 1997).
On the transcriptional level, T-cap transcripts are abundantly expressed in heart and skeletal muscles (Valle et al., 1997). At day 9.5 pc, the distribution of T-cap transcripts in somites and in the developing heart closely resembles that of titin (Fig. 5 for T-cap; Kolmerer et al., 1996 for titin). Future studies will be needed to address the significance of T-cap transcripts in the otic vesicle during early development, and if the initial appearance of titin transcripts at 8.0 pc (Kolmerer et al., 1996) precedes that of T-cap.
To elucidate further the functional significance of titin's association with the Z-line components, we overexpressed the NH2-terminal domains Z1 and Z2 of titin and their binding partner, T-cap, in primary cultures of cardiac myocytes. Using either microinjection or transfection techniques, overexpression of either molecule resulted in severe myofibril disruption. One explanation for this phenomenon is that a dominant-negative phenotype occurred. That is, overexpressed titin Z1-Z2 fragments competed for the T-cap binding site of endogenous titin, and overexpressed T-cap competed for the titin binding site on endogenous T-cap. These events might result in inhibiting endogenous titin and/or T-cap from assembling and stabilizing Z-discs.
The results from our titin Z1-Z2 and T-cap overexpression studies suggest that the association of titin filaments with Z-discs is critical for the assembly and maintenance of myofibril structure. Disruption of myofibrils has also been reported as a result of overexpression of (a) the first 362 amino acids of titin (zeugmatin) in cardiac and skeletal muscle cultures (Turnacioglu et al., 1997); (b) the entire Z-disc region of titin in the myogenic cell line Hsk btsA58 (Peckham et al., 1997); and (c) a COOH-terminal truncated fragment of α-actinin (thus, missing it's binding site for titin) in skeletal muscle cultures (Schultheiss et al., 1992). Thus, the association of titin filaments with Z-discs (i.e., via its interaction with T-cap and α-actinin) is critical for the assembly and maintenance of myofibril structure.
It is not difficult to understand why disrupting the interaction of titin with α-actinin would result in Z-line disruption, since it is likely that the interaction of these two proteins (together with the thin filaments) contributes greatly to the structural continuity of the sarcomere. It is more difficult to understand why overexpression of the 19-kD T-cap, which interacts with the most NH2-terminal region of titin, located at the periphery of the Z-line, is also required for Z-line/sarcomere maintenance. A highly speculative idea is that perhaps T-cap acts as a “bolt,” functioning to anchor the giant titin filament at its NH2 terminus. Future studies will be focused on exploring further the interaction of titin with T-cap, e.g., determining how the interaction of T-cap with titin is regulated.
In summary, we report here that the NH2 terminus of titin spans the Z-line. So far, using different regions of Z-disc titin as baits in yeast two-hybrid studies, two ligands for this region of titin have been identified: α-actinin (Ohtsuka et al., 1997; Sorimachi et al., 1997; Young et al., 1998) and T-cap (this study). Studies in live myocytes demonstrate that all components of this network, including the NH2 terminus of titin, COOH terminus of α-actinin, and T-cap are required for Z-line structure. Since the amount of titin filament overlap coincides with Z-line width, and many studies have demonstrated that α-actinin and Z-disc titin are important in myofibril formation, it appears likely that the NH2-terminal region of titin is intimately involved in the assembly of Z-lines during myogenesis.
Acknowledgments
The authors would like to thank Dr. Naoji Toyota for generously providing pCMVmyc.
This work was supported by grants from the Human Frontier Science Programme (S. Labeit, H. Sorimachi, and C.C. Gregorio), National Institutes of Health (NIH) HL57461 (C.C. Gregorio), NIH AR42652 (H. Granzier), the Deutsche Forschungsgemeinschaft (La668/2-3), and the “Forschungsfond für Klinische Medizin Mannheim” (S. Labeit). The Leica confocal microscope used in this study is supported by the NIEHS Southwest Environmental Health Science Grant ES-06694.
Abbreviations used in this paper
- 3-AT
3-amino-1, 2, 4-triazole
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- pc
post coitum
- RT-PCR
reverse transcription–polymerase chain reaction
- T-cap
titin-cap
Note Added in Proof
T-cap (telethonin) was shown to be phosphorylated by titin's M-line catalytic serine/threonine kinase domain in vitro. Therefore, by a T-cap (telethonin)-dependent mechanism, the Z-disc and the M-line ends of the titin molecule may communicate with each other (Mayans, O., P. van der Ven, M. Wilm, A. Mues, P. Young, D.O. Fürst, M. Wilmanns, and M. Gautel. 1998. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. In press).
Footnotes
Address all correspondence to Carol C. Gregorio, Ph.D., Department of Cell Biology and Anatomy, LSN441, University of Arizona, 1501 N. Campbell Avenue, Tucson, AZ 85724. Tel.: (520) 626-8113. Fax: (520) 626-2097. E-mail: gregorio@u.arizona.edu
References
- Beggs AH, Byers TJ, Knoll JHM, Boyce FM, Bruns GAP, Kunkel LM. Cloning and characterization of two human skeletal muscle α-actinin genes located on chromosomes 1 and 11. J Biol Chem. 1992;267:9281–9288. [PubMed] [Google Scholar]
- Conlon RA, Herrmann BG. Detection of messenger RNA by in situ hybridization to post-implantation embryo whole mounts. Methods Enzymol. 1993;225:373–383. doi: 10.1016/0076-6879(93)25026-x. [DOI] [PubMed] [Google Scholar]
- Dabiri GA, Turnacioglu KK, Sanger JM, Sanger JW. Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc Natl Acad Sci USA. 1997;94:9493–9498. doi: 10.1073/pnas.94.17.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri, G.A., K. Turnacioglu, J.C. Ayoob, J.M. Sanger, and J.W. Sanger. 1998. Transfections of primary muscle cell cultures with plasmids coding for GFP linked to full length and truncated muscle proteins. Methods Cell Biol. In press. [DOI] [PubMed]
- Eilertsen KJ, Kazmierski ST, Keller TCS., III Cellular titin localization in stress fibers and interaction with myosin II filaments in vitro. J Cell Biol. 1994;126:1201–1210. doi: 10.1083/jcb.126.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Fischman DA. Molecular analysis of protein assembly in muscle development. Science. 1991;251:1039–1044. doi: 10.1126/science.1998120. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton AB, Alftine C. Organization of protein and mRNA for titin and other myofibril components during myofibrillogenesis in cultured chicken skeletal muscle. Cell Struct Funct. 1997;22:51–58. doi: 10.1247/csf.22.51. [DOI] [PubMed] [Google Scholar]
- Funatsu T, Higuchi H, Ishiwata S. Elastic filaments in skeletal muscle revealed by selective removal of thin filaments with plasma gelsolin. J Cell Biol. 1990;110:53–62. doi: 10.1083/jcb.110.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst DO, Osborn M, Nave R, Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J Cell Biol. 1988;106:1563–1572. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst DO, Osborn M, Weber K. Myogenesis in the mouse embryo: differential onset of expression of myogenic proteins and the involvement of titin in myofibril assembly. J Cell Biol. 1989a;109:517–527. doi: 10.1083/jcb.109.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst DO, Nave R, Osborn M, Weber K. Repetitive titin epitopes with a 42 nm spacing coincide in relative position with known A band striations also identified by major myosin-associated proteins. An immunoelectron-microscopical study on myofibrils. J Cell Sci. 1989b;94:119–125. doi: 10.1242/jcs.94.1.119. [DOI] [PubMed] [Google Scholar]
- Gautel M, Goulding D, Bullard B, Weber K, Fürst DO. The central Z-disk region of titin is assembled from a novel repeat in variable copy numbers. J Cell Sci. 1996;109:2747–2754. doi: 10.1242/jcs.109.11.2747. [DOI] [PubMed] [Google Scholar]
- Granzier HL, Irving TC. Passive tension in cardiac muscle: contribution of collagen, titin, microtubules and intermediate filaments. Biophys J. 1995;68:1027–1044. doi: 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier H, Kellermayer M, Trombitas K. Titin elasticity and mechanism of passive force development in rat cardiac myocytes probed by thin-filament extraction. Biophys J. 1997;73:2043–2053. doi: 10.1016/S0006-3495(97)78234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Fowler VM. Mechanisms of thin filament assembly in embryonic chick cardiac myocytes: tropomodulin requires tropomyosin for assembly. J Cell Biol. 1995;129:683–695. doi: 10.1083/jcb.129.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Repasky EA, Fowler VM, Black JD. Dynamic properties of ankyrin in T lymphocytes: colocalization with spectrin and protein kinase C β. J Cell Biol. 1994;125:345–358. doi: 10.1083/jcb.125.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Weber A, Bondad M, Pennise CR, Fowler VM. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377:83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- Handel SE, Greaser ML, Schultz E, Wang S-M, Bulinski JC, Lin JJ-C, Lessard JL. Chicken cardiac myofibrillogenesis studied with antibodies specific for titin and the muscle and nonmuscle isoforms of actin and tropomyosin. Cell Tissue Res. 1991;263:419–430. doi: 10.1007/BF00327276. [DOI] [PubMed] [Google Scholar]
- Holtzer H, Hijikata T, Lin ZX, Zhang ZQ, Holtzer S, Protasi F, Frazini-Armstrong C, Sweeney HL. Independent assembly of 1.6 μm long bipolar MHC filaments and I-Z-I bodies. Cell Struct Funct. 1997;22:83–93. doi: 10.1247/csf.22.83. [DOI] [PubMed] [Google Scholar]
- Horowits R, Kempner ES, Bisher ME, Podolski RJ. A physiological role for titin and nebulin in skeletal muscle. Nature. 1986;323:160–164. doi: 10.1038/323160a0. [DOI] [PubMed] [Google Scholar]
- Houmeida A, Holt J, Tskhovrebova L, Trinick J. Studies of the interaction between titin and myosin. J Cell Biol. 1995;131:1471–1481. doi: 10.1083/jcb.131.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller TCS. Molecular bungees. Nature. 1997;387:233–235. doi: 10.1038/387233a0. [DOI] [PubMed] [Google Scholar]
- Kolmerer B, Olivieri N, Witt CC, Herrmann BG, Labeit S. Genomic organization of the M line titin and its tissue-specific expression in two distinct isoforms. J Mol Biol. 1996;256:556–563. doi: 10.1006/jmbi.1996.0108. [DOI] [PubMed] [Google Scholar]
- Labeit S, Gautel M, Lakey A, Trinick J. Towards a molecular understanding of titin. EMBO (Eur Mol Biol Organ) J. 1992;11:1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B. Titins, giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B, Linke WA. The giant protein titin: emerging roles in physiology and pathophysiology. Circ Res. 1997;80:290–294. doi: 10.1161/01.res.80.2.290. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Linke WA, Ivemeyer M, Olivieri N, Kolmerer B, Rüegg JC, Labeit S. Towards a molecular understanding of the elasticity of titin. J Mol Biol. 1996;261:62–71. doi: 10.1006/jmbi.1996.0441. [DOI] [PubMed] [Google Scholar]
- Linke WA, Ivemeyer M, Labeit S, Hinssen H, Ruegg JC, Gautel M. Actin-titin interaction in cardiac myofibrils: probing a physiological role. Biophys J. 1997;73:905–919. doi: 10.1016/S0006-3495(97)78123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther PK, Munro PMG, Squire JM. Muscle ultrastructure in the teleost fish. Micron. 1995;26:431–459. [Google Scholar]
- MacDonald RJ, Swift GH, Przybyla AE, Chirgwin JM. Isolation of RNA using guanidinium salts. Methods Enzymol. 1987;152:219–226. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]
- Maruyama K. Connectin/titin, giant elastic protein of muscle. FASEB (Fed Am Soc Exp Biol) J. 1997;11:341–345. doi: 10.1096/fasebj.11.5.9141500. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Matsubara R, Natori Y, Nonomura S, Kimura S, Ohashi K, Murakami F, Handa S, Eguchi G. Connectin, an elastic protein of muscle. J Biochem. 1977;82:317–337. [PubMed] [Google Scholar]
- Maruyama K, Kimura S, Yamamoto K, Wakabayashi T, Suzuki T. Connectin causes aggregation of myosin rods but not myosin heads. Biomed Res. 1985a;6:423–427. [Google Scholar]
- Maruyama K, Yoshioka T, Higuchi H, Ohashi K, Kimura S, Natori R. Connectin filaments link thick filaments and Z lines in frog skeletal muscle as revealed by immunoelectron microscopy. J Cell Biol. 1985b;101:2167–2172. doi: 10.1083/jcb.101.6.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WMJ, Gautel M, Steiner F, van der Ven PFM, Weber K, Fürst DO. The structure of the sarcomeric M band: localization of defined domains of myomesin, M protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy. J Cell Biol. 1996;134:1441–1453. doi: 10.1083/jcb.134.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka H, Yajima H, Kimura S, Maruyama K. Binding of the N terminal fragment of connectin/titin to α-actinin as revealed by yeast two-hybrid systems. FEBS Lett. 1997;401:65–67. doi: 10.1016/s0014-5793(96)01432-9. [DOI] [PubMed] [Google Scholar]
- Peckham M, Young P, Gautel M. Constitutive and variable regions of Z-disk titin/connectin in myofibril formation: a dominant-negative screen. Cell Struct Funct. 1997;22:95–101. doi: 10.1247/csf.22.95. [DOI] [PubMed] [Google Scholar]
- Rhee D, Sanger JM, Sanger JW. The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil Cytoskel. 1994;28:1–24. doi: 10.1002/cm.970280102. [DOI] [PubMed] [Google Scholar]
- Rowe RWD. The ultrastructure of Z disks from white, intermediate, and red fibers of mammalian striated muscle. J Cell Biol. 1973;57:261–277. doi: 10.1083/jcb.57.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki RK, Scharf SJ, Faloona F, Mullis GT, Erlich HA. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Schroeter JP, Bretaudiere J-P, Sass RL, Goldstein MA. Three-dimensional structure of the Z band in a normal mammalian skeletal muscle. J Cell Biol. 1996;133:571–583. doi: 10.1083/jcb.133.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss T, Choi J, Lin Z, Cohen-Gould L, Fischman D, Holtzer H. A sarcomeric α-actinin truncated at the carboxyl end induces the breakdown of stress fibers in PtK2 cell and the formation of nemaline-like bodies and breakdown of myofibrils in myotubes. Proc Natl Acad Sci USA. 1992;89:9282–9286. doi: 10.1073/pnas.89.19.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Komiyama M, Begum S, Maruyama K. Development of connectin/titin and nebulin in striated muscles of chicken. Adv Biophys. 1996;33:223–234. [PubMed] [Google Scholar]
- Sorimachi H, Freiburg A, Kolmerer B, Ishiura S, Stier G, Gregorio CC, Labeit D, Linke WA, Suzuki K, Labeit S. Tissue-specific expression and α-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J Mol Biol. 1997;270:688–695. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- Squire JM. Architecture and function in the muscle sarcomere. Curr Opin Struct Biol. 1997;7:247–257. doi: 10.1016/s0959-440x(97)80033-4. [DOI] [PubMed] [Google Scholar]
- Stromer MH, Goll DE. Studies on purified α-actinin. J Mol Biol. 1972;67:489–494. doi: 10.1016/0022-2836(72)90465-2. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffat BA. RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1991;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT, Maher PA. Immunocytochemical studies of cardiac myofibrillogenesis in early chick embryos. I. Presence of immunofluorescent titin spots in premyofibrillar stages. J Cell Biol. 1987a;105:2781–2793. doi: 10.1083/jcb.105.6.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT, Maher PA. Immunocytochemical studies of cardiac myofibrillogenesis in early chick embryos. II. Generation of α-actinin dots within titin spots at the time of the first myofibril formation. J Cell Biol. 1987b;105:2795–2801. doi: 10.1083/jcb.105.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinick J. Cytoskeleton: titin as a scaffold and spring. Curr Biol. 1996;6:258–260. doi: 10.1016/s0960-9822(02)00472-4. [DOI] [PubMed] [Google Scholar]
- Trombitas K, Granzier H. Actin removal from cardiac myocytes shows that near the Z-line titin attaches to actin while under tension. Am J Physiol. 1997;273:C662–C670. doi: 10.1152/ajpcell.1997.273.2.C662. [DOI] [PubMed] [Google Scholar]
- Trombitas K, Jin J-P, Granzier H. The mechanically active domain of titin in cardiac muscle. Circ Res. 1995;77:856–861. doi: 10.1161/01.res.77.4.856. [DOI] [PubMed] [Google Scholar]
- Trombitas K, Greaser M, Labeit S, Jin J-P, Kellermayer M, Helmes M, Granzier H. Titin extensibility in situ: entropic elasticity of permanently folded and permanently unfolded molecular segments. J Cell Biol. 1998;140:853–859. doi: 10.1083/jcb.140.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnacioglu KK, Mittal B, Dabiri GA, Sanger JM, Sanger JW. An N-terminal fragment of titin coupled to green fluorescent protein localizes to the Z-bands in living muscle cells: overexpression leads to myofibril disassembly. Mol Biol Cell. 1997;8:705–717. doi: 10.1091/mbc.8.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle G, Faulkner G, De Antoni A, Pacchioni B, Pallavicini A, Pandolofo D, Tiso N, Toppo S, Trevisan S, Lanfranchi G. Telethonin, a novel sarcomeric protein of heart and skeletal muscle. FEBS Lett. 1997;415:163–168. doi: 10.1016/s0014-5793(97)01108-3. [DOI] [PubMed] [Google Scholar]
- Vigoreaux JO. The muscle Z-band: lessons in stress management. J Muscle Res Cell Motil. 1994;15:237–255. doi: 10.1007/BF00123477. [DOI] [PubMed] [Google Scholar]
- Wang K. Titin/connectin and nebulin: giant protein rulers of muscle structure and function. Adv Biophys. 1996;33:123–134. [PubMed] [Google Scholar]
- Wang K, McClure J, Tu A. Titin: major myofibrillar components of striated muscle. Proc Natl Acad Sci USA. 1979;76:3698–3702. doi: 10.1073/pnas.76.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting A, Wardale J, Trinick J. Does titin regulate the length of muscle thick filaments? . J Mol Biol. 1989;205:263–268. doi: 10.1016/0022-2836(89)90381-1. [DOI] [PubMed] [Google Scholar]
- Yajima H, Ohtsuka H, Kawamura Y, Kume H, Maruyama T, Abe H, Kimura S, Maruyama K. A 11.5 kb 5′-terminal cDNA sequence of chicken breast muscle connectin/titin reveals its Z line binding region. Biochem Biophys Res Commun. 1996;223:160–164. doi: 10.1006/bbrc.1996.0862. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Izumimoto M, Robson RM, Stromer MH. Fine structure of wide and narrow vertebrate muscle Z-lines. A proposed model and computer simulation of Z-line architecture. J Mol Biol. 1985;184:621–644. doi: 10.1016/0022-2836(85)90308-0. [DOI] [PubMed] [Google Scholar]
- Young P, Ferguson C, Bañuelos S, Gautel M. Molecular structure of the sarcomeric Z-disk: two types of titin interactions lead to an asymmetrical sorting of α-actinin. EMBO (Eur Mol Biol Organ) J. 1998;17:1614–1624. doi: 10.1093/emboj/17.6.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]